ABSTRACT
Long non-coding RNA nuclear paraspeckle assembly transcript 1 (NEAT1) is a novel pro-inflammatory factor in severe human diseases. Since inflammatory plays important roles in periodontitis progression, we aimed to explore the role of NEAT1 in chronic periodontitis (CP) in vitro. We established a periodontitis cell model was established by Porphyromonas gingivalis lipopolysaccharide (Pg-LPS)-induced periodontal ligament cells (PDLCs). Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed to detect the expression of NEAT1, microRNA (miR)-200c-3p, and tumor necrosis factor receptor-associated factor 6 (TRAF6). Cell viability, inflammatory factors, and protein expression of Bcl-2, Bax, and TRAF6 were analyzed by MTT, enzyme-linked immunosorbent assay, and Western blot. The target relationships among NEAT1, miR-200c-3p, and TRAF6 were predicted by the StarBase/TargetScan software, and further validated by dual-luciferase reporter assay. In this research, NEAT1 is up-regulated in CP tissues and periodontitis model group. Silencing of NEAT1 and over-expression of miR-200c-3p enhanced cell viability and repressed apoptosis in the periodontitis model group. NEAT1 targets miR-200c-3p, and miR-200c-3p further targets TRAF6. MiR-200c-3p inhibitor or over-expression of TRAF6 reversed the promoting effect of NEAT1 knockdown on cell viability, and the inhibiting effects on inflammatory cytokines and cell apoptosis. Consequently, the silencing of NEAT1 inhibits inflammation and apoptosis via targeting miR-200c-3p/TRAF6 axis, thereby contributing to alleviate the progression of CP. This finding could provide an underlying target for the treatment of CP.
KEYWORDS: NEAT1, miR-200c-3p, TRAF6, chronic periodontitis, in vitro
Introduction
Chronic periodontitis (CP) is a common dental inflammatory disease [1] primarily caused by Porphyromonas gingivalis (P. gingivalis), and its virulence factor lipopolysaccharide (LPS) is the main pathogenic factor of periodontitis [2,3]. Its pathological features include increased subgingival pathogens and connective tissues injuries surrounding the teeth [4,5]. Inflammation of periodontal ligament cells (PDLCs) is important for periodontal ligament tissues regeneration [6,7]. Therefore, this study places emphasis on the regulatory mechanisms underlying the pathogenesis of periodontitis based on Pg-LPS-induced PDLCs inflammatory damage.
Long non-coding RNAs (lncRNAs) have no protein-coding ability with 200 nucleotides in length [8] and are closely associated with inflammatory-related diseases, such as CP [9,10]. Previous studies have found that over-expression of taurine-up-regulated gene 1 (TUG1) facilitates the proliferation of LPS-induced PDLCs and restrains cell apoptosis [11]. Chen et al. suggested that the up-regulation of lncRNA FGD5-antisense RNA 1 (AS1) in LPS-induced PDLCs could protect against periodontitis [12]. On the other hand, silencing of metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) attenuates the inflammatory response in LPS-induced human gingival fibroblasts (HGFs) [13]. These findings imply that the abnormal expression of lncRNAs influences the progression of CP. lncRNA nuclear paraspeckle assembly transcript 1 (NEAT1) modulates inflammation in several diseases, including asthma [14], chronic obstructive pulmonary disease (COPD) [15], diabetic nephropathy [16], and sepsis [17–19]. Importantly, a study has reported dysregulation of lncRNA NEAT1 in gingival tissues or blood samples of patients with periodontitis in comparison with healthy subjects [20]. Huang et al. indicated that the level of lncRNA NEAT1 was up-regulated in PDLCs subjected to compressive force by qRT-PCR and RNA sequencing assay [21]. However, studies of CP involving NEAT1 are restricted to the level of tissues. Therefore, the mechanism of NEAT1 involved in CP in cellular level needs to be further elucidated.
Some microRNAs (miRs) have anti-inflammation roles in CP. For instance, miR-146a or miR-210 over-expression decreases the secretion of IL-1β and IL-6 in LPS-induced periodontal ligament fibroblasts [22,23]. Additionally, miR-200c-3p is reported to exert an anti-inflammatory role in pre-osteoblasts and HGFs, eventually attenuating the development of periodontitis [24,25]. Notably, it remains unclear whether miR-200c-3p is modulated by NEAT1 to exert its anti-inflammatory function in CP pathogenesis.
Tumor necrosis factor receptor-associated factor (TRAF) is an oncogene in the pathogenesis of several human cancers, including colorectal [26], gastric [27], breast [28], and prostate [29] cancers. In recent years, the pro-inflammatory role of TRAF6 has attracted increasing attention [30,31]. TRAF6 is regulated by lncRNA MIAT aggravates the inflammatory response in LPS-induced septic cardiomyopathy [30]. Silencing of TRAF6 has a nephroprotective effect on LPS-induced acute renal injury by suppressing inflammation [31]. P22077 could inhibit inflammation and reduce the lung injury by promoting TRAF6 degradation in LPS-induced endotoxemia mice [32]. The down-regulation of TRAF6 has a suppressive effect on inflammation in PDLCs induced by P. gingivalis [33]. Furthermore, TRAF6 is also negatively regulated by miR-146a in CP [22]. Nevertheless, the interactions among TRAF6, NEAT1, and miR-200c-3p axis in the pathogenesis of CP are relatively unknown.
In this research, we employed CP tissues and LPS-induced PDLCs in vitro to determine whether NEAT1 is involved in the regulation of CP and what is the underlying mechanism in the progression of CP. The result showed that NEAT1 was up-regulated in CP tissues and model group. The silencing of NEAT1 could protect the PDLCs against LPS-induced inflammation and apoptosis by targeting miR-200c-3p/TRAF6 axis, thereby contributing to alleviate the progression of CP. The results may provide a novel insight for the pathophysiology mechanism of CP and may provide support for NEAT1 in the clinical applications of CP therapy.
Materials and methods
Tissues collection
In total, 28 patients with CP without other diseases were selected from 2017 to 2018 in our hospital. Simultaneously, 20 healthy volunteers undergoing a physical examination were recruited. Gingiva tissues were obtained through operation from CP patients, followed by an original ineffectual nonsurgical scaling and root planning in accordance with the established professional and required oral hygiene of patients. Gingival tissues were also procured through crown-lengthening procedures from healthy individuals with the following inclusion criteria: clinical attachment loss <4 mm, probing depth (PD) <4 mm, and no alveolar bone destruction at radiographic level [12]. All the collected gingival tissue samples were frozen in liquid nitrogen and then stored immediately at −80°C for further experiments. The procedures were conducted based on the Declaration of Helsinki and obtained the approval of our hospital’s ethics committee. Each participant has obtained informed consent.
Isolation, culture, and transfection of human PDLCs
PDLCs were isolated from the healthy periodontal ligament in the middle third of the periodontal ligament root of the third molars as previously described [34]. The cells were cultured in Dulbecco’s Modified of Eagle’s Medium (DMEM). Immunohistochemistry and Alizarin red staining were used to identify the PDLCs as previously described [34]. Immunocytochemistry showed that cells with a spindle shape were positive in vimentin and negative in keratin. Alizarin red staining revealed that the cells could osteogenically differentiate. Consequently, these cells were identified as PDLCs. PDLCs in the third generation were used in the next experiments [35]. PDLCs were then divided into a control group and a model group. PDLCs induced by Pg-LPS (10 μg/mL) were served as the periodontitis model group [36]. These cells served as the control (without LPS treatment). shRNA-negative control (sh-NC), sh-NEAT1, miR-200c-3p inhibitor, miR-200c-3p mimics, miR-NC, and over-expression TRAF6 vector (pcDNA-TRAF6) were co-transfected with LPS-induced PDLCs for 48 h. PDLCs were collected to perform further experiments.
Quantitative real-time PCR qRT-PCR
The total RNA from PDLCs was extracted using RNAiso Plus (Takara, Tokyo, Japan). RNA was reversed transcribed into cDNA using M-MLV Reverse Transcriptase kit (Sigma-Aldrich). qRT-PCR was performed using a SYBR® Green PCR Kit (Sigma-Aldrich) according to the instructions of the manufacturer. GAPDH or U6 was used for the normalization of the mRNA expression of NEAT1, TRAF6, and miR-200c-3p.
Western blotting assay
The protein samples were extracted using RIPA lysis buffer (Solarbio, Beijing, China). The proteins were then separated by 10% SDS-PAGE and then transferred onto polyvinylidene difluoride (PVDF). The membranes were blocked by 5% skim milk at 25°C for 1 h. Afterward, the membranes were incubated with primary antibodies, including TRAF6 (1:1000, Abcam), Bax (1:1000, Abcam), Bcl-2 (1:1000, Abcam), and β-actin (1:1000, Abcam) at 4°C overnight. Afterward, the membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibody at 25°C for 1 h. Blots were visualized and analyzed using a chemiluminescence system (Bio-Rad, CA, USA). β-actin was employed as a protein loading control.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay
The LPS-induced PDLCs were seeded in 96-well plate and incubated for 24 h. Thereafter, 5 mg/mL of MTT was added to incubate for another 2 h at 37°C with 5% CO2. Dimethyl sulfoxide was added to terminate reactions. The viability was analyzed by a microplate reader (Thermo Fisher Scientific) at OD 450.
Enzyme-linked immunosorbent assay (ELISA)
Culture media was collected from each group, including the control, Model, Model + sh-NEAT1/NC, Model + miR-200c-3p mimics/NC, Model + sh-NEAT1 + miR-200c-3p inhibitor, and Model + sh-NEAT1 + pcDNA-TRAF6. The levels of tumor necrosis factor (TNF)-α, interleukin (IL)-6, and IL-1β were detected by ELISA kits according to the manufacturer’s guidelines (Eusebio, Shanghai, China). The absorbance was detected at 450 nm by a microplate reader (Bio-Rad).
Dual luciferase reporter (DLR) assay
Target-prediction analyses for the targets miRNAs of NEAT1 and miRNA targets sites were formed using TargetScan (http://www.targetscan.org//) and StarBase (https://starbase.sysu.edu.cn/index.php). The 3’-UTR fragment of NEAT1 or TRAF6 which contained binding sites of miR-200c-3p was introduced into a pGL3-promotor vector to construct NEAT1 wt (mut) or TRAF6 wt (mut). Next, the above reporter vectors along with miR-NC/miR-200c-3p mimics were transfected into PDLCs via Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. After transfection for 48 h, relative luciferase activity was assessed with a Dual-Glo Luciferase assay kit (Promega, Madison, WI, USA).
Statistical analysis
Data were expressed as the mean ± SD and analyzed using SPSS 20.0 (SPSS; Chicago, USA). Student’s t-test was used to analyze the differences between the two groups. One-way ANOVA followed by Tukey post-hoc tests and two-way ANOVA was used for comparisons between two groups or among multiple groups. P< 0.05 was considered to be a statistically significant difference.
Results
In this study, we established a periodontitis cell model by Pg-LPS-induced PDLCs. We determined the levels of TNF-α, IL-1β, and IL-6, as well as protein expression of Bcl-2, Bax, and TLR4 by ELISA and Western blot. The expression of NEAT1, miR-200c-3p, and TRAF6 was detected by qRT-PCR. The results showed that the silencing of NEAT1 could protect the PDLCs against LPS-induced inflammation and apoptosis by targeting miR-200c-3p/TRAF6 axis, thereby contributing to alleviate the progression of CP.
Knockdown of NEAT1 inhibits inflammatory responses in model group
The expression of NEAT1 was initially determined by qRT-PCR, and we found that NEAT1 was highly expressed in CP tissues (Figure 1a, p < 0.01) and the model group compared with respective controls (Figure 1b, p < 0.01). qRT-PCR assay showed that the expression of NEAT1 was decreased in sh-NEAT1 group compared with sh-NC group (Figure 1c, p < 0.01), suggesting the successful transfection of sh-NEAT1. The viability of PDLCs was decreased after LPS stimulation, while sh-NEAT1 reversed the inhibitory effect of LPS on cell viability (Figure 1d, p < 0.01). Thereafter, the influences of NEAT1 silencing on inflammatory responses were researched. As illustrated in Figure 1e-g, the levels of IL-6, IL-1β, and TNF-α were distinctly promoted in model groups. However, the promoting roles of LPS in these inflammatory cytokines were reversed by sh-NEAT1 (P < 0.01). Western blotting assay uncovered that the LPS treatment had a promoting effect on Bax level, and a suppressive effect on Bcl-2 protein expression (Figure 1h, p < 0.01). As expected, the transfection of sh-NEAT1 significantly reversed the effects of LPS on Bax and Bcl-2 (Figure 1h, p < 0.01). LPS was also found to dramatically elevate the Bax/Bcl-2 ratio, which was reversed by the transfection of sh-NEAT1 (Figure 1i, p < 0.01).
Figure 1.
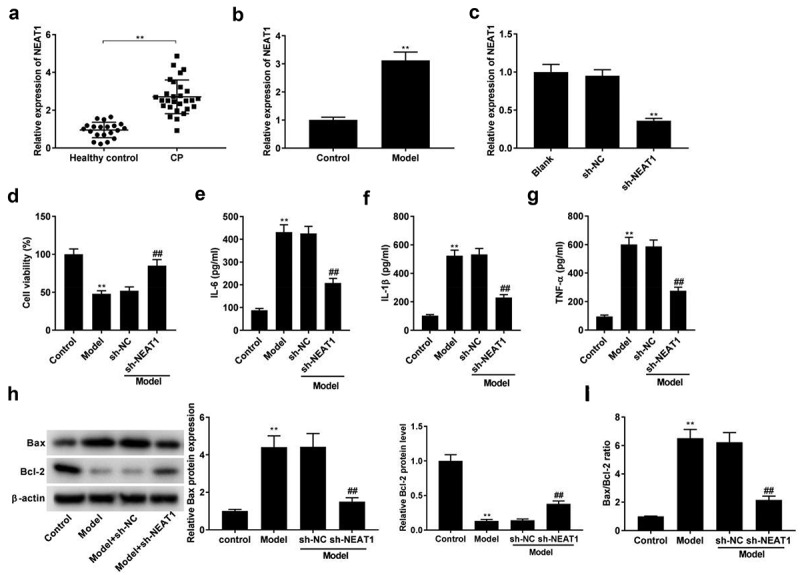
Knockdown of NEAT1 inhibits inflammation in model group. (a) NEAT1 expression in healthy control individuals and CP tissues. **P < 0.01 vs. healthy control. (b) NEAT1 expression in model group. **P< 0.01 vs. Control. (c) The transfection efficiency of NEAT1 was detected by qRT-PCR. **P< 0.01 vs. sh-NC. (d-g) Cell viability, and the levels of IL-6, IL-1β, and TNF-α in control, model, model + sh-NC, and model + sh-NEAT1 groups. **P< 0.01 vs. Control. ##P < 0.01 vs. model + sh-NC. (h) Relative protein expression of Bax and Bcl-2 was detected by Western blot. **P< 0.01 vs. Control. ##P < 0.01 vs. model + sh-NC. (i) The ratio of Bax/Bcl-2 in control, model, model + sh-NC, and model + sh-NEAT1 groups. **P< 0.01 vs. Control. ##P < 0.01 vs. model + sh-NC.
NEAT1 targets miR-200c-3p
StarBase was used to predict the relationship between NEAT1 and miR-200c-3p, and the binding site is shown in Figure 2a. We found that NEAT1 knockdown elevated miR-200c-3p expression (Figure 2b, p < 0.01). In addition, miR-200c-3p mimics co-transfected with NEAT1 wt remarkably declined luciferase activity in PDLCs (Figure 2c, p < 0.01), which confirmed that NEAT1 directly targeted miR-200c-3p.
Figure 2.
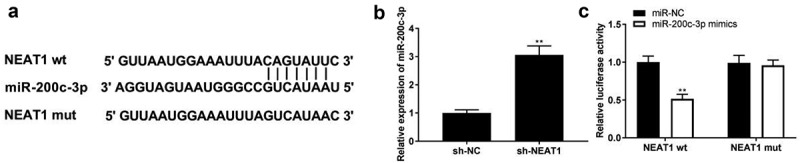
NEAT1 targets miR-200c-3p. (a) The binding site of NEAT1 and miR-200c-3p was predicted by TargetScan. (b) The expression of miR-200c-3p was detected by qRT-PCR. **P < 0.01 vs. sh-NC. (c) The interaction between NEAT1 and miR-200c-3p was determined by DLR assay. **P < 0.01 vs. miR-NC.
Over-expression of miR-200c-3p represses inflammation of in model group
In order to explore the function of miR-200c-3p in CP, the expression of miR-200c-3p was detected by qRT-PCR, and we found that miR-200c-3p was down-expressed in CP tissues and model group (Figure 3a-b, P < 0.01). Then, miR-200c-3p mimics were successfully transfected into model group to explore the function of miR-200c-3p on CP progression in vitro (Figure 3c, p < 0.01). As presented in Figure 3d-g, miR-200c-3p mimics remarkably facilitated the viability (P < 0.01) and repressed the levels of IL-6, IL-1β, and TNF-α (P< 0.01). In addition, miR-200c-3p mimics down-regulated the expression of Bax and up-regulated Bcl-2 expression, as well as decreased Bax/Bcl-2 ratio (Figure 3h-i, P < 0.01).
Figure 3.
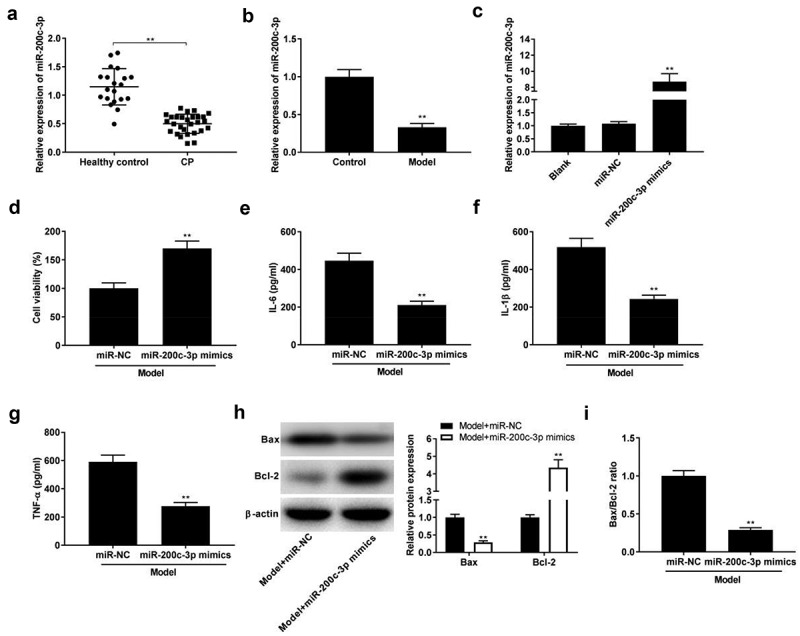
Over-expression of miR-200c-3p inhibits inflammatory response in model group. (a) The expression of miR-200c-3p in healthy control individuals and CP tissues. **P < 0.01 vs. healthy control. (b) The expression of miR-200c-3p in model group. **P< 0.01 vs. control. (c) The expression of miR-200c-3p was detected by qRT-PCR after transfection of miR-NC and miR-200c-3p mimics. **P < 0.01 vs. miR-NC. (d-g) Cell viability and the levels of IL-6, IL-1β, and TNF-α in model + miR-NC and model + miR-200c-3p mimics groups. **P< 0.01 vs. miR-NC. (h-i) Relative protein expression of Bax and Bcl-2, and the ratio of Bax/Bcl-2 in model + miR-NC and model + miR-200c-3p mimics groups. **P < 0.01 vs. miR-NC.
TRAF6 is a target of miR-200c-3p
The binding site between miR-200c-3p and TRAF6 was predicted using TargetScan software, and a binding site is shown in Figure 4a. Western blot assay showed that TRAF6 expression was significantly decreased by the transfection of miR-200c-3p mimics (Figure 4b, p< 0.01). The DLR assay indicated that luciferase activity significantly decreased in PDLCs co-transfected with TRAF6-wt and miR-200c-3p mimics by contrast to in the TRAF6-wt + miR-NC group (Figure 4c, p< 0.01).
Figure 4.
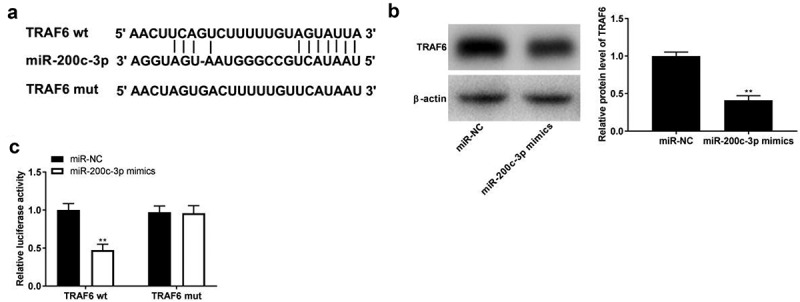
TRAF6 is a target of miR-200c-3p. (a) The binding sites of TRAF6 and miR-200c-3p were predicted by StarBase software. (b) The expression of TRAF6 was detected by Western blot. **P < 0.01 vs. miR-NC. (c) Target relationship of TRAF6 and miR-200c-3p. **P < 0.01 vs. miR-NC.
Silencing of NEAT1 inhibits inflammation and apoptosis by targeting miR-200c-3p/TRAF6 axis
The expression of TRAF6 in CP tissues and model group was visibly up-regulated compared with respective controls (Figure 5a-b, P < 0.01). At the same time, we analyzed the protein level of TRAF6 in model group. The results showed that protein expression of TRAF6 was down-regulated by sh-NEAT1, while miR-200c-3p inhibitor partly reversed the effect of sh-NEAT1 on protein expression of TRAF6 in model group (Figure 5c, p < 0.01). Subsequently, feedback verification experiments were performed to investigate the interactions among NEAT1, miR-200c-3p, and TRAF6 on CP progression in vitro. As illustrated in Figure 5d, the transfection of miR-200-3p inhibitor and pcDNA-TRAF6 reversed the promoting effect of sh-NEAT1 on viability in model groups (Figure 5d, p < 0.01). The anti-inflammatory effects of sh-NEATT were markedly reversed by the transfection of miR-200-3p inhibitor and pcDNA-TRAF6 in model groups (Figure 5e-g, P < 0.01). Moreover, miR-200-3p inhibitor and pcDNA-TRAF6 also reversed the effects of sh-NEATT on the expression of Bcl-2 and Bax, and the ratio of Bax/Bcl-2 in model groups (Figure 5h-i, P < 0.01).
Figure 5.
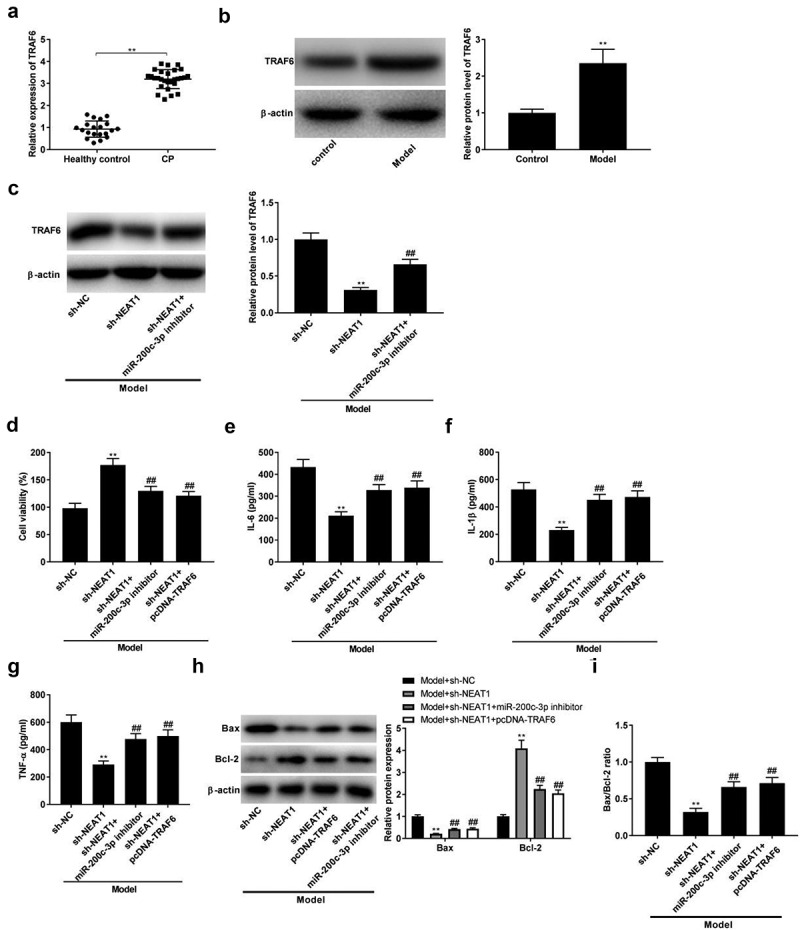
Silencing of NEAT1 inhibits inflammation and apoptosis by targeting miR-200c-3p/TRAF6 axis. (a) TRAF6 expression in healthy control individuals and CP tissues. **P < 0.01 vs. healthy control. (b) TRAF6 expression in control and model groups. **P< 0.01 vs. control. (c) The expression of TRAF6 was determined by Western blot after transfection of sh-NEAT1/sh-NEAT1 + miR-200c-3p inhibitor. **P < 0.01 vs. sh-NC. ##P < 0.01, vs. sh-NEAT1. (d-g) Cell viability and the levels of IL-6, IL-1β, and TNF-α in model + sh-NC, model + sh-NEAT1, model + sh-NEAT1 + miR-200c-3p inhibitor, and model + sh-NEAT1 + pcDNA-TRAF6 groups. **P < 0.01 vs. sh-NC. ##P < 0.01, vs. sh-NEAT1. (h-i) Relative protein expression of Bax and Bcl-2, and the ratio of Bax/Bcl-2 in Model + sh-NC, model + sh-NEAT1, model + sh-NEAT1 + miR-200c-3p inhibitor, and model + sh-NEAT1 + pcDNA-TRAF6 groups. **P < 0.01 vs. sh-NC. ##P < 0.01, vs. sh-NEAT1.
Discussion
A study showed that Pg-LPS inhibited cell viability and triggered inflammation of PDLCs [36]. Consistent with this study, our result showed that LPS stimulation reduced the viability of PDLCs and increased the levels of inflammatory factors (IL-6, IL-1β, and TNF-α) in model groups. Furthermore, LncRNA NEAT1 plays a crucial role in several inflammation diseases [14,16,18,37]. The up-regulation of NEAT1 occurs in the tissues of patients with asthma, COPD, and acute kidney injury [14,16,38]. In this study, we found that NEAT1 was significantly up-expressed in CP tissues. Consistent with our results, Sayad et al. also indicated that NEAT1 expression in CP tissues is dramatically elevated [20]. Additionally, we showed that the expression of NEAT1 was up-regulated in model group, suggesting that NEAT1 may play a vital role in CP. To further explore the exact role of NEAT1 in CP, relevant in vitro experiments were performed in model groups. We demonstrated that the suppression of NEAT1 facilitates viability and inhibits apoptosis and inflammation in model group. In line with these results, Yi et al. discovered that knockdown of NEAT1 could promote cell viability, and suppress inflammatory factors and cell apoptosis in sepsis-induced acute kidney injury [38]. Therefore, we speculated that the silencing of NEAT1 might be a suppressor in the occurrence and development of CP.
miRNAs act as suppressors to participate in the inflammation reaction of CP in vitro or in vivo [23,39,40]. A decreased expression of miR-335-5p has been found in periodontitis tissues [40], and miR-210 is low-expressed not only in CP tissues but also in model group [23]. Additionally, miR-218 is minimally expressed both in CP tissues and periodontal ligament progenitor cells [39]. In this study, miR-200c-3p was reduced in CP tissues and PDLCs, and over-expression of miR-200c-3p reduced the levels of inflammatory factors in model group, which was consistent with previous studies [24,25]. Additionally, in the present study, we demonstrated that the up-regulation of miR-200c-3p promoted cell viability and inhibited apoptosis in model group. More importantly, miR-200c-3p was found to be up-regulated by sh-NEAT1, and a target of NEAT1. Our feedback verification experiments suggested that inhibition of miR-200c-3p reversed the effects of sh-NEAT1 on cell viability, cell apoptosis, and inflammatory factors in PDLCs. The above data indicated that silencing of NEAT1 inhibited apoptosis and inflammation model groups by up-regulating miR-200c-3p expression, thereby contributing to alleviate the progression of CP.
TRAF6 is involved in the regulation of CP [33,41] and is highly expressed in LPS-induced PDLCs [41]. LPS stimulation elevates TRAF6 expression in PDLCs [33]. Similarly, we found that the protein level of TRAF6 was up-regulated in the model group, and an increased expression of TRAF6 in CP tissues was observed. The result indicated that TRAF6 may be closely related to the progression of CP. Simultaneously, we verified that TRAF6 was a target gene of miR-200c-3p and silencing of NEAT1 down-regulated TRAF6 expression. The results of the feedback verification experiment verified that the over-expression of TRAF6 reversed the anti-inflammation and anti-apoptosis effects, as well as the promoting effect on the cell viability of sh-NEAT1. In other words, we believed that silencing NEAT1 alleviated the progression of CP via regulation of the miR-200c-3p/TRAF6 axis.
Conclusions
In conclusion, NEAT1 was significantly highly expressed in CP tissues and LPS-induced PDLCs. Silencing of NEAT1 markedly suppressed the inflammatory response and apoptosis via the miR-200c-3p/TRAF6 axis in the model group. Accordingly, NEAT may act as a potential therapeutic target for CP therapy in clinical applications. In addition, we failed to verify the NEAT1/miR-200c-3p/TRAF6 axis in vivo, and this may be a limitation of this study. We will elucidate this in the future.
Funding Statement
The author(s) reported that there is no funding associated with the work featured in this article.
Highlights
NEAT1 is up-regulated in CP tissues and model group.
NEAT1 targets miR-200c-3p in model group.
TRAF6 is a target of miR-200c-3p.
Silencing of NEAT1 inhibits inflammation and apoptosis by targeting miR-200c-3p/TRAF6 axis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- [1].Hajishengallis G. Immunomicrobial pathogenesis of periodontitis: keystones, pathobionts, and host response. Trends Immunol. 2014;35:3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Jain S, Darveau RP. Contribution of Porphyromonas gingivalis lipopolysaccharide to periodontitis. Periodontol 2000. 2010;54:53–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Poole S, Singhrao SK, Kesavalu L, et al. Determining the presence of periodontopathic virulence factors in short-term postmortem Alzheimer’s disease brain tissue. J Alzheimers Dis. 2013;36:665–677. [DOI] [PubMed] [Google Scholar]
- [4].Ratheesh V, Subramanian S, Prakash PSG, et al. Evaluation of association of vitamin D receptor genetic polymorphism with severe chronic periodontitis in an ethnic Tamilian population. Genet Test Mol Biomarkers. 2018;22:615–621. [DOI] [PubMed] [Google Scholar]
- [5].Van Dyke TE. The management of inflammation in periodontal disease. J Periodontol. 2008;79:1601–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Oates TW, Rouse CA, Cochran DL. Mitogenic effects of growth factors on human periodontal ligament cells in vitro. J Periodontol. 1993;64:142–148. [DOI] [PubMed] [Google Scholar]
- [7].Kanzaki H, Chiba M, Shimizu Y, et al. Periodontal ligament cells under mechanical stress induce osteoclastogenesis by receptor activator of nuclear factor kappaB ligand up-regulation via prostaglandin E2 synthesis. J Bone Miner Res. 2002;17:210–220. [DOI] [PubMed] [Google Scholar]
- [8].Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. [DOI] [PubMed] [Google Scholar]
- [9].Zhang P, Sun Y, Peng R, et al. Long non-coding RNA Rpph1 promotes inflammation and proliferation of mesangial cells in diabetic nephropathy via an interaction with Gal-3. Cell Death Dis. 2019;10:526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ma M, Pei Y, Wang X, et al. LncRNA XIST mediates bovine mammary epithelial cell inflammatory response via NF-κB/NLRP3 inflammasome pathway. Cell Prolif. 2019;52:e12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Han Y, Wang F, Shao L, et al. LncRNA TUG1 mediates lipopolysaccharide-induced proliferative inhibition and apoptosis of human periodontal ligament cells by sponging miR-132. Acta Biochim Biophys Sin (Shanghai). 2019;51:1208–1215. [DOI] [PubMed] [Google Scholar]
- [12].Chen H, Lan Z, Li Q, et al. Abnormal expression of long noncoding RNA FGD5-AS1 affects the development of periodontitis through regulating miR-142-3p/SOCS6/NF-κB pathway. Artif Cells Nanomed Biotechnol. 2019;47:2098–2106. [DOI] [PubMed] [Google Scholar]
- [13].Li J, Wang M, Song L, et al. LncRNA MALAT1 regulates inflammatory cytokine production in lipopolysaccharide-stimulated human gingival fibroblasts through sponging miR-20a and activating TLR4 pathway. J Periodontal Res. 2020;55:182–190. [DOI] [PubMed] [Google Scholar]
- [14].Li X, Ye S, Lu Y. Long non-coding RNA NEAT1 overexpression associates with increased exacerbation risk, severity, and inflammation, as well as decreased lung function through the interaction with microRNA-124 in asthma. J Clin Lab Anal. 2020;34:e23023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ming X, Duan W, Yi W. Long non-coding RNA NEAT1 predicts elevated chronic obstructive pulmonary disease (COPD) susceptibility and acute exacerbation risk, and correlates with higher disease severity, inflammation, and lower miR-193a in COPD patients. Int J Clin Exp Pathol. 2019;12:2837–2848. [PMC free article] [PubMed] [Google Scholar]
- [16].Ma J, Zhao N, Du L, et al. Downregulation of lncRNA NEAT1 inhibits mouse mesangial cell proliferation, fibrosis, and inflammation but promotes apoptosis in diabetic nephropathy. Int J Clin Exp Pathol. 2019;12:1174–1183. [PMC free article] [PubMed] [Google Scholar]
- [17].Wang W, Guo ZH. Downregulation of lncRNA NEAT1 ameliorates LPS-induced inflammatory responses by promoting macrophage M2 polarization via miR-125a-5p/TRAF6/TAK1 axis. Inflammation. 2020;43:1548–1560. [DOI] [PubMed] [Google Scholar]
- [18].Xia D, Yao R, Zhou P, et al. LncRNA NEAT1 reversed the hindering effects of miR-495-3p/STAT3 axis and miR-211/PI3K/AKT axis on sepsis-relevant inflammation. Mol Immunol. 2020;117:168–179. [DOI] [PubMed] [Google Scholar]
- [19].Zhang CC, Niu F. LncRNA NEAT1 promotes inflammatory response in sepsis-induced liver injury via the Let-7a/TLR4 axis. Int Immunopharmacol. 2019;75:105731. [DOI] [PubMed] [Google Scholar]
- [20].Sayad A, Mirzajani S, Gholami L, et al. Emerging role of long non-coding RNAs in the pathogenesis of periodontitis. Biomed Pharmacothe. 2020;129:110362. [DOI] [PubMed] [Google Scholar]
- [21].Huang Y, Zhang Y, Li X, et al. The long non-coding RNA landscape of periodontal ligament stem cells subjected to compressive force. Eur J Orthod. 2019;41:333–342. [DOI] [PubMed] [Google Scholar]
- [22].Tang L, Li X, Bai Y, et al. MicroRNA-146a negatively regulates the inflammatory response to porphyromonas gingivalis in human periodontal ligament fibroblasts via TRAF6/p38 pathway. J Periodontol. 2019;90:391–399. [DOI] [PubMed] [Google Scholar]
- [23].Jia S, Yang X, Yang X, et al. MicroRNA-210 protects against periodontitis through targeting HIF-3α and inhibiting p38MAPK/NF-κB pathway. Artif Cells Nanomed Biotechnol. 2020;48:129–136. [DOI] [PubMed] [Google Scholar]
- [24].Hong L, Sharp T, Khorsand B, et al. MicroRNA-200c represses IL-6, IL-8, and CCL-5 expression and enhances osteogenic differentiation. PloS One. 2016;11:e0160915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Akkouch A, Zhu M, Romero-Bustillos M, et al. MicroRNA-200c attenuates periodontitis by modulating proinflammatory and osteoclastogenic mediators. Stem Cells Dev. 2019;28:1026–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Liao X, Zhan W, Zhang J, et al. Long noncoding RNA LINC01234 promoted cell proliferation and invasion via miR-1284/TRAF6 axis in colorectal cancer. J Cell Biochem. 2020;121:4295–4309. [DOI] [PubMed] [Google Scholar]
- [27].Yang M, Jin M, Li K, et al. TRAF6 promotes gastric cancer cell self-renewal, proliferation, and migration. Stem Cells Int. 2020;2020:3296192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bishop RT, Marino S, Carrasco G, et al. Combined administration of a small-molecule inhibitor of TRAF6 and Docetaxel reduces breast cancer skeletal metastasis and osteolysis. Cancer Lett. 2020;488:27–39. [DOI] [PubMed] [Google Scholar]
- [29].Khusbu FY, Zhou X, Roy M, et al. Resveratrol induces depletion of TRAF6 and suppresses prostate cancer cell proliferation and migration. Int J Biochem Cell Biol. 2020;118:105644. [DOI] [PubMed] [Google Scholar]
- [30].Xing PC, An P, Hu GY, et al. LncRNA MIAT promotes inflammation and oxidative stress in sepsis-induced cardiac injury by targeting miR-330-5p/TRAF6/NF-κB axis. Biochem Genet. 2020;58:783–800. [DOI] [PubMed] [Google Scholar]
- [31].Chen X, Zhao Y, Xu J, et al. The nephroprotective effect of TNF receptor-associated factor 6 (TRAF6) blockade on LPS-induced acute renal injury through the inhibition of inflammation and oxidative stress. Med Sci Monit. 2020;26:e919698. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [32].Zhao XB, Ji FY, Li HR, et al. P22077 inhibits LPS-induced inflammatory response by promoting K48-linked ubiquitination and degradation of TRAF6. Aging (Albany NY). 2020;12:10969–10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Tang L, Zhou XD, Wang Q, et al. TNF receptor-associated factor 6 suppression inhibits inflammatory response to Porphyromonas gingivalis in human periodontal ligament cells. Quintessence Int (Berlin, Germany: 1985). 2011;42:787–796. [PubMed] [Google Scholar]
- [34].Li C, Li C, Yue J, et al. miR-21 and miR-101 regulate PLAP-1 expression in periodontal ligament cells. Mol Med Rep. 2012;5:1340–1346. [DOI] [PubMed] [Google Scholar]
- [35].Chen Q, Cao M, Ge H. Knockdown of MALAT1 inhibits the progression of chronic periodontitis via targeting miR-769-5p/HIF3A axis. Biomed Res Int. 2021;2021:8899863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chen W, Su J, Cai S, et al. Cullin3 aggravates the inflammatory response of periodontal ligament stem cells via regulation of SHH signaling and Nrf2. Bioengineered. 2021;12:3089–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Liu W, Konermann A, Guo T, et al. Canonical Wnt signaling differently modulates osteogenic differentiation of mesenchymal stem cells derived from bone marrow and from periodontal ligament under inflammatory conditions. Biochim Biophys Acta. 2014;1840:1125–1134. [DOI] [PubMed] [Google Scholar]
- [38].Chen Y, Qiu J, Chen B, et al. RETRACTED: long non-coding RNA NEAT1 plays an important role in sepsis-induced acute kidney injury by targeting miR-204 and modulating the NF-κB pathway. Int Immunopharmacol. 2018;59:252–260. [DOI] [PubMed] [Google Scholar]
- [39].Guo J, Zeng X, Miao J, et al. MiRNA-218 regulates osteoclast differentiation and inflammation response in periodontitis rats through Mmp9. Cell Microbiol. 2019;21:e12979. [DOI] [PubMed] [Google Scholar]
- [40].Lian J, Wu X, Liu Y, et al. Potential roles of miR-335-5p on pathogenesis of experimental periodontitis. J Periodontal Res. 2020;55:191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Tang L, Zhou XD, Wang Q, et al. Expression of TRAF6 and pro-inflammatory cytokines through activation of TLR2, TLR4, NOD1, and NOD2 in human periodontal ligament fibroblasts. Arch Oral Biol. 2011;56:1064–1072. [DOI] [PubMed] [Google Scholar]


