Abstract
Ubiquitination is a post-translational modification involving the reversible attachment of the small protein ubiquitin to a target protein. Ubiquitination is involved in numerous cellular processes, including the membrane trafficking of cargo proteins. However, the ubiquitination of the trafficking machinery components and their involvement in environmental responses are not well understood. Here, we report that the Arabidopsis thaliana trans-Golgi network/early endosome localized SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) protein SYP61 interacts with the transmembrane ubiquitin ligase ATL31, a key regulator of resistance to disrupted carbon (C)/nitrogen/(N)-nutrient conditions. SYP61 is a key component of membrane trafficking in Arabidopsis. The subcellular localization of ATL31 was disrupted in knockdown mutants of SYP61, and the insensitivity of ATL31-overexpressing plants to high C/low N-stress was repressed in these mutants, suggesting that SYP61 and ATL31 cooperatively function in plant responses to nutrient stress. SYP61 is ubiquitinated in plants, and its ubiquitination level is upregulated under low C/high N-nutrient conditions. These findings provide important insights into the ubiquitin signaling and membrane trafficking machinery in plants.
The TGN/EE SNARE protein SYP61 plays a critical role in plant carbon/nitrogen nutrient stress responses and is ubiquitinated in response to low carbon/high nitrogen conditions.
IN A NUTSHELL.
Background: Ubiquitination is a post-translational modification that functions in numerous cellular processes by serving as a molecular signal, such as protein degradation, membrane trafficking, and complex formation. Although numerous reports describe the ubiquitination of cargo proteins, the ubiquitination of components of the trafficking machinery is not well understood. We previously identified the transmembrane ubiquitin ligase ATL31 and demonstrated that it regulates carbon (C)/nitrogen (N)-nutrient stress responses in Arabidopsis. As ATL31 requires its transmembrane domain for this function, we characterized the subcellular localization of ATL31 and determined that it interacts with the trans-Golgi network/early endosome-localized SNARE protein SYP61.
Question: SNARE proteins are key regulators of membrane trafficking. However, their roles in plant nutrient responses were not well known. Also, the ubiquitination of SNARE proteins was not well characterized. Therefore, we investigated whether the SNARE protein SYP61 is involved in plant C/N-nutrient responses and is ubiquitinated.
Findings: We found that SYP61 is required for the proper subcellular localization of ATL31 and its function in high C/low N-nutrient stress responses. In addition, we discovered that SYP61 is transiently ubiquitinated in response to low C/high N-nutrient conditions. Our results demonstrate the importance of membrane trafficking regulation in plant nutrient responses, and they suggest that SNARE proteins might be regulated by ubiquitination in response to nutrient conditions.
Next steps: We are currently trying to reveal the downstream effects of SNARE ubiquitination. It would also be interesting to investigate how common this modification is among SNARE proteins. Further experiments including analysis of ubiquitin-deficient mutants of SNARE proteins will help us understand the dynamics of the posttranslational regulation of the membrane trafficking machinery.
Introduction
Ubiquitination plays critical roles in regulating the activities of various proteins and enables flexible responses to the changing environment (Hershko and Ciechanover, 1998; Oh et al., 2018). Ubiquitin is an 8.5-kDa protein attaches to lysine (K) residues of its target proteins by ubiquitin ligases. Ubiquitin can form chains via its seven K residues and one N-terminal methionine residue. Therefore, there are several types of topologically different ubiquitin chains, which modulate distinct cellular processes (Oh et al., 2018). For example, proteins modified by compact structured K48- and K11-linked ubiquitin chains undergo proteasomal degradation, whereas more flexible K63-linked ubiquitination is involved in several processes, including vacuolar targeting, endocytosis, DNA repair, and signal activation (Callis, 2014; Isono and Kalinowska, 2017; Oh et al., 2018; Romero-Barrios and Vert, 2018).
Ubiquitination is involved in regulating the membrane trafficking of cargo proteins during plant responses to environmental stress. For example, multiple mono-ubiquitination of the plant metal transporter IRON-REGULATED TRANSPORTER1 (IRT1) is required for its constitutive turnover from the plasma membrane to the trans-Golgi network/early endosome (TGN/EE); the extension of ubiquitination into K63-linked chains in the presence of excess amounts of noniron metal ions leads to vacuolar degradation (Barberon et al., 2011; Dubeaux et al., 2018). Ubiquitination of other plasma membrane localized proteins, including the brassinosteroid receptor BRASSINOSTEROID INSENSITIVE1 (BRI1) and the auxin efflux carrier PIN FORMED2 (PIN2), also triggers endocytosis and vacuolar targeting (Leitner et al., 2012; Korbei et al., 2013; Martins et al., 2015; Zhou et al., 2018). Ubiquitination of the boron transceptor REQUIRES HIGH BORON1 (BOR1) is required for its vacuolar degradation under high boron concentrations (Kasai et al., 2011; Yoshinari et al., 2021). In addition, mono-ubiquitination of the membrane-associated receptor-like cytoplasmic kinase BOTRYTIS-INDUCED KINASE1 (BIK1) triggers its endocytosis and plays important roles in the activation of plant immunity (Ma et al., 2020). Deubiquitination reactions are also important for cargo protein trafficking via the endosomal sorting complex required for transport (ESCRT) system (Isono and Kalinowska, 2017). Although these findings describe the regulation of the trafficking of cargo proteins, less is known about the ubiquitination of the trafficking machinery itself.
Soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins are key regulators of membrane trafficking that are conserved among eukaryotes (Jahn and Scheller, 2006; Lipka et al., 2007; Wickner and Schekman, 2008). These proteins mediate vesicle fusion by forming SNARE complexes consisting of four different types of SNARE motifs: Qa-, Qb-, Qc-, and R-SNARE. The Arabidopsis thaliana genome encodes more than 60 SNARE proteins (Sanderfoot et al., 2000; Uemura et al., 2004; Lipka et al., 2007; Sanderfoot, 2007). Each SNARE protein localizes to specific membranes and has specific SNARE partners in the cell (Pratelli et al., 2004; Uemura et al., 2004; Jahn and Scheller, 2006; Fujiwara et al., 2014). Some SNARE proteins have several different SNARE partners and mediate different trafficking pathways. For example, the TGN/EE localized Qc-SNARE protein SYNTAXIN OF PLANTS61 (SYP61) mediates vacuolar trafficking with SYP41 (Qa-SNARE), VPS TEN INTERACTING12 (VTI12, Qb-SNARE), and the Sec1/Munc18 (SM) family protein VACUOLAR PROTEIN SORTING45 (VPS45; Zouhar et al., 2009; Kim and Bassham, 2011). SYP61 also mediates retrograde and anterograde trafficking of the aquaporin PLANT PLASMA MEMBRANE INTRINSIC PROTEINS 2;7 (PIP2;7), possibly with SYP121 (Qa-SNARE; Hachez et al., 2014). SYP61 mediates the exocytotic trafficking of cell wall components as well (Drakakaki et al., 2012; Gendre et al., 2013). SNARE proteins play important roles in various plant physiological responses. For example, a knockdown mutant of SYP61 is hypersensitive to salt and osmotic stress (Zhu et al., 2002).
Sugar (carbon, C) and nitrogen (N) are essential components of organisms whose relative availability (C/N-nutrient balance) affects many aspects of plant physiology. For example, excess sugar with limited N during the early post-germination stage inhibits seedling growth and leads to the accumulation of anthocyanin, whereas high CO2/low N conditions during maturity promote the progression of senescence in Arabidopsis (Coruzzi and Zhou, 2001; Martin et al., 2002; Aoyama et al., 2014a, 2014b). These high C/low N-nutrient stress responses are rescued by either reducing the amount of carbon sources or enhancing the availability of nitrogen.
We previously reported that the T6P (Trehalose 6-Phosphate)-SnRK1 (SNF1-related protein kinase 1 complex) module mediates C/N-nutrient signaling in Arabidopsis (Li et al., 2020). T6P is an intermediate metabolite of trehalose biosynthesis that inhibits the activity of SnRK1 by directly binding to the SnRK1α subunit (Zhai et al., 2018). We also identified the membrane-localized ubiquitin ligase ARABIDOPSIS TOXICOS EN LEVADURA 31 (ATL31) as a downstream target of the SnRK1-regulated kinases CBL-INTERACTING PROTEIN KINASE7, 12, and 14 (CIPK7/12/14; Yasuda et al., 2017; Li et al., 2020). ATL31 enhances plant tolerance to high C/low N-nutrient stress by targeting 14-3-3 proteins for proteasomal degradation by binding to them via CIPK7/12/14-mediated phosphorylation (Sato et al., 2009, 2011; Yasuda et al., 2014, 2017).
In this study, we identified SYP61 as an interactor of ATL31. We demonstrate that SYP61 is required for the proper localization of ATL31 and for ATL31-mediated C/N-nutrient stress responses. We also demonstrate that SYP61 is ubiquitinated in plants and that its ubiquitination level is regulated in response to C/N-nutrient availability, suggesting that SNARE proteins might be regulated at the post-translational level in response to nutrient conditions.
Results
ATL31 localizes to the plasma membrane and endosomal compartments
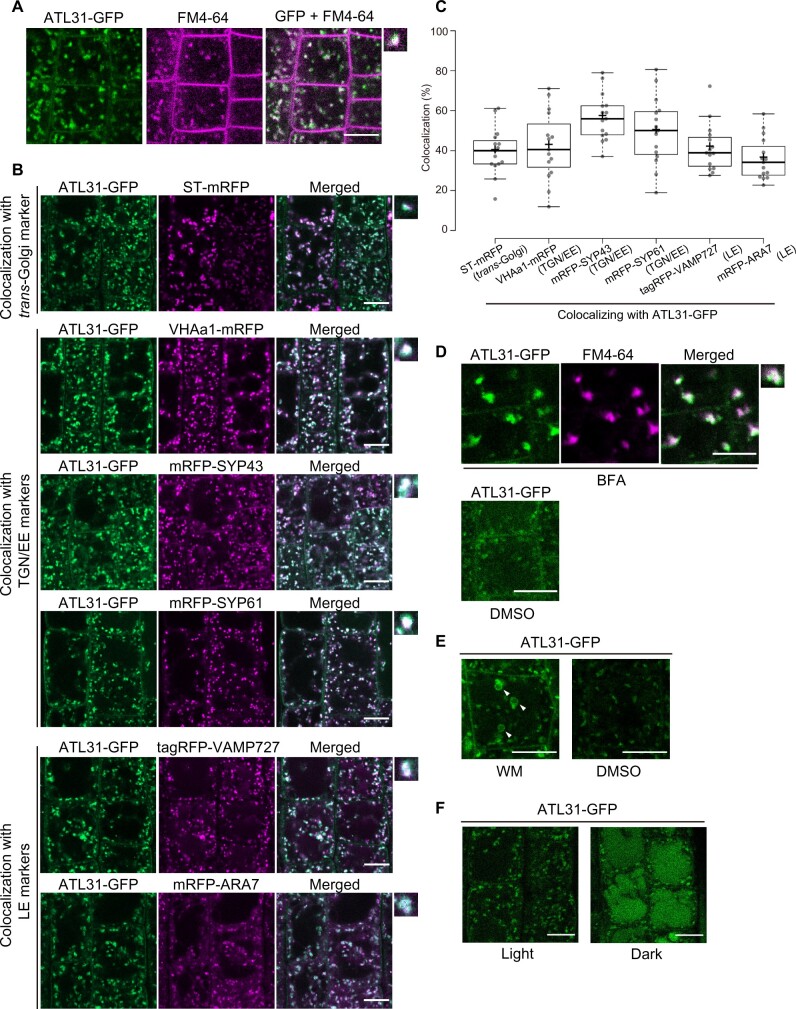
We previously showed that the N-terminal transmembrane domain of ATL31 is required for the role of this ubiquitin ligase in C/N-nutrient stress responses (Sato et al., 2009). To characterize the subcellular localization of ATL31, we performed confocal microscopy observations of Arabidopsis plants expressing green fluorescent protein-tagged ATL31 (ATL31-GFP). In root tip epidermal cells, ATL31-GFP signals were detected at intracellular punctate structures and the plasma membrane (Figure 1A). The GFP signals colocalized with the internalized FM4–64 dye in <20 min after staining, suggesting that ATL31 localizes to the TGN/EE (Figure 1A).
Figure 1.
ATL31 localizes to the plasma membrane and endosomal compartments. A, Representative confocal images of FM4–64-stained Arabidopsis root epidermal cells expressing ATL31-GFP. The root was stained with 2-µM FM4–64 for 2 min and washed with medium without dye. The photograph was taken 18 min after staining. Bar = 10 µm. B, Representative confocal images of Arabidopsis root epidermal cells co-expressing ATL31-GFP with ST-mRFP (trans-Golgi), mRFP-SYP43 (TGN/EE), mRFP-SYP61 (TGN/EE), VHAa1-mRFP (TGN/EE), mRFP-ARA7 (LE), and tagRFP-VAMP727 (LE; F1 generation). Small panels on the right show enlarged views of the dot-like structures. Bars = 10 µm. C, Quantification of the data in (B). Percentage of dot-like structures of ATL31-GFP colocalizing with the indicated markers. The center–center distances of the nearest dot-like structures were calculated using the ImageJ plugin DiAna, and endosomes at a distance closer than 0.2 µm (approximate theoretical resolution limit of the confocal microscope) were considered to be colocalizing. n = 15 cell slices from five roots were analyzed. In the box plots, center line, median; box limits, lower and upper quartiles; +, mean; dots, individual data points; whiskers, highest and lowest data points (the whiskers extend to data points that are less than 1.5× interquartile range (IQR) away from the 1st and 3rd quartile.). D, Representative confocal images of Arabidopsis root epidermal cells expressing ATL31-GFP treated with 50-µM BFA or DMSO for 30 min. Cells were stained with 5-µM FM4–64 15 min before BFA treatment. The small panel on the right shows and enlarged view of BFA bodies. Bars = 10 µm. E, Representative confocal images of Arabidopsis root epidermal cells expressing ATL31-GFP treated with 33-µM WM or DMSO for 1 h. Arrowheads indicate the ring-like structures representing vacuolized late endosomal compartments. Bars = 10 µm. F, Representative confocal images of Arabidopsis root epidermal cells expressing ATL31-GFP after 18 h of light or dark treatment. Bars = 10 µm.
To further explore the identity of these dot-like structures, we performed colocalization analysis with monomeric red fluorescent protein (mRFP)- or tagRFP-tagged endomembrane organelle markers in transgenic Arabidopsis plants. ATL31-GFP colocalized with the TGN/EE markers mRFP-SYP43, mRFP-SYP61, and VHAa1-mRFP (Figure 1, B and C). We also observed the colocalization of ATL31-GFP with the trans-Golgi marker ST-mRFP (cytoplasmic and transmembrane domains of a rat sialyl transferase) and the late endosome (LE) markers mRFP-ARA7 and tagRFP-VAMP727 (Figure 1, B and C). Since ST often colocalizes with part of the TGN/EE called Golgi-associated-TGN (GA-TGN; Uemura et al., 2014; Shimizu et al., 2021), the colocalization of ATL31-GFP with ST-mRFP suggests it might be localized to the GA-TGN.
To confirm the localization of ATL31 at the TGN/EE and the LE, we tested the sensitivity of ATL31-GFP to the commonly used membrane trafficking inhibitors brefeldin A (BFA) and wortmannin (WM). BFA inhibits the BFA-sensitive ADP-ribosylation factors guanine nucleotide exchange factors and generates large aggregations of endosomal compartments known as BFA bodies (Geldner et al., 2003; Grebe et al., 2003; Dettmer et al., 2006; Robinson et al., 2008). A 30-min treatment with 50-μM BFA induced the aggregation of ATL31-GFP, with fluorescent signals overlapping FM4–64 containing BFA bodies (Figure 1D). WM is an inhibitor of phosphoinositide 3-kinase (PI3K) and PI4K that causes the swelling and vacuolization of late endosomal compartments (Jaillais et al., 2008; Wang et al., 2009). After 1 h of 33-μM WM treatment, ATL31-GFP signals were observed as small ring-like structures (Figure 1E). These pharmacological analyses further support the notion that ATL31 localizes to the TGN/EE and the LE.
Membrane-localized proteins often undergo vacuolar degradation, which can be visualized as fluorescent signals of tagged GFP in the vacuole after dark treatment (Tamura et al., 2003). After 18 h of dark treatment, GFP fluorescence was observed in vacuoles in addition to the intracellular punctuate structures (Figure 1F), whereas mRFP-SYP61 was not observed in the vacuole (Supplemental Figure S1). The vacuolar targeting of ATL31 was confirmed by treatment with the vacuolar ATPase inhibitor Concanamycin A (ConcA; Dröse et al., 1993; Matsuoka et al., 1997; Dettmer et al., 2006; Supplemental Figure S2). After 3 h of ConcA treatment, ATL31-GFP was observed in abnormal patches, and the ratio of full-length ATL31-GFP to free GFP protein was enhanced (Supplemental Figure S2). These results suggest that ATL31-GFP is subjected to vacuolar degradation in Arabidopsis cells.
Because ATL31 is a C/N-nutrient response regulator, we also examined the subcellular localization of ATL31 under different C/N-nutrient conditions. Transgenic Arabidopsis seedlings expressing ATL31-GFP with the TGN/EE marker VHAa1-mRFP or the LE marker tagRFP-VAMP727 were treated with liquid medium containing 0-mM glucose (Glc)/60-mM N, 100-mM Glc/30-mM N, or 200-mM Glc/0.3-mM N for 90–110 min (Supplemental Figure S3). The colocalization of ATL31 with VHAa1-mRFP was not dependent on C/N-nutrient conditions, possibly because TGN/EE is a trafficking hub that harbors both exocytic and endocytic pathways (Uemura et al., 2014, 2019; Shimizu et al., 2021). On the other hand, fewer dot-like structures of ATL31-GFP colocalized with tagRFP-VAMP727 under higher C/lower N conditions. We previously reported that ATL31 is stabilized under higher C/lower N conditions (Yasuda et al., 2017). The reduced colocalization of ATL31-GFP with the LE marker suggests that the vacuolar targeting of ATL31 decreased under these conditions.
Altogether, these results indicate that ATL31 localizes to multiple organelles including the plasma membrane, TGN/EE, and LE and is degraded in the vacuole. The observation that the subcellular localization of ATL31 is affected by C/N-nutrient conditions points to the relevance of the regulation of ATL31 trafficking to its function.
ATL31 physically interacts with the TGN/EE SNARE SYP61
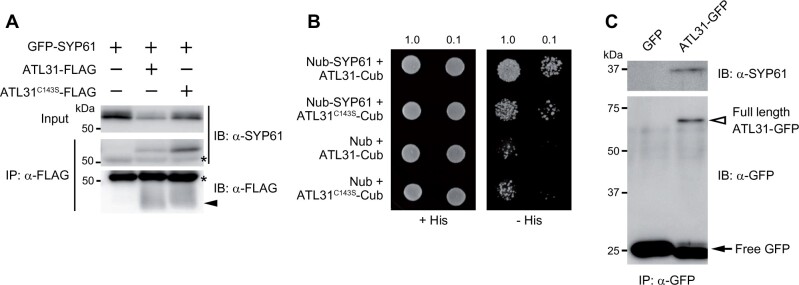
SYP61 is a Qc-SNARE with three N-terminal helical domains and a C-terminal transmembrane region that mediates membrane trafficking at the TGN/EE in Arabidopsis (Sanderfoot et al., 2001; Uemura et al., 2004). In a proteomic analysis of the SYP61 compartment, ATL6, the closest homolog of ATL31, was identified (Drakakaki et al., 2012). SYP61 mediates the secretion of cell wall components (Drakakaki et al., 2012; Wilkop et al., 2019), and ATL31 is thought to be involved in the accumulation of cell wall components (Maekawa et al., 2014). This functional relevance motivated us to evaluate the interaction between ATL31 and SYP61. We transiently co-expressed GFP-tagged SYP61 with FLAG-tagged ATL31 or the catalytically inactive form of ATL31 (with a point-mutation at the conserved cysteine-143 residue in the RING domain to serine [ATL31C143S]) in Nicotiana benthamiana leaves and subjected the proteins to a co-immunoprecipitation assay. SYP61 was co-immunoprecipitated with both intact ATL31 and ATL31C143S (Figure 2A). Their physical interactions were confirmed by split ubiquitin yeast two-hybrid analysis (Figure 2B). In addition, immunoblotting with anti-SYP61 antibody detected endogenous SYP61 in the immunoprecipitate of ATL31-GFP from transgenic Arabidopsis plants (Figure 2C). These results demonstrate that ATL31 physically interacts with SYP61 in planta.
Figure 2.
ATL31 interacts with SYP61. A, Co-immunoprecipitation of GFP-SYP61 (56 kDa) with ATL31-FLAG or ATL31C143S-FLAG (43 kDa, indicated by arrowhead). Proteins were expressed in N. benthamiana leaves and immunoprecipitated with anti-FLAG antibody beads, followed by immunoblotting with anti-SYP61 and anti-FLAG antibodies. Asterisks, nonspecific bands and IgG heavy chain. B, Split ubiquitin yeast-two hybrid assays of ATL31-Cub and ATL31C143S-Cub with Nub-SYP61 or empty vector. Yeast cultures were diluted 1- or 10-fold (indicated by 1.0 or 0.1, respectively) and grown on solid medium with or without histidine (His). Cub, C-terminal half of ubiquitin; Nub, N-terminal half of ubiquitin. C, Co-immunoprecipitation of endogenous SYP61 with ATL31-GFP in Arabidopsis. Proteins were extracted from Arabidopsis plants expressing GFP (27 kDa, indicated by arrow) or ATL31-GFP (71 kDa, indicated by open arrowhead) and subjected to immunoprecipitation with anti-GFP antibody beads, followed by immunoblotting with anti-SYP61 and anti-GFP antibodies.
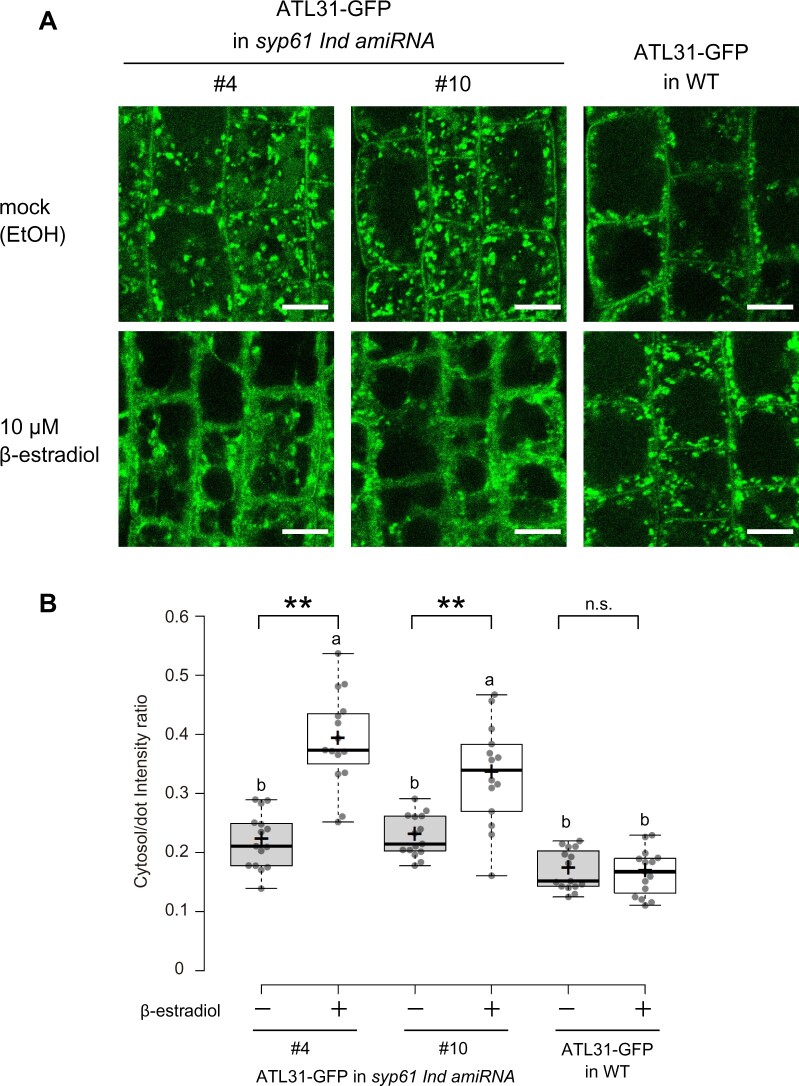
Induced knockdown of SYP61 affects the subcellular localization of ATL31
As SYP61 is a regulator of membrane trafficking, we observed the subcellular localization of ATL31 in Arabidopsis syp61 mutants. We generated β-estradiol-inducible knockdown mutants using an artificial SYP61 microRNA (syp61 Ind amiRNA) and crossed the two independent transformants with ATL31-GFP-expressing plants. We confirmed that the knockdown of SYP61 was successfully induced in the presence of 10-µM β-estradiol, while ATL31 was constitutively overexpressed in these plants (Supplemental Figure S4). We observed seedlings grown on medium containing 10-µM β-estradiol or ethanol (mock) by confocal laser-scanning microscopy (Figure 3). In the syp61 knockdown background, the fluorescent signals of ATL31-GFP were more dispersed in the cytosol compared to the wild-type background (Figure 3A). Quantification showed that the mean intensity ratio of signals in the cytosol to the dot-like structures was significantly enhanced in the mutant (Figure 3B). This abnormal pattern change upon β-estradiol treatment was not observed in the wild-type background, suggesting that this phenotype was caused by the decreased expression of SYP61 and was not a side-effect of β-estradiol treatment. These results indicate that SYP61 is required for the proper localization of ATL31.
Figure 3.
Subcellular localization of ATL31 is affected by knockdown of SYP61. A, Representative confocal images of Arabidopsis root epidermal cells expressing ATL31-GFP in the syp61 inducible knockdown mutant (syp61 Ind amiRNA; F3 generation, homo) or wild-type (WT) background. Plants were grown for 7 days on medium containing ethanol (EtOH) or 10-µM β-estradiol. Bars = 10 µm. The induction of SYP61 knockdown by β-estradiol in the lines is confirmed in Supplemental Figure S4. B, Quantification of the intensity ratios of ATL31-GFP in the cytosol and dot-like structures. The intracellular region excluding the dot-like structures was considered to be cytosol. n = 14–15 cell slices from five roots were analyzed. **P <0.01 by two-tailed Welch’s t test. NS, not significant. Different letters indicate significant difference, as determined by post hoc Tukey honestly significant difference test (P <0.05). In the box plots, center line, median; box limits, lower and upper quartiles; +, mean; dots, individual data points; whiskers, highest and lowest data points (the whiskers extend to data points that are less than 1.5× IQR away from the 1st and 3rd quartile).
SYP61 is required for the plant response to disrupted C/N-nutrient balance
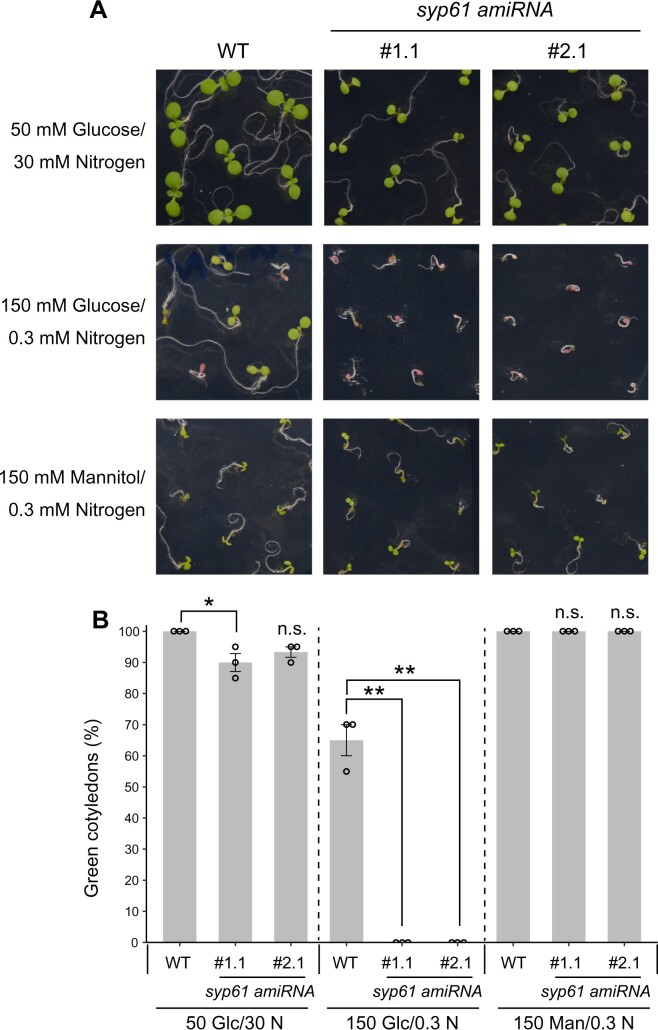
We then examined whether SYP61 is involved in ATL31-mediated high C/low N-nutrient stress responses. First, we tested the high C/low N stress tolerance of syp61 mutants. Two independent lines of knockdown mutant plants constitutively expressing the artificial SYP61 microRNA (syp61 amiRNA; Supplemental Figure S5) showed significant hypersensitivity to 150-mM Glc/0.3-mM N conditions, with no green cotyledons, whereas 65% of wild-type seedlings showed expanded green cotyledons under these conditions (Figure 4). We also performed the assay using osm1, a previously reported syp61 mutant (Zhu et al., 2002). In the osm1 mutant, the expression pattern of SYP61 is altered due to a T-DNA insertion upstream of the translation start site of SYP61 (Zhu et al., 2002). Because this mutant was established in the Arabidopsis C24 accession background, which is more resistant to high C/low N-nutrient stress than the Col-0 accession, we examined the osm1 phenotype in medium containing more glucose than the medium used for mutants in the Col-0 background. In line with the results for the syp61 amiRNA mutants, the percentage of green cotyledons was significantly lower for osm1 (10%) than for C24 wild-type (27%) under 300-mM Glc/0.3-mM N stress conditions (Supplemental Figure S6). All wild-type and syp61 seedlings showed green cotyledons in mannitol medium, confirming that the effects of C/N-nutrient stress are distinct from those of osmotic stress (Figure 4;Supplemental Figure S6).
Figure 4.
syp61 knockdown mutants are hypersensitive to C/N-nutrient stress. A, Representative images of WT (Col-0) and syp61 constitutive knockdown mutant (syp61 amiRNA) seedlings grown for 8 days on 50-mM glucose/30-mM nitrogen, 150-mM glucose/0.3-mM nitrogen, or 150-mM mannitol/0.3-mM nitrogen. B, Percentage of seedlings with green cotyledons after 8 days of growth. Glc, glucose; N, nitrogen; Man, mannitol; unit, mM. Data are mean ± sem (n = 3 independent batches of plants). Statistical significance was determined by two-tailed Dunnett’s test (*P <0.05, **P <0.01). NS, not significant.
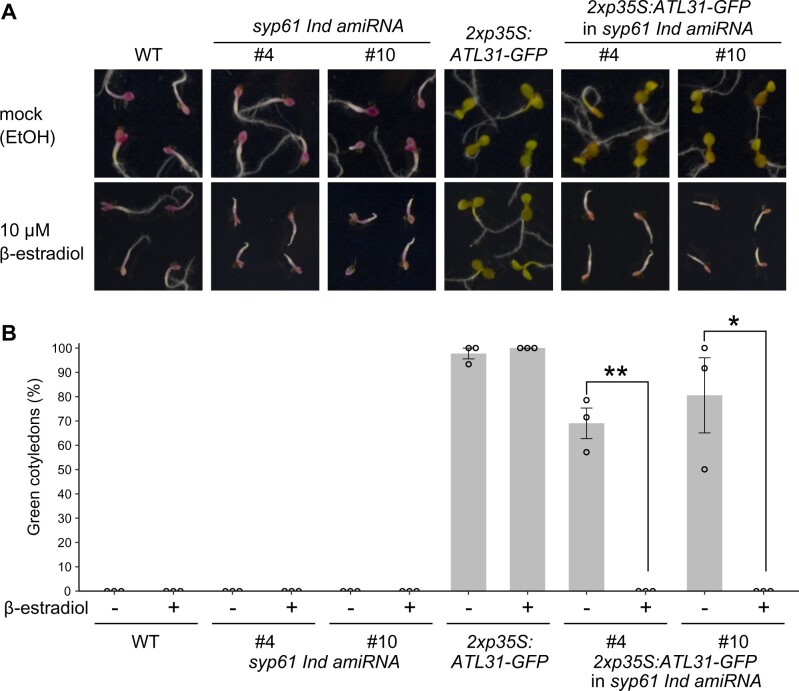
Next, to examine the relevance of SYP61 to ATL31 function, we tested the C/N-nutrient stress responses of Arabidopsis plants overexpressing ATL31 in the syp61 background, which were the same plant lines used for the localization analysis. The progeny of ATL31-overexpressing and syp61 Ind amiRNA plants were grown on high C/low N medium containing 300-mM Glc/0.3-mM N with 10-µM β-estradiol or ethanol (mock; Figure 5;Supplemental Figure S4). On mock medium, the ATL31-overexpressing and progeny plants exhibited an insensitive phenotype. In contrast, the post-germination growth of the progeny plants was inhibited on β-estradiol-containing nutrient stress medium, and the phenotype of the syp61 knockdown mutant was not rescued by the overexpression of ATL31. These results demonstrate that SYP61 plays an essential role in plant adaptation to C/N-nutrient conditions and is crucial for ATL31-mediated insensitivity to C/N-nutrient stress
Figure 5.
Overexpressing ATL31 does not rescue the hypersensitivity of SYP61 knockdown mutants to C/N-nutrient stress. A, Representative images of WT (Col-0), ATL31-overexpressing (2xp35S:ATL31-GFP), syp61 inducible knockdown mutant (syp61 Ind amiRNA) plants, and their F1 progeny grown for 12 days on 300-mM glucose/0.3-mM nitrogen with ethanol or β-estradiol. B, Percentage of seedlings with green cotyledons after 12 days of growth on 300-mM glucose/0.3-mM nitrogen with ethanol (−) or 10-µM β-estradiol (+).Data are mean ± sem (n = 3 independent batches of plants). Statistical significance was determined by two-tailed Welch’s t test (*P <0.05, **P <0.01).
SYP61 protein is ubiquitinated in plant cells
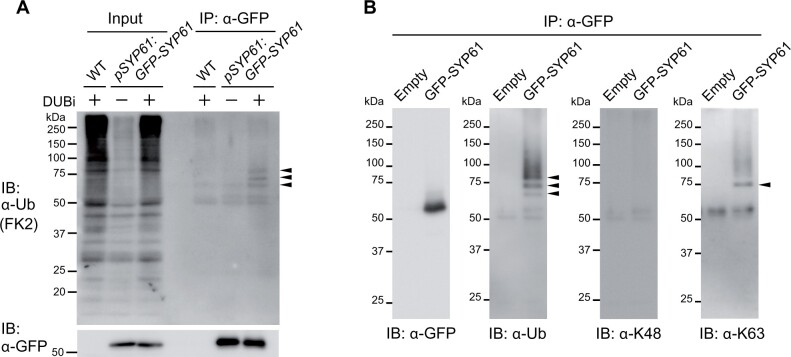
Because ATL31 is a ubiquitin ligase, we also examined the possibility of the ubiquitin modification on SYP61. First, we extracted proteins from transgenic Arabidopsis plants expressing GFP-fused SYP61 under the control of the native promoter (pSYP61:GFP-SYP61). GFP-SYP61 was purified by immunoprecipitation in the absence or presence of inhibitors of deubiquitination enzymes (DUBi). Immunoblotting with anti-GFP antibody detected a signal corresponding to intact GFP-SYP61, whereas immunoblotting with anti-ubiquitin antibody yielded sharp bands of higher molecular weight, which were enhanced by DUBi treatment (Figure 6A). Because the predicted molecular size of GFP-SYP61 in this construct is ∼55 kDa and that of ubiquitin is 8.5 kDa, the lowest band in this blot is presumed to be mono-ubiquitinated GFP-SYP61, and the second and the third bands may represent di- and tri-ubiquitinated GFP-SYP61. We confirmed the ubiquitination of SYP61 by immunoblotting using FLAG-tag fused SYP61 (FLAG-SYP61), and FLAG-GFP as a control (Supplemental Figure S7). Based on their molecular weights, the bands detected with ubiquitin antibody were likely ubiquitinated SYP61, not SYP61 fused with GFP, FLAG-tag, or other interactors. To further assess the biochemical characteristics of SYP61 ubiquitination, we analyzed the ubiquitin chain type. The second band of ubiquitinated SYP61, which is slightly smaller than 75 kDa, was also bound by the antibody specific for K63-linked, but not K48-linked, ubiquitination (Figure 6B). These findings indicate that SYP61 is ubiquitinated in plants and that some pools of SYP61 are modified by K63-type ubiquitination.
Figure 6.
SYP61 is ubiquitinated in plants. A, Immunoblot analysis to detect ubiquitinated SYP61 in Arabidopsis. Proteins were extracted from WT or GFP-SYP61 (55 kDa)-expressing Arabidopsis plants (pSYP61:GFP-SYP61) in the presence or absence of DUBi. Proteins were immunoprecipitated with anti-GFP antibody beads and detected by anti-ubiquitin (FK2) or anti-GFP antibodies. Arrowheads indicate ubiquitinated GFP-SYP61. B, Immunoblot analysis to identify the chain type of SYP61 ubiquitination. GFP-SYP61 (56 kDa) was transiently expressed in N. benthamiana leaves. Extracted proteins were immunoprecipitated with anti-GFP antibody beads and detected with anti-GFP, anti-ubiquitin (FK2), anti-K48-linked ubiquitin (Apu2), and anti-K63-linked ubiquitin (Apu3) antibodies. Arrowheads indicate ubiquitinated GFP-SYP61. The molecular weight of GFP-SYP61 transiently expressed in N. benthamiana is higher than that of GFP-SYP61 in stably transformed Arabidopsis plants, as it contains a longer linker.
ATL31 is able to mediate SYP61 ubiquitination
To investigate whether ATL31 can mediate K63-linked ubiquitination, we performed an in vitro ubiquitination reaction with recombinant ATL31 (MBP-ATL31) and quantitatively determined the chain type of the ubiquitination on MBP-ATL31 by parallel reaction monitoring (PRM), a type of targeted tandem mass spectrometry, using Ub-absolute quantification (Ub-AQUA) peptides (Kirkpatrick et al., 2005; Tsuchiya et al., 2017). Mass spectrometry showed that ATL31 catalyzes ubiquitin chains with K63 linkage, as well as other linkage types, such as K11 and K48 (Supplemental Figure S8).
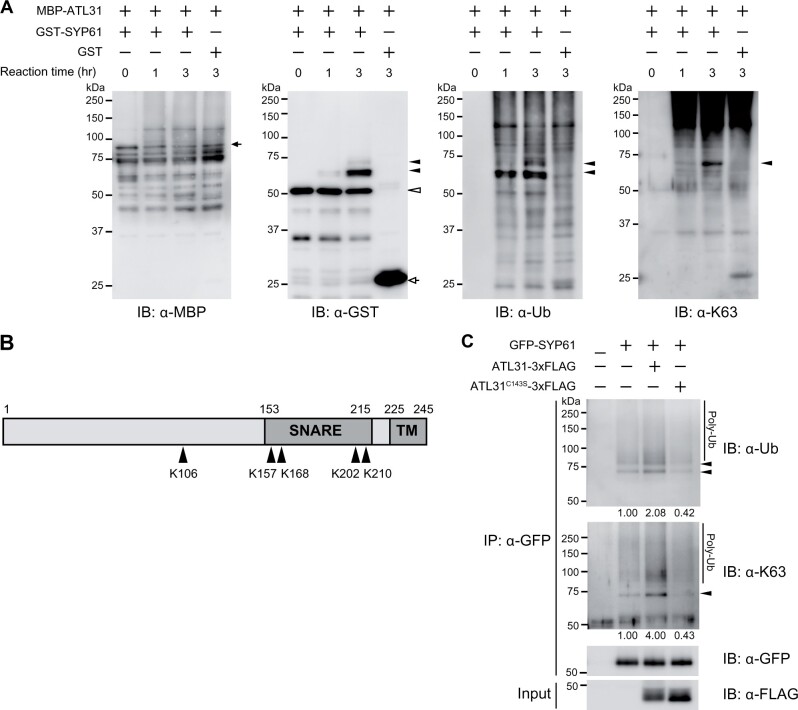
To examine the direct involvement of ATL31 in SYP61 ubiquitination, we performed in vitro ubiquitination assays with MBP-tag fused ATL31 (MBP-ATL31) and GST-tag-fused SYP61 (GST-SYP61) recombinant proteins. Incubation of GST-SYP61 with MBP-ATL31 for 1 or 3 h, followed by immunoblotting with either anti-GST or anti-ubiquitin antibody, revealed the time-associated shifting of the GST-SYP61 bands (Figure 7A). This ubiquitination signal was not detected after incubation with GST protein, indicating that SYP61 was ubiquitinated by ATL31 in vitro. The molecular size of the bands suggested that SYP61 was ubiquitinated with at least one molecule (mono-ubiquitin) but could also be ubiquitinated with two molecules. SYP61 bound to two ubiquitins was also detected with anti-K63 ubiquitination antibody (Figure 7A), indicating that ATL31 can catalyze the attachment of a K63-linked ubiquitin chain to SYP61.
Figure 7.
ATL31 ubiquitinates SYP61 in vitro and in planta. A, In vitro ubiquitination assay of SYP61 by ATL31. GST-SYP61 or GST was incubated with MBP-ATL31 in the presence of ubiquitin, E1, E2, and ATP for the indicated time. Immunoblot analysis was performed with anti-MBP, anti-GST, anti-ubiquitin (FK2), and anti-K63-linked ubiquitin (Apu3) antibodies. Closed arrow, MBP-ATL31 (81 kDa); open arrow, GST (28 kDa); open arrowhead, GST-SYP61 (53 kDa); closed arrowheads, ubiquitinated SYP61. B, Schematic diagram of the primary structure of SYP61 protein. Arrowheads indicate the ATL31-catalyzed ubiquitination sites of SYP61, as determined by mass spectrometry analysis following an in vitro ubiquitination assay as in A. Numbers indicate the amino acid positions. SNARE, SNARE domain; TM, transmembrane domain. The original data are shown in Supplemental Table S1. C, Immunoblot analysis to examine SYP61 ubiquitination by ATL31 in plants. GFP-SYP61 (56 kDa) was transiently expressed alone or with ATL31-3xFLAG or ATL31C143S-3xFLAG (45 kDa) in N. benthamiana leaves. Extracted proteins were immunoprecipitated with anti-GFP antibody beads and detected with anti-GFP, anti-ubiquitin (FK2), and anti-K63-linked ubiquitin (Apu3) antibodies. Arrowheads indicate ubiquitinated GFP-SYP61. The numbers show the relative intensities of the di-Ub bands (the bands indicated by the lower arrowhead in the anti-ubiquitin blot) normalized by the band intensity of GFP-SYP61.
As upshifted bands of ubiquitinated GST-SYP61 were also observed with the ubiquitin protein without lysine 63 (Ub-K63R), ATL31 can also catalyze other types of ubiquitination, such as multiple-mono or other chain types (Supplemental Figure S9A). Nevertheless, the anti-K63 ubiquitination antibody did not detect the di-ubiquitination of GST-SYP61 with Ub-K63R, whereas the band was detected with the K63-only ubiquitin mutant, which can only form K63-linked chains, further confirming the existence of the K63-linked chain (Supplemental Figure S9, A and B). Mass spectrometry showed that five lysine residues in SYP61, including four in the SNARE domain, were ubiquitinated by ATL31 in vitro (Figure 7B;Supplemental Table S1).
We also confirmed that ATL31 ubiquitinates SYP61 in plant cells. We expressed GFP-SYP61 alone or with 3xFLAG-tagged ATL31 (ATL31-3xFLAG) or ATL31C143S (ATL31C143S-3xFLAG) in N. benthamiana leaves by agroinfiltration. The co-expression of ATL31-3xFLAG enhanced the ubiquitination of SYP61 including the K63-linked chain (Figure 7C). The enhanced signal was not observed with the catalytically inactive form of ATL31, indicating that ATL31 enhances the ubiquitination of SYP61 via its ubiquitination activity.
Ubiquitination of SYP61 is affected by C/N-nutrient availability
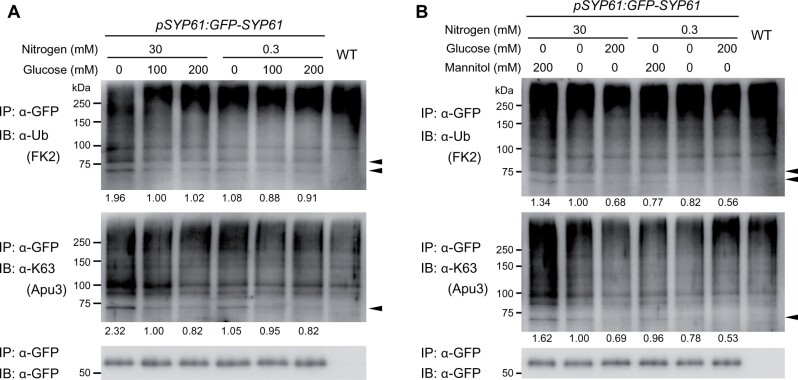
Finally, we investigated the relevance of SYP61 ubiquitination in plant C/N-nutrient responses (Figure 8A). To examine the effects of gradually changing C/N-nutrient conditions, transgenic Arabidopsis pSYP61:GFP-SYP61 plants were grown in liquid medium containing 100-mM Glc/30-mM N for 10 days and treated with six different types of medium for 3 h: medium containing 0-, 100-, or 200-mM Glc and 30- or 0.3-mM N. Interestingly, the signal of ubiquitinated SYP61 was highest in samples treated with 0-mM Glc/30-mM N. The difference in the signal of K63-linked ubiquitin was even more pronounced. The signal was higher in 0-mM Glc-treated samples than under the other glucose conditions with the same nitrogen status. Comparing the two 0-mM Glc-treated samples under different N conditions, the sample treated with 30-mM N showed higher band intensity than the sample treated with 0.3-mM N. The overall ubiquitination profiles did not show this tendency, suggesting that the ubiquitination of SYP61 is specifically regulated by these conditions (Supplemental Figure S10). The ubiquitination signals of SYP61 were not reduced by the addition of mannitol instead of glucose, which confirmed that the observed difference was not due to osmotic conditions (Figure 8B).
Figure 8.
Ubiquitination of SYP61 is affected by C/N-nutrient availability. A and B, Immunoblot analysis to examine the effect of C/N-nutrient availability on SYP61 ubiquitination. GFP-SYP61 (55 kDa)-expressing Arabidopsis plants (pSYP61:GFP-SYP61) were grown for 10 days in liquid medium containing 100-mM glucose/30-mM nitrogen and treated with medium containing the indicated concentrations of nitrogen and glucose (A), or nitrogen, glucose, and mannitol (B) for 3 h. Extracted proteins were immunoprecipitated with anti-GFP antibody beads and detected with anti-GFP, anti-ubiquitin (FK2), and anti-K63-linked ubiquitin (Apu3) antibodies. Arrowheads indicate ubiquitinated GFP-SYP61. The numbers indicate the relative intensities of the di-Ub bands (the bands indicated by the lower arrowhead in the anti-ubiquitin blot) normalized by the band intensity of GFP-SYP61.
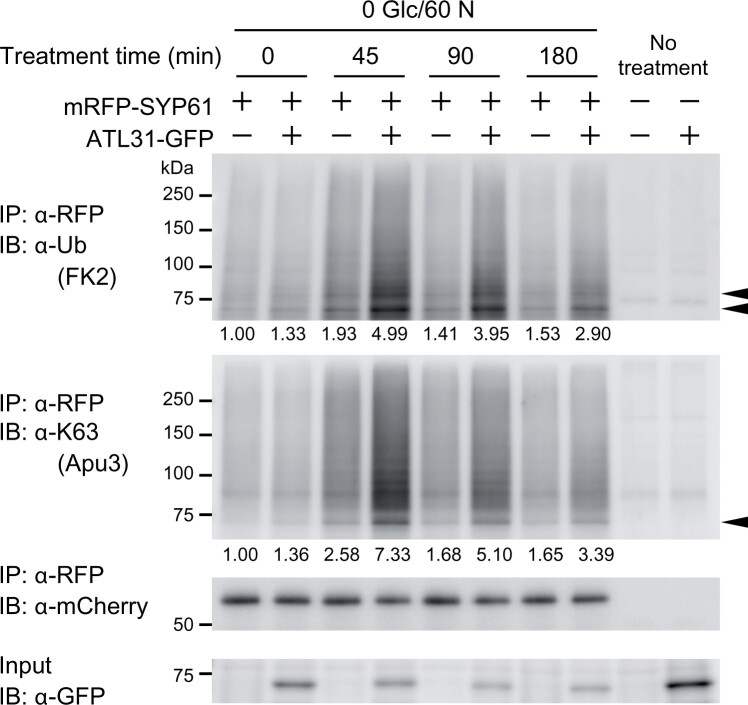
As the ubiquitination of SYP61 was upregulated under lower C/higher N conditions, we used 0-mM Glc/60-mM N low C/high N treatment in the following experiments. To test if ATL31 mediates the low C/high N-induced ubiquitination of SYP61, we performed an experiment with transgenic Arabidopsis plants expressing mRFP-SYP61 in the ATL31-overexpressing background (pSYP61:mRFP-SYP61 in the 2xp35S:ATL31-GFP background). We grew the plants in liquid medium containing 100-mM Glc/30-mM N for 11 days and treated them with low C/high N medium containing 0-mM Glc and 60-mM N for 0, 45, 90, or 180 min (Figure 9). The levels of total and K63-linked ubiquitination of mRFP-SYP61 were highest 45 min after treatment and gradually decreased in the wild-type background. The tendency was the same in the ATL31-overexpressing background, but the amplitude was much higher. We also performed this experiment using transgenic plants expressing GFP-SYP61 in the ATL31 and ATL6 double knockout mutant background (pSYP61:GFP-SYP61 in the atl31-1 atl6-1 background), as ATL6 is the closest homolog of ATL31, to confirm the necessity of these proteins (Supplemental Figure S11). However, SYP61 ubiquitination was not abolished in the mutant background, and these plants responded to the C/N-nutrient treatment like plants in the wild-type background. Thus, ATL31 is not the only ubiquitin ligase that functions in this process.
Figure 9.
Overexpressing ATL31 enhances low C/high N-nutrient-responsive ubiquitination of SYP61. Immunoblot analysis to examine the effect of ATL31 on SYP61 ubiquitination in low C/high N-nutrient condition. Arabidopsis plants expressing mRFP-SYP61 (53 kDa; pSYP61:mRFP-SYP61) in the WT or ATL31-GFP-overexpressing background (2xp35S:ATL31-GFP) were grown for 11 days in liquid medium containing 100-mM glucose/30-mM nitrogen and treated with medium containing 0-mM glucose/60-mM nitrogen for the indicated time. WT and 2xp35S:ATL31-GFP plants were used as negative controls. Extracted proteins were immunoprecipitated with anti-RFP antibody beads and detected with anti-mCherry, anti-ubiquitin (FK2), and anti-K63-linked ubiquitin (Apu3) antibodies. ATL31-GFP in the crude extract was detected with anti-GFP antibody. Arrowheads indicate ubiquitinated mRFP-SYP61. The numbers represent the relative intensities of the di-Ub bands (the bands indicated by the lower arrowhead in the anti-ubiquitin blot) normalized by the band intensity of mRFP-SYP61. Glc, glucose; N, nitrogen; unit, mM.
To test the possibility that the ubiquitination of SYP61 enhances its internalization from the plasma membrane, we quantified the amount of mRFP-SYP61 in the plasma membrane using the plant materials and C/N-nutrient conditions mentioned above (Supplemental Figure S12). The proportion of plasma membrane-localized mRFP-SYP61 decreased with decreasing C/increasing N conditions. However, the amount of plasma membrane-localized SYP61 did not decrease in response to ATL31 overexpression. Therefore, the ATL31-mediated ubiquitination of SYP61 does not likely serve as an endocytosis signal.
Discussion
The SNARE protein SYP61 mediates post-Golgi membrane trafficking in Arabidopsis (Sanderfoot et al., 2001; Uemura et al., 2004). In this study, we identified SYP61 as an interactor of the ubiquitin ligase ATL31. Our results suggest that the cooperative function of SYP61 with ATL31, including the proper regulation of the trafficking of ATL31 by SYP61, is important for plant adaptation to high C/low N-nutrient stress conditions. Because SYP61 is a Qc-SNARE, the suppressed expression of SYP61 protein might have caused the low efficiency of membrane fusion at the TGN/EE in transgenic Arabidopsis, resulting in the dispersion of ATL31-GFP-positive compartments.
We also determined that SYP61 is subjected to K63-linked ubiquitination in plant cells. Furthermore, low C/high N-nutrient conditions enhance the ubiquitination of SYP61, and ATL31 promotes this process. ATL31 catalyzes the mono- and K63-linked ubiquitination of SYP61 in vitro and in plant cells, although it is not the only ubiquitin ligase with this activity. The ubiquitination of SYP61 detected in this study might be of multiple types with different molecular meanings. Our mass spectrometry analysis identified several sites of SYP61 that were directly ubiquitinated by ATL31 in vitro. In the in planta assays, while ubiquitination bands of higher molecular weight than di-Ub bands were detected by anti-ubiquitination antibody, the bands were not clearly detected by a K63-chain-specific antibody. Thus, the bands that appeared above di-Ub in the gel might contain SYP61 with non-K63-linked chains or with multiple mono-ubiquitination. These observations point to the heterogeneity of the ubiquitination of SYP61.
Both the physiological and molecular functions of SYP61 ubiquitination remain open questions, but several conclusions can be drawn from our results. First, it is not likely that ATL31 targets SYP61 to enhance plant growth under high C/low N-nutrient conditions, because these conditions downregulated SYP61 ubiquitination. Second, the ATL31-mediated ubiquitination of SYP61 does not likely serve as an endocytosis signal, while the possibility that some form of low C/high N-mediated ubiquitination plays a role in the endocytosis pathway cannot be excluded. Because both ATL31 and SYP61 colocalized with TGN/EE and LE markers, it would be interesting to examine if ATL31-mediated ubiquitination modifies the SYP61-mediated trafficking pathway in these compartments.
A recent report indicates that the TGN/EE marked by SYP61 harbors both a secretory-trafficking zone with the R-SNARE VAMP721 and a vacuolar-targeting zone with VAMP727 (Shimizu et al., 2021). In response to flg22 treatment, the proportion of the hybrid compartment of the TGN/EE and LE marked by SYP61 and ARA7/RABF2b increases temporarily to mediate the endocytic trafficking of FLS2 (Choi et al., 2013). SYP61 localizes not only to the TGN/EE, but also to the plasma membrane and the vacuolar membranes under specific conditions (Gendre et al., 2013; Hachez et al., 2014; Rosquete et al., 2019; Heinze et al., 2020). This dynamic and seemingly undiscriminating behavior of SYP61 suggests that as-yet-unknown post-translational mechanisms are involved in regulating SYP61 localization and function in response to environmental stimuli and that ubiquitination might be involved in the process.
The few studies on the ubiquitination of SNARE proteins reported to date have revealed the involvement of this process in several different molecular pathways. In yeast, ubiquitination of Tlg1, the SYP61 ortholog, and a Qa-SNARE Pep12 serves as a signal for vacuolar degradation through multivesicular bodies (Reggiori and Pelham, 2002; Valdez-Taubas and Pelham, 2005). Poly-ubiquitination of the Rat Qa-SNARE Syntaxin1 induces its proteasomal degradation (Chin et al., 2002). On the other hand, mono-ubiquitination of the Qa-SNARE Syntaxin3 triggers nondegradative endocytosis in human epithelial cells (Giovannone et al., 2017). Also, K63-linked ubiquitination of Snc1, an yeast R-SNARE that mediates trafficking between the plasma membrane and the TGN, functions as a nondegradative trafficking signal that facilitates its recycling back from the endocytic pathway to the exocytic pathway (Xu et al., 2017). The mono-ubiquitination and subsequent deubiquitination of the human Qa-SNARE Syntaxin5 serve as a gate for the time-specific formation of SNARE complexes to regulate Golgi membrane fusion during the cell cycle (Huang et al., 2016). K63-linked ubiquitination of the human R-SNARE Sec22b is also thought to be involved in SNARE complex formation during interactions with the bacterial pathogen Legionella pneumophila (Kitao et al., 2020). In plants, the plasma membrane-localized Qa-SNARE SYP122 was identified in a K63-linked ubiquitinome analysis in Arabidopsis (Johnson and Vert, 2016), although its upstream and downstream molecular effects have not been characterized. SNARE ubiquitination appears to function as various molecular signals in a wide range of species.
Our results also point to the multifunctionality of ATL31. Our mass spectrometry (MS)-based analysis indicated that ATL31 has the ability to catalyze multiple ubiquitin chain types. As ubiquitination chains are formed by the attachment of a donor ubiquitin molecule onto an acceptor ubiquitin molecule, the ubiquitin chain type is directly determined by the positioning of the donor and acceptor ubiquitin molecules (Deol et al., 2019). Therefore, when a ubiquitination chain forms on a ubiquitin molecule that is already bound to the target, all of the proteins in the complex serve as determinants of the chain type. For a RING-type ubiquitin ligase like ATL31, the major determinants will be E2, the ubiquitin ligase itself, and the target proteins (Deol et al., 2019; Romero-Barrios et al., 2019).
The ubiquitination types catalyzed by ATL31 in plant cell require further investigation. While we used human UbcH5a as the E2 in our in vitro assay, the Arabidopsis genome contains 37 E2 genes (Callis, 1995). The types of ubiquitin chains that form via ATL31 will be affected by which E2 protein it interacts with. It is also possible that other co-factors and the post-translational modification of ATL31 have effects on this process. Nevertheless, our data demonstrate the potential of ATL31 to catalyze these multiple types of ubiquitination, suggesting that ATL31 might mediate multiple cellular signals via the ubiquitination of distinct target proteins. We previously demonstrated that ATL31 targets 14-3-3 proteins for proteasomal degradation via phosphorylation by CIPK7/12/14 kinases under high C/low N-nutrient stress conditions (Sato et al., 2011; Yasuda et al., 2014, 2017). Thus, the activity of ATL31, including the target interactions and possibly the type of ubiquitin linkages, appears to be tightly regulated in response to environmental conditions. The ubiquitination of SYP61 by ATL31 might also be activated by as-yet-unknown upstream signals.
In addition to C/N-nutrient responses, ATL31 and SYP61 might jointly function in other environmental responses. In addition to high C/low N-nutrient stress responses, ATL31 positively regulates plant resistance to pathogen attack and possibly to salt stress (Maekawa et al., 2012, 2014; Peng et al., 2014; Wibowo et al., 2016). ATL31 enhances callose deposition during plant immunity (Maekawa et al., 2012, 2014). ATL31 targets fungal penetration sites in association with SYP121/PEN1 and accelerates the formation of rigid cell walls called papillae (Maekawa et al., 2014), suggesting that ATL31 might mediate the vesicle trafficking of cell wall material and/or the related enzymes. SYP61 is also involved in salt-stress tolerance and cell wall secretion. The osm1 mutant is hypersensitive to the salt and osmotic stress (Zhu et al., 2002) and shows disrupted localization of the plasma membrane aquaporin PIP2;7 (Hachez et al., 2014). Proteomic analysis of the SYP61 compartment identified cell wall-related enzymes as potential cargos (Drakakaki et al., 2012), and a recent glycomic analysis indicated that SYP61 also mediates the trafficking of cell wall materials, such as polysaccharides and glycoproteins (Wilkop et al., 2019). In addition to the overlap in the subcellular localizations of ATL31 and SYP61 and their physical interaction, these functional similarities of ATL31 and SYP61 point to their possible cooperation in diverse physiological processes in plants.
In this study, we demonstrated the importance of the regulation of membrane trafficking by a SNARE protein for plant nutrient responses. Our data also suggest the possibility that SNARE proteins are regulated via ubiquitination in response to nutrient conditions. Further studies should reveal the downstream effects of SNARE ubiquitination, as well as how common this modification is among SNARE proteins. It will also be interesting to identify additional cargos whose localizations are regulated in response to C/N-nutrient conditions. Our findings provide important insights into the membrane trafficking machinery and ubiquitin signaling during plant nutrient responses.
Materials and methods
Plant material and growth conditions
The Arabidopsis thaliana ecotypes Columbia-0 (Col-0) and C24 were used in this study. The T-DNA insertion mutant osm1 (Zhu et al., 2002) was kindly provided by Dr Jianhua Zhu (University of Maryland). The T-DNA insertion mutants atl31-1 and atl6-1 were characterized previously (Sato et al., 2009). Transgenic Arabidopsis plants, 2xp35S:ATL31-GFP, pSYP61:GFP-SYP61, p35S:ST-mRFP, pSYP61:mRFP-SYP61, pSYP43:mRFP-SYP43, pVHAa1:VHAa1-mRFP, pARA7:mRFP-ARA7, and pVAMP727:tagRFP-VAMP727 have been described (Fecht-Bartenbach et al., 2007; Sato et al., 2009; Uemura et al., 2012; Inada et al., 2016; Shimizu et al., 2021). pVHAa1:VHAa1-mRFP was kindly provided by Dr Karin Schumacher (Heidelberg University, Germany). The SYP61 amiRNA constitutively expressing or inducible lines were generated as described below. Co-expression lines were generated by crossing, and F1 or homozygous lines of the F3 generation were used for microscopic observation and carbon/nitrogen-nutrient response analysis. For seed production, Arabidopsis seeds were surface-sterilized and sown on 1× Murashige and Skoog (MS) medium supplemented with 1% sucrose, vitamins, and 0.4% gellan gum (pH 5.7). Following incubation in the dark at 4°C for 2–4 days to synchronize germination, the plants were grown under a 16-h light/8-h dark cycle at 22°C on the plates for several weeks and transferred to pots with soil containing compost and vermiculate at a ratio of 1:6. The fluorescent lamps TOSHIBA FL20SW and NEC FL40SW were used, and the photosynthetic photon flux density was ∼40 to 50 µmol m−2 s−1. Tap water was used for watering. Nicotiana benthamiana plants were used for transient protein expression. Surface-sterilized seeds were sown on 1× MS medium supplemented with 1% sucrose, vitamins, and 0.4% gellan gum (pH 5.7), grown under a 16-h light/8-h dark cycle at 22°C on the plates for 2 weeks, and transferred to pots with soil containing compost and vermiculate at a ratio of 1:6. Tap water was used for watering, and the plants were supplied with 1/1,000 diluted Hyponex (HYPONeX JAPAN CORP, Japan) once a week.
For microscopy imaging of Arabidopsis root cells, seeds were grown vertically on modified MS medium containing 100-mM glucose, 30-mM nitrogen (10-mM NH4NO3 and 10-mM KNO3), vitamins, and 0.8% agar (pH 5.7). Modified MS medium was prepared as previously described (Sato et al., 2009; Huarancca Reyes et al., 2018). For vacuolar accumulation analysis, 2xp35S:ATL31-GFP seedlings were wrapped in a double layer of aluminum foil 18 h before observation, and 6-day-old plants were observed. For microscopy of syp61-inducible amiRNA lines, plants were grown for 7 days on modified MS medium containing 50-mM glucose, 30-mM nitrogen (10-mM NH4NO3 and 10-mM KNO3), vitamins, and 0.8% agar (pH 5.7) supplied with 10 µM of β-estradiol or an equal amount of ethanol.
To detect the ubiquitination of SYP61 in Arabidopsis, seeds were grown in liquid 1× MS medium supplemented with 1% sucrose and vitamins (pH 5.7) for 10 days under constant light with shaking at 70 rpm.
To test the effect of C/N-nutrient conditions on SYP61 ubiquitination, seeds were grown in modified MS liquid medium containing 100-mM glucose, 30-mM nitrogen (10-mM NH4NO3 and 10-mM KNO3), vitamins, and 2-mM MES (pH 5.7) for 10 or 11 days under constant light with shaking at 70 rpm and treated with MS medium containing different concentrations of glucose, nitrogen, and mannitol for the indicated time.
Plant transformation
The artificial microRNA (amiRNA) was designed to target SYP61 using the online WMD interface (http://wmd2.weigelworld.org;Schwab et al., 2006; Ossowski et al., 2008). The amiRNA construct was amplified by polymerase chain reaction (PCR) using the primers listed in Supplemental Table S2 and the plasmid pRS300 (Ossowski et al., 2008) as template. The PCR product was subcloned into pENTR/D-TOPO and transferred to the binary vector pGWB402-omega for the constitutive knockdown lines (Nakagawa et al., 2007) or the pMDC7B vector (Curtis and Grossniklaus, 2003) for the β-estradiol-inducible lines by LR recombination according to the manufacturer’s protocol (Invitrogen). To generate transgenic Arabidopsis plants, these constructs were introduced into Agrobacterium tumefaciens GV3101 (pMP90) by electroporation, followed by transformation of Arabidopsis plants (Col-0) by the floral dip method (Clough and Bent, 1998).
Transient expression in N. benthamiana
For plasmid construction, the coding sequences of each gene were amplified by PCR and cloned into vector pENTR/D-TOPO (Life Technologies). These fragments were subsequently transferred to destination vectors using the Gateway system according to the manufacturer’s protocol (Invitrogen). For immunoprecipitation assays, the coding sequences of ATL31 and ATL31C143S were subcloned into the pGWB11 destination vector, and SYP61 (At1g28490) was subcloned into the pGWB6 and pGWB12 destination vectors (Nakagawa et al., 2007). For in vivo ubiquitination analysis, the coding sequences of ATL31 and ATL31C143S were transferred into the pAMPAT-GW-3xFLAG destination vector (Yamada et al., 2016), and using them as templates, the sequences from ATL31 or ATL31C143S to 3xFLAG were subcloned into the pGWB502-Ω destination vector (Nakagawa et al., 2007). The primers are listed in Supplemental Table S3. All constructs were introduced into Agrobacterium tumefaciens strain GV3101 (pMP90) by electroporation. Agrobacterium tumefaciens cells containing the constructs were grown in 2xYT liquid medium at 28°C overnight with shaking, and the cells were resuspended in infiltration buffer (10-mM MES, 10-mM MgCl2, and 450-µM acetosyringone, pH 5.6). The suspension mixtures were infiltrated into the leaves of 5- to 6-week-old N. benthamiana plants using a needleless syringe. Agrobacterium tumefaciens carrying the p19 suppressor was co-infiltrated with the all samples (Takeda et al., 2002). After 3 days, the leaves were harvested and immediately frozen in liquid nitrogen.
Immunoblotting analysis of ATL31-GFP following ConcA treatment
The 2xp35S:ATL31-GFP transgenic plants were grown in modified MS liquid medium containing 100-mM glucose, 30-mM nitrogen (10-mM NH4NO3 and 10-mM KNO3), vitamins, and 2-mM MES (pH 5.7) for 10 days under constant light with shaking at 70 rpm, followed by treatment with medium containing 1-µM ConcA (SIGMA-ALDRICH, C9705) or the same amount of DMSO (1:200 dilution) for 3 h. The plants were frozen in liquid nitrogen and ground to a powder with stainless steel beads. Approximately 6-µL/mg-FW of 1× SDS-sample buffer was directly added to the samples. 1× SDS was prepared by diluting 2× solution (125-mM Tris–HCl [pH 6.8], 4% SDS, 20% glycerol, 10% 2-mercaptoethanol, 0.02% bromophenol blue) in protein extraction buffer (50-mM Tris–HCl [pH 7.5], 0.5% Triton X-100, 150-mM NaCl, 10% glycerol, 1-mM EDTA, pH 7.5). The extracts were incubated at 55°C for 40 min, and the debris was removed by two rounds of centrifugation at 20,000g for 5 min each time. Proteins were separated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) and detected by immunoblotting with anti-GFP (MBL, 598) and anti-alpha-Tubulin (DM1A; Calbiochem, CP06-100UGCN). CBB staining was performed using AE-1340 EzStain AQua (ATTO, 2332370) according to the manufacturer’s protocol. The band intensity of full-length ATL31-GFP and free GFP were quantified using ImageJ software (NIH, Bethesda, MD, USA), and the ratio was calculated.
Immunoprecipitation
The plant materials were frozen in liquid nitrogen, ground to a powder with a mortar and pestle, and the expressed proteins extracted using protein extraction buffer (50-mM Tris–HCl [pH 7.5], 0.5% Triton X-100, 150-mM NaCl, 10% glycerol, 1-mM ethylenediaminetetraacetic acid (EDTA)) supplemented with 10-µM MG132 and Complete Protease Inhibitor Mixture (Roche Applied Science). To detect ubiquitination, the deubiquitination inhibitors 20-mM N-ethylmaleimide and 2-mM 1,10-phenanthroline were added to the samples. To test the in planta ubiquitination of SYP61 by ATL31, and the effect of C/N-nutrient conditions, 500 nM of MLN-7243 (also known as TAK-243; Active Biochem, A-1384) was also added as an E1 inhibitor to avoid additional ubiquitination during the extraction steps. The mixtures were centrifuged at 20,000g for 5 min at 4°C and the supernatants were collected. This step was repeated twice, and the proteins were then immunoprecipitated with anti-FLAG M2 affinity gel (Sigma-Aldrich, M8823), anti-GFP mAb-magnetic agarose (MBL, D153-10), anti-GFP mAb-Magnetic Beads (MBL, D153-11), or anti-RFP mAb-Magnetic Beads (MBL, M165-11). D153-11 was used to examine the ubiquitination of GFP-SYP61 in response to C/N-nutrient conditions, and D153-10 was used for other experiments to immunoprecipitate GFP-tagged proteins. The extracts and beads were mixed, incubated with rotation at 4°C for 1 h, and washed more than three times with extraction buffer without inhibitors. To detect FLAG-SYP61 ubiquitination, proteins were eluted with 150 µg·mL−1 3× FLAG peptide (Sigma-Aldrich, F4799) in extraction buffer, followed by precipitation in cold acetone, resuspension in SDS sample buffer (62.5-mM Tris–HCl [pH 6.8]), 2% SDS, 10% glycerol, 5% 2-mercaptoethanol, 0.01% bromophenol blue), and incubation at 90°C for 5 min. In other experiments, proteins were eluted with SDS sample buffer at 90°C for 5 min or 55°C for 30 min.
Proteins were separated by SDS–PAGE and detected by immunoblotting with anti-FLAG (SIGMA-ALDRICH, F1804-200UG), anti-SYP61, anti-GFP (MBL, 598), anti-mCherry (Clontech, 632543), anti-ubiquitin (FK2; Wako, 302-06751), anti-K48 ubiquitination (Apu2; Millipore, 05-1307), and anti-K63 ubiquitination (Apu3; Millipore, 05-1308) antibodies. Anti-SYP61 antibody was generated in a rabbit by immunization with recombinant GST-SYP61 as described below. The band intensity was quantified using ImageJ software.
Anti-SYP61 antibody
GST-SYP61 (1–218) was expressed in Escherichia coli strain BL21 (DE3) using the pGEX 6P-1 vector (GE Healthcare). The cells expressing fusion proteins were collected, resuspended in BugBuster Protein Extraction Reagent (MercK), and centrifuged at 10,000g for 20 min. The supernatants were loaded onto a glutathione–Sepharose 4B column (GE Healthcare) and washed with washing buffer (phosphate-buffered saline (PBS) + 1% Triton), and the fusion proteins were eluted with elution buffer (50-mM reduced glutathione, 50-mM Tris, pH 8.0, 150-mM NaCl, and 0.1% mercaptoethanol). Purified GST-SYP61 was used as an antigen to raise the anti-SYP61 polyclonal antibody. The antibody was purified by protein G affinity column chromatography (GE Healthcare; Supplemental Figure S13).
Split ubiquitin yeast two-hybrid assay
All assays were performed using the yeast strain L40ccua (MATa his3Δ200 trp1-901 leu2-3,112 LYS2::(lexAop)4-HIS3 ura3::(lexAop)8-lacZ ADE2::(lexAop)8-URA3 gal80 canR cyh2R). The full-length coding sequences of ATL31 and ATL31C143S were subcloned into the destination vector pMetYC_GW, containing the C-terminal half of ubiquitin, and the full-length coding sequence of SYP61 was subcloned into the destination vector pNX32_GW, containing the N-terminal half of ubiquitin (Obrdlik et al., 2004). The empty vector pNX32_GW was used as a negative control. The constructs were transfected into yeast using a Frozen-EZ Yeast Transformation II Kit (Zymo Research) according to the manufacturer’s protocol, with growth assessed as described in the Yeast Protocols Handbook (Clontech).
Recombinant protein expression and purification for the in vitro ubiquitination assay
GST-tagged truncated SYP61, which included amino acid residues 1–218, was expressed in E. coli strain BL21(DE3) pLysS (Novagen) and purified with Glutathione-Sepharose 4B (GE Healthcare, 17-0756-01) following the manufacturer’s protocol.
In vitro ubiquitination assay
In vitro ubiquitination assays were performed as described (Sato et al., 2009) with some modifications. For quantitative analysis of ubiquitin chain type with AQUA peptide, 500 ng of MBP-ATL31 was incubated for 0–3 h at 30°C in 30 µL of reaction mixture containing 50-ng E1 (Wako, 219-01111), 50-ng UbcH5a for E2 (Wako, 215-01191), 4-µg ubiquitin (Sigma-Aldrich, U6253), 25-µM MG132, 40-mM Tris–HCl (pH 7.5), 5-mM MgCl2, 2-mM ATP, and 2-mM dithiothreitol (DTT). For ubiquitination analysis of GST-SYP61, 500 ng of GST-SYP61 or GST was incubated with 500 ng of MBP-ATL31 for 0–3 h at 30°C in 30 µL of reaction mixture containing 50-ng E1 (Wako, 219-01111), 51-ng UbcH5a for E2 (Wako, 215-01191), 4-µg ubiquitin (Sigma-Aldrich, U6253), 25-µM MG132, 40-mM Tris–HCl (pH 7.5), 5-mM MgCl2, 2-mM ATP, and 2-mM DTT. For the ubiquitination assays with mutated ubiquitins, 500 ng of GST-SYP61 or GST was incubated with 500 ng of MBP-ATL31 for 0–1 h at 30°C in 30 µL of reaction mixture containing 200-nM E1 (R&D Systems, E-305-025), 250-ng UbcH5a for E2 (Wako, 215-01191), 6-µg ubiquitin proteins (native ubiquitin: Ubiquitin human, U-100H, Boston Biochem; K63R ubiquitin: rhUbiquitin K63R, UM-K63R, Boston Biochem; K63 only ubiquitin: rhUbiquitin K63 only, UM-K630, Boston Biochem), 25-µM MG132, 40-mM Tris–HCl (pH 7.5), 5-mM MgCl2, 2-mM ATP, and 2-mM DTT. Reactions were stopped by adding the same amount of 2× SDS sample buffer (125-mM Tris–HCl [pH 6.8], 4% SDS, 20% glycerol, 10% 2-mercaptoethanol, 0.02% bromophenol blue). Proteins were separated by SDS–PAGE and detected by immunoblotting using anti-MBP (New England BioLabs, E8032S), anti-GST (MBL, M071-3), anti-ubiquitin (FK2; Wako, 302-06751), anti-K48 ubiquitination (Apu2; Millipore, 05-1307), and anti-K63 ubiquitination (Apu3; Millipore, 05-1308) antibodies.
Mass spectrometry analysis of ubiquitination sites
Ubiquitinated GST-SYP61 proteins were separated by SDS–PAGE and stained with SYPRO Ruby (Lonza, Switzerland) as described in the manufacturer’s protocol. Gel pieces containing ubiquitinated GST-SYP61 were excised from the gel, dehydrated with 100% acetonitrile, and incubated in 10-mM dithiothreitol and 50-mM ammonium bicarbonate for 45 min at 56°C with shaking. The gel pieces were subsequently incubated in 55-mM chloroacetamide/50-mM ammonium bicarbonate for 30 min at room temperature, washed with 25-mM ammonium bicarbonate, and dehydrated with 100% acetonitrile. The dried gels were incubated for 16 h at 37°C in 50-mM ammonium bicarbonate containing sequence grade modified trypsin (Trypsin Gold; Promega, USA). The digested peptides were eluted from the gels with 50% acetonitrile (v/v)/5% formic acid (v/v) and dried using an evaporator. The peptides were dissolved in 2% acetonitrile (v/v)/0.1% formic acid (v/v) and filtered through Ultrafree-MC Centrifugal Filters (polyvinylidene fluoride (PVDF) 0.45 μm; Millipore, USA). Peptides were identified using an EASY-nLC 1000 liquid chromatograph coupled to an Orbitrap Mass Spectrometer (Thermo Scientific, USA), followed by assessment using the SEQUEST algorithm embedded in Proteome Discoverer 1.4 software (Thermo Scientific, USA) against TAIR10 (http://www.arabidopsis.org/index.jsp), as described (Lu et al., 2016).
Quantitative analysis of ubiquitin chain type with AQUA peptide
Tandem mass spectrometry (MS/MS)-based AQUA of ubiquitin peptides by PRM was performed as described previously (Tsuchiya et al., 2017). Proteins were separated by a short run (1 cm) on 4%–12% Nu-PAGE gels, and the gel region corresponding to molecular weight >75 kDa was subjected to trypsinization. Trypsinized peptides were extracted, spiked with ubiquitin AQUA peptides, and analyzed in targeted MS/MS mode on a Q Exactive mass spectrometer coupled to an EASY-nLC 1000 (Thermo Scientific). Raw data were processed using PinPoint software version 1.3 (Thermo Scientific).
Gene expression analysis
Total RNA was isolated from 7-day-old Arabidopsis seedlings using TRIzol reagent (Invitrogen) and treated with RQ1 RNase-free DNase (Promega) according to the manufacturer’s protocols. cDNA was synthesized using oligo(dT) primer (Promega) with 18S rRNA-specific primers and ReverTraAce reverse transcriptase (Toyobo). Quantitative real-time PCR (qRT-PCR) analysis was performed using SYBR premix Ex Taq (TaKaRa) and Mx3000P (Agilent Technologies) with MxPro Software version 4.10 (Agilent Technologies) according to the manufacturer’s protocol. The primers are listed in Supplemental Table S4.
Carbon/nitrogen-nutrient response analysis
Seeds of each genotype were surface-sterilized and sown on modified MS-based solid medium containing 0.4% gellan gum, vitamins, glucose, mannitol, and nitrogen (KNO3 and NH4NO3 were used at a 1:1 molar ratio) at the concentration indicated in the figures. The medium was prepared as described (Sato et al., 2009; Huarancca Reyes et al., 2018). Ten micromolar of β-estradiol or an equal amount of ethanol was added to the medium for the experiment with syp61 inducible amiRNA lines. The seeds were incubated in the dark at 4°C for 2 days to synchronize germination and grown under a 16-h light/8-h dark cycle at 22°C. The percentage of germinated seeds with green cotyledons was calculated.
Chemical treatment of samples for microscopy
Five- or 6-day-old seedlings were incubated in modified liquid MS medium with 100-mM glucose/30-mM nitrogen containing 50-μM BFA or 33-μM WM. Fluorescence was monitored by confocal microscopy 30 min and 1 h later. For BFA treatment, seedlings were pre-incubated in 5-μM FM4–64 dye for 15 min and transferred to BFA-containing medium.
For ConcA treatment, 7-day-old seedlings were incubated in modified liquid MS medium with 100-mM glucose/30-mM nitrogen containing 1-µM ConcA (SIGMA-ALDRICH, C9705) or the same amount of DMSO (1:200 dilution) and observed by confocal microscopy 3 h later.
FM4–64 internalization assay
Seven-day-old seedlings were pulse-stained with 2-µM FM4–64 dye in modified liquid MS medium containing 100-mM glucose/30-mM nitrogen for 2 min, washed twice with dye-free medium, and observed by microscopy.
C/N-nutrient treatment of samples for microscopy
For colocalization analysis of ATL31-GFP and the organelle markers, 6-day-old seedlings were treated with modified liquid MS medium containing the indicated amounts of glucose and nitrogen and observed 90–110 min after treatment. To analyze the plasma membrane localization of mRFP-SYP61, 5-day-old plants were subjected to overnight treatment, and 6-day-old seedlings were observed.
Confocal laser-scanning microscopy
Confocal laser-scanning microscopy was performed using a Zeiss LSM510 microscope equipped with a C-Apochromat 40×/1.20 W Korr UV-VIS-IR lens, an LSM980 microscope with a Plan-Apochromat 63×/NA 1.4 Oil objective lens, and a Nikon Ti-E inverted microscope equipped with Plan Apo λ 60× Oil lens and a Nikon A1Rsi spectral imaging confocal scanning system. For observation under the Zeiss LSM510 microscope, GFP fluorescence was excited by a 488 nm laser and detected using a 505–550 nm band-pass emission filter, whereas RFP fluorescence was excited by a 561-nm laser and detected using a 575–615 nm band-pass emission filter. FM4–64 fluorescence was excited by a 561-nm DPSS laser and detected using a 575-nm long-pass emission filter. For observation under the Zeiss LSM980 microscope, fluorescence was excited by a 488-nm laser for GFP and 543 nm for RFP, and the fluorescence emissions were detected at 490–543 nm for GFP and 596–694 nm for RFP. For observation with the Nikon microscope system, GFP fluorescence was excited by a 488-nm laser and detected using a 500–550-nm band-pass emission filter, and RFP and FM4–64 fluorescence were excited by a 561-nm laser and detected using a 570–620 nm band-pass emission filter. Images were processed using ImageJ software.
Colocalization analysis
Object-based distance analysis was performed using the ImageJ plugin DiAna (Gilles et al., 2017). To remove the noise, a median filter of 2 pxl-radius was utilized before analysis. To analyze untreated plants, the dot-like structures were segmented by DiAna-segmentation with the following parameters: spot segmentation; RadXY, 3; RadZ, 1; Noise, 50; Radius Max (pxl), 7; sd value, 1.4; volume min (pxl), 4; volume max (pxl), 200; exclude objects on XY edges, yes; seed threshold, 3,000 for ATL31-GFP, 2,000 for ST-mRFP, 4,000 for VHAa1-mRFP, 3,000 for mRFP-SYP43, 2,800 for mRFP-SYP61, 3,000 for tagRFP-VAMP727, and 3,500 for mRFP-ARA7. To analyze plants following C/N-nutrient liquid medium treatments, the following parameters were used: spot segmentation; RadXY, 3; RadZ, 1; Noise, 50; Radius Max (pxl), 7; sd value, 1.4; volume min (pxl), 4; volume max (pxl), 200; exclude objects on XY edges, yes; seed threshold, 5,500 for ATL31-GFP, 4500 for VHAa1-mRFP and tagRFP-VAMP727. The segmented objects with signal at the plasma membrane and the outside of the cell of interest were removed manually. After segmentation, the distance of the centroids of the closest neighbor was measured by DiAna-analysis. The objects with a center–center distance shorter than the theoretical resolution were considered to be colocalizing. The optical resolution was considered to be 0.2 µm (according to the Rayleigh criterion, the theoretical lateral resolution was calculated with the following formula: 0.51λ/NA (λ, excitation wavelength; NA, numerical aperture of the objective lens) = 0.51*543/1.40 = 197.8 nm < 0.2 µm). n = 15 cell slices from five roots were analyzed.
Analysis of the dot/cytosol intensity ratio of ATL31-GFP
ImageJ was used to analyze the dot/cytosol intensity ratio of ATL31-GFP. A median filter of 2 pxl-radius was used to remove the noise. The dot-like structures of ATL31-GFP were segmented by DiAna with the following parameters: spot segmentation; RadXY, 3; RadZ, 1; Noise, 50; Radius Max (pxl), 7; sd value, 1.4; volume min (pxl), 4; volume max (pxl), 200; exclude objects on XY edges, yes; seed threshold, 16,000. The intracellular region excluding the dot-like structures was considered to be cytosol in this analysis. The original images were used for measurement. The ratio of the mean intensity of the cytosol to the total area of the dot-like structures was calculated. n = 15 cell slices from five roots were analyzed.
Analysis of the PM/dot intensity ratio of mRFP-SYP61
ImageJ was used to analyze the PM/Dot intensity ratio of mRFP-SYP61. A median filter of 2 pxl-radius was used to remove the noise. The dot-like structures of mRFP-SYP61 were segmented by DiAna with the following parameters: iterative thresholding, volume min (pxl), 10; volume max (pxl), 200; min threshold, 200; STEP value, 50. The plasma membrane was selected manually using the selection brush tool. The original images were used for measurement. The ratio of the mean intensity of the plasma membrane to the dot-like structures was calculated. The mean intensity of the dot-like structures was calculated by dividing the total intensity of the dot-like structures by their total area. n = 20 cell slices from five roots were analyzed.
Statistical analysis
All values shown in bar graphs are mean ± standard deviation (sd) or standard error of the mean (sem) as stated. P-values were calculated by two-tailed Welch’s t test for two-group comparisons, by two-tailed Dunnett’s test for multiple-group comparisons with a single control, and by one-way analysis of variance (ANOVA) followed by Tukey’s honestly significant difference test for other multiple-group comparisons. Statistical significance was set based on P-values. ns, P > 0.05; *P < 0.05; **P < 0.01. Detailed ANOVA and t test results are described in Supplemental Data Set S1.
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ATL31, At5g27420; SYP61, AT1g28490; SYP43, AT3G05710; VHAa1, At2g28520; ARA7, AT4G19640; VAMP727, AT3G54300.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. ATL31-GFP accumulates in the vacuole after 18-h dark treatment, whereas mRFP-SYP61 does not.
Supplemental Figure S2. ATL31-GFP is sensitive to Concanamycin A.
Supplemental Figure S3. Subcellular localization of ATL31 in response to C/N-nutrient conditions.
Supplemental Figure S4. Knockdown of SYP61 is induced under β-estradiol treatment, while ATL31 is constitutively overexpressed in the progeny of ATL31-overexpressing and syp61 inducible knockdown lines.
Supplemental Figure S5. SYP61 expression is reduced in syp61 amiRNA constitutive knockdown plants.
Supplemental Figure S6. The osm1 mutant is hypersensitive to C/N-nutrient stress.
Supplemental Figure S7. SYP61 is ubiquitinated in plants.
Supplemental Figure S8. ATL31 has the ability to catalyze several types of ubiquitination.
Supplemental Figure S9. ATL31 ubiquitinates SYP61 in vitro.
Supplemental Figure S10. The overall ubiquitination profile under different C/N-nutrient conditions.
Supplemental Figure S11. Ubiquitination of SYP61 in the atl31 atl6 double knockout mutant.
Supplemental Figure S12. The plasma membrane localization of SYP61 in response to C/N-nutrient conditions in different genetic backgrounds.
Supplemental Figure S13. Recognition of SYP61 by anti-SYP61 antibody.
Supplemental Table S1. Ubiquitinated SYP61 peptides detected by mass spectrometry analysis.
Supplemental Table S2. Primer sequences used for syp61 amiRNA construction.
Supplemental Table S3. Primer sequences used for plasmid construction.
Supplemental Table S4. Primer sequences used for expression analysis.
Supplemental Data Set S1. ANOVA and t test results.
Supplementary Material
Acknowledgments
We thank Drs Tsuyoshi Nakagawa (Shimane University, Japan) and Yoshihisa Ueno (Ryukoku University, Japan) for kindly providing the Gateway destination vectors used in this study. We also thank Drs Bernhard Grimm (Humboldt University, Germany) and Ryouich Tanaka (Hokkaido University, Japan) for kindly providing materials for split ubiquitin yeast two-hybrid assays. The osm1 mutant seeds were kindly provided by Dr Jianhua Zhu (University of Maryland, USA). We thank Drs Yoko Ito (Ochanomizu University, Japan), Emi Nakamura Ito (Ochanomizu University, Japan), Kazuo Ebine (National Institute for Basic Biology, Japan), Ooi-Kock Teh (Hokkaido University, Japan) and the Nikon Imaging Center, Hokkaido University for kind support in microscope analysis, and the Instrumental Analysis Division, Global Facility Center, Creative Research Institution, Hokkaido University for mass spectrometry analysis.
Funding
Y.H. was supported by Japan Society for the Promotion of Science (JSPS) Research Fellowship for Young Scientists [Grant reference number: 17J06430]. T.H.R. was supported by MEXT Honors Scholarship for Privately Financed International Students. R.P.B. was funded by grant from Knut and Alice Wallenberg Foundation. This work was supported by the MEXT KAKENHI [Grant No. JP18H05275 to A.N., Nos. JP15H01167 and JP21H02150 to J.Y., Nos. JP20K05949 and JP21H05644 to T.S., and JP16H06280], JST (CREST [Grant No. JPMJCR20E5 to T.U.]), Sumitomo Foundation (Grant for Basic Science Research Project to T.U.), the NOASTEC foundation (to T.S.), and Hokkaido University (Hokkaido University Young Scientist Support Program to T.S.).
Conflict of interest statement. The authors declare that they have no conflict of interest.
Y.H. and T.S. designed the experiments. Y.H., T.H.R., T.U., A.F., Y.L., Y.M., Y.S., S.M., S.Y., K.M., and Y.F. performed the experiments. A.B. generated the constitutive and inducible SYP61 knockdown lines. Y.H., Y.S., K.T., A.N., J.T., R.P.B., and T.S. analyzed the data. Y.H. and T.S. wrote the article, and J.Y., Y.S., J.T., and R.P.B. edited it. All the authors approved the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Takeo Sato (t-satou@sci.hokudai.ac.jp).
References
- Aoyama S, Huarancca Reyes T, Guglielminetti L, Lu Y, Morita Y, Sato T, Yamaguchi J (2014a) Ubiquitin ligase ATL31 functions in leaf senescence in response to the balance between atmospheric CO2 and nitrogen availability in Arabidopsis. Plant Cell Physiol 55: 293–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama S, Lu Y, Yamaguchi J, Sato T (2014b) Regulation of senescence under elevated atmospheric CO2 via ubiquitin modification. Plant Signal Behav 9: e28839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberon M, Zelazny E, Robert S, Conéjéro G, Curie C, Friml J, Vert G (2011) Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc Natl Acad Sci USA 108: E450–E458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis J (1995) Regulation of protein degradation. Plant Cell 7: 845–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callis J (2014) The ubiquitination machinery of the ubiquitin system. Arab B 12: e0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin LS, Vavalle JP, Li A (2002) Staring, a novel E3 ubiquitin-protein ligase that targets syntaxin 1 for degradation. J Biol Chem 277: 35071–35079 [DOI] [PubMed] [Google Scholar]
- Choi S, Tamaki T, Ebine K, Uemura T, Ueda T, Nakano A (2013) RABA members act in distinct steps of subcellular trafficking of the FLAGELLIN SENSING2 receptor. Plant Cell 25: 1174–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coruzzi GM, Zhou L (2001) Carbon and nitrogen sensing and signaling in plants: emerging “matrix effects”. Curr Opin Plant Biol 4: 247–253 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Breakthr Technol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deol KK, Lorenz S, Strieter ER (2019) Enzymatic logic of ubiquitin chain assembly. Front Physiol 10: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J, Hong-Hermesdorf A, Stierhof Y-D, Schumacher K (2006) Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakakaki G, van de Ven W, Pan S, Miao Y, Wang J, Keinath NF, Weatherly B, Jiang L, Schumacher K, Hicks G, et al. (2012). Isolation and proteomic analysis of the SYP61 compartment reveal its role in exocytic trafficking in Arabidopsis. Cell Res 22: 413–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dröse S, Bindseil KU, Bowman EJ, Siebers A, Zeeck A, Altendorf K (1993) Inhibitory effect of modified bafilomycins and concanamycins on P- and V-type adenosinetriphosphatases. Biochemistry 32: 3902–3906 [DOI] [PubMed] [Google Scholar]
- Dubeaux G, Neveu J, Zelazny E, Vert G (2018) Metal sensing by the IRT1 transporter-receptor orchestrates its own degradation and plant metal nutrition. Mol Cell 69: 953–964.e5 [DOI] [PubMed] [Google Scholar]
- Fujiwara M, Uemura T, Ebine K, Nishimori Y, Ueda T, Nakano A, Sato MH, Fukao Y (2014) Interactomics of Qa-SNARE in Arabidopsis thaliana. Plant Cell Physiol 55: 781–789 [DOI] [PubMed] [Google Scholar]
- Geldner N, Anders N, Wolters H, Keicher J, Kornberger W, Muller P, Delbarre A, Ueda T, Nakano A, Jürgens G (2003) The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112: 219–230 [DOI] [PubMed] [Google Scholar]
- Gendre D, McFarlane HE, Johnson E, Mouille G, Sjödin A, Oh J, Levesque-Tremblay G, Watanabe Y, Samuels L, Bhalerao RP (2013) Trans-Golgi network localized ECHIDNA/Ypt interacting protein complex is required for the secretion of cell wall polysaccharides in Arabidopsis. Plant Cell 25: 2633–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilles J-F, Dos Santos M, Boudier T, Bolte S, Heck N (2017) DiAna, an ImageJ tool for object-based 3D co-localization and distance analysis. Methods 115: 55–64 [DOI] [PubMed] [Google Scholar]
- Giovannone AJ, Reales E, Bhattaram P, Fraile-Ramos A, Weimbs T (2017) Mono-ubiquitination of syntaxin 3 leads to retrieval from the basolateral plasma membrane and facilitates cargo recruitment to exosomes. Mol Biol Cell 28: 2843–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebe M, Xu J, Möbius W, Ueda T, Nakano A, Geuze HJ, Rook MB, Scheres B (2003) Arabidopsis sterol endocytosis involves actin-mediated trafficking via ARA6-positive early endosomes. Curr Biol 13: 1378–1387 [DOI] [PubMed] [Google Scholar]
- Hachez C, Laloux T, Reinhardt H, Cavez D, Degand H, Grefen C, De Rycke R, Inzé D, Blatt MR, Russinova E, et al. (2014). Arabidopsis SNAREs SYP61 and SYP121 coordinate the trafficking of plasma membrane aquaporin PIP2;7 to modulate the cell membrane water permeability. Plant Cell 26: 3132–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinze L, Freimuth N, Rößling A, Hahnke R, Riebschläger S, Fröhlich A, Sampathkumar A, McFarlane HE, Sauer M (2020) EPSIN1 and MTV1 define functionally overlapping but molecularly distinct trans-Golgi network subdomains in Arabidopsis. Proc Natl Acad Sci USA 117: 25880–25889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Huang S, Tang D, Wang Y (2016) Monoubiquitination of syntaxin 5 regulates Golgi membrane dynamics during the cell cycle. Dev Cell 38: 73–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarancca Reyes T, Scartazza A, Pompeiano A, Ciurli A, Lu Y, Guglielminetti L, Yamaguchi J (2018) Nitrate reductase modulation in response to changes in C/N balance and nitrogen source in Arabidopsis. Plant Cell Physiol 59: 1248–1254 [DOI] [PubMed] [Google Scholar]
- Inada N, Betsuyaku S, Shimada TL, Ebine K, Ito E, Kutsuna N, Hasezawa S, Takano Y, Fukuda H, Nakano A, et al. (2016). Modulation of plant RAB GTPase-mediated membrane trafficking pathway at the interface between plants and obligate biotrophic pathogens. Plant Cell Physiol 57: 1854–1864 [DOI] [PubMed] [Google Scholar]
- Isono E, Kalinowska K (2017) ESCRT-dependent degradation of ubiquitylated plasma membrane proteins in plants. Curr Opin Plant Biol 40: 49–55 [DOI] [PubMed] [Google Scholar]
- Jahn R, Scheller RH (2006) SNAREs–engines for membrane fusion. Nat Rev Mol Cell Biol 7: 631–643 [DOI] [PubMed] [Google Scholar]
- Jaillais Y, Fobis-Loisy I, Miège C, Gaude T (2008) Evidence for a sorting endosome in Arabidopsis root cells. Plant J 53: 237–247 [DOI] [PubMed] [Google Scholar]
- Johnson A, Vert G (2016) Unraveling K63 polyubiquitination networks by sensor-based proteomics. Plant Physiol 171: 1808–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai K, Takano J, Miwa K, Toyoda A, Fujiwara T (2011) High boron-induced ubiquitination regulates vacuolar sorting of the BOR1 borate transporter in Arabidopsis thaliana. J Biol Chem 286: 6175–6183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-J, Bassham DC (2011) TNO1 is involved in salt tolerance and vacuolar trafficking in Arabidopsis. Plant Physiol 156: 514–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick DS, Gerber SA, Gygi SP (2005) The absolute quantification strategy: a general procedure for the quantification of proteins and post-translational modifications. Methods 35: 265–273 [DOI] [PubMed] [Google Scholar]
- Kitao T, Taguchi K, Seto S, Arasaki K, Ando H, Nagai H, Kubori T (2020) Legionella manipulates non-canonical SNARE pairing using a bacterial deubiquitinase. Cell Rep 32: 108107. [DOI] [PubMed] [Google Scholar]
- Korbei B, Moulinier-Anzola J, De-Araujo L, Lucyshyn D, Retzer K, Khan MA, Luschnig C (2013) Arabidopsis TOL proteins act as gatekeepers for vacuolar sorting of PIN2 plasma membrane protein. Curr Biol 23: 2500–2505 [DOI] [PubMed] [Google Scholar]
- Leitner J, Petrasek J, Tomanov K, Retzer K, Parezova M, Korbei B, Bachmair A, Zazimalova E, Luschnig C (2012) Lysine63-linked ubiquitylation of PIN2 auxin carrier protein governs hormonally controlled adaptation of Arabidopsis root growth. Proc Natl Acad Sci USA 109: 8322–8327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Sanagi M, Lu Y, Nomura Y, Stolze SC, Yasuda S, Saijo Y, Schulze WX, Feil R, Stitt M, et al. (2020) Protein phosphorylation dynamics under carbon/nitrogen-nutrient stress and identification of a cell death-related receptor-like kinase in Arabidopsis. Front Plant Sci 11: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka V, Kwon C, Panstruga R (2007) SNARE-ware: the role of SNARE-domain proteins in plant biology. Annu Rev Cell Dev Biol 23: 147–174 [DOI] [PubMed] [Google Scholar]
- Lu Y, Yasuda S, Li X, Fukao Y, Tohge T, Fernie AR, Matsukura C, Ezura H, Sato T, Yamaguchi J (2016) Characterization of ubiquitin ligase SlATL31 and proteomic analysis of 14-3-3 targets in tomato fruit tissue (Solanum lycopersicum L.). J Proteomics 143: 254–264 [DOI] [PubMed] [Google Scholar]
- Ma X, Claus LAN, Leslie ME, Tao K, Wu Z, Liu J, Yu X, Li B, Zhou J, Savatin DV, et al. (2020) Ligand-induced monoubiquitination of BIK1 regulates plant immunity. Nature 581: 199–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa S, Inada N, Yasuda S, Fukao Y, Fujiwara M, Sato T, Yamaguchi J (2014) The carbon/nitrogen regulator ARABIDOPSIS TOXICOS EN LEVADURA31 controls papilla formation in response to powdery mildew fungi penetration by interacting with SYNTAXIN OF PLANTS121 in Arabidopsis. Plant Physiol 164: 879–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa S, Sato T, Asada Y, Yasuda S, Yoshida M, Chiba Y, Yamaguchi J (2012) The Arabidopsis ubiquitin ligases ATL31 and ATL6 control the defense response as well as the carbon/nitrogen response. Plant Mol Biol 79: 217–227 [DOI] [PubMed] [Google Scholar]
- Martin T, Oswald O, Graham IA (2002) Arabidopsis seedling growth, storage lipid mobilization, and photosynthetic gene expression are regulated by carbon:nitrogen availability. Plant Physiol 128: 472–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins S, Dohmann EMN, Cayrel A, Johnson A, Fischer W, Pojer F, Satiat-Jeunemaître B, Jaillais Y, Chory J, Geldner N, et al. (2015). Internalization and vacuolar targeting of the brassinosteroid hormone receptor BRI1 are regulated by ubiquitination. Nat Commun 6: 6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka K, Higuchi T, Maeshima M, Nakamura K (1997) A vacuolar-type H+-ATPase in a nonvacuolar organelle is required for the sorting of soluble vacuolar protein precursors in tobacco cells. Plant Cell 9: 533–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Obrdlik P, El-Bakkoury M, Hamacher T, Cappellaro C, Vilarino C, Fleischer C, Ellerbrok H, Kamuzinzi R, Ledent V, Blaudez D, et al. (2004) K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc Natl Acad Sci USA 101: 12242–12247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Akopian D, Rape M (2018) Principles of ubiquitin-dependent signaling. Annu Rev Cell Dev Biol 34: 137–162 [DOI] [PubMed] [Google Scholar]
- Ossowski S, Schwab R, Weigel D (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53: 674–690 [DOI] [PubMed] [Google Scholar]
- Peng J, Li Z, Wen X, Li W, Shi H, Yang L, Zhu H, Guo H (2014) Salt-induced stabilization of EIN3/EIL1 confers salinity tolerance by deterring ROS accumulation in Arabidopsis. PLoS Genet 10: e1004664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli R, Sutter JU, Blatt MR (2004) A new catch in the SNARE. Trends Plant Sci 9: 187–195 [DOI] [PubMed] [Google Scholar]
- Reggiori F, Pelham HRB (2002) A transmembrane ubiquitin ligase required to sort membrane proteins into multivesicular bodies. Nat Cell Biol 4: 117–123 [DOI] [PubMed] [Google Scholar]
- Robinson DG, Jiang L, Schumacher K (2008) The endosomal system of plants: charting new and familiar territories. Plant Physiol 147: 1482–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Barrios N, Monachello D, Dolde U, Wong A, San Clemente H, Cayrel A, Johnson A, Lurin C, Vert G (2019) Advanced cataloging of lysine-63 polyubiquitin networks by genomic, interactome, and sensor-based proteomic analyses. Plant Cell 32: 123–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Barrios N, Vert G (2018) Proteasome-independent functions of lysine-63 polyubiquitination in plants. New Phytol 217: 995–1011 [DOI] [PubMed] [Google Scholar]
- Rosquete MR, Worden N, Ren G, Sinclair RM, Pfleger S, Salemi M, Phinney BS, Domozych D, Wilkop T, Drakakaki G (2019) AtTRAPPC11/ROG2: a role for TRAPPs in maintenance of the plant trans-Golgi network/early endosome organization and function. Plant Cell 31: 1879–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot A (2007) Increases in the number of SNARE genes parallels the rise of multicellularity among the green plants. Plant Physiol 144: 6–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot AA, Assaad FF, Raikhel NV (2000) The Arabidopsis genome. An abundance of soluble N-ethylmaleimide-sensitive factor adaptor protein receptors. Plant Physiol 124: 1558–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot AA, Kovaleva V, Bassham DC, Raikhel NV (2001) Interactions between syntaxins identify at least five SNARE complexes within the Golgi/prevacuolar system of the Arabidopsis cell. Mol Biol Cell 12: 3733–3743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Maekawa S, Yasuda S, Sonoda Y, Katoh E, Ichikawa T, Nakazawa M, Seki M, Shinozaki K, Matsui M, et al. (2009) CNI1/ATL31, a RING-type ubiquitin ligase that functions in the carbon/nitrogen response for growth phase transition in Arabidopsis seedlings. Plant J 60: 852–864 [DOI] [PubMed] [Google Scholar]
- Sato T, Maekawa S, Yasuda S, Domeki Y, Sueyoshi K, Fujiwara M, Fukao Y, Goto DB, Yamaguchi J (2011) Identification of 14-3-3 proteins as a target of ATL31 ubiquitin ligase, a regulator of the C/N response in Arabidopsis. Plant J 68: 137–146 [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y, Takagi J, Ito E, Ito Y, Ebine K, Komatsu Y, Goto Y, Sato M, Toyooka K, Ueda T, et al. (2021) Cargo sorting zones in the trans-Golgi network visualized by super-resolution confocal live imaging microscopy in plants. Nat Commun 12: 1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda A, Sugiyama K, Nagano H, Mori M, Kaido M, Mise K, Tsuda S, Okuno T (2002) Identification of a novel RNA silencing suppressor, NSs protein of Tomato spotted wilt virus. FEBS Lett 532: 75–79 [DOI] [PubMed] [Google Scholar]
- Tamura K, Shimada T, Ono E, Tanaka Y, Nagatani A, Higashi S-I, Watanabe M, Nishimura M, Hara-Nishimura I (2003) Why green fluorescent fusion proteins have not been observed in the vacuoles of higher plants. Plant J 35: 545–555 [DOI] [PubMed] [Google Scholar]
- Tsuchiya H, Ohtake F, Arai N, Kaiho A, Yasuda S, Tanaka K, Saeki Y (2017) In vivo ubiquitin linkage-type analysis reveals that the Cdc48-Rad23/Dsk2 axis contributes to K48-linked chain specificity of the proteasome. Mol Cell 66: 488–502.e7 [DOI] [PubMed] [Google Scholar]
- Uemura T, Nakano RT, Takagi J, Wang Y, Kramer K, Finkemeier I, Nakagami H, Tsuda K, Ueda T, Schulze-Lefert P, et al. (2019). A Golgi-released subpopulation of the trans-Golgi network mediates protein secretion in Arabidopsis. Plant Physiol 179: 519–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Suda Y, Ueda T, Nakano A (2014) Dynamic behavior of the trans-Golgi network in root tissues of Arabidopsis revealed by super-resolution live imaging. Plant Cell Physiol 55: 694–703 [DOI] [PubMed] [Google Scholar]
- Uemura T, Ueda T, Nakano A (2012) The physiological role of SYP4 in the salinity and osmotic stress tolerances. Plant Signal Behav 7: 1118–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Ueda T, Ohniwa RL, Nakano A, Takeyasu K, Sato MH (2004) Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct Funct 29: 49–65 [DOI] [PubMed] [Google Scholar]
- Valdez-Taubas J, Pelham H (2005) Swf1-dependent palmitoylation of the SNARE Tlg1 prevents its ubiquitination and degradation. EMBO J 24: 2524–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von der Fecht-Bartenbach J, Bogner M, Krebs M, Stierhof Y-D, Schumacher K, Ludewig U (2007) Function of the anion transporter AtCLC-d in the trans-Golgi network. Plant J 50: 466–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Cai Y, Miao Y, Lam SK, Jiang L (2009) Wortmannin induces homotypic fusion of plant prevacuolar compartments. J Exp Bot 60: 3075–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibowo A, Becker C, Marconi G, Durr J, Price J, Hagmann J, Papareddy R, Putra H, Kageyama J, Becker J, et al. (2016) Hyperosmotic stress memory in Arabidopsis is mediated by distinct epigenetically labile sites in the genome and is restricted in the male germline by DNA glycosylase activity. eLife 5: 1–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner W, Schekman R (2008) Membrane fusion. Nat Struct Mol Biol 15: 658–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkop T, Pattathil S, Ren G, Davis DJ, Bao W, Duan D, Peralta AG, Domozych DS, Hahn MG, Drakakaki G (2019) A hybrid approach enabling large-scale glycomic analysis of post-Golgi vesicles reveals a transport route for polysaccharides. Plant Cell 31: 627–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Hankins HM, MacDonald C, Erlinger SJ, Frazier MN, Diab NS, Piper RC, Jackson LP, MacGurn JA, Graham TR (2017) COPI mediates recycling of an exocytic SNARE by recognition of a ubiquitin sorting signal. eLife 6: 1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Yamashita-Yamada M, Hirase T, Fujiwara T, Tsuda K, Hiruma K, Saijo Y (2016) Danger peptide receptor signaling in plants ensures basal immunity upon pathogen-induced depletion of BAK1. EMBO J 35: 46–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda S, Aoyama S, Hasegawa Y, Sato T, Yamaguchi J (2017) Arabidopsis CBL-interacting protein kinases regulate carbon/nitrogen-nutrient response by phosphorylating ubiquitin ligase ATL31. Mol Plant 10: 605–618 [DOI] [PubMed] [Google Scholar]
- Yasuda S, Sato T, Maekawa S, Aoyama S, Fukao Y, Yamaguchi J (2014) Phosphorylation of Arabidopsis ubiquitin ligase ATL31 is critical for plant carbon/nitrogen nutrient balance response and controls the stability of 14-3-3 proteins. J Biol Chem 289: 15179–15193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinari A, Hosokawa T, Beier MP, Oshima K, Ogino Y, Hori C, Takasuka TE, Fukao Y, Fujiwara T, Takano J (2021) Transport-coupled ubiquitination of the borate transporter BOR1 for its boron-dependent degradation. Plant Cell 33: 420–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Z, Keereetaweep J, Liu H, Feil R, Lunn JE, Shanklin J (2018) Trehalose 6-phosphate positively regulates fatty acid synthesis by stabilizing WRINKLED1. Plant Cell 30: 2616–2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liu D, Wang P, Ma X, Lin W, Chen S, Mishev K, Lu D, Kumar R, Vanhoutte I, et al. (2018) Regulation of Arabidopsis brassinosteroid receptor BRI1 endocytosis and degradation by plant U-box PUB12/PUB13-mediated ubiquitination. Proc Natl Acad Sci USA 115: E1906–E1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Gong Z, Zhang C, Song C, Damsz B, Inan G, Koiwa H, Zhu J, Hasegawa PM, Bressan RA (2002) OSM1/SYP61: a syntaxin protein in Arabidopsis controls abscisic acid-mediated and non-abscisic acid-mediated responses to abiotic stress. Plant Cell 14: 3009–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouhar J, Rojo E, Bassham DC (2009) AtVPS45 is a positive regulator of the SYP41/SYP61/VTI12 SNARE complex involved in trafficking of vacuolar cargo. Plant Physiol 149: 1668–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.