ABSTRACT
Viruses have evolved diverse strategies to hijack the cellular gene expression system for their replication. The poly(A) binding proteins (PABPs), a family of critical gene expression factors, are viruses’ common targets. PABPs act not only as a translation factor but also as a key factor of mRNA metabolism. During viral infections, the activities of PABPs are manipulated by various viruses, subverting the host translation machinery or evading the cellular antiviral defense mechanism. Viruses harness PABPs by modifying their stability, complex formation with other translation initiation factors, or subcellular localization to promote viral mRNAs translation while shutting off or competing with host protein synthesis. For the past decade, many studies have demonstrated the PABPs’ roles during viral infection. This review summarizes a comprehensive perspective of PABPs’ roles during viral infection and how viruses evade host antiviral defense through the manipulations of PABPs.
KEYWORDS: viral infection, host defense, poly(A)-binding protein (PABP)
INTRODUCTION
Whether DNA or RNA viruses, no known virus encodes all apparatus required to synthesize viral proteins. Viruses compete with the host translational factory to translate their viral mRNAs. This situation inspires viruses to develop strategies to hijack the cellular gene expression system to promote their propagation in host cells. Viruses usually manipulate the translational apparatus by host cell shutoff, inhibiting host protein synthesis for viral replication. Few other viruses finish their propagation in host cells more quietly by shutting down only a small part of the host translational system or facilitating virus replication with ongoing host protein synthesis. Although viruses regulate cellular translation at different molecular levels, the important and early step to interfere with translation is to disrupt translation initiation machinery on polyadenylated mRNAs. The translation machinery contains eukaryotic initiation factors (eIFs), poly(A) binding proteins (PABPs), and other cellular factors (Fig. 1B). eIF4F is a protein complex that binds to the 5′ cap of a mature mRNA. The eIF4F complex consists of eIF4E (a small cap-binding protein), eIF4A (a DEAD box RNA helicase), and the central factor eIF4G (a large scaffold protein) (1). eIF4G anchors the 43S pre-initiation complex (consisting of eIF3, eIF1, eIF1A, eIF2-GTP-tRNAiMet, eIF5, and the 40S subunit of the ribosome) to the 5′-end of the mRNA through the interaction with eIF3 (1). PABPs bind to the 3′-end poly(A) tail of mRNA and interact with the eIF4G protein on a 5′ cap, thereby stabilizing the structure of mRNA by forming a head-to-tail loop and promoting the recruitment of ribosome subunits for translation initiations (1, 2). PABPs are general viral targets, and the modification of PABP activities allows viruses to hinder the host translation initiations (3). To date, numerous studies have demonstrated that viruses target the translation initiation factor PABPs. Consequently, how viruses subvert PABPs during viral infections needs to be updated despite a review of PABP as a viral target being published more than a decade ago (3).
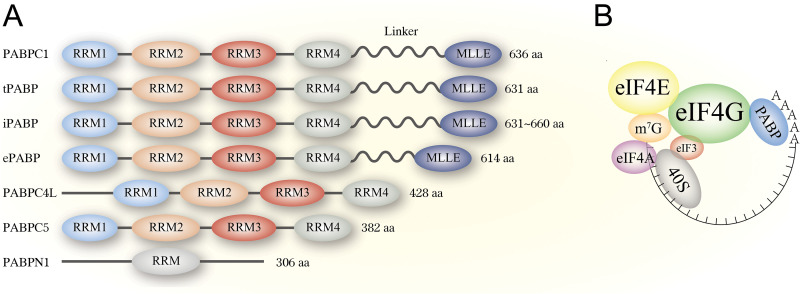
FIG 1.
The structure of PABPs and translation initiation complex in uninfected cells. (A) The structure of different human PABPs. PABP comprises N-terminus RRM domains, C-terminus MLLE domain, and a proline-rich linker sequence connect these two parts. PABPC5 and PABPN1 do not have MLLE and the linker region. (B) The translation initiation complex in uninfected cells. PABPs bind to poly(A) tail eukaryotic cellular mRNA, and they guide 3′-end of mRNA to its 5′ cap through interaction with eIF4G. eIF4G is a bridge between the PABP-poly(A) tail and 5′ cap, forming a head-to-tail loop and recruiting ribosome 40S subunit for further translation initiations.
RNA-binding proteins (RBPs) regulate numerous post-transcriptional processes by binding to intended RNAs (4). A clear-identified and well-conserved group of RBPs is the PABPs subfamily. PABPs are a cluster of proteins that interact with polyadenylated mRNAs in eukaryotic cells. It was first discovered to be bound to the polyadenylated region of eukaryotic messenger RNAs (mRNAs) in the 1970s (5–7). In the early days, PABPs were mainly supposed to protect mRNA stability from deadenylation of the 3′-end poly(A) tail. In addition to its RNA-binding capabilities, PABPs are also critical mediators of gene expression at the level of RNA metabolism (8). Nevertheless, most studies claim the critical role of PABPs in enhancing the cellular translation initiation, which is PABPs binds poly(A) tail to 5′ cap of mRNA to facilitate the efficiency of translation initiation or regulates the length of 3′ poly(A) tail (PAT) for different translation purposes (8–10).
Moreover, PABPs act as enhancing factors of mRNA translation initiation and as translation factors themselves. They function as scaffold proteins, supporting protein-protein interactions with multiple translation factors, thus promoting translation initiation and regulating gene expression (11). To date, PABPs have been shown to take part in all the metabolic pathways of the mRNA, including mRNA polyadenylation, mRNA deadenylation, mRNA export, mRNA surveillance, mRNA translation and stability, mRNA decay, microRNA-associated regulation, and regulation of expression during development and so on (11–15). These metabolic pathways are good routines for the viruses to utilize in their infection strategies. Targeting PABPs by both DNA and RNA viruses again highlights the role of PABPs as central regulators of gene expression. This review updates an integrative account of the significance of PABP during viral infections.
THE CLASSIFICATION OF PABPS
There are significant differences in PABPs family proteins among different species from yeast to humans. Different species have different numbers of PABP proteins. For example, dicot Arabidopsis thaliana contains eight PABPs (16, 17), Xenopus laevis contains three PABPs (18–20), and Mus musculus contains two PABPs (21, 22). In humans, seven PABPs have been discovered. In irrespective of species, PABPs can be classified according to their distribution in cells. PABPs include cytoplasmic PABPs, nuclear PABP, embryonic PABP, and the X chromosome-encoded protein. Cytoplasmic PABPs (PABPC) includes PABPC1 (also called PABP1), tPABP (testis-specific PABP, also called PABPC2 or PABPC3), iPABP (inducible PABP, also called PABPC4 or PABP4), and PABPC4L; embryonic PABP is called ePABP or PABPC1L, and X chromosome-encoded protein is named PABPC5; and the nuclear PABP is named PABPN1 (11, 23, 24). Nevertheless, the most well-studied PABPs during viral infections are PABP1. Only PABP1 was reported in virus infections. This review focus on how PABP1 (refer to PABPs in the following text) is manipulated during viral infections.
THE STRUCTURE OF PABPS
Cytoplasmic PABPs bind specifically to the poly(A) tail of eukaryotic mRNAs via its RNA-recognition motifs (RRMs), which have a lower affinity for poly(U) and poly(G), and no detectable binding to poly(C) (25–28). The structures of PABPs are highly conserved, with its N-terminus containing four non-identical RRMs, and its C-terminus consisting of a highly conserved carboxy-terminal helical domain named MLLE, previously known as PABC (Fig. 1A) (11, 29, 30). Proline-rich linker sequences connect the four RRM domains to the MLLE domain (31, 32). Despite that the RRM motifs are conserved for RNA recognition, their binding abilities to RNA and proteins appear to be specifically different (27, 28). For instance, RRM 1 and 2 exhibits highly poly(A) binding abilities to eIF4G and PABP-interacting protein 1 (Paip1), while RRM 3 and 4 provide lower binding affinity to poly(A) while binding to AU-rich RNA and mediate protein-protein interactions (25, 33, 34). Multiple genes encoding PABPs are found in metazoans and plant species.
Furthermore, not all PABPs encode four RRM domains. In plants like A. thaliana, only two of the eight PABP genes encode proteins containing four RRM domains(17). In plants, such as PABPs in N. tabacum, only one of the three expressed PABP genes have four RRMs (17). In contrast to RRMs roles, the MLLE is not required for RNA recognition but is necessary for interaction with initiation factors through its binding site, such as the release factor eRF3, translation repressor Paip1/Paip2. A 15-residue sequence “PABP interaction motif,” termed PAM2, is critical for this process. This oligomerization controls mRNA translation, deadenylation, and regulates PABPs binding ability to poly(A) tail and eIF4G (35–40).
VIRUSES USURP PABP FUNCTIONS FOR VIRAL INFECTIONS
Viruses have long been reported to shut off host protein synthesis, usually disrupting the translation initiation machinery. PABPs are general translation initiation factors targeted by viruses to promote viral propagation and/or thwart cell gene expression during infections. In eukaryotic cells, PABPs bind to the 3′-end poly (A) tail and indirectly bind to the 5′-end cap complex to promote efficient translation initiations (1). However, some viruses do not contain a canonical 5′-end cap and/or 3′-end poly(A) tail. Hence, viruses develop strategies to manipulate PABPs for viral replication and protein synthesis. Depending on distinct structures of viral mRNAs, different RNA viruses and DNA viruses favor different mechanisms to manipulate PABPs. In reported cases, viruses could commandeer PABPs to hinder the efficient host translation initiation through following ways, such as cleavage of PABPs by viral proteases, regulation of eIF4F-PABP translational complex, relocalization of cytoplasmic PABPs into the nucleus, interfering with PABPs containing stress granules (SGs), expressing viral translation factor analogs (3, 8, 41). The genomic structure of RNA viruses containing both canonical 5′-end cap and 3′-end poly(A) tail would prefer to use a cap-dependent translation initiation method to translate their mRNA while simultaneously shutting off the cellular and viral mRNA translation. In this situation, viruses would compensate their translation by distinct means to escape the whole translation shutoff. The more specific mechanism of how viruses escape their shutoff is summarized and discussed in each virus in this review. Coincidentally, DNA viruses transcripts are similar to host mRNAs, using a cap-dependent strategy. The genomic structure of RNA viruses lacking a 5′-end cap usually bears an internal ribosomal entry site (IRES) in the 5′ untranslated region (5'UTR). They utilize cap-independent translation to promote viral mRNA translation and inhibit the host protein synthesis by degrading PABPs or eIF4F complex. When RNA viruses have a genomic structure without a classic poly(A) tail, they usually express PABPs surrogates in enhancing viral mRNA translation initiation by binding to viral 3′-end tail and redistributing the cytoplasmic PABPs into the nucleus. Although there seems to be a rule to follow by deciphering the relationship between the viral mRNA structures and their strategies in usurping PABPs, the underlying mechanisms that each virus subverts PABPs are distinct. In this regard, more studies are warranted to draw a map about how viruses hinder the host PABPs.
Cap-dependent RNA viruses.
(i) Coronaviridae.
The coronavirus (CoV) contains a single-stranded, positive-sense RNA genome, which resembles the structure of most eukaryotic cellular mRNA. CoV genomic RNA contains a cap structure at the 5′-end, a poly(A) tail at the 3′-end, and untranslated regions (UTRs) (42). During CoV infections, knockdown of PABPs could attenuate virus propagations. For instance, transmissible gastroenteritis virus (TEGV) is a member of the Coronaviridae family, and it was reported that PABPs play significant roles in virus RNA synthesis, possibly by binding to the TEGV genome through its 3′ end poly(A) tail (43) However, a new report showed that in bovine coronavirus (BCoV), it is the viral nucleocapsids (N) protein, not PABPs involving in RNA synthesis by binding to both 5′- and 3′-ends (44). Even though PABP is important for CoV replication (43, 45), it still unclear how PABPs influence virus RNA synthesis during the CoV life cycle.
Moreover, CoV competitively utilized PABPs as translation initiation factors with the host for viral mRNA translation. CoV could decrease the translation of host mRNA by its nucleocapsid (N) protein. For example, BCoV N protein competes with PABP to interact with poly(A) tail and eIF4E to decrease translation initiation efficiency of virus and host both in vitro and in vivo (46). This finding suggests that CoV can suppress host mRNA translation by competitively inhibiting the interaction between PABP and host mRNAs' poly(A) tails. Even this process could also harm viral mRNAs translation. However, N protein also binds to viral replicase proteins, which might be an important way to rescue the disturbance of viral mRNAs. Once viral replicase protein is synthesized, it associates with N protein and guides the N protein-associated viral mRNAs to the replication complex at the endoplasmic reticulum (ER) to promote viral mRNA synthesis (47). This process may compensate for viral mRNA quantity for later translation events.
A recent study demonstrated that CoV could manipulate PABPs in a different strategy. SARS-unique domain (SUD) is a non-conserved part of non-structural protein 3 (NSP3) shared by SARS-CoV-2 and SARS-CoV (48–50). Brunn et al. showed that SUD promoted PABP-interacting protein 1 (Paip1) and PABPs interactions by binding to 40S/80S ribosome, and Paip1 interacts with the RRM domains of PABPs to stimulate translation, thus increasing both viral and host mRNAs translation efficiency (36, 40, 51–53). However, viruses hinder host translation by specifically degrading host mRNAs and blocking them from binding to the 40S ribosome by viral protein NSP1 (54–56). Consequently, only viral but not host translation is enhanced by SARS-CoV-2 and SARS-CoV SUD (51).
In conclusion, the CoVs either evade cellular shutoff by compensating viral mRNA levels or blocking host mRNA translation by viral protein after stimulating a high activity of the translation system.
(ii) Arteriviridae.
Porcine reproductive and respiratory syndrome virus (PRRSV) is a single-stranded positive-sense RNA virus containing a 5′-end cap, 3′-end poly(A) tail, and UTRs. Depending on the similar viral mRNA structures, PRRSV and CoV may use similar strategies to manipulate PABPs. However, this hypothesis is not verified as limited studies reported manipulating PABPs in PRRSV. Jiang et al. found that PRRSV N protein was responsible for redistributing cytoplasmic PABPs into the infected nuclei through the interaction with the C-terminal half of PABPs (57). Moreover, knockdown of PABPs significantly influences viral RNA synthesis and virus titers, suggesting that the mRNA translation or replication of PRRSV is closely related to PABPs (57). The underlying mechanisms of how PRRSV N protein or other viral proteins manipulate PABPs need further investigation.
(iii) Togaviridae.
The Rubella virus (RV) belongs to the Rubivirus genus of the Togaviridae family. Its genomic structure is a positive-strand, capped, and polyadenylated RNA. Like other viruses with similar RNA structures, such as PRRSV, RV would redistribute cytoplasmic PABPs into the nucleus (58). This phenomenon is also observed in rotavirus and herpes simplex virus (HSV), which have classic 5′-end cap mRNA. Notably, RV capsid protein binds to the C-terminal region of PABPs and co-localizes it to the perinuclear region of the cell in vitro and in vivo (58). PABPs levels increased in RV infected cells, but its activity in translation may be blocked, causing the inhibition of translation; however, this situation was alleviated by excess PABPs (58).
Nevertheless, it is still unclear whether the interactions between RV capsid protein and PABPs are associated with viral mRNA translation by restricting PABPs in the nucleus or hijacking PABPs to inhibit cellular protein synthesis. Furthermore, it was suggested that by sequestration of PABPs, the viral capsids could increase the efficiency of viral RNA packaging into capsids instead of viral RNA translation (58). If so, more nucleocapsid formation-related identifications need to be performed to verify this hypothesis (59).
(iv) Orthomyxoviridae.
Influenza A contains a single-stranded, negative-sense RNA genome. Its genome is both capped and polyadenylated with 5′-UTR. Influenza A encodes a non-structural protein 1 (NS1) that binds to the 5′-UTR region of viral mRNA and functions in viral replication and protein synthesis (60, 61). NS1 also interacts with PABPs and eIF4G on its different binding domains (59). It does not evict PABPs from the eIF4G as relative amounts of eIF4G, and PABPs proteins are co-purified with NS1. NSP1 specifically accelerates PABPs and eIF4G to form a complex with it, and this complex is beneficial for viral translation initiation (62). This mechanism of manipulating PABPs is very different from other positive-sense RNA viruses.
(v) Rhabdoviridae.
Some viruses induce SGs formation in infected cells, such as the rabies virus (RABV), the typical species of the Lyssavirus genus within the Rhabdoviridae family (63). The viral genome is a single-stranded, negative-sense RNA consisting of a 5′-end cap and 3′-end poly(A) tail (64). No report shows how RABV manipulates PABPs in a cap-dependent way. However, RABV might exploit PABPs to disrupt the formation of SGs. PABP-specific SGs are among the major distinguishable RNA particles in host cells (41, 65). It is an important component of host antiviral defense. Viral infection exerts pressure on the host cells to induce the formation of SGs (41). However, viruses must face the adverse environment from host immunity and take various strategies to block the formation of SGs and promote its infection and proliferation effectively. RABV mRNAs synthesize in cytoplasmic viral factories, known as Negri bodies (NBs), closely related to SGs (63). This close correlation of viral mRNAs accumulating around SGs reveals material communications between them and may inhibit the antiviral function of SGs (63). Nevertheless, it is unclear how the virus targets PABPs in RABV induced SGs, and the relationship between PABP-specific SGs and viral infections needs further explorations generally.
Cap-independent RNA viruses.
(i) Picornaviridae.
The genera Enterovirus, Rhinovirus, Hepatovirus, Parechovirus, Cardiovirus, and Aphthovirus belong to the Picornaviridae family. Picornavirus contains an uncapped and polyadenylated positive-strand RNA genome with UTRs. Lacking the 5′-end cap, they employ an IRES located in the 5′-UTR instead to initiate the translation of viral mRNAs (66). As a 5′-end cap is required in host cellular translation initiation, in this regard, IRES serves as a 5′-end cap-like element in picornavirus mRNA translation initiation. Picornavirus exhibits alternative translation control strategies based on the IRES structure of the viral genome.
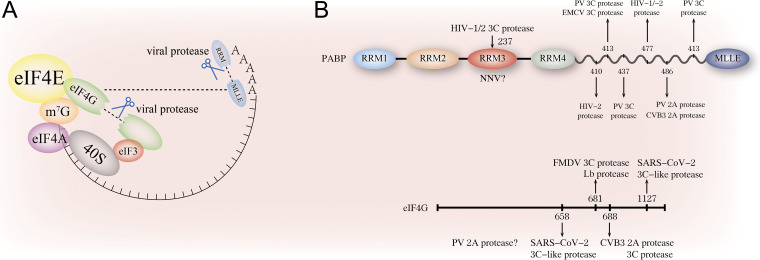
RNA viruses of this family have been extensively responsible for PABPs and eIF4G cleavage by the viral proteases, facilitating viral translation by inhibiting cellular mRNA translation (Fig. 2). 3C is a viral cysteine protease that cleaves the proline-rich linker of the PABPs and eIF4G structure (67). The 3C proteases of coxsackievirus (CV), poliovirus (PV), encephalomyocarditis virus (EMCV), and duck hepatitis A virus (DHAV) target PABPs to destabilize PABP-poly(A) association (68–71). These 3C proteases separate the N-terminus of PABPs from its C-terminus by cleaving its flexible linker. Even the recognition sites vary (Fig. 2B). PV 3C cleaves PABPs at Q537/G538, Q437/G438, and Q413/T414, while the cleavage sites on PABPs by EMCV and DHAV 3C are WTAQ437/G438 and Q367/G368, respectively (70, 72, 73). PABPs are also degraded by PV and coxsackievirus B3 (CVB3) 2A proteases between amino acids 486 and 487. Even this cleavage is not as efficient as 3C protease does (67, 70). Moreover, PV 2A protease would cut eIF4G, which evicts the N-terminus of eIF4G from the C-terminus, destabilizes the PABP-eIF4G interaction, and causes impaired cellular translation initiation (74).
FIG 2.
Cleavage of PABPs or eIF4G by viral proteases. (A) Cleavage of PABPs and eIF4G on the translation initiation complex by viral proteases. Viral 2A, 3C, and other viral proteases will cut PABPs from their RRM domains or proline-rich linker sequences when host cells are infected. This cleavage will lose the interaction between PABPs and eIF4G, leading to inhibition of cellular translation. Besides, viral protease (2A/3C/Lb proteases) will cleave eIF4G, separating its N-terminus from C-terminus, thus destabilizing PABPs interactions with poly(A) or disrupting eIF4G interaction with other translation initiation factors. (B) Schematic of the cleavage sites on PABPs and eIF4G by different viral proteases. The cleavage site of PV protease on eIF4G and NNV protease on PABPs is not reported yet. Thus, '”?'” mark is used to indicate the unfound cleavage site.
Picornaviruses escape from their shutoff through a cap-independent initiation way. PV mRNA is translated using an IRES instead of a 5′-end cap, which does not require intact eIF4G (74–76). Intriguingly, the truncated eIF4G works more efficiently on the viral IRES-driven than the host cap-driven translation (77). PABPs might promote PV mRNA translation without eIF4G as it interacts with other eIFs. Although 3C protease mediates the proteolysis of PABPs, leading to the inhibition of host translation, there are still some uncleaved PABPs. These uncleaved PABPs promote PV mRNAs translation (75, 78), and picornavirus could evade the complete shutoff by utilizing the uncleaved PABPs.
Concerning the cleavage of eIF4G, it is mediated by viral 2A, 3C protease, or other viral proteases. For example, foot-and-mouth disease virus (FMDV) 3C protease and leader (Lb) protease induce the cleavage of eIF4G at C-terminus residue 681/682 (79, 80). CVB3 2A proteases degrade eIF4G at 688/689, and 3C also cleaves eIF4G even with no evidence showing the cleavage site (81). SARS-CoV-2 3C-like protease, also called the main (M) protease or non-structural G protein 5 (NSP5), induces the cleavage of eIF4G at two sites LQ658/GI659 and LQ1127/QA1128, even the role of host cell shutoff behind this cleavage is not elucidated (82).
FMDV Lb protease also cleaves G3BP1 and G3BP2, the scaffold protein in the stress granules (SGs), to disturb the formation of SGs (83). G3BP1 interacts directly with the viral IRES region and negatively regulates viral translation. FMDV 3C protease cleaves G3BP1 at amino acid residue 284/285 to release the IRES site and destroy the G3BP1 role to activate innate immunity (84). Another picornavirus CVB3 3C protease cleave G3BP1 at amino acid residue 325/326, inhibiting SG formation and promoting CVB3 replication (85). In addition, Feline calicivirus (FCV) NS6 protease was reported to cleave G3BP1 to inhibit SG formation (86).
(ii) Caliciviridae.
The family of Caliciviridae consists of Vesivirus, Lagovirus, Norovirus, Sapovirus, and Nebovirus genera. The genome structure is highly conserved with a positive-sense, single-stranded RNA. However, Calicivirus RNA contains a 3′-end poly(A) tail and short UTRs without a canonical 5′-end cap (87). Calicivirus genome is different but closely related with picornaviruses. Calicivirus does not contain IRES elements. Instead, the 5′-end covalently links with a small virus-encoded protein (VPg), which would interact with the classic translation initiation factors for translation (88). FCV and norovirus (NV) express 3C-like protease because it shares similarity to picornavirus 3C protease in the active site region. Viral 3C-like protease also inhibits cellular translation by the cleavage of PABPs. FCV and norovirus cleave PABPs within the proline-rich linker as well (89).
(iii) Retroviridae.
Human immunodeficiency virus (HIV) is Lentivirus belonging to the Retroviridae family. Retrovirus genomic RNAs contain 5′-end cap, 3′-end poly(A) tail, and UTRs regions, resembling host cellular transcripts. HIV mRNA utilizes cap-dependent translation initiation. However, HIV mRNA also contains the IRES region, which uses IRES-dependent translation initiation (90). Retroviruses like HIV, simian immunodeficiency virus (SIV), or Moloney murine leukemia virus (MMLV) contain the IRES region (91–93), which may let the virus take advantage of cap-independent translation, even without intact eIF4G. Like picornavirus or calicivirus, which do not contain a classic 5′-end cap but an IRES part, HIV-1 and HIV-2 encode proteases to degrade eIF4G and PABPs (94).HIV-1 and HIV-2 proteases cleave PABPs at amino acid residue 237/238 and 477/478, while HIV-2 additionally cleaves at 410/411 (95). The reason may be based on the IRES region located in the 5′-UTR. Although these viral IRESs contain diverse sequences, many have similar secondary structures and facilitate translation through similar mechanisms (96).
Overall, these studies demonstrated that cytolytic RNA viruses, including the Piconaviridae, Calicivirade, and Retroviridae, downregulate host metabolism mostly by drastically degrading cellular translation factors, such as PABPs and eIF4G, while viral translation continues via a cap-independent mechanism, which ensures that the translational machinery stays available only to viral mRNAs.
Non-polyadenylated RNA viruses.
(i) Reoviridae.
Rotavirus is a double-stranded RNA virus belonging to the Reoviridae family. The rotavirus mRNAs are capped but not polyadenylated. The rotavirus mRNA structure is quite different from the host mRNA, and the rotavirus uses distinct translation logic from the cap-dependent translation initiation. It is well known that the PABP-eIF4G complex is efficient for the translation initiation of cellular polyadenylated mRNAs, and rotavirus dispatched a viral non-structural protein NSP3 to compete with PABPs from binding to eIF4G (Fig. 3) (97). Consequently, rotavirus utilizes NSP3 to mediate the shutdown of host protein synthesis by suppressing the translation of cellular mRNAs. Poncet et al. found out that in infected cells, NSP3 is genetically linked to GACC-tailed mRNA, not poly(A)-tailed mRNA, competing with host for efficient translation initiation (98, 99). Notably, the 3′-end of rotavirus mRNA contains a conserved binding site (GACC motif) for NSP3 (99). The homodimerized NSP3 recognizes the specific motif and interacts with eIF4G to enhance viral mRNA translation, thus escaping from the host shutoff (100).
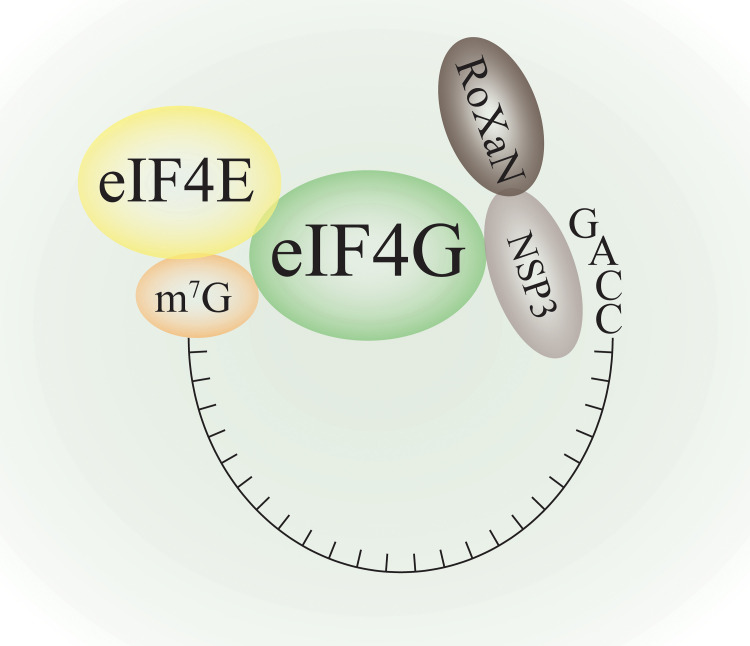
FIG 3.
Displacement of PABPs by viral structure protein. In RV infected cells, viral protein NSP3 surrogate PABPs by binding to viral 3′-end tail (GACC tail instead of AAAA tail in RV mRNAs). NSP3 and RoXaN are two RV proteins that bind to eIF4G to promote viral mRNAs translation.
Rotavirus NPS3 usurps PABPs by working as a surrogate of PABPs to evict them from the translation initiation complex and redistributing the cytoplasmic PABPs into the nucleus. This redirect of PABPs requires the interaction of NSP3 with eIF4G and RoXaN (rotavirus X protein associated with NSP3) (101, 102). Another research showed that the nuclear localization of PABPs by NSP3 upon rotavirus infection simultaneously happens with the altered splicing of the stress-related transcription factor XBP1, and the eIF4G-binding domain of NSP3 is required for this process (103). XBP1 is a cellular factor involved in innate immunity and stress response (104). During infections, it could be possible that NSP3 evicts PABPs from eIF4G change the conformation of PABPs or eIF4G, thus exposing the nuclear import signal of cytoplasmic PABPs (103), and PABPs are relocalized into the nuclear to interact with XBP1 pre-RNA, causing retention of transcripts. Alter splicing of XBP1 could be a specific host response to rotavirus infection or virus cause nuclear localization of PABPs. Despite that NSP3 displaces PABPs' roles in host protein translation initiation, the function or mechanism of rotavirus NSP3 needs to be elucidated for further understanding.
(ii) Nodaviridae.
Nervous necrosis virus (NNV) is classified as the Betanodavirus genus of the Nodaviridae family. NNV has a positive, single-stranded RNA containing a 5′-end cap but lacks a 3′-end poly(A) tail. It is a pathogen causing viral nervous necrosis disease in marine fish, larvae, and juveniles (105). Chang et al. reported that NNV infections induced host shutoff by degrading host cytoplasmic PABP sand translocating it into the nucleus (106). The NNV sequestered PABPs in the infected nucleus via interaction with its coat protein (106). This interaction requires the N-terminal shell domain of NNV coat protein and proline-rich linker region of PABPs (106). This mechanism of host shutoff is also seen with RV (58). Moreover, PABPs were degraded up to the RRM3 domain at a later infection stage through the ubiquitin-proteasome system, distinct from viruses like picornaviruses, calicivirus, or other viruses retrovirus, which carry proteolytic enzymes to degrade PABPs. It is a relatively new mechanism found in manipulating PABPs during viral infection.
Plant RNA viruses.
The studies between plant viruses and PABPs are limited. Despite some of the PABPs structures in plant cells may be distinct from human or animal cells, it was targeted by plant viruses to affect efficient translation initiation. In some plant RNA viruses, their viral mRNAs lack both 5′-end cap and/or 3′-end poly(A) tail. Its mRNAs have cap-independent translation enhancer elements (CITEs) located in the 3′ UTR, mediating their translation. For instance, in red clover necrotic mosaic virus (RCNMV), A-rich sequence (ARS) and 3′-end CITEs are substitutes for the 3′-end poly(A) tail and the 5′-end cap of eukaryotic mRNAs for translation initiation (107). PABPs bind to ARS in the viral 3′ UTR to facilitate translation of uncapped nonpolyadenylated viral mRNAs (107). Some plant viruses use IRES for translation initiation, like the picornavirus or calicivirus. However, unlike many IRES elements, the crucifer-infecting tobamovirus (CrTMV) contains a polypurine tract rather than a pyrimidine-rich tract for PABPs binding activities (108). For example, PABPs interact with the polypurine tract of CrTMV IRES in the presence of poly(A) tail to enhance IRES activity (108). In tobacco etch virus (TEV), PABPs play important roles in the kinetics and stability of eIF4F binding to IRES, thus facilitating viral mRNAs translation initiations (109). VPg of turnip mosaic virus (TuMV) plays an important role in the cap-independent initiation of viral protein synthesis by interacting with PABPs and eIF4B to eIF4F, which resembles what happens in calicivirus (110, 111).
DNA viruses.
RNA viruses manipulate PABPs in diverse manners as the structures of viral mRNAs are diverse. DNA viruses have double-stranded DNA, and their viral transcripts are both capped and polyadenylated; therefore, they are more likely to employ the canonical cap-dependent translation initiation way for their replications.
Poxviridae.
Poxvirus genera is a large, complex, enveloped virus belonging to the Poxviridae family. Unlike herpesviruses, which take advantage of the host DNA and RNA polymerases, poxvirus could replicate in the cytosol as it encodes itself DNA and RNA polymerases. Poxvirus replicates in the cytoplasmic compartments known as DNA factories, containing viral mRNA, RNA-binding proteins, ribosomal proteins, virally transcription factors, and cellular translation initiation factors eIF4E and eIF4G (112). Poxvirus and herpesvirus mRNAs are translated via the classic cellular cap-dependent route. Vaccinia virus (VV), a model of the poxvirus, utilized its viral factors to inactivate the eIF4E binding protein (4E-BP) (113). 4E-BP is a translational repressor that sequesters eIF4E from its association with eIF4G, a core bridge between PABPs and eIF4F complex (114). VV negatively regulate eIF4F to suppress cellular cap-dependent mechanism to translate their mRNAs by destroying 4E-BP (113). Moreover, in VV infected cells, eIF4E, eIF4G, and PABPs are redistributed to the distinct viral replication compartments in the cytoplasm (113). VV hijacks of the host translation apparatus to DNA factories enhance the efficiency of viral replication and thwart cellular protein synthesis.
Herpesviridae.
Herpesviridae family comprises three subfamilies, including the Alphaherpesvirinae, Betaherpesvirinae, and Gammaherpesvirinae. Alphaherpesvirus and betaherpesvirus are represented by herpes simplex virus 1 (HSV-1) and human cytomegalovirus (HCMV), respectively, whereas Kaposi’s sarcoma-associated herpesvirus (KSHV) and Epstein-Barr virus (EBV) are two models of a gammaherpesvirus. HSV-1 and KSHV induce host shutoff gene expression while HCMV does not. Besides this, they develop both similar and distinct mechanisms to ensure efficient viral mRNAs translation initiation.
Alphaherpesvirus.
PABPs could shuttle between the cytoplasm and the nucleus under the assist of many viruses even without a typical nuclear export or import signal. For instance, PABPs are associated with HSV-1 ICP27 protein. HSV-1 immediately early gene-encoded protein ICP27 is a nucleo-cytoplasmic shuttling protein involved in transcriptional and posttranscriptional regulation of viral and cellular gene expression (115). Gromeier and colleagues investigated the significance of ICP27-PABP interaction. They reported that in HSV-1 ICP27 alone expressed HeLa cells, PABPs were largely relocated into the nucleus without affecting the expression levels and integrity of PABPs or their associating translation initiation factors (116). Moreover, the interaction of ICP27 and UL47 decreased PABPs interactions to their physiological binding partner eIF4G and Paip2 (116). It is unknown whether the dissociation of eIF4G or Paip2 from PABPs could also result from PABPs redistribution to the nucleus, causing fewer cytoplasmic PABPs ready for efficient translational initiation.
Another group investigated the ICP27-null mutant and discovered that ICP27 does not influence the relocalization of PABPs into the nucleus, suggesting that other HSV-1 encoded viral factors may influence PABPs relocalization (117). It is argued that redistribution of PABPs into the nucleus was vhs dependent (118). Again, more work needs to be done on understanding how HSV-1 manipulates PABPs during infection. Although both ICP27 and UL47 are RNA-binding proteins, abundant nuclear PABPs seem more convenient for interacting with viral mRNA poly(A) tails in the nucleus and facilitating viral mRNAs export from the nuclei. These findings suggest that HSV-1 targets PABPs by changing its subcellular localization and disrupting its binding to the translational initiation complex. Gray and colleagues found that HSV-1 ICP27 also form a complex with PABPs and eIF4G to promote small ribosomal subunit recruitment for viral mRNAs translation purposes independently (119), suggesting PABP-eIF4G interaction is not only targeted for mRNA repression but also activation. Besides, similar to VV, HSV-1 infection also enhances eIF4F assembly by phosphorylation and proteasome-mediated degradation of the 4E-BP, thus negatively regulating eIF4F complex formation (120), which is similar to what happens in VV infected cells. Consequently, viruses may also target PABPs to affect viral or cellular mRNA processing and transport in addition to mRNA translation initiation. More viral and cellular factors related to this process await investigations to propose a clear and intact model to decipher how ICP27 or other associated proteins commandeer PABPs or other translation initiation factors during HSV-1 infections.
HSV-2 is another alphaherpesvirus that might utilize PABPs during infections. However, the interaction between PABPs and HSV-2 encoding factors is not clear. Viral protein vhs is required to disrupt SGs, but the abundance of PABPs is not changed in SGs of infected cells (121). Whether PABPs or their binding partner were affected by vhs or other viral factors is not clear, and future investigations are needed to explore if vhs interact with PABPs to disrupt SGs integrity.
Betaherpesvirus.
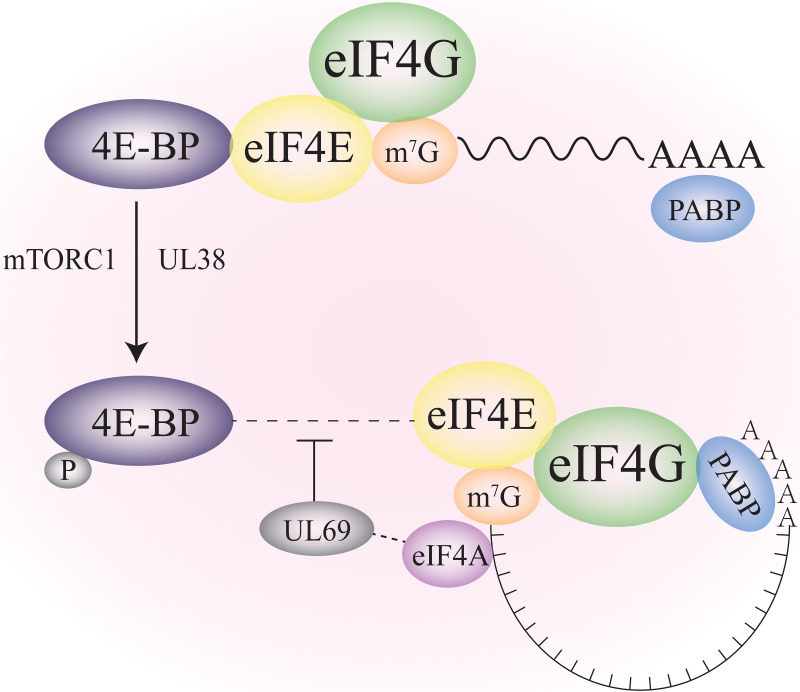
In HSV-1, HCMV, and KSHV-infected cells, viral mRNAs compete with host mRNAs for translation by stimulating the assembly of cellular translation initiation factors eIF4F (122, 123). However, only HCMV increases host eIF4F components (eIF4E, eIF4G, and eIF4A) and cytoplasmic PABPs levels, and this process requires the inactivation of cellular translational repressor 4E-BP (124). HSV-1 and KSHV exclude PABPs from the eIF4F complex by restricting them in the nucleus (116, 125). By contrast, HCMV manipulates PABPs in a cytoplasmic-dependent manner. PABPs in HCMV infected cells aggregate in the cytoplasm and are sensitive to rapamycin complex 1 (mTORC1)-mediated inhibition of 4E-BP phosphorylation (Fig. 4) (124). HCMV UL38 preserves the role of mTORC1 activity to inactivate 4E-BP. mTORC1 kinase mediates the inactivation of 4E-BP, the inactivated 4E-BP resulted in the release of eIF4E, leading to more efficient eIF4E-eIF4G interaction for more efficient translation initiation (122, 126, 127). Finally, the accumulated PABPs in the cytoplasm are ready to bind to eIF4F and promote efficient translation of viral mRNAs. Moreover, UL38 is an HCMV encoded target of mTORC1 activator, which translationally increases PABPs synthesis by regulating terminal oligopyrimidine (TOP) elements found on PABP mRNA 5′ UTR (127). The HCMV-induced PABPs accumulation promotes eIF4F assembly, thus increasing host translation initiation factors to foster viral protein synthesis and stimulate virus replication (127). Another HCMV UL69 protein also antagonizes the active state of 4E-BP by a different mechanism. UL69 interacts directly with eIF4A, excluding 4E-BP from the cap-binding complex and PABPs (128). In the meantime, the host is trying to restrict HCMV by increasing Paip2 levels (129). Paip2 is a gene expression repressor that is tightly regulated with PABPs. Despite the cellular Paip2 functioning as an innate defense to impede virus propagation, HCMV could sufficiently increase PABP levels to overcome its repression caused by Paip2 (129).VV, HSV-1, and HCMV can inactivate 4E-BP to promote the initiation of viral mRNA, even exploiting different mechanisms. Thus, depending on these discoveries, it is still worth studying how other HSV-1 and HCMV proteins manipulate PABPs and propose a working model of different herpesviruses, which might be good antiviral targets for the future.
FIG 4.
Negative regulation of PABPs by HCMV proteins. 4E-BP, a translation inhibitor binding to eIF4E, regulates cellular translation initiation. HCMV excludes 4E-BP from the translation initiation complex to promote viral mRNA translation. HCMV UL38 disassociates 4E-BP from eIF4E by enhancing its phosphorylation through the mTORC1 signal, thus relieving the inhibition of translation initiations. UL38 also increases PABPs levels in the cytoplasm, assisting viral mRNA translations. HCMV UL69 excludes 4E-BP from cap-binding complex and PABPs by indirectly interacting with eIF4A.
Gammaherpesvirus.
Redistribution of PABPs into the nucleus is, at least in part, a typical method utilized by viruses to induce host shutoff. In KSHV, viral protein K10/10.1 may target PABPs for their redistribution into the infected nuclei, which would mediate host translation defects. However, the importance and mechanism of this interaction are not elucidated due to the unknown function of the K10/10.1 protein in KSHV (130). Additionally, KSHV ORF37 encodes a protein named SOX which strongly induces a host shutoff (131, 132). KSHV SOX was shown to possess both endonuclease and exonuclease activity and facilitates host shutoff by promoting cytoplasmic mRNA degradation, nuclear mRNA hyperadenylation, and their nuclear retention (133–137), causing retaining of host mRNAs in infected nuclei and mediating mRNA export defects (125, 137); The nuclear import signals of PABPs are masked when bind to RNA in the cytoplasm, and SOX is sufficient to mediate nuclear relocalization of PABPs by degrading mRNAs on PABPs, resulting in RNA-free PABPs imported into the nucleus (138).
Moreover, during KSHV lytic infections, an intronless viral long noncoding (lnc) RNA called polyadenylated nuclear (PAN) RNA is transcribed at high levels (139). SOX upregulates the accumulation of PAN RNA in the nucleus further to restrict PABPs in the nucleus (139). These measures taken by SOX are consistent with the host shutoff effect. In addition to that, PAN RNA might promote viral gene expression at the late stage of infections as knockdown of PAN RNA is associated with the reduced amount of virus released into cell culture media (139).
ORF57 induces the translocation of PABPs from the cytoplasm to the nucleus, similar to SOX protein (140). Moreover, KSHV ORF57 is responsible for the stability of viral PAN RNA structures (140, 141). PABPs inhibit PAN RNA accumulation by destabilizing its structure in the absence of ORF57. However, the N-terminus of ORF57 interacts with the RRM of PAPBs, colocalizing PABPs with nuclear PAN RNA and alleviating the negative effect of PAPBs on PAN (140). As the relocating nuclear PABPs interrupt mRNA nuclear export and cause subsequent hyperventilation, one of the functions of ORF57 is probably to prevent viral RNA from hyperadenylation in the nucleus.
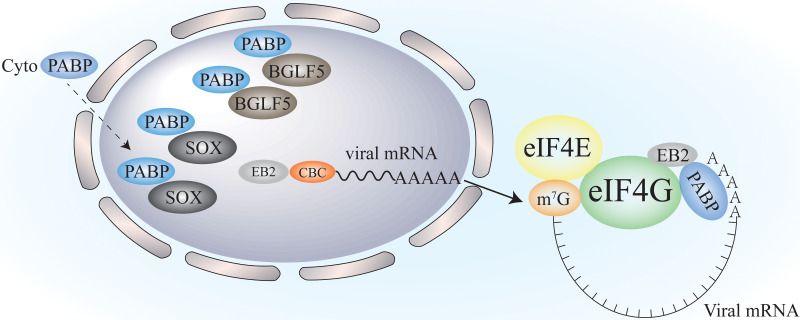
KSHV expressed another protein, ORF10, which directly targeted the nuclear export pathway by interacting with Rae1 (142), a highly conserved eukaryotic cellular export factor to inhibit nuclear mRNA exit. Export of mRNA-ribonucleoprotein (mRNP) complex requires the transit of them through the nucleopore complex (NPC), and Rae1 was involved in mRNA export by interacting with Nup98 at NPC, disrupting Rae1 and Nup98 complex working in nuclear mRNA export (133, 142). EBV and murine gamma-HV 68 (MHV68) encode homologs of KSHV SOX: BGLF5 and muSOX, respectively (143). KSHV SOX roles are mimicked by these two homologs suggesting the universal roles of KSHV SOX and the possible representative mechanism model in gammaherpesviruses (Fig. 5).
FIG 5.
Redistribution of cytoplasmic PABPs into the infected nucleus. In herpesviruses infected cells, viral proteins such as ICP27 and UL47 in HSV-1, SOX in KSHV, and BGLF5 in EBV are required to redistribute of PABPs into the nucleus. This process inhibits cellular translation by decreasing cytoplasmic PABPs. In addition, EBV EB2 protein promotes viral mRNA export from the nucleus and associate with PABP-eIF4F to promote translation initiation of viral mRNAs.
Epstein-Barr virus (EBV) encodes an RNA-binding protein EB2 to facilitate translation initiation of viral mRNA through association with PABP-eIF4F (144). EB2 protein is encoded to promote the recruitment of viral mRNAs in the cytoplasm. Most of the viral genes of herpesvirus are intronless, and herpesvirus requires a specific mechanism of mRNA export and translation for efficient expression of their non-intron mRNAs. EBV EB2 can shuttle between the nucleus and the cytoplasm and enhance the nuclear export of intronless viral mRNAs (145). Gruffat et al. provided a model showing how EB2 of EBV facilitates the efficient translation of viral mRNAs (144). They reported that EB2 (Fig. 5) first binds to mRNA's cap-binding complex (CBC) in the nucleus. After the formation of mature mRNAs, mRNPs are transported into the cytoplasm by CBC (144). The mature viral mRNAs then interacted with the N-terminus of eIF4G and PABPs to facilitate translation initiation in the cytoplasm (144). HSV-1 ICP27, HCMV UL69, and KSHV ORF57 are homologs of EBV EB2. They play important roles in viral mRNA translation, and their mechanisms are different and not fully explored. For example, KSHV ORF57 proteins recruited a cellular factor PYM to promote viral mRNAs translation, and the interaction between ORF57 and PYM links the 40S subunit of ribosomes and initiation factors (146). ICP27 of HSV-1 and UL69 from HCMV were both claimed to target PABPs and eIF4A/eIF4G to facilitate translation initiation as described above (119, 128, 147, 148). The Herpesviridae could relocate PABPs into the nucleus. However, it looks like only gammaherpesvirus would accelerate host mRNA decay to promote this process. The mechanism models of how the Herpesviridae target PABPs await further clarifications.
Summary and future perspectives.
The host mRNA translation apparatus has long been recognized as a base to attack during viral infections. PABPs are the number one targets as they play significant roles in translation initiation. Manipulating PABPs would impede host translation efficiency and hijack them to work for viral mRNA synthesis simultaneously. In general, there are a few ways to control PABPs activities.
Firstly, PABPs are cleaved by viral proteases. Enterovirus (PV and CV), Hepatovirus (HAV and HCV), Calicivirus (FCV and NV), Lentiviruses (HIV-1 and HIV-2) encoding proteases mediate proteolysis of PABPs and/or the eIF4G, which directly decrease the translation initiation efficiency. In addition, in PV and CV, PABPs were observed to facilitate viral RNA replication directly via an unknown mechanism. Secondly, PABPs are displaced from the translation initiation complex. For example, rotavirus NSP3 displaces PABPs in the interaction with eIF4G and evicts PABPs to mediate host shutoff. Thirdly, a virus may target PABPs for their presence in virus-induced host SGs via an unclear mechanism. More work is needed to understand how the virus regulates PABPs in SGs. Last but not least, PABPs are redistributed from the cytoplasm into the nucleus to shut off host translation initiation. Different viruses could use different strategies to achieve the same aim. For example, rotavirus and herpesvirus use different mechanisms to restrict PABPs in the nucleus.
Although similar measures are taken to target PABPs, the underlying mechanisms are disparate, and virus replication would be promoted differently (see Table 1). Most investigations on PABPs focus on it as an initiation factor, but it has additional roles in RNA metabolism, such as RNA splicing, RNA decay, and others. Whether these additional ways are served as targets for viral manipulations remains future investigations. Besides, are there more viral and cellular factors involved? More specifically, an overview map of how viruses target PABPs needs to be elucidated. In the meantime, the cytoplasmic PABPs are the most studied at present. Other members of the PABPs family are less explored during viral infections. The specific functions of other PABP family members are still vague and need to be further clarified during viral infection. According to the current research results, the roles of PABPs were investigated in cell lines and may vary from case to case. Organoids are recommended to mimic viral infections in actual circumstances to improve the consistency of the virology study and warranty fidelity.
TABLE 1.
Manipulation of PABPs by viruses
| Genome | Virus family | Virus | Mechanism |
|---|---|---|---|
| dsDNA | Herpesviridae | HSV-1 | Relocalization of PABPs into the nucleus and disrupt PABPs binding to eIF4G and Paip2 by ICP27 and UL47 interaction; promote viral mRNAs translation by ICP27-PABP-eIF4G complex |
| HCMV | Increase cytoplasmic PABPs levels and induce their accumulation to promote eIF4F assembly; UL38 and UL69 exclude 4E-BP from the cap-binding complex and PABPs to promote viral mRNAs translation initiation | ||
| KSHV | Induces host shutoff and relocalization of PABPs into the nucleus by SOX; ORF57 and PYM interaction link 40S subunit to facilitate viral mRNAs translation; ORF57 prevent viral RNA from hyperadenylation by interacting with PABPs to stabilize viral PAN RNA | ||
| EBV | BGLF5 redistribute PABPs into the nucleus; EB2 associate with PABP-eIF4F to promote translation initiation of viral mRNAs | ||
| (+)ssRNA | Picornaviridae | PV | Cleavage of eIF4G and PABPs by protease 2A and 3C |
| CV | Cleavage of PABPs by protease 3C and may increase PABPs levels by manipulating vimentin rearrangements |
||
| EMCV | Cleavage of PABPs between Q437 and G438 sites by protease 3C | ||
| Caliciviridae | FCV NV |
Cleavage of PABPs by 3C-like protease Cleavage of PABPs by 3C-like protease |
|
| Togaviridae | RV | Redistribute PABPs into the nucleus | |
| Flaviviridae | DENV | Recruit PABPs to viral 3′-UTR to activate its translation | |
| HCV | Cleavage of PABPs by protease 3C | ||
| HAV | Cleavage of PABPs by protease 3C | ||
| DHAV | Cleavage of PABPs between Q367 and G368 sites by protease 3C | ||
| Ateriviridae | PRRSV | N protein recruits PABPs accumulate at perinuclear sites of viral synthesis and sequesters it to inhibit cellular protein synthesis | |
| (-)ssRNA | Coronaviridae | TEGV | PABPs bind to viral genome poly(A) tail to increase virus propagation |
| BCoV | Inhibit interaction efficiency between poly(A) and eIF4F by N protein | ||
| Rhabdoviridae | RABV | May modify PABP containing SGs to promote viral replication | |
| dsRNA | Reoviridae | Rotavirus | NSP3 is a surrogate of PABPs that link RV mRNA to eIF4G for efficient translation initiation; redistributing PABPs into the nucleus needs NSP3/RoXaN interaction |
PABPs have roles in RNA metabolism, including cellular mRNA decay, mRNA deadenylation, mRNA export, mRNA surveillance. PABPs may regulate viral mRNAs identified by the innate immune system through pattern recognition receptors (PRRs) (149). How PABPs mediate, viral mRNAs metabolism is unrevealed. It reminds us that viruses may target PABPs to metabolize viral RNAs to escape PRRs arrest, suggesting PABPs’ possible roles in innate immunity, an undiscovered and promising area during viral infections.
Although the importance of PABPs during viral infections has been concerned in the last decades, their function in infected cells is not fully addressed in many aspects. With the progress of cell research technology, different views on the roles of PABPs during viral infection and the whole organism may appear. Here, we have outlined and discussed the current knowledge of the PABPs during viral infection and provided information for future study of PABPs.
ACKNOWLEDGMENTS
Our work is supported by the Natural Sciences Foundation of Inner Mongolia Autonomous Region of China grant 2021BS03013, the Inner Mongolia University High-level Talents Award 10000-21311201/057. We apologize to colleagues whose work could not be cited because of length restrictions.
Contributor Information
Chunfu Zheng, Email: zheng.alan@hotmail.com.
Rozanne M. Sandri-Goldin, University of California, Irvine
REFERENCES
- 1.Pestova TV, Lorsch JR, Hellen CUT. 2007. The mechanism of translation initiation in eukaryotes, p 87–128. In Mathews MB, Sonenberg N, Hershey JWB (ed), Translational control in biology and medicine. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, NY. [Google Scholar]
- 2.Kahvejian A, Roy G, Sonenberg N. 2001. The mRNA closed-loop model: the function of PABP and PABP-interacting proteins in mRNA translation. Cold Spring Harbor Symp Quant Biol 66:293–300. 10.1101/sqb.2001.66.293. [DOI] [PubMed] [Google Scholar]
- 3.Smith RW, Gray NK. 2010. Poly(A)-binding protein (PABP): a common viral target. Biochem J 426:1–12. 10.1042/BJ20091571. [DOI] [PubMed] [Google Scholar]
- 4.Nostrand E, Freese P, Pratt GA, Wang X, Wei X, Blue SM, Dominguez D, Cody N, Olson S, Sundararaman B. 2017. A large-scale binding and functional map of human RNA binding proteins. Nature 583:711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morris RJ, Mcsherry E, Sebastian A. 1973. An adenylic acid-rich sequence in messenger RNA of HeLa cells and its possible relationship to reiterated sites in DNA. Harvard Business Rev 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blobel G. 1973. A protein of molecular weight 78,000 bound to the polyadenylate region of eukaryotic messenger RNAs. Proc Natl Acad Sci USA 70:924–928. 10.1073/pnas.70.3.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edmonds M, Vaughan MH, Nakazato H. 1971. Polyadenylic acid sequences in the heterogeneous nuclear RNA and rapidly-labeled polyribosomal RNA of HeLa cells: possible evidence for a precursor relationship. Proc Natl Acad Sci USA 68:1336–1340. 10.1073/pnas.68.6.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goss DJ, Kleiman FE. 2013. Poly(A) binding proteins: are they all created equal? Wiley Interdiscip Rev RNA 4:167–179. 10.1002/wrna.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicholson AL, Pasquinelli AE. 2019. Tales of detailed poly(a) tails. Trends Cell Biol 29:191–200. 10.1016/j.tcb.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laishram RS. 2014. Poly(A) polymerase (PAP) diversity in gene expression - Star-PAP vs canonical PAP. FEBS Lett 588:2185–2197. 10.1016/j.febslet.2014.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mangus DA, Evans MC, Jacobson A. 2003. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol 4:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Funakoshi Y, Doi Y, Hosoda N, Uchida N, Osawa M, Shimada I, Tsujimoto M, Suzuki T, Katada T, Hoshino S-i. 2007. Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev 21:3135–3148. 10.1101/gad.1597707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eliseeva IA, Lyabin DN, Ovchinnikov LP. 2013. Poly(A)-binding proteins: structure, domain organization, and activity regulation. Biochemistry (Mosc) 78:1377–1391. 10.1134/S0006297913130014. [DOI] [PubMed] [Google Scholar]
- 14.Smith RW, Blee TK, Gray NK. 2014. Poly(A)-binding proteins are required for diverse biological processes in metazoans. Biochem Soc Trans 42:1229–1237. 10.1042/BST20140111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorgoni B, Richardson WA, Burgess HM, Anderson RC, Wilkie GS, Gautier P, Martins J, Brook M, Sheets M, Gray NK. 2011. Poly(A)-binding proteins are functionally distinct and have essential roles during vertebrate development. Proc Natl Acad Sci USA 108:7844–7849. 10.1073/pnas.1017664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallie DR, Liu R. 2014. Phylogenetic analysis reveals dynamic evolution of the poly(A)-binding protein gene family in plants. BMC Evol Biol 14:238. 10.1186/s12862-014-0238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belostotsky DA. 2003. Unexpected complexity of poly(A)-binding protein gene families in flowering plants: three conserved lineages that are at least 200 million years old and possible auto- and cross-regulation. Genetics 163:311–319. 10.1093/genetics/163.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cosson B, Braun F, Paillard L, Blackshear P, Osborne HB. 2004. Identification of a novel Xenopus laevis poly (A) binding protein. Biol Cell 96:519–519. 10.1016/j.biolcel.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 19.A BC, A AC, A RLG, B JM, C SC, C GZ, A MP. 2002. Characterization of the poly(A) binding proteins expressed during oogenesis and early development of Xenopus laevis. Biology of the Cell 94:217–231. [DOI] [PubMed] [Google Scholar]
- 20.Voeltz GK, Ongkasuwan J, Standart N, Steitz JA. 2001. A novel embryonic poly(A) binding protein, ePAB, regulates mRNA deadenylation in Xenopus egg extracts. Genes Dev 15:774–788. 10.1101/gad.872201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu W, Kwon Y, Oko R, Hermo L, Hecht NB. 1995. Poly (A) binding protein is bound to both stored and polysomal mRNAs in the mammalian testis. Mol Reprod Dev 40:273–285. 10.1002/mrd.1080400303. [DOI] [PubMed] [Google Scholar]
- 22.Kleene KC, Wang MY, Cutler M, Hall C, Shih D. 1994. Developmental expression of poly(A) binding protein mRNAs during spermatogenesis in the mouse. Mol Reprod Dev 39:355–364. 10.1002/mrd.1080390403. [DOI] [PubMed] [Google Scholar]
- 23.Kühn U, Wahle E. 2004. Structure and function of poly(A) binding proteins. Biochim Biophys Acta 1678:67–84. 10.1016/j.bbaexp.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 24.Yang H, Duckett CS, Lindsten T. 1995. iPABP, an inducible poly(A)-binding protein detected in activated human T cells. Mol Cell Biol 15:6770–6776. 10.1128/MCB.15.12.6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kühn U, Pieler T. 1996. XenopusPoly(A) binding protein: functional domains in RNA binding and protein – protein interaction. J Mol Biol 256:20–30. 10.1006/jmbi.1996.0065. [DOI] [PubMed] [Google Scholar]
- 26.Burd CG, Matunis EL, Dreyfuss G. 1991. The multiple RNA-binding domains of the mRNA poly(A)-binding protein have different RNA-binding activities. Mol Cell Biol 11:3419–3424. 10.1128/mcb.11.7.3419-3424.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sachs AB, Bond MW, Kornberg RD. 1986. A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: domain structure and expression. Cell 45:827–835. 10.1016/0092-8674(86)90557-x. [DOI] [PubMed] [Google Scholar]
- 28.Adam SA, Nakagawa T, Swanson MS, Woodruff TK, Dreyfuss G. 1986. mRNA polyadenylate-binding protein: gene isolation and sequencing and identification of a ribonucleoprotein consensus sequence. Mol Cell Biol 6:2932–2943. 10.1128/mcb.6.8.2932-2943.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorgoni B, Gray NK. 2004. The roles of cytoplasmic poly(A)-binding proteins in regulating gene expression: a developmental perspective. Brief Funct Genomic Proteomic 2:125–141. 10.1093/bfgp/3.2.125. [DOI] [PubMed] [Google Scholar]
- 30.Kozlov G, Safaee N, Rosenauer A, Gehring K. 2010. Structural basis of binding of P-body-associated proteins GW182 and ataxin-2 by the Mlle domain of poly(A)-binding protein. J Biol Chem 285:13599–13606. 10.1074/jbc.M109.089540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gray NK, Hrabálková L, Scanlon JP, Smith R. 2015. Poly(A)-binding proteins and mRNA localization: who rules the roost? Biochem Soc Trans 43:1277–1284. 10.1042/BST20150171. [DOI] [PubMed] [Google Scholar]
- 32.Kozlov G, Siddiqui N, Coillet-Matillon S, Trempe JF, Ekiel I, Sprules T, Gehring K. 2002. Solution structure of the orphan PABC domain from Saccharomyces cerevisiae poly(A)-binding protein. J Biol Chem 277:22822–22828. 10.1074/jbc.M201230200. [DOI] [PubMed] [Google Scholar]
- 33.Burgess HM, Gray NK. 2010. mRNA-specific regulation of translation by poly(A)-binding proteins. Biochem Soc Trans 38:1517–1522. 10.1042/BST0381517. [DOI] [PubMed] [Google Scholar]
- 34.Brook M, Smith JW, Gray NK. 2009. The DAZL and PABP families: RNA-binding proteins with interrelated roles in translational control in oocytes. Reproduction 137:595–617. 10.1530/REP-08-0524. [DOI] [PubMed] [Google Scholar]
- 35.Roy G. 2008. Characterization of PABP-interacting proteins 1 and 2. Universitaire Pers Maastricht. [Google Scholar]
- 36.Derry MC, Yanagiya A, Martineau Y, Sonenberg N. 2006. Regulation of poly(A)-binding protein through PABP-interacting proteins. Cold Spring Harbor Symp Quant Biol 71:537–543. 10.1101/sqb.2006.71.061. [DOI] [PubMed] [Google Scholar]
- 37.Khaleghpour K, Svitkin YV, Craig AW, DeMaria CT, Deo RC, Burley SK, Sonenberg N. 2001. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol Cell 7:205–216. 10.1016/s1097-2765(01)00168-x. [DOI] [PubMed] [Google Scholar]
- 38.Kozlov G, Ménade M, Rosenauer A, Long N, Gehring K. 2010. Molecular determinants of PAM2 recognition by the MLLE domain of poly(A)-binding protein. J Mol Biol 397:397–407. 10.1016/j.jmb.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 39.Guennadi K, Kalle G, Petri K. 2010. Molecular basis of eRF3 recognition by the MLLE domain of poly(A)-binding protein. PLoS One 5:e10169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roy G, De Crescenzo G, Khaleghpour K, Kahvejian A, O'Connor-McCourt M, Sonenberg N. 2002. Paip1 interacts with poly(A) binding protein through two independent binding motifs. Mol Cell Biol 22:3769–3782. 10.1128/MCB.22.11.3769-3782.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Q, Sharma NR, Zheng ZM, Chen M. 2019. Viral regulation of RNA granules in infected cells. Virol Sin 34:175–191. 10.1007/s12250-019-00122-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lai MM, Cavanagh D. 1997. The molecular biology of coronaviruses. Adv Virus Res 48:1–100. 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galán C, Sola I, Nogales A, Thomas B, Akoulitchev A, Enjuanes L, Almazán F. 2009. Host cell proteins interacting with the 3' end of TGEV coronavirus genome influence virus replication. Virology 391:304–314. 10.1016/j.virol.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lo CY, Tsai TL, Lin CN, Lin CH, Wu HY. 2019. Interaction of coronavirus nucleocapsid protein with the 5'- and 3'-ends of the coronavirus genome is involved in genome circularization and negative-strand RNA synthesis. FEBS J 286:3222–3239. 10.1111/febs.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spagnolo JF, Hogue BG. 2000. Host protein interactions with the 3' end of bovine coronavirus RNA and the requirement of the poly(A) tail for coronavirus defective genome replication. J Virol 74:5053–5065. 10.1128/jvi.74.11.5053-5065.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsai TL, Lin CH, Lin CN, Lo CY, Wu HY. 2018. Interplay between the poly(A) tail, poly(A)-binding protein, and coronavirus nucleocapsid protein regulates gene expression of coronavirus and the host cell. J Virol 92. 10.1128/JVI.01162-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Madhugiri R, Fricke M, Marz M, Ziebuhr J. 2016. Coronavirus cis-acting RNA elements. PLoS Adv Virus Res. 96:127–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu B, Zeng LP, Yang XL, Ge XY, Zhang W, Li B, Xie JZ, Shen XR, Zhang YZ, Wang N, Luo DS, Zheng XS, Wang MN, Daszak P, Wang LF, Cui J, Shi ZL. 2017. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog 13:e1006698. 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang XL, Hu B, Wang B, Wang MN, Zhang Q, Zhang W, Wu LJ, Ge XY, Zhang YZ, Daszak P, Wang LF, Shi ZL. 2015. Isolation and characterization of a novel bat coronavirus closely related to the direct progenitor of severe acute respiratory syndrome coronavirus. J Virol 90:3253–3256. 10.1128/JVI.02582-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, Poon LL, Guan Y, Rozanov M, Spaan WJ, Gorbalenya AE. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol 331:991–1004. 10.1016/s0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lei J, Ma-Lauer Y, Han Y, Thoms M, Buschauer R, Jores J, Thiel V, Beckmann R, Deng W, Leonhardt H, Hilgenfeld R, von Brunn A. 2021. The SARS-unique domain (SUD) of SARS-CoV and SARS-CoV-2 interacts with human Paip1 to enhance viral RNA translation. EMBO J 40:e102277. 10.15252/embj.2019102277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pfefferle S, Schöpf J, Kögl M, Friedel CC, Müller MA, Carbajo-Lozoya J, Stellberger T, von Dall'Armi E, Herzog P, Kallies S, Niemeyer D, Ditt V, Kuri T, Züst R, Pumpor K, Hilgenfeld R, Schwarz F, Zimmer R, Steffen I, Weber F, Thiel V, Herrler G, Thiel HJ, Schwegmann-Wessels C, Pöhlmann S, Haas J, Drosten C, von Brunn A. 2011. The SARS-coronavirus-host interactome: identification of cyclophilins as target for pan-coronavirus inhibitors. PLoS Pathog 7:e1002331. 10.1371/journal.ppat.1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kozlov G, Trempe JF, Khaleghpour K, Kahvejian A, Ekiel I, Gehring K. 2001. Structure and function of the C-terminal PABC domain of human poly(A)-binding protein. Proc Natl Acad Sci USA 98:4409–4413. 10.1073/pnas.071024998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanaka T, Kamitani W, DeDiego ML, Enjuanes L, Matsuura Y. 2012. Severe acute respiratory syndrome coronavirus nsp1 facilitates efficient propagation in cells through a specific translational shutoff of host mRNA. J Virol 86:11128–11137. 10.1128/JVI.01700-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thoms M, Buschauer R, Ameismeier M, Koepke L, Denk T, Hirschenberger M, Kratzat H, Hayn M, Mackens-Kiani T, Cheng J, Straub JH, Stürzel CM, Fröhlich T, Berninghausen O, Becker T, Kirchhoff F, Sparrer KMJ, Beckmann R. 2020. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science 369:1249–1255. 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schubert K, Karousis ED, Jomaa A, Scaiola A, Echeverria B, Gurzeler LA, Leibundgut M, Thiel V, Mühlemann O, Ban N. 2020. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat Struct Mol Biol 27:959–966. 10.1038/s41594-020-0511-8. [DOI] [PubMed] [Google Scholar]
- 57.Wang X, Bai J, Zhang L, Wang X, Li Y, Jiang P. 2012. Poly(A)-binding protein interacts with the nucleocapsid protein of porcine reproductive and respiratory syndrome virus and participates in viral replication. Antiviral Res 96:315–323. 10.1016/j.antiviral.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 58.Ilkow CS, Mancinelli V, Beatch MD, Hobman TC. 2008. Rubella virus capsid protein interacts with poly(a)-binding protein and inhibits translation. J Virol 82:4284–4294. 10.1128/JVI.02732-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aragón T, de la Luna S, Novoa I, Carrasco L, Ortín J, Nieto A. 2000. Eukaryotic translation initiation factor 4GI is a cellular target for NS1 protein, a translational activator of influenza virus. Mol Cell Biol 20:6259–6268. 10.1128/MCB.20.17.6259-6268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hale BG, Randall RE, Ortín J, Jackson D. 2008. The multifunctional NS1 protein of influenza A viruses. J Gen Virol 89:2359–2376. 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- 61.de la Luna S, Fortes P, Beloso A, Ortín J. 1995. Influenza virus NS1 protein enhances the rate of translation initiation of viral mRNAs. J Virol 69:2427–2433. 10.1128/JVI.69.4.2427-2433.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burgui I, Aragón T, Ortín J, Nieto A. 2003. PABP1 and eIF4GI associate with influenza virus NS1 protein in viral mRNA translation initiation complexes. J Gen Virol 84:3263–3274. 10.1099/vir.0.19487-0. [DOI] [PubMed] [Google Scholar]
- 63.Nikolic J, Civas A, Lama Z, Lagaudrière-Gesbert C, Blondel D. 2016. Rabies Virus Infection Induces the Formation of Stress Granules Closely Connected to the Viral Factories. PLoS Pathog 12:e1005942. 10.1371/journal.ppat.1005942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ogino M, Ito N, Sugiyama M, Ogino T. 2016. The Rabies Virus L Protein Catalyzes mRNA Capping with GDP Polyribonucleotidyltransferase Activity. Viruses 8:144. 10.3390/v8050144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ma S, Bhattacharjee RB, Bag J. 2009. Expression of poly(A)-binding protein is upregulated during recovery from heat shock in HeLa cells. FEBS J 276:552–570. 10.1111/j.1742-4658.2008.06803.x. [DOI] [PubMed] [Google Scholar]
- 66.Martínez-Salas E, Francisco-Velilla R, Fernandez-Chamorro J, Lozano G, Diaz-Toledano R. 2015. Picornavirus IRES elements: RNA structure and host protein interactions. Virus Res 206:62–73. 10.1016/j.virusres.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 67.Joachims M, Breugel PCV, Lloyd RE. 1999. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J Virol 73:718–727. 10.1128/JVI.73.1.718-727.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bo Z, Graziella M, Verena GM, Yuri K. 2007. Poly(A) binding protein, C-terminally truncated by the hepatitis A virus proteinase 3C, inhibits viral translation. Nucleic Acids Res 35:5975–5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kerekatte V, Keiper BD, Badorff C, Cai A, Knowlton KU, Rhoads RE. 1999. Cleavage of poly(A)-binding protein by coxsackievirus 2A protease in vitro and in vivo: another mechanism for host protein synthesis shutoff? J Virol 73:709–717. 10.1128/JVI.73.1.709-717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kuyumcu-Martinez NM, Joachims M, Lloyd RE. 2002. Efficient cleavage of ribosome-associated poly(A)-binding protein by enterovirus 3C protease. J Virol 76:2062–2074. 10.1128/jvi.76.5.2062-2074.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kuyumcu-Martinez NM, Van Eden ME, Younan P, Lloyd RE. 2004. Cleavage of poly(A)-binding protein by poliovirus 3C protease inhibits host cell translation: a novel mechanism for host translation shutoff. Mol Cell Biol 24:1779–1790. 10.1128/MCB.24.4.1779-1790.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun D, Wang M, Wen X, Cheng A, Jia R, Sun K, Yang Q, Wu Y, Zhu D, Chen S, Liu M, Zhao X, Chen X. 2017. Cleavage of poly(A)-binding protein by duck hepatitis A virus 3C protease. Sci Rep 7:16261. 10.1038/s41598-017-16484-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kobayashi M, Arias C, Garabedian A, Palmenberg AC, Mohr I. 2012. Site-specific cleavage of the host poly(A) binding protein by the encephalomyocarditis virus 3C proteinase stimulates viral replication. J Virol 86:10686–10694. 10.1128/JVI.00896-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lamphear BJ, Kirchweger R, Skern T, Rhoads RE. 1995. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem 270:21975–21983. 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 75.Svitkin YV, Costa-Mattioli M, Herdy B, Perreault S, Sonenberg N. 2007. Stimulation of picornavirus replication by the poly(A) tail in a cell-free extract is largely independent of the poly(A) binding protein (PABP). RNA 13:2330–2340. 10.1261/rna.606407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bergamini G, Preiss T, Hentze MW. 2000. Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA 6:1781–1790. 10.1017/s1355838200001679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ohlmann T, Rau M, Morley SJ, Pain VM. 1995. Proteolytic cleavage of initiation factor eIF-4 gamma in the reticulocyte lysate inhibits translation of capped mRNAs but enhances that of uncapped mRNAs. Nucleic Acids Res 23:334–340. 10.1093/nar/23.3.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Herold J, Andino R. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol Cell 7:581–591. 10.1016/s1097-2765(01)00205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Belsham GJ, McInerney GM, Ross-Smith N. 2000. Foot-and-mouth disease virus 3C protease induces cleavage of translation initiation factors eIF4A and eIF4G within infected cells. J Virol 74:272–280. 10.1128/jvi.74.1.272-280.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kirchweger R, Ziegler E, Lamphear BJ, Waters D, Liebig HD, Sommergruber W, Sobrino F, Hohenadl C, Blaas D, Rhoads RE. 1994. Foot-and-mouth disease virus leader proteinase: purification of the Lb form and determination of its cleavage site on eIF-4 gamma. J Virol 68:5677–5684. 10.1128/JVI.68.9.5677-5684.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chau DH, Yuan J, Zhang H, Cheung P, Lim T, Liu Z, Sall A, Yang D. 2007. Coxsackievirus B3 proteases 2A and 3C induce apoptotic cell death through mitochondrial injury and cleavage of eIF4GI but not DAP5/p97/NAT1. Apoptosis 12:513–524. 10.1007/s10495-006-0013-0. [DOI] [PubMed] [Google Scholar]
- 82.Koudelka T, Boger J, Henkel A, Schönherr R, Krantz S, Fuchs S, Rodríguez E, Redecke L, Tholey A. 2021. N-terminomics for the identification of in vitro substrates and cleavage site specificity of the SARS-CoV-2 main protease. Proteomics 21:e2000246. 10.1002/pmic.202000246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Visser LJ, Medina GN, Rabouw HH, de Groot RJ, Langereis MA, de Los Santos T, van Kuppeveld FJM. 2019. Foot-and-mouth disease virus leader protease cleaves G3BP1 and G3BP2 and inhibits stress granule formation. J Virol 93. 10.1128/JVI.00922-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ye X, Pan T, Wang D, Fang L, Ma J, Zhu X, Shi Y, Zhang K, Zheng H, Chen H, Li K, Xiao S. 2018. Foot-and-mouth disease virus counteracts on internal ribosome entry site suppression by G3BP1 and inhibits G3BP1-mediated stress granule assembly via post-translational mechanisms. Front Immunol 9:1142. 10.3389/fimmu.2018.01142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fung G, Ng CS, Zhang J, Shi J, Wong J, Piesik P, Han L, Chu F, Jagdeo J, Jan E, Fujita T, Luo H. 2013. Production of a dominant-negative fragment due to G3BP1 cleavage contributes to the disruption of mitochondria-associated protective stress granules during CVB3 infection. PLoS One 8:e79546. 10.1371/journal.pone.0079546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Humoud MN, Doyle N, Royall E, Willcocks MM, Sorgeloos F, van Kuppeveld F, Roberts LO, Goodfellow IG, Langereis MA, Locker N. 2016. Feline calicivirus infection disrupts assembly of cytoplasmic stress granules and induces G3BP1 cleavage. J Virol 90:6489–6501. 10.1128/JVI.00647-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Alhatlani B, Vashist S, Goodfellow I. 2015. Functions of the 5' and 3' ends of calicivirus genomes. Virus Res 206:134–143. 10.1016/j.virusres.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Goodfellow I. 2011. The genome-linked protein VPg of vertebrate viruses - a multifaceted protein. Curr Opin Virol 1:355–362. 10.1016/j.coviro.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kuyumcu-Martinez M, Belliot G, Sosnovtsev SV, Chang KO, Green KY, Lloyd RE. 2004. Calicivirus 3C-like proteinase inhibits cellular translation by cleavage of poly(A)-binding protein. J Virol 78:8172–8182. 10.1128/JVI.78.15.8172-8182.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Monette A, Valiente-Echeverría F, Rivero M, Cohen ÉA, Lopez-Lastra M, Mouland AJ. 2013. Dual mechanisms of translation initiation of the full-length HIV-1 mRNA contribute to gag synthesis. PLoS One 8:e68108. 10.1371/journal.pone.0068108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Berlioz C, Darlix JL. 1995. An internal ribosomal entry mechanism promotes translation of murine leukemia virus gag polyprotein precursors. J Virol 69:2214–2222. 10.1128/JVI.69.4.2214-2222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ohlmann T, Lopez-Lastra M, Darlix JL. 2000. An internal ribosome entry segment promotes translation of the simian immunodeficiency virus genomic RNA. J Biol Chem 275:11899–11906. 10.1074/jbc.275.16.11899. [DOI] [PubMed] [Google Scholar]
- 93.Buck CB, Shen X, Egan MA, Pierson TC, Walker CM, Siliciano RF. 2001. The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J Virol 75:181–191. 10.1128/JVI.75.1.181-191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Castelló A, Franco D, Moral-López P, Berlanga JJ, Alvarez E, Wimmer E, Carrasco L. 2009. HIV- 1 protease inhibits Cap- and poly(A)-dependent translation upon eIF4GI and PABP cleavage. PLoS One 4:e7997. 10.1371/journal.pone.0007997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Alvarez E, Castelló A, Menéndez-Arias L, Carrasco L. 2006. HIV protease cleaves poly(A)-binding protein. Biochem J 396:219–226. 10.1042/BJ20060108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang Y, Wang Z. 2019. IRES-mediated cap-independent translation, a path leading to hidden proteome. J Mol Cell Biol 11:911–919. 10.1093/jmcb/mjz091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gratia M, Sarot E, Vende P, Charpilienne A, Baron CH, Duarte M, Pyronnet S, Poncet D. 2015. Rotavirus NSP3 is a translational surrogate of the poly(A) binding protein-poly(A) complex. J Virol 89:8773–8782. 10.1128/JVI.01402-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Poncet D. 2003. Translation of rotavirus mRNAs in infected cell. Perspectives in Medical Virology 9:185–205. 10.1016/S0168-7069(03)09012-8. [DOI] [Google Scholar]
- 99.Gratia M, Vende P, Charpilienne A, Baron HC, Laroche C, Sarot E, Pyronnet S, Duarte M, Poncet D. 2016. Challenging the roles of NSP3 and untranslated regions in rotavirus mRNA translation. PLoS One 11:e0145998. 10.1371/journal.pone.0145998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Deo RC, Groft CM, Rajashankar KR, Burley SK. 2002. Recognition of the rotavirus mRNA 3' consensus by an asymmetric NSP3 homodimer. Cell 108:71–81. 10.1016/s0092-8674(01)00632-8. [DOI] [PubMed] [Google Scholar]
- 101.Arnold MM, Brownback CS, Taraporewala ZF, Patton JT. 2012. Rotavirus variant replicates efficiently although encoding an aberrant NSP3 that fails to induce nuclear localization of poly(A)-binding protein. J Gen Virol 93:1483–1494. 10.1099/vir.0.041830-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Harb M, Becker MM, Vitour D, Baron CH, Vende P, Brown SC, Bolte S, Arold ST, Poncet D. 2008. Nuclear localization of cytoplasmic poly(A)-binding protein upon rotavirus infection involves the interaction of NSP3 with eIF4G and RoXaN. J Virol 82:11283–11293. 10.1128/JVI.00872-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Duarte M, Vende P, Charpilienne A, Gratia M, Laroche C, Poncet D. 2019. Rotavirus infection alters splicing of the stress-related transcription factor XBP1. J Virol 93. 10.1128/JVI.01739-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415:92–96. 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 105.Bandín I, Souto S. 2020. Betanodavirus and VER disease: A 30-year research review. Pathogens 9:106. 10.3390/pathogens9020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cheng C-A, Luo J-M, Chiang M-H, Fang K-Y, Li C-H, Chen C-W, Wang Y-S, Chang C-Y, Ou J-HJ. 2021. Nervous necrosis virus coat protein mediates host translation shutoff through nuclear translocalization and degradation of polyadenylate binding protein. J Virol 95:e02364-20. 10.1128/JVI.02364-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Iwakawa HO, Tajima Y, Taniguchi T, Kaido M, Mise K, Tomari Y, Taniguchi H, Okuno T. 2012. Poly(A)-binding protein facilitates translation of an uncapped/nonpolyadenylated viral RNA by binding to the 3' untranslated region. J Virol 86:7836–7849. 10.1128/JVI.00538-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Marom L, Hen-Avivi S, Pinchasi D, Chekanova JA, Belostotsky DA, Elroy-Stein O. 2009. Diverse poly(A) binding proteins mediate internal translational initiation by a plant viral IRES. RNA Biol 6:446–454. 10.4161/rna.6.4.8951. [DOI] [PubMed] [Google Scholar]
- 109.Yumak H, Khan MA, Goss DJ. 2010. Poly(A) tail affects equilibrium and thermodynamic behavior of tobacco etch virus mRNA with translation initiation factors eIF4F, eIF4B and PABP. Biochim Biophys Acta 1799:653–658. 10.1016/j.bbagrm.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 110.Khan MA, Goss DJ. 2018. PABP and eIF4B effects binding affinity and kinetic rates of genome linked viral protein (VPg) with eukaryotic initiation factor 4F. FASEB J 32. 10.1096/fasebj.2018.32.1_supplement.652.19. [DOI] [Google Scholar]
- 111.Khan MA, Goss DJ. 2019. Poly (A) binding protein enhances the binding affinity of potyvirus VPg to eukaryotic initiation factor eIF4F and activates in vitro translation. Int J Biol Macromol 121:947–955. 10.1016/j.ijbiomac.2018.10.135. [DOI] [PubMed] [Google Scholar]
- 112.Katsafanas GC, Moss B. 2007. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe 2:221–228. 10.1016/j.chom.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Walsh D, Arias C, Perez C, Halladin D, Escandon M, Ueda T, Watanabe-Fukunaga R, Fukunaga R, Mohr I. 2008. Eukaryotic translation initiation factor 4F architectural alterations accompany translation initiation factor redistribution in poxvirus-infected cells. Mol Cell Biol 28:2648–2658. 10.1128/MCB.01631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Moerke NJ, Aktas H, Chen H, Cantel S, Reibarkh MY, Fahmy A, Gross JD, Degterev A, Yuan J, Chorev M, Halperin JA, Wagner G. 2007. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell 128:257–267. 10.1016/j.cell.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 115.Sandri-Goldin RM. 2011. The many roles of the highly interactive HSV protein ICP27, a key regulator of infection. Future Microbiol 6:1261–1277. 10.2217/fmb.11.119. [DOI] [PubMed] [Google Scholar]
- 116.Dobrikova E, Shveygert M, Walters R, Gromeier M. 2010. Herpes simplex virus proteins ICP27 and UL47 associate with polyadenylate-binding protein and control its subcellular distribution. J Virol 84:270–279. 10.1128/JVI.01740-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Salaun C, MacDonald AI, Larralde O, Howard L, Lochtie K, Burgess HM, Brook M, Malik P, Gray NK, Graham SV. 2010. Poly(A)-binding protein 1 partially relocalizes to the nucleus during herpes simplex virus type 1 infection in an ICP27-independent manner and does not inhibit virus replication. J Virol 84:8539–8548. 10.1128/JVI.00668-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Elliott G, Pheasant K, Ebert-Keel K, Stylianou J, Franklyn A, Jones J. 2018. Multiple posttranscriptional strategies to regulate the herpes simplex virus 1 vhs endoribonuclease. J Virol 92:e00818-18. 10.1128/JVI.00818-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Smith RWP, Anderson RC, Larralde O, Smith JWS, Gorgoni B, Richardson WA, Malik P, Graham SV, Gray NK. 2017. Viral and cellular mRNA-specific activators harness PABP and eIF4G to promote translation initiation downstream of cap binding. Proc Natl Acad Sci USA 114:6310–6315. 10.1073/pnas.1610417114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Walsh D, Mohr I. 2006. Assembly of an active translation initiation factor complex by a viral protein. Genes Dev 20:461–472. 10.1101/gad.1375006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Finnen RL, Pangka KR, Banfield BW. 2012. Herpes simplex virus 2 infection impacts stress granule accumulation. J Virol 86:8119–8130. 10.1128/JVI.00313-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Walsh D, Perez C, Notary J, Mohr I. 2005. Regulation of the translation initiation factor eIF4F by multiple mechanisms in human cytomegalovirus-infected cells. J Virol 79:8057–8064. 10.1128/JVI.79.13.8057-8064.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Arias C, Walsh D, Harbell J, Wilson AC, Mohr I. 2009. Activation of host translational control pathways by a viral developmental switch. PLoS Pathog 5:e1000334. 10.1371/journal.ppat.1000334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Perez C, McKinney C, Chulunbaatar U, Mohr I. 2011. Translational control of the abundance of cytoplasmic poly(A) binding protein in human cytomegalovirus-infected cells. J Virol 85:156–164. 10.1128/JVI.01778-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lee YJ, Glaunsinger BA, Sugden B. 2009. Aberrant herpesvirus-induced polyadenylation correlates with cellular messenger RNA destruction. PLoS Biol 7:e1000107. 10.1371/journal.pbio.1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kudchodkar SB, Yu Y, Maguire TG, Alwine JC. 2004. Human cytomegalovirus infection induces rapamycin-insensitive phosphorylation of downstream effectors of mTOR kinase. J Virol 78:11030–11039. 10.1128/JVI.78.20.11030-11039.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.McKinney C, Perez C, Mohr I. 2012. Poly(A) binding protein abundance regulates eukaryotic translation initiation factor 4F assembly in human cytomegalovirus-infected cells. Proc Natl Acad Sci USA 109:5627–5632. 10.1073/pnas.1202829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Aoyagi M, Gaspar M, Shenk TE. 2010. Human cytomegalovirus UL69 protein facilitates translation by associating with the mRNA cap-binding complex and excluding 4EBP1. Proc Natl Acad Sci USA 107:2640–2645. 10.1073/pnas.0914856107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.McKinney C, Yu D, Mohr I. 2013. A new role for the cellular PABP repressor Paip2 as an innate restriction factor capable of limiting productive cytomegalovirus replication. Genes Dev 27:1809–1820. 10.1101/gad.221341.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Katano SH. 2006. Expression of Kaposi's sarcoma-associated herpesvirus-encoded K10/10.1 protein in tissues and its interaction with poly(A)-binding protein. Virology 352:100–109. [DOI] [PubMed] [Google Scholar]
- 131.Glaunsinger B, and, Ganem D. 2004. Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Mol Cell 13:713–723. 10.1016/S1097-2765(04)00091-7. [DOI] [PubMed] [Google Scholar]
- 132.Lee H, Patschull AOM, Bagnéris C, Ryan H, Sanderson CM, Ebrahimi B, Nobeli I, Barrett TE. 2017. KSHV SOX mediated host shutoff: the molecular mechanism underlying mRNA transcript processing. Nucleic Acids Res 45:4756–4767. 10.1093/nar/gkw1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Pardamean CI, Wu TT. 2021. Inhibition of host gene expression by KSHV: sabotaging mRNA stability and nuclear export. Front Cell Infect Microbiol 11:648055. 10.3389/fcimb.2021.648055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mendez AS, Vogt C, Bohne J, Glaunsinger BA. 2018. Site specific target binding controls RNA cleavage efficiency by the Kaposi's sarcoma-associated herpesvirus endonuclease SOX. Nucleic Acids Res 46:11968–11979. 10.1093/nar/gky932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Uppal T, Meyer D, Agarwal A, Verma SC. 2019. The DNase activity of Kaposi's sarcoma-associated herpesvirus SOX protein serves an important role in viral genome processing during lytic replication. J Virol 93. 10.1128/JVI.00977-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Glaunsinger B, Ganem D. 2004. Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Mol Cell 5:713–723. [DOI] [PubMed] [Google Scholar]
- 137.Kumar GR, Glaunsinger BA. 2010. Nuclear import of cytoplasmic poly(A) binding protein restricts gene expression via hyperadenylation and nuclear retention of mRNA. Mol Cell Biol 30:4996–5008. 10.1128/MCB.00600-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kumar GR, Shum L, Glaunsinger BA. 2011. Importin alpha-mediated nuclear import of cytoplasmic poly(A) binding protein occurs as a direct consequence of cytoplasmic mRNA depletion. Mol Cell Biol 31:3113–3125. 10.1128/MCB.05402-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Borah S, Darricarrère N, Darnell A, Myoung J, Steitz JA, Damania B. 2011. A viral nuclear noncoding RNA binds re-localized poly(A) binding protein and is required for late KSHV gene expression. PLoS Pathog 7:e1002300. 10.1371/journal.ppat.1002300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Massimelli MJ, Majerciak V, Kruhlak M, Zheng ZM. 2013. Interplay between polyadenylate-binding protein 1 and Kaposi's sarcoma-associated herpesvirus ORF57 in accumulation of polyadenylated nuclear RNA, a viral long noncoding RNA. J Virol 87:243–256. 10.1128/JVI.01693-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Majerciak V, Zheng ZM. 2015. KSHV ORF57, a protein of many faces. Viruses 7:604–633. 10.3390/v7020604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gong D, Kim YH, Xiao Y, Du Y, Xie Y, Lee KK, Feng J, Farhat N, Zhao D, Shu S, Dai X, Chanda SK, Rana TM, Krogan NJ, Sun R, Wu TT. 2016. A herpesvirus protein selectively inhibits cellular mRNA nuclear export. Cell Host Microbe 20:642–653. 10.1016/j.chom.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Covarrubias S, Richner JM, Clyde K, Lee YJ, Glaunsinger BA. 2009. Host shutoff is a conserved phenotype of gammaherpesvirus infection and is orchestrated exclusively from the cytoplasm. J Virol 83:9554–9566. 10.1128/JVI.01051-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mure F, Panthu B, Zanella-Cléon I, Delolme F, Manet E, Ohlmann T, Gruffat H. 2018. Epstein-Barr virus protein EB2 stimulates translation initiation of mRNAs through direct interactions with both poly(A)-binding protein and eukaryotic initiation factor 4G. J Virol 92. 10.1128/JVI.01917-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hiriart E, Farjot G, Gruffat H, Nguyen MV, Sergeant A, Manet E. 2003. A novel nuclear export signal and a REF interaction domain both promote mRNA export by the Epstein-Barr virus EB2 protein. J Biol Chem 278:335–342. 10.1074/jbc.M208656200. [DOI] [PubMed] [Google Scholar]
- 146.Boyne JR, Jackson BR, Taylor A, Macnab SA, Whitehouse A. 2010. Kaposi's sarcoma-associated herpesvirus ORF57 protein interacts with PYM to enhance translation of viral intronless mRNAs. EMBO J 29:1851–1864. 10.1038/emboj.2010.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Larralde O, Smith RW, Wilkie GS, Malik P, Gray NK, Clements JB. 2006. Direct stimulation of translation by the multifunctional herpesvirus ICP27 protein. J Virol 80:1588–1591. 10.1128/JVI.80.3.1588-1591.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fontaine-Rodriguez EC, Knipe DM. 2008. Herpes simplex virus ICP27 increases translation of a subset of viral late mRNAs. J Virol 82:3538–3545. 10.1128/JVI.02395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mogensen TH. 2009. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev 22:240–273. 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]