Abstract
Hyperactive signal transducer and activator of transcription 3 (STAT3) signaling is frequently detected in human triple-negative breast cancer (TNBC) and gastric cancer, leading to uncontrolled tumor growth, resistance to chemotherapy, and poor prognosis. Thus, inhibition of STAT3 signaling is a promising therapeutic approach for both TNBC and gastric cancer, which have high incidences and mortality and limited effective therapeutic approaches. Here, we report a small molecule, WZ-2-033, capable of inhibiting STAT3 activation and dimerization and STAT3-related malignant transformation. We present in vitro evidence from surface plasmon resonance analysis that WZ-2-033 interacts with the STAT3 protein and from confocal imaging that WZ-2-033 disrupts HA-STAT3 and Flag-STAT3 dimerization in intact cells. WZ-2-033 suppresses STAT3-DNA-binding activity but has no effect on STAT5-DNA binding. WZ-2-033 inhibits the phosphorylation and nuclear accumulation of pY705-STAT3 and consequently suppresses STAT3-dependent transcriptional activity and the expression of STAT3 downstream genes. Moreover, WZ-2-033 significantly inhibited the proliferation, colony survival, migration, and invasion of TNBC cells and gastric cancer cells with aberrant STAT3 activation. Furthermore, administration of WZ-2-033 in vivo induced a significant antitumor response in mouse models of TNBC and gastric cancer that correlated with the inhibition of constitutively active STAT3 and the suppression of known STAT3 downstream genes. Thus, our study provides a novel STAT3 inhibitor with significant antitumor activity in human TNBC and gastric cancer harboring persistently active STAT3.
Keywords: STAT3 inhibitor, WZ-2-033, TNBC, gastric cancer
Introduction
Signal transducer and activator of transcription 3 (STAT3) is an important regulator of multiple cellular processes, including proliferation, survival, differentiation, apoptosis, immune function, and angiogenesis [1–3]. STAT3 signaling is strictly regulated in normal cells in response to stimulation with diverse cytokines (i.e., IL-6) and growth factors (i.e., EGF) [4–6]. However, hyperactivated STAT3 signaling leads to uncontrolled tumor growth and survival, resistance to chemotherapy, and poor prognosis in many types of cancer [3, 7, 8].
Breast cancer is a cancer with high incidence and mortality, and triple-negative breast cancer (TNBC) is more aggressive and lethal than other types of breast cancer [9, 10]. Many studies have proven that aberrant STAT3 plays an important role in TNBC. It regulates the expression of genes related to proliferation and antiapoptotic mechanisms (Bcl-2, Bcl-xL, survivin, c-Myc, etc.) [11, 12], invasion and migration (MMP-2/9, twist, vimentin, MEK-5, etc.), angiogenesis (VEGF, HIF-1α, HGF, etc.) [13–17], drug resistance (TNFR1, NF-κB, Oct-4, etc.) [18–20], and immunosuppression (IL-6/10, TGF-β, PD-L1, etc.) [21–24], leading to the tumorigenesis, malignant progression, and poor prognosis of TNBC.
On the other hand, gastric cancer (GC) is one of the most common gastrointestinal cancers and has a high incidence rate and mortality rate [25]. Early GC can be treated by surgery [26]. However, most patients are diagnosed in the late stage, and there are limited effective therapeutic approaches. Clinical studies have confirmed that STAT3 is hyperactivated in GC and closely associated with the proliferation, survival, angiogenesis, immune escape, and poor prognosis [27–29]. Therefore, the development of effective anticancer drugs targeting STAT3 is urgently needed for GC therapy.
STAT3 has been recognized as an attractive and potent target for the therapy of both TNBC and GC. Several STAT3 inhibitors have been developed and have demonstrated potential effects on TNBC and GC in vivo and in vitro [27, 30–33]. Among them, OPB-51602, napabucasin, AZD9150, and STAT3 decoys have even entered clinical trials and achieved certain efficacy in diverse human cancers, including TNBC and GC [27, 34]. However, poor specificity, weak effects, low oral bioavailability (niclosamide [35]), poor solubility (C188-9 [36]), and structural instability (BP-1-102 [37]) have prevented the further clinical application of the existing STAT3 inhibitors. None of the STAT3 inhibitors have been approved for TNBC and GC therapy in the clinic. Therefore, it is urgent to develop novel STAT3 inhibitors with higher potency, less toxicity, higher selectivity, and better stability for the therapy of TNBC and GC.
Although several approaches have been proposed to develop novel STAT3 inhibitors [37–42], targeting the STAT3 Src-homology 2 (SH2) domain and disrupting STAT3 dimerization are still some of the most important strategies. The SH2 domain in the STAT3 protein plays a pivotal role in the signaling cascade. Targeting the STAT3 SH2 domain prevents the dimerization and transcriptional activity of STAT3 [32, 36]. Here, we describe a novel STAT3 inhibitor, WZ-2-033, that can disrupt STAT3 dimerization, selectively inhibit STAT3 activation, further suppress the proliferation, migration, and invasion of TNBC and GC cells, and significantly inhibit the growth of human TNBC and GC cell xenografts in a mouse model. Further studies revealed that active STAT3 and downstream genes are also suppressed by WZ-2-033 in tumor tissue. Thus, our study provides a novel STAT3 inhibitor with significant antitumor activity and explores its pharmacological mechanisms.
Materials and methods
Chemical methods and synthesis
The chemical synthesis methods and routes are described in detail in the Supplementary Information.
Cell lines and cell culture
HEK-293T cells, human TNBC cells (MDA-MB-231, MDA-MB231-4175, and HCC70 cells), and human GC cells (AGS and MGC-803 cells) were obtained from American Type Culture Collection (VA, USA). HEK-293T, MDA-MB-231, and MDA-MB231-4175 cells were cultured in Dulbecco’s modified Eagle’s medium containing 10% (v/v) fetal bovine serum (FBS). HCC70, AGS, and MGC-803 cells were cultured in Roswell Park Memorial Institute 1640 (RPMI-1640) medium containing 10% (v/v) FBS. In addition, 50 μg/mL penicillin and 50 μg/mL streptomycin (Gibco, NY, USA) were added during culture. All cells were incubated at 37 °C in an incubator containing 5% CO2.
Reagents and antibodies
WZ-2-033 was dissolved in sterile dimethyl sulfoxide (DMSO; MP Biomedicals, 196055, CA, USA) and stored at −20 °C. Primary antibodies against phosphorylated STAT3 (Y705, #9145; S727, #9134), c-Myc (#5605), Mcl-1 (#4572), HA-tag (#3742), cleaved-caspase 7 (#9491), and β-actin (#3700) and fluorescently labeled secondary Alexa Fluor® 488 Conjuate2 (#4412) and Alexa Fluor® 555 Conjuate2 (#4409) antibodies were purchased from Cell Signaling Technology, Inc. (MA, USA). An antibody against STAT3 (10253-2-AP) was purchased from ProteinTech Group, Inc. (Wuhan, China). The antibody targeting Bcl-xL (sc-8392) was purchased from Santa Cruz Biotechnology (TX, USA). The monoclonal anti-FLAG® M2 antibody was purchased from Sigma-Aldrich (MO, USA). The secondary antibodies goat anti-rabbit IgG H&L (HRP) (ab6721) and goat anti-mouse IgG H&L (HRP) (ab6789) were purchased from Abcam (Cambridge, UK).
Plasmids and molecular cloning
The STAT3 reporter gene pGL3-STAT3 includes seven copies of the Stat3-specific binding sequence (AATCCCAGAA) in the C-reactive protein gene promoter. The Flag-STAT3 plasmid expressing Flag-tagged STAT3 protein was purchased from FulenGen (EX-Z2385-M35, Guangzhou, China). The HA-STAT3 plasmid was constructed based on Flag-STAT3. The new construct was confirmed by sequencing. STAT3C is a recombinant plasmid with cysteine residues substituted for specific amino acids in STAT3 (A662C and N664C) to render the molecule constitutively active.
Molecular docking
The Maestro 11.1 software was utilized to dock the STAT3 protein (PDB ID: 1BG1) and the small molecule. First, the structure of the STAT3 protein was optimized using the “Protein Preparation Wizard” module, including removal of water molecules, hydrogenation, minimization of energy, and treatment of metal ions and disulfide bonds. The “Lig Prep” module was used to optimize the small molecule, including the formation of a 3D structure and low-energy conformation. Next, the SH2 domain and transactivation domain (TAD) of the STAT3 protein were extracted as active pocket centers, and pocket grid points were generated using the “Receptor Grid Generation” module in the software. Finally, we used the “Ligand Docking” module to simulate the interactions between the STAT3 protein and the small molecule. In addition, we also used the “Protein Surface Analyzer” module to analyze the STAT3 protein structure according to electronegativity and label it with different colors.
Bioinformatics analysis
The gene expression profile datasets GSE19826, GSE14548, and GSE6861 were downloaded from the GEO database. The Gene Expression Profiling Interactive Analysis website (http://gepia.cancer-pku.cn/) was employed to determine the relationship of relevant genes with clinical survival.
Cellular thermal shift assay (CETSA)
To determine the target engagement of STAT3 in compound with cells, AGS cells were treated with WZ-2-033 or DMSO for 1 h when the cells were 70%–80% confluent. The cells were harvested and suspended in 1 mL PBS (containing PMSF and phosphatase inhibitor cocktail) and subsequently divided into 0.2 mL PCR tubes with 100 μL per tube. Each tube was heated to the designated temperature point for 2 min, tubes were removed and cooled to room temperature immediately after heating, and then, tubes were frozen in liquid nitrogen immediately. To lyse cells, the tubes were subjected to three freeze and thaw cycles in liquid nitrogen. The cell lysates were centrifuged at 20,000 × g for 20 min at 4 °C. Then, the cell lysates were subsequently analyzed by Western blotting.
Surface plasmon resonance (SPR) analysis
The SPR assay was performed using Biacore 8K (GE Healthcare, GA, USA) and Biacore Insight Evaluation Software. Purified STAT3 protein (0.17 mg/mL) without Tris and glycerin was dissolved in PBS and injected onto the CM5 chip (GE Healthcare, GA, USA). Different concentrations of WZ-2-033 were dissolved in running buffer (filtered 1×PBS, 0.01% DMSO) and passed over the chip to produce diverse response signals. The binding affinity (KD) was evaluated using Biacore Insight Evaluation Software.
Cell viability assay
Cell counting kit-8 (Bimake, B34302, Shanghai, China) was employed to measure the viability of the cells treated with WZ-2-033. TNBC cells (MDA-MB-231, MDA-MB-231-4175, and HCC70 cells) and GC cells (AGS and MGC-803 cells) were seeded in 96-well plates at a density of 1 × 103 ~ 5 × 103 cells per well and cultured in an incubator overnight. Then, the cells were administered different concentrations of WZ-2-033 or vehicle (medium containing 0.1% DMSO) for 72 h. Subsequently, a 1/10 volume of CCK-8 was added to each well of cells and incubated for 0.5–4 h at 37 °C. Finally, we used a microplate reader (FLUOstar Omega ACU, Offenburg, Germany) to examine the absorbance (optical density, OD) of the cells at 450 nm. Cell viability was calculated as follows: cell viability (%) = (ODcompound − ODblank)/(ODvehicle − ODblank) × 100%, where “blank” indicates medium only.
Colony formation assay
In this experiment, TNBC cells (MDA-MB-231 and MDA-MB231-4175 cells) and GC cells (AGS and MGC-803 cells) were seeded in six-well plates at a density of 0.5 × 103 ~ 1 × 103 cells per well. Then, the cells were treated with vehicle (0.1% DMSO) and various concentrations of WZ-2-033. The medium of the cells was changed, and the indicated concentration of WZ-2-033 was added to the cells every 2–3 days until colonies were visible in the vehicle control wells. After 10–15 days, the cells were fixed with 4% paraformaldehyde (Beyotime, P0099, Shanghai, China) and then stained with crystal violet (Beyotime, C0121, Shanghai, China). Finally, images of the cells were captured with a camera and analyzed by ImageJ software. All experiments were performed in triplicate.
Wounding healing assay
A wound healing assay was performed to measure the effect of WZ-2-033 on cell migration. The cells were seeded in six-well plates at a density of ~0.8 × 106 ~ 1.2 × 106 cells per well. When the cell fusion rate was 100%, the cells were scratched with pipette tips. Subsequently, the indicated concentration of WZ-2-033 was added to the cells until the vehicle cells migrated into the scratched area. Images of the scratched area of the cells before and after WZ-2-033 treatment were captured with a microscope. All experiments were performed in triplicate.
Transwell assay for cancer cell invasion
Twenty-four-well Transwell® plates with 8.0-µm-pore polycarbonate filters (Corning, 3422-ND) were utilized for the invasion assay. MDA-MB-231, HCC70, and MGC-803 cells were seeded in the top chamber of the insert at a density of 1.5 × 104 ~ 3 × 104 cells/well, treated with different concentrations of WZ-2-033, and cultured with serum-free medium. The bottom chamber of the insert was supplemented with medium containing 20% FBS as an attractant. The cells in the top chamber were carefully removed with a cotton swab after 12 h, and the cells in the bottom chamber were fixed with 4% paraformaldehyde for 15 min and stained with crystal violet for 30 min. Finally, the invaded cells were photographed and counted.
Flow cytometry analysis of apoptosis
A BestBio Annexin V-FITC/PI Cell Death Detection Kit (BB-4101-3) was employed to detect the apoptosis of the cells treated with WZ-2-033. The cells were treated with the indicated concentration of WZ-2-033 for 48 h. Then, the cells were digested with trypsin without EDTA, washed with cold PBS twice and resuspended in 400 µL 1× binding buffer. The cell suspension was incubated with Annexin V-FITC (5 µL) for 5 min and PI (10 µL) for 15 min at 4 °C in the dark. The samples were assessed within 1 h by flow cytometry. The data were analyzed by FlowJo 7.6 software.
RNA interference
RNA interference was performed as previously described [38]. AGS cells were seeded into six-well plates and incubated at 37 °C for 18–24 h. When the cells reached 50%–60% confluency, they were transfected with negative control siRNA or siRNA targeting STAT3 according to the DharmaFECT (T-2001-03, Dharmacon, USA) protocol. Then, the culture medium was replaced with fresh culture medium 6 h later, and the cells were cultured for 48 or 72 h. For the STAT3 interference experiments, the sequences of the two siRNAs targeting STAT3 are as follows:
STAT3-1: forward: 5′-UCCAGUUUCUUAAUUUGUUGACGGGUC-3′, reverse: 5′-GACCCGUCAACAAAUUAAGAAACUGGA-3′;
STAT3-1: forward: 5′-AUAGUCCUAUCUUCUAUUUGGAUGUCA-3′, reverse: 5′-UGACAUCCAAAUAGAAGAUAGGACUAU-3′.
A nonspecific oligonucleotide lacking complementary to any human gene was used as a negative control. All of the above siRNAs were synthesized by Sangon Biotech (Guangdong, China).
Western blot and immunoprecipitation
The cells treated with WZ-2-033 were lysed with RIPA buffer (50 mM Tris, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS sodium orthovanadate, sodium fluoride, EDTA, and leupeptin, pH 7.4) supplemented with 1 mM PMSF (Beyotime, ST506, Shanghai, China) and 1% (v/v) phosphatase inhibitor cocktail (Bimake, B15001, Shanghai, China). The cell lysates containing 20–50 μg proteins were denatured by boiling, analyzed by electrophoresis on SDS-PAGE gels, and transferred onto polyvinylidene difluoride membranes (Merck Millipore, Darmstadt, Germany). Then, the membranes were blocked with 5% bovine serum albumin (Beyotime, ST023, Shanghai, China) at room temperature for 1 h. After that, the membranes were incubated with the corresponding primary antibodies at 4 °C overnight and then incubated with the corresponding secondary antibodies for 1 h at room temperature. Finally, an ECL kit (Tanon, 180-5001, Shanghai, China) was employed to detect the expression level of the proteins following the manufacturer’s protocol.
Immunofluorescence
For colocalization analysis, HEK-293T cells were seeded in confocal dishes and cotransfected with HA-STAT3 and Flag-STAT3 plasmids. After the HA-STAT3 and Flag-STAT3 proteins were stably expressed in HEK-293T cells, various concentrations of WZ-2-033 were added to the cells and incubated for 24 h. The cells were stimulated with 100 ng/mL human IL-6 (PeproTech, 200-06-20) for 30 min before testing. Subsequently, the samples were fixed with 4% paraformaldehyde, permeabilized with 0.3% Triton X-100, blocked with goat serum at room temperature for 1 h, and incubated with anti-HA-tag and anti-Flag-tag antibodies at 4 °C overnight. The cells were incubated with Alexa Fluor 555-conjugated and Alexa Fluor 488-conjugated secondary antibodies for Flag-STAT3 and HA-STAT3 detection, respectively, for 1 h at room temperature in the dark. The cell nuclei were stained with DAPI (Yeasen, 40728ES10) in the dark for 10 min. Finally, the specimens were observed and imaged with a FV3000 laser scanning confocal microscope (Olympus, Tokyo, Japan). To determine the cellular location of the pY705-STAT3 protein, cells treated with WZ-2-033 were fixed with 4% paraformaldehyde, permeabilized with 0.3% Triton X-100, blocked with goat serum at room temperature for 1 h, incubated with anti-pY705-STAT3 antibody at 4 °C overnight, and incubated with the Alexa Fluor 488-conjugated fluorescently labeled secondary antibody for 1 h in the dark. The subsequent experiments were the same as above.
Coimmunoprecipitation
For coimmunoprecipitation experiments, 293T cells were prepared and treated as described for the immunofluorescence experiments. Subsequently, the cells were lysed with IP RIPA buffer (Beyotime, Shanghai, China) supplemented with protease inhibitors (Beyotime, Shanghai, China) and phosphatase inhibitors (Bimake, America), and lysates were obtained. Half of the protein lysate was cultured with anti-Flag Affinity beads (B23102, Bimake, USA) overnight at 4 °C. The beads were washed with PBST three times, 1× loading buffer was added, and the proteins were denatured by heating for 5 min at 100 °C. Then, the proteins were resolved by SDS-PAGE, transferred to PVDF membranes, and analyzed by immunoblotting.
Electrophoretic mobility shift assay (EMSA)
The nuclear proteins of MDA-MB-231 cells treated with different concentrations of WZ-2-033 were extracted using a Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime, P0027, Shanghai, China) following the manufacturer’s protocol. The nuclear extracts were incubated with biotin-labeled STAT3 (Beyotime, GS083B, Shanghai, China) or biotin-labeled STAT5 (Beyotime, GS085B, Shanghai, China) probes before EMSA. Subsequently, the DNA-protein complexes were separated in 6% native polyacrylamide gels and transferred onto positively charged nylon membranes (Merck Millipore, Darmstadt, Germany). The membranes were analyzed by a Chemiluminescent Biotin-labeled Nucleic Acid Detection Kit (Beyotime, D3308, Shanghai, China) following the manufacturer’s instructions.
Dual-luciferase reporter gene assay
HEK-293T cells were cotransfected with 50 ng pGL3-STAT3 carrying a reporter gene, 50 ng STAT3C and 40 ng Renilla luciferase plasmid as an internal control. After the cells stably expressed the STAT3 reporter gene, various concentrations of WZ-2-033 were added to the cells. The cells were lysed, and luciferase activity was detected by a Dual-Luciferase Reporter Gene Assay Kit (Beyotime, RG027, Shanghai, China) following the manufacturer’s protocol within 24 h.
Animal experiments
The animal procedures were approved by the Research Ethics Committee of Sun Yat-sen University (SYSU-IACUC-2020-000478 and SYSU-IACUC-2020-000479) and complied with the Guide for the Care and Use of Laboratory Animals. Mice were euthanized when they met the institutional euthanasia criteria for tumor size and overall health. Four-week-old nude mice (male or female, weighing 16–17 g, SPF grade, certification no. SYXK(Guangdong) 2016-0112) were obtained from GemPharmatech Co., Ltd. (Nanjing, China). For the subcutaneous implantation model, nude mice were injected subcutaneously with MGC-803 cells (3 × 106) or MDA-MB-231-4175 cells (5 × 106). When tumor volumes reached ~100 mm3, mice were randomly distributed into groups of six mice. The mice were treated by intraperitoneal injection (i.p.) with 5 or 15 mg/kg/d WZ-2-033 or vehicle. During the treatment, tumor size was measured with a caliper every 3 days. Tumor volume (V) was calculated with the formula V = A × B2 × π/6, where A and B stand for the longest and shortest diameters, respectively. After 21 days, tumor tissues were harvested and fixed with 4% paraformaldehyde fixation solution for paraffin-embedded sectioning.
Results
Discovery of the novel potent STAT3 inhibitor WZ-2-033
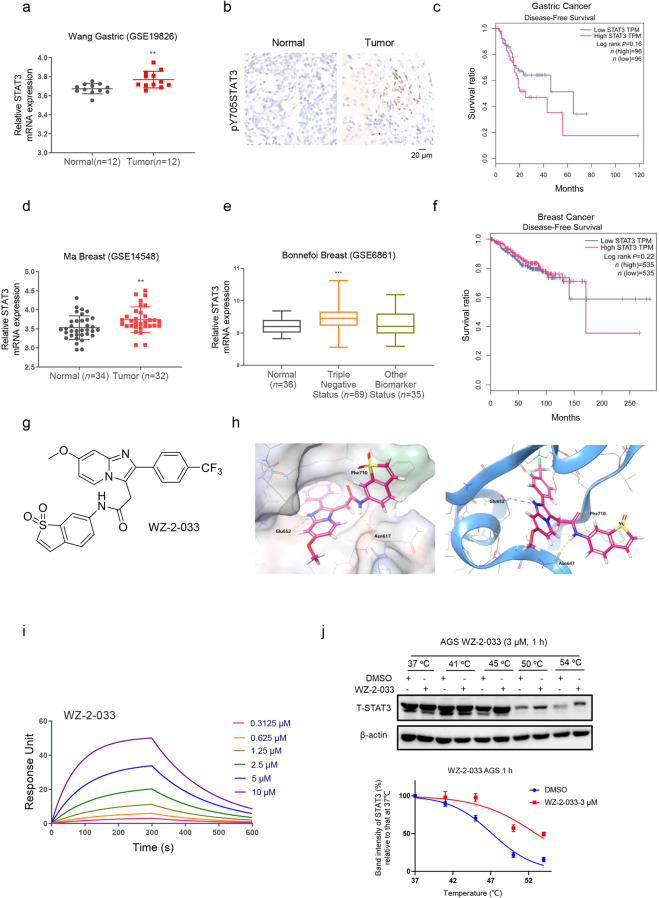
To investigate the role of STAT3 in GC and breast cancer, we analyzed GEO datasets of gastric tumors (GSE19826), breast tumors (GSE14548), and corresponding normal tissues. We found that STAT3 was indeed upregulated in gastric tumors (Fig. 1a) compared with normal tissue, as well as in breast tumors (Fig. 1d). We further confirmed the upregulation of STAT3 in TNBC (ER−/PR−/Her2−) (Fig. 1e), suggesting that STAT3 may play an important role in aggressive breast cancer. JAK tyrosine kinases phosphorylate the Tyr705 residue of STAT3 and activate STAT3, suggesting that the protein expression of pY705-STAT3 is very important. Thus, we performed an immunohistochemistry (IHC) assay to examine the protein expression of pY705-STAT3 in gastric tumor tissue. Consistently, the pY705-STAT3 protein was significantly overexpressed in gastric tumor tissues compared to adjacent tissues (Fig. 1b). In addition, upregulation of STAT3 was significantly associated with lower survival rates in patients with GC (Fig. 1c) and patients with breast cancer (Fig. 1f). This finding indicates that STAT3 is specifically upregulated in GC and breast cancer and closely associated with the occurrence and development of these two cancers.
Fig. 1. Discovery of the novel STAT3 inhibitor WZ-2-033.
a STAT3 expression level in normal and gastric tumor tissues (normal, n = 12; tumor, n = 12). **P ≤ 0.01 vs normal. b IHC analysis of STAT3 expression in gastric tumor tissues and corresponding normal tissues. Scale bar = 20 μm. c Disease-free survival analysis of gastric cancer patients (n = 96) from TCGA with high or low STAT3 expression levels. d STAT3 expression levels in normal and breast tumor tissues (normal, n = 34; tumor, n = 32). **P ≤ 0.01 vs normal. e Expression of STAT3 in molecular subtypes of breast tumors. Triple-negative status: ER−/PR−/Her2−. ***P ≤ 0.001. f Disease-free survival analysis of breast cancer patients (n = 535) from TCGA with high or low STAT3 expression levels. g Chemical structure of WZ-2-033. h Molecular docking of WZ-2-033 and the STAT3 protein. The red molecule represents WZ-2-033. WZ-2-033 occupies subpockets in the SH2 domain of STAT3. i SPR analysis of the interaction between WZ-2-033 and STAT3. j Western blot (upper) and melt curve (lower) analyses of the STAT3 protein in AGS cells after treatment with WZ-2-033 (3 μM) for 1 h at the indicated CETSA temperatures.
Therefore, we combined synthetic chemistry, gene reporter assays, and high-throughput screening to establish an in-house library of small molecules to identify potential STAT3 inhibitors. WZ-2-033 was identified as a potent STAT3 inhibitor from this library (Fig. 1g). To explore the binding model of WZ-2-033 with the target protein STAT3, we employed Maestro 11.1 to simulate the interaction between WZ-2-033 and STAT3. As shown in Fig. 1h, WZ-2-033 bound to the SH2 domain of the STAT3 protein and formed hydrogen bonds with Glu652, Asn647, and Phe710. In addition, WZ-2-033 also formed a salt bridge with Glu652, which is more stable than hydrogen bonds. SPR analysis was further utilized to explore the interaction between WZ-2-033 and STAT3. WZ-2-033 bound to the wild-type STAT3 protein with a KD of 7.40 μM (Fig. 1i). Furthermore, we performed a CETSA to examine the interaction between WZ-2-033 and the STAT3 protein. As shown in Fig. 1j, STAT3 was more stabilized in WZ-2-033-treated (3 μM, 1 h) cells than DMSO-treated cells, suggesting the binding of WZ-2-033 to STAT3.
WZ-2-033 disrupted STAT3 dimerization and significantly inhibited STAT3 signaling
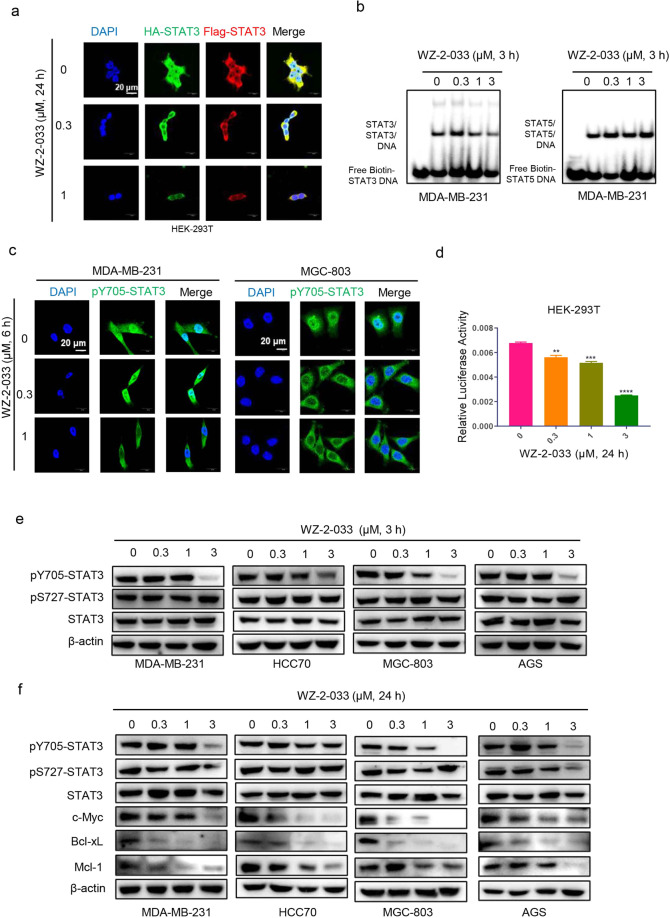
STAT3 contains six domains, including the N-terminal domain, coiled-coil domain, DNA-binding domain, linker domain, SH2 domain, and TAD. The SH2 domain is crucial for STAT3 dimerization, which is required for STAT3 nuclear translocation, DNA binding, and transcriptional activation [39–41]. Therefore, targeting the STAT3 SH2 domain is an attractive strategy to disrupt STAT3 signaling. STAT3 dimerization occurs through binding of two reciprocal phosphotyrosine-SH2 domains. To examine the effect of WZ-2-033 on STAT3 dimerization, we generated a cell model by cotransfecting HA-tagged STAT3 and Flag-tagged STAT3 into the HEK-293T cell line and measured STAT3–STAT3 dimerization directly in intact cells. HEK-293T cells cotransfected with Flag-STAT3 and HA-STAT3 were treated with WZ-2-033 for 24 h and 50 ng/mL EGF for 30 min before harvesting. As shown in Fig. 2a, the colocalization (yellow) of HA-STAT3 (green) and Flag-STAT3 (red) was significantly reduced by WZ-2-033 in a concentration-dependent manner, which indicated that WZ-2-033 inhibits the dimerization of HA-STAT3 and Flag-STAT3. The coimmunoprecipitation study results in Fig. S2 also demonstrated that STAT3 dimerization was disrupted by WZ-2-033.
Fig. 2. WZ-2-033 disrupted STAT3 dimerization and inhibited STAT3 signaling.
a HEK-293T cells stably expressing HA-STAT3 (green) and Flag-STAT3 (red) proteins were treated with the indicated concentrations of WZ-2-033 for 24 h and assessed by immunofluorescence analysis. DAPI nuclear staining is shown in blue. Scale bar = 20 μm. b EMSA analysis: The nuclear extracts of MDA-MB-231 cells treated with WZ-2-033 were incubated with a biotin-STAT3 or biotin-STAT5 probe and subjected to EMSA. c Cellular localization of pY705-STAT3 (green) in the cells treated with WZ-2-033 for 6 h. DAPI nuclear staining is shown in blue. Scale bar = 20 μm. d STAT3C, pGL3-STAT3-promoter, and Renilla luciferase (as a reference) plasmids were transiently cotransfected into HEK-293T cells. The luciferase activity of the cells after treatment with WZ-2-033 for 24 h was detected as described in the methods. e The phosphorylation level (pY705 and pS727) and expression level of STAT3 in TNBC and gastric cancer cells were measured by Western blot after treatment with WZ-2-033 for 3 h. f The expression levels of c-Myc, Bcl-xL, and Mcl-1 in TNBC and gastric cancer cells were measured by Western blot after treatment with WZ-2-033 for 24 h.
Furthermore, an EMSA was carried out to examine the effect of WZ-2-033 on STAT3-DNA-binding activity. Nuclear protein was harvested from MDA-MB-231 cells treated with WZ-2-033 at the indicated concentrations for 3 h. Then, the nuclear extracts were incubated with biotin-labeled STAT3 or biotin-labeled STAT5 for EMSA. As shown in Fig. 2b, WZ-2-033 inhibited STAT3-DNA-binding activity but had no obvious effect on STAT5-DNA-binding activity, which means that WZ-2-033 preferentially suppressed STAT3-DNA binding. Activated STAT3 monomer proteins form a dimer in the cytoplasm and then translocate to the nucleus. We observed that the level of tyrosine-phosphorylated STAT3 (pY705-STAT3, green) was significantly decreased in the nuclei of human breast cancer and GC cells treated with WZ-2-033, as assessed with laser confocal microscopy (Fig. 2c). As a transcription factor, STAT3 binds to specific DNA response elements and regulates the expression of target genes. We further examined whether WZ-2-033 suppresses STAT3-dependent transcriptional activation through a luciferase gene reporter assay. HEK-293T cells were transiently cotransfected with pGL3-STAT3 (a STAT3-responsive promoter-firefly luciferase reporter), STAT3C (to induce constitutive activation of STAT3), and Renilla luciferase reporter (for normalization) plasmids. Compared to those in the control group, the cells treated with WZ-2-033 had lower luciferase activity (Fig. 2d), which implies that WZ-2-033 significantly inhibits STAT3 transcriptional activity. To further investigate whether WZ-2-033 exerts anticancer activity by regulating the intracellular STAT3 signaling pathway, we examined the effect of WZ-2-033 on the level of pSTAT3. The tyrosine (Y705) phosphorylation level of STAT3 was reduced in response to WZ-2-033 treatment, while the serine (S727) phosphorylation level and total protein level of STAT3 did not show obvious changes, as shown in Fig. 2e. Subsequently, the expression of STAT3 downstream genes, including Bcl-2, Bcl-xL, and c-Myc, was inhibited by WZ-2-033 in a dose-dependent manner in TNBC and GC cells (Fig. 2f).
Taken together, the above results indicate that WZ-2-033 inhibits STAT3 dimerization and STAT3 Y705 phosphorylation, which in turn affects STAT3 nuclear translocation and DNA binding, resulting in the downregulation of transcriptional activity and subsequent inhibition of the expression of downstream genes, such as c-Myc, Bcl-xL, and Mcl-1, in breast cancer and GC cells.
WZ-2-033 strongly suppressed the proliferation, colony survival, migration, and invasion of TNBC and GC cells with aberrantly active STAT3
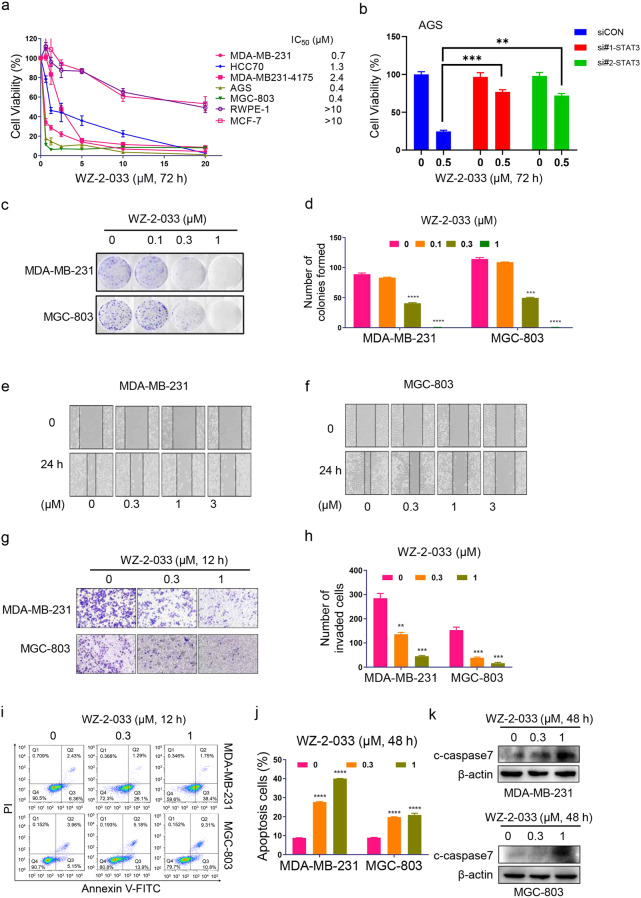
Abnormal activation of STAT3 promotes the transcription and expression of genes related to tumor proliferation, survival, migration, invasion, apoptosis, and angiogenesis, leading to malignant progression of tumors, and these characteristics make STAT3 a potential target for cancer therapy [42, 43]. To evaluate the effect of WZ-2-033 on cell proliferation, human breast and GC cells harboring aberrantly activated STAT3 were treated with different concentrations of WZ-2-033 for 72 h and subsequently examined with a CCK-8 kit. The breast cancer cell line MCF-7, which has lower STAT3 activity [37, 44], and the normal epithelial cell line RWPE-1 were also examined to evaluate the selectivity of WZ-2-033. As shown in Fig. 3a, WZ-2-033 significantly inhibited cancer cell proliferation in a dose-dependent manner. The IC50 values of WZ-2-033 in the malignant breast cancer cell lines MDA-MB-231, HCC70, and MDA-MB231-4175 were 0.7, 1.3, and 1.3 μM, respectively. Compared with the human breast cancer cells, human GC cells (AGS and MGC-803 cells) were more sensitive, with an IC50 of 0.4 μM (Fig. 3a). The decrease in viability in breast cancer MCF-7 cells and normal epithelial RWPE-1 cells which have lower STAT3 activity, induced by WZ-2-033 was much weaker than that in cancer cells with constitutive STAT3 activity (Fig. 3a). To further identify the selectivity of WZ-2-033, we knocked down STAT3 in AGS cells and found that the inhibition of cancer cell proliferation induced by WZ-2-033 was STAT3 dependent (Fig. 3b and Fig. S1a, b). Subsequently, a colony formation assay was performed to examine the effect of WZ-2-033 on cancer cell survival. The colony number of the cells was significantly decreased even at 0.3 μM (Fig. 3c, d and Figs. S1c-d) in breast cancer and GC cells.
Fig. 3. Antitumor effects of WZ-2-033 in malignant TNBC and gastric cancer cells with aberrantly active STAT3 in vitro.
a, b Cancer cells harboring constitutively activated STAT3 were treated with 0–20 μM WZ-2-033 for 72 h and then subjected to cell viability assays with a CCK-8 kit. MCF-7 cells, which have lower STAT3 activity, and normal epithelial RWPE-1 cells were also examined by CCK-8 assay. **P < 0.01; ***P < 0.001 vs siCON. c, d Colony formation assays: the designated cells were administered the indicated concentrations of WZ-2-033 and grown until colonies were visible, at which point they were stained with crystal violet (c). The number of colonies formed was determined and analyzed (d). Values are presented as the mean ± SEM. n = 3. ***P < 0.001; **** P < 0.0001 vs control. e, f Cells were scratched with pipette tips when the cell confluency was 100% and then WZ-2-033 was added to the cells, the images were taken again in 24 h. g, h The effect of WZ-2-033 on cell invasion was measured by transwell analysis. The cells were stained with crystal violet (g) and analyzed (h) as described in the “Materials and methods” section. Values are presented as the mean ± SEM. n = 3. **P < 0.01; ***P < 0.001 vs control. i, j Flow cytometry analysis was performed to detect the apoptosis of the cells treated with 0–1 μM WZ-2-033 for 48 h (i). The percentage of apoptotic cells was calculated and analyzed (j). Values are presented as the mean ± SEM. n = 3. ****P < 0.0001 vs control. k Cleaved caspase 7 (c-caspase 7) immunoblots for MDA-MB-231 and MGC-803 cells treated with WZ-2-033 for 48 h.
Metastasis is one of the main factors leading to poor prognosis and drug resistance [45–47]. To evaluate the effect on tumor metastasis, we measured cell migration and invasion through wound healing and transwell assays in TNBC and GC cells. As shown in Fig. 3e, f and Fig. S1e, f, WZ-2-033 inhibited the migration of TNBC and GC cells in a dose-dependent manner. The transwell cell invasion assay results suggested that the invasion of breast cancer and GC cells was suppressed by 0.3 μM WZ-2-033 (Fig. 3g, h and Fig. S1g, h). Taken together, these results show that WZ-2-033 significantly suppressed the migration and invasion of human breast and GC cells.
Furthermore, we performed flow cytometry analysis to examine whether the proliferative inhibition induced by WZ-2-033 was because of apoptosis. As shown in Fig. 3i, j, 0.3 μM WZ-2-033 significantly induced apoptosis in MDA-MB-231, MGC-803, and AGS cells, with apoptotic cell percentages of 27.7%, 19.8%, and 7.4%, respectively (Fig. 3i, j and Fig. S1i, j). Moreover, cleavage of caspase 7, one of the hallmarks of apoptosis, was induced by WZ-2-033 in MDA-MB-231 and MGC-803 cells, as shown in Fig. 3k.
WZ-2-033 inhibited human GC cell xenografts
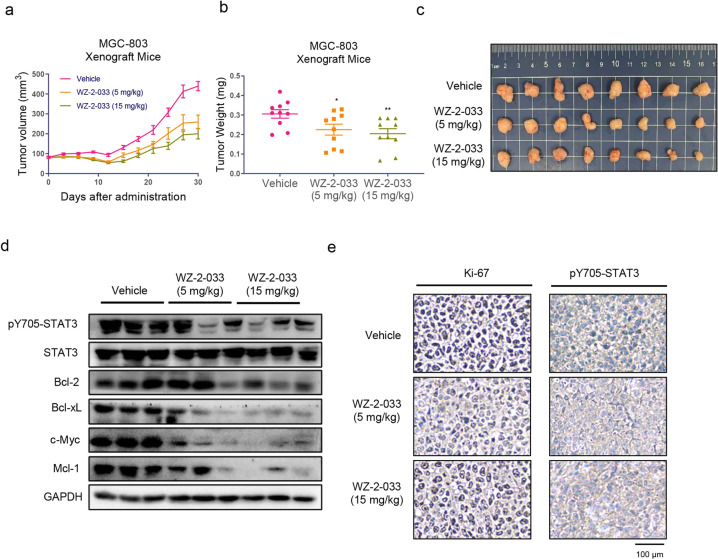
To further demonstrate the therapeutic efficacy of WZ-2-033, subcutaneous xenografts of MG-803 human GC cells in a mouse model were utilized to evaluate antitumor effects. As shown in Fig. 4a–c, the growth of human GC xenografts following 30 days of treatment with 5 mg/kg or 15 mg/kg WZ-2-033 per day by i.p. injection was significantly inhibited. The tumor volumes and tumor weights were decreased dose dependently in the 5 and 15 mg/kg groups (Fig. 4a, b). In addition, there were no obvious changes in body weights or signs of toxicity, such as loss of appetite, decreased activity, or lethargy during treatment (Fig. S3a–c).
Fig. 4. Antitumor effect of WZ-2-033 in the MGC-803 mouse xenograft model.
a MGC-803 xenograft mice were administered 5 or 15 mg/kg WZ-2-033 or vehicle (15% Cremophor EL in PBS) via i.p. injection per day. The tumor size was measured every 3 days. Values are presented as the mean ± SEM. n = 10. b Tumor weight after 30 days of administration. Values are presented as the mean ± SEM. n = 10. *P < 0.05; **P < 0.01 vs the vehicle group. c Representative images of gastric cancer xenografts. d pY705-STAT3, STAT3, Bcl-2, Bcl-xL, c-myc, and Mcl-1 immunoblots for tumor tissue lysates from three different vehicle- and WZ-2-033-treated mice. e IHC for Ki-67 and pY705-STAT3 in the tumor tissues of vehicle- and WZ-2-033-treated mice. Scale bar = 100 μm
The tumor mitotic index (Ki-67) and STAT3 signaling in tumor tissues were further evaluated by IHC and Western blotting. As shown in Fig. 4e, the expression of Ki-67 was significantly reduced by WZ-2-033 compared with the vehicle control, indicating that tumor proliferation and progression were suppressed by WZ-2-033 administration. IHC analysis of tumor xenografts showed that the level of pY705-STAT3 was decreased in the WZ-2-033-treated group (Fig. 4e). The expression of pY705-STAT3 and the STAT3 downstream genes Bcl-2, c-Myc, Mcl-1, and Bcl-xL were also examined by Western blotting. WZ-2-033 significantly suppressed the level of pY705-STAT3 and subsequently downregulated the expression of STAT3-targeting genes in human GC cell xenografts, as shown in Fig. 4d. Taken together, these results suggest that WZ-2-033 significantly suppressed excessive STAT3 signaling and gastric tumor growth in mouse models.
WZ-2-033 inhibited human TNBC cell xenografts
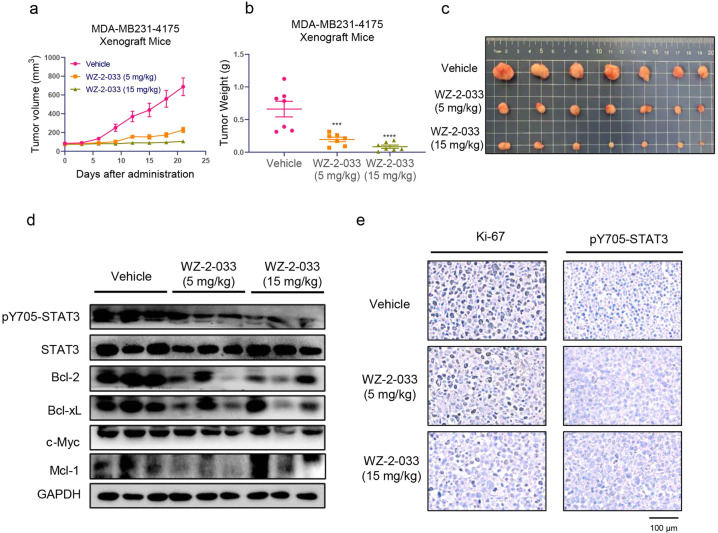
Next, we evaluated the therapeutic efficacy of WZ-2-033 in breast cancer using subcutaneous mouse xenografts of MDA-MB231-4175 human breast cancer cells (MDA-MB231-4175). Tumor-bearing mice were administered WZ-2-033 at doses of 5 and 15 mg/kg per day for 21 consecutive days. Tumor volume and body weight were measured every 3 days. WZ-2-033 demonstrated significant antitumor efficacy with a tumor growth inhibition (TGI) rate greater than 85% with the high dosage. The low-dosage group also showed effective antitumor activity with a TGI of 70.5%. TNBC appears to be more sensitive to WZ-2-033 than GC. Similar to the results in the GC model, there were no obvious changes in body weights or signs of toxicity, such as loss of decreased appetite, decreased activity, or lethargy, during treatment (Fig. S4a–c). The expression of pY705-STAT3 and the STAT3 downstream genes Bcl-2, c-Myc, Mcl-1, and Bcl-xL was also examined by Western blotting (Fig. 5d). WZ-2-033 significantly suppressed the level of pY705-STAT3 and subsequently downregulated the expression of STAT3-targeted genes in human GC cell xenografts, as shown in Fig. 5d, e.
Fig. 5. Antitumor effect of WZ-2-033 in a mouse xenograft model of MDA-MB231-4175 cells.
a MDA-MB231-4175 xenograft mice were administered 5 or 15 mg/kg WZ-2-033 or vehicle (15% Cremophor EL in PBS) via i.p. injection every day. Tumor sizes were measured every 3 days. Values are presented as the mean ± SEM. n = 10. b Tumor weight after 21 days of administration. Values are presented as the mean ± SEM. n = 10. *P < 0.05; **P < 0.01 vs the vehicle group. c Representative images of breast cancer xenografts. d pY705-STAT3, STAT3, Bcl-2, Bcl-xL, c-Myc, and Mcl-1 immunoblots for tumor tissue lysates from three different vehicle- and WZ-2-033-treated mice. e IHC analysis for Ki-67 and pY705-STAT3 in tumor tissues from vehicle- and WZ-2-033-treated mice. Scale bar = 100 μm
Discussion
Normal STAT3 signaling is tightly controlled in standard cellular responses, while constitutive STAT3 activation occurs frequently in a variety of human cancers. Hyperactive STAT3 signaling is frequently detected in breast cancer and GC cell lines, tumor models, and patient samples [30]. Aberrant STAT3 activation promotes cancer cell proliferation and survival and enhances resistance to chemotherapy and targeted therapy. Although these two types of cancer feature high incidence and mortality, limited effective therapeutic approaches are clinically available. Therefore, the development of new targeted cancer drugs, including novel direct STAT3 inhibitors, will benefit breast cancer and gastric patients harboring constitutive STAT3 activation.
Given the importance of STAT3 in cancer initiation and progression, many efforts have been made to develop specific and potent STAT3 inhibitors in the past two decades, especially inhibitors targeting STAT3 dimerization. Nonetheless, these inhibitors have demonstrated moderate inhibitory potency in cell and mouse models, possibly because dimerization of activated STAT3 proteins is a type of protein–protein interaction (PPI). The binding interfaces of PPIs, such as the binding site in the STAT3 SH2 domain for the pY-peptide, generally spread over large, relatively flat surfaces and lack well-defined, deep binding pockets. Therefore, these sites are difficult to target with small molecules. OPB-51602 and OPB-31121 are promising STAT3 inhibitors that have reached early-phase clinical trials and have shown promising therapeutic efficacy [48, 49]. However, most reported inhibitors still suffer from multiple challenges, such as low specificity, weak binding affinity, low oral bioavailability, poor solubility, structural instability, unfavorable PK profiles, and potential severe toxicities. The potent STAT3 degrader SD-36 was developed based on the proteolysis targeting chimera concept, utilizing a ligand for the cereblon/cullin 4A E3 ligase and a peptidomimetic STAT3 SH2 domain inhibitor, and SD-36 was recently reported to induce potent antitumor activity in blood cancers [50]. However, none of the STAT3 inhibitors have been approved for cancer therapy in the clinic. Therefore, discovering new STAT3 inhibitors with higher potency and excellent drug properties is urgently needed.
To this end, we synthesized and screened a series of small molecules targeting STAT3. WZ-2-033 was identified from an in-house library. We simulated the interaction between WZ-2-033 and STAT3 with docking software and found that it bound to the SH2 domain of the STAT3 protein. Subsequently, we employed SPR analysis to examine the affinity between WZ-2-033 and the STAT3 protein. Moreover, CETSA analysis was further utilized to confirm the interaction between WZ-2-033 and STAT3. We also generated a cell line that could be used to measure direct STAT3–STAT3 dimerization in intact cells and revealed that WZ-2-033 inhibited the colocalization of HA-STAT3 and Flag-STAT3 through confocal imaging. As a transcription factor, STAT3 binds to specific DNA sequences and regulates the expression of target genes. The EMSA analysis demonstrated that WZ-2-033 selectively inhibited STAT3-DNA binding without obvious effects on STAT5-DNA binding. Luciferase reporter studies showed that WZ-2-033 dose dependently inhibited STAT3-dependent transcriptional activity. Confocal imaging showed that the accumulation of pY705-STAT3 in the nucleus was downregulated by WZ-2-033. Moreover, WZ-2-033 reduced Y705 phosphorylation of STAT3 and downregulated the expression of STAT3 target genes, including c-Myc, Bcl-xL, and Mcl-1, in breast cancer and GC cell lines. However, although we determined that WZ-2-033 interacts with STAT3 protein and modulates STAT3 signaling using multiple approaches, without cocrystal structure evidence, it is difficult to identify the direct binding sites of WZ-2-033 within the STAT3 protein. Further structural biology studies will reveal more information on the detailed effects of WZ-2-033 on STAT3 signaling.
Next, we evaluated the antitumor effects of WZ-2-033 in cell and mouse models. WZ-2-033 significantly inhibited the proliferation of breast cancer and GC cell lines with constitutive STAT3 activity. Cancer cells with lower STAT3 activation, such as MCF-7 cells, were not sensitive to WZ-2-033 treatment. WZ-2-033 also strongly suppressed the survival, migration, and invasion of breast cancer and GC cells with aberrantly active STAT3 and induced cell apoptosis. Moreover, WZ-2-033 significantly inhibited the growth of subcutaneous mouse xenografts of breast cancer cell lines and GC cell lines harboring aberrantly active STAT3. The level of pY705-STAT3 in tumor tissue was significantly inhibited by WZ-2-033 according to Western blotting and IHC analyses. Furthermore, we found that the expression of STAT3 downstream genes, including c-Myc, Bcl-2, and Bcl-xL, in tumor tissue was also downregulated by WZ-2-033. Thus, WZ-2-033 effectively inhibits STAT3 signaling with potent antitumor activity in vitro and in vivo. In addition to its critical roles in the proliferation, invasion, and metastasis of human cancer cells, STAT3 also plays an important role in regulating the tumor microenvironment and immune response. STAT3 can bind to the PD-L1 promoter and regulate PD-L1 expression [21–24]. We did not illustrate the effect of WZ-2-033 as a STAT3 inhibitor on the immune response in an animal model. Future studies will elucidate the detailed mechanism of WZ-2-033 antitumor effects and its role in immunotherapy.
In the present study, we demonstrated that WZ-2-033, as a novel STAT3 inhibitor, effectively inhibited STAT3 signaling with potent antitumor activity in human TNBC and GC in vitro and in vivo. The pharmacological mechanisms were assessed in multiple ways. WZ-2-033 is a potential candidate worthy of further development for clinical application, particularly in human TNBC and GC, which lack effective therapeutic approaches.
Supplementary information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81973359 and 21977128), Guangdong Basic and Applied Basic Research Foundation (2019A1515011215), and Guangzhou Basic and Applied Basic Research Foundation (202002030408 and 202103000097). Grants from the Fundamental Research Funds for the Central Universities, Sun Yat-sen University (2021qntd44), Jilin Province Science and Technology Development Project (20190304046YY and 20200404105YY), National Engineering and Technology Research Center for New Drug Druggability Evaluation (Seed Program of Guangdong Province, 2017B090903004), National Major Special Projects for the Creation and Manufacture of New Drugs (2019ZX09301104), Key-Area Research and Development Program of Guangdong Province (2020B1111110003), and Guangdong Provincial Key Laboratory of Construction Foundation (2019B030301005) are also appreciated.
Author contributions
XLZ, YXW, and PQL designed the research and supervised the whole study and YZ, SMOY, and JSM performed the studies in the cell models. SS conducted the animal studies. LD and QYH synthesized the compound. KZ and XMQ contributed to data analysis and technology support.
Data availability
All data generated or analyzed during this study are included in this published article.
Competing interests
The authors declare no competing interests.
Ethics statement
The animal procedures were approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University (SYSU-IACUC-2020-000478 and SYSU-IACUC-2020-000479) and followed the Guide for the Care and Use of Laboratory Animals.
Footnotes
These authors contributed equally: Yan Zhong, Lin Deng, Shuo Shi
Contributor Information
Pei-qing Liu, Email: liupq@mail.sysu.edu.cn.
Yuan-xiang Wang, Email: wangyx95@mail.sysu.edu.cn.
Xiao-lei Zhang, Email: zhangxlei5@mail.sysu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41401-021-00718-0.
References
- 1.Bromberg J, Darnell J. The role of STATs in transcriptional control and their impact on cellular function. Oncogene. 2000;19:2468–73. doi: 10.1038/sj.onc.1203476. [DOI] [PubMed] [Google Scholar]
- 2.Levy DE, C-k Lee. What does Stat3 do? J Clin Invest. 2002;109:1143–8. doi: 10.1172/JCI15650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu H, Jove R. The STATs of cancer–new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 4.Heinrich P, Behrmann I, Müller-Newen G, Schaper F, Graeve L. Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem J. 1998;334:297–314. doi: 10.1042/bj3340297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, van Boxel-Dezaire AH, Cheon H, Yang J, Stark GR. STAT3 activation in response to IL-6 is prolonged by the binding of IL-6 receptor to EGF receptor. Proc Natl Acad Sci USA. 2013;110:16975–80. doi: 10.1073/pnas.1315862110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan J, Zhang F, Niu R. Multiple regulation pathways and pivotal biological functions of STAT3 in cancer. Sci Rep. 2015;5:17663. doi: 10.1038/srep17663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yue P, Turkson J. Targeting STAT3 in cancer: how successful are we? Expert Opin Investig Drugs. 2009;18:45–56. doi: 10.1517/13543780802565791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao C, Li H, Lin HJ, Yang S, Lin J, Liang G, et al. Feedback activation of STAT3 as a cancer drug-resistance mechanism. Trends Pharmacol Sci. 2016;37:47–61. doi: 10.1016/j.tips.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Hubalek M, Czech T, Muller H. Biological subtypes of triple-negative breast cancer. Breast Care. 2017;12:8–14. doi: 10.1159/000455820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar P, Aggarwal R. An overview of triple-negative breast cancer. Arch Gynecol Obstet. 2016;293:247–69. doi: 10.1007/s00404-015-3859-y. [DOI] [PubMed] [Google Scholar]
- 11.Diaz N, Minton S, Cox C, Bowman T, Gritsko T, Garcia R, et al. Activation of stat3 in primary tumors from high-risk breast cancer patients is associated with elevated levels of activated SRC and survivin expression. Clin Cancer Res. 2006;12:20–8. doi: 10.1158/1078-0432.CCR-04-1749. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Shen Y, Wang S, Shen Q, Zhou X. The role of STAT3 in leading the crosstalk between human cancers and the immune system. Cancer Lett. 2018;415:117–28. doi: 10.1016/j.canlet.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang F, Wang Z, Fan Y, Xu Q, Ji W, Tian R, et al. Elevated STAT3 signaling-mediated upregulation of MMP-2/9 confers enhanced invasion ability in multidrug-resistant breast cancer cells. Int J Mol Sci. 2015;16:24772–90. doi: 10.3390/ijms161024772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McDaniel J, Varley K, Gertz J, Savic D, Roberts B, Bailey S, et al. Genomic regulation of invasion by STAT3 in triple negative breast cancer. Oncotarget. 2017;8:8226–38. doi: 10.18632/oncotarget.14153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie Q, Yang Z, Huang X, Zhang Z, Li J, Ju J, et al. Ilamycin C induces apoptosis and inhibits migration and invasion in triple-negative breast cancer by suppressing IL-6/STAT3 pathway. J Hematol Oncol. 2019;12:60. doi: 10.1186/s13045-019-0744-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamran MZ, Patil P, Gude RP. Role of STAT3 in cancer metastasis and translational advances. Biomed Res Int. 2013;2013:421821. doi: 10.1155/2013/421821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu F, Zhang H, Song H. Upregulation of MEK5 by Stat3 promotes breast cancer cell invasion and metastasis. Oncol Rep. 2017;37:83–90. doi: 10.3892/or.2016.5256. [DOI] [PubMed] [Google Scholar]
- 18.Cheng CC, Shi LH, Wang XJ, Wang SX, Wan XQ, Liu SR, et al. Stat3/Oct-4/c-Myc signal circuit for regulating stemness-mediated doxorubicin resistance of triple-negative breast cancer cells and inhibitory effects of WP1066. Int J Oncol. 2018;53:339–48. doi: 10.3892/ijo.2018.4399. [DOI] [PubMed] [Google Scholar]
- 19.Egusquiaguirre SP, Yeh JE, Walker SR, Liu S, Frank DA. The STAT3 target gene TNFRSF1A modulates the NF-kappaB pathway in breast cancer cells. Neoplasia. 2018;20:489–98. doi: 10.1016/j.neo.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuo WY, Hwu L, Wu CY, Lee JS, Chang CW, Liu RS. STAT3/NF-kappaB-regulated lentiviral TK/GCV suicide gene therapy for cisplatin-resistant triple-negative breast cancer. Theranostics. 2017;7:647–63. doi: 10.7150/thno.16827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen M, Pockaj B, Andreozzi M, Barrett MT, Krishna S, Eaton S, et al. JAK2 and PD-L1 amplification enhance the dynamic expression of PD-L1 in triple-negative breast cancer. Clin Breast Cancer. 2018;18:e1205–15. doi: 10.1016/j.clbc.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 22.Kitamura H, Ohno Y, Toyoshima Y, Ohtake J, Homma S, Kawamura H, et al. Interleukin-6/STAT3 signaling as a promising target to improve the efficacy of cancer immunotherapy. Cancer Sci. 2017;108:1947–52. doi: 10.1111/cas.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Yang Y, Wang H, Wang B, Zhao K, Jiang W, et al. Syntenin1/MDA-9 (SDCBP) induces immune evasion in triple-negative breast cancer by upregulating PD-L1. Breast Cancer Res Treat. 2018;171:345–57. doi: 10.1007/s10549-018-4833-8. [DOI] [PubMed] [Google Scholar]
- 24.Sasidharan Nair V, Toor S, Ali B, Elkord E. Dual inhibition of STAT1 and STAT3 activation downregulates expression of PD-L1 in human breast cancer cells. Expert Opin Ther Targets. 2018;22:547–57. doi: 10.1080/14728222.2018.1471137. [DOI] [PubMed] [Google Scholar]
- 25.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 26.Nomura S, Kaminishi M. Surgical treatment of early gastric cancer. Digest Surg. 2007;24:96–100. doi: 10.1159/000101895. [DOI] [PubMed] [Google Scholar]
- 27.Giraud AS, Menheniott TR, Judd LM. Targeting STAT3 in gastric cancer. Expert Opin Ther Targets. 2012;16:889–901. doi: 10.1517/14728222.2012.709238. [DOI] [PubMed] [Google Scholar]
- 28.Kim DY, Cha ST, Ahn DH, Kang HY, Kwon CI, Ko KH, et al. STAT3 expression in gastric cancer indicates a poor prognosis. J Gastroenterol Hepatol. 2009;24:646–51. doi: 10.1111/j.1440-1746.2008.05671.x. [DOI] [PubMed] [Google Scholar]
- 29.Li MX, Bi XY, Huang Z, Zhao JJ, Han Y, Li ZY, et al. Prognostic role of phospho-STAT3 in patients with cancers of the digestive system: a systematic review and meta-analysis. PLoS One. 2015;10:e0127356. doi: 10.1371/journal.pone.0127356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu J, Yang S, Zhou T, Si Y, Xiang Y, Ke W, et al. Cucurbitacin B inhibits gastric cancer progression by suppressing STAT3 activity. Arch Biochem Biophys. 2020;684:108314. doi: 10.1016/j.abb.2020.108314. [DOI] [PubMed] [Google Scholar]
- 31.Cheong JH, Hong SY, Zheng Y, Noh SH. Eupatilin inhibits gastric cancer cell growth by blocking STAT3-mediated VEGF expression. J Gastric Cancer. 2011;11:16–22. doi: 10.5230/jgc.2011.11.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dai X, Yin C, Guo G, Zhang Y, Zhao C, Qian J, et al. Schisandrin B exhibits potent anticancer activity in triple negative breast cancer by inhibiting STAT3. Toxicol Appl Pharmacol. 2018;358:110–9. doi: 10.1016/j.taap.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 33.Yang B, Shen JW, Zhou DH, Zhao YP, Wang WQ, Zhu Y, et al. Precise discovery of a STAT3 inhibitor from Eupatorium lindleyanum and evaluation of its activity of anti-triple-negative breast cancer. Nat Prod Res. 2019;33:477–85. doi: 10.1080/14786419.2017.1396596. [DOI] [PubMed] [Google Scholar]
- 34.Qin JJ, Yan L, Zhang J, Zhang WD. STAT3 as a potential therapeutic target in triple negative breast cancer: a systematic review. J Exp Clin Cancer Res. 2019;38:195. doi: 10.1186/s13046-019-1206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren X, Duan L, He Q, Zhang Z, Zhou Y, Wu D, et al. Identification of niclosamide as a new small-molecule inhibitor of the STAT3 signaling pathway. ACS Med Chem Lett. 2010;1:454–9. doi: 10.1021/ml100146z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Di JX, Zhang HY. C188-9, a small-molecule STAT3 inhibitor, exerts an antitumor effect on head and neck squamous cell carcinoma. Anticancer Drugs. 2019;30:846–53. doi: 10.1097/CAD.0000000000000783. [DOI] [PubMed] [Google Scholar]
- 37.Zhang X, Yue P, Page BD, Li T, Zhao W, Namanja AT, et al. Orally bioavailable small-molecule inhibitor of transcription factor Stat3 regresses human breast and lung cancer xenografts. Proc Natl Acad Sci USA. 2012;109:9623–8. doi: 10.1073/pnas.1121606109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng Q, Dong H, Mo J, Zhang Y, Huang J, Ouyang S, et al. A novel STAT3 inhibitor W2014-S regresses human non-small cell lung cancer xenografts and sensitizes EGFR-TKI acquired resistance. Theranostics. 2021;11:824–40. doi: 10.7150/thno.49600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang G, Yan H, Ye S, Tong C, Ying QL. STAT3 phosphorylation at tyrosine 705 and serine 727 differentially regulates mouse ESC fates. Stem Cells. 2014;32:1149–60. doi: 10.1002/stem.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ren Z, Mao X, Mertens C, Krishnaraj R, Qin J, Mandal PK, et al. Crystal structure of unphosphorylated STAT3 core fragment. Biochem Biophys Res Commun. 2008;374:1–5. doi: 10.1016/j.bbrc.2008.04.049. [DOI] [PubMed] [Google Scholar]
- 41.Sgrignani J, Garofalo M, Matkovic M, Merulla J, Catapano CV, Cavalli A. Structural biology of STAT3 and its implications for anticancer therapies development. Int J Mol Sci. 2018;19:1591. doi: 10.3390/ijms19061591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Furtek S, Backos D, Matheson C, Reigan P. Strategies and approaches of targeting STAT3 for cancer treatment. ACS Chem Biol. 2016;11:308–18. doi: 10.1021/acschembio.5b00945. [DOI] [PubMed] [Google Scholar]
- 43.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14:736–46. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- 44.Zhang X, Sun Y, Pireddu R, Yang H, Urlam MK, Lawrence HR, et al. A novel inhibitor of STAT3 homodimerization selectively suppresses STAT3 activity and malignant transformation. Cancer Res. 2013;73:1922–33. doi: 10.1158/0008-5472.CAN-12-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tseng LM, Hsu NC, Chen SC, Lu YS, Lin CH, Chang DY, et al. Distant metastasis in triple-negative breast cancer. Neoplasma. 2013;60:290–4. doi: 10.4149/neo_2013_038. [DOI] [PubMed] [Google Scholar]
- 46.Aysola K, Desai A, Welch C, Xu J, Qin Y, Reddy V, et al. Triple negative breast cancer—an overview. Hereditary Genet. 2013;2013:001. doi: 10.4172/2161-1041.S2-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yakata Y, Nakayama T, Yoshizaki A, Kusaba T, Inoue K, Sekine I. Expression of p-STAT3 in human gastric carcinoma: significant correlation in tumour invasion and prognosis. Int J Oncol. 2007;30:437–42. [PubMed] [Google Scholar]
- 48.Wong AL, Soo RA, Tan DS, Lee SC, Lim JS, Marban PC, et al. Phase I and biomarker study of OPB-51602, a novel signal transducer and activator of transcription (STAT) 3 inhibitor, in patients with refractory solid malignancies. Ann Oncol. 2015;26:998–1005. doi: 10.1093/annonc/mdv026. [DOI] [PubMed] [Google Scholar]
- 49.Okusaka T, Ueno H, Ikeda M, Mitsunaga S, Ozaka M, Ishii H, et al. Phase 1 and pharmacological trial of OPB-31121, a signal transducer and activator of transcription-3 inhibitor, in patients with advanced hepatocellular carcinoma. Hepatol Res. 2015;45:1283–91. doi: 10.1111/hepr.12504. [DOI] [PubMed] [Google Scholar]
- 50.Bai L, Zhou H, Xu R, Zhao Y, Chinnaswamy K, McEachern D, et al. A potent and selective small-molecule degrader of STAT3 achieves complete tumor regression in vivo. Cancer Cell. 2019;36:498–511. doi: 10.1016/j.ccell.2019.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.