Abstract
Background:
Ischemic heart disease following the obstruction of coronary vessels leads to the death of cardiac tissue and the formation of a fibrotic scar. In contrast to adult mammals, zebrafish can regenerate their heart after injury, enabling the study of the underlying mechanisms. One of the earliest responses following cardiac injury in adult zebrafish is coronary revascularization. Defects in this process lead to impaired cardiomyocyte repopulation and scarring. Hence, identifying and investigating factors that promote coronary revascularization holds great therapeutic potential.
Methods:
We used wholemount imaging, immunohistochemistry and histology to assess various aspects of zebrafish cardiac regeneration. Deep transcriptomic analysis allowed us to identify targets and potential effectors of Vegfc signaling. We employed newly generated loss- and gain-of-function genetic tools to investigate the role of Emilin2a and Cxcl8a-Cxcr1 signaling in cardiac regeneration.
Results:
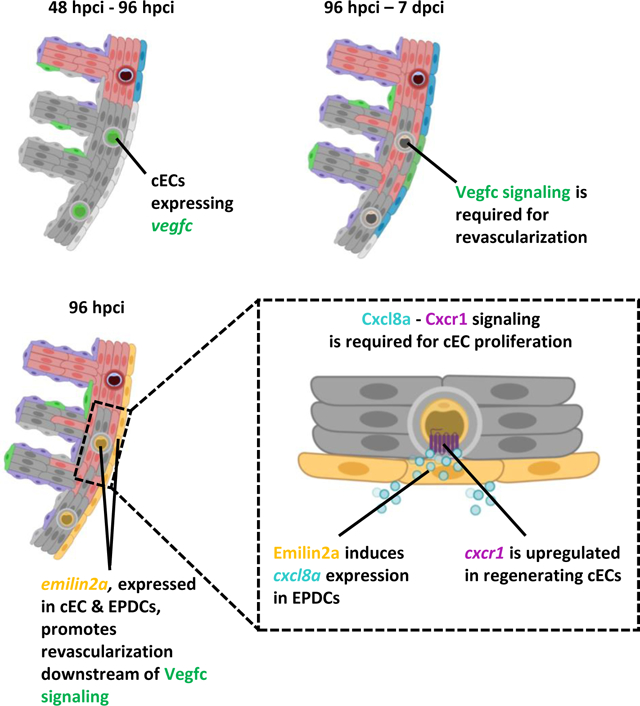
We first show that regenerating coronary endothelial cells upregulate vegfc upon cardiac injury in adult zebrafish, and that Vegfc signaling is required for their proliferation during regeneration. Notably, blocking Vegfc signaling also significantly reduces cardiomyocyte dedifferentiation and proliferation. Using transcriptomic analyses, we identified emilin2a as an effector of Vegfc signaling, and found that manipulation of emilin2a expression can modulate coronary revascularization as well as cardiomyocyte proliferation. Mechanistically, Emilin2a induces the expression of the chemokine gene cxcl8a in epicardium-derived cells, while cxcr1, the Cxcl8a receptor gene, is expressed in coronary endothelial cells. We further show that Cxcl8a-Cxcr1 signaling is also required for coronary endothelial cell proliferation during cardiac regeneration.
Conclusions:
These data show that after cardiac injury, coronary endothelial cells upregulate vegfc to promote coronary network re-establishment and cardiac regeneration. Mechanistically, Vegfc signaling upregulates epicardial emilin2a and cxcl8a expression to promote cardiac regeneration. These studies aid in understanding the mechanisms underlying coronary revascularization in zebrafish, with potential therapeutic implications to enhance revascularization and regeneration in injured human hearts.
Graphical Abstract
INTRODUCTION
Ischemic heart disease arises from reduced blood flow to the heart caused by narrowed coronary arteries. Blockage of coronary arteries results in myocardial infarction (MI), leading to the death of downstream tissues. These events lead to cardiac fibrosis and remodeling1, thereby imposing a major medical challenge given the inability of the adult mammalian heart to regenerate2. Following MI in humans, new capillaries sprout from pre-existing ones via angiogenesis to help re-oxygenate the injured tissue3,4. In addition, epicardial collaterals develop through the enlargement and widening of preformed ones to help the ischemic area in a process termed arteriogenesis4. Patients in which collateral arteries form to a larger extent exhibit improved outcome following MI in comparison to those with a lesser extent of collateral formation5. These observations have motivated researchers to develop angiogenic and arteriogenic therapies to treat MI; however, clinical trials have thus far proven ineffective6. Therefore, understanding the mechanisms that regulate coronary network re-establishment in a regenerative setting might ultimately lead to beneficial clinical outcomes. Over the last two decades, the zebrafish (Danio rerio) has been used extensively as a model organism to study cardiac regeneration, owing to the remarkable ability of its heart to regenerate7,8,9,10. Recent studies have shown that coronary revascularization of the injured tissue is essential for a complete regenerative response11. Following cardiac injury in zebrafish, coronary endothelial cells rapidly enter the cell cycle and revascularize the injured tissue11. Regenerating coronaries sprout from pre-existing vessels superficially, as well as intraventricularly, forming a scaffold for cardiomyocytes (CMs) to repopulate the injured tissue12. Blocking this process leads to reduced CM proliferation and repopulation of the injured tissue, as well as increased scarring11,12. Given the importance of coronary revascularization, identifying molecules that aid this process holds great therapeutic potential.
Recent studies have highlighted the importance of angiocrine factors in tissue homeostasis and repair13. Angiocrine molecules are secreted by endothelial cells and can have various effects on neighboring tissues12. Amongst the many angiocrine molecules, vascular endothelial growth factor C (Vegfc) displays a significant upregulation in its gene expression levels following both ventricular cryoinjury and resection14,15, and intraperitoneal administration of VEGFC can improve cardiac output and performance following MI in adult mouse hearts16. Vegfc regulates lymphangiogenesis during mouse and zebrafish development17,18,19,20,21 as well as in the adult zebrafish heart22,23,24. VEGFC also regulates blood vessel angiogenesis in different developmental settings25,26,27,28,29. For instance, addition of VEGFC stimulates angiogenic sprouting in chick embryos and mouse corneas25. In zebrafish, knockdown of vegfc using morpholinos reduces intersegmental vessel sprouting30,31. Conversely, overexpressing a constitutively active form of human VEGFC in zebrafish embryos led to intersegmental vessel hypersprouting, further highlighting a role for vegfc in regulating blood vessel angiogenesis32. Recent studies have also suggested an important role for VEGFC in regulating coronary development27,33. Cardiac ventricles from Vegfc mutant mice display a significant reduction in the coverage and branching of subepicardial coronaries27,33. Moreover, cohort studies on patients with coronary artery disease further indicate that patients with high levels of circulating VEGFC exhibit a better prognosis28. Furthermore, intramyocardial administration of VEGFC promoted the formation of collateral arteries following MI in pigs26. However, the mechanisms by which VEGFC signaling induces coronary revascularization or collateral formation following cardiac injury remain elusive.
Here, we show that after cardiac injury in adult zebrafish, Vegfc signaling promotes coronary endothelial cell (cEC) and CM proliferation by positively regulating the expression of emilin2a, a gene encoding an extracellular matrix (ECM) component. Using tissue specific gain-of-function tools, we show that Emilin2a is also a pro-regenerative factor that induces cEC proliferation and scar resolution. Mechanistically, Emilin2a induces the expression of the chemokine gene cxcl8a in epicardium-derived cells (EPDCs), while its receptor cxcr1 is expressed in regenerating cECs. Our results further indicate that Cxcl8a-Cxcr1 signaling is likewise required for coronary revascularization and scar resolution. Overall, these results highlight the role of the angiocrine molecule Vegfc in regulating matrisome-associated factors to create a more permissive environment for cardiac regeneration in adult zebrafish.
METHODS
All supporting data are available within the article and its Supplemental Materials. Detailed materials and methods can be found in the Supplemental Materials. Please see the Major Resources Table in the Supplemental Materials.
DATA AVAILABILITY
RNAseq data from this study have been deposited in the Gene Expression Omnibus (GEO) database under accession number GSE168175.
RESULTS
vegfc expression is upregulated in coronary endothelial cells after cardiac injury in zebrafish
During zebrafish embryogenesis, vegfc is expressed in the dorsal aorta and it regulates developmental angiogenesis51,52. In adult zebrafish, vegfc expression is induced after cardiac injury14,15. To gain insight into the temporal pattern of vegfc expression during cardiac regeneration, we performed RT-qPCR analyses on zebrafish ventricles at 48 and 96 (hpci) as well as at 7 days post cryoinjury (dpci). We observed that vegfc was upregulated at these time points when compared with sham operated ventricles, showing the highest expression levels at 96 hpci (Figure 1A), which coincides with the peak of cEC proliferation12. To identify which cell type(s) express vegfc during cardiac regeneration, we performed in situ hybridization on wild-type (WT) ventricles and observed vegfc expression at 96 hpci in the periphery of the injured tissue, where regenerating coronaries and EPDCs are located (Figure 1B). To better define the source of vegfc expression during cardiac regeneration, we performed RT-qPCR analyses on sorted cECs and EPDCs using the Tg(−0.8flt1:RFP) and TgBAC(tcf21:mCherry) lines, respectively. These analyses showed an upregulation in vegfc expression in sorted cECs at 96 hpci in comparison with sham operated hearts (96 hps) (Figure 1C), but not in sorted EPDCs (Figure S1A and S1B). We also analyzed vegfc expression in sorted adult CMs using the Tg(myl7:DsRed) line, and observed very low expression before and after cardiac injury (Figure S1C). Altogether, these results indicate that vegfc expression is upregulated by cECs during zebrafish cardiac regeneration.
Figure 1: Vegfc signaling promotes coronary endothelial cell proliferation after cardiac injury in zebrafish.
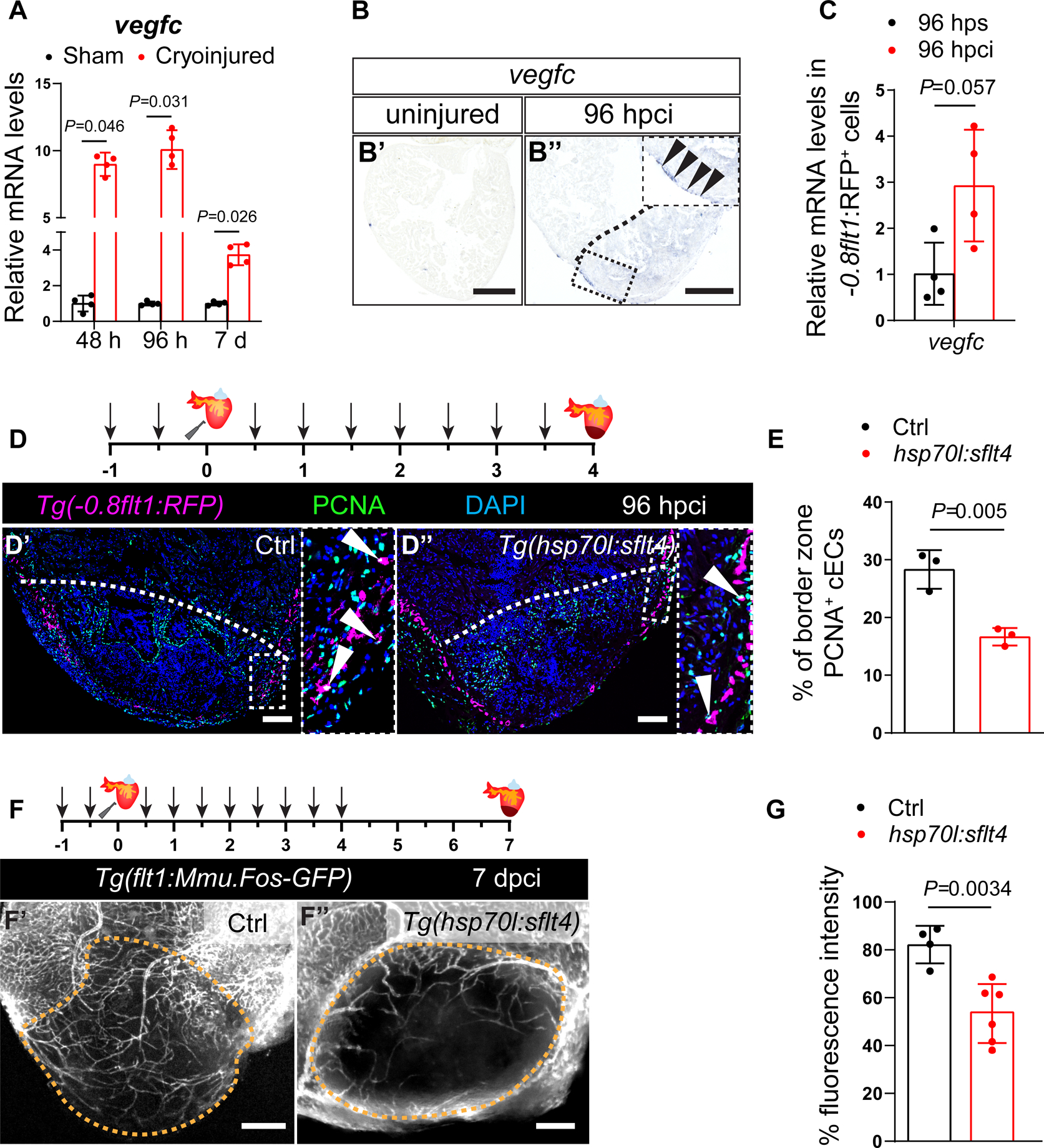
A. RT-qPCR analysis of vegfc mRNA levels at 48 and 96 hpci and 7 dpci in the injured tissue normalized to sham-operated hearts (n=4); h: hpci, d: dpci. B. in situ hybridization for vegfc expression on sections of uninjured (B’) and 96 hpci (B”) hearts. Arrowheads point to vegfc expression. C. RT-qPCR analysis of vegfc mRNA levels at 96 hpci normalized to sham-operated hearts (96 hps) in sorted −0.8flt1:RFP+ cells (cECs) (n=4). D. Schematic representation of heat shock treatments (arrows) and cardiac cryoinjury. Immunostaining of sections of cryoinjured hearts of sibling Tg(0.8flt1:RFP) (Ctrl) (D’) and Tg(hsp70l:sflt4); Tg(−0.8flt1:RFP) (D”) zebrafish at 96 hpci; sections stained for RFP (coronaries, magenta), PCNA (proliferation marker, green), and DNA (DAPI, blue). Arrowheads point to PCNA+ cECs. E. Percentage of PCNA+ cECs in the border zone of Tg(−0.8flt1RFP) (Ctrl) (n=3) and Tg(hsp70l:sflt4); Tg(−0.8flt1:RFP) (n=3) ventricles at 96 hpci. F. Schematic representation of heat shock treatments (arrows) and cardiac cryoinjury. Wholemount images of cryoinjured hearts of sibling Tg(flt1:Mmu.Fos-GFP) (Ctrl) (F’) and Tg(hsp70l:sflt4); Tg(flt1:Mmu.Fos-GFP) (F”) zebrafish at 7 dpci. G. Percentage of GFP fluorescence intensity in the injured tissue of Tg(flt1:Mmu.Fos-GFP) (Ctrl) (n=4) and Tg(hsp70l:sflt4); Tg(flt1:Mmu.Fos-GFP) (n=6) ventricles at 7 dpci. Black (B), white (D), and orange (F) dotted lines delineate the injured tissue. Statistical tests: Non-parametric Mann-Whitney test (A,C), Student’s t-test (E,G). Scale bars: 100 μm (B,D,F). Ct values of RT-qPCR data are listed in table S3.
Vegfc signaling promotes coronary endothelial cell proliferation after cardiac cryoinjury in zebrafish
vegfc upregulation in cECs after cardiac injury suggests a role during regeneration. To test this hypothesis, we utilized a Tg(hsp70l:sflt4) line. Upon heat shock, the expression of the soluble receptor sFlt4 is induced, which acts as a decoy to block Vegfc signaling29. Together with this line, and in order to visualize coronaries, we used the Tg(−0.8flt1:RFP) and Tg(flt1:Mmu.Fos-GFP) lines, which specifically label cECs in the zebrafish ventricle (Figure S1D). After optimization of the heat shock regimen (Figure S1E), we performed daily heat shocks on Tg(hsp70:sflt4); Tg(−0.8flt1:RFP) and Tg(−0.8flt1:RFP) (controls), and analyzed their ventricles (Figure 1D). We quantified cEC proliferation at 96 hpci and found that blocking Vegfc signaling resulted in a significant reduction in cEC proliferation (Figures 1D and 1E); it also resulted in reduced coronary vessel coverage of the injured tissue at 7 dpci (Figures 1F and 1G), a stage when the injured tissue is extensively repopulated by regenerating coronaries in control zebrafish11. In addition, we observed a decrease in the expression levels of cxcr4a and apln (Figure S1F), two genes that have been shown to regulate coronary regeneration in zebrafish12.
During cardiac regeneration, CMs dedifferentiate and proliferate to replenish the injured tissue53,54,55,56, and this process is regulated in part by cECs11,12. Hence, we reasoned that altering revascularization by blocking Vegfc signaling could lead to defects in CM regeneration. To test this possibility, we performed heat shock treatments on cryoinjured Tg(hsp70l:sflt4) and non-transgenic siblings until 96 hpci, when revascularization is very active, and analyzed CM dedifferentiation and proliferation at 7 dpci. We found that blocking Vegfc signaling resulted in a reduction in both CM dedifferentiation and proliferation in the injury border zone (Figures 2A–D and S1G). To gain more insight into these CM phenotypes, we analyzed the expression of vegfr1, vegfr2 and vegfr3 in sorted CMs and observed very low or no detectable mRNA levels before or after injury (Figures S1H and S1I), indicating that they are unlikely due to reduced Vegfc signaling in CMs. Next, to test whether reducing coronary revascularization by blocking Vegfc signaling impacts scar resolution, we induced the expression of sflt4 until 96 hpci, when cEC proliferation is at its peak12, and analyzed the scar area at 30 and 90 dpci. We found that ventricles in which Vegfc signaling was blocked displayed a significantly larger scar area when compared with non-transgenic siblings subjected to the same treatment at both 30 (Figures S1J and S1K) and 90 (Figures 2E and 2F) dpci, in line with recent findings using vegfc+/um18 zebrafish22 as well as Tg(hsp70l:sflt4) zebrafish23.
Figure 2: Blocking Vegfc signaling leads to reduced cardiomyocyte dedifferentiation and proliferation after cardiac injury in zebrafish.
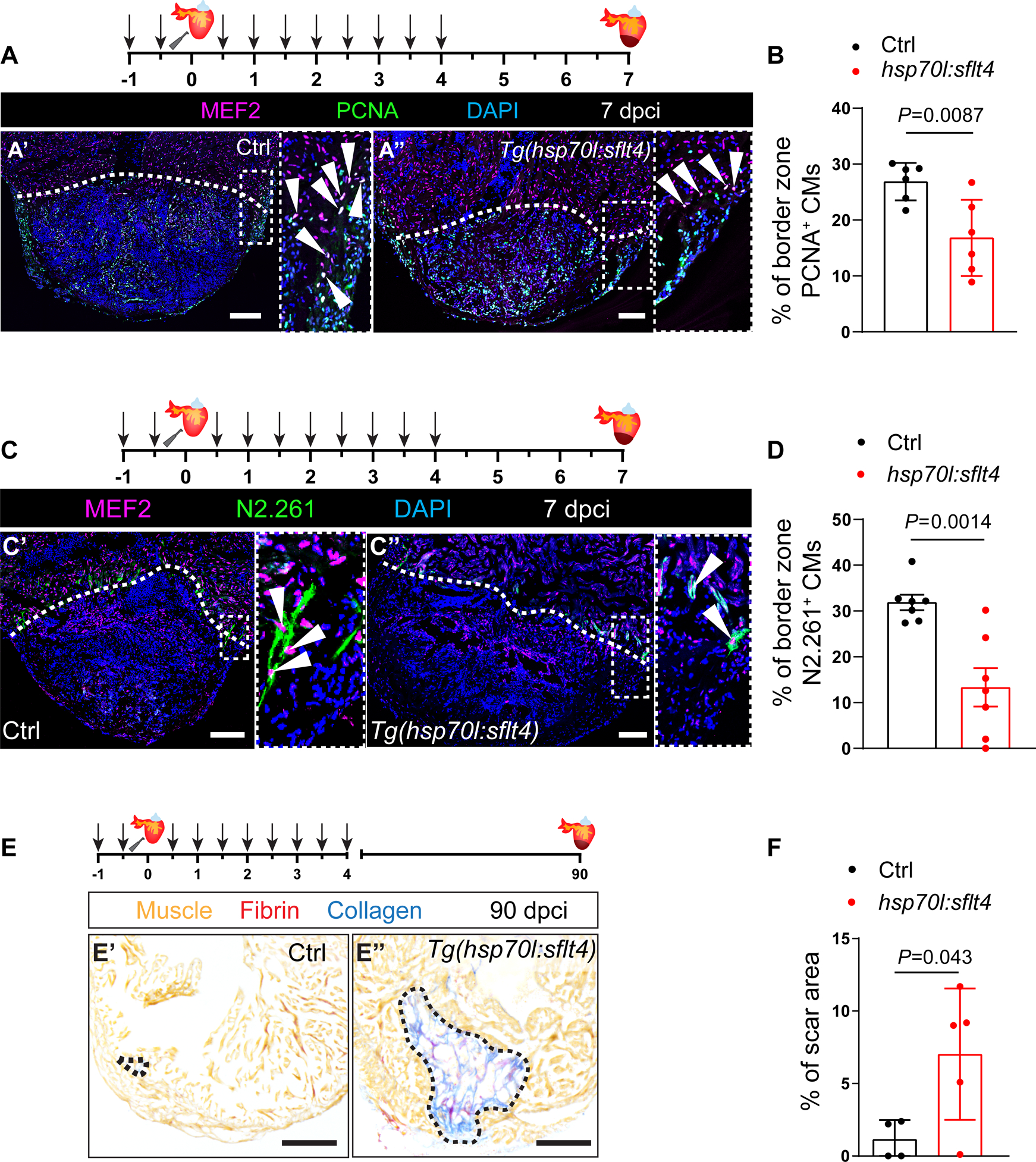
A. Schematic representation of heat shock treatments (arrows) and cardiac cryoinjury. Immunostaining of sections of cryoinjured hearts of non-transgenic sibling (Ctrl) (A’) and Tg(hsp70l:sflt4) (A”) zebrafish at 7 dpci; sections stained for MEF2 (CMs, magenta), PCNA (proliferation marker, green), and DNA (DAPI, blue). Arrowheads point to PCNA+ CMs. B. Percentage of PCNA+ CMs in the border zone of non-transgenic sibling (Ctrl) (n=6) and Tg(hsp70l:sflt4) (n=6) ventricles at 7 dpci. C. Schematic representation of heat shock treatments (arrows) and cardiac cryoinjury. Immunostaining of sections of cryoinjured hearts of non-transgenic sibling (Ctrl) (C’) and Tg(hsp70l:sflt4) (C”) zebrafish at 7 dpci; sections stained for MEF2 (CMs, magenta), N2.261 (embryonic myosin heavy chain, green), and DNA (DAPI, blue). Arrowheads point to N2.261+ CMs. D. Percentage of N2.261+ CMs in the border zone of non-transgenic sibling (Ctrl) (n=7) and Tg(hsp70l:sflt4) (n=7) ventricles at 7 dpci. E. Schematic representation of heat shock treatments (arrows) and cardiac cryoinjury. AFOG staining of sections of non-transgenic sibling (Ctrl) (E’) and Tg(hsp70l:sflt4) (E”) ventricles at 90 dpci. Orange, Muscle; red, Fibrin; blue, Collagen. F. Percentage of scar area relative to ventricular area in non-transgenic sibling (Ctrl) (n=4) and Tg(hsp70l:sflt4) (n=5) ventricles at 90 dpci. White (A,C), and black (E) dotted lines delineate the injured tissue. Statistical test: Student’s t-test (B,D,F). Scale bars: 100 μm (A,C,E).
Vegfc regulates lymphangiogenesis during development21,57 and cardiac regeneration in zebrafish22,23. To rule out that the regeneration phenotypes observed upon global Vegfc signaling interference are due to lymphatic defects, we performed injuries on 3 months old (~27–28 mm in length) zebrafish, in which, based on reporter expression, lymphatics are only observed in the bulbus arteriosus and not yet present in the ventricle22,23. To further test for the presence of lymphatic vessels at these stages and during regeneration, we performed intramyocardial injections of Qdots which are taken up and cleared by lymphatics23. These experiments were performed in animals carrying the Tg(lyve1b:DsRed) transgene which marks lymphatic vessels. Using this approach, we confirmed the absence of lymphatics in untouched 3 months old zebrafish ventricles and during regeneration experiments in WT animals as well as after blockade of Vegfc signaling (Figures S2A and S2B). Similar to our previous experiments with older (i.e., 8 months old) animals (Figures 1D and 1E), we observed a significant decrease in cEC proliferation at 96 hpci in 3 months old Tg(hsp70l:sflt4) zebrafish when compared with controls (Figures S2C and S2D). These data indicate that reduced revascularization upon blocking Vegfc signaling is independent of the presence or absence of ventricular lymphatics. However, in order to work with zebrafish devoid of ventricular lymphatics, we used animals 27–28 mm in length for the rest of the study.
Since Flt4 (Vegfr3) is the main receptor for both Vegfc and Vegfd58,59, it is possible that the cardiac regeneration defects in Tg(hsp70l:sflt4) zebrafish are at least partially due to alterations in Vegfd signaling. To test this possibility, we performed cryoinjuries on WT, hypomorphic vegfc−/− 32 and vegfd−/− 22 zebrafish, and analyzed cEC proliferation in injured ventricles at 96 hpci. Notably, cEC proliferation was reduced in vegfc−/− but not vegfd−/− ventricles (Figures S2E and S2F), consistent with previous findings that vegfd−/− zebrafish do not exhibit cardiac regeneration defects22,24. These data indicate that defects observed in cEC regeneration upon sflt4 overexpression are due to blocked Vegfc signaling, although one cannot completely rule out an effect on other Vegf signaling pathways. Overall, these findings indicate that blocking Vegfc signaling impairs coronary revascularization as well as CM dedifferentiation and proliferation during cardiac regeneration in zebrafish.
Vegfc signaling induces expression of the extracellular matrix gene emilin2a to promote coronary revascularization
To gain mechanistic insight into how Vegfc signaling regulates coronary regeneration, we performed transcriptomic analyses of cryoinjured ventricles after blocking Vegfc signaling using the Tg(hsp70l:sflt4) line and non-transgenic siblings as controls. We extracted injured ventricles at 24 hpci, and after comparing their transcriptome (Figure 3A), cross-referenced the significantly downregulated genes with those upregulated during WT cardiac regeneration in zebrafish, from a published dataset15 (Figure S3A). We identified 10 candidate genes (Figure S3A). Next, we performed RT-qPCR analysis to check their expression levels in WT and Tg(hsp70l:sflt4) ventricles at 24 hpci (time point when the RNAseq was performed) and 96 hpci (time point when we observed the reduced cEC phenotype) (Figures S3B and S3C). Amongst these candidate genes, emilin2a was the only one to display a consistent and significant downregulation when blocking Vegfc signaling (Figures 3B, S3B and S3C). To test whether emilin2a is a target of Vegfc signaling, we injected vegfc mRNA into one-cell stage WT embryos and found that emilin2a expression was significantly increased at 48 hpf (Figure S3D). As Vegfc signaling regulates endothelial sprouting51,52, we reasoned that the changes in emilin2a expression could occur as a consequence of reduced revascularization. To test this possibility, we analyzed emilin2a expression in ventricles from Tg(hsp70l:dn-vegfaa) zebrafish, a heat shock inducible transgenic line expressing a dominant negative form of Vegfaa that can block revascularization11,34. We performed 1 hour heat shocks every day for 3 days on WT, Tg(hsp70l:sflt4) and Tg(hsp70l:dn-vegfaa) zebrafish, and analyzed emilin2a expression by RT-qPCR on uninjured ventricles. While emilin2a expression was significantly downregulated in Tg(hsp70l:sflt4) ventricles, it remained unchanged in Tg(hsp70l:dn-vegfaa) ventricles (Figure S3E), further indicating that emilin2a is a target of Vegfc signaling, and not merely an endothelial marker. To explore the possibility that the VEGFC-EMILIN2 axis is also at play in human endothelial cells, we knocked down VEGFC in cultured human umbilical vein endothelial cells (HUVECs) and observed a significant reduction in EMILIN2 expression (Figure 3C). Altogether, these results indicate that EMILIN2 is an effector of VEGFC signaling in zebrafish as well as in human ECs.
Figure 3: Vegfc signaling stimulates emilin2a expression to promote coronary endothelial cell proliferation in regenerating zebrafish hearts.
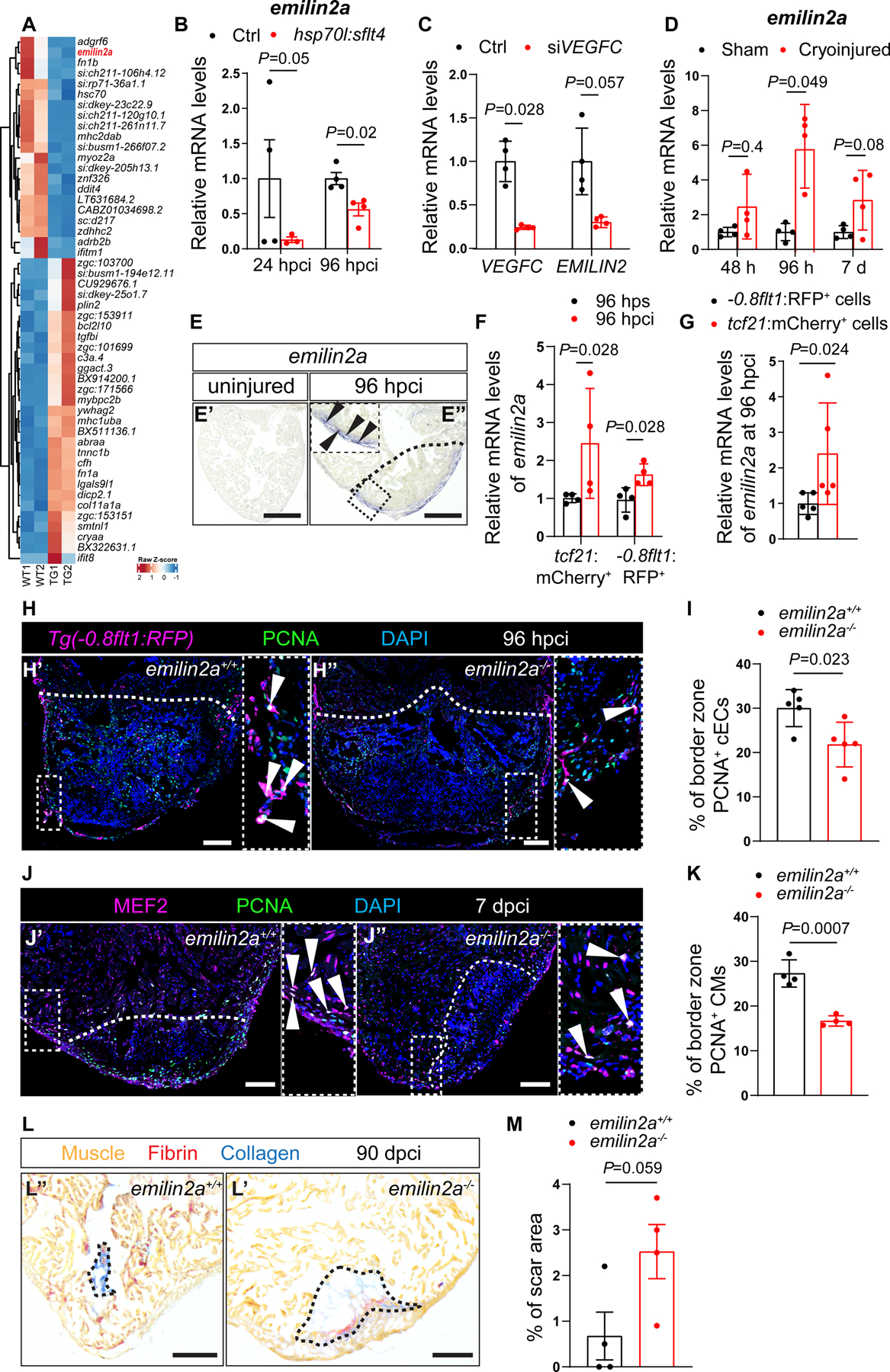
A. Heat map showing differentially expressed genes in non-transgenic sibling (WT) versus Tg(hsp70l:sflt4) hearts at 24 hpci. Genes are broadly divided into two clusters: one that contains genes that are upregulated in Tg(hsp70l:sflt4) ventricles and the other that contains genes that are downregulated in Tg(hsp70l:sflt4) ventricles. B. RT-qPCR analysis of emilin2a mRNA levels at 24 and 96 hpci in Tg(hsp70l:sflt4) ventricles normalized to non-transgenic sibling (Ctrl) hearts (n=4). C. RT-qPCR analysis of VEGFC and EMILIN2 mRNA levels in HUVECs after siVEGFC treatment (n=4) in comparison with scrambled control (n=4). D. RT-qPCR analysis of emilin2a mRNA levels at 48 and 96 hpci and 7 dpci in injured tissue normalized to sham-operated hearts (n=4); h: hpci, d: dpci. E. in situ hybridization for emilin2a expression on sections of uninjured (E’) and 96 hpci (E”) hearts. Arrowheads point to emilin2a expression. F. RT-qPCR analysis of emilin2a mRNA levels in sorted tcf21:mCherry+ cells (EPDCs) (n=4) and sorted −0.8flt1:RFP+ cells (cECs) (n=4) at 96 hpci normalized to 96 hps. G. RT-qPCR analysis of emilin2a mRNA levels in sorted tcf21:mCherry+ cells (EPDCs) (n=5) normalized to sorted −0.8flt1:RFP+ cells (cECs) (n=5) at 96 hpci. H. Immunostaining of sections of cryoinjured hearts of Tg(flt1:Mmu.Fos-GFP); emilin2a+/+ (H’) and Tg(flt1:Mmu.Fos-GFP); emilin2a−/− (H”) sibling zebrafish at 96 hpci; sections stained for GFP (coronaries, magenta), PCNA (proliferation marker, green), and DNA (DAPI, blue). Arrowheads point to PCNA+ cECs. I. Percentage of PCNA+ cECs in the border zone of Tg(flt1:Mmu.Fos-GFP); emilin2a+/+ (n=5) and Tg(flt1:Mmu.Fos-GFP); emilin2a−/− (n=5) ventricles at 96 hpci. J. Immunostaining of sections of cryoinjured hearts of emilin2a+/+ (J’) and emilin2a−/− (J”) sibling zebrafish at 7 dpci; sections stained for MEF2 (CMs, magenta), PCNA (proliferation marker, green), and DNA (DAPI, blue). Arrowheads point to PCNA+ CMs. K. Percentage of PCNA+ CMs in the border zone of emilin2a+/+ (n=4) and emilin2a−/− (n=4) ventricles at 7 dpci. L. AFOG staining of sections of emilin2a+/+ (L’) and emilin2a−/−(L”) ventricles at 90 dpci. Orange, Muscle; red, Fibrin; blue, Collagen. M. Percentage of scar area relative to ventricular area in emilin2a+/+ (n=4) and emilin2a−/− (n=4) ventricles at 90 dpci. Black (E,L), and white (H,J) dotted lines delineate the injured tissue. Statistical tests: Non-parametric Mann-Whitney test (B,C,D,F,G), Student’s t-test (I,K,M). Scale Bars: 100 μm (E,H,J,L). Ct values of RT-qPCR data are listed in table S3.
EMILIN2 is a glycoprotein that belongs to the EMI domain endowed (EDEN) superfamily of proteins which consists of EMILIN1, EMILIN2, MMRN1 and MMRN260. It has previously been shown that emilin2a is expressed in the dorsal aorta of zebrafish embryos at 24 hpf 61; however, its role in heart regeneration remains unknown.
We next analyzed the expression pattern of emilin2a at different time points after cryoinjury and found that emilin2a was strongly upregulated in the injured tissue at 96 hpci (Figure 3D), which coincides with the peak of vegfc expression (Figure 1A) and cEC proliferation12. Using in situ hybridization on ventricular sections, we observed emilin2a expression in the periphery of the injured tissue at 96 hpci, similar to vegfc expression, while no expression was detectable in uninjured ventricles (Figure 3E). We further analyzed this expression by RT-qPCR on sorted cECs (−0.8flt1:RFP+) and sorted EPDCs (tcf21:mCherry+) and found emilin2a expression induced in both cell types after injury (Figure 3F), while very low expression was observed in CMs before and after cardiac injury (Figure S3F). We also compared emilin2a expression between both cells types (EPDCs and cECs) and found that EPDCs expressed higher levels than cECs at 96 hpci (Figure 3G).
Previous studies have shown that stimulating HUVECs with recombinant EMILIN2 enhanced their migration in a scratch assay62. We also observed a significant upregulation of EMILIN2 at 6 hours after scratch (Figure S3G), when cells are actively migrating. Furthermore, the loss of EMILIN2 significantly reduced microvessel sprouting from cultured mouse aortic rings, suggesting an angiogenic role63,64. These data led us to hypothesize that Emilin2a regulates different aspects of revascularization during cardiac regeneration in zebrafish. To test this hypothesis, we generated an emilin2a mutant allele using the CRISPR/Cas9 technology. The zebrafish genome contains two emilin2 genes, emilin2a and emilin2b61. Because of the existence of emilin2b in the zebrafish genome, we generated a full locus deletion (FLD) allele in order to avoid possible transcriptional adaptation65,66 (Figures S4A–S4E). emilin2a FLD mutants lack emilin2a expression (Figure S4F) and display no obvious defects as larvae (Figure S4G). Furthermore, the gross morphology and coronary coverage of emilin2a−/− adult ventricles were comparable with those of emilin2a+/+ ventricles when uninjured (Figure S4H). Upon cryoinjury, emilin2a−/− ventricles displayed a significant reduction in cEC proliferation at 96 hpci (Figures 3H and 3I) and impaired coronary vessel coverage at 7 dpci (Figure S5A and S5B), while devoid of lymphatics (Figure S5C). In addition, emilin2a−/− ventricles exhibited a significant reduction in CM proliferation at 7 dpci (Figures 3J and 3K) and displayed a larger scar at 30 (Figure S5D and S5E) and 90 (Figures 3L and 3M) dpci. Altogether, these results indicate that emilin2a is required for cEC proliferation and scar resolution during cardiac regeneration in zebrafish.
Previously published transcriptomic analyses have shown that, unlike in zebrafish, emilin2a is not upregulated following cryoinjury in medaka (Oryzias latipes)15, a teleost species that cannot regenerate its heart and lacks a coronary network67 (Figure S5F). Furthermore, our data showing that emilin2a mutant ventricles retain a larger fibrotic scar at 90 dpci as well as the HUVEC data mentioned earlier62 (Figure S3G) led us to hypothesize that Emilin2a can enhance cardiac regeneration. To test this hypothesis, we generated a transgenic zebrafish line expressing emilin2a under a heat shock promoter. We quantified cEC proliferation at 96 hpci in Tg(hsp70l:emilin2a); Tg(−0.8flt1:RFP) ventricles in comparison with controls (i.e., Tg(−0.8flt1:RFP) zebrafish subjected to the same treatment) (Figure 4A). We found that overexpressing emilin2a significantly increased cEC proliferation at 96 hpci (Figure 4A and 4B) and marginally increased coronary vessel coverage at 7 dpci (Figure S6A and S6B), despite the absence of ventricular lymphatics (Figure S6C). Moreover, overexpressing emilin2a resulted in an increase in CM proliferation at 7 dpci (Figure 4C and 4D) and a reduced scar area at 30 dpci (Figure 4E and 4F), in comparison with non-transgenic siblings.
Figure 4: emilin2a overexpression increases coronary endothelial cell proliferation and can rescue Vegfc signaling block in regenerating zebrafish hearts.
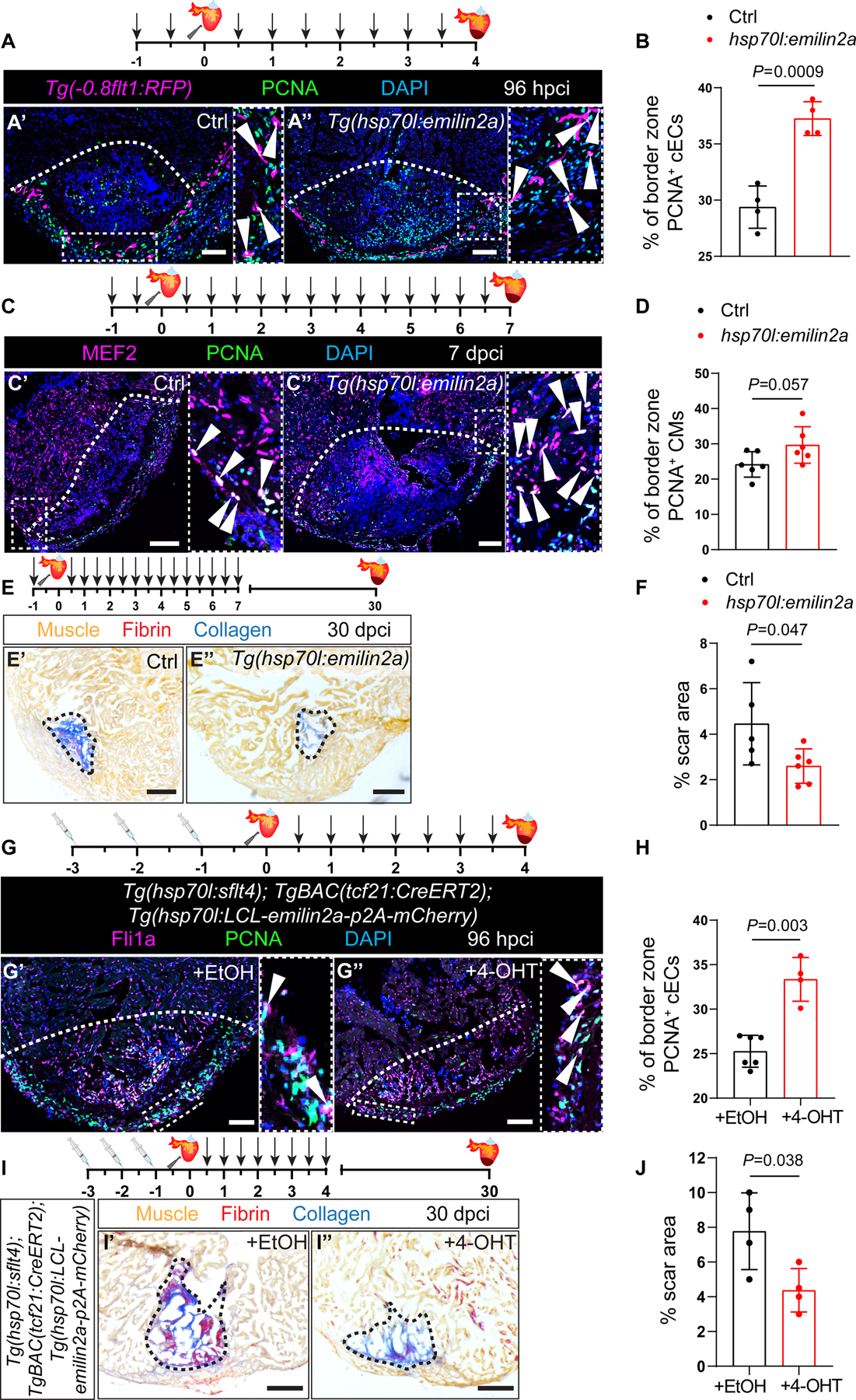
A. Schematic representation of heat shock treatments (arrows) and cardiac cryoinjury. Immunostaining of sections of cryoinjured hearts of Tg(0.8flt1:RFP) (Ctrl) (A’) and Tg(hsp70l:emilin2a); Tg(−0.8flt1:RFP) (A”) sibling zebrafish at 96 hpci; sections stained for RFP (coronaries, magenta), PCNA (proliferation marker, green), and DNA (DAPI, blue). Arrowheads point to PCNA+ cECs. B. Percentage of PCNA+ cECs in the border zone of Tg(−0.8flt1RFP) (Ctrl) (n=4) and Tg(hsp70l:emilin2a); Tg(−0.8flt1:RFP) ventricles (n=4) at 96 hpci. C. Schematic representation of heat shock treatments (arrows) and cardiac cryoinjury. Immunostaining of sections of cryoinjured hearts of non-transgenic sibling (Ctrl) (C’) and Tg(hsp70l:emilin2a) (C”) zebrafish at 7 dpci; sections stained for MEF2 (CMs, magenta), PCNA (proliferation marker, green), and DNA (DAPI, blue). Arrowheads point to PCNA+ CMs. D. Percentage of PCNA+ CMs in the border zone of non-transgenic sibling (Ctrl) (n=6) and Tg(hsp70l:emilin2a) (n=6) ventricles at 7 dpci. E. Schematic representation of heat shock treatments (arrows) and cardiac cryoinjury. AFOG staining of sections of non-transgenic sibling (Ctrl) (E’) and Tg(hsp70l:emilin2a) (E”) ventricles at 30 dpci. Orange, Muscle; red, Fibrin; blue, Collagen. F. Percentage of scar area relative to ventricular area in non-transgenic sibling (Ctrl) (n=5) and Tg(hsp70l:emilin2a) (n=6) ventricles at 30 dpci. G. Schematic representation of Ethanol (EtOH) or 4-hydroxytamoxifen (4-OHT) injections followed by cardiac cryoinjury and heat shock treatments (arrows). Immunostaining of sections of cryoinjured hearts of Tg(hsp70l:sflt4); TgBAC(tcf21:CreERT2); Tg(hsp70l:LCL-emilin2a-p2A-mCherry) zebrafish injected with EtOH (G’) or 4-OHT (G”) at 96 hpci; sections stained for Fli1a (endothelial cells, magenta), PCNA (proliferation marker, white), and DNA (DAPI, blue). Arrowheads point to PCNA+ cECs. H. Percentage of PCNA+ cECs in the border zone of Tg(hsp70l:sflt4); TgBAC(tcf21:CreERT2); Tg(hsp70l:LCL-emilin2a-mCherry) at 96 hpci injected with EtOH (n=6) or 4-OHT (n=4). I. Schematic representation of Ethanol (EtOH) or 4-hydroxytamoxifen (4-OHT) injections followed by cardiac cryoinjury and heat shock treatments (arrows). AFOG staining of sections of cryoinjured hearts of Tg(hsp70l:sflt4); TgBAC(tcf21:CreERT2); Tg(hsp70l:LCL-emilin2a-p2A-mCherry) zebrafish injected with EtOH (I’) or 4-OHT (I”) at 30 dpci. Orange, Muscle; red, Fibrin; blue, Collagen. J. Percentage of scar area relative to ventricular area in Tg(hsp70l:sflt4); TgBAC(tcf21:CreERT2); Tg(hsp70l:LCL-emilin2a-mCherry) at 30 dpci injected with EtOH (n=4) or 4-OHT (n=4). White (A,C,G), and Black (E,I) dotted lines delineate the injured tissue. Statistical test: Student’s t-test (B,D,F,H,J). Scale Bars: 100 μm (A,C,G), 200 μm (E,I).
In view of these results, we reasoned that the coronary revascularization phenotype in Tg(hsp70l:sflt4) zebrafish hearts might be rescued by overexpressing emilin2a. We utilized the HOT-CRE system68 and generated a Tg(hsp70l:LCL-emilin2a-p2A-mCherry) zebrafish line, which in combination with the TgBAC(tcf21:CreERT2) line38, allows spatial and temporal control of emilin2a expression in EPDCs (Figure S6D–S6G). Next, we overexpressed emilin2a in EPDCs while blocking Vegfc signaling and observed a significant increase in cEC proliferation in comparison with hearts in which emilin2a expression was not induced (Figures 4G and 4H). Notably, we also observed an increase in CM proliferation (Figure S6H and S6I) and a significant decrease in scar area at 30 dpci (Figures 4I and 4J). Altogether, these findings indicate that emilin2a, whose expression is stimulated by Vegfc signaling in EPDCs and cECs, is required for, and can enhance, heart regeneration.
emilin2a induces cxcl8a expression in epicardium-derived cells
Previous studies indicate that in HUVECs and human gastric tumors, EMILIN2 exerts its angiogenic role via CXCL862,64, a chemokine that signals through the G-protein coupled receptors CXCR1 and CXCR2 to regulate various aspects of wound healing69. Notably, other studies have highlighted the ability of CXCL8 to act as a pro-angiogenic factor in in vitro settings70,71,72. Thus, Emilin2a might induce cxcl8a expression during cardiac regeneration in zebrafish. To test this possibility, we first analyzed cxcl8a expression levels in emilin2a−/− ventricles at 96 hpci and observed a significant reduction when compared with emilin2a+/+ ventricles (Figure 5A). Moreover, we observed cxcl8a upregulation in Tg(hsp70l:emilin2a) ventricles at 96 hpci when compared with non-transgenic siblings (Figure S7A). To further test whether cxcl8a is downstream of Vegfc signaling, we analyzed cxcl8a expression after blocking Vegfc signaling and found a significant downregulation in Tg(hsp70l:sflt4) ventricles at 96 hpci (Figure S7B). Conversely, injecting vegfc mRNA into one-cell stage zebrafish embryos resulted in an increase in cxcl8a expression (Figure S7C). In the regenerating heart, we observed cxcl8a expression in cells covering the injured tissue (Figure 5B). To determine the source of this expression, we performed RT-qPCR analyses on sorted EPDCs and sorted cECs, and found increased cxcl8a expression in regenerating EPDCs (Figure 5C), but not cECs, at 96 hpci when compared with sham operated controls (Figures S7D and S7E). Overall, these findings indicate that downstream of Vegfc signaling, the ECM protein Emilin2a induces cxcl8a expression, and that EPDCs are a source of Cxcl8a during zebrafish cardiac regeneration.
Figure 5: Emilin2a induces cxcl8a expression in EPDCs to promote coronary endothelial cell proliferation after cardiac injury in zebrafish.
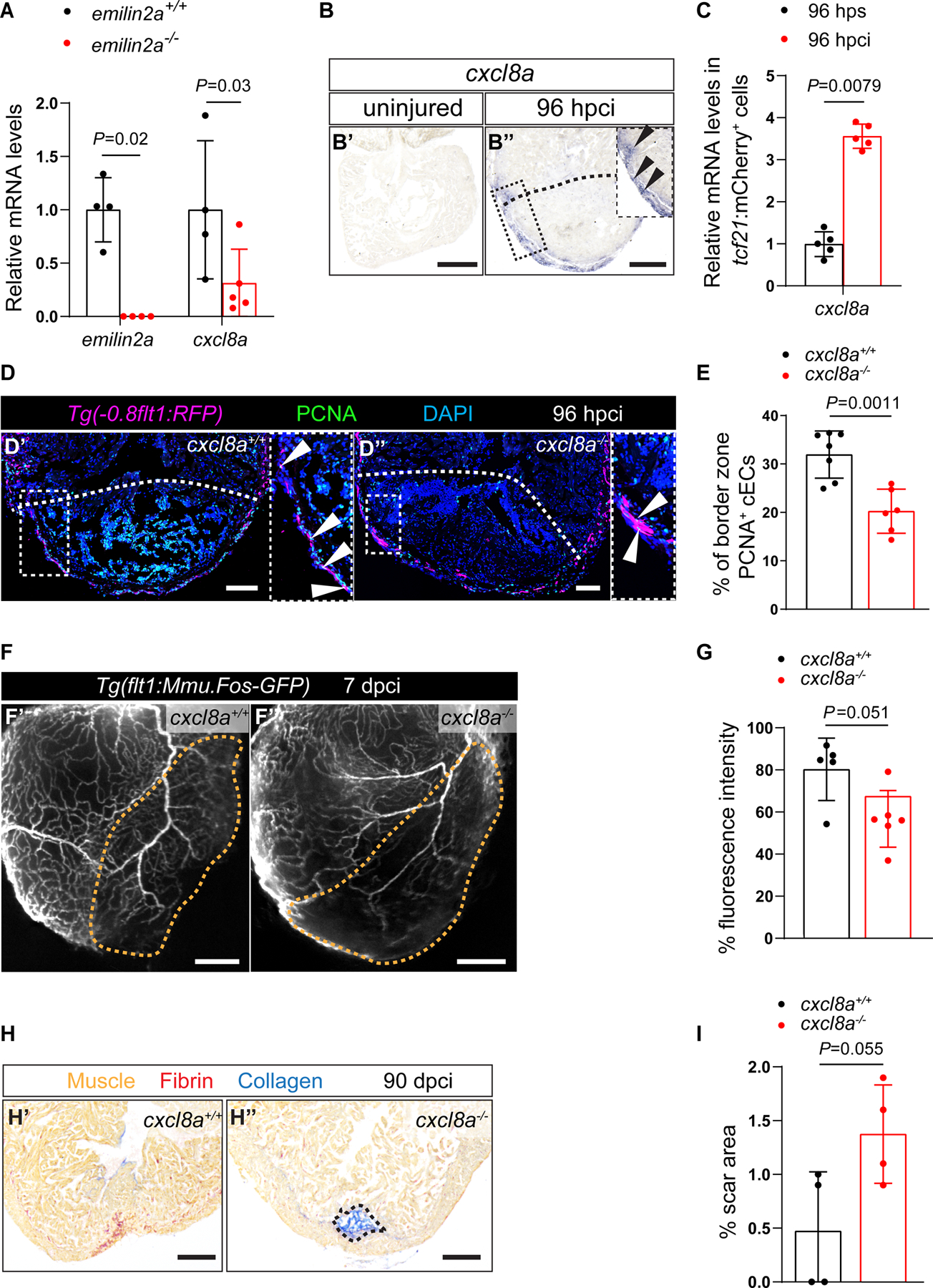
A. RT-qPCR analysis of emilin2a and cxcl8a mRNA levels in emilin2a−/− ventricles normalized to emilin2a+/+ ventricles at 96 hpci (n=4–5). B. in situ hybridization for cxcl8a expression on sections of uninjured (B’) and 96 hpci (B”) hearts. Arrowheads point to cxcl8a expression. C. RT-qPCR analysis of cxcl8a mRNA levels in sorted tcf21:mCherry+ (EPDCs) (n=5) at 96 hpci normalized to 96 hps. D. Immunostaining of sections of cryoinjured hearts of Tg(flt1:Mmu.Fos-GFP); cxcl8a+/+ (D’) and Tg(flt1:Mmu.Fos-GFP); cxcl8a−/− (D”) sibling zebrafish at 96 hpci; sections stained for GFP (coronaries, magenta), PCNA (proliferation marker, green), and DNA (DAPI, blue). Arrowheads point to PCNA+ cECs. E. Percentage of PCNA+ cECs in the border zone of cxcl8a+/+ (n=7) and cxcl8a−/− (n=6) ventricles at 96 hpci. F. Wholemount images of ventricles of Tg(flt1:Mmu.Fos-GFP); cxcl8a+/+ (F’) and Tg(flt1:Mmu.Fos-GFP); cxcl8a−/− (F”) sibling zebrafish at 7 dpci. G. Percentage of GFP fluorescence intensity in the injured tissue of Tg(flt1:Mmu.Fos-GFP); cxcl8a+/+ (n=5) and Tg(flt1:Mmu.Fos-GFP); cxcl8a−/− (n=6) ventricles at 7 dpci. H. AFOG staining of sections of cxcl8a+/+ (H’) and cxcl8a−/− (H”) ventricles at 90 dpci. Orange, Muscle; red, Fibrin; blue, Collagen. I. Percentage of scar area relative to ventricular area in cxcl8a+/+ (n=4) and cxcl8a−/− (n=4) ventricles at 90 dpci. Black (B,H), white (D) and orange (F) dotted lines delineate the injured tissue. Statistical tests: Non-parametric Mann-Whitney test (A,C,G), Student’s t-test (E,I). Scale Bars: 200 μm (B,F,H), 100 μm (D). Ct values of RT-qPCR data are listed in table S3.
Cxcl8a-Cxcr1 signaling promotes coronary revascularization.
In view of the injury-induced upregulation of cxcl8a expression, we wanted to test whether Cxcl8a plays an important role during cardiac regeneration, and thus generated cxcl8a mutants using the CRISPR/Cas9 technology (Figures S8A and S8B). In silico analysis predicts that the protein encoded by the mutant transcript lacks the C-terminal 31 amino acids which are essential for Cxcl8a dimerization and receptor binding73. We analyzed cEC proliferation in Tg(flt1:Mmu.Fos-GFP); cxcl8a+/+ and Tg(flt1:Mmu.Fos-GFP); cxcl8a−/− siblings at 96 hpci, and found a significant reduction in the mutants (Figures 5D and 5E). Moreover, cxcl8a−/− ventricles displayed a significant reduction in coronary vessel coverage at 7 dpci (Figures 5F and 5G), while devoid of lymphatics (Figure S8C). In addition, cxcl8a−/− ventricles displayed a marginally larger scar at 30 (Figures S8D and S8E), and 90 (Figures 5H and 5I) dpci. Altogether, these data indicate that Cxcl8a plays an important role in coronary revascularization during cardiac regeneration in zebrafish.
In view of the reduced cEC proliferation phenotype in cxcl8a mutants, we reasoned that its receptor(s) might be expressed in cECs during heart regeneration. Indeed, we detected cxcr1 expression at the periphery of the injured tissue at 96 hpci by in situ hybridization (Figure 6A). We also observed a significant upregulation of cxcr1 expression in sorted cECs at 96 hpci, while no cxcr2 expression was detected (Figure 6B). Importantly, pharmacological inhibition (with SB225002)74 of Cxcr1 reduced coronary vessel coverage at 7 dpci (Figures S8F and S8G). Furthermore, in order to investigate the role of cxcr1 during cardiac regeneration, we performed cryoinjuries on cxcr1−/− ventricles and observed a significant reduction in cEC proliferation at 96 hpci (Figures 6C and 6D), as well as defective coronary vessel coverage at 7 dpci (Figures 6E and 6F). Next, we checked whether cxcr1−/− ventricles were able to regenerate. Although they displayed similar scar size as cxcr1+/+ sibling controls at 30 dpci (Figures S8H and S8I), cxcr1−/− ventricles displayed significantly larger scars at 90 dpci (Figures 6G and 6H). Altogether, these findings suggest that Cxcl8a signals through Cxcr1 in regenerating cECs to promote revascularization and support cardiac regeneration in zebrafish.
Figure 6: Cxcl8a-Cxcr1 signaling promotes coronary endothelial cell proliferation after cardiac injury in zebrafish.
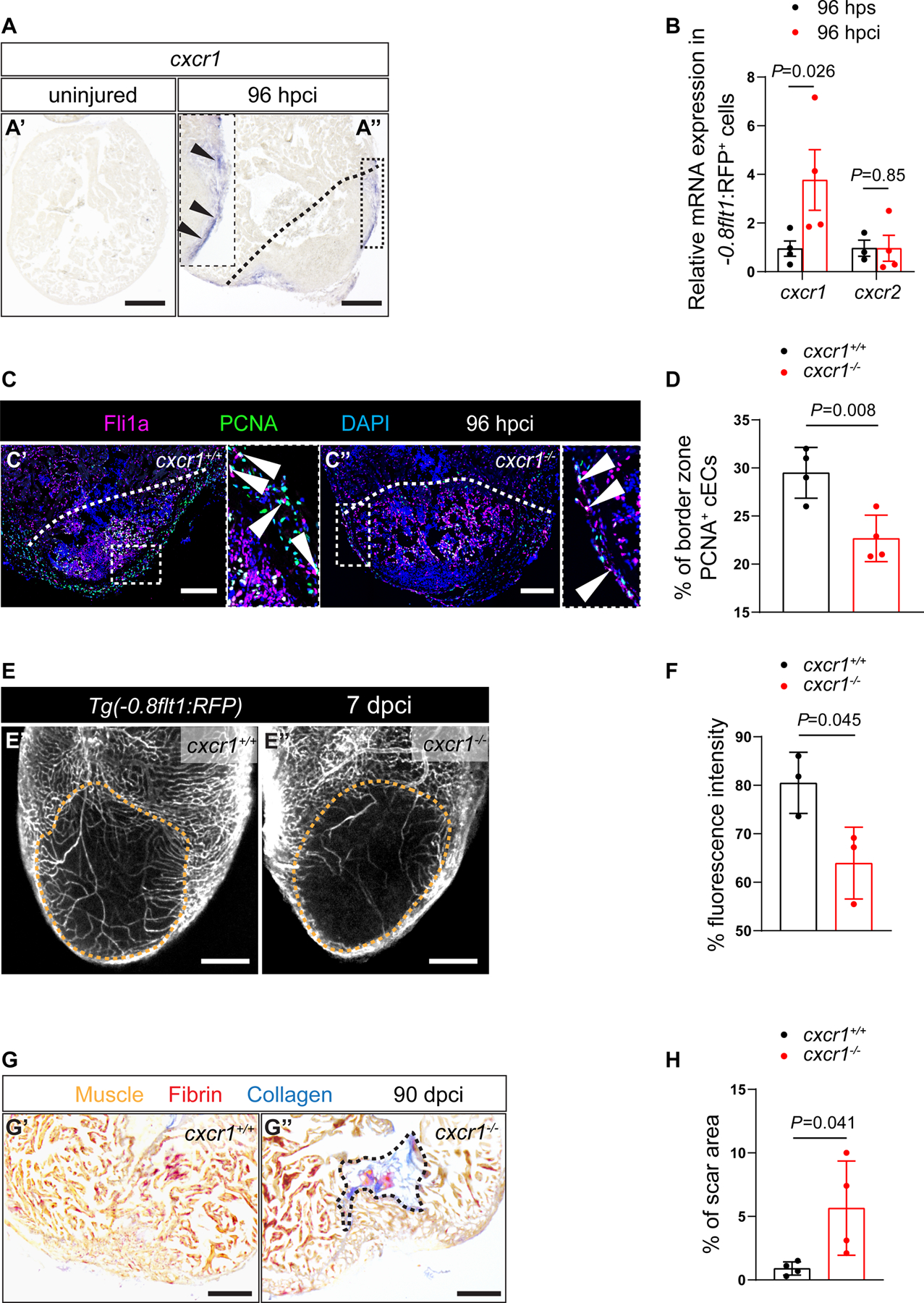
A. in situ hybridization for cxcr1 expression on sections of uninjured (A’) and 96 hpci (A”) hearts. Arrowheads point to cxcr1 expression. B. RT-qPCR analysis of cxcr1 and cxcr2 mRNA levels in sorted −0.8flt1:RFP+ cells (cECs) (n=3–4) at 96 hpci normalized to 96 hps. C. Immunostaining of sections of cryoinjured hearts of cxcr1+/+ (C’) and cxcr1−/− (C”) sibling zebrafish at 96 hpci; sections stained for Fli1a (endothelial cells, magenta), PCNA (proliferation marker, green), and DNA (DAPI, blue). Arrowheads point to PCNA+ cECs. D. Percentage of PCNA+ cECs in the border zone of cxcr1+/+ (n=4) and cxcr1−/− (n=4) ventricles at 96 hpci. E. Wholemount images of ventricles of Tg(−0.8flt1:RFP); cxcr1+/+ (E’) and Tg(−0.8flt1:RFP); cxcr1−/− (E”) sibling zebrafish at 7 dpci. F. Percentage of RFP fluorescence intensity in the injured tissue of Tg(−0.8flt1:RFP); cxcr1+/+ (n=3) and Tg(−0.8flt1:RFP); cxcr1−/− (n=3) ventricles at 7 dpci. G. AFOG staining of sections of cxcr1+/+ (G’) and cxcr1−/− (G”) ventricles at 90 dpci. Orange, Muscle; red, Fibrin; blue, Collagen. H. Percentage of scar area relative to ventricular area in cxcr1+/+ (n=4) and cxcr1−/− (n=4) ventricles at 90 dpci. Black (A,G), white (C) and orange (E) dotted lines delineate the injured tissue. Statistical tests: Non-parametric Mann-Whitney test (B), Student’s t-test (D,F,H). Scale Bars: 200 μm (A,E,G), 100 μm (C). Ct values of RT-qPCR data are listed in table S3.
DISCUSSION
Following cardiac injury, the zebrafish heart mounts a fast angiogenic response that is essential for the regenerative process11. Here, we show that the regenerating coronary endothelium upregulates vegfc which is required for coronary revascularization after cardiac injury, independent of the presence or absence of lymphatics in the ventricle. We further show that reducing coronary revascularization by blocking Vegfc signaling impaired key aspects of cardiac regeneration including CM dedifferentiation and proliferation, as well as scar resolution. Mechanistically, Vegfc signaling induces emilin2a and cxcl8a expression, mostly in EPDCs, to promote coronary revascularization during cardiac regeneration in zebrafish.
One of the factors contributing to the inability of the adult mammalian heart to regenerate is its poor revacsularization75. In contrast, models with cardiac regenerative capacity like the zebrafish display a robust and fast angiogenic response to cardiac injury that supports heart regeneration11. Supporting this notion, perturbing coronary revascularization in regenerating zebrafish hearts impairs CM proliferation and prevents scar resolution11. On the other hand, inducing collateral artery formation following MI in adult mice results in a significant improvement in cardiac function76. Similarly, stabilizing vessel-like structures in the non-regenerative medaka heart following cardiac injury reduces scarring15. Therefore, improving revascularization in non-regenerating hearts should stimulate their regenerative response, and a better understanding of the mechanisms underlying revascularization in the regenerating zebrafish heart may help identify new therapeutic avenues.
Our data show that blocking Vegfc signaling results in reduced coronary revascularization, reduced CM proliferation, and the retention of a larger scar. Coronaries provide a scaffold for CMs during cardiac regeneration as well as during development12. Therefore, we hypothesize that the reduced CM dedifferentiation and proliferation phenotypes observed upon blocking Vegfc signaling occur as a consequence of reduced revascularization; however, tissue specific loss- and gain-of-function approaches will be needed to test this hypothesis. Our data show that Vegfc is a crucial angiocrine factor secreted by cECs to promote cardiac regeneration. Recent studies have emphasized how endothelial cells in different organs play important roles in regulating tissue homeostasis and repair by secreting angiocrine factors13. For instance, liver sinusoidal ECs govern the balance between liver regeneration and fibrosis by the preferential activation of the CXCR7 and CXCR4 receptors, respectively77. Similarly, following pneumonectomy, pulmonary capillary ECs secrete angiocrine factors, such as MMP14, that promote alveolar epithelial regeneration78. Importantly, it has been shown that EC-derived VEGFC stimulates the proliferation of neural stem cells during development and regeneration79,80. Likewise, our study illustrates the angiocrine role of Vegfc in the context of zebrafish cardiac regeneration to promote the proliferation of cECs and subsequently CMs. We further describe a mechanism by which Vegfc signaling is required for cardiac regeneration by stimulating the expression of the ECM gene emilin2a. During zebrafish development, Vegfc acts in both an autocrine and paracrine manner to induce venous as well as lymphatic sprouting by inducing cell cycle arrest via Vegfr332,81. Similarly, our data showing emilin2a expression by cECs and EPDCs during cardiac regeneration suggest that Vegfc acts in both an autocrine and paracrine manner to promote emilin2a expression. Importantly, we found that global overexpression of emilin2a stimulates coronary revascularization and reduces scarring after cardiac injury. Previous studies have shown that emilin2a expression is not induced following cardiac injury in non-regenerative organisms such as medaka15 and adult mice82. These observations, as well as our data highlighting the importance of emilin2a in promoting cardiac regeneration in zebrafish, indicate that Emilin2a enhance the regeneration process. Furthermore, we show that the regulation of emilin2a expression by Vegfc signaling is also present in human endothelial cells, encouraging future investigations into the role of VEGFC and EMILIN2 in promoting collateral formation in patients after MI.
Our data reveal how alterations in the ECM molecule Emilin2a affect coronary revascularization and cardiac regeneration in zebrafish. Indeed, recent studies have emphasized the role of the ECM in regulating different processes essential for cardiac regeneration including angiogenesis83,84,85. Interestingly, we found a number of ECM genes downregulated upon blocking Vegfc signaling (Table S4), including fibronectin 1b (fn1b) which has been previously implicated in angiogenesis86,87,88 as well as in zebrafish cardiac regeneration89. We speculate that Vegfc signaling alters the expression of several ECM genes, thereby creating a microenvironment that facilitates coronary revascularization following cardiac injury. Further studies are needed to explore the potential of various ECM proteins in promoting cardiac regeneration.
To better understand how Emilin2a exerts its role as a pro-regenerative factor, we identified the pro-inflammatory chemokine Cxcl8a as a potential target. These results are in line with data showing that EMILIN2 induces CXCL8 expression to promote the migration of HUVECs58. Our data reveal that cxcl8a is expressed in EPDCs in the injured tissue while cxcr1 is expressed in regenerating cECs, suggesting an interaction between these cell types to regulate coronary revascularization. Recent studies have shown that EPDCs are a source of important signals that regulate coronary regeneration in zebrafish12 and mouse90. CXCL8 has been shown in different settings to act as a pro-angiogenic factor via the activation of the G-protein coupled receptors CXCR1/270,71,72. Our data showing reduced cEC proliferation in both cxcl8a and cxcr1 mutants suggest that in the context of cardiac regeneration, Cxcl8a mediates its angiogenic role by activating Cxcr1 expressed by cECs. Accordingly, previous reports have implicated CXCR1 in promoting the proliferation of HUVECs in culture and endothelial cells in chick embryos, via different signaling pathways including Rho, Rac and MAPK activation91. Here, we provide evidence for a cellular crosstalk regulated by Cxcl8a-Cxcr1 signaling to promote cEC proliferation and cardiac regeneration.
Overall, our data highlight the importance of Vegfc signaling in promoting cardiac regeneration in zebrafish. We propose that Vegfc signaling induces the expression of the ECM component Emilin2a and chemokine Cxcl8a, as well as other factors, to provide a milieu that promotes cardiac regeneration. Our findings, along with data showing the importance of VEGFC signaling in promoting coronary development in mammals33,38, lay a strong foundation to investigate the potential of a VEGFC-EMILIN2-CXCL8 signaling axis to enhance collateral formation and thereby improve cardiac function following myocardial infarction in mammals.
Supplementary Material
NOVELTY AND SIGNIFICANCE.
What is known?
The adult zebrafish heart mounts a fast angiogenic response following cardiac injury, and blocking this process impedes cardiac regeneration.
vegfc expression is upregulated following cardiac injury in adult zebrafish
Vegfc signaling is required for lymphangiogenesis during development and cardiac regeneration.
What new information does this article contribute?
Vegfc signaling is required for coronary revascularization during cardiac regeneration in adult zebrafish.
We identify two new effectors of Vegfc signaling: the genes encoding the extracellular matrix protein Emilin2a and the chemokine Cxcl8a, and show that they are both required for coronary revascularization during cardiac regeneration in adult zebrafish.
Vegfc signaling promotes the expression of emilin2a, which in turn promotes cxcl8a expression in epicardium-derived cells to orchestrate a coronary-epicardium cross-talk necessary for cardiac regeneration.
Ischemic heart disease is a leading cause of death worldwide due to the inability of the adult mammalian heart to replace lost tissue. However, patients with improved collateral artery formation display improved clinical prognosis. Hence, identifying angiogenic factors in a regenerative setting holds promising therapeutic potential. We show that vegfc expression is upregulated in regenerating coronaries following cardiac injury in adult zebrafish and that Vegfc signaling is required for coronary revascularization. To understand the underlying mechanisms, we performed transcriptomic analysis and identified the extracellular matrix gene emilin2a and the chemokine gene cxcl8a as targets of Vegfc signaling. Mechanistically, we show that Emilin2a promotes the expression of cxcl8a in epicardium-derived cells to induce coronary revascularization and cardiac regeneration in adult zebrafish. These findings reveal an angiocrine role for Vegfc in promoting the expression of several factors that provide a milieu suitable for coronary revascularization and cardiac regeneration.
ACKNOWLEDGEMENTS
We thank Khrievono Kikhi and Ann Atzberger for assistance with cell sorting, Radhan Ramadass for help with imaging, Srinivas Allanki, Chi-Chung Wu, Arica Beisaw, Gülsüm Kayman-Kürekçi and Stéphanie Larrivée Vanier for helpful discussions and comments on the manuscript.
SOURCE OF FUNDING
H.E-S was funded by a Boehringer Ingelheim Fonds PhD fellowship. B.Y and W.C were funded by NIH 1R01DK117147. R.M-J is currently supported by the Canadian Institutes of Health Research (PJT-178037) and a FRQS Junior-1 award. Work in the Stainier lab was supported in part by funds from the Max Planck Society, the DFG (project number 394046768) SFB1366/project A4, and the Leducq Foundation.
NON-STANDARD ABBREVIATIONS AND ACRONYMS
- MI
myocardial infarction
- cECs
coronary endothelial cells
- EPDCs
epicardium-derived cells
- CMs
cardiomyocytes
- ECM
extracellular matrix
- hpci
hours post cryoinjury
- dpci
days post cryoinjury
- 4-OHT
4-hydroxytamoxifen
- EtOH
ethanol
- HS
heat shock
Footnotes
REFERENCES
- 1.Pfeffer MA, Braunwald E. Ventricular remodeling after myocardial infarction: Experimental observations and clinical implications. Circulation 1990;81:1161–1172. doi: 10.1161/01.CIR.81.4.1161. [DOI] [PubMed] [Google Scholar]
- 2.Talman V, Ruskoaho H. Cardiac fibrosis in myocardial infarction—from repair and remodeling to regeneration. Cell Tissue Res 2016;365:563–581. doi: 10.1007/s00441-016-2431-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Habib GB, Heibig J, Forman SA, Brown BG, Roberts R, Terrin ML, et al. Influence of coronary collateral vessels on myocardial infarct size in humans. Results of Phase I thrombolysis in myocardial infarction (TIMI) trial. Circulation 1991;83:739–746. doi: 10.1161/01.CIR.83.3.739. [DOI] [PubMed] [Google Scholar]
- 4.Seiler C, Stoller M, Pitt B, Meier P. The human coronary collateral circulation: Development and clinical importance. Eur Heart J 2013;34:2674–2682. doi: 10.1093/eurheartj/eht195. [DOI] [PubMed] [Google Scholar]
- 5.Sabia PJ, Powers ER, Ragosta M, Sarembock IJ, Burwell LR, Kaul S. An Association between Collateral Blood Flow and Myocardial Viability in Patients with Recent Myocardial Infarction. N Engl J Med 1992;327:1825–1831. doi: 10.1056/nejm199212243272601. [DOI] [PubMed] [Google Scholar]
- 6.Robich MP, Chu LM, Oyamada S, Sodha NR, Sellke FW. Myocardial therapeutic angiogenesis: A review of the state of development and future obstacles. Expert Rev Cardiovasc Ther 2011;9:1469–1479. doi: 10.1586/erc.11.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science (80- ) 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 8.Chablais F, Veit J, Rainer G, Jaźwińska A. The zebrafish heart regenerates after cryoinjury-induced myocardial infarction. BMC Dev Biol 2011;11:21. doi: 10.1186/1471-213X-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.González-Rosa JM, Martín V, Peralta M, Torres M, Mercader N. Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development 2011;138:1663–1674. doi: 10.1242/dev.060897. [DOI] [PubMed] [Google Scholar]
- 10.Schnabel K, Wu CC, Kurth T, Weidinger G. Regeneration of cryoinjury induced necrotic heart lesions in zebrafish is associated with epicardial activation and cardiomyocyte proliferation. PLoS One 2011;6. doi: 10.1371/journal.pone.0018503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marín-Juez R, Marass M, Gauvrit S, Rossi A, Lai S-L, Materna SC, et al. Fast revascularization of the injured area is essential to support zebrafish heart regeneration. Proc Natl Acad Sci U S A 2016;113:11237–11242. doi: 10.1073/pnas.1605431113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marín-Juez R, El-Sammak H, Helker CSM, Kamezaki A, Mullapuli ST, Bibli SI, et al. Coronary Revascularization During Heart Regeneration Is Regulated by Epicardial and Endocardial Cues and Forms a Scaffold for Cardiomyocyte Repopulation. Dev Cell 2019;51:503–515.e4. doi: 10.1016/j.devcel.2019.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rafii S, Butler JM, Ding B Sen. Angiocrine functions of organ-specific endothelial cells. Nature 2016;529:316–325. doi: 10.1038/nature17040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lien CL, Schebesta M, Makino S, Weber GJ, Keating MT. Gene expression analysis of zebrafish heart regeneration. PLoS Biol 2006;4:1386–1396. doi: 10.1371/journal.pbio.0040260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai SL, Marín-Juez R, Moura PL, Kuenne C, Lai JKH, Tsedeke AT, et al. Reciprocal analyses in zebrafish and medaka reveal that harnessing the immune response promotes cardiac regeneration. Elife 2017;6. doi: 10.7554/eLife.25605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klotz L, Norman S, Vieira JM, Masters M, Rohling M, Dubé KN, et al. Cardiac lymphatics are heterogeneous in origin and respond to injury. Nature 2015;522:62–67. doi: 10.1038/nature14483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh SJ, Jeltsch MM, Birkenhäger R, McCarthy JEG, Weich HA, Christ B, et al. VEGF and VEGF-C: Specific induction of angiogenesis and lymphangiogenesis in the differentiated avian chorioallantoic membrane. Dev Biol 1997;188:96–109. doi: 10.1006/dbio.1997.8639. [DOI] [PubMed] [Google Scholar]
- 18.Secker GA, Harvey NL. VEGFR signaling during lymphatic vascular development: From progenitor cells to functional vessels. Dev Dyn 2015;244:323–331. doi: 10.1002/dvdy.24227. [DOI] [PubMed] [Google Scholar]
- 19.Karaman S, Leppänen VM, Alitalo K. Vascular endothelial growth factor signaling in development and disease. Dev 2018;145. doi: 10.1242/dev.151019. [DOI] [PubMed] [Google Scholar]
- 20.Rauniyar K, Jha SK, Jeltsch M. Biology of vascular endothelial growth factor C in the morphogenesis of lymphatic vessels. Front Bioeng Biotechnol 2018;6. doi: 10.3389/fbioe.2018.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Küchler AM, Gjini E, Peterson-Maduro J, Cancilla B, Wolburg H, Schulte-Merker S. Development of the Zebrafish Lymphatic System Requires Vegfc Signaling. Curr Biol 2006;16:1244–1248. doi: 10.1016/j.cub.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 22.Gancz D, Raftrey BC, Perlmoter G, Marín-Juez R, Semo J, Matsuoka RL, et al. Distinct origins and molecular mechanisms contribute to lymphatic formation during cardiac growth and regeneration. Elife 2019;8. doi: 10.7554/eLife.44153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrison MR, Feng X, Mo G, Aguayo A, Villafuerte J, Yoshida T, et al. Late developing cardiac lymphatic vasculature supports adult zebrafish heart function and regeneration. Elife 2019;8. doi: 10.7554/eLife.42762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vivien CJ, Pichol-Thievend C, Sim CB, Smith JB, Bower NI, Hogan BM, et al. Vegfc/d-dependent regulation of the lymphatic vasculature during cardiac regeneration is influenced by injury context. Npj Regen Med 2019;4. doi: 10.1038/s41536-019-0079-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao Y, Linden P, Farnebo J, Cao R, Eriksson A, Kumar V, et al. Vascular endothelial growth factor C induces angiogenesis in vivo. Proc Natl Acad Sci U S A 1998;95:14389–14394. doi: 10.1073/pnas.95.24.14389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pätilä T, Ikonen T, Rutanen J, Ahonen A, Lommi J, Lappalainen K, et al. Vascular endothelial growth factor C-induced collateral formation in a model of myocardial ischemia. J Hear Lung Transplant 2006;25:206–213. doi: 10.1016/j.healun.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 27.Chen HI, Poduri A, Numi H, Kivela R, Saharinen P, McKay AS, et al. VEGF-C and aortic cardiomyocytes guide coronary artery stem development. J Clin Invest 2014;124:4899–4914. doi: 10.1172/JCI77483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wada H, Suzuki M, Matsuda M, Ajiro Y, Shinozaki T, Sakagami S, et al. VEGF-C and mortality in patients with suspected or known coronary artery disease. J Am Heart Assoc 2018;7. doi: 10.1161/JAHA.118.010355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuoka RL, Marass M, Avdesh A, Helker CSM, Maischein HM, Grosse AS, et al. Radial glia regulate vascular patterning around the developing spinal cord. Elife 2016;5. doi: 10.7554/eLife.20253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Covassin LD, Villefranc JA, Kacergis MC, Weinstein BM, Lawson ND. Distinct genetic interactions between multiple Vegf receptors are required for development of different blood vessel types in zebrafish. Proc Natl Acad Sci U S A 2006;103:6554–6559. doi: 10.1073/pnas.0506886103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Villefranc JA, Nicoli S, Bentley K, Jeltsch M, Zarkada G, Moore JC, et al. A truncation allele in vascular endothelial growth factor c reveals distinct modes of signaling during lymphatic and vascular development. Development 2013;140:1497–1506. doi: 10.1242/dev.084152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Guen L, Karpanen T, Schulte D, Harris NC, Koltowska K, Roukens G, et al. Ccbe1 regulates Vegfc-mediated induction of Vegfr3 signaling during embryonic lymphangiogenesis. Development 2014;141:1239–1249. doi: 10.1242/dev.100495. [DOI] [PubMed] [Google Scholar]
- 33.Chen HI, Sharma B, Akerberg BN, Numi HJ, Kivelä R, Saharinen P, et al. The sinus venosus contributes to coronary vasculature through VEGFC-stimulated angiogenesis. Dev 2014;141:4500–4512. doi: 10.1242/dev.113639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rossi A, Gauvrit S, Marass M, Pan L, Moens CB, Stainier DYR. Regulation of Vegf signaling by natural and synthetic ligands. Blood 2016;128:2359–2366. doi: 10.1182/blood-2016-04-711192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bussmann J, Bos FL, Urasaki A, Kawakami K, Duckers HJ, Schulte-Merker S. Arteries provide essential guidance cues for lymphatic endothelial cells in the zebrafish trunk. Development 2010;137:2653–2657. doi: 10.1242/dev.048207. [DOI] [PubMed] [Google Scholar]
- 36.Chi NC, Shaw RM, Jungblut B, Huisken J, Ferrer T, Arnaout R, et al. Genetic and physiologic dissection of the vertebrate cardiac conduction system. PLoS Biol 2008;6:1006–1019. doi: 10.1371/journal.pbio.0060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, Cao J, Dickson AL, Poss KD. Epicardial regeneration is guided by cardiac outflow tract and Hedgehog signalling. Nature 2015;522:226–230. doi: 10.1038/nature14325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kikuchi K, Gupta V, Wang J, Holdway JE, Wills AA, Fang Y, et al. Tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development 2011;138:2895–2902. doi: 10.1242/dev.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okuda KS, Astin JW, Misa JP, Flores MV., Crosier KE, Crosier PS. Lyve1 expression reveals novel lymphatic vessels and new mechanisms for lymphatic vessel development in zebrafish. Dev 2012;139:2381–2391. doi: 10.1242/dev.077701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nicenboim J, Malkinson G, Lupo T, Asaf L, Sela Y, Mayseless O, et al. Lymphatic vessels arise from specialized angioblasts within a venous niche. Nature 2015;522:56–61. doi: 10.1038/nature14425. [DOI] [PubMed] [Google Scholar]
- 41.Kettleborough RNW, Busch-Nentwich EM, Harvey SA, Dooley CM, De Bruijn E, Van Eeden F, et al. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature 2013;496:494–497. doi: 10.1038/nature11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerri C, Marín-Juez R, Marass M, Marks A, Maischein HM, Stainier DYR. Hif-1α regulates macrophage-endothelial interactions during blood vessel development in zebrafish. Nat Commun 2017;8. doi: 10.1038/ncomms15492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 44.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liao Y, Smyth GK, Shi W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 47.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koth J, Wang X, Killen AC, Stockdale WT, Potts HG, Jefferson A, et al. Runx1 promotes scar deposition and inhibits myocardial proliferation and survival during zebrafish heart regeneration. Dev 2020;147. doi: 10.1242/DEV.186569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lowe V, Wisniewski L, Sayers J, Evans I, Frankel P, Mercader-Huber N, et al. Neuropilin 1 mediates epicardial activation and revascularization in the regenerating zebrafish heart. Dev 2019;146. doi: 10.1242/dev.174482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ogawa M, Geng FS, Humphreys DT, Kristianto E, Sheng DZ, Hui SP, et al. Krüppel-like factor 1 is a core cardiomyogenic trigger in zebrafish. Science (80- ) 2021;372. doi: 10.1126/science.abe2762. [DOI] [PubMed] [Google Scholar]
- 51.Hogan BM, Herpers R, Witte M, Heloterä H, Alitalo K, Duckers HJ, et al. Vegfc/Flt4 signalling is suppressed by Dll4 in developing zebrafish intersegmental arteries. Development 2009;136:4001–4009. doi: 10.1242/dev.039990. [DOI] [PubMed] [Google Scholar]
- 52.Hogan BM, Bos FL, Bussmann J, Witte M, Chi NC, Duckers HJ, et al. Ccbe1 is required for embryonic lymphangiogenesis and venous sprouting. Nat Genet 2009;41:396–398. doi: 10.1038/ng.321. [DOI] [PubMed] [Google Scholar]
- 53.Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, et al. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morikawa Y, Zhang M, Heallen T, Leach J, Tao G, Xiao Y, et al. Actin cytoskeletal remodeling with protrusion formation is essential for heart regeneration in Hippo-deficient mice. Sci Signal 2015;8. doi: 10.1126/scisignal.2005781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beisaw A, Kuenne C, Guenther S, Dallmann J, Wu CC, Bentsen M, et al. AP-1 Contributes to Chromatin Accessibility to Promote Sarcomere Disassembly and Cardiomyocyte Protrusion during Zebrafish Heart Regeneration. Circ Res 2020;126:1760–1778. doi: 10.1161/CIRCRESAHA.119.316167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsedeke AT, Allanki S, Gentile A, Jimenez-Amilburu V, Rasouli SJ, Guenther S, et al. Cardiomyocyte heterogeneity during zebrafish development and regeneration. Dev Biol 2021;476:259–271. doi: 10.1016/j.ydbio.2021.03.014. [DOI] [PubMed] [Google Scholar]
- 57.Yaniv K, Isogai S, Castranova D, Dye L, Hitomi J, Weinstein BM. Live imaging of lymphatic development in the zebrafish. Nat Med 2006;12:711–716. doi: 10.1038/nm1427. [DOI] [PubMed] [Google Scholar]
- 58.Jeltsch M, Kaipainen A, Joukov V, Meng X, Lakso M, Rauvala H, et al. Hyperplasia of lymphatic vessels in VEGF-C transgenic mice. Science (80- ) 1997;276:1423–1425. doi: 10.1126/science.276.5317.1423. [DOI] [PubMed] [Google Scholar]
- 59.Stacker SA, Stenvers K, Caesar C, Vitali A, Domagala T, Nice E, et al. Biosynthesis of vascular endothelial growth factor-D involves proteolytic processing which generates non-covalent homodimers. J Biol Chem 1999;274:32127–32136. doi: 10.1074/jbc.274.45.32127. [DOI] [PubMed] [Google Scholar]
- 60.Colombatti A, Spessotto P, Doliana R, Mongiat M, Bressan GM, Esposito G. The EMILIN/multimerin family. Front Immunol 2012;2. doi: 10.3389/fimmu.2011.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Milanetto M, Tiso N, Braghetta P, Volpin D, Argenton F, Bonaldo P. Emilin genes are duplicated and dynamically expressed during zebrafish embryonic development. Dev Dyn 2008;237:222–232. doi: 10.1002/dvdy.21402. [DOI] [PubMed] [Google Scholar]
- 62.Paulitti A, Andreuzzi E, Bizzotto D, Pellicani R, Tarticchio G, Marastoni S, et al. The ablation of the matricellular protein EMILIN2 causes defective vascularization due to impaired EGFR-dependent IL-8 production affecting tumor growth. Oncogene 2018;37:3399–3414. doi: 10.1038/s41388-017-0107-x. [DOI] [PubMed] [Google Scholar]
- 63.Mongiat M, Marastoni S, Ligresti G, Lorenzon E, Schiappacassi M, Perris R, et al. The extracellular matrix glycoprotein elastin microfibril interface located protein 2: A dual role in the tumor microenvironment. Neoplasia 2010;12:294–304. doi: 10.1593/neo.91930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Andreuzzi E, Fejza A, Capuano A, Poletto E, Pivetta E, Doliana R, et al. Deregulated expression of Elastin Microfibril Interfacer 2 (EMILIN2) in gastric cancer affects tumor growth and angiogenesis. Matrix Biol Plus 2020;6–7. doi: 10.1016/j.mbplus.2020.100029. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 65.Rossi A, Kontarakis Z, Gerri C, Nolte H, Hölper S, Krüger M, et al. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 2015;524:230–233. doi: 10.1038/nature14580. [DOI] [PubMed] [Google Scholar]
- 66.El-Brolosy MA, Kontarakis Z, Rossi A, Kuenne C, Günther S, Fukuda N, et al. Genetic compensation triggered by mutant mRNA degradation. Nature 2019;568:193–197. doi: 10.1038/s41586-019-1064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ito K, Morioka M, Kimura S, Tasaki M, Inohaya K, Kudo A. Differential reparative phenotypes between zebrafish and medaka after cardiac injury. Dev Dyn 2014;243:1106–1115. doi: 10.1002/dvdy.24154. [DOI] [PubMed] [Google Scholar]
- 68.Hesselson D, Anderson RM, Beinat M, Stainier DYR. Distinct populations of quiescent and proliferative pancreatic β-cells identified by HOTcre mediated labeling. Proc Natl Acad Sci U S A 2009;106:14896–14901. doi: 10.1073/pnas.0906348106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ha H, Debnath B, Neamati N. Role of the CXCL8-CXCR1/2 axis in cancer and inflammatory diseases. Theranostics 2017;7:1543–1588. doi: 10.7150/thno.15625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Strieter RM, Kunkel SL, Elner VM, Martonyi CL, Koch AE, Polverini PJ, et al. Interleukin-8: A corneal factor that induces neovascularization. Am J Pathol 1992;141:1279–1284. [PMC free article] [PubMed] [Google Scholar]
- 71.Heidemann J, Ogawa H, Dwinell MB, Rafiee P, Maaser C, Gockel HR, et al. Angiogenic effects of interleukin 8 (CXCL8) in human intestinal microvascular endothelial cells are mediated by CXCR2. J Biol Chem 2003;278:8508–8515. doi: 10.1074/jbc.M208231200. [DOI] [PubMed] [Google Scholar]
- 72.Li A, Dubey S, Varney ML, Dave BJ, Singh RK. IL-8 Directly Enhanced Endothelial Cell Survival, Proliferation, and Matrix Metalloproteinases Production and Regulated Angiogenesis. J Immunol 2003;170:3369–3376. doi: 10.4049/jimmunol.170.6.3369. [DOI] [PubMed] [Google Scholar]
- 73.Clark-Lewis I, Schumacher C, Baggiolini M, Moser B. Structure-Activity Relationships of Interleukin-8 Determined Using Chemically Synthesized Analogs. J Biol Chem 1991;266:23128–23134. [PubMed] [Google Scholar]
- 74.White JR, Lee JM, Young PR, Hertzberg RP, Jurewicz AJ, Chaikin MA, et al. Identification of a potent, selective non-peptide CXCR2 antagonist that inhibits interleukin-8-induced neutrophil migration. J Biol Chem 1998;273:10095–10098. doi: 10.1074/jbc.273.17.10095. [DOI] [PubMed] [Google Scholar]
- 75.Kocijan T, Rehman M, Colliva A, Groppa E, Leban M, Vodret S, et al. Genetic lineage tracing reveals poor angiogenic potential of cardiac endothelial cells. Cardiovasc Res 2021;117:256–270. doi: 10.1093/cvr/cvaa012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Das S, Goldstone AB, Wang H, Farry J, D’Amato G, Paulsen MJ, et al. A Unique Collateral Artery Development Program Promotes Neonatal Heart Regeneration. Cell 2019. doi: 10.1016/j.cell.2018.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sen Ding B, Cao Z, Lis R, Nolan DJ, Guo P, Simons M, et al. Divergent angiocrine signals from vascular niche balance liver regeneration and fibrosis. Nature 2014;505:97–102. doi: 10.1038/nature12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sen Ding B, Nolan DJ, Guo P, Babazadeh AO, Cao Z, Rosenwaks Z, et al. Endothelial-derived angiocrine signals induce and sustain regenerative lung alveolarization. Cell 2011;147:539–553. doi: 10.1016/j.cell.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Le Bras B, Barallobre MJ, Homman-Ludiye J, Ny A, Wyns S, Tammela T, et al. VEGF-C is a trophic factor for neural progenitors in the vertebrate embryonic brain. Nat Neurosci 2006;9:340–348. doi: 10.1038/nn1646. [DOI] [PubMed] [Google Scholar]
- 80.Han J, Calvo CF, Kang TH, Baker KL, Park JH, Parras C, et al. Vascular Endothelial Growth Factor Receptor 3 Controls Neural Stem Cell Activation in Mice and Humans. Cell Rep 2015;10:1158–1172. doi: 10.1016/j.celrep.2015.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jerafi-Vider A, Bassi I, Moshe N, Tevet Y, Hen G, Splittstoesser D, et al. VEGFC/FLT4-induced cell-cycle arrest mediates sprouting and differentiation of venous and lymphatic endothelial cells. Cell Rep 2021;35. doi: 10.1016/j.celrep.2021.109255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zangrando J, Zhang L, Vausort M, Maskali F, Marie PY, Wagner DR, et al. Identification of candidate long non-coding RNAs in response to myocardial infarction. BMC Genomics 2014;15. doi: 10.1186/1471-2164-15-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu CC, Jeratsch S, Graumann J, Stainier DYR. Modulation of Mammalian Cardiomyocyte Cytokinesis by the Extracellular Matrix. Circ Res 2020;127:896–907. doi: 10.1161/CIRCRESAHA.119.316303. [DOI] [PubMed] [Google Scholar]
- 84.Bassat E, Mutlak YE, Genzelinakh A, Shadrin IY, Baruch Umansky K, Yifa O, et al. The extracellular matrix protein agrin promotes heart regeneration in mice. Nature 2017;547:179–184. doi: 10.1038/nature22978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mongiat M, Andreuzzi E, Tarticchio G, Paulitti A. Extracellular matrix, a hard player in angiogenesis. Int J Mol Sci 2016;17. doi: 10.3390/ijms17111822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Astrof S, Hynes RO. Fibronectins in vascular morphogenesis. Angiogenesis 2009;12:165–175. doi: 10.1007/s10456-009-9136-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou X, Rowe RG, Hiraoka N, George JP, Wirtz D, Mosher DF, et al. Fibronectin fibrillogenesis regulates three-dimensional neovessel formation. Genes Dev 2008;22:1231–1243. doi: 10.1101/gad.1643308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chiu CH, Chou CW, Takada S, Liu YW. Development and fibronectin signaling requirements of the Zebrafish interrenal vessel. PLoS One 2012;7. doi: 10.1371/journal.pone.0043040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang J, Karra R, Dickson AL, Poss KD. Fibronectin is deposited by injury-activated epicardial cells and is necessary for zebrafish heart regeneration. Dev Biol 2013;382:427–435. doi: 10.1016/j.ydbio.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhou B, Honor LB, He H, Qing M, Oh JH, Butterfield C, et al. Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. J Clin Invest 2011;121:1894–1904. doi: 10.1172/JCI45529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li A, Varney ML, Valasek J, Godfrey M, Dave BJ, Singh RK. Autocrine role of interleukin-8 in induction of endothelial cell proliferation, survival, migration and MMP-2 production and angiogenesis. Angiogenesis 2005;8:63–71. doi: 10.1007/s10456-005-5208-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNAseq data from this study have been deposited in the Gene Expression Omnibus (GEO) database under accession number GSE168175.


