Abstract
Purpose:
Develop a quantitative image analysis method to characterize the heterogeneous patterns of nodule components for the classification of pathological categories of nodules.
Materials and Methods:
With IRB approval and permission of the National Lung Screening Trial (NLST) project, 103 subjects with low dose CT (LDCT) were used in this study. We developed a radiomic quantitative CT attenuation distribution descriptor (qADD) to characterize the heterogeneous patterns of nodule components and a hybrid model (qADD+) that combined qADD with subject demographic data and radiologist-provided nodule descriptors to differentiate aggressive tumors from indolent tumors or benign nodules with pathological categorization as reference standard. The classification performances of qADD and qADD+ were evaluated and compared to the Brock and the Mayo Clinic Models by analysis of the area under the receiver operating characteristic curve (AUC).
Results:
The radiomic features were consistently selected into qADDs to differentiate pathological invasive nodules from (1) preinvasive nodules, (2) benign nodules, and (3) the group of preinvasive and benign nodules, achieving test AUCs of 0.847±0.002, 0.842±0.002 and 0.810±0.001, respectively. The qADD+ obtained test AUCs of 0.867±0.002, 0.888±0.001 and 0.852±0.001, respectively, which were higher than both the Brock and the Mayo Clinic Models.
Conclusion:
The pathologic invasiveness of lung tumors could be categorized according to the CT attenuation distribution patterns of the nodule components manifested on LDCT images, and the majority of invasive lung cancers could be identified at baseline LDCT scans.
Keywords: radiomic, lung nodule, pathologic categorization, LDCT, lung cancer screening
Introduction
Lung cancer usually manifests as noncalcified nodules with solid and subsolid (part- and non-solid) composition on CT images. Histologically, lung cancer is classified into the categories of invasive (INV) carcinoma and pre-invasive (Pre-INV) carcinoma (including adenocarcinoma in situ (AIS) and minimally invasive adenocarcinoma (MIA)), representing a histological spectrum of carcinomas ranged from indolent to aggressive tumors (1, 2). Both AIS and MIA can achieve excellent (nearly 100%) postsurgery 5-year survival, whereas INV has worse prognosis (2). Studies of radiologic- pathologic correlation suggest that the degree of pathologic invasive growth in carcinoma can be quantified according to the proportion of the solid component of nodules on CT (3, 4). However, the visual classification of solid and subsolid nodules, and the outlining of the nodules with manual or semi-automated method have been reported to have large inter- and intra-observer variability among radiologists (5–9). The inconsistency may cause inaccurate and subjective assessment of nodule composition (10, 11). This study is to 1) develop a quantitative CT attenuation distribution descriptor (qADD) based method to characterize the heterogeneous patterns of nodule components, 2) investigate its capability for the classification of pathological categories of pulmonary nodules, and 3) compare to the Brock (12) and the Mayo Clinic Models (13).
Materials and Methods
Study population
With Institutional Review Board approval and permission of the National Lung Screening Trial (NLST) project, 103 subjects with positive baseline low dose CT (LDCT) scans were randomly selected from the NLST dataset. The image acquisition techniques were 80–120 kVp, 40–120 mAs, and reconstructed at 1–2.5 mm slice interval. Forty-nine of the 103 subjects (47.6%) were women (median age 62 years; range 55–73 years) and 54 were men (median age 60 years; range 55–74 years). A total of 166 nodules with size < 20 mm found by the NLST readers were included. Table 1 shows the categories of pathologically diagnosed invasive, preinvasive and benign nodules. Eighty-nine nodules were pathologically diagnosed as lung cancer in 53 subjects who underwent biopsy, of which 45 and 44 were invasive and preinvasive, respectively. Seventy-seven nodules were determined to be benign in 50 subjects by biopsy or 5-year follow-up exams.
Table 1.
the categories of pathologically diagnosed invasive, preinvasive and benign nodules
| Invasive (n=45) | Preinvasive (n=44) | Benign (n=77) | ||
| acinar | 17 | Adenocarcinomas in situ | 35 | 77 |
| papillary | 7 | Atypical adenomatous hyperplasia | 7 | |
| large cell carcinomas | 7 | Squamous cell carcinomas in situ | 2 | |
| squamous cell | 7 | |||
| mixed subtype | 7 |
Model development and evaluation
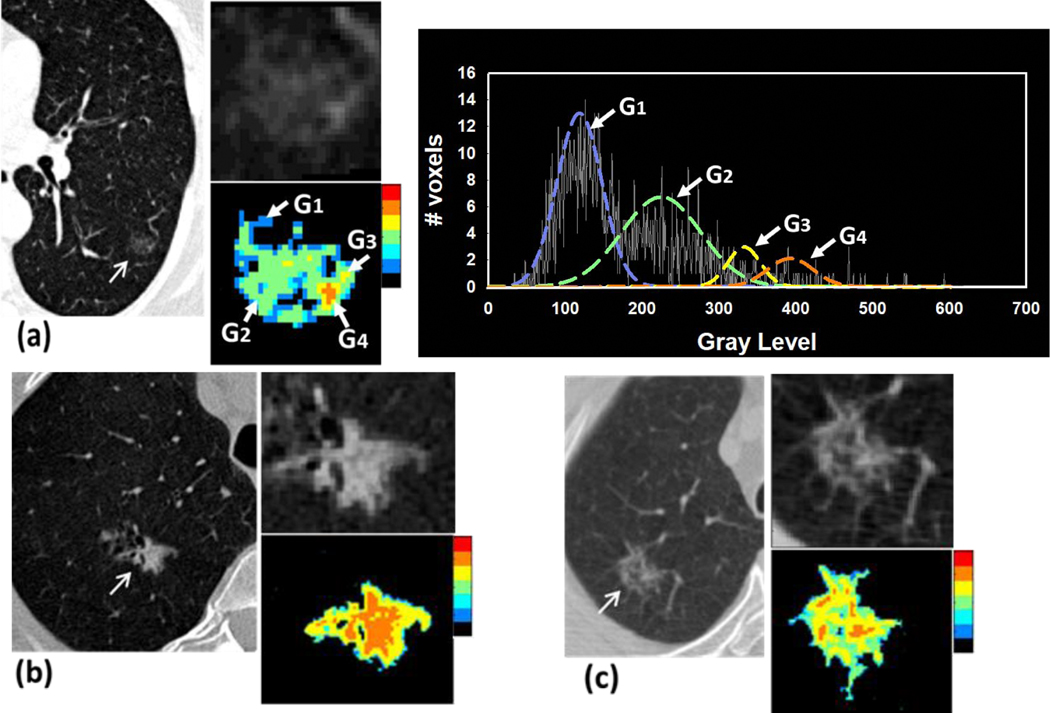
We developed a new 3D adaptive multi-component Expectation-Maximization (EM) analysis (AMEA) method (14, 15) to extract the volumes of the entire nodule, the solid and subsolid components, and the lung parenchymal region surrounding the nodule. The EM algorithm (16) is an iterative method to find maximum likelihood or maximum a posteriori (MAP) estimation of model parameters. Our AMEA method is fully automated after the location of the nodule is marked by a single point. The EM analysis was performed in a cubical volume of interest (VOI) with side-length of 32 mm centered at the point and enclosing the nodule. We assumed up to 6 regions (e.g., air, solid or subsolid regions, etc.) being segmented in the nodule VOI as a priori knowledge. Assuming the CT values of each region followed a Gaussian mixture distribution, 6 initial Gaussians with equal variances were distributed evenly across the CT value histogram of the VOI. The EM algorithm then iteratively computed MAP estimates to fit a Gaussian model to each component. After the iteration reached convergence, any two adjacent fitted Gaussians were merged into one Gaussian when the difference of their means (mi) was less than one of their standard deviations (σi ), i.e., |m1-m2| < σ1 or σ2, where i =1, 2. Finally, all segmented components, excluding the background, constituted the nodule volume. A rim-like surrounding lung parenchyma region was then obtained by expanding the nodule volume by 3D morphological dilation with a 5-mm-diameter rolling ball structuring element. Fig.1(a) shows an example to illustrate the fitting of the Gaussian model by EM algorithm, in which the 6 fitted Gaussian models were merged into 4, indicating that 4 nodule components were segmented.
Fig.1.
Examples of automated segmentation of three types of nodules using the AMEA method. The regions of the segmented nodule components are rendered by a color heat map where the region with high CT values is mapped to hot colors and low CT values to cold colors. (a) a pure non solid nodule located at the left lower lobe, measuring 15×12 mm in diameters with poorly defined margins, pathologically diagnosed as adenocarcinoma in situ. The histogram of the nodule VOI excluding zero gray level illustrates that the 4 segmented nodule components including ground glass tissue and small portion of solid component were represented by 4 EM fitted Gaussians (G1 – G4). (b) a subsolid nodule located at the right lower lobe, measuring 15×10 mm in diameters, with spiculated margins, pathologically diagnosed as invasive adenocarcinoma, (c) a non-solid nodule containing small solid components located at right upper lobe, measuring 9×7 mm in diameters with poorly defined margins, pathologically diagnosed as adenocarcinoma in situ.
After AMEA segmentation, for each screen-detected nodule, we extracted 11 features to describe the size, the CT attenuation distributions of solid and subsolid components of the nodule, and the surrounding lung parenchyma region:
Volume of the entire nodule (V-Nodule)
Percent volume of solid (%-Solid) and subsolid (%-SS) components within entire nodule
Mean and standard deviation of CT attenuation of solid (μ-Solid and σ-Solid) and subsolid (μ-SS and σ-SS) component
Entropy (17) of CT attenuation of entire nodule (S-Nodule) and lung parenchyma (S-LP)
Density of solid (ρ-Solid = %-Solid * μ-Solid), and subsolid (ρ-SS = %-SS * μ-SS) components.
The support vector machine (SVM) (18) is a supervised machine learning algorithm by finding a decision boundary to separate the two classes of data. We trained three SVM classifiers with linear kernel separately as a quantitative CT attenuation distribution descriptor (qADD) to differentiate invasive cancers from 1) preinvasive nodules, 2) benign nodules, and 3) the group of both preinvasive and benign nodules. For each task, we also built two hybrid models, qADD+ and qADD++ that combined radiomic qADD features with NLST documented demographic factors (Table 2, f1 to f12), excluding and including clinical risk factors (Table 2, f13 to f21), respectively. The least absolute shrinkage and selection operator (LASSO) method (19) with 5-fold cross-validation was used to select the effective features for each task.
Table 2.
NLST documented demographic factors ( f1 to f12 )and clinical radiologic risk factors ( f13 to f21 )
| Variable | Subcategory | *Group-1 | *Group-2 | *Group-3 | *Group-4 | ** P 1 | ** P 2 | ** P 3 |
|---|---|---|---|---|---|---|---|---|
| Per-Subject | N=33 | N=20 | N=50 | N=70 | ||||
|
| ||||||||
| Gender (f1) | female | 18 | 10 | 21 | 31 | |||
| Male | 15 | 10 | 29 | 39 | 1.000 | 0.736 | 0.400 | |
| Age (f2) | 62.8±5.7 | 64.6±5.1 | 60.6±5.2 | 61.7±5.4 | 0.516 | 0.142 | 0.348 | |
| Race (f3) | White | 32 | 20 | 48 | 68 | |||
| other | 1 | 0 | 2 | 2 | 1.000 | 1.000 | 0.999 | |
| Ethnicity (f4) | Hispanic/Latino | 0 | 0 | 2 | 2 | |||
| other | 33 | 20 | 48 | 68 | 1.000 | 1.000 | 0.999 | |
| Packs/yr (f5) | 61.5±20.5 | 65.3±29.6 | 61.6±39.3 | 62.7±36.6 | 1.000 | 1.000 | 0.834 | |
| Smoke | Starting Age (f6) | 16.5±3.6 | 16.4±3.1 | 16.5±3.8 | 16.5±3.6 | 1.000 | 1.000 | 0.939 |
| Years (f7) | 43.0±5.6 | 43.3±6.7 | 37.5±7.5 | 39.2±7.7 | 1.000 | 0.0006 | 0.005 | |
| BMI (f8) | 26.2±3.7 | 27.0±4.4 | 27.5±4.9 | 27.4±4.7 | 1.000 | 0.302 | 0.168 | |
| Emphysema (f9) | 5 | 1 | 3 | 4 | 0.144 | 0.370 | 0.071 | |
| Medical | COPD (f10) | 1 | 0 | 2 | 2 | 1.000 | 1.000 | 0.540 |
| History | Emphysema/COPD | 6 | 1 | 4 | 5 | 0.466 | 0.370 | 0.168 |
| (E-COPD) (f11) | ||||||||
| Family (f12) | 7 | 5 | 11 | 16 | 1.000 | 1.000 | 0.810 | |
| Per-Nodule | N=45 | N=44 | N=77 | N=121 | ||||
|
| ||||||||
| Upper-Lobe (f13) | 27 | 32 | 34 | 66 | 0.526 | 0.266 | 0.599 | |
| Attenuation | Soft (f14) | 31 | 5 | 48 | 53 | <0.001 | 1.000 | <0.001 |
| GGO (f15) | 5 | 29 | 17 | 46 | <0.001 | 0.301 | <0.001 | |
| Mix (f16) | 9 | 10 | 12 | 22 | 0.800 | 1.000 | 0.824 | |
| Size (f17) | 12.1±5.6 | 11.4±6.2 | 7.3±3.0 | 8.8±4.8 | 1.000 | <0.001 | <0.001 | |
| Margin | Spiculated (f18) | 18 | 1 | 5 | 6 | <0.001 | <0.001 | <0.001 |
| Smooth (f19) | 11 | 14 | 52 | 66 | 0.972 | <0.001 | <0.001 | |
| Poor (f20) | 12 | 23 | 13 | 36 | 0.012 | 0.612 | 0.596 | |
| Undetermined (f21) | 4 | 6 | 7 | 13 | 1.000 | 1.000 | 0.999 | |
Group-1, 2, 3, 4 are Invasive, Preinvasive, Benign, and group of Preinvasive and Benign nodules, respectively.
P1, P2, P3: P-value of the differences between Group-1 and Group-2, Group-3 and Group-4, respectively. The values of P1, P2 were corrected for Bonferroni multiple comparisons (N=2).
-Age, Smoke history, BMI and Size are shown as mean±standard deviation; the P values of their differences are calculated by Student’s t-test. The integers are the number of the subjects or nodules and compared by Fisher Exact Test (P value).
Statistical Analysis
The documented demographic data of the subjects and the distribution of nodule size, margin and attenuation categories, were summarized by descriptive statistics and bar chart. The Student’s t- test and Fisher exact test (20) for independence were used to compare their differences between invasive, preinvasive and benign nodules. The qADD, qADD+ and qADD++ were trained and validated with 5-fold cross-validation resampling method. The classification performances of the qADDs were evaluated and compared to the Brock model (21) and the Mayo clinic model (13) by analysis of the area under the receiver operating characteristic (ROC) curve (AUC) (22). The ROC curves were compared with the method of DeLong et al. (23). For subjects with more than one nodule, clustered ROC data analysis was used to account for the intra-subject correlation between different nodules within the same subject (24). The Bonferroni correction (25) was used to adjust the P-values for multiple comparison of invasive vs preinvasive and benign nodules. P-values less than 0.05 after adjustment were considered statistically significant. The ROC analysis, SVM and LASSO methods, and other statistical analyses_were performed by using the R software packages (version 3.5.1; http://www.r-project.org/).
Results
Per-Subject analysis: NLST-risk factors
The distribution of the NLST-documented subject demographic data was summarized in Table 2. The Student’s t-test shows that the ages of the subjects with invasive nodules were not significantly different from those with preinvasive (P=0.516) or benign nodules (P=0.142), and the group of subjects who had preinvasive and benign nodule (P=0.348). Only the years of smoking for subjects with invasive nodules (n=33, years 43.0±5.6) were significantly different from those with benign nodules (n=50, years 37.5±7.5) and the groups of subjects with preinvasive or benign nodules (n=70, years 39.2.0±7.7) (P < 0.05 by Student’s t-test). The differences of all other risk factors were not significant for subjects who had invasive, preinvasive or benign nodules.
Per-Nodule analysis: radiologist-provided radiologic descriptors, Brock and Mayo Clinic Models
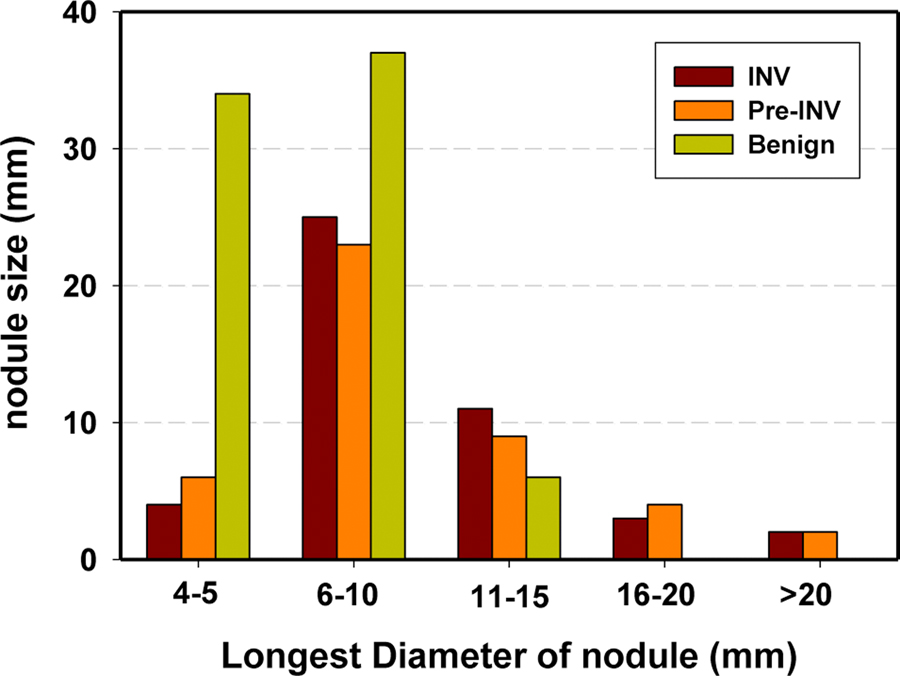
Table 2 also summarizes radiologic risk factors provided by NLST radiologists for the 166 nodules. Fig. 2 shows the bar chart of distribution of nodule sizes measured as the longest diameter (mm) by NLST radiologists. The mean size of the 89 (45+44) malignant invasive and preinvasive nodules (11.7±5.9 mm) was significantly larger than that of the 77 benign nodules (7.3±3.0 mm) (P <0.001 by Student’s t-test). Among the 89 malignant nodules, there was extensive overlap between the size of invasive nodules (12.1±5.6 mm) and that of preinvasive nodules (11.4±6.2 mm) (P =1.000). On the other hand, the mean size of the invasive nodules is significantly larger than that of the 121 (44+77) preinvasive and benign nodules (8.8±4.8 mm) and that of the 77 benign nodules (7.3±3.0 mm), respectively (P <0.001).
Fig.2.
Distribution of the longest diameters of the lung nodules in the data set as measured by NLST radiologists on the axial slices of the CT examinations.
The Fisher exact test indicates that a larger number of invasive nodules had spiculated margins than that of the preinvasive and benign nodules (18 vs 1+5) (P<0.05), and a larger number of benign nodules exhibited smooth margins than invasive (52 vs 11) and preinvasive nodules (52 vs 14) (P<0.05). The NLST radiologists described the majority of invasive (31/45=68.9%) and benign nodules (48/77=62.3%) as homogenous soft tissue attenuation, while more than half of the preinvasive nodules (29/44=65.9%) had non-solid/ground glass attenuation.
We applied the NLST-documented subject demographic data, clinical risk factors and radiologic descriptors to the Brock and the Mayo Clinic Models for differentiating the invasive nodules from the preinvasive, the benign, and the group of preinvasive and benign nodules. Table 3 shows that the Brock models achieved a test AUC of 0.741±0.033, 0.855±0.031 and 0.843±0.031, respectively. The corresponding test AUCs achieved by the Mayo Clinic model were 0.693±0.030, 0.821±0.032 and 0.771±0.032 for the three classification tasks, respectively.
Table 3.
Test AUC achieved by qADD, the combination of qADD and subject demographics and risk factors, and excluding (qADD+) or including radiologic descriptors (qADD++), the Brock and Mayo Clinic Models for differentiating invasive nodules from the preinvasive nodules, Benign nodules and the group of Pre-INV and Benign nodules.
| Task | Invasive vs Preinvasive | Invasive vs Benign | Invasive vs Preinvasive &Benign | |
|---|---|---|---|---|
|
| ||||
| qADD | AUC | 0.847±0.002 | 0.842±0.002 | 0.810±0.001 |
| Selected features | μ-Solid, σ-Solid, μ-SS, ρ-SS | σ-Solid, μ-SS, S-Nodule, S-LP | σ-Solid, μ-SS, S-LP | |
|
| ||||
| AUC | 0.867 ±0.002 | 0.888±0.001 | 0.852±0.001 | |
| qADD+ | Selected features | μ-Solid, σ-Solid, μ-SS, ρ-SS, E-COPD | σ-Solid, μ-SS, S-Nodule, S-LP, Gender, Smoke-years, Upper-lobe | σ-Solid, μ-SS, S-LP, ρ-SS, Gender, Smoke-years, E-COPD |
| vs qADD | P = 0.208 | P =0.089 | P =0.280 | |
|
| ||||
| qADD++ | AUC | 0.901±0.001 | 0.909±0.001 | 0.877±0.001 |
| Selected featu s | μ-Solid, σ-Solid, μ-SS, ρ-SS, E-COPD, GGO, Spiculated | σ-Solid, μ-SS, S-Nodule, S-LP, Smoke-years, E- COPD, Size, Spiculated | σ-Solid, μ-SS, S-LP, ρ-SS, Smoke-years, E-COPD, Size | |
| vs qADD | P = 0.046 | P =0.027 | P =0.069 | |
| vs qADD+ | P = 0.102 | P =0.159 | P =0.096 | |
|
| ||||
| AUC | 0.741±0.033 | 0.855±0.031 | 0.843±0.031 | |
| Risk factors | Age, Gender, Family, Emphysema, Size, Attenuation, Upper-lobe, Spiculated. | |||
| Brock Model | vs qADD | P=0.121 | P=0.185 | P=0.469 |
| vs qADD+ | P=0.039 | P =0.167 | P=0.770 | |
|
| ||||
| Mayo Clinic Model | AUC | 0.693±0.030 | 0.821±0.032 | 0.771±0.032 |
| Risk factors | Age, Smoker, Non-Lung-Cancer, Size, Upper-lobe, Spiculated. | |||
| vs qADD | P =0.033 | P =0.681 | P =0.523 | |
| vs qADD+ | P =0.014 | P =0.042 | P =0.028 | |
| vs Brock | P=0.242 | P=0.341 | P=0.012 | |
qADD for categorization of pathologic subtypes of nodules
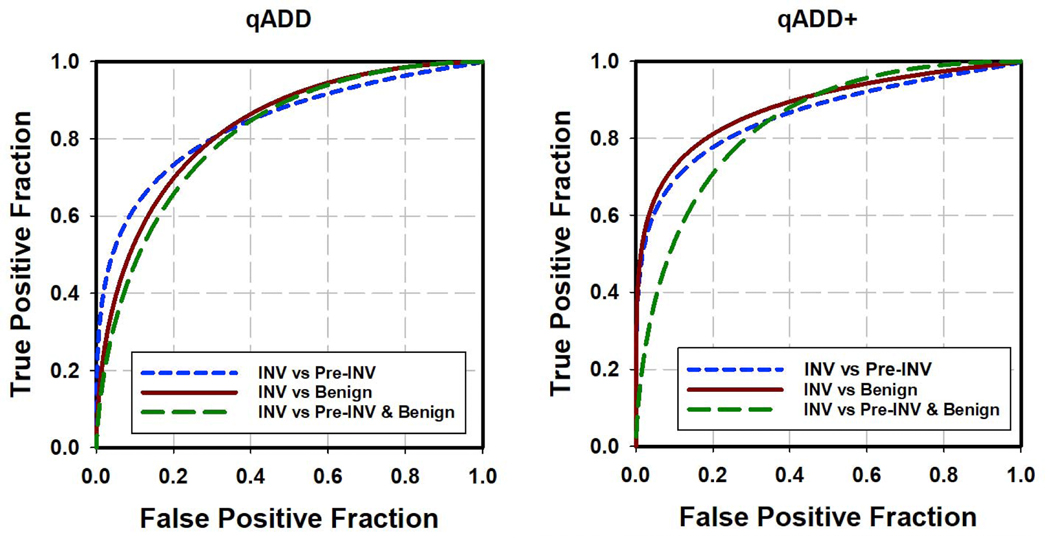
With the LASSO feature selection method, 4, 4, and 3 effective features were selected from the 11 radiomic features for the three classification tasks of differentiating the invasive nodules from the preinvasive, the benign, and the group of preinvasive and benign nodules, respectively. The selected features were combined within each task to generate three qADDs by three SVM classifiers, respectively. The selected features and classification results are listed in Table 3. The test ROC curves of the three qADDs obtained from the 5-fold cross-validation method are shown in Fig. 3. The corresponding AUCs were 0.847±0.002, 0.842±0.002, and 0.810±0.001 for the three classification tasks, respectively.
Fig.3.
Test ROC curves for classification of different types of nodules by qADD (left) and qADD+ that combined NLST risk factors with qADD (right).
qADD+ and qADD++ : added value of subject demographic and risk factors
We added NLST-documented subject risk factors (12 factors (f1 to f12) shown in Table 2) to the 11 radiomic features to form a new feature pool. Among those 23 features, 5, 7, and 7 features, respectively, were identified to be effective by the LASSO feature selection method for distinguishing the invasive nodules from the preinvasive, the benign, and the group of benign and preinvasive nodules. The comparison of the AUCs between the classifiers with and without adding the subject data to the qADD is summarized in Table 3. The same 4, 4 and 3 radiomic features selected for the qADD classifiers were consistently selected for the corresponding qADD+ classifiers, respectively. The qADD+ for differentiating the invasive from benign nodules achieved higher AUC of 0.888 by selecting the subject gender, smoking-years, and nodule location (upper-lobe or not) as additional features. The qADD+ classifier for differentiating the invasive from the preinvasive selected the feature of emphysema or COPD (E-COPD) history and improved the AUC from 0.847 to 0.867. The qADD+ classifier for differentiating the invasive from the group of preinvasive and benign nodules selected the additional features of gender, smoke-years and E-COPD and improved the AUC from 0.810 to 0.852. However, none of the improvement reached statistical significance.
We also formed another new feature pool of 32 features by adding the 9 radiologic descriptors provided by NLST radiologists (f13 to f21) together with 12 NLST factors (f1 to f12) to the 11 radiomic features. Three qADD++ classifiers were built with 7, 8 and 7 LASSO-selected features for the same three tasks to distinguish invasive nodules (Table 3). The same 4, 4 and 3 radiomic features selected for the qADD and qADD+ classifiers were also consistently selected by the qADD++ classifiers. Compared with qADD+, the descriptors of GGO, spiculated and the nodule size provided by the radiologists as additional features slightly improved the AUC from 0.867 to 0.901, from 0.888 to 0.909, and from 0.852 to 0.877, for differentiating invasive from preinvasive, benign and the group of preinvasive and benign nodules, respectively, but none of the improvement reached statistical significance.
Comparing qADD and qADD+ to two clinical models (Table 3), the test AUC achieved by the qADD+ was significantly (P<0.05) higher than those of the Mayo Clinic Models in all three classification tasks. Without the additional factors, the test AUC of qADD was significantly higher than that of Mayo Clinic Model for differentiating the invasive from preinvasive (P<0.05).
Discussions
Biologically, the pulmonary tumor cells initially grow along the alveolar lining with minimal thickening of the alveolar septa. As the number of tumor cells increases, the alveolar walls become thickened and collapse. The nodule becomes denser due to the alveoli invaded and replaced by cells, which appear on CT images as subsolid or completely solid. Several radiology-pathology correlation studies (2–4, 6, 26, 27) found that the degree of tumor invasion to the alveoli seen with microscopic histology assessment is correlated with the size of solid components manifested on macroscopic CT images. The presence of a solid component in the nodule that is measurable with CT images depends on the amount of tumor cell invasion to the alveoli. For lung cancer screening with LDCT, although the image quality of LDCT is sufficient for detecting nodules, the increased image noise degrades the visibility of the nodule margins and solid components, thereby affecting the classification of benign and malignant nodules as well as the degree of tumor cell invasion. Our AMEA method has the advantage of being more consistent than manual segmentation. It not only can segment multiple regions of interest allowing quantitative analysis but also facilitates direct visualization of nodule component structures with a color heat map (Fig.1). Thus may be useful for monitoring the changes of nodule components during follow-up CT scans.
Unlike other radiomics methods extracting hundreds of features that may contain a significant amount of noise and highly correlated features (28, 29) for a specific classification task, we extracted only 11 features that were designed to characterize the volumes and CT value distribution of AMEA- segmented nodule components. The results of our qADD classifiers showed that several effective features were selected consistently by the LASSO method and achieved high accuracies for different classification tasks (Table 3). The features of σ-Solid and μ-SS were designed to quantify the variation in attenuation of the solid portion and the mean attenuation of the ground-glass area of a nodule that may characterize the degree of tumor cell invasion to the alveoli manifested on CT images; both were selected for all three classification tasks. The entropy-based feature S-LP was designed to characterize the inhomogeneity of CT attenuation in the lung parenchyma surrounding the nodule that may be caused by tumor angiogenesis extending to the lung parenchyma. Our results indicated that this feature played an important role in distinguishing invasive nodules from the benign and the group of preinvasive and benign nodules. The NLST-factors, including nodule characteristics and subject demographic data, provide varied degrees of correlation with cancer (Tables 2 & 3).
A number of studies (6, 30–33) demonstrated that the nodule size is highly predictive of the risk of malignancy. Our data set shows that, the number of benign nodules is significantly larger than that of invasive and preinvasive malignant nodules for the nodules <5 mm in diameter (Fig.2). It is worth noting that, none of our size-related radiomic features such as %-Solid, %-SS and V-Nodule were selected as effective features for differentiating invasive nodules from other types of nodules (Table 3). Table 2 and Table 3 show that the radiologic descriptors of nodule attenuation (soft (f14), and GGO (f15)) can be a useful factor to differentiate invasive nodules from preinvasive nodules, and the spiculated margin (f18) to differentiate invasive nodules from preinvasive and benign nodules. The major difference between the two clinic models is that the Brock model includes the attenuation descriptor of solid or subsolid nodule. Table 3 shows the Brock model achieved higher AUCs than the Mayo model for the three classification tasks. The comparison of qADD+ to qADD++ (Table 3) show that adding the radiologic descriptors did not significantly improve the AUC for the three classification tasks. The subjective attenuation descriptors might correlate with some of our radiomic features that were designed to characterize nodule attenuation distribution patterns.
There are several limitations in our pilot study. First, the relatively small sample size might not be representative of a general lung cancer screening population, although the subject cases in our study were basically random samples from the national multicenter prospective NLST study. It is important to further validate the performance with a large independent data set, and investigate the predictive values of incorporating the quantitative descriptors of nodule components with other risk factors (i.e., occupation, environmental exposure, etc.). Further independent cohort studies are also needed to validate the qADD approach to improving the baseline interpretation of nodule aggressiveness, as well as the management of indeterminate nodules. Second, due to the limited sample size available, there were not enough samples to further divide the data into subsets and study the effects of the CT acquisition or reconstruction parameters (e.g., kVp, mAs, slice thickness, etc) on the performance of our methods, which will be of interest in future studies. Third, we did not directly evaluate the segmentation accuracy of our AMEA method because radiologists’ segmentation has large variabilities. Instead of attempting to obtain gold standard to evaluate the accuracy of our AMEA method for nodule component segmentation, we used task-driven methodology to evaluate the performance of our nodule classification method, in which radiomic features were extracted from the segmented nodule components. The high classification accuracy indicated that the nodule components obtained from the AMEA segmentation correlated with the invasiveness of the nodule, regardless of whether the segmentation agreed with manual segmentations.
In conclusion, our study demonstrated the feasibility of estimating the pathologic invasiveness of lung cancers using our qADD approach to characterize the CT attenuation distribution patterns of the nodule components manifested on LDCT images, and that the majority of invasive lung nodules could be identified early before treatment, thus has potential to reduce over-diagnosis and over- treatment of indolent lung cancer.
Highlights.
EM analysis is feasible to extract the volume of the nodule and its solid and subsolid components.
Radiomics have potentials to quantify CT attenuation distribution patterns of nodule components.
The pathologic invasiveness of lung tumors could be categorized by radiomic descriptors.
Clinical risk factors add discriminative value to radiomics in differentiating nodule subtypes.
The Brock and Mayo Clinic Models are less accurate than the quantitative approach.
Acknowledgements
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number U01CA216459. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Credit Author Statement
Chuan Zhou: Conceptualization, Methodology, Software, Writing- Original draft preparation. Heang-Ping Chan: Conceptualization, Writing- Validation. Aamer Chughtai: Data Curation. Lubomir M. Hadjiiski: Methodology. Ella A. Kazerooni: Editing. Jun Wei: Methodology
Conflict of interest
The authors and authors’ institutions have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kerr KM. Pulmonary preinvasive neoplasia. J Clin Pathol. 2001;54(4):257–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Travis WD, Brambilla E, Noguchi M, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol. 2011;6(2):244–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee HY, Choi YL, Lee KS, et al. Pure ground-glass opacity neoplastic lung nodules: histopathology, imaging, and management. AJR Am J Roentgenol. 2014;202(3):W224–33. [DOI] [PubMed] [Google Scholar]
- 4.Isaka T, Yokose T, Ito H, et al. Comparison between CT tumor size and pathological tumor size in frozen section examinations of lung adenocarcinoma. Lung Cancer. 2014;85(1):40–6. [DOI] [PubMed] [Google Scholar]
- 5.Naidich DP, Bankier AA, MacMahon H, et al. Recommendations for the management of subsolid pulmonary nodules detected at CT: a statement from the Fleischner Society. Radiology. 2013;266(1):304–17. [DOI] [PubMed] [Google Scholar]
- 6.Ridge CA, Yildirim A, Boiselle PM, et al. Differentiating between Subsolid and Solid Pulmonary Nodules at CT: Inter- and Intraobserver Agreement between Experienced Thoracic Radiologists. Radiology. 2016;278(3):888–96. [DOI] [PubMed] [Google Scholar]
- 7.van Riel SJ, Sanchez CI, Bankier AA, et al. Observer Variability for Classification of Pulmonary Nodules on Low-Dose CT Images and Its Effect on Nodule Management. Radiology. 2015;277(3):863–71. [DOI] [PubMed] [Google Scholar]
- 8.Chen PA, Huang EP, Shih LY, et al. Qualitative CT Criterion for Subsolid Nodule Subclassification: Improving Interobserver Agreement and Pathologic Correlation in the Adenocarcinoma Spectrum. Acad Radiol. 2018;25(11):1439–45. [DOI] [PubMed] [Google Scholar]
- 9.Armato SG, McNitt-Gray MF, Reeves AP, et al. The Lung Image Database Consortium (LIDC): An evaluation of radiologist variability in the identification of lung nodules on CT scans. Academic Radiology. 2007;14(11):1409–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lassen BC, Jacobs C, Kuhnigk JM, van Ginneken B, van Rikxoort EM. Robust semi-automatic segmentation of pulmonary subsolid nodules in chest computed tomography scans. Phys Med Biol. 2015;60(3):1307–23. [DOI] [PubMed] [Google Scholar]
- 11.Chae HD, Park CM, Park SJ, Lee SM, Kim KG, Goo JM. Computerized texture analysis of persistent part-solid ground-glass nodules: differentiation of preinvasive lesions from invasive pulmonary adenocarcinomas. Radiology. 2014; 273(1):285–93. [DOI] [PubMed] [Google Scholar]
- 12.McWilliams A, Tammemagi MC, Mayo JR, et al. Probability of cancer in pulmonary nodules detected on first screening CT. N Engl J Med. 2013;369(10):910–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swensen SJ, Silverstein MD, Ilstrup DM, Schleck CD, Edell ES. The probability of malignancy in solitary pulmonary nodules. Application to small radiologically indeterminate nodules. Archives of internal medicine. 1997; 157(8):849–55. [PubMed] [Google Scholar]
- 14.Zhou C, Chan HP, Sahiner B, et al. Automatic multiscale enhancement and hierarchical segmentation of pulmonary vessels in CT pulmonary angiography (CTPA) images for CAD applications. Medical physics. 2007;34(12):4567–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou C, Chan H-P, Wei J, Hadjiiski LM, Chughtai A, Kazerooni EA. Quantitative analysis of CT attenuation distribution patterns of nodule components for pathologic categorization of lung nodules. Proc SPIE 2017; 10134:1013422–6. [Google Scholar]
- 16.Dempster NM, Laird AP, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. JRStatist Soc B. 1977; 39:185–97. [Google Scholar]
- 17.Haralick RM, Shanmugam K, Dinstein I. Texture features for image classification. IEEE Transactions on Systems, Man, and Cybernetics. 1973; SMC-3:610–21. [Google Scholar]
- 18.Chapelle O, Haffner P, Vapnik VN. Support vector machines for histogram-based image classification. IEEE Transactions on Neural Networks. 1999; 10(5):1055–64. [DOI] [PubMed] [Google Scholar]
- 19.Tibshirani R. Regression shrinkage and selection via the Lasso. Journal of the Royal Statistical Society Series B-Methodological. 1996; 58(1):267–88. [Google Scholar]
- 20.Bewick V, Cheek L, Ball J. Statistics review 8: Qualitative data - tests of association. Critical care (London, England). 2004;8(1):46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McWilliams A, Tammemagi MC, Mayo JR, et al. Probability of Cancer in Pulmonary Nodules Detected on First Screening CT. 2013; 369(10):910–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swets JA. ROC analysis applied to the evaluation of medical imaging techniques. Invest Radiology. 1979; 14:109–21. [DOI] [PubMed] [Google Scholar]
- 23.Delong ER, Delong DM, Clarkepearson DI. Comparing the areas under 2 or more correlated receiver operating characteristic curves - A nonparametric approach. Biometrics. 1988;44(3):837–45. [PubMed] [Google Scholar]
- 24.Obuchowski NA. Nonparametric analysis of clustered ROC curve data. Biometrics. 1997; 53(2):567–78. [PubMed] [Google Scholar]
- 25.Bland JM, Altman DG. Statistics notes: Multiple significance tests: the Bonferroni method. BMJ. 1995; 310(6973):170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noguchi M, Morikawa A, Kawasaki M, et al. Small adenocarcinoma of the lung. Histologic characteristics and prognosis. Cancer. 1995; 75(12):2844–52. [DOI] [PubMed] [Google Scholar]
- 27.Thunnissen E, Beasley MB, Borczuk AC, et al. Reproducibility of histopathological subtypes and invasion in pulmonary adenocarcinoma. An international interobserver study. Mod Pathol. 2012; 25(12):1574–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nature communications. 2014; 5:4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oakden-Rayner L, Carneiro G, Bessen T, Nascimento JC, Bradley AP, Palmer LJ. Precision Radiology: Predicting longevity using feature engineering and deep learning methods in a radiomics framework. Scientific reports. 2017; 7(1):1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petkovska I, Brown MS, Goldin JG, et al. The effect of lung volume on nodule size on CT. Academic Radiology. 2007; 14(4):476–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrou M, Quint LE, Nan B, Baker LH. Pulmonary Nodule Volumetric Measurement Variability as a Function of CT Slice Thickness and Nodule Morphology. Am J Roentgenol. 2007; 188(2):306–12. [DOI] [PubMed] [Google Scholar]
- 32.Way TW, Chan H-P, Goodsitt MM, et al. Effect of CT scanning parameters on volumetric measurements of pulmonary nodules by 3D active contour segmentation: a phantom study. Physics in Medicine and Biology. 2008; 53(5):1295–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee KH, Goo JM, Park SJ, et al. Correlation between the size of the solid component on thin-section CT and the invasive component on pathology in small lung adenocarcinomas manifesting as ground-glass nodules. J Thorac Oncol. 2014; 9(1):74–82. [DOI] [PubMed] [Google Scholar]