Abstract
Recent studies have sparked debate over whether catalytical reactions enhance the diffusion coefficients D of enzymes. Through high statistics of the transient (600 µs) displacements of unhindered single molecules freely diffusing in common buffers, we here quantify D for four enzymes under catalytic turnovers. We thus formulate how ~±1% precisions may be achieved for D, and show no changes in diffusivity for catalase, urease, aldolase, and alkaline phosphatase under the application of wide concentration ranges of substrates. Our single-molecule approach thus overcomes potential limitations and artifacts underscored by recent studies to show no enhanced diffusion in enzymatic reactions.
Graphical Abstract
INTRODUCTION
The recent reporting of accelerated diffusion of enzymes in catalytic reactions has generated keen research interest and debate.1–8 Measurements based on fluorescence correlation spectroscopy (FCS) have documented substantial (>~30%) increases in the diffusion coefficient D for multiple enzymes under catalytic turnovers.1,2,9–16 Several recent studies, however, emphasize FCS artifacts3,17,18 and report contradicting results. Dynamic light scattering (DLS) experiments, with low sensitivity for the very small sizes of proteins, also yield inconsistent results either supporting14,16 or refuting19 substrate-enhanced diffusion. Likewise, different nuclear magnetic resonance (NMR) experiments have separately reported the presence or absence of reaction-enhanced D for enzymes and small molecules,20–23 with unsettled debate on data interpretation.8,24,25
Single-molecule experiments26–28 provide potential means to achieve precise D measurements by probing one molecule at a time. However, with expected D~30-50 µm2/s,6,29 the fast motions of typical enzymes make it difficult to detect freely diffusing single molecules. Using reducing-oxidizing chemicals that suppress fluorophore blinking, a recent study achieved long-time anti-Brownian electrokinetic (ABEL) single-molecule trapping of dye-labeled alkaline phosphatase (ALP) to show no change in D with the addition of its substrate.18 Meanwhile, single-molecule tracking has been reported for dye-labeled urease and ALP that were substantially slowed down by using viscous reagents as methylcellulose30,31 and glycerol18,31 or by tethering to supported lipid bilayers,31 which yielded contrasting results of no change versus up to 3-fold increases in D upon substrate addition. Thus, it remains difficult to quantify D through single-molecule measurements for molecules freely diffusing in regular buffers, whereas the addition of blinking-suppressing chemicals and viscous reagents complicates data interpretation.
We recently developed single-molecule displacement/diffusivity mapping (SMdM),32,33 a single-molecule imaging-based super-resolution method34–36 capable of determining molecular diffusivity. In SMdM, stroboscopic excitation pulses of τ <1 ms duration reduce motion-blur to capture snapshots of fast-moving molecules in the wide field. The excitation pulses are applied as pairs across tandem camera frames with a fixed Δt <~1 ms interval (Figure 1a), so that images captured in the two frames (Figure 1b) can be compared to determine the transient displacements d of the observed molecules within the Δt time window. Repeating this tandem excitation scheme then enables the accumulation of d over time for D-value determination.
Figure 1.
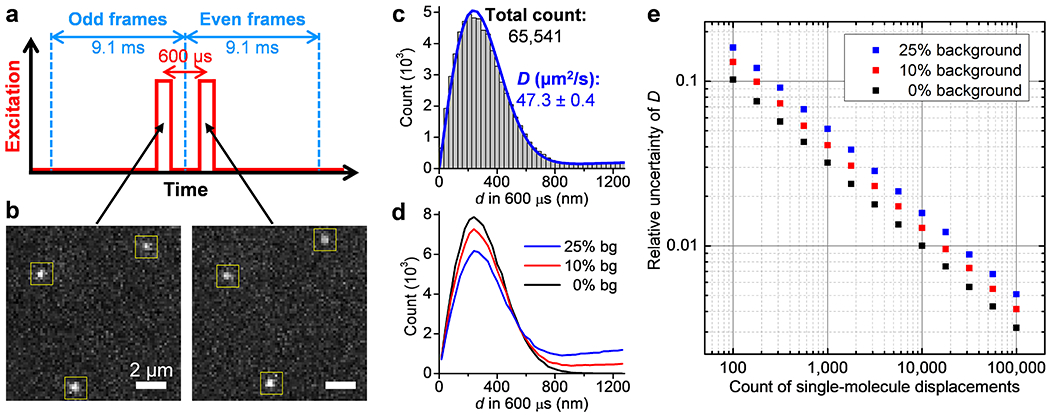
High-throughput statistics of unhindered single-molecule displacements. (a) Schematics: tandem excitation pulses of 300 µs duration are applied at Δt = 600 µs center-to-center separations across paired frames of the recording EM-CCD camera. (b) Example single-molecule images captured in the paired frames, for Cy3B-labeled catalase freely diffusing in the PBS buffer, which enabled the determination of the transient displacements d of the 3 molecules marked by the yellow boxes in the Δt = 600 µs time window. (c) Histogram: distribution of the 65,541 transient single-molecule displacements collected in 9 min by repeating the tandem excitation scheme 30,000 times. Blue line: MLE yielding a diffusion coefficient D of 47.3±0.4 µm2/s (95% confidence interval). (d) Example distributions of 100,000 simulated displacements, of which 0%, 10%, and 25% are backgrounds from molecules randomly entered the vicinity while the rest are 600-µs displacements with D = 50 µm2/s. (e) Relative uncertainty of D (σD/D) from MLE results of the simulated data at different levels of backgrounds, as a function of the counts of displacements. The standard deviations σD were calculated from the differences between 5,000 rounds of simulations under each condition.
Whereas we previously focused on the high spatial resolution of SMdM to study intracellular diffusion heterogeneities,32,33 here we harness its capability for high single-molecule statistics to enable the precise quantification of D for enzymes freely diffusing in regular buffers. We thus formulate how ~±1% precisions may be achieved for D, and then utilize this capability to show no changes in diffusivity for four enzymes under wide concentration ranges of substrates.
RESULTS AND DISCUSSION
To optimize SMdM for the ~60% higher D versus the ~20-30 µm2/s values in our previous work on intracellular diffusion,32 we accordingly reduced the excitation pulse duration τ and center-to-center separation Δt to 300 and 600 µs, respectively (Figure 1a and Figure S1). The use of Cy3B,37 a bright synthetic dye comparable in size to those used in previous enzyme-diffusion studies, to label the enzymes enabled single-molecule detection with good signals (Figure 1b). By running the recording EM-CCD camera at 110 frames per second, ~104 pairs of tandem frames were executed in minutes, from which >5×104 single-molecule displacements were typically accumulated (Figure 1c). Maximum likelihood estimation (MLE) with a predefined model based on normal diffusion plus background32 (Methods) yielded good fits, from which D values were obtained with their 95% confidence intervals, e.g., D = 47.3±0.4 µm2/s for Cy3B-labeled catalase in phosphate-buffered saline (PBS) (Figure 1c). We also analyzed the single-molecule displacements in two dimensions,33 and found the diffusion isotropic, with identical D values calculated along different directions (Figure S2). As we varied Δt between 400 µs and 1 ms, we further observed a linear dependence for the MLE-extracted mean squared displacements (Figure S2). Thus, SMdM characterized normal (Gaussian) diffusion in our experiments.
To define how the achievable precision in D depends on the count of single-molecule displacements N, we simulated 600-µs displacements d for molecules diffusing at D = 50 µm2/s. Different levels of backgrounds were further included, i.e., assuming 0%, 10%, or 25% of the total counts were due to extraneous molecules that randomly entered the vicinity during Δt (Figure 1d). MLE of the simulated data showed that with zero background, the relative uncertainty of D (σD/D) was ~1/√N, so that 10% and 1% uncertainties were achieved with 100 and 10,000 counts of d, respectively (Figure 1e). This result is expected: each displacement provides an independent reporting of diffusivity, so the signal-to-noise ratio improves as ~√N. At background levels of 10% and 25%, 1% uncertainty in D was achieved with ~20,000 and ~30,000 counts of d, respectively. As our experimental data contained <10% background (compare Figure 1c and Figure 1d), ~0.5% relative uncertainty (hence an ~±1% bracket for the 95% confidence interval) was thus obtained (Figure 1e) with the typical >50,000 counts of d in our experiments, in agreement with our above MLE results on the experimental data (Figure 1c). We further compared experimental results from different days, and observed D values in the range of 46.3–48.1 µm2/s for Cy3B-labeled catalase in PBS (Figure S2). The ~±2% variations between runs are larger than the above MLE-deduced ~±1% bracket from the single-molecule statistics alone, and may be attributed to uncontrolled experimental factors. In particular, an ~2% increase in D is expected per 1 K temperature increase, mainly attributable to changes in water viscosity.6
To verify whether the above high precisions would enable the experimental detection of small differences in D, we examined the diffusion of Cy3B-labeled catalase in PBS buffers containing varying amounts of glycerol. The addition of 5%, 10%, and 15% weight percentages of glycerol led to substantial shifts in the measured d distribution (Figure 2a), with MLE yielding ~10%, ~21%, and ~30% decreases in D, respectively (Figure 2b). Two batches of samples measured a month apart yielded consistent D values, as well as good matches to the trend predicted by the glycerol concentration-dependent viscosity38 (Figure 2b). Among these results, the 2.5% glycerol sample showed ~5% reduction in D and was well differentiated from the glycerol-free sample (Figure 2b). Whereas it is difficult to produce substantially higher D with the same enzyme, we employed SMdM to measure the diffusion coefficients of 13 different protein samples in a wide 14–545 kDa size range in PBS and Tris buffers (Figure 2c and Table S1). We thus obtained results in agreement with that is predicted by the Young-Carroad-Bell model,6,29 including for the fast-diffusing small proteins as RNase A and lysozyme at >100 µm2/s. Thus, SMdM is well poised for detecting D differences over a broad range.
Figure 2.
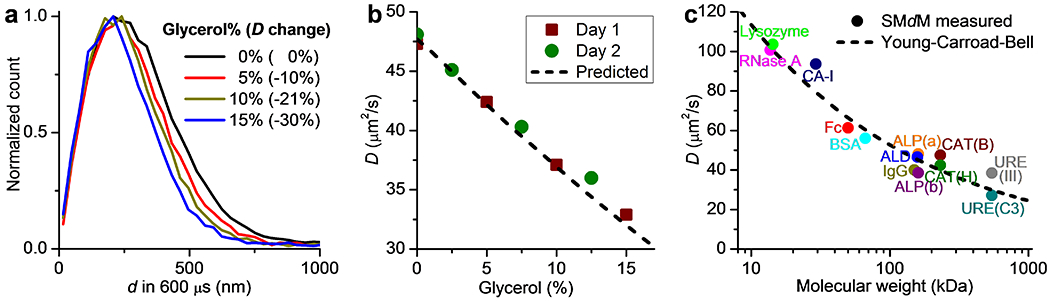
Precise determination of diffusivity via single-molecule statistics. (a) Normalized count distributions of the measured 600-µs single-molecule displacements for Cy3B-labeled catalase diffusing in PBS buffers containing 0%, 5%, 10%, and 15% glycerol. (b) The measured D as a function of the glycerol percentage, for two batches of samples measured a month apart (squares and circles; 95% confidence intervals are comparable to the symbol sizes), compared to that is predicted by scaling the 0% glycerol value with the known glycerol concentration-dependent viscosity (dash line). (c) SMdM-measured D values of 13 different protein samples of varying sizes in PBS and Tris buffers. Dash line: expected values at 20 °C according to the Young-Carroad-Bell model. See protein sample details in Table S1.
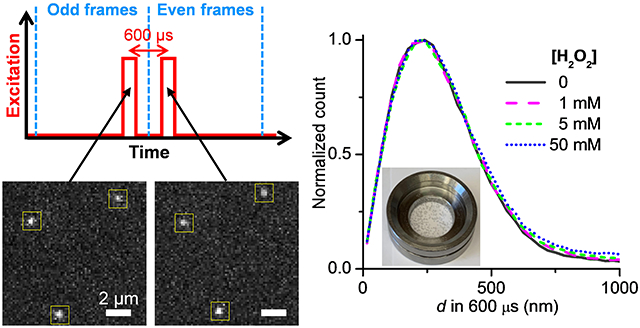
For catalytic reactions, we started with catalase in the presence of its substrate H2O2, for which system FCS has reported ~30-40% increase in D with ~25 mM H2O2.2,9,11 SMdM statistics showed that the addition of 1, 5, and 50 mM H2O2 did not noticeably shift the single-molecule displacement distribution (Figure 3a). The catalytic reaction generated oxygen bubbles, which were more notable for samples with higher H2O2 concentrations (Figure 3a inset). The bubbles scattered light and increased the fluorescence background to slightly reduce our capability to track molecules across the tandem frames, and so led to a moderately increased tail in the d distribution (Figure 3a). Nonetheless, MLE yielded good fits (Figure 3b), and the resultant D values showed minimal variations in the range of 46.5–48.0 µm2/s between the different substrate concentrations (Figure 3c). We also conducted SMdM experiments at a longer pulse separation of Δt = 1 ms, as well as at increased single-molecule densities for higher backgrounds in the d distribution, and similarly observed unvaried D independent of the presence of the H2O2 substrate (Figure S3). These results contrast with the ~30–40% substrate-induced D enhancements reported in previous FCS studies.2,9,11
Figure 3.
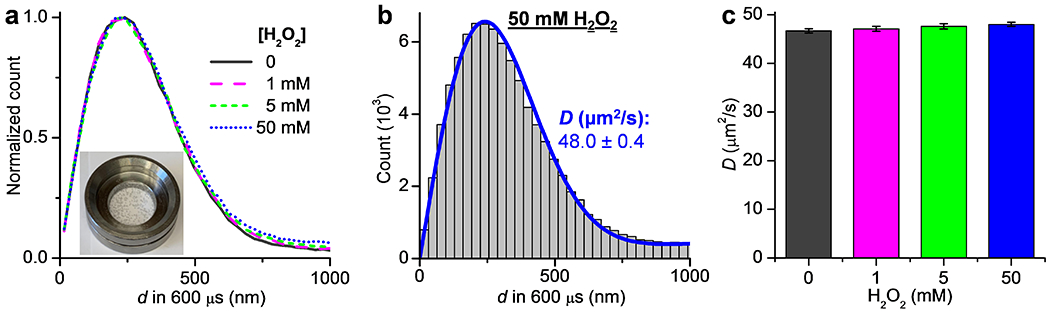
No substantial change in single-molecule displacements observed for catalase under turnover. (a) Normalized count distributions of the measured 600-µs single-molecule displacements for Cy3B-labeled catalase diffusing in PBS buffers with H2O2 added at 0, 1, 5, and 50 mM. Inset: photo of a sample with 50 mM H2O2, showing the generation of bubbles from the catalytic reaction. (b) Histogram: the count distribution with the 50 mM H2O2 solution. Blue line: MLE yielding D = 48.0±0.4 µm2/s (95% confidence interval). (c) MLE-determined D at the different substrate concentrations. Error bars: 95% confidence intervals of MLE.
We next examined urease, for which previous FCS experiments have reported up to ~40% D enhancement in the presence of 10-100 mM of its substrate urea.1,2,12,14–16 Moreover, ~3-fold substrate-induced D enhancement is recently reported for single urease molecules diffusing in the highly crowded or membrane-tethered systems at orders of magnitudes lower D values.30,31 SMdM showed virtually identical distributions for the 600-µs single-molecule displacements of urease in PBS with and without the addition of 0.1-100 mM of urea (Figure 4a). MLE hence yielded comparable D values of 26.4-27.3 µm2/s for the five conditions with no noticeable trend on the substrate concentration (Figure 4b).
Figure 4.
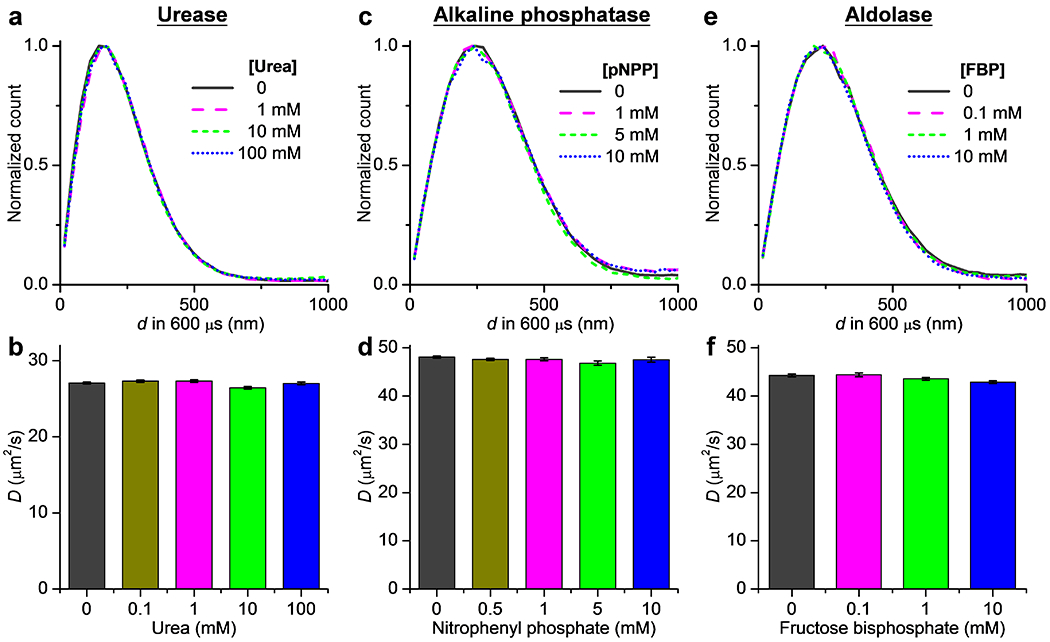
Results on urease, alkaline phosphatase, and aldolase also show no changes in single-molecule displacements under catalytic turnover. (a) Normalized distributions of the measured 600-µs single-molecule displacements for Cy3B-labeled urease in PBS, with and without the addition of urea at different concentrations. (b) MLE-determined D values at different urea concentrations. Error bars: 95% confidence intervals of MLE. (c,d) Similar to that in (a,b), but for Cy3B-labeled alkaline phosphatase in 0.1 M Tris-HCl (pH = 9.8) with the addition of 1 mM MgCl2 and 20 µM ZnCl2, with p-nitrophenyl phosphate (pNPP) as the substrate. (e,f) Similar to that in (a,b), but for Cy3B-labeled aldolase in 0.1 M HEPES (pH = 7.4), with fructose 1,6-bisphosphate (FBP) as the substrate.
For ALP, FCS has initially reported up to an 80% increase in D with the addition of 2.6 mM of its substrate p-nitrophenyl phosphate (pNPP).2 However, recent FCS experiments,3,16,18 as well as single-molecule trapping in blinking-suppressed buffers and tracking in glycerol-slowed solutions18 indicate no substrate-induced D enhancement. For ALP in its working buffer of 100 mM Tris-HCl (pH = 9.8) containing 1 mM MgCl2 and 20 µM ZnCl2, SMdM showed no substantial changes in single-molecule displacements with the addition of 0.5–10 mM pNPP (Figure 4c). Accordingly, the resultant D values of 46.8–48.1 µm2/s indicated no enhanced diffusion (Figure 4d).
We further examined aldolase, for which previous FCS experiments have either reported ~30% increase10 or no change14,16 of D in the presence of ~1 mM of its substrate, fructose 1,6-bisphosphate (FBP), whereas DLS14,19 and NMR21 experiments have indicated no D enhancement over broad substrate ranges. SMdM of aldolase in a HEPES buffer (pH = 7.4) showed no substantial changes in single-molecule displacements with and without the addition of 0.1-10 mM of FBP (Figure 4e), and hence no enhanced diffusion of D values in the range of 42.9-44.4 µm2/s (Figure 4f).
To verify that the enzymes were active throughout our experiments, we performed standard enzyme activity assays under identical conditions as in SMdM. We thus showed that the enzymes were active, comparable to that reported in previous studies (Figure S4). Using these results, we further calculated the expected substrate concentrations and reaction rates as a function of time (Figure S5). Changes in both parameters were small over 10 min. Indeed, as SMdM is based on single-molecule imaging, its ~200 pM working enzyme concentration is an order of magnitude lower than that is typically used in FCS,2,16 and so substrate consumption during measurement is less concerning. Following recent discussions that D may increase linearly with the free energy released in the enzyme-catalyzed reaction,16 we also calculated the expected Gibbs energy dissipation rates for the different systems (Figure S5). The resultant values for urease and aldolase were comparable to the study,16 which would predict >10% reaction-induced diffusion enhancement for the former. For catalase, the calculated Gibbs energy dissipation rates were one order of magnitude higher than the highest values discussed in the study.16 Our observation of no enhanced diffusion for both systems thus does not support the exergonic argument.
CONCLUSION
Together, with the high precision achieved through the high single-molecule statistics of SMdM, our results consistently showed unvarying D for four enzymes that have been reported as exhibiting enhanced diffusion under catalytic turnovers. These results, together with recent experiments that reexamine artifacts due to photophysical processes as fluorescence quenching and blinking,3,17,18 underscore the difficulties with quantifying D through FCS. Although blinking-suppressing buffers partly alleviate these issues while also enabling single-molecule trapping for D measurements,18 the addition of reducing and oxidizing chemicals complicates data interpretation. Meanwhile, the high D values of enzymes have largely limited single-molecule tracking to systems in which viscous reagents as methylcellulose30,31 and glycerol18,31 are added to slow down molecular motions.
In contrast, with SMdM, here we were able to work with the diffusion of enzymes in regular buffers without the addition of extraneous components that may potentially affect the system: As SMdM captured and accumulated the transient (600 µs) displacements of single molecules, it removed the needs to establish long-term fluorophore photostability or to impede diffusion to permit multi-frame tracking. Whereas in this study we have employed MLE to extract D values based on normal (Gaussian) diffusion, the highly invariant distributions of single-molecule displacements we observed under different substrate concentrations suggest no changes in diffusivity independent of the specific model. The sub-millisecond motion-detection time window of SMdM is comparable to that of FCS, which has been the primary method used in previous studies to advocate enhanced diffusion in the presence of substrates. For the four enzymes examined, this time window should accommodate multiple turnovers for catalase, urease, and ALP, but not aldolase (based on reaction rates in Figure S5). Our results thus support recent analysis that reaction-facilitated diffusion vanishes at the molecular scale,8 and call for the reexamination of factors that may have inadvertently affected previous measurements, including both photophysical processes3,17,18 and the dissociation of enzyme subunits.3,14 The application of the tools developed in this work to other systems in which precise D values are important, potentially including the even faster diffusion of smaller molecules, awaits future efforts.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge support by the National Science Foundation (CHE-1554717), the National Institute of General Medical Sciences of the National Institutes of Health (DP2GM132681), the Beckman Young Investigator Program, the Packard Fellowships for Science and Engineering, and the Heising-Simons Faculty Fellows award, to K.X. K.X. is a Chan Zuckerberg Biohub investigator.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Materials and methods; Table of SMdM-determined D values for different proteins; Timing waveforms used in this work; Additional characterizations of single-molecule displacements from SMdM; SMdM results at different pulse separations and single-molecule densities; Enzyme activity assays; Simulated substrate concentrations, reaction rates, and Gibbs energy dissipation rates in our SMdM experiments (PDF)
REFERENCES
- (1).Muddana HS; Sengupta S; Mallouk TE; Sen A; Butler PJ Substrate catalysis enhances single-enzyme diffusion. J. Am. Chem. Soc 2010, 132, 2110–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Riedel C; Gabizon R; Wilson CAM; Hamadani K; Tsekouras K; Marqusee S; Presse S; Bustamante C The heat released during catalytic turnover enhances the diffusion of an enzyme. Nature 2015, 517, 227–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Gunther JP; Borsch M; Fischer P Diffusion Measurements of Swimming Enzymes with Fluorescence Correlation Spectroscopy. Acc. Chem. Res 2018, 51, 1911–1920. [DOI] [PubMed] [Google Scholar]
- (4).Agudo-Canalejo J; Adeleke-Larodo T; Illien P; Golestanian R Enhanced Diffusion and Chemotaxis at the Nanoscale. Acc. Chem. Res 2018, 51, 2365–2372. [DOI] [PubMed] [Google Scholar]
- (5).Zhao X; Gentile K; Mohajerani F; Sen A Powering motion with enzymes. Acc. Chem. Res 2018, 51, 2373–2381. [DOI] [PubMed] [Google Scholar]
- (6).Zhang YF; Hess H Enhanced diffusion of catalytically active enzymes. ACS Cent. Sci 2019, 5, 939–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Feng M; Gilson MK Enhanced Diffusion and Chemotaxis of Enzymes. Annu. Rev. Biophys 2020, 49, 87–105. [DOI] [PubMed] [Google Scholar]
- (8).Zhang Y; Hess H Chemically-powered swimming and diffusion in the microscopic world. Nature Rev. Chem 2021, 5, 500–510. [DOI] [PubMed] [Google Scholar]
- (9).Sengupta S; Dey KK; Muddana HS; Tabouillot T; Ibele ME; Butler PJ; Sen A Enzyme Molecules as Nanomotors. J. Am. Chem. Soc 2013, 135, 1406–1414. [DOI] [PubMed] [Google Scholar]
- (10).Illien P; Zhao X; Dey KK; Butler PJ; Sen A; Golestanian R Exothermicity Is Not a Necessary Condition for Enhanced Diffusion of Enzymes. Nano Lett. 2017, 17, 4415–4420. [DOI] [PubMed] [Google Scholar]
- (11).Jiang L; Santiago I; Foord J Observation of nanoimpact events of catalase on diamond ultramicroelectrodes by direct electron transfer. Chem. Comm 2017, 53, 8332–8335. [DOI] [PubMed] [Google Scholar]
- (12).Jee AY; Dutta S; Cho YK; Tlusty T; Granick S Enzyme leaps fuel antichemotaxis. Proc. Natl. Acad. Sci. U. S. A 2018, 115, 14–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Jee AY; Cho YK; Granick S; Tlusty T Catalytic enzymes are active matter. Proc. Natl. Acad. Sci. U. S. A 2018, 115, E10812–E10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Jee AY; Chen K; Tlusty T; Zhao J; Granick S Enhanced diffusion and oligomeric enzyme dissociation. J. Am. Chem. Soc 2019, 141, 20062–20068. [DOI] [PubMed] [Google Scholar]
- (15).Ghosh S; Mohajerani F; Son S; Velegol D; Butler PJ; Sen A Motility of enzyme-powered vesicles. Nano Lett. 2019, 19, 6019–6026. [DOI] [PubMed] [Google Scholar]
- (16).Jee AY; Tlusty T; Granick S Master curve of boosted diffusion for 10 catalytic enzymes. Proc. Natl. Acad. Sci. U. S. A 2020, 117, 29435–29441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Kandula HN; Jee AY; Granick S Robustness of FCS (Fluorescence Correlation Spectroscopy) with Quenchers Present. J. Phys. Chem. A 2019, 123, 10184–10189. [DOI] [PubMed] [Google Scholar]
- (18).Chen Z; Shaw A; Wilson H; Woringer M; Darzacq X; Marqusee S; Wang Q; Bustamante C Single-molecule diffusometry reveals no catalysis-induced diffusion enhancement of alkaline phosphatase as proposed by FCS experiments. Proc. Natl. Acad. Sci. U. S. A 2020, 117, 21328–21335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Zhang YF; Armstrong MJ; Kazeruni NMB; Hess H Aldolase Does Not Show Enhanced Diffusion in Dynamic Light Scattering Experiments. Nano Lett. 2018, 18, 8025–8029. [DOI] [PubMed] [Google Scholar]
- (20).Pavlick RA; Dey KK; Sirjoosingh A; Benesi A; Sen A A catalytically driven organometallic molecular motor. Nanoscale 2013, 5, 1301–1304. [DOI] [PubMed] [Google Scholar]
- (21).Gunther JP; Majer G; Fischer P Absolute diffusion measurements of active enzyme solutions by NMR. J. Chem. Phys 2019, 150, 124201. [DOI] [PubMed] [Google Scholar]
- (22).MacDonald TSC; Price WS; Astumian RD; Beves JE Enhanced Diffusion of Molecular Catalysts is Due to Convection. Angew. Chem.-Int. Edit 2019, 58, 18864–18867. [DOI] [PubMed] [Google Scholar]
- (23).Wang H; Park M; Dong R; Kim J; Cho Y-K; Tlusty T; Granick S Boosted molecular mobility during common chemical reactions. Science 2020, 369, 537–541. [DOI] [PubMed] [Google Scholar]
- (24).Günther J-P; Fillbrook LL; MacDonald TSC; Majer G; Price WS; Fischer P; Beves JE Comment on “Boosted molecular mobility during common chemical reactions”. Science 2021, 371, eabe8322. [DOI] [PubMed] [Google Scholar]
- (25).Wang H; Park M; Dong R; Kim J; Cho Y-K; Tlusty T; Granick S Response to Comment on “Boosted molecular mobility during common chemical reactions”. Science 2021, 371, eabe8678. [DOI] [PubMed] [Google Scholar]
- (26).Ha T Single-molecule methods leap ahead. Nat. Methods 2014, 11, 1015–1018. [DOI] [PubMed] [Google Scholar]
- (27).Moerner WE; Shechtman Y; Wang Q Single-molecule spectroscopy and imaging over the decades. Faraday Discuss 2015, 184, 9–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Shen H; Tauzin LJ; Baiyasi R; Wang WX; Moringo N; Shuang B; Landes CF Single particle tracking: from theory to biophysical applications. Chem. Rev 2017, 117, 7331–7376. [DOI] [PubMed] [Google Scholar]
- (29).Young ME; Carroad PA; Bell RL Estimation of diffusion coefficients of proteins. Biotechnol. Bioeng 1980, 22, 947–955. [Google Scholar]
- (30).Xu MQ; Ross JL; Valdez L; Sen A Direct Single Molecule Imaging of Enhanced Enzyme Diffusion. Phys. Rev. Lett 2019, 123, 128101. [DOI] [PubMed] [Google Scholar]
- (31).Xu M; Rogers WB; Ahmed WW; Ross JL Comparison of different approaches to single-molecule imaging of enhanced enzyme diffusion. Dec 31, 2020. arXiv:2012.15424. (accessed 2022–02-25). [Google Scholar]
- (32).Xiang L; Chen K; Yan R; Li W; Xu K Single-molecule displacement mapping unveils nanoscale heterogeneities in intracellular diffusivity. Nat. Methods 2020, 17, 524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Yan R; Chen K; Xu K Probing nanoscale diffusional heterogeneities in cellular membranes through multidimensional single-molecule and super-resolution microscopy. J. Am. Chem. Soc 2020, 142, 18866–18873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Xu K; Shim S-H; Zhuang X Super-resolution imaging through stochastic switching and localization of single molecules: an overview. In Far-Field Optical Nanoscopy; Tinnefeld P, Eggeling C, Hell SW, Eds.; Springer: Berlin, 2015; pp 27–64. [Google Scholar]
- (35).Möckl L; Moerner WE Super-resolution microscopy with single molecules in biology and beyond-essentials, current trends, and future challenges. J. Am. Chem. Soc 2020, 142, 17828–17844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Xiang L; Chen K; Xu K Single molecules are your quanta: A bottom-up approach toward multidimensional super-resolution microscopy. ACS Nano 2021, 15, 12483–12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Cooper M; Ebner A; Briggs M; Burrows M; Gardner N; Richardson R; West R Cy3B™: Improving the Performance of Cyanine Dyes. J. Fluorescence 2004, 14, 145–150. [DOI] [PubMed] [Google Scholar]
- (38).Sheely ML Glycerol viscosity tables. Ind. Eng. Chem 1932, 24, 1060–1064. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


