Abstract
Background
The consensus molecular subtypes (CMSs) of colorectal cancer (CRC) capture tumor heterogeneity at the gene-expression level. Currently, a restricted number of molecular features are used to guide treatment for CRC. We summarize the evidence on the clinical value of the CMSs.
Methods
We systematically identified studies in Medline and Embase that evaluated the prognostic and predictive value of CMSs in CRC patients. A random-effect meta-analysis was performed on prognostic data. Predictive data were summarized.
Results
In local disease, CMS4 tumors were associated with worse overall survival (OS) compared with CMS1 (hazard ratio [HR] = 3.28, 95% confidence interval = 1.27 to 8.47) and CMS2 cancers (HR = 2.60, 95% confidence interval = 1.93 to 3.50). In metastatic disease, CMS1 consistently had worse survival than CMS2-4 (OS HR range = 0.33-0.55; progression-free survival HR range = 0.53-0.89). Adjuvant chemotherapy in stage II and III CRC was most beneficial for OS in CMS2 and CMS3 (HR range = 0.16-0.45) and not effective in CMS4 tumors. In metastatic CMS4 cancers, an irinotecan-based regimen improved outcome compared with oxaliplatin (HR range = 0.31-0.72). The addition of bevacizumab seemed beneficial in CMS1, and anti-epidermal growth factor receptor therapy improved outcome for KRAS wild-type CMS2 patients.
Conclusions
The CMS classification holds clear potential for clinical use in predicting both prognosis and response to systemic therapy, which seems to be independent of the classifier used. Prospective studies are warranted to support implementation of the CMS taxonomy in clinical practice.
Colorectal cancer (CRC) is the third-most common cancer and the second-leading cause of cancer deaths worldwide (1). CRC is a highly heterogeneous disease with respect to clinical and biological features, resulting in striking differences in disease progression and treatment response (2). This diversity of CRC complicates estimation of prognosis and the optimal timing and selection of treatment regimens for individual patients. Currently, mainly pathological staging by tumor node metastasis (TNM), sidedness, and a few molecular markers, including mismatch repair (MMR) status and RAS and BRAF mutation status, are regularly used in clinical practice to select patients for specific therapies. Most recently, the BEACON regimen (encorafenib [BRAF-inhibitor], binimetinib [MEK-inhibitor], and cetuximab [anti-EGFR]) was found to be effective for BRAF mutant metastatic CRC (mCRC), and immunotherapy (pembrolizumab; anti-PD-1) statistically significantly prolonged progression-free survival (PFS) in the first-line setting of patients with microsatellite instable (MSI) tumors (3,4). However, a clinically significant number of patients within the currently used subgroups for treatment selection do not benefit from the available regimens. For instance, 59% of RAS/BRAF wild-type (wt) patients with left-sided tumors, for whom anti-EGFR treatment is an option, do not respond (stable or progressive disease) (5). Therefore, additional molecular tumor characteristics are needed to further personalize therapies and prevent both under- and overtreatment of CRC patients.
The consensus molecular subtype (CMS) classification is a thoroughly studied and robust stratification strategy for CRC (6). The CMS taxonomy was principally founded on the basis of differences in tumor biology rather than clinical outcomes, thereby capturing the intrinsic biomolecular heterogeneity of CRC. Based on differential gene expression in tumor tissue, comprising both cancer cells as well as the microenvironment, CRC can be divided into 4 subtypes (CMS1-4). CMS1 is the immunogenic subtype, enriched for MSI tumors and BRAF mutations; CMS2 has epithelial characteristics with marked WNT and MYC signaling and high CIN; CMS3 also has epithelial features but less CIN, is enriched for KRAS mutations, and presents with evident metabolic dysregulation; and CMS4 is the mesenchymal subtype with prominent transforming growth factor bèta (TGF-β) activation, stromal invasion, angiogenesis, and an inflammatory, immunosuppressive phenotype (6,7).
Although the CMS taxonomy provides valuable insight into tumor biology and could be used to guide drug development, the direct clinical utility of the CMS classification lies in the possibility to estimate survival (prognostic value) and select patients for both adjuvant or palliative chemotherapy and currently used targeted agents (predictive value). The poor prognostic value of CMS4 and the relatively favorable prognosis of CMS1 and CMS2 in nonmetastatic disease have been demonstrated before (6), although no aggregated evidence of different survival measures per CMS was previously reported. We argue that molecular stratification of CRC patients in clinical trials should be encouraged and that the path towards clinical implementation should be investigated. We therefore conducted a systematic review and meta-analysis to evaluate the current state of the evidence on the prognostic and predictive potential of the CMSs to guide future treatment strategies. In both local and metastatic disease, we compared the CMSs for overall survival (OS), PFS, relapse-free survival (RFS), and survival after relapse (SAR), and we assessed benefit of (adjuvant) chemotherapy regimens and targeted agents in each subtype.
Methods
Study Protocol and Objectives
The PROSPERO international prospective register of systematic reviews was consulted before the start of the study, and no ongoing reviews with similar scope were identified. Our study protocol was submitted to PROSPERO before the start of data extraction and is accessible via the PROSPERO database (PROSPERO identifier CRD42020165483) (8). Reporting is in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines (9).
The primary objectives of this study were to evaluate the prognostic and predictive impact of the CMS classification in CRC patients, thereby investigating their relevance for use in clinical practice.
Search Strategy and Study Selection
We conducted a literature search in Medline and Embase to identify original studies and conference abstracts covering the prognostic and predictive value of the CMSs in CRC between 2011 and 2020. No restrictions on study design were set. Key words consisted of “colorectal cancer” and “consensus molecular subtypes.” The initial search was conducted in December 2019 and repeated in December 2020.
Eligible studies contained data on the prognostic or predictive value of the CMSs for CRC of any stage. Included studies used either CMS labels or 1 of the 6 transcriptomic classifications at the foundation of the CMSs, from which the CMS labels could be deduced (10-15). All methods of CMS labeling were considered eligible. Both study selection based on title and abstract and full-text screening were performed independently by 2 reviewers (S.H. and T.B.) using Rayyan software, a web and mobile application for systematic reviews (16). After removal of blinding, the selected studies were discussed and inclusion of the studies was consensus-based without disagreements between authors.
Quality Assessment and Data Collection
The Prediction model Risk Of Bias ASsessment Tool was used to assess the risk of bias of the included full-text studies (17). Two reviewers (S.H. and T.B.) independently completed the tool for all studies, and the final assessment was consensus-based. No disagreements occurred.
Abstracts, full-text manuscripts, and supplementary materials were screened for data of interest. Relevant missing data were requested from the authors. Data were extracted by 2 independent reviewers (S.H. and T.B.), and inconsistencies were resolved by consensus. General study characteristics were retrieved, such as study design, Gene Expression Omnibus accession number of used preexisting cohorts, number of included patients with CMS label, CRC stage, methods of CMS labeling, and distribution of CMSs across the study populations. Outcome data of interest included median absolute OS (mOS) and RFS (mRFS); hazard ratios (HRs) for OS, PFS, RFS, and SAR from Cox regression analyses; and response rates (RRs) for systemic therapies, all stratified for CMSs and disease stage. Overlap between studies was accounted for by assessing used cohorts and types of comparisons and analyses performed. When considerable overlap between studies existed, the study with the most robust data on the clinical value was included.
Statistical Analyses
For the prognostic data, meta-analyses and posthoc subgroup and sensitivity analyses were performed using the R packages “meta,” “metafor,” and “dmetar” (18). For calculating both the pooled mOS times per CMS in mCRC and the CMS comparisons per survival outcome in local and metastatic disease, random-effect inverse variance meta-analyses were performed. The DerSimonian-Laird τ2-estimator was used for pooling hazard ratios, whereas the Restricted Maximum-Likelihood τ2-estimator was used for pooling median survival times (19). Additional t tests were performed for pairwise CMS comparisons of pooled mOS times. Data on the predictive value of the CMSs were limited and heterogeneous in terms of treatment strategies and could therefore not be pooled, but were summarized individually.
Between-study heterogeneity in all meta-analyses was assessed with the Higgins I2 index and the Cochran’s Q-test (20). Heterogeneity was defined as either an I2 index of at least 50% or a statistically significant Q-test. Sources of heterogeneity were explored by identifying outliers and influential studies (21). The relative contribution of individual studies to heterogeneity compared with their impact on the pooled effect size was visualized with Baujat plots (data not shown) (22). Sensitivity analyses were performed to assess changes in heterogeneity and effect sizes after exclusion of studies that imposed heterogeneity. These studies were not excluded from the final analyses if no statistically significant change of the pooled effects occurred.
To explore the impact on our findings of varying CMS classification methods used in the included articles, we performed subgroup and sensitivity analyses. We compared phenotypic (immunohistochemical [IHC]) classification vs RNA-based classification, single-group classification vs direct CMS subtyping, and 3 methods of RNA-based CMS classification. Subgroup analyses were only performed if at least 2 cohorts with the same classification method could be included per subgroup; otherwise, sensitivity analyses were run. Subgroup analyses were performed using a random-effect model to pool the effect sizes within the subgroups and a fixed-effect (plural) model to compare the subgroups (mixed-effect model) (23).
Publication bias was assessed with funnel plots and quantified using the Egger’s test (24,25).
All statistical analyses were performed with R software version 3.6.1 or SPSS version 26. Absolute survival data were presented as medians with 95% confidence interval (CI) and response data as percentages of the total. Survival comparisons were described with hazard ratio, 95% confidence interval, and P value and depicted with forest plots. Missing confidence intervals and P values of hazard ratios were calculated using previously described methods (26,27). Distribution of CMS labels in the local vs metastatic setting was assessed with the χ2 statistic and Pearson residuals. P values were 2-sided, and a P value of less than .05 was considered statistically significant. Multiple testing correction, using the Benjamini-Hochberg procedure, was applied to P values from our primary analyses, with an alpha value of .05. We corrected P values per independent hypothesis, that is, for the analyses into local CRC, mCRC, and mOS per CMS. We used the function “mt.rawp2adjp()” from the R package “multtest” to adjust raw P values according to the classical Benjamini-Hochberg procedure (28).
Results
Selection of Eligible Studies
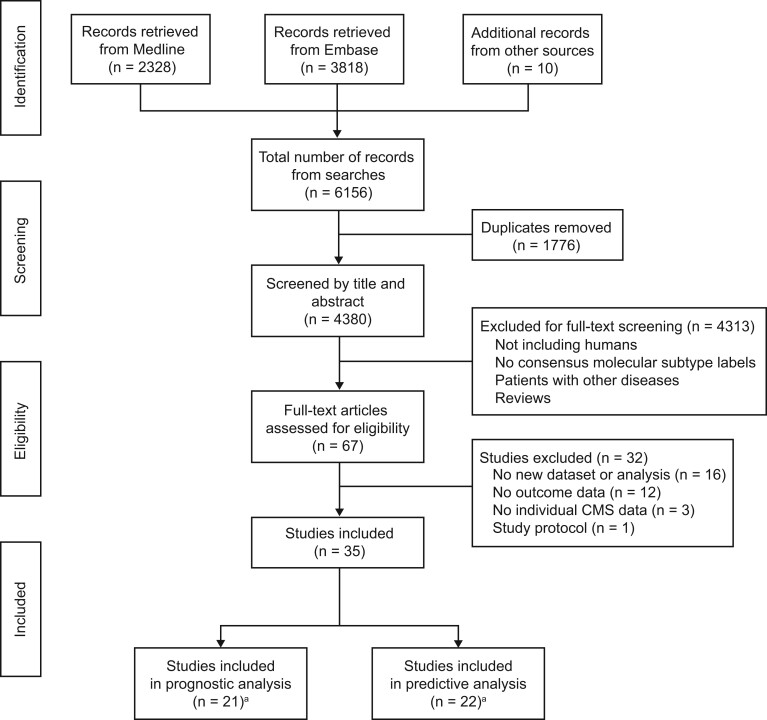
The initial search yielded 4380 studies, of which 67 studies were deemed potentially relevant for further review. After full-text screening, 32 additional studies were excluded. The majority of the exclusions (n = 16) were due to considerable overlap in (publicly available) cohorts. In case different studies used similar cohorts, we included those with the most extensive analysis on the prognostic or predictive value of the CMSs (10,13,14,29–41). Other reasons for exclusion after full-text screening were no outcome data of interest reported (n = 12) (7,42–50), no data on the individual CMSs reported (n = 3) (51–53), or a study protocol (n = 1) (54) (Figure 1).
Figure 1.
Flowchart of study identification and selection process. aEight studies were used for both prognostic and predictive analysis.
Among the 35 eligible studies were 27 full-text articles (6,11,12,15,55–77) and 8 conference abstracts (78–85). Prognostic data were extracted from 21 studies, of which 2 were conference abstracts (6,11,15,57,58,60,61,63,64,66–73,75,77,80,81). The predictive value of the CMSs was assessed in 16 studies and 6 conference abstracts (12,15,55–57,59,62,64–67,69,72–74,76,78,79,82–85).
Overview of Study Characteristics and Risk of Bias
The median size of the study populations with CMS or CMS-related labels was 237 (interquartile range = 113-748) (Table 1; Supplementary Table 1 and 2, available online). The majority of the studies were performed in mCRC patients, and most studies reported RNA-based CMS labels derived from the primary tumor. OS and RFS were the most frequently reported outcome data. Most predictive studies reported on adjuvant chemotherapy or (the addition of) bevacizumab and cetuximab.
Table 1.
Overview of study characteristicsa
| Characteristic | Studies, No. (%) | Prognostic, No. (%) | Predictive, No. (%) |
|---|---|---|---|
| Total No. | 35 | 21 | 22 |
| Articles | |||
| Full text | 27 (77.1) | 19 (90.5) | 16 (72.7) |
| Abstracts | 8 (22.9) | 2 (9.5) | 6 (27.3) |
| Study design | |||
| Retrospective | 31 (88.6) | 20 (95.2) | 19 (86.4) |
| Prospective | 4 (11.4) | 1 (4.8) | 3 (13.6) |
| Disease stages in survival analysis | |||
| II and III | 7 (20.0) | 5 (23.8) | 4 (18.2) |
| III | 3 (8.6) | 1 (4.8) | 2 (9.1) |
| IV | 17 (48.6) | 9 (42.9) | 13 (59.1) |
| I-IV | 3 (8.6) | 1 (4.8) | 2 (9.1) |
| Other | 5 (14.3) | 5 (23.8) | 1 (4.5) |
| Transcriptomic classification | |||
| Consensus molecular subtypes (CMS) | 30 (85.7) | 18 (85.7) | 18 (81.8) |
| CMS R package | 16 (45.7) | 11 (52.4) | 9 (40.9) |
| NanoString | 8 (22.6) | 4 (19.0) | 5 (22.7) |
| Immunohistochemistry | 4 (11.4) | 2 (9.5) | 3 (13.6) |
| Other | 2 (5.7) | 1 (4.8) | 1 (4.5) |
| Sadanandam subtypes | 2 (5.7) | 1 (4.8) | 2 (9.1) |
| De Sousa e Melo subtypes | 1 (2.9) | 0 (0.0) | 1 (4.5) |
| Budinska subtypes | 1 (2.9) | 1 (4.8) | 0 (0.0) |
| Roepman subtypes | 1 (2.9) | 1 (4.8) | 1 (4.5) |
| Survival outcomes | |||
| Overall | 26 (74.3) | 16 (76.2) | 17 (77.3) |
| Relapse free | 13 (37.1) | 10 (47.6) | 5 (22.7) |
| Progression free | 12 (34.3) | 7 (33.3) | 10 (45.5) |
| Survival after relapse | 3 (8.6) | 3 (14.3) | 0 (0.0) |
| Systemic treatment | |||
| Adjuvant chemotherapy | 6 (17.1) | 3 (14.3) | 6 (27.3) |
| Targeted agents | 11 (31.4) | 4 (19.0) | 11 (50.0) |
| First-line chemotherapy metastatic colorectal cancer | 5 (14.3) | 2 (9.5) | 5 (22.7) |
| Immunotherapy | 2 (5.7) | 0 (0.0) | 2 (9.1) |
| No. of classified samples | |||
| Median | 237 | 237 | 273 |
| Interquartile range | 113-748 | 113-765 | 168-660 |
The percentages were calculated using the total number in the first row for each column.
In general, studies were of high quality as scored with the Prediction model Risk Of Bias ASsessment Tool (Supplementary Table 3, available online). Risk of bias was rated as high in 5 studies, which was commonly due to small sample sizes. Applicability was a concern for 14 studies, which was invariably related to a custom method of CMS labeling or highly specific study populations. When outcome definitions or methods of CMS labeling were lacking, applicability was deemed unclear (n = 3).
CMS Distribution
First, we evaluated the relationship between cancer stage and the distribution of reported CMS labels. We observed a statistically significant shift in the CMS distribution between the local and metastatic disease setting (χ2 = 304.65, P < .001) (Supplementary Table 4, available online). CMS1 was less abundant in the metastatic compared with the localized setting (9.9% vs 18.7%), and conversely an increase in CMS4 tumors was found in metastatic disease (36.1% vs 25.6%). These findings are in line with those in earlier individual studies (6,39).
Prognostic Value of the CMSs
Local disease . For clinicians and CRC patients, the CMSs could be a valuable tool to estimate disease prognosis. Two studies reported mRFS measures per CMS class for stage II and III patients, predominantly treated with adjuvant chemotherapy, which ranged from 120 to 122 months for CMS1, 114 to 123 months for CMS2, 107 to 115 months for CMS3, and 66 to 120 months in CMS4 (58,72).
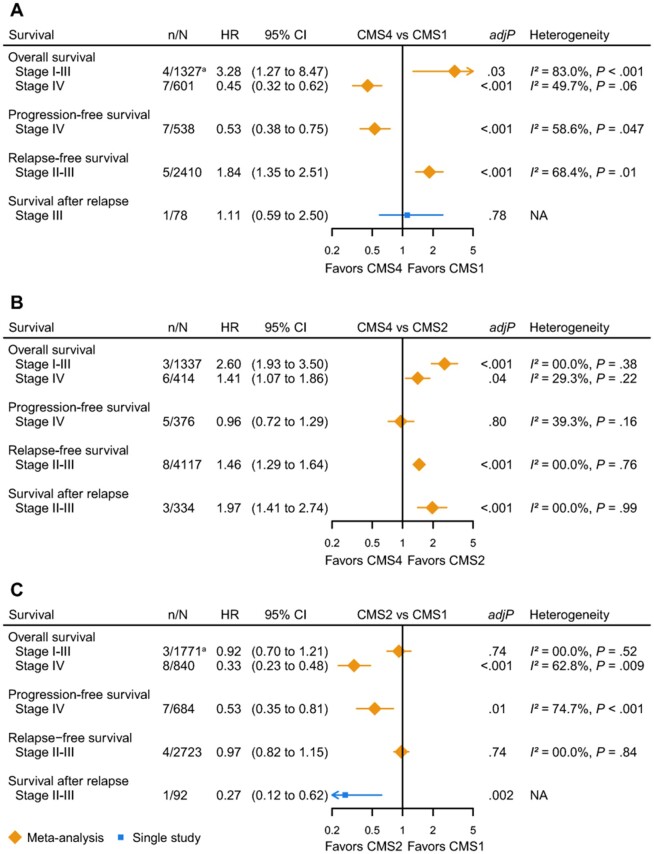
The meta-analyses into the prognostic value of the CMSs in local disease included patients from 11 cohorts. We found worse OS for CMS4 compared with CMS1 (stage I-III: HR = 3.28, 95% CI = 1.27 to 8.47; number of included cohorts [n] = 4) (15,61,66,70) and for CMS4 compared with CMS2 (stage I-III: HR = 2.60, 95% CI = 1.93 to 3.50; n = 3) (11,70). The same pattern was observed for the RFS and SAR (Figure 2, A and B) (11,58,70–72,75). To strengthen these findings, we performed 2 secondary meta-analyses into OS and RFS for stage I-III CMS4 vs the other subtypes, which confirmed the worse survival for CMS4 tumors [OS: HR = 1.67, 95% CI = 1.34 to 2.06, n = 4 (15,31,82,83); RFS: HR = 1.49, 95% CI = 1.11 to 2.00, n = 3 (29,58,60)](data not graphically shown). Dismal OS and RFS for stage II and III CMS4 tumors were confirmed in 4 other studies (60,63,71,82), with 1 study reporting the lowest 5-year lymph node metastasis-free and RFS for T1 CMS4 tumors (66.7%) (60). This is most probably due to the invasive and mesenchymal properties of CMS4 tumors, resulting in early metastasis (12,39). For CMS3, there is a worse OS compared with CMS2, but better OS than CMS1, although both are not statistically significant. RFS of CMS3 did not differ from CMS1, CMS2, and CMS4 (Supplementary Figure 1, available online) (11,58,61,72,75).
Figure 2.
Forest plots of pooled and single hazard ratios (HRs) for different survival outcomes per pairwise consensus molecular subtype (CMS) comparison in colorectal cancer. A) CMS4 vs CMS1. B) CMS4 vs CMS2. C) CMS2 vs CMS1. Number of cohorts (n) and total number of included patients (N) per meta-analysis indicated with n/N; because several studies described more cohorts, the number of cohorts n does not reflect the number of studies. adjP indicates significance for the random-effect model (corrected for multiple testing). Heterogeneity depicted as I2 index and Cochran’s Q-test P value. All tests were 2-sided. aEstimate. adjP = adjusted P value; CI = confidence interval; NA = not applicable.
Three (out of 10) conducted pairwise meta-analyses for local disease suffered from between-study heterogeneity; differences in sample sizes appeared to explain heterogeneity, because exclusion of these studies reduced heterogeneity without statistically significantly changing the pooled effect sizes (Supplementary Table 5; Supplementary Figure 2, available online).
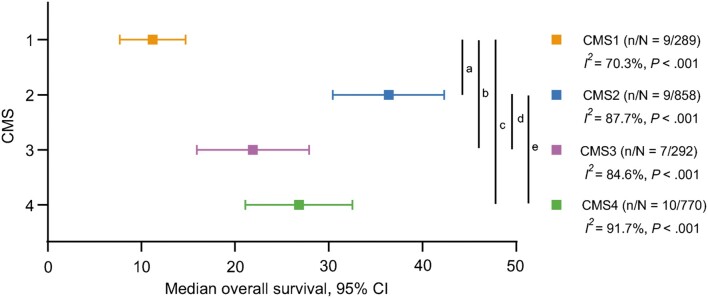
Metastatic disease. In synchronous or metachronous mCRC, the pooled mOS differed across the CMSs (Figure 3; Supplementary Table 6, available online). Interestingly, CMS1 had the poorest mOS of 11.2 months (6,64,67–70,73), which was statistically significantly worse compared with CMS2 (36.4 months; CMS2 vs CMS1, P < .001) (6,64,67–70,73,80), CMS3 (21.9 months; CMS3 vs CMS1, P = .02) (6,64,67–69,73), and CMS4 (26.8 months; CMS4 vs CMS1, P = .001) (6,64,67–70,73,80). CMS2 had the most favorable OS, which was statistically significantly better than CMS3 (P = .009) and CMS4 (P = .04). The mOS of CMS3 and CMS4 was comparable (P = .27). Heterogeneity was present in these meta-analyses and was most likely caused by differences in treatment regimens between the cohorts (anti-EGFR treated vs first-line chemotherapy). Exclusion of heterogeneous studies did not statistically significantly alter the pooled survival times of the CMSs (Supplementary Table 7, available online).
Figure 3.
Median overall survival in metastatic disease per consensus molecular subtype (CMS). Vertical lines indicate statistically significant comparisons (t test). aP less than .001, bP = .02, cP = .001, dP = .009, eP = .04 (corrected for multiple testing). Number of cohorts (n) and total number of included patients (N) per meta-analysis indicated with n/N; because several studies described more cohorts, the number of cohorts n does not reflect the number of studies. Heterogeneity depicted as I2 index and Cochran’s Q-test P value. All tests were 2-sided.
Pairwise CMS comparisons in the metastatic setting included patients from 9 unique cohorts and showed that CMS1 consistently had worse OS and PFS than CMS2, CMS3, and CMS4 (OS HR range = 0.33-0.55; PFS HR range = 0.53-0.89) (57,64,67,69,70,81) (Figure 2, A and C; Supplementary Figure 1, A, available online). This is in line with the reported mOS times. The mesenchymal CMS4 subtype was associated with worse OS compared with the epithelial CMS2 subtype (HR = 1.41, 95% CI = 1.07 to 1.86, n = 6) (57,69,70,77), but PFS did not differ (Figure 2, B) (57,69,70). CMS3 did not show statistically significant differences in OS and PFS compared with CMS2 and CMS4, although PFS tended to be worse for CMS3 compared with CMS4 (57,69) (Supplementary Figure 1, available online). Heterogeneity was detected in 5 out of 12 meta-analyses, which could partly be explained by differences in cohort sizes, and exclusion of these studies did not change the summary effect sizes (Supplementary Table 5, available online).
Predictive Value of the CMSs
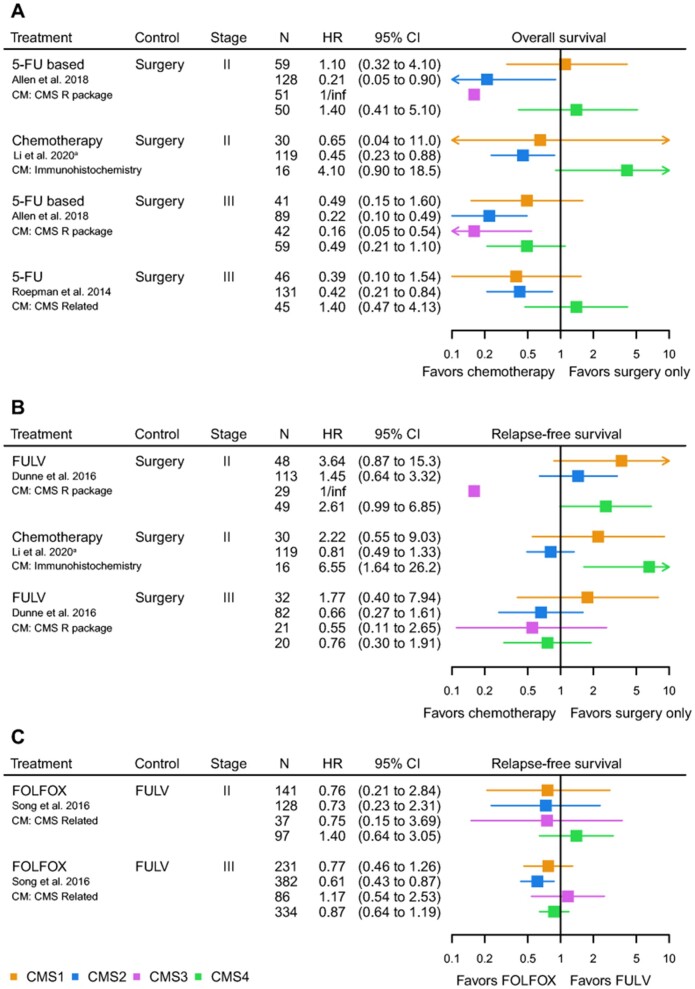
Local disease. Currently, patient selection for adjuvant chemotherapy is suboptimal, leading to over- and undertreatment in a large proportion of patients (2). Differences in tumor biology, as captured by the CMSs, could serve as predictors for treatment benefit. Because data on the predictive value of the CMSs were rather heterogeneous in terms of received treatment regimens, we summarized the results of each study individually. Adjuvant chemotherapy, compared with surgery alone, improved OS for both stage II and III in CMS2 and CMS3 (HR range = 0.16-0.45) (Figure 4, A) (15,55,66). Adjuvant chemotherapy was not clearly associated with better survival in CMS1 and CMS4 tumors (Figure 4, A and B) (59). With respect to combination regimens for adjuvant treatment, the addition of oxaliplatin to 5-fluorouracil increased RFS in stage III CMS2 patients in 1 study (HR = 0.61, 95% CI = 0.43 to 0.87) (Figure 4, C) (72), whereas CMS4 patients seemed not to benefit from this regimen in another study (83). The difference in treatment efficacy between CMS2 and CMS3 vs CMS1 and CMS4 likely results from intrinsic molecular differences between epithelial vs mesenchymal and immunogenic tumors.
Figure 4.
Forest plots of hazard ratios (HRs) for the predictive value of the consensus molecular subtypes (CMSs) for adjuvant chemotherapy in colorectal cancer. A) Overall survival for adjuvant chemotherapy vs surgery alone. B) Relapse-free survival for adjuvant chemotherapy vs surgery alone. C) Relapse-free survival for oxaliplatin plus 5-fluorouracil and leucovorin vs 5-fluorouracil plus leucovorin. aImmunohistochemistry classification: CMS2 and CMS3 depicted as CMS2. 5-FU = 5-fluorouracil; FOLFOX = 5-fluorouracil, leucovorin, and oxaliplatin; FULV = 5-fluorouracil and leucovorin.
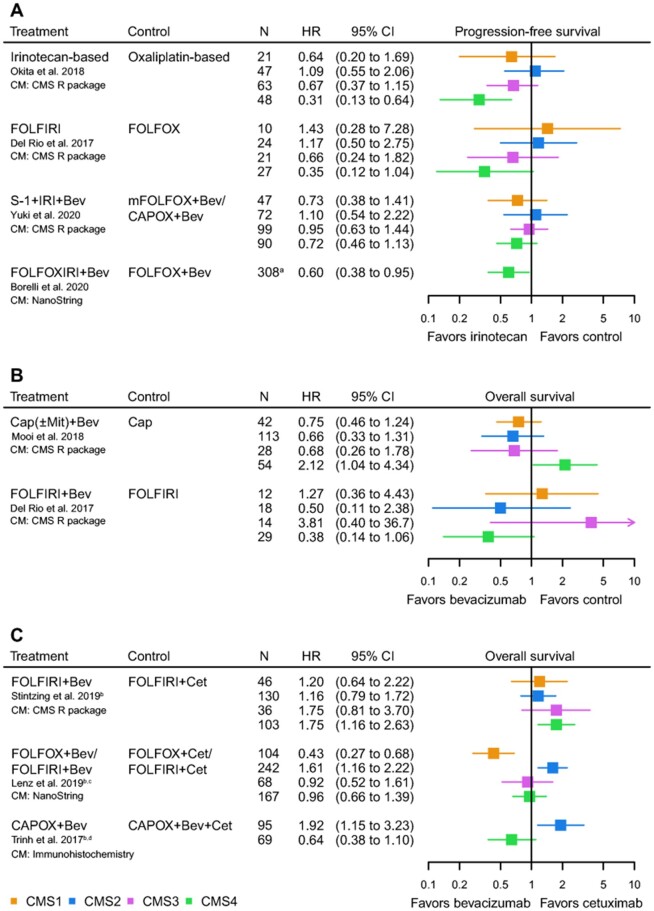
Metastatic disease. All predictive studies in mCRC assessed first-line treatment regimens, except for 2 studies and 1 conference abstract reporting on more recently developed targeted therapies in the second line (62,65,84). Strikingly, all studies uniformly showed that an irinotecan backbone increased both PFS and OS in patients with CMS4 tumors (PFS HR range = 0.31-0.72; OS HR range = 0.43-0.45) (Figure 5, A; Supplementary Figure 3, A, available online) (57,69,78,85). One abstract reported an increased OS for oxaliplatin- compared with irinotecan-based treatment in CMS4 patients (HR = 0.42, 95% CI = 0.17 to 1.00) (79). However, irinotecan-based therapy was given to a minority of patients compared with oxaliplatin-based chemotherapy (19.7% vs 80.3%), which might have influenced the results. For CMS1, a slight increase of survival with an irinotecan-based regimen was also observed (Figure 5, A; Supplementary Figure 3, A, available online). These findings are of importance considering that the first-choice regimen in the first-line setting often is oxaliplatin based, and, consequently, a large group of CMS1 and CMS4 mCRC patients may receive suboptimal first-line treatment.
Figure 5.
Forest plots of hazard ratios (HRs) for the predictive value of the consensus molecular subtypes (CMSs) for first-line systemic therapy in metastatic colorectal cancer. A) Progression-free survival for an irinotecan backbone vs control regimen. B) Overall survival for the addition of bevacizumab vs control regimen. C) Overall survival for the addition of bevacizumab vs cetuximab. aTotal number of patients, no information on the number of CMS4 patients. bKRAS wild-type population. c75.2% FOLFOX/24.8% FOLFIRI. dImmunohistochemistry classification: CMS2 and CMS3 depicted as CMS2. Bev = bevacizumab; Cap = capecitabine; CAPOX = capecitabine and oxaliplatin; Cet = cetuximab; CM = classification method; FOLFIRI = 5-fluorouracil, leucovorin, and irinotecan; FOLFOX = 5-fluorouracil, leucovorin, and oxaliplatin; mFOLFOX = modified FOLFOX; FOLFOXIRI, 5-fluorouracil, leucovorin, oxaliplatin and irinotecan; IRI = irinotecan; Mit = mitomycin; N = number of patients.
Regarding targeted therapies, a PFS benefit from adding bevacizumab to capecitabine in both CMS2 and CMS3 was seen (HR = 0.44 [95% CI = 0.29 to 0.68] and HR = 0.35 [95% CI = 0.14 to 0.86], respectively) (Supplementary Figure 3, B, available online). For OS, the same trend was observed (Figure 5, B) (67). Adding bevacizumab to FOLFIRI (but not to capecitabine) compared with FOLFIRI monotherapy was associated with better OS (HR = 0.38, 95% CI = 0.14 to 1.06) and PFS in CMS4 (Figure 5, A; Supplementary Figure 3, B, available online) (57,67). However, when comparing FOLFIRI combined with either bevacizumab or cetuximab in CMS4 patients, bevacizumab was associated with inferior outcomes compared with cetuximab for both OS (HR = 1.75, 95% CI = 1.16 to 2.63) and PFS (Figure 5, C; Supplementary Figure 3, C, available online) (73). The combination of bevacizumab with FOLFOX appeared more effective than cetuximab plus FOLFOX in CMS1 for both OS (HR = 0.43, 95% CI = 0.27 to 0.68) and PFS (Figure 5, C; Supplementary Figure 3, C, available online) (64).
The addition of cetuximab to either FOLFOX or CAPOX-bevacizumab was beneficial in KRAS wt CMS2 patients for OS (FOLFOX: HR = 1.61, 95% CI = 1.16 to 2.22; CAPOX-bevacizumab: HR = 1.92, 95% CI = 1.15 to 3.23) and PFS (Figure 5, C; Supplementary Figure 3, C, available online) (64,74). One abstract also showed an increased OS for anti-EGFR in CMS2 and 3 compared with bevacizumab (HR = 0.67, 95% CI = 0.42 to 1.06) (79). Furthermore, evidence for anti-EGFR benefit in CMS2 was observed in terms of RECIST-defined tumor response rates, showing a very high disease control rate for both CMS2 (92.3%) and CMS4 (81.8%) with anti-EGFR therapy (±80.0% combined with irinotecan and ±20.0% monotherapy) (56,69,76). Additionally, clinical response and disease-free survival of cetuximab monotherapy were found to be better for KRAS wt CMS2 patients (12).
Two articles and 1 conference abstract reported on targeted therapies as second-line regimens for mCRC. One study showed an increase in PFS when CMS4 tumors were treated with nintedanib (triple angiokinase inhibitor) compared with placebo (HR = 0.60, 95% CI = 0.40 to 0.91); no effect was seen in CMS2 tumors. Unclassified patients had both an increased OS (HR = 0.29, 95% CI = 0.14 to 0.52) and PFS (HR = 0.20, 95% CI = 0.08 to 0.49) (65). The combination of napabucasin (blocks STAT3; downregulates IDO1 and PD-L1) and pembrolizumab showed a response rate of 33.0% for CMS1 and CMS4 tumors (62). The third study showed no clear benefit for CMS4 from bintrafusp alfa, targeting both TGF-β and PD-L1, with or without radiotherapy (84).
Altogether, these data indicate that CMS2 and CMS3 could predict benefit from adjuvant chemotherapy. In mCRC, CMS4 has predictive value for the use of first-line irinotecan-based chemotherapy. Also, combining an irinotecan backbone with bevacizumab or cetuximab (for KRAS wt patients) is favorable in CMS4. CMS1 appears to be predictive for the addition of bevacizumab, and CMS2 has potential predictive value for the use of cetuximab in KRAS wt patients. Immunotherapy might be an effective treatment for CMS1 mCRC.
Impact of CMS Classification Methods
Differences in CMS classification methods could affect the estimated prognostic and predictive values of the individual CMSs as determined in our study. The majority of included studies used gene expression data and the CMS R package (n = 15). Other classification methods were NanoString-based (n = 7), IHC-based (n = 4), the CMScaller (n = 1), the multinomial elastic net classifier (n = 1), or single-group classification from which the CMSs could be deduced (n = 4).
We previously described a shift in distribution of CMS labels between the local and metastatic setting, and this remained the same when excluding those studies using classification methods with only 3 CMS classes (IHC, Roepman, and De Sousa subtypes) (Supplementary Table 8, available online). Studies in which the distribution of CMS labels was markedly altered are indicated in Supplementary Tables 1 and 2. Most differences were due to the usage of the IHC CMS classifier because this cannot detect CMS3 labels, or due to a specific subgroup of patients (eg, T1 tumors or a RAS wt cohort). Furthermore, it was recognized that specific classification methods are associated with slightly different CMS distributions (Supplementary Figure 4, available online). In the local setting, more CMS3 and unknown labels were observed when using the CMS R package but reduced CMS4 labels. The IHC method yielded more CMS2 and reduced unknown labels. In the metastatic setting, NanoString classifiers detected less CMS3, and the CMS R package detected less CMS2 and CMS4 labels.
To explore the effect of the different classification methods on the prognostic value of the CMSs, we looked into 3 comparisons: transcriptomic vs IHC-based labeling, single-group transcriptomic labels as surrogates for CMS labels vs direct CMS labeling, and CMS R package classification vs other RNA-based methods. Two analyses included IHC-based CMS labels next to RNA-based CMS labels (Supplementary Tables 9 and 10, available online). In sensitivity analyses, no major changes in (direction of) the pooled effect sizes occurred after exclusion of IHC-based data (Supplementary Table 10, available online). Nine meta-analyses included single-group transcriptomic labels (Budinska, Roepman, and Sadanandam) as surrogates for CMS labels. For both Budinska and Roepman labels, neither subgroup nor sensitivity analyses showed a statistically significant difference in prognostic value compared with CMS labels (Supplementary Tables 11 and 12, available online). Two subgroup analyses for Sadanandam labels also showed similar prognostic value compared with CMS classes, but a third subgroup analysis found a statistically significant difference between these taxonomies (CMS4 vs CMS1, RFS local) (Supplementary Table 11, available online). However, the direction of both effect sizes was similar, and therefore our conclusions were not affected. Furthermore, subgroup and sensitivity analyses showed no statistically significant differences between the different RNA-based CMS classification methods, except for 1 subgroup analysis into NanoString-based classification vs the CMS R package (CMS2 mOS) (Supplementary Tables 13-15, available online). This again had no effect on the interpretation of our findings.
Regarding the predictive value, 3 different CMS classification methods were used in the studies investigating localized disease [the CMS R package (55), single-group classification (15), and the IHC-classifier (66)]. The OS benefit of adjuvant chemotherapy compared with surgery alone for CMS2 and CMS3 was consistent across the 4 cohorts despite using different classifiers. As for stage II CMS4, no clear benefit from adjuvant chemotherapy was detected by using both the CMS R package and the IHC classifier (55,59). In the metastatic setting, the beneficial effect of irinotecan-based chemotherapy for CMS4 compared with oxaliplatin-based chemotherapy was mainly based on CMS labels from the CMS R package (3 studies), although the same effect was found with a NanoString classifier (78). The favorable effect of cetuximab combined with an oxaliplatin-based backbone for CMS2 patients was shown with both NanoString-based and IHC-based CMS classification (64,74). Other supporting evidence for the benefit of anti-EGFR therapy compared with the addition of bevacizumab in CMS2 patients was shown with the IHC classifier (79), but also with NanoString-based classification (56), the CMS R package (69), and the CMScaller (76).
Altogether, it seems that the prognostic and predictive value of the CMSs are robust and remain unaffected by the classification method used. The possibility to use a variety of CMS classification methods with largely similar CMS detection and related clinical associations could be a major advantage for future implementation of the CMSs in the clinic.
Publication Bias
In the majority of 21 out of 26 meta-analyses, publication bias did not play a statistically significant role, as assessed in funnel plots and quantified with the Egger’s test (Supplementary Figures 5-7, available online). In 5 meta-analyses into mCRC, that is, OS and PFS of CMS4 vs CMS1, OS of CMS2 vs CMS1, OS of CMS3 vs CMS1, and absolute mOS of CMS1, publication bias could have influenced our outcomes, warranting careful interpretation of these specific results. However, in general, the role of publication bias in our meta-analyses seemed limited.
Discussion
To our knowledge, this is the first systematic review and meta-analysis providing a comprehensive overview of the prognostic and predictive significance of the CMS classification (6). Our study underlines the additive value of molecular CMS classification and its potential for patient stratification in clinical trials and future clinical use, both with regards to estimating disease prognosis and selecting patients for specific treatment regimens. These benefits seem not to be restricted to a specific classification method but appear to be rather universal for a variety of molecular classifiers, depending on the materials and resources available.
We demonstrated with aggregated data the unfavorable prognostic value of CMS4 in local disease compared with CMS1 and CMS2. As an example, 1 of the included studies looked into a relatively large cohort of 860 stage III patients and identified that CMS4 performed statistically significantly worse than CMS1 and CMS2 with regards to OS (70). In mCRC, we found that CMS1 was associated with worse survival compared with the other subtypes. Compared with our pooled data, the largest included study in the metastatic setting investigated a cohort of 581 patients and found an mOS of 15 months for CMS1 and a statistically significantly worse outcome compared with CMS2 and CMS4 (64). This is in line with published data showing worse survival for metastasized BRAF mutated and MSI tumors (86–88). Because both MSI and BRAF mutations are typical properties of the CMS1 subtype, they might explain the worse survival of metastatic CMS1 tumors as identified in our study. In both local and metastatic disease, we identified CMS2 as the subtype with the most favorable prognosis.
Our findings provide insight into the variable efficacy of systemic therapies in CRC, currently leading to substantial over- and undertreatment in a large proportion of patients. In local disease, CMS2 and CMS3 predict benefit from adjuvant chemotherapy in contrast to the mesenchymal and immune subtypes. The lack of fluoropyrimidine-monotherapy benefit for defective MMR CMS1 tumors could be explained by the fact that a sufficient MMR machinery is required for FU-induced cell death (89–91). CMS4 tumors possibly behave more like the early-dissemination model instead of the classical linear-progression model (92,93). Therefore, a large proportion of patients diagnosed with local disease might already have (yet undetectable) micro-metastases and, consequently, lack benefit from adjuvant chemotherapy. The predictive relevance of the CMSs for neoadjuvant chemotherapy in resectable stage II and III colon cancer will be further evaluated in the CONNECTION II trial (94).
In CMS4 metastatic disease, studies showed better efficacy of an irinotecan-based first-line regimen compared with an oxaliplatin-containing regimen. One of these is the recently published retrospective analysis of the phase III TRIBE2 study on the superiority of bevacizumab combined with triplet chemotherapy (FOLFOXIRI) compared with doublets. Adding irinotecan to doublet therapy with bevacizumab showed a statistically significant benefit for CMS4 tumors compared with doublet therapy with bevacizumab (78). The phase III TRICOLORE trial also found the best effectivity of an irinotecan-based regimen (S-1/irinotecan/bevacizumab) for CMS4 compared with an oxaliplatin backbone combined with bevacizumab (85). Currently, standard first-line regimens in mCRC are often oxaliplatin based, which stresses the relevance of stratifying patients according to molecular features to prevent suboptimal treatment. In addition, the majority of peritoneal metastases are of the CMS4 subtype. Therefore, intraperitoneal irinotecan-based treatment holds great promise in light of our findings and will be assessed in the phase I INTERACT trial (95,96). CMS4 mCRC also predicted benefit from irinotecan combined with cetuximab (in KRAS wt tumors) and bevacizumab (in KRAS mutant tumors), and thus, combinations of these targeted agents with an irinotecan backbone could serve as preferred first-line options in CMS4 mCRC patients.
CMS1 tumors likely benefit most from immunotherapy in metastatic disease. The available studies using CMS1 instead of MSI are sparse, but based on the clear efficacy in these studies we assume that immunotherapy will be the treatment of choice (4,97,98). As second-line regimen for CMS1 tumors, a chemotherapy backbone combined with bevacizumab could be an effective alternative, as was shown in the retrospective analysis of a phase III Cancer and Leukemia Group B (CALGB) and Southwest Oncology Group (SWOG) alliance trial that assessed the relative efficacy of cetuximab vs bevacizumab when added to standard FOLFOX or FOLFIRI-based first-line chemotherapy (Figure 5C) (64). The preference for bevacizumab rather than cetuximab in CMS1 could be explained by the important role of T-lymphocytes (abundant in CMS1 tumors) in vessel normalization induced by blockade of angiogenesis. This results in reduced tumor hypoxia and vessel leakage, increasing influx of immune effector cells and chemotherapy into the tumor parenchyma (7,99,100).
Cetuximab benefit was most evident in patients with CMS2 KRAS wt mCRC. It was previously shown that CMS2 tumors are predominantly left-sided, and specifically these tumors are characterized by an activated EGFR pathway, rendering them susceptible for cetuximab treatment (64,101). Previous studies have also shown that combined APC and TP53 mutations are related to cetuximab sensitivity, and these mutations are enriched in CMS2 patients (75.0% vs 17.0 to 41.0% in CMS1-4) (102,103).
An important and ongoing topic of discussion is the additive value of the CMSs compared with standardly used TNM staging, MMR, and gene mutation status. Regarding the latter, a strong CMS-dependent prognostic impact of BRAFV600E and KRAS mutations in MSS tumors has been demonstrated, with the strongest prognostic value of BRAFV600E mutations in CMS1 tumors and KRAS mutations in CMS2 and CMS3 tumors (104). With respect to TNM staging, we recently reported that gene signatures of advanced-stage CRC were highly correlated with CMS4 and identified a CMS4-subgroup within high-risk stage II patients with worse survival than the total group of stage II patients, stressing the additive prognostic value of transcriptomic subtypes to TNM staging (39). Furthermore, a retrospective study in early-stage CRC demonstrated that tumor microenvironment features, such as cancer-associated-fibroblasts (CAFs) and CD3+ and CD8+ T-cell infiltration, are particularly prognostic compared with T and N stage; MMR, BRAF, and KRAS mutational status; and CMS4 score (58). Because microenvironment features, including CAFs and immune cell infiltration, are intrinsic properties of the CMSs, this suggests that transcriptomic profiling might be more accurate in prognosis prediction compared with TNM staging. This, however, requires further refinement into more specific prognostic subgroups that go beyond sole CMS labeling. For example, next to CAF and CytoLym scores, several studies within the established CMSs have shown the favorable prognostic value of immune markers, such as expression of chemokine-like factor in CMS1 (59), immunoscore-like metagenes within CMS2 and CMS3 (36), and both specific transcriptomic immune subtypes and immune checkpoint metagenes in CMS3 (36,105). For CMS1 and CMS3, deregulated Bcl2-dependent apoptosis was a poor prognostic marker (106), whereas in CMS4 factors related to the antioxidant response and oxidative DNA damage conferred poor prognosis(107).
As for predicting treatment benefit, the additive value of transcriptomic subtyping to current standard of care is promising, providing a full overview of molecular targets and exploitable tumor vulnerabilities for each tumor, enabling better selection for commonly used chemotherapeutic regimens and bringing newly discovered targeted agents within reach for each patient. For example, in preclinical models, CMS1 and CMS4 predicted response to heat shock protein 90 inhibitors (31), but also response to triple therapy with trametinib (MEK1/2-inhibitor), neratinib (pan-ERBB inhibitor), and trastuzumab (HER2-inhibitor) (108). On the other hand, CMS2 and CMS3 were more sensitive to aurora kinase inhibitors (10). These studies point out differences in treatment sensitivity between CMS1 and CMS4 (mesenchymal or immune) vs CMS2 and CMS3 (epithelial) tumors.
So far, clinical implementation of molecular classification systems has proven to be challenging. Extensive genomic and transcriptomic profiling of tumors is time-consuming and costly, and its cost-effectiveness has yet to be demonstrated. Furthermore, RNA from formalin-fixed paraffin-embedded (FFPE) tissue, which is widely used in hospitals, is vulnerable and generally of low quality. The CMS classification must be optimized for use on this material, and a standardized pipeline needs to be developed to determine individual patients’ subtypes while accounting for intra-tumor heterogeneity. We showed that the value of CMS classification is not restricted to a specific classification method but appears to be rather universal for a variety of molecular classifiers, which could enhance clinical implementation. Further advancements in this field have been made; Morris et al. (68) most recently developed a 100-gene FFPE NanoString classifier with high classification accuracy (>80.0% compared with the original CMS classifier) and validated it in a Clinical Laboratory Improvement Amendments (CLIA) certified laboratory. Similarly, for Sadanandam subtypes, a 38-gene NanoString CRCA classifier was developed that showed high concordance between classification of FFPE and fresh frozen material, and with CMS subtypes (75.0%) (109). Alternatively, image-based transcriptomic classification could be further explored, because this circumvents the above-mentioned issues and seems promising in first attempts(110). Additional studies that address these issues are warranted to pursue successful implementation of molecular classification systems in the clinic.
The limitations of our systematic review and meta-analysis arise mainly from the retrospective nature of the included studies. Survival data were either incomplete or lacking, and therefore median survival per CMS in the curative setting and some pairwise CMS comparisons per survival outcome could not be pooled. The number of studies included in the meta-analyses of localized CRC was limited and more often had an applicability concern in our risk of bias assessment, which warrants careful interpretation of these results. Using data from retrospective cohorts with differences in samples sizes, CMS classification methods and treatments, imposed heterogeneity on our meta-analyses. Most pooled cohorts used for the prognostic meta-analyses included patients who were treated with adjuvant chemotherapy in the curative setting and first-line chemotherapy in the metastatic setting. However, in the curative setting, we could not completely rule out differences in received treatment, which has affected the pooled estimates. However, sensitivity analyses showed no statistically significant change of effect sizes after exclusion of cohorts that imposed heterogeneity, indicating robustness of our meta-analyses (Supplementary Table 5, available online). Due to limited data on the predictive value of the CMSs, a traditional meta-analysis was not feasible. Consequently, although insightful, the results of the predictive potential of the CMSs are preliminary, and these findings should be validated in future randomized trials. Lastly, publication bias, as observed in funnel plots and quantified with the Egger’s test, was a minor concern for the majority of our meta-analyses (Supplementary Figures 5-7, available online).
Importantly, the varying CMS classification methods used in the included studies have an influence on the CMS distribution of patient cohorts. Labeling of individual samples can vary by methods of normalization of the raw data, settings of the specific classifier (eg, SSP vs RF in CMS R package), and preselection of specific patient groups. Therefore, we thoroughly explored the effect of different CMS labeling methods on our pooled prognostic outcomes, and we could not identify any difference between the CMS classifiers that would change our findings. The same holds true for the summarized predictive value of the CMSs. Although CMS labeling varies to some extent between the classifiers, it seems that the subtypes are sufficiently different to be detected, regardless of classification method and preselection of cohorts.
This comprehensive systematic review and meta-analysis of the CMS classification demonstrates that it is a robust framework for both prognostication and predicting treatment benefit in a biology-driven, patient-centered fashion. Future studies should include molecular subtyping in their trial design to validate these associations. In addition, refinement of the subtype classification using prognostic biomarkers may lead to an even more thorough understanding of the different subtypes. Once the CMSs are sufficiently validated in prospective clinical trials, in addition to TNM staging, they could have great impact on improving and tailoring treatment strategies for the individual CRC patient.
Funding
This work is supported by Oncode Institute, the Dutch Cancer Society (KWF 10529), the European Research Council (ERG-StG 638193), ZonMw (Vidi 016.156.308), and Innovatiefonds Zorgverzekeraars (B17-140) to LV. LV is a New York Stem Cell Foundation—Robertson Investigator.
Notes
Role of the funder: The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosures: LV received speaker and consultancy fees from Genentech, Bayer, Servier, Boehringer-Ingelheim, MSD and Pierre Fabre. LV received unrestricted grants from Novartis, Servier, Roche and Roche Diagnostics. LV declares ongoing collaborations with Firalis, LeadPharma and Genentech. DS, TB and SH have no competing interests to declare.
Author contributions: SH, TB, DS and LV designed the study. SH and TB identified eligible studies, assessed quality of the included studies, extracted data and performed data analyses. SH, TB, DS and LV wrote and revised the manuscript.
Acknowledgements: We would like to express our gratitude to René Spijker, who assisted in performing our literature search and selection, to Bart Ferwerda and Tom van den Bosch for statistical support on the meta-analyses. We would also like to thank the authors from the original studies for providing additional information upon request.
Data Availability
Data used in this systematic review and meta-analysis could be provided upon request, only with permission of the authors of the original studies.
Supplementary Material
Contributor Information
Sanne ten Hoorn, Amsterdam University Medical Centers (UMC), University of Amsterdam, Lab. for Experimental Oncology and Radiobiology, Center for Experimental and Molecular Medicine, Cancer Center Amsterdam, Amsterdam, the Netherlands; Oncode Institute, Amsterdam UMC, Amsterdam, the Netherlands.
Tim R de Back, Amsterdam University Medical Centers (UMC), University of Amsterdam, Lab. for Experimental Oncology and Radiobiology, Center for Experimental and Molecular Medicine, Cancer Center Amsterdam, Amsterdam, the Netherlands; Oncode Institute, Amsterdam UMC, Amsterdam, the Netherlands.
Dirkje W Sommeijer, Amsterdam University Medical Centers (UMC), University of Amsterdam, Lab. for Experimental Oncology and Radiobiology, Center for Experimental and Molecular Medicine, Cancer Center Amsterdam, Amsterdam, the Netherlands; Department of Medical Oncology, Amsterdam UMC, University of Amsterdam, Amsterdam, the Netherlands; Flevohospital, Department of Internal Medicine, Almere, the Netherlands.
Louis Vermeulen, Amsterdam University Medical Centers (UMC), University of Amsterdam, Lab. for Experimental Oncology and Radiobiology, Center for Experimental and Molecular Medicine, Cancer Center Amsterdam, Amsterdam, the Netherlands; Oncode Institute, Amsterdam UMC, Amsterdam, the Netherlands; Department of Medical Oncology, Amsterdam UMC, University of Amsterdam, Amsterdam, the Netherlands.
References
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 2. Punt CJ, Koopman M, Vermeulen L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat Rev Clin Oncol. 2017;14(4):235–246. [DOI] [PubMed] [Google Scholar]
- 3. Kopetz S, Grothey A, Yaeger R, et al. Encorafenib, binimetinib, and cetuximab in BRAF V600E-mutated colorectal cancer. N Engl J Med. 2019;381(17):1632–1643. [DOI] [PubMed] [Google Scholar]
- 4. Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Moretto R, Cremolini C, Rossini D, et al. Location of primary tumor and benefit from anti-epidermal growth factor receptor monoclonal antibodies in patients with RAS and BRAF wild-type metastatic colorectal cancer. Oncologist. 2016;21(8):988–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21(11):1350–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Becht E, de Reynies A, Giraldo NA, et al. Immune and stromal classification of colorectal cancer is associated with molecular subtypes and relevant for precision immunotherapy. Clin Cancer Res. 2016;22(16):4057–4066. [DOI] [PubMed] [Google Scholar]
- 8.PROSPERO: International Prospective Register of Systematic Reviews. Registered 28-04-2020 [CRD42020165483]. https://www.crd.york.ac.uk/prospero/display_record.php?RecordID=165483.
- 9. Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. 2009;339:b2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schlicker A, Beran G, Chresta CM, et al. Subtypes of primary colorectal tumors correlate with response to targeted treatment in colorectal cell lines. BMC Med Genom. 2012;5:66. [Electronic Resource] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Budinska E, Popovici V, Tejpar S, et al. Gene expression patterns unveil a new level of molecular heterogeneity in colorectal cancer. J Pathol. 2013;231(1):63–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. De Sousa EMF, Wang X, Jansen M, et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med. 2013;19(5):614–618. [DOI] [PubMed] [Google Scholar]
- 13. Marisa L, de Reynies A, Duval A, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10(5):e1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sadanandam A, Lyssiotis CA, Homicsko K, et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19(5):619–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Roepman P, Schlicker A, Tabernero J, et al. Colorectal cancer intrinsic subtypes predict chemotherapy benefit, deficient mismatch repair and epithelial-to-mesenchymal transition. Int J Cancer. 2014;134(3):552–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ouzzani M, Hammady H, Fedorowicz Z, Elmagarmid A. Rayyan-a web and mobile app for systematic reviews. Syst Rev. 2016;5(1):210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wolff RF, Moons KGM, Riley RD, et al. PROBAST: a tool to assess the risk of bias and applicability of prediction model studies. Ann Intern Med. 2019;170(1):51–58. [DOI] [PubMed] [Google Scholar]
- 18. Harrer M, Cuijpers P, Furukawa TA, Ebert DD. 2019. Doing meta-analysis in R: a hands-on guide. https://bookdown.org/MathiasHarrer/Doing_Meta_Analysis_in_R/. Accessed June 22, 2020.
- 19. Veroniki AA, Jackson D, Viechtbauer W, et al. Methods to estimate the between-study variance and its uncertainty in meta-analysis. Res Synth Methods. 2016;7(1):55–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–1558. [DOI] [PubMed] [Google Scholar]
- 21. Viechtbauer W, Cheung MW. Outlier and influence diagnostics for meta-analysis. Res Synth Methods. 2010;1(2):112–125. [DOI] [PubMed] [Google Scholar]
- 22. Baujat B, Mahe C, Pignon JP, Hill C. A graphical method for exploring heterogeneity in meta-analyses: application to a meta-analysis of 65 trials. Stat Med. 2002;21(18):2641–2652. [DOI] [PubMed] [Google Scholar]
- 23. Borenstein M, Higgins JP. Meta-analysis and subgroups. Prev Sci. 2013;14(2):134–143. [DOI] [PubMed] [Google Scholar]
- 24. Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sterne JA, Egger M. Funnel plots for detecting bias in meta-analysis: guidelines on choice of axis. J Clin Epidemiol. 2001;54(10):1046–1055. [DOI] [PubMed] [Google Scholar]
- 26. Altman DG, Bland JM. How to obtain the confidence interval from a P value. BMJ. 2011;343:d2090. [DOI] [PubMed] [Google Scholar]
- 27. Altman DG, Bland JM. How to obtain the P value from a confidence interval. BMJ. 2011;343:d2304. [DOI] [PubMed] [Google Scholar]
- 28. Pollard KS, Gilbert HN, Ge Y, Taylor SE, Dudoit S. Multtest: resampling-based multiple hypothesis testing. R Package Version 2. 2021;46. Available at: Multtest.pdf (bioconductor.org). 2021. Accessed February 20, 2021. [Google Scholar]
- 29. Bramsen JB, Rasmussen MH, Ongen H, et al. Molecular-subtype-specific biomarkers improve prediction of prognosis in colorectal cancer. Cell Reports. 2017;19(6):1268–1280. [DOI] [PubMed] [Google Scholar]
- 30. Pilati C, Taieb J, Balogoun R, Marisa L, de Reynies A, Laurent-Puig P. CDX2 prognostic value in stage II/III resected colon cancer is related to CMS classification. Ann Oncol. 2017;28(5):1032–1035. [DOI] [PubMed] [Google Scholar]
- 31. Sveen A, Bruun J, Eide PW, et al. Colorectal cancer consensus molecular subtypes translated to preclinical models uncover potentially targetable cancer cell dependencies. Clin Cancer Res. 2018;24(4):794–806. [DOI] [PubMed] [Google Scholar]
- 32. Sveen A, Johannessen B, Tengs T, et al. Multilevel genomics of colorectal cancers with microsatellite instability-clinical impact of JAK1 mutations and consensus molecular subtype 1. Genome Med. 2017;9(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aderka D, Stintzing S, Heinemann V. Explaining the unexplainable: discrepancies in results from the CALGB/SWOG 80405 and FIRE-3 studies. Lancet Oncol. 2019;20(5):e274–e283. [DOI] [PubMed] [Google Scholar]
- 34. Fessler E, Jansen M, De Sousa EMF, et al. A multidimensional network approach reveals microRNAs as determinants of the mesenchymal colorectal cancer subtype. Oncogene. 2016;35(46):6026–6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim SR, Song N, Yothers G, et al. Tumour sidedness and intrinsic subtypes in patients with stage II/III colon cancer: analysis of NSABP C-07 (NRG Oncology). Br J Cancer. 2018;118(5):629–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marisa L, Svrcek M, Collura A, et al. The balance between cytotoxic T-cell lymphocytes and immune checkpoint expression in the prognosis of colon tumors. J Natl Cancer Inst. 2018;110(1):01. [DOI] [PubMed] [Google Scholar]
- 37. Sztupinszki Z, Gyorffy B. Colon cancer subtypes: concordance, effect on survival and selection of the most representative preclinical models. Sci Rep. 2016;6:37169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Eide PW, Eilertsen IA, Sveen A, Lothe RA. Long noncoding RNA MIR31HG is a bona fide prognostic marker with colorectal cancer cell-intrinsic properties. Int J Cancer. 2019;144(11):2843–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Coebergh Van Den Braak RRJ, Ten Hoorn S, Sieuwerts AM, et al. Interconnectivity between molecular subtypes and tumor stage in colorectal cancer. BMC Cancer. 2020;20(1):850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Matsuyama T, Kandimalla R, Ishikawa T, et al. A novel mesenchymal-associated transcriptomic signature for risk-stratification and therapeutic response prediction in colorectal cancer. Int J Cancer. 2020;147(11):3250–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang B, Wang L, Liu Z, Shao B, Jiang W, Shu P. Integrated analysis identifies an immune-based prognostic signature for the mesenchymal identity in colorectal cancer. Medicine. 2020;99(25):e20617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Berg KCG, Sveen A, Holand M, et al. Gene expression profiles of CMS2-epithelial/canonical colorectal cancers are largely driven by DNA copy number gains. Oncogene. 2019;38(33):6109–6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Khan M, Loree JM, Advani SM, et al. Prognostic implications of mucinous differentiation in metastatic colorectal carcinoma can be explained by distinct molecular and clinicopathologic characteristics. Clin Colorectal Canc. 2018;17(4):e699–e709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim S-K, Kim S-Y, Kim CW, et al. A prognostic index based on an eleven gene signature to predict systemic recurrences in colorectal cancer. Exp Mol Med. 2019;51(10):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lal N, White BS, Goussous G, et al. KRAS mutation and consensus molecular subtypes 2 and 3 are independently associated with reduced immune infiltration and reactivity in colorectal cancer. Clin Cancer Res. 2018;24(1):224–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schirripa M, Biason P, Lonardi S, et al. Class 1, 2, and 3 BRAF-mutated metastatic colorectal cancer: a detailed clinical, pathologic, and molecular characterization. Clin Cancer Res. 2019;25(13):3954–3961. [DOI] [PubMed] [Google Scholar]
- 47. Sirinukunwattana K, Domingo E, Richman SD, et al. Image-based consensus molecular subtype (imCMS) classification of colorectal cancer using deep learning. Gut. 2021;70(3):544–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Trinh A, Ladrach C, Dawson HE, et al. Tumour budding is associated with the mesenchymal colon cancer subtype and RAS/RAF mutations: a study of 1320 colorectal cancers with Consensus Molecular Subgroup (CMS) data. Br J Cancer. 2018;119(10):1244–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vellinga TT, Kranenburg O, Frenkel N, et al. Lymphangiogenic gene expression is associated with lymph node recurrence and poor prognosis after partial hepatectomy for colorectal liver metastasis. Ann Surg. 2017;266(5):765–771. [DOI] [PubMed] [Google Scholar]
- 50. Xiong Y, Deng Y, Wang K, et al. Profiles of alternative splicing in colorectal cancer and their clinical significance: a study based on large-scale sequencing data. EBioMedicine. 2018;36:183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kim WG, Kim JY, Park DY. Simple classifiers for molecular subtypes of colorectal cancer. Arab J Gastroenterol. 2017;18(4):191–200. [DOI] [PubMed] [Google Scholar]
- 52. Purcell RV, Schmeier S, Lau YC, Pearson JF, Frizelle FA. Molecular subtyping improves prognostication of stage 2 colorectal cancer. BMC Cancer. 2019;19(1):1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Trumpi K, Ubink I, Trinh A, et al. Neoadjuvant chemotherapy affects molecular classification of colorectal tumors. Oncogenesis. 2017;6(7):e357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ubink I, Bloemendal HJ, Elias SG, et al. Imatinib treatment of poor prognosis mesenchymal-type primary colon cancer: a proof-of-concept study in the preoperative window period (ImPACCT). BMC Cancer. 2017;17(1):282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Allen WL, Dunne PD, McDade S, et al. Transcriptional subtyping and CD8 immunohistochemistry identifies poor prognosis stage II/III colorectal cancer patients who benefit from adjuvant chemotherapy. J Clin Oncol Precis Oncol. 2018;(2):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cremolini C, Benelli M, Fontana E, et al. Benefit from anti-EGFRs in RAS and BRAF wild-type metastatic transverse colon cancer: a clinical and molecular proof of concept study. ESMO Open. 2019;4(2):e000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Del Rio M, Mollevi C, Bibeau F, et al. Molecular subtypes of metastatic colorectal cancer are associated with patient response to irinotecan-based therapies. Eur J Cancer. 2017;76:68–75. [DOI] [PubMed] [Google Scholar]
- 58. Dienstmann R, Villacampa G, Sveen A, et al. Relative contribution of clinicopathological variables, genomic markers, transcriptomic subtyping and microenvironment features for outcome prediction in stage II/III colorectal cancer. Ann Oncol. 2019;30(10):1622–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dunne PD, O’Reilly PG, Coleman HG, et al. Stratified analysis reveals chemokine-like factor (CKLF) as a potential prognostic marker in the MSI-immune consensus molecular subtype CMS1 of colorectal cancer. Oncotarget. 2016;7(24):36632–36644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Haasnoot KJC, Backes Y, Moons LMG, et al. ; on behalf of the Dutch T1 CRC Working Group. Associations of non-pedunculated T1 colorectal adenocarcinoma outcome with consensus molecular subtypes, immunoscore, and microsatellite status: a multicenter case-cohort study. Mod Pathol. 2020;33(12):2626–2636. [DOI] [PubMed] [Google Scholar]
- 61. Jary M, Hasanova R, Vienot A, et al. Molecular description of ANGPT2 associated colorectal carcinoma. Int J Cancer. 2020;147(7):2007–2018. [DOI] [PubMed] [Google Scholar]
- 62. Kawazoe A, Kuboki Y, Shinozaki E, et al. Multicenter phase I/II trial of napabucasin and pembrolizumab in patients with metastatic colorectal cancer (EPOC1503/SCOOP Trial). Clin Cancer Res. 2020;26(22):5887–5894. [DOI] [PubMed] [Google Scholar]
- 63. Kwon Y, Park M, Jang M, et al. Prognosis of stage III colorectal carcinomas with FOLFOX adjuvant chemotherapy can be predicted by molecular subtype. Oncotarget. 2017;8(24):39367–39381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lenz H-J, Ou F-S, Venook AP, et al. Impact of consensus molecular subtype on survival in patients with metastatic colorectal cancer: results from CALGB/SWOG 80405 (alliance). J Clin Oncol. 2019;37(22):1876–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lenz HJ, Argiles G, Yoshino T, et al. Association of consensus molecular subtypes and molecular markers with clinical outcomes in patients with metastatic colorectal cancer: biomarker analyses from LUME-colon 1. Clin Colorectal Canc. 2021;20(1):84:–95.. [DOI] [PubMed] [Google Scholar]
- 66. Li Y, Yao Q, Zhang L, et al. Immunohistochemistry-based consensus molecular subtypes as a prognostic and predictive biomarker for adjuvant chemotherapy in patients with stage II colorectal cancer. Oncologist. 2020;25(12):e1968–e1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mooi JK, Wirapati P, Asher R, et al. The prognostic impact of consensus molecular subtypes (CMS) and its predictive effects for bevacizumab benefit in metastatic colorectal cancer: molecular analysis of the AGITG MAX clinical trial. Ann Oncol. 2018;29(11):2240–2246. [DOI] [PubMed] [Google Scholar]
- 68. Morris JS, Luthra R, Liu Y, et al. Development and validation of a gene signature classifier for consensus molecular subtyping of colorectal carcinoma in a CLIA-certified setting. Clin Cancer Res. 2021;27(1):120–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Okita A, Takahashi S, Ouchi K, et al. Consensus molecular subtypes classification of colorectal cancer as a predictive factor for chemotherapeutic efficacy against metastatic colorectal cancer. Oncotarget. 2018;9(27):18698–18711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Piskol R, Huw L, Sergin I, et al. A clinically applicable gene-expression classifier reveals intrinsic and extrinsic contributions to consensus molecular subtypes in primary and metastatic colon cancer. Clin Cancer Res. 2019;25(14):4431–4442. [DOI] [PubMed] [Google Scholar]
- 71. Shinto E, Oki E, Shimokawa M, et al. A validation study for recurrence risk stratification of stage II colon cancer using the 55-gene classifier. Oncology. 2020;98(8):534–538. [DOI] [PubMed] [Google Scholar]
- 72. Song N, Pogue-Geile KL, Gavin PG, et al. Clinical outcome from oxaliplatin treatment in stage II/III colon cancer according to intrinsic subtypes: secondary analysis of NSABP C-07/NRG oncology randomized clinical trial. JAMA Oncol. 2016;2(9):1162–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Stintzing S, Wirapati P, Lenz HJ, et al. Consensus molecular subgroups (CMS) of colorectal cancer (CRC) and first-line efficacy of FOLFIRI plus cetuximab or bevacizumab in the FIRE3 (AIO KRK-0306) trial. Ann Oncol. 2019;30(11):1796–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Trinh A, Trumpi K, De Sousa EMF, et al. Practical and robust identification of molecular subtypes in colorectal cancer by immunohistochemistry. Clin Cancer Res. 2017;23(2):387–398. [DOI] [PubMed] [Google Scholar]
- 75. Williams DS, Mouradov D, Jorissen RN, et al. Lymphocytic response to tumour and deficient DNA mismatch repair identify subtypes of stage II/III colorectal cancer associated with patient outcomes. Gut. 2019;68(3):465–474. [DOI] [PubMed] [Google Scholar]
- 76. Woolston A, Khan K, Spain G, et al. Genomic and transcriptomic determinants of therapy resistance and immune landscape evolution during anti-EGFR treatment in colorectal cancer. Cancer Cell. 2019;36(1):35–50.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Schlicker A, Ellappalayam A, Beumer IJ, et al. Investigating the concordance in molecular subtypes of primary colorectal tumors and their matched synchronous liver metastasis. Int J Cancer. 2020. doi:10.1002/ijc.33003. [DOI] [PubMed] [Google Scholar]
- 78. Borelli B, Fontana E, Giordano M, et al. Consensus molecular subtypes and CRC assigner classifications in metastatic colorectal cancer (mCRC): prognostic and predictive impact in the TRIBE2 study. J Clin Oncol. 2020;38(suppl 15):4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gomez OH, Soto VH, Machado I, et al. Prognostic and predictive role of consensus molecular subtypes (CMS) determined by immunohistochemistry in metastatic colorectal cancer (mCRC). Ann Oncol. 2020;31:S442–S3. [Google Scholar]
- 80. Lam M, Marie PK, Sarshekeh AM, et al. Consensus molecular subtypes (CMS) as a marker for treatment and disease biology in metastatic colorectal cancer (CRC). J Clin Oncol. 2020;38(suppl 15):4089. [Google Scholar]
- 81. Lee MS, Selitsky SR, Parker JS, et al. Association of consensus molecular subtypes (CMS) with time to progression (TTP), progression free survival (PFS), and overall survival (OS) with second-line FOLFIRI +/- regorafenib in metastatic colorectal cancer (mCRC). J Clin Oncol. 2019;37(suppl 4):597. [Google Scholar]
- 82. Marisa L, Ayadi M, Balogoun R, et al. Clinical utility of colon cancer molecular subtypes: validation of two main colorectal molecular classifications on the PETACC-8 phase III trial cohort. J Clin Oncol. 2017;35(suppl 15):3509. [Google Scholar]
- 83. Pogue-Geile KL, Andre T, Song N, et al. Association of colon cancer (CC) molecular signatures with prognosis and oxaliplatin prediction-benefit in the MOSAIC Trial (Multicenter International Study of Oxaliplatin/5-FU-LV in the Adjuvant Treatment of Colon Cancer). J Clin Oncol. 2019;37(suppl 15):3503. [Google Scholar]
- 84. Sarshekeh AM, Lam M, Zorrilla IR, et al. Consensus molecular subtype (CMS) as a novel integral biomarker in colorectal cancer: a phase II trial of bintrafusp alfa in CMS4 metastatic CRC. J Clin Oncol. 2020;38(suppl 15):4084. [Google Scholar]
- 85. Yuki S, Gamoh M, Denda T, et al. Analysis of consensus molecular subtypes (CMS) classification in the TRICOLORE trial: a randomized phase III trial of S-1 and irinotecan (IRI) plus bevacizumab (Bmab) versus mFOLFOX6 or CapeOX plus Bmab as first-line treatment for metastatic colorectal cancer (mCRC). J Clin Oncol. 2020;38(suppl 4):169.31536446 [Google Scholar]
- 86. Aasebo KO, Dragomir A, Sundstrom M, et al. Consequences of a high incidence of microsatellite instability and BRAF-mutated tumors: a population-based cohort of metastatic colorectal cancer patients. Cancer Med. 2019;8(7):3623–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Venderbosch S, Nagtegaal ID, Maughan TS, et al. Mismatch repair status and BRAF mutation status in metastatic colorectal cancer patients: a pooled analysis of the CAIRO, CAIRO2, COIN, and FOCUS studies. Clin Cancer Res. 2014;20(20):5322–5330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tran B, Kopetz S, Tie J, et al. Impact of BRAF mutation and microsatellite instability on the pattern of metastatic spread and prognosis in metastatic colorectal cancer. Cancer. 2011;117(20):4623–4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Des Guetz G, Schischmanoff O, Nicolas P, Perret GY, Morere JF, Uzzan B. Does microsatellite instability predict the efficacy of adjuvant chemotherapy in colorectal cancer? A systematic review with meta-analysis. Eur J Cancer. 2009;45(10):1890–1896. [DOI] [PubMed] [Google Scholar]
- 90. Sargent DJ, Marsoni S, Monges G, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28(20):3219–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Tajima A, Hess MT, Cabrera BL, Kolodner RD, Carethers JM. The mismatch repair complex hMutS alpha recognizes 5-fluorouracil-modified DNA: implications for chemosensitivity and resistance. Gastroenterology. 2004;127(6):1678–1684. [DOI] [PubMed] [Google Scholar]
- 92. Ghajar CM, Bissell MJ. Metastasis: pathways of parallel progression. Nature. 2016;540(7634):528–529. [DOI] [PubMed] [Google Scholar]
- 93. Naxerova K, Reiter JG, Brachtel E, et al. Origins of lymphatic and distant metastases in human colorectal cancer. Science. 2017;357(6346):55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. van den Berg I, van de Weerd S, Roodhart JML, et al. Improving clinical management of colon cancer through CONNECTION, a nation-wide colon cancer registry and stratification effort (CONNECTION II trial): rationale and protocol of a single arm intervention study. BMC Cancer. 2020;20(1):776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. de Boer NL, Brandt-Kerkhof ARM, Madsen EVE, et al. Concomitant intraperitoneal and systemic chemotherapy for extensive peritoneal metastases of colorectal origin: protocol of the multicentre, open-label, phase I, dose-escalation INTERACT trial. BMJ Open. 2019;9(12):e034508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ubink I, van Eden WJ, Snaebjornsson P, et al. Histopathological and molecular classification of colorectal cancer and corresponding peritoneal metastases. Br J Surg. 2018;105(2):e204–e211. [DOI] [PubMed] [Google Scholar]
- 97. Overman MJ, McDermott R, Leach JL, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18(9):1182–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Overman MJ, Lonardi S, Wong KYM, et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J Clin Oncol. 2018;36(8):773–779. [DOI] [PubMed] [Google Scholar]
- 99. Tian L, Goldstein A, Wang H, et al. Mutual regulation of tumour vessel normalization and immunostimulatory reprogramming. Nature. 2017;544(7649):250–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Hamzah J, Jugold M, Kiessling F, et al. Vascular normalization in Rgs5-deficient tumours promotes immune destruction. Nature. 2008;453(7193):410–414. [DOI] [PubMed] [Google Scholar]
- 101. Arnold D, Lueza B, Douillard JY, et al. Prognostic and predictive value of primary tumour side in patients with RAS wild-type metastatic colorectal cancer treated with chemotherapy and EGFR directed antibodies in six randomized trials. Ann Oncol. 2017;28(8):1713–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Yang M, Schell MJ, Loboda A, et al. Repurposing EGFR inhibitor utility in colorectal cancer in mutant APC and TP53 subpopulations. Cancer Epidemiol Biomarkers Prev. 2019;28(7):1141–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Thota R, Yang M, Davis T, Schell MJ, Pledger WJ, Yeatman TJ. APC and TP53 as potential biomarkers for EGFR sensitivity in colorectal cancer. J Clin Oncol. 2020;38(suppl 15):4094. [Google Scholar]
- 104. Smeby J, Sveen A, Merok MA, et al. CMS-dependent prognostic impact of KRAS and BRAFV600E mutations in primary colorectal cancer. Ann Oncol. 2018;29(5):1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Soldevilla B, Carretero-Puche C, Gomez-Lopez G, et al. The correlation between immune subtypes and consensus molecular subtypes in colorectal cancer identifies novel tumour microenvironment profiles, with prognostic and therapeutic implications. Eur J Cancer (Oxford, England: 1990). 2019;123:118–129. [DOI] [PubMed] [Google Scholar]
- 106. Lindner AU, Salvucci M, Morgan C, et al. BCL-2 system analysis identifies high-risk colorectal cancer patients. Gut. 2017;66(12):2141–2148. [DOI] [PubMed] [Google Scholar]
- 107. van der Waals LM, Jongen JMJ, Elias SG, et al. Increased levels of oxidative damage in liver metastases compared with corresponding primary colorectal tumors: association with molecular subtype and prior treatment. Am J Pathol. 2018;188(10):2369–2377. [DOI] [PubMed] [Google Scholar]
- 108. Pal R, Wei N, Song N, et al. Molecular subtypes of colorectal cancer in pre-clinical models show differential response to targeted therapies: treatment implications beyond KRAS mutations. PLoS ONE [Electronic Resource. 2018;13(8):e0200836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Ragulan C, Eason K, Fontana E, et al. Analytical validation of multiplex biomarker assay to stratify colorectal cancer into molecular subtypes. Sci Rep. 2019;9(1):7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Sirinukunwattana K, Domingo E, Richman SD, et al. Image-based consensus molecular subtype (imCMS) classification of colorectal cancer using deep learning. Gut. 2021;70(3):544–554. doi:10.1136/gutjnl-2019-319866. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data used in this systematic review and meta-analysis could be provided upon request, only with permission of the authors of the original studies.