Abstract
Over 50% of all cancer patients are treated with radiation therapy (RT). However, RT is often insufficient as a monotherapy and requires a non-toxic radiosensitizer. Squalene epoxidase (SQLE) controls cholesterol biosynthesis by converting squalene to 2,3-oxidosqualene. Given that SQLE is frequently overexpressed in human cancer, this study investigated the importance of SQLE in breast cancer (BC) and non-small cell lung cancer (NSCLC), two cancers often treated with RT. SQLE-positive immunohistochemical staining was observed in 68% of BC and 56% of NSCLC specimens versus 15% and 25% in normal breast and lung tissue, respectively. Importantly, SQLE expression was an independent predictor of poor prognosis, and pharmacological inhibition of SQLE enhanced breast and lung cancer cell radiosensitivity. In addition, SQLE inhibition enhanced sensitivity to PARP inhibition. Inhibition of SQLE interrupted homologous recombination by suppressing ATM activity via the translational upregulation of wild-type p53-induced phosphatase (WIP1), regardless of the p53 status. SQLE inhibition and subsequent squalene accumulation promoted this upregulation by triggering the endoplasmic reticulum (ER) stress response. Collectively, these results identify a novel tumor-specific radiosensitizer by revealing unrecognized crosstalk between squalene metabolites, ER stress, and the DNA damage response (DDR). Although SQLE inhibitors have been used as antifungal agents in the clinic, they have not yet been used as antitumor agents. Repurposing existing SQLE-inhibiting drugs may provide new cancer treatments.
Keywords: squalene epoxidase, radiation therapy, endoplasmic reticulum stress, DNA damage response
INTRODUCTION
The cure of solid tumors requires effective local control. Radiotherapy (RT) is an important treatment for more than 50% of all cancer patients (1). Although there is a genetic basis for variations in radiosensitivity and resistance (2), there is a lack of precision medicine-based strategies for the large majority of RT patients, resulting in toxicity and over- or under-treatment. In addition, it is difficult to leave the tissue surrounding tumors unharmed during RT because of the nature of radiation beam delivery, significantly limiting the doses that can be administered. Thus, RT is often an insufficient therapy on its own, emphasizing the need for radiosensitizers.
The proteins required for cholesterol biosynthesis can be upregulated to meet the high demand of tumor metabolism. Squalene epoxidase (squalene monooxygenase, SQLE) controls cholesterol biosynthesis by converting squalene to 2, 3-oxidosqualene. SQLE is likely an oncogene as it promotes oncogenic signaling (3–7). Indeed, frequent SQLE amplification and differential expression in broad types of cancer, especially breast cancer (BC), have been reported (8–11). Given that the correlation between gene transcripts and protein abundance in tumors can be low, and small-molecule SQLE inhibitors target protein, not mRNA, it is important to investigate SQLE protein expression in cancer and evaluate its significance in patient prognosis.
SQLE is a limiting factor for cholesterol biosynthesis but fewer studies have focused on targeting SQLE to treat cholesterol-related diseases. Nevertheless, inhibitors targeting SQLE (e.g., terbinafine [TF]) have been approved for antifungal therapy which can be quickly translated into cancer therapy. Inhibitors targeting mammalian SQLE (e.g., NB-598) are currently under development. However, a recent study suggested that SQLE inhibition could cause normal tissue toxicity at efficacious doses against tumors in vivo (12), limiting the use of NB-598 as monotherapy. Given that synergistic combination therapy is often advantageous over monotherapeutic approaches in cancer treatment, pre-clinical testing for new targeted therapy must be done in the context of existing standard-of-care therapy to facilitate entry of these agents into clinical trials and increase their potential for success. Therefore, it is important to study the efficacy of SQLE inhibition in combination with standard therapy at non-toxic doses.
Ionizing radiation (IR), used in RT, activates the ataxia-telangiectasia mutated (ATM) kinase, which responds to the presence of DNA double-strand breaks (DSBs) by activating the cell cycle checkpoint and DNA repair mechanisms (13). One such mechanism is homologous recombination (HR), a major pathway for repairing DSBs and stalled or collapsed replication forks. Thus, ATM is important for HR regulation during replication stress (RS) and fixing IR-induced DSBs in cancer cells (14–16). ATM activity is negatively regulated by the nuclear protein phosphatase known as wild-type p53-induced phosphatase (WIP1) (17). Following DNA damage, increased WIP1 expression reduces the activation of ATM-dependent signaling (18). Thus, WIP1 plays a role in the DNA damage response (DDR) by dephosphorylating DDR proteins (19). It is well established that a p53-dependent mechanism upregulates WIP1 after DNA damage (18,19).
Protein synthesis, folding, and major steps of cholesterol synthesis occur in the endoplasmic reticulum (ER). ER stress results from excess levels of unfolded or misfolded proteins and toxic lipids in the ER (20). This accumulation activates the ER stress response, also called the unfolded protein response (UPR) (20,21). The UPR cause a general decrease in protein translation but upregulates the translation of certain individual protein (22). In addition, lipid droplets (LDs), which are organelles that store lipids for energy and protect cells from lipotoxicity, can also form following ER stress (20).
Here, we reveal high SQLE protein expression in patient BC and non-small cell lung cancer (NSCLC) samples compared to normal adjacent breast and lung tissue. Targeting SQLE enhanced sensitivity of cancer cells to radiotherapy by decreasing ATM activity and subsequent HR reduction through the squalene accumulation-induced ER stress response and the associated p53- independent increase in WIP1 translation. Interestingly, loss of the SQLE activity had no impact on cholesterol levels; however, it dramatically increased the levels of the intermediate metabolite squalene. SQLE inhibition induced radiosensitivity, and the effects on the WIP1-ATM axis and ER stress were dependent on squalene. Our study identified a novel radiosensitizer and unrecognized crosstalk between squalene, ER stress, and DDR. Our study has the potential to repurpose a clinically available SQLE inhibitor for combination therapy with RT.
MATERIALS AND METHODS
Clinical samples and SQLE immunohistochemistry
The retrospective BC cohort consisted of 442 BC individuals with primary early-stage BC (stages I-III) with complete data available. One group of 294 patients was recruited at the First Affiliated Hospital of Sun Yat-Sen University (Guangzhou, China) from 2004 to 2008, with a median of 127 months (19–171 months) follow-up. The data cutoff for the OS and DFS analysis was September 28, 2018. Another group of 148 patients with complete clinical and pathologic variables and 125 paired ANTs was recruited from 2014 to 2018. The study was approved by the Ethics Committee of the First Affiliated Hospital of Sun Yat-Sen University and conducted following the Declaration of Helsinki and Good Clinical Practice guidelines. All patients gave written informed consent for the analysis of tumor samples for biomarker assessment. Inclusive criteria were described previously (23,24). Assignment of BC subtype and tissue microarray construction were previously described (23,24). Additional information was included in Supplemental Materials and Methods.
Lung cancer TMA cohorts were purchased from Shanghai OUTDO Co with full follow-up data (First cohort: HLugA180Su06, HLugS180Su01) and Wuhan Service Bio without follow-up data (Second cohort: LAC-1401, LAC-1403). The retrospective cohort consisted of 179 individuals with early-stage I–III lung cancer enrolled in our study from July 1, 2004 to September 8, 2009. The samples included 98 adenocarcinomas, 81 squamous carcinomas, and 170 paired adjacent normal lung or bronchus tissues. The secondary cohort consisted of 125 lung cancer tumors from patients with stage I–III disease, including 60 adenocarcinomas, 65 squamous carcinomas, and 33 paired adjacent normal lung or bronchus tissues. SQLE immunohistochemistry was carried out as previously reported (24). IgG-rabbit polyclonal antibody against SQLE (1:50 dilution; 12544-1-AP, Proteintech) was used for SQLE detection by IHC. This antibody has been used and validated for IHC staining in previous studies (25,26).
Cell lines, plasmids, inhibitors, and transfection
The breast (MCF-7, BT-549, HCC1143, HCC-38, and MDA-MB-231) and lung (H1299, H1437) cancer cell lines were purchased from ATCC and authenticated via STR profiling by the MCIC Genomics core at Ohio State University. MCF-7 and H1299 were cultured in DMEM medium (Hyclone), and HCC1437, BT-549, HCC1143, and HCC-38 were grown in RIPM 1640 medium (Hyclone). All media were supplemented with 10% fetal bovine serum (FBS; Gibco). All cells were grown in a humidified atmosphere containing 5% CO2 at 37°C. All cell lines were free of mycoplasma, as determined using the LookOut Mycoplasma PCR Detection Kit (MP0035, Sigma). The shRNAs were purchased from Sigma-Aldrich. The SQLE inhibitors terbinafine (A8533) and NB-598 A3645) and PARP inhibitor AZD2281 (olaparib, A4154) were obtained from APExBIO Technology. The squalene synthase inhibitor TAK-475 (SML-2168) was purchased from MilliporeSigma. The ATM kinase inhibitor KU-55933 (S1092) was obtained from Selleck Chemicals. All chemicals were dissolved in dimethyl sulfoxide (DMSO). Transfection was performed with Lipofectamine 2000 using the manufacturer’s recommendations (Invitrogen).
MTT, colony formation, and comet assays
The MTT, colony formation, and comet assays were performed as previously described (27).
Homologous recombination assay
Cycloheximide assay
The cycloheximide assay was performed as previously published (27)
Squalene and cholesterol detection by liquid chromatography-mass spectrometry (LC-MS)
The cells were extracted with methanol, dried using a SpeedVac, and re-extracted with 400 μL of chloroform:methanol (2:1) containing 1 ppm of heavy-labeled 5C13-cholesterol and was detected by LC-MS (see supplementary data for details).
Tumor xenograft studies
All animal procedures were approved by the Institutional Animal Care and Use Committee of the Ohio State University (OSU) (Columbus, OH). Male and female athymic nude mice (Strain code 553, NCI Frederick; 4–5 weeks of age) were obtained from the OSU Target Validation Shared Resource. The mice were bred at the University Laboratory Animal Resources of OSU and housed five per cage in conventional barrier conditions with a 12 h light/dark cycle at 22°C, with free access to water and food. NCI-H1299 cells (5×105 cells in 100 μL of sterile PBS with 50% Matrigel (Corning) were subcutaneously injected into both mouse flanks. Tumors measurements were performed every two days using digital calipers, and the tumor volume (mm3) was calculated using the formula: volume = (width)2 × length/2. When tumor volume reached 100 mm3, mice (n = 10) received the following agents either alone or in combination: vehicle (DMSO) or 60 mg/kg TF daily from days 1 to 14 via intraperitoneal injection; local IR (2 Gy) to the tumor on days 5 to 9, with the body shielded. The endpoint for the survival study was when the tumor volume reached five times the volume measured on day 0 or when the end of the follow-up period was reached. IR was administered using an RS-2000 irradiator (Rad Source Company, USA) in the Small Animal Imaging Core of OSU (Columbus, OH). All animal procedures are adherence to the NIH Guide for the Care and Use of Laboratory Animals,
Serum and organ collection and pathological analysis
See supplementary data for details
Analysis of polysome-associated WIP1 mRNA expression
Sucrose density gradient centrifugation was employed to separate the ribosome fractions following treatment of cells with drugs. Fifteen minutes before collection, cycloheximide (100 μg/mL) was added to the culture medium. Cells were washed in ice-cold PBS containing 100 μg/mL cycloheximide, and harvested in polysome lysis buffer (5 mM Tris-HCl, pH7.5, 2.5 mM MgCl2, 1.5 mM KCl, 2 mM DTT, 0.5% Triton X-100, 0.5% sodium deoxycholate, 100 μg/mL cycloheximide, 200 U/mL RNAsin, 0.2 mg/mL heparin, and protease inhibitors). Cells were incubated on ice for 15 min and then centrifuged at 10,000 × g for 10 min at 4 °C. The supernatant (4 mg protein) was layered on a pre-chilled 10–50% linear sucrose gradient preparing in the gradient buffer (5 mM Tris-HCl, pH7.5, 2.5 mM MgCl2, 1.5 mM KCl, 2 mM DTT, 100 μg/mL cycloheximide, 40 U/mL RNAsin, 0.1 mg/ml heparin, and protease inhibitors), and then centrifuged in a Thermo Fisher Scientific TH-641 rotor at 250,000 × g for 2.5 h at 4 °C. Gradients were fractionated while monitoring absorbance at A254 with a Density Gradient Fractionation System (Brandel, Gaithersburg, MD, USA). RNA was isolated from gradient fractions using TRIzol Reagent (Invitrogen) according to the manufacturer’s protocol. The RNA was dissolved in 20 μL of nuclease-free water. RNA concentration was determined using a Nanodrop, and the quality of RNA was evaluated on a 1% agarose gel. The purified RNA (1 μg) was reverse transcribed to cDNA with the Roche Transcriptor First Strand cDNA Synthesis Kit and then diluted 1:5 with nuclease-free water. The qRT-PCR was performed using a pair of gene-specific primers. The percent distribution of the mRNAs across the fractions was calculated using the cycle threshold (CT) values.
Statistics
Statistical analysis was performed using IBM SPSS 23.0 (Armonk, NY, USA) and GraphPad Prism 7.0 (San Diego, CA, USA). Associations between SQLE expression and clinical or prognostic factors were evaluated by the chi-square test and logistic regression. Comparisons of SQLE IHC scores were performed using the Mann–Whitney test. Survival was determined using the Kaplan–Meier method, and differences between groups were tested using the log-rank test. Univariate and multivariate analyses were performed with the Cox proportional hazard regression model. All in vitro experiments were performed with biological triplicates and reproduced at least twice. The mean differences between groups were compared using the Student’s t-test (two-tailed, two groups) or ANOVA (more than 2 groups). Additional comparisons were done with the Bonferroni’s post-hoc test. Correlations were evaluated with the Spearman rank correlation. Statistically significance was defined as P < 0.05. The asterisks define the degree of significance as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Differences that were not significant are indicated by ns.
RESULTS
SQLE expression and poor prognosis in BC and NSCLC
SQLE protein expression was examined by IHC in 442 BC tissue patient samples which were from two cohort of BC as described in our previous publication (24) including 36 ductal carcinomas in situ (DCIS) and 406 invasive ductal carcinomas (IDC). It was found that 67.6% of the samples were SQLE-positive compared to only 15.2% of the 125 adjacent normal tissues (ANTs) (Fig. 1A, P < 0.001). The median SQLE immunoreactive score (IRS) was significantly higher for the BC samples than ANTs (Fig. 1B, P < 0.001). Representative SQLE-staining images of a matched BC and ANT are shown (Fig. 1C). Although a relatively high positive staining rate was observed for the triple negative BC (TNBC) subtype (70.7%), the differences in SQLE protein expression between each subtype did not reach statistical significance (Fig. 1D).
Figure 1. SQLE is overexpressed and correlated with poor survival in BC and NSCLC.
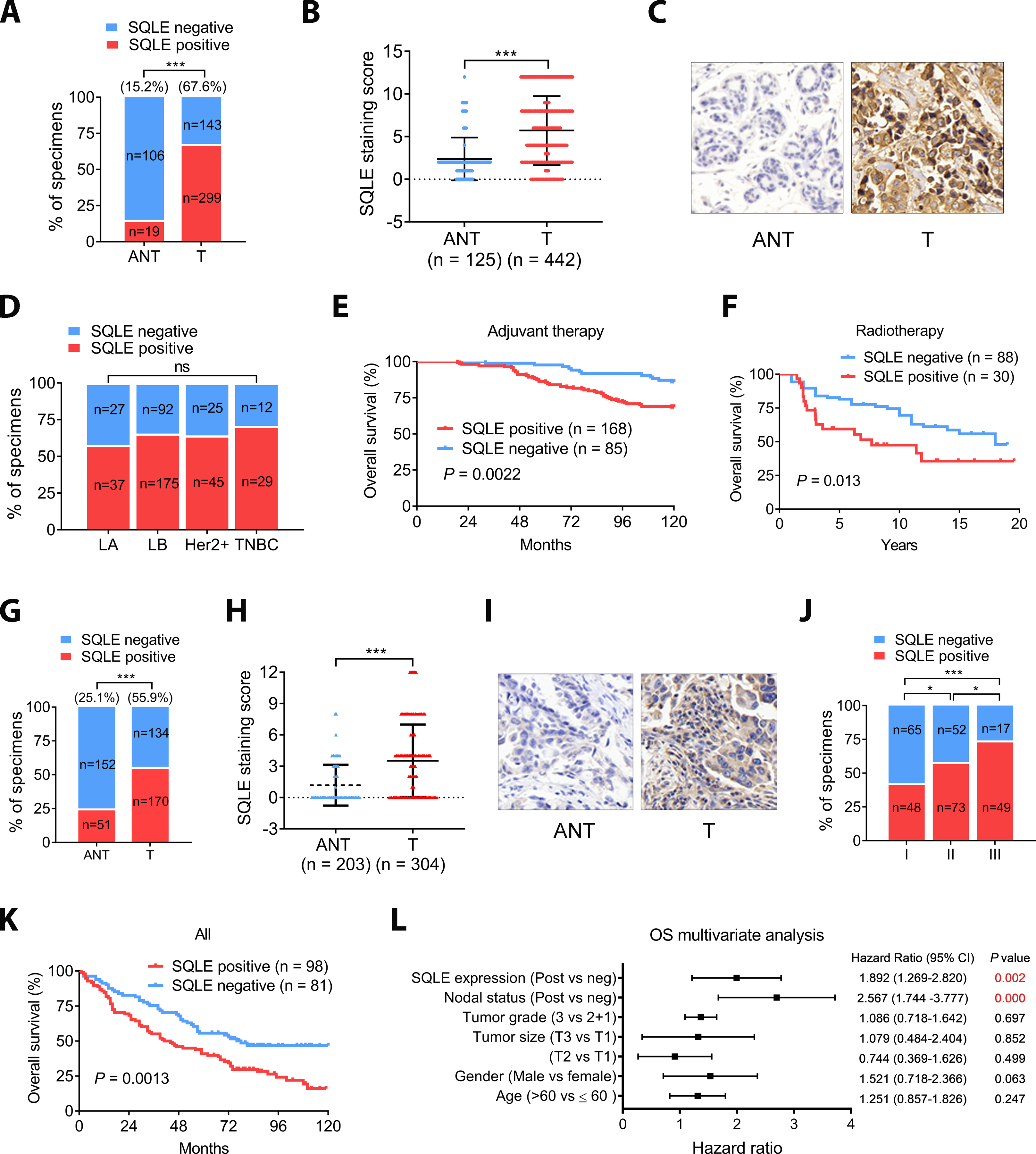
A, SQLE expression was elevated in BC. B, The median SQLE IHC score (Immunoreactive score, IRS) of the BC tumors was significantly higher than that of the ANTs. IRS was determined from the staining intensity (SI) and percentage of positive cells (PP): IRS = SI × PP. An IRS ≥ 3 score was classified as SQLE positive. Groups were compared with the Mann–Whitney test. C, Representative IHC images of matched BC and ANT tissue. Magnification 40×. D, Association of SLQE expression and BC subtypes. E, High SQLE expression correlates with poor survival in a BC population on adjuvant therapy. Kaplan-Meier survival analysis of our cohorts. F, TCGA data analysis showed that high SQLE expression correlates with poor survival in a BC population administered radiation therapy (GSE10374). G, SQLE expression was elevated in NSCLC. SQLE protein expression in NSCLC and ANT was determined using IHC. H, The median SQLE staining of NSCLC tumors was significantly higher than in the ANTs. I, Representative IHC images of NSCLC tumors with more than or less than 3-year survival. Magnification 40×. J, The percentage of positive SQLE staining in NSCLC tumors of different TNM stages. K, High SQLE expression correlated with poor survival in NSCLC. L, Multivariate cox regression analysis of OS in our cohort of lung cancer. Data in (A, D, G and H) was evaluated by chi-square test and in (B and H) was evaluated by Mann-Whitney test. Survival in (E, F and K) were determined using the Kaplan-Meier method, and differences between groups were tested using the log-rank test. Multivariate analysis in (L) was performed with the cox proportional hazard regression model.*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Kaplan-Meier analysis indicated that the 10-year overall survival (OS) and disease-free survival (DFS) were 74.5% and 67.1%, respectively, for the 294 individuals in the first cohort with complete follow-up data. The OS of all individuals was significantly shorter for those patients with SQLE-positive tumors than those that were negative (Supplemental Fig. S1A, 67.83% vs. 85.26%). Individuals with SQLE-positive tumors also had significantly worse DFS than those with SQLE-negative BC (Supplemental Fig. S1B, 59.3% vs. 83.16%). In the subset of BC patients who received adjuvant cancer therapy, SQLE expression was associated with poor prognosis (Fig. 1E). Although the two BC cohorts in our study had no information on RT status, SQLE expression might also be associated with poor prognosis for patients that receive RT because there is a strong correlation between responses to IR and chemotherapy (2). Indeed, the analysis of BC cohort GSE10374 that had RT as a therapy component suggested that SQLE expression shortened recurrence-free survival after RT (Fig. 1F). Moreover, high SQLE expression was previously shown to confer therapeutic radioresistance to pancreas cancer cells (30). We also determined the predictive value of SQLE expression. For the first cohort (294 cases), nodal-positive status and positive SQLE expression were identified as significant predictors of poor OS and DFS by both univariate (Supplemental Fig. S1C) and multivariate (Supplemental Fig. S1D) analysis.
SQLE protein expression was also observed in ~56% (170/304) of the NSCLC samples analyzed compared to only 25.1% (51/203) of normal lung tissue controls (Fig. 1G). Of note, although the rate of positive staining in NSCLC was high, the intensity was generally lower than in BC (Fig. 1H and I, vs Fig. 1B and C). SQLE protein expression was associated with tumor grade and poor prognosis in NSCLC (Fig. 1J and K) and was identified as predictors of poor OS in NSCLC by multivariate analysis (Fig. 1L). Together, our retrospective clinical study suggested that SQLE protein expression is high in BC and NSCLC and associated with poor prognosis. SQLE was an independent predictor of poor prognosis in our BC cohort. SQLE expression also predicted the outcome of BC that received adjuvant therapy and radiotherapy.
SQLE inhibition leading to enhancement of radiosensitivity
Given the significant association between SQLE expression and poor prognosis, especially in a subgroup of BC patients who received RT (Fig. 1F), we focused on targeting SQLE with TF combined with RT. Because ideal radiosensitizers should have no toxicity alone but enhance radiosensitivity, we evaluated TF in combination with RT at a concentration that had no significant impact on cell growth by itself. The IC50s for TF in the MCF-7 and HCC-38 BC cell lines that carry multiple copies of the SQLE gene were 70 μM and 163 μM, respectively (Fig. 2A). Its IC50 for the NSCLC cell line H1299 was 90 μM (Fig. 2A). Therefore, we choose 50 μM for the combination experiments with RT for all three cell lines since this concentration had little or no impact on cell growth (Fig. 2A, Fig. S2). TF increased the radiosensitivity of all three cell lines based on colony formation (Fig. 2B). We also used the linear-quadratic (LQ) model to fit each survival curve of Fig. 2B (Supplemental Fig. S3). SQLE inhibition decreased the α/β values of the survival curve, further suggesting increased radiosensitivity mediated by TF. Similarly, sub-IC50 concentrations of NB-598 (2 μM, 8 μM) significantly increased the sensitivity of the three cell lines to RT (Supplemental Fig. S4A and B). In contrast, SQLE inhibition had no impact on the radiosensitivity of MDA-MB-231 TNBC cells, which do not express SQLE (Supplemental Fig. S4C, (8)). Lastly, SQLE knockdown by shRNA resulted in enhanced radiosensitivity (Supplemental Fig. S5), consistent with the results obtained using SQLE inhibitors. This result confirmed the specificity of the SQLE inhibitors.
Figure 2. SQLE inhibition enhanced radiosensitivity in vitro and in vivo.
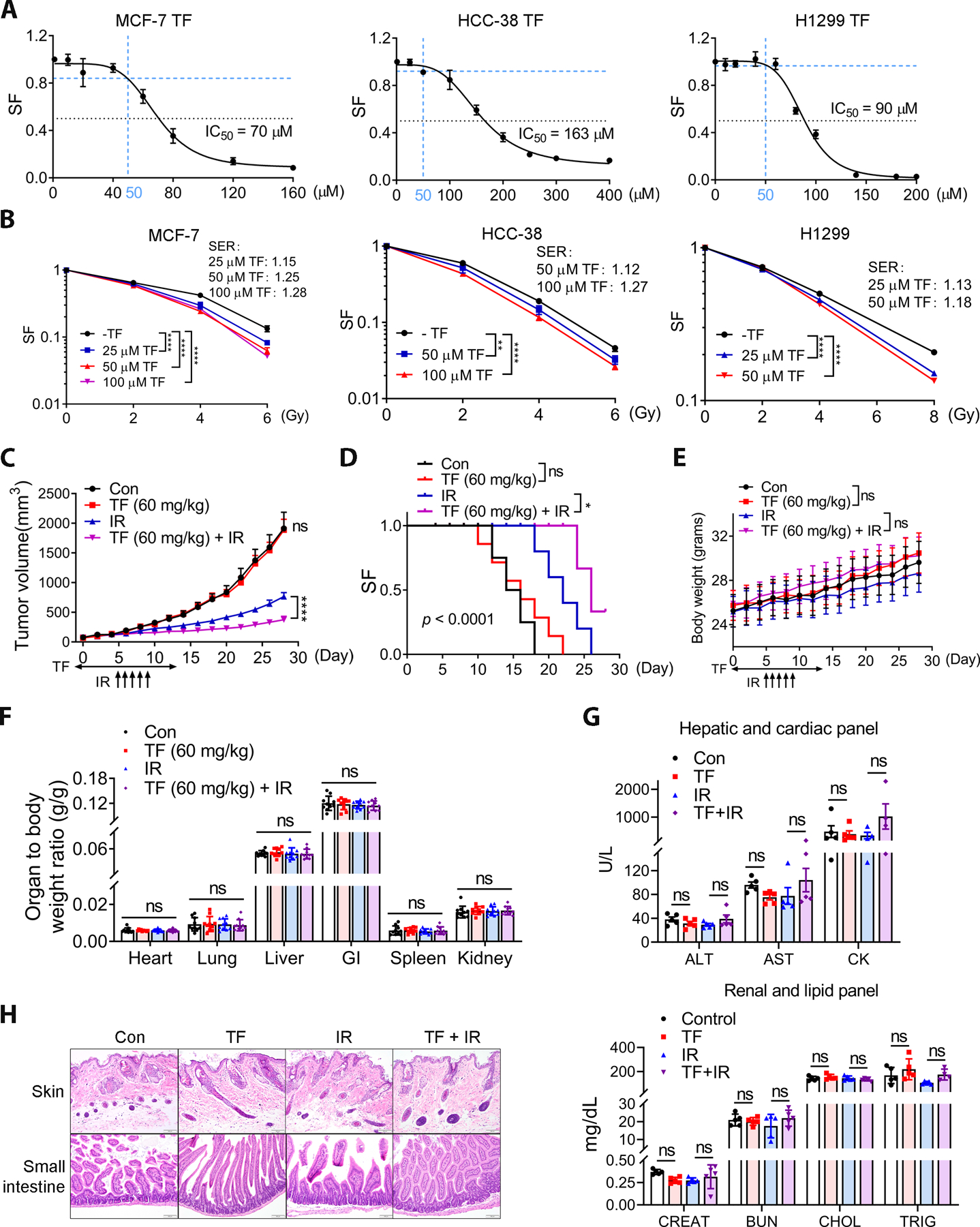
A, The IC50s of TF in the BC cell lines (MCF-7 and HCC-38) and also NSCLC H1299 (24 h treatment). Blue dash lines indicate the dose of 50 μM TF that did not significantly inhibit cell viability and was used for the colony formation assay. SF, Survival fraction. B, TF sensitized BC and NSCLC to ionizing radiation (IR) in the in vitro colony formation assay. Sensitizer enhanced ratio (SER) was showed in each cell lines. TF was given 24 h prior to irradiation. Data in (B) are presented as mean ± SD. Statistical significance was evaluated by two-way ANOVA and the Bonferroni post-hoc test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. C and D, TF enhanced IR efficacy. Vehicle (DMSO) or SQLE inhibitor TF (60 mg/kg) were administered by intraperitoneal injection daily for two weeks. Local fractionated x-ray radiation (2 Gy) was administered for five days, beginning on day 5. Data are presented as the mean tumor volume ± SEM. Statistical significance was evaluated by one-way ANOVA and the Bonferroni post-hoc test; ****, P < 0.0001 (C). Kaplan-Meier analysis was used for overall survival, and the statistical significance was examined using the Mantel-Cox test; *, P < 0.05 (D). E, SQLE inhibition had no impact on body weight. One-way ANOVA did not show significant differences between groups. F, SQLE inhibition did not affect organ weights (one-way ANOVA). G, Representative serum biochemistry profiling indicated no toxicity to the liver (ALT and AST), heart (CK), kidneys (CREAT and BUN) or lipid metabolism (CHOL and TRIG) in the TF-treated or TF combined IR groups. For one-way ANOVA, ns, not significant; ALT, alanine aminotransferase; AST, aspartate aminotransferase; CK, creatinine kinase; CREAT, creatinine; BUN, blood urea nitrogen; CHOL, cholesterol; TRIG, triglyceride. H, Representative histological images of skin and intestine. Magnification, 40×.
We evaluated the therapeutic potential of targeting SQLE with TF in combination with RT using in vivo model of H1299 cells (Fig. 2C). Local fractionated IR (2 Gy × 5 days) began on day 5 after intraperitoneal treatment with 60 mg/kg TF. SQLE inhibition did not affect tumor growth as a single agent; however, it significantly improved the efficacy of RT (Fig. 2C). As a result, SQLE inhibition significantly improved the survival of tumor-bearing mice exposed to IR compared to the control group (Fig. 2D). Importantly, no significant body weight or organ weight changes or altered serum chemistry across the experiment groups (Fig. 2E–G, and Supplemental Fig. S6A). Finally, no abnormalities were observed in the histology of the gastrointestinal tract or skin as the result of SQLE inhibition (Fig. 2H and Supplemental Fig. S6B). Thus, SQLE inhibition by TF could sensitize NSCLC xenografts to RT at doses that did not promote normal tissue toxicity.
SQLE inhibition and HR and RT-induced DSB repair
PARP inhibitors specifically kill cells with defective HR (31). SQLE inhibition with TF or NB-598 reduced the survival of MCF-7 and HCC-38 cells treated with the PARP inhibitor olaparib (AZD2281) (Fig. 3A and B), suggesting that SQLE inhibition affected HR. Using a HR reporter assay (28,29), we found that SQLE inhibition by TF or NB598 significantly decreased the percentage of I-SceI-induced GFP-positive cells compared to untreated MCF-7 and H1299 cells (Fig. 3C and Supplemental Fig. S7A). An almost identical cell cycle profile was observed in cells with or without SQLE inhibition (Supplemental Fig. S7B and C), indicating that impaired HR in the context of SQLE inhibition was not the consequence of alterations in the proportions of cells in S and G2/M.
Figure 3. SQLE inhibition impaired HR activity and delayed RT-induced DSB repair.
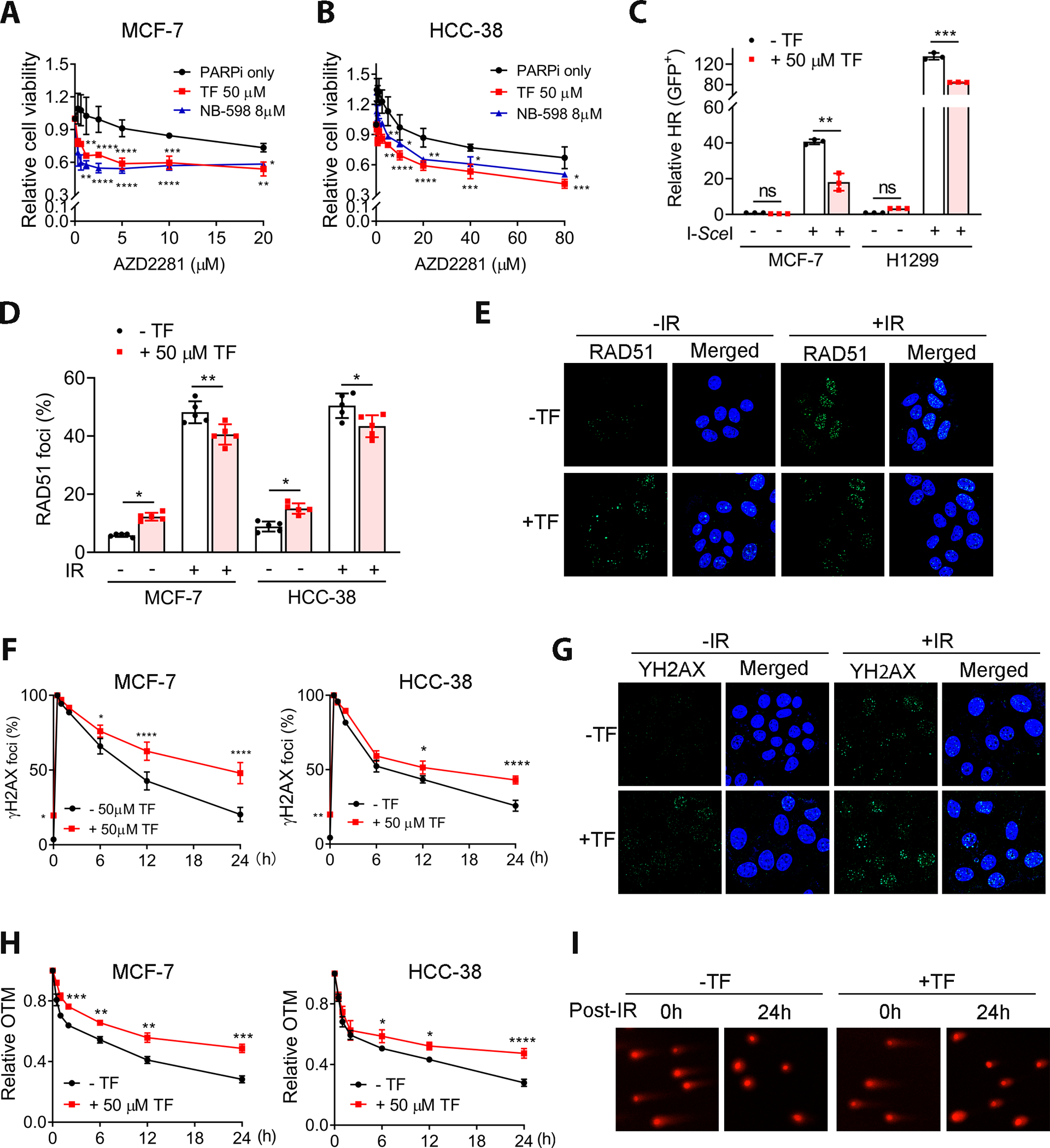
A and B, SQLE inhibition by TF or NB-598 promoted sensitivity to PARP inhibitor (AZD2281) in MCF-7 and HCC-38 cell lines. C, SQLE inhibition impaired HR. The percentage of I-SceI-induced GFP-positive cells was significantly decreased in MCF-7 and H1299 cells treated with 50 μM TF. All data are normalized with negative control. D, SQLE inhibition by TF abrogated IR-induced RAD51 foci formation in MCF-7 and HCC-38 cells, demonstrated by IF. The percentage of cells with positive RAD51 foci were presented. The cells with ≥10 foci were counted as positive. E, Representative IF images of IR-induced Rad51 foci in MCF-7 treated with or without 50 μM TF (magnification, 63× oil. IR: 4 Gy). F, SQLE inhibition led to the retention of IR-induced γ-H2AX in TF-treated MCF-7 and HCC-38 cells (IR, 4 Gy). The percentage of cells with positive γ-H2AX foci were presented. The cells with ≥10 foci were counted as positive. G, Representative γ-H2AX foci in MCF-7 cells (magnification, 63× oil). H, SQLE inhibition by TF promoted IR-induced DSBs by the comet assay (IR, 4 Gy). OTM, olive tail moment. I, Representative comet assay images used to quantify the olive tail moment in (H) (magnification, 20×). TF was given 24 h prior to irradiation (D-I). Data are presented as mean ± SD. Statistical significance was evaluated by one-way (C and D) or two-way (A, B, F and H) ANOVA and Bonferroni’s post-hoc test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Consistent with the reduced HR frequency, TF reduced IR-induced RAD51 foci formation (Fig. 3D and E). Furthermore, cells treated with TF had higher levels of γ-H2AX foci, a marker for DSB, following IR (Fig. 3F and G), indicating that SQLE inhibition could cause the retention of unrepaired radiation-induced DSBs due to an HR defect. Using the comet assay, we determined that the rate of IR-induced DSB repair in control cells was faster than in SQLE-inhibited cells (Fig. 3H and I), indicating reduced DNA repair in SQLE-inhibited cells. Similar results were found with MCF-7 and HCC-38 cells after NB-598 treatment (Supplemental Fig. S8A–D). In contrast, SQLE inhibition had no impact on the γ H2AX foci and comet tail in MDA-MB-231 cells (Supplemental Fig. S8E and F). Notably, we cannot exclude the possibility that the comet assay measured DNA fragmentation in dead cells, but not DNA repair kinetics. However, even the DNA fragmentation in dead cells was involved, this result still supports our hypothesis because SQLE inhibition leads to impaired DSB repair and the retention of IR-induced DNA damage, resulting in more cell death.
Although HR is defective in SQLE-inhibited cells, SQLE inhibition did not affect the IR-induced G2/M phase checkpoint (Supplemental Fig. S9A and B). Therefore, SQLE inhibition impaired HR activity, delaying DSB repair in SQLE-expressing cells following RT.
In addition to the repair of IR-induced DSBs, HR is important for the reactivation of stalled replication forks and HR-deficient cells display spontaneous replication stress (RS) (32). SQLE inhibition alone increased spontaneous γ-H2AX foci (Fig. 3F and G). Increased spontaneous RS/DSBs can trigger RAD51 foci formation. Indeed, SQLE inhibition increased RAD51 foci in the absence of IR (Fig. 3D and E). Our data are consistent with previous studies suggesting that although ATM is required for DSB repair by promoting HR in mitotically dividing cells, increased recombination might result from an increased number of lesions due to defective DNA damage signaling or repair in the absence of ATM (33,34). We speculate that the increased DSBs in SQLE-inhibited cells are related to RS. Indeed, SQLE inhibition increased p-RPA2 and p-CHK1, markers of RS in breast and lung cancer cells (Supplemental Fig. S10A and B). No obvious increase in ROS levels was observed in SQLE-depleted cells, whereas H2O2 treatment (positive control) led to a significant increase in ROS in BC (Supplemental Fig. S10C). Taken together, these results suggest that SQLE inhibition impairs HR, leading to DSB accumulation after IR and increased spontaneous RS.
p53-independent upregulation of WIP1 following SQLE inhibition
A deficiency in ATM activity enhances radiosensitivity by interrupting HR (35). It has been suggested that squalene-treated cells decreased ATM activity and increased WIP1 expression (36). Since SQLE inhibition has a potential to increase substrate squalene, we next determined if SQLE inhibition alters axis of WIP1-ATM. First, we analyzed the IR-induced phosphorylation of ATM on Ser1981, a marker for ATM activity. SQLE inhibition by TF or NB598 impaired IR-induced p-ATM (S1981) protein levels and foci formation in MCF-7 and HCC-38 cells (Fig. 4A and B). In support of the hypothesis that ATM activity is impaired, p-KAP1 that is a well-known downstream factor and readout of ATM activity is also reduced to SQLE inhibited cells (Supplemental Fig. S11A). In addition, the SQLE inhibition impaired p-ATM (S1981) foci at different time points post IR (Fig. 4C and D). Of note, a slight increase in p-ATM (S1981) was observed in the cells treated with TF alone (Fig. 4A and B). This effect was associated with increased spontaneous RS and DSBs (Fig. 3). Even when p-ATM (S1981) activity was suppressed in SQLE-inhibited cells, the level of spontaneous DSBs resulting from HR deficiency led to increased p-ATM (S1981) levels because of the increase in spontaneous DSBs, a major inducer of p-ATM (S1981). A similar result was observed in cells treated with NB-598 (Supplemental Fig. S11B). Moreover, SQLE inhibition-induced reduction in HR was dependent on ATM activity, and ATM inhibition abrogated TF- and NB-598-induced HR defects (Fig. 4E and Supplemental Fig. S11C) and SQLE inhibition-induced radio sensitivity (Supplemental Fig. S11D).
Figure 4. SQLE inhibition impaired the WIP1-ATM axis.
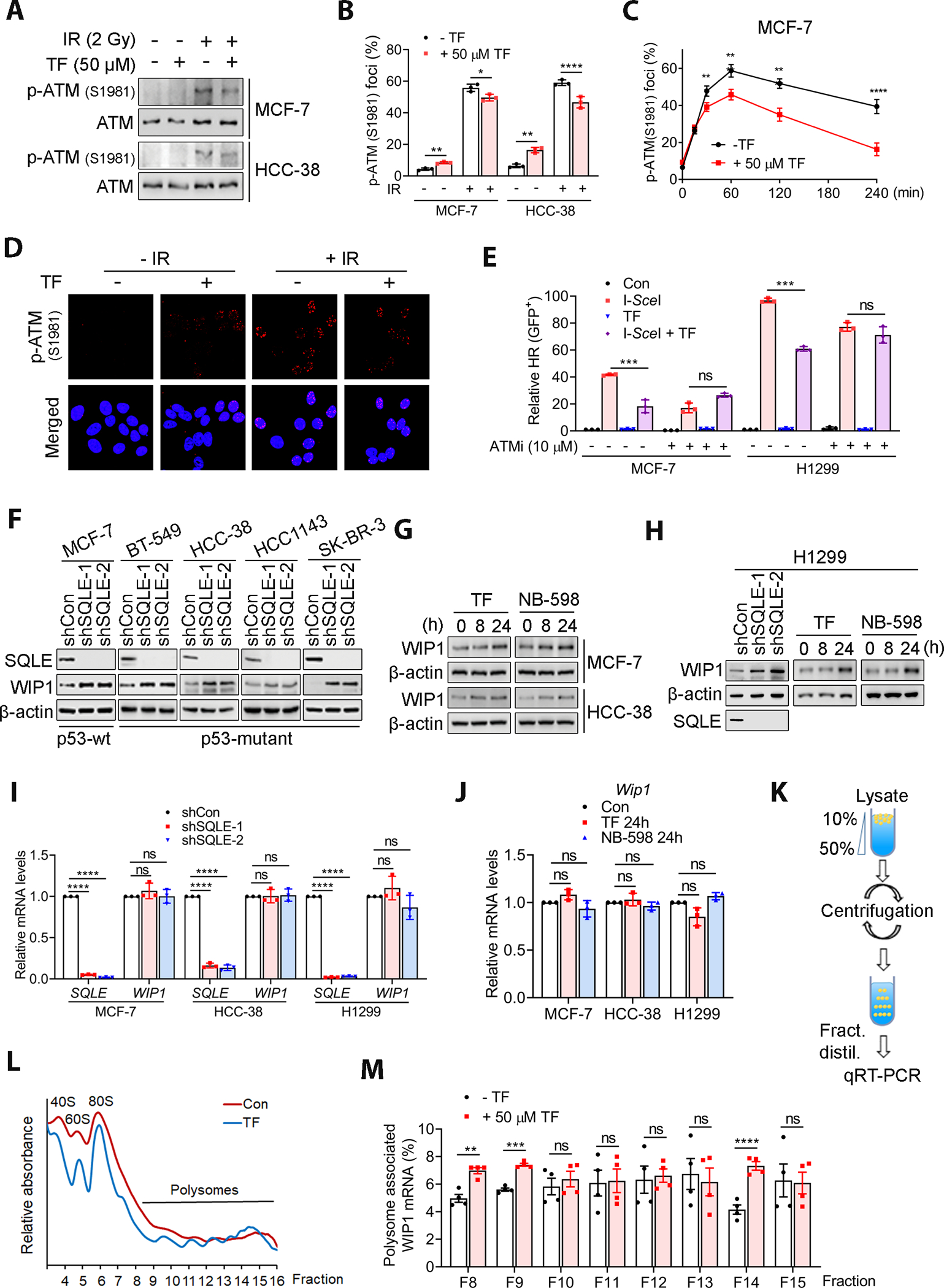
A and B, SQLE inhibition led to a decrease in IR-induced p-ATM (S1981) levels and phosphor-ATM-1981 foci (30 min post-2Gy IR) in MCF-7 and HCC-38 cells, as detected by WB and immunofluorescence staining. TF was given 24 h prior to irradiation. The percentage of cells with positive p-ATM (S1981) foci were presented. The cells with ≥10 foci were counted as positive. C, SQLE inhibition by TF decreased IR-induced p-ATM (S1981) levels at different time points after IR, as measured by immunofluorescence staining of MCF-7 cells. TF was given 24 h prior to irradiation. D, Representative immunofluorescence images of p-ATM (S1981) foci (magnification, 63× oil). E, ATM inhibition (10 μM) abrogated the TF-induced reduction in HR activity in MCF-7 and H1299 cells, as detected by the HR-DrGFP reporter assay. F-H, SQLE knockdown or pharmaceutical inhibition increased WIP1 protein expression in cells with or without wild-type. I and J, SQLE inhibition by SQLE knockdown or inhibitors did not affect WIP1 mRNA expression. K, Schematic representation of sucrose gradient preparation, polysome fractionation, and fraction analysis. L, Polysomes profiling. M, SQLE inhibition by TF increased polysome-associated WIP1 mRNA in MCF-7 cells. WIP1 mRNA was detected by qRT-PCR. The TF-treated MCF-7 cells showed significant increments in the WIP1 mRNA-associated polysomes. TF was given 24 h prior to irradiation (A-D). Data are presented as the mean ± SD in (B, C, E, I and J) and as the mean ± SEM in (M). All data in (I and J) are normalized with shCon or Con group. Statistical significance was determined by two-way (C) or one-way ANOVA (B, E, I and J) followed by Bonferroni’s post-hoc analysis for multiple comparisons. The Student’s t-test was used for the data in (M) for each group comparison. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
WIP1 negatively regulates ATM, and WIP1 overexpression reduces p-ATM (S1981) (17). SQLE inhibition by SQLE shRNA knockdown or chemical inhibitors increased WIP1 expression in BC and H1299 cells with wild-type or mutant p53 (Fig. 4F–H). In addition, WIP1 knockdown abrogates the SQLE inhibition-induced radio sensitivity (Supplemental Fig. S12). The impact of SQLE inhibition on WIP1 expression was independent of WIP1 transcription and p53 because WIP1 mRNA was unchanged in cells with our without SQLE inhibition, regardless of their p53 status (Fig. 4I and J, and Supplemental Fig. S13A). DNA damage upregulates WIP1 in a p53-dependent manner (18). Although SQLE inhibition induced RS and DSBs (Supplemental Fig. S10A and B), these effects might not be responsible for the SQLE inhibition-induced WIP1 increase because this increase occurred in both p53-WT and p53-deficient cells. In addition, SQLE inhibition did not affect IR-induced upregulation of WIP1 transcription. Consistent with a previous report (18), we saw IR-induced WIP1 transcript upregulation in p53 wild-type cells but not P53-deficient cells (Supplemental Fig. S13B). However, SQLE inhibition had no impact on the IR-induced upregulation of WIP1. Therefore, the SQLE inhibition-induced increase in WIP1 differed from the DNA damage-induced, p53-dependent WIP1 transcript regulation.
The increased WIP1 expression by SQLE inhibition was not associated with the change in the level of protein stability as analyzed by protein degradation rate (Supplemental Fig. S13C). Thus, we hypothesized that SQLE inhibition likely regulated WIP1 expression at the level of its mRNA translation. Given that mRNAs are actively translated in polysomes, we determined whether polysome-associated WIP1 mRNA levels were increased in SQLE-inhibited cells. The polysomal fractions were separated by sucrose density gradient centrifugation (Fig. 4K). Remarkably, WIP1 mRNA translation was enhanced in TF-treated MCF-7 cells compared to the control cells, as revealed by a shift in WIP1 mRNA association towards more actively translating polysomal fractions (Fig. 4L and M). These results suggested that SQLE inhibition impaired ATM activity by upregulating WIP1 expression regardless of the p53 status, and SQLE inhibition-induced WIP1 expression was controlled at the level of translation.
Squalene-dependent alterations in the WIP1-ATM axis and radiosensitivity
We next investigated why WIP1 expression increased in SQLE-inhibited cells. It has been suggested that squalene-treated cells have increased WIP1 expression (36). Thus, SQLE inhibition likely induces WIP1 expression as a result of squalene accumulation. Indeed, squalene levels markedly increased in BC and H1299 cells when SQLE was downregulated by shRNAs or inhibited by TF or NB-598 (Fig. 5A and B). In addition, SQLE inhibition increased SQLE expression (Fig. 5C), which is consistent with a recent study showing that SQLE inhibition stabilizes SQLE protein because squalene accumulation, most likely in the ER, prevents SQLE protein degradation (37). Increases in squalene accumulation and SQLE expression indicated that SQLE activity was inhibited by these inhibitors. In support of the hypothesis that SQLE inhibition-induced WIP1 expression is dependent on squalene, depleting FDFT1 (farnesyl-diphosphate farnesyltransferase), the enzyme responsible for squalene synthesis, by shRNA (FDFT1 shRNA) or pharmaceutical inhibitor (TAK-475) abrogated the WIP1 expression induced by SQLE inhibition (Fig. 5D–G). Moreover, TAK-475 abolished the SQLE inhibition-induced reduction in IR-induced p-ATM (S1981) foci and I-SceI-induced HR (Fig. 5H and I, and Supplemental Fig. S14A) and SQLE inhibition-induced radiosensitivity in MCF-7 cells (Fig. 5J). Similar results were obtained with HCC-38 cells (Fig. 5K and Supplemental Fig. S14B). Squalene depletion by TAK-475 blocked the induction of p-RPA2 and γ-H2AX by SQLE inhibition (Supplemental Fig. S14C and D) and FDFT1 knockdown (Supplemental Fig. S14E and F) in MCF-7 and HCC-38 cells. A similar result was obtained with H1299 cells (Supplemental Fig. S14G). These results were consistent with the requirement of squalene in the SQLE inhibition-induced reduction in HR. Thus, SQLE inhibition-induced squalene accumulation is required for WIP1 expression, reduced ATM activity, HR impairment, and increased radiosensitivity.
Figure 5. SQLE inhibition-induced alterations to the WIP1-ATM axis and radiosensitivity was dependent on squalene.
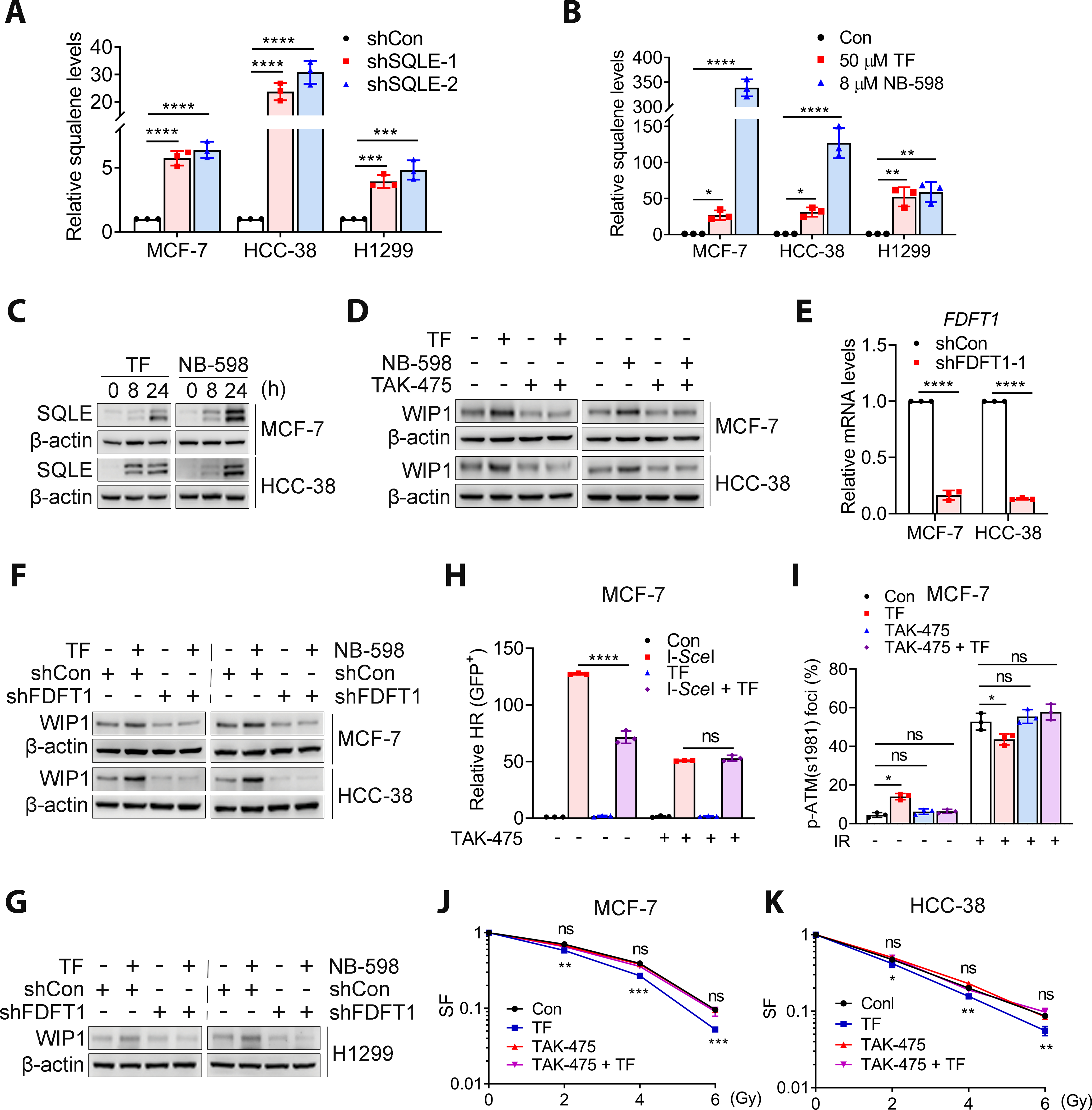
A and B, SQLE inhibition led to increased squalene levels in BC and lung cancer cells. Squalene levels were detected by LC-MS and normalized to control or shCon values. C, SQLE inhibition by 50 μM TF or 8 μM NB-598 increased SQLE protein in MCF-7 and HCC-38 cells. D, Blocking squalene synthesis for 48 h with TAK-475 (10 μM) reversed the effects of TF (50 μM) and NB-598 (8 μM) effects on WIP1 expression in MCF-7 and HCC-38 cells. E, Relative mRNA level of FDFT1 in MCF-7 and HCC-38 with or without FDFT1 knockdown. F and G, Squalene depletion by FDFT1 knockdown abrogated the increased WIP1 levels induced by TF (50 μM) or NB-598 (8 μM) in MCF-7, HCC-38, and H1299 cells. H, TAK-475 (10 μM) for 48 h restored the SQLE inhibition (50 μM TF)-induced reduction in HR in MCF-7 cells. All data are normalized with negative control. I, TAK-475 (10 μM) restored the SQLE inhibition (50 μM TF)-induced reduction in IR-induced p-ATM (S1981) foci in MCF-7 cells. J and K, TAK-475 (10 μM) eliminated SQLE inhibition-induced radiosensitivity in MCF-7 cells or HCC-38 cells. TF was given 24 h prior to irradiation (I-K). Data are presented as the mean ± SD. All data in (A, B, E and H) are normalized with shCon or Con group. Statistical significance was evaluated with one-way (A, B, E, H and I) or two-way (J and K) ANOVA and Bonferroni’s post-hoc test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
Of note, although the increase in squalene levels was much more profound in NB-598-treated cells compared to TF-treated cells (Fig. 5B), the magnitude of the enhanced radiosensitivity was almost similar (Fig. 2). We inferred that SQLE inhibition-induced squalene was likely stored in a nontoxic form by the formation of LDs (38,39), organelles that can prevent the toxicity of squalene. Indeed, LD formation was more pronounced in cells treated with NB-598 compared to those treated with TF (Supplemental Fig. S15A). The difference in the LDs in NB-598-treated cells might be underestimated because the results are presented as the number of LD per cell, and the size of the LDs was not taken into account. In fact, the size and intensity of the LDs were bigger in NB-598-treated cells in addition to the number of LD (Supplemental Fig. S15B).
The SQLE inhibition-induced WIP1 increase and enhanced radiosensitivity might also be associated with altered cholesterol levels because of the role of SQLE in cholesterol biosynthesis. Surprisingly, SQLE inhibition by shRNA knockdown or pharmacological inhibitor did not affect the total cholesterol levels in BC and lung cancer cells (Supplemental Fig. S16A). This result was confirmed using the Amplex Red Cholesterol Assay (Supplemental Fig. S16B). Normal cells meet their cholesterol needs through de novo synthesis and its uptake. SQLE inhibition likely suppresses endogenous cholesterol biosynthesis; however, the cells can still take up cholesterol from the complete medium, compensating for the loss of endogenous cholesterol biosynthesis caused by SQLE inhibition. Indeed, SQLE inhibition decreased cholesterol levels when cells were grown in lipoprotein-depleted serum (LPDS) medium (Supplemental Fig. S16C). Similarly, SQLE inhibition had no impact on cholesterol levels in H1299 cells cultured in full medium (Supplemental Fig. S16A and B) but decreased cholesterol levels for the cells grown in LPDS medium (Supplemental Fig. S16D). Together, these results suggested that SQLE inhibition caused squalene accumulation, rendering cells vulnerable to RT by impairing HR via upregulating WIP1 and subsequently reducing ATM activity.
SQLE inhibition and ER stress
The last question we addressed was how SQLE inhibition-induced squalene accumulation upregulated WIP1 translation. Squalene binds to the N-100 domain of SQLE, a region located in the ER (40,41). Squalene resides in the ER membrane and LDs, organelles formed by budding off from the ER, thought to be an important mechanism to reduce ER stress. Given that SQLE inhibition increased LD formation, we hypothesized that SQLE inhibition elevated WIP1 translation by triggering the ER stress response/UPR as a result of squalene accumulation. Three major ER stress sensors exist in mammalian cells, PERK, IRE1, and ATF6. The ER chaperone GRP78 is a firmly established regulator and downstream effector of these three sensors. SQLE inhibition by shRNA knockdown (Fig. 6A) or treatment with TF or NB-598 (Fig. 6B) increased GRP78 expression in both MCF-7 and HCC-38 cells as early as 8 hr. Indeed, squalene accumulation was also observed at this time point (Fig. S17). In addition, SQLE inhibition increased GRP78 mRNA levels (Fig. 6C and D). A similar result was observed with H1299 cells (Fig. 6E–G). Remarkably, TAK-475 treatment or FDFT1 knockdown abrogated the GRP78 expression triggered by SQLE inhibition in both breast and lung cancer cells (Fig. 6H–J). Thus, SQLE inhibition could induce the ER stress response and require squalene for this response. SQLE inhibition might also lead to squalene accumulation and ER stress and then subsequent WIP1 expression.
Figure 6. SQLE inhibition-induced ER stress was dependent on squalene.
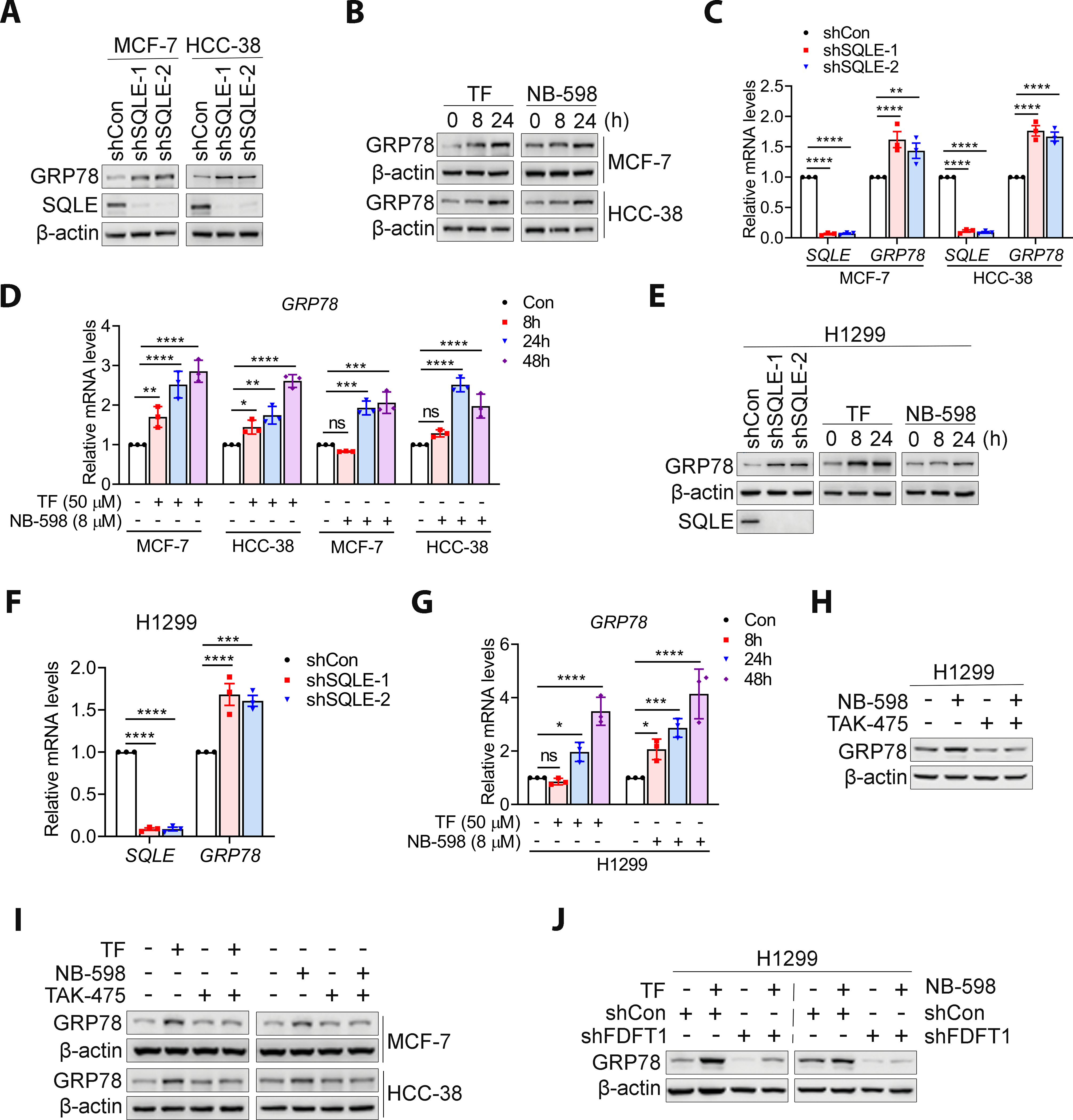
A, SQLE knockdown increased GRP78 expression in MCF-7 and HCC-38 cells. B, SQLE inhibition by TF (50 μM) or NB-598 (8 μM) increased GRP78 expression, as detected by western blotting. C, SQLE knockdown increased GRP78 transcript levels in MCF-7 and HCC-38 cells, as detected by qRT-PCR. D, SQLE inhibition by TF (50 μM) or NB-598 (8 μM) increased GRP78 mRNA levels in MCF-7 and HCC-38 cells. E and F, SQLE inhibition by shRNA knockdown or TF (50 μM) and NB-598 (8 μM) triggered GRP78 protein and transcript expression in H1299 cells. G, SQLE inhibition by TF (50 μM) or NB-598 (8 μM) increased GRP78 transcript levels in H1299 cells, as detected by qRT-PCR. H-J, Squalene was required for SQLE inhibition-induced GRP78 expression. Squalene depletion by the 10 μM TAK-475 inhibitor for 48 h or FDFT1 knockdown abrogated the effects of TF (50 μM) and NB-598 (8 μM) on GRP78 expression in BC and H1299 cells. Data are presented as the mean ± SD. All data are normalized with shCon or Con group. Statistical significance was evaluated with one-way ANOVA (C, D, F and G) and Bonferroni’s post-hoc test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
SQLE inhibition and PERK-eIF2α pathway
To further decipher the ER stress responses that contribute to WIP1 expression, we evaluated the PERK activation that is well-known for its role in inhibiting general protein translation but promoting the specific translation of stress response transcripts through the phosphorylation of eIF2α (42). For instance, ATF4 is synthesized by the PERK- eIF2α pathway during ER stress and induces the transcription of CHOP. SQLE knockdown (Fig. 7A) and pharmacological inhibition (Fig. 7B) increased p-eIF2α in MCF-7 and HCC-38 cells. Furthermore, increased ATF4 protein (Fig. 7A and B) and CHOP transcription (Fig. 7C) was observed, suggesting that the PERK- eIF2α mediated translation was activated by SQLE inhibition. Most importantly, eIF2α depletion abrogated SQLE inhibition-induced WIP1 protein expression (Fig. 7D). Therefore, SQLE inhibition-induced PERK-eIF2α pathway activation is likely required for WIP1 expression. Similar results obtained in BC cell are also observed in H1299 cells (Fig. 7E–H). Taken together, SQLE inhibition could increase. WIP1 translation through activation of PERK-eIF2α pathways in breast and lung cancers. The working model was presented in Fig. 7I.
Figure 7. SQLE inhibition activated the PERK- eIF2α pathway.
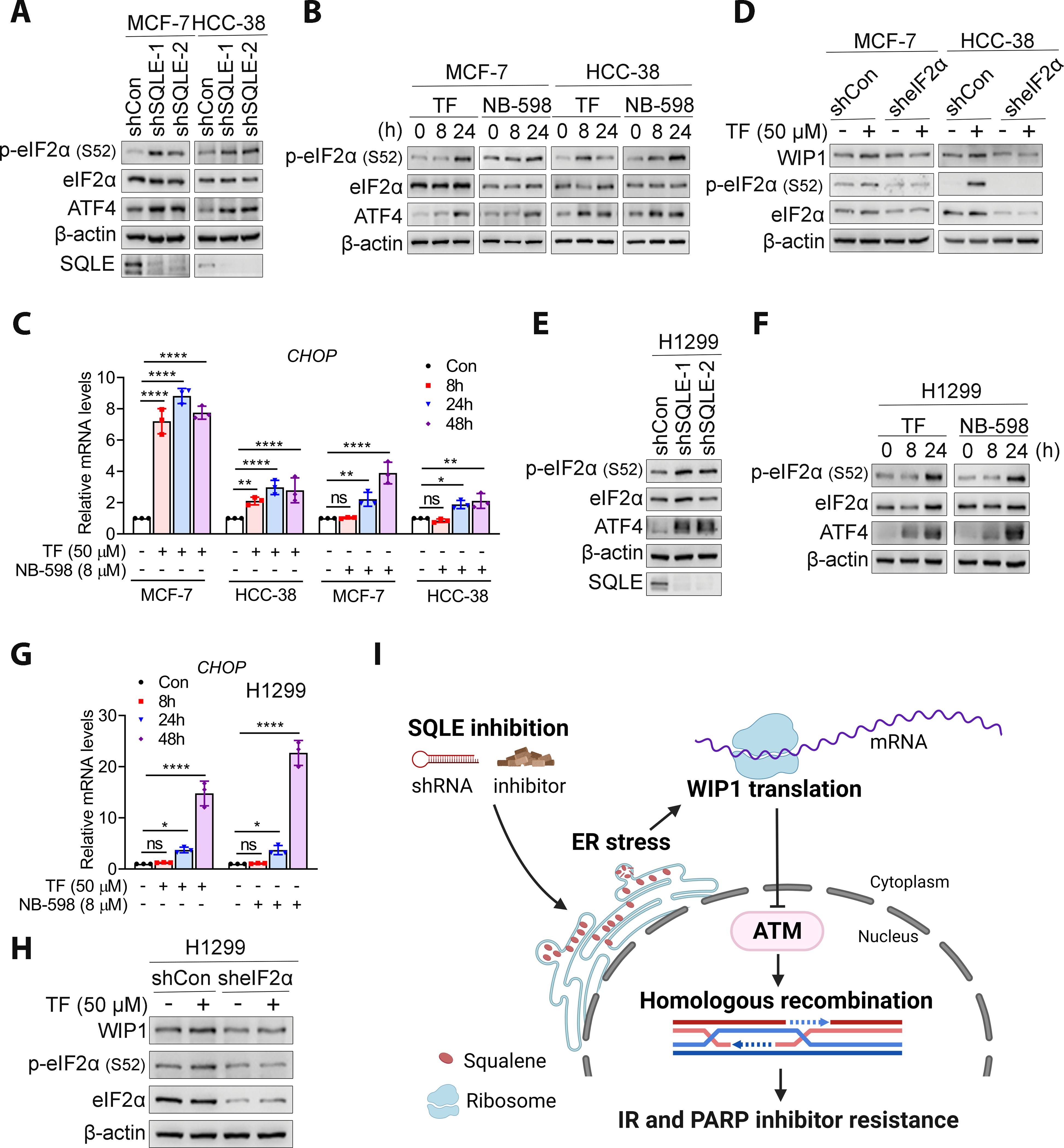
A and B, SQLE inhibition by knockdown or inhibitors (50 μM TF and 8 μM NB-598) increased p-eIF2α and ATF4 levels in MCF-7 and HCC-38 cell lines. C, CHOP transcription was upregulated by 50 μM TF or 8 μM NB-598 in MCF-7 and HCC-38 cells. All data are normalized with Con group. D, eIF2α downregulation abrogated TF-induced WIP1 expression in MCF-7 and HCC-38 cells. E and F, SQLE inhibition promoted the expression of p-eIF2a and ATF4 in H1299 cell. G, CHOP expression was upregulated following the treatment of 50 μM TF or 8 μM NB-598 in H1299 cell. H, eIF2α knockdown eliminated TF-induced WIP1 expression in H1299 cells. I, A schematic model for radiosensitization by SQLE inhibition. Figure created using BioRender (https://biorender.com/). The data in C and G are presented as mean ± SD. Statistical significance was evaluated with one-way ANOVA and Bonferroni’s post-hoc test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
DISCUSSION
Our retrospective clinical study demonstrated that SQLE expression was elevated in BC and NSCLC and an independent predictor of poor prognosis. In this study, we found that 67.6% of BC tumors were SQLE-positive compared to 15.2% of adjacent normal breast tissue. A previous study revealed that 62% of BC have high SQLE gene expression due to the gaining of an SQLE locus (8). Thus, it is likely that increased copy number or gene amplification contribute to SQLE expression. However, we cannot exclude other mechanisms that likely contribute to increased SQLE expression in BC as methylation and other posttranslational mechanisms might be involved in the upregulated SQLE expression in human cancers. Our results are consistent with recently published data showing that the SQLE expression is associated with breast tumor progression and poor prognosis (43).
We determined that SQLE expression was associated with significantly worse outcomes, which is in line with the prognostic value of SQLE mRNA and protein in BC (8,43). This result is not limited to BC because were obtained similar results with NSCLC. Our retrospective study and TCGA analysis suggested that SQLE or SQLE expression is associated with poor prognosis in a subgroup of BC that received adjuvant therapy, RT, or both. For estrogen receptor-positive BC, high SQLE expression may be responsible for the resistance associated with endocrine therapy (44) Thus, although the mechanisms by which SQLE expression contributes to poor prognosis remain unknown, high SQLE expression might contribute to poor prognosis by causing resistance to standard BC treatment, including RT. Given that paraffin-embedded tissue is routinely archived for clinical diagnostics and our study suggest that high SQLE expression occurs at 67.6% that is at a similar rate to high SQLE mRNA expression (8), detection of SQLE expression by IHC could be a potential readout for SQLE amplification and a predictor of radioresistance.
In the clinic, the therapeutic ratio compares the dose of a therapy that causes the desired amount of tumor control to that which causes toxicity. RT is dose-limiting because of the potential toxicity to normal tissue. Radiosensitizers must increase the efficacy of RT against the tumor and the probability of tumor cure without causing toxicity for them to be clinically useful in improving the therapeutic ratio. Our study has provided evidence that SQLE inhibitors are novel radiosensitizer candidates that fit these criteria. SQLE is highly expressed in various human tumors (e.g., BC) compared to the other 13 genes required for cholesterol biosynthesis, including HMGCR, the target of statin (45). Our results indicated that SQLE is highly expressed in BC and lung cancer, and SQLE inhibition could sensitize SQLE-expressing cells to RT but had no impact on cancer cells that lacked SQLE expression. Interestingly, hypoxic tumors are often radioresistant, and SQLE gene amplification is associated with hypoxic environments (11), making targeting SQLE to enhance radiosensitivity even more interesting. SQLE inhibition combined with RT should be considered as a new approach for treating this subset of cancer with the possibility of maximizing the response and minimizing toxicity. Given that enhanced sensitivity to PARP inhibitors was observed in SQLE-inhibited cells, SQLE inhibition may not be limited to a combination with RT. In addition, given that the process to bring new cancer treatments to market is slow and expensive, an affordable and safe treatment approach is to repurpose available licensed non-cancer drugs as new anticancer treatments. Thus, repurposing existing SQLE inhibitors (e.g., TF) provides a cost-efficient way to increase the treatment options for BC and lung cancer patients
SQLE inhibition sensitizes the cells to RT via disrupting HR by regulating WIP1-ATM axis. SQLE inhibition induced squalene accumulation is important for this regulation. Squalene may serve as a natural antioxidant in the skin; however, its role in cancer therapy is not clear. Squalene is a lipophilic metabolite present in ER membranes and LDs. Although squalene levels are very low to undetectable, SQLE inhibition increases its levels. Our study demonstrated that SQLE inhibition in human cancer cells renders the cells sensitive to RT by increasing squalene and disrupting HR due to dysregulation of the WIP1-ATM pathway without altering cholesterol levels. Interestingly, the current reports on the role of squalene in antitumor activity are inconsistent. For instance, squalene accumulation induced by SQLE inhibition is toxic to some small cell lung cancer (SCLC) cell lines (46); however, squalene protected ALK+ anaplastic large cell lymphoma cells from lipid oxidation-induced death (39). A recent study demonstrated that decreased SQLE due to increased cholesterol promoted tumor growth and metastasis, independent of squalene (47). Thus, from an antitumor perspective, the effect of squalene on cancer therapy can be suppressive, promoting, or inert. Thus, the effect of squalene is cell context-dependent.
The ER is the cellular site for protein folding, protein modifications, and lipid metabolism. Unfolded or misfolded protein and toxic lipid accumulation cause ER stress and subsequent activation of the ER stress response and LD formation (20,21). The ER stress response restores homeostasis by modulating LD formation (reduces lipid toxicity) and the protein load and folding capacity of the ER. Squalene is found in the middle of a lipid bilayer and is stored in the ER and plasma membrane (48). Squalene can alter the structure and properties of artificial membranes (49,50). Thus, it is likely that its accumulation triggers ER stress. Indeed, SQLE inhibition triggers ER stress. LD formation is an important mechanism for reducing ER stress because toxic lipids can be stored in the LD (20). Our finding that LD levels increase in SQLE-inhibited cells further supports the hypothesis that SQLE inhibition triggers ER stress. Translation initiation is the rate-limiting step in translation and is the primary target of regulation under stress, including ER stress. Our study demonstrated that SQLE inhibition-induced WIP1 expression occurs through translation, perhaps at the translation initiation steps. Several mechanisms trigger protein translation initiation under ER stress. The best-characterized mechanism is the PERK-eIF2α pathway. When PERK phosphorylates eIF2α, it prevents GTP binding, which interferes with the initiation of mRNA translation and reduces general protein synthesis. However, some proteins can elude this mechanism and translation of the transcript occurs. Our study suggest that the PERK branch activation likely is important for SQLE inhibition-induced WIP1 expression. However, we cannot exclude the possibility that translation regulation by other ER stress response branch might also involves the WIP1 translation and that SQLE inhibition might also affect RT-induced ER stress.
Overall, our work has provided for the first time a novel perspective on how SQLE inhibition combined with RT acts as antitumor therapy without increased toxicity. We propose a hypothetical model to illustrate the role of SQLE in cancer prognosis and how targeting SQLE improves treatment response, particularly for RT (Fig. 7I). According to this model, SQLE upregulation is associated with poor prognosis and resistance to cancer therapy, including RT. Targeting SQLE leads to squalene accumulation and subsequent ER stress that triggers WIP1 translation. Under such conditions, when cells are exposed to IR, RT-induced DSBs cannot be sufficiently repaired by HR due to ATM inhibition, rendering the cells sensitive to RT. However, we cannot exclude the possibility that other ATM functions might also be involved in SQLE inhibition-induced radiosensitization. The knowledge obtained from our studies is valuable for understanding why the inhibition of an enzyme required for cholesterol synthesis increased radiosensitivity.
There were several limitations to our study. For the retrospective study, our cohort sizes were relatively small, especially for individual BC subtypes. Additional large-scale studies are necessary to verify the prognostic value of SQLE expression in BC subtypes, including TNBC. For the preclinical studies, although we provide evidence supporting the hypothesis that SQLE inhibition-induced squalene accumulation triggers ER stress and subsequent WIP1 expression, more mechanistic studies are needed to clarify whether and how WIP1 is involved in the ER stress response and how squalene triggers ER stress by disrupting the properties of the ER membrane. Lastly, our findings were mainly performed in cell lines and mice. Studies in human cancer patients would be worthwhile.
Our results suggest alterations in the activity of the metabolic enzyme SQLE influence metabolite intermediates, altering non-metabolic cellular functions. The results of our study have significant implications for cancer therapy by introducing a novel radiosensitizer for treating SQLE-expressing BC and NSCLC. In addition, our study provides significant advances in translating the use of SQLE inhibitors to the clinical setting. Given that SQLE overexpression exists in a wide range of human cancers, repurposing existing SQLE inhibitors offers a cost-efficient way to increase cancer patient treatment options.
Supplementary Material
Significance:
Squalene epoxidase inhibitors are novel tumor-specific radiosensitizers that promote ER stress and suppress homologous recombination, providing a new potential therapeutic approach to enhance radiotherapy efficacy.
Acknowledgments:
We thank Dr. Feng Geng from Dr. Deliang Guo’s lab for assisting with cholesterol detection assay and lipid droplet staining and quantification. We thank Dr. Linlin Yang from Dr. Terence M. Williams’s lab for assisting with the mouse radiation. We thanks pathologist Dr. Yu Wang from Dr. Allan Tsung’s lab for providing pathological assessment of mouse stomach tissues and taking representative images. This work was partially funded by the National Institutes of Health (NIH)/National Cancer Institute (NCI) (R21CA226317, R21 CA241242, R01CA240374, and R01CA249198), the America Lung Cancer Association, and the Breast Cancer Alliance and the Ohio State University James Comprehensive Cancer Intramural Research Program (Pelotonia) (to J. Zhang). Pathology services were provided by the Comparative Pathology & Digital Imaging Shared Resource, Department of Veterinary Biosciences and the Comprehensive Cancer Center, The Ohio State University (Columbus, OH) and supported in part by grant P30 CA16058 (NCI). The project was also supported by the National Center for Advancing Translational Sciences (UL1TR002733). The content is solely the authors’ responsibility and does not necessarily represent the official views of the National Center for Advancing Translational Sciences or the NIH. A startup fund from The First Affiliated Hospital of Sun Yat-sen University to Z. Ma.
Footnotes
Competing interests: The authors declare no potential conflicts of interest.
References
- 1.Baskar R, Lee KA, Yeo R, Yeoh KW. Cancer and radiation therapy: current advances and future directions. Int J Med Sci 2012;9:193–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yard BD, Adams DJ, Chie EK, Tamayo P, Battaglia JS, Gopal P, et al. A genetic basis for the variation in the vulnerability of cancer to DNA damage. Nature communications 2016;7:11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sui Z, Zhou J, Cheng Z, Lu P. Squalene epoxidase (SQLE) promotes the growth and migration of the hepatocellular carcinoma cells. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine 2015;36:6173–9 [DOI] [PubMed] [Google Scholar]
- 4.Ge H, Zhao Y, Shi X, Tan Z, Chi X, He M, et al. Squalene epoxidase promotes the proliferation and metastasis of lung squamous cell carcinoma cells though extracellular signal-regulated kinase signaling. Thorac Cancer 2019;10:428–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polycarpou-Schwarz M, Gross M, Mestdagh P, Schott J, Grund SE, Hildenbrand C, et al. The cancer-associated microprotein CASIMO1 controls cell proliferation and interacts with squalene epoxidase modulating lipid droplet formation. Oncogene 2018 [DOI] [PubMed] [Google Scholar]
- 6.Liu D, Wong CC, Fu L, Chen H, Zhao L, Li C, et al. Squalene epoxidase drives NAFLD-induced hepatocellular carcinoma and is a pharmaceutical target. Science translational medicine 2018;10. [DOI] [PubMed] [Google Scholar]
- 7.Li L, Zhang Q, Wang X, Li Y, Xie H, Chen X. Squalene epoxidase-induced cholesteryl ester accumulation promotes nasopharyngeal carcinoma development by activating PI3K/AKT signaling. Cancer Sci 2020;111:2275–83 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Brown DN, Caffa I, Cirmena G, Piras D, Garuti A, Gallo M, et al. Squalene epoxidase is a bona fide oncogene by amplification with clinical relevance in breast cancer. Scientific reports 2016;6:19435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A, et al. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proceedings of the National Academy of Sciences of the United States of America 2003;100:10393–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helms MW, Kemming D, Pospisil H, Vogt U, Buerger H, Korsching E, et al. Squalene epoxidase, located on chromosome 8q24.1, is upregulated in 8q+ breast cancer and indicates poor clinical outcome in stage I and II disease. Br J Cancer 2008;99:774–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haider S, McIntyre A, van Stiphout RG, Winchester LM, Wigfield S, Harris AL, et al. Genomic alterations underlie a pan-cancer metabolic shift associated with tumour hypoxia. Genome Biol 2016;17:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagaraja R, Olaharski A, Narayanaswamy R, Mahoney C, Pirman D, Gross S, et al. Preclinical toxicology profile of squalene epoxidase inhibitors. Toxicol Appl Pharmacol 2020;401:115103. [DOI] [PubMed] [Google Scholar]
- 13.Shiloh Y The ATM-mediated DNA-damage response: taking shape. Trends Biochem Sci 2006;31:402–10 [DOI] [PubMed] [Google Scholar]
- 14.Golding SE, Rosenberg E, Khalil A, McEwen A, Holmes M, Neill S, et al. Double strand break repair by homologous recombination is regulated by cell cycle-independent signaling via ATM in human glioma cells. The Journal of biological chemistry 2004;279:15402–10 [DOI] [PubMed] [Google Scholar]
- 15.Morrison C, Sonoda E, Takao N, Shinohara A, Yamamoto K, Takeda S. The controlling role of ATM in homologous recombinational repair of DNA damage. The EMBO journal 2000;19:463–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nature cell biology 2006;8:37–45 [DOI] [PubMed] [Google Scholar]
- 17.Shreeram S, Demidov ON, Hee WK, Yamaguchi H, Onishi N, Kek C, et al. Wip1 phosphatase modulates ATM-dependent signaling pathways. Molecular cell 2006;23:757–64 [DOI] [PubMed] [Google Scholar]
- 18.Fiscella M, Zhang H, Fan S, Sakaguchi K, Shen S, Mercer WE, et al. Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proceedings of the National Academy of Sciences of the United States of America 1997;94:6048–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu X, Nguyen TA, Moon SH, Darlington Y, Sommer M, Donehower LA. The type 2C phosphatase Wip1: an oncogenic regulator of tumor suppressor and DNA damage response pathways. Cancer metastasis reviews 2008;27:123–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hapala I, Marza E, Ferreira T. Is fat so bad? Modulation of endoplasmic reticulum stress by lipid droplet formation. Biol Cell 2011;103:271–85 [DOI] [PubMed] [Google Scholar]
- 21.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 2007;8:519–29 [DOI] [PubMed] [Google Scholar]
- 22.Ryoo HD, Vasudevan D. Two distinct nodes of translational inhibition in the Integrated Stress Response. BMB Rep 2017;50:539–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang X, Pan Y, Qiu Z, Du Z, Zhang Y, Fa P, et al. RNF126 as a Biomarker of a Poor Prognosis in Invasive Breast Cancer and CHEK1 Inhibitor Efficacy in Breast Cancer Cells. Clinical cancer research : an official journal of the American Association for Cancer Research 2018;24:1629–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wan L, Liu T, Hong Z, Pan Y, Sizemore ST, Zhang J, et al. NEDD4 expression is associated with breast cancer progression and is predictive of a poor prognosis. Breast Cancer Res 2019;21:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, Fang L, Liu W. High SQLE Expression and Gene Amplification Correlates with Poor Prognosis in Head and Neck Squamous Cell Carcinoma. Cancer Manag Res 2021;13:4709–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He L, Li H, Pan C, Hua Y, Peng J, Zhou Z, et al. Squalene epoxidase promotes colorectal cancer cell proliferation through accumulating calcitriol and activating CYP24A1-mediated MAPK signaling. Cancer Commun (Lond) 2021;41:726–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiu Z, Fa P, Liu T, Prasad CB, Ma S, Hong Z, et al. A genome-wide pooled shRNA screen identifies PPP2R2A as a predictive biomarker for the response to ATR and CHK1 inhibitors. Cancer research 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J, Ma Z, Treszezamsky A, Powell SN. MDC1 interacts with Rad51 and facilitates homologous recombination. Nature structural & molecular biology 2005;12:902–9 [DOI] [PubMed] [Google Scholar]
- 29.Shi W, Ma Z, Willers H, Akhtar K, Scott SP, Zhang J, et al. Disassembly of MDC1 foci is controlled by ubiquitin-proteasome-dependent degradation. The Journal of biological chemistry 2008;283:31608–16 [DOI] [PubMed] [Google Scholar]
- 30.Souchek JJ, Baine MJ, Lin C, Rachagani S, Gupta S, Kaur S, et al. Unbiased analysis of pancreatic cancer radiation resistance reveals cholesterol biosynthesis as a novel target for radiosensitisation. Brit J Cancer 2014;111:1139–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lord CJ, Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science 2017;355:1152–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daboussi F, Courbet S, Benhamou S, Kannouche P, Zdzienicka MZ, Debatisse M, et al. A homologous recombination defect affects replication-fork progression in mammalian cells. J Cell Sci 2008;121:162–6 [DOI] [PubMed] [Google Scholar]
- 33.Meyn MS. High spontaneous intrachromosomal recombination rates in ataxia-telangiectasia. Science 1993;260:1327–30 [DOI] [PubMed] [Google Scholar]
- 34.Bishop AJ, Barlow C, Wynshaw-Boris AJ, Schiestl RH. Atm deficiency causes an increased frequency of intrachromosomal homologous recombination in mice. Cancer research 2000;60:395–9 [PubMed] [Google Scholar]
- 35.Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene 2007;26:7741–8 [DOI] [PubMed] [Google Scholar]
- 36.Tatewaki N, Konishi T, Nakajima Y, Nishida M, Saito M, Eitsuka T, et al. Squalene Inhibits ATM-Dependent Signaling in gammaIR-Induced DNA Damage Response through Induction of Wip1 Phosphatase. PloS one 2016;11:e0147570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshioka H, Coates HW, Chua NK, Hashimoto Y, Brown AJ, Ohgane K. A key mammalian cholesterol synthesis enzyme, squalene monooxygenase, is allosterically stabilized by its substrate. Proceedings of the National Academy of Sciences of the United States of America 2020;117:7150–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ta MT, Kapterian TS, Fei W, Du X, Brown AJ, Dawes IW, et al. Accumulation of squalene is associated with the clustering of lipid droplets. The FEBS journal 2012;279:4231–44 [DOI] [PubMed] [Google Scholar]
- 39.Garcia-Bermudez J, Baudrier L, Bayraktar EC, Shen Y, La K, Guarecuco R, et al. Squalene accumulation in cholesterol auxotrophic lymphomas prevents oxidative cell death. Nature 2019;567:118–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chan TA. 14-3-3Sigma is required to prevent mitotic catastrophe after DNA damage. Nature (London);401:616–20 [DOI] [PubMed] [Google Scholar]
- 41.Nathan JA. Squalene and cholesterol in the balance at the ER membrane. Proceedings of the National Academy of Sciences of the United States of America 2020;117:8228–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Molecular cell 2000;6:1099–108 [DOI] [PubMed] [Google Scholar]
- 43.Kim NI, Park MH, Kweon SS, Cho N, Lee JS. Squalene epoxidase expression is associated with breast tumor progression and with a poor prognosis in breast cancer. Oncol Lett 2021;21:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simigdala N, Gao Q, Pancholi S, Roberg-Larsen H, Zvelebil M, Ribas R, et al. Cholesterol biosynthesis pathway as a novel mechanism of resistance to estrogen deprivation in estrogen receptor-positive breast cancer. Breast Cancer Res 2016;18:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu H, Zhou S, Tang Q, Xia H, Bi F. Cholesterol metabolism: New functions and therapeutic approaches in cancer. Biochim Biophys Acta Rev Cancer 2020;1874:188394. [DOI] [PubMed] [Google Scholar]
- 46.Mahoney CE, Pirman D, Chubukov V, Sleger T, Hayes S, Fan ZP, et al. A chemical biology screen identifies a vulnerability of neuroendocrine cancer cells to SQLE inhibition. Nature communications 2019;10:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jun SY, Brown AJ, Chua NK, Yoon JY, Lee JJ, Yang JO, et al. Reduction of Squalene Epoxidase by Cholesterol Accumulation Accelerates Colorectal Cancer Progression and Metastasis. Gastroenterology 2020 [DOI] [PubMed] [Google Scholar]
- 48.Spanova M, Czabany T, Zellnig G, Leitner E, Hapala I, Daum G. Effect of lipid particle biogenesis on the subcellular distribution of squalene in the yeast Saccharomyces cerevisiae. The Journal of biological chemistry 2010;285:6127–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lohner K, Degovics G, Laggner P, Gnamusch E, Paltauf F. Squalene promotes the formation of non-bilayer structures in phospholipid model membranes. Biochimica et biophysica acta 1993;1152:69–77 [DOI] [PubMed] [Google Scholar]
- 50.Hauss T, Dante S, Dencher NA, Haines TH. Squalane is in the midplane of the lipid bilayer: implications for its function as a proton permeability barrier. Biochimica et biophysica acta 2002;1556:149–54 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


