Abstract
Erythroblastic island (EBI), composed of a central macrophage surrounded by developing erythroid cells, is a structure found in hematopoietic tissues such as fetal liver and bone marrow. It is the first described hematopoietic niche that predominantly supports erythropoiesis. Although it is well accepted that EBIs and EBI macrophage play important roles during erythropoiesis, the mechanisms by which they support erythropoiesis remain largely unclear due to our inability to identify and isolate EBI macrophages. Earlier efforts to identify surface markers for EBI macrophages have focused on the adhesion molecules which are involved in macrophage's interaction with erythroblasts. These include EMP, Vcam1, CD169, CD163, and αV integrin. Findings from these earlier studies suggested that combination of Vcam1, CD169, and mouse macrophage surface marker F4/80 can be used to define mouse EBI macrophage. We found that not all F4/80+Vcam1+CD169+ macrophages are EBI macrophages. Instead, we discovered that EBI macrophages are characterized by the expression of Epor in both mouse and man. RNA-seq analyses of the newly identified EBI macrophages revealed that EBI macrophages have involved specialized function in supporting erythropoiesis. Our findings provide foundation for future studies. Here we will review current knowledge of EBI macrophages and discuss future perspectives.
Keywords: Epor, Erythroblast islands, Erythroblastic island macrophages, Erythropoiesis
1. INTRODUCTION
Erythropoiesis is a complex process by which mature red cells are generated from hematopoietic stem cells and can be subdivided into three stages: early stage erythropoiesis, terminal erythroid differentiation, and reticulocyte maturation. Defects at any stage during this process can lead to disordered erythropoiesis.1–3 These include Cooley anemia,4,5 congenital dyserythropoietic anemia,6–8 Diamond–Blackfan anemia,9,10 malarial anemia,11–13 myelodysplastic syndromes,14–16 and polycythemia vesa.17–20 Thus, a better and detailed understanding of the process of erythropoiesis is of both biological and clinical significance. Previous studies on the regulations of erythropoiesis have been primarily focused on cell intrinsic factor of erythroid cells, such as erythropoiesis master regulator transcription factors GATA binding protein 1 (GATA1) and Krueppel-like factor 1 (KLF1) as well as erythropoietin (EPO) and its receptor (EPOR) mediated signal transduction.21–25
In addition to cell intrinsic regulations, erythropoiesis is also influenced by niche and microenvironment. It has been well established that erythropoiesis occurs at the erythroblastic island (EBI) which is composed of a central macrophage surrounded by developing erythroblasts. EBI was first defined in 1958 by Marcel Bessis, based on the analysis of transmission electron micrographs of sections of bone marrow.26 Based on the structural observations, Bessis et al made a number of interesting inferences concerning the role of central macrophages. It was suggested that the macrophage functions as a “nurse” cell, providing iron to developing erythroblasts for heme synthesis.27 The study of EBI was extended in 1970s by reconstruction of three-dimensional scale models of bone marrow.28 Later studies further confirmed the existence of EBIs in the mammal bone marrow (BM), spleen and fetal liver (FL), but not in yolk sac.29,30 It was proposed that the macrophages in the center of EBIs not only transport iron to erythroblasts for the synthesis of hemoglobin, they also secrete many cytokines to support the survival and differentiation of erythroblasts,29,31,32 as well as act as phagocytes to engulf and degrade erythroblast nuclei during nuclear extrusion.33–35 Despite extensive studies on EBIs over the past few decades, the identity of EBI macrophage has remained elusive until very recently.32
The lack of inability to identify and isolate EBI macrophages prospectively has hindered the investigation on EBI macrophages. Very recently, our group discovered that EBI macrophages are characterized by the expression of Epor in both mouse and human. We further performed RNA-seq on the newly identified EBI macrophages. Bioinformatic analyses revealed that EBI macrophages have involved specialized function in supporting erythropoiesis and suggested potential underlying molecular mechanisms. These findings provide solid foundation for many future studies.32 In this review, we will provide an overview of identification and characterization of EBI macrophage. We will also discuss future perspectives.
2. IDENTIFICATION OF EBI MACROPHAGE
2.1. Adhesion molecules involved in EBI formation
EBI formation involves the interaction between EBI macrophages and erythroblasts. Therefore, earlier efforts to identify surface markers for EBI macrophages were focused on the adhesion molecules. Erythroblast macrophage protein (EMP) was the first molecule identified on both erythroblast and EBI macrophage that mediates the association between macrophages and erythroblast attachments through hemophilic interaction.36 In EBI cultures, absence of Emp leads to aberrant erythropoiesis and increased levels of apoptosis, suggesting that the direct association between the EBI macrophage and the erythroblasts is essential for erythroid maturation and prevention of cell death.37 However, a recent study demonstrated that EMP expressed by macrophages but not by erythroblasts is important for the EBI formation.38
The other receptor/counter receptor identified as mediating cell–cell interactions within EBI were VCAM1 in EBI macrophages and α4β1 integrin in erythroblasts.39 The biological significance of this interaction is underlined in the experiment in which the formation of EBI was disrupted by the application of antibodies against either α4β1 integrin or VCAM-1.39 This receptor/counter receptor interaction also contributes to the integrity of EBIs during primitive erythropoiesis.40,41 However, the lack of phenotypic changes of Vcam1−/− mice questions the physiological role of Vcam1 in erythropoiesis.38
In addition, the adhesion molecule αV integrin expressed on macrophage and ICAM4 expressed on erythroblasts also plays critical roles in maintaining EBI integrity, disrupting the binding of αV integrin, and ICAM4 leads to decreasing number of EBI.42,43
The other adhesion molecule expressed by EBI macrophage is CD169. It has been shown that CD169 localizes at the site of macrophage/erythroblast contact in EBIs.44–47 A recent study shows that depletion of CD169+ macrophages in mice leads to impaired erythropoiesis in vivo.48 It should be noted that the counter receptor for CD169 on erythroblasts has yet to be identified.
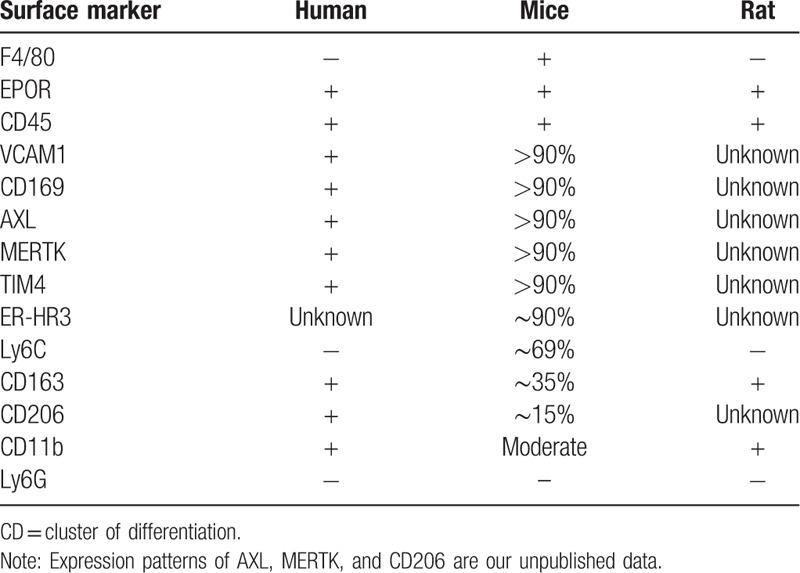
A number of additional macrophage surface proteins have been identified as receptors for erythroblasts. One macrophage adhesion glycoprotein with an unidentified erythroid binding partner is CD163 (formerly called ED2 antigen), which functions as a receptor for hemoglobin–haptoglobin complexes and is involved in the clearance of free hemoglobin.49 CD163 contains an erythroblast adhesion motif as well, which mediates binding of macrophages to erythroid precursors facilitating erythroblast expansion and survival.50 Other molecules that have been reported to be expressed on EBI macrophages are lectin-like sheep erythrocyte receptor, erythroblast receptor (EbR), ED2, CD206, ER-HR3, Ly6G, tyrosine kinase receptor Mertk, and Axl.51–53
2.2. Use of imaging flow cytometry to characterize EBI macrophages
In 2017, Seu et al used a novel method, multispectral imaging flow cytometry (IFC), to characterize EBI macrophages. IFC combines the high-throughput advantage of flow cytometry with the morphological and fluorescence features derived from microscopy.54 This method provides quantitative analysis of EBIs, as well as structural and morphological details of the EBI macrophages and associated cells. Using IFC, Seu et al showed that consistent with previous findings, F4/80, VCAM1, and CD169 are expressed on EBI macrophages. However, CD163 is not expressed on mouse EBI macrophages even though it is expressed on rat EBI macrophages. Intriguingly, CD11b is not expressed on EBI macrophages in mice, although it is abundantly expressed on myeloid cells within the islands.
2.3. EBI macrophages are characterized by the expression of Epor
Based on previous findings, it is thought that combination of F4/80, Vcam1, and CD169 can be used to define EBI macrophage. However, by examining the ratio of F4/80+Vcam1+CD169+ macrophages and erythroblasts, we found that the ratio of F4/80+Vcam1+CD169+ versus erythroblasts in mouse bone marrow is only about 1:2.6. It has been reported that the number of erythroblasts per island ranges from 10 cells observed in tissue sections from rat femur to 5 to >30 erythroblasts in islands harvested from human BM. Thus, our findings strongly suggest that it is unlikely all F4/80+VCAM1+CD169+ macrophages are EBI macrophages.
Given the facts Epo/Epor is essential for erythropoiesis and that Epor is expressed in numerous non-erythroid cells including macrophages,55–58 we hypothesized that Epor is expressed on EBI macrophages so that Epo can act on both erythroid cells and EBI macrophages to ensure efficient red cell production. To test this, we first used Epor-eGFP knockin mouse model.32 We show that a subpopulation of macrophages in mouse bone marrow and fetal live express Epor. Importantly, the EBIs are predominantly formed by the Epor+ macrophages in both mouse and man. The representative images of EBI are shown as Figure 1. Furthermore, using flow cytometry and IFC, we characterized the surface expression of previously described EBI macrophages. These results along with previous findings are summarized in Table 1. Notably, although CD163 is a marker for human and rat EBI macrophages, it is only expressed on about 35% of mouse EBI macrophages, indicating differences among species.32
Figure 1.
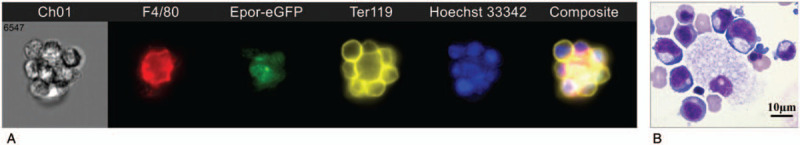
Images of EBI. (A) Native mouse bone marrow EBI revealed by imaging flow cytometry. F4/80: mouse macrophage marker; Ter119, mouse erythroid cell marker. (B) Cytospin image of in vitro formed human EBI.
Table 1.
Expression of surface markers on EBI macrophages in human, mouse and rat.
3. FUTURE PERSPECTIVE
Since the discovery of the EBIs more than 60 years, the major gap in the understanding of EBI has been what is the identity of EBI macrophage? Our very recent identification of the EBI macrophages and transcriptomic analyses of the EBI macrophages not only fill this important gap but also offer the opportunity to begin to fill the gaps in mechanistic understanding of the roles of EBI macrophages in normal as well as disordered erythropoiesis. Specifically, with the finding that EBI macrophages are characterized by the expression of Epor, the question is what is the role of Epo/Epor in EBI macrophages? Another question is that although it has long been postulated that EBI macrophages could provide nutrients locally to the surrounding erythroid cells, so far there was no study to demonstrate this is the case. With the iron being the most needed nutrient for erythropoiesis, do EBI macrophages provide iron locally to the developing erythroid cells? Our finding that molecules involved in iron recycle such as PS receptor Tim4, tyrosine kinase MerTK, Axl, heme oxygenase HO-1, iron exporter ferroportin, and iron transporter transferrin are abundantly expressed in EBI macrophages strongly suggest that EBI macrophages could provide iron locally to support erythropoiesis.32 Similarly, it has been thought that EBI macrophages may secret cytokines to promote erythropoiesis. However, it remains unknown which erythropoiesis-promoting cytokines they secret. Our finding that erythropoiesis-promoting cytokine Igf1 is expressed in EBI macrophages but not non-EBI macrophages raises the question of whether local secretion of Igf1 by EBI macrophages contributes to erythropoiesis.
Other knowledge gaps include but are not limited to: first, erythroblast enucleation is the last step of terminal erythroid maturation. Do EBI macrophages promote enucleation? If so, does Epo play a role?; second, one function of EBI macrophage is to engulf extruded nuclei. Does Epo enhance engulfment of nuclei by EBI macrophages? If so, what are the underlying molecular pathways?; third, in addition to iron, do EBI macrophages provide other nutrients to developing erythroid cells? If so, what are these nutrients and what are the routes these nutrients are transferred from EBI macrophages to erythroid cells?; fourth, fetal liver erythropoiesis is different from adult BM erythropoiesis in that FL erythropoiesis is a kind of stress erythropoiesis. Do EBI macrophages contribute to the differences between FL and BM erythropoiesis. If so, to what extent and how? Our ability to identify and isolate EBI macrophages as well as the RNA-seq database we have generated provide the foundation for many future studies to fill these gaps.
REFERENCES
- [1].Yan H, Wang Y, Qu X, et al. Distinct roles for TET family proteins in regulating human erythropoiesis. Blood 2017;129(14):2002–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Qu X, Zhang S, Wang S, et al. TET2 deficiency leads to stem cell factor dependent clonal expansion of dysfunctional erythroid progenitors. Blood 2018;132(22): doi: 10.1182/blood-2018-05-853291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Huang Y, Hale J, Wang Y, et al. SF3B1 deficiency impairs human erythropoiesis via activation of p53 pathway: implications for understanding of ineffective erythropoiesis in MDS. J Hematol Oncol 2018;11(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ginzburg Y, Rivella S. β-Thalassemia: a model for elucidating the dynamic regulation of ineffective erythropoiesis and iron metabolism. Blood 2011;118(16):4321–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Finch CA, Sturgeon P. Erythrokinetics in Cooley's anemia. Blood 1957;12(1):64–73. [PubMed] [Google Scholar]
- [6].Heimpel H, Schwarz K, Ebnöther M, et al. Congenital dyserythropoietic anemia type I (CDA I): molecular genetics, clinical appearance, and prognosis based on long-term observation. Blood 2006;107(1):334–340. [DOI] [PubMed] [Google Scholar]
- [7].Schwarz K, Iolascon A, Verissimo F, et al. Mutations affecting the secretory COPII coat component SEC23B cause congenital dyserythropoietic anemia type II. Nat Genet 2009;41(8):936–940. [DOI] [PubMed] [Google Scholar]
- [8].Wickramasinghe SN, Wood WG. Advances in the understanding of the congenital dyserythropoietic anaemias. Br J Haematol 2005;131(4):431–446. [DOI] [PubMed] [Google Scholar]
- [9].Diamond LK, Wang WC, Alter BP. Congenital hypoplastic anemia. Adv Pediatr 1976;22:349–378. [PubMed] [Google Scholar]
- [10].Lipton JM, Ellis SR. Diamond-Blackfan anemia: diagnosis, treatment, and molecular pathogenesis. Hematol Oncol Clin North Am 2009;23(2):261–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Haldar K, Mohandas N. Malaria, erythrocytic infection, and anemia. Hematol Am Soc Hematol Educ Progr 2009;2009(1):87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Casals-Pascual C, Kai O, Cheung JOP, et al. Suppression of erythropoiesis in malarial anemia is associated with hemozoin in vitro and in vivo. Blood 2006;108(8):2569–2577. [DOI] [PubMed] [Google Scholar]
- [13].Chang K-H, Tam M, Stevenson MM. Inappropriately low reticulocytosis in severe malarial anemia correlates with suppression in the development of late erythroid precursors. Blood 2004;103(10):3727–3735. [DOI] [PubMed] [Google Scholar]
- [14].Ebert BL, Galili N, Tamayo P, et al. An erythroid differentiation signature predicts response to lenalidomide in myelodysplastic syndrome. PLoS Med 2008;5(2):e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].List A, Dewald G, Bennett J, et al. Lenalidomide in the myelodysplastic syndrome with chromosome 5q deletion. N Engl J Med 2006;355(14):1456–1465. [DOI] [PubMed] [Google Scholar]
- [16].Nimer SD. Myelodysplastic syndromes. Blood 2008;111(10):4841–4851. [DOI] [PubMed] [Google Scholar]
- [17].James C, Ugo V, Le Couédic J-P, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 2005;434(7037):1144–1148. [DOI] [PubMed] [Google Scholar]
- [18].Guglielmelli P, Tozzi L, Bogani C, et al. Overexpression of microRNA-16-2 contributes to the abnormal erythropoiesis in polycythemia vera. Blood 2011;117(25):6923–6927. [DOI] [PubMed] [Google Scholar]
- [19].Hsia CC, Xenocostas A. Polycythemia vera with pancytopenia and red cell fragmentation. Blood 2011;118(18):4769. [DOI] [PubMed] [Google Scholar]
- [20].Spivak JL. Polycythaemia vera. Hematology 2013;18(4):244–245. [DOI] [PubMed] [Google Scholar]
- [21].Koury MJ, Bondurant MC. Erythropoietin retards DNA breakdown and prevents programmed death in erythroid progenitor cells. Science 1990;248(4953):378–381. [DOI] [PubMed] [Google Scholar]
- [22].Kieran MW, Perkins AC, Orkin SH, Zon LI. Thrombopoietin rescues in vitro erythroid colony formation from mouse embryos lacking the erythropoietin receptor. Proc Natl Acad Sci U S A 1996;93(17):9126–9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wu H, Liu X, Jaenisch R, Lodish HF. Generation of committed erythroid BFU-E and CFU-E progenitors does not require erythropoietin or the erythropoietin receptor. Cell 1995;83(1):59–67. [DOI] [PubMed] [Google Scholar]
- [24].Siatecka M, Bieker JJ. The multifunctional role of EKLF/KLF1 during erythropoiesis. Blood 2011;118(8):2044–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pevny L, Lin CS, D’Agati V, et al. Development of hematopoietic cells lacking transcription factor GATA-1. Development 1995;121(1):163–172. [DOI] [PubMed] [Google Scholar]
- [26].Bessis M. Erythroblastic island, functional unity of bone marrow. Rev Hematol 1958;13(1):8–11. [PubMed] [Google Scholar]
- [27].Bessis M, Breton-Gorius J. The erythroblastic islet and the rhopheocytosis of ferritin in inflammation. Nouv Rev Fr Hematol 1961;1:569–582. [PubMed] [Google Scholar]
- [28].Mohandas N, Prenant M. Three-dimensional model of bone marrow. Blood 1978;51(4):633–643. [PubMed] [Google Scholar]
- [29].Chasis JA, Mohandas N. Erythroblastic islands: niches for erythropoiesis. Blood 2008;112(3):470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ferkowicz MJ, Yoder MC. Blood island formation: longstanding observations and modern interpretations. Exp Hematol 2005;33(9):1041–1047. [DOI] [PubMed] [Google Scholar]
- [31].Sawada K, Krantz SB, Dessypris EN, Koury ST, Sawyer ST. Human colony-forming units-erythroid do not require accessory cells, but do require direct interaction with insulin-like growth factor I and/or insulin for erythroid development. J Clin Invest 1989;83(5):1701–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Li W, Wang Y, Zhao H, et al. Identification and transcriptome analysis of erythroblastic island macrophages. Blood 2019;doi: 10.1182/blood.2019000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bessis M, Breton-Gorius J. Iron metabolism in the bone marrow as seen by electron microscopy: a critical review. Blood 1962;19:635–663. [PubMed] [Google Scholar]
- [34].Leimberg MJ, Prus E, Konijn AM, Fibach E. Macrophages function as a ferritin iron source for cultured human erythroid precursors. J Cell Biochem 2008;103(4):1211–1218. [DOI] [PubMed] [Google Scholar]
- [35].Manwani D, Bieker JJ. The erythroblastic island. Curr Top Dev Biol 2008;82:23–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hanspal M, Hanspal JS. The association of erythroblasts with macrophages promotes erythroid proliferation and maturation: a 30-kD heparin-binding protein is involved in this contact. Blood 1994;84(10):3494–3504. [PubMed] [Google Scholar]
- [37].Hanspal M, Smockova Y, Uong Q. Molecular identification and functional characterization of a novel protein that mediates the attachment of erythroblasts to macrophages. Blood 1998;92(8):2940–2950. [PubMed] [Google Scholar]
- [38].Wei Q, Boulais PE, Zhang D, et al. Maea expressed by macrophages, but not erythroblasts, maintains postnatal murine bone marrow erythroblastic islands. Blood 2019;doi: 10.1182/blood-2018-11-888180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sadahira Y, Yoshino T, Monobe Y. Very late activation antigen 4-vascular cell adhesion molecule 1 interaction is involved in the formation of erythroblastic islands. J Exp Med 1995;181(1):411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kingsley PD, Malik J, Fantauzzo KA, Palis J. Yolk sac-derived primitive erythroblasts enucleate during mammalian embryogenesis. Blood 2004;104(1):19–25. [DOI] [PubMed] [Google Scholar]
- [41].McGrath KE, Kingsley PD, Koniski AD, et al. Enucleation of primitive erythroid cells generates a transient population of “pyrenocytes” in the mammalian fetus. Blood 2008;111(4):2409–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Mankelow TJ, Spring FA, Parsons SF, et al. Identification of critical amino-acid residues on the erythroid intercellular adhesion molecule-4 (ICAM-4) mediating adhesion to alpha V integrins. Blood 2004;103(4):1503–1508. [DOI] [PubMed] [Google Scholar]
- [43].Lee G, Lo A, Short SA, et al. Targeted gene deletion demonstrates that the cell adhesion molecule ICAM-4 is critical for erythroblastic island formation. Blood 2006;108(6):2064–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Crocker PR, Gordon S. Properties and distribution of a lectin-like hemagglutinin differentially expressed by murine stromal tissue macrophages. J Exp Med 1986;164(6):1862–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Crocker PR, Gordon S. Mouse macrophage hemagglutinin (sheep erythrocyte receptor) with specificity for sialylated glycoconjugates characterized by a monoclonal antibody. J Exp Med 1989;169(4):1333–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Morris L, Crocker PR, Fraser I, Hill M, Gordon S. Expression of a divalent cation-dependent erythroblast adhesion receptor by stromal macrophages from murine bone marrow. J Cell Sci 1991;99(Pt 1):141–147. [DOI] [PubMed] [Google Scholar]
- [47].Crocker PR, Werb Z, Gordon S, Bainton DF. Ultrastructural localization of a macrophage-restricted sialic acid binding hemagglutinin, SER, in macrophage-hematopoietic cell clusters. Blood 1990;76(6):1131–1138. [PubMed] [Google Scholar]
- [48].Chow A, Huggins M, Ahmed J, et al. CD169+ macrophages provide a niche promoting erythropoiesis under homeostasis and stress. Nat Med 2013;19(4):429–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kristiansen M, Graversen JH, Jacobsen C, et al. Identification of the haemoglobin scavenger receptor. Nature 2001;409(6817):198–201. [DOI] [PubMed] [Google Scholar]
- [50].Fabriek BO, Polfliet MMJ, Vloet RPM, et al. The macrophage CD163 surface glycoprotein is an erythroblast adhesion receptor. Blood 2007;109(12):5223–5229. [DOI] [PubMed] [Google Scholar]
- [51].Barbé E, Huitinga I, Döpp EA, Bauer J, Dijkstra CD. A novel bone marrow frozen section assay for studying hematopoietic interactions in situ: the role of stromal bone marrow macrophages in erythroblast binding. J Cell Sci 1996;109(Pt 1):2937–2945. [DOI] [PubMed] [Google Scholar]
- [52].Jacobsen RN, Forristal CE, Raggatt LJ, et al. Mobilization with granulocyte colony-stimulating factor blocks medullar erythropoiesis by depleting F4/80+VCAM1+CD169+ER-HR3+Ly6G+ erythroid island macrophages in the mouse. Exp Hematol 2014;42(7):547–561. e4. [DOI] [PubMed] [Google Scholar]
- [53].Heideveld E, Hampton-O’neil LA, Cross SJ, et al. Glucocorticoids induce differentiation of monocytes towards macrophages that share functional and phenotypical aspects with erythroblastic Island macrophages. Haematologica 2018;103(3):395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Seu KG, Papoin J, Fessler R, et al. Unraveling macrophage heterogeneity in erythroblastic islands. Front Immunol 2017;8:1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ott C, Martens H, Hassouna I, et al. Widespread expression of erythropoietin receptor in brain and its induction by injury. Mol Med 2015;21(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].von Salisch S, Klar M, Thurisch B, Bungert J, Dame C. Gata4 and Sp1 regulate expression of the erythropoietin receptor in cardiomyocytes. J Cell Mol Med 2011;15(9):1963–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Westenfelder C, Baranowski RL. Erythropoietin stimulates proliferation of human renal carcinoma cells. Kidney Int 2000;58(2):647–657. [DOI] [PubMed] [Google Scholar]
- [58].Lopez TV, Lappin TRJ, Maxwell P, et al. Autocrine/paracrine erythropoietin signalling promotes JAK/STAT-dependent proliferation of human cervical cancer cells. Int J Cancer 2011;129(11):2566–2576. [DOI] [PubMed] [Google Scholar]


