Abstract
Stomata play important roles in gas and water exchange in leaves. The morphological features of stomata and pavement cells are highly plastic and are regulated during development. However, it is very laborious and time-consuming to collect accurate quantitative data from the leaf surface by manual phenotyping. Here, we introduce LeafNet, a tool that automatically localizes stomata, segments pavement cells (to prepare them for quantification), and reports multiple morphological parameters for a variety of leaf epidermal images, especially bright-field microscopy images. LeafNet employs a hierarchical strategy to identify stomata using a deep convolutional network and then segments pavement cells on stomata-masked images using a region merging method. LeafNet achieved promising performance on test images for quantifying different phenotypes of individual stomata and pavement cells compared with six currently available tools, including StomataCounter, Cellpose, PlantSeg, and PaCeQuant. LeafNet shows great flexibility, and we improved its ability to analyze bright-field images from a broad range of species as well as confocal images using transfer learning. Large-scale images of leaves can be efficiently processed in batch mode and interactively inspected with a graphic user interface or a web server (https://leafnet.whu.edu.cn/). The functionalities of LeafNet could easily be extended and will enhance the efficiency and productivity of leaf phenotyping for many plant biologists.
LeafNet is an accurate, robust, automatic, and high-throughput analytical tool for identifying and quantifying morphological features of both stomata and pavement cells in leaf epidermal images.
IN A NUTSHELL.
Background: Stomata are microscopic openings in the leaf epidermis that allow for oxygen and carbon dioxide exchange between a plant and the atmosphere. The phenotypes of stomata and the surrounding pavement cells, including their numbers, sizes, and other morphological features, are important for understanding their functions and regulation. Traditional phenotyping generally depends on laborious and time-consuming manual work by specialists in plant biology. In recent years, several programs were independently developed to detect stomata or to segment pavement cells. However, the accuracy of these methods for analyzing light microscopy images from a broad range of species is limited.
Question: We aimed to develop an automatic tool to accurately identify and quantify different features of stomata and pavement cells at the same time using light microscopy images to facilitate plant biology studies.
Findings: We introduce LeafNet, a tool that can automatically localize stomata, segment pavement cells, and report multiple morphological parameters for a variety of leaf epidermal images, especially images generated by bright-field microscopy. We employed a hierarchical strategy to identify stomata using a deep convolutional network and then segment pavement cells on stomata-masked image using a region merging method. LeafNet achieved promising performance on test images when quantifying different phenotypes of individual stomata and pavement cells in a comparison with six currently available tools. LeafNet shows great flexibility, and we further expanded its ability to analyze bright-field images from various species as well as confocal images using transfer learning. Users can install LeafNet locally via the conda package or directly use it in the webserver at https://leafnet.whu.edu.cn/.
Next steps: We believe that the plant community needs more well-labeled datasets for training and testing. We think that pavement cell segmentation and stoma detection could be better solved with a single joint deep learning model, with more datasets released by researchers in the future.
Introduction
Stomata are microscopic openings in the epidermis of leaves, stems, and other plant organs that allow for oxygen and carbon dioxide exchange between a plant and the atmosphere as well as for water loss by transpiration (Zoulias et al., 2018). In general, each stoma contains a pair of specialized guard cells. In many plants, two or more subsidiary or accessory cells that are adjacent to guard cells cooperatively regulate stomatal aperture. Stomatal function is essential for photosynthesis and respiration, which are critical for plant survival in the terrestrial environment and for plant productivity (Qi and Torii, 2018). Thus, stomata biology has attracted the interest of many plant researchers over the years.
Recent changes in climate, including elevated CO2 levels, high temperatures, and drought, have significantly influenced the ecosystem structure and the productivity of global agriculture (Engineer et al., 2016; Xu et al., 2016). High-throughput leaf thermal imaging has identified multiple mutants in the CO2 response (Hashimoto et al., 2006). To optimize the regulatory functions of stomata in response to the changing environment, the generation, development, and patterning of stomata and pavement cells are regulated by the complex interplay between internal developmental programs and various environmental cues (Casson and Hetherington, 2010). However, the irregularity of plant epidermal cells makes quantitative analysis difficult and inefficient. Hence, there is an urgent need for high-throughput technologies for screening large populations of genetic materials to identify regulators of stomatal development.
Due to the irregularity of epidermal cells surrounding stomata, traditional phenotyping of stomata and pavement cells in images generally depends on laborious, time-consuming manual work by specialists in plant biology. Fetter et al. developed StomataCounter to identify and count stomata in scanning electron microscopy (SEM) images and differential interference contrast (DIC) images using deep convolutional neural networks (CNNs; Fetter et al., 2019), but this technique has reduced accuracy for bright-field images. Several tools have recently been developed for the segmentation of pavement cells (to prepare them for quantification), including PaCeQuant (Möller et al., 2017), PlantSeg (Wolny et al., 2020), and Cellpose (Stringer et al., 2021). PlantSeg and PaCeQuant can generate accurate segmentation for confocal and light sheet images, but their accuracy is limited using bright-field images taken under a light microscope. Cellpose performs well with convex polygon-like cells, but fails with puzzle-shaped cells. MorphoGraphX is a 3D image analysis tool that can be used to collect accurate leaf epidermal information from confocal or light sheet images (Barbier de Reuille et al., 2015); however, collecting large numbers of 3D images is expensive and time-consuming compared with 2D bright-field images. Therefore, an automatic tool is needed to accurately identify stomata and pavement cells simultaneously in bright-field images taken under a light microscope.
Here, we present an accurate, robust, automatic, high-throughput analytical tool called LeafNet for identifying and quantifying different features of both stomata and pavement cells in light microscopy images for plant biology studies.
Results
Hierarchical strategy for segmenting stomata and pavement cells
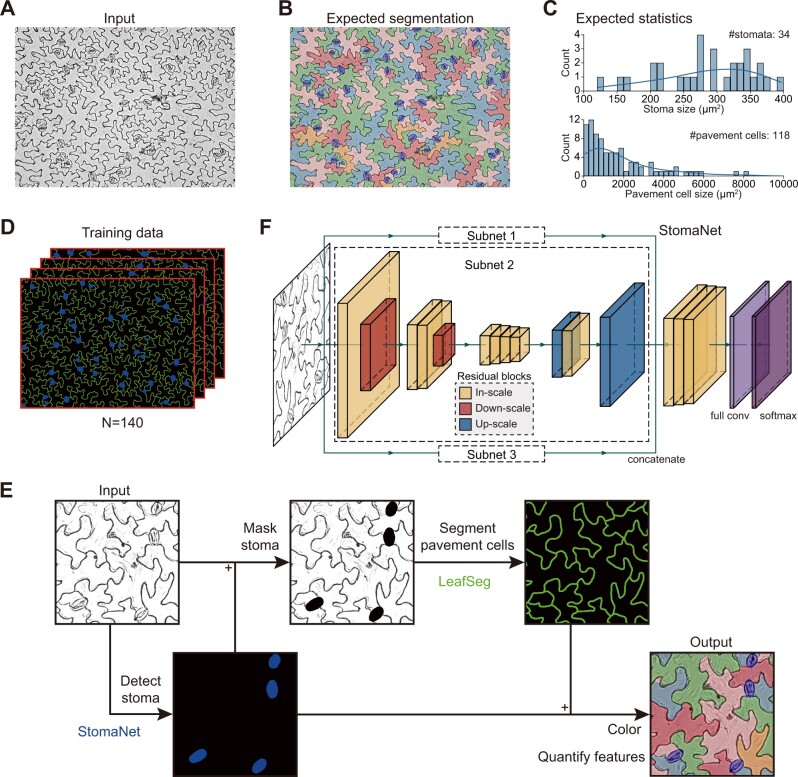
In representative bright-field images of Arabidopsis thaliana leaf epidermis obtained under a light microscope (Figure 1A and Supplemental Figure S1A), the stomata appear ellipse-like, whereas the pavement cells are extremely irregular in shape. Furthermore, the experimental process of creating the images can generate various types of noise, which makes it difficult to perform accurate segmentation and to quantify their numbers and other features. Manual segmentation can be accurate (Figure 1B), but it is very laborious and time-consuming to label the boundaries of individual stomata and the puzzle-shaped contours of pavement cells. Besides segmentation, it is important to characterize the general features of stomata and pavement cells, such as count and size (Figure 1C), for a large number of leaves from the same or different genotypes.
Figure 1.
Hierarchical strategy of LeafNet to segment stomata and pavement cells. A, Representative bright-field image of stomata and pavement cells. B, Representative result from manual segmentation of the input image in (A). Stomata are labeled in blue, and pavement cells are filled with different colors. C, Expected statistics from the segmentation in (B) on the size distribution for stomata (top) and for pavement cells (bottom). D, Training data prepared from manual segmentation. The stomata are shown in blue, and the borders of pavement cells are labeled in green. E, Hierarchical strategy and workflow of LeafNet with the StomaNet module for detecting stomata, and the LeafSeg module for segmenting pavement cells on stoma-masked input. A graphical illustration of each step is shown in Supplemental Figure S1. F, Graphical illustration of the deep residual neural network for the StomaNet module. This module is composed of three subnets with in-scale (orange), down-scale (red), and up-scale (blue) residual blocks.
To solve these problems more efficiently, we built an automatic tool for identifying stomata using a deep-learning approach, segmenting pavement cells with a region merging algorithm, and quantifying their features, including count, size, and length-to-width ratio for stomata and 28 different morphological parameters (e.g. size, perimeter, circularity, lobe count, and so on) for pavement cells. We initially manually annotated 140 images with fine segmentation of stomata and pavement cells by labeling pavement cell walls in green, stomata in blue, and the background in black (Figure 1D). All of these images with manual annotations are available at the LeafNet website (https://leafnet.whu.edu.cn/suppdata/).
We developed the LeafNet program, which employs a hierarchical strategy to sequentially identify stomata and pavement cells (Figure 1E). We first trained a stoma detector, the StomaNet (Supplemental Figure S1B) module, based on a deep residual network (Figure 1F and Supplemental Figure S2) to identify reliable stomata in the input image. We then masked the stomata out of the original image by coloring them black. We built the LeafSeg module to reliably identify pavement cell borders using a region merging algorithm (Supplemental Figure S1C; see “Methods” section for details). Finally, we merged the stomata and pavement cell borders. Combining the two modules, LeafNet can generate pixel-wise segmentation of the input image (Supplemental Figure S1D) and then collect different morphological features of stomata and pavement cells (Figure 1E).
LeafNet shows good performance for the segmentation of stomata and pavement cells
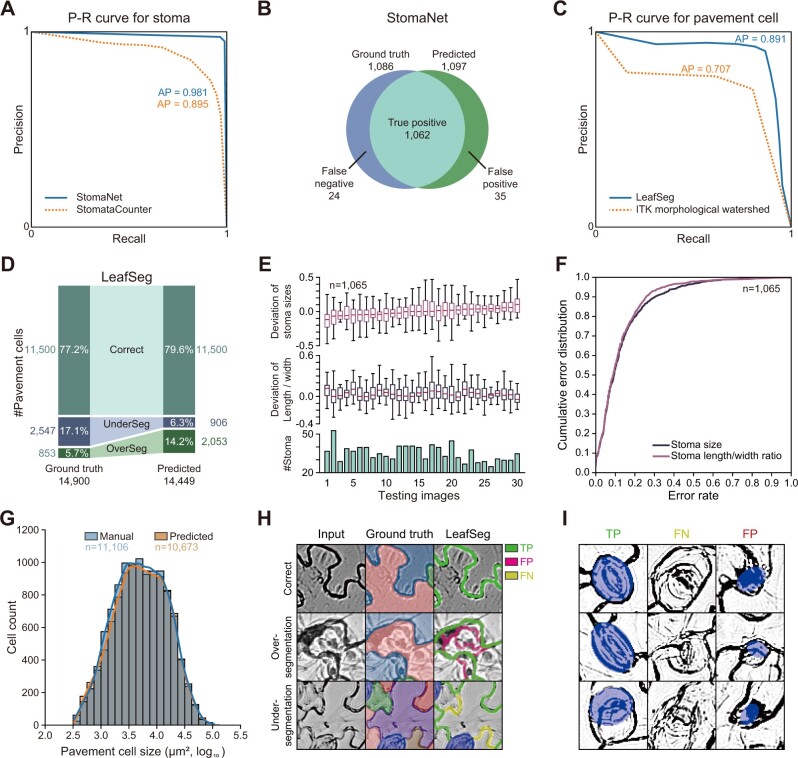
To evaluate the performance of LeafNet for detecting stomata, we manually labeled 30 images with 1,086 complete stomata as a test set for stomata detection. The stomata detecting module StomaNet achieved an average precision (AP) of 98.1%, while StomataCounter (Fetter et al., 2019) reached 89.5% precision as our baseline (Figure 2A). As a result, of the 1,086 ground truth stomata (using manual labeling as the gold standard), StomaNet successfully detected 1,062 stomata (missing 24) and falsely detected 35 stomata (Figure 2B).
Figure 2.
Performance of LeafNet for the recognition and quantification of stomata and pavement cells. A, Precision-recall curve of StomaNet (blue) and StomataCounter (orange dotted line) for counting stomata in 30 testing images. The thresholds were evaluated from 0.1 to 0.9 to calculate AP. B, Venn diagram showing the performance of StomaNet for detecting stomata in the testing dataset using default settings. C, Precision-recall curve for segmenting pavement cells with LeafSeg (blue) and the ITK morphological watershed algorithm (orange). D, Performance of LeafSeg for segmenting pavement cells in the testing dataset using default settings. The numbers and percentages of correct, under-segmented (UnderSeg), and over-segmented (OverSeg) cells are shown. E, Deviation from the ground truth of the quantification of size (top) and length/width ratio (middle) for stomata in each image. The per-image counts of stomata are shown as a bar graph (bottom). F, Cumulative distribution of the size deviation (blue) and length/width ratio deviation (red) for all stomata (n = 1,065) in the testing dataset. G, Distribution of the pavement cell sizes obtained by manual annotation (blue) and LeafSeg prediction (orange). H, Representative examples of LeafSeg predictions in three cases: correct (top row), over-segmentation (middle row), and under-segmentation (bottom row). The middle column (Ground truth) shows the correct segmentation of cells filled with different colors. In the right column, the TP, FP, and FN borders are shown in green, magenta, and yellow, respectively. I, Representative stomata correctly identified (TP), missed (FN), and falsely identified (FP) by StomaNet.
To compare LeafSeg with a baseline method, the morphological watershed algorithm (which operates on the topographic surface of an image gradient), we performed baseline analysis using Insight Toolkit (ITK), a general image processing library widely used for biological images (McCormick et al., 2014). We added another optional module, ITK morphological watershed, into LeafNet by calling the ITK morphological watershed algorithm for pavement cell segmentation (see “Methods” section for details). The ITK morphological watershed module is interchangeable with LeafSeg in LeafNet. For our 140 manually labeled images, the LeafSeg module of LeafNet achieved an AP of 89.1% for segmenting ∼14,900 pavement cells, while ITK morphological watershed achieved an AP of 70.7% (Figure 2C). In detail, 79.6% pavement cells were correctly predicted, 6.3% cells resulted from under-segmentation (multiple ground truth cells were merged into one cell), and 14.2% cells resulted from over-segmentation (one ground truth cell was split into multiple cells) (Figure 2D).
To further evaluate the ability of LeafNet to quantify different epidermal cell characteristics through pixel-wise segmentation, we compared the predicted stoma sizes, length/width ratios, and pavement cell sizes with the ground truth. The average deviations of stoma sizes were in the range of −13.3% to 13.9% by image, while the average deviations of length/width ratios ranged from −0.1% to 13.2% (Figure 2E). The MAE metrics (mean absolute error) of stoma size and length/width ratio reached 12.6% and 12.4%, respectively. The cumulative error distribution showed that most stomata were correctly predicted, with ∼10% of stomata having large errors in size or length/width ratio (Figure 2F). Although under-segmented and over-segmented pavement cells existed, the difference in pavement cell sizes between the predictions and ground truth was not significant (P-value as 0.48 from two-tailed t test on ∼11,000 complete pavement cells in images) (Figure 2G).
Moreover, we analyzed the success and failure of LeafNet for individual cases. The LeafSeg module of pavement cell segmentation is noise-tolerant (Figure 2H, top row), but occasionally failed when using images with strong noise (Figure 2H, middle row) or with multiple breaks in short borders (Figure 2H, bottom row). As shown in the three representative examples (Figure 2I, left column), the StomaNet module faithfully captured stomata in most cases. The presence of fuzzy contours (Figure 2I, middle column) or ellipse-like cell walls and noise (Figure 2I, right column) seldom prevented StomaNet from correctly identifying stomata.
Taken together, LeafNet performed satisfactorily in identifying stomata and pavement cells and quantifying their biological features.
Comparison of LeafNet with StomataCounter and PaCeQuant
For stoma detection, StomataCounter was developed to automatically count stomata from micrographs of the leaf epidermis (Fetter et al., 2019). This program performed well for SEM and DIC images, but it had limited capacity to detect stomata in our dataset with bright-field images. In our testing dataset used in Figure 2, StomataCounter only reached an AP of 89.5% (Figure 2A); it successfully detected 918 of 1,086 true stomata (missed 168 stomata) but falsely detected 157 stomata with the best threshold. In addition, this tool failed to predict the contours of the stomata and as a result could not quantify their sizes.
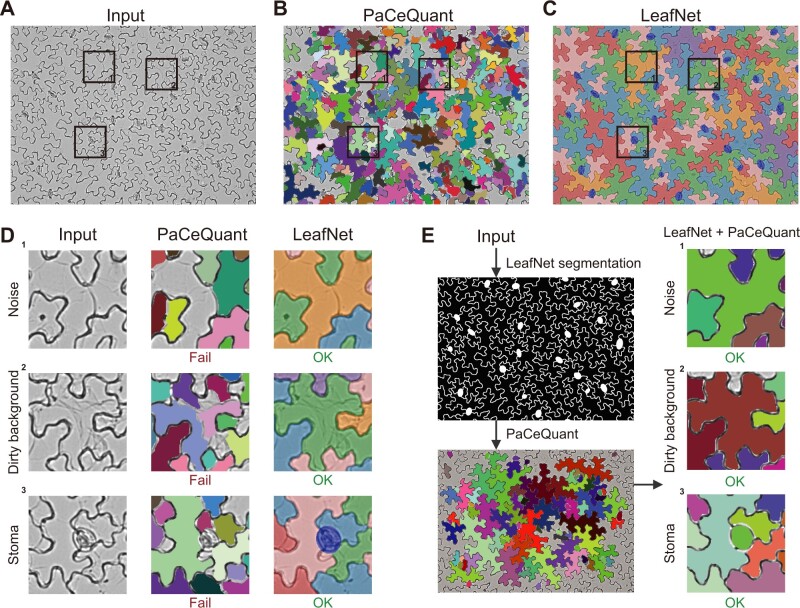
For pavement cell segmentation, we first tried PaCeQuant, a recently developed tool for pavement cell segmentation and morphological analysis of fluorescence microscopy images (Möller et al., 2017). This tool also performed well for 2D images converted from confocal images by maximum intensity z-projection (Supplemental Figure S3A). When our dataset (representative image in Figure 3A) was examined with PaCeQuant, almost all pavement cells were over-segmented into tiny areas, and no reasonable results were generated (Figure 3B) in contrast to LeafNet (Figure 3C), suggesting that PaCeQuant is not suitable for analyzing bright-field images. We further examined the results from the PaCeQuant output in detail and concluded that PaCeQuant is very sensitive to various types of noise in bright-field images, such as dots and lines (Figure 3D, top row) or a dirty background (Figure 3D, middle row). The missing feature of PaCeQuant to detect stomata also affected its performance to segment cells adjacent to those stomata (Figure 3D, bottom row), whereas the accuracy of LeafNet in segmenting pavement cells is substantially enhanced by masking stomata before performing pavement cell segmentation.
Figure 3.
Comparison of LeafNet with PaCeQuant. A, Representative raw input image with three areas highlighted in boxes. B, Results of segmentation of the image in (A) using the PaCeQuant program with the default configuration. Individual cells are filled with different colors. C, Results of segmentation of the image in (A) using LeafNet with the default configuration. Stomata are colored in blue, and neighboring pavement cells are filled with different colors. D, Zoom-in views of the three representative areas in the image in (A) showing typical noise and difficulties encountered in light microscope images. The raw input (left), segmented cells from PaCeQuant (middle), and those from LeafNet (right) are filled with different colors as in (B) and (C). E, Segmentation results of PaCeQuant using LeafNet’s segmentation as input (left). The combination of LeafNet and PaCeQuant achieved good results for the three representative areas, highlighting the advantages of LeafNet’s tolerance to various types of noise.
To further verify that the poor results of PaCeQuant were due to its low tolerance to noise, we used the segmentation from LeafNet to generate input images without these types of noise for PaCeQuant. In this case, PaCeQuant successfully segmented the pavement cells (Figure 3E, left), as shown in the three areas in Figure 3E (right), in contrast to when PaCeQuant was directly applied to the bright-field images (Figure 3D, middle column). These results indicate that LeafNet performs well in tolerating various types of noise in bright-field images and that its hierarchical strategy is effective for avoiding the interference from stomata during pavement cell segmentation.
To perform further morphological analysis, we implemented a script to parse the annotation image generated by LeafNet and to directly feed the pavement cell segmentation results into PaCeQuant without calling its own segmentation function. The combination of LeafNet’s cell segmentation and PaCeQuant’s feature extraction enabled us to obtain 28 morphological parameters such as perimeter, circularity, lobe counts, and so on in bright-field images. These quantification results were then visualized within the segmentation (Supplemental Figure S4). In addition, the annotation image from LeafNet can be manually corrected using GNU Image Manipulation Program (GIMP) or Photoshop before extracting the morphological features (see “Methods” section for details).
Quantitative evaluation of pavement cell segmentation using LeafNet and other programs
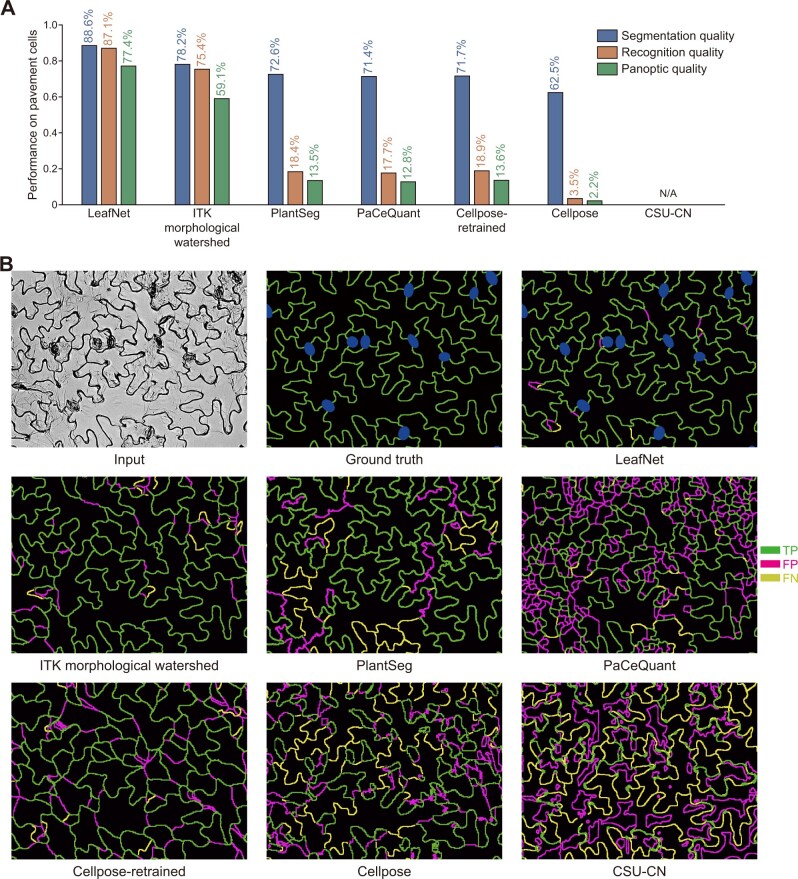
We quantitatively evaluated LeafNet using three different metrics: recognition quality (RQ; evaluating LeafNet’s ability to correctly report pavement cells from an image), segmentation quality (SQ, evaluating LeafNet’s ability to closely match the predicted borders with cell walls), and panoptic quality (PQ; the product of SQ and RQ; see “Methods” section for details). Meanwhile, we used the same dataset to investigate the performance of ITK morphological watershed and several recently developed programs, including PaCeQuant (Möller et al., 2017), PlantSeg (Wolny et al., 2020), Cellpose (Stringer et al., 2021), and CSU-CN from Cell Segmentation Benchmark. As expected, LeafNet performed much better than PaCeQuant, especially for RQ and PQ (Figure 4A), with a representative example shown in Figure 4B. Consistent with the results of our comparison based on AP (Figure 2C), the performance of ITK morphological watershed was worse than that of LeafNet with the LeafSeg module for these three metrics (Figure 4, A and B).
Figure 4.
Quantitative evaluation of the performance of LeafNet and related tools for pavement cell segmentation. A, Performance of different tools for pavement cell segmentation in bright-field images of Arabidopsis leaves using three different metrics. B, Representative examples of segmentation results using different tools. The TP, FP, and FN borders are shown in green, magenta, and yellow, respectively.
PlantSeg, which contains a pretrained model to segment 2D images, performed well on 2D images converted from confocal images by maximum intensity z-projection (Supplemental Figure S3B). However, PlantSeg’s performance on our dataset was worse than LeafNet’s, probably because the pretrained model was not trained for bright-field images (Figure 4, A and B). Please note that PlantSeg has multiple algorithm options for pavement cell segmentation, and the results are based on the default GASP algorithm.
Cellpose accurately segmented convex polygon-like cells in bright-field images (Supplemental Figure S3C). On our dataset of bright-field images from Arabidopsis leaves, Cellpose did not perform well with its pretrained cyto model. We retrained a new model called Cellpose-retrained with the images from our dataset. Cellpose-retrained showed improvement over Cellpose; however, its performance was still worse than LeafNet’s (Figure 4, A and B), probably because the Cellpose algorithm does not support nonconvex-shaped cells.
We chose the CSU-CN method as a representative tool from the Cell Segmentation Benchmark in Cell Tracking Challenge (http://celltrackingchallenge.net/) because it achieved the highest score on the Fluo-N2DH-GOWT1 dataset, which contains high-contrast bright cells separated by a dark background, similar to our images. CSU-CN accurately segmented the image from the testing set Fluo-N2DH-GOWT1 (Supplemental Figure S3D). However, CSU-CN failed to segment our bright-field images of Arabidopsis leaves (Figure 4, A and B), likely because all of its training data were morphologically different from our images.
These results indicate that LeafNet well tolerates various types of noise when bright-field images are used, and its hierarchical strategy is effective for avoiding the interference from stomata during pavement cell segmentation.
Extension of LeafNet to confocal images
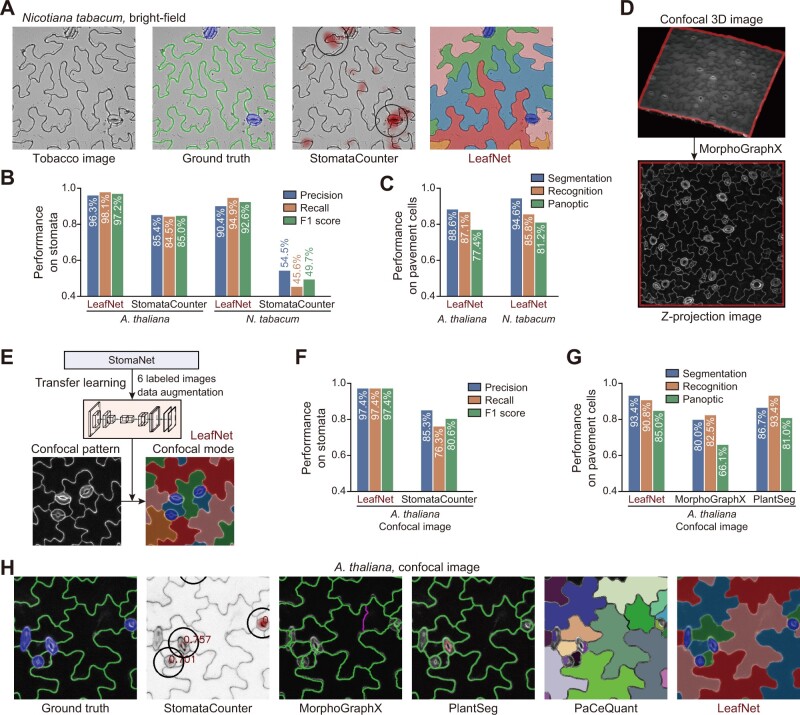
To explore LeafNet’s flexibility and broad utility, we first examined its performance using bright-field images from a different plant species, tobacco (Nicotiana tabacum). As expected, as shown by a representative example in Figure 5A, LeafNet can faithfully identify the two stomata and segment pavement cells, as in the ground truth image (manual labeling as the gold standard), while StomataCounter can only count stomata without giving exact borders. For our dataset of 14 images from N. tabacum, there were 79 stomata and 285 pavement cells based on ground truth labeling. LeafNet successfully detected 75 and falsely detected 8 stomata, while StomataCounter successfully detected only 36 and falsely detected 30 stomata. The overall performance of LeafNet (F1 score 92.6%) was much better than that of StomataCounter (F1 score 49.7%) (Figure 5B). The performance of StomataCounter for N. tabacum was much worse than that for A. thaliana, while LeafNet’s performance showed a small loss (Figure 5B; ˂5% in F1 score). On tobacco pavement cells, LeafNet achieved a good panoptic score that appeared to be slightly better than that for A. thaliana (Figure 5C), pointing to LeafNet’s good adaptability to species with similar cell morphology.
Figure 5.
Extension of LeafNet to analyze species with similar morphology as well as confocal images. A, Representative example of LeafNet results for a bright-field image of N. tabacum. Raw image, ground truth labeling, StomataCounter results, and LeafNet results are shown from left to right. B, Performance of LeafNet for stoma detection in A. thaliana and N. tabacum bright-field images compared with StomataCounter. C, Performance of LeafNet for segmenting pavement cells in A. thaliana and N. tabacum bright-field images using three different metrics. D, Representative example of preprocessing a confocal 3D image to a z-projection image using MorphoGraphX. E, Extension of LeafNet to analyze z-projection image using transfer learning based on limited number of newly labeled data to identify stomata with its confocal mode. F, Performance of LeafNet for detecting stomata in A. thaliana confocal images compared with StomataCounter. G, Performance of LeafNet for pavement cell segmentation in A. thaliana confocal images compared with MorphoGraphX and PlantSeg for three different metrics. H, Representative outputs of five programs using confocal images. StomataCounter, PlantSeg, PaCeQuant, and LeafNet use max intensity z-projection images as input, while MorphoGraphX takes 3D image stacks as input.
Next, we extended LeafNet to analyze confocal images. Confocal imaging can generate a 3D stack of images of leaf tissue, as shown in Figure 5D (top panel) (Erguvan et al., 2019). As LeafNet is designed to analyze 2D bright-field images, we established a pipeline to convert 3D image stacks from confocal or light sheet microscopes to a maximum intensity z-projection 2D image using MorphoGraphX (Barbier de Reuille et al., 2015) and ImageJ (Schindelin et al., 2012; Figure 5D bottom, Supplemental Figure S5A, and see “Methods” section for details). We noticed that LeafNet’s default mode accurately segmented pavement cells but had difficulty in precisely identifying stomata, probably because new confocal patterns were not observed in bright-field images. Thus, we trained a StomaNet confocal model using transfer learning on a confocal dataset of Arabidopsis leaves (Erguvan et al., 2019) after preprocessing them into 2D images and manually labeling all stomata and pavement cells (Figure 5E).
We compared LeafNet with four other state-of-the-art programs for analyzing confocal images, including StomataCounter for stoma detection and PlantSeg, PaCeQuant, and MorphoGraphX for pavement cell segmentation (Figure 5, F–H and Supplemental Figure S5B). PlantSeg and PaCeQuant perform segmentation on maximum intensity z-projections of image stacks in a similar manner to LeafNet, while MorphoGraphX performs 2.5D surface segmentation on the original image stacks, and we converted the results to 2D segmentation with z-projection, allowing us to compare the results with other tools (see “Methods” section for details).
For the 38 ground truth stomata, LeafNet successfully detected 37 and falsely detected 1 stoma, while StomataCounter successfully detected 29 and falsely detected 5 stomata: the performance scores are summarized in Figure 5F. For the 92 ground truth pavement cells, LeafNet achieved a panoptic score of 85.0%, while MorphoGraphX obtained a score of 66.1% and PlantSeg obtained a score of 81.0%. PaCeQuant accurately identified most pavement cells but falsely recognized stomata as 74 pavement cells. As PaCeQuant rejects pavement cells that are adjacent to image edges, we only quantified the performance of MorphoGrahX and PlantSeg to compared with LeafNet (Figure 5G). LeafNet achieved the highest accuracy in terms of both stomata (LeafNet F1 score of 0.97 versus 0.80 for StomataCounter) and pavement cells (LeafNet F1 score of 0.85 versus 0.66 for MorphoGraphX and 0.81 for PlantSeg). In the representative patch in Figure 5H (see complete image in Supplemental Figure S5B), StomataCounter missed true stoma, MorphoGraphX, and PlantSeg mis-segmented pavement cells, and PaCeQuant frequently miscalled one or more pavement cells in one stoma.
In summary, LeatNet outperforms the state-of-the-art programs in handling confocal images and has good adaptivity to different species, pointing to its flexibility and potential broad utility.
Extension of LeafNet to a wide range of species
To further examine the capability of LeafNet to analyze images from a broad range of species and images obtained using different micrographic methods, we performed systematic comparisons of datasets with a variety of images.
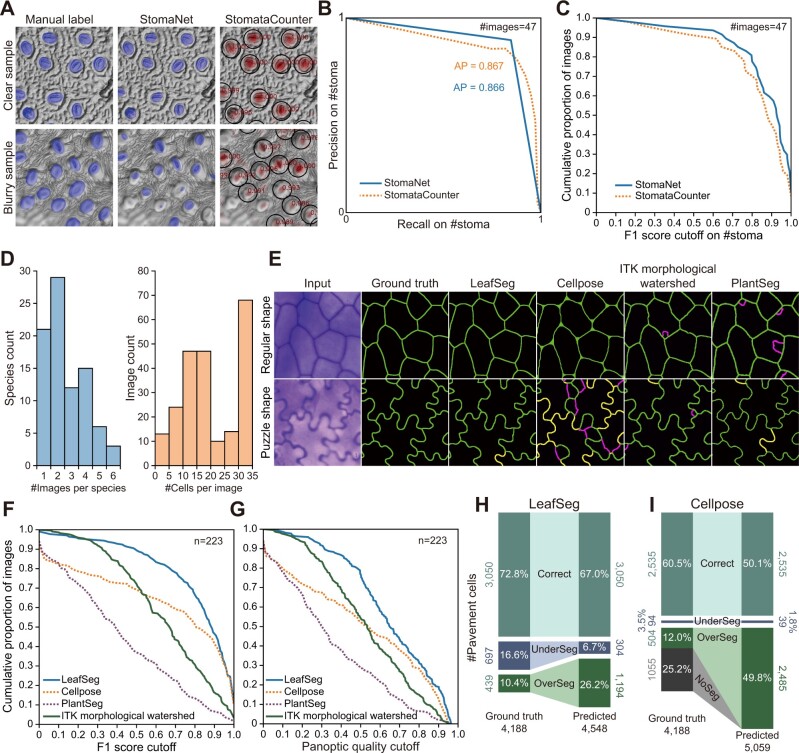
For stoma detection, we trained another universal model in StomaNet using the same deep learning network structure with the training data used by StomataCounter (Fetter et al., 2019). The training data contain ˃4,000 leaf epidermal images from more than 600 species taken by DIC microscopy, SEM, and bright-field microscopy; however, no label information is available for this dataset (https://datadryad.org/stash/dataset/doi:10.5061/dryad.kh2gv5f). We used StomataCounter’s prediction on 960 images for rough training and then manually labeled 140 randomly selected images for further training. The StomaNetUniversal model has a similar performance to StomataCounter on 47 manually labeled testing images randomly selected from the testing set (Figure 6A). StomaNet achieved an AP of 86.6%, similar to that of StomataCounter (86.7% AP) (Figure 6B).
Figure 6.
Evaluation of complex datasets using LeafNet and other tools. A, Representative examples of the results of StomaNet and StomataCounter for evaluating StomataCounter’s dataset. StomaNet and StomataCounter both achieved high quality with stomata with clear boundaries (top) and suffered performance loss with blurry images (bottom). B, Precision-recall curve of StomaNet (blue) and StomataCounter (orange dotted line) for counting stomata in 30 testing images. The thresholds were evaluated from 0.1 to 0.9 to calculate AP. C, Cumulative distribution of F1 scores for stoma detection in all images of the stoma detection testing dataset (n = 47). D, The distribution of species counts by the number of images per species (left) and the distribution of image counts by the labeled cell counts per image (right). This pavement cell data set contains 4,188 cells in 223 images from 86 species. E, Representative examples of the segmentation results from different programs for regularly shaped (top) and puzzle-shaped cells (bottom). F and G, Cumulative distribution of F1 scores (F) and PQ scores (G) of pavement cell segmentation from different programs using the pavement cell segmentation testing dataset (n = 223). H and I, Performance of LeafSeg (H) and Cellpose (I) in segmenting pavement cells for a testing dataset from a wide range of species. The numbers and percentages of correct, under-segmented, and over-segmented cells are shown in the comparison of predictions to ground truth.
For pavement cell segmentation, we used a dataset containing leaf epidermal images from different species with stained cell walls (Vőfély et al., 2019). The authors indicated that it is very difficult to perform automatic segmentations on this dataset due to various image defects, which prompted them to manually track the boundaries of pavement cells. The boundaries for each pavement cell were individually saved as coordinates relative to the cell center, but their positions in the original image are not available. We successfully mapped 4,188 pavement cells to 223 leaf epidermal images from 86 different species as our testing dataset (Figure 6D; see Supplemental Figure S6, A and B for three representative examples), which are provided in LeafNet’s website (https://leafnet.whu.edu.cn/suppdata/).
Based on this large testing dataset, we evaluated the LeafSeg module of LeafNet compared to other existing methods including ITK morphological watershed, PlantSeg, PaCeQuant, and Cellpose. LeafSeg and Cellpose were more robust to various types of noise in regularly shaped cells comparing to the other programs (Figure 6E top and Supplemental Figure S6, C and D left), and they both achieved F1 scores >0.95 and PQ scores >0.75 for ˃30% of the images (Figure 6, F and G). However, Cellpose showed reduced performance for images with puzzle-shaped pavement cells or with uneven lighting, which had little impact on LeafSeg’s performance (Figure 6, E–G and Supplemental Figure S6, C and D middle and right). PlantSeg and ITK morphological watershed showed similar performance for images with both regularly shaped and puzzle-shaped cells (Figure 6E and Supplemental Figure S6, E and F), but their overall performance scores were worse than that of LeafSeg (Figure 6, F–G). We also tested CSU-CN in this dataset, but the results were unusable (Supplemental Figure S6G).
Overall, for this complex dataset, LeafSeg had the best performance, achieving an average F1 score of 0.74 and a PQ score of 0.64, while ITK morphological watershed obtained scores of 0.64 and 0.52, Cellpose obtained scores of 0.58 and 0.50, and PlantSeg obtained scores of 0.40 and 0.35, respectively. We then examined the predicted pavement cells compared with the ground truth for LeafSeg and Cellpose, which generated more acceptable results (F1 score > 0.95 and PQ > 0.75) than PlantSeg and ITK morphological watershed. Of the 4,188 manually labeled pavement cells, LeafSeg correctly segmented 3,050 cells (72.8%), under-segmented 697 cells (16.6%) into 304 cells, and over-segmented 439 cells (10.4%) into 1,194 cells (Figure 6H). Cellpose correctly segmented 2,535 pavement cells (60.5%), under-segmented 94 cells (3.5%) into 39 cells, over-segmented 504 cells (12.0%) into 2,485 cells, and reported 1,055 cells as background (Figure 6I). These results are consistent with the finding that Cellpose performed worse than LeafSeg based on F1 score and PQ.
In summary, these results suggest that the performance of StomaNet for stoma counting is similar to that of StomataCounter based on the 140 manually multi-species training data and show that StomaNet can obtain accurate boundaries for stomata with clear signals. For pavement cell segmentation, LeafSeg showed the best performance among the tools examined on a large set of images with various defects from different species. By combining StomaNet and LeafSeg, LeafNet can be extended to a wide range of species and to images taken using different methods.
LeafNet detects significant biological differences
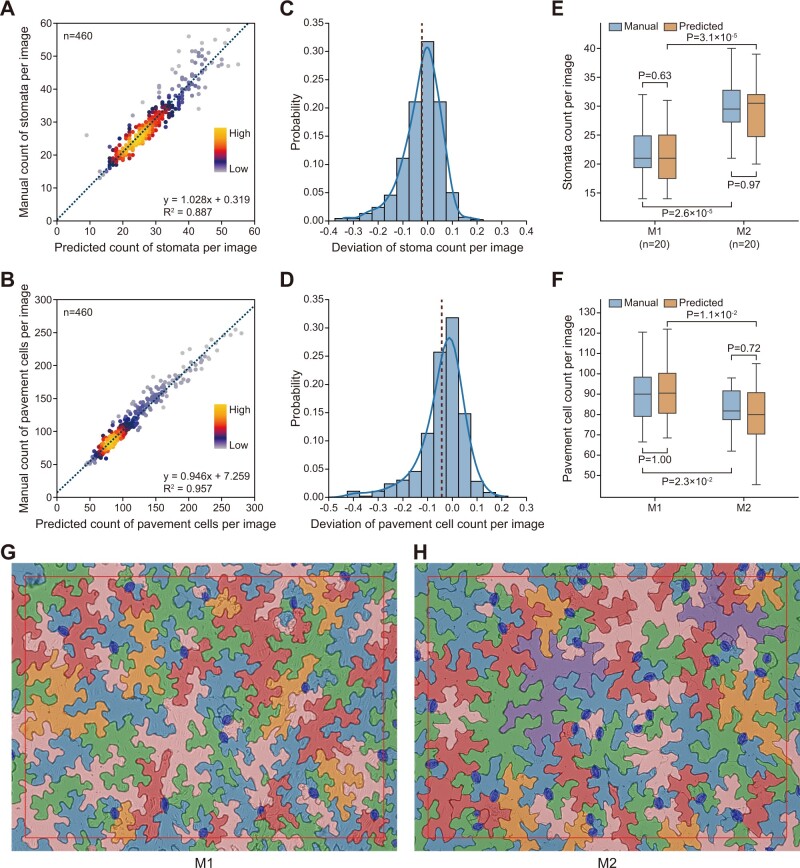
Next, we applied LeafNet to evaluate its ability to automatically analyze large-scale microscopy image datasets and to assess its difference from manual labeling using statistics. In total, we analyzed 460 images using LeafNet, manually inspected the segmentations of stomata and pavement cells, and recorded the correct counts as ground truth. We compared and evaluated the differences between the predicted and manual results. The predicted counts of stomata and pavement cells had good linear relationships with the manual counts (Figure 7, A and B), and the deviation of counts by image showed a tight distribution centered around 0 (Figure 7, C and D), with a mean absolute error of 5.80% and 5.45% for stomata and pavement cells, respectively.
Figure 7.
Application of LeafNet for evaluating large-scale microscope images. A and B, Linear regression between LeafNet’s results and manual counting of stomata (A) and pavement cells (B). The point densities are represented by heatmap with KDE smoothing. C and D, The deviation of stomata (C) and pavement cell (D) counts per image from LeafNet’s results versus manual annotation. E and F, LeafNet’s performance in quantifying the counts of stomata (E) and pavement cells (F) in leaves from two different genotypes: M1 and M2. P-values are based on paired two-tailed t test. G–H, Representative examples of LeafNet predictions using images from genotype M1 (G) and M2 (H).
Furthermore, we tested whether LeafNet can detect statistically significant differences in the densities of stomata and pavement cells between two different Arabidopsis genotypes in the Columbia-0 (Col-0) genetic background (M1 and M2). M1 is Pro35S:PIF4 (expressing PHYTOCHROME INTERACTING FACTOR4 under the control of the 35S promoter), and M2 is the wild-type control (Col-0). We compared the counts of stomata and pavement cells generated by LeafNet with those obtained from a manual annotation of 40 images. As shown in the representative examples for stomata (Figure 7E) and for pavement cells (Figure 7F), we observed consistent results between the predicted (blue) and manual (orange) results for nonsignificant differences in M1 or in M2 (not significant P-value), and significant differences between M1 and M2, from paired t tests.
These results suggest that LeafNet is a useful tool for plant biologists to process large numbers of images and quantify the biological differences in a reliable manner.
Using LeafNet in the CLI, GUI, and web server
To make LeafNet widely accessible to different users, we designed a standalone program that can be run on most computer systems. We also developed a graphic user interface (GUI; Supplemental Figure S7A) and a web server (Supplemental Figure S7B) for users without Linux experience, and a command-line interface (CLI) for experienced users with servers (Supplemental Figure S7C). The web server is hosted at https://leafnet.whu.edu.cn/. We have also created two Conda packages for both CPU and GPU environments, and thus users can easily install LeafNet with one command “conda install -c anaconda -c conda-forge -c zhouyulab leafnet(-gpu)” in Linux, Mac OS, or Windows systems.
LeafNet can use images generated from bright-field, confocal z-projection, and other imaging methods as input by using different modules. During preprocessing, Peeled denoiser works with bright-field images from peeled leaf epidermal (e.g. Figure 2A) and with confocal images (e.g. Figure 5D), while Stained denoiser works with leaf epidermal images with stained cell walls (e.g. Supplemental Figure S6A) and is recommended for other types of images. For stoma detection, StomaNet is trained for bright-field images from Arabidopsis by peeling off leaf epidermis, StomaNetConfocal is trained for confocal images from Arabidopsis, and StomaNetUniversal is trained to detect stomata in a wide range of species.
LeafNet can generate three types of output results. The first type is a preview image provided for visualizing the segmentation, which is shown on the original image with cells of different colors and stomata marked in blue. The second type is an annotation image provided for further analysis, which uses green lines to label pavement cell walls and labels stomata with blue ellipses. The third type is a statistical text with quantified data, including morphological features of the leaf epidermal image, such as the counts and sizes of stomata and pavement cells (Supplemental Figure S8, A–C). All three output results are available in the CLI, GUI, and web server.
In addition, users can use LeafNet in annotation mode and manually correct the output. Users can load the annotation image from LeafNet together with the input image into GIMP or Photoshop and then correct the annotation image for further analysis (see “Methods” section for details). Existing epidermal image processing pipelines could also benefit from LeafNet by simplifying the image annotation procedure, as the annotation image generated by LeafNet could easily be handled by other tools. Supplemental Figure S4 shows a feasible scenario in which manual correction can be performed on the annotation image from LeafNet and the corrected image can be fed into PaCeQuant to extract morphological information.
We also provide training utilities to extend LeafNet for other types of images. Advanced users can perform transfer learning to improve the stoma detection model, and our investigations and codes provide an exemplar workflow. New types of stomata for other species can be analyzed using newly labeled images with transfer learning in a similar manner (Figure 5E). Meanwhile, LeafNet’s segmentation can be used as a good starting point for further manual correction to efficiently construct training data.
Discussion
Here we introduce LeafNet, a fully automatic program capable of precisely detecting stomata and segmenting pavement cells. We devised a hierarchical strategy to accurately identify stomata first and then segment pavement cells in stomata-masked images. By incorporating two modules, StomaNet and LeafSeg, LeafNet sequentially conquers the two challenges of precisely detecting stomata and segmenting pavement cells while avoiding the interference between these two types of objects with different characteristics. The StomaNet module accurately segments stomata using a deep neural network. The LeafSeg module tolerates various types of noise and puzzle-shaped cell shapes, exhibiting acceptable performance on bright-field images using a region merging algorithm. LeafNet adapts to images from a broad range of species and outperforms several state-of-the-art programs, enabling biologists to perform fast and simple experiments using easily accessible bright-field microscopy, SEM, or confocal microscopy.
Object detection and cell segmentation are classic tasks in biological image processing, and many tools have been built for different scenarios. At the level of stomata detection, StomataCounter (Fetter et al., 2019) performed well on different types of images. StomaNet achieved higher accuracy than StomataCounter for bright-field images from Arabidopsis or species with similar morphology using 140 training images. StomaNetUniversal achieved similar accuracy to StomataCounter on images from a wide range of species using 960 roughly labeled and 140 manually labeled training images, in contrast to StomataCounter’s much larger training data of 4,618 images, suggesting that StomaNet requires less training data than StomataCounter and that StomaNetUniversal has the potential to evolve once more training data have been introduced. Moreover, StomaNet’s ability to accurately segment the borders of stomata enables LeafNet to mask stomata, which prevents the stomata from interfering with subsequent pavement cell segmentation.
For pavement cell segmentation, we tested three nondeep learning-based tools including MorphoGraphX (Barbier de Reuille et al., 2015), PaCeQuant (Möller et al., 2017), and ITK morphological watershed (McCormick et al., 2014), and three recently developed deep learning-based tools including PlantSeg (Wolny et al., 2020), Cellpose (Stringer et al., 2021), and CSU-CN (from Cell Tracking Challenge), based on a systematic evaluation on our own and published datasets. MorphoGraphX has been used to perform surface segmentation on confocal image stacks (Sapala et al., 2018). However, its performance was slightly worse than that of LeafNet and PlantSeg (Figure 5, G and H), and it could not process bright-field images. PaCeQuant could not tolerate the noise, uneven lighting, and inconsistent border signals in bright-field images. ITK morphological watershed showed better tolerance to image defects, but it did not perform as well as LeafSeg (Figures 4 and 6). PlantSeg, Cellpose, and CSU-CN achieved state-of-the-art performance on their own preferred input images (Supplemental Figure S3, B–D), but their performance on bright-field leaf epidermal images was worse than that of LeafSeg, especially on puzzle-shaped pavement cells (Figures 4 and 6). Based on the results for these representative tools, we conclude that the segmentation of pavement cells in bright-field images is challenging and that LeafSeg represents a significant advancement: it is well-adapted to this task and performs even better than the three deep learning-based methods examined.
To explore the possibility of using a CNN to enhance cell wall signals as in PlantSeg, we retrained a PlantSeg CNN model with 100 training images from our dataset in Figure 4. We applied our Stained Denoiser before training and prediction to improve its generalization ability. We evaluated the performance of this retrained network (named CNNwall) on 40 other images from the same dataset. With CNNwall enhancement, PlantSeg’s PQ increased from 13.5% to 70.4% on testing images, outperforming ITK morphological watershed (59.1%), and LeafSeg’s PQ increased from 77.4% to 81.4% (Supplemental Figure S9A).
We further tested CNNwall’s broad utility using the Vofely dataset (Vőfély et al., 2019) used in Figure 6, D–I. PlantSeg’s PQ increased from 34.5% to 55.7%, outperforming ITK morphological watershed (52.9%), suggesting that CNNwall is better adapted to various input images than PlantSeg’s original model (Supplemental Figure S9, B and C). However, LeafSeg’s PQ dropped from 64.5% to 56.5%, a value similar to PlantSeg’s (55.7%) (Supplemental Figure S9, B and C). As illustrated in the exemplar images, we reasoned that although CNNwall could enhance the signals of cell walls (Supplemental Figure S9D), it could also introduce extra artifacts when input images are different from those in the training dataset, which would impair cell segmentation (Supplemental Figure S9E). Consistently, the differences in pavement cells in a wide range of species and using different imaging methods had a huge impact to the performance of previously reported deep learning-based cell segmentation methods (Supplemental Figure S3, B–D and Supplemental Figure S6, D, E, and G). Therefore, a much larger dataset may be required to train a universal deep learning-based pavement cell segmentation model than to train a good stoma detection model. Considering that manually labeling pavement cell boundaries take more time than labeling stomata, we currently provide CNNwall-enhanced LeafSeg as an optional method and provide the original LeafSeg as our default universal method for pavement cell segmentation.
Nevertheless, we believe that pavement cell segmentation and stoma detection could be better solved using a single joint deep learning model in the future. Many articles reported to date by the plant community only contain morphological information but do not provide manually corrected segmentation. Platforms such as Cell Tracking Challenge (Ulman et al., 2017) provide different types of images from cultivated cell lines with pixel-wise labels for researchers and programmers to test and compare their methods, but there no such platform is currently available for pavement cell segmentation and stoma detection. We believe that the plant community needs more well-labeled datasets, and thus we shared all our training datasets, testing datasets, and the results from LeafNet and existing tools in the Download page of the LeafNet web server. In the future, as more datasets are released by researchers, a deep learning-based universal model could be created to segment pavement cells and stomata simultaneously with better accuracy and to further enhance the performance of automatic morphological analysis tools on bright-field images.
The LeafNet program, the associated web server, strategy, and codes are provided for the plant community with the potential to replace manual work, enhance productivity, and increase reproducibility. We have shown that LeafNet is flexible and can be extended to different species or confocal images, and we anticipate that it will be useful for a broad range of researchers interested in quantifying stomata and pavement cells.
Methods
Plant culture
All A. thaliana plants used in this study were of the Col-0 genetic background. The seeds were sterilized with 75% ethanol for 2 min and 2% sodium hypochlorite solution for 15 min. The seeds were sown in Petri dishes containing 0.5 strength Murashige and Skoog medium solidified with agar and placed at 4°C for 3 days in complete darkness, followed by growth under short days (10-h light/14-h dark) or long days (14-h light/10-h dark) at 22°C. Seedlings were grown under white fluorescent light at a light intensity of 100 μmol photons/m2/s.
Image collection
The light microscope images in this study were taken using a modified method as described (Engineer et al., 2014). Briefly, plant tissues (leaves) were sampled throughout plant growth. To obtain epidermal peels, glass slides (CITOGLAS 9821) were sprayed with Hollister Medical Adhesive (3.8 oz. Spray HH7730), and the abaxial epidermal surfaces of leaves from independent seedlings were gently pressed onto the slides. The mesophyll tissues were removed from the slides with a blade, and the epidermal peels were imaged under a Leica DMi8 microscope at 200× magnification.
Manual labeling to create the training dataset
We used the GIMP program to annotate stomata and pavement cells in input images. Briefly, we created a new annotation layer on top of the sample layer and set its opacity to 50%. We manually labeled the boundaries of pavement cells with a green line with 100 hardness and filled the stomata with blue coloring on the annotation layer. We then set the opacity of the annotation layer back to 100% and set its background to black. Finally, we removed the sample layer, flattened the image, and saved it as an annotation figure. The manual annotations were validated by one or more other annotators independently.
LeafNet workflow
The LeafNet workflow consists of image preprocessing, the StomaNet module, and the LeafSeg module.
Preprocessing
Images taken under a light microscope can be noisy and must be denoised before training or prediction. LeafNet has two different preprocessing modules. For our images of the epidermal surface peeled from leaves, we used the Peeled Denoiser based on the noise reduction function from the generic graphics library GEGL. Preprocessing involves the following steps: (1) resize the image to the resolution of the trained model in PIL with Image.ANTIALIAS; (2) invert the image only if it is a fluorescence image; (3) separate the image into dark and bright parts with Otsu threshold; (4) perform an adaptive linear transformation on the bright part to scale the mean of pixel gray scale to 200; (5) merge the dark part and transform the bright part; and (6) denoise the image with the noise reduction function from GEGL.
For other types of images, we used the Stained Denoiser. This denoiser is better adapted to different types of images and is recommended for most scenarios. This denoiser involves the following steps: (1–2) the same steps as for the Peeled Denoiser; (3) apply a median filter to the image; (4) apply a high-pass filter to the image; (5) perform adaptive area normalization on the image; and (6) apply mean curvature blur on the image.
The StomaNet module for detecting stomata
StomaNet input
As we used valid convolution and transpose convolution without padding, a fixed network size is needed, as the layer output size must be a positive integer. Therefore, the sliding window method is used for network input, including the following steps: (1) As the network output is smaller than the input, denoised images are copy padded with OpenCV using the border reflect method and (2) the sliding window method is used to split the input images into smaller images as network input. The input images are broken into patches whose sizes match the network input (186 for StomaNet) with a stride of network output (104 for StomaNet).
StomaNet network architecture
StomaNet is a deep residual network inspired by ISL (Christiansen et al., 2018) built with TensorFlow and Keras. The network comprises three sub-networks (called towers) of different input sizes to collect information at different scales. The towers are composed of residual blocks. Residual blocks are sub-networks of the structure shown in Supplemental Figure S2A. Three types of residual blocks are used in the network: down-scale, in-scale, and up-scale, the parameters of which are shown in Supplemental Figure S2B.
Residual blocks consist of two parts. The first part directly copies the data to output, forming a residual connection. Up sampling, cropping, and average pooling are used in different types of residual blocks to keep the data in the same shape with convolution filters. The second part consists of two convolution layers, which form an encoder-decoder structure. Input is batch normalized and activated by ReLU and Tanh (concatenated), encoded by a convolution layer called Conv2D expand. The output of the encoder is concatenated with the max pooling result of block input, batch normalized, and activated by ReLU and Tanh (concatenated) again. The 1*1 convolution is used as a decoder to reduce the count of filters to make it match the block input. The results of the two steps are added to generate the output of a residual block.
StomaNet output
The output of the network are images that represent the pixel-wise plausibility of stomata in the area (104 for StomaNet) of the center of the input (186 for StomaNet). As the input is generated by the sliding window approach, output images are stacked, generating a full-sized probability heatmap of stomata.
Training samples
For StomaNet, we manually labeled stomata in blue (0,0,255) in raw images from Arabidopsis using GIMP software, and saved the annotated images with the same name and resolution as the original image in a label folder. A 4 px Gaussian Blur was applied to stomata labels. The labeled images were broken into patches using the sliding-window method (the step length equals the size of the network output in generating nonoverlapping results) and split into a training set and validation set. An additional 15 negative images (including five photographs, five instances of random noise, and five different cells) without any stomata were added to the training set to prevent overfitting. For StomaNet confocal mode, we labeled six additional 2D images transformed from 3D confocal images for transfer learning. For StomaNetUniversal model, we manually labeled 140 images of diverse species using the same method.
Model training
StomaNet was built to be trainable with devices accessible to most researchers in an acceptable timeframe. We trained StomaNet with 4 Tesla K40m in approximately 2 h, and it could be trained with a GTX 1060 6G in ∼8 h. It is possible to train StomaNet with cheaper video gaming cards, but the batch size should be decreased according to the VRAM of the card used. We used Nadam optimizer with a batch size of 25 for StomaNet.
Transfer learning
Transfer learning is implemented by using another pretrained model’s weights as initial weights and then applying the above training procedure to fine-tune the model. We did not freeze the weights of any layers in the initial model, as low-level features also vary using different imaging methods. The StomaNetConfocal model was trained with six manually labeled images based on StomaNet’s model. The StomaNetUniversal model was trained with 140 manually labeled images based on an initial model trained on 960 images with rough labels from StomataCounter’s prediction.
Prediction
The prediction of stomata involves three steps: (1) stack results from the network to produce a heatmap with the same size as the input image; (2) assign the score as weight for the pixels in the heatmap with scores >0.5 and perform DBSCAN (eps = 10, minimum samples = 40) on these pixels to generate clusters; and (3) perform PCA on each cluster and use the two principal components to describe the stoma as an ellipse. The stoma center is the weighted average of all pixels, and stoma size is the count of all pixels in the cluster multiplied by the size correction ratio (0.85 for StomaNet). The stoma length/width ratio is computed as (PC1/PC2)0.5, and stoma angle is the angle of PC1.
The LeafSeg module for segmenting pavement cells
After detecting stomata in the input image, the LeafSeg module segments pavement cells using a region merging algorithm as follows: (1) mask stomata in black (grayscale = 0) with the length and width multiplied by 1.5 and copy the masked image; (2) perform a median blur on the masked image; (3) binarize the blurred image with Otsu threshold and copy it; (4) skeletonize the image, remove isolated skeletons <64 px in size, and dilate the remaining skeletons by 4 px to obtain smooth borders; (5) perform a Euclidean distance transform on the border image; (6) perform the watershed algorithm on the masked image from step 1, with the peaks in the distance image from step 5 as different labels; (7) obtain border dots from watershed areas, and define the border score as the average value of the binarized image from step 3 under the border; and (8) merge the areas based on border score. An area can only merge with one other area (with the lowest score) at a time. Border score is recalculated after each merge. The final borders are used as the boundaries of pavement cells for counting and statistical analysis.
The borders of an image (50 px wide after resizing) are considered invalid areas, which are marked with a red line in the segmentation results. If the stoma center is located in a valid area, the stoma is counted as 1 stoma; otherwise, it is not counted in the final statistical result. Pavement cells covering the edge of a valid area are counted as 0.5 pavement cell and are not included in other statistical results such as cell size.
Metrics for evaluating pavement cell segmentation
To evaluate pavement cell segmentation, three metrics were used: RQ, SQ, and PQ. The PQ is the product of SQ and RQ, which are defined as follows: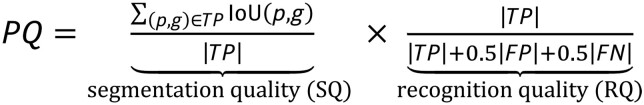 where TP represents true positives; FP represents false positives; FN represents false negatives; p and g represent prediction and ground truth; IoU stands for Intersection over Union; and |x| represents the number of x.
where TP represents true positives; FP represents false positives; FN represents false negatives; p and g represent prediction and ground truth; IoU stands for Intersection over Union; and |x| represents the number of x.
2D segmentation with ITK morphological watershed, PlantSeg, PaCeQuant, and Cellpose
For ITK morphological watershed, we applied the following steps for a fair comparison with LeafNet: (1) preprocess the image with Stained Denoiser, as described in the LeafNet workflow; (2) perform ITK morphological watershed on the preprocessed image, try different segmentation levels, and choose the one with the best PQ; (3) remove the areas darker than the Otsu threshold of the full image to remove cell walls; and (4) perform watershed with the remaining areas as labels to fill in the removed areas.
For PlantSeg, we used the confocal_2D_unet_bce_dice_ds3x model for boundary detection, applied rescaling based on the image resolution, and used the GASP algorithm for segmentation. We tried different Under-/Over-segmentation factors and CNN prediction thresholds and chose the values with the best PQ, and used the default values for the other parameters.
For PaCeQuant and Cellpose, we only set one parameter based on the input image resolution and used default values for other parameters.
Preprocessing of 3D images to 2D images
MorphoGraphX is used to generate 2D images from 3D image stacks via the following processes: (1) use Stack->Canvas->ReverseAxes to reverse z-axis when the image stack is upside down; (2) use Stack->Filters->Gaussian Blur Stack to denoise the image by 1 μm; (3) use Stack->Multi-stack->Copy Work to Main Stack to save the denoised stack; (4) use Stack->Morphology->Edge Detect to create a solid shape; (5) use Mesh->Creation->Marching Cube Surface to create a mesh surface; (6) use Mesh->Structure->Subdivide and Mesh->Structure->Smooth Mesh to smooth the mesh; (7) use Stack->Multi-stack->Copy Main to Work Stack to load the input signal; (8) use Stack->Mesh Interaction->Annihilate(minDist = 6 μm, maxDist = 8 μm) to remove the surface; and (9) save the work stack, and use ImageJ to generate a maximum intensity z-projection image.
Surface segmentation with MorphoGraphX
To segment 3D image stacks with MorphoGraphX, we first used the above steps 1–6 to create a mesh surface and then performed the following operations: (1) use Stack->Multi-stack->Copy Main to Work stack to load input signal; (2) use Mesh->Signal->Project Signal (minDist = 6 μm, maxDist = 8 μm) to project the signal onto the mesh surface; and (3) use Mesh->Segmentation->Auto-Segmentation (blur for seeding = 5 μm, radius for auto seeding = 5 μm, blur for cell outlines = 1 μm, normalize radius = 20 μm, border distance = 0.5 μm, merge threshold = 1.5 for input signal) to segment the mesh surface.
Prediction of stomata using StomataCounter
To predict stomata with StomataCounter, we used the model named sc_feb2019.caffemodel, set the scale parameter as 2 for our regular (A. thaliana, bright-field) and confocal (A. thaliana, confocal) dataset, and set the scale as 1 for the N. tabacum, bright-field dataset. Confocal maximum intensity z-projection images should be inverted for better performance. We tried different values for the threshold parameter in console mode to call stoma and used the one with the best F1 score (1.625 for the regular dataset, 2.25 for the N. tabacum dataset, and 0.3 for the confocal dataset).
Manual correction procedure with LeafNet
All results of LeafNet reported in this article are original output without any correction. We added this section for users to improve the segmentation results. The manual correction procedure involves the following steps: (1) use LeafNet to generate a segmentation image; (2) load the sample image and annotation image into GIMP or Photoshop; (3) place the annotation image in a layer above the sample image; (4) set the opacity of the annotation layer to 50%; (5) correct pavement cell boundaries with 100 hardness and in green (0,255,0) on the annotation layer, and correct stomata with 100 hardness and in blue (0,0,255) on the annotation layer; (6) set the opacity of the annotation layer back to 100%; (7) set the background of the image to black (0,0,0); (8) remove the sample image layer; and (9) flatten the image, and save the corrected annotation.
Quantification and statistical analysis
The quantifications of stomata and pavement cells are from LeafNet version 1.0. To test for the differences between manual and LeafNet predictions across different genotypes (Figure 6, E and F), we used the paired two-tailed t test.
Data and code availability
Source code and released LeafNet packages are available in the GitHub repository: https://github.com/zhouyulab/leafnet, and the web application is available at https://leafnet.whu.edu.cn/. The training data and the results from all the analysis, as well as detailed configurations to run these tools, are available at https://leafnet.whu.edu.cn/suppdata/.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Graphical representation of the LeafNet workflow.
Supplemental Figure S2. Detailed structure of StomaNet.
Supplemental Figure S3. Representative examples of segmentation results from four programs using their preferred images.
Supplemental Figure S4. Integration of LeafNet with PaCeQuant.
Supplemental Figure S5. Preprocessing of a 3D image and representative results from five programs.
Supplemental Figure S6. Representative examples of segmentation results from five tools using complex datasets.
Supplemental Figure S7. User interfaces for LeafNet.
Supplemental Figure S8. Sample output for LeafNet.
Supplemental Figure S9. Performance of CNNwall enhancement for pavement cell segmentation.
Supplementary Material
Acknowledgments
The authors thank the undergraduates in Class 4 of Biological Science in Grade 2019 from College of Life Science at Wuhan University, especially Keyu Lu, Dongzi Liu, Yuxuan Yang, Liran Mao, Yinghao Sun, Miao Lou, Zhiming He, and Shilong Zhao for their help in manual annotation of data.
Funding
This study was supported by grants from the National Natural Science Foundation of China (31922039 to Y.Z. and 31830057 to Y.X.Z.), the National Key R&D Program of China (2017YFA0504400 to Y.Z.), the Natural Science Foundation of Hubei Province, China (2020CFA057 to Y.Z.), the Chinese Scholarship Council (predoctoral fellowships to C.Z.), and the Ghent University “Bijzondere Onderzoekfonds” (BOF/CHN/010 to C.Z.). Part of computation in this work was done on the supercomputing system in the Supercomputing Center of Wuhan University.
Conflict of interest statement: None declared.
Contributor Information
Shaopeng Li, College of Life Sciences, Wuhan University, Wuhan 430072, China.
Linmao Li, College of Life Sciences, Wuhan University, Wuhan 430072, China.
Weiliang Fan, College of Life Sciences, Wuhan University, Wuhan 430072, China.
Suping Ma, College of Life Sciences, Wuhan University, Wuhan 430072, China.
Cheng Zhang, Department of Plant Biotechnology and Bioinformatics, Ghent University, Ghent 9052, Belgium; Center for Plant Systems Biology, VIB, Ghent 9052, Belgium.
Jang Chol Kim, College of Life Sciences, Wuhan University, Wuhan 430072, China.
Kun Wang, College of Life Sciences, Wuhan University, Wuhan 430072, China.
Eugenia Russinova, Department of Plant Biotechnology and Bioinformatics, Ghent University, Ghent 9052, Belgium; Center for Plant Systems Biology, VIB, Ghent 9052, Belgium.
Yuxian Zhu, College of Life Sciences, Wuhan University, Wuhan 430072, China; Institute for Advanced Studies, Wuhan University, Wuhan 430072, China.
Yu Zhou, College of Life Sciences, Wuhan University, Wuhan 430072, China; Institute for Advanced Studies, Wuhan University, Wuhan 430072, China; State Key Laboratory of Virology, Wuhan University, Wuhan 430072, China; Frontier Science Center for Immunology and Metabolism, Wuhan University, Wuhan 430072, China.
Y.Z., Y.X.Z., and K.W. designed the research. L.L. performed the experiments. L.L., S.L., S.M., and J.C.K. generated manually labeled data. S.L., Y.Z., and C.Z. developed the computational method. S.L. implemented the standalone tool and W.F. implemented the web server. Y.Z., Y.X.Z., and E.R. wrote the paper with inputs from S.L. and L.L. All authors discussed the results and approved the manuscript.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) are: Yuxian Zhu (zhuyx@whu.edu.cn) and Yu Zhou (yu.zhou@whu.edu.cn).
References
- Barbier de Reuille P, Routier-Kierzkowska AL, Kierzkowski D, Bassel GW, Schüpbach T, Tauriello G, Bajpai N, Strauss S, Weber A, Kiss A, et al. (2015) MorphoGraphX: A platform for quantifying morphogenesis in 4D. eLife 4: e05864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson SA, Hetherington AM (2010) Environmental regulation of stomatal development. Curr Opin Plant Biol 13: 90–95 [DOI] [PubMed] [Google Scholar]
- Christiansen EM, Yang SJ, Ando DM, Javaherian A, Skibinski G, Lipnick S, Mount E, O’Neil A, Shah K, Lee AK, et al. (2018) In silico labeling: predicting fluorescent labels in unlabeled images. Cell 173: 792–803.e19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engineer CB, Ghassemian M, Anderson JC, Peck SC, Hu H, Schroeder JI (2014) Carbonic anhydrases, EPF2 and a novel protease mediate CO2 control of stomatal development. Nature 513: 246–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engineer CB, Hashimoto-Sugimoto M, Negi J, Israelsson-Nordström M, Azoulay-Shemer T, Rappel WJ, Iba K, Schroeder JI (2016) CO2 sensing and CO2 regulation of stomatal conductance: Advances and open questions. Trends Plant Sci 21: 16–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erguvan Ö, Louveaux M, Hamant O, Verger S (2019) ImageJ SurfCut: A user-friendly pipeline for high-throughput extraction of cell contours from 3D image stacks. BMC Biol 17: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetter KC, Eberhardt S, Barclay RS, Wing S, Keller SR (2019) StomataCounter: A neural network for automatic stomata identification and counting. New Phytol 223: 1671–1681 [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Negi J, Young J, Israelsson M, Schroeder JI, Iba K (2006) Arabidopsis HT1 kinase controls stomatal movements in response to CO2. Nat Cell Biol 8: 391–397 [DOI] [PubMed] [Google Scholar]
- McCormick M, Liu X, Jomier J, Marion C, Ibanez L (2014) ITK: Enabling reproducible research and open science. Front Neuroinformatics 8: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller B, Poeschl Y, Plötner R, Bürstenbinder K (2017) PaCeQuant: A tool for high-throughput quantification of pavement cell shape characteristics. Plant Physiol 175: 998–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Torii KU (2018) Hormonal and environmental signals guiding stomatal development. BMC Biol 16: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapala A, Runions A, Routier-Kierzkowska AL, Das Gupta M, Hong L, Hofhuis H, Verger S, Mosca G, Li CB, Hay A, et al. (2018) Why plants make puzzle cells, and how their shape emerges. eLife 7: e32794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: An open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer C, Wang T, Michaelos M, Pachitariu M (2021) Cellpose: A generalist algorithm for cellular segmentation. Nat Methods 18: 100–106 [DOI] [PubMed] [Google Scholar]
- Ulman V, Maška M, Magnusson KEG, Ronneberger O, Haubold C, Harder N, Matula P, Matula P, Svoboda D, Radojevic M, et al. (2017) An objective comparison of cell-tracking algorithms. Nat Methods 14: 1141–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vőfély RV, Gallagher J, Pisano GD, Bartlett M, Braybrook SA (2019) Of puzzles and pavements: A quantitative exploration of leaf epidermal cell shape. New Phytol 221: 540–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolny A, Cerrone L, Vijayan A, Tofanelli R, Barro AV, Louveaux M, Wenzl C, Strauss S, Wilson-Sánchez D, Lymbouridou R, et al. (2020) Accurate and versatile 3D segmentation of plant tissues at cellular resolution. eLife 9: e57613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Jiang Y, Jia B, Zhou G (2016) Elevated-CO2 response of stomata and its dependence on environmental factors. Front Plant Sci 7: 657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoulias N, Harrison EL, Casson SA, Gray JE (2018) Molecular control of stomatal development. Biochem J 475: 441–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source code and released LeafNet packages are available in the GitHub repository: https://github.com/zhouyulab/leafnet, and the web application is available at https://leafnet.whu.edu.cn/. The training data and the results from all the analysis, as well as detailed configurations to run these tools, are available at https://leafnet.whu.edu.cn/suppdata/.