Abstract
Significance:
Reactive oxygen species (ROS) are well known to promote innate immune responses during and in the absence of microbial infections. However, excessive or prolonged exposure to ROS provokes innate immune signaling dysfunction and contributes to the pathogenesis of many autoimmune diseases. The relatively high basal expression of pattern recognition receptors (PRRs) in innate immune cells renders them prone to activation in response to minor intrinsic or extrinsic ROS misbalances in the absence of pathogens.
Critical Issues:
A prominent source of ROS are mitochondria, which are also major inter-organelle hubs for innate immunity activation, since most PRRs and downstream receptor molecules are directly located either at mitochondria or at mitochondria-associated membranes. Due to their ancestral bacterial origin, mitochondria can also act as quasi-intrinsic self-microbes that mimic a pathogen invasion and become a source of danger-associated molecular patterns (DAMPs) that triggers innate immunity from within.
Recent Advances:
The release of mitochondrial DAMPs correlates with mitochondrial metabolism changes and increased generation of ROS, which can lead to the oxidative modification of DAMPs. Recent studies suggest that ROS-modified mitochondrial DAMPs possess increased, persistent immunogenicity.
Future Directions:
Herein, we discuss how mitochondrial DAMP release and oxidation activates PRRs, changes cellular metabolism, and causes innate immune response dysfunction by promoting systemic inflammation, thereby contributing to the onset or progression of autoimmune diseases. The future goal is to understand what the tipping point for DAMPs is to become oxidized, and whether this is a road without return. Antioxid. Redox Signal. 36, 441–461.
Keywords: mitochondria, damage-associated molecular pattern (DAMP), redox, reactive oxygen species (ROS), mitochondrial ROS (mtROS), innate immunity, oxidized mitochondrial DNA (mtDNA), oxidized mitochondrial RNA (mtRNA), neutrophil extracellular trap (NET), stimulator of interferon genes (STING), cyclic GMP-AMP synthase (cGAS), transcription factor A, mitochondrial TFAM, N-formyl peptides (NFPs), Carbamoyl phosphate synthetase-1 (CPS1), cytochrome C, cardiolipin (CL), oxidized cardiolipin, mitochondrial antiviral signaling (MAVS) protein, MAVS oligomerization, ATP, oxidized ATP, succinate, autoimmunity, systemic lupus erythematosus (SLE), metabolism
Introduction
Essentially all living organisms, from simple prokaryotes to multicellular eukaryotes, rely for their entire life on their innate immune system for protection against microbial infections (46, 80). In the past, innate immunity was deemed simple and unsophisticated compared with the specific and long-lasting responses of the adaptive immune system. However, it has since become clear that innate immunity depends on vastly diverse, immunologically active preexisting proteins, not just immune cells, to trigger an effective host response to a pathogen and to prime adaptive immunity (76). Further, it is now recognized that innate immune responses are not only accountable for inducing chronic inflammation and autoimmunity, but they are as well the direct cause of cell, tissue, and organ damage that is generally associated with autoimmunity (153).
Pattern recognition receptors (PRRs) are the key players for the induction of innate immunity and can be divided into several classes. Starting with the extracellular space, plasma membrane-positioned type 1 transmembrane proteins, known as Toll-like receptors (TLRs), recognize hydrophobic lipids and proteins exposed on the surface of pathogens (91). Within the cell, endosomal TLRs have the distinct ability to recognize the genetic material of pathogens (e.g., DNA, RNA). Alike is another set of cytoplasmic PRRs, known as retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), which are able to recognize specific patterns within the genomic material of pathogens, such as conserved motifs from different RNA-viruses or RNA-intermediates from DNA-viral replication (114).
Pathogen-associated DNA is recognized by other distinct cytosolic sensors localized freely in the cytoplasm, such as cyclic guanosine monophosphate-adenosine monophosphate (cGAMP) synthase (cGAS) (170, 197); interferon-gamma-inducible protein 16 (IFI16), a member of the pyrin and HIN domain-containing (PHYIN) protein family (181); Z-DNA-binding protein 1 (ZBP1), also known as DNA-dependent activator of interferon-regulatory factors (DAI) (172); or the absent in melanoma 2 (AIM2) protein, which is not only a DNA sensor, but also reportedly an inflammasome activator and inducer of pyroptosis, an inflammatory form of cell death (53, 71).
In addition to inducing proinflammatory cytokines postengagement, PRRs can mobilize adaptive immune responses by increasing major histocompatibility complex class II expression and by inducing the expression of costimulatory molecules, such as cluster of differentiation (CD) −40, −80, and −86 on antigen-presenting cells (177). Further, PRRs may recognize endogenous motifs that are known as danger-associated molecular patterns (DAMPs) of “self”-derived material origin, which comprise cellular or organelle components rather than a pathogen-associated “nonself” structure of motif (Table 1). During cellular stress or injury, otherwise spatially confined components can relocate between the sub-cellular compartments and/or be chemically modified to become mimics of “nonself” motifs that typically induce immunity (111).
Table 1.
Intra- and Extracellular Pattern Recognition Receptors for Mitochondria-Derived Danger-Associated Molecular Patterns
| mtDAMP | PRRs of mtDAMPs |
|
|---|---|---|
| Cytoplasmic | Extracellular | |

|
cGAS (170, 197), STING (176), TLR9 (2, 99, 169). | RAGE when in complex with TFAM (81). |

|
RIG-I (10), MDA5 (45, 132), PKR (88), TLR 3, 7, 8 (55, 86). | Not known |

|
Not known | RAGE, TLR9 (82). |

|
Not known | FPR (26, 115). |

|
Not known | Possibly specific macrophage receptor (135). |

|
TBK1, IRF3/7, NF-kB (23, 142, 162). | Not known |

|
IP3R-NLRP3? | Serum LPG (36). |

|
NLRP3 (77). | P-type ATPase transmembrane lipid pump (ATP8b1) (147), CD36 (9). |
|
NLRP3 (63). | Purinergic P2 receptor (32, 50). |
|
PHD enzyme (160). | GPR91 (105, 152, 178). |
CD, cluster of differentiation; cGAMP, cyclic guanosine-adenosine-monophosphate; cGAS, cGAMP synthase; FPR, formyl peptide receptor; GPR91, G protein-coupled receptor 91; IP3R, inositol-1,4,5-triphosphate receptor; LPG, leucine-rich α-2-glycoprotein-1; mtDAMP, mitochondrial DAMP; NLRP3, NOD-, LRR- and pyrin domain-containing protein 3; PHD, prolyl hydroxylase domain; PKR, protein kinase R; PRRs, pattern recognition receptors; RAGE, receptor for advanced glycation endproducts; RIG-I, retinoic acid-inducible gene I; STING, stimulator of interferon genes; TFAM, mitochondrial transcription factor A; TLR, Toll-like receptor.
Reactive oxygen species (ROS) are generated in all cell types, and under normal physiologic conditions limited ROS production is known to be overall beneficial and necessary to maintain biological functions of the cell (165). This includes gene expression, molecular signaling between organelles, control of the balance between the cell's proliferation and death, and response to microbial infections (126). Increased and/or chronic, potentially harmful levels of ROS occur during tissue injury, microbial infection, or exposure to environmental factors, such as ionizing radiation, ultraviolet (UV) irradiation associated with sun exposure, or heavy metal ions present in drinking water or overall increased air pollution (184) (Fig. 1).
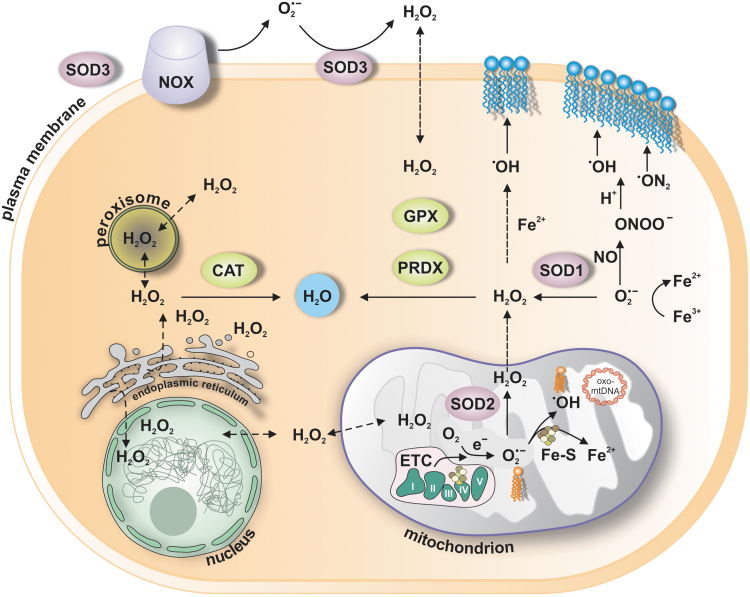
FIG. 1.
Major sites in the cell, where key ROS are produced. ROS comprise oxygen-containing reactive species, including superoxide anion (O2˙−), H2O2, ˙OH), NO, and ONOO−, among others. Production of ˙OH is shown to occur in mitochondria or cytoplasm and will ensue when H2O2 reacts with Fe2+ in Fenton reaction or when O2˙− is reducing Fe3+ to Fe2+. Reaction of NO with O2˙− is shown to lead to the formation of ONOO−, which is known to induce nitration of proteins, or alike ˙OH oxidation of lipids (blue at the plasma membrane or orange—cardiolipin in the mitochondria). Conversion of O2˙− to H2O2 is either spontaneous or catalyzed by SODs (SOD1, SOD2, SOD3), which are localized to the cytoplasm, mitochondria, plasma membrane, and/or extracellular space, respectively. There are several mechanisms for the removal of H2O2 from each site in the cell. The major contributors include PRDX and GPX, which are specifically expressed either in the cytoplasm or in mitochondria, and peroxisome-bound CAT, all of which convert H2O2 to water and molecular oxygen. CAT, catalase; H2O2, hydrogen peroxide; ˙OH, hydroxyl radical; GPX, glutathione peroxidases; NO, nitric oxide; NOX, NADPH oxidase; ONOO−, peroxynitrite; PRDX, peroxiredoxins; ROS, reactive oxygen species; SODs, superoxide dismutases.
Chronically increased ROS may lead to the accumulation and release of DAMPs, which can continuously activate the innate immune system (74). Studies of innate immunity activation in the absence of pathogens suggest that the various forms of ROS can be considered as DAMPs on their own, leading to the increased secretion of proinflammatory cytokines, changes in cellular metabolism, and induction of inflammation that could contribute to the development of autoimmune diseases (23, 148).
The ROS still posit a conundrum, because their increased presence in one cell type might not correspond to the effect it would have in a different cell type, or even in the same cell under analogous physiological conditions, as such an effect depends on the cell localization in the tissue, cellular density, molecular interactions, and timespan of the events (6). The activity of ROS, although when it was determined to be at the same concentration, was shown to depend not only on a level of multitude of relating cellular antioxidants, but it was also suggested to depend on the density of the cytoplasmic milieu, which might control the rate of ROS or antioxidant diffusion, respectively (192, 198).
Further, latest findings suggest that the mobility of different types of ROS within one cell can be important in distributing oxidative stress at the tissue or even entire organ level (116). The role of ROS in becoming a DAMP and the role of ROS-mediated modifications of DAMPs in the onset or potential progressive acceleration of autoimmune disease development are just at the beginning of being evaluated.
The term ROS comprises a heterogenous group of various oxygen-containing molecules (Fig. 1), produced from molecular oxygen, which can participate in electron transfer processes (56). The reactivity of different ROS molecules differs by at least 11 orders of magnitude respective to their kinetic interaction rate with a given substrate (125). However, the type of ROS and the level of relative concentration that is necessary to promote specific DAMP accumulation and oxidation, which in turn leads to changes in cellular metabolism to promote chronic inflammation and development of autoimmunity, is not known.
Eighty five percent to 90% of cellular oxygen is metabolized in mitochondria (Fig. 1), where superoxide anion (O2˙−) is produced after the interaction of a single electron with molecular oxygen at the electron transport chain (ETC) (56). The O2˙− can also be produced in the cytoplasm and at the plasma membrane (Fig. 1), where the catalytic action of many cytoplasmic enzymes has been shown to involve O2˙− (165). The capacity of O2˙− to become an efficient DAMP modifier is limited by its very short lifetime, caused by spontaneous or enzymatic dismutation to less reactive hydrogen peroxide (H2O2) (56). The rate of spontaneous O2˙− dismutation is fast and the enzymatic reaction catalyzed by cytoplasmic, mitochondrial, and extracellular compartmental-specific superoxide dismutases (SODs) is at least 103 times faster and close to the rate of free diffusion (56).
Hence, O2˙− is not able to travel far from where it is produced, or passage across cellular membranes, therefore unlikely being directly responsible for DAMP modification (56). The immediate targets of the negatively charged O2˙− in the mitochondria are positively charged iron/sulfur (Fe-S) centers in proteins, such as tricarboxylic acid (TCA) cycle enzyme aconitase or succinate dehydrogenase (SDH), both of which were shown to be more sensitive to oxidation by O2˙− than by H2O2 (27). The interaction of O2˙− with aconitase occurs in the mitochondrial matrix and was indicated to lead to the release of hydroxyl radical (HO˙) (182) (Fig. 1).
HO· is one of the strongest oxidants known, and because of its very low activation energy and quick diffusion has been suggested to be responsible for mitochondrial DNA (mtDNA) oxidations (137, 190). The protonated form of O2˙−, perhydroxyl radical (HOO˙), on the other hand, was suggested to be responsible for modification of mitochondrial phospholipids, for example cardiolipin (20). Another strong ROS, which can modify proteins and is responsible for lipid peroxidation, is peroxynitrite (ONOO−) (167). ONOO− is a product of near-diffusion-controlled reaction of O2˙− with nitric oxide (NO), which is on its own capable of oxidizing biomolecules (12, 167).
H2O2, although it is not a free radical like O2˙− or HO· and is less redox active because of its high activation energy, is relatively longer lived and was suggested to disseminate through membranes by passive diffusion (165) (Fig. 1). H2O2 has augmented reactivity toward cysteine residues in certain proteins, depending on the structure and environment, providing a basis for selectivity and specificity of H2O2 under physiological conditions (191, 195). Under oxidative distress conditions, when the H2O2 concentration increases in the cell from 1–10 to ∼100 nM and higher, nonspecific oxidation of proteins and damage to DNA and other biomolecules was suggested to occur (165) (Fig. 2).
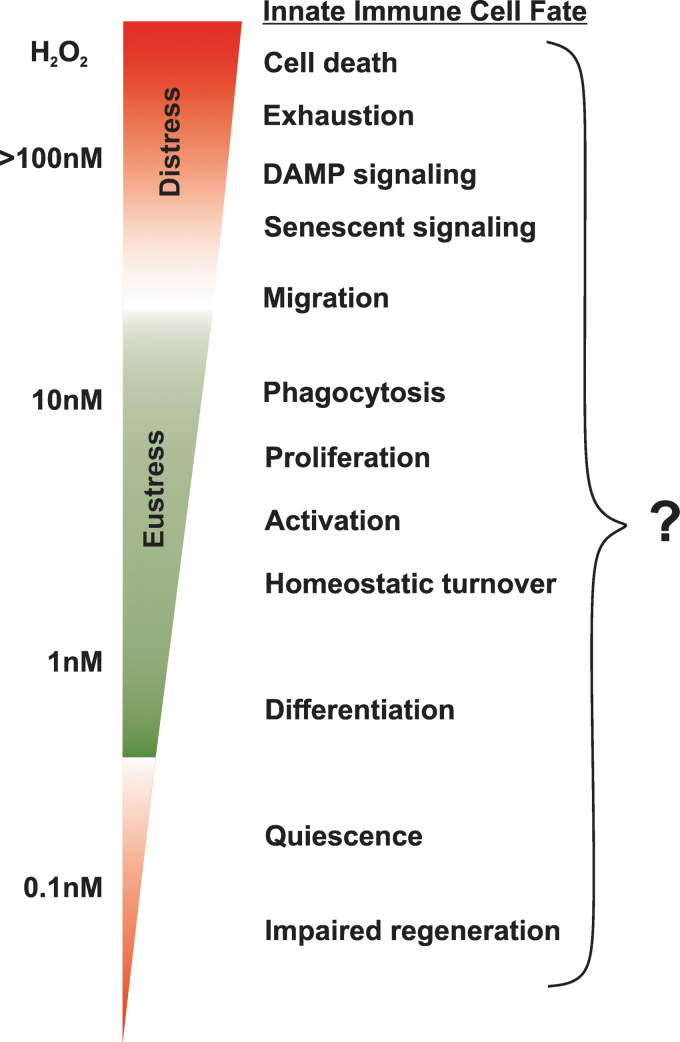
FIG. 2.
Intracellular concentration range of H2O2 in the cell and simplified associated stages of innate immune cell physiology. The exact level of either type of ROS at each stage of life and function of innate immune cells like, for example, plasmacytoid dendritic cells, which express a high level of PRRs, is not known and does not have to be necessary linear as presented. Thus far, limited published evidence suggests that myeloid cell-derived ROS externally can regulate myeloid progenitor proliferation and differentiation in bacteria-induced or sterile inflammation (164, 200). DAMP, danger-associated molecular pattern; PRRs, pattern recognition receptors.
It should be noted that the overall H2O2 concentration in the mitochondria was estimated to be at least two to five times higher than that detected in the cytoplasm, but lower than this was observed in the lumen of the endoplasmic reticulum (ER; ∼700 nM), where the formation of each disulfide bridge during the physiological folding and maturation of proteins leads to the generation of one molecule of H2O2 (165). The exact concentration of ROS in mitochondria to induce DAMP formation in immune cells, which in turn has a higher capacity to induce DAMP-dependent signaling, is not known and will most probably fluctuate with every organ (Fig. 2).
Future studies will need to focus on evaluating how innate immune cells respond to different levels of extracellular molecular oxygen in a tissue-specific context, simply because of their highly migratory nature. Only under this consideration we will be able to better understand the necessary conditions for the generation and release of DAMP and its associated pathologies.
The aim of this review is to present a comprehensive summary of how ROS modify mitochondrial components and become DAMPs on release, and how these intrinsic “self”-targets influence metabolic reprogramming, which ultimately contributes to the onset or promotes the progression of various autoimmune diseases.
Genomic Material: mtDNA and mtRNA
Mitochondrial DNA
As bacterial descendants, mitochondria possess plenty of DNA motifs that resemble pathogen-associated molecular patterns (PAMPs); hence, mtDNA is often considered to be the prototypical mitochondrial DAMP. Similar to bacterial DNA, mtDNA is circular in structure and arranged in nucleoids, which, on average, contain one copy apiece of mtDNA and mitochondrial transcription factor A (TFAM) (Fig. 3). TFAM is responsible for inducing mtDNA transcription and is also known to regulate mtDNA metabolism by directing mtDNA to lysosomes for degradation (95). Due to its localization in the mitochondrial matrix and thus in close vicinity to the ETC—a major source of ROS—mtDNA is known to be vulnerable to oxidation (194) (Fig. 3).
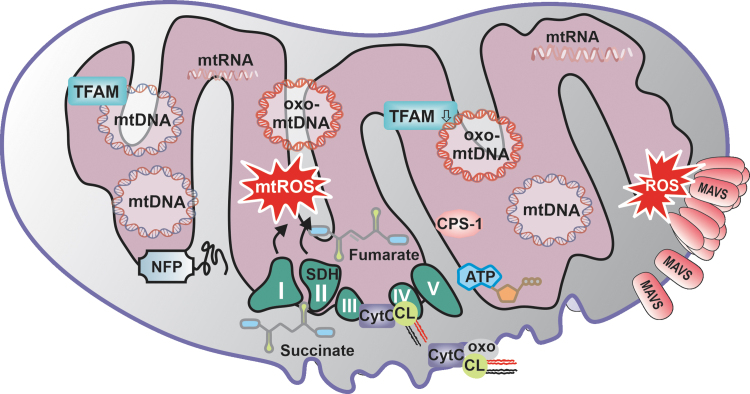
FIG. 3.
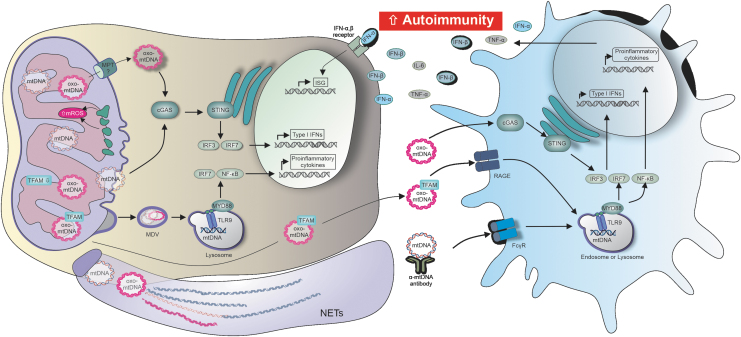
Mitochondrial components as DAMPS. The mitochondrial generation of ROS can lead to the oxidation of already existing DAMPs, such as mitochondrial nucleic acids. mtDNA and mtRNA are per se immunogenic, and it is plausible that oxidation of these molecules may augment their immunogenicity. Although mtDNA is known to be vulnerable to oxidation, emerging studies suggested only recently that mtRNA oxidation plays a significant role in the development of various diseases. Whether mtRNA exists in a stable oxidized form, and whether it can translocate to the cytoplasm to become a PRR substrate, remains unknown. However, it is feasible that oxidized mtRNA can, like oxo-mtDNA, mimic viral infection and drive the activation of antiviral signaling pathways. Moreover, multiple mitochondrial proteins such as TFAM, NFP, or CPS-1 were reported as DAMP on translocation to the cytoplasm or extracellular space, but the role of protein oxidation was only recently elucidated for a few proteins, such as MAVS or NFP. A further important group of mitochondrial DAMPs that are sensitive to oxidation are lipids such as cardiolipin, which was linked to autoimmune diseases through the presence of specific anti-cardiolipin antibodies in individuals with SLE or anti-phospholipid syndrome. Lastly, certain mitochondrial metabolites and even mtROS are now considered to play a role as DAMPS through accumulation, and nonselective oxidative crosslinking or modulation of cellular motifs. CPS-1, carbamoyl phosphate synthetase-1; MAVS, mitochondrial-antiviral signaling; mtDNA, mitochondrial DNA; mtRNA, mitochondrial RNA; mtROS, mitochondrial ROS; NFP, N-formyl peptide; oxo-mtDNA, oxidized mitochondrial DNA; SLE, systemic lupus erythematosus; TFAM, mitochondrial transcription factor A.
Murine studies have shown that intra-articular injection of mtDNA purified from cell extracts, but not DNA isolated from the nucleus, causes increased localized inflammation, suggesting a major difference in the capacity of the two DNA to induce inflammation (37). This observation was confirmed with the injection of a synthetic, sequence-analogue DNA with and without oxidized nucleotides, which when oxidized lead to the secretion of proinflammatory cytokines or inflammation (37) (Figs. 3 and 4). mtDNA can simulate several PRRs, including cytosolic cGAS, which promotes type I interferon (IFN) responses, and AIM2, which leads to the formation of inflammasome and endosomal TLR9 (150, 189, 196).
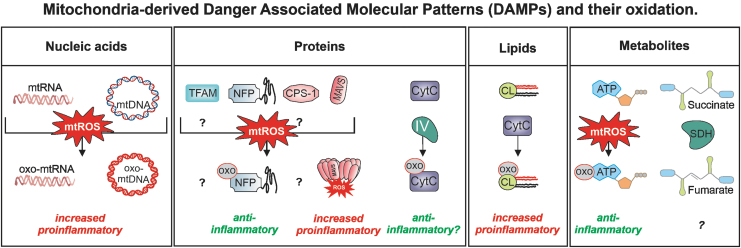
FIG. 4.
Oxidation of mitochondrial DAMPs and its consequences. Oxidized mitochondrial DAMPs can gain access to the cytoplasm of innate immune cells and subsequently activate PRRs. What level of oxidation of mitochondrial DAMPs defines their pro- or anti-inflammatory properties was—as of now—not studied and might depend on the organ that the particular innate immune cell will be infiltrating. CytC, cytochrome C.
The release of mtDNA into the cytoplasm acts as a danger signal and can occur during infection, cell death, or in response to the exposure to diverse environmental factors (Figs. 4 and 5). In fact, it has been suggested that the release of mtDNA into the cytoplasm during infection strengthens a cell's antiviral response by activating the cGAS pathway and prompting the secretion of antiviral type I IFNs and proinflammatory cytokines, which could prevent the spread of the virus to neighboring cells.
FIG. 5.
Oxidized and nonoxidized mitochondrial nucleic acids induce proinflammatory cytokine secretion, and contribute to the onset and progression of autoimmunity, through multiple distinct pathways. mtDNA and mtRNA were shown to be per se immunogenic by acting as DMAPs on multiple pathways, such as cGAS/STING or TLR-mediated signaling. Oxidized and nonoxidized mtDNA also play an important role in NETosis, and mtDNA complexed with the transcription factor TFAM showed an augmented immunogenicity. Mitochondrial nucleic acids that reach the extracellular space induce cytokine secretion in bystander cells. Systemic or chronic inflammation is induced through sustained cytokine secretion, and quite possibly a major contributing factor to the onset or progression of autoimmune diseases such as SLE. cGAMP, cyclic guanosine-adenosine-monophosphate; cGAS, cGAMP synthase; Fcγ, constant fragment gamma receptor; ISG, interferon-stimulated gene; RAGE, receptor for advanced glycation endproducts; STING, stimulator of interferon genes; TLR, Toll-like receptor.
For example, several herpes viruses have acquired the ability to diminish mtDNA release as a defense mechanism by directly degrading mtDNA, which results in the complete loss of mtDNA from infected cells (39). Interestingly, it has been reported that mtDNA depletion does not affect viral replication; however, to date, it has yet to be investigated as to what impact virus-mitigated loss of mtDNA has on the metabolism of a cell affected by infection (49). mtDNA release is associated not only with DNA virus infection, but also with small RNA viruses that lack DNA or DNA intermediates in their genome, such as dengue virus (DENV) (159).
During DENV infection, it has been shown that cytosolic mtDNA interacts directly with cGAS in an oxidized state (oxo-mtDNA), and is further released from the cell, where it can interact with TLR9 and promote antiviral immunity (2, 99, 169). Interestingly, inflammatory stimulation of bystander cells with interleukin (IL)-1β, which is secreted in response to viral infection, also leads to the release of mtDNA into the cytoplasm and cGAS activation (Fig. 5). However, this mtDNA-cGAS interaction occurs without apparent loss of mitochondrial mass and increase in mitochondrial membrane potential (Δψm), and absence of cytochrome C release, which indicates lack of cell death induction (1).
It is currently not known whether there is a difference in the efficiency or kinetics of cGAS or TLR9 stimulation between mtDNA and oxo-mtDNA and whether there are differences between mtDNA and oxo-mtDNA in bystander cell activation, nor how it influences bystander cells' metabolism or inflammatory responses (Fig. 2). mtDNA-TLR9 interaction is known to occur not only after viral infection but it is also implicated in the progression of autoimmune diseases such as rheumatoid arthritis (RA), atherosclerosis, diseases associated with sterile inflammation, steatohepatitis, or acute liver injury (57, 188, 196).
Although identified under many different clinical conditions, the exact molecular mechanism of mtDNA interaction with TLR9 is still unclear due to the controversy that surrounds how to determine the precise degree of methylation of CpG motifs in mtDNA (70, 138). mtDNA is also being discussed to play a role in regulating hyperglycemia during type 2 diabetes, where circulating and cell-free mtDNA was suggested to contribute to AIM2 inflammasome activation (7).
The rate and level of mtDNA release is now being evaluated in many other than autoimmune diseases and it is becoming clear that mtDNA has a longer lifetime in circulation, even in healthy individuals, as compared with DNA originating from the nucleus (112, 179). To date, most studies have focused on mtDNA's extracellular secretion from neutrophils, which extrude fibrous networks known as neutrophil extracellular traps (NETs)—easily detectable by fluorescence microscopy—that contain mtDNA in response to microbial activation (22). In addition, it was more recently reported that not only neutrophils but also macrophages and monocytes can release extracellular traps with nuclear DNA; however, the presence of mtDNA was not investigated (163).
The NET formation can also occur spontaneously in the absence of microbial infection and was associated with a variety of autoimmune diseases, such as atherosclerosis, psoriasis, RA, gout, anti-neutrophil cytoplasmic antibodies-associated vasculitis, or systemic lupus erythematosus (SLE) (101). It has been suggested that NET formation and release induces changes in cellular metabolism, associated with an increase in glycolytic adenosine triphosphate (ATP) production and ROS-regulated dynamics of cytoskeleton (5) (Figs. 5 and 6).
FIG. 6.
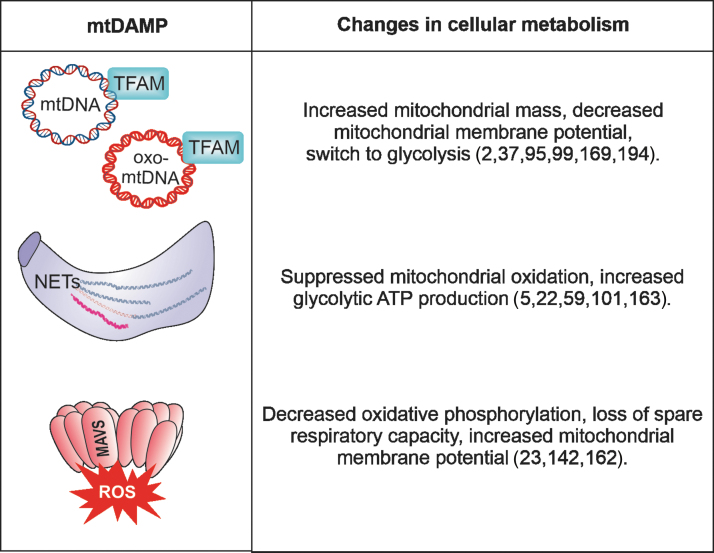
Mitochondrial DAMPs and their impact on cellular metabolism. The release of various mitochondrial DAMPs was shown to induce metabolic reprogramming. The individual outcome, for example a shunt from oxidative phosphorylation to glycolysis, depends on the nature of the DAMP, the cell type, and the tissue milieu.
Recent evidence indicates that activated eosinophils can also release extracellular traps (EET), however the exact molecular mechanism of EET formation remains unknown (72, 166). It is also unclear whether EETs are performed in eosinophils and subsequently released, as recent studies suggest that the kinetics of mtDNA release from eosinophils differs from EET formation (60). Whether extracellular EET signaling can affect cellular metabolism of bystander cells is not known.
Dissemination of mtDNA may occur through the direct fusion of mitochondrial membranes with the cell membrane; when this occurs, the levels of oxo-mtDNA have been shown to be high (24, 106). oxo-mtDNA released from neutrophils in SLE patients was reported to interact with TLR9 on plasmacytoid dendritic cells (pDCs), drive type I IFN production, and have increased immunostimulatory potency compared with nonoxidized mtDNA (24, 139). The exact mechanism responsible for the increased interferogenicity of oxo-mtDNA is not known. One study suggests that oxidized DNA, regardless of mitochondrial or nuclear origin, is less susceptible to 3′ repair exonuclease 1 (TREX1)-mediated degradation (59).
Other studies were unable to show differences between oxo-mtDNA and nonoxidized mtDNA regarding susceptibility to degradation by extracellular deoxyribonucleases or differences in the quantitative uptake, intracellular compartmentalization, or stability within pDCs (139). This supports the idea that there may be a unique TLR9-oxo-mtDNA interaction, or a recruitment of an as-of-yet-unidentified co-receptor that facilitates the interaction and determines the specificity (139).
Mitochondrial RNA
During mitochondrial gene expression, both strands of mtDNA are simultaneously transcribed in reverse, resulting in the formation of lengthy polycistronic mitochondrial RNA (mtRNA) transcripts (171) (Fig. 3). These are then processed by endoribonucleases, yielding mature messenger RNAs (mRNAs), ribosomal RNAs (rRNAs), transfer RNAs (tRNAs), and large amounts of complementary, noncoding RNA from the opposite strand of mRNA and rRNA (4). The untranslated light strand is under physiological conditions quickly degraded in mitochondria by polynucleotide phosphorylase (PNPase) and further un-winded in the cytoplasm by the helicase HSuv3 to become a substrate for cytoplasmic RNases (17, 96).
PNPase localization at the inter-mitochondrial membrane space ensures that any mtRNA of pathogen-resembling structure, such as long double stranded (ds) RNA, are detected before escaping into the cytoplasm to become a DAMP. In fact, antisense mtRNA, if not degraded but accumulated in mitochondria, was shown to form long dsRNA helices analogous to viral genomes, which can specifically be recognized by the antiviral RLR-family member melanoma differentiation gene 5 (MDA5) helicase, causing type I IFNs and proinflammatory cytokines secretion (45, 132).
Notably, patients carrying mutations in the PNPase-encoding PNPT1 gene—whose depletion is embryonically lethal—display mitochondrial dsRNA buildup linked with upregulation of type I IFN-stimulated genes and other proinflammatory markers frequently found in SLE patients (45). ds-mtRNA can also be recognized by the protein kinase R (PKR) (88): A PKR ds-mtRNA complex was shown to induce type I IFN secretion, and to influence eukaryotic initiation factor 2 alpha's (eIF2α) phosphorylation, respectively suggesting that indirectly may impact translation and cell signaling (88). Interestingly, mtRNA can be interferogenic not only because of the formation of dsRNA stretches but can also be mistaken for viral RNA because of the specific structure of the 5′-end untranslated regions.
Since mtRNA is not capped at the 5′-end like other cellular mRNAs found in the cytoplasm, it can bind to RLR-helicases such as RIG-I, which then induces type I IFN secretion (10). Finally, similar to bacterial RNA, mtRNA contains less of naturally occurring chemically modified nucleotides, which were shown to affect primary, secondary, and tertiary structures of cytoplasmic RNA and suggested to render them inefficient to interact, for example with TLR 3, 7, and 8 (55, 86).
Emerging studies suggest that RNA oxidation plays a significant role in the development of various diseases (30). Whether mtRNA exists in an oxidized form, and whether it could reach the cytoplasm to become a PRR substrate, remains unknown; to-date, no peer-reviewed reports address a role for oxidized mtRNA. It is also not known whether the escape of mtRNA, or potentially its oxidized form, into the cytoplasm could lead to changes in cellular metabolism, such as those that occur through translational or cell-cycle interference (193), which could be expected because of the interaction with PKR and its ribosomal localization.
Mitochondrial Proteins: TFAM, N-Formyl Peptides, Carbamoyl Phosphate Synthetase-1, Cytochrome C
Mitochondrial transcription factor A
TFAM is an indispensable element of the mtDNA transcription machinery (51). TFAM has nonspecific DNA-binding properties that are similar to family members of the high mobility group (HMG) proteins, which interact with nuclear DNA and are known as nucleus-derived DAMPs (134) (Fig. 2). However, the similarity between TFAM and HMGs is only structural, as the amino acid sequences differ substantially (134). Similar to HMGs, TFAM can be released from a variety of immune and nonimmune cells during cellular stress, alone or in complex with mtDNA, which were shown to stimulate dendritic cells and macrophages, causing a dose-dependent increase in tumor necrosis factor (TNF)-α secretion (81, 84, 157).
Interestingly, exogenous HMGs are stronger immunogenic factors when associated with nuclear DNA (151). Whether the same signal amplification applies as well to TFAM and mtDNA has yet to be investigated in detail. Injection of purified glutathione S-transferase-tagged TFAM, but not the tag alone, was shown to upregulate circulating levels of proinflammatory cytokines, increase neutrophil infiltration of the lungs, and cause organ injury in healthy animals, all of which suggest that extracellular TFAM can trigger inflammation in the absence of mtDNA (31).
Further, TFAM augmented TNF-α secretion in response to cytosine and guanine with phosphodiester backbone (CpGA) DNA, which were then in complex recognized extracellularly by the receptor for advanced glycation endproducts (RAGE) (Fig. 5), an immunoglobulin superfamily transmembrane receptor, and TLR9 (82). Interestingly, TFAM was also shown to be recognized by brain microglia and proposed in the human brain to have intercellular signaling function, which causes pro-inflammatory and cytotoxic responses (104).
Recent data suggest that even though TFAM transcript levels increase in the nucleus and cytoplasm during sepsis, the levels of intra-mitochondrial TFAM protein and its level of interaction with mitochondrial transcription factor 2B (a key for mtDNA transcription initiation) do not (145). The exact mechanism underlying spatial misplacement of TFAM during sepsis is unknown. One possible explanation is that TFAM may either be post-translationally modified, and thus unable to enter mitochondria, or it may nonspecifically interact with other proteins or nuclear DNA, and subsequently be extruded from the cell as a DAMP.
In fact, it has been shown that ubiquitination of TFAM blocks its ability to enter mitochondria, resulting in dysfunction of retinal cells, and a subsequent loss of sight (156). In addition to ubiquitination, TFAM can also be acetylated and phosphorylated (89). Phosphorylation of human TFAM in mitochondria is vital for the maintenance and expression of the mitochondrial genome, and it has been also shown to occur within other HMGs by cAMP-dependent protein kinase (PKA) and result in the rapid and selective degradation of TFAM (107).
Interestingly, the PKA-dependent phosphorylation of Saccharomyces cerevisiae's TFAM analogue appears to be tied in to metabolic processes, since TFAM's affinity to mtDNA under conditions of glucose repression was shown to be decreased (33). Moreover, TFAM can be phosphorylated by extracellular signal-regulated protein kinases (ERKs), which translocate to the mitochondria (44, 185). Although this phosphorylation reduces TFAM binding to transcription initiation sites, it does not decrease binding to nonspecific-mtDNA sequences (44, 185). Although phosphorylation and other modifications have been well described, it has not yet been established whether human TFAM can be modified by ROS at the two cysteines that are encoded in its sequence, either in mitochondria, in the cytoplasm, or in extracellular space (120).
In contrast, ROS modification was observed in HMG homologues, where three conserved cysteines were found in an oxidized state on translocation from the nucleus into the cytoplasm or extracellular space (109). One set of cysteines in the N-terminus was shown to form intramolecular bridges, thus altering the structure of the protein and causing at least a 10-fold decrease in its affinity for DNA. On the other hand, the oxidation of cysteine positioned in the middle of the protein was shown to be responsible for its translocation (109, 136). Nuclear HMGs have been also shown to become immune-silent after caspase-dependent oxidation (87).
Based on TFAM's localization in the middle of mitochondria, it is unlikely that oxidation could be selective enough to regulate TFAM, like it was observed for HMGs, to promote the immune function associated with tissue regeneration, chemotaxis, and inflammation (174). Therefore, it is not surprising that TFAM levels are responsive to more selective redox modifiers such as hydrogen sulfide, which was shown to not directly modify TFAM per se, but to sulfhydrate interferon regulatory factor 1 (IRF-1), which inhibits the methylation of the TFAMs gene promoter and represses its expression (103).
Somewhat surprisingly, TFAM expression levels are not solely contributors to the development of danger in the cell; increased expression of TFAM can also facilitate cardioprotection, prevent skeletal muscle atrophy, or positively impact mtDNA copy number, as seen in a rat model of diabetic neuropathy (29, 97). In conclusion, TFAM can have a pleiotropic effect on cellular function, and simply associating its increased qualitative expression with cellular stress in a given organism should be approached with caution. This is especially true, considering that the impact of TFAM modification associated with oxidative stress and danger signals secondary to posttranslational phosphorylation or other modification that originates from mitochondria had not yet been carefully addressed in any autoimmune disease.
N-formyl peptides
N-formyl peptides (NFPs) are derivatives of mitochondrial and/or bacterial translation (158). Mitochondria, similar to bacteria, start protein synthesis with an N-formylmethionine residue; after the release of mitochondrial proteins into extracellular space, this residue can function as a chemoattractant that is sensed by high-affinity formyl peptide receptors (FPRs) (26, 115). Interestingly, these FPRs are not only found to be expressed on immune cells such as neutrophils, monocytes, and dendritic cells, but as well on cells of the nervous system, endothelial cells, and hepatocytes (94). Mitochondrial NFPs attract and activate various immune cells that result, for example, in the secretion of proinflammatory signals such as matrix metalloproteinase-8 (MMP-8) and IL-8, but also attract thrombocytes, thereby potentially triggering unwanted blood coagulation (43, 146).
The oxidation of NFPs is so far not widely studied; however, early studies have shown that human neutrophils can induce oxidation of the synthetic chemotactic peptide N-formyl methionyl-leucyl-phenylalanine by the myeloperoxidase system (180) (Fig. 4). Oxidation was proposed to diminish NFPs' chemotactic activity, which may be one of the mechanisms by which neutrophils modulate the inflammatory process. The ratio of oxidized to nonoxidized mitochondrial NFPs released from the cell under different stress conditions has not yet been investigated, but it would be interesting to determine whether possible differences in NFP oxidation under different types of cell stress and death exist. Overall, this implies that oxidation and thereby inactivation of humoral mediators plays a significant pathophysiologic role as a negative feedback inflammatory control mechanism (34).
Carbamoyl phosphate synthetase-1
Carbamoyl phosphate synthetase-1 (CPS-1) is a 165-kDa protein that is translated as a proenzyme in the cytoplasm and, after transfer into the mitochondrial matrix, is cleaved to yield the mature protein, which is responsible for generating the precursor of citrulline (80a) (Figs. 3 and 4). The expression of CPS-1 is mostly restricted to the liver, where it constitutes up to 15%–20% of total hepatic mitochondrial protein; it is characterized by a long half-life of ∼8 days (35). More recent work described CPS-1 expression also in intestinal enterocytes and intestinal mucosa, where it appears to contribute to pyrimidine biosynthesis and citrulline/arginine production (61).
A possible role for CPS-1 as an extracellular DAMP molecule triggering innate immune responses has only recently been considered. Early findings showed that CPS-1 can be released by the liver into circulation after lipopolysaccharide (LPS) stimulation, although pro- or anti-inflammatory consequences of this process were not evaluated (168). The release of CPS-1 after LPS stimulation shows, however, interesting kinetics, as compared with the release of acute inflammation cytokines such as TNF-α (168). Unlike TNF-α, which can be detected several minutes after LPS stimulation in circulation and can return to basal level within a few hours, extracellular CPS-1 levels have been shown to increase with a delay of several hours and to remain sustained for extended periods of time (168).
The release of CPS-1 into circulation has also been observed after induction of cecal ligation and puncture sepsis, where it is proposed to be associated with increased mitochondria depletion due to increased lysosomal clearance rather than cell death (3). Interestingly, increased circulating CPS-1 has also been associated with mitochondrial morphology abnormalities in the liver (42). A later study showed that the liver also releases CPS-1 into circulation during Fas-ligand-induced apoptosis; CPS-1 has under these conditions a rather shorter half-life of merely 2 h, suggesting that its lifetime in circulation may directly be dependent on necrotic, apoptotic, or cell-death-independent release from mitochondria and the cell (187).
However, besides the mere observation of the highly variable stability of CPS-1, the underlying molecular mechanism is not known. Of note, recent in vivo studies suggest a CPS-1 release during acute liver injury into the blood, followed by a rapid uptake by monocytes, which become as a result anti-inflammatory, and homing to the liver (135). CPS-1's function in monocytes may be independent of its enzymatic ability to produce carbamoyl phosphate (135). This would suggest that CPS-1, instead of acting as a DAMP, would have protective and anti-inflammatory properties, although it is not clear what precise mechanism would underlie this monocyte/macrophage activation. At present, no information is available about CPS-1 stress-related modifications or what impact released CPS-1 may have on cell metabolism, especially in monocytes.
Cytochrome C and cardiolipin
Cytochrome C and cardiolipin release into circulation is fairly well documented in patients who have SLE, liver injury, suffered a myocardial infarction, or are undergoing chemotherapy (11, 78, 110, 117, 144). Cytochrome C's role as an intra- and extracellular danger signal is not as well understood as its canonical function in the ETC at mitochondria, where it interchanges from a reduced to an oxidized form by transferring electrons from complex III to complex IV (64) (Fig. 3). Besides its functional role in mitochondria, cytochrome C is structurally essential for the proper association of the entire respiratory supercomplexes (140). In addition, cytochrome C is also responsible for the import of cysteine-rich proteins into the mitochondrial intermembrane space via sulfhydryl oxidase-mediated redox reactions (15).
Cytochrome C escapes from mitochondria on detection of proapoptotic stimuli such as DNA damage, metabolic stress, accumulation of unfolded proteins, chemical triggers such as H2O2, etoposide, actinomycin D, or exposure to UV light (62). The mechanisms of cytochrome C release from mitochondria are still controversial, as it seems it can occur either via nonspecific pores in the outer mitochondrial membrane (OMM), via “apoptotic pores” formed by the B cell lymphoma protein-2 (Bcl-2) family of proteins, or via mitochondrial porins, which are formed by the voltage-dependent anion channel (VDAC) protein (62).
Altogether, this suggests that cytochrome C escape from mitochondria is cell-type specific and depends not on one but diverse apoptotic triggers. Several studies now suggest that cytochrome C release from mitochondria is a gradual process that may also take place under nonapoptotic conditions, without mitochondrial membrane permeabilization (47).
Interestingly, under homeostatic conditions and at maximal mitochondrial respiration, not all cytochrome C is bound to the inner mitochondrial membrane (IMM), but at least 20% can be also found firmly attached via electrostatic and hydrophobic interactions to the cytosolic interface (58). Some studies suggest that after release into the cytoplasm, cytochrome C migrates into the nucleus, where it interacts with a series of chromatin-binding factors, thereby changing its function from a “killer” to a “healer” by regulating the dynamics of damaged chromatin (64).
The best described nonapoptotic cytoplasmic receptor for cytochrome C is inositol-1,4,5-trisphosphate receptor (IP3R), a member of ion channels, which facilitate the release of calcium from the ER into the cytosol by the binding of IP3 (14). Cytochrome C was shown to bind to IP3R, before apoptosis induction and independently of caspase activation, and to promote calcium release, which was suggested in a feedback loop to promote mitochondrial depolarization and augment cytochrome C release (131). Interestingly, the role of cytochrome C binding to IP3R and increased calcium release from the ER was not studied in the context of NOD-, LRR-, and pyrin domain-containing protein 3 (NLRP3) activation, which is well documented to occur after calcium influx in the cytoplasm (123).
As it is not known which form of cytochrome C binds to IP3R, whether it being oxidized or nonoxidized, post-translationally modified, or in a complex with another compound, differences in cytochrome C's redox state may offer an explanation that yet has to be answered. Cytochrome C in its reduced form in neurons is not able to interact with proapoptotic proteins to form the apoptosome (183). Early studies suggested that cytochrome C, when exogenously added to cytoplasmic extracts, is rapidly reduced—within less than 5 min—and that only oxidized species can activate caspases (67).
In line with this observation, later studies that were performed in a cell-free system suggested that the presence of cytochrome C reductases, such as glutathione, L-cysteine, ascorbate, or N-acetylcysteine, prevents the formation of the apoptosome (133).
In contrast, more recent studies suggest that the presence of the cytochrome C reductant tetramethyl-p-phenylenediamine (TMPD) does not entirely eliminate cytochrome C activity in the induction of apoptosome and activation of caspase 3, as it was studied in cell-free extracts isolated from either murine macrophage J774 cells or human HeLa cells (18). Also, recent studies using an in vitro purified system and cytochrome C point mutation variants, which can accommodate different oxidation states, suggest that the redox state of cytochrome C is irrelevant for the induction of the apoptosome complex (113).
Cytochrome C's role is, therefore, complex and should not be examined in an isolated state. Physiologically, cytochrome C is noncovalently attached to the mitochondrial membrane via interaction with acidic phospholipids primarily found in the IMM, such as cardiolipin (75). Under physiological conditions, cardiolipin is equally distributed between the inner and outer leaflet of the IMM and not present at the OMM (83). Cytochrome C interaction with cardiolipin leads to peroxidation and subsequent relocation of cardiolipin to the OMM, which is associated with the OMM permeabilization and release of pro-apoptotic factors (83, 130).
Although the role of oxidized cardiolipin as a DAMP signal has not been widely investigated, it is known that anti-cardiolipin antibodies are present in SLE patients and individuals with antiphospholipid syndrome (141). Anticardiolipin antibodies after the interaction with their target antigen beta-2-glycoprotein-I, an anionic phospholipid-binding serum protein, can bind to TLRs on monocytes, macrophages, and platelets, which subsequently leads to their activation and pro-inflammatory responses (48, 122, 155). Another example of the pathologic role of cardiolipin was shown in lung pathology associated with pneumonias, where increased extracellular cardiolipin was associated with lung malfunction (147).
In this study, cardiolipin was shown to interact with the P-type ATPase transmembrane lipid pump (ATP8b1) and internalized into lung epithelium; however, the fate of the cardiolipin after uptake or proinflammatory signaling was not investigated (147). In vitro, experiments suggest that lack of cardiolipin impairs NLRP3 inflammasome activation and cannot be rescued by other phospholipids, as it was tested in cell-free extracts by measuring caspase-1 activity (77). A contradictory, beneficial function was identified for extracellular cardiolipin, when bound to mitochondria and possibly cytochrome C.
Extracellular mitochondria coated with both oxidized and nonoxidized forms of cardiolipin have been shown to be efficiently engulfed by professional phagocytes through the receptor CD36, causing an attenuation of innate immunity responses (9). Further, extracellular cytochrome C has been shown to interact with serum leucine-rich α-2-glycoprotein-1 (LPG), and this interaction was shown, instead of being pathologic, to protect lymphocytes from cell death (36).
The role of cardiolipin in the assumption of distinct metabolic programs and mitochondrial function in immune cells was only just recently investigated. Cardiolipin was shown to be necessarily present in the cell before stimulation, which leads to immune memory development (41). In conclusion, DAMP properties of cytochrome C and its binding cofactor cardiolipin suggest that a cytoplasmic localization favors the development of a proinflammatory phenotype (Figs. 3 and 4). In contrast, cytochrome C and cardiolipin in the extracellular space were thus far shown to be recognized by sensors that trigger an anti-inflammatory response. Future research should also focus on potential differences of oxidized versus nonoxidized forms of cytochrome C and cardiolipin in innate immune cells to precisely dissect their pro-apoptotic and/or pro-inflammatory functions.
Mitochondrial antiviral signaling protein
Mitochondrial antiviral signaling (MAVS) protein is located downstream of PRRs of the RIG-I-like helicases family, which are activated by either viral RNA recognition during infection, or self-RNA interaction that was, for example, released by mitochondria (161) (Figs. 3 and 4). MAVS has been demonstrated by our group to be activated by oxidative stress alone, independent of viral infection and upstream RLR interaction (23) (Fig. 6). In fact, chemically generated oxidative stress in the presence or absence of pan- or mitochondria-specific ROS scavengers demonstrated that specifically mitochondrial ROS (mtROS) lead to the formation of a disulfide bond between a highly conserved cysteine at position 79 (C79) in neighboring MAVS molecules.
This disulfide-dependent dimer formation triggers further MAVS oligomerization, functional formation of the MAVS signaling complex with its downstream proteins, and secretion of proinflammatory cytokines such as type I IFN and IL-6 (23). In contrast, inhibition of mtROS by the mitochondria-targeted antioxidant MitoQ inhibited MAVS aggregation and type I IFN secretion. Interestingly, these stable MAVS oligomers could be detected in immune cells and in the circulation of SLE patients (23, 162). The observation that overall disease intensity in SLE patients carrying an MAVS-mutation of cysteine 79 to phenylalanine (C79F) is significantly reduced substantiates the notion that mtROS-dependent MAVS oligomerization drives type I IFN production (23, 142).
Of note, this C79F variation is found naturally in 30% of the healthy sub-Saharan population; at this geographical location, the C79F variant appears to be a natural adaptation that offers some protection from ROS-driven MAVS-oligomerization due to continuous exposure to UV-irradiation (142). Taken together, these findings suggest that mtROS modified MAVS in SLE patients serves as a DAMP molecule and contributes to the type I IFN signature characteristic of the disease (23). Of particular interest, mtROS-dependent MAVS oligomers have been shown to not only affect the proinflammatory state of the cell, but also to cause mitochondrial hyperpolarization, reduced spare respiratory capacity, and decreased ATP production (23).
Mitochondria-associated membrane PRRs: cGAS and stimulator of interferon genes
Another example of direct ROS modulation of a PRR has been observed for the cytosolic DNA sensor cGAS and its interacting partner stimulator of interferon genes (STING) (176). Exposure of murine bone marrow-derived macrophages (BMDMs) to vitamin K3 (menadione) can lead to the oxidation of STING at cysteine 147 (176). Contrary to MAVS, oxidation of STING did not lead to accelerated signaling and secretion of proinflammatory cytokines; in fact, there was an opposite effect, as STING signaling was suppressed. STING is not the only example for a PRR to be directly and/or specifically modified by mtROS, especially during microbial infections. For example, during infection with Listeria monocytogenes, specifically mtROS, but not cytoplasmic ROS, was shown to induce disulfide bridges in the NF-κB essential modulator (NEMO) (69).
It was further demonstrated that mtROS-dependent dimerization of NEMO led to the secretion of proinflammatory cytokines such as IL-1β, TNF α, and IL-6, and induced macrophage polarization (69). mtROS was also implicated in the activation of the inflammasome, a supramolecular signaling complex that converts inactive pro-IL-1β into its active pro-inflammatory form (IL-1β) (199). The exact role of mtROS versus cytoplasmic ROS in activation of inflammasome is still quite controversial. Interestingly, NRLP3, one of the key proteins forming a DAMP- but not PAMP-specific inflammasome, has been shown to contain a highly conserved and redox-sensitive disulfide bond (8).
It would be valuable to evaluate the role of this disulfide bridge formation in the presence of MitoPQ, a mitochondria-specific redox cycler, which increases superoxide production within the mitochondrial matrix (149). Additional research will be needed to determine how this spatiotemporal action of mtROS occurs and whether mtROS plays a more direct role in the activation of nonmitochondrial PRR rather than via the ancillary proteins in the downstream complex.
Metabolites: ATP, succinate, mtROS
Adenosine triphosphate
ATP is a vital mitochondrial metabolite that fulfills multiple roles as an energy molecule, a phosphate donor, and a signaling molecule inside the cells. Intracellularly generated ATP is under oxidizing conditions largely produced in mitochondria, whereas cytoplasmic glycolysis is the most important extra-mitochondrial source of ATP. ATP that is present in the extracellular space is now recognized as a compound that not only can change different cellular functions associated with cellular metabolism but also can play a key role in promoting an innate immunity response during bacterial infections, in addition to becoming a DAMP signal by itself (65).
The release of ATP is now known not to be a random process that is associated with gross leakage of other cellular content after cell rupture, but rather being specifically controlled by pannexin 1 plasma membrane channels and recognized by purinergic P2 receptors as a “find me” signal on phagocytes (32, 50).
Despite ATP's essential part in regulating a proper immune response during microbial infection or cancer, extracellular ATP has been shown be a sensor in its own right that induces the release of pro-allergic mediators and contributes to chronic inflammation in lungs under certain conditions (73, 93). Extracellular ATP can elicit NLRP3 inflammasome activation in the presence or absence of secondary PAMPs or DAMPs (63). Extracellular ATP that interacts with P2 receptor on DCs promotes T cell priming, possibly setting a chronic inflammatory state, as it was, for example, described in inflammatory bowel disease (98).
Whether stress-released ATP is oxidized is not known. Very early studies indicate that oxidized ATP is an inhibitor of purinergic macrophage receptors and rather attenuates proinflammatory signaling (13, 124). Follow-up studies on the enzymes responsible for nucleotide pool recycling and DNA repair uncovered further that the influence of free nucleotides' oxidation is surprisingly larger than that of direct oxidation of DNA (128). Interestingly, a so-called sanitizing enzyme for the oxidized nucleotide pool, mutator T homolog 1 (MTH1), also known as nudix hydrolase 1 (NUDT1), was shown to efficiently hydrolyze not only free not-incorporated-in-DNA deoxy-triphosphate purines, but also purine ribonucleoside triphosphates such as 2-OH-ATP, 8-oxo-ATP, and 8-oxo-GTP (129, 154).
This suggests that oxidized ATP could exist in the pool of free nucleotides released from the cell and disturb signal transduction or metabolic function, where ATP is an essential mediator (Fig. 4). Twelve modified variants of endogenous NTPs were recently discovered in various eukaryotic cells, and NUDT1 was shown to colocalize to mitochondria (79, 85). Whether oxidized ATP would pose as a relatively stronger or weaker DAMP is presently unknown; interestingly, a polymorphism of the NUDT1 gene that results in a less stable variant of the protein may be associated with the development of type 1 diabetes in the female population (119).
Succinate
Succinate appears in the TCA cycle as a metabolic intermediate. It is oxidized by SDH into fumarate in complex II of the ETC in mitochondria. Recent studies suggests that succinate, besides being a metabolite, can act as a DAMP in the cytoplasm and in the extracellular space (118, 127). The multifaceted function for succinate as a DAMP, aside from its classical role as mitochondrial “fuel,” can only be understood when the role of succinate in mtROS homeostasis under different conditions is considered. Under saturating levels of succinate, when the function of either complex I or III is not pharmacologically inhibited, the flow of electrons is slow, and all adenosine diphosphate (ADP) is converted to ATP (so-called state 4 respiration).
In this case, succinate can reverse electron transfer (RET) through complex I when Δψm increases (90). RET then leads to the generation of mtROS at a very high rate, increased Δψm, and a slow flow of electrons (28, 92). Under RET conditions, succinate in the cytoplasm was demonstrated to inhibit the activity of prolyl hydroxylase domain (PHD) enzyme by direct interaction. In turn, the succinate-mediated inhibition of PHD leads to stabilization of the hypoxia-inducible factor 1α (HIF-1α), which further interacts with HIF response elements (HREs) of proinflammatory genes such as IL-1β and the HREs of genes responsible for the induction of glycolysis (160).
Of note, succinate-dependent HIF-1α stabilization can occur both under hypoxia, when oxygen levels are low and hence the activity of SDH dampened, and during normoxia, when SDH is unable to convert succinate to fumarate, for example due to genetic aberrations (21, 175) (Fig. 4). Stabilization of HIF-1α can also occur independently from succinate's interaction with PHD, as HIF-1α levels were shown to be increased by direct action of mtROS (66, 143). It has been suggested that mtROS-mediated stabilization of HIF-1α may be the dominant mechanism in most cells, whereas succinate-mediated activation of HIF-1α occurs specifically in inflammatory macrophages (118, 127).
It is noteworthy that, in addition to a reduction in the function of SDH or RET, there are several other ways in which accumulation of succinate can occur in a cell (118). For example, succinate levels can rise as a result of increased glutamine metabolism through anaplerosis of α-ketoglutarate through the TCA cycle and γ-aminobutyric acid (GABA) and its transporters, which, in turn, generates succinate by way of the so called “GABA-shunt” or “glyoxylate shunt,” which leads to the conversion of isocitrate to succinate by isocitrate lyase (175, 186).
Although the impact of succinate's accumulation under different conditions and its function as a DAMP are still being precisely determined, the general consensus is that the pathological outcome of succinate buildup will depend on the accumulation mechanism, the level of mtROS released into the cytoplasm, and the cell's oxygen supply. A consequence of cytoplasmic succinate accumulation, aside from the possibility that it will engage with mitochondria, mtROS, SDH, or other processes, is its release into the extracellular space. Once there, it interacts with G protein-coupled receptor 91 (GPR91), likewise known as succinate receptor 1 (SUCNR1) (68).
GPR91 is strongly expressed on dendritic cells and was shown to play an important role in adipose, hepatic, renal, splenic, and retinal tissues, and associated tissue-specific diseases (105, 152, 178). It has also been suggested that extracellular succinate activates adaptive immunity in the gut via Tuft cells, which express high levels of GPR91 (102). In SLE patients, succinate has been shown to support the expansion and metabolism of a specific subtype of T-follicular helper cells (Tfh cells), which provide B cell help (25).
Interestingly, similar T cells can be also generated from naive CD4+ T cells that have been exposed to pDC activated by oxo-mt DNA (24). Although here the discussion posits succinate as a DAMP, it is also important to mention that itaconate, another mitochondrial metabolite that acts as an endogenous SDH inhibitor, has recently been shown to regulate succinate levels and function and mitochondrial respiration, as well as reduce inflammatory cytokine production (40, 100). In SLE patients, it has been observed that 4-octyl itaconate has inhibited proinflammatory cytokine production in peripheral blood mononuclear cells (173). Lastly, it has been proposed that succinate and its dehydrogenase may not only promote mtROS production but also control mitochondrial lipid peroxidation, a DAMP signal in its own right as discussed for cytochrome C—cardiolipin interaction (16).
Mitochondrial ROS
mtROS has the ability to directly activate PRRs, thus bypassing any interaction with other DAMPs to induce a downstream signaling cascade. Therefore, mtROS per se qualifies as a DAMP. Under normal physiological conditions, mitochondria contribute at least one-third of total cellular H2O2 production (52) through at least 11 distinct sites within the mitochondrial ETC that generate mtROS in the form of H2O2 or superoxide anions O2·−. Superoxide anions are rapidly converted to H2O2 by manganese SOD that resides in high concentrations within the mitochondrial matrix (38).
The sites with the utmost capacity to generate ROS are associated with respiratory chain's complex I, which releases mtROS into the matrix, and complex III, which is able to disseminate superoxide toward either side of the inner mitochondrial membrane (19). Theoretically, superoxide in the intermembrane space has easier access to cytosol than matrix-localized O2·−, because it needs to cross only the OMM, rather than both IMM and OMM (121). Thus, by the time superoxide could reach the cytoplasm, it is mostly converted to H2O2 by mitochondrial SODs.
Although less reactive than the superoxide anion, H2O2 can oxidize any given PRRs thiol group of cysteines to form sulfenic acid, thereby acting as a quasi-cytoplasmic PRR ligand. This cysteine-oxidation can then lead to glutathionylation, the formation of intra- or intercellular crosslinking disulfide bonds with an adjacent thiol, or a reaction with an amide group to form sulfenyl amide (54).
In summary, in this review, we discussed how mitochondria-derived DAMPs, which undergo oxidation, start forming an independent, strongly interferogenic family of ligands for PRR, which are associated with chronic changes in cell metabolism and are associated with the development of autoimmunity (Fig. 5).
Author's Contributions
All the authors contributed equally to this review.
Abbreviations Used
- Δψm
mitochondrial membrane potential
- ADP
adenosine diphosphate
- AIM2
absent in melanoma 2
- ATP
adenosine triphosphate
- Bcl-2
B cell lymphoma protein-2
- BMDMs
bone marrow-derived macrophages
- CAT
catalase
- CD
cluster of differentiation
- cGAMP
cyclic guanosine-adenosine-monophosphate
- cGAS
cGAMP synthase
- CpGA
cytosine and guanine with phosphodiester backbone
- CPS-1
carbamoyl phosphate synthetase-1
- CytC
cytochrome C
- DAI
DNA-dependent activator of interferon-regulatory factor
- DAMP
danger-associated molecular pattern
- DENV
dengue virus
- ds
double stranded
- EET
eosinophile extracellular trap
- eIF2α
eukaryotic initiation factor 2 alpha
- ER
endoplasmic reticulum
- ERK
extracellular signal-regulated protein kinase
- ETC
electron transport chain
- Fcγ
constant fragment gamma receptor
- Fe-S
iron-sulfur
- FPR
formyl peptide receptor
- GABA
γ-aminobutyric acid
- GPR91
G protein-coupled receptor 91
- GPX
glutathione peroxidase
- H2O2
hydrogen peroxide
- HIF
hypoxia-inducible factor
- HIN
hematopoietic expression, interferon-inducible nature, and nuclear localization
- HMG
high mobility group
- HO˙
hydroxyl radical
- HOO˙
perhydroxyl radical
- HRE
HIF response element
- IFI16
interferon-gamma-inducible protein 16
- IFN
interferon
- IL
interleukin
- IMM
inner mitochondrial membrane
- IP3R
inositol-1,4,5-trisphosphate receptor
- IRF
interferon-regulatory factor
- ISG
interferon-stimulated gene
- LPG
leucine-rich α-2-glycoprotein-1
- LPS
lipopolysaccharide
- LRR
leucine-rich repeat
- MAVS
mitochondrial-antiviral signaling
- MDA5
melanoma differentiation gene 5
- MMP-8
matrix metalloproteinase-8
- mRNAs
messenger RNAs
- mtDAMP
mitochondrial DAMP
- mtDNA
mitochondrial DNA
- MTH1
mutator T homolog 1
- mtRNA
mitochondrial RNA
- mtROS
mitochondrial ROS
- NEMO
NF-κB essential modulator
- NET
neutrophil extracellular trap
- NFP
N-formyl peptide
- NLRP3
NOD-, LRR- and pyrin domain-containing protein 3
- NO
nitric oxide
- NOD
Nucleotide-binding oligomerization domain
- NOX
NADPH oxidase
- NUDT1
nudix hydrolase 1
- O2˙−
superoxide anion
- OMM
outer mitochondrial membrane
- ONOO·
peroxynitrite
- oxo-mtDNA
oxidized mitochondrial DNA
- PAMP
pathogen-associated molecular pattern
- pDC
plasmacytoid dendritic cell
- PHD
prolyl hydroxylase domain
- PHYIN
pyrin and HIN domain-containing
- PKA
cAMP-dependent protein kinase
- PKR
protein kinase R
- PNPase
polynucleotide phosphorylase
- PRDX
peroxiredoxins
- PRR
pattern recognition receptor
- RA
rheumatoid arthritis
- RAGE
receptor for advanced glycation endproducts
- RET
reverse electron transfer
- RIG-I
retinoic acid-inducible gene I
- RLR
RIG-I-like receptor
- ROS
reactive oxygen species
- rRNAs
ribosomal RNAs
- SDH
succinate dehydrogenase
- SLE
systemic lupus erythematosus
- SOD
superoxide dismutase
- STING
stimulator of interferon genes
- SUCNR1
succinate receptor 1
- TCA
tricarboxylic acid
- TFAM
mitochondrial transcription factor A
- Tfh cells
T-follicular helper cells
- TLR
Toll-like receptor
- TMPD
tetramethyl-p-phenylenediamine
- TNF
tumor necrosis factor
- TREX1
3′ repair exonuclease 1
- tRNAs
transfer RNAs
- UV
ultraviolet
- VDAC
voltage-dependent anion channel
- ZBP1
Z-DNA-binding protein 1
Author Disclosure Statement
No competing financial interests exist.
Funding Information
The authors were supported by grants NIH/NIAMS R01AR073282, NIH/NIAID R21AI128510, and NIH/NIAMS R21AR069830 (I.A.B.-K.) and the Harold S. Geneen Charitable Trust Awards Program for Coronary Heart Disease Research (A.K.).
References
- 1. Aarreberg LD, Esser-Nobis K, Driscoll C, Shuvarikov A, Roby JA, and Gale M Jr. Interleukin-1beta induces mtDNA release to activate innate immune signaling via cGAS-STING. Mol Cell 74: 801–815.e6, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aguirre S, Luthra P, Sanchez-Aparicio MT, Maestre AM, Patel J, Lamothe F, Fredericks AC, Tripathi S, Zhu T, Pintado-Silva J, Webb LG, Bernal-Rubio D, Solovyov A, Greenbaum B, Simon V, Basler CF, Mulder LC, Garcia-Sastre A, and Fernandez-Sesma A. Dengue virus NS2B protein targets cGAS for degradation and prevents mitochondrial DNA sensing during infection. Nat Microbiol 2: 17037, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aki T, Unuma K, and Uemura K. Emerging roles of mitochondria and autophagy in liver injury during sepsis. Cell Stress 1: 79–89, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aloni Y and Attardi G. Symmetrical in vivo transcription of mitochondrial DNA in HeLa cells. Proc Natl Acad Sci U S A 68: 1757–1761, 1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Amini P, Stojkov D, Felser A, Jackson CB, Courage C, Schaller A, Gelman L, Soriano ME, Nuoffer JM, Scorrano L, Benarafa C, Yousefi S, and Simon HU. Neutrophil extracellular trap formation requires OPA1-dependent glycolytic ATP production. Nat Commun 9: 2958, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Auten RL and Davis JM. Oxygen toxicity and reactive oxygen species: the devil is in the details. Pediatr Res 66: 121–127, 2009. [DOI] [PubMed] [Google Scholar]
- 7. Bae JH, Jo SI, Kim SJ, Lee JM, Jeong JH, Kang JS, Cho NJ, Kim SS, Lee EY, and Moon JS. Circulating cell-free mtDNA contributes to AIM2 inflammasome-mediated chronic inflammation in patients with type 2 diabetes. Cells 8: 328, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bae JY and Park HH. Crystal structure of NALP3 protein pyrin domain (PYD) and its implications in inflammasome assembly. J Biol Chem 286: 39528–39536, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Balasubramanian K, Maeda A, Lee JS, Mohammadyani D, Dar HH, Jiang JF, St Croix CM, Watkins S, Tyurin VA, Tyurina YY, Kloditz K, Polimova A, Kapralova VI, Xiong Z, Ray P, Klein-Seetharaman J, Mallampalli RK, Bayir H, Fadeel B, and Kagan VE. Dichotomous roles for externalized cardiolipin in extracellular signaling: promotion of phagocytosis and attenuation of innate immunity. Sci Signal 8: ra95, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barchiesi A and Vascotto C. Transcription, processing, and decay of mitochondrial RNA in health and disease. Int J Mol Sci 20: 2221, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barczyk K, Kreuter M, Pryjma J, Booy EP, Maddika S, Ghavami S, Berdel WE, Roth J, and Los M. Serum cytochrome c indicates in vivo apoptosis and can serve as a prognostic marker during cancer therapy. Int J Cancer 116: 167–173, 2005. [DOI] [PubMed] [Google Scholar]
- 12. Beckman JS, Beckman TW, Chen J, Marshall PA, and Freeman BA. Apparent hydroxyl radical production by peroxynitrite: implications for endothelial injury from nitric oxide and superoxide. Proc Natl Acad Sci U S A 87: 1620–1624, 1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Beigi RD, Kertesy SB, Aquilina G, and Dubyak GR. Oxidized ATP (oATP) attenuates proinflammatory signaling via P2 receptor-independent mechanisms. Br J Pharmacol 140: 507–519, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Berridge MJ. Inositol trisphosphate and calcium signalling. Nature 361: 315–325, 1993. [DOI] [PubMed] [Google Scholar]
- 15. Bihlmaier K, Mesecke N, Terziyska N, Bien M, Hell K, and Herrmann JM. The disulfide relay system of mitochondria is connected to the respiratory chain. J Cell Biol 179: 389–395, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bindoli A, Cavallini L, and Jocelyn P. Mitochondrial lipid peroxidation by cumene hydroperoxide and its prevention by succinate. Biochim Biophys Acta 681: 496–503, 1982. [DOI] [PubMed] [Google Scholar]
- 17. Borowski LS, Dziembowski A, Hejnowicz MS, Stepien PP, and Szczesny RJ. Human mitochondrial RNA decay mediated by PNPase-hSuv3 complex takes place in distinct foci. Nucleic Acids Res 41: 1223–1240, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Borutaite V and Brown GC. Mitochondrial regulation of caspase activation by cytochrome oxidase and tetramethylphenylenediamine via cytosolic cytochrome c redox state. J Biol Chem 282: 31124–31130, 2007. [DOI] [PubMed] [Google Scholar]
- 19. Brand MD. Mitochondrial generation of superoxide and hydrogen peroxide as the source of mitochondrial redox signaling. Free Radic Biol Med 100: 14–31, 2016. [DOI] [PubMed] [Google Scholar]
- 20. Brand MD. Riding the tiger—physiological and pathological effects of superoxide and hydrogen peroxide generated in the mitochondrial matrix. Crit Rev Biochem Mol Biol 55: 592–661, 2020. [DOI] [PubMed] [Google Scholar]
- 21. Briere JJ, Favier J, Benit P, El Ghouzzi V, Lorenzato A, Rabier D, Di Renzo MF, Gimenez-Roqueplo AP, and Rustin P. Mitochondrial succinate is instrumental for HIF1alpha nuclear translocation in SDHA-mutant fibroblasts under normoxic conditions. Hum Mol Genet 14: 3263–3269, 2005. [DOI] [PubMed] [Google Scholar]
- 22. Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, and Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science 303: 1532–1535, 2004. [DOI] [PubMed] [Google Scholar]
- 23. Buskiewicz I, Montgomery T, Yasewicz EC, Huber SA, Murphy MP, Hartley RC, Kelly R, Crow MK, Perl A, Budd RC, and Koenig A. Reactive oxygen species induce virus-independent MAVS oligomerization in systemic lupus erythematosus. Sci Signal 2016 Nov 29;9(456) ra115, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Caielli S, Athale S, Domic B, Murat E, Chandra M, Banchereau R, Baisch J, Phelps K, Clayton S, Gong M, Wright T, Punaro M, Palucka K, Guiducci C, Banchereau J, and Pascual V. Oxidized mitochondrial nucleoids released by neutrophils drive type I interferon production in human lupus. J Exp Med 213: 697–713, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Caielli S, Veiga DT, Balasubramanian P, Athale S, Domic B, Murat E, Banchereau R, Xu Z, Chandra M, Chung CH, Walters L, Baisch J, Wright T, Punaro M, Nassi L, Stewart K, Fuller J, Ucar D, Ueno H, Zhou J, Banchereau J, and Pascual V. A CD4(+) T cell population expanded in lupus blood provides B cell help through interleukin-10 and succinate. Nat Med 25: 75–81, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Carp H. Mitochondrial N-formylmethionyl proteins as chemoattractants for neutrophils. J Exp Med 155: 264–275, 1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Castro L, Tortora V, Mansilla S, and Radi R. Aconitases: non-redox iron-sulfur proteins sensitive to reactive species. Acc Chem Res 52: 2609–2619, 2019. [DOI] [PubMed] [Google Scholar]
- 28. Chance B and Hollunger G. The interaction of energy and electron transfer reactions in mitochondria. I. General properties and nature of the products of succinate-linked reduction of pyridine nucleotide. J Biol Chem 236: 1534–1543, 1961. [PubMed] [Google Scholar]
- 29. Chandrasekaran K, Muragundla A, Demarest TG, Choi J, Sagi AR, Najimi N, Kumar P, Singh A, Ho CY, Fiskum G, Koch LG, Britton SL, and Russell JW. mGluR2/3 activation of the SIRT1 axis preserves mitochondrial function in diabetic neuropathy. Ann Clin Transl Neurol 4: 844–858, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chao MR, Evans MD, Hu CW, Ji Y, Moller P, Rossner P, and Cooke MS. Biomarkers of nucleic acid oxidation—a summary state-of-the-art. Redox Biol 42: 101872, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chaung WW, Wu R, Ji Y, Dong W, and Wang P. Mitochondrial transcription factor A is a proinflammatory mediator in hemorrhagic shock. Int J Mol Med 30: 199–203, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chekeni FB, Elliott MR, Sandilos JK, Walk SF, Kinchen JM, Lazarowski ER, Armstrong AJ, Penuela S, Laird DW, Salvesen GS, Isakson BE, Bayliss DA, and Ravichandran KS. Pannexin 1 channels mediate ‘find-me’ signal release and membrane permeability during apoptosis. Nature 467: 863–867, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cho JH, Lee YK, and Chae CB. The modulation of the biological activities of mitochondrial histone Abf2p by yeast PKA and its possible role in the regulation of mitochondrial DNA content during glucose repression. Biochim Biophys Acta 1522: 175–186, 2001. [DOI] [PubMed] [Google Scholar]
- 34. Clark RA. Chemotactic factors trigger their own oxidative inactivation by human neutrophils. J Immunol 129: 2725–2728, 1982. [PubMed] [Google Scholar]
- 35. Clarke S. A major polypeptide component of rat liver mitochondria: carbamyl phosphate synthetase. J Biol Chem 251: 950–961, 1976. [PubMed] [Google Scholar]
- 36. Codina R, Vanasse A, Kelekar A, Vezys V, and Jemmerson R. Cytochrome c-induced lymphocyte death from the outside in: inhibition by serum leucine-rich alpha-2-glycoprotein-1. Apoptosis 15: 139–152, 2010. [DOI] [PubMed] [Google Scholar]
- 37. Collins LV, Hajizadeh S, Holme E, Jonsson IM, and Tarkowski A. Endogenously oxidized mitochondrial DNA induces in vivo and in vitro inflammatory responses. J Leukoc Biol 75: 995–1000, 2004. [DOI] [PubMed] [Google Scholar]
- 38. Collins Y, Chouchani ET, James AM, Menger KE, Cocheme HM, and Murphy MP. Mitochondrial redox signalling at a glance. J Cell Sci 125: 801–806, 2012. [DOI] [PubMed] [Google Scholar]
- 39. Corcoran JA, Saffran HA, Duguay BA, and Smiley JR. Herpes simplex virus UL12.5 targets mitochondria through a mitochondrial localization sequence proximal to the N terminus. J Virol 83: 2601–2610, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cordes T, Wallace M, Michelucci A, Divakaruni AS, Sapcariu SC, Sousa C, Koseki H, Cabrales P, Murphy AN, Hiller K, and Metallo CM. Immunoresponsive gene 1 and itaconate inhibit succinate dehydrogenase to modulate intracellular succinate levels. J Biol Chem 291: 14274–14284, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Corrado M, Edwards-Hicks J, Villa M, Flachsmann LJ, Sanin DE, Jacobs M, Baixauli F, Stanczak M, Anderson E, Azuma M, Quintana A, Curtis JD, Clapes T, Grzes KM, Kabat AM, Kyle R, Patterson AE, Geltink RK, Amulic B, Steward CG, Strathdee D, Trompouki E, O'Sullivan D, Pearce EJ, and Pearce EL. Dynamic cardiolipin synthesis is required for CD8(+) T cell immunity. Cell Metab 32: 981–995.e7, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Crouser ED, Julian MW, Huff JE, Struck J, and Cook CH. Carbamoyl phosphate synthase-1: a marker of mitochondrial damage and depletion in the liver during sepsis. Crit Care Med 34: 2439–2446, 2006. [DOI] [PubMed] [Google Scholar]
- 43. Czapiga M, Gao JL, Kirk A, and Lekstrom-Himes J. Human platelets exhibit chemotaxis using functional N-formyl peptide receptors. Exp Hematol 33: 73–84, 2005. [DOI] [PubMed] [Google Scholar]
- 44. Dagda RK, Zhu J, Kulich SM, and Chu CT. Mitochondrially localized ERK2 regulates mitophagy and autophagic cell stress: implications for Parkinson's disease. Autophagy 4: 770–782, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dhir A, Dhir S, Borowski LS, Jimenez L, Teitell M, Rotig A, Crow YJ, Rice GI, Duffy D, Tamby C, Nojima T, Munnich A, Schiff M, de Almeida CR, Rehwinkel J, Dziembowski A, Szczesny RJ, and Proudfoot NJ. Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature 560: 238–242, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dimitriu T, Szczelkun MD, and Westra ER. Evolutionary ecology and interplay of prokaryotic innate and adaptive immune systems. Curr Biol 30: R1189–R1202, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Doran E and Halestrap AP. Cytochrome c release from isolated rat liver mitochondria can occur independently of outer-membrane rupture: possible role of contact sites. Biochem J 348 Pt 2: 343–350, 2000. [PMC free article] [PubMed] [Google Scholar]
- 48. Doring Y, Hurst J, Lorenz M, Prinz N, Clemens N, Drechsler MD, Bauer S, Chapman J, Shoenfeld Y, Blank M, Lackner KJ, and von Landenberg P. Human antiphospholipid antibodies induce TNFalpha in monocytes via Toll-like receptor 8. Immunobiology 215: 230–241, 2010. [DOI] [PubMed] [Google Scholar]
- 49. Duguay BA, Saffran HA, Ponomarev A, Duley SA, Eaton HE, and Smiley JR. Elimination of mitochondrial DNA is not required for herpes simplex virus 1 replication. J Virol 88: 2967–2976, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Elliott MR, Chekeni FB, Trampont PC, Lazarowski ER, Kadl A, Walk SF, Park D, Woodson RI, Ostankovich M, Sharma P, Lysiak JJ, Harden TK, Leitinger N, and Ravichandran KS. Nucleotides released by apoptotic cells act as a find-me signal to promote phagocytic clearance. Nature 461: 282–286, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Falkenberg M, Larsson NG, and Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem 76: 679–699, 2007. [DOI] [PubMed] [Google Scholar]
- 52. Fang J, Wong HS, and Brand MD. Production of superoxide and hydrogen peroxide in the mitochondrial matrix is dominated by site IQ of complex I in diverse cell lines. Redox Biol 37: 101722, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fernandes-Alnemri T, Yu JW, Datta P, Wu J, and Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458: 509–513, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Finkel T. From sulfenylation to sulfhydration: what a thiolate needs to tolerate. Sci Signal 5: pe10, 2012. [DOI] [PubMed] [Google Scholar]
- 55. Freund I, Eigenbrod T, Helm M, and Dalpke AH. RNA modifications modulate activation of innate toll-like receptors. Genes (Basel) 10: 92, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fridovich I. Superoxide anion radical (O2−.), superoxide dismutases, and related matters. J Biol Chem 272: 18515–18517, 1997. [DOI] [PubMed] [Google Scholar]
- 57. Garcia-Martinez I, Santoro N, Chen Y, Hoque R, Ouyang X, Caprio S, Shlomchik MJ, Coffman RL, Candia A, and Mehal WZ. Hepatocyte mitochondrial DNA drives nonalcoholic steatohepatitis by activation of TLR9. J Clin Invest 126: 859–864, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Garrido C, Galluzzi L, Brunet M, Puig PE, Didelot C, and Kroemer G. Mechanisms of cytochrome c release from mitochondria. Cell Death Differ 13: 1423–1433, 2006. [DOI] [PubMed] [Google Scholar]
- 59. Gehrke N, Mertens C, Zillinger T, Wenzel J, Bald T, Zahn S, Tuting T, Hartmann G, and Barchet W. Oxidative damage of DNA confers resistance to cytosolic nuclease TREX1 degradation and potentiates STING-dependent immune sensing. Immunity 39: 482–495, 2013. [DOI] [PubMed] [Google Scholar]
- 60. Germic N, Fettrelet T, Stojkov D, Hosseini A, Horn MP, Karaulov A, Simon D, Yousefi S, and Simon HU. The release kinetics of eosinophil peroxidase and mitochondrial dna is different in association with eosinophil extracellular trap formation. Cells 10: 306, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ginguay A and De Bandt JP. Citrulline production and protein homeostasis. Curr Opin Clin Nutr Metab Care 22: 371–376, 2019. [DOI] [PubMed] [Google Scholar]
- 62. Gogvadze V, Orrenius S, and Zhivotovsky B. Multiple pathways of cytochrome c release from mitochondria in apoptosis. Biochim Biophys Acta 1757: 639–647, 2006. [DOI] [PubMed] [Google Scholar]
- 63. Gombault A, Baron L, and Couillin I. ATP release and purinergic signaling in NLRP3 inflammasome activation. Front Immunol 3: 414, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gonzalez-Arzola K, Velazquez-Cruz A, Guerra-Castellano A, Casado-Combreras MA, Perez-Mejias G, Diaz-Quintana A, Diaz-Moreno I, and De la Rosa MA. New moonlighting functions of mitochondrial cytochrome c in the cytoplasm and nucleus. FEBS Lett 593: 3101–3119, 2019. [DOI] [PubMed] [Google Scholar]
- 65. Grazioli S and Pugin J. Mitochondrial damage-associated molecular patterns: from inflammatory signaling to human diseases. Front Immunol 9: 832, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Guzy RD, Sharma B, Bell E, Chandel NS, and Schumacker PT. Loss of the SdhB, but Not the SdhA, subunit of complex II triggers reactive oxygen species-dependent hypoxia-inducible factor activation and tumorigenesis. Mol Cell Biol 28: 718–731, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hampton MB, Zhivotovsky B, Slater AF, Burgess DH, and Orrenius S. Importance of the redox state of cytochrome c during caspase activation in cytosolic extracts. Biochem J 329 (Pt 1): 95–99, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. He W, Miao FJ, Lin DC, Schwandner RT, Wang Z, Gao J, Chen JL, Tian H, and Ling L. Citric acid cycle intermediates as ligands for orphan G-protein-coupled receptors. Nature 429: 188–193, 2004. [DOI] [PubMed] [Google Scholar]
- 69. Herb M, Gluschko A, Wiegmann K, Farid A, Wolf A, Utermohlen O, Krut O, Kronke M, and Schramm M. Mitochondrial reactive oxygen species enable proinflammatory signaling through disulfide linkage of NEMO. Sci Signal 12: eaar5926, 2019. [DOI] [PubMed] [Google Scholar]
- 70. Hong EE, Okitsu CY, Smith AD, and Hsieh CL. Regionally specific and genome-wide analyses conclusively demonstrate the absence of CpG methylation in human mitochondrial DNA. Mol Cell Biol 33: 2683–2690, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, and Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458: 514–518, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hwang CS, Park SC, Cho HJ, Park DJ, Yoon JH, and Kim CH. Eosinophil extracellular trap formation is closely associated with disease severity in chronic rhinosinusitis regardless of nasal polyp status. Sci Rep 9: 8061, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Idzko M, Hammad H, van Nimwegen M, Kool M, Willart MA, Muskens F, Hoogsteden HC, Luttmann W, Ferrari D, Di Virgilio F, Virchow JC Jr., and Lambrecht BN. Extracellular ATP triggers and maintains asthmatic airway inflammation by activating dendritic cells. Nat Med 13: 913–919, 2007. [DOI] [PubMed] [Google Scholar]
- 74. Itagaki K, Rica I, Konecna B, Kim HI, Park J, Kaczmarek E, and Hauser CJ. Role of mitochondria-derived danger signals released after injury in systemic inflammation and sepsis. Antioxid Redox Signal 35: 1273–1290, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Iverson SL and Orrenius S. The cardiolipin-cytochrome c interaction and the mitochondrial regulation of apoptosis. Arch Biochem Biophys 423: 37–46, 2004. [DOI] [PubMed] [Google Scholar]
- 76. Iwasaki A and Medzhitov R. Control of adaptive immunity by the innate immune system. Nat Immunol 16: 343–353, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Iyer SS, He Q, Janczy JR, Elliott EI, Zhong Z, Olivier AK, Sadler JJ, Knepper-Adrian V, Han R, Qiao L, Eisenbarth SC, Nauseef WM, Cassel SL, and Sutterwala FS. Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity 39: 311–323, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jemmerson R, LaPlante B, and Treeful A. Release of intact, monomeric cytochrome c from apoptotic and necrotic cells. Cell Death Differ 9: 538–548, 2002. [DOI] [PubMed] [Google Scholar]
- 79. Jiang HP, Xiong J, Liu FL, Ma CJ, Tang XL, Yuan BF, and Feng YQ. Modified nucleoside triphosphates exist in mammals. Chem Sci 9: 4160–4167, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Jones JD, Vance RE, and Dangl JL. Intracellular innate immune surveillance devices in plants and animals. Science 354: aaf6395, 2016. [DOI] [PubMed] [Google Scholar]
- 80a. Jones ME, Spector L, and Lipmann F. Carbamyl phosphate, the carbamyl donor in enzymatic citrulline synthesis. J Am Chem Soc 77: 819–820, 1955. [Google Scholar]
- 81. Julian MW, Shao G, Bao S, Knoell DL, Papenfuss TL, VanGundy ZC, and Crouser ED. Mitochondrial transcription factor A serves as a danger signal by augmenting plasmacytoid dendritic cell responses to DNA. J Immunol 189: 433–443, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Julian MW, Shao G, Vangundy ZC, Papenfuss TL, and Crouser ED. Mitochondrial transcription factor A, an endogenous danger signal, promotes TNFalpha release via RAGE- and TLR9-responsive plasmacytoid dendritic cells. PLoS One 8: e72354, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kagan VE, Tyurin VA, Jiang J, Tyurina YY, Ritov VB, Amoscato AA, Osipov AN, Belikova NA, Kapralov AA, Kini V, Vlasova, II, Zhao Q, Zou M, Di P, Svistunenko DA, Kurnikov IV, and Borisenko GG. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat Chem Biol 1: 223–232, 2005. [DOI] [PubMed] [Google Scholar]
- 84. Kang D, Kim SH, and Hamasaki N. Mitochondrial transcription factor A (TFAM): roles in maintenance of mtDNA and cellular functions. Mitochondrion 7: 39–44, 2007. [DOI] [PubMed] [Google Scholar]
- 85. Kang D, Nishida J, Iyama A, Nakabeppu Y, Furuichi M, Fujiwara T, Sekiguchi M, and Takeshige K. Intracellular localization of 8-oxo-dGTPase in human cells, with special reference to the role of the enzyme in mitochondria. J Biol Chem 270: 14659–14665, 1995. [DOI] [PubMed] [Google Scholar]
- 86. Kariko K, Buckstein M, Ni H, and Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23: 165–175, 2005. [DOI] [PubMed] [Google Scholar]
- 87. Kazama H, Ricci JE, Herndon JM, Hoppe G, Green DR, and Ferguson TA. Induction of immunological tolerance by apoptotic cells requires caspase-dependent oxidation of high-mobility group box-1 protein. Immunity 29: 21–32, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kim Y, Park J, Kim S, Kim M, Kang MG, Kwak C, Kang M, Kim B, Rhee HW, and Kim VN. PKR senses nuclear and mitochondrial signals by interacting with endogenous double-stranded RNAs. Mol Cell 71: 1051–1063.e6, 2018. [DOI] [PubMed] [Google Scholar]
- 89. King GA, Hashemi Shabestari M, Taris KH, Pandey AK, Venkatesh S, Thilagavathi J, Singh K, Krishna Koppisetti R, Temiakov D, Roos WH, Suzuki CK, and Wuite GJL. Acetylation and phosphorylation of human TFAM regulate TFAM-DNA interactions via contrasting mechanisms. Nucleic Acids Res 46: 3633–3642, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Komlodi T, Geibl FF, Sassani M, Ambrus A, and Tretter L. Membrane potential and delta pH dependency of reverse electron transport-associated hydrogen peroxide production in brain and heart mitochondria. J Bioenerg Biomembr 50: 355–365, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kopp EB and Medzhitov R. The Toll-receptor family and control of innate immunity. Curr Opin Immunol 11: 13–18, 1999. [DOI] [PubMed] [Google Scholar]
- 92. Korshunov SS, Skulachev VP, and Starkov AA. High protonic potential actuates a mechanism of production of reactive oxygen species in mitochondria. FEBS Lett 416: 15–18, 1997. [DOI] [PubMed] [Google Scholar]
- 93. Kouzaki H, Iijima K, Kobayashi T, O'Grady SM, and Kita H. The danger signal, extracellular ATP, is a sensor for an airborne allergen and triggers IL-33 release and innate Th2-type responses. J Immunol 186: 4375–4387, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Krysko DV, Agostinis P, Krysko O, Garg AD, Bachert C, Lambrecht BN, and Vandenabeele P. Emerging role of damage-associated molecular patterns derived from mitochondria in inflammation. Trends Immunol 32: 157–164, 2011. [DOI] [PubMed] [Google Scholar]
- 95. Kukat C, Wurm CA, Spahr H, Falkenberg M, Larsson NG, and Jakobs S. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc Natl Acad Sci U S A 108: 13534–13539, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Kummer E and Ban N. Mechanisms and regulation of protein synthesis in mitochondria. Nat Rev Mol Cell Biol 22: 307–325, 2021. [DOI] [PubMed] [Google Scholar]
- 97. Kunkel GH, Chaturvedi P, Thelian N, Nair R, and Tyagi SC. Mechanisms of TFAM-mediated cardiomyocyte protection. Can J Physiol Pharmacol 96: 173–181, 2018. [DOI] [PubMed] [Google Scholar]
- 98. Kurashima Y, Amiya T, Nochi T, Fujisawa K, Haraguchi T, Iba H, Tsutsui H, Sato S, Nakajima S, Iijima H, Kubo M, Kunisawa J, and Kiyono H. Extracellular ATP mediates mast cell-dependent intestinal inflammation through P2X7 purinoceptors. Nat Commun 3: 1034, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Lai JH, Wang MY, Huang CY, Wu CH, Hung LF, Yang CY, Ke PY, Luo SF, Liu SJ, and Ho LJ. Infection with the dengue RNA virus activates TLR9 signaling in human dendritic cells. EMBO Rep 19: e46182, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lampropoulou V, Sergushichev A, Bambouskova M, Nair S, Vincent EE, Loginicheva E, Cervantes-Barragan L, Ma X, Huang SC, Griss T, Weinheimer CJ, Khader S, Randolph GJ, Pearce EJ, Jones RG, Diwan A, Diamond MS, and Artyomov MN. Itaconate links inhibition of succinate dehydrogenase with macrophage metabolic remodeling and regulation of inflammation. Cell Metab 24: 158–166, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Lee KH, Kronbichler A, Park DD, Park Y, Moon H, Kim H, Choi JH, Choi Y, Shim S, Lyu IS, Yun BH, Han Y, Lee D, Lee SY, Yoo BH, Lee KH, Kim TL, Kim H, Shim JS, Nam W, So H, Choi S, Lee S, and Shin JI. Neutrophil extracellular traps (NETs) in autoimmune diseases: a comprehensive review. Autoimmun Rev 16: 1160–1173, 2017. [DOI] [PubMed] [Google Scholar]
- 102. Lei W, Ren W, Ohmoto M, Urban JF Jr., Matsumoto I, Margolskee RF, and Jiang P.. Activation of intestinal tuft cell-expressed Sucnr1 triggers type 2 immunity in the mouse small intestine. Proc Natl Acad Sci U S A 115: 5552–5557, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Li S and Yang G. Hydrogen sulfide maintains mitochondrial DNA replication via demethylation of TFAM. Antioxid Redox Signal 23: 630–642, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Little JP, Simtchouk S, Schindler SM, Villanueva EB, Gill NE, Walker DG, Wolthers KR, and Klegeris A. Mitochondrial transcription factor A (Tfam) is a pro-inflammatory extracellular signaling molecule recognized by brain microglia. Mol Cell Neurosci 60: 88–96, 2014. [DOI] [PubMed] [Google Scholar]
- 105. Littlewood-Evans A, Sarret S, Apfel V, Loesle P, Dawson J, Zhang J, Muller A, Tigani B, Kneuer R, Patel S, Valeaux S, Gommermann N, Rubic-Schneider T, Junt T, and Carballido JM. GPR91 senses extracellular succinate released from inflammatory macrophages and exacerbates rheumatoid arthritis. J Exp Med 213: 1655–1662, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Lood C, Blanco LP, Purmalek MM, Carmona-Rivera C, De Ravin SS, Smith CK, Malech HL, Ledbetter JA, Elkon KB, and Kaplan MJ. Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nat Med 22: 146–153, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Lu B, Lee J, Nie X, Li M, Morozov YI, Venkatesh S, Bogenhagen DF, Temiakov D, and Suzuki CK. Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease. Mol Cell 49: 121–132, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. This reference has been deleted.
- 109. Malarkey CS and Churchill ME. The high mobility group box: the ultimate utility player of a cell. Trends Biochem Sci 37: 553–562, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Mamula MJ, Jemmerson R, and Hardin JA. The specificity of human anti-cytochrome c autoantibodies that arise in autoimmune disease. J Immunol 144: 1835–1840, 1990. [PubMed] [Google Scholar]
- 111. Matzinger P. Tolerance, danger, and the extended family. Annu Rev Immunol 12: 991–1045, 1994. [DOI] [PubMed] [Google Scholar]
- 112. Meddeb R, Dache ZAA, Thezenas S, Otandault A, Tanos R, Pastor B, Sanchez C, Azzi J, Tousch G, Azan S, Mollevi C, Adenis A, El Messaoudi S, Blache P, and Thierry AR. Quantifying circulating cell-free DNA in humans. Sci Rep 9: 5220, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Mendez DL, Akey IV, Akey CW, and Kranz RG. Oxidized or reduced cytochrome c and axial ligand variants all form the apoptosome in vitro. Biochemistry 56: 2766–2769, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Meylan E, Tschopp J, and Karin M. Intracellular pattern recognition receptors in the host response. Nature 442: 39–44, 2006. [DOI] [PubMed] [Google Scholar]
- 115. Migeotte I, Communi D, and Parmentier M. Formyl peptide receptors: a promiscuous subfamily of G protein-coupled receptors controlling immune responses. Cytokine Growth Factor Rev 17: 501–519, 2006. [DOI] [PubMed] [Google Scholar]
- 116. Millare B, O'Rourke B, and Trayanova N. Hydrogen peroxide diffusion and scavenging shapes mitochondrial network instability and failure by sensitizing ROS-induced ROS release. Sci Rep 10: 15758, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Miller TJ, Knapton A, Adeyemo O, Noory L, Weaver J, and Hanig JP. Cytochrome c: a non-invasive biomarker of drug-induced liver injury. J Appl Toxicol 28: 815–828, 2008. [DOI] [PubMed] [Google Scholar]
- 118. Mills E and O'Neill LA. Succinate: a metabolic signal in inflammation. Trends Cell Biol 24: 313–320, 2014. [DOI] [PubMed] [Google Scholar]
- 119. Miyako K, Kohno H, Ihara K, Kuromaru R, Matsuura N, and Hara T. Association study of human MTH1 gene polymorphisms with type 1 diabetes mellitus. Endocr J 51: 493–498, 2004. [DOI] [PubMed] [Google Scholar]
- 120. Morozov YI, Agaronyan K, Cheung AC, Anikin M, Cramer P, and Temiakov D. A novel intermediate in transcription initiation by human mitochondrial RNA polymerase. Nucleic Acids Res 42: 3884–3893, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Muller FL, Liu Y, and Van Remmen H. Complex III releases superoxide to both sides of the inner mitochondrial membrane. J Biol Chem 279: 49064–49073, 2004. [DOI] [PubMed] [Google Scholar]
- 122. Muller-Calleja N and Lackner KJ. Mechanisms of cellular activation in the antiphospholipid syndrome. Semin Thromb Hemost 44: 483–492, 2018. [DOI] [PubMed] [Google Scholar]
- 123. Murakami T, Ockinger J, Yu J, Byles V, McColl A, Hofer AM, and Horng T. Critical role for calcium mobilization in activation of the NLRP3 inflammasome. Proc Natl Acad Sci U S A 109: 11282–11287, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Murgia M, Hanau S, Pizzo P, Rippa M, and Di Virgilio F. Oxidized ATP. An irreversible inhibitor of the macrophage purinergic P2Z receptor. J Biol Chem 268: 8199–8203, 1993. [PubMed] [Google Scholar]
- 125. Murphy MP. How mitochondria produce reactive oxygen species. Biochem J 417: 1–13, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Murphy MP, Holmgren A, Larsson NG, Halliwell B, Chang CJ, Kalyanaraman B, Rhee SG, Thornalley PJ, Partridge L, Gems D, Nystrom T, Belousov V, Schumacker PT, and Winterbourn CC. Unraveling the biological roles of reactive oxygen species. Cell Metab 13: 361–366, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Murphy MP, and O'Neill LAJ. Krebs cycle reimagined: the emerging roles of succinate and itaconate as signal transducers. Cell 174: 780–784, 2018. [DOI] [PubMed] [Google Scholar]
- 128. Nakabeppu Y, Behmanesh M, Ymaguchi H, Tsuchimoto D, and Kunihiko S. Prevention of the mutagenecity and cytotoxicity of oxidized purine nucleotides. In: Oxidative Damage to Nucleic Acids, 2007-08-28. Landes Bioscience, Austin, Texas, USA, 2007, pp. 40–53. [Google Scholar]
- 129. Nakabeppu Y, Oka S, Sheng Z, Tsuchimoto D, and Sakumi K. Programmed cell death triggered by nucleotide pool damage and its prevention by MutT homolog-1 (MTH1) with oxidized purine nucleoside triphosphatase. Mutat Res 703: 51–58, 2010. [DOI] [PubMed] [Google Scholar]
- 130. Nakagawa Y. Initiation of apoptotic signal by the peroxidation of cardiolipin of mitochondria. Ann N Y Acad Sci 1011: 177–184, 2004. [DOI] [PubMed] [Google Scholar]
- 131. Orrenius S, Zhivotovsky B, and Nicotera P. Regulation of cell death: the calcium-apoptosis link. Nat Rev Mol Cell Biol 4: 552–565, 2003. [DOI] [PubMed] [Google Scholar]
- 132. Pajak A, Laine I, Clemente P, El-Fissi N, Schober FA, Maffezzini C, Calvo-Garrido J, Wibom R, Filograna R, Dhir A, Wedell A, Freyer C, and Wredenberg A. Defects of mitochondrial RNA turnover lead to the accumulation of double-stranded RNA in vivo. PLoS Genet 15: e1008240, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Pan Z, Voehringer DW, and Meyn RE. Analysis of redox regulation of cytochrome c-induced apoptosis in a cell-free system. Cell Death Differ 6: 683–688, 1999. [DOI] [PubMed] [Google Scholar]
- 134. Parisi MA and Clayton DA. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science 252: 965–969, 1991. [DOI] [PubMed] [Google Scholar]
- 135. Park MJ, D'Alecy LG, Anderson MA, Basrur V, Feng Y, Brady GF, Kim DI, Wu J, Nesvizhskii AI, Lahann J, Lukacs NW, Fontana RJ, and Omary MB. Constitutive release of CPS1 in bile and its role as a protective cytokine during acute liver injury. Proc Natl Acad Sci U S A 116: 9125–9134, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Park S and Lippard SJ. Redox state-dependent interaction of HMGB1 and cisplatin-modified DNA. Biochemistry 50: 2567–2574, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Patergnani S, Bouhamida E, Leo S, Pinton P, and Rimessi A. Mitochondrial oxidative stress and “mito-inflammation”: actors in the diseases. Biomedicines 9: 216, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Patil V, Cuenin C, Chung F, Aguilera JRR, Fernandez-Jimenez N, Romero-Garmendia I, Bilbao JR, Cahais V, Rothwell J, and Herceg Z. Human mitochondrial DNA is extensively methylated in a non-CpG context. Nucleic Acids Res 47: 10072–10085, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Pazmandi K, Agod Z, Kumar BV, Szabo A, Fekete T, Sogor V, Veres A, Boldogh I, Rajnavolgyi E, Lanyi A, and Bacsi A. Oxidative modification enhances the immunostimulatory effects of extracellular mitochondrial DNA on plasmacytoid dendritic cells. Free Radic Biol Med 77: 281–290, 2014. [DOI] [PubMed] [Google Scholar]
- 140. Perez-Mejias G, Guerra-Castellano A, Diaz-Quintana A, De la Rosa MA, and Diaz-Moreno I. Cytochrome c: surfing off of the mitochondrial membrane on the tops of complexes III and IV. Comput Struct Biotechnol J 17: 654–660, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Pizzuto M and Pelegrin P. Cardiolipin in immune signaling and cell death. Trends Cell Biol 30: 892–903, 2020. [DOI] [PubMed] [Google Scholar]
- 142. Pothlichet J, Niewold TB, Vitour D, Solhonne B, Crow MK, and Si-Tahar M. A loss-of-function variant of the antiviral molecule MAVS is associated with a subset of systemic lupus patients. EMBO Mol Med 3: 142–152, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Quinlan CL, Orr AL, Perevoshchikova IV, Treberg JR, Ackrell BA, and Brand MD. Mitochondrial complex II can generate reactive oxygen species at high rates in both the forward and reverse reactions. J Biol Chem 287: 27255–27264, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Radhakrishnan J, Wang S, Ayoub IM, Kolarova JD, Levine RF, and Gazmuri RJ. Circulating levels of cytochrome c after resuscitation from cardiac arrest: a marker of mitochondrial injury and predictor of survival. Am J Physiol Heart Circ Physiol 292: H767–H775, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Rahmel T, Marko B, Nowak H, Bergmann L, Thon P, Rump K, Kreimendahl S, Rassow J, Peters J, Singer M, Adamzik M, and Koos B. Mitochondrial dysfunction in sepsis is associated with diminished intramitochondrial TFAM despite its increased cellular expression. Sci Rep 10: 21029, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Raoof M, Zhang Q, Itagaki K, and Hauser CJ. Mitochondrial peptides are potent immune activators that activate human neutrophils via FPR-1. J Trauma 68: 1328–1332; discussion 133 2–1334, 2010. [DOI] [PubMed] [Google Scholar]
- 147. Ray NB, Durairaj L, Chen BB, McVerry BJ, Ryan AJ, Donahoe M, Waltenbaugh AK, O'Donnell CP, Henderson FC, Etscheidt CA, McCoy DM, Agassandian M, Hayes-Rowan EC, Coon TA, Butler PL, Gakhar L, Mathur SN, Sieren JC, Tyurina YY, Kagan VE, McLennan G, and Mallampalli RK. Dynamic regulation of cardiolipin by the lipid pump Atp8b1 determines the severity of lung injury in experimental pneumonia. Nat Med 16: 1120–1127, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Rivera-Serrano EE, DeAngelis N, and Sherry B. Spontaneous activation of a MAVS-dependent antiviral signaling pathway determines high basal interferon-beta expression in cardiac myocytes. J Mol Cell Cardiol 111: 102–113, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Robb EL, Gawel JM, Aksentijevic D, Cocheme HM, Stewart TS, Shchepinova MM, Qiang H, Prime TA, Bright TP, James AM, Shattock MJ, Senn HM, Hartley RC, and Murphy MP. Selective superoxide generation within mitochondria by the targeted redox cycler MitoParaquat. Free Radic Biol Med 89: 883–894, 2015. [DOI] [PubMed] [Google Scholar]
- 150. Rongvaux A, Jackson R, Harman CC, Li T, West AP, de Zoete MR, Wu Y, Yordy B, Lakhani SA, Kuan CY, Taniguchi T, Shadel GS, Chen ZJ, Iwasaki A, and Flavell RA. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell 159: 1563–1577, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Rouhiainen A, Tumova S, Valmu L, Kalkkinen N, and Rauvala H. Pivotal advance: analysis of proinflammatory activity of highly purified eukaryotic recombinant HMGB1 (amphoterin). J Leukoc Biol 81: 49–58, 2007. [DOI] [PubMed] [Google Scholar]
- 152. Rubic T, Lametschwandtner G, Jost S, Hinteregger S, Kund J, Carballido-Perrig N, Schwarzler C, Junt T, Voshol H, Meingassner JG, Mao X, Werner G, Rot A, and Carballido JM. Triggering the succinate receptor GPR91 on dendritic cells enhances immunity. Nat Immunol 9: 1261–1269, 2008. [DOI] [PubMed] [Google Scholar]
- 153. Saferding V and Bluml S. Innate immunity as the trigger of systemic autoimmune diseases. J Autoimmun 110: 102382, 2020. [DOI] [PubMed] [Google Scholar]
- 154. Sakumi K, Furuichi M, Tsuzuki T, Kakuma T, Kawabata S, Maki H, and Sekiguchi M. Cloning and expression of cDNA for a human enzyme that hydrolyzes 8-oxo-dGTP, a mutagenic substrate for DNA synthesis. J Biol Chem 268: 23524–23530, 1993. [PubMed] [Google Scholar]
- 155. Salem D, Subang R, Laplante P, Levine JS, and Rauch J. The dual role of innate immunity in antiphospholipid syndrome and systemic lupus erythematosus. Lupus 23: 1327–1331, 2014. [DOI] [PubMed] [Google Scholar]
- 156. Santos JM, Mishra M, and Kowluru RA. Posttranslational modification of mitochondrial transcription factor A in impaired mitochondria biogenesis: implications in diabetic retinopathy and metabolic memory phenomenon. Exp Eye Res 121: 168–177, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Scaffidi P, Misteli T, and Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 418: 191–195, 2002. [DOI] [PubMed] [Google Scholar]
- 158. Schiffmann E, Corcoran BA, and Wahl SM. N-formylmethionyl peptides as chemoattractants for leucocytes. Proc Natl Acad Sci U S A 72: 1059–1062, 1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, Dabelic R, Manicassamy B, Aitchison JD, Aderem A, Elliott RM, Garcia-Sastre A, Racaniello V, Snijder EJ, Yokoyama WM, Diamond MS, Virgin HW, and Rice CM. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505: 691–695, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, and Gottlieb E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell 7: 77–85, 2005. [DOI] [PubMed] [Google Scholar]
- 161. Seth RB, Sun L, Ea CK, and Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122: 669–682, 2005. [DOI] [PubMed] [Google Scholar]
- 162. Shao WH, Shu DH, Zhen Y, Hilliard B, Priest SO, Cesaroni M, Ting JP, and Cohen PL. Prion-like aggregation of mitochondrial antiviral signaling protein in lupus patients is associated with increased levels of type I interferon. Arthritis Rheumatol 68: 2697–2707, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Shen F, Tang X, Cheng W, Wang Y, Wang C, Shi X, An Y, Zhang Q, Liu M, Liu B, and Yu L. Fosfomycin enhances phagocyte-mediated killing of Staphylococcus aureus by extracellular traps and reactive oxygen species. Sci Rep 6: 19262, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Sheng KC, Pietersz GA, Tang CK, Ramsland PA, and Apostolopoulos V. Reactive oxygen species level defines two functionally distinctive stages of inflammatory dendritic cell development from mouse bone marrow. J Immunol 184: 2863–2872, 2010. [DOI] [PubMed] [Google Scholar]
- 165. Sies H and Jones DP. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat Rev Mol Cell Biol 21: 363–383, 2020. [DOI] [PubMed] [Google Scholar]
- 166. Silva JDC, Thompson-Souza GA, Barroso MV, Neves JS, and Figueiredo RT. Neutrophil and eosinophil DNA extracellular trap formation: lessons from pathogenic fungi. Front Microbiol 12: 634043, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Speckmann B, Steinbrenner H, Grune T, and Klotz LO. Peroxynitrite: from interception to signaling. Arch Biochem Biophys 595: 153–160, 2016. [DOI] [PubMed] [Google Scholar]
- 168. Struck J, Uhlein M, Morgenthaler NG, Furst W, Hoflich C, Bahrami S, Bergmann A, Volk HD, and Redl H. Release of the mitochondrial enzyme carbamoyl phosphate synthase under septic conditions. Shock 23: 533–538, 2005. [PubMed] [Google Scholar]
- 169. Sun B, Sundstrom KB, Chew JJ, Bist P, Gan ES, Tan HC, Goh KC, Chawla T, Tang CK, and Ooi EE. Dengue virus activates cGAS through the release of mitochondrial DNA. Sci Rep 7: 3594, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Sun L, Wu J, Du F, Chen X, and Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339: 786–791, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Taanman JW. The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta 1410: 103–123, 1999. [DOI] [PubMed] [Google Scholar]
- 172. Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, Ohba Y, and Taniguchi T. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448: 501–505, 2007. [DOI] [PubMed] [Google Scholar]
- 173. Tang C, Wang X, Xie Y, Cai X, Yu N, Hu Y, and Zheng Z. 4-Octyl itaconate activates Nrf2 signaling to inhibit pro-inflammatory cytokine production in peripheral blood mononuclear cells of systemic lupus erythematosus patients. Cell Physiol Biochem 51: 979–990, 2018. [DOI] [PubMed] [Google Scholar]
- 174. Tang D, Kang R, Zeh HJ, 3rd, and Lotze MT. High-mobility group box 1, oxidative stress, and disease. Antioxid Redox Signal 14: 1315–1335, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, Zheng L, Gardet A, Tong Z, Jany SS, Corr SC, Haneklaus M, Caffrey BE, Pierce K, Walmsley S, Beasley FC, Cummins E, Nizet V, Whyte M, Taylor CT, Lin H, Masters SL, Gottlieb E, Kelly VP, Clish C, Auron PE, Xavier RJ, and O'Neill LA. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature 496: 238–242, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Tao L, Lemoff A, Wang G, Zarek C, Lowe A, Yan N, and Reese TA. Reactive oxygen species oxidize STING and suppress interferon production. Elife 9: e57837, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Thompson MR, Kaminski JJ, Kurt-Jones EA, and Fitzgerald KA. Pattern recognition receptors and the innate immune response to viral infection. Viruses 3: 920–940, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Toma I, Kang JJ, Sipos A, Vargas S, Bansal E, Hanner F, Meer E, and Peti-Peterdi J. Succinate receptor GPR91 provides a direct link between high glucose levels and renin release in murine and rabbit kidney. J Clin Invest 118: 2526–2534, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Trumpff C, Michelson J, Lagranha CJ, Taleon V, Karan KR, Sturm G, Lindqvist D, Fernstrom J, Moser D, Kaufman BA, and Picard M. Stress and circulating cell-free mitochondrial DNA: a systematic review of human studies, physiological considerations, and technical recommendations. Mitochondrion 59: 225–245, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Tsan MF and Denison RC. Oxidation of n-formyl methionyl chemotactic peptide by human neutrophils. J Immunol 126: 1387–1389, 1981. [PubMed] [Google Scholar]
- 181. Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, and Bowie AG. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 11: 997–1004, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Vasquez-Vivar J, Kalyanaraman B, and Kennedy MC. Mitochondrial aconitase is a source of hydroxyl radical. An electron spin resonance investigation. J Biol Chem 275: 14064–14069, 2000. [DOI] [PubMed] [Google Scholar]
- 183. Vaughn AE and Deshmukh M. Glucose metabolism inhibits apoptosis in neurons and cancer cells by redox inactivation of cytochrome c. Nat Cell Biol 10: 1477–1483, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Vojdani A, Pollard KM, and Campbell AW. Environmental triggers and autoimmunity. Autoimmune Dis 2014: 798029, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Wang KZ, Zhu J, Dagda RK, Uechi G, Cherra SJ, 3rd, Gusdon AM, Balasubramani M, and Chu CT. ERK-mediated phosphorylation of TFAM downregulates mitochondrial transcription: implications for Parkinson's disease. Mitochondrion 17: 132–140, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Wayne LG and Lin KY. Glyoxylate metabolism and adaptation of Mycobacterium tuberculosis to survival under anaerobic conditions. Infect Immun 37: 1042–1049, 1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Weerasinghe SV, Jang YJ, Fontana RJ, and Omary MB. Carbamoyl phosphate synthetase-1 is a rapid turnover biomarker in mouse and human acute liver injury. Am J Physiol Gastrointest Liver Physiol 307: G355–G364, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. West AP and Shadel GS. Mitochondrial DNA in innate immune responses and inflammatory pathology. Nat Rev Immunol 17: 363–375, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. White MJ, McArthur K, Metcalf D, Lane RM, Cambier JC, Herold MJ, van Delft MF, Bedoui S, Lessene G, Ritchie ME, Huang DC, and Kile BT. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell 159: 1549–1562, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Winterbourn CC. The biological chemistry of hydrogen peroxide. Methods Enzymol 528: 3–25, 2013. [DOI] [PubMed] [Google Scholar]
- 191. Winterbourn CC. Biological production, detection, and fate of hydrogen peroxide. Antioxid Redox Signal 29: 541–551, 2018. [DOI] [PubMed] [Google Scholar]
- 192. Wong F, Stokes JM, Cervantes B, Penkov S, Friedrichs J, Renner LD, and Collins JJ. Cytoplasmic condensation induced by membrane damage is associated with antibiotic lethality. Nat Commun 12: 2321, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Xiong W, Jiao Y, Huang W, Ma M, Yu M, Cui Q, and Tan D. Regulation of the cell cycle via mitochondrial gene expression and energy metabolism in HeLa cells. Acta Biochim Biophys Sin (Shanghai) 44: 347–358, 2012. [DOI] [PubMed] [Google Scholar]
- 194. Yakes FM and Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci U S A 94: 514–519, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Zeida A, Trujillo M, Ferrer-Sueta G, Denicola A, Estrin DA, and Radi R. Catalysis of peroxide reduction by fast reacting protein thiols. Chem Rev 119: 10829–10855, 2019. [DOI] [PubMed] [Google Scholar]
- 196. Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, Brohi K, Itagaki K, and Hauser CJ. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature 464: 104–107, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, and Chen ZJ. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell 51: 226–235, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Zhou L, Aon MA, Almas T, Cortassa S, Winslow RL, and O'Rourke B. A reaction-diffusion model of ROS-induced ROS release in a mitochondrial network. PLoS Comput Biol 6: e1000657, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Zhou R, Yazdi AS, Menu P, and Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature 469: 221–225, 2011. [DOI] [PubMed] [Google Scholar]
- 200. Zhu H, Kwak HJ, Liu P, Bajrami B, Xu Y, Park SY, Nombela-Arrieta C, Mondal S, Kambara H, Yu H, Chai L, Silberstein LE, Cheng T, and Luo HR. Reactive oxygen species-producing myeloid cells act as a bone marrow niche for sterile inflammation-induced reactive granulopoiesis. J Immunol 198: 2854–2864, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]