Abstract
Autism Spectrum Disorder (ASD) diagnosis is still based on behavioral criteria through a lengthy and time-consuming process. Much effort is being made to identify brain imaging biomarkers and develop tools that could facilitate its diagnosis. In particular, using Machine Learning classifiers based on resting-state fMRI (rs-fMRI) data is promising, but there is an ongoing need for further research on their accuracy and reliability. Therefore, we conducted a systematic review and meta-analysis to summarize the available evidence in the literature so far. A bivariate random-effects meta-analytic model was implemented to investigate the sensitivity and specificity across the 55 studies that offered sufficient information for quantitative analysis. Our results indicated overall summary sensitivity and specificity estimates of 73.8% and 74.8%, respectively. SVM stood out as the most used classifier, presenting summary estimates above 76%. Studies with bigger samples tended to obtain worse accuracies, except in the subgroup analysis for ANN classifiers. The use of other brain imaging or phenotypic data to complement rs-fMRI information seems promising, achieving higher sensitivities when compared to rs-fMRI data alone (84.7% versus 72.8%). Finally, our analysis showed AUC values between acceptable and excellent. Still, given the many limitations indicated in our study, further well-designed studies are warranted to extend the potential use of those classification algorithms to clinical settings.
Subject terms: Diagnostic markers, Biomarkers, Diseases, Neurology, Mathematics and computing
Introduction
Autism Spectrum Disorder (ASD) is a life-long neurodevelopmental condition associated with the atypical development of the brain. Individuals in this group, in general, present a slow development in certain activities when compared to individuals of Typical Development (TD)—such as speech, motor coordination, and social activities—and difficulties communicating and relating to others1,2.
Despite being considered a neurological disorder, the diagnosis of ASD remains exclusively based on behavioral criteria3. This may be due to the great heterogeneity within the population, possibly reflecting an enormous amount of different neurodevelopmental etiologies4,5.
Typically identified in early childhood, ASD’s development is believed to have genetic and environmental roots6,7. According to recent publications8,9, the former accounts for approximately 80% of the cases. Also, epidemiological studies suggest an increase in its global prevalence in recent years, and a systematic review published in 2012 estimated it to be about 0.62%10.
The impact of this condition on the quality of life extends beyond the affected individual to the entire family. For example, parents of children with ASD report higher stress levels than parents of children with other disabilities11. Also, the majority of researches regarding autism is based on data from high-income countries. This creates inequities across the world in access to services and supports12.
On the other hand, given the brain’s plasticity during the first years of life, early detection paired with early treatment would have considerably stronger benefits than later treatments13,14.
The gold standard diagnosis of ASD is based on a differential diagnostic examination by an experienced clinician, frequently supported by tools such as the Autism Diagnostic Interview-Revised (ADI-R)15—a standardized caregiver interview—and the Autism Diagnostic Observation Schedule (ADOS/-2)16—a semi-structured standardized observation for individuals with suspected ASD. Therefore, this is a long and time-consuming process that requires a multi-disciplinary team to assess information from various sources17,18.
In recent years, Machine Learning (ML) classifiers have been increasingly applied to neuroimaging data to diagnose psychiatric disorders, including ASD. Those classification methods hold the promise of facilitating and speeding up the diagnostic process19,20.
Throughout the different types of neuroimaging data, the resting-state functional Magnetic Resonance Imaging (rs-fMRI) is increasingly used to investigate neural connectivity and identify biomarkers of psychiatric disorders. It is based on spontaneous fluctuations in the Blood Oxygenation Level-Dependent (BOLD) signal obtained through a non-invasive and relatively fast acquisition process. Also, the rs-fMRI is task-free—requiring no active and focused participation of the patient—and the data can be easily combined to generate large databases19,21,22.
We can highlight the Autism Brain Imaging Data Exchange (ABIDE) as one example of such databases. Together, the first23 and second24 versions of the repository (ABIDE I and ABIDE II) aggregate rs-fMRI and corresponding structural data of more than 2000 individuals with ASD and of TD collected across more than 24 international brain imaging laboratories.
Studies using rs-fMRI data have revealed brain functional connectivity patterns that could serve as biomarkers for classifying depression25, Parkinson’s disease26, Attention Deficit Hyperactivity Disorder27, ASD28, and even age29. However, the reproducibility and generalizability of these approaches in research or clinical settings are debatable. There are many potential sources of variation across studies, and its effect on diagnosis and biomarker extraction is still poorly understood22,30.
Therefore, we conducted a systematic review and meta-analysis of studies that used ML classifiers based on rs-fMRI data to distinguish patients with ASD from individuals of TD. We aimed to critically review the current literature on this area based on the following research questions:
Which ML techniques are used to classify ASD and TD individuals based on rs-fMRI?
What are the results obtained by the studies using these approaches?
Which methodological differences are associated with the performance measures obtained throughout the publications?
The approaches are robust enough to be applied in a clinical setting?
What are the aspects that still need to be investigated?
Results
We searched for articles using four digital libraries and a backward snowballing approach31—i.e., looking for new relevant articles in the references of the selected ones. Three authors conducted a selection process based on specific inclusion and exclusion criteria. Articles were pre-selected if at least one author concluded they should be, and the final selected papers were defined by consensus.
Data extraction was performed on the selected articles using a standardized data extraction sheet, considering only one result per independent sample. Publications with enough information to obtain measures of True Positive (TP), True Negative (TN), False Positive (FP), and False Negative (FN) were included in the meta-analysis. Also, the Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2)32 was applied to assess studies’ methodological quality. More details of our methodology can be found in “Methods” section.
The following subsections present the characteristics of the studies selected for (i) the systematic review and (ii) the meta-analysis, and the results of (iii) the quality assessment and (iv) the quantitative meta-analysis.
General study characteristics
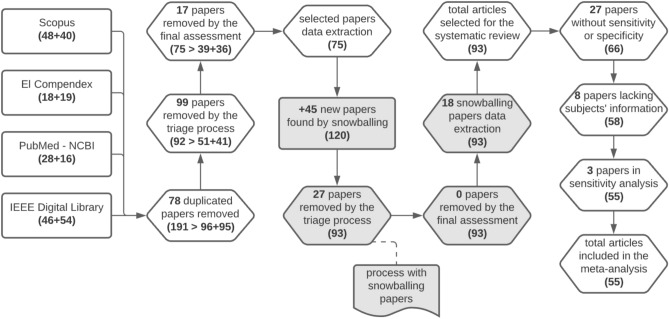
A total of 935,19,22,30,33–121 studies were selected for the systematic review. Figure 1 summarizes our selection methodology and the details according to the screening stage. Also, the final publications identified and selected can be found as Supplementary Tables S1–S3.
Figure 1.
Screening and selection of studies according to inclusion and exclusion criteria at different stages of the meta-analysis. The numbers between parentheses indicate the total of articles remaining after each step. The numbers separated by indicate the count of articles from the first and second search, respectively. Created with Lucidchart Free https://www.lucidchart.com.
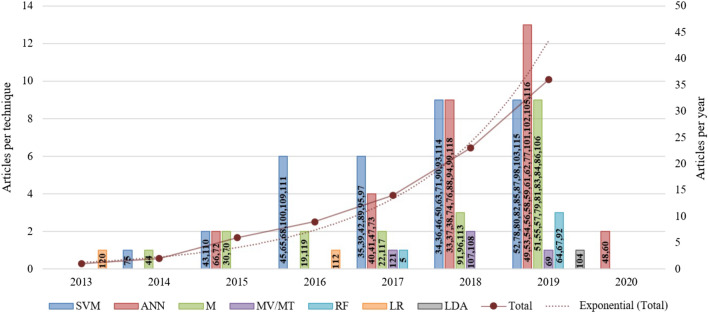
All the 93 studies were published between 2013 and 2020 and used samples that varied from 24 to 2352 individuals. The most commonly applied ML techniques for classification were Support Vector Machine (SVM, ) and Artificial Neural Network (ANN, ), followed by studies that used more than one technique (M, ). Figure 2 shows the distribution of the selected articles by year and ML technique used, whereas Table 1 shows the general characteristics of the included studies.
Figure 2.
Distribution of the selected studies by year of publication and type of ML technique used (MV/MT multiview/multitask learning, RF Random Forest, LR Logistic Regression; LDA Linear Discriminant Analysis). The numbers inside the bars indicate each article. Created with Microsoft Excel 2019.
Table 1.
General characteristics of the studies selected in the systematic review (SR) and the studies and samples included in the meta-analysis (MA).
| Characteristics | Studies (SR) | Studies (MA) | Samples (MA) |
|---|---|---|---|
| Total | 93 | 55 | 132 |
| ML technique | |||
| SVM | 33 | 27 | 54 |
| L-SVM | − | 14 | 33 |
| Other | − | 13 | 21 |
| ANN | 30 | 13 | 44 |
| CNN | − | 5 | 16 |
| Other | − | 8 | 28 |
| M | 19 | 2 | 2 |
| MV/MT | 4 | 3 | 15 |
| RF | 4 | 2 | 2 |
| LR | 2 | 3 | 4 |
| LDA | 1 | 2 | 8 |
| Ridge | − | 1 | 1 |
| XGB | − | 1 | 1 |
| Affine | − | 1 | 1 |
| Dataset | |||
| ABIDE (any version) | 79 | 45 | 121 |
| ABIDE without version | 34 | 21 | 41 |
| ABIDE I − preprocessed | 34 | 19 | 54 |
| ABIDE I + ABIDE II | 7 | 5 | 26 |
| ABIDE I | 2 | 0 | 0 |
| ABIDE II | 2 | 0 | 0 |
| UMCD | 3 | 2 | 2 |
| NDAR | 3 | 2 | 2 |
| Own sample | 3 | 4 | 4 |
| Own sample + ABIDE | 3 | 2 | 2 |
| Others | 2 | 1 | 1 |
| Type of data | |||
| Only rs-fMRI | 73 | 49 | 114 |
| rs-fMRI plus other types of brain imaging data | 11 | 3 | 14 |
| rs-fMRI plus phenotypic information | 9 | 3 | 4 |
| Sex of the subjects | |||
| Males and females | 62 | 37 | 80 |
| Not enough information | 26 | 13 | 44 |
| Only males | 5 | 6 | 8 |
| Age of the subjects | |||
| Both above and below 18 y.o. | 42 | 26 | 62 |
| Not enough information | 28 | 10 | 25 |
| Below 18 y.o. | 20 | 20 | 39 |
| Above 18 y.o. | 3 | 4 | 6 |
| FIQ of the subjects | |||
| Not enough information | − | 33 | 90 |
| Both high- and low-functioning | − | 14 | 30 |
| Only high-functioning | − | 8 | 12 |
Note that for the dataset, sex, and age of the subjects, the sum of the column Studies (MA) is greater than 55 due to articles with multiple samples included in different categories.
L-SVM Linear SVM, CNN Convolutional Neural Network, Ridge Ridge classifier, XGB Extreme Gradient Boosting, Affine Affine-Invariant, y.o. years old, FIQ Full Intelligence Quotient.
Significance values are given in italics.
Almost 85% of the studies () extracted their samples from versions of the ABIDE, specially ABIDE I preprocessed () or ABIDE without specifying the version (). The other articles used data from the UCLA Multimodal Connectivity Database122 (UMCD, ), the National Database of Autism Research (NDAR, ) [http://ndar.nih.gov], own samples (), own samples and ABIDE (), others ().
The majority of the studies () used only rs-fMRI data for classification. Beyond that, some studies used other types of brain imaging data () or phenotypic data ().
Regarding the subjects’ characteristics, we found studies that included both males and females (), only male subjects (), and studies that did not present enough information regarding the sex of the selected individuals (). Furthermore, there were samples with subjects both above and below 18 years old (), only below 18 (), only above 18 (), and studies without enough information ().
Studies included in the meta-analysis
From the 93 studies selected for the systematic review, 2733–59 did not report any data regarding sensitivity or specificity and were excluded from the meta-analysis. Five articles60–64 did not present the exact number of TD and ASD subjects on the sample or the test set, making it impossible to calculate the measures necessary for the meta-analysis. Two articles65,66 defined specific sample percentages as training or test sets and performed some random trials. Thus, it was impossible to determine the exact number of subjects in the test set nor the proportion of ASD and TD subjects. In another study67, seven subjects had corrupted rs-fMRI imaging files, so they were included in the structural MRI (sMRI) analysis and excluded from fMRI analysis and sMRI-fMRI modalities fusion. However, the study did not inform from which group (ASD or TD) those subjects were.
Two articles68,69 presented their results through bar charts without showing the exact sensitivity and specificity values, so we decided not to include them in the meta-analysis. In another publication5, an RF was used as the classifier, and sensitivity and specificity measures were presented only for the external validation data-set. The main results from the article were obtained through the out-of-bag (OOB) error—by subsampling with replacement to create training samples for each tree, the excluded data are used for testing, and the mean of the results generates the OOB error—but only the accuracy was reported. Since the results from the validation data-set and the ones obtained using the OOB error presented high variation, we decided not to use the results from this article. However, those three articles were included in a sensitivity analysis to assess their effect on the meta-analysis results.
Finally, 55 studies19,22,30,70–121—published between 2013 and 2019—provided sufficient data for a quantitative meta-analysis. Note that the information presented in the rest of this section is related to the main results extracted from those studies, as described in “Data extraction” section.
Since some studies comprise multiple samples, a total of 132 independent samples were extracted, with sensitivity and specificity ranging from 37.5% to 100% and 20% to 100%, respectively. About 85% of the studies extracted their samples from versions of the ABIDE, corresponding to 93% of the samples. A coupled forest plot of sensitivity and specificity for all the samples included in the meta-analysis can be found as Supplementary Fig. S1. Table 1 also presents the general characteristics of the studies and samples included.
From all samples, 123 used ABIDE data in the test sets. To better understand their overlap, we calculated how many samples used each ABIDE site. Sixteen of them did not present enough information to define which sites they came from. ABIDE I was used in 100 samples, while ABIDE II was used in nine. Note that some samples comprised both versions of the database and, for this analysis, we defined the ABIDE version according to the sites’ names presented if the articles did not directly specify which ABIDE they used.
In the case of ABIDE I, each site was used from 13 to 33 times throughout the samples, with a mean of 19.2 samples using each site. Besides, 60 samples used only one site, 16 samples used two sites, and seven samples used all ABIDE I sites. As for ABIDE II, ignoring the sites that were not used, each site was used from one to three times, with a mean of 1.5 samples using each site. In this case, six samples used only one site.
It is worth noticing that a site being used in 33 samples does not mean a complete overlap 33 times since each sample used a different number of subjects from each site. Even knowing the number of subjects used from each site in each sample and the total number of subjects available in each site, one cannot assess the extent of the overlap once it is not known which specific subjects were used. In fact, only two of the articles22,96 presented which subjects from ABIDE were used in their studies.
Beyond that, less than 50% of the studies ( articles/61 samples) presented the mean age, almost 24% () had not enough information regarding the sex of the subjects, and only 20% () presented the mean FIQ of their ASD and TD samples. Also, only 1 of the samples112 used in the meta-analysis—a multi-site dataset from Japan—was not from North America or Europe.
Most of the articles defined Regions of Interest (ROIs) of the brain to reduce the dimensionality of their features using a priori atlases or combinations of them (), especially the Automated Anatomical Labelling123 in versions of 90 (AAL90, ) and 116 ROIs (AAL116, ) and the Craddock atlas with 200 ROIs124 (CC200, ).
Throughout the studies, there were various approaches to extract features from the data, from different points of view, and in varying levels of complexity. However, the most used type of features consisted of estimating functional connectivity (FC) patterns19 using the Pearson Correlation (PC) between all pairs of averaged time-series. Those studies used the PC either on its original version () or normalized by Fisher transformation ().
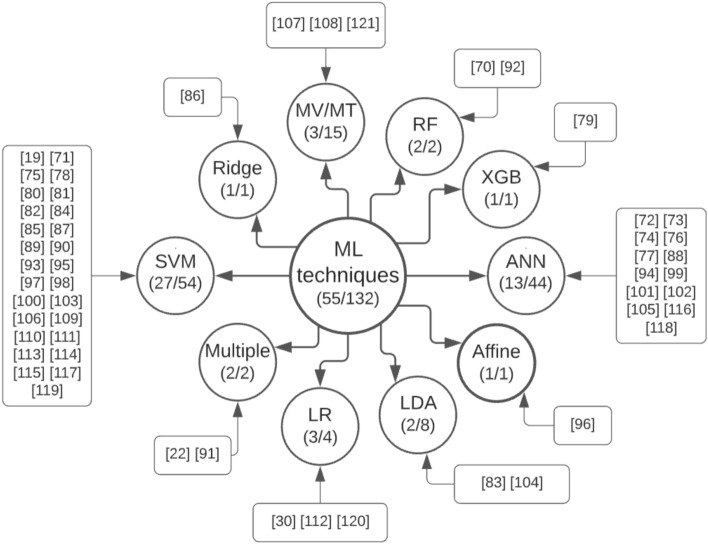
According to the main results, the techniques used for classification are presented through a conceptual map in Fig. 3.
Figure 3.
Conceptual map of ML techniques used throughout the articles selected for meta-analysis (number of articles/number of samples). Created with Lucidchart Free https://www.lucidchart.com.
Quality assessment
The QUADAS-2 is a tool designed to assess the quality of primary diagnostic accuracy studies, evaluating if systematic flaws or limitations might distort their results—i.e., if there is a Risk of Bias (RoB)—and if these studies apply to the review’s research question32.
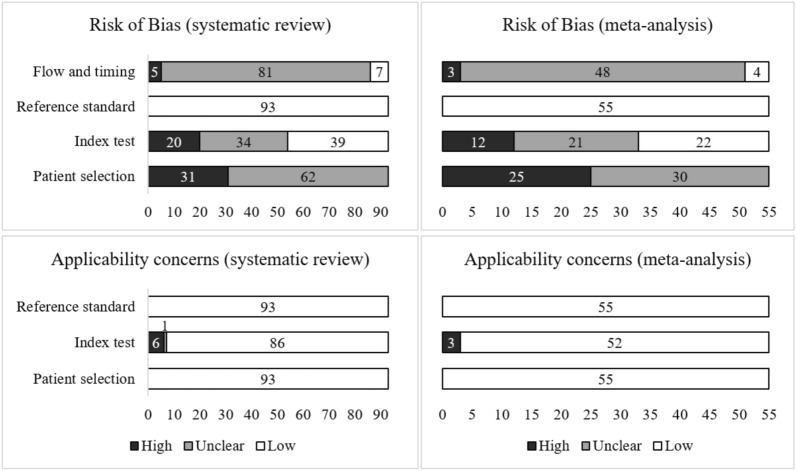
Figure 4 shows the distribution of the results of QUADAS-2 considering all the publications selected for the systematic review or the studies included in the meta-analysis (see Supplementary Table S4). The only difference in the results between the two applications of the tool was in the RoB by index test. Since some of the results in the articles did not have enough information to be used in the meta-analysis, three studies70,71,117 showed low RoB by index test when assessed as a whole, but when considering only the results used for the meta-analysis they presented unclear RoB.
Figure 4.
Risk of bias and applicability concerns by domain in QUADAS-2 for all the studies selected for the systematic review (left) and considering only the ones included in the meta-analysis (right). Created with Microsoft Excel 2019.
We can highlight that none of the studies was considered to have a low RoB by patient selection domain. Most of them had an unclear RoB (62 out of 93 in the first and 30 out of 55 in the second application) since they used databases such as the ABIDE and did not present details regarding the recruitment of the subjects nor sufficient information of the characteristics of the subjects selected. The remaining articles were shown to have a high RoB due mainly to the selection of subjects in restricted intervals of age and intelligence quotient (IQ) or the exclusion of female subjects.
The great majority of the studies were considered to have an unclear RoB by flow and timing domain (81 and 48 studies) mainly because they did not present the interval between the application of the index test and the reference standard nor sufficient information to conclude if all subjects received the same reference standard.
All of the articles were shown to have a low RoB by reference standard domain given that we considered the reference standards used in databases such as the ABIDE as reliable even if the article did not present exactly what reference standards were used. For the same reason, all of the studies were assessed to have low concerns regarding applicability by the same domain. We highlight that the reference standards used throughout the studies were similar to those used in ABIDE23: combining clinical judgment and diagnostic instruments; clinical judgment only; or diagnostic tools only.
More than half of the articles were considered to have an unclear or high RoB (54 and 33) by the index test domain. Also, most of the studies (86 and 52) were assessed to have low concerns regarding applicability by the same domain. Finally, all of the articles were shown to have low concerns regarding applicability by the patient selection domain.
According to the first application of the QUADAS-2 tool, the RoB was judged as high in at least one category in 42 studies, and 12 studies presented a high RoB in at least two domains. Finally, according to the second analysis, the RoB was judged as high in at least one category in 29 studies, and 9 studies presented a high RoB in at least two domains.
Diagnostic accuracy
As commented before, there is a literature limitation regarding sample overlap. This should be taken in consideration while analyzing the quantitative results presented in this section. The implications and other considerations regarding this issue can be found in “Limitations” section.
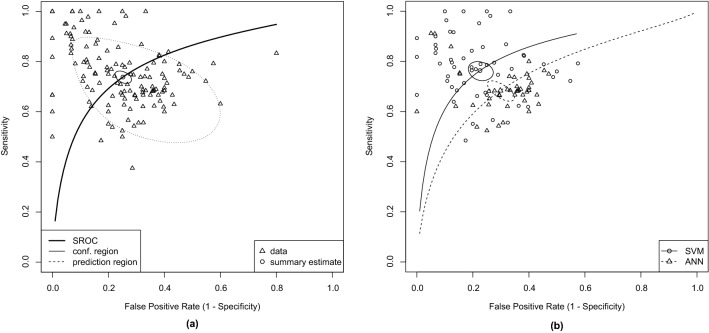
Using the bivariate model, machine learning-based classifiers separated ASD from TD with a sensitivity of 73.8% (95% confidence interval (CI) 71.8–75.8%), a specificity of 74.8% (95% CI 72.3–77.1%), and area under the curve (AUC)/partial AUC (pAUC) of 0.803/0.765. A Summary Receiver Operating Characteristic (SROC) curve of the included studies—along with the estimated summary point, confidence region, and prediction region—is presented in Fig. 5a. Of the 132 samples, 40 were outside the 95% predictive region of the SROC curve, indicating heterogeneity. All the results obtained with the main analysis can be found in Supplementary Table S5.
Figure 5.
SROC curves of all the included studies with summary estimate (a) and the studies using SVM and ANN with their summary estimates and confidence region (b). Created with R Statistics125 version 4.1.1 using the package mada126 version 0.5.10.
Regression with year of publication did not affect sensitivity () or specificity (), even segregating per type of ML technique.
ML technique and sample size
Considering the type of ML technique, only SVM and ANN classification tools were used in 5 or more articles. When analyzing the SVM studies, we obtained a sensitivity of 76.3% (95% CI 73.2–79.2%), a specificity of 77.5% (95% CI 73.7–80.8%), and AUC/pAUC of 0.832/0.748. The ANN studies had a sensitivity of 68.4% (95% CI 65–71.5%), a specificity of 70.2% (95% CI 66.2–73.9%), and AUC/pAUC of 0.743/0.582. The SROC curves for the studies using SVM and ANN are presented in Fig. 5b. We performed a subgroup analysis and found a significant difference between the sensitivities () and the specificities (). However, after correction for multiple comparisons, only the difference in sensitivities remained significant.
We also analyzed the subtype of ML technique. For SVM, only L-SVM was used in five or more articles - sensitivity of 73.9% (95% CI 70.2-77.2%), specificity of 77.5% (95% CI 73.3–81.2%), and AUC/pAUC of 0.813/0.708—whereas for ANN the same happened with CNN - sensitivity of 66.7% (95% CI 63.3–69.9%), specificity of 70.1% (95% CI 66.3–73.7%), and AUC/pAUC of 0.732/0.565. Thus, we compared L-SVM with other types of SVM and CNN with other types of ANN. The comparison showed no effect on sensitivity or specificity (all ). Finally, the comparison with L-SVM against CNN indicated higher sensitivity () and specificity () in L-SVM studies. However, neither of those effects survived multiple comparisons correction.
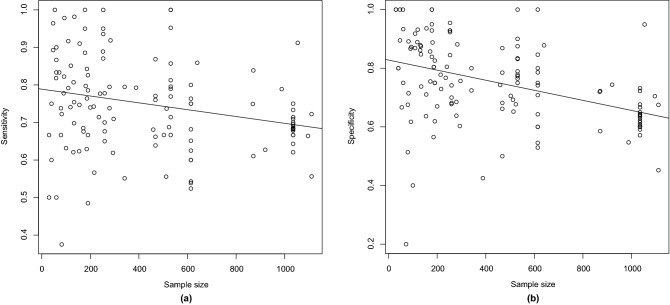
Regression with sample size as moderator showed a significant effect on both sensitivity () and specificity () when analyzing all the samples together, even considering multiple comparisons correction. Figure 6 shows the linear regression models with sample size predicting sensitivity and specificity and indicates that bigger sample sizes tend to obtain worse accuracies.
Figure 6.
Linear regression models with sample size predicting sensitivity (a) and specificity (b) for all the studies. Created with R Statistics125 version 4.1.1 using the package mada126 version 0.5.10.
However, the same analysis segregating the studies per type of ML technique used indicated a significant effect on specificities () and no impact on sensitivities () for SVM studies, with worse specificities in studies with larger samples. Also, no significant effect was found for the ANN studies (all ).
Subjects characteristics
No significant effects of sex (only males against males and females studies) or FIQ (neither considering the mean FIQ nor comparing studies that used only high-functioning subjects with the ones that used high- and low-functioning subjects) on sensitivity or specificity (all ) were observed.
Regression with the mean age of the subjects did not affect sensitivity or specificity (all considering ASD or TD groups). Comparison between samples with subjects under 18 years old and samples composed of individuals both under and above that age showed a significant difference between the specificities () but no effect on sensitivity (), indicating higher specificity in studies that used only subjects under 18 y.o. (77.6—95% CI 73–81.6%—versus 70.5—95% CI 66.6–74.1%). This effect, however, did not survive multiple comparisons correction. Segregating per type of ML technique, only SVM had enough studies (17 studies and 37 samples) to conduct the analysis. Still, the results did not show any effect on sensitivity () or specificity (). Sensitivity analysis considering other age thresholds (19, 20, 21) yielded the same conclusions (see Supplementary Table S6).
Sources of the samples
Subgroup analysis considering the database or source of the sample (ABIDE without version—comprising the studies that did not specify which version of ABIDE they were using—ABIDE I preprocessed or ABIDE I + ABIDE II) indicated a significant effect on the sensitivity when comparing ABIDE without version with ABIDE I preprocessed () or ABIDE I + ABIDE II (). In both cases, the ABIDE without version group presented higher sensitivity (77.1%—95% CI 73.2–80.6%—versus 72%—95% CI 69–74.9%—and 69.2%—95% CI 65.8–72.4%—respectively). Nevertheless, this effect did not survive multiple comparisons. All the other analyses indicated no significant impact on sensitivity or specificity (all ). The same analysis with SVM samples (ABIDE without version or ABIDE I preprocessed) did not indicate any effect on sensitivity () or specificity ().
We conducted another analysis comparing the studies that used any version of ABIDE with studies that used databases or samples other than ABIDE (Own sample, NDAR, UMCD). The regression indicated higher sensitivity () and specificity () in studies that used databases or samples other than ABIDE (Sensitivity: 81.8%—95% CI 73.4–88.1%—versus 73.2%—95% CI 71.1–75.2%; Specificity: 83%—95% CI 72.5–90%—versus 74.1%—95% CI 71.6–76.5%), but the effects did not survive multiple comparisons.
Features definition
Subgroup analysis considering type of data (only rs-fMRI or rs-fMRI plus other data types) showed a significant difference between the sensitivities () and the specificities (), indicating better results in studies that used other types of data together with rs-fMRI (Sensitivity: 84.7%—95% CI 78.5–89.4%— versus 72.8%—95% CI 70.6–74.8%; Specificity: 81%—95% CI 74.1–86.3%—versus 73.9%—95% CI 71.3–76.4%). However, after correction for multiple comparisons, only the difference on sensitivities remained significant.
Subgroup analysis considering the atlas used (AAL90, AAL116 or CC200), indicated that: studies using AAL90 obtained better specificity (74.9%—95% CI 68.7–80.1%; ) than studies using CC200 (64.4%—95% CI 60.7–67.9%) but no significant effect was observed on the sensitivity (); studies using AAL116 obtained better sensitivity (77.7%—95% CI 73.7–81.2%; ) and specificity (78.2%—95% CI 72.8–82.9%; ) than studies using CC200 (Sensitivity: 68%—95% CI 65.4–70.4%); there was no significant effect on sensitivity () or specificity () between studies using AAL 116 or 90. Figure 7 shows the SROC curves for the studies using AAL90, AAL116 or CC200. All those effects remained significant after multiple comparisons correction.
Figure 7.
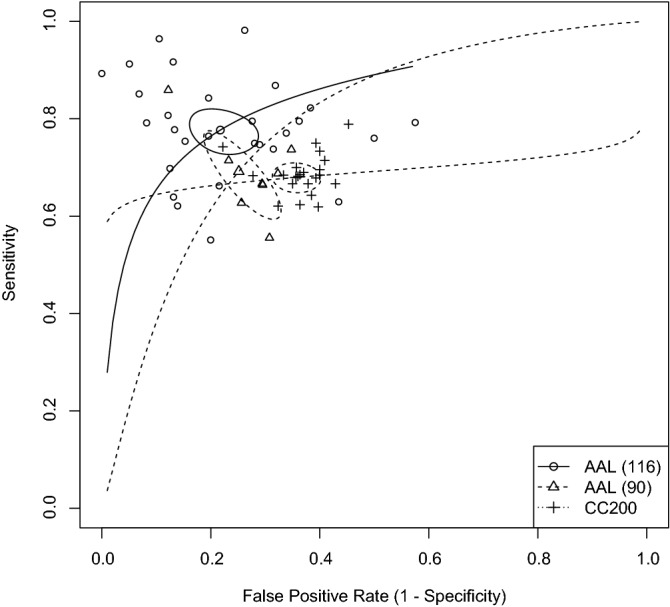
SROC curves of the studies using AAL90, AAL116, or CC200 with their summary estimates and confidence regions. Created with R Statistics125 version 4.1.1 using the package mada126 version 0.5.10.
Regression considering the number of ROIs used showed significant effects on sensitivity () and specificity (). When segregating per ML technique, there was a significant effect on sensitivity () for the SVM studies but no effect on specificity () whereas for the ANN studies there was a significant effect on specificity () but no effect on sensitivity (). For all the significant effects, the linear regression models indicated lower values of sensitivity/specificity as the number of regions increased. None of these effects survived multiple comparisons correction.
Subgroup analysis using the type of feature as moderator (PC, PC Fisher-transformed or others), indicated that: studies using PC (Fisher-transformed) obtained better sensitivity (76.7%—95% CI 71.3–81.3%; ) and specificity (81%—95% CI 75.6–85.4%; ) than studies using PC (Sensitivity: 68.9%—95% CI 66.8–70.9%; Specificity: 68.3%—95% CI 64.3–72.1%); similarly, studies using other features obtained better sensitivity (73.5%—95% CI 70.6–76.2%; ) and specificity (74.7%—95% CI 71–78%; ) than studies using PC; there was no significant effect on sensitivity () or specificity () between studies using PC (Fisher-transformed) and other features. Only the effects of the first analysis survived multiple comparisons.
QUADAS-2 analyses
Subgroup analysis considering the number of domains with low RoB in QUADAS-2 results (one or two) showed a significant difference between the specificities () but no effect on sensitivity (), indicating higher specificity in studies that had only one domain with a low risk of bias (78.4%—95% CI 75.5–81.1%—versus 69.6%—95% CI 65.9–73%).
Analysis considering the RoB of the index test obtained from QUADAS-2 (high, unclear or low) indicated that: studies with unclear RoB obtained better sensitivity (80.5%—95% CI 76.1–84.2%; ) than studies with high RoB (70.1%—95% CI 66.3–73.5%) but no significant effect was observed on the specificity (); studies with high RoB obtained better specificity (76.6%— 95% CI 73.2–79.8%; —did not survive multiple comparisons) than studies with low RoB (69.9%—95% CI 66.2–73.3%) but no significant effect was observed on the sensitivity (); studies with unclear RoB obtained better sensitivity (—did not survive multiple comparisons) and specificity (81.3%—95% CI 76.9–85%; ) than studies with low RoB (Sensitivity: 72.1%—95% CI 69.6–74.4%).
We conducted another analysis splitting the studies with low RoB between the ones that performed a temporal (using data from newly recruited subjects) or geographic (using data collected by independent investigators at a different site) validation (5 articles and 35 samples) and the ones that performed a split-sample validation127. The conclusions were the same, except for the comparison between the studies with high RoB and the ones with low RoB (split-sample) that did not indicate a significant effect on sensitivity () or specificity (). Also, there was no effect on sensitivity () or specificity () when comparing the studies that used a split-sample validation and the ones that used a temporal or geographic validation.
Sensitivity analyses
Sensitivity analysis including three more articles5,68,69—four new samples—that were initially excluded from the meta-analysis (as cited in “Studies included in the meta-analysis” section) indicated no significant change in overall sensitivity (73.7%—95% CI 71.7–75.6%) or specificity (75%— 95% CI 72.7–77.2%). All the results obtained with this sensitivity analysis can be found in Supplementary Table S7.
We repeated all the tests that the addition of those articles could impact. In general, their influence on the outcome was minor, but the conclusions from the meta-regression differed in three analyses. The regression with type of data as moderator showed the same conclusion for the sensitivities () but no significant effect between the specificities (). The regression considering the database or source of the sample indicated no significant effect on sensitivity when comparing ABIDE with ABIDE I preprocessed () or ABIDE I + ABIDE II (). Regression with atlas as moderator showed a significant effect on sensitivity () between studies using AAL 116 or 90, indicating higher sensitivity in studies that used the AAL 116 (78%—95% CI 74.1–81.4%—versus 69.2%—95% CI 61.4-76%).
Discussion
ML techniques and sample size
As shown in Fig. 2, the number of publications in the area has been increasing following an exponential trend, and SVM was the most used classification technique, especially until 2018. Also, from the 54 samples that used SVM for classification, 33 of them used an L-SVM. The inherent characteristics of SVMs help them generalize well and deal with noisy, correlated features and high-dimensional data sets128,129.
However, we can observe an increasing number of studies using ANN techniques, making it the second most used technique in absolute numbers and the first when considering only articles published in 2019. Many ANN classifiers used deep-learning methods, especially the Convolutional Neural Network, corresponding to more than 35% of the ANN techniques.
Despite that, analysis considering the type of classification algorithm used indicated better results for SVM against ANN, especially on sensitivity. Also, the analysis comparing L-SVM and CNN resulted in the same conclusions (although not surviving multiple comparisons). In all those cases, the difference was about seven percentage points.
More complex models—such as deep-learning ones—tend to be more powerful but generally have more hyperparameters. Therefore, they are potentially more capable of explaining noise in the data and overfitting130. Also, it is not clear that those complex models always provide a significant advantage in practical performance. However, this can reflect the small number of samples available instead of indicating an absence of complicated relationships between features131.
We conducted a regression analysis to investigate the effect of the sample size on the results. Considering all the samples selected for the meta-analysis, we found worse results on both sensitivity and specificity by increasing the sample size. This same trend was also observed on specificities when considering only the SVM studies.
However, using only the ANN studies, we could not find any significant effect. Since a great part of the ANN methods were also deep-learning methods and more complex models may demand larger samples to avoid overfitting, our analysis suggests that ANN techniques may have an advantage when dealing with larger samples.
Subjects characteristics
Several studies indicate gender, age, and IQ differences in autistic symptoms and impairments. For example, boys with ASD showed more restricted and repetitive behaviors than girls with ASD132,133; significant though modest effects of IQ and age indicated increased Autism severity with decreasing IQ and age134; also, lower socio-communicative symptoms were found in older compared to younger individuals132.
It is well known that ASD shows an imbalanced male–female ratio, and recent studies suggest values between 2:1 and 5:1135–137. There is also evidence that this ratio is lower in individuals with lower IQ135,138. Since most autism studies tend to follow this ratio or include only male participants, the underrepresentation of females may have led to an understanding of the disorder biased toward males139. Females are generally diagnosed later, and even with similar levels of severity of autistic traits, males are more likely to receive a diagnosis140–142.
Still, early detection and treatment would enormously benefit the individuals within the spectrum13,14. Therefore, it is essential to understand how those variables may affect classification accuracy to obtain a clinically useful ML diagnostic tool. In our analysis, we could not find any significant effect of the sex of the subjects or their FIQ on sensitivities or specificities. However, we must highlight some issues.
The regression considering the sex of the subjects compared the articles that used only male subjects with the ones without a sex restriction—whose samples were composed of males and females. Also, there were ten times fewer samples in the former compared to the latter subgroup. From the articles selected in the systematic review, only two42,50 performed tests considering different categories of gender. They obtained higher classification accuracies for females than males, even though the number of training samples for females was significantly lower.
Regarding the FIQ, we performed tests considering the mean FIQ and compared the samples composed of high-functioning subjects against those with both high- and low-functioning subjects. However, only 20% and 40% of the studies were included on those tests, respectively.
Regression with the mean age of the subjects did not affect sensitivity or specificity, but less than half of the articles presented enough information to be included in this analysis. On the other hand, analysis considering an adulthood threshold indicated higher specificity in studies that used only subjects under 18 y.o. when compared to studies without this restriction. This result is in accordance with some of the selected studies42,65,66, in which the adulthood segregation of the sample improved classification performance. However, the effect observed did not survive multiple comparisons, and the same analysis using only the SVM studies showed no significant effects (although it included almost three times fewer samples).
Analyzing the ABIDE—the most used database—we found a low proportion of child subjects: the ABIDE I do not include individuals below 7 years old whereas ABIDE II do not include individuals below 5 years old; in addition, from an analysis of the subjects that were available in both databases, less than 20% were below 10 years old (about 100 subjects in each one). The lack of younger individuals in those studies raises some questions. Those classifiers may be, in fact, detecting the consequences in terms of brain circuitry alterations of living with ASD instead of identifying the true roots of the disorder30,143.
As we can see, there is a lack of information regarding the characteristics of the subjects and samples included in many of the studies. Aggravating the problem, variables such as IQ, symptom severity, and handedness are missing for some sites of ABIDE70.
Sources of the samples
As a heterogeneous and complex disorder, any ASD cohort is likely composed of ill-understood subtypes with different brain features. The use of large samples, such as provided by the ABIDE, can be helpful to address those issues70. Studies based on smaller datasets from a single site are composed of more homogeneous participants, reducing the generalizability of those models. Therefore, large multi-site datasets are needed to include a greater diversity of participants and obtain more reliable, robust diagnostic systems that generalize better to new data, revealing common features that contribute to classification109,119,144,145.
On the other hand, the massive use of a single database, as we saw in this study with ABIDE, end up limiting the interpretation and generalization of the analyses and results obtained.
We conducted analyses comparing the different versions of ABIDE used throughout the studies. At first, we found a significant effect on sensitivities, but this effect was not present in the sensitivity analysis or considering only the SVM studies. It is noteworthy that the ABIDE without version group is composed of samples from the ABIDE I, ABIDE I preprocessed, or ABIDE II, but the studies did not specify which version they were using.
When comparing the studies that used any version of ABIDE (121 samples) with studies that used databases or samples other than ABIDE (9 samples), the analysis indicated higher sensitivity and specificity in the studies of the first subgroup. This may reflect the greater size and diversity of the ABIDE compared to the other sources. However, the effect did not survive multiple comparisons, and we highlight the imbalance between those subgroups.
It is also vital to notice that, while most individuals with ASD live in low- and middle-income countries10,12, all of the samples used in this meta-analysis were from high-income countries, and only one of them was not from North America or Europe.
Therefore, it is of utmost importance that more diverse datasets and studies be created and conducted. The applicability of the results obtained so far needs to be tested and confirmed across different cultures and social classes. Also, larger and more diverse samples would allow studies using restricted samples (such as low-motion data or samples composed entirely of female subjects) to obtain more reliable and robust results by selecting a bigger number of participants.
Features definition
Even though the focus of our research was the ML diagnostic tools that used rs-fMRI for classification, some of the selected studies used other types of data together with rs-fMRI aiming to obtain better results by complementing the information available. In general, we found two types of complementary data: phenotypic information such as age and sex; other brain imaging data, specially sMRI. As there were not enough articles in each of these subgroups, we compared the studies that used only rs-fMRI data with those that used any other type of data together with rs-fMRI. Our results indicated a higher specificity in the latter case.
Different brain images provide different views of the same brain and may reveal hidden evidence of ASD that is not available by using a single imaging modality121. However, we must highlight that investigation of the effect of combining different types of data in the classification is not the main objective of this study.
Most of the studies defined ROIs using a priori atlases. However, those atlases are often selected arbitrarily in the rs-fMRI community33. Therefore, we conducted a subgroup analysis with the atlas used as moderator.
Both versions of the AAL obtained significantly better results than the CC200, but there was no significant difference between them. The sensitivity analysis, however, indicated better sensitivity for studies using the AAL116. We must also highlight that, in both cases, the p-values of the comparison between the versions of the AAL were close to the threshold of 0.05, and those results should be taken with extra caution.
We made a regression analysis considering the number of ROIs used throughout the studies. Our results indicated smaller accuracies as the number of regions used increased—more specifically, worse sensitivities for the SVM studies and worse specificities for the ANN studies. This may be because more ROIs generally result in more features available. Using a large number of features relative to the number of data samples can cause classifiers to overfit19. However, the effect did not survive multiple comparisons.
The variety of choices in data processing adds to the variability of the results obtained in the studies of the field22,146. Therefore, we conducted a subgroup analysis considering the type of feature used. Our results indicated a significant advantage on sensitivity and specificity to the studies using the Fisher-transformed version of the PC against the studies using it without modifications. The studies using other features showed the same benefit against the PC ones (although not surviving multiple comparisons). Finally, even though the studies using the PC Fisher-transformed obtained better summary estimates for sensitivity and specificity, their comparison against the studies using other features did not indicate any significant effect—which is not a surprise considering the great variety in the latter group.
QUADAS-2 analyses
Bias and variation are often present in diagnostic test accuracy studies. Therefore, they need to be detected and assessed to understand the validity of the meta-analytic results obtained147,148. Through QUADAS-2 application (see Fig. 4), we found many studies with high RoB on the patient selection domain, basically due to the selection of subjects in restricted intervals of age and IQ or the exclusion of female subjects. The effect of those variables on the classification results was already discussed in “Subjects characteristics” section.
Beyond that, we can see that many studies were assessed to have an unclear RoB, especially on the patient selection and flow and timing domains. This reinforces the necessity to present more detailed information regarding the characteristics of the subjects and samples included in the publications.
We conducted some analysis using the QUADAS-2 results. The first one considered the number of domains with low RoB in each study and indicated higher specificity in studies with only one low RoB domain. We also performed a subgroup analysis using the index test domain results. As expected, studies with high or unclear RoB obtained significantly better results than studies with low RoB. We also found better outcomes for the studies with unclear RoB in comparison to those with high RoB. We suppose that a significant part of those studies assessed as unclear should have been assessed as high, but there was not enough information to conclude that. This also indicates a bias of overestimation—at least for the specificities—on the studies that do not apply their best models to independent sets after testing different numbers of features or atlases.
For the last analysis, we separated the low RoB category into the articles that performed a temporal or geographic validation and those using a split-sample validation127. Even though the latter obtained better summary estimates than the former, there was no significant effect between their sensitivities or specificities.
From the clinical standpoint, complete external validation (temporal or geographic) is preferred. Split-sample validation would not accurately assess the generalizability of a model. In contrast, geographic validation is helpful for this purpose since it may be performed with different technical parameters at different sites127. Our analysis did not indicate a significant difference between the results using each of these types of validation. However, we must highlight that only 5 articles with low RoB by the index test domain performed a complete external validation, and it is still the more reliable approach to assess generalizability.
Clinical validity
At present, ASD diagnosis is based on behavioral criteria, being vulnerable to subjectivity and interpretative bias. Also, less experienced clinicians seem to have more problems with the challenges of this complex diagnostic process17,149.
The diagnostic utility and discriminative ability of the ADOS-G and the ADI-R were assessed using a clinical population of children150. The results indicated approximately 75% of agreement with the qualified multidisciplinary team diagnoses, and most inconsistencies were false positives. The accuracy and validity of the ADOS-2 and ADI-R in diagnosing ASD in adults without an intellectual disability were also evaluated149. The original algorithm of ADOS-2 Module 4 obtained 85.9% and 82.9%, whereas its revised algorithm obtained 87.2% and 74.3% of sensitivity and specificity, respectively. On the other hand, the ADI-R got 43.1% of sensitivity and 94.7% of specificity.
Considering all the studies selected for the meta-analysis, we found summary sensitivity and specificity of 73.8% and 74.8%, respectively, for the ASD diagnosis using rs-fMRI and ML classifiers. Also, the AUC/pAUC of 0.803/0.765 indicates values between acceptable and excellent151 (0.5: no discrimination; 0.7–0.79: acceptable; 0.8–0.89: excellent; 0.9 outstanding). If we look at the analysis considering only the SVM studies, those results were even better, with sensitivity and specificity above 76% and AUC of 0.832. Also, we found acceptable AUC values within the articles that presented lower RoB and, therefore, more reliable results.
Even though these results seem promising and somewhat close to the ones obtained with diagnostic tools currently used for ASD, there is a long journey ahead before those ML algorithms could be used in clinical practice. First of all, the limitation found in the literature regarding the almost exclusive use of ABIDE as a source of samples also limit the interpretability of the quantitative results in this meta-analysis.
Beyond that, the articles included in our analysis presented a great variety of features extracted and selected, classifiers used, and validation approaches applied. Thus, the summary estimates that we obtained show the overall potential of those procedures but do not indicate a specific one to be used in clinical practice. It would even be possible to use different classifiers for subjects with different characteristics—such as sex and age—similarly to what happens with the modules of ADOS-2.
Taking the variety of neurodevelopmental etiologies believed to exist within the ASD population, there may not be an exceptional biomarker to diagnose the disorder4,5. Perhaps the classifiers must consider different biomarkers for different etiologies, partitioning the ASD into more than a single class152.
The decision on ASD diagnosis concerns the relationship established by the individual with their environment, which can lead to significant barriers to their quality of life and not to a specific condition in itself153. Therefore, a binary classifier of ASD vs. non-ASD might not be clinically helpful by not considering the environmental factors or other similarly presenting conditions. Still, the margin of doubt regarding the impairment on quality of life is limited to milder ranges of the disorder, reducing the probability that the individual does not meet the diagnostic criteria as the autistic traits accumulate. Suppose these classifiers reliably demonstrate their consistency with formal clinical diagnostic. In that case, we could take advantage of these more efficient and possibly more disseminated and democratic tools to build a scenario of access to diagnosis to all those who need the resulting social support.
Another issue to consider is the difficulty of many children and low-functioning individuals with ASD to tolerate fMRI scans, which may explain their underrepresentation within the samples analyzed. However, the application of fMRI during natural sleep can help to overcome this limitation154,155. The only article in this review that used subjects below five years of age89 applied this methodology to obtain data from 6-month-old infants, with classification results above 80%.
Many questions need to be assessed to define the clinical validity of those procedures. It includes the underrepresentation of females in research and clinical practice, the effects of subject’s IQ and age, the lack of such information in many studies, and the necessity of larger and more diverse samples to confirm the generalizability of the classification tools.
Limitations
Some limitations must be considered. The biggest one is the sample overlap between the studies, especially considering the lack of information on the patient selection process and the large number of studies that used the ABIDE database. Sample overlap induces a correlation structure among empirical outcomes, which, if not accounted for, can harm the statistical properties of meta-analysis methods and result in higher rates of false positives156. Thus, it is not clear to which extent this overlap could bias the results obtained. Furthermore, due to the tremendous heterogeneity of ASD, this high degree of overlap may limit the interpretability and generalizability of our analysis.
Despite that, we highlight that all the significant results obtained in our analyses were reasonable and in line with the literature, as we discussed in the previous subsections. Also, we clearly stated this limitation throughout the study and hope that it serves as a guide to future works, eventually reaching a state where more robust analyses can be done.
Considering the significant heterogeneity within the selected publications, the summary estimates obtained through the meta-analysis have to be interpreted with caution and in light of the methodologic quality of the studies, as previously discussed. Most studies provided only limited information regarding the patients samples and their clinical characteristics. However, detailed information about the participants’ disease status, symptoms, current medication, history of interventions, or comorbidities is crucial for evaluating the potential of the proposed models to be applied in clinical practice70,157. Thus, the impact of those variables on classification accuracy needs to be better investigated.
The studies included in our analysis identified ASD-distinctive brain patterns as compared to healthy volunteers. Nevertheless, it is critical to investigate the patterns of brain abnormalities that differentiate between different psychiatric disorders. Also, the results obtained in this meta-analysis do not apply to individuals below five years of age since almost none of the studies included individuals with such low age.
In addition, some methodological steps were not investigated in our analyses, such as the data preprocessing and feature selection procedures. Those aspects still need to be assessed to define their effects on classification accuracy.
Recommendations
Based on our results, we recommend that future studies obtain their features using the PC Fisher-transformed instead of using it without modifications. Also, the AAL116 seems to be a good choice of atlas, and we encourage studies to explore other types of data to complement rs-fMRI.
In the face of large samples, ANN techniques seem to have an advantage compared to SVM. However, considering the limitations of our study and the other methods not analyzed, we think it is a bit premature to recommend any of these techniques. For example, two articles70,92 using RF obtained results around 90%, and it would be interesting to include this technique in future analyses.
It would be of great value for future publications in this field to apply the following best practices, when possible: report, at least, measures of sensitivity and specificity; present detailed information of the subjects and samples used (sex, age, IQ, number of TD and ASD subjects, etc.); inform the reference standard used and how the patients were selected, even if the sample came from an existing database; after conducting all the tests, apply the best model to an independent sample, preferably using a temporal or geographic validation; use more diverse samples, especially from low- and middle-income countries; if the samples came from an existing database, specify which subjects were included.
Conclusions
We performed a comprehensive analysis on the literature of ML classifiers using rs-fMRI data for ASD diagnosis, indicating promising pathways and questions to be addressed. Our results showed overall sensitivity and specificity estimates of around 75%. We found better accuracy for SVM classifiers, but ANN techniques may have an advantage in dealing with larger samples. Also, the use of other types of data to complement rs-fMRI information seems to be promising.
To the best of our knowledge, this is the first meta-analysis focused on the topic, and we reiterate the availability of all extracted data. However, given the many limitations indicated in our study and the poor methodological quality found in a great part of the selected articles, further well-designed studies are warranted to extend the potential use of those classification algorithms to clinical settings, and the quantitative meta-analytical results presented here should be taken with caution.
Methods
Search strategy
The articles used in this review were found through four digital libraries: Scopus, El Compendex, PubMed—NCBI, and IEEE Xplore. Considering that the El Compendex and PubMed NCBI libraries resulted in many duplicated articles (approximately 73% and 91% of the articles, respectively), we decided not to include other libraries in the search and find out more studies through the snowballing.
The search expression was iteratively defined using keywords considered appropriate. We analyzed the titles and abstracts of the publications found through the searches to determine whether they were related or not to the purpose of this study. Based on that, we refined the search expression and obtained the final version presented below:
We started using other expressions related to ASD as defined by the Diagnostic and Statistical Manual of mental disorders 4th edition (DSM-IV)158 to possibly include articles published before 2013—when the DSM-V3 was first published. These expressions were: “Pervasive Development Disorders”; “PDD”; “Autistic Disorder”; “Asperger’s Disorder”; “Asperger”; “Childhood Disintegrative Disorder”; “PDD-NOS”. However, the addition of these terms only resulted in two new articles that were not related to the purpose of this study. Therefore, we decided to simplify the expression by removing those terms.
The search was carried out in two parts. First, we searched for articles published between January 1, 2010, and December 7, 2018, the date of the last search conducted. The string was applied directly in the digital libraries El Compendex and PubMed. The advanced mode was used for Scopus, and the search was specified for title, abstract, and keywords. Likewise, the advanced mode was used for IEEE Xplore, but the search was specified for full text. The start date was defined considering that, during the tests with the string, only one article published before 2010 was found, and it did not fulfill the criteria to be included in this study. Furthermore, the use of the snowballing technique should retrieve the most relevant papers published before this date.
After the first search, the development of the study took longer than expected. Therefore, a second search was performed to keep the study updated. We searched for articles published between December 7, 2018, and April 3, 2020, the last search date. The string was applied to the digital libraries using the same process as in the first search. The only exception was the IEEE Xplore, for which we used the command search instead of the advanced mode, and the search was specified for full text and metadata.
Study selection
First, a triage process was applied to the non-duplicate publications. Three authors (C.P.S., E.A.C., I.D.R.) submitted each paper to a selection based on specific inclusion and exclusion criteria previously defined (see Supplementary Table S8). However, some exclusion criteria needed to be created or adjusted during the selection for better classification.
Generally speaking, we included publications that used ML techniques to classify subjects between ASD and TD based only on rs-fMRI or based on rs-fMRI together with other types of data. Guidelines for applying ML techniques in the classification of brain images were included if they presented classification results regarding rs-fMRI and ASD. Also, publications focused on distinguishing ASD from other disorders were included if they classified ASD vs. TD.
The criteria were applied based on the abstracts of the studies. When it was not sufficient, a superficial reading of the entire article was carried out—it is worth noting that this was conducted only for a pre-selection of the articles. The papers were selected if at least one of the researchers concluded it should be. Then, the same three researchers performed a new assessment to confirm the selection. In this step, each paper selected was read carefully to determine if it fulfilled three requirements: (1) used rs-fMRI data; (2) performed a classification between ASD and TD; (3) the classification was performed using an ML technique. If at least one of those requirements was not fulfilled, the article was excluded from the study.
Data extraction
Three authors (C.P.S., E.A.C., I.D.R.) used a standardized data extraction sheet to collect data from all included studies (see Supplementary Table S4 online). We extracted the source and type of the data, sample size, if the study included both males and females, average age and FIQ of the subjects, preprocessing steps, feature extraction and selection procedures, the validation process, classifiers used, outcomes reported, main results (accuracy, sensitivity, specificity, and measures of TP, TN, FP, and FN), other tests performed, and important brain areas.
We extracted/calculated only one result from each independent sample in a study. Since the majority of the publications presented multiple results from different tests, the main results were selected according to the following criteria: results from the classification method proposed in the article were prioritized; results presenting enough information to conduct the meta-analysis (measures of TP, TN, FP, and FN, number of ASD and TD subjects in the test set) were prioritized; results using only rs-fMRI data were prioritized; results using a hold-out test set, an inter-site (leave-one-site-out) approach or a train/validation/test procedure were prioritized; tests using larger samples were prioritized; if the study presented results using different numbers of folds for the cross-validation, tenfold was prioritized (the most common approach); finally, the results with higher accuracy were prioritized.
Snowballing
Snowballing means systematically searching for primary studies based on references to and from other studies. Since we limited our research to the date of the last search conducted, we only performed a backward snowballing31. The goal was to broaden the scope of this work and include the maximum number of related articles, especially those before 2010, if any.
As the selected articles were analyzed, we looked for references that could be included in this systematic review according to the inclusion/exclusion criteria. It resulted in many duplicated articles, so we decided not to re-apply the snowballing technique. Also, the new articles found went through the same selection process presented before.
Quality assessment
One author (C.P.S.) assessed methodological quality using the QUADAS-232—the currently recommended tool for a systematic review of diagnostic accuracy studies147,159.
QUADAS-2 assesses study quality in four key domains: patient selection, index test, reference standard, and flow and timing. All the domains are assessed in terms of RoB, and the first three are also evaluated in terms of concerns about applicability (the concern that a study does not match the review question)32.
The tool was tailored by two authors (C.P.S., E.A.C.). After defining the signaling questions and review-specific guidance, both authors applied the tool using five articles. The answers to the signaling questions and the risks of bias/applicability were compared, and any disagreement was discussed to reach a consensus. We maintained the core signaling questions for each domain as defined by the QUADAS-232 except for the index test domain, for which we defined the level of RoB by reviewing the validation process used to obtain the classification accuracy.
Studies using a nested cross-validation procedure or a hold-out set for testing the proposed classification algorithms were assessed as having low RoB. If a study presented their results per number of features, per atlas used or similar, but the best model was not applied to an independent set, there was a high RoB. Studies using a cross-validation scheme without providing any further information were considered as having an unclear RoB. The applicability concerns of the same domain were based on the type of data used. Studies using other data types beyond rs-fMRI were assessed as having high concerns unless the data used would be available in a real application (e.g., age or gender).
The QUADAS-2 tool was applied two times. In the first, all articles were assessed as a whole. In the second, only the papers selected for the meta-analysis were evaluated, considering the main results (as defined in “Data extraction” section) used for the statistical analysis. In both cases, the information used to reach the judgment of each of the domains was recorded to make the rating transparent and facilitate discussion.
Statistical analysis
Studies were eligible for inclusion in the quantitative meta-analysis if TP, TN, FP, and FN measures were available or if the data allowed for their calculation. Therefore, we excluded studies that: did not report sensitivity/specificity (nor equivalent metrics); did not present enough information regarding the number of TD and ASD subjects on the test set. We also chose not to include articles with results reported only through bar charts nor RF studies without enough information on their OOB results (see “General study characteristics” section for more details). However, those three articles were included in a sensitivity analysis. The TP/TN/FP/FN values were extracted or calculated from each independent sample in a study according to the criteria defined in “Data extraction” section.
To avoid bias, handling sample overlap between the studies is necessary, possibly excluding samples with considerable overlap. However, the majority of the studies selected in this review extracted their samples from the ABIDE database. Thereby, we have a lot of potential overlapping samples. At the same time, there is little information concerning the exact individuals used in each study to conclude the real extent of the overlap. Excluding all the potential overlapping samples would make it difficult to perform a meta-analysis since only a few results would remain. Furthermore, we can consider that the studies vary considerably regarding characteristics such as the preprocessing, features, and classification techniques used. Thus,this overlap could not be accounted for and we decided to use all the results regardless of it.
The statistical analysis was performed using the open-source package mada126 version 0.5.10 in R Statistics125 version 4.1.1. A coupled forest plot of sensitivity and specificity was created using RevMan version 5.3160. SROC curves, summary estimates of sensitivity and specificity, and the corresponding 95% CIs were calculated by the bivariate model of Reitsma et al.161. Prediction region, AUC, and pAUC were also obtained. Studies that were visually deviant from the 95% prediction region on the SROC curves were considered heterogeneous162.
Subgroup analysis and bivariate meta-regression with potential covariables were performed to reduce any heterogeneity noted between the studies. The ML technique used, year of publication, sample size, type of data, source of the sample, atlas used, number of ROIs, QUADAS-2 results, type of features, and sex, IQ, and age of the subjects were investigated. Knowing that the bivariate model has five parameters162, we considered the minimum number of studies to justify a separate meta-analysis. All tests were based on a 2-sided significance level of .
Since we conducted many tests of significance, we applied the Bonferroni correction163 to account for multiple comparisons. We considered different corrections for different families of tests164. Therefore, for subgroup analysis (30 tests) the corrected significance level is while for meta-regression (13 tests) the corrected significance level is .
In sensitivity analysis, three studies that were initially excluded from the meta-analysis were included to verify the robustness of the results. Also, we investigated the effect of the age of the subjects considering different adulthood thresholds (18–21 years old).
Publication bias was not assessed in our analysis, as there are currently no statistically adequate models in the field of meta-analysis of DTA studies, and further research is required162.
Supplementary Information
Acknowledgements
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brazil (CAPES) - Finance Code 001, and the Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG) (APQ-01565-18).
Author contributions
C.P.S. designed this study. C.P.S., E.A.C., and I.D.R. were involved in the selection and data extraction processes. C.P.S. performed all the statistical analysis. C.P.S. analyzed the results. C.P.S. drafted this manuscript. E.A.C., I.D.R., G.S.B., A.D.S., and L.L.B. reviewed and suggested modifications and increments for this work. G.S.B. and A.D.S. mentored the development of this study.
Data availability
The datasets analyzed during the current study are available in the ABIDE I and ABIDE II repositories, https://fcon_1000.projects.nitrc.org/indi/abide/.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-09821-6.
References
- 1.Rapin I, Tuchman RF. Autism: Definition, neurobiology, screening, diagnosis. Pediatr. Clin. N. Am. 2008;55:1129–1146. doi: 10.1016/j.pcl.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 2.Hahler E-M, Elsabbagh M. Autism: A global perspective. Curr. Dev. Disord. Rep. 2015;2:58–64. doi: 10.1007/s40474-014-0033-3. [DOI] [Google Scholar]
- 3.American Psychiatric Pub . Diagnostic and Statistical Manual of Mental Disorders. 5. American Psychiatric Pub; 2013. [Google Scholar]
- 4.Geschwind DH, State MW. Gene hunting in autism spectrum disorder: On the path to precision medicine. The Lancet Neurol. 2015;14:1109–1120. doi: 10.1016/S1474-4422(15)00044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jahedi A, Nasamran CA, Faires B, Fan J, Müller R-A. Distributed intrinsic functional connectivity patterns predict diagnostic status in large autism cohort. Brain Connect. 2017;7:515–525. doi: 10.1089/brain.2017.0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang C, Geng H, Liu W, Zhang G. Prenatal, perinatal, and postnatal factors associated with autism: A meta-analysis. Medicine. 2017;96:e6696. doi: 10.1097/MD.0000000000006696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hertz-Picciotto I, et al. The charge study: An epidemiologic investigation of genetic and environmental factors contributing to autism. Environ. Health Perspect. 2006;114:1119–1125. doi: 10.1289/ehp.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sandin S, et al. The heritability of autism spectrum disorder. JAMA. 2017;318:1182–1184. doi: 10.1001/jama.2017.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carvalho EA, Santana CP, Rodrigues ID, Lacerda L, Bastos GS. Hidden Markov models to estimate the probability of having autistic children. IEEE Access. 2020;8:99540–99551. doi: 10.1109/ACCESS.2020.2997334. [DOI] [Google Scholar]
- 10.Elsabbagh M, et al. Global prevalence of autism and other pervasive developmental disorders. Autism Res. 2012;5:160–179. doi: 10.1002/aur.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayes SA, Watson SL. The impact of parenting stress: A meta-analysis of studies comparing the experience of parenting stress in parents of children with and without autism spectrum disorder. J. Autism Dev. Disord. 2013;43:629–642. doi: 10.1007/s10803-012-1604-y. [DOI] [PubMed] [Google Scholar]
- 12.Durkin MS, et al. Autism screening and diagnosis in low resource settings: challenges and opportunities to enhance research and services worldwide. Autism Res. 2015;8:473–476. doi: 10.1002/aur.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Webb SJ, Jones EJ, Kelly J, Dawson G. The motivation for very early intervention for infants at high risk for autism spectrum disorders. Int. J. Speech Lang. Pathol. 2014;16:36–42. doi: 10.3109/17549507.2013.861018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogers SJ, et al. Autism treatment in the first year of life: A pilot study of infant start, a parent-implemented intervention for symptomatic infants. J. Autism Dev. Disord. 2014;44:2981–2995. doi: 10.1007/s10803-014-2202-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rutter M, LeCouteur A, Lord C. Autism Diagnostic Interview Revised (adi-r) Western Psychological Services; 2003. [Google Scholar]
- 16.Lord C, et al. Autism Diagnostic Observation Schedule, (ados-2) Modules 1–4. Western Psychological Services; 2012. [Google Scholar]
- 17.Kamp-Becker I, et al. Diagnostic accuracy of the ados and ados-2 in clinical practice. Eur. Child Adolesc. Psychiatry. 2018;27:1193–1207. doi: 10.1007/s00787-018-1143-y. [DOI] [PubMed] [Google Scholar]
- 18.Falkmer T, Anderson K, Falkmer M, Horlin C. Diagnostic procedures in autism spectrum disorders: A systematic literature review. Eur. Child Adolesc. Psychiatry. 2013;22:329–340. doi: 10.1007/s00787-013-0375-0. [DOI] [PubMed] [Google Scholar]
- 19.Kassraian-Fard P, Matthis C, Balsters JH, Maathuis MH, Wenderoth N. Promises, pitfalls, and basic guidelines for applying machine learning classifiers to psychiatric imaging data, with autism as an example. Front. Psych. 2016;7:177. doi: 10.3389/fpsyt.2016.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Litjens G, et al. A survey on deep learning in medical image analysis. Med. Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 21.Matthews PM, Jezzard P. Functional magnetic resonance imaging. J. Neurol. Neurosurg. Psychiatry. 2004;75:6–12. [PMC free article] [PubMed] [Google Scholar]
- 22.Abraham A, et al. Deriving reproducible biomarkers from multi-site resting-state data: An autism-based example. Neuroimage. 2017;147:736–745. doi: 10.1016/j.neuroimage.2016.10.045. [DOI] [PubMed] [Google Scholar]
- 23.Di Martino A, et al. The autism brain imaging data exchange: Towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol. Psychiatry. 2014;19:659–667. doi: 10.1038/mp.2013.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Martino A, et al. Enhancing studies of the connectome in autism using the autism brain imaging data exchange II. Sci. Data. 2017;4:1–15. doi: 10.1038/sdata.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sundermann B, Beverborg MO, Pfleiderer B. Toward literature-based feature selection for diagnostic classification: A meta-analysis of resting-state fmri in depression. Front. Hum. Neurosci. 2014;8:692. doi: 10.3389/fnhum.2014.00692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tahmasian M, et al. A systematic review on the applications of resting-state fmri in Parkinson’s disease: Does dopamine replacement therapy play a role? Cortex. 2015;73:80–105. doi: 10.1016/j.cortex.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 27.Milham MP, Fair D, Mennes M, Mostofsky SH, et al. The adhd-200 consortium: A model to advance the translational potential of neuroimaging in clinical neuroscience. Front. Syst. Neurosci. 2012;6:62. doi: 10.3389/fnsys.2012.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hull JV, et al. Resting-state functional connectivity in autism spectrum disorders: A review. Front. Psychiatry. 2017;7:205. doi: 10.3389/fpsyt.2016.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dosenbach NU, et al. Prediction of individual brain maturity using fmri. Science. 2010;329:1358–1361. doi: 10.1126/science.1194144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plitt M, Barnes KA, Martin A. Functional connectivity classification of autism identifies highly predictive brain features but falls short of biomarker standards. NeuroImage Clin. 2015;7:359–366. doi: 10.1016/j.nicl.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wohlin C, et al. Experimentation in Software Engineering. Springer; 2012. [Google Scholar]
- 32.Whiting PF, et al. Quadas-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011;155:529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 33.Khosla, M., Jamison, K., Kuceyeski, A. & Sabuncu, M. 3d convolutional neural networks for classification of functional connectomes. Preprint at http://arxiv.org/abs/1806.04209 (2018). [DOI] [PMC free article] [PubMed]
- 34.Bi X-A, et al. Analysis of asperger syndrome using genetic-evolutionary random support vector machine cluster. Front. Physiol. 2018;9:1646. doi: 10.3389/fphys.2018.01646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crimi, A., Dodero, L., Murino, V. & Sona, D. Case-control discrimination through effective brain connectivity. In 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), 970–973. 10.1109/ISBI.2017.7950677 (2017).
- 36.Bi X-A, Wang Y, Shu Q, Sun Q, Xu Q. Classification of autism spectrum disorder using random support vector machine cluster. Front. Genet. 2018;9:18. doi: 10.3389/fgene.2018.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao, D. & Lu, H. Classify autism and control based on deep learning and community structure on resting-state fmri. In 2018 Tenth International Conference on Advanced Computational Intelligence (ICACI), 289–294 (IEEE, 2018).
- 38.Dvornek, N. C., Ventola, P. & Duncan, J. S. Combining phenotypic and resting-state fmri data for autism classification with recurrent neural networks. In 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), 725–728 (IEEE, 2018). [DOI] [PMC free article] [PubMed]
- 39.Chaitra, N. & Vijaya, P. A. Comparing univalent and bivalent brain functional connectivity measures using machine learning. In 2017 Fourth International Conference on Signal Processing, Communication and Networking (ICSCN), 1–5. 10.1109/ICSCN.2017.8085741 (2017).
- 40.Guo X, et al. Diagnosing autism spectrum disorder from brain resting-state functional connectivity patterns using a deep neural network with a novel feature selection method. Front. Neurosci. 2017;11:460. doi: 10.3389/fnins.2017.00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dvornek, N. C., Ventola, P., Pelphrey, K. A. & Duncan, J. S. Identifying autism from resting-state fmri using long short-term memory networks. In International Workshop on Machine Learning in Medical Imaging, 362–370 (Springer, 2017). [DOI] [PMC free article] [PubMed]
- 42.Subbaraju V, Suresh MB, Sundaram S, Narasimhan S. Identifying differences in brain activities and an accurate detection of autism spectrum disorder using resting state functional-magnetic resonance imaging: A spatial filtering approach. Med. Image Anal. 2017;35:375–389. doi: 10.1016/j.media.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 43.Dodero, L., Minh, H. Q., Biagio, M. S., Murino, V. & Sona, D. Kernel-based classification for brain connectivity graphs on the riemannian manifold of positive definite matrices. In 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), 42–45. 10.1109/ISBI.2015.7163812 (2015).
- 44.Zhou Y, Yu F, Duong T. Multiparametric mri characterization and prediction in autism spectrum disorder using graph theory and machine learning. PLoS ONE. 2014;9:e90405. doi: 10.1371/journal.pone.0090405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu, Y. et al. Reveal consistent spatial-temporal patterns from dynamic functional connectivity for autism spectrum disorder identification. In International Conference on Medical Image Computing and Computer-Assisted Intervention, 106–114 (Springer, 2016). [DOI] [PMC free article] [PubMed]
- 46.Sartipi, S., Kalbkhani, H. & Shayesteh, M. G. Ripplet ii transform and higher order cumulants from r-fmri data for diagnosis of autism. In 2017 10th International Conference on Electrical and Electronics Engineering (ELECO), 557–560 (IEEE, 2017).
- 47.Parisot, S. et al. Spectral graph convolutions for population-based disease prediction. In International Conference on Medical Image Computing and Computer-Assisted Intervention, 177–185 (Springer, 2017).
- 48.Gupta S, Rajapakse JC, Welsch RE, Initiative ADN, et al. Ambivert degree identifies crucial brain functional hubs and improves detection of alzheimer’s disease and autism spectrum disorder. NeuroImage Clin. 2020;25:102186. doi: 10.1016/j.nicl.2020.102186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khosla M, Jamison K, Kuceyeski A, Sabuncu MR. Ensemble learning with 3d convolutional neural networks for functional connectome-based prediction. Neuroimage. 2019;199:651–662. doi: 10.1016/j.neuroimage.2019.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sartipi, S., Shayesteh, M. G. & Kalbkhani, H. Diagnosing of autism spectrum disorder based on garch variance series for rs-fmri data. In 2018 9th International Symposium on Telecommunications (IST), 86–90 (IEEE, 2018).
- 51.DSouza, A. M., Abidin, A. Z. & Wismüller, A. Classification of autism spectrum disorder from resting-state fmri with mutual connectivity analysis. In Medical Imaging 2019: Biomedical Applications in Molecular, Structural, and Functional Imaging, vol. 10953, 109531D (International Society for Optics and Photonics, 2019).
- 52.Bi X-A, et al. The genetic-evolutionary random support vector machine cluster analysis in autism spectrum disorder. IEEE Access. 2019;7:30527–30535. doi: 10.1109/ACCESS.2019.2902889. [DOI] [Google Scholar]
- 53.El-Gazzar, A. et al. A hybrid 3dcnn and 3dc-lstm based model for 4d spatio-temporal fmri data: An abide autism classification study. In OR 2.0 Context-Aware Operating Theaters and Machine Learning in Clinical Neuroimaging, 95–102 (Springer, 2019).
- 54.Zhang, M. et al. Comparison of neural networks’ performance in early screening of autism spectrum disorders under two mri principles. In 2019 International Conference on Networking and Network Applications (NaNA), 338–343 (IEEE, 2019).
- 55.Mostafa S, Tang L, Wu F-X. Diagnosis of autism spectrum disorder based on eigenvalues of brain networks. IEEE Access. 2019;7:128474–128486. doi: 10.1109/ACCESS.2019.2940198. [DOI] [Google Scholar]
- 56.Zhao, Y., Dai, H., Zhang, W., Ge, F. & Liu, T. Two-stage spatial temporal deep learning framework for functional brain network modeling. In 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), 1576–1580 (IEEE, 2019).
- 57.Mellema, C., Treacher, A., Nguyen, K. & Montillo, A. Multiple deep learning architectures achieve superior performance diagnosing autism spectrum disorder using features previously extracted from structural and functional mri. In 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), 1891–1895 (IEEE, 2019). [DOI] [PMC free article] [PubMed]
- 58.Anirudh, R. & Thiagarajan, J. J. Bootstrapping graph convolutional neural networks for autism spectrum disorder classification. In ICASSP 2019-2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), 3197–3201 (IEEE, 2019).
- 59.Bengs, M., Gessert, N. & Schlaefer, A. 4d spatio-temporal deep learning with 4d fmri data for autism spectrum disorder classification. Preprint at http://arxiv.org/abs/2004.10165 (2020).
- 60.Sherkatghanad Z, et al. Automated detection of autism spectrum disorder using a convolutional neural network. Front. Neurosci. 2019;13:1325. doi: 10.3389/fnins.2019.01325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sairam K, Naren J, Vithya G, Srivathsan S. Computer aided system for autism spectrum disorder using deep learning methods. Int. J. Psychosoc. Rehabil. 2019;23:01. [Google Scholar]
- 62.Rajesh G, Pannirselvam S. Lucid ant colony optimization based denoiser for effective autism spectrum disorder classification. Int. J. Adv. Sci. Technol. 2019;28:865–876. [Google Scholar]
- 63.Bhaumik R, Pradhan A, Das S, Bhaumik DK. Predicting autism spectrum disorder using domain-adaptive cross-site evaluation. Neuroinformatics. 2018;16:197–205. doi: 10.1007/s12021-018-9366-0. [DOI] [PubMed] [Google Scholar]
- 64.Li, J., Ji, J., Liang, Y., Zhang, X. & Wang, Z. Deep forest with cross-shaped window scanning mechanism to extract topological features. In 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), 688–691 (IEEE, 2019).
- 65.Mahanand, B. S., Vigneshwaran, S., Suresh, S. & Sundararajan, N. An enhanced effect-size thresholding method for the diagnosis of autism spectrum disorder using resting state functional mri. In 2016 Second International Conference on Cognitive Computing and Information Processing (CCIP), 1–6. 10.1109/CCIP.2016.7802874 (2016).
- 66.Vigneshwaran, S., Mahanand, B. S., Suresh, S. & Sundararajan, N. Using regional homogeneity from functional mri for diagnosis of asd among males. In 2015 International Joint Conference on Neural Networks (IJCNN), 1–8. 10.1109/IJCNN.2015.7280562 (2015).
- 67.Dekhil O, et al. A personalized autism diagnosis cad system using a fusion of structural mri and resting-state functional mri data. Front. Psychiatry. 2019;10:392. doi: 10.3389/fpsyt.2019.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zu, C. et al. Identifying high order brain connectome biomarkers via learning on hypergraph. In International Workshop on Machine Learning in Medical Imaging, 1–9 (Springer, 2016). [DOI] [PMC free article] [PubMed]
- 69.Huang, F. et al. Multi-template based auto-weighted adaptive structural learning for asd diagnosis. In International Workshop on Machine Learning in Medical Imaging, 516–524 (Springer, 2019).
- 70.Chen CP, et al. Diagnostic classification of intrinsic functional connectivity highlights somatosensory, default mode, and visual regions in autism. NeuroImage Clin. 2015;8:238–245. doi: 10.1016/j.nicl.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dekhil, O. et al. Using resting state functional mri to build a personalized autism diagnosis system. In Biomedical Imaging (ISBI 2018), 2018 IEEE 15th International Symposium on, 1381–1385 (IEEE, 2018). [DOI] [PMC free article] [PubMed]
- 72.Iidaka T. Resting state functional magnetic resonance imaging and neural network classified autism and control. Cortex. 2015;63:55–67. doi: 10.1016/j.cortex.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 73.Kam T-E, Suk H-I, Lee S-W. Multiple functional networks modeling for autism spectrum disorder diagnosis. Hum. Brain Mapp. 2017;38:5804–5821. doi: 10.1002/hbm.23769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li H, Parikh NA, He L. A novel transfer learning approach to enhance deep neural network classification of brain functional connectomes. Front. Neurosci. 2018;12:491. doi: 10.3389/fnins.2018.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Price, T., Wee, C.-Y., Gao, W. & Shen, D. Multiple-network classification of childhood autism using functional connectivity dynamics. In International Conference on Medical Image Computing and Computer-Assisted Intervention, 177–184 (Springer, 2014). [DOI] [PubMed]
- 76.Zhao, Y., Ge, F., Zhang, S. & Liu, T. 3d deep convolutional neural network revealed the value of brain network overlap in differentiating autism spectrum disorder from healthy controls. In International Conference on Medical Image Computing and Computer-Assisted Intervention, 172–180 (Springer, 2018).
- 77.Aghdam MA, Sharifi A, Pedram MM. Diagnosis of autism spectrum disorders in young children based on resting-state functional magnetic resonance imaging data using convolutional neural networks. J. Digit. Imaging. 2019;32:899–918. doi: 10.1007/s10278-019-00196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brahim, A., El Hassani, M. H. & Farrugia, N. Classification of autism spectrum disorder through the graph fourier transform of fmri temporal signals projected on structural connectome. In International Conference on Computer Analysis of Images and Patterns, 45–55 (Springer, 2019).
- 79.Dammu, P. S. & Bapi, R. S. Employing temporal properties of brain activity for classifying autism using machine learning. In International Conference on Pattern Recognition and Machine Intelligence, 193–200 (Springer, 2019).
- 80.Kazeminejad A, Sotero RC. Topological properties of resting-state fmri functional networks improve machine learning-based autism classification. Front. Neurosci. 2019;12:1018. doi: 10.3389/fnins.2018.01018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lanka P, et al. Supervised machine learning for diagnostic classification from large-scale neuroimaging datasets. Brain Imaging Behav. 2019;14:1–39. doi: 10.1007/s11682-019-00191-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Spera G, et al. Evaluation of altered functional connections in male children with autism spectrum disorders on multiple-site data optimized with machine learning. Front. Psychiatry. 2019;10:620. doi: 10.3389/fpsyt.2019.00620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martial EET, Hu L, Yuqing S. Characterising and predicting autism spectrum disorder by performing resting-state functional network community pattern analysis. Front. Hum. Neurosci. 2019;13:203. doi: 10.3389/fnhum.2019.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang C, Xiao Z, Wu J. Functional connectivity-based classification of autism and control using svm-rfecv on rs-fmri data. Phys. Med. 2019;65:99–105. doi: 10.1016/j.ejmp.2019.08.010. [DOI] [PubMed] [Google Scholar]
- 85.Wang, J. et al. Interpretable feature learning using multi-output Takagi-Sugeno-Kang fuzzy system for multi-center asd diagnosis. In International Conference on Medical Image Computing and Computer-Assisted Intervention, 790–798 (Springer, 2019).
- 86.Yang, X., Islam, M. S. & Khaled, A. A. Functional connectivity magnetic resonance imaging classification of autism spectrum disorder using the multisite abide dataset. In 2019 IEEE EMBS International Conference on Biomedical & Health Informatics (BHI), 1–4 (IEEE, 2019).
- 87.Yuan, D., Zhu, L. & Huang, H. Prediction of autism spectrum disorder based on imbalanced resting-state fmri data using clustering oversampling. In Tenth International Conference on Signal Processing Systems, vol. 11071, 110710W (International Society for Optics and Photonics, 2019).
- 88.Aghdam MA, Sharifi A, Pedram MM. Combination of rs-fmri and smri data to discriminate autism spectrum disorders in young children using deep belief network. J. Dig. Imaging. 2018;31:1–9. doi: 10.1007/s10278-017-0006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Emerson RW, et al. Functional neuroimaging of high-risk 6-month-old infants predicts a diagnosis of autism at 24 months of age. Sci. Transl. Med. 2017;9:2882. doi: 10.1126/scitranslmed.aag2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sen B, Borle NC, Greiner R, Brown MR. A general prediction model for the detection of adhd and autism using structural and functional mri. PLoS ONE. 2018;13:e0194856. doi: 10.1371/journal.pone.0194856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tolan, E. & Isik, Z. Graph theory based classification of brain connectivity network for autism spectrum disorder. In International Conference on Bioinformatics and Biomedical Engineering, 520–530 (Springer, 2018).
- 92.Eill A, et al. Functional connectivities are more informative than anatomical variables in diagnostic classification of autism. Brain Connect. 2019;9:604–612. doi: 10.1089/brain.2019.0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mastrovito D, Hanson C, Hanson SJ. Differences in atypical resting-state effective connectivity distinguish autism from schizophrenia. NeuroImage Clin. 2018;18:367–376. doi: 10.1016/j.nicl.2018.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Heinsfeld AS, Franco AR, Craddock RC, Buchweitz A, Meneguzzi F. Identification of autism spectrum disorder using deep learning and the abide dataset. NeuroImage Clin. 2018;17:16–23. doi: 10.1016/j.nicl.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jun, E. & Suk, H.-I. Region-wise stochastic pattern modeling for autism spectrum disorder identification and temporal dynamics analysis. In International Workshop on Connectomics in Neuroimaging, 143–151 (Springer, 2017).
- 96.Wong, E., Anderson, J. S., Zielinski, B. A. & Fletcher, P. T. Riemannian regression and classification models of brain networks applied to autism. In International Workshop on Connectomics in Neuroimaging, 78–87 (Springer, 2018). [DOI] [PMC free article] [PubMed]
- 97.Zhu, Y., Zhu, X., Kim, M., Yan, J. & Wu, G. A tensor statistical model for quantifying dynamic functional connectivity. In International Conference on Information Processing in Medical Imaging, 398–410 (Springer, 2017). [DOI] [PMC free article] [PubMed]
- 98.Ren, Y. & Wang, S. Exploring functional connectivity biomarker in autism using group-wise sparse representation. In Multimodal Brain Image Analysis and Mathematical Foundations of Computational Anatomy, 21–29 (Springer, 2019).
- 99.Dekhil, O. et al. Identifying personalized autism related impairments using resting functional mri and ados reports. In International Conference on Medical Image Computing and Computer-Assisted Intervention, 240–248 (Springer, 2018).
- 100.Ren, Y. et al. Identifying autism biomarkers in default mode network using sparse representation of resting-state fmri data. In 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), 1278–1281. 10.1109/ISBI.2016.7493500 (2016).
- 101.Chen, Z., Ji, J. & Liang, Y. Convolutional neural network with an element-wise filter to classify dynamic functional connectivity. In 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), 643–646 (IEEE, 2019).
- 102.El Gazzar, A., Cerliani, L., van Wingen, G. & Thomas, R. M. Simple 1-d convolutional networks for resting-state fmri based classification in autism. In 2019 International Joint Conference on Neural Networks (IJCNN), 1–6 (IEEE, 2019).
- 103.Huang, F. et al. Sparse low-rank constrained adaptive structure learning using multi-template for autism spectrum disorder diagnosis. In 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), 1555–1558 (IEEE, 2019).
- 104.Sidhu G. Locally linear embedding and fmri feature selection in psychiatric classification. IEEE J. Transl. Eng. Health Med. 2019;7:1–11. doi: 10.1109/JTEHM.2019.2936348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang C, Xiao Z, Wang B, Wu J. Identification of autism based on svm-rfe and stacked sparse auto-encoder. IEEE Access. 2019;7:118030–118036. doi: 10.1109/ACCESS.2019.2936639. [DOI] [Google Scholar]
- 106.Wang M, et al. Identifying autism spectrum disorder with multi-site fmri via low-rank domain adaptation. IEEE Trans. Med. Imaging. 2019;39:644–655. doi: 10.1109/TMI.2019.2933160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang J, et al. Sparse multiview task-centralized ensemble learning for asd diagnosis based on age- and sex-related functional connectivity patterns. IEEE Trans. Cybern. 2018;48:1–14. doi: 10.1109/TCYB.2018.2839693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhou D, Wang J, Jiang B, Guo H, Li Y. Multi-task multi-view learning based on cooperative multi-objective optimization. IEEE Access. 2018;6:19465–19477. doi: 10.1109/ACCESS.2017.2777888. [DOI] [Google Scholar]
- 109.Chen H, et al. Multivariate classification of autism spectrum disorder using frequency-specific resting-state functional connectivity—A multi-center study. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2016;64:1–9. doi: 10.1016/j.pnpbp.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 110.Dodero, L., Sambataro, F., Murino, V. & Sona, D. Kernel-based analysis of functional brain connectivity on grassmann manifold. In International Conference on Medical Image Computing and Computer-Assisted Intervention, 604–611 (Springer, 2015).
- 111.Wee C-Y, Yap P-T, Shen D. Diagnosis of autism spectrum disorders using temporally distinct resting-state functional connectivity networks. CNS Neurosci. Therap. 2016;22:212–219. doi: 10.1111/cns.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yahata N, et al. A small number of abnormal brain connections predicts adult autism spectrum disorder. Nat. Commun. 2016;7:11254. doi: 10.1038/ncomms11254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bernas A, Aldenkamp AP, Zinger S. Wavelet coherence-based classifier: A resting-state functional mri study on neurodynamics in adolescents with high-functioning autism. Comput. Methods Progr. Biomed. 2018;154:143–151. doi: 10.1016/j.cmpb.2017.11.017. [DOI] [PubMed] [Google Scholar]
- 114.Huang H, et al. Enhancing the representation of functional connectivity networks by fusing multi-view information for autism spectrum disorder diagnosis. Hum. Brain Mapp. 2019;40:833–854. doi: 10.1002/hbm.24415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kazeminejad A, Sotero RC. The importance of anti-correlations in graph theory based classification of autism spectrum disorder. BioRxiv. 2019 doi: 10.1101/557512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Saeed F, Eslami T, Mirjalili V, Fong A, Laird A. Asd-diagnet: A hybrid learning approach for detection of autism spectrum disorder using fmri data. Front. Neuroinform. 2019;13:70. doi: 10.3389/fninf.2019.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tejwani, R., Liska, A., You, H., Reinen, J. & Das, P. Autism classification using brain functional connectivity dynamics and machine learning. Preprint at http://arxiv.org/abs/1712.08041 (2017).
- 118.Xing, X., Ji, J. & Yao, Y. Convolutional neural network with element-wise filters to extract hierarchical topological features for brain networks. In 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), 780–783 (IEEE, 2018).
- 119.Ghiassian S, Greiner R, Jin P, Brown MR. Using functional or structural magnetic resonance images and personal characteristic data to identify adhd and autism. PLoS ONE. 2016;11:e0166934. doi: 10.1371/journal.pone.0166934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Uddin LQ, et al. Salience network-based classification and prediction of symptom severity in children with autism. JAMA Psychiatry. 2013;70:869–879. doi: 10.1001/jamapsychiatry.2013.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang J, et al. Multi-task diagnosis for autism spectrum disorders using multi-modality features: A multi-center study. Hum. Brain Mapp. 2017;38:3081–3097. doi: 10.1002/hbm.23575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Brown JA, Rudie JD, Bandrowski A, Van Horn JD, Bookheimer SY. The ucla multimodal connectivity database: A web-based platform for brain connectivity matrix sharing and analysis. Front. Neuroinform. 2012;6:28. doi: 10.3389/fninf.2012.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tzourio-Mazoyer N, et al. Automated anatomical labeling of activations in spm using a macroscopic anatomical parcellation of the mni mri single-subject brain. Neuroimage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- 124.Craddock RC, James GA, Holtzheimer PE, III, Hu XP, Mayberg HS. A whole brain fmri atlas generated via spatially constrained spectral clustering. Hum. Brain Mapp. 2012;33:1914–1928. doi: 10.1002/hbm.21333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Team, R. C. R: A Language and Environment for Statistical Computing. (R Foundation for Statistical Computing, 2012). https://www.r-project.org/.
- 126.Doebler P, Holling H. Meta-analysis of diagnostic accuracy with mada. R Packag. 2015;1:15. doi: 10.1007/s11336-014-9430-0. [DOI] [PubMed] [Google Scholar]
- 127.Park SH, Han K. Methodologic guide for evaluating clinical performance and effect of artificial intelligence technology for medical diagnosis and prediction. Radiology. 2018;286:800–809. doi: 10.1148/radiol.2017171920. [DOI] [PubMed] [Google Scholar]
- 128.Russell SJ, Norvig P. Artificial Intelligence: A Modern Approach. Pearson Education Limited; 2016. [Google Scholar]
- 129.James G, Witten D, Hastie T, Tibshirani R. An Introduction to Statistical Learning. Springer; 2013. [Google Scholar]
- 130.Arbabshirani MR, Plis S, Sui J, Calhoun VD. Single subject prediction of brain disorders in neuroimaging: Promises and pitfalls. Neuroimage. 2017;145:137–165. doi: 10.1016/j.neuroimage.2016.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pereira F, Mitchell T, Botvinick M. Machine learning classifiers and fmri: A tutorial overview. Neuroimage. 2009;45:S199–S209. doi: 10.1016/j.neuroimage.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tillmann J, et al. Evaluating sex and age differences in adi-r and ados scores in a large european multi-site sample of individuals with autism spectrum disorder. J. Autism Dev. Disord. 2018;48:2490–2505. doi: 10.1007/s10803-018-3510-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Van Wijngaarden-Cremers PJ, et al. Gender and age differences in the core triad of impairments in autism spectrum disorders: A systematic review and meta-analysis. J. Autism Dev. Disord. 2014;44:627–635. doi: 10.1007/s10803-013-1913-9. [DOI] [PubMed] [Google Scholar]
- 134.Mayes SD, Calhoun SL. Impact of iq, age, ses, gender, and race on autistic symptoms. Res. Autism Spectrum Disord. 2011;5:749–757. doi: 10.1016/j.rasd.2010.09.002. [DOI] [Google Scholar]
- 135.Loomes R, Hull L, Mandy WPL. What is the male-to-female ratio in autism spectrum disorder? A systematic review and meta-analysis. J. Am. Acad. Child Adolesc. Psychiatry. 2017;56:466–474. doi: 10.1016/j.jaac.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 136.Maenner MJ, Shaw KA, Baio J, et al. Prevalence of autism spectrum disorder among children aged 8 years-autism and developmental disabilities monitoring network, 11 sites, united states, 2016. MMWR Surveill. Summ. 2020;69:1. doi: 10.15585/mmwr.ss6904a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jensen CM, Steinhausen H-C, Lauritsen MB. Time trends over 16 years in incidence-rates of autism spectrum disorders across the lifespan based on nationwide danish register data. J. Autism Dev. Disord. 2014;44:1808–1818. doi: 10.1007/s10803-014-2053-6. [DOI] [PubMed] [Google Scholar]
- 138.Fombonne E. Epidemiology of pervasive developmental disorders. Pediatr. Res. 2009;65:591–598. doi: 10.1203/PDR.0b013e31819e7203. [DOI] [PubMed] [Google Scholar]
- 139.Lai M-C, Lombardo MV, Auyeung B, Chakrabarti B, Baron-Cohen S. Sex/gender differences and autism: Setting the scene for future research. J. Am. Acad. Child Adolesc. Psychiatry. 2015;54:11–24. doi: 10.1016/j.jaac.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Russell G, Steer C, Golding J. Social and demographic factors that influence the diagnosis of autistic spectrum disorders. Soc. Psychiatry Psychiatr. Epidemiol. 2011;46:1283–1293. doi: 10.1007/s00127-010-0294-z. [DOI] [PubMed] [Google Scholar]
- 141.Giarelli E, et al. Sex differences in the evaluation and diagnosis of autism spectrum disorders among children. Disabil. Health J. 2010;3:107–116. doi: 10.1016/j.dhjo.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Begeer S, et al. Sex differences in the timing of identification among children and adults with autism spectrum disorders. J. Autism Dev. Disord. 2013;43:1151–1156. doi: 10.1007/s10803-012-1656-z. [DOI] [PubMed] [Google Scholar]
- 143.Karmiloff-Smith A. Challenging the use of adult neuropsychological models for explaining neurodevelopmental disorders: Developed versus develop ing brains: The 40th sir frederick bartlett lecture. Q. J. Exp. Psychol. 2013;66:1–14. doi: 10.1080/17470218.2012.744424. [DOI] [PubMed] [Google Scholar]
- 144.Segall JM, et al. Voxel-based morphometric multisite collaborative study on schizophrenia. Schizophr. Bull. 2009;35:82–95. doi: 10.1093/schbul/sbn150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Biswal BB, et al. Toward discovery science of human brain function. Proc. Natl. Acad. Sci. 2010;107:4734–4739. doi: 10.1073/pnas.0911855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Carp J. The secret lives of experiments: Methods reporting in the fmri literature. Neuroimage. 2012;63:289–300. doi: 10.1016/j.neuroimage.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 147.Kim KW, Lee J, Choi SH, Huh J, Park SH. Systematic review and meta-analysis of studies evaluating diagnostic test accuracy: A practical review for clinical researchers-part I. General guidance and tips. Korean J. Radiol. 2015;16:1175–1187. doi: 10.3348/kjr.2015.16.6.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Jones CM, Ashrafian H, Darzi A, Athanasiou T. Guidelines for diagnostic tests and diagnostic accuracy in surgical research. J. Investig. Surg. 2010;23:57–65. doi: 10.3109/08941930903469508. [DOI] [PubMed] [Google Scholar]
- 149.Fusar-Poli L, et al. Diagnosing asd in adults without id: Accuracy of the ados-2 and the adi-r. J. Autism Dev. Disord. 2017;47:3370–3379. doi: 10.1007/s10803-017-3258-2. [DOI] [PubMed] [Google Scholar]
- 150.Mazefsky CA, Oswald DP. The discriminative ability and diagnostic utility of the ados-g, adi-r, and gars for children in a clinical setting. Autism. 2006;10:533–549. doi: 10.1177/1362361306068505. [DOI] [PubMed] [Google Scholar]
- 151.Hosmer DW, Jr, Lemeshow S, Sturdivant RX. Applied Logistic Regression. Wiley; 2013. [Google Scholar]
- 152.Iakoucheva LM, Muotri AR, Sebat J. Getting to the cores of autism. Cell. 2019;178:1287–1298. doi: 10.1016/j.cell.2019.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wheelwright S, Auyeung B, Allison C, Baron-Cohen S. Defining the broader, medium and narrow autism phenotype among parents using the autism spectrum quotient (aq) Mol. Autism. 2010;1:1–9. doi: 10.1186/2040-2392-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pierce K. Early functional brain development in autism and the promise of sleep fmri. Brain Res. 2011;1380:162–174. doi: 10.1016/j.brainres.2010.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Graham AM, et al. The potential of infant fmri research and the study of early life stress as a promising exemplar. Dev. Cogn. Neurosci. 2015;12:12–39. doi: 10.1016/j.dcn.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bom PR, Rachinger H. A generalized-weights solution to sample overlap in meta-analysis. Res. Synth. Methods. 2020;11:812–832. doi: 10.1002/jrsm.1441. [DOI] [PubMed] [Google Scholar]
- 157.Devillé WL, et al. Conducting systematic reviews of diagnostic studies: Didactic guidelines. BMC Med. Res. Methodol. 2002;2:9. doi: 10.1186/1471-2288-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.American Psychiatric Pub . Diagnostic and Statistical Manual of Mental Disorders. 4. American Psychiatric Pub; 2000. [Google Scholar]
- 159.Biondi-Zoccai G. Diagnostic Meta-Analysis: A Useful Tool for Clinical Decision-Making. Springer; 2018. [Google Scholar]
- 160.Community, C. Review Manager (revman). Version 5.3. https://community.cochrane.org/help/tools-and-software/revman-5/revman-5-download (Accessed 6 January 2020).
- 161.Reitsma JB, et al. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J. Clin. Epidemiol. 2005;58:982–990. doi: 10.1016/j.jclinepi.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 162.Macaskill, P., Gatsonis, C., Deeks, J., Harbord, R. & Takwoingi, Y. Chapter 10: Analysing and presenting results. In Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy Version, Vol. 1 (2010).
- 163.Dunn OJ. Multiple comparisons among means. J. Am. Stat. Assoc. 1961;56:52–64. doi: 10.1080/01621459.1961.10482090. [DOI] [Google Scholar]
- 164.Polanin JR, Pigott TD. The use of meta-analytic statistical significance testing. Res. Synth. Methods. 2015;6:63–73. doi: 10.1002/jrsm.1124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed during the current study are available in the ABIDE I and ABIDE II repositories, https://fcon_1000.projects.nitrc.org/indi/abide/.