Abstract
Overarching themes in the terminal differentiation of the enteric nervous system, an autonomously acting unit of animal nervous systems, have so far eluded discovery. We describe here the overall regulatory logic of enteric nervous system differentiation of the nematode Caenorhabditis elegans that resides within the foregut (pharynx) of the worm. A C. elegans homolog of the Drosophila Sine oculis homeobox gene, ceh-34, is expressed in all 14 classes of interconnected pharyngeal neurons from their birth throughout their life time, but in no other neuron type of the entire animal. Constitutive and temporally controlled ceh-34 removal shows that ceh-34 is required to initiate and maintain the neuron type-specific terminal differentiation program of all pharyngeal neuron classes, including their circuit assembly. Through additional genetic loss of function analysis, we show that within each pharyngeal neuron class, ceh-34 cooperates with different homeodomain transcription factors to individuate distinct pharyngeal neuron classes. Our analysis underscores the critical role of homeobox genes in neuronal identity specification and links them to the control of neuronal circuit assembly of the enteric nervous system. Together with the pharyngeal nervous system simplicity as well as its specification by a Sine oculis homolog, our findings invite speculations about the early evolution of nervous systems.
Research organism: C. elegans
Introduction
Across animal phylogeny, enteric nervous systems constitute a self-contained, autonomously acting neuronal network that detects physiological conditions to control the peristaltic movement of food through the digestive tract (Ayali, 2004; Copenhaver, 2007; Fung and Vanden Berghe, 2020; Hartenstein, 1997; Laranjeira and Pachnis, 2009). Because of structural and functional autonomy, the enteric nervous system has been referred to as a ‘second brain’ (Gershon, 1998). In mammals, the enteric nervous system is composed of around 20 different neuron types, categorized into intrinsic sensory, inter- or motor neurons, which line the interior lumen of different sections of the digestive system (Drokhlyansky et al., 2020; Furness, 2000). While progress has been made in understanding early developmental patterning events that establish the fate of neurons in the enteric nervous system of mammals (Nagy and Goldstein, 2017; Sasselli et al., 2012), fish (Ganz, 2018), and flies (Copenhaver, 2007; Myers et al., 2018), much less is known about terminal differentiation programs of enteric neurons, both in vertebrate and invertebrate models (Copenhaver, 2007; Hao and Young, 2009; Memic et al., 2018; Morarach et al., 2021; Rao and Gershon, 2018). Specifically, it has remained unclear as to whether there are common unifying themes in how enteric neurons acquire their terminally differentiated state. This is particularly interesting from an evolutionary standpoint. The function of enteric neurons in controlling feeding behavior is an ancient one that may precede the evolution of the bilaterian central nervous system (Cook et al., 2020; Furness and Stebbing, 2018; Gilbert, 2019; Koizumi, 2007). Understanding how enteric neurons acquire their terminal features may therefore provide novel insights into nervous system evolution.
The nematode Caenorhabditis elegans contains an autonomously acting nervous system in its foregut, the pharynx, composed of 20 synaptically interconnected neurons that fall into 14 anatomically distinct classes (Albertson and Thomson, 1976; Cook et al., 2020; Mango, 2007; Figure 1). Due to its association with the digestive tract of the worm, the pharyngeal nervous system can be considered to be the enteric nervous system of C. elegans. Apart from this anatomical association, the pharyngeal nervous system shares functional features of enteric nervous systems of more complex animals. It is required for movement of food through the digestive tract of the worm and functions in an entirely autonomous manner, even if removed from the rest of the animal (Albertson and Thomson, 1976; Avery, 2012; Cook et al., 2020; Mango, 2007). Like other enteric nervous systems, the nematode pharyngeal nervous system constitutes a non-centralized neuronal network isolated from the rest of the nervous system and, in rough analogy to the vagus nerve, is connected to the remainder of the nervous system through a single nerve fiber, that of the bilateral RIP neuron pair (Albertson and Thomson, 1976; Cook et al., 2020; Cook et al., 2019; White et al., 1986). Like vertebrate enteric neurons (Fung and Vanden Berghe, 2020), pharyngeal neurons have sensory, inter- and motorneuron function (Avery and Horvitz, 1989; Cook et al., 2020; Trojanowski et al., 2014).
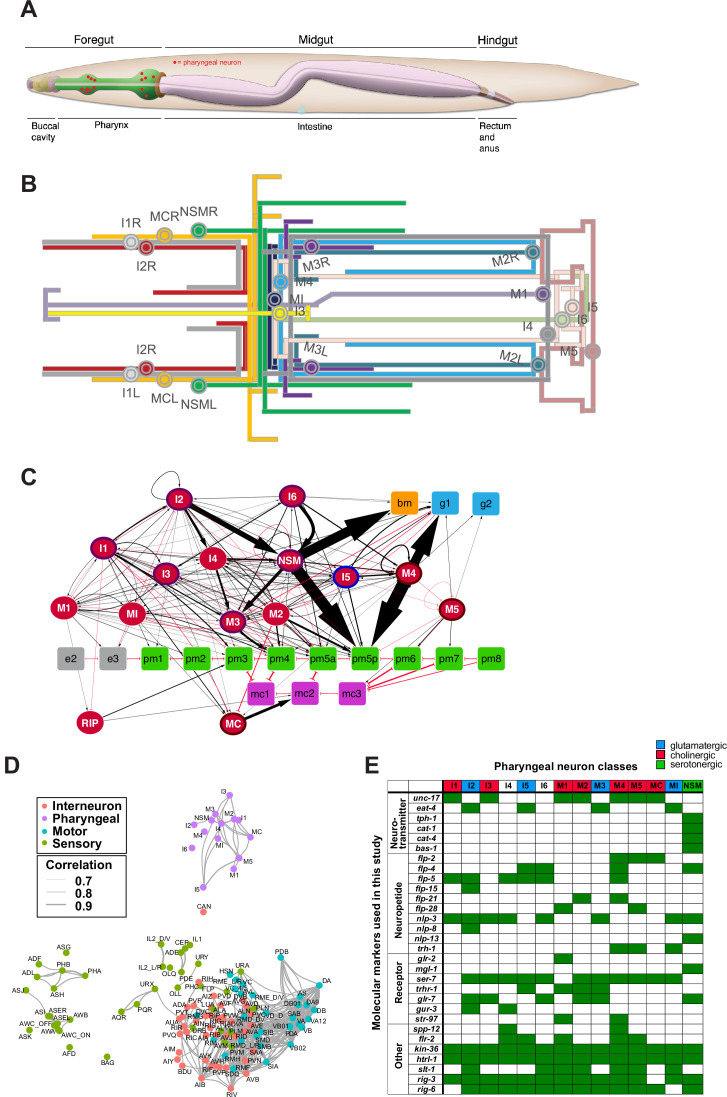
Figure 1. The pharyngeal nervous system of Caenorhabditis elegans.
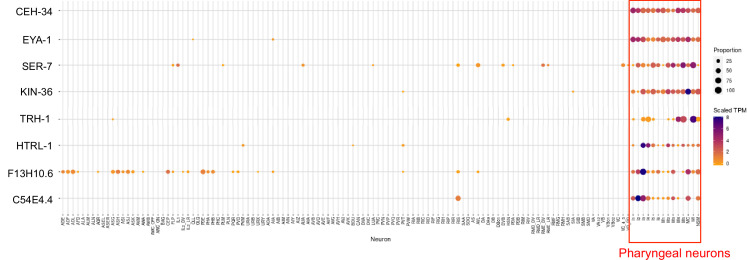
(A) Overview of the C. elegans alimentary system from Wormbook (Hall and Altun, 2007), with neuronal cell bodies in the pharynx added in red. (B) Projection patterns of pharyngeal neurons within the pharynx displayed in the format of a subway map (kindly provided by SJ Cook). (C) Full connectome of pharyngeal nervous system, adapted from Cook et al., 2020. Square nodes are end organs, including muscle (green), marginal cells (fuchsia), gland cells (blue), epithelial cells (gray), and basement membrane (orange). Neurons are red ellipses. Neurons with outlines have either apical (purple), unexposed (brown), or embedded (blue) sensory endings. Directed chemical edges and undirected gap junction edges are represented by black arrows and red lines, respectively. The line width is proportional to the anatomical strength of that connection (# serial sections). The pharyngeal nervous system is connected to the rest of the nervous system through a single neuron pair (RIP). (D) Single cell transcriptome similarity between neuron types classes with widths of edges indicating strengths of similarity (Pearson correlation coefficients > 0.7), showing that pharyngeal neurons are more similar to each other than to other neurons in the C. elegans nervous system. Reproduced from Taylor et al., 2021. (E) Molecular markers used in this study for cell fate analysis. See Supplementary file 1 for information on reporter constructs.
© 2020, John Wily and Sons
Panel C is adapted from Cook et al., 2020 with permission from John Wily and Sons. It is not covered by the CC-BY 4.0 licence and further reproduction of this panel would need permission from the copyright holder.
© 2021, Elsevier
Panel D is reproduced from Figure 2i in Taylor et al., 2021 with permission from Elsevier. It is not covered by the CC-BY 4.0 licence and further reproduction of this panel would need permission from the copyright holder.
Our recent re-analysis of pharyngeal nervous system anatomy has shown that rather than segregating these functions over distinct neurons as vertebrates do (Fung and Vanden Berghe, 2020), most pharyngeal neurons each combine sensory, inter- and motorneuron function, that is, are polymodal (Cook et al., 2020). The 14 distinct pharyngeal neuron types are defined by their unique anatomy, that is, axonal projections, morphology, synaptic connectivity (Figure 1B and C; Cook et al., 2020), and unique functional features (Avery and Horvitz, 1989; Trojanowski et al., 2014). This anatomical and functional classification has recently been further extended by the description of their unique molecular fingerprint, determined by scRNA transcriptomic analysis (Taylor et al., 2021; Figure 1D). For example, each pharyngeal neuron class is uniquely defined by characteristic signatures of neurotransmitter systems, from acetylcholine (ACh), glutamate (Glu) to serotonin, and by unique combinations of neuropeptide-encoding genes (Figure 1E). While pharyngeal neurons are clearly different from one another, scRNA profiling has shown that their molecular signatures are more similar to each other than to other neurons in the nervous system (Taylor et al., 2021; Figure 1D).
One fascinating aspect of the C. elegans pharyngeal nervous system is that it has an appearance of what one could imagine an ancestral, primitive nervous system to have looked like. Primitive nervous systems are generally thought to have emerged in the context of monolayers of epithelial cells, with individual cells in such layers specializing into primitive sensory motor-type neurons (Arendt, 2008; Mackie, 1970; Varoqueaux and Fasshauer, 2017). These diffusely organized neurons may have sensed the environment and relayed such sensory information to the other primitive cell type thought to have arisen early in evolution, namely, contractile ‘myoepithelial’ cells that were able to generate motion. The organization of the C. elegans pharynx reveals some striking parallels to such a presumptive primitive nervous system: It is also essentially a single monolayer of cells that is organized into a tubular structure and the vast majority of constituent cells are myoepithelial cells (pharyngeal muscle) and polymodal, interconnected sensory/motor neurons (Cook et al., 2020; Mango, 2007; Portereiko and Mango, 2001). Pharyngeal neurons combine sensory, inter- and motor neuron features and are also not localized to ganglia but rather diffusely localized, resembling the architecture of more ancient nerve nets (Albertson and Thomson, 1976; Watanabe et al., 2009). The interconnectivity of pharyngeal neurons also displays less selectivity than non-pharyngeal neurons, such that mere physical proximity is an almost sufficient criterion for connectivity (Cook et al., 2020). Moreover, pharyngeal neurons are closely related to non-neuronal pharyngeal cells by lineage; for example, some muscle and neurons derive from a common mother cell (Sulston et al., 1983). The idea of enteric neurons being reflective of an early, primitive state of the nervous system has also been brought forward in the context of comparing enteric nervous systems from widely divergent species (Furness and Stebbing, 2018; Gilbert, 2019). Specifically, these authors argued that the nerve net-like hydra nervous system displays features of the vertebrate enteric nervous system.
The self-contained and hypothetically primitive state of the C. elegans enteric nervous system, with all its unique features, encouraged us to use this system as a model to probe several concepts of neuronal identity specification that have emerged from the centralized, non-pharyngeal nervous system of C. elegans: (1) The first is the concept of terminal selectors, transcription factors that act in a master-regulatory manner to coordinately control the many identity features of a terminally differentiating neuron (Hobert, 2016). Are members of terminal gene batteries in each pharyngeal neuron also controlled in a coordinated manner, via terminal selectors? One study in the NSM neurons provided some limited evidence in this regard (Zhang et al., 2014), but how broadly this applies throughout the pharyngeal nervous system was less clear. (2) Second, homeodomain transcription factors have a predominant role as terminal selectors of neuronal identity in the non-pharyngeal nervous system (Hobert, 2021; Reilly et al., 2020). A recent cataloguing of the expression patterns of all homeodomain proteins in the C. elegans genome revealed that each one of the C. elegans’ 118 neuron classes, including the pharyngeal neurons, display a unique combinatorial signature of homeodomain protein expression (Reilly et al., 2020). Are all pharyngeal neurons indeed specified by homeodomain protein combinations? (3) Lastly, there is evidence from both C. elegans (Berghoff et al., 2021; Pereira et al., 2015) and other systems (Brunet and Pattyn, 2002) that synaptically connected neurons are often specified by the same transcription factor, suggesting that such transcription factors may be involved in assembling neurons in functional circuitry. We have termed such transcription factors ‘circuit organizers’ (Berghoff et al., 2021; Pereira et al., 2015) and sought to test whether the isolated pharyngeal circuitry is similarly specified by a circuit organizer transcription factor.
In this paper, we show that all three predictions are fulfilled in the context of the nematode’s pharyngeal/enteric nervous system. We show that a C. elegans ortholog of the Sine oculis/Six1/Six2 homeobox gene, ceh-34, is expressed in all pharyngeal neurons from their birth throughout their life time. Its expression is induced by the foregut organ selector gene pha-4/FoxA. We demonstrate that ceh-34 initiates and maintains the terminally differentiated state of all synaptically connected pharyngeal neurons, that ceh-34 is required to assemble and maintain pharyngeal neuron architecture and that in distinct pharyngeal neuron types, ceh-34 cooperates with distinct homeobox genes to specify and maintain their respective identity. Taken together, our studies further substantiate overarching themes of nervous system development and potentially provide insights into the evolution of nervous systems.
Results
Expression of paralogous genes ceh-34 and ceh-33, the two C. elegans Sine oculis/Six1/2 orthologs
Genome sequence mining revealed several C. elegans homologs of the Sine oculis/Six family of homeodomain proteins (Ruvkun and Hobert, 1998) whose founding member was first identified in Drosophila for its role in eye patterning (Cheyette et al., 1994). This specific homeodomain transcription factor family is characterized by the presence of a conserved domain, located N-terminally to the DNA binding homeodomain, the ~150 amino acid-long SIX domain, involved in both protein-DNA, as well as protein-protein interactions (Kumar, 2009; Patrick et al., 2013). C. elegans Six-type homeodomain proteins fall into several, phylogenetically conserved families, the Sine oculis/Six1/2, the Six4/5, and the Six3/6 subfamily (Dozier et al., 2001; Kumar, 2009). We focus here on the Sine oculis subfamily.
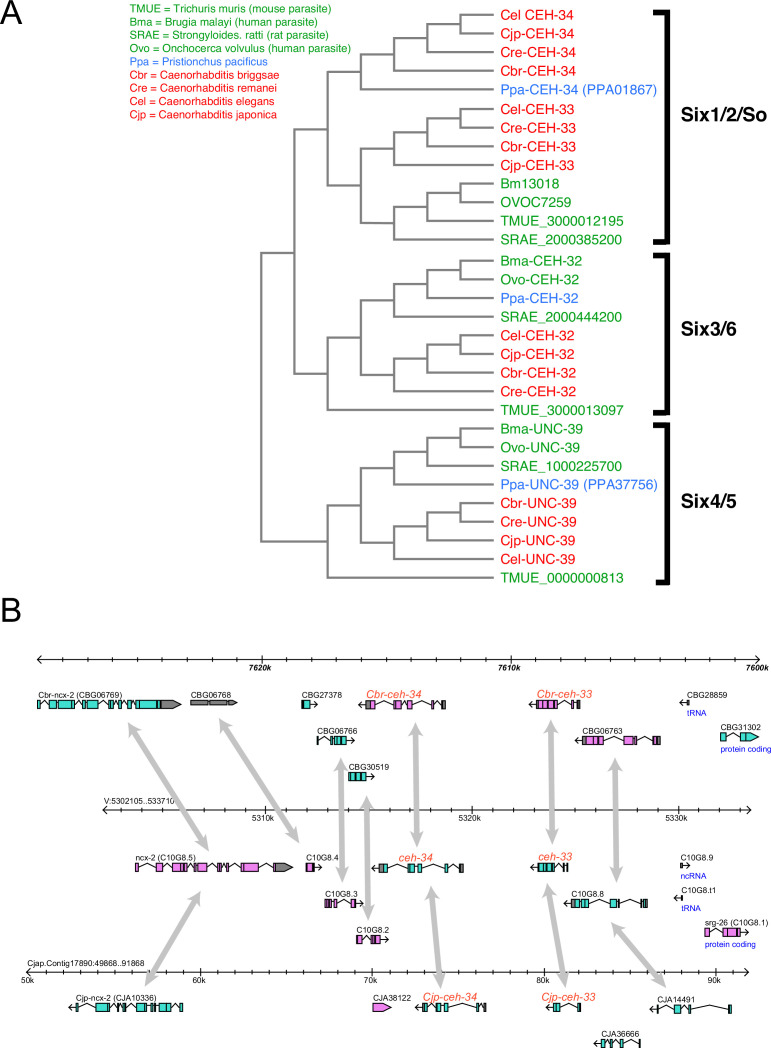
Through the analysis of multiple nematode genome sequences, we found that nematodes generally contain a single ortholog of the Sine oculis/Six1/2 subfamily of SIX homeodomain proteins, but that this locus has duplicated in the Caenorhabditis genus into two immediately adjacent paralogs, ceh-33 and ceh-34 (Figure 2, Figure 2—figure supplement 1). Using a fosmid-based reporter in which the ceh-33 locus is tagged with gfp, we found that the CEH-33 protein shows no expression in the nervous system within or outside the pharynx at any developmental stage. The only observed expression was in a subset of head muscle cells (Figure 2B).
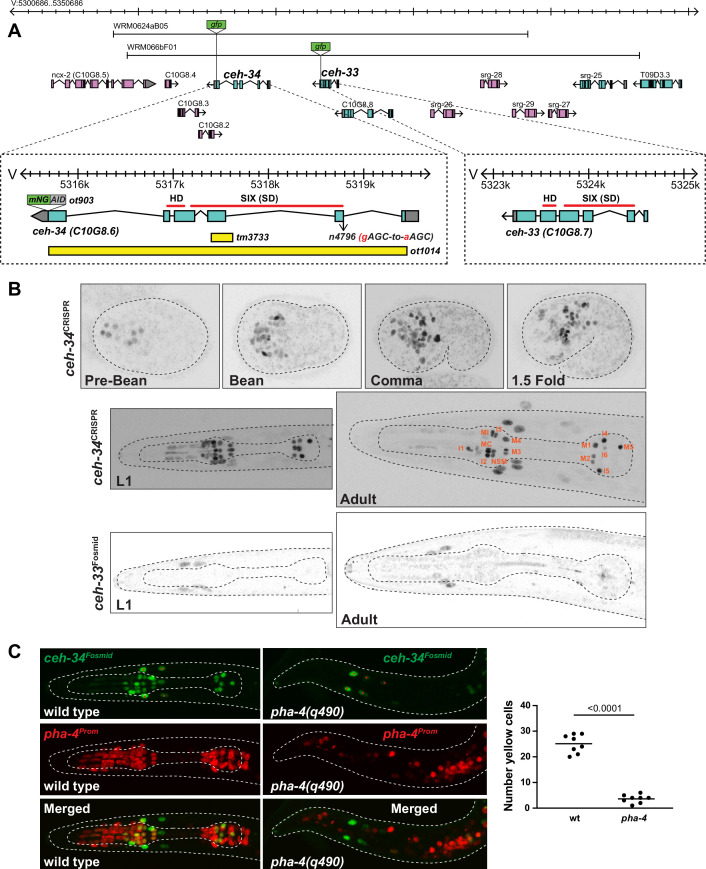
Figure 2. ceh-34 is expressed in all pharyngeal neurons.
(A) ceh-33 and ceh-34 loci showing different alleles and fosmid reporters used in this study. (B) Expression of the ceh-34 CRISPR/Cas-9-engineered reporter allele ot903 over the course of development. ceh-33 fosmid reporter (wgIs575) shows expression in a subset of head muscle cells. (C) Pharynx organ selector pha-4 controls ceh-34 expression (as analyzed with the wgIs524 transgene). Animals were scored at the L1 stage. Presumptive ‘pharyngeal cells’ in pha-4 mutant are marked with a red pha-4 promoter fusion (stIs10077). Cells co-expressing ceh-34 and pha-4 were counted (yellow cells). ceh-34 expression in head muscle cells, marked with red asterisk, is not affected since they do not express pha-4.
Figure 2—figure supplement 1. Evolution of Sine oculis orthologs in nematodes.
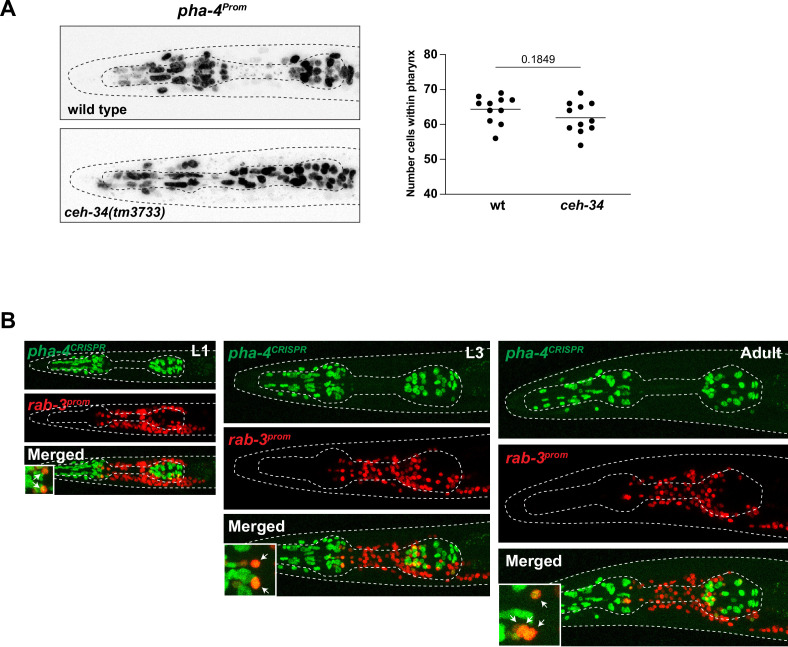
Figure 2—figure supplement 2. Expression of the organ selector pha-4.
A previously described reporter construct that contains the coding region and 3.8 kb of promoter region of the neighboring ceh-34 gene showed expression in all pharyngeal neurons (Hirose et al., 2010). A fosmid-based reporter shows the same expression pattern (Reilly et al., 2020). To further confirm this strikingly selective pattern of neuronal expression and also to examine expression at different developmental stages with the best possible reagent, we used the CRISPR/Cas9 system to engineer a reporter allele of ceh-34. This reporter shows the same expression as the fosmid-based reporter in all pharyngeal neurons, but no other neurons (Figure 2B). The ceh-34 reporter is turned on in the embryo at around the time of birth of pharyngeal neurons and is maintained in all pharyngeal neurons throughout all larval and adult stages (Figure 2B).
The remarkable restriction of ceh-34 expression within the nervous system to all pharyngeal neurons prompted us to ask whether the Forkhead transcription factor pha-4, an organ selector gene involved in early patterning of the pharynx (Gaudet and Mango, 2002; Horner et al., 1998; Kalb et al., 1998; Mango et al., 1994), is required for ceh-34 expression. Crossing a ceh-34 reporter into pha-4(q490) mutant animals, we indeed observed a loss of ceh-34 expression (Figure 2C). Conversely, ceh-34 does not affect pha-4 expression (Figure 2—figure supplement 2A). The regulation of ceh-34 by pha-4 mirrors the effects that pha-4 has on the expression of other transcription factors that control terminal differentiation of other tissue types in the pharynx, for example, the ceh-22/NKX homeobox gene that specifies pharyngeal muscle differentiation (Vilimas et al., 2004), or the hlh-6 bHLH gene that specifies pharyngeal gland differentiation (Smit et al., 2008). We furthermore note that a pha-4 reporter allele that we generated through CRISPR/Cas9 genome engineering is continuously expressed throughout the entire pharyngeal nervous system, at all postembryonic stages (Figure 2—figure supplement 2B), raising the possibility that pha-4 may not only initiate, but also maintain ceh-34 expression.
ceh-34 controls the expression of diverse neurotransmitter signaling pathways in pharyngeal neurons
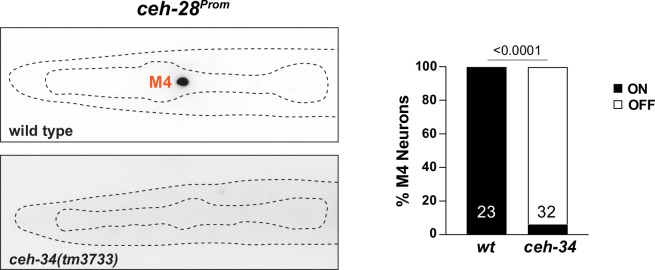
To begin to assess the function of ceh-34 in enteric nervous system differentiation, we used CRISPR/Cas9 engineering to generate a null allele of the ceh-34 locus, ot1014, in which the entire ceh-34 locus is deleted (Figure 2A). Previously described ceh-34 alleles include a hypomorphic splice site allele, n4796, and a small deletion allele, tm3733 (Amin et al., 2009; Hirose et al., 2010). In our ensuing mutant analysis, we found ot1014 to be phenotypically indistinguishable from the tm3733 deletion allele and we therefore used both alleles interchangeably. Both the ot1014 and tm3733 alleles result in a completely penetrant early larval arrest phenotype, as expected from loss of pharynx function and resulting inability to feed.
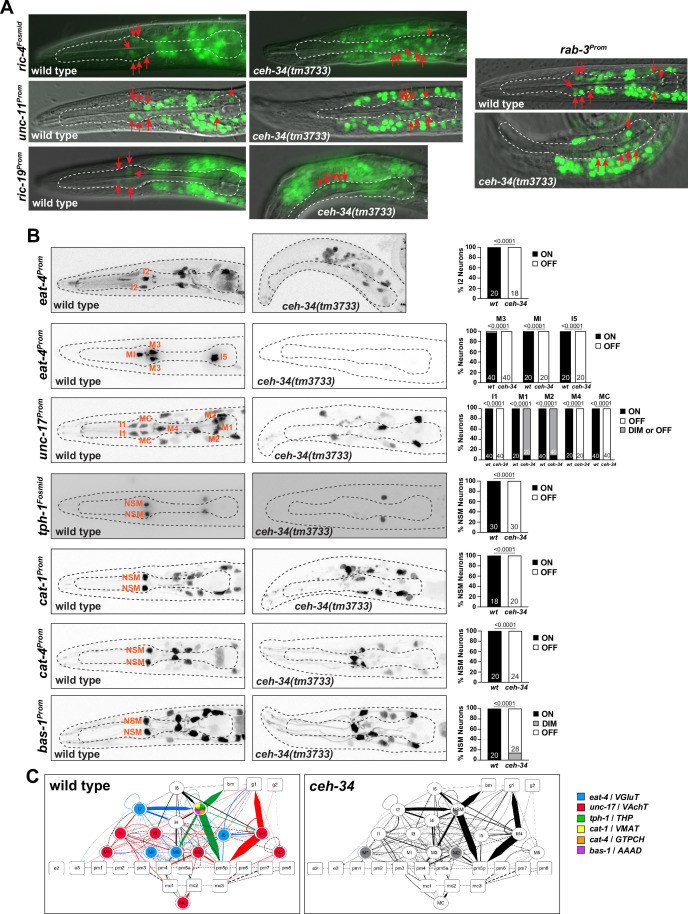
We first asked whether ceh-34 is required for the generation of pharyngeal neurons. Using a pha-4 reporter that labels all pharyngeal cells, we observed no obvious differences in the number of pharyngeal cells in ceh-34 mutants (Figure 2—figure supplement 2A). Moreover, the expression of the pan-neuronal genes ric-4/SNAP25, ric-19/ICA1, rab-3/RAB3, and unc-11/AP180 is unaffected (Figure 3A), indicating that pharyngeal neurons are generated and properly execute a generic neuronal differentiation program.
Figure 3. Pharyngeal neurons are generated in ceh-34 mutants but lose their neurotransmitter identity.
(A) Pictures at the L1 stage showing expression of pan-neuronal reporter transgenes that monitor ric-4 (otIs350), ric-19 (otIs380), unc-11 (otIs620), and rab-3 (otIs291) expression. A single focal plane with a subset of pharyngeal neurons marked with red arrows is shown for clarity. (B) ceh-34 affects the expression of neurotransmitter identity genes. Glutamatergic, cholinergic, and serotonergic identity is lost. Reporter transgenes used are eat-4 (otIs487, otIs558), unc-17 (otIs661), tph-1 (otIs517), cat-1 (otIs221), cat-4 (otIs225), and bas-1 (otIs226). Animals were scored at the L1 stage. Statistical analysis was performed using Fisher’s exact test or chi-square test. N is indicated within each bar and represents number of neurons scored. (C) Circuit diagram summarizing the effect of ceh-34 on neurotransmitter identity. Nodes are colored to illustrate neurotransmitter identity gene expression. Nodes lose coloring when expression is affected in ceh-34 mutants (gray indicates partial effect). Edges are colored if the source neuron expresses either eat-4 (glutamatergic), unc-17 (cholinergic), or tph-1 (serotonergic). Edges lose coloring when expression of these genes is affected in the source neuron in ceh-34 mutants (irrespective of whether the effect is partial or total). Note that in this and ensuing circuit diagrams, the existence of gray edges does not indicate whether those edges are generated properly in ceh-34 mutants. Directed edges (arrows) represent chemical synapses. Undirected edges (dashed lines) represent electrical synapses. The width of the edge is proportional to the weight of the connection (the number of serial section electron micrographs where a synaptic specialization is observed).
We next assessed the effect of ceh-34 on a large collection of neuron type-specific molecular identity features (illustrated in Figure 1E). To this end, we first focused on the signaling capacities of all pharyngeal neurons. In the vertebrate enteric nervous system, each individual neuron class is distinguished by its unique set of signaling molecules, from classic neurotransmitters to neuropeptides (Morarach et al., 2021). The same applies to all neuron classes in the pharyngeal/enteric nervous system of C. elegans (Horvitz et al., 1982; Pereira et al., 2015; Serrano-Saiz et al., 2013; Taylor et al., 2021). We first considered the three classic neurotransmitters ACh, Glu, and serotonin which are employed in C. elegans much like in the enteric nervous system of other species: 7 of the 14 pharyngeal neuron classes use ACh as neurotransmitter, 4 use Glu, and 1 uses serotonin (NSM) (Horvitz et al., 1982; Pereira et al., 2015; Serrano-Saiz et al., 2013). Of those neurotransmitters, serotonin is perhaps the best studied neurotransmitter both in the vertebrate enteric nervous system (Gershon, 2013) and in the nematode pharyngeal nervous system (Horvitz et al., 1982; Ishita et al., 2020; Song and Avery, 2013). Acquisition of ACh, Glu, and serotonin neurotransmitter identity features can be visualized through the expression of a number of enzymes and transporters: unc-17/VAChT for cholinergic identity, eat-4/VGluT for glutamatergic identity, and tph-1/TPH, cat-1/VMAT, bas-1/AAAD, and cat-4/GCH for serotonergic identity. We examined the expression of all these markers, using various reporter genes, in all pharyngeal neurons of ceh-34 mutant animals and found that the seven cholinergic, four glutamatergic, and single serotonergic neuron classes fail to acquire their respective neurotransmitter identity (Figure 3B). We schematize these results in the context of a pharyngeal circuit diagram to illustrate the systemic nature of neurotransmission defects in ceh-34 mutants (Figure 3C).
We note that while in most cases, the effect of ceh-34 on expression of neurotransmitter systems (as well as other identity markers described in ensuing sections) is fully penetrant and fully expressive, in some cases reporter expression is diminished, but not completely eliminated. At the end of this paper, we describe cofactors for ceh-34 which may be partially able to compensate for loss of ceh-34 function.
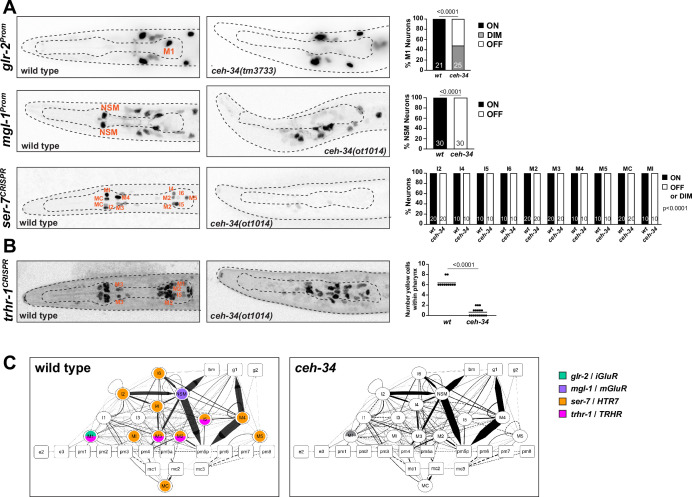
As in vertebrate enteric nervous systems (Drokhlyansky et al., 2020), pharyngeal neurons also display highly patterned expression of various neurotransmitter receptors (Taylor et al., 2021). We analyzed the ceh-34-dependence of three representative receptors, the serotonin receptor ser-7, the ortholog of vertebrate HTR7, as well as a metabotropic and an ionotropic Glu receptor, mgl-1 and glr-2. Each of these receptors is expressed in specific subsets of pharyngeal neurons and ser-7 is known to control feeding behavior (Hobson et al., 2006; Song and Avery, 2012; Song and Avery, 2013). Using a combination of reporter transgenes and CRISPR/Cas9-engineered gfp reporter alleles, we found that the expression of mgl-1, glr-2, and ser-7 is strongly affected in different pharyngeal neuron types, if not entirely abrogated upon removal of ceh-34 (Figure 4A–C).
Figure 4. ceh-34 affects the expression of receptors for neurotransmitters and neuropeptides.
(A) Representative pictures and quantification showing neurotransmitter receptor expression loss in ceh-34 mutants. Reporter genes used are transgenes glr-2 (ivIs26), mgl-1 (otIs341), and a CRISPR/Cas9-engineered reporter allele for ser-7 (syb4502). Animals were scored at the L1 stage. Statistical analysis was performed using Fisher’s exact test or chi-square test. N is indicated within each bar and represents number of neurons scored. (B) Representative pictures and quantification showing neuropeptide receptor expression loss in ceh-34 mutants. Reporter gene used is the CRISPR/Cas9-engineered reporter allele trhr-1 (syb4453). Animals were scored at the L1 stage, with a red pan-neuronal marker (otIs355) in the background to facilitate scoring. Number of “yellow“ cells (overlap of red pan-neuronal marker and green reporter) within the pharynx were counted. Statistical analysis was performed using unpaired t-test. (C) Circuit diagram summarizing the effect of ceh-34 on neurotransmitter and neuropeptide receptor expression. Nodes lose coloring when expression is affected in ceh-34 mutants (gray indicates partial effect; ser-7 and trhr-1 are colored white in all neurons since identity of neurons with partial effect is not known). See legend to Figure 3 for more information on circuit diagram features.
ceh-34 controls diverse neuropeptidergic identities of pharyngeal neurons
The function of vertebrate enteric nervous systems is modulated by a number of prominent neuropeptidergic signaling systems (Abot et al., 2018; Llewellyn-Smith, 1989). In some cases, both neuropeptide and receptor are expressed in the vertebrate enteric nervous system, while in others, the peptide is produced elsewhere but acts on neuropeptide receptors located in the enteric nervous system. Likewise, the C. elegans pharyngeal nervous system expresses a great diversity of neuropeptides and neuropeptide receptors, including homologs of neuropeptide signaling systems that function in the vertebrate enteric nervous system (Taylor et al., 2021). In fact, each pharyngeal neuron expresses a unique combination of neuropeptides and their receptor proteins (Taylor et al., 2021; Figure 5).
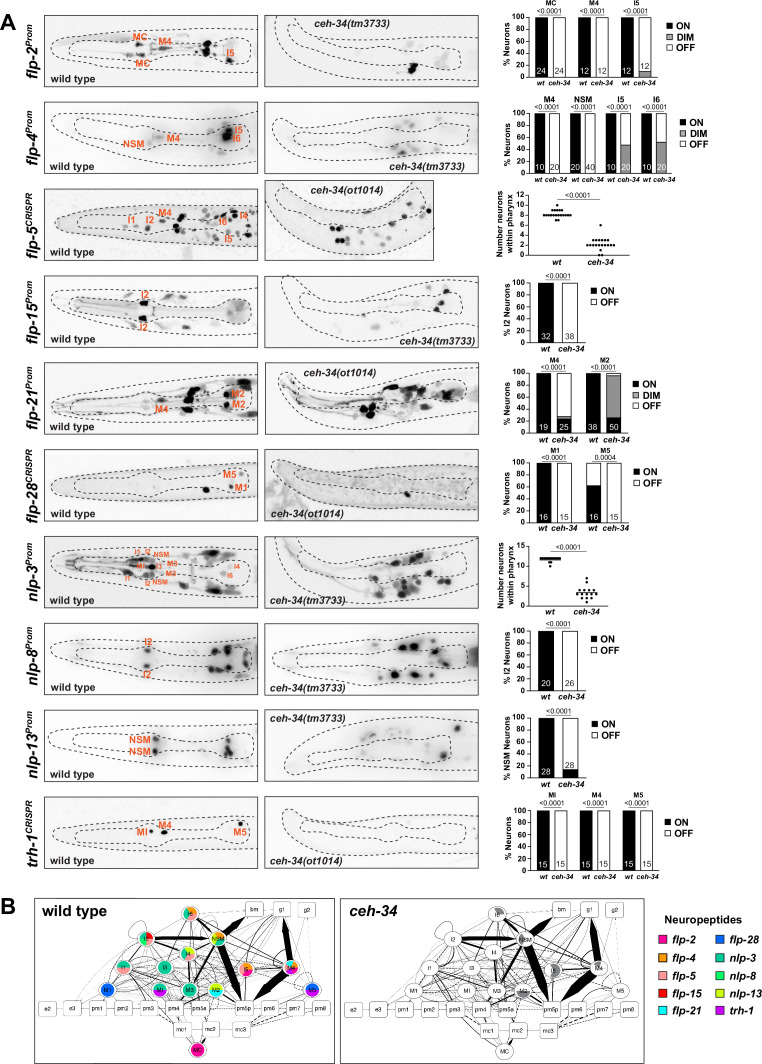
Figure 5. ceh-34 affects neuropeptidergic identity of pharyngeal neurons.
(A) Representative pictures and quantification showing expression of 10 different neuropeptides is affected in ceh-34 mutants. Reporter genes used are transgenic reporters for flp-2 (ynIs57), flp-4 (ynIs30), flp-15 (ynIs45), flp-21 (ynIs80), nlp-3 (otIs695), nlp-8 (otIs711), and nlp-13 (otIs742) and CRISPR/Cas9-engineered reporter alleles for flp-5 (syb4513), flp-28 (syb3207), and trh-1 (syb4421). Animals were scored at the L1 stage. Statistical analysis was performed using Fisher’s exact test, chi-square test, or unpaired t-test. N is indicated within each bar and represents number of neurons scored. (B) Circuit diagram summarizing the effect of ceh-34 on neuropeptide expression. Nodes lose coloring when neuropeptide expression is affected in ceh-34 mutants (gray indicates partial effect; flp-5 and nlp-3 are colored white in all neurons since identity of neurons with partial effect is not known). See legend to Figure 3 for more information on circuit diagram features.
To test whether ceh-34 affects the neuron type-specific expression of various neuropeptidergic signaling systems, we first examined the expression of the phylogenetically conservedthyrotropin-releasing hormone (TRH) signaling axis that is important in stimulating gastrointestinal motility in vertebrates (Abot et al., 2018). C. elegans homologs of either the thyrotropin-releasing hormone (TRH-1) or its receptor (TRHR-1) are expressed in the pharyngeal nervous system (Hunt-Newbury et al., 2007; Van Sinay et al., 2017), a notion we confirmed and extended with CRISPR/Cas9-engineered reporter alleles, showing that trh-1 is expressed in the MI, M4, and M5 neurons and trhr-1 in the M1, M2, M3, and I5 neurons (Figure 4 and Figure 5). We found that the expression of the trh-1 and trhr-1 reporter alleles in these pharyngeal neurons is strongly affected in ceh-34 mutants (Figure 4 and Figure 5A).
In addition to this deeply conserved neuropeptidergic system, we also tested the expression of a cohort of neuropeptides from the FMRFamide family (flp-2, flp-4, flp-5, flp-15, flp-21, flp-28) and other miscellaneous neuropeptides (nlp-3, nlp-8, nlp-13). The expression of these nine neuropeptides is neuron type-specific, but in aggregate they cover the entire pharyngeal nervous system, often with unique cell type-specific combinations (schematized in Figure 5B). We found that the expression of all of these nine neuropeptides, analyzed with either reporter transgenes or CRISPR/Cas9 genome-engineered gfp reporter alleles, is affected in ceh-34 mutants (Figure 5A). We schematize these results again in the context of a pharyngeal circuit diagram (Figure 5B). Together with our analysis of classic neurotransmitters (ACh, Glu, serotonin) and receptors, we conclude that ceh-34 is required to endow pharyngeal neurons with their neuron type-specific arsenal of signaling molecules and, hence, that ceh-34 is a critical specifier of several key aspects of pharyngeal neuron identity and function.
ceh-34 is required for sensory receptor expression in the pharyngeal nervous system
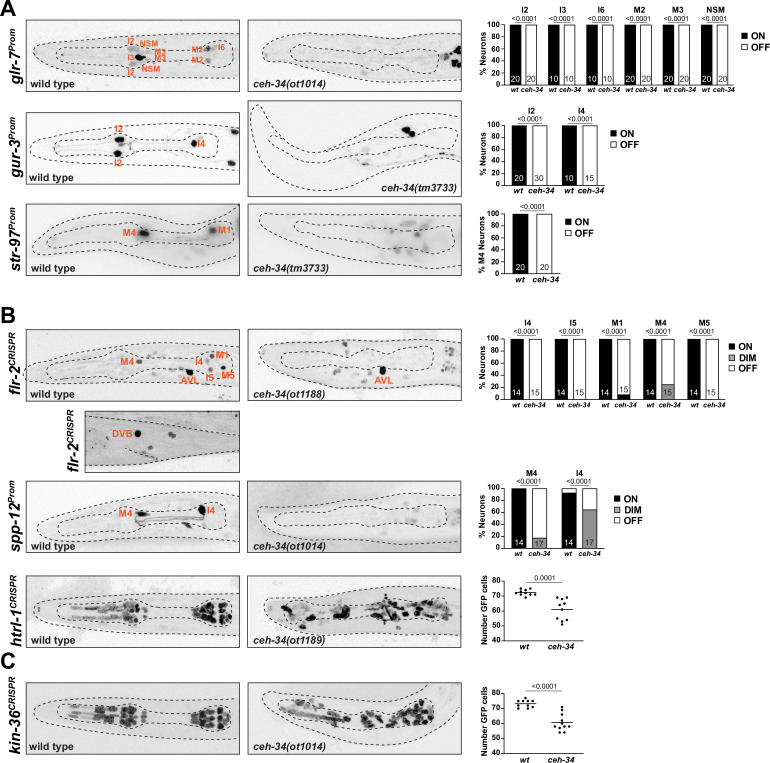
We sought to extend our analysis of ceh-34 mutants by examining the expression of other molecular features of pharyngeal neurons. Like neurons in the vertebrate enteric nervous system, many of the pharyngeal neurons are likely internal sensory neurons that perceive sensory information to modulate peristaltic movements of the alimentary tract (Cook et al., 2020). While the sensory apparatus of pharyngeal neurons is not well understood, there are several candidate sensory receptors expressed in pharyngeal neurons. A gustatory receptor family member, gur-3, a possible light receptor (Bhatla and Horvitz, 2015), is expressed in two pharyngeal neuron classes and its expression is lost in ceh-34 mutants (Figure 6A). Pharyngeal neurons also express the two sole members of the ionotropic sensory receptor family (Croset et al., 2010), encoded by glr-7 and glr-8 in C. elegans (Brockie et al., 2001; Hobert, 2013). We examined expression of glr-7 expression, normally observed in six pharyngeal neuron classes (I2, I3, I6, M2, M3, and NSM), in ceh-34 mutants and found its expression to be completely lost (Figure 6A). Finally, we analyzed the expression of str-97, a putative chemosensory receptor of the GPCR family which we found to be expressed in several pharyngeal neurons (Vidal et al., 2018; Figure 6A). We found that ceh-34 is required for proper str-97 expression (Figure 6A).
Figure 6. ceh-34 affects other identity features of pharyngeal neurons.
(A) Representative pictures and quantification showing sensory receptor expression loss in ceh-34 mutants. Reporter transgenes used are glr-7 (otIs809), gur-3 (nIs780), and str-97 (otIs716). str-97 (otIs716) is expressed in M1 in adult animals, but also in M4 in first larval stage animals. Expression in M4 is lost in ceh-34 mutant, but expression in M1 could not be reliably scored. Animals were scored at the L1 stage. Statistical analysis was performed using Fisher’s exact test. N is indicated within each bar and represents number of neurons scored. (B) Representative pictures and quantification showing effect of ceh-34 on antimicrobial defense genes. Reporter genes used are spp-12 (otIs868) and CRISPR/Cas9-engineered reporter alleles for flr-2 (syb4861) and htrl-1 (syb4895). Animals were scored at the L1 stage. Statistical analysis was performed using Fisher’s exact test, chi-square test, or unpaired t-test. N is indicated within each bar and represents number of neurons scored. (C) Representative pictures and quantification showing effect of ceh-34 on pan-pharyngeal genes. Reporter gene is the CRISPR/Cas9-engineered reporter allele kin-36 (syb4677). Animals were scored at the L1 stage. Statistical analysis was performed using unpaired t-test.
Figure 6—figure supplement 1. Summary of previous single cell transcriptomic analysis of genes broadly expressed in pharyngeal neurons.
Figure 6—figure supplement 2. ceh-34 affects ceh-28 expression.
ceh-34 is required for the expression of antimicrobial defense machinery
One deeply conserved feature of the gut and its associated nervous system is its engagement in antimicrobial defense, either directly through the release of antimicrobial peptides or through the employment of signaling systems that activate the immune system (Klimovich and Bosch, 2018; Muniz et al., 2012). Similar defense strategies operate in C. elegans (Dierking et al., 2016). Aside from the epithelial cells of the intestine, the pharyngeal nervous system appears to play a direct role in these microbial control mechanisms, as inferred by the pharyngeal neuron expression of specific proteins implicated in antimicrobial defense. For example, pharyngeal neurons express a hormone, FLR-2, homologous to glycoprotein hormone alpha subunit, that signals to the intestine to orchestrate antimicrobial defense (Oishi et al., 2009). Pharyngeal neurons also secrete pore-forming polypeptides that directly kill bacteria, such as the SPP-12 protein (Hoeckendorf et al., 2012), as well as a defensin-type antimicrobial peptide, ABF-2 and fungal-induced peptides (FIPR proteins; Kato et al., 2002; Taylor et al., 2021). Our scRNA transcriptome analysis (Taylor et al., 2021) revealed pharyngeal neuron expression of another saposin-related secreted protein, which we named htrl-1 (see Materials and methods). We visualized expression of these signaling molecules, using a promoter fusion for spp-12 (Hoeckendorf et al., 2012) and CRISPR/Cas9-engineered gfp reporter alleles for flr-2 and htrl-1 (Figure 6B). We found that the spp-12 reporter is expressed in I4 and M4 neurons, the flr-2::SL2::gfp::h2b reporter allele is expressed in five pharyngeal neuron classes (I4, I5, M1, M4, and M5), and the htrl-1::SL2::gfp::h2b reporter allele is expressed in all pharyngeal neurons (and in all other pharyngeal cells, but nowhere outside the pharynx at the L1 stage; Figure 6B). A notable feature of the extrapharyngeal expression of the flr-2 reporter allele is expression in the AVL and DVB neurons (Figure 6B), the only extrapharyngeal neurons of the C. elegans nervous system that innervate gut tissue (not the foregut, but the midgut; White et al., 1986). Crossing these reporters into a ceh-34 mutant background, we found that expression of all three genes (spp-12, flr-2, htrl-1) is severely reduced or eliminated in pharyngeal neurons (Figure 6B).
ceh-34 is required for the expression of pan-pharyngeal nervous system genes
In addition to investigating the ceh-34-dependence of genes that fall into specific functional categories, we also sought to capitalize on the recently released single cell transcriptome profiling of the entire C. elegans nervous system that included the entire pharyngeal nervous system (Taylor et al., 2021). This analysis had shown that the molecular signatures of pharyngeal neurons are more similar to each other than to other neurons in the nervous system (Taylor et al., 2021; Figure 1D). This pattern is driven, in part, by a number of previously entirely uncharacterized genes with very broad, if not pan-pharyngeal nervous system expression (but no expression in non-pharyngeal neurons), including the above-mentioned saposin-related htrl-1 gene, small cell surface proteins (e.g. C54E4.4) and a novel receptor tyrosine kinase, which we named kin-36 (Figure 6—figure supplement 1). To confirm this expression pattern, we tagged the kin-36 locus with a gfp::H2B::SL2 cassette at its 5’ end, using CRISPR/Cas9 genome engineering. We found that kin-36 indeed displays pan-pharyngeal neuron expression; expression is also observed in all other pharyngeal cell types, but no cell types outside the pharynx, except some unidentified vulval cells (Figure 6C). Crossing the kin-36 reporter allele into ceh-34 mutants, we observed what appears to be selective loss of kin-36 expression from many, albeit not all pharyngeal neurons, a similar effect to what we observed for htrl-1 (Figure 6B and C).
We conclude that ceh-34 is required for the adoption of a broad palette of individual molecular features of all pharyngeal neurons, consistent with a role as a terminal selector of neuronal identity of all pharyngeal neurons. Like other terminal selectors, ceh-34 only affects neuron type-specific features, but not features that are expressed by all neurons throughout the nervous system.
ceh-34 is continuously required to maintain the differentiated and functional state of enteric neurons
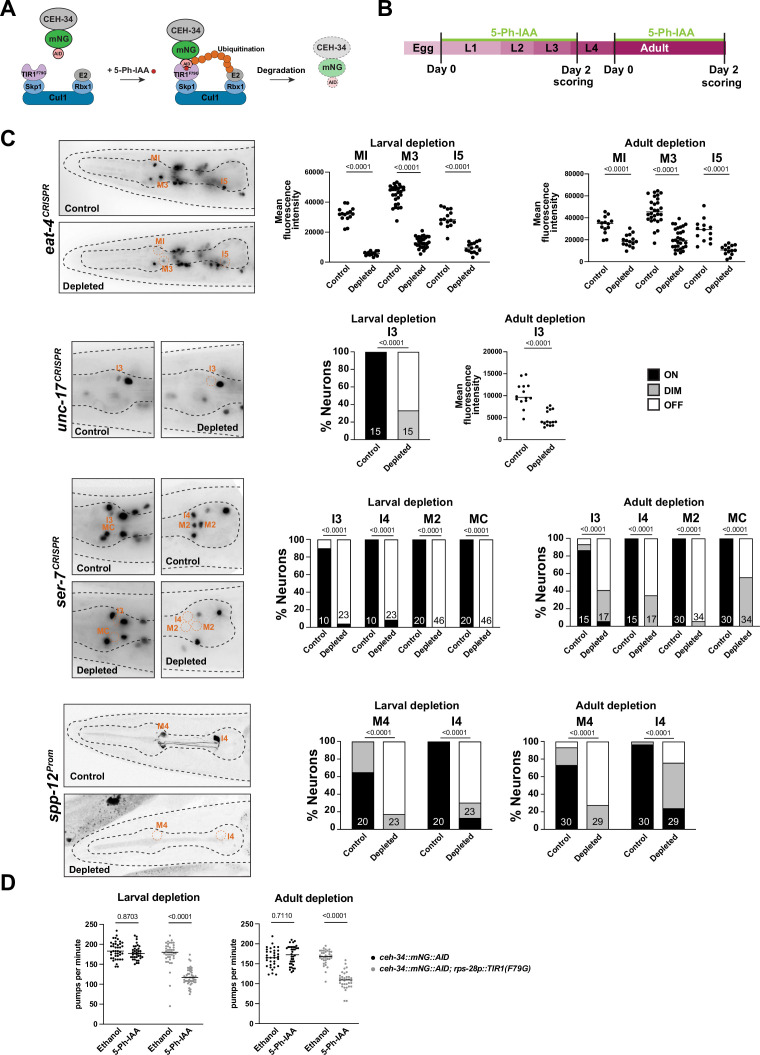
The effect of ceh-34 on terminal marker expression and its continuous expression throughout the life of all pharyngeal neurons suggests that, like other terminal selectors in the non-pharyngeal nervous system, ceh-34 may not only initiate but also maintain the terminally differentiated state. To test this possibility, we generated a conditional ceh-34 allele that allowed us to deplete CEH-34 protein postdevelopmentally. To this end, we inserted an auxin-inducible degron (AID) (Zhang et al., 2015) into the ceh-34 locus using CRISPR/Cas9 genome engineering. Together with a ubiquitously expressed AtTIR1F79G ubiquitin ligase that recognizes the degron (rps-28 driver; cshIs140; Hills-Muckey et al., 2022), this approach allows for temporal depletion of CEH-34 protein through addition of an auxin derivative (5-Ph-IAA) to the worm diet (Figure 7A and B). Postembryonic 5-Ph-IAA addition at either larval or adult stages resulted in downregulation of four tested markers for the differentiated state of different pharyngeal neuron classes, eat-4/VGluT, unc-17/VAChT, ser-7, and spp-12 (Figure 7B and C). These effects are not as strong as in null mutants, but this is likely due to incomplete CEH-34 protein depletion, since constitutive 5-Ph-IAA exposure from parental stages throughout all developmental stages also produces only limited defects in expression of these markers.
Figure 7. ceh-34 is continuously required to maintain gene expression and function of pharyngeal neurons.
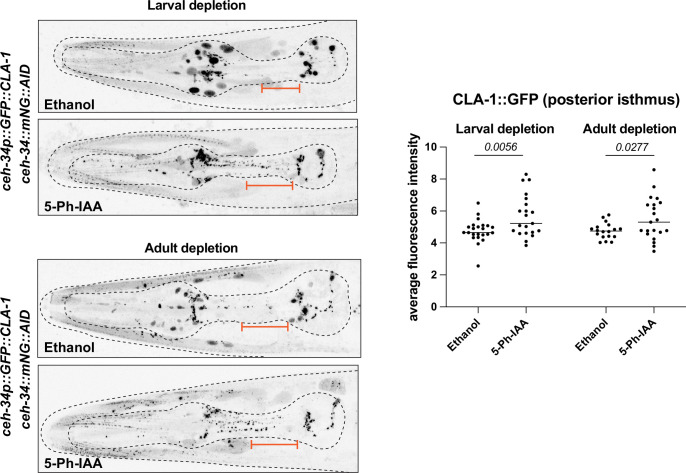
(A) Schematic of the AIDv2 system (Hills-Muckey et al., 2022). Skp1, Cul1, Rbx1, and E2 are phylogenetically conserved components of the E3 ligase complex. TIR1F79G is a modified plant-specific substrate-recognizing subunit of the E3 ligase complex. In the presence of the auxin analog (5-Ph-IAA), the enzyme TIR1F79G binds to the AID fused to a protein of interest, leading to ubiquitination and proteasomal degradation of the targeted protein. (B) Schematic depicting the 5-Ph-IAA treatment. Synchronized populations of worms at the L1 and young adult stage were transferred onto 5-Ph-IAA-coated plates and scored 48 hr later. Worms were expressing TIR1F79G ubiquitously under the rps-28 promoter (cshIs140). The ceh-34 locus was tagged with mNG::AID (ot903). (C) ceh-34 is required for maintained expression of identity genes. Reporter genes used are spp-12 (otIs868) and CRISPR/Cas-9-engineered reporter alleles for eat-4 (syb4257), unc-17 (syb4491), and ser-7 (syb4502). Since ceh-34::mNG::AID (ot903) and reporter genes scored are all fluorescent green, and this was obscuring the scoring on ethanol conditions, the control conditions are reporter genes on their own treated with 5-Ph-IAA. Representative pictures of larval depletion are shown on the left and quantification for larval and adult depletion is shown on the right. Quantification is only shown for neurons that were affected. Neurons unaffected by temporally controlled 5-Ph-IAA addition are also unaffected by constitute 5-Ph-IAA addition, indicating an inability to completely deplete CEH-34 protein. Statistical analysis was performed using unpaired t-test, Fisher’s exact test, or chi-square test. N is indicated within each bar and represents number of neurons scored. (D) ceh-34 is required for maintained pharyngeal function. Larval and adult ceh-34 depletion results in decreased pharyngeal pumping. Statistical analysis was performed using two-way ANOVA.
We also assessed the functional consequences of CEH-34 protein depletion in adult animals, as well as larval stage animals. Using again the AID approach, we found that postembryonic CEH-34 depletion at either larval or adult stages results in substantial defects in pharyngeal pumping, as expected from a disruption of enteric nervous system function (Figure 7D). We conclude that ceh-34 is required to maintain differentiated features of pharyngeal neurons and therefore fulfills another key criterion to classify as a terminal selector of pharyngeal neuron identity.
Pharyngeal nervous system architecture is severely disorganized in ceh-34 mutants
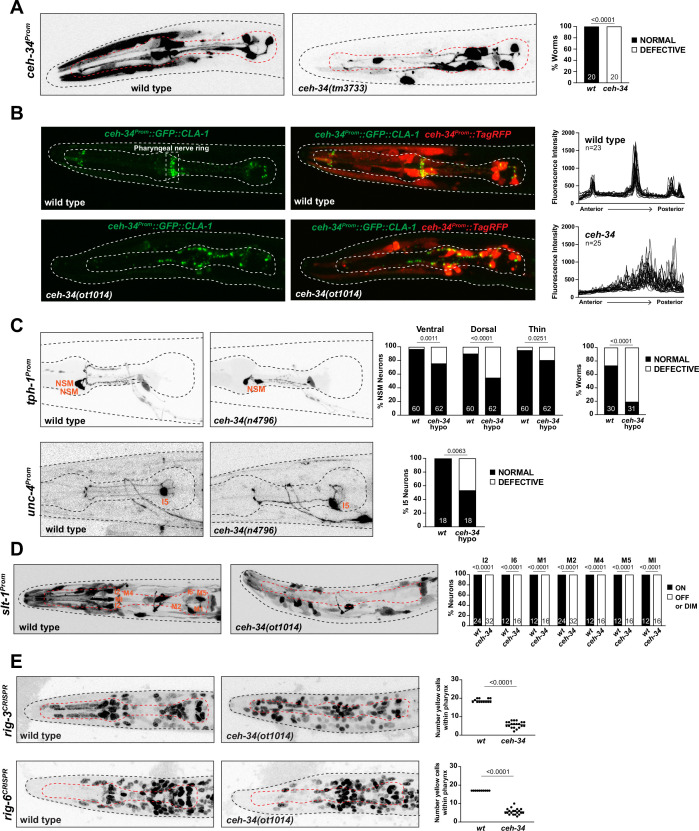
We further extended our analysis of ceh-34 function by analyzing the anatomy of pharyngeal neuron circuitry in ceh-34 mutants. This analysis is particularly important in light of the observation that, both in C. elegans (Berghoff et al., 2021; Pereira et al., 2015) and in vertebrates (Brunet and Pattyn, 2002), several instances have been described in which synaptically interconnected, but otherwise distinct neurons express the same transcription factor. Such observation suggests that these transcription factors may have a role in assembling neurons into functional circuitry. ceh-34 represents a particularly extreme version of this scenario, because all ceh-34(+) neurons are heavily synaptically interconnected and only make a single robust synaptic contact to the rest of the C. elegans nervous system (Figure 1C; Cook et al., 2020). To assess whether ceh-34 not only specifies terminal molecular properties of pharyngeal neurons, but also organizes overall circuit architecture, we examined pharyngeal nervous system architecture using fluorescent reporter constructs. Such analysis is complicated by the fact that genes that are selectively expressed in ceh-34(+) neurons, and hence could serve as drivers for a fluorescent reporter, are turned off in ceh-34 null mutant animals, thereby preventing an easy visualization of individual axonal tracts or synaptic contacts. However, we found that the ceh-34 promoter itself is still expressed until the first larval stage in ceh-34 null mutants, when these animals arrest development. This ceh-34 promoter transgene (otIs762) reveals that although their identity is not properly specified, as described above, pharyngeal neurons retain the capability to grow neuronal projections in ceh-34 mutants; however, axonal tracts are severely disorganized (Figure 8A).
Figure 8. ceh-34 affects the assembly of pharyngeal circuitry.
(A) ceh-34 null mutants display disorganized axodendritic projections. Axodendritic projections were scored as a whole rather than by individual neuron because with all the pharyngeal neurons being labeled it was difficult to assign specific projections to individual neurons. Projections were classified as defective only when obviously deviating from the wild-type path. Representative pictures and quantification are shown. Reporter gene is ceh-34 (otIs762). Animals were scored at the L1 stage. Statistical analysis was performed using Fisher’s exact test. N is indicated within each bar and represents number of worms. (B) ceh-34 null mutants show disorganized pharyngeal nerve ring presynaptic specializations as visualized with CLA-1 puncta. Representative pictures are shown. Quantification (right panels) shows GFP fluorescent intensity profiles along the anterior posterior axis. Reporter gene is otIs785. Animals were scored at the L1 stage. (C) ceh-34(n4796) hypomorph mutants show axonal defects in NSM (top panel) and I5 (bottom panel). Representative pictures and quantification are shown. For NSM, the ventral, dorsal, and thin projection (not visible in picture) were scored separately (graph on the left) and then data was pulled together to indicate the percentage of worms showing any defect (graph on the right). Reporter genes used are tph-1 (zdIs13) and unc-4 (otEx7503). Animals were scored at the L4 stage. Statistical analysis was performed using Fisher’s exact test. N is indicated within each bar and represents number of neurons or number of worms. (D) ceh-34 affects expression of the axon guidance cue slt-1 (kyIs174). Representative pictures and quantification are shown. Animals were scored at the L1 stage. Statistical analysis was performed using Fisher’s exact test. N is indicated within each bar and represents number of neurons scored. (E) ceh-34 affects expression of CRISPR/Cas9-engineered gfp reporter alleles of rig-3 (syb4763) and rig-6 (syb4729), two Ig superfamily members. rig-3 and rig-6 are expressed in almost all pharyngeal neurons plus many other cells within and outside the pharynx. Worms were scored with a red pan-neuronal marker (otIs355) or a red ceh-34promoter fusion (stIs10447) in the background to facilitate scoring. Number of yellow cells were counted within the pharynx. Animals were scored at the L1 stage. Statistical analysis was performed using unpaired t-test.
Figure 8—figure supplement 1. ceh-34 is required to maintain synapse organization in the pharyngeal nervous system.
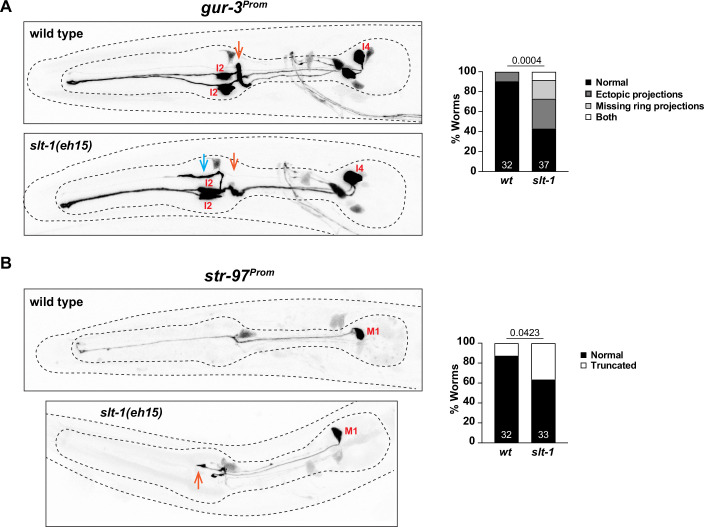
Figure 8—figure supplement 2. Axonal defects in slt-1 mutants.
We also expressed the cytoplasmically localized TagRFP reporter together with a synaptically localized, GFP-tagged CLA-1/Clarinet protein (a synaptic active zone marker; Xuan et al., 2017) under control of the ceh-34 promoter. In wild-type animals, this transgene (otIs785) reveals (1) the axonal tracts of the pharyngeal nervous system and (2) synaptic structures that are strongly enriched in the pharyngeal nerve ring (Figure 8B). In ceh-34 null mutants, we observed not only a disruption of axonal tract anatomy, but a severe disorganization of presynaptic clusters throughout the entire pharyngeal nervous system (Figure 8B).
Using the CLA-1 synaptic marker, we also asked whether CEH-34 is continuously required to maintain synaptic architecture postembryonically (i.e. after the time when synaptic connections initially form). To this end, we again made use of the AID system and removed CEH-34::mNG::AID either throughout larval stages or in the adult stage. In both cases, we found that such depletion resulted in synaptic clusters becoming disorganized, such that ectopic presynaptic clusters form at ectopic locations in the isthmus of the pharynx (Figure 8—figure supplement 1).
Lastly, we made use of a mild ceh-34 hypomorphic allele, n4796 (Hirose et al., 2010). In ceh-34(n4796) animals, the expression of many molecular markers for individual pharyngeal neurons are not affected, allowing to visualize their morphology. Focusing on two neuron types, NSM and I5, we found that in both types, specific axons branches fail to form in ceh-34(n4796) animals (Figure 8C). We conclude that ceh-34 is required to establish and maintain proper pharyngeal nervous system architecture.
ceh-34 affects the expression of molecules involved in proper wiring of the pharyngeal nervous system
To further explore how ceh-34 may affect pharyngeal nervous system architecture, we considered the expression of molecules with potential or explicitly demonstrated roles in axon guidance, fasciculation, and/or synapse formation. A genetic analysis of axon guidance and circuit formation has only been conducted in a small number of pharyngeal neurons, mainly the M2 and NSM neuron classes (Pilon, 2008). In both neuron classes, the SLT-1 axon guidance cue, the C. elegans ortholog of Slit, has been found to be required for proper axon guidance (Axäng et al., 2008; Rauthan et al., 2007). We extended this phenotypic characterization, finding that other pharyngeal neurons also display axon pathfinding defects in slt-1 mutants (Figure 8—figure supplement 2). To examine potential links between slt-1 and ceh-34, we made use of a promoter::gfp fusion that captures the entire upstream intergenic region of the slt-1 locus (Hao et al., 2001) and which shows selective expression in seven pharyngeal neuron classes (I2, I6, M1, M2, M4, M5, and MI) at the first larval stage. We found that slt-1 expression is strongly affected in ceh-34 mutants (Figure 8D).
Effects of ceh-34 on molecules potentially involved in circuit formation are not restricted to slt-1. Two Ig superfamily members, rig-3 and rig-6 (the sole C. elegans ortholog of contactin), have previously been implicated in axon outgrowth and synapse function in the C. elegans nervous system (Babu et al., 2011; Bhardwaj et al., 2020; Katidou et al., 2013; Kim and Emmons, 2017) and promoter fusion transgenes have indicated their expression in pharyngeal neurons (Schwarz et al., 2009). We used CRISPR/Cas9 to tag both loci with gfp and found that both genes are broadly expressed in many pharyngeal neurons (Figures 1E and 8E). ceh-34 affects the pharyngeal neuron expression of both rig-3 and rig-6 reporter alleles (Figure 8E).
The Six homeodomain cofactor, Eyes absent/Eya, shows limited cooperation with ceh-34
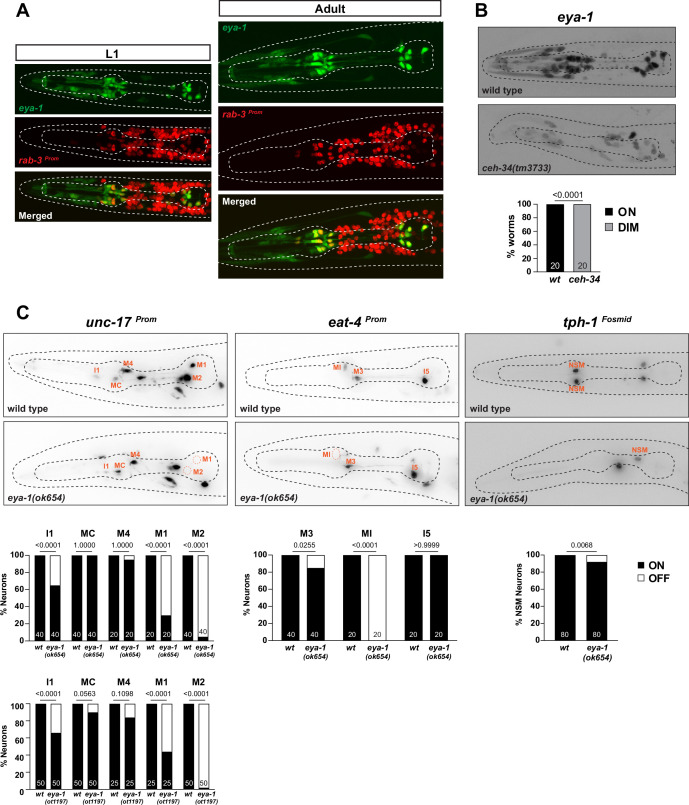
To gain further insights into how CEH-34 patterns the identity of a wide array of distinct pharyngeal neuron types, we considered the involvement of cell type-specific cofactors. As a first step, we considered the EYes Absent/EYA protein, a phylogenetically conserved transcriptional co-activator of specific subsets of Six homeodomain proteins (Ohto et al., 1999; Patrick et al., 2013; Tadjuidje and Hegde, 2013). The C. elegans ortholog of Eyes absent, called eya-1 (Furuya et al., 2005), also directly physically interacts with CEH-34 protein (Amin et al., 2009; Hirose et al., 2010). We first examined eya-1 expression in pharyngeal neurons using a genomic fragment that contains the entire, gfp-tagged eya-1 locus (Furuya et al., 2005). We observed expression in all pharyngeal neurons throughout all larval and adult stages, a phenocopy of the ceh-34 expression pattern (Figure 9A), that is also corroborated by our recent scRNA analysis (Figure 6—figure supplement 1; Taylor et al., 2021). Moreover, we found that eya-1 expression requires ceh-34 function, suggesting that ceh-34 acts in a feedforward configuration to induce its own transcriptional cofactor (Figure 9B).
Figure 9. Limited involvement of eya-1 in pharyngeal neuron identity specification.
(A) eya-1 is expressed in all pharyngeal neurons throughout the life of the worm. Images of L1 and adult worms showing co-localization of eya-1 expression (nIs352) with the pan-neuronal gene rab-3 (otIs355) in pharyngeal neurons. (B) eya-1 expression is regulated by ceh-34. Representative pictures and quantification are shown. Reporter gene is eya-1 (nIs352). Animals were scored at the L1 stage. Statistical analysis was performed using Fisher’s exact test. N is indicated within each bar and represents number of worms scored. (C) eya-1 mutant animals show defects in neurotransmitter identity specification. Representative images and quantification are shown for unc-17 (otIs661), eat-4 (otIs487), and tph-1 (otIs517). Animals were scored at the L4 stage. Statistical analysis was performed using Fisher’s exact test. N is indicated within each bar and represents number of neurons scored.
We analyzed the function of eya-1 in the context of pharyngeal neuron specification. Animals that carry a deletion of a part of the eya-1 locus, ok654, display pharyngeal neuron specification defects, albeit much milder than those observed in ceh-34 null mutant animals (Figure 9C). The larval arrest phenotype of ceh-34 null mutants is also more penetrant than that of eya-1 mutants, which are very slow growing, but still homozygous viable (Furuya et al., 2005). To exclude the possibility that the ok654 allele is not a null allele, we used CRISPR/Cas9 to generate a null allele in which the entire locus is deleted. Animals carrying this deletion allele (ot1197) display phenotypes that are indistinguishable from those of ok654 animals. They are still homozygous viable, albeit as slowly growing as ok654 animals, and they display very similar, limited neuronal cell fate marker defects (Figure 9C). Given the milder spectrum of eya-1 defects compared to ceh-34 null mutants, we conclude that ceh-34 may be able to partly function without eya-1.
In other organisms, Dachshund proteins are components of Sine oculis/Eya complexes in several cellular contexts (Hanson, 2001). However, the sole C. elegans ortholog of Dachshund is not expressed in pharyngeal neurons (Colosimo et al., 2004; Taylor et al., 2021) and dac-1 null mutants also do not display the larval growth/arrest phenotype characteristic of ceh-34 and eya-1 mutants (Colosimo et al., 2004). While these observation do not entirely rule out a function for DAC-1 in pharyngeal neurons, it appears unlikely that DAC-1 is an essential cofactor of CEH-34.
ceh-34 cooperates with a multitude of other homeobox genes to specify distinct pharyngeal neuron types
How does ceh-34 activate distinct genes in different pharyngeal neuron types? One obvious possibility is that ceh-34 cooperates with neuron type-specific cofactors in neuron type-specific terminal selector complexes to drive specific fates. As candidates for such cofactors, we considered homeobox genes, for two reasons: (1) like any other neuron in the C. elegans nervous system, each individual pharyngeal neuron expresses a unique combination of homeobox genes, in addition to pan-pharyngeal ceh-34 (Reilly et al., 2020); (2) previous studies had already implicated a few homeobox genes in controlling some select functional or molecular aspects of individual pharyngeal neurons (Aspöck et al., 2003; Feng and Hope, 2013; Mörck et al., 2004; Ramakrishnan and Okkema, 2014; Ray et al., 2008; Zhang et al., 2014). For example, the homeobox gene ceh-2, the C. elegans ortholog of vertebrate EMX and Drosophila Ems, is required for proper function of the M3 neuron, but effects of ceh-2 on molecular aspects of M3 neuron differentiation had not been reported (Aspöck et al., 2003). We therefore set out to analyze homeobox gene function throughout the pharyngeal nervous system and to examine potential interactions of ceh-34 with other homeobox genes.
NSM neurons
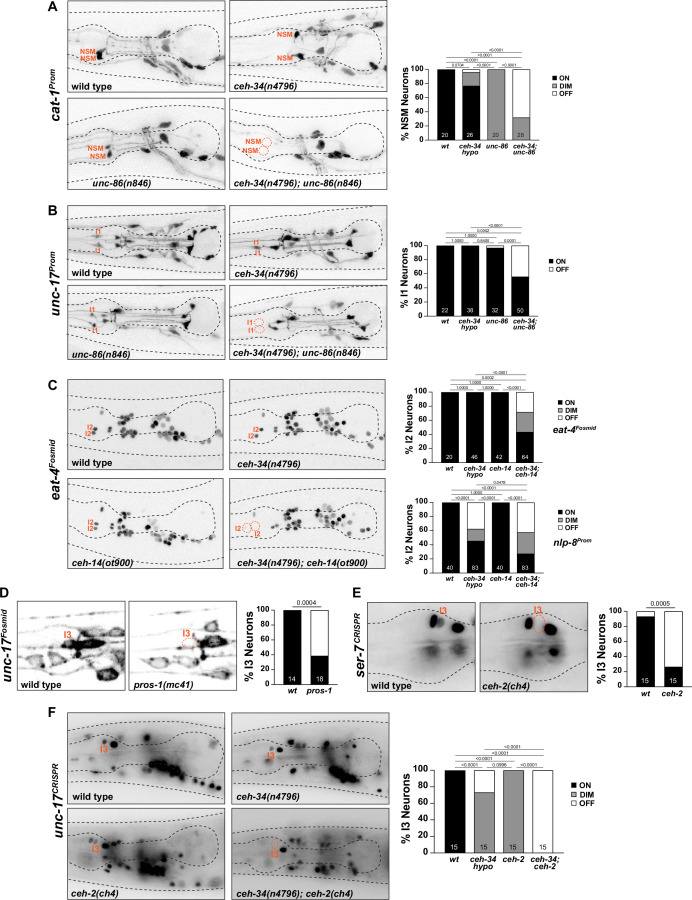
To ask whether distinct pharyngeal neuron type-specific homeobox genes cooperate with ceh-34 in distinct neuronal cell types throughout the pharyngeal nervous system, we made use of a hypomorphic ceh-34 allele, n4796 (Hirose et al., 2010). Unlike the ceh-34 null allele, which results in very strong expression defects of all NSM molecular markers (Figures 3—6), the n4796 allele displays only subtle if any marker expression effects on its own (Figure 10A). However, when combined with a mutant allele of unc-86, a POU homeobox gene that affects many but not all NSM marker genes (Zhang et al., 2014), strong synergistic differentiation defects of NSM are observed (Figure 10A). This genetic interaction mirrors the synergistic effects of unc-86 and the LIM homeobox gene ttx-3, another regulator of NSM differentiation (Zhang et al., 2014).
Figure 10. ceh-34 cooperates with homeobox genes to specify distinct pharyngeal neuron types.
(A) unc-86 and ceh-34 synergistically affect NSM differentiation. Representative images and quantification are shown. Reporter gene used is cat-1 (otIs224). Animals were scored at the L4 stage. Statistical analysis was performed using Fisher’s exact test. p-values were adjusted with the Holm-Sidak correction for multiple comparisons. N is indicated within each bar and represents number of neurons scored. (B) unc-86 and ceh-34 show synergistic defects in I1 neuron differentiation. Representative images and quantification are shown. Reporter gene used is unc-17 (otIs661). Animals were scored at the L4 stage. Statistical analysis was performed using Fisher’s exact test. p-values were adjusted with the Holm-Sidak correction for multiple comparisons. N is indicated within each bar and represents number of neurons scored. (C) ceh-14 and ceh-34 show synergistic effects on I2 neuron differentiation. Representative images and quantification are shown for eat-4 (otIs518). Bottom graph shows quantification for nlp-8 (otIs711). Animals were scored at the L4 stage. Statistical analysis was performed using Fisher’s exact test or chi-square test. p-values were adjusted with the Holm-Sidak correction for multiple comparisons. N is indicated within each bar and represents number of neurons scored. (D) pros-1 affects I3 neuron differentiation. Representative images and quantification are shown. Reporter gene is unc-17 (otIs576). Animals were scored at the L1 stage. Statistical analysis was performed using Fisher’s exact test. N is indicated within each bar and represents number of neurons scored. (E) ceh-2 affects I3 neuron differentiation. Representative images and quantification are shown. Reporter gene used is a CRISPR/Cas9-enginereed allele for ser-7 (syb4502). Animals were scored at the L4 stage. Statistical analysis was performed using Fisher’s exact test. N is indicated within each bar and represents number of neurons scored. (F) ceh-2 and ceh-34 show synergistic defects in I3 neuron differentiation. Representative images and quantification are shown. Reporter gene used is CRISPR/Cas9-engineered allele for unc-17 (syb4491). Animals were scored at the L4 stage. Statistical analysis was performed using Fisher’s exact test. p-values were adjusted with the Holm-Sidak correction for multiple comparisons. N is indicated within each bar and represents number of neurons scored.
I1 neurons
A similar genetic interaction between ceh-34 and unc-86 is observed in the cholinergic I1 neuron pair, the only other pharyngeal neuron class that also co-expresses ceh-34 and unc-86 (Baumeister et al., 1996; Serrano-Saiz et al., 2018), but which does not express ttx-3. While cholinergic identity, visualized via unc-17/VAChT expression, is not affected in unc-86(n846) single mutants, a combination of the ceh-34(n4796) hypomorphic allele with the unc-86(n846) mutation results in I1 losing its cholinergic identity (Figure 10B).
I2 neurons
Synergistic interactions are also observed in the glutamatergic I2 neuron pair. Like the I1 neurons, no identity regulators were previously known for this neuron class. Our homeobox gene expression atlas (Reilly et al., 2020) showed that the I2 neuron expresses the LIM homeobox gene ceh-14, the C. elegans ortholog of vertebrate Lhx3/4. No other pharyngeal neuron expresses ceh-14. While ceh-14 single null mutants show no effect on eat-4/VGluT expression (the marker of glutamatergic identity), in combination with the ceh-34(n4796) hypomorphic allele, a strong synergistic effect on glutamatergic identity acquisition is observed in I2 (Figure 10C). Another molecular marker for I2 identity, the neuropeptide nlp-8, is also synergistically regulated by ceh-34 and ceh-14 (Figure 10C).
I3 neuron
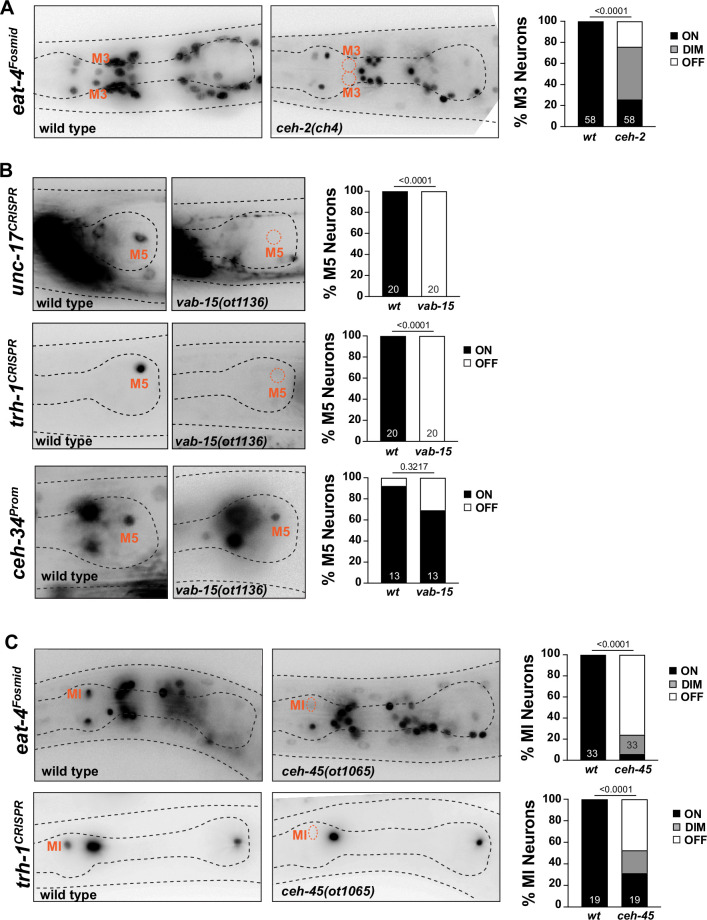
The previously unstudied cholinergic I3 neuron class expresses, in addition to ceh-34, the C. elegans ortholog of Empty spiracles/EMX, ceh-2 (Aspöck et al., 2003; Reilly et al., 2020), as well as the Prospero ortholog pros-1, whose function in the nervous system has not previously been examined. We find that in pros-1 null mutant animals, cholinergic identity of I3, measured with an unc-17/VAChT reporter transgene, is not properly acquired (Figure 10D). The ceh-2(ch4) null allele alone also shows a reduction of unc-17/VAChT expression (Figure 10F), as well as a reduction of the serotonin receptor ser-7 expression (Figure 10E). In combination with the ceh-34(n4796) hypomorphic allele, unc-17/VAChT expression, and, hence, cholinergic identity of I3, is eliminated in ceh-2 mutant animals (Figure 10F).
M3 neuron
Apart from expression in I3, the EMX ortholog ceh-2 is also expressed in the glutamatergic M3 neurons and is required for proper M3 function (Aspöck et al., 2003), but molecular correlates for this functional defect have not previously been identified. We found that in ceh-2 single mutants, expression of the eat-4/VGluT identity marker is affected in M3 (Figure 11A).
Figure 11. Other homeobox genes involved in specifying distinct pharyngeal neuron types.
(A) ceh-2 affects M3 neuron differentiation. Representative images and quantification are shown. Reporter gene is eat-4 (otIs388). Animals scored at the L4 stage. Statistical analysis was performed using chi-square test. N is indicated within each bar and represents number of neurons scored. (B) vab-15 affects M5 neuron differentiation. Representative images and quantification are shown. Reporter genes used are ceh-34 (stIs10447) and CRISPR/Cas9-engineered alleles for unc-17 (ot907) and trh-1 (syb4421). Animals were scored at the L4 stage. Statistical analysis was performed using Fisher’s exact test. N is indicated within each bar and represents number of neurons scored. (C) ceh-45 affects MI neuron differentiation. Representative images and quantification are shown. Reporter genes used are eat-4 (otIs388) and CRISPR/Cas9-engineered allele for trh-1 (syb4421). Animals were scored at the L4 stage. Statistical analysis was performed using Fisher’s exact test. N is indicated within each bar and represents number of neurons scored.
Figure 11—figure supplement 1. Homeobox mutant alleles.
Figure 11—figure supplement 2. Homeobox misexpression experiments.
M4 neuron
In the cholinergic M4 neuron, the ceh-28 and zag-1 homeobox genes have previously been shown to each regulate subsets of M4 identity features (Ramakrishnan and Okkema, 2014). Both zag-1 and ceh-28 affected flp-2 expression, but only zag-1, and not ceh-28, was found to affect ser-7 expression (Ramakrishnan and Okkema, 2014). In contrast, ceh-28 but not zag-1 affected flp-5 expression and neither zag-1 nor ceh-28 affected unc-17 or flp-21 expression (Ramakrishnan and Okkema, 2014). ceh-34 null mutants show effects on the expression of all the tested ceh-28 or zag-1-dependent (ser-7, flp-2, and flp-5) or -independent markers (unc-17, flp-21; Figures 3—5), indicating that ceh-34 may collaborate with these homeobox genes to control distinct subsets of M4 differentiation markers. ceh-34 also affects ceh-28 expression (Figure 6—figure supplement 2).
M5 neuron
The cholinergic M5 neuron expresses, in addition to ceh-34, the Msh/Msx ortholog vab-15 (Reilly et al., 2020). Since only a hypomorphic allele of vab-15 was previously available (Du and Chalfie, 2001), we generated a molecular null allele, ot1136, through CRISPR/Cas9 genome engineering (Figure 11—figure supplement 1A). In contrast to the hypomorphic allele, these null mutant animals are very slow growing, but still homozygous viable. We found a complete loss of expression of the cholinergic marker unc-17/VAChT, as well as the neuropeptidergic marker, trh-1, in vab-15(ot1136) animals (Figure 11B). Complete loss of marker expression is not an indicator of failure of this neuron to be generated, since crossing a ceh-34 marker into vab-15 null mutants revealed the presence and normal ceh-34 expression of M5 (Figure 11B).
MI neuron
In our previous genome-wide analysis of homeobox gene expression (Reilly et al., 2020), we had shown that the glutamatergic MI neuron expresses the sole worm ortholog of the Goosecoid homeobox gene, ceh-45. Embryonically, ceh-45 is expressed in multiple pharyngeal tissues (Ma et al., 2021), but its expression resolves to exclusive expression in the MI and I1 neurons (Reilly et al., 2020). ceh-45 had not previously been functionally characterized. We examined ceh-45 function by generating a null allele using CRISPR/Cas9 genome engineering, ot1065 (Figure 11—figure supplement 1A). ceh-45 null mutant animals display partially penetrant embryonic lethality, with escapers being slow growing. Mirroring the ceh-34 defects, we found that glutamatergic identity specification of MI (as assessed by eat-4/VGluT expression) is strongly affected in surviving ceh-45 null mutant animals (Figure 11C). Similarly, expression of the neuropeptide trh-1 is also affected in MI (Figure 11C).
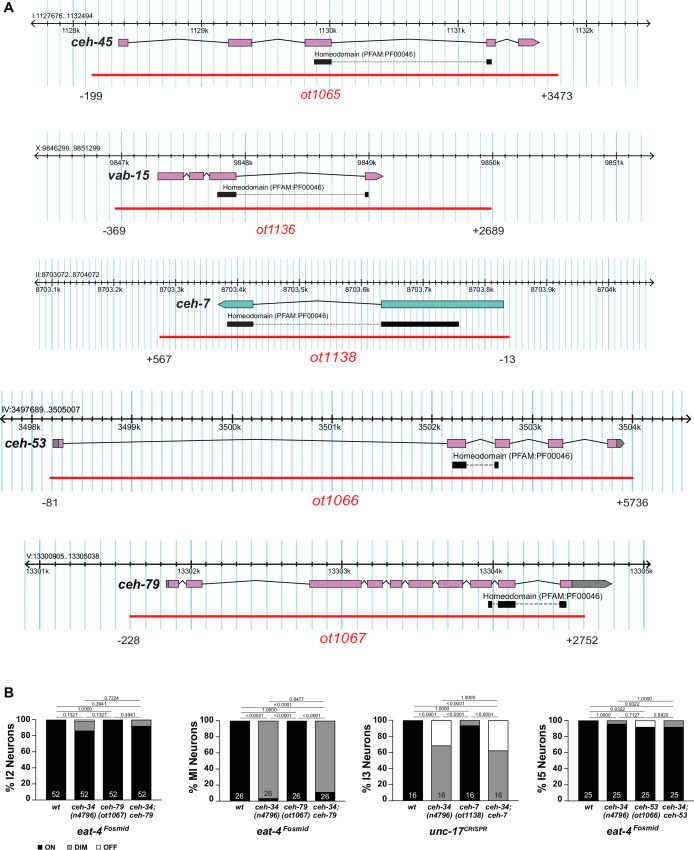
In our search for potential ceh-34 cofactors, we also considered three divergent, non-conserved homeobox genes, ceh-7, ceh-53, and ceh-79 that display expression in subsets of pharyngeal neurons (Reilly et al., 2020). We generated null alleles for these three genes using CRISPR/Cas9 genome engineering (Figure 11—figure supplement 1A). However, we observed no eat-4/VGluT or unc-17/VAChT expression defects in either of these mutant strains, either alone or in combination with the ceh-34 hypomorphic allele n4796 (Figure 11—figure supplement 1B).
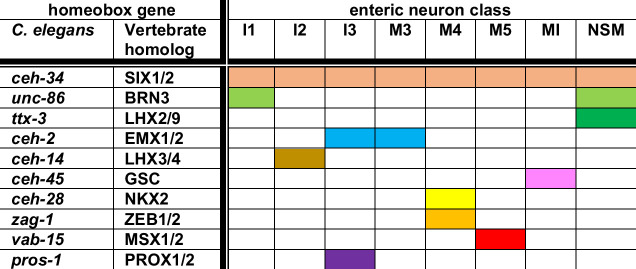
In conclusion, nine phylogenetically conserved homeobox genes appear to collaborate with ceh-34 in 8 of the 14 pharyngeal neuron classes to specify their proper identity (summarized in Figure 12). Since the remaining six classes also express specific combinations of homeobox genes (Reilly et al., 2020), we anticipate that future analysis will likely reveal homeobox codes throughout the entire pharyngeal nervous system.
Figure 12. Summary of homeobox gene codes involved in pharyngeal neuron identity specification.
Shown here are homeobox genes for which an involvement in pharyngeal neuron differentiation has been shown, as well as the 8 (of a total of 14) pharyngeal neuron classes for which a homeobox regulator besides ceh-34 has been identified to date. Each neuron expresses additional homeobox genes (resulting in neuron type-specific combination of homeobox genes; Reilly et al., 2020), but the function of these additional genes remains to be characterized.
Discussion
We have identified here common, overarching themes in the differentiation of the pharyngeal nervous system, the enteric nervous system of the nematode C. elegans. A number of previous studies have identified transcription factors involved in regulating specific differentiation aspects of a small subset of pharyngeal neurons (Aspöck et al., 2003; Feng and Hope, 2013; Mörck et al., 2004; Ramakrishnan and Okkema, 2014; Rauthan et al., 2007; Ray et al., 2008; Zhang et al., 2014), yet no common theme emerged from these studies. We have shown here that a Sine oculis ortholog, ceh-34, orchestrates the terminal differentiation program of all pharyngeal neurons. CEH-34 appears to act as a terminal selector of pharyngeal neuron identity, as inferred from its requirement to initiate terminal neuronal differentiation programs in all pharyngeal neurons (without affecting pan-neuronal identity, a unifying trait of all terminal selectors), as well as its continuous role in maintaining the differentiated state (another defining trait of terminal selectors). The aspects of the differentiation program that we consider here, and found to be under control of ceh-34, include anatomical (axon outgrowth and synapse formation), molecular, and functional features. Molecular and functional features affected by ceh-34 range from neuron-neuron communication to presumptive sensory functions to the intriguing function of enteric neurons as potential regulators of microbial colonization. There is good reason to believe that CEH-34 controls these diverse phenotypic identity features in a direct manner, that is, it may not act through intermediary factors. CEH-34 is among the many C. elegans transcription factors whose binding sites have been determined in vitro through protein binding microarrays (Narasimhan et al., 2015) and a phylogenetic footprinting pipeline reveals that these motifs are significantly enriched in the single cell transcriptome of most pharyngeal neuron classes (Glenwinkel et al., 2021). This is in accordance with CEH-34 being a shared terminal selector component of all pharyngeal neuron classes.
Within the nervous system, the selectivity of ceh-34 expression in all pharyngeal neurons is remarkable – based on extensive gene expression pattern analysis, including recent scRNA data, there is no other transcription factor that so selectively and comprehensively defines all pharyngeal, but no non-pharyngeal neuronal cell types. We found that the key determinant of this expression is the organ selector PHA-4 (Gaudet and Mango, 2002; Horner et al., 1998; Kalb et al., 1998; Mango et al., 1994), which is expressed earlier in development to act both as a pioneer factor (Hsu et al., 2015) and to induce the expression of a number of different terminal selectors for different tissue types within the foregut – the ceh-34 gene for all neurons (this paper), the bHLH transcription factor hlh-6 for pharyngeal gland cells (Smit et al., 2008) and the Nk-type homeobox gene ceh-22 for pharyngeal muscle identity (Vilimas et al., 2004). Given the continuous expression of PHA-4 throughout postembryonic life, it is conceivable that PHA-4 acts in a regulatory feedforward motif configuration, where it not only induces tissue-type terminal selectors (ceh-34, hlh-6, ceh-22), but then also collaborates with them to induce and maintain terminal differentiation batteries.
Our work indicates that CEH-34 is a shared component of neuron type-specific terminal selector complexes, such that CEH-34 interacts with a distinct set of at least eight homeodomain cofactors to impose unique features in distinct pharyngeal neuron types. As is the case for CEH-34 target sites, the in vitro-determined binding sites for several of these homeodomain cofactors display a phylogenetically conserved enrichment in the respective neuron type-specific gene batteries (Glenwinkel et al., 2021). For example, CEH-34 and UNC-86 binding sites are co-enriched in the I1 neuronal transcriptome, CEH-34 and CEH-14 binding sites in the I2 neuronal transcriptome, CEH-34 and CEH-2 in the I3 transcriptome, CEH-34 and CEH-45 in the MI transcriptome, and VAB-15 and CEH-34 in the M5 transcriptome (Glenwinkel et al., 2021). The interactions of CEH-34 with its various collaborating factors is likely highly dependent on the cis-regulatory architecture of individual target genes. We infer this from ceh-34 mutant phenotypes, which, depending on target gene, can be fully or partially penetrant (i.e. not all animals affected), or fully or partially expressive (i.e. ‘dimming’ of target gene expression), or a combination of both. Depending on the number, affinity, and arrangement of binding sites for individual factors, CEH-34 and its individual cofactors may each have a more or less pronounced role in the regulation of individual target genes.
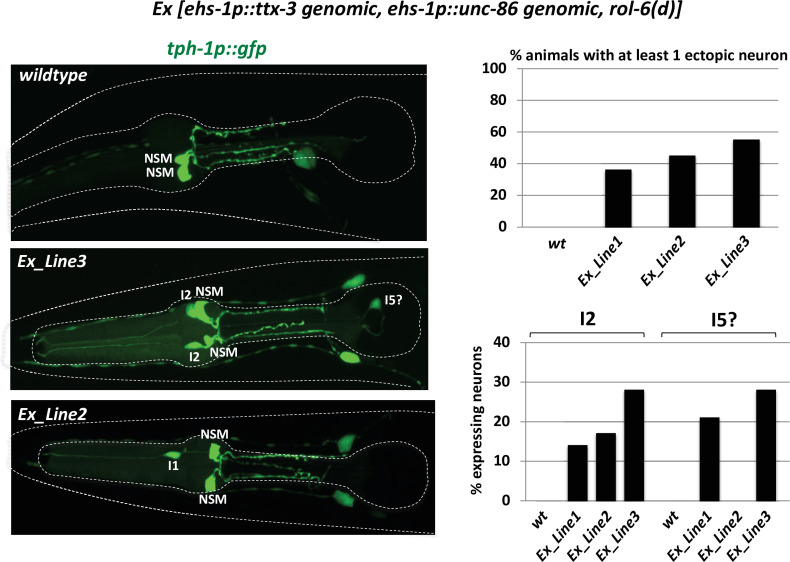
We found that the ectopic expression of pharyngeal homeobox genes reveal a limited capacity to respecify identity features of pharyngeal neuron (Figure 11—figure supplement 2). This is a likely reflection of our incomplete knowledge of the entire set of collaborating factors and possibly also a reflection of the difficulties associated with overriding endogenous terminal differentiation programs by ectopic expression of drivers of alternative fates (Patel and Hobert, 2017).
Taken together, our findings not only reveal a common terminal selector-based regulatory logic for how a self-contained, enteric nervous system acquires its terminal differentiated state. They also corroborate two themes that have emerged from recent studies, primarily in C. elegans (and also emerging in other systems):
Homeobox genes – and more specifically, combinatorial codes of homeobox genes – are prominently employed in neuron identity specification throughout all neurons of the nervous system, as exemplified here in the context of the enteric nervous system, the so-called ‘second brain’ of animals (Gershon, 1998).
A number of identity-specifying terminal selectors, such as CEH-34, are expressed in synaptically connected neurons, suggesting they may specify the assembly of neurons into functional circuitry. It is presently unclear how prominent such a connectivity theme is. We observed a few cases of such putative ‘circuit organizer’ transcription factors in the non-pharyngeal nervous system (Berghoff et al., 2021; Pereira et al., 2015) and there are some striking potential examples in vertebrates (Brunet and Pattyn, 2002; Dauger et al., 2003; Ha and Dougherty, 2018; Ruiz-Reig et al., 2019; Sokolowski et al., 2015). This present study provides an extreme example of this. An entire set of synaptically connected neurons (the worm’s enteric nervous system) is specified by a single transcription factor (CEH-34), which apparently helps these neurons to become assembled into functional circuitry. In each pharyngeal neuron type, CEH-34 pairs up with different homeodomain proteins to diversify pharyngeal neurons into distinct identities. Following Dobzhansky’s dictum that ‘nothing in biology makes sense except in the light of evolution’ (Dobzhansky, 1964), we speculate that the pharyngeal nervous system may have derived from a homogenous set of interconnected, identical neurons, all specified by ceh-34, which may have regulated a homophilic adhesion molecule that functionally linked these ancestral neurons. The more complex, present-day circuitry may have evolved through the eventual partnering of CEH-34 protein with distinct sets of homeodomain proteins that diversified neuronal identities, connectivity, and function in the pharyngeal nervous system.
Arguing for a conserved function of Six homeodomain factors in enteric nervous system differentiation is the observation that in flies, the Sine oculis paralog Optix is indeed expressed in the frontal ganglion (Seo et al., 1999), which constitutes the nervous system of insect foreguts (Hartenstein, 1997). In the context of studying Sine oculis function in the Drosophila corpus cardiacum, it was also noted that the entire stomatogastric ganglion, that is, the entire enteric nervous system (of which the frontal ganglion is a part) does not form in Sine oculis mutants (De Velasco et al., 2004). In sea urchin, pharyngeal neurons also express, and require for their proper development, the Sine oculis paralog Six3 (Wei et al., 2011). Other than an early report of Six2 expression in the mouse foregut region (Ohto et al., 1998), the expression and function of Sine oculis orthologs in vertebrate enteric nervous systems has, to our knowledge, not yet been examined.
Notably, another homeobox gene appears to have a critical and very broad function in vertebrate enteric nervous system development that is akin to the broadness of ceh-34 function in C. elegans. The paired-type homeobox gene Phox2b is expressed in enteric nervous system precursors and required early in development for the generation of all enteric ganglia (Pattyn et al., 1999; Tiveron et al., 1996). While Phox2b is also continuously expressed throughout the adult enteric nervous system (Corpening et al., 2008; Drokhlyansky et al., 2020; Morarach et al., 2021), its function in terminal differentiation and perhaps even maintenance of enteric neurons identity remains to be examined, for example, via temporally controlled, postdevelopmental knock-out in juvenile or adult stage animals. If the analogy to ceh-34 holds, the enteric neurons of such animals may lose their differentiated state. Remarkably, additional homeobox genes have recently been noted to show highly selective expression patterns within the vertebrate enteric nervous system, effectively discriminating distinct neuronal subtypes (Memic et al., 2018). One of them, the Meis ortholog Pbx3, has been confirmed to play an important role in the postmitotic specification of distinct enteric neuron types (Morarach et al., 2021). Hence, it appears that the overall logic of a pan-enteric homeobox gene, cooperating with cell type-specific homeobox genes, may be conserved from worms to vertebrates.
Another evolutionary perspective of our findings considers the origins of the enteric nervous system, and maybe nervous systems as a whole. Based on a number of anatomical and functional features, it has been proposed that enteric nervous systems preceded, and then paralleled the emergence of centralized nervous systems of bilaterian animals (Furness and Stebbing, 2018; Gilbert, 2019; Klimovich and Bosch, 2018). This argument is bolstered by considering a number of features of the enteric, that is, pharyngeal nervous system of C. elegans: (1) its polymodality (sensory + inter + motor neuron) of most pharyngeal neurons, (2) its innervation of what is essentially a single sheath of myoepithelial cells, a proposed feature of primitive nervous systems (Mackie, 1970), (3) its simple immune functions (also thought to be a feature of primitive neurons, e.g. in hydra; Klimovich et al., 2020; Klimovich and Bosch, 2018), and (4) the relatively indiscriminate synaptic cross-innervation patterns among pharyngeal neurons (Cook et al., 2020). If pharyngeal neurons indeed resemble a more primitive, ancestral state of neurons, our observation that CEH-34 acts as a terminal selector in these neurons would point to the ancient nature of (1) a terminal selector-type logic of neuronal identity specification and (2) the deployment of a homeobox gene in such function. Sine oculis homologs appear to be employed broadly in sensory neuron specification across animal phylogeny, even in the most basal metazoan (Jacobs et al., 2007). CEH-34/Sine oculis may represent an ancestral determinant of neuronal cell types.
Materials and methods
C. elegans strains
Worms were grown at 20°C on nematode growth media (NGM) plates seeded with E. coli (OP50) bacteria as a food source. The wild-type strain used is Bristol N2. A complete list of strains used in this study can be found in Supplementary file 1.
Generation of deletion alleles
Mutant alleles for the ceh-34, ceh-45, vab-15, ceh-7, ceh-53, ceh-79, and eya-1 genes (schematized in Figure 2 and Figure 11—figure supplement 1) were generated by CRISPR/Cas9 genome engineering as described (Dokshin et al., 2018). A deletion of the full locus was generated using two crRNAs and an ssODN donor. Sequences are as follows:
ceh-34(ot1014): crRNAs (cgacaagaggacgacgctct and ttattctaatggtcttgagg), ssODN (gcgacattcactgggggacgacaagaggacgacgccaagaccattagaataacttttaactatatttttg).
ceh-34(ot1188) and ceh-34(ot1189) were generated the same way as ceh-34(ot1014) and are molecularly identical. The difference is that ot1188 was generated in the background of flr-2(syb4861) and ot1189 was generated in the background of htrl-1(syb4895) because these loci are very closely linked to ceh-34.
ceh-45(ot1065): crRNAs (taggccaccgatacaagcag and tccgccagagaccggtcggg), ssODN (aactgaaattcgaaattctaggccaccgatacaaggaccggtctctggcggattactgtagccgtttggg).
vab-15(ot1136): crRNAs (ggtcaacacatctgcttata and ttgtgaaaagcgtaatactt), ssODN (agcgcgtggtgttatattggtcaacacatctgctttattacgcttttcacaatattttatggactaacca).
ceh-7(ot1138): crRNAs (ccccttgtactgacaattga and tgatcaggaatttgctctcg), ssODN (cgaaacgaaaacgggcggccccttgtactgacaatgagcaaattcctgatcatctgacacttttccagac).
ceh-53(ot1066): crRNAs (gcggcgcttccgggactctg and gaaatcaggggcaaacttgg), ssODN (gctccatcagaaaaaggggcggcgcttccgggactagtttgcccctgatttcgaatatttatgtgaaaaa).
ceh-79(ot1067): crRNAs (aagaagaaccgacgaaccca and cacccccgaactgtgttcac), ssODN (aactcctgtctctccttcgatgatcttttccatggcactggacacatatctttaacttttccgatgtgta).
eya-1(ot1197): crRNAs (ttttgtacgagtgactcagt and acacctgtatctctgcgggg), ssODN (cggtcgtcagattggtagccctccaaaatcccactcgcagagatacaggtgttcaaaatcggggtgaaga).
With the exception of ceh-34(ot1014), ceh-34(ot1188) and ceh-34(ot1189) animals, all other null mutant alleles are homozygous viable. ceh-45(ot1065) and vab-15(ot1136) are slow growing and at least ceh-45(ot1065) animals also display a partially penetrant embryonic lethality.
Generation of reporter knock-ins
The ceh-34 locus was tagged with mNG::3xFLAG::AID to generate ceh-34(ot903). The AID sequence was amplified and inserted into the pDD268 vector (mNG::SEC::3xFLAG) (Dickinson et al., 2015) to generate the plasmid pUA77 (ccdB::mNG::SEC::3xFLAG::AID ccdB; Aghayeva et al., 2021). The construct contains a self-excising drug selection cassette (SEC) and was used for SEC-mediated CRISPR insertion of mNG::3xFLAG::AID right before the stop codon of ceh-34 as described in Dickinson et al., 2015. The guide RNA used targets the following sequence: ttattctaatggtcttgagg.
The pha-4 locus was tagged with gfp at its 3’end to generate pha-4(ot946), using Cas9 protein, tracrRNA, and crRNA from IDT, as previously described (Dokshin et al., 2018). One crRNA (attggagatttataggttgg) and an asymmetric double-stranded gfp-loxP-3xFLAG cassette, amplified from a plasmid, were used to insert the fluorescent tag at the C-terminal.
Reporter alleles for flp-5(syb4513), flp-28(syb3207), flr-2(syb4861), htrl-1(syb4895), ser-7(syb4502), rig-3(syb4763), rig-6(syb4729), trh-1(syb4421), and trhr-1(syb4453) were generated by CRISPR/Cas9 to insert an SL2::GFP::H2B cassette at the C-terminus of the respective gene. For the unc-17(syb4491) allele T2A::GFP::H2B was inserted at the C-terminus. For the kin-36(syb4677) locus, a GFP::HIS::SL2 sequence was inserted at the N-terminus. These strains were generated by Sunybiotech and are listed in Supplementary file 1.
Generation of transgenic reporter strains
To generate otIs762(ceh-34prom::TagRFP) a 3720 bp PCR fragment containing the whole ceh-34 intergenic region plus the first 2 exons and 2 introns was amplified from N2 genomic DNA and cloned into a TagRFP vector using Gibson Assembly (NEBuilder HiFi DNA Assembly Master Mix, Catalog # E2621L). The following primers were used: aatgaaataagcttgcatgcctgcaTGTTTATTTTCTATGTAATTTCTAATAAAGTCCC and cccggggatcctctagagtcgacctgcaCTGAAAGTTGAAATATAGAATTTTTAATTTTTTTTTTTTG. The resulting construct was injected as a simple extrachromosomal array (50 ng/µl) into pha-1(e2123) animals, using a pha-1 rescuing plasmid (pBX, 50 ng/µl) as co-injection marker. A representative line was integrated into the genome with gamma irradiation and backcrossed four times.
To generate otIs785(ceh-34prom::GFP::CLA-1), a 3720 bp PCR fragment containing the whole ceh-34 intergenic region plus the first 2 exons and 2 introns was amplified from N2 genomic DNA and cloned into PK065 (kindly shared by Peri Kurshan) using Gibson Assembly (NEBuilder HiFi DNA Assembly Master Mix, Catalog # E2621L). The following primers were used: gattacgccaagcttgcatgcTGTTTATTTTCTATGTAATTTCTAATAAAGTC and gttcttctcctttactcatcccgggCTGAAAGTTGAAATATAGAATTTTTAATTTTTTTTTTTTG. The resulting construct was injected at 7 ng/µl together with ceh-34prom::TagRFP (50 ng/µl) (see above) and rol-6(su1006) as a co-injection marker. A representative line was integrated into the genome with gamma irradiation and backcrossed four times.
Choice of pharyngeal fate markers
Most fate markers used in this paper were previously described. For several of those, we used previous expression patterns (based on transgenic reporter fusions) as an impetus to then generate reporter alleles by CRISPR/Cas9 genome engineering (e.g. flp-5, ser-7; all listed in previous sections). One case warrants specific emphasis: scRNA has shown that the T11F9.12 gene is expressed exclusively in most, if not all pharyngeal neurons, a notion we confirmed with a CRISPR/Cas9 genome-engineered reporter allele. T11F9.12 encodes for a relatively large (736aa), secreted and nematode-specific protein that contains a Pfam-annoted domain (Htrl domain; PF09612) that is, outside nematodes, only found in a bacterial protein HtrL. This bacterial protein currently has no assigned function but is found in a region of LPS core biosynthesis genes which are involved in bacterial immune defense (Bertani and Ruiz, 2018). Interestingly, hidden Markov model-based searches in the Panther database reveal a sequence pattern (PTHR21579) along the entire T11F9.12 protein that is otherwise only found in C. elegans saposin proteins, which are bona fide immune effector proteins (Bányai and Patthy, 1998; Hoeckendorf et al., 2012; Oishi et al., 2009). We named this protein HTRL-1.
Temporally controlled CEH-34 protein degradation
We used conditional protein depletion with a modified auxin-inducible degradation system (C.e.AIDv2; Hills-Muckey et al., 2022). AID-tagged proteins are conditionally degraded when exposed to 5-Ph-IAA in the presence of AtTIR1F79G. To generate the experimental strain, the conditional allele ceh-34(ot903[ceh-34::mNG::AID]) was crossed with cshIs140[rps-28p::TIR1(F79G)], which expresses AtTIR1F79G ubiquitously. The synthetic auxin analog 5-Ph-IAA was purchased from BioAcademia (#30-003-10) and dissolved in ethanol (EtOH) to prepare 100 mM stock solutions. NGM agar plates with fully grown OP50 bacterial lawn were coated with the 5-Ph-IAA stock solution to a final concentration of 200 μM and allowed to dry overnight at room temperature. To induce protein degradation, synchronized L1 or young adult worms were transferred onto 5-Ph-IAA -coated plates and kept at 20°C. As a control, worms were transferred onto EtOH-coated plates instead. 5-Ph-IAA solutions and experimental plates were shielded from light.
Microscopy and image analysis
Worms were anesthetized using 100 mM sodium azide (NaN3) and mounted on 5% agarose pads on glass slides. Z-stack images (each ~0.7 µm thick) were acquired using a Zeiss confocal microscope (LSM880) or Zeiss compound microscope (Imager Z2) with the ZEN software. Maximum intensity projections of 2–30 slices were generated with the ImageJ software (Schindelin et al., 2012).
Reporter gene expression in different neurons was visualized in wild-type and mutant animals and usually assigned to one of the following categories: ‘on’ (fluorescence levels comparable to wild-type animals), ‘dim’ (fluorescence still detectable but much dimmer than wild-type animals), or ‘off’ (fluorescence not detectable). In cases were fluorescence levels were variable between animals and difference with wild type was not obvious mean fluorescence intensity in each neuron was measured with the ImageJ software.
Acknowledgements
We thank Chi Chen for generating transgenic lines, Seth Taylor and Michael Gershon for discussion, Kelly Liu, Robert Horvitz, Peri Kurshan, and Matthias Leippe for providing reagents, Steven Cook for providing illustrations and Michael Gershon and members of the Hobert lab for comments on the manuscript. This work was funded by NIH R21NS106843, NIHR01NS039996, and the Howard Hughes Medical Institute.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Oliver Hobert, Email: or38@columbia.edu.
Douglas Portman, University of Rochester, United States.
Piali Sengupta, Brandeis University, United States.
Funding Information
This paper was supported by the following grants:
Howard Hughes Medical Institute to Oliver Hobert.
National Institutes of Health R01 NS039996 to Oliver Hobert.
National Institutes of Health R21NS106843 to Oliver Hobert.
Additional information
Competing interests
No competing interests declared.
No competing interests declared.
Author contributions
Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing - review and editing.
Formal analysis, Investigation.
Conceptualization, Data curation, Formal analysis, Investigation, Visualization, Writing - review and editing.
Formal analysis, Investigation, Visualization.
Formal analysis, Investigation, Visualization.
Formal analysis, Investigation, Visualization.
Conceptualization, Funding acquisition, Project administration, Supervision, Writing - original draft.
Additional files
This file provides a list of all Caenorhabditis elegans strains used in this study.
Data availability
All data generated or analysed during this study are included in the manuscript and supporting files.
References
- Abot A, Cani PD, Knauf C. Impact of Intestinal Peptides on the Enteric Nervous System: Novel Approaches to Control Glucose Metabolism and Food Intake. Frontiers in Endocrinology. 2018;9:328. doi: 10.3389/fendo.2018.00328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghayeva U, Bhattacharya A, Sural S, Jaeger E, Churgin M, Fang-Yen C, Hobert O. DAF-16/FoxO and DAF-12/VDR CControl Cellular Plasticity Both Cell-Autonomously and via Interorgan SignalingControl Cellular Plasticity Both Cell-Autonomously and via Interorgan Signaling. bioRxiv. 2021 doi: 10.1101/2020.12.15.422925. [DOI] [PMC free article] [PubMed]
- Albertson DG, Thomson JN. The pharynx of Caenorhabditis elegans. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 1976;275:299–325. doi: 10.1098/rstb.1976.0085. [DOI] [PubMed] [Google Scholar]
- Amin NM, Lim SE, Shi H, Chan TL, Liu J. A conserved Six-Eya cassette acts downstream of Wnt signaling to direct non-myogenic versus myogenic fates in the C. elegans postembryonic mesoderm. Developmental Biology. 2009;331:350–360. doi: 10.1016/j.ydbio.2009.05.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt D. The evolution of cell types in animals: emerging principles from molecular studies. Nature Reviews. Genetics. 2008;9:868–882. doi: 10.1038/nrg2416. [DOI] [PubMed] [Google Scholar]
- Aspöck G, Ruvkun G, Bürglin TR. The Caenorhabditis elegans ems class homeobox gene ceh-2 is required for M3 pharynx motoneuron function. Development (Cambridge, England) 2003;130:3369–3378. doi: 10.1242/dev.00551. [DOI] [PubMed] [Google Scholar]
- Avery L, Horvitz HR. Pharyngeal pumping continues after laser killing of the pharyngeal nervous system of C. elegans. Neuron. 1989;3:473–485. doi: 10.1016/0896-6273(89)90206-7. [DOI] [PubMed] [Google Scholar]
- Avery L. C. Elegans Feeding. WormBook; 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axäng C, Rauthan M, Hall DH, Pilon M. Developmental genetics of the C. elegans pharyngeal neurons NSML and NSMR. BMC Developmental Biology. 2008;8:38. doi: 10.1186/1471-213X-8-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayali A. The insect frontal ganglion and stomatogastric pattern generator networks. Neuro-Signals. 2004;13:20–36. doi: 10.1159/000076156. [DOI] [PubMed] [Google Scholar]
- Babu K, Hu Z, Chien SC, Garriga G, Kaplan JM. The immunoglobulin super family protein RIG-3 prevents synaptic potentiation and regulates Wnt signaling. Neuron. 2011;71:103–116. doi: 10.1016/j.neuron.2011.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bányai L, Patthy L. Amoebapore homologs of Caenorhabditis elegans. Biochimica et Biophysica Acta. 1998;1429:259–264. doi: 10.1016/s0167-4838(98)00237-4. [DOI] [PubMed] [Google Scholar]
- Baumeister R, Liu Y, Ruvkun G. Lineage-specific regulators couple cell lineage asymmetry to the transcription of the Caenorhabditis elegans POU gene unc-86 during neurogenesis. Genes & Development. 1996;10:1395–1410. doi: 10.1101/gad.10.11.1395. [DOI] [PubMed] [Google Scholar]
- Berghoff EG, Glenwinkel L, Bhattacharya A, Sun H, Varol E, Mohammadi N, Antone A, Feng Y, Nguyen K, Cook SJ, Wood JF, Masoudi N, Cros CC, Ramadan YH, Ferkey DM, Hall DH, Hobert O. The Prop1-like homeobox gene unc-42 specifies the identity of synaptically connected neurons. eLife. 2021;10:e64903. doi: 10.7554/eLife.64903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani B, Ruiz N. Function and Biogenesis of Lipopolysaccharides. EcoSal Plus. 2018;8:ESP-0001-2018. doi: 10.1128/ecosalplus.ESP-0001-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj A, Pandey P, Babu K. Control of locomotory behavio Locomotory Behavior of Caenorhabditis elegans by the immunoglobulin superfamily proteinImmunoglobulin Superfamily Protein RIG-3. Genetics. 2020;214:135–145. doi: 10.1534/genetics.119.302872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatla N, Horvitz HR. Light and hydrogen peroxide inhibit C. elegans Feeding through gustatory receptor orthologs and pharyngeal neurons. Neuron. 2015;85:804–818. doi: 10.1016/j.neuron.2014.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockie PJ, Madsen DM, Zheng Y, Mellem J, Maricq AV. Differential expression of glutamate receptor subunits in the nervous system of Caenorhabditis elegans and their regulation by the homeodomain protein UNC-42. The Journal of Neuroscience. 2001;21:1510–1522. doi: 10.1523/JNEUROSCI.21-05-01510.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet JF, Pattyn A. Phox2 genes - from patterning to connectivity. Current Opinion in Genetics & Development. 2002;12:435–440. doi: 10.1016/s0959-437x(02)00322-2. [DOI] [PubMed] [Google Scholar]
- Cheyette BN, Green PJ, Martin K, Garren H, Hartenstein V, Zipursky SL. The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron. 1994;12:977–996. doi: 10.1016/0896-6273(94)90308-5. [DOI] [PubMed] [Google Scholar]
- Colosimo ME, Brown A, Mukhopadhyay S, Gabel C, Lanjuin AE, Samuel ADT, Sengupta P. Identification of thermosensory and olfactory neuron-specific genes via expression profiling of single neuron types. Current Biology. 2004;14:2245–2251. doi: 10.1016/j.cub.2004.12.030. [DOI] [PubMed] [Google Scholar]
- Cook SJ, Jarrell TA, Brittin CA, Wang Y, Bloniarz AE, Yakovlev MA, Nguyen KCQ, Tang LTH, Bayer EA, Duerr JS, Bülow HE, Hobert O, Hall DH, Emmons SW. Whole-animal connectomes of both Caenorhabditis elegans sexes. Nature. 2019;571:63–71. doi: 10.1038/s41586-019-1352-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook SJ, Crouse CM, Yemini E, Hall DH, Emmons SW, Hobert O. The connectome of the Caenorhabditis elegans pharynx. The Journal of Comparative Neurology. 2020;528:2767–2784. doi: 10.1002/cne.24932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver PF. How to innervate a simple gut: familiar themes and unique aspects in the formation of the insect enteric nervous system. Developmental Dynamics. 2007;236:1841–1864. doi: 10.1002/dvdy.21138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpening JC, Cantrell VA, Deal KK, Southard-Smith EM. A Histone2BCerulean BAC transgene identifies differential expression of Phox2b in migrating enteric neural crest derivatives and enteric glia. Developmental Dynamics. 2008;237:1119–1132. doi: 10.1002/dvdy.21498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croset V, Rytz R, Cummins SF, Budd A, Brawand D, Kaessmann H, Gibson TJ, Benton R. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLOS Genetics. 2010;6:e1001064. doi: 10.1371/journal.pgen.1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauger S, Pattyn A, Lofaso F, Gaultier C, Goridis C, Gallego J, Brunet JF. Phox2b controls the development of peripheral chemoreceptors and afferent visceral pathways. Development (Cambridge, England) 2003;130:6635–6642. doi: 10.1242/dev.00866. [DOI] [PubMed] [Google Scholar]
- De Velasco B, Shen J, Go S, Hartenstein V. Embryonic development of the Drosophila corpus cardiacum, a neuroendocrine gland with similarity to the vertebrate pituitary, is controlled by sine oculis and glass. Developmental Biology. 2004;274:280–294. doi: 10.1016/j.ydbio.2004.07.015. [DOI] [PubMed] [Google Scholar]
- Dickinson DJ, Pani AM, Heppert JK, Higgins CD, Goldstein B. Streamlined Genome Engineering with a Self-Excising Drug Selection Cassette. Genetics. 2015;200:1035–1049. doi: 10.1534/genetics.115.178335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierking K, Yang W, Schulenburg H. Antimicrobial effectors in the nematode Caenorhabditis elegans: an outgroup to the Arthropoda. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 2016;371:20150299. doi: 10.1098/rstb.2015.0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. Biology, molecular and organismic. American Zoologist. 1964;4:443–452. doi: 10.1093/icb/4.4.443. [DOI] [PubMed] [Google Scholar]
- Dokshin GA, Ghanta KS, Piscopo KM, Mello CC. Robust Genome Editing with Short Single-Stranded and Long, Partially Single-Stranded DNA Donors in Caenorhabditis elegans. Genetics. 2018;210:781–787. doi: 10.1534/genetics.118.301532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dozier C, Kagoshima H, Niklaus G, Cassata G, Bürglin TR. The Caenorhabditis elegans Six/sine oculis class homeobox gene ceh-32 is required for head morphogenesis. Developmental Biology. 2001;236:289–303. doi: 10.1006/dbio.2001.0325. [DOI] [PubMed] [Google Scholar]
- Drokhlyansky E, Smillie CS, Van Wittenberghe N, Ericsson M, Griffin GK, Eraslan G, Dionne D, Cuoco MS, Goder-Reiser MN, Sharova T, Kuksenko O, Aguirre AJ, Boland GM, Graham D, Rozenblatt-Rosen O, Xavier RJ, Regev A. The Human and Mouse Enteric Nervous System at Single-Cell Resolution. Cell. 2020;182:1606–1622. doi: 10.1016/j.cell.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Chalfie M. Genes regulating touch cell development in Caenorhabditis elegans. Genetics. 2001;158:197–207. doi: 10.1093/genetics/158.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Hope IA. The Caenorhabditis elegans homeobox gene ceh-19 is required for MC motorneuron function. Genesis (New York) 2013;51:163–178. doi: 10.1002/dvg.22365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung C, Vanden Berghe P. Functional circuits and signal processing in the enteric nervous system. Cellular and Molecular Life Sciences. 2020;77:4505–4522. doi: 10.1007/s00018-020-03543-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furness JB. Types of neurons in the enteric nervous system. Journal of the Autonomic Nervous System. 2000;81:87–96. doi: 10.1016/s0165-1838(00)00127-2. [DOI] [PubMed] [Google Scholar]
- Furness JB, Stebbing MJ. The first brain: Species comparisons and evolutionary implications for the enteric and central nervous systems. Neurogastroenterology and Motility. 2018;30:nmo.13234. doi: 10.1111/nmo.13234. [DOI] [PubMed] [Google Scholar]
- Furuya M, Qadota H, Chisholm AD, Sugimoto A. The C. elegans eyes absent ortholog EYA-1 is required for tissue differentiation and plays partially redundant roles with PAX-6. Developmental Biology. 2005;286:452–463. doi: 10.1016/j.ydbio.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Ganz J. Gut feelings: Studying enteric nervous system development, function, and disease in the zebrafish model system. Developmental Dynamics. 2018;247:268–278. doi: 10.1002/dvdy.24597. [DOI] [PubMed] [Google Scholar]
- Gaudet J, Mango SE. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science (New York, N.Y.) 2002;295:821–825. doi: 10.1126/science.1065175. [DOI] [PubMed] [Google Scholar]
- Gershon MD. The Second Brain. HarperCollins; 1998. [Google Scholar]
- Gershon MD. 5-Hydroxytryptamine (serotonin) in the gastrointestinal tract. Current Opinion in Endocrinology, Diabetes, and Obesity. 2013;20:14–21. doi: 10.1097/MED.0b013e32835bc703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SF. Evolutionary transitions revisited: Holobiont evo-devo. Journal of Experimental Zoology. Part B, Molecular and Developmental Evolution. 2019;332:307–314. doi: 10.1002/jez.b.22903. [DOI] [PubMed] [Google Scholar]
- Glenwinkel L, Taylor SR, Langebeck-Jensen K, Pereira L, Reilly MB, Basavaraju M, Rafi I, Yemini E, Pocock R, Sestan N, Hammarlund M, Miller DM, Hobert O. In silico analysis of the transcriptional regulatory logic of neuronal identity specification throughout the C. elegans nervous system. eLife. 2021;10:e64906. doi: 10.7554/eLife.64906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha NT, Dougherty KJ. Spinal Shox2 interneuron interconnectivity related to function and development. eLife. 2018;7:e42519. doi: 10.7554/eLife.42519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DH, Altun Z. C. Elegans Atlas. Cold Spring Harbor Laboratory Press; 2007. [Google Scholar]
- Hanson IM. Mammalian homologues of the Drosophila eye specification genes. Seminars in Cell & Developmental Biology. 2001;12:475–484. doi: 10.1006/scdb.2001.0271. [DOI] [PubMed] [Google Scholar]
- Hao JC, Yu TW, Fujisawa K, Culotti JG, Gengyo-Ando K, Mitani S, Moulder G, Barstead R, Tessier-Lavigne M, Bargmann CI. C. elegans slit acts in midline, dorsal-ventral, and anterior-posterior guidance via the SAX-3/Robo receptor. Neuron. 2001;32:25–38. doi: 10.1016/s0896-6273(01)00448-2. [DOI] [PubMed] [Google Scholar]
- Hao MM, Young HM. Development of enteric neuron diversity. Journal of Cellular and Molecular Medicine. 2009;13:1193–1210. doi: 10.1111/j.1582-4934.2009.00813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstein V. Development of the insect stomatogastric nervous system. Trends in Neurosciences. 1997;20:421–427. doi: 10.1016/s0166-2236(97)01066-7. [DOI] [PubMed] [Google Scholar]
- Hills-Muckey K, Martinez MAQ, Stec N, Hebbar S, Saldanha J, Medwig-Kinney TN, Moore FEQ, Ivanova M, Morao A, Ward JD, Moss EG, Ercan S, Zinovyeva AY, Matus DQ, Hammell CM. An engineered, orthogonal auxin analog/AtTIR1(F79G) pairing improves both specificity and efficacy of the auxin degradation system in Caenorhabditis elegans. Genetics. 2022;220:iyab174. doi: 10.1093/genetics/iyab174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T, Galvin BD, Horvitz HR. Six and Eya promote apoptosis through direct transcriptional activation of the proapoptotic BH3-only gene egl-1 in Caenorhabditis elegans. PNAS. 2010;107:15479–15484. doi: 10.1073/pnas.1010023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. The neuronal genome of Caenorhabditis elegans. WormBook: The Online Review of C. Elegans Biology. 2013;1:1–106. doi: 10.1895/wormbook.1.161.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. Terminal Selectors of Neuronal Identity. Current Topics in Developmental Biology. 2016;116:455–475. doi: 10.1016/bs.ctdb.2015.12.007. [DOI] [PubMed] [Google Scholar]
- Hobert O. Homeobox genes and the specification of neuronal identity. Nature Reviews. Neuroscience. 2021;22:627–636. doi: 10.1038/s41583-021-00497-x. [DOI] [PubMed] [Google Scholar]
- Hobson RJ, Hapiak VM, Xiao H, Buehrer KL, Komuniecki PR, Komuniecki RW. SER-7, a Caenorhabditis elegans 5-HT7-like receptor, is essential for the 5-HT stimulation of pharyngeal pumping and egg laying. Genetics. 2006;172:159–169. doi: 10.1534/genetics.105.044495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeckendorf A, Stanisak M, Leippe M. The saposin-like protein SPP-12 is an antimicrobial polypeptide in the pharyngeal neurons of Caenorhabditis elegans and participates in defence against a natural bacterial pathogen. The Biochemical Journal. 2012;445:205–212. doi: 10.1042/BJ20112102. [DOI] [PubMed] [Google Scholar]
- Horner MA, Quintin S, Domeier ME, Kimble J, Labouesse M, Mango SE. pha-4, an HNF-3 homolog, specifies pharyngeal organ identity in Caenorhabditis elegans. Genes & Development. 1998;12:1947–1952. doi: 10.1101/gad.12.13.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvitz HR, Chalfie M, Trent C, Sulston JE, Evans PD. Serotonin and octopamine in the nematode Caenorhabditis elegans. Science (New York, N.Y.) 1982;216:1012–1014. doi: 10.1126/science.6805073. [DOI] [PubMed] [Google Scholar]
- Hsu HT, Chen HM, Yang Z, Wang J, Lee NK, Burger A, Zaret K, Liu T, Levine E, Mango SE. TRANSCRIPTION. Recruitment of RNA polymerase II by the pioneer TRANSCRIPTION factor PHA-4. Science (New York, N.Y.) 2015;348:1372–1376. doi: 10.1126/science.aab1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt-Newbury R, Viveiros R, Johnsen R, Mah A, Anastas D, Fang L, Halfnight E, Lee D, Lin J, Lorch A, McKay S, Okada HM, Pan J, Schulz AK, Tu D, Wong K, Zhao Z, Alexeyenko A, Burglin T, Sonnhammer E, Schnabel R, Jones SJ, Marra MA, Baillie DL, Moerman DG. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLOS Biology. 2007;5:e237. doi: 10.1371/journal.pbio.0050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishita Y, Chihara T, Okumura M. Serotonergic modulation of feeding behavior in Caenorhabditis elegans and other related nematodes. Neuroscience Research. 2020;154:9–19. doi: 10.1016/j.neures.2019.04.006. [DOI] [PubMed] [Google Scholar]
- Jacobs DK, Nakanishi N, Yuan D, Camara A, Nichols SA, Hartenstein V. Evolution of sensory structures in basal metazoa. Integrative and Comparative Biology. 2007;47:712–723. doi: 10.1093/icb/icm094. [DOI] [PubMed] [Google Scholar]
- Kalb JM, Lau KK, Goszczynski B, Fukushige T, Moons D, Okkema PG, McGhee JD. pha-4 is Ce-fkh-1, a fork head/HNF-3alpha,beta,gamma homolog that functions in organogenesis of the C. elegans pharynx. Development (Cambridge, England) 1998;125:2171–2180. doi: 10.1242/dev.125.12.2171. [DOI] [PubMed] [Google Scholar]
- Katidou M, Tavernarakis N, Karagogeos D. The contactin RIG-6 mediates neuronal and non-neuronal cell migration in Caenorhabditis elegans. Developmental Biology. 2013;373:184–195. doi: 10.1016/j.ydbio.2012.10.027. [DOI] [PubMed] [Google Scholar]
- Kato Y, Aizawa T, Hoshino H, Kawano K, Nitta K, Zhang H. abf-1 and abf-2, ASABF-type antimicrobial peptide genes in Caenorhabditis elegans. The Biochemical Journal. 2002;361:221–230. doi: 10.1042/0264-6021:3610221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Emmons SW. Multiple conserved cell adhesion protein interactions mediate neural wiring of a sensory circuit in C. elegans. eLife. 2017;6:e29257. doi: 10.7554/eLife.29257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimovich AV, Bosch TCG. Rethinking the Role of the Nervous System: Lessons From the Hydra Holobiont. BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology. 2018;40:201800060. doi: 10.1002/bies.201800060. [DOI] [PubMed] [Google Scholar]
- Klimovich A, Giacomello S, Björklund Å, Faure L, Kaucka M, Giez C, Murillo-Rincon AP, Matt AS, Willoweit-Ohl D, Crupi G, de Anda J, Wong GCL, D’Amato M, Adameyko I, Bosch TCG. Prototypical pacemaker neurons interact with the resident microbiota. PNAS. 2020;117:17854–17863. doi: 10.1073/pnas.1920469117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi O. Nerve ring of the hypostome in hydra: is it an origin of the central nervous system of bilaterian animals? Brain, Behavior and Evolution. 2007;69:151–159. doi: 10.1159/000095204. [DOI] [PubMed] [Google Scholar]
- Kumar JP. The sine oculis homeobox (SIX) family of transcription factors as regulators of development and disease. Cellular and Molecular Life Sciences. 2009;66:565–583. doi: 10.1007/s00018-008-8335-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laranjeira C, Pachnis V. Enteric nervous system development: Recent progress and future challenges. Autonomic Neuroscience. 2009;151:61–69. doi: 10.1016/j.autneu.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Llewellyn-Smith IJ. In: In Regulatory Peptides. Polak JM, editor. Basel: Birkhäuser Basel; 1989. Neuropeptides and the microcircuitry of the enteric nervous system; pp. 247–265. [Google Scholar]
- Ma X, Zhao Z, Xiao L, Xu W, Kou Y, Zhang Y, Wu G, Wang Y, Du Z. A 4D single-cell protein atlas of transcription factors delineates spatiotemporal patterning during embryogenesis. Nature Methods. 2021;18:893–902. doi: 10.1038/s41592-021-01216-1. [DOI] [PubMed] [Google Scholar]
- Mackie GO. In: Neuroid Conduction and the Evolution of Conducting Tissues. Mackie GO, editor. The University of Chicago Press; 1970. Q Rev Biol; pp. 319–332. [DOI] [PubMed] [Google Scholar]
- Mango SE, Lambie EJ, Kimble J. The pha-4 gene is required to generate the pharyngeal primordium of Caenorhabditis elegans. Development (Cambridge, England) 1994;120:3019–3031. doi: 10.1242/dev.120.10.3019. [DOI] [PubMed] [Google Scholar]
- Mango SE. In: The C. Elegans Pharynx: A Model for Organogenesis. Mango SE, editor. Oxford University Press; 2007. WormBook: The Online Review of C. Elegans Biology; pp. 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memic F, Knoflach V, Morarach K, Sadler R, Laranjeira C, Hjerling-Leffler J, Sundström E, Pachnis V, Marklund U. Transcription and Signaling Regulators in Developing Neuronal Subtypes of Mouse and Human Enteric Nervous System. Gastroenterology. 2018;154:624–636. doi: 10.1053/j.gastro.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morarach K, Mikhailova A, Knoflach V, Memic F, Kumar R, Li W, Ernfors P, Marklund U. Diversification of molecularly defined myenteric neuron classes revealed by single-cell RNA sequencing. Nature Neuroscience. 2021;24:34–46. doi: 10.1038/s41593-020-00736-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mörck C, Rauthan M, Wågberg F, Pilon M. pha-2 encodes the C. elegans ortholog of the homeodomain protein HEX and is required for the formation of the pharyngeal isthmus. Developmental Biology. 2004;272:403–418. doi: 10.1016/j.ydbio.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Muniz LR, Knosp C, Yeretssian G. Intestinal antimicrobial peptides during homeostasis, infection, and disease. Frontiers in Immunology. 2012;3:310. doi: 10.3389/fimmu.2012.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers L, Perera H, Alvarado MG, Kidd T. The Drosophila Ret gene functions in the stomatogastric nervous system with the Maverick TGFbeta ligand and the Gfrl co-receptor. Development (Cambridge, England) 2018;145:dev.157446. doi: 10.1242/dev.157446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy N, Goldstein AM. Enteric nervous system development: A crest cell’s journey from neural tube to colon. Seminars in Cell & Developmental Biology. 2017;66:94–106. doi: 10.1016/j.semcdb.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan K, Lambert SA, Yang AWH, Riddell J, Mnaimneh S, Zheng H, Albu M, Najafabadi HS, Reece-Hoyes JS, Fuxman Bass JI, Walhout AJM, Weirauch MT, Hughes TR. Mapping and analysis of Caenorhabditis elegans transcription factor sequence specificities. eLife. 2015;4:e06967. doi: 10.7554/eLife.06967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto H, Takizawa T, Saito T, Kobayashi M, Ikeda K, Kawakami K. Tissue and developmental distribution of Six family gene products. The International Journal of Developmental Biology. 1998;42:141–148. [PubMed] [Google Scholar]
- Ohto H, Kamada S, Tago K, Tominaga SI, Ozaki H, Sato S, Kawakami K. Cooperation of six and eya in activation of their target genes through nuclear translocation of Eya. Molecular and Cellular Biology. 1999;19:6815–6824. doi: 10.1128/MCB.19.10.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi A, Gengyo-Ando K, Mitani S, Mohri-Shiomi A, Kimura KD, Ishihara T, Katsura I. FLR-2, the glycoprotein hormone alpha subunit, is involved in the neural control of intestinal functions in Caenorhabditis elegans. Genes to Cells. 2009;14:1141–1154. doi: 10.1111/j.1365-2443.2009.01341.x. [DOI] [PubMed] [Google Scholar]
- Patel T, Hobert O. Coordinated control of terminal differentiation and restriction of cellular plasticity. eLife. 2017;6:e24100. doi: 10.7554/eLife.24100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick AN, Cabrera JH, Smith AL, Chen XS, Ford HL, Zhao R. Structure-function analyses of the human SIX1-EYA2 complex reveal insights into metastasis and BOR syndrome. Nature Structural & Molecular Biology. 2013;20:447–453. doi: 10.1038/nsmb.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattyn A, Morin X, Cremer H, Goridis C, Brunet JF. The homeobox gene Phox2b is essential for the development of autonomic neural crest derivatives. Nature. 1999;399:366–370. doi: 10.1038/20700. [DOI] [PubMed] [Google Scholar]
- Pereira L, Kratsios P, Serrano-Saiz E, Sheftel H, Mayo AE, Hall DH, White JG, LeBoeuf B, Garcia LR, Alon U, Hobert O. A cellular and regulatory map of the cholinergic nervous system of C. elegans. eLife. 2015;4:e12432. doi: 10.7554/eLife.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon M. Fishing lines, time-delayed guideposts, and other tricks used by developing pharyngeal neurons in Caenorhabditis elegans. Developmental Dynamics. 2008;237:2073–2080. doi: 10.1002/dvdy.21636. [DOI] [PubMed] [Google Scholar]
- Portereiko MF, Mango SE. Early morphogenesis of the Caenorhabditis elegans pharynx. Developmental Biology. 2001;233:482–494. doi: 10.1006/dbio.2001.0235. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan K, Okkema PG. Regulation of C. elegans neuronal differentiation by the ZEB-family factor ZAG-1 and the NK-2 homeodomain factor CEH-28. PLOS ONE. 2014;9:e113893. doi: 10.1371/journal.pone.0113893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao M, Gershon MD. Enteric nervous system development: what could possibly go wrong? Nature Reviews. Neuroscience. 2018;19:552–565. doi: 10.1038/s41583-018-0041-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauthan M, Mörck C, Pilon M. The C. elegans M3 neuron guides the growth cone of its sister cell M2 via the Krüppel-like zinc finger protein MNM-2. Developmental Biology. 2007;311:185–199. doi: 10.1016/j.ydbio.2007.08.037. [DOI] [PubMed] [Google Scholar]
- Ray P, Schnabel R, Okkema PG. Behavioral and synaptic defects in C. elegans lacking the NK-2 homeobox gene ceh-28. Developmental Neurobiology. 2008;68:421–433. doi: 10.1002/dneu.20599. [DOI] [PubMed] [Google Scholar]
- Reilly MB, Cros C, Varol E, Yemini E, Hobert O. Unique homeobox codes delineate all the neuron classes of C. elegans. Nature. 2020;584:595–601. doi: 10.1038/s41586-020-2618-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Reig N, Rakotobe M, Bethus I, Le Menn G, Huditz H-I, Marie H, Lamonerie T, D’Autréaux F. Developmental Requirement of Homeoprotein Otx2 for Specific Habenulo-Interpeduncular Subcircuits. The Journal of Neuroscience. 2019;39:1005–1019. doi: 10.1523/JNEUROSCI.1818-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvkun G, Hobert O. The taxonomy of developmental control in Caenorhabditis elegans. Science (New York, N.Y.) 1998;282:2033–2041. doi: 10.1126/science.282.5396.2033. [DOI] [PubMed] [Google Scholar]
- Sasselli V, Pachnis V, Burns AJ. The enteric nervous system. Developmental Biology. 2012;366:64–73. doi: 10.1016/j.ydbio.2012.01.012. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nature Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz V, Pan J, Voltmer-Irsch S, Hutter H. IgCAMs redundantly control axon navigation in Caenorhabditis elegans. Neural Development. 2009;4:13. doi: 10.1186/1749-8104-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HC, Curtiss J, Mlodzik M, Fjose A. Six class homeobox genes in Drosophila belong to three distinct families and are involved in head development. Mechanisms of Development. 1999;83:127–139. doi: 10.1016/s0925-4773(99)00045-3. [DOI] [PubMed] [Google Scholar]
- Serrano-Saiz E, Poole RJ, Felton T, Zhang F, De La Cruz ED, Hobert O. Modular control of glutamatergic neuronal identity in C. elegans by distinct homeodomain proteins. Cell. 2013;155:659–673. doi: 10.1016/j.cell.2013.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Saiz E, Leyva-Díaz E, De La Cruz E, Hobert O. BRN3-type POU Homeobox Genes Maintain the Identity of Mature Postmitotic Neurons in Nematodes and Mice. Current Biology. 2018;28:2813–2823. doi: 10.1016/j.cub.2018.06.045. [DOI] [PubMed] [Google Scholar]
- Smit RB, Schnabel R, Gaudet J. The HLH-6 transcription factor regulates C. elegans pharyngeal gland development and function. PLOS Genetics. 2008;4:e1000222. doi: 10.1371/journal.pgen.1000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolowski K, Esumi S, Hirata T, Kamal Y, Tran T, Lam A, Oboti L, Brighthaupt S-C, Zaghlula M, Martinez J, Ghimbovschi S, Knoblach S, Pierani A, Tamamaki N, Shah NM, Jones KS, Corbin JG. Specification of select hypothalamic circuits and innate behaviors by the embryonic patterning gene dbx1. Neuron. 2015;86:403–416. doi: 10.1016/j.neuron.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B, Avery L. Serotonin activates overall feeding by activating two separate neural pathways in Caenorhabditis elegans. The Journal of Neuroscience. 2012;32:1920–1931. doi: 10.1523/JNEUROSCI.2064-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song BM, Avery L. The pharynx of the nematode C. elegans: A model system for the study of motor control. Worm. 2013;2:e21833. doi: 10.4161/worm.21833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Developmental Biology. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Tadjuidje E, Hegde RS. The Eyes Absent proteins in development and disease. Cellular and Molecular Life Sciences. 2013;70:1897–1913. doi: 10.1007/s00018-012-1144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SR, Santpere G, Weinreb A, Barrett A, Reilly MB, Xu C, Varol E, Oikonomou P, Glenwinkel L, McWhirter R, Poff A, Basavaraju M, Rafi I, Yemini E, Cook SJ, Abrams A, Vidal B, Cros C, Tavazoie S, Sestan N, Hammarlund M, Hobert O, Miller DM. Molecular topography of an entire nervous system. Cell. 2021;184:4329–4347. doi: 10.1016/j.cell.2021.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiveron MC, Hirsch MR, Brunet JF. The expression pattern of the transcription factor Phox2 delineates synaptic pathways of the autonomic nervous system. The Journal of Neuroscience. 1996;16:7649–7660. doi: 10.1523/JNEUROSCI.16-23-07649.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojanowski NF, Padovan-Merhar O, Raizen DM, Fang-Yen C. Neural and genetic degeneracy underlies Caenorhabditis elegans feeding behavior. Journal of Neurophysiology. 2014;112:951–961. doi: 10.1152/jn.00150.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Sinay E, Mirabeau O, Depuydt G, Van Hiel MB, Peymen K, Watteyne J, Zels S, Schoofs L, Beets I. Evolutionarily conserved TRH neuropeptide pathway regulates growth in Caenorhabditis elegans. PNAS. 2017;114:E4065–E4074. doi: 10.1073/pnas.1617392114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varoqueaux F, Fasshauer D. Getting Nervous: An Evolutionary Overhaul for Communication. Annual Review of Genetics. 2017;51:455–476. doi: 10.1146/annurev-genet-120116-024648. [DOI] [PubMed] [Google Scholar]
- Vidal B, Aghayeva U, Sun H, Wang C, Glenwinkel L, Bayer EA, Hobert O. An atlas of Caenorhabditis elegans chemoreceptor expression. PLOS Biology. 2018;16:e2004218. doi: 10.1371/journal.pbio.2004218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilimas T, Abraham A, Okkema PG. An early pharyngeal muscle enhancer from the Caenorhabditis elegans ceh-22 gene is targeted by the Forkhead factor PHA-4. Developmental Biology. 2004;266:388–398. doi: 10.1016/j.ydbio.2003.10.015. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Fujisawa T, Holstein TW. Cnidarians and the evolutionary origin of the nervous system. Development, Growth & Differentiation. 2009;51:167–183. doi: 10.1111/j.1440-169X.2009.01103.x. [DOI] [PubMed] [Google Scholar]
- Wei Z, Angerer RC, Angerer LM. Direct development of neurons within foregut endoderm of sea urchin embryos. PNAS. 2011;108:9143–9147. doi: 10.1073/pnas.1018513108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Xuan Z, Manning L, Nelson J, Richmond JE, Colón-Ramos DA, Shen K, Kurshan PT. Clarinet (CLA-1), a novel active zone protein required for synaptic vesicle clustering and release. eLife. 2017;6:e29276. doi: 10.7554/eLife.29276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Bhattacharya A, Nelson JC, Abe N, Gordon P, Lloret-Fernandez C, Maicas M, Flames N, Mann RS, Colón-Ramos DA, Hobert O. The LIM and POU homeobox genes ttx-3 and unc-86 act as terminal selectors in distinct cholinergic and serotonergic neuron types. Development (Cambridge, England) 2014;141:422–435. doi: 10.1242/dev.099721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ward JD, Cheng Z, Dernburg AF. The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development (Cambridge, England) 2015;142:4374–4384. doi: 10.1242/dev.129635. [DOI] [PMC free article] [PubMed] [Google Scholar]