Abstract
We previously demonstrated that pentamidine, which has been clinically used against Pneumocystis carinii, inhibits in vitro a group I intron ribozyme from that organism. Another fungal pathogen, Candida albicans, also harbors a group I intron ribozyme (Ca.LSU) in the essential rRNA genes in almost half of the clinical isolates analyzed. To determine whether pentamidine inhibits Ca.LSU in vitro and in cells, phylogenetically closely related intron-containing (4-1) and intronless (62-1) strains were studied. Splicing in vitro of the Ca.LSU group I intron ribozyme was completely inhibited by pentamidine at 200 μM. On rich glucose medium, the intron-containing strain was more sensitive to growth inhibition by pentamidine than was the intronless strain, as measured by disk or broth microdilution assays. On rich glycerol medium, they were equally susceptible to pentamidine. At pentamidine levels selectively inhibiting the intron-containing strain (1 μM) in glucose liquid cultures, inhibition of splicing and rRNA maturation was detected by quantitative reverse transcription-PCR within 1 min with a 10- to 15-fold accumulation of precursor rRNA. No comparable effect was seen in the intronless strain. These results correlate the cellular splicing inhibition of Ca.LSU with the growth inhibition of strain 4-1 harboring Ca.LSU. Broth microdilution assays of 13 Candida strains showed that intron-containing strains were generally more susceptible to pentamidine than the intronless strains. Our data suggest that ribozymes found in pathogenic microorganisms but absent in mammals may be targets for antimicrobial therapy.
The widespread use of broad-spectrum antibacterial agents as well as immunosuppressive treatments has increased the incidence of fungal infections (16). In the immunocompromised population, opportunistic fungal infections have become a major cause of morbidity and mortality (7). Candida albicans, normally a commensal fungus in humans, can cause mild, superficial infections in an immunocompetent host but is capable of causing life-threatening systemic disseminated disease in a compromised patient. In human immunodeficiency virus (HIV)-infected patients, mucosal candidiasis has a reported incidence of more than 90% and oropharyngeal candidiasis, which is the most common fungal infection in these patients, has been used to predict the development of AIDS in HIV-1-infected patients (8, 16). Furthermore, the emerging problem of increasing azole-resistant C. albicans isolates (7, 17, 19, 34) and the fact that azoles and other major classes of antifungal drugs target the ergosterol biosynthesis pathway raise the possibility of the simultaneous appearance of resistance to multiple agents (7, 12). Therefore, the search for novel antifungal targets has become an important priority because intolerance, resistance, toxicity, and multiple-drug interactions among the current antimicrobial agents are major issues yet to be resolved (14).
C. albicans harbors a self-splicing group I intron (Ca.LSU) in the nuclear rRNA genes in about 40% of all isolates so far analyzed (30). In Candida dubliniensis a similar ribozyme has been found in every isolate tested (2). Other pathogens harboring nuclear group I introns include Pneumocystis carinii (22, 39) and Naegleria (10) and Acanthamoeba species (11). The presence of group I intron ribozymes in essential genes of pathogens but not in the human genome suggests that splicing inhibitors can potentially act as antimicrobial agents (21–24, 27, 29, 30, 35, 46–49). Consistent with these expectations, Mercure et al. (30) demonstrated a differential susceptibility to 5-fluorocytosine between an intron-containing C. albicans strain and a closely related intronless strain. The higher susceptibility of the intron-containing strain appears to be due to the inhibitory effect of biosynthetic incorporation of this base analog into the group I intron ribozyme. Pentamidine, a proven in vitro inhibitor of group I intron ribozymes from P. carinii (24), has been shown to inhibit the growth of a DNA virus harboring such a ribozyme in an essential gene without detectable growth inhibition of its algal host (51). The observation that Saccharomyces cerevisiae, which generally contains group I and group II introns in its mitochondrial genome, is more susceptible to pentamidine when grown on a nonfermentable glycerol carbon source (25; Y. Zhang, A. Bell, P. S. Perlman, and M. J. Leibowitz, submitted for publication) is consistent with the possibility that these ribozymes are targets for this antimicrobial action. Zhang et al. (submitted) confirmed the specific inhibitory effect of pentamidine on S. cerevisiae mitochondrial intron splicing in vivo and in vitro. The failure of ribozyme introns to be removed from rRNA precursors can be expected to be deleterious to the cell. Introduction of a splicing-deficient mutant group I intron from the Tetrahymena thermophila 25S rRNA gene into the 23S rRNA gene of Escherichia coli resulted in nonfunctional ribosomes which failed to assemble normally, indicating the potential antimicrobial activity of splicing inhibitors (31).
The aim of this study was to determine whether pentamidine, a known inhibitor of group I intron splicing in vitro, inhibits a C. albicans nuclear group I intron ribozyme in the 25S rRNA gene, denoted Ca.LSU, and whether this inhibition in cells results in anticandidal activity. We have found, both in vitro and in cultured C. albicans cells, that this ribozyme activity is inhibited by pentamidine. Furthermore, the cellular inhibition of splicing correlates with differential inhibition of growth of an intron-containing strain compared to a closely related intronless strain, suggesting that ribozyme inhibition is a possible mechanism of action of antifungal agents.
MATERIALS AND METHODS
Antimicrobial agents.
Pentamidine isethionate, tetracycline, and amphotericin B were from Sigma (St. Louis, Mo.). The pentamidine analogs BBE [trans-1,2-bis(5-amidino-2-benzimidazolyl)ethene] and DIMP {1,3-di[4-(2-imidazolinyl)-2-methoxyphenoxy]propane} and propamidine were kindly provided by R. R. Tidwell (University of North Carolina). Amphotericin B was dissolved in dimethyl sulfoxide. Tetracycline and pentamidine and its analogs were dissolved in sterile water.
C. albicans strains and culture media.
Clinically isolated C. albicans strains 4-1 and 62-1 were kindly provided by G. Lemay (University of Montreal, Montreal, Canada). C. albicans 4-1, but not strain 62-1, bears the Ca.LSU self-splicing group I intron in the 25S ribosomal DNA (rDNA) (30). These two strains are closely related based on genetic sequence analysis of variable regions V2 and V3 of the genes encoding 25S rRNA (2). Ten C. albicans clinical isolates, previously tested for their susceptibility to pentamidine (40), were kindly provided by G. St-Germain (Laboratoire de Santé Publique du Québec, Quebec, Canada). C. albicans ATCC 90028, obtained from the American Type Culture Collection (Manassas, Va.), was used as a control for drug susceptibility assays. These strains were maintained on YPD medium plates (1% yeast extract, 2% peptone, 2% dextrose, 2% agar). The MICs of pentamidine and amphotericin B were determined in YPD and YPG (1% yeast extract, 2% peptone, 3% glycerol) media. Other media used included SD (6.7% yeast nitrogen base without amino acids, 2% dextrose), and RPMI 1640, buffered to pH 7.0 with MOPS (morpholinepropanesulfonic acid) (37).
Oligonucleotide primers.
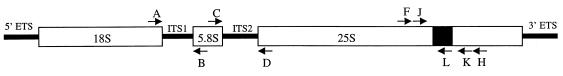
The sequence of each oligonucleotide is shown in Table 1. The location of each primer used for PCR of rDNA and for reverse transcription (RT)-PCR of rRNA and precursors is depicted in Fig. 1. The primer pairs were A-B for the internal transcribed spacer 1 (ITS1) (248-bp product), C-D for the ITS2 (207-bp product), F-H for the in vitro transcription DNA template (735-bp product), and J-K (256- and 635-bp intronless and intron-containing products, respectively) or J-L (465-bp product) for the 25S rRNA or rDNA containing the Ca.LSU intron.
TABLE 1.
Oligonucleotides used
| Designation | Sequence (5′ to 3′)a | 5′ coordinate | Accession no. |
|---|---|---|---|
| A | CCGAGAAGCTGGTCAAAC | 1703 (+); 18S | M60302 |
| B | ACCAAGAGATCCGTTGTTG | 26 (−); 5.8S | X71088 |
| C | GCATGCCTGTTTGAGCGTC | 136 (+); 5.8S | X71088 |
| D | CAGCGGGTAGTCCTACCTG | 72 (−); 25S | X70659 |
| F | ATTTAGGTGACACTATAGAATTAAAACATAGCATTGTGATG | 2158 (+); 25S | X70659 |
| H | CCGAAGCTCCCACTTATTC | 2495 (−); 25S | X70659 |
| J | CAATGTGATTTCTGCCCAGTG | 2201 (+); 25S | X70659 |
| K | CACAATGTCAAACTAGAGTCAAG | 2457 (−); 25S | X70659 |
| L | CATTGCTCCAAGAAATCGC | 379 (−); Ca.LSU intron | X74272 |
| 10816 | GGTGATGGTGTTACTCACG | 2150 (+); actin | X16377 |
| 10473 | GCTTCCAAACCTAAATCAGC | 2496 (−); actin | X16377 |
| Oligo(dT) | T18 |
Underlined sequence in primer F is a promoter sequence from bacteriophage SP6; the 5′ coordinate shown is for nucleotide 19 of this sequence.
FIG. 1.
Structure of the rRNA gene operon and location of each PCR and RNA RT-PCR primer pair. The RT-PCR primer pairs were A-B for ITS1, C-D for ITS2, J-L for the intron in precursor rRNA, and J-K for colony PCR of Candida cells. The PCR primer pair for the amplification of the DNA template used for the SP6 in vitro transcription of precursor RNA was F-H. The open rectangles represent the mature rRNAs; the solid square represents the Ca.LSU intron; the thick lines represent the ETSs and ITSs (not proportional to length, with ITS being shorter and ETS longer than indicated).
Splicing assays and kinetic analysis.
In vitro splicing reactions were performed as previously described (24) using in vitro-transcribed rRNA precursor derived from the genome of C. albicans strain 4-1. Primers F and H were used to PCR amplify a 735-bp fragment from the 25S rRNA gene (including a 17-bp SP6 promoter region derived from primer F) containing the complete group I intron (379 nucleotides) and partial flanking exon regions (130 and 209 nucleotides; 5′ and 3′, respectively) using Taq DNA polymerase (Qiagen, Valencia, Calif.). The amplification conditions were 2 cycles of 94°C for 30 s, 50°C for 1 min, and 72°C for 45 s, followed by 30 cycles of 94°C for 10 s, 53°C for 1 min, and 72°C for 45 s, followed by a 5-min incubation at 72°C. In vitro transcription of this PCR product DNA to produce a 718-nucleotide E1-I-E2 precursor RNA (E1 indicates a 5′ exon fragment, I indicates an intron, and E2 indicates a 3′ exon fragment) was carried out using SP6 RNA polymerase with the Riboprobe System II (Promega, Madison, Wis.) in the presence of 3.4 μM [α-32P]UTP (50 μCi; NEN, Boston, Mass.), under the recommended conditions. After 40 min at 37°C, 1 U of DNase (RNase-free; RQ1; Promega) was added and the DNA template was digested for 15 min at 37°C. Transcripts were purified with the RNeasy kit (Qiagen), and the full-length precursor was purified by preparative polyacrylamide gel electrophoresis (PAGE; 5% acrylamide, 0.1% bisacrylamide) run in the presence of 8 M urea, followed by electroelution (Elu-Trap; Schleicher and Schuell, Keene, N.H.) and ethanol precipitation.
Splicing reactions were carried out in the presence of purified precursor RNA, 50 mM Tris-HCl [pH 7.5], 1.25 mM magnesium chloride, 0.4 mM spermidine, 4 U of RNasin (Promega), and 10 μM GTP at 37°C for 10 min, followed by gel electrophoresis as described above. The gels were dried, and the precursor, intermediates, and products of the splicing reaction were visualized by autoradiography and quantitated using a densitometer (GS-670; Bio-Rad, Hercules, Calif.) or a phosphorimager (GS-525; Bio-Rad). The extent of the splicing reaction was calculated by the percentage of radioactivity in the products (I plus E1-E2) and intermediates (I-E2 plus E1) relative to total radioactivity (P plus I-E2 plus E1 plus I plus E1-E2). Kinetic analysis to determine the constant of inhibition values for compounds inhibiting the splicing reaction were derived from data expressed in Hanes-Woolf plots (50), as previously described (24).
Drug susceptibility testing.
Determination of the pentamidine MIC80 (lowest concentration that produced an 80% growth decrease compared with a drug-free control) and the amphotericin B MIC100 (lowest concentration that completely inhibited growth) for C. albicans strains 4-1 and 62-1 in YPD and YPG media was performed by a modified microdilution method (37). C. albicans control strain ATCC 90028 was tested in the standard RPMI 1640 medium buffered to pH 7.0 with MOPS. Briefly, flat-bottom 96-well microtiter plates were prepared by making twofold serial dilutions (in duplicate or triplicate) of pentamidine ranging from 100 to 0.19 μM and of amphotericin B ranging from 16 to 0.313 μg/ml. The final volume of the drug solution was 0.1 ml. Each strain inoculum (0.1 ml) was prepared from colonies (≥1 mm in diameter) on YPD medium plates grown for 42 h at 30°C in 0.85% saline by using a 0.5 McFarland turbidity standard and then diluted in each culture medium to 0.5 × 103 to 2.5 × 103 CFU/ml. MIC endpoints were determined after incubation for 24 and 48 h at 35°C by spectrophotometry at 540 nm after the plates were gently agitated to homogenize the cultures. The average background optical density (OD) of the inoculum-free wells (medium-only control) was subtracted from the average of the OD of each experimental group (in duplicate). These values were divided by the average of the OD of the drug-free control wells to calculate the growth percentage of experimental wells relative to the drug-free-growth positive control. All 13 C. albicans isolates at hand were tested simultaneously for susceptibility to pentamidine in YPD medium at 35°C for 24 h as described above, except that the inoculum colonies (<1 mm in diameter) used were from YPD medium plates grown at 35°C for 24 h.
Disk assay.
The susceptibilities of C. albicans strains 4-1 and 62-1 to pentamidine on YPD medium plates were measured by a disk assay. Cells grown at 30°C for 24 to 48 h on YPD medium plates were suspended in sterile water at 1.25 × 107/ml, and 0.4 ml of the cell suspension was spread circularly from the center out using a glass pipette as the plate was continuously rotated on a turntable. After the cell lawn was allowed to dry briefly, five 3MM paper disks (6-mm diameter) (Whatman, Maidstone, England) were placed on the plate and 10 μl of pentamidine (200, 20, 2, or 0.2 nmol) or sterile water was spotted on each disk. Zones of inhibition were measured after 4 days of incubation at 30°C. An inhibition zone 8 mm in diameter was the smallest that could be accurately measured.
Preparation of C. albicans total RNA.
Ten milliliters of SD medium was inoculated with a single colony of C. albicans 4-1 or 62-1 from a YPD plate, and the colony was grown overnight at 28°C at 250 rpm. Fresh 100-ml YPD cultures were inoculated at an OD at 600 nm (OD600) of 0.1, and growth was continued. At an OD600 of 0.4 to 0.6 (mid-log phase), pentamidine isethionate dissolved in water was added to a 1-μM final concentration, and at various times 1-ml aliquots were aseptically transferred to 1.5-ml microcentrifuge tubes. Cell pellets were collected by a brief spin to remove the medium and were immediately frozen in a dry ice-ethanol bath. A modified version of a hot phenol-chloroform protocol (36) was used to extract RNA from the frozen cell pellets. The modifications included cell breakage with glass beads by vortexing each microcentrifuge tube three times for 10 s during a 12-min incubation in a water bath at 65°C and two reextractions of the organic phase plus interphase for 1 min with 0.1 ml of AE buffer (36). All three aqueous phases were pooled before the final extraction with AE-buffered phenol-chloroform. Every RNA preparation was treated with RQ1 DNase I (Promega) and purified with the RNeasy kit RNA clean-up protocol (Qiagen). The RNA quantity and quality were checked by spectrophotometry (OD260 and OD260/OD280 ratio) and by agarose gel electrophoresis. Alternatively, RNA was extracted from cells grown in YPD (20-ml cultures) to an OD600 of 0.5 at 35°C following a protocol for the extraction of precursor rRNA and preribosomes (18). This RNA preparation was used for the competitive RT-PCR analysis.
RT-PCR.
RT-PCR of each RNA template was carried out using the Titan One Tube RT-PCR system (Boehringer Mannheim, Indianapolis, Ind.). RT was performed by a 30-min incubation at 50°C for rRNA templates or 55°C for the actin mRNA template, followed by a 2.5-min incubation at 94°C to inactivate the reverse transcriptase. PCR amplification conditions for the resulting cDNAs were as follows. For the 25S intron (primer pair J-L) and actin mRNA control [primer pair 10816-10473 and oligo(dT)], the PCR conditions were 5 cycles of 94°C for 30 s, 52°C for 30 s, and 68°C for 45 s, followed by 25 cycles of 94°C for 30 s, 54 to 55°C for 30 s, and 68°C for 1 min. For ITS1 (primer pair A-B), the conditions were 5 cycles of 94°C for 30 s, 49°C for 30 s, and 68°C for 30 s, followed by 25 cycles of 94°C for 30 s, 52°C for 30 s, and 68°C for 45 s. For ITS2 (primer pair C-D), the conditions were 5 cycles of 94°C for 30 s, 56°C for 30 s, and 68°C for 45 s, followed by 25 cycles of 94°C for 30 s, 58°C for 30 s, and 68°C for 1 min. All reactions were followed by a 5-min incubation at 68°C. All cDNA PCR products were resolved in 5% PAGE gels, and the bands were visualized by SybrGreen I staining (FMC, Rockland, Maine). Photographs of gels were scanned with a densitometer, and each band was quantitated with Molecular Analyst/PC software (Bio-Rad).
Preparation of competitor RNA.
Competitor RNA refers to the in vitro-synthesized RNA used as an internal control in the quantitation of precursor rRNA by RT-PCR. Primers F and L (within the 25S rRNA exon 1 and the 3′ end of the Ca.LSU group I intron, respectively) were used to amplify a fragment from the 735-bp PCR product generated with primers F and H. This fragment was blunt ended and phosphorylated using bacteriophage T4 DNA polymerase (GIBCO BRL) and polynucleotide kinase (Pharmacia Biotech, Piscataway, N.J.) under the recommended conditions. The fragment was ligated with T4 DNA ligase (Rapid DNA ligation kit; Boehringer Mannheim) to vector plasmid pUC19 that had been linearized with SmaI (GIBCO BRL), and dephosphorylated with calf intestine alkaline phosphatase (Boehringer Mannheim). To produce a 50-bp deletion within the intron sequence of this construct, a double digestion with StuI (Boehringer Mannheim) and MscI (New England Biolabs, Beverly, Mass.) followed by ligation was performed. In vitro transcription of this deletion construct to synthesize competitor RNA transcript was carried out using SP6 RNA polymerase and the Riboprobe Gemini System II (Promega) as recommended by the manufacturer.
Quantitative competitive RT-PCR.
Competitive RT-PCR of the target and competitor RNA was carried out using the Titan One Tube RT-PCR System. RT and PCR conditions were identical to those of the noncompetitive RT-PCR with the exception that each reaction mixture contained a constant amount of competitor RNA (determined by comparison with standards on agarose electrophoretic gels stained with ethidium bromide) and varying amounts of total (target) RNA. The cDNA PCR products were resolved in a 5% PAGE gel, and the bands were visualized by ethidium bromide or SybrGreen I (FMC) staining. The gel images were then scanned using a densitometer (GS-670; Bio-Rad), and each band was quantitated using the Molecular Analyst/PC software. Quantitative analysis of the band measurements was done as described previously (52).
Colony PCR.
Detection of Ca.LSU in 13 C. albicans clinical isolates was performed by PCR amplification with primers J and K using whole cells from single colonies grown on YPD plates at 35°C. An initial incubation at 95°C with the cells in the reaction mixture was followed by 3 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 45 s and 22 cycles of 94°C for 30 s, 59°C for 30 s, and 72°C for 1 min. This reaction was followed by a 5-min incubation at 72°C. The PCR products were resolved in a 1% agarose gel.
RESULTS
In vitro splicing inhibition assays.
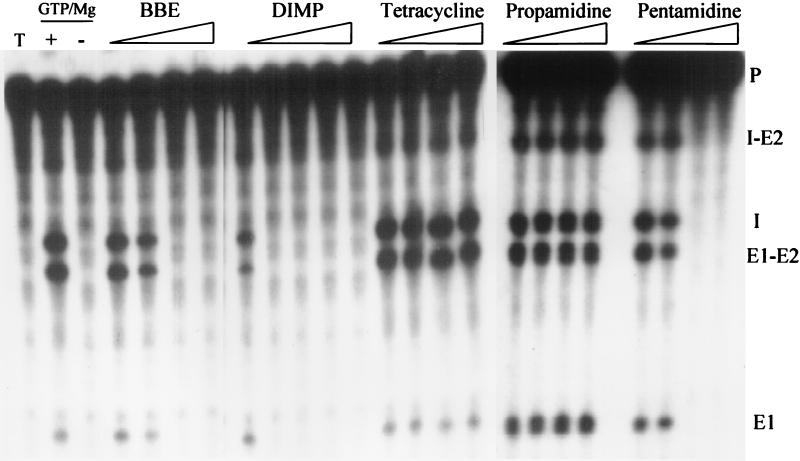
We have analyzed the C. albicans 4-1 strain 25S rDNA ribozyme Ca.LSU (30) for its sensitivity to known group I intron splicing inhibitors and other chemical agents, based on protocols used for in vitro characterization of the P. carinii rDNA group I intron ribozyme Pc1.LSU (24). Like other group I intron ribozymes, Ca.LSU requires magnesium and GTP for activity (30). The optimal magnesium chloride concentration at 37°C and ≤7.8 μM GTP was determined to be 1.25 mM, and the splicing reaction was in a linear range for up to 12 min (data not shown). The calculated Michaelis constant for GTP was 2.5 μM, which is comparable to 3 μM for the P. carinii ribozyme Pc1.LSU (24) and far below the intracellular GTP concentration in S. cerevisiae. Some agents known to inhibit the in vitro splicing of Pc1.LSU were tested for the ability to inhibit splicing of Ca.LSU precursor RNA under standardized conditions described in Materials and Methods. As shown in Fig. 2, pentamidine, BBE, and DIMP completely inhibited the Ca.LSU intron splicing reaction at 200, 0.5, and 10 μM, respectively. These values are 1.25-, 5-, and 6-fold lower than the levels required to completely inhibit splicing of Pc1.LSU (24). On the other hand, tetracycline, which inhibited Pc1.LSU in vitro splicing at 50 μM, failed to inhibit Ca.LSU intron in vitro splicing at 300 μM (data not shown). Propamidine at up to 1.0 mM failed to inhibit splicing of either intron. Hanes-Woolf plots for the Ca.LSU intron in vitro-splicing inhibitors pentamidine, BBE, and DIMP indicated noncompetitive inhibition (data not shown), as previously described for Pc1.LSU (24).
FIG. 2.
Inhibition of Ca.LSU intron splicing by pentamidine analogs. Reactions were run under standard conditions as indicated in Materials and Methods. The first lane (T) shows precursor RNA only, the second lane (+) shows the complete reaction without drugs, and the third lane (−) shows the reaction in the absence of magnesium and GTP. The experimental compounds are present in a complete splicing reaction at the following concentrations (from left to right): 0.125, 0.25, 0.5, or 0.75 μM BBE; 5, 10, 20, 40, or 60 μM DIMP; 5, 10, 30, or 60 μM tetracycline; 0.5, 0.75, 1.0, or 1.25 μM propamidine; and 100, 150, 200, or 250 μM pentamidine. The bands are labeled as follows: P, precursor RNA; I-E2, G intron-3′ exon intermediate; I, excised linear G intron; E1-E2, reaction product consisting of ligated exon fragments; E1, 5′ exon fragment intermediate.
Broth dilution drug susceptibility testing.
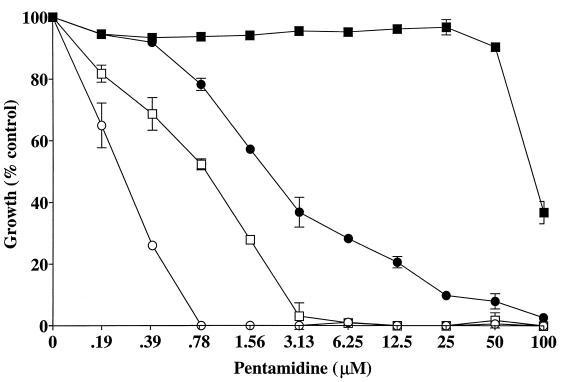
As shown in Table 2, strains 4-1 (intron containing) and 62-1 (intronless) were similarly susceptible to amphotericin B and pentamidine in YPG medium. On the other hand, while they were similarly susceptible to amphotericin B in YPD medium, there was a ≥16-fold increase in susceptibility to pentamidine of the intron-containing strain 4-1 relative to strain 62-1. Note that the pentamidine sensitivities of the two strains were greater on YPG medium than on YPD medium and there was relatively less difference between these strains on YPG medium than on YPD medium. These results suggest that the two strains may share a common mitochondrial pentamidine-sensitive target, as has been reported for S. cerevisiae (25). In each experiment, the MIC100 of amphotericin B for the reference strain ATCC 90028 (in RPMI 1640) was within the expected range (0.25 to 1.0 μg/ml). The differential susceptibility to pentamidine after 24 h in YPD and YPG media is graphically presented in Fig. 3, indicating that low concentrations of pentamidine selectively inhibit the intron-containing strain. This differential susceptibility was confirmed in the cultures (at 35°C and 250 rpm) used for the extraction of RNA from each strain and by disk assay performed on YPD plates (data not shown). Under these conditions, 1 μM pentamidine added during logarithmic phase (mid-log) slowed growth of the intron-containing 4-1 strain but not of the intronless 62-1 strain (data not shown).
TABLE 2.
MICs of pentamidine and amphotericin B against C. albicans intron-containing strain 4-1 and intronless strain 62-1
| Strain | Antibiotic | MICa after 24 h
|
|
|---|---|---|---|
| YPG | YPD | ||
| 4-1 | Pentamidine (μM) | 0.78 | 12.5 |
| Amphotericin B (μg/ml) | 0.5 | 1.0 | |
| 62-1 | Pentamidine (μM) | 3.13 | >100 |
| Amphotericin B (μg/ml) | 0.25 | 1.0 | |
The 80% and 100% inhibition endpoints were used for pentamidine and amphotericin B, respectively.
FIG. 3.
Comparison of the intron-containing (4-1) and intronless (62-1) strain susceptibility to pentamidine. The 24-h values were determined by a broth microdilution assay in YPD or YPG medium. ●, strain 4-1 in YPD medium; ○, strain 4-1 in YPG medium; ■, strain 62-1 in YPD medium; □, strain 62-1 in YPG medium. The error bars indicate standard deviations.
Effect of pentamidine on rRNA processing.
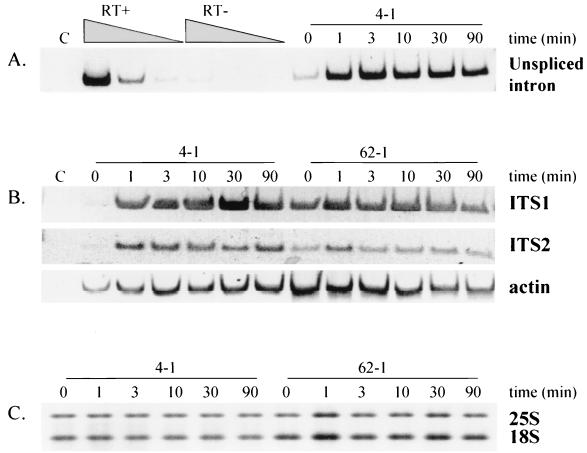
As demonstrated in Fig. 4 and 5A, treatment of the intron-containing strain 4-1 with 1 μM pentamidine in YPD medium, which resulted in a reduced rate of growth, produced an increase in detectable intron-containing rRNA precursor in 1 min. This pentamidine-dependent accumulation of putative pre-rRNA transcript was detected by nonquantitative as well as by competitive quantitative RT-PCR (52), using the same set of primers with different RNA preparations from similarly treated cells. By quantitative RT-PCR the level of intron-containing target RNA transcript (copies/ng of RNA) was determined to be 15.6 in untreated cells and 222.5 in pentamidine-treated cells (1 μM for 1 min), indicating a 14.3-fold increase after 1 min of pentamidine treatment (Fig. 4). Visual inspection (Fig. 5A) and densitometry (data not shown) of RT-PCR images for the nonquantitative RT-PCR experiment indicated that the increase in intron-containing RNA was obvious at 1 min and sustained over 90 min. The relative differences were similar whether the RT-PCR analysis was quantitative or nonquantitative, supporting the use of nonquantitative RT-PCR for additional analysis. RT-PCR of ITS1 and ITS2-containing RNAs showed that both ITS regions rapidly accumulated in unprocessed form after pentamidine treatment of the intron-containing strain 4-1, while no similar changes were seen in the intronless strain 62-1 (Fig. 5B). It is noteworthy that the linear range for RT-PCR amplification of both the intron and the ITS1-containing precursor was at RNA template dilutions of 1:103 to 1:105; for ITS2-containing RNAs, the linear range was at a 100-fold greater dilution (1:105 to 1:107 dilution). This difference suggests that more ITS2-containing RNA is present in cells than RNA containing the intron or ITS1. Note that pentamidine treatment resulted in no comparable changes in RT-PCR-detectable actin mRNA (Fig. 5B) or levels of ethidium bromide stain-detected mature rRNA (Fig. 5C) over 90 min of treatment.
FIG. 4.
Quantitation of the accumulation of the intron-containing precursor rRNA. Strain 4-1 was grown to logarithmic phase in YPD medium. Aliquots were taken before (pentamidine −) and 1 min after (pentamidine +) addition of 1 M pentamidine. Total RNA was extracted and subjected to quantitative competitive RT-PCR with primers J and L using a constant amount of in vitro-transcribed competitor RNA (1.016 × 10−17 g) and three different dilutions of total RNA, followed by PAGE analysis. The target and competitor represent amplification products of the cellular and synthetic RNA transcripts, respectively.
FIG. 5.
Effect of pentamidine on rRNA processing. Strains 4-1 (intron containing) and 62-1 (intronless) were grown to logarithmic phase in YPD. Aliquots were taken at the indicated times after pentamidine addition (all panels); RNA was extracted and subjected to RT-PCR, followed by PAGE analysis (A and B). (A) Amplification (at an RNA dilution of 1:104) using primers derived from the intron (L) and 5′ exon (J) of precursor RNA from strain 4-1 at the indicated times after addition of 1 μM pentamidine. RT+, dose response for RNA template from untreated 4-1 cells (extracted total RNA at 0.1 μg/μl) with serial dilutions from 1:103 to 1:105; RT−, identical reactions run using heat-inactivated reverse transcriptase. (B) Amplification using primer pairs A and B flanking ITS1 at an RNA dilution of 1:104 (ITS1), primers C and D flanking ITS2 at an RNA dilution of 1:106 (ITS2), and primers [oligo(dT) and 10816/10473] for actin at an RNA dilution of 1:10 (actin). (C) Mature 25S and 18S rRNAs were subjected to agarose gel electrophoresis and visualized by ethidium bromide staining (0.5 μg of the extracted total RNA at 0.1 μg/μl). In panels A and B, the lanes marked C are negative controls (no template). Time (in minutes) represents the time when the RNA was extracted, before (0 min) or after (1 to 90 min) adding 1 μM pentamidine to a mid-log-phase culture. The same RNA preparations were used in all panels.
Intron presence correlates with pentamidine sensitivity.
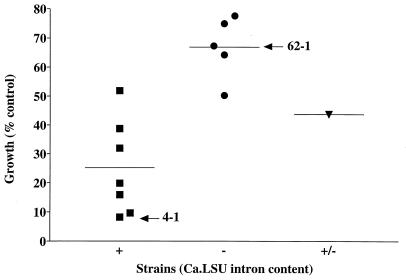
Thirteen clinical isolates of C. albicans were tested for the presence of introns in their 25S rRNA genes by PCR using primers J and K and were tested for their growth sensitivity to pentamidine in YPD medium. Figure 6 shows that seven intron-containing strains appeared to be relatively more sensitive to pentamidine than five intronless strains. We noted that one strain (provided by G. St-Germain) appeared to contain both intron-containing and intronless rRNA genes by PCR analysis. This result was obtained on all single colonies isolated after repeated restreaking, suggesting that this strain does harbor two distinct rRNA gene alleles. The data in Fig. 6 show that this strain had an intermediate level of pentamidine sensitivity. Although solubility problems prevented accurate pentamidine MIC80 determinations for the more resistant strains among those tested here, in these experiments the MIC80s for four of seven intron-containing strains, but none of the intronless or the mixed-genotype strains, were ≤100 μM.
FIG. 6.
Pentamidine sensitivity correlates with the presence of intron Ca.LSU. Thirteen clinical isolates were tested for growth inhibition of a standard inoculum by 0.1 mM pentamidine during 24 h of growth at 35°C in YPD medium. PCR with primers flanking the intron revealed that seven strains (including 4-1) harbored (+) the intron (■), five strains (including 62-1) lacked (−) the intron (●), and one strain yielded PCR products indicating a mixture (+/−) of intron-containing and intronless rRNA genes (▾). The last strain yielded the same pattern after repeated single-colony isolations, indicating that it is not due to a mixed population of cells. The horizontal lines indicate the average percent growth in the presence of pentamidine for each set of strains.
DISCUSSION
Inhibition of the in vitro splicing of group I intron ribozymes by various antibiotics (22, 24, 35, 46–49) suggests that these ribozymes, which have not yet been found in the human genome, may be potential therapeutic targets for antimicrobial agents. We have confirmed that the Ca.LSU intron of C. albicans shows approximately the same sensitivity to pentamidine in vitro as does the P. carinii intron Pc1.LSU. This inhibition was also observed in culture at concentrations of pentamidine that inhibit the growth of an intron-containing, but not an intronless, strain of C. albicans. The correlation of in vitro and in-culture effects of pentamidine in this system suggests that ribozyme-catalyzed splicing might be a target for the action of this drug on C. albicans.
In vitro splicing by Ca.LSU was inhibited by pentamidine, with complete inhibition seen at 200 μM pentamidine, close to the 250 μM level required to inhibit Pc1.LSU (24). The pentamidine analogs BBE and DIMP were also modestly more potent inhibitors of Ca.LSU than of Pc1.LSU, and in both systems pentamidine and its analogs inhibited the ribozyme in a noncompetitive manner relative to the guanosine substrate, as indicated graphically on Hanes-Woolf plots (data not shown). Despite the small quantitative differences between the sensitivities of the two ribozymes to pentamidine and its analogs, pentamidine sensitivity seems to be a property of multiple group I introns, including the intron encoded by the DNA genome of an algal virus (51). On the other hand, tetracycline, also a noncompetitive inhibitor of Pc1.LSU (24), failed to inhibit Ca.LSU at a sixfold-higher molar concentration.
The availability of intronless (62-1) and intron-containing (4-1) strains of C. albicans which are closely related phylogenetically based on genomic sequence analysis (2), suggested that we could use these strains to study the effects of pentamidine on splicing in cultured cells and the effects of intron presence on pentamidine sensitivity, despite the lack of truly isogenic strains for this study. The two strains also showed similar sensitivities to amphotericin B.
When the MIC was determined after 24 h of incubations, intron-containing strain 4-1 was only four times more sensitive to pentamidine than intronless strain 62-1 in YPG and at least 16 times more sensitive on YPD medium. Table 2 and Fig. 3 show that the sensitivities of both strains to pentamidine on rich medium was greater with glycerol than with glucose as a carbon source. This result is similar to the finding that S. cerevisiae is more sensitive to pentamidine under nonfermentative growth conditions, suggesting a mitochondrial target for this agent (25). Therefore, the relatively greater sensitivity of both strains of C. albicans to pentamidine on glycerol-containing media is consistent with these strains sharing a target for pentamidine action resembling the mitochondrial target suggested for S. cerevisiae. The nuclear group I intron ribozyme could be the target for the differential effect of pentamidine on the two strains grown on glucose.
Addition of a low concentration of pentamidine (1 μM), which inhibited growth in YPD medium of intron-containing strain 4-1 but not intronless strain 62-1, resulted in the rapid accumulation of intron-containing rRNA species in the intron-containing strain (Fig. 4 and 5A). In strain 4-1, rRNA precursors containing ITS1 and ITS2 also accumulated with a similar time course after the addition of pentamidine, while no similar effects were seen on the levels of actin mRNA or mature rRNA; no effects on rRNA precursor levels were seen in the intronless strain 62-1. This result indicates that pentamidine does not inhibit rRNA maturation in strain 62-1, suggesting that the effects on rRNA processing in strain 4-1 might be secondary to inhibition of processing of the group I intron. The order of cleavages in the maturation of rRNA from the primary transcript has been determined in S. cerevisiae (45) and presumably resembles that in C. albicans, except that in the latter organism, in strains (such as 4-1) which contain a group I intron, that intron must be removed. In T. thermophila, removal of a similar group I intron in the 25S rRNA gene is one of the first steps in rRNA processing (3, 6, 13). The results in this model organism, and the results of our experiments, suggest that intron splicing precedes or is simultaneous with cleavage in the ITS1 region, which in S. cerevisiae is coupled to 3′ external transcribed spacer (ETS) processing and is believed to occur cotranscriptionally (1). This cleavage in ITS1 and the 3′ ETS, along with cleavage in the 5′ ETS, is the earliest step in the maturation of rRNA in S. cerevisiae (33), prior to steps removing ITS2. The results of RT-PCR analysis of the rRNA precursors indicate that in both Candida strains, total cellular RNA as a template needed to be diluted about 100-fold more to show a linear response for amplification of ITS2-containing precursors than to demonstrate precursors containing ITS1 and Ca.LSU (Fig. 5). If it is assumed that the RT and PCR amplification efficiencies of all of these RNA species are similar, then this implies that the level of precursors containing intact ITS2 is greater than the level of those containing intact ITS1 or Ca.LSU. Furthermore, in the intronless strain 62-1 without pentamidine treatment, precursors containing intact ITS2 are more readily detected than in the intron-containing strain 4-1; the levels in strain 4-1 rapidly increase in response to pentamidine treatment, while those in strain 62-1 remain unchanged. These results suggest that in the intronless strain, the step(s) removing the ITS2 is rate limiting, so without treatment, ITS2-containing precursors are readily detected by RT-PCR. On the other hand, such ITS2-containing precursors are much less detectable in the untreated intron-containing strain, in which an earlier step (such as intron removal) may be rate limiting. Failure to observe changes in ITS1- or ITS2-containing precursors in the intronless strain with pentamidine treatment suggests that this agent does not act directly on removal of either ITS in rRNA maturation.
In S. cerevisiae, and probably in other organisms as well, processing of rRNA (44) is tightly regulated, so if any step is blocked, processing is shut down and accumulated precursors and/or intermediates or aberrantly processed molecules are subsequently degraded (1, 13, 15, 20, 28). This regulation ensures that only functional rRNA is assembled into ribosomes. Our results indicate that when the early intron removal step is inhibited by pentamidine, very early precursors of rRNA (containing the intron and both ITS regions) accumulate rapidly due to the ordered sequence of processing steps. Later intermediates also transiently accumulate in this strain, such as those containing ITS2 but not ITS1 (see above). This effect may be due to regulatory processes that inhibit all later steps in the maturation pathway when an early step (intron removal) is blocked. This is consistent with the finding that insertion of an unspliceable mutant intron derived from Pp.LSU3 into the rRNA gene of S. cerevisiae results in accumulation of two ITS2-containing 27S rRNA precursor species (21).
The potential coupling of rRNA maturation with intron processing is indicated by the observation that when the T. thermophila intron was introduced into a genetically marked plasmid-expressed rDNA gene in Schizosaccharomyces pombe, intron removal from the 25S rRNA precursor was detectable but the genetically marked 5.8S rRNA and its 7S immediate precursor containing the marked sequence were undetectable (13). This result suggests that perturbation of rRNA maturation, even by insertion of a removable intron, can result in failure to process rRNA correctly. On the other hand, such tight regulation does not occur in all situations, since when a mutant intron incapable of splicing was introduced into the 23S rRNA gene of E. coli, rRNA containing the unspliced intron was assembled into 50S subunit particles which were defective in functional association with 30S subunits (31).
Sequence similarities between rRNA genes of strains 4-1 and 62-1 indicate their phylogenetic relatedness (2), although they are not isogenic. On the other hand, no such estimation of similarity among the other strains tested can be made. Therefore the differences in pentamidine sensitivity seen in Fig. 6 can be due to many factors, including permeability, metabolism, or efflux of the drug. However, it is striking that the intron-containing strains tend to be more pentamidine sensitive than the intronless strains.
The PCR test for the presence of introns in the 25S rRNA gene revealed that one isolate contained two alleles of this gene, including both intron-containing and intronless gene copies. Such heterozygous strains of C. albicans have previously been reported (4, 26). Further studies of the genetic structure of the heterozygous rRNA genes in this C. albicans isolate are in progress.
The results presented here indicate that pentamidine, which acts as a noncompetitive inhibitor of group I intron ribozymes, can inhibit splicing of the Ca.LSU group I intron in cells of C. albicans and that this inhibition occurs under conditions in which an intron-containing strain is more sensitive to pentamidine than an apparently closely related intronless strain. This inhibition has proven useful as a tool to study the rRNA maturation pathway in this organism. The in vivo inhibition of intron splicing supports the hypothesis that ribozyme inhibitors might represent a new class of antimicrobial agents acting on this RNA target (22–24, 27, 29, 30, 35, 46–49). It is not known if variation in the presence of an intron in the 25S rRNA gene of different isolates of C. albicans (2, 30) is responsible for the reported variation in the MIC of pentamidine for various isolates of this organism (40). Since inhibition of group I intron splicing (Fig. 4 and 5) occurs at much lower pentamidine levels in cultured cells than in vitro (Fig. 2), pentamidine might be concentrated at the site where splicing occurs in living cells.
It is noteworthy that successful treatment with pentamidine of some serious C. albicans infections has been reported (41–43). Although pentamidine has been more widely used to treat P. carinii than C. albicans infections (32), its target in P. carinii remains incompletely defined (9). Pentamidine and a series of analogs have been shown to inhibit the growth of C. albicans in culture (5, 40). Topoisomerases have been proposed as a target of pentamidine (9). However, it has recently been shown that the cloned topoisomerase I from P. carinii is not inhibited by pentamidine, nor does pentamidine enhance topoisomerase I cleavage of linear DNA, a hallmark of topoisomerase I poisons. These results agree with in vivo results indicating that in yeast cells in which P. carinii topoisomerase I has been engineered to replace the endogenous enzyme, topoisomerase I is not the molecular target of pentamidine (R. T. Van Dross and M. M. Sanders, submitted for publication). In a C. albicans strain with a nuclear group I intron ribozyme grown on glucose, inhibition of ribozyme catalysis appears to be the most sensitive target of pentamidine action, although at high concentrations pentamidine acts equivalently on strains with and without this intron, indicating a second, less sensitive target; this might be similar to the mitochondrial target identified in S. cerevisiae (25), which lacks nuclear group I introns. The greater pentamidine sensitivity of intron-containing and intronless strains grown on glycerol supports the existence of a common mitochondrial target, whose nature is undefined at present. The lack of a convenient axenic culture system for P. carinii (38) has thus far prevented in vivo tests of nuclear group I introns as the targets of pentamidine action in this organism.
The results presented here demonstrate that group I intron ribozymes can be targets for antimicrobial action against pathogenic microorganisms. Although pentamidine may act on other targets, it clearly can inhibit group I intron catalysis in vitro and in living cells of C. albicans, and the presence of the Ca.LSU intron correlates with increased growth sensitivity to pentamidine.
ACKNOWLEDGMENTS
We thank G. Lemay (University of Montreal, Montreal, Canada) and G. St-Germain (Laboratoire de Santé Publique du Québec, Quebec, Canada) for C. albicans strains vital to this work, R. R. Tidwell (University of North Carolina) for providing BBE and DIMP, and R. Van Dross and M. Sanders (UMDNJ-Robert Wood Johnson Medical School) for sharing their data before publication. We also thank M. Sanders, T. Goss Kinzy and D. Pilch, of this institution, for helpful advice on the preparation of the manuscript, and E. C. Cultrone for help with the graphical data.
This work was supported by the NIH through a grant (1RO1 GM53815 to M.J.L.) and an Individual National Research Service Award (1F32 GM18912 to K.E.M.).
REFERENCES
- 1.Allmang C, Tollervey D. The role of the 3′ external transcribed spacer in yeast pre-rRNA processing. J Mol Biol. 1998;278:67–78. doi: 10.1006/jmbi.1998.1693. [DOI] [PubMed] [Google Scholar]
- 2.Boucher H, Mercure S, Montplaisir S, Lemay G. A novel group I intron in Candida dubliniensis is homologous to a Candida albicans intron. Gene. 1996;180:189–196. doi: 10.1016/s0378-1119(96)00453-2. [DOI] [PubMed] [Google Scholar]
- 3.Cech T R, Rio D C. Localization of transcribed regions on extrachromosomal ribosomal RNA genes of Tetrahymena thermophila by R-loop mapping. Proc Natl Acad Sci USA. 1979;76:5051–5055. doi: 10.1073/pnas.76.10.5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clemons K V, Feroze F, Holmberg K, Stevens D A. Comparative analysis of genetic variability among Candida albicans isolates from different geographic locales by three genotypic methods. J Clin Microbiol. 1997;358:1332–1336. doi: 10.1128/jcm.35.6.1332-1336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Del Poeta M, Schell W A, Dykstra C C, Jones S, Tidwell R R, Czarny A, Bajic M, Kumar A, Boykin D, Perfect J R. Structure-in vitro activity relationships of pentamidine analogues and dication-substituted bis-benzimidazoles as new antifungal agents. Antimicrob Agents Chemother. 1998;42:2495–2502. doi: 10.1128/aac.42.10.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Din N, Engberg J. The intervening sequence in the 26S rRNA coding region of T. thermophila is transcribed within the largest stable precursor for rRNA. Cell. 1979;18:525–532. doi: 10.1016/0092-8674(79)90069-2. [DOI] [PubMed] [Google Scholar]
- 7.Dixon D M, McNeil M M, Cohen M L, Gellin B G, LaMontagne J R. Fungal infections: a growing threat. Public Health Rep. 1996;111:226–235. [PMC free article] [PubMed] [Google Scholar]
- 8.Dodd C L, Greenspan D, Katz M H, Westenhouse J L, Feigal D W, Greenspan J S. Oral candidiasis in HIV infection: pseudomembranous and erythematous candidiasis show similar rates of progression to AIDS. AIDS. 1991;5:1339–1343. [PubMed] [Google Scholar]
- 9.Dykstra C C, McClernon D R, Elwell L P, Tidwell R R. Selective inhibition of topoisomerases from Pneumocystis carinii compared with that of topoisomerases from mammalian cells. Antimicrob Agents Chemother. 1994;38:1890–1898. doi: 10.1128/aac.38.9.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Embley T M, Dyal P, Kilvington S. A group I intron in the small subunit ribosomal RNA gene from Naegleria andersoni ssp. andersoni strain PPMF-6. Nucleic Acids Res. 1992;20:6411. doi: 10.1093/nar/20.23.6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gast R J, Fuerst P A, Byers T J. Discovery of group I introns in the nuclear small subunit ribosomal RNA genes of Acanthamoeba. Nucleic Acids Res. 1994;22:592–596. doi: 10.1093/nar/22.4.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Georgopapadakou N H, Walsh T J. Human mycoses: drugs and targets for emerging pathogens. Science. 1994;264:371–373. doi: 10.1126/science.8153622. [DOI] [PubMed] [Google Scholar]
- 13.Good L, Elela S A, Nazar R N. Tetrahymena ribozyme disrupts rRNA processing in yeast. J Biol Chem. 1994;269:22169–22172. [PubMed] [Google Scholar]
- 14.Graybill J R. The future of antifungal therapy. Clin Infect Dis. 1996;22(Suppl. 2):S166–S178. doi: 10.1093/clinids/22.supplement_2.s166. [DOI] [PubMed] [Google Scholar]
- 15.Hitchen J, Ivakine E, Melekhovets Y F, Lalev A, Nazar R N. Structural features in the 3′ external transcribed spacer affecting intragenic processing of the yeast rRNA. J Mol Biol. 1997;274:481–490. doi: 10.1006/jmbi.1997.1376. [DOI] [PubMed] [Google Scholar]
- 16.Hood S, Denning D W. Treatment of fungal infection in AIDS. J Antimicrob Chemother. 1996;37(Suppl. B):71–85. doi: 10.1093/jac/37.suppl_b.71. [DOI] [PubMed] [Google Scholar]
- 17.Johnson E M, Warnock D W, Luker J, Porter S R, Scully C. Emergence of azole drug resistance in Candida species from HIV-infected patients receiving prolonged fluconazole therapy for oral candidiosis. J Antimicrob Chemother. 1995;35:103–114. doi: 10.1093/jac/35.1.103. [DOI] [PubMed] [Google Scholar]
- 18.Klootwijk J, Planta R J. Isolation and characterization of yeast ribosomal RNA precursors and preribosomes. Methods Enzymol. 1989;180:96–109. doi: 10.1016/0076-6879(89)80095-3. [DOI] [PubMed] [Google Scholar]
- 19.Law D, Moore C B, Wardle H M, Ganguli L A, Keaney M G L, Denning D W. High prevalence of antifungal resistance in Candida spp. from patients with AIDS. J Antimicrob Chemother. 1994;34:659–668. doi: 10.1093/jac/34.5.659. [DOI] [PubMed] [Google Scholar]
- 20.Lee Y, Melekhovets Y F, Nazar R N. Termination as a factor in “quality control” during ribosome biogenesis. J Biol Chem. 1995;270:28003–28005. doi: 10.1074/jbc.270.47.28003. [DOI] [PubMed] [Google Scholar]
- 21.Lin J, Vogt V M. I-Ppo I, the endonuclease encoded by the group I intron PpLSU3, is expressed from an RNA polymerase I transcript. Mol Cell Biol. 1998;188:5809–5817. doi: 10.1128/mcb.18.10.5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Leibowitz M J. Variation and in vitro splicing of group I introns in rRNA genes of Pneumocystis carinii. Nucleic Acids Res. 1993;21:2415–2421. doi: 10.1093/nar/21.10.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, Leibowitz M J. Bidirectional effectors of a group I intron ribozyme. Nucleic Acids Res. 1995;23:1284–1291. doi: 10.1093/nar/23.8.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Tidwell R R, Leibowitz M J. Inhibition of in vitro splicing of a group I intron of Pneumocystis carinii. J Eukaryot Microbiol. 1994;41:31–38. doi: 10.1111/j.1550-7408.1994.tb05931.x. [DOI] [PubMed] [Google Scholar]
- 25.Ludewig G, Williams J M, Li Y, Staben C. Effects of pentamidine isethionate on Saccharomyces cerevisiae. Antimicrob Agents Chemother. 1994;38:1123–1128. doi: 10.1128/aac.38.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCullough M J, Clemons K V, Stevens D A. Molecular and phenotypic characterization of genotypic Candida albicans subgroups and comparison with Candida dubliniensis and Candida stellatoidea. J Clin Microbiol. 1999;37:417–421. doi: 10.1128/jcm.37.2.417-421.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mei H Y, Cui M, Sutton S T, Truong H N, Chung F Z, Czarnik A W. Inhibition of self-splicing group I intron RNA: high throughput screening assays. Nucleic Acids Res. 1996;24:5051–5053. doi: 10.1093/nar/24.24.5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melekhovets Y F, Good L, Elela S A, Nazar R N. Intragenic processing in yeast rRNA is dependent on the 3′ external transcribed spacer. J Mol Biol. 1994;239:170–180. doi: 10.1006/jmbi.1994.1361. [DOI] [PubMed] [Google Scholar]
- 29.Mercure S, Cousineau L, Montplaisir S, Belhumeur P, Lemay G. Expression of a reporter gene interrupted by the Candida albicans group I intron is inhibited by base analogs. Nucleic Acids Res. 1997;25:431–437. doi: 10.1093/nar/25.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mercure S, Montplaisir S, Lemay G. Correlation between the presence of a self-splicing intron in the 25S rDNA of C. albicans and strains susceptibility to 5-fluorocytosine. Nucleic Acids Res. 1993;21:6020–6027. doi: 10.1093/nar/21.25.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nikolcheva T, Woodson S A. Association of a group I intron with its splice junction in 50S ribosomes: implications for intron toxicity. RNA. 1997;3:1016–1027. [PMC free article] [PubMed] [Google Scholar]
- 32.Queener S F. New drug developments for opportunistic infections in immunosuppressed patients: Pneumocystis carinii. J Med Chem. 1995;38:4739–4759. doi: 10.1021/jm00024a001. [DOI] [PubMed] [Google Scholar]
- 33.Raue H A, Planta R J. The pathway to maturity: processing of ribosomal RNA in Saccharomyces cerevisiae. Gene Expr. 1995;5:71–77. [PMC free article] [PubMed] [Google Scholar]
- 34.Rex J H, Rinaldi G, Pfaller M A. Resistance of Candida species to fluconazole. Antimicrob Agents Chemother. 1995;39:1–8. doi: 10.1128/aac.39.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rogers J, Davies J. The pseudodisaccharides: a novel class of group I intron splicing inhibitors. Nucleic Acids Res. 1994;22:4983–4988. doi: 10.1093/nar/22.23.4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitt M E, Brown T A, Trumpower B L. A rapid and simple method for the preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sewell D L, Pfaller M A, Barry A L. Comparison of broth macrodilution, broth microdilution, and E test antifungal susceptibility tests for fluconazole. J Clin Microbiol. 1994;328:2099–2102. doi: 10.1128/jcm.32.9.2099-2102.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sloand E, Laughon B, Armstrong M, Bartlett M S, Blumenfeld W, Cushion M, Kalica A, Kovacs J A, Martin W, Pitt E, Pesanti E L, Richards F, Rose R, Walzer P D. The challenge of Pneumocystis carinii culture. J Eukaryot Microbiol. 1993;40:188–195. doi: 10.1111/j.1550-7408.1993.tb04902.x. [DOI] [PubMed] [Google Scholar]
- 39.Sogin M L, Edman J C. A self-splicing intron in the small subunit rRNA gene of Pneumocystis carinii. Nucleic Acids Res. 1989;17:5349–5359. doi: 10.1093/nar/17.13.5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.St-Germain G. Effects of pentamidine alone and in combination with ketoconazole or itraconazole on the growth of Candida albicans. Antimicrob Agents Chemother. 1990;34:2304–2306. doi: 10.1128/aac.34.12.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stenderup A. Effect of diamidines on Candida albicans in vitro. Acta Pathol Microbiol Scand. 1955;36:361–364. doi: 10.1111/j.1699-0463.1955.tb04630.x. [DOI] [PubMed] [Google Scholar]
- 42.Stenderup A, Bichel J, Kissmeyer-Nielsen F. Moniliasis treated with pentamidine. Lancet. 1956;i:20–21. doi: 10.1016/s0140-6736(56)91856-6. [DOI] [PubMed] [Google Scholar]
- 43.Tóth G, Miklós G, Kerekes A, Katona J, Szarvas D. Pentamidin-Behandlung der kongenitalen generalisierten Candidiasis. Acta Paediatr Acad Sci Hung. 1977;18:207–211. [PubMed] [Google Scholar]
- 44.Udem S A, Warner J R. Ribosomal RNA synthesis in Saccharomyces cerevisiae. J Mol Biol. 1972;65:227–242. doi: 10.1016/0022-2836(72)90279-3. [DOI] [PubMed] [Google Scholar]
- 45.Venema J, Tollervey D. Processing of pre-ribosomal RNA in Saccharomyces cerevisiae. Yeast. 1995;11:1629–1650. doi: 10.1002/yea.320111607. [DOI] [PubMed] [Google Scholar]
- 46.von Ahsen U, Schroeder R. Streptomycin inhibits splicing of group I introns by competition with the guanosine substrate. Nucleic Acids Res. 1991;19:2261–2265. doi: 10.1093/nar/19.9.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.von Ahsen U, Davies J, Schroeder R. Antibiotic inhibition of group I ribozyme function. Nature. 1991;368:368–370. doi: 10.1038/353368a0. [DOI] [PubMed] [Google Scholar]
- 48.von Ahsen U, Davies J, Schroeder R. Non-competitive inhibition of a group I intron RNA self-splicing by aminoglycoside antibiotics. J Mol Biol. 1992;226:935–941. doi: 10.1016/0022-2836(92)91043-o. [DOI] [PubMed] [Google Scholar]
- 49.Wank H, Rogers J, Davies J, Schroeder R. Peptide antibiotics of the tuberactinomycin family as inhibitors of group I intron RNA splicing. J Mol Biol. 1994;236:1001–1010. doi: 10.1016/0022-2836(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 50.Wilkinson G N. Statistical estimation in enzyme kinetics. Biochem J. 1961;80:324–332. doi: 10.1042/bj0800324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamada T, Tamura K, Aimi T, Songsri P. Self-splicing group I introns in eukaryotic viruses. Nucleic Acids Res. 1994;22:2532–2537. doi: 10.1093/nar/22.13.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang J, Desai M, Ozanne S E, Doherty C, Hales C N, Byrne C D. Two variants of quantitative reverse transcriptase PCR used to show differential expression of α-, β- and γ-fibrinogen genes in rat liver lobes. Biochem J. 1997;321:769–775. doi: 10.1042/bj3210769. [DOI] [PMC free article] [PubMed] [Google Scholar]