Abstract
Squalene synthase (farnesyl-diphosphate farnesyltransferase, EC 2.5.1.21) is the first committed enzyme of the sterol biosynthesis pathway. Inhibitors of this enzyme have been intensively studied as potential antifungal agents. To assess the effect of deactivating squalene synthase on the growth of fungi in mice, we isolated the squalene synthase (ERG9) gene from the pathogenic fungus Candida glabrata and generated strains in which the CgERG9 gene was under the control of the tetracycline-regulatable promoter. Depletion of the ERG9 gene by doxycycline (DOX), a derivative of tetracycline, decreased the cell viability in laboratory media, whereas it did not affect cell growth in mice at all. The growth defect caused by DOX in laboratory media was suppressed by the addition of serum. Analyses of the sterol composition of the restored cells in serum-containing media suggest that the defect of ergosterol biosynthesis can be complemented by the incorporation of exogenous cholesterol into the cells. Thus, deactivation of squalene synthase did not affect fungal growth in mice, presumably because the cells were able to incorporate cholesterol from the serum. These results showed that squalene synthase could not be a suitable target of antifungals for the treatment of C. glabrata infection.
The incidence of life-threatening fungal infections has been increasing, particularly among patients who are immunocompromised by human immunodeficiency virus infection and among those who are receiving immunosuppressive therapy for organ transplantation or chemotherapy for cancer. Current therapies against such infections rely on two main groups of drugs: polyenes, which disrupt membrane function, and azoles, which inhibit the synthesis of ergosterol (4, 5). The toxicity of polyenes, however, and the rapid appearance of azole-resistant strains have motivated us to discover and develop new antifungal drugs.
Sterol biosynthesis inhibitors are widely used not only in the medical field, including as treatments for systemic fungal infections, but also in the agricultural field. They are roughly classified into three groups according to their target molecules. One is the group of sterol 14α-demethylase inhibitors, including azoles, which lead to cell death by accumulation of the aberrant sterol 14α-methylergosta-8,24(28)-dien-3β,6α-diol (14, 15). Morpholines and piperidines constitute a second group of inhibitors that inhibit sterol Δ14-reductase and Δ8→7-isomerase. Treatment of fungal infections with these compounds results in fungal growth inhibition by accumulation of Δ8,14-sterol and a corresponding depletion of ergosterol (16). The third group, including allylamines, inhibits squalene epoxidase and causes depletion of ergosterol (30). Thus, sterol biosynthesis inhibitors have been well characterized. Nevertheless, two sterol biosynthesis inhibitor groups other than azoles have limited usefulness against fungal infections due to host toxicity for morpholines and undesirable pharmacokinetics for allylamines. Therefore, two approaches are thought to be necessary for the further development of sterol biosynthesis inhibitors as antifungal agents; one is to seek additional potential sites that are yet to be fully investigated. Another is to investigate the molecular responses of species resistant to such compounds.
Candida glabrata causes not only mucocutaneous but also deep-seated infections in transplant recipients and immunosuppressed patients (10, 31, 32). Recent data indicate a population shift to non-Candida albicans species, including C. glabrata, Candida krusei, and Candida tropicalis, whereas C. albicans is mainly isolated from immunocompromised patients (3). Among infections with Candida species other than C. Albicans, the incidence of C. glabrata infection has been increasing, mostly in conjunction with the use of azole antifungals (24–26). Furthermore, this organism always grows as a haploid yeast cell, which makes genetic manipulation of organisms such as Saccharomyces cerevisiae easy. Furthermore, functional analysis of a gene, including in a host, can be performed by using a tetracycline-regulatable expression system (23). Thus, C. glabrata is able to be an attractive experimental model to understand molecular responses of Candida species to antifungal compounds.
Squalene synthase (Erg9p) is the first enzyme branched out from other farnesyl pyrophosphate (FPP) derivatives, ubiquinones, dolichols, heme, and C15- or C20-isoprenoid chains. Loss of its function in an ortholog of S. cerevisiae leads to cell death (13) due to the defect of ergosterol biosynthesis. Therefore, inhibitors of this enzyme as candidates of antifungal compounds have been intensively pursued. The essential nature, however, of the ERG9 gene in C. glabrata has not been reported. To address this, we first isolated the ERG9 gene from this organism and investigated its essentiality, including in a host, by regulating its expression with a tetracycline-regulatable system (23). Contrary to our expectation based on its essentiality in vitro, depletion of the squalene synthase gene did not block the growth of C. glabrata in mice. We also showed several lines of evidence supporting the ability of C. glabrata to incorporate exogenous sterol from serum in aerobic conditions, suggesting that host cholesterol complements the defect of ergosterol biosynthesis in C. glabrata. These findings would provide new insight for exploiting target-driven drug discovery programs.
MATERIALS AND METHODS
Strains, growth media, and transformation.
The C. glabrata strains used in this study were ATCC 2001, ACG4 (his3 trp1 PScADH1::tetR::GAL4AD::TRP1), (23), and ERG9-controllable strain 97SQS (which is the same as ACG4 but 97t::ERG9 HIS3 mutant). The C. glabrata strains were grown at 37°C on yeast extract-peptone-dextrose (YEPD) complex medium containing 2% glucose, 2% Bacto Peptone (Difco Laboratories), and 1% yeast extract (Difco). YEPD agar plates contained 2% agar (Difco) as a supplement. Yeast nitrogen base (0.67% [Difco]) with 2% glucose and 2% agar (Difco) with appropriate amino acids and bases was used as the selective medium after transformation of ACG4. Yeast transformations were carried out by the modified lithium acetate method (6, 12). Escherichia coli DH5α was used as the host strain for all plasmid constructions and was grown on standard media.
Construction of plasmids and strains.
A genomic library was constructed by ligating 5- to 10-kb fragments of C. glabrata genomic DNA, which had been partially digested with Sau3AI, into the BamHI site of pRS415 (Stratagene). Genomic DNA was extracted from C. glabrata ATCC 2001 as described by Rose et al. (28). The construction of the plasmid p97ERG9 was generated by introducing region A (nucleotides [nt] −503 to −133) and region B (nt −6 to 314) of ERG9 into SacII/XbaI sites and EcoRI/SalI sites of p97CGH, respectively (23). Region A (nt −503 to −133) or region B (nt −6 to 314) of CgERG9 was amplified by PCR with the primer pair ERG9AF (5′-CAGTCTCCGCGGCCACAATGGACTCCGGG-3′) and ERG9AR (5′-ACAGCATCTAGAGGACTTCGAAGTTTATGCTC-3′) or primer pair ERG9BF (5′-AAAAATGAATTCATAACCATGGGTAAAGTACTTG-3′) and ERG9BR (5′-GGAGTCGTCGACACGCAACACTTTGACCTTC-3′), respectively. To replace the endogenous promoter with the tetracycline-regulatable promoter (97t) by homologous recombination, p97ERG9 linearized with SacII/SalI was used to transform ACG4, resulting in strain 97SQS.
Cloning and DNA sequencing of the C. glabrata ERG9 gene.
An approximately 0.4-kb fragment of the CgERG9 gene was amplified from C. glabrata genomic DNA by PCR with the pair of primers ERG9-2 (5′-TAYTGYCAYTAYGTIGCIGGIYTIGTIGG-3′) and ERG9-4RV (5′-ATIGCCATIACYTGIGGDATIGCRCARAA-3′), which were designed based on the sequence of the conserved region among S. cerevisiae, C. albicans, Schizosaccharomyces pombe, humans, and Arabidopsis thaliana (Fig. 1). The amplified fragment was cloned in pT7blue (Novagen), and its sequence was confirmed by the Sanger dideoxy chain termination method with M13 universal and reverse primers by using an ABI 377 sequencer. The full-length ERG9 gene was identified from the genomic library by screening with a 350-bp region of the central portion of this fragment as the probe, which was radiolabeled by the random priming method by using [α-32P]dCTP. The DNA probe was amplified by PCR with primers ERG9FW (5′-TGAATTGATTGTCCTTGCAGG-3′) and ERG9RV (5′-TGAGGATTGCTCGTGGATTG-3′). The DNA sequencing of the C. glabrata ERG9 gene was performed by the Sanger dideoxy chain termination method with M13 universal and reverse primers and synthetic oligonucleotides complemented to specific regions of CgERG9 using an ABI 377 sequencer.
FIG. 1.
Nucleotide and deduced amino acid sequence of CgERG9. Nucleotides and amino acids are numbered to the left and right. The CgERG9 open reading frame (ORF) begins at the base +1 and extends 1,329 nt to the stop codon beginning at position 1330. The region amplified by PCR with degenerate primers is boxed. Asterisk indicates termination of protein synthesis.
Investigation of the number of viable cells in several culture media.
Approximately 106 97SQS and ATCC 2001 cells were separately inoculated into YEPD medium with or without 10 μg of doxycycline (DOX) per ml. After 14 h of incubation at 37°C, the optical density at 660 nm was determined. The number of viable cells was determined by counting the number of colonies on an agar plate in which 20 μl of diluted cultures had been spread after incubation for 24 h at 37°C. For the time course experiments, approximately 105 97SQS cells were inoculated and cultured in YEPD medium at 37°C with or without 10 μg of DOX per ml. Their growth was monitored by measuring the optical density at 660 nm at indicated times after adding DOX. The number of viable cells was also determined as described above. For permeability experiments, approximately 103 ATCC 2001 and 97SQS cells were inoculated and cultured in YEPD medium at 37°C for 14 h, with or without 10 μg of DOX per ml, in the presence of the indicated concentrations of human serum (Irvine Scientific), human lipoprotein (Sigma), human lipoprotein-deficient serum (Sigma), 50 μg of squalene per ml (Sigma), and 25 μg of each of three sterols per ml (Sigma).
Determination of the number of viable C. glabrata cells in mice.
To generate immunocompromised mice, male CD-1 mice were treated as described previously (23). Each mouse was intravenously inoculated with 105 ATCC 2001 and 97SQS cells after having been given 5% of sucrose solution with or without 2 mg of DOX per ml as drinking water starting 2 days before the infection. On the indicated days, the mouse kidneys were removed and homogenized. The homogenates were spread onto YEPD plates containing penicillin G (200 U/ml) and streptomycin (200 μg/ml). The number of colonies that had appeared after culturing the cells for 24 h at 37°C was counted.
Measurement of squalene synthase activity.
Approximately 106 cells/ml were inoculated into YEPD medium or YEPD medium containing 5% (vol/vol) human serum. The cells were harvested after 8 h of incubation at 37°C with or without 10 μg of DOX per ml. Microsomal fractions were prepared from them as described previously (11). The harvested cells were suspended with buffer S (50 mM Tris-HCl [pH 7.5], 1 mM EDTA, 1 mM β-mercaptoethanol, 1 M sucrose, 1 mM phenylmethylsulfonyl fluoride). The suspension was vortexed with glass beads, and its extract was clarified by centrifugation. The supernatant was then centrifuged at 100,000 × g for 1 h. After removing the supernatant, the pellet was suspended with buffer A (25 mM Tris-HCl [pH 7.5], 0.5 mM EDTA, 1 mM β-mercaptoethanol, 30% [vol/vol] glycerol), and its extract was clarified by centrifugation. Protein concentrations of these microsome fractions were determined with a bicinchoninic acid protein assay kit (Pierce). The same volume of premix solution (400 mM phosphate-buffered saline, 10 mM MgCl2, 2 mM NADPH, 7.5 μM FPP, containing 3H-labeled FPP) was added to each microsomal fraction. After 1 h of incubation at 30°C, the reaction was terminated by adding 1/5 volume of ice-cold stop solution (0.5 M EDTA, 5% [wt/vol] KOH). These samples were washed with ice-cold water, filtered, and then dried. After that, the count was measured with a scintillation counter (TOP count; Amersham-Pharmacia).
Analyses of sterol composition of C. glabrata cells.
To prepare samples for analyzing sterol content, we modified the method described previously (8). Approximately 106 cells per ml were inoculated and cultured in YEPD medium or YEPD medium containing 10% (vol/vol) human serum at 37°C. The cells were harvested after 8 h of incubation with or without 10 μg of DOX per ml. They were washed with sterile water and lyophilized. The lipid fraction of each sample was extracted with 3 ml of CH2Cl2-MeOH (2:1) solution at 70°C. After drying, the lipid fractions were saponified with 6% (wt/vol) methanolic KOH for 1 h at 90°C. An equal volume of water was added to the saponified samples. After removal of the unsaponified fractions with 3 volumes of hexane, each sample was acetylated with toluene-Ac2O-pyridine (1:2:1 vol/vol/vol) for 16 h at room temperature and then evaporated; sterol compositions were analyzed with gas chromatograph-mass spectrometry (JEOL). We used 4,4-diphenyl-1-benzyl-piperidine as the internal control for the analyses. When the mass spectrum was measured, the chamber temperature was 200°C and ionization voltage was 30 eV. The gas chromatography was performed at 250°C with a 2% OV-17 (5 mm by 1.5 m) column, and its flow rate was 30 ml/min.
Nucleotide sequence accession numbers.
The DDBJ/EMBL/GenBank accession number for the sequence reported in this paper is AB009978 for CgERG9. The GenBank accession numbers for the previously determined nucleotide sequences of ERG9 of S. cerevisiae, C. albicans, S. pombe, humans, and A. thaliana are M63979, D89610, L06071, X69141, and D29017, respectively.
RESULTS
Isolation of the C. glabrata squalene synthase (ERG9) gene.
To isolate the C. glabrata ERG9 gene, we designed degenerate primers corresponding to the amino acid sequence in the highly conserved regions among five other species: S. cerevisiae (2, 13), C. albicans (N. Ishii, unpublished data), S. pombe, humans (27), and A. thaliana (22) (Fig. 1 and Materials and Methods). With these primers, we reproducibly amplified an approximately 400-bp fragment from C. glabrata genomic DNA (data not shown). The DNA sequence of this fragment was capable of encoding a polypeptide that was homologous to the other five species (Fig. 2). The central portion of this PCR fragment was used as a probe to obtain clones from the C. glabrata genomic library. Thirteen hybridization-positive clones were obtained from approximately 100,000 colonies, and their DNA sequences were determined by primer walking.
FIG. 2.
Alignment of deduced amino acid sequences for squalene synthases. Cg, C. glabrata; Sc, S. cerevisiae; Ca, C. albicans; Sp, S. pombe; Hs, human; and At, A. thaliana. Asterisks indicate amino acid residues conserved in the six species. Dots show conserved similar amino acid residues in the six species. Predicted kinase motifs are boxed. The arrows indicate the region that was used for designing the degenerated primers.
This C. glabrata open reading frame predicted a protein of 443 amino acids, and the sequence has 71.3% identity to S. cerevisiae Erg9p, 53.1% identity to C. albicans Erg9p, 37.7% identity to S. pombe Erg9p, 37.7% identity to human Erg9p, and 33.4% identity to A. thaliana Erg9p. Thus, the C. glabrata protein resembled most of the S. cerevisiae protein. Furthermore, C. glabrata Erg9p conserves the squalene and phytoene sequence motifs, Y(C/S/A)XXVA(G/A)XVG (positions 178 to 187) and (L/I/V/M)GXXXQXXNIXRD(L/I/V/M/F/Y)XX(D/E) (positions 217 to 231) (Fig. 1 and 2). Of numerous potential sites for phosphorylation present in the C. glabrata protein, two phosphorylation sites by casein kinase II and a single phosphorylation site by protein kinase C were also highly conserved. Concerning the sites for casein kinase II, positions 80 to 83 in C. glabrata were conserved in all six species' enzymes, and positions 171 to 174 in C. glabrata were conserved in all four yeast species. As to the site of protein kinase C phosphorylation, positions 49 to 51 in C. glabrata were conserved in five species, excluding the A. thaliana protein (Fig. 2). The hydrophobicity plot for the primary sequence of the C. glabrata protein exhibited a pattern similar to those of the other species (data not shown).
Investigation of the importance of the ERG9 gene for cell growth in laboratory media and in mice.
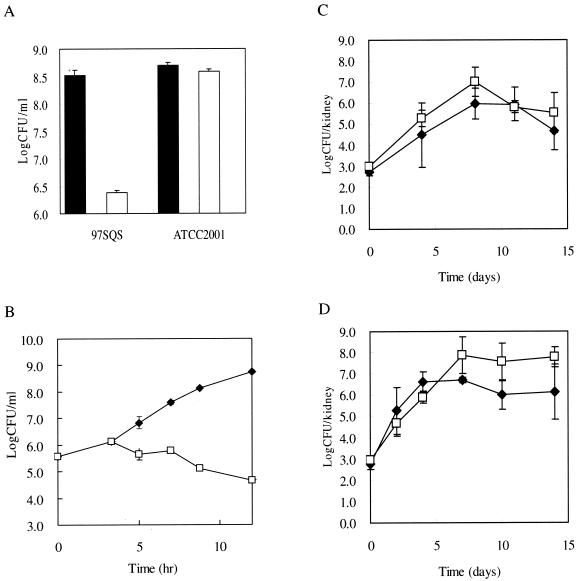
To assess the importance of squalene synthase for the fungal growth in mice, we generated ERG9-controllable strain 97SQS, in which the CgERG9 gene was controlled by the tetracycline-regulatable promoter 97t (23). The corrected replacement of the endogenous ERG9 promoter with 97t was confirmed by PCR diagnosis and Southern blot analysis (data not shown). First of all, we investigated whether or not depletion of the CgERG9 gene by DOX affects the growth of 97SQS cells. When 97SQS cells were cultured for 14 h in YEPD medium with DOX, a severe growth defect was observed (Fig. 3A). The same result was obtained with a defined medium (data not shown). We also investigated squalene synthase activity of 97SQS and ATCC 2001 (wild-type strain) after 8 h of incubation with or without DOX. As shown in Table 1, squalene synthase activity of 97SQS was almost the same as that of the wild type in the absence of DOX, and the addition of DOX almost completely inhibited its activity. We next determined the number of viable cells at various time points after the addition of DOX. A reduction in the number of viable cells was observed 5 h after adding DOX (Fig. 3B), suggesting that the lack of squalene synthase activity caused rapid cell death. Furthermore, we investigated the importance of the enzyme for growth in mice by counting the number of surviving cells in mice treated with DOX or left untreated. Surprisingly, the 97SQS cells in the mice treated with DOX could proliferate as normally as those in the untreated mice. The results were almost the same as those obtained when wild-type cells were inoculated into mice. In contrast, a severe growth defect was observed when other controllable strains, such as 99TEF3, in which the TEF3 gene was controlled by a tetracycline-regulatable promoter, 99t (23), were inoculated into DOX-treated mice (data not shown).
FIG. 3.
Viability of the ERG9-controllable strains in laboratory media and in mice. (A) The numbers of viable cells for 97SQS and ATCC 2001. Here, 105 97SQS and ATCC 2001 cells were inoculated into YEPD medium. After culturing in the absence (closed bars) or presence (open bars) of 10 μg of DOX per ml for 14 h, CFU was determined by counting the number of colonies on YEPD agar plates where 20 μl of diluted cultures had been spread. Each value is based on three independent experiments. (B) Time course for the number of viable 97SQS cells cultured with or without 10 μg of DOX per ml. For each strain, 104 cells were inoculated into YEPD medium and cultured in the absence (closed diamonds) or presence (open squares) of 10 μg of DOX per ml for the indicated times. Each line represents the average for three independent experiments. The error bars are not shown where the symbol is larger than the error bar. (C) Effects of DOX on the survival of ATCC 2001 in mouse kidneys. The mice were sacrificed, and the ATCC 2001 cells in their kidneys were recovered. Closed diamonds, the number of cells recovered from untreated mice; open squares, the number of cells recovered from DOX-treated mice. Each line represents the average for the number of cells recovered from five mice. Repeated experiments showed the same results. (D) Effects of DOX on the survival of 97SQS in mouse kidneys. The mice were sacrificed and the 97SQS cells in their kidneys were recovered. Closed diamonds, the number of cells recovered from untreated mice; open squares, the number of cells recovered from treated mice. Each line represents the average for the number of cells recovered from five mice. Repeated experiments showed the same results.
TABLE 1.
Effect of DOX and human serum on squalene synthase activity in ATCC 2001 and 97SQS cellsa
| Strain | Medium supplementation
|
Squalene synthase-specific activity (pmol/h/mg of protein) | |
|---|---|---|---|
| DOX (μg/ml) | Human serum (%)b | ||
| ATCC 2001 | 0 | 0 | 1,635 ± 174 |
| 10 | 0 | 1,589 ± 170 | |
| 0 | 10 | 441 ± 130 | |
| 97SQS | 0 | 0 | 1,680 ± 166 |
| 10 | 0 | 16 ± 9 | |
| 0 | 10 | 714 ± 115 | |
| 10 | 10 | 12 ± 6 | |
Values are means ± standard deviations (three independent samples per group).
Volume per volume.
Serum suppresses the growth defect of 97SQS cells treated with DOX.
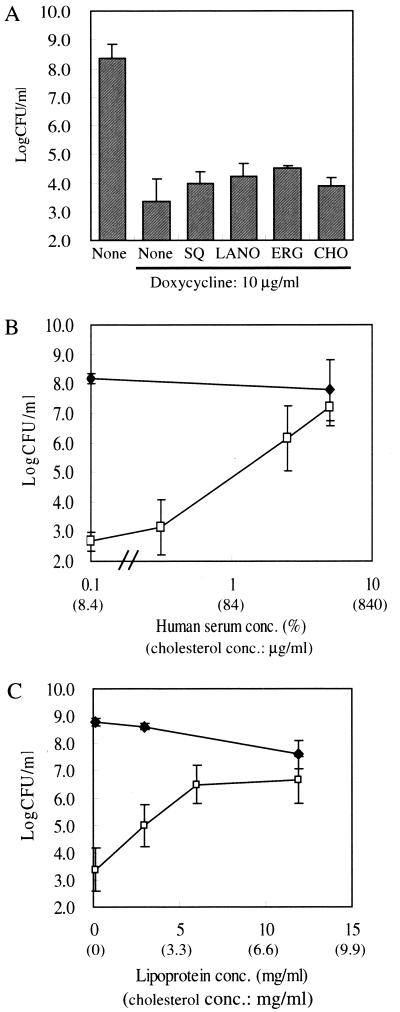
In yeast cells, including C. glabrata, the sterol biosynthesis pathway is strictly dependent on oxygen. Therefore, sterol auxotrophs normally appear only under laboratory anaerobic conditions, unless the sterol defect is accompanied by heme deficiency (20). ATCC 2001, 97SQS, and its parent strain, ACG4, had heme competency because they could grow on nonfermentable media (data not shown). It has been reported that the concentration of oxygen in an animal body is 10 to 50 mm Hg (1). Admittedly, that figure is much lower than laboratory aerobic conditions; however, this value is not low enough to induce oxygen-dependent gene expression (1). Furthermore, 10 μg of cholesterol per ml was sufficient for the wild-type S. cerevisiae strain treated with lovastatin or fenpropimorph to grow under aerobic conditions (19, 21). We, therefore, examined the permeability of the cells for lipids in aerobic conditions to address how the 97SQS cells were able to maintain normal growth in the mice treated with DOX. When 97SQS cells were inoculated into DOX-containing YEPD medium, the growth defect still occurred in the presence of 25-μg/ml lanosterol, ergosterol, and cholesterol and in the presence of 50-μg/ml squalene (Fig. 4A). On the other hand, the addition of human serum, which contains a high concentration of cholesterol (5.5 mg per ml of serum), to YEPD medium could suppress the growth defect of the 97SQS caused by DOX. The numbers of viable cells were in correlation with the concentration of human serum below 5% (vol/vol) (Fig. 4B). Moreover, we could obtain similar results when the sera of other species, such as calf or mouse, were added to YEPD medium (data not shown). To exclude the possibility that serum affects the binding of DOX to the fusion transactivator tetR-GAL4AD, we investigated the activity of squalene synthase in DOX-treated 97SQS cells in the presence of human serum. As shown in Table 1, the squalene synthase activity of 97SQS cells was almost completely depressed in the media containing human serum and DOX. Furthermore, human serum did not affect the DOX-dependent growth defect of other controllable strains such as 99TEF3 (data not shown). Thus, the growth defect of 97SQS by DOX would be suppressed in the presence of serum. We then examined whether the addition of lipoprotein to media could suppress the growth defect of 97SQS cells treated with DOX, because almost all cholesterol exists as a lipoprotein in serum. The addition of lipoprotein could suppress the growth defect by DOX (Fig. 4C). Nevertheless, the suppression was not observed in the medium containing lipoprotein-deficient serum (data not shown).
FIG. 4.
Effect of added lipids, serum, and lipoprotein on growth of 97SQS. (A) Effect of added sterols and squalene on the growth of 97SQS. The 97SQS cells (103) were inoculated into YEPD medium containing 50 μg of squalene (SQ) per ml, 25 μg of lanosterol (LANO) per ml, 25 μg of ergosterol (ERG) per ml, and 25 μg of cholesterol (CHO) per ml. They were then cultured in the absence and presence of 10 μg of DOX per ml for 14 h at 37°C. Each value is based on three independent experiments. (B) Effect of added serum on the growth of 97SQS cells. For both cell lines, 103 cells were inoculated into YEPD medium containing the indicated concentrations of serum and were cultured in the absence (closed symbols) and presence (open symbols) of 10 μg of DOX per ml for 14 h at 37°C. The amount of cholesterol contained in the media is indicated in parentheses on the x axis. Each line represents the average of three independent experiments. The error bars are not shown where the symbol is larger than the error bar. (C) Effect of added lipoprotein on the growth of 97SQS cells. The 97SQS cells (103) were inoculated into YEPD medium containing the indicated concentrations (wt/vol) of lipoprotein and were cultured in the absence (closed diamonds) or presence (open squares) of 10 μg of DOX per ml for 14 h at 37°C. The amount of cholesterol contained in the media is indicated in parentheses on the x axis. Each line represents the average of three independent experiments.
Effect of serum on sterol composition in C. glabrata cells.
To confirm that cholesterol would suppress the DOX-induced growth defect in the 97SQS cells, we analyzed the sterol composition of the cells. As shown in Table 2, deactivation of squalene synthase by DOX affected only sterol biosynthesis and resulted in reduced amounts of ergosterol in the cells without the accumulation of aberrant sterols. Reduction of ergosterol correlated with a loss of viability in the 97SQS cells cultured with YEPD medium containing only DOX. On the other hand, when 97SQS cells were cultured in the presence of human serum and DOX, the cholesterol fraction was detected, and their growth defect was also suppressed. Interestingly, a cholesterol fraction was detected in all the cells cultured with human serum. This is the first demonstration that wild-type yeast cells can incorporate sterols from growth media under aerobic conditions. The amount of cholesterol in the 97SQS cells cultured with DOX and serum was higher than that in cultures with only serum. By investigating the effect of DOX on the sterol content of 97SQS cells cultured with serum, we found that the reduced amount of ergosterol was almost the same as the increased amount of cholesterol in the 97SQS cells cultured with DOX and serum, implying that incorporated cholesterol could be compensating for decreased ergosterol. Thus, these results suggest that incorporated cholesterol complements the defect of ergosterol biosynthesis and results in recovering cell growth.
TABLE 2.
Effect of DOX and human serum on viability and sterol composition of ATCC 2001 and 97SQS cellsa
| Strain | Medium supplementation
|
Viability (log CFU/ml) | Sterol content (μg/g [dry/wt] of cellsb
|
||||
|---|---|---|---|---|---|---|---|
| DOX (μg/ml) | Human serum (%) | Cholesterol | Ergosterol | Lanosterol | Total | ||
| ATCC 2001 | 0 | 0 | 9.00 ± 0.04 | 0d | 276.0 ± 3.0 | 4.75 ± 0.6 | 280.7 ± 2.4 |
| 10 | 0 | NAc | 0d | 287.6 ± 17.4 | 5.9 ± 0.4 | 293.4 ± 17.6 | |
| 0 | 10 | 8.89 ± 0.03 | 975.4 ± 50.1 | 218.4 ± 6.9 | 8.1 ± 1.0 | 1,201.85 ± 44.2 | |
| 97SQS | 0 | 0 | 8.77 ± 0.04 | 0d | 378.4 ± 20.3 | 9.5 ± 0.5 | 387.8 ± 20.8 |
| 10 | 0 | 6.67 ± 0.03 | 0d | 70.7 ± 8.7 | UDe | 70.7 ± 8.7 | |
| 0 | 10 | 8.82 ± 0.04 | 409.6 ± 1.7 | 239.9 ± 29.5 | 4.8 ± 3.3 | 654.3 ± 26.3 | |
| 10 | 10 | 8.33 ± 0.07 | 667.3 ± 22.9 | 92.9 ± 9.6 | 0.3 ± 0.2 | 667.3 ± 22.9 | |
Values are means ± standard deviations (three independent samples per group).
4,4-Diphenyl-1-benzyl-piperidine was used as the internal standard in these experiments.
NA, not assayed.
No cholesterol detected.
UD, under the detection limit.
DISCUSSION
To estimate the efficacy of squalene synthase inhibitors for the therapy of C. glabrata infections, we investigated the importance of the squalene synthase gene for the growth of fungi in mice by using the C. glabrata tetracycline-regulatable expression system (23). Herein, we report interesting evidence that the CgERG9 gene is essential for fungal growth in laboratory medium; however, it is not required at all for fungal growth in mice. We also found that erg9-deficient cells could grow in the media containing serum and that the ability to incorporate exogenous cholesterol can suppress their growth defect. Taken together, these results suggest that this ability would also support growth of the erg9-deficient cells in mice. Inhibitors of squalene synthase are, therefore, not useful for the treatment of C. glabrata infections.
Both in S. cerevisiae and in mammalian cells, squalene synthase activity is regulated transcriptionally and posttranslationally to maintain the cellular sterol content (7, 17, 27). Squalene synthase activity was decreased both in ATCC 2001 (endogenous ERG9 promoter) and 97SQS (the artificial promoter 97t) cultured with serum-containing media (in the absence of DOX), presumably due to the ability of both cells to incorporate cholesterol from serum. These findings suggest that the activity can be regulated posttranslationally. The predicted phosphorylation sites, which are conserved in squalene synthases, might participate in this regulation, although discussion and further studies are required to verify this. In addition, the squalene synthase activity of wild-type ATCC 2001 cells was more strictly repressed than that of 97SQS cells cultured with serum, implying that squalene synthase was also regulated transcriptionally.
As shown in Fig. 4 and Table 2, C. glabrata cells can take up sterol in an oxygen-independent manner. The ATCC 2001 and 97SQS cells still incorporated exogenous cholesterol in the presence of serum, even though the cells generated amounts of ergosterol similar to those obtained when they were cultured under normal conditions (in the absence of serum). From our data, it is difficult to determine how cholesterol was incorporated and whether or not the sterol uptake of normal cells is governed by the same mechanism as in cells lacking ergosterol. Nevertheless, these results allow us to speculate that ergosterol is the major sterol used for maintenance of membrane integrity for this fungal cell proliferation in mice and that incorporated cholesterol may have an important role(s) in maintaining fungal growth in the presence of serum and in the animal body. Furthermore, when the amount of cholesterol in media containing lipoprotein and the amount of cholesterol in media containing human serum are compared, these results suggest that a larger amount of cholesterol is necessary to rescue the growth in the media containing lipoprotein, implying that sterol uptake, at least in ergosterol-deficient cells, could be enhanced by some factor(s) in serum other than lipoprotein.
Numerous sterol biosynthesis inhibitors have been developed as antifungal agents. It has been reported, however, that these agents have limited usefulness for infections by fungi, including Candida species. It has been reported that allylamines, which are squalene epoxidase inhibitors, have been marketed as oral and topical treatments for dermatophytosis (29). The limited usefulness of squalene epoxidase (ERG1) inhibitors against Candida species has been thought to be due to difficulties in crossing the plasma membrane (30). In C. glabrata infection, this defect could be also explained on the basis of our finding; incorporation of cholesterol could rescue growth by inhibiting squalene epoxidase, as is observed in erg9-deficient cells. On the other hand, morpholines and piperidines, which inhibit sterol Δ14-reductase and Δ8→7-isomerase, are also limited for use against superficial fungal infections. These drugs cause fungicidal effects by the intracellular accumulation of Δ8,14-sterol and depletion of ergosterol (9). It has been reported, however, that Δ8,14-sterol did not block the growth of S. cerevisiae growing in ergosterol-containing medium under anaerobic conditions (18). Our findings suggest that treatment of C. glabrata infections with these drugs could only result in accumulation of Δ8,14-sterol. Thus, the sterol uptake ability in ergosterol-deficient cells could explain the limited usefulness of squalene epoxidase inhibitors and inhibitors of sterol Δ14-reductase and Δ8→7-isomerase for C. glabrata infections. Therefore, the sterol uptake observed in our study is an important factor to estimate the efficacy of known sterol biosynthesis inhibitors in the treatment of C. glabrata infections. Alternatively, sterol uptake inhibitors would have potency as antifungal drugs for the therapy of C. glabrata infection, especially if they were administered simultaneously with known sterol biosynthesis inhibitors, such as squalene epoxidase inhibitors and inhibitors of sterol Δ14-reductase and Δ8→7-isomerase. We believe that our findings and hypothesis for the limited usefulness of squalene epoxidase inhibitors or sterol Δ14-reductase and Δ8→7-isomerase inhibitors against C. glabrata infections apply to infections by other fungi that have activity for uptake of in-host cholesterol.
ACKNOWLEDGMENTS
We thank N. Ishii for kindly providing the degenerated primers; M. Aoki, H. Shirai, H. Kato, and T. Tsukuda for their advice on the experiments; S. B. Inoue and S. Nagahashi for helpful discussions; and S. B. Miwa and F. Ford for critical reading of the manuscript.
REFERENCES
- 1.Bunn H F, Poyton R O. Oxygen sensing and molecular adaptation to hypoxia. Physiol Rev. 1996;76:839–885. doi: 10.1152/physrev.1996.76.3.839. [DOI] [PubMed] [Google Scholar]
- 2.Fegueur M, Richard L, Charles A D, Karst F. Isolation and primary structure of the ERG9 gene of Saccharomyces cerevisiae encoding squalene synthetase. Curr Genet. 1991;20:365–372. doi: 10.1007/BF00317063. [DOI] [PubMed] [Google Scholar]
- 3.Fidel P L, Jr, Vazquez J A, Sobel J D. Candida glabrata: review of epidemiology, pathogenesis, and clinical disease with comparison to C. albicans. Clin Microbiol Rev. 1999;12:80–96. doi: 10.1128/cmr.12.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallis H A, Drew R H, Pickard W W. Amphotericin B: 30 years of clinical experience. Rev Infect Dis. 1990;12:308–329. doi: 10.1093/clinids/12.2.308. [DOI] [PubMed] [Google Scholar]
- 5.Georgopapadakou N H, Walsh T J. Antifungal agents: chemotherapeutic targets and immunologic strategies. Antimicrob Agents Chemother. 1996;40:279–291. doi: 10.1128/aac.40.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gietz R D, Schiestl R H, Willems A R, Woods R A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 7.Goldstein J L, Brown M S. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 8.Grandmougin-Ferjani A, Dalpé Y, Hartmann M-A, Laruelle F, Sancholle M. Sterol distribution in arbuscular mycorrhizal fungi. Phytochemistry. 1999;50:1027–1031. [Google Scholar]
- 9.Hartman P G, Polak A. The action of amorolfine: from molecule to cell. In: Rippon J W, Fromtling R A, editors. Cutaneous antifungal agents. New York, N.Y: Marcel Dekker; 1993. pp. 27–36. [Google Scholar]
- 10.Hickey W F, Sommerville L H, Schoen F J. Disseminated Candida glabrata: report of a uniquely severe infection and a literature review. Am J Clin Pathol. 1983;80:724–727. doi: 10.1093/ajcp/80.5.724. [DOI] [PubMed] [Google Scholar]
- 11.Inoue S B, Takewaki N, Takasuka T, Mio T, Adachi M, Fujii Y, Miyamoto C, Arisawa M, Furuichi Y, Watanabe T. Characterization and gene cloning of 1,3-β-d-glucan synthase from Saccharomyces cerevisiae. Eur J Biochem. 1995;231:845–854. doi: 10.1111/j.1432-1033.1995.tb20770.x. [DOI] [PubMed] [Google Scholar]
- 12.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jennings S M, Tsay Y H, Fisch T M, Robinson G W. Molecular cloning and characterization of the yeast gene for squalene synthetase. Proc Natl Acad Sci USA. 1991;88:6038–6042. doi: 10.1073/pnas.88.14.6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly S L, Arnodi A, Kelly D E. Molecular genetic analysis of azole antifungal mode of action. Biochem Soc Trans. 1993;21:1034–1038. doi: 10.1042/bst0211034. [DOI] [PubMed] [Google Scholar]
- 15.Kelly S L, Lamb D C, Corran A J, Baldwin B C, Kelly D E. Mode of action and resistance to azole antifungals associated with the formation of 14α-methylergosta-8,24(28)-dien-3β,6α-diol. Biochem Biophys Res Commun. 1995;207:910–915. doi: 10.1006/bbrc.1995.1272. [DOI] [PubMed] [Google Scholar]
- 16.Kelly D E, Rose M E, Kelly S L. Investigation of the role of sterol delta 8→7-isomerase in the sensitivity of Saccharomyces cerevisiae to fenpropimorph. FEMS Microbiol Lett. 1994;122:223–226. doi: 10.1111/j.1574-6968.1994.tb07171.x. [DOI] [PubMed] [Google Scholar]
- 17.Kennedy M A, Barbuch R, Bard M. Transcriptional regulation of the squalene synthase gene (ERG9) in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 1999;1445:110–122. doi: 10.1016/s0167-4781(99)00035-4. [DOI] [PubMed] [Google Scholar]
- 18.Lees N D, Skaggs B, Kirsch D R, Bard M. Cloning of the late genes in the ergosterol biosynthetic pathway of Saccharomyces cerevisiae. Lipids. 1995;30:221–226. doi: 10.1007/BF02537824. [DOI] [PubMed] [Google Scholar]
- 19.Lorenz R T, Parks L W. Effects of lovastatin (mevinolin) on sterol levels and on activity of azoles in Saccharomyces cerevisiae. Antimicrob Agents Chemother. 1990;34:1660–1665. doi: 10.1128/aac.34.9.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lorenz R T, Parks L W. Involvement of heme components in sterol metabolism of Saccharomyces cerevisiae. Lipids. 1991;26:598–603. doi: 10.1007/BF02536423. [DOI] [PubMed] [Google Scholar]
- 21.Lorenz R T, Parks L W. Physiological effects of fenpropimorph on wild-type Saccharomyces cerevisiae and fenpropimorph-resistant mutants. Antimicrob Agents Chemother. 1991;35:1532–1537. doi: 10.1128/aac.35.8.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakashima T, Inoue T, Oka A, Nishino T, Osumi T, Hata S. Cloning, expression, and characterization of cDNAs encoding Arabidopsis thaliana squalene synthase. Proc Natl Acad Sci USA. 1995;92:2328–2332. doi: 10.1073/pnas.92.6.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakayama H, Izuta M, Nagahashi S, Sihta E Y, Sato Y, Yamazaki T, Arisawa M, Kitada K. A controllable gene-expression system for the pathogenic fungus Candida glabrata. Microbiology. 1998;144:2407–2415. doi: 10.1099/00221287-144-9-2407. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen M H, Peacock J E, Morris A J, Tanner D C, Nguyen M L, Snydman D R, Wagener M M, Rinaldi M G, Yu V L. The changing face of candidemia: emergence of non-Candida albicans species and antifungal resistance. Am J Med. 1996;100:617–623. doi: 10.1016/s0002-9343(95)00010-0. [DOI] [PubMed] [Google Scholar]
- 25.Pfaller M A, Jones R N, Messer S A, Edmond M B, Wenzel R P. National surveillance of nosocomial blood stream infection due to species of Candida other than Candida albicans: frequency of occurrence and antifungal susceptibility in the SCOPE program. Diagn Microbiol Infect Dis. 1998;31:327–332. doi: 10.1016/s0732-8893(97)00240-x. [DOI] [PubMed] [Google Scholar]
- 26.Pfaller M A, Messer S A, Houston A, Rangel-Frausto M S, Wiblin T, Blumberg H M, Edwards J E, Jarvis W, Martin M A, Neu H C, Saiman L, Patterson J E, Dibb J C, Roldan C M, Rinaldi M G, Wenzel R P. National epidemiology of mycoses survey: a multicenter study of strain variation and antifungal susceptibility among isolates of Candida species. Diagn Microbiol Infect Dis. 1998;31:281–296. doi: 10.1016/s0732-8893(97)00245-9. [DOI] [PubMed] [Google Scholar]
- 27.Robinson G W, Tsay Y H, Kienzle B K, Smith-Monroy C A, Bishop R W. Conservation between human and fungal squalene synthetases: similarities in structure, function, and regulation. Mol Cell Biol. 1993;13:2706–2717. doi: 10.1128/mcb.13.5.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rose M D, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. pp. 128–129. [Google Scholar]
- 29.Ryder N S, Stuetz A, Nussbaumer P. Squalene epoxidase inhibitors. Structural determinants for activity and selectivity of allylamines and related compounds. In: Nes W D, Parish E J, Trzaskos J M, editors. Regulation of isopentenoid metabolism. Washington, D.C.: American Chemical Society; 1992. pp. 192–204. [Google Scholar]
- 30.Vanden Bossche H, Warnock D W, Dupont B, Kerridge D, Sen Gupta S, Improvisi L, Marichal P, Odds F C, Provost F, Ronin O. Mechanisms and clinical impact of antifungal drug resistance. J Med Vet Mycol. 1994;32(Suppl. 1):189–202. doi: 10.1080/02681219480000821. [DOI] [PubMed] [Google Scholar]
- 31.Wingard J R, Merz W G, Rinaldi M G, Miller C B, Karp J E, Saral R. Association of Torulopsis glabrata infections with fluconazole prophylaxis in neutropenic bone marrow transplant patients. Antimicrob Agents Chemother. 1993;37:1847–1849. doi: 10.1128/aac.37.9.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wingard J R. Importance of Candida species other than C. albicans as pathogens in oncology patients. Clin Infect Dis. 1995;20:115–125. doi: 10.1093/clinids/20.1.115. [DOI] [PubMed] [Google Scholar]