Significance Statement
The mechanisms regulating the urinary excretion of uromodulin remain mostly unknown. A meta-GWAS conducted in 29,315 individuals from 13 cohorts identified two novel, genome-wide significant loci, KRT40 and WDR72, in addition to the previously known UMOD-PDILT locus, to be associated with urinary uromodulin. KRT40 colocalizes with uromodulin in TAL cells and functional studies showed that its expression affects the processing and apical excretion of uromodulin. WDR72, which does not colocalize with uromodulin, has been associated with kidney function, urinary acidification, and kidney stones. These studies provide novel insights into the biology of uromodulin and keratins and into the influence of the UMOD-PDILT locus on kidney function.
Keywords: cytokeratin, thick ascending limb, Tamm-Horsfall protein, loop of Henle, KRT40, WDR72
Abstract
Background
Uromodulin, the most abundant protein excreted in normal urine, plays major roles in kidney physiology and disease. The mechanisms regulating the urinary excretion of uromodulin remain essentially unknown.
Methods
We conducted a meta-analysis of genome-wide association studies for raw (uUMOD) and indexed to creatinine (uUCR) urinary levels of uromodulin in 29,315 individuals of European ancestry from 13 cohorts. We tested the distribution of candidate genes in kidney segments and investigated the effects of keratin-40 (KRT40) on uromodulin processing.
Results
Two genome-wide significant signals were identified for uUMOD: a novel locus (P 1.24E–08) over the KRT40 gene coding for KRT40, a type 1 keratin expressed in the kidney, and the UMOD-PDILT locus (P 2.17E–88), with two independent sets of single nucleotide polymorphisms spread over UMOD and PDILT. Two genome-wide significant signals for uUCR were identified at the UMOD-PDILT locus and at the novel WDR72 locus previously associated with kidney function. The effect sizes for rs8067385, the index single nucleotide polymorphism in the KRT40 locus, were similar for both uUMOD and uUCR. KRT40 colocalized with uromodulin and modulating its expression in thick ascending limb (TAL) cells affected uromodulin processing and excretion.
Conclusions
Common variants in KRT40, WDR72, UMOD, and PDILT associate with the levels of uromodulin in urine. The expression of KRT40 affects uromodulin processing in TAL cells. These results, although limited by lack of replication, provide insights into the biology of uromodulin, the role of keratins in the kidney, and the influence of the UMOD-PDILT locus on kidney function.
Uromodulin (UMOD, previously known as Tamm-Horsfall protein) is the most abundant protein excreted in the normal urine. This kidney-specific protein is essentially produced by the cells lining the thick ascending limb (TAL) of the loop of Henle, and, to a much smaller extent, the initial segment of the distal convoluted tubule (DCT).1 As a typical glycosylphosphatidylinositol-anchored protein, UMOD matures along the secretory pathway, becoming heavily glycosylated and sorted to the apical plasma membrane, where it is cleaved by the serine protease hepsin.2 Once in the lumen, UMOD monomers assemble into homopolymeric filaments, which encapsulate and aggregate uropathogens, such as type-1 fimbriated Escherichia coli, promoting their clearance in the urine.3,4 At the level of tubular cells, UMOD regulates apical transport systems operating in the TAL and in the DCT,5,6 modulating salt reabsorption and blood pressure control.7,8
Multilevel evidence supports the role of UMOD, the gene coding for UMOD, in a spectrum of kidney disorders. Rare missense mutations of UMOD are the most common cause of autosomal dominant tubulointerstitial kidney disease (ADTKD), a disease entity characterized by tubular damage and interstitial fibrosis in the absence of glomerular lesions, progressing to kidney failure.9 ADTKD-UMOD is caused by a toxic gain of function mechanism, with accumulation of intracellular aggregates of mutant UMOD in the TAL, leading to tissue damage and kidney fibrosis.9–12 In parallel, genome-wide association studies (GWAS) have consistently associated the UMOD locus with the eGFR and the risk for developing CKD in the general population.13,14 Remarkably, the UMOD locus has a relatively large effect size on eGFR and CKD risk, consistent across most ethnic groups studied so far. The top GWAS risk variants map in the same linkage disequilibrium (LD) block encompassing the promoter of UMOD, and they are associated with an increased expression of UMOD.7,15
Although the roles of UMOD are increasingly recognized, our knowledge about the mechanisms regulating its production in tubular cells and its excretion into the urine remains limited. Recent studies have shown that the excretion of UMOD is functionally linked with the activity of transport processes operating in the TAL.16–18 Using a meta-GWAS approach performed on 10,884 individuals of European descent from six cohorts, we previously identified common variants within the promoter of UMOD as the single genome-wide significant locus associated with the levels of UMOD in urine.19 The top UMOD promoter variant, rs12917707, was associated in a dose-dependent fashion with the urinary levels of UMOD (uUMOD),19 confirming the biologic link between these variants and the expression of UMOD in kidney and urine.
To gain further insights into the factors regulating the production and excretion of UMOD, and increase the statistical power to detect novel loci associated with urinary excretion of UMOD, we conducted a meta-GWAS in 29,315 individuals from 13 cohorts of European ancestry. We performed detailed expression studies and investigated the biologic relevance of the novel genome-wide significant KRT40 locus in the processing of UMOD.
Materials and Methods
Cohorts
The concentrations of urinary UMOD and creatinine were measured in 29,315 individuals of European ancestry in 13 cohorts from both urban and isolate communities: CARTaGENE, CoLaus, CROATIA-Korcula, CROATIA-Split, CROATIA-Vis, Framingham Heart Study (FHS), Genetic and Phenotypic Determinants of Blood Pressure and other Cardiovascular Risk Factors, German Chronic Kidney Disease (GCKD), Generation Scotland: Scottish Family Health Study (GS:SFHS), INGI-Carlantino, INGI-Val Borbera, Lothian Birth Cohort 1936 (LBC1936), and Viking Health Study-Shetland (VIKING). Written informed consent was provided by all participants. The characteristics of each cohort are summarized in Supplemental Appendix 1, Supplemental Methods, and Supplemental References, and the study sample characteristics for uUMOD measurement are detailed in Supplemental Table 1. UMOD and UMOD indexed to creatinine (uUCR) were inverse-normal transformed before adjusting for age, sex, and relatedness. Each study conducted linear regression analysis of the residuals of uUMOD and uUCR from genotyped and either 1000G Phase 3, HapMap, or Haplotype Reference Consortium imputed single nucleotide polymorphisms (SNPs) using an additive model with appropriate statistical software (Supplemental Table 2).20–23 The Hg19 genome build was used for the reference panel. The genetic kinship matrix fitted for all family-based cohorts was calculated using the “ibs” function in GenABEL/ProbABEL,22 and GWAS was performed using various software listed in Supplemental Table 2.
UMOD and Creatinine Measurement
Spot urine samples were collected and were frozen and stored before analysis (Supplemental Table 1). Urinary UMOD levels were measured using a well-established ELISA as described previously.24 Human UMOD (AG 733; EMD Millipore, Temecula, CA) was used to determine the standard curve. For the capture antibody, a sheep anti-human UMOD antibody was used (K90071C; Meridian Life Science, Memphis, TN), a mouse monoclonal anti-human UMOD antibody (CL 1032A; Cedarlane Laboratories, Burlington, NC) was used as the primary antibody, and a goat anti-mouse IgG (H+L) horseradish peroxidase–conjugated protein (172.1011; BioRad Laboratories, Inc., Hercules, CA) as the secondary antibody. The detection range for the assay was between 3.9 ng/ml and 500 ng/ml (Supplemental Table 1). Urinary UMOD levels in samples from FHS were measured by the Rules-Based Medicine array (Rules-Based Medicine, Inc., Austin, TX) using immunoassay with a bead Luminex platform.19 UMOD levels were expressed as uUMOD (μg/ml) or uUCR (mg/g creatinine).
Heritability
Estimates of heritability of urinary UMOD in family-based cohorts (GS:SFHS, VIKING, CROATIA-Korcula, CROATIA-Split, CROATIA-Vis, INGI-Carlantino, INGI-Val Borbera, and FHS) were derived from the analysis of the polygenic model in GenABEL software,22 with age and sex as covariates.
Statistical Analyses (GWAS)
The GWAS summary output files from the 13 cohorts were processed for quality control using the EasyQC package,25 which filtered out SNPs with an effect allele frequency of <0.01 and imputation quality of <0.4. Furthermore, we excluded SNPs with missing values in each cohort, carried out allele and marker name harmonization to the 1000 genomes reference panel, and performed frequency checks to output a clean and harmonized GWAS file for each of the cohorts, which were subsequently used for meta-analyses. The inverse-variance weighted fixed-effects method implemented in METAL (v2011–03–25),26 software was used to conduct meta-analyses of uUMOD and uUCR levels. A genomic control coefficient was computed for each cohort and was used to correct P values for any cryptic relatedness or population stratification by METAL. SNPs that were present in at least two cohorts were used to identify genome-wide significant and suggestive loci and to create Manhattan plots. Meta-analysis using sample sizes and P values was also calculated using METAL to account for any heterogeneity. Meta-analysis on urinary UMOD values adjusted for age and sex were compared against meta-analysis on urinary UMOD using age, sex, and eGFR as covariates in seven available cohorts (GS:SFHS, VIKING, CROATIA-Korcula, CROATIA-Split, CROATIA-Vis, GCKD and CoLaus, n=20,620), to account for kidney function. eGFR was calculated using the CKD Epidemiology Collaboration equation.
A threshold of P≤5.0E–08 was used to determine significant associations after a second correction for calculated genomic inflation factor (ʎ) from the meta-analysis. Annotation of association results was performed using Haploreg,27 SNiPa,28 and the UCSC genome browser.29 A suggestive threshold was defined as P<1.0E–05. LocusZoom was used to create regional association maps, thus identifying other genome-wide significant and suggestive SNPs in LD with the SNP of interest and other genes within each locus.30 Spearman’s rank correlation was used to compare the meta-GWAS effect sizes of SNP of interest, in association with uUMOD and uUCR. Genome-wide significant SNPs and genes were investigated for the presence of candidate expression quantitative trait loci (eQTLs) using the Genotype-Tissue Expression (GTEx) database v8.31 Versatile A Gene-based Association Study (VEGAS2) was performed on summary metadata.32 SNPs with the lowest P value in novel genome-wide significant loci were queried for association with eGFR, urinary creatinine, and plasma creatinine levels using UK Biobank GWAS summary statistics in the Global Biobank Engine.33–35
Conditional Analyses
The GCTA COJO Slct algorithm was applied on summary metadata to select independently associated SNPs using a stepwise model selection procedure.36 Default P value cutoff of 5.0E–08 and collinearity cutoff of 0.9 were used. The reference panel used was Phase 3 1000G (Europeans only). Conditional analysis on the SNPs identified as independent by GCTA COJO in UMOD and PDILT was carried out on uUMOD and uUCR in the GS:SFHS cohort.
Candidate Gene Analyses
A list of genes expressed in the TAL and associated with rare monogenic disorders affecting the TAL or with ADTKD was compiled (Supplemental Table 3).9,37 All SNPs in each gene were queried in the meta-analysis results and a gene-specific threshold was calculated as previously described.19 The gene region was defined as ±1 kb from gene start and end position as listed in Ensembl for each of the genes. For each gene, region-specific multiple testing was carried out by calculating a gene-specific threshold, namely, 0.05 divided by the number of found LD blocks at r2=0.2. Variants that had a P value lower than gene-specific thresholds were declared significant. The 1000G dataset and r2 value of 0.1 was used to define LD blocks. SNPs with minor allele frequency <0.01 and imputation quality <0.4 were removed before analysis.
To determine if there was a stronger association between genes expressed in the TAL compared with other segments, a simple gene query was carried out using 10 genes expressed in specific segments (Supplemental Table 4) and the genes that were close to significance in the previous meta-GWAS for UMOD (SORL1, CAB39, FAM83A, and MARCH1).19 As above, the gene region was defined as ±1 kb from the gene start and end position.
Mouse Kidney Samples and Microdissection of Nephron Segments
C57BL/6J mice were housed in a light- and temperature-controlled environment with ad libitum access to tap water and standard chow (Diet AO3, SAFE; 25/18 GR Mucedola Srl, Settimo Milanese, Italy). Mice were sacrificed by cervical dislocation after anesthesia with isoflurane (Minrad International Inc., Orchard Park, NY, USA) for kidney collection. One kidney was used to obtain material for the primary mouse TAL (mTAL) cell culture, whereas the other was further processed for histologic analyses. Kidney tubule segments were prepared from mouse kidneys briefly digested with Liberase (Roche, Basel, Switzerland), with manual isolation of the segments according to morphologic criteria as described.38 The samples were lysed in either Radioimmunoprecipitation assay buffer (RIPA buffer) for immunoblotting experiments or in the RNA lysis buffer from the RNAqueous Total RNA Isolation Kit (Invitrogen, Carlsbad, CA, USA) for transcript analysis. Quantitative RT-PCR was performed on pools of approximately 70 samples for each isolated fraction. The experiments were performed in accordance with the ethical guidelines at University of Zurich (Zurich, Switzerland) and the legislation of animal care and experimentation of Canton Zurich, Switzerland (Gesundheitsdirektion Veterinäramt; protocol ZH049/17).
Isolation, Culture, and Treatment of Primary mTAL Cells
Primary cultures of mTAL cells were prepared from kidneys of C57/BL6J mice and validated as previously described.38 Briefly, TALs were isolated under a light microscope on the basis of morphologic characteristics and cultured on permeable filter supports (Transwell-COL, pore size 0.4 µm, Corning Costar, USA) containing a DMEM:F12-based medium for 7–10 days in a humidified chamber at 37 °C and 5% CO2 until confluent monolayers were formed. For Krt40 silencing, an adenovirus expressing a short hairpin RNA against mouse Krt40 (Ad-shKrt40; targeting sequence: GGATGAGATGCGATGTCAATA, Vector Biolabs) and a scramble (Ad-GFP-U6-scrmbl-shRNA) control were used. The transduction protocol was performed as previously described.39 Cells were plated on filters and transduction was performed when they reached approximately 70%–80% of confluence (24 hours after plating). Cells were subsequently incubated overnight at 37°C with culture medium containing the virus at the appropriate concentration (0.2125 × 109 plaque forming units (PFU)/ml). Culture medium was changed every day and the cells were collected for analysis after 5 days of serum-free conditions, to ensure the best compromise between UMOD expression and the knockdown in KRT40.38
Histologic Analysis and Immunostaining
Kidneys were fixed overnight at 4 °C in 4% formaldehyde (Sigma-Aldrich), dehydrated, and subsequently embedded in paraffin. Paraffin blocks were cut into 5 µm-thick sections, deparaffinized in xylene, and rehydrated in decreasing ethanol concentrations. The sections were incubated with sheep anti-UMOD (1:300; Meridian Life Science Inc., Cincinnati, OH, USA), rabbit anti-KRT40 (1:50–1:100, LifeSpan Bioscience), rabbit anti-WDR72 (1:500; Sigma-Aldrich), goat anti-AQP2 (1:400, SantaCruz Biotechnology), and rabbit anti-KRT39 (1:50, ThermoFisher Scientific) overnight at 4°C. After washing steps, sections were incubated with the appropriate Alexa Fluor–labeled secondary antibody (1:400, Life Technologies, Carlsbad, CA) for 1 hour at room temperature. The use of human kidney biopsies has been approved by the UCLouvain Ethical Review Board. Monolayers of mTAL cells on polytetrafluoroethylene filters were fixed for 10 minutes in 4% formaldehyde (Sigma-Aldrich), permeabilized for 30 minutes using 0.5% saponin (Sigma-Aldrich), and blocked in 3% BSA (Sigma-Aldrich). Immunostaining procedures were similar to those used for kidney sections. After the last washing step, filters were cut and mounted on a glass slide using Prolong Gold Anti-fade reagent containing DAPI (Invitrogen Corp., Waltham, MA), and viewed under a confocal microscope (Leica Microsystems GmbH, Wetzlar, Germany) using a ×63 1.4 NA oil immersion objective.
Protein Samples Preparation and Immunoblotting
mTAL cells were lysed in ice-cold RIPA buffer (Sigma-Aldrich, St. Louis, MO, USA) containing protease (Roche) and phosphatase (PhosSTOP, Sigma) inhibitors. Samples (20 μg/lane) were thawed on ice, diluted in Laemmli sample buffer (BioRad), separated on a 7.5% SDS-PAGE gel, and blotted on PVDF membranes. After blocking with 5% nonfat milk (BioRad) in PBS, the membranes were incubated overnight at 4°C with primary antibody. Blots were subsequently washed and incubated with peroxidase-conjugated secondary antibodies, washed again, and visualized by Immun-Star enhanced chemiluminescence (BioRad). Quantitative analysis was performed by scanning the blots and measuring the relative density of each band normalized to β-actin by using ImageJ software.
RNA Isolation and Quantitative RT-PCR
Total RNA was extracted from mTAL cells with RNAqueousR kit (Invitrogen, Carlsbad, CA). One μg of RNA was used to perform the reverse transcription reaction with iScript TM cDNA Synthesis Kit (BioRad). Changes in target genes mRNA levels were determined by relative quantitative RT-PCR with a CFX96TM Real-Time PCR Detection System (BioRad) using iQTM SYBR Green Supermix (BioRad). The analyses were performed in duplicate with 100 nM of both sense and antisense primers in a final volume of 20 µL using iQTM SYBR Green Supermix (BioRad). Specific primers were designed using Primer3 (Supplemental Table 5). PCR conditions were 95°C for 3 minutes followed by 40 cycles of 15 seconds at 95°C, and 30 seconds at 60°C. The PCR products were sequenced with the BigDye terminator kit (Perkin Elmer Applied Biosystems) using ABI3100 capillary sequencer (Perkin Elmer Applied Biosystems). The efficiency of each set of primers was determined by dilution curves (Supplemental Table 5). The relative changes in targeted genes over Gapdh mRNAs were calculated using the 2-ΔΔCt formula.
Antibodies
The following primary antibodies were used: sheep anti-UMOD (K90071C, Meridian Life Science Inc., Cincinnati, OH, USA; 1:500 for western blot (WB) and 1:300 for immunofluorescence (IF)), mouse anti-UMOD (CL1032A; Cedarlane Laboratories, for ELISA), mouse anti-hair cortex cytokeratin (ab16113, Abcam, 1:1000 for WB, 1:100 for IF), rabbit anti-KRT40 (LS-C400568, Life Span Bio Science, 1:50 for IF), mouse anti-β actin (A5441, Sigma-Aldrich; 1:10,000 for WB), and rabbit anti-WDR72 (HPA057410, Sigma-Aldrich, 1:500 for IF).
Statistical Analysis (Experimental Studies)
The quantitative data were expressed as mean±SEM. Normality of the sample distribution was assessed using Shapiro–Wilk test, whereas the F test was used to check equality of the variance in the experimental groups. The difference between experimental groups were evaluated using unpaired, two-tailed t test or Mann–Whitney test in the case of non-normal distribution. The sample size (n) of each experimental group is described in the corresponding figure legends. All results are representative of more than three independent experiments. GraphPad Prism software was used for all statistical analysis. Statistical significance was set as P<0.05.
Information on the datasets and summary statistics are available in the Edinburgh Datashare repository, under the link https://doi.org/10.7488/ds/3012 created on April 7, 2021.
Results
Summary Data and Heritability
The summary data for the 29,315 participants from the 13 cohorts with uUMOD levels are shown in Table 1. The λ values in individual cohorts ranged from 0.99 to 1.04 for uUMOD and 0.99–1.05 for uUCR. Detailed information on cohorts, samples, assays, genotyping, and imputation platforms are given in Supplemental Appendix 1 and Supplemental Tables 1 and 2. The estimated heritabilities for uUMOD in the family-based cohorts ranged from 11% (CROATIA-Vis) to 45% (GS:SFHS) (Supplemental Table 6). All genome-wide significant loci associated with uUMOD and uUCR in the overall population are listed in Table 2.
Table 1.
Main characteristics and UMOD levels in the 13 study cohorts
| Study Cohort | Population Type | Sample Size (n) | Women (n, %) | Age (yr) | BMI (kg/m2) | eGFRcreat (ml/min per 1.73 m2) | uUMOD (ug/ml) | uUCR (mg/g creat) |
|---|---|---|---|---|---|---|---|---|
| CARTaGENE | Nonisolate urban | 675 | 344 (51.1) | 53.5±8.7 | 27.0±5.2 | 90±14.0 | 14.0 (3.32–78.0) | 23.4 (3.84–88.1) |
| CoLaus | Nonisolate urban | 5112 | 2627 (51.7) | 54.1±10.9 | 25.9±4.6 | 89.5±19.9 | 25.9 (4.93–73.7) | 18.3 (3.69–46.7) |
| CROATIA-Korcula | Isolate | 1687 | 1125 (66.7) | 55.2±15.2 | 27.7±4.4 | 83.2±23.2 | 14.2 (2.47–112) | 15.2 (1.54–80.1) |
| CROATIA-Split | Nonisolate urban | 487 | 275 (56.5) | 49.5±14.7 | 27.0±4.2 | 85.1±21.9 | 28.5 (5.05–89.0) | 20.6 (3.51–48.9) |
| CROATIA-Vis | Isolate | 200 | 120 (60) | 53.8±12.4 | 27.4±3.8 | 84.9±22.7 | 36.3 (4.75–111) | 43.6 (4.81–95.2) |
| FHS | Nonisolate urban | 2643 | 1404 (53.2) | 58.4±9.6 | 27.9±5.1 | 87.5±25.0 | 7.43 (0.30–33.1) | 9.52 (0.26–27.0) |
| GAPP | Nonisolate urban | 1518 | 815 (53.7) | 35.5±5.3 | 24.5±3.7 | 109±12.2 | 19.4 (6.09–117) | 4.40 (0.78–14.3) |
| GCKD | Nonisolate | 4716 | 1855 (39.3) | 60.0±12.0 | 29.8±5.9 | 49.6±18.1 | 9.71 (1.34–25.8) | 12.2 (2.20–30.4) |
| GS:SFHS | Nonisolate urban | 7652 | 4588 (60) | 51.3±13.5 | 26.6±5.1 | 98.9±20.9 | 8.50 (1.67–53.1) | 15.7 (1.20–57.1) |
| INGI-CARL | Isolate | 337 | 191 (56.9) | 48.1±20.4 | 26.7±5.9 | 101.5±44.2 | 5.05 (0.98–21.2) | 6.69 (1.41–30.5) |
| LBC1936 | Nonisolate urban | 661 | 317 (48) | 72.7±0.7 | 28.0±4.3 | 63.2±5.75 | 15.3 (3.09–56.2) | 16.3 (4.16–46.8) |
| INGI-VB | Isolate | 1538 | 858 (55.8) | 54.5±18.0 | 25.9±4.5 | 90.1±22.4 | 6.74 (1.52–30.4) | 7.91 (1.54–34.4) |
| VIKING | Isolate | 2089 | 1254 (60) | 49.9±15.2 | 27.4±4.9 | 94.1±24.1 | 8.46 (2.36–37.5) | 6.82 (1.57–59.3) |
Data are presented as mean±SD or median (5th percentile, 95th percentile) for continuous variables; and n (%) for categorical variables. Median and percentile range were used for uUMOD and uUCR as the original distribution was not normally distributed, which is why we rank transformed the phenotype before running GWAS. BMI, body mass index; eGFRcreat, eGFR on the basis of plasma creatinine, using the Chronic Kidney Disease Epidemiology Collaboration equation; GAPP, Genetic and Phenotypic Determinants of Blood Pressure and other Cardiovascular Risk Factors; INGI-CARL, INGI-Carlantino; INGI-VB, INGI-Val Borbera.
Table 2.
Genome-wide significant loci associated with urinary UMOD (raw and indexed to creatinine) levels in the overall population
| Marker Name | EA | NEA | EAF | Effect | SE | P Value | Imputation Quality | CHR | BP | ID | Gene |
|---|---|---|---|---|---|---|---|---|---|---|---|
| uUMOD | |||||||||||
| 16:20357281 | t | c | 0.153 | −0.2344 | 0.012 | 2.17E–88 | 0.95 | 16 | 20357281 | rs12934455 | Intron 5 of UMOD |
| 16:20392332 | a | g | 0.190 | −0.2154 | 0.011 | 5.33E–79 | 0.84 | 16 | 20392332 | rs77924615 | Intron 3 of PDILT |
| 17:39135505 | c | g | 0.284 | −0.0537 | 0.009 | 1.24E–08 | 0.94 | 17 | 39135505 | rs8067385 | Intron 7 of KRT40 |
| uUCR | |||||||||||
| 16:20359831 | t | c | 0.167 | −0.2550 | 0.011 | 3.86E–118 | 0.97 | 16 | 20359831 | rs13335818 | Exon 3 of UMOD |
| 16:20392332 | a | g | 0.190 | −0.2351 | 0.011 | 1.27E–97 | 0.84 | 16 | 20392332 | rs77924615 | Intron 3 of PDILT |
| 15:53879241 | g | t | 0.474 | −0.0502 | 0.009 | 1.65E–08 | 0.99 | 15 | 53879241 | rs9672398 | Intron 18 of WDR72 |
The overall population includes 29,315 participants from 13 cohorts. BP, base position; CHR, chromosome; EA, effect allele; NEA, noneffect allele; EAF, effect allele frequency; CHR, chromosome; ID, SNP identification.
Meta-GWAS for Raw Urinary UMOD Levels
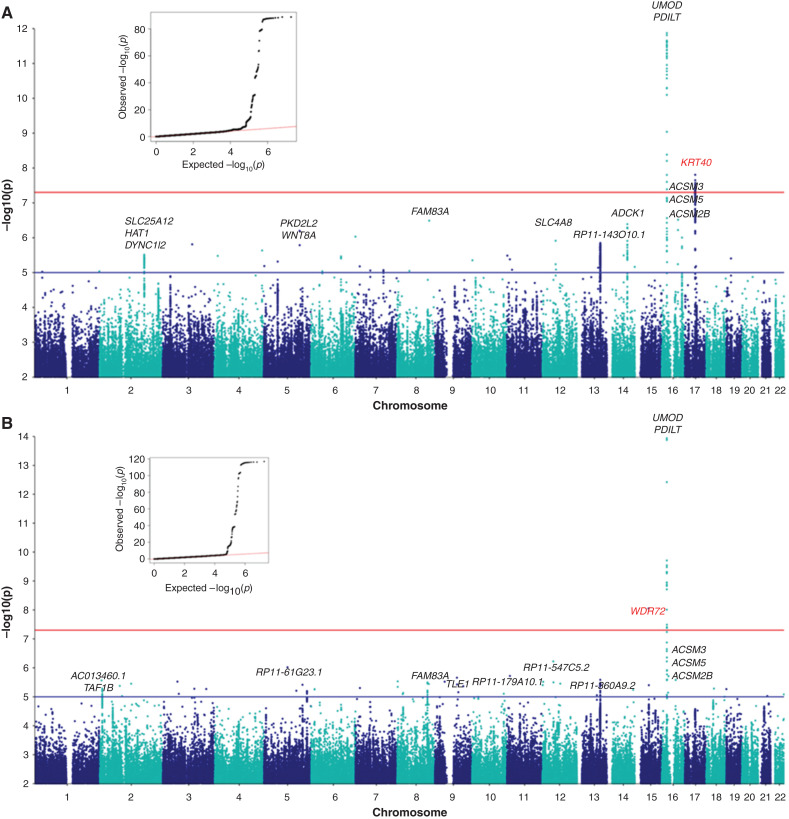
Meta-analysis of uUMOD levels in 29,315 individuals resulted in the identification of two genome-wide significant signals. A novel, genome-wide significant locus (P value 1.24E–08) was identified on chromosome 17 spanning the KRT40 gene. A major signal (P value 2.17E–88) was present on chromosome 16, corresponding to the previously described UMOD-PDILT locus (Figure 1A). The quantile-quantile plot of the –log10 observed versus expected P values for raw UMOD had a ʎ value of 1.00, indicating there is no significant genomic inflation (Figure 1A, inset). The full Manhattan plot without y-axis cutoff is shown in Supplemental Figure 1A.
Figure 1.
Genetic loci associated with raw uUMOD levels and UMOD indexed to creatinine (uUCR). Manhattan plot of meta-GWAS showing –log10 P values (y-axis cutoff at 1.0E–12 and 1.0E–14) in 13 cohorts. The blue line is at 1E–05 “suggestive” level and the red line is at the commonly used 5E–08 threshold for significance in GWAS. (A) Two genome-wide significant loci are associated with uUMOD: the first on chromosome 16 near the UMOD and PDILT genes, with the lowest P value (2.17E–88) at rs12934455; the second on chromosome 17, with the lowest P value rs806738 (1.24E–08) identified in and near the KRT40 gene. The quantile-quantile (QQ) plot of observed versus expected –log10 P values of the meta-GWAS is shown at the top left-hand corner. (B) Two genome-wide significant loci are associated with uUCR: the first on chromosome 16 spanning the UMOD gene, with the SNP with the lowest P value being a synonymous UMOD variant (rs13335818, P value 3.86E–118). A second genome-wide significant locus was detected on chromosome 15 within the WDR72 gene with rs9672398 showing the strongest association (P value 1.65E–08).
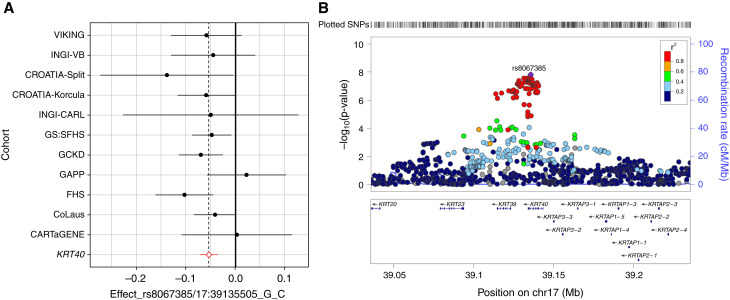
Association of uUMOD levels and genotypes at rs8067385, the SNP with the lowest P value (1.24E–08) within the KRT40 locus, is shown in Tables 3 and 4. The effect size of the SNP on uUMOD levels was consistent in direction and similar in magnitude across most of the cohorts, as evidenced in a Forest plot (Figure 2A). The minor allele, C, of rs8067385 was associated with lower uUMOD levels in most of the cohorts and had an average standardized effect size of -0.05 and a standard error of 0.009 (Tables 3 and 4; Figure 2A), which explained 0.1% of the variance observed. The regional association plot (Figure 2B) showed all of the genome-wide significant SNPs in high LD with rs8067385 over the KRT40 region. SNPs in each locus with a P value <1.0E–05 are listed in Supplemental Table 7. All KRT40 SNPs with P value <1.0E–05 that were in high LD (r2 ≥ 0.8) with rs8067385 are listed in Supplemental Table 8 and annotated using SNiPa. The exonic variants rs9908304 and rs721958 are predicted to be damaging/deleterious by SIFT and PolyPhen2 (Supplemental Table 8). KRT40 encodes keratin-40 (KRT40), a type I keratin expressed in the kidney. The 17q21.2 region also includes type I keratin genes KRT39 and KRT23.
Table 3.
Association of rs8067385 genotypes (KRT40 locus) with urinary UMOD levels in the 13 study cohorts
| uUMOD (ug/ml) | Sample Size (n) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cohorts | GG | GC | CC | GG | GC | CC | Total Sample Size (n) | P Value in Each Cohort | EAF (C) | Imputation Quality | Beta | SE |
| CARTaGENE | 23.99±27.53 | 27.67±36 | 25.63±31.47 | 345 | 264 | 65 | 674 | 9.58E–01 | 0.293 | 0.992 | 0.003 | 0.057 |
| CoLaus | 31.93±28.59 | 30.52±27.14 | 30.24±23.18 | 2564 | 2090 | 458 | 5112 | 5.91E–02 | 0.297 | 0.969 | −0.041 | 0.022 |
| CROATIA-Korcula | 33.54±42.37 | 31.42±36.44 | 28.49±33.29 | 728 | 741 | 218 | 1687 | 4.71E–02 | 0.345 | 0.904 | −0.059 | 0.030 |
| CROATIA-Split | 34.08±25.57 | 36.56±35.50 | 32.15±24.34 | 204 | 232 | 51 | 487 | 4.35E–02 | 0.289 | 0.881 | −0.139 | 0.069 |
| CROATIA-Vis | 45.22±34.22 | 39.26±32.95 | 49.07±27.78 | 111 | 74 | 14 | 199 | 8.68E–01 | 0.263 | 0.894 | −0.020 | 0.118 |
| FHS | 11.65±13.39 | 10.73±12.27 | 9.14±9.23 | 1318 | 1076 | 246 | 2640 | 6.20E–04 | 0.295 | 0.976 | −0.103 | 0.030 |
| GAPP | 33.51±36.78 | 33.00±35.09 | 35.39±36.66 | 817 | 580 | 121 | 1518 | 5.80E–01 | 0.271 | 0.978 | 0.023 | 0.041 |
| GCKD | 10.2±13.22 | 9.29±12.66 | 8.69±8.86 | 2463 | 1862 | 391 | 4716 | 2.07E–03 | 0.286 | 0.993 | −0.070 | 0.023 |
| GS:SFHS | 16.25±22.05 | 15.59±22.25 | 13.88±17.84 | 4402 | 2816 | 433 | 7651 | 1.83E–02 | 0.240 | 0.951 | −0.048 | 0.020 |
| INGI-CARL | 7.30±7.20 | 8.13±14.15 | 7.31±6.83 | 159 | 152 | 26 | 337 | 5.84E–01 | 0.306 | 0.891 | −0.05 | 0.09 |
| LBC1936a | NA | NA | NA | NA | NA | NA | 661 | NA | NA | NA | NA | NA |
| INGI-VB | 10.55±11.35 | 10.47±10.85 | 9.65±10.89 | 761 | 640 | 137 | 1538 | 3.04E–01 | 0.306 | 0.873 | −0.045 | 0.043 |
| VIKING | 13.57±12.24 | 13.76±27.65 | 12.42±10.43 | 1134 | 813 | 142 | 2089 | 1.10E–01 | 0.261 | 0.988 | −0.058 | 0.037 |
EAF, effect allele frequency; GAPP, Genetic and Phenotypic Determinants of Blood Pressure and other Cardiovascular Risk Factors; INGI-CARL, INGI-Carlantino; INGI-VB, INGI-Val Borbera; NA, not available.
The SNP was not present in LBC1936 for meta-analysis.
Table 4.
The average effect size of the SNP
| CHR | BP | ID | Gene | Allele1 | Allele2 | Freq1 | Effect | SE | P Value | Het I Sq | Het P Value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 17 | 39135505 | rs8067385 | intron of KRT40 | c | g | 0.284 | −0.054 | 0.009 | 1.24E–08 | 0 | 0.557 |
CHR, chromosome; BP, base position; ID, SNP identification; Freq1, Frequency of the allele 1; Het I Sq, heterogeneity I square; Het P value; heterogeneity P value.
Figure 2.
Effect size of rs8067385 and regional association plot of KRT40 locus for raw urinary UMOD levels. (A) Forest plot showing effect sizes of rs8067385 (top SNP in KRT40 locus) on uUMOD meta-analyses in the 13 cohorts. The red diamond represents the average effect size of -0.054 and a standard error of 0.009 of the minor, C allele of rs8067385 in association with uUMOD. Information on this SNP was not available in the GWAS for the LBC1936 cohort. Effect sizes are shown for cohorts with at least 10 individuals for each of the genotypes of rs8067385. (B) Regional association plot of the KRT40 locus for uUMOD meta-analysis in 13 cohorts. The genome-wide significant locus spans over KRT40 and KRT39 genes, whereas the top rs8067385 variant (purple diamond) is above KRT40. Each dot represents a SNP; the color code refers to the LD toward the top SNP. Red dot represents high LD with the top SNP.
Association of uUMOD levels and genotypes at rs12934455, the SNP with the lowest P value (2.17E–88) within the UMOD-PDILT locus, is shown in Tables 5 and 6. The average standardized effect size was −0.23 with a standard error of 0.01, which explains 1.4% of the variance observed. The Forest plot, showing effect sizes of the minor allele of rs12934455 consistent in direction for all cohorts, and the regional association plot of the UMOD-PDILT locus are shown in Supplemental Figure 2. The rs12934455 SNP had a heterogeneity I Sq value of 68.7, likely due to the lower imputation quality for that variant in CoLaus and FHS cohorts. Indeed, heterogeneity for that SNP was not significant (Het I Sq=45.2, Het P value=0.06) when meta-analysis of uUMOD was carried out excluding CoLaus and FHS. The UMOD-PDILT locus has been consistently associated with kidney function and levels of urinary UMOD in previous GWAS.14,19,40
Table 5.
Association of rs12934455 genotypes (UMOD-PDILT locus) with urinary UMOD levels in the 13 study cohorts
| uUMOD (ug/ml) | Sample Size (n) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cohorts | CC | CT | TT | CC | CT | TT | Total Sample Size (n) | P Value in Each Cohort | EAF (T) | Imputation Quality | Beta | SE |
| CARTaGENE | 25.97±32.08 | 24±29.24 | 8.5±5.73 | 473 | 189 | 12 | 674 | 3.33E-02 | 0.158 | 0.997 | −0.159 | 0.075 |
| CoLaus | 33.20±29.41 | 27.39±22.67 | 20.43±15.82 | 3544 | 1408 | 160 | 5112 | 7.85E-28 | 0.141 | 0.748 | −0.349 | 0.032 |
| CROATIA-Korcula | 29.72±32.85 | 25.06±28.14 | 20.98±25.82 | 1194 | 405 | 28 | 1627 | 7.72E-05 | 0.140 | 0.972 | −0.158 | 0.040 |
| CROATIA-Split | 38.40±33.23 | 28.32±21.95 | 15.52±9.03 | 349 | 124 | 14 | 487 | 2.42E-06 | 0.149 | 0.966 | −0.371 | 0.079 |
| CROATIA-Vis | 45.84±35.48 | 32.86±20.18 | 52.17 | 159 | 40 | 1 | 200 | 2.20E-02 | 0.104 | 0.923 | −0.403 | 0.176 |
| FHS | 12.22±13.72 | 8.87±9.62 | 5.04±5.98 | 1796 | 775 | 72 | 2643 | 9.87E-16 | 0.172 | 0.880 | −0.313 | 0.039 |
| GAPP | 34.46±37.44 | 31.49±32.81 | 26.51±30.55 | 1071 | 412 | 35 | 1518 | 6.75E-03 | 0.159 | 0.978 | −0.136 | 0.050 |
| GCKD | 10.16±12.62 | 8.63±13.26 | 6.36±7.5 | 3510 | 1095 | 111 | 4716 | 1.36E-14 | 0.140 | 0.992 | −0.225 | 0.029 |
| GS:SFHS | 16.94±23.16 | 14.16±19.19 | 8.47±10.63 | 5286 | 2142 | 224 | 7652 | 1.24E-16 | 0.169 | 0.992 | −0.186 | 0.022 |
| INGI-CARL | 8.69±12.73 | 6.25±6.01 | 2.96±2.63 | 217 | 105 | 15 | 337 | 3.16E-04 | 0.198 | 0.965 | −0.338 | 0.094 |
| LBC1936 | 23.04±23.37 | 20.38±22.64 | 10.55±8.29 | 455 | 190 | 16 | 661 | 5.49E-04 | 0.164 | 0.944 | −0.263 | 0.076 |
| INGI-VB | 11.11±11.50 | 8.78±9.83 | 4.87±4.31 | 1150 | 355 | 33 | 1538 | 1.37E-08 | 0.136 | 0.959 | −0.306 | 0.054 |
| VIKING | 14.19±21.83 | 11.88±10.26 | 8.84±7.18 | 1578 | 472 | 39 | 2089 | 9.89E-05 | 0.131 | 0.998 | −0.181 | 0.047 |
EAF, effect allele frequency; GAPP, Genetic and Phenotypic Determinants of Blood Pressure and other Cardiovascular Risk Factors; INGI-CARL, INGI-Carlantino; INGI-VB, INGI-Val Borbera.
Table 6.
The average effect size of the SNP
| CHR | BP | ID | Gene | Allele1 | Allele2 | Freq1 | Effect | SE | P Value | Het I Sq | Het P Value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 16 | 20357281 | rs12934455 | intron of UMOD | t | c | 0.153 | −0.234 | 0.012 | 2.17E-88 | 68.7 | 0.0001 |
CHR, chromosome; BP, base position; ID, SNP identification; Freq1, Frequency of the allele 1; Het I Sq, heterogeneity I square; Het P value; heterogeneity P value.
Meta-GWAS for Urinary UMOD Levels Indexed to Creatinine
The meta-GWAS for uUCR yielded two genome-wide significant signals: the known UMOD-PDILT locus on chromosome 16, and a novel locus on chromosome 15 over the WDR72 gene (Figure 1B). The minor G allele of rs9672398 in WDR72 is associated with lower levels of uUCR in most of the cohorts (Supplemental Figure 3), with an average standardized effect size of −0.05 and a standard error of 0.0089 (Supplemental Table 9), which explains 0.1% of the variance observed. The regional association plot for the chromosome 15 signal revealed that only one SNP, rs9672398, reached genome-wide significance (P value 1.65E–08) but was in LD with several other SNPs over WDR72 (Supplemental Figure 3). The association of uUMOD did not reach genome-wide significance (P value 3.336E–05). WDR72 is highly expressed in the kidney. By GWAS, common variants in WDR72 have been associated with kidney function and CKD,14,41 urine pH,42 and kidney stones.42,43
The rs13335818 SNP with the lowest P value (3.86E–118) was found within the UMOD-PDILT locus, and association to uUCR is shown in Supplemental Table 10 and Supplemental Figure 4. The regional association plot showed that rs13335818 is in high LD (r2=0.94) with the top rs12934455, identified in meta-analysis of uUMOD levels. As for the raw UMOD level, the genome-wide signal included independent variants on UMOD (rs13335818) and PDILT (rs77924615), respectively (Supplemental Figure 4). SNPs from each locus with P<1E–05 for uUCR are shown in Supplemental Table 11.
The combined variance of rs12934455 (UMOD) and rs8067385 (KRT40) for uUMOD is 1.5% and for uUCR is 1.9%. Similar associations for uUMOD and uUCR were seen when meta-analysis was conducted using sample size and P values (Supplemental Figure 5). Spearman’s rank correlation of effect sizes (P=0.02) of the most significant KRT40 SNP, rs8067385, in association with uUMOD and uUCR indicated the effect sizes between the two traits were similar in each of the cohorts (Supplemental Figure 6). Analysis of the candidate genes associated with raw and indexed urinary UMOD levels revealed a number of genes expressed in the TAL, with encoded proteins playing important roles in cell homeostasis, mitochondrial function, transport, and inflammatory signaling (Supplemental Table 12).
Meta-GWAS for Urinary UMOD Levels Normalized for eGFR
As some cohorts included individuals with CKD, potentially influencing the levels of UMOD in urine,16 we repeated the meta-analysis using UMOD normalized for eGFR, sex, and age in seven cohorts with available information. The UMOD-PDILT locus remained at genome-wide significance and the KRT40 locus at a suggestive threshold (Supplemental Table 13) whereas the WDR72 locus did not (Supplemental Table 14).
Effect of UMOD Genotype on Urinary Levels of UMOD
The minor, T alleles of the UMOD-PDILT variants rs12934455 and rs13335818 are associated with significantly lower levels of urinary UMOD, either raw or indexed to creatinine. For each trait, the homozygous TT carriers showed approximately 50% lower levels compared with the homozygous carriers of the reference, C allele (Supplemental Figure 7).
Associations of the New Loci with Creatinine and eGFR in UK Biobank
To evaluate the role of indexing urinary levels of UMOD to creatinine, we investigated potential associations of the new genome-wide significant loci for plasma and urinary creatinine and derived eGFR in the UK Biobank summary statistics database “Global Biobank Engine.”34 These analyses revealed that rs9672398 in WDR72 (index SNP for uUCR) was significantly associated with eGFR and plasma creatinine, but not with urinary creatinine levels, whereas the rs8067385 SNP in KRT40 (index for uUMOD) was not associated with any trait related to creatinine (Table 7).
Table 7.
Association of the top variants in KRT40 and WDR72 with eGFR and plasma and urinary creatinine levels in UK Biobank
| GWAS | Gene | SNP | POS | A1 | A2 | N | AF1 | BETA | SE | P value | Imputation quality |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Plasma creatinine | KRT40 | rs8067385 | 39135505 | G | C | 408181 | 0.751 | 0.006 | 0.003 | 1.55E-01 | 0.994 |
| WDR72 | rs9672398 | 53879241 | T | G | 408181 | 0.536 | 0.020 | 0.002 | 2.51E-16 | 0.999 | |
| Urinary creatinine | KRT40 | rs8067385 | 39135505 | G | C | 408181 | 0.751 | 0.001 | 0.003 | 6.75E-01 | 0.994 |
| WDR72 | rs9672398 | 53879241 | T | G | 408181 | 0.536 | 0.005 | 0.002 | 2.45E-02 | 0.999 | |
| eGFR | KRT40 | rs8067385 | 39135505 | G | C | 408181 | 0.751 | −0.006 | 0.003 | 4.16E-02 | 0.994 |
| WDR72 | rs9672398 | 53879241 | T | G | 408181 | 0.536 | −0.020 | 0.002 | 2.05E-16 | 0.999 |
Plasma and urine creatinine and eGFR were all obtained from UK Biobank summary statistics. POS, position, A1, allele 1; A2, allele 2; N, sample size number; AF1, allele 1 frequency.
Conditional Analysis
Implementing the GCTA COJO -Slct function for conditional analysis on summary statistics from meta-analysis resulted in identification of two independent loci in chromosome 16, one in UMOD (rs13335818) and one in the upstream gene, PDILT (rs11864909). A conditional GWAS using either of these two variants as covariates on the basis of individual-level genotype data from our largest cohort (GS:SFHS) confirmed that both signals remained genome-wide significant, indicating the loci are independent in the Scottish “healthy” population (Supplemental Figure 8).
Additional Genome-wide Analyses
VEGAS2 is a gene-based association method that uses GWAS summary data and the associated P values and a simulation approach to calculate gene-based empirical association P values. The method tests for enrichment of multiple SNPs associated with the disease/trait that individually have a too modest effect on the phenotype to reach genome-wide significance using a per-SNP test.32 The VEGAS analysis identified the region of chromosome 16 containing UMOD and PDILT and chromosome 17 containing KRT40 as statistically significant regions (P value 1.24E–08) for uUMOD, with the top UMOD SNP from the meta-analysis (rs12934455, P value 2.17E–88) being the main contributor to the finding. For uUCR, only the top SNPs in UMOD and PDILT reached significance, with KRT40 being the third most significant result (P value 3.60E–05). The top five results from VEGAS2 are included in Supplemental Table 15.
Candidate Gene and Sensitivity Analyses
As UMOD is essentially produced in the TAL cells, we tested whether common variants within genes causing Mendelian disorders affecting the TAL may also influence the urinary excretion of UMOD. SLC12A1, KCNJ1, CLDN19, HNF1B, and MUC1 showed at least one SNP with a P value below the gene-specific threshold associated with uUMOD and/or uUCR (Supplemental Figure 9; Supplemental Table 16). We should note the association for genes expressed in the TAL was not stronger than that identified for genes expressed in other nephron segments, when queried against the meta-GWAS results (Supplemental Table 17). The suggestive loci (SORL1, CAB39, FAM83A, and MARCH1) identified in the first meta-GWAS for UMOD,19 also did not reach the suggestive threshold (Supplemental Table 17).
The effect sizes of the most significant SNP within UMOD and PDILT are similar, as shown in Supplemental Table 18. The sensitivity analysis removing the FHS cohort, which measured UMOD using a distinct immunoassay, showed the UMOD/PDILT locus remained genome-wide significant and both the KRT40 and WDR72 loci remained within the suggestive threshold limit, despite the reduced sample size (Supplemental Table 19).
Segmental Distribution of KRT40 and WDR72 in relation to UMOD
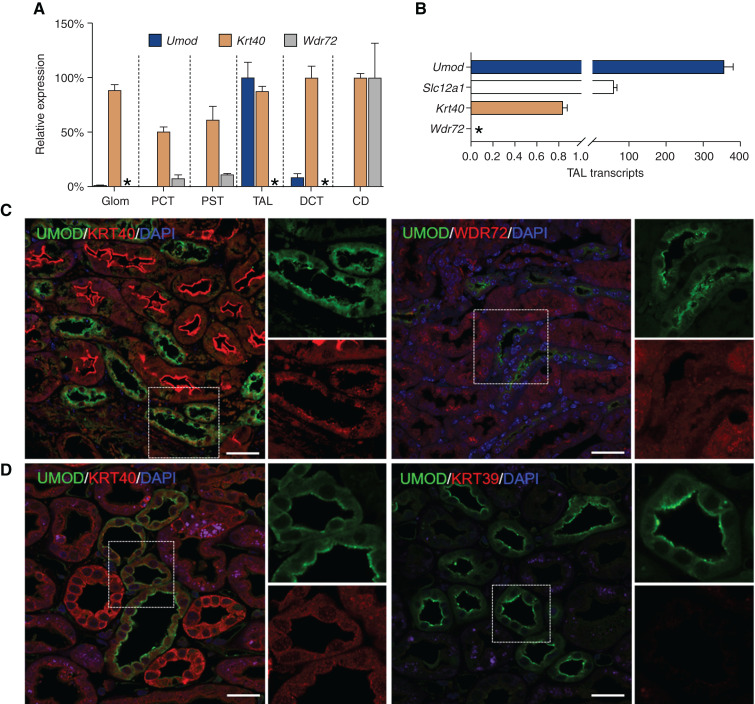
To substantiate the biologic relevance of the newly identified KRT40 and WDR72 loci, we first evaluated the segmental distribution of KRT40, WDR72, and UMOD in mouse kidney (Figure 3). KRT40 is widely distributed in the kidney, more abundant in distal tubular segments and overlapping with UMOD in the TAL—both at the mRNA and protein levels (Figure 3, A–C). In isolated TAL segments, the expression of KRT40 was at least two orders of magnitude lower than that of UMOD and NKCC2 (Slc12a1) (Figure 3B). In situ hybridization evidenced a weak, selective expression of Krt40 in Umod-positive segments of the mouse kidney, with no signal for Krt39 (Supplemental Figure 10). Immunostaining confirmed a signal for KRT40 in UMOD-positive tubules, particularly at the apical pole of cells lining the TAL, whereas no colocalization between UMOD and WDR72 was observed (Figure 3C). Both KRT40 and WDR72 were detected in AQP2-positive segments of the mouse kidney (Supplemental Figure 11). In the human kidney, KRT40 was detected in both UMOD-positive and negative tubules, whereas KRT39 did not show any signal (Figure 3D).
Figure 3.
Segmental distribution of UMOD, KRT40, and WDR72 in the mouse kidney. (A) The mRNA levels of Umod, Krt40, and Wdr72 in isolated mouse nephron segments were analyzed by SYBR green quantitative PCR. Quantification of targeted genes was done in comparison with Gapdh, which was used as housekeeping gene (n=4 pools for each segment). The nephron segments were validated by enrichment in specific markers.6,18 (B) Relative expression of Krt40, Wdr72, Slc12a1, and Umod transcript levels in isolated TALs from C57BL/6J mice as assessed by SYBR green quantitative PCR. Values are expressed as 2^(CtGapdh- CtGene of interest) × 10^2. Bars indicate average±SEM n=4 TAL fractions. Asterisk (*), not detected (A and B). (C) Representative immunofluorescence staining for UMOD (UMOD, green) and KRT40 or WDR72 (red) on paraffin-embedded kidney sections from wild-type mice, showing colocalization of the UMOD and KRT40 signals in the TAL. No staining for WDR72 is detected in UMOD-positive segments. Nuclei are counterstained with DAPI (blue). Scale bar: 25 µm. (D) Representative immunofluorescence staining for UMOD (UMOD, green) and KRT40 or KRT39 (red) on paraffin-embedded kidney sections from a normal human kidney. KRT40 is localized in both UMOD-positive and negative tubules, whereas no signal for KRT39 is detected. Nuclei are counterstained with DAPI (blue). Scale bar: 25 µm.
Modulation of KRT40 Expression Influences UMOD Excretion by mTAL Cells
The codistribution of KRT40 and UMOD led us to test whether the level of KRT40 expression may modulate the processing and excretion of UMOD by TAL cells. This hypothesis was supported by the existence of at least two exonic variants in high LD with the index KRT40 variant rs8067385, predicted to be damaging/deleterious by SIFT and PolyPhen2 (Supplemental Table 8). Furthermore, in the GTEx portal, the minor, C allele of rs8067385 is associated with a significant, dose-dependent decrease in the expression of KRT40 in a variety of epithelial tissues including the testis, pancreas, esophagus, and colon (no eQTL data for kidney medulla tissue available) (Supplemental Figure 12).
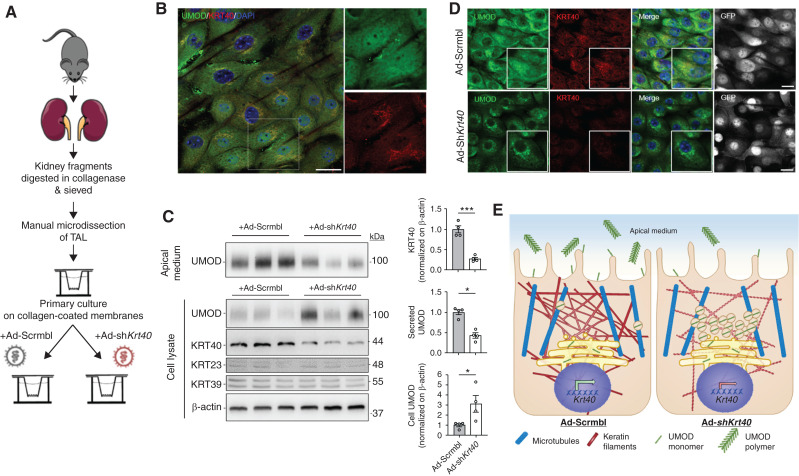
Characterization of mTAL cells verified that KRT40 and UMOD were both endogenously expressed (Figure 4, A and B). Transduction of mTAL cells with an adenovirus expressing a short hairpin RNA against mouse Krt40 (Ad-shKrt40) induced a specific silencing of KRT40, compared with cells treated with a scramble adenovirus (Ad-Scrmbl) (Figure 4C). In these conditions, the downregulation of KRT40 was reflected by a significant accumulation of UMOD and a sharp decrease in the amount of excreted UMOD in the apical medium of mTAL cells (Figure 4C). Confocal microscopy indicated the silencing of KRT40 in mTAL cells resulted in perinuclear accumulation of UMOD, contrasting with the control signal in cells transduced with Ad-Scrmbl (Figure 4D). The trafficking defect induced by KRT40 downregulation was confirmed by Z-stack image analysis, with a perinuclear staining for UMOD contrasting with the diffuse signal observed in control conditions (Supplemental Figure 13A). The silencing of KRT40 had also an effect on the trafficking of ROMK (Supplemental Figure 13B), but did not modify the expression of TAL genes including Slc12a1, Kcnj1, Hnf1b, and Muc1 in mTAL cells (data not shown). Taken together, these data suggest the expression of the cytokeratin KRT40 regulates the processing and excretion of UMOD in TAL cells (Figure 4E).
Figure 4.
Effect of KRT40 modulation on UMOD processing in mTAL cells. (A) Schematic diagram illustrating the protocol to generate differentiated primary cell cultures (mTAL cells) from mouse kidney.38 (B) Representative immunofluorescence staining for UMOD (UMOD, green) and KRT40 (red) on mTAL cells. Nuclei are counterstained with DAPI (blue). Scale bar: 25 μm. (C) Representative Western blot of secreted (apical medium) and cellular UMOD in mTAL cells. The apical medium and whole cell lysates were collected 5 days after treatment with Ad-shKrt40 or Ad-Scrmbl. Krt40 downregulation resulted in an increase of intracellular UMOD, and a reduced release in the apical medium. β-actin was used as a loading control. Densitometry analysis for KRT40, secreted and cellular UMOD signals are shown relative to Ad-Scrmbl. Bars indicate mean±SEM. Unpaired two-tailed t test (KRT40) or Mann–Whitney test (cellular and secreted UMOD), *P < 0.05; ***P < 0.001, n=4. (D) Representative immunofluorescence staining for UMOD (UMOD, green) and KRT40 (red) on mTAL cells after transduction with Ad-shKrt40. Accumulation of UMOD is observed in the perinuclear compartment of Krt40 silenced cells. Nuclei are counterstained with DAPI (blue). Both adenoviral vectors express GFP (gray). Scale bar: 25 µm. (E) Model showing the potential link between variants in KRT40 and the excretion of UMOD. Specific KRT40 variants (e.g., the minor, C allele of rs8067385) may affect the expression of KRT40 in TAL cells, affecting the cytoskeleton, and altering the processing and apical excretion of UMOD in the urine.
Discussion
To gain novel insights into the mechanisms regulating UMOD excretion, we performed a meta-GWAS on urinary UMOD levels in 29,315 individuals of European ancestry, three times more than in our previous analysis.19 We identified two novel, genome-wide significant loci, KRT40 and WDR72, in addition to the previously known UMOD-PDILT locus to be associated with uUCR and uUMOD. Mechanistic studies in primary mTAL cells demonstrated that modulating the expression of KRT40 affects the processing and apical excretion of UMOD. These studies provide insights into the biology of UMOD and keratins, and into the links between the UMOD-PDILT locus and kidney function.
The UMOD-PDILT locus has been consistently among the strongest associated loci with eGFR and CKD.14,40 The relevance of the UMOD variants, which are associated with the levels of UMOD in the kidney and urine, is immediate because the gene is kidney specific and involved in a spectrum of kidney diseases.1,7,40 In our meta-analysis, the variant showing the strongest association with uUCR is rs13335818 (P value 3.86E–118), a synonymous variant within UMOD, in high LD (r2 = 0.98) with the top SNP in our previous study, rs12917707, and with UMOD promoter variants associated with eGFR and CKD and with expression of UMOD.7,40 In a previous study of genetic associations with urinary UMOD levels,14 two independently associated variants in the UMOD-PDILT locus were identified in conditional analyses: rs77924615, mapping into an intron of the upstream gene PDILT, and rs34262842, mapping to an intron of UMOD. Similarly, our conditional analysis in a large cohort with individual-level genotype data identified two independent loci in that region, one in UMOD (SNP rs13335818 in high LD with rs34262842, r2=0.94) and one in PDILT (rs11864909, in almost complete LD with rs77924615, r2=0.98).
The lead SNP in PDILT from our meta-analysis, rs77924615, had the strongest association with CKD and eGFR in the GWAS performed by the CKDGen Consortium.14 Of interest, the intronic PDILT rs77924615 maps to open chromatin regions identified from various kidney cell types. Because PDILT is not expressed in the human kidney and rs77924615 was significantly associated with both differential expression of UMOD in kidney tissue and urine UMOD levels (obtained in the GCKD cohort), it was considered a regulatory SNP.14 Collectively, these results substantiate the independent association between UMOD and PDILT variants and the levels of UMOD in urine. A recent Mendelian randomization study clarified the causality between uUMOD levels and kidney function in individuals of European descent: genetically driven levels of UMOD have a direct, causal, and adverse effect on kidney function outcome in the general population.44
The KRT40 locus on chromosome 17 is a novel, genome-wide significant locus associated with UMOD levels in the urine. The association at the KRT40 locus is on the basis of multiple genome-wide significantly associated SNPs in high LD with the index SNP, rs8067385. Individuals homozygous for the minor, C allele of rs8067385 had lower levels of uUMOD compared with individuals homozygous for the G allele. Of note, the effect sizes of rs8067385 for uUMOD and uUCR are only marginally different, as indicated by Spearman’s rank correlation analysis. The KRT40 signal remained above the suggestive threshold (P<5.17E–07) in the meta-analysis on urinary UMOD corrected for eGFR, and in the VEGAS analysis. In contrast, the rs8067385 SNP in KRT40 was not significantly associated with eGFR and plasma or urinary creatinine levels in the UK Biobank. Together, these results support the value of the KRT40 signal in relation to urinary UMOD levels.
KRT40 encodes KRT40, a type I keratin that belongs to the family of intermediate filament-forming keratins that form the cytoskeleton in epithelial cells. Types I and II keratins form obligate heterodimers and are regulated in a pairwise fashion in epithelia, depending on the tissue, the differentiation state, and the biologic context.45 KRT40 belongs to a cluster of type I KRT genes located on chromosome 17q21.2, close to KRT39. As cytoskeletal proteins, keratins are involved in maintaining the physical integrity, mechanical stability, and shape of epithelial cells. They are also important for intracellular organization and transport within cells, for example, trafficking of proteins to the plasma membrane.46 Keratins are considered as cytoprotective, undergoing dynamic upregulation in disease states, and potentially affecting migration, growth, proliferation, and protein synthesis.47 Several inherited keratinopathies (e.g., skin disorders) have been reported, but none involving KRT40.
Little is known about the role of keratins in the kidney. Recent studies evidenced robust changes in the expression and subcellular localization of KRT7–8 and KRT18–19 in response to kidney stress, with KRT18 in urine being a potential biomarker for tubular cell injury.48 Our studies in mouse and human kidney reveal that KRT40 is weakly expressed in the TAL, where it colocalizes with UMOD. The prediction of missense variants in KRT40 in LD with rs8067385 and the association in GTEx of the minor allele of rs8067385 with a decreased expression of KRT40 in epithelial tissues suggest a possible loss of function of the KRT40 variant. We tested this hypothesis in the mTAL cells, which endogenously express both KRT40 and UMOD. Specific silencing of KRT40 in mTAL cells was reflected by a sharp decrease in the apical excretion of UMOD, causing the intracellular accumulation of the protein. The silencing was also reflected by altered apical targeting of ROMK in these cells. That altered expression of KRT40 affects UMOD and ROMK processing in TAL cells may suggest a role of KRT40 on the polarized sorting of proteins to the apical membrane, affecting the release of UMOD in urine (Figure 4E).
We detected a genome-wide significant association between variants in WDR72 and the urinary UMOD level indexed to creatinine (uUCR). WDR72 encodes a protein with eight WD40 (or β-transducin) repeats, which fold to form two circular, β-propeller structures, and an α-solenoid tail at the C-terminus. This combination of domains is conserved among membrane-coating proteins, which serve as a docking site for protein–protein interactions and stabilize membrane curvature.49 WDR72 is highly expressed in the kidney, although we found it clustered in the collecting ducts. Recessive mutations in WDR72 have been associated with amelogenesis imperfecta,50 and distal renal tubular acidosis.51,52 By GWAS, variants in WDR72 have been associated with kidney function and CKD,14,41,53 urine pH,42 and risk of kidney stones.42,43 In CKDGen, the index SNP at WDR72 was associated with blood urea nitrogen.14 Variants in WDR72 are strongly associated with eGFR on the basis of serum creatinine or cystatin C and with BUN.41 The fact that rs9672398 is significantly associated with eGFR and plasma creatinine levels in UK Biobank, that the WDR72 locus does not reach any suggestive threshold in the meta-analysis using UMOD normalized for eGFR, and that WDR72 does not colocalize with UMOD suggest the WDR72 signal, only detected for uUCR, is most likely related to its effect on eGFR.
Although limited by power, our candidate gene analysis revealed that a few common variants in genes causing rare Mendelian disorders targeting the TAL are weakly associated with UMOD levels. These results support the functional interactions operating in TAL cells, including the transcription factor HNF1-β, known to be an essential transcriptional regulator of UMOD,1,9 and ROMK, which directly regulates processing and release of UMOD by TAL cells.17 Analysis of the candidate genes from the loci associated with uUMOD and uUCR with a suggestive P value (<1.0E–05) revealed a number of genes expressed in the TAL/DCT, with encoded proteins playing roles in cell homeostasis, mitochondrial function and transport. Future studies will address the relevance and biologic mechanisms that underlie these genetic associations.
Our study combines the advantages of the largest to date meta-GWAS on urinary UMOD, measured with a robust assay in various types of cohorts, and complemented with detailed expression studies in mouse and human kidneys, and functional investigations in TAL cells. Limitations of this study include the availability of data only for individuals of European descent and the lack of replication due to limited availability of additional cohorts with available uUMOD measurements. We noted some variability of UMOD levels that were measured in different cohorts, even when using the same assay and apparently unrelated to sample processing and/or storage conditions.19,24 Variations in the physiology excretion of UMOD have been reported, potentially linked to dietary habits, tubular transport activities, or level of residual kidney function.1,16
Common, independent variants in KRT40, UMOD, and PDILT influence the levels of UMOD in urine. The expression of the type I keratin KRT40 affects UMOD processing in TAL cells. These results advance our understanding of the biology of UMOD, the role of keratins in the kidney and substantiate the association of UMOD-PDILT variants with kidney function.
Disclosures
A. Köttgen reports receiving honoraria from Sanofi Genzyme; reports being a scientific advisor or member of the American Kidney Fund, American Journal of Kidney Diseases, Journal of the American Society of Nephrology, Kidney International, and Nature Reviews Nephrology. C. Black reports having other interests/relationships through an honorary contract with the National Health Service. D. Conen reports consultancy agreements with Roche Diagnostics; reports receiving research funding from the Canadian Institutes of Health Research; and reports receiving honoraria from Bristol Myers Squibb/Pfizer. E. Wuehl reports being a scientific advisor to or member of the Alnylam Pharmaceuticals Advisory Board, Editorial Board Member of the Journal of Hypertension and Pediatric Nephrology, Executive Board Member of the German Hypertension League (Deutsche Hochdruckliga), and Vice-Chair COST Action HyperChildNET (EU Programme Horizon 2020). F. Madore reports receiving research funding from AstraZeneca, Bayer, Boehringer Ingelheim, GlaxoSmithKline, and Janssen; and reports being a scientific advisor or membership as Associate Editor for the Canadian Journal of Kidney Health and Disease. F. Schaefer reports having consultancy agreements with Akebia, Amgen, Alexion, Alnylam, Astellas, AstraZeneca, Bayer, Boehringer Ingelheim, Fresenius Medical Care, Otsuka, Roche, and Relypsa; reports receiving research funding from Fresenius Medical Care; reports receiving honoraria from Amgen, Gilead, Otsuka, Relypsa, and Roche; and reports being a scientific advisor or member of the Scientific Advisory Board activities for Alexion and Otsuka. M. Bochud reports receiving research funding from Merck Sharp & Dohme; reports receiving honoraria from various Swiss Federal Agencies (Swiss Federal Office of Public Health, Swiss Federal Office of Food Security and Veterinary Affairs); reports being a scientific advisor or member of scientific journals, such as Nutrients and Hypertension, Member of the Council of the Swiss Society of Public Health Plus, Member of the Council of the The National Institute for Cancer Epidemiology and Registration (NICER) Foundation (cancer epidemiology in Switzerland), representative of the University of Lausanne at the Swiss Academy of Medical Sciences; and reports other interests/relationships as a member of the Swiss Federal Commission on Nutrition, the Swiss Society of Hypertension, the Swiss Society of Nephrology, the Swiss Society of Nutrition, the Swiss Society of Public Health Plus. O. Devuyst reports having consultancy agreements with Alnylam, Galapagos, Otsuka Pharmaceuticals, and Sanofi; reports receiving research funding from Otsuka Pharmaceuticals and Roche; reports being a scientific advisor or member of the editorial board of CJASN, Kidney International, Nephrology Dialysis Transplantation, Pflügers Archiv, Peritoneal Dialysis International, and Orphanet Journal of Rare Diseases. K. Eckardt reports consultancy agreements with Akebia, AstraZeneca, Bayer, Boehringer Ingelheim, Genzyme, Otsuka, Travere, and Vifor; research funding from Amgen, AstraZeneca, Bayer, Evotec, Fresenius, Genzyme, Shire, and Vifor; honoraria from Akebia, AstraZeneca, Bayer, Boehringer Ingelheim, Genzyme, Otsuka, Travere, and Vifor; and advisory or leadership role with KI and BMJ (editorial boards). All remaining authors have nothing to disclose.
Funding
This work was supported by the Swiss National Science Foundation (project grant 310030_189044), the University Research Priority Program Innovative Therapies in Rare Diseases of the University of Zurich (UZH), the Swiss National Centre of Competence in Research, Kidney Control of Homeostasis (Kidney.CH), and the TrainCKDis project, funded by the European Union’s Horizon 2020 research and innovation programme under Marie Skłodowska-Curie grant agreement 860977 (to O. Devuyst) and by an Medical Research Council University Unit Programme grant MC_UU_00007/10 (QTL in Health and Disease) (to C. Hayward). CARTaGENE is supported by the Kidney Foundation of Canada and the Fonds de la Recherche du Québec-Santé. CoLaus received financial contributions from GlaxoSmithKline, the Faculty of Biology and Medicine of Lausanne, and the Swiss National Science Foundation (33CSCO-122661, 3200BO-111361/2, 3100AO-116323/1, and 310000-112552). The CROATIA_Vis, CROATIA_Korcula, and CROATIA_Split studies were funded by European Commission Framework 6 project EUROSPAN (contract LSHG-CT-2006-018947) and Republic of Croatia Ministry of Science, Education and Sports research grants (108-1080315-0302). The FHS is supported by the National Heart, Lung, and Blood Institute (FHS contract N01-HC-25195). The Genetic and Phenotypic Determinants of Blood Pressure and other Cardiovascular Risk Factors study was supported by the Liechtenstein Government, the Swiss Heart Foundation, the Swiss Society of Hypertension, the University Hospital Basel, the Hanela Foundation, the Mach-Gaensslen Foundation, Schiller AG, and Novartis. The GCKD study was supported by the German Ministry of Education and Research (Bundesministerium für Bildung und Forschung, grants FKZ 01ER 0804, 01ER 0818, 01ER 0819, 01ER 0820 and 01ER 0821) and the KfH Foundation for Preventive Medicine (Kuratorium für Heimdialyse und Nierentransplantation e.V.–Stiftung Präventivmedizin) and corporate sponsors (www.gckd.org). The work of A. Kottgen and M. Wuttke is funded by the Deutsche Forschungsgemeinschaft (German Research Foundation) Project-ID 431984000– SFB 1453. GS:SFHS received core support from the Chief Scientist Office of the Scottish Government Health Directorates (CZD/16/6) and the Scottish Funding Council (HR03006). INGI-Carlantino and INGI-Val Borbera were supported by Italian Ministry of Health grants RC 35/17 and D70-RESRICGIROTTO. The LBC1936 is supported by Age UK (Disconnected Mind project) and the Medical Research Council (MR/M01311/1, MR/K026992/1). The Viking Health Study– Shetland was supported by Medical Research Council University Unit Programme (MC_UU_00007/10, QTL in Health and Disease).
Supplementary Material
Acknowledgments
We are grateful for the willingness of the patients to participate in the study and CRediT Taxonomy. O. Devuyst and C. Hayward conceptualized the study; S. Aeschbacher, P. Awadalla, C. Black, S. Bergmann, M. Bochud, V. Bruat, A. Campbell, H. Campbell, M. Cocca, M. Concas, D. Conen, T. Corre, I. Deary, O. Devuyst, K.-U. Eckardt, G. Girotto, S. Harris, C. Hayward, J. Huffman, C. Joseph, I. Kolcic, A. Köttgen, F. Madore, M. Mariniello, J. Marten, M. Olden, E. Olinger, O. Polasek, D. Porteous, A. Robino, F. Schaefer, G. Schiano, S. Thèriault, S. Troyanov, S. Ulivi, J. Wilson, E. Wühl, M. Wuttke, an A. Yoshifuji were responsible for the data curation; S. Aeschbacher, P. Awadalla, C. Black, S. Bergmann, M. Bochud, V. Bruat, M. Cocca, M. Concas, D. Conen, T. Corre, I. Deary, O. Devuyst, K.-U. Eckardt, G. Girotto, S. Harris, C. Hayward, J. Huffman, C. Joseph, A. Köttgen, F. Madore, M. Mariniello, J. Marten, M. Olden, E. Olinger, A. Richmond, A. Robino, C. Sala, G. Schiano, S. Thèriault, S. Troyanov, S. Ulivi, J. Wilson, E. Wühl, M. Wuttke, and A. Yoshifuji were responsible for the formal analysis; H. Campbell, O. Devuyst, C. Hayward, and S. Troyanov were responsible for the funding acquisition; Y. Cheng, O. Devuyst, C. Hayward, C. Joseph, J. Lake, M. Mariniello, G. Schiano, S. Troyanov, and A. Yoshifuji were responsible for the investigation; T. Corre, O. Devuyst, C. Hayward, C. Joseph, J. Lake, M. Mariniello, J. Marten, M. Olden, G. Schiano, and A. Yoshifuji were responsible for the methodology; A. Campbell, H. Campbell, O. Devuyst, S. Harris, C. Hayward, and D. Porteous were responsible for the project administration; S. Aeschbacher, P. Awadalla, C. Black, S. Bergmann, M. Bochud, V. Bruat, A. Campbell, H. Campbell, Y. Cheng, M. Cocca, M. Concas, D. Conen, T. Corre, I. Deary, O. Devuyst, K.-U. Eckardt, G. Girotto, S. Harris, C. Hayward, J. Huffman, A. Köttgen, F. Madore, M. Olden, E. Olinger, O. Polasek, D. Porteous, A. Robino, C. Sala, F. Schaefer, S. Thèriault, S. Troyanov, S. Ulivi, J. Wilson, E. Wühl, and M. Wuttke were responsible for the resources; O. Devuyst provided supervision; O. Devuyst, C. Hayward, S. Troyanov, and A. Yoshifuji were responsible for the validation; O. Devuyst, C. Hayward, C. Joseph, J. Lake, M. Mariniello, and G. Schiano were responsible for the visualization; O. Devuyst, C. Hayward, C. Joseph, and A. Yoshifuji wrote the original draft; S. Aeschbacher, P. Awadalla, C. Black, S. Bergmann, M. Bochud, V. Bruat, A. Campbell, H. Campbell, Y. Cheng, M. Cocca, M. Concas, D. Conen, T. Corre, I. Deary, O. Devuyst, K.-U. Eckardt, G. Girotto, I. Kolcic, A. Köttgen, A. Robino, S. Harris, C. Hayward, J. Huffman, C. Joseph, J. Lake, F. Madore, M. Mariniello, J. Marten, M. Olden, E. Olinger, O. Polasek, D. Porteous, A. Richmond, C. Sala, F. Schaefer, G. Schiano, S. Thèriault, S. Troyanov, S. Ulivi, J. Wilson, E. Wühl, M. Wuttke, and A. Yoshifuji reviewed and edited the manuscript. We deeply acknowledge Professor John Starr (1960–2018), founding Director of the Alzheimer Scotland Dementia Research Centre, University of Edinburgh, Edinburgh, United Kingdom, who contributed in the early phase of this study. The help of Larissa Govers and Huguette Debaix (UZH, Zurich) is deeply appreciated. We thank Aleksander Edelman (Institut Necker Enfants Malades, Paris, France) for fruitful discussions on keratins. CARTaGENE: We thank the dedicated team at CARTaGENE for their diligent help. CoLaus: The computations for CoLaus imputation were performed in part at the Vital-IT center for high-performance computing of the Swiss Institute of Bioinformatics. M. Bochud is supported by the Swiss School of Public Health Plus. CROATIA-Korcula, CROATIA-Split, CROATIA-Vis: We would like to acknowledge the staff of several institutions in Croatia that supported the field work, including but not limited, to the University of Split and Zagreb Medical Schools, Institute for Anthropological Research in Zagreb, and the Croatian Institute for Public Health. We also thank all of the participants from the islands of Vis and Korcula and the city of Split. FHS: This research was conducted in part using data and resources from the FHS of the National Institutes of Health National Heart Lung and Blood Institute (NHLBI) and Boston University School of Medicine. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. GCKD: Genotyping was supported by Bayer Pharma AG. The GCKD study was/is funded by grants from the Federal Ministry of Education and Research (BMBF, grant number 01ER0804) and the KfH Foundation for Preventive Medicine. We are grateful for the willingness of the patients to participate in the GCKD study. The enormous effort of the study personnel of the various regional centers is highly appreciated. We thank the many nephrologists who provide routine care for the patients and collaborate with the GCKD study. GS:SFHS: We are grateful to all of the families who took part, the general practitioners and the Scottish School of Primary Care for their help in recruiting them, and the whole Generation Scotland team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, healthcare assistants, and nurses. GS:SFHS is supported by the Wellcome Trust (216767/Z/19/Z). Genotyping of the GS:SFHS samples was carried out by the Genetics Core Laboratory at the Edinburgh Clinical Research Facility, University of Edinburgh, Scotland and was funded by the Medical Research Council UK and the Wellcome Trust (Wellcome Trust Strategic Award “STratifying Resilience and Depression Longitudinally” Reference 104036/Z/14/Z). INGI-Carlantino and INGI-Val Borbera: We would like to thank the people of Carlantino and of the Val Borbera Valley for the everlasting support. LBC: The authors thank all LBC study participants and research team members who have contributed, and continue to contribute, to ongoing LBC studies. Genotyping was funded by the Biotechnology and Biological Sciences Research Council (BB/F019394/1). VIKING: We would like to acknowledge the invaluable contributions of the research nurses in Shetland, the administrative team in Edinburgh and the people of Shetland. We thank the UK Biobank Resource, approved under application 19655. The authors would like to thank the Rivas lab for making the Global Biobank Engine resource available. DNA extractions and genotyping were performed at the Edinburgh Clinical Research Facility, University of Edinburgh. The GTEx Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by National Cancer Institute, National Human Genome Research Institute, National Heart, Lung, and Blood Institute, National Institute on Drug Abuse, National Institute of Mental Health, and National Institute of Neurological Disorders and Stroke. The data used for the analyses described in this manuscript were obtained from the GTEx Portal (V8) on December 14, 2020.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
See related editorial, “Urine Uromodulin and Genetics of its Variation,” on pages 461–462.
Data Sharing Statement
Information on the datasets and summary statistics are available in the Edinburgh Datashare repository, under the link https://doi.org/10.7488/ds/3012 created on April 7, 2021 with additional material under the link https://doi.org/10.7488/ds/3262 created on 16th December 2021. For the purpose of open access, the author has applied a Creative Commons Attribution (CC BY) licence to any Author Accepted Manuscript version arising from this submission.
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2021040491/-/DCSupplemental.
Supplemental Table 1. Study sample characteristics for UMOD measurement.
Supplemental Table 2. Genotyping and imputation platforms.
Supplemental Table 3. List of candidate genes associated with rare Mendelian disorders affecting the TAL.
Supplemental Table 4. List of genes associated with diseases in specific kidney segments and candidate genes previously published by Olden et al. (2014).
Supplemental Table 5. Primers for quantitative RT-PCR analyses.
Supplemental Table 6. Heritability estimates for family-based cohorts.
Supplemental Table 7. Most significant SNP from each locus with P value <1E–05 from urinary UMOD (uUMOD) meta-analysis.
Supplemental Table 8. List of KRT40 variants, in high LD with rs8067385, and with P value <1E–05 in association with uUMOD.
Supplemental Table 9. Association of rs9672398 (WDR72 locus) with urinary UMOD levels indexed to creatinine.
Supplemental Table 10. Association of rs13335818 (UMOD-PDILT locus) with urinary UMOD levels indexed to creatinine.
Supplemental Table 11. Most significant SNP from each locus with P value <1E–05 from urinary UMOD indexed to creatinine (uUCR) meta-analysis.
Supplemental Table 12. Encoded protein, expression, and disease association for candidate genes (P value <1E-05) at urinary UMOD (uUMOD)-associated and indexed urinary UMOD (uUCR)-associated variants.
Supplemental Table 13. Effect size and P values of rs8067385 from meta-analysis of uUMOD concentration and uUMOD_eGFR using seven cohorts.
Supplemental Table 14. Effect size and P values of rs9672398 from meta-analysis of uUMOD concentration and uUMOD_eGFR using seven cohorts.
Supplemental Table 15. VEGAS2 results for uUMOD and uUCR meta-analysis.
Supplemental Table 16. Candidate gene analysis for uUMOD and uUCR levels.
Supplemental Table 17. Look-up analyses of genes associated with specific segments of the kidney and candidate genes previously published by Olden et al. (2014).
Supplemental Table 18. Effect sizes of the most significant SNP in UMOD and PDILT in association with uUMOD and uUCR.
Supplemental Table 19. Effect sizes of the most significant SNP in UMOD, KRT40 and WDR72 in association with uUMOD and uUCR in 12 cohorts (FHS excluded).
Supplemental Appendix 1. Summary characteristics of the study cohorts.
Supplemental Figure 1. Genetic loci associated with uUMOD and uUCR.
Supplemental Figure 2. Effect size of rs12934455 and regional association plot of UMOD-PDILT locus from raw UMOD levels.
Supplemental Figure 3. Effect size of rs9672398 and regional association plot of WDR72 locus for uUCR meta-analysis.
Supplemental Figure 4. Effect size of rs13335818 and regional association plot of UMOD-PDILT locus from uUCR meta-analysis.
Supplemental Figure 5. Manhattan plot of meta-GWAS of uUMOD and uUCR using sample size and P values for analysis of the 13 study cohorts.
Supplemental Figure 6. Forest plot showing effect sizes of rs8067385 (KRT40 locus) on uUMOD and uUCR meta-analysis in the 13 cohorts.
Supplemental Figure 7. Effect of UMOD genotype on urinary UMOD (uUMOD and uUCR) levels.
Supplemental Figure 8. Manhattan plot showing GWAS results in association with uUMOD and uUMOD conditioned for rs12934455 or for rs11864909 using GS:SFHS.
Supplemental Figure 9. Candidate genes influencing the urinary excretion of UMOD.
Supplemental Figure 10. In situ hybridization for Umod, Krt40 and Krt39 on mouse kidney.
Supplemental Figure 11. Immunofluorescence staining for AQP2 and KRT40 or WDR72 on mouse kidney.
Supplemental Figure 12. eQTL data for the KRT40 variant rs8067385.
Supplemental Figure 13. UMOD (Z-stack) and ROMK distribution in mTAL cells following KRT40 knockdown.
References
- 1.Devuyst O, Olinger E, Rampoldi L: Uromodulin: From physiology to rare and complex kidney disorders. Nat Rev Nephrol 13: 525–544, 2017 [DOI] [PubMed] [Google Scholar]
- 2.Brunati M, Perucca S, Han L, Cattaneo A, Consolato F, Andolfo A, et al. : The serine protease hepsin mediates urinary secretion and polymerisation of Zona Pellucida domain protein uromodulin. eLife 4: e08887, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiss GL, Stanisich JJ, Sauer MM, Lin CW, Eras J, Zyla DS, et al. : Architecture and function of human uromodulin filaments in urinary tract infections. Science 369: 1005–1010, 2020 [DOI] [PubMed] [Google Scholar]
- 4.Stanisich JJ, Zyla DS, Afanasyev P, Xu J, Kipp A, Olinger E, et al. : The cryo-EM structure of the human uromodulin filament core reveals a unique assembly mechanism. eLife 9: e60265, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mutig K, Kahl T, Saritas T, Godes M, Persson P, Bates J, et al. : Activation of the bumetanide-sensitive Na+,K+,2Cl- cotransporter (NKCC2) is facilitated by Tamm-Horsfall protein in a chloride-sensitive manner. J Biol Chem 286: 30200–30210, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tokonami N, Takata T, Beyeler J, Ehrbar I, Yoshifuji A, Christensen EI, et al. : Uromodulin is expressed in the distal convoluted tubule, where it is critical for regulation of the sodium chloride cotransporter NCC. Kidney Int 94: 701–715, 2018 [DOI] [PubMed] [Google Scholar]
- 7.Trudu M, Janas S, Lanzani C, Debaix H, Schaeffer C, Ikehata M, et al. ; SKIPOGH team : Common noncoding UMOD gene variants induce salt-sensitive hypertension and kidney damage by increasing uromodulin expression. Nat Med 19: 1655–1660, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graham LA, Padmanabhan S, Fraser NJ, Kumar S, Bates JM, Raffi HS, et al. : Validation of uromodulin as a candidate gene for human essential hypertension. Hypertension 63: 551–558, 2014 [DOI] [PubMed] [Google Scholar]
- 9.Devuyst O, Olinger E, Weber S, Eckardt KU, Kmoch S, Rampoldi L, et al. : Autosomal dominant tubulointerstitial kidney disease. Nat Rev Dis Primers 5: 60, 2019 [DOI] [PubMed] [Google Scholar]
- 10.Dahan K, Devuyst O, Smaers M, Vertommen D, Loute G, Poux JM, et al. : A cluster of mutations in the UMOD gene causes familial juvenile hyperuricemic nephropathy with abnormal expression of uromodulin. J Am Soc Nephrol 14: 2883–2893, 2003 [DOI] [PubMed] [Google Scholar]
- 11.Bernascone I, Janas S, Ikehata M, Trudu M, Corbelli A, Schaeffer C, et al. : A transgenic mouse model for uromodulin-associated kidney diseases shows specific tubulo-interstitial damage, urinary concentrating defect and renal failure. Hum Mol Genet 19: 2998–3010, 2010 [DOI] [PubMed] [Google Scholar]
- 12.Piret SE, Olinger E, Reed AAC, Nesbit MA, Hough TA, Bentley L, et al. : A mouse model for inherited renal fibrosis associated with endoplasmic reticulum stress. Dis Model Mech 10: 773–786, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Köttgen A, Glazer NL, Dehghan A, Hwang SJ, Katz R, Li M, et al. : Multiple loci associated with indices of renal function and chronic kidney disease. Nat Genet 41: 712–717, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wuttke M, Li Y, Li M, Sieber KB, Feitosa MF, Gorski M, et al. ; Lifelines Cohort Study; V. A. Million Veteran Program : A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat Genet 51: 957–972, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Köttgen A, Hwang SJ, Larson MG, Van Eyk JE, Fu Q, Benjamin EJ, et al. : Uromodulin levels associate with a common UMOD variant and risk for incident CKD. J Am Soc Nephrol 21: 337–344, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pruijm M, Ponte B, Ackermann D, Paccaud F, Guessous I, Ehret G, et al. : Associations of urinary uromodulin with clinical characteristics and markers of tubular function in the general population. Clin J Am Soc Nephrol 11: 70–80, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schiano G, Glaudemans B, Olinger E, Goelz N, Müller M, Loffing-Cueni D, et al. : The urinary excretion of uromodulin is regulated by the potassium channel ROMK. Sci Rep 9: 19517, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tokonami N, Olinger E, Debaix H, Houillier P, Devuyst O: The excretion of uromodulin is modulated by the calcium-sensing receptor. Kidney Int 94: 882–886, 2018 [DOI] [PubMed] [Google Scholar]
- 19.Olden M, Corre T, Hayward C, Toniolo D, Ulivi S, Gasparini P, et al. : Common variants in UMOD associate with urinary uromodulin levels: A meta-analysis. J Am Soc Nephrol 25: 1869–1882, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.1000 Genomes Project Consortium , Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, et al. : A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, et al. ; Haplotype Reference Consortium : A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 48: 1279–1283, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM: GenABEL: An R library for genome-wide association analysis. Bioinformatics 23: 1294–1296, 2007 [DOI] [PubMed] [Google Scholar]
- 23.Haller T, Kals M, Esko T, Mägi R, Fischer K: RegScan: A GWAS tool for quick estimation of allele effects on continuous traits and their combinations. Brief Bioinform 16: 39–44, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Youhanna S, Weber J, Beaujean V, Glaudemans B, Sobek J, Devuyst O: Determination of uromodulin in human urine: Influence of storage and processing. Nephrol Dial Transplant 29: 136–145, 2014 [DOI] [PubMed] [Google Scholar]
- 25.Winkler TW, Day FR, Croteau-Chonka DC, Wood AR, Locke AE, Mägi R, et al. ; Genetic Investigation of Anthropometric Traits (GIANT) Consortium : Quality control and conduct of genome-wide association meta-analyses. Nat Protoc 9: 1192–1212, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willer CJ, Li Y, Abecasis GR: METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26: 2190–2191, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ward LD, Kellis M: HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 40: D930–D934, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arnold M, Raffler J, Pfeufer A, Suhre K, Kastenmüller G: SNiPA: An interactive, genetic variant-centered annotation browser. Bioinformatics 31: 1334–1336, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. : The human genome browser at UCSC. Genome Res 12: 996–1006, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. : LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics 26: 2336–2337, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carithers LJ, Ardlie K, Barcus M, Branton PA, Britton A, Buia SA, et al. ; GTEx Consortium : A novel approach to high-quality postmortem tissue procurement: The GTEx Project. Biopreserv Biobank 13: 311–319, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mishra A, Macgregor S: VEGAS2: Software for more flexible gene-based testing. Twin Res Hum Genet 18: 86–91, 2015 [DOI] [PubMed] [Google Scholar]
- 33.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. : UK biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 12: e1001779, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Global Biobank Engine : Stanford, CA. Available at: https://biobankengine.stanford.edu/. Accessed March 20th 2021.
- 35.McInnes G, Tanigawa Y, DeBoever C, Lavertu A, Olivieri JE, Aguirre M, et al. : Global Biobank Engine: Enabling genotype-phenotype browsing for biobank summary statistics. Bioinformatics 35: 2495–2497, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J, Ferreira T, Morris AP, Medland SE, Madden PA, Heath AC, et al. ; Genetic Investigation of ANthropometric Traits (GIANT) Consortium; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium : Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet 44: 369–375, S1–S3, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Wijst J, Belge H, Bindels RJM, Devuyst O: Learning physiology from inherited kidney disorders. Physiol Rev 99: 1575–1653, 2019 [DOI] [PubMed] [Google Scholar]
- 38.Glaudemans B, Terryn S, Gölz N, Brunati M, Cattaneo A, Bachi A, et al. : A primary culture system of mouse thick ascending limb cells with preserved function and uromodulin processing. Pflugers Arch 466: 343–356, 2014 [DOI] [PubMed] [Google Scholar]
- 39.Festa BP, Chen Z, Berquez M, Debaix H, Tokonami N, Prange JA, et al. : Impaired autophagy bridges lysosomal storage disease and epithelial dysfunction in the kidney. Nat Commun 9: 161, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Devuyst O, Pattaro C: The UMOD locus: Insights into the pathogenesis and prognosis of kidney disease. J Am Soc Nephrol 29: 713–726, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stanzick KJ, Li Y, Schlosser P, Gorski M, Wuttke M, Thomas LF, et al. ; VA Million Veteran Program : Discovery and prioritization of variants and genes for kidney function in >1.2 million individuals. Nat Commun 12: 4350, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benonisdottir S, Kristjansson RP, Oddsson A, Steinthorsdottir V, Mikaelsdottir E, Kehr B, et al. : Sequence variants associating with urinary biomarkers. Hum Mol Genet 28: 1199–1211, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Howles SA, Wiberg A, Goldsworthy M, Bayliss AL, Gluck AK, Ng M, et al. : Genetic variants of calcium and vitamin D metabolism in kidney stone disease. Nat Commun 10: 5175, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ponte B, Sadler MC, Olinger E, Vollenweider P, Bochud M, Padmanabhan S, et al. : Mendelian randomization to assess causality between uromodulin, blood pressure and chronic kidney disease. Kidney Int 100: 1282–1291, 2021 [DOI] [PubMed] [Google Scholar]
- 45.Jacob JT, Coulombe PA, Kwan R, Omary MB: Types I and II keratin intermediate filaments. Cold Spring Harb Perspect Biol 10: a018275, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davezac N, Tondelier D, Lipecka J, Fanen P, Demaugre F, Debski J, et al. : Global proteomic approach unmasks involvement of keratins 8 and 18 in the delivery of cystic fibrosis transmembrane conductance regulator (CFTR)/deltaF508-CFTR to the plasma membrane. Proteomics 4: 3833–3844, 2004 [DOI] [PubMed] [Google Scholar]
- 47.Toivola DM, Boor P, Alam C, Strnad P: Keratins in health and disease. Curr Opin Cell Biol 32: 73–81, 2015 [DOI] [PubMed] [Google Scholar]
- 48.Djudjaj S, Papasotiriou M, Bülow RD, Wagnerova A, Lindenmeyer MT, Cohen CD, et al. : Keratins are novel markers of renal epithelial cell injury. Kidney Int 89: 792–808, 2016 [DOI] [PubMed] [Google Scholar]
- 49.Katsura KA, Horst JA, Chandra D, Le TQ, Nakano Y, Zhang Y, et al. : WDR72 models of structure and function: A stage-specific regulator of enamel mineralization. Matrix Biol 38: 48–58, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.El-Sayed W, Parry DA, Shore RC, Ahmed M, Jafri H, Rashid Y, et al. : Mutations in the beta propeller WDR72 cause autosomal-recessive hypomaturation amelogenesis imperfecta. Am J Hum Genet 85: 699–705, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rungroj N, Nettuwakul C, Sawasdee N, Sangnual S, Deejai N, Misgar RA, et al. : Distal renal tubular acidosis caused by tryptophan-aspartate repeat domain 72 (WDR72) mutations. Clin Genet 94: 409–418, 2018 [DOI] [PubMed] [Google Scholar]
- 52.Khandelwal P, Mahesh V, Mathur VP, Raut S, Geetha TS, Nair S, et al. : Phenotypic variability in distal acidification defects associated with WDR72 mutations. Pediatr Nephrol 36: 881–887, 2021 [DOI] [PubMed] [Google Scholar]
- 53.Köttgen A, Pattaro C, Böger CA, Fuchsberger C, Olden M, Glazer NL, et al. : New loci associated with kidney function and chronic kidney disease. Nat Genet 42: 376–384, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.