Abstract
When exposed to desiccation stress, extremotolerant organisms from all domains of life produce protective disordered proteins with the potential to inform the design of excipients for formulating biologics and industrial enzymes. However, the mechanism(s) of desiccation protection remain largely unknown. To investigate the role of water sorption in desiccation protection, we use thermogravimetric analysis to study water adsorption by two desiccation‐tolerance proteins, cytosolic abundant heat soluble protein D from tardigrades and late embryogenesis abundant protein 4 from the anhydrobiotic midge Polypedilum vanderplanki, and, as a control, the globular B1 domain of staphylococcal protein G. All samples adsorb similar amounts of water, suggesting that modulated water retention is not responsible for dehydration protection by desiccation‐tolerance proteins.
Keywords: desiccation, disordered proteins, tardigrades, thermogravimetric analysis
1. INTRODUCTION
Water is essential to biology, 1 , 2 but selected organisms from all domains of life can survive anhydrobiosis, a state in which cellular water is less than 5% w/w compared to typical values of 60–70%. 3 Although the mechanism(s) of anhydrobiosis remain largely unknown, many resistant organisms express small, disordered proteins upon desiccation. 4 , 5
Drying protein‐based drugs (biologics) and industrial enzymes can increase their stability and thereby avoid challenges associated with refrigerated transport and storage (the so‐called cold chain) that hinder their use. 6 , 7 , 8 Unfortunately, many proteins do not withstand dehydration because of water's crucial role in globular protein structure and function. 9 , 10 Proteins can be formulated with protective molecules called excipients prior to drying, 8 , 11 but the process of choosing an effective excipient is empirical, and formulation often fails because we know so little about dehydration protection. 8 , 11 , 12
Uncovering the mechanisms by which desiccation‐tolerance molecules protect proteins will advance our understanding of anhydrobiosis and facilitate the logical choice and design of excipients, making enzymes and life‐saving biologics more affordable and accessible. 13
Hypotheses about the mechanism of protein dehydration protection have been proposed. In the preferential hydration hypothesis, a hydration layer is maintained by protectants that trap or crowd water at the hydrophilic surface of client proteins. 14 Using this model, one might predict that desiccation‐tolerance molecules are more hygroscopic than those unrelated to desiccation tolerance, holding on to water that then interacts with the client protein surface.
In the water replacement hypothesis, protectants provide H‐bonds to client proteins that are usually made by water. 12 , 15 Furthermore, water can accelerate the chemical degradation of proteins such that desiccation protection might involve inhibiting water's adverse effects. 12 , 17 , 18 These ideas suggest that desiccation‐tolerance molecules limit degradation by replacing water, which means they perhaps bind less water than molecules unrelated to desiccation tolerance. In summary, the amount of retained water by desiccation‐tolerance proteins might be skewed in either direction compared to proteins not related to desiccation tolerance.
Here, we use thermogravimetric analysis (TGA) to quantify water sorption by two desiccation‐tolerance proteins, one from a tardigrade and another from a midge. Tardigrades are microscopic animals many species of which survive desiccation and/or other extreme stresses. 19 , 20 , 21 Cytosolic abundant heat soluble (CAHS) proteins are unique to tardigrades, necessary for their desiccation survival, intrinsically disordered, and protect heterologously expressing cells and enzymes from desiccation damage. 22 , 23 , 24 They also form reversible, concentration‐dependent hydrogels. 25 , 26 We tested CAHS D from the tardigrade Hypsibius exemplaris. The midge Polypedilum vanderplanki survives multiple cycles of complete dehydration and accumulates late embryogenesis abundant (LEA) proteins upon desiccation. 27 LEA proteins, also disordered, are involved in desiccation tolerance of many plants and animals, and inhibit protein aggregation. 27 , 28 , 29 , 30 We tested LEA protein 4 from P. vanderplanki (PvLEA4). 27 As a control we tested a globular protein not implicated in desiccation tolerance, the B1 domain of staphylococcal protein G (GB1). 31
2. RESULTS
To determine if the mechanism of desiccation protection is related to water adsorption, we performed TGA on lyophilized samples of GB1, CAHS D, and PvLEA4. Immediately after 24 hr of lyophilization, samples possess 10–11% water by mass (Figure 1), which is less than one surface layer of water for GB1 (Table S1). Upon exposure to 75% relative humidity, the water content sharply increases before plateauing by 24 hr. PvLEA4 samples plateau with a water content ~19%, CAHS D ~16%, and GB1 ~18%, which amounts to slightly more than one layer on the surface of GB1, similar to other results (Table S1). 32 , 33 From 2 hr on, the pattern from highest to lowest water content is PvLEA4, GB1, CAHS D (Figure 1). However, the water content of PvLEA4 and CAHS D samples is within the uncertainty for GB1 at each timepoint except 4 hr, where the water content of PvLEA4 is higher than that of GB1 (Figure 1). After exposure to 75% relative humidity for 72 h, all samples adsorb similar amounts of water, showing a total water content of ~17% (Figure 1). These results are comparable to estimations of water sorption based on the hydrophilic groups in a dry protein; the procedure of Leeder and Watt 34 , 35 predicts equilibrium water contents for our proteins of ~19–21%, not far from our experimental results (Table S2, Figure 1).
FIGURE 1.
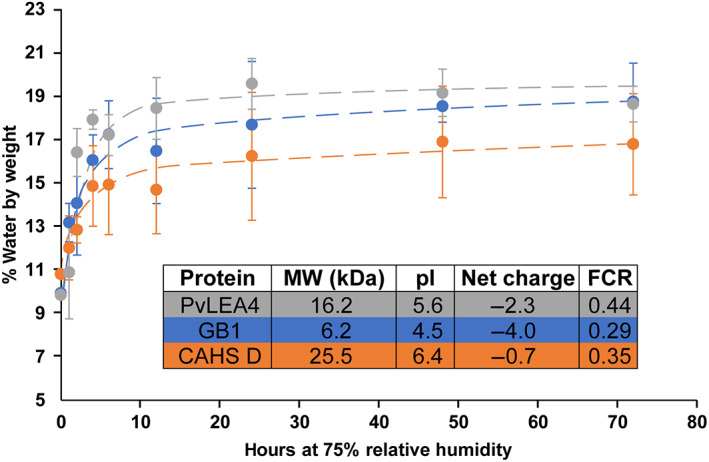
Water content, in percent weight of solid protein sample, as determined by TGA while heating at 4°C/min. Samples comprising 2 mg of GB1, CAHS D, or PvLEA4 in 650 μl 1.5 mM 4‐(2‐hydroxyethyl)‐1‐piperazineethanesulfonic acid, pH 6.5 were lyophilized for 24 hr, then exposed to 75% relative humidity at room temperature. Error bars represent the SD from three independent experiments. Molecular weight (MW), isoelectric point (pI), net charge calculated at pH 6.5, and FCRs are shown in the inset. CAHS D, cytosolic abundant heat‐soluble protein D; FCRs, fraction of charged residues; GB1, B1 domain of staphylococcal protein G; PvLEA4, LEA protein 4 from P. vanderplanki; TGA, thermogravimetric analysis
3. DISCUSSION
The observation that PvLEA4 and CAHS D adsorb neither more nor less water than GB1 (Figure 1) and other globular proteins at 75% relative humidity (e.g., lysozyme, ribonuclease, chymotrypsinogen, bovine somatotropin) 34 , 36 shows that neither elevated nor depressed sorption is a distinguishing feature of desiccation‐tolerance proteins. In the dry state, a globular protein comprises an ensemble of partially folded conformations, 32 , 37 which suggests that the vapor‐accessible surface of a dry globular protein is more comparable to that of a dry disordered protein than it is to the surface of a globular protein in solution.
Although water coordination to client proteins may be an aspect of their mechanism, simple accumulation is not sufficient to explain the function of desiccation‐tolerance proteins. Similarly, although desiccation‐tolerance proteins may replace water H‐bonds to client proteins while combating plasticizing and degradative effects, they do not adsorb less (or more) water than other proteins. We have also examined gelatin, a disordered protein unrelated to desiccation tolerance. A gelatin‐GB1 mixture adsorbs a similar amount of water as mixtures of CAHS D and GB1 or PvLEA4 and GB1. 33 In the same residue‐level study of dehydration protection, we show that water content of client protein/protectant protein mixtures does not correlate with dehydration protection. 33
These data also suggest that the ability of CAHS D to form hydrogels 25 , 26 is not explained by hygroscopicity, because CAHS D adsorbs no more water than nongelling proteins (GB1, PvLEA4, etc.).
Neither charge, fraction of charged residues, amino acid composition, nor hydrophobicity completely explain the amounts of retained water (Figure 1, Tables S3 and S4). However, PvLEA4, the protein with the highest fraction of charged residues, on average retains more water than GB1 or CAHS D. This observation suggests that more charged proteins adsorb more water, in agreement with studies showing that hydrophilic groups in a dry protein correlate with sorption capacity. 34 , 35 , 38 , 39
In summary, desiccation‐tolerance proteins adsorb water similarly to globular proteins, suggesting that modulated water retention does not explain desiccation protection. In vivo, other molecules may modulate hydration near desiccation‐tolerance proteins, but desiccation‐tolerance proteins themselves do not necessarily bind water differently than other proteins. Investigating what properties are particular to desiccation‐tolerance proteins will reveal their protective mechanism(s), allowing rational design of excipients to make protein products more affordable and accessible.
4. MATERIALS AND METHODS
4.1. Materials
Ampicillin, kanamycin sulfate, and 4‐(2‐hydroxyethyl)‐1‐piperazineethanesulfonic acid (HEPES) (Thermo Fisher) were used without further purification. H2O with a resistivity >17 MΩ cm−1 was used to prepare buffers. A constant relative humidity of 75 ± 5% as measured by a digital hygrometer (Fisherbrand TraceableGO™ Bluetooth datalogging digital hygrometer) was created by sealing a 0.5‐L chamber containing 200 ml of H2O saturated with sodium chloride (Thermo Fisher). 40
4.2. Protein expression and purification
The pET11a plasmid (Novagen) containing the gene for the T2Q variant of GB1 was provided by Leonard D. Spicer's laboratory at Duke University (Durham, North Carolina). This variant, which we call GB1, was chosen because the mutation prevents N‐terminal deamidation. 41 The pET28b plasmid containing the gene for CAHS D was engineered as described. 22 The pET28b plasmid containing the gene for PvLEA4 fused to an N‐terminal hexahistadine (His‐) tag and a TEV protease cleavage site was ordered from Gene Universal Inc. Vectors were transformed into Agilent BL21 Gold (DE3) Escherichia coli as described. 22
A single colony was used to inoculate 100 ml of Luria‐Bertani broth (Fisher, 10 g/l tryptone, 5 g/l yeast extract, 5 g/l NaCl) supplemented with the antibiotic ampicillin (GB1) or kanamycin (CAHS D and PvLEA4) to a final concentration of 60 μg/ml. The culture was shaken at 37°C overnight (New Brunswick Scientific I26 incubator, 225 rpm). Ten milliliters of the overnight culture were used to inoculate 1 l of antibiotic‐supplemented LB. One‐liter cultures were shaken at 37°C until they reached an optical density at 600 nm of 0.6–0.8, at which point protein expression was induced by adding isopropyl β‐d‐1‐thiogalactopyranoside (1 mM final concentration). Three hours after induction, cells were harvested via centrifugation at 4,000g. The cell pellet from each culture was resuspended in 10 ml of 20 mM Tris, pH 7.5, and stored at −20°C.
Cell pellets from GB1 expression were lysed by sonication (500‐W dismembrator 1/8‐in. tip, 15% amplitude [Fisher Scientific]) for 8 min using a 2 s on/1 s off duty cycle, and then GB1 was purified as described. 42 CAHS D was purified as described. 23 PvLEA4 was purified as described. 33 Purified proteins were exchanged into H2O by dialysis (ThermoScientific Snakeskin™ dialysis tubing, 3,500 Da molecular weight cutoff), and divided into 2 mg aliquots. Aliquots were flash‐frozen, lyophilized, and stored at −20°C. Purity was confirmed by observation of a single band on sodium dodecylsufate polyacrylamide gel electrophoresis and by quadrupole time‐of‐flight mass spectrometry (ThermoScientific, Q Exactive HF‐X) in the UNC Mass Spectrometry Chemical Research and Teaching Core Laboratory.
4.3. Thermogravimetric analysis
Aliquots of purified, lyophilized protein were resuspended in 650 μl of 1.5 mM HEPES buffer, pH 6.5, flash‐frozen, and lyophilized (LABCONCO FreeZone 1 Liter Benchtop Freeze Dry System) for 24 hr. Samples were then placed, without caps, in a chamber with a controlled relative humidity of 75 ± 5%, created as described above. Individual tubes were removed after 0, 1, 2, 4, 6, 12, 24, 48, and 72 hr, and protein samples were loaded into a TA Instruments model 550 thermogravimetric analyzer on an open Pt pan and heated from 25 to 175°C at a rate of 4°C/min under a N2(g) sample purge of 60 ml/min and a balance purge of 40 ml/min. The well‐defined mass loss ending around 125°C was used to quantify H2O content. 43 , 44 Thermograms were analyzed using Trios V5.1.0.56403 software.
CONFLICT OF INTEREST
The authors declare no potential conflict of interest.
AUTHOR CONTRIBUTIONS
Julia A. Brom: Conceptualization (lead); data curation (lead); formal analysis (lead); investigation (lead); methodology (lead); validation (lead); visualization (lead); writing—review and editing (lead). Gary J. Pielak: Conceptualization (lead); project administration (lead); writing—review and editing (equal).
Supporting information
Tables S1‐S4 H2O content and amino acid compositions
ACKNOWLEDGMENTS
This research was supported by NIH grant R01GM127291 to Gary J. Pielak. The authors thank Samantha Stadmiller for GB1 preparation. The authors thank Brandie Ehrmann from the UNC Chemistry Mass Spectrometry Core Laboratory for equipment maintenance and advice, Elizabeth Pielak for comments on the manuscript, and the Pielak Laboratory for helpful discussion.
Brom JA, Pielak GJ. Desiccation‐tolerance and globular proteins adsorb similar amounts of water. Protein Science. 2022;31(5):e4288. 10.1002/pro.4288
Review Editor: John Kuriyan
Funding information National Institutes of Health, Grant/Award Number: R01GM127291
REFERENCES
- 1. Ball P. Water is an active matrix of life for cell and molecular biology. Proc Natl Acad Sci USA. 2017;114:13327–13335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tanford C. The hydrophobic effect and the organization of living matter. Science. 1978;200:1012–1018. [DOI] [PubMed] [Google Scholar]
- 3. Crowe JH, Hoekstra FA, Crowe LM. Anhydrobiosis. Annu Rev Physiol. 1992;54:579–599. [DOI] [PubMed] [Google Scholar]
- 4. Boothby TC, Pielak GJ. Intrinsically disordered proteins and desiccation tolerance: Elucidating functional and mechanistic underpinnings of anhydrobiosis. Bioessays. 2017;39:1700119. [DOI] [PubMed] [Google Scholar]
- 5. Hoekstra FA, Golovina EA, Buitink J. Mechanisms of plant desiccation tolerance. Trends Plant Sci. 2001;6:431–438. [DOI] [PubMed] [Google Scholar]
- 6. Morrow T, Felcone LH. Defining the difference: What makes biologics unique. Biotechnol Healthc. 2004;1:24–29. [PMC free article] [PubMed] [Google Scholar]
- 7. Frokjaer S, Otzen DE. Protein drug stability: A formulation challenge. Nat Rev Drug Discov. 2005;4:298–306. [DOI] [PubMed] [Google Scholar]
- 8. Piszkiewicz S, Pielak GJ. Protecting enzymes from stress‐induced inactivation. Biochemistry. 2019;58:3825–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bellissent‐Funel MC, Hassanali A, Havenith M, et al. Water determines the structure and dynamics of proteins. Chem Rev. 2016;116:7673–7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang W. Lyophilization and development of solid protein pharmaceuticals. Int J Pharm. 2000;203:1–60. [DOI] [PubMed] [Google Scholar]
- 11. Bjelosevic M, Zvonar Pobirk A, Planinsek O, Ahlin Grabnar P. Excipients in freeze‐dried biopharmaceuticals: Contributions toward formulation stability and lyophilisation cycle optimisation. Int J Pharm. 2020;576:1–12. [DOI] [PubMed] [Google Scholar]
- 12. Mensink MA, Frijlink HW, van der Voort MK, Hinrichs WL. How sugars protect proteins in the solid state and during drying (review): Mechanisms of stabilization in relation to stress conditions. Eur J Pharm Biopharm. 2017;114:288–295. [DOI] [PubMed] [Google Scholar]
- 13. Hill AB, Kilgore C, McGlynn M, Jones CH. Improving global vaccine accessibility. Curr Opin Biotechnol. 2016;42:67–73. [DOI] [PubMed] [Google Scholar]
- 14. Hesgrove C, Boothby TC. The biology of tardigrade disordered proteins in extreme stress tolerance. Cell Commun Signal. 2020;18:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Crowe JH, Clegg JS, Crowe LM. Anhydrobiosis: The water replacement hypothesis. In: Reid DS, editor. The properties of water in foods ISOPOW 6. Boston, MA: Springer US, 1998; p. 440–455. [Google Scholar]
- 16. Liu WR, Langer R, Klibanov AM. Moisture‐induced aggregation of lyophilized proteins in the solid state. Biotechnol Bioeng. 1991;37:177–184. [DOI] [PubMed] [Google Scholar]
- 17. Lai MC, Topp EM. Solid‐state chemical stability of proteins and peptides. J Pharm Sci. 1999;88:489–500. [DOI] [PubMed] [Google Scholar]
- 18. Ahlneck C, Zografi G. The molecular basis of moisture effects on the physical and chemical stability of drugs in the solid state. Int J Pharm. 1990;62:87–95. [Google Scholar]
- 19. Rebecchi L, Guidetti R, Borsari S, Altiero T, Bertolani R. Dynamics of long‐term anhydrobiotic survival of lichen‐dwelling tardigrades. Hydrobiologia. 2006;558:23–30. [Google Scholar]
- 20. Jonsson KI, Rabbow E, Schill RO, Harms‐Ringdahl M, Rettberg P. Tardigrades survive exposure to space in low earth orbit. Curr Biol. 2008;18:R729–R731. [DOI] [PubMed] [Google Scholar]
- 21. Hengherr S, Brmmer F, Schill RO. Anhydrobiosis in tardigrades and its effects on longevity traits. J Zool. 2008;275:216–220. [Google Scholar]
- 22. Boothby TC, Tapia H, Brozena AH, Piszkiewicz S, Smith AE, Giovannini I, Rebecchi L, Pielak GJ, Koshland D, Goldstein B (2017) Tardigrades use intrinsically disordered proteins to survive desiccation. Mol Cell 65: 975–984 e975, 975, 984.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Piszkiewicz S, Gunn KH, Warmuth O, et al. Protecting activity of desiccated enzymes. Protein Sci. 2019;28:941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yamaguchi A, Tanaka S, Yamaguchi S, et al. Two novel heat‐soluble protein families abundantly expressed in an anhydrobiotic tardigrade. PLoS One. 2012;7:e44209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Malki A, Teulon JM, Camacho‐Zarco AR, et al. Intrinsically disordered tardigrade proteins self‐assemble into fibrous gels in response to environmental stress. Angew Chem Int Ed. 2021;134:e20210996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yagi‐Utsumi M, Aoki K, Watanabe H, et al. Desiccation‐induced fibrous condensation of CAHS protein from an anhydrobiotic tardigrade. Sci Rep. 2021;11:21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hatanaka R, Hagiwara‐Komoda Y, Furuki T, et al. An abundant LEA protein in the anhydrobiotic midge, PvLEA4, acts as a molecular shield by limiting growth of aggregating protein particles. Insect Biochem Mol Biol. 2013;43:1055–1067. [DOI] [PubMed] [Google Scholar]
- 28. Wise MJ, Tunnacliffe A. POPP the question: What do LEA proteins do? Trends Plant Sci. 2004;9:13–17. [DOI] [PubMed] [Google Scholar]
- 29. Battaglia M, Olvera‐Carrillo Y, Garciarrubio A, Campos F, Covarrubias AA. The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 2008;148:6–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hand SC, Menze MA, Toner M, Boswell L, Moore D. LEA proteins during water stress: Not just for plants anymore. Annu Rev Physiol. 2011;73:115–134. [DOI] [PubMed] [Google Scholar]
- 31. Gronenborn AM, Filpula DR, Essig NZ, et al. A novel, highly stable fold of the immunoglobulin binding domain of streptococcal protein g. Science. 1991;253:657–661. [DOI] [PubMed] [Google Scholar]
- 32. Crilly CJ, Brom JA, Kowalewski ME, Piszkiewicz S, Pielak GJ. Dried protein structure revealed at the residue level by liquid‐observed vapor exchange NMR. Biochemistry. 2021;60:152–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Crilly CJ, Brom JA, Warmuth O, Esterly HJ, Pielak GJ. Protection by desiccation‐tolerance proteins probed at the residue level. Protein Sci. 2022;31:396–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Leeder JD, Watt IC. The stoichiometry of water sorption by proteins. J Colloid Interface Sci. 1974;48:339–344. [Google Scholar]
- 35. Watt IC, Leeder JD. 24—Stoichiometric analysis of the wool‐water isotherm. J Text Inst. 1968;59:353–364. [Google Scholar]
- 36. Hageman MJ. The role of moisture in protein stability. Drug Dev Ind Pharm. 2008;14:2047–2070. [Google Scholar]
- 37. Crilly CJ, Eicher JE, Warmuth O, Atkin JM, Pielak GJ. Water's variable role in protein stability uncovered by liquid‐observed vapor exchange NMR. Biochemistry. 2021;60:3041–3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Leeder JD, Watt IC. The role of amino groups in water absorption by keratin. J Phys Chem. 1965;69:3280–3284. [DOI] [PubMed] [Google Scholar]
- 39. Watt IC, Leeder JD. Role of carboxyl groups in water absorption by keratin. J Appl Chem. 1968;18:1–4. [DOI] [PubMed] [Google Scholar]
- 40. Rockland LB. Saturated salt solutions for static control of relative humidity between 5° and 40°C. Anal Chem. 1960;32:1375–1376. [Google Scholar]
- 41. Smith CK, Withka JM, Regan L. A thermodynamic scale for the beta‐sheet forming tendencies of the amino acids. Biochemistry. 1994;33:5510–5517. [DOI] [PubMed] [Google Scholar]
- 42. Monteith WB, Pielak GJ. Residue level quantification of protein stability in living cells. Proc Natl Acad Sci USA. 2014;111:11335–11340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. May JC, Grim E, Wheeler RM, Westy J. Determination of residual moisture in freeze‐dried viral vaccines: Karl fischer, gravimetric and thermogravimetric methodologies. J Biol Stand. 1982;10:249–259. [DOI] [PubMed] [Google Scholar]
- 44. May JC, Wheeler RM, Grim E. The gravimetric method for the determination of residual moisture in freeze‐dried biological products. Cryobiology. 1989;26:277–284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1‐S4 H2O content and amino acid compositions


