Abstract
The emergence of the SARS-CoV-2 Omicron variant is dominant in many countries worldwide. The high number of spike mutations is responsible for the broad immune evasion from existing vaccines and antibody drugs. To understand this, we first present the cryo-electron microscopy structure of ACE2-bound SARS-CoV-2 Omicron spike. Comparison to previous spike antibody structures explains how Omicron escapes these therapeutics. Secondly, we report structures of Omicron, Delta, and wild-type spikes bound to a patient-derived Fab antibody fragment (510A5), which provides direct evidence where antibody binding is greatly attenuated by the Omicron mutations, freeing spike to bind ACE2. Together with biochemical binding and 510A5 neutralization assays, our work establishes principles of binding required for neutralization and clearly illustrates how the mutations lead to antibody evasion yet retain strong ACE2 interactions. Structural information on spike with both bound and unbound antibodies collectively elucidates potential strategies for generation of therapeutic antibodies.
Keywords: SARS-CoV-2, Omicron, spike, neutralization antibody, cryo-EM structures, immune evasion
Graphical abstract
Guo et al. report the cryo-EM structures of ACE2-bound SARS-CoV-2 Omicron spike and human antibody (510A5)-bound spikes of Omicron, Delta, and WT. The structures clearly illustrate how Omicron mutations lead to antibody evasion yet retain strong ACE2 interactions.
Introduction
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants associated with increased virulence or infectivity that occurred during the late months of 2020 has caused massive outbreaks in different regions (Tao et al., 2021; Wu et al., 2020; Zhou et al., 2020). The Delta variant (lineage B.1.617.2, first detected in India), which has become a dominant strain around the world since mid-2021 (Del Rio et al., 2021), has been gradually surpassed by the fast-spreading B.1.1.529 variant that was first identified on November 9, 2021. Due to the high level of genetic mutations, particularly in spike, the B.1.1.529 variant was classified as a variant of concern (VOC) and designated as Omicron by the World Health Organization on November 26, 2021. The US Centers for Disease Control and Prevention (Christensen et al., 2022) estimated 95.4% of COVID-19 cases between December 26, 2021, and January 1, 2022, were Omicron, barely one month after the first case was reported in the United States.
The epidemiology and transmission dynamics studies suggests that the Omicron variant spreads more rapidly and has a higher rate of asymptomatic infection than other VOCs (Christensen et al., 2022; Garrett et al., 2022). Of note, groups from different research institutions found that Omicron reduces or abrogates neutralization titers by sera from Pfizer-BioNTech or AstraZeneca vaccines and convalescent patients (Cao et al., 2021; Carreno et al., 2021; Dejnirattisai et al., 2022; Garcia-Beltran et al., 2022; Hoffmann et al., 2022; Planas et al., 2021). Additionally, a panel of the potent receptor-binding domain (RBD)-directed monoclonal antibodies (mAbs) were shown to completely or partially lose neutralizing activity against Omicron (Cameroni et al., 2021; Liu et al., 2021a). Unlike the Delta variant, Omicron has attenuated viral fusogenicity and relatively low pathogenicity, which may explain the fact that Omicron patients were younger with milder clinical symptoms (Halfmann et al., 2022; Maslo et al., 2021; Shuai et al., 2022; Suzuki et al., 2022). The immune evasion attributed to the high number of spike mutations undoubtedly correlates with Omicron’s rapid dissemination. However, these antigenic changes are not sufficient to explain the hyper-transmissibility of Omicron. Although accumulating evidence indicates that the Omicron variant replicates more readily in human bronchus tissue than lung tissue (McMahan et al., 2022; Peacock et al., 2022), the underlying mechanisms remain unclear.
SARS-CoV-2 uses its homotrimeric glycosylated spike protein to enter host cells (Walls et al., 2020). Spike is composed of two functional fragments S1 and S2 after proteolytic cleavage by furin-like proteases. The subunit S1 contains the N-terminal domain (NTD, residues 14–305), which recognizes attachment factors (Wang et al., 2021b), and the RBD (residues 328–531), which is responsible for engagement with the host receptor angiotensin-converting enzyme 2 (ACE2) (Walls et al., 2020). The RBD is the most important target for neutralizing antibodies elicited by either vaccination or viral infection (Piccoli et al., 2020; Wec et al., 2020).
Human neutralizing monoclonal antibodies (NAbs) targeting the RBD can be categorized into four classes according to their binding region as well as their neutralizing mechanisms (Barnes et al., 2020; Liu et al., 2021a). Class 1 and 2 include the most potent antibodies that bind to the RBD with epitopes that overlap or are close to RBD residues K417, E484, and N501. Class 3 NAbs bind outside the ACE2-binding site, comprising residues 440-450, and result in a relatively condensed epitope on RBD. Class 4 NAbs also do not overlap with the ACE2 binding site, and their highly conserved cryptic epitope can only be accessed when at least two RBDs on the spike protein are in the “up” state (Tortorici et al., 2021; Yuan et al., 2020). Several studies have shown that most of the substitutions that contribute to decreased antibody binding and immune escape are located on the RBD, which indicates the immunodominance of RBD (Harvey et al., 2021).
Compared to the original wild-type (WT) strain of SARS-CoV-2 (strain Wuhan-Hu-1) (Wu et al., 2020), the Omicron variant has an unusually high 60 mutations, 37 of which are in spike (Figure 1 A). 15 amino acid substitutions are located on the RBD (Viana et al., 2022). Some of these mutations are shared by previously circulating VOCs and have been proved to be associated with increased replication and transmission. For instance, the globally prevalent spike D614G mutant is responsible for enhanced binding to ACE2 and increased replication in animal models of infection (Weissman et al., 2021; Zhou et al., 2021). The N501Y substitution, present in the Alpha, Beta, and Gamma variants, increases ACE2 affinity and exhibits adaptive fitness gains for transmission (Liu et al., 2021b). Other mutants including K417N and E484A/K reduce the neutralization potency of vaccinated or convalescent plasma (Wang et al., 2021a, 2021c). Hence, understanding how these concerning mutations affect Omicron’s biological function and how SARS-CoV-2 establishes a balance between transmissibility, virulence, and immune escape is urgently needed.
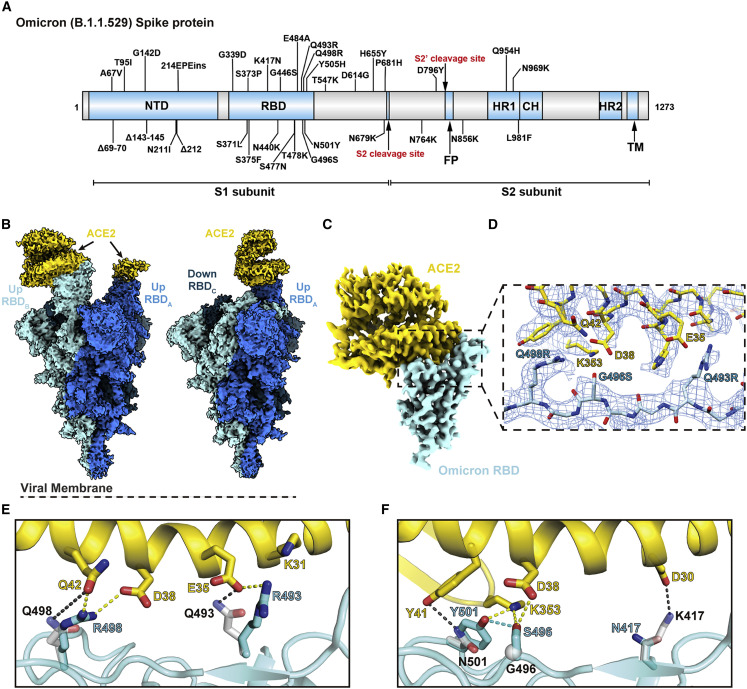
Figure 1.
Cryo-EM structure of the SARS-CoV-2 Omicron spike in complex with ACE2
(A) An overview diagram of the domain arrangement of the Omicron spike protein.
(B) Cryo-EM structures of Omicron spike with double- (left) and single (right)-bound ACE2, with three protomers shown in different colors.
(C) Cryo-EM map of the Omicron spike RBD in complex with human ACE2 after local refinement.
(D) Cryo-EM density at the Omicron RBD-ACE2 interface. The fitted atomic model is shown as sticks with oxygen colored in red, nitrogen colored in blue, and sulfurs colored in yellow.
(E and F) Comparison of the RBD-ACE2 interface between WT and the Omicron variant. Residues forming hydrogen bonds and ionic interactions are shown as sticks. Yellow and black dashed lines represent interactions between ACE2 and Omicron or WT RBDs, respectively. The hydrogen bond formed between Omicron RBD mutations G496S and N501Y is shown as a blue dashed line.
See also Figures S1 and S2; Table S1.
In this study, we present the cryo-electron microscopy (cryo-EM) structure of the Omicron spike complexed with ACE2 to analyze how Omicron can evade certain NAbs while maintaining RBD affinities to the receptors through new interactions. We also determined the cryo-EM structures of Omicron, WT, and Delta spikes in complex with one particular anti-WT/Beta RBD NAb to demonstrate how Omicron specifically evades the antibody neutralization. Our structural data are likely to contribute to the understanding for the rapid spread of the Omicron variant and help guide strategies for the development of broadly neutralizing antibodies.
Results
Cryo-EM structure determination of Omicron spike in complex with ACE2
The recombinant spike trimers including WT, Delta (B.1.617.2), and Omicron variants were expressed and purified to homogeneity. The properties of proteins were compared by using differential scanning fluorimetry (DSF) (Figure S1). The rapid DSF assay measures the inflection temperature (Ti) indicating the transition of the folding state of a protein, to demonstrate the thermal stability of the sample (Figure S1J). Ti of the Omicron spike shifted to lower temperatures compared to the WT and Delta, suggesting reduced protein stability.
To understand how Omicron spike engages ACE2, we analyzed the complex by cryo-EM. During preparation of the EM grids, the Omicron spikes appeared unstable compared to the WT and Delta spike trimers, as there was a much lower proportion of intact particles valid for further data collection (Figure S2A), which is consistent with the DSF results. We observed that the majority of classes containing ACE2 are bound to one or two RBDs from the spike trimer in the “up” conformation, with the other RBD in the “down” conformation (Figures 1B and S2B). No particles of spike trimer lacking ACE2 could be classified. The final asymmetric cryo-EM reconstruction of the Omicron spike-ACE2 complex was determined to an overall resolution of 2.8 Å (Figures S2B–S2G, Table S1). Local refinement resulted in clearly observed side chains of the ACE2-RBD interacting residues and reliable model building (Figures 1C–1F).
The overall architecture of the Omicron spike ECD is similar to WT and all earlier variants. The cryo-EM density of the Omicron spike clearly shows many of the mutations present in this variant (Figure 2 A). Consistent with recent reports during our manuscript preparation (Cui et al., 2022; Mannar et al., 2022; McCallum et al., 2022; Ni et al., 2021), the mutations are distributed mostly on the surface and some in the interior of the Omicron spike. The 15 RBD mutations are majorly clustered at one side of the domain, and some are involved in RBD-targeting antibody evasion (Figure 2A).
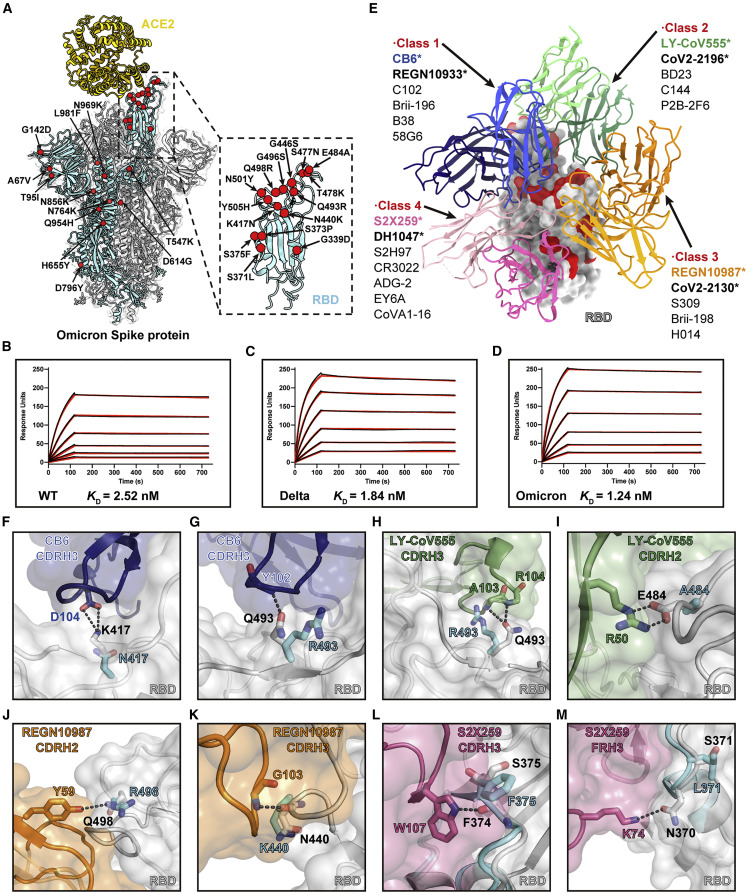
Figure 2.
Analysis of the Omicron spike RBD mutations in ACE binding and antibody evasion
(A) Model of Omicron spike protein with Cα of all mutated residues shown as red spheres.
(B–D) SPR analysis of the WT (B), Delta (C), and Omicron (D) spike protein affinities for human ACE2. The dissociation constant (KD) indicates mean ± SD of three independent replicates. Raw data is colored in black, and the red lines represent the fit to the raw data.
(E) Structural depiction of summarized Abs binding the RBD epitopes. A representative Ab from each class is shown. The Abs reported to be escaped by the Omicron variants are marked with stars.
(F–M) Structural details of the interface between neutralizing antibody Fabs and RBD. The Omicron RBD mutations S371L (M), S375F (L), K417N (F), N440K (K), E484A (I), Q493R (G and H), and Q498R (J) are likely to introduce steric clashes and/or abolish the specific hydrogen bonds.
Structural analysis reveals the molecular mechanism of Omicron mutations evading antibody recognition while retaining efficient ACE2 binding
The interface between ACE2 and the receptor-binding motif (RBM) on RBD has been extensively studied, which mainly involves polar interactions, particularly between ACE2 D30 and RBD K417, ACE2 K31/E35, and RBD Q493, and between ACE2 D38/Q42/K353 and RBD G496/Q498/N501 (Figure 1D). The ACE2 residues mostly lie on a long helix at the interface, with their side chains moving freely to interact with the RBD residues at the loop regions, most of which are not conserved in different variants, such as K417N, Q493R, G496S, Q498R, and N501Y in Omicron RBD (Figures 1E and 1F).
The absence of a strictly fit interface between ACE2 and RBD allows for higher tolerance to mutations. For example, in the Omicron RBD, the Q493R mutation forms a new salt bridge to ACE2 E35 (Figure 1E). Q498R forms a hydrogen bond and a new salt bridge with ACE2 Q42 and D38, respectively (Figure 1E). G496S adds new hydrogen bonds to ACE2 K353 and D38 at the interface and also stabilizes N501Y by a hydrogen bond (Figure 1F). We confirmed the importance of these interactions by surface plasmon resonance (SPR) assays using spike trimers of WT, Delta, and Omicron variants to ACE2. Unlike the reported scenario for other variants such as Alpha with a much lower dissociation constant (k d) (Han et al., 2021; Tanaka et al., 2021), here all three spikes assayed showed approximately similar binding affinity (Figures 2B–2D), which agrees with other previous studies (Cao et al., 2021; Mannar et al., 2022; Ni et al., 2021).
The spike RBD is the major target for NAbs from convalescent plasma from individuals recovering from SARS-CoV-2 (Hoffmann et al., 2022). Among the many mAbs in various development stages, current therapeutic antibodies bind to the RBD and effectively neutralize SARS-CoV-2 (Barnes et al., 2020; Greaney et al., 2021; Kim et al., 2021; Wheatley et al., 2021). We analyzed these antibodies with published structures, including those used as single agents or combination therapies (CB6, the parent mAb of JS016 etesevimab [Du et al., 2021]; COV2-2196 and COV2-2130, the parent mAbs of AZD8895 and AZD1061 [Zost et al., 2020]; LY-CoV555 [Starr et al., 2021b], REGN10933 and REGN10987 [Baum et al., 2020], S2X259 [Tortorici et al., 2021], etc.) by superposing the Fab-RBD structures on that of the Omicron RBD. The mutations clearly cluster at the NAb epitope sites (Figure 2E).
To explain the significantly reduced binding affinity and potency of NAbs against RBD, we analyzed those Omicron mutations involved in ACE2 binding. These antibodies bind using heavy chain complementarity determining regions that form a snug fit to RBD, such as CDRH3 loops in CB6 (Figures 2F and 2G), CDRH2 and CDRH3 loops in LY-CoV555 (Figures 2H and 2I), and CDRH2 and CDRH3 loops in REGN-10987 (Figures 2J and 2K). Mutations on the interface are likely to introduce steric clashes and/or abolish specific hydrogen bonds. K417N could be responsible for reducing the activity of CB6 (class 1) due to the lost hydrogen bonds to D104 in CDRH3 (Figure 2F). Q493R could also contribute to the disruption of CB6 binding due to the large side chain of arginine clashing with the closely embedded CDRH3 and failing to form a hydrogen bond with the mainchain NH group of Y102 (Figure 2G). A similar situation could also be observed in LY-CoV555 (class 2) where Q493R loses the hydrogen bonds to the main-chain NH groups of A103 and R104 in CDRH3 (Figure 2H). The previously known E484A mutation (Laurini et al., 2021; Shruti et al., 2021) also abolishes the hydrogen bond to the side chain of R50 in CDRH2 (Figure 2I). Q498R could account for the escape against REGN10987 (class 3) as the mutation disrupts the well-fitting contact and the hydrogen bonds between RBD Q498 and Y59 in CDRH2 (Figure 2J). N440K additionally contributes to the resistance to REGN10987 by steric hindrance and the loss of bonding to the mainchain NH group of G103 in CDRH3 (Figure 2K). For S2X259 (class 4), the mutations from serine to other bulkier side chains such as S375F and S371L cause a conformational change to disorder the interactions mediated by neighboring residues (Figures 2L and 2M). Due to indirect interference, the potency of the antibody in class 4 was reported to be reduced but not eliminated (Cameroni et al., 2022; Liu et al., 2021a). These structural analyses reveal that specific antibody recognition to RBD epitopes appears strict and less tolerable to RBD mutations compared to receptor engagement. These analyses are verified by several research groups that report RBD-targeting neutralizing antibodies showed abolished or impaired potency against the Omicron variant, including antibodies of all four classes (Barnes et al., 2020; Liu et al., 2021a; Ni et al., 2021).
To sum up, we show that Omicron RBD mutations effectively interrupt the snug fit of the Fab binding regions to RBD. Remarkably, these mutations form new hydrogen bonds and salt bridges with ACE2 without disrupting the binding interface, showing that the mutations have dual roles. However, due to the large variety of antibody interactions and the approach angles in each class, the selected antibodies from different classes may not represent the entire classes of NAbs. This analysis may only explain the escape mechanism for these specific antibodies.
Cryo-EM structures of Omicron spike in complex with antibody 510A5 against WT and Beta show how Omicron maintains ACE2 binding
In our previous work, we have reported one potent RBD-targeting mAb derived from a patient’s serum with remarkable neutralizing efficacy against the WT and Beta variants, named 510A5 (Li et al., 2021). In both the spike ECD and RBD binding assays, 510A5 showed a significant reduction in the binding affinity for Omicron compared to WT and Delta (Figures 3A and 3B). Using a pseudovirus neutralization assay, we probed whether 510A5 could also block entry of Omicron and Delta pseudoviruses into ACE2-expressing host cells and found that 510A5 failed to efficiently cross-neutralize Omicron, in contrast to its retained potency against Delta (Figure 3C). Loss of RBD antibody binding due to attenuated affinity could majorly result in the loss of Omicron neutralization activity of 510A5.
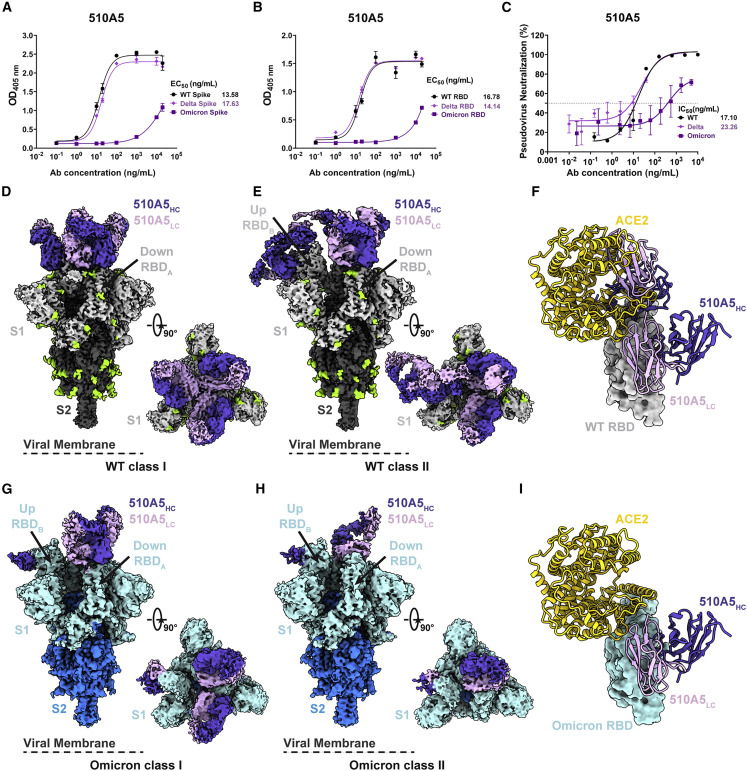
Figure 3.
Cryo-EM structures of 510A5 mAbs binding to SARS-CoV-2 WT, Delta, and Omicron spike protein
(A and B) The binding capabilities of 510A5 against spike protein (A) and RBD protein (B) of WT, Delta, or Omicron measured by ELISA. Data are presented as mean values ± SEM of three independent experiments.
(C) The neutralizing potencies of the 510A5 against WT, Delta, and Omicron measured by pseudovirus neutralization assay. Data are presented as mean values ± SEM of three independent experiments. Dashed line indicates a 50% reduction in viral neutralization.
(D and E) Cryo-EM densities for the 510A5 Fab-WT spike complex observed in two classes. (D) WT class I, 3.2 Å, revealing binding of 510A5 to RBDs in the “3 down” state; (E) WT class II, 3.4 Å, revealing binding of 510A5 to RBDs in “1 up, 2 down” state.
(F) Superposition of the local refined RBD-ACE2 model to that of WT RBD-510A5 model shows binding of two Fabs on both epitopes 1 and 3 completely blocks the ACE2 binding.
(G and H) Cryo-EM densities for the 510A5 Fab-Omicron spike complex observed in two classes with RBDs in a “1 up, 2 down” state. (G) Omicron class I, 3.7 Å, three Fabs bound; (H) Omicron class II, 3.7 Å, two Fabs bound.
(I) Superposition of the local refined Omicron RBD-ACE2 model to that of the 510A5-Omicron RBD model shows no steric hindrance between 510A5 Fab and ACE2.
See also Figures S3 and S4; Table S2.
To understand the structural basis of how 510A5 fails to neutralize only Omicron, we solved cryo-EM structures of the 510A5 Fab in complex with either WT (Figures S3A and S3B), Delta (Figures S4A and S4C), or Omicron (Figures S4B and S4D) spike ECD trimers. All spike trimers in the cryo-EM reconstruction were decorated with 510A5 Fabs, including the Omicron spike, albeit with different conformational states and Fab recognition modes.
For Fab-bound WT spike complexes, two major classes of particles could be identified, which we name WT class I and class II, at a global resolution of 3.2 Å and 3.4 Å, respectively (Figures S3C–S3F). In WT class I, three Fabs bind to three “down” RBDs, preserving the 3-fold symmetric conformation (Figure 3D). WT class II revealed an asymmetric conformation with four Fabs bound to the “1 up, 2 down” RBD conformation, with two Fabs attached to one up RBD and two Fabs bound to the other two down RBDs (Figure 3E). The up RBD occupied by two Fabs simultaneously occludes the ACE2 binding site (Figure 3F). In the Fab-bound Delta spike complexes, only one dominant particle class was observed with a global resolution of 3.6 Å, which is identical to WT class I (Figures S4E and S4H). In both WT class I and Delta, the Fabs lock spike in the “three down” RBD conformation, which would also contribute to prohibiting ACE2 binding.
Despite the low binding affinity, we were able to capture a structure with 510A5 Fabs binding to Omicron spike. In the scenario where the Fab binds Omicron, all the spike trimers adopted the “1 up, 2 down” conformation, and no particles with the “three down” RBD conformation could be classified (Figures 3G and 3H). Omicron spike relieves the conformational restriction caused by 510A5 binding. Two particle classes (Omicron class I and class II) could be obtained with three or two Fabs bound, respectively, at a global resolution of 3.7 Å (Figures S4F–S4J). In both classes, there was only one Fab bound to the up RBD, and other Fabs bound with the down RBDs. Superimposition of the up RBDs in the structures of Fab-Omicron spike and ACE2-Omicron spike complexes suggested no significant overlap between the Fab and ACE2 (although only a few minor clashes may occur between side chains), which could also be one of the possible explanations for the loss of 510A5 potency to Omicron when the Fab is able to bind spike (Figure 3I).
To validate the neutralizing mechanism of 510A5 we proposed based on our cryo-EM structures, we investigated the affinities between 510A5 and either WT spike or RBD using bio-layer interferometry (BLI) (Figure S5A). The affinity (K D values) of 510A5 for WT spike was more than 200 times higher than RBD. The off-rates (k d values) of 510A5 for WT spike and RBD exhibited more than a 1000-fold difference. The kinetic differences between 510A5 and either WT spike or RBD support our structural observation that the conformation of RBD in the context of spike, and likely avidity due to trimerization, play a role in 510A5 affinity. In contrast, the BLI data of 510A5 binding to the Omicron spike showed that the affinity is 2000 times weaker than WT spike, and there is a 7000-fold faster off-rate than the WT spike (Figure S5B). This is consistent with our ELISA experiments that the binding of 510A5 to Omicron spike is significantly reduced compared to the WT. Next, we compared the ability of 510A5 to compete with ACE2 for binding to the WT or Omicron spike. In our BLI assay, 510A5 exhibited strong inhibition of ACE2-WT spike binding (Figure S5C) as we reported previously (Li et al., 2021), while it failed to efficiently block the interaction between ACE2 and Omicron spike (Figure S5D). The fast-off rate characterization of 510A5 binding to Omicron spike should contribute mainly to the lack of competition with ACE2.
In the scenario where 510A5 does bind to Omicron, our structural studies reveal that Omicron spike presents distinct conformations and Fab recognition modes that lead to an exposed RBM and potentiated ACE2 binding. The fast-off rate and low binding affinity together with these structural differences may explain the loss of neutralization activity of 510A5 against Omicron but not other variants.
Omicron mutations greatly disrupt 510A5 binding and allow for spike to bind ACE2
To understand how Omicron, but not WT, spike can expose an up RBD in the presence of 510A5, we examined interfaces between the Fab variable domains and the RBD of WT and Omicron, respectively. In both WT and Omicron classes, we identified two epitopes (1 and 2) for 510A5 bound to the down configuration of RBD, as one Fab could span two RBDs, so one RBD could associate simultaneously with two Fabs (Figures 4A–4C). One Fab is associated with a down RBD through epitope 1, mainly via residues from CDRH1 and CDRH3 to RBD residues T345, R346, D442, K444, V445, and Y451 (Figures 4D and 4I). A small region of Fab light chain complementarity determining regions 1 and 3 (CDRL1 and CDRL3) also contacts RBD residues of N439, N440, and T500 (Figures 4D and 4I). These surface residues were fully solvent exposed even when the RBD was in the down conformation. In any particle class containing “down” RBDs, irrespective of whether they are WT, Delta, or Omicron spike, 510A5 engaged epitope 1 with a buried surface area of 719 Å2 (Figures 4D and 4G). Based on the location of epitope 1 and the binding mode of 510A5 to RBD, we infer 510A5 as a class 3 NAb. Epitope 2, comprised of RBD residues Y449, Q498, T500, and N501, bound light chain framework regions 1 and 3 (FRL1 and FRL3) on the adjacent down RBD, which contributed an additional 272 Å2 (Figures 4E and 4J).
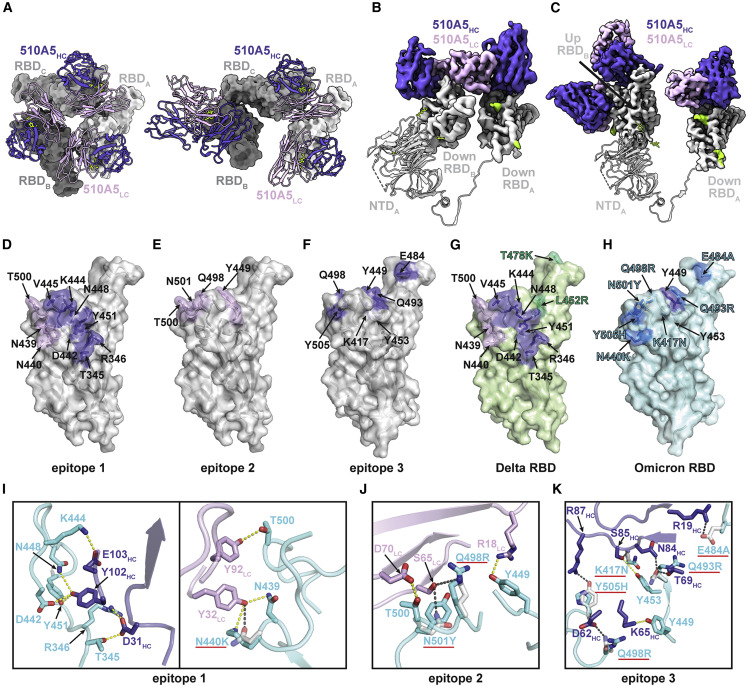
Figure 4.
Structural mechanism of Omicron spike escaping 510A5
(A) Left, a close-up view of 510A5 light-chain-mediated contacts on adjacent protomer “down” RBD, preserving a 3-fold symmetric conformation in WT class I. Right, a close-up view of an asymmetric conformation with four Fabs bound to the “1 up, 2 down” RBD conformation in WT class II.
(B) 510A5 shares inter-protomer contacts via binding to an adjacent down RBD to form the effective epitope 1 in all five classes of complexes observed.
(C) An extra 510A5 Fab binds to the up RBD to form epitope 3 in WT class II.
(D–F) Surface representations of 510A5 epitope 1 (purple and pink, D), epitope 2 (pink, E) and epitope 3 (purple, F) on the WT RBD surface (gray). RBD epitope residues (defined as residues forming potential hydrogen bonds with 510A5 Fab residues) are labeled in black.
(G) Surface representations of 510A5 epitope 1 (purple and pink) on the Delta RBD surface (green). RBD epitope residues are labeled in black. Two Delta mutations are labeled in green.
(H) Surface representations of the Omicron RBD (blue). Mutations on the Omicron RBD surface region corresponding to the WT epitope 3 are labeled in blue.
(I and J) Composite model of 510A5 Fab-WT RBD overlaid with the 510A5 Fab-Omicron RBD model. Potential hydrogen bonds on epitope 1 (I) and epitope 2 (J) are illustrated by dashed yellow (Omicron) or black (WT) lines, respectively. Omicron mutations are highlighted with red underlines.
(K) Composite model of 510A5 Fab-WT RBD overlaid with the Omicron RBD model. Potential hydrogen bonds are illustrated by dashed yellow (Omicron) or black (WT) lines, respectively. Omicron mutations (highlighted with red underlines) disrupt numerous interactions to 510A5 and exclude the epitope 3.
See also Figure S5.
In WT class I and Delta, three Fabs coordinate in turns to lock all three RBDs in the down conformation (Figure 4A). Here, the receptor-blocking activities of 510A5 are straightforward because it inhibits receptor recognition by preventing any up RBDs and subsequent exposure of the ACE2 binding site. We noticed that there is one Omicron mutation (N440K) in epitope 1, which could disrupt the local interactions between 510A5 and RBD. In the structure, it shows that there might be extra space for the larger lysin side chain, which could potentially tolerate the mutation to some extent. Since N440K is the only Omicron mutation on epitope 1 and is only involved in the interactions with the light chain of 510A5, it might explain the residual binding of 510A5 to epitope 1 of Omicron RBD (Figures 4H and 4I). Moreover, Q498R and N501Y in the Omicron variant were positioned at the center of epitope 2 of 510A5 and would weaken interactions and/or clash with S65 from the FRL3 and 510A5 binding from this region of RBD. This would explain the absence of the “three down” RBD conformation for Fab-Omicron spike complexes (Figures 4H and 4J).
In WT class II and two Omicron classes, 510A5 still occupied RBD epitope 1 in the up conformation. The up conformation of RBD exposed extra space for ACE2 binding (Figures 3G–3I). In WT class II, we observed another Fab-RBD interacting interface on the up conformation of RBD, named epitope 3 with a buried surface area of 670 Å2 and comprising RBD residues K417, Y449, Y453, E484, Q493, Q498, and Y505 (Figure 4F). The interface features extensive hydrogen bonding and salt bridge interactions between heavy chain framework regions 1 and 3 (FRH1 and FRH3) of 510A5 and the RBD residues (Figures 4F and 4K). The interface partially overlapped with the RBD-ACE2 binding surface. Thus, binding of two Fabs to the RBD in the up conformation to both epitopes 1 and 3 completely blocks the binding of ACE2, which could also contribute to the high potency of 510A5 to WT SARS-CoV-2. Our competitive ELISA and SPR assays support this mechanism where 510A5 also directly inhibits the interaction of WT RBD and ACE2 (Li et al., 2021). However, Omicron mutations in epitope 3, such as K417N, E484A, Q493R, Q498R, and Y505H, would disrupt interactions to Fab heavy chain residues R19, D62, R87, N84, and S85, and this would exclude binding of the second Fab to epitope 3 (Figures 4H and 4K). Structural analysis reveals that Delta RBD mutations (T487K and L452R) would lead to minimal disturbance to epitopes 1, 2, and 3 (Figure 4G), which is consistent with our observation of the potent neutralization activity of 510A5 against Delta.
In all, the structure of Omicron spike in complex with an antibody with minimal neutralizing activities has provided direct evidence captured by structural studies to uncover the molecular mechanism of antibody escape by Omicron.
Discussion
The high level of mutations in the Omicron variant have led to serious concerns over vaccine failure, immune escape, and increased transmissibility that have not been previously reported in any other VOC of SARS-CoV-2. Although many mutations emerged in Omicron, especially clustered in RBD, and most of them were not observed in the previous variants, the cryo-EM structures we determined and those reported by other groups recently demonstrate no large conformational changes (Cui et al., 2022; Mannar et al., 2022; Ni et al., 2021). It is of great interest and value to see the impact and details of mutations on ACE2 binding and antibody interactions to give a molecular explanation for the sweeping immune evasion of Omicron.
Here we present the cryo-EM structure of Omicron spike in complex with ACE2. The mutations on RBD contribute collectively to ACE2 binding, with some such as K417N inhibiting binding, while others such as N501Y enhance affinity. We speculate that the many newly emerged mutations subsequently result in retained ability to efficiently engage ACE2 while escaping the recognition by neutralizing antibodies.
To achieve this, viruses seem to evolve a distinct structural framework for the two types of binding activity. The ACE2 engagement involves a less compact interaction mediated by a long helical region in ACE2, leaving adequate space for side-chain interactions between RBD and ACE2. Such structural arrangement allows for high tolerance to mutations to compensate for the otherwise lost interactions or risk of steric hindrance. In contrast, a snug fit formed between Fabs and RBD leads to specific interactions between antibodies and RBD, with a consequence of tighter binding affinities for antibodies (Li et al., 2021; Wang et al., 2020) and competitive binding to ACE2. This rigid conformational feature results in a fastidious nature for antibodies toward RBD mutations. Thus, Omicron takes advantage of its malleable ACE2 binding to harbor mutations in spike that escape the strict antibody epitopes while simultaneously generating new ACE2 interactions to retain infectivity. Structural superposition of the Fab-WT RBD structures for several antibodies to that of the Omicron RBD depicts the potential influence of the RBD mutations on antibody binding. Due to the plasticity of the antigen surface, more quantitative studies are needed to further elucidate how individual mutations drive Omicron to escape.
In addition, we present the cryo-EM structure of one patient-derived SARS-CoV-2 NAb 510A5, which fails to neutralize the Omicron variant. It provides direct structural evidence for Omicron antibody evasion. The action modes of RBD-directed antibodies tested in different labs could be most generally described as two types: (1) blocking ACE2-RBD interactions through competitive binding to prevent viral entry and (2) targeting to RBD without occluding receptor binding. Some of the latter participate in conformational restrictions that prevent the binding of ACE2 and/or virus fusion. In our work, the antibody 510A5 functions in WT, Delta, and Beta variants, using both of the modes with up to four Fabs bound to the spike trimer, on three epitopes. We classified distinct conformational states for Omicron in complex with 510A5 in contrast to those observed for the WT and Delta. Our structures highlight the effect of N501Y, Q498R, Q493R, Y505H, K417N, and E484A on Fab interactions. We noted that although there are a few structures reported having two Fab molecules bind to the same protomer RBD, such as C144, C002, S2M11, 2-4, and others (Barnes et al., 2020; Liu et al., 2020; Tortorici et al., 2020), it is uncommon to observe either this situation in most other reported antibody-spike structures or several FR residues involved in interaction with RBD. Our structural data allows for unambiguous modeling of the interface between RBD and 510A5 in the WT structure (Figures S4I and S4J), enabling confident structural interpretation given the observed specific interactions. However, nonspecific binding may also exist to contribute to the contact between RBD and the Fab molecule given that some FR residues are involved. In summary, no structures of spike in complex with an antibody escaped by Omicron have been reported previously in the published work. Our findings provide the molecular basis for antibody evasion of Omicron due to extensive mutations that emerged on RBD while retaining the ability for ACE2 binding and high transmissibility.
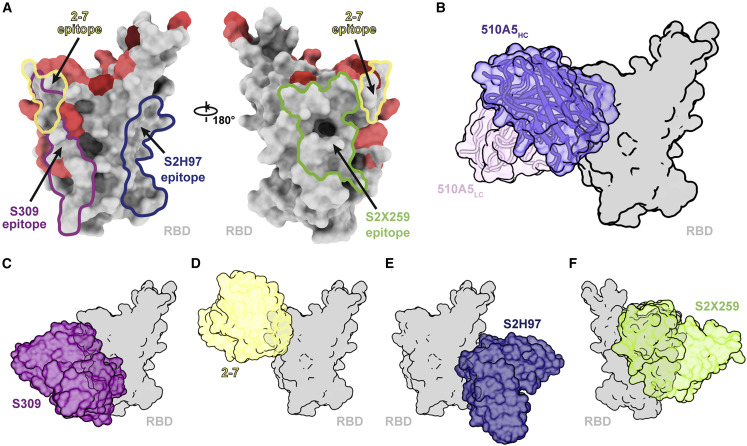
The strategy for 510A5 neutralization and Omicron evasion revealed from our structural studies shows that binding alone is insufficient for neutralization as reported previously in several other cases (Beaudoin-Bussières et al., 2022; Brouwer Philip et al., 2020; Ilinykh et al., 2020). We identified key epitopes of spike that result in 510A5 inactivation. When composing a mutational landscape of RBD colored with varied mutation frequencies observed in all the variants (Figure 5 A), most high-frequency mutations localize to regions related to the receptor binding site, tending to complicate drug and vaccine development. As reported recently, some existing therapeutic antibodies preserve their protection against Omicron, such as S309 (class 3, the parent mAb of VIR-7831 [sotrovimab]), S2H97 (class 4), S2X259 (class 4), and COV2-2196/COV2-2130 combination (class 2 and 3, the parent mAbs of AZD8895 [tixagevimab] and AZD1061 [cilgavimab]) (Pinto et al., 2020; Starr et al., 2021a; Tortorici et al., 2021; Zost et al., 2020). We analyzed the epitopes and the action modes of these antibodies and found that binding regions for class 3 and class 4 antibodies have fewer Omicron mutations (Figures 5C–5F). Therapies targeting these most highly conserved regions could be broadly and potently effective. Indeed, S309 of class 3 that retained substantial inhibitory activity against Omicron was originally discovered during SARS-CoV-1 natural infection and does not bind the ACE2-binding site (Pinto et al., 2020). 510A5 also acts similar to class 3 antibody epitope regions on Omicron RBD (Figure 5B). The dual-mode of action observed for 510A5 against WT RBD highlights the importance of antibody combination therapies. Though we noticed that 510A5 epitopes 2 and 3 are escaped by Omicron mutants, it shows the potential for increased neutralization when combined with antibodies in class 1 or 2 that block ACE2. As reported previously, S309 has shown enhanced SARS-CoV-2 neutralization when in combination with other antibodies (McCallum et al., 2022; Pinto et al., 2020). Targeting these epitopes simultaneously through a cocktail of antibodies may yield a broad SARS-CoV-2 therapy against Omicron and future variants, which recent studies have shown that the COV2-2196/COV2-2130 combination preserved considerable potency against Omicron (Liu et al., 2021a; Zost et al., 2020). The Omicron variant promotes the development of new interventions focusing on more conserved viral elements.
Figure 5.
Potential epitope regions on the spike RBD for effective antibody design
(A) Epitope footprints of four RBD-directed antibodies. The RBD is shown as surface, with mutations within the Omicron RBD highlighted from dark red to light red according to the mutation frequency among different variants so far. Different epitopes are labeled by color.
(B) Surface representation of RBD in complex with 510A5 Fab.
(C–F) Surface models of RBD in complex with four representative antibodies that remain some neutralizing ability against the Omicron variant. RBD is colored in gray, and the four Fab fragments are colored as follows: S309, purple (C); 2-7, yellow (D); S2H97, navy (E); S2X259, green (F).
Given the slippery nature of the spike-ACE2 interface, which is highly tolerant to mutations, strategies of combination therapies with antibodies targeting different spike epitopes are likely to be greatly beneficial and of high value.
Limitations of the study
The cryo-EM reconstructions of the Omicron spike trimer solved in this study using engineered stabilized constructs maintain a stable and active conformation for receptor and antibody recognition. Future studies on Omicron spike properties with in situ virion structures could provide more insights into the receptor binding and antibody evasion processes. In addition, the molecular mechanism of Omicron mutations evading antibody recognition is studied exclusively from structural alignments and immunological data reported in recent research. Moreover, the authentic virus infection assay, as well as the animal experiments need to be further studied since the pseudovirus system only represents the influence of spike protein, while other viral proteins could also contribute to infectivity and evasion.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| 510A5 | (Li et al., 2021) | N/A |
| Bacterial and virus strains | ||
| E. coli DH5α Competent Cells | WEIDI | Cat# DL1001 |
| Chemicals, peptides, and recombinant proteins | ||
| Trypsin | OXOID | Lot# 2862082 |
| Yeast Extract | OXOID | Lot# 2831838-02 |
| HEPES | VETEC | Lot# WXBD4400V |
| TRIS | VETEC | Lot# WXBD0198V |
| His-tagged SARS-CoV-2 Omicron variant Spike ECD | This paper | N/A |
| His-tagged SARS-CoV-2 WT Spike ECD | This paper | N/A |
| His-tagged SARS-CoV-2 Delta variant Spike ECD | This paper | N/A |
| His-tagged human ACE2 PD | This paper | N/A |
| Polyethylenimine | Polysciences | Cat# 24765-2 |
| Deposited data | ||
| Cryo-EM structure of SARS-CoV-2 WT Spike in complex with three 510A5 Fabs | This paper | EMDB: EMD-32739 PDB: 7WS0 |
| Cryo-EM structure of SARS-CoV-2 WT Spike in complex with four 510A5 Fabs | This paper | EMDB: EMD-32740 PDB: 7WS1 |
| Cryo-EM structure of SARS-CoV-2 WT down RBD in complex with one 510A5 Fab local refinement | This paper | EMDB: EMD-32741 PDB: 7WS2 |
| Cryo-EM structure of SARS-CoV-2 WT up RBD in complex with two 510A5 Fabs local refinement | This paper | EMDB: EMD-32749 PDB: 7WS7 |
| Cryo-EM structure of SARS-CoV-2 Delta Spike in complex with three 510A5 Fabs | This paper | EMDB: EMD-32742 PDB: 7WS3 |
| Cryo-EM structure of SARS-CoV-2 Omicron Spike in complex with two 510A5 Fabs | This paper | EMDB: EMD-32743 PDB: 7WS4 |
| Cryo-EM structure of SARS-CoV-2 Omicron Spike in complex with three 510A5 Fabs | This paper | EMDB: EMD-32744 PDB: 7WS5 |
| Cryo-EM structure of SARS-CoV-2 Omicron down RBD in complex with one 510A5 Fab local refinement | This paper | EMDB: EMD-32748 PDB: 7WS6 |
| Cryo-EM structure of SARS-CoV-2 Omicron Spike in complex with two ACE2 | This paper | EMDB: EMD-32750 PDB: 7WS8 |
| Cryo-EM structure of SARS-CoV-2 Omicron Spike in complex with one ACE2 | This paper | EMDB: EMD-32751 PDB: 7WS9 |
| Cryo-EM structure of SARS-CoV-2 Omicron up RBD in complex with ACE2 local refinement | This paper | EMDB: EMD-32752 PDB: 7WSA |
| Experimental models: Cell lines | ||
| FreeStyle 293 cells | ThermoFisher Scientific | Cat# R79007 |
| Recombinant DNA | ||
| Vector: pCAG-human ACE2 PD | This paper | N/A |
| Vector: pcDNA3.1-SARS-CoV-2 Omicron Spike ECD | This paper | N/A |
| Vector: pcDNA3.1-SARS-CoV-2 WT Spike ECD | This paper | N/A |
| Vector: pcDNA3.1-SARS-CoV-2 Delta Spike ECD | This paper | N/A |
| Software and algorithms | ||
| SerialEM | (Mastronarde, 2005) | https://bio3d.colorado.edu/SerialEM |
| MotionCor2 | (Zheng et al., 2017) | https://emcore.ucsf.edu/ucsf-software |
| RELION 3.0 | (Scheres, 2012) | https://www3.mrc-lmb.cam.ac.uk/relion |
| cryoSPARC v2.14.2 | (Punjani et al., 2017) | https://cryosparc.com/ |
| Coot 0.9.6 | (Emsley et al., 2010) | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| PHENIX | (Liebschner et al., 2019) | http://www.phenix-online.org/ |
| UCSF Chimera | (Pettersen et al., 2004) | https://www.cgl.ucsf.edu/chimera |
| UCSF ChimeraX | (Pettersen et al., 2021) | https://www.cgl.ucsf.edu/chimeraX |
| Pymol | Schrodinger, LLC | https://pymol.org/2/ |
| GraphPad Prism 8 | GraphPad | https://www.graphpad.com/ |
| Other | ||
| Ni Sepharose 6 Fast Flow | GE Healthcare | Lot# 10289601 |
| Superdex 200 Increase 10/300 column | GE Healthcare | Lot# 10246673 |
| Superose 6 Increase 10/300 column | GE Healthcare | Lot# 10290586 |
| 30 kDa cutoff concentrators | Millipore | REF# UFC903096 |
| 100 kDa cutoff concentrators | Millipore | REF# UFC910096 |
| R1.2/1.3 300 mesh Cu holey carbon grids | Quantifoil | Cat# Q3100CR-1.3 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Xiaoyun Ji (xiaoyun.ji@nju.edu.cn).
Materials availability
Bacterial strains and Antibodies were obtained from the commercial or academic sources described in the STAR Methods key resources table.
Reagents from this study are available upon request.
Experimental models and subject details
E. coli DH5α competent cells used in this study were from WEIDI. Cells were grown in LB medium.
FreeStyle 293-F cells (ThermoFisher Scientific, Cat# R79007) were maintained in OPM-293 CD05 Medium (OPM Biosciences) at 37°C with 5% CO2 before and after transfection.
Method details
Protein expression and purification
The gene encoding residues 1-1208 of SARS-CoV-2 Omicron Spike with 6P and furin mutations was synthesized, fused to a T4 fibritin trimerization motif, an HRV-3C protease cleavage site, a Twin-Strep-tag, and an 8×His-tag at the C terminus according to previous researches (Li et al., 2021; Mannar et al., 2022), and then cloned into the pcDNA3.1 vector. For the Spike protein production, the expression vector was transiently transfected into FreeStyle 293-F cells using PEI. Four days after transfection, the Omicron Spike protein was purified from filtered cell supernatants by affinity and gel filtration chromatography (Figure S1C). The production of the SARS-CoV-2 WT or Delta Spike protein was described previously (Li et al., 2021) consistent with this study (Figures S1A and S1B).
The gene encoding human ACE2 PD domain was cloned into a pCAG vector with an HRV-3C cleavage site and an 8×His-Tag at the end. The ACE2 protein was expressed by transient transfection of FreeStyle 293-F cells and purified by affinity and gel filtration chromatography.
Thermostability assays
The thermal stability was performed using Tycho NT.6 (NanoTemper Technologies). The protein unfolding profile was monitored using the intrinsic fluorescence at 350 and 330 nm with a temperature gradient from 35 to 95°C at a rate of 30 K/min. Data analysis was performed using the internal evaluation features of the Tycho instrument.
Cryo-EM sample preparation and data collection
Purified SARS-CoV-2 Omicron Spike protein was diluted to a concentration of 1.2 mg/mL and was incubated with ACE2 at a molar ratio of 1:3. Purified SARS-CoV-2 WT, Delta and Omicron Spike protein were diluted to a concentration of 1.6 mg/mL and were incubated with 510A5 Fab at a molar ratio of 1:3, respectively. To prevent aggregation during vitrification, 0.01% (w/v) n-dodecyl β-D-maltoside (DDM) was added to the sample before plunge freezing. The mixture sample was applied onto an H2/O2 glow-discharged, 300-mesh Quantifoil R1.2/1.3 copper grid. The grid was then blotted for 2.5 s with a blot force of −1 at 8°C and 100% humidity and plunge-frozen in liquid ethane using a Vitrobot Mark IV (ThermoFisher Scientific). Cryo-EM datasets were collected at a 300 kV Titan Krios microscope (ThermoFisher Scientific) equipped with a K3 detector (Gatan). The exposure time was set to 2.4 s for the WT and Delta datasets, and 2.0 s for the Omicron dataset, with a total accumulated dose of 60 electrons per Å2. Micrographs were collected using SerialEM (Mastronarde, 2005). The statistics of cryo-EM data collection can be found in Tables S1 and S2.
Cryo-EM data processing
All dose-fractioned images were motion-corrected and dose-weighted by MotionCor2 software (Zheng et al., 2017) and their contrast transfer functions were estimated by cryoSPARC patch CTF estimation (Punjani et al., 2017). The following particle picking, extraction, 2D classification, Ab-Initio reconstruction, 3D classification, 3D refinements and local resolution estimation were all carried out in cryoSPARC. For the dataset of the SARS-CoV-2 Omicron Spike-ACE2 complex, the final 3D reconstructions were obtained using non-uniform refinement with C1 symmetry, achieving a resolution of 3.0 Å for double-bound ACE2 and 2.8 Å for single-bound ACE2. For the dataset of the SARS-CoV-2 WT Spike-510A5 Fab complex, the final 3D reconstructions were obtained using non-uniform refinement, achieving a resolution of 3.2 Å for 3-Fabs-bound class, 3.4 Å for 4-Fabs-bound class and 3.3 Å for apo class. For the dataset of the SARS-CoV-2 Delta Spike-510A5 Fab complex, the final 3D reconstructions were obtained using non-uniform refinement, achieving a resolution of 3.6 Å for the 3-Fabs-bound class. For the dataset of the SARS-CoV-2 Omicron Spike-510A5 Fab complex, the final 3D reconstructions were obtained using non-uniform refinement, achieving a resolution of 3.7 Å for both 3 Fabs-bound and 2 Fabs-bound classes.
To improve the resolution for the binding interface, a local refinement focusing on the RBD-ACE2 region was carried out, achieving a 3.0 Å map representing the RBD-ACE2 interface. Similarly, the local resolution between 510A5 Fab variable domains and WT or Omicron RBD were improved to 3.4 Å or 3.9 Å, respectively.
The full cryo-EM data processing workflows are described in Figures S2–S4.
Model building and refinement
The recently reported structural model PDB entry 7T9K (Mannar et al., 2022) was used as an initial template for model building of the Omicron Spike trimer and ACE2. PDB entry 7K43 was used as an initial template to build the WT Spike trimer. The 510A5 Fab model was predicted using Phyre2 (Kelley et al., 2015). Manual and automated model building were iteratively performed using real-space refinement in Phenix 1.19 (Liebschner et al., 2019) and Coot 0.9.6 (Emsley et al., 2010), respectively. The SARS-CoV-2 Delta and Omicron Spike-510A5 Fab complex models were placed and rigid-body fitted into cryo-EM electron density maps using the already built WT Spike-510A5 Fab structure, respectively. The statistics of model validation can be found in Tables S1 and S2.
Surface plasmon resonance (SPR) experiments
SPR experiments were performed on the Biacore 8K instrument. Recombinant human ACE2-Fc was immobilized using the Series S Sensor protein A chip (Cytiva). Increasing concentrations (1.5625 nM, 3.125 nM, 6.25 nM, 12.5 nM, 25 nM, 50 nM) of various Spike ECD were flowed over the surface for single-cycle kinetic experiments. The surface was regenerated in 10mM glycine pH 1.0. The experiments were performed at 25°C, using a running buffer containing 10mM HEPES, 150mM NaCl, and 0.05% v/v Surfactant P20.
ELISA experiments
2 μg/mL of the recombinant Spike or RBD proteins derived from SARS-CoV-2 WT, Delta or Omicron (Sino Biological) were added to the 384-well plates (Corning) at 4°C overnight. Plates were blocked with blocking buffer (PBS containing 5% BSA) at 37°C for 1 h. Serially diluted 510A5 was added to the plates and incubated at 37°C for 45 min. Plates were washed with PBS, 0.05% Tween-20 (PBST) and ALP-conjugated Goat (Fab')2 Anti-Human IgG (Fab')2 (Abcam, Ab98532) was added into each well and incubated at 37°C for 30 min. Lastly, the PNPP substrate was added, and absorbance was measured at 405 nm by a microplate reader (ThermoFisher).
Production of pseudovirus bearing spike protein
pVSVG expressing SARS-CoV-2 Spike protein was constructed using the packaging plasmid (VSV-G pseudotyped ΔG-luciferase). It encoded either the Spike protein of SARS-CoV-2 WT, Delta or Omicron. Lenti-X293T cells were grown to 80% confluency before transfection with VSV-G pseudotyped ΔG-luciferase, pWPXL and pSFAX2. These cells were cultured overnight at 37°C with 5% CO2. DMEM supplemented with 5% fetal bovine serum and 100 IU/mL of penicillin and 100 μg/mL of streptomycin was added to the inoculated cells, which were cultured overnight for 72 h. The supernatant was harvested, filtered by 0.45 μm filter and centrifugated at 300 g for 10 min to collect the supernatant, then aliquoted and storied at −80°C.
Pseudovirus neutralization assay
Serially diluted mAbs (30 μL) were incubated with the same volume of the Lenti-X293T cell supernatants containing pseudovirus for 1 h at 37°C. These pseudovirus-antibody mixtures were added to ACE2 expressing Lenti-X293T cells (293T/ACE2). After 6 h, the wells were removed with supernatant and eluted with fresh cell culture mediums. The luciferase activities of infected 293T/ACE2 cells were detected by the One-Lumi luciferase reporter assay kit (Beyotime, RG055M). The IC50 of the evaluated mAbs were tested by the Varioskan LUX Microplate Spectrophotometer (ThermoFisher), and calculated by a nonlinear logistic regression using GraphPad Prism 8.0.
Bio-layer interferometry assays
Bio-layer interferometry assays were conducted on Octet® K2 Protein Analysis System (Fortebio). Protein biotinylation was performed using the EZ-link NHS-PEO Solid Phase Biotinylation Kit (Pierce) and purified using MINI Dialysis Unit (ThermoFisher). After baseline adsorption of nonspecific binding, SA biosensors (Fortebio) were immersed with biotinylated WT and Omicron RBD to capture RBD to 0.3 nm and 1 nm, then sensors were immersed in kinetics buffer (0.02% Tween-20, 1 mg/mL BSA in PBS) to the baseline. After association with different concentrations of 510A5 for 180 s, disassociation was conducted for 300 s. For the detection of the affinity of 510A5 with Spike protein, biotinylated 510A5 Fab was captured to 0.3 nm. Different concentrations of WT and Omicron Spike were conducted for 180 s, and disassociation was conducted for 300 s. For ACE2 binding competition experiments, biotinylated WT or Omicron Spike protein was loaded at 1 μg/mL to 3 nm in kinetics buffer onto SA biosensors. Association of mAbs was performed in kinetics buffer at 20 μg/mL for 300 s, and then ACE2-Flag-His was loaded for 300 s at 40 μg/mL in kinetics buffer. Data were recorded and analyzed using Octet BLI Discovery (12.0).
Quantification and statistical analysis
Quantification and statistical analyses employed in this publication pertain to the analysis on electron microscopy data and the determination of structures by electron microscopy. In Figures S2–S4, the resolution estimations of cryo-EM density maps are based on the 0.143 Fourier Shell Correlation (FSC) criterion (Chen et al., 2013; Rosenthal and Henderson, 2003).
Surface plasmon resonance assays and bio-layer interferometry assays were performed in triplicates. Neutralization assays were performed in biological triplicates.
Acknowledgments
We thank Dr. Lei Sun from The Fifth People’s Hospital of Shanghai, Fudan University, for the optimization guidance of spike protein expression and purification. We thank Dr. Yan Sun from Ruijin Hospital affiliated to Shanghai Jiao Tong University School of Medicine, for SPR data collection and analysis. We thank Dr. Kailin Yang for his valuable discussions. We thank the Bio-Electron Microscopy Facility of ShanghaiTech University, and we are grateful to Dr. Qianqian Sun, Zhihui Zhang and Yaping Wang for their help with cryo-EM technical support. This work was supported by funds from the National Key Research and Development Program of China (2018YFA0507100, 2016YFD0500300 to X.J.), National Natural Science Foundation of China (grant No. 81871639 to X.J.), National Key R&D Program of China (grant No. 2020YFA0707502 to H.Y.), Lingang Laboratory (grant No. LG202101-01-07 to H.Y.), Science and Technology Commission of Shanghai Municipality (grant No. YDZX20213100001556 and 20XD1422900 to H.Y.), National Natural Science Foundation of China (grant No. 92169109 to H.Y.), Shanghai Municipal Science and Technology Major Project (grant no. ZD2021CY001 to H.Y.), the program for Innovative Talents and Entrepreneur in Jiangsu, and the Fundamental Research Funds for the Central Universities.
Author contributions
X.J., H.Y., and Z.R. conceived and designed the project; H.G., X.J., and H.Y. designed the experiments; H.G., L.Z., Y.L., and Y.L. cloned, expressed, and purified proteins; A.J. and A.H. supervised antibody production and purification; H.G. and Y.L. performed SPR assay; F.L., S.S., and T.L. performed ELISA and BLI assays for the binding ability and the affinity of NAbs; F.L. performed BLI assay for the competition of 510A5 and ACE2 for WT or Omicron spike; T.L. conducted the pseudovirus neutralization assays; T.Y. was responsible for the thermostability assays of the spike variants; H.G., Y.G., and Y.L. collected the cryo-EM data; H.G. and Y.G. processed cryo-EM data; H.G. and Y.G. built and refined the structure model; X.J., H.Y., and H.Z. analyzed and discussed the data; H.G., T.L., H.N., H.Y., and X.J. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: April 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110770.
Supplemental information
Data and code availability
-
•
Cryo-EM structure coordinates and electron density maps for the SARS-CoV-2 WT, Delta and Omicron Spike and 510A5 Fab complexes, and the SARS-CoV-2 Omicron Spike and ACE2 complexes have been deposited with the Protein Data Bank and Electron Microscopy Data Bank and are publicly available as of the date of publication. Accession numbers are listed here and also in the key resources table: WT Spike in complex with three 510A5 Fabs: PDB 7WS0, EMD-32739; WT Spike in complex with four 510A5 Fabs: PDB 7WS1, EMD-32740; WT down RBD in complex with one 510A5 Fab local refinement: PDB 7WS2, EMD-32741; WT up RBD in complex with two 510A5 Fabs local refinement: PDB 7WS7, EMD-32749; Delta Spike in complex with three 510A5 Fabs: PDB 7WS3, EMD-32742; Omicron Spike in complex with two 510A5 Fabs: PDB 7WS4, EMD-32743; Omicron Spike in complex with three 510A5 Fabs: PDB 7WS5, EMD-32744; Omicron down RBD in complex with one 510A5 Fab local refinement: PDB 7WS6, EMD-32748; Omicron Spike in complex with two ACE2: PDB 7WS8, EMD-32750; Omicron Spike in complex with one ACE2: PDB 7WS9, EMD-32751; Omicron RBD in complex with ACE2 local refine: PDB 7WSA, EMD-32752.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.M.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Ajithdoss D., Copin R., Zhou A., Lanza K., Negron N., Ni M., Wei Y., Mohammadi K., Musser B., et al. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science. 2020;370:1110–1115. doi: 10.1126/science.abe2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin-Bussières G., Chen Y., Ullah I., Prévost J., Tolbert W.D., Symmes K., Ding S., Benlarbi M., Gong S.Y., Tauzin A., et al. A Fc-enhanced NTD-binding non-neutralizing antibody delays virus spread and synergizes with a nAb to protect mice from lethal SARS-CoV-2 infection. Cell Rep. 2022;38:110368. doi: 10.1016/j.celrep.2022.110368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer Philip J.M., Caniels Tom G., van der Straten K., Snitselaar Jonne L., Aldon Y., Bangaru S., Torres Jonathan L., Okba Nisreen M.A., Claireaux M., Kerster G., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K., Pinto D., VanBlargan L.A., De Marco A., di Iulio J., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2021;602:664–670. doi: 10.1038/d41586-021-03825-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K., Pinto D., VanBlargan L.A., De Marco A., di Iulio J., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602:664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2021;602:657–663. doi: 10.1038/d41586-021-03796-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreno J.M., Alshammary H., Tcheou J., Singh G., Raskin A., Kawabata H., Sominsky L., Clark J., Adelsberg D.C., Bielak D., et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature. 2021;602:682–688. doi: 10.1038/d41586-021-03846-z. [DOI] [PubMed] [Google Scholar]
- Chen S., McMullan G., Faruqi A.R., Murshudov G.N., Short J.M., Scheres S.H.W., Henderson R. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen P.A., Olsen R.J., Long S.W., Snehal R., Davis J.J., Saavedra M.O., Reppond K., Shyer M.N., Cambric J., Gadd R., et al. Signals of significantly increased vaccine breakthrough, decreased hospitalization rates, and less severe disease in patients with COVID-19 caused by the Omicron variant of SARS-CoV-2 in Houston, Texas. medRxiv. 2022 doi: 10.1101/2021.12.30.21268560. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z., Liu P., Wang N., Wang L., Fan K., Zhu Q., Wang K., Chen R., Feng R., Jia Z., Yang M., Xu G., Zhu B., Fu W., Chu T., Feng L., Wang Y., Pei X., Yang P., Xie X.S., Cao L., Cao Y., Wang X. Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron. Cell. 2022;185:860–871.e13. doi: 10.1016/j.cell.2022.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., Huo J., Zhou D., Zahradník J., Supasa P., Liu C., Duyvesteyn H.M.E., Ginn H.M., Mentzer A.J., Tuekprakhon A., et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185:467–484.e15. doi: 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Rio C., Malani P.N., Omer S.B. Confronting the Delta variant of SARS-CoV-2, summer 2021. JAMA. 2021;326:1001–1002. doi: 10.1001/jama.2021.14811. [DOI] [PubMed] [Google Scholar]
- Du Y., Shi R., Zhang Y., Duan X., Li L., Zhang J., Wang F., Zhang R., Shen H., Wang Y., et al. A broadly neutralizing humanized ACE2-targeting antibody against SARS-CoV-2 variants. Nat. Commun. 2021;12:5000. doi: 10.1038/s41467-021-25331-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. Section D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., St Denis K.J., Hoelzemer A., Lam E.C., Nitido A.D., Sheehan M.L., Berrios C., Ofoman O., Chang C.C., Hauser B.M., et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2022;185:457–466.e4. doi: 10.1016/j.cell.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett N., Tapley A., Andriesen J., Seocharan I., Fisher L.H., Bunts L., Espy N., Wallis C.L., Randhawa A.K., Ketter N., et al. High rate of asymptomatic carriage associated with variant strain Omicron. medRxiv. 2022 doi: 10.1101/2021.12.20.21268130. Preprint at. [DOI] [Google Scholar]
- Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H.D., et al. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29:44–57.e9. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann P.J., Iida S., Iwatsuki-Horimoto K., Maemura T., Kiso M., Scheaffer S.M., Darling T.L., Joshi A., Loeber S., Singh G., et al. Consortium Mount Sinai Pathogen Surveillance PSP study group SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature. 2022;603:687–692. doi: 10.1038/s41586-022-04441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han P., Su C., Zhang Y., Bai C., Zheng A., Qiao C., Wang Q., Niu S., Chen Q., Zhang Y., et al. Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants. Nat. Commun. 2021;12:6103. doi: 10.1038/s41467-021-26401-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Peacock S.J., Robertson D.L., COVID-19 Genomics UK COG-UK Consortium SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Krüger N., Schulz S., Cossmann A., Rocha C., Kempf A., Nehlmeier I., Graichen L., Moldenhauer A.-S., Winkler M.S., Lier M., Dopfer-Jablonka A., Jack H.M., Behrens G.M., Pohlmann S. The Omicron variant is highly resistant against antibody-mediated neutralization: implications for control of the COVID-19 pandemic. Cell. 2022;185:447–456.e11. doi: 10.1016/j.cell.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilinykh P.A., Huang K., Santos R.I., Gilchuk P., Gunn B.M., Karim M.M., Liang J., Fouch M.E., Davidson E., Parekh D.V., et al. Non-neutralizing antibodies from a marburg infection survivor mediate protection by fc-effector functions and by enhancing efficacy of other antibodies. Cell Host Microbe. 2020;27:976–991.e11. doi: 10.1016/j.chom.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C., Ryu D.-K., Lee J., Kim Y.-I., Seo J.-M., Kim Y.-G., Jeong J.-H., Kim M., Kim J.-I., Kim P., et al. A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein. Nat. Commun. 2021;12:288. doi: 10.1038/s41467-020-20602-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurini E., Marson D., Aulic S., Fermeglia A., Pricl S. Molecular rationale for SARS-CoV-2 spike circulating mutations able to escape bamlanivimab and etesevimab monoclonal antibodies. Sci. Rep. 2021;11:20274. doi: 10.1038/s41598-021-99827-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Han X., Gu C., Guo H., Zhang H., Wang Y., Hu C., Wang K., Liu F., Luo F., et al. Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants. Nat. Commun. 2021;12:6304. doi: 10.1038/s41467-021-26539-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D., Afonine P.V., Baker M.L., Bunkoczi G., Chen V.B., Croll T.I., Hintze B., Hung L.-W., Jain S., McCoy A.J., et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. Section D. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Iketani S., Guo Y., Chan J.F., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2021;602:676–681. doi: 10.1038/d41586-021-03826-3. [DOI] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F.W., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Liu Y., Liu J., Plante K.S., Plante J.A., Xie X., Zhang X., Ku Z., An Z., Scharton D., Schindewolf C., et al. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature. 2021;602:294–299. doi: 10.1038/s41586-021-04245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannar D., Saville James W., Zhu X., Srivastava Shanti S., Berezuk Alison M., Tuttle Katharine S., Marquez A.C., Sekirov I., Subramaniam S. SARS-CoV-2 Omicron variant: antibody evasion and cryo-EM structure of spike protein–ACE2 complex. Science. 2022;375:760–764. doi: 10.1126/science.abn7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslo C., Friedland R., Toubkin M., Laubscher A., Akaloo T., Kama B. Characteristics and outcomes of hospitalized patients in South Africa during the COVID-19 Omicron wave compared with previous waves. JAMA. 2022;327:583. doi: 10.1001/jama.2021.24868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- McCallum M., Czudnochowski N., Rosen Laura E., Zepeda Samantha K., Bowen John E., Walls Alexandra C., Hauser K., Joshi A., Stewart C., Dillen Josh R., et al. Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement. Science. 2022;375:864–868. doi: 10.1126/science.abn8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahan K., Giffin V., Tostanoski L.H., Chung B., Siamatu M., Suthar M.S., Halfmann P., Kawaoka Y., Piedra-Mora C., Martinot A.J., et al. Reduced pathogenicity of the SARS-CoV-2 Omicron variant in hamsters. bioRxiv. 2022 doi: 10.1101/2022.01.02.474743. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni D., Lau K., Turelli P., Raclot C., Beckert B., Nazarov S., Pojer F., Myasnikov A., Stahlberg H., Trono D. Structural analysis of the Spike of the Omicron SARS-COV-2 variant by cryo-EM and implications for immune evasion. bioRxiv. 2021 doi: 10.1101/2021.12.27.474250. Preprint at. [DOI] [Google Scholar]
- Peacock T.P., Brown J.C., Zhou J., Thakur N., Newman J., Kugathasan R., Sukhova K., Kaforou M., Bailey D., Barclay W.S. The SARS-CoV-2 variant, Omicron, shows rapid replication in human primary nasal epithelial cultures and efficiently uses the endosomal route of entry. bioRxiv. 2022 doi: 10.1101/2021.12.31.474653. Preprint at. [DOI] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera-A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., Ferrin T.E. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Snell G., Walls A.C., Veesler D., Beltramello M., Silacci-Fregni C., Pinto D., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042.e1021. doi: 10.2210/pdb7jxe/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J., Bolland W.H., Porrot F., Staropoli I., Lemoine F., et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2021;602:671–675. doi: 10.1038/d41586-021-03827-2. [DOI] [PubMed] [Google Scholar]
- Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Rosenthal P.B., Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Scheres S.H.W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shruti K., Céline G., Lucas F., Mark B.S., Gunter B., Amadou D., Nancy A., Joses H., Raphael T.C.L., Winston Y., et al. Perspectives: GISAID’s role in pandemic response. China CDC Weekly. 2021;3:1049–1051. doi: 10.46234/ccdcw2021.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai H., Chan J.F.-W., Hu B., Chai Y., Yuen T.T.-T., Yin F., Huang X., Yoon C., Hu J.-C., Liu H., et al. Attenuated Replication and Pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature. 2022;603:693–699. doi: 10.1038/s41586-022-04442-5. [DOI] [PubMed] [Google Scholar]
- Starr T.N., Czudnochowski N., Liu Z., Zatta F., Park Y.-J., Addetia A., Pinto D., Beltramello M., Hernandez P., Greaney A.J., et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature. 2021;597:97–102. doi: 10.1038/s41586-021-03807-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Dingens A.S., Bloom J.D. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. Cell Rep. Med. 2021;2:100255. doi: 10.1016/j.xcrm.2021.100255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R., Yamasoba D., Kimura I., Wang L., Kishimoto M., Ito J., Morioka Y., Nao N., Nasser H., Uriu K., et al. The Genotype to Phenotype Japan G2P-Japan Consortium Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature. 2022;603:700–705. doi: 10.1038/s41586-022-04462-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Hirayama A., Nagai H., Shirai C., Takahashi Y., Shinomiya H., Taniguchi C., Ogata T. Increased transmissibility of the SARS-CoV-2 Alpha variant in a Japanese Population. Int. J. Environ. Res. Public Health. 2021;18:7752. doi: 10.3390/ijerph18157752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao K., Tzou P.L., Nouhin J., Gupta R.K., de Oliveira T., Kosakovsky Pond S.L., Fera D., Shafer R.W. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 2021;22:757–773. doi: 10.1038/s41576-021-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Beltramello M., Lempp Florian A., Pinto D., Dang Ha V., Rosen Laura E., McCallum M., Bowen J., Minola A., Jaconi S., et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370:950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Czudnochowski N., Starr T.N., Marzi R., Walls A.C., Zatta F., Bowen J.E., Jaconi S., Di Iulio J., Wang Z., et al. Broad sarbecovirus neutralization by a human monoclonal antibody. Nature. 2021;597:103–108. doi: 10.1038/s41586-021-03817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana R., Moyo S., Amoako D.G., Tegally H., Scheepers C., Althaus C.L., Anyaneji U.J., Bester P.A., Boni M.F., Chand M., et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022;603:679–688. doi: 10.1038/d41586-021-03832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A.D.M.E., van Kuppeveld F.J.M., Haagmans B.L., Grosveld F., Bosch B.J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Zhang Q., Ge J., Ren W., Zhang R., Lan J., Ju B., Su B., Yu F., Chen P., et al. Analysis of SARS-CoV-2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species. Immunity. 2021;54:1611–1621.e5. doi: 10.1016/j.immuni.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Qiu Z., Hou Y., Deng X., Xu W., Zheng T., Wu P., Xie S., Bian W., Zhang C., et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021;31:126–140. doi: 10.1038/s41422-020-00460-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wec A.Z., Wrapp D., Herbert A.S., Maurer D.P., Haslwanter D., Sakharkar M., Jangra R.K., Dieterle M.E., Lilov A., Huang D., et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman D., Alameh M.G., de Silva T., Collini P., Hornsby H., Brown R., LaBranche C.C., Edwards R.J., Sutherland L., Santra S., et al. D614G spike mutation increases SARS CoV-2 susceptibility to neutralization. Cell Host Microbe. 2021;29:23–31.e4. doi: 10.1016/j.chom.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley A.K., Pymm P., Esterbauer R., Dietrich M.H., Lee W.S., Drew D., Kelly H.G., Chan L.-J., Mordant F.L., Black K.A., et al. Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain. Cell Rep. 2021;37:109822. doi: 10.1016/j.celrep.2021.109822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S.Q., Palovcak E., Armache J.-P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Thao T.T.N., Hoffmann D., Taddeo A., Ebert N., Labroussaa F., Pohlmann A., King J., Steiner S., Kelly J.N., et al. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature. 2021;592:122–127. doi: 10.1038/s41586-021-03361-1. [DOI] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Cryo-EM structure coordinates and electron density maps for the SARS-CoV-2 WT, Delta and Omicron Spike and 510A5 Fab complexes, and the SARS-CoV-2 Omicron Spike and ACE2 complexes have been deposited with the Protein Data Bank and Electron Microscopy Data Bank and are publicly available as of the date of publication. Accession numbers are listed here and also in the key resources table: WT Spike in complex with three 510A5 Fabs: PDB 7WS0, EMD-32739; WT Spike in complex with four 510A5 Fabs: PDB 7WS1, EMD-32740; WT down RBD in complex with one 510A5 Fab local refinement: PDB 7WS2, EMD-32741; WT up RBD in complex with two 510A5 Fabs local refinement: PDB 7WS7, EMD-32749; Delta Spike in complex with three 510A5 Fabs: PDB 7WS3, EMD-32742; Omicron Spike in complex with two 510A5 Fabs: PDB 7WS4, EMD-32743; Omicron Spike in complex with three 510A5 Fabs: PDB 7WS5, EMD-32744; Omicron down RBD in complex with one 510A5 Fab local refinement: PDB 7WS6, EMD-32748; Omicron Spike in complex with two ACE2: PDB 7WS8, EMD-32750; Omicron Spike in complex with one ACE2: PDB 7WS9, EMD-32751; Omicron RBD in complex with ACE2 local refine: PDB 7WSA, EMD-32752.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.