The introduction of the fluoroquinolones (FQs) in the 1980s provided clinicians with a class of broad-spectrum agents applicable to a range of gram-negative infections including urinary tract infections, gastrointestinal infections, respiratory tract infections, sexually transmitted diseases, bone and joint infections, and infections of the skin and soft tissue (reviewed in references 10 and 43). Targeted microorganisms include the family Enterobacteriaceae, Haemophilus spp., Neisseria spp., and Moraxella spp., which are highly susceptible to these agents, as well as important nosocomial pathogens such as Pseudomonas aeruginosa and Acinetobacter spp. FQs are less active but still clinically useful against Legionella spp.
Given this broad spectrum of activity, it is unfortunate that resistance to FQs has increased in a number of gram-negative organisms, most notably in P. aeruginosa but in virtually all organisms where FQs have been employed (1, 43). Resistance is due usually to mutations in the genes for the bacterial targets of the FQs (DNA gyrase [GyrA] and topoisomerase IV [ParC]) or to active efflux of the agents via antibiotic efflux pumps (59). This review focuses on efflux mechanisms of FQ resistance, their distribution and clinical significance in gram-negative pathogens, the possible natural function(s) of these, and, finally, the potential therapeutic value of efflux pump inhibitors.
ANTIBIOTIC EFFLUX
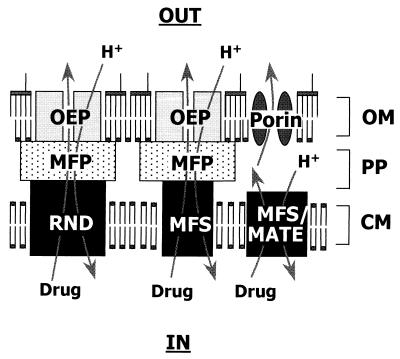
Efflux as a mechanism of antibiotic resistance was first reported in the early 1980s, for tetracycline, by two groups of researchers (11, 85). Since then, efflux-mediated resistance to several antimicrobial agents, including FQs, has been reported in a variety of bacterial species, and a number of efflux determinants have been cloned and sequenced (109) (Table 1). Bacterial antimicrobial efflux transporters have generally been grouped into four superfamilies, primarily on the basis of amino acid sequence homology. These include the major facilitator superfamily (MFS) (108), the ATP-binding cassette family (137), the resistance-nodulation-division (RND) family (97, 121), and the small multidrug resistance (SMR) protein family (110). Recently, a fifth family, referred to as the multidrug and toxic compound extrusion (MATE) family, has been identified (13). Antibiotic efflux pumps fall into the RND, MFS, or MATE groups (Fig. 1) and utilize the energy of the proton motive force to export antibiotics from the cell (97, 108, 109). RND family transporters are unique to gram-negative bacteria and typically work in conjunction with a periplasmic membrane fusion protein (MFP) (26, 121) (also called a periplasmic efflux protein [54]) and an outer membrane protein (97) (also called outer membrane [OM] efflux protein [OEP] [54]). This organization provides for efflux of antibiotics across both membranes of the typical gram-negative organism.
TABLE 1.
FQ efflux systems of gram-negative bacteria
| Organism | Efflux componenta
|
Regulatory gene(s) | Expressionb | Substrates | Referencesc | ||||
|---|---|---|---|---|---|---|---|---|---|
| MFP | RND | OEP | MFS | MATE | |||||
| B. fragilis | NorA?e | ?e | wt +; mutant ++ | Antibiotics, dyes | 88 | ||||
| B. cepacia | CeoA | CeoB | OpcM | ? | wt −; mutant + | Antibiotics | 15, 16 | ||
| C. jejuni | ? | ? | ? | ? | ? | ? | ? | ? | 20 |
| C. freundii | ? | ? | ? | ? | ? | ? | ? | ? | 94, 134a |
| E. aerogenes | ? | ? | ? | ? | ? | ? | ? | ? | 77 |
| E. coli | AcrA | AcrB | TolC | acrR, marA, robA, soxS | wt +; marR ++; acrR ++ | Antibiotics, dyes, disinfectants, detergents, solvents | 22, 24, 44, 74, 90 | ||
| AcrE | AcrF | ? | acrS | wt −; mutant + | Antibiotics, dyes, detergents | 76 | |||
| YdhE | ? | ? | Antibiotics, dyes, lipophilic cations | 92 | |||||
| MdfA | ? | ? | Antibiotics, dyes, lipophilic cations | 28 | |||||
| H. influenzae | AcrA | AcrB | ? | ? | wt + | Antibiotics, dyes, detergents | 123 | ||
| K. pneumoniae | ? | ? | ? | ? | ? | ? | ? | ? | 25, 79 |
| N. gonorrhoeae | MtrC | MtrD | MtrE | mtrR | wt +; mtrR ++ | Antibiotics, detergents, lipids, antimicrobial peptides | 40 | ||
| P. vulgaris | ? | ? | ? | ? | ? | ? | ? | ? | 49 |
| P. aeruginosa | MexA | MexB | OprM | mexR | wt +; nalB +++; nalCd ++ | Antibiotics, dyes, solvents, detergents, homoserine lactones, disinfectants | 18, 19, 33, 39, 67, 86, 114, 115, 117, 120, 145 | ||
| MexC | MexD | OprJ | nfxB | wt −; nfxB ++ | Antibiotics, dyes, detergents, solvents | 113 | |||
| MexE | MexF | OprN | mexT | wt −; nfxCd ++ | Antibiotics, solvents | 61 | |||
| MexX (AmrA) | MexY (AmrB) | OprM | mexZ (amrR) | wt + | Antibiotics, dyes | 3, 87, 139 | |||
| S. enterica serovar Typhimurium | AcrA | AcrB | ? | ? | wt +; mutant ++ | Antibiotics, dyes, detergents | 37, 65, 99 | ||
| S. dysenteriae | ? | ? | ? | ? | ? | ? | ? | ? | 2, 36 |
| S. maltophilia | SmeA | SmeB | SmeC | smeRS | ? | Antibiotics | 6, 14, 9; Li et al., unpublished data | ||
| V. parahaemolyticus | NorM | ? | ? | Antibiotics, dyes, lipophilic cations | 92 | ||||
In organisms in which FQ efflux systems have been identified, components are identified as members of the single-component MATE or MFS group of efflux pumps or as members of the three-component RND-MFP-OEP group. In some instances the OEP component has yet to be confirmed.
wt +, efflux system is expressed in wild-type cells (under laboratory growth conditions); mutant ++, expression is enhanced in resistant strains; wt −, efflux system is not expressed in wild-type; mutant +, efflux system is expressed in resistant strains. In instances where the nature of the mutation leading to enhanced efflux gene expression is known, the gene is indicated along with the relative level of gene expression (++, somewhat enhanced; +++, very enhanced).
Where the identity of an FQ efflux system has yet to be made, references supporting the existence of FQ efflux mechanisms are supplied. Where FQ efflux systems have been identified, the more general references in support of efflux are indicated in boldface type opposite the organism name, and references pertaining to specific FQ efflux systems are indicated in lightface type opposite the relevant efflux system. Only the initial description of the latter is referenced here. Details of the regulation and substrate specificity are cited in the text.
The nalC and nfxC genes have not yet been identified.
?, uncertain.
FIG. 1.
Schematic demonstrating the organization and operation of antimicrobial efflux pumps of gram-negative bacteria. Although some MFS pumps work in conjunction with MFP and OEP counterparts, FQ efflux via a MFS-MFP-OEP tripartite pump has yet to be demonstrated. Abbreviations: PP, periplasmic space; CM, cytoplasmic membrane.
FQ EFFLUX IN GRAM-NEGATIVE BACTERIA
FQ resistance attributable to efflux has been reported in a number of gram-negative organisms including Burkholderia cepacia, Campylobacter jejuni, Citrobacter freundii, Enterobacter aerogenes, Escherichia coli, Klebsiella pneumoniae, Proteus vulgaris, P. aeruginosa, Salmonella enterica serovar Typhimurium, Shigella dysentariae, Stenotrophomonas maltophilia, Vibrio parahaemolyticus, and the anaerobe Bacteroides fragilis (Table 1). In most instances efflux was identified as the resistance mechanism because of an observed increase in FQ accumulation in FQ-resistant strains that was, when examined, compromised upon the addition of an energy inhibitor such as carbonyl cyanide m-chlorophenylhydrazone (CCCP). Unfortunately, a CCCP-promoted increase in FQ accumulation has been observed in the absence of an efflux mechanism (35, 100). As such, demonstration of CCCP-enhanced FQ accumulation in bacterial cells alone is insufficient to support the existence of an FQ efflux mechanism. The demonstration of FQ efflux, directly, or of reduced FQ accumulation in FQ-resistant relative to -susceptible strains is necessary before claims of an efflux mechanism can be made.
An interesting feature of strains expressing efflux-mediated quinolone resistance is their cross-resistance to a number of structurally unrelated antimicrobial agents (6, 15, 16, 18–20, 24, 25, 33, 36, 44, 66, 67, 77, 82, 99, 112, 117, 120, 123, 125, 149). This is due to the broad substrate specificity of the FQ efflux systems, which are capable of accommodating a variety of clinically relevant antimicrobial agents in addition to FQs. As such, FQ use and the attendant development of FQ resistance threatens to increase the incidence of multidrug resistance (MDR) in a number of important human pathogens. This is especially true for an organism like P. aeruginosa, where efflux-mediated resistance to FQs seems to predominate as a mechanism of resistance to these agents (50, 51, 62, 144).
RND-Type Efflux Systems
P. aeruginosa.
Organisms with known FQ efflux systems of the MFP-RND-OEP type are highlighted in Table 1. In P. aeruginosa, four FQ-MDR efflux systems have been described to date, although numerous homologues are identifiable in the recently completed genome sequence (http://www.pseudomonas.com). The first to be reported, encoded by the mexAB-oprM operon (39, 69, 114, 115), is expressed constitutively in wild-type cells cultivated under usual laboratory conditions, where it contributes to intrinsic resistance to quinolones and other antibiotics (60, 116, 131). The system is also hyperexpressed in so-called nalB mutants, which display elevated resistance to FQs and a variety of other antimicrobials (60, 82, 83, 116, 117). nalB strains carry mutations in a gene, mexR, which occurs immediately upstream of the efflux operon and encodes a repressor of mexAB-oprM expression (53, 116, 122, 132, 152). MexAB-OprM hyperexpression independent of mutations in mexR and the mexR mexAB-oprM intergenic region have also recently been described (132, 152). Dubbed nalC mutants (132), these presumably carry a mutation in a hitherto unidentified regulator of mexAB-oprM expression. The MexAB-OprM system is also growth phase regulated, its expression increasing in late log phase (30). Thus, this FQ-MDR efflux system is highly regulated in P. aeruginosa.
The MexCD-OprJ (113) and MexEF-OprN (61) systems are apparently not expressed in wild-type cells under normal laboratory conditions (45, 61, 131) but are hyperexpressed in nfxB (42, 83, 113) and nfxC (33, 61, 83) mutants, respectively. NfxB mutants carry mutations in a gene, nfxB (105, 106), which is located upstream of the efflux genes and encodes a repressor of mexCD-oprJ expression (113). Two classes of nfxB mutants have been described, expressing moderate (type A) or high (type B) levels of the efflux system, with resistance levels correlating with efflux gene expression (81). The nature of mutations leading to MexEF-OprN hyperexpression in nfxC strains has yet to be elucidated. MexEF-OprN hyperexpression is, however, dependent upon the mexT gene, which is located upstream of mexEF-oprN and encodes a positive regulator of mexEF-oprN expression (63, 102). Unlike the aforementioned efflux operons, the recently described mexXY system (also called amrA [139]) lacks a linked OM gene (87), reminiscent of the acrAB FQ-MDR efflux operon of E. coli (see below). Still, MexXY appears to utilize the product of the oprM gene as its OM constituent (3, 87), consistent with an earlier observation that OprM is functional in efflux-mediated MDR in the absence of MexAB (151). Given that the OM efflux proteins are not functional in the absence of RND-MFP counterparts (i.e., these pumps operate as tripartite pumps only) (141), this was interpreted as an indication that OprM functioned as the OM efflux component for additional efflux systems (151). Again, this was reminiscent of E. coli, in which the TolC OM component of the AcrAB-TolC FQ-MDR efflux system (see below) also functioned as the OM constituent of several other three-component pumps involved in the export of uncouplers (97), colicin V (46) and hemolysin (138). MexXY-OprM-mediated resistance to FQs has only been demonstrated with the cloned genes in E. coli (87) and P. aeruginosa (3). Thus, it is not clear to what extent the chromosomal mexXY genes promote FQ resistance in P. aeruginosa. Inactivation of chromosomal mexXY enhances the organism's susceptibility to several antibiotics, including aminoglycosides, but not to FQs (3), and FQ or MDR mutants hyperexpressing this system have yet to be described. A gene, mexZ (also called amrR [139]), has been identified upstream of mexXY and apparently encodes a repressor of mexXY (amrAB) expression (3, 139).
These three-component efflux systems include an inner membrane, RND-type presumed drug-proton antiporter (MexB, MexD, MexF, or MexY); a periplasmic link or membrane fusion protein (MexA, MexC, MexE, or MexX); and an OM, presumed channel-forming protein (OprM, OprJ, or OprN). The inner membrane and OM components function in the export of antibiotics across the corresponding membrane, while the MFP apparently couples antibiotic export across both membranes by bringing these export components (or the membranes [147]) into close apposition (76, 97). Although drug-proton antiport has not been demonstrated for the RND pump components of P. aeruginosa, their amino acid sequence homology to a heavy metal-proton antiporter in Alcaligenes eutrophus (Ralstonia eutropha) (95) and an MDR-type drug-proton antiporter in E. coli (147) and their inhibition by uncouplers such as CCCP (69) favor this mechanism of action. The MFP components, though periplasmic, are anchored to the cytoplasmic membrane, probably via the acyl moiety of these presumed lipoproteins. The MexA MFP has, in fact, been shown to be acylated, although this acylation appears to be unnecessary for its activity (142). Channel-forming activity has yet to be demonstrated for the OEP components, which appear to be distinct from the porin class of OM channel-forming proteins (54). Moreover, the OEP components contain so-called lipoprotein boxes, typically sites of acylation in lipoproteins, at their N termini, and indeed, acylation of OprM has been demonstrated (N. Bianco and K. Poole, unpublished data). The observation that tonB mutants are compromised with respect to MexAB-OprM-mediated resistance suggests that OprM, at least, is a “corked” channel whose opening is dependent upon energy and the TonB protein (150).
FQ-MDR clinical isolates of the nalB (53, 152), nfxB (51, 53), and nfxC (34) type have been described, although clinical FQ-resistant strains typically display target site mutations (17, 93, 143). It is unlikely, however, that efflux mechanisms were assessed in many of these strains. Still, a recent report indicates that hyperexpression of MexCD-OprJ or MexEF-OprN is the predominant mechanism of FQ resistance in strains isolated from the lungs of cystic fibrosis patients (52). The presence of both efflux and gyrase or topoisomerase mutations in the same strain has, for example, been reported in a number of clinical strains, particularly those exhibiting high-level FQ resistance (18, 47, 53, 145, 148). Given that high-level FQ resistance is invariably associated with multiple mutations (53), it is likely that efflux is a significant contributing factor in many instances. Interestingly, selection of FQ-resistant strains in vitro at FQ concentrations two to four times the MIC in a recent study yielded efflux mutants in >90% of the cases (62), a trend noted previously for the FQs (50, 51, 144) and distinct from the pattern seen for nalidixic acid-resistant strains (where gyrase mutations predominated) (144). This has obvious implications vis-à-vis FQ selection of MDR strains in vivo.
E. coli and the Enterobacteriaceae.
The predominant FQ efflux system of E. coli is encoded by the acrAB-tolC genes (32, 74, 76). The AcrAB proteins are highly homologous to the Mex proteins of P. aeruginosa, while TolC has limited homology to the OEPs of P. aeruginosa. TolC has, however, been shown to form channels in planar lipid bilayer membranes (12), consistent with a role in the export of antibiotics across the OM of E. coli. As with the FQ efflux mechanisms of P. aeruginosa, this system is broadly specific and accommodates a number of clinically relevant antimicrobials in addition to FQs (74). This system is expressed in wild-type cells under normal laboratory growth conditions, where it provides for intrinsic resistance, and its hyperexpression in mutants results in elevated resistance to FQs and other agents (74, 97). A related MDR efflux system, encoded by the acrEF (76) (previously called envCD [58]) genes is not expressed in wild-type cells (76) but does accommodate the same antibiotics, including FQs, as does AcrAB (J. Hwang and H. Nikaido, personal communication). Acylation of EnvC-AcrE has been demonstrated (127) although this does not appear to be essential for the function of the MFPs (146). Homologues of these systems have been described in both S. enterica serovar Typhimurium (37, 65, 99) and Haemophilus influenzae (123), although an OM constituent has yet to be confirmed. While the S. enterica serovar Typhimurium counterpart has a demonstrated role in efflux and FQ resistance (99), the H. influenzae AcrAB homologue does not (123). Still, the latter organism possesses an OM of comparatively high permeability (124), and it is known that the OM barrier is an important determinant of resistance mediated by FQ-MDR efflux systems (i.e., they function synergistically) (41, 76, 98). Despite the presence of highly homologous FQ-MDR efflux systems in P. aeruginosa (MexAB-OprM) and E. coli (AcrAB-TolC), for example, the former provides resistance to a broader range of compounds, in particular small hydrophilic antibiotics (131). Moreover, expression of MexAB-OprM in E. coli affords resistance to a narrower range of antibiotics than it does in P. aeruginosa (130). This is apparently because the OM of E. coli is more permeable to these agents than is P. aeruginosa's (96), and their rate of influx, then, is sufficient to overcome the effects of efflux (76). Similarly, disruption of the OM in P. aeruginosa compromises antibiotic resistance mediated by the MexAB-OprM pump (70a, 98). Thus, while the AcrAB system of H. influenzae may well pump FQs, the rather permeable OM of this organism (i.e., enhanced influx) would likely, as H. Nikaido has stated (123), negate this effect. In this vein, Neisseria gonorrhoeae also possesses a homologue of the AcrAB-MexAB FQ-MDR efflux systems, MtrCDE (40), that functions as an antibiotic efflux system but does not provide resistance to FQs. Again, the OM of this organism is apparently more permeable than that of E. coli (and P. aeruginosa) (123).
Expression of acrAB is governed primarily by AcrR, the product of a repressor gene located immediately adjacent to the efflux genes (73), and MarA, a positive regulator encoded by the marA gene of the marRAB operon (4). AcrR is a member of a family of repressor proteins that includes AcrS, which apparently regulates acrEF expression (76), and MtrR, which regulates mtrCDE expression (40). Mutations in acrR (73) or mtrR (40) produce modest increases in efflux gene expression and, thus, modest increases in resistance to substrate antibiotics. The marRAB operon, the so-called mar locus, has long been known to be the site of mutations in E. coli that produce a multiple antibiotic resistance (Mar) phenotype (4). The marR gene encodes a repressor of marRAB expression (and is the site of mutation in most mar strains [4]), while the marA gene product activates a variety of genes associated with resistance to antibiotics and oxygen stress (4). Still, the predominant locus responsible for the MDR of mar mutants is acrAB. This was aptly demonstrated by observations that expression of acrAB is increased in marR mutants (which show elevated marA expression) (75) and the multiple antibiotic resistance of marR mutants is compromised in strains with acrAB deletions (107). Moreover, the cloned marA gene enhances expression of both acrA and tolC (9). Two additional genes, robA and soxS, also increase acrA and tolC expression (9), highlighting the complexity of acrAB-tolC regulation in E. coli. Recently, a null mutation in the mppA gene encoding a periplasmic murein peptide-binding protein was shown to increase MarA production and, concomitantly, the antibiotic resistance of E. coli (68). Still, the mechanism by which MppA influences marA expression remains to be elucidated.
Intriguingly, MarA homologues have been identified in several bacteria, including serovar Typhimurium (21, 133), Shigella spp., Klebsiella spp., C. freundii, Hafnia alvei, Enterobacter spp. (21) and P. vulgaris (48), with marA expression associated with a mar phenotype in Enterobacter agglomerans and S. typhimurium (21). A Mar phenotype has also been reported in P. aeruginosa (148), although a mar locus has yet to be identified in this organism. It is possible, therefore, that these organisms possess FQ-MDR transporters of the AcrAB type. The observation that FQ resistance is increased in K. pneumoniae upon exposure to salicylate (27), a known inducer of the mar operon (23), also suggests that this organism possesses an AcrAB-type efflux system.
The clinical relevance of AcrAB-mediated resistance to FQs or any antibiotic is unclear, although the presence of mar mutations in clinical FQ-resistant strains (78, 104) and the observation that mar mutants are less sensitive to the bactericidal effects of FQs (38) and mutate more readily to high-level FQ resistance (4) suggest it may be important. Moreover, clinical strains of E. coli resistant to FQs and exhibiting decreased accumulation of ciprofloxacin have been described, many of which show very marked increases in ciprofloxacin accumulation upon treatment with CCCP (31). These strains also carry mutations in gyrA and parC, indicating, again, that FQ resistance often results from a combination of efflux and target site mutations (see also reference 56).
Burkholderia spp.
First identified as a determinant of chloramphenicol resistance in a clinical strain (15), the ceoAB-opcM operon (GenBank accession number U97042) of B. cepacia encodes an MDR efflux pump which accommodates and, thus, provides resistance to FQs (16; J. L. Burns, C. Ptritzlaff, J. Barry, M. Charon, and M. Cieri, Abstr. 98th Gen. Meet. Am. Soc. Microbiol., abstr. V-108, 1998). Associated with the MDR of a clinical strain, it is unclear if this homologue of the RND-MFP-OEP efflux systems of P. aeruginosa is only expressed in mutant strains (like the nfxB and nfxC mutants of P. aeruginosa) or is expressed constitutively and, thus, contributes to the well-known intrinsic resistance of this organism to many antibiotics. A regulatory gene has not been reported for this efflux system, although salicylate induction of FQ-MDR in B. cepacia (14) is suggestive of a mar locus in this organism (see below). An RND-MFP-OEP homologue, AmrAB-OprA, has recently been described in Burkholderia pseudomallei (91). Also an MDR transporter, this system exports and provides resistance to aminoglycosides and macrolides but not FQs.
S. maltophilia.
FQ selection of MDR strains of S. maltophilia (then called Xanthamonas maltophilia), reminiscent of FQ selection of efflux mutants in P. aeruginosa (see above), has been seen, although an efflux mechanism was not originally implicated (66). More recently, FQ-MDR strains have been described which appear to possess efflux mechanisms responsible for the FQ-MDR (6, 149). Of significance, several clinical MDR strains of S. maltophilia expressing homologues of the MexAB-OprM efflux system of P. aeruginosa have been reported (149). In fact, a MexAB-OprM-like efflux system, encoded by the smeABC operon (GenBank accession number AF173226), has been discovered recently in this organism and shown to accommodate FQs as well as other antibiotics (X.-Z. Li, L. Zhang, and K. Poole, unpublished data).
Genome projects.
Homologues of the RND-MFP-type MDR systems with and without linked OEP genes are identifiable in the genomes of many bacteria, including Rickettsia prowazekii (7) and Helicobacter pylori (5, 136), where a hefABC operon has been identified by Bina and Hancock (accession number AF059041); as well as the cyanobacterium Synechocystis sp. (55) and Rhodobacter capsulatus (64). BLAST searching of the unfinished genome sequences available on-line (http://www.ncbi.nlm.nih.gov/BLAST/unfinishedgenome.html) also revealed numerous homologues in S. enterica serovar Typhi (four), Bordetella pertussis (four), Yersinia pestis (five), C. jejuni (one), and Vibrio cholerae (two). Whether any of these function in antibiotic efflux remains to be determined. Still, the possibility exists, at least, that FQ-MDR efflux systems are widespread in gram-negative bacteria, where they may play a significant role in resistance to this important class of antibiotic.
MFS- and MATE-Type Efflux Systems
The most common example of an MFS antibiotic efflux system in gram-negative bacteria is that encoded by the various tet genes associated with tetracycline efflux and resistance (119). Members of this family that efflux FQs are, in contrast, rare in gram-negative bacteria, including only the MdfA transporter of E. coli (28), although it appears to be a more effective pump for nonantibiotics. Originally described as MFS transporters, the NorM pump of V. parahaemolyticus (92) and its E. coli homologue YdhE (92) now appear to be members of the newly reported MATE family of transporters (13). These pumps facilitate resistance to multiple agents, including antibiotics and nonantibiotics, and appear to be better exporters of FQs than is MdfA. In the case of all three of the aforementioned transporters, however, efflux and antibiotic resistance were assessed using genes cloned on plasmids and expressed in E. coli. Thus, the relevance of the chromosomal counterparts to FQ resistance is uncertain. As with MFS-type exporters involved in FQ resistance in gram-positive bacteria FQ resistance attributable to NorM and YdhE is limited to the more hydrophilic FQs such as ciprofloxacin and norfloxacin (92).
FQ EFFLUX SYSTEMS EXHIBIT BROAD SUBSTRATE SPECIFICITY
The most striking feature of the RND-MFP-OEP MDR efflux systems of gram-negative bacteria is their incredibly broad substrate specificity, encompassing a variety of structurally unrelated antimicrobial agents, including clinically relevant antibiotics, dyes (28, 40, 74, 88, 92, 101, 123, 134), detergents (40, 74, 130, 135), disinfectants (84, 89), antimicrobial peptides (129), organic solvents (8, 57, 70, 71, 140), inhibitors of fatty acid synthesis (126), and homoserine lactones involved in bacterial cell-to-cell signalling (29, 111). The binding of multiple structurally varied substrates is uncommon in biology, and how this is achieved in gram-negative FQ-MDR transporters is as yet unknown. This broad substrate specificity contrasts with other examples of antibiotic efflux systems, which tend to be agent or class specific (e.g., the tetracycline [tet] [119] efflux systems). Interestingly, too, the FQ-MDR efflux systems are invariably chromosomally encoded and conserved in both sensitive and resistant strains, with resistance usually resulting from mutational upregulation of the efflux genes. The recently described plasmid-borne FQ resistance determinant (80) appears not to involve efflux, although the mechanism of resistance has yet to be elucidated (G. A. Jacoby, personal communication). Again, this contrasts with the tetracycline efflux systems, which are generally plasmid-borne or transposon-encoded (119). This suggests that FQ-MDR efflux systems are an intrinsic part of the gram-negative bacterium and function independently of antibiotic efflux and resistance, while the others function uniquely in antibiotic efflux and resistance and their acquisition from outside sources provides for antibiotic resistance.
NATURAL FUNCTION OF FQ-MDR EFFLUX SYSTEMS
The natural role of FQ-MDR efflux systems is the subject of some debate, with support for export of and, thus, protection from exogenous antimicrobial agents available in some instances. The inducibility of the E. coli AcrAB system by toxic fatty acids (75) and the demonstrated role of AcrAB in the export of and resistance to bile salts (135) are consistent with a role for AcrAB in protecting the cell from the action of these agents in the gut (75). A protective function is also likely attributable to the MtrCDE system, which provides for resistance to fecal lipids in rectal isolates of N. gonorrhoeae (128) and, probably, bile salts known to bathe mucous membranes (40). Still, in none of these cases are antibiotics the intended substrate. The fact, too, that most pump genes have linked regulatory genes indicates that the efflux systems are highly regulated and, thus, likely respond to something environmental or cell associated. Although some cell-derived compounds have been identified as substrates for these efflux systems, including homoserine lactone autoinducers in P. aeruginosa (MexAB-OprM) (29, 111) and indole (a precursor of tryptophan) in E. coli (AcrEF) (K. Sato, K. Shibayama, T. Horii, Y. Arakawa, and M. Ohta, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C-126, 1998), it is far from clear that these are the intended substrates. MexEF-OprN expression in P. aeruginosa, for example, is associated with a decrease in pyocyanin production (61), a phenotype also seen in nalB strains and attributed to homoserine lactone efflux by MexAB-OprM (29). Thus, homoserine lactones may be just another in a line of shared substrates for these highly accommodating MDR efflux systems in P. aeruginosa (K. Sato et al. 38th ICAAC).
THERAPEUTIC VALUE OF EFFLUX PUMP INHIBITORS
Given the contribution of FQ-MDR efflux systems to low- and high-level FQ resistance in clinical strains of a variety of important human pathogens, it seems logical that they be targets for therapeutic intervention. Indeed, a variety of genetic and inhibitor studies have confirmed the usefulness of pump inactivation in increasing bacterial susceptibility to FQs (and other antibiotics) and preventing emergence of FQ resistance. Mutants of P. aeruginosa with deletions of the genes coding for the three best-characterized FQ-MDR efflux systems of this organism were markedly FQ hypersusceptible, and FQ-resistant derivatives of this deletion strain could not be selected in vitro at clinically relevant concentrations of FQ (72). Moreover, elimination of the FQ-MDR efflux systems in this organism compromised resistance mediated by gyrA mutations (72; K. Poole, unpublished data). Similarly, loss of acrAB in E. coli rendered topoisomerase mutations generally inconsequential as regards clinical FQ resistance (103). Recently, the first examples of broad-spectrum efflux pump inhibitors of the Mex efflux system of P. aeruginosa have been reported (118; O. Lomovskaya, K. Hoshino, H. Ishida, A. Lee, M. Warren, and J. Galazzo, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. F-1264, 1999). These potentiate the activity of a number of antibiotics, including FQs, in vitro (118; Lomovskaya et al., 39th ICAAC) and in animal models of P. aeruginosa infection (D. Griffith, O. Lomovskaya, V. Lee, and M. Dudley, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. F-1268, 1999) and appear also to work in a variety of other gram-negative pathogens (J. Blais, D. Cho, K. Tangen, C. Ford, A. Lee, O. Lomovskaya, and S. Chamberland, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. F-1266, 1999). These inhibitors were effective at reversing acquired FQ resistance attributable to efflux or target site mutations (Lomovskaya et al., 39th ICAAC). Moreover, these inhibitors markedly decreased the frequency with which highly FQ-resistant strains could be selected in vitro (Lomovskaya et al., 39th ICAAC).
CONCLUSIONS
Efflux mechanisms of FQ resistance are widely distributed among important gram-negative human pathogens, where they appear to be components of the normal genetic complement of these bacteria. The identification, too, of several homologues of the RND-MFP-OEP-type efflux systems in the genome sequences of a number of these organisms, in which efflux-mediated FQ resistance has yet to be studied, suggests that the potential for efflux to contribute to FQ resistance is particularly great in gram-negative bacteria. Although there is much debate and uncertainty regarding the natural function of these plentiful FQ-MDR efflux systems, it is clear that they are important contributors to intrinsic and acquired FQ resistance. Inhibition of efflux systems would seem, therefore, a prudent approach to combating and/or preventing FQ resistance. Given the high degree homology of these efflux systems, it should be possible to identify broad-spectrum inhibitors, and indeed, preliminary work with P. aeruginosa efflux pump inhibitors seems to bear this out.
ACKNOWLEDGMENTS
Work on FQ/MDR efflux systems carried out in the author's laboratory was generously supported by the Canadian Cystic Fibrosis Foundation (CCFF) and the Canadian Bacterial Diseases Network, one of the Networks of Centers of Excellence. K.P. is a CCFF Martha Morton Scholar.
I thank R. Srikumar and G. McKay for critically reading the manuscript.
ADDENDUM IN PROOF
Recently, the crystal structure of the TolC OM channel of the AcrAB-TolC efflux system of E. coli was reported (V. Koronakis, A. Sharff, E. Koronakis, B. Luisi, and C. Hughes, Nature 405:914–919, 2000). Also, channel-forming activity has now been demonstrated for the OprM component of the MexAB-OprM efflux system of P. aeruginosa (K. K. Y. Wong and R. E. W. Hancock, J. Bacteriol. 182:2402–2410, 2000), although the observed channel size is less than what would be needed to accommodate the various known substrates of this efflux system. Thus, it is still likely that TonB is necessary to mediate channel opening.
REFERENCES
- 1.Acar J F. Trends in bacterial resistance to fluoroquinolones. Clin Infect Dis. 1997;24(Suppl. 1):S67–73. doi: 10.1093/clinids/24.supplement_1.s67. [DOI] [PubMed] [Google Scholar]
- 2.Ahamed J, Gangopadhyay J, Kundu M. Mechanisms of quinolone resistance in clinical isolates of Shigella dysenteriae. Antimicrob Agents Chemother. 1999;43:2333–2334. doi: 10.1128/aac.43.9.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aires J R, Köhler T, Nikaido H, Plésiat P. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob Agents Chemother. 1999;43:2624–2628. doi: 10.1128/aac.43.11.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alekshun M N, Levy S B. The mar regulon: multiple resistance to antibiotics and other toxic chemicals. Trends Microbiol. 1999;7:410–413. doi: 10.1016/s0966-842x(99)01589-9. [DOI] [PubMed] [Google Scholar]
- 5.Alm R A, Ling L S, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 6.Alonso A, Martinez J L. Multiple antibiotic resistance in Stenotrophomonas maltophilia. Antimicrob Agents Chemother. 1997;41:1140–1142. doi: 10.1128/aac.41.5.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersson S G, Zomorodipour A, Andersson J O, Sicheritz-Ponten T, Alsmark U C, Podowski R M, Naslund A K, Eriksson A S, Winkler H H, Kurland C G. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 8.Aono R. Improvement of organic solvent tolerance level of Escherichia coli by overexpression of stress-responsive genes. Extremophiles. 1998;2:239–248. doi: 10.1007/s007920050066. [DOI] [PubMed] [Google Scholar]
- 9.Aono R, Tsukagoshi N, Yamamoto M. Involvement of outer membrane protein TolC, a possible member of the mar-sox regulon, in maintenance and improvement of organic solvent tolerance of Escherichia coli K-12. J Bacteriol. 1998;180:938–944. doi: 10.1128/jb.180.4.938-944.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ball P. The quinolones: history and overview. In: Andriole V T, editor. The quinolones. 2nd ed. San Diego, Calif: Academic Press; 1998. pp. 1–28. [Google Scholar]
- 11.Ball P R, Shales S W, Chopra I. Plasmid-mediated tetracycline resistance in Escherichia coli involves increased efflux of the antibiotic. Biochem Biophys Res Commun. 1980;93:74–81. doi: 10.1016/s0006-291x(80)80247-6. [DOI] [PubMed] [Google Scholar]
- 12.Benz R, Maier E, Gentshev I. TolC of Escherichia coli functions as an outer membrane channel. J Bacteriol. 1993;178:5803–5805. doi: 10.1016/s0934-8840(11)80836-4. [DOI] [PubMed] [Google Scholar]
- 13.Brown M H, Paulsen I T, Skurray R A. The multidrug efflux protein NorM is a prototype of a new family of transporters. Mol Microbiol. 1999;31:393–395. doi: 10.1046/j.1365-2958.1999.01162.x. [DOI] [PubMed] [Google Scholar]
- 14.Burns J L, Clark D K. Salicylate-inducible antibiotic resistance in Pseudomonas cepacia associated with absence of a pore-forming outer membrane protein. Antimicrob Agents Chemother. 1992;36:2280–2285. doi: 10.1128/aac.36.10.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burns J L, Hedin L A, Lien D M. Chloramphenicol resistance in Pseudomonas cepacia because of decreased permeability. Antimicrob Agents Chemother. 1989;33:136–141. doi: 10.1128/aac.33.2.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burns J L, Wadsworth C D, Barry J J, Goodall C P. Nucleotide sequence analysis of a gene from Burkholderia (Pseudomonas) cepacia encoding an outer membrane lipoprotein involved in multiple antibiotic resistance. Antimicrob Agents Chemother. 1996;40:307–313. doi: 10.1128/aac.40.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cambau E, Perani E, Dib C, Petinon C, Trias J, Jarlier V. Role of mutations in DNA gyrase genes in ciprofloxacin resistance of Pseudomonas aeruginosa susceptible or resistant to imipenem. Antimicrob Agents Chemother. 1995;39:2248–2252. doi: 10.1128/aac.39.10.2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Celesk R A, Robillard R A. Factors influencing the accumulation of ciprofloxacin in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1989;33:1921–1926. doi: 10.1128/aac.33.11.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chamberland S, Bayer A S, Schollaardt T, Wong S A, Bryan L E. Characterization of mechanisms of quinolone resistance in Pseudomonas aeruginosa strains isolated in vitro and in vivo during experimental endocarditis. Antimicrob Agents Chemother. 1989;33:624–634. doi: 10.1128/aac.33.5.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charvalos E, Tselentis Y, Michea Hamzehpour M, Koehler T, Pechere J-C. Evidence for an efflux pump in multidrug-resistant Campylobacter jejuni. Antimicrob Agents Chemother. 1995;39:2019–2022. doi: 10.1128/aac.39.9.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen S H, Yan W, Levy S B. A multidrug resistance regulatory chromosomal locus is widespread among enteric bacteria. J Infect Dis. 1993;168:484–488. doi: 10.1093/infdis/168.2.484. [DOI] [PubMed] [Google Scholar]
- 22.Cohen S P, Hooper D C, Wolfson J S, Souza K S, McMurry L M, Levy S B. Endogenous active efflux of norfloxacin in susceptible Escherichia coli. Antimicrob Agents Chemother. 1988;32:1187–1191. doi: 10.1128/aac.32.8.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen S P, Levy S B, Foulds J, Rosner J L. Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J Bacteriol. 1993;175:7856–7862. doi: 10.1128/jb.175.24.7856-7862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen S P, McMurry L M, Hooper D C, Wolfson J S, Levy S B. Cross-resistance to fluoroquinolones in multiple-antibiotic-resistance (Mar) Escherichia coli selected by tetracycline resistance: decreased drug accumulation asociated with membrane changes in addition to OmpF reduction. Antimicrob Agents Chemother. 1989;33:1318–1325. doi: 10.1128/aac.33.8.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deguchi T, Kawamura T, Yasuda M, Nakano M, Fukuda H, Kato H, Kato N, Okanao Y, Kawada Y. In vivo selection of Klebsiella pneumoniae strains with enhanced quinolone resistance during fluoroquinolone treatment of urinary tract infections. Antimicrob Agents Chemother. 1997;41:1609–1611. doi: 10.1128/aac.41.7.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dinh T, Paulsen I T, Saier M H., Jr A family of extracytoplasmic proteins that allow transport of large molecules across the outer membranes of gram-negative bacteria. J Bacteriol. 1994;176:3825–3831. doi: 10.1128/jb.176.13.3825-3831.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Domenico P, Hopkins T, Cunha B A. The effect of sodium salicylate on antibiotic susceptibility and synergy in Klebsiella pneumoniae. J Antimicrob Chemother. 1990;26:343–351. doi: 10.1093/jac/26.3.343. [DOI] [PubMed] [Google Scholar]
- 28.Edgar R, Bibi E. MdfA, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition. J Bacteriol. 1997;179:2274–2280. doi: 10.1128/jb.179.7.2274-2280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evans K, Passador L, Srikumar R, Tsang E, Nezezon J, Poole K. Influence of the MexAB-OprM multidrug efflux system on quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1998;180:5443–5447. doi: 10.1128/jb.180.20.5443-5447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans K, Poole K. The MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa is growth-phase regulated. FEMS Microbiol Lett. 1999;173:35–39. doi: 10.1111/j.1574-6968.1999.tb13481.x. [DOI] [PubMed] [Google Scholar]
- 31.Everett M J, Jin Y F, Ricci V, Piddock L J. Contributions of individual mechanisms to fluoroquinolone resistance in 36 Escherichia coli strains isolated from humans and animals. Antimicrob Agents Chemother. 1996;40:2380–2386. doi: 10.1128/aac.40.10.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fralick J A. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J Bacteriol. 1996;178:5803–5805. doi: 10.1128/jb.178.19.5803-5805.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fukuda H, Hosaka M, Hirai K, Iyobe S. New norfloxacin resistance gene in Pseudomonas aeruginosa PAO. Antimicrob Agents Chemother. 1990;34:1757–1761. doi: 10.1128/aac.34.9.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fukuda H, Hosaka M, Iyobe S, Gotoh N, Nishino T, Hirai K. nfxC-type quinolone resistance in a clinical isolate of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:790–792. doi: 10.1128/AAC.39.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Furet Y X, Deshusses J, Pechere J C. Transport of pefloxacin across the bacterial cytoplasmic membrane in quinolone-susceptible Staphylococcus aureus. Antimicrob Agents Chemother. 1992;36:2506–2511. doi: 10.1128/aac.36.11.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghosh A S, Ahamed J, Chauhan K K, Kundu M. Involvement of an efflux system in high-level fluoroquinolone resistance of Shigella dysenteriae. Biochem Biophys Res Commun. 1998;242:54–56. doi: 10.1006/bbrc.1997.7902. [DOI] [PubMed] [Google Scholar]
- 37.Giraud E, Cloeckaert A, Kerboeuf D, Chaslus-Dancla E. Evidence for active efflux as the primary mechanism of resistance to ciprofloxacin in Salmonella enterica serovar Typhimurium. Antimicrob Agents Chemother. 2000;44:1223–1228. doi: 10.1128/aac.44.5.1223-1228.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldman J D, White D G, Levy S B. Multiple antibiotic resistance (mar) locus protects Escherichia coli from rapid cell killing by fluoroquinolones. Antimicrob Agents Chemother. 1996;40:1266–1269. doi: 10.1128/aac.40.5.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gotoh N, Tsujimoto H, Poole K, Yamagishi J-I, Nishino T. The outer membrane protein OprM of Pseudomonas aeruginosa is encoded by oprK of the mexA-mexB-oprK multidrug resistance operon. Antimicrob Agents Chemother. 1995;39:2567–2569. doi: 10.1128/aac.39.11.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hagman K E, Pan W, Spratt B G, Balthazar J T, Judd R C, Shafer W M. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology. 1995;141:611–622. doi: 10.1099/13500872-141-3-611. [DOI] [PubMed] [Google Scholar]
- 41.Hancock R E W. The bacterial outer membrane as a drug barrier. Trends Microbiol. 1997;5:37–42. doi: 10.1016/S0966-842X(97)81773-8. [DOI] [PubMed] [Google Scholar]
- 42.Hirai K, Suzue S, Irikura T, Iyobe S, Mitsuhashi S. Mutations producing resistance to norfloxacin in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1987;31:582–586. doi: 10.1128/aac.31.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hooper D C. Clinical applications of quinolones. Biochim Biophys Acta. 1998;1400:45–61. doi: 10.1016/s0167-4781(98)00127-4. [DOI] [PubMed] [Google Scholar]
- 44.Hooper D C, Wolfson J S, Souza K S, Ng E Y, McHugh G L, Swartz M N. Mechanisms of quinolone resistance in Escherichia coli: characterization of nfxB and cfxB, two mutant resistance loci decreasing norfloxacin accumulation. Antimicrob Agents Chemother. 1989;33:283–290. doi: 10.1128/aac.33.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hosaka M, Gotoh N, Nishino T. Purification of a 54-kilodalton protein (OprJ) produced in NfxB mutants of Pseudomonas aeruginosa and production of a monoclonal antibody specific to OprJ. Antimicrob Agents Chemother. 1995;39:1731–1735. doi: 10.1128/aac.39.8.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hwang J, Zhong X, Tai P C. Interactions of dedicated export membrane proteins of the colicin V secretion system: CvaA, a member of the membrane fusion protein family, interacts with CvaB and TolC. J Bacteriol. 1997;179:6264–6270. doi: 10.1128/jb.179.20.6264-6270.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Irikura T, Iyobe S, Mitsuhashi S. New quinolone resistance in clinical isolates of Pseudomonas aeruginosa. Rev Infect Dis. 1989;11(Suppl. 5):S972–973. [Google Scholar]
- 48.Ishida H, Fuziwara H, Kaibori Y, Horiuchi T, Sato K, Osada Y. Cloning of multidrug resistance gene pqrA from Proteus vulgaris. Antimicrob Agents Chemother. 1995;39:453–457. doi: 10.1128/aac.39.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ishii H, Sato K, Hoshino K, Sato M, Yamaguchi A, Sawai T, Osada Y. Active efflux of ofloxacin by a highly quinolone-resistant strain of Proteus vulgaris. J Antimicrob Chemother. 1991;28:827–836. doi: 10.1093/jac/28.6.827. [DOI] [PubMed] [Google Scholar]
- 50.Iyobe S, Hirai K, Hashimoto H. Drug resistance of Pseudomonas aeruginosa with special reference to new quinolones. Chemotherapy (Basel) 1991;44:209–214. doi: 10.1159/000420316. [DOI] [PubMed] [Google Scholar]
- 51.Jakics E B, Iyobe S, Hirai K, Fukuda H, Hashimoto H. Occurrence of the nfxB type mutation in clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992;36:2562–2565. doi: 10.1128/aac.36.11.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jalal S, Ciofu O, Hoiby N, Gotoh N, Wretlind B. Molecular mechanisms of fluoroquinolone resistance in Pseudomonas aeruginosa isolates from cystic fibrosis. Antimicrob Agents Chemother. 2000;44:710–712. doi: 10.1128/aac.44.3.710-712.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jalal S, Wretlind B. Mechanisms of quinolone resistance in clinical strains of Pseudomonas aeruginosa. Microbiol Drug Resist. 1998;4:257–261. doi: 10.1089/mdr.1998.4.257. [DOI] [PubMed] [Google Scholar]
- 54.Johnson J M, Church G M. Alignment and structure prediction of divergent protein families: periplasmic and outer membrane proteins of bacterial efflux pumps. J Mol Biol. 1999;287:695–715. doi: 10.1006/jmbi.1999.2630. [DOI] [PubMed] [Google Scholar]
- 55.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 56.Kern W V, Oethinger M, Jellen-Ritter A S, Levy S B. Non-target gene mutations in the development of fluoroquinolone resistance in Escherichia coli. Antimicrob Agents Chemother. 2000;44:814–820. doi: 10.1128/aac.44.4.814-820.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kieboom J, Dennis J J, Zylstra G J, de Bont J A. Active efflux of organic solvents by Pseudomonas putida S12 is induced by solvents. J Bacteriol. 1998;180:6769–6772. doi: 10.1128/jb.180.24.6769-6772.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klein J R, Henrich B, Plapp R. Molecular analysis and nucleotide sequence of the envCD operon of Escherichia coli. Mol Gen Genet. 1991;230:230–240. doi: 10.1007/BF00290673. [DOI] [PubMed] [Google Scholar]
- 59.Köhler T, Pechere J-C. Bacterial resistance to quinolones: mechanisms and clinical implications. In: Andriole V T, editor. The quinolones. London, United Kingdom: Academic Press; 1998. pp. 117–142. [Google Scholar]
- 60.Köhler T, Kok M, Michea-Hamzehpour M, Plesiat P, Gotoh N, Nishino T, Kocjanici Curty L, Pechere J-C. Multidrug efflux in intrinsic resistance to trimethoprim and sulfamethoxazole in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1996;40:2288–2290. doi: 10.1128/aac.40.10.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Köhler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty L K, Pechere J-C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 62.Köhler T, Michea-Hamzehpour M, Plesiat P, Kahr A-L, Pechere J-C. Differential selection of multidrug efflux systems by quinolones in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;41:2540–2543. doi: 10.1128/aac.41.11.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Köhler T, Epp S F, Curty L K, Pechère J-C. Characterization of MexT, the regulator of the MexE-MexF-OprN multidrug efflux system of Pseudomonas aeruginosa. J Bacteriol. 1999;181:6300–6305. doi: 10.1128/jb.181.20.6300-6305.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kumar V, Fonstein M, Haselkorn R. Bacterium genome sequence. Nature. 1996;381:653–654. doi: 10.1038/381653a0. [DOI] [PubMed] [Google Scholar]
- 65.Lacroix F J C, Cloeckaert A, Grépinet O, Pinault C, Popoff M Y, Waxin H, Pardon P. Salmonella typhimurium acrB-like gene: identification and role in resistance to biliary salts and detergents and in murine infection. FEMS Microbiol Lett. 1996;135:161–167. doi: 10.1111/j.1574-6968.1996.tb07983.x. [DOI] [PubMed] [Google Scholar]
- 66.Lecso-Bornet M, Pierre J, Sarkis-Karam D, Lubera S, Bergogne-Berezin E. Susceptibility of Xanthamonas maltophilia to six quinolones and study of outer membrane proteins in resistant mutants selected in vitro. Antimicrob Agents Chemother. 1992;36:669–671. doi: 10.1128/aac.36.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Legakis N J, Tzouvelekis L S, Makris A, Kotsifaki H. Outer membrane alterations in multiresistant mutants of Pseudomonas aeruginosa selected by ciprofloxacin. Antimicrob Agents Chemother. 1989;33:124–127. doi: 10.1128/aac.33.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li H, Park J T. The periplasmic murein peptide-binding protein MppA is a negative regulator of multiple antibiotic resistance in Escherichia coli. J Bacteriol. 1999;181:4842–4847. doi: 10.1128/jb.181.16.4842-4847.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li X-Z, Nikaido H, Poole K. Role of MexA-MexB-OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1948–1953. doi: 10.1128/aac.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li X-Z, Zhang L, Poole K. Role of the multidrug efflux systems of Pseudomonas aeruginosa in organic solvent tolerance. J Bacteriol. 1998;180:2987–2991. doi: 10.1128/jb.180.11.2987-2991.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70a.Li X-Z, Zhang L, Poole K. Interplay between the MexAB-OprM multidrug efflux system and the outer membrane barrier in the multiple antibiotic resistance of Pseudomonas aeruginosa. J Antimicrob Chemother. 2000;45:433–436. doi: 10.1093/jac/45.4.433. [DOI] [PubMed] [Google Scholar]
- 71.Li X Z, Poole K. Organic solvent-tolerant mutants of Pseudomonas aeruginosa display multiple antibiotic resistance. Can J Microbiol. 1999;45:18–22. doi: 10.1139/cjm-45-1-18. [DOI] [PubMed] [Google Scholar]
- 72.Lomovskaya O, Lee A, Hoshino K, Ishida H, Mistry A, Warren M S, Boyer E, Chamberland S, Lee V J. Use of a genetic approach to evaluate the consequences of inhibition of efflux pumps in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43:1340–1346. doi: 10.1128/aac.43.6.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ma D, Alberti M, Lynch C, Nikaido H, Hearst J E. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol Microbiol. 1996;19:101–112. doi: 10.1046/j.1365-2958.1996.357881.x. [DOI] [PubMed] [Google Scholar]
- 74.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst J E. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J Bacteriol. 1993;175:6299–6313. doi: 10.1128/jb.175.19.6299-6313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst J E. Genes acrA and acrB encode a stress-induced system of Escherichia coli. Mol Microbiol. 1995;16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 76.Ma D, Cook D N, Hearst J E, Nikaido H. Efflux pumps and drug resistance in Gram-negative bacteria. Trends Microbiol. 1994;2:489–493. doi: 10.1016/0966-842x(94)90654-8. [DOI] [PubMed] [Google Scholar]
- 77.Mallea M, Chevalier J, Bornet C, Eyraud A, Davin-Regli A, Bollet C, Pagès J-M. Porin alteration and active efflux: two in vivo drug resistance strategies used by Enterobacter aerogenes. Microbiology. 1998;144:3003–3009. doi: 10.1099/00221287-144-11-3003. [DOI] [PubMed] [Google Scholar]
- 78.Maneewannakul K, Levy S B. Identification for mar mutants among quinolone-resistant clinical isolates of Escherichia coli. Antimicrob Agents Chemother. 1996;40:1695–1698. doi: 10.1128/aac.40.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martínez-Martínez L, García I, Ballesta S, Benedí V J, Hernández-Allés S, Pascual A. Energy-dependent accumulation of fluoroquinolones in quinolone-resistant Klebsiella pneumoniae strains. Antimicrob Agents Chemother. 1998;42:1850–1852. doi: 10.1128/aac.42.7.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martínez-Martínez L, Pascual A, Jacoby G A. Quinolone resistance from a transferable plasmid. Lancet. 1998;351:797–799. doi: 10.1016/S0140-6736(97)07322-4. [DOI] [PubMed] [Google Scholar]
- 81.Masuda N, Gotoh N, Ohya S, Nishino T. Quantitative correlation between susceptibility and OprJ production in NfxB mutants of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1996;40:909–913. doi: 10.1128/aac.40.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Masuda N, Ohya S. Cross-resistance to meropenem, cephems, and quinolones in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992;36:1847–1851. doi: 10.1128/aac.36.9.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Masuda N, Sakagawa E, Ohya S. Outer membrane proteins responsible for multiple drug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:645–649. doi: 10.1128/AAC.39.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McMurry L M, Oethinger M, Levy S B. Overexpression of marA, soxS, or acrAB produces resistance to triclosan in laboratory and clinical strains of Escherichia coli. FEMS Microbiol Lett. 1998;166:305–309. doi: 10.1111/j.1574-6968.1998.tb13905.x. [DOI] [PubMed] [Google Scholar]
- 85.McMurry L M, Petrucci R E, Jr, Levy S B. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc Natl Acad Sci USA. 1980;77:3974–3977. doi: 10.1073/pnas.77.7.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Michea-Hamzehpour M, Lucain C, Pechere J-C. Resistance to pefloxacin in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1991;35:512–518. doi: 10.1128/aac.35.3.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mine T, Morita Y, Kataoka A, Mitzushima T, Tsuchiya T. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43:415–417. doi: 10.1128/aac.43.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Miyamae S, Nikaido H, Tanaka Y, Yoshimura F. Active efflux of norfloxacin by Bacteroides fragilis. Antimicrob Agents Chemother. 1998;42:2119–2121. doi: 10.1128/aac.42.8.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moken M C, McMurry L M, Levy S B. Selection of multiple-antibiotic-resistant (mar) mutants of Escherichia coli by using the disinfectant pine oil: roles of the mar and acrAB loci. Antimicrob Agents Chemother. 1997;41:2770–2772. doi: 10.1128/aac.41.12.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Moniot-Ville N, Moreau J G, Acar J F, Collatz E, Gutmann L. Mechanisms of quinolone resistance in a clinical isolate of Escherichia coli highly resistant to fluoroquinolones but susceptible to nalidixic acid. Antimicrob Agents Chemother. 1991;35:519–523. doi: 10.1128/aac.35.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Moore R A, DeShazer D, Reckseidler S, Weissman A, Woods D E. Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob Agents Chemother. 1999;43:465–470. doi: 10.1128/aac.43.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Morita Y, Kodama K, Shiota S, Mine T, Kataoka A, Mizushima T, Tsuchiya T. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob Agents Chemother. 1998;42:1778–1782. doi: 10.1128/aac.42.7.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mouneimne H, Robert J, Jarlier V, Cambau E. Type II topoisomerase mutations in ciprofloxacin-resistant strains of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43:62–66. doi: 10.1128/aac.43.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Navia M M, Ruiz J, Ribera A, de Anta M T, Vila J. Analysis of the mechanisms of quinolone resistance in clinical isolates of Citrobacter freundii. J Antimicrob Chemother. 1999;44:743–748. doi: 10.1093/jac/44.6.743. [DOI] [PubMed] [Google Scholar]
- 95.Nies D H. The cobalt, zinc, and cadmium efflux system CzcABC from Alcaligenes eutrophus functions as a cation-proton antiporter in Escherichia coli. J Bacteriol. 1995;177:2707–2712. doi: 10.1128/jb.177.10.2707-2712.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nikaido H. Outer membrane barrier as a mechanism of antimicrobial resistance. Antimicrob Agents Chemother. 1989;33:1831–1836. doi: 10.1128/aac.33.11.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nikaido H. Antibiotic resistance caused by gram-negative multidrug efflux pumps. Clin Infect Dis. 1998;27(Suppl. 1):S32–41. doi: 10.1086/514920. [DOI] [PubMed] [Google Scholar]
- 98.Nikaido H. The role of outer membrane and efflux pumps in the resistance of gram-negative bacteria. Can we improve drug access? Drug Res Update. 1998;1:93–98. doi: 10.1016/s1368-7646(98)80023-x. [DOI] [PubMed] [Google Scholar]
- 99.Nikaido H, Basina M, Nguyen V, Rosenberg E Y. Multidrug efflux pump AcrAB of Salmonella typhimurium excretes only those β-lactam antibiotics containing lipophilic side chains. J Bacteriol. 1998;180:4686–4692. doi: 10.1128/jb.180.17.4686-4692.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nikaido H, Thanassi D G. Penetration of lipophilic agents with multiple protonation sites into bacterial cells: tetracyclines and fluoroquinolones as examples. Antimicrob Agents Chemother. 1993;37:1393–1399. doi: 10.1128/aac.37.7.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ocaktan A, Yoneyama H, Nakae T. Use of fluorescence probes to monitor function of the subunit proteins of the MexA-MexB-OprM drug extrusion machinery in Pseudomonas aeruginosa. J Biol Chem. 1997;272:21964–21969. doi: 10.1074/jbc.272.35.21964. [DOI] [PubMed] [Google Scholar]
- 102.Ochs M M, McCusker M P, Bains M, Hancock R E. Negative regulation of the Pseudomonas aeruginosa outer membrane porin OprD selective for imipenem and basic amino acids. Antimicrob Agents Chemother. 1999;43:1085–1090. doi: 10.1128/aac.43.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Oethinger M, Kern W V, Jellen-Ritter A S, McMurry L M, Levy S B. Ineffectiveness of topoisomerase mutations in mediating clinically significant fluoroquinolone resistance in Escherichia coli in the absence of the AcrAB efflux pump. Antimicrob Agents Chemother. 2000;44:10–13. doi: 10.1128/aac.44.1.10-13.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Oethinger M, Podglajen I, Kern W V, Levy S B. Overexpression of the marA or soxS regulatory gene in clinical topoisomerase mutants of Escherichia coli. Antimicrob Agents Chemother. 1998;42:2089–2094. doi: 10.1128/aac.42.8.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Okazaki T, Hirai K. Cloning and nucleotide sequence of the Pseudomonas aeruginosa nfxB gene, conferring resistance to new quinolones. FEMS Microbiol Lett. 1992;97:197–202. doi: 10.1016/0378-1097(92)90386-3. [DOI] [PubMed] [Google Scholar]
- 106.Okazaki T, Iyobe S, Hashimoto H, Hirai K. Cloning and characterization of a DNA fragment that complements the nfxB mutation in Pseudomonas aeruginosa PAO. FEMS Microbiol Lett. 1991;79:31–36. doi: 10.1016/0378-1097(91)90522-c. [DOI] [PubMed] [Google Scholar]
- 107.Okusu H, Ma D, Nikaido H. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J Bacteriol. 1996;178:306–308. doi: 10.1128/jb.178.1.306-308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pao S S, Paulsen I T, Saier M H., Jr Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Paulsen I T, Skurray R A, Tam R, Saier M H, Jr, Turner R J, Weiner J H, Goldberg E B, Grinius L L. The SMR family: a novel family of multidrug efflux protein involved with the efflux of lipophilic drugs. Mol Microbiol. 1996;19:1175. doi: 10.1111/j.1365-2958.1996.tb02462.x. [DOI] [PubMed] [Google Scholar]
- 111.Pearson J P, Van Delden C, Iglewski B H. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J Bacteriol. 1999;181:1203–1210. doi: 10.1128/jb.181.4.1203-1210.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Piddock L J V, Hall M C, Bellido F, Bains M, Hancock R E W. A pleiotropic, posttherapy, enoxacin-resistant mutant of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992;36:1057–1061. doi: 10.1128/aac.36.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, Yamasaki T, Neshat S, Yamagishi J-I, Li X-Z, Nishino T. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB multidrug resistant strains of Pseudomonas aeruginosa. Mol Microbiol. 1996;21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 114.Poole K, Heinrichs D E, Neshat S. Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa: regulation by iron and possible involvement in the secretion of the siderophore pyoverdine. Mol Microbiol. 1993;10:529–544. doi: 10.1111/j.1365-2958.1993.tb00925.x. [DOI] [PubMed] [Google Scholar]
- 115.Poole K, Krebes K, McNally C, Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Poole K, Tetro K, Zhao Q, Neshat S, Heinrichs D, Bianco N. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob Agents Chemother. 1996;40:2021–2028. doi: 10.1128/aac.40.9.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rella M, Haas D. Resistance of Pseudomonas aeruginosa PAO to nalidixic acid and low levels of β-lactam antibiotics: mapping of chromosomal genes. Antimicrob Agents Chemother. 1982;22:242–249. doi: 10.1128/aac.22.2.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Renau T E, Leger R, Flamme E M, Sangalang J, She M W, Yen R, Gannon C L, Griffith D, Chamberland S, Lomovskaya O, Hecker S J, Lee V J, Ohta T, Nakayama K. Inhibitors of efflux pumps in Pseudomonas aeruginosa potentiate the activity of the fluoroquinolone antibacterial levofloxacin. J Med Chem. 1999;42:4928–4931. doi: 10.1021/jm9904598. [DOI] [PubMed] [Google Scholar]
- 119.Roberts M C. Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol Lett. 1996;19:1–24. doi: 10.1111/j.1574-6976.1996.tb00251.x. [DOI] [PubMed] [Google Scholar]
- 120.Robillard R A, Scarpa A L. Genetic and physiological characterization of ciprofloxacin resistance in Pseudomonas aeruginosa PAO. Antimicrob Agents Chemother. 1988;32:535–539. doi: 10.1128/aac.32.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Saier M H, Tam R, Reizer A, Reizer J. Two novel families of bacterial membrane proteins concerned with nodulation, cell division and transport. Mol Microbiol. 1994;11:841–847. doi: 10.1111/j.1365-2958.1994.tb00362.x. [DOI] [PubMed] [Google Scholar]
- 122.Saito K, Yoneyama H, Nakae T. nalB-type mutations causing the overexpression of the MexAB-OprM efflux pump are located in the mexR gene of the Pseudomonas aeruginosa chromosome. FEMS Microbiol Lett. 1999;179:67–72. doi: 10.1111/j.1574-6968.1999.tb08709.x. [DOI] [PubMed] [Google Scholar]
- 123.Sanchez L, Pan W, Vinas M, Nikaido H. The acrAB homolog of Haemophilus influenzae codes for a functional multidrug efflux pump. J Bacteriol. 1997;179:6855–6857. doi: 10.1128/jb.179.21.6855-6857.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sanchez L, Puig M, Fuste C, Loren J G, Vinas M. Outer membrane permeability of non-typable Haemophilus influenzae. J Antimicrob Chemother. 1996;37:341–344. doi: 10.1093/jac/37.2.341. [DOI] [PubMed] [Google Scholar]
- 125.Sanders C C, Sanders W E, Jr, Goering R V, Werner V. Selection of multiple antibiotic resistance by quinolones, β-lactams, and aminoglycosides with special reference to cross-resistance between unrelated drug classes. Antimicrob Agents Chemother. 1984;26:797–801. doi: 10.1128/aac.26.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schweizer H P. Intrinsic resistance to inhibitors of fatty acid biosynthesis in Pseudomonas aeruginosa is due to efflux: application of a novel technique for generation of unmarked chromosomal mutations for the study of efflux systems. Antimicrob Agents Chemother. 1998;42:394–398. doi: 10.1128/aac.42.2.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Seiffer D, Klein J R, Plapp R. EnvC, a new lipoprotein of the cytoplasmic membrane of Escherichia coli. FEMS Microbiol Lett. 1993;107:175–178. doi: 10.1111/j.1574-6968.1993.tb06026.x. [DOI] [PubMed] [Google Scholar]
- 128.Shafer W M, Balthazar J T, Hagman K E, Morse S A. Missense mutations that alter the DNA-binding domain of the MtrR protein occur frequently in rectal isolates of Nesseria gonorrhoeae that are resistant to fecal lipids. Microbiology. 1995;141:907–911. doi: 10.1099/13500872-141-4-907. [DOI] [PubMed] [Google Scholar]
- 129.Shafer W M, Qu X-D, Waring A J, Lehrer R I. Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc Natl Acad Sci USA. 1998;95:1829–1833. doi: 10.1073/pnas.95.4.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Srikumar R, Kon T, Gotoh N, Poole K. Expression of Pseudomonas aeruginosa multidrug efflux pumps MexA-MexB-OprM and MexC-MexD-OprJ in a multidrug-sensitive Escherichia coli strain. Antimicrob Agents Chemother. 1998;42:65–71. doi: 10.1128/aac.42.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Srikumar R, Li X-Z, Gotoh N, Poole K. The inner membrane efflux components are responsible for the β-lactam specificity of multidrug efflux pumps in Pseudomonas aeruginosa. J Bacteriol. 1997;179:7875–7881. doi: 10.1128/jb.179.24.7875-7881.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Srikumar R, Paul C J, Poole K. Influence of mutations in the mexR repressor gene on expression of the MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa. J Bacteriol. 2000;182:1410–1414. doi: 10.1128/jb.182.5.1410-1414.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sulavik M C, Dazer M, Miller P F. The Salmonella typhimurium mar locus: molecular and genetic analyses and assessment of its role in virulence. J Bacteriol. 1997;179:1857–1866. doi: 10.1128/jb.179.6.1857-1866.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Takiff H G, Cimino M, Musso M C, Weisbrod T, Martinez R, Delgado M B, Salazar L, Bloom B R, Jacobs W R., Jr Efflux pump of the proton antiporter family confers low-level fluoroquinolone resistance in Mycobacterium smegmatis. Proc Natl Acad Sci USA. 1996;93:362–366. doi: 10.1073/pnas.93.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134a.Tavío M M, Vila J, Ruiz J, Amicosante G, Franceschini N, Martín-Sánchez A M, de Anta M T J. In vitro selected fluoroquinolone-resistant mutants of Citrobacter freundii: analysis of the quinolone resistance acquisition. J Antimicrob Chemother. 2000;45:521–524. doi: 10.1093/jac/45.4.521. [DOI] [PubMed] [Google Scholar]
- 135.Thanassi D G, Cheng L W, Nikaido H. Active efflux of bile salts in Escherichia coli. J Bacteriol. 1997;179:2512–2518. doi: 10.1128/jb.179.8.2512-2518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzegerald L M, Lee N, Adams M D, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 137.van Veen H W, Konings W N. The ABC family of multidrug transporters in microorganisms. Biochim Biophys Acta. 1998;1365:31–36. doi: 10.1016/s0005-2728(98)00039-5. [DOI] [PubMed] [Google Scholar]
- 138.Wandersman C, Delepelaire P. TolC, an Escherichia coli outer membrane protein required for hemolysin secretion. Proc Natl Acad Sci USA. 1990;87:4776–4780. doi: 10.1073/pnas.87.12.4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Westbrock-Wadman S, Sherman D R, Hickey M J, Coulter S N, Zhu Y Q, Warrener P, Nguyen L Y, Shawar R M, Folger K R, Stover C K. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob Agents Chemother. 1999;43:2975–2983. doi: 10.1128/aac.43.12.2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.White D G, Goldman J D, Demple B, Levy S B. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J Bacteriol. 1997;179:6122–6126. doi: 10.1128/jb.179.19.6122-6126.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wong K K Y, Poole K, Gotoh N, Hancock R E W. Influence of OprM expression on multiple antibiotic resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;41:2009–2012. doi: 10.1128/aac.41.9.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yoneyama H, Maseda H, Kamiguchi H, Nakae T. Function of the membrane fusion protein, MexA, of the MexAB-OprM efflux pump in Pseudomonas aeruginosa without an anchoring membrane. J Biol Chem. 2000;275:4628–4634. doi: 10.1074/jbc.275.7.4628. [DOI] [PubMed] [Google Scholar]
- 143.Yonezawa M, Takahata M, Matsubara N, Watanabe Y, Narita H. DNA gyrase gyrA mutations in quinolone-resistant clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1970–1972. doi: 10.1128/aac.39.9.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yoshida H, Nakamura M, Bogaki M, Nakamura S. Proportion of DNA gyrase mutants among quinolone-resistant strains of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1990;34:1273–1275. doi: 10.1128/aac.34.6.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yoshida T, Muratani T, Iyobe S, Mitsuhashi S. Mechanisms of high-level resistance to quinolones in urinary tract isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1994;38:1466–1469. doi: 10.1128/aac.38.7.1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zgurskaya H I, Nikaido H. AcrA is a highly asymmetric protein capable of spanning the periplasm. J Mol Biol. 1999;285:409–420. doi: 10.1006/jmbi.1998.2313. [DOI] [PubMed] [Google Scholar]
- 147.Zgurskaya H I, Nikaido H. Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc Natl Acad Sci USA. 1999;96:7190–7195. doi: 10.1073/pnas.96.13.7190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhanel G G, Karlowsky J A, Saunders M H, Davidson R J, Hoban D J, Hancock R E, McLean I, Nicolle L E. Development of multiple-antibiotic-resistant (Mar) mutants of Pseudomonas aeruginosa after serial exposure to fluoroquinolones. Antimicrob Agents Chemother. 1995;39:489–495. doi: 10.1128/aac.39.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhang L, Li X-Z, Poole K. Multiple antibiotic resistance in Stenotrophomonas maltophilia: involvement of a multidrug efflux system. Antimicrob Agents Chemother. 2000;44:287–293. doi: 10.1128/aac.44.2.287-293.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhao Q, Li X-Z, Mistry A, Srikumar R, Zhang L, Lomovskaya O, Poole K. Influence of the TonB energy-coupling protein on efflux-mediated multidrug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1998;42:2225–2231. doi: 10.1128/aac.42.9.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhao Q, Li X-Z, Srikumar R, Poole K. Contribution of outer membrane efflux protein OprM to antibiotic resistance in Pseudomonas aeruginosa independent of MexAB. Antimicrob Agents Chemother. 1998;42:1682–1688. doi: 10.1128/aac.42.7.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ziha-Zarifi I, Llanes C, Koehler T, Pechere J-C, Plesiat P. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa overexpressing the active efflux system MexA-MexB-OprM. Antimicrob Agents Chemother. 1999;43:287–291. doi: 10.1128/aac.43.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]