Abstract
Despite their importance in disease and evolution, highly identical segmental duplications (SDs) are among the last regions of the human reference genome (GRCh38) to be fully sequenced. Using a complete telomere-to-telomere human genome (T2T-CHM13), we present a comprehensive view of human SD organization. SDs account for nearly one-third of the additional sequence increasing the genome-wide estimate from 5.4% to 7.0% (218 Mbp). An analysis of 268 human genomes shows that 91% of the previously unresolved T2T-CHM13 SD sequence (68.3 Mbp) better represents human copy number variation. Comparing long-read assemblies from human (n=12) and nonhuman primate (n=5) genomes, we systematically reconstruct the evolution and structural haplotype diversity of biomedically relevant and duplicated genes. This analysis reveals patterns of structural heterozygosity and evolutionary differences in SD organization between humans and other primates.
STRUCTURED ABSTRACT
Introduction:
Large high-identity duplicated sequences, termed segmental duplications (SDs), are frequently the last regions of genomes to be sequenced and assembled. While the human reference genome provided a road map of the SD landscape, more than 50% of the remaining gaps correspond to regions of complex SDs.
Rationale:
SDs are major sources of evolutionary gene innovations and contribute disproportionately to genetic variation within and between ape species. A complete human genome (T2T-CHM13) has the potential to identify genes and uncover patterns of human genetic variation.
Results:
We identified 51 Mbp of additional human SD in T2T-CHM13 and now estimate that 7% of the human genome consists of SDs (218 Mbp of 3.1 Gbp). SDs make up two-thirds (45.0/68.1 Mbp) of acrocentric short arms and are the largest SDs in the human genome (Fig. 0a). Additionally, 54% of acrocentric SDs are copy number variable or map to different chromosomes among six individuals examined. A detailed comparison between the current reference genome (GRCh38) and T2T-CHM13 for SD content identifies 81 Mbp of previously unresolved or structurally variable SDs. Short-read whole-genome sequence data from a diversity panel of 268 humans shows that human copy number is nine times (59.26/6.55 Mbp) more likely to match T2T-CHM13 rather than GRCh38, including 119 protein-coding genes (Fig. 0b). Using long-read-sequencing data from 25 human haplotypes, we investigated patterns of human genetic variation identifying significant increases in structural and single-nucleotide diversity. We identified gene-rich regions (e.g., TBC1D3) that vary by hundreds of kilobase pairs and gene copy number between individuals showing some of the highest genome-wide structural heterozygosity (85–90%). Our analysis identified 182 candidate protein-encoding genes as well as the complete sequence for structurally variable gene models that were previously unresolved. Among these is the complete gene structure of lipoprotein A (LPA) including the expanded Kringle IV repeat domain. Reduced copies of this domain are among the strongest genetic associations with cardiovascular disease, especially among African Americans, and sequencing of multiple human haplotypes not only identified copy number variation but also other forms of rare coding variation potentially relevant to disease risk. Finally, we compared global methylation and expression patterns between duplicated and unique genes. Transcriptionally inactive duplicate genes are more likely to map to hypomethylated genomic regions; however, specifically over the transcription start site we observe an increase in methylation suggesting that as many as two-thirds of duplicated genes are epigenetically silenced. Additionally, SD genes show a high degree of concordance between methylation profiles and transcription levels allowing us to define the actively transcribed members of high-identity gene families that are otherwise indistinguishable by coding sequence.
Conclusion:
A complete human genome provides a more comprehensive understanding of the organization, expression, and regulation of duplicated genes. Our analysis reveals underappreciated patterns of human genetic diversity and suggests unique features of methylation and gene regulation. This resource will serve as a critical baseline for improved gene annotation, genotyping, and novel associations for some of the most dynamic regions of our genome.
INTRODUCTION
Genomic duplications have long been recognized as important sources of structural change and gene innovation (1, 2). In humans, the most recent and highly identical sequences (>90% and >1 kbp), referred to as segmental duplications (SDs) (3), promote meiotic unequal crossover events contributing to recurrent rearrangements associated with ~5% of developmental delay and autism (4). These same SDs are reservoirs for human-specific genes important in increasing synaptic density and the expansion of the frontal cortex since humans diverged from other ape lineages (5–8). SDs are also ~10-fold enriched for normal copy number variation although most of this genetic diversity has yet to be fully characterized or associated with human phenotypes (9, 10). Their length (frequently >100 kbp), sequence identity, and extensive structural diversity among human haplotypes have hampered our ability to characterize these regions at a genomic level. This is because sequence reads have been insufficiently long and human haplotypes too structurally diverse to resolve duplicate copies or distinguish allelic variants.
One of the first human whole-genome sequence (WGS) assembly drafts created with Sanger sequencing technology was almost devoid of SDs and their underlying genes (11, 12). Similarly, BAC-based approaches to assemble the human genome from different haplotypes led to many misjoins creating de facto gaps that took years to resolve (13). While combining WGS and BAC-based data for early sequencing of human genomes provided a road map of the SD landscape (14), more than 50% of the gaps within the human reference genome have corresponded to regions of complex SDs.
The development of genomic resources (15–17), including BAC libraries and long-read sequence data from complete hydatidiform moles (which represent a single human haplotype), was motivated in large part by efforts to resolve the organization of these regions and concomitantly complete the human reference genome. CHM13 has the advantage of being from a single haplotype and from predominantly a single ancestral group (European, (18) in contrast to the GRCh38 reference, which is a composite representation of multiple human haplotypes and ancestries (19). These resources, combined with advances in long-read technologies, have produced the gapless human genome assembly T2T-CHM13 (20). Here, we use this genome assembly to present a complete view of SDs in a human genome and highlight their importance in advancing our understanding of human genetic diversity, evolution, and disease.
RESULTS
SD content and organization.
We characterized the SD content of the T2T-CHM13 v1.0 assembly using sequence read-depth and pairwise sequence alignments (>90% and >1 kbp) (21). Our analysis of the assembly identifies 208 Mbp of nonredundant segmentally duplicated sequence within chromosome-level scaffolds (including 15.6 Mbp of SD located on chrY, which is included from GRCh38), compared to just 167 Mbp in the current reference (GRCh38) (Table 1 and Fig. 1). This raises the percent estimate of the human genome that is segmentally duplicated from 5.4% to 6.7%. However, five SD-related gaps remained in the initial assembly of the female CHM13 genome (T2T-CHM13 v1.0). Each corresponded to a cluster of tandemly repeated rDNA genes on each acrocentric chromosome where we confirm long-read sequence pileups consistent with unresolved SDs. The estimated amount of missing rDNA sequence was calculated by Nurk et al. using both digital droplet PCR (22) and a whole-genome Illumina coverage analysis (20). Assuming a canonical repeat length of 45 kbp for the rDNA molecule (23, 24), the total amount of missing sequences was approximated at ~10 Mbp and ~200 copies of unresolved rDNA sequence (20). These findings are consistent with the subsequent specialized assembly of the rDNA released as part of the T2T-CHM13 v1.1 assembly. Including this estimate, the overall SD content of the human genome is now 7.0% (6.7% not including rDNA; see Table 1 for statistics breakdown by SD type) and is likely to increase as more complete genomes of diverse origins are sequenced and assembled.
Table 1:
Summary statistics of segmental duplications in T2T-CHM13 and GRCh38
| Assembly | Gbp | % SD | SD (Mbp) | # SDs | inter (Mbp) | # inter | intra (Mbp) | # intra | acro (Mbp) | # acro | peri (Mbp) | # peri | telo (Mbp) | # telo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||
| T2T CHM13 v1.0* | 3.114 | 6.665 | 207.563 | 41289 | 121.113 | 30484 | 142.958 | 10805 | 35.106 | 13264 | 88.606 | 24985 | 10.975 | 4998 |
| GRCh38 | 3.114 | 5.372 | 167.297 | 24280 | 83.556 | 16348 | 120.710 | 7932 | 6.624 | 1407 | 53.944 | 10606 | 8.926 | 1529 |
| Difference | 0.000 | 1.293 | 40.266 | 17009 | 37.556 | 14136 | 22.248 | 2873 | 28.482 | 11857 | 34.662 | 14379 | 2.049 | 3469 |
| previously unresolved or structurally variable | 0.240 | 33.885 | 81.338 | 25161 | 61.873 | 20579 | 54.932 | 4582 | 35.039 | 13258 | 54.037 | 19607 | 5.616 | 4005 |
| T2T CHM13 v1.0* + rDNA estimate | 3.114 | 6.987 | 217.598 | 66042 | 131.148 | 49213 | 152.993 | 16829 | 45.141 | 38017 | 98.641 | 49738 | 10.975 | 4998 |
The version of T2T-CHM13 v1.0 used included chrY from GRCh38
Mbp: the number of nonredundant Mbp of SD; peri: within 5 Mbp of the heterochromatin surrounding the centromere; telo: within 500 kbp of the telomere; acro: within the short arms of the acrocentric chromosomes.
Difference: SD content difference between T2T-CHM13 v1.0 and GRCh38.
Previously unresolved or structurally variable: Sequence in T2T-CHM13 that does not have 1 Mbp of synteny with GRCh38.
GRCh38 contains 149,690,719 of gap sequence included in the reported # of Gbp.
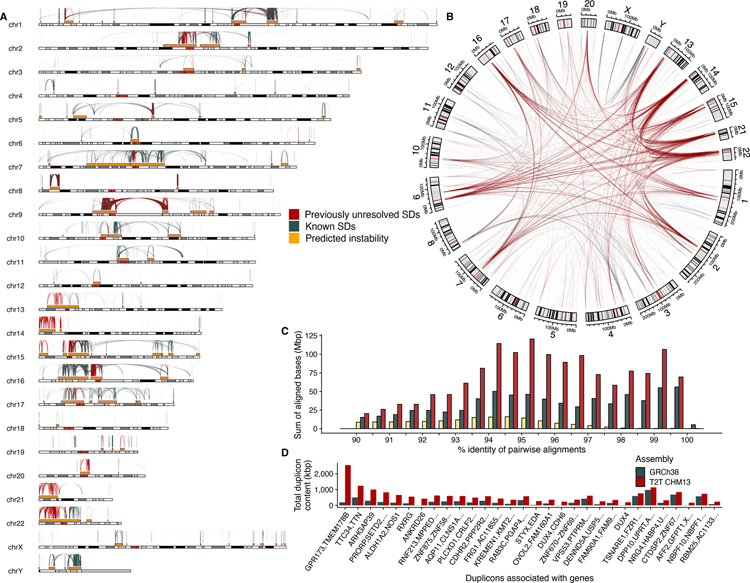
Fig. 1. Segmental duplication (SD) content of the T2T-CHM13 genome.
A) The pattern of previously unresolved or structurally variant intrachromosomal duplications in T2T-CHM13 (red) compared to known duplications in GRCh38 (blue-gray). These predict hotspots of genomic instability (gold) flanked by large (>10 kbp), high-identity (>95%) interspersed (>50 kbp) SDs. B) Circos plot highlighting previously unresolved interchromosomal SDs (red) shows the preponderance of previously unresolved SDs mapping to pericentromeric and acrocentric regions. C) A histogram comparing SD content in different human reference genomes. The sum of bases in pairwise SD alignments stratified by their percent identity for the Celera (yellow, Sanger-based), GRCh38 (blue-gray, BAC-based), and T2T-CHM13 (red, long-read) assemblies. D) The 30 genic duplicons (ancestral repeat units) with the greatest copy number difference between GRCh38 and T2T-CHM13 as determined by DupMasker (table S2). All of the 30 largest differences are increased in T2T-CHM13.
One-third (81.3 Mbp, 25) of SD sequence in T2T-CHM13 is wholly uncharacterized in GRCh38 (16.5 Mbp) or differs in copy number and structure (64.8 Mbp, 25). Most of these involve large, high-identity SDs. For example, there is a 70% increase (41,285/24,280) in the number of SD pairs and a doubling of the number of bases in pairwise alignments with greater than 95% identity (Fig. 1C). Among these previously unresolved or variable SDs, 13,258 (35.0 Mbp) map to the acrocentric short arms of chromosomes 13, 14, 15, 21, and 22 (Fig. 1B and Table 1), which are unassembled in the GRCh38. These SDs do not correspond to rDNA duplications but represent other segments predominantly shared among acrocentric (n=5,332 alignments) and non-acrocentric chromosomes (n= 5,500 alignments, table S1). In particular, the pericentromeric regions of chromosomes 1, 3, 4, 7, 9, 16 and 20 show the most extensive SD homology with acrocentric DNA (Fig. 1B). Non-rDNA acrocentric SDs are 1.75-fold longer than all other SDs (N50: 74,704 vs. 42,842) – significantly longer (p-value < 0.01, one-sided Wilcoxon rank-sum test) than any other defined SD category in the human genome [intrachromosomal, interchromosomal, pericentromeric, and telomeric (fig. S1)].
We annotated all T2T-CHM13 SDs using DupMasker (26), which defines ancestral evolutionary units of duplication on the basis of mammalian outgroups and a repeat graph (27). Focusing on duplicons that carry genes or duplicated portions of genes, we identify 30 duplicons that show the greatest copy number change between T2T-CHM13 and GRCh38. These 30 genic SDs represent regions where gene annotation is most likely to change; all predicted differences favor an increase in copy number for the T2T-CHM13 assembly (Fig. 1D and table S2).
We also compared the number of SDs more directly by defining syntenic regions (5 Mbp) between GRCh38 and T2T-CHM13 (25). Of the 15 windows with the largest increase, nine mapped to the acrocentric short arms while six were in pericentromeric regions (fig. S1 and table S3). In particular, the intervals between the centromeric satellite and secondary constrictions (qh regions) on chromosomes 1, 9, and 16 show a 4.6-fold increase in the number of SDs (5,254/1,141) and the most differences in organization compared to GRCh38. SDs in these regions are almost exclusively interchromosomal and depleted for intrachromosomal duplications (figs. S2 and S3).
Validation and heteromorphic variation.
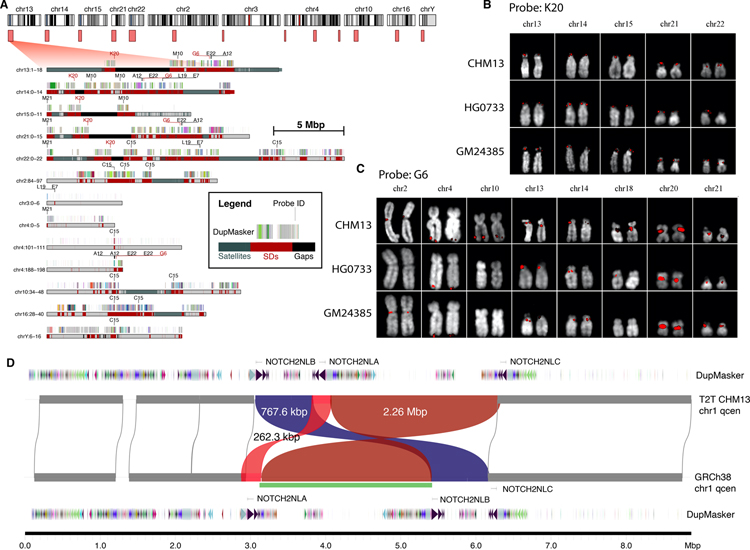
Because the acrocentric short arms as well as the qh regions on chromosomes 1, 9, and 16 were either previously unresolved or showed the most significant differences in terms of SD content, we focused first on validating their organization. We mapped available end-sequence data from a human fosmid genome library (28) to the T2T-CHM13 assembly and selected nine distinct clones as probes (Fig. 2A) to confirm the patterns of high-identity (>95%) SDs (25). All 30 of the distinct duplication predictions from T2T-CHM13 SDs were corroborated by FISH against chromosomal metaphases of the CHM13 cell line (Fig. 2, B and C, and table S4).
Fig. 2. Validation of previously unresolved SDs in T2T-CHM13 and heteromorphic variation.
A) Ideogram (top) depicts large SD regions (light red boxes) present in T2T-CHM13 but absent from the current reference human genome (GRCh38). An expanded view of the duplication (red) and satellite organization (blue) are depicted below showing the location of fosmid FISH probes (e.g., C15) and SD organization compared to ancestral duplicon segments (multi-colored bars) (see inset). B,C) FISH signals (red) shown on extracted metaphase for two probes and three human cell lines. Probe K20 shows a fixed signal (except for one heterozygous signal), and G6 is heteromorphic among humans (see table S4 and fig. S4 for complete description for all nine probes). D) Inversion polymorphism (green bar) between T2T-CHM13 and GRCh38 in the pericentromeric chromosome 1q region. The inversion (green bar) identified by Strand-seq (32) is confirmed in the assembly; however, the sequence-resolved assembly shows a more complex structure including two inversions (red) and one reordered segment (blue) mapping near the NOTCH2NL human-specific duplications.
Interestingly, FISH also revealed nine additional signals not originally predicted by our SD analysis (fig. S4). However, we were able to identify lower identity duplications confirming seven of these sites leading to an overall concordance of 95% (37/39) between FISH and the T2T-CHM13 SD assembly content. We extended this analysis to five additional human cell lines of diploid origin because both pericentromeric and acrocentric portions of chromosomes have been shown to be cytogenetically heteromorphic (29–31). In total, we identified 61 distinct cytogenetic locations of which 28 (46%) were fixed while 33 (54%) were variable in their presence or absence on specific homologues (both acrocentric and pericentromeric regions of the human genome) (fig. S4). Of the 61 FISH signals, all but six were observed in more than one of the six human cell lines indicating that such heteromorphic variation is common and prevalent.
We found a correlation (Pearson’s correlation coefficient, r=0.96) between genome-wide copy number variation from the assembly and Illumina read-depth data generated from the same CHM13 source (25). Because SDs frequently map to the breakpoints of inversion polymorphisms (28, 32, 33), we validated 65 inversions relative to GRCh38 with Strand-seq analysis of the CHM13 assembly (figs. S5 and S6, (25). While 32 of these represent known human polymorphisms, 33 have not been observed in six previously analyzed human genomes (32). However, by analysis of Strand-seq data from one additional human haplotype (CHM1), we further confirmed 30 of these inversions (i.e., present in CHM1 and CHM13) suggesting that at least 95.4% (62/65) represent true large-scale human inversion polymorphisms (fig. S5). Consistent with previous literature (34), inversions associated with SDs (n=30) are significantly longer than those not associated with SDs (p-value < 0.01, one-sided Wilcoxon rank-sum test) and are polymorphic among humans (fig. S6). One striking example is an inversion polymorphism mapping to chromosome 1q21. It is a complex event consisting of two inversions (262.3 kbp, 2.26 Mbp) originally predicted by Sanders and colleagues (33) but our sequence analysis shows a relocation of 767.6 kbp of genic sequence (Fig. 2D). The large inversion (chr1:146,350,000–148,610,000) is flanked by the core duplicon, the NPBF gene family, and in combination with the other rearrangements changes the order of human-specific genes NOTCH2NLA, B and C, which have been implicated in the expansion of the frontal cortex (8, 35). As a final test, we resolved this region in eight additional human haplotypes (25)—all of which support the T2T-CHM13 configuration with one exception (CHM1), which is consistent with the GRCh38 configuration (fig. S7).
Single-nucleotide and copy number variation within SDs.
The high quality and single haplotype nature of both the T2T-CHM13 and GRCh38 reference genomes provides us an opportunity to compare the genome-wide pattern of single-nucleotide variation in regions that have been typically excluded from most analyses due to their repetitive nature. We aligned GRCh38 to T2T-CHM13 and retained only regions deemed to be “syntenic” on the basis of an unambiguous one-to-one correspondence between both reference genomes and at least 1 Mbp of aligned sequence (25).
Most unique regions of the genome (2,693 Mbp) could be compared, while only 60% (124 Mbp) of the SDs within T2T-CHM13 exhibited a clear orthologous relationship between the two human reference genomes. As expected, the X chromosome and the region corresponding to the major histocompatibility complex (MHC) are the least and most divergent, respectively (Fig. 3A), due to the slower rate of evolution for the female X and the deep coalescence of MHC.
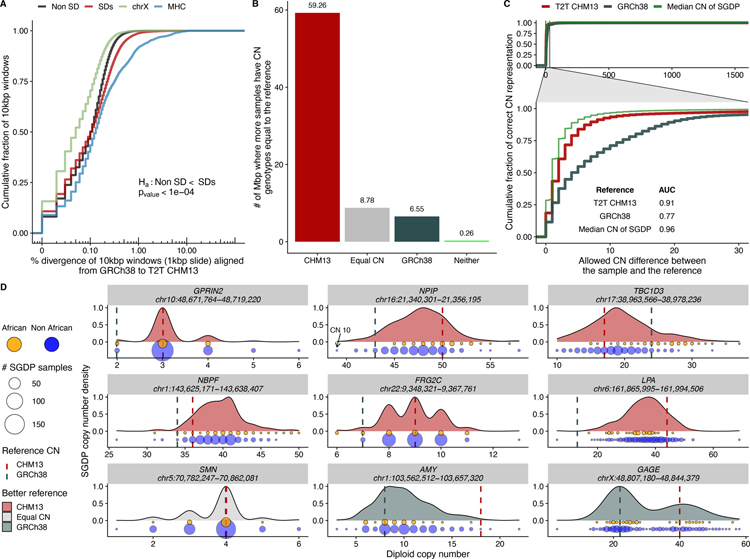
Fig. 3. SD single-nucleotide and copy number variation.
A) Sequence divergence (% in 10 kbp bins) based on syntenic alignments between GRCh38 and T2T-CHM13 for SDs (red), and unique genomic regions (black). SD regions show significantly more divergence when compared to unique sequence (black) and chromosome X (blue) but less than the MHC regions (green). B) Copy number of SD regions that are previously unresolved or structurally different in T2T-CHM13 compared to GRCh38 based on 268 human genomes from the Simons Genome Diversity Project (SGDP). The histogram shows the number of Mbp where more samples support the copy number of the given assembly [T2T-CHM13 (red), GRCh38 (blue), neither (green), or both equally (equal copy number)]. C) Empirical cumulative distribution showing how many samples genotype correctly with either GRCh38 or T2T-CHM13 as a function of the allowed difference between sample and reference copy number. The inset shows the area under the curve (AUC) calculation for both references allowing a maximum copy number difference of 30. The green curve shows an in silico reference made using the median copy number of the SGDP samples at each site. D) Genic copy number variation. Copy number variation in nine gene families are shown (generated with SGDP) and distribution is colored according to which reference better reflects the median copy number; GRCh38 generally underestimates copy number (vertical lines) and Africans (orange) tend to show higher copy number than non-Africans (blue); circle size indicates # of samples.
Of note, SD sequences are significantly more diverged than unique sequences (p-value < 0.001, one-sided Mann-Whitney U test) (fig. S8). Comparing only syntenic regions (25) between GRCh38 and T2T-CHM13, we estimate the single-nucleotide variant (SNV) density to be 0.95 SNVs/kbp for unique regions of the genome when compared to SD regions where density rises to 1.47 SNVs/kbp (table S5). This 50% increase could be due to an increased mutation rate of SDs (e.g., due to the action of interlocus gene conversion), or a deeper average coalescence of duplicated sequences. Another possible explanation for this observation is erroneous alignment of paralogous instead of allelic sequence; however, we believe this is unlikely given the requirement of at least 1 Mbp of continuous, one-to-one, best alignment between GRCh38 and T2T-CHM13 (25).
As part of this analysis, we also identified regions that structurally differ or are absent from GRCh38 when compared to the T2T-CHM13 assembly. Using 1 Mbp LASTZ alignments (25), we identified 126 non-syntenic regions for a total of 240 Mbp (N50 length of 12.7 Mbp; fig. S9). Of these, 33.9% (81.34/240 Mbp) overlap SD regions. Using sequence read-depth (25) from 268 human genomes (Simons Genome Diversity Project or SGDP), we compared the copy number of both T2T-CHM13 and GRCh38 (36) successfully genotyping 1,292 distinct copy number variable regions (74.85 Mbp). We find that CHM13 approximates the median (+/−2 s.d.) human copy number from SGDP for 94% of bases (70.6 Mbp) in contrast to GRCh38 where 57% of bases (42.8 Mbp) meet this metric (fig. S10). In particular, human copy number is nine times (59.26/6.55 Mbp) more likely to more closely match the CHM13 copy number rather than GRCh38 in non-syntenic SD regions (Fig. 3B). Thus, CHM13 is a better predictor (AUC 0.91) than GRCh38 (AUC 0.77) of human copy number variation and better approximates an in silico human reference constructed with the median copy number of the SGDP samples at every site (AUC 0.96, Fig. 3C). In non-syntenic SD regions GRCh38 tends to underestimate normal human copy number (by on average 9.2 copies or a median of 3.0 copies).
We identify 119 protein-encoding genes where CHM13 copy number better represents the true human copy number state. In contrast, only 65 genes are better represented by GRCh38 (table S6). These include both biomedically important genes relevant to disease risk (LPA and MUC3A) (37–43) as well as gene families implicated in the expansion of the human brain during human evolution (TBC1D3, NPIP, NPBF) (Fig. 3D and table S6) (7, 44–46). In T2T-CHM13, for example, there are additional copies of NPIP, NPBF, and GOLGA that are absent from GRCh38—each of these has been described as core duplicons responsible for the expansion of interspersed duplications in the human genome (27) as well as the emergence of human-specific gene families.
Interestingly, African genomes tend to have overall a higher copy number status when compared to non-African genomes. In particular, TBC1D3 shows ~7 fewer copies in non-Africans when compared to Africans (p-value < 1e-12). These findings suggest that higher copy number is likely ancestral (table S7) and CHM13, once again, better captures that diversity. Despite its primarily European origin, our results show that the more complete genome assembly serves as a better reference for copy number variation irrespective of population group (fig. S11).
Structural variation and massive evolutionary changes in the human lineage.
Advances in long-read genome assembly (47, 48) enable sequence resolution of complex structural variation associated with SDs at the haplotype level (49). We generated or used existing high-fidelity (HiFi) sequence data from 12 human and 5 nonhuman primate genomes to understand both the structural diversity and evolution of specific SD regions. To guide the selection of candidate regions for analysis, we constructed a hifiasm genome assembly of chimpanzee (Clint), compared it to the T2T-CHM13 assembly, and searched for regions of significant structural difference between the lineages. We focused first on the largest regions of insertion on the human lineage before sub-selecting those regions that contain genes of biomedical or evolutionary significance (tables S8 and S9). We restricted the analysis to insertions >50 kbp in length and selected 10 loci for a more detailed analysis, including genes associated with the expansion of the human frontal cortex (tables S8–S9 and fig. S12). Assemblies of additional haplotypes recapitulated the structural organization of T2T-CHM13 for eight of the 10 loci whereas evidence for the structural organization of GRCh38 was only found in five of the 10 loci (25). Overall, 73% of human haplotype assemblies were successfully reconstructed (table S8); however, the fraction of human haplotypes resolved at each locus varied considerably depending on the size and complexity of the region (fig. S13). For example, in the case of the 8.9 Mbp region corresponding to NOTCH2NL and SRGAP2B/2D, we recovered only 37.5% of human haplotypes (table S8 and fig. S7). Similarly, we resolved only six haplotypes (from a potential of 24 haplotypes) for the 3.4 Mbp region harboring the SMN1 and SMN2 loci (fig. S14).
Among haplotypes that could be resolved, we find a high degree of structural heterozygosity among human genomes (25) with 249 kbp differing on average when compared to T2T-CHM13 (table S10). In some cases, the structural changes are simple, such as ~12 kbp insertion or deletion of CYP2D6, which contributes to differential drug metabolism activity as well as other human disease susceptibilities (50–56) (fig. S15). In other cases, the patterns of structural variation are complex involving hundreds of kilobase pairs of inserted or deleted gene-rich sequence along with large-scale inversion events that alter gene order for specific human haplotypes (see ARGHAP11A/B; fig. S16 and NOTCH2NLA/B; fig. S7). Furthermore, the spinal muscular atrophy (SMA) locus containing SMN1 and SMN2—one of the most difficult regions to finish as part of the Human Genome Project on chromosome 5 (57)—shows a unique structure for all seven assembled haplotypes including GRCh38. Some haplotypes not only show increases in SMN2 copy number (fig. S14), a genetic modifier of SMA (58), but also potential functional differences in the organization and composition of SMN2. Since SMN2 serves as a target for small-molecule drug therapy improving splice-site efficiency compensating for the loss SMN1 in SMA patients (59), this level of sequence resolution suggests practical utility for disease risk assessment and treatment of patients.
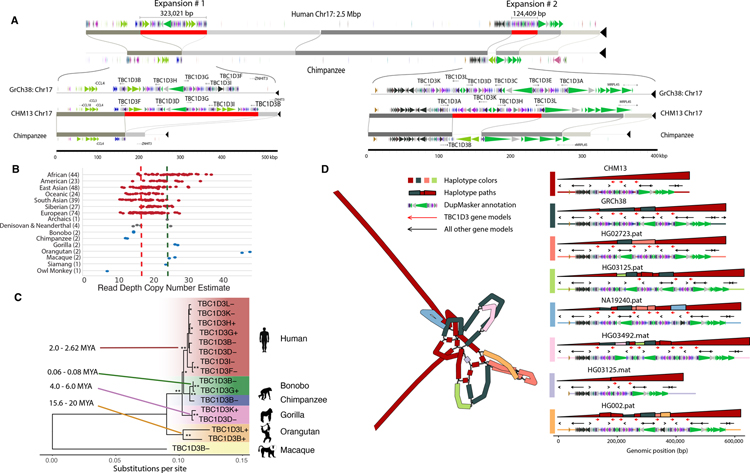
Of particular interest is the TBC1D3 gene family (44) (Fig. 4 and figs. S17–S18) whose protein products modulate epidermal growth factor receptor signaling and trafficking (60) and whose duplication in humans has been associated with expansion of the human prefrontal cortex as evidenced by mouse transgenic experiments (7). A comparison to chimpanzee (Fig. 4A) shows two massive genomic expansions in the human lineage (323.0 and 124.4 kbp). Both the high sequence identity (99.6%) and sequence read-depth comparisons of TBC1D3 copy number are consistent with expansion occurring in the human lineage after divergence from chimpanzee (Fig. 4B).
Fig. 4. Human-specific expansion of TBC1D3 compared to nonhuman primates.
A) Regions of homology between human T2T-CHM13’s chromosome 17 (top) and a HiFi assembly of the chimpanzee genome (bottom). Red blocks represent regions of human-specific expansion, including TBC1D3 duplications. Colored arrows above and below the homologous sequence represent unique ancestral units (duplicons) identified by DupMasker. Inset plots for both expansion sites are included below with the gene models identified by Liftoff (94). B) Copy number (diploid) estimates from an Illumina read-depth analysis of SGDP, ancient hominids, and nonhuman primates for a TBC1D3 paralog (table S14). Copy number estimates include pseudogenes (5) not included in the phylogeny, explaining the higher counts observed. The T2T-CHM13 copy number and GRCh38 copy number are represented by the red and blue lines, respectively. C) Phylogeny of TBC1D3 copies at these two expansion sites as well as nonhuman primate copies. Single asterisks at nodes indicate bootstrap values greater than or equal to 70%, while double asterisks indicate 100%. The data illustrate a human-specific expansion, as well as several independent expansions in the macaque, gorilla, and orangutan. Using macaque sequence as an outgroup, we estimate the human-specific expansion to be ~2.3 million years ago (MYA). D) Variation in human haplotypes across the first TBC1D3 expansion site: a graph representation (rGFA, left) of the locus where colors indicate the source genome for the sequence, and on the right the path for each haplotype-resolved assembly through the graph. The top row for each haplotype composed of large polygons represents an alignment comparing the haplotype-resolved sequence (horizontal) against the graph (vertical), and color represents the source haplotype for the vertical sequence. For example, a single large red triangle indicates there is a one-to-one alignment between CHM13 and the haplotype. Structural variants can be identified from discontinuities in height (deletion), changes between colors (insertion), or changes in the direction of the polygon (inversion). Below is shown the gene of interest (red arrow) and other genic content in the region (black arrow). Colored bars show ancestral duplication segments (duplicons) that compose the larger duplication blocks.
We extended this analysis to other nonhuman primates by generating HiFi assemblies for bonobo, gorilla, orangutan, and macaque. We identified TBC1D3 homologues in each species and constructed a maximum likelihood phylogeny using intronic or noncoding sequence flanking the gene (Fig. 4C). The analysis reveals recurrent and independent expansions of TBC1D3 in the orangutan, gorilla, and macaque species at different time points during primate evolution with the most recent expansions occurring 2 and 2.6 million years ago. However, these estimates assume that there has not been significant interlocus gene conversion, which may not be the case.
Complete sequencing of human TBC1D3 haplotypes reveals remarkable structural diversity (Fig. 4D) with TBC1D3 copy number ranging from three to fourteen TBC1D3 copies at expansion site #1, and two to nine copies at expansion site #2. In total, approximately one-third of human expansion site #2 shows large-scale structural variation and we identify >1.8 Mbp of duplicated sequence and >650 kbp of inverted sequence across the 18 haplotypes (including GRCh38). We estimate the structural heterozygosity of this locus to be 90.1% with 14 of 18 haplotypes showing structurally distinct duplication configurations (fig. S18). Similarly, TBC1D3 expansion site #1 is 87.6% heterozygous with 14 of 22 of the haplotypes displaying unique structures corresponding to copy number differences in the TBC1D3 gene family (fig. S17). Using orthogonal Oxford Nanopore Technologies (ONT) ultra-long-read sequencing, we validated these complex patterns of structural variation in a subset of the samples investigated here (25) (figs. S19 and S20). To better represent the structural genetic variation at this locus, we used a graph-based representation (61), which identified two TBC1D3 genes as common among all human haplotypes examined thus far (TBC1D3B at site #1 and TBC1D3A at site #2).
Additional gene models and variable duplicate genes.
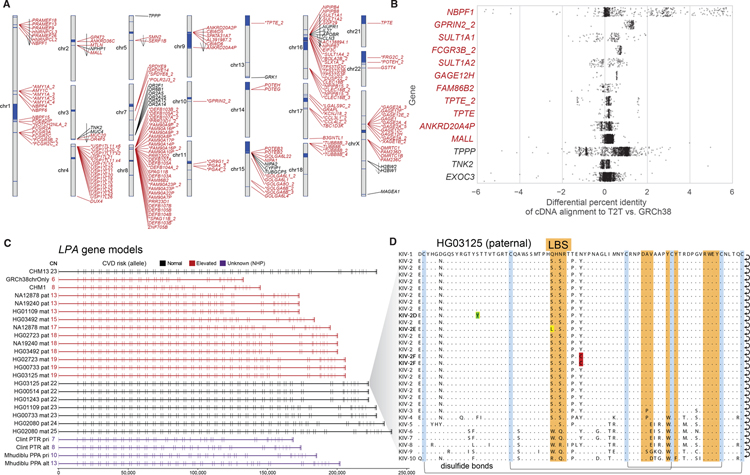
We identified 182 candidate previously unresolved or non-syntenic genes (25) in the T2T-CHM13 genome assembly (compared to GRCh38) with open reading frames and multiple exons (table S11). Of these 91% (166) corresponded to SD gene families (Fig. 5A). Many of these represent expanded tandem duplications (e.g., GAGE gene family members on the X chromosome) or large interspersed duplications (e.g., beta-defensin locus) adding additional copies of nearly identical genes to the human genome (Fig. 5A).
Fig. 5. Genic variation in previously unresolved SD regions of T2T-CHM13.
A) Ideogram showing the previously unresolved or non-syntenic gene models (open reading frames [ORFs] with >200 bp of coding sequence and multiple exons) in the T2T-CHM13 assembly as predicted by Liftoff. Previously unresolved genes mapping to SDs (red) are indicated with an asterisk if predicted to be an expansion in the gene family relative to GRCh38 (25). Arrows indicate inverted regions. Most unique genes mapping to non-syntenic regions (black) are the result of an inversion (arrow). B) Percent improvement in mapping of CHM13 Iso-Seq reads in candidate duplicated genes (red) mapping to non-syntenic regions of the T2T-CHM13 assembly. Positive values identify Iso-Seq reads aligning better to T2T-CHM13 than GRCh38. C) Gene models of LPA with ORF generated from haplotype-resolved HiFi assemblies. The double-exon repeat in these gene models encode for the Kringle IV subtype 2 domain of the LPA protein. Highlighted in red are haplotypes with reduced Kringle IV subtype 2 repeats predicted to increase risk of cardiovascular disease. D) Amino acid variation in the Kringle IV subtype 2 repeat in the paternal haplotype of HG01325 identifies a previously unknown set of amino acid substitutions including rare variants: Ser42Leu in the active site, Ser24Tyr and Tyr49Cys.
We searched for evidence that these copy number polymorphic or structurally variant regions were transcribed by aligning long-read transcript sequencing data and searching for perfect matches (25). We constructed a database of 44.2 million full-length cDNA transcripts derived from 31 human tissue samples and compared them to both the GRCh38 and T2T-CHM13 human genome references. For those 182 previously unresolved protein-coding genes where an unambiguous assignment could be made, 36% (65/182, >20 Iso-Seq reads) were confirmed as expressed with 23 showing the majority of reads mapping better to T2T-CHM13 when compared to GRCh38 (Fig. 5B).
Overall, across the entire genome, 12% of full-length transcripts exhibit at least 0.2% higher alignment identity when mapped against CHM13, while 8% align better to GRCh38. These results are consistent with the notion that the T2T-CHM13 is more complete, but that both assemblies are, in some cases, capturing different structurally variant haplotypes associated with genes. In addition to entirely new genes, we identify several gene models that were previously incomplete—many of which encode proteins with large tandem repeat domains (ZNF, LPA, Mucin; Fig. 5C). Among these is the complete gene structure of the Kringle IV domain of the lipoprotein A gene. Reduced copies of this domain are among the strongest genetic associations with cardiovascular disease, especially among African Americans (37–40, 62). Sequencing of multiple human haplotypes not only identified length variation but also other forms of rare coding variants potentially relevant to disease risk (Fig. 5D).
SD methylation and transcription.
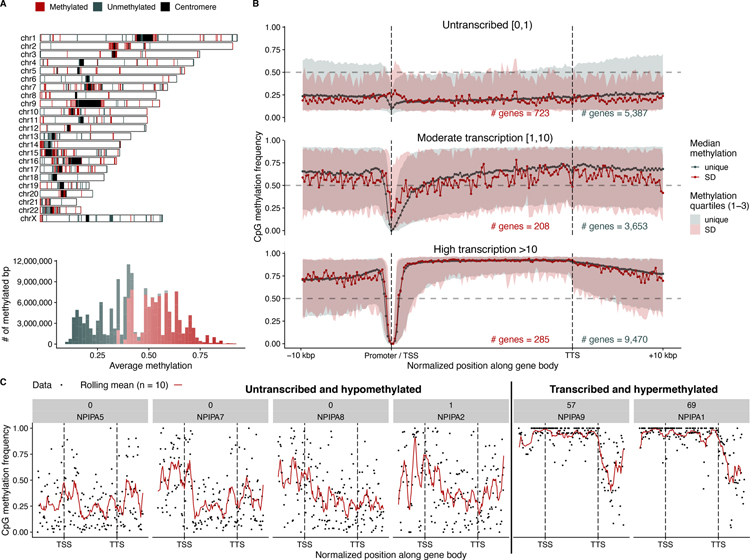
Since methylation is an important consideration in regulating gene transcription, we took advantage of the signal inherent in ultra-long-read ONT data (63–65) to investigate the CpG methylation status of SD genes within the CHM13 genome (25). Using hierarchical clustering, we find that SD blocks are generally either methylated or unmethylated as an entire block; (fig. S21 and Fig. 6A). Specifically, we find that 452 SD blocks flanked (127.7 Mbp) by unique sequences are hypermethylated in contrast to 222 hypomethylated SD blocks (52.1 Mbp). Methylation status does not appear to be driven by genomic location, e.g., proximity to the centromeres, acrocentric short arms, or telomeres (Fig. 6A).
Fig. 6. SD methylation and gene transcription.
A) Methylated (red) or unmethylated (blue-gray) SD blocks in the CHM13 genome based on processing ONT data. The histogram shows the distribution of average methylation across these regions. B) Median methylation signal of SD (red) and unique (blue-gray) genes stratified by their Iso-Seq expression levels in CHM13. The filled intervals represent the 25 and 75 quartiles of the observed data. Vertical lines indicate the position of the transcription start site (TSS) and the transcription termination site (TTS). C) Methylation signal across the recently duplicated NPIPA gene family in CHM13, showing increased methylation in transcriptionally active copies. Black points are individual methylation calls, and the red line is a rolling mean across 10 methylation sites. The labels in gray show the number of CHM13 Iso-Seq transcripts and the gene name.
Using full-length transcript data from CHM13, we compared methylation and transcription status of duplicated genes (25). If we stratify genes by their number of full-length transcripts, we observe distinct methylation patterns for transcribed and non-transcribed SD genes (Fig. 6B). For highly transcribed SD genes (genes without at least one exon overlapping with SD sequence) and unique genes, the gene body and flanking sequence are generally hypermethylated with a dramatic dip near the transcription start site (TSS)/promoter (66). In contrast, non-transcribed genes show moderate to low methylation across the gene body and flanking sequence.
Restricting the analysis to genes mapping within SDs, we find that transcriptionally silenced duplicate genes are more likely (10,000 permutations, p=0.0018) to map to hypomethylated regions of SD sequence (Fig. 6A) when compared to transcribed duplicate genes. Additionally, in untranscribed SD genes we observe a statistically significant (p-value < 0.001, one-sided Mann-Whitney U test) increase in TSS methylation (6.6% increase) when compared to unique genes where the TSS is more likely to be depleted for methylation (8.2% decrease).
One important consideration in this analysis is the presence of a CpG island within 1500 bp of the promoter (67). In our analysis of CHM13, for example, unexpressed unique genes have a low CpG count, consistent with a lack of CpG islands (fig. S22). If we repeat the same analysis on SD genes, we find that the unexpressed SD genes exist with and without CpG islands (fig. S22). In total, these observations suggest a process of epigenetic silencing for a subset of duplicate genes through general demethylation of the gene body but hypermethylation of promoter regions. Based on these observed signatures, we investigated whether these epigenetic features coincided with actively transcribed members of duplicate gene families.
We investigated a recently duplicated hominid gene family (NPIPA) (68) where sufficient paralogous sequence differences exist to unambiguously assign full-length transcripts to specific loci. While promoter/TSS signatures are less evident at the individual gene level, the gene body methylation signal appears diagnostic (Fig. 6C). NPIPA1 and NPIPA9, for example, are the most transcriptionally active and show demonstrably distinct methylation patterns providing an epigenetic signature to distinguish transcriptionally active loci associated with high-identity gene families that are otherwise largely indistinguishable. We show this trend also holds for other high-copy number gene families (fig. S23).
DISCUSSION
This work provides a comprehensive view of the organization of SDs in the human genome. The T2T-CHM13 reference adds a chromosome’s worth (81 Mbp) of SDs increasing the human genome average from 5.4% to 7.0% nearly doubling the number of SD pairwise relationships (24 vs. 41 thousand) and, as a result, predicts regions of genomic instability due to their potential to drive unequal crossing-over events during meiosis.
By every metric, T2T-CHM13 improves our representation of the structure of the human genome. This includes sequence-based organization of the short arms of chromosomes 13, 14, 15, 20 and 21 where we find that SDs account for more sequence (34.6 Mbp) than either heterochromatic satellite (26.7 Mbp) or rDNA (10 Mbp). Acrocentric SDs are almost twice as large when compared to non-acrocentric regions likely due to ectopic exchange events occurring among the short arms, which associate more frequently during the formation of the nucleolus (69).
Notably, nearly half of the acrocentric SDs involve duplications with non-acrocentric pericentromeric regions of chromosomes 1, 3, 4, 7, 9, 16, and 20. These duplicated islands of euchromatic-like sequences within acrocentric DNA are much more extensive than previously thought but have been shown to be transcriptionally active (70). While the underlying mechanism for their formation is unknown, it is noteworthy that three of the non-acrocentric regions have large secondary constriction sites (chromosomes 1q, 9q, and 16q) composed almost entirely of heterochromatic satellites (HSAT2 & 3) (fig. S2). These particular SD blocks, thus, are bracketed by large tracts of heterochromatic satellites and such configurations may make them particularly prone to double-strand breakage events (71) promoting interchromosomal duplications (fig. S3) between acrocentric and non-acrocentric chromosomes.
The T2T-CHM13 reference, along with resources from other human genomes, provides a baseline for investigating more complex forms of human genetic variation. For example, this complete reference sequence facilitates the design of sequence-anchored probes to systematically discover and characterize SD heteromorphic variation where chromosome organization differs among individuals (Fig. 2). Such chromosomal heteromorphisms have been traditionally investigated cytogenetically and are thought to be clinically benign (29–31). However, recent work indicates that these large-scale variants associate with infertility by increasing sperm aneuploidy, decreasing rates of embryonic cleavage (IVF), and increasing miscarriages (72–79). Distinguishing between fixed and heteromorphic acrocentric SDs will facilitate such research as well as the characterization of breakpoints associated with Robertstonian translocations—the most common form of human translocation (80).
At a finer-grained level, the T2T-CHM13 reference and the use of long reads from other human genomes provides access to other complex forms of variation involving duplicated gene families. Short-read copy number variation analyses and single-nucleotide polymorphism microarrays have long predicted that SDs are enriched 10-fold for copy number variation but the structural differences underlying these regions as well as their functional consequences have remained elusive (10, 81). We reveal elevated levels of human genetic variation in genes important for neurodevelopment (TBC1D3) and human disease (LPA, SMN). Even between just two genomes (GRCh38 and CHM13), we find that 37% (81 Mbp) of SD bases are uncharacterized or structurally variable and this predicts 182 copy number variable genes between two human haplotypes (table S6).
In cases such as TBC1D3, we find that most human haplotypes vary (64–78%). Different humans carry different complements and arrangements of the TBC1D3 gene family. The potential ramifications of this dramatic expansion in humans versus chimpanzees and of such high structural heterozygosity among humans are intriguing given the gene’s purported role in expansion of the frontal cortex (7). Similarly, we were able to reconstruct the complete structure of the LPA gene model in multiple human genotypes. While this is only a single gene, variability in the tandemly repeated 5.2 kbp protein-encoding Kringle IV domain underlies one of the most significant genetic risk factors for cardiovascular disease. Sequence resolution of the structural variation, as well as underlying amino-acid differences, allow us to predict previously uncharacterized risk alleles for disease (Fig. 5). Sequence-resolved structural variation improves genotyping and tests of selection (49, 82, 83) providing a path forward for understanding the disease and evolutionary implications of these complex forms of genetic variation.
Finally, and perhaps most importantly, the T2T-CHM13 reference coupled with other long-read datasets enables genome-wide functional characterization of recently duplicated genes. Both gene annotation and large-scale efforts to characterize the regulatory landscape of the human genome have typically excluded repetitive regions, including the 859 human genes mapping to high-identity SDs (84, 85). This is because the underlying short-read sequencing limits conventional RNA-seq or Chip-seq data from being assigned unambiguously to specific duplicated genes.
In this study, we generated long-read full-length transcript data (Iso-Seq) with long-read methylation data from ONT sequencing of the same genome to simultaneously investigate epigenetic and transcriptional data against a fully assembled reference genome. The long-read data from the same haploid source facilitated the unambiguous assignment of these functional readouts allowing us to correlate methylation and transcript abundance. Our initial analyses suggest that a large fraction of duplicate genes are in fact epigenetically silenced (characterized by hypermethylation of the promoter and hypomethylation of the gene body) and that this epigenetic mark may be used to predict actively transcribed loci even when genes are virtually identical (Fig. 6 and fig. S23). While more human genomes and diverse tissues need to be interrogated to assess the significance of this observation, it is clear that phased genome assemblies (49) with long-read functional readouts such as methylation (65), transcription, or Fiber-seq (86, 87) provide a powerful approach to understanding the regulatory landscape of duplicated and copy number polymorphic genes in the human genome.
There are several remaining challenges. First, not all human haplotypes corresponding to specific duplicated regions could be fully sequence resolved with automated assembly of long-read HiFi sequencing technology. The majority of the 250 unresolved regions of phased human genomes generated solely with HiFi long reads correspond to some of the largest and most variable duplicated regions of the human genome (49). For example, only 25% of SMN1/SMN2 haplotypes were fully resolved by HiFi assembly and unresolved loci are predicted to carry some of the most complex structural variation patterns. In comparison, the T2T-CHM13 assembly used both accurate HiFi and ultra-long ONT data, and future assembly methods that combine these technologies will likely be critical for diploid T2T assembly and the complete characterization of SD haplotypes (18, 88).
Another important challenge going forward will be how to accurately represent these more complex forms of human genetic variation, including functional annotation, where linear representations may be insufficient. While a more complex pangenome reference graph could overcome these limitations, it is unclear how this will be achieved in practice or how it will be adopted by the genomics and clinical communities. This highlights the importance of not only the construction of a pangenome reference but the necessary tools that will distinguish paralogous and orthologous sequences within duplications to allow for comparison between haplotypes with different SD architectures. The work currently underway by the Human Pangenome Reference Consortium (HPRC), Human Genome Structural Variation Consortium (HGSVC), and Telomere-to-Telomere (T2T) Consortium will be key to developing these methods and completing our understanding of SDs and their role in human genetic variation.
METHODS SUMMARY
SDs in T2T-CHM13 were identified using SEDEF (21) after repeat making with Tandem Repeats Finder (TRF) (89) and RepeatMasker (90). Syntenic one-to-one alignments were determined using halSynteny (91). Copy number prediction based on short-read data was performed with WSSD (3, 14) and mrsFAST (92), and regions of comparable copy number were determined with the changepoint package in R (93). To generate gene annotations we used the tools Liftoff (94) and GffRead (95). Fosmid probes were selected from the ABC10 library (28, 96) and two-color FISH was performed to experimentally validate acrocentric SDs (97–99). All assemblies (table S12) with the exception of T2T-CHM13 and GRCh38 were assembled with hifiasm v0.12 (47) using default parameters. Assembly validation of TBC1D3 was performed using sample-matched ONT data by checking the consistency of read alignments to the assemblies (100, 101). Phylogenetic analysis of TBC1D3 was performed with MAFFT and RAxML (102–105). Assembling pangenome graphs for select loci was performed with minigraph (61). Methylation analysis was performed using the methods and data described in Gershman et al. (106) using Winnowmap2 and Nanopolish for mapping and methylation calling (65, 107). Data visualization and figures [with the exception of Miropeats (108)] were primarily made in R making use of GenomicRanges (109), Tidyverse (110), karyoploteR (111), and circlize (112). Pipelines used for large-scale data analysis were constructed with Snakemake (113–115). Detailed descriptions of materials and methods are available in the supplementary materials (25).
Supplementary Material
Fig. 0. More complete segmental duplication content improves genotyping.
a) ~10-fold increase in the number of large (>10 kbp) acrocentric segmental duplications (red) in T2T-CHM13 (right) compared to GRCh38 (left). b) Read-depth genotyping of short-read Illumina whole-genome sequence from a human diversity panel (n=268) better matches T2T-CHM13 (red) when compared to GRCh38 (blue) irrespective of human population group considered.
ACKNOWLEDGEMENTS
The authors thank T. Brown and A. Lo for help in editing this manuscript.
Funding:
This work was supported, in part, by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health (S.N., S.K., and A.M.P.), grants from the U.S. National Institutes of Health (NIH grants 5R01HG002385, 5U01HG010971, and 1U01HG010973 to E.E.E.; 1R01HG011274 to K.H.M.; 5R01HG009190 to W.T.; and U41HG007234 to M.D.), and a grant from Futuro in Ricerca (2010-RBFR103CE3 to M.V.). E.E.E. is an investigator of the Howard Hughes Medical Institute.
Competing interests: E.E.E. is an SAB member of Variant Bio, Inc; S.K. and K.H.M. received travel funds to speak at events hosted by Oxford Nanopore Technologies; W.T. has licensed two patents to Oxford Nanopore Technologies (US 8748091 and 8394584).
Footnotes
Data materials and availability: CHM13hTERT cells were obtained for research use via a material transfer agreement with the University of Pittsburg. PacBio HiFi data has been deposited into NCBI Sequence Read Archive (SRA) under the following accessions: SRX7897688, SRX7897687, SRX7897686, and SRX7897685 for CHM13; SRR14407677 and SRR14407676 for CHM1; SRR10382244, SRR10382245, SRR10382248 and SRR10382249 for HG002; PRJNA540705 for NA12878; PRJEB36100 for HG00733 and HG00514; ERX4787609, ERX4787607, ERX4787606, ERX4782632, and ERX4781730 for NA19240; PRJNA701308 for HG01109, HG01243, HG02080, HG02723, HG03125, and HG03492; and PRJNA659034 and PRJNA691628 for all nonhuman primate samples. The human lymphoblastoid cell lines GM24385, GM19240, HG00514, and HG00733 used in the FISH experiments were obtained from Coriell. The T2T-CHM13 v1.0 assembly can be found on NCBI (GCA_009914755.2) and all associated read data were uploaded on SRA under the BioProject identifier PRJNA686988. Table S13 contains the accession information for all Iso-Seq data used in the paper. The canonical rDNA unit used to estimate copy number can be found on the NCBI nucleotide repository (KY962518.1). Human and nonhuman primate genome assemblies, SD annotations, methylation data, and Liftoff gene models can be found on Zenodo (DOI: 10.5281/zenodo.4721956, 116). Code for Snakemake pipelines, data analysis, and figure generation are also available on Zenodo (10.5281/zenodo.5498993 and 10.5281/zenodo.5498988, 117, 118).
LIST OF SUPPLEMENTARY CONTENT
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
REFERENCES AND NOTES
- 1.Ohno Wolf, Atkin, Evolution from fish to mammals by gene duplication. Hereditas. 59, 169–187 (1968). [DOI] [PubMed] [Google Scholar]
- 2.Ohno, Evolution by Gene Duplication (Springer Science & Business Media, 1970). [Google Scholar]
- 3.Bailey Yavor, Massa Trask, Eichler, Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 11, 1005–1017 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper Coe, Girirajan Rosenfeld, Vu, et al. , A copy number variation morbidity map of developmental delay. Nat. Genet. 43, 838–846 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dennis Nuttle, Sudmant Antonacci, Graves, et al. , Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell. 149, 912–922 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Florio Heide, Pinson Brandl, Albert, et al. , Evolution and cell-type specificity of human-specific genes preferentially expressed in progenitors of fetal neocortex. Elife. 7, e32332 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ju Hou, Sheng Wu, Zhou, et al. , The hominoid-specific gene TBC1D3 promotes generation of basal neural progenitors and induces cortical folding in mice. Elife. 5 (2016), doi: 10.7554/eLife.18197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fiddes Lodewijk, Mooring Bosworth, Ewing, et al. , Human-specific NOTCH2NL genes affect notch signaling and cortical neurogenesis. Cell. 173, 1356–1369.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sudmant Kitzman, Antonacci Alkan, Malig, et al. , Diversity of human copy number. Science. 11184, 2–7 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sudmant Mallick, Nelson Hormozdiari, Krumm, et al. , Global diversity, population stratification, and selection of human copy-number variation. Science. 349 (2015), doi: 10.1126/science.aab3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venter Adams, Myers Li, Mural, et al. , The sequence of the human genome. Science. 291, 1304–1351 (2001). [DOI] [PubMed] [Google Scholar]
- 12.She Jiang, Clark Liu, Cheng, et al. , Shotgun sequence assembly and recent segmental duplications within the human genome. Nature. 431, 927–930 (2004). [DOI] [PubMed] [Google Scholar]
- 13.IHGSC, Initial sequencing and analysis of the human genome. Nature. 409, 860–921 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Bailey Gu, Clark Reinert, Samonte, et al. , Recent segmental duplications in the human genome. Science. 297, 1003–1007 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Eichler Surti, Ophoff, Proposal for Construction a Human Haploid BAC library from Hydatidiform Mole Source Material (2002).
- 16.Fredman White, Potter Eichler, Dunnen Den, et al. , Complex SNP-related sequence variation in segmental genome duplications. Nat. Genet. 36, 861–866 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Chaisson Huddleston, Dennis Sudmant, Malig, et al. , Resolving the complexity of the human genome using single-molecule sequencing. Nature. 517, 608–611 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miga Koren, Rhie Vollger, Gershman, et al. , Telomere-to-telomere assembly of a complete human X chromosome. Nature. 585, 79–84 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green Krause, Briggs Maricic, Stenzel, et al. , A draft sequence of the Neandertal genome. Science. 328, 710–722 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nurk Koren, Rhie Rautiainen, Bzikadze, et al. , The complete sequence of a human genome. bioRxiv (2021), p. 2021.05.26.445798,, doi: 10.1101/2021.05.26.445798. [DOI] [Google Scholar]
- 21.Numanagic Gökkaya, Zhang Berger, Alkan, et al. , Fast characterization of segmental duplications in genome assemblies. Bioinformatics. 34, i706–i714 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bell Usher, McCarroll, Analyzing Copy Number Variation with Droplet Digital PCR. Methods Mol. Biol. 1768, 143–160 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez Sylvester, Complete sequence of the 43-kb human ribosomal DNA repeat: analysis of the intergenic spacer. Genomics. 27, 320–328 (1995). [DOI] [PubMed] [Google Scholar]
- 24.Kim Dilthey, Nagaraja Lee, Koren, et al. , Variation in human chromosome 21 ribosomal RNA genes characterized by TAR cloning and long-read sequencing. Nucleic Acids Res. 46, 6712–6725 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Supplementary materials and methods for segmental duplications and their variation in a complete human genome. [DOI] [PMC free article] [PubMed]
- 26.Jiang Hubley, Smit Eichler, DupMasker: a tool for annotating primate segmental duplications. Genome Res. 18, 1362–1368 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang Tang, Ventura Cardone, Marques-Bonet, et al. , Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat. Genet. 39, 1361–1368 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Kidd Cooper, Donahue Hayden, Sampas, et al. , Mapping and sequencing of structural variation from eight human genomes. Nature. 453, 56–64 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhasin, Human population cytogenetics: A review. Int. J. Hum. Genet. 5, 83–152 (2005). [Google Scholar]
- 30.Hsu Benn, Tannenbaum Perlis, Carlson, Chromosomal polymorphisms of 1, 9, 16, and Y in 4 major ethnic groups: a large prenatal study. Am. J. Med. Genet. 26, 95–101 (1987). [DOI] [PubMed] [Google Scholar]
- 31.Barber, Euchromatic heteromorphism or duplication without phenotypic effect? Prenat. Diagn. 14 (1994), pp. 323–324. [DOI] [PubMed] [Google Scholar]
- 32.Chaisson Sanders, Zhao Malhotra, Porubsky, et al. , Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat. Commun. 10, 1784 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanders Hills, Porubský Guryev, Falconer, et al. , Characterizing polymorphic inversions in human genomes by single-cell sequencing. Genome Res. 26, 1575–1587 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Porubsky Sanders, Höps Hsieh, Sulovari, et al. , Recurrent inversion toggling and great ape genome evolution. Nat. Genet. 52, 849–858 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki Gacquer, Heurck Van, Kumar Wojno, et al. , Human-Specific NOTCH2NL Genes Expand Cortical Neurogenesis through Delta/Notch Regulation. Cell. 173, 1370–1384.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mallick Li, Lipson Mathieson, Gymrek, et al. , The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature. 538, 201–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clarke Peden, Hopewell Kyriakou, Goel, et al. , Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N. Engl. J. Med. 361, 2518–2528 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Coassin Schönherr, Weissensteiner Erhart, Forer, et al. , A comprehensive map of single-base polymorphisms in the hypervariable LPA kringle IV type 2 copy number variation region. J. Lipid Res. 60, 186–199 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kronenberg Utermann, Lipoprotein(a): resurrected by genetics. J. Intern. Med. 273, 6–30 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Schmidt Noureen, Kronenberg Utermann, Structure, function, and genetics of lipoprotein (a). J. Lipid Res. 57, 1339–1359 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gum Hicks, Swallow Lagace, Byrd, et al. , Molecular cloning of cDNAs derived from a novel human intestinal mucin gene. Biochem. Biophys. Res. Commun. 171, 407–415 (1990). [DOI] [PubMed] [Google Scholar]
- 42.Pratt Crawley, Hicks Ho, Nash, et al. , Multiple transcripts of MUC3: evidence for two genes, MUC3A and MUC3B. Biochem. Biophys. Res. Commun. 275, 916–923 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Kyo Muto, Nagawa Lathrop, Nakamura, Associations of distinct variants of the intestinal mucin gene MUC3A with ulcerative colitis and Crohn’s disease. J. Hum. Genet. 46, 5–20 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Paulding Ruvolo, Haber, The Tre2 (USP6) oncogene is a hominoid-specific gene. Proc. Natl. Acad. Sci. U. S. A. 100, 2507–2511 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cantsilieris Sunkin, Johnson Anaclerio, Huddleston, et al. , An evolutionary driver of interspersed segmental duplications in primates. Genome Biol. 21, 202 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marques-Bonet Eichler, The evolution of human segmental duplications and the core duplicon hypothesis. Cold Spring Harb. Symp. Quant. Biol. 74, 355–362 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng Concepcion, Feng Zhang, Li, Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods. 18, 170–175 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nurk Walenz, Rhie Vollger, Logsdon, et al. , HiCanu: accurate assembly of segmental duplications, satellites, and allelic variants from high-fidelity long reads. Genome Res. 30, 1291–1305 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ebert Audano, Zhu Rodriguez-Martin, Porubsky, et al. , Haplotype-resolved diverse human genomes and integrated analysis of structural variation. Science (2021), doi: 10.1126/science.abf7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bertilsson Dahl, Dalén Al-Shurbaji, Molecular genetics of CYP2D6: clinical relevance with focus on psychotropic drugs. Br. J. Clin. Pharmacol. 53, 111–122 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hammer Sjöqvist, Plasma levels of monomethylated tricyclic antidepressants during treatment with imipramine-like compounds. Life Sciences. 6 (1967), pp. 1895–1903. [DOI] [PubMed] [Google Scholar]
- 52.Alexanderson Evans, Sjöqvist, Steady-state plasma levels of nortriptyline in twins: influence of genetic factors and drug therapy. Br. Med. J. 4, 764–768 (1969). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skoda Gonzalez, Demierre Meyer, Two mutant alleles of the human cytochrome P-450db1 gene (P450C2D1) associated with genetically deficient metabolism of debrisoquine and other drugs. Proc. Natl. Acad. Sci. U. S. A. 85, 5240–5243 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dahl Johansson, Palmertz Ingelman-Sundberg, Sjöqvist, Analysis of the CYP2D6 gene in relation to debrisoquin and desipramine hydroxylation in a Swedish population. Clin. Pharmacol. Ther. 51, 12–17 (1992). [DOI] [PubMed] [Google Scholar]
- 55.Gaedigk Blum, Gaedigk Eichelbaum, Meyer, Deletion of the entire cytochrome P450 CYP2D6 gene as a cause of impaired drug metabolism in poor metabolizers of the debrisoquine/sparteine polymorphism. Am. J. Hum. Genet. 48, 943–950 (1991). [PMC free article] [PubMed] [Google Scholar]
- 56.Johansson Lundqvist, Bertilsson Dahl, Sjöqvist, et al. , Inherited amplification of an active gene in the cytochrome P450 CYP2D locus as a cause of ultrarapid metabolism of debrisoquine. Proc. Natl. Acad. Sci. U. S. A. 90, 11825–11829 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmutz Martin, Terry Couronne, Grimwood, et al. , The DNA sequence and comparative analysis of human chromosome 5. Nature. 431, 268–274 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Butchbach, Copy Number Variations in the Survival Motor Neuron Genes: Implications for Spinal Muscular Atrophy and Other Neurodegenerative Diseases. Front Mol Biosci. 3, 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bebee Thomas, Bebee, Splicing regulation of the Survival Motor Neuron genes and implications for treatment of spinal muscular atrophy. Frontiers in Bioscience. 15 (2010), p. 1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wainszelbaum Charron, Kong Kirkpatrick, Srikanth, et al. , The hominoid-specific oncogene TBC1D3 activates Ras and modulates epidermal growth factor receptor signaling and trafficking. J. Biol. Chem. 283, 13233–13242 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Feng, Chu, The design and construction of reference pangenome graphs with minigraph. Genome Biol. 21, 265 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kronenberg, Human Genetics and the Causal Role of Lipoprotein(a) for Various Diseases. Cardiovasc. Drugs Ther. 30, 87–100 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Quick Loman, Duraffour Simpson, Severi, et al. , Real-time, portable genome sequencing for Ebola surveillance. Nature. 530, 228–232 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Loman Quick, Simpson, A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat. Methods. 12, 733–735 (2015). [DOI] [PubMed] [Google Scholar]
- 65.Simpson Workman, Zuzarte David, Dursi, et al. , Detecting DNA cytosine methylation using nanopore sequencing. Nat. Methods. 14, 407–410 (2017). [DOI] [PubMed] [Google Scholar]
- 66.Ball Li, Gao Lee, LeProust, et al. , Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 27, 361–368 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saxonov Berg, Brutlag, A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. U. S. A. 103, 1412–1417 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Johnson Viggiano, Bailey Abdul-Rauf, Goodwin, et al. , Positive selection of a gene family during the emergence of humans and African apes. Nature. 413, 514–519 (2001). [DOI] [PubMed] [Google Scholar]
- 69.Arnheim Nei, Koehn, Evolution of genes and proteins. Sinauer, Sunderland, MA, 38–61 (1983). [Google Scholar]
- 70.Lyle Prandini, Osoegawa ten Hallers, Humphray, et al. , Islands of euchromatin-like sequence and expressed polymorphic sequences within the short arm of human chromosome 21. Genome Res. 17, 1690–1696 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Luke Verma, Conte Mathews, Molecular characterization of the secondary constriction region (qh) of human chromosome 9 with pericentric inversion. J. Cell Sci. 103 ( Pt 4), 919–923 (1992). [DOI] [PubMed] [Google Scholar]
- 72.Barber Zhang, Friend Collins, Maloney, et al. , Duplications of proximal 16q flanked by heterochromatin are not euchromatic variants and show no evidence of heterochromatic position effect. Cytogenet. Genome Res. 114, 351–358 (2006). [DOI] [PubMed] [Google Scholar]
- 73.Sahin Yilmaz, Yuregir Bulakbasi, Ozer, et al. , Chromosome heteromorphisms: an impact on infertility. J. Assist. Reprod. Genet. 25, 191–195 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Codina-Pascual Navarro, Oliver-Bonet Kraus, Speicher, et al. , Behaviour of human heterochromatic regions during the synapsis of homologous chromosomes. Hum. Reprod. 21, 1490–1497 (2006). [DOI] [PubMed] [Google Scholar]
- 75.Caglayan Ozyazgan, Demiryilmaz Ozgun, Are heterochromatin polymorphisms associated with recurrent miscarriage? J. Obstet. Gynaecol. Res. 36, 774–776 (2010). [DOI] [PubMed] [Google Scholar]
- 76.Madon Athalye, Parikh, Polymorphic variants on chromosomes probably play a significant role in infertility. Reprod. Biomed. Online. 11, 726–732 (2005). [DOI] [PubMed] [Google Scholar]
- 77.Minocherhomji Athalye, Madon Kulkarni, Uttamchandani, et al. , A case-control study identifying chromosomal polymorphic variations as forms of epigenetic alterations associated with the infertility phenotype. Fertil. Steril. 92, 88–95 (2009). [DOI] [PubMed] [Google Scholar]
- 78.Hong Zhou, Tao Wang, Zhao, Do polymorphic variants of chromosomes affect the outcome of in vitro fertilization and embryo transfer treatment? Hum. Reprod. 26, 933–940 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kalantari Sepehri, Behjati Ashtiani, Akbari, Chromosomal studies in infertile men. Tsitol. Genet. 35, 50–54 (2001). [PubMed] [Google Scholar]
- 80.Wilch Morton, Historical and Clinical Perspectives on Chromosomal Translocations. Adv. Exp. Med. Biol. 1044, 1–14 (2018). [DOI] [PubMed] [Google Scholar]
- 81.Locke Segraves, Nicholls Schwartz, Pinkel, et al. , BAC microarray analysis of 15q11-q13 rearrangements and the impact of segmental duplications. J. Med. Genet. 41, 175–182 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ebler Clarke, Rausch Audano, Houwaart, et al. , Pangenome-based genome inference. Cold Spring Harbor Laboratory (2020), p. 2020.11.11.378133,, doi: 10.1101/2020.11.11.378133. [DOI] [Google Scholar]
- 83.Hsieh Vollger, Dang Porubsky, Baker, et al. , Adaptive archaic introgression of copy number variants and the discovery of previously unknown human genes. Science. 366 (2019), doi: 10.1126/science.aax2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.GTEx Consortium, Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group, Statistical Methods groups—Analysis Working Group, Enhancing GTEx (eGTEx) groups, NIH Common Fund, et al. , Genetic effects on gene expression across human tissues. Nature. 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dougherty Underwood, Nelson Tseng, Munson, et al. , Transcriptional fates of human-specific segmental duplications in brain. Genome Res. 28, 1566–1576 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stergachis Debo, Haugen Churchman, Stamatoyannopoulos, Single-molecule regulatory architectures captured by chromatin fiber sequencing. Science. 368, 1449–1454 (2020). [DOI] [PubMed] [Google Scholar]
- 87.Abdulhay McNally, Hsieh Kasinathan, Keith, et al. , Massively multiplex single-molecule oligonucleosome footprinting. Elife. 9 (2020), doi: 10.7554/eLife.59404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Logsdon Vollger, Hsieh Mao, Liskovykh, et al. , The structure, function and evolution of a complete human chromosome 8. Nature. 593, 101–107 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Benson, Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Smit Hubley, Green, RepeatMasker (1996).
- 91.Krasheninnikova Diekhans, Armstrong Dievskii, Paten, et al. , halSynteny: a fast, easy-to-use conserved synteny block construction method for multiple whole-genome alignments. Gigascience. 9 (2020), doi: 10.1093/gigascience/giaa047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hach Hormozdiari, Alkan Hormozdiari, Birol, et al. , MrsFAST: A cache-oblivious algorithm for short-read mapping. Nat. Methods. 7, 576–577 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Killick Haynes, Eckley Fearnhead, Lee, Package ‘changepoint.’ R package version 0. 4. −2011. - http://cran.rproject.org/web/packages/changepoint/index.html (2016) (available at https://cran.r-project.org/web/packages/changepoint/changepoint.pdf). [Google Scholar]
- 94.Shumate Salzberg, Liftoff: accurate mapping of gene annotations. Bioinformatics (2020), doi: 10.1093/bioinformatics/btaa1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pertea Pertea, GFF Utilities: GffRead and GffCompare. F1000Res 9 (2020), doi: 10.12688/f1000research.23297.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Altschul Gish, Miller Myers, Lipman, Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 97.Cardone Alonso, Pazienza Ventura, Montemurro, et al. , Independent centromere formation in a capricious, gene-free domain of chromosome 13q21 in Old World monkeys and pigs. Genome Biol. 7, R91 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sanders Falconer, Hills Spierings, Lansdorp, Single-cell template strand sequencing by Strand-seq enables the characterization of individual homologs. Nat. Protoc. 12, 1151–1176 (2017). [DOI] [PubMed] [Google Scholar]
- 99.Standing Committee on Human Cytogenetic Nomenclature, ISCN 1995: An International System for Human Cytogenetic Nomenclature (1995) : Recommendations of the International Standing Committee on Human Cytogenetic Nomenclature, Memphis, Tennessee, USA, October 9–13, 1994 (Karger Medical and Scientific Publishers, 1995). [Google Scholar]
- 100.Quinlan Hall, BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li, Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 34, 3094–3100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Katoh Misawa, Kuma Miyata, MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stamatakis, RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Berger Munson, A novel randomized iterative strategy for aligning multiple protein sequences. Comput. Appl. Biosci. 7, 479–484 (1991). [DOI] [PubMed] [Google Scholar]
- 105.Gotoh, Optimal alignment between groups of sequences and its application to multiple sequence alignment. Comput. Appl. Biosci. 9, 361–370 (1993). [DOI] [PubMed] [Google Scholar]
- 106.Gershman Sauria, Hook Hoyt, Razaghi, et al. , Epigenetic Patterns in a Complete Human Genome. bioRxiv (2021), p. 2021.05.26.443420,, doi: 10.1101/2021.05.26.443420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jain Rhie, Zhang Chu, Walenz, et al. , Weighted minimizer sampling improves long read mapping. Bioinformatics. 36, i111–i118 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Parsons, Miropeats: graphical DNA sequence comparisons. Comput. Appl. Biosci, 615–619 (1995). [DOI] [PubMed] [Google Scholar]
- 109.Lawrence Huber, Pagès Aboyoun, Carlson, et al. , Software for computing and annotating genomic ranges. PLoS Comput. Biol. 9, e1003118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wickham Averick, Bryan Chang, McGowan, et al. , Welcome to the tidyverse. J. Open Source Softw. 4, 1686 (2019). [Google Scholar]
- 111.Gel Serra, karyoploteR: an R/Bioconductor package to plot customizable genomes displaying arbitrary data. Bioinformatics. 33, 3088–3090 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gu Gu, Eils Schlesner, Brors, circlize Implements and enhances circular visualization in R. Bioinformatics. 30, 2811–2812 (2014). [DOI] [PubMed] [Google Scholar]
- 113.Köster Rahmann, Snakemake—a scalable bioinformatics workflow engine. Bioinformatics. 28, 2520–2522 (2012). [DOI] [PubMed] [Google Scholar]
- 114.Köster Rahmann, Snakemake-a scalable bioinformatics workflow engine. Bioinformatics. 34, 3600 (2018). [DOI] [PubMed] [Google Scholar]
- 115.Mölder Jablonski, Letcher Hall, Tomkins-Tinch, et al. , Sustainable data analysis with Snakemake. F1000Res. 10, 33 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vollger, Assemblies and data generated for “SEGMENTAL DUPLICATIONS AND THEIR VARIATION IN A COMPLETE HUMAN GENOME” (2021), doi: 10.5281/ZENODO.4721956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vollger, mrvollger/Data-Analysis-for-SDs-in-T2T-CHM13: (2021; https://zenodo.org/record/5498994).
- 118.Vollger, mrvollger/assembly_workflows: (2021; https://zenodo.org/record/5498989).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.