Abstract
Human immunodeficiency virus type 1 (HIV-1) resistance to antiretroviral drugs is the main cause of patient treatment failure. Despite the problems associated with interpretation of HIV-1 resistance testing, resistance monitoring should help in the rational design of initial or rescue antiretroviral therapies. It has previously been shown that the activity of the HIV-1 protease can be monitored by using a bacteriophage lambda-based genetic assay. This genetic screening system is based on the bacteriophage lambda regulatory circuit in which the viral repressor cI is specifically cleaved to initiate the lysogenic to lytic switch. We have adapted this simple lambda-based genetic assay for the analysis of the activities and phenotypes of different HIV-1 proteases. Lambda phages that encode HIV-1 proteases either from laboratory strains (strain HXB2) or from clinical samples are inhibited in a dose-dependent manner by the HIV-1 protease inhibitors indinavir, ritonavir, saquinavir, and nelfinavir. Distinct susceptibilities to different drugs were also detected among phages that encode HIV-1 proteases carrying different resistance mutations, further demonstrating the specificity of this assay. Differences in proteolytic processing activity can also be directly monitored with this genetic screen system since two phage populations compete in culture with each other until one phage outgrows the other. In summary, we present here a simple, safe, and rapid genetic screening system that may be used to predict the activities and phenotypes of HIV-1 proteases in the course of viral infection and antiretroviral therapy. This assay responds appropriately to well-known HIV-1 protease inhibitors and can be used to search for new protease inhibitors.
Human immunodeficiency virus (HIV) type 1 (HIV-1) protease is essential in the replication and maturation of the virus because it processes gag (p55) and gag-pol (p160) polyprotein products into functional core and viral enzymes (4). Several inhibitors of the HIV-1 protease have become available for the treatment of HIV-1-infected patients (14). Early antiviral therapy, mainly directed against the reverse transcriptase, showed limited antiviral activity. However, treatment with antiretroviral combination therapy that includes at least a protease inhibitor has made possible the reduction of plasma virus levels to below the limit of detection (9, 11, 14). Nevertheless, suboptimal therapies that fail to achieve a complete and sustained suppression of virus replication lead to the selection of drug-resistant virus mutants (5, 21, 29). Moreover, there is an overlap of resistance-conferring mutations among most of the available protease inhibitors (1). A latent reservoir of replication-competent virus remains in CD4 T cells (3, 7, 30), and the persistence of HIV-1 replication has also been detected in some patients with sustained suppression of viremia (8, 15, 17, 32), suggesting that drug-resistant viruses are expected to emerge even in patients with undetectable virus in their plasma. Faster and simpler assays for the phenotypic detection of drug resistance and cross-resistance may facilitate the study of the increasing number of patients bearing HIV-1 strains resistant to protease inhibitors. New drug-screening systems might also be important in the search for new anti-HIV-1 drugs.
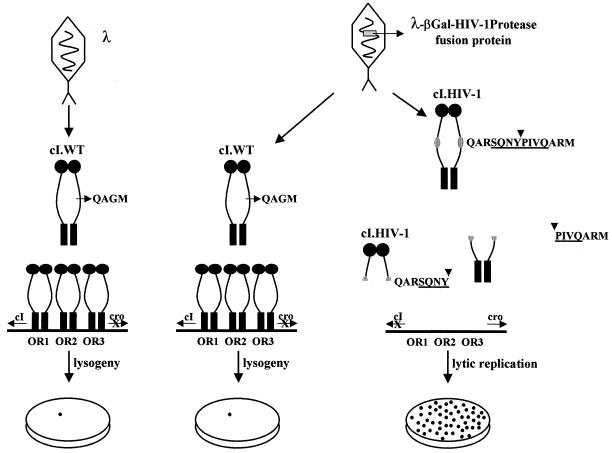
It has previously been demonstrated that a bacteriophage lambda-based genetic screen can be used to monitor the activity of the HIV-1 protease (28). This genetic screening system is based on the bacteriophage lambda cI-cro regulatory circuit in which the viral repressor cI is specifically cleaved to initiate the lysogenic to lytic switch (26). An endogenous bacterial protease, RecA, cleaves the lambda repressor cI at a specific protein region, avoiding an efficient DNA-cI complex and switching on the viral lytic genes that will produce the phage progeny. When a recombinant cI repressor containing a specific HIV-1 protease cleavage site was tested in vivo it was found to be resistant to RecA cleavage. Interestingly, the introduction of an HIV-1 protease in a wild-type phage will cleave this mutant cI repressor containing a specific HIV-1 protease cleavage site, allowing the phage to go into the lytic replication cycle (Fig. 1) (28).
FIG. 1.
Bacteriophage lambda-based genetic screen for characterization of the activity and phenotype of HIV-1 protease. This genetic screening system is based on the bacteriophage lambda cI-cro regulatory circuit in which the viral repressor cI is specifically cleaved to initiate the lysogenic to lytic switch. When phages that contain a specific HIV-1 protease (λ-HIV-1p) infect E. coli cells that express the recombinant cI.HIV-1 repressor, the infection results in lytic replication. Phages that lack HIV-1 proteases are not able to replicate in cells that express the cI.HIV-1 repressor. Likewise, phages that contain HIV-1 proteases cannot replicate in cells that express the wild-type cI repressor. The cI.HIV-1 repressor contains the matrix-capsid (p17/p24) gag cleavage site sequence shown here. The structure and construction of cI.HIV-1 repressors are described by Sices and Kristie (28). WT, wild type; βGal, β-galactosidase.
The complexity and time-consuming nature of the current ex vivo or in vitro protocols for the characterization of HIV-1 protease activity prompted us to explore this bacteriophage lambda-based genetic screening system as a simple alternative approach to the characterization of HIV-1 protease enzymatic activity and phenotype. Here, we demonstrate that lambda phages that encode HIV-1 proteases either from HIV-1 laboratory strains or from clinical samples are inhibited in a dose-dependent manner by four different HIV-1 protease inhibitors. Furthermore, differences in proteolytic processing activity among mutant enzymes could be also monitored with this simple genetic screening system.
MATERIALS AND METHODS
HIV-1 proteases.
Viral RNA was isolated from an HIV-1 HXB2 strain stock (obtained through the Medical Research Council AIDS Reagent Program, London, United Kingdom) cultured in Sup-T1 cells and from six plasma samples obtained from three HIV-1-infected patients. These three patients were selected for this study because they had failed therapy with different antiprotease regimens. Table 1 shows the clinical characteristics and the antiprotease drug experiences of these patients and for the two samples analyzed for each patient. RNA was extracted from a volume of 140 μl of plasma or the supernatant of a Sup-T1 cell culture with the QIAamp blood kit (Qiagen). After viral RNA isolation, 10 μl of resuspended RNA was reverse transcribed and was amplified with the Titan one-tube reverse transcription (RT)-PCR System (Boehringer Mannheim) according to the manufacturer's instructions. Briefly, the RT-PCR mixture contained 10 pmol of the protease oligonucleotides 5′prot1 (sense) (5′-AGC TAA TTT TTT AGG GAA GAT CTG GCC TTC C-3′; HXB2 positions 2077 to 2108 [22]) and 3′prot1 (antisense) (5′-GCA CCT ACT GGA GTA TTG TAT GGA TTT TCA GG′-3; HXB2 positions 2703 to 2733), 200 μM deoxynucleoside triphosphates, 1.5 mM MgCl2, RT-PCR buffer, 5 mM dithiothreitol, 5 U of RNase inhibitor, 1 μl of enzyme mix (avian myeloblastosis virus and Expand High Fidelity PCR System) in a total reaction volume of 50 μl. The samples were incubated for 30 min at 50°C; then one cycle of denaturation at 94°C for 2 min; then 10 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 68°C for 1 min; and then 25 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 68°C for 1 min plus 5 s for each cycle. A final extension at 68°C for 7 min was added to the last cycle. A 5-μl aliquot was again amplified in a 100-μl reaction mixture containing 10 pmol of the protease oligonucleotides HIVproL (sense) (5′-GGG GAA TTC TAA GGC CAG GGA ATT TTC TTC-3′; HXB2 positions 2117 to 2136) and HIVproR (antisense) (5′-GGG GAA TTC AAA GGC CAT CCA TTC CTG GC-3′; HXB2 positions 2587 to 2603) (underscores indicate an EcoRI restriction site), 200 μM deoxynucleoside triphosphates, 1.5 mM MgCl2, PCR buffer (50 mM KCl, 10 mM Tris-Cl [pH 8.3]), and 0.5 U of Taq DNA polymerase (Perkin-Elmer). Cycling parameters were one cycle of denaturation at 94°C for 2 min and then 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min. This was followed by a 7-min incubation at 72°C. By following the above protocol, the sequence amplified included the full-length HIV-1 protease plus 44 amino acids upstream and 18 amino acids downstream of the protease-coding region.
TABLE 1.
Clinical characteristics and treatment histories of the three study patients
| Patient | Sample no. | No. of CD4+ cells/μla | No. of HIV-1 RNA copies/mlb | Protease inhibitor treatment history |
|---|---|---|---|---|
| IRLL | 95 | 54 | 140,000 | Naive |
| 98 | 6 | 363,344 | 11 mo of IND, 19 mo of RIT, 19 mo of SQV | |
| JAD | 93 | 128 | 76,823 | Naive |
| 98 | 71 | 11,798 | 24 mo of RIT, 10 mo of SQV, 4 mo of IND | |
| JGR | 95 | 630 | 81,771 | Naive |
| 97 | 327 | 1,996 | 3 mo of RIT, 10 mo of IND |
CD4 T-cell counts were measured by flow-activated cytometric assay and are expressed as the number per microliter of blood.
Plasma HIV-1 RNA levels were determined by using the Amplicor Monitor assay (Roche Molecular Systems, Inc.).
Construction of recombinant phages.
The HIV-1 protease PCR products were digested with EcoRI, ligated to EcoRI-digested bacteriophage lambda ZapII (Stratagene), and packaged in vitro. Phage stocks were prepared from the correct insert orientation. Phage DNA sequencing confirmed that there were no major differences in the protease amino acid sequences between the RT-PCR-amplified HIV-1 RNA and the recombinant phages (data not shown).
Lambda-based genetic assay.
Escherichia coli JM109 containing plasmid p2X-cI.HIV was transformed with plasmid pcI.HIV-cro as described by Sices and Kristie (28). Briefly, 200 μl (108 cells) of the resulting strain was infected with 107 PFU of phages containing the different HIV-1 proteases (λ-HIV-1p) for 15 min at 37°C, washed with 1 ml of 10 mM MgSO4, and resuspended in LB medium containing 12.5 μg of tetracycline per ml, 0.2% maltose, 10 mM MgSO4, 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and the appropriate drug concentration (see Fig. 4) in a final volume of 1 ml. At 3 h postinfection at 37°C, aliquots of the cultures were coplated with E. coli XL-1 Blue cells in top agar containing 12.5 μg of tetracycline per ml, 0.2% maltose, 0.1 mM IPTG, and the indicated drug concentration for 6 h (see Fig. 4). The competitive replication assay between phages carrying different HIV-1 proteases was carried out by coinfecting 108 pcI.HIV-cro cells with 107 PFU of λ-HIV-1pJAD93 and 107 PFU of λ-HIV-1pJAD98. After one cycle of selection, additional cycles of selective growth were done by resuspending 1/10 (100 μl) of the infected cells with a fresh aliquot (108 cells) of pcI.HIV-cro cells and cycling was continued as described above. After three selective cycles phage DNA from 5 μl of culture supernatant was directly amplified by PCR with T3 and T7 oligonucleotides. PCR conditions were the same as those described above for the HIVproL and HIVproR oligonucleotides. PCR products were purified by using the Qiaquick spin PCR purification kit (Qiagen). Sequencing reactions were carried out with the ABI PRISM dRhodamine Terminator Cycle Sequencing kit (Applied Biosystems) and T3 and T7 oligonucleotides. The products of the reactions were then analyzed on an Applied Biosystems 310 sequencer. Sequence editing was performed with the Sequence Navigator program (Applied Biosystems). The Factura DNA analysis software package (Applied Biosystems) was used to quantify the proportion of individual nucleotide mixture ratios during three cycles of competitive phage selective growth (see Fig. 5). At the relevant nucleotide positions, for example, protease codon 82, the peaks heights of the nucleotide bases representing the wild-type and mutant coding sequences were compared.
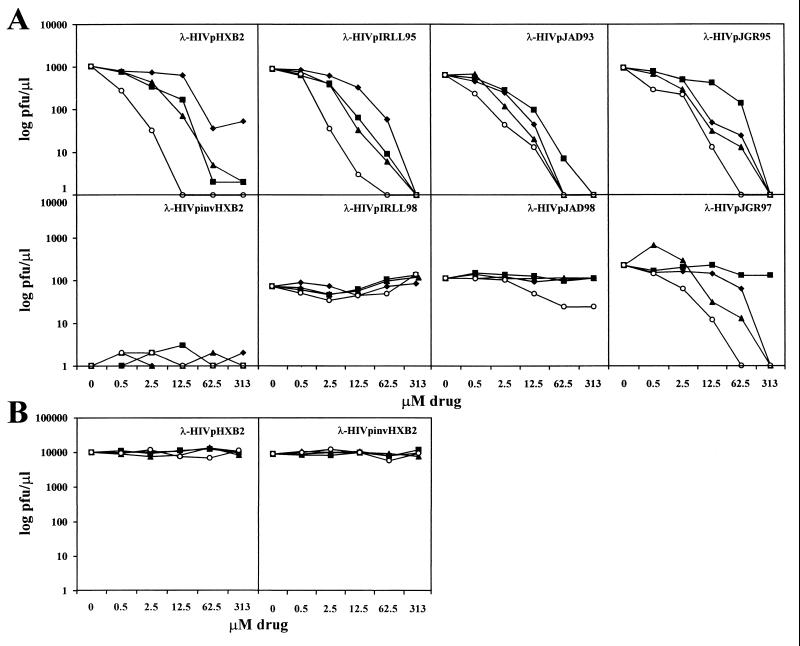
FIG. 4.
(A) Inhibition of seven different λ-HIV-1 proteases in the presence of different protease inhibitors. Cells transformed with the cI.HIV-1-cro construct were infected with λ-HIV-1 proteases in the absence or presence of different concentrations of the HIV-1 protease inhibitors IND (⧫), RIT (■), SQV (▴), and NFV (○). At 3 h postinfection, the titer of the resulting phage was determined. Each datum point is the average of two independent experiments. (B) Control cells that did not express the cI.HIV-1-cro construct were infected with λ-HIV-1pHXB2 or λ-HIV-1pinvHXB2 phages in the presence of different concentrations of the four HIV-1 protease inhibitors listed above. Drugs did not have any effect on the growth of λ-HIV-1 proteases in these control cells.
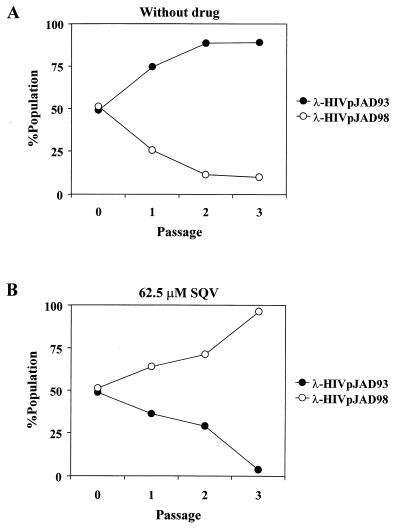
FIG. 5.
Competitive replication assay between phages carrying patient sample JAD93 (λ-HIV-1pJAD93) and JAD98 (λ-HIV-1pJAD98) proteases in the absence (A) or presence of SQV at a concentration of 62.5 μM (B). Data were generated on the basis of the relative peak heights in electropherograms produced directly from the DNA sequence of the phage genome at the different passages. For example, equal peak heights for both wild-type and mutant bases indicate a 50:50 ratio. Each datum point is the average of two independent experiments.
Ex vivo drug susceptibility testing.
Peripheral blood mononuclear cells from patient IRLL isolates 95 and 98 were cocultured with phytohemagglutinin (Sigma)-interleukin 2 (Boehringer Inghelheim)-stimulated peripheral blood mononuclear cells from an HIV-seronegative blood donor. When the HIV-1 p24 antigen concentration in the culture surpassed 20 ng/ml, the supernatants were harvested. Titration of these two virus stocks was performed in MT-4 cells. The HXB2 control HIV-1 isolate and the MT4-4 cells were obtained from the AIDS Reagent Project (Medical Research Council). HXB2 virus was propagated and titrated in MT4-4 cells. Patient and HXB2 isolates were tested in triplicate with indinavir (IND; Merck & Co, West Point, Pa.), ritonavir (RIT; Abbott Laboratories, Park Road, Ill.), saquinavir (SQV; Roche Laboratories, London, United Kingdom), and nelfinavir (NFV; Agouron Pharmaceuticals, San Diego, Calif.). Anti-HIV-1 activity measurements in MT-4 cells were based on the viabilities of cells that had been infected or not infected with HIV-1 (multiplicity of infection, 0.003) and exposed to various concentrations of the drug. After the MT-4 cells were allowed to proliferate for 5 days, the number of viable cells was quantified by a tetrazolium-based colorimetric method (MTT method) as described elsewhere (24).
RESULTS
In order to assess if the bacteriophage lambda-based genetic assay can be used to monitor the activities of HIV-1 proteases either from laboratory strains or from clinical samples, RT-PCR-amplified DNAs encompassing the full-length protease-coding sequence from the HXB2 laboratory virus strain and from six plasma samples from three HIV-1-infected patients were cloned in a lambda phage to generate a β-galactosidase–HIV-1 protease fusion protein (Fig. 1). The amino acid sequences of the seven HIV-1 proteases analyzed in this study are shown in Fig. 2. E. coli cells containing the HIV-1 cI repressor (cI.HIV-1-cro) construct (Fig. 1) were then infected with phages encoding the seven different β-galactosidase–HIV-1 protease fusion proteins (λ-HIV-1p). Wild-type lambda phage (λ) and a phage carrying an inverted HXB2 protease (λ-HIV-1pinvHXB2) were used as controls.
FIG. 2.
Deduced amino acid sequence alignment of the seven HIV-1 proteases analyzed in the present study. Amino acid changes relative to the clade B consensus sequence (22) are indicated. Dots indicate amino acid identity. The sampling time points (years) are indicated for the three patients.
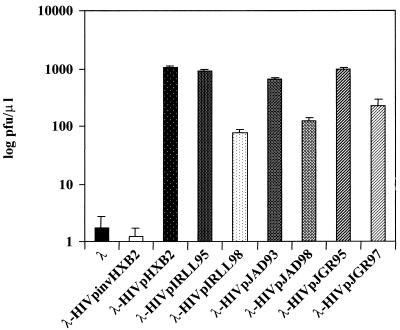
As shown in Fig. 3, the seven λ-HIV-1p proteases replicated 100 to 1,000 times more efficiently than λ-HIV-1pinvHXB2 or a wild-type λ phage in cells expressing the cI.HIV-1-cro construct. This result demonstrates that the activities of HIV-1 proteases from different sources can be monitored by using this simple bacteriophage lambda-based genetic assay. Furthermore, the low deviations observed after four independent activity determinations for each sample (Fig. 3) showed the high degree of reproducibility of this system. No significant differences in replicative capacities were observed among the four phages carrying wild-type proteases (λ-HIV-1pHXB2, -IRLL95, -JAD93, and -JGR95) (Fig. 3). Interestingly, those phages containing proteases with genotypic resistance to anti-HIV-1 protease drugs (λ-HIV-1pIRLL98, -JAD98, and -JGR97) (Fig. 2) replicated up to nine times less efficiently than naive λ-HIV-1p (Fig. 3).
FIG. 3.
Selective growth of λ-HIV-1 proteases in cells expressing the cI.HIV-1-cro construct. Cells expressing the cI.HIV-1-cro construct were infected with different λ-HIV-1 proteases or control λ-HIV-1pinvHXB2 and λ phages. As shown, selection in cI.HIV-1-cro cells resulted in the replication of λ-HIV-1 proteases, whereas the replication of λ-HIV-1pinvHXB2 or λ phages was severely compromised. Each datum point is the average of four independent experiments.
Measurement of susceptibilities of recombinant phages carrying different HIV-1 proteases to protease inhibitors.
The compounds IND, RIT, SQV, and NFV were used to evaluate whether λ-HIV-1 proteases were sensitive to different well-characterized HIV-1 protease inhibitors. Infection of E. coli cells that expressed cI.HIV-1-cro with λ-HIV-1p was carried out in the presence of various micromolar concentrations of protease inhibitors. As shown in Fig. 4A, the growth of the four drug-naive λ-HIV-1p proteases (HXB2, IRLL95, JAD93, and JGR95) was inhibited in a dose-dependent manner by the four protease inhibitors used. The inhibition of λ-HIV-1p was observed either with proteases from a laboratory strain (HXB2) or with proteases isolated from three different HIV-1-infected patients. Similar to the results shown in Fig. 3, the control λ-HIV-1pinvHXB2 did not replicate in the presence of the cI.HIV-1-cro construct in either the absence or the presence of each of the four different drugs (Fig. 4A). In addition, these four drugs did not affect the growth of λ-HIV-1pHXB2 or the λ-HIV-1pinvHXB2 when control cells that did not express the cI.HIV-1 construct were used (Fig. 4B). Thus, cellular metabolism was unaffected by the micromolar concentrations of the four drugs used in this study.
By using the data shown in Fig. 4A, the mean 50% inhibitory concentration (IC50s) for the different λ-HIV-1p and protease inhibitors were calculated. The results are given in Table 2. For all wild-type samples analyzed (λ-HIV-1pHXB2, -IRLL95, -JAD93, and -JGR95), comparable IC50s were obtained with RIT and SQV, with values ranging between 1.28 and 3.73 and 1.16 and 1.88 μM, respectively. NFV was the most potent inhibitor, with IC50s ranging between 0.17 and 0.30 μM. Three of the four phage samples showed a sensitivity to IND that was similar to that obtained with RIT and SQV (IC50 range, 1.43 to 6.31 μM), and one phage (λ-HIV-1pHXB2) was moderately resistant to IND (IC50, 17.5 μM) (Table 2).
TABLE 2.
Phenotypes and genotypes of recombinant phages containing different HIV-1 proteases
| Patient sample | Protease mutational patterna | IC50 (μM) (fold increase)b
|
|||
|---|---|---|---|---|---|
| IND | RIT | SQV | NFV | ||
| λ-HIVpHXB2 | 37S | 17.5 ± 2.3 | 1.28 ± 0.8 | 1.69 ± 0.2 | 0.30 ± 0.2 |
| λ-HIVpIRLL95 | 35D, 63P, 77I | 6.31 ± 7.2 | 1.78 ± 1.5 | 1.88 ± 1.4 | 0.17 ± 0.5 |
| λ-HIVpIRLL98 | 10I, 35D, 37D, 48V, 54V, 63P, 71V, 82A, 90M, 93L | >313 (>50) | >313 (>176) | >313 (>166) | >313 (>1,043) |
| λ-HIVpJAD93 | 63P | 1.43 ± 0.9 | 1.97 ± 1.5 | 1.38 ± 0.1 | 0.35 ± 0.2 |
| λ-HIVpJAD98 | 10I, 14V, 33F, 36M, 37C, 54V, 63P, 67F, 71V, 72M, 73S, 77I, 82A, 84V, 90M | >313 (>219) | >313 (>159) | >313 (>227) | 10.13 ± 6.8 (29) |
| λ-HIVpJGR95 | 14V, 20R, 63P, 64V, 77I | 2.76 ± 0.4 | 3.73 ± 0.8 | 1.16 ± 0.1 | 0.28 ± 0.0 |
| λ-HIVpJGR97 | 14V, 20R, 32I, 63P, 64V, 71V, 82A | 23.37 ± 1.1 (8) | >313 (>84) | 0.93 ± 0.4 (0) | 1.3 ± 0.3 (5) |
Amino acid changes relative to the clade B consensus sequence. Boldface indicates critical resistance-associated mutations.
Fold increase in the IC50 relative to the IC50 for the wild-type patient. IC50s were derived from two separate determinations and are expressed as the mean ± standard deviation.
Taken together, these results demonstrate that the lambda-based genetic screening assay can be used not only to monitor the activity of an HIV-1 protease but also to measure the extent of inhibition by different anti-HIV-1 protease drugs.
Phages carrying mutated HIV-1 proteases are resistant to different protease inhibitors.
To further demonstrate the specificity of this bacteriophage lambda-based genetic system, phages containing proteases with genotypic resistance to anti-HIV-1-protease drugs (λ-HIV-1pIRLL98, -JAD98, and -JGR97) (Fig. 2) and obtained from three patients who failed treatment with different antiprotease drugs (see Materials and Methods and Table 1) were propagated in the presence of four protease inhibitors (Fig. 4A). The sample λ-HIV-1pIRLL98, which had the mutations 48V, 82A, and 90M, all of which are critical in the development of resistance to anti-HIV-1 protease inhibitors, was highly resistant to the four drugs tested (Fig. 4A and Table 2). Similarly, the sample λ-HIV-1pJAD98, with critical mutations at residues 82A, 84V, and 90M, was highly resistant to IND (>219-fold), RIT (>159-fold), and SQV (>227-fold) but was moderately sensitive to NFV (IC50, 10.13 μM). In contrast, the phage in sample λ-HIV-1pJGR98, which had only one critical mutation, 82A, was highly resistant to RIT (>84-fold), moderately sensitive to IND (IC50, 23.37 μM; eightfold resistant), and highly sensitive to SQV and NFV (IC50s, 0.93 and 1.30 μM, respectively) (Fig. 4A and Table 2). Therefore, the resistance to the drugs shown by phages containing mutated HIV-1 proteases further demonstrates that the actions of the four protease inhibitors used in this study are specific to the HIV-1 protease. In addition, these results also indicate that this bacteriophage lambda-based genetic system is able to detect a broader phenotypic resistance to the different protease inhibitors in phages carrying larger numbers of amino acid changes at positions critical to the development of resistance to anti-HIV-1 protease drugs.
Competitive replication assay between phages carrying different HIV-1 proteases in the presence and absence of drug pressure.
As mentioned above, phages containing proteases with genotypic resistance to anti-HIV-1 protease drugs (λ-HIV-1pIRLL98, -JAD98, and -JGR97) replicated up to nine times less efficiently than their wild-type λ-HIV-1p counterparts (Fig. 3), suggesting that these mutated proteases had less efficient enzymatic activity than wild-type HIV-1 proteases. In order to confirm the relative differences in fitness observed between wild-type and mutated proteases, we used a competitive phage replication assay. Cells carrying the cI.HIV-1-cro construct were coinfected with the same amount (PFU) of λ-HIV-1pJAD93 and λ-HIV-1pJAD98. After three rounds of infection-selection, the phage DNAs present in the culture supernatants from the three infection cycles were amplified by PCR. The sequences of the former PCR products showed that after three cycles of culture competition 90% of the phage present was wild-type λ-HIV-1pJAD93 (Fig. 5A). This result further demonstrates the higher replication capacity of this phage compared to that of mutated phage λ-HIV-1pJAD98. As expected, when competition with the former phage was carried out in the presence of SQV, the wild-type phage was outgrown by mutated λ-HIV-1pJAD98 phage (Fig. 5B). Hence, the bacteriophage lambda-based genetic system also allows a direct comparison of the relative fitness and degree of drug resistance of phages carrying different drug-resistant HIV-1 proteases.
Comparison of protease inhibitor susceptibilities of recombinant phages carrying HIV-1 proteases with ex vivo HIV-1 replication assays.
To assess whether the drug susceptibility data obtained by the bacteriophage lambda-based genetic assay was equivalent to that obtained by standard ex vivo HIV-1 replication assays, the susceptibilities to the four protease inhibitors were determined in MT-4 cells with the MTT-based assay for HXB2 and patient viruses IRLL95 and IRLL98. The results given in Table 3 showed that for viruses in naive samples (λ-HIV-1pHXB2 and -IRLL95) IND, SQV, and NFV had similar IC50s, ranging between 2.6 and 20.0 nM for IND, 4.3 and 9.8 nM for SQV, and 1.9 and 8.4 nM for NFV. In contrast, RIT was a less potent inhibitor, with IC50s ranging between 116 and 169 nM. Similar to the results obtained with the lambda-based genetic assay, the virus isolated from the mutated sample IRLL98 was found to be highly resistant to RIT, SQV, and NFV and to a lesser extent to IND (Table 3). Although some differences in drug susceptibility were detected between the ex vivo assay and the lambda-based genetic system, that is, a lower sensitivity to RIT was detected by the ex vivo assay (see Table 2 and 3), these results illustrate that protease amino acid substitutions that resulted in IND, RIT, SQV, and NFV resistance in intact virus also appear to lower the sensitivity of the HIV-1 protease to the four drugs in the bacteriophage lambda-based assay.
TABLE 3.
Sensitivities of HIV-1HXB2, -IRLL95, and -IRLL98 to protease inhibitors
| Patient sample | Protease mutational patterna | IC50b (nM) (fold increase)c
|
|||
|---|---|---|---|---|---|
| IND | RIT | SQV | NFV | ||
| HXB2 | 37S | 20 ± 11.1 | 169 ± 89.8 | 9.8 ± 4.6 | 8.4 ± 5.8 |
| IRLL95 | 35D, 63P, 77I | 2.6 ± 9.8 | 116 ± 18.3 | 4.3 ± 2.0 | 1.9 ± 8.2 |
| IRLL98 | 10I, 35D, 37D, 48V, 54V, 63P, 71V, 82A, 90M, 93L | 30 ± 3.2 (11.5) | >500 (>4.5) | >500 (>116) | 96 ± 38.2 (50.5) |
Boldface indicates critical resistance-associated mutations.
IC50s were measured by the MTT assay.
Fold increase in the IC50 relative to the IC50 for the patient wild-type. IC50s were derived from two separate determinations and are expressed as the mean ± standard deviation.
DISCUSSION
In the present study we demonstrate that the bacteriophage lambda-based genetic screening assay (28) can be used not only to monitor the activities of different HIV-1 proteases but also to measure the extent of inhibition by different anti-HIV-1 protease drugs. Moreover, the phenotypes of phages containing mutated HIV-1 proteases can also be quantitatively evaluated by this assay, further increasing the utility of this lambda-based system. Protease, a key enzyme in the replication and maturation of HIV-1, has become a major target for antiviral therapy (1). Although encouraging clinical results have been obtained with anteretroviral combination therapy (23), the development of drug resistance during therapy remains a major cause of treatment failure (25). Fast and sensitive genotypic and phenotypic monitoring of resistance could improve HIV-1 therapeutics. By rapidly identifying drug resistance in infected patients, the best treatment regimens against susceptible or resistant viral populations could be designed. The rapid, simple, and safe lambda-based genetic screening system presented here will expand the biochemical approaches for the phenotypic detection of resistance to protease inhibitors. Additionally, the lambda-based assay may be also appropriate for the identification of new HIV-1 protease inhibitors because it is inexpensive, safe, and easy enough for the screening of large numbers of different drugs and replicates containing various concentrations of inhibitor.
The bacteriophage lambda-based genetic screening assay differs from current ex vivo methods for the detection of phenotypic resistance to HIV-1 protease inhibitors in that it needs micromolar concentrations of drug (Table 2) to inhibit 50% of phage replicative events (IC50), whereas the IC50s calculated by the ex vivo assays are in the nanomolar range for the four protease inhibitors tested (Table 3) (12, 13, 27). A possible explanation might be the differences in cell uptake and/or metabolism between bacteria and human eucaryotic cells. Nevertheless, the results shown in Fig. 4 for phages carrying either wild-type or mutated proteases demonstrate that it can be used to monitor HIV-1 protease inhibition with micromolar concentrations of drug. Phages containing proteases with different patterns of genotypic resistance (λ-HIV-1pIRLL98, -JAD98, and -JGR97) and that were obtained from patients failing different antiretroviral treatments were compared. Differences in their phenotypic resistance to the four protease inhibitors could be monitored (Table 1 and Fig. 4A). Thus, phages containing the JGR97 protease that have only one substitution (82A) critical to the development of current protease drug resistance were phenotypically resistant to RIT and IND (>84- and 8-fold, respectively) but were susceptible to SQV and were moderately sensitive to NFV (Table 2). These findings are in agreement with those of previous studies carried out by using ex vivo assays, in which the 82A mutation was implicated in cross-resistance to IND and RIT, but in the absence of other critical mutations, viruses carrying this substitution were still sensitive to SQV and NFV (13, 16, 21, 29). At the time when sample JGR97 was taken, patient JGR had begun a new treatment regimen that included SQV and NFV plus two nucleoside analogues. Interestingly, the plasma viral load in this patient (<20 copies/ml) had remained undetectable after more than 2 years of treatment with this antiretroviral therapy (data not shown). Likewise, the results obtained in the present study with phage λ-HIV-1pIRLL98, the one with three substitutions that are critical to the development of phenotypic protease inhibitor resistance (48V, 82A and 90M) (Fig. 2), confirmed previous reports that revealed the loss of susceptibility to IND, RIT, SQV, and NFV of this mutant with multiple mutations (13, 27). Patient IRLL and patient JAD did not achieve viral RNA suppression in plasma after treatment with several drug combinations, stressing the dangers of selecting suboptimal sequential antiretroviral therapies for HIV-1-infected patients (2). Overall, these findings confirm the clinical management benefits of HIV-1 resistance testing. Because some quantitative differences were observed when the bacteriophage lambda-based genetic screening system was compared to eucaryotic cell culture assays (compare Tables 2 and 3), the lambda-based assay can be seen as a complement to current viral propagation assays for the monitoring of drug sensitivities. One advantage of the lambda-based assay is that current ex vivo methods rely on human cell culture systems infected with live virus. These methods are time-consuming and require the use of biosafety precautions. The gene for HIV-1 protease can be amplified from the blood of infected patients, ligated to predigested lambda DNA, and packaged in vitro in 24 h. The lambda-based genetic screen for the determination of the HIV-1 protease phenotype needs only 12 additional h (see Materials and Methods). Therefore, the genetic screening assay described here can easily be performed in 2 to 3 working days. In contrast, current cell culture systems used to detect phenotypic resistance to HIV-1 inhibitors, such as the recombinant virus assay (13) or the coculture of infected peripheral blood mononuclear cells with seronegative donor peripheral blood mononuclear cells (24), require at least 3 to 4 weeks for virus isolation and phenotypic characterization. By taking advantage of the genetics of bacteriophage lambda, an additional important aspect of this genetic assay is that it allows the rapid detection of functional minority viral populations. The selection of different phage clones (Fig. 1) and reexamination of their phenotypes and genotypes might facilitate the identification of minority strains of resistant viruses. Since multiple-drug-resistant mutants were not commonly found in bulk sequences from previously treated patients who failed antiretroviral combination therapy (19), detection of minor populations of resistant virus in those patients may improve the utility of genotypic and phenotypic monitoring of resistance for clinical management.
Differences in HIV-1 protease activity can also be measured with the bacteriophage lambda-based genetic screening system. The activities of the protease inhibitor-resistant phages λ-HIV-1pIRLL98, λ-HIV-1pJAD98, and λ-HIV-1pJGR97 are reduced nine-, five-, and fourfold, respectively, in comparison to the activities of the phages containing naive proteases (Fig. 3). It is noteworthy that λ-HIV-1pIRLL98, the phage most resistant to the four protease inhibitors tested, was the phage that showed the lowest level of protease activity (Fig. 3). Several protease inhibitor resistance mutations, alone or in combination, can reduce the replicative capacity of HIV-1 (6, 10, 18, 20, 31). In addition to the data provided by the independent determination of the protease activity of each λ-HIV-1p (Fig. 3), the system described here also allows direct comparison of relative HIV-1 protease fitness. The relative fitness of two phages that differ in their HIV-1 protease genotypes can be directly compared since two phage populations in a culture compete with each other until one phage outgrows the other (Fig. 5). We have found that in the absence of protease inhibitors the mutated protease obtained from patient JAD after 2.5 years of protease inhibitor therapy (Table 1) showed a lower level of fitness than the baseline wild-type virus (Fig. 5A). In the presence of drug, however, λ-HIV-1pJAD98 outgrew λ-HIV-1pJAD93 (Fig. 5B), confirming the replication advantage of the mutant phage under drug pressure. The relative fitness in the absence of drugs, as measured here, can be associated with the catalytic efficiency of the mutant enzyme. Estimation of the effect of drug resistance on viral fitness may have clinical relevance because it can affect viral pathogenesis and transmissibility. The potential ability to rapidly analyze the relative fitness of drug-resistant proteases in this system may aid in predicting the viral genotypic and phenotypic changes in the course of HIV-1 infection and antiviral therapy.
Finally, another application of the bacteriophage lambda-based genetic screening system is the characterization of an in vitro-generated library of mutated HIV-1 proteases. The analysis of a large collection of mutant viral proteases will ease studies on structure-function relationships, viral genetic evolution, and novel drug-resistant mutants. Since this system has the potential to isolate enzymes with low levels of catalytic activity (28), coupling of random sequences with positive genetic selection will allow the study of functional mutants which are unlikely to be isolated from HIV-1-infected patients. The former mutants may be of interest in the characterization of catalytic properties of proteases in the absence or the presence of drugs (M. Cabana and M.-A. Martínez, unpublished data). Identification and characterization of new residues implicated in protease inhibitor resistance from a random library of HIV-1 protease mutants may also be useful in the design and screening of new antiprotease inhibitors.
In summary, the bacteriophage lambda-based genetic screening system described here may be of use in predicting the activities and phenotypes of HIV-1 proteases in the course of HIV-1 infection and antiretroviral therapy. Characterization of mutant HIV-1 proteases and the search for new inhibitors can be also be facilitated by this genetic screening system.
ACKNOWLEDGMENTS
We thank H. J. Sices and T. M. Kristie for the cI.HIV-cro constructs and for helping us in setting up the bacteriophage lambda-based genetic system.
This work was supported by the irsiCaixa Foundation and by grants from the Spanish Fondo de Investigación Sanitaria 98/0054-03 and 98/0868. M. A. Martínez visited T. M. Kristie's laboratory at the National Institutes of Health (Bethesda, Md.) with support from a fellowship from the Human Frontier Science Program (Strasbourg, France).
REFERENCES
- 1.Boden D, Markowitz M. Resistance to human immunodeficiency virus type 1 protease inhibitors. Antimicrob Agents Chemother. 1998;42:2775–2783. doi: 10.1128/aac.42.11.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cabana M, Clotet B, Martinez M A. Emergence and genetic evolution of HIV-1 variants with mutations conferring resistance to multiple reverse transcriptase and protease inhibitors. J Med Virol. 1999;59:480–490. doi: 10.1002/(sici)1096-9071(199912)59:4<480::aid-jmv10>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 3.Chun T W, Stuyver L, Mizell S B, Ehler L A, Mican J A, Baseler M, Lloyd A L, Nowak M A, Fauci A S. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci USA. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coffin J M, Hughes H S, Varmus H E. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 5.Condra J H, Schleif W A, Blahy O M, Gabryelski L J, Graham D J, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M, et al. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;374:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 6.Croteau G, Doyon L, Thibeault D, McKercher G, Pilote L, Lamarre D. Impaired fitness of human immunodeficiency virus type 1 variants with high-level resistance to protease inhibitors. J Virol. 1997;71:1089–1096. doi: 10.1128/jvi.71.2.1089-1096.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finzi D, Hermankova M, Pierson T, Carruth L M, Buck C, Chaisson R E, Quinn T C, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho D D, Richman D D, Siliciano R F. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 8.Furtado M R, Callaway D S, Phair J P, Kunstman K J, Stanton J L, Macken C A, Perelson A S, Wolinsky S M. Persistence of HIV-1 transcription in peripheral-blood mononuclear cells in patients receiving potent antiretroviral therapy. N Engl J Med. 1999;340:1614–1622. doi: 10.1056/NEJM199905273402102. [DOI] [PubMed] [Google Scholar]
- 9.Gulick R M, Mellors J W, Havlir D, Eron J J, Gonzalez C, McMahon D, Richman D D, Valentine F T, Jonas L, Meibohm A, Emini E A, Chodakewitz J A. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N Engl J Med. 1997;337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 10.Gulnik S V, Suvorov L I, Liu B, Yu B, Anderson B, Mitsuya H, Erickson J W. Kinetic characterization and cross-resistance patterns of HIV-1 protease mutants selected under drug pressure. Biochemistry. 1995;34:9282–9287. doi: 10.1021/bi00029a002. [DOI] [PubMed] [Google Scholar]
- 11.Hammer S M, Squires K E, Hughes M D, Grimes J M, Demeter L M, Currier J S, Eron J J, Jr, Feinberg J E, Balfour H H, Jr, Deyton L R, Chodakewitz J A, Fischl M A. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. AIDS Clinical Trials Group 320 Study Team. N Engl J Med. 1997;337:725–733. doi: 10.1056/NEJM199709113371101. [DOI] [PubMed] [Google Scholar]
- 12.Hecht F M, Grant R M, Petropoulos C J, Dillon B, Chesney M A, Tian H, Hellmann N S, Bandrapalli N I, Digilio L, Branson B, Kahn J O. Sexual transmission of an HIV-1 variant resistant to multiple reverse-transcriptase and protease inhibitors. N Engl J Med. 1998;339:307–311. doi: 10.1056/NEJM199807303390504. [DOI] [PubMed] [Google Scholar]
- 13.Hertogs K, de Bethune M P, Miller V, Ivens T, Schel P, Van Cauwenberge A, Van Den Eynde C, Van Gerwen V, Azijn H, Van Houtte M, Peeters F, Staszewski S, Conant M, Bloor S, Kemp S, Larder B, Pauwels R. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob Agents Chemother. 1998;42:269–276. doi: 10.1128/aac.42.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirsch M S, Conway B, D'Aquila R T, Johnson V A, Brun-Vezinet F, Clotet B, Demeter L M, Hammer S M, Jacobsen D M, Kuritzkes D R, Loveday C, Mellors J W, Vella S, Richman D D. Antiretroviral drug resistance testing in adults with HIV infection: implications for clinical management. International AIDS Society—USA Panel. JAMA. 1998;279:1984–1991. doi: 10.1001/jama.279.24.1984. [DOI] [PubMed] [Google Scholar]
- 15.Ibanez A, Puig T, Elias J, Clotet B, Ruiz L, Martinez M A. Quantification of integrated and total HIV-1 DNA after long-term highly active antiretroviral therapy in HIV-1-infected patients. AIDS. 1999;13:1045–1049. doi: 10.1097/00002030-199906180-00007. [DOI] [PubMed] [Google Scholar]
- 16.Markowitz M, Mo H, Kempf D J, Norbeck D W, Bhat T N, Erickson J W, Ho D D. Selection and analysis of human immunodeficiency virus type 1 variants with increased resistance to ABT-538, a novel protease inhibitor. J Virol. 1995;69:701–706. doi: 10.1128/jvi.69.2.701-706.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez M A, Cabana M, Ibanez A, Clotet B, Arno A, Ruiz L. Human immunodeficiency virus type 1 genetic evolution in patients with prolonged suppression of plasma viremia. Virology. 1999;256:180–187. doi: 10.1006/viro.1999.9601. [DOI] [PubMed] [Google Scholar]
- 18.Martinez-Picado J, Savara A V, Sutton L, D'Aquila R T. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J Virol. 1999;73:3744–3752. doi: 10.1128/jvi.73.5.3744-3752.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez-Picado J, Sutton L, De Pasquale M P, Savara A V, D'Aquila R T. Human immunodeficiency virus type 1 cloning vectors for antiretroviral resistance testing. J Clin Microbiol. 1999;37:2943–2951. doi: 10.1128/jcm.37.9.2943-2951.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maschera B, Darby G, Palu G, Wright L L, Tisdale M, Myers R, Blair E D, Furfine E S. Human immunodeficiency virus. Mutations in the viral protease that confer resistance to saquinavir increase the dissociation rate constant of the protease-saquinavir complex. J Biol Chem. 1996;271:33231–33235. doi: 10.1074/jbc.271.52.33231. [DOI] [PubMed] [Google Scholar]
- 21.Molla A, Korneyeva M, Gao Q, Vasavanonda S, Schipper P J, Mo H M, Markowitz M, Chernyavskiy T, Niu P, Lyons N, Hsu A, Granneman G R, Ho D D, Boucher C A, Leonard J M, Norbeck D W, Kempf D J. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat Med. 1996;2:760–766. doi: 10.1038/nm0796-760. [DOI] [PubMed] [Google Scholar]
- 22.Myers G, Foley B, Mellors J W, Korber B, Jeang K T, Wain-Hobson S. Human retroviruses and AIDS database. Los Alamos, N.Mex: Los Alamos National Laboratory; 1996. [Google Scholar]
- 23.Palella F J, Jr, Delaney K M, Moorman A C, Loveless M O, Fuhrer J, Satten G A, Aschman D J, Holmberg S D. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med. 1998;338:853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 24.Pauwels R, Balzarini J, Baba M, Snoeck R, Schols D, Herdewijn P, Desmyter J, De Clercq E. Rapid and automated tetrazolium-based colorimetric assay for the detection of anti-HIV compounds. J Virol Methods. 1988;20:309–321. doi: 10.1016/0166-0934(88)90134-6. [DOI] [PubMed] [Google Scholar]
- 25.Perrin L, Telenti A. HIV treatment failure: testing for HIV resistance in clinical practice. Science. 1998;280:1871–1873. doi: 10.1126/science.280.5371.1871. [DOI] [PubMed] [Google Scholar]
- 26.Ptashne M. A genetic switch. Cambridge, Mass: Cell Press; 1986. [Google Scholar]
- 27.Shafer R W, Winters M A, Palmer S, Merigan T C. Multiple concurrent reverse transcriptase and protease mutations and multidrug resistance of HIV-1 isolates from heavily treated patients. Ann Intern Med. 1998;128:906–911. doi: 10.7326/0003-4819-128-11-199806010-00008. [DOI] [PubMed] [Google Scholar]
- 28.Sices H J, Kristie T M. A genetic screen for the isolation and characterization of site-specific proteases. Proc Natl Acad Sci USA. 1998;95:2828–2833. doi: 10.1073/pnas.95.6.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tisdale M, Myers R E, Maschera B, Parry N R, Oliver N M, Blair E D. Cross-resistance analysis of human immunodeficiency virus type 1 variants individually selected for resistance to five different protease inhibitors. Antimicrob Agents Chemother. 1995;39:1704–1710. doi: 10.1128/aac.39.8.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong J K, Hezareh M, Gunthard H F, Havlir D V, Ignacio C C, Spina C A, Richman D D. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 31.Zennou V, Mammano F, Paulous S, Mathez D, Clavel F. Loss of viral fitness associated with multiple Gag and Gag-Pol processing defects in human immunodeficiency virus type 1 variants selected for resistance to protease inhibitors in vivo. J Virol. 1998;72:3300–3306. doi: 10.1128/jvi.72.4.3300-3306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang L, Ramratnam B, Tenner-Racz K, He Y, Vesanen M, Lewin S, Talal A, Racz P, Perelson A S, Korber B T, Markowitz M, Ho D D. Quantifying residual HIV-1 replication in patients receiving combination antiretroviral therapy. N Engl J Med. 1999;340:1605–1613. doi: 10.1056/NEJM199905273402101. [DOI] [PubMed] [Google Scholar]