Abstract
Concatemers of d(TCCC) that were first detected through their association with deletions at the RACK7 locus, are widespread throughout the human genome. Circular dichroism spectra show that d(GGGA)n sequences form G-quadruplexes when n > 3, while i-motif structures form at d(TCCC)n sequences at neutral pH when n ≥ 7 in vitro. In the PC3 cell line, deletions are observed only when the d(TCCC)n variant is long enough to form significant levels of unresolved i-motif structure at neutral pH. The presence of an unresolved i-motif at a representative d(TCCC)n element at RACK7 was suggested by experiments showing that that the region containing the d(TCCC)9 element was susceptible to bisulfite attack in native DNA and that d(TCCC)9 oligo formed an i-motif structure at neutral pH. This in turn suggested that that the i-motif present at this site in native DNA must be susceptible to bisulfite mediated deamination even though it is a closed structure. Bisulfite deamination of the i-motif structure in the model oligodeoxynucleotide was confirmed using mass spectrometry analysis. We conclude that while G-quadruplex formation may contribute to spontaneous mutation at these sites, deletions actually require the potential for i-motif to form and remain unresolved at neutral pH.
INTRODUCTION
Regions of GC Skew in eukaryotic genomes, where dG residues are generally confined to one strand and dC residues are confined to the other, are prone to the formation of several types of non-B DNA structure. Among the more common of these are regions that conform to a general sequence motif: d(G3+N1–7G3+N1–7G3+N1–7G3+). This motif has been used to detect genomic sites with potential to fold into G-quadruplex structures on the G-rich strand in regions of GC Skew (1). Since G-quadruplex structures are strongly stabilized by the presence of potassium (2), they are favored under physiological conditions. Nevertheless other alternative DNA structures such as triplexes (3) and i-motifs (4) can also form at sites conforming to this general sequence motif.
Genomic sequence comparisons across Eukarya show that sequences conforming to this motif have increased in frequency during the evolution of ever more complex eukaryotic organisms (5,6) suggesting that they are under positive selective pressure and may serve one or more biological functions. Since they are often present in promotor regions (7) there has been considerable interest in the possibility that they play roles in transcription and other epigenetic phenomena. Evidence in support of these ideas has been obtained in a number of studies. Triplex formation at the MYC promoter has been implicated in transcription regulation at this gene (3), and i-motif formation at the BCL2 gene has been shown to play a role in transcription at that locus (8). Moreover, formation of stable structures at these sequences has been shown to promote methylation in their vicinity (9) and disrupt the formation of repressive chromatin structures (10). G-quadruplexes, for example may influence DNA methylation patterns via the direct inhibition of DNA methyltransferase I (11). It has also been shown that G-quadruplex formation on the G-rich strand in a region of GC Skew may indirectly influence DNA methylation patterning by stabilizing R-loop RNA:DNA hybrids on the C-rich DNA strand, since RNA:DNA hybrids have also been shown in vitro to inhibit DNA methyltransferase I (12,13).
In normal cells, G-quadruplex formation is likely to be transient, given the constellation of G-quadruplex resolvases known to be expressed in normal mammalian cells. Among these are the RecQ helicases encoded by the BLM (14) and WRN genes (15) and the helicase encoded by the Fanconi Anemia locus FANCJ (16). Further, during DNA replication in normal cells the Y-polymerase family genes (REV1, POLK and POLH) express polymerases capable of replication through G-quadruplex DNA. Rev1 actually disrupts G residues in the quadruplex and inserts a dC residue on the nascent strand (17) while, Pol κ and Pol η also appear to function in replicating through G-quadruplex forming sequences (18). Even so, replication through extended regions of G-quadruplex potential like the d(GGC)n repeat at FRAXA in disease carriers appears to be able to induce replication fork collapse and chromosome breaks and subsequent deletion and insertion prone repair (19).
In the most extensive study of quadruplex-linked replication impediments to date, the dog-1 mutation in Caenorhabditis elegans was shown to promote genome wide deletions confined to oligo dG sequences (20,21). The mutation at dog-1 knocks out the C. elegans homolog of human FANCJ, which is known to resolve G-quadruplex sequences (16), thus implicating unresolved G-quadruplex sequences as replication impediments at multiple genomic locations. The deletions in C. elegans were shown to occur 5′- to the oligo dG strand (i.e. the G-rich strand), and often deleted much of the oligo dG element itself at each site (20). Deletion frequency also increased with the length of the oligo dG element (20).
Interestingly, the oligo dC strand has been shown to form an i-motif quadruplex at neutral pH in vitro (22). These sequences form optimally when they conform to a (4n– 1) length rule for n > 2 (22). That is to say, the potential for i-motif structure at pH 7 in vitro is greatest at lengths of 11, 15, 19 and 23 dC residues, since the pHT (i.e. the pH at which half the population of sequence adopts an i-motif) is 7.2 at sequence lengths conforming to this (4n –1) rule. Data from the dog-1 experiments shows that at lengths of 15 and 19 the deletion frequency appears to be suppressed but not at sequences of greater length (20). For short sequences of this type this is consistent with evidence showing that G-quadruplex and i-motif formation at shorter sequences can be mutually exclusive due to steric hindrance while other, often longer sequences, permit the formation of both structures offset from one another (23–25). Taken together, this suggests that i-motif formation in oligo dC at lengths of 15 and 19 suppresses G-quadruplex formation in C. elegans and thus hinders dog-1 induced deletions, while at longer d(G:C) sequences the formation of i-motifs does not prevent formation of G-quadruplexes resulting in dog-1 induced deletions. For this interpretation to be correct, C. elegans must either retain a repair system that resolves i-motif structures, or i-motif structures do not form replication impediments in C. elegans.
In previous work, we devised a method designed to clone sequences from stalled replication forks at sites in DNA where DNA polymerase and CMG helicase uncoupling is present prior to DNA isolation (Supplementary Figure S1). Double strand fragments originating near a putative CMG helicase halt site were often recovered associated with a non-B structure forming element generally detected within 1800 bp of the cloned sequence (26). One of the sequences detected in that screen, d(TCCC)9, was shown to be in an open conformation in PC3 cells as judged by bisulfite accessibility in native chromosomal DNA, and was characterized by short deletions in the PC3 prostate tumor cell line (26). Since the sequence was capable of forming an i-motif at neutral pH in vitro, this raised the possibility that the i-motif structure may form a replication impediment that is not suppressed in human prostate cancer cells. The absence of a functional p53 gene in the PC3 cell line is consistent with the possibility that i-motif suppression by DNA repair systems could be defective in this repair compromised cell line.
In this report we determined the tendency of the d(TCCC)n sequences to from i-motif structures and the tendency of the d(GGGA)n complementary sequence to form G-quadruplex at neutral pH in vitro. Since the d(TCCC)n element occurs with different lengths at multiple chromosomal locations in the human genome we compared these in vitro results with the frequency of deletions at sites ranging from d(TCCC)5 to d(TCCC)15 observed in the PC3 cell line. We found that G-quadruplex formation in vitro was spontaneous at physiological pH over the range tested d(GGGA)4 to d(GGGA)14 while i-motif formation at neutral pH increased over the range d(TCCC)5 to d(TCCC)15. Deletion frequency in the PC3 cell line reached 100% of cloned representatives when the pHT observed in vitro for i-motif formation exceeded pH 7. Moreover, deletions were confined 5′- to the d(TCCC)n element (i.e. only on the C-rich strand) in each gene examined.
MATERIALS AND METHODS
Cell culture
Cell culture methods and preparation of DNA from the PC3 cell line were as previously described (27).
DNA isolation
DNA isolation methods have been previously described (27).
PCR methods
All PCR Primers were obtained from Integrated DNA Technologies (San Diego, CA).
For the region from the TAFA5 gene: The forward primer sequence was 5′-GCACTCAGCTCTCTGCTGTCCTTGCCTC-3′, the reverse primer sequence was: 5′-AGACCCTGATCCAGCCAGCCAAGCCCTC-3′. PCR reactions contained 12.5 μl 2X KAPA2G Fast Multiplex mix (Kapa Biosystems, Wilmington, MA), 1.25 μl of 10 μM stock of each primer (500 nM final concentration), 100 ng target DNA, made up to a final volume of 25 μl with Molecular Biology Water (Sigma). PCR cycling conditions were 95°C for 5 min, followed by 35 cycles of 95°C for 15 s, 56°C for 30 s, 72°C for 30 s followed by a final extension at 72°C for 26 s.
For the region from the BCR gene: The forward primer sequence was 5′-TTGCTCCTCTTATTTCCTAGCAGCAGATGT-3′, the reverse primer sequence was: 5′-CAGTAGGTGTCACATCAACTTCAAATGAAC-3′. PCR reactions contained 12.5 μl 2X KAPA2G Fast Multiplex mix (Kapa Biosystems, Wilmington, MA), 0.55 μl of 9 μM stock of each primer (198 nM final concentration), 200 ng target DNA, made up to a final volume of 25 μl with Molecular Biology Water (Sigma). PCR cycling conditions were 95°C for 5 min, followed by 35 cycles of 95°C for 15 s, 56°C for 30 s, 72°C for 30 s followed by a final extension at 72°C for 26 s.
For the region from the ABL1gene: The forward primer sequence was 5′-ATATACTTAGAGGTGGGCTGGTAAAG-3′, the reverse primer sequence was: 5′-CCATGTTAGGTCAGGTGCTCATGTCT-3′. PCR reactions contained 12.5 μl 2X KAPA2G Fast Multiplex mix (Kapa Biosystems, Wilmington, MA), 1.25 μl of 10 μM stock of each primer (500 nM final concentration), 100 ng target DNA, made up to a final volume of 25 μl with Molecular Biology Water (Sigma). PCR cycling conditions were 95°C for 3 min, followed by 35 cycles of 95°C for 15 s, 53°C for 30 s, 72°C for 30 s followed by a final extension at 72°C for 26 s.
For the region from the RACK 7 gene: The forward primer sequence was 5′-GGTTGGCCAGTTGTGTACCTCACAGTGG-3′; and the reverse primer sequence was 5′-GCTCCAGCCTGGGCAACAAAGCAACAGT-3′. PCR reactions contained 12.5 μl 2X KAPA2G Fast Multiplex mix (Kapa Biosystems, Wilmington, MA), 0.55 μl 9 μM stock of each primer (198 nM final concentration), 200 ng target DNA, made up to a final volume of 25 μl with Molecular Biology Water (Sigma). PCR cycling conditions were 95°C for 3 min, followed by 35 cycles of 95°C for 1 min, 60°C for 30 s, 72°C for 30 s followed by a final extension at 72°C for 26 s.
For the region from the PLA2G2A gene: The forward primer sequence was 5′-TAGAGGTGATGGTTGTACAACACTGTGAAT-3′; and the reverse primer sequence was 5′-AGAGCAAGACCCTGTCTTATTGTTGAACCG-3′. PCR reactions contained 12.5 μl 2X KAPA2G Fast Multiplex mix (Kapa Biosystems, Wilmington, MA), 0.55 μl of 9 μM stock of each primer (198 nM final concentration), 200 ng target DNA, made up to a final volume of 25 μl with Molecular Biology Water (Sigma). PCR cycling conditions were 95°C for 3 min, followed by 35 cycles of 95°C for 1 min, 63°C for 30 s, 72°C for 30 s followed by a final extension at 72°C for 26 s.
Bisulfite mediated deamination of native genomic DNA
Genomic DNA isolated from the PC3 cell line was treated with bisulfite using the EZ methylation Kit (Zymo Research, Irvine CA). For these experiments, DNA was added to the conversion reagent without M-dilution buffer (a denaturant) or heat treatment in order to preserve the native structure present in the isolated DNA. The reaction mixture was incubated in the dark at 37°C for 16 h. Thereafter, the kit protocol including the desulfonation step was carried out in order to recover DNA for subsequent PCR amplification of the selected region in the RACK7 gene.
In vitro analysis of d(TCCC)n and d(GGGA)n oligodeoxynucleotides
Oligonucleotides
were purchased from IDT (San Diego, CA) or Eurogentec (Liege, Belgium) and were HPLC purified. Solid DNA samples were initially dissolved as a stock solution in purified water (1 mM); further dilutions were carried out in the respective sodium cacodylate buffer. Samples were thermally annealed in a heat block at 95°C for 5 min and cooled slowly to room temperature overnight.
UV absorption spectroscopy
UV spectroscopy experiments were performed on a Cary 60 UV–Vis spectrometer (Agilent Technologies) and recorded using low volume masked quartz cuvettes (1 cm path length). ODNs were diluted to 2.5 μM in buffer at the desired pH. Samples (200 μl) were transferred to a cuvette and stoppered to reduce evaporation of the sample. The absorbance of the ODN was measured at 295 nm as the temperature of the sample was held for 10 min at 4°C, heated to 95°C at a rate of 0.5°C per min, then held at 95°C for 10 min before the process was reversed; each melting/annealing process was repeated three times. Data were recorded every 1°C during both melting and annealing and melting temperatures (Tm) and annealing temperatures (Ta) were determined using the first derivative method. This analysis was performed independently on each of the three melting curves collected for each ODN and the values presented are the average and standard deviation of these. TDS were calculated by subtracting the spectrum of the folded structure between 220 and 320 nm at 4°C from that of the unfolded structure at 95°C. The data was normalized and maximum change in absorption was set to +1 as previously described (28).
Circular dichroism
CD spectra were recorded on a Jasco J-810 spectropolarimeter using a 1 mm path length quartz cuvette. ODNs were diluted to 10 μM (total volume: 200 μl) in buffer at pH increments of 0.25 pH units from 5.0 to 8.0. The scans were recorded at room temperature between 200 and 320 nm. Data pitch was set to 0.5 nm and measurements were taken at a scanning speed of 200 nm/min, response time of 1 s, bandwidth of 2 nm and 100 mdeg sensitivity; each spectrum was the average of three scans. Samples containing only buffer were also scanned according to these parameters to allow for blank subtraction. Transitional pH (pHT) for each i-motif was calculated from the inflection point of fitted ellipticity at 288 nm.
Frontier orbital calculations
As previously described (29), model compounds were used in electronic structure calculations. Models of C, C+, C:G and C:C+ in single stranded, Watson–Crick paired duplex or i-motif structures in DNA, were 1-methyl-cytosine,1-methyl-N3-protonated-cytosine, 1-methly-cytosine Watson–Crick base paired with 1-methyl guanine, and 1–methyl-cytosine base paired with 1-methyl-N3-protonated-cytosine respectively. Electronic structure calculations for each of the above compounds and the HSO3– anion were carried out in Spartan’18 (Wavefunction, Irvine, CA). Preliminary geometries obtained with molecular mechanics were submitted for equilibrium geometry calculation with Parametric Method 3. The resulting geometry was submitted for ab initio equilibrium geometry calculation using the Hartree–Fock level of theory with a final geometry optimization using the 6-31G* basis set. All molecular orbital parameters and surfaces were computed with this basis set.
Mass spectrometry analysis of (TCCC)9
Bisulfite mediated deamination of the (TCCC)9 oligodeoxynucleotide was carried out at 37°C and pH 5.3, where more than 95% of this oligodeoxynucleotide adopts an i-motif structure (see Table 2), using the procedure described above for non-denatured genomic DNA. Bisulfite treated DNA recovered from 0.5, 1 or 2 μg of input oligodeoxynucleotide was submitted to Novatia, LLC (Newtown, PA) for High Resolution Liquid Chromatography Mass Spectroscopic analysis. dC→dU conversions were detected as 1 Da increases in mass of the reference mass of the (TCCC)9 oligodeoxynucleotide. Deconvolution reports like the representative report given in Supplementary Data Set 1 were used to determine average frequencies for each level of conversion from 1 dU/36mer to 27 dU/36mer.
Table 2.
UV melting and annealing temperatures of d(TCCC) repeats determined by the first derivative method. DNA at 2.5 μM in 10 mM sodium cacodylate with 100 mM KCl at pH 5.3. Values are the mean from triplicate experiments with standard deviations
| Sequence** | T m (°C) | T a (°C) | Hysteresis (°C) |
|---|---|---|---|
| d(TCCC)5 | 50.2 ± 3.5 | 45.8 ± 3.5 | 4.3 ± 0.5 |
| d(TCCC)9 | 66.7 ± 2.0 | 62.0 ± 2.0 | 4.6 ± 0.5 |
RESULTS
Bisulfite treatment of native genomic DNA
At pH 5.3 and 37°C isolated DNA is primarily in the B-form DNA conformation with Watson-Crick base pairs. Biologically formed non-B structures can be expected to persist under these conditions, provided that they are not maintained by constraints present in chromatin (30,31). Cytosine residues that are Watson-Crick base paired in the B-DNA duplex are not susceptible to attack by the bisulfite nucleophile, which has been shown experimentally (31,32) and is confirmed by the frontier orbital energy of the C:G base pair model (Figure 1), where the lowest unoccupied molecular orbital (LUMO) of the base pair electrophile (centered on the cytosine moiety at C6) is very high relative to the highest occupied molecular orbital (HOMO) energy of the HSO3− nucleophile. Consequently, for the regions comprising d(TCCC)5 and d(TCCC)9 depicted in Figure 3, bisulfite mediated deamination in native DNA at pH 5.3 and 37°C shows that these regions are in non-B structures on some chromosomes in PC3 cells. Deletions were also detected at the 5′ end of the d(TCCC)5 and d(TCCC)9 elements. Moreover, deamination and deletion at d(TCCC)9 was more extensive than it was at d(TCCC)5. Only two forms capable of reaction with the bisulfite nucleophile are possible at these sites: an unpaired and disordered loop, or an i-motif. Since pH 5.3 is near the pKa of N3 of cytosine, should dissociation of a biologically formed i-motif occur during DNA isolation result in an unpaired loop, both the protonated and unprotonated forms of deoxycytosine would be present. While the frontier orbital picture virtually precludes nucleophilic attack by the HSO3− anion on dC, given the high energy of its LUMO, the protonated dC+ residue is expected to be readily attacked as shown both by experiment (32,33) and by the proximity to its LUMO energy relative to the energy of the HOMO of HSO3− (Figure 1).
Figure 1.
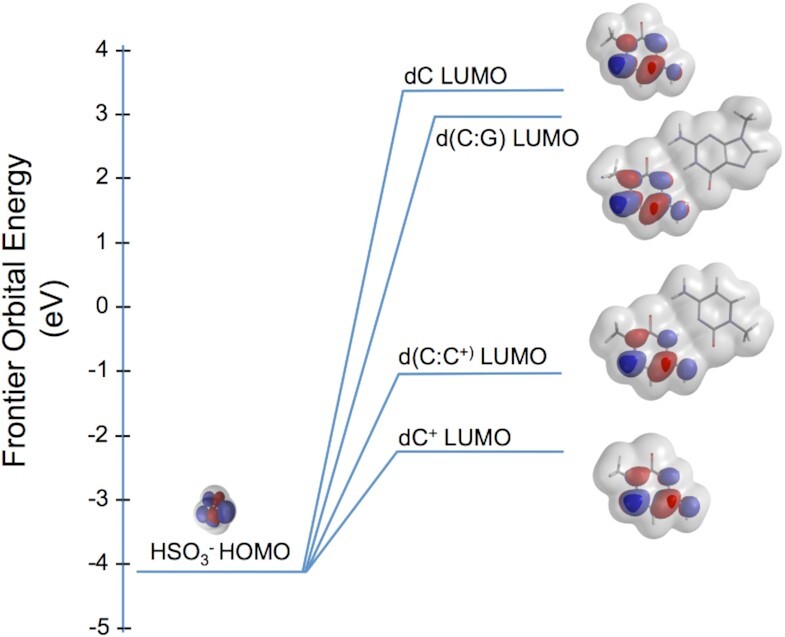
Frontier orbitals in the reaction between the bisulfite anion and cytosine in nucleic acids. The equilibrium geometry highest occupied molecular orbital (HOMO) for the bisulfite anion (lower left) is shown at its energy in eV under the transparent electron density that envelops of the molecule. Similar equilibrium geometry calculations, all at the Hartree-Fock 6–31G* level of theory, are depicted for the lowest unoccupied molecular orbital (LUMO) of models of cytosine in its several possible configurations in DNA. In each model the deoxyribose moiety at N1 of cytosine or guanine is modeled by a simple methyl group. Hence, the bases used in the electronic structure calculations are 1-methyl-cytosine, 1-methyl_cytoisne+ or 1-methyl-guanine. The geometry of the LUMO’s identify C6 of cytosine as the point of nucleophilic attack by the anion (blue center above the transparent electron density) in each model, and order the reactivity of each based on the LUMO energies in eV: dC+>d(C:C+)>>d(C:G)>dC.
Figure 3.
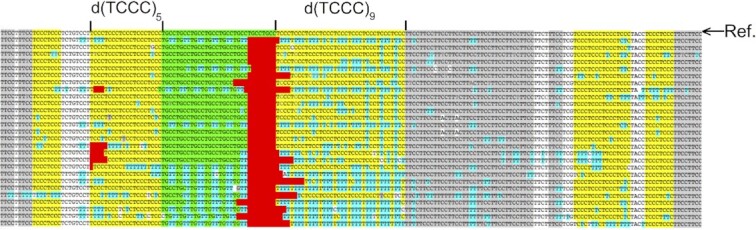
Bisulfite mediated deamination of native DNA in the region of RACK7 containing d(TCCC)5 and d(TCCC)9. After bisulfite treatment of native DNA at pH 5.3 and 37°C, representative cloned isolates of the region were sequenced and the sequences aligned under the genomic reference sequence from the GRCh38.p12 primary assembly of the human genome (marked Ref. in the figure). d(TCCC)n elements are depicted in yellow. The alignment shows that deletions of various length have occurred in different cells at the 5′ends of both d(TCCC)5 and d(TCCC)9. However, both the level of deamination (blue) and the level of deletion (red) are much more extensive at d(TCCC)9 than in d(TCCC)5.
Although one might expect that an intact i-motif structure might resist bisulfite attack given the results with duplex DNA, the energy of the LUMO of the d(C:C+) base pair electrophile (centered over C6 of the dC+ moiety) in the i-motif (Figure 1) is also close to that of the nucleophile and therefore should be more readily attacked than either unprotonated dC or the C:G base pair. These considerations and the frontier orbital analysis above suggested that although the i-motif is a closed structure, it can be attacked by bisulfite. To directly confirm this possibility we treated d(TCCC)9 with bisulfite using the same procedure used for genomic DNA (i.e. pH 5.3 and 37°C) where it is almost exclusively in the i-motif form (Table 2). High resolution liquid chromatography mass spectrometry analysis showed that significant conversion of dC to dU occurred during the bisulfite reaction even though the i-motif structure was the dominant form in solution with a Tm of 66.7°C (Table 2). In these experiments intact 36mers accounted for 92% of the deconvoluted mass spectrum. A representative deconvolution report is given as Supplementary Data set 1. While the data clearly show significant conversion ranging from 1/27 to 27/27 cytosines converted. The frequency distribution for the recovered species was approximately random and peaks at about nine dC→dU conversions per 36mer (Figure 2). Unfortunately, sequence assignments are not possible with this method.
Figure 2.
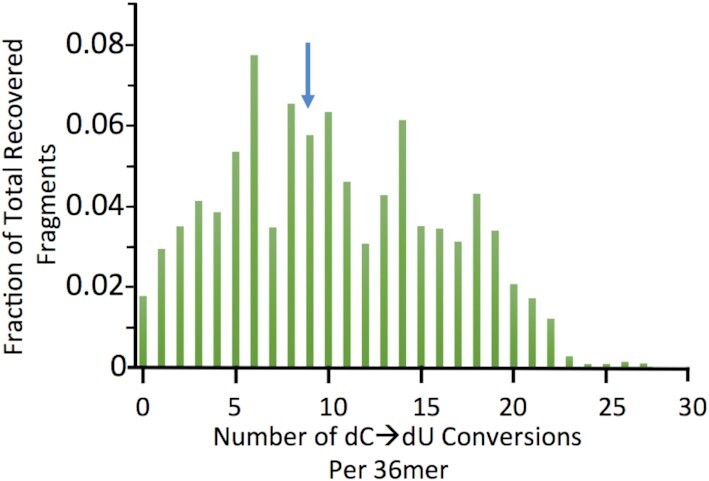
Frequency distribution for recovered 36mers after bisulfite treatment at 37°C and pH 5.3. Bisulfite treated DNA at 0.5, 1 and 2 μg input oligodeoxynucleotide DNA was recovered after high resolution liquid chromatography. Mass spectrometry showed that it is partially converted with a range of dU/36mer deaminations that are distributed from 0 to 27 dU deaminations in individual 36mers distinguished based on mass. The frequency of each species is plotted versus the number of dUs present in each 36mer as the average for the three input levels. The Distribution is centered on approximately 9dU/36mer (blue arrow). 92% of fragments in the spectrum had masses ≥ that of the d(TCCC)9 36mer.
The sequencing results given in Figure 3 show aligned exemplars originating from individual chromosomes. In spite of the deletions observed in many of the sequences, both the d(TCCC)5 and d(TCCC)9 regions are generally complete. The C→T conversion frequency in d(TCCC)5 was 10.4% while the conversion frequency in d(TCCC)9 is 37.4%. Further, deletions were confined to the 5′- end at both d(TCCC)5 and d(TCCC)9 with a frequency of 18.5% and 100% of the cloned sequences respectively.
In vitro analysis of d(TCCC)n and d(GGGA)n model oligodeoxynucleotides
Given that the observed bisulfite mediated deamination was more extensive at d(TCCC)9 than at d(TCCC)5 we asked if the potential for i-motif structure formation at neutral pH was a function of length as seen in other i-motif forming sequences (22). Circular dichroism (CD) spectra exhibit well-characterized and easily distinguishable signatures for both the i-motif structure and the G-quadruplex structure for single-stranded oligodeoxynucleotide sequences. G-quadruplex sequences form spontaneously at neutral pH, however, i-motif structure formation is enhanced at low pH because protonation of N3 of cytosine increases the formation of the C:C+ base pair in parallel DNA strands. However, many i-motif structures are known to form effectively at neutral pH (4), suggesting that successive stacking interactions in longer i-motif forming sequences may also enhance i-motif formation.
We determined titration profiles for i-motif formation using CD spectra at different pHs for the d(TCCC)n element with n ranging from 4 to 15. Data for the full analyses for the length series d(TCCC)4 to d(TCCC)15 is given in the Supplementary Figure S7A–I, and is summarized in Table 1. The input concentration of 10 μM oligodeoxynucleotide was chosen to permit detection at a concentration that favors unimolecular folding. The pHT (i.e. the transition pH values at which equal concentration of i-motif and single strands are present in solution) for each sequence was seen to increase with increasing length. The pHT values steadily increase with length up to a plateau with pHT reaching neutral pH at lengths greater than n = 8 with corresponding average Tm values of 41°C (Table 1 and Supplementary Figure S8). In contrast, the equivalent G-rich sequences formed stable G-quadruplexes over the whole length range. When we performed analyses at physiological mimicking potassium cation concentrations (100 mM KCl), the Tm values were all >90°C, or near the limit of detection. Reduction of the KCl levels to 20 mM still gave average melting temperatures of >70°C for every sequence (Table 1 and Supplementary Figures S5A–H and Supplementary Table S1). This gives us confidence that in physiological conditions, these G-quadruplexes are highly stable, regardless of sequence length. Thermal difference spectra for G-quadruplex formation are given in Supplementary Figure S6.
Table 1.
UV melting and annealing temperatures of d(TCCC) repeats determined by the first derivative method. DNA at 2.5 μM in 10 mM sodium cacodylate with 100 mM KCl at pH 6.5. Values are the mean from triplicate experiments with standard deviations. Transitional pH values determined by CD, with standard errors from the curve fit*
| Sequence* | T m (°C) | T a (°C) | Hysteresis (°C) | pHT |
|---|---|---|---|---|
| d(TCCC)4 | 15.6 ± 1.0 | 13.8 ± 1.2 | 1.8 ± 2.1 | 6.6 ± 0.01 |
| d(TCCC)5 | 28.3 ± 0.6 | 26.0 ± 0.6 | 2.4 ± 1.0 | 6.7 ± 0.04 |
| d(TCCC)6 | 31.4 ± 0.6 | 29.2 ± 0.0 | 2.2 ± 0.6 | 6.8 ± 0.02 |
| d(TCCC)7 | 32.8 ± 1.0 | 29.1 ± 0.0 | 3.7 ± 1.0 | 6.8 ± 0.03 |
| d(TCCC)8 | 40.7 ± 0.6 | 30.7 ± 0.6 | 10.0 ± 0.0 | 6.9 ± 0.01 |
| d(TCCC)9 | 40.7 ± 0.6 | 30.3 ± 0.6 | 10.4 ± 0.6 | 7.1 ± 0.04 |
| d(TCCC)12 | 40.2 ± 0.6 | 30.5 ± 0.0 | 9.8 ± 0.6 | 7.2 ± 0.08 |
| d(TCCC)14 | 42.0 ± 4.0 | 30.4 ± 0.0 | 11.6 ± 4.0 | 7.1 ± 0.07 |
| d(TCCC)15 | 40.2 ± 4.0 | 23.2 ± 2.0 | 16.0 ± 2.0 | 7.1 ± 0.08 |
The pH profiles for d(TCCC)5 and d(TCCC)9 are depicted in Figure 4. Both sequences have significant potential for the formation of i-motif structures at physiological pH, with the pHT values of 6.7 for d(TCCC)5 and 7.1 for d(TCCC)9. Since the pH of the bisulfite reaction is very close to the pKa of the N3 of cytosine, protonated cytosines are expected to be present during the reaction should a biologically formed i-motif dissociate into a loop. However, the CD titration profiles of each sequence suggest that at pH 5.3 a biologically formed i-motif would be preserved as the dominant form in solution during the bisulfite reaction and hence the primary substrate for the deamination reaction (Figure 4). Further, we assessed the stability of each of these sequences (d(TCCC)5 and d(TCCC)9) at pH 5.3 and, as expected, the melting temperatures were high (Supplementary Figure S4A and B). Tm profiles were determined for each d(TCCC)n oligodeoxynucleotide at pH 6.7 and are given in Supplementary Figure S2A–I. Thermal difference spectra for i-motif formation are given in Supplementary Figure S3. At pH 6.5, the Tm value for d(TCCC)5 was found to be 28.3 ± 0.6°C and that for d(TCCC)9 was 40.7 ± 0.6°C (Table 1). However, at pH 5.3 the Tm value for d(TCCC)5 was found to be 50.2 ± 3.5°C and that for d(TCCC)9 was 66.7 ± 2.0°C (Table 2). This indicates that, under the conditions of the bisulfite reaction (pH 5.3 and 37°C), both these two regions would remain almost exclusively in the i-motif conformation in those chromosomes that had formed an i-motif biologically.
Figure 4.
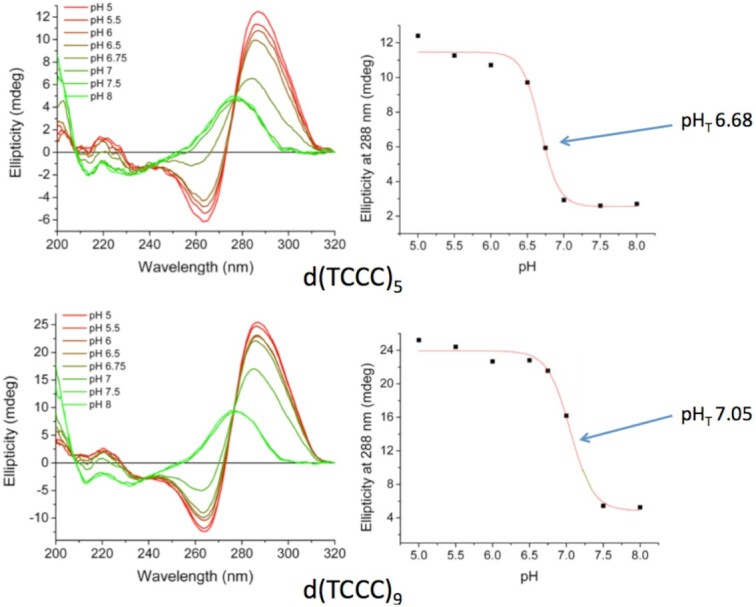
Titration curves for oligodeoxynucleotides with d(TCCC)5 and d(TCCC)9 repeats. CD spectra for each repeat over the titration range pH 5 to pH 8. In each titration, the characteristics i-motif signature emerge as pH is decreased, with the full signature emerging at pH 5 for d(TCCC)5 and at pH 5.5 for d(TCCC)9. Transition pH (pHT) values show that a significant fraction of each sequence would be present as i-motif near physiological pH.
Deletion Frequencies in the PC3 cell line
The d(TCCC)n repetitive element occurs in multiple locations across the whole human genome. Of these, there are about 220 distinct locations were d(TCCC)n for n ≥ 9. We used PCR to isolate fragments spanning sequences of various lengths from the genomic DNA of the PC3 cell line as previously described (26). Isolated fragments were cloned and sequenced also as described previously (26). Data in Figure 5 shows the results obtained for the HAFA5 gene (d(TCCC)5 element), the BCR gene (d(TCCC)7 element), the RACK7 gene (d(TCCC)9 element), the ABL1 gene (d(TCCC)11 element), and the PLA2G2A gene (d(TCCC)15 element). The sequences were aligned as previously described against the genomic sequence (GRCh38.p12 Primary Assembly) of the human genome (26). As depicted in Figure 5, deletions (highlighted in red) are uniformly associated with the 5′-end of the genomic sequence of the C-rich strand. Deletions are absent from cloned representatives of the HAFA5 site, with a single deletion recovered at the BCR site. However, the RACK7, ABL1 and PLA2G2A showed significant deletions at each cloned representative. The data is summarized in Figure 6. The potential formation of G-quadruplex structures is not correlated with deletion frequency at all, since in the absence of an i-motif (i.e. as a single-stranded oligodeoxynucleotide), G-quadruplex structures were observed to form in vitro at physiological pH at each length tested (Supplementary Figure S9). Moreover, even at sub-physiological potassium levels (20 mM KCl) all the melting temperatures for the G-quadruplex forming sequences were above 70°C (see Supplementary Figure S5A–H and Supplementary Table S1). Thermal difference spectra for G-quadruplex formation are given in Supplementary Figure S6. Conversely, we observe a strong correlation between the potential for i-motif formation in vitro and the frequency of deletion in PC3, since 100% of the recovered clones showed a deletion when the pHT of the relevant i-motif forming sequence observed in vitro is above pH 7.0.
Figure 5.
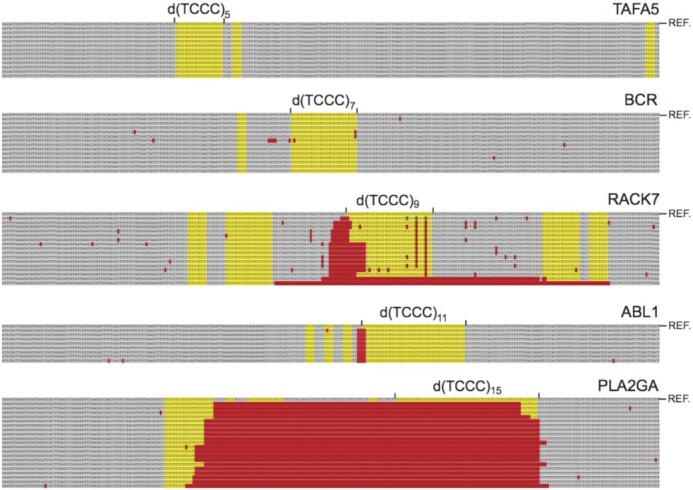
Deletions at multiple genomic locations in cultured human prostate cancer cells. Deleted sequences (red) are located near the 5′- end of the C-rich strand of the d(TCCC)n repeat and often extend into the repeat itself. Deletions were not observed for d(TCCC)5 in the HAFA5 gene, and were very infrequent for d(TCCC)7 in the BCR gene, however for the d(TCCC)9 in the RACK7 gene, d(TCCC)11 in the ABL1 gene and d(TCCC)15 in the PLA2GA2 gene each cloned representative carried a deletion.
Figure 6.
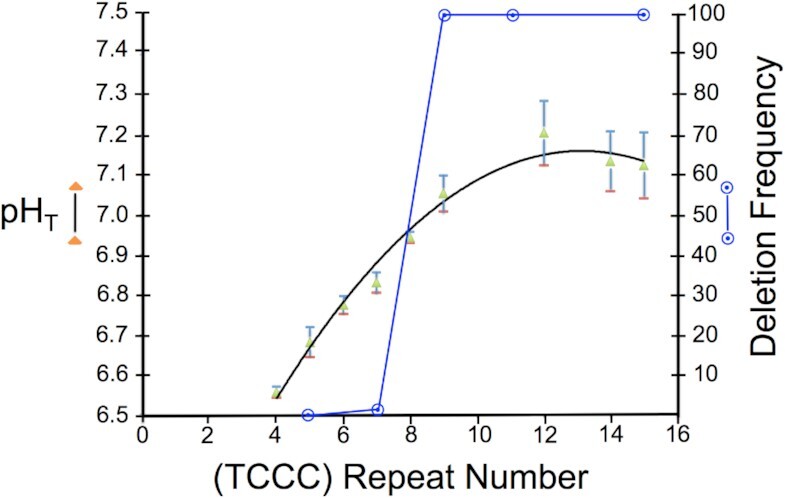
Length dependence of i-motif formation at neutral pH correlated with deletion frequency. pHT value: closed triangles (green) ± standard deviation determined from the titration curve fit. Fraction of cloned sequences with a deletion: open circles (blue).
DISCUSSION
While G-quadruplex and i-motif structures can both form at neutral pH, as noted here for the d(TCCC)n and as reported for dCn (22), i-motif structure formation at neutral pH can depend dramatically on sequence length and composition. Consequently, the correlation between deletion frequency and d(TCCC)n folding at neutral pH (Figure 6) provides strong evidence that i-motif structures are linked to deletions at d(TCCC)n elements in the human genome. This correlation suggests that i-motif structures form in vivo in the repair compromised PC3 cell line. Further, the in vitro findings of pHT’s near 7.1 and a melting temperatures above 40°C, for each sequence length associated with a significant deletion frequency in PC3 (Supplementary Figures S2A-I, S7A-I and S8), indicate that the i-motif structure, once formed, is expected maintain a structural disruption of duplex DNA structure at 37°C and physiological pH at d(TCCC)n with n ≥ 9.
Even so, we can only speculate on the process that leads to the extrusion of G-quadruplexes and i-motifs biologically. Evidence suggesting roles in transcription require complex biological processes that elicit these structures (7,8,11,34). Such systems might alter local topology in order to extrude quadruplexes and it is known that transcription itself can extrude a G-quadruplex at a site of RNA:DNA hybrids during R-Loop formation (35). However, i-motif structures have not been found associated with R-Loops, since RNA:DNA hybrids appear to form exclusively on the C-rich strand in regions of GC-Skew.
That one or both forms are in fact extruded so as to elicit replication stress is strongly suggested by the data in C. elegans (20,21,36) and our own data on the human PC3 cell line (26). In C. elegans dog-1 mutant worms, unresolved G-quadruplex structures cause replication stress and replication fork stalling and collapse (37). Since replication fork stalling can lead to HMG helicase uncoupling (38), once a replication fork stalls and the HMG helicase uncouples (38) the two strands will be separated and both strands can form non-B structures as topological constraints outside the region are relieved. In previous work we used a cloning procedure (see Supplementary Figure 1) designed to clone sequences near uncoupled HMG halt sites (26). Clones isolated from PC3 cell line DNA using that procedure were most often within 5 kb of a G-quadruplex motif (26). The d(TCCC)9 site in RACK7 was identified with this cloning procedure suggesting that the clone came from a site undergoing replication stress and HMG helicase uncoupling (26).
Our previous work on the RACK7 repeat in PC3 cells also showed that d(TCCC)9 sequence at this site was heavily deaminated by bisulfite under non-denaturing conditions (26), demonstrating that it is not paired with its complementary strand (31). However, the data presented here suggests that the i-motif structure can even survive after DNA isolation, as has been shown for other non-B structures (30,31). Importantly, the frequency of deamination at the d(TCCC)5 element in the region of 10% compared to the frequency of 37% for d(TCCC)9 correlates with the relative lower stability of d(TCCC)5 (pHT 6.7 and Tm = 28°C) compared to d(TCCC)9 (pHT 7.1 and Tm = 41°C) under physiological conditions. We considered the possibility that the region around d(TCCC)9 was simply single-stranded and the i-motif might not be present given the possibility that, if it were present, it might behave like native duplex DNA and would resist nucleophilic attack by bisulfite. However, careful analysis of the features of the bisulfite reaction (Figure 1) and pH and melting profiles (Table 2 and Figure 4) suggested that: the i-motif itself is the mostly likely initial substrate for nucleophilic attack by the bisulfite anion at pH 5.3 and 37°C (Figure 3). This suggestion was confirmed by high resolution liquid chromatography mass spectrometry of the bisulfite treated d(TCCC)9 36mer (Figure 2) at 37°C and pH 5.3. After bisulfite treatment, between 1/27→27/27 dC→dU conversions were observed in intact 36mers in a roughly random distribution (Figure 2). While the method does not allow sequence assignments for the conversions, the approximate peak in the frequency distribution (Figure 2) at 9 conversions/ 36mer is consistent with the frequency of 10.8 conversions/36mer seen in bisulfite treated genomic DNA (Figure 3).
Further, the roughly random distribution of frequencies is consistent with the frontier orbital picture (Figure 1) since in the initial stages of the reaction, cytosines exposed in a loop would be expected to be only marginally more reactive than those in a closed i-motif. As the reaction progressed, bisulfite addition to the i-motif structure is expected to destabilize the structure by neutralizing the charge on the protonated cytosine and introducing sp3 carbons at both C6 and C5. Such destabilization is expected to permit further bisulfite addition at exposed dC+ residues in unpaired single-strands.
While the mass spectrometry data does not address the nature of the structure of the i-motif that forms in d(TCCC)9, for a sequence of this type with 27 dC residues, two forms seem plausible. In the first form the sequence can be viewed as two i-motif 16mers linked as follows: d(TCCC)4–d(TCCC)–d(TCCC)4 (Figure 7A). In the second a single 32mer forms the i-motif linked as follows: d(TCCC)–d(TCCC)8. Although our data do not prove that the structure is more like the second structure (Figure 7B) one expects the Tm for the tandem structure (Figure 7A) to be very close to that observed for d(TCCC)4 at neutral pH i.e. 15.6°C as opposed to the observed value for d(TCCC)9 at neutral pH of 40.7°C (Table 1A). In both models (Figure 7) a single dT residue is exposed in the loops of each i-motifs. Additional stability in a longer structure, like that depicted in Figure 7B, may be achieved in part by the additional stacking energy and in part by the increase in pKa associated with additional base pairing (39,40).
Figure 7.
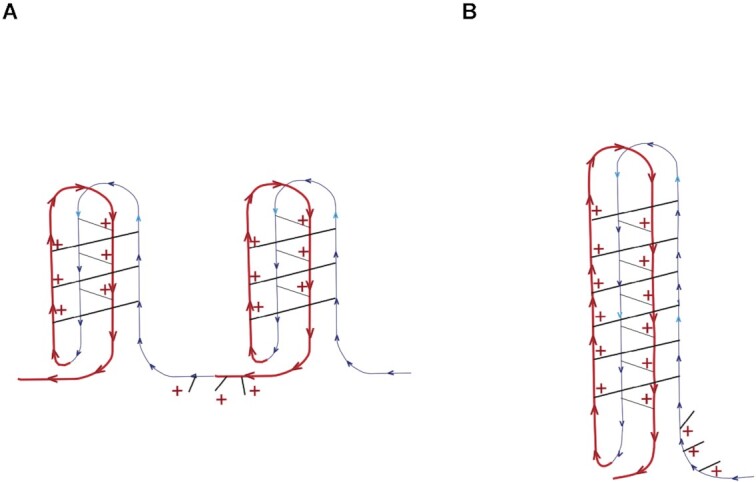
Possible structures for d(TCCC)9. (A) A Tandem Structure Composed of two linked i-motifs formed from two d(TCCC)4 elements linked by a single d(TCCC): d(TCCC)4–d(TCCC)–d(TCCC)4. (B) A single d(TCCC)9 i-motif. In each structure the loops contain a single dT residue.
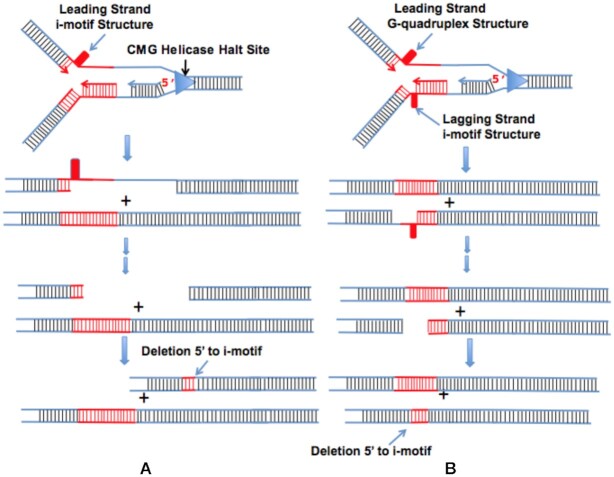
In addition to these findings it is important to note that independent of d(TCCC) repeat length, all of the deletions we observe, mapped near the 5′ end of C-rich strand of the d(TCCC)n element and often extended into the element itself (Figure 3). The opposite orientation for deletions has been clearly shown for dog-1 mutations in C. elegans (20,21). There, deletions map 5′- to the G-rich strand and G-quadruplex provides the impetus for deletion. Hence, the orientation of the human deletions reported here implicates i-motif formation during or prior to DNA replication in the genesis of deletions at d(TCCC)n sequences.
By analogy with the multiple repair systems known to play roles in suppressing G-quadruplex formation, it seems likely that similar suppression systems for i-motif formation may exist, given the association of the i-motif with deletions as described here. However, the efficiency of such systems remains unknown. In this regard, it must be noted that the PC3 cell line has been in long-term cell culture, hence the deletions we see may also indicate a simple stochastic accumulation of deletions representing the overall frequency of i-motif occurrence and the efficiency of suppression at each replication cycle. In this regard, it is interesting to note that if the in vitro pHT values for d(TCCC)n sequences for n ≥ 9 reflect a high rate of i-motif formation at these sites, and the overall efficiency of an i-motif suppression system translates to a probability of remaining unsuppressed in vivo during replication of only 0.05, then the probability that an i-motif would impair replication and induce DNA damage on average at least once in 25 replication cycles is 0.72.
Models of the deletion process induced by the dog-1 mutation in C. elegans suggest that poorly suppressed G-quadruplexes provide a replication impediment that creates a gap beyond the impediment that survives replication and leads to deletions in a subsequent replication cycle that map to 5′ end of the impediment (21,37). Our findings in the human PC3 cell line for i-motif impediments formed at the d(TCCC)n elements are completely consistent with this model (Figure 8). The data presented here and in (26) would support HMG helicase uncoupling and uncoupled replication followed by leading strand repriming to produce a gap (41) near an i-motif leading strand impediment (Figure 8A). A transient G-quadruplex that led to HMG helicase uncoupling and uncoupled replication and the formation of an i-motif on the lagging strand would leave a gap in the lagging strand 3′ to the i-motif (Figure 8B). Such a gap would explain why bisulfite can attack the region 3′ to d(TCCC)9 element in RACK7 (Figure 3). Given that the dog-1 mutation impairs G-quadruplex suppression and results in deletions mapping 5′ to end of the G-quadruplex in the dGn elements in C. elegans, models like that shown in Figure 8A in which the leading strand impediment is an unresolved G-quadruplex are ruled out since they require that the deletions occur 5′ to the G-quadruplex impediment on the G-rich strand and 3′ on the C-rich strand, because all of the deletions we observe are 5′- to the i-motif/G-quadruplex forming sequence on the C-rich strand. Given the the ability of the d(GGGA)n motifs to form G-quadruplexes in vitro over the range of lengths n > 3 we conclude that while G-quadruplex formation at these sites may be a necessary or contributing condition for these deletions, i-motif formation a under physiological conditions is required for deletions to occur.
Figure 8.
i-Motif induced deletion models. i-Motifs can form on either the leading or lagging strand depending on the direction of replication. When replication completes, either by restarting replication beyond the leading strand impediment or by replication from a distant replication origin two types of structure are created (20,21,36). A: When an i-motif forms the leading strand impediment and survives after replication deletions occur 5′ to the i-motif on the C-rich strand often extending into the repeat sequence itself. B: When a G-quadruplex forms a transient leading strand impediment and an i-motif forms on the lagging strand, deletions again occur near the i-motif often extending into the repeat sequence itself. Note that when only G-quadruplex survives replication as is the case in dog-1 mutant C. elegans deletions will occur 5′ to the G-quadruplex on the G-rich strand (20,21,36) often extending into the repeat itself.
Finally, given the dog-1 precedent indicating that loss of a G-quadruplex resolvase results in deletions at sites that can form G-quadruplexes and i-motifs it is reasonable to speculate that the failure of an unknown i-motif suppression system could result in the deletions in the deletions observed here in the PC3 cell line. Given the absence of a functional p53 gene in the PC3 cell line, failure to suppress i-motif formation may be linked to p53, however, direct evidence for this suggestion is currently unavailable.
Supplementary Material
ACKNOWLEDGEMENTS
Author contributions: M.M., R.V.C. and M.A.S.A. conducted experiments, F.P. helped with experiments, Z.A.E.W. designed and conducted experiments, S.S.S. designed and conducted experiments and wrote the paper.
Contributor Information
Marianna Martella, Judy and Bernard Briskin Center for Multiple Myeloma Research, City of Hope, 1500 E. Duarte Rd., Duarte, CA 91010, USA.
Flavia Pichiorri, Judy and Bernard Briskin Center for Multiple Myeloma Research, City of Hope, 1500 E. Duarte Rd., Duarte, CA 91010, USA.
Rupesh V Chikhale, School of Pharmacy, University of East Anglia, Norwich Research Park, Norwich NR4 7TJ, UK; UCL School of Pharmacy, 29-39 Brunswick Square, London WC1N 1AX, UK.
Mahmoud A S Abdelhamid, School of Pharmacy, University of East Anglia, Norwich Research Park, Norwich NR4 7TJ, UK.
Zoë A E Waller, School of Pharmacy, University of East Anglia, Norwich Research Park, Norwich NR4 7TJ, UK; UCL School of Pharmacy, 29-39 Brunswick Square, London WC1N 1AX, UK.
Steven S Smith, Department of Hematologic Malignancies Translational Science, City of Hope, 1500 E. Duarte Rd., Duarte, CA 91010, USA; Beckman Research Institute of the City of Hope, 1500 E. Duarte Rd., Duarte, CA 91010, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Funding
Biotechnology and Biological Sciences Research Council [BB/L02229X/1 to Z.A.E.W, 5R01-CA102521 to S.S.S.] from the U.S. National Cancer Institute of the National Institutes of Health; Ensign Foundation; Diabetes UK [18/0005820 to R.V.C.]; research reported in this publication also included work performed in the Integrative Genomics and Bioinformatics Core supported by the National Cancer Institute of the National Institutes of Health [P30CA033572]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Funding for open access charge: Institutional Funds.
Conflict of interest statement. None declared.
REFERENCES
- 1. Huppert J.L., Balasubramanian S.. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005; 33:2908–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sundquist W.I., Klug A.. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989; 342:825–829. [DOI] [PubMed] [Google Scholar]
- 3. Del Mundo I.M.A., Zewail-Foote M., Kerwin S.M., Vasquez K.M.. Alternative DNA structure formation in the mutagenic human c-MYC promoter. Nucleic Acids Res. 2017; 45:4929–4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Day H.A., Pavlou P., Waller Z.A.. i-Motif DNA: structure, stability and targeting with ligands. Bioorg. Med. Chem. 2014; 22:4407–4418. [DOI] [PubMed] [Google Scholar]
- 5. Smith S.S. Evolutionary expansion of structurally complex DNA sequences. Cancer Genomics Proteomics. 2010; 7:207–215. [PubMed] [Google Scholar]
- 6. Wu F., Niu K., Cui Y., Li C., Lyu M., Ren Y., Chen Y., Deng H., Huang L., Zheng S.et al.. Genome-wide analysis of DNA G-quadruplex motifs across 37 species provides insights into G4 evolution. Commun Biol. 2021; 4:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huppert J.L., Balasubramanian S.. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007; 35:406–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kendrick S., Kang H.J., Alam M.P., Madathil M.M., Agrawal P., Gokhale V., Yang D., Hecht S.M., Hurley L.H.. The dynamic character of the BCL2 promoter i-motif provides a mechanism for modulation of gene expression by compounds that bind selectively to the alternative DNA hairpin structure. J. Am. Chem. Soc. 2014; 136:4161–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clark J., Smith S.S.. Secondary structure at a hot spot for DNA methylation in DNA from human breast cancers. Cancer Genomics Proteomics. 2008; 5:241–251. [PMC free article] [PubMed] [Google Scholar]
- 10. Sarkies P., Reams C., Simpson L.J., Sale J.E.. Epigenetic instability due to defective replication of structured DNA. Mol. Cell. 2010; 40:703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mao S.Q., Ghanbarian A.T., Spiegel J., Martinez Cuesta S., Beraldi D., Di Antonio M., Marsico G., Hansel-Hertsch R., Tannahill D., Balasubramanian S.. DNA G-quadruplex structures mold the DNA methylome. Nat. Struct. Mol. Biol. 2018; 25:951–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Smith S.S. Stalling of DNA methyltransferase in chromosome stability and chromosome remodelling (Review). Int. J. Mol. Med. 1998; 1:147–156. [PubMed] [Google Scholar]
- 13. Ginno P.A., Lott P.L., Christensen H.C., Korf I., Chedin F.. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell. 2012; 45:814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun H., Karow J.K., Hickson I.D., Maizels N.. The bloom's syndrome helicase unwinds G4 DNA. J. Biol. Chem. 1998; 273:27587–27592. [DOI] [PubMed] [Google Scholar]
- 15. Fry M., Loeb L.A.. Human werner syndrome DNA helicase unwinds tetrahelical structures of the fragile x syndrome repeat sequence d(CGG)n. J. Biol. Chem. 1999; 274:12797–12802. [DOI] [PubMed] [Google Scholar]
- 16. Wu Y., Shin-ya K., Brosh R.M. Jr. FANCJ helicase defective in fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol. Cell. Biol. 2008; 28:4116–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eddy S., Ketkar A., Zafar M.K., Maddukuri L., Choi J.Y., Eoff R.L.. Human rev1 polymerase disrupts G-quadruplex DNA. Nucleic Acids Res. 2014; 42:3272–3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Betous R., Rey L., Wang G., Pillaire M.J., Puget N., Selves J., Biard D.S., Shin-ya K., Vasquez K.M., Cazaux C.et al.. Role of TLS DNA polymerases eta and kappa in processing naturally occurring structured DNA in human cells. Mol. Carcinog. 2009; 48:369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kononenko A.V., Ebersole T., Vasquez K.M., Mirkin S.M.. Mechanisms of genetic instability caused by (CGG)n repeats in an experimental mammalian system. Nat. Struct. Mol. Biol. 2018; 25:669–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kruisselbrink E., Guryev V., Brouwer K., Pontier D.B., Cuppen E., Tijsterman M.. Mutagenic capacity of endogenous G4 DNA underlies genome instability in FANCJ-defective c. elegans. Curr. Biol. 2008; 18:900–905. [DOI] [PubMed] [Google Scholar]
- 21. Koole W., van Schendel R., Karambelas A.E., van Heteren J.T., Okihara K.L., Tijsterman M.. A polymerase Theta-dependent repair pathway suppresses extensive genomic instability at endogenous G4 DNA sites. Nat. Commun. 2014; 5:3216. [DOI] [PubMed] [Google Scholar]
- 22. Fleming A.M., Ding Y., Rogers R.A., Zhu J., Zhu J., Burton A.D., Carlisle C.B., Burrows C.J.. 4n-1 is a "Sweet spot" in DNA i-Motif folding of 2′-Deoxycytidine homopolymers. Nat. Commun. 2017; 139:4682–4689. [DOI] [PubMed] [Google Scholar]
- 23. Cui Y., Kong D., Ghimire C., Xu C., Mao H.. Mutually exclusive formation of G-Quadruplex and i-Motif is a general phenomenon governed by steric hindrance in duplex DNA. Biochemistry. 2016; 55:2291–2299. [DOI] [PubMed] [Google Scholar]
- 24. King J.J., Irving K.L., Evans C.W., Chikhale R.V., Becker R., Morris C.J., Pena Martinez C.D., Schofield P., Christ D., Hurley L.H.et al.. DNA G-quadruplex and i-Motif structure formation is interdependent in human cells. J. Am. Chem. Soc. 2020; 142:20600–20604. [DOI] [PubMed] [Google Scholar]
- 25. Wolski P., Nieszporek K., Panczyk T.. G-Quadruplex and I-Motif structures within the telomeric DNA duplex. A molecular dynamics analysis of protonation states as factors affecting their stability. J. Phys. Chem. B. 2019; 123:468–479. [DOI] [PubMed] [Google Scholar]
- 26. Amparo C., Clark J., Bedell V., Murata-Collins J.L., Martella M., Pichiorri F., Warner E.F., Abdelhamid M.A.S., Waller Z.A.E., Smith S.S.. Duplex DNA from sites of helicase-polymerase uncoupling links Non-B DNA structure formation to replicative stress. Cancer Genomics Proteomics. 2020; 17:101–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Munson K., Clark J., Lamparska-Kupsik K., Smith S.S.. Recovery of bisulfite-converted genomic sequences in the methylation-sensitive QPCR. Nucleic Acids Res. 2007; 35:2893–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mergny J.L., Li J., Lacroix L., Amrane S., Chaires J.B.. Thermal difference spectra: a specific signature for nucleic acid structures. Nucleic. Acids. Res. 2005; 33:e138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Clark J., Shevchuk T., Kho M.R., Smith S.S.. Methods for the design and analysis of oligodeoxynucleotide-based DNA (cytosine-5) methyltransferase inhibitors. Anal. Biochem. 2003; 321:50–64. [DOI] [PubMed] [Google Scholar]
- 30. Raghavan S.C., Swanson P.C., Wu X., Hsieh C.L., Lieber M.R.. A non-B-DNA structure at the bcl-2 major breakpoint region is cleaved by the RAG complex. Nature. 2004; 428:88–93. [DOI] [PubMed] [Google Scholar]
- 31. Raghavan S.C., Tsai A., Hsieh C.L., Lieber M.R.. Analysis of non-B DNA structure at chromosomal sites in the mammalian genome. Methods Enzymol. 2006; 409:301–316. [DOI] [PubMed] [Google Scholar]
- 32. Shapiro R., Braverman B., Louis J.B., Servis R.E.. Nucleic acid reactivity and conformation. II. Reaction of cytosine and uracil with sodium bisulfite. J. Biol. Chem. 1973; 248:4060–4064. [PubMed] [Google Scholar]
- 33. Hayatsu H., Wataya Y., Kazushige K.. The addition of sodium bisulfite to uracil and to cytosine. J. Am. Chem. Soc. 1970; 92:724–726. [DOI] [PubMed] [Google Scholar]
- 34. Guo K., Gokhale V., Hurley L.H., Sun D. Intramolecularly folded G-quadruplex and i-motif structures in the proximal promoter of the vascular endothelial growth factor gene. Nucleic Acids Res. 2008; 36:4598–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Duquette M.L., Pham P., Goodman M.F., Maizels N.. AID binds to transcription-induced structures in c-MYC that map to regions associated with translocation and hypermutation. Oncogene. 2005; 24:5791–5798. [DOI] [PubMed] [Google Scholar]
- 36. Lemmens B., van Schendel R., Tijsterman M.. Mutagenic consequences of a single G-quadruplex demonstrate mitotic inheritance of DNA replication fork barriers. Nat. Commun. 2015; 6:8909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. van Kregten M., Tijsterman M.. The repair of G-quadruplex-induced DNA damage. Exp. Cell. Res. 2014; 15:178–183. [DOI] [PubMed] [Google Scholar]
- 38. Taylor M.R.G., Yeeles J.T.P.. Dynamics of replication fork progression following helicase-polymerase uncoupling in eukaryotes. J. Mol. Biol. 2019; 431:2040–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wright E.P., Huppert J.L., Waller Z.A.E.. Identification of multiple genomic DNA sequences which form i-motif structures at neutral pH. Nucleic Acids Res. 2017; 45:13095–13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Skolakova P., Renciuk D., Palacky J., Krafcik D., Dvorakova Z., Kejnovska I., Bednarova K., Vorlickova M.. Systematic investigation of sequence requirements for DNA i-motif formation. Nucleic Acids Res. 2019; 47:2177–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yeeles J.T.P., Poli J., Marians K.J., Pasero P.. Rescuing stalled or damaged replication forks. Cold Spring Harb. Perspect. Biol. 2013; 5:a012815. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.