Abstract
Purpose
Monocarboxylate transporter 4 (MCT4) is an important component of cancer cell glycolytic metabolism. It has been confirmed that MCT4 is highly expressed in hepatocellular carcinoma (HCC) cells and tissues and is significantly associated with poor prognosis of HCC patients. However, research on its downstream molecules that affect HCC is still insufficient. The aim of current research was to investigate the MCT downstream molecule and its role of in HCC development.
Patients and Methods
After MCT4 expression was knocked down by RNA interference, RNA sequencing and quantitative real-time PCR were used to screen for differentially expressed genes in an HCC cell line (HCCLM3). Immunohistochemistry in HCC tissue microarray was carried out to evaluate the Trafficking Protein Particle Complex Subunit 5 (TRAPPC5) expression. Cell proliferation, migration, and invasion were evaluated by CCK-8 assay, colony formation assay, transwell and wound-healing test, respectively. Xenograft experiment was employed to investigate the function of TRAPPC5 on tumor growth in vivo. Related signaling pathway proteins were evaluated by Western blot.
Results
TRAPPC5 expression was significantly downregulated after knocking down of MCT4 in HCCLM3. TRAPPC5 was highly expressed in HCC tissues, and it could enhance the proliferation, migration, invasion and epithelial–mesenchymal transition (EMT) process of HCC cells. In vivo experiment showed that TRAPPC5 could promote HCC tumorigenesis.
Conclusion
In the process of MCT4 affecting the progression of HCC, TRAPPC5 is one of the most important related molecules. TRAPPC5 suppression could significantly reduce HCC cell proliferation, migration and invasion and could serve as a therapeutic target in HCC.
Keywords: MCT4, hepatocellular carcinoma, TRAPPC5, RNA sequencing, tissue microarrays
Introduction
Hepatocellular carcinoma (HCC) accounts for approximately 80% of primary liver malignancies, and is considered the sixth most common cancer and the third leading cause of cancer deaths worldwide.1,2 Most cases are treated with surgical resection, percutaneous ablation and transarterial chemoembolization (TACE) depends on tumor burden, location and comorbidities. Although there have been significant advances in targeted drugs (antiangiogenic tyrosine kinase inhibitors) and immunotherapy (anti-PD1 agents) in recent years, the 5-year average survival rate of HCC patients is still less than 20%.3,4 In addition to conventional clinicopathological factors such as lymph node metastasis, PNI, pathological grade, and staging, many molecules such as AFP, HSP90 and GPC-3 have been widely studied and applied as prognostic biomarkers involved in tumorigenesis and development,5,6 more studies is needed to help providing effective treatment targets and prognostic markers.
Monocarboxylate transporter 4 (MCT4) is an effective lactic acid transporter and an important component of cancer cell glycolytic metabolism. And lactate efflux results in an acidic tumor microenvironment, which contributes to tumor invasion, metastasis, immunosurveillance avoidance, and therapeutic resistance.7 Overexpression of MCT4 has been reported associated with poor survival of head and neck cancer,8 Adrenocortical carcinoma,9 lung cancer,10 Glioblastoma,11 breast cancer12 and colorectal cancer.13 In our previous study, it has been confirmed that MCT4 is highly expressed in HCC cells and tissues, promotes the proliferation, invasion and metastasis of tumor cells, and is significantly associated with poor prognosis of HCC patients. MCT4 might be a therapeutic target for HCC.14 However, research on its downstream molecules that affect HCC is still insufficient. In this study, MCT4 expression in HCC cells was knocked down by RNA interference, Trafficking Protein Particle Complex Subunit 5 (TRAPPC5) gene was screened out by high-throughput sequencing (RNA sequencing), and its role in HCC cell proliferation and metastasis was studied. Our results demonstrated that high expression of TRAPPC5 promotes the proliferation, invasion and metastasis of HCC cells for the first time.
Materials and Methods
Cell Culture
HCCLM3 cells and BEL-7404 cell lines were purchased from the cell bank of the Shanghai Chinese Academy of Sciences (Shanghai, China). They were cultured routinely at 37°C and 5% CO2 in DMEM medium (Gibco, Grand Island, NY, USA) plus 10% fetal bovine serum (FBS; Gibco) in an incubator at 37 °C containing 5% CO2, with a serum concentration of 10%, a penicillin concentration of 100 U/mL, and a streptomycin 0.1 mg/mL; When the cells enter the logarithmic growth phase, use a centrifuge at 900 rpm for 3 minutes to adjust the cell density and plant them in a six-well plate for subsequent experiments. When the cell density in the well plate reaches about 80%, the cells are processed for subsequent experiments.
Cell Transfection
For expression silencing, small interfering RNA (siRNA) was utilized. In brief, siRNA targeting TRAPPC5 (si-TRAPPC5) and siRNA negative control (si-NC) were constructed by Oligobi (Beijing, China). SiRNA were transfected into cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to detailed usage instructions.
Fluorescence Quantitative PCR Detection of Interference Effect
RNA extraction kit (Kwbio, Taizhou, China) was used to extract total RNA from cells, and after reverse transcription to form cDNA, real-time fluorescent quantitative PCR (HEHUIBIO, Suzhou, China) was used to detect the interference effect.
Cell Counting Kit-8 (CCK-8) Assay
HCCLM3 cells and BEL-7404 cells were plated in 96-well plates (1500 cells/well) with different transfection. After culturing for 24, 48 and 72 h, cells were exposed to 10 μL CCK-8 reagent (MCE, Monmouth Junction, NJ, USA) and incubate in a 37°C incubator for 1.5h. The absorbance at 450 nm of HCC cells at different time points was examined using a microplate reader (Tecan, Switzerland). Each sample was in triplicate, and three independent experiments were conducted.
Colony Formation Assay
HCC cells were cultured in a 60 mm dish containing 5 mL of 37°C pre-warmed culture medium at a gradient density of 500 cells per dish with different transfection. The plates were incubated in a cell incubator at 37°C, 5% CO2 and saturated humidity for 2 weeks. Formed colonies were fixed using methanol and then stained using 0.1% crystal violet followed by photograph under a microscope (Olympus, Tokyo, Japan). Each sample was in triplicate, and three independent experiments were conducted.
Transwell Migration and Invasion Experiment
Cell migration was tested using transwell chambers (Costar, Corning, NY, USA). Cell invasion was analyzed with Matrigel-coated transwell chambers. For transwell assay, HCCLM3 cells and BEL-7404 cells (1 × 104 for migration assay, 5×104 for invasion assay) with different transfections in serum-free DMEM were plated into the superior chambers, the lower chamber was supplemented with 500μL DMEM with 10% serum. After 24h, wipe off the cells and Matrigel in the upper chamber with a cotton swab. Fix with paraformaldehyde for 15 minutes. After staining with 0.1% crystal violet for 5 minutes and washing with PBS, cut the basement membrane and place it on a glass slide for observation, taking pictures and counting and examined under a microscope (× 100 magnification, Olympus, Tokyo, Japan) with 6 random fields.
Wound Healing Assay
Cells with various transfections were seeded into 6-well plates at a density of 5×104 cells/well and cultured in DMEM with 10% FBS until a cell monolayer was formed. Cells were scratched with a 10 μL pipette tip. Wash the cells three times with PBS before adding serum-free media. Place it in a 37°C 5% CO2 incubator to culture. After 24 hours of culture, the migration distance of the cell scratch region was measured using a microscope. (× 100 magnification, Olympus, Tokyo, Japan).
HCC Tissue Microarray and Immunohistochemical Staining
A tissue microarray (Cat No. HLivH060CS01) including 20 HCC tissue specimens, 20 corresponding adjacent tissues specimens was obtained from Shanghai Outdo Biotechnology (Shanghai, China). The human tissue experiments were approved by Ethics Committee of Shandong Provincial Hospital, all methods were carried out in accordance with relevant guidelines and regulations. IHC staining was carried out to measure TRAPPC5 protein level in HCC tissues following the instructions from the manufacturer using an anti- TRAPPC5 antibody (1:500, DF9965, Affinity). To summarize, the paraffin-embedded sections were first deparaffinized. After that, citrate buffer (pH 6.0) was used for antigen retrieval, and 0.3% H2O2 was used to inhibit endogenous peroxidase activity. The sections were treated with the primary and secondary antibodies listed. DAB was used to label the nuclei. Each HCC sample was evaluated based on the staining intensity and the percentage of cells with positive staining. Then apply the DensitoQuant software in the Quant center to automatically identify and set all dark brown on the tissue section as strong positive, brownish yellow as moderately positive, light yellow as weakly positive, and blue nuclei as negative. Then identify and analyze the areas of strong positives, moderate positives, weak positives and negatives for each tissue point (unit: pixels), percentage of positives, and get the finally H-score. Paired t-test was used to compare the CDT1 expression in HCC tissues and the paired non-tumor tissues.
Western Blot
Total proteins were extracted in RIPA Lysis Buffer (Beyotime) and quantitated by a protein quantitative kit (Beyotime). 20 μg/lane Protein were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred onto polyvinylidene difluoride (PVDF) membranes (Bio-Rad) and incubated with 5% non-fat milk. Subsequently, the membranes containing protein bands were probed with the primary antibodies (Supplementary Table 1) at 4 °C overnight and subsequently treated with the secondary antibodies (Supplementary Table 2). The protein bands were imaged using ECL reagent (Amyjet Scientific, Wuhan, China), the blots were visualized by ChemiDoc XRS+ system (Bio-rad Laboratories, Richmond, California, USA) and analyzed Image J v1.8 software (NIH, Bethesda, MD, USA). Each sample was in triplicate, and three independent experiments were conducted.
Xenograft Experiment
In total 20 BALB/c nude mice (female, 5-week-old, 18–20g) were purchased from Beijing Laboratory Animal Center (Beijing, China) and housed under specific pathogen-free conditions. The animal experiments were approved by Ethics Committee of Shandong Provincial Hospital, all methods were carried out in accordance with relevant guidelines and regulations. This study was carried out in compliance with the ARRIVE guidelines. Mice were randomly divided into 2 groups (n = 10 per group). Lentiviruses expressing siRNA-TRAPPC5 and the negative control were supplied by Genechem Co. LTD. (Shanghai, China). Stable HCCLM3 cells were established by transducing with different lentiviruses, selected by puromycin. Stable HCCLM3 cells (1 × 106 /mouse) were hypodermically inoculated into the right flanks of the mice. Tumor volume was estimated every week. 3 weeks later, sacrificed to collect tumor tissues and weighed.
RNA Sequencing in HCC Cell Line
Total RNA was extracted from HCCLM3 cells transfected with siRNA (HCCLM3-S3) and sent to BGI (Shenzhen, China) for RNA sequencing. Raw reads were edited to remove low-quality bases and adaptor sequences (default parameters) using fastp, HISTAT2 was used to map reads to the GRCh38 genome. After creating the BAM file, featureCounts was used to calculate the read counts for each gene. Reads per kilobase per million (RPKM) values were obtained using gene counts. The data was analyzed using R software and a low-stringency filtering technique of a 1.5-fold change in the expression level following MAS5 normalization of all datasets.
Gene Expression Profiling Interactive Analysis (GEPIA)
GEPIA is a newly developed interactive web server for analyzing the RNA sequencing expression data from the TCGA and the GTEx projects (http://gepia.cancer-pku.cn/).15 In this study, we used GEPIA to investigated the expression of TRAPPC5 in HCC tissues and patients at different stages, and eventually obtained a survival analysis.
Statistical Analysis
Use SPSS18.0 statistical analysis software to analyze the experimental data. Measurement data are expressed as mean±standard deviation (Mean±SD), the comparison between the two groups was performed by t test, and P<0.05 was considered statistically significant. The experimental results were expressed as mean ± standard deviation.16 Statistical analysis was performed using SPSS18.0 statistical analysis software and GraphPad Prism 7.0 software (Graph Pad, La Jolla, CA, USA). Data analysis between two or more groups was conducted via Student’s t-test. Statistics with P<0.05 was considered to be statistically significant.
Results
TRAPPC5 Was Identified as MCT4 Downstream Gene via RNA Sequencing in MCT4 Knocking Down HCC Cell Line (HCCLM3)
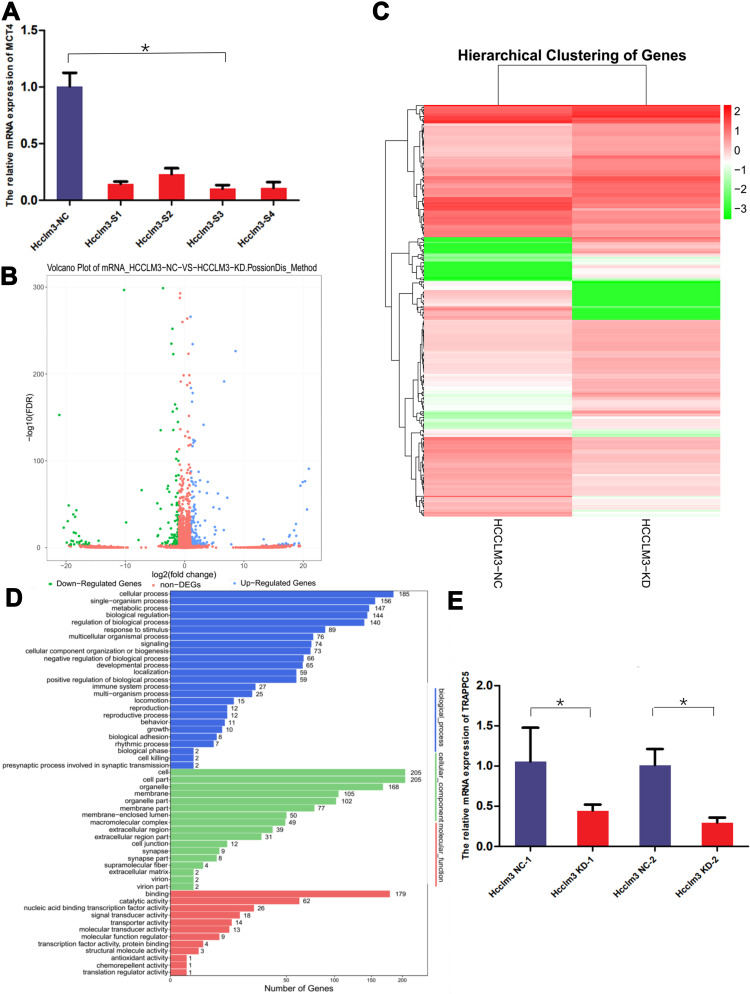
To study the related molecules of MCT4 in HCC cells, we induced RNA sequencing of HCCLM3 cell line after knockdown MCT4 by transfection of siRNA. Four siRNAs (HCCLM3-S1-4) of MCT4 (Table 1) were induced in transfection and the HCCLM3-S3 was chosen because of its highest efficiency verified by qPCR (Figure 1A). Then the mRNA sequencing of HCCLM3 cells was performed after knockdown of MCT4. According to the sequencing results, TRAPPC5 whose expression decreased the most after knocking down of MCT4 in HCCLM3 cells was screened out for database screening (Figure 1B–D) and verified by qPCR (Figure 1E).
Table 1.
The Sequence of siRNAs Used to Knockdown MCT4
| Target | Sequence (5’-3’) |
|---|---|
| si-MCT4-1 | CCATCTGCTTTGCCATCTT |
| si-MCT4-2 | GCTGTGGTAACTACCAAAT |
| si-MCT4-3 | GCAGGCTGGTTATATGATT |
| si-MCT4-4 | CCGGTTTGGGTTCTGCTTT |
| si-NC | TTCTCCGAACGTGTCACGT |
Figure 1.
TRAPPC5 was identified as MCT4 downstream gene via RNA sequencing in MCT4 knocking down HCC cell line (HCCLM3). (A) HCCLM3-S3 (siRNA of MCT4) was chosen to knock down MCT4 expression because of its highest efficiency verified by qPCR. (B) Volcano plot demonstrating differentially expressed genes between the knockdown and control group. (C) Heat-map displayed the genes of differential expression in MCT4 knocking down and control group. (D) Functional categorization by Gene Ontology (GO) analysis of the differential expression genes between knockdown and control group. (E) QPCR test showed that TRAPPC5 is downregulated in MCT4 knockdown group. *P < 0.05.
TRAPPC5 is Overexpressed in HCC Tissue Microarrays
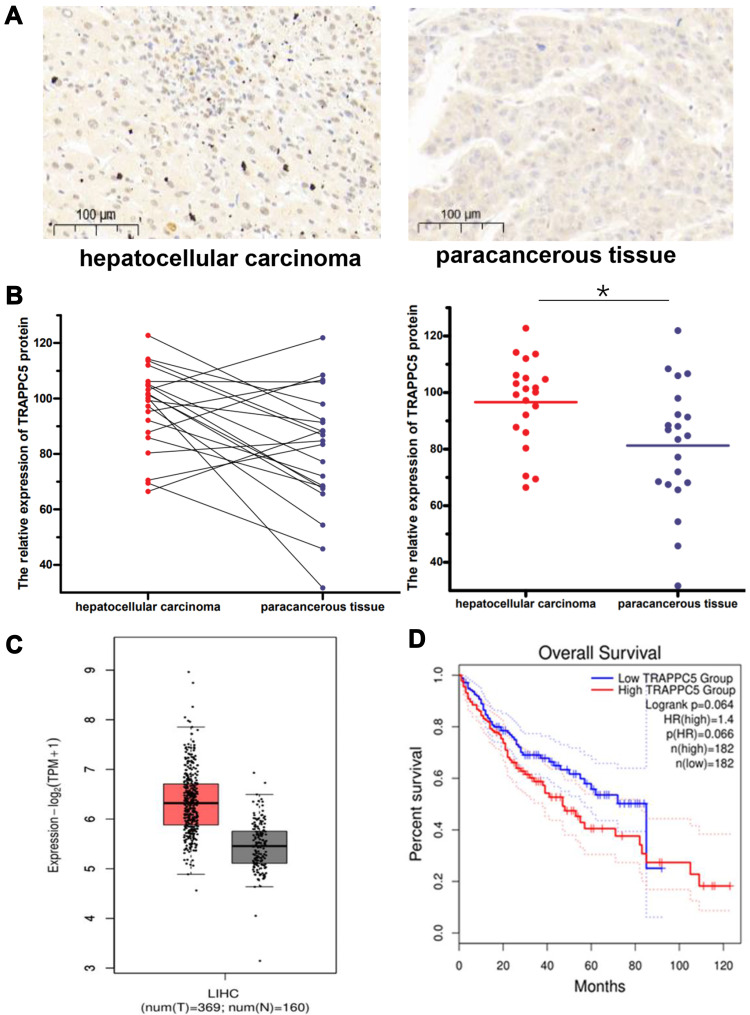
We detected TRAPPC5 protein expression by immunohistochemistry in commercial tissue microarrays which containing cancer tissues of patients with HCC and paracancerous tissues (n=21). TRAPPC5 protein was detected in cancer tissues in higher levels, whereas paracancerous tissues expressed much lower (Figure 2A and B). Furthermore, we used the gene expression profiling interactive analysis (GEPIA, http://gepia.cancer-pku.cn/index.html) to investigated the expression of TRAPPC5 in HCC tissues and patients at different stages, and eventually obtained a survival analysis. The database showed that TRAPPC5 is highly expressed in HCC tissues and has a poor prognosis (Figure 2C and D).These findings indicated expression of TRAPPC5 may contribute to the development of HCC.
Figure 2.
TRAPPC5 is overexpressed in HCC tissue microarrays and indicated poor prognosis. (A, B) TRAPPC5 is overexpressed in HCC tissue microarrays (n=21). TRAPPC5 protein was detected in cancer tissues in higher levels, whereas paracancerous tissues expressed much lower. Scale bars, 100 μm (magnification, 100×). (C) TRAPPC5 is highly expressed in HCC tissues (n=369) compared with that in normal tissues (n=160) using the GEPIA database. (D) Kaplan-Meier analysis of overall survival showed that patients with overexpression of TRAPPC5 have a poor prognosis (n=182) using the GEPIA database. *P < 0.05.
TRAPPC5 Knockdown Inhibited HCC Cell Proliferation, Migration, and Invasion
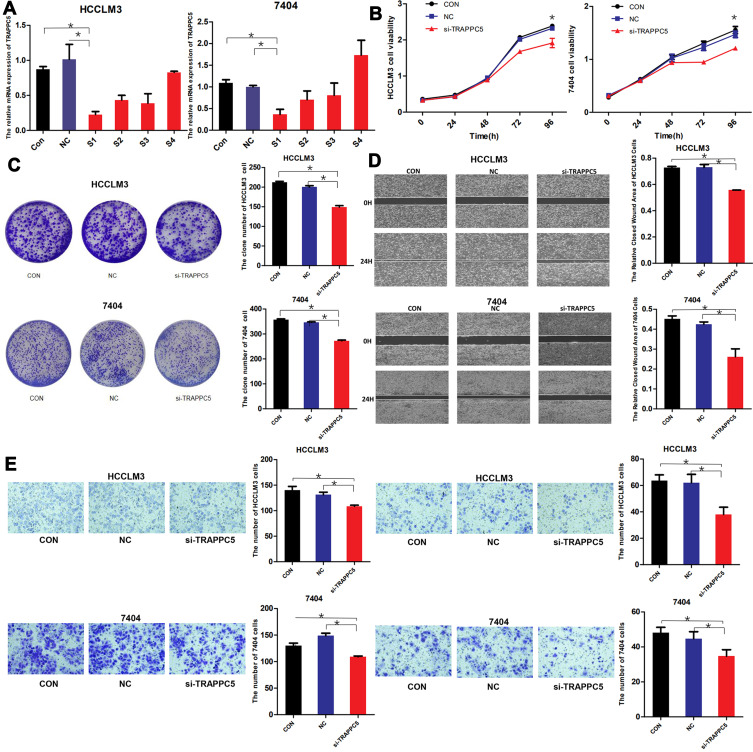
Knocking down of TRAPPC5 by siRNA was conducted to explore the role of TRAPPC5 in HCC cells. The down-regulation effect of four siRNAs (Table 2) were tested by qPCR and si-TRAPPC5-1 (S1) was selected for further research. After the transfection of si-TRAPPC5 (S1), the expression of TRAPPC5 was significantly declined in HCCLM3 and BEL-7404 cells (Figure 3A). TRAPPC5 downregulation significantly inhibited HCC cell proliferation as analyzed by CCK-8 assay and colony formation. (Figure 3B and C). TRAPPC5 downregulation also suppressed HCC cell invasion and migration by wound healing assay and transwell assay (Figure 3D and E). These data suggested that TRAPPC5 downregulation inhibited malignant behaviors of HCC in vitro.
Table 2.
The Sequence of siRNAs Used to Knockdown TRAPPC5
| Target | Sequence (5’-3’) |
|---|---|
| si-TRAPPC5-1 | GCACCACGCTCATGATCAA |
| si-TRAPPC5-2 | CGCGCACCTTCTACATCAT |
| si-TRAPPC5-3 | GCTCATCAACACCTACATC |
| si-TRAPPC5-4 | GCCGGAAGTTTGAGACCAA |
| si-NC | TTCTCCGAACGTGTCACGT |
Figure 3.
TRAPPC5 knockdown inhibited HCC cell proliferation, migration, and invasion. (A) The down-regulation effect of four siRNAs (S1-4) in two HCC cell lines (HCCLM3 and BEL-7404) were tested by qPCR and S1 was selected for further research. (B, C) TRAPPC5 downregulation significantly inhibited HCC cell proliferation as analyzed by CCK-8 assay and colony formation. (D, E) TRAPPC5 downregulation also suppressed HCC cell invasion and migration by wound healing assay and transwell assay. *P < 0.05.
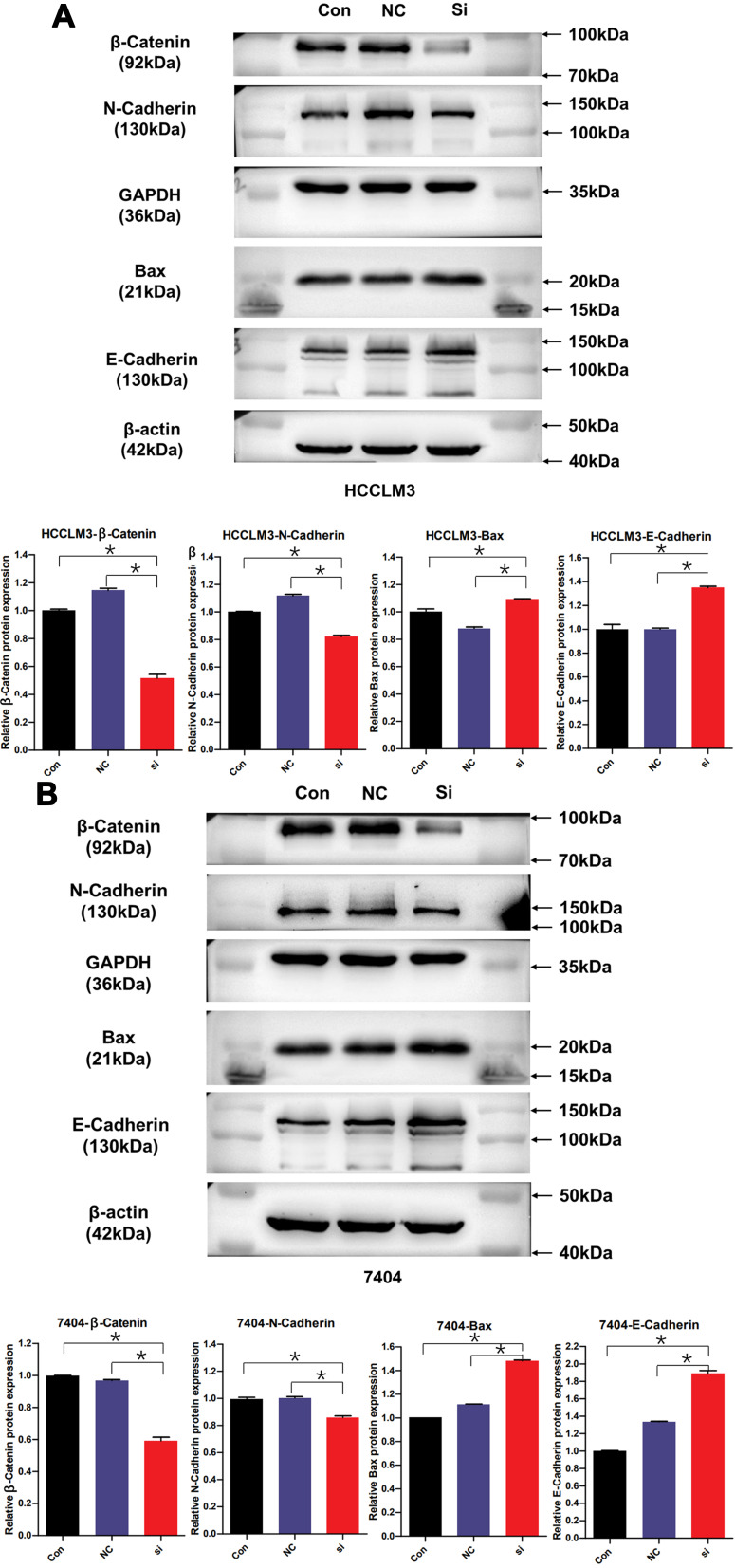
TRAPPC5 Knockdown Affected the Expression of Bax and EMT-Related Proteins in HCC Cell Lines
The Western blot analysis showed that in HCCLM3 and BEL-7404 cells, TRAPPC5 knockdown decreased the expression of Bax, EMT-related proteins N-cadherin and β-catenin while increasing the expression of E-cadherin. GAPDH and β-actin was used as a loading control (Figure 4A and B).
Figure 4.
TRAPPC5 knockdown affected the expression of Bax and EMT-related proteins in HCC cell lines. (A, B) The Western blot analysis of the protein levels of β-Catenin, Bax and EMT-related proteins (N-Cadherin and E-Cadherin) in HCCLM3 and BEL-7404 cells transfected with si-TRAPPC5 or controls. GAPDH and β-actin was used as a loading control. *P < 0.05.
TRAPPC5 Knockdown Repressed Tumor Growth in vivo
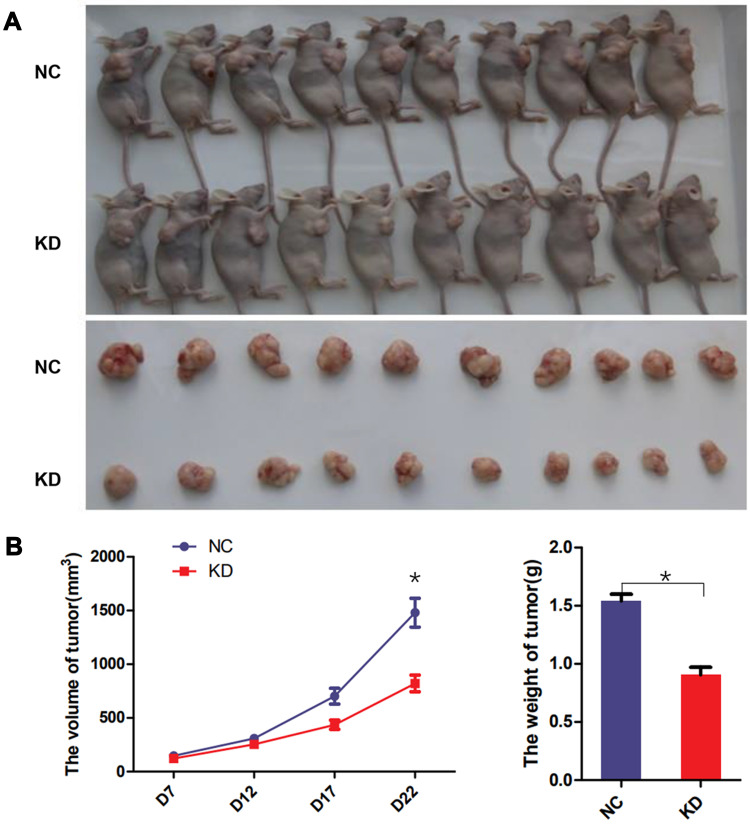
To investigate the effects of TRAPPC5 in vivo, HCCLM3 stable cells with TRAPPC5 knockdown and negative control were used for the tumorigenic test. The result showed the tumor growth rate of nude mice in the TRAPPC5 downregulated group was significantly slower than that in the control group. The tumor’s final size and weight of TRAPPC5 downregulated group were significantly smaller than in NC group (Figure 5A and B).
Figure 5.
TRAPPC5 knockdown repressed tumor growth in vivo. (A, B) Stable HCCLM3 cells (1 × 106 /mouse) were hypodermically inoculated into the right flanks of the mice. Tumor volume was estimated every week. Three weeks later, the mice sacrificed to collect tumor tissues and weighed. *P < 0.05.
Discussion
In our previous study, we have demonstrated that the expression profiles of MCT4 in HCC patients are closely correlated with AFP levels and tumor size. The amount of MCT4 expression can predict survival and TACE treatment response for HCC patients. The inhibition of MCT4 can induce inactivation of HIF-1α and inhibit phosphorylation of AKT. MCT4 may be a potential therapeutic target for the treatment of HCC.14 However, there is still no research to reveal its downstream molecules and mechanism of action by mRNA sequencing. In this study, we used siRNA to knockdown the expression of MCT4 in the HCC cell line HCCLM3, and further performed the mRNA sequencing, initially screened the molecules with significant differential expression. TRAPPC5 whose expression decreased most obviously was selected as the downstream gene of MCT4 for experiments. Tissue array immunohistochemistry of HCC specimens removed from the patient showed that the expression of TRAPPC5 in cancer was significantly higher than that in adjacent cancer tissues. Through in vivo and in vitro experiments, TRAPPC5 was discovered to accelerate HCC cell proliferation, migration, invasion, Epithelial–mesenchymal transition (EMT) process and tumorigenesis.
TRAPPC5 is a subunit of the transport protein particle (TRAPP) complex which is important for complex integrity and anterograde membrane transport from the endoplasmic reticulum (ER) to the ER-Golgi intermediate compartment. The collection of secretory cargo from the endoplasmic reticulum lumen initiates the transport of proteins and lipids between the endoplasmic reticulum and the Golgi apparatus. Following that, transport carriers bud from this membrane and are carried to, and then fuse with, the Golgi.17 Because of the Golgi’s ability to process and traffic a large portion of the proteome as well as regulate multiple processes in the cell such as mitosis, apoptosis, and migration, the dysregulation of ER–Golgi vesicular transport is associated with several pathologies such as Parkinson’s and Alzheimer’s disease,18 as well as cancer.19 Yoon et al showed that USO1 isoforms differentially promote liver cancer progression by dysregulating the ER–Golgi network. ERK and GRASP65 were found to be involved in the USO1-T-mediated Golgi dysfunction.20 In another study, Golgi compaction was found in cells that had completed an Epithelial to Mesenchymal transition (EMT), driven by the scaffolding protein PAQR11.21 Experimental evidence also indicates that ER and Golgi apparatus can activate both pro-survival (recovery) mechanisms as well as cell suicide programs if the stress-signaling threshold is exceeded.22 For cancer therapy, ER–Golgi vesicular transport has been identified as potential drug targets for therapeutic interventions.23 The successful targeting of the ER–Golgi pathway with brefeldin A (BFA) in fludarabine-resistant CLL cells had provided a novel approach to eradicating cancerous B-cells regardless of p53 status and pathological overexpression of Bcl-2, Bcl-XL, Mcl-1, and XIAP proteins.24 Aside from B-CLL cells, BFA has been shown to induce apoptosis in multiple myeloma, Jurkat, HeLa, leukemia, colon, prostate, and adenoid cystic sarcoma cells.25 TRAPP complexes, a key for transport through the Golgi, was predicted to be similarly affected. TRAPP subunit mutations have been linked to a variety of diseases in humans.26 Recent studies revealed that trafficking protein particle subunit 4 (TRAPPC4), another core subunit of TRAPP complex, promotes colorectal carcinogenesis by activating Wnt signaling and contributes to the aggressiveness of gastric cancer through controlling the transduction of extracellular signal-regulated protein kinase (ERK) signaling.27,28 Beside that a recent study revealed that TRAPPC4 can regulate the intracellular trafficking of PD-L1 and antitumor immunity by acting as a scaffold for PD-L1 and RAB11 to coordinate RAB11-mediated recycling of PD-L1 and protecting PD-L1 from lysosomal degradation, ultimately promoting immune evasion and tumor progression.29
According to previous research, MCT4 played a role in regulation of cargo export and sorting at the trans-Golgi network.30 For our study, MCT4 may dysregulating the ER–Golgi network by affecting the expression of TRAPPC5. And TRAPPC5 promoted liver cancer progression and associated with EMT. However, the apparent differentiation of human malignant tumor associated with different TRAPP subunits including TRAPPC5 remained unclear.
Our research is the first time to use RNA sequencing technology to screen MCT4 related molecules and describe the relationship between TRAPPC5 expression and HCC malignant behavior. TRAPPC5 may be a potential therapeutic target for the treatment of HCC.
Conclusion
In the process of MCT4 affecting the progression of HCC, TRAPPC5 is one of the most important related molecules. TRAPPC5 suppression could significantly reduce HCC cell proliferation, migration and invasion and could serve as a therapeutic target in HCC.
Acknowledgments
This work is supported by the Natural Science Foundation of China Youth Project (Grant No. 81802379) and Jinan Science and Technology Development Project (Grant No. 201805029) and Natural Science Foundation of Shandong Province, China (Grant No.ZR2020QH039).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Colombet M, Soerjomataram I, et al. Cancer statistics for the year 2020: an overview. Int J Cancer. 2021. doi: 10.1002/ijc.33588 [DOI] [PubMed] [Google Scholar]
- 3.Vogel A, Cervantes A, Chau I, et al. Hepatocellular carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2019;30:871–873. doi: 10.1093/annonc/mdy510 [DOI] [PubMed] [Google Scholar]
- 4.Sangro B, Sarobe P, Hervas-Stubbs S, Melero I. Advances in immunotherapy for hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2021;18:525–543. doi: 10.1038/s41575-021-00438-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nouri-Vaskeh M, Alizadeh L, Hajiasgharzadeh K, et al. The role of HSP90 molecular chaperones in hepatocellular carcinoma. J Cell Physiol. 2020;235:9110–9120. doi: 10.1002/jcp.29776 [DOI] [PubMed] [Google Scholar]
- 6.Han HH, Qiu YJ, Shi YY, et al. Glypican-3-targeted precision diagnosis of hepatocellular carcinoma on clinical sections with a supramolecular 2D imaging probe. Theranostics. 2018;8:3268–3274. doi: 10.7150/thno.24711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Felmlee MA, Jones RS, Rodriguez-Cruz V, Follman KE, Morris ME. Monocarboxylate Transporters (SLC16): function, Regulation, and Role in Health and Disease. Pharmacol Rev. 2020;72:466–485. doi: 10.1124/pr.119.018762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curry JM, Tuluc M, Whitaker-Menezes D, et al. Cancer metabolism, stemness and tumor recurrence: MCT1 and MCT4 are functional biomarkers of metabolic symbiosis in head and neck cancer. Cell Cycle. 2013;12:1371–1384. doi: 10.4161/cc.24092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pinheiro C, Granja S, Longatto-Filho A, et al. Metabolic reprogramming: a new relevant pathway in adult adrenocortical tumors. Oncotarget. 2015;6:44403–44421. doi: 10.18632/oncotarget.5623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Izumi H, Takahashi M, Uramoto H, et al. Monocarboxylate transporters 1 and 4 are involved in the invasion activity of human lung cancer cells. Cancer Sci. 2011;102:1007–1013. doi: 10.1111/j.1349-7006.2011.01908.x [DOI] [PubMed] [Google Scholar]
- 11.Kubelt C, Peters S, Ahmeti H, et al. Intratumoral Distribution of Lactate and the Monocarboxylate Transporters 1 and 4 in Human Glioblastoma Multiforme and Their Relationships to Tumor Progression-Associated Markers. Int J Mol Sci. 2020;21. doi: 10.3390/ijms21176254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lisanti MP, Sotgia F, Pestell RG, Howell A, Martinez-Outschoorn UE. Stromal glycolysis and MCT4 are hallmarks of DCIS progression to invasive breast cancer. Cell Cycle. 2013;12:2935–2936. doi: 10.4161/cc.26280 [DOI] [PubMed] [Google Scholar]
- 13.Gotanda Y, Akagi Y, Kawahara A, et al. Expression of monocarboxylate transporter (MCT)-4 in colorectal cancer and its role: MCT4 contributes to the growth of colorectal cancer with vascular endothelial growth factor. Anticancer Res. 2013;33:2941–2947. [PubMed] [Google Scholar]
- 14.Gao HJ, Zhao MC, Zhang YJ, et al. Monocarboxylate transporter 4 predicts poor prognosis in hepatocellular carcinoma and is associated with cell proliferation and migration. J Cancer Res Clin Oncol. 2015;141:1151–1162. doi: 10.1007/s00432-014-1888-8 [DOI] [PubMed] [Google Scholar]
- 15.Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodrigues IS, Pereira LMG, Lisboa J, et al. Author Correction: involvement of Hsp90 and cyclophilins in intoxication by AIP56, a metalloprotease toxin from Photobacterium damselae subsp. piscicida. Sci Rep. 2020;10:1266. doi: 10.1038/s41598-020-58152-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watson P, Stephens DJ. ER-to-Golgi transport: form and formation of vesicular and tubular carriers. Biochim Biophys Acta. 2005;1744:304–315. doi: 10.1016/j.bbamcr.2005.03.003 [DOI] [PubMed] [Google Scholar]
- 18.Martinez-Menarguez JA, Tomas M, Martinez-Martinez N, Martinez-Alonso E. Golgi Fragmentation in Neurodegenerative Diseases: is There a Common Cause? Cells. 2019;8:242. doi: 10.3390/cells8070748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howley BV, Howe PH. Metastasis-associated upregulation of ER-Golgi trafficking kinetics: regulation of cancer progression via the Golgi apparatus. Oncoscience. 2018;5:142–143. doi: 10.18632/oncoscience.426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoon S, Choi JH, Shah M, et al. USO1 isoforms differentially promote liver cancer progression by dysregulating the ER-Golgi network. Carcinogenesis. 2021;42:1208–1220. doi: 10.1093/carcin/bgab067 [DOI] [PubMed] [Google Scholar]
- 21.Tan X, Banerjee P, Guo HF, et al. Epithelial-to-mesenchymal transition drives a pro-metastatic Golgi compaction process through scaffolding protein PAQR11. J Clin Invest. 2017;127:117–131. doi: 10.1172/JCI88736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hicks SW, Machamer CE. Golgi structure in stress sensing and apoptosis. Biochim Biophys Acta. 2005;1744:406–414. doi: 10.1016/j.bbamcr.2005.03.002 [DOI] [PubMed] [Google Scholar]
- 23.Broker LE, Kruyt FA, Giaccone G. Cell death independent of caspases: a review. Clin Cancer Res. 2005;11:3155–3162. doi: 10.1158/1078-0432.CCR-04-2223 [DOI] [PubMed] [Google Scholar]
- 24.Carew JS, Nawrocki ST, Krupnik YV, et al. Targeting endoplasmic reticulum protein transport: a novel strategy to kill malignant B cells and overcome fludarabine resistance in CLL. Blood. 2006;107:222–231. doi: 10.1182/blood-2005-05-1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wlodkowic D, Skommer J, McGuinness D, Hillier C, Darzynkiewicz Z. ER-Golgi network–a future target for anti-cancer therapy. Leuk Res. 2009;33:1440–1447. doi: 10.1016/j.leukres.2009.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim JJ, Lipatova Z, Segev N. TRAPP Complexes in Secretion and Autophagy. Front Cell Dev Biol. 2016;4:20. doi: 10.3389/fcell.2016.00020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kong X, Qian J, Chen LS, et al. Synbindin in extracellular signal-regulated protein kinase spatial regulation and gastric cancer aggressiveness. J Natl Cancer Inst. 2013;105:1738–1749. doi: 10.1093/jnci/djt271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ai L, Ren Y, Li Y, et al. Synbindin deficiency inhibits colon carcinogenesis by attenuating Wnt cascade and balancing gut microbiome. Int J Cancer. 2019;145:206–220. doi: 10.1002/ijc.32074 [DOI] [PubMed] [Google Scholar]
- 29.Ren Y, Qian Y, Ai L, et al. TRAPPC4 regulates the intracellular trafficking of PD-L1 and antitumor immunity. Nat Commun. 2021;12:5405. doi: 10.1038/s41467-021-25662-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castorino JJ, Deborde S, Deora A, et al. Basolateral sorting signals regulating tissue-specific polarity of heteromeric monocarboxylate transporters in epithelia. Traffic. 2011;12:483–498. doi: 10.1111/j.1600-0854.2010.01155.x [DOI] [PMC free article] [PubMed] [Google Scholar]