Abstract
Protein ubiquitination is a dynamic and reversible post‐translational modification that controls diverse cellular processes in eukaryotes. Ubiquitin‐dependent internalization, recycling, and degradation are important mechanisms that regulate the activity and the abundance of plasma membrane (PM)‐localized proteins. In plants, although several ubiquitin ligases are implicated in these processes, no deubiquitinating enzymes (DUBs), have been identified that directly remove ubiquitin from membrane proteins and limit their vacuolar degradation. Here, we discover two DUB proteins, UBP12 and UBP13, that directly target the PM‐localized brassinosteroid (BR) receptor BR INSENSITIVE1 (BRI1) in Arabidopsis. BRI1 protein abundance is decreased in the ubp12i/ubp13 double mutant that displayed severe growth defects and reduced sensitivity to BRs. UBP13 directly interacts with and effectively removes K63‐linked polyubiquitin chains from BRI1, thereby negatively modulating its vacuolar targeting and degradation. Our study reveals that UBP12 and UBP13 play crucial roles in governing BRI1 abundance and BR signaling activity to regulate plant growth.
Keywords: endocytosis, membrane protein, phytohormone, receptor, ubiquitination
Subject Categories: Plant Biology, Post-translational Modifications & Proteolysis, Signal Transduction
The deubiquitinating enzymes UBP12 and UBP13 act as negative regulators of brassinosteroid (BR) receptor BRI1 degradation in the vacuole through the deubiquitination of BRI1, thus playing positive roles in BR‐responsive plant growth.

Introduction
Protein ubiquitination is a critical post‐translational modification that regulates multiple cellular processes and signaling pathways in eukaryotic cells (Hershko & Ciechanover, 1998; Swatek & Komander, 2016). Ubiquitin is a 76‐amino‐acid polypeptide, of which the carboxyl terminal glycine (G) can be covalently attached to the target protein through the sequential action of an enzymatic cascade, comprising a ubiquitin‐activating enzyme (E1), a ubiquitin‐conjugating enzyme (E2), and a ubiquitin ligase (E3). The versatility of ubiquitin‐dependent signaling is imposed by the diversity of polyubiquitin chains, that is, the substrate‐attached ubiquitin can be further ubiquitinated on its seven internal lysine (K) residues or on the N‐terminus encompassing complex topologies. Different ubiquitin chains direct distinct outcomes for the substrate. The prevalent examples are K48‐ and K63‐linked polyubiquitin chains, which mainly regulate proteasomal degradation and intracellular trafficking, respectively (Oh et al, 2018).
The antagonistic process to ubiquitination is accomplished by the action of deubiquitinating enzymes (DUBs) (Komander et al, 2009; Mevissen & Komander, 2017). Two of the Arabidopsis thaliana DUBs, UBP12 and UBP13, that belong to the family of ubiquitin‐binding or ubiquitin‐specific proteases (UBPs or USPs in mammals) (Hu et al, 2005; Wu et al, 2019) and share a high sequence homology (91%) act redundantly in multiple plant physiology aspects. They regulate immune response (Ewan et al, 2011), circadian rhythm and flowering time (Cui et al, 2013; Lee et al, 2019), root meristem maintenance (An et al, 2018), jasmonic acid (JA) signaling (Jeong et al, 2017), and leaf development and senescence (Park et al, 2019; Vanhaeren et al, 2020). UBP12 is reported to cleave the K48‐linked polyubiquitin chains (Ewan et al, 2011) and both UBP12 and UBP13 can directly deubiquitinate the transcription factors MYC2 and ORESARA1 (ORE1), thus preventing them from proteasomal degradation (Jeong et al, 2017; Park et al, 2019). The deubiquitinating activity of UBP12 and UBP13 against K63‐linked polyubiquitin chains remains elusive. Previously, the ubp12/ubp13 double‐null mutant has been reported to be seedling lethal and a weak double mutant displayed a dwarf phenotype (Cui et al, 2013), but the detailed mechanism and deubiquitination targets causing these phenotypes are largely unknown.
Brassinosteroids (BRs) are steroidal phytohormones that are essential for growth and development (Clouse, 2011; Nolan et al, 2020). BRs are perceived by the plasma membrane (PM)‐localized receptor BR INSENSITIVE1 (BRI1) in the apoplast and the activation of BRI1 triggers its dimerization with the co‐receptor BRI1‐ASSOCIATED KINASE1 (BAK1). The receptor complex is further activated through auto‐ and trans‐phosphorylation that initiates a phosphorylation/dephosphorylation cascade, which ultimately leads to the dephosphorylation and accumulation of BRASSINAZOLE RESISTANT1 (BZR1) and BR‐INSENSITIVE‐EMS‐SUPPRESSOR1 (BES1)/BZR2 transcription factors in the nucleus to regulate gene expression (Wang et al, 2002; Shimada et al, 2015). After ligand binding, BRI1 is internalized for signaling attenuation by reducing the number of receptors present on the cell surface; therefore, BRI1 and BRs provide the model for receptor–ligand pairs to understand the interplay between receptor‐mediated endocytosis and signaling in plants (Irani et al, 2012). BRI1 dynamically undergoes internalization from the PM to the trans‐Golgi network/early endosomes (TGN/EEs), from which BRI1 can then be recycled back to the PM or targeted to the vacuole for degradation via the late endosome/multivesicular bodies (LE/MVBs) (Russinova et al, 2004; Geldner et al, 2007; Irani et al, 2012). BRI1 is modified by K63‐linked polyubiquitin chains that enhance its internalization and serve as a sorting signal for BRI1 vacuolar degradation, thereby BRI1 ubiquitination negatively regulates BR signaling activity (Martins et al, 2015). BR perception promotes BRI1 phosphorylation of and association with the plant U‐box ubiquitin ligase PUB13, which further ubiquitinates BRI1 leading to the internalization and degradation of the receptor with down‐regulation of BR response as a consequence (Zhou et al, 2018). However, until now, DUB proteins that can mediate BRI1 deubiquitination have not been described.
Here, we demonstrate that UBP12 and UBP13 function as key negative regulators of BRI1 degradation. UBP13 directly interacts with BRI1 and removes K63‐linked polyubiquitin chains. The polyubiquitinated form of BRI1 is increased in the knockdown ubp12i/ubp13 double mutant, whereas the total BRI1 protein levels decreased. Ubiquitination‐deficient BRI1 rescued the dwarf phenotype of the ubp12i/ubp13 mutant. We further show that UBP12 and UBP13 limit the BRI1 degradation in the vacuole. These findings provide new insight into the regulatory mechanism of BRI1 degradation and plant growth via deubiquitination.
Results and Discussion
ubp12i/ubp13 double mutant is severely dwarfed and shows reduced sensitivity to BRs
Previously, the ubp12/ubp13 double mutant of null alleles had been shown to be not viable, whereas growth of a weak double mutant was stunted (Cui et al, 2013). To gain more insights of UBP12 and UBP13 in plant development, we generated a dexamethasone (DEX)‐inducible RNAi UBP12 silencing transgenic plants in the T‐DNA insertional knockout ubp13‐1 mutant background (ubp12i/ubp13). Two independent lines (#1‐7 and #12‐8) were selected for further analysis (Appendix Fig S1). In these two lines, the expression level of UBP12 was reduced to 30–40% of that in mock‐treated plants upon DEX induction (Appendix Fig S1B). Whereas the ubp12i/ubp13 plants resembled the wild‐type under mock conditions, they were severely impaired in growth after DEX induction and exhibited a dwarf phenotype, including short petioles, round and inwardly curved true leaves, and small compact rosettes (Appendix Fig S1A), similar to that of mutants defective in either BR biosynthesis or signaling. To examine the involvement of UBP12 and UBP13 in BR responses, we analyzed the root phenotypes of ubp12i/ubp13 plants along with the BR receptor mutant (the bri1‐null mutant). Compared to the wild‐type plants, both bri1‐null and ubp12i/ubp13 plants displayed severe defects in root development, including a decreased primary root length and a drastically reduced lateral root density, the latter was even more prominent in the ubp12i/ubp13 double mutant than that in the bri1‐null mutant (Fig 1A and Appendix Fig S1C). We then examined whether the exogenously applied brassinolide (BL), the most active BR, restores the dwarf phenotype of ubp12i/ubp13. Upon BL application, wild‐type plants showed BL‐induced root curling and bending petioles, whereas exogenous BL had little effect on the ubp12i/ubp13 dwarf phenotype similar to that of the bri1‐null mutant (Fig 1A). Next, we measured the hypocotyl length of light‐grown seedlings in the presence of increasing BL concentrations and found that ubp12i/ubp13 plants were less sensitive to BL at 10–1,000 nM (Fig 1B and C). To investigate whether the observed phenotypes in ubp12i/ubp13 plants resulted from a perturbed BR signaling, we monitored the BES1 dephosphorylation status, which is a well‐established BR signaling activation readout (Yin et al, 2002; Yu et al, 2011). Consistent with the observed reduction in BR sensitivity upon exogenous BL treatment in the light, the accumulation of dephosphorylated BES1 was lower in the ubp12i/ubp13 plants than that in the wild‐type (Fig 1D and E). We further analyzed the response to exogenous BL of dark‐grown hypocotyls of ubp12i/ubp13 plants and found that both lines exhibited a reduced sensitivity to BL, although to a lesser extent than the bri1‐null mutant (Appendix Fig S2B–E). To exclude that the possible variance in DEX‐induced RNAi might lead to individual variation in BR responses, we also generated transgenic plants constitutively expressing UBP13 under the control of a CaMV 35S promoter (35S:UBP13) and studied their BR responses. As expected, 35S:UBP13 plants were more sensitive to BL, supporting the UBP12 and UBP13 roles in BR signaling (Appendix Fig S2A–C). Altogether, these results suggest that UBP12 and UBP13 positively regulate BR signaling in both seedlings and mature plants. The differences in BR sensitivity of ubp12i/ubp13 mutant plants, where it was only partially reduced at a young seedling stage contrasting the pronounced insensitive phenotype of the ubp12i/ubp13 mature plants, indicate that such regulation may depend on the growth stage. Local BR signaling activities are differentially regulated in each developmental zone in Arabidopsis roots (Chaiwanon & Wang, 2015; Vukašinović et al, 2021), it is possible that UBP12 and UBP13 contribute to regulate such developmental stage‐specific BR responses.
Figure 1. Loss of UBP12 and UBP13 caused growth defects and decreased BR sensitivity in Arabidopsis .
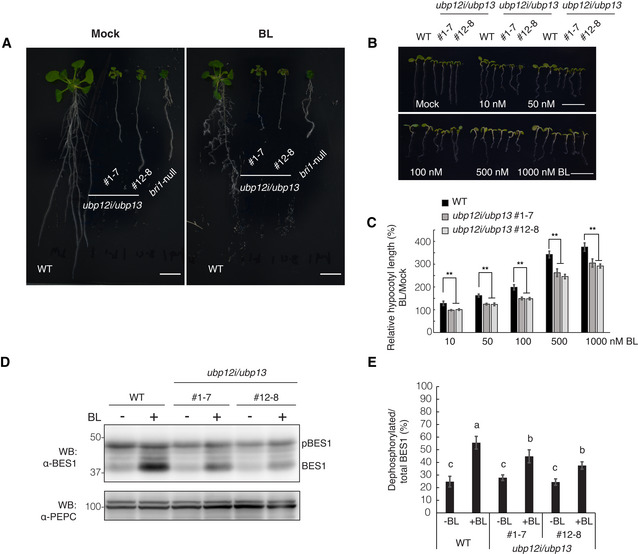
- Growth phenotypes of the wild‐type (WT, Col‐0), bri1‐null mutant, and two independent ubp12i/ubp13 double mutant lines grown on 1/2MS medium supplemented with 10 μM DEX (DEX medium) for 10 days and transferred to DEX medium containing 1 μM BL or mock solution (0.1% [v/v] EtOH) for additionally 8 days. Scale bars: 10 mm.
- Phenotype of the WT and ubp12i/ubp13 double mutant grown on DEX medium in the presence of increasing concentrations of BL in the light for 6 days. Scale bars: 10 mm.
- Hypocotyl length relative to the mock control of seedlings (B). Experiments were done in triplicate (n > 15 seedlings for each line).
- Western blot (WB) analysis of BES1 dephosphorylation after BL treatment with the α‐BES1 antibody. Total proteins were isolated from 6‐day‐old seedlings grown on DEX medium in the light, either in the presence of 100 nM BL or mock. PEPC was used as a loading control.
- Percentage of dephosphorylated BES1 relative to the total BES1 from (D) (n = 6 biological replicates).
Data information: Data are presented as means ± SD (C and E). **P < 0.01 (one‐way ANOVA and post hoc Tukey's test) compared with WT under each condition (C). Different letters indicate statistically significant differences at P < 0.01 (one‐way ANOVA and post‐ hoc Tukey's test) (E).
UBP12 and UBP13 modulate the BRI1 protein abundance in Arabidopsis
The dwarf phenotype and the BR hyposensitivity of the ubp12i/ubp13 plants hinted at the possibility that UBP12 and UBP13 target BR signaling components for deubiquitination. As BRI1 is polyubiquitinated, in turn regulating its endocytosis and protein abundance (Martins et al, 2015; Zhou et al, 2018), we hypothesized that BRI1 is the deubiquitination target of UBP12 and UBP13. To examine this possibility, we assessed the BRI1 protein amounts in ubp12i/ubp13 plants. The BRI1 protein level was dramatically lower in ubp12i/ubp13 plants under DEX treatment than that under mock treatment or that of wild‐type plants (Fig 2A). The expression level of BRI1 was not reduced in the ubp12i/ubp13 plants, suggesting that UBP12 and UBP13 regulate the protein levels of BRI1 post‐translationally (Appendix Fig S3A). We then assessed the BRI1 protein stability in ubp12i/ubp13 plants in the presence of the protein synthesis inhibitor cycloheximide (CHX). In wild‐type plants, the half‐life of BRI1 was around 5 h, as previously reported (Geldner et al, 2007), whereas ubp12i/ubp13 decreased the BRI1 half‐life to less than 5 h (Appendix Fig S4). To further understand how UBP12 and UBP13 affect the BRI1 protein, we analyzed the effect of the vacuolar ATPase inhibitor concanamycin A (ConcA), which prevents the degradation of proteins targeted to the vacuole (Takano et al, 2005). After exposure to 1 µM ConcA for 4 h, the reduced BRI1 protein level was partly restored in the ubp12i/ubp13 plants (Fig 2B). Taken together, these results imply that UBP12 and UBP13 are positive regulators of the BRI1 protein stability, probably limiting its vacuolar degradation.
Figure 2. UBP12 and UBP13 negatively regulated BRI1 protein degradation.
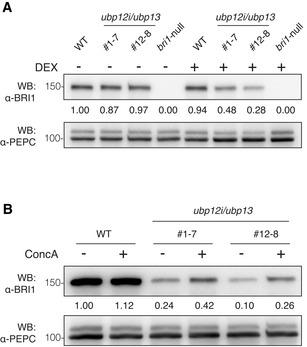
- Western blot analysis of BRI1 protein amount in ubp12i/ubp13 plants with the α‐BRI1 antibody. Total proteins were isolated from 18‐day‐old plants grown on 1/2MS medium supplemented with 10 μM DEX (+) or not (−).
- Western blot analysis of BRI1 protein level in concanamycin A (ConcA)‐treated plants. Eighteen‐day‐old plants were grown on DEX medium and exposed to 1 μM ConcA for 4 h before total protein isolation.
Data information: PEPC was used as a loading control. The values shown above each lane indicate the abundance of the BRI1 protein relative to that of WT without DEX treatment (A) or to that of WT without ConcA treatment (B).
UBP13 interacts with and deubiquitinates BRI1
To examine whether UBP12 and UBP13 directly regulate BRI1 amounts, we tested the interaction between UBP13 and BRI1. The split‐ubiquitin yeast two‐hybrid assays in which full‐length BRI1 and UBP13 were fused to the C‐terminal (BRI1‐Cub) and the N‐terminal half of ubiquitin (UBP13‐Nub), respectively, revealed that UBP13 could bind to BRI1 in yeast (Fig 3A). Next, we tested whether UBP13 interacted with BRI1 in plant cells by co‐immunoprecipitation and western blot analysis. The FLAG‐tagged UBP13 (FLAG‐UBP13) and Myc‐tagged BRI1 (BRI1‐Myc) were transiently co‐expressed in Nicotiana benthamiana leaves and FLAG‐UBP13 was co‐immunoprecipitated with BRI1‐Myc (Fig 3B), indicating a physical interaction between UBP13 and BRI1 in planta.
Figure 3. UBP13 interacts with and deubiquitinates BRI1.
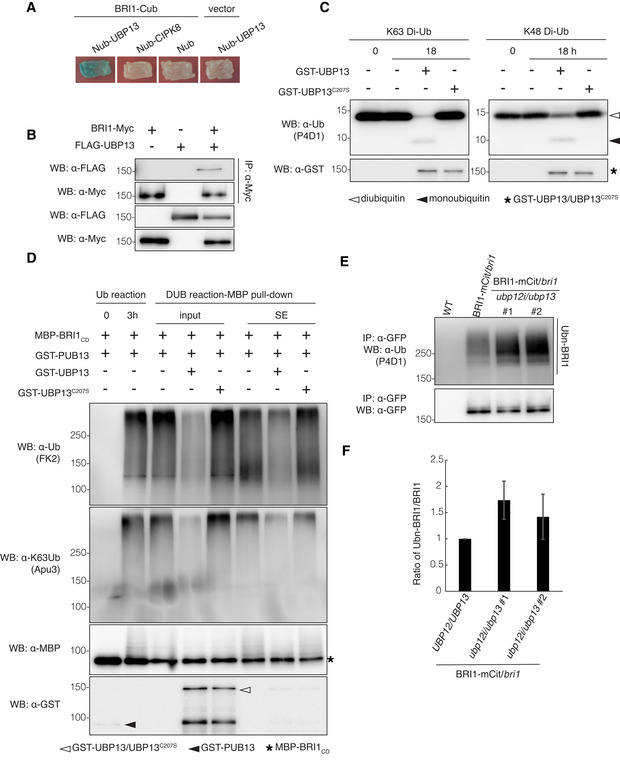
- Split‐ubiquitin yeast two‐hybrid assay revealing the UBP13 and BRI1 interactions. The indicated constructs were co‐transformed into yeast cells. BRI1‐Cub with Nub, Nub‐CIPK8, and Nub‐UBP13 with an empty vector were used as negative controls. Transformants were streaked onto solidified medium supplemented with X‐gal. Blue patch indicates positive interaction. Cub, the C‐terminal half of ubiquitin; Nub, the N‐terminal half of ubiquitin.
- UBP13 interaction with BRI1 in plants. BRI1‐Myc and FLAG‐UBP13 were co‐expressed in Nicotiana benthamiana leaves and co‐immunoprecipitation was done on solubilized microsomal proteins with α‐Myc antibody beads. The association of BRI1‐UBP13 was detected by western blot with an α‐FLAG antibody after α‐Myc IP (Top).
- Deubiquitinating activity of UBP13 against K48‐ and K63‐linked ubiquitination. Diubiquitins linked through K48 or K63 (K48 Di‐Ub or K63 Di‐Ub) were incubated alone or with GST‐UBP13 or GST‐UBP13C207S for 18 h. Deubiquitination of the diubiquitins was analyzed by western blot with the α‐ubiquitin (α‐Ub, P4D1) antibody. The presence of GST‐UBP13/UBP13C207S recombinant proteins was confirmed by the α‐GST antibody.
- UBP13 deubiquitination of K63‐ubiquitinated BRI1 in vitro. Polyubiquitinated MBP‐BRI1CD was generated by incubation with GST‐PUB13 (Ub reaction) and then with GST‐UBP13 or GST‐UBP13C207S (DUB reaction‐MBP pull‐down, input). After in vitro deubiquitination, MBP‐BRI1CD was purified by an MBP pull‐down to analyze the BRI1CD ubiquitination (DUB reaction‐MBP pull‐down, SE). The ubiquitinated proteins were detected by western blot with an α‐ubiquitin antibody (FK2). Ubiquitin chain specificity was detected with an α‐K63Ub antibody (Apu3). The presence of recombinant proteins was confirmed by the α‐GST and α‐MBP antibodies.
- Western blot analysis of BRI1 ubiquitination on plants expressing pBRI1:BRI1‐mCitrine complementing the bri1 mutant in either the UBP12/UBP13 (BRI1‐mCit/bri1) or ubp12i/ubp13 (BRI1‐mCit/bri1/ubp12i/ubp13) background. BRI1‐mCitrine proteins were isolated from 18‐day‐old plants grown on DEX medium and then immunoprecipitated with α‐GFP antibody beads from solubilized microsomal proteins. Ubiquitinated BRI1 and basal BRI1 proteins were detected by the α‐ubiquitin (P4D1) and α‐GFP antibodies, respectively.
- Quantification of BRI1 ubiquitination profiles from (E) (n = 3 biological replicates). The ubiquitinated BRI1 fraction was normalized to the total immunoprecipitated BRI1 detected by the α‐GFP antibody.
Data information: Data in (F) are presented as means ± SD.
UBP12 had previously been shown to possess deubiquitinating enzymatic activity toward the K48‐linked polyubiquitination in vitro (Ewan et al, 2011), both UBP12 and UBP13 modulated proteasomal degradation in plants, implying that they cleave K48‐linked polyubiquitin chains in vivo (Jeong et al, 2017; Park et al, 2019). On the contrary, BRI1 is modified by the K63‐linked polyubiquitin chains in vivo (Martins et al, 2015). Therefore, we tested the deubiquitinating activity of UBP13 against the K63‐linked polyubiquitination and its ability to deubiquitinate BRI1. An in vitro deubiquitination assay was carried out with K48‐ or K63‐linked diubiquitins (K48 Di‐Ub and K63 Di‐Ub) as substrates to examine the chain‐specific deubiquitinating activity of UBP13. We prepared the recombinant glutathione S‐transferase‐UBP13 fusion protein (GST‐UBP13) and its catalytically inactive mutant (GST‐UBP13C207S), in which the conserved cysteine of the enzymatically active site was substituted by serine (Ewan et al, 2011; Cui et al, 2013). GST‐UBP13, but not GST‐UBP13C207S, cleaved both K48‐ or K63‐linked diubiquitins, indicating that UBP13 possesses deubiquitinating activities against not only K48‐ but also K63‐linked polyubiquitin chains (Fig 3C). To examine whether UBP13 deubiquitinated K63‐ubiquitinated BRI1 in vitro, we used ubiquitinated BRI1 produced by PUB13 as DUB substrate. PUB13 had been shown to polyubiquitinate BRI1 both in vitro and in vivo (Zhou et al, 2018). Although PUB13 might plausibly catalyze K63‐linked ubiquitin signals on BRI1, the configuration of such a ubiquitination has not been demonstrated experimentally. By using lysine (K)‐to‐arginine (R) ubiquitin variants, we first validated that PUB13 had both K63 and K48 ubiquitination activities (Appendix Fig S5A and B). To confirm that PUB13 catalyzed K63 polyubiquitination on BRI1, we incubated the maltose‐binding protein (MBP)‐tagged cytosolic domain of BRI1 (MBP‐BRI1CD) with GST‐fused PUB13 (GST‐PUB13) and native or K63R/K48R ubiquitins. The reaction co‐incubated with native ubiquitin analyzed with a K63‐linked polyubiquitin chain‐specific antibody revealed that GST‐PUB13 mediated K63‐linked polyubiquitination toward MBP‐BRI1CD. Moreover, BRI1 ubiquitination was clearly reduced, although not diminished with the K63R ubiquitin variant, suggesting that GST‐PUB13 catalyzes at least K63‐linked polyubiquitin chains on MBP‐BRI1CD in vitro (Appendix Fig S5C and D). The ubiquitination reaction containing a mixture of polyubiquitinated MBP‐BRI1CD and GST‐PUB13 was terminated by adding an E1 inhibitor, and in vitro deubiquitination assays were done by co‐incubating with GST‐UBP13 or GST‐UBP13C207S. MBP‐BRI1CD proteins pulled down by amylose resin were subjected to western blot analysis with the chain‐type specific ubiquitin antibody. The MBP‐BRI1CD ubiquitination level was clearly reduced by co‐incubation with GST‐UBP13, but not with GST‐UBP13C207S, indicating that UBP13 possesses the deubiquitinating activity toward K63‐linked BRI1 polyubiquitination in vitro (Fig 3D).
As a further confirmation of this observation in vivo, Arabidopsis transgenic plants, expressing the BRI1‐mCitrine fusion driven by the endogenous BRI1 promoter in the bri1‐null mutant, were crossed into the ubp12i/ubp13 plants (BRI1‐mCit/bri1/ubp12i/ubp13) and the suppression of the UBP12 expression was successfully induced by DEX (Appendix Fig S3A). Using the microsomal protein fraction from crude extracts, BRI1‐mCitrine proteins were immunoprecipitated with anti‐GFP antibodies and subjected to western blot analysis. A high molecular mass smear above 170 kDa was detected in the immunoprecipitates from plants expressing BRI1‐mCitrine, corresponding to the polyubiquitinated BRI1‐mCitrine proteins (Fig 3E). Notably, the level of polyubiquitinated BRI1‐mCitrine proteins was markedly higher in BRI1‐mCit/bri1/ubp12i/ubp13 than that in BRI1‐mCit/bri1 plants (Fig 3E and F). Additionally, when polyubiquitinated BRI1‐mCitrine proteins were incubated with GST‐UBP13 or GST‐UBP13C207S, the resulted ubiquitination levels were reduced to nearly 50% when incubated with native UBP13 compared with the UBP13C207S inactive control (Appendix Fig S6). Taken together, the data indicated that UBP13 associates with BRI1 and deubiquitinates BRI1 removing K63‐linked polyubiquitin chains.
Expression of ubiquitination‐deficient BRI1 restored BRI1 protein levels and partly the growth defects of the ubp12i/ubp13 mutant
The BRI1‐mCit/bri1/ubp12i/ubp13 transgenic plants were dwarfed compared with BRI1‐mCit/bri1 and wild‐type plants, although did not completely phenocopy ubp12i/ubp13 (Fig 4A). They displayed elongated petioles, larger leaf areas, longer primary roots, and an increased lateral root density compared with ubp12i/ubp13 plants (Fig 4A, Appendix Figs S3B and S7). We evaluated the BRI1‐mCitrine protein amounts in these transgenic plants, which were notably lower in BRI1‐mCit/bri1/ubp12i/ubp13 plants than those in BRI1‐mCit/bri1 plants (Fig 4B). To further dissect the relationship among UBP12, UBP13, and BRI1, we analyzed the phenotype of BRI125KR‐mCit/bri1/ubp12i/ubp13 plants, namely ubp12i/ubp13 Arabidopsis plants, expressing a functional but ubiquitination‐deficient BRI1 (BRI125KR). As reported previously, the BRI125KR‐mCitrine mutant protein is more stable at the cell surface due to compromised BRI1 ubiquitination (Martins et al, 2015). As expected, the ubp12i/ubp13 dwarf phenotype was dramatically restored by the BRI125KR‐mCitrine transgene expression (Fig 4A, Appendix Figs S3B and S7). Accordingly, the protein amounts of BRI125KR‐mCitrine remained similar in BRI125KR‐mCit/bri1/ubp12i/ubp13 and BRI125KR‐mCit/bri1 plants (Fig 4B). BR responses were also assessed in these lines by measuring the length of light‐grown hypocotyls. Whereas the hypocotyl elongation was reduced in ubp12i/ubp13 lines in wild‐type and BRI1‐mCit/bri1 backgrounds, the BL‐induced hypocotyl elongation was unaffected by ubp12i/ubp13 in BRI125KR‐mCit/bri1 plants (Fig 4C, Appendix Fig S7A), correlating with the BL‐treated phenotype. Indeed, exogenous BL failed to restore the dwarf phenotype of BRI‐mCit/bri1/ubp12i/ubp13 plants and they showed straight or slightly curled petioles. In contrast, the BRI125KR‐mCit/bri1/ubp12i/ubp13 plants displayed highly curled petioles in response to BL (Appendix Fig S7B). These results suggest that the enhanced ubiquitin‐dependent degradation of BRI1 protein in the ubp12i/ubp13 mutant partly contributes to its dwarf phenotype. The data support a critical role for UBP12 and UBP13 as DUBs by counteracting BRI1 polyubiquitination for its protein accumulation to regulate BR‐responsive plant growth. The growth phenotype of the ubp12i/ubp13 mutant was significantly, however, not fully restored by the expression of BRI125KR proteins, suggesting that, besides BRI1, UBP12 and UBP13 might deubiquitinate other targets to regulate plant growth (Fig 4A, Appendix Figs S3B and S7). Two BR‐responsive transcription factors BZR1 and BES1 are also under the regulation of ubiquitin‐mediated 26S proteasomal degradation or selective autophagy (Nolan et al, 2020). An early proteomics analysis had revealed that UBP12 and UBP13 were BZR1‐interacting proteins and proved the interaction between UBP12 and BZR1 in the nucleus of N. benthamiana (Wang et al, 2013). Whether UBP12 and UB13 deubiquitinate BZR1 to regulate the BR signaling awaits further research and validation.
Figure 4. The ubiquitination‐deficient BRI1 restores the growth defects of ubp12i/ubp13 .
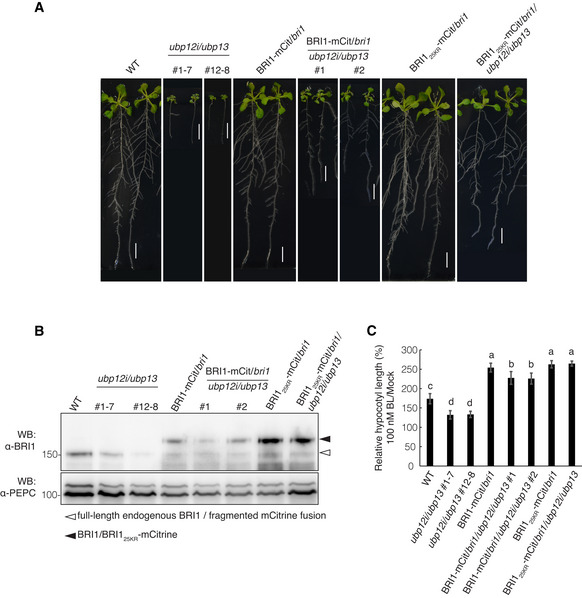
- Growth phenotype comparison of BRI1‐mCit/bri1/ubp12i/ubp13 and BRI125KR‐mCit/bri1/ubp12i/ubp13. The indicated transgenic lines were grown on DEX medium for 18 days. Scale bars: 10 mm.
- Western blot analysis monitoring the BRI1 protein accumulation in BRI125KR‐mCit/bri1/ubp12i/ubp13 plants with the α‐BRI1 antibody. Total proteins were isolated from the roots of the lines indicated (A). PEPC was used as a loading control.
- Hypocotyl length relative to the mock control. Seedlings of the indicated lines were grown on DEX medium containing 100 nM BL or mock in the light for 6 days (Appendix Fig S7A). Experiments were done in triplicate (n > 15 seedlings for each line).
Data information: Data in (C) are presented as means ± SD. Different letters indicate statistically significant differences at P < 0.05 (one‐way ANOVA and post hoc Tukey's test).
Loss of UBP12 and UBP13 accelerated the vacuolar targeting of BRI1
Brassinosteroid signaling primarily depends on the PM pool of BRI1 (Irani et al, 2012) and ubiquitination can enhance the internalization of BRI1, targeting it to the vacuole for degradation, thus playing a negative role in BR signaling (Martins et al, 2015; Zhou et al, 2018). The ubiquitination of BRI1 is considered to be BR dependent because BL promotes the association between BRI1 and PUB13 and BRI1 kinase activity is required for the direct phosphorylation of PUB13 (Zhou et al, 2018). Given that BRI1 ubiquitination had increased, while the protein levels were drastically reduced in the ubp12i/ubp13 plants and the ubp12i/ubp13 plants were hyposensitive to BR, we hypothesize that loss of UBP12 and UBP13 would promote the vacuolar degradation of BRI1. To test this assumption, we evaluated the vacuolar accumulation of BRI1‐mCitrine or BRI125KR‐mCitrine in the ubp12i/ubp13 background using quantitative microscopy. Seedlings were transferred to dark conditions in the presence of CHX for inhibition of de novo protein synthesis before imaging. By measuring the fluorescence intensity of BRI1‐mCitrine, we observed that in BRI1‐mCit/bri1/ubp12i/ubp13 transgenic plants the relative fluorescence intensity ratio of the PM‐to‐cytoplasm‐localized BRI1 was lower than that of the BRI1‐mCit/bri1 control (Fig 5A and B). On the contrary, the BRI125KR‐mCitrine proteins were mostly located at the PM and the BRI125KR‐mCit/bri1 and BRI125KR‐mCit/bri1/ubp12i/ubp13 plants did not significantly differ (Fig 5C and D).
Figure 5. The vacuolar targeting of BRI1 is accelerated in ubp12i/ubp13 .
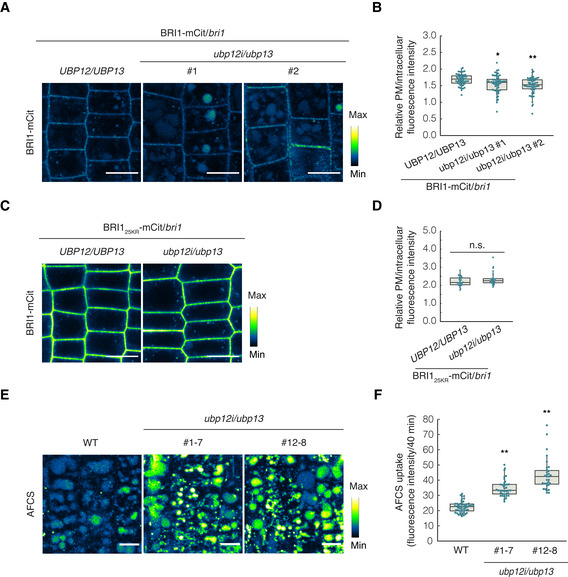
-
A–DAnalysis of the BRI1 vacuolar targeting on Arabidopsis seedlings expressing BRI1 (A) or BRI125KR (C) tagged with mCitrine (mCit) in either bri1/UBP12/UBP13 or bri1/ubp12i/ubp13 background. Six‐day‐old seedlings were grown on DEX medium, and kept in the dark for 2 h in the presence of 50 μM CHX before imaging. Confocal images of epidermal cells from root meristem are shown. Scale bars: 10 μm. Measurements of the relative PM/intracellular BRI1‐mCit fluorescence intensity. For each line, at least 55 and 25 cells from over 25 and 15 seedlings were measured in (B) and (D), respectively.
-
EPulse‐chase AFCS uptake experiments in WT and ubp12i/13 plants. Six‐day‐old seedlings were grown on DEX medium and pulsed with 40 µM AFCS for 40 min followed by a 30‐min chase before imaging. Scale bars: 10 μm.
-
FQuantification of AFCS fluorescence intensity in (E) (n > 30 ROIs from 7 to 9 seedlings for each line).
Data information: Measurements shown in box plots are the first and third quartiles and are split by the medians, whiskers extending 1.5‐fold interquartile range beyond the box (B, D, and F). Data points are plotted as dots. n.s., not significant, *P < 0.05, **P < 0.01 (one‐way ANOVA and post hoc Tukey's test).
We next used the bioactive fluorescent BR, Alexa Fluor 647‐catasterone (AFCS), which visualizes the endocytosis of BRI1‐BR complexes (Irani et al, 2012) to evaluate the impact of the loss of UBP12 and UBP13 on endogenous BRI1. We analyzed the uptake of AFCS in wild‐type and ubp12i/ubp13 plants by quantifying the vacuolar accumulation of the dye after a 30‐min chase. The signal intensity was strikingly higher in ubp12i/13 plants, suggesting that the loss of UBP12 and UBP13 facilitated AFCS uptake and BRI1 endocytosis from the PM (Fig 5E and F). Taken together, our data indicate that UBP12 and UBP13 negatively regulate the internalization and vacuolar targeting of BRI1 through direct removal of its ubiquitin modifications.
Plant DUBs have been demonstrated to facilitate the down‐regulation of receptors via membrane trafficking. In Arabidopsis, the associated molecule with the SH3 domain of STAM3 (AMSH3) protein is the DUB that interacts with components of the endosomal sorting complex required for transport (ESCRT) and is essential for degradation of ubiquitinated membrane proteins (Isono et al, 2010; Katsiarimpa et al, 2011, 2014). However, plant DUBs that can negatively regulate membrane protein vacuolar degradation via counteracting its ubiquitination have not been described yet. UBP12 and UBP13 were reported to maintain the protein stability of another receptor‐like kinase root meristem growth factor 1 receptor (RGFR1) during root development, however, through the 26S proteasomal pathway (An et al, 2018). Here, we showed that UBP12 and UBP13 directly associate with BRI1 and function to negatively modulate its vacuolar degradation via the K63‐linked ubiquitin signal, thereby they control the BRI1 protein levels to fine‐tune plant sensitivity to BRs that is required for optimal growth. Our findings provide evidence that plant DUBs act in opposition during intracellular trafficking. In mammals, deubiquitination has been demonstrated to play a dual role in the regulation of intracellular trafficking (Clague & Urbé, 2006; Millard & Wood, 2006). DUBs facilitate endocytosis and vacuolar degradation of PM receptors by recruiting the components of clathrin‐mediated endocytosis (Jaworski et al, 2014). Nevertheless, DUBs also prevent internalization of the receptors from the PM or promote endosomal recycling via direct ubiquitin removal, hence limiting vacuolar degradation (McCann et al, 2016). As the acidic environment of the early endosomes in Arabidopsis might preclude ligand dissociation, the ligand‐bound BRI1 recycling is questioned (Luo et al, 2015). Further studies are needed to determine whether deubiquitination is required to retain BRI1 at the PM and to recycle ligand‐free BRI1 back to the PM.
Materials and Methods
Plant material and growth conditions
Arabidopsis thaliana (L.) Heynh., accession Columbia‐0 (Col‐0), was used as the wild‐type in all experiments. The bri1‐null T‐DNA knockout mutant (GABI_134E10) and transgenic lines pBRI1:BRI1‐mCitrine/bri1 (BRI1‐mCit/bri1) and pBRI1:BRI125KR‐mCitrine/bri1 (BRI125KR‐mCit/bri1) have been described previously (Jaillais et al, 2011; Belkhadir et al, 2012; Martins et al, 2015; Zhou et al, 2018). To generate the dexamethasone (DEX)‐inducible ubp12i/ubp13 double‐knockdown mutant, the pOpOff2 RNAi system was used (Wielopolska et al, 2005). A UBP12‐coding sequence fragment (400 bp; positions 650–1049) was cloned into the pENTR/D‐TOPO vector and transferred to the pOpOff2 destination vector with the Gateway System (Invitrogen). The expression construct was then introduced into the ubp13‐1 (SALK_128312) T‐DNA knockout mutant by Agrobacterium tumefaciens–mediated transformation (Clough & Bent, 1998). The transgenic mutant lines BRI1‐mCit/bri1/ubp12i/ubp13 and BRI125KR‐mCit/bri1/ubp12i/ubp13 were generated by crossing BRI1‐mCit/bri1 or BRI125KR‐mCit/bri1 into the ubp12i/ubp13 double mutant. To generate the 35S:UBP13 lines, the full‐length coding sequence of UBP13 was cloned into the pEarleygate202 (Earley et al, 2006) destination vector to produce the N‐terminally FLAG‐fused construct under the control of a Cauliflower mosaic virus (CaMV)35S promoter and then introduced into wild‐type Arabidopsis plants as described above. Primers used to generate the abovementioned constructs are listed in Appendix Table S1.
Arabidopsis seeds were surface sterilized and sown on plates with half‐strength Murashige and Skoog medium (1/2MS, 1% [w/v] sucrose, 0.4% [w/v] gellan gum, pH 5.7). For the expression induction of the UBP12 RNAi constructs, DEX was supplemented in the medium at a final concentration of 10 μM. For the BL response assays, BL concentrates were added to the medium with a final EtOH concentration of 0.1% (v/v). After stratification at 4°C in the dark for 2–4 days, the plants were grown under a 16‐h light/8‐h dark photoperiod at 22°C, unless otherwise indicated.
Chemical treatments
DEX (Wako; 10 mM stock in EtOH), BL (Cayman; 5 mM stock in EtOH), ConcA (Sigma‐Aldrich; 1 mM stock in dimethyl sulfoxide [DMSO]), and CHX (Sigma‐Aldrich; 50 mM or 200 mM stock in EtOH) were used at the concentrations indicated in the figure legends.
Hypocotyl and root growth assays
Imbibed seeds were stratified at 4°C for 4 days to synchronize seed germination between different genotypes. In the hypocotyl growth assay for the BL response in the dark, after stratification, seeds were exposed to light for over 6 h and then kept in the dark for 6 days, whereas for the BL response in the light, the light intensity was approximately 75 μmol/m2/s. Seedlings were imaged and hypocotyl/primary root lengths were measured with Fiji/ImageJ (National Institutes of Health).
Western blot analysis and immunoprecipitation
For BES1 dephosphorylation and BRI1 protein accumulation analyses, plant samples were homogenized in liquid nitrogen and total proteins were extracted with denaturing buffer (20 mM Tris‐HCl [pH 7.5], 150 mM NaCl, 1% [w/v] SDS, 100 mM DTT, and EDTA‐free protease inhibitor mixture complete [Roche]). For immunodetection, α‐BES1 (1/5,000) (Yu et al, 2011) and α‐BRI1 (1/5,000; Agrisera) antibodies were used as primary antibodies followed by the incubation with a secondary α‐rabbit (1/5,000; CST) antibody conjugated to horseradish peroxidase (HRP). Equal loading was verified with an α‐phosphoenolpyruvate carboxylase (PEPC) (1/1,000; Agrisera) antibody. The band intensity was quantified with Fiji/imageJ.
For in vivo BRI1 deubiquitination analysis, solubilized microsomal proteins were immunoprecipated. Samples (2 g) were ground in liquid nitrogen and resuspended in 4 ml of ice‐cold sucrose buffer (100 mM Tris‐HCl [pH 7.5], 810 mM sucrose, 5% [v/v] glycerol, 10 mM EDTA [pH 8.0], 10 mM EGTA [pH 8.0], 5 mM KCl, 10 mM N‐ethylmaleimide [NEM, Wako], 1 mM 1,10‐phenanthroline [1,10‐PT; Wako], 1 μM MLN7243 [Active Biochem], 10 μM MG132, EDTA‐free protease inhibitor mixture complete [Roche], and PhosSTOP phosphatase inhibitor cocktail [Roche]). Samples were clarified by centrifugation twice (5,000 g for 10 min and then for 5 min) and filtered through two layers of Miracloth (Millipore). Microsomes were pelleted from the homogenate by ultracentrifugation at 100,000 g for 90 min. Pellets were resuspended in 1 ml of immunoprecipitation buffer (25 mM Tris‐HCl [pH 7.5], 150 mM NaCl, 0.1% [w/v] SDS, 10 mM NEM, 1 mM 1,10‐PT, 1 μM MLN7243, 10 μM MG132, EDTA‐free protease inhibitor mixture complete, and PhosSTOP phosphatase inhibitor cocktail [Roche]), left on a rotating wheel for 30 min at 4°C. Non‐resuspended material was removed by centrifugation (20,000 g for 10 min). Supernatants enriched with microsomal proteins were incubated with α‐GFP mAb‐magnetic beads (MBL, D153‐11) for 1 h at 4°C with gentle rocking. For immunodetection, α‐Ub (P4D1, 1/2500; Santa Cruz) and α‐GFP (1/2,000; MBL) antibodies were used. For quantification of BRI1 ubiquitination, the ratio of normalized immunoprecipitation signal intensity obtained with α‐GFP and α‐Ub antibodies was determined with Fiji/ImageJ.
Transient expression in N. benthamiana
For the co‐immunoprecipitation assay, the full‐length coding sequence of BRI1 was cloned into the pGWB17 (Nakagawa et al, 2007) destination vector as a C‐terminally Myc‐fused construct with the primers listed in Appendix Table S1. pGWB17‐BRI1 and pEarleygate202‐UBP13 were introduced into the A. tumefaciens strain GV3101 (pMP90) by electroporation. Overnight cultures of Agrobacterium containing the expression constructs were pelleted and resuspended in the infiltration buffer (10 mM MES, 10 mM MgCl2, and 450 μM acetosyringone, pH 5.6). The suspension mixtures were infiltrated into 4‐week‐old N. benthamiana leaves with a syringe without needle. The p19 suppressor‐carrying Agrobacterium was co‐infiltrated with all samples (Takeda et al, 2002).
Co‐immunoprecipitation assay
Co‐immunoprecipitation was carried out on solubilized microsomal proteins derived from 1.5 g infiltrated N. benthamiana leaf extracts. Microsomes were pelleted as described above and the pellets were resuspended in 1 ml of co‐immunoprecipitation buffer (50 mM Tris‐HCl [pH 7.5], 10% [v/v] glycerol, 150 mM NaCl, 0.1% [v/v] Triton X‐100, 1 mM EDTA, 10 mM NEM, 1 mM 1,10‐PT, 1 μM MLN7243, 10 μM MG132, EDTA‐free protease inhibitor mixture complete [Roche], and PhosSTOP phosphatase inhibitor cocktail [Roche]), kept on a rotating wheel for 30 min at 4°C. Non‐resuspended material was removed by centrifugation (20,000 g for 10 min). The supernatants enriched with the microsomal proteins were incubated with α‐cMyc antibody beads (Wako, 10D11) for 1 h at 4°C under gentle rocking. Immunoprecipitates were analyzed by western blot with α‐FLAG (1/1,000; MBL) and α‐Myc (1/1,000; Wako) antibodies.
Split‐ubiquitin yeast two‐hybrid assay
Full‐length coding sequences of BRI1, UBP13, and calcineurin B‐like (CBL)‐interacting protein kinase 8 (CIPK8) were cloned into the pMetYC_GW or pNX32_GW destination vectors (Obrdlik et al, 2004). Constructs were transformed into the yeast strain L40ccua (MATa his3Δ200 trp1‐901 leu2‐3112 LYS2::(lexAop)4‐HIS3 ura3::(lexAop)8‐lacZ ADE2::(lexAop)8‐URA3 gal80 canR cyh2R ) with a Frozen‐EZ Yeast Transformation II Kit (Zymo Research) according to the manufacturer's protocol. Yeast cultures were handled and β‐galactosidase assays were done as described in the Yeast Protocols Handbook (Clontech).
In vitro ubiquitination and deubiquitination assays
Full‐length coding sequences of UBP13 and its deubiquitination‐inactive mutant UBP13C207S, harboring a point mutation of Cys207 to Ser207 generated by site‐directed mutagenesis, were subcloned into the pYU1274 destination vector with N‐terminal GST‐tag (kindly provided by Dr. Yoshihisa Ueno, Ryukoku University, Kyoto, Japan) with primers listed in Appendix Table S1. Constructs expressing MBP‐BRI1CD (Liu et al, 2020) and GST‐PUB13 have been described previously (Zhou et al, 2018). All constructs were transformed into the Escherichia coli strain BL21(DE3) pLysS (Novagen) to produce recombinant proteins induced by 0.1–0.2 mM isopropyl β‐d‐thiogalactopyranoside. The produced GST‐UBP13, GST‐UBP13C207S, MBP‐BRI1CD, and GST‐PUB13 recombinant proteins were purified with Glutathione Sepharose 4B (GE Healthcare) or amylose resin (New England BioLabs) according to the manufacturers' instructions.
For the in vitro chain‐specific deubiquitination assay, 500 ng K48‐ or K63‐diubiquitin (Boston Biochem) were incubated with 1 μg GST‐UBP13 or GST‐UBP13C207S in 30 μl of deubiquitination buffer (40 mM Tris‐HCl [pH 7.5], 5 mM MgCl2, 2 mM DTT, and 150 mM NaCl) for 0 or 18 h at 30°C. Assay reactions were stopped by addition of an equal volume of 2× sample buffer (125 mM Tris‐HCl [pH 6.8], 20% [v/v] glycerol, 4% [w/v] SDS, 10% [v/v] β‐mercaptoethanol, and 0.02% [w/v] bromophenol blue).
The in vitro ubiquitination assay was carried out as described previously (Sato et al, 2009) with minor modifications. In brief, 83.3 nM human E1 (UBE1; Boston Biochem), 250 ng E2 (UbcH5a; Wako), 4 μg ubiquitin (Sigma‐Aldrich), 150 ng GST‐PUB13, and 500 ng MBP‐BRI1CD in 30 μl of ubiquitination buffer (40 mM Tris‐HCl [pH 7.5], 5 mM MgCl2, 2 mM ATP, and 2 mM DTT) were incubated for 3 h at 30°C. The reaction was terminated by incubation with 1 μM E1 inhibitor (MLN7243) for 30 min at 30°C. After ubiquitination had been stopped, the reaction was supplemented with 150 mM NaCl and separately incubated with 1 μg GST‐UBP13 or GST‐UBP13C207S for 18 h at 30°C. As a mock control, Milli‐Q water (Millipore) was used. After the in vitro deubiquitination, the subsequent reactions were incubated with 10 μl of amylose resin in 1 ml of pull‐down buffer (40 mM Tris‐HCl [pH 7.5], 5 mM MgCl2, 2 mM DTT, 150 mM NaCl, and 1% [v/v] Triton X‐100) for 2 h at 4°C under gentle rocking to purify the polyubiquitinated MBP‐BRI1CD proteins that were eluted by 1× SDS sample buffer.
For the in vitro ubiquitination assays with ubiquitin variants, PUB13 auto‐ubiquitination was done by incubating 500 ng GST‐PUB13 with 4 μg native ubiquitins or lysine‐to‐arginine single mutated ubiquitins (Ubiquitin human, U‐100H; rhUbiquitin K63R, UM‐K63R; rhUbiquitin K48R, UM‐K48R; Boston Biochem) for 1 h at 30°C, or by incubating 1 μg GST‐PUB13 with 6 μg ubiquitin mutants retaining only one lysine residue (rhUbiquitin K63 only, UM‐K630; rhUbiquitin K48 only, UM‐K480; Boston Biochem) for 3 h at 30°C in 30 μl of ubiquitination buffer containing 83.3 or 200 nM human E1 (UBE1; Boston Biochem) and 250 ng E2 (UbcH5a; Wako). To confirm BRI1 was K63 ubiquitinated by PUB13, 500 ng MBP‐BRI1CD was incubated with 250 ng GST‐PUB13 in 30 μl of ubiquitination buffer containing 83.3 nM human E1 (UBE1; Boston Biochem) and 200 nM E2 (UbcH5a; Boston Biochem) for 30 min at 30°C. Ubiquitination was stopped by incubation with the E1 inhibitor and the polyubiquitinated MBP‐BRI1CD was purified by MBP pull‐down.
Protein ubiquitination or deubiquitination was examined by western blot analyses after SDS‐PAGE with α‐ubiquitin (P4D1 [Santa Cruz] or FK2 [Wako]), α‐K63Ub (Apu3; Millipore), and α‐K48Ub (Apu2; Millipore) antibodies. The α‐GST (MBL) and α‐MBP (New England BioLabs) antibodies were used to confirm the presence of recombinant proteins.
Cell‐free deubiquitination assay
Polyubiquitinated BRI1‐mCit was purified from 18‐day‐old BRI1‐mCit/bri1 plants by immunoprecipitation with α‐GFP mAb‐magnetic beads on solubilized microsomal proteins as described above. Bead‐bound Ubn‐BRI1‐mCit was resuspended in 30 μl of deubiquitination buffer, and separately incubated with 2 μg GST‐UBP13 or GST‐UBP13C207S for 10 h at 30°C. After incubation, Ubn‐BRI1‐mCit proteins were eluted from the beads by adding 1× SDS sample buffer and analyzed by western blot.
Gene expression analysis
Total RNA was isolated from plant samples by means of the TRIzol reagent (Invitrogen) and treated with RQ1 RNase‐free DNase (Promega), according to the manufacturers' protocols. First‐strand cDNA was synthesized with oligo(dT) primers (Promega) and ReverTraAce reverse transcriptase (TOYOBO) and subjected to qRT‐PCR analysis on an Mx3000P system (Agilent Technologies) with TB Green Premix EX Taq (TaKaRa) as described by the manufacturers.
Confocal microscopy and image analysis
To analyze the BRI1 vacuolar targeting, Arabidopsis roots were mounted in water and imaged by a confocal laser‐scanning microscope (LSM980; Zeiss) equipped with 63× oil immersion objective. The excitation/emission wavelengths used were 514 nm/516–605 nm. The imaging position was kept consistently in the same region of the meristematic zone of the root tip, 10–15 cells above the quiescent center. Images were converted to 8 bit in Fiji/ImageJ for the BRI1‐mCitrine fluorescence signal intensity measurement. Regions of interest (ROIs) were selected based on the PM or cytosol localization. Histograms listing all intensity values per ROI were generated and the average intensity of the 100 pixels with the highest signal was used for calculations.
The AFCS uptake assay was done as previously described (Irani et al, 2014) with modifications. Six‐day‐old seedlings grown on DEX medium were transferred to 200 µl of liquid 1/2MS containing 10 µM DEX (liquid DEX medium) on a piece of parafilm placed in a Petri plate, humidified with wet laboratory wipes for 10 min. The medium was replaced with liquid DEX medium supplemented with 40 µM AFCS for 40 min (pulse). Seedlings were washed six times and chased for 30 min to ensure complete ligand uptake into the vacuole. Epidermal cells of the root meristematic zone were imaged with a SP8X laser scanning confocal microscope (Leica) equipped with an HC PL APO CS2 40×/1.10 water‐corrected objective and 2× digital zoom. The excitation wavelength was 633 nm by white light laser. Emission was detected at 650–700 nm by hybrid detectors (Leica). For the measurement of the fluorescence signal in vacuoles, stacks of four to six slices of 1.5 μm were obtained. Image analysis and signal quantification were carried out with the Fiji/ImageJ software. Four to six slices covering the epidermal cell layer were merged with maximum projection. The images were processed with a Gaussian blur filter (1) and subtraction background (100). Rectangular ROIs were determined, avoiding regions that were overstained, and the signal intensity was quantified in seven to nine seedlings per genotype.
Quantification and statistical analysis
The P‐values were calculated with one‐way analysis of variance (ANOVA) and post hoc Tukey's test for multiple‐group comparisons. Values shown in bar graphs are mean ± SD and measurements shown in box plots display the first and third quartiles, split by the medians with whiskers extending 1.5‐fold the interquartile range beyond the box. Data points are plotted as dots. Statistical significance was set based on P‐values. n.s., not significant (P > 0.05); *P < 0.05; **P < 0.01.
Author contributions
Yongming Luo: Conceptualization; Data curation; Investigation; Writing—original draft. Junpei Takagi: Data curation; Investigation; Writing—review and editing. Lucas Alves Neubus Claus: Data curation; Investigation; Writing—review and editing. Chao Zhang: Writing—review and editing. Shigetaka Yasuda: Data curation; Investigation. Yoko Hasegawa: Data curation; Investigation. Junji Yamaguchi: Writing—review and editing. Libo Shan: Data curation; Investigation; Writing—review and editing. Eugenia Russinova: Conceptualization; Data curation; Investigation; Writing—original draft. Takeo Sato: Conceptualization; Data curation; Investigation; Writing—original draft.
In addition to the CRediT author contributions listed above, the contributions in detail are:
YL, ER, and TS designed research; YL, JT, LANC, SY, YH, LS, ER, and TS performed research and analyzed data; YL, ER, and TS wrote the manuscript with the help of JT, LANC, CZ, JY, and LS.
Disclosure and competing interests statement
The authors declare no conflict of interest.
Supporting information
Appendix
Acknowledgements
We thank Dr. Tsuyoshi Nakagawa (Shimane University, Matsue, Japan) and Dr. Yoshihisa Ueno (Ryukoku University, Kyoto, Japan) for providing the Gateway destination vectors, Dr. Bernhard Grimm (Humboldt University, Berlin, Germany) and Dr. Ryouich Tanaka (Hokkaido University, Sapporo, Japan) for providing materials for the split‐ubiquitin yeast two‐hybrid assays, Dr. Yanhai Yin (Iowa State University, Ames, IA, USA) for providing the BES1 antibody, Dr. Gregory Vert (Université de Toulouse, Toulouse, France) for providing the BRI1‐mCit/bri1 and BRI125KR‐mCit/bri1 transgenic lines, and Martine De Cock (Ghent University) for help in preparing the manuscript. This work was supported by Grants‐in‐Aid for Scientific Research from the Japan Society for the Promotion of Science (JSPS) [No. JP20K05949 and JP21H05644 to T.S. and No. JP21H02150 to J.Y.], Hokkaido University Young Scientist Support Program to T.S., and National Institute of Health [R01GM097247] to L.S. Y.L. and C.Z. are supported by the JSPS Research Fellowships for Young Scientists and a China Scholarship Council fellowship, respectively.
EMBO reports (2022) 23: e53354.
Data availability
This study includes no data deposited in external repositories.
References
- An Z, Liu Y, Ou Y, Li J, Zhang B, Sun D, Sun Y, Tang W (2018) Regulation of the stability of RGF1 receptor by the ubiquitin‐specific proteases UBP12/UBP13 is critical for root meristem maintenance. Proc Natl Acad Sci USA 115: 1123–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir Y, Jaillais Y, Epple P, Balsemão‐Pires E, Dangl JL, Chory J (2012) Brassinosteroids modulate the efficiency of plant immune responses to microbe‐associated molecular patterns. Proc Natl Acad Sci USA 109: 297–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaiwanon J, Wang Z‐Y (2015) Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in Arabidopsis roots. Curr Biol 25: 1031–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clague MJ, Urbé S (2006) Endocytosis: the DUB version. Trends Cell Biol 16: 551–559 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Clouse SD (2011) Brassinosteroids. Arabidopsis Book 9: e0151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X, Lu F, Li Y, Xue Y, Kang Y, Zhang S, Qiu QI, Cui X, Zheng S, Liu B et al (2013) Ubiquitin‐specific proteases UBP12 and UBP13 act in circadian clock and photoperiodic flowering regulation in Arabidopsis . Plant Physiol 162: 897–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway‐compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Ewan R, Pangestuti R, Thornber S, Craig A, Carr C, O'Donnell L, Zhang C, Sadanandom A (2011) Deubiquitinating enzymes AtUBP12 and AtUBP13 and their tobacco homologue NtUBP12 are negative regulators of plant immunity. New Phytol 191: 92–106 [DOI] [PubMed] [Google Scholar]
- Geldner N, Hyman DL, Wang X, Schumacher K, Chory J (2007) Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev 21: 1598–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Hu M, Li P, Song L, Jeffrey PD, Chenova TA, Wilkinson KD, Cohen RE, Shi Y (2005) Structure and mechanisms of the proteasome‐associated deubiquitinating enzyme USP14. EMBO J 24: 3747–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irani NG, Di Rubbo S, Mylle E, Van den Begin J, Schneider‐Pizoń J, Hniliková J, Šíša M, Buyst D, Vilarrasa‐Blasi J, Szatmári A‐M et al (2012) Fluorescent castasterone reveals BRI1 signaling from the plasma membrane. Nat Chem Biol 8: 583–589 [DOI] [PubMed] [Google Scholar]
- Irani NG, Di Rubbo S, Russinova E (2014) In vivo imaging of brassinosteroid endocytosis in Arabidopsis . Methods Mol Biol 1209: 107–117 [DOI] [PubMed] [Google Scholar]
- Isono E, Katsiarimpa A, Müller IK, Anzenberger F, Stierhof Y‐D, Geldner N, Chory J, Schwechheimer C (2010) The deubiquitinating enzyme AMSH3 is required for intracellular trafficking and vacuole biogenesis in Arabidopsis thaliana . Plant Cell 22: 1826–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillais Y, Belkhadir Y, Balsemão‐Pires E, Dangl JL, Chory J (2011) Extracellular leucine‐rich repeats as a platform for receptor/coreceptor complex formation. Proc Natl Acad Sci USA 108: 8503–8507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski J, de la Vega M, Fletcher SJ, McFarlane C, Greene MK, Smyth AW, Van Schaeybroeck S, Johnston JA, Scott CJ, Rappoport JZ et al (2014) USP17 is required for clathrin mediated endocytosis of epidermal growth factor receptor. Oncotarget 5: 6964–6975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong JS, Jung C, Seo JS, Kim J‐K, Chua N‐H (2017) The Deubiquitinating enzymes UBP12 and UBP13 positively regulate MYC2 levels in jasmonate responses. Plant Cell 29: 1406–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsiarimpa A, Anzenberger F, Schlager N, Neubert S, Hauser M‐T, Schwechheimer C, Isono E (2011) The Arabidopsis deubiquitinating enzyme AMSH3 interacts with ESCRT‐III subunits and regulates their localization. Plant Cell 23: 3026–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsiarimpa A, Muñoz A, Kalinowska K, Uemura T, Rojo E, Isono E (2014) The ESCRT‐III‐interacting deubiquitinating enzyme AMSH3 is essential for degradation of ubiquitinated membrane proteins in Arabidopsis thaliana . Plant Cell Physiol 55: 727–736 [DOI] [PubMed] [Google Scholar]
- Komander D, Clague MJ, Urbé S (2009) Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol 10: 550–563 [DOI] [PubMed] [Google Scholar]
- Lee C‐M, Li M‐W, Feke A, Liu W, Saffer AM, Gendron JM (2019) GIGANTEA recruits the UBP12 and UBP13 deubiquitylases to regulate accumulation of the ZTL photoreceptor complex. Nat Commun 10: 3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Kumar R, Claus LAN, Johnson AJ, Siao W, Vanhoutte I, Wang P, Bender KW, Yperman K, Martins S et al (2020) Endocytosis of BRASSINOSTEROID INSENSITIVE1 is partly driven by a canonical Tyr‐based motif. Plant Cell 32: 3598–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo YU, Scholl S, Doering A, Zhang YI, Irani NG, Di Rubbo S, Neumetzler L, Krishnamoorthy P, Van Houtte I, Mylle E et al (2015) V‐ATPase activity in the TGN/EE is required for exocytosis and recycling in Arabidopsis . Nat Plants 1: 15094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins S, Dohmann EMN, Cayrel A, Johnson A, Fischer W, Pojer F, Satiat‐Jeunemaître B, Jaillais Y, Chory J, Geldner N et al (2015) Internalization and vacuolar targeting of the brassinosteroid hormone receptor BRI1 are regulated by ubiquitination. Nat Commun 6: 6151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann AP, Scott CJ, Van Schaeybroeck S, Burrows JF (2016) Deubiquitylating enzymes in receptor endocytosis and trafficking. Biochem J 473: 4507–4525 [DOI] [PubMed] [Google Scholar]
- Mevissen TET, Komander D (2017) Mechanisms of deubiquitinase specificity and regulation. Annu Rev Biochem 86: 159–192 [DOI] [PubMed] [Google Scholar]
- Millard SM, Wood SA (2006) Riding the DUBway: regulation of protein trafficking by deubiquitylating enzymes. J Cell Biol 173: 463–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nolan TM, Vukašinović N, Liu D, Russinova E, Yin Y (2020) Brassinosteroids: multidimensional regulators of plant growth, development, and stress responses. Plant Cell 32: 295–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obrdlik P, El‐Bakkoury M, Hamacher T, Cappellaro C, Vilarino C, Fleischer C, Ellerbrok H, Kamuzinzi R, Ledent V, Blaudez D et al (2004) K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc Natl Acad Sci USA 101: 12242–12247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Akopian D, Rape M (2018) Principles of ubiquitin‐dependent signaling. Annu Rev Cell Dev Biol 34: 137–162 [DOI] [PubMed] [Google Scholar]
- Park S‐H, Jeong JS, Seo JS, Park BS, Chua N‐H (2019) Arabidopsis ubiquitin‐specific proteases UBP12 and UBP13 shape ORE1 levels during leaf senescence induced by nitrogen deficiency. New Phytol 223: 1447–1460 [DOI] [PubMed] [Google Scholar]
- Russinova E, Borst J‐W, Kwaaitaal M, Caño‐Delgado A, Yin Y, Chory J, de Vries SC (2004) Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16: 3216–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Maekawa S, Yasuda S, Sonoda Y, Katoh E, Ichikawa T, Nakazawa M, Seki M, Shinozaki K, Matsui M et al (2009) CNI1/ATL31, a RING‐type ubiquitin ligase that functions in the carbon/nitrogen response for growth phase transition in Arabidopsis seedlings. Plant J 60: 852–864 [DOI] [PubMed] [Google Scholar]
- Shimada S, Komatsu T, Yamagami A, Nakazawa M, Matsui M, Kawaide H, Natsume M, Osada H, Asami T, Nakano T (2015) Formation and dissociation of the BSS1 protein complex regulates plant development via brassinosteroid signaling. Plant Cell 27: 375–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swatek KN, Komander D (2016) Ubiquitin modifications. Cell Res 26: 399–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano J, Miwa K, Yuan L, von Wirén N, Fujiwara T (2005) Endocytosis and degradation of BOR1, a boron transporter of Arabidopsis thaliana, regulated by boron availability. Proc Natl Acad Sci USA 102: 12276–12281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda A, Sugiyama K, Nagano H, Mori M, Kaido M, Mise K, Tsuda S, Okuno T (2002) Identification of a novel RNA silencing suppressor, NSs protein of Tomato spotted wilt virus . FEBS Lett 532: 75–79 [DOI] [PubMed] [Google Scholar]
- Vanhaeren H, Chen Y, Vermeersch M, De Milde L, De Vleeschhauwer V, Natran A, Persiau G, Eeckhout D, De Jaeger G, Gevaert K et al (2020) UBP12 and UBP13 negatively regulate the activity of the ubiquitin‐dependent peptidases DA1, DAR1 and DAR2. eLife 9: e52276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukašinović N, Wang Y, Vanhoutte I, Fendrych M, Guo B, Kvasnica M, Jiroutová P, Oklestkova J, Strnad M, Russinova E (2021) Local brassinosteroid biosynthesis enables optimal root growth. Nat Plants 7: 619–632 [DOI] [PubMed] [Google Scholar]
- Wang Z‐Y, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T et al (2002) Nuclear‐localized BZR1 mediates brassinosteroid‐induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2: 505–513 [DOI] [PubMed] [Google Scholar]
- Wang C, Shang J‐X, Chen Q‐X, Oses‐Prieto JA, Bai M‐Y, Yang Y, Yuan M, Zhang Y‐L, Mu C‐C, Deng Z et al (2013) Identification of BZR1‐interacting proteins as potential components of the brassinosteroid signaling pathway in Arabidopsis through tandem affinity purification. Mol Cell Proteomics 12: 3653–3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielopolska A, Townley H, Moore I, Waterhouse P, Helliwell C (2005) A high‐throughput inducible RNAi vector for plants. Plant Biotechnol J 3: 583–590 [DOI] [PubMed] [Google Scholar]
- Wu R, Zheng W, Tan J, Sammer R, Du L, Lu C (2019) Protein partners of plant ubiquitin‐specific proteases (UBPs). Plant Physiol Biochem 145: 227–236 [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang Z‐Y, Mora‐Garcia S, Li J, Yoshida S, Asami T, Chory J (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191 [DOI] [PubMed] [Google Scholar]
- Yu X, Li L, Zola J, Aluru M, Ye H, Foudree A, Guo H, Anderson S, Aluru S, Liu P et al (2011) A brassinosteroid transcriptional network revealed by genome‐wide identification of BESI target genes in Arabidopsis thaliana . Plant J 65: 634–646 [DOI] [PubMed] [Google Scholar]
- Zhou J, Liu D, Wang P, Ma X, Lin W, Chen S, Mishev K, Lu D, Kumar R, Vanhoutte I et al (2018) Regulation of Arabidopsis brassinosteroid receptor BRI1 endocytosis and degradation by plant U‐box PUB12/PUB13‐mediated ubiquitination. Proc Natl Acad Sci USA 115: E1906–E1915 [DOI] [PMC free article] [PubMed] [Google Scholar]


