SUMMARY
Alternative splicing generates distinct mRNA variants and is essential for development, homeostasis, and renewal. Proteins of the serine/arginine (SR)-rich splicing factor family are major splicing regulators that are broadly required for organ development as well as cell and organism viability. However, how these proteins support adult organ function remains largely unknown. Here, we used the continuously growing mouse incisor as a model to dissect the functions of the prototypical SR-family protein SRSF1 during tissue homeostasis and renewal. We identified an SRSF1-governed alternative splicing network that is specifically required for dental proliferation and survival of progenitors but dispensable for the viability of differentiated cells. We also observed a similar progenitor-specific role of SRSF1 in the small intestinal epithelium, indicating a conserved function of SRSF1 across adult epithelial tissues. Thus, our findings define a regulatory mechanism by which SRSF1 specifically controls progenitor-specific alternative splicing events to support adult tissue homeostasis and renewal.
Keywords: Alternative splicing, splicing factor, progenitor, tissue homeostasis, tissue renewal, mouse, incisor, intestine
eTOC Blurb:
Cycling progenitors generate the terminally differentiated cells needed to maintain a functional organ. Yu et al. demonstrate in two self-renewing tissues, the ever-growing mouse incisor and the small intestine, that epithelial progenitors are maintained through proper splicing of several targets of the splice factor SRSF1.
Graphical Abstract

INTRODUCTION
Alternative splicing generates mRNA variants through selective inclusion or exclusion of exons or introns during pre-mRNA processing. This process is the primary mechanism to increase the diversity of protein isoforms that are structurally and functionally specialized in different tissues to ensure normal development (reviewed in Jin et al., 2018). Over the past decade, transcriptomic analyses performed in individual organs or whole organisms have identified distinct clusters of alternative splicing events that are required in various cell populations as well as at different developmental stages to support cell viability and tissue development (Salomonis et al., 2010; Graveley et al., 2011; Zhang et al., 2014). However, the spatial and temporal regulation of alternative splicing events by splicing regulators remains relatively unexplored. One mechanism for precisely controlling alternative splicing is via specific expression of splicing regulators in certain cell types (Hayakawa-Yano et al., 2017) or at specific developmental stages (Kalsotra et al., 2008). However, because many splicing regulators are broadly expressed (Kalsotra et al., 2008; Yeo et al., 2008; Damianov and Black. 2010), additional mechanisms that confer specificity on these important RNA binding proteins must exist.
Among the principal regulators of alternative splicing are members of the serine/arginine (SR)-rich splicing factor family, which bind to splicing enhancer sequences to promote alternative splicing (reviewed in Shepard and Hertel, 2009). The SR splicing factor family has 12 members in humans that share a conserved SR domain (reviewed in Manley and Krainer, 2010). Many SR splicing factors have been reported to be essential for development. Germline deletion of these factors leads to embryonic lethality or perinatal death (Jumaa et al., 1999; Ding et al., 2004; Xu et al., 2005; Feng et al., 2009), and tissue-specific deletion of SR genes causes organ failure, resulting in growth retardation and often lethality (Xu et al., 2005; Cheng et al., 2016). Despite the indispensable role of SR splicing factors during early development, their functions in adult tissues are largely unknown. With more than 95% of genes undergoing alternative splicing in adult tissues (Pan et al., 2008; Wang et al., 2008), many important questions remain unanswered, including how SR splicing factors maintain tissue homeostasis and renewal, as well as their roles in various cell types, including stem cells, transit-amplifying progenitors and differentiated cells.
We have turned to the continuously growing adult mouse incisor as a model to study the cellular function of SR splicing factors. The constant wear on the incisor as the animal gnaws on food is balanced by rapid replacement of cells and mineralized tissue thanks to a pool of adult stem cells. Therefore, like other high turnover epithelial organs, such as the intestine, the balance between cell proliferation and differentiation must be maintained to ensure proper homeostasis and renewal. An epithelial structure located at the proximal end of the adult mouse incisor called the labial cervical loop (laCL) houses a group of proliferating progenitors that produce multiple cell populations to support the continuously growing incisors throughout the animal’s lifetime (Fig. 1A) (reviewed in Yu and Klein, 2020). Dental progenitors reside in the inner enamel epithelium (IEE), giving rise to multiple cell lineages, including those in the stellate reticulum and the outer enamel epithelium (OEE) regions at the proximal end of laCL. IEE progenitors also contribute to the differentiation and maturation of enamel-secreting ameloblasts and to the formation of a single layer of stratum intermedium cells immediately adjacent to the ameloblasts.
Figure 1. Deleting Srsf1 specifically disrupts the function of dental progenitors in the adult mouse laCL.
(A) Schematic diagram of the labial cervical loop (laCL), located at the proximal mouse incisor. Dental progenitors residing in the proliferating inner enamel epithelium (IEE) drive the formation and differentiation of a variety of cell types (red arrow lines), including the enamel-secreting ameloblasts (AMB), stellate intermedium (SI) cells and other cell populations that reside in outer enamel epithelium (OEE) and stellate reticulum (SR). Dental mesenchyme-derived odontoblasts (ODB) produce dentin at the lingual surface of enamel. liCL, lingual cervical loop. Light green dashed line with arrowheads marks the IEE region. Dark green dashed line with arrowheads marks the mature region.
(B and C) Immunostaining of SRSF1 (B) and SRSF3 (C) in the laCL.
(D) Timeline depicting tamoxifen (Tam)-induced Cre activation (black arrowheads) and sample collection (blue arrowheads).
(E and F) BrdU/Survivin co-labelling in the control (E) and K14CreER;Srsf1fl/fl (F) laCL. White dashed lines with arrowheads outline the reduced region of proliferation in K14CreER;Srsf1fl/fl (F), compared to control (E).
(G and H) TUNEL staining in the control (G) and K14CreER;Srsf1fl/fl (H) laCL. Increased cell death (red arrowheads) can be detected specifically in the proliferating IEE upon Srsf1 deletion (H).
(I and J) Co-labelling of TUNEL and immunostaining of AMBN in control (I) and K14CreER;Srsf1fl/fl (J) laCL. Normal initiation and pattern of AMBN expression (green arrowheads) in both control (I) and K14CreER;Srsf1fl/fl (J). Increased cell death (red arrowheads) detected specifically in the proliferating IEE and no TUNEL+ cells detected in the mature region with AMBN expression upon Srsf1 deletion (J). Images were stitched manually, selecting a common feature appearing in the overlapping region. A black background was applied to the top edges of the image to make up the gaps due to image stitching.
(K-N) Quantification of the number of BrdU+ cells in dental epithelium (K) and mesenchyme adjacent to IEE (L), TUNEL+ cells in the proliferating IEE region (M) and mature region (N) in the control (Con), K14CreER;Srsf1fl/fl (Srsf1cKO) and K14CreER;Srsf3fl/fl (Srsf3cKO) laCL. All quantitative data are shown as mean ± SD (*P < 0.05, ** P < 0.001 and *** P <0.001).
(O-Q) H&E staining of the control (O), K14CreER;Srsf1fl/fl (P) and K14CreER;Srsf3fl/fl (Q) laCL 6 days after Tamoxifen treatment. Black dashed lines with arrowheads outline the region of tissue damage in (P) and (Q) upon Srsf1 (P) and Srsf3 (Q) deletion, respectively. Images were stitched manually, selecting a common feature appearing in the overlapping region.
Dashed lines outline laCL. Representative images and quantitative data are shown. Scale bar: 50 μm. See also Figures S1, S2, S3, and S7.
Here, we identify a regulatory mechanism by which the ubiquitously expressed SRSF1 protein specifically controls progenitor-specific alternative splicing events to maintain adult dental tissue homeostasis and renewal. Misregulation of the SRSF1-dependent splicing network activates the p53/p21 pathway, which arrests the cell cycle and impairs cell survival in the adult mouse laCL. Notably, a similar progenitor-specific function of SRSF1 is also observed in the small intestine epithelium, indicating a conserved role of SR splicing factors in regulating alternative splicing in adult epithelial tissues.
RESULTS
SRSF1 is required for maintenance of dental progenitors but dispensable in differentiated cell populations in the adult mouse incisor
We found that 11 SR genes were expressed in the adult mouse laCL (Fig. S1A). In addition, our previous analysis suggested that Srsf3 is highly expressed in the proliferating IEE cells of the laCL (Seidel et al., 2017), pointing to a role for SR splicing factors in proliferative cells in this region. Furthermore, extensive autoregulation and cross-regulation have been reported among different SR splicing factors, especially between SRSF1 and SRSF3 (Ni et al., 2007; Gonçalves et al., 2009). Therefore, to better understand the role of SR splicing factors in adult tissue, we set out to study the cellular and regulatory mechanism of SRSF1 and SRSF3 in regulating adult mouse incisor homeostasis and renewal. We first performed immunostaining with amplification (Fig. 1B, C and Fig. S1B, C) and quantitative RNAscope in situ hybridization (Fig. S1D–E”‘) to determine at high resolution the expression patterns of SRSF1 and SRSF3 in the adult mouse laCL. These analyses demonstrated that both SR splicing factors were ubiquitously expressed in the wild-type laCL.
To determine the cellular functions of SR splicing factors in the dental epithelium, we genetically deleted Srsf1 or Srsf3 in the adult incisor epithelium by crossing Srsf1 (Srsf1fl/fl) (Xu et al., 2005) or Srsf3 (Srsf3fl/fl) (Jumaa et al., 1999) conditional alleles with an inducible recombinase expressed in the epithelium, Keratin 14CreER (K14CreER) (Li et al., 2000). After two consecutive doses of tamoxifen injection (Fig. 1D), Cre recombinase was induced in the incisor epithelium of K14CreER;Srsf1fl/fl and K14CreER;Srsf3fl/fl conditional knockout mutants, and this resulted in a loss of SRSF1 or SRSF3 protein, respectively, shown by immunostaining (Fig. S2A–D” and Fig. S3A, B).
The impact upon deletion of SR genes was apparent two days after tamoxifen injection. Both proliferation and survival of dental progenitors in the K14CreER;Srsf1fl/fl and K14CreER;Srsf3fl/fl mutants were strongly disrupted. The number of 5-bromo-2’-deoxyuridine (BrdU+) cells was significantly reduced in the Survivin co-labeled proliferating IEE region (Fig. 1E, F, K and Fig. S3C, F). A decrease in BrdU+ progenitors was also observed in the dental mesenchyme adjacent to IEE in both mutants (Fig. 1E, F, L and Fig. S3C, F), most likely resulting from mesenchymal-epithelial cross-talk. In addition, TUNEL+ apoptotic cells were increased in the proliferating IEE region (Fig. 1G–J, M and Fig. S3D, G). In the K14CreER;Srsf3fl/fl mutant laCL, cell death was also detected in the differentiating and differentiated regions, including pre-ameloblasts and ameloblasts (Fig. 1N and Fig. S3D, G). In contrast, very low numbers of TUNEL+ cells were observed in the mature region of K14CreER;Srsf1fl/fl dental epithelium (Fig. 1I, J, N). Furthermore, whereas the expression of the mature ameloblast marker ameloblastin (AMBN) was almost undetectable in K14CreER;Srsf3fl/fl mutant dental epithelium (Fig. S3E, H), the expression pattern of AMBN remained similar in both control and K14CreER;Srsf1fl/fl mutant (Fig. 1I, J). Disruption of the ameloblast layer with Srsf3 deletion resulted in lack of enamel formation (Fig. S3K), compared to the orderly aligned ameloblasts and enamel mineralization in control (Fig. S3I) and K14CreER;Srsf1fl/fl mutant (Fig. S3J). These data suggest that SRSF1 and SRSF3 have intrinsically different roles in regulating ameloblast maturation.
Six days after tamoxifen injection, the K14CreER;Srsf3fl/fl mutant epithelium exhibited a much more severe loss of dental epithelial tissue (Fig. 1P) compared to the K14CreER;Srsf1fl/fl mutants (Fig. 1O), consistent with the larger number of affected cells in the K14CreER;Srsf3fl/fl mutants. Because of the rapid tissue turnover, the IEE had almost fully recovered in both mutants over this time period. As a result, the regenerated IEE cells pushed the earlier affected tissue to a more distal region. (Fig. 1P, Q). Both K14CreER;Srsf1fl/fl and K14CreER;Srsf3fl/fl mutant animals ceased eating and became moribund seven days after Cre induction, potentially due to the requirement for SR proteins in other epithelial organs, which precluded assessment of longer-term consequences of Srsf1 or Srsf3 deletions.
These findings indicate that, despite both SR proteins showing ubiquitous expression patterns, they have distinct roles. SRSF3 is required in various cell types regardless of their cycling status. On the other hand, SRSF1 displays a progenitor-specific function, and it is dispensable in differentiated cells. Given the specific role of SRSF1 in regulating incisor progenitors that are required for continual tissue renewal, we next focused on studying the function of SRSF1 and understanding its progenitor-specific regulatory role.
SRSF1 regulates alternative splicing events related to maintaining the survival and proliferation of dental progenitors
To detect alternative splicing events governed by the main splicing regulator SRSF1, we performed bulk RNA-Seq of dissected adult dental epithelium from 3 control and 4 K14CreER;Srsf1fl/fl mutant mice. Using the JuncBASE (junction based analysis of splicing events) tool (Brooks et al., 2011), we evaluated and classified alternative splicing events with a false discovery rate lower than 0.1. We identified 170 alternative splicing events strongly associated with Srsf1 deletion with the absolute “percent spliced in” value (|PSI|) higher than 10% (Fig. 2A, Table S1). Almost half of these SRSF1-regulated alternative splicing events corresponded to cassette exons (CA-exons), also known as exon skipping (Fig. 2A). The pervasive effect of Srsf1 deletion on CA-exons splicing was further supported by a skew toward p-values that were lower than expected (Fig. 2B). A larger proportion of the CA-exon events identified had ΔPSI lower than −10%, indicating that Srsf1 mutation was more associated with the skipping of exons rather than inclusion (Fig. 2C). All 170 alternative splicing events were plotted in a heatmap (Fig. 2D), revealing a trend toward a higher exon inclusion rate in control samples, or in other words, more exon skipping events in K14CreER;Srsf1fl/fl mutants. Together, these data indicate that CA-exon events were highly prevalent with the deletion of Srsf1.
Figure 2. SRSF1 governs a large number of alternative splicing events in the adult mouse laCL.
(A) Categories of alternative splicing events significantly associated with Srsf1 deletion versus control adult mouse laCL from bulk RNA-Seq.
(B) Q-Q plot analysis of cassette exon (CA-exon) splicing events.
(C) Volcano plot analysis of alternative splicing events upon deletion of Srsf1. Red dashed/dotted lines indicate a large magnitude of splicing change percent spliced in |ΔPSI| >= 10% (top) or a significant p-value (bottom).
(D) Heatmap of up- and downregulated alternative splicing events between the control and K14CreER;Srsf1fl/fl (Srsf1 mutant) laCL. The genes that contain splicing events that are highly likely to be directly regulated by SRSF1 are labeled and highlighted in black. Red or blue color indicates up or downregulation of each splicing event, respectively. See also Figure S4.
To identify direct binding targets of SRSF1, we compared published cross-linking immunoprecipitation (CLIP) tags (Sanford et al., 2008; Pandit et al., 2013; Anczuków et al., 2015) with the bulk RNA-Seq alternative spliced gene list (Table S1). As a result, 52 overlapping targets were found as putative alternatively spliced binding targets of SRSF1 (Table S2). To predict the biological impact of alternatively spliced targets upon Srsf1 deletion, we analyzed the topological correlation between alternatively spliced SRSF1 binding targets (Table S2) and differentially expressed genes in Srsf1 mutants (Table S3) using STRING plot (Fig. S4A). 10 highlighted putative SRSF1 targets were identified, grouping into different clusters (Fig. S4A). The rankings of |ΔPSI| of these highlighted targets are shown in the heatmap (Fig. 2D).
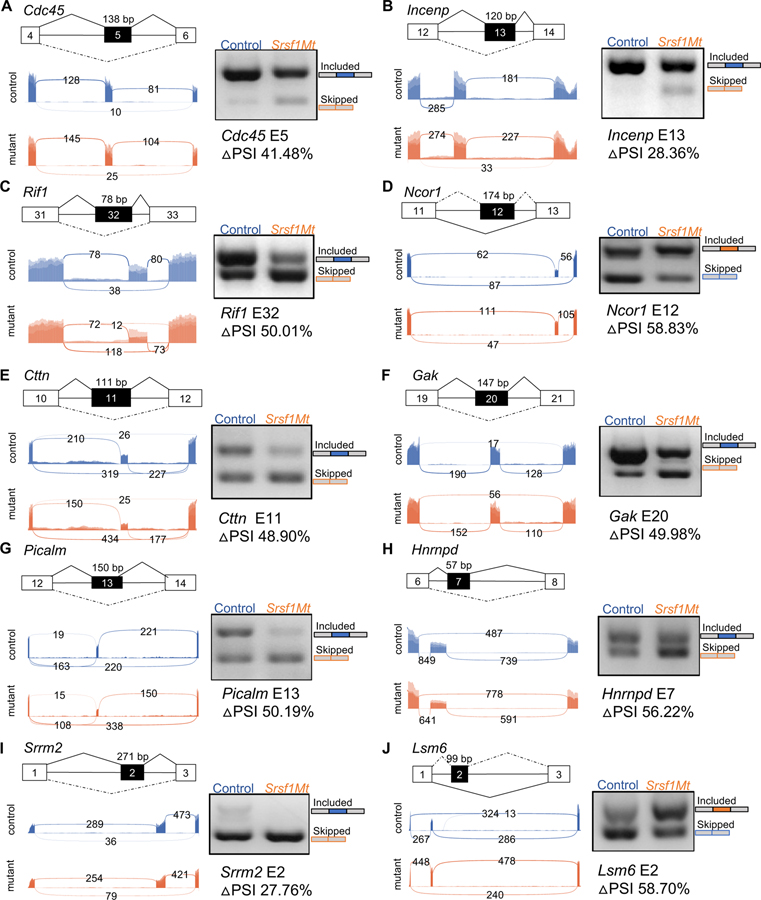
Next, we assessed the accuracy of splicing changes from the RNA-Seq data. Then, we quantitatively visualized sequencing reads aligned to gene annotations in control and Srsf1 mutants with Sashimi plots (Fig. 3). To further validate the PSI of 10 SRSF1 binding targets, we performed reverse transcriptase PCR (RT-PCR) in triplicate, using primers flanking the shaded CA-exons to amplify both isoforms that included and excluded the exon (Fig. 3). All 10 targets showed significant downregulation of their full-length transcripts and upregulation of their truncated transcripts upon Srsf1 deletion (Fig. 3), consistent with the quantification of the Sashimi plots (Fig. S5).
Figure 3. Deletion of Srsf1 alters alternative splicing of its binding targets.
(A-J) SRSF1-regulated CA-exons events of Cdc45 (A), Incenp (B), Rif1 (C), Ncor1 (D), Cttn (E), Gak (F), Picalm (G), Hnrnpd (H), Srrm2 (I) and Lsm6 (J) in the mouse incisor laCL. For each gene, exons and introns are indicated in scaled diagrams. CA-exons are shaded. Included exons transcripts are spliced with solid lines and skipped transcripts are spliced with dashed lines. Exon number and the size of CA-exons are labeled in and on top of the diagram, respectively. Sashimi plots (below the scaled diagrams) were used to quantitatively visualize splice junctions in control and Srsf1 mutants (Srsf1 Mt). Sashimi plots are depicted under the gene structure with each peak representing read coverage of each exon. RT-PCR validation of each gene is shown at the right side of the panel, with representative gel images and quantification of the ΔPSI expression in Srsf1 mutants. The representative cropped gel images are shown from 3 biological replicates. All quantitative data are shown as mean ± SD (*P < 0.05 and ** P < 0.001). See also Figure S5.
To determine the biological functions altered by Srsf1 deletion, the STRING analysis was cross-referenced with Gene Ontology (GO) clusters. GO analysis was performed on the list of SRSF1 translational genes (Table S3) using the DAVID database for functional annotation under the category of “Biological Process”. Genes clustered into two main terms: 1) cell death and DNA damage or 2) cell cycling (Fig. S4B). These assignments were consistent with our observation that cell death was increased, and proliferation was reduced, following Srsf1 deletion. The biological functions of targets located within the main branch in the STRING plot (Fig. S4A, blue-labeled targets), namely Cdc45, Incenp, Rif1, and Ncor1, were closely related to cell survival and cycling, as indicated in the GO clusters. In contrast, the yellow-labeled SRSF1 targets, which were reported as genes functioning during cell adhesion and RNA splicing, were not directly associated with the two main biological GO clusters. In addition, they were located outside of the main topological branch in the STRING plot (Fig. S4A), suggesting they are less likely to be responsible for gene expression or histological changes after Srsf1 deletion. Because we observed significant changes in cell proliferation and survival upon Srsf1 deletion, we proceeded to analyze the roles of Cdc45, Incenp, Rif1, and Ncor1 in maintaining dental tissue homeostasis and supporting tissue renewal.
To address how the ubiquitously expressed SRSF1 protein can regulate distinct functions in specific cell types, we analyzed the expression of its four putative binding targets related to cell survival and cycling in the adult laCL. All four targets were expressed at significantly higher levels in the proliferating IEE (Fig. 4A–H) compared to the other regions of the dental epithelium. Among them, CDC45 and INCENP were most strongly enriched in the IEE (Fig. 4A–B’, E, F), whereas RIF1 and NCOR1 were mainly expressed in the IEE and in the laCL (Fig. 4C–D’, G–H). Since we observed some background staining using the INCENP, RIF1, and NCOR1 antibodies (Fig. 4B–D’), we also performed in situ hybridization to confirm the IEE-enriched expression of Incenp, Rif1, and Ncor1 in the adult laCL (Fig. S6A–C”). Together, our findings suggest that the progenitor-specific phenotype of SRSF1 arises because it controls alternative splicing of progenitor-enriched targets.
Figure 4. SRSF1 binding targets are highly expressed in proliferating IEE and crypts in the laCL and the small intestine, respectively.
(A, B, C, and D) Immunostaining of CDC45 (A), INCENP (B), RIF1 (C) and NCOR1 (D) in the laCL. Red dashed lines with arrowheads mark higher expression regions of the proteins. INCENP antibody staining showed strong background in ameloblasts, making it hard to quantify in AMB region.
(A’, B’, C’, and D’) Greyscales of protein expression of CDC45 (A’), INCENP (B’), RIF1 (C’) and NCOR1 (D’) in the laCL. White dashed lines with arrowheads mark higher expression regions of the proteins.
(E-H) Quantification of regional protein expression of CDC45 (E), INCENP (F), RIF1 (G) and NCOR1 (H).
OEE, outer enamel epithelium. SR, stellate reticulum. IEE, inner enamel epithelium. pre-AMB, pre-ameloblast. AMB, ameloblast.
Dashed lines outline laCL. Images were stitched manually, selecting a common feature appearing in the overlapping region.
(I-L) Immunostaining of CDC45 (I), INCENP (J), RIF1 (K) and NCOR1 (L) in the small intestine. Red dashed lines with arrowheads mark the villus region. White dashed lines with arrowheads mark the crypt region. Dashed lines outline small intestine crypts. INCENP antibody showed strong background, therefore in situ hybridization data was added in Figure S6 (A-A”).
(M-P) Quantification of regional protein expression of CDC45 (M), INCENP (N), RIF1 (O), and NCOR1 (P).
Representative images and quantitative data are shown. All quantitative data are shown as mean ± SD (*P < 0.05, ** P < 0.001, *** P <0.001 and n.s. not significant). Scale bar represents 50 μm in A-D’; 20 μm in I-L. See also Figure S6.
SRSF1 plays an essential role in maintaining progenitors but is dispensable in differentiated cells in the adult mouse small intestine
Our data suggested that SRSF1 plays a specific role in splicing key targets required for dental progenitor function. Given that in vivo studies have shown that SRSF1 is required to maintain normal development of various organs at different stages (Xu et al., 2005; Kanadia et al., 2008; Katsuyama et al., 2019), we therefore considered whether the regulation of progenitor function by SRSF1 was conserved across different tissues. To address this question, we tested whether SRSF1 serves a similar progenitor-specific function in another highly regenerative epithelium, the adult mouse small intestine. In the intestinal crypts, lysozyme-secreting differentiated Paneth cells are intermingled between Lgr5+ intestinal stem cells, which divide daily to give rise to transit-amplifying (T-A) progenitors (Barker et al., 2007). T-A cells proliferate and contribute to various secretory and absorptive cell lineages in the villi (reviewed in Zwick et al., 2019). To determine whether SRSF1 also has a distinct cellular function in different intestinal cell types, we deleted Srsf1 in the adult mouse small intestine by crossing an Srsf1 conditional allele (Srsf1fl/fl) (Xu et al., 2005) with the pan-intestinal epithelial VillinCreER (Marjou et al., 2004).
As in the adult mouse incisor epithelium, SRSF1 was also expressed ubiquitously in the small intestinal epithelium throughout the crypt-villus axis, suggesting a potential role in intestinal epithelial cell dynamics (Fig. 5A). Three days after two consecutive tamoxifen injections, SRSF1 expression was reduced significantly in the epithelium of both crypts and villi (Fig. 5B). As determined by both TUNEL assay (Fig. 5C, F) and cleaved caspase 3 (CASP3) immunostaining (Fig. 5D, H), increased cell death was observed specifically in the intestinal crypts upon Srsf1 deletion compared to controls (Fig. 5E, G). TUNEL+ cells were found among the BrdU+ progenitor-enriched region in the intestinal crypts in VillinCreER;Srsf1fl/fl mutants, without overlapping with the Paneth cells at the crypt base (Fig. 5I, J). Correspondingly, the number of BrdU+ cells was reduced (Fig. 5I–K). Neither mature Paneth cells in the crypt (Fig. 5I, J, L), nor the number of differentiated goblet cells in the villi (Fig. 5M) were obviously affected by the loss of SRSF1. Taken together, our results demonstrate that SRSF1 plays a critical role in maintaining the cycling population of T-A cells in intestinal crypts of adult mice, whereas it is not required for the survival of differentiated cells in either crypts or villi. These data indicate a similar progenitor-specific function of SRSF1 in another epithelial tissue distinct from the adult mouse incisor. Interestingly, the expression of the intestinal stem cell marker, OLFM4, was also decreased in the VillinCreER;Srsf1fl/fl compared to control (Fig. 5O, N), suggesting that there might be feedback from progenitors to stem cells in adult mouse intestinal epithelium.
Figure 5. SRSF1 is required for maintaining the survival of intestinal progenitors, while it is dispensable for the differentiated cell populations.
(A and B) Immunostaining of SRSF1 in control (A) and VillinCreER;Srsf1fl/fl (B) the mouse small intestine. Significantly reduced expression of SRSF1 upon intestinal epithelial deletion of Srsf1 (B). Images were stitched manually, selecting a common feature appearing in the overlapping region.
(C and D) Quantification of the number of TUNEL+ cells (C) and CASP3+ cells (D) in the crypts and villi, respectively.
(E-H) Detection of cell apoptosis using TUNEL assay (E-F) and immunostaining of CASP3 (G-H) in control (E and G) and VillinCreER;Srsf1fl/fl (F and H) small intestine. Increased apoptotic cells (red arrowheads) can be detected in Srsf1 mutants (F and H).
(I and J) Triple-labelling TUNEL (red) with immunostaining of Paneth cell marker, Lysozyme (green) and BrdU (grey) in control (I) and VillinCreER;Srsf1fl/fl (J) adult mouse small intestine. Increased cell death (red arrowheads) can be detected specifically in the proliferating region enriched in BrdU+ (grey) cells (J). The number of Paneth cells (green) shows no difference upon Srsf1 deletion in the small intestine (J), compared to control (I).
(K-M) Number of BrdU+ cells (K), Paneth cells (L) in the crypt, and goblet cells (M) per crypt-villus unit in control and VillinCreER;Srsf1fl/fl small intestine. All quantitative data are shown as mean ± SD (*** P <0.001 and n.s. not significant).
(N and O) Immunostaining of OLFM4 in control (N) and VillinCreER;Srsf1fl/fl (O) the mouse small intestine. Significantly reduced expression of OLFM4 upon Srsf1 deletion (O).
Dashed lines outline small intestine crypts. Representative images and quantitative data are shown. Scale bar represents 50 μm in A and B, E-H, N and O; 20 μm in I and J.
To determine whether these same alternatively spliced putative binding targets of SRSF1 can be identified in the intestinal epithelium, we used fluorescence activated cell sorting (FACS) to purify crypt cells and performed RT-PCR for validation. Similar changes in splicing events of all four genes were observed in VillinCreER;Srsf1fl/fl mutants, showing a significant downregulation of their full-length transcripts and upregulation of their truncated transcripts upon Srsf1 deletion (Fig. S6D, E). In addition, we also analyzed the expression of these four putative binding targets in the intestinal epithelium. CDC45, INCENP, RIF1, and NCOR1 were all expressed at significantly higher levels in crypt cells (Fig. 4I–P) compared to villus cells, providing another piece of evidence in a different epithelial system to support the hypothesis that the progenitor-specific phenotype of SRSF1 arises because it controls alternative splicing of progenitor-enriched targets.
Srsf1 mutation activates the p53/p21 pathway, impairing cellular function of both dental and intestinal progenitors
The functions of all four SRSF1 binding targets have been linked to the p53/p21 pathway via various in vitro studies (Battaglia et al., 2010; Datta et al., 2017; Sun et al., 2019; Eke et al., 2020). Given that defects in cell proliferation and cell survival were two main consequences of Srsf1 deletion in the adult mouse laCL and small intestine, and changes in the p53 pathway are key to both cell cycling and apoptosis (reviewed in Bartek and Lukas, 2001; reviewed in Harris and Levine, 2005), we hypothesized that SRSF1 might control the activity of P53 and P21 through the regulation of the splice variants of Cdc45, Incenp, Rif1, and Ncor1. Therefore, we next analyzed the expression of P53 and P21 in the K14CreER;Srsf1fl/fl and VillinCreER;Srsf1fl/fl mutants.
In both adult mouse laCL and small intestine crypts, P53 was not detectable in the controls (Fig. 6A, A’, F), whereas its immunostaining signal was upregulated in IEE progenitors in K14CreER;Srsf1fl/fl mutants (Fig. 6B, B’, E) and in the intestinal crypt progenitors in VillinCreER;Srsf1fl/fl mutants (Fig. 6G, J). The immunostaining of the p53-dependent cell-cycle regulator P21 was also increased in the similar IEE region in laCLs (Fig. 6C–E), as well as in the intestinal crypts (Fig. 6H–J) upon Srsf1 deletion. Together, these data suggest that misregulation of alternative splicing events due to loss of SRSF1 activates P53 to stabilize P21, which then inhibits the activity of cyclin-dependent kinase to arrest cell cycle progression. In turn, inappropriate activation of the p53/p21 pathway stimulates apoptosis.
Figure 6. Deletion of Srsf1 activates p53/p21 pathway, leading to impaired function of both dental and intestinal progenitors.
(A-B’) Immunostaining of P53 in the control (A) and K14CreER;Srsf1fl/fl (B) laCL. Enlarged images of boxed regions are displayed in (A’) and (B’), respectively.
(C-D’) Immunostaining of P21 in the control (C) and K14CreER;Srsf1fl/fl (D) laCL. Enlarged images of boxed regions are displayed in (C’) and (D’), respectively.
(E) Quantification of regional P21 and P53 expression in the IEE.
(F and G) Immunostaining of P53 in the control (F) and VillinCreER;Srsf1fl/fl (G) small intestine.
(H and I) Immunostaining of P21 in the control (H) and VillinCreER;Srsf1fl/fl (I) small intestine.
(J) Quantification of regional P21 and P53 expression in the small intestine.
Dashed lines outline laCL and small intestine crypts. Representative images and quantitative data are shown. All quantitative data are shown as mean ± SD (*** P <0.001). Scale bar: 50 μm.
AMO-mediated exon exclusion of targeted genes recapitulates loss of Srsf1
Although traditionally considered as a splicing regulator, SRSF1 has been shown to regulate other cellular functions outside of alternative splicing (Das et al., 2014). To understand whether the observed activation of p53/p21 is due to mis-splicing of the identified targets or other SRSF1-dependent functions, we reasoned that inducing the specific mis-splicing events without affecting Srsf1 levels could help identify how SRSF1 is acting. We tested this approach using antisense morpholino oligonucleotides (AMOs), splice-switching molecules that bind to a reverse complementary sequence of a pre-mRNA target. Steric blocking of SRSF1’s ability to bind and promote exon-inclusion results in an exon skipping event. This allows for the following possible splice site to be used, thus mimicking the exon skipping event observed in the Srsf1 mutant (Figure 7A). Primary dental epithelial cells harvested from the progenitor-rich cervical loop region (Chavez et al., 2014) and primary intestinal cells harvested from organoid crypts (Thorne et al., 2018) were cultured and transfected with a control AMO or an AMO targeting Cdc45 exon (E)5, Incenp1 E13, or Rif1 E32 for 48 hours. Each target-specific AMO induced the respective exon skipping event observed in vivo without affecting the levels of Srsf1, as observed by RT-PCR (Figure 7B and 7F). Furthermore, immunostaining showed increased activation of both P53 and P21 with the target-specific AMO for both primary dental epithelial cells or intestinal epithelial cells (Figure 7C–E and G–I). These data suggest that a major role of SRSF1 in the incisor is as a splicing regulator of these target genes, which are essential for the survival of highly proliferative progenitor cells.
Figure 7. AMO-mediated exon exclusion of targeted genes recapitulates loss of Srsf1 phenotype.
(A) Schematic of AMO-mediated exon skipping event.
(B) Representative RT-PCR of control and target gene AMO transfections in primary dental epithelial cells and resulting ΔPSI upon transfection.
(C) Representative immunocytochemistry staining for P21 (magenta) and P53 (blue) in control AMO and individual target gene AMOs treated primary dental epithelial cells as marked by KRT14-mGFP (green). ΔE denotes the exon skipped upon AMO transfection.
(D-E) Quantification of regional P21 and P53 expression, respectively, in primary dental epithelial cells.
(F) Representative RT-PCR of control AMO and target gene AMO transfections in primary intestinal epithelial cells and resulting ΔPSI upon transfection.
(G) Representative immunocytochemistry staining for P21(magenta) and P53 (blue) in control AMO and individual target gene AMOs treated primary intestinal epithelial cells as marked by LGR5-mGFP (green). ΔE denotes the exon skipped upon AMO transfection.
(H-I) Quantification of regional P21 and P53 expression, respectively, in primary intestinal epithelial cells.
All quantitative data are shown as mean ± SD (*** P <0.001). Scale bar: 25 μm.
DISCUSSION
To maintain tissue homeostasis and regenerative capability, proliferative progenitors must generate various cell populations. Among the mechanisms contributing to the regulation of self-renewal, proliferation, and differentiation of progenitor cells, alternative splicing can play a significant role in producing the complex repertoire of protein isoforms required from embryonic stages to adulthood, as well as under health and disease conditions. In this study, using the adult mouse incisor and small intestine as two fast-cycling in vivo model systems, we identified a mechanism by which the ubiquitously expressed SRSF1 splicing factor governs alternative splicing in a progenitor-specific manner. Loss of SRSF1 function resulted in misregulation of alternative splicing events, thus activating p53/p21 signaling and leading to cell cycle arrest and apoptosis of progenitors. Our findings thus reveal an intricate alternative splicing regulatory system in rapidly renewing tissues.
Progenitor-specific alternative splicing regulation
Previously reported progenitor-specific function of splicing regulators mainly focused on identifying pro-pluripotent splicing events using pre-enriched human/mouse embryonic stem cells or mouse neural stem cells (Atlasi et al., 2008; Hayakawa-Yano et al., 2017). However, many splicing regulators exhibit broad expression patterns (Kalsotra et al., 2008; Yeo et al., 2008; Damianov and Black. 2010), pointing to a need to look beyond stem cells to reveal distinct cellular functions of splicing regulators at the tissue or organ level. In this study, by interrogating cells at various stages of differentiation, our findings identified a key role of SRSF1 in specifically maintaining progenitor proliferation and survival during homeostatic turnover in adult epithelial tissues. Interestingly, instead of having a regionally restricted expression pattern, the master splicing regulator SRSF1 was expressed ubiquitously and yet specifically governed the splicing of genes enriched in the progenitors in the adult mouse laCL. Even though SRSF1 was also expressed in other more differentiated cell types, it was dispensable for their function and survival. To rule out the possibility that the dental progenitor-specific defects observed in K14CreER;Srsf1fl/fl mutant laCL were indirectly introduced from potential disruption in other cell types in which K14CreER was activated, we further included ShhCreER;Srsf1fl/fl to allow more restricted deletion of Srsf1 in lower IEE in the incisor cervical loops (Fig. S7). In this knockout system, we observed similar disrupted function specifically in dental progenitors (Fig. S7E–L), as well as similar changes in alternative splicing of 4 SRSF1 targets (Fig. S7M). These data support the notion that the splicing defects were caused by a specific impact on dental progenitors upon Srsf1 deletion.
In adult organs that require rapid cell turnover to counterbalance constant tissue loss, stem cells and progenitors play an essential role in ensuring these systems’ normal function. In recent years, numerous studies have identified cell plasticity in committed cell types. When high levels of cell death occur in the progenitor pool upon acute tissue injury, differentiated cells can undergo dedifferentiation or transdifferentiation to replenish the loss in the progenitor niche to repair the tissue (reviewed in Varga and Greten, 2017; reviewed in McKinley et al., 2020). For example, in the small intestine, cellular plasticity has been observed in numerous immature and committed cell types, which can dedifferentiate to restore intestinal homeostasis upon ablation of Lgr5+ intestinal stem cells (Jadhav et al., 2017; Yan et al., 2017). Therefore, maintaining ubiquitous expression of SRSF1 might be essential for the dedifferentiation or transdifferentiation of cells to quickly restore progenitor cells and support tissue injury repair.
In vivo analysis of the cellular mechanism of alternative splicing regulation during adult tissue homeostasis and renewal
Studies performed in human or mouse embryonic stem cells, as well as in cancer cells, have provided important information about how alternative splicing events change in response to different stages or under disease conditions (Salomonis et al., 2010; Xu et al., 2018; reviewed in Qi et al., 2020). However, whether all in vitro data are representative or able to recapitulate in vivo conditions remains debatable. For example, cross-regulation and antagonism between SRSF1 and SRSF3 have been reported through in vitro studies (Jumaa et al., 1997; Gonçalves et al., 2009; Sun et al., 2010). In this study, we did not observe a strong inter-regulatory function between these two SR proteins in the adult mouse laCL. Although 60% of alternative splicing events regulated by SRSF3 corresponded to CA-exons (Fig. S3L), similar to SRSF1 (Fig. 2A), only three overlapping genes were governed by SRSF1 and SRSF3, and no antagonistic regulation was identified (data not shown). A slight down-regulation of RNA expression of Srsf3 was detected in the K14CreER;Srsf1fl/fl mutant laCL (Fig. S3M). However, due to cell heterogeneity in the dental tissue, no solid conclusion can be drawn regarding whether Srsf1 and Srsf3 can compensate for each other in distinct cell types. Compared to the relatively simpler in vitro culture systems that have one or a small number of cell types, single cell RNA Seq will be needed to answer this question in the incisor laCL. It will be important to consider tissue-specific differences or potential variations between in vitro and in vivo studies in future work.
Furthermore, cellular responses were observed in vivo upon Srsf1 deletion in laCLs that could not be detected through in vitro analysis. Consistent with mGFP expression reporting Cre-mediated recombination throughout the K14CreER;Srsf1fl/fl mutant laCL, the expression of SRSF1 in IEE cells, SI cells, pre-ameloblasts, and ameloblasts was almost undetectable one to two days after tamoxifen injection (1–2 dpi), whereas SRSF1 was still highly expressed in cells in the stellate reticulum and OEE regions (Fig. S2B–B”, F, G). SRSF1 expression started to decrease in the stellate reticulum and OEE at 3 dpi, and the reduction became evident at 4 dpi (Fig. S2H, I). Recovery of SRSF1 expression in the IEE initiated at around 4 dpi (Fig. S2I). By 5 dpi, a more pronounced expression of SRSF1 was observed in the IEE (Fig. S2J). These data together suggest that the half-life of SRSF1 varies in different cell types in the adult mouse laCL, which could inform further understanding of the cellular regulation of alternative splicing during injury repair.
Together, our findings define a mechanism by which SRSF1-dependent alternative splicing events regulate homeostasis in adult tissues, revealing a paradigm for cell-type specific splicing factor function. We identified a conserved mechanism of SRSF1 function in both the incisor and small intestine, suggesting a fundamental role of this splicing factor during development and posing new questions for future research into the regulation of alternative splicing in tissue regeneration and injury repair.
LIMITATIONS OF THE STUDY
A major limitation of conducting in vivo analysis on alternative splicing regulation is the difficulty in obtaining sufficient protein. For example, more than 250 mice (500 laCLs) of relatively similar age would be needed to obtain enough SRSF1 protein to perform RNA immunoprecipitation, such as CLIP. Therefore, we cross-referenced to published CLIP data from in vitro culture of HEK293T cells, mouse embryo fibroblasts, MCF-10A cells, and HeLa cells (Sanford et al., 2008; Pandit et al., 2013; Anczuków et al., 2015). Next, the alternatively spliced transcripts of all 10 putative binding targets (Table S2) were validated through RT-PCR, strongly suggesting that they were directly regulated by SRSF1. Thus, immediate splicing responses were recorded one day after tamoxifen injection. Using AMOs, we validated the effects of the particular exon exclusion for Cdc45, Incenp1, and Rif1 in the activation of p53/p21. However, inducing an exon inclusion event as seen in Ncor1 is much more challenging, given the frequency of sequences involved in splicing found within the large intronic regions. Nevertheless, the robust activation of P53 and P21 upon the exon skipping events in Cdc45, Incenp1, and Rif1 gives us confidence to suggest important roles for these targets in the proper maintenance of adult epithelial progenitors.
STAR Methods
Resource availability
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Ophir Klein (Ophir.Klein@ucsf.edu). All unique/stable reagents generated in this study are available by contacting the Lead Contact, but we may require a completed Materials Transfer Agreement if there is potential for commercial application.
Experimental models and subject details
Animals
K14CreER (Li et al., 2000), R26mT/mG (Muzumdar et al., 2007), VillinCreER (Marjou et al., 2004) and ShhCreER (Harfe et al., 2004), as well as conditional alleles of Srsf1fl/fl (Xu et al., 2005) and Srsf3fl/fl (Jumaa et al., 1999) mice were housed and genotyped as previously published. Both male and female adult mutant and Cre-negative littermate control mice at 8–10 weeks of age were used for experiments. All experimental procedures involving mice were approved by the Institutional Animal Care and Use Committee (IACUC) and Laboratory Animal Resource Center (LARC) at the University of California, San Francisco, and the mice were handled in accordance with the principles and procedures of the Guide for the Care and Use of Laboratory Animals under the approved protocol AN180876.
Method Details
Tamoxifen injection
Tamoxifen (Sigma T5648) was prepared in corn oil at the concentration of 25 mg/ml. Both control and mutant mice, were injected intraperitoneally with 2.5 mg tamoxifen per day per 20 g body weight for two successive days to induce Cre-mediated gene deletion. Animals were euthanized two to six days after tamoxifen injection.
BrdU injection
5-bromo-2’-deoxyuridine (BrdU) (Sigma B9285) was prepared in D-PBS (w/o calcium and magnesium salts) at the concentration of 10 mg/ml. Both control and mutant mice were injected intraperitoneally with a single dose (1 mg) per 20g body weight. Mice were euthanized 30 minutes after BrdU injection. The number of BrdU positive cells was counted for quantitative analyses.
Tissue preparation
Lower mandible
Adult mice were euthanized and perfused with 4% paraformaldehyde (PFA) in PBS. Lower mandibles were dissected from control, K14CreER;Srsf1fl/fl and K14CreER;Srsf3fl/lf mutants, and post-fixed in 4% PFA overnight at 4°C. Mandibles were decalcified with 0.5M EDTA for 2 weeks, dehydrated in 70% EtOH, embedded in paraffin and sectioned at 6 μm.
Small intestine
Adult control and VillinCreER;Srsf1fl/fl mutants were euthanized and perfused with 4% PFA in PBS. The first 10 cm of small intestinal tissues were perfusion-fixed with 4% PFA and post-fixed in 4% PFA overnight at 4°C. Tissues were then dehydrated in 70% EtOH, embedded in paraffin and sectioned at 6 μm.
Immunofluorescence assays
Immunofluorescence staining was performed as previously described (Hu et al., 2017). Additionally, for staining of CASP3, CDC45, NCOR1, NICD, OLFM4, P53, P21, RIF1, SRSF1 and SRSF3, primary antibodies were detected by biotinylated secondary antibodies, and then sequentially amplified using VECTASTAIN Elite ABC HRP Kit (Vector Laboratories) and Tyramide Signal Amplification (Perkin Elmer). All images were acquired with a Leica TCS SP8 X confocal microscope.
TUNEL assay
TUNEL assay was performed using the In Situ Cell Death Detection Kit, TMR red (Roche), according to the manufacturer’s instructions. The number of TUNEL positive nuclei was counted for quantitative analyses.
Hematoxylin & Eosin (H&E) staining
Adult mouse mandibles were dissected and fixed in 4% PFA overnight at 4°C. Samples were washed, decalcified for 2 weeks and then dehydrated in 70% EtOH. Mandibles were embedded in paraffin and sectioned at 6 μm. H&E staining was performed as previously described (Hu et al., 2017).
Bulk RNA-Seq and analysis
3 control and 4 K14CreER;Srsf1fl/fl mutant mice were used for RNA extraction. Each RNA sample was isolated from 2 laCLs using the RNeasy mini kit (QIAGEN 74104) according to the manufacturer’s instructions. RNA quality was assessed using a Pico Chip on an Agilent 2100 Bioanalyzer (Agilent Technologies). Samples with RNA-integrity (RIN) scores > 8.0 were used by the UCSF Functional Genomics Core. Bulk RNA-Seq was performed on Illumina HiSeq4000 system according to the manufacturer’s instructions with paired-end 100bp sequencing type. For each condition, RNA-Seq libraries were prepared with the TruSeq mRNA Kit. RNA-Seq reads were mapped to the reference mouse genome (GRCm38.78). Kallisto (0.44.0) was used to count the number of reads aligned to each transcript. Differential expression analysis was performed using the DESeq2 (v1.16.1) to identify genes that were significantly up- or down-regulated in the Srsf1 mutant laCL (n=4), relative to controls (n=3). The p-values were adjusted to account for multiple testing with the false discovery rate (FDR) < 0.1. The bulk RNA-Seq data reported in this study will be uploaded upon acceptance of the paper.
Bulk RNA-Seq alternative splicing analysis.
JuncBASE (Brooks et al., 2011) was used to identify and quantify AS events. The length-normalized counts for each AS inclusion or exclusion event in each of the sample was obtained from the last step of JuncBASE. The counts were adapted to a format amenable for DRIMSeq (Nowicka and Robinson, 2016), a statistical testing framework. DRIMSeq was used to assign p-values to each AS event. Q-Q plots and volcano plots were output from JuncBASE. The heatmap was plotted using a custom script written with the matplotlib package in python. Sashimi plots for quantitative visualization of sequencing reads aligned to gene annotations were generated using ggsashimi (Garrido-Martin et al., 2018). To identify the potential binding targets of SRSF1, the output list of alternative spliced events from JuncBASE was overlapped with published CLIP tags from 4 papers (Sanford et al., 2008; Pandit et al., 2013; Anczuków et al., 2015), respectively using Venn diagram. 52 overlapping targets were found as putative alternatively spliced binding targets of SRSF1, shown in Table S2. Venn diagrams were made using the matplotlib venn package in python.
Reverse transcription PCR (RT-PCR)
LaCL samples were isolated as previously described (Chavez et al., 2014). DAPIlowEpCAM+CD44+ small intestine crypt epithelial cells were obtained through flow cytometry as previously described (Nusse et al., 2018). RNA samples obtained from control and K14CreER;Srsf1fl/fl mutant laCL 1 day after tamoxifen injection, as well as control and VillinCreER;Srsf1fl/fl mutant small intestine crypt epithelial cells 2 days after tamoxifen injection were isolated using GenCatch™ Total RNA miniprep kit (Epoch Life Science 1660050). RNA samples were then reverse-transcribed to cDNA using SensiFAST™ cDNA Synthesis Kit (BioLine BIO-65053). Both steps were performed according to the manufacturer’s instructions. RT-PCR were performed using iTAQ CYBR green (Biorad) with the following conditions: 5 minutes at 95°C and 35 cycles of amplification (30 seconds at 95°C, 50 seconds at 60°C and 1 minute at 72°C). The relative changes of transcript expression between controls and Srsf1 mutants were analyzed by ImageJ as previously described (Yu et al., 2017). In brief, band intensities were first converted into histograms, from which the area under the curve can be measured using the Wand tool and the relative expression between control and mutant samples were calculated. Percent spliced in changes (ΔPSI) were calculated using the band intensity of the inclusion event divided by the band intensity of the inclusion event plus band intensity of the exclusion (i.e., inclusion event/(inclusion + exclusion event)).The primer sets used are listed in the key resouces table. All measurements were normalized to GAPDH.
Key Resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit Anti-Ameloblastin (M-300) antibody | Santa Cruz Biotechnology | Cat# sc-50534, RRID:AB_2226393 |
| Rat Anti-BrdU antibody | Abcam | Cat# ab6326, RRID:AB_305426 |
| Rabbit Anti-Caspase-3, phospho (Cleaved Asp175) antibody | Cell Signaling Technology | Cat# 8202, RRID:AB_1658166 |
| Rabbit Anti-Survivin antibody | Cell Signaling Technology | Cat# 2808, RRID:AB_2063948 |
| Rabbit Anti-Phospho-Histone H3 antibody | Cell Signaling Technology | Cat# 9706, RRID:AB_331748 |
| Rabbit Anti-Cleaved Notch1 (Val1744) (D3B8) antibody | Cell Signaling Technology | Cat# 4147, RRID:AB_2153348 |
| Rabbit Anti-Olfm4 (D6Y5A) XP® antibody | Cell Signaling Technology | Cat# 39141, RRID:AB_2650511 |
| Rabbit Anti-Cdc45 (D7G6) antibody | Cell Signaling Technology | Cat# 11881, RRID:AB_2715569 |
| Mouse Anti-P21 antibody | BD Biosciences | Cat# 556431, RRID:AB_396415 |
| Mouse Anti-Incenp antibody (B-4) | Santa Cruz Biotechnology | Cat# sc-376514, RRID:AB_11149761 |
| Mouse Anti-Rif1 antibody (B-4) | Santa Cruz Biotechnology | Cat# Sc-515573 |
| Rabbit Anti-P53 antibody | Proteintech | Cat# 10442–1-AP, RRID:AB_2206609 |
| Rabbit Anti-SRSF1 antibody | Abcam | Cat# ab38017, RRID:AB_882519 |
| Rabbit Anti-SRSF3 antibody | ProSci | Cat# 8179 |
| Chicken Anti- Green Fluorescent Protein (GFP) Antibody | Aves Lab | Cat# GFP-1020, RRID:AB_10000240 |
| Chemicals, peptides, and recombinant proteins | ||
| Control AMO | GeneTools Inc. | Stand. Control Oligo Cat# (PCO-100) |
| Oligo: mCdc45 E5 ACTGAAAAACAGGACGAGGCACTGT | GeneTools Inc. | Custom Oligo Cat# CO-300 |
| Oligo: Incenp E13 TGTCCCCACCTAGTCGCACCTGTTG | GeneTools Inc. | Custom Oligo Cat# CO-300 |
| Oligo: Rif1 E32 GGTATTATGCTAGACAGAAGTAGA |
GeneTools Inc. | Custom Oligo Cat# CO-300 |
| Endo-Porter (PEG) | GeneTools Inc. | Cat# OT-EP-PEG-1 |
| Critical commercial assays | ||
| RNAscope 2.5 HD Red detection kit | ACD | Cat#322350 |
| In Situ Cell Death Detection Kit, TMR red | Roche | Cat# 12156792910 |
| TSA Cyanine 3 Tyramide Reagent Pack | Perkin Elmer | Cat# AT704B001EA |
| VECTASTAIN Elite ABC HRP Kit (Peroxidase, Standard) | Vector Laboratories | Cat # PK-6100 |
| Deposited Data | ||
| Raw and processed RNAseq data | This paper | GEO: GSE193492 |
| Oligonucleotides | ||
| Primer: Srsf1fl/fl Forward GGGACTAATGTGGGAAGAATG Primer: Srsf1fl/fl Reverse AACCTAAACTATTGCTCCCATCTG |
IDT | Xu et al., 2005 |
| Primer: Srsr31fl/fl Forward GCGCAGGTACTTGAGAGA Primer: Srsf3fl/fl Reverse CCCTTTTATTGGTCAGTGA |
IDT | Jumaa et al., 1999 |
| Primer: CreER Forward TGGAGATCTTCGACATGCTG Primer: CreER Reverse CACGTTCTTGCACTTCATGC |
IDT | JAX |
| Primer: Cdc45 Forward TGTGTGCTTGCAAGATCCTC Primer: Cdc45 Reverse ACCCATCACCGTCACTGTCT |
IDT | This Paper |
| Primer: Cttn Forward AGAGAAGCACGAATCCCAGA Primer: Cttn Reverse TCTTGTCCATCCGATCC |
IDT | This Paper |
| Primer: Gak Forward GGAAGAGCAGCAGGACATTC Primer: Gak Reverse GTAGCCCCAGGAGATCAACA |
IDT | This Paper |
| Primer: GAPDH Forward CATGGCCTTCCGTGTTCCTA Primer: GAPDH Reverse CCTGCTTCACCACCTTCTTGAT |
IDT | This Paper |
| Primer: Hnrnpd Forward GACGCCAGTAAGAACGAGGA Primer: Hnrnpd Reverse TCAGGTGTGTCTGGAGAAAGG |
IDT | This Paper |
| Primer: Incenp Forward CGTGAGAGGGTGGAACAGAT Primer: Incenp Reverse CTGCTCCTTCCGTTCCTGTT |
IDT | This Paper |
| Primer: Lsm6 Forward TGTGAGCGTTCTGGATCCG Primer: Lsm6 Reverse GGATGAACGCATCTCCGTACT |
IDT | This Paper |
| Primer: Ncor1 Forward GCTCTTTACCAACGGCACAT Primer: Ncor1 Reverse GTTGGTCGTGTTGTTGGAAG |
IDT | This Paper |
| Primer: Picalm Forward CTCAGCAGGGGGAATAATGA Primer: Picalm Reverse AGAATGTGGCTGTGCAACTG |
IDT | This Paper |
| Primer: Rif1 Forward GTGTCTCGTTTGCAGATCCA Primer: Rif1 Reverse GCGTACTCAAATCCCCAATG |
IDT | This Paper |
| Primer: Srrm2 Forward ACCTCCCTGTTTGACAGTCG Primer: Srrm2 Reverse GGGAGGCTCAGGAGCTATTT |
IDT | This Paper |
| Software and algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| ImageJ Colocalization plugin | Image J | https://imagej.nih.gov/ij/plugins/colocalization.html |
| SPRING Viewer | Klein Lab | https://github.com/AllonKleinLab/SPRING_de |
| Prism 9 | Graphpad software, Inc | N/A |
| Adobe illustrator 2020 | Adobe Computer software company | N/A |
Quantification and statistical analysis
Statistical analysis
All of the data points are biological replicates and were replicated at least three times. Bar charts indicate the mean of samples and error bars represent mean ± SD (standard deviation). P value were derived from unpaired two tail Student’s t tests, assuming equal variance (*P < 0.05, ** P < 0.001, *** P <0.001 and n.s. not significant).
ImageJ image analysis
Colocalization of SRSF1, SRSF3, CDC45, RIF1, NCOR1 with DAPI was measured using the ImageJ plugin “Colocalization”. Percentage of the immunostained area was detected as previously described in Hu et al., 2017.
Data and Code Availability
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
This manuscript did not generate new code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Supplementary Material
Table S1. List of alternative splicing events strongly associated with Srsf1 mutation |ΔPSI| > 10%, Related to Figure 2.
Table S2. List of putative alternative spliced binding targets of SRSF1, Related to Figure 2.
Table S3. List of differentially expressed genes after deletion of Srsf1, Related to Figure 2 and Figure 3.
Highlights:
SRSF1 governs epithelial progenitor homeostasis.
Mis-splicing of SRSF1 targets leads to epithelial progenitor cell death.
Cdc45, Incenp, Rif1, and Ncor1 are important targets of SRSF1.
ACKNOWLEDGMENTS
We thank A. Rathnayake, B. Hoehn, E. Sandoval and S. Alto for technical assistance, Drs. J. Hu, K. McKinley, P. Marangoni, R. Zwick, T. Huycke, A. Sharir, members of the Klein, Floor, Brooks and Goodwin labs for helpful discussions, Dr. K. Seidel for importing and maintaining the Srsf1fl/fl and Srsf3fl/fl mouse lines at the beginning of the project, and the UCSF Functional Genomics Core for help with RNA-Seq. This work was funded by NIDCR R01-DE024988 and R35-DE026602 to O.D.K.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
INCLUSION AND DIVERSITY
The sex balance of our samples was included as part of our experimental designs. Our laboratories take pride in fostering a diverse and inclusive environment, and we strive to make equitable opportunities for all our trainees. The authors of this paper identify as members of one or more of the following communities: AAPI, Black, Differently Able, Latinx, and LGBTQ+. Also, in this study, one or more of the authors received support from a program designed to increase minority representation.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Anczuków O, Akerman M, Cléry A, Wu J, Shen C, Shirole NH, Raimer A, Sun S, Jensen MA, Hua Y, and Allain FHT (2015). SRSF1-regulated alternative splicing in breast cancer. Molecular cell, 60(1), pp.105–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlasi Y, Mowla SJ, Ziaee SA, Gokhale PJ, and Andrews PW (2008). OCT4 spliced variants are differentially expressed in human pluripotent and nonpluripotent cells. Stem cells, 26(12), pp.3068–3074. [DOI] [PubMed] [Google Scholar]
- Barker N, Van Es JH, Kuipers J, Kujala P, Van Den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, and Clevers H (2007). Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature, 449(7165), pp.1003–1007. [DOI] [PubMed] [Google Scholar]
- Bartek J, and Lukas J (2001). Mammalian G1-and S-phase checkpoints in response to DNA damage. Current opinion in cell biology, 13(6), pp.738–747. [DOI] [PubMed] [Google Scholar]
- Battaglia S, Maguire O, Thorne JL, Hornung LB, Doig CL, Liu S, Sucheston LE, Bianchi A, Khanim FL, Gommersall LM, and Coulter HS (2010). Elevated NCOR1 disrupts PPARα/γ signaling in prostate cancer and forms a targetable epigenetic lesion. Carcinogenesis, 31(9), pp.1650–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks AN, Yang L, Duff MO, Hansen KD, Park JW, Dudoit S, Brenner SE, and Graveley BR (2011). Conservation of an RNA regulatory map between Drosophila and mammals. Genome research, 21(2), pp.193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez MG, Hu J, Seidel K, Li C, Jheon A, Naveau A, Horst O, and Klein OD (2014). Isolation and culture of dental epithelial stem cells from the adult mouse incisor. JoVE (Journal of Visualized Experiments), (87), p.e51266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Luo C, Wu W, Xie Z, Fu X, and Feng Y (2016). Liver-specific deletion of SRSF2 caused acute liver failure and early death in mice. Molecular and cellular biology, 36(11), pp.1628–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Krainer AR. (2014) Emerging functions of SRSF1, splicing factor and oncoprotein, in RNA metabolism and cancer. Mol Cancer Res,12(9), pp.1195–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damianov A, and Black DL (2010). Autoregulation of Fox protein expression to produce dominant negative splicing factors. Rna, 16(2), pp.405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A, Ghatak D, Das S, Banerjee T, Paul A, Butti R, Gorain M, Ghuwalewala S, Roychowdhury A, Alam SK, and Das P (2017). p53 gain-of-function mutations increase Cdc7-dependent replication initiation. EMBO reports, 18(11), pp.2030–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding JH, Xu X, Yang D, Chu PH, Dalton ND, Ye Z, Yeakley JM, Cheng H, Xiao RP, Ross J, and Chen J (2004). Dilated cardiomyopathy caused by tissue-specific ablation of SC35 in the heart. The EMBO journal, 23(4), pp.885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eke I, Zong D, Aryankalayil MJ, Sandfort V, Bylicky MA, Rath BH, Graves EE, Nussenzweig A, and Coleman CN (2020). 53BP1/RIF1 signaling promotes cell survival after multifractionated radiotherapy. Nucleic Acids Research, 48(3), pp.1314–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Valley MT, Lazar J, Yang AL, Bronson RT, Firestein S, Coetzee WA, and Manley JL (2009). SRp38 regulates alternative splicing and is required for Ca2+ handling in the embryonic heart. Developmental cell, 16(4), pp.528–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido-Martín D, Palumbo E, Guigó R, and Breschi A (2018). ggsashimi: Sashimi plot revised for browser-and annotation-independent splicing visualization. PLoS computational biology, 14(8), p.e1006360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves V, Matos P, and Jordan P (2009). Antagonistic SR proteins regulate alternative splicing of tumor-related Rac1b downstream of the PI3-kinase and Wnt pathways. Human molecular genetics, 18(19), pp.3696–3707. [DOI] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, and Brown JB (2011). The developmental transcriptome of Drosophila melanogaster. Nature, 471(7339), pp.473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa-Yano Y, Suyama S, Nogami M, Yugami M, Koya I, Furukawa T, Zhou L, Abe M, Sakimura K, Takebayashi H, and Nakanishi A (2017). An RNA-binding protein, Qki5, regulates embryonic neural stem cells through pre-mRNA processing in cell adhesion signaling. Genes & development, 31(18), pp.1910–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, Scherz PJ, Nissim S, Tian H, McMahon AP and Tabin CJ (2004). Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell, 118(4), pp.517–528. [DOI] [PubMed] [Google Scholar]
- Harris SL, and Levine AJ (2005). The p53 pathway: positive and negative feedback loops. Oncogene, 24(17), pp.2899–2908. [DOI] [PubMed] [Google Scholar]
- Hu JKH, Du W, Shelton SJ, Oldham MC, DiPersio CM, and Klein OD (2017). An FAK-YAP-mTOR signaling axis regulates stem cell-based tissue renewal in mice. Cell stem cell, 21(1), pp.91–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Wang K, and Li M (2019). Detecting differential alternative splicing events in scRNA-seq with or without UMIs. bioRxiv, p.738997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav U, Saxena M, O’Neill NK, Saadatpour A, Yuan GC, Herbert Z, Murata K, and Shivdasani RA (2017). Dynamic reorganization of chromatin accessibility signatures during dedifferentiation of secretory precursors into Lgr5+ intestinal stem cells. Cell Stem Cell, 21(1), pp.65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Dong H, Shi Y, and Bian L (2018). Mutually exclusive alternative splicing of pre-mRNAs. Wiley Interdisciplinary Reviews: RNA, 9(3), p.e1468. [DOI] [PubMed] [Google Scholar]
- Jumaa H, and Nielsen PJ (1997). The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. The EMBO journal, 16(16), pp.5077–5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumaa H, Wei G, and Nielsen PJ (1999). Blastocyst formation is blocked in mouse embryos lacking the splicing factor SRp20. Current Biology, 9(16), pp.899–902. [DOI] [PubMed] [Google Scholar]
- Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, and Cooper TA (2008). A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proceedings of the National Academy of Sciences, 105(51), pp.20333–20338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanadia RN, Clark VE, Punzo C, Trimarchi JM, and Cepko CL (2008). Temporal requirement of the alternative-splicing factor Sfrs1 for the survival of retinal neurons. Development, 135(23), pp.3923–3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuyama T, Li H, Comte D, Tsokos GC, and Moulton VR (2019). Splicing factor SRSF1 controls T cell hyperactivity and systemic autoimmunity. The Journal of clinical investigation, 129(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Indra AK, Warot X, Brocard J, Messaddeq N, Kato S, Metzger D, and Chambon P (2000). Skin abnormalities generated by temporally controlled RXRα mutations in mouse epidermis. Nature, 407(6804), pp.633–636. [DOI] [PubMed] [Google Scholar]
- Manley JL, and Krainer AR (2010). A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes & development, 24(11), pp.1073–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Marjou F, Janssen KP, Hung-Junn Chang B, Li M, Hindie V, Chan L, Louvard D, Chambon P, Metzger D, and Robine S (2004). Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. genesis, 39(3), pp.186–193. [DOI] [PubMed] [Google Scholar]
- McKinley KL, Castillo-Azofeifa D, and Klein OD Tools and concepts for interrogating and defining cellular identity. Cell Stem Cell, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, and Luo L (2007). A global double-fluorescent Cre reporter mouse. genesis, 45(9), pp.593–605. [DOI] [PubMed] [Google Scholar]
- Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O’Brien G, Shiue L, Clark TA, Blume JE, and Ares M (2007). Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes & development, 21(6), pp.708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowicka M, and Robinson MD (2016). DRIMSeq: a Dirichlet-multinomial framework for multivariate count outcomes in genomics. F1000Research, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse YM, Savage AK, Marangoni P, Rosendahl-Huber AK, Landman TA, de Sauvage FJ, Locksley RM and Klein OD (2018). Parasitic helminths induce fetal-like reversion in the intestinal stem cell niche. Nature, 559(7712), pp.109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, and Blencowe BJ (2008). Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nature genetics, 40(12), p.1413. [DOI] [PubMed] [Google Scholar]
- Pandit S, Zhou Y, Shiue L, Coutinho-Mansfield G, Li H, Qiu J, Huang J, Yeo GW, Ares M Jr, and Fu XD (2013). Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Molecular cell, 50(2), pp.223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F, Li Y, Yang X, Wu YP, Lin LJ, and Liu XM (2020). Significance of alternative splicing in cancer cells. Chinese Medical Journal, 133(2), p.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomonis N, Schlieve CR, Pereira L, Wahlquist C, Colas A, Zambon AC, Vranizan K, Spindler MJ, Pico AR, Cline MS, and Clark TA (2010). Alternative splicing regulates mouse embryonic stem cell pluripotency and differentiation. Proceedings of the National Academy of Sciences, 107(23), pp.10514–10519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JR, Coutinho P, Hackett JA, Wang X, Ranahan W, and Caceres JF (2008). Identification of nuclear and cytoplasmic mRNA targets for the shuttling protein SF2/ASF. PLoS One, 3(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel K, Ahn CP, Lyons D, Nee A, Ting K, Brownell I, Cao T, Carano RA, Curran T, Schober M, et al. (2010). Hedgehog signaling regulates the generation of ameloblast progenitors in the continuously growing mouse incisor. Development, 137, pp.3753–3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel K, Marangoni P, Tang C, Houshmand B, Du W, Maas RL, Murray S, Oldham MC, and Klein OD (2017). Resolving stem and progenitor cells in the adult mouse incisor through gene co-expression analysis. Elife, 6, p.e24712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharir A, Marangoni P, Zilionis R, Wan M, Wald T, Hu JK, Kawaguchi K, Castillo-Azofeifa D, Epstein L, Harrington K, and Pagella P (2019). A large pool of actively cycling progenitors orchestrates self-renewal and injury repair of an ectodermal appendage. Nature cell biology, 21(9), pp.1102–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard PJ, and Hertel KJ (2009). The SR protein family. Genome biology, 10(10), p.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Zhang Z, Sinha R, Karni R, and Krainer AR (2010). SF2/ASF autoregulation involves multiple layers of post-transcriptional and translational control. Nature structural & molecular biology, 17(3), p.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Veschi V, Bagchi S, Xu M, Mendoza A, Liu Z, and Thiele CJ (2019). Targeting the Chromosomal Passenger Complex Subunit INCENP Induces Polyploidization, Apoptosis, and Senescence in Neuroblastoma. Cancer research, 79(19), pp.4937–4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne CA, Chen IW, Sanman LE, Cobb MH, Wu LF, Altschuler SJ. (2018) Enteroid Monolayers Reveal an Autonomous WNT and BMP Circuit Controlling Intestinal Epithelial Growth and Organization. Dev cell, 44(5), pp. 624–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga J, and Greten FR (2017). Cell plasticity in epithelial homeostasis and tumorigenesis. Nature cell biology, 19(10), pp.1133–1141. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, and Burge CB (2008). Alternative isoform regulation in human tissue transcriptomes. Nature, 456(7221), pp.470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Yang D, Ding JH, Wang W, Chu PH, Dalton ND, Wang HY, Bermingham JR Jr, Ye Z, Liu F, and Rosenfeld MG (2005). ASF/SF2-regulated CaMKIIδ alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell, 120(1), pp.59–72. [DOI] [PubMed] [Google Scholar]
- Xu Y, Zhao W, Olson SD, Prabhakara KS, and Zhou X (2018). Alternative splicing links histone modifications to stem cell fate decision. Genome biology, 19(1), pp.1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan KS, Gevaert O, Zheng GX, Anchang B, Probert CS, Larkin KA, Davies PS, Cheng ZF, Kaddis JS, Han A, and Roelf K (2017). Intestinal enteroendocrine lineage cells possess homeostatic and injury-inducible stem cell activity. Cell stem cell, 21(1), pp.78–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanger K, Zong Y, Maggs LR, Shapira SN, Maddipati R, Aiello NM, Thung SN, Wells RG, Greenbaum LE, and Stanger BZ (2013). Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes & development, 27(7), pp.719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, and Gage FH (2009). An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nature structural & molecular biology, 16(2), p.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, and Klein OD (2020). Molecular and cellular mechanisms of tooth development, homeostasis and repair. Development, 147(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Graf M, Renn J, Schartl M, Larionova D, Huysseune A, Witten PE, and Winkler C (2017). A vertebrate-specific and essential role for osterix in osteogenesis revealed by gene knockout in the teleost medaka. Development, 144(2), pp.265–271. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, and Deng S (2014). An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. Journal of Neuroscience, 34(36), pp.11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick RK, Ohlstein B, and Klein OD (2019). Intestinal renewal across the animal kingdom: comparing stem cell activity in mouse and Drosophila. American Journal of Physiology-Gastrointestinal and Liver Physiology, 316(3), pp.G313–G322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of alternative splicing events strongly associated with Srsf1 mutation |ΔPSI| > 10%, Related to Figure 2.
Table S2. List of putative alternative spliced binding targets of SRSF1, Related to Figure 2.
Table S3. List of differentially expressed genes after deletion of Srsf1, Related to Figure 2 and Figure 3.
Data Availability Statement
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
This manuscript did not generate new code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.