Abstract
Bacillus subtilis spores are encased in two concentric shells: an outer proteinaceous “coat” and an inner peptidoglycan “cortex,” separated by a membrane. Cortex assembly depends on coat assembly initiation, but how cells achieve this coordination across the membrane is unclear. Here, we report that the protein SpoVID monitors the polymerization state of the coat basement layer via an extension to a functional intracellular LysM domain that arrests sporulation when coat assembly is initiated improperly. Whereas extracellular LysM domains bind mature peptidoglycan, SpoVID LysM binds to the membrane-bound lipid II peptidoglycan precursor. We propose that improper coat assembly exposes the SpoVID LysM domain, which then sequesters lipid II and prevents cortex assembly. SpoVID defines a widespread group of firmicute proteins with a characteristic N-terminal domain and C-terminal peptidoglycan-binding domains that might combine coat and cortex assembly roles to mediate a developmental checkpoint linking the morphogenesis of two spatially separated supramolecular structures.
In brief
The coordinated assembly of cellular structures is a hallmark of development. Delerue et al. identify a bacterial sporulation protein that sequesters a precursor molecule to halt assembly of one supramolecular structure until it detects that construction of another structure in a different cellular compartment has been initiated successfully.
Graphical Abstract
INTRODUCTION
Checkpoints are regulatory mechanisms that exist during growth and development that ensure the orderly execution of events during the cell cycle or a developmental program (Hartwell and Weinert, 1989). Unlike events that are intrinsically linked, checkpoints enforce the dependency of an event on a previous event correctly occurring. Therefore, if a dependency of one event on another may be removed by mutation, it is likely under the control of a checkpoint. As a result, checkpoints guard critical transitions in a cell by ensuring that a previous phase is complete and error free before the system is permitted to move forward in the program (Khodjakov and Rieder, 2009; Nasmyth, 1996). Checkpoints consist of two parts: a surveillance mechanism that monitors the completion of a phase and a regulatory mechanism that permits the second phase to be initiated.
Bacterial spore formation (sporulation) is a relatively simple developmental program in which a cell that normally grows by binary fission to create two identical progeny instead differentiates into a dormant cell type termed an “endospore” (or simply a “spore”) (Higgins and Dworkin, 2012; Riley et al., 2020; Shen et al., 2019; Tan and Ramamurthi, 2014). Spores are highly resistant to environmental insults and thus represent one of the hardiest life forms on the planet (Setlow, 2014). This process has been best studied in the model organism Bacillus subtilis, a soil bacterium in which sporulation is triggered in response to nutrient deprivation. Sporulation in B. subtilis is initiated with the ramped activation of a master regulator (Fujita et al., 2005) that permits one final round of DNA replication and cell division (Rahn-Lee et al., 2009; Wagner et al., 2009), after which the cell divides asymmetrically to produce two genetically identical but morphologically dissimilar daughter cells that will display different cell fates: a smaller “forespore” that will eventually become the mature spore and a larger “mother cell” that is destined to lyse (Figure 1A). After asymmetric division, the sporangium commits to completing the sporulation program, which takes ~6–8 h to complete, even if favorable growth conditions resume. Expectedly, then, the asymmetric division is governed by multiple checkpoints that ensure ploidy and chromosomal integrity before the cell irreversibly enters the sporulation program (Bejerano-Sagie et al., 2006; Burkholder et al., 2001; Veening et al., 2009). After asymmetric division, the mother cell swallows the forespore such that the forespore resides as a double membrane-bound cell in the mother cell cytosol. Next, the mother cell nurtures the forespore by sequentially constructing two concentric shells that encase the forespore: first, a proteinaceous outer shell termed the “coat,” composed of ~80 proteins, and second, an inner peptidoglycan “cortex” that is constructed between the two membranes of the forespore (Figures 1G–1J). Finally, after forespore maturation, the mother cell lyses, thereby releasing the forespore. The coat and cortex of the released spore, therefore, represent a unique bacterial cell surface (Driks and Eichenberger, 2016; Henriques and Moran, 2007; McKenney et al., 2013). Although the construction of both structures occurs well after the commitment step, their construction was recently discovered to be under surveillance by at least two independent quality control systems that ensure the removal of defective spores from the population (Ebmeier et al., 2012; Ramírez-Guadiana et al., 2017; Tan et al., 2015).
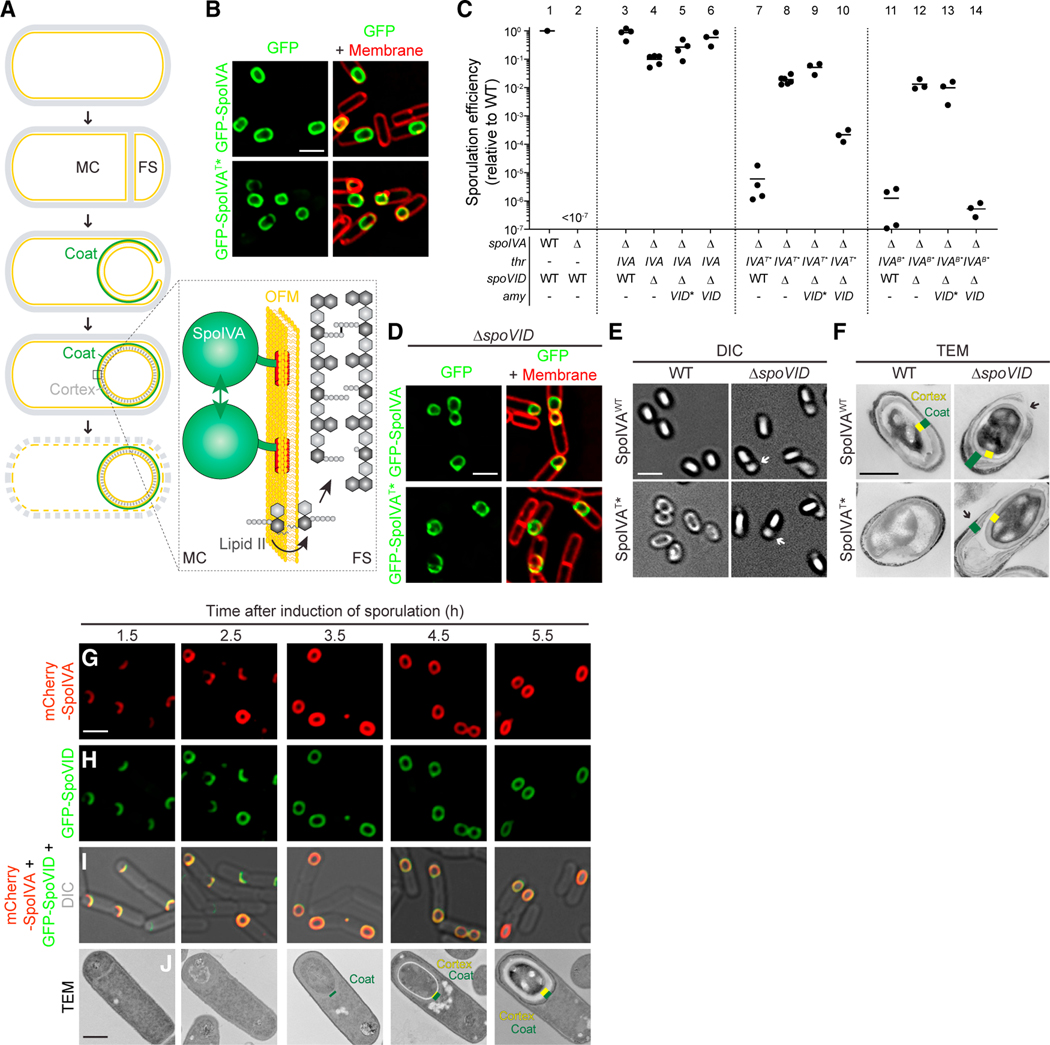
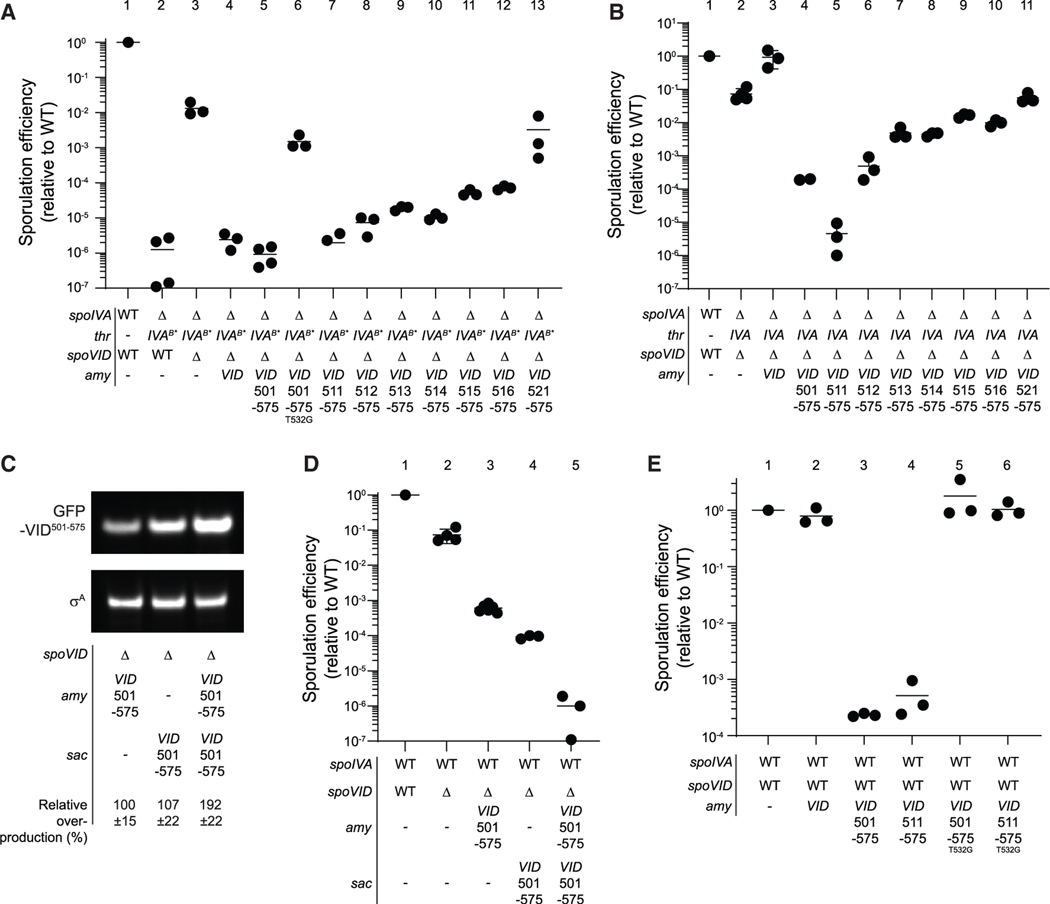
Figure 1. Disrupting spoVID suppresses spore envelope assembly defects caused by mis-assembly of the spore coat basement layer.
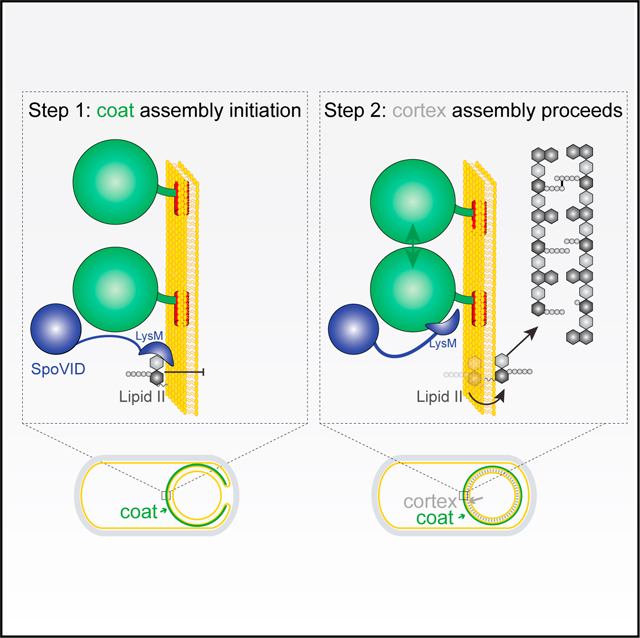
(A) Schematic of sporulation in B. subtilis. Sporulation initiates when the rod-shaped progenitor cell (top) asymmetrically divides, producing unequal progeny: a larger mother cell (MC) and smaller forespore (FS). The mother cell then engulfs the forespore (middle panel) and produces ~80 proteins that deposit around the outer forespore membrane to form the coat (green). The forespore is bounded by two membranes: the outer forespore membrane (OFM) originally derived from the mother cell and the inner forespore membrane (IFM), both depicted in yellow. Once coat assembly initiates successfully, the peptidoglycan cortex (gray dashes, fourth panel) assembles between the two membranes surrounding the forespore. Ultimately, the mother cell lyses (bottom), releasing the mature, dormant spore. Membranes depicted in yellow; cell wall depicted in gray. Inset: the basement layer of the spore coat is built with SpoIVA (green), which polymerizes on the MC face of the OFM (yellow). The lipid II peptidoglycan precursor (gray) is also made in the MC, inserts into the OFM, then flips to the intermembrane space, and is incorporated into the assembling cortex.
(B) Subcellular localization of GFP-SpoIVA (top panels) or GFP-SpoIVAT*, a variant that fails to polymerize (bottom panels) in sporulating B. subtilis (strains JPC156 and JPC243) 3 h after induction of sporulation. Left: fluorescence from GFP; right: overlay, fluorescence from GFP and membranes visualized with FM4–64.
(C) Sporulation efficiencies, determined as resistance to heat, relative to WT (PY79). Strain genotypes at spoIVA and spoVID loci are indicated below the graph; thr and amy are ectopic chromosomal loci used to complement spoIVA and spoVID deletions, respectively, with different alleles of those genes. Symbols are independent cultures; bars represent mean values. Strains used: PY79, KP73, KR394, JB171, JB175, JB174, JPC221, JB168, JB177, JB177, JB176, JB103, JB280, JB294, and JB293.
(D) Subcellular localization of GFP-SpoIVA (top panels) or GFP-SpoIVAT* (bottom panels) in the absence of spoVID in sporulating B. subtilis (strains TD300 and TD302) 3 h after induction of sporulation. Size bar: 2 mm.
(E and F) Differential interference contrast (DIC) light microscopy (E; size bar: 2 mm) or transmission electron microscopy (TEM) images (F; size bar: 500 nm) of released spores harboring WT (top) or T* alleles of spoIVA in WT (left) or ΔspoVID (right) strains (strains KR394, JPC221, JB171, JB168). Yellow indicates cortex; green indicates coat.
(G–I) Subcellular localization of (G) mCherry-SpoIVA and (H) GFP-SpoVID in sporulating WT cells (strain TD1074) imaged at the indicated times after induction of sporulation. (I) Overlay of mCherry and GFP fluorescence; size bar: 2 μm. (J) TEM of a representative WT sporulating cell (strain PY79) harvested at indicated time points after induction of sporulation. Yellow indicates cortex; green indicates coat; size bar: 500 nm. Strain genotypes are listed in STAR Methods (see also Figure S1A).
Coat assembly is initiated when a small 26-amino-acid protein produced in the mother cell localizes to the forespore surface (Gill et al., 2015; Kim et al., 2017; Ramamurthi et al., 2009) and recruits a structural protein (Peluso et al., 2019; Ramamurthi et al., 2006; Wu et al., 2015), also produced in the mother cell, termed “SpoIVA” (Price and Losick, 1999; Roels et al., 1992; Stevens et al., 1992) (Figure 1G). SpoIVA is an unusual cytoskeletal ATPase that uses the energy released by ATP hydrolysis to polymerize irreversibly on the forespore surface, thereby constructing a platform, or “basement layer,” atop which the rest of the spore coat assembles (Castaing et al., 2013; Ramamurthi and Losick, 2008; Updegrove et al., 2021) (Figure 1A, inset). Subsequent assembly of the peptidoglycan cortex is similarly largely mediated by the mother cell, analogous to the assembly of the bacterial cell wall during normal growth. Soluble peptidoglycan precursors are made in the mother cell cytosol and culminate in the synthesis of the membrane-bound lipid II precursor that is subsequently flipped to the intermembrane space surrounding the forespore, whereupon the peptidoglycan components of lipid II are incorporated into the assembling cortex (Popham and Bernhards, 2015) (Figure 1A, inset, and Figure 1J). Cortex assembly was known to initiate only after coat assembly started (Figures 1G–1J), but surprisingly, several early observations of sporulation mutants using electron microscopy also revealed that improper initiation of coat assembly actually prevented cortex assembly, even though both structures are separated by a membrane (Coote, 1972; Imae and Strominger, 1976; Piggot and Coote, 1976). This suggested the existence of a checkpoint that monitors the morphogenesis of the spore coat. Subsequent investigations revealed that successful tethering and construction of the basement layer of the coat was required to permit cortex assembly (Levin et al., 1993; Roels et al., 1992; Stevens et al., 1992), but despite decades of study, the mechanism that ensures the orchestrated assembly of both spatially separated structures remained unknown.
Here, we report that the mother cell-produced SpoVID protein functions as the primary checkpoint that links the assembly of the coat and cortex. SpoVID is a well-studied protein that is implicated in tethering the outer layers of the coat to the platform created by SpoIVA (Beall et al., 1993; de Francesco et al., 2012; Müllerová et al., 2009; Nunes et al., 2018; Ozin et al., 2000; Qiao et al., 2013; Wang et al., 2009) (Figure 1H), but we discovered an additional negative regulatory role for SpoVID that is manifested when the coat fails to form properly. The C terminus of SpoVID harbors a LysM domain (Costa et al., 2006) that is usually found in cell surface proteins and is implicated in binding N-acetyl-glucosamine (GlcNAc) present in the cell walls of plants, fungi, and bacteria (chitin or peptidoglycan, for example) (Bateman and Bycroft, 2000; Ponting et al., 1999). The presence of this domain in SpoVID has been puzzling because mature peptidoglycan is absent in the cytosol of the mother cell (Figure 1A). An N-terminal extension to this domain was implicated in binding to SpoIVA (McKenney and Eichenberger, 2012; Nunes et al., 2018; Wang et al., 2009), but previous studies reported that the LysM domain of SpoVID is likely not functional because it failed to bind mature cortex in vitro, thereby increasing the mystery regarding the function of this domain (Pereira et al., 2019). We found that SpoVID harbors a functional LysM domain that binds the lipid II peptidoglycan precursor, which contains GlcNAc and is present in the mother cell cytosol. Additionally, we report that SpoVID preferentially binds to polymerized SpoIVA compared with unpolymerized SpoIVA, which may be characteristic of either a mis-assembled spore coat or an immature spore coat. We propose a model in which mis-assembly of the spore coat liberates the C-terminus of SpoVID, which then sequesters lipid II and prevents cortex assembly, specifically in cells that fail to assemble the coat basement layer. The SpoVID checkpoint therefore represents a mechanism that directly monitors a physical characteristic of the developing cell surface to link the morphogenesis of two spatially separated supramolecular structures via a transcription-independent, small-molecule-sensing mechanism.
RESULTS
Deletion of spoVID restores cortex assembly caused by ATPase-defective SpoIVA variants
Deletion of spoIVA not only results in mis-assembly of the spore coat but also prevents initiation of cortex assembly, even though both structures are spatially separated by the outer forespore membrane (Roels et al., 1992). As a result, cells harboring a deletion of spoIVA exhibit a >107-fold decrease in the production of heat-resistant spores compared with the wild type (Figure 1C, lanes 1 and 2). To investigate the pathway that links cortex morphogenesis to the initiation of coat assembly, we sought to employ a genetic approach. However, ΔspoIVA cells display pleiotropic sporulation defects (Roels et al., 1992), and mutations that bypass the SpoIVA requirement for sporulation have not been reported. We therefore examined well-characterized SpoIVA variants that displayed a specific assembly defect. The ATP-binding active site of SpoIVA harbors two motifs that are essential for ATP hydrolysis: a Walker B motif that helps coordinate a Mg2+ ion and activates a water molecule for nucleophilic attack on the γ-phosphate during ATP hydrolysis and a Sensor threonine (“Sensor T”) residue that detects the γ-phosphoryl of the bound ATP to trigger hydrolysis (Castaing et al., 2013; Leipe et al., 2003). Disruption of either motif in SpoIVA specifically abolishes ATP hydrolysis but not ATP binding, consequently abrogating SpoIVA polymerization (Castaing et al., 2013). When fused to GFP, the SpoIVA variant harboring a Sensor T disruption (SpoIVAT*) displayed a relatively minor subcellular localization defect compared with GFP-SpoIVA (Figure 1B) that causes improper coat assembly (Castaing et al., 2013). When viewed under a light microscope, cells harboring SpoIVAT* failed to produce so-called “phase-bright” spores and instead released phase-gray spores, indicating that the spores failed to achieve and maintain a dehydrated spore core (Figure 1E, left panel). This is likely due to the absence of a cortex that is visible via transmission electron microscopy of negative stained thin sections of mature WT B. subtilis spores but not in cells producing SpoIVAT* (Figure 1F, left panel). As a result of the cortex assembly defect, cells harboring either the spoIVAT* or spoIVAB* (defective Walker B motif) allele as the only copy of spoIVA exhibited a 10−5–10−6-fold reduction in sporulation efficiency, despite producing similar levels of protein as WT SpoIVA (Figure 1C, lanes 7 and 11, and Figure S1A). We took advantage of this phenotype to isolate spontaneously arising mutants that corrected the sporulation defect caused by SpoIVAT*. Cells were grown in sporulation medium and nonsporulating cells, and poorly formed spores were subsequently killed by exposure to 80°C for 20 min. Surviving cells were enriched by repeatedly diluting the resulting culture into fresh sporulation media where they could germinate and re-sporulate. Whole-genome sequencing of spoIVAT*-expressing cells displaying enhanced sporulation efficiency revealed an extragenic mutation in the spoVID gene wherein a single nucleotide transition from cytosine to thymidine in codon 546 specifying Gln changed the position to a premature stop codon, presumably truncating the C-terminal 30 amino acids of SpoVID (Figure 2A).
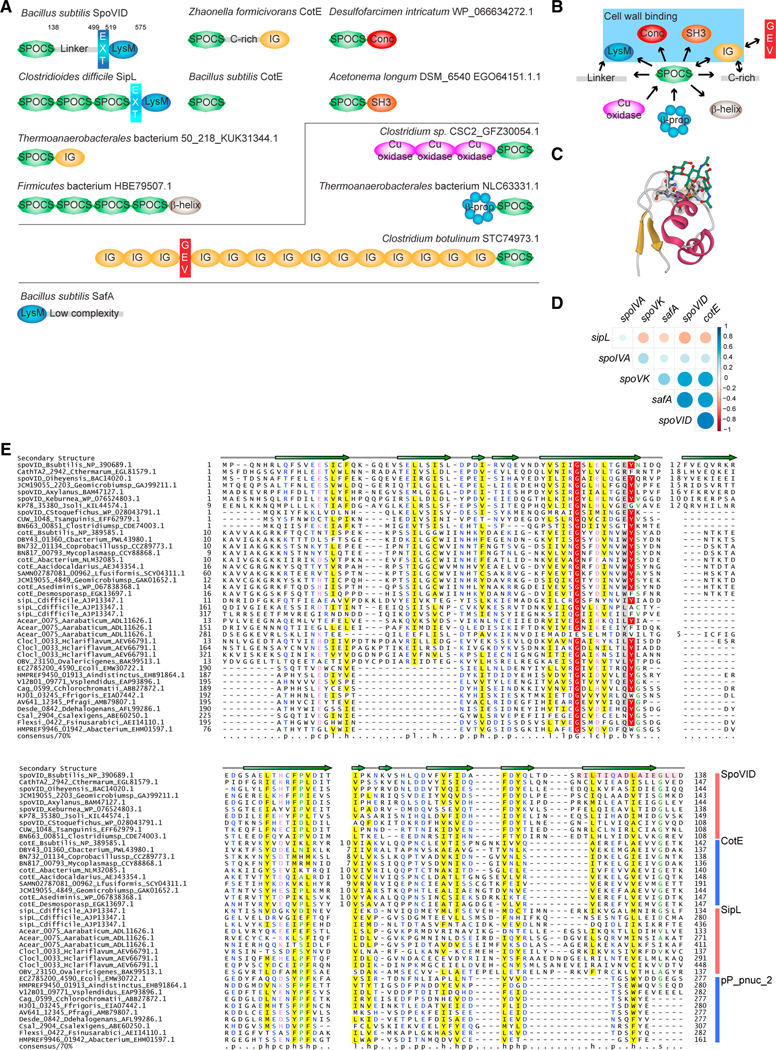
Figure 2. SpoVID domain architecture and phylogeny reveal a superfamily of coat assembly proteins.
(A) Domain architectures of SPOCS (SpoVID-CotE-SipL) domain-containing proteins. Domain architectures are labeled with organism name and a representative gene name (whenever a known gene name is present) or accession. GEV, GEVED; conc, Concanavalin-like; β-prop, beta propeller; EXT, N-terminal extension in LysM domains found in SpoVID and SipL (difference in color denotes a difference in the nature of the extension). Lines delineate proteins harboring N-terminal or C-terminal SPOCS domain(s). For B. subtilis, numbers indicate residue positions that mark C-terminal boundaries of each domain. Also depicted is B. subtilis SafA, a sporulation protein harboring a LysM domain that interacts with SpoVID but does not contain a SPOCS domain. Domains are not drawn to scale.
(B) A simple network is depicted to summarize observed domain arrangements. Arrows represent amino acid sequence direction, from N terminus to C terminus.Blue box indicates domains associated with SPOCS domain that have been implicated in binding peptidoglycan.
(C) The first LysM domain of the NlpC/P60 family peptidoglycan D-L endopeptidase bound to three GlcNAc moieties (green) (PDB: 4UZ3). The threonine facing the reader (circled in yellow) is the equivalent of T532 in SpoVID. The remaining conserved residues of the binding pocket are shown in ball and stick and Gaussian volume representations. Gold, β-strands; maroon, α-helices; green, Glc-Nac moieties; red, O; blue, N; black, C.
(D) Correlation matrix of SpoIVA, SpoVK, SafA, and SPOCS domain-containing proteins SpoVID, CotE, and sipL/DUF3794. Presence or absence of the each of these protein in a given genome is correlated. Positive correlations are displayed in shades of blue and negative correlations in shades of red color. Color intensity and the size of the circle are proportional to the correlation coefficient.
(E) Sequence alignment of the SPOCS domain. Representative set of SPOCS domains from SpoVID, CotE, SipL, and pp_pnuc_2 families is shown in multiple sequence alignment. The encasement region of SpoVID is shown with a pink box. The predicted secondary structure is shown above the alignment with the arrows representing β-strands. Inserts are shown with numbers. The 70% consensus shown below the alignment was derived for the SPOCS domain using the following amino acid classes: hydrophobic (h: ALICVMYFW, yellow), small (s: ACDGNPSTV, green), polar (p: CDEHKNQRST, blue) and its charged subset (c: DEHKR, pink), and big (b: FILMQRWYEK; gray). Numbers at the beginning and end are the start and end of the sequence for the SPOCS domain in a given protein. Numbers in the middle are the length of nonconserved inserts. Aacidocaldarius, Alicyclobacillus acidocaldarius; Aarabaticum, Acetohalobium arabaticum; Abacterium, Acetobacteraceae AT-5844; Abacterium, Acholeplasmataceae bacterium; Aindistinctus, Alistipes indistinctus; Asediminis, Amphibacillus sediminis; Axylanus, Amphibacillus xylanus; Bsubtilis, Bacillus subtilis; Cstoquefichus, Candidatus stoquefichus massiliensis; Cbacterium, Clostridiaceae bacterium; Cchlorochromatii, Chlorobium chlorochromatii; Cdifficile, Clostridioides difficile; Clostridiumsp, Clostridium sp. CAG:451; Coprobacillussp, Coprobacillus sp. CAG:605; Csalexigens, Chromohalobacter salexigens; Cthermarum, Caldalkalibacillus thermarum; Ddehalogenans, Desulfitobacterium dehalogenans; E. coli, Escherichia coli; Ffrigoris, Flavobacterium frigoris; Fsinusarabici, Flexistipes sinusarabici; Geomicrobiumsp, Geomicrobium sp. JCM19055; Hclariflavum, Hungateiclostridium clariflavum DSM19732; Jsoli, Jeotgalibacillus soli; Keburnea, Kroppenstedtia eburnea; Lfusiformis, Lysinibacillus fusiformis; Mycoplasmasp, Mycoplasma sp. CAG:956; Oiheyensis, Oceanobacillus iheyensis; Ovalericigenes, Oscillibacter valericigenes; Pfragi, Pseudomonas fragi; Tsanguinis, Turicibacter sanguinis; Vsplendidus, Vibrio splendidus.
To test whether this extragenic suppressor mutation resulted in a loss of function in SpoVID, we examined the sporulation efficiencies of cells harboring either spoIVAT* or spoIVAB* in the absence of spoVID. Cells harboring a deletion of spoVID in the presence of WT SpoIVA displayed a relatively minor sporulation defect that was ~10-fold lower than WT (Figure 1C, lane 4; Beall et al., 1993). In cells harboring either spoIVAT* or spoIVAB*, spoVID deletion improved sporulation efficiency to ~10−1–10−2 (Figure 1C, lanes 8 and 12); reintroducing spoVID at an ectopic chromosomal locus (amy) in these cells resulted in lower sporulation efficiency, indicating that SpoVID negatively regulates sporulation in the presence of these defective spoIVA alleles. We next tested the effect of removing SpoVID on coat and cortex assembly. The absence of SpoVID did not affect GFP-SpoIVA localization (Wang et al., 2009), nor did it appear to correct the minor mis-localization defect of GFP-SpoIVAT* (Figure 1D). SpoVID, like SpoIVA, is a spore coat protein that has been implicated in the encasement step of sporulation; thus, the absence of SpoVID results in a loosely attached coat (Beall et al., 1993; Wang et al., 2009). Accordingly, when viewed by light microscopy, deletion of spoVID in an otherwise wild-type strain produced phase-bright spores, many of which displayed an appendage that was likely a loosely attached coat (Figure 1E, arrows) (Beall et al., 1993). Deletion of spoVID in cells producing SpoIVAT* also displayed the loose-coat phenotype, but unlike in the presence of SpoVID, these released spores were phase bright, consistent with the heat-resistant phenotype of this strain. Examination of these strains by electron microscopy (Figure 1F) confirmed the presence of a loosely attached coat (green, arrows) and the presence of a cortex (yellow) upon deletion of spoVID, despite these strains producing a SpoIVA variant that could not properly assemble the coat. Taken together, these data suggest that SpoVID, in addition to its previously described role in positively influencing the encasement step (de Francesco et al., 2012; Nunes et al., 2018; Wang et al., 2009), plays a negative regulatory role during sporulation. Removing SpoVID could permit cells to proceed through the sporulation program to produce a cortex despite displaying coat basement layer assembly defects.
The domain architecture and phyletic pattern of SpoVID reveal connections to murein biogenesis and define a superfamily of coat assembly-related domains
The C-terminus of SpoVID harbors a single LysM domain (Figure 2A), a small conserved globular domain that is widely found in carbohydrate-binding proteins from both bacteria and eukaryotes (Buist et al., 2008). In bacteria, LysM domains have been shown to bind peptidoglycan, while in eukaryotes the usual substrate is chitin, the common denominator being the GlcNAc moieties in these molecules. Most bacterial LysM domains are extracellular proteins as indicated by their signal peptide or lipobox anchor and are often fused to other domains that either bind or enzymatically act on murein and other extracellular biopolymers. SpoVID lacks a signal peptide, indicating that its LysM domain paradoxically acts in the intracellular compartment, which is not known to contain mature peptidoglycan. Reciprocal BLAST searches and sequence alignments showed that the LysM domain of SpoVID is most closely related to LysM domains found in SipL, a protein from Clostridium difficile proposed to play a comparable role to SpoVID in that organism (Putnam et al., 2013; Touchette et al., 2019), and B. subtilis SafA, which has been shown to bind SpoVID (Ozin et al., 2001). Notably, like SpoVID, both proteins lack signal peptides (Figure 2A). This implies that the action of multiple firmicute LysM domains in the intracellular compartment might be an important aspect of spore biogenesis.
We then mapped the conserved residues and the predicted secondary structure of the LysM domain of SpoVID onto the co-crystal structure of the same domain from the Thermus thermophilus NlpC/P60 family peptidoglycan D-L endopeptidase N-acetyl-chitohexaose (Wong et al., 2015) (Figure 2C). This showed that the core features of the SpoVID LysM domain are conserved, including the N- and C-terminal β-strands that form a two-stranded sheet within which a central helical hairpin forming the sugar-binding interface is inserted. This mapping showed that four key residues (T532, E558, L559, and K560) are conserved, which predicts that SpoVID LysM should bind a sugar moiety, despite its reported inability to do so (Pereira et al., 2019). Moreover, unlike several LysM domains that occur as multiple tandem copies, there is only one such domain in SpoVID, raising the possibility that it might specialize in binding non-polymeric peptidoglycan precursors.
In addition to the LysM domain, SpoVID contains a previously uncharacterized globular domain at the N terminus that is linked to the former via a flexible linker (predicted to be low complexity and hence disordered), which greatly varies in length between SpoVID orthologs. Using a multiple sequence alignment (MSA) of this N-terminal domain, we performed transitive profile–profile searches that recovered with significant probabilities (Figure 2E): (1) the so-called DUF3794, a Pfam model corresponding to a domain found in three copies in the C. difficile SipL protein N-terminal to the LysM domain (Figure 2A) (Putnam et al., 2013), (2) the family of firmicute proteins defined by the B. subtilis CotE protein that interacts with SpoVID (Zheng et al., 1988), and (3) the pP_pnuc_2, a domain we had earlier identified in prokaryotic Piwi-containing anti-phage immunity systems that recruit effector nucleases (Burroughs et al., 2014) (Figure 2E). We accordingly name this superfamily of homologous domains as SPOCS (for SpoVID, CotE, SipL). The conservation pattern of residues extracted from the MSA, secondary structure inference using the Jnet program (Cole et al., 2008; Drozdetskiy et al., 2015), and deep learning-based contact prediction using the RaptorX algorithm (Xu, 2019) strongly suggest a β-strand-rich sandwich fold with long twisted strands for the SPOCS domain (Figure 2E). Consistent with this, the HHpred searches against the PDB database as well as model prediction with RaptorX suggested that the structural fold of the SPOCS domain has features resembling the lipopolysaccharide-binding protein LBP (Eckert et al., 2013), raising the possibility that it might bind a lipid moiety. The previously mapped “encasement region” of SpoVID maps to the terminal strand of the SPOCS domain (Figure 2E), suggesting that it might additionally facilitate homo- and hetero-typic protein–protein interactions with partners such as CotE (also containing a SPOCS domain) and SafA.
We found that the SPOCS domain is present in diverse architectures, but one widespread theme that stands out (including in SpoVID) combines one or more N-terminal copies of this domain with a C-terminal murein-binding domain separated by a flexible linker (Figures 2A and 2B). The C-terminal murein-binding domain might either recognize sugars (e.g., LysM and concanavalin) or peptides (e.g., SH3). Notably, other than the pP_pnuc_2 clade (not discussed here), all versions of the SPOCS domain are found in firmicutes that are known or predicted to sporulate (Figure 2E). Further, its presence in interacting coat assembly proteins (e.g., CotE) suggests that the SPOCS domain might be a key determinant in recruiting proteins for coat assembly. The architectures with C-terminal peptidoglycan-binding domains separated by a linker (SpoVID included) also predict that these proteins might play a broader role in both coat and cortex assembly via their functionally distinct N- and C-terminal domains.
Examination of the phyletic patterns of multiple sporulation proteins involved in coat and cortex assembly (Figure 2D) indicates that SpoIVA is the only coat protein that positively correlates with all of them. This is consistent with SpoIVA being the conserved component of the basement layer across all sporulating firmicutes. Notably, SpoVID is positively correlated with SpoVK, SafA, and CotE, indicating that it forms a specific functional module with those proteins. However, SpoVID and its above partners are negatively correlated with SipL-related proteins with the DUF3794-clade SPOCS domain. This raises the possibility that SipL-related proteins (Putnam et al., 2013) define an alternative pathway that takes the place of the SpoVID-centric system in a subset of the sporulating firmicutes such as C. difficile.
Inactivation of SpoVID LysM domain restores cortex assembly defect caused by defective SpoIVA variants
Like SpoIVA, SpoVID is produced in the mother cell under the control of the σE transcription factor (Beall et al., 1993). Localization of SpoVID to the forespore surface is directly dependent on SpoIVA and involves amino acids 500–524 of SpoVID, previously termed “region A.” It maps to the conserved α-helix just N-terminal to the LysM domain that we term the “LysM extension” (Figure 2A) (Costa et al., 2006; Wang et al., 2009). In the absence of SpoIVA, GFP-SpoVID mislocalized in the mother cell cytosol (Figures 3A, 3B, 3D, and 3E) (Wang et al., 2009), but GFP-SpoVID localized properly in cells producing SpoIVAT* (Figures 3C and 3F). Immunoblot analysis indicated that similar amounts of SpoVID were detected in the presence of various spoIVA mutant alleles (Figure S1B). SpoVID interacts with at least two other coat proteins, CotE and SafA, via the C-terminal region of the SPOCS domain, thereby tethering the outer layers of the coat to the basement layer of the coat formed by SpoIVA as it migrates around the engulfing forespore in a process termed “encasement” (de Francesco et al., 2012; McKenney and Eichenberger, 2012; Nunes et al., 2018; Ozin et al., 2000; Ozin et al., 2001) (deletion of safA or cotE did not suppress the sporulation defect caused by spoIVAB* or spoIVAT*, though). SpoVID presents a conundrum as, unlike typical peptidoglycan-binding proteins, it is situated in the mother cell cytosol where there is no mature peptidoglycan; thus, its function during sporulation has been mysterious (Pereira et al., 2019).
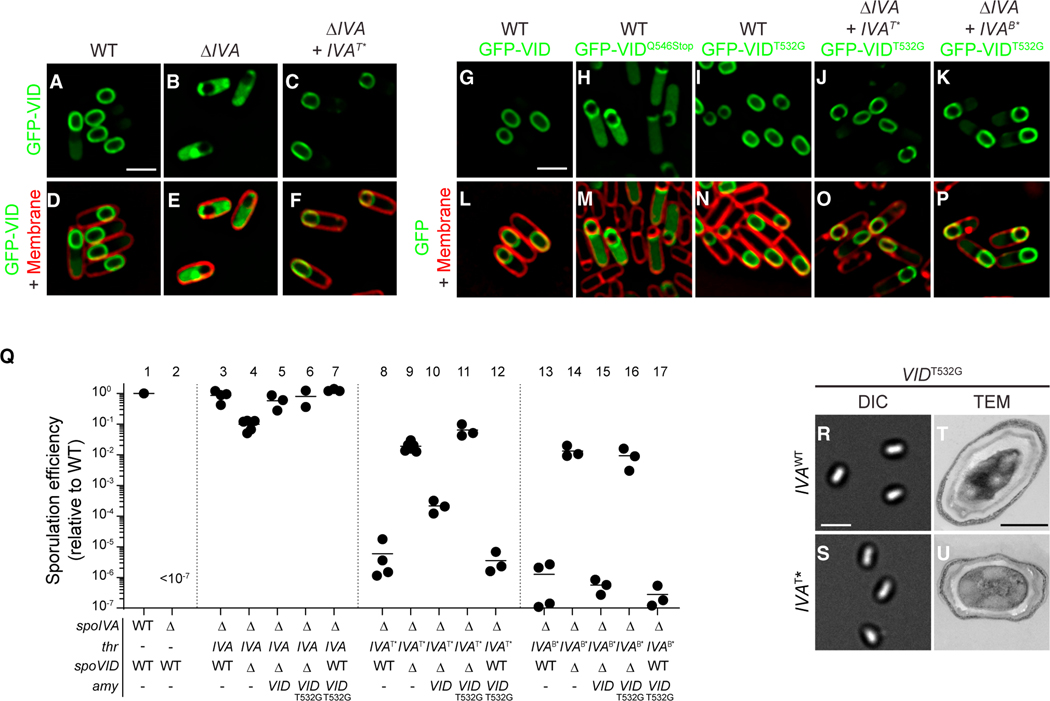
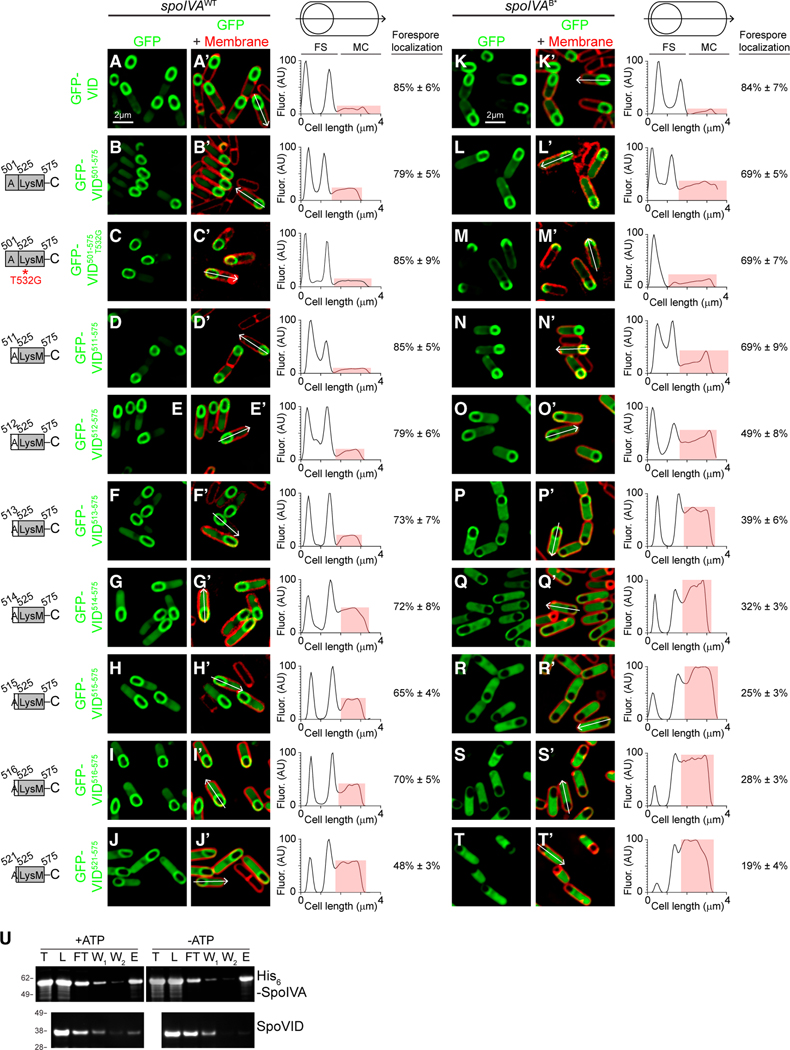
Figure 3. Disrupting the LysM domain of SpoVID suppresses coat assembly defects.
(A–P) Subcellular localization of (A–F) GFP-SpoVID in WT (A and D), absence of spoIVA (B and E), or presence of spoIVAT* (C and F); (G, P) GFP-SpoVID, (G and L) GFP-SpoVIDQ546Stop, and (H and M) GFP-SpoVIDT532G in otherwise WT background; and GFP-SpoVIDT532G in the presence of (I and N) spoIVA, (J and O) spoIVAT*, or (K and P) spoIVAB* cells 3.5 h after induction of sporulation (strains TD139, TD172, TD144, TD139, TD154, TD251, TD954, and TD953). (A–C and G–K) Fluorescence from GFP; (D–F and L–P) overlay of fluorescence from GFP and membranes stained with FM4–64; size bar: 2 μm.
(Q) Sporulation efficiencies, determined as resistance to heat, relative to WT (PY79). Strain genotypes at spoIVA and spoVID loci are indicated below the graph; thr and amy are ectopic chromosomal loci used to complement spoIVA and spoVID deletions, respectively, with indicated alleles of those genes. Symbols are independent cultures; bars represent mean values. Strains used: PY79, KP73, KR394, JB171, JB174, JB281, TD1139, JPC221, JB168, JB176, JB242, TD1140, JB103, JB280, JB293, JB295, and TD1141.
(R–U) Morphologies of spores harboring (R and T) WT or (S and U) T* alleles of spoIVA in the presence of spoVIDT532G as visualized using (R–S; size bar: 2 μm) DIC light microscopy or (T and U; size bar: 500 nm) TEM (strains JB281 and JB242) (see also Figures S1B and S1C).
Since the suppressor mutation in spoVID truncated the LysM domain, we sought to test the effect of specifically inactivating the LysM domain without interfering with either the encasement function or localization of SpoVID. Removing the C-terminal 30 residues of SpoVID resulted in partial mis-localization of the protein: a population of GFP-SpoVIDQ546STOP localized to the mother cell cytosol, and the population that did localize to the forespore surface failed to uniformly coat the forespore and instead formed caps on the mother cell-distal and -proximal poles of the forespore (Figures 3H and 3M). We next utilized the above predictions of the sugar-binding interface to make targeted mutations in the LysM domain that would specifically abrogate its ability to bind substrate. Substituting one of the predicted sugar-binding residues, T532, with Gly resulted in proper localization of GFP-SpoVIDT532G around the forespore (Figures 3I–3K and 3N–3P). Complementation of the spoVID deletion mutant with spoVIDT532G resulted in near wild-type levels of sporulation and phase-bright spores without a poorly attached coat (Figures 3Q, lanes 5 and 6, 3R, and 3T), suggesting that the encasement function of SpoVID was largely unaffected by this substitution. However, similar to the deletion of spoVID, SpoVIDT532G suppressed the sporulation defect of strains harboring spoIVAT* or spoIVAB* (Figure 3Q, lanes 11 and 16, respectively). Immunoblotting cell extracts of sporulating B. subtilis strains indicated that SpoVIDT532G was produced at similar levels in the spoIVA mutant strains as in the wild-type strain (Figure S1C). Concomitantly, strains harboring SpoVIDT532G and SpoIVAT* were able to produce phase-bright forespores (Figure 3S) and elaborate a cortex, as detected by transmission electron microscopy (Figure 3U). Consistent with the T532G substitution resulting in a loss of function, in a merodiploid strain that also produced WT SpoVID, SpoVIDT532G was recessive in correcting the sporulation defect of SpoIVAT* and SpoIVAB* (Figure 3Q, lanes 12 and 17).
Taken together, we conclude that the T532G substitution in the LysM domain of SpoVID specifically inactivated its quality control function of arresting cortex assembly in cells that misassemble the spore coat.
SpoVID harbors a functional LysM domain that binds to the lipid II peptidoglycan precursor
In addition to the lack of mature peptidoglycan in the mother cell cytosol, the activity of the LysM domain of SpoVID has been puzzling because it evidently failed to bind purified peptidoglycan in vitro (Pereira et al., 2019), leading to the hypothesis that it may only function as a protein–protein interaction domain. However, it has been proposed that positive charges in LysM domains play an important role in substrate binding, and therefore, in vitro binding assays should be performed at a pH that is near the isoelectric point of a particular LysM domain (Visweswaran et al., 2011). The isoelectric point of SpoVID residues 525–575 is 4.8, and previous binding studies using purified SpoVID were performed at near-neutral pH. To test whether the LysM domain of SpoVID can bind peptidoglycan under a different buffer condition, we first purified the C terminus of SpoVID (residues 501–575, which comprise the LysM domain) fused to GFP, incubated it at acidic pH (5.5) with purified peptidoglycan from B. subtilis, and measured sedimentation of the protein by immunoblotting in the presence of increasing amounts of the insoluble peptidoglycan upon centrifugation (Yamamoto et al., 2008). At this pH, we observed that the fraction of GFP-SpoVID501–575 in the pellet fraction increased in the presence of increasing amounts of purified peptidoglycan, which was accompanied by a concomitant decrease in the supernatant fraction, indicating that GFP-SpoVID501–575 interacts with purified peptidoglycan (Figure 4A). In contrast, the sedimentation of GFP-SpoVID501–545, which corresponds to the suppressor variant of SpoVID that harbors a truncated LysM domain (Figure 1C, VID*; the T532G variant was not used because its purification yield was poor), and purified GFP were diminished (Figures 4B–4D).
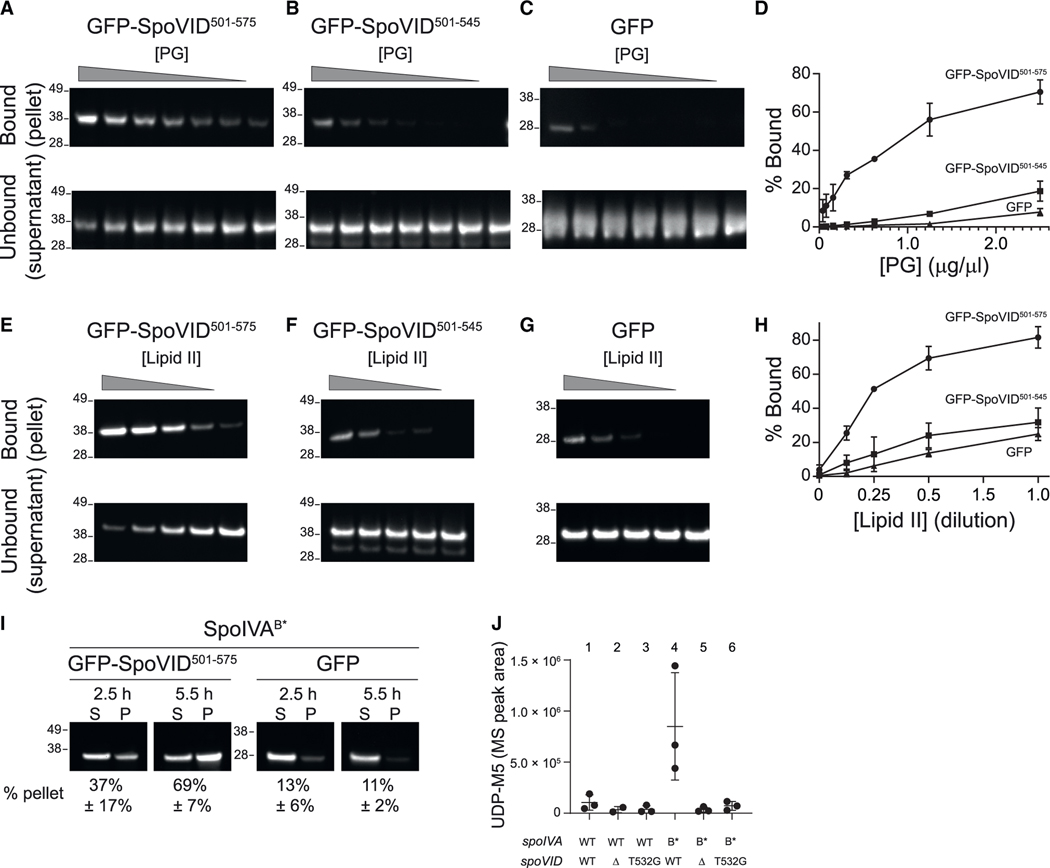
Figure 4. The LysM domain of SpoVID binds to lipid II.
(A–H) Immunoblot analysis of purified (A and E) GFP-SpoVID501–575 or (B and F) GFP-SpoVID501–545 or (C and G) GFP, detected with anti-GFP antiserum, incubated with (A–C) purified mature peptidoglycan (PG) at decreasing concentrations (denoted by triangle), or (E–G) purified lipid II at decreasing concentrations (denoted by triangle) and centrifuged to separate insoluble PG-bound or lipid II-bound protein in the pellet (top) from unbound supernatant (bottom). (D and H) Quantification of fraction of GFP-SpoVID501–575, GFP-SpoVID501–545 or GFP in the bound (pellet) fraction as a function of concentration of purified (D) PG and (H) lipid II. Symbols represent mean values (n = 3); errors are SD. (I) Immunoblot analysis of cell extracts fractionated into insoluble pellet (P) or soluble supernatant (S) fractions by ultracentrifugation, harvested from sporulating cells harboring SpoIVAB* producing either (left) GFP-SpoVID501–575 or (right) GFP, at indicated time points after induction of sporulation, detected with anti-GFP antiserum. Quantification of fraction of GFP-SpoVID501–575 or GFP in the pellet fraction is indicated below each blot (mean values, n = 3 independent cultures; errors are SD). Relative mobility of molecular weight markers (kDa) indicated to the left of each blot. (J) Accumulation of peptidoglycan precursor Park’s nucleotide (UDP-M5) in various strains of sporulating B. subtilis harboring the indicated alleles of spoIVA and spoVID harvested 5.5 h after the induction of sporulation (symbols are independent samples; bars represent mean value; errors are SD). Strains used: KR394, JB171, JB281, JB103, JB280, and JB295.
Although the LysM domain of SpoVID appeared functional in vitro, the physical separation of cytosolic SpoVID and mature peptidoglycan in the intermembrane space of the forespore would prevent such an interaction in vivo. Based on this and domain architectural considerations (see above), we hypothesized that the SpoVID LysM domain may bind to a GlcNAc-containing peptidoglycan precursor in the mother cell cytosol. Such interactions are not usually studied since proteins harboring LysM domains are typically on the cell surface in the same compartment as mature peptidoglycan; nonetheless, soluble fragments of mature peptidoglycan containing GlcNAc have been shown to interact with LysM-containing proteins (Mesnage et al., 2014). Only two peptidoglycan precursors harbor a GlcNAc moiety: soluble UDP-GlcNAc and insoluble lipid II, the final peptidoglycan precursor that is found on the forespore surface (where SpoVID localizes) immediately before it is flipped to the intermembrane space of the forespore where it integrates into the assembling cortex (Ducret and Grangeasse, 2021; Gifford and Meyer, 2015; Meeske et al., 2015). To test whether the SpoVID LysM domain could bind lipid II, we repeated the sedimentation assay using purified insoluble lipid II as the substrate instead of mature peptidoglycan. With increasing amounts of lipid II, the fraction of GFP-SpoVID501–575 increased in the pellet fraction and decreased in the supernatant fraction (Figure 4E). In contrast, the sedimentation of GFP-SpoVID501–545 and GFP was diminished (Figures 4F–4H). Consistent with this in vitro activity, we observed that GFP-SpoVID501–575, but not GFP, was largely in the insoluble fraction of cell extracts prepared from sporulating cells that harbored SpoIVAB* (Figure 4I), consistent with an in vivo interaction between the otherwise soluble LysM domain of SpoVID and a membrane-associated component of the cell extract such as lipid II. The insolubility of polymerized WT SpoIVA, which interacts with GFP-SpoVID501–575, prevented us from performing a similar centrifugation experiment with extracts prepared from cells harboring WT SpoIVA. Nonetheless, these data indicated that SpoVID harbors a functional LysM domain that is capable of specifically binding the GlcNAc-containing peptidoglycan precursor lipid II, and that this interaction may occur in vivo in cells that mis-assemble the basement layer of the coat.
Another in vivo prediction of SpoVID binding lipid II, consistent with a negative regulatory activity of SpoVID, would be the accumulation of peptidoglycan precursors in the mother cell cytosol, specifically when coat assembly (and therefore cortex assembly) is disrupted. This accumulation of precursors is similar to what occurs when cells are treated with cell wall-disrupting antibiotics such as vancomycin (Ling et al., 2015) or when cortex assembly is genetically disrupted during sporulation (Vasudevan et al., 2007). To test this prediction, we extracted total peptidoglycan precursors from sporulating B. subtilis cells and examined the accumulation of Park’s nucleotide (UDP-N-acetylmuramyl pentapeptide), the penultimate precursor of lipid II. Cells harboring wild-type SpoIVA in the presence or absence of SpoVID or in the presence of SpoVIDT532G did not accumulate this intermediate, consistent with the ability of these cells to assemble a cortex (Figure 4J, lanes 1–3). However, cells harboring SpoIVAB* accumulated increased amounts of Park’s nucleotide, suggesting a block in cortex assembly upon disrupting coat assembly (Figure 4J, lane 4) and was similar to the accumulation of Park’s nucleotide observed when several genes encoding for cortex biosynthesis were deleted (Vasudevan et al., 2007). This accumulation in Park’s nucleotide was alleviated upon deletion of SpoVID or in the presence of SpoVIDT532G (Figure 4J, lanes 5 and 6), which was consistent with the ability of these strains to form a cortex despite the inability to correctly form the basement layer of the coat.
The intracellular copy number of lipid II during vegetative growth in B. subtilis has been estimated to be ~18,000, with ~15,000 of those located in the outer leaflet of the plasma membrane and ~3,000 in the inner leaflet (Piepenbreier et al., 2019). To compare the relative abundance of SpoVID relative to lipid II, we immunoblotted cell extracts with anti-SpoVID antiserum and compared the intensity of those blots with that of purified SpoVID of known concentration and divided that total by the number of sporulating cells. This approach revealed that 5.5 h after induction of sporulation, cells harbored 650,000 ± 250,000 copies of SpoVID (Table S1). By comparison, we previously reported that sporulating B. subtilis cells produce a similar number (270,000 ± 90,000) of SpoIVA molecules 4 h after sporulation induction (Peluso et al., 2019). Moreover, assuming a similar intracellular lipid II copy number during sporulation for assembling a smaller cell wall (the cortex) over the course of several hours, these data suggest that the intracellular SpoVID molecules easily outnumber the amount of lipid II. Taken together, these data are thus far consistent with a model in which, upon sensing a defect in spore coat assembly, the LysM domain of SpoVID binds and sequesters lipid II, thereby preventing assembly of the cortex.
The C terminus of SpoVID harbors a sporulation-inhibitory activity
To test the quality control function of SpoVID LysM domain, we tested if GFP-SpoVID501–575 could complement the negative regulatory function of SpoVID in a spoVID deletion mutant. The sporulation defect caused by spoIVAB* may be suppressed by deletion of spoVID (Figures 1C and 5A, lanes 2 and 3). Complementation of this strain with full-length GFP-SpoVID restored the defect, as did complementation with GFP-SpoVID501–575 (but not GFP-SpoVID501–575(T532G)) (Figure 5A, lanes 4–6). This indicates that the C-terminal fragment of SpoVID retains its quality control activity in a manner dependent on a functional LysM domain. Further, this activity is distinct from the coat encasement function that maps to the N-terminal SPOCS domain of SpoVID.
Figure 5. The carboxy terminus of SpoVID is sufficient for sporulation-inhibitory activity and is dominant-negative effect.
(A and B) Sporulation efficiencies of various strains, determined as resistance to heat, relative to WT (PY79). Strain genotypes at spoIVA and spoVID loci are indicated below the graph; thr and amy are ectopic chromosomal loci used to complement spoIVA and spoVID deletions, respectively, with different alleles of those genes. Strains used: PY79, JB103, JB280, TD139, TD931, TD955, TD996, TD998, TD999, TD1000, TD1001, TD1002, TD997, JB171, TD139, TD931, TD996, TD998, TD999, TD1000, TD1001, TD1002, and TD997.
(C) Representative immunoblot of cell extracts of sporulating B. subtilis cultures harboring a deletion of spoVID and expressing gfp-spoVID501–575 ectopically from either the amy (left), sac (center) loci, or from both amy and sac loci (right). Strain genotypes and relative overproduction of GFP-SpoVID501–575 as determined by quantification of immunoblots from three independent cultures are indicated below (errors are SD). Strains used: TD1070, TD1085, and TD1086.
(D) Sporulation efficiencies of WT, ΔspoVID, or strains in (C) producing GFP-SpoVID501–575. All symbols are independent cultures; bars represent mean values. Strains used: PY79, TD100, TD1070, TD1085, and TD1086.
(E) Sporulation efficiencies of strains harboring native copies of spoIVA and spoVID and expressing ectopically either GFP-SpoVID501–575 or GFP-SpoVID511–575, or GFP-SpoVID501–575 or GFP-SpoVID511–575 harboring a disruption in the LysM domain (T532G). Strains used: PY79, JB217, TD1029, TD1031, TD1030, and TD1032 (see also Figure S1D).
We next sequentially removed N-terminal residues from this SpoVID fragment and examined the resulting effect on sporulation efficiency. The deletion of 10 residues, which truncated the LysM extension, did not affect the function of GFP-SpoVID511–575 (Figure 5A, lane 7). However, sequential deletion of single amino-terminal residues from positions 512–516, which were produced at similar levels as GFP-SpoVID501–575 (Figure S1D), resulted in incremental defects in the quality control function of SpoVID, as evidenced by the progressively increasing ability of cells producing SpoIVAB* to sporulate in the presence of these GFP-SpoVID truncations (Figure 5A, lanes 7–12). GFP-SpoVID521–575, which is missing most of the LysM extension, inactivated the negative regulatory activity of SpoVID (Figure 5A, lane 13), similar to the level resulting from disrupting the LysM domain (Figure 5A, lane 6).
We next tested if producing the C terminus of SpoVID alone would result in unregulated negative activity in cells that properly initiate spore coat assembly. Complementation of the spoVID deletion with full-length gfp-spoVID restored the ~10-fold sporulation defect (Figure 5B, lanes 1–3); however, complementing the spoVID deletion mutant with GFP-SpoVID501–575 instead resulted in an additional ~1000-fold reduction in sporulation efficiency, even though the cells produced WT SpoIVA (Figure 5B, lane 4). This inhibitory activity was largely retained upon further deletion of 11 additional residues such that GFP-SpoVID512–575 inhibited sporulation in cells that properly initiated coat assembly (Figure 5B, lanes 5 and 6). Further truncations exhibited intermediate levels of inhibition (Figure 5B, lanes 7–10), but GFP-SpoVID521–575 did not inhibit sporulation more than the spoVID deletion (Figure 5B, lane 11). These data were therefore consistent with the notion that the C terminus of SpoVID was not only sufficient for the negative regulatory role of SpoVID in the context of a mis-assembling spore coat, but that it could inhibit sporulation in an uncontrolled fashion even in the presence of a properly assembling coat basement layer.
To test this uncontrolled inhibitory activity further, we tested if increasing the production of the C terminus of SpoVID would result in increased inhibitory activity. To this end, we expressed gfp-spoVID501–575 from two different chromosomal loci (amy or sac) in the absence of the native copy of spoVID in an otherwise WT cell. Expression of this construct from either locus produced roughly equal amounts of protein, as detected by immunoblotting (Figure 5C, lanes 1 and 2) and reduced sporulation efficiency ~103–104-fold (Figure 5D, lanes 3 and 4). However, producing the construct from both loci simultaneously approximately doubled the protein level (Figure 5C, lane 3) and further reduced sporulation efficiency to ~10−6 relative to WT (Figure 5D, lane 5), indicating that the inhibitory effect of the C terminus of SpoVID in cells that properly assemble the coat is dose dependent.
These results suggest that the C-terminal fragment of SpoVID could exert a dominant-negative activity in a merodiploid strain harboring the native copy of spoVID in cells that assemble the spore coat properly. Co-expressing a second copy of full-length gfp-spoVID from an ectopic chromosomal locus did not affect the sporulation efficiency of otherwise wild-type cells (Figure 5E, lane 2). However, expressing gfp-spoVID501–575 or gfpspoVID511–575 resulted in a ~5000-fold decrease in sporulation efficiency (Figure 5E, lanes 3 and 4), indicating that the C-terminal fragment of SpoVID, in the absence of the rest of the protein, harbors an activity that is dominant over the full-length SpoVID. As a control, producing either GFP-SpoVID501–575 or GFPSpoVID511–575 harboring the T532G substitution did not result in sporulation inhibition (Figure 5E, lanes 5 and 6).
The C terminus of SpoVID detects the polymerization state of the spore coat basement layer
We wondered how the negative regulatory function of the LysM domain of SpoVID could be exerted only when the spore coat basement layer mis-assembles. To investigate this, we examined if the various SpoVID truncations described above would differentially localize when the basement layer of the spore coat assembles or mis-assembles. GFP-SpoVID501–575, which includes the entire LysM domain and the helical extension, localized to the forespore surface, both in WT cells and in cells harboring spoIVAB* that mis-assembles the coat (Figures 6B– 6B′ and 5L–5L′). This pattern was similar to the localization pattern of full-length GFP-SpoVID (Figures 6A–6A′ and 4K– 4K′) (Wang et al., 2009). GFP-SpoVID501–575 harboring the T532G substitution also localized properly in WT cells (Figures 6C–6C′). However, in the presence of spoIVAB*, although this construct localized to the forespore, GFP-SpoVID501–575(T532G) largely accumulated at the mother cell-distal pole of the forespore (Figures 6M–6M′), suggesting that the LysM region plays a subtle role in the proper localization of SpoVID. Deletion of a further 10 residues did not affect localization GFP-SpoVID511–575 (Figures 6D–6D′ and 6N–6N′) in either strain background. However, deletion of an additional residue resulted in an increased mis-localization of GFP-SpoVID512–575 specifically in cells harboring SpoIVAB* but not in WT cells (Figures 6E–E′ and 6O–O′; compare the cytosolic signal intensity, shaded in red, of the line scan to the right of the micrographs), as evidenced by the increased GFP signal intensity in the mother cell cytosol. This pattern of preferential increased cytosolic mis-localization in cells harboring SpoIVAB* continued as we examined sequential truncations in the LysM extension (Figures 6F–6I′ and 6P–6S′). Deletion of almost the entire LysM extension resulted in the mis-localization of greater than 80% of GFP-SpoVID521–575 in the mother cell cytosol in cells harboring SpoIVAB*, but only 50% of GFP-SpoVID521–575 in the mother cell cytosol of WT cells (Figures 6J–6J′ and 6T– 6T′). This suggests that the SpoVID LysM domain and its α-helical extension could directly sense the assembly state of the basement layer of the spore coat and that it may become liberated when the basement layer mis-assembles.
Figure 6. The carboxy terminus of SpoVID can detect the polymerization state of SpoIVA.
(A–T′) Subcellular localization of indicated sequential truncations of SpoVID C-terminal fragments fused to GFP in sporulating cells 3.5 h after induction of sporulation in the presence of (A–J′) WT or (K–T′) B* variant of SpoIVA. A–T: fluorescence from GFP; A′–T′: overlay of fluorescence from GFP and membranes stained with FM4–64. Arrows indicate linescan used to quantify GFP fluorescence intensity along an individual sporangium; quantification of the linescan of the single indicated cell shown to the right (red highlights GFP fluorescence intensity localized in the mother cell cytosol), along with mean fraction of forespore-localized fluorescence intensity (n = 10 cells; errors are SD). Size bar: 2 mm. Strains used: TD139, TD931, TD955, TD996, TD998, TD999, TD1000, TD1001, TD1002, TD997, TD142, TD933, TD957, TD961, TD979, TD980, TD981, TD982, TD983, and TD962. (U) Representative immunoblot of SpoIVA and GFP-SpoVID501–575 with polymerized SpoIVA (+ATP) or nonpolymerized SpoIVA (−ATP). Total amount of SpoIVA before polymerization (T), proteins loaded onto the affinity column after SpoIVA polymerization and SpoVID interaction (L), column flow through (FT), first and second column washes (W1, W2), proteins eluted from the column (E).
To directly test whether the C terminus of SpoVID preferentially binds to polymerized SpoIVA, we first overproduced and purified His6-tagged SpoIVA from Escherichia coli and incubated it in the presence (or absence) of ATP for 4 h to induce polymerization (Castaing et al., 2013; Updegrove et al., 2021). Next, we briefly incubated this reaction with purified GFP-SpoVID501–575 that did not contain a His6 affinity tag, purified His6SpoIVA using Ni2+-affinity chromatography and examined the copurification of GFP-SpoVID501–575 with either the polymerized or unpolymerized SpoIVA by immunoblotting. After incubation in the presence or absence of ATP, we recovered nearly all the SpoIVA that we had initially purified (Figure 6U; compare lanes “T,” total, with “L,” load in the SpoIVA blot). We detected a small amount of SpoIVA in the column flow through (Figure 6U; “FT”) and, after two washes (Figure 6U; “W1” and “W2”), recovered similar amounts of polymerized and unpolymerized SpoIVA in the elution (Figure 6U; “E”). When we examined the copurification of GFP-SpoVID501–575, we observed that the amount of GFP-SpoVID501–575 in the elution was 7.0-fold (±2.2, n = 3) higher in the presence of polymerized SpoIVA than unpolymerized SpoIVA. Taken together, these data indicate that proper localization of the SpoVID LysM domain at the forespore surface is required for its quality control function. More importantly, the LysM domain of SpoVID distinguishes between polymerized (+ATP) and unpolymerized (ATP) states of SpoIVA by dissociating from SpoIVA when SpoIVA fails to assemble properly.
These data are consistent with a model in which the C terminus of SpoVID can detect the polymerization state of SpoIVA and at least partially disengage from SpoIVA when SpoIVA fails to polymerize. We propose that the disengaged LysM domain, provided it remains localized near the forespore surface, can inhibit the progression of sporulation by sequestering lipid II, thereby preventing cortex assembly, specifically in cells that fail to assemble the spore coat.
DISCUSSION
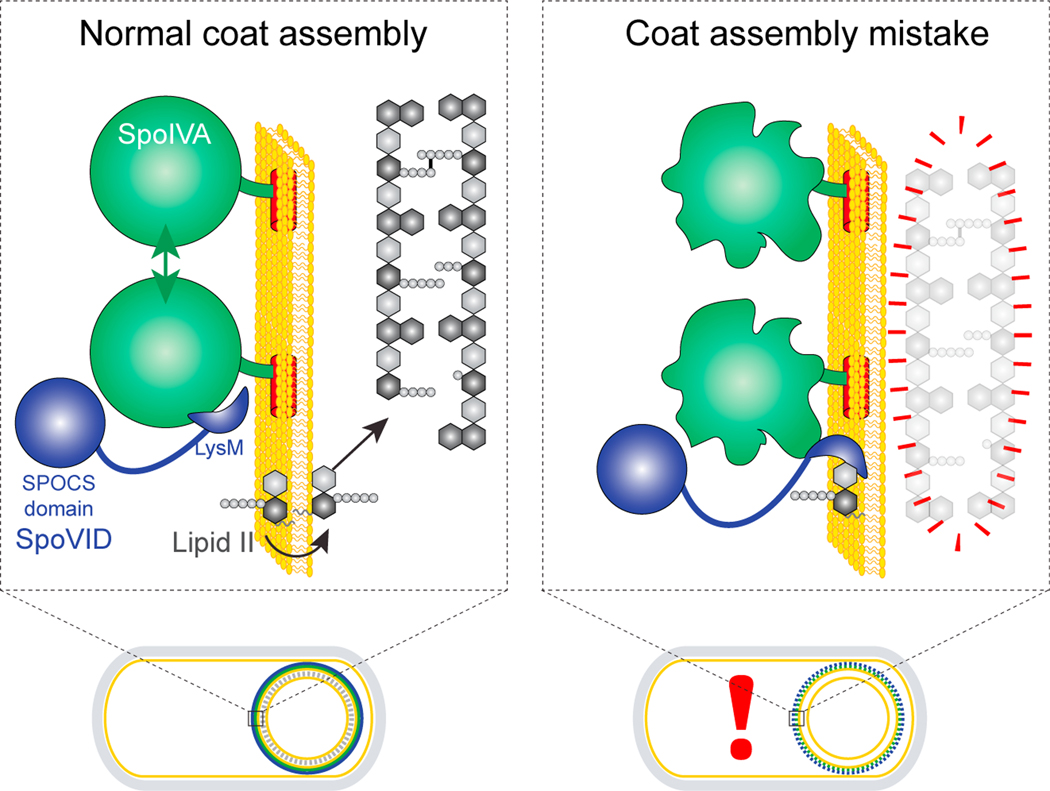
Here, we have investigated a mechanism by which the morphogenesis of two static supramolecular structures (the bacterial spore coat and cortex), spatially separated by a membrane, can be coupled during development. Our proposed mechanism also describes how the cell physically monitors the assembly status of one of those structures (the coat) and couples its morphogenesis with the assembly of the other (the cortex). In our model, the SpoVID protein, which was identified in a genetic selection to uncouple cortex assembly from coat assembly, is the central regulator in this orchestration (Figure 7, blue). SpoIVA is a conserved sporulation protein (Galperin et al., 2012) that polymerizes around the developing forespore in an ATP hydrolysis-dependent manner (Ramamurthi and Losick, 2008) to form the basement layer of the spore coat (Figure 7, green). SpoIVA directly or indirectly recruits several coat proteins, among which is SpoVID, which has been previously implicated in tethering other coat proteins to the platform structure created by polymerized SpoIVA (Driks and Eichenberger, 2016). SpoVID performs its tethering function via its N-terminal SPOCS domain, which we have identified in this report as a conserved domain in multiple firmicute-specific spore coat proteins. In our model, SpoVID has an additional, negative, regulatory role: to inhibit sporulation by halting cortex assembly when SpoIVA assembles improperly. We propose that SpoVID primarily achieves this function via its cytosolic C-terminal LysM domain that binds to GlcNAc moieties. (Ponting et al., 1999). SpoVID harbors an N-terminal α-helical extension to the LysM domain, and residues in this region have previously been implicated in binding to SpoIVA (Costa et al., 2006). We propose that the LysM domain of SpoVID and its extension have a dual function: to selectively bind to SpoIVA when SpoIVA polymerizes properly (thereby either sterically hindering the sugar-binding site of the LysM domain or driving a conformational change in the LysM domain that prevents substrate binding) and to disengage from SpoIVA when SpoIVA fails to polymerize and to instead bind to the cell wall precursor lipid II, thereby sequestering it and preventing lipid II from flipping across the outer forespore membrane and incorporating into the growing cortex cell wall (Figure 7).
Figure 7. Model for the SpoVID-mediated orchestrated assembly of coat and cortex during sporulation.
Depicted are a wild-type sporulating bacterium (left) and a mutant bacterium that mis-assembles the spore coat (right). An expansion of the spore envelope (spore coat basement layer, outer forespore membrane, and cortex) is depicted above each cell. When SpoIVA (green) assembles properly in a wildtype cell, the LysM domain of SpoVID (blue) is occluded. Lipid II (gray) is produced in the mother cell and is permitted to flip to the intermembrane space to incorporate into the growing cortex. When SpoIVA mis-assembles (right), the LysM domain of SpoVID is liberated and sequesters lipid II, preventing it from being flipped into the intermembrane space and thereby disrupting cortex assembly.
Our model is supported by several lines of genetic, biochemical, and cytological evidence. First, we demonstrated that the LysM domain of SpoVID is functional. The presence of this domain in a cytosolic protein has been historically puzzling due to the absence of peptidoglycan in this compartment and attempts at demonstrating the functionality of this domain in SpoVID were previously unsuccessful (Nunes et al., 2018). In our buffer conditions (which may possibly mimic an underappreciated in vivo microenvironment of the forespore surface), optimized for the unique amino acid composition of the LysM domain of SpoVID, we demonstrated that SpoVID could bind in vitro to lipid II, the final precursor of peptidoglycan biosynthesis and a molecule that is located in the same cellular compartment as SpoVID. To our knowledge, this is the first example of a biological role for an intracellular LysM domain binding to a peptidoglycan precursor instead of mature peptidoglycan (Masser et al., 2021). Accordingly, mutations in the LysM domain predicted to abrogate LysM function uncoupled cortex assembly from improper coat morphogenesis in vivo and permitted the sporulation program to proceed despite the presence of a defective coat. Consistent with a negative regulatory function of this domain, we observed that the penultimate precursor of lipid II, Park’s nucleotide, accumulated in cells that mis-assembled the coat but failed to accumulate this precursor once the LysM domain was disrupted. Second, the LysM domain of SpoVID functioned independently in this negative regulatory role but did so in an uncontrolled fashion. Producing just the C terminus of SpoVID successfully inhibited cortex assembly in mutant cells that misassembled the coat but also inhibited sporulation in cells that properly initiated coat assembly. This result predicts that an interplay with the N-terminal SPOCS domain of SpoVID might be required to regulate the activity of the LysM domain. Indeed, producing the SpoVID LysM fragment displayed a dominant-negative phenotype in merodiploid strains harboring a wildtype copy of spoVID, and increasing production of the LysM domain alone exhibited a dose-dependent inhibition of sporulation when the coat properly assembled. The results are collectively consistent with a simple model wherein the LysM domain directly binds to free lipid II and sequesters it, although it is formally possible that the LysM domain binds lipid II in the context of another lipid II-binding protein, such as a protein in the peptidoglycan biosynthetic pathway, to exert its inhibitory activity. Third, the LysM domain of SpoVID preferentially bound to polymerized SpoIVA in vitro. In vivo, this discriminatory binding was revealed by the mis-localization of the SpoVID LysM fragment fused to GFP in cells harboring a polymerization-defective allele of spoIVA but not in otherwise wild-type cells in which coat assembly proceeded normally. In summary, these data revealed that SpoVID is a bifunctional protein whose (positive) encasement function during sporulation may be uncoupled from its (negative) regulatory role that monitors the progress of the assembly of the spore coat basement layer.
Although it is not immediately clear why the assemblies of the coat and cortex are coupled, the phenomenon appears to be broadly conserved in other sporulating firmicutes. In Clostridium difficile, coat and cortex morphogenesis are not as stringently linked as they are in B. subtilis (Touchette et al., 2019), but deleting spoIVA or sipL (which encodes a protein homologous to SpoVID) nonetheless results in some cortex assembly defects, indicating an evolutionarily conserved advantage during sporulation in orchestrating coat and cortex assembly. Since coat assembly occurs at a late stage during sporulation after the program is irreversible (Dworkin and Losick, 2005), the accumulation of mutations that subtly abrogate coat assembly could result in the propagation of mutations that could ultimately undermine the fidelity of the sporulation program in progeny cells. As such, SpoVID may represent a mechanism that ensures the faithful assembly of a static structure wherein a cell may abort the pathway rather than continuing with mistakes, akin to other late-stages porulation quality control pathways described previously (Decker and Ramamurthi, 2017; Ramírez-Guadiana et al., 2017; Tan et al., 2015). A key feature of our model is that the inhibition by SpoVID should be reversible: once SpoIVA polymerizes, the LysM domain would be occluded, upon which cortex assembly could proceed. Thus, in a wild-type cell, SpoVID likely represents a timing mechanism that ensures that cortex assembly does not begin until coat assembly properly initiates. Could the linked assembly of both structures occur in a similar manner in C. difficile? C. difficile contains a protein termed SipL that not only contains a C-terminal LysM domain (Putnam et al., 2013) but, according to our analysis, also harbors three N-terminal SPOCS domains, thereby suggesting a degree of functional equivalence. Indeed, SipL was reported to preferentially interact with wild-type SpoIVA in an in vivo copurification experiment compared with a putative ATPase-defective variant of SpoIVA (Benito de la Puebla et al., 2020; Putnam et al., 2013), which suggests a conserved mechanism by which both species may use a similar protein to orchestrate coat and cortex assembly during sporulation.
Myriad biological processes involve the orchestrated construction of large static structures, and mechanisms have evolved to monitor intermediate steps to ensure the correct, ordered assembly of the entire structure. For example, phage heads and viral capsids typically require a scaffolding protein that ensures the templated assembly of the head or capsid, without which capsid proteins typically assembly into aberrant, irregular structures. Additionally, a portal protein, required for DNA entry, assembles at the base of the assembling capsid that ensures DNA is packaged only after capsid assembly is completed (Aksyuk and Rossmann, 2011). In this scenario, the stepwise association of key regulatory proteins to the assembling structure ensures the orderly self-assembly of the entire structure. During bacterial flagellar assembly, ordered assembly of the flagellum is instead primarily dictated by coupling gene expression to key morphogenetic events (Chilcott and Hughes, 2000). For example, the transcription of genes encoding structural elements that form the bulk of the extracellular portion of the flagellum is repressed until the basal body of the flagellum is constructed. This link is achieved via a transcriptional repressor that is itself a substrate for export by the basal body but does not itself assemble into the flagellum. In this way, once construction of the basal body is completed, the transcriptional repressor is secreted, which then permits the production of factors that make up the final part of the flagellum. A distinguishing feature of the checkpoint mechanism we have proposed is that it physically monitors the state of a particular event (in this case, spore cell surface assembly in the mother cell cytosol) and initiates a response that is manifested in a different compartment (the intermembrane space surrounding the forespore). Thus, the case of the ordered assembly of the coat, then the cortex, applies not simply to two parts of the same ultrastructure but instead to two different supramolecular structures in two different subcellular locations, without invoking transcriptional control. This phenomenon of physically monitoring protein polymerization is reminiscent of the taxol response in eukaryotes (Kelling et al., 2003), wherein disrupting microtubule dynamics results in the arrest of mitosis. This indicates that a mechanism exists in that system whereby cells physically monitor a disrupted microtubule and arrest progression through the cell cycle (Joglekar and Kukreja, 2017; Long et al., 2019; Salmon and Bloom, 2017). By comparison, the coupled morphogenesis of the coat and cortex during sporulation involves directly monitoring the polymerization state of the unusual cytoskeletal protein SpoIVA and either permitting or prohibiting progression through the sporulation program. Thus, the ability of cells to directly monitor the assembly of supramolecular structures and trigger a response appears to have arisen in disparate scenarios.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and fulfilled by the Lead Contact, Kumaran S. Ramamurthi (ramamurthiks@mail.nih.gov).
Materials availability
All bacterial strains and antisera raised against B. subtilis proteins used herein are freely available upon request.
Data and code availability
Microscopy data can be shared upon request.
No original code was developed in this study.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
METHOD DETAILS
Strain construction
Strains are otherwise isogenic derivatives of B. subtilis PY79 (Youngman et al., 1984). Genes of interest were PCR amplified to include their native promoter and cloned into integration vectors pDG1662 (for insertion into the amyE locus), pDG1731(thrC locus) or pSac-Cm (sacA locus) (Guérout-Fleury et al., 1996; Middleton and Hofmeister, 2004) containing the appropriate inserts as templates. Site-directed mutagenesis was achieved using QuikChange kit (Agilent). All plasmids were integrated into the B. subtilis chromosome by double recombination at the specified ectopic locus. Plasmid construction was verified by DNA sequencing.
General methods
To determine sporulation efficiencies, cells were grown and induced to sporulate in Difco Sporulation Medium (KD Medical) for at least 24 h; nonsporulating cells and defective spores were killed by exposure to 80° C for 20 min. Cultures were then serially diluted and colony-forming units (cfu) that survived were determined and reported relative to the cfu obtained in a parallel culture of the WT PY79 strain. Spontaneous suppressor mutant was isolated by enriching for colonies that grew after multiple rounds of heat treatment and growth; mutation was identified by whole genome sequencing as described previously [Tan]. SpoVID, SpoIVA and σA levels were determined by immunoblotting cell extracts prepared as described previously (Ramamurthi and Losick, 2008) using rabbit antiserum raised against purified SpoIVA, SpoVID or σA (Covance) as primary antibody and a goat Starbright Blue 700 (Bio-Rad) as secondary antibody. Bands were quantified, as needed, using Fiji software (NIH).
Sequence analysis
Homologs were identified using sequence profile searches performed with the PSI-BLAST program (Altschul and Koonin, 1998). The searches were run against the non-redundant (NR) protein database of National Center for Biotechnology Information (NCBI) or the same database compressed by clustering at 50% (nr50) or a custom database of 4210 complete prokaryotic proteomes. Profile-profile searches were conducted using the HHpred program (Söding, 2005; Söding et al., 2005) with multiple alignments to derive the query hidden Markov model augmented by hits against the nr50 or nr70 databases. They were run against 1) PDB; 2) Pfam; 3) An inhouse collection of profiles; the significance of the hits were assessed using the probability percentage and p-value of the HHpred hits. Deep-learning-based contact prediction for fold inference was performed using the RaptorX algorithm (Xu, 2019). Domain architectures were obtained using a combination of profile searches against Pfam and in-house profiles. Detection of homologs for phyletic pattern correlation analysis was done on a curated set of 4210 complete prokaryotic proteomes. The correlation analysis was done in the R language using the cor() function with the “pearson” method to compute correlation. The plot was drawn using the R Corrplot library (Wei and Simko, 2021). Multiple sequence alignment was built using MUSCLE (Edgar, 2004) or KALIGN programs (Lassmann et al., 2009; Lassmann and Sonnhammer, 2005) and manually corrected based on secondary structure. Protein secondary structure was predicted using a multiple alignment as the input for the Jnet program (Cole et al., 2008; Cuff and Barton, 2000; Drozdetskiy et al., 2015) and single sequence as input for the Deep-Learning based secondary structure inference from the RaptorX algorithm (Xu, 2019). Structural visualization of the pdb files were carried out using the Mol*viewer (Sehnal et al., 2021).
Epifluorescence microscopy
Cells were induced to sporulate by resuspension in SM medium (Sterlini and Mandelstam, 1969). At various time points, 100 μl of culture was harvested and resuspended in 10 μl SM medium containing 5 μg·mL-1 fluorescent dye FM4–64 to visualize membranes, and placed on a glass- bottomed culture dish (Mattek) 3 μl was spotted on a glass bottom culture dish (Mattek) and covered with a 1% agarose pad made with SM medium. Cells were viewed at room temperature with a DeltaVision Core microscope system (Applied Precision/GE Healthcare). Seven planes were acquired every 200 nm, and the data were deconvolved using SoftWorx software (Eswara et al., 2018). Linescan analyses were performed using Fiji.
Transmission electron microscopy
Cells were allowed to sporulate in DSM for greater than 24 hours. Cells were harvested by centrifugation, washed with water, and resuspended in 20% metrizoic acid (Sigma S4506). Resuspended cells were then added to the top of a step gradient of five different metrizoic acid concentrations (70%, 60%, 50%, 40% and 30%) and centrifuged at 40,000 x g for 60 min at 4C (Fukushima et al., 2004). Spores were found in the middle layers and were collected, washed with water, and fixed in 4% formaldehyde, 2% glutaraldehyde in 0.1 M cacodylate buffer, post fixed using a 1% osmium tetroxide solution, then dehydrated sequentially in 35%, 50%, 75%, 95% and 100% ethanol followed by 100% propylene oxide. Cells were infiltrated in an equal volume of 100% propylene oxide and epoxy resin overnight and embedded in pure resin the following day. The epoxy resin was cured at 55 °C for 48 h. The cured block was thin-sectioned and stained in uranyl acetate and lead citrate. The sample was imaged with a Hitachi H7600 TEM equipped with a CCD camera (Ebmeier et al., 2012).
Protein purification
For copurification experiments, GFP-SpoVID501−575-FLAG was cloned into pET28a vector to create pTD354. Overnight cultures of E. coli BL21(DE3) harboring either pTD354 or pKR145 (expressing his6-spoIVA (Ramamurthi and Losick, 2008)) grown in LB medium supplemented with 50 μg/ml kanamycin for plasmid maintenance were diluted 1:50 into 500 ml fresh LB/Kanamycin and grown for 2 h at 37 °C at 250 rpm. Cells were induced to produce the proteins by addition of 1 μM final concentration IPTG and grown for an additional 3 h at 37 °C at 250 rpm. Cells were harvested by centrifugation, resuspended in buffer A (50 mM Tris at pH 7.5, 150 mM NaCl), and disrupted using a French pressure cell at ca. 20,000 psi. Cell lysate was cleared by ultracentrifugation at ~100 k × g for 30 min at 4 C. Supernatant containing His6-SpoIVA was removed and placed on Ni2+-NTA agarose beads (Qiagen), washed with buffer A containing 20 mM imidazole, and eluted with buffer A containing 250 mM imidazole and additionally purified using anion exchange chromatography (MonoQ, Cytivia) using a step gradient of NaCl. Protein peak at 400 mM NaCl was collected and stored at −80 °C. Supernatant containing GFP-SpoVID501−575-FLAG was purified using magnetic agarose anti-DYKDDDDK beads (Pierce) in buffer A, washed with buffer A, and eluted using 150 mg/ml 3X DYKDDDDK peptide. Peak fractions were collected and additionally purified using size exclusion chromatography (Superdex 75 Increase, Cytiva) and stored at 4° C for a maximum of 4 days. For peptidoglycan and Lipid II binding experiments, gfp-spoVID501-575-his6, gfp-spoVID501-545-his6, and gfp-his6 were cloned into pET28a vector to create pTD352, pTD354, and pKR168, respectively. Cells harboring these plasmids were induced and purified using Ni2+-affinity chromatography as described above and additionally purified using size exclusion chromatography (Superdex 75 Increase, Cytiva).
Peptidoglycan and lipid II binding assays
Peptidoglycan was purified from B. subtilis largely as described previously (Alvarez et al., 2016). 1 L of B. subtilis culture was pelleted, resuspended in 5 mL PBS, and added dropwise to an equal volume of boiling 10% SDS and boiled with stirring for 2 h. The insoluble material was pelleted by centrifugation in a benchtop ultracentrifuge (153,000 × g for 15 min at 20 °C) and washed three times in water to remove the SDS. Final pellets were resuspended in 1 mL water; 200 mg glass beads (0.1 mm) were added and cells were disrupted by homogenization twice for 90s by bead beating. The insoluble fraction was pelleted by ultracentrifugation as before, and the pellet was resuspended in 1 mL 100 mM Tris-HCl at pH 7.5. 10 μL of 10 mg/ml ɑ-amylase (Sigma) was added and the reaction was incubated for 2 h at 37° C with shaking to remove excess polysaccharides. A further 1 ml of 100 mM Tris-HCl pH 7.5 was added along with 40 μL 1 M MgSO4, 2 μL of RNase A (10 mg/mL, Thermo) and 1 mL of DNase I (1000 U/mL, Thermo); the reaction was incubated with shaking at 37C for a further 2 h. Next, 50 μL of 50 mM CaCl2 and 100 μL of trypsin solution (2.5 mg/mL, Sigma) were added and the mixture was incubated overnight at 37°C with stirring. The next day, the trypsin was inactivated by adding SDS to 1% and boiling the samples for 10 min. The insoluble peptidoglycan was pelleted by ultracentrifugation, resuspended in 1 mL 8 M LiCl and incubated for 10 min at 37 °C to remove bound proteins. The peptidoglycan was then pelleted again, resuspended in 1 mL 100 mM EDTA and incubated for 10 min at 37 °C. Next, the peptidoglycan was pelleted and resuspended in 3 mL acetone:H2O (1:2) and then washed twice in water. The final pellet was resuspended in 300 mL water, dried under vacuum, and resuspended in 1 mL cold 47% HF. The mixture was transferred to a 15 mL tube and stirred for 48 h at 4 C. The peptidoglycan was pelleted by ultracentrifugation and washed four times in water, then dried by lyophilization. Peptidoglycan binding assays were based on the assay by Yamamoto et al. (Yamamoto et al., 2008). Briefly, 100 nM of purified GFP-SpoVID501−575 GFP-SpoVID501−545 or GFP were incubated 30 min at 37 °C in presence of 2.5, 1.25, 0.625, 0.312, 0.156, 0.08, 0 mg/ml of purified peptidoglycan in 100 ml total reaction buffer (50 mM Sodium Acetate at pH 5.5, 100 mM NaCl, 0.02% Tween 20). Samples were then centrifugated at 15,000 3 g for 2 min at room temperature. 75 ml supernatant was removed and prepared for immunoblotting by adding 25 ml 4X sample buffer. To equalize concentrations between pellet and supernatant, the pellet was resuspended in 133 ul 1x sample buffer. Equal volumes of pellet and supernatant fractions were separated and examined by immunoblotting using anti-GFP antiserum. Lipid II was purified from B. subtilis essentially as described previously (Qiao et al., 2017). 1 L B. subtilis culture was pelleted and resuspended in 5 mL PBS, then added to 40 mL chloroform:methanol (1:2) and stirred for 1 h. The mixture was centrifuged at 4000 × g at 4 °C for 10 min and the supernatant was transferred to a new flask containing 24 ml chloroform and 18 ml PBS. The mixture was stirred for 1 h, centrifuged at 4000 × g at 4 °C for 10 min and the tubes were stored at 4 °C overnight to allow the interphase (a white layer between the organic and aqueous phases) to settle. The next day, the interphase was removed and dried under vacuum. The dried interphase was dissolved in 15 ml organic solvent (6 M pyridium acetate:n-butanol (1:2)), transferred to a separating funnel and 15 ml of aqueous solvent (water saturated with n-butanol) was added to wash the organic phase. The organic phase was removed and stored, while the aqueous phase was washed with a further 10 ml organic solvent and 4 ml aqueous solvent. The organic phase was removed and combined with the previous one, then washed 3 times with 10 ml aqueous solvent. The final organic fraction was dried under vacuum, resuspended in 1 μl DMSO, dried under vacuum again, and resuspended in 100 mL DMSO. To confirm the presence of lipid II, a 1 mL aliquot was delipidated by hydrolysis in 10 mM ammonium acetate pH 4.2 with boiling for 30 min, before being dried under vacuum and resuspended in 15 μL water for MS analysis. 50 nM of purified GFP-SpoVID501−575, GFP-SpoVID501-545, or GFP were incubated for 30 min at 37 °C in presence of 2-fold decreasing concentrations of purified Lipid II in 100ul reaction buffer [50 mM Sodium Acetate at pH 5.2, 100 mM NaCl, 0.02% Tween 20, 2% DMSO]. Samples were then centrifugated at 15,000 × g for 2 minutes at room temperature. The pellet and the supernatant were separated and prepared for immunoblotting as described. Quantification of the bound fraction (pellet) was determined using Fiji software.
Peptidoglycan accumulation
Strains KR394, JB171, JB281, JB103, JB280, JB295 were induced to sporulate by resuspension. Cells were harvested at t = 5.5 h and washed three times in 2 ml ice cold 0.9% NaCl. The final pellets were resuspended in 100 μL of 0.9% NaCl and boiled for 5 minutes to lyse the cells and extract Park’s nucleotide, before centrifugation at 21,000 × g for 5 min to pellet the insoluble material. The resulting supernatant was filtered through a 0.22 mm pore-size filter and used for LC-MS analysis. Detection and quantification of Park’s nucleotide was performed on a UPLC system (Waters) equipped with an ACQUITY UPLC BEH C18 column (130Å pore size, 1.7 μm particle size, 2.1 mm x 150 mm, Waters) coupled to a Xevo G2-XS QTOF mass spectrometer (Waters). Chromatographic separation of the soluble fraction was achieved using a linear gradient from 0.1% formic acid in water to 0.1% formic acid in acetonitrile over 18 minutes at 45 °C. The QTOF instrument was operated in positive ionisation mode and detection of UDP-M5 was performed in the untargeted MSe mode. The MS parameters were set as follows: capillary voltage 3 kV, source temperature 120 C, desolvation temperature 350 C, sample cone voltage 40 V, cone gas flow 100 L h−1 and desolvation gas flow 500 L h-1. Data acquisition and processing was performed using the UNIFI software (Waters). To quantify the Park’s nucleotide, its calculated [M+2H]2+ ion of m/z 597.68 was extracted from the total ion chromatogram, and the corresponding peak in the resulting extracted ion chromatogram was integrated to give a peak area
GFP-SpoVID501−575 copurification assay
4 μM of purified SpoIVA were incubated in 120 μL buffer B (50 mM Tris-HCl pH 7.5, 400 mM NaCl) in the presence or absence of 4 mM ATP and 5mM MgCl2 and incubated at 37 °C for 4 h. 20 μl were removed and saved for analysis (T, total). 500 nM final concentration of purified GFP-SpoVID501−575 was added to the remaining 100 μL of each sample and incubated at room temperature for 15 min. 20 μl were removed and saved for analysis (L, load). The remaining 80 μL were incubated with Ni-NTA agarose beads for 5 min at room temperature and centrifuged to separate unbound material, of which 20 μl were collected (FT, flow through) for analysis. Beads were washed twice (W1-W2) with 80 μl of buffer B and eluted with 80 μl of buffer B containing 250 mM imidazole after incubating for 2 min at room temperature. Samples were analyzed by immunoblotting using anti-SpoIVA and anti-GFP antiserum.
Supplementary Material
KEY RESOURCES TABLE
| Bacillus subtilis strains used in this study | ||
|---|---|---|
|
| ||
| Strain | Genotype or description | Reference |
|
| ||
| PY79 | Prototrophic derivative of B. subtilis 168 | Youngman et al. (1984) |
| JB103 | ΔspoIVA::neo thrC::spoIVAD97A spec | This study |
| JB168 | ΔspoIVA::erm thrC::spoIVAT70A,T71A spec ΔspoVID::neo | This study |
| JB171 | ΔspoIVA::erm DspoVID::neo thrC::spoIVA spec | This study |
| JB174 | ΔspoIVA::erm DspoVID::neo thrC::spoIVA spec amyE::spoVID cat | This study |
| JB175 |
ΔspoIVA::erm thrC::spoIVA spec ΔspoVID::neo amyE::spoVID1−545 cat |
This study |
| JB176 | ΔspoIVA::erm ΔspoVID::neo thrC::spoIVAT70A,T71A spec amyE::spoVID cat | This study |
| JB177 |
ΔspoIVA::erm ΔspoVID::neo thrC::spoIVAT70A,T71A
spec amyE:: spoVID1−545 cat |
This study |
| JB217 | amyE::gfp-spoVID cat | This study |
| JB242 |
amyE::spoVIDT532G
cat ΔspoIVA::erm ΔspoVID::neo thrC:: spoIVAT70A,T71A spec |
This study |
| JB280 |
ΔspoIVA::erm thrC:: spoIVAD97A
spec ΔspoVID::neo |
This study |
| JB281 |
amyE::spoVIDT532G
cat ΔspoIVA::erm ΔspoVID::neo thrC::spoIVA spec |
This study |
| JB293 |
amyE::spoVID cat ΔspoIVA::erm ΔspoVID::neo thrC:: spoIVAD97A spec |
This study |
| JB294 | ΔspoIVA::erm thrC:: spoIVAD97A ΔspoVID::neo spec amyE::spoVIDQ546stop cat | This study |
| JB295 |
amyE::spoVIDT532G
cat ΔspoIVA::erm ΔspoVID::neo thrC::spoIVAD97A spec |
This study |
| JPC156 | ΔspoIVA::neo amyE::spoIVA cat thrC::GFPspoIVA spec | Castaing et al. (2013) |
| JPC221 | ΔspoIVA::neo thrC:: spoIVAT70A,T71A spec | Castaing et al. (2013) |
| JPC243 | ΔspoIVA::neo amyE:: spoIVAT70A,T71A cat thrC::GFP-spoIVAT70A,T71A spec | Castaing et al. (2013) |
| KP73 | ΔspoIVA::kan | Price and Losick (1999) |
| KR394 | ΔspoIVA::neo thrC::spoIVA spec | Ramamurthi and Losick (2008) |
| TD100 | ΔspoVID::neo | This study |
| TD1000 | ΔspoIVA::erm thrC::spoIVA spec ΔspoVID::neo amyE::GFPspoVID514−575 cat | This study |
| TD1001 | ΔspoIVA::erm thrC::spoIVA spec ΔspoVID::neo amyE::GFPspoVID515−575 cat | This study |
| TD1002 | ΔspoIVA::erm thrC::spoIVA spec ΔspoVID::neo amyE::GFPspoVID564−575 cat | This study |
| TD1029 | amyE::GFP-spoVID501−575 cat | This study |
| TD1030 | amyE:: GFP-spoVID501-575(T532G) cat | This study |
| TD1031 | amyE::GFP-spoVID511−575 cat | This study |
| TD1032 | amyE:: GFP-spoVID511−575(T532G) cat | This study |
| TD1056 |
ΔspoIVA::erm ΔspoVID::neo thrC:: spoIVAD97A spec amyE::PspoVID-FLAG-GFP cat |
This study |
| TD1070 | ΔspoVID::neo amyE::GFPspoVID501−575 cat | This study |
| TD1074 | ΔspoIVA::erm thrC::spoIVA spec ΔspoVID::neo amyE::gfp-spoVID cat pyrD::mcherry-spoIVA cat::tet | This study |
| TD1085 | ΔspoVID::neo sacA::GFP- spoVID501−575 cat::tet | This study |
| TD1086 | ΔspoVID::neo amyE:: GFP- spoVID501−575 cat sacA::GFP- spoVID501−575 cat::tet | This study |
| TD1139 | ΔspoIVA::neo thrC::spoIVA spec amyE::spoVIDT532G cat | This study |
| TD1140 | ΔspoIVA::erm thrC::spoIVAT70A,T71A spec amyE::spoVIDT532G cat | This study |
| TD1141 | ΔspoIVA::erm thrC::spoIVAD97A spec amyE::spoVIDT532G cat | This study |
| TD139 |
ΔspoIVA::erm thrC::spoIVA spec
ΔspoVID::neo amyE::gfp-spoVID cat |
This study |
| TD142 |
ΔspoIVA::erm thrC::spoIVAD97A
spec ΔspoVID::neo amyE::gfp-spoVID cat |
This study |
| TD144 |
ΔspoIVA::erm thrC::spoIVAT70A,T71A
spec ΔspoVID::neo amyE::gfp-spoVID cat |
This study |
| TD154 | ΔspoIVA::erm thrC::spoIVA spec ΔspoVID::neo amyE::gfpspoVIDQ546Stop cat | This study |
| TD172 | ΔspoIVA::erm ΔspoVID::neo amyE::gfpspoVID cat | This study |
| TD251 |
ΔspoIVA::erm thrC::spoIVA spec ΔspoVID::neo amyE::gfp-spoVIDT532G cat |
This study |
| TD300 |
ΔspoIVA::neo amyE::spoIVA cat
ΔspoVID::erm thrC::GFP-spoIVA spec |
This study |
| TD302 | ΔspoIVA::neo amyE:: spoIVAT70A,T71A cat ΔspoVID::erm thrC::GFPspoIVAT70A,T71A spec | This study |
| TD931 |
ΔspoIVA::erm thrC::spoIVA spec
ΔspoVID::neo amyE::GFP-spoVID 501−575 |
This study |
| TD933 |
ΔspoIVA::erm thrC:: spoIVAD97A
spec ΔspoVID::neo amyE::gfp-spoVID501−575 |
This study |
| TD953 |
ΔspoIVA::erm DspoVID::neo thrC:: spoIVAD97A spec amyE::gfpspoVIDT532G cat |
This study |
| TD954 |
ΔspoIVA::erm thrC:: spoIVAT70A,T71A
spec ΔspoVID::neo amyE::gfp- spoVIDT532G cat |
This study |
| TD955 | ΔspoIVA::erm DspoVID::neo thrC::spoIVA spec amyE::gfp-spoVID501−575(T532G) cat | This study |
| TD957 | ΔspoIVA::erm ΔspoVID::neo thrC::spoIVAD97A spec amyE::gfpspoVID501–575(T532G) cat | This study |
| TD961 |
ΔspoIVA::erm thrC::spoIVAD97A
spec ΔspoVID::neo amyE::gfp-spoVID511−575 |
This study |
| TD962 |
ΔspoIVA::erm thrC::spoIVAD97A
spec ΔspoVID::neo amyE::gfp-spoVID521−575 |
This study |
| TD979 |
ΔspoIVA::erm thrC:: spoIVAD97A
spec ΔspoVID::neo amyE::gfp-spoVID512−575 |
This study |
| TD980 |
ΔspoIVA::erm thrC:: spoIVAD97A
spec ΔspoVID::neo amyE::gfp-spoVID513−575 |
This study |
| TD981 |
ΔspoIVA::erm thrC:: spoIVAD97A
spec ΔspoVID::neo amyE::gfp-spoVID514−575 |
This study |
| TD982 |
ΔspoIVA::erm thrC:: spoIVAD97A
spec ΔspoVID::neo amyE::gfp-spoVID515−575 |
This study |
| TD983 |
ΔspoIVA::erm thrC:: spoIVAD97A
spec ΔspoVID::neo amyE:: gfp-spoVID516−575 |
This study |
| TD996 |
ΔspoIVA::erm thrC::spoIVA spec
ΔspoVID::neo amyE::gfp-spoVID 511−575 |
This study |
| TD997 |
ΔspoIVA::erm thrC::spoIVA spec
ΔspoVID::neo amyE::gfp-spoVID 521−575 |
This study |
| TD998 |
ΔspoIVA::erm thrC::spoIVA spec
ΔspoVID::neo amyE::gfp-spoVID 512−575 |
This study |
| TD999 |
ΔspoIVA::erm thrC::spoIVA spec
ΔspoVID::neo amyE::gfp-spoVID 513−575 |
This study |
|
| ||
| Escherichia coli strains used in this study | ||
|
| ||
| Strain | Genotype and description | Reference |
|
| ||
| BL21(DE3) | pKR168 (pET28a backbone, PT7-GFP-His6) | Ramamurthi and Losick (2008) |
| BL21(DE3) |
pTD352 (pET28a backbone, P
T7
-GFP-
SpoVID501–575-His6) |
This study |
| BL21(DE3) | pTD354 (pET28a backbone, P T7 -GFPspoVID 501−575 -FLAG) | This study |
| BL21(DE3) | pTD436 (pET28a backbone, P T7 - GFPspoVID501–545-His6) | This study |
|
| ||
| Item | Source | Catalog number |
|
| ||
| Antisera | Purified antigen used | |
|
| ||
| Rabbit anti-GFP | Ramamurthi lab; Covance/LabCorp | Purified GFP-His6 |
| Rabbit anti-SpoIVA | Ramamurthi lab; Covance/LabCorp | Purified His6-SpoIVA |
| Rabbit anti-SpoVID | Ramamurthi lab; Covance/LabCorp | Purified His6-SpoVID |
|
| ||
| Microscopy | ||
|
| ||
| FM4–64 | Thermo Fisher Scientific | T13320 |
| 35 mm Dish, No. 1.5 Coverslip | Mat Tek | P35GC-1.5–14-C |
|
| ||
| Protein purification and detection | ||
|
| ||
| Ni-NTA agarose beads | Qiagen | 30250 |
| Pierce Anti-DYKDDDDK Magnetic Agarose | Thermo Fisher Scientific | A36798 |
| QuikChange Lightning Site-Directed Mutagenesis Kit |
Agilent | 210519 |
| StarBright Blue 700 Goat Anti-Rabbit IgG | BioRad | 12004161 |
Highlights.
During sporulation, cortex assembly depends on successful coat assembly initiation
SpoVID directly measures the successful polymerization of the coat basement layer
SpoVID harbors a cytosolic LysM domain that binds lipid II to halt cortex assembly
The SpoVID checkpoint ensures orchestrated assembly of the spore envelope
ACKNOWLEDGMENTS
We thank members of KSR lab for suggestions and comments on the manuscript; S. Gottesman, S. Wickner, G. Storz, M. Maurizi, A. Khare, and A. Kelly for discussions; I.-L. Wu and J. Barriga for strains; K. Nagashima and Z. Wang of the Electron Microscopy Laboratory of CCR for TEM sample preparation and imaging; and the CCR Genomics Core Facility for whole genome sequencing. This work was funded by National Institutes of Health (NIH) grant R01GM138630 (D.L.P.), the Swedish Research Council, the Laboratory of Molecular Infection Medicine Sweden (MIMS), Umeå University, the Knut and Alice Wallenberg Foundation (KAW), the Kempe Foundation (F.C.), the Intramural Research Program of the NIH, the National Cancer Institute, the Center for Cancer Research (K.S.R.), and the National Library of Medicine (L.A.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.devcel.2021.12.021.
REFERENCES
- Aksyuk AA, and Rossmann MG (2011). Bacteriophage assembly. Viruses 3, 172–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, and Koonin EV (1998). Iterated profile searches with PSIBLAST–a tool for discovery in protein databases. Trends Biochem. Sci. 23, 444–447. [DOI] [PubMed] [Google Scholar]
- Alvarez L, Hernandez SB, de Pedro MA, and Cava F. (2016). Ultra-sensitive, high-resolution liquid chromatography methods for the high-throughput quantitative analysis of bacterial cell wall chemistry and structure. Methods Mol. Biol. 1440, 11–27. [DOI] [PubMed] [Google Scholar]
- Bateman A, and Bycroft M. (2000). The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J. Mol. Biol. 299, 1113–1119. [DOI] [PubMed] [Google Scholar]
- Beall B, Driks A, Losick R, and Moran CP Jr. (1993). Cloning and characterization of a gene required for assembly of the Bacillus subtilis spore coat. J. Bacteriol. 175, 1705–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano-Sagie M, Oppenheimer-Shaanan Y, Berlatzky I, Rouvinski A, Meyerovich M, and Ben-Yehuda S. (2006). A checkpoint protein that scans the chromosome for damage at the start of sporulation in Bacillus subtilis. Cell 125, 679–690. [DOI] [PubMed] [Google Scholar]
- Benito de la Puebla H, Giacalone D, Cooper A, and Shen A. (2020). Role of SpoIVA ATPase motifs during Clostridioides difficile sporulation. J. Bacteriol.202, e00387–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buist G, Steen A, Kok J, and Kuipers OP (2008). LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol. Microbiol. 68, 838–847. [DOI] [PubMed] [Google Scholar]
- Burkholder WF, Kurtser I, and Grossman AD (2001). Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell 104, 269–279. [DOI] [PubMed] [Google Scholar]
- Burroughs AM, Ando Y, and Aravind L. (2014). New perspectives on the diversification of the RNA interference system: insights from comparative genomics and small RNA sequencing. Wiley Interdiscip. Rev. RNA 5, 141–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaing JP, Nagy A, Anantharaman V, Aravind L, and Ramamurthi KS (2013). ATP hydrolysis by a domain related to translation factor GTPases drives polymerization of a static bacterial morphogenetic protein. Proc. Natl.Acad. Sci. USA 110, E151–E160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilcott GS, and Hughes KT (2000). Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64, 694–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C, Barber JD, and Barton GJ (2008). The Jpred 3 secondary structure prediction server. Nucleic Acids Res 36, W197–W201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coote JG (1972). Sporulation in Bacillus subtilis. Characterization of oligosporogenous mutants and comparison of their phenotypes with those of asporogenous mutants. J. Gen. Microbiol. 71, 1–15. [DOI] [PubMed] [Google Scholar]
- Costa T, Isidro AL, Moran CP Jr., and Henriques AO (2006). Interaction between coat morphogenetic proteins SafA and SpoVID. J. Bacteriol. 188, 7731–7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuff JA, and Barton GJ (2000). Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins 40, 502–511. [DOI] [PubMed] [Google Scholar]
- de Francesco M, Jacobs JZ, Nunes F, Serrano M, McKenney PT, Chua MH, Henriques AO, and Eichenberger P. (2012). Physical interaction between coat morphogenetic proteins SpoVID and CotE is necessary for spore encasement in Bacillus subtilis. J. Bacteriol. 194, 4941–4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker AR, and Ramamurthi KS (2017). Cell death pathway that monitors spore morphogenesis. Trends Microbiol 25, 637–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driks A, and Eichenberger P. (2016). The spore coat. Microbiol. Spectr. 4. [DOI] [PubMed] [Google Scholar]
- Drozdetskiy A, Cole C, Procter J, and Barton GJ (2015). JPred4: A protein secondary structure prediction server. Nucleic Acids Res 43, W389–W394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducret A, and Grangeasse C. (2021). Recent progress in our understanding of peptidoglycan assembly in Firmicutes. Curr. Opin. Microbiol. 60, 44–50. [DOI] [PubMed] [Google Scholar]
- Dworkin J, and Losick R. (2005). Developmental commitment in a bacterium.Cell 121, 401–409. [DOI] [PubMed] [Google Scholar]
- Ebmeier SE, Tan IS, Clapham KR, and Ramamurthi KS (2012). Small proteins link coat and cortex assembly during sporulation in Bacillus subtilis.Mol. Microbiol. 84, 682–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert JK, Kim YJ, Kim JI, Gu€rtler K, Oh DY, Sur S, Lundvall L, Hamann L, van der Ploeg A, Pickkers P, et al. (2013). The crystal structure of lipopolysaccharide binding protein reveals the location of a frequent mutation that impairs innate immunity. Immunity 39, 647–660. [DOI] [PubMed] [Google Scholar]
- Edgar RC (2004). MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswara PJ, Brzozowski RS, Viola MG, Graham G, Spanoudis C, Trebino C, Jha J, Aubee JI, Thompson KM, Camberg JL, et al. (2018). An essential Staphylococcus aureus cell division protein directly regulates FtsZ dynamics. eLife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Gonzá lez-Pastor JE, and Losick R. (2005). High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J. Bacteriol. 187, 1357–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima T, Tanabe T, Yamamoto H, Hosoya S, Sato T, Yoshikawa H, and Sekiguchi J. (2004). Characterization of a polysaccharide deacetylase gene homologue (pdaB) on sporulation of Bacillus subtilis. J. Biochem. 136, 283–291. [DOI] [PubMed] [Google Scholar]
- Galperin MY, Mekhedov SL, Puigbo P, Smirnov S, Wolf YI, andRigden DJ (2012). Genomic determinants of sporulation in Bacilli and Clostridia: Towards the minimal set of sporulation-specific genes. Environ.Microbiol. 14, 2870–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford SM, and Meyer P. (2015). Enzyme function is regulated by its localization. Comput. Biol. Chem. 59, 113–122. [DOI] [PubMed] [Google Scholar]
- Gill RL Jr., Castaing JP, Hsin J, Tan IS, Wang X, Huang KC, Tian F, and Ramamurthi KS (2015). Structural basis for the geometry-driven localization of a small protein. Proc. Natl. Acad. Sci. U. S. A. 112, E1908–E1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gué rout-Fleury AM, Frandsen N, and Stragier P. (1996). Plasmids for ectopic integration in Bacillus subtilis. Gene 180, 57–61. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, and Weinert TA (1989). Checkpoints: Controls that ensure the order of cell cycle events. Science 246, 629–634. [DOI] [PubMed] [Google Scholar]
- Henriques AO, and Moran CP Jr. (2007). Structure, assembly, and function of the spore surface layers. Annu. Rev. Microbiol. 61, 555–588. [DOI] [PubMed] [Google Scholar]
- Higgins D, and Dworkin J. (2012). Recent progress in Bacillus subtilis sporulation. FEMS Microbiol. Rev. 36, 131–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imae Y, and Strominger JL (1976). Cortex content of asporogenous mutants of Bacillus subtilis. J. Bacteriol. 126, 914–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP, and Kukreja AA (2017). How kinetochore architecture shapes the mechanisms of its function. Curr. Biol. 27, R816–R824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelling J, Sullivan K, Wilson L, and Jordan MA (2003). Suppression of centromere dynamics by taxol in living osteosarcoma cells. Cancer Res 63, 2794–2801. [PubMed] [Google Scholar]
- Khodjakov A, and Rieder CL (2009). The nature of cell-cycle checkpoints:Facts and fallacies. J. Biol. 8, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EY, Tyndall ER, Huang KC, Tian F, and Ramamurthi KS (2017). Dash-and-recruit mechanism drives membrane curvature recognition by the small bacterial protein SpoVM. Cell Syst 5, 518–526, e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassmann T, Frings O, and Sonnhammer EL (2009). Kalign2: High-performance multiple alignment of protein and nucleotide sequences allowing external features. Nucleic Acids Res 37, 858–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassmann T, and Sonnhammer EL (2005). Kalign–An accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics 6, 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe DD, Koonin EV, and Aravind L. (2003). Evolution and classification of P-loop kinases and related proteins. J. Mol. Biol. 333, 781–815. [DOI] [PubMed] [Google Scholar]
- Levin PA, Fan N, Ricca E, Driks A, Losick R, and Cutting S. (1993). An unusually small gene required for sporulation by Bacillus subtilis. Mol.Microbiol. 9, 761–771. [DOI] [PubMed] [Google Scholar]
- Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Scha TF, Hughes DE, Epstein S, et al. (2015). A new antibiotic kills pathogens without detectable resistance. Nature 517, 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long AF, Kuhn J, and Dumont S. (2019). The mammalian kinetochoremicrotubule interface: Robust mechanics and computation with many microtubules. Curr. Opin. Cell Biol. 60, 60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masser EA, Burby PE, Hawkins WD, Gustafson BR, Lenhart JS, and Simmons LA (2021). DNA damage checkpoint activation affects peptidoglycan synthesis and late divisome components in Bacillus subtilis. Mol.Microbiol. 116, 707–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney PT, Driks A, and Eichenberger P. (2013). The Bacillus subtilis endospore: Assembly and functions of the multilayered coat. Nat. Rev.Microbiol. 11, 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney PT, and Eichenberger P. (2012). Dynamics of spore coat morphogenesis in Bacillus subtilis. Mol. Microbiol. 83, 245–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeske AJ, Sham LT, Kimsey H, Koo BM, Gross CA, Bernhardt TG, and Rudner DZ (2015). MurJ and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 112, 6437–6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnage S, Dellarole M, Baxter NJ, Rouget JB, Dimitrov JD, Wang N, Fujimoto Y, Hounslow AM, Lacroix-Desmazes S, Fukase K, et al. (2014). Molecular basis for bacterial peptidoglycan recognition by LysM domains. Nat. Commun. 5, 4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton R, and Hofmeister A. (2004). New shuttle vectors for ectopic insertion of genes into Bacillus subtilis. Plasmid 51, 238–245. [DOI] [PubMed] [Google Scholar]
- Mu€llerova D, Krajcí´kova D, and Bara k, I. (2009). Interactions between Bacillus subtilis early spore coat morphogenetic proteins. FEMS Microbiol.Lett. 299, 74–85. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. (1996). Viewpoint: Putting the cell cycle in order. Science 274, 1643–1645. [DOI] [PubMed] [Google Scholar]
- Nunes F, Fernandes C, Freitas C, Marini E, Serrano M, Moran CP Jr., Eichenberger P, and Henriques AO (2018). SpoVID functions as a noncompetitive hub that connects the modules for assembly of the inner and outer spore coat layers in Bacillus subtilis. Mol. Microbiol. 110, 576–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozin AJ, Henriques AO, Yi H, and Moran CP Jr. (2000). Morphogenetic proteins SpoVID and SafA form a complex during assembly of the Bacillus subtilis spore coat. J. Bacteriol. 182, 1828–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozin AJ, Samford CS, Henriques AO, and Moran CP Jr. (2001). SpoVID guides SafA to the spore coat in Bacillus subtilis. J. Bacteriol. 183, 3041–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluso EA, Updegrove TB, Chen J, Shroff H, and Ramamurthi KS (2019). A 2-dimensional ratchet model describes assembly initiation of a specialized bacterial cell surface. Proc. Natl. Acad. Sci. U. S. A. 116, 21789–21799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira FC, Nunes F, Cruz F, Fernandes C, Isidro AL, Lousa D, Soares CM, Moran CP Jr., Henriques AO, and Serrano M. (2019). A LysM domain intervenes in sequential protein-protein and protein-peptidoglycan interactions important for spore coat assembly in Bacillus subtilis. J. Bacteriol. 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepenbreier H, Diehl A, and Fritz G. (2019). Minimal exposure of lipid II cycle intermediates triggers cell wall antibiotic resistance. Nat. Commun.10, 2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot PJ, and Coote JG (1976). Genetic aspects of bacterial endospore formation. Bacteriol. Rev. 40, 908–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Aravind L, Schultz J, Bork P, and Koonin EV (1999). Eukaryotic signalling domain homologues in archaea and bacteria. Ancient ancestry and horizontal gene transfer. J. Mol. Biol. 289, 729–745. [DOI] [PubMed] [Google Scholar]
- Popham DL, and Bernhards CB (2015). Spore peptidoglycan. Microbiol. Spectr. 3, TBS-0005–2012. [DOI] [PubMed] [Google Scholar]
- Price KD, and Losick R. (1999). A four-dimensional view of assembly of a morphogenetic protein during sporulation in Bacillus subtilis. J. Bacteriol.181, 781–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam EE, Nock AM, Lawley TD, and Shen A. (2013). SpoIVA and SipL are Clostridium difficile spore morphogenetic proteins. J. Bacteriol. 195, 1214–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Krajcikova D, Xing C, Lu B, Hao J, Ke X, Wang H, Barak I, and Tang J. (2013). Study of the interactions between the key spore coat morphogenetic proteins CotE and SpoVID. J. Struct. Biol. 181, 128–135. [DOI] [PubMed] [Google Scholar]
- Qiao Y, Srisuknimit V, Rubino F, Schaefer K, Ruiz N, Walker S, and Kahne D. (2017). Lipid II overproduction allows direct assay of transpeptidase inhibition by beta-lactams. Nat. Chem. Biol. 13, 793–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahn-Lee L, Gorbatyuk B, Skovgaard O, and Losick R. (2009). The conserved sporulation protein YneE inhibits DNA replication in Bacillus subtilis. J. Bacteriol. 191, 3736–3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi KS, Clapham KR, and Losick R. (2006). Peptide anchoring spore coat assembly to the outer forespore membrane in Bacillus subtilis.Mol. Microbiol. 62, 1547–1557. [DOI] [PubMed] [Google Scholar]
- Ramamurthi KS, Lecuyer S, Stone HA, and Losick R. (2009). Geometric cue for protein localization in a bacterium. Science 323, 1354–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi KS, and Losick R. (2008). ATP-driven self-assembly of a morphogenetic protein in Bacillus subtilis. Mol. Cell 31, 406–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Guadiana FH, Meeske AJ, Wang X, Rodrigues CDA, and Rudner DZ (2017). The Bacillus subtilis germinant receptor GerA triggers premature germination in response to morphological defects during sporulation.Mol. Microbiol. 105, 689–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley EP, Schwarz C, Derman AI, and Lopez-Garrido J. (2020).Milestones in Bacillus subtilis sporulation research. Microb. Cell 8, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roels S, Driks A, and Losick R. (1992). Characterization of spoIVA, a sporulation gene involved in coat morphogenesis in Bacillus subtilis. J. Bacteriol.174, 575–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon ED, and Bloom K. (2017). Tension sensors reveal how the kinetochore shares its load. BioEssays 39, 1600216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehnal D, Bittrich S, Deshpande M, Svobodova R, Berka K, Bazgier V, Velankar S, Burley SK, Koca J, and Rose AS (2021). Mol* Viewer: modern web app for 3D visualization and analysis of large biomolecular structures.Nucleic Acids Res 49, W431–W437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow P. (2014). Spore resistance properties. Microbiol. Spectr. 2, 2–5. [DOI] [PubMed] [Google Scholar]
- Shen A, Edwards AN, Sarker MR, and Paredes-Sabja D. (2019). Sporulation and germination in clostridial pathogens. Microbiol. Spectr. 7. 10.1128/microbiolspec.GPP3-0017-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sö ding J. (2005). Protein homology detection by HMM-HMM comparison.Bioinformatics 21, 951–960. [DOI] [PubMed] [Google Scholar]
- Sö ding J, Biegert A, and Lupas AN (2005). The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33, W244–W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterlini JM, and Mandelstam J. (1969). Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem. J. 113, 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens CM, Daniel R, Illing N, and Errington J. (1992). Characterization of a sporulation gene, spoIVA, involved in spore coat morphogenesis in Bacillus subtilis. J. Bacteriol. 174, 586–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan IS, and Ramamurthi KS (2014). Spore formation in Bacillus subtilis.Environ. Microbiol. Rep. 6, 212–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan IS, Weiss CA, Popham DL, and Ramamurthi KS (2015). A qualitycontrol mechanism removes unfit cells from a population of sporulating bacteria. Dev. Cell 34, 682–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchette MH, Benito de la Puebla H, Ravichandran, and Shen A. (2019). SpoIVA-SipL complex formation is essential for Clostridioides difficile spore assembly. J. Bacteriol. 201, e00042–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegrove TB, Harke J, Anantharaman V, Yang J, Gopalan N, Wu D, Piszczek G, Stevenson DM, Amador-Noguez D, Wang JD, et al. (2021). Reformulation of an extant ATPase active site to mimic ancestral GTPase activity reveals a nucleotide base requirement for function. eLife 10, e65845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan P, Weaver A, Reichert ED, Linnstaedt SD, and Popham DL (2007). Spore cortex formation in Bacillus subtilis is regulated by accumulation of peptidoglycan precursors under the control of sigma K. Mol. Microbiol. 65, 1582–1594. [DOI] [PubMed] [Google Scholar]
- Veening JW, Murray H, and Errington J. (2009). A mechanism for cell cycle regulation of sporulation initiation in Bacillus subtilis. Genes Dev 23, 1959–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visweswaran GR, Dijkstra BW, and Kok J. (2011). Murein and pseudomurein cell wall binding domains of bacteria and archaea–a comparative view.Appl. Microbiol. Biotechnol. 92, 921–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JK, Marquis KA, and Rudner DZ (2009). SirA enforces diploidy by inhibiting the replication initiator DnaA during spore formation in Bacillus subtilis. Mol. Microbiol. 73, 963–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KH, Isidro AL, Domingues L, Eskandarian HA, McKenney PT, Drew K, Grabowski P, Chua MH, Barry SN, Guan M, et al. (2009). The coat morphogenetic protein SpoVID is necessary for spore encasement in Bacillus subtilis. Mol. Microbiol. 74, 634–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T, and Simko V. (2021). R package ‘corrplot’: Visualization of a Correlation Matrix. (Version 0.92), https://github.com/taiyun/corrplot.
- Wong JE, Midtgaard SR, Gysel K, Thygesen MB, Sørensen KK, Jensen KJ, Stougaard J, Thirup S, and Blaise M. (2015). An intermolecular binding mechanism involving multiple LysM domains mediates carbohydrate recognition by an endopeptidase. Acta Crystallogr. D Biol. Crystallogr. 71, 592–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu IL, Narayan K, Castaing JP, Tian F, Subramaniam S, and Ramamurthi KS (2015). A versatile nano display platform from bacterial spore coat proteins. Nat. Commun. 6, 6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. (2019). Distance-based protein folding powered by deep learning. Proc.Natl. Acad. Sci. U. S. A. 116, 16856–16865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Miyake Y, Hisaoka M, Kurosawa S, and Sekiguchi J. (2008). The major and minor wall teichoic acids prevent the sidewall localization of vegetative DL-endopeptidase LytF in Bacillus subtilis. Mol. Microbiol.70, 297–310. [DOI] [PubMed] [Google Scholar]
- Youngman P, Perkins JB, and Losick R. (1984). Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposonborne erm gene. Plasmid 12, 1–9. [DOI] [PubMed] [Google Scholar]
- Zheng LB, Donovan WP, Fitz-James PC, and Losick R. (1988). Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Dev 2, 1047–1054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microscopy data can be shared upon request.
No original code was developed in this study.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.