Gao et al. show that an endosomal population carrying the TORC1 signaling complex, which they term signaling endosomes (SEs), requires the HOPS tethering complex and MVB biogenesis for their formation and identity and for efficient TORC1 signaling.
Abstract
The endomembrane system of eukaryotic cells is essential for cellular homeostasis during growth and proliferation. Previous work showed that a central regulator of growth, namely the target of rapamycin complex 1 (TORC1), binds both membranes of vacuoles and signaling endosomes (SEs) that are distinct from multivesicular bodies (MVBs). Interestingly, the endosomal TORC1, which binds membranes in part via the EGO complex, critically defines vacuole integrity. Here, we demonstrate that SEs form at a branch point of the biosynthetic and endocytic pathways toward the vacuole and depend on MVB biogenesis. Importantly, function of the HOPS tethering complex is essential to maintain the identity of SEs and proper endosomal and vacuolar TORC1 activities. In HOPS mutants, the EGO complex redistributed to the Golgi, which resulted in a partial mislocalization of TORC1. Our study uncovers that SE function requires a functional HOPS complex and MVBs, suggesting a tight link between trafficking and signaling along the endolysosomal pathway.
Introduction
The endocytic pathway connects the plasma membrane to the endolysosomal compartment with its early and late endosome (EE and LE) and the lytic lysosome, where proteins are selectively degraded (Huotari and Helenius, 2011; Langemeyer et al., 2018). This connection allows a constant adjustment of the plasma membrane protein and lipid content in response to environmental cues or metabolic needs. Consequently, proteins are continuously surveyed and are selectively removed by endocytosis if they are bound to a ligand or cargo (Sardana and Emr, 2021). During endocytosis, internalized proteins are packaged into small vesicles, which are first delivered to the EE. Here, some proteins release their cargo and are sorted via the recycling endosome to the plasma membrane, whereas others are transferred from the EE to the LE (Huotari and Helenius, 2011). This process requires both maturation of EE to LE and multiple fusion events among EEs and LEs (Zeigerer et al., 2012). To allow membrane protein degradation, endosomal sorting complex required for transport (ESCRT) complexes sort these proteins into intraluminal vesicles (ILVs; Zhen et al., 2021). Consequently, maturation changes the tubular EE into a spherical structure with multiple ILVs. Mature LEs, now also called multivesicular bodies (MVBs), finally fuse with the lysosome to allow protein degradation for reuse of nutrients (Sardana and Emr, 2021; Huotari and Helenius, 2011).
Rab GTPases are crucial regulators of membrane trafficking, docking, and fusion events (Barr, 2013; Wandinger-Ness and Zerial, 2014; Hutagalung and Novick, 2011; Goody et al., 2017). All Rabs can bind to both GTP and GDP. For activation, a guanine nucleotide exchange factor (GEF) promotes loading of the Rab with GTP as a prerequisite for its ability to bind to effector proteins. Inactivation of the Rab requires a GTPase activating protein, which allows extraction of the Rab-GDP by the chaperone GDP dissociation inhibitor. In the endolysosomal system, Rab5 functions on EEs and interacts with the effector tethering complex class C core vacuole/endosome tethering (CORVET), among others, to promote EE fusion (Balderhaar and Ungermann, 2013; Balderhaar et al., 2013). During endosome maturation, Rab5 recruits and activates the Mon1–Ccz1 GEF complex, which in turn activates Rab7 on LEs (Nordmann et al., 2010; Langemeyer et al., 2020). In yeast, the Rab7-homolog Ypt7 then recruits the heterohexameric homotypic fusion and vacuole protein sorting (HOPS) complex (Wurmser et al., 2000; Bröcker et al., 2012; Seals et al., 2000). HOPS has two binding sites for Ypt7 and bridges LEs and vacuoles to promote the assembly of SNAREs from both organelles, and thus drive fusion (Wickner and Rizo, 2017; Mima and Wickner, 2009; Baker et al., 2015; Bröcker et al., 2012; van der Beek et al., 2019). Importantly, HOPS also supports fusion of autophagosomes and AP-3 vesicles with the yeast vacuole (Schoppe et al., 2020; Gao et al., 2018; van der Beek et al., 2019; Cabrera et al., 2010).
Endosomal maturation is accompanied by changes in lipid composition, most prominently in phosphoinositides. EEs are marked by phosphatidylinositol-3-phosphate (PI3P), which is generated by the Vps34 PI-3 kinase complex (Schu et al., 1993), whose activity is promoted by Rab5 (Tremel et al., 2021). At LEs, PI3P is further phosphorylated by the only PI3P 5-kinase Fab1 (PIKfyve in metazoans; Hasegawa et al., 2017; Ho et al., 2012). Both lipid kinases function as part of large complexes and localize to multiple membranes of the endolysosomal system. Proteins can specifically bind to the phosphorylated inositol head group, often with coinciding binding to Rab GTPases or other membrane proteins (Balla, 2013). Consequently, changes in Rab composition and PIPs also result in a change in the general membrane composition of maturing organelles.
The endolysosomal system of yeast seems to be less complex than the mammalian system (Day et al., 2018). It thus came as a surprise when signaling endosomes (SEs) were described as a novel endosomal population distinct from MVBs in yeast (Hatakeyama and Virgilio, 2019b; Hatakeyama et al., 2019; Hatakeyama and Virgilio, 2019a). Like vacuoles, these endosomes harbor the highly conserved target of rapamycin complex 1 (TORC1) kinase and its regulatory exit from G0 (EGO) complex (Chen et al., 2021; Hatakeyama et al., 2019), named Rag–Ragulator complex in metazoans (Kanarek et al., 2020). Interestingly, SEs lack the ESCRT-IV ATPase Vps4 required for ILV formation (Babst et al., 1998) but contain a population of the Fab1 lipid kinase (Chen et al., 2021). We recently showed that Fab1 is a substrate of the TORC1 complex and that Fab1 phosphorylation promotes its localization to SEs in addition to its localization to MVBs and vacuoles (Chen et al., 2021). This suggests that the activity of TORC1 controls Fab1 and thus the biogenesis of SEs. However, the exact link between MVBs and SEs remains unresolved.
One additional marker protein found on SEs is the I-BAR protein Ivy1 (Chen et al., 2021). Ivy1 is an effector of Ypt7, binds PI3P, and can inhibit Fab1 (Numrich et al., 2015; Malia et al., 2018; Lazar et al., 2002). It also dynamically relocalizes from puncta to vacuoles and vacuolar microdomains in response to nutrient starvation or cellular stress (Numrich et al., 2015; Zweytick et al., 2014; Varlakhanova et al., 2018a; Ishii et al., 2019). We showed before that a fraction of Ivy1 colocalizes with the EGO complex (EGOC) in endosomal dots, which are distinct from MVBs, as they lack Vps4 (Chen et al., 2021; Hatakeyama et al., 2019). Here, we focus on the biogenesis of SEs as a novel endosomal population. Our data reveal that SEs harbor not just a pool of TORC1 and the EGOC, but also the Rab7-like Ypt7 and the Rab5-like Vps21. Importantly, both ESCRTs and HOPS are important to maintain the identity of SEs. Our data suggest that SEs are dynamic structures that form at an interface between the endocytic pathway and the Golgi by continuous fission and fusion processes.
Results
SEs and MVBs are distinct endosomal populations
We previously showed that Ivy1, Fab1, and TORC1 localize to the vacuole and to endosomal dots proximal to the vacuole, which we coined SEs (Hatakeyama et al., 2019; Chen et al., 2021). To understand the dynamics and function of SEs in the context of the endosomal pathway, we wondered whether we could take Ivy1 as a reference marker of SEs given that the protein, like the other described and above-mentioned SE marker proteins, localizes dynamically to endosomes and vacuoles (Malia et al., 2018; Varlakhanova et al., 2018a; Numrich et al., 2015; Chen et al., 2021). However, this dynamic localization also applies to all other marker proteins of SEs and MVBs. We thus reasoned that the analysis of the relative localization of these markers to each other should reveal, how SEs form or maintain their identity.
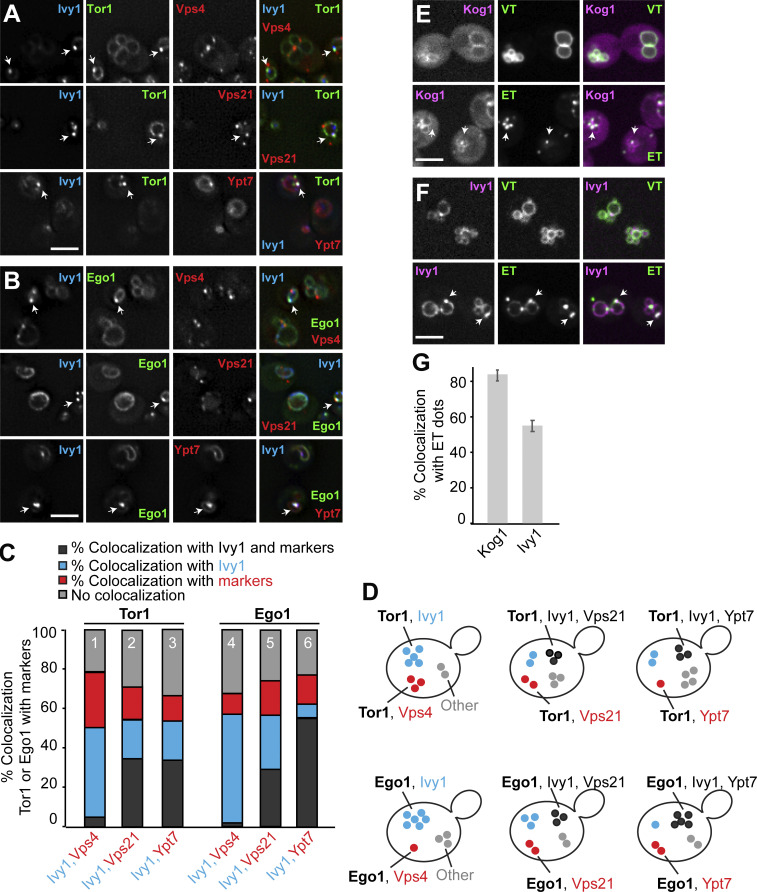
As a start, we analyzed the localization of Ivy1, marked C-terminally with a Halo tag, relative to several endosomal markers and signaling proteins using three-color imaging. As a marker of the TORC1 complex, we selected the catalytic subunit Tor1, which is preferentially found on endosomes (and vacuolar membranes) if N-terminally tagged with GFP (Hatakeyama et al., 2019; Chen et al., 2021). As a marker of the EGOC, we selected its myristoylated subunit Ego1. We colocalized these with endosomal and vacuolar markers: mCherry-tagged Vps4 (a subunit of ESCRT complex), Vps21 (a Rab5-like protein at endosomes), or Ypt7 (a Rab7-like protein at LEs; Fig. 1, A and B). We then scored the level of triple colocalization (dark gray), dual colocalization (red, blue), or no colocalization (light gray; Fig. 1, C and D).
Figure 1.
Ivy1 localizes to SEs. (A and B) Localization of Tor1 or Ego1 relative to Ivy1 and Vps4, Vps21, or Ypt7. Cells expressing mGFP-tagged Tor1 or Ego1 with Halo-tagged Ivy1 and mCherry-tagged Vps4, Vps21, or Ypt7 were grown in synthetic complete medium. The cells were incubated with the Janelia fluor 646 Halo-tag ligand for 1 h and washed eight times before imaging. The cells were analyzed by fluorescence microscopy and are shown as individual slices. Scale bar, 5 µm. Arrows show colocalizing dots. (C) Quantification of Tor1 or Ego1 dots colocalizing with Ivy1 and/or Vps4, Vps21, or Ypt7 puncta (from A and B). Cells (n ≥ 150), Tor1 dots (n ≥ 100), Ego1 dots (n ≥ 100), Ivy1 dots (n ≥ 150), Vps4 dots (n ≥ 500), Vps21 dots (n ≥ 500), and Ypt7 dots (n ≥ 100) from three independent experiments were quantified by ImageJ. (D) Schematic representation showing the observed populations of fluorescent mGFP-Tor1 and Ego1-mGFP dots in cells. Dark gray dots indicate the fraction of Tor1 (top) or Ego1 (bottom) colocalizing with Ivy1 and Vps4 (left), Vps21 (middle), or Ypt7 (right) puncta. Blue spots indicate the fraction of Tor1 or Ego1 colocalizing just with Ivy1. Red dots indicate the fraction of Tor1 or Ego1 colocalizing with just Vps4 (left), just Vps21 (middle), or just Ypt7 (right). Light gray dots indicate the fraction of Tor1 or Ego1 without any colocalization with the selected markers. (E and F) Localization of Kog1 or Ivy1 relative to VT and ET reporters. Cells expressing mCherry-tagged Kog1 or mScarlet-tagged Ivy1 were transformed with ET (FYVE-GFP-Sch9C-term) or VT (Sch9C-term-GFP-Pho8N-term) reporters. The cells were grown in a synthetic medium and analyzed by fluorescence microscopy and are shown as individual slices. Scale bar, 5 µm. Arrows show colocalizing dots. (G) Quantification of the number of Kog1 or Ivy1 dots colocalizing with ET-positive dots. Cells (n ≥ 150), Kog1 dots (n ≥ 100), Ivy1 dots (n ≥ 150), and ET dots (n ≥ 200) were quantified by ImageJ. Error bars represent SD of three independent experiments.
Our analysis revealed that Ivy1 colocalized well with Tor1 and Ego1 (40–50%, blue fraction and dark gray, columns 1 and 4). A large fraction of Ivy1/Tor1- and Ivy1/Ego1-positive structures were also positive for Vps21 and even more for Ypt7, in the case of Ego1 (dark gray, columns 2, 3, 5, and 6). Importantly, Ivy1/Tor1- or Ivy1/Ego1-positive structures hardly overlapped with Vps4 (dark gray part, columns 1 and 3), suggesting that Ivy1 marks a fraction of endosomes that are distinct from MVBs (Hatakeyama and Virgilio, 2019b; Hatakeyama et al., 2019). We noticed in addition that a fraction of Tor1 (28 ± 0.5%) and Ego1 (10 ± 2.6%) also colocalized with Vps4 (red part, columns 1 and 4). For some Tor1 and Ego1 dots (20–30%), no colocalization was found (light gray part of columns). However, we also noticed that there were more Ivy1 dots than Tor1 or Ego1 dots. Moreover, Vps4 dots were clearly more abundant than Ivy1 dots (Fig. S1 A). As Vps4 marks ESCRT III–positive LEs and endosomal TORC1 (ET) phosphorylates Vps27 as an ESCRT-0 subunit (Hatakeyama et al., 2019; Hatakeyama and Virgilio, 2019a, b; Lahiri and Klionsky, 2019), we decided to focus on the Ivy1-positive endosomal population. We reasoned that this is likely an endosomal pool where signaling via TORC1 occurs, preventing ESCRT function (Hatakeyama et al., 2019).
Figure S1.
Relocalization of SE-specific proteins to class E compartments of vps4 mutant cells. (A) Quantification of Ivy1, Tor1, Ego1, and Vps4 dots per cell. Cells (n ≥ 200), Ivy1 dots (n ≥ 200), Tor1 dots (n ≥ 100), Ego1 dots (n ≥ 100), and Vps4 dots (n ≥ 600) were analyzed. Error bars represent SD of three independent experiments. (B) Quantification of endosomal protein puncta relative to Ivy1 (related to Fig. 2 C). Cells (n ≥ 150), Ivy1 dots (n ≥ 150), Vps4 dots (n ≥ 300), Vps8 dots (n ≥ 50), Vps21 dots (n ≥ 200), Ypt7 dots (n ≥ 150), Ego1 dots (n ≥ 50), Gtr2 dots (n ≥ 50), Kog1 dots (n ≥ 50), or Fab1 dots (n ≥ 50) were quantified by ImageJ and are shown as the percentage of endosomal markers that show Ivy1-positive signals. Error bars represent SD of three independent experiments. (C) Quantification of endosomal protein puncta colocalizing with Ivy1 in wild-type (wt) and vps4∆ mutant (related to Fig. 3 D). Cells (n ≥ 200), Ivy1 dots (n ≥ 200), Vps4 dots (n ≥ 400), Vps21 dots (n ≥ 300), Ypt7 dots (n ≥ 100), Ego1 dots (n ≥ 50), Kog1 dots (n ≥ 100), or Fab1 dots (n ≥ 50) were quantified by ImageJ and are expressed as percentage of endosomal markers that show Ivy1-positive signals. Error bars represent SD of three independent experiments. ns, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01 (Student’s t test). (D) Localization of Ivy1 relative to Fab1 in vps4 ts cells. vps4 ts cells expressing mCherry-tagged Ivy1 and mNeon-tagged Fab1 were grown at 24°C in a synthetic medium, and then shifted or not to 37°C for 1 h. Arrows show colocalizing dots. (E) Quantification of mean fluorescent intensity of Ivy1-mGFP from C. Cells (n ≥ 200) and Ivy1 dots (n ≥ 200) were quantified by ImageJ. Error bars represent SD of three independent experiments. ***, P ≤ 0.001 (Student’s t test). (F) Percentage of Ivy1 colocalizing with Fab1 puncta (upper panel) and percentage of Fab1 puncta colocalizing with Ivy1 (lower panel). Cells (n ≥ 200), Ivy1 dots (n ≥ 200), and Fab1 dots (n ≥ 50) were quantified by ImageJ. Error bars represent SD of three independent experiments. ns, P > 0.05 (Student’s t test). (G) Localization of Ivy1 relative to Cps1. Wild-type and vps4Δ cells expressing mCherry-tagged Ivy1 and GFP-tagged Cps1 were grown in a synthetic medium and analyzed by fluorescence microscopy. Individual slices are shown. Scale bar, 5 µm. Arrows show colocalizing dots. (H) Quantification of Ivy1 colocalizing with Cps1-positive puncta. Cells (n ≥ 150), Ivy1 dots (n ≥ 150), and Cps1 dots (n ≥ 50) were quantified by ImageJ. Error bars represent SD of three independent experiments. **, P ≤ 0.01 (Student’s t test).
We previously established reporter constructs to determine ET and vacuolar TORC1 (VT) activities (Hatakeyama et al., 2019). They consist of fusion proteins, which target a truncated form of the TORC1 substrate Sch9 to either endosomes or the vacuole (Hatakeyama et al., 2019). To confirm whether we could take Ivy1 as an apparent marker of SEs, we colocalized mScarlet-tagged Ivy1 or mCherry-tagged Kog1 (a TORC1 subunit) with GFP-tagged ET and VT. More than 50% (52.9 ± 2.8%) of Ivy1 and 80% (83.33 ± 1.6%) of Kog1 colocalized with ET (Fig. 1, E–G). In addition, VT colocalized with both proteins, indicating that Ivy1 has two populations that overlap with TORC1, SEs, and the vacuole (Fig. 1, E–G).
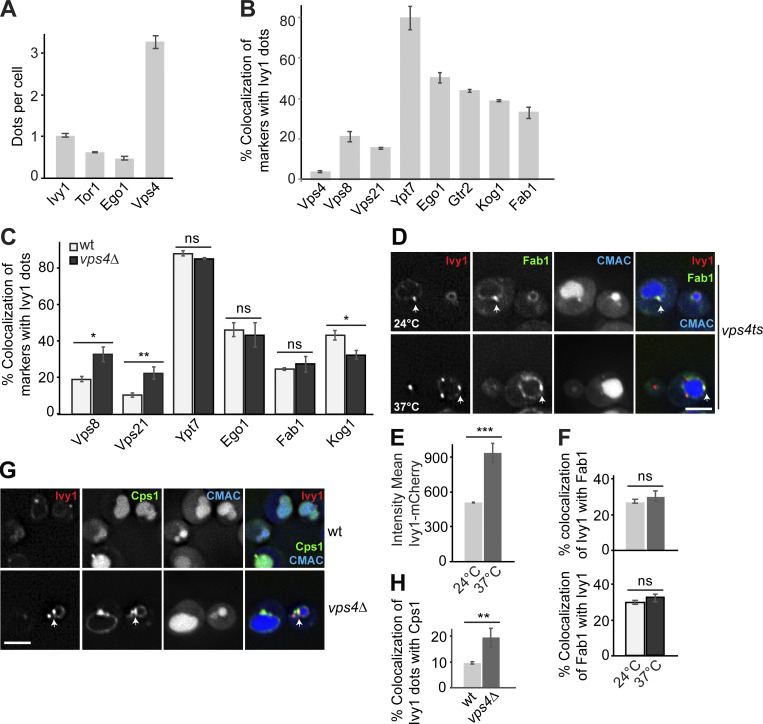
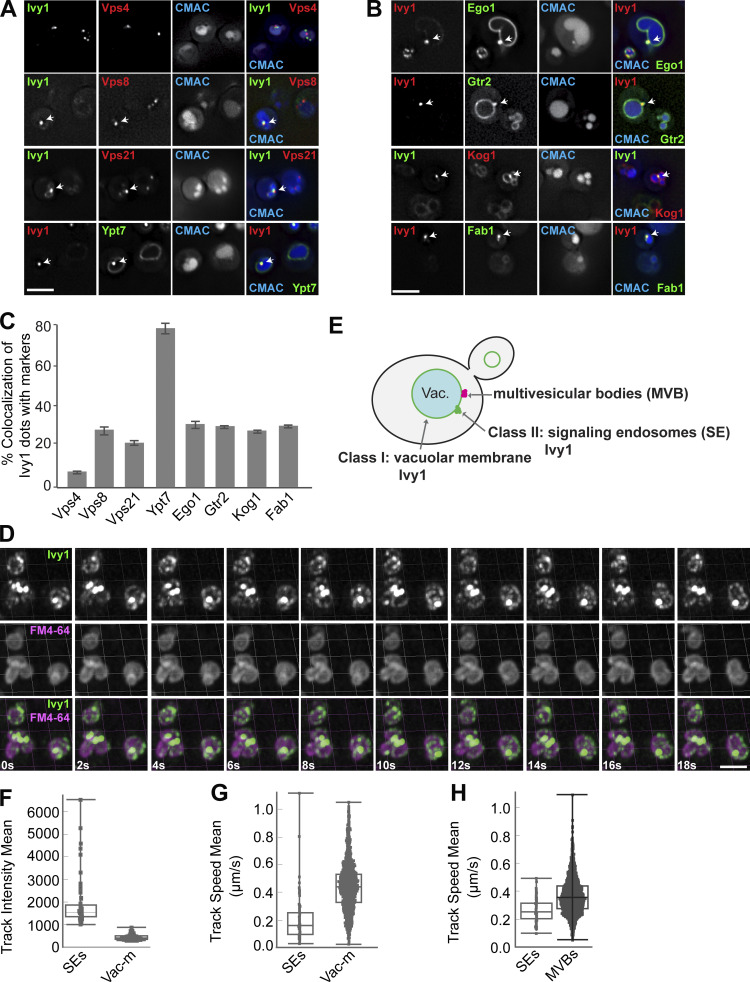
To further determine the identity of Ivy1 dots, we colocalized C-terminally mGFP or mCherry-tagged Ivy1 with functionally tagged markers of the endosome or vacuole carrying the other fluorophore. In particular, we analyzed Vps4, Vps8 (a subunit of CORVET complex at EEs), Vps21, Ypt7, Ego1, Gtr2 (a subunit of the heterodimeric Rag GTPase module that controls TORC1 at SEs), Kog1, and Fab1 (a PI3P 5-kinase). In agreement with our previous studies (Chen et al., 2021; Malia et al., 2018; Numrich et al., 2015), we observed that Ivy1 strongly colocalized with Ypt7, and a fraction of Ivy1 colocalized with Vps8, Vps21, Ego1, Gtr2, Kog1, and Fab1 (Fig. 2, A–C). As observed before (Fig. 1), Ivy1 localized only very weakly with Vps4 (Fig. 2, A and C). In a reverse quantification, we noticed that Ego1, Gtr2, and Kog1 showed increased colocalization with Ivy1 and almost no colocalization with Vps4, while Vps21 and Vps8 showed less colocalization with Ivy1 (Fig. S1 B). This is because Ego1, Gtr2, and Kog1 form fewer dots than Ivy1, whereas Vps21 dots are more abundant. Thus, we hereafter took Ivy1 as a reference marker protein to study SEs in more detail.
Figure 2.
Ivy1-positive structures mark SEs that are distinct from MVBs. (A and B) Localization of Ivy1-positive dots relative to endosomal proteins. Cells expressing mGFP-tagged Ivy1 and mCherry-tagged Vps4, Vps8, Vps21, and Kog1 or expressing mCherry-tagged Ivy1 and GFP-tagged Ypt7, Ego1, Gtr2, and mNeon-tagged Fab1 were grown in a synthetic medium. Vacuoles were stained with CMAC. The cells were analyzed by fluorescence microscopy, and individual slices are shown. Scale bar, 5 µm. Arrows show colocalizing dots. (C) Quantification of Ivy1 dots colocalizing with endosomal proteins. Cells (n ≥ 150), Ivy1 dots (n ≥ 150), Vps4 dots (n ≥ 300), Vps8 dots (n ≥ 50), Vps21 dots (n ≥ 200), Ypt7 dots (n ≥ 150), Ego1 dots (n ≥ 50), Gtr2 dots (n ≥ 50), Kog1 dots (n ≥ 50), or Fab1 dots (n ≥ 50) were quantified by ImageJ. Error bars represent SD of three independent experiments. (D) Ivy1 localization by LLSM after 3D deconvolution (Video 1). Cells expressing mGFP-tagged Ivy1 were grown in synthetic medium. Vacuoles were stained with FM4-64 and visualized relative to Ivy1-mGFP by Imaris. Scale bar, 5 μm. 200–500 cells were analyzed in each independent experiment. (E) Schematic model showing the location of fluorescent Ivy1-mGFP expressed in yeast cells. The green ring corresponds to Ivy1 localization on the vacuolar membrane (class I), green spots indicate SEs (class II), and magenta spots are MVBs. (F) Fluorescence intensity distribution of all tracked spots for Ivy1-mGFP from Video 1. (G) Speed distribution based on trajectory displacements per time point of all tracks for Ivy1-mGFP from Video 1. The data were analyzed as in F. (H) Speed distribution for Ivy1-mCherry (SEs) and Vps4-HA-mGFP (MVBs) from Video 2. The data were analyzed as in F.
Ivy1- and Vps4-positive endosomes differ in their mobility relative to the vacuole
To determine the dynamics of Ivy1-positive structures, we took advantage of lattice light-sheet microscopy (LLSM) to trace Ivy1-mGFP, which enabled us to follow the molecular events in living cells with high spatiotemporal resolution and utmost detection efficiency of lowest signals (Chen et al., 2014). Intriguingly, we detected two classes of fluorescent signals of Ivy1-mGFP (Fig. 2 E): hyperdynamic Ivy1 signals at the vacuolar membrane (class I) and rather immobile Ivy1 dots next to the vacuole (class II; Fig. 2, D–G; and Video 1). Ivy1 signals on the vacuole membrane were dim but mobile, whereas Ivy1 dots were rather bright (Fig. 2, F and G). To compare the dynamics of Ivy1-positive endosomal compartments and MVBs in the cells, we monitored Ivy1-mCherry relative to Vps4-HA-mGFP by LLSM and observed that Ivy1 puncta moved much slower than Vps4-positive dots (Fig. 2 H and Video 2). This suggests that SEs and MVBs differ not only in some key proteins, but also in their relative mobility at the vacuole.
Video 1.
Ivy1 localization relative to vacuoles by LLSM analysis after 3D deconvolution. Related to Fig. 2 D. Cells expressing mGFP-tagged Ivy1 with FM4-64-stained vacuoles were imaged for 1 min. The video shows 3D renderings (maximum-intensity projection mode in Imaris) of a deconvolved 3D time-lapse stack (see Materials and methods for details). The video was recorded at 8 frames/s; the original images were taken at 1.2-s time intervals.
Video 2.
Ivy1 and Vps4 localization by LLSM analysis after 3D deconvolution. Related to Fig. 2 H. Cells expressing mCherry-tagged Ivy1 and HA-mGFP-tagged Vps4 were imaged for 1 min. The video shows 3D renderings (maximum-intensity projection mode in Imaris) of a deconvolved 3D time-lapse stack (see Materials and methods for details). The video was recorded at 8 frames/s; the original images were taken at 8-s time intervals.
SE identity depends on MVB biogenesis
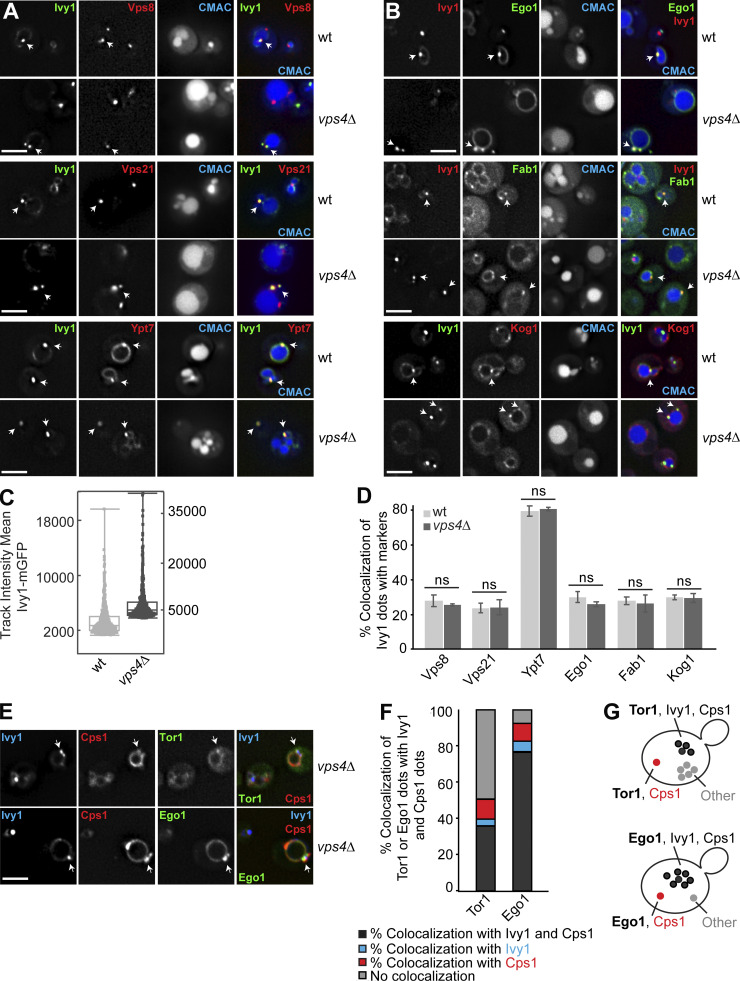
Because SEs also carry endosomal proteins such as the CORVET subunits Vps8 and Vps21, and also the Rab7-like Ypt7 (Figs. 1 and 2), we wondered whether impaired MVB biogenesis would affect SE identity. We therefore analyzed the localization of Ivy1-mGFP in wild-type and vps4Δ cells. Loss of ESCRT proteins results in the accumulation of multilamellar structures, called the class E compartment, next to the vacuole, where all endosomal proteins accumulate (Raymond et al., 1992; Rieder et al., 1996; Babst et al., 1998; Adell et al., 2017; Russell et al., 2012). When we analyzed Ivy1 in vps4∆ cells, the protein strongly accumulated in bright puncta next to the vacuole, and most cells lost the vacuolar localization of Ivy1 (Fig. 3, A and B; and Videos 3 and 4); however, Ivy1 dots were still positive for Vps8, Vps21, Ypt7, Ego1, Fab1, and Kog1 (Fig. 3, A and B; and Fig. S1 C). The same observation was made upon inactivation of Vps4 in a vps4 temperature-sensitive (ts) strain (Fig. S1 D; Babst et al., 1997). At permissive temperature (24°C), mCherry-tagged Ivy1 and mNeon-tagged Fab1 (one of the proteins we tested for colocalization with Ivy1 in vps4∆ cells) partially colocalized in dots and at the vacuolar membrane. However, when shifted to the nonpermissive temperature (37°C), Ivy1 strongly accumulated as in the vps4∆ cells in dots (Fig. S1, E and F), which were partially positive for Fab1 and likely correspond to class E compartments (Adell et al., 2017; Fig. S1, D–F).
Figure 3.
SE identity depends on MVB functionality. (A and B) Localization of Ivy1 relative to endosomal proteins in ESCRT mutants. Wild-type or vps4Δ cells expressing mGFP-tagged Ivy1 and mCherry-tagged Vps8, Vps21, Ypt7, and Kog1 or expressing mCherry-tagged Ivy1, GFP-tagged Ego1, and mNeon-tagged Fab1 were grown in a synthetic medium. Vacuoles were stained with CMAC. The cells were analyzed by fluorescence microscopy and are shown as individual slices. Scale bar, 5 µm. Arrows show colocalizing dots. (C) 3D track of the mean fluorescence intensity of Ivy1-mGFP from Videos 3 and 4. Wild-type or vps4Δ cells expressing mGFP-tagged Ivy1 were grown in a synthetic medium. Vacuoles were stained with CMAC, and the data were analyzed as in Fig. 2 F. (D) Quantification related to A and B. Cells (n ≥ 200), Ivy1 dots (n ≥ 200), Vps4 dots (n ≥ 400), Vps21 dots (n ≥ 300), Ypt7 dots (n ≥ 100), Ego1 dots (n ≥ 50), Kog1 dots (n ≥ 100), or Fab1 dots (n ≥ 50) were quantified by ImageJ. Error bars represent SD of three independent experiments. ns, P > 0.05 (Student’s t test). (E) Localization of Tor1 or Ego1 relative to Ivy1 and Cps1 in vps4∆ cells. Cells expressing mGFP-tagged Tor1 or Ego1 with HaloTag-Ivy1 and mCherry-Cps1 were grown in a synthetic medium. The cells were incubated with the Janelia fluor 646 HaloTag ligand for 1 h and washed eight times before imaging. The cells were analyzed by fluorescence microscopy and are shown as individual slices. Scale bar, 5 µm. Arrows show colocalizing dots. (F) Quantification of Tor1 or Ego1 dots colocalizing with Ivy1 and/or Cps1 from E. Cells (n ≥ 200), Tor1 dots (n ≥ 100), Ego1 dots (n ≥ 100), Ivy1 dots (n ≥ 200), and Cps1 dots (n ≥ 200) from three independent experiments were quantified by ImageJ. (G) Schematic model showing the different populations of mGFP-Tor1 and Ego1-mGFP dots analyzed in F. Dark gray spots indicate Tor1 or Ego1 dots colocalizing with both Ivy1 and Cps1. Blue and red spots indicate Tor1 or Ego1 dots colocalizing with just Ivy1 or just Cps1, respectively. Light gray spots indicate Tor1 or Ego1 dots that show colocalization with neither Ivy1 nor Cps1.
Video 4.
Ivy1 localization relative to vacuoles in vps4∆ cells by LLSM analysis after 3D deconvolution. Related to Fig. 3 C. vps4∆ cells expressing mGFP-tagged Ivy1 with CMAC-stained vacuoles were imaged for 1 min. The video shows 3D renderings (maximum-intensity projection mode in Imaris) of a deconvolved 3D time-lapse stack (see Materials and methods for details). The video was recorded at 8 frames/s; the original images were taken at 1.1-s time intervals.
To confirm that Ivy1 was indeed present on class E compartments in vps4 mutant cells, we analyzed the colocalization of mCherry-tagged Ivy1 with GFP-tagged ESCRT substrate carboxypeptidase S (Cps1) in wild-type and vps4∆ cells. Upon deletion of vps4, Ivy1 colocalized more strongly with Cps1, which accumulated at class E compartments (Fig. S1, G and H). To further test if SEs remain as distinct endosomes in vps4∆ cells, we analyzed the colocalization of mGFP-tagged Tor1 or Ego1 with Halo-tagged Ivy1 and mCherry-tagged Cps1. We observed that 35.85 ± 0.9% of the Tor1 dots and 76.27 ± 4.2% of the Ego1 dots colocalized with Ivy1 and Cps1 (Fig. 3 F, dark gray). There was almost no colocalization of Tor1 or Ego1 with just Ivy1 (blue) and very little with just Cps1 (red), suggesting that SEs were lost at the expense of the expanded class E compartments of the ESCRT mutant (Fig. 3, E–G). We thus conclude that the maintenance of SEs as an endosomal population is directly linked to the biogenesis of MVBs.
HOPS function is required to maintain SE number and endolysosomal trafficking
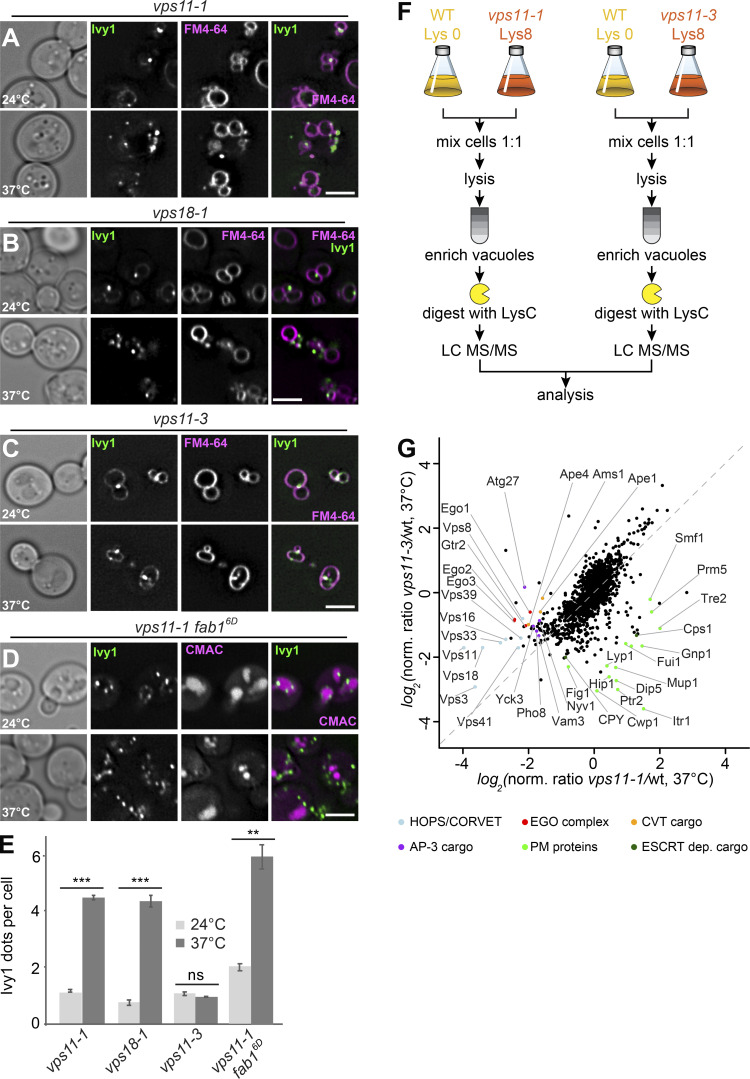
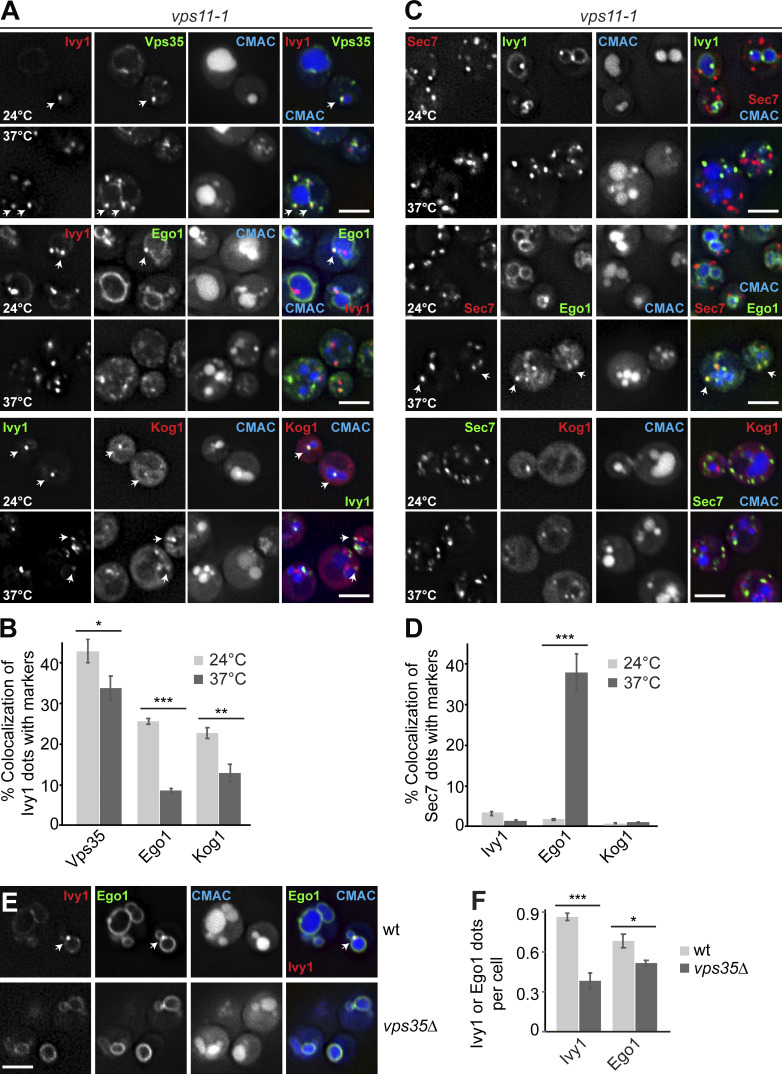
The biogenesis of the LE depends on multiple fusion events at EEs and LEs as a prerequisite for MVB formation (Zeigerer et al., 2012). EE fusion requires the CORVET tethering complex, whereas the fusion of MVBs with vacuoles depends on HOPS (van der Beek et al., 2019; Balderhaar and Ungermann, 2013). We therefore asked whether HOPS or CORVET was required to maintain the identity of SEs. In a previous study, two temperature-sensitive alleles for Vps11 were identified that disable HOPS (vps11-1) or CORVET (vps11-3) function, whereas a vps18-1 mutant specifically impairs HOPS (Robinson et al., 1991; Peterson and Emr, 2001). All mutants are functional at the permissive temperature of 24°C but at 37°C show a protein-sorting defect toward the vacuole and a partial growth defect (Peterson and Emr, 2001). We therefore tagged Ivy1 with mGFP in these strains and analyzed its localization relative to FM4-64–stained vacuoles at permissive (24°C) or restrictive (37°C) temperature. Both vps11-1 and vps18-1 cells strongly accumulated Ivy1-mGFP in fourfold more dots proximal to the vacuole at the restrictive temperature (Fig. 4, A, B, and E), whereas the vps11-3 mutant had no effect on Ivy1 localization (Fig. 4, C and E). This indicates that the inactivation of HOPS, but not of CORVET, affects the number of observed Ivy1-positive structures in the cell. We previously showed that Ivy1 accumulates at SEs in cells expressing a phosphomimetic Fab16D allele (Chen et al., 2021). We thus wondered whether the number of Ivy1 dots would increase in a vps11-1 fab16D double mutant. This was indeed observed (Fig. 4, D and E), suggesting that impairment of HOPS and Fab1 both affect the formation of SEs independently.
Figure 4.
Ivy1-positive structures accumulate in HOPS mutants. (A–C) Localization of Ivy1 relative to the vacuole. Selected ts strains (vps11-1, vps18-1, and vps11-3) expressing mGFP-tagged Ivy1 were grown at 24°C in a synthetic medium, and then shifted to 24 or 37°C for 1 h. Vacuoles were stained with FM4-64. The cells were analyzed by fluorescence microscopy and are shown as individual slices. Scale bar, 5 μm. (D) vps11-1 fab16D cells expressing mGFP-tagged Ivy1 were stained with CMAC and analyzed as before. (E) Quantification of Ivy1 dots per cell in the indicated mutant strains grown at 24 and 37°C. Cells (n ≥ 150) and Ivy1 dots (n ≥ 200) were analyzed. Error bars represent SD of three independent experiments. ns, P > 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 (Student’s t test). (F) Design of the experimental procedure to determine vacuolar proteomics. (G) Vacuolar proteomic analysis. Wild-type (wt) and vps11-1 or vps11-3 cells were grown in light lysine (wild-type cells) or heavy lysine (mutant cells) containing SILAC medium as described in Materials and methods and incubated at 37°C for 1 h. The vacuoles were isolated and analyzed by mass spectrometry. Intensities of identified proteins are plotted in normalized heavy over light SILAC ratios. Selected vacuolar proteins are marked. CVT, cytoplasm-to-vacuole targeting; PM, plasma membrane.
As the vps11-1 mutant caused a strong increase in Ivy1-positive dots, we asked whether we could see general changes in the endosomal and vacuolar proteome due to HOPS inactivation. We therefore turned to a recently established method of stable isotope labeling by/with amino acids in cell culture (SILAC)–based vacuolar proteomics, which allows the identification of all vacuolar proteins in comparative analyses (Eising et al., 2019). We reasoned that mutants impaired in HOPS (vps11-1) or CORVET (vps11-3) should differ in their vacuolar proteome at the restrictive temperature and thus reveal impaired cargo trafficking. We therefore isolated vacuoles from light labeled wild-type cells and compared them either to vacuoles from heavy labeled vps11-1 or vacuoles from heavy labeled vps11-3 cells (Fig. 4 F). We plotted the ratios of vps11-1 over wild-type on the x axis against the ratios of vps11-3 over wild-type on the y axis. This analysis revealed that all subunits of HOPS and CORVET were affected in both mutants (light blue dots). The effect of the vps11-1 mutant appears to be stronger than that of the vps11-3 mutant. However, we were able to identify clear differences regarding the vacuolar proteome of both analyzed mutants. As expected, the vps11-1 mutation affected the abundance of EGOC subunits (red dots), cargoes of the autophagy-related cytoplasm-to-vacuole targeting pathway (orange dots), and AP-3 cargo proteins (purple dots). In contrast, the vps11-3 mutant mostly affected the abundance of proteins following the endolysosomal pathway, especially plasma membrane proteins (light green dots) and vacuolar hydrolases such as CPY and Cps1 (dark green dots; Fig. 4 G). Together, these findings indicate that the CORVET-specific vps11-3 allele impairs endocytosis, whereas the vps11-1 allele affects HOPS function, which is required for all fusion events at LEs and vacuoles (Wickner and Rizo, 2017; van der Beek et al., 2019; Peterson and Emr, 2001). Vacuolar proteomics of temperature-sensitive alleles can thus recapitulate the affected trafficking defects (Lin et al., 2008; Cabrera et al., 2013; Markgraf et al., 2009; Peplowska et al., 2007).
HOPS is required to maintain identities of endosomal structures
Vacuolar proteomics can reveal the overall changes in protein abundance on vacuoles and associated compartments yet cannot resolve how a HOPS mutant affects the relative distribution of endosomal proteins at SEs, MVBs, and vacuoles. We therefore colocalized both Vps4 and Ivy1 with several endosomal and Golgi markers relative to the vacuole in vps11-1 mutant at the permissive and restrictive temperatures. For this, we used tagged constructs with mCherry or mGFP fluorophores that maintain the functionality of the proteins (Numrich et al., 2015; Adell et al., 2017).
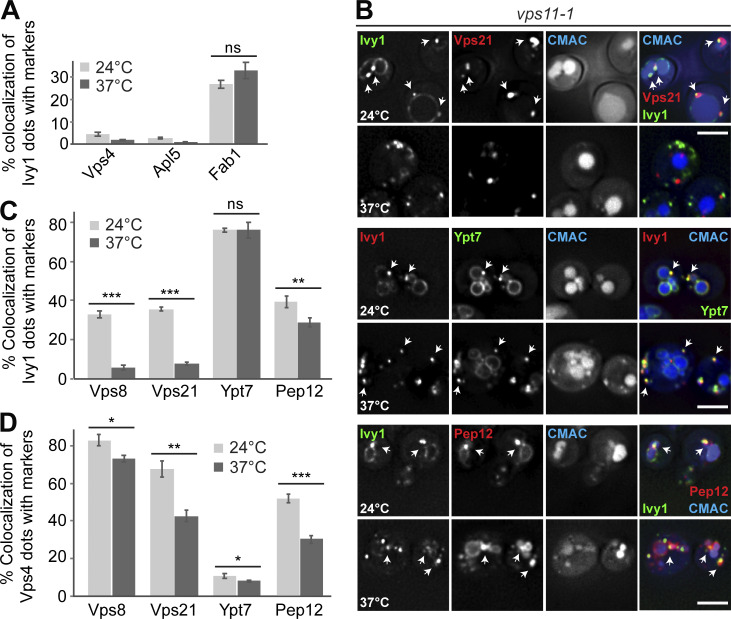
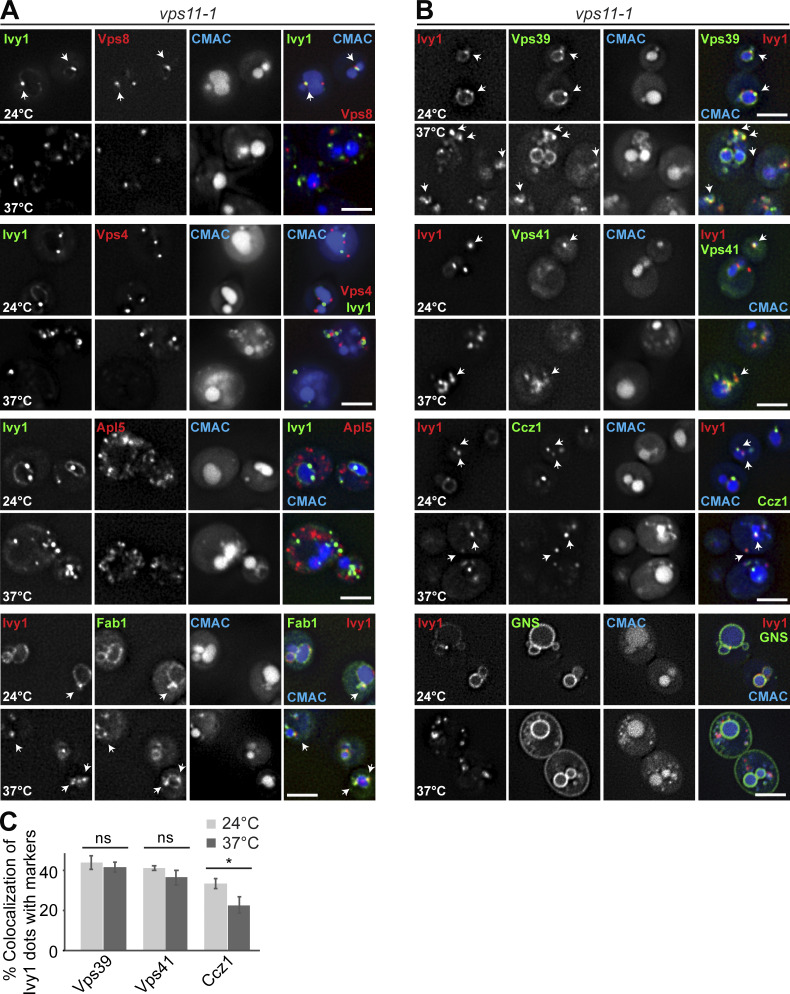
We initially focused on Ivy1 as a protein found at SEs. At the permissive and restrictive temperatures (24°C and 37°C, respectively), Ivy1 dots were still strongly positive for Ypt7 and also, to a large extent (26.8 ± 1.8%, 32.74 ± 3.7%), Fab1, the SNARE Pep12 (40.75 ± 3.2%, 29.67 ± 2.4%), the Ypt7 GEF Ccz1 (33.28 ± 2.5%, 22.66 ± 4%), and the HOPS subunits Vps39 (43.7 ± 3.5%, 41.52 ± 2.5%) and Vps41 (41.1 ± 1.2%, 36.32 ± 1.6%; Fig. 5, A–C; and Fig. S2, A–C). We also did not detect an increase in Ivy1 colocalization with Vps4 or the AP-3 marker Apl5 (Figs. 5 A and S2 A).
Figure 5.
Ivy1 structures lose EE marker proteins upon HOPS inactivation. (A) Quantification of Ivy1 dots colocalizing with Vps4, Apl5, or Fab1 puncta in vps11-1 mutant cells grown at 24 or 37°C (related to Fig. S2 A). Cells (n ≥ 150), Ivy1 dots (n ≥ 200), Vps4 dots (n ≥ 400), Apl5 dots (n ≥ 500), or Fab1 dots (n ≥ 50) were quantified by ImageJ. Error bars represent SD of three independent experiments. ns, P > 0.05 (Student’s t test). (B) Localization of Ivy1 relative to the endosomal Rabs Vps21 and Ypt7 or the SNARE Pep12. vps11-1 ts cells expressing mGFP-tagged Ivy1 and mCherry-tagged Vps21, Pep12 or expressing mScarlet-tagged Ivy1 and mGFP-tagged Ypt7 were grown at 24°C in a synthetic medium, and then shifted to 24 or 37°C for 1 h. Vacuoles were stained with CMAC. The cells were analyzed by fluorescence microscopy, and individual slices are shown. Scale bar, 5 µm. Arrows show colocalizing dots. (C) Quantification of Ivy1 dots that colocalize with Vps8, Vps21, Ypt7, or Pep12 puncta (related to Fig. S2 A). Cells (n ≥ 150), Ivy1 dots (n ≥ 150), Vps8 dots (n ≥ 100), Vps21 dots (n ≥ 300), Ypt7 dots (n ≥ 100), or Pep12 dots (n ≥ 150) were quantified by ImageJ. Error bars represent SD of three independent experiments. ns, P > 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 (Student’s t test). (D) Quantification of Vps4 dots that colocalize with Vps8, Vps21, Ypt7, or Pep12 puncta (related to Fig. S3 A). Cells (n ≥ 150) and dots of Vps4 (n ≥ 400), Vps8 (n ≥ 100), Vps21 (n ≥ 300), Ypt7 (n ≥ 100), or Pep12 (n ≥ 150) were quantified by ImageJ. Error bars represent SD of three independent experiments. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 (Student’s t test).
Figure S2.
Analysis of endosomal and vacuolar markers relative to Ivy1 in HOPS mutant cells. (A) Localization of Ivy1 relative to Vps8, Vps4, Apl5, and Fab1 (quantification is shown in Fig. 5, A and C). vps11-1 ts cells expressing mGFP-tagged Ivy1 and mCherry-tagged Vps8, Vps4, and Apl5 or mScarlet-tagged Ivy1 and mNeon-tagged Fab1 were grown at 24°C in a synthetic medium, and then shifted or not to 37°C for 1 h. Vacuoles were stained with CMAC. The cells were analyzed by fluorescence microscopy, and individual slices are shown. Arrows show colocalizing dots. (B) Localization of Ivy1 relative to Vps39, Vps41, Ccz1, and the artificial cargo GNS. vps11-1 cells expressing mScarlet-tagged Ivy1; mNeon-tagged Vps39, Vps41, or Ccz1; or GFP-tagged GNS were grown and analyzed as in A. Scale bar, 5 µm. Arrows show colocalizing dots. (C) Quantification of Ivy1 colocalizing with Vps39, Vps41, and Ccz1 puncta. Cells (n ≥ 200), Ivy1 dots (n ≥ 250), Vps39 dots (n ≥ 100), Vps41 dots (n ≥ 100), or Ccz1 dots (n ≥ 200) were quantified by ImageJ. Error bars correspond to SD of three independent experiments. ns, P > 0.05; *, P ≤ 0.05 (Student’s t test).
This picture changed when we analyzed early endosomal markers. Both the Rab5-like Vps21 and the CORVET subunit Vps8 colocalized with Ivy1 at the permissive temperature, as in wild-type (Fig. 5, B and C). However, at the restrictive temperature, Vps8- and Vps21-positive dots colocalized significantly less with Ivy1 (Fig. 5, B and C). This suggests that loss of HOPS function results in a change of surface composition of Ivy1-marked structures, and thus likely in the entire SE pool. Importantly, Ivy1-positive dots were still positive for the endosomal SNARE Pep12, Fab1, and Ypt7 but lost the early endosomal markers Vps21 and Vps8 (Fig. 5 C).
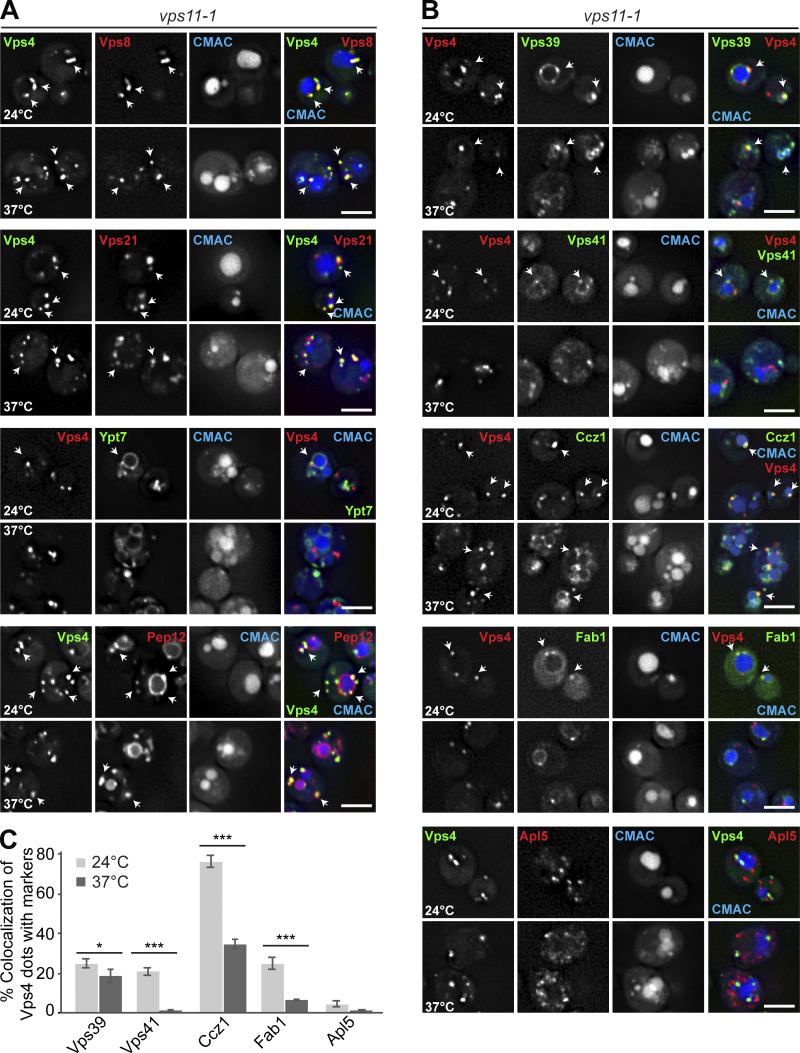
To determine whether HOPS inactivation also changed the late endosomal identity, we traced the colocalization of Vps4 with the same markers. Vps4 largely colocalized with Vps8 (82.7 ± 2.85%), Vps21 (67.75 ± 4.4%), and Pep12 (73.44 ± 5.7%), which decreased by 10–30% upon HOPS inactivation (Fig. 5 D). Interestingly, Vps4 also colocalized well with the subunit of the Ypt7 GEF-subunit, Ccz1 (75.95 ± 3%), and to a lesser degree with Vps39 (24.95 ± 2%) and Fab1 (25.04 ± 2.7%), while only little Ypt7 (10.7 ± 1.1%) was found at these structures (Fig. 5 D and Fig. S3, A–C). At the restrictive temperature, the colocalization of Vps4 with Ccz1, Vps41, or Fab1 decreased strongly, suggesting that the composition of Vps4-positive LEs also changes upon HOPS inactivation (Fig. S3, B and C).
Figure S3.
Analysis of proteins found on Vps4-positive LEs in HOPS mutant cells. (A) Localization of the MVB marker Vps4 relative to endosomal proteins. vps11-1 ts cells expressing mGFP-tagged Vps4 and mCherry-tagged Vps8, Vps21, and Pep12 or mCherry-tagged Vps4, and mGFP-tagged Ypt7 were grown at 24°C in a synthetic medium, and then shifted or not to 37°C for 1 h. Vacuoles were stained with CMAC. The cells were analyzed by fluorescence microscopy and are shown via individual slices. Scale bar, 5 µm. Quantification is shown in Fig. 5 D. Arrows show colocalizing dots. (B) Localization of Vps4 relative to Vps39, Vps41, Ccz1, Fab1, and Apl5. vps11-1 cells expressing mCherry-tagged Vps4 and mNeon-tagged Vps39, Vps41, Ccz1, and Fab1 or mGFP-tagged Vps4 and 3xmCherry-tagged Apl5 were grown at 24°C in a synthetic medium, and then shifted or not to 37°C for 1 h. Vacuoles were stained with CMAC. Cells were analyzed by fluorescence microscopy, and individual slices are shown. Scale bar, 5 µm. Arrows show colocalizing dots. (C) Quantification. Cells (n ≥ 150), Vps4 dots (n ≥ 400), Vps39 dots (n ≥ 100), Vps41 dots (n ≥ 100), Ccz1 dots (n ≥ 200), or Fab1 dots (n ≥ 50) were quantified by ImageJ. Error bars represent SD of three independent experiments. *, P ≤ 0.05; ***, P ≤ 0.001 (Student’s t test).
HOPS and retromer function maintain SE identity
Given that both Ivy1- and Vps4-positive endosomes seem to require HOPS to maintain their identity, we asked whether Ego1 and TORC1 would remain at endosomes if HOPS is inactivated. At the permissive temperature, Ego1 and Kog1 colocalized with Ivy1 as in wild-type (Fig. 6, A and B; and Fig. 2 C). However, at the restrictive temperature, colocalization between Ego1 or Kog1 dots and Ivy1 was largely lost (Fig. 6, A and B). As Ego1 reaches the vacuole surface by the AP-3 pathway, we tested if some Ivy1 or Ego1 was found also on the Golgi (marked by Sec7) but did not detect any overlap at the permissive temperature (Fig. 6, C and D). Surprisingly, at the restrictive temperature, we observed that Ego1 now colocalized with the Golgi marker Sec7 (Fig. 6, C and D). The number of Sec7 dots stayed the same, however, indicating that HOPS inactivation did not affect Golgi function per se. For Kog1, we did not detect colocalization with Sec7 and currently do not know the identity of the remaining dots (Fig. 6, C and D). These data suggest that loss of HOPS results in an accumulation of the EGOC at the Golgi, while TORC1 is found elsewhere.
Figure 6.
HOPS and retromer mutants differentially affect Ego1 localization away from SEs. (A) Ivy1 structures lose signaling complexes upon HOPS inactivation. vps11-1 cells expressing mScarlet-tagged Ivy1 and mGFP-tagged Vps35 or Ego1 or expressing mGFP-tagged Ivy1 and mCherry-tagged Kog1 were grown at 24°C in a synthetic medium and then shifted to 24 or 37°C for 1 h. Vacuoles were stained with CMAC. The cells were analyzed by fluorescence microscopy, and individual slices are shown. Arrows show colocalizing dots. (B) Quantification of Ivy1 dots colocalizing with Vps35, Ego1, or Kog1 puncta. Cells (n ≥ 200), Ivy1 dots (n ≥ 200), Vps35 dots (n ≥ 150), Ego1 dots (n ≥ 150), or Kog1 dots (n ≥ 100) were quantified by ImageJ. Error bars represent SD of three independent experiments. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001 (Student’s t test). (C) Localization of Sec7 relative to Ivy1, Ego1, or Kog1. vps11-1 cells expressing mScarlet-tagged Sec7 and mGFP-tagged Ivy1, Ego1, or mGFP-tagged Sec7 and mCherry-tagged Kog1 were grown and analyzed as in A. Arrows show colocalizing dots. (D) Percentage of Sec7 dots colocalizing with Ivy1, Ego1, or Kog1 puncta. Cells (n ≥ 200) and Ivy1 (n ≥ 200), Sec7 (n ≥ 350), Ego1 (n ≥ 150), or Kog1 (n ≥ 100) dots were quantified by ImageJ. Error bars represent SD of three independent experiments. ***, P ≤ 0.001 (Student’s t test). (E) Localization of Ivy1 relative to Ego1 in wild-type (wt) and retromer mutant. Wild-type or vps35∆ mutant expressing mCherry-tagged Ivy1 and mGFP-tagged Ego1 were grown in a synthetic medium. Vacuoles were stained with CMAC. The cells were analyzed by fluorescence microscopy, and individual slices are shown. Scale bar, 5 µm. Arrows show colocalizing dots. (F) Quantification of Ivy1 and Ego1 dots per wild-type or vps35Δ cell. Cells (n ≥ 200), Ivy1 dots (n ≥ 200), and Ego1 dots (n ≥ 200) were quantified. Error bars represent SD of three independent experiments. *, P ≤ 0.05; ***, P ≤ 0.001 (Student’s t test).
Previous work in mammalian cells showed that retromer plays a critical role in TORC1 signaling by controlling a Rab7 GTPase-activating protein and thus Rab7 levels at lysosomes (Kvainickas et al., 2019). A possible explanation for the Golgi localization of Ego1 is that Ego1 became a substrate of retromer, which is found on SEs (Fig. 6, A and B). We thus analyzed the localization of Ivy1 relative to Ego1 in vps35∆ cells and observed a loss of both Ivy1 and Ego1 dots at the expense of vacuolar localization. This suggests that the localization of the EGO and Tor complexes to SEs requires both retromer and HOPS function (Fig. 6, E and F).
Because Ego1 reaches the vacuolar surface via the AP-3 pathway, we further tested whether other AP-3 cargoes also accumulate at the Golgi if HOPS is inactivated. We therefore monitored AP-3–dependent trafficking of the artificial cargo GNS to the vacuole (Reggiori et al., 2000). This GNS cargo consists of the N-terminally tagged cytosolic part of the vacuolar SNARE Nyv1, a bona fide AP-3 cargo (Wen et al., 2006), linked to the longer transmembrane domain of Snc1. If the AP-3 pathway is defective, then GNS is rerouted via the plasma membrane to the vacuole (Reggiori et al., 2000). We therefore followed GNS in the vps11-1 mutant and observed that it localized to the vacuole at the permissive temperature. At the restrictive temperature, GNS also stained the plasma membrane, indicating an AP-3 defect (Fig. S2 B). This indicates that the inactivation of HOPS does not result in a general rerouting of AP-3 cargoes to the Golgi or its retention. It suggests rather that the localization of the EGO and TOR complexes to SEs is determined by dynamic fission and fusion processes that require a functional retromer and the HOPS complex.
HOPS function is required for TORC1 signaling
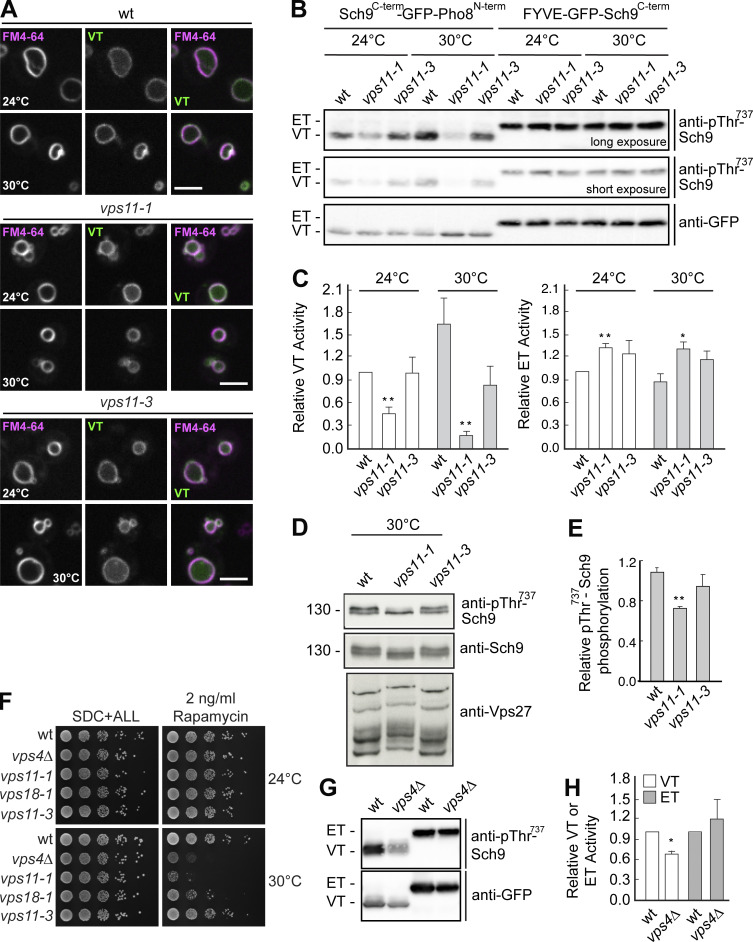
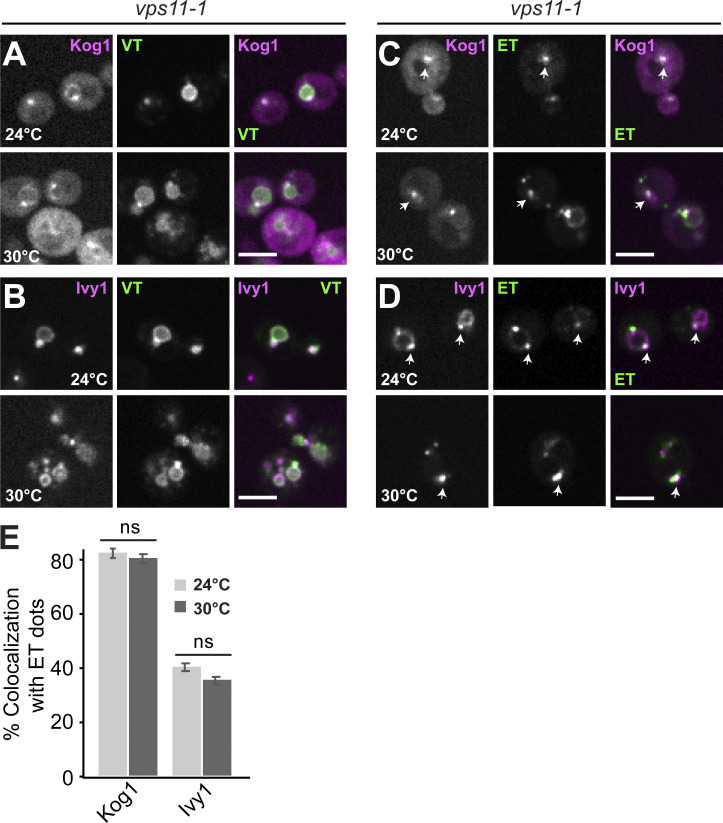
As a complete impairment of HOPS function results in the redistribution of Ego1 from SEs and other locations to the Golgi, we expected an alteration in vacuolar and ET activities. Using our previously described reporter system to measure VT and ET activities (Hatakeyama et al., 2019), we independently confirmed this expectation. These reporters are found in wild-type cells at SEs and vacuoles (Fig. 1, E–G). As TORC1 activity is temperature sensitive, we here used temperatures between 24 and 30°C. Under these conditions, the VT reporter arrived successfully at the vacuole, whereas the ET reporter colocalized with Kog1 and Ego1 (Fig. 1, E–G; Fig. 7 A; and Fig. S4, A–E).
Figure 7.
HOPS mutants affect TORC1 activity. (A) Localization of VT relative to the vacuole. Wild-type (wt), vps11-1, and vps11-3 cells were transformed with the VT (Sch9C-term-GFP-Pho8N-term) reporter and grown in a synthetic medium at 24 or 30°C. Vacuoles were stained with FM4-64. The cells were analyzed by fluorescence microscopy and are shown as individual slices. Scale bar, 5 μm. (B) The vps11-1 allele causes changes in both VT and ET activities. Strains with the indicated genotypes were transformed with ET (FYVE-GFP-Sch9C-term) or VT (Sch9C-term-GFP-Pho8N-term) reporters and grown exponentially at 24 or 30°C on SDC + all medium. To measure ET/VT activities (Hatakeyama et al., 2019), proteins were extracted and run on SDS-PAGE, and the phosphorylation levels of the ET/VT reporters were detected by immunoblotting using phospho-specific anti-Sch9-pThr737 antibodies. ET/VT input levels were detected with anti-GFP antibodies. Different exposures are shown to better visualize the effects on ET and VT. (C) Quantifications of the ET/VT assays in A. Significance was determined with a two-tailed Student’s t test (**, P < 0.005; *, P < 0.05). (D) Phosphorylation states of vacuolar Sch9 and endosomal Vps27. Wild-type, vps11-1, and vps11-3 were grown in synthetic complete medium. Corresponding cells extracts were run on 7.5 and 9% SDS-PAGE and probed with phosphospecific Thr737 Sch9 and anti-Sch9 antibodies or run on a 6% gel containing 50 μM Mn2+-Phos-tag and probed with anti-Vps27 antibodies. (E) Quantifications of the Sch9 Thr737 phosphorylation assayed in D. Error bars represent SD of three independent experiments. **, P ≤ 0.01 (Student’s t test). (F) Growth of wild-type, vps4Δ, vps11-1, vps18-1, and vps11-3 on rapamycin-containing plates. The cells were grown in synthetic medium, spotted onto plates containing SDC + all with or without 2 ng/ml rapamycin, and grown at either 24 or 30°C for 2–5 d. (G) VPS4 deletion affects vacuolar but not ET activity. Wild-type and vps4Δ cells were transformed with ET (FYVE-GFP-Sch9C-term) or VT (Sch9C-term-GFP-Pho8N-term) reporters and grown exponentially at 30°C in a synthetic medium. ET/VT activities were assessed as in B. (H) Quantifications of the ET/VT assay in G. Significance was determined with a two-tailed Student’s t test (*, P ≤ 0.05). Source data are available for this figure: SourceData F7.
Figure S4.
Analysis of the localization of TORC1 reporters in the vps11-1 mutant. (A–D) Localization of Kog1 and Ivy1 relative to VT and ET reporters. vps11-1 cells expressing Kog1-mCherry or Ivy1-mScarlet were transformed with VT (Sch9C-term-GFP-Pho8N-term) or ET (FYVE-GFP-Sch9C-term) reporters, grown exponentially at 24 or 30°C in a synthetic medium, and analyzed by fluorescence microscopy. Individual slices are shown. Scale bar, 5 µm. Arrows show colocalizing dots. (E) Quantification of Kog1 or Ivy1 colocalizing with ET dots. Cells (n ≥ 150), Kog1 dots (n ≥ 100), Ivy1 dots (n ≥ 150), and ET dots (n ≥ 200) were quantified by ImageJ. Error bars represent SD of three independent experiments.
We then determined VT and ET activities. Cells containing the vps11-1 allele, but not vps11-3 cells, exhibited a significant reduction in VT activity at 24°C that became even more pronounced at 30°C, while the ET activities were even slightly, but significantly, increased in vps11-1, but not vps11-3 cells (Fig. 7, B and C). As additional readouts, we followed the phosphorylation of Sch9 and Vps27, which are substrates of VT and ET, respectively (Hatakeyama et al., 2019). We detected less Sch9 phosphorylation (assayed by immunoblot analyses using phosphospecific antibodies that target the TORC1 residue T737 in Sch9) and more Vps27 phosphorylation (assayed by a slower electrophoretic migration in Phos-tag gel analyses) in vps11-1 but not in vps11-3 cells (Fig. 7, D and E; Hatakeyama et al., 2019). This shows that the impairment of HOPS function, in parallel to affecting the endosomal localization of various proteins, significantly disturbs the partitioning of TORC1 signaling between endosomes and vacuolar membranes.
Because VT primarily defines rapamycin-sensitive growth through its vacuolar target Sch9 (Urban et al., 2007; Hatakeyama et al., 2019), these data explain why a moderate reduction in HOPS function at semipermissive temperatures (in vps11-1 and vps18-1 cells), but not a reduction in CORVET function (in vps11-3 cells), resulted in rapamycin-sensitive growth (Fig. 7, B–E). Notably, in line with its rapamycin-sensitive growth at 30°C (Fig. 7 F), the vps4∆ strain also exhibited significantly lower VT, but not ET, activity (Fig. 7, G and H). As vps4∆ strains have no AP-3–sorting defect (Babst et al., 1997), this corroborates our conclusion above that defective MVB biogenesis has a significant impact on TORC1 signaling. Taken together, HOPS is needed to maintain proper TORC1 activities at SEs and vacuoles, which is also in agreement with previous genetic analyses (Kingsbury et al., 2014; Hatakeyama et al., 2019; Zurita-Martinez et al., 2007).
HOPS is required for reformation of Ivy1-positive structures next to the vacuoles
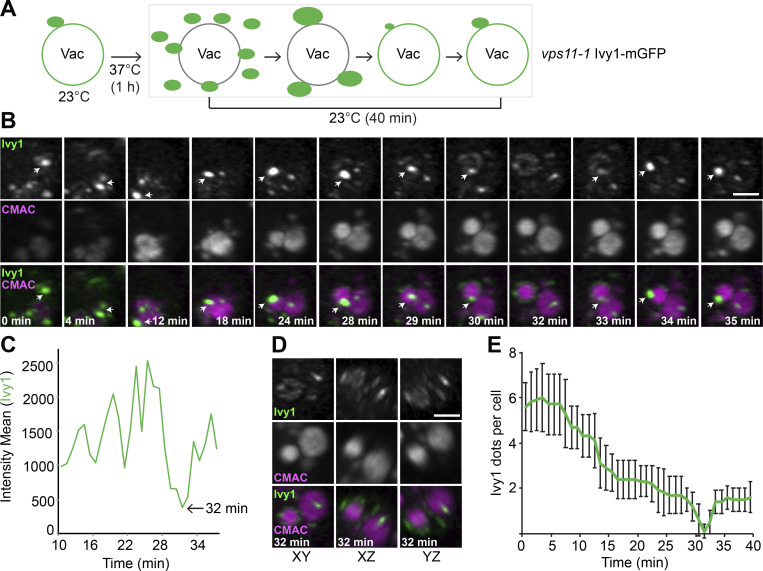
As HOPS inactivation causes an accumulation of Ivy1-positive structures owing to a possible fusion defect, we wondered whether we could observe recovery of Ivy1-positive structures with vacuoles by shifting vps11-1 mutant cells back to the permissive temperature (Fig. 8 A). To monitor this, we shifted cells to 37°C to accumulate Ivy1-mGFP dots and then traced Ivy1 dots by LLSM after cells were exposed to the permissive temperature (Fig. 8 B). Over the first 30 min, we observed that the number of Ivy1 puncta strongly decreased at the expense of one large dot (Fig. 8 B). At 32 min, this bright dot suddenly disappeared (Fig. 8 C), and Ivy1 fluorescence thereafter was equally distributed over the entire vacuole surface (Fig. 8, B and D; and Videos 5 and 6). After this event, Ivy1 dots then reappeared proximal to the vacuole, suggesting either reformation of SEs or relocalization of Ivy1 (Fig. 8 E). These data suggest that Ivy1-positive structures may initially undergo homotypic fusion before fusing with the vacuole or MVB, and all fusion or reformation events are HOPS dependent. During this process, they most likely also acquire the EGO and TORC1 signaling complexes, resulting in the reformation of SEs.
Figure 8.
HOPS function is required for SE recovery. (A) Schematic diagram of the method for monitoring SEs fusion with vacuoles (Vac). (B and C) Tracing of SEs by LLSM during recovery of vps11-1 mutant cells (from Video 5). vps11-1 cells expressing mGFP-tagged Ivy1 were grown at 24°C in a synthetic medium, incubated at 37°C for 1 h, and then tracked and analyzed by LLSM at 24°C. Vacuoles were stained with CMAC. The 3D stacks were cropped by Imaris after deconvolution, and the different channels were split by ImageJ. Scale bar, 2 µm. Imaris-defined xy, xz, and yz planes of the 32-min time point are shown. The analyzed dot is indicated in B by white arrows. Ivy1 fluorescent intensity was analyzed by vantage time plots in Imaris, and the plot statistic values were measured by surpass objects with spots (C). (D) 3D view of Ivy1 after dots dispersion. The image was extracted from Video 5, and all views are shown. (E) Quantification of Ivy1 dots in Video 6. The numbers of Ivy1 dots were counted manually and analyzed by ImageJ. Error bars represent SD of three independent experiments.
Video 5.
Ivy1 localization relative to vacuoles in vps11-1 cells by LLSM analysis after 3D deconvolution. Related to Fig. 8 B. vps11-1 cells expressing mGFP-tagged Ivy1 with CMAC-stained vacuoles were imaged for 40 min. The video shows cropped 3D renderings (maximum-intensity projection mode in Imaris) from Video 6 of a deconvolved 3D time-lapse stack (see Materials and methods for details). The video was recorded at 10 frames/s; the original images were taken in 60-s time intervals.
Video 6.
Ivy1 localization in vps11-1 cells by LLSM analysis after 3D deconvolution. Related to Fig. 8 E. vps11-1 cells expressing mGFP-tagged Ivy1 were imaged for 40 min (CMAC staining is not shown). The video shows 3D renderings (maximum-intensity projection mode in Imaris) of a deconvolved 3D time-lapse stack (see Materials and methods for details). The video was recorded at 10 frames/s; the original images were taken in 60-s time intervals.
Endosomal cargo can pass through Ivy1-positive structures
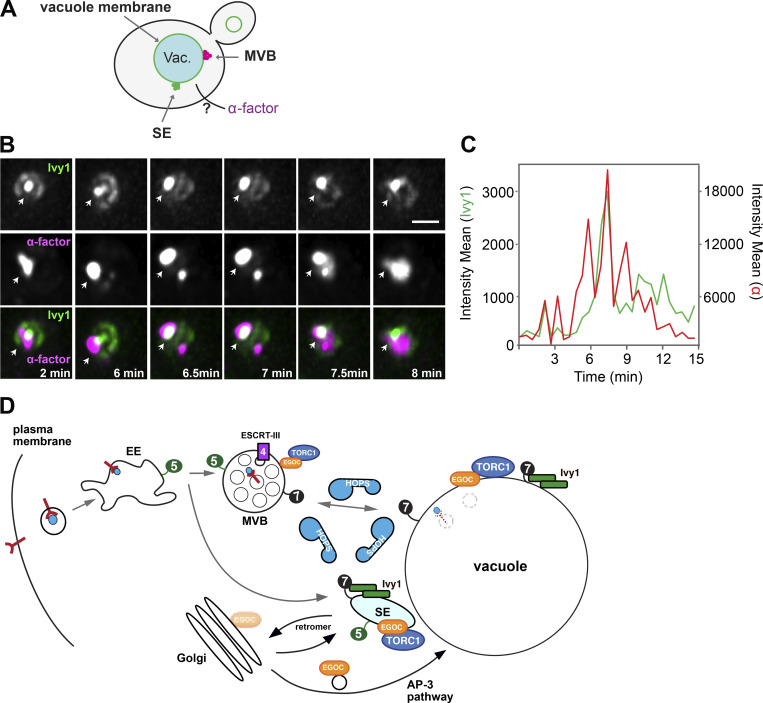
Previous analyses suggested that SEs have a key function in ET activity to control protein synthesis, macroautophagy, and ESCRT-mediated microautophagy at the vacuole (Hatakeyama et al., 2019; Hatakeyama and Virgilio, 2019b; Lahiri and Klionsky, 2019). However, it has not been resolved how ET may sense the nutrient status of the cell. We considered the possibility that SEs are part of the endocytic pathway and may thus detect the flux of cargo or possibly receptor proteins. To test if Ivy1-positive SEs are connected to the endocytic pathway, we monitored the trafficking of Cy5-labeled α-factor via its pheromone receptor Ste2 from the plasma membrane through the endocytic pathway to the vacuole (Arlt et al., 2015; Day et al., 2018). For this, α-factor was added to cells expressing mGFP-tagged Ivy1, and both signals were recorded by 3D LLSM over time. Because of the time needed between α-factor addition to cells and their mounting at the LLSM stage, we observed events only at the Ivy1-decorated SEs. We observed α-factor and Ivy1 in the same structure over time, followed by the appearance of α-factor in the vacuole lumen (Video 7 and Fig. 9, B and C), suggesting that endosomal cargo can pass through endosomal structures marked by Ivy1.
Video 7.
Ivy1 localization relative to α-factor by LLSM analysis after 3D deconvolution. Related to Fig. 9, B and C. Cells expressing mGFP-tagged Ivy1 and treated with fluorescent α-factor were imaged for 12 min. The video shows 3D renderings (maximum-intensity projection mode in Imaris) of a deconvolved 3D time-lapse stack (see Materials and methods for details). The video was recorded by 8 frames/s, the original images were taken at 30-s time intervals.
Figure 9.
Plasma membrane–derived cargo can pass through the SEs. (A) Schematic model of α-factor uptake by yeast cells. Green dots refer to SE, green ring to vacuoles (Vac.), and magenta dots to MVB. (B) Trafficking of α-factor relative to SEs. Selected time points from LLSM image (Video 7) after 3D deconvolution are shown. Cells expressing mGFP-tagged Ivy1 were grown in a synthetic medium, cooled to 4°C to block endocytosis, and treated with fluorescent α-factor for 15 min at 4°C. After mounting, α-factor was tracked by LLSM at 23°C. Indicated time points refer to the time interval after 5 min when cells were shifted to 23°C. (C) 3D track mean fluorescence intensities of Ivy1-mGFP and α-factor from Video 7. Respective Ivy1 and α-factor fluorescence intensities were analyzed by vantage time plots in Imaris, the plots statistics values measured by surpass objects with spots. The analyzed dots were indicated in B by white arrows. The experiment was done three times with similar observations. (D) Working model of the SE function in endolysosomal trafficking. Endocytic transport of a plasma membrane protein (red) bound to cargo (blue) occurs via the EE and MVB toward the vacuole. SEs are shown at the interface between the EE and the Golgi. Rab5 (5, green) and Rab7 (7, black) indicate membrane identity of each compartment. Vps4 (4, pink) is present on MVBs, Ivy1 on SEs and the vacuole, where the two pools of EGOC (a substrate of the AP-3 pathway) and TORC1 are also observed. A fraction of EGOC and TORC1 also resides on MVBs. HOPS promotes fusion between these compartments. For details, see text.
Discussion
SEs are a novel endosomal population in yeast, which harbor ET (Hatakeyama and Virgilio, 2019b; Lahiri and Klionsky, 2019; Hatakeyama et al., 2019; Chen et al., 2021). Here, we set out to determine the identity of SEs and their link to the endocytic pathway using Ivy1 as a reference marker. We show that these SEs are distinct structures with slower mobility than MVBs that localize close to the vacuole, yet are tightly connected to MVB biogenesis. If the ESCRT-IV protein Vps4 is lacking or impaired, Tor1, Ego1, and Ivy1 as proteins on SEs shift largely to class E compartments as shown for many other endosomal proteins (Russell et al., 2012). Using a HOPS-inactivating vps11-1 allele (Peterson and Emr, 2001), we uncover that most of the Ivy1-positive structures remain endosomal but lose SE-specific signaling markers such as the TORC1 subunit Kog1 and Ego1, which is then found at the Golgi (Fig. 6). Once HOPS is reactivated, these Ivy1-positive structures reform into a punctum next to the vacuole, suggesting that they are reformed SEs. In agreement with the signaling function of SEs, HOPS inactivation impairs VT activity even at semipermissive temperatures and slightly enhances ET activity (Fig. 7). This is explained by the depletion of Ego1 (and hence EGOC; Nicastro et al., 2017) at vacuolar membranes and its clustering at SEs and the Golgi compartment. Overall, we reveal that SEs form at a branch between endocytosis and MVB biogenesis, thus linking signaling to protein trafficking (Fig. 9 D).
SEs as an endosomal population have escaped attention in previous studies. One reason for this could be that SEs harbor basically all endosomal markers, such as the Rab5-like Vps21, CORVET, or the SNARE Pep12. Also, the ESCRT-0 subunit Vps27 is present on SEs (Hatakeyama et al., 2019), and Vps27 has been taken as a bona fide marker of endosomes in many studies (Kama et al., 2011; Dobzinski et al., 2015; Kanneganti et al., 2011; MacDonald et al., 2012; Bilodeau et al., 2003; Katzmann et al., 2003; Curwin et al., 2009). However, as shown here and before (Chen et al., 2021), SEs, which we follow using Ivy1 as a reference marker, lack the ESCRT-IV subunit Vps4 and are thus unable to form ILVs, yet all SE-specific markers accumulate at class E compartments if VPS4 is deleted (Fig. 3). This shows that the function of the SE as a dynamic endosomal compartment requires functional MVBs. Even the CORVET subunit Vps8 as an effector of the Rab5-like Vps21 may not be the best marker to trace endosomes in general and thus follow their fusion with the vacuole (Day et al., 2018; Casler and Glick, 2020).
We realize that the endosome population in the cell is heterogeneous and that various stages of endosome maturation exist, which creates a challenge to trace SEs. We used mainly Ivy1 as a reference marker, although are aware that not all Ivy1 dots localize to SEs. Ivy1 is an IBAR protein with a preference for negative curvature that binds both PI3P and Ypt7, can inhibit Fab1 function, and is found in endosomal dots and on the vacuole, similarly to Tor1 and Ego1 (Lazar et al., 2002; Numrich et al., 2015; Malia et al., 2018). Ivy1 also relocalizes from dots to the vacuole in response to changes in amino acids and is found in vacuolar domains after long starvation (Sullivan et al., 2019; Varlakhanova et al., 2018b, a; Murley et al., 2017; Toulmay and Prinz, 2012; Numrich et al., 2015). We are aware that Ivy1 as a peripheral membrane protein may relocalize from endosomes to vacuoles without membrane fusion. However, we followed Ivy1 under normal growth conditions and observed clear colocalization of Tor1 or Ego1, together with Ivy1 and endosomal proteins such as Vps21, but also Ypt7 (Fig. 1, A–D). Importantly, Ivy1 dots also colocalized well with ET (Fig. 1, F and G). Moreover, Ivy1 structures accumulate if HOPS is impaired, suggesting that they require HOPS to fuse with MVBs and the vacuole. These data indicate that Ivy1 dots correspond largely to SEs, possibly at different stages of their maturation (Fig. 1, A–D). In addition to the more static Ivy1-positive dots (which we here consider SEs), we observed a rather mobile Ivy1 fraction on vacuoles (Fig. 2, F and G), which was also observed when we reactivated HOPS and traced the vacuolar pool of Ivy1 on the vacuole over time (Fig. 8). We consider it likely that these mobile dots on vacuoles correspond to individual Ivy1 molecules, but not endosomes. In comparison to Ivy1 dots, MVBs (as monitored by Vps4 mobility) are more mobile (Fig. 2 H), though we do not know the reason for this difference in mobility presently.
Several studies have used ultrastructural analyses to dissect the yeast endocytic pathway by following the endocytic uptake of nanogold particles (Griffith and Reggiori, 2009; Prescianotto-Baschong and Riezman, 2002). Here, tubular intermediates appeared, which were interpreted as EEs. As we found colocalization of Ivy1 with α-factor as an endocytic cargo, we speculate that the biogenesis of SEs and TORC1 signaling is linked to nutrient transporter shuttling (Fig. 9 D). How such a link between trafficking and signaling may work is presently unclear. We favor a model where nutrient transporters themselves either activate TORC1 or bring along signaling molecules. This would allow TORC1 to translate trafficking of nutrient transporters into the metabolic state of the cell. If TORC1 is then active, it may phosphorylate several proteins such as the Fab1 complex (Chen et al., 2021), which may stabilize the SE, affecting signaling and thus growth.
Our data uncover a key role of the HOPS complex in maintaining SE identities. HOPS is a tethering complex that binds SNAREs and promotes fusion of Ypt7-positive membranes (Wickner and Rizo, 2017; Zick and Wickner, 2016; Mima and Wickner, 2009; Bröcker et al., 2012; Lürick et al., 2017; Ho and Stroupe, 2015). The two vps11 alleles clearly affect HOPS and CORVET differently (Peterson and Emr, 2001). The vps11-1 allele affects primarily HOPS and thus fusion events at the vacuole but still allows CORVET-dependent endocytosis, whereas the vps11-3 allele blocks the latter process without interfering with fusion at the vacuole. In agreement, vacuolar proteomics clearly show that the vps11-3 mutant strongly blocks endocytosis, whereas the vps11-1 analysis shows that both HOPS and the EGOC are lost from vacuolar fractions at the restrictive temperature (Fig. 4, F and G). If SEs then harbor both HOPS and Ypt7, why do not they fuse with the vacuole? We speculate that signaling at the SE may also block the fusion machinery, by phosphorylation of Vps27 or other proteins (Fig. 7, C and D; Hatakeyama et al., 2019; Hatakeyama and Virgilio, 2019a). In this case, loss of signaling may revert this process and promote fusion of SEs with MVBs or the vacuole. We are currently testing this hypothesis.
SEs are possibly also connected to the AP-3 pathway (Nagano et al., 2019; Toshima et al., 2014) and thus may exist at a branch between the biosynthetic sorting pathway to the vacuole and the endocytic pathway (Fig. 9 D). This would explain why the EGOC, an identified substrate of the AP-3 pathway (Hatakeyama et al., 2019), localizes to SEs and vacuoles. This localization may be far more dynamic than anticipated as Ego1 (and likely the entire EGOC) appears at the Golgi if HOPS has been inactivated. Furthermore, Ego1 and Ivy1 dots are reduced in retromer mutant, suggesting a role of sorting nexins or retromer in retrograde transport at SEs. Localization of EGOC and TORC1 to SEs may thus require a balance between the HOPS-dependent fusion and a retromer-dependent recycling pathway.
In summary, our data reveal that SEs are tightly connected to the biogenesis of LEs in yeast, are linked to the endocytic pathway, and may thus receive signal input for ET activity for their stabilization as an endosomal structure. We uncover a key role of the HOPS complex in keeping SE identities, suggesting that fusion regulation may be part of the signaling cascade. Future studies need to dissect how TORC1 or other signaling complexes promote SE formation, sense endocytic trafficking, and thus translate this into metabolic adjustments.
Materials and methods
Yeast strains and molecular biology
Strains used in this study are listed in Table S1. Deletions and tagging of genes in the cells were done by PCR-based homologous recombination with corresponding primers and templates (Janke et al., 2004; Puig et al., 1998). Mutations in Fab1 were generated by a CRISPR-Cas9 approach (Generoso et al., 2016). Vps4-mCherry has an HA-tag as a spacer before the mCherry tag, which maintains protein functionality (Adell et al., 2017). Plasmids are listed in Table S2. Primers are listed in Table S3.
Fluorescence microscopy
Yeast cells were grown in a synthetic complete medium (yeast nitrogen base without amino acids and with ammonium sulfate) containing 2% glucose to log phase at 30°C. Selective ts strains were cultured in a synthetic complete medium at 24°C to log phase and then shifted to 37°C for 1 h. Cells were imaged on a DeltaVision Elite imaging system based on an inverted microscope with 100×, NA 1.49 objectives, an sCMOS camera (PCO), and an Insight SSI (TM) illumination system. Stacks of six to eight images with 0.2–0.35 µm spacing were taken, and images were deconvolved using SoftWoRx software (Applied Precision). To analyze the localization of ET or VT relative to the vacuole, Ivy1, and Kog1, images were captured with an inverted spinning-disk confocal microscope (Nikon Ti-E; VisiScope CSU-W1) that was equipped with a Photometrics pco.edge 4.2 sCMOS camera and a 100×, NA 1.3 oil-immersion Nikon CFI series objective (Egg).
Real-time 3D LLSM and image processing
Wild-type cells expressing Ivy1-mGFP were grown in a synthetic complete medium to log phase, and vacuoles were stained with FM4-64 or 7-amino-4-chlormethylcumarin (CMAC; Videos 1, 2, 3, and 4). vps11-1 ts cells expressing Ivy1-mGFP were grown in synthetic complete medium at 24°C to a log phase, incubated at 37°C for 1 h, and vacuoles were stained with CMAC. For α-factor uptake, cells expressing Ivy1-mGFP were grown in synthetic complete medium to a log phase, incubated on ice for 15 min, and then 2.5 μM labeled α-factor was added to the cells for additional 15-min incubation on ice.
Video 3.
Ivy1 localization relative to vacuoles by LLSM analysis after 3D deconvolution. Related to Fig. 3 C. Cells expressing mGFP-tagged Ivy1 with CMAC-stained vacuoles were imaged for 1 min. The video shows 3D renderings (maximum-intensity projection mode in Imaris) of a deconvolved 3D time-lapse stack (see Materials and methods for details). The video was recorded at 8 frames/s; the original images were taken at 1.1-s time intervals.
5 μl of cells were spotted on the top of 5-mm round glass coverslips (11888372; Thermo Fisher Scientific) coated with concanavalin A for 5 min at room temperature to make them adhere. They were then mounted on a sample holder specially designed for LLSM, which was an exact home-built clone of the original design by the Eric Betzig group (Chen et al., 2014). The holder was inserted into a sample bath containing synthetic complete medium at room temperature (25°C). A two-channel image stack was acquired in sample scan mode through a fixed light sheet with a step size of 500 nm, which is equivalent to a ∼271-nm slicing with respect to the z-axis considering the sample scan angle of 32.8°. We used a dithered square lattice pattern generated by multiple Bessel beams, with inner and outer numerical apertures of the excitation objective of 0.48 and 0.55, respectively. Each 3D image stack (512 × 320 × 150 voxels) contained 50–100 cells and was imaged at 30–40 frames. For time-lapse videos, we recorded protein dynamics with a full 3D stack every 1 s for a total time of 1 min (60 time points). For vps11-1 cell recovery assay, we recorded a full 3D stack every 1 min for a total time of 40 min (40 time points). For α-factor uptake assay, we recorded a full 3D stack every 30 s for a total time of 15 min (30 time points). The different channels were sequentially excited using a 405-nm laser (LBX-405; Oxxius) for CMAC, a 488-nm laser (2RU-VFL-P-300-488-B1R; MPB Communications) for GFP (Ivy1 or Vps4), and a 560-nm laser (2RU-VFL-P-500-560-B1R; MPB Communications) for mCherry (Ivy1). Fluorescence was detected by an sCMOS camera (ORCAFlash 4.0; Hamamatsu) using a quadband emission filter (446/523/600/677 HC Quadband; Semrock), with an exposure time of 13.2 ms for monitoring protein dynamics in each channel and an exposure time of 23.2 ms for the vps11-1 cell recovery and α-factor uptake assays. The final pixel size in the image is 104.5 nm. The raw data were further processed using an open-source LLSM postprocessing utility called LLSpy v0.4.9 (https://github.com/tlambert03/LLSpy) for deskewing, deconvolution, 3D stack rotation, and rescaling. Deconvolution was performed using experimental point spread functions and is based on the Richardson–Lucy algorithm using 10 iterations. Finally, image data were analyzed using spot detection and tracking in Imaris 9.5 (Bitplane). By using the built-in spot detection routine, single fluorescent signals were classified as diffraction-limited ellipsoids with a diameter of 250 nm in x and y directions and 600 nm in z direction. These spots were tracked with the built-in autoregressive motion model using a maximum single-step displacement of 1.2 µm and a maximum gap size of 0 time points. Only trajectories >2.5 s were considered for further analysis. Mean track intensity and speed box plots were generated by the Imaris built-in Vantage plot tool. Box plots show minimum (Q0 percentile) and maximum (Q4 percentile) values, the box defines data points within Q1 and Q3 percentiles, and the line defines the median (Q2 percentile).
ET/VT assay to determine TORC1 activity
Wild-type, vps11-1, and vps11-3 cells were transformed either with the ET reporter (FYVE-GFP-Sch9C-term) harboring plasmid pRS425-VAC8p-EEA1(human)1257-1411-GFP-SCH9709-824 or the VT reporter (Sch9C-term-GFP-Pho8N-term) harboring plasmid pRS426-PRC1p-SCH9709-824-GFP-PHO81-63. The cells were grown at 24 or 30°C in a synthetic complete medium (2% glucose, yeast nitrogen base, ammonium sulfate, and all amino acids) until mid-log phase, and 10 ml of cell culture was mixed with TCA at a final concentration of 6%. After centrifugation, the pellet was washed with cold acetone and dried in a speed-vac. The pellet was resuspended in lysis buffer (50 mM Tris-HCl, pH 7.5, 5 mM EDTA, 6 M urea, and 1% SDS), the amount being proportional to the OD600 of the original cell culture. Proteins were extracted by agitation in a Precellys machine after the addition of glass beads. After the addition of 2× Laemmli buffer (350 mM Tris-HCl, pH 6.8, 30% glycerol, 600 mM DTT, 10% SDS, and 0.02% bromophenol blue), the mix was boiled at 98°C for 5 min. The analysis was carried out by SDS-PAGE using phosphospecific rabbit anti-Sch9-pThr737 (custom made), goat anti-Sch9 (custom made), and mouse anti-GFP (11814460001; Roche) antibodies. Band intensities were quantified using ImageJ software. For Vps27 phosphorylation state analysis, EDTA-free protein extracts were run on a 6% gel containing 50 μM Mn2+-Phos-tag and probed with rabbit anti-Vps27 antibodies (custom made).
Vacuole isolation and proteomics
Wild-type and ts mutant cells were grown in 500 ml synthetic medium with 30 mg/l normal lysine or 30 mg/l heavy lysine (L-lysine 13C615N2; Cambridge Isotope Laboratories) at 23°C and then incubated at 37°C for 1 h to an OD600 of ∼0.8–1.0, respectively. The vacuole isolation assay was performed as described before (Gao et al., 2018). Isolated vacuoles were precipitated with 20% TCA, incubated on ice for 20 min, and resuspended in ice-cold acetone twice by sonication. The vacuole precipitate was further purified using PreOmics IST kit for the final mass spectrometry measurements.
Reverse-phase chromatography was performed on a Thermo Ultimate 3000 RSLCnano system connected to a Q Exactive-Plus mass spectrometer (Thermo Fisher Scientific). Peptides were separated and eluted as described previously (Eising et al., 2019). The MS results were analyzed by MaxQuant (v1.6.14.0, www.maxquant.org/) as described before (Fröhlich et al., 2013), and the plots were performed with the R software package (www.r-project.org/).
Online supplemental material
Fig. S1 shows the relocalization of SE-specific proteins in vps4 mutants and supports Fig. 3. Fig. S2 shows the localization of endosomal and vacuolar proteins relative to Ivy1 in HOPS mutant strains and supports Fig. 5. Fig. S3 shows analysis of proteins on Vps4-positive endosomes in HOPS mutants cells in support of Figs. 5 and 6. Fig. S4 shows the localization of TORC1 reporters in the vps11 mutant and supports Fig. 7. Videos 1 and 2 show the mobility of Ivy1 relative to FM4-64 stained vacuoles and Vps4 (related to Fig. 2, D and H). Video 3 shows the mobility of Ivy1 relative to CMAC stained vacuolesis (related to Fig. 3 C). Video 4 shows the Ivy1 localization relative to vacuoles in the vps4 mutant (related to Fig. 3 C). Videos 5 and 6 show Ivy1 localization in vps11-1 mutants (related to Fig. 8, B and E). Video 7 shows Ivy1 localization relative to endocytosed α-factor and is related to Fig. 9, B and C. Table S1 lists all yeast strains used in this study. Table S2 lists the plasmids used. Table S3 lists the primers used. Data S1 shows quantification of Fig. 1, A, B, E, and F, and correlates to Fig. 1, C and G. Data S2 shows quantification of Fig. 2, A and B, and correlates to Fig. 2 C. Data S3 shows quantification of Fig. 3, A, B, and E, and correlates to Fig. 3, D and F. Data S4 shows quantification of Fig. 4, A–D, and correlates to Fig. 4 F. Data S5 shows quantification of Fig. S2 A, Fig. 5B, and Fig. S3 A, and correlates to Fig. 5, A, C, and D. Data S6 shows quantification of Fig. 6, A, C, and E, correlates to Fig. 6 B, D, and F. Data S7 shows quantification of Fig. 7, B, D, and G, and correlates to Fig. 6, C, E, and H. Data S8 shows quantification of Fig. 8 B and correlates to Fig. 8 E. Data S9 shows quantification of Fig. 9 B and correlates to Fig. 9 C. Data S10 shows quantification of Fig. 1, A and B; reverse quantification of Fig. 2, A and B, and Fig. 3, A and B; quantification of Fig. S1, D and G; and correlates to Fig. S1, A–C, E, F, and H. Data S11 shows quantification of Fig. S2 B and correlates to Fig. S2 C. Data S12 shows quantification of Fig. S3 B and correlates to Fig. S3 C.
Supplementary Material
lists yeast strains used in this study.
lists plasmids used in this study.
lists primers used in this study.
contains original blots for Fig. 7.
shows quantification of Fig. 1, A, B, E, and F, and correlates to Fig. 1, C and G.
shows quantification of Fig. 2, A and B, and correlates to Fig. 2 C.
shows quantification of Fig. 3, A, B, and E, and correlates to Fig. 3, D and F.
shows quantification of Fig. 4, A–D , and correlates to Fig. 4 F.
shows quantification of Fig. S2 A, Fig. 5B, and Fig. S3 A, and correlates to Fig. 5, A, C, and D.
shows quantification of Fig. 6, A, C, and E, correlates to Fig. 6 B, D, and F.
shows quantification of Fig. 7, B, D, and G, and correlates to Fig. 6, C, E, and H.
shows quantification of Fig. 8 B and correlates to Fig. 8 E.
shows quantification of Fig. 9 B and correlates to Fig. 9 C.
shows quantification of Fig. 1, A and B; reverse quantification of Fig. 2, A and B, and Fig. 3, A and B; quantification of Fig. S1, D and G; and correlates to Fig. S1, A–C, E, F, and H.
shows quantification of Fig. S2 B and correlates to Fig. S2 C.
shows quantification of Fig. S3 B and correlates to Fig. S3 C.
Acknowledgments
We thank Scott Emr (Cornell University, Ithaca, NY), David Teis (University of Innsbruck, Innsbruck, Austria), Fulvio Reggiori (University of Groningen, Groningen, Netherlands), and Markus Babst (University of Utah, Salt Lake City, UT) for strains and advice; Lars Langemeyer, Ayelén Gonzaléz Montoro, and all members of the Ungermann lab for feedback; and Kathrin Auffarth and Angela Perz for expert technical assistance.
This work was funded by the Deutsche Forschungsgemeinschaft (UN111/10-2 to C. Ungermann), imaging facility iBiOs (PI 405/14-1 to J. Piehler and R. Kurre), the Sonderforschungsbereich 944 (project P11 to C. Ungermann, project P20 to F. Fröhlich, P8 to J. Piehler, and Z to R. Kurre), and the Swiss National Science Foundation (310030_184671 to C. De Virgilio).
Author contributions: J. Gao performed all localization and trafficking experiments with support from S. Grziwa, Z. Chen, and M.-P. Péli-Gulli. R. Nicastro, M.-P. Péli-Gulli, and C. De Virgilio analyzed TORC1 activity. LLSM analysis was done by R. Kurre and J. Piehler. All mass spectrometry analyses were done by F. Fröhlich. C. Ungermann supervised the study and wrote the manuscript together with J. Gao, with the support of all authors.
References
- Adell, M.A.Y., Migliano S.M., Upadhyayula S., Bykov Y.S., Sprenger S., Pakdel M., Vogel G.F., Jih G., Skillern W., Behrouzi R., et al. 2017. Recruitment dynamics of ESCRT-III and Vps4 to endosomes and implications for reverse membrane budding. eLife. 6:e31652. 10.7554/elife.31652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt, H., Auffarth K., Kurre R., Lisse D., Piehler J., and Ungermann C.. 2015. Spatiotemporal dynamics of membrane remodeling and fusion proteins during endocytic transport. Mol. Biol. Cell. 26:1357–1370. 10.1091/mbc.e14-08-1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst, M., Sato T.K., Banta L.M., and Emr S.D.. 1997. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 16:1820–1831. 10.1093/emboj/16.8.1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst, M., Wendland B., Estepa E.J., and Emr S.D.. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17:2982–2993. 10.1093/emboj/17.11.2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, R.W., Jeffrey P.D., Zick M., Phillips B.P., Wickner W.T., and Hughson F.M.. 2015. A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science. 349:1111–1114. 10.1126/science.aac7906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balderhaar, H.J., Lachmann J., Yavavli E., Bröcker C., Lürick A., and Ungermann C.. 2013. The CORVET complex promotes tethering and fusion of Rab5/Vps21-positive membranes. Proc. Natl. Acad. Sci. USA. 110:3823–3828. 10.1073/pnas.1221785110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balderhaar, H.J., and Ungermann C.. 2013. CORVET and HOPS tethering complexes: Coordinators of endosome and lysosome fusion. J. Cell Sci. 126:1307–1316. 10.1242/jcs.107805 [DOI] [PubMed] [Google Scholar]
- Balla, T. 2013. Phosphoinositides: Tiny lipids with giant impact on cell regulation. Physiol. Rev. 93:1019–1137. 10.1152/physrev.00028.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr, F.A. 2013. Rab GTPases and membrane identity: Causal or inconsequential? J. Cell Biol. 202:191–199. 10.1083/jcb.201306010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau, P.S., Winistorfer S.C., Kearney W.R., Robertson A.D., and Piper R.C.. 2003. Vps27-Hse1 and ESCRT-I complexes cooperate to increase efficiency of sorting ubiquitinated proteins at the endosome. J. Cell Biol. 163:237–243. 10.1083/jcb.200305007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröcker, C., Kuhlee A., Gatsogiannis C., Balderhaar H.J., Hönscher C., Engelbrecht-Vandré S., Ungermann C., and Raunser S.. 2012. Molecular architecture of the multisubunit homotypic fusion and vacuole protein sorting (HOPS) tethering complex. Proc. Natl. Acad. Sci. USA. 109:1991–1996. 10.1073/pnas.1117797109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera, M., Arlt H., Epp N., Lachmann J., Griffith J., Perz A., Reggiori F., and Ungermann C.. 2013. Functional separation of endosomal fusion factors and the class C core vacuole/endosome tethering (CORVET) complex in endosome biogenesis. J. Biol. Chem. 288:5166–5175. 10.1074/jbc.m112.431536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera, M., Langemeyer L., Mari M., Rethmeier R., Orban I., Perz A., Bröcker C., Griffith J., Klose D., Steinhoff H.-J., et al. 2010. Phosphorylation of a membrane curvature–sensing motif switches function of the HOPS subunit Vps41 in membrane tethering. J. Cell Biol. 191:845–859. 10.1083/jcb.201004092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casler, J.C., and Glick B.S.. 2020. A microscopy-based kinetic analysis of yeast vacuolar protein sorting. eLife. 9. e56844. 10.7554/elife.56844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, B.-C., Legant W.R., Wang K., Shao L., Milkie D.E., Davidson M.W., Janetopoulos C., Wu X.S., Hammer J.A., Liu Z., et al. 2014. Lattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal resolution. Science. 346:1257998. 10.1126/science.1257998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z., Malia P.C., Hatakeyama R., Nicastro R., Hu Z., Péli-Gulli M.-P., Gao J., Nishimura T., Eskes E., Stefan C.J., et al. 2021. TORC1 determines Fab1 lipid kinase function at signaling endosomes and vacuoles. Curr. Biol. 31:297–309.e8. 10.1016/j.cub.2020.10.026 [DOI] [PubMed] [Google Scholar]
- Curwin, A.J., Fairn G.D., and McMaster C.R.. 2009. Phospholipid transfer protein Sec14 is required for trafficking from endosomes and regulates distinct trans-Golgi export pathways. J. Biol. Chem. 284:7364–7375. 10.1074/jbc.m808732200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day, K.J., Casler J.C., and Glick B.S.. 2018. Budding yeast has a minimal endomembrane system. Dev. Cell. 44:56–72.e4. 10.1016/j.devcel.2017.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzinski, N., Chuartzman S.G., Kama R., Schuldiner M., and Gerst J.E.. 2015. Starvation-dependent regulation of Golgi quality control links the TOR signaling and vacuolar protein sorting pathways. Cell Rep. 12:1876–1886. 10.1016/j.celrep.2015.08.026 [DOI] [PubMed] [Google Scholar]
- Eising, S., Thiele L., and Fröhlich F.. 2019. A systematic approach to identify recycling endocytic cargo depending on the GARP complex. eLife. 8. e42837. 10.7554/elife.42837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich, F., Christiano R., and Walther T.C.. 2013. Native SILAC: Metabolic labeling of proteins in prototroph microorganisms based on lysine synthesis regulation. Mol. Cell Proteom. 12:1995–2005. 10.1074/mcp.m112.025742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, J., Reggiori F., and Ungermann C.. 2018. A novel in vitro assay reveals SNARE topology and the role of Ykt6 in autophagosome fusion with vacuoles. J. Cell Biol. 217:3670–3682. 10.1083/jcb.201804039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Generoso, W.C., Gottardi M., Oreb M., and Boles E.. 2016. Simplified CRISPR-Cas genome editing for Saccharomyces cerevisiae. J. Microbiol. Meth. 127:203–205. 10.1016/j.mimet.2016.06.020 [DOI] [PubMed] [Google Scholar]
- Goody, R.S., Müller M.P., and Wu Y.-W.. 2017. Mechanisms of action of Rab proteins, key regulators of intracellular vesicular transport. Biol. Chem. 398:565–575. 10.1515/hsz-2016-0274 [DOI] [PubMed] [Google Scholar]
- Griffith, J., and Reggiori F.. 2009. Ultrastructural analysis of nanogold-labeled endocytic compartments of yeast Saccharomyces cerevisiae using a cryosectioning procedure. J. Histochem. Cytochem. 57:801–809. 10.1369/jhc.2009.952952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa, J., Strunk B.S., and Weisman L.S.. 2017. PI5P and PI(3,5)P2: Minor, but essential phosphoinositides. Cell Struct. Funct. 42:17003. 10.1247/csf.17003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama, R., Péli-Gulli M.-P., Hu Z., Jaquenoud M., Osuna G.M.G., Sardu A., Dengjel J., and Virgilio C.D.. 2019. Spatially distinct pools of TORC1 balance protein homeostasis. Mol. Cell. 73:325–338.e8. 10.1016/j.molcel.2018.10.040 [DOI] [PubMed] [Google Scholar]
- Hatakeyama, R., and Virgilio C.D.. 2019a. TORC1 specifically inhibits microautophagy through ESCRT-0. Curr. Genet. 65:1243–1249. 10.1007/s00294-019-00982-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama, R., and Virgilio C.D.. 2019b. A spatially and functionally distinct pool of TORC1 defines signaling endosomes in yeast. Autophagy. 15:915–916. 10.1080/15548627.2019.1580107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, C.Y., Alghamdi T.A., and Botelho R.J.. 2012. Phosphatidylinositol-3,5-Bisphosphate: No longer the poor PIP2. Traffic. 13:1–8. 10.1111/j.1600-0854.2011.01246.x [DOI] [PubMed] [Google Scholar]
- Ho, R., and Stroupe C.. 2015. The HOPS/class C Vps complex tethers membranes by binding to one Rab GTPase in each apposed membrane. Mol. Biol. Cell. 26:2655–2663. 10.1091/mbc.e14-04-0922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huotari, J., and Helenius A.. 2011. Endosome maturation. EMBO J. 30:3481–3500. 10.1038/emboj.2011.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutagalung, A.H., and Novick P.J.. 2011. Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 91:119–149. 10.1152/physrev.00059.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii, A., Kurokawa K., Hotta M., Yoshizaki S., Kurita M., Koyama A., Nakano A., and Kimura Y.. 2019. Role of Atg8 in the regulation of vacuolar membrane invagination. Sci. Rep. 9:14828. 10.1038/s41598-019-51254-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke, C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., and Knop M.. 2004. A versatile toolbox for PCR-based tagging of yeast genes: New fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 21:947–962. 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- Kama, R., Kanneganti V., Ungermann C., and Gerst J.E.. 2011. The yeast Batten disease orthologue Btn1 controls endosome–Golgi retrograde transport via SNARE assembly. J. Cell Biol. 195:203–215. 10.1083/jcb.201102115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanarek, N., Petrova B., and Sabatini D.M.. 2020. Dietary modifications for enhanced cancer therapy. Nature. 579:507–517. 10.1038/s41586-020-2124-0 [DOI] [PubMed] [Google Scholar]
- Kanneganti, V., Kama R., and Gerst J.E.. 2011. Btn3 is a negative regulator of Btn2-mediated endosomal protein trafficking and prion curing in yeast. Mol. Biol. Cell. 22:1648–1663. 10.1091/mbc.e10-11-0878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann, D.J., Stefan C.J., Babst M., and Emr S.D.. 2003. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J. Cell Biol. 162:413–423. 10.1083/jcb.200302136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury, J.M., Sen N.D., Maeda T., Heitman J., and Cardenas M.E.. 2014. Endolysosomal membrane trafficking complexes drive nutrient-dependent TORC1 signaling to control cell growth in Saccharomyces cerevisiae. Genetics. 196:1077–1089. 10.1534/genetics.114.161646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvainickas, A., Nägele H., Qi W., Dokládal L., Jimenez-Orgaz A., Stehl L., Gangurde D., Zhao Q., Hu Z., Dengjel J., et al. 2019. Retromer and TBC1D5 maintain late endosomal RAB7 domains to enable amino acid-induced mTORC1 signaling. J. Cell Biol. 218:3019–3038. 10.1083/jcb.201812110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri, V., and Klionsky D.J.. 2019. Spatially distinct pools of TORC1 balance protein homeostasis. Autophagy. 15:561–564. 10.1080/15548627.2019.1575162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langemeyer, L., Borchers A.-C., Herrmann E., Füllbrunn N., Han Y., Perz A., Auffarth K., Kümmel D., and Ungermann C.. 2020. A conserved and regulated mechanism drives endosomal Rab transition. eLife. 9. e56090. 10.7554/elife.56090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langemeyer, L., Fröhlich F., and Ungermann C.. 2018. Rab GTPase function in endosome and lysosome biogenesis. Trends Cell Biol. 28:957–970. 10.1016/j.tcb.2018.06.007 [DOI] [PubMed] [Google Scholar]
- Lazar, T., Scheglmann D., and Gallwitz D.. 2002. A novel phospholipid-binding protein from the yeast Saccharomyces cerevisiae with dual binding specificities for the transport GTPase Ypt7p and the Sec1-related Vps33p. Eur. J. Cell Biol. 81:635–646. 10.1078/0171-9335-00290 [DOI] [PubMed] [Google Scholar]
- Lin, C.H., MacGurn J.A., Chu T., Stefan C.J., and Emr S.D.. 2008. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell. 135:714–725. 10.1016/j.cell.2008.09.025 [DOI] [PubMed] [Google Scholar]
- Lürick, A., Gao J., Kuhlee A., Yavavli E., Langemeyer L., Perz A., Raunser S., and Ungermann C.. 2017. Multivalent Rab interactions determine tether-mediated membrane fusion. Mol. Biol. Cell. 28:322–332. 10.1091/mbc.e16-11-0764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald, C., Buchkovich N.J., Stringer D.K., Emr S.D., and Piper R.C.. 2012. Cargo ubiquitination is essential for multivesicular body intralumenal vesicle formation. EMBO Rep. 13:331–338. 10.1038/embor.2012.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malia, P., Numrich J., Nishimura T., Montoro A.G., Stefan C.J., and Ungermann C.. 2018. Control of vacuole membrane homeostasis by a resident PI-3,5-kinase inhibitor. Proc. Natl. Acad. Sci. USA. 115:201722517. 10.1073/pnas.1722517115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markgraf, D.F., Ahnert F., Arlt H., Mari M., Peplowska K., Epp N., Griffith J., Reggiori F., and Ungermann C.. 2009. The CORVET subunit Vps8 cooperates with the Rab5 homolog Vps21 to induce clustering of late endosomal compartments. Mol. Biol. Cell. 20:5276–5289. 10.1091/mbc.e09-06-0521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mima, J., and Wickner W.. 2009. Phosphoinositides and SNARE chaperones synergistically assemble and remodel SNARE complexes for membrane fusion. Proc. Natl. Acad. Sci. USA. 106:16191–16196. 10.1073/pnas.0908694106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murley, A., Yamada J., Niles B.J., Toulmay A., Prinz W.A., Powers T., and Nunnari J.. 2017. Sterol transporters at membrane contact sites regulate TORC1 and TORC2 signaling. J. Cell Biol. 216:2679–2689. 10.1083/jcb.201610032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano, M., Toshima J.Y., Siekhaus D.E., and Toshima J.. 2019. Rab5-mediated endosome formation is regulated at the trans-Golgi network. Commun. Biol. 2:419. 10.1038/s42003-019-0670-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro, R., Sardu A., Panchaud N., and Virgilio C.D.. 2017. The architecture of the Rag GTPase signaling network. Biomol. 7:48. 10.3390/biom7030048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann, M., Cabrera M., Perz A., Bröcker C., Ostrowicz C., Engelbrecht-Vandré S., and Ungermann C.. 2010. The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr. Biol. 20:1654–1659. 10.1016/j.cub.2010.08.002 [DOI] [PubMed] [Google Scholar]
- Numrich, J., Péli-Gulli M.-P., Arlt H., Sardu A., Griffith J., Levine T., Engelbrecht-Vandré S., Reggiori F., Virgilio C.D., and Ungermann C.. 2015. The I-BAR protein Ivy1 is an effector of the Rab7 GTPase Ypt7 involved in vacuole membrane homeostasis. J. Cell Sci. 128:2278–2292. 10.1242/jcs.164905 [DOI] [PubMed] [Google Scholar]
- Peplowska, K., Markgraf D.F., Ostrowicz C.W., Bange G., and Ungermann C.. 2007. The CORVET tethering complex interacts with the yeast Rab5 homolog Vps21 and is involved in Endo-lysosomal biogenesis. Dev. Cell. 12:739–750. 10.1016/j.devcel.2007.03.006 [DOI] [PubMed] [Google Scholar]
- Peterson, M.R., and Emr S.D.. 2001. The class C Vps complex functions at multiple stages of the vacuolar transport pathway. Traffic. 2:476–486. 10.1034/j.1600-0854.2001.20705.x [DOI] [PubMed] [Google Scholar]
- Prescianotto-Baschong, C., and Riezman H.. 2002. Ordering of compartments in the yeast endocytic pathway. Traffic. 3:37–49. 10.1034/j.1600-0854.2002.30106.x [DOI] [PubMed] [Google Scholar]
- Puig, O., Rutz B., Luukkonen B., Kandels-Lewis S., Bragado-Nilsson E., and Seraphin B.. 1998. New constructs and strategies for efficient PCR-based gene manipulations in yeast. Yeast. 14:1139–1146. [DOI] [PubMed] [Google Scholar]
- Raymond, C., Howald-Stevenson I., Vater C., and Stevens T.. 1992. Morphological classification of the yeast vacuolar protein sorting mutants: Evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell. 3:1389–1402. 10.1091/mbc.3.12.1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori, F., Black M., and Pelham H.. 2000. Polar transmembrane domains target proteins to the interior of the yeast vacuole. Mol. Biol. Cell. 11:3737–3749. 10.1091/mbc.11.11.3737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder, S.E., Banta L.M., Kohrer K., McCaffery J.M., and Emr S.D.. 1996. Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol. Biol. Cell. 7:985–999. 10.1091/mbc.7.6.985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, J., Graham T., and Emr S.. 1991. A putative zinc finger protein, Saccharomyces cerevisiae Vps18p, affects late Golgi functions required for vacuolar protein sorting and efficient alpha-factor prohormone maturation. Mol. Cell Biol. 11:5813–5824. 10.1128/mbc.11.12.5813-5824.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, M.R.G., Shideler T., Nickerson D.P., West M., and Odorizzi G.. 2012. Class E compartments form in response to ESCRT dysfunction in yeast due to hyperactivity of the Vps21 Rab GTPase. J. Cell Sci. 125:5208–5220. 10.1242/jcs.111310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardana, R., and Emr S.D.. 2021. Membrane protein quality control mechanisms in the Endo-lysosome system. Trends Cell Biol. 31:269–283. 10.1016/j.tcb.2020.11.011 [DOI] [PubMed] [Google Scholar]
- Schoppe, J., Mari M., Yavavli E., Auffarth K., Cabrera M., Walter S., Fröhlich F., and Ungermann C.. 2020. AP-3 vesicle uncoating occurs after HOPS-dependent vacuole tethering. EMBO J. 39:e2020105117. 10.15252/embj.2020105117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schu, P., Takegawa K., Fry M., Stack J., Waterfield M., and Emr S.. 1993. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 260:88–91. 10.1126/science.8385367 [DOI] [PubMed] [Google Scholar]
- Seals, D.F., Eitzen G., Margolis N., Wickner W.T., and Price A.. 2000. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc. Natl. Acad. Sci. USA. 97:9402–9407. 10.1073/pnas.97.17.9402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, A., Wallace R.L., Wellington R., Luo X., and Capaldi A.P.. 2019. Multilayered regulation of TORC1-body formation in budding yeast. Mol. Biol. Cell. 30:400–410. 10.1091/mbc.e18-05-0297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toshima, J.Y., Nishinoaki S., Sato Y., Yamamoto W., Furukawa D., Siekhaus D.E., Sawaguchi A., and Toshima J.. 2014. Bifurcation of the endocytic pathway into Rab5-dependent and -independent transport to the vacuole. Nat. Commun. 5:1–11. 10.1038/ncomms4498 [DOI] [PubMed] [Google Scholar]
- Toulmay, A., and Prinz W.A.. 2012. A conserved membrane-binding domain targets proteins to organelle contact sites. J. Cell Sci. 125:49–58. 10.1242/jcs.085118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremel, S., Ohashi Y., Morado D.R., Bertram J., Perisic O., Brandt L.T.L., von Wrisberg M.-K., Chen Z.A., Maslen S.L., Kovtun O., et al. 2021. Structural basis for VPS34 kinase activation by Rab1 and Rab5 on membranes. Nat. Commun. 12:1564. 10.1038/s41467-021-21695-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban, J., Soulard A., Huber A., Lippman S., Mukhopadhyay D., Deloche O., Wanke V., Anrather D., Ammerer G., Riezman H., et al. 2007. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell. 26:663–674. 10.1016/j.molcel.2007.04.020 [DOI] [PubMed] [Google Scholar]
- van der Beek, J., Jonker C., van der Welle R., Liv N., and Klumperman J.. 2019. CORVET, CHEVI and HOPS: Multisubunit tethers of the endo-lysosomal system in health and disease. J. Cell Sci. 132:jcs189134. 10.1242/jcs.189134 [DOI] [PubMed] [Google Scholar]
- Varlakhanova, N.V., Tornabene B.A., and Ford M.G.J.. 2018a. Ivy1 is a negative regulator of Gtr-dependent TORC1 activation. J. Cell Sci. 131:jcs218305. 10.1242/jcs.218305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varlakhanova, N.V., Tornabene B.A., and Ford M.G.J.. 2018b. Feedback regulation of TORC1 by its downstream effectors Npr1 and Par32. Mol. Biol. Cell. 29:2751–2765. 10.1091/mbc.e18-03-0158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandinger-Ness, A., and Zerial M.. 2014. Rab proteins and the compartmentalization of the endosomal system. CSH Persp. Biol. 6:a022616. 10.1101/cshperspect.a022616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, W., Chen L., Wu H., Sun X., Zhang M., and Banfield D.. 2006. Identification of the yeast R-SNARE Nyv1p as a novel longin domain-containing protein. Mol. Biol. Cell. 17:4282–4299. 10.1091/mbc.e06-02-0128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner, W., and Rizo J.. 2017. A cascade of multiple proteins and lipids catalyzes membrane fusion. Mol. Biol. Cell. 28:707–711. 10.1091/mbc.e16-07-0517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmser, A.E., Sato T.K., and Emr S.D.. 2000. New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J. Cell Biol. 151:551–562. 10.1083/jcb.151.3.551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeigerer, A., Gilleron J., Bogorad R.L., Marsico G., Nonaka H., Seifert S., Epstein-Barash H., Kuchimanchi S., Peng C.G., Ruda V.M., et al. 2012. Rab5 is necessary for the biogenesis of the endolysosomal system in vivo. Nature. 485:465–470. 10.1038/nature11133 [DOI] [PubMed] [Google Scholar]
- Zhen, Y., Radulovic M., Vietri M., and Stenmark H.. 2021. Sealing holes in cellular membranes. EMBO J. 40:e106922. 10.15252/embj.2020106922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zick, M., and Wickner W.. 2016. Improved reconstitution of yeast vacuole fusion with physiological SNARE concentrations reveals an asymmetric Rab(GTP) requirement. Mol. Biol. Cell. 27:2590–2597. 10.1091/mbc.e16-04-0230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurita-Martinez, S.A., Puria R., Pan X., Boeke J.D., and Cardenas M.E.. 2007. Efficient Tor signaling requires a functional class C Vps protein complex in Saccharomyces cerevisiae. Genetics. 176:2139–2150. 10.1534/genetics.107.072835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweytick, D., Leitner E., Kohlwein S.D., Yu C., Rothblatt J., and Daum G.. 2014. Contribution of Are1p and Are2p to steryl ester synthesis in the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 267:1075–1082. 10.1046/j.1432-1327.2000.01103.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists yeast strains used in this study.
lists plasmids used in this study.
lists primers used in this study.
contains original blots for Fig. 7.
shows quantification of Fig. 1, A, B, E, and F, and correlates to Fig. 1, C and G.
shows quantification of Fig. 2, A and B, and correlates to Fig. 2 C.
shows quantification of Fig. 3, A, B, and E, and correlates to Fig. 3, D and F.
shows quantification of Fig. 4, A–D , and correlates to Fig. 4 F.
shows quantification of Fig. S2 A, Fig. 5B, and Fig. S3 A, and correlates to Fig. 5, A, C, and D.
shows quantification of Fig. 6, A, C, and E, correlates to Fig. 6 B, D, and F.
shows quantification of Fig. 7, B, D, and G, and correlates to Fig. 6, C, E, and H.
shows quantification of Fig. 8 B and correlates to Fig. 8 E.
shows quantification of Fig. 9 B and correlates to Fig. 9 C.
shows quantification of Fig. 1, A and B; reverse quantification of Fig. 2, A and B, and Fig. 3, A and B; quantification of Fig. S1, D and G; and correlates to Fig. S1, A–C, E, F, and H.
shows quantification of Fig. S2 B and correlates to Fig. S2 C.
shows quantification of Fig. S3 B and correlates to Fig. S3 C.