Summary
Radial progenitor development and function lay the foundation for the construction of the cerebral cortex. Radial glial scaffold, through its functions as a source of neurogenic progenitors and neuronal migration guide, is thought to provide a template for the formation of the cerebral cortex. Emerging evidence challenges this limited view. Intriguingly, radial glial scaffold may also play a role in axonal growth, guidance, and neuronal connectivity. Radial glial cells not only facilitate the generation, placement, and allocation of neurons in the cortex but also regulate how they wire up. The organization and function of radial glial cells may thus be a unifying feature of the developing cortex that helps to precisely coordinate the right patterns of neurogenesis, neuronal placement, and connectivity necessary for the emergence of a functional cerebral cortex. This perspective critically explores this emerging view and its impact in the context of human brain development and disorders.
Keywords: Radial glia, Corticogenesis, Cortical connectivity, Cortical circuits, Neurodevelopmental disorders, Brain evolution
In Brief
Radial glial progenitors serve as an instructive matrix to coordinate the generation, placement, and connectivity of appropriate numbers and types of neurons in the developing cerebral cortex. Here Casingal et al explore this concept in the context of cortical development and neurodevelopmental disorders.
Introduction
Neurogenic and gliogenic functions of molecularly diverse radial progenitors enable the generation of appropriate number and types of neurons and glia during brain development (Borell & Reillo, 2012; Florio & Huttner, 2014; Hansen et al., 2010; Hartfuss et al., 2001; Lui et al., 2011; Noctor et al., 2001; Rakic 1988, 2009; Molnár et al., 2019). Radial glia’s simultaneous role in the guidance of neuronal migration leads to the allocation of the right number and types of neurons at the right time into different areas of the cerebral cortex (Rakic, 1988). The molecularly and areally diverse radial glia’s ability to direct or guide early axon/dendrite development of the newly arrived neurons in the cortical plate may then lead to the formation of distinct patterns of cortical neuronal connectivity (Henrikson & Vaughn, 1974; Kaur et al., 2020; Norris & Kalil, 1991; Xu et al., 2015; Okada et al., 2017; Varadarajan et al., 2017). Radial progenitor organization imparts essential modularity to how cortical neurons organize and connect in the emerging neocortex (Rakic, 1988, 2000, 2007, 2009; Nakagawa et al., 2019). These findings suggest that radial glia’s singular ability to coordinate neurogenesis, neuronal placement (i.e., laminar, columnar, and radial units), and neuronal connectivity during development underlies cortical circuit formation. Here, we explore this concept and the current knowledge of each aspect of radial glial function during the emergence of the cerebral cortex with an emphasis on how they may inform our path ahead for cortical development and associated disorders.
Development and Neurogenic Functions of Radial Progenitors
Neocortical development relies on the timely generation of appropriate number of neuronal and glial subtypes by radial glial cells (RGCs [also referred to as radial progenitors]) and their derivatives, intermediate progenitors (Borell & Reillo, 2012; Florio & Huttner, 2014; Götz et al., 2016; Haydar et al., 2000; Lui et al., 2011; Rakic, 2009). During the early stages of corticogenesis, polarized neuroepithelial cells (NECs), the predecessors of RGCs in the telencephalon, undergo interkinetic nuclear migration (IKN) and divide symmetrically to expand the neuroepithelial precursor pool. At the onset of neurogenesis, patterning morphogens from organizers surrounding the neural tube facilitate the transformation of NECs into ventricular radial glial cells (Haubensak et al., 2004). Loss of ZO1+ tight junctions and waning of FGF10 expression in NECs characterize this transition. Like NECs, RGCs maintain contact with both the apical and basal surfaces of the developing cerebral wall, but RGCs have a uniquely long, slim, basal process that spans the entire cerebral wall (Figure 1A). Maintenance of this apicobasal polarity is vital for RGC development and functions (Yokota et al., 2009; Weimer et al., 2009). Apical endfeet accumulation Numb and Numbl (numb-like) facilitate the basolateral insertion of cadherins in the apical adherens junctions, necessary to organize the apical end of RGCs (Kadowaki et al., 2007; Rasin et al., 2007). Further, polarized mRNA transport and local translation of signaling and cytoskeletal regulators, such as FMRP and Kif26a, at the RGC endfeet help maintain the basal process growth (Pilaz et al., 2016). Polarized transport of adhesion proteins like GPR88 to the endfeet and molecular signals derived from nascent CSF (e.g., LIFR signaling) further facilitate RGC organization (Chau et al., 2015; Nakagawa et al., 2019; Ferent et al., 2020). The combined activity of these mechanisms promotes the apicobasal architecture of RGCs and enables radial glia to maintain their contact with both ventricular and pial surfaces as the cerebral wall thickness expands during embryonic development.
Figure 1. Radial progenitors in the developing cerebral cortex.
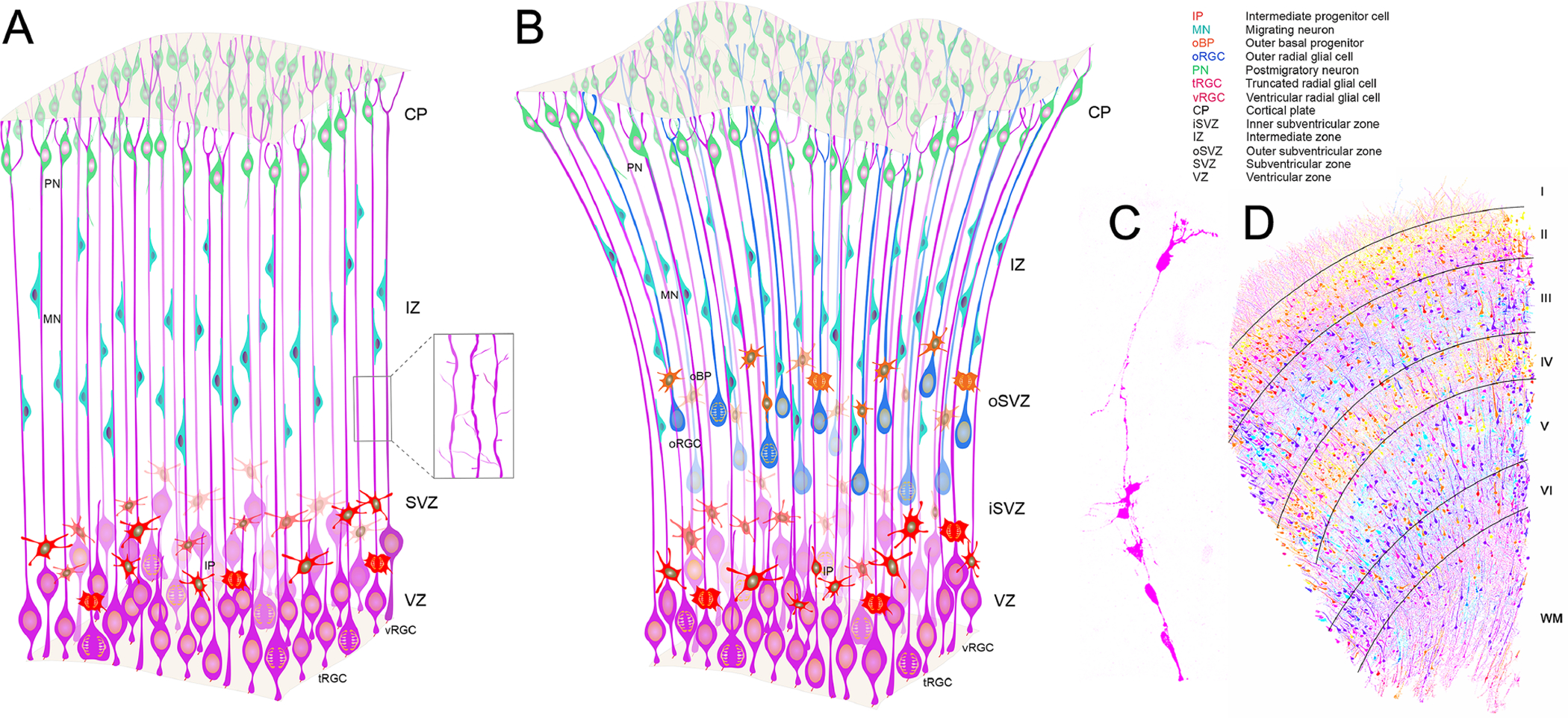
(A, B) Major radial glial types and other progenitors in the embryonic neocortex of mice (A) and humans (B). Boxed area (A) illustrates extensive inter-radial glial interactions all along basal processes. (C) MADM (Mosaic Analysis with Double Markers)-based labeling of a single radial unit containing a mother RGC and its progenies in E16 mouse cortex (modified from Nakagawa et al, 2019). Please note that in contrast to the cylindrically shaped radial units in mice, human radial units are more conically shaped due to the existence of oSVZ and abundance of oRGCs (Borrell, 2019). (D) Laminar and area-specific allocation of distinct types and numbers of neurons in human cortex resulting from embryonic radial glial development, organization, and function (modified from Shapson-Coe et al, 2021). Radial glial interactions with axons, interneurons, vasculature, and glia (astrocytes, microglia, and ependymal cells) are not illustrated. I-VI; cortical layers; WM, white matter.
The early RGCs divide symmetrically in the ventricular zone (VZ) to expand the RGC pool. The RGCs then divide asymmetrically, producing a self-renewing ventricular radial glial cell (vRGC; also called apical RGC [aRGC]) or an intermediate progenitor (IP). A balance in the energy metabolism of progenitor subtypes, particularly that of glycolysis versus oxidative phosphorylation (OXPHOS), likely regulated by angiogenesis based oxygen availability in the germinal zones, is a crucial regulator of progenitor differentiation (Khacho et al., 2016; Namba et al., 2021). For example, mouse vRGCs require higher glycolysis and lower OXPHOS, whereas IPs rely on lower glycolysis and higher OXPHOS. The IPs delaminate to the subventricular zone (SVZ). Newborn neurons migrate to the cortical plate using the basal processes of RGCs as a migratory scaffold (Rakic, 1978; Schmechel & Rakic, 1979a, 1979b). The vRGCs continue to divide asymmetrically, increasing the number of neurons while maintaining the vRGC pool in the VZ. In contrast, IPs rapidly divide symmetrically in the SVZ to expand the IP pool or to generate two neurons, hence, amplifying cortical neuronal production (Haubensak et al., 2004; Miyata et al., 2004; Noctor et al., 2004; Lui et al., 2011; Borell & Reillo, 2012; Florio & Huttner, 2014). In addition to vRGCs and IPs, the embryonic mouse neocortex includes other less abundant types of progenitors that have originated from the RGCs. These include the short neural precursors, which lack a long, radial basal process, but can directly produce postmitotic neurons (Stancik et al., 2010), neurogenic outer radial glial cells (Wang et al., 2011), and a subset of bipolar RGCs, which divide in sub-apical locations to generate IPs (Pilz et al., 2013). As neurogenesis decrease, gliogenic RGC divisions give rise to astrocytes, oligodendrocyte precursors (OPCs), or ependymal cells. Early born neurons are known to secrete cytokines such as cardiotrophin-1, which can act as a feedback mechanism to trigger the onset of neurogenic to gliogenic transition of radial progenitors (Barnabè-Heider et al., 2005). The remainder of RGCs, during late embryonic stages or soon after birth, lose their ventricular and pial attachments and transform into multipolar astrocytes (Bayraktar et al., 2015; Kriegstein & Alvarez-Bulla, 2009; Schmechel & Rakic, 1979a).
In contrast to mice, a specialized progenitor zone, outer subventricular zone (oSVZ), forms around gestational week (GW) 12 in humans. oSVZ contains progenitors derived from ventricular RGCs which have lost their apical contacts (Fietz et al., 2010; Hansen et al., 2010; LaMonica et al., 2013; Pollen et al., 2015; Reillo et al., 2011; Smart et al., 2002; Thomsen et al., 2015; Figure 1B). Daughter cells from oblique divisions of ventricular RGCs inherit the basal process but are unable to generate an apical endfoot via an apical adhesion plaque, thus leading to the formation of outer radial glial cells (oRGCs) (Fujita et al., 2020). These outer radial glial cells (also called basal radial glial cells [bRGCs]) are molecularly distinct, lack apical processes, and extend a radial process towards pia. oRGCs undergo asymmetric divisions where they self-renew and generate daughter IPs (i.e., basal intermediate progenitors). During this process, they undergo characteristically rapid “mitotic somatic translocation” (MST) along the basal process towards the pial surface just before division (Hansen et al., 2010; Ostrem et al., 2014). Human-specific expression of ARHGAP11B and post-translational modification of proteins via palmitoylation facilitates full level of oRGC proliferation in the human fetal neocortex (Florio et al., 2015; Heide et al., 2020; Kalebic et al., 2019). For example, human-specific expression of a morpho-regulatory protein PALMD, which upon palmitoylation associates with the plasma membrane to increase oRGC process formation necessary for interactions with the extracellular matrix and appropriate levels of oRGC expansion in the human embryonic neocortex (Kalebic et al., 2019; Namba et al., 2021). The proliferation of oRGCs is thought to underlie the massive expansion of cortical area and upper layer neurons in humans (Borrell, 2019; Nowakowski et al., 2016; Stahl et al., 2013; L. Wang et al., 2016). By early-mid neurogenesis (17 GW) oRGCs are the main contributors to the radial scaffold in the human embryonic cortex (Nowakowski et al., 2016). Some of the ventricular RGCs maintain apical attachments, but their basal processes no longer reach the pial surface, thus giving rise to truncated radial glia (around GW 14–14.5) in humans that are capable of neurogenesis, but are unable to support radial migration of neurons (LaMonica et al., 2013; Nowakowski et al., 2016).
Radial Glial guided Neuronal Placement and Allocation
Neurons, once generated from radial glia or IPs, migrate hundreds of cell soma size distances to their targeted laminar and areal locations. Neuronal cell types, numbers, and density across different layers and cortical areas vary (Figure 1D). These differences underly the functional specification of cortical areas and associated circuits (Brodmann, 2006; DeFelipe et al., 2002; Gouwens et al., 2019; Hodge et al., 2019; Molnàr & Rockland, 2020). For example, the visual cortex is characterized by a richly sub layered layer IV, whereas layer IV is barely detectable in the motor cortex. Proper acquisition of neuronal numbers and position is achieved through dynamic regulation of neurogenic progenitor proliferation and a process of active migration of newborn neurons away from the ventricular and subventricular zones to their final laminar destination in the cortical plate (Rakic, 1972; Rakic, 1988; Haydar et al., 2000). Early generated cortical projection neurons migrate radially using somal translocation, whereas later generated neurons arrive at the cortical plate using radial glia-guided locomotion (Hatten & Mason, 1990; Miyata et al., 2001; Morest & Silver, 2003; Nadarajah & Parnavelas, 2002; Noctor et al., 2001; O’Rourke et al., 1992; Rakic, 1972). Cortical interneurons, on the other hand, migrate tangentially into the cerebral wall from the ganglionic eminence before radial glial guided migration to the cortical plate (Anderson et al., 1997; Nadarajah & Parnavelas, 2002; Yokota, Gashghaei, et al., 2007). Comparative analysis of radial glial cell surface cues involved in neuronal migration or neuron- radial glial interactions indicates that the radial glial strands (i.e., basal processes) spanning the cerebral wall present an instructive pathway for neuronal migration, enabling a complex orchestration of changes in cell-cell recognition, cell adhesion, and cell motility events (Evsyukova et al., 2013). Temporal and spatial changes in the distribution of cell surface cues along radial glial guides actively instruct neuronal migration and placement during corticogenesis. For example, radial glial surface based anti-adhesive molecule, sparc-like 1 (SC1), localized in the segment of radial glia spanning the upper cortical plate (CP), where neurons terminate their migration, functions as a cue for cessation of neuronal migration by enabling neuronal detachment from radial glial guides at their final destination. SC1 reduces the adhesivity of neurons to the radial glial cell surface, thus causing them to detach and begin their aggregation into specific layers of the cerebral cortex. In addition to radial glial cues such as SC1, RGC induced changes in membrane trafficking and degradation of neuronal cell adhesion molecules such as N-cadherin may also provide a ‘release’ signal that initiates the conversion from the migratory to post-migratory state in the developing cortex (Gongidi et al., 2004; Kawauchi et al., 2010; Yokota et al., 2007).
Following detachment from radial glial guides, cohorts of excitatory projection neurons begin to assemble into their respective layers. Formation of cortical circuits involves laminar and areal specific placement of both excitatory and inhibitory neuronal subtypes. Live imaging of interneurons and radial glia in the developing cerebral cortex suggests that radial glial cells can repulse, attract, or allow oriented migration of interneuronal subsets in the developing cortex (Yokota, Gashghaei, et al., 2007). Tangentially oriented streams of interneurons in the marginal zone, intermediate zone, and ventricular zones invade the dorsal cortex from the subpallium. Local radial glial cells, by their ability to repulse, attract, stall, or permit the continued migration of distinct subsets of interneurons, may play a guiding role in the targeting of appropriate subsets of interneurons into correct areal locations (Yokota, Gashghaei, et al., 2007). Once the interneurons invade the cortex, differential interactions or specific positional affinities between interneurons and areally distinct radial glial scaffold may alter the tangential migratory trajectory of interneurons, and thus facilitating interneuronal positioning within distinct areal and laminar domains of the developing cerebral cortex. For example, as the interneurons migrating in the marginal zone stream reach their areal destinations, they descend into the underlying cortex to assume their position as Martinotti type interneurons (Lim et al., 2018). The identity of secreted or surface-bound radial glial signals during these interactions remains unclear. Nevertheless, the ability of radial glia to influence the migration of both excitatory and inhibitory neurons suggest that radial glia may provide an integrated, structural and molecular guidance matrix to coordinate the placement of both interneurons and excitatory neurons in the developing cerebral cortex, a requisite step in the formation of cortical circuits.
Radial Glia, Neuronal Connectivity, and Circuit Formation
Radial glial cell’s role in sculpting the dendritic and axonal growth and connectivity of neurons it has guided into position in cortex is surprisingly underappreciated. As neurons reach their target location, they extend a single axon and multiple dendrites, necessary for the formation of functional cortical circuitry. Radial glial scaffold affects the initiation of axons in excitatory neurons. During this process, N-cadherin-mediated radial glia–neuron interaction induces active RhoA at the neuronal contact site (i.e., in the leading process) and active Rac1 in the opposing, trailing neurite. RhoA–Rho-kinase signaling in the leading neurite inhibits axon formation, whereas Rac1 induction in the trailing neurite promotes elongation and axon formation, thus enabling radial glia to facilitate the axon-dendrite growth of newly arrived neurons in the cortical plate (Xu et al., 2015). In vitro studies have also identified radial glia secreted factors such as Meteorin as facilitators of axonal extension (Nishino et al., 2004).
Callosal axons, as they extend to connect with neurons in reciprocal regions of the contralateral cortical hemisphere, are intimately apposed to RG fibers, and thus RG scaffold is thought to guide callosal axons towards their cortical targets (Norris & Kalil, 1991). Further, Fgfr1-mediated somal translocation of radial glial cells from the dorsomedial ventricular zone (glial wedge region) to the subpial region of the dorsomedial pallium is critical for the callosal axons to cross the midline (Smith et al., 2006).
Corticospinal axons are guided to their subcerebral targets by the radial glial scaffold at the striato-pallidal junction. Perturbing the organization of these RG guides, which depends on a signaling interaction between an atypical Rho-GTPase, RND3 (RhoE), and its binding partner ARHGAP35 (P190A), a Rho GTPase-activating protein, results in the misrouting of corticospinal axons. In the absence of an organized RG scaffold at the striato-pallidal junction, these cortical axons do not enter nor form the internal capsule after coursing through the striatum, remain stuck at the striato-pallidal junction, and eventually misroute through the globus pallidus to aberrantly terminate near the surface of the ventral pallidum, instead of in the spinal cord (Kaur et al., 2020). Importantly, radial glial cells express and transport essential corticospinal axon growth and guidance regulators in a topographically selective manner. For example, secretion of the endosulfatases Sulf1 and Sulf2, by the radial glia of the third ventricle, is necessary for the extracellular removal of 6-O-sulfate groups from heparan sulfate (HS) and appropriate presentation of HS binding Slit 2 to developing corticospinal axons. Deletion of Sulf1 and Sulf2 in radial glia disrupts corticospinal tract formation and guidance (Okada et al., 2017; Aizawa et al., 2020).
Radial glia during late embryonic stages, particularly the ones transforming to become astrocytes, are thought to facilitate dendritic branching and growth (Crandall et al., 1990; Wallace & Withers, 2018), though the initial dendritic outgrowth of cortical neurons appears to not occur in alignment with radial glial fibers (Kostović, Sedmak, et al., 2019). However, neonatal radial glia may influence the refinement of dendritic patterning in functional cortical circuits. Whisker barrels in the somatosensory (S1) cortex of mice form from a seemingly homogeneous neuropil around first postnatal week. In the mature barrel cortex, the number of radial dendrites in the barrel walls is nearly twice that of the barrel hollows. Intriguingly, MAP2+ dendrites and RC2+ radial glial fibers are denser near the walls of barrels than near the hollows (centers) of barrels, suggesting that radial glial fibers may help differentially pattern radial dendrites in the layer IV of the barrel field. Further, apical dendrites of cortical neurons can bundle within cortical columns (Molnàr & Rockland, 2020). Whether this is in part due to early axo-dendritic polarity induced by radial glia on their neuronal progeny remains unclear.
Migration and axon targeting programs are also coupled in interneurons as they dive into different cortical layers (Lim et al., 2018). Whether radial glia, guiding the migration of interneurons during the final stages of their journey into the cortex, also impact the appropriate axon-dendrite initiation in these neurons remains enigmatic. Determining if radial glial guides can coordinately affect the final stages of migration and neurite growth of interneurons, as they do for excitatory neurons, will be highly informative. It will help establish if and how radial glia function to optimize and coordinate the assembly of excitatory/inhibitory circuits in the cerebral cortex.
In humans, between 11.5–13GW, thalamocortical (TH-C) axons traverse the periventricular area and enter the cortical intermediate zone in a radiating fashion, reminiscent of association with radial glial fibers (Krsnik et al., 2017). This early invasion of thalamocortical projections into cortical neuroepithelium before the onset of neurogenesis suggests a potentially influential role for these afferents in cortical progenitor behavior (Marin-Padilla, 1983). TH-C axons can release FGF to influence RGC proliferation (Dehay et al., 2001). As they wait to enter the cortical plate, between 16–20GW, TH-C axons interdigitate with callosal axons which are known to align with RG fibers (Krsnik et al., 2017). Furthermore, diffusion MR imaging suggests that corticofugal projection axons are positioned closer to the ventricles when compared to corticopetal (i.e., thalamocortical) fibers, which run more superficially (Vasung et al., 2017). These invading callosal, corticofugal, and corticopetal axonal fibers, traversing in radial, sagittal, and transverse directions, delaminate the SVZ into multiple cell bands, thus forming a multilaminar axonal-cellular compartment (MACC), below the subplate, in the ventricular zone. This meshwork of progenitors, radial glial fibers, and axons form a cellular grid work in the ventricular niche that may enable radial glia to simultaneously modulate neurogenesis, neuronal migration, and axonal growth in and around the proliferative zone during human cortical development (Išasegi, Žunić et al., 2018; Kostović, Sedmak, et al., 2019; Rakic, 1988, 2007; Figure 2).
Figure 2. Progenitor niche of the human fetal cerebral cortex.

Cellular diversity in a segment of the ventricular and outer subventricular regions of the early to mid-fetal (~16–20GW) dorsal cortex.
Radial Unit Hypothesis and Cortical Development
Radial glia’s role as neuronal progenitors and guides of neuronal migration is the basis for the “radial unit hypothesis”, an enduring conceptual framework for cerebral cortical formation (Figure 1 C; Rakic, 1988; Noctor et al., 2001; Hartfuss et al., 2001). According to this hypothesis, “the ependymal layer of the embryonic cerebral ventricle consists of proliferative units that provide a proto-map of prospective cytoarchitectonic areas. The output of the proliferative units is translated via glial guides to the expanding cortex in the form of ontogenetic columns, whose final number for each area can be modified through interaction with afferent input”. Consistent with this hypothesis, cortical expansion in evolution is accomplished primarily by expanding cortical surface area. In contrast, cortical thickness changes minimally (Molnàr & Rockland, 2020). The cerebral cortical surface area ratio in mouse, macaque, and human is ~1:100:1000, whereas the average cortical thickness ratio is similar in all three species (Molnàr & Rockland, 2020; Rakic, 2009).
Evolutionary expansion of radial unit numbers as the basis for cortical surface expansion is an attractive preposition worth further exploration. Importantly, genetic variants associated with the cortical global surface area (and the surface area of multiple sub regions of the cortex [caudal anterior cingulate, entorhinal, lateral occipital, lingual, and pericalcarine]), but not the average thickness across the entire cortex, are significantly enriched in human cortical progenitors, perhaps indicative of the significance of expansion of radial progenitor pool for the increased cortical surface area in humans (Liang et al., 2021). Lineage tracing studies indicate that the pyramidal neuronal progeny of a single radial glial cell in the dorsal telencephalon is organized into defined radial clusters of sibling excitatory neurons (Kornack & Rakic, 1995; Nakagawa et al., 2019; Noctor et al., 2001). For example, neuronal siblings derived from E13 radial progenitors in mice span 150–250 μm in the horizontal axis and contribute to all cortical layers generated after E13 (Costa et al., 2009). On average, a neurogenic RGC produces 8–9 neurons (Gao et al., 2014). Excitatory neurons generated from the same radial progenitor not only keep spatial relationships but also display preferential synaptic connections. Multiplication of the ontogenetic columns of radial units that can form minicolumns or columns of neurons may thus underlie the increased cerebral cortical complexity during evolution (Rakic, 1988, 2009), though the precise number of radial units that make up an anatomical or functional mini column or column in mature cortex remains to be established.
Regional Diversity of Radial Progenitors, Inter RGC Interactions, and Cortical Organization
The areal diversity of cortical neurons and cortical circuit organization germinates from the molecular diversity in radial progenitors. Analysis of the expression patterns of 234 ventricular zone-specific genes (www.eurexpress.org) reveals strong regional diversity in the molecular characteristics of radial glial progenitors (Figure 3A–B). Transcriptional priming, reflected by the expression of untranslated mRNAs in radial progenitors, facilitates radial progenitor diversity (Li et al.,2020). This regional diversity in radial progenitors may initiate distinct patterns of intrinsic gene regulatory programs in them. Thus providing a matrix to dynamically coordinate the neurogenic, neuronal migration, and neuronal growth influencing functions of radial glia necessary to generate distinct cortical functional domains (Figure 3C–D). Examination of how perturbing this regional molecular diversity of RGCs affects the generation, placement, allocation, and connectivity of specific neurons in distinct cerebral cortical regions will help to firmly establish if radial glia are the guiding elements of the developing cerebral cortex that coordinate the right patterns of neurogenesis, placement, and connectivity necessary for the emergence of a functional cerebral cortex. Supporting this possibility, perturbing transcription factors such as Emx1, Emx2, Pax6, Sp8, and Lhx2, which are preferentially expressed in distinct RGCs, lead to changes in the cortical area maps (O’Leary et al., 2007).
Figure 3. Radial progenitor diversity and its impact on the construction of the cerebral cortex.

(A) Molecular and areal diversity of radial progenitors. Different patterns of radial progenitor gene expression in the ventricular zone (arrow) across the developing mouse cerebral cortex (E16; from www.eurexpress.org). A, anterior; P, posterior; D, dorsal; V, ventral. (B) Different gene expression patterns were superimposed to illustrate the molecular diversity of radial progenitors in distinct cortical ventricular zone regions. (C) Molecularly distinct radial glial cells (purple) give rise to intermediate progenitors (yellow) and neurons. Neurons (blue) use RGC basal processes as migratory guides. RGCneuron interactions facilitate axon-dendrite growth (red, green) as they settle into distinct layers in the cortical plate. Migrating neurons are illustrated in shades of blue. Upper and lower layer neurons are red and green, respectively. (D) The function of molecularly distinct radial glial cells (purple-orange) helps integrate neuronal genesis, migration, placement, and connectivity in distinct domains of the developing cerebral cortex. Axons initially align with radial glial guides and strategically placed radial glia along the route (gray) further guide the targeting of major cortical axonal tracts. Callosal axons are illustrated in green and corticospinal axons are in red.
Molecularly distinct radial glia appear to provide a surprisingly dynamic scaffold for the generation, guidance, and growth of neurons during cortical development. Live imaging of radial glial populations indicates that radial glial basal processes within the scaffold actively extend, retract, or interact with each other (Yokota et al., 2010). Adjacent radial glial cells extend filopodial or spine-like protrusions all along their processes to contact each other. RGCs can also extend longer processes (10–40 μm) that can span the width of several radial glial cells, enabling interactions between non-adjacent radial glia (Figure 1 A [inset]). Further, the radial glial endfeet are not stable structures attached to the pial basement membrane, rather, they are highly motile, constantly remodel, continuously probe adjacent radial glia, and may modulate the migratory behavior of neurons passing through the marginal zone. These extensive inter-radial glial interactions appear to occur concurrently as radial glia proliferate and support neuronal migration. Neurons can also sequentially migrate along adjacent radial glial fibers (Gertz & Kriegstein, 2015). In the ventricular zone, a trigger radial glial cell releasing ATP can initiate calcium waves in a cohort of radial glia cells surrounding it. Spontaneous calcium waves propagating through clusters of radial glia may modulate the proliferation of cohorts of radial glia, leading to the production of isochronic neurons destined to the same cortical layer or column (Weissman et al., 2004). Furthermore, calcium waves in radial glial clusters may also promote the release of neurotransmitters (e.g., glutamate or GABA), which in turn can modulate or trigger RG-guided neuronal migration of these isochronic neurons (Marín & Rubenstein, 2003). The dynamic interactions between cohorts of molecularly distinct radial glia all along their length may further coordinate neuronal generation, migration, and growth, and thus promote and refine the emergence of ‘radial unit’ like organization and connectivity of the cerebral cortex (Rakic, 1988).
Transcriptomic and Epigenomic Analysis of Radial Glial Diversity and Function
What mechanisms regulate the cellular and functional diversity of radial progenitors? Recent single-cell RNA sequencing (scRNA-seq) and transposase-accessible chromatin using sequencing (scATAC-seq) studies in human and mouse cortices have established transcriptional heterogeneity of progenitors and their progenies, resulting in the identification of different cell type taxonomies, lineage trajectories, and spatial reconstruction of gene expression profiles during cortical development (DiBella et al., 2021; Kang et al., 2011; Loo et al., 2019; Li et al., 2018; Li et al., 2020; Nowakowski et al., 2017; Song et al., 2020; Trevino et al., 2021; Telley et al., 2019; X. Wang et al., 2011; Zhong et al., 2018).
Single-cell RNA-sequencing across multiple regions of the developing human brain (GW6–10) has identified nine different radial progenitor populations in the telencephalon (Eze et al., 2021). 3/4th of these progenitors expressed cortical areal identity. RNA-Velocity analysis suggests lineage relationships between many of these progenitor clusters. This molecular and spatial atlas of the early embryonic human brain demonstrates that RG cell types are organized in a hierarchical manner, exhibiting varying degrees of relatedness with each other and their progeny. Further, chief regulators of cell fate transition of these progenitor clusters are enriched for genes associated with various neurodevelopmental and psychiatric disorders (e.g., autism, schizophrenia, cortical malformations). Two of these progenitor clusters, marked either by C1orf61 or ID4 expression, are enriched in the developing human, but not mouse cortex. Other human-specific radial glial clusters (e.g., Sox2 only bRGCs) have also been identified (Li et al., 2020). The unique presence of these progenitor clusters may enable accelerated expansion of progenitors and neurons in the human cortex when compared to mice (Eze et al., 2021). Further, elucidation of spatially resolved single cell transcriptomes (Rodriques et al., 2019; Kanton et al., 2019; Li et al., 2020) of these RG subtypes across the entire range of human cortical development will be highly informative in revealing how these cell types contribute to areal and circuit diversity in the neocortex.
Signaling pathways embedded within the RG transcriptome display heterogeneous activation during radial glial diversification (Nowakowski et al., 2017). Several scRNA-seq studies show dynamic expression of genes that are associated with LIFR/STAT3, mTOR, Wnt, FGF, and Notch signaling pathways in cortical progenitors (Eze et al., 2021; Micali et al., 2020; Pollen et al., 2015; Suzuki et al., 2018). LIFR and its co-receptor IL6ST (GP130) are selectively enriched in oRGCs but not in vRGCs (Pollen et al., 2015). Further, oRGCs show enriched expression of several regulators of the mTOR signaling pathway and increased phosphorylation of the S6 ribosomal protein, when compared to vRGCs, tRGCs, and IPs (Nowakowski et al., 2017). Dysregulation of mTOR signaling specifically affects oRG cellular morphology, migration, and mitotic behavior, but not proliferation or cell fate (Andrews et al., 2020).
Temporal analysis of radial progenitor molecular states in mice revealed that a core set of evolutionarily conserved, temporally patterned genes that sequentially unfold during development, drive the differentiation of radial progenitors and their ability to generate appropriate type and number of cortical neurons with proper patterns of connectivity (DiBella et al., 2021; Telley et al., 2019; Li et al., 2020). Initially, radial progenitors express mainly genes that are DNA replication/cell cycle, chromatin organization, or cell-death related, necessary for the control of self-renewal and pool size. As development proceeds, expression of genes for membrane receptors, lipid metabolism, cell-cell signaling-related proteins, and excitability-related proteins facilitate the ability of progenitors to respond to environmental signals. This is reflected by the gradual lengthening of G1 phase of cell cycle necessary for environment sensing, fate decision, and increased competence in progenitors’ ability to respond to environmental stimuli such as glutamate or Ca2+. Importantly, radial progenitors begin to acquire neuronal transcription features and express neuronal differentiation (e.g., axonogenesis, dendrite development) and function (e.g., synaptogenesis, neurotransmission) related genes as corticogenesis proceeds and neurogenesis accelerates. Such temporally patterned genes are transmitted from apical progenitors to their neuronal progeny, essentially giving them molecular birthmarks or seeds of identity that are further refined into final neuronal identity and connectivity by cell-extrinsic input (DiBella et al., 2021; Telley et al., 2019; Li et al., 2020). For example, receptors such as NTRK3, a TrkC receptor critical for neuronal survival, are colocalized with SOX2-positive early radial progenitor population and shift from progenitor to neuronal expression as neurogenesis unfolds (Eze et al., 2021). Progenitors may be epigenetically primed for future cell fates by the stage-specific presence and activity of super-interactive promoters or genes with predictive chromatin (i.e., genes with large numbers of nearby accessible enhancers) enriched for binding of lineage-defining transcription factors (DiBella et al., 2021; Song et al., 2020; Trevino et al., 2021). This evolutionarily conserved program is defined by the progressive loss of radial progenitor–derived molecular birthmarks and the gradual prominence of input-dependent transcriptional programs, epigenetically modulated in part by the Polycomb repressor complex PRC2. Consistent with this, the loss of PRC2 leads to atypical acceleration of the sequence of radial progenitor’s neurogenic differentiation program (Telley et al., 2019).
In addition to the transcriptome, the epigenome has also been intensively investigated to understand the sequential specification of RGCs and their role in the generation of diverse cortical cell types (DiBella et al., 2021; Song et al., 2020; Trevino et al., 2021). The identity of each RGC subtype is associated with unique transcriptional signatures shaped by epigenetic modifications. Analysis of global changes in the DNA methylome during cortical development showed three waves of regulation: the first wave is demethylation of neuron-specific gene promoters during the transition from NECs to RGCs/IPs; the second step is demethylation of glial gene promoters, which occurs during the differentiation of RGCs/IPs into potential astrocytes and oligodendrocytes; and the third step is de novo methylation of neuronal genes in glial cells to inhibit the expression of neuronal genes in glia (Sanosaka et al., 2017). Further, promoters of both neurogenic and gliogenic genes undergo various histone modifications to ensure the sequential production of different cell types at the appropriate stages of cortical development (Hirabayashi & Gotoh, 2010). For example, methylation on H3K4 and H3K36 usually activates gene transcription critical for neurogenesis (Bernstein et al., 2005). In contrast, the loss of ASH2L, a COMPASS family histone methyltransferase co-factor, in RGCs impairs H3K4me3 levels, disrupts transcriptional activation of Wnt-β catenin signaling, and inhibits progenitor proliferation and neurogenesis (L. Li et al., 2019).
Approaches such as Paired-seq (parallel analysis of individual cells for RNA expression and DNA accessibility by sequencing), SNARE-seq (single-nucleus chromatin accessibility and mRNA expression sequencing), and other combinations of scRNA-seq and scATAC-seq, for simultaneous profiling of transcriptome and accessible chromatin in millions of fetal forebrain cells, are enabling us to infer the transcriptome and open chromatin landscapes in cortical progenitor cell types, examine how gene expression patterns defining these cell types are transcriptionally regulated, and characterize the cellular lineage trajectories of these cells within the developing cortex (Chen et al., 2019; DiBella et al., 2021; Trevino et al., 2021; Zhu et al., 2019). Linking chromatin accessibility to the corresponding transcriptome suggests that accessible sites in RGCs are enriched for binding motifs of transcription factors (TFs) such as Sox2 and LHX2, regulators of neurogenesis and gliogenesis. Similarly, open sites in radial glia that are not in cell cycle are enriched for Wnt signaling pathway components. Pseudotime trajectories of progenitors to cortical excitatory neuron gene expression profiles and the trajectories of accessibility profiles show corresponding directional changes in promoter accessibility and gene expression levels. For example, Sox6, a transcription factor needed for the maintenance of neural progenitors showed a decline along neuronal differentiation, whereas Khdrbs2 (SLM1), encoding an RNA-binding alternative splicing regulator, and its target Nrxn1 (a synaptic receptor) showed a corresponding directional rise along neurogenesis (Chen et al., 2019).
Furthermore, Methyl-HiC or single-nucleus methyl-3C sequencing assays that can capture both the chromatin organization and DNA methylome in a cell may help reveal how coordination of DNA methylation between distal genomic segments that are in spatial proximity in the nucleus facilitates the chromatin architecture and DNA methylome necessary for RG cell-type-specific functions in the developing cortex (Li et al., 2019). Detecting genome-wide RG type specific chromatin conformation and differential DNA methylation will help identify the type of interactions between epigenetic mechanisms necessary to regulate gene expression underlying RG cell functions in the developing cortex (Albert et al., 2017; Liang et al., 2021; Song et al., 2020). For example, human RGCs are enriched, relative to other cortical cell types, for chromatin interactivity of Notch signaling genes and human gained enhancers, which are critical for the expansion of RGCs (Song et al., 2020). Identifying the RG-specific overlap between chromatin interactions and differentially methylated regions (DMRs), across cortical development, will help construct the evolution of RG cell type specific chromatin conformation maps during cortical development (Armand et al., 2021; Lee et al., 2019; Song et al., 2020).
Development and refinement of new approaches to comprehensively identify unique cellular signatures of radial progenitors, such as single-nucleus methylCytosine, Chromatin accessibility and Transcriptome sequencing (snmC2T-seq), will facilitate parallel analyses of the methylome, transcriptome, chromatin accessibility and conformation in these RG cell types. Identifying the precise connections between epigenome and transcriptome in specific progenitor types and reconstructing regulatory lineages for these cell populations throughout cortical development will be necessary to fully identify their functional impact on the emergence of areal identity and circuit specification in human cortex. Further, identification of enriched genetic risk for neurodevelopmental disorders (NDD) within these unique RG cellular signatures will enable one to predict with temporal and spatial specificity the progenitor cell types with causal roles in neurodevelopmental diseases (Di Bella et al., 2021; Liang et al., 2021; Song et al., 2020; Trevino et al., 2021). Combining such approaches with high-throughput functional screens (e.g., Perturb-Seq; Jin et al., 2021) and spatial transcriptomics can yield a rich canvass of downstream cellular and molecular effects of NDD risk genes in radial progenitors.
Imaging and Genetics of Human Brain Disorders of Radial Progenitor Development and Function
Despite the insights gained from genetic model systems, our understanding of radial progenitor-guided human brain development and malformations is imprecise. Combined application of high resolution human fetal brain imaging and genetic dissection of neurodevelopmental disorders provides an effective window into the in utero dynamics of radial guided cortical development. Diffusion tensor imaging (DTI) of fetal human brains indicates a correlation between DTI fractional anisotropy (FA) and histology (Huang et al, 2012). High FA values and organized radial orientation of diffusion tensor primary eigenvectors correlate with organized radial glial scaffold in histological images (Ouyang et al., 2019; Takahashi et al., 2011; Vasung, Charvet, et al., 2019; Figure 4A–B). Heterogeneous nature of FA maps in 13–21GW human fetal brains likely indicates the regional differences in radial glial development and function (Huang et al., 2013). The radial coherence of telencephalic cerebral wall, reflecting the radial orientation of radial glial scaffold and associated migrating neurons and axons, is prominent in GW 18–30 human brains when radial glial guided cortical development is unfolding (Khan et al., 2019; Kostović et al., 2021; Kostović, Sedmak, et al., 2019; Takahashi et al., 2012; Vasung et al., 2017; Vasung, Charvet, et al., 2019; Xu et al., 2015). Disruption of the radial organization of radial glial scaffold results in FA decrease (Ouyang et al., 2019). Further, transformation of radial glia into astrocytes correlates with the emergence of cortico-cortico and subcortical tracts in high-angular resolution diffusion (HARDI) tractography (Ouyang et al., 2019; Vasung, Charvet, et al., 2019). Radial organization diminishes first in dorsal parieto-occipital regions and then in ventral frontotemporal regions. Crest of gyri tends to have longer persistence of radial organization than the depths of sulci (Takahashi et., 2012). Developmental stage-specific appearance (during GW13–21) and gradual disappearance (around GW21–39) of radial coherence provide a time window in which detection of changes in normal cortical patterns of radial coherence may provide the earliest indication of radial progenitor malfunction (Khan et al., 2019; Vasung, Charvet, et al., 2019).
Figure 4. Human fetal imaging of radial glial scaffold.
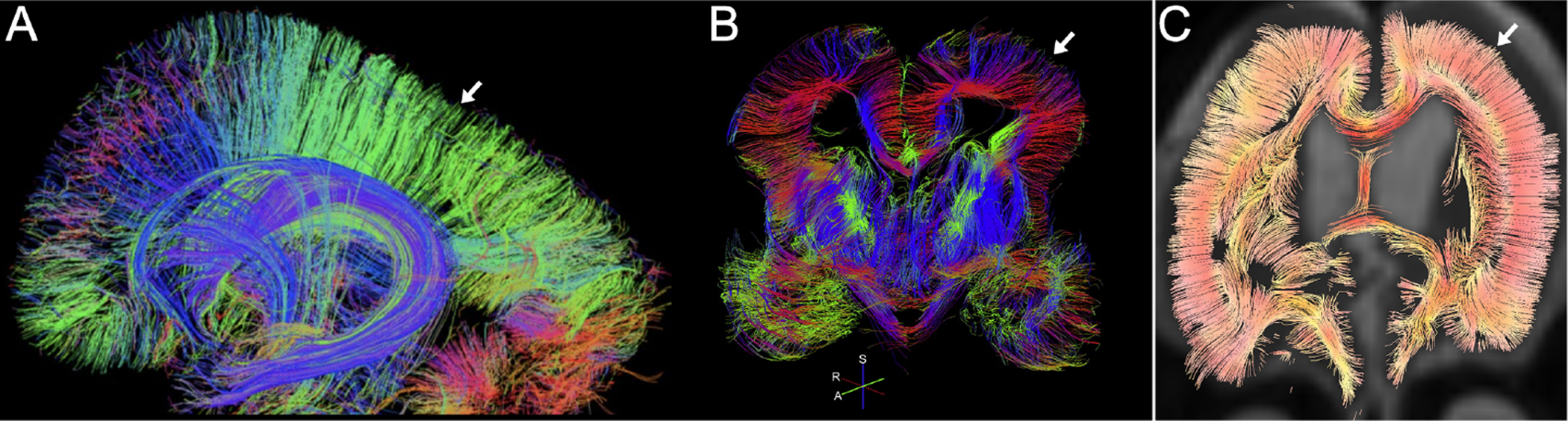
Diffusion-weighted MRI tractography of radially coherent structures (arrow) indicative of radial glial scaffold in GW18 (sagittal, A) and GW19 (coronal, B) human fetal cortex. (C) Persistence of these radial structures (arrow) in a one-year-old patient with lissencephaly, indicative of aberrant radial glial development. R, right; A, anterior; S, superior. Copied from Takahashi et al, 2012 (A), Kolasinski et al, 2013 (B), and Kamiya et al, 2014 (C).
Intriguingly, genes that are expressed in embryonic RGCs (e.g., ADCY8, ITGA9) show a strong correlation between gene expression and FA ratio changes, suggestive of their influence on regional growth and connectivity patterns of the human embryonic cortex (Huang et al., 2013). In Down syndrome (DS) fetal brains, characterized by simplified gyri and reduced cortical surface area, correlative histological and MRI studies indicate that reduced Sox2+ radial progenitors may correspond to reduced and altered diffusion MRI radial coherence structures in oSVZ and IZ (Baburamani et al., 2020). Consistent with these observations, postnatal MRI studies of DS patients demonstrate irregular cortical thickness (Levman et al., 2019). Neuroimaging studies of microcephalic, megalencephalic, or lissencepahlic patients, where radial glia mediated neurogenesis or neuronal migration is affected, indicate abnormal axonal fiber tract organization of affected neurons (Kamiya et al., 2014; Poretti et al., 2013; Vasung, Rezayev, et al., 2019). Mutation in TUBA1A, a gene expressed in radial progenitors, leads to microcephaly, lissencephaly, and associated intractable epilepsy and developmental delay. DTI imaging at post-birth (~1 year) indicates the persistence of radially coherent structures that are normally evident only during embryonic gestational weeks (15–28), indicating disrupted radial scaffold/unit formation and function in these patients (Kamiya et al., 2014; Figure 4C). These studies highlight the potential value of non-invasive imaging methods in predicting changes in radial glial guided cortical development and subsequent neurological consequences. Further, preterm birth-related alterations in cortical morphometry and microstructure captured by multimodal MRI correlate with gene expression changes underlying radial progenitor differentiation and neuronal migration (Ball et al., 2020). While the in vivo MRI characteristics (e.g., FA and signal intensity values) change across the developing cerebral wall domains (e.g., VZ, IZ, SP, CP) and with gestational age, ex vivo MRI and corresponding histological characterization of human fetal brains are essential to establish imaging landmarks characteristic of radial glia scaffold, radial neuronal migration, neuronal lamination, and radial glial associated growth of axons/dendrites in cortex (Kolasinski et al., 2013; Takahashi et al., 2011; Vasung, Charvet, et al., 2019; Vasung, Rezayev, et al., 2019; Vasung, Turk, et al., 2019). In particular, light sheet imaging of immunostained and cleared human fetal brains may provide high resolution 3D images of cellular dynamics in the developing cortex (Belle et al., 2017). Combining such approaches with comprehensive spatiotemporal gene expression and progenitor subtype maps of the developing fetal human cortex and AI based deep-learning models of pattern recognition may help establish a reference map of MRI landmarks underlying RG guided cortical development in utero during human gestation. It will help assess the trajectories of normal cortical development as well as disorders arising from radial glial malfunction in real-time in humans.
Cortical surface area expansion and cortical thickness variability associated with autism spectrum disorder (ASD), as well as the brain overgrowth and macrocephaly linked to a subset of genetically-defined ASD patients, suggest that disrupted radial progenitor function and the resultant changes in local cortical connectivity may underly the ASD pathophysiology in these patients (Bernier et al., 2014; Ciarrusta et al., 2020; Fang et al., 2014; Hazlett et al., 2017; Levman et al., 2019; Marchetto et al., 2017; Mariani et al., 2015; Ohta et al., 2016; O’Reilly et al., 2017). Several ASD risk genes (e.g., TCFL2) are enriched in oRGC (Polioudakis et al., 2019; Pollen et al., 2015; Satterstrom et al., 2020; Treviono et al., 2021). Furthermore, ASD-associated mutations in genes such as MEMO1 disrupt radial glial tiling and results in focal disruption of cortical columnar and laminar architecture. Disrupted laminar architecture in Memo1 cKO cortex resembles patch-like, focal neuronal disorganization found in prefrontal, temporal, and occipital neocortical regions in children with autism (Stoner et al., 2014). These regions mediate communication, emotional, social, and language functions that are perturbed in autism. Measuring MRI radial coherence changes in utero in subjects with a family history of ASD may therefore help predict ASD diagnostic outcome at 24 months, thus enabling early presymptomatic intervention in ASD risk children. Together, these studies indicate the impact of radial progenitor organization and function on cortical circuit formation relevant to ASD.
Common risk variants for multiple neuropsychiatric disorders, including ASD, schizophrenia, major depressive disorder (MDD), neuroticism, and depressive symptoms, are enriched in regulatory elements that are active during midgestation, particularly during radial progenitor development and differentiation in cortex. Integrated examination of chromatin accessibility Quantitative Trait Loci (caQTLs) and brain relevant genome wide association data indicates that noncoding risk alleles disrupt transcription factor binding within progenitor-type-specific regulatory elements (REs), hence leading to disruptions in gene expression and downstream disease risk effects. Analysis of caQTLs in cultured human neural progenitor cells and their differentiated neuronal progeny from a large number of donors (87) suggest that a subset of MDD risk variants are enriched in progenitor specific regulatory elements and affect chromatin accessibility and gene expression within progenitors, but not in neurons. For example, a SNP strongly associated with risk for MDD disrupts the motifs of multiple TFs, including ETV1, and is associated with differences in chromatin accessibility specifically in progenitors. These observations suggest that genetic variants that disrupt human progenitor REs contribute to risk for neuropsychiatric disorders, perhaps by affecting transcriptional regulation of radial glia mediated functions that are necessary for circuit formation (Liang et al., 2021). Developing a patient cell derived human cortical organoid system that has consistent high fidelity to all stages of RGC development in the human fetal cortex (Eze et al., 2021) will vastly facilitate this effort.
Opportunities and Outlook
Collective evidence on radial glial development, diversity, and functions imply that radial glia serve as a coordinator of neurogenesis, neuronal placement, and connectivity in the developing cerebral cortex. Yet, several major challenges remain as indicated earlier and below.
Detailed ultrastructural reconstruction of radial glial morphology and their cellular interactions, particularly during the late stages of cortical development, is lacking. Whether they form functional associations with immature dendritic spines, growing vasculature, newly formed astrocytes, or microglia remain underexplored (Bjornsson et al., 2015). Such interactions with their local cellular environment could allow them to be receptive to signals from neurons, glia, and vasculature, which may then modulate RG fate and functions, thus reciprocally sculpt the emerging neural circuitry in the neonatal cortex.
An expanding array of diverse radial glial identities is being defined by combined transcriptomic and epigenomic approaches (Di Bella et al., 2021; Eze et al., 2020; Li et al., 2020; Song et al., 2019; Trevino et al., 2021). Characterizing the morphological and topographic attributes of these progenitors in the developing cortical progenitor niche will be necessary to fully evaluate their functional significance in the generation of cortical neuronal, astrocyte, and oligodendroglial diversity.
A major unanswered question is how the integrative function of radial glia leads to the assembly of local cortical circuits. While the radial allocation of clonally related neurons from a progenitor is clear, transfer of neurons from the parental to the neighboring radial glial fibers can occur (Gertz & Kriegstein, 2015; Kornack & Rakic, 1995; Noctor et al., 2001; Tan & Breen, 1993) and this mixing may serve to introduce functional diversity between adjacent radial units. Further, how radial units of excitatory neurons recruit appropriate types and number of interneurons into neuronal columns is a mystery. This is particularly intriguing since, unlike excitatory neurons, clonally related or molecularly distinct interneurons can spread widely across cortical functional area boundaries (Mayer et al., 2016; Saunders et al., 2018; Tasic et al., 2018). Extensive radial movement of interneurons also occurs in the early neonatal human frontal lobe (Parades et al., 2016). Radial migration of interneurons, after they invade the dorsal cortex, along radial glial guides, might facilitate coordination between interneurons and projection neurons in a radial column. Such RG-guided targeting of interneurons may enable appropriate layer-specific matching between excitatory projection neurons and interneuron subtypes in a cortical column (Hevner et al., 2004; Lodato et al., 2011; Wester et al., 2019). Thus testing if radial glial guide-dependent repositioning of specific projection neurons can also reroute corresponding interneurons in a radial glia-dependent manner will help establish if the assembly of local cortical circuits depends on radial glial scaffold functions.
The significance of radial glial guided columnar organization for the functional organization of the adult cortex remains enigmatic. It was suggested that “neurons within a given column are stereotypically interconnected in the vertical dimension, share extrinsic connectivity, and hence act as basic functional units subserving a set of common static and dynamic cortical operations that include not only sensory and motor areas but also association areas subserving the highest cognitive functions” (Jones & Rakic, 2010). Combined application of lineage tracing methods that can differentially label individual radial units with targeted mapping of neurons activated by a specific stimulus (e.g., TRAP method) can help us understand if and how radial units form the basic information processing units of the cerebral cortex (Contreras et al., 2021; Guenthner et al., 2013; Lui et al., 2021; Nakagawa et al., 2019). Such studies will be necessary to establish if neuronal ensembles emanating from embryonic radial units are functionally relevant in the mature cortex. If they are not, it will suggest that the early cortical neuronal organization (genesis, placement, and connectivity) engendered by diverse radial progenitors is merely the initial blueprint of cortical organization, which is then modified and eventually erased by neuronal activity based mechanisms (Blakemore & Cooper, 1970; Cadwell et al., 2019; Rakic, 1988).
The main conceptual framework of corticogenesis, the radial unit hypothesis, posits that proliferative radial glial units provide a proto-map of prospective cytoarchitectonic areas. The neuronal output of these progenitors is translated via radial glial guides to the expanding cortex in the form of radial columns, whose final number for each cortical area is thought to be modified through interaction with incoming afferent input. Perhaps, radial glia’s emerging role in axonal growth and guidance necessitates a revamping of this foundational hypothesis. Radial progenitors not only guide the formation of radial units of neurons that underlie the laminar and columnar organization of the cerebral cortex, but also facilitate the appropriate growth of neurons within the radial units towards their appropriate targets. In this sense, radial glia serves to integrate neurogenesis, neuronal migration, neuronal placement, and neuronal growth that underlies neural circuit formation in the cerebral cortex. Radial units form the basis for cortical microcircuits and underlie the modular, evolutionary expansion of the primate cortex (Rakic, 1988, 2000, 2007, 2009; Molnár et al, 2019; Molnár and Rockland, 2020). Inclusion of an expanded, integrative role of radial glia in radial unit generation and connectivity may better guide the ongoing efforts to piece together how progenitor dynamics coordinate formation and connectivity of the neocortex across brain evolution.
Acknowledgements
This research was supported by NIH grants R35NS116859 and HD098657 to E.S.A.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- Aizawa S, Okada T, Keino-Masu K, Doan TH, Koganezawa T, Akiyama M, Tamaoka A, & Masu M (2020). Abnormal pyramidal decussation and bilateral projection of the corticospinal tract axons in mice lacking the heparan sulfate endosulfatases, Sulf1 and Sulf2. Frontiers in molecular neuroscience, 12, 333. 10.3389/fnmol.2019.00333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert M, Kalebic N, Florio M, Lakshmanaperumal N, Haffner C, Brandl H, Henry I, & Huttner WB (2017). Epigenome profiling and editing of neocortical progenitor cells during development. The EMBO journal, 36(17), 2642–2658. 10.15252/embj.201796764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SA, Eisenstat DD, Shi L, & Rubenstein JL (1997). Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science (New York, N.Y.), 278(5337), 474–476. 10.1126/science.278.5337.474 [DOI] [PubMed] [Google Scholar]
- Andrews MG, Subramanian L, & Kriegstein AR (2020). mTOR signaling regulates the morphology and migration of outer radial glia in developing human cortex. eLife, 9, e58737. 10.7554/eLife.5873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armand EJ, Li J, Xie F, Luo C, & Mukamel EA (2021). Single-cell sequencing of brain cell transcriptomes and epigenomes. Neuron, 109(1), 11–26. 10.1016/j.neuron.2020.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baburamani AA et al. (2020). Assessment of radial glia in the frontal lobe of fetuses with Down syndrome. Acta Neuropathologica Communications, 3, 1–17. 10.1186/s40478-020-01015-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball G et al. (2020). Cortical morphology at birth reflects spatiotemporal patterns of gene expression in the fetal human brain. In PLoS Biology (Vol. 18, Issue 11). 10.1371/journal.pbio.3000976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnabè-Heider F, Wasylnka JA, Fernandes KJL, Porsche C, Sendtner M, Kaplan DR, & Miller FD (2005). Evidence that embryonic neurons regulate the onset of cortical gliogenesis. Neuron, 48, 253–265. 10.1016/j.neuron.2005.08.037 [DOI] [PubMed] [Google Scholar]
- Bayraktar OA, Fuentealba LC, Alvarez-Buylla A, & Rowitch DH (2015). Astrocyte development and heterogeneity. Cold Spring Harbor Perspectives in Biology, 7(1). 10.1101/cshperspect.a020362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belle M, Godefroy D, Couly G, Malone SA, Collier F, Giacobini P, & Chédotal A (2017). Tridimensional visualization and analysis of early human development. Cell, 169(1), 161–173. e12. 10.1016/j.cell.2017.03.008 [DOI] [PubMed] [Google Scholar]
- Bernier R et al. (2014). Disruptive CHD8 mutations define a subtype of autism early in development. Cell, 158(2), 263–276. 10.1016/j.cell.2014.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE et al. (2005). Genomic maps and comparative analysis of histone modifications in human and mouse. Cell, 120(2), 169–181. 10.1016/j.cell.2005.01.001 [DOI] [PubMed] [Google Scholar]
- Bjornsson CS, Apostolopoulou M, Tian Y, & Temple S (2015). It takes a village: constructing the neurogenic niche. Developmental cell, 32(4), 435–446. 10.1016/j.devcel.2015.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakemore C, & Cooper GF (1970). Development of the brain depends on the visual environment. Nature, 228, 1969–1970. [DOI] [PubMed] [Google Scholar]
- Borrell V (2019). Recent advances in understanding neocortical development. F1000Research, 8, 1–9. 10.12688/f1000research.20332.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrell V, & Reillo I (2012). Emerging roles of neural stem cells in cerebral cortex development and evolution. Developmental neurobiology, 72(7), 955–971. 10.1002/dneu.22013 [DOI] [PubMed] [Google Scholar]
- Brodmann K (2006). Brodmann’s Localisation in the Cerebral Cortex (Garey LJ (ed.)). Springer US. [Google Scholar]
- Cadwell CR, Bhaduri A, Mostajo-radji MA, Keefe MG, & Nowakowski TJ (2019). Development and arealization of the cerebral cortex. Neuron, 103(6), 980–1004. 10.1016/j.neuron.2019.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau KF, Springel MW, Broadbelt KG, Park HY, Topal S, Lun MP, Mullan H, Maynard T, Steen H, LaMantia AS, & Lehtinen MK (2015). Progressive Differentiation and Instructive Capacities of Amniotic Fluid and Cerebrospinal Fluid Proteomes following Neural Tube Closure. Developmental cell, 35(6), 789–802. 10.1016/j.devcel.2015.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Lake BB, & Zhang K (2019). High-throughput sequencing of the transcriptome and chromatin accessibility in the same cell. Nature Biotechnology, 37(12), 1452–1457. 10.1038/s41587-019-0290-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciarrusta J et al. (2020). Emerging functional connectivity differences in newborn infants vulnerable to autism spectrum disorders. Translational Psychiatry, 10(1). 10.1038/s41398-020-0805-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras X et al. (2021). A genome-wide library of MADM mice for single-cell genetic mosaic analysis. Cell Reports, 35(122). 10.1016/j.celrep.2021.109274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa MR, Bucholz O, Schroeder T, & Götz M (2009). Late origin of glia-restricted progenitors in the developing mouse cerebral cortex. Cerebral Cortex, 19(July), i135–i143. 10.1093/cercor/bhp046 [DOI] [PubMed] [Google Scholar]
- Crandall JE, Mlsson J-P, & Butler D (1990). The development of radial glia and radial dendrites during barrel formation in mouse somatosensory cortex. Developmental Brain Research, 55, 87–94. 10.1016/01653806(90)90108-b [DOI] [PubMed] [Google Scholar]
- DeFelipe J, Alonso-Nanclares L, & Arellano JI (2002). Microstructure of the neocortex: Comparative aspects. Journal of Neurocytology, 31, 299–316. 10.1023/a:1024130211265 [DOI] [PubMed] [Google Scholar]
- Dehay C, Savatier P, Kennedy H, & Vision C (2001). Cell-cycle kinetics of neocortical precursors are influenced by embryonic thalamic axons. Journal of Neuroscience, 21(1), 201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bella DJ et al. (2021). Molecular logic of cellular diversification in the mouse cerebral cortex. Nature, 595(7868), 554–559. 10.1038/s41586-021-03670-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evsyukova I, Plestant C, & Anton ES (2013). Integrative mechanisms of oriented neuronal migration in the developing brain. Annual Review of Cell and Developmental Biology, 29, 299–353. 10.1146/annurev-cellbio-101512-122400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eze UC, Bhaduri A, Haeussler M, Nowakowski TJ, & Kriegstein AR (2021). Single-cell atlas of early human brain development highlights heterogeneity of human neuroepithelial cells and early radial glia. Nature Neuroscience, 24(4), 584–594. 10.1038/s41593-020-00794-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W-Q, Chen W-W, Jiang L, Liu K, Yung W-H, Fu A, & Ip N (2014). Overproduction of upper-layer neurons in the neocortex leads to autism-like features in mice. Cell Reports, 9(5), 1635–1643. 10.1016/j.celrep.2014.11.003 [DOI] [PubMed] [Google Scholar]
- Ferent J, Zaidi D, & Francis F (2020). Extracellular Control of Radial Glia Proliferation and Scaffolding During Cortical Development and Pathology. Frontiers in cell and developmental biology, 8, 578341. 10.3389/fcell.2020.578341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fietz SA et al. (2010). OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nature Neuroscience, 13(6), 690–699. 10.1038/nn.2553 [DOI] [PubMed] [Google Scholar]
- Florio M, & Huttner WB (2014). Neural progenitors, neurogenesis and the evolution of the neocortex. Development (Cambridge, England), 141(11), 2182–2194. 10.1242/dev.090571 [DOI] [PubMed] [Google Scholar]
- Florio M et al. (2015). Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science (New York, N.Y.), 347(6229), 1465–1470. 10.1126/science.aaa1975 [DOI] [PubMed] [Google Scholar]
- Fujita I et al. (2020). Endfoot regeneration restricts radial glial state and prevents translocation into the outer subventricular zone in early mammalian brain development. Nature Cell Biology, 22(1), 26–37. 10.1038/s41556-019-0436-9 [DOI] [PubMed] [Google Scholar]
- Gao P et al. (2014). Deterministic progenitor behavior and unitary production of neurons in the neocortex. Cell, 159(4), 775–788. 10.1016/j.cell.2014.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz CC, & Kriegstein AR (2015). Neuronal migration dynamics in the developing ferret cortex. Journal of Neuroscience, 35(42), 14307–14315. 10.1523/JNEUROSCI.2198-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gongidi V, Ring C, Moody M, Brekken R, Sage EH, Rakic P, Anton ES, Hill C, & Carolina N (2004). SPARC-like 1 regulates the terminal phase of radial glia-guided migration in the cerebral cortex. Neuron, 41, 57–69. 10.1016/S0896-6273(03)00818-3 [DOI] [PubMed] [Google Scholar]
- Götz M, Nakafuku M, & Petrik D (2016). Neurogenesis in the developing and adult brain—similarities and key differences. Cold Spring Harbor Perspectives in Biology, 8(7), 1–23. 10.1101/cshperspect.a018853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouwens NW et al. (2019). Classification of electrophysiological and morphological neuron types in the mouse visual cortex. Nature Neuroscience, 22(July), 1182–1195. 10.1038/s41593-019-0417-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenthner CJ, Miyamichi K, Yang HH, Heller HC, & Luo L (2013). Permanent genetic access to transiently active neurons via TRAP : Targeted recombination in active populations. Neuron, 78(5), 773–784. 10.1016/j.neuron.2013.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Parker PRL, & Kriegstein AR (2010). Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature, 464(7288), 554–561. 10.1038/nature08845 [DOI] [PubMed] [Google Scholar]
- Hartfuss E, Galli R, Heins N, & Götz M (2001). Characterization of CNS precursor subtypes and radial glia. Developmental biology, 229(1), 15–30. 10.1006/dbio.2000.9962 [DOI] [PubMed] [Google Scholar]
- Hatten M, & Mason C (1990). Mechanisms of glial-guided neuronal migration in vitro and in vivo. Experimentia, 46(9), 907–916. 10.1007/BF01939383 [DOI] [PubMed] [Google Scholar]
- Haubensak W, Attardo A, Denk W, & Huttner WB (2004). Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: A major site of neurogenesis. Proceedings of the National Academy of Sciences of the United States of America, 101(9), 3196–3201. 10.1073/pnas.0308600100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydar TF, Wang F, Schwartz ML, & Rakic P (2000). Differential modulation of proliferation in the neocortical ventricular and subventricular zones. Journal of Neuroscience, 20(15), 5764–5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazlett HC et al. (2017). Early brain development in infants at high risk for autism spectrum disorder. Nature, 542, 348–351. 10.1038/nature21369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heide M, Haffner C, Murayama A, Kurotaki Y, Shinohara H, Okano H, Sasaki E, & Huttner WB (2020). Human-specific ARHGAP11B increases size and folding of primate neocortex in the fetal marmoset. Science (New York, N.Y.), 369(6503), 546–550. 10.1126/science.abb2401 [DOI] [PubMed] [Google Scholar]
- Henrikson CK, & Vaughn JE (1974). Fine structural relationships between neurites and radial glial processes in developing mouse spinal cord. Journal of Neurocytology, 675, 659–675. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Daza RA, Englund C, Kohtz J, & Fink A (2004). Postnatal shifts of interneuron position in the neocortex of normal and reeler mice: Evidence for inward radial migration. Neuroscience, 124(3), 605–618. 10.1016/j.neuroscience.2003.11.033 [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y, & Gotoh Y (2010). Epigenetic control of neural precursor cell fate during development. Nature Reviews Neuroscience, 11(6), 377–388. 10.1038/nrn2810 [DOI] [PubMed] [Google Scholar]
- Hodge RD et al. (2019). Conserved cell types with divergent features in human versus mouse cortex. Nature, 573, 61–68. 10.1038/s41586-0191506-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Jeon T, Sedmak G, Pletikos M, Vasung L, Xu X, & Yarowsky P (2013). Coupling diffusion imaging with histological and gene expression analysis to examine the dynamics of cortical areas across the fetal period of human brain development. Cerebral Cortex, 23, 2620–2631. 10.1093/cercor/bhs241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Išasegi Žunić, I., Milan R, Željka K, Marko R, Vesna B, & Kostović I (2018). Interactive histogenesis of axonal strata and proliferative zones in the human fetal cerebral wall. Brain Structure and Function, 223(9), 3919–3943. 10.1007/s00429-018-1721-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X et al. (2020). In vivo Perturb-Seq reveals neuronal and glial abnormalities associated with autism risk genes. Science (New York, N.Y.), 370(6520), eaaz6063. 10.1126/science.aaz6063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EG, & Rakic P (2010). Radial columns in cortical architecture : It is the composition that counts. Cerebral Cortex, 20(October), 2261–2264. 10.1093/cercor/bhq127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki M, Nakamura S, Machon O, Krauss S, Radice GL, & Takeichi M (2007). N-cadherin mediates cortical organization in the mouse brain. Developmental biology, 304(1), 22–33. 10.1016/j.ydbio.2006.12.014 [DOI] [PubMed] [Google Scholar]
- Kalebic N et al. (2019). Neocortical expansion due to increased proliferation of basal progenitors Is linked to changes in their morphology. Cell Stem Cell, 24(4), 535–550. e9. 10.1016/j.stem.2019.02.017 [DOI] [PubMed] [Google Scholar]
- Kamiya K, Tanaka F, & Ikeno M (2014). DTI tractography of lissencephaly caused by TUBA1A mutation. Neurological Sciences, 35, 801–803. 10.1007/s10072-014-1662-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ et al. (2011). Spatio-temporal transcriptome of the human brain. Nature, 478(7370), 483–489. 10.1038/nature10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanton S et al. (2019). Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature, 574(7778), 418–422. 10.1038/s41586-019-1654-9 [DOI] [PubMed] [Google Scholar]
- Kaur N et al. (2020). Neural stem cells direct axon guidance via their radial fiber scaffold. Neuron, 107, 1197–1211. 10.1016/j.neuron.2020.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi T, Sekine K, Shikanai M, Chihama K, Tomita K, Kubo K, Nakajima K, Nabeshima Y, & Hoshino M (2010). Rab GTPases-dependent endocytic pathways regulate neuronal migration and maturation through N-cadherin trafficking. Neuron, 67(4), 588–602. 10.1016/j.neuron.2010.07.007 [DOI] [PubMed] [Google Scholar]
- Khacho M et al. (2016). Mitochondrial dynamics impacts stem cell identity and fate decisions by regulating a nuclear transcriptional program. Stem Cell, 19(2), 232–247. 10.1016/j.stem.2016.04.015 [DOI] [PubMed] [Google Scholar]
- Khan S, Vasung L, Marami B, Rollins CK, Afacan O, Ortinau CM, Yang E, Warfield SK, & Gholipour A (2019). Fetal brain growth portrayed by a spatiotemporal diffusion tensor MRI atlas computed from in utero images. NeuroImage, 185(October 2017), 593–608. 10.1016/j.neuroimage.2018.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolasinski J, Takahashi E, Stevens AA, Benner T, Fischl B, Zöllei L, & Grant PE (2013). Radial and tangential neuronal migration pathways in the human fetal brain : Anatomically distinct patterns of diffusion MRI coherence. NeuroImage, 79, 412–422. 10.1016/j.neuroimage.2013.04.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornack DR, & Rakic P (1995). Radial and horizontal deployment of clonally related cells in the primate neocortex : relationship to distinct mitotic lineages. Neuron, 15, 311–321. [DOI] [PubMed] [Google Scholar]
- Kostović I, Rados M, Kostović-Srzentić M, & Željka K (2021). Fundamentals of the development of connectivity in the human fetal brain in late gestation : From 24 weeks gestational age to term. Journal of Neuropathology and Experimental Neurology, 80(5), 393–414. 10.1093/jnen/nlab024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostović I, Sedmak G, & Judas M (2019). Neural histology and neurogenesis of the human fetal and infant brain. NeuroImage, 188, 743–773. 10.1016/j.neuroimage.2018.12.043 [DOI] [PubMed] [Google Scholar]
- Kriegstein Arnold R., Alvarez-Bulla A (2009). The glial nature of embryonic and adult neural stem cells. Annual Reviews of Neuroscience, 32, 149–184. 10.1146/annurev.neuro.051508.135600.The [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krsnik Ž, Majic V, Vasung L, Huang H, & Kostović I (2017). Growth of thalamocortical fibers to the somatosensory cortex in the human fetal brain. Frontiers in Neuroscience, 11(233). 10.3389/fnins.2017.00233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMonica BE, Lui JH, Hansen DV, & Kriegstein AR (2013). Mitotic spindle orientation predicts outer radial glial cell generation in human neocortex. Nature Communications, 4. 10.1038/ncomms2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DS et al. (2019). Simultaneous profiling of 3D genome structure and DNA methylation in single human cells. Nature Methods, 16(10), 999–1006. 10.1038/s41592-019-0547-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levman J, Macdonald A, Baumer N, Macdonald P, Stewart N, Lim A, Cogger L, Shiohama T, & Takahashi E (2019). Structural magnetic resonance imaging demonstrates abnormal cortical thickness in Down syndrome: Newborns to young adults. NeuroImage: Clinical, 23(October 2018), 101874. 10.1016/j.nicl.2019.101874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M et al. (2018). Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science (New York, N.Y.), 362(6420), eaat7615. 10.1126/science.aat7615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Liu Y, Zhang Y, Kubo N, Yu M, Fang R, Kellis M, & Ren B (2019). Joint profiling of DNA methylation and chromatin architecture in single cells. Nature Methods, 16(10), 991–993. 10.1038/s41592-019-0502-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L et al. (2019). The COMPASS family protein ASH2L mediates corticogenesis via transcriptional regulation of Wnt signaling. Cell Reports, 28(3), 698–711. e5. 10.1016/j.celrep.2019.06.055 [DOI] [PubMed] [Google Scholar]
- Li Z, Tyler WA, Zeldich E, Santpere Baró G, Okamoto M, Gao T, Li M, Sestan N, & Haydar TF (2020). Transcriptional priming as a conserved mechanism of lineage diversification in the developing mouse and human neocortex. Science advances, 6(45), eabd2068. 10.1126/sciadv.abd2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D et al. (2021). Cell-type-specific effects of genetic variation on chromatin accessibility during human neuronal differentiation. Nature Neuroscience. 10.1038/s41593-021-00858-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L, Llorca A, Mi D, & Marin O (2018). Development and functional diversification of cortical interneurons. Neuron, 100(2), 294–313. 10.1016/j.neuron.2018.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato S, Rouaux C, Quast KB, Studer M, Hensch TK, & Arlotta P (2011). Excitatory projection neuron subtypes differentially control the distribution of local inhibitory interneurons in the cerebral cortex. Neuron, 69(4), 763–779. 10.1016/j.neuron.2011.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo L, Simon JM, Xing L, McCoy ES, Niehaus JK, Guo J, Anton ES, & Zylka MJ (2019). Single-cell transcriptomic analysis of mouse neocortical development. Nature Communications, 10(1), 1–11. 10.1038/s41467-018-08079-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JH, Hansen DV, & Kriegstein AR (2011). Development and evolution of the human neocortex. Cell, 146(1), 18–36. 10.1016/j.cell.2011.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JH et al. (2021). Differential encoding in prefrontal cortex projection neuron classes across cognitive tasks. Cell, 184(2), 489–506. e26. 10.1016/j.cell.2020.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto MC et al. (2017). Altered proliferation and networks in neural cells derived from idiopathic autistic individuals. Molecular Psychiatry, 22(6), 820–835. 10.1038/mp.2016.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani J et al. (2015). FOXG1-dependent dysregulation of GABA/glutamate neuron differentiation in autism spectrum disorders. Cell, 162(2), 375–390. 10.1016/j.cell.2015.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Padilla M (1983). Structural organization of the human cerebral cortex prior to the appearance of the cortical plate. Anatomy and Embryology, 168, 21–40. [DOI] [PubMed] [Google Scholar]
- Marín O, & Rubenstein JLR (2003). Cell migration in the forebrain. Annual Reviews of Neuroscience, 26, 441–483. 10.1146/annurev.neuro.26.041002.131058 [DOI] [PubMed] [Google Scholar]
- Mayer C, Bandler RC, Mayer C, Bandler RC, & Fishell G (2016). Lineage is a poor predictor of interneuron positioning within the forebrain. Neuron, 92, 45–51. 10.1016/j.neuron.2016.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micali N et al. (2020). Variation of human neural stem cells generating organizer states in vitro before committing to cortical excitatory or inhibitory neuronal fates. Cell Reports, 31(5). 10.1016/j.celrep.2020.107599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Okano H, & Ogawa M (2001). Asymmetric inheritance of radial glial fibers by cortical neurons. Neuron, 31, 727–741. [DOI] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Saito K, Kawano M, Muto T, & Ogawa M (2004). Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development, 131(13), 3133–3145. 10.1242/dev.01173 [DOI] [PubMed] [Google Scholar]
- Molnár Z et al. (2019). New insights into the development of the human cerebral cortex. Journal of anatomy, 235(3), 432–451. 10.1111/joa.13055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnàr Z, & Rockland KS (2020). Cortical columns. In Rubenstein JLR, Chen B, Rakic P, & Kwan KY (Eds.), Neural Circuit and Cognitive Development (2nd ed., pp. 103–126). Academic Press. 10.1016/B978-0-12-814411-4.00005-6 [DOI] [Google Scholar]
- Morest DK, & Silver J (2003). Precursors of neurons, neuroglia, and ependymal cells in the CNS: what are they? Where are they from? How do they get where they are going?. Glia, 43(1), 6–18. 10.1002/glia.10238 [DOI] [PubMed] [Google Scholar]
- Nadarajah B, & Parnavelas JG (2002). Modes of neuronal migration in the developing cerebral cortex. Nature Reviews Neuroscience, 3(June). 10.1038/nrn845 [DOI] [PubMed] [Google Scholar]
- Nakagawa N et al. (2019). Memo1-mediated tiling of radial glial cells facilitates cerebral cortical development. Neuron, 103(5), 836–852. e5. 10.1016/j.neuron.2019.05.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba T, Nardelli J, Gressens P, & Huttner WB (2021). Metabolic regulation of neocortical expansion in development and evolution. Neuron, 109(3), 408–419. 10.1016/j.neuron.2020.11.014 [DOI] [PubMed] [Google Scholar]
- Nishino J, Yamashita K, Hashiguchi H, Fujii H, & Shimazaki T (2004). Meteorin: a secreted protein that regulates glial cell differentiation and promotes axonal extension. EMBO Journal, 23(9), 1998–2008. 10.1038/sj.emboj.7600202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Dammerman RS, & Kriegstein AR (2001). Neurons derived from radial glial cells establish radial units in neocortex. Nature, 409(February), 714–720. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeño V, Ivic L, & Kriegstein AR (2004). Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nature Neuroscience, 7(2), 136–144. 10.1038/nn1172 [DOI] [PubMed] [Google Scholar]
- Norris CR, & Kalil K (1991). Guidance of callosal axons by radial glia in the developing cerebral cortex. Journal of Neuroscience, 11(11), 3481–3492. 10.1523/JNEUROSCI.11-11-03481.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski TJ et al. (2017). Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science, 358(6368), 1318–1323. 10.1126/science.aap8809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski TJ, Pollen AA, Sandoval-Espinosa C, & Kriegstein AR (2016). Transformation of the radial glia scaffold demarcates two stages of human cerebral cortex development. Neuron, 91(6), 1219–1227. 10.1016/j.neuron.2016.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary D, Chou S, & Sahara S (2007). Area patterning of the mammalian cortex. Neuron, 56(2), 252–269. 10.1016/j.neuron.2007.10.010 [DOI] [PubMed] [Google Scholar]
- O’Reilly C, Lewis JD, & Elsabbagh M (2017). Is functional brain connectivity atypical in autism ? A systematic review of EEG and MEG studies. PLoS ONE, 12(5), e0175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke NA, Dailey ME, Smith SJ, & Mcconnell SK (1992). Diverse migratory pathways in the developing cerebral cortex. Science, 258(5080), 1992. 10.1126/science.1411527 [DOI] [PubMed] [Google Scholar]
- Ohta H, Nordahl CW, Iosif A-M, Lee A, Rogers S, & Amaral DG (2016). Increased surface area, but not cortical thickness, in a subset of young boys with autism spectrum disorder. Autism Research, 9(2), 232–248. 10.1002/aur.1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Keino-Masu K, Nagamine S, Kametani F, Ohto T, Hasegawa M, van Kuppevelt TH, Kunita S, Takahashi S, & Masu M (2017). Desulfation of heparan sulfate by Sulf1 and Sulf2 is required for corticospinal tract formation. Scientific reports, 7(1), 13847. 10.1038/s41598-017-14185-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrem BEL, Lui JH, Gertz CC, & Kriegstein AR (2014). Control of outer radial glial stem cell mitosis in the human brain. Cell Reports, 8(3), 656–664. 10.1016/j.celrep.2014.06.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang M, Dubois J, Yu Q, Mukherjee P, & Huang H (2019). Delineation of early brain development from fetuses to infants with diffusion MRI and beyond. NeuroImage, 185(October 2017), 836–850. 10.1016/j.neuroimage.2018.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes MF et al. (2016). Extensive migration of young neurons into the infant human frontal lobe. Science (New York, N.Y.), 354(6308), aaf7073. 10.1126/science.aaf7073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilaz LJ, Lennox AL, Rouanet JP, & Silver DL (2016). Dynamic mRNA Transport and Local Translation in Radial Glial Progenitors of the Developing Brain. Current biology : CB, 26(24), 3383–3392. 10.1016/j.cub.2016.10.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilz GA et al. (2013). Amplification of progenitors in the mammalian telencephalon includes a new radial glial cell type. Nature Communications, 4, 1–11. 10.1038/ncomms3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polioudakis D et al. (2019). A single-cell transcriptomic atlas of human neocortical development during mid-gestation. Neuron, 103(5), 785–801. e8. 10.1016/j.neuron.2019.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen AA et al. (2015). Molecular identity of human outer radial glia during cortical development. Cell, 163(1), 55–67. 10.1016/j.cell.2015.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poretti A, Meoded A, & Rossi A (2013). Diffusion tensor imaging and fiber tractography in brain malformations. Pediatric Radiology, 23, 28–54. 10.1007/s00247-012-2428-9 [DOI] [PubMed] [Google Scholar]
- Rakic P (1972). Mode of cell migration to the su perficial layers of fetal monkey neocortex. Journal of Comparative Neurology, 145, 61–83. 10.1002/cne.901450105 [DOI] [PubMed] [Google Scholar]
- Rakic P (1978). Neuronal migration and contact guidance in the primate telencephalon. Postgraduate Medical Journal, 54(Suppl 1), 25–40. [PubMed] [Google Scholar]
- Rakic P (1988). Specification of cerebral cortical areas. Science, 241(4862), 170–176. 10.1126/science.3291116 [DOI] [PubMed] [Google Scholar]
- Rakic P (2000). Radial unit hypothesis of neocortical expansion. Novartis Foundation Symposium, 228(May), 30–45. 10.1002/0470846631.ch3 [DOI] [PubMed] [Google Scholar]
- Rakic P (2007). The radial edifice of cortical architecture : From neuronal silhouettes to genetic engineering. Brain Research Reviews, 55, 204–219. 10.1016/j.brainresrev.2007.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P (2009). Evolution of the neocortex: A perspective from developmental biology. Nature Reviews Neuroscience, 10(10), 724–735. 10.1038/nrn2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasin MR et al. (2007). Numb and Numbl are required for maintenance of cadherin-based adhesion and polarity of neural progenitors. Nature neuroscience, 10(7), 819–827. 10.1038/nn1924 [DOI] [PubMed] [Google Scholar]
- Reillo I, De Juan Romero C, García-Cabezas MÁ, & Borrell V (2011). A role for intermediate radial glia in the tangential expansion of the mammalian cerebral cortex. Cerebral Cortex, 21(7), 1674–1694. 10.1093/cercor/bhq238 [DOI] [PubMed] [Google Scholar]
- Rodriques SG, Stickels RR, Goeva A, Martin CA, Murray E, Vanderburg CR, Welch J, Chen LM, Chen F, & Macosko EZ (2019). Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science, 363(6434), 1463–1467. 10.1126/science.aaw1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanosaka T et al. (2017). DNA methylome analysis identifies transcription factor-based epigenomic signatures of multilineage competence in neural Stem/progenitor cells. Cell Reports, 20(12), 2992–3003. 10.1016/j.celrep.2017.08.086 [DOI] [PubMed] [Google Scholar]
- Satterstrom FK et al. (2020). Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell, 180(3), 568–584. e23. 10.1016/j.cell.2019.12.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A et al. (2018). Molecular diversity and specializations among the cells of the adult mouse brain. Cell, 174(4), 1015–1030. e16. 10.1016/j.cell.2018.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmechel DE, & Rakic P (1979a). A golgi study of radial glial cells in developing monkey telencephalon : Morphogenesis and transformation into astrocytes. Anatomy and Embryology, 152, 115–152. [DOI] [PubMed] [Google Scholar]
- Schmechel DE, & Rakic P (1979b). Arrested proliferation of radial glial cells during midgestation in rhesus monkey. Nature, 277(5694), 303–305. 10.1038/277303a0 [DOI] [PubMed] [Google Scholar]
- Shapson-Coe A et al. (2021). A connectomic study of a petascale fragment of human cerebral cortex. bioRxiv, 2021.05.29.446289; doi: 10.1101/2021.05.29.446289 [DOI] [Google Scholar]
- Smart IH, Dehay C, Giroud P, Berland M, & Kennedy H (2002). Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cerebral cortex (New York, N.Y. : 1991), 12(1), 37–53. 10.1093/cercor/12.1.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KM, Ohkubo Y, Maragnoli ME, Rasin M-R, Schwartz ML, Nenad S, & Vaccarino FM (2006). Midline radial glia translocation and corpus callosum formation require FGF signaling. Nature Neuroscience, 9(6), 787–797. 10.1038/nn1705 [DOI] [PubMed] [Google Scholar]
- Song M et al. (2020). Cell-type-specific 3D epigenomes in the developing human cortex. Nature, 587(7835), 644–649. 10.1038/s41586-020-2825-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl R et al. (2013). Trnp1 regulates expansion and folding of the mammalian cerebral cortex by control of radial glial fate. Cell, 153(3), 535–549. 10.1016/j.cell.2013.03.027 [DOI] [PubMed] [Google Scholar]
- Stancik EK, Navarro-Quiroga I, Sellke R, & Haydar TF (2010). Heterogeneity in ventricular zone neural precursors contributes to neuronal fate diversity in the postnatal neocortex. Journal of Neuroscience, 30(20), 7028–7036. 10.1523/JNEUROSCI.6131-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoner R, Chow ML, Boyle MP, Sunkin SM, Mouton PR, Roy S, Wynshaw-Boris A, Colamarino SA, Lein ES, & Courchesne E (2014). Patches of disorganization in the neocortex of children with autism. The New England Journal of Medicine, 370(13), 1209–1219. 10.1056/NEJMoa1307491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki IK et al. (2018). Human-specific NOTCH2NL genes expand cortical neurogenesis through delta/notch regulation. Cell, 173(6), 1370–1384. e16. 10.1016/j.cell.2018.03.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi E, Dai G, Rosen GD, Wang R, Ohki K, Folkerth RD, Galaburda AM, Wedeen VJ, & Grant PE (2011). Developing neocortex organization and connectivity in cats revealed by direct correlation of diffusion tractography and histology. Cerebral Cortex, 21. 10.1093/cercor/bhq084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi E, Folkerth RD, Galaburda AM, & Grant PE (2012). Emerging cerebral connectivity in the human fetal brain : An MR tractography study. Cerebral Cortex, 22(2), 455–464. 10.1093/cercor/bhr126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S, & Breen S (1993). Radial mosaicism and tangential cell dispersion both contribute to mouse neocortical development. Nature, 362(April), 638–640. [DOI] [PubMed] [Google Scholar]
- Tasic B et al. (2018). Shared and distinct transcriptomic cell types across neocortical areas. Nature, 563(7729), 72–78. 10.1038/s41586-0180-654-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telley L et al. (2019). Temporal patterning of apical progenitors and their daughter neurons in the developing neocortex. Science, 364(6440). 10.1126/science.aav2522 [DOI] [PubMed] [Google Scholar]
- Thomsen ER et al. (2015). Fixed single-cell transcriptomic characterization of human radial glial diversity. Nature Methods, 13(1), 87–93. 10.1038/nmeth.3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevino AE et al. (2021). Chromatin and gene-regulatory dynamics of the developing human cerebral cortex at single-cell resolution. Cell, 184(19), 5053–5069. e23. 10.1016/j.cell.2021.07.039 [DOI] [PubMed] [Google Scholar]
- Varadarajan SG, Kong JH, Phan KD, Kao TJ, Panaitof SC, Cardin J, Eltzschig H, Kania A, Novitch BG, & Butler SJ (2017). Netrin1 Produced by Neural Progenitors, Not Floor Plate Cells, Is Required for Axon Guidance in the Spinal Cord. Neuron, 94(4), 790–799. e3. 10.1016/j.neuron.2017.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasung L, Charvet CJ, Shiohama T, Gagoski B, Levman J, & Takahashi E (2019). Ex vivo fetal brain MRI : Recent advances, challenges, and future directions. NeuroImage, 195(November 2017), 23–37. 10.1016/j.neuroimage.2019.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasung L, Raguz M, Kostović I, & Takahashi E (2017). Spatiotemporal relationship of brain pathways during human fetal development using high-angular resolution diffusion MR imaging and histology. Frontiers in Neuroscience, 11(JUL), 1–16. 10.3389/fnins.2017.00348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasung L et al. (2019). Structural and diffusion MRI analyses with histological observations in patients with lissencephaly. Frontiers in Cell and Developmental Biology, 7(124), 1–10. 10.3389/fcell.2019.00124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasung L, Turk EA, Ferradal SL, Sutin J, Stout JN, Ahtam B, Lin P, & Grant PE (2019). Exploring early human brain development with structural and physiological neuroimaging. NeuroImage, 187, 226–254. 10.1016/j.neuroimage.2018.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace CS, & Withers GS (2018). Evidence that astroglia influence dendrite morphogenesis and synaptogenesis independently in the vertebrate central nervous system. In Astrocyte: Physiology and Pathology (pp. 35–49). 10.5772/intechopen.73891 [DOI] [Google Scholar]
- Wang L, Hou S, & Han YG (2016). Hedgehog signaling promotes basal progenitor expansion and the growth and folding of the neocortex. Nature Neuroscience, 19(7), 888–896. 10.1038/nn.4307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tsai JW, Lamonica B, & Kriegstein AR (2011). A new subtype of progenitor cell in the mouse embryonic neocortex. Nature Neuroscience, 14(5), 555–562. 10.1038/nn.2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer JM, Yokota Y, Stanco A, Stumpo DJ, Blackshear PJ, & Anton ES (2009). MARCKS modulates radial progenitor placement, proliferation and organization in the developing cerebral cortex. Development (Cambridge, England), 136(17), 2965–2975. 10.1242/dev.036616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman TA, Riquelme PA, Ivic L, Flint AC, & Kriegstein AR (2004). Calcium waves propagate through radial glial cells and modulate proliferation in the developing neocortex. Neuron, 43, 647–661. 10.1016/j.neuron.2004.08.015 [DOI] [PubMed] [Google Scholar]
- Wester J et al. (2019). Neocortical projection neurons instruct inhibitory interneuron circuit development in a lineage-dependent manner. Neuron, 102(5), 960–975. e6. 10.1016/j.neuron.2019.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Funahashi Y, Watanabe T, Takano T, Nakamuta S, Namba T, & Kaibuchi K (2015). Radial glial cell – neuron interaction directs axon formation at the opposite side of the neuron from the contact site. Journal of Neuroscience, 35(43), 14517–14532. 10.1523/JNEUROSCI.1266-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota Y, Eom T, Stanco A, Kim W, Rao S, Snider WD, & Anton ES (2010). Cdc42 and Gsk3 modulate the dynamics of radial glial growth, inter-radial glial interactions and polarity in the developing cerebral cortex. Development, 137, 4101–4110. 10.1242/dev.048637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota Y, Gashghaei HT, Han C, Watson H, Campbell KJ, & Anton ES (2007). Radial glial dependent and iondependent dynamics of interneuronal migration in the developing cerebral cortex. PLoS ONE, 2(8), e794. 10.1371/journal.pone.0000794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota Y, Ring C, Cheung R, Pevny L, & Anton ES (2007). Nap1-regulated neuronal cytoskeletal dynamics is essential for the final differentiation of neurons in cerebral cortex. Neuron, 54(3), 429–445. 10.1016/j.neuron.2007.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota Y, Kim WY, Chen Y, Wang X, Stanco A, Komuro Y, Snider W, & Anton ES (2009). The adenomatous polyposis coli protein is an essential regulator of radial glial polarity and construction of the cerebral cortex. Neuron, 61(1), 42–56. 10.1016/j.neuron.2008.10.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S et al. (2018). A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature, 555(7697), 524–528. 10.1038/nature25980 [DOI] [PubMed] [Google Scholar]
- Zhu C, Yu M, Huang H, Juric I, Abnousi A, Hu R, Lucero J, Behrens MM, Hu M, & Ren B (2019). An ultra high-throughput method for single-cell joint analysis of open chromatin and transcriptome. Nature Structural and Molecular Biology, 26(11), 1063–1070. 10.1038/s41594-019-0323-x [DOI] [PMC free article] [PubMed] [Google Scholar]


