Abstract
Epigenetic pharmacotherapies have emerged as a promising treatment option for substance use disorder (SUD) due to their ability to reverse maladaptive transcriptional and behavioral responses to drugs of abuse. In particular, inhibitors of bromodomain and extra terminal domain (BET) reader proteins have been shown to reduce cocaine- and opioid-seeking behaviors in rodents. However, only pan-BET inhibitors, small molecules that bind to both bromodomains (BD1 and BD2) with all BET proteins, have been investigated in animal models of SUD. Given the potential side effects associated with pan-BET inhibitors, safer and more selective strategies are needed to advance BET therapeutics as a potential treatment for SUD. Here, we show that RVX-208, a clinically tested, BD2-selective BET inhibitor, dose-dependently reduced cocaine conditioned place preference in male and female mice, similar to the pan-BET inhibitor JQ1. In other behavioral experiments, RVX-208 treatment did not alter distance traveled, anxiety-like behavior, or novel object recognition memory. At the transcriptional level, RVX-208 attenuated the expression of multiple cocaine-induced genes in the nucleus accumbens in a sex-dependent manner. RVX-208 produced a distinct transcriptional response in stimulated primary neurons compared to JQ1 but had little effect on gene expression in non-stimulated neurons. Together, these data indicate that targeting domain-specific BET mechanisms may be an effective and safer strategy to reduce cocaine-induced neurobehavioral adaptations.
Keywords: epigenetics, bromodomain, BET, BRD4, cocaine, addiction, substance use disorder
Graphical Abtract
Introduction
Repeated drug use alters histone acetylation processes in reward-related brain regions, which contributes, in part, to the molecular maladaptations involved in drug intake and relapse (Rogge and Wood, 2013). While writers (e.g., histone acetyltransferases, HATs) and erasers (e.g., histone deacetylases, HDACs) of histone acetylation have well-established roles in animal models of substance use disorder (SUD) (Kennedy et al., 2013; Malvaez et al., 2013, 2011; Rogge et al., 2013), readers of histone acetylation, called bromodomains, have also emerged as potential therapeutic targets (Egervari et al., 2017; Guo et al., 2020; Sartor et al., 2015). Specifically, pharmacological inhibition of bromodomain and extra terminal domain (BET) reader proteins (BRD2, BRD3, BRD4 and BRDT) has been shown to normalize transcriptional aberrations and behavioral symptoms in a wide range of disease models, including SUD (For review see: Singh and Sartor, 2020; Zaware and Zhou, 2017). Recognizing the growing success and usefulness of BET inhibitors, many large and small pharmaceutical companies have developed new and more selective BET therapeutics that are being tested in a variety of preclinical and clinical studies (Lu et al., 2020; Sartor, 2019).
Each BET protein contains highly conserved, tandem bromodomains (BD1 and BD2) that have been successfully targeted by small molecule inhibitors (Wu et al., 2019). First-generation BET inhibitors, also referred to as pan-BET inhibitors (e.g., JQ1, IBET151, IBET762), displayed similar affinity for BD1 and BD2 within all BET proteins and have been shown to alter disease-associated gene expression in multiple cell types by blocking BET bromodomain interactions with acetylated histones (Dawson et al., 2011; Filippakopoulos et al., 2010; Huang et al., 2009; Nicodeme et al., 2010). In preclinical SUD models, the pan-BET inhibitor, JQ1, was found to attenuate behavioral and transcriptional responses to cocaine, amphetamine, nicotine, and heroin in mice and rats (Babigian et al., 2021; Egervari et al., 2017; Guo et al., 2020; Sartor et al., 2015). However, our understanding of bromodomain-specific BET mechanisms in drug-seeking behaviors remains limited, as JQ1 blocks both bromodomains (BD1 and BD2) within all BET proteins. Additionally, pan-BET inhibitors similar to JQ1 are known to have side effects in humans that may limit their clinical applications for SUD (Amorim et al., 2016; Berthon et al., 2016).
To reduce potential toxic liabilities associated with pan-BET inhibitors, more recent efforts have focused on developing domain-selective BET inhibitors that target specific bromodomains within BET proteins (Petretich et al., 2020). RVX-208 (also called Apabetalone and RVX000222) was one of the first small molecule BET inhibitors to display preferential binding to the BD2 domain within BET proteins (Picaud et al., 2013). In cell lines, RVX-208 only modestly affected BET-dependent gene expression and produced a unique transcriptional outcome compared to JQ1 (Picaud et al., 2013). More recent studies indicate that RVX-208 may offer therapeutic benefits in multiple disease models by blocking the transcriptional responses to inflammatory stimuli (Tsujikawa et al., 2019; Wasiak et al., 2020). In humans, RVX-208 was found to be safe and well tolerated, and it is currently being investigated in Phase II/III clinical trials for the treatment of acute coronary syndromes, atherosclerosis, and Alzheimer’s disease (McNeill, 2010; Nikolic et al., 2015). Given that RVX-208 is a clinically tested, potentially safe, BD2-selective BET inhibitor, we sought to investigate its effects on cocaine-induced behavior and transcriptional activity. Here, we found that RVX-208 dose-dependently reduced cocaine conditioned place preference in male and female mice without altering locomotor activity, thigmotaxis, and novel object recognition learning. At the transcriptional level, RVX-208 reduced cocaine-induced gene expression in the nucleus accumbens (NAc) and produced a disparate transcriptional response in primary neurons compared to JQ1. Together, these data indicate that the BET BD2 plays an important role in the behavioral and transcriptional responses to cocaine and that BD2 BET inhibition has limited effects in non-stimulated neurons and other behaviors.
Materials and Methods
Drug treatments:
For in vitro studies, JQ1 and RVX-208 (Cayman chemicals) were dissolved in dimethyl sulfoxide (DMSO) and administered at 0.1% v/v. In in vivo studies, JQ1 and RVX-208 were dissolved in 10% DMSO and 5% Tween 80 (v/v) and then diluted with sterile saline. RVX-208, JQ1, and vehicle were administered by an intraperitoneal (i.p.) injection at a volume of 0.1–0.15 mL. Cocaine HCl (10 mg/kg, National Institutes of Health’s National Institute on Drug Abuse Drug Supply Program) was dissolved in 0.9% sterile saline and administered by an i.p. injection.
Animals:
Male and female C57BL/6 mice (10–12 weeks old; Charles River Laboratories) were housed 4 mice per cage under a reverse 12 h/12 h light/dark cycle and had ad libitum access to food and water. For primary neuron experiments, a pregnant female rat (Sprague Dawley; Charles River Laboratories) carrying embryonic day 16 (E16) pups was single housed for two days with ad libitum access to food and water. Animals were housed in a humidity- and temperature-controlled and AAALAC-accredited animal facility at the University of Connecticut. Experiments were approved by the Institutional Animal Care and Use Committee and conducted according to guidelines established by the U.S. Public Health Service Policy on Humane Care and Use of Laboratory Animals.
Primary neurons:
For primary cortical neuron experiments, we used similar methods described in BrainBits® primary neuron culture protocol (https://www.brainbitsllc.com/primary-neuron-plating-protocol/). Briefly, brain collection and dissection were carried out under sterile conditions. Pregnant female rats were euthanized in a carbon dioxide chamber. Embryos (E18) were removed, and brains were quickly collected and placed in cold 0.1 M PBS with 0.1% glucose wt./vol. For precise identification of the brain structures, the dissections were performed under a stereo microscope. The meninges and blood vessels were carefully removed, and the cortices were carefully separated from the brain. Frontal cortices were collected, cut into smaller pieces, and digested in papain for 10 min at 37°C (2 mg/ml papain solution in Hibernate-E minus CaCl2, BrainBits LLC). Papain solution was removed, and the tissue was washed three times with Hibernate-E solution followed by 15-minute incubation in Hibernate-E solution and DNAse (1% wt/vol.) at room temperature. Cells were mechanically dissociated with a fire-polished pasteur pipette, resuspended in NbActive4 media (BrainBits LLC), and plated at a density of ~400,000 cells on a poly-L-ornithine (100 μg/ml) and laminin (15 μg/ml) coated 12-well plates. Neurons were maintained at 5% CO2 and 37° C for 12 days in vitro (DIV), and half the media was replaced with fresh media every three days. The use of anti-mitotic compounds to suppress glia growth is known to have toxic side effects in neurons and were not used (Meberg and Miller, 2003). In some experiments, neurons were treated with DMSO (vehicle), RVX-208 (100 nM and 500 nM) and JQ1 (100 nM and 500 nM) for 1 h, 2 h and 3 h. In other experiments, neurons received a 2 h cotreatment of AMPA (20 μM) or BDNF (30 ng/mL) with DMSO, RVX-208 (500 nM), or JQ1 (500 nM). These time points were selected to capture the early effects of BET inhibition and are based on previous studies using pan-BET inhibitors (Sartor et al., 2015; Sullivan et al., 2015). BDNF and AMPA stimulation were used because previous reports indicate that BET protein activity is regulated by glutamate and tyrosine receptor kinase B (TrKB) receptor signaling (Korb et al., 2015; Sullivan et al., 2015), and both receptor signaling pathways have been implicated in cocaine-induced neuroplasticity (Bowers et al., 2010; Li and Wolf, 2015). After treatment, neurons were collected for RNA extraction as described below.
RT-qPCR:
In in vivo studies, mice received an i.p. injection of vehicle or RVX-208 (50 mg/kg) 5 minutes before an i.p. injection of saline or cocaine (10 mg/kg). The nucleus accumbens (NAc) was collected and homogenized in TRIzol 2 hours after the last injection, similar to previous experiments (Sartor et al., 2015). In cell culture experiments, the media was aspirated and 1 ml of TRIzol was added to each well. RNEasy Mini Kit (Qiagen) was used for RNA extraction according to the manufacturer’s instructions. RNA was reverse transcribed using High-Capacity cDNA Reverse Transcription Kit (Thermo Scientific). The genes selected for measurement were based on validated targets of pan-BET inhibitors and/or genes involved in cocaine-induced neuroplasticity (Korb et al., 2015; Sartor et al., 2015; Sullivan et al., 2015). Using validated TaqMan primer probes for Bdnf (Thermo Scientific, Rn02531967_s1, Mm04230607_s1), Gria1 (Rn06323759_m1, Mm00433752_m1), Arc (Rn00571208_g1, Mm01204954_g1), Nr4a1 (Rn01533237_m1, Mm01300401_m1), c-fos (Rn02396759_m1, Mm00487425_m1), and Fosb (Mm00500401_m1), cDNA was run in triplicate and analyzed using the 2−ΔΔCT method with β-actin (Rn00667869_m1, Mm02619580_g1) as a normalization control. Fosb levels were extremely low in non-stimulated primary neurons (RT values > 35 or undetectable), and these data were not reported.
Conditioned place preference (CPP):
The CPP apparatus (Med Associates Inc.) consisted of two compartments with distinct contextual cues on each side: black walls and grid rod-style floors in one compartment and white walls mesh style floors in the other compartment. The compartments were separated by a divider with a small door that could be opened or closed. In a pretest, drug-free acclimation session, mice were allowed to freely explore both compartments for 15 min via an opening in the partition. The time spent on each side of the CPP chamber was automatically recorded by infrared photobeam detectors on the chamber walls. Mice that exhibited a strong bias for one side of the chamber during the pretest (> 65% of time spent on one side) were excluded from further testing. For the next three consecutive days, animals were conditioned as previously described (Sartor et al., 2015; Sartor and Aston-Jones, 2012). During the 30-minute conditioning sessions, one side of the chamber was paired with an i.p. saline injection and the other side was paired with a cocaine injection (10 mg/kg, i.p.). Conditioning occurred in morning and afternoon sessions in a counterbalanced design (at least 4 h apart). Five minutes before each cocaine conditioning session, mice received an i.p. injection of vehicle, JQ1 (50 mg/kg) or RVX-208 (10, 25, or 50 mg/kg). One day after the last conditioning session, mice received a drug-free CPP test where they had free access to each side of the chamber. The preference score was calculated by subtracting the time spent on the cocaine-paired side during the posttest from the time spent on the same side during the pretest.
Open field:
The open field (Ugo Basile) consists of a 46 × 46 cm chamber with opaque gray walls. Baseline behavior (distance traveled and time spent in the inner and outer zone) was measured in a 30-minute, drug-free habituated test. Treatments were assigned so that baseline measurements did not differ between groups. The next day, mice were injected with vehicle or RVX-208 (50 mg/kg, i.p.) and were placed in the chamber 5 minutes post-injection. Distance traveled and time spent in the inner and outer zone were measured for 30 minutes using EthoVision tracking software (Noldus).
Novel object recognition (NOR):
On days 1–3, mice were allowed to freely explore a clean mouse home cage (without objects) for 5 min per day. On day 4, mice were exposed to two identical objects in the cage for 10 min. After the test, mice were immediately injected with vehicle or RVX-208 (50 mg/kg) and returned to the colony room. Previous studies using this approach showed that JQ1 reduced NOR (Korb et al., 2015). The next day, mice were tested in a 10 min retention trial with one familiar object and one novel object. The objects consisted of a stack of Lego Duplo bricks and a 100 ml glass beaker. The location of the objects was counterbalanced for left and right positions in each treatment group. The objects were thoroughly cleaned with 70% ethanol after each session. Total time spent exploring and facing each object within 2.5 cm was calculated by Ethovision tracking software. Results were presented as time exploring the objects and discrimination index: [(novel object investigation time minus familiar object investigation time)/(total investigation time of both objects) * 100].
Data analysis:
Statistical analyses were performed with GraphPad Prism 7.0 software. Mean values from behavioral studies and Rq values from qRT-PCR experiments were compared between groups using one-way, two-way or three-way analysis of variance (ANOVA). When comparing multiple groups, a Bonferroni correction test was utilized. Data are expressed as means ±SEM, and the level of significance was set at P < 0.05.
Results
The effects of RVX-208 on gene expression in primary neurons
In in vitro experiments, primary neurons were treated with vehicle, RVX-208 (100 nM or 500 nM), or JQ1 (100 nM or 500 nM) and gene expression was measured after 1, 2, and 3 h of treatment (Figures 1A–E). Using a two-way ANOVA, significant treatment, time, and interaction effects were observed in Bdnf expression (Treatment: F (4, 28) = 22.95, P < 0.0001; Time: F (2, 56) = 33.66, P < 0.0001; Interaction: F (8, 56) = 6.95, P < 0.0001) and c-fos expression (Treatment: F (4, 28) = 26.24, P < 0.0001; Time: F (2, 56) = 8.57, P = 0.0006; Interaction: F (8, 56) = 2.75, P = 0.012) (Figure 1A and 1D). Significant treatment and interaction effects were detected in Arc expression, but no time effect was found (Treatment: F (4, 28) = 34.22, P < 0.0001; Time: F (2, 56) = 0.92, P = 0.404; Interaction: F (8, 56) = 2.61, P = 0.017) (Figure 1B). A significant treatment effect was observed in Nr4a1 expression but time and interaction effects were not detected (Treatment: F (4, 28) = 55.36, P < 0.0001; Time: F (2, 56) = 0.36, P = 0.675; Interaction: F (8, 56) = 1.71, P = 0.117) (Figure 1C). No significant treatment or interaction effects on Gria1 expression were found, but a time effect was observed (Treatment: F (4, 28) = 0.594, P = 0.670; Time: F (2, 56) = 8.26, P = 0.0007; Interaction: F (8, 56) = 1.77, P = 0.103) (Figure 1E). Bonferroni post hoc analysis revealed that RVX-208 had minimal effect on gene expression compared to vehicle, with the only significant change occurring in Bdnf expression after 2 h of treatment (Figure 1A). JQ1, on the other hand, altered the expression of most genes examined compared to vehicle, including: a decrease in Bdnf at 1, 2 and 3 h timepoints (Figures 1A); a decrease in Arc at 1, 2 and 3 h timepoints (Figures 1B); a decrease in Nr4a1 at 1, 2 and 3 h timepoints (Figures 1C); and an increase in c-fos at the 2 and 3 h timepoints (Figure 1D).
Figure 1: RVX-208 has limited effects in non-stimulated primary neurons.
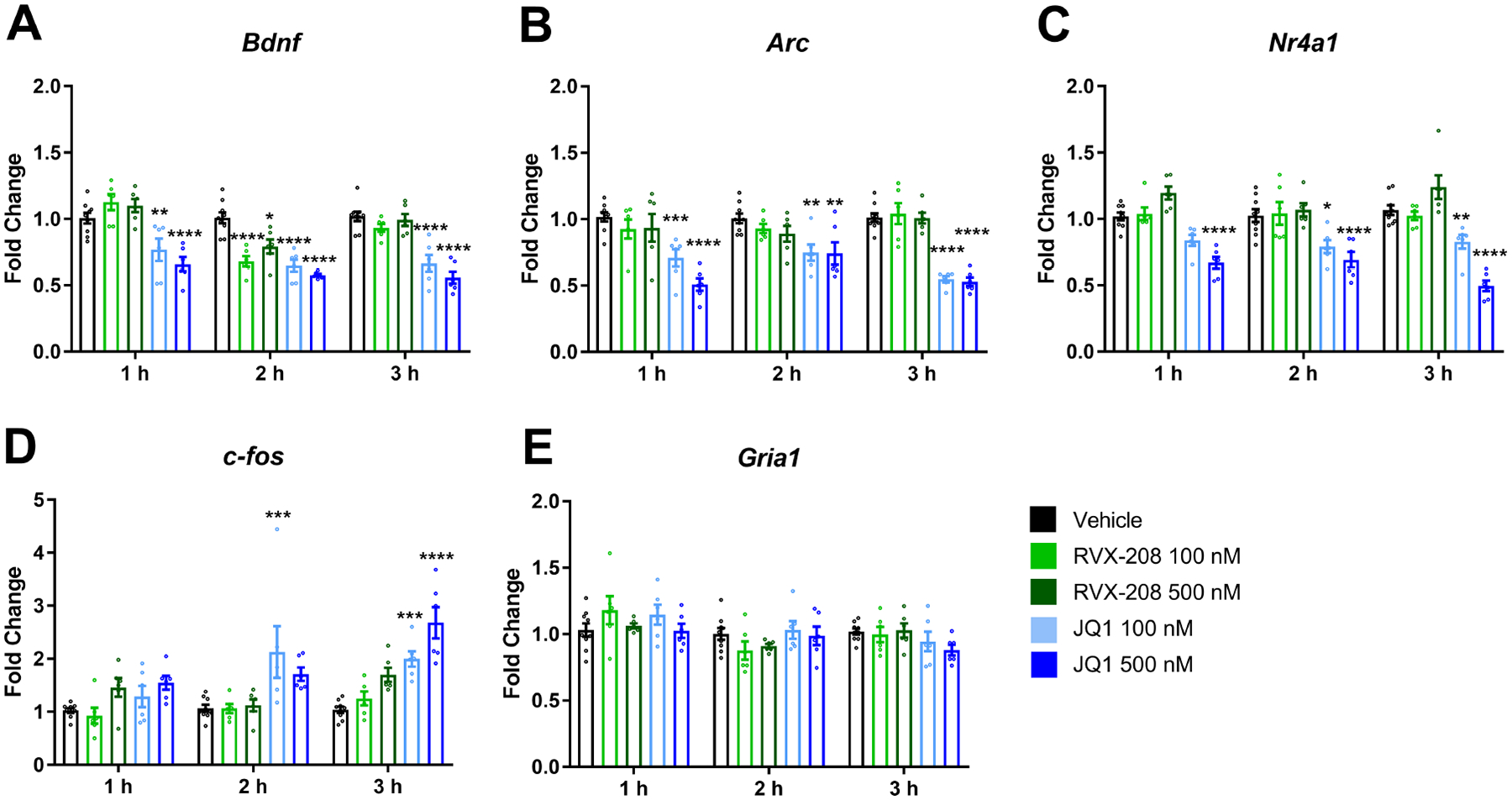
Primary neurons were treated with vehicle (n = 9), RVX-208 (n = 6), or JQ1 (n = 6) (100 nM and 500 nM) for 1, 2, and 3 hours. Following treatment, (A) Bdnf, (B) Arc, (C) Nr4a1, (D) c-fos, and (E) Gria1 expression levels were measured via RT-qPCR. Using Bonferroni post hoc test, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 indicate a significant difference from vehicle at the same timepoint. Data are mean ±SEM.
The effects of RVX-208 on gene expression in stimulated primary neurons
Previous experiments indicate that the BET BD2 domain is responsible for stimulus-induced gene expression in cell lines (Gilan et al., 2020). To examine if RVX-208 alters gene expression in stimulated neurons, we co-treated neurons for 2 h with vehicle, RVX-208 (500 nM), or JQ1 (500 nM) plus BDNF (Figure 2) or AMPA (Figure 3). In BDNF and AMPA stimulated neurons, all genes examined, except Gria1, were elevated by more than 5-fold compared to untreated neurons (baseline) (Figures 2 and 3). In BDNF simulated neurons, RVX-208 and JQ1 significantly reduced the expression of Bdnf (One-way ANOVA main effect: F(3, 20) = 44.69, P < 0.0001 followed by Bonferroni post-hoc test, P = 0.025 for BDNF vs. RVX-208 + BDNF, P = 0.022 for Vehicle + BDNF vs. RVX-208 + BDNF, P < 0.0001 for BDNF vs. JQ1 + BDNF, P < 0.0001 for Vehicle + BDNF vs. JQ1 + BDNF) (Figure 2A). Arc expression was reduced by RVX-208 but not by JQ1 (One-way ANOVA main effect: F(3, 20) = 10.65, P = 0.0002 followed by Bonferroni post-hoc test, P = 0.044 for BDNF vs. JQ1 + BDNF, P = 0.0086 for Vehicle + BDNF vs. RVX-208 + BDNF) (Figure 2B). Nr4a1 was reduced by RVX-208 but unchanged by JQ1 (One-way ANOVA main effect: F(3, 20) = 8.56, P = 0.0007 followed by Bonferroni post-hoc test, P = 0.0007 for BDNF vs. RVX-208 + BDNF, P = 0.0042 for vehicle + BDNF vs. RVX-208 + BDNF) (Figure 2C), and c-fos was significantly elevated by JQ1 but unchanged RVX-208 in BDNF stimulated neurons (One-way ANOVA main effect: F(3, 20) = 111.0, P < 0.0001 followed by Bonferroni post-hoc test, P < 0.0001 for BDNF vs. JQ1 + BDNF, P < 0.0001 for vehicle + BDNF vs. JQ1 + BDNF) (Figure 2D). Gria1 was not altered by BDNF stimulation or RVX-208 and JQ1 treatments (One-way ANOVA main effect: F(3, 20) = 0.923, P = 0.448) (Figure 2E).
Figure 2: RVX-208 alters the expression of multiple genes in BDNF stimulated primary neurons.
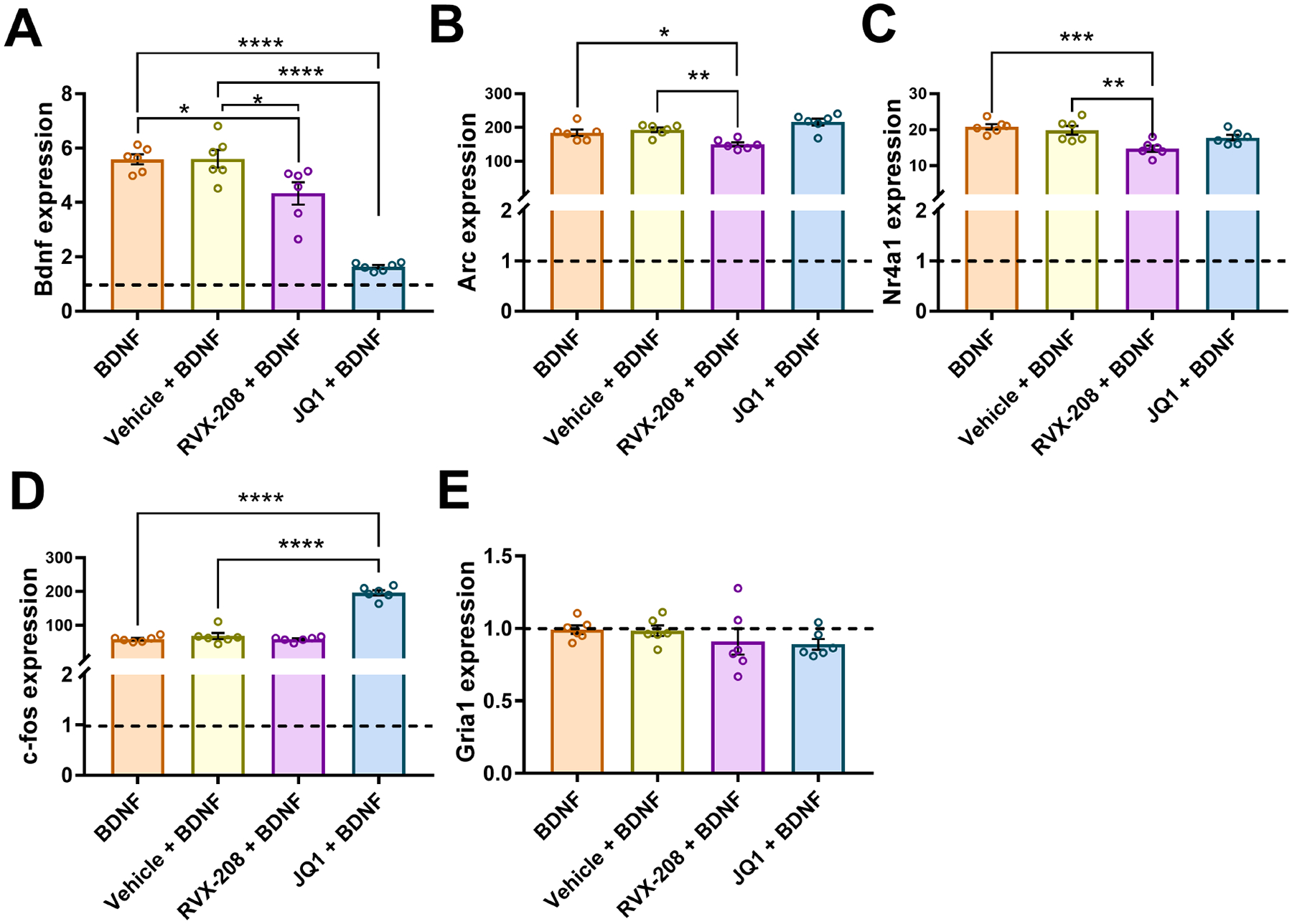
Primary neurons were treated with BDNF, vehicle + BDNF, RVX-208 + BDNF or JQ1 + BDNF for 2 hours. The expression of (A) Bdnf, (B) Arc, (C) Nr4a1, (D) c-fos, and (E) Gria1 was measured via RT-qPCR. Horizontal dotted line indicates baseline expression of untreated neurons. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 indicate a significant difference compared to BDNF or Vehicle + BDNF using Bonferroni post hoc test. Data are mean ±SEM, n = 6 per treatment.
Figure 3: RVX-208 has no effect on gene expression in AMPA stimulated primary neurons.
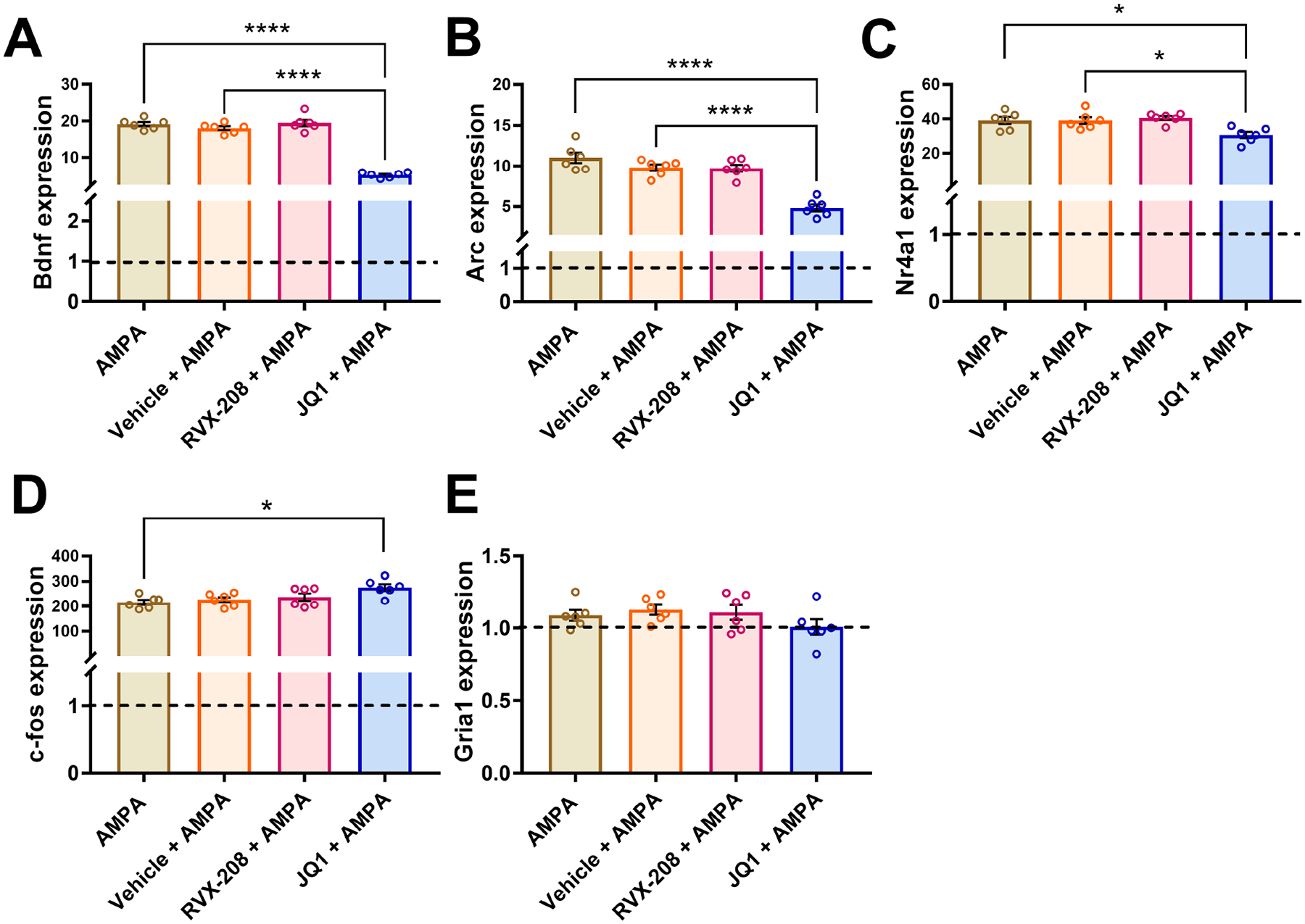
Primary neurons were treated with AMPA, vehicle + AMPA, RVX-208 + AMPA, or JQ1 + AMPA for 2 hours. The expression of (A) Bdnf, (B) Arc, (C) Nr4a1, (D) c-fos, and (E) Gria1 was measured via RT-qPCR. Horizontal dotted line indicates baseline expression of untreated neurons. *P < 0.05 and ****P < 0.0001 indicate a significant difference compared to AMPA or Vehicle + AMPA using Bonferroni post hoc test. Data are mean ±SEM, n = 6 per treatment.
In AMPA stimulated neurons, RVX-208 did not affect the expression of the genes measured (P values > 0.05) (Figure 3). JQ1, on the other hand, decreased Bdnf (One-way ANOVA main effect: F(3, 20) = 127.5, P < 0.0001 followed by Bonferroni post-hoc test, P < 0.0001 for AMPA vs. JQ1 + AMPA, P < 0.0001 for vehicle + AMPA vs. JQ1 + AMPA) (Figure 3A), Arc (One-way ANOVA main effect: F(3, 20) = 31.37, P < 0.0001 followed by Bonferroni post-hoc test, P < 0.0001 for AMPA vs. JQ1 + AMPA, P < 0.0001 for vehicle + AMPA vs. JQ1 + AMPA) (Figure 3B), and Nr4a1 in AMPA stimulated neurons (One-way ANOVA main effect: F(3, 20) = 6.15, P = 0.004 followed by Bonferroni post-hoc test, P = 0.019 for AMPA vs. JQ1 + AMPA, P = 0.02 for vehicle + AMPA vs. JQ1 + AMPA (Figure 3C). Expression of c-fos was increased by JQ1 in AMPA stimulated neurons (One-way ANOVA main effect: F(3, 20) = 4.32, P = 0.017 followed by Bonferroni post-hoc test, P = 0.016 for AMPA vs. JQ1 + AMPA) (Figure 3D), and expression of Gria1 was unchanged by AMPA, RVX-208, and JQ1 (F values < 1.4, P values > 0.05) (Figure 3E).
Effects of RVX-208 on cocaine-induced gene expression in the NAc
To examine the effects of RVX-208 on cocaine-induced gene expression, male and female mice were treated with vehicle or RVX-208 (50 mg/kg) prior to an injection of cocaine (10 mg/kg) or saline (Figure 4). In the NAc, vehicle/cocaine significantly increased the expression of Arc in males but not females when compared to vehicle/saline (Two-way ANOVA treatment effect: F (3, 56) = 17.57 P < 0.0001; sex effect: F (1, 56) = 0.3769, P = 0.5417; interaction effect: F (3, 56) = 4.105, P = 0.012) (Figure 4B). Nr4a1 was increased by cocaine in both sexes (Two-way ANOVA treatment effect: F (3, 56) = 19.45, P < 0.0001; sex effect: F (1, 56) = 4.278, P = 0.043; interaction effect: F (3, 56) = 0.460, P = 0.71) (Figure 4C), while c-fos (Two-way ANOVA treatment effect: F (3, 56) = 17.78, P < 0.0001, sex effect: F (1, 56) = 3.155, P = 0.081; interaction effect: F (3, 56) = 1.61, P = 0.198) (Figure 4D) and Fosb (Two-way ANOVA treatment effect: F (3, 56) = 27.1, P < 0.0001; sex effect: F (1, 56) = 17.3, P = 0.0001; interaction effect: F (3, 56) = 7.28, P = 0.0003) were increased by cocaine in males but not females ((Figure 4E). Treatments had no effect on Gria1 expression (Two-way ANOVA F values < 0.8, P values > 0.05) (Figure 4F). Although a statistically significant treatment effect was observed in Bdnf expression (Two-way ANOVA treatment effect: F (3, 56) = 5.622, P = 0.002; sex effect: F (1, 56) = 0.9583, P = 0.33; interaction effect: F (3, 56) = 1.519, P = 0.22), no significant changes were observed via Bonferroni post hoc comparisons (P values > 0.05) (Figure 4A). When comparing vehicle/cocaine to RVX-208/cocaine, RVX-208 attenuated cocaine-induced gene expression of Arc in males (P < 0.0001), Nr4a1 in males and females (P = 0.04 for males and P = 0.0006 for females), c-fos in males (P < 0.0001), and Fosb in males (P < 0.0001) (Figures 4B–E). When comparing sex differences in vehicle/cocaine treated mice, expression of Fosb was significantly higher in males compared to females (P < 0.0001). There were no significant changes in gene expression when comparing vehicle/saline to RVX-208/saline treated mice (P values > 0.05).
Figure 4: RVX-208 alters cocaine-induced gene expression in the NAc.
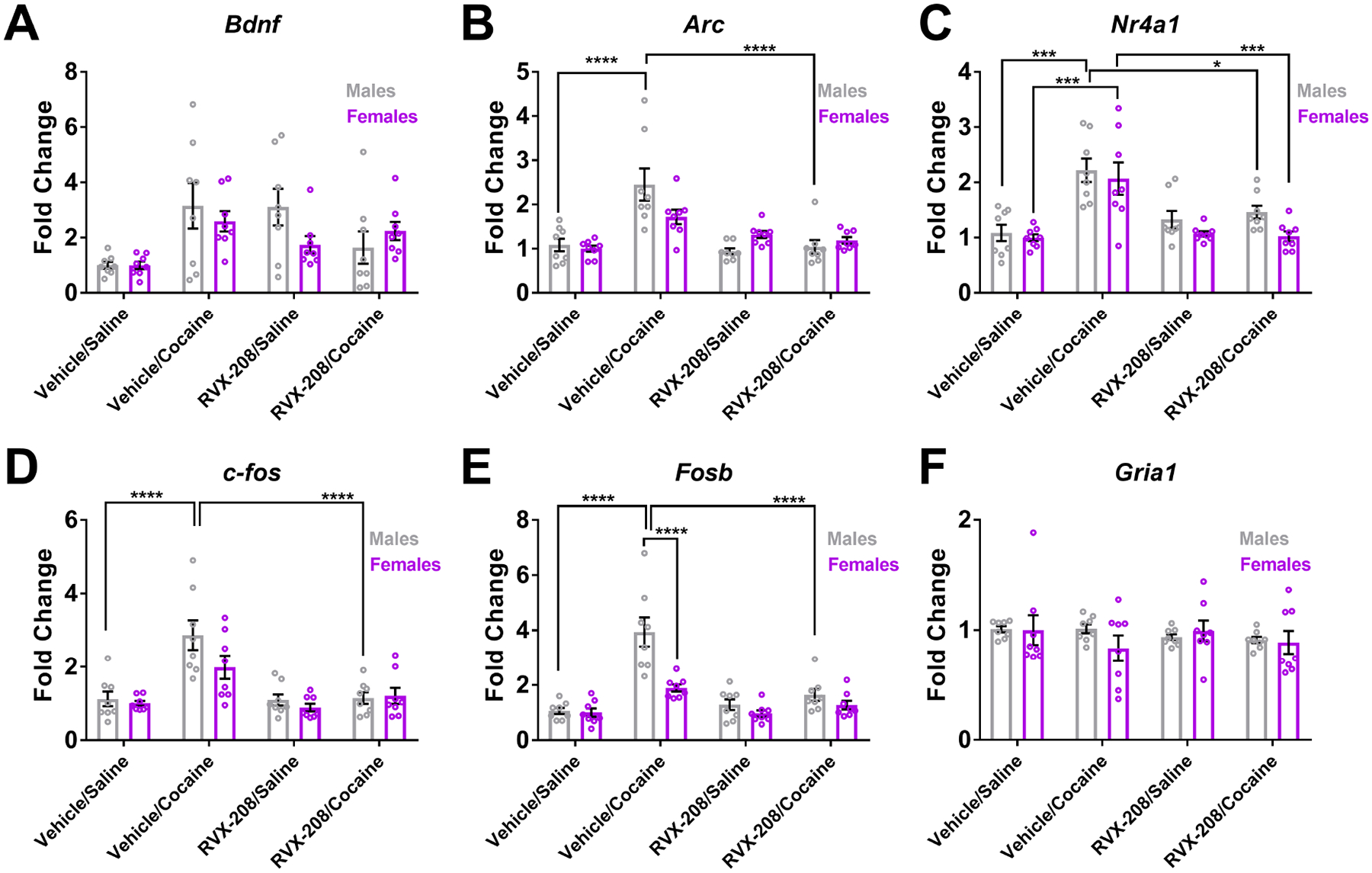
Male and female mice were treated with vehicle/saline, vehicle/cocaine, RVX-208/saline, or RVX-208/cocaine. Following treatment, (A) Bdnf, (B) Arc (C), Nr4a1 (D), c-fos, (E) Fosb, and (F) Gria1 expression levels were measured via RT-qPCR. *P < 0.05, ***P < 0.001, and ****P < 0.0001 indicate a significant difference using Bonferroni post hoc test. Data are mean ±SEM, n = 8 per treatment.
Effects of RVX-208 on cocaine conditioned place preference and other behaviors
To examine the effects of RVX-208 on cocaine-induced behavior, male and female mice were injected with vehicle, RVX-208, or JQ1 prior to each cocaine conditioning session. RVX-208 dose-dependently reduced the acquisition of cocaine conditioned place preference (CPP) compared to vehicle treated male and female mice (Two-way ANOVA treatment effect: F (4, 77) = 13.28, P < 0.0001; sex effect: F (1, 77) = 4.031, P = 0.048; interaction effect: F (4, 77) = 0.7318, P = 0.57) (Figure 5A). In post-hoc analysis, RVX-208 significantly reduced cocaine CPP in females at 10, 25, and 50 mg/kg compared to vehicle, and in males RVX-208 significantly reduced CPP at 25 and 50 mg/kg but not 10 mg/kg. Consistent with previous results in male mice, JQ1 (50 mg/kg) attenuated acquisition of cocaine CPP compared to vehicle in male and female mice (Figure 5A). In other mice, an acute injection of RVX-208 (50 mg/kg) did not alter distance traveled in males and females, but distance traveled was higher in females vs. males, similar to previous reports (Borbélyová et al., 2019) (Two-way ANOVA treatment effect: F (1, 28) = 0.01. P = 0.92; sex effect: F (1, 28) = 19.14, P = 0.0002; interaction effect: F (1, 28) = 0.333, P = 0.57) (Figure 5B). Time spent in the inner and outer zones in an open field was not altered by RVX-208 (Three-way ANOVA treatment effect: F (1, 56) < 0.001, P = 0.99; sex effect: F (1, 56) < 0.001, P = 0.99; zone effect: F (1, 56) = 666, P < 0.0001) (Figure 5C). In the novel object recognition (NOR) task, time exploring the objects during acquisition was not statistically different between treatments or sexes (Two-way ANOVA F values < 0.5, P values > 0.05) (Figure 5D). During the NOR test, vehicle and RVX-208 treated mice showed an increase in time exploring the novel object (NO) compared to the familiar object (FO) (Three-way ANOVA effect on object exploration: F (1, 72) = 31.13, P < 0.0001; sex effect: F (1, 72) = 5.46, P=0.022; treatment effect: F (1, 72) < 0.001, P = 0.99) (Figure 5E). Finally, no significant treatment effect on novel object discrimination index (DI) was observed (Two-way ANOVA F values < 0.9, P values > 0.05) (Figure 5F).
Figure 5. Effects of RVX-208 on behavior.
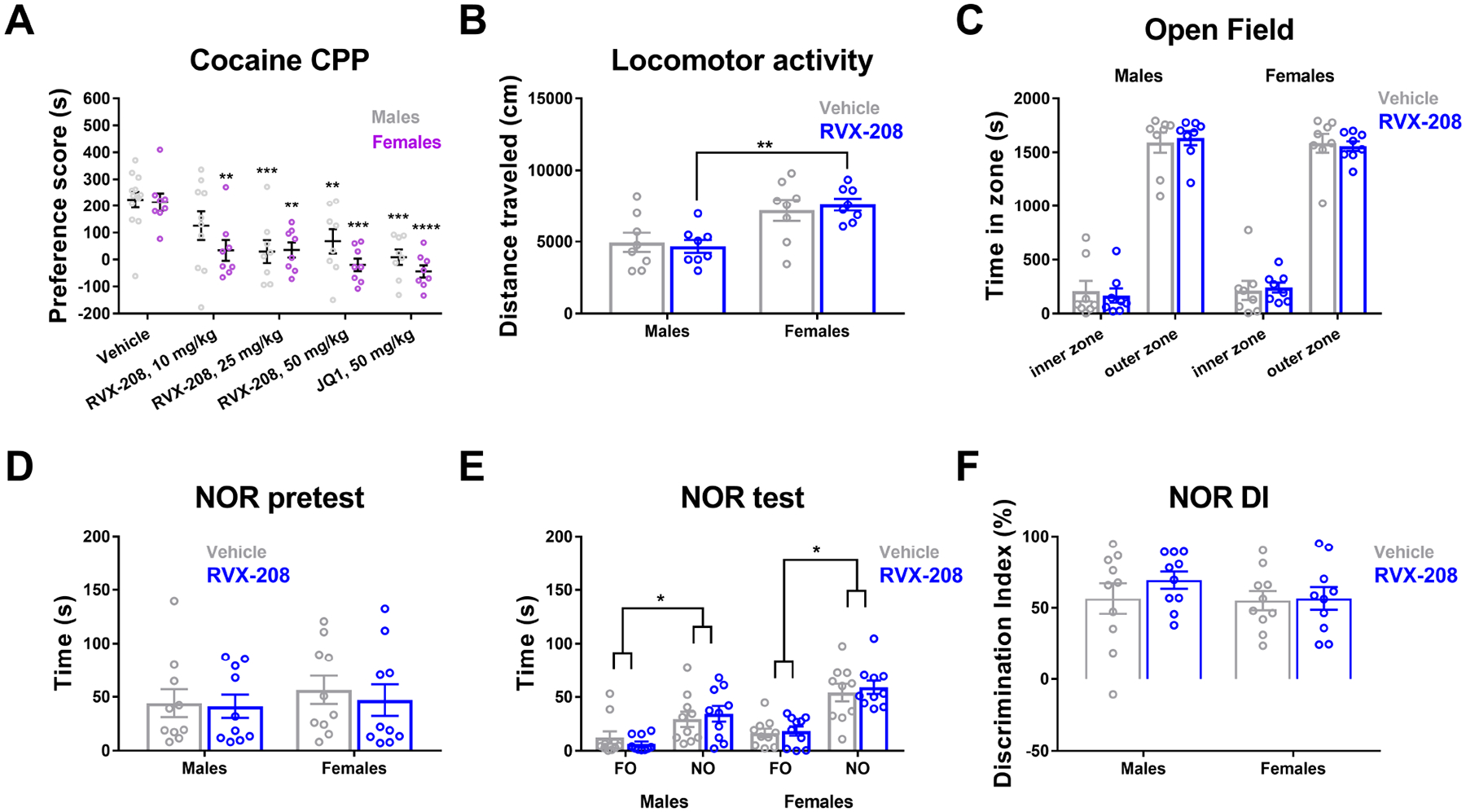
(A) Dose-dependent reduction of cocaine conditioned place preference (CPP) by RVX-208 compared to vehicle in male and female mice (n = 8–13). (B) The effects of RVX-208 (50 mg/kg) on locomotor activity and (C) time spent in the inner and outer zones in open field in male and female mice (n = 8). (D) Time spent exploring objects during novel object recognition (NOR) pretest in male and female mice (n = 10). (E) Time spent exploring the novel object (NO) and familiar object (FO) during the NOR test in vehicle and RVX-208 (50 mg/kg) treated male and female mice (n = 10). (F) NOR discrimination index (DI) values for vehicle and RVX-208 treated male and female mice (n = 10). *P < 0.05, **P < 0.01, ***P < 0.001 indicate a significant difference using Bonferroni post hoc test. Data are mean ±SEM.
Discussion
A role for BD2-selective BET inhibition in cocaine-induced behavioral and transcriptional responses
In previous studies, we showed that BET protein activity and expression are altered by cocaine use, and BET bromodomain inhibition attenuated behavioral and transcriptional responses to cocaine (Sartor et al., 2015). Consistent with these initial findings, more recent studies have confirmed that BET inhibitors reduce other types of drug-seeking behaviors (Egervari et al., 2017; Sartor et al., 2019; Guo et al., 2020; Babigian et al., 2021). To date, however, all addiction-related BET experiments have been conducted in males using the pan-BET inhibitor JQ1, a tool compound that is not suitable for clinical testing due to its poor pharmacokinetic properties (Tanaka et al., 2016). In addition to having side effects that may restrict their clinical potential as a SUD treatment, pan-BET inhibitors similar to JQ1 block both bromodomains within all BET proteins, restricting our understanding of domain-specific BET mechanisms in drug-seeking behaviors. In the pursuit of more selective and safer therapeutic approaches for SUD, here, we report that RVX-208, a clinically tested, BD2-selective BET inhibitor, attenuated cocaine-conditioned responses in male and female mice without altering distance traveled and anxiety-like behavior in an open field. In previous studies, JQ1 reduced novel object recognition (Korb et al., 2015), an indication that pan-BET inhibitors may have undesirable effects on learning and memory. Distinct from these previous results, we found that RVX-208 did not alter novel object recognition, supporting the concept that BD2-selective BET inhibitors have fewer side effects compared to pan-BET inhibitors.
At the transcriptional level, RVX-208 attenuated cocaine-induced expression of Arc, Nr4a1, c-fos, and Fosb in the NAc, genes that play key roles in cocaine-induced neuroplasticity (Bannon et al., 2002; Carpenter et al., 2020; Cates et al., 2019; Hope, 1998; Penrod et al., 2020; Salery et al., 2017; Walker et al., 2018; Xu, 2008). The significant effects were mainly observed in males, as cocaine-induced gene expression in the NAc was generally lower in females compared to males. However, similar trends in treatment-induced gene expression were observed between sexes. Importantly, RVX-208 treatment without cocaine had no significant effect on baseline gene expression in the NAc compared to vehicle-treated mice. These results are in contrast to previous studies that showed pan-BET inhibitors (JQ1 and I-BET858) altered baseline expression of multiple neuroplasticity-associated genes in the NAc and primary neurons (Korb et al., 2015; Sartor et al., 2015; Sullivan et al., 2015). Our current results are in line with previous reports demonstrating that BET BD2 is involved in stimulus-induced gene expression, whereas the BD1 domain (a target of pan-BET inhibitors) maintains steady-state gene expression (Gilan et al., 2020).
Similar to our in vivo findings, RVX-208 had little effect on gene expression in non-stimulated primary neurons, but RVX-208 did reduce Bdnf, Arc and Nr4a1 expression in BDNF, but not AMPA, stimulated neurons. JQ1, on the other hand, produced a sustained and robust change in many of the genes measured in non-stimulated and stimulated neurons. The limited effects of RVX-208 on AMPA-induced gene expression indicates that BD2-mediated transcriptional responses may differ depending on the stimulus used. Consistent with this idea, the effects of BD2-selective inhibition on gene expression also varied depending on stimulus applied in immunoinflammatory disease models (Gilan et al., 2020). Importantly, as we only examined a few target genes in the current study, it is possible that RVX-208 alters the expression of AMPA-induced genes that were not measured.
When comparing RVX-208 to JQ1, a difference in c-fos and Fosb expression was also observed. In the current studies, JQ1 increased c-fos expression in stimulated and non-stimulated neurons, and in previous studies, JQ1 and I-BET858 elevated c-fos and Fosb in the NAc and primary neurons (Sartor et al., 2015; Sullivan et al., 2015). In the current studies, RVX-208 reduced c-fos and Fosb in the NAc of cocaine-treated male mice and had no effect on c-fos in stimulated and non-stimulated primary neurons. These data indicate that inhibition of the BD1 domain via pan-BET inhibition may be responsible for the observed elevation of c-fos and Fosb. A more recent study has confirmed that that inhibition of BD1, but not the BD2, BET domain is responsible for the induction of c-fos in neurons (Jones-Tabah et al., 2022). Together, these data reveal that BET BD2 regulates gene expression in stimulated neurons and brain tissue, but unlike pan-BET inhibition, BET BD2 inhibition has minimal impact on baseline expression.
The limited effects on baseline gene expression in non-stimulated cells is consistent with the improved safety profile of BD2-selective inhibitors compared to pan-BET inhibitors. For example, in multiple clinical trials, RVX-208 was shown to be well tolerated in patients without causing thrombocytopenia and gastrointestinal toxicity (Nicholls et al., 2011; Tsujikawa et al., 2019), which are dose-limiting side effects reported in humans treated with pan-BET inhibitors (Amorim et al., 2016; Berthon et al., 2016). In preclinical and clinical studies, RVX-208’s primary indication has been for the treatment of cardiovascular and metabolic diseases (e.g., atherosclerosis, hypertension, hyperlipidemia) by elevating the expression of apolipoprotein AI (ApoA-I) and high-density lipoprotein (HDL) and reducing the expression of inflammation factors (Gilham et al., 2016; Tsujikawa et al., 2019). Interestingly, chronic cocaine use is also known to cause deleterious effects on the cardiovascular system, and cocaine users have an increased risk of developing coronary atherosclerosis (Bachi et al., 2017). Thus, beyond reducing drug-seeking behaviors, RVX-208 may be beneficial to the cardiovascular health of patients suffering from cocaine use disorder.
Regulation of stimulus-dependent transcription by the BET BD2 domain
Since the development of RVX-208, more selective BD2 BET inhibitors have been reported (Faivre et al., 2020; Gilan et al., 2020). For example, potent and highly selective BD1 (GSK778) and BD2 BET (GSK046) inhibitors were recently identified (Gilan et al., 2020). GSK046 displayed >300-fold selectivity for BD2 over BD1, whereas GSK778 showed ≥130-fold selectivity for BD1 over BD2. The BD1-selective inhibitor GSK778 exhibited similar transcriptional effects compared to pan-BET inhibitors in cancer cells, consistent with previous studies showing that BD1 plays the dominant role in maintaining established transcriptional programs (Picaud et al., 2013). However, distinct from BD1-selective and pan-BET inhibitors, the BD2-selective inhibitor GSK046 primarily affected stimulus-induced gene expression and caused limited disruptions in cell proliferation, cell viability, and BRD4 chromatin binding. In SUD-related research, future studies examining potential differences between modestly selective (RVX-208) versus highly selective (GSK046) BD2 BET inhibitors in cocaine-induced transcriptional and behavioral responses would be of high interest and could potentially guide BET drug development efforts for cocaine use disorder.
Biochemical analysis of the BET protein, BRD4, also indicates that the BD2 domain is involved in stimulus-dependent transcriptional regulation (Chiang, 2016). For example, when the flanking regions of BD2 are dephosphorylated by protein phosphatase 2 (PP2A), the BD2 domain on BRD4 is sterically impeded, preventing it from interacting with acetylated histones and non-histone proteins (Wu et al., 2013). When these regions are phosphorylated by casein kinase 2 (CK2), protein kinase A (PKA), and potentially other kinases, BRD4 undergoes a conformational change that uncovers BD2 and promotes its interactions with other proteins. Though the upstream receptor signaling mechanisms responsible for BRD4 phosphorylation have yet to be fully explored, cocaine and BDNF stimulation were found to increase BRD4 phosphorylation levels in the NAc and primary neurons, respectively (Guo et al., 2020; Korb et al., 2015). The data reported here also suggest that upstream signaling mechanisms following BDNF and cocaine, but not AMPA, stimulation regulate transcription of multiple BET-targeted genes in a BD2-dependent manner.
Outlook of BD2-selective BET inhibitors for the treatment of cocaine use disorder.
Identifying safe and effective treatments for cocaine addiction is an urgent need as there are currently no FDA-approved medications for cocaine use disorder. Developing a brand-new drug treatment, however, takes an enormous amount of time, effort, and money due to years of preclinical and clinical testing for safety and efficacy (Morgan et al., 2011). RVX-208 has been shown to be extremely well tolerated by patients, with safety data now surpassing 2,700 patient-years (Tsujikawa et al., 2019). To date, the only dose-limiting side effect observed with RVX-208 is a reversible and transient elevation in liver enzymes (alanine aminotransferases/aspartate aminotransferases) in a small percentage of patients (Nicholls et al., 2011). With a well-established safety profile in humans, RVX-208 has the potential to make a rapid impact as an epigenetic pharmacotherapy for cocaine use disorder. However, before it can be considered a promising treatment option, RVX-208 must be tested in more sophisticated and translatable models of cocaine use disorder (e.g., short-, long-, and intermittent-access cocaine self-administration and reinstatement/behavioral economic procedures). Additional behavioral tests in female rodents are also needed, as this is the first SUD-related study to report BET inhibitor effects in female rodents. Finally, while we have identified a few target genes that potentially mediate RVX-208’s effects on cocaine-seeking behavior, future transcriptomic and cell type-specific studies are necessary to fully elucidate the mechanisms by which RVX-208 regulates cocaine-induced neurobehavioral adaptations. Ongoing efforts to address these questions will determine the therapeutic potential of domain-selective BET inhibitors for the treatment of substance use disorder.
Highlights:
RVX-208 is a clinically tested BD2-selective BET inhibitor with limited side effects.
RVX-208 reduced cocaine conditioned responding without affecting other behaviors.
RVX-208 attenuated cocaine-induced gene expression in the nucleus accumbens.
In primary neurons, RVX-208 produced a unique transcriptional profile compared to JQ1.
Acknowledgements:
This work was supported by National Institute on Drug Abuse grant R00DA040744.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Statement
Author contributions: G.C.S. and M.B.S. designed research; G.C.S., M.B.S., and C.J.B. performed research; G.C.S. and M.B.S. analyzed data; G.C.S. and M.B.S. wrote the paper.
References
- Amorim S, Stathis A, Gleeson M, Iyengar S, Magarotto V, Leleu X, Morschhauser F, Karlin L, Broussais F, Rezai K, Herait P, Kahatt C, Lokiec F, Salles G, Facon T, Palumbo A, Cunningham D, Zucca E, Thieblemont C, 2016. Bromodomain inhibitor OTX015 in patients with lymphoma or multiple myeloma: a dose-escalation, open-label, pharmacokinetic, phase 1 study. Lancet Haematol 3, e196–204. 10.1016/S2352-3026(16)00021-1 [DOI] [PubMed] [Google Scholar]
- Babigian CJ, Wiedner HJ, Wahlestedt C, Sartor GC, 2021. JQ1 attenuates psychostimulant- but not opioid-induced conditioned place preference. Behavioural Brain Research in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachi K, Mani V, Jeyachandran D, Fayad ZA, Goldstein RZ, Alia-Klein N, 2017. Vascular disease in cocaine addiction. Atherosclerosis 262, 154–162. 10.1016/j.atherosclerosis.2017.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannon MJ, Pruetz B, Manning-Bog AB, Whitty CJ, Michelhaugh SK, Sacchetti P, Granneman JG, Mash DC, Schmidt CJ, 2002. Decreased expression of the transcription factor NURR1 in dopamine neurons of cocaine abusers. Proc Natl Acad Sci U S A 99, 6382–6385. 10.1073/pnas.092654299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthon C, Raffoux E, Thomas X, Vey N, Gomez-Roca C, Yee K, Taussig DC, Rezai K, Roumier C, Herait P, Kahatt C, Quesnel B, Michallet M, Recher C, Lokiec F, Preudhomme C, Dombret H, 2016. Bromodomain inhibitor OTX015 in patients with acute leukaemia: A dose-escalation, phase 1 study. The Lancet Haematology 3, e186–e195. 10.1016/S2352-3026(15)00247-1 [DOI] [PubMed] [Google Scholar]
- Borbélyová V, Janišová K, Mysliveček J, Riljak V, 2019. Sex-related differences in locomotion and climbing of C57Bl/6NTac mice in a novel environment. Physiol Res 68, S353–S359. 10.33549/physiolres.934348 [DOI] [PubMed] [Google Scholar]
- Bowers MS, Chen BT, Bonci A, 2010. AMPA receptor synaptic plasticity induced by psychostimulants: the past, present, and therapeutic future. Neuron 67, 11–24. 10.1016/j.neuron.2010.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter MD, Hu Q, Bond AM, Lombroso SI, Czarnecki KS, Lim CJ, Song H, Wimmer ME, Pierce RC, Heller EA, 2020. Nr4a1 suppresses cocaine-induced behavior via epigenetic regulation of homeostatic target genes. Nat Commun 11, 504. 10.1038/s41467-020-14331-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cates HM, Lardner CK, Bagot RC, Neve RL, Nestler EJ, 2019. Fosb Induction in Nucleus Accumbens by Cocaine Is Regulated by E2F3a. eNeuro 6, ENEURO.0325–18.2019. 10.1523/ENEURO.0325-18.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C-M, 2016. Phospho-BRD4: transcription plasticity and drug targeting. Drug Discov Today Technol 19, 17–22. 10.1016/j.ddtec.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J-H, Park J-Y, Park SP, Lee H, Han S, Park KH, Suh YH, 2016. Regulation of mGluR7 trafficking by SUMOylation in neurons. Neuropharmacology 102, 229–235. 10.1016/j.neuropharm.2015.11.021 [DOI] [PubMed] [Google Scholar]
- Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan W-I, Robson SC, Chung C, Hopf C, Savitski MM, Huthmacher C, Gudgin E, Lugo D, Beinke S, Chapman TD, Roberts EJ, Soden PE, Auger KR, Mirguet O, Doehner K, Delwel R, Burnett AK, Jeffrey P, Drewes G, Lee K, Huntly BJP, Kouzarides T, 2011. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478, 529–533. 10.1038/nature10509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egervari G, Landry J, Callens J, Fullard JF, Roussos P, Keller E, Hurd YL, 2017. Striatal H3K27 Acetylation Linked to Glutamatergic Gene Dysregulation in Human Heroin Abusers Holds Promise as Therapeutic Target. Biol Psychiatry 81, 585–594. 10.1016/j.biopsych.2016.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faivre EJ, McDaniel KF, Albert DH, Mantena SR, Plotnik JP, Wilcox D, Zhang L, Bui MH, Sheppard GS, Wang L, Sehgal V, Lin X, Huang X, Lu X, Uziel T, Hessler P, Lam LT, Bellin RJ, Mehta G, Fidanze S, Pratt JK, Liu D, Hasvold LA, Sun C, Panchal SC, Nicolette JJ, Fossey SL, Park CH, Longenecker K, Bigelow L, Torrent M, Rosenberg SH, Kati WM, Shen Y, 2020. Selective inhibition of the BD2 bromodomain of BET proteins in prostate cancer. Nature 578, 306–310. 10.1038/s41586-020-1930-8 [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE, 2010. Selective inhibition of BET bromodomains. Nature 468, 1067–1073. 10.1038/nature09504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilan O, Rioja I, Knezevic K, Bell MJ, Yeung MM, Harker NR, Lam EYN, Chung C, Bamborough P, Petretich M, Urh M, Atkinson SJ, Bassil AK, Roberts EJ, Vassiliadis D, Burr ML, Preston AGS, Wellaway C, Werner T, Gray JR, Michon A-M, Gobbetti T, Kumar V, Soden PE, Haynes A, Vappiani J, Tough DF, Taylor S, Dawson S-J, Bantscheff M, Lindon M, Drewes G, Demont EH, Daniels DL, Grandi P, Prinjha RK, Dawson MA, 2020. Selective targeting of BD1 and BD2 of the BET proteins in cancer and immunoinflammation. Science 368, 387–394. 10.1126/science.aaz8455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilham D, Wasiak S, Tsujikawa LM, Halliday C, Norek K, Patel RG, Kulikowski E, Johansson J, Sweeney M, Wong NCW, Gordon A, McLure K, Young P, 2016. RVX-208, a BET-inhibitor for treating atherosclerotic cardiovascular disease, raises ApoA-I/HDL and represses pathways that contribute to cardiovascular disease. Atherosclerosis 247, 48–57. 10.1016/j.atherosclerosis.2016.01.036 [DOI] [PubMed] [Google Scholar]
- Guo W, Long H, Bu Q, Zhao Y, Wang H, Tian J, Cen X, 2020. Role of BRD4 phosphorylation in the nucleus accumbens in relapse to cocaine-seeking behavior in mice. Addict Biol 25, e12808. 10.1111/adb.12808 [DOI] [PubMed] [Google Scholar]
- Hope BT, 1998. Cocaine and the AP-1 transcription factor complex. Ann N Y Acad Sci 844, 1–6. [PubMed] [Google Scholar]
- Huang B, Yang X-D, Zhou M-M, Ozato K, Chen L-F, 2009. Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol Cell Biol 29, 1375–1387. 10.1128/MCB.01365-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Tabah J, Martin RD, Chen JJ, Tanny JC, Clarke PBS, Hébert TE, 2022. A role for BET proteins in regulating basal, dopamine-induced and cAMP/PKA-dependent transcription in rat striatal neurons. Cell Signal 91, 110226. 10.1016/j.cellsig.2021.110226 [DOI] [PubMed] [Google Scholar]
- Kennedy PJ, Feng J, Robison AJ, Maze I, Badimon A, Mouzon E, Chaudhury D, Damez-Werno DM, Haggarty SJ, Han M-H, Bassel-Duby R, Olson EN, Nestler EJ, 2013. Class I HDAC inhibition blocks cocaine-induced plasticity by targeted changes in histone methylation. Nat Neurosci 16, 434–440. 10.1038/nn.3354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korb E, Herre M, Zucker-Scharff I, Darnell RB, Allis CD, 2015. BET protein Brd4 activates transcription in neurons and BET inhibitor Jq1 blocks memory in mice. Nat Neurosci 18, 1464–1473. 10.1038/nn.4095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wolf ME, 2015. Multiple faces of BDNF in cocaine addiction. Behav Brain Res 279, 240–254. 10.1016/j.bbr.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Lu W, Luo C, 2020. A patent review of BRD4 inhibitors (2013–2019). Expert Opin Ther Pat 30, 57–81. 10.1080/13543776.2020.1702645 [DOI] [PubMed] [Google Scholar]
- Malvaez M, McQuown SC, Rogge GA, Astarabadi M, Jacques V, Carreiro S, Rusche JR, Wood MA, 2013. HDAC3-selective inhibitor enhances extinction of cocaine-seeking behavior in a persistent manner. Proc Natl Acad Sci U S A 110, 2647–2652. 10.1073/pnas.1213364110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvaez M, Mhillaj E, Matheos DP, Palmery M, Wood MA, 2011. CBP in the nucleus accumbens regulates cocaine-induced histone acetylation and is critical for cocaine-associated behaviors. J Neurosci 31, 16941–16948. 10.1523/JNEUROSCI.2747-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill E, 2010. RVX-208, a stimulator of apolipoprotein AI gene expression for the treatment of cardiovascular diseases. Curr Opin Investig Drugs 11, 357–364. [PubMed] [Google Scholar]
- Meberg PJ, Miller MW, 2003. Culturing hippocampal and cortical neurons. Methods Cell Biol 71, 111–127. 10.1016/s0091-679x(03)01007-0 [DOI] [PubMed] [Google Scholar]
- Morgan S, Grootendorst P, Lexchin J, Cunningham C, Greyson D, 2011. The cost of drug development: a systematic review. Health Policy 100, 4–17. 10.1016/j.healthpol.2010.12.002 [DOI] [PubMed] [Google Scholar]
- Nicholls SJ, Gordon A, Johansson J, Wolski K, Ballantyne CM, Kastelein JJP, Taylor A, Borgman M, Nissen SE, 2011. Efficacy and safety of a novel oral inducer of apolipoprotein a-I synthesis in statin-treated patients with stable coronary artery disease a randomized controlled trial. J Am Coll Cardiol 57, 1111–1119. 10.1016/j.jacc.2010.11.015 [DOI] [PubMed] [Google Scholar]
- Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung C-W, Chandwani R, Marazzi I, Wilson P, Coste H, White J, Kirilovsky J, Rice CM, Lora JM, Prinjha RK, Lee K, Tarakhovsky A, 2010. Suppression of inflammation by a synthetic histone mimic. Nature 468, 1119–1123. 10.1038/nature09589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolic D, Rizzo M, Mikhailidis DP, Wong NC, Banach M, 2015. An evaluation of RVX-208 for the treatment of atherosclerosis. Expert Opin Investig Drugs 24, 1389–1398. 10.1517/13543784.2015.1083010 [DOI] [PubMed] [Google Scholar]
- Penrod RD, Thomsen M, Taniguchi M, Guo Y, Cowan CW, Smith LN, 2020. The activity-regulated cytoskeleton-associated protein, Arc/Arg3.1, influences mouse cocaine self-administration. Pharmacol Biochem Behav 188, 172818. 10.1016/j.pbb.2019.172818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petretich M, Demont EH, Grandi P, 2020. Domain-selective targeting of BET proteins in cancer and immunological diseases. Curr Opin Chem Biol 57, 184–193. 10.1016/j.cbpa.2020.02.003 [DOI] [PubMed] [Google Scholar]
- Picaud S, Wells C, Felletar I, Brotherton D, Martin S, Savitsky P, Diez-Dacal B, Philpott M, Bountra C, Lingard H, Fedorov O, Müller S, Brennan PE, Knapp S, Filippakopoulos P, 2013. RVX-208, an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain. Proc Natl Acad Sci U S A 110, 19754–19759. 10.1073/pnas.1310658110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogge GA, Singh H, Dang R, Wood MA, 2013. HDAC3 is a negative regulator of cocaine-context-associated memory formation. J Neurosci 33, 6623–6632. 10.1523/JNEUROSCI.4472-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogge GA, Wood MA, 2013. The role of histone acetylation in cocaine-induced neural plasticity and behavior. Neuropsychopharmacology 38, 94–110. 10.1038/npp.2012.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salery M, Dos Santos M, Saint-Jour E, Moumné L, Pagès C, Kappès V, Parnaudeau S, Caboche J, Vanhoutte P, 2017. Activity-Regulated Cytoskeleton-Associated Protein Accumulates in the Nucleus in Response to Cocaine and Acts as a Brake on Chromatin Remodeling and Long-Term Behavioral Alterations. Biol Psychiatry 81, 573–584. 10.1016/j.biopsych.2016.05.025 [DOI] [PubMed] [Google Scholar]
- Sartor GC, 2019. Epigenetic pharmacotherapy for substance use disorder. Biochemical Pharmacology 168, 269–274. 10.1016/j.bcp.2019.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor GC, Aston-Jones GS, 2012. A septal-hypothalamic pathway drives orexin neurons, which is necessary for conditioned cocaine preference. J Neurosci 32, 4623–4631. 10.1523/JNEUROSCI.4561-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor GC, Malvezzi AM, Kumar A, Andrade NS, Wiedner HJ, Vilca SJ, Janczura KJ, Bagheri A, Al-Ali H, Powell SK, Brown PT, Volmar CH, Foster TC, Zeier Z, Wahlestedt C, 2019. Enhancement of BDNF Expression and Memory by HDAC Inhibition Requires BET Bromodomain Reader Proteins. J Neurosci 39, 612–626. 10.1523/JNEUROSCI.1604-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor GC, Powell SK, Brothers SP, Wahlestedt C, 2015. Epigenetic Readers of Lysine Acetylation Regulate Cocaine-Induced Plasticity. J Neurosci 35, 15062–15072. 10.1523/JNEUROSCI.0826-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh MB, Sartor GC, 2020. BET bromodomains as novel epigenetic targets for brain health and disease. Neuropharmacology 181, 108306. 10.1016/j.neuropharm.2020.108306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Gu X, Zhang Z, Li W, Wang X, 2020. Retinoic acid receptor gamma is targeted by microRNA-124 and inhibits neurite outgrowth. Neuropharmacology 163, 107657. 10.1016/j.neuropharm.2019.05.034 [DOI] [PubMed] [Google Scholar]
- Sullivan JM, Badimon A, Schaefer U, Ayata P, Gray J, Chung C, von Schimmelmann M, Zhang F, Garton N, Smithers N, Lewis H, Tarakhovsky A, Prinjha RK, Schaefer A, 2015. Autism-like syndrome is induced by pharmacological suppression of BET proteins in young mice. J Exp Med 212, 1771–1781. 10.1084/jem.20151271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Roberts JM, Seo H-S, Souza A, Paulk J, Scott TG, DeAngelo SL, Dhe-Paganon S, Bradner JE, 2016. Design and characterization of bivalent BET inhibitors. Nature Chemical Biology 12, 1089–1096. 10.1038/nchembio.2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujikawa LM, Fu L, Das S, Halliday C, Rakai BD, Stotz SC, Sarsons CD, Gilham D, Daze E, Wasiak S, Studer D, Rinker KD, Sweeney M, Johansson JO, Wong NCW, Kulikowski E, 2019. Apabetalone (RVX-208) reduces vascular inflammation in vitro and in CVD patients by a BET-dependent epigenetic mechanism. Clin Epigenetics 11, 102. 10.1186/s13148-019-0696-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DM, Cates HM, Loh Y-HE, Purushothaman I, Ramakrishnan A, Cahill KM, Lardner CK, Godino A, Kronman HG, Rabkin J, Lorsch ZS, Mews P, Doyle MA, Feng J, Labonté B, Koo JW, Bagot RC, Logan RW, Seney ML, Calipari ES, Shen L, Nestler EJ, 2018. Cocaine Self-administration Alters Transcriptome-wide Responses in the Brain’s Reward Circuitry. Biological Psychiatry, Cocaine Addiction, Obsessive-Compulsive Disorder, and Corticostriatal Plasticity 84, 867–880. 10.1016/j.biopsych.2018.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasiak S, Dzobo KE, Rakai BD, Kaiser Y, Versloot M, Bahjat M, Stotz SC, Fu L, Sweeney M, Johansson JO, Wong NCW, Stroes ESG, Kroon J, Kulikowski E, 2020. BET protein inhibitor apabetalone (RVX-208) suppresses pro-inflammatory hyper-activation of monocytes from patients with cardiovascular disease and type 2 diabetes. Clin Epigenetics 12, 166. 10.1186/s13148-020-00943-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Heidenreich D, Zhou S, Ackloo S, Krämer A, Nakka K, Lima-Fernandes E, Deblois G, Duan S, Vellanki RN, Li F, Vedadi M, Dilworth J, Lupien M, Brennan PE, Arrowsmith CH, Müller S, Fedorov O, Filippakopoulos P, Knapp S, 2019. A chemical toolbox for the study of bromodomains and epigenetic signaling. Nat Commun 10, 1915. 10.1038/s41467-019-09672-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S-Y, Lee A-Y, Lai H-T, Zhang H, Chiang C-M, 2013. Phospho Switch Triggers Brd4 Chromatin Binding and Activator Recruitment for Gene-Specific Targeting. Molecular Cell 49, 843–857. 10.1016/j.molcel.2012.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, 2008. c-Fos is an intracellular regulator of cocaine-induced long-term changes. Ann N Y Acad Sci 1139, 1–9. 10.1196/annals.1432.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaware N, Zhou M-M, 2017. Chemical modulators for epigenome reader domains as emerging epigenetic therapies for cancer and inflammation. Curr Opin Chem Biol 39, 116–125. 10.1016/j.cbpa.2017.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]


