Abstract
Objective
After deploying a clinical prediction model, subsequently collected data can be used to fine-tune its predictions and adapt to temporal shifts. Because model updating carries risks of over-updating/fitting, we study online methods with performance guarantees.
Materials and Methods
We introduce 2 procedures for continual recalibration or revision of an underlying prediction model: Bayesian logistic regression (BLR) and a Markov variant that explicitly models distribution shifts (MarBLR). We perform empirical evaluation via simulations and a real-world study predicting Chronic Obstructive Pulmonary Disease (COPD) risk. We derive “Type I and II” regret bounds, which guarantee the procedures are noninferior to a static model and competitive with an oracle logistic reviser in terms of the average loss.
Results
Both procedures consistently outperformed the static model and other online logistic revision methods. In simulations, the average estimated calibration index (aECI) of the original model was 0.828 (95%CI, 0.818–0.938). Online recalibration using BLR and MarBLR improved the aECI towards the ideal value of zero, attaining 0.265 (95%CI, 0.230–0.300) and 0.241 (95%CI, 0.216–0.266), respectively. When performing more extensive logistic model revisions, BLR and MarBLR increased the average area under the receiver-operating characteristic curve (aAUC) from 0.767 (95%CI, 0.765–0.769) to 0.800 (95%CI, 0.798–0.802) and 0.799 (95%CI, 0.797–0.801), respectively, in stationary settings and protected against substantial model decay. In the COPD study, BLR and MarBLR dynamically combined the original model with a continually refitted gradient boosted tree to achieve aAUCs of 0.924 (95%CI, 0.913–0.935) and 0.925 (95%CI, 0.914–0.935), compared to the static model’s aAUC of 0.904 (95%CI, 0.892–0.916).
Discussion
Despite its simplicity, BLR is highly competitive with MarBLR. MarBLR outperforms BLR when its prior better reflects the data.
Conclusions
BLR and MarBLR can improve the transportability of clinical prediction models and maintain their performance over time.
Keywords: model recalibration, machine learning, clinical prediction models, Bayesian model updating
BACKGROUND
A growing number of prediction models have been validated and approved as clinical decision support systems and medical diagnostic devices.1 Models that have been successfully deployed need to be regularly monitored and updated over time, because locked algorithms are known to decay in performance due to changes in clinical practice patterns, patient case mix, measurement procedures, and more.2–4
With the expansion of electronic health record systems, we have a unique opportunity to embed models that continuously learn and evolve by analyzing streaming medical data, which are often referred to as online learning or continual learning systems.5–8 Online learning systems not only have the potential to protect against the consequences of distributional shifts over time, but they may also improve prediction performance, for example, by increasing precision of their estimates or personalizing predictions to local medical practices.9,10 Nevertheless, there are major technical challenges in developing reliable online learning systems,8 so establishing their safety and effectiveness is of utmost importance.11
In this study, we focus our attention on online model revision for risk prediction models, in which data are revealed in a sequential manner and the goal of the online model is to dynamically predict the probability of having or developing a disease (or outcome) given forecasted scores from an underlying model. We build on the common practice of using logistic regression to recalibrate and/or revise forecasted scores from an underlying prediction model on an initial dataset2 and extend it to the online setting with streaming labelled data (Figure 1). In the simplest case, the revisions only depend on the forecasted score, which is also known as online model recalibration. In more complex situations, the updated prediction can depend on both the forecasted score and other patient variables. We are particularly interested in procedures that can safely update “black-box” models such as gradient boosted trees and neural networks, which have achieved unprecedented success by capturing nonlinearities and interactions in the data. Simply refitting black-box models on accumulating data may carry risks because the refitted version is not guaranteed to outperform the original model.12 However, we can analyze the theoretical properties of logistic model revision, even for an underlying black-box prediction model.
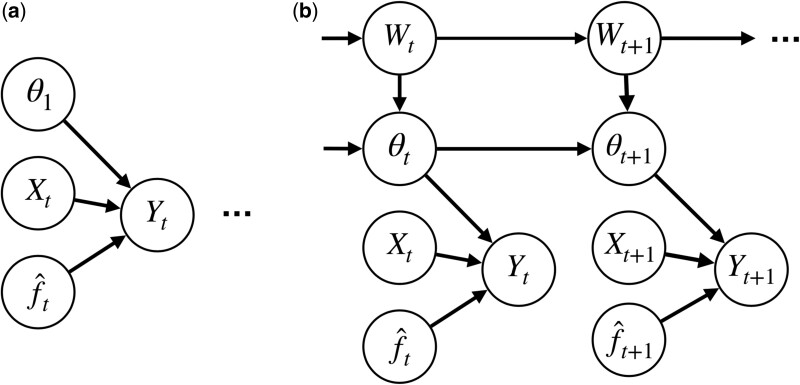
Figure 1.
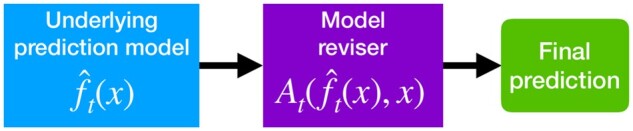
Given a patient with variables, the model reviser wraps around an underlying machine learning model to predict the true probability of having or developing a disease (or outcome). The focus of this work is the design of an online model reviser.
Methods for continually updating clinical prediction models have traditionally relied on dynamic Bayesian models13,14 or online hypothesis testing.15 However, these methods do not provide theoretical guarantees under model misspecification or distributional shifts. More recently, game-theoretic online learning methods have been applied to the problem of online model recalibration, which provide performance guarantees that bound its cumulative loss relative to some oracle procedure (also known as “regret”). Kuleshov et al16 use a nonparametric binning technique, but this method converges slowly in practice and cannot be used to revise a model with respect to patient variables. Davis et al17 apply Adam to the related problem of estimating dynamic calibration curves18; however, recent theoretical results show that the optimal regret bound for online logistic regression is, in fact, achieved by Bayesian model updating.19,20
OBJECTIVE
In this work, we develop online revision methods for “black-box” models that may be locked or evolving over time and provide theoretical guarantees without making any assumptions about the data distribution or the quality of the underlying model. We investigate online model revision using Bayesian logistic regression (BLR) and BLR with a Markov prior that explicitly models distribution shifts (MarBLR). To quantify the safety and effectiveness of the proposed online model revisers, we introduce the notions of Type I and II regret. We derive regret bounds for BLR and MarBLR, which provides a recipe for selecting a Bayesian prior that satisfies desired performance guarantees. In simulation studies, we evaluate BLR and MarBLR for online model recalibration and revision and as wrappers for black-box refitting procedures. We then apply the online updating procedures to Chronic Obstructive Pulmonary Disease (COPD) risk prediction in a retrospective dataset from 2012 to 2020. Code is available at http://github.com/jjfeng/bayesian_model_revision.
MATERIALS AND METHODS
Because the nature of future distribution shifts is typically unknown, we study the safety and effectiveness of an online model reviser in the presence of arbitrary distribution shifts, following the game-theoretic online learning literature. This general framework allows us to study, for example, distribution shifts induced by the deployment of the machine learning (ML) model itself, which has been raised as a concern in a number of recent works.21,22
A framework for evaluating online model revision algorithms
Denote patient variables with and binary outcomes with . Suppose the streaming data are received at discrete times . At time , we observe a new observation . Let denote the underlying prediction model. Let be the model revision that outputs a probability. The initial clinical prediction model is defined by the composition . If is well-calibrated, we may simply define to be the identity function; otherwise, should be estimated on an initial recalibration dataset.2 Let be any sequence of times in which the model revision is updated. In certain cases, one may observe a batch of observations at each time point instead. We discuss how the theoretical framework and results need to be adjusted to handle batched data in the Supplementary Appendix.
The online learning procedure can be described as follows. For time steps :
Patient is revealed. The online reviser deploys modification for model and releases a prediction for the patient.
We observe the respective outcome
The evolving model selects The online reviser selects . The next observation is acquired, but not revealed to the model yet.
In this setup, we do not make any assumptions about how the data are generated or the reliability of the underlying model.
For the theoretical analyses, we quantify the performance of the online model reviser by its average over the entire time period, that is, where denotes the negative log likelihood for the outcome and predicted probability . This is a direct extension of offline logistic regression—which is usually fit using maximum likelihood estimation—to the online setting.
For an online model reviser to be safe, it should be, at the very least, noninferior to locking the original model. Drawing analogy to the hypothesis testing literature, locking the model can be viewed as the “null” hypothesis and using the online model reviser as the “alternative.” Type I error is then the incorrect rejection of the null. Combining this with the notion of regret from the online learning literature, we define Type I regret as the average increase in the loss when using the online reviser instead of the original model, that is
Type I regret spans the nonnegative real values, where a smaller value is better. A safe online model reviser should control its value below some pre-specified noninferiority margin . Over-zealous model updating (also known as over-updating2) tends to inflate Type I regret. Nevertheless, it is not enough to solely control Type I regret because locking the original model perfectly controls Type I regret without offering any protection against distribution shifts.
In addition, we quantify the effectiveness of an online model reviser by comparing its performance to that achieved by the best sequence of model revisions in retrospect. More specifically, if one had access to observations for times , there is some oracle sequence of model revisions , restricted to update times , that minimizes the average loss. We refer to an oracle as static when is the empty set (ie the model revision sequence is the optimal constant sequence) and dynamic otherwise. We define Type II -regret as the average performance difference between the online reviser and this dynamic oracle, that is
Type II regret is large when we fail to update the model fast enough. It is especially large when we make a Type II error—when we fail to reject the “null” hypothesis—and do not update the model at all. Similar to Type I regret, Type II regret spans the nonnegative real values where small values are better.
Our aim is to design online model revisers that minimize Type II regret while controlling Type I regret, regardless of how the data distribution and underlying prediction model change over time. There is a trade-off between Type I and II regret, since increasing the frequency and magnitude of the model revision updates typically increase Type I regret but decrease Type II regret.
Bayesian logistic revision and a Markov variant
BLR and MarBLR perform inference for logistic model revisers of the form
where is some basis expansion of the score from the underlying model and patient variables. Let denote the logistic revision parameters at time . After receiving a new labeled observation, we update the posterior for the model revision parameters according to Bayes’ theorem. We predict the probability that for a patient variables using the posterior mean of .
BLR defines a Gaussian prior over the model revision parameters and assumes the parameters are fixed over the entire time period, that is, the true do not have time dependence. Because BLR is an over-simplification of the data, we also consider a generalization of BLR called MarBLR that allows for updates to the revision parameters over time (Figure 2). In particular, MarBLR supposes the revision parameters need updating at each time point with a prior probability of and evolves according to the Gaussian random walk , where are independent random vectors and is a binary random variable with success probability . Thus, is the prior probability for how frequently the model revision needs to be updated and is our prior regarding the magnitude of these updates. In the theoretical analyses, we analyze MarBLR with . In practice, we choose where is the posterior covariance matrix of the at time . MarBLR reduces to BLR when or .
Figure 2.
BLR and its Markov variant MarBLR update the deployed model revision at each time point per the evolving Bayesian posterior. Theoretical guarantees for BLR and MarBLR hold under misspecification of the logistic model and/or priors. A, Bayesian logistic revision (BLR) estimates the model revision parameters for the underlying prediction model with the simplifying assumption that the data are independently and identically distributed for a constant set of model revision parameters over all time points . B, MarBLR defines a prior over revision parameter sequences that change over time. It assumes that the revision parameters change with probability at each time , as modeled by a binary random variable . It supposes that changes in the model revision parameters follow a Gaussian prior centered at zero.
We derive Type I and II regret bounds for BLR and MarBLR by extending,19 which only compared BLR to a static oracle. We generalize the results to handle dynamic oracles and the more general MarBLR procedure. The Type I regret bounds hold as long as BLR and MarBLR are able to revert to the original model. Thus, one should always choose the basis expansion to include scores from both the original and evolving models and , respectively.
Bayesian inference for MarBLR requires marginalizing over possible update times. Enumerating all possible shift times is computationally intractable and because the posterior does not have a closed form, we use instead a Laplace approximation of the logistic posterior23 and perform Kalman filtering with collapsing13,24 (see Section A in the Supplementary Appendix).
Empirical analyses
Simulation settings
We assess the performance of BLR and MarBLR in 3 simulation studies with increasingly complex model revisions. We generate 10-dimensional patient variables using a multivariate normal distribution and binary outcomes using a logistic model. We introduce distribution shifts by perturbing coefficients of this model. The underlying prediction model is a gradient-boosted tree (GBT). To ensure that the initial prediction model is well-calibrated, we fit on 100 observations held out from the original training data. Thereafter, we observe a single subject at each time point and run the procedure for time steps. For each simulated condition, we perform 50 replicates to estimate standard errors. (Additional simulation details are in the Supplementary Appendix.)
Scenario 1 is on online recalibration of a locked underlying model in the presence of temporal shifts as well as recalibration across patient subgroups to see if these methods can improve algorithmic fairness. Our motivation is based on recent works that highlight disparities in the performance of ML models when patient populations are heterogeneous and unbalanced.25 We define patient subgroups A and B with 20% and 80% prevalence, respectively. We simulate an initial distribution shift in each subgroup and introduce 1 and 2 subsequent shifts in subgroups A and B, respectively. We fit a univariate logistic recalibration that ignores subgroup status as well as a subgroup-aware recalibration using interaction terms between the forecasted score and subgroup status.
Scenario 2 is on learning logistic model revisions where the inputs are the forecasted score from a locked underlying model and 10 patient variables, that is
where is the intercept, is the coefficient for the locked model, and is a vector of coefficients associated with the patient variables. We consider 3 types of data distributions: independent and identically distributed (IID) data after an initial distribution shift (Initial Shift), cyclical variation (Cyclical), and repeated dataset shifts that lead to gradual performance decay of the original model (Decay). The initial GBT is trained on 300 observations.
Scenario 3 performs online ensembling of the original model and a black-box refitting procedure . The logistic revision is of the form
where and are the coefficients for the locked and evolving models, respectively. We use the Initial Shift and Decay data settings from scenario 2 and the same initial model. We simulate a reliable black-box refitting procedure by refitting on all available data (All-Refit). To test if the online model reviser can protect against black-box refitting procedures with unreliable performance, we simulate an evolving model that refits on the most recent 75 observations up to time and then suddenly refits using only the most recent 30 observations thereafter(Subset-Refit). While this refitting procedure is unlikely to be used in practice, it lets us simulate how the online model reviser might respond to sudden deterioration in the evolving model.
Performance metrics
For baseline comparison, we lock the original model, perform online logistic revision using Adam, and cumulative logistic regression (CumulativeLR) as suggested in.19 Briefly, Adam performs a gradient-based update to the current logistic revision parameters and CumulativeLR refits logistic revision parameters by minimizing with respect to all prior observations. BLR is similar to CumulativeLR in nature but appropriately integrates over uncertainty using a Bayesian framework, and MarBLR additionally models distribution shifts. We evaluate the methods in terms of the expected negative log likelihood (NLL), estimated calibration index (ECI),26 and area under the receiver-operating characteristic curve (AUC), when appropriate. Note that a model is better if it has smaller NLL and ECI and bigger AUC, where the minimum ECI is zero and the maximum AUC is 1. We use aNLL, aECI, and aAUC to denote their average value over the time period.
Hyperparameter selection
We select the Gaussian prior for BLR and MarBLR such that its mean is the estimated logistic revision parameters on the initial recalibration dataset and its covariance is a scaled version of the standard error matrix to achieve the desired Type I regret control. In the third simulation, we must construct an initial recalibration dataset such that the fixed and evolving models are not exactly the same. We do this by representing the evolving model with a model that was trained on 90% of the original training data. The Gaussian prior at is then centered at the estimated revision parameters with the constraint that the coefficient for the evolving model is zero and the prior covariance is a scaled version of the Hessian matrix.
The priors for BLR and MarBLR are selected such that their Type I regret is no more than 5% of the initial loss of the original locked model in the first and third scenarios. Because the second scenario considers higher dimensional model revisions and Type I regret scales with the dimensionality of the problem (see Theoretical Results in the following section), we use a looser bound of 10% in this setting. Because the regret bounds for Adam and CumulativeLR are too wide to be meaningful, these methods are run without any Type I regret control.
COPD dataset
We analyzed 108 002 in-patient admissions to UCSF Health from June 15, 2012 to December 1, 2020, of which 2756 admissions resulted in a primary or secondary diagnosis of COPD (based on ICD-9 and ICD-10 codes). We ordered observations by their admission dates. There are a total of 36 predictors available, including age, history of smoking, history of COPD, active outpatient medications prior to emergency department (ED) presentation, and medications administered in the ED prior to the point of admission. The initial model is fit using a gradient boosted tree on the first 2500 observations, of which 92 are positive cases. For initial model recalibration, we use the 1000 immediately following observations, of which 52 are positive cases.
We apply BLR and MarBLR for online model recalibration, logistic model revision, and online ensembling, using the same procedures described for the simulations. Logistic model revision was restricted to 3 predictors based on clinical knowledge: age, history of COPD, and history of smoking. In the online ensembling experiment, we refit the gradient boosted tree on all prior data every 270 observations (across 400 time points). We selected the hyperparameters for BLR and MarBLR such that the regret bound was no more than 5% of the initially estimated loss for the locked model. For computational speed, we run BLR and MarBLR on batches of observations.
We split the data into 4 time periods with an equal number of observations. We evaluate the deployed models using walk-forward testing, that is, the forecasted probability was compared to the observed outcome at each time point, and estimated the AUC, ECI, and NLL for each time period. We average these performance metrics across the time periods to calculate aAUC, aECI, and aNLL. Confidence intervals are constructed using 200 bootstrap replicates.
Computation
All empirical studies were performed on an Intel Gold 6240 CPU. The computation time for each experiment was no more than thirty minutes, except for the online ensembling procedure for the COPD data analysis. This particular experiment took around 3 h, where the bulk of the time was spent on refitting a GBT each time new data was collected.
RESULTS
Theoretical results
We derived Type I and II regret bounds for BLR and MarBLR, which give us the theoretical guarantees regarding the safety and effectiveness of the 2 procedures in the presence of distribution shifts. The regret bounds provide guidance for choosing between BLR and MarBLR as well as their hyperparameters. The results are finite-sample, do not assume that the Bayesian modeling assumptions are correct, and hold even if the data are adversarially chosen. A summary of the regret bounds is shown in Table 1. Theorems and proofs are provided in the Supplementary Appendix. Below, we highlight the trade-off between Type I and II regret as we vary hyperparameters in the 2 procedures.
Table 1.
Overview of theoretical results.
| Type I regret bound | Type II -regret bound | |
| BLR | ||
| MarBLR |
Note: The regret bounds are displayed in asymptotic notation. is the best locked model revision sequence in retrospect and is the best dynamic model revision sequence with revision times in retrospect. Symbol meanings: =dimension of logistic revision parameters, =total number of time steps, =initial logistic parameters, =inflation factor for MarBLR posterior, =update probability in MarBLR, =MarBLR prior over update times.
Type I regret for BLR converges at the rate of , where d denotes the dimension of the logistic revision parameter, and T denotes the total number of time steps. This is best currently known rate for online logistic regression.20 Because this regret bound converges quickly to zero as T increases, it can be used to meaningfully control Type I regret on realistic time horizons. Although not shown in Table 1, the Type I regret for BLR decreases to zero as we shrink the prior covariance matrix and concentrate the prior around locking the original model (and its revision).
In comparison, the Type I regret bound for MarBLR includes an additional error term of that corresponds to the prior probability of revision sequences that change over time. As we increase and , MarBLR prior puts less weight on the original model and searches over more dynamic revision sequences. Because this additional error term does not go to zero as increases, so one needs to choose the MarBLR hyperparameters with care. This illustrates how Type I regret increases when we try to estimate more complex model revision sequences using MarBLR.
Type II regret for MarBLR is plus the distance between the dynamic oracle and the prior. If the true sequence of revision update times is known, selecting in MarBLR minimizes the bound on the average expected loss. On the other hand, BLR assumes the oracle model revision sequence is static and sets . Consequently, its Type II regret bound incurs an additional term that quantifies the error in approximating the dynamic oracle with a locked oracle .
To summarize, BLR has smaller Type I but larger Type II regret bounds than MarBLR for the same Gaussian prior at time . We can further fine-tune the Type I regret control achieved by the 2 methods by selecting more or less diffuse priors.
Simulation studies
Scenario 1: online recalibration of a locked underlying model
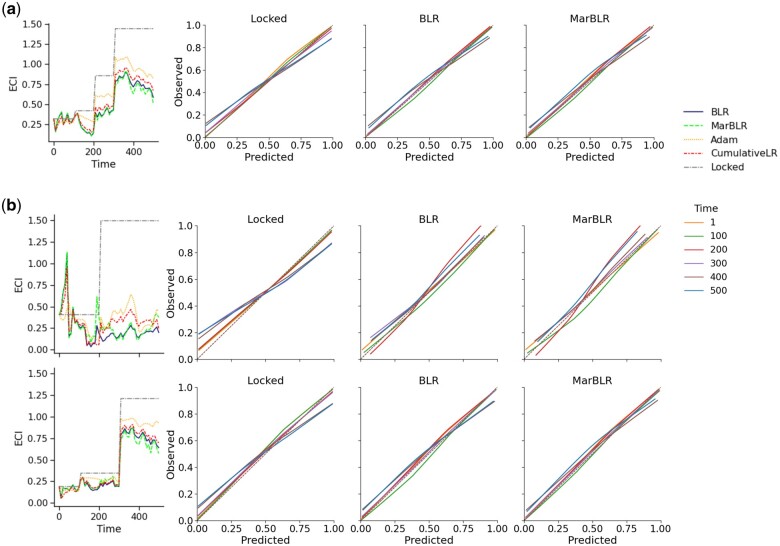
By design, the online recalibration methods aim to minimize the aNLL with respect to the general population. Indeed, we find that all the online recalibration methods significantly improved aNLL and aECI compared to the locked model, with BLR and MarBLR achieving the smallest values (Table 2, Figure 3). Online recalibration also improved aNLL and aECI within each subpopulation, but different methods achieved different levels of calibration of across the subgroups. In general, we find that that subgroup-aware recalibration leads to more similar aECI between the subpopulations than univariate recalibration. For instance, MarBLR achieved aECIs of 0.590 (95% CI, 0.543–0.637) and 0.176 (95% CI, 0.158–0.193) in subgroups A and B, respectively, when performing univariate recalibration. In contrast, the aECIs for subgroups A and B are 0.498 (95% CI, 0.429–0.566) and 0.233 (95% CI, 0.208–0.258), respectively, when performing subgroup-aware recalibration. Finally, we note that there is a spike in the ECI at early time points but quickly disappears as data accumulates.
Table 2.
Average performance of online logistic recalibration methods of a fixed underlying prediction model and simulated patients subgroups A and B with prevalence 20% and 80%, respectively (scenario 1).
| Average ECI (aECI) |
Average NLL (aNLL) |
|||||
|---|---|---|---|---|---|---|
| Subgroup | A | B | Combined | A | B | Combined |
| Locked | 1.371 (0.009) | 0.612 (0.005) | 0.910 (0.005) | 0.661 (0.001) | 0.559 (0.000) | 0.580 (0.000) |
| Online univariate logistic recalibration | ||||||
| MarBLR | 0.590 (0.024) | 0.176 (0.009) | 0.301 (0.016) | 0.622 (0.001) | 0.538 (0.000) | 0.555 (0.001) |
| BLR | 0.655 (0.027) | 0.199 (0.012) | 0.346 (0.019) | 0.624 (0.001) | 0.539 (0.001) | 0.556 (0.001) |
| Adam | 0.878 (0.018) | 0.273 (0.012) | 0.494 (0.014) | 0.635 (0.001) | 0.543 (0.001) | 0.561 (0.001) |
| CumulativeLR | 0.745 (0.025) | 0.225 (0.013) | 0.403 (0.019) | 0.629 (0.001) | 0.541 (0.001) | 0.558 (0.001) |
| Online subgroup-aware logistic recalibration | ||||||
| MarBLR | 0.498 (0.035) | 0.233 (0.013) | 0.241 (0.013) | 0.616 (0.001) | 0.541 (0.001) | 0.556 (0.001) |
| BLR | 0.465 (0.037) | 0.270 (0.016) | 0.265 (0.018) | 0.615 (0.002) | 0.542 (0.001) | 0.557 (0.001) |
| Adam | 0.439 (0.025) | 0.426 (0.011) | 0.320 (0.016) | 0.614 (0.001) | 0.550 (0.001) | 0.562 (0.001) |
| CumulativeLR | 0.449 (0.040) | 0.303 (0.017) | 0.311 (0.020) | 0.616 (0.002) | 0.544 (0.001) | 0.558 (0.001) |
Note: Methods include Bayesian logistic revision (BLR) and its Markov variant (MarBLR), Adam, cumulative refitting of a logistic regression model (CumulativeLR), and locking the original model (Locked). Standard errors over 50 replicates are shown in parentheses.
Abbreviations: aECI: average estimated calibration index; aNLL: average negative log likelihood.
Figure 3.
Results from online model recalibration of a fixed underlying prediction model in a patient population with patient subgroups A and B with prevalence 20% and 80% (scenario 1). Left: Estimated calibration index (ECI) at each time point. Right: Calibration curves for the original model and the revised versions from BLR and MarBLR. The ideal calibration curve is the identity function, which has an ECI of zero. A, Univariate recalibration; calibration measured with respect to the general population. B, Subgroup-aware recalibration; calibration measured with respect to subgroups A (top) and B (bottom).
Scenario 2: online logistic revision of a locked underlying model
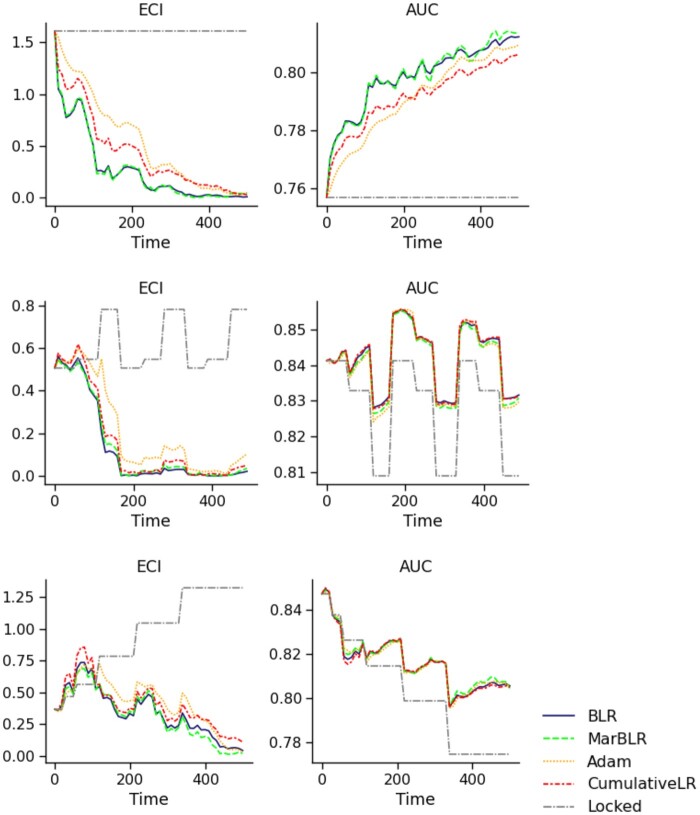
BLR and MarBLR learned beneficial logistic revisions faster than the other online methods across all data settings (Table 3, Figure 4), improving in both model discrimination and calibration over the locked model. BLR and MarBLR significantly improved model discrimination in the setting with IID data after an initial shift. We also observed improvements in model discrimination when there were cyclical distribution shifts, but to a lesser extent. In the Decay data setting, the locked model had an initial AUC of 0.85 and achieved an average AUC of 0.803 (95% CI, 0.801–0.805). In contrast, BLR and MarBLR slowed down the performance decay, achieving an aAUC of 0.819 (95% CI, 0.817–0.821).
Table 3.
Performance of online logistic revision of fixed underlying prediction model with respect to the forecasted score and patient variables (scenario 2).
| aAUC | aECI | aNLL | |
|---|---|---|---|
| Initial Shift | |||
| MarBLR | 0.799 (0.001) | 0.288 (0.012) | 0.554 (0.001) |
| BLR | 0.800 (0.001) | 0.278 (0.013) | 0.553 (0.001) |
| Adam | 0.795 (0.001) | 0.576 (0.014) | 0.574 (0.001) |
| CumulativeLR | 0.797 (0.001) | 0.417 (0.017) | 0.563 (0.001) |
| Locked | 0.767 (0.001) | 1.637 (0.008) | 0.661 (0.001) |
| Cyclical | |||
| MarBLR | 0.834 (0.001) | 0.211 (0.011) | 0.510 (0.001) |
| BLR | 0.834 (0.001) | 0.202 (0.012) | 0.509 (0.001) |
| Adam | 0.834 (0.001) | 0.293 (0.011) | 0.514 (0.001) |
| CumulativeLR | 0.834 (0.001) | 0.240 (0.013) | 0.511 (0.001) |
| Locked | 0.826 (0.001) | 0.605 (0.002) | 0.542 (0.000) |
| Decay | |||
| MarBLR | 0.819 (0.001) | 0.281 (0.015) | 0.532 (0.001) |
| BLR | 0.819 (0.001) | 0.277 (0.017) | 0.532 (0.001) |
| Adam | 0.820 (0.001) | 0.379 (0.015) | 0.537 (0.001) |
| CumulativeLR | 0.819 (0.001) | 0.337 (0.018) | 0.535 (0.001) |
| Locked | 0.803 (0.001) | 0.979 (0.002) | 0.588 (0.000) |
Note: Simulated data settings include IID data after an initial shift (Initial Shift), nonstationary data that cycles between 3 distributions (Cyclical), nonstationary data where the performance of the original model decays over time (Decay). Methods include Bayesian logistic revision (BLR) and its Markov variant MarBLR, Adam, cumulative refitting of a logistic regression model (CumulativeLR) and locking the original model (Locked). Standard errors over 50 replicates are shown in parentheses
Abbreviations: aAUC: area under the receiver operating characteristic curve; aECI: average estimated calibration index, aNLL: average negative log likelihood.
Figure 4.
Results from online logistic revision of a fixed underlying model with respect to the forecasted score and ten patient variables (scenario 2), in terms of the estimated calibration index (ECI, left) and AUC (right). Data are simulated to be stationary over time after an initial shift (Initial Shift, top), shift in a cyclical fashion (Cyclical, middle), and shift such that the original model decays in performance over time (Decay, bottom). All online logistic revision methods outperformed locking the original model in terms of the average ECI and AUC, with BLR and MarBLR performing the best. Note that the revised models were worse than the original model briefly in the Decay setting.
Scenario 3: online ensembling of a locked and continuously refitted black-box model
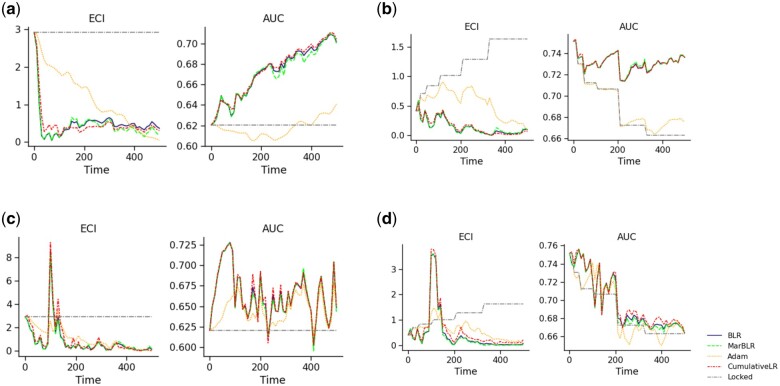
CumulativeLR, BLR, and MarBLR achieved the top performance across the different model refitting procedures and data distributions (Table 4, Figure 5). In All-Refit, the evolving model had better performance than the original model so the online revisers learned to place more weight on the evolving model, thereby improving model calibration and discrimination. In Subset-Refit, we simulated an unreliable evolving black-box model to test how the online revisers respond to sudden model deterioration. All the online model revisers drop in performance when the evolving model suddenly deteriorates, but they recover over time, some faster than others.
Table 4.
Performance of online logistic revision as a wrapper for a continually refitted gradient boosted tree (scenario 3).
| aAUC | aECI | aNLL | |
|---|---|---|---|
| Initial Shift, All-Refit | |||
| MarBLR | 0.689 (0.002) | 0.452 (0.024) | 0.646 (0.001) |
| BLR | 0.689 (0.002) | 0.482 (0.027) | 0.647 (0.001) |
| Adam | 0.621 (0.001) | 1.085 (0.029) | 0.702 (0.001) |
| CumulativeLR | 0.690 (0.002) | 0.448 (0.021) | 0.646 (0.001) |
| Locked | 0.624 (0.001) | 2.695 (0.017) | 0.762 (0.001) |
| Initial Shift, Subset-Refit | |||
| MarBLR | 0.658 (0.001) | 0.707 (0.033) | 0.671 (0.001) |
| BLR | 0.658 (0.001) | 0.696 (0.035) | 0.671 (0.001) |
| Adam | 0.643 (0.001) | 1.128 (0.035) | 0.695 (0.001) |
| CumulativeLR | 0.659 (0.001) | 0.787 (0.040) | 0.675 (0.001) |
| Locked | 0.624 (0.001) | 2.695 (0.017) | 0.762 (0.001) |
| Decay, All-Refit | |||
| MarBLR | 0.726 (0.001) | 0.238 (0.014) | 0.617 (0.001) |
| BLR | 0.725 (0.001) | 0.224 (0.014) | 0.617 (0.001) |
| Adam | 0.689 (0.001) | 0.514 (0.019) | 0.652 (0.001) |
| CumulativeLR | 0.727 (0.001) | 0.211 (0.014) | 0.616 (0.001) |
| Locked | 0.688 (0.001) | 1.218 (0.007) | 0.679 (0.000) |
| Decay, Subset-Refit | |||
| MarBLR | 0.703 (0.001) | 0.456 (0.029) | 0.639 (0.001) |
| BLR | 0.704 (0.001) | 0.450 (0.031) | 0.639 (0.001) |
| Adam | 0.698 (0.001) | 0.647 (0.026) | 0.650 (0.001) |
| CumulativeLR | 0.705 (0.001) | 0.533 (0.034) | 0.642 (0.001) |
| Locked | 0.688 (0.001) | 1.218 (0.007) | 0.679 (0.000) |
Note: Refitting procedures include refitting on all available data (All-Refit) and refitting on the most recent window of data (Subset-Refit). Methods include Bayesian logistic revision (BLR) and its Markov variant MarBLR, Adam, cumulative refitting of a logistic regression model (CumulativeLR), and locking the original model (Locked). Standard errors over 50 replicates are shown in parentheses.
Abbreviations: aAUC: area under the receiver operating characteristic curve; aECI: average estimated calibration index; aNLL: average negative log likelihood.
Figure 5.
Model calibration and discrimination (left and right panels, respectively) from online ensembling of the original model with an underlying prediction model that is continually refitted over time (scenario 3). Data are simulated to be stationary over time after an initial shift (Initial Shift) and nonstationary such that the original model decays in performance over time (Decay). Underlying prediction model is updated by continually refitting on all previous data (All-Refit) or refit on the most recent subset of data (Subset-Refit). Note that Subset-Refit simulates a sudden drop in performance for the continually refitted model at time t = 100 and, consequently, across all online ensembling procedures. BLR and MarBLR recover from this sudden performance decay and achieve better performance than locking the original model in terms of the average ECI and AUC. A, Initial Shift, All-Refit. B, Decay, All-Refit. C, Initial Shift, Subset-Refit. D, Decay, Subset-Refit.
We can gain more insight into the operating characteristics of BLR and MarBLR by visualizing how the logistic revision coefficients change over time (Supplementary Appendix Figure A.1). For All-Refit, the 2 methods gradually increased the importance of the evolving model and decreased the importance of the locked model. By the end of the time period, both methods assigned higher importance to the evolving model. As expected, this switch in model importance occurs earlier in the nonstationary setting. For Subset-Refit, BLR and MarBLR increase the coefficient of the evolving model during the time period when it was a good predictor of the outcome (t < 100). Once the evolving model decayed in prediction accuracy, its coefficient starts to decrease towards zero.
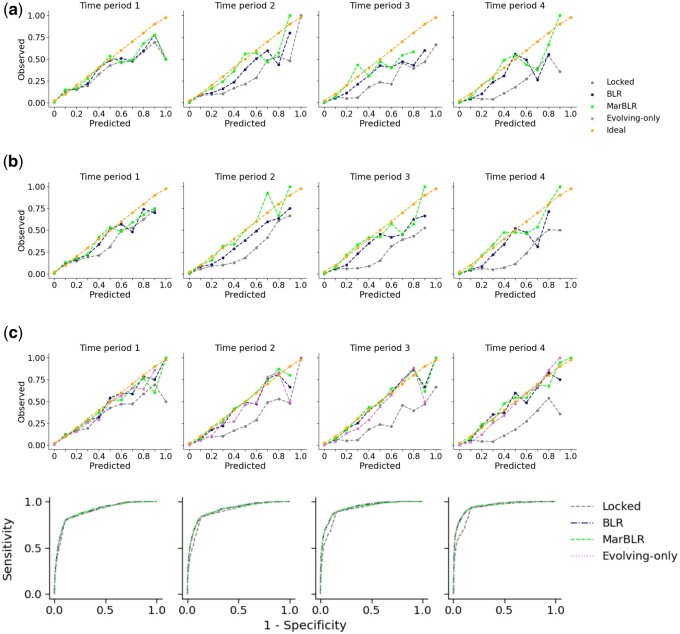
COPD case study
Due to real-world temporal shifts, the original model decayed in calibration (Table 5, Figure 6). Online logistic recalibration gradually decreased the forecasted score, which reflects the general downward trend in COPD diagnosis rates. It improved the aECI from 1.526 (95% CI, 1.371–1.680) to 0.693 (95% CI, 0.624–0.762) with BLR and 0.450 (95% CI, 0.411–0.490) with MarBLR. The performance of online logistic revision was similar to online logistic recalibration. We observe significant improvements when BLR and MarBLR ensemble the original and continually refitted GBTs. Using MarBLR, we achieve an aAUC of 0.925 (95% CI, 0.914–0.935) compared to the locked model’s aAUC of 0.904 (95% CI, 0.892–0.916). Although the initial coefficient for the refitted model is set to zero, BLR and MarBLR increased the weight of the refitted model and decreased that of the original model. Finally, BLR and MarBLR had similar AUC as the continually refitted model but were better calibrated. Perhaps even more importantly, BLR and MarBLR offer safety guarantees whereas the continual refitting procedure does not.
Table 5.
Results from COPD risk prediction task using Bayesian logistic revision (BLR) and its Markov variant MarBLR.
| aAUC | aECI | aNLL | |
|---|---|---|---|
| Locked | 0.904 (0.892,0.916) | 1.526 (1.371,1.680) | 0.099 (0.097,0.101) |
| Online logistic recalibration | |||
| MarBLR | 0.907 (0.895,0.919) | 0.450 (0.411,0.490) | 0.079 (0.077,0.082) |
| BLR | 0.906 (0.894,0.918) | 0.693 (0.624,0.762) | 0.082 (0.080,0.085) |
| Online logistic revision | |||
| MarBLR | 0.909 (0.897,0.921) | 0.460 (0.427,0.493) | 0.078 (0.075,0.080) |
| BLR | 0.907 (0.894,0.919) | 0.708 (0.647,0.769) | 0.082 (0.079,0.084) |
| Online ensembling of the original and continually refitted models | |||
| MarBLR | 0.925 (0.914,0.935) | 0.482 (0.450,0.514) | 0.072 (0.070,0.074) |
| BLR | 0.924 (0.913,0.935) | 0.429 (0.401,0.458) | 0.071 (0.069,0.073) |
| Continual Refit Only | 0.924 (0.914, 0.935) | 0.657 (0.624, 0.690) | 0.078 (0.076, 0.080) |
Note. 95% bootstrap confidence intervals are shown in parentheses.
Abbreviations: aAUC: average area under the receiver operating characteristic curve; aECI: average estimated calibration index; aNLL: average negative log likelihood.
Figure 6.
Results from online logistic recalibration and revision for a fixed COPD risk prediction model (A and B, respectively) and online reweighting for fixed and continually refitted (evolving) COPD risk prediction models (C) using BLR and MarBLR. Calibration curves are estimated across 4 time periods that divide the full dataset into equal lengths. The receiver operating characteristic (ROC) curve is shown for the online ensembling approach because the ROC curves did not change significantly for (A) or (B). A, Online recalibration of a fixed prediction model. B, Online logistic revision with respect to a fixed prediction model and patient variables. C, Online ensembling of the original and continually refitted prediction models.
DISCUSSION
Performance degradation of clinical ML algorithms occur for a variety of reasons, such as abrupt system-wide changes in the record keeping system,27 changes in the event rate and patient case mix,28 and changes in clinical order patterns.29 A growing number of papers have highlighted the need for regular monitoring and updating of clinical prediction algorithms,3,30 but there are currently no online procedures with practical performance guarantees. Instead, much of the literature has focused on learning 1-time model updates,31–33 which have inflated error rates when applied repeatedly over time. Dynamic model updating has also been suggested for clinical prediction models,14,34,35 but theoretical guarantees in the presence of model misspecification and dataset shifts have been lacking.
In this work, we show that online model revision by BLR or MarBLR is a promising solution that both exhibits strong empirical performance and provides theoretical guarantees. We derived Type I regret bounds that guarantee the online revision methods will be noninferior to locking the original model in the presence of arbitrary distribution shifts and Type II regret bounds that guarantee the methods quickly learn beneficial revisions. Our data analyses verified these results: BLR and MarBLR consistently outperformed locking the original model by slowing down performance decay in nonstationary settings and gradually improved overall performance in stationary settings. For example, the risk predictions from the locked model became increasingly inflated in the COPD dataset; by the end of the 8-year time span, a risk prediction of 0.8 from the locked model corresponded to an observed event rate of 0.5. Although other online methods helped protect against distribution shifts, their theoretical guarantees are much weaker and they tended to learn good model revisions more slowly.
The key difference between BLR and MarBLR is that the former assumes the oracle sequence of model revisions is static, whereas the latter allows for dynamic sequences. The theoretical results highlight a tradeoff between the 2 procedures: BLR incurs higher bias because of its simplifying assumptions, whereas MarBLR is more sensitive to sampling noise because it searches over a richer class of model revision sequences. In practice, we find that BLR is highly competitive with MarBLR. Although MarBLR tended to outperform BLR in settings with more severe distribution shifts, the performance differences were negligible in most, if not all, cases. Given that BLR is a simpler procedure involving fewer hyperparameters, we believe BLR is sufficient in most settings. Moreover, BLR can be implemented using standard software packages for Bayesian inference.36,37
We highlight that MarBLR and BLR only provide theoretical guarantees in terms of the average negative log likelihood. As seen in the simulations, MarBLR and BLR may perform worse than the original model for a few time points because of small sample sizes early on, or because the continual refitting procedure for the underlying prediction model suddenly introduces a bad update. This serves as a point of caution. Many online learning methods, including ours, do not guarantee that the deployed model will outperform the static model at all time points. Nevertheless, MarBLR and BLR are guaranteed to recover from sudden performance decay such that the average performance compares favorably to locking the original model. Analogously, MarBLR and BLR are not guaranteed to outperform the original model within a particular patient subgroup if the online revision procedure trains on data from the general population. Although the results from the first simulation are promising, we can only control Type I regret if separate instances of MarBLR/BLR are deployed within each patient subgroup. Future work should evaluate the algorithmic fairness of MarBLR and BLR in a wide variety of settings and introduce any necessary extensions to ensure fairness will be maintained over time. This paper has only explored the simplest setting with 2 patient subgroups defined a priori; in practice, one may be interested in many more subgroups and may even want to preserve fairness across subgroups that have yet to be defined clinically.
A key assumption in this work is that one can measure gold standard outcomes and perform an unbiased evaluation of the underlying prediction model. One must verify these assumptions hold, at least approximately, before applying MarBLR and BLR. In our case study, we considered a risk prediction model for predicting COPD diagnosis. This model may influence clinical decision making and perturb the distribution of both the covariates and the outcome, thereby complicating model evaluation and updating. This issue called “performative prediction”38 can be even more severe in high acuity settings where ML recommendations are designed to change clinical workflows. Nevertheless, one may be able to obtain more accurate patient outcomes by incorporating a lag time, during which missed diagnoses are corrected. Another option is to consider the counterfactual framework and define the true outcome as the outcome that would have been observed, had the ML prediction not been made available to the clinician. Two avenues for identifying this true outcome are to either randomize a subset of patients to receive no ML prediction or use a causal inference/missing data framework. Both options come with substantial challenges and require further research.39
In this paper, we require the user to decide the class of logistic revisions upfront. That is, they must decide between online model recalibration versus revision; if they choose the latter, they must decide which variables to include and whether to incorporate a continually refitted model. Because future data distributions are unknown, it is difficult to anticipate which type of model revision will lead to best online performance. As seen in the COPD case study, the logistic model revisions with respect to the selected patient variables did not improve model discrimination but incorporating a continually refitted model did. Instead of requiring a model revision class to be selected upfront, future extensions of MarBLR/BLR may be able to incorporate model selection using a hierarchical modeling approach such as in McCormick et al.14 In addition, new biomarkers will be discovered as the biomedical field continues to advance. An important use case is to let the model developer add these newly discovered biomarkers to the online model revision procedure.
Finally, a limitation of the current work is that the derived regret bounds scale linearly with the number of variables used during online model revision. As such, MarBLR and BLR may not provide meaningful safety guarantees in high-dimensional settings. Because many modern ML algorithms analyze a large number of variables, future work should look to refine our regret bounds by characterizing, say, the L1-norm or sparsity of the oracle model revision.20
CONCLUSION
Our theoretical and empirical results support the use of online model revision by BLR or MarBLR over other online methods for regular monitoring and updating of clinical prediction algorithms when performance drift is of concern.
FUNDING
This work was supported by the Food and Drug Administration (FDA) of the U.S. Department of Health and Human Services (HHS) as part of a financial assistance award Center of Excellence in Regulatory Science and Innovation grant to University of California, San Francisco (UCSF) and Stanford University, U01FD005978 totaling $79 250 with 100% funded by FDA/HHS. The contents are those of the author(s) and do not necessarily represent the official views of, nor an endorsement, by FDA/HHS, or the U.S. Government.
AUTHOR CONTRIBUTIONS
Conceptualization: JF, AG, BS, RP. Manuscript drafting and/or editing: JF, AG, BS, RP. Development and analysis of methodology: JF, AG. Empirical investigation and validation: JF, AG.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
This work was greatly improved by helpful suggestions and feedback from Pang Wei Koh and Andrew Magee. We thank Ari Hoffman, Mark Pletcher, and Jennifer Creasman for sharing the COPD dataset.
CONFLICT OF INTEREST STATEMENT
None declared.
DATA AVAILABILITY
Simulation data can be reproduced using code available at http://github.com/jjfeng/bayesian_model_revision. The COPD dataset cannot be shared publicly due to patient privacy. The data will be shared on reasonable request to the corresponding author.
Contributor Information
Jean Feng, Department of Epidemiology and Biostatistics, University of California, San Francisco, San Francisco, California, USA.
Alexej Gossmann, CDRH-Center for Devices and Radiological Health, Food and Drug Administration, Silver Spring, Maryland, USA.
Berkman Sahiner, CDRH-Center for Devices and Radiological Health, Food and Drug Administration, Silver Spring, Maryland, USA.
Romain Pirracchio, Department of Anesthesia and Perioperative Care, University of California, San Francisco, San Francisco, California, USA.
References
- 1. Benjamens S, Dhunnoo P, Meskó B. The state of artificial intelligence-based FDA-approved medical devices and algorithms: an online database. NPJ Digit Med 2020; 3 (1): 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Steyerberg EW. Clinical Prediction Models: A Practical Approach to Development, Validation, and Updating. New York, NY: Springer; 2009. [Google Scholar]
- 3. Pirracchio R, Ranzani OT. Recalibrating our prediction models in the ICU: time to move from the abacus to the computer. Intensive Care Med 2014; 40 (3): 438–41. [DOI] [PubMed] [Google Scholar]
- 4. Amarasingham R, Patzer RE, Huesch M, et al. Implementing electronic health care predictive analytics: considerations and challenges. Health Aff (Millwood) 2014; 33 (7): 1148–54. [DOI] [PubMed] [Google Scholar]
- 5. Thrun S. Lifelong learning algorithms. In: Thrun S, Pratt L, eds. Learning to Learn. Boston, MA: Springer US; 1998: 181–209. [Google Scholar]
- 6. Cesa-Bianchi N, Lugosi G. Prediction, Learning, and Games. New York, NY: Cambridge University Press; 2006. [Google Scholar]
- 7. Baweja C, Glocker B, Kamnitsas K. Towards continual learning in medical imaging. In: Medical Imaging meets NIPS Workshop, 32nd Conference on Neural Information Processing Systems (NIPS2018); 2018; Montréal, Canada. [Google Scholar]
- 8. Lee CS, Lee AY. Clinical applications of continual learning machine learning. Lancet Digital Health 2020; 2 (6): e279–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Janssen KJM, Moons KGM, Kalkman CJ, et al. Updating methods improved the performance of a clinical prediction model in new patients. J Clin Epidemiol 2008; 61 (1): 76–86. [DOI] [PubMed] [Google Scholar]
- 10. Strobl AN, Vickers AJ, Van Calster B, et al. Improving patient prostate cancer risk assessment: moving from static, globally-applied to dynamic, practice-specific risk calculators. J Biomed Inform 2015; 56: 87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.U.S. Food and Drug Administration. Proposed regulatory framework for modifications to artificial intelligence/machine learning (AI/ML)-based software as a medical device (SaMD): discussion paper and request for feedback; 2019. https://www.fda.gov/media/122535/download. Accessed August 31, 2021.
- 12. Viering T, Mey A, Loog M. Open Problem: Monotonicity of Learning In: Beygelzimer A, Hsu D, eds. Proceedings of the Thirty-Second Conference on Learning Theory. Phoenix, USA: PMLR; 2019: 3198–201. [Google Scholar]
- 13. West M, Harrison J. Bayesian Forecasting and Dynamic Models. New York, NY: Springer; 1997. [Google Scholar]
- 14. McCormick TH, Raftery AE, Madigan D, et al. Dynamic logistic regression and dynamic model averaging for binary classification. Biometrics 2012; 68 (1): 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Feng J, Emerson S, Simon N. Approval policies for modifications to machine learning-based software as a medical device: a study of bio-creep. Biometrics 2020; 77 (1): 31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kuleshov V, Ermon S. Estimating uncertainty online against an adversary. In: AAAI; 2017. [Google Scholar]
- 17. Davis SE, Greevy RA Jr, Lasko TA, et al. Detection of calibration drift in clinical prediction models to inform model updating. J Biomed Inform 2020; 112: 103611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kingma DP, Ba J. Adam: a method for stochastic optimization. In: Bengio Y, LeCun Y, eds. 3rd International Conference for Learning Representations; 2015; San Diego, CA. [Google Scholar]
- 19. Kakade SM, Ng A. Online bounds for Bayesian algorithms. In: Saul L, Weiss Y, Bottou L, eds. Advances in Neural Information Processing Systems. MIT Press; 2005: 641–8. [Google Scholar]
- 20. Shamir GI. Logistic regression regret: what’s the catch? arXiv [cs.LG]; 2020. http://arxiv.org/abs/2002.02950
- 21. Lum K, Isaac W. To predict and serve? Significance 2016; 13 (5): 14–9. [Google Scholar]
- 22. Ensign D, Friedler SA, Neville S, et al. Runaway feedback loops in predictive policing. In: Friedler SA, Wilson C, eds. Proceedings of the 1st Conference on Fairness, Accountability and Transparency. New York, NY, USA: PMLR; 2018: 160–71. [Google Scholar]
- 23. Lewis SM, Raftery AE. Estimating Bayes factors via posterior simulation with the laplace—metropolis estimator. J Am Stat Assoc 1997; 92: 648–55. [Google Scholar]
- 24. Gordon K, Smith AFM. Modeling and monitoring biomedical time series. J Am Stat Assoc 1990; 85: 328–37. [Google Scholar]
- 25. Chouldechova A, Roth A. The frontiers of fairness in machine learning. arXiv [cs.LG]; 2018. http://arxiv.org/abs/1810.08810. Accessed August 31, 2021.
- 26. Van Hoorde K, Van Huffel S, Timmerman D, et al. A spline-based tool to assess and visualize the calibration of multiclass risk predictions. J Biomed Inform 2015; 54: 283–93. [DOI] [PubMed] [Google Scholar]
- 27. Nestor B, McDermott MBA, Boag W, et al. Feature robustness in non-stationary health records: caveats to deployable model performance in common clinical machine learning tasks. Mach Learn Healthcare 2019; 106: 381–405. [Google Scholar]
- 28. Davis SE, Lasko TA, Chen G, et al. Calibration drift in regression and machine learning models for acute kidney injury. J Am Med Inform Assoc 2017; 24 (6): 1052–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen JH, Alagappan M, Goldstein MK, et al. Decaying relevance of clinical data towards future decisions in data-driven inpatient clinical order sets. Int J Med Inform 2017; 102: 71–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Saria S, Subbaswamy A. Tutorial: safe and reliable machine learning. In: Proceedings of the Conference on Fairness, Accountability, and Transparency. New York, NY: Association for Computing Machinery; 2019; Atlanta, Georgia.
- 31. Davis SE, Greevy RA, Fonnesbeck C, et al. A nonparametric updating method to correct clinical prediction model drift. J Am Med Inform Assoc 2019; 26 (12): 1448–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vergouwe Y, Nieboer D, Oostenbrink R, et al. A closed testing procedure to select an appropriate method for updating prediction models. Stat Med 2017; 36 (28): 4529–39. [DOI] [PubMed] [Google Scholar]
- 33. Steyerberg EW, Borsboom GJJM, van Houwelingen HC, et al. Validation and updating of predictive logistic regression models: a study on sample size and shrinkage. Stat Med 2004; 23 (16): 2567–86. [DOI] [PubMed] [Google Scholar]
- 34. Su T-L, Jaki T, Hickey GL, et al. A review of statistical updating methods for clinical prediction models. Stat Methods Med Res 2018; 27 (1): 185–97. [DOI] [PubMed] [Google Scholar]
- 35. Raftery AE, Kárný M, Ettler P. Online prediction under model uncertainty via dynamic model averaging: application to a cold rolling mill. Technometrics 2010; 52 (1): 52–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stan Development Team. Stan modeling language users guide and reference manual; 2021. https://mc-stan.org/. Accessed August 31, 2021.
- 37. Salvatier J, Wiecki TV, Fonnesbeck C. Probabilistic programming in Python using PyMC3. PeerJ Comput Sci 2016; 2: e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Perdomo J, Zrnic T, Mendler-Dünner C, et al. Performative prediction. In: Iii HD, Singh A, eds. Proceedings of the 37th International Conference on Machine Learning. PMLR; 2020: 7599–609. [Google Scholar]
- 39. Liley J, Emerson S, Mateen B, et al. Model updating after interventions paradoxically introduces bias. In: Banerjee A, Fukumizu K, eds. International Conference on Artificial Intelligence and Statistics; PMLR; 2021;130:3916–24. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Simulation data can be reproduced using code available at http://github.com/jjfeng/bayesian_model_revision. The COPD dataset cannot be shared publicly due to patient privacy. The data will be shared on reasonable request to the corresponding author.