Abstract
Non-healing wounds in Type 2 Diabetes (T2D) patients represent the most common cause of amputation in the US, with an associated 5-year mortality of nearly 50%. Our lab has examined tissue from both T2D murine models and human wounds in order to explore mechanisms contributing to impaired wound healing. Current published data in the field point to macrophage function serving a pivotal role in orchestrating appropriate wound healing. Wound macrophages in mice and patients with T2D are characterized by a persistent inflammatory state; however, the mechanisms that control this persistent inflammatory state are unknown. Current literature demonstrates that gene regulation through histone modifications, DNA modifications, and microRNA can influence macrophage plasticity during wound healing. Further, accumulating studies reveal the importance of cells such as adipocytes, infiltrating immune cells (PMNs and T cells), and keratinocytes secrete factors that may help drive macrophage polarization. This review will examine the role of macrophages in the wound healing process, along with their function and interactions with other cells, and how it is perturbed in T2D. We also explore epigenetic factors that regulate macrophage polarization in wounds, while highlighting the emerging role of other cell types that may influence macrophage phenotype following tissue injury.
Keywords: Diabetes, Epigenetics, Macrophage, Wound, Inflammation, Phenotype
1. Introduction
Around 34.1 million adults in the US have diabetes, and the incidence has been increasing by 5% per year [1]. These patients develop several complications, with impaired lower extremity wound healing being one of the most common causes of hospitalization. Standard therapy for these wounds often leads to amputation, as over 70% of diabetic wounds fail to heal with the current standard of care [1]. The significant morbidity following amputation in these patients leads to an associated 50% mortality at five years [2]. Current literature supports a role for decreased microcirculation and peripheral neuropathy in impaired diabetic wound healing [3]. Additionally, chronic inflammation plays a vital role in this process, with macrophage functional impairment serving a crucial role in the hyper-inflammatory phenotype seen in non-healing diabetic wounds [4].
Wound healing is a complex process regulated by multiple cell types. Our lab and others have demonstrated that macrophages are critical for this healing process. Macrophages’ plasticity allows them to be pro-inflammatory initially and then transition to an anti-inflammatory phenotype to perform functions vital for tissue repair [5,6]. Diabetic wounds exhibit a chronic inflammatory state where imbalances in macrophage phenotype prevent wound inflammation resolution [7]. Molecular mechanisms that control macrophage phenotype are not well understood. However, recent studies by our lab and others have suggested epigenetics plays a critical role in controlling macrophage plasticity following tissue injury [8]. Current literature demonstrates that gene regulation through histone modifications, DNA modifications, and microRNA can influence macrophage plasticity during wound healing [8,9]. Further, there is increasing evidence that cells such as adipocytes, infiltrating immune cells (PMNs and T cells), and keratinocytes secrete factors that may help drive macrophage polarization during wound healing [10–14].
This review will examine the role of macrophages in the wound healing process, their function and interactions with other cells, along with how these functions are disturbed in T2D. It will also explore epigenetic factors that regulate macrophage polarization in diabetic wounds and highlight emerging evidence on the role of other immune cells and structural cells that may influence macrophage phenotype following tissue injury.
2. Phases of wound healing
Dermal wound healing is an intricate process encompassing four classically described phases of wound healing: hemostasis, inflammation, proliferation, and remodeling [15]. Under normal conditions, these phases occur in an overlapping, linear fashion. The hemostasis phase is initiated following injury; this involves the aggregation of platelets and solubilized coagulant proteins in the blood to form a clot at sites of active bleeding. These platelets degranulate to release factors critical for the initial inflammatory phase, such as P-selectin, allowing for neutrophil adherence to the wound [16].
Early in the inflammatory phase, neutrophils are the primary cell type within the dermal wound. They release antimicrobial peptides (e.g., LL37), reactive oxygen species (ROS), and neutrophil extracellular traps (NETs). Additionally, neutrophils secrete chemokines to attract monocytes/macrophages [17,18]. These monocytes/macrophages, in addition to tissue-resident macrophages, then become the dominant cell type in the inflammatory phase and are critical in regulating this inflammation [7,19–21]. Initially, macrophages are pro-inflammatory or M1-like, producing cytokines such as IL-12, IL-1β, IL-6, TNFα, and iNOS. During normal wound repair, anti-inflammatory or M2-like macrophage subtypes become dominant over time to trigger the proliferative phase.
During the proliferative phase, fibroblasts and keratinocytes are stimulated by growth factors secreted by macrophages and other cells, to re-epithelialize and form granulation tissue within the wound; this is via the secretion of collagen and other extracellular matrices (ECM) proteins. Macrophages play a critical role stimulating these fibroblasts, this is demonstrated by the ablation of macrophages leading to a significant delay in the appearance of fibroblast during the proliferative phase of wound healing [22]. Interestingly, recent single-cell analysis has also revealed that in a murine skin wounding model, about 6–11% of fibroblasts are myeloid-derived, demonstrating the important role and plasticity of myeloid cells through the stages of wound healing [23].
Lastly, the remodeling phase is the longest phase of wound healing in which type III collagen is replaced with type I collagen for higher tensile strength. Depending on the depth, mechanism of injury, and species, dermal structures such as hair follicles and pre-injury tensile strength do not entirely recover [24].
3. Monocytes/macrophages during wound healing
3.1. Monocytes
There are two types of circulating monocytes, classical (CD16−/Ly6Chi) and nonclassical monocytes (CD16+/Ly6CLo). In response to initial signals of Damage Associated Molecular Patterns (DAMPs) or Pathogen Associated Molecular Patterns (PAMPs), classical monocytes (CD16−/Ly6CHi) extravasate at the site of injury and immediately release inflammatory cytokines and chemokines to recruit more myeloid cells [25]. These infiltrating myeloid cells perform phagocytosis, efferocytosis, and autophagy function to ‘clean’ the wound, priming it for the placement of granulation tissue and proliferation. They also massively proliferate within the wound and bone marrow, peaking in murine dermal skin wound models on day six following injury in a highly conserved fashion [26,27]. Following proliferation, these monocytes will differentiate into ‘M1’ inflammatory macrophages and undergo in situ differentiation to Ly6CLo monocyte/M2-like ‘anti-inflammatory’ macrophages and are the primary source of cells within the wound [28].
In contrast to classical monocytes, nonclassical monocytes (CD16+/Ly6CLo) are ‘anti-inflammatory’ and express VEGF, TGFβ, and IL-10. They are biased progenitors of ‘M2’ macrophages [29], which are key promoters of the proliferative and remodeling phases of wound healing. However, fewer of these monocytes are circulating; thus, they are not the major contributors to myeloid-function within the wound healing setting.
3.2. Macrophages phenotypes in wound healing
Macrophages play a critical role in normal wound healing. Indeed, macrophage-specific ablation resulted in delayed re-epithelialization and exhibited a reduction in the secretion of VEGF and TGF-β1, making wounds less conducive to angiogenesis and cell proliferation [30,31]. Macrophages are classically identified based on in vitro studies into subsets based on their markers, phenotype, and role. These in vitro phenotypes are the ‘M1’ or “classically-activated” macrophage and the ‘M2’ or “alternatively-activated” macrophage. ‘M1’ macrophages (CD86+) are the primary players in pathogen destruction, secretion of inflammatory cytokines, and driving a Th1-type response in wound healing. Meanwhile, ‘M2’ macrophages (CD206+) are associated with critical aspects of wound healing, including angiogenesis, ECM remodeling, production of anti-inflammatory cytokines, and inflammation resolution [32]. However, more recent work has identified multiple unique subtypes that are host and context dependent – and demonstrate that a ‘spectrum’ of macrophage phenotypes exist, especially in the immunomodulatory phenotypes, rather than a dichotomous M1 and M2 polarization seen with acute in vitro stimulation [33]. Further, the in vitro ‘M1’ and ‘M2’ phenotype classification omits the role and context of the in vivo wound on influencing macrophage phenotypes. In vivo, heterogenous individual cells exhibit varying characteristics of traditional M1/M2 phenotypes on a spectrum [33,34]. These subtypes and others’ specific roles continue to be discovered, especially in how they function in tissues in vivo. As a whole, in the physiologic dermal wound, in vivo macrophages dynamically change their phenotypes from pro-inflammatory to reparative over time, rather than a dichotomous switch between M1 and M2 [23,28]. One recent study shows that wound angiogenesis in murine and zebrafish models, is dependent on pro-inflammatory TNFα-secreting macrophages[35], highlighting the complexity of the wound microenvironment. Further, specific macrophage subtypes continue to be discovered. For example, one recent study established two novel in vivo macrophage subtypes (CX3CR1Hi vs. CX3CR1Med/Lo) and showed how a loss of CX3CR1Hi macrophages in T2D contributes to poor wound healing and a pro-inflammatory state [36]. Kinetic regulation of these subtypes is critical for physiologic maturation of the dermal wound. Recent work examining in vivo macrophage transcriptional networks showed that the Runx2 locus had strong control over the transition from pro-inflammatory in vivo macrophages to reparative phenotypes in a murine ear wound model [37]. Loss of function of Runx2 led to persistence of pro-inflammatory phenotypes and delayed wound healing [37]. This highlights the importance of managing in vivo macrophage phenotypes in a kinetic fashion to progress the wound through the stages of wound healing, and how the artificial in vitro M1/M2 dichotomy, although easy to describe, does not apply perfectly to the wound.
Another active area of research focuses on differences between tissue-resident macrophages vs. monocyte-derived macrophages in the context of wound healing. Dermal macrophages are essential for promoting the initial inflammatory response to DAMPs/PAMPs in wounds [38,39]. They are quickly outnumbered by monocyte-derived macrophages during wound repair, but upon resolution, they will self-renew to return the environment to homeostasis.
3.3. Macrophage dysregulation in type II diabetic wounds
Wound macrophage dysregulation in T2D is characterized by the decreased host response to pathogens in the wound [40], sustained presence of inflammatory phenotypes [28], increased number of monocytes/macrophages at late time points in wounds [41], and a dampened initial inflammatory response [42].
Diabetic wounds are known to have enhanced susceptibility to bacterial infections [40], and this is in part due to a dysfunctional macrophage host response in diabetes [43]. Diabetic wounds are colonized in a polymicrobial fashion, and Staphylococcus aureus is typically present [44]. Peritoneal macrophages isolated from diabetic mice demonstrated impaired phagocytosis of fluorescently labeled S. Aureus compared to controls [43]. Furthermore, bone marrow-derived macrophages (BMDMs) cultured in high glucose media exhibited decreased phagocytic ability and were less bactericidal than controls [43]. They also have a dampened inflammatory response to lipopolysaccharide (LPS), a component of the gram-negative bacteria cell wall, despite their increased basal inflammatory activity [45]. The interplay between the microbiota colonizing wounds and macrophages likely plays a role in altering their phenotype and subsequent wound repair ability [44]; thus, this deserves further study, especially on identifying the specific epigenetic changes on macrophages exposed to various microbiomes. Hyperglycemic conditions have been shown to impair macrophage efferocytosis. In particular, macrophage clearing of apoptotic neutrophils and necrotic cells in the tissue is dampened by local hyperglycemia in the form of advanced glycosylated end products via the RAGE receptor [46,47]. This dysregulated macrophage function contributes to the well-described pro-inflammatory diabetic wound milieu via failure to remove pathogens and inflammatory apoptotic neutrophils.
Increased persistence of M1-type (Ly6CHi) monocytes/macrophages in diabetic wounds following the monocytes initial recruitment phase is well established [28]. Several studies have demonstrated their role in contributing to delayed healing, showing that reversal of this persistent inflammatory phenotype improves wound healing. One study demonstrated that administration of anti-monocyte chemoattractant protein-1 (MCP-1) to wounds prevented this influx of late inflammatory monocyte/macrophages (Ly6CHi) and enhanced wound healing in diabetic mice [28]. Our group has recently illustrated this via single-cell RNA sequencing of human wound skin, demonstrating increased NFκB–mediated inflammation in diabetic wounds compared to control; this was in part due to increased cyclooxygenase 2/prostaglandin E2 (COX-2/PGE2). Subsequent inhibition of the COX-2/PGE2 pathway genetically (Cox2fl/fl Lyz2Cre+) in mice or with macrophage-specific nanotherapy targeting COX-2 in murine tissue macrophages reversed the inflammatory macrophage phenotype and improved wound repair [48]. Another study reversed epigenetic modifications priming NFkB-mediated inflammation in diabetic macrophages and subsequently improved wound healing [11,49]. The wealth of translational studies showing improved diabetic healing with reversal of prolonged macrophage-mediated inflammation underscores their important role during in vivo wound repair.
There is also mounting evidence involving regulation in the timing of macrophage presence in diabetic wounds; however, whether M1 macrophages are increased or decreased initially following an injury is unclear. Several studies using an unsplinted wound model demonstrate an increased presence of inflammatory or ‘M1’ macrophages at early time points in diabetic wounds [27,50]. In contrast, using a splinted wound model in Leprdb/db mice, Yan et al. showed a decreased presence of both ‘M1’ and ‘M2’ macrophages at early time points when the wounds were less than 20% healed by size [51]. This difference in the presence of ‘M1’ macrophages early in the diabetic wound may be due to the ‘chronic’ splinted wound model used by Yan et al., compared to an acute unsplinted model used by the first groups. Unsplinted wounded models can lead to healing via contracture instead of secondary intention and granulation, seen in the splinted model. One study highlights the importance of the initial tissue macrophage response early after injury. Increasing ‘M2’ macrophage phenotypes in unsplinted diabetic wounds for the first three days leads to increased keratinocyte migration, increased granulation tissue formation, increased angiogenesis, and faster-wound healing [52]. In fact, our group has also found delayed epigenetic regulation of inflammation by Mixed lineage leukemia 1 (MLL1) in T2D leads to a decrease in inflammatory macrophages early following injury and impaired diabetic wound healing [42]. Thus, the effects here lend credence to the significance of a dampened initial macrophage response to tissue injury in diabetes contributing to poor wound healing. However, the exact nature of macrophage dysregulation and timing in early wound healing merits further investigation, and is likely model dependent. This could be achieved by directly comparing in vivo macrophage phenotypes between various sized murine skin wounds and comparing splinted vs. unsplinted wounds at multiple time points. Further, linking murine findings with human data from diabetic wounds is critical to establish clinical relevance of differing models.
The changes contributing in part to macrophage dysregulation in the wound stem from alterations in hematopoietic stem cells in the bone marrow. One study showed that, following adoptive transfer, there was dysregulation of both ‘M1’ and ‘M2’ macrophages early on in the wound course and a subsequent preference to ‘M1’ macrophages later [51]. Epigenetic changes to hematopoietic stem cells are conserved, contributing to M1 and M2 macrophage dysregulation and impaired wound healing [27,50].
As single-cell RNA sequencing becomes more accessible and the technical challenges performing this in human diabetic wounds is overcome, more targets affecting macrophage plasticity and phenotype in diabetes will be discovered [53]. Furthermore, the regulators of macrophage phenotypes – transcriptional networks, epigenetic modifications, and other wound cells in the wound microenvironment will need to be analyzed and offer exciting opportunities for potential therapeutic targets for management of diabetic wounds.
4. Epigenetic modifications of macrophages in type 2 diabetic wounds
Epigenetic modifications regulate monocyte/macrophage phenotype during wound healing by altering the chromatin to activate or silence a gene without changing the genetic code [8,54–56]. Studies demonstrating the importance of epigenetic regulation in macrophage polarization are summarized in Table 1. There are three main types of epigenetic modifications: histone modification, DNA modification, and ATP-dependent remodeling [57], with the former two being more studied in wound healing. Additionally, miRNA can change the macrophage inflammatory profile by regulating gene expression (Fig. 1).
Table 1.
Epigenetic enzymes that regulate macrophage polarization
| Histone methyltransferases | ||
|---|---|---|
| Name | Function | Upstream regulators |
| MLL1 | ||
| SMYD3 |
|
|
| SMYD2 |
|
|
| SMYD5 |
|
|
| SETDB2 |
|
|
| SETDB1 |
|
|
| SET7 |
|
|
| PRMT1 |
|
|
| Histone demethylases | ||
| JMJD3 |
|
|
| UTX |
|
|
| JMJD1A |
|
|
| Histone acetyltransferases | ||
| MOF |
|
|
| SIRT1 |
|
|
| SIRT6 |
|
|
| Histone deacteylases | ||
| HDAC6 |
|
|
| HDAC2 |
|
|
| HDAC3 |
|
|
| DNA methyltransferases | ||
| DNMT1 | ||
| DNMT3t |
|
|
Fig. 1.
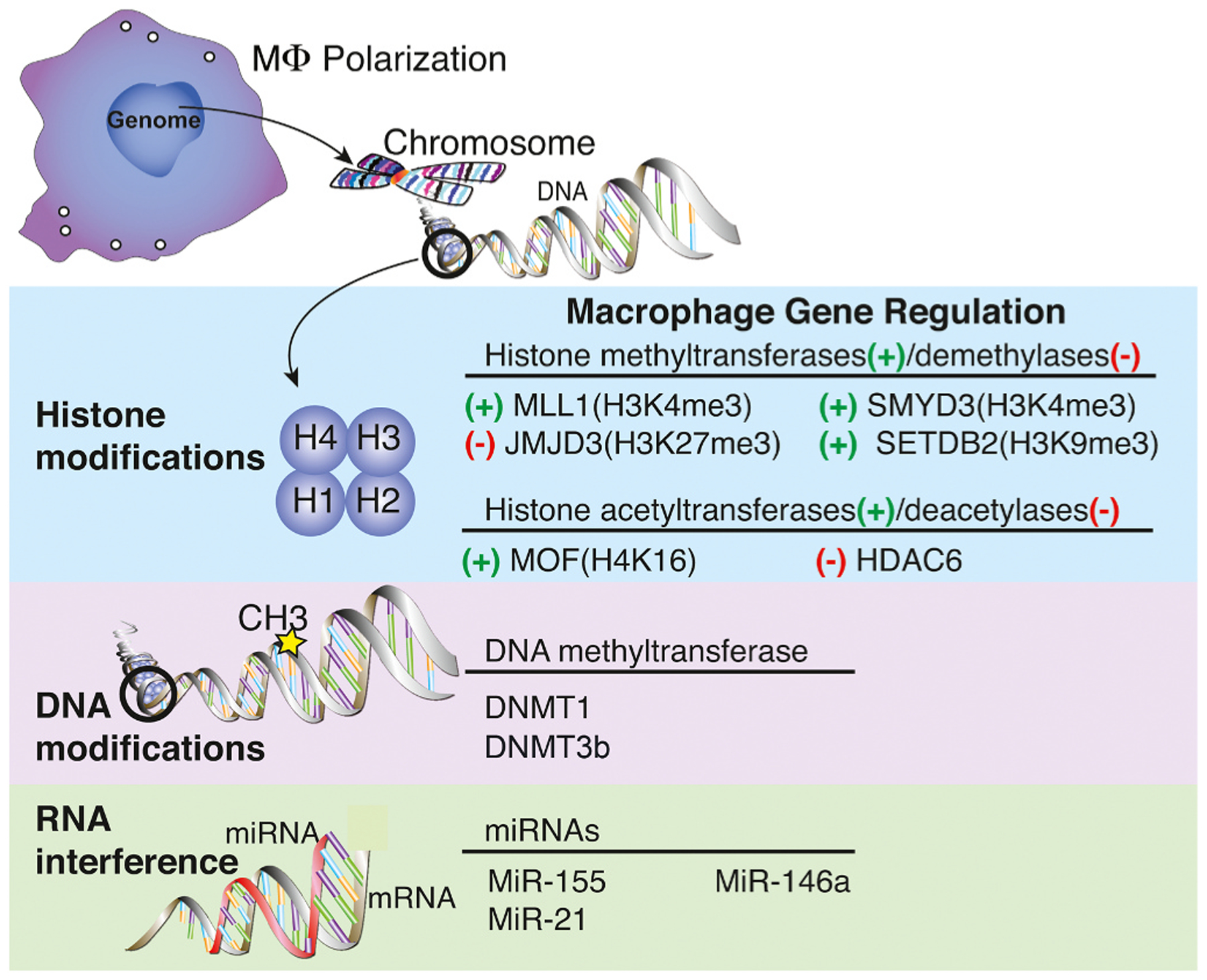
Gene regulation enzymes associated with inducing macrophage polarization. Histone modifications, DNA modifications, and microRNAs associated with influencing macrophage phenotype. Histone deacetylase (HDAC); Jumonji domain-containing protein (JMJD); Mixed-lineage leukemia (MLL); SET domain bifurcated (SETDB); SET and MYN domain (SMYD); Males-absent on the first (MOF); DNA methyltransferases (DNMT).
4.1. Histone modifications
Current literature demonstrates a role for histone modification in regulating macrophage polarization. In eukaryotes, DNA and histones form units called nucleosomes [58]. When DNA is tightly wrapped around histones (heterochromatin), transcription is inhibited by blocking transcriptional machinery access. However, when DNA-histone interactions are relaxed (euchromatin), transcriptional proteins can bind to allow transcription to occur [58–60]. Histones have an N-terminal “tail” with lysine (K) residues that histone-modifying enzymes can methylate and acetylate to direct formation of heterochromatin and euchromatin [59].
Histone methylation and demethylation are the most highly studied histone modifications and play a role in macrophage polarization. Methylation of a histone activates or represses transcription depending on the number of methyl groups added and their location on the histone tail. For example, the tri-methylation of lysine 4 on histone 3 (H3K4me3) causes the chromatin to open, promoting transcription. On the other hand, the tri-methylation of lysine 27 (H3K27me3) or lysine 9 (H3K9me3) is associated with promoting heterochromatin formation [14,56,60,61]. These histone methylation marks play a part in regulating the macrophage switch during wound healing [14,56].
Histone methyltransferases (HMTs) and Histone demethylases (HDMs) respectively achieve methylation and demethylation of a histone. Several HMTs have been ascribed with roles in macrophage polarization during wound healing. One methyltransferase necessary for catalyzing H3K4me3 deposition driving macrophage polarization is MLL1. Our lab has shown that MLL1 is associated with promoting pro-inflammatory gene expression in macrophages during the inflammatory phase of wound healing. Use of an MLL1 inhibitor or myeloid specific deletion of MLL1 delayed wound healing and decreased pro-inflammatory cytokine production [42]. We further demonstrate that monocytes isolated from T2D mice exhibited impaired MLL1 expression early during wound healing, followed by overexpression in the latter phase [42]. Additionally, we show in T2D patients and mice that MLL1 regulates macrophage phenotype in part through TLR4 receptor signaling, as myeloid-specific knockout of MLL reduced TLR4 expression and improved wound healing [62,63]. Conversely, SMYD3, another H3K4me3 methyltransferase, seems to regulate M2-like polarization [64]. We have recently shown that SETDB2, a H3K9me3 methyltransferase, negatively regulates pro-inflammatory genes, inducing a tissue repair macrophage phenotype. Further, we demonstrate that SETDB2 expression is regulated by IFN-β [14]. Not only do HMTs play a role in macrophage polarization in wound repair, but HDMs also control this process. The H3K27 demethylase Jumonji domain-containing protein 3 (JMJD3) has roles in activating both pro-and anti-inflammatory macrophage phenotypes depending on the environment/tissue [56,65,66]. Our lab and others have shown that in T2D murine models, wound macrophages exhibit decreased H3K27me3 expression mediated by JMJD3 release of H3K27me3 to promote inflammatory gene expression [49,67]. JMJD3 expression has also been shown to increase in response to inflammatory stimuli such as LPS and IL-4 [67]. These data indicate that histone methylation and demethylation regulated by HMTs and HDMs influence macrophage polarization following injury.
Histone acetylation and deacetylation are other mechanisms important in macrophage polarization during wound healing. Acetylation of the lysine residue on the histone tail interferes with the interaction between the DNA and histone, promoting transcriptional activation. Histone acetyltransferases (HATs) carry out the acetyl group’s transfer from acetyl CoA to the lysine residue [68]. Our lab has shown that Males-absent on the first (MOF), a histone acetyltransferase, is elevated in T2D mice and is regulated by TNF-alpha stimulation [69]. The primary substrate for MOF activity, H4K16, has increased deposition associated with promoters of inflammatory genes of diabetic macrophages [69]. Others have demonstrated that histone deacetylase 6 (HDAC6) influences macrophage production of IL-1β, while inhibiting IL-10 production, under hyperglycemic conditions [70]. Though histone acetylation can induce macrophage polarization, further studies into upstream regulation are needed to elucidate this mechanism fully. Overall, since histone modifications demonstrate an important role in the macrophage “switch” required for wound healing, targeting these modifications directly or indirectly may prove useful in improving wound healing.
4.2. DNA methylation
DNA methylation is predominantly associated with transcriptional repression. It is characterized by the transfer of a methyl group to the cytosine ring of DNA by DNA methyltransferases (DNMTs), which occurs at clusters of CpG islands. Methylation of CpG islands within the promoter can directly silence transcription by impeding transcription factor binding [71]. DNMT1 can regulate macrophage and bone marrow progenitor cells’ inflammatory profile towards an ‘M1’ phenotype [51, 72]. DNMT1 inhibition by 5-aza-2’-deoxycytidine promoted ‘M2’ macrophage formation and suppressed inflammation in bone marrow-derived macrophages (BMDMs) [72]. This inhibition of DNMT1 also resulted in protection against obesity-induced inflammation and insulin resistance [72]. Further, DNMT1 is elevated in BMDMs from type 2 diabetic murine models and promotes a pro-inflammatory macrophage phenotype. Indeed, the knockout of DNMT1 improved wound healing in these mice [51]. DNMT3b has also been shown to regulate macrophage polarization towards a ‘M1’ phenotype [73]. The expression of DNMT3b is elevated in macrophages isolated from diet-induced obese mice compared to control [73]. Additionally, knockout of DNMT3b in macrophages in vitro, induced polarization towards a ‘M2’ phenotype [73]. While DNMT3b may regulate macrophage polarization, how this contributes to wound repair is unclear.
4.3. miRNA regulation
Research recently has shown microRNA as an additional regulator in gene expression during macrophage polarization [9]. One group demonstrated that microRNA-146a (MiR-146a) expression is elevated in ‘M2’ macrophages but attenuated in ‘M1’ macrophages [74]. MiR-146a exhibits protective effects on macrophages by inhibiting the activation and secretion of pro-inflammatory cytokines [74,75]. Meanwhile, microRNA- 155 (miR-155) expression promotes an ‘M1’ macrophage phenotype by repressing negative regulators of inflammatory cytokine expression [76]. Additionally, overexpression of microRNA-21 (miR-21) in diabetic wound macrophages was associated with the upregulation of pro-inflammatory genes such as IL-1β and TNFα [77]. Similarly, long noncoding RNA may also demonstrate a role in regulating macrophage polarization [78], though further studies are needed to elucidate the mechanism fully.
5. Cellular regulation of macrophage phenotype in wounds
Current research has demonstrated that the pro-inflammatory microenvironment in diabetic wounds contributes to the persistent ‘M1’ macrophage phenotype exhibited following injury. In particular, our lab and others have shown that blocking or inhibiting cytokines (e.g., IL-1β) and chemokines (MCP-1) elevated in the wound microenvironment can reduce the overabundance of inflammatory macrophages in diabetic wounds [4,28]. Additionally, keratinocytes, adipocytes, T cells, and neutrophils exhibit altered phenotypes in diabetic wounds [79–83]. The factors secreted by these cells have been suggested to be involved in macrophage polarization (Fig. 2).
Fig. 2.
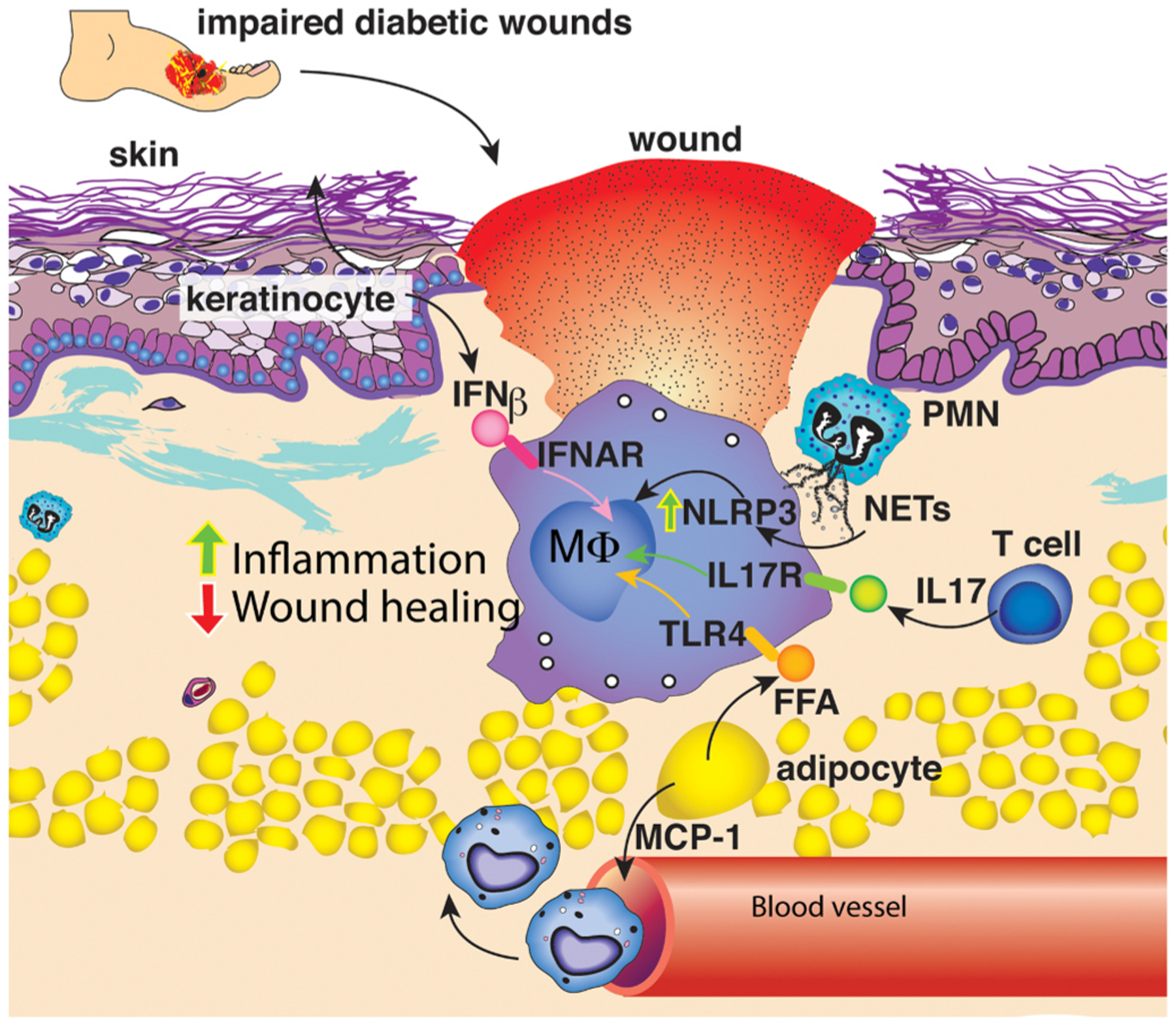
Cellular regulation of macrophage phenotype in the wound microenvironment. Following injury infiltrating immune cells and structural cells within the wound microenvironment secrete factors that influence macrophage polarization.
5.1. Adipocytes
Adipocytes in obese patients and mice release MCP-1, TNF, and free fatty acids [10]. Specifically, dermal adipocytes can release palmitic acid and oleic acid, and fatty acid levels are elevated in T2D patients [84–87]. Our lab and others have shown that fatty acids can manipulate the macrophage inflammatory profile [10,11]. In particular, we demonstrate that palmitate can increase JMJD3 expression in macrophages in a TLR4/MyD88-dependent manner, releasing H3K27me3 deposition on the promoters of inflammatory genes [11]. Fatty acids can also increase the expression of SIRT3, a primary mitochondrial deacetylase in macrophages [88]. We found that SIRT3 negatively regulates inflammatory cytokine production in macrophages and is important for normal wound healing. In diabetic wounds, macrophages exhibit attenuated SIRT3 expression in a fatty acid-binding protein (FABP4)--dependent manner [89]. However, the mechanism by which FABP4 is regulated in diabetic wounds is unclear. Together these studies suggest following injury, adipocyte secretion of fatty acids may manipulate macrophage phenotype through inducing epigenetic modification.
5.2. Infiltrating immune cells
Infiltrating immune cells into the skin is also associated with manipulating macrophage phenotype during wound healing. Neutrophils are among the first cells entering the wound, secreting neutrophil extracellular traps (NETs). When healthy and diabetic skin was compared, diabetic patients exhibited increased NETosis. NETosis is associated with activation of NLRP3 inflammasome and IL-1β production by macrophages [12]. Within the wound microenvironment, research suggests lymphocytes also contribute to macrophage polarization [90]. Increased T cells are present in diabetic wounds, particularly gamma delta and Th17 cells [13]. In diabetic wounds, IL-17, a cytokine produced mainly by Th17 cells, demonstrates a role in macrophage polarization as knockout of IL-17 improved wound healing in a diabetic mouse (db/db) model by reducing ‘M1’ macrophages and maintaining ‘M2’ macrophages [91]. Further studies are warranted to identify the specific signaling pathways or epigenetic enzymes that are driving Th17 polarization, and whether there are viable therapeutic targets to improve wound healing by preventing excessive Th17 polarization in wound T cells.
5.3. Keratinocytes
Keratinocytes not only function as a barrier in the skin, but they are also primed to respond to environmental stimuli and serve as the first link in cutaneous immunity through the release of cytokines/chemokines. During chronic skin inflammation, keratinocytes secrete NFκB regulated cytokines such as IL-18 and IL-6, in addition to type I IFNs. Although other chronic inflammation skin models demonstrate keratinocyte production of type I IFNs influences immune cell inflammatory profiles [92], less is known about their role in tissue repair. Diabetic studies to date have focused on IFN-β, demonstrating that administration of IFN-β to high-fat-diet (HFD) mice has been shown to attenuate inflammation in adipose tissue, reduce weight gain, and improve glucose tolerance [93]. We found that knockout of the type I IFN receptor impairs wound healing; this impaired wound healing is associated with an increased inflammatory macrophage phenotype [14]. Additionally, we demonstrate that IFN-β induces Setdb2 expression in macrophages resulting in the turning off of inflammatory genes; however, in diabetic wounds, this IFN-β/Setdb2 axis is impaired [14]. These data suggest a role for IFN-β in regulating macrophage phenotype during wound healing. As keratinocytes are known for type I IFN production, further investigation is needed to understand how keratinocytes may contribute to macrophage polarization to lead to novel topical therapies.
6. Conclusion
Dysregulation of diabetic macrophages leads to heightened inflammation and poor wound healing in diabetic wounds. Here, we have described the epigenetic changes and regulations by other cell types in the wound microenvironment, leading to these pathologic changes. However, there remains significant questions about the interactions between macrophages and their environment, especially with other immune cells and structural cells. In particular, very little is known about the interplay between keratinocyte cytokine production on macrophage polarization during wound healing, and filling in this gap in knowledge may lead to the development of potential topical therapies. Further studies investigating the therapeutic potential of targeting epigenetic enzymes such as MLL1 or MOF in a local, cell-specific manner are also warranted. Given that the current standard of care leaves 70% of diabetic wounds unhealed [94], it is critical to investigate the therapeutic potential of these recently discovered pathways triggering pathologic inflammation.
Acknowledgments
All authors contributed to the writing and editing of this publication. We would like to thank Robin G. Kunkel, University of Michigan, for her work on the figures.
The lab is supported by National Institutes of Health (NIH)R01-DK124290. Dr. Wolf is supported by the University of Michigan Institutional Research and Academic Career Develoment Awards (IRACDA) program NIH/NIGMS grant: K12 GM111725.
Footnotes
Declarations of interest
None.
References
- [1].Centers for Disease Control and Prevention, National Diabetes Statistics Report U. S. Dept of Health and Human Services, Centers for Disease Control and Prevention, 2020. [Google Scholar]
- [2].Lamont P, Franklyn K, Rayman G, Boulton AJ, Update on the diabetic foot 2012: the 14th biennial Malvern Diabetic Foot Conference, J. Low. Extrem. Wounds 12 (1) (2013) 71–75. [DOI] [PubMed] [Google Scholar]
- [3].Ackermann PW, Hart DA, Influence of comorbidities: neuropathy, vasculopathy, and diabetes on healing response quality, Adv. Wound Care (N. Rochelle) 2 (8) (2013) 410–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Barman PK, Koh TJ, Macrophage dysregulation and impaired skin wound healing in diabetes, Front Cell Dev. Biol 8 (2020) 528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wynn TA, Vannella KM, Macrophages in tissue repair, regeneration, and fibrosis, Immunity 44 (3) (2016) 450–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Italiani P, Boraschi D, From monocytes to M1/M2 macrophages: phenotypical vs. functional differentiation, Front Immunol. 5 (2014) 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Boniakowski AE, Kimball AS, Jacobs BN, Kunkel SL, Gallagher KA, Macrophage-mediated inflammation in normal and diabetic wound healing, J. Immunol 199 (1) (2017) 17–24. [DOI] [PubMed] [Google Scholar]
- [8].den Dekker A, Davis FM, Kunkel SL, Gallagher KA, Targeting epigenetic mechanisms in diabetic wound healing, Transl. Res 204 (2019) 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Self-Fordham JB, Naqvi AR, Uttamani JR, Kulkarni V, Nares S, MicroRNA: dynamic regulators of macrophage polarization and plasticity, Front Immunol. 8 (2017) 1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Olefsky JM, Glass CK, Macrophages, inflammation, and insulin resistance, Annu. Rev. Physiol 72 (2010) 219–246. [DOI] [PubMed] [Google Scholar]
- [11].Davis FM, d enDekker A, Joshi AD, Wolf SJ, Audu C, Melvin WJ, Mangum K, Riordan MO, Kunkel SL, Gallagher KA, Palmitate-TLR4 signaling regulates the histone demethylase, JMJD3, in macrophages and impairs diabetic wound healing, Eur. J. Immunol 50 (2020) 1929–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lee MKS, Sreejit G, Nagareddy PR, Murphy AJ, Attack of the NETs! NETosis primes IL-1beta-mediated inflammation in diabetic foot ulcers, Clin. Sci. (London) 134 (12) (2020) 1399–1401. [DOI] [PubMed] [Google Scholar]
- [13].Lee J, Rodero MP, Patel J, Moi D, Mazzieri R, Khosrotehrani K, Interleukin-23 regulates interleukin-17 expression in wounds, and its inhibition accelerates diabetic wound healing through the alteration of macrophage polarization, FASEB J. 32 (4) (2018) 2086–2094. [DOI] [PubMed] [Google Scholar]
- [14].Kimball AS, Davis FM, denDekker A, Joshi AD, Schaller MA, Bermick J, Xing X, Burant CF, Obi AT, Nysz D, Robinson S, Allen R, Lukacs NW, Henke PK, Gudjonsson JE, Moore BB, Kunkel SL, Gallagher KA, The histone methyltransferase Setdb2 modulates macrophage phenotype and uric acid production in diabetic wound repair, Immunity 51 (2) (2019) 258–271, e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Diegelmann RF, Evans MC, Wound healing: an overview of acute, fibrotic and delayed healing, Front Biosci. 9 (2004) 283–289. [DOI] [PubMed] [Google Scholar]
- [16].Zarbock A, Polanowska-Grabowska RK, Ley K, Platelet-neutrophil-interactions: linking hemostasis and inflammation, Blood Rev. 21 (2) (2007) 99–111. [DOI] [PubMed] [Google Scholar]
- [17].Ridiandries A, Tan JTM, Bursill CA, The role of chemokines in wound healing, Int J. Mol. Sci 19 (10) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Martins-Green M, Petreaca M, Wang L, Chemokines and their receptors are key players in the orchestra that regulates wound healing, Adv. Wound Care (N. Rochelle) 2 (7) (2013) 327–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhang X, Mosser DM, Macrophage activation by endogenous danger signals, J. Pathol 214 (2) (2008) 161–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Koh TJ, DiPietro LA, Inflammation and wound healing: the role of the macrophage, Expert Rev. Mol. Med 13 (2011) 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Eming SA, Wynn TA, Martin P, Inflammation and metabolism in tissue repair and regeneration, Science 356 (6342) (2017) 1026–1030. [DOI] [PubMed] [Google Scholar]
- [22].Leibovich SJ, Ross R, The role of the macrophage in wound repair. A study with hydrocortisone and antimacrophage serum, Am. J. Pathol 78 (1) (1975) 71–100. [PMC free article] [PubMed] [Google Scholar]
- [23].Guerrero-Juarez CF, Dedhia PH, Jin S, Ruiz-Vega R, Ma D, Liu Y, Yamaga K, Shestova O, Gay DL, Yang Z, Kessenbrock K, Nie Q, Pear WS, Cotsarelis G, Plikus MV, Single-cell analysis reveals fibroblast heterogeneity and myeloid-derived adipocyte progenitors in murine skin wounds, Nat. Commun 10 (1) (2019) 650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Levenson SM, Geever EF, Crowley LV, Oates JF 3rd, Berard CW, Rosen H, The healing of rat skin wounds, Ann. Surg 161 (1965) 293–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kono H, Rock KL, How dying cells alert the immune system to danger, Nat. Rev. Immunol 8 (4) (2008) 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Pang J, Urao N, Koh TJ, Proliferation of Ly6C+ monocytes/macrophages contributes to their accumulation in mouse skin wounds, J. Leukoc. Biol 107 (4) (2020) 551–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Barman PK, Pang J, Urao N, Koh TJ, Skin wounding-induced monocyte expansion in mice is not abrogated by IL-1 receptor 1 deficiency, J. Immunol 202 (9) (2019) 2720–2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kimball A, Schaller M, Joshi A, Davis FM, denDekker A, Boniakowski A, Bermick J, Obi A, Moore B, Henke PK, Kunkel SL, Gallagher KA, Ly6C(Hi) blood monocyte/macrophage drive chronic inflammation and impair wound healing in diabetes mellitus, Arterioscler. Thromb. Vasc. Biol 38 (5) (2018) 1102–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Olingy CE, San Emeterio CL, Ogle ME, Krieger JR, Bruce AC, Pfau DD, Jordan BT, Peirce SM, Botchwey EA, Non-classical monocytes are biased progenitors of wound healing macrophages during soft tissue injury, Sci. Rep 7 (1) (2017) 447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Mirza R, DiPietro LA, Koh TJ, Selective and specific macrophage ablation is detrimental to wound healing in mice, Am. J. Pathol 175 (6) (2009) 2454–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lucas T, Waisman A, Ranjan R, Roes J, Krieg T, Muller W, Roers A, Eming SA, Differential roles of macrophages in diverse phases of skin repair, J. Immunol 184 (7) (2010) 3964–3977. [DOI] [PubMed] [Google Scholar]
- [32].Ferrante CJ, Leibovich SJ, Regulation of macrophage polarization and wound healing, Adv. Wound Care (N. Rochelle) 1 (1) (2012) 10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Xue J, Schmidt SV, Sander J, Draffehn A, Krebs W, Quester I, De Nardo D, Gohel TD, Emde M, Schmidleithner L, Ganesan H, Nino-Castro A, Mallmann MR, Labzin L, Theis H, Kraut M, Beyer M, Latz E, Freeman TC, Ulas T, Schultze JL, Transcriptome-based network analysis reveals a spectrum model of human macrophage activation, Immunity 40 (2) (2014) 274–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ogle ME, Segar CE, Sridhar S, Botchwey EA, Monocytes and macrophages in tissue repair: implications for immunoregenerative biomaterial design, Exp. Biol. Med (Maywood) 241 (10) (2016) 1084–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Gurevich DB, Severn CE, Twomey C, Greenhough A, Cash J, Toye AM, Mellor H, Martin P, Live imaging of wound angiogenesis reveals macrophage orchestrated vessel sprouting and regression, EMBO J. 37 (13) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Burgess M, Wicks K, Gardasevic M, Mace KA, Cx3CR1 expression identifies distinct macrophage populations that contribute differentially to inflammation and repair, Immunohorizons 3 (7) (2019) 262–273. [DOI] [PubMed] [Google Scholar]
- [37].Bagnati M, Moreno-Moral A, Ko JH, Nicod J, Harmston N, Imprialou M, Game L, Gil J, Petretto E, Behmoaras J, Systems genetics identifies a macrophage cholesterol network associated with physiological wound healing, JCI Insight 4 (2) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Malissen B, Tamoutounour S, Henri S, The origins and functions of dendritic cells and macrophages in the skin, Nat. Rev. Immunol 14 (6) (2014) 417–428. [DOI] [PubMed] [Google Scholar]
- [39].Minutti CM, Knipper JA, Allen JE, Zaiss DM, Tissue-specific contribution of macrophages to wound healing, Semin. Cell Dev. Biol 61 (2017) 3–11. [DOI] [PubMed] [Google Scholar]
- [40].Hirsch T, Spielmann M, Zuhaili B, Koehler T, Fossum M, Steinau HU, Yao F, Steinstraesser L, Onderdonk AB, Eriksson E, Enhanced susceptibility to infections in a diabetic wound healing model, BMC Surg. 8 (2008) 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Mirza R, Koh TJ, Dysregulation of monocyte/macrophage phenotype in wounds of diabetic mice, Cytokine 56 (2) (2011) 256–264. [DOI] [PubMed] [Google Scholar]
- [42].Kimball AS, Joshi A, Ft Carson W, Boniakowski AE, Schaller M, Allen R, Bermick J, Davis FM, Henke PK, Burant CF, Kunkel SL, Gallagher KA, The histone methyltransferase MLL1 directs macrophage-mediated inflammation in wound healing and is altered in a murine model of obesity and type 2, Diabetes 66 (9) (2017) 2459–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Pavlou S, Lindsay J, Ingram R, Xu H, Chen M, Sustained high glucose exposure sensitizes macrophage responses to cytokine stimuli but reduces their phagocytic activity, BMC Immunol. 19 (1) (2018) 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Deusenbery CB, Kalan L, Meisel JS, Gardner SE, Grice EA, Spiller KL, Human macrophage response to microbial supernatants from diabetic foot ulcers, Wound Repair Regen. 27 (6) (2019) 598–608. [DOI] [PubMed] [Google Scholar]
- [45].Canedo-Dorantes L, Canedo-Ayala M, Skin acute wound healing: a comprehensive review, Int J. Inflamm 2019 (2019), 3706315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Friggeri A, Banerjee S, Biswas S, de Freitas A, Liu G, Bierhaus A, Abraham E, Participation of the receptor for advanced glycation end products in efferocytosis, J. Immunol 186 (11) (2011) 6191–6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].He M, Kubo H, Morimoto K, Fujino N, Suzuki T, Takahasi T, Yamada M, Yamaya M, Maekawa T, Yamamoto Y, Yamamoto H, Receptor for advanced glycation end products binds to phosphatidylserine and assists in the clearance of apoptotic cells, EMBO Rep. 12 (4) (2011) 358–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Davis FM, Tsoi LC, Wasikowski R, denDekker A, Joshi A, Wilke C, Deng H, Wolf S, Obi A, Huang S, Billi AC, Robinson S, Lipinski J, Melvin WJ, Audu CO, Weidinger S, Kunkel SL, Smith A, Gudjonsson JE, Moore BB, Gallagher KA, Epigenetic regulation of the PGE2 pathway modulates macrophage phenotype in normal and pathologic wound repair, JCI Insight 5 (17) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Gallagher KA, Joshi A, Carson WF, Schaller M, Allen R, Mukerjee S, Kittan N, Feldman EL, Henke PK, Hogaboam C, Burant CF, Kunkel SL, Epigenetic changes in bone marrow progenitor cells influence the inflammatory phenotype and alter wound healing in type 2 diabetes, Diabetes 64 (4) (2015) 1420–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Barman PK, Urao N, Koh TJ, Diabetes induces myeloid bias in bone marrow progenitors associated with enhanced wound macrophage accumulation and impaired healing, J. Pathol 249 (4) (2019) 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Yan J, Tie G, Wang S, Tutto A, DeMarco N, Khair L, Fazzio TG, Messina LM, Diabetes impairs wound healing by Dnmt1-dependent dysregulation of hematopoietic stem cells differentiation towards macrophages, Nat. Commun 9 (1) (2018) 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kaymakcalan OE, Abadeer A, Goldufsky JW, Galili U, Karinja SJ, Dong X, Jin JL, Samadi A, Spector JA, Topical alpha-gal nanoparticles accelerate diabetic wound healing, Exp. Dermatol 29 (4) (2020) 404–413. [DOI] [PubMed] [Google Scholar]
- [53].Januszyk M, Chen K, Henn D, Foster DS, Borrelli MR, Bonham CA, Sivaraj D, Wagh D, Longaker MT, Wan DC, Gurtner GC, Characterization of diabetic and non-diabetic foot ulcers using single-cell RNA-sequencing, Micro (Basel) 11 (9) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Barrero MJ, Boue S, Izpisua JC, Belmonte, epigenetic mechanisms that regulate cell identity, Cell Stem Cell 7 (5) (2010) 565–570. [DOI] [PubMed] [Google Scholar]
- [55].Ft Carson W, Cavassani KA, Soares EM, Hirai S, Kittan NA, Schaller MA, Scola MM, Joshi A, Matsukawa A, Aronoff DM, Johnson CN, Dou Y, Gallagher KA, Kunkel SL, The STAT4/MLL1 epigenetic axis regulates the antimicrobial functions of murine macrophages, J. Immunol 199 (5) (2017) 1865–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Ishii M, Wen H, Corsa CA, Liu T, Coelho AL, Allen RM, Ft Carson W, Cavassani KA, Li X, Lukacs NW, Hogaboam CM, Dou Y, Kunkel SL, Epigenetic regulation of the alternatively activated macrophage phenotype, Blood 114 (15) (2009) 3244–3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Portela A, Esteller M, Epigenetic modifications and human disease, Nat. Biotechnol 28 (10) (2010) 1057–1068. [DOI] [PubMed] [Google Scholar]
- [58].Allshire RC, Madhani HD, Ten principles of heterochromatin formation and function, Nat. Rev. Mol. Cell Biol 19 (4) (2018) 229–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Strahl BD, Allis CD, The language of covalent histone modifications, Nature 403 (6765) (2000) 41–45. [DOI] [PubMed] [Google Scholar]
- [60].Jenuwein T, Allis CD, Translating the histone code, Science 293 (5532) (2001) 1074–1080. [DOI] [PubMed] [Google Scholar]
- [61].Ferrari KJ, Scelfo A, Jammula S, Cuomo A, Barozzi I, Stutzer A, Fischle W, Bonaldi T, Pasini D, Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity, Mol. Cell 53 (1) (2014) 49–62. [DOI] [PubMed] [Google Scholar]
- [62].Davis FM, d enDekker A, Kimball A, Joshi AD, El Azzouny M, Wolf SJ, Obi AT, Lipinski J, Gudjonsson JE, Xing X, Plazyo O, Audu C, Melvin WJ, Singer K, Henke PK, Moore BB, Burant C, Kunkel SL, Gallagher KA, Epigenetic regulation of TLR4 in diabetic macrophages modulates immunometabolism and wound repair, J. Immunol 204 (9) (2020) 2503–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Davis FM, Kimball A, denDekker A, Joshi AD, Boniakowski AE, Nysz D, Allen RM, Obi A, Singer K, Henke PK, Moore BB, Kunkel SL, Gallagher KA, Histone methylation directs myeloid TLR4 expression and regulates wound healing following cutaneous tissue injury, J. Immunol 202 (6) (2019) 1777–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Kittan NA, Allen RM, Dhaliwal A, Cavassani KA, Schaller M, Gallagher KA, Ft Carson W, Mukherjee S, Grembecka J, Cierpicki T, Jarai G, Westwick J, Kunkel SL, Hogaboam CM, Cytokine induced phenotypic and epigenetic signatures are key to establishing specific macrophage phenotypes, PLoS One 8 (10) (2013) 78045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, Kumagai Y, Miyake T, Matsushita K, Okazaki T, Saitoh T, Honma K, Matsuyama T, Yui K, Tsujimura T, Standley DM, Nakanishi K, Nakai K, Akira S, The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection, Nat. Immunol 11 (10) (2010) 936–944. [DOI] [PubMed] [Google Scholar]
- [66].Kruidenier L, Chung CW, Cheng Z, Liddle J, Che K, Joberty G, Bantscheff M, Bountra C, Bridges A, Diallo H, Eberhard D, Hutchinson S, Jones E, Katso R, Leveridge M, Mander PK, Mosley J, Ramirez-Molina C, Rowland P, Schofield CJ, Sheppard RJ, Smith JE, Swales C, Tanner R, Thomas P, Tumber A, Drewes G, Oppermann U, Patel DJ, Lee K, Wilson DM, A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response, Nature 488 (7411) (2012) 404–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G, The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing, Cell 130 (6) (2007) 1083–1094. [DOI] [PubMed] [Google Scholar]
- [68].Grunstein M, Histone acetylation in chromatin structure and transcription, Nature 389 (6649) (1997) 349–352. [DOI] [PubMed] [Google Scholar]
- [69].d enDekker AD, Davis FM, Joshi AD, Wolf SJ, Allen R, Lipinski J, Nguyen B, Kirma J, Nycz D, Bermick J, Moore BB, Gudjonsson JE, Kunkel SL, Gallagher KA, TNF-alpha regulates diabetic macrophage function through the histone acetyltransferase MOF, JCI Insight 5 (5) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Karnam K, Sedmaki K, Sharma P, Routholla G, Goli S, Ghosh B, Venuganti VVK, Kulkarni OP, HDAC6 inhibitor accelerates wound healing by inhibiting tubulin mediated IL-1β secretion in diabetic mice, Biochim. Biophys. Acta Mol. Basis Dis 1866 (11) (2020), 165903. [DOI] [PubMed] [Google Scholar]
- [71].Li E, Zhang Y, DNA methylation in mammals, Cold Spring Harb. Perspect. Biol 6 (5) (2014), 019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Wang X, Cao Q, Yu L, Shi H, Xue B, Shi H, Epigenetic regulation of macrophage polarization and inflammation by DNA methylation in obesity, JCI Insight 1 (19) (2016) 87748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Yang X, Wang X, Liu D, Yu L, Xue B, Shi H, Epigenetic regulation of macrophage polarization by DNA methyltransferase 3b, Mol. Endocrinol 28 (4) (2014) 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Huang C, Liu XJ, QunZhou, Xie J, Ma TT, Meng XM, Li J, MiR-146a modulates macrophage polarization by inhibiting Notch1 pathway in RAW264.7 macrophages, Int Immunopharmacol. 32 (2016) 46–54. [DOI] [PubMed] [Google Scholar]
- [75].Li D, Duan M, Feng Y, Geng L, Li X, Zhang W, MiR-146a modulates macrophage polarization in systemic juvenile idiopathic arthritis by targeting INHBA, Mol. Immunol 77 (2016) 205–212. [DOI] [PubMed] [Google Scholar]
- [76].O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D, MicroRNA-155 is induced during the macrophage inflammatory response, Proc. Natl. Acad. Sci. USA 104 (5) (2007) 1604–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Liechty C, Hu J, Zhang L, Liechty KW, Xu J, Role of microRNA-21 and its underlying mechanisms in inflammatory responses in diabetic wounds, Int J. Mol. Sci 21 (9) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Hu J, Zhang L, Liechty C, Zgheib C, Hodges MM, Liechty KW, Xu J, Long noncoding RNA GAS5 regulates macrophage polarization and diabetic wound healing, J. Investig. Dermatol 140 (8) (2020) 1629–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Mirza RE, Fang MM, Ennis WJ, Koh TJ, Blocking interleukin-1beta induces a healing-associated wound macrophage phenotype and improves healing in type 2 diabetes, Diabetes 62 (7) (2013) 2579–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Mirza RE, Fang MM, Weinheimer-Haus EM, Ennis WJ, Koh TJ, Sustained inflammasome activity in macrophages impairs wound healing in type 2 diabetic humans and mice, Diabetes 63 (3) (2014) 1103–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Spravchikov N, Sizyakov G, Gartsbein M, Accili D, Tennenbaum T, Wertheimer E, Glucose effects on skin keratinocytes: implications for diabetes skin complications, Diabetes 50 (7) (2001) 1627–1635. [DOI] [PubMed] [Google Scholar]
- [82].Hirota T, Levy JH, Iba T, The influence of hyperglycemia on neutrophil extracellular trap formation and endothelial glycocalyx damage in a mouse model of type 2 diabetes, Microcirculation 27 (5) (2020) 12617. [DOI] [PubMed] [Google Scholar]
- [83].Martinez N, Vallerskog T, West K, Nunes-Alves C, Lee J, Martens GW, Behar SM, Kornfeld H, Chromatin decondensation and T cell hyperresponsiveness in diabetes-associated hyperglycemia, J. Immunol 193 (9) (2014) 4457–4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Passos ME, Alves HH, Momesso CM, Faria FG, Murata G, Cury-Boaventura MF, Hatanaka E, Massao-Hirabara S, Gorjao R, Differential effects of palmitoleic acid on human lymphocyte proliferation and function, Lipids Health Dis. 15 (1) (2016) 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Souza CO, Teixeira AA, Biondo LA, Silveira LS, Calder PC, Rosa JC, Neto, palmitoleic acid reduces the inflammation in LPS-stimulated macrophages by inhibition of NFkappaB, independently of PPARs, Clin. Exp. Pharm. Physiol 44 (5) (2017) 566–575. [DOI] [PubMed] [Google Scholar]
- [86].Camell C, Smith CW, Dietary oleic acid increases m2 macrophages in the mesenteric adipose tissue, PLoS One 8 (9) (2013) 75147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].I.S.S. A, A.B. C, J.S. A, Changes in plasma free fatty acids associated with type-2 diabetes, Nutrients 11 (9) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Xu H, Hertzel AV, Steen KA, Bernlohr DA, Loss of fatty acid binding protein 4/aP2 reduces macrophage inflammation through activation of SIRT3, Mol. Endocrinol 30 (3) (2016) 325–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Boniakowski AM, d enDekker AD, Davis FM, Joshi A, Kimball AS, Schaller M, Allen R, Bermick J, Nycz D, Skinner ME, Robinson S, Obi AT, Moore BB, Gudjonsson JE, Lombard D, Kunkel SL, Gallagher KA, SIRT3 regulates macrophage-mediated inflammation in diabetic wound repair, J. Investig. Dermatol 139 (12) (2019) 2528–2537, e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Seraphim PM, Leal EC, Moura J, Goncalves P, Goncalves JP, Carvalho E, Lack of lymphocytes impairs macrophage polarization and angiogenesis in diabetic wound healing, Life Sci. 254 (2020), 117813. [DOI] [PubMed] [Google Scholar]
- [91].Rodero MP, Hodgson SS, Hollier B, Combadiere C, Khosrotehrani K, Reduced Il17a expression distinguishes a Ly6c(lo)MHCII(hi) macrophage population promoting wound healing, J. Investig. Dermatol 133 (3) (2013) 783–792. [DOI] [PubMed] [Google Scholar]
- [92].Jiang Y, Tsoi LC, Billi AC, Ward NL, Harms PW, Zeng C, Maverakis E, Kahlenberg JM, Gudjonsson JE, Cytokinocytes: the diverse contribution of keratinocytes to immune responses in skin, JCI Insight 5 (20) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Alsaggar M, Mills M, Liu D, Interferon beta overexpression attenuates adipose tissue inflammation and high-fat diet-induced obesity and maintains glucose homeostasis, Gene Ther. 24 (1) (2017) 60–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Boulton AJ, Vileikyte L, Ragnarson-Tennvall G, Apelqvist J, The global burden of diabetic foot disease, Lancet 366 (9498) (2005) 1719–1724. [DOI] [PubMed] [Google Scholar]
- [95].Xu G, Liu G, Xiong S, Liu H, Chen X, Zheng B, The histone methyltransferase Smyd2 is a negative regulator of macrophage activation by suppressing interleukin 6 (IL-6) and tumor necrosis factor alpha (TNF-alpha) production, J. Biol. Chem 290 (9) (2015) 5414–5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Stender JD, Pascual G, Liu W, Kaikkonen MU, Do K, Spann NJ, Boutros M, Perrimon N, Rosenfeld MG, Glass CK, Control of proinflammatory gene programs by regulated trimethylation and demethylation of histone H4K20, Mol. Cell 48 (1) (2012) 28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Hachiya R, Shiihashi T, Shirakawa I, Iwasaki Y, Matsumura Y, Oishi Y, Nakayama Y, Miyamoto Y, Manabe I, Ochi K, Tanaka M, Goda N, Sakai J, Suganami T, Ogawa Y, The H3K9 methyltransferase Setdb1 regulates TLR4-mediated inflammatory responses in macrophages, Sci. Rep 6 (2016) 28845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Li Y, Reddy MA, Miao F, Shanmugam N, Yee JK, Hawkins D, Ren B, Natarajan R, Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-kappaB-dependent inflammatory genes. Relevance to diabetes and inflammation, J. Biol. Chem 283 (39) (2008) 26771–26781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Hassa PO, Covic M, Bedford MT, Hottiger MO, Protein arginine methyltransferase 1 coactivates NF-kappaB-dependent gene expression synergistically with CARM1 and PARP1, J. Mol. Biol 377 (3) (2008) 668–678. [DOI] [PubMed] [Google Scholar]
- [100].Tausendschon M, Dehne N, Brune B, Hypoxia causes epigenetic gene regulation in macrophages by attenuating Jumonji histone demethylase activity, Cytokine 53 (2) (2011) 256–262. [DOI] [PubMed] [Google Scholar]
- [101].Yoshizaki T, Schenk S, Imamura T, Babendure JL, Sonoda N, Bae EJ, Oh DY, Lu M, Milne JC, Westphal C, Bandyopadhyay G, Olefsky JM, SIRT1 inhibits inflammatory pathways in macrophages and modulates insulin sensitivity, Am. J. Physiol. Endocrinol. Metab 298 (3) (2010) E419–E428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Ji L, Chen Y, Wang H, Zhang W, He L, Wu J, Liu Y, Overexpression of Sirt6 promotes M2 macrophage transformation, alleviating renal injury in diabetic nephropathy, Int. J. Oncol 55 (1) (2019) 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Wu C, Li A, Hu J, Kang J, Histone deacetylase 2 is essential for LPS-induced inflammatory responses in macrophages, Immunol. Cell Biol 97 (1) (2019) 72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Ghiboub M, Zhao J, Li Yim AYF, Schilderink R, Verseijden C, van Hamersveld PHP, Duarte JM, Hakvoort TBM, Admiraal I, Harker NR, Tough DF, Henneman P, de Winther MPJ, de Jonge WJ, HDAC3 mediates the inflammatory response and LPS tolerance in human monocytes and macrophages, Front Immunol. 11 (2020), 550769. [DOI] [PMC free article] [PubMed] [Google Scholar]


