Abstract
Norovirus is the leading cause of epidemic and endemic acute gastroenteritis worldwide and the most frequent cause of foodborne illness in the United States. There is no specific treatment for norovirus infections and therapeutic interventions are based on alleviating symptoms and limiting viral transmission. The immune response to norovirus is not completely understood and mechanistic studies have been hindered by lack of a robust cell culture system. In recent years, the human intestinal enteroid/human intestinal organoid system (HIE/HIO) has enabled successful human norovirus replication. Cells derived from HIE have also successfully been subjected to genetic manipulation using viral vectors as well as CRISPR/Cas9 technology, thereby allowing studies to identify antiviral signaling pathways important in controlling norovirus infection. RNA sequencing using HIE cells has been used to investigate the transcriptional landscape during norovirus infection and to identify antiviral genes important in infection. Other cell culture platforms such as the microfluidics-based gut-on-chip technology in combination with the HIE/HIO system also have the potential to address fundamental questions on innate immunity to human norovirus. In this review, we highlight the recent advances in understanding the innate immune response to human norovirus infections in the HIE system, including the application of advanced molecular technologies that have become available in recent years such as the CRISPR/Cas9 and RNA sequencing, as well as the potential application of single cell transcriptomics, viral proteomics, and gut-on-a-chip technology to further elucidate innate immunity to norovirus.
Keywords: enteroids, innate immunity, norovirus
INTRODUCTION
Norovirus is the leading cause of epidemic and endemic acute gastroenteritis worldwide and the most frequent cause of foodborne illness in the United States [1]. Norovirus gastroenteritis causes approximately 21 million cases of illness in the United States and over 600 million cases worldwide every year [2]. Although the disease is typically self-limiting, some cases result in severe illness, with an estimated 56 000 to 71 000 hospitalizations and 570 to 800 norovirus related deaths reported in the United States each year [2]. Human norovirus spreads primarily via the oral-faecal route or through contact with contaminated food, water, or surfaces. As few as 18–1018 genome equivalents are sufficient to cause infection [3–5], and an estimated 50% human infectious dose (HID50) ranges between 1320 and 2800 genome equivalents [5]. Norovirus disease is characterized by stomach pain, nausea, explosive vomiting and diarrhoea within 12–48 h of exposure [6]. In immune-competent individuals these symptoms typically resolve within 1–3 days although the virus might be detectable for several weeks [7]. There is no specific treatment for norovirus infection and therapeutic interventions are based on alleviating symptoms [8]. Outbreak management relies heavily on early identification of cases, isolation of infected individuals, and strict disinfection and decontamination protocols [7, 8]. Several norovirus vaccine candidates are under development, four of which have undergone clinical trials [9]; however, none have currently been licensed. In this review, we will summarize the current information on the innate immune response to norovirus infection in human intestinal enteroids (HIE) and advanced molecular technologies that may help to better understand the molecular mechanisms that regulate norovirus replication allowing the development of effective prophylactic and therapeutic interventions.
Human volunteer challenge studies have provided important information on aspects of virus infection such as the environmental conditions that affect virus stability [10], the HID50 [3, 5], host genetic factors that govern susceptibility to infection [11], as well as insights on the immune response during infection [12–14]. Several studies have shown association between susceptibility to norovirus infection and expression of a functional FUT2 gene [11, 15–17]. FUT2 encodes α–1,2-fucosyltransferase, an enzyme that is important for expression of human blood group antigen (HBGA) molecules on mucosal surfaces. α–1,2-fucosyltransferase transfers a second fucose molecule to the H blood group antigen precursor, thereby generating H antigen [17, 18]. Norovirus infection requires HBGA which function as binding ligands to facilitate virus attachment to cells [19]. Individuals that express a functional FUT2 gene are termed secretor-positive and are therefore susceptible to norovirus infection [17]. Secretor-negative individuals lack functional FUT2 alleles, consequently, they do not express H-antigen structures on their mucosa and are resistant to infection by most norovirus strains, including the globally predominant genotype GII.4 [20], although exceptions have been reported [6, 17].
Human volunteer challenge studies have also given initial insights into the antiviral response that restricts norovirus infections. Analysis of serum collected during the first 4 days after infection in two human volunteer challenge studies showed that norovirus induces T-helper 1 and T-helper 2 (Th1 and Th2) cytokines, chemokines, and inflammatory cytokines including IFN-γ, IL-6, IL-8, IL-12p70, MCP-1 and TNF-α, as part of the acute response, with peak detection at 2 days post-infection [12]. Assessment of norovirus-specific antibody responses during infection using saliva collected from elderly individuals in 43 long-term care facilities showed that virus-specific salivary IgA titers increase beginning at 5 days after symptom onset, with peak titers at 14 days [21]. Together, these studies show that both the innate and adaptive immune responses are important for controlling norovirus infection [22, 23]. What remains to be elucidated are the specific molecular mechanisms and signalling pathways involved in the antiviral response against human norovirus.
Models for norovirus infection
For many years mechanistic studies were hampered by lack of a robust cell culture system. Efforts to grow norovirus in a number of well-established cell lines, including primary kidney cell lines, primary intestinal cell lines, and colon carcinoma cell lines, failed [24]. Despite the established tropism of murine norovirus (MNV) for innate immune cells, efforts to replicate human norovirus in the same cell types derived from peripheral blood mononuclear cells (PBMCs) also failed [25]. It has been reported that human noroviruses are capable of replicating in human B cells [26, 27]. Attempts to obtain sustained norovirus replication in B cells using unfiltered, unprocessed stool as inoculum showed that bacterial surface expressed human blood group antigens (HBGA) are important factors for successful virus replication [26]. Additionally, the addition of HBGA-expressing Enterobacter cloacae to the cell culture could restore the infectivity of filtered human norovirus positive stool filtrate, whereas a non-HBGA-expressing bacterium could not [26]. BJAB and Raji B cell lines initially showed promise for replicating human norovirus however these studies have been shown to be difficult to reproduce [26, 27].
Several replicon models that stably express human norovirus RNA have been developed as tools to facilitate studying the immune response to norovirus infection [28–30]. A number of animal models have also been utilized to study human norovirus including non-human primates [31, 32], gnotobiotic pigs [33, 34] and humanized mice [35]. Each of these models had limitations including low levels of virus replication. Recently, zebrafish larvae have been reported as a robust model for human norovirus infection [36]. However, this model does not necessarily represent cells in the human gut and the ensuing innate immune responses after a norovirus infection. Despite the above-mentioned efforts, what remains unclear are the antiviral proteins that specifically restrict virus replication as well as the molecules that recognize norovirus to initiate the antiviral signal pathways. Until recently, much of our understanding of the molecular mechanisms involved in norovirus pathogenesis and immune response has been derived from transformed cell lines and infection with human norovirus surrogate viruses such as feline calicivirus, porcine calicivirus, MNV and Tulane virus [37–39] (Fig. 1).
Fig. 1.
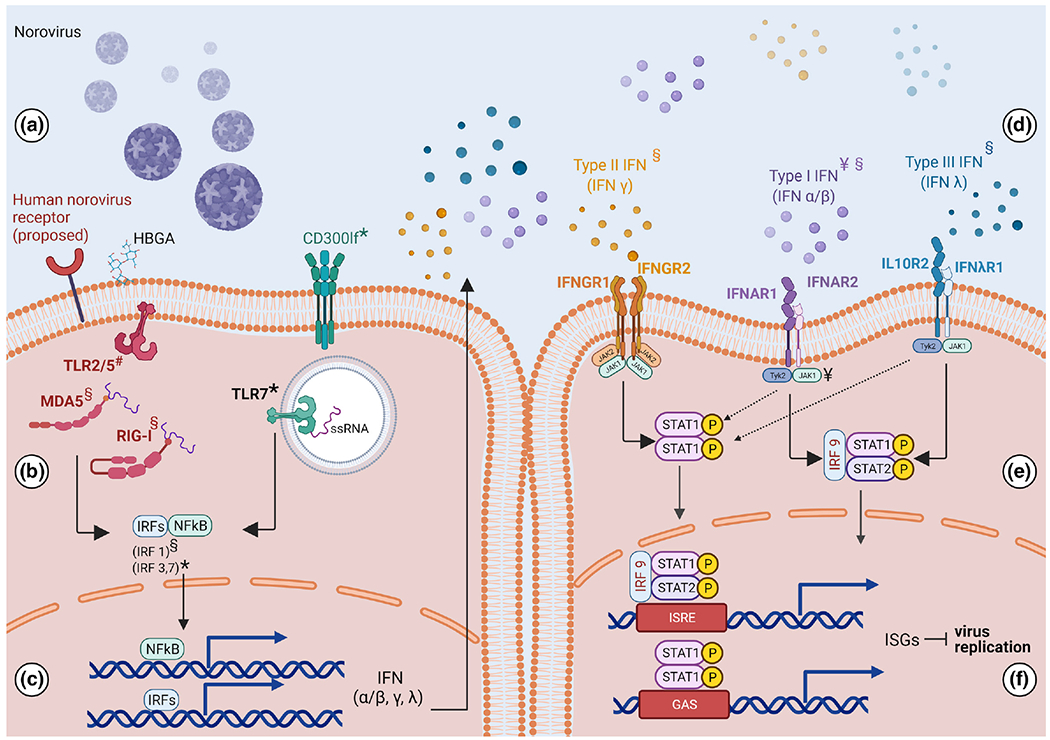
The antiviral response in norovirus-infected cells. Norovirus infection induces an antiviral response that restricts virus replication. a. Human norovirus attachment is facilitated by human blood group antigens (HBGA) and a receptor that is yet to be identified. In mice the receptor is CD300lf [66]. b. Viral entry results in sensing of the virus and viral components by molecular sensors. The molecular sensors that are triggered during human norovirus infection have not been identified, however human norovirus virus-like particles (VLPs) have been shown to trigger TLR2 and TLR5 (70). MDA5 and RIG-I are also involved in the response against human norovirus [102] and TLR7 has been shown to trigger an antiviral response that protects against murine norovirus (MNV) [103]. c. Virus sensing results in activation of transcription factors including NFkB [103] and interferon regulatory factors IRF1(102), IRF3 and IRF7(3) which facilitate transcription of genes encoding type I, type II and type III interferon. d. Interferon is secreted and engages corresponding receptors on the cell surface, resulting in phosphorylation and activation of STAT1/2 through the JAK/STAT pathway [55]. e. STAT1/2 translocate to the nucleus and facilitate upregulation of hundreds of antiviral interferon stimulated genes (ISGs). f. The proteins encoded by antiviral ISGs restrict virus replication. This figure summarizes the immune components involved in the antiviral response to norovirus infection and includes findings from human norovirus infection as well as its surrogates: ¥human norovirus infection in human intestinal enteroids, *murine norovirus, # human norovirus virus like particles (VLPs), §human norovirus replicon. Created with BioRender.com (accessed on 12 October 2021).
In recent years, technical advances in the culture of primary human intestinal epithelial cells using intestinal 3D organoid cultures [40] along with availability of advanced molecular technologies, have revolutionized approaches to study the immune response in norovirus infection. Techniques such as gene manipulation, bulk and single-cell RNA sequencing (scRNA-seq), proteomics [41–43], as well as gut-on-a-chip technology, which mimic the intestinal physiological environment [40–42], have the potential to be applied to human norovirus studies in order to move the field forward (Fig. 2).
Fig. 2.
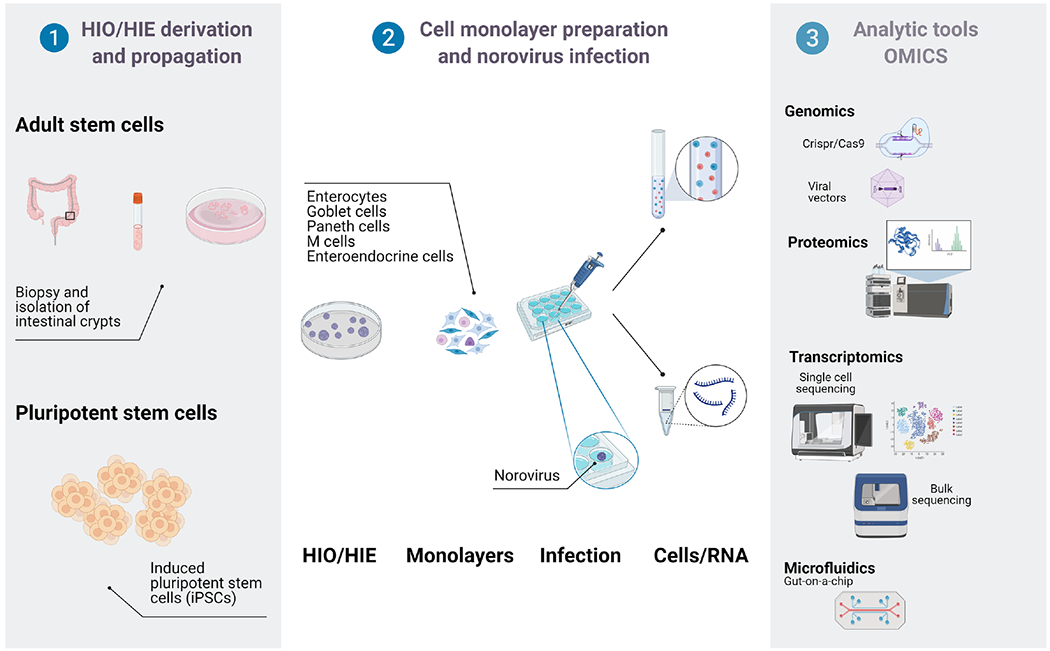
Application of human intestinal enteroids/organoids (HIE/HIO) in understanding the immune response to human norovirus infection. HIE/HIO can be applied in multiple ways to investigate the immune response to norovirus infection and determine host antiviral factors that restrict norovirus replication. HIE and HIO are derived either from adult intestinal biopsies, or adult induced pluripotent cells (iPSCs). Once HIE/HIO are propagated and differentiated, they can be infected with human norovirus. The cells can also be genetically manipulated using CRISPR/Cas9 or viral vectors to create mutant cell lines that can also be used in norovirus studies. Cells and RNA derived from infected HIE/HIO can be analysed for transcriptional changes using RNA sequencing. Similarly, the cells can also be subjected to proteomics-based analyses to investigate effects of norovirus infection on expression. Another potential application of HIE/HIO is measuring metabolites during norovirus infection using gut-on a chip technology. Created with BioRender.com (accessed on 12 October 2021).
Human intestinal enteroid/organoid cultures and norovirus tropism
HIEs and organoid cultures [40, 44–47] have created platforms to study the cellular processes and signalling pathways involved in restricting replication of enteric viruses including human norovirus [48, 49]. Human intestinal organoids (HIO) and HIE are three-dimensional (3D) cultures containing multiple intestinal cell types that are derived from Lgr5+ intestinal stem cells [44] (Fig. 2). HIO contain a mesenchymal niche and are derived from embryonic or pluripotent stem cells (iPSCs) [50], whereas HIE are derived from adult stem cells isolated from intestinal biopsies [40, 51, 52]. These stem cells are propagated in a 3D format supported by Matrigel which allows assembly of the cells into organoid structures that retain cellular composition and physiological functions of the intestinal epithelium [53]. Additionally, adult stem cells are intrinsically programmed with their location-specific function [54], and the differentiated cells that are derived from these stem cells retain an immune profile akin to that of the cells in the corresponding intestinal segment [55]. For replication of human norovirus infection in vitro, these 3D HIE cultures are dissociated and plated as monolayers which are then utilized in a wide variety of studies [56] (Fig. 2).
Successful norovirus replication in vitro using monolayers of HIEs was first reported in 2016 [57] and later confirmed by several other laboratories [58–62]. The cell types found in enteroid cultures include enterocytes, goblet, enteroendocrine, and Paneth cells [53]. Human norovirus potentially replicates in multiple cells types including enterocytes and enteroendocrine cells (EECs) [63, 64]. Presence of human norovirus in enterocytes was first discovered by histological comparison of tissue biopsies from infected and uninfected immunocompromised transplant patients which showed presence of the major capsid protein VP1 in enterocytes from infected individuals [63]. Interestingly, the VP1 expression was also detected in other cells types including, macrophages, T cells and dendritic cells, however, non-structural proteins RdRp and VPg were detected along with VP1 only in enterocytes [63]. Recently human norovirus has been shown to replicate in enteroendocrine epithelial cells (EEC) [64]. Immunohistochemical staining of tissue from the jejunum and ileum of a paediatric intestinal transplant recipient with severe gastroenteritis showed the presence of human norovirus VP1 protein in EEC. Confocal fluorescence microscopy showing colocalization of positive and negative sense human norovirus RNA with the EEC marker (chromogranin A -CgA), confirmed active norovirus replication in this cell type in vivo [64, 65].
The use of commercial media has further optimized human norovirus replication in HIE yielding higher levels of virus replication compared to home-made conditioned media [61]. However, not all norovirus strains can replicate in HIEs [57, 58] with success rate of samples with high viral load as low as 20% [58]. GII.4 viruses demonstrate higher replication levels compared to other genotypes such as GII.3 [57, 58]. A potential explanation for this difference could be that norovirus strains respond differently to the antiviral mechanisms employed by the host cell to restrict virus replication. Other components of the complex intestinal environment, such as the intestinal microbiome and M cells, may also play a role in the strain-specific differences in virus replication [61]. The absence of these components potentially represent major drawbacks of the HIE system.
Human norovirus replication is enhanced or depends on the inclusion of bile acids in the cell culture media [57, 66]. However, the requirement for bile is strain dependent as inclusion is critical for replication of GI.1, GII.1, GII.3, GII.6, and GII.17 strains whereas GII.4 virus replication occurs without supplementation, but is enhanced by bile [61]. This breakthrough has cleared the way for other lines of research including investigation of the antiviral mechanisms that restrict virus replication.
Genetic manipulation of enteroids
Several research groups have begun exploring whether cells derived from HIE and HIO are amendable to genetic manipulation. Using CRISPR/Cas9 technology, a knockout cell line for the FUT2 gene was created [16], FUT2 encodes an enzyme that affects HBGA expression in intestinal epithelial cells and susceptibility to human norovirus infection [17]. This FUT2 knockout cell line demonstrated diminished replication of GII.4, GII.17, and GI.1 viruses. Further, norovirus replication was shown in secretor-negative J4 cells by knocking in the FUT2 gene, thereby demonstrating that FUT2 expression is necessary and sufficient for norovirus replication in HIEs. While the role of FUT2 has been established epidemiologically, these knockout HIE cell lines provided the genetic basis for this observation [16].
In another study which explored the role of interferon signalling in norovirus infection, lentiviral vectors were used to express proteins that antagonize interferon signalling thereby generating intestinal organoid lines incapable of interferon signalling [59]. Specifically, lentiviral vectors were used to express bovine viral diarrhoea virus NPro or parainfluenza virus type 5 (PIV5) V proteins. BVDF Npro blocks IFN production by degrading interferon regulatory factor 3 (IRF3) whereas the PIV5 V protein compromises IFN production and signalling by targeting key molecules such as STAT1, melanoma differentiation-associated protein 5 (MDA5), and LGP2 for degradation [59]. This study demonstrated that enteroid cells are robust enough for transfection and have the potential to be genetically modified to create cells that reliably sustain norovirus replication. Importantly, it showed that the enteroid cells can be modified to increase virus yield by disrupting the interferon signalling pathway, as demonstrated by a 33-fold increase in norovirus GII.3 replication in BVDF Npro-expressing cells compared to control (nontransduced) cells, and a six-fold increase in PIV5 V protein-expressing cells compared to control cells.
What is currently known about innate immune response to norovirus infection?
While the receptor for human norovirus is yet to be found, CD300lf (Fig. 1a) has been identified as the primary receptor for MNV [19, 67–69]. Following viral entry, virus particles are recognized by molecular sensors on the plasma membrane, in endosomes, and in the cytosol which trigger induction of an antiviral or inflammatory response [70] (Fig. 1b). The specific sensors that recognize human norovirus have yet to be identified; however, a recent study showed that Toll-like receptors (TLR) 2 and 5 are activated by norovirus virus-like particles (VLPs) [71] (Fig. 1b). Using a TLR2-transfected HEK293 responder cell line, the authors demonstrated that norovirus VLPs can attach to TLR2. Using a TLR5 expressing cell line with an NF-kB-luciferase cassette, they also demonstrated that norovirus VLPs attachment to TLR5 can induce NF-κB driven inflammatory signalling (Fig. 1b, c). These results suggest that TLR 2 and 5 may be involved in recognition of human norovirus leading to induction of an inflammatory response upon infection. Whether this observation can be recapitulated in a physiologically relevant system such as enterocytes derived from HIE requires additional studies.
An early study utilizing 293FT cells transfected with stool-isolated human norovirus RNA showed that while these cells were capable of replicating norovirus RNA, a robust type I interferon response is not induced [72], which was in contrast with the prominent role of type I interferon in the restriction of MNV replication in macrophages and dendritic cells [73–75]. It has now been demonstrated that human noroviruses indeed induce a robust innate immune response chiefly orchestrated by type I and type III interferon [59, 76, 77] (Fig. 1d). Studies with MNV have further dissected this pathway to reveal pivotal roles for transcription factors STAT1 and 2 as well as interferon regulatory factors (IRF) 1, 3 and 7 [59, 78, 79] (Fig. 1b, e). MDA5, another molecular sensor that recognizes single stranded RNA, is also thought to be involved [80, 81]. Using the Norwalk replicon system, it was shown that MDA5 activation by human norovirus RNA results in activation of the JAK-STAT pathway which leads to production of interferon, a cytokine response that induces an antiviral state in infected cells and surrounding uninfected cells [81]. Fig. 1 summarizes what is currently known regarding the antiviral response to norovirus infection. Although some factors involved in the antiviral response to MNV are included, the immune response to MNV infection was out of the scope of this review and has been covered extensively elsewhere [76, 82].
HIE and the innate immune response to human norovirus infection
Interferon (IFN) is a major component of the antiviral response that is induced upon norovirus infection [75]. To investigate this, HIE cells that had been treated with exogenous IFN (type I IFN [IFNα1 and IFNβ1] or type III IFN [IFNλ1, IFNλ2, and IFNλ3]) were infected with norovirus GII.3 or GII.4. Both strains showed reduction in replication suggesting that GII.4 and GII.3 norovirus strains are sensitive to IFN [77]. Consistent with this finding, when enteroid-derived IFN-receptor-knockout cell lines were infected with GII.3 and GII.4 strains, both strains showed higher levels of replication compared to infection in wild-type cells [77]. However, GII.3 virus replication was rescued to a greater extent than GII.4, suggesting that GII.3 infected cells are more susceptible to IFN restriction [77]. This was further confirmed using transcriptome analysis which demonstrated that human norovirus elicits a predominantly type III IFN response, and that GII.3 strains induced a more robust IFN-stimulated gene response compared to GII.4 strains [77].
Using a specific Janus kinase 1 (JAK1)/JAK2 inhibitor Ruxolitinib (Rux) to disrupt IFN signalling downstream of the IFN-receptor prior to infection of duodenal IECs with GII.3 or GII.4 strains of norovirus resulted in an increase in GII.4 virus replication. This further demonstrated the importance of IFN signalling in restricting virus replication in HIE [59]. Altogether, these studies clearly highlight the benefit of using HIE in understanding the role of IFN in the antiviral response against human norovirus.
Interferon stimulated genes that restrict human norovirus infection
The IFN signalling pathway is a cytokine-based response that results in restriction of virus growth in infected cells and upregulation of antiviral genes in surrounding uninfected cells. IFN secreted from virus-infected cells functions in an autocrine and paracrine manner to engage the IFN-receptors on the cell surface and activate JAK kinases and phosphorylation of STAT1/2 which facilitate upregulation of hundreds of interferon stimulated genes (ISG) (Fig. 1e, f). These ISGs encode effectors of the antiviral response, which antagonize virus replication [83, 84](Fig. 1f).
Little is known about specific antiviral genes that restrict norovirus replication in enteroid/organoid-derived cells. To investigate this, monolayers from two organoid-derived cell lines (terminal ileum organoids) were infected with GII.4 viruses [59] and using RNA-sequencing, 162 genes were found to be differentially regulated in one cell line, and 70 genes were differentially regulated in another cell line [59]. A majority of these were ISGs, demonstrating that human norovirus induces a robust ISG response. The highly upregulated genes included IFI44L, OAS2, OASL, MX-1 and ISG15 which have shown antiviral activity against several viruses including, Zika virus, respiratory syncytial virus, and influenza [85–89].
In another study, transcriptome analysis of two enteroid cell lines using RNA-sequencing also demonstrated a robust transcriptional response 72 h after infection. Additionally, this study found that diverse type I (IFN β) and type III (IFN λ) IFN-driven responses were induced [90]. The use of HIE/HIO has significantly advanced the identification of potential antiviral genes that control norovirus infection. However, much work remains to be done to fully understand the molecular mechanisms by which the antiviral proteins restrict virus replication.
FUTURE PERSPECTIVES
A major caveat of using this enteroid/organoid system is that the gene expression changes that have been found using transcriptomic analysis represent changes in a bulk population of cells. Norovirus replicates in enterocytes, however the proportion of enterocytes that are infected and sustain virus replication is unclear. Furthermore, the extent to which other cellular types in these culture systems can sustain norovirus replication is not known. Other important information for the development of targeted therapies that could mitigate norovirus replication such as the contribution of each cell type to the immune response during infection, remains unclear. Several recent technologies, such as single cell transcriptomics [41, 42, 91], viral proteomics [92, 93] and gut on a chip system [94–96] are promising approaches to further dissect the innate immune response to norovirus.
Single-cell RNA-sequencing has also led to the discovery of rare intestinal cell types [97], and the capability of norovirus to infect these rare cell types is yet to be clarified.
A crucial component of the innate immune response to enteric pathogens are Microfold cells (M cells) [98]. M cells are unique as they function as a first line of defence in an innate immune capacity but also bridge the innate immune response and adaptive immune response by functioning as antigen presenting cells that facilitate the production of antibodies to protect from subsequent infection. Because of their important role in the intestine during the immune response against murine norovirus infection [99, 100], it may be important to develop culture conditions that support differentiation of M cells from enteroid monolayers [101] in order to ascertain their role in human norovirus infection. By using single cell approaches, the transcriptional response in each cell type can be determined to understand the individual contribution of each cell type to the response [102].
Along with single cell transcriptomics, advancements in mass spectrometry and high-throughput cell imaging allow large-scale surveys at protein level [103]. Mass spectrometry proteomics approaches are frequently employed to study cell-viral interactions, how viruses affect cellular signalling pathways, and which cellular proteins are crucial for viral persistence [92, 104–107]. Every virus encodes proteins that manipulate key cellular pathways to promote viral replication and evade the host immune response [92]. Data from a proteomics study on HIE has confirmed that Paneth and goblet cells generated from intestinal stem cells in vitro share features typical of these cell types observed in vivo further confirming that HIE are useful models to investigate normal and disease processes in the intestine [108]. Applying viral proteomics to norovirus infections in enterocytes/HIE will help understand cellular responses during viral pathogenesis as well as in identifying diagnostic and therapeutic targets against human norovirus. Combining data generated by transcriptomics and viral proteomics methods [93] will allow a more comprehensive understanding of the regulatory network driving the human host response to norovirus infection (Fig. 2).
The knowledge derived from using the HIE/HIO system to understand the innate immune response to norovirus infection can be applied to other systems such as the recently developed gut-on-a-chip system which is an innovative in vitro platform for studying gut physiology [94–96]. This technology attempts to mimic the complexity and physiology of native tissues in vitro using cells grown in a series of chambers and maintained in culture medium under conditions that maintain physiological function of the tissue from which the cells were derived [94]. Compared to static cell culture, gut-on-a-chip technology allows the cells to be maintained under mechanically active conditions, and small amounts of the media can be continuously sampled for metabolites, cytokines, or even virus production [94, 95, 109].
Recent work in which this technology was used to study Coxsackie B virus 1 (CVB1) replication in CaCo2 cells showed that gut-on-a-chip has the potential to be applied to norovirus studies [110]. In this study, CaCo2 cells were seeded in a gut-on-a-chip device containing two hollow microchannels separated by a porous membrane. Six days after seeding the cells were polarized and CVB1 was injected into the device allowing for infection on the apical side of the cell monolayers. Media was collected and virus replication and cytokine production (IP-10 and IL-8) were detectable at 24 h post-infection [110]. Differentiated organoid cells also have the potential to be maintained in this microenvironment [111], which could expand the scope of the studies done with organoid cells in norovirus infection.
CONCLUSIONS
HIE/HIO are non-transformed cell culture models that contain multiple intestinal epithelial cell types that comprise the intestinal epithelium. HIEs provide an excellent platform to study human norovirus replication, which until recently has been a major hurdle in advancing human norovirus research. Fundamental questions and challenges can now be addressed to further our understanding of norovirus infection. Remaining questions in the field include identifying the human norovirus receptor that facilitates virus entry, the molecular sensors that trigger an antiviral response once infection has been established as well as the antiviral genes most relevant for restricting norovirus infection.
Another major challenge in studying norovirus replication and innate immune response in differentiated HIEs is the relatively low success for virus replication and difficulty in passaging the viruses. To overcome this, detailed understanding of the complexity of virus host interactions is required. Use of recently described molecular approaches such as RNAseq analysis and CRISPR/Cas9 modification of HIEs have identified many potential immune targets involved in norovirus replication opening opportunities to study the innate immune response after human norovirus infection.
Going forward, advanced technologies such as single cell transcriptomics, viral proteomics and gut on a chip technology may help to better understand the molecular mechanisms that regulate norovirus replication allowing the development of effective prophylactic and therapeutic interventions.
Funding information
This work received no specific grant from any funding agency.
Abbreviations:
- EEC
enteroendocrine epithelial cell
- FUT2
α-1,2-fucosyltransferase
- HBGA
human blood group antigen
- HIE
human intestinal enteroids
- HIO
human intestinal organoids
- IFN
interferon
- ISG
interferon stimulated gene
- JAK
Janus kinase
- MDA5
melanoma differentiation-associated protein 5
- MNV
murine norovirus
- PBMC
peripheral blood mononuclear cell
- PIV5
parainfluenza virus type 5
- RIG-I
retinoic acid-inducible gene 1
- STAT
signal transducer and activator of transcription
- TLR
toll-like receptor
- VLP
virus-like particle
- VP1
virus protein 1
- VPg
viral protein genome-linked
Footnotes
Conflicts of interest
The authors declare that there are no conflicts of interest.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
References
- 1.Ahmed SM, Hall AJ, Robinson AE, Verhoef L, Premkumar P, et al. Global prevalence of norovirus in cases of gastroenteritis: a systematic review and meta-analysis. Lancet infect Dis 2014;14:725–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hall AJ, Lopman BA, Payne DC, Patel MM, Gastañaduy PA, et al. Norovirus disease in the United States. Emerg infect Dis 2013;19:1198–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teunis PFM, Moe CL, Liu P, Miller SE, Lindesmith L, et al. Norwalk virus: how infectious is it? J Med Virol 2008;80:1468–1476. [DOI] [PubMed] [Google Scholar]
- 4.Vinjé J Advances in laboratory methods for detection and typing of norovirus. J Clin Microbiol 2015;53:373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atmar RL, Opekun AR, Gilger MA, Estes MK, Crawford SE, et al. Determination of the 50% human infectious dose for Norwalk virus. J Infect Dis 2014;209:1016–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donaldson EF, Lindesmith LC, Lobue AD, Baric RS. Norovirus pathogenesis: mechanisms of persistence and immune evasion in human populations. Immunol Rev 2008;225:190–211. [DOI] [PubMed] [Google Scholar]
- 7.Division of Viral Diseases NCfI, Respiratory Diseases CfDC, Prevention. Updated norovirus outbreak management and disease prevention guidelines. MMWR Recomm Rep 2011;60:1–18. [PubMed] [Google Scholar]
- 8.Barclay L, Park GW, Vega E, Hall A, Parashar U, et al. Infection control for norovirus. Clin Microbiol Infect 2014;20:731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cates JE, Vinjé J, Parashar U, Hall AJ. Recent advances in human norovirus research and implications for candidate vaccines. Expert Rev Vaccines 2020;19:539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seitz SR, Leon JS, Schwab KJ, Lyon GM, Dowd M, et al. Norovirus infectivity in humans and persistence in water. Appl Environ Microbiol 2011;77:6884–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindesmith L, Moe C, Marionneau S, Ruvoen N, Jiang X, Human susceptibility and resistance to Norwalk virus infection. Nat Med 2003;9:548–553. [DOI] [PubMed] [Google Scholar]
- 12.Newman KL, Moe CL, Kirby AE, Flanders WD, Parkos CA, Human norovirus infection and the acute serum cytokine response. Clin Exp Immunol 2015;182:195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernstein DI, Atmar RL, Lyon GM, Treanor JJ, Chen WH, et al. Norovirus vaccine against experimental human GII.4 virus illness: a challenge study in healthy adults. J Infect Dis 2015;211:870–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atmar RL, Bernstein DI, Lyon GM, Treanor JJ, Al-Ibrahim MS, Serological correlates of protection against a GII.4 norovirus. Clin Vaccine Immunol 2015;22:923–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Currier RL, Payne DC, Staat MA, Selvarangan R, Shirley SH, Innate susceptibility to norovirus infections influenced by FUT2 genotype in a United States pediatric population. Clin Infect Dis 2015;60:1631–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haga K, Ettayebi K, Tenge VR, Karandikar UC, Lewis MA, Genetic manipulation of human intestinal enteroids demonstrates the necessity of a functional fucosyltransferase 2 gene for secretor-dependent human norovirus infection. mBio 2020;11 :e00251–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nordgren J, Svensson L. Genetic susceptibility to human norovirus infection: an update. Viruses 2019;11:E226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindesmith LC, Brewer-Jensen PD, Mallory ML, Jensen K, Yount BL, et al. Virus-host interactions between nonsecretors and human norovirus. Cell Mol Gastroenterol Hepatol 2020;10:245–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graziano VR, Wei J, Wilen CB. Norovirus attachment and entry. Viruses 2019;11:E495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kendra JA, Tohma K, Ford-Siltz LA, Lepore CJ, Parra GI. Antigenic cartography reveals complexities of genetic determinants that lead to antigenic differences among pandemic GII.4 noroviruses. Proc Natl Acad Sci U S A. 2021;118:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Costantini VP, Cooper EM, Hardaker HL, Lee LE, DeBess EE, et al. Humoral and mucosal immune responses to human norovirus in the elderly. J Infect Dis 2020;221:1864–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reeck A, Kavanagh O, Estes MK, Opekun AR, Gilger MA, et al. Serological correlate of protection against norovirus-induced gastroenteritis. J Infect Dis 2010;202:1212–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindesmith LC, Donaldson E, Leon J, Moe CL, Frelinger JA, et al. Heterotypic humoral and cellular immune responses following Norwalk virus infection. J Virol 2010;84:1800–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duizer E, Schwab KJ, Neill FH, Atmar RL, Koopmans MPG, et al. Laboratory efforts to cultivate noroviruses. J Gen Virol 2004;85:79–87. [DOI] [PubMed] [Google Scholar]
- 25.Lay MK, Atmar RL, Guix S, Bharadwaj U, He H, et al. Norwalk virus does not replicate in human macrophages or dendritic cells derived from the peripheral blood of susceptible humans. Virology 2010;406:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones MK, Grau KR, Costantini V, Kolawole AO, de Graaf M, et al. Human norovirus culture in B cells. Nat Protoc 2015;10:1939–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones MK, Watanabe M, Zhu S, Graves CL, Keyes LR, et al. Enteric bacteria promote human and mouse norovirus infection of B cells. Science 2014;346:755–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang KO, Sosnovtsev SV, Belliot G, King AD, Green KY. Stable expression of a Norwalk virus RNA replicon in a human hepatoma cell line. Virology 2006;353:463–473. [DOI] [PubMed] [Google Scholar]
- 29.Katayama K, Murakami K, Sharp TM, Guix S, Oka T, et al. Plasmid-based human norovirus reverse genetics system produces reporter-tagged progeny virus containing infectious genomic RNA. Proc Natl Acad Sci U S A 2014;111:E4043–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arthur SE, Sorgeloos F, Hosmillo M, Goodfellow IG. Epigenetic Suppression of Interferon Lambda Receptor Expression Leads to Enhanced Human Norovirus Replication In Vitro. mBio 2019; 10 :e02155–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rockx BHG, Bogers WMJM, Heeney JL, van Amerongen G, Koopmans MPG. Experimental norovirus infections in non-human primates. J Med Virol 2005;75:313–320. [DOI] [PubMed] [Google Scholar]
- 32.Bok K, Parra GI, Mitra T, Abente E, Shaver CK, et al. Chimpanzees as an animal model for human norovirus infection and vaccine development. Proc Natl Acad Sci U S A 2011;108:325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheetham S, Souza M, Meulia T, Grimes S, Han MG, et al. Pathogenesis of a genogroup II human norovirus in gnotobiotic pigs. J Virol 2006;80:10372–10381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park B-J, Jung S-T, Choi C-S, Myoung J, Ahn H-S, et al. Pathogenesis of human norovirus genogroup II genotype 4 in post-weaning gnotobiotic pigs. J Microbiol Biotechnol 2018;28:2133–2140. [DOI] [PubMed] [Google Scholar]
- 35.Taube S, Kolawole AO, Höhne M, Wilkinson JE, Handley SA, et al. A mouse model for human norovirus. mBio 2013;4:e00450–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Dycke J, Ny A, Conceição-Neto N, Maes J, Hosmillo M, et al. A robust human norovirus replication model in zebrafish larvae. PLoS Pathog 2019;15:e1008009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wobus CE, Thackray LB, Virgin HW 4th. Murine norovirus: a model system to study norovirus biology and pathogenesis. J Virol 2006;80:5104–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ha S, Choi IS, Choi C, Myoung J. Infection models of human norovirus: challenges and recent progress. Arch Virol 2016;161:779–788. [DOI] [PubMed] [Google Scholar]
- 39.Vashist S, Bailey D, Putics A, Goodfellow I. Model systems for the study of human norovirus biology. Future Virol 2009;4:353–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sato T, Stange DE, Ferrante M, Vries RGJ, Van Es JH, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 2011;141:1762–1772. [DOI] [PubMed] [Google Scholar]
- 41.Hedlund E, Deng Q. Single-cell RNA sequencing: technical advancements and biological applications. Mol Aspects Med 2018;59:36–46. [DOI] [PubMed] [Google Scholar]
- 42.Kulkarni A, Anderson AG, Merullo DP, Konopka G. Beyond bulk: a review of single cell transcriptomics methodologies and applications. Curr Opin Biotechnol 2019;58:129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hrdlickova R, Toloue M, Tian B. RNA-Seq methods for transcriptome analysis. Wiley Interdiscip Rev RNA 2017;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramani S, Crawford SE, Blutt SE, Estes MK. Human organoid cultures: transformative new tools for human virus studies. Curr Opin Virol 2018;29:79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.In JG, Foulke-Abel J, Estes MK, Zachos NC, Kovbasnjuk O, et al. Human mini-guts: new insights into intestinal physiology and host-pathogen interactions. Nat Rev Gastroenterol Hepatol 2016;13:633–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Almeqdadi M, Mana MD, Roper J, Yilmaz ÖH. Gut organoids: mini-tissues in culture to study intestinal physiology and disease. Am J Physiol Cell Physiol 2019;317:C405–C419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009;459:262–265. [DOI] [PubMed] [Google Scholar]
- 48.Kolawole AO, Wobus CE. Gastrointestinal organoid technology advances studies of enteric virus biology. PLoS Pathog 2020;16:e1008212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crawford SE, Ramani S, Blutt SE, Estes MK. Organoids to Dissect Gastrointestinal Virus-Host Interactions: What Have We Learned? Viruses 2021;13:999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pleguezuelos-Manzano C, Puschhof J, van den Brink S, Geurts V, Beumer J, et al. Establishment and Culture of Human Intestinal Organoids Derived from Adult Stem Cells. Curr Protoc Immunol 2020;130:e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mahe MM, Sundaram N, Watson CL, Shroyer NF, Helmrath MA. Establishment of human epithelial enteroids and colonoids from whole tissue and biopsy. J Vis Exp 2015;2015:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stelzner M, Helmrath M, Dunn JCY, Henning SJ, Houchen CW, et al. A nomenclature for intestinal in vitro cultures. Am J Physiol Gastrointest Liver Physiol 2012;302:G1359–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zachos NC, Kovbasnjuk O, Foulke-Abel J, In J, Blutt SE, et al. Human Enteroids/Colonoids and Intestinal Organoids Functionally Recapitulate Normal Intestinal Physiology and Pathophysiology. J Biol Chem 2016;291:3759–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Middendorp S, Schneeberger K, Wiegerinck CL, Mokry M, Akkerman RDL, et al. Adult stem cells in the small intestine are intrinsically programmed with their location-specific function. Stem Cells 2014;32:1083–1091. [DOI] [PubMed] [Google Scholar]
- 55.Kayisoglu O, Weiss F, Niklas C, Pierotti I, Pompaiah M, et al. Location-specific cell identity rather than exposure to GI microbiota defines many innate immune signalling cascades in the gut epithelium. Gut 2021;70:687–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meneses AMC, Schneeberger K, Kruitwagen HS, Penning LC, van Steenbeek FG, et al. Intestinal Organoids-Current and Future Applications. Vet Sci 2016;3:E31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, et al. Replication of human noroviruses in stem cell-derived human enteroids. Science 2016;353:1387–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Costantini V, Morantz EK, Browne H, Ettayebi K, Zeng X-L, et al. Human Norovirus Replication in Human Intestinal Enteroids as Model to Evaluate Virus Inactivation. Emerg Infect Dis 2018;24:1453–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hosmillo M, Chaudhry Y, Nayak K, Sorgeloos F, Koo B-K, et al. Norovirus replication in human intestinal epithelial cells is restricted by the interferon-induced JAK/STAT signaling pathway and RNA polymerase II-mediated transcriptional responses. mBio 2020; 11 :e00215–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chan MC-W, Cheung SKC, Mohammad KN, Chan JCM, Estes MK, et al. Use of human intestinal enteroids to detect human norovirus infectivity. Emerg Infect Dis 2019;25:1730–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ettayebi K, Tenge VR, Cortes-Penfield NW, Crawford SE, Neill FH, et al. New insights and enhanced human norovirus cultivation in human intestinal enteroids. mSphere 2021;6:e01136–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Randazzo W, Costantini V, Morantz EK, Vinjé J. Human intestinal enteroids to evaluate human norovirus GII.4 inactivation by aged-green tea. Front Microbiol 2020;11:1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karandikar UC, Crawford SE, Ajami NJ, Murakami K, Kou B, et al. Detection of human norovirus in intestinal biopsies from immunocompromised transplant patients. J Gen Virol 2016;97:2291–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Green KY, Kaufman SS, Nagata BM, Chaimongkol N, Kim DY, et al. Human norovirus targets enteroendocrine epithelial cells in the small intestine. Nat Commun 2020;11:2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goldspink DA, Reimann F, Gribble FM. Models and tools for studying enteroendocrine cells. Endocrinology 2018;159:3874–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Murakami K, Tenge VR, Karandikar UC, Lin S-C, Ramani S, et al. Bile acids and ceramide overcome the entry restriction for GII.3 human norovirus replication in human intestinal enteroids. Proc Natl Acad Sci U S A 2020;117:1700–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Graziano VR, Walker FC, Kennedy EA, Wei J, Ettayebi K, et al. CD300lf is the primary physiologic receptor of murine norovirus but not human norovirus. PLoS Pathog 2020;16:e1008242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haga K, Fujimoto A, Takai-Todaka R, Miki M, Doan YH, et al. Functional receptor molecules CD300lf and CD300ld within the CD300 family enable murine noroviruses to infect cells. Proc Natl Acad Sci U S A 2016;113:E6248–E6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Orchard RC, Wilen CB, Doench JG, Baldridge MT, McCune BT, et al. Discovery of a proteinaceous cellular receptor for a norovirus. Science 2016;353:933–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jensen S, Thomsen AR. Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J Virol 2012;86:2900–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ponterio E, Mariotti S, Tabolacci C, Ruggeri FM, Nisini R. Virus like particles of GII.4 norovirus bind Toll Like Receptors 2 and 5. Immunol Lett 2019;215:40–44. [DOI] [PubMed] [Google Scholar]
- 72.Qu L, Murakami K, Broughman JR, Lay MK, Guix S, et al. Replication of human norovirus RNA in mammalian cells reveals lack of interferon response. J Virol 2016;90:8906–8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wobus CE, Karst SM, Thackray LB, Chang K-O, Sosnovtsev SV, et al. Replication of norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol 2004;2:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nice TJ, Osborne LC, Tomov VT, Artis D, Wherry EJ, et al. Type I interferon receptor deficiency in dendritic cells facilitates systemic murine norovirus persistence despite enhanced adaptive immunity. PLoS Pathog 2016;12:e1005684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ingle H, Peterson ST, Baldridge MT. Distinct effects of type I and III interferons on enteric viruses. Viruses 2018;10:E46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sarvestani ST, Cotton B, Fritzlar S, O’Donnell TB, Mackenzie JM. Norovirus infection: replication, manipulation of host, and interaction with the host immune response. J Interferon Cytokine Res 2016;36:215–225. [DOI] [PubMed] [Google Scholar]
- 77.Lin S-C, Qu L, Ettayebi K, Crawford SE, Blutt SE, et al. Human norovirus exhibits strain-specific sensitivity to host interferon pathways in human intestinal enteroids. Proc Natl Acad Sci U S A 2020;117:23782–23793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thackray LB, Duan E, Lazear HM, Kambal A, Schreiber RD, et al. Critical role for interferon regulatory factor 3 (IRF-3) and IRF-7 in type I interferon-mediated control of murine norovirus replication. J Virol 2012;86:13515–13523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Peterson ST, Kennedy EA, Brigleb PH, Taylor GM, Urbanek K, et al. Disruption of type III interferon (IFN) genes Ifnl2 and Ifnl3 recapitulates loss of the type III IFN receptor in the mucosal antiviral response. J Virol 2019;93:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.MacDuff DA, Baldridge MT, Qaqish AM, Nice TJ, Darbandi AD, et al. HOIL1 is essential for the induction of type I and III interferons by MDA5 and regulates persistent murine norovirus infection. J Virol 2018;92:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li Y, Yu P, Qu C, Li P, Li Y, et al. MDA5 against enteric viruses through induction of interferon-like response partially via the JAK-STAT cascade. Antiviral Res 2020;176:104743. [DOI] [PubMed] [Google Scholar]
- 82.Newman KL, Leon JS. Norovirus immunology: of mice and mechanisms. Eur J Immunol 2015;45:2742–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hemann EA, Gale M, Savan R. Interferon lambda genetics and biology in regulation of viral control. Front Immunol 2017;8:1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mesev EV, LeDesma RA, Ploss A. Decoding type I and III interferon signaling during viral infection. Nat Microbiol 2019;4:914–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liao X, Xie H, Li S, Ye H, Li S, et al. 2’, 5’-oligoadenylate synthetase 2 (OAS2) inhibits zika virus replication through activation of type I IFN signaling pathway. Viruses 2020;12:E418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Verhelst J, Hulpiau P, Saelens X. Mx proteins:antiviral gatekeepers that restrain the uninvited. Microbiol Mol Biol Rev 2013;77:551–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhu J, Ghosh A, Sarkar SN. OASL-a new player in controlling antiviral innate immunity. Curr Opin Virol 2015;12:15–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Perng YC, Lenschow DJ. ISG15 in antiviral immunity and beyond. Nat Rev Microbiol 2018;16:423–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Busse DC, Habgood-Coote D, Clare S, Brandt C, Bassano I, et al. Interferon-induced protein 44 and interferon-induced protein 44-like restrict replication of respiratory syncytial virus. J Virol 2020;94:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lin L, Han J, Yan T, Li L, Li J, et al. Replication and transcriptionomic analysis of human noroviruses in human intestinal enteroids. Am J Transl Res 2019;11:3365–3374. [PMC free article] [PubMed] [Google Scholar]
- 91.Paolillo C, Londin E, Fortina P. Single-cell genomics. Clin Chem 2019;65:972–985. [DOI] [PubMed] [Google Scholar]
- 92.McBride AA. The promise of proteomics in the study of oncogenic viruses. Mol Cell Proteomics 2017;16:S65–S74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jean Beltran PM, Federspiel JD, Sheng X, Cristea IM. Proteomics and integrative omic approaches for understanding host-pathogen interactions and infectious diseases. Mol Syst Biol 2017;13:922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ashammakhi N, Nasiri R, Barros NR de, Tebon P, Thakor J, et al. Gut-on-a-chip: current progress and future opportunities. Biomaterials 2020;255:120196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bein A, Shin W, Jalili-Firoozinezhad S, Park MH, Sontheimer-Phelps A, et al. Microfluidic organ-on-a-chip models of human intestine. Cell Mol Gastroenterol Hepatol 2018;5:659–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xiang Y, Wen H, Yu Y, Li M, Fu X, et al. Gut-on-chip: recreating human intestine in vitro. J Tissue Eng 2020;11:2041731420965318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Grün D, Lyubimova A, Kester L, Wiebrands K,Basak O, et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature 2015;525:251–255. [DOI] [PubMed] [Google Scholar]
- 98.Corr SC, Gahan CCGM, Hill C. M-cells: origin, morphology and role in mucosal immunity and microbial pathogenesis. FEMS Immunol Med Microbiol 2008;52:2–12. [DOI] [PubMed] [Google Scholar]
- 99.Grau KR, Roth AN, Zhu S, Hernandez A, Colliou N, et al. The major targets of acute norovirus infection are immune cells in the gut-associated lymphoid tissue. Nat Microbiol 2017;2:1586–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Karst SM, Wobus CE. A working model of how noroviruses infect the intestine. PLoS Pathog 2015;11:e1004626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fasciano AC, Blutt SE, Estes MK, Mecsas J. Induced differentiation ofDifferentiation of M cell-like cells in human stem cell-derived ileal enteroid monolayersCell-like Cells in Human Stem Cell-derived Ileal Enteroid Monolayers. J Vis Exp 2019;2019:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mead BE, Ordovas-Montanes J, Braun AP, Levy LE, Bhargava P, et al. Harnessing single-cell genomics to improve the physiological fidelity of organoid-derived cell types. BMC Biol 2018;16:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet 2012;13:227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Muller M, Demeret C. The HPV E2-host protein-protein interactions: a complex hijacking of the cellular network. Open Virol J 2012;6:173–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Malik-Soni N, Frappier L. Proteomic profiling of EBNA1-host protein interactions in latent and lytic epstein-barr virus infections. J Virol 2012;86:6999–7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gan J, Wang C, Jin Y, Guo Y, Xu F, et al. Proteomic profiling identifies the SIM-associated complex of KSHV-encoded LANA. Proteomics 2015;15:2023–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sperk M, van Domselaar R, Rodriguez JE, Mikaeloff F, Sá Vinhas B, et al. Utility of proteomics in emerging and re-emerging infectious diseases caused by RNA viruses. J Proteome Res 2020;19:4259–4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Luu L, Matthews ZJ, Armstrong SD, Powell PP, Wileman T, et al. Proteomic profiling of enteroid cultures skewed toward development of specific epithelial lineages. Proteomics 2018;18:e1800132. [DOI] [PubMed] [Google Scholar]
- 109.Kim HJ, Ingber DE. Gut-on-a-chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr Biol (Camb) 2013;5:1130–1140. [DOI] [PubMed] [Google Scholar]
- 110.Villenave R, Wales SQ, Hamkins-Indik T, Papafragkou E, Weaver JC, et al. Human gut-on-a-chip supports polarized infection of coxsackie B1 virus in vitro. PLoS One 2017;12:e0169412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yu F, Hunziker W, Choudhury D. Engineering microfluidic organoid-on-a-chip platforms. Micromachines (Basel) 2019;10:E165. [DOI] [PMC free article] [PubMed] [Google Scholar]


