Abstract
BACKGROUND:
Extended red blood cell (RBC) antigen matching is recommended to limit alloimmunization in patients with sickle cell disease (SCD). DNA-based testing to predict blood group phenotypes has enhanced availability of antigen-negative donor units and improved typing of transfused patients, but replacement of routine serologic typing for non-ABO antigens with molecular typing for patients has not been reported.
STUDY DESIGNS AND METHODS:
This study compared the historical RBC antigen phenotypes obtained by hemagglutination methods with genotype predictions in 494 patients with SCD. For discrepant results, repeat serologic testing was performed and/or investigated by gene sequencing for silent or variant alleles.
RESULTS:
Seventy-one typing discrepancies were identified among 6360 antigen comparisons (1.1%). New specimens for repeat serologic testing were obtained for 66 discrepancies and retyping agreed with the genotype in 64 cases. One repeat Jk(b−) serologic phenotype, predicted Jk(b+) by genotype, was found by direct sequencing of JK to be a silenced allele, and one N typing discrepancy remains under investigation. Fifteen false-negative serologic results were associated with alleles encoding weak antigens or single-dose Fyb expression.
CONCLUSIONS:
DNA-based RBC typing provided improved accuracy and expanded information on RBC antigens compared to hemagglutination methods, leading to its implementation as the primary method for extended RBC typing for patients with SCD at our institution.
Red blood cell (RBC) transfusions are an essential treatment for patients with sickle cell disease (SCD) but alloimmunization to RBC antigens remains a major complication (reviewed in Chou et al.1 and Yazdanbaksh et al.2). Differences in antigen prevalence among patients of African descent and donors of European background provide one major explanation for high alloimmunization rates in patients with SCD in Europe and the United States. RH heterogeneity resulting in altered Rh antigen expression on patient RBCs is an additional risk factor.3–5 Pretransfusion determination of extended Rh (CcEe) and K antigen phenotypes is necessary for prophylactic matching strategies that have been implemented to reduce alloimmunization for chronically transfused patients. Transfusion with donor units negative for C, E, and K for patients who lack these antigens is effective in reducing alloimmunization.6,7 More than 80% of comprehensive sickle cell centers obtain a pretransfusion extended RBC phenotype (Rh, Kell, Kidd, Duffy, Lewis, and MNS systems) to provide C-, E-, and K-matched RBCs and to guide new antibody evaluations,8 but this practice is not universal standard of care.9
RBC phenotyping by hemagglutination has been the gold standard, but is labor-intensive and hampered by subjectivity in interpreting agglutination reactions and transcription errors when manually transcribing results. Moreover, patients who have been recently transfused or have a positive direct antiglobulin test may not be accurately typed. Testing is also limited by lack of availability of reagents for a number of clinically significant antigens.10
DNA-based assays targeting single-nucleotide polymorphisms (SNPs) associated with blood group antigen expression offer an alternative.11–14 Genotyping methods provide information on RBC antigens for which standardized serologic typing reagents are not available and are amenable to high-throughput testing with automated computerized interpretation. Here we compared RBC antigen phenotypes determined by single-nucleotide polymorphism analysis with serologic testing for 13 routinely tested RBC antigens and report the prevalence of 35 antigens predicted by DNA in 494 patients with SCD.The aim of this study was to determine the accuracy and any potential benefits of RBC typing with DNA methods to replace extended serologic antigen typing for patients with SCD.
MATERIALS AND METHODS
This is a retrospective study of RBC antigen phenotypes and genotypes of patients with SCD at The Children’s Hospital of Philadelphia performed under a protocol approved by the institutional review board. RBC genotyping was performed for 494 subjects between 2008 and 2014 by human erythrocyte antigen (HEA) BeadChip DNA array (Bioarray/Immucor, Warren, NJ) to determine polymorphisms associated with 35 antigens in 11 blood group systems (Rh, Kell, Kidd, Duffy, MNS, Dombrock, Lutheran, Landsteiner-Wiener, Diego, Colton, and Scianna). DNA-based typing results were electronically imported into a study database (Filemaker, Inc., Santa Clara, CA) and compared to serologic data.
Historic serologic RBC antigen phenotypes, performed by standard manual tube hemagglutination methods, were ascertained from blood bank records. Per institution protocol, the phenotype is performed once on a pretransfusion sample and includes C, c, E, e, K, Fya, Fyb, Jka, Jkb, Lea, Leb, M, N, S, s, and P1 antigens. The RBC phenotype is typically obtained at age 1 year or at the first clinic visit upon transfer of care from another institution. The historic serologic phenotype was compared to the genotype result, with the exception of Lea, Leb, and P1, which are not included on the HEA BeadChip. Twenty-four patients had no serologic phenotype recorded for one or more antigens with a total of 62 antigens missing serologic data; thus, a total of 6360 serologic and genotype antigen results were compared.
To resolve discordant results between antigen type predicted by DNA and historical serologic phenotype, repeat serologic typing was performed on a new sample as discrepancies were identified. For patients transfused in the preceding three months, autologous RBCs for serologic retype were isolated by a hypotonic saline wash procedure (n = 25).15 Samples remaining discordant after repeat serologic typing were tested with multiple commercial reagents and gene sequencing was performed to investigate for a silent or variant allele. Statistical analysis of antigen frequency was performed using bivariate comparisons of categorical variables using a two-tailed chi-square test with Yates’ correction or a Fisher’s exact test.
RESULTS
The 494 subjects included hemoglobin (Hb)SS (67.2%), HbSC (22.4%), HbSβ thalassemia (9.5%), or HbSβvariant (0.9%) genotypes. A total of 135 patients received chronic transfusions (27.3%), 190 episodic transfusions (38.5%), and 169 (34.2%) had not been transfused. A total of 236 patients (47.8%) were female and 258 (52.2%) were male. The median patient age was 13.6 years (range, 1–43 years). The prevalence of 35 RBC antigens predicted from the genotype in this cohort of patients was similar to historical values determined by serology for blacks16 (Fig. 1A, Table 1, and Table S1 [available as supporting information in the online version of this paper]). As expected, the prevalence of C, E, and K antigens in patients with SCD was low and there were significant differences in the prevalence of Jkb (p < 0.0001), Fya (p < 0.0001), Fyb (p < 0.0001), and S (p < 0.0001) compared to Caucasians. Twenty-five percent of patients expressed V, 28% VS, and 17% Jsa, which are low-frequency antigens in all but African ethnic groups.
Fig. 1.
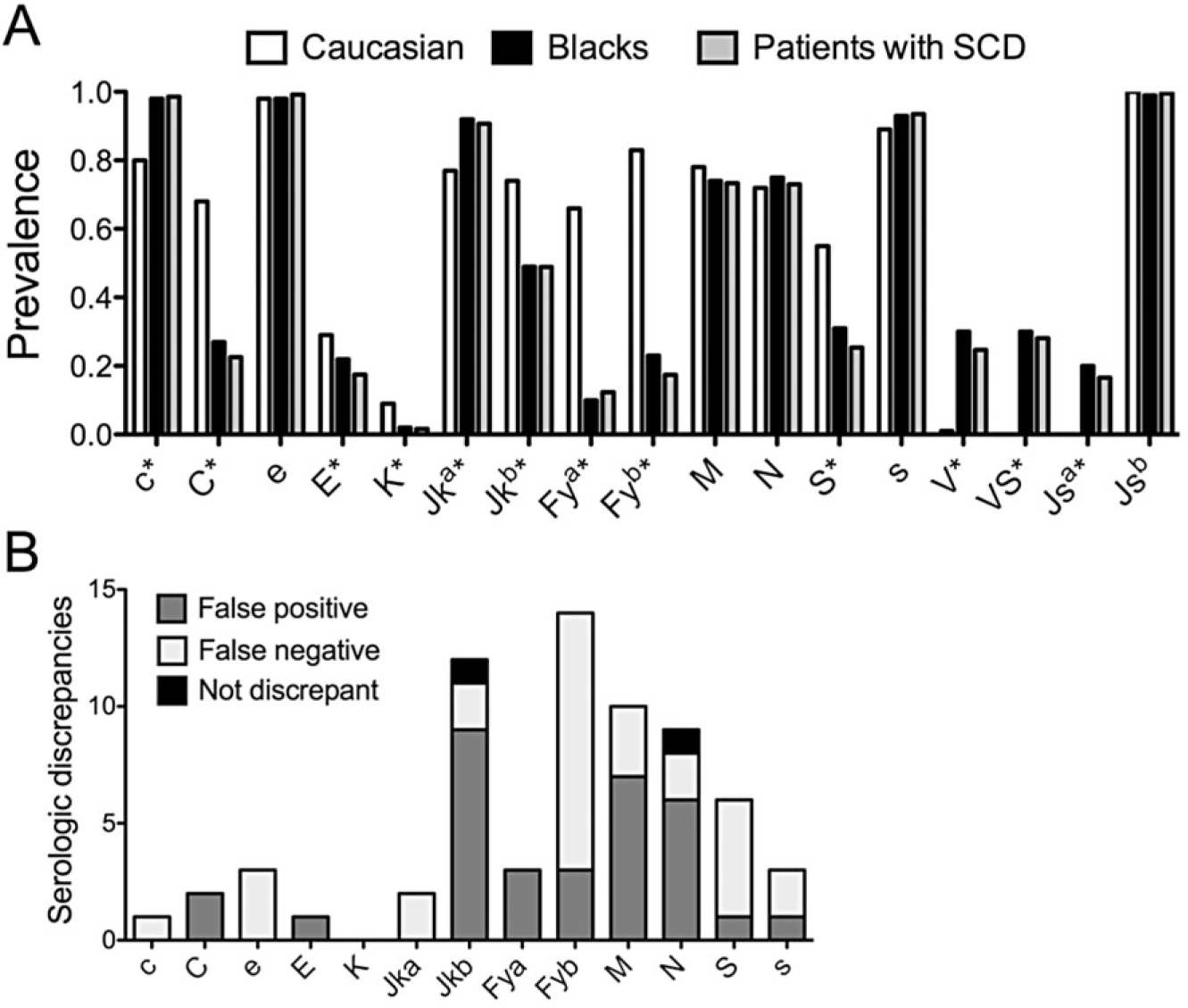
Comparison of DNA-based RBC typing with serologic typing. (A) Prevalence of RBC antigens predicted by genotyping in a cohort of patients with SCD (n = 494) compared to reported prevalence determined by serologic typing for black and Caucasian populations in Reid et al.16 *Antigens for which the frequency between Caucasians and patients with SCD had a p value of less than 0.0001. (B) Seventy-one total discrepancies out of 6360 antigen result comparisons between serologic and genotype predicted results. Bars indicate number of serologic discrepancies per antigen for which repeat serologic testing confirmed the genotype result in all but two cases. Not discrepant indicates the two repeat serology results that were consistent with the historical serologic record.
TABLE 1.
Prevalence of 23 antigens not routinely available by serologic testing*
| Patients with SCD |
Prevalence |
||||
|---|---|---|---|---|---|
| Antigen | Positive | Negative | Patients with SCD | Blacks | Caucasian |
|
| |||||
| hrB† | 436 | 54‡ | 0.883 | 0.960 | 1.000 |
| U | 489§ | 5 | 0.990 | 0.990 | 0.999 |
| Joa | 491 | 3 | 0.994 | 0.990 | 1.000 |
| Hy | 493 | 1 | 0.998 | 0.990 | 1.000 |
| V† | 122 | 372 | 0.247 | 0.300 | 0.010 |
| VS† | 139 | 355 | 0.281 | 0.300 | 0.0001 |
| Jsa† | 82 | 412 | 0.166 | 0.200 | 0.0001 |
| Jsb | 492 | 2 | 0.996 | 0.990 | 1.000 |
| k | 494 | 0 | 1.000 | 1.000 | 0.998 |
| Kpa | 0 | 494 | 0.000 | 0.0001 | 0.020 |
| Kpb | 494 | 0 | 1.000 | 1.000 | 1.000 |
| Lua∥ | 18 | 464 | 0.037 | 0.050 | 0.080 |
| Lub∥ | 485 | 0 | 1.000 | 0.998 | 0.998 |
| Dia∥ | 0 | 493 | 0.000 | 0.0001 | 0.0001 |
| Dib∥ | 493 | 0 | 1.000 | 1.000 | 1.000 |
| Coa | 494 | 0 | 1.000 | 0.999 | 0.999 |
| Cob† | 8 | 486 | 0.016 | 0.100 | 0.100 |
| Doa† | 214 | 280 | 0.433 | 0.550 | 0.670 |
| Dob† | 460 | 34 | 0.931 | 0.890 | 0.820 |
| LWa | 494 | 0 | 1.000 | 1.000 | 1.000 |
| LWb | 0 | 494 | 0.000 | NA | 0.010 |
| Sc1 | 494 | 0 | 1.000 | 0.990 | 0.990 |
| Sc2 | 0 | 494 | 0.000 | NA | 0.010 |
Number of patients with SCD (n = 494) who genotyped positive or negative for each RBC antigen. Antigen prevalence among this cohort was compared to reported prevalence for blacks and Caucasians determined by serological typing in Reid et al.16 with the exception of hrB (authors’ experience).
Antigens for which the frequency between Caucasians and patients with SCD had a p value of less than 0.0001.
Does not include four individuals predicted to be hrB– associated with DcE/DcE (R2R2).
Includes three U variants.
No results were determined for 12 Lua, nine Lub, one Dia, and one Dib due to low signal intensity on the HEA BeadChip.
NA = not available.
Historical serologic RBC antigen phenotypes were compared to DNA-based types. We identified 71 typing discrepancies among 6360 antigen comparisons (1.1%) in 57 patients (11.5%). Of these 71 discordant antigen types, 34 serologic types were performed before 2008, and 37 were obtained between 2008 and 2014. Serologic typing was performed by manual tube method and there have been no notable changes in serologic typing reagents during this time frame. One antigen typing discrepancy was found in 47 patients, two discrepancies in seven, three in two, and four in one patient. Antigen results most often discrepant included 16 Fyb, 13 Jkb, 10 M, 10 N, and seven S (Fig. 1B). New specimens for repeat serologic typing were obtained for 66 of 71 discrepancies. Two patients were unavailable to obtain a repeat specimen. Serologic retyping was not performed for three discrepancies: two Fy(b−) historic serologic results were explained by inheritance of FY*X (265C>T missense mutation)17 encoding very weak antigen expression often not detected by serologic reagents and one weak e antigen expression associated with RHCE*ceJAL in trans to RHCE*cE18 revealed by high resolution RH genotyping (Table S2, available as supporting information in the online version of this paper). Retyping agreed with the genotype in 64 cases (Table S2). Many discrepancies were associated with samples with single-dose Fyb expression that were not originally detected as positive; 86% of Fyb discrepancies reacted weak+ to 2 + on the repeat serologic testing (Table S2). Direct sequencing revealed one Jka discrepancy explained by a JK*A nucleotide 130G>A change19 associated with weak antigen expression. Overall, discrepant results were equally associated with false-positive (n = 34) and false-negative (n = 33) historical serologic types.
Only two repeat serology results were consistent with historical serologic records (Table S2). One Jk(b−) serologic phenotype, predicted to be Jk(b+) by genotype, was found by direct sequencing of JK to be a silenced allele (JK*B 191G>A).20 A serologic N+ type predicted N− by genotype is under investigation. Thus, the concordance rate between DNA-based testing and serology confirmation was 0.9997, with two true genotype-phenotype discrepancies in 6360 total antigens.
DNA-based testing predicted expression for 35 antigens that can guide antibody evaluations and choice of donor units. In these 494 subjects, 66 lack or have altered high prevalence antigens: 54 hrB–, five U–, three U variant, three Jo(a−), and one Hy– (Table 1). The assay also identifies the T>C substitution in the FY erythroid promoter that disrupts binding of the GATA1 erythroid transcription factor and results in RBC-specific loss of Fyb antigen expression.21 The RBC genotype of 410 patients (83%) was predicted to be Fyb–, but 404 (98.5%) were positive for the GATA mutation and, thus, not at risk for producing anti-Fyb.
Patients with SCD commonly inherit RH variant alleles that result in RBCs that lack common or carry novel Rh antigenic epitopes.3–5 DNA-based testing identified 122 V+ (24.6%) and 139 VS+ (28.1%) individuals (Table 1). Fifty-four patients (11.0%) were homozygous for the 733C>G change (predicting V/VS+) associated with loss of the high-prevalence hrB antigen. The presence of markers for a V–VS+ phenotype identified the potential for a hybrid RHD*DIIIa-CE(4–7)-D gene that does not encode D but encodes partial C antigen in 30 patients. High-resolution RH genotyping confirmed RHD*DIIIa-CE(4–7)-D alleles in 23 of the 30 individuals (77%). The RBCs serologically type as C+ but these patients can develop anti-C if exposed to conventional C antigen.22 Providing C– RBCs to these individuals minimizes anti-C alloimmunization.
DISCUSSION
The field of transfusion medicine has over a decade of experience with RBC genotyping. However, elimination of serologic testing and implementation of DNA-based typing for non-ABO blood groups, without confirmatory serologic testing, requires acceptance of practice change. Many studies in blood donors demonstrate that genotyping is reliable and correlates with serologic typing.10,23 Of the 71 serologic antigen types that were discordant with the genotype, 34 were performed before 2008 and 37 were typed between 2008 and 2014. Serologic typing was performed by manual tube method for all samples, and the authors are not aware of any major changes in commercial serologic typing reagents during this time period. The erroneous serologic results revealed in this study likely have multiple etiologies inherent to all manual test methods. These include recording and/or processing errors with manual typing, testing performed inadvertently on a posttransfusion specimen, or an antigen that was expressed weakly and not detected by serologic reagents.
In this study, the concordance rate between DNA-based testing and serologic confirmation was 0.9997, similar to a 0.9995 concordance rate observed in a comparison of 356 reagent donor RBCs analyzed by hemagglutination and genotype using the same high-throughput DNA platform used here.24 Fifty-seven patients (11.5%) had at least one serologic discrepancy noted, compared to studies in multiply transfused patients with thalassemia and SCD that reported serologic and DNA-predicted phenotype discrepancies in 36% to 51% of individuals.25,26 The lower rate of discordant results in this study likely reflects the policy of obtaining an extended RBC phenotype by age 1 year before any transfusion and systematically obtaining the RBC phenotype after a hypotonic saline wash if a patient was recently transfused.
Based on the data reported here, RBC genotyping has replaced serologic typing for blood groups other than ABO, RhD, and Lewis at our institution. This change of practice does require recognition of some limitations of DNA-based testing. The genotype predicts phenotype and does not directly detect antigen expression. High-throughput genotyping platforms identify the most common polymorphisms and do not detect all variants or rare silenced alleles that result in false-positive types. For the Rh system, genotype prediction of common C/c and E/e antigens correlated well with the patients’ serologic results, but high-resolution RH genotyping is required to distinguish most Rh variants.3–5 From a cost perspective, an extended RBC antigen genotype (35 antigens) costs approximately 15% less than a serologic phenotype (15 antigens) when performed by our reference laboratories.
Efforts to prevent alloimmunization in patients with SCD have focused on prophylactic antigen matching. Antigen matching programs for Rh and K have decreased alloimmunization in patients with SCD, but these strategies have not been as effective as predicted.3,5,6,27 Individuals with variant Rh antigens continue to form antibodies against the Rh blood group system despite receiving Rh-matched transfusions.3,5 Extended matching for Kidd, Duffy, and S antigens can further minimize alloantibody formation, but is limited by donor availability and cost. RBC genotyping of both patients and donors may have the potential to improve RBC antigen matching. DNA-based typing provides data on significantly more antigens than routine serologic typing, including the clinically relevant U, Doa, Dob, partial C, and hrB antigens. Providing antigen-negative units as a prophylactic prevention strategy for patients who lack high-prevalence antigens (hrB, U, Joa, Hy) could improve transfusion safety but would require a large increase in minority donations and donor genotyping. DNA-based screening of donors can help expand antigen-negative unit inventories, particularly with strategies to increase donations from minority populations.
Major challenges for mass scale genotyping of donors and patients are data handling and integration of information technology systems between reference molecular laboratories, blood donor centers, and hospital transfusion services. Electronic importing of DNA-predicted RBC phenotypes from the molecular laboratories to hospital transfusion services would be ideal. A data clearinghouse is also necessary to efficiently match donors and patients using RBC genotype information along with ABO status that currently remains dependent on serologic methods. Numerous blood bank, reference laboratory, and blood donor center information systems currently exist. Thus, a concerted effort among providers to build a shared information system is crucial and necessitates uniform standards for antigen nomenclature, data representation, and interfacing.
Implementation of RBC genotyping for patient care requires acceptance of molecular typing without serologic confirmation, which represents a major practice change for transfusion services. This large cohort study demonstrates that use of a DNA-based array to predict RBC antigen phenotype is highly reliable in patients with SCD, including those who are chronically transfused. High alloimmunization rates despite prophylactic Rh and K matching in this patient population suggest that more precise matching is necessary.3,7,27 Based on these findings, our institution has adopted molecular typing as the primary method to predict RBC antigen phenotypes outside the ABO, RhD, and Lewis systems. RBC genotyping can provide antigen status on many clinically significant antigens for which serologic reagents are limited or not available, which can expedite new antibody evaluations and potentially improve RBC matching. Blood donation centers are readily adapting DNA-based screening of donors to efficiently manage their inventories. Now, the field of transfusion medicine is poised to integrate RBC genotyping of both patients and donors into routine clinical practice, which can particularly benefit individuals with SCD.
Supplementary Material
Table S1. Red blood cell antigen prevalence. Antigen prevalence among this cohort was compared to reported prevalence for Blacks and Caucasians determined by serological typing in.16 Statistical analysis of frequency was performed using bivariate comparisons of categorical variables using a two-tailed chi-square test with Yates’ correction or a Fisher’s exact test. NA, not available.
Table S2. Seventy-one discrepancies between historical RBC antigen phenotype determined by serology compared to genotype predicted phenotype. UPIN, unique patient identifier number; NP, confirmatory serology testing not performed because discrepancy explained by inheritance of alleles encoding weak antigen expression; NA, repeat specimen not available; w+, weak positive agglutination; 1+, 1+ agglutination; 2+, 2+ agglutination; *, strength of agglutination not available.
ACKNOWLEDGMENTS
We thank the staff at the blood bank at the Children’s Hospital of Philadelphia and the Immunohematology and Genomics Laboratory at New York Blood Center. STC and CMW designed the study; and JC, DFF, TJ, SV, CMW, and STC performed the research, analyzed the data, and wrote the paper.
This work was supported by the Innovations in Clinical Research Award from the Doris Duke Charitable Foundation Grant 2011097 to STC and CMW.
ABBREVIATIONS:
- HEA
human erythrocyte antigen
- SCD
sickle cell disease
Footnotes
CONFLICT OF INTEREST
The authors have disclosed no conflicts of interest.
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article at the publisher’s Web site:
REFERENCES
- 1.Chou ST, Liem RI, Thompson AA. Challenges of alloimmunization in patients with haemoglobinopathies. Br J Haematol 2012;159:394–404. [DOI] [PubMed] [Google Scholar]
- 2.Yazdanbakhsh K, Ware RE, Noizat-Pirenne F. Red blood cell alloimmunization in sickle cell disease: pathophysiology, risk factors, and transfusion management. Blood 2012; 120:528–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chou ST, Jackson T, Vege S, et al. High prevalence of red blood alloimmunization in sickle cell disease despite transfusion from Rh-matched minority donors. Blood 2013;122: 1062–71. [DOI] [PubMed] [Google Scholar]
- 4.Noizat-Pirenne F, Tournamille C. Relevance of RH variants in transfusion of sickle cell patients. Transfus Clin Biol 2011;18:527–35. [DOI] [PubMed] [Google Scholar]
- 5.Silvy M, Tournamille C, Babinet J, et al. Red blood cell immunization in sickle cell disease: evidence of a large responder group and a low rate of anti Rh linked to partial Rh phenotype. Haematologica 2014;99:e115–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vichinsky EP, Earles A, Johnson RA, et al. Alloimmunization in sickle cell anemia and transfusion of racially unmatched blood. N Engl J Med 1990;322: 1617–21. [DOI] [PubMed] [Google Scholar]
- 7.O’Suoji C, Liem RI, Mack AK, et al. Alloimmunization in sickle cell anemia in the era of extended red cell typing. Pediatr Blood Cancer 2013;60:1487–91. [DOI] [PubMed] [Google Scholar]
- 8.Afenyi-Annan A, Willis MS, Konrad TR, et al. Blood bank management of sickle cell patients at comprehensive sickle cell centers. Transfusion 2007;47:2089–97. [DOI] [PubMed] [Google Scholar]
- 9.Osby M, Shulman IA. Phenotype matching of donor red blood cell units for nonalloimmunized sickle cell disease patients: a survey of 1182 North American laboratories. Arch Pathol Lab Med 2005;129:190–3. [DOI] [PubMed] [Google Scholar]
- 10.Jungbauer C, Hobel CM, Schwartz DW, et al. High-throughput multiplex PCR genotyping for 35 red blood cell antigens in blood donors. Vox Sang 2012;102:234–42. [DOI] [PubMed] [Google Scholar]
- 11.Hashmi G, Shariff T, Seul M, et al. A flexible array format for large-scale, rapid blood group DNA typing. Transfusion 2005;45:680–8. [DOI] [PubMed] [Google Scholar]
- 12.Denomme GA, Van Oene M. High-throughput multiplex single-nucleotide polymorphism analysis for red cell and platelet antigen genotypes. Transfusion 2005;45:660–6. [DOI] [PubMed] [Google Scholar]
- 13.Westhoff CM. Molecular testing for transfusion medicine. Curr Opin Hematol 2006;13:471–5. [DOI] [PubMed] [Google Scholar]
- 14.Avent ND, Martinez A, Flegel WA, et al. The Bloodgen Project of the European Union, 2003–2009. Transfus Med Hemother 2009;36:162–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown DJ. A rapid method for harvesting autologous red cells from patients with hemoglobin S disease. Transfusion 1988;28:21–3. [DOI] [PubMed] [Google Scholar]
- 16.Reid ME, Lomas-Francis C, Olsson ML. The blood group antigen facts book. 3rd ed San DIego (CA): Elsevier Academic Press; 2012. [Google Scholar]
- 17.Tournamille C, Le Van Kim C, Gane P, et al. Arg89Cys substitution results in very low membrane expression of the Duffy antigen/receptor for chemokines in Fy(x) individuals. Blood 1998;92:2147–56. [PubMed] [Google Scholar]
- 18.Westhoff CM, Vege S, Wylie DE, et al. The JAL antigen (RH48) is the result of a change in RHCE that encodes Arg114Trp. Transfusion 2009;49:725–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wester ES, Storry JR, Olsson ML. Characterization of Jk(a+(weak)): a new blood group phenotype associated with an altered JK*01 allele. Transfusion 2011;51: 380–92. [DOI] [PubMed] [Google Scholar]
- 20.Billingsley KL, Posadas JB, Moulds JM, et al. A novel JK null allele associated with typing discrepancies among African Americans. Immunohematology 2013;29: 145–8. [PubMed] [Google Scholar]
- 21.Tournamille C, Colin Y, Cartron JP, et al. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat Genet 1995;10:224–8. [DOI] [PubMed] [Google Scholar]
- 22.Tournamille C, Meunier-Costes N, Costes B, et al. Partial C antigen in sickle cell disease patients: clinical relevance and prevention of alloimmunization. Transfusion 2010;50: 13–9. [DOI] [PubMed] [Google Scholar]
- 23.Hashmi G, Shariff T, Zhang Y, et al. Determination of 24 minor red blood cell antigens for more than 2000 blood donors by high-throughput DNA analysis. Transfusion 2007;47:736–47. [DOI] [PubMed] [Google Scholar]
- 24.Kappler-Gratias S, Peyrard T, Beolet M, et al. Blood group genotyping by high-throughput DNA analysis applied to 356 reagent red blood cell samples. Transfusion 2011;51: 36–42. [DOI] [PubMed] [Google Scholar]
- 25.Ribeiro KR, Guarnieri MH, da Costa DC, et al. DNA array analysis for red blood cell antigens facilitates the transfusion support with antigen-matched blood in patients with sickle cell disease. Vox Sang 2009;97:147–52. [DOI] [PubMed] [Google Scholar]
- 26.Bakanay SM, Ozturk A, Ileri T, et al. Blood group genotyping in multi-transfused patients. Transfus Apher Sci 2013;48:257–61. [DOI] [PubMed] [Google Scholar]
- 27.Miller ST, Kim HY, Weiner DL, et al. Red blood cell alloimmunization in sickle cell disease: prevalence in 2010. Transfusion 2013;53:704–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Red blood cell antigen prevalence. Antigen prevalence among this cohort was compared to reported prevalence for Blacks and Caucasians determined by serological typing in.16 Statistical analysis of frequency was performed using bivariate comparisons of categorical variables using a two-tailed chi-square test with Yates’ correction or a Fisher’s exact test. NA, not available.
Table S2. Seventy-one discrepancies between historical RBC antigen phenotype determined by serology compared to genotype predicted phenotype. UPIN, unique patient identifier number; NP, confirmatory serology testing not performed because discrepancy explained by inheritance of alleles encoding weak antigen expression; NA, repeat specimen not available; w+, weak positive agglutination; 1+, 1+ agglutination; 2+, 2+ agglutination; *, strength of agglutination not available.


