Abstract
Background
The skin is the largest organ in the human body and serves as a multilayered protective shield from the environment as well as a sensor and thermal regulator. However, despite its importance, many details about skin structure and function at the molecular level remain incompletely understood. Recent advances in liquid chromatography tandem mass spectrometry (LC-MS/MS) proteomics have enabled the quantification and characterization of the proteomes of a number of clinical samples, including normal and diseased skin.
Summary
Here, we review the current state of the art in proteomic analysis of the skin. We provide a brief overview of the technique and skin sample collection methodologies as well as a number of recent examples to illustrate the utility of this strategy for advancing a broader understanding of the pathology of diseases as well as new therapeutic options.
Key Messages
Proteomic studies of healthy skin and skin diseases can identify potential molecular biomarkers for improved diagnosis and patient stratification as well as potential targets for drug development. Collectively, efforts such as the Human Skinatlas offer improved opportunities for enhancing clinical practice and patient outcomes.
Keywords: Proteomics, Label-free mass spectrometry, Melanoma, Psoriasis, Eczema
Introduction
In medicine, including dermatology, we have traditionally relied on patients' medical history and examination for the diagnosis and choice of treatments. However, over the last two decades, medical practice and clinical research have undergone a remarkable transformation fueled by technological breakthroughs, most notably in DNA sequencing. Decreasing sequencing costs and increasing speed and accuracy have moved genetics and genomics into mainstream medicine. This has allowed the medical community to begin to envision the future of personalized and precision medicine where each patient would receive care appropriate to their individual physiology (physiome). Although the knowledge of individuals' genetic makeup is a critical first step towards this goal, the full understanding and evaluation of individual physiology would benefit from knowledge of the proteome, the list and quantities of all the proteins present, as well as other readouts of physiology. This review will focus on proteomics, more specifically the recent advances in proteomic analysis of the skin. We will first introduce the technology of proteomics, especially liquid chromatography tandem mass spectrometry (LC-MS/MS)-based proteomics, as a clinically relevant strategy to diagnose and characterize disease as well as monitor response to treatment. We will provide comments on current state-of-the-art methodology for sample collection and analysis, exemplified by the recently published “proteomic Skinatlas” (http://skin.science) and discuss several examples of how MS-based proteomics can be used in the context of precision dermatology.
MS-Based Proteomics in Clinical Research and Practice
Proteomics is the study of proteins encoded by the genome of an organism and expressed at a given state [1, 2, 3]. The identification of the protein composition of clinical samples provides the opportunity to characterize diseases at a deeper level and leads to a broader understanding of the pathology of skin diseases; it also promotes the development and evaluation of new therapeutic options. Given that proteins carry out cellular and tissue function, they more accurately define the disease state than genomics alone [4]. Additionally, in contrast to whole-genome analysis, the proteome is highly dynamic [3], as the expression of proteins reflects a balance of linked processes, from the transcription, processing, and degradation of mRNAs to the translation, localization, modification, and programed destruction of the proteins [5, 6]. Therefore, just knowing the identity of genes and/or the level at which they are transcribed, the transcriptome cannot fully predict protein abundance and function [5, 6]. More importantly, proteins are subject to an array of posttranslational modifications, which introduce different types of moieties into proteins, ranging from small chemical groups, such as a phosphate, one of the most common modifications in cellular signal transduction, to attachment of whole proteins, e.g., ubiquitin, in a process that regulates protein degradation and turnover. Understanding protein composition structure and function may therefore also require a detailed analysis of protein properties such as posttranslational modifications, protein localization, protein synthesis and turnover rate, protein interaction, and tissue distribution, in addition to the relative or absolute abundance of proteins [3, 4, 7].
Currently, most clinical research and practice relies on antibody-based methods, such as immunohistochemistry and enzyme-linked immunosorbent assay (ELISA), for protein detection and quantification [4, 8, 9]. Although mostly reliable and widely implemented, these methods are restricted to individual and already known proteins, and therefore provide a limited amount of information [3, 4, 10]. Implementing MS-based proteomics in a clinical setting would open opportunities to provide a much deeper view of the proteins that are present and in what quantities in a given sample at a given time. The fast-paced technological developments in the field of MS-based techniques have already resulted in clinical applications such as bacterial and fungal phenotyping, newborn screenings, and urine toxicology screenings [11]. Moreover, MS-based proteomic research is poised to advance discoveries in clinical medicine, including subclassification of diseases, identification of targets for treatment, prediction of treatment response, and prognosis [10]. For example, quantitative MS-based protein analysis on formalin-fixed paraffin-embedded (FFPE) tissue samples enabled classification of the large B-cell lymphoma subtypes according to the cell of origin [12]. In another example, the strategy was used to stratify patients with non-small-cell lung cancer and enabled selection of patients for treatment with a specific agent, the oral receptor tyrosine kinase inhibitor crizotinib [13], and it has shown promise for the development of new diagnostic tests [10, 13]. Interestingly, a proteomic study of medulloblastoma identified substantial differences at the protein level, but not at the genomic and transcriptomic levels, enabling subsequent classification into subgroups with a potential for developing future treatment strategies [14].
These examples only scratch the surface of the wide range of MS-based proteomics applications and the way this level of granular insight could empower clinicians. For the purpose of this review, we will focus on label-free MS-based proteomics in the context of dermatology. In brief, “label-free” methods refer to a subset of quantitative MS-based proteomic strategies that do not require the use of isotope labeling to identify and quantify proteins present in the sample, making them suitable for clinical implementation. Moreover, we have found label-free MS proteomics to be fast and to have wider dynamic range and simple sample preparation, while producing reliable results and deep proteome coverage [3, 15].
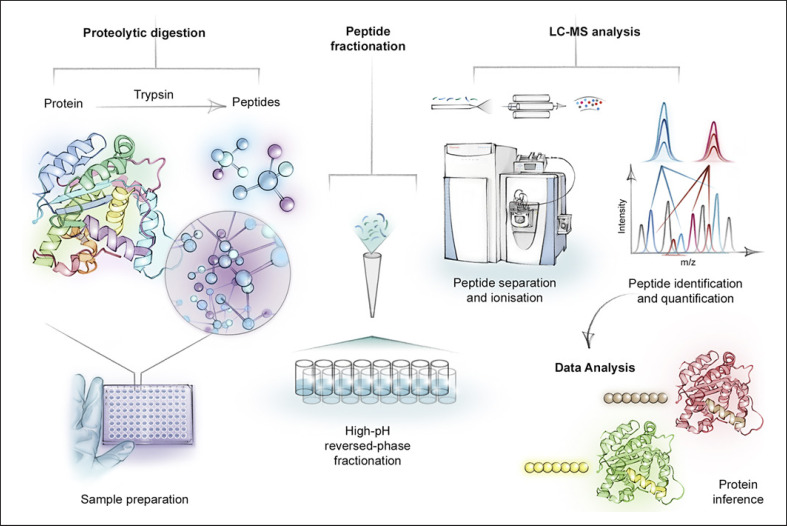
The LC-MS/MS pipeline includes several preparation steps, an LC analytical step, and two MS analysis steps. The process also involves the use of proteolytic enzymes to specifically break down proteins into smaller fragments, namely peptides (Fig. 1). The peptide mixture is first separated by reverse-phase LC, based on hydrophobicity, and then electrosprayed as ionized peptide ions that are transported into the vacuum of a mass spectrometer [16]. In the mass spectrometer the peptides are fragmented to generate MS/MS spectra, adding another dimension of information to identify and quantify the peptides [3, 17]. Proteomics experiments can be performed in “shotgun” or targeted modes. In the former as many peptides as possible are analyzed whereas in the latter just a few peptides of interest are repeatedly analyzed. Label-free shotgun proteomics can similarly be performed in two ways: (1) Data-dependent acquisition, whereby the first MS step selects the most abundant peptides, followed by fragmentation and second MS analysis of those fragments (tandem MS). (2) More consistent coverage of the proteome is achieved by scanning across the entire range of m/z values in the first MS step in data-independent acquisition. The information from these experiments is fed into a bioinformatics data processing pipeline where peptide fragments are matched to a proteomic database using statistical methods that rigorously control the false discovery rates in peptide and protein identification [3]. When followed by quantitative proteomic algorithms, this workflow results in an inventory of protein abundances in complex samples, such as skin tissue, without the need to identify the proteins in advance [3]. For more about label-free MS-based proteomics and other aspects of proteomic research, we refer the reader to several recent reviews that include a critical assessment of this strategy [3, 18].
Fig. 1.
Bottom-up proteomics workflow begins with sample preparation. In the first step, samples are homogenized and proteins are extracted and digested by a sequence-specific enzyme such as trypsin. In some cases, samples can be fractionated (through high-pH reversed-phase fractionation) to produce less complex samples. The cleaved and fractionated peptides are separated by reverse-phase liquid chromatography and electrosprayed as ionized peptides into the tandem mass spectrometer. In the mass spectrometer the peptide mixture undergoes tandem mass spectrometry fragmentation and sequence data are obtained. Proteins are identified and quantified by peptides that have unique sequences with the help of bioinformatic software.
Skin Sample Collection and Preparation for MS-Based Proteomic Analysis
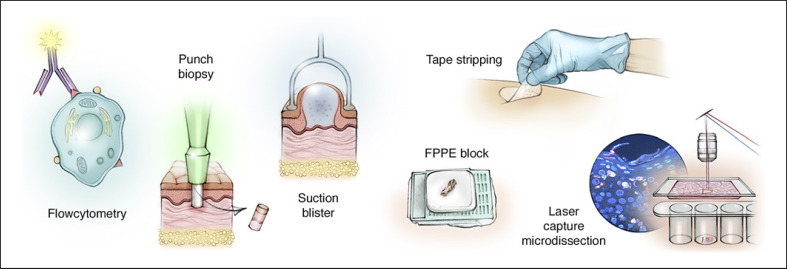
The collection and initial processing of the protein-containing material is critical in every branch of proteomics. In the context of dermatology, there are several specific challenges and corresponding types of skin sampling techniques that enable MS-based proteomic studies (Fig. 2). Skin biopsies are extensively used for histological and immunological examination to diagnose skin diseases [19, 20]. The biopsies are often FFPE-stained due to the feasibility for long preservation [10]. Both FFPE and snap-frozen biopsies have been proven useful for proteomic investigations with LC-MS/MS-based protein research (Fig. 2) [21, 22].
Fig. 2.
In dermatology, several types of skin sampling techniques enable mass spectrometry-based proteomic studies. In the clinical setting, the skin biopsies are often preserved as formalin-fixed paraffin-embedded samples and these samples can be analyzed whole or more selectively through the use of laser capture microdissection. Mass spectrometry-based proteomic analysis can also be performed on skin layers produced by suction blisters. Individual cells or subpopulations of cells can be collected by flow cytometry or tape stripping.
In addition to whole skin samples, there are multiple ways to sample portions of the skin. Tape stripping has the advantage of being easy to use and is noninvasive [19, 20]. However, tape stripping is limited to studies of the stratum corneum and cannot be used for protein analysis of the deeper part of the epidermis and dermis [20]. Suction blisters is a sampling technique that allows access to the deeper layers of the epidermis and uses a vacuum pump to detach the epidermis from the dermis [23]. The technique is time-consuming but associated with little pain and is less invasive than punch biopsies. There are reliable methods for protein analysis of the blister fluid or cells [23, 24]. Cells from the suction blisters or single-cell suspensions can be sorted using cytofluorimetric analyses and subsequently analyzed by MS-based proteomics. Recent technological advances have led to ultra-high sensitivity where in-depth characterization of cellular function of individual cells is attainable [25].
Collected skin samples can be further processed using laser capture microdissection, which is useful for proteomic analyses of specific cells or areas of interest within the skin [26, 27]. Laser capture microdissection can be applied to several preparations, such as heterogenous tissue sections, cytological preparations, or live cell cultures [26, 28]. The advantage of laser capture microdissection is its precision [29]. Laser capture microdissection can be limited due to its reliance on fragility and visual discrimination of target cells and due to other sample preparation challenges related to tissue staining and fixation techniques [26, 28, 30]. Some of these challenges can be addressed by combining artificial intelligence-driven image analysis of cellular phenotypes with automated single-cell laser microdissection and ultra-high-sensitivity MS. This novel technique, called deep visual proteomics, links protein abundance to complex cellular or even subcellular phenotypes while preserving spatial context [31].
After collecting the clinical samples, the selected tissue sections are processed to extract the proteins for subsequent proteomic analysis [32]. Samples that include all layers of the skin are challenging to work with due to the dominance of structural proteins such as keratins and collagens in some of them that contribute to the resilience of the skin. In addition, these high-abundant structural proteins result in a high dynamic range (the difference between most abundant and least abundant proteins), which makes it difficult to detect low-abundant proteins in the same samples. To extract proteins, the skin can be disintegrated by homogenization and subsequently digested enzymatically [3, 17] before the sample is advanced through the LC-MS/MS workflow described above. In general, the most important aspects of skin sample collection and preparation are strategic planning to determine which layers of the skin or skin-associated cells are of interest and to choose the appropriate sampling technique to ensure best performance of the subsequent proteomics runs, as well as improve reproducibility.
Label-Free LC-MS/MS Proteomics in Dermatological Studies
In dermatology, clinicians have for decades been using knowledge of histological anatomy and relying on visual structural characterization of tissue samples, often combined with immunohistochemistry to support the diagnostic process [33]. A multitude of proteins have been described and characterized in this way [34]. Recently, the advent of proteomics has opened opportunities to analyze skin samples in new ways to identify additional biomarkers that can guide clinicians with the diagnosis, prognosis, and treatment of dermatological conditions. Below we summarize some of the recent examples of proteomic studies done on both healthy and diseased human skin samples. Due to lack of space, murine studies and studies of skin-associated cell cultures or immune cells will not be discussed here.
Proteomic Analysis of Healthy Human Skin
Human skin is a complex organ with functionally distinct layers and cell types as well as high abundance of extracellular matrix. As mentioned, its complexity results in difficulties in separating and homogenizing the tissue for mass spectrometric analyses, and the dominance of structural proteins further complicates the process. Consequently, until recently, prior studies have only identified a relatively small fraction of the expressed skin proteome [21, 22, 33, 35, 36, 37, 38, 39]. For example, Mikesh et al. [33] analyzed the composition in three skin layers from two anatomical regions, resulting in characterization of about 200 proteins and providing insight into varying protein composition depending on skin layer and skin region. However, this represents only a narrow snapshot of the skin, given the low number of characterized proteins and the limited number of skin layers and regions sampled.
To fill this knowledge gap, we have recently created a comprehensive quantitative proteomic atlas of healthy human skin using MS-based proteomics with an advanced sampling strategy that accounts for the cellular complexity of the skin. This resulted in a spatially resolved proteomes atlas of skin layers and skin-associated immune cells including 10,701 proteins [40]. This rich resource for the community provides the deepest proteomic coverage of the main skin layers and cell types. The study comprises separate proteomic analyses of four skin layers (stratum corneum, inner epidermis, dermis, subcutis) as well as four skin cell subsets (fibroblasts, keratinocytes, melanocytes, endothelial cells) and five different skin-associated immune cells (macrophages, dendritic cells, mast cells, epidermal and dermal T cells). Collectively, these data should enable community-wide efforts to better understand the healthy human skin. To enable broad and unrestricted use of this resource and to accelerate the pace of discovery in this area, we have made the data publicly available at https://skin.science.
Proteomic Studies of Malignant Melanomas
Genetic and molecular characterization of melanoma has recently led to the development of targeted treatments, such as immunotherapy and treatments targeting the BRAF/MEK pathway. For patients with metastatic melanoma, these treatments have fundamentally changed the prognosis of disease [32, 38, 41]. MS-based proteomic analyses have enabled further characterization of melanoma tumor biology, including the molecular alterations associated with different stages of disease and mutational status. For example, a proof-of-principle proteomic study of primary melanomas revealed that the cellular proteome exhibits mutational status-dependent differences [38]. While wild-type BRAF tumors exhibited increased expression of proteins involved in melanogenesis and extracellular matrix formation, mutant BRAF primary tumors showed decreased expression of proteins with antiproliferative and tumor suppressor functions as well as proteins involved in fatty acid metabolism [38]. These insights suggest that proteomic analysis may reveal molecular signatures of different disease subtypes that could be used to diagnose, stage, and stratify patients.
Additionally, a major attraction of proteomics would be the identification of biomarkers in skin diseases such as melanomas. Studies have examined metastatic melanoma alone [42], melanoma in situ compared to invasive melanoma [43], melanocytic nevus compared to metastatic melanoma [44], primary melanoma compared to metastatic melanoma [45], as well as progression of disease [32, 46], identifying potential biomarkers for metastatic disease [42, 44, 45, 46]. Moreover, they reflected the complex pattern of progression on a molecular level, with upregulated proteins covering a wide variety of biological functions such as motility, adhesion, migration and tumor progression, apoptosis, and proliferation [42, 44, 45]. Although the sample size in some of these studies was small, the results affirm the potential of MS for new biological discoveries and development of targeted treatments.
Proteomic Studies of Cutaneous Squamous Cell Carcinoma
Cutaneous squamous cell carcinoma (SCC) as well as the premalignant lesions actinic keratosis (AK) and Bowen's disease (BD) are caused predominantly by exposure to ultraviolet radiation and derived from an uncontrolled growth of keratinocytes [47, 48, 49]. Until recently, the molecular transformation of normal keratinocytes to malignant lesions had not been characterized at the proteomic level. Several recent studies have now provided insights into proteomic changes that are associated with this transformation [19, 21, 37, 50]. Altered expression levels of proteins associated with inflammation and angiogenesis, cell proliferation, apoptosis, migration, tumor suppression, and terminal epithelial differentiation have been observed in SCC [21, 50]. In BD, changes in proteins associated with cell cycle arrest, apoptosis, and repair mechanism have been found, while in AK, the most significant upregulated pathways include apoptosis and RNA translation [21, 37]. The higher activation of DNA repair pathways seen in BD could possibly be associated with the low transformation rate of BD to SCC [21]. Identification of many common protein changes and the disruption of similar biological processes (cell-cell adhesion, cytoskeleton organization, regulation of cell proliferation, damage response, apoptosis, extracellular matrix organization, and angiogenesis) between AK, BD, and SCC lesions support the notion that AK and BD are precursor lesions of SCC [21].
MS-based proteomics has also been used to study disease progression by analyzing highly, moderately, and poorly differentiated SCC. Proteins known to be associated with cell cycle progression as well as cell proliferation and differentiation in several cancer types were increased in highly differentiated SCC, suggesting a potential role of these proteins in SCC differentiation and malignancy [37]. For poorly differentiated SCC, this study found oncogenic and tumor-suppressive proteins known to be associated with other poorly differentiated cancers, such as advanced stages of colorectal and metastatic gallbladder cancer (GOSR1), increased invasiveness in colorectal and renal cell carcinoma (AACS), and poor differentiation in colorectal cancer and invasive metastatic melanoma (BGN) [37].
The same disrupted tumorigenic pathways were identified in a proteomic analysis of tape-stripped stratum corneum when compared to punch biopsies of AK lesions. The total number of proteins detected in tape-stripped AK lesions was, as expected, smaller than the total number of proteins detected in FFPE samples from AK lesions [19]. Molecular pathway analysis of the tape-stripped stratum corneum samples from AK lesions revealed equally disrupted tumorigenic pathways as those identified in the biopsies from AK lesions, indicating that tape stripping of superficial skin lesions might be an appropriate, alternative sampling method for protein analysis of the upper part of the skin with LC-MS/MS [19].
Proteomic Studies of Hand Eczema
The etiology of hand eczema includes both exogenous and endogenous factors [51]. Amongst the exogenous factors are irritants and mechanical or other irritation of the skin, while endogenous variables are related to impaired skin barrier function [51]. Proteomic analysis of skin biopsies from patients with chronic hand eczema and healthy controls identified 185 differentially regulated proteins [52]. The protein changes included downregulation of filaggrin, filaggrin 2, and hornerin, confirming the disturbance in the skin barrier in chronic hand eczema. The S100 family members S100A7, S100A8, and S100A9 were among the markedly upregulated proteins in chronic hand eczema. These proteins are prevalent in skin and are upregulated in response to loss of epidermal barrier integrity. They serve as a defense mechanism against microbial infections and seem to be involved in the sensitization phase of allergic contact dermatitis [53]. These results shed light on the regulation of the epidermal barrier in chronic hand eczema at the protein level. More studies are needed to further investigate the multifactorial pathogenesis of chronic hand eczema, including the role of environmental exposure, irritant damage, and allergen exposure.
Proteomic Studies of Ichthyosis
Ichthyosis is a genetic disease clinically characterized by scaling of the skin [54]. In a proteomic study comparing ichthyosis vulgaris, lamellar ichthyosis, X-linked ichthyosis, and atopic dermatitis (AD) to healthy skin, the protein compositions of stratum corneum from the multiple anatomical areas were analyzed [54]. Comparison of the ichthyosis subtypes revealed that phenotypic severity was associated with increased alterations of keratins and junctional proteins and to changes in the cornified envelope of the skin barrier. Both findings have been validated in another study [36]. Studying protein composition in different genetic subtypes of ichthyosis may help to identify underlying pathogenic pathways as well as individuals with both the causal gene defect and concomitant altered downstream effects of the different mutations [54].
Proteomic Studies of Psoriasis
Genome-wide association studies have recently provided insight into the pathogenesis of psoriasis [55, 56]. Biological therapies targeting TNFα- and IL-23/Th17-related pathways are the most effective treatments to date [55, 56]. However, a subset of patients present with treatment-resistant psoriasis, highlighting the need for identifying additional factors involved in the pathogenesis that can be targeted for treatment [55, 56].
Comparative proteomic analysis of lesional psoriatic and healthy skin identified 249 differentially expressed proteins [57]. They represent pathways involved in cell proliferation and development, with 23% of them in broad biological processes known to be dysregulated in psoriasis, such as development and innate immune system [57]. When the proteome of lesional skin was compared to that of either nonlesional or healthy skin, 44 proteins exhibited altered expression, with half of them not previously associated with psoriasis. Those with decreased expression were found to be involved in apoptosis, signaling, endothelial cell proliferation, migration, cell-cell interaction, and extracellular matrix interaction. Although the total number of proteins quantified in this study is unknown, the findings illustrate the potential of proteomic studies to find proteins that have not been associated with psoriasis.
Analysis of differential expression patterns at both the transcriptome as well as the proteome level was performed on lesional and nonlesional skin from 14 patients with psoriasis [58]. A moderate association was observed between the transcriptome level and the proteome level. In psoriatic lesions, ribosomal proteins and elongation factors were more abundant, corresponding to accelerated epidermal turnover, proliferation of keratinocytes, and abnormal differentiation of basal keratinocytes characteristic for psoriasis [58].
Taken together, the studies discussed here illustrate how implementing MS-based proteomics in clinical research and practice of dermatology can complement, improve, and accelerate current routinely used strategies to diagnose and treat skin diseases.
Proteomic Studies of AD
AD is a multifactorial disease with complex genetic and environmental susceptibility factors as well as multiple clinical phenotypes. Proteomic studies of AD have mainly focused on identifying different inflammatory and barrier biomarkers. Amongst these, the fatty acid-binding protein e-fabp, which functions as a key regulator of inflammatory and metabolic signaling pathways, has been identified as being involved in the pathogenesis of AD [59, 60]. Studies of protein changes in tape-stripped lesional AD skin compared to nonlesional AD skin and skin of healthy subjects have identified several differentially expressed proteins related to skin barrier function [59, 60, 61, 62, 63]. In one study, elevated expression levels of a group of 45 proteins were positively correlated to transepidermal water loss in patients with AD and food allergies compared to patients with AD but without allergic sensitization [64]. This group of proteins includes keratin-intermediate filaments, proteins associated with inflammatory response, glycolysis, and oxidative stress response. The raised levels of these proteins found in AD patients with allergic sensitization to peanuts are therefore suggested to be involved in skin barrier function, and the altered expression an indication of the increased penetration of allergens through the skin barrier [64]. Furthermore, lower levels of enzymes involved in the generation of moisturizing factors have been found in AD lesional skin [60]. These studies have provided new insights into the predisposition to dry skin and skin infections within this patient group due to the increased penetration of microbes [60].
In addition to the current studies that characterize the changes in the skin of patients with AD, MS-based proteomics can be used to characterize clinical phenotypes of AD in the future.
Current Challenges and Future Perspective on Proteomic Research in Dermatology
Over the last decades, proteomic research using MS-based proteomics has evolved into a powerful technology. It supports the analysis of many aspects of proteins, including quantity and their state of modification. It has the capacity to rapidly identify and quantify thousands of proteins from small amounts of tissue or even just a few cells. This review illustrates the technological progress that has been made in the last decade. The major remaining challenges to proteomic studies are the complexity of the tissues that we investigate and, especially for skin, the high dynamic range due to abundant structural proteins. In addition, every project generates vast quantities of data that require extensive data processing, especially if the projects integrate multiple “omics” data. There are continuous efforts to improve techniques and instrumentation, resulting in a fast pace of technological developments that are useful for clinical samples. In particular, the ability of MS-based proteomics to analyze FFPE samples offers many possibilities to advance translational dermatological research, including retrospective analyses of archival clinical samples obtained during sensibly designed prospective clinical trials. These could help to identify mechanisms responsible for disease progression and therapy resistance. Label-free proteomic quantification offers potential insight into clinical phenotypes, so-called proteotypes, in patients that are clinically well characterized. Knowledge of disease-related proteotypes could provide a better understanding of pathological and physiological changes in skin diseases and potentially lead to more targeted treatment approaches in dermatology. As methods improve, posttranslational modification research will add another layer of information and potentially serve as a biomarker for skin disease, especially in cancer, to identify posttranslational modification signatures that could be correlated with disease progression and that may help to stratify patients. Through MS-based proteomic analyses we can investigate the association between genetic loci, biological pathways, and disease phenotypes and supplement the current gene-centric approach to precision medicine.
Key Message
High-resolution MS-based proteomics enables identification and quantification of the nearly complete proteomes of skin diseases.
Conflict of Interest Statement
The authors declare no competing financial interests.
Funding Sources
B. Dyring-Andersen is supported by grants from the Leo Foundation, the Novo Nordisk Foundation, the Lundbeck Foundation, the Aage Bangs Foundation, the Johan Boserup and Lise Boserup Foundation, and the A.P. Møller Foundation for the Advancement of Medical Sciences.
Author Contributions
Conceptualization: B. Dyring-Andersen. Data collection: G. Fredman, B. Dyring-Andersen. First draft of the manuscript: G. Fredman. Interpretation and critical revision of the manuscript: B. Dyring-Andersen, G. Fredman, M. Mann, L. Skov.
Acknowledgment
We thank Juliet Percival for creating the illustrations for this paper.
References
- 1.Altelaar AF, Munoz J, Heck AJ. Next-generation proteomics: towards an integrative view of proteome dynamics. Nat Rev Genet. 2013 Jan;14((1)):35–48. doi: 10.1038/nrg3356. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein AM. Changing paradigms in dermatology: proteomics: a new approach to skin disease. Clin Dermatol. 2003 Oct;21((5)):370–4. doi: 10.1016/j.clindermatol.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 3.Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016 Sep;537((7620)):347–55. doi: 10.1038/nature19949. [DOI] [PubMed] [Google Scholar]
- 4.Mann M, Kulak NA, Nagaraj N, Cox J. The coming age of complete, accurate, and ubiquitous proteomes. Mol Cell. 2013 Feb;49((4)):583–90. doi: 10.1016/j.molcel.2013.01.029. [DOI] [PubMed] [Google Scholar]
- 5.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012 Mar;13((4)):227–32. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Mol Biosyst. 2009 Dec;5((12)):1512–26. doi: 10.1039/b908315d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larance M, Lamond AI. Multidimensional proteomics for cell biology. Nat Rev Mol Cell Biol. 2015 May;16((5)):269–80. doi: 10.1038/nrm3970. [DOI] [PubMed] [Google Scholar]
- 8.Li H, Brewer G, Ongo G, Normandeau F, Omeroglu A, Juncker D. Immunohistochemistry microarrays. Anal Chem. 2017 Sep;89((17)):8620–5. doi: 10.1021/acs.analchem.7b00807. [DOI] [PubMed] [Google Scholar]
- 9.Lin AV. Direct ELISA. Methods Mol Biol. 2015;1318:61–7. doi: 10.1007/978-1-4939-2742-5_6. [DOI] [PubMed] [Google Scholar]
- 10.Doll S, Gnad F, Mann M. The case for proteomics and phospho-proteomics in personalized cancer medicine. Proteomics Clin Appl. 2019;13((2)):e1800113. doi: 10.1002/prca.201800113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sabbagh B, Mindt S, Neumaier M, Findeisen P. Clinical applications of MS-based protein quantification. Proteomics Clin Appl. 2016 Apr;10((4)):323–45. doi: 10.1002/prca.201500116. [DOI] [PubMed] [Google Scholar]
- 12.Deeb SJ, Tyanova S, Hummel M, Schmidt-Supprian M, Cox J, Mann M. Machine Learning-based classification of diffuse large B-cell lymphoma patients by their protein expression profiles. Mol Cell Proteomics. 2015 Nov;14((11)):2947–60. doi: 10.1074/mcp.M115.050245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007 Dec;131((6)):1190–203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 14.Archer TC, Ehrenberger T, Mundt F, Gold MP, Krug K, Mah CK, et al. Proteomics, post-translational modifications, and integrative analyses reveal molecular heterogeneity within medulloblastoma subgroups. Cancer Cell. 2018 Sep;34((3)):396–410.e8. doi: 10.1016/j.ccell.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kulak NA, Geyer PE, Mann M. Loss-less nano-fractionator for high sensitivity, high coverage proteomics. Mol Cell Proteomics. 2017;16((4)):694–705. doi: 10.1074/mcp.O116.065136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sinha A, Mann M. A beginner's guide to mass spectrometry-based proteomics. Biochem (Lond) 2020;42((5)):64–9. [Google Scholar]
- 17.Hammers CM, Tang HY, Chen J, Emtenani S, Zheng Q, Stanley JR. Research techniques made simple: mass spectrometry for analysis of proteins in dermatological research. J Invest Dermatol. 2018 Jun;138((6)):1236–42. doi: 10.1016/j.jid.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richards AL, Merrill AE, Coon JJ. Proteome sequencing goes deep. Curr Opin Chem Biol. 2015 Feb;24:11–7. doi: 10.1016/j.cbpa.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Azimi A, Ali M, Kaufman KL, Mann GJ, Fernandez-Penas P. Tape stripped stratum corneum samples prove to be suitable for comprehensive proteomic investigation of actinic keratosis. Proteomics Clin Appl. 2019 May;13((3)):e1800084. doi: 10.1002/prca.201800084. [DOI] [PubMed] [Google Scholar]
- 20.Clausen ML, Slotved HC, Krogfelt KA, Agner T. Tape stripping technique for stratum corneum protein analysis. Sci Rep. 2016 Jan;6((1)):19918. doi: 10.1038/srep19918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Azimi A, Kaufman KL, Ali M, Arthur J, Kossard S, Fernandez-Penas P. Differential proteomic analysis of actinic keratosis, Bowen's disease and cutaneous squamous cell carcinoma by label-free LC-MS/MS. J Dermatol Sci. 2018 Jul;91((1)):69–78. doi: 10.1016/j.jdermsci.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 22.Lerche CM, Olsen P, Nissen CV, Philipsen PA, Wulf HC. A novel LC-MS/MS method to quantify eumelanin and pheomelanin and their relation to UVR sensitivity − a study on human skin biopsies. Pigment Cell Melanoma Res. 2019 Nov;32((6)):809–16. doi: 10.1111/pcmr.12805. [DOI] [PubMed] [Google Scholar]
- 23.Svoboda M, Hlobilová M, Marešová M, Sochorová M, Kováčik A, Vávrová K, et al. Comparison of suction blistering and tape stripping for analysis of epidermal genes, proteins and lipids. Arch Dermatol Res. 2017 Nov;309((9)):757–65. doi: 10.1007/s00403-017-1776-6. [DOI] [PubMed] [Google Scholar]
- 24.Macdonald N, Cumberbatch M, Singh M, Moggs JG, Orphanides G, Dearman RJ, et al. Proteomic analysis of suction blister fluid isolated from human skin. Clin Exp Dermatol. 2006 May;31((3)):445–8. doi: 10.1111/j.1365-2230.2006.02078.x. [DOI] [PubMed] [Google Scholar]
- 25.Brunner AD, Thielert M, Vasilopoulou CG, Ammar C, Coscia F, Mund A, et al. Ultra-high sensitivity mass spectrometry quantifies single-cell proteome changes upon perturbation. bioRxiv. doi: 10.15252/msb.202110798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Gonzalez E, McGee JS. Research techniques made simple: laser capture microdissection in cutaneous research. J Invest Dermatol. 2016 Oct;136((10)):e99–103. doi: 10.1016/j.jid.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 27.Taverna D, Pollins AC, Nanney LB, Sindona G, Caprioli RM. Histology-guided protein digestion/extraction from formalin-fixed and paraffin-embedded pressure ulcer biopsies. Exp Dermatol. 2016 Feb;25((2)):143–6. doi: 10.1111/exd.12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Espina V, Wulfkuhle JD, Calvert VS, VanMeter A, Zhou W, Coukos G, et al. Laser-capture microdissection. Nat Protoc. 2006 Aug;1((2)):586–603. doi: 10.1038/nprot.2006.85. [DOI] [PubMed] [Google Scholar]
- 29.Datta S, Malhotra L, Dickerson R, Chaffee S, Sen CK, Roy S. Laser capture microdissection: big data from small samples. Histol Histopathol. 2015 Nov;30((11)):1255–69. doi: 10.14670/HH-11-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fend F, Specht K, Kremer M, Quintanilla-Martínez L. Laser capture microdissection in pathology. Methods Enzymol. 2002;356:196–206. doi: 10.1016/s0076-6879(02)56933-0. [DOI] [PubMed] [Google Scholar]
- 31.Mund A, Coscia F, Hollandi R, Kovács F, Kriston A, Brunner AD, et al. AI-driven deep visual proteomics defines cell identity and heterogeneity. bioRxiv. doi: 10.1038/s41587-022-01302-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gil J, Betancourt LH, Pla I, Sanchez A, Appelqvist R, Miliotis T, et al. Clinical protein science in translational medicine targeting malignant melanoma. Cell Biol Toxicol. 2019 Aug;35((4)):293–332. doi: 10.1007/s10565-019-09468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mikesh LM, Aramadhaka LR, Moskaluk C, Zigrino P, Mauch C, Fox JW. Proteomic anatomy of human skin. J Proteomics. 2013 Jun;84:190–200. doi: 10.1016/j.jprot.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 34.Edqvist PH, Fagerberg L, Hallström BM, Danielsson A, Edlund K, Uhlén M, et al. Expression of human skin-specific genes defined by transcriptomics and antibody-based profiling. J Histochem Cytochem. 2015;63((2)):129–41. doi: 10.1369/0022155414562646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newton VL, Riba-Garcia I, Griffiths CEM, Rawlings AV, Voegeli R, Unwin RD, et al. Mass spectrometry-based proteomics reveals the distinct nature of the skin proteomes of photoaged compared to intrinsically aged skin. Int J Cosmet Sci. 2019 Apr;41((2)):118–31. doi: 10.1111/ics.12513. [DOI] [PubMed] [Google Scholar]
- 36.Karim N, Durbin-Johnson B, Rocke DM, Salemi M, Phinney BS, Naeem M, et al. Proteomic manifestations of genetic defects in autosomal recessive congenital ichthyosis. J Proteomics. 2019 Jun;201:104–9. doi: 10.1016/j.jprot.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 37.Azimi A, Yang P, Ali M, Howard V, Mann GJ, Kaufman KL, et al. Data independent acquisition proteomic analysis can discriminate between actinic keratosis, Bowen's disease, and cutaneous squamous cell carcinoma. J Invest Dermatol. 2020 Jan;140((1)):212–22.e11. doi: 10.1016/j.jid.2019.06.128. [DOI] [PubMed] [Google Scholar]
- 38.Trilla-Fuertes L, Gámez-Pozo A, Prado-Vázquez G, Zapater-Moros A, Díaz-Almirón M, Fortes C, et al. Melanoma proteomics suggests functional differences related to mutational status. Sci Rep. 2019 May;9((1)):7217. doi: 10.1038/s41598-019-43512-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma J, Xu S, Wang X, Zhang J, Wang Y, Liu M, et al. Noninvasive analysis of skin proteins in healthy Chinese subjects using an Orbitrap Fusion Tribrid mass spectrometer. Skin Res Technol. 2019 Jul;25((4)):424–33. doi: 10.1111/srt.12668. [DOI] [PubMed] [Google Scholar]
- 40.Dyring-Andersen B, Løvendorf MB, Coscia F, Santos A, Møller LBP, Colaço AR, et al. Spatially and cell-type resolved quantitative proteomic atlas of healthy human skin. Nat Commun. 2020 Nov;11((1)):5587. doi: 10.1038/s41467-020-19383-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Owens B. Melanoma. Nature. 2014 Nov;515((7527)):S109. doi: 10.1038/515S109a. [DOI] [PubMed] [Google Scholar]
- 42.Rezaul K, Murphy M, Lundgren DH, Wilson L, Han DK. Combined mass spectrometry- and immunohistochemistry-based approach to determine protein expression in archival melanoma − proof of principle. Pigment Cell Melanoma Res. 2010 Dec;23((6)):849–52. doi: 10.1111/j.1755-148X.2010.00774.x. [DOI] [PubMed] [Google Scholar]
- 43.Dowling P, Moran B, McAuley E, Meleady P, Henry M, Clynes M, et al. Quantitative label-free mass spectrometry analysis of formalin-fixed, paraffin-embedded tissue representing the invasive cutaneous malignant melanoma proteome. Oncol Lett. 2016 Nov;12((5)):3296–304. doi: 10.3892/ol.2016.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Byrum S, Avaritt NL, Mackintosh SG, Munkberg JM, Badgwell BD, Cheung WL, et al. A quantitative proteomic analysis of FFPE melanoma. J Cutan Pathol. 2011 Nov;38((11)):933–6. doi: 10.1111/j.1600-0560.2011.01761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang SK, Darfler MM, Nicholl MB, You J, Bemis KG, Tegeler TJ, et al. LC/MS-based quantitative proteomic analysis of paraffin-embedded archival melanomas reveals potential proteomic biomarkers associated with metastasis. PLoS One. 2009;4((2)):e4430. doi: 10.1371/journal.pone.0004430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Byrum SD, Larson SK, Avaritt NL, Moreland LE, Mackintosh SG, Cheung WL, et al. Quantitative proteomics identifies activation of hallmark pathways of cancer in patient melanoma. J Proteomics Bioinform. 2013 Mar;6((3)):43–50. doi: 10.4172/jpb.1000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lomas A, Leonardi-Bee J, Bath-Hextall F. A systematic review of worldwide incidence of nonmelanoma skin cancer. Br J Dermatol. 2012 May;166((5)):1069–80. doi: 10.1111/j.1365-2133.2012.10830.x. [DOI] [PubMed] [Google Scholar]
- 48.Alam M, Armstrong A, Kim JYS, Kozlow JH, Mittal B, Moyer J, et al. Guidelines of care for the management of cutaneous squamous cell carcinoma. J Am Acad Dermatol. 2018 Mar;78((3)):560–78. doi: 10.1016/j.jaad.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stang A, Khil L, Kajüter H, Pandeya N, Schmults CD, Ruiz ES, et al. Incidence and mortality for cutaneous squamous cell carcinoma: comparison across three continents. J Eur Acad Dermatol Venereol. 2019 Dec;33((Suppl 8)):6–10. doi: 10.1111/jdv.15967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Azimi A, Kaufman KL, Ali M, Kossard S, Fernandez-Penas P. In silico analysis validates proteomic findings of formalin-fixed paraffin embedded cutaneous squamous cell carcinoma tissue. Cancer Genomics Proteomics. 2016 Oct;13((6)):453–65. doi: 10.21873/cgp.20008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Agner T, Elsner P. Hand eczema: epidemiology, prognosis and prevention. J Eur Acad Dermatol Venereol. 2020 Jan;34((Suppl 1)):4–12. doi: 10.1111/jdv.16061. [DOI] [PubMed] [Google Scholar]
- 52.Molin S, Merl J, Dietrich KA, Regauer M, Flaig M, Letulé V, et al. The hand eczema proteome: imbalance of epidermal barrier proteins. Br J Dermatol. 2015 Apr;172((4)):994–1001. doi: 10.1111/bjd.13418. [DOI] [PubMed] [Google Scholar]
- 53.Petersen B, Wolf M, Austermann J, van Lent P, Foell D, Ahlmann M, et al. The alarmin Mrp8/14 as regulator of the adaptive immune response during allergic contact dermatitis. EMBO J. 2013 Jan;32((1)):100–11. doi: 10.1038/emboj.2012.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rice RH, Bradshaw KM, Durbin-Johnson BP, Rocke DM, Eigenheer RA, Phinney BS, et al. Distinguishing ichthyoses by protein profiling. PLoS One. 2013;8((10)):e75355. doi: 10.1371/journal.pone.0075355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boehncke WH, Brembilla NC. Unmet needs in the field of psoriasis: pathogenesis and treatment. Clin Rev Allergy Immunol. 2018 Dec;55((3)):295–311. doi: 10.1007/s12016-017-8634-3. [DOI] [PubMed] [Google Scholar]
- 56.Conrad C, Gilliet M. Psoriasis: from pathogenesis to targeted therapies. Clin Rev Allergy Immunol. 2018 Feb;54((1)):102–13. doi: 10.1007/s12016-018-8668-1. [DOI] [PubMed] [Google Scholar]
- 57.Szél E, Bozó R, Hunyadi-Gulyás É, Manczinger M, Szabó K, Kemény L, et al. Comprehensive proteomic analysis reveals intermediate stage of non-lesional psoriatic skin and points out the importance of proteins outside this trend. Sci Rep. 2019 Aug;9((1)):11382. doi: 10.1038/s41598-019-47774-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swindell WR, Remmer HA, Sarkar MK, Xing X, Barnes DH, Wolterink L, et al. Proteogenomic analysis of psoriasis reveals discordant and concordant changes in mRNA and protein abundance. Genome Med. 2015 Dec;7((1)):86. doi: 10.1186/s13073-015-0208-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Broccardo CJ, Mahaffey SB, Strand M, Reisdorph NA, Leung DY Peeling off the layers: skin taping and a novel proteomics approach to study atopic dermatitis. J Allergy Clin Immunol. 2009 Nov;124((5)):1113–5.e1–11. doi: 10.1016/j.jaci.2009.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Broccardo CJ, Mahaffey S, Schwarz J, Wruck L, David G, Schlievert PM, et al. Comparative proteomic profiling of patients with atopic dermatitis based on history of eczema herpeticum infection and Staphylococcus aureus colonization. J Allergy Clin Immunol. 2011 Jan;127((1)):186–11. doi: 10.1016/j.jaci.2010.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sakabe J, Kamiya K, Yamaguchi H, Ikeya S, Suzuki T, Aoshima M, et al. Proteome analysis of stratum corneum from atopic dermatitis patients by hybrid quadrupole-orbitrap mass spectrometer. J Allergy Clin Immunol. 2014 Oct;134((4)):957–60.e8. doi: 10.1016/j.jaci.2014.07.054. [DOI] [PubMed] [Google Scholar]
- 62.Winget JM, Finlay D, Mills KJ, Huggins T, Bascom C, Isfort RJ, et al. Quantitative proteomic analysis of stratum corneum dysfunction in adult chronic atopic dermatitis. J Invest Dermatol. 2016;136((8)):1732–5. doi: 10.1016/j.jid.2016.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu M, Zhang J, Wang Y, Xin C, Ma J, Xu S, et al. Non-invasive proteome-wide quantification of skin barrier-related proteins using label-free LC-MS/MS analysis. Mol Med Rep. 2020 May;21((5)):2227–35. doi: 10.3892/mmr.2020.11020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goleva E, Calatroni A, LeBeau P, Berdyshev E, Taylor P, Kreimer S, et al. Skin tape proteomics identifies pathways associated with transepidermal water loss and allergen polysensitization in atopic dermatitis. J Allergy Clin Immunol. 2020 Dec;146((6)):1367–78. doi: 10.1016/j.jaci.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]