Abstract
The effect of quinolones on the inhibition of DNA synthesis in Staphylococcus aureus was examined by using single resistance mutations in parC or gyrA to distinguish action against gyrase or topoisomerase IV, respectively. Norfloxacin preferentially attacked topoisomerase IV and blocked DNA synthesis slowly, while nalidixic acid targeted gyrase and inhibited replication rapidly. Ciprofloxacin exhibited an intermediate response, consistent with both enzymes being targeted. The absence of RecA had little influence on target choice by this assay, indicating that differences in rebound (repair) DNA synthesis were not responsible for the results. At saturating drug concentrations, norfloxacin and a gyrA mutant were used to show that topoisomerase IV-norfloxacin-cleaved DNA complexes are distributed on the S. aureus chromosome at intervals of about 30 kbp. If cleaved complexes block DNA replication, as indicated by previous work, such close spacing of topoisomerase-quinolone-DNA complexes should block replication rapidly (replication forks are likely to encounter a cleaved complex within a minute). Thus, the slow inhibition of DNA synthesis at growth-inhibitory concentrations suggests that a subset of more distantly distributed complexes is physiologically relevant for drug action and is unlikely to be located immediately in front of the DNA replication fork.
The fluoroquinolones are potent antibacterial agents that have DNA gyrase and DNA topoisomerase IV as their intracellular targets (reviewed in references 10, 13, and 14). The compounds have had good success against gram-negative bacteria, but resistance of gram-positive pathogens, such as Staphylococcus aureus, has become a problem (5). This difference is probably due in part to the sensitivity of gyrase from S. aureus being much lower than that of gyrase from gram-negative species such as Escherichia coli (4). Indeed, sensitivity is even lower than that of topoisomerase IV, making the latter enzyme the primary target of the commonly used compounds ciprofloxacin (16, 17, 29) and levofloxacin (1, 18, 22). Studies of quinolone action against topoisomerase IV in E. coli (26, 27) suggest that the chromosomal location of this enzyme, which is thought to be behind replication forks (26), may exacerbate the ineffectiveness of quinolones with organisms in which topoisomerase IV is the primary target. How topoisomerase IV in S. aureus responds to quinolone attack has not been studied, and so it is unclear whether work with E. coli can be generalized.
The key step in quinolone action is trapping gyrase or topoisomerase IV on DNA as ternary drug-enzyme-DNA complexes (reviewed in reference 10). The complexes block replication fork movement (23), explaining the well-known ability of the quinolones to inhibit DNA synthesis (8). Inhibition of DNA synthesis correlates well with inhibition of growth as measured by MIC (7); consequently, the MIC has been taken as a measure of complex formation (40). In gyrase-containing complexes, the DNA is broken and treatment with ionic detergents releases chromosomal DNA fragments (35). Measurement of fragment size provides an estimate of the distribution of cleaved complexes on chromosomal DNA (35).
In the present work we examined the ability of several quinolones to inhibit DNA synthesis in S. aureus. Mutations in gyrA (gyrase) or parC (also called grlA in S. aureus) (topoisomerase IV) were used to direct the compounds toward one target or the other. The compounds tested showed a clear preference: norfloxacin preferentially attacked topoisomerase IV, while nalidixic acid preferred gyrase. Even though nalidixic acid is much less potent than norfloxacin, it inhibited DNA synthesis more rapidly. When the distribution of norfloxacin-topoisomerase IV-cleaved DNA complexes was measured, they were found at 30-kbp intervals, about three times more often than gyrase complexes on the E. coli chromosome (35). The slower inhibition of DNA synthesis, despite the close spacing of complexes, raises questions about bacterial chromosome structure. We also found that the MIC of norfloxacin was subsaturating with respect to topoisomerase IV complex formation on the chromosome. This finding suggests that the inhibition of DNA synthesis and cell growth is caused by a subset of drug-enzyme-DNA complexes or possibly by complexes in which single-strand rather than double-strand DNA breaks occurred.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The S. aureus strains used in this study are described in Table 1. These strains were grown at 37°C with shaking in Trypticase soy broth or CY liquid medium (30), unless otherwise specified. To measure the inhibition of DNA synthesis, the bacteria were grown in minimal medium as described by Wilkinson (38), using glucose as a carbon source without additional components. For recA-deficient strains, erythromycin was added at 5 μg/ml.
TABLE 1.
S. aureus strains used
| Strain | Genotype | MIC (μg/ml) of:
|
Source or reference | ||
|---|---|---|---|---|---|
| Norfloxacin | Nalidixic acid | Ciprofloxacin | |||
| ISP794 | 8325 pig-131 | 0.5 | 64.0 | 0.25 | 36 |
| MT5 | ISP794 gyrB142 (Ser102,Ile144) | 0.5 | 64.0 | 0.25 | 37 |
| SS1 | ISP794 gyrB142 (Ser102, Ile144) gyrA (Leu84) | 0.5 | 256.0 | 0.25 | 3 |
| MT5224c4 | ISP794 grlA542 (Phe80) | 8.0 | 64.0 | 2.0 | 37 |
| EN1252a | ISP794 gyrB142 (Ser102, Ile144) gyrA (Leu84) grlA542 (Phe80) | 64.0 | 256.0 | 32.0 | 3 |
| ISP2272 | 8325-4 uvs-568Ω(Tn551)1074 | 2 | |||
| BF10 | ISP794 uvs-568Ω(Tn551)1074 | This study: ISP2272 DNA × ISP794 | |||
| BF11 | MT5224c4 uvs-568Ω(Tn551)1074 | This study: ISP2272 DNA × MT5224c4 | |||
| BF12 | SS1 uvs-568Ω(Tn551)1074 | This study: ISP2272 DNA × SS1 | |||
Construction of recA-deficient derivatives.
Chromosomal DNA of strain ISP2272, which carries the uvs-568 mutation closely linked to the transposon Tn551 (2), was obtained by the procedure of Stahl and Pattee (36). This DNA was then introduced into competent cells of strains ISP794, MT5224c4, and SS1 as previously described (36), followed by the selection of transformants on agar containing 5 μg of erythromycin/ml. This procedure produced strains BF10, BF11, and BF12, respectively (Table 1). Two findings verified that these strains lacked recA. First, the MIC of ethylmethane sulfonate for strains BF10, BF11, and BF12 was only 25% of that for parental strains. Second, strains BF10, BF11, and BF12 exhibited a threefold increase in sensitivity to ultraviolet light relative to the parental strains.
Measurement of DNA synthesis.
The rate of DNA synthesis was determined at 3- to 5-min intervals by transferring 200 μl of an exponentially growing culture of S. aureus to tubes containing 1 μCi of [3H]thymidine. After incubation at 37°C for 2 min, the incorporation of radioactivity was terminated by the addition of 1 ml of 10% trichloroacetic acid. Acid precipitates were processed and counted on filters as previously described (15).
Sucrose density gradient analysis.
Sedimentation analysis was performed as previously described for E. coli (6, 11, 35). S. aureus was grown to mid-log phase, and DNA was radioactively labeled by additional growth for 60 min in [3H]thymidine (30 μCi/ml; Amersham). The cells were then treated with various concentrations of norfloxacin for 20 min, after which they were chilled on ice and harvested by centrifugation. The protoplasts were prepared by incubation in SSTB buffer (50 mM Tris-HCl [pH 7.6], 50 mM NaCl, 20% [wt/vol] sucrose) plus 400 μg of lysostaphin/ml and norfloxacin on ice for 20 min. The protoplasts were lysed by gentle mixing with 5 volumes of preheated (60°C) lysis buffer (1.2% sodium dodecyl sulfate, 25 mM EDTA). DNA was gently loaded onto 5 to 20% (wt/vol) sucrose density gradients containing 0.1 M NaCl, 0.05 M sodium phosphate buffer (pH 6.8), and 0.5% sodium dodecyl sulfate. Centrifugation was performed with a Beckman SW50.1 rotor at 23°C. The gradients were fractionated from the bottom of tubes, and DNA was precipitated on paper filters by using 10% trichloroacetic acid followed by washes with 1 N HCl, water, and ethanol. The amount of radioactivity on the filters was determined by liquid scintillation spectrometry. 14C-labeled bacteriophage T4B DNA was prepared as previously described (11).
RESULTS
Selectivity of ciprofloxacin, norfloxacin, and nalidixic acid for type II topoisomerases with respect to the inhibition of DNA synthesis.
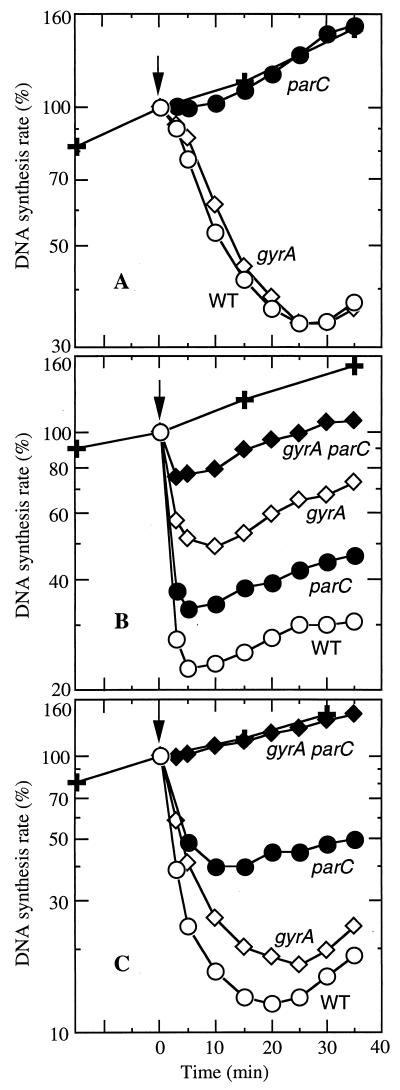
As a test for quinolone target specificity during exponential growth in liquid medium, we compared the inhibition of DNA synthesis by norfloxacin, nalidixic acid, and ciprofloxacin in quinolone-resistant gyrA and parC mutants of S. aureus (for MICs, see Table 1). Norfloxacin inhibited DNA synthesis gradually for both wild-type and gyrA (Leu84) strains (Fig. 1A), as expected from a prior study with E. coli in which the selective use of resistance mutations was thought to leave topoisomerase IV as the target (27). A parC mutation that conferred norfloxacin resistance abolished the inhibition of DNA synthesis, while a gyrA (Leu84) allele had almost no effect (Fig. 1A).
FIG. 1.
Quinolone-mediated inhibition of DNA synthesis in S. aureus topoisomerase mutants. Strains ISP794 (wild type [WT]), MT5224c4 (parC [Phe80]), SS1 (gyrA [Leu84]), and EN1252a (gyrA [Leu84] parC [Phe80]) were grown to mid-log phase in minimal medium (38). Quinolone was added at zero time (arrows), and the rate of DNA synthesis was determined every 3 to 5 min. For an untreated culture (✚), determinations were every 15 min. The relative rate of DNA synthesis is expressed as a percentage of the untreated control at time zero. (A) Norfloxacin, 0.25 μg/ml; (B) nalidixic acid, 25 μg/ml; (C) ciprofloxacin, 0.25 μg/ml. Data are the means of at least two determinations.
Nalidixic acid, a less potent quinolone, inhibited synthesis rapidly at 25 μg/ml (Fig. 1B), even though the extent of inhibition was similar to that observed with norfloxacin. In contrast to the findings with norfloxacin, there was a limited reduction of inhibition in strain MT5224c4 (parC [Phe80]) and a more extensive reduction in strain SS1 (gyrA [Leu84]). The most extensive reduction was observed with strain EN1252a (gyrA [Leu84] parC [Phe80]) (Fig. 1B). We tested two additional concentrations of nalidixic acid (14 and 50 μg/ml), and a similar pattern of inhibition was observed (data not shown). These findings indicate that at these concentrations of nalidixic acid, interactions with both topoisomerases contribute to the inhibition of DNA synthesis but interaction with DNA gyrase dominates.
Ciprofloxacin behavior was opposite to that of nalidixic acid, with inhibition of DNA synthesis reduced more in a parC (Phe80) mutant than in a gyrA (Leu84) mutant (Fig. 1C). In this case, interaction with topoisomerase IV was a major contributor to the inhibition of DNA synthesis. The inhibition of DNA synthesis was completely abolished, however, only in EN1252a (gyrA [Leu84] parC [Phe80]), indicating that interactions with both enzymes contributed to the kinetics of inhibition of DNA synthesis with ciprofloxacin. In an effort to identify a more selective concentration at which partial inhibition attributable to DNA gyrase was undetectable, we tested a threefold-lower concentration of ciprofloxacin (0.075 μg/ml). The same pattern of inhibition with the parent and mutant strains was seen (data not shown). Thus, ciprofloxacin, in the concentration ranges studied, inhibits DNA synthesis to a considerable extent through interaction with topoisomerase IV, but interactions with both topoisomerase IV and DNA gyrase contribute to the rapidity of inhibition of DNA synthesis. Moreover, inhibition by ciprofloxacin is slower than that by nalidixic acid even when the total extent of inhibition is greater (compare Fig. 1B and C). Differences in the kinetics of quinolone-mediated inhibition of DNA synthesis identify norfloxacin as more selective than nalidixic acid or ciprofloxacin for generating topoisomerase IV-quinolone cleavage complexes in S. aureus during short-term treatment.
Some of the quinolone experiments described above utilized strain SS1 (gyrA [Leu84]), which contained a closely linked coumarin-resistant gyrB mutation used for strain construction (3). As a control, we compared the inhibition of DNA synthesis by norfloxacin, nalidixic acid, and ciprofloxacin in the otherwise isogenic wild-type strain ISP794 and its coumarin-resistant gyrB mutant MT5. No difference was observed in the rates of inhibition (data not shown). Thus, the gyrB mutation itself does not affect the rates of inhibition of DNA synthesis by the quinolones tested and probably had no effect on inhibition in strain SS1.
Rates of inhibition of DNA synthesis resulting from quinolone target selectivity.
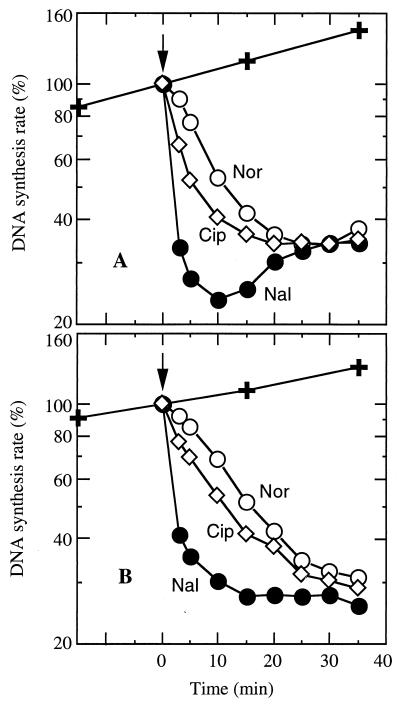
In order to compare directly the kinetics of inhibition of DNA synthesis mediated by different quinolones in the wild-type strain, concentrations of quinolones were adjusted to generate a plateau inhibition of DNA synthesis at approximately 30% of control values. The DNA synthesis rate was determined with additional samples taken 3 min after addition of the drug (Fig. 2A). Nalidixic acid produced the most rapid inhibition (67% at 3 min), followed by ciprofloxacin (34%) and norfloxacin (10%). Norfloxacin inhibited synthesis at less than 10% of the rate seen with nalidixic acid, i.e., norfloxacin required 36 to 40 min to produce the same level of inhibition as nalidixic acid at 3 min.
FIG. 2.
Quinolone-mediated inhibition of DNA synthesis in wild-type S. aureus. Inhibition of DNA synthesis was determined as described for Fig. 1. The relative rate of DNA synthesis is expressed as a percentage of the untreated control at time zero. ✚, untreated control; Nor, norfloxacin (0.25 μg/ml); Nal, nalidixic acid (22 μg/ml); Cip, ciprofloxacin (0.13 μg/ml). (A) Strain ISP794 (recA+); (B) strain BF10 (recA deficient). Data are the means of at least two determinations.
Repair DNA synthesis, which is thought to contribute to the plateau and rebound of DNA synthesis that is apparent 30 min or more after addition of a quinolone (15), might also have undetected effects on early kinetics. Thus, we determined the early kinetics of inhibition of DNA synthesis by using a recA-deficient strain (BF10) at the quinolone concentrations used with the wild-type cells (Fig. 2B). As expected, for BF10 the plateau of inhibition was less pronounced and no rebound DNA synthesis was seen. Furthermore, the same relative pattern of kinetics of inhibition by the three quinolones occurred: nalidixic acid (59% inhibition at 3 min), ciprofloxacin (23%), and norfloxacin (8%).
Altering the kinetics of DNA synthesis inhibition.
Depending on the intrinsic differences in the sensitivities of wild-type and mutant gyrase and topoisomerase IV for a given quinolone, conditions may also be adjusted to make one enzyme or the other the primary target. For example, at the lowest growth-inhibitory concentrations of many quinolones for wild-type S. aureus, interactions with topoisomerase IV may predominate; in contrast, the higher quinolone concentrations required to inhibit growth of a resistant parC mutant strain may result in effects due to drug interactions with DNA gyrase.
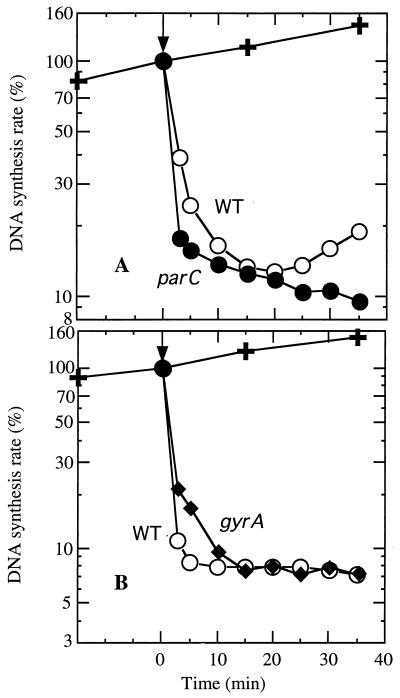
To confirm that a low rate of inhibition of DNA synthesis is due to interaction with topoisomerase IV and that a higher rate of inhibition is due to interaction with DNA gyrase, we used the two quinolones that interacted with both topoisomerases but with differing relative selectivities (ciprofloxacin and nalidixic acid). For these experiments we reasoned that for strains with resistance mutations in the primary enzyme target, use of higher quinolone concentrations at a level corresponding to the MIC for the mutant would increase drug interaction with the secondary enzyme target and thereby alter the pattern of kinetics of inhibition of DNA synthesis. These results could then be compared with the pattern of inhibition for the parent wild-type strain at its MIC, a concentration at which interaction with the primary enzyme target predominates.
For ciprofloxacin, we determined the inhibition of DNA synthesis of ISP794 (recA+ gyrA+ parC+) at the MIC (0.25 μg/ml) (19) compared to that of MT5224c4 (recA+ parC [Phe80]) at the MIC (1 μg/ml) (Fig. 3A). These concentrations also generated a similar plateau of inhibition at 30 min. The rate of inhibition of DNA synthesis for ISP794 (61% inhibition at 3 min) was lower than that for MT5224c4 (82% inhibition). When the recA-deficient derivatives of ISP794 (BF10) and MT5224c4 (BF11) were compared using the same concentrations of ciprofloxacin as for the recA+ strains, a similar but more pronounced pattern was seen, with more rapid inhibition in the parC mutant, in which interaction of the drug was with gyrase (70 versus 36% inhibition at 3 min).
FIG. 3.
Altering the kinetics of DNA synthesis inhibition. (A) Ciprofloxacin inhibition of DNA synthesis in wild-type (WT) (0.25 μg/ml) and parC mutant (1 μg/ml) S. aureus strains; (B) nalidixic acid inhibition of DNA synthesis in wild-type (WT) (60 μg/ml) and gyrA mutant (175 μg/ml) S. aureus strains. Inhibition of DNA synthesis was determined as described for Fig. 1. The relative rate of DNA synthesis is expressed as a percentage of the untreated control at time zero. ✚, untreated control; WT, ISP794 (recA+ gyrA+ parC+); parC, MT5224c4 (recA+ parC [Phe80]); gyrA, SS1 (recA+ gyrA [Leu84]). Data are the means of at least two determinations.
For experiments with nalidixic acid, as for those with ciprofloxacin, the drug concentrations chosen were at the respective MICs of nalidixic acid for the strains used (ISP794 [60 μg/ml] and SS1 gyrA [Leu84] [175 μg/ml]) in order to promote interaction with the primary target (gyrase) in the wild-type strain and interaction with the secondary target (topoisomerase IV) in the mutant strain and to generate a similar plateau of inhibition for the two kinetic curves. For the recA+ strains, the initial rate of inhibition of DNA synthesis at 3 min was slightly lower for SS1 (recA+ gyrA [Leu84] parC+) (79%) than for ISP794 (recA+ gyrA+ parC+) (89%). This lower rate persisted until the plateau level of inhibition was reached, despite the use of higher concentrations of nalidixic acid for strain SS1 (Fig. 3B). These data are consistent with nalidixic acid attacking topoisomerase IV in gyrA mutants.
Chromosomal distribution of topoisomerase IV-DNA-quinolone complexes.
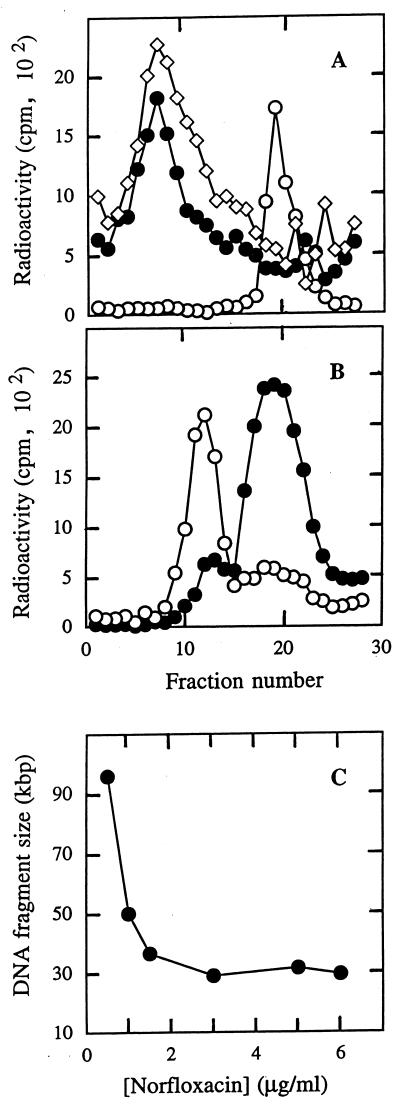
To measure the size of DNA fragments generated by quinolone action, it was necessary to develop cell lysis and DNA handling procedures that themselves introduce few DNA breaks. Standard protocols used for preparing S. aureus protoplasts, which involve long periods of incubation with lysostaphin (35), produced small DNA fragments (data not shown), presumably due to the release of cellular nucleases. The modification of buffer conditions, lysostaphin concentration, and incubation temperature allowed us to recover large DNA molecules from cells that were not treated with norfloxacin (Fig. 4A; identical sedimentation profiles were obtained with the DNA from untreated cultures of ISP794, SS1, and EN1252a [data not shown]). When S. aureus strain SS1 gyrA (Leu84) was treated with 3 μg of norfloxacin/ml for 20 min, chromosomal DNA was cleaved to a size much smaller than that of bacteriophage T4 DNA (Fig. 4B). To ensure that the norfloxacin concentration was saturating, a series of cultures was treated with increasing concentrations of norfloxacin and DNA sedimentation was measured as for Fig. 4B (under the conditions used, the sedimentation rate was almost linear with distance sedimented, allowing the use of a single molecular-weight marker to calibrate the gradients, as described in reference 11). Number-average molecular weight was calculated (11) and plotted against norfloxacin concentration (Fig. 4C). Since the sedimentation coefficient of DNA fragments of this size is unaffected by rotor speed under the conditions used (33) and since varying the DNA concentration had no effect on the DNA sedimentation coefficient at saturating norfloxacin concentrations (data not shown), no additional correction for these effects was required. Moreover, norfloxacin treatment of a gyrA parC double mutant, under conditions found to be saturating for strain SS1 gyrA (Leu84), caused no detectable breaks (Fig. 4A). Consequently, topoisomerase IV was the drug target in the gyrA mutant. To show that the formation of complexes of norfloxacin, topoisomerase IV, and DNA was complete by the 20-min exposure used above, we performed a time-course experiment using strain SS1 (gyrA [Leu84]) exposed to 3 μg of norfloxacin/ml. Entrapment of topoisomerase IV-DNA complexes by norfloxacin was nearly complete by 5 min and was fully saturated by 20 min (data not shown). These controls allow us to conclude from the data shown in Fig. 4C that under saturating conditions, norfloxacin produces breaks at intervals of about 30 kbp, which correspond to about 100 breaks (topoisomerase IV-DNA interactions) per 2,900-kbp chromosome.
FIG. 4.
Sedimentation of DNA fragments obtained from norfloxacin-treated S. aureus. [3H]DNA was obtained from extracts of gently lysed S. aureus and sedimented into sucrose density gradients by centrifugation with a Beckman SW50.1 rotor at 7,100 rpm at 23°C. (A) DNA from strain SS1 (gyrA [Leu84]) (●) without quinolone treatment or strain EN1252a (gyrA [Leu84] parC [Phe80]) (◊), treated with 5 μg of norfloxacin/ml, was sedimented in separate gradients for 17 h. Each gradient contained 14C-labeled bacteriophage T4B DNA (molecular weight = 1.32 × 108 daltons) (○) as a sedimentation marker. Sedimentation is from right to left. (B) DNA from strain SS1 (gyrA [Leu84]) (●), treated with 3 μg of norfloxacin/ml for 20 min, was centrifuged as for panel A for 40 h. ○, 14C-labeled T4B DNA. For both panels A and B each experiment was repeated at least once, and sedimentation profiles similar to those shown were obtained. (C) Effects of norfloxacin concentration on DNA size. Strain SS1 (gyrA [Leu84]) was treated with the indicated concentrations of norfloxacin, cells were gently lysed, and DNA size was determined by sedimentation as shown in panel B. Number-average molecular weight was calculated as previously described (11).
DISCUSSION
The work described above continues our characterization of fluoroquinolone action against S. aureus (9, 19, 20, 29, 39). In this organism, topoisomerase IV is the primary target of most fluoroquinolones (16, 17, 29), and so its sensitivity is a key aspect of drug action. As with gyrase, the physiological observations appear to derive from the formation of quinolone-topoisomerase IV-DNA complexes. We used resistance alleles and drug-target preferences to direct norfloxacin to topoisomerase IV and nalidixic acid to gyrase. We then found that the attack of topoisomerase IV inhibited DNA replication much more slowly than the attack of gyrase (Fig. 1 and 2). A similar phenomenon has been seen with E. coli (27). With that organism, slow inhibition is likely to be due in part to topoisomerase IV functioning behind replication forks (27; reviewed in reference 10), while rapid inhibition is likely to derive from action on gyrase ahead of forks (12).
The data presented above allow us to discern a relationship among complex formation, inhibition of DNA synthesis, and inhibition of growth, as measured by the MIC. A good correlation has been found between the inhibition of DNA synthesis and the MIC in E. coli (7). Other data show correlations between the MIC and the 50% inhibitory concentration, an in vitro measurement of drug activity against purified gyrase (24). In that case, MICs are somewhat lower than 50% inhibitory concentrations (24). Our measurement of complex formation, which we determined from DNA fragment sizes (Fig. 4), showed that the norfloxacin concentration at which all of the complexes have been created (1.5 μg/ml) (Fig. 4C) is about three times greater than the MIC. At half this concentration of norfloxacin, the DNA fragments were larger and more heterogeneous. Thus, growth-inhibitory concentrations of norfloxacin are subsaturating with respect to double-strand breaks. It appears that for S. aureus topoisomerase IV-DNA-drug complexes, as is the case with E. coli gyrase-DNA-drug complexes (35), a subset of complexes may be sufficient to inhibit cell growth and block DNA synthesis. It is also possible that the production of single-strand DNA breaks (not measured by the methods employed here) rather than of double-strand breaks correlates best with the inhibition of DNA synthesis and the MIC. For S. aureus, quinolone concentrations at the MIC caused about 75% inhibition of DNA synthesis (Fig. 1 and 2). For the oxolinic acid attack of gyrase in E. coli, this level of inhibition causes a fivefold-greater accumulation of single-strand DNA nicks than of double-strand DNA breaks (35).
Early measurements of DNA fragmentation caused by quinolones showed that gyrase complexes in E. coli are distributed at 100-kbp intervals (35), which corresponds to once per topological domain (34). In the present study, comparable measurements using norfloxacin to form topoisomerase IV-containing complexes revealed a distribution of one per 30 kbp (Fig. 4C). Based on estimated rates of replication fork progression, norfloxacin-topoisomerase IV-DNA complexes should be encountered by a replication fork on average in less than 1 min. If topoisomerase IV functions behind replication forks, as suggested by the slow inhibition of DNA synthesis in S. aureus (Fig. 2) and by several types of experiments with E. coli (26, 27), it is difficult to explain how the complexes can be distributed at such close intervals without blocking replication quickly. We have postulated previously that gyrase acts at two levels on the chromosome, one in association with replication forks and one scattered over the chromosome (12). If both are equally susceptible to quinolones, then the sensitivity of the distributed complexes will reflect that of complexes associated with the replication fork. Thus, for gyrase only a subset of all complexes is relevant for the inhibition of DNA synthesis and bacterial growth. The same appears to be true for topoisomerase IV in S. aureus. In the case of gyrase these key complexes are likely to be ahead of replication forks, a circumstance that promotes a rapid collision with the fork. In contrast, the subset of topoisomerase IV-DNA complexes formed under conditions that slowly inhibit DNA synthesis must be more distantly distributed than all possible complexes and are unlikely to be positioned directly ahead of replication forks (26). Resolving the paradox of how the inhibition of DNA synthesis can be slow when complexes are closely spaced could provide new insights into bacterial chromosome structure.
The effect of a recA deficiency during an attack of topoisomerase IV by quinolones was similar to that observed when gyrase was the primary target: little effect is exerted on the inhibition of DNA synthesis, but rebound synthesis is blocked (15). The absence of recA is likely to affect lethal action, which we have postulated involves the release of double-strand breaks from quinolone-enzyme-DNA complexes (6). These considerations fit with the observation that recA deficiencies have a major impact on survival in the presence of quinolones but little effect on the inhibition of DNA synthesis and presumably the MIC (15, 28). Thus, the killing of cells but not the blocking of DNA replication may be affected by the repair of DNA double-strand breaks.
Quinolone structure determines whether gyrase or topoisomerase IV is the preferred target (31). The relative target selection of other quinolones in S. aureus needs further study, but some compounds, such as gatifloxacin, that target gyrase in Streptococcus pneumoniae (21) appear to target topoisomerase IV in S. aureus (25). Thus, relative target preference cannot necessarily be generalized even within gram-positive species.
The compounds in the present study differ in ways that do not allow the assignment of target preference to particular chemical groups. But nalidixic acid and norfloxacin do set structural boundaries for future studies aimed at finding highly effective compounds that attack both targets equally. Such a compound would be highly desirable because if it can attack both topoisomerase IV and gyrase simultaneously, two mutations, one in each target, would be required for a cell to become resistant. Such an event is expected to occur rarely, and so the compound would restrict selection of resistance. A precedent for this concept has been found with C-8-methoxy fluoroquinolones with S. aureus (9) and with a C-8-chlorine compound tested against Streptococcus pneumoniae (32).
ACKNOWLEDGMENTS
We thank Marila Genarro, Samuel Kayman, and Steve Calderwood for critical comments on the manuscript and Ken Bayles for providing strain ISP2272.
The work was supported by grants AI35257 (to K.D.) and AI23988 (to D.C.H.) from the National Institutes of Health.
REFERENCES
- 1.Anderson V E, Zaniewski R P, Kaczmarek F S, Gootz T D, Osheroff N. Action of quinolones against Staphylococcus aureus topoisomerase IV: basis for DNA cleavage enhancement. Biochemistry. 2000;39:2726–2732. doi: 10.1021/bi992302n. [DOI] [PubMed] [Google Scholar]
- 2.Bayles K W, Brunskill E W, Iandolo J J, Hruska L L, Huang S, Pattee P A, Smiley B K, Yasbin R E. A genetic and molecular characterization of the recA gene from Staphylococcus aureus. Gene. 1994;147:13–20. doi: 10.1016/0378-1119(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 3.Bisognano C, Vaudaux P E, Lew D P, Ng E Y W, Hooper D C. Increased expression of fibronectin-binding proteins by fluoroquinolone-resistant Staphylococcus aureus exposed to subinhibitory levels of ciprofloxacin. Antimicrob Agents Chemother. 1997;41:906–913. doi: 10.1128/aac.41.5.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanche F, Cameron B, Bernard F-X, Maton L, Manse B, Ferrero L, Ratet N, Lecoq C, Goniot A, Bish D, Crouzet J. Differential behaviors of Staphylococcus aureus and Escherichia coli type II DNA topoisomerases. Antimicrob Agents Chemother. 1996;40:2714–2720. doi: 10.1128/aac.40.12.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blumberg H M, Rimland D, Carroll D J, Terry P, Wachsmuth I K. Rapid development of ciprofloxacin resistance in methicillin-susceptible and -resistant Staphylococcus aureus. J Infect Dis. 1991;163:1279–1285. doi: 10.1093/infdis/163.6.1279. [DOI] [PubMed] [Google Scholar]
- 6.Chen C R, Malik M, Snyder M, Drlica K. DNA gyrase and topoisomerase IV on the bacterial chromosome: quinolone-induced DNA cleavage. J Mol Biol. 1996;258:627–637. doi: 10.1006/jmbi.1996.0274. [DOI] [PubMed] [Google Scholar]
- 7.Chow R T, Dougherty T J, Fraimow H S, Bellin E Y, Miller M H. Association between early inhibition of DNA synthesis and the MICs and MBCs of carboxyquinolone antimicrobial agents for wild-type and mutant [gyrA nfxB(ompF) acrA] Escherichia coli K-12. Antimicrob Agents Chemother. 1988;32:1113–1118. doi: 10.1128/aac.32.8.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deitz W H, Cook T M, Goss W A. Mechanism of action of nalidixic acid on Escherichia coli. III. Conditions required for lethality. J Bacteriol. 1966;91:768–773. doi: 10.1128/jb.91.2.768-773.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong Y, Zhao X, Domagala J, Drlica K. Effect of fluoroquinolone concentration on selection of resistant mutants of Mycobacterium bovis BCG and Staphylococcus aureus. Antimicrob Agents Chemother. 1999;43:1756–1758. doi: 10.1128/aac.43.7.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drlica K. Mechanism of fluoroquinolone action. Curr Opin Microbiol. 1999;2:504–508. doi: 10.1016/s1369-5274(99)00008-9. [DOI] [PubMed] [Google Scholar]
- 11.Drlica K, Chen C R, Kayman S. Sedimentation analysis of bacterial nucleoid structure. Methods Mol Biol. 1999;94:87–98. doi: 10.1385/1-59259-259-7:87. [DOI] [PubMed] [Google Scholar]
- 12.Drlica K, Manes S H, Engle E C. DNA gyrase on the bacterial chromosome: possibility of two levels of action. Proc Natl Acad Sci USA. 1980;77:6879–6883. doi: 10.1073/pnas.77.11.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drlica K, Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drlica K, Zhao X. DNA topoisomerase IV as a quinolone target. Curr Opin Antiinfect Investig Drugs. 1999;1:435–442. [Google Scholar]
- 15.Engle E C, Manes S H, Drlica K. Differential effects of antibiotics inhibiting gyrase. J Bacteriol. 1982;149:92–98. doi: 10.1128/jb.149.1.92-98.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrero L, Cameron B, Manse B, Lagneaux D, Crouzet J, Famechon A, Blanche F. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target of fluoroquinolones. Mol Microbiol. 1994;13:641–653. doi: 10.1111/j.1365-2958.1994.tb00458.x. [DOI] [PubMed] [Google Scholar]
- 17.Ferrero L, Cameron B, Crouzet J. Analysis of gyrA and grlA mutations in stepwise-selected ciprofloxacin-resistant mutants of Staphylococcus aureus. Antimicrob Agents Chemother. 1995;39:1554–1558. doi: 10.1128/aac.39.7.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fitzgibbon J E, John J F, Delucia J L, Dubin D T. Topoisomerase mutations in trovafloxacin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1998;42:2122–2124. doi: 10.1128/aac.42.8.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fournier B, Hooper D C. Mutations in topoisomerase IV and DNA gyrase of Staphylococcus aureus: novel pleiotropic effects on quinolone and coumarin activity. Antimicrob Agents Chemother. 1998;42:121–128. doi: 10.1128/aac.42.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fournier B, Hooper D C. Effects of mutations in GrlA of topoisomerase IV from Staphylococcus aureus on quinolone and coumarin activity. Antimicrob Agents Chemother. 1998;42:2109–2112. doi: 10.1128/aac.42.8.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fukada H, Hiramatsu K. Primary targets of fluoroquinolones in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1999;43:410–412. doi: 10.1128/aac.43.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gootz T D, Zaniewski R P, Haskell S L, Kaczmarek F S, Maurice A E. Activities of trovafloxacin compared with those of other fluoroquinolones against purified topoisomerases and gyrA and grlA mutants of Staphylococcus aureus. Antimicrob Agents Chemother. 1999;43:1845–1855. doi: 10.1128/aac.43.8.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiasa H, Yousef D O, Marians K J. DNA strand cleavage is required for replication fork arrest by a frozen topoisomerase-quinolone-DNA ternary complex. J Biol Chem. 1996;271:26424–26429. doi: 10.1074/jbc.271.42.26424. [DOI] [PubMed] [Google Scholar]
- 24.Hooper D C, Wolfson J S, Ng E Y, Swartz M N. Mechanisms of action of and resistance to ciprofloxacin. Am J Med. 1987;82:12–20. [PubMed] [Google Scholar]
- 25.Ince D, Aras R, Hooper D C. Mechanisms and frequency of resistance to gatifloxacin in comparison with ciprofloxacin in Staphylococcus aureus. Drugs. 1999;58:134–135. doi: 10.1128/AAC.45.10.2755-2764.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khodursky A B, Cozzarelli N R. The mechanism of inhibition of topoisomerase IV by quinolone antibacterials. J Biol Chem. 1998;273:27668–27677. doi: 10.1074/jbc.273.42.27668. [DOI] [PubMed] [Google Scholar]
- 27.Khodursky A B, Zechiedrich E L, Cozzarelli N R. Topoisomerase IV is a target of quinolones in Escherichia coli. Proc Natl Acad Sci USA. 1995;92:11801–11805. doi: 10.1073/pnas.92.25.11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDaniel L S, Rogers L H, Hill W E. Survival of recombination-deficient mutants of Escherichia coli during incubation with nalidixic acid. J Bacteriol. 1978;134:1195–1198. doi: 10.1128/jb.134.3.1195-1198.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ng E Y, Trucksis M, Hooper D C. Quinolone resistance mutations in topoisomerase IV: relationship to the flqA locus and genetic evidence that topoisomerase IV is the primary target and DNA gyrase is the secondary target of fluoroquinolones in Staphylococcus aureus. Antimicrob Agents Chemother. 1996;40:1881–1888. doi: 10.1128/aac.40.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Novick R P, Brodsky R J. Studies on plasmid replication: plasmid incompatibility and establishment in Staphylococcus aureus. J Mol Biol. 1972;68:285–302. doi: 10.1016/0022-2836(72)90214-8. [DOI] [PubMed] [Google Scholar]
- 31.Pan X-S, Fisher L M. Targeting of DNA gyrase in Streptococcus pneumoniae by sparfloxacin: selective targeting of gyrase or topoisomerase IV by quinolones. Antimicrob Agents Chemother. 1997;41:471–474. doi: 10.1128/aac.41.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan X-S, Fisher L M. DNA gyrase and topoisomerase IV are dual targets of clinafloxacin action in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1998;42:2810–2816. doi: 10.1128/aac.42.11.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rubenstein I, Leighton S B. The influence of rotor speed on the sedimentation behavior in sucrose gradients of high molecular weight DNA's. Biophys Chem. 1974;1:292–299. doi: 10.1016/0301-4622(74)80015-3. [DOI] [PubMed] [Google Scholar]
- 34.Sinden R R, Carlson J O, Pettijohn D E. Torsional tension in the DNA double helix measured with trimethylpsoralen in living Escherichia coli cells. Cell. 1980;21:773–783. doi: 10.1016/0092-8674(80)90440-7. [DOI] [PubMed] [Google Scholar]
- 35.Snyder M, Drlica K. DNA gyrase on the bacterial chromosome: DNA cleavage induced by oxolinic acid. J Mol Biol. 1979;131:287–302. doi: 10.1016/0022-2836(79)90077-9. [DOI] [PubMed] [Google Scholar]
- 36.Stahl M L, Pattee P A. Confirmation of protoplast fusion-derived linkages in Staphylococcus aureus by transformation with protoplast DNA. J Bacteriol. 1983;154:406–412. doi: 10.1128/jb.154.1.406-412.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trucksis M, Wolfson J S, Hooper D C. A novel locus conferring fluoroquinolone resistance in Staphylococcus aureus. J Bacteriol. 1991;173:5854–5860. doi: 10.1128/jb.173.18.5854-5860.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilkinson B J. Biology. In: Crossley K B, Archer G L, editors. The staphylococci in human disease. New York, N.Y: Churchill Livingstone; 1997. pp. 1–38. [Google Scholar]
- 39.Zhao X, Wang J-Y, Xu C, Dong Y, Zhou J, Domagala J, Drlica K. Killing of Staphylococcus aureus by C-8-methoxy fluoroquinolones. Antimicrob Agents Chemother. 1998;42:956–958. doi: 10.1128/aac.42.4.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao X, Xu C, Domagala J, Drlica K. DNA topoisomerase targets of the fluoroquinolones: a strategy for avoiding bacterial resistance. Proc Natl Acad Sci USA. 1997;94:13991–13996. doi: 10.1073/pnas.94.25.13991. [DOI] [PMC free article] [PubMed] [Google Scholar]