Abstract
Background
Since the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 2019, viral variants with greater transmissibility or immune-evasion properties have arisen, which could jeopardize recently deployed vaccine- and antibody-based countermeasures.
Methods
Here, we evaluated in mice and hamsters the efficacy of a pre-clinical version of the Moderna mRNA vaccine (mRNA-1273) and the Johnson & Johnson recombinant adenoviral-vectored vaccine (Ad26.COV2.S) against the B.1.621 (Mu) variant of SARS-CoV-2, which contains spike mutations T95I, Y144S, Y145N, R346K, E484K, N501Y, D614G, P681H, and D950N.
Findings
Immunization of 129S2 and K18-human ACE2 transgenic mice with the mRNA-1273 vaccine protected against weight loss, lung infection, and lung pathology after challenge with the B.1.621 or WA1/2020 N501Y/D614G SARS-CoV-2 strain. Similarly, immunization of 129S2 mice and Syrian hamsters with a high dose of Ad26.COV2.S reduced lung infection after B.1.621 virus challenge.
Conclusions
Thus, immunity induced by the mRNA-1273 or Ad26.COV2.S vaccine can protect against the B.1.621 variant of SARS-CoV-2 in multiple animal models.
Funding
This study was supported by the NIH (R01 AI157155 and U01 AI151810), NIAID Centers of Excellence for Influenza Research and Response [CEIRR] contracts 75N93021C00014 and 75N93021C00016, and the Collaborative Influenza Vaccine Innovation Centers [CIVIC] contract 75N93019C00051. It was also supported, in part, by the National Institutes of Allergy and Infectious Diseases Center for Research on Influenza Pathogenesis (HHSN272201400008C) and the Japan Program for Infectious Diseases Research and Infrastructure (JP21wm0125002) from the Japan Agency for Medical Research and Development (AMED).
Keywords: mRNA-1273, SARS-CoV-2, Ad26.COV2.S, in vivo vaccine efficacy studies, Syrian hamster, mouse
Graphical abstract
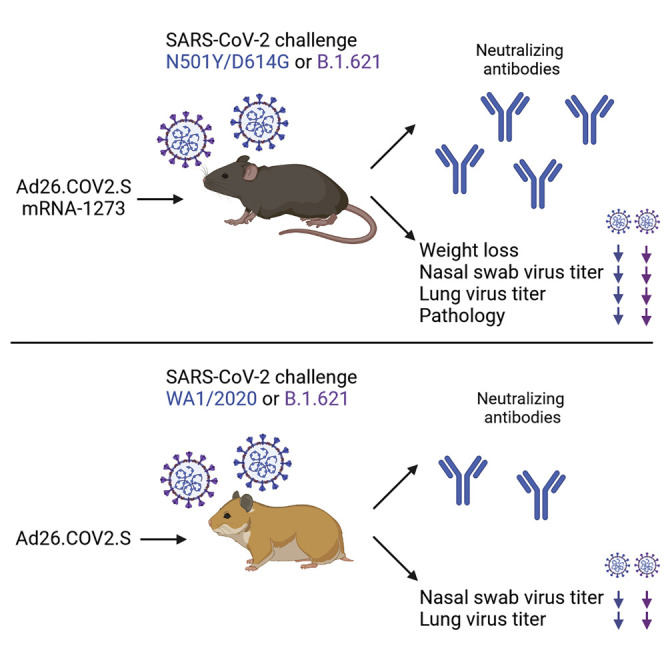
Context and significance
New variants of SARS-CoV-2 have emerged that are more resistant to neutralizing antibodies. Prior to Omicron, the Mu variant (B.1.621) was one of the more neutralizing-antibody-resistant variants of SARS-CoV-2. Here we evaluated the efficacy of two vaccines for protection against the Mu variant in mice and hamsters. Immunization with the mRNA-1273 or Ad26.COV2.S vaccine induced serum neutralizing antibody responses and protected the animals against disease after challenge with B.1.621. Thus, immunity induced by these vaccines can protect against the B.1.621 variant of SARS-CoV-2 in multiple animal models.
Darling et al. show that the Ad26.COV2.S and mRNA-1273 vaccines induce neutralizing antibodies against the B.1.621 variant of SARS-CoV-2 in mice and Syrian hamsters and protect against a B.1.621 virus challenge.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the etiological agent of coronavirus disease 2019 (COVID-19), has caused hundreds of millions of infections worldwide, with more than 5 million deaths. Vaccines targeting the SARS-CoV-2 spike protein were developed within 1 year of the start of the pandemic. Several of these (mRNA and adenoviral vectored) are remarkably effective in protecting against severe COVID-19, with efficacy rates ranging from 75% to 95% depending on the vaccine and age of the individual.1, 2, 3 Vaccines also protect against infection, and likely transmission, albeit at lower rates of 50%–70%.4 , 5 The emergence of several SARS-CoV-2 variants with amino-acid substitutions in the spike protein has jeopardized the efficacy of current vaccines to protect against infection and disease. These variants can be more transmissible and also evade serum neutralizing antibodies. As an example, while the Delta variant (B.1.617.2) had no appreciable effect on vaccine efficacy against hospitalization, vaccine-mediated protection against infection was markedly reduced.1, 2, 3, 4, 5 Thus, evaluating vaccine efficacy against emerging variants of SARS-CoV-2 is important for deciding when to administer booster shots and determining if and when mono- or multivalent vaccines with variant spike antigens are needed.
After the emergence of the first D614G variant, several variants of concern (VOCs) or interest (VOIs) arose, including B.1.1.7 (Alpha), B.1.351 (Beta), B.1.1.28 (Gamma), B.1.617.2 (Delta), and, more recently, B.1.1.529 (Omicron). Isolates from these lineages showed increased resistance to neutralizing antibodies and enhanced transmissibility compared with the antecedent SARS-CoV-2 strains.6, 7, 8 Beyond these major VOCs, other variants have emerged. The B.1.621 (Mu) variant was first detected in Colombia in January of 2021 and since then has spread to 51 countries including the United States, Japan, and the United Kingdom. The spike protein of B.1.621 varies at nine positions compared with the original SARS-CoV-2 isolate: T38I, Y144T, Y145S, R346K, E484K, N501Y, D614G, P681H, and D950N.9 The E484K mutation, also found in the B.1.351 (Beta) and P.1 (Gamma) variants, is predicted to reduce serum neutralizing antibody titers against this virus. The R346K mutation, first identified in B.1.621 and more recently in a subset of B.1.1.529 (Omicron), is considered a key mutation that confers resistance to serum antibodies from convalescent and vaccinated individuals10 and class 2 neutralizing monoclonal antibodies (mAbs).11 In serum from individuals immunized with Ad26.COV2.S, mRNA-1273, or BNT162b2 vaccines, difference in neutralization comparing historical and B.1.621 SARS-CoV-2 ranged from 2.1- to 8.7-fold depending on the study population and vaccine.12, 13, 14, 15, 16 A similar difference (4.7- to 12-fold) in neutralization was observed in convalescent sera from previously infected individuals.13 , 14 However, the impact of the spike mutations in B.1.621 on the protective efficacy of vaccines in vivo remains unknown. Here, we evaluated the immunogenicity and efficacy of two vaccines, mRNA-1273 (Moderna) and Ad26.COV2.S (Johnson & Johnson), against the B.1.621 variant of SARS-CoV-2. We show that immunity induced by mRNA-1273 and Ad26.COV2.S protects mice and hamsters from challenge with the B.1.621 variant of SARS-CoV-2.
Results
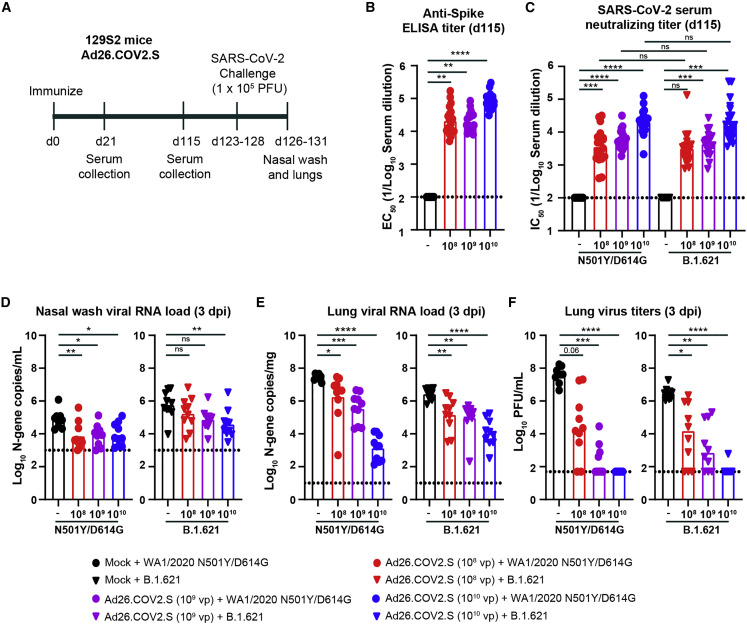
Immunogenicity and protection by Ad26.COV2.S vaccine against B.1.621 challenge in 129S2 mice
Groups of 5- to 6-week-old male 129S2 mice were immunized once with 108, 109, or 1010 virus particles of fresh or freeze-thawed Ad26.COV2.S vaccine (Figure 1A). Serum was collected 21 and 115 days later, and antibody responses were evaluated by ELISA for spike (S)-specific antibody responses. As expected, serum from control mice that received PBS did not bind to the S protein by ELISA (Figure 1B). In comparison, serum collected from mice 115 days after immunization with a 108, 109, or 1010 dose of Ad26.COV2.S contained anti-S-specific antibodies with geometric mean titers (GMTs) of 1:21,087, 1:19,870, and 1:80,599, respectively (Figure 1B). No differences in GMTs were observed between animals immunized with the fresh or freeze-thawed Ad26.COV2.S vaccine (Figures S1A and S1B). The serum anti-S antibody response was higher in animals that received the 1010 dose (p < 0.001; Figure 1B) than those immunized with the 108 or 109 dose of vaccine. A comparison of the anti-S response between 21 and 115 days after immunization revealed an approximately 3-fold increase in anti-S response over time (Figure S1C). Serum samples also were tested for neutralization of SARS-CoV-2 by focus reduction neutralization test (FRNT). Whereas serum from the control animals did not neutralize WA1/2020 N501Y/D614G or B.1.621 (Figure 1C), serum from mice immunized with the 108, 109, or 1010 dose of Ad26.COV2.S did (WA1/2020 N501Y/D614G: GMTs of 1:3,602, 1:6,071, and 1:20,592, respectively, and B.1.621: GMTs of 1:3,172, 1:4,460, and 1:20,520, respectively) (Figures 1C and S2A). No significant difference (p > 0.5) in serum neutralization was observed between the WA1/2020 N501Y/D614G and B.1.621 strains. Also, no significant difference (p > 0.05) was observed in a pairwise comparison of serum neutralization titers against WA1/2020 N501Y/D614G and B.1.621 (Figure S1D).
Figure 1.
Ad26.COV2.S vaccine protects 129S2 mice against challenge with WA1/2020 N501Y/D614G and B.1.621
(A) Experimental setup.
(B) Serum anti-S protein antibody response (EC50) in control mice (black symbols) and mice immunized with 108 (red symbols), 109 (purple symbols), or 1010 (blue symbols) of Ad26.COV2.S (∗∗∗∗p < 0.0001, ∗∗∗p < 0.001 by non-parametric one-way ANOVA with a Dunn’s post-test).
(C) Serum neutralizing titer (IC50) against WA1/2020 N501Y/D614G (circles) or B.1.621 (triangles) measured by FRNT from 129S2 mice immunized once with 108 (red symbols), 109 (purple symbols), or 1010 (blue symbols) of Ad26.COV2.S (∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ns = not significant by non-parametric one-way ANOVA with a Dunn’s post-test).
(D–F) 129S2 mice were challenged with 105 PFU of the WA1/2020 N501Y/D614G (circles) or B.1.621 (triangles) variant of SARS-CoV-2, and nasal washes (D) and lungs (E and F) were collected for analysis of viral RNA levels by qRT-PCR (D) and infectious virus by plaque assay (E and F) (∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, ns = not significant by one-way ANOVA with a Dunnett’s [D and E] or Dunn’s [F] post-test).
(B–F) Bars indicate the geometric mean values, and dotted lines are the limits of detection (LODs) of the assays. Animals at the limit of detection are arbitrarily assigned this value. These values are combined with those having values above the limit to determine the GMT. The results are from two independent experiments, and each symbol represents an individual animal.
See also Figure S1.
Next, we challenged the Ad26.COV2.S-immunized 129S2 mice with 105 plaque-forming units (PFUs) of WA1/2020 N501Y/D614G, or B.1.621 virus and 3 days later collected nasal washes and the left lung lobe for viral-burden analysis. We used 129S2 mice for these studies because these animals are permissive to infection by some SARS-CoV-2 variants (e.g., B.1.1.7, B.1.1.28, and B.1.351) or mouse-adapted or engineered strains17, 18, 19 that encode an N501Y mutation, which enables engagement of murine ACE2.20 Infection of 129S2 mice with SARS-CoV-2 results in mild to moderate lung infection and clinical disease with subsequent recovery.17 , 19 In the nasal wash of control animals challenged with WA1/2020 N501Y/D614G, we detected ∼105 copies of the N gene transcript/mL (Figure 1D). Immunization with the 108, 109, or 1010 dose of Ad26.COV2.S reduced the viral RNA levels by 10-, 9-, and 8-fold (p < 0.01, 0.05, and 0.05, respectively; Figure 1D). After challenge with B.1.621, we measured ∼106 copies of N gene transcript/mL in the nasal wash, and this was reduced 3-, 8-, and 15-fold (p < 0.01 for 1010 dose) in animals immunized with the 108, 109, or 1010 dose of Ad26.COV2.S, respectively.
Viral RNA levels also were quantified in the left lung lobe at 3 days post infection (dpi). In the control groups challenged with the WA1/2020 N501Y/D614G virus, ∼107 N gene transcript copies/mg and ∼107 PFU/mL were measured in lung homogenates (Figures 1E and 1F). Immunization with the 108, 109, or 1010 dose of Ad26.COV2.S reduced the N gene copy number by 15-, 80-, and 20,000-fold (p < 0.05, 0.001, and 0.0001, respectively) and infectious virus levels by 103-, 105-, and 106-fold (p = 0.06, <0.001, and <0.0001, respectively) (Figures 1E and 1F). A second cohort of Ad26.COV2.S-immunized animals were challenged with the B.1.621 strain of SARS-CoV-2. In the control groups, we detected ∼106 N gene transcript copies/mg and ∼106 PFU/mL of infectious virus of lung homogenate (Figures 1E and 1F). Immunization with the 108, 109, or 1010 dose of Ad26.COV2.S reduced the viral RNA levels by 18-, 30-, and 250-fold (p < 0.01, 0.01, and 0.0001, respectively) and infectious virus burden by 350-, 2,000-, and 50,000-fold (p < 0.05, 0.01, and 0.0001, respectively) (Figures 1E and 1F).
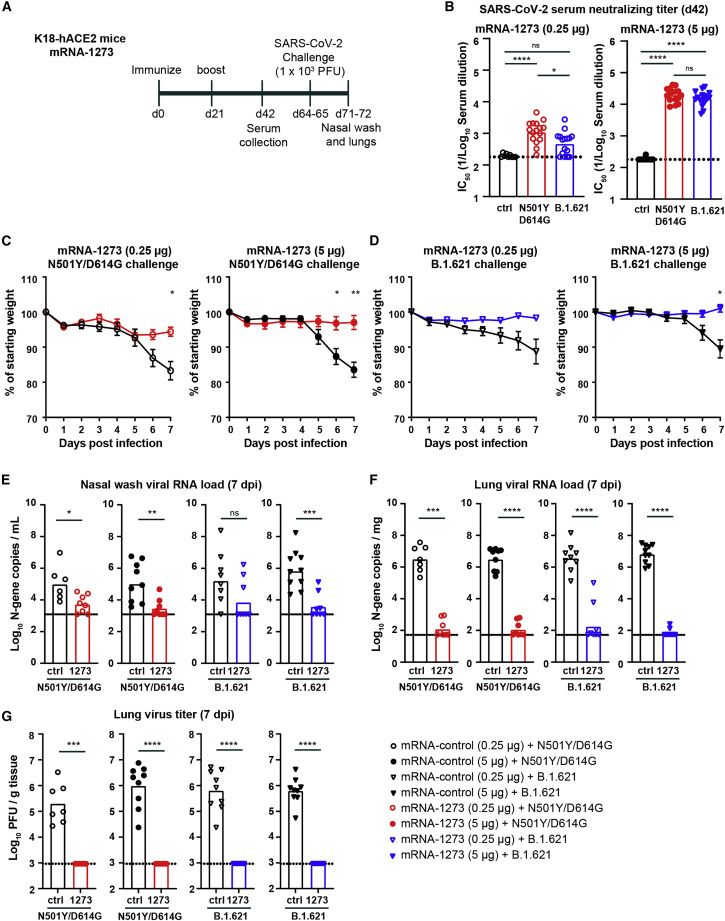
Immunogenicity and protection by mRNA-1273 against B.1.621 challenge in K18-hACE2 mice
Next, we evaluated the efficacy of pre-clinical non-good manufacturing practice (GMP) lots of the Moderna mRNA-1273 vaccine encoding a sequence-optimized 2 proline-stabilized S protein of Wuhan-1 for protection against B.1.621 in K18-hACE2 transgenic mice. Groups of 7- to 8-week-old female mice were immunized and boosted via an intramuscular route with a low (0.25 μg) or high (5 μg) dose of the mRNA-1273 or a control mRNA vaccine (mRNA-control; Figure 2A). Serum was collected 21 days after the second immunization, and inhibitory antibody responses were evaluated by FRNT against WA1/2020 N501Y/D614G and B.1.621. As expected, serum from mice immunized with the control vaccine did not inhibit virus infection (Figure 2B). In contrast, serum from mice immunized with a 0.25 μg dose of mRNA-1273 neutralized the infectious virus with GMTs of 1:1,125 and 1:434 for WA1/2020 N501Y/D614G and B.1.621 viruses, respectively. Serum from mice immunized with 5 μg mRNA-1273 showed greater neutralizing activity against WA1/2020 N501Y/D614G and B.1.621, with GMTs of 1:19,751 and 1:15,130, respectively. A statistical difference in serum GMTs was observed between WA1/2020 N501Y/D614G and B.1.621 for the low dose (p < 0.05), but not the high dose (p > 0.5), vaccine. However, a pairwise comparison showed a 1.3- to 2.3-fold difference in serum neutralizing antibody titers between WA1/2020 N501Y/D614G and B.1.621 for the high- (p < 0.05) and low- (p < 0.01) dose mRNA-1273-immunized mice (Figure S1E).
Figure 2.
mRNA-1273 protects K18-hACE2 transgenic mice against challenge with N501Y/D614G and B.1.621
(A) Experimental setup.
(B) Serum neutralizing titer (IC50) against WA1/2020 N501Y/D614G (red circles) or B.1.621 (blue triangles) from K18-hACE2 mice immunized twice with 0.25 (open symbols) or 5 μg (closed symbols) of mRNA-1273 or mRNA control vaccine (∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 by non-parametric one-way ANOVA with a Dunn’s post-test).
(C and D) Mean ± SEM of weight loss/gain in SARS-CoV-2-challenged mice that were immunized twice with 0.25 (open symbols, left in C and D) or 5 μg (closed symbols, right in C and D) of mRNA-1273 or mRNA control vaccine (∗∗p < 0.01, ∗p < 0.05, ns = not significant by two-way ANOVA).
(E–G) K18-hACE2 mice were challenged with 103 PFU of the WA1/2020 N501Y/D614G (red circles) or B.1.621 (blue triangles), and nasal washes (E) and lungs (F and G) were collected for analysis of viral RNA levels by qRT-PCR (E) and infectious virus by plaque assay (F and G) (∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, ns = not significant by Mann-Whitney test).
(B and E–G) Bars indicate the geometric mean values, and dotted lines are the LODs of the assays. Animals at the limit of detection are arbitrarily assigned this value. These values are combined with those having values above the limit to determine the GMT. The results are from two independent experiments, and each symbol represents an individual animal.
See also Figures S1 and S2.
We next evaluated the protective effect of the mRNA-1273 vaccine in K18-hACE2 mice. Mice immunized with mRNA-1273 or control mRNA vaccine were challenged via an intranasal route with 103 PFU of WA1/2020 N501Y/D614G or B.1.621 virus, and body weight was recorded for 7 days before a nasal wash and the left lung lobe were collected for viral-burden analysis. In the control groups challenged with the WA1/2020 N501Y/D614G virus, substantial weight loss was observed at 6 and 7 dpi. Immunization with 0.25 or 5 μg of mRNA-1273 significantly prevented weight loss (p < 0.05 and 0.01, respectively; Figure 2C). Similar protection against weight loss after challenge with B.1.621 was observed in the groups immunized with 0.25 (p = 0.19) or 5 μg (p < 0.05) of mRNA-1273 (Figure 2D).
We quantified the amount of viral RNA in the nasal wash at 7 dpi. In control-vaccinated animals challenged with WA1/2020 N501Y/D614G, we detected ∼105 copies of the SARS-CoV-2 N gene transcript/mL in the nasal wash (Figure 2E). Immunization with mRNA-1273 reduced the levels of WA1/2020 N501Y/D614G RNA by 20- to 45-fold for the 0.25 (p < 0.05) and 5 μg (p < 0.01) doses, respectively (Figure 2E). In the groups vaccinated with the control mRNA and challenged with the B.1.621 virus, we detected 105–106 copies of the N gene transcript/mL in the nasal wash (Figure 2E). Immunization with 0.25 (p = 0.1) or 5 μg (p < 0.001) of mRNA-1273 reduced the N gene copy number by ∼25- and 200-fold, respectively (Figure 2E).
The amount of virus in lung homogenates was also quantified. In the control group challenged with WA1/2020 N501Y/D614G, ∼106 copies of the N transcript/mg and 105–106 PFU/g tissue were detected (Figures 2F and 2G). Immunization with 0.25 μg of mRNA-1273 resulted in markedly reduced (∼20,000-fold, p < 0.0001) levels of viral RNA and no detectable infectious virus titer in the lung (Figures 2F and 2G). Similar results were seen in mice immunized with 5 μg of mRNA-1273 (Figures 2F and 2G). A second group of immunized animals was challenged with B.1.621. In the control-mRNA-vaccinated groups, we detected high levels of viral RNA (N gene, >106 copies/mg) and infectious virus (6 × 105 PFU/g) in lung homogenates (Figures 2F and 2G). Immunization with either dose of the mRNA-1273 vaccine significantly reduced levels of B.1.621 viral RNA and infectious virus in the lung (∼20,000-fold, p < 0.0001; Figures 2F and 2G).
K18-hACE2 mice vaccinated with 0.25 or 5 μg of mRNA-1273 also had markedly reduced, if not absent, lung pathology at 7 dpi compared with the control mRNA-vaccinated and challenged animals (Figure S2). Lung sections obtained from mice immunized with the control mRNA vaccine and challenged with B.1.621 showed severe pneumonia characterized by immune cell infiltration, alveolar space consolidation, vascular congestion, and interstitial edema. In comparison, mice immunized with either dose of mRNA-1273 did not show evidence of lung pathology. Overall, these data indicate that the mRNA-1273 vaccine protects against the B.1.621 variant of SARS-CoV-2 in K18-hACE2 mice.
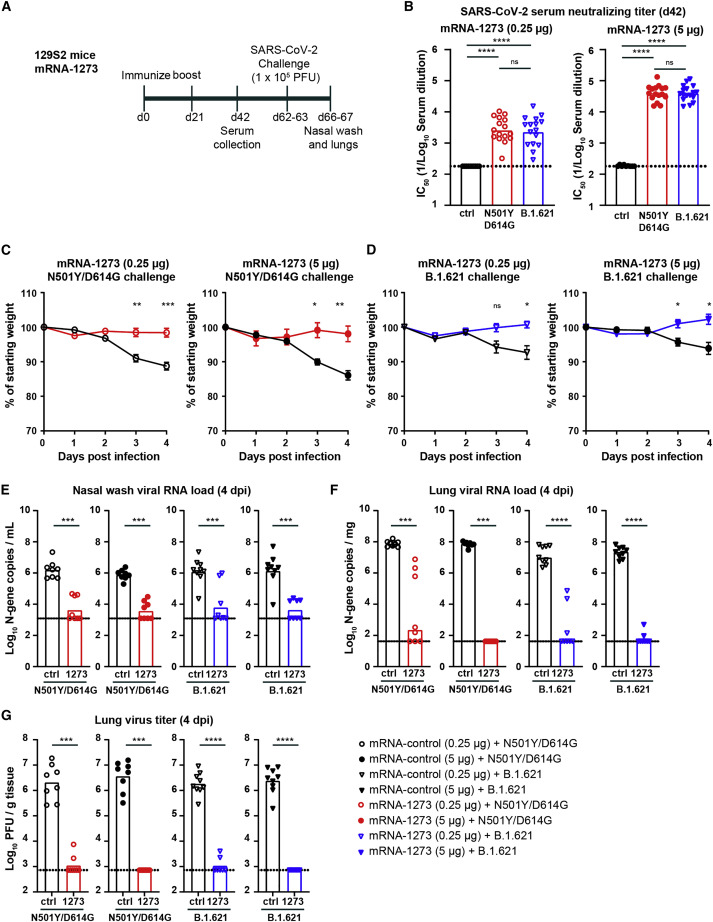
Immunogenicity and protection by mRNA-1273 vaccine against B.1.621 challenge in 129S2 mice
To corroborate our findings, we also tested the mRNA-1273 vaccine in immunocompetent 129S2 mice. Groups of 7- to 8-week-old female mice were immunized and boosted via an intramuscular route with 0.25 or 5 μg of mRNA-1273 or control mRNA vaccine (Figure 3A). Serum was collected 21 days after the second immunization, and antibody responses were evaluated by FRNT. As expected, serum from 129S2 mice immunized with the control mRNA vaccine did not neutralize virus infection (Figure 3B). In contrast, serum from mice immunized with 5 μg of mRNA-1273 robustly neutralized infection with GMTs of 1:40,066 and 1:38,675 for WA1/2020 N501Y/D614G and B.1.621, respectively. Sera from mice immunized with the 0.25 μg dose of mRNA-1273 inhibited infection of WA1/2020 N501Y/D614G and B.1.621 to a lesser extent, with GMTs of 1:2,665 and 1:2,407, respectively. No significant difference (p > 0.05) in vaccine-induced GMTs were observed between the WA1/2020 N501Y/D614G and B.1.621 viruses (Figure 3B). Also, no significant difference (p > 0.5) was observed in a pairwise comparison of the neutralization titers against WA1/2020 N501Y/D614G and B.1.621 in serum from 129S2 mice immunized with high- and low-dose mRNA-1273 vaccine (Figure S1F).
Figure 3.
mRNA-1273 protects 129S2 mice against challenge with WA1/2020 N501Y/D614G and B.1.621
(A) Experimental setup.
(B) Serum neutralizing titer (IC50) against WA1/2020 N501Y/D614G (red circles) or B.1.621 (blue triangles) from 129S2 mice immunized twice with 0.25 (open symbols) or 5 μg (closed symbols) of mRNA-1273 or mRNA control vaccine (∗∗∗∗p < 0.0001, ∗∗∗p < 0.001 by non-parametric one-way ANOVA with a Dunn’s post-test).
(C and D) Mean ± SEM of weight loss/gain in SARS-CoV-2-challenged mice that were immunized twice with 0.25 (open symbols, left in C and D) or 5 μg (closed symbols, right in C and D) of mRNA-1273 or mRNA control vaccine (∗∗p < 0.01, ∗p < 0.05, ns = not significant by two-way ANOVA).
(E–G) 129S2 mice were challenged with 105 PFU of WA1/2020 N501Y/D614G (red symbols) or B.1.621 (blue symbols), and nasal washes (E) and lungs (F and G) were evaluated for viral RNA levels by qRT-PCR (E) and infectious virus by plaque assay (F and G) (∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, ns = not significant by Mann-Whitney test).
(B and E–G) Bars indicate the geometric mean values, and dotted lines are the LODs of the assays. Animals at the limit of detection are arbitrarily assigned this value. These values are combined with those having values above the limit to determine the GMT. The results are from two independent experiments, and each symbol represents an individual animal.
See also Figures S1 and S3.
We next challenged immunized 129S2 mice with 105 PFU of WA1/2020 N501Y/D614G or B.1.621 via an intranasal route. Weights were recorded for 4 days before the animals were necropsied, and nasal washes and the left lung lobe were collected for viral-burden analysis. Control animals challenged with the WA1/2020 N501Y/D614G virus lost ∼10% of their starting body weight by 4 dpi (Figure 3C). Immunization with 0.25 and 5 μg of mRNA-1273 reduced the weight loss (p < 0.001 and 0.01, respectively; Figure 3C). Inoculation of control-vaccinated 129S2 mice with B.1.621 resulted in ∼7% weight loss at 4 dpi (Figure 3D), and immunization with 0.25 or 5 μg of mRNA-1273 prevented this weight loss (p < 0.05).
In animals vaccinated with the control mRNA and challenged with the WA1/2020 N501Y/D614G virus, we detected ∼106 copies/mL of N gene transcript (Figure 3E) in the nasal wash at 4 dpi. Immunization with 0.25 or 5 μg of mRNA-1273 reduced WA1/2020 N501Y/D614G viral RNA levels in the nasal wash (∼200-fold, p < 0.001). In mice vaccinated with the control mRNA vaccine and challenged with B.1.621, we detected ∼106 N gene transcript copies/mL in the nasal wash (Figure 3E). Immunization with 0.25 or 5 μg of mRNA-1273 reduced the viral RNA levels substantially (∼200-fold, p < 0.001; Figure 3E). Viral burden also was measured in lung homogenates of these same animals. In the control mRNA group challenged with the WA1/2020 N501Y/D614G virus, we detected ∼108 copies/mg of N gene transcript (Figure 3F) and >106 PFU/g of infectious virus (Figure 3G) in lung homogenates. Immunization with 0.25 μg of mRNA-1273 effectively reduced the viral RNA (106-fold, p < 0.001; Figure 3F) and infectious virus levels (1,000-fold, p < 0.001; Figure 3G), although some animals showed breakthrough infection. Immunization with a 5 μg dose of mRNA-1273 also reduced the levels of viral RNA (106-fold, p < 0.001) and infectious virus (1,000-fold, p < 0.001; Figures 3F and 3G). Importantly, no breakthrough infection was detected. A second group of immunized animals were challenged with the B.1.621 virus. In the control-mRNA-vaccinated groups, we detected in the lung homogenates ∼107 copies/mg of viral RNA transcript (Figure 3F) and ∼106 PFU/g of infectious virus (Figure 3E). Immunization with 0.25 or 5 μg doses of mRNA-1273 significantly reduced B.1.621 viral RNA (105-fold, p < 0.0001) and infectious virus (1,000-fold, p < 0.0001) levels in the lung (Figures 3F and 3G) to a similar degree after WA1/2020 N501Y/D614G challenge.
The reduction in viral burden in the lungs of mice immunized with 0.25 or 5 μg of mRNA-1273 and challenged with WA1/2020 N501Y/D614G or B.1.621 corresponded with an absence of lung pathology at 4 dpi (Figure S3). Lung sections from mice immunized with a control mRNA and challenged with WA1/2020 N501Y/D614G or B.1.621 showed evidence of immune cell infiltration and mild tissue injury characterized by vascular congestion and interstitial edema. Immunization with 0.25 and 5 μg of mRNA-1273 prevented lung pathology after challenge with both the WA1/2020 N501Y/D614G and B.1.621 viruses. Overall, these data indicate that the mRNA-1273 vaccine protects against the B.1.621 variant of SARS-CoV-2 in non-transgenic immunocompetent 129S2 mice.
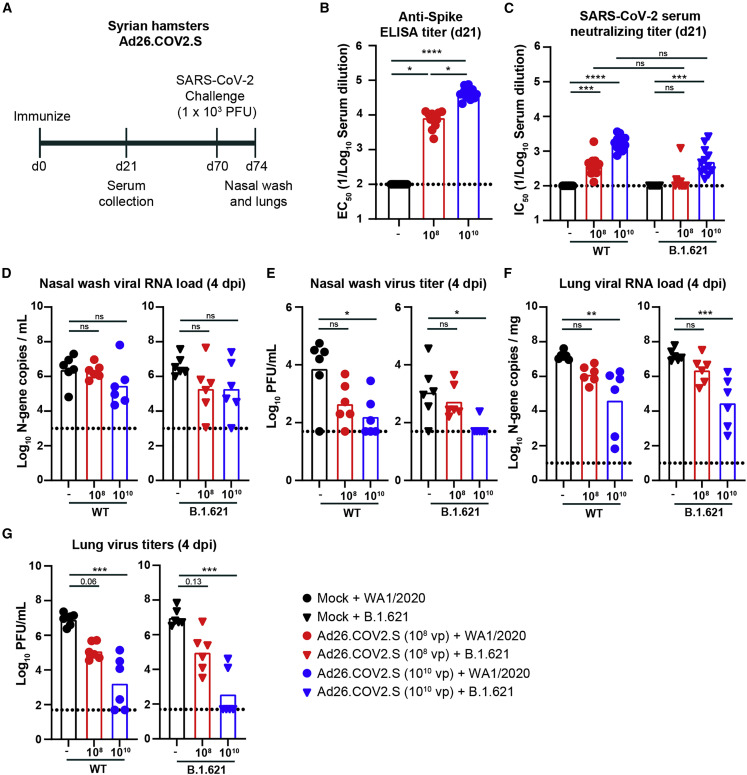
Immunogenicity and protection by Ad26.COV2.S vaccine against B.1.621 challenge in Syrian hamsters
We next evaluated the efficacy of a single dose of the Ad26.COV2.S vaccine in Syrian hamsters. Syrian hamsters are naturally susceptible to SARS-CoV-2 and considered an excellent model for COVID-19.19 , 21, 22, 23, 24 Groups of 5- to 6-week-old male hamsters were immunized once with the 108 or 1010 dose of the Ad26.COV2.S vaccine (Figure 4A). Serum was collected 21 days later, and antibody responses were evaluated by ELISA and FRNT. As expected, serum from control hamsters that received PBS did not bind the viral S protein (Figure 4B). However, sera collected from hamsters immunized with the 108 or 1010 dose of Ad26.COV2.S contained anti-S antibodies with GMTs of 1:7,506, and 1:40,913, respectively (Figure 4B). The serum anti-S antibody response was higher in animals receiving the 1010 vaccine dose (p < 0.05; Figure 4B). Serum samples were tested for neutralization of SARS-CoV-2 by FRNT. Serum from hamsters immunized with the 108 or 1010 dose of Ad26.COV2.S neutralized WA1/2020, with GMTs of 1:381 and 1:1,692, respectively (Figure 4C), and B.1.621, with GMTs of 1:136 and 1:501, respectively (Figure 4C). No difference in GMTs was observed with respect to neutralization of WA1/2020 and B.1.621 variants after immunization with the 108 (p = 0.06) or 1010 (p = 0.6) dose of Ad26.COV2.S (Figure 4C). A pairwise comparison found a ∼3-fold reduction in serum neutralizing antibody titers between WA1/2020 and B.1.621 for the high- (p < 0.0001) and low- (p < 0.001) dose Ad26-COV2.S-immunized hamsters (Figure S1G).
Figure 4.
Ad26.COV2.S vaccine protects Syrian hamsters against challenge with WA1/2020 and B.1.621
(A) Experimental setup.
(B) Serum anti-S protein antibody response (EC50) in control hamsters (black symbols) and hamsters immunized with 108 (red symbols) or 1010 (blue symbols) of Ad26.COV2.S (∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ns = not significant by non-parametric one-way ANOVA with a Dunn’s post-test).
(C) Serum neutralizing titer (IC50) against WA1/2020 (circles) or B.1.621 (triangles) from hamsters immunized once with 108 (red symbols) or 1010 (blue symbols) of Ad26.COV2.S (∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ns = not significant by non-parametric one-way ANOVA with a Dunn’s post-test).
(D–G) Syrian hamsters were challenged with 103 PFU of the WA1/2020 (circles) or B.1.621 (triangles), and nasal washes (D and E) and lungs (F and G) were evaluated for viral RNA levels by qRT-PCR (D and F) and infectious virus by plaque assay (E and G) (∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, ns = not significant by one-way ANOVA with a Dunnett’s [D and F] or Dunn’s [E and G] post-test).
(B–G) Bars indicate the geometric mean values, and dotted lines are the LODs of the assays. Animals at the limit of detection are arbitrarily assigned this value. These values are combined with those having values above the limit to determine the GMT. The results are from one experiment, and each symbol represents an individual animal.
See also Figures S1 and S4.
Seventy days after immunization, the hamsters were challenged via an intranasal route with 103 PFU of WA1/2020 or B.1.621. Four days later, the animals were necropsied, and a nasal wash and the left lung lobe were collected for viral-burden analysis. In the nasal wash of control animals challenged with the WA1/2020 virus, we detected ∼106 copies of the N gene transcript/mL of nasal wash (Figure 4D) and ∼104 PFU/mL of infectious virus (Figure 4E). Immunization with the 108 or 1010 dose of Ad26.COV2.S did not reduce the viral RNA levels in the nasal wash (Figure 4D), although the infectious virus titer was decreased by 16- and 46-fold (p > 0.05 and <0.05, respectively; Figure 4E). Upon challenge of hamsters with B.1.621, we detected ∼106 N gene copies/mL (Figure 4D) and ∼103 PFU/mL of infectious virus (Figure 4E) in the nasal wash. Immunization with the 108 or 1010 dose of Ad26.COV.2 reduced the N gene copy number 19-fold (p > 0.05; Figure 4D). In comparison, the infectious virus titer was reduced by 2- and 17-fold (p > 0.05 and < 0.05 for the 108 and 1010 doses, respectively; Figure 4E)
We also measured the viral burden in lung homogenates of hamsters. In the control group challenged with WA1/2020, the N gene copy number was ∼107 copies/mg (Figure 4F) and the infectious virus levels were ∼107 PFU/mL (Figure 4G). Immunization with the 108 or 1010 dose of Ad26.COV2.S reduced the viral RNA (11- and 360-fold, p > 0.05 and < 0.01) and infectious virus (66- and 4,800-fold, p = 0.06 and < 0.001) levels (Figures 4F and 4G). After B.1.621 challenge of the control group, we detected ∼107 N gene copies/mg of tissue (Figure 4F) and ∼107 PFU/mL of infectious virus in the lung (Figure 4G). Immunization with the 108 or 1010 dose of Ad26.COV2.S reduced the viral RNA (7- and 540-fold, p > 0.05 and < 0.05) and infectious virus (100- and 25,500-fold, p = 0.13 and < 0.001) levels (Figures 4F and 4G). A comparison between neutralization titers (half maximal effective concentration [EC50] values) and infectious virus titers in the lungs in animals challenged with WA1/2020 revealed a significant correlation (R2 = 0.67, p = 0.0012; Figure S4). In contrast, this association was not clearly observed in hamsters challenged with B.1.621 (R2 = 0.24, p = 0.11; Figure S4), suggesting that other immune factors (non-neutralizing antibodies, T cells, or anamnestic B cell responses) likely contribute more to vaccine-induced protection against variant SARS-CoV-2 viruses.
Discussion
In this study, we evaluated the efficacy of two vaccines, mRNA-1273 and Ad26.COV2.S, against the B.1.621 variant of SARS-CoV-2 in three pre-clinical models: 129S2-immuno-competed mice, K18-hACE2 transgenic mice, and Syrian hamsters. The mRNA-1273 vaccine induced high levels of neutralizing antibodies against WA1/2020 N501Y/D614G and B.1.621 viruses, and this response was associated with robust protection from an intranasal challenge. Immunization of 129S2 mice or Syrian hamsters with different doses of Ad26.COV2.S induced moderate to high serum neutralizing antibody responses against the B.1.621 virus. However, only the high dose (1010 virus particles) Ad26.COV2.S reduced virus titers substantially.
Our studies provide a comparison of the immunogenicity and efficacy of the Ad26.COV2.S and mRNA-1273 vaccines in 129S2 mice. The serum neutralizing antibody titer was similar after one dose of Ad26.COV2.S or two doses of mRNA-1273 in129S2 mice, and this was true for both the high-dose (5 μg versus 1010 virus particles) and the low-dose (0.25 μg versus 108 virus particles) vaccine regimens. Despite the similarity in neutralization titer, the mRNA-1273 vaccine more effectively reduced viral load in the lungs of WA1/2020 N501Y/D614G- or B.1.621-challenged animals than the Ad26.COV2.S vaccine. In mice immunized with a low dose of the mRNA-1273 vaccine, approximately 20% of the animals showed evidence of breakthrough infections after challenge (Figure 3G). In contrast, 75% of the animals that received a low dose of Ad26.COV2.S and were challenged with WA1/2020 N501Y/D614G or B.1.621 showed virus breakthrough despite relatively equivalent levels of serum neutralizing antibody titers at the time of challenge (Figure 1F). One explanation for this difference could that the mRNA vaccine requires two doses, while the Ad26.COV2.S vaccine was only given once. Another possibility is the time between the last immunization and the virus challenge, which is 41 and 115 days for the mRNA-1273- and Ad26.COV2.S-immunized animals, respectively. Differences in the glycosylation pattern or the immunoglobulin G (IgG) subclass of antibodies between anti-S antibodies induced by mRNA-1273 and Ad26.COV2.S induced could contribute to differences in protection, as seen in non-human primates and humans.25 , 26 It is also possible that mRNA-1273 vaccine induced a better anamnestic B or T cell response in 129S2 mice compared with the Ad26.COV2.S vaccine at the time of virus challenge.
The antibody response after vaccination varied between the mouse and hamster models. Immunization of mice with 5 μg of mRNA-1273 or 1010 Ad26.COV2.S induced serum neutralizing antibody responses with a GMT of >10,000 in both 129S2 and K18-hACE2 mice. In contrast, in Syrian hamsters, the GMT against WA1/2020 was ∼10-fold lower than in mice yet still severalfold higher than that observed in humans vaccinated with one dose of Ad26.COV2.S.27, 28, 29, 30 The reason for this difference in vaccine response between mice and hamsters remains unknown. It is possible that the hamster immune response targets different epitopes on the S protein. Alternatively, the S protein contains fewer T cell epitopes for hamsters compared with mouse, although that seems unlikely given the size of the antigen. The Syrian hamster also may be more tolerogenic, perhaps due to its complex microbiome as opposed to the SPF microbiome of 129S2 and K18 TG mice. Mice that received the microbiome from pet stores or from field mice had blunted vaccine responses compared with laboratory-housed mice in pathogen-free facilities.31 It is also possible that the differences in antibody response after immunization are impacted by differential immunological maturity between 7- and 8-week-old mice and 5- to 6-week-old hamsters. Additional studies are required to elucidate the causes for this difference, but in general, the magnitude of the antibody response in Syrian hamsters is more similar to the human antibody response after vaccination.
The B.1.621 (Mu) variant of SARS-CoV-2 has R346K and E484K mutations in the receptor-binding domain of the S protein and is believed to be more resistant to virus neutralization by serum antibodies compared with the historical SARS-CoV-2 virus. In sera from vaccinated or infected individuals, the fold difference in neutralization between the D614G (B.1) variant of SARS-CoV-2 and B.1.621 was between 2- and 12-fold.12, 13, 14, 15, 16 , 32 In Syrian hamsters, we observed a ∼3-fold decrease (p < 0.001) in serum neutralization titers between the WA1/2020 and B.1.621 virus. In K18-hACE2 mice immunized with mRNA-1273, the difference in serum neutralization titers between WA1/2020 N501Y/D614G and B.1.621 was 1.3- to 2.3-fold. In contrast, no difference in neutralization titers between WA1/2020 N501Y/D614G and B.1.621 was observed in 129S2 mice immunized with mRNA-1273 or Ad26.COV2.S, suggesting mouse-strain and species-specific differences in the antibody response. Another reason may be the N501Y mutation in WA1/2020 N501Y/D614G, which has been shown to reduce the neutralization titer of serum and certain mAbs compared with WA1/2020 and WA1/2020 D614G, respectively.6 , 33 We used the WA1/2020 N501Y/D614G virus because the N501Y mutation was required for virus infection in immunocompetent 129S2 mice lacking hACE2 expression. Finally, it is possible that the insertion of a threonine at positions 144–145 in our particular B.1.621 isolate reduced the resistance to serum neutralizing antibodies.
We observed no difference in efficacy of the mRNA and adenoviral-vectored vaccine to protect against B.1.621 in three different animal models. Near full protection, defined by undetectable levels of SARS-CoV-2 viral RNA and infectious virus, plus the absence of immunopathology in the vaccinated animals, was observed against both B.1.621 and the control WA1/2020 virus in mice immunized with mRNA-1273. While immunization with lower doses of Ad26.COV2.S offered only partial protection, we did not observe a difference in virus titer or frequency of breakthrough infection between B.1.621 and control virus. This suggests that a 2- to 3-fold reduction in serum neutralization titers has limited impact on mRNA and adenoviral vaccine protection against the variant B.1.621 virus.
Limitations of the study
We note several limitations of our study. (1) We did not evaluate the effects of the vaccine on the transmission of SARS-CoV-2 in Syrian hamsters, which may be an important measure of vaccine protection. (2) We used lower doses of vaccine to mimic suboptimal and possibly waning immunity. Studies that directly compare the quality of a waning immune response with that of a low-dose-vaccine-induced immune response are needed. (3) The challenge dose of SARS-CoV-2 used in our hamster model (103 PFU) is several orders of magnitude higher that the minimal infectious dose (5 PFU).34 While this creates a very robust virus challenge model, it could underestimate the protective effects of vaccines. (4) We did not establish correlates of immune protection. We noted that lower serum antibody neutralization titers were associated with high viral loads and infectious virus titers in the Ad26.COV2.S-immunized animals, as well as breakthrough infections in some of the mRNA-1273 vaccinated mice, albeit this did not explain all breakthrough infections. A more detailed analysis of T cell and non-neutralizing antibody responses coupled with even lower vaccine doses may be needed to fully establish a correlate of protection against breakthrough infection. (5) We did not evaluate each vaccine in male and female mice and hamsters. Due to the number of variables (vaccines and doses) and viruses (WA1/2020 and B.1.621 variants), testing male and female animals in each experiment was impractical. Instead, we opted to test different sexes in separate cohorts for each rodent model to address whether sex impacts the efficacy of the vaccine against the B.1.621 variant.
Overall, our studies demonstrate that the Moderna mRNA-1273 and Johnson & Johnson Ad26.COV2.S vaccines authorized for emergency use are immunogenic in mice and Syrian hamsters and protect against the B.1.621 (Mu) variant of SARS-CoV-2 without substantial loss of potency.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-mouse IgG | Southern Biotec | Cat #1030-05; RRID: AB_2619742 |
| anti-hamster-IgG(H+L)-HRP | Southern Biotech | Cat#6061-05; RRID: AB_2796135 |
| SARS2-2 | Diamond Laboratory | N/A |
| SARS2-71 | Diamond Laboratory | N/A |
| SARS2-11 | Diamond Laboratory | N/A |
| SARS2-16 | Diamond Laboratory | N/A |
| SARS2-31 | Diamond Laboratory | N/A |
| SARS2-38 | Diamond Laboratory | N/A |
| SARS2-57 | Diamond Laboratory | N/A |
| anti-mouse IgG | Sigma | Cat # A8924; RRID: AB_258426 |
| Bacterial and virus strains | ||
| SARS-CoV-2 (strain 2019 n-CoV/USA_WA1/2020) | CDC/BEI Resources | Cat#NR52281 |
| SARS-CoV-2 (strain hCoV-19/USA/WI-UW-4340/2021) | This paper (Kawoaka Laboratory) | N/A |
| SARS-CoV-2 N501Y/D614G | Pei-Yong Shi Laboratory | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant Spike protein of SARS-CoV-2 | Hassan et al35 | N/A |
| TMB substrate | Vector laboratories | Cat#SK4400 |
| Critical commercial assays | ||
| RNA isolation kit | Omega Bio-Tek | Cat#R6834-01 |
| TaqMan™ RNA-to-CT 1-Step Kit | Thermo Scientific | Cat#4352042 |
| MagMAX MirVana Total RNA isolation kit | Applied Biosystems | A27828 |
| Experimental models: Cell lines | ||
| Vero-hTMPRSS2 | Chen et al6 | N/A |
| Vero-hACE2-hTMPRSS2 | Chen et al6 | N/A |
| Experimental models: Organisms/strains | ||
| LVG Golden Syrian Hamster | Charles Rivers Laboratories | Crl:LVG(SYR) |
| K18-hACE2 transgenic mice | Jackson Laboratories | Cat # 34860 |
| 129S2/SvPasCrl | Charles Rivers Laboratories | Cat # 287 |
| Oligonucleotides | ||
| SARS-CoV-2 N F: 5′-GACCCCAAAATCAGCGAAAT-3′ | Integrated DNA technologies | N/A |
| SARS-CoV-2 N R: 5′-TCTGGTTACTGCCAGTTGAATCTG-3′ | Integrated DNA technologies | N/A |
| SARS-CoV-2 N Probe: 5'-/56-FAM/ACCCCGCATTACGTTTGGTGGACC/3IABkFQ/-3′ | Integrated DNA technologies | N/A |
| SARS-CoV-2 N F: 5′-ATGCTGCAATCGTGCTACAA-3′ | Integrated DNA technologies | N/A |
| SARS-CoV-2 N R: 5′-GACTGCCGCCTCTGCTC-3′ | Integrated DNA technologies | N/A |
| SARS-CoV-2 N Probe: 5'-/56-FAM/TCAAGGAACAACATTGCCAA/3IABkFQ/-3′ | Integrated DNA technologies | N/A |
| Software and algorithms | ||
| GraphPad Prism | GraphPad | v9.3 (www.graphpad.com) |
| Nanozoomer Digital Pathology | Hamamatsu | v2 (https://www.hamamatsu.com/us/en/product/type/U12388-01/index.html) |
| BioSpot analyzer | Cellular Technology Limited | N/A |
| AxioImager Z2 system | Zeiss | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Adrianus C.M. Boon (jboon@wustl.edu).
Materials availability
All requests for resources and reagents should be directed to the lead contact. This includes viruses, vaccines, and primer-probe sets. All reagents will be made available on request after completion of a Materials Transfer Agreement.
Data and code availability
All data supporting the findings of this study are available within the paper and are available from the corresponding author upon request. This paper does not include original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Cells and viruses
Vero cells expressing human ACE2 and TMPRSS2 (Vero-hACE2-hTMPRSS2,6 , 36 gift from Adrian Creanga and Barney Graham, NIH) were cultured at 37°C in Dulbecco’s Modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 10 mM HEPES (pH 7.3), 100 U/mL of Penicillin-Streptomycin, and 10 μg/mL of puromycin. Vero cells expressing TMPRSS2 (Vero-hTMPRSS2)36 were cultured at 37°C in Dulbecco’s Modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 10 mM HEPES (pH 7.3), 100 U/mL of Penicillin-Streptomycin, and 5 μg/mL of blasticidin.
SARS-CoV-2 (strain 2019-nCoV/USA-WA1/2020) was obtained from the US Centers for Disease Control (CDC) and propagated on Vero-hTMPRSS2 cells. The B.1.621 variant of SARS-CoV-2 (strain hCoV-19/USA/WI-UW-4340/2021) was obtained from a nasal swab isolate and propagated on Vero-hTMPRSS2 cells. Recombinant SARS-CoV-2 with a N501Y and D614G mutations in the S protein of SARS-CoV-2 has been published previously6 and was propagated on Vero-hTMPRSS2 cells. The virus stocks were subjected to next-generation sequencing, and the S protein sequences were identical to the original isolates. The infectious virus titer was determined by plaque or focus-forming assay on Vero-hACE2-hTMPRSS2 or Vero-hTMPRSS2 cells.
Pre-clinical vaccine mRNA and lipid nanoparticle production process
A sequence-optimized mRNA encoding prefusion-stabilized Wuhan-Hu-1 (mRNA-1273) SARS-CoV-2 S-2P protein was synthesized in vitro using an optimized T7 RNA polymerase-mediated transcription reaction with complete replacement of uridine by N1m-pseudouridine.37 The reaction included a DNA template containing the immunogen open-reading frame flanked by 5′ untranslated region (UTR) and 3′ UTR sequences and was terminated by an encoded polyA tail. After transcription, the cap-1 structure was added to the 5′ end using the vaccinia virus capping enzyme (New England Biolabs) and vaccinia virus 2′-O-methyltransferase (New England Biolabs). The mRNA was purified by oligo-dT affinity purification, buffer exchanged by tangential flow filtration into sodium acetate, pH 5.0, sterile filtered, and kept frozen at −20°C until further use.
The mRNA was encapsulated in a lipid nanoparticle through a modified ethanol-drop nanoprecipitation process described previously.38 Ionizable, structural, helper, and polyethylene glycol lipids were briefly mixed with mRNA in an acetate buffer, pH 5.0, at a ratio of 2.5:1 (lipid:mRNA). The mixture was neutralized with Tris-HCl, pH 7.5, sucrose was added as a cryoprotectant, and the final solution was sterile-filtered. Vials were filled with formulated lipid nanonparticle and stored frozen at −20°C until further use. The vaccine product underwent analytical characterization, which included the determination of particle size and polydispersity, encapsulation, mRNA purity, double-stranded RNA content, osmolality, pH, endotoxin, and bioburden, and the material was deemed acceptable for in vivo study.
Recombinant proteins
Recombinant S, was expressed as previously described.39 Briefly, a mammalian cell codon-optimized nucleotide sequence coding for soluble S (GenBank: MN908947.3, amino acids 1-1,213) modified to remove the polybasic cleavage site (RRAR to A), but introducing two stabilizing mutations (K986P and V987P, wild-type numbering) and a C-terminal thrombin cleavage site, T4 foldon trimerization domain, and a 6xHIS tag were cloned into mammalian expression vector pCAGGS.40 Recombinant S was produced in Expi293F cells (ThermoFisher, Cat #A14527) by transfection with purified DNA using the ExpiFectamine 293 Transfection Kit (ThermoFisher, Cat #A14524). Supernatants from transfected cells were harvested 4 days post-transfection, and recombinant proteins were purified using Ni-NTA agarose (ThermoScientific, Cat #88222), then buffer exchanged into phosphate buffered saline (PBS) and concentrated using Amicon Ultracel centrifugal filters (EMD Millipore, UFC903024).
Mouse experiments
Animal studies were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee at the Washington University School of Medicine (assurance number A3381–01). Seven-to-nine week old male 129S2 (strain: 129S2/SvPasCrl, Cat # 287) or female K18-hACE2 transgenic mice (strain: 2B6.Cg-Tg(K18-ACE2)2Prlmn/J, Cat # 34860) were obtained from Charles River Laboratories and Jackson Laboratories, respectively and housed at Washington University. Animals were housed in groups and fed standard chow diet.
Some of the animals were vaccinated via intramuscular (IM) route with 108, 109, or 1010 viral particles of fresh or freeze-thawed Ad26.COV2.S in 100 μL of phosphate buffered saline (PBS). The freeze-thawed vaccine was stored at -80°C prior to thawing at room temperature. No difference in serum antibody responses were detected between the fresh and freeze-thawed Ad26.COV2.S vaccine (Figures S1A and S1B). Control animals for the adenoviral vaccine received PBS alone. Twenty-one days and 115 days later, serum samples were obtained, and used for ELISA and virus neutralization assays. This later time point was selected to mimic potential waning immunity. Separately, 129S2 mice and K18-hACE2 mice were immunized and boosted with 0.25 or 5 μg of mRNA-1273 or a control mRNA (mRNA-control) vaccine at three week intervals. Twenty-one days after the second immunization, serum was obtained and used for virus neutralization assays. These two mouse models were selected because of their different characteristics: (a) K18-hACE2 mice are among the most susceptible model for SARS-CoV-2 infection and accounts for the SARS-CoV-2 spike protein-hACE2 entry pathways and the impact of vaccine induced antibodies on this interaction; and (b) 129 mice were used because in contrast to BALB/c and C57BL/6 mice, in 6 to 8 week-old animals, they lose weight upon challenge with 105 PFU of SARS-CoV-2.
Following transfer to the enhanced Biosafety level 3 laboratory, the animals were challenged via intranasal route with 103 or 105 PFU of the SARS-CoV-2 N501Y/D614G or B.1.621 variant. This differences in challenge dose reflects the differences in susceptibility to SARS-CoV-2 between the 129S2 (105 PFU) and K18-hACE2 transgenic mice (103 PFU). Animal weights were measured daily for the duration of the experiment. At different time points after challenge, the animals were necropsied, and their lungs were collected for virological and histological analysis. These time points for necropsies and tissue collections were based on optimized ones that were established in the Boon and Diamond laboratories.19 , 21 , 22 , 24 , 41 , 42 These time points coincided with maximal infection, pathological findings, and weight loss. The left lobe was homogenized in 1.0 mL of Dulbecco’s Modified Eagle Medium (DMEM), clarified by centrifugation (1,000 x g for 5 min) and used for viral titer analysis by quantitative RT-PCR (RT-qPCR) using primers and probes targeting the N gene, and by plaque assay. A nasal wash also was collected, by inoculating 1.0 mL of PBS with 0.1% bovine serum albumin into one nostril and collecting the wash from the other nostril (Figures 1 and 4). Alternatively, 0.5 mL of PBS with 0.1% bovine serum albumin was flushed through the nasal cavity after dissecting off the lower jaw (Figures 2 and 3). The nasal wash was clarified by centrifugation (2,000 x g for 10 min) and used for viral titer analysis by RT-qPCR using primers and probes targeting the N gene, and by plaque assay.
Hamster experiments
Animal studies were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee at the Washington University School of Medicine (assurance number A3381–01). Five-week old male hamsters were obtained from Charles River Laboratories and housed at Washington University. Five days after arrival, the animals were immunized via intramuscular injection with 108 of 1010 viral particles of freeze-thawed Ad26.COV2.S in 100 μL of PBS. Control animals received PBS alone. Serum samples were obtained 21 days later, and six weeks after that the animals were transferred to the enhanced Biosafety level 3 laboratory. This time point was selected to assess responses in the setting of possible waning immunity. One day later, the animals were challenged via intranasal route with 103 PFU of WA1/2020 or B.1.621 variant. Syrian hamsters are highly susceptible to SARS-CoV-2 with an estimated infectious dose of 5 virus particles.34 Therefore, we chose a challenge dose that is approximately 100-fold higher to evaluate vaccine efficacy in this model. Animal weights were measured daily for the duration of the experiment. Four days after challenge, the animals were necropsied, and their lungs were collected for virological and histological analysis. The left lobe was homogenized in 1.0 mL of DMEM, clarified by centrifugation (1,000 x g for 5 min) and used for viral titer analysis by quantitative RT-PCR using primers and probes targeting the N gene, and by plaque assay. A nasal wash was also collected, by inoculating 1.0 mL of PBS with 0.1% bovine serum albumin into one nostril and collecting the wash from the other nostril. The nasal wash was clarified by centrifugation (2,000 x g for 10 min) and used for viral titer analysis by quantitative RT-PCR using primers and probes targeting the N gene, and by plaque assay.
Method details
Focus reduction neutralization titer assay (FRNT)
Serial dilutions of serum samples were incubated with 102 focus-forming units (FFU) of different strains of SARS-CoV-2 for 1 h at 37°C. Antibody-virus complexes were added to Vero-hTMPRSS2 cell monolayers in 96-well plates and incubated at 37°C for 1 h. Subsequently, cells were overlaid with 1% (w/v) methylcellulose in Eagle’s Minimal Essential medium (MEM, Thermo Fisher Scientific). Plates were harvested 30 h later by removing overlays and fixed with 4% paraformaldehyde (PFA) in PBS for 20 min at room temperature. Plates were washed and sequentially incubated with an oligoclonal pool of SARS2-2, SARS2-11, SARS2-16, SARS2-31, SARS2-38, SARS2-57, and SARS2-71 43 anti-S protein antibodies and HRP-conjugated goat anti-mouse IgG (Sigma Cat # A8924) in PBS supplemented with 0.1% saponin and 0.1% bovine serum albumin. SARS-CoV-2-infected cell foci were visualized using TrueBlue peroxidase substrate (KPL) and quantitated on an ImmunoSpot microanalyzer (Cellular Technologies).
Virus titration assays
Plaque assays were performed on Vero-hACE2-hTRMPSS2 cells in 24-well plates. Lung tissue homogenates or nasal washes were diluted serially by 10-fold, starting at 1:10, in cell infection medium (DMEM + 2% FBS + 100 U/mL of penicillin-streptomycin). Two hundred and fifty microliters of the diluted virus were added to a single well per dilution per sample. After 1 h at 37°C, the inoculum was aspirated, the cells were washed with PBS, and a 1% methylcellulose overlay in MEM supplemented with 2% FBS was added. Seventy-two hours after virus inoculation, the cells were fixed with 4% formalin, and the monolayer was stained with crystal violet (0.5% w/v in 25% methanol in water) for 1 h at 20°C. The number of plaques were counted and used to calculate the plaque forming units/mL (PFU/mL).
To quantify viral load in lung tissue homogenates and nasal washes, RNA was extracted from 100 μL samples using E.Z.N.A.® Total RNA Kit I (Omega) and eluted with 50 μL of water. Four microliters RNA was used for real-time RT-qPCR to detect and quantify N gene of SARS-CoV-2 using TaqMan™ RNA-to-CT 1-Step Kit (Thermo Fisher Scientific) as described44 using the following primers and probes: Forward: GACCCCAAAATCAGCGAAAT; Reverse: TCTGGTTACTGCCAGTTGAATCTG; Probe: ACCCCGCATTACGTTTGGTGGACC; 5′Dye/3′Quencher: 6-FAM/ZEN/IBFQ. Viral RNA was expressed as N gene copy numbers per mg for lung tissue homogenates or mL for nasal swabs and nasal washes, based on a standard included in the assay, which was created via in vitro transcription of a synthetic DNA molecule containing the target region of the N gene.
Histology
The lungs from SARS-CoV-2 infected and control mice were inflated and fixed in 10% formalin for seven days. Lungs were embedded in paraffin and sectioned before hematoxylin and eosin staining. Lung slides were scanned using the Hamamatsu NanoZoomer slide scanning system and head sections were imaged using the Zeiss AxioImager Z2 system.
ELISA
Ninety-six-well microtiter plates (Nunc MaxiSorp; ThermoFisher Scientific) were coated with 100 μL of recombinant SARS-CoV-2 S protein (Wuhan strain) at a concentration of 1 μg/mL in PBS (Gibco) at 4°C overnight; negative control wells were coated with 1 μg/mL of BSA (Sigma). Plates were blocked for 1.5 h at room temperature with 280 μL of blocking solution (PBS supplemented with 0.05% Tween-20 (Sigma) and 10% FBS (Corning)). Serum from mice and hamsters were diluted serially in blocking solution, starting at 1:100 dilution and incubated for 1.5 h at room temperature. The plates were washed three times with T-PBS (1X PBS supplemented with 0.05% Tween-20), and 100 μL of goat anti-mouse IgG (Southern Biotech Cat #1030-05) diluted 1:2,000 in blocking solution or 100 μL of HRP-conjugated anti-hamster IgG(H+L) antibody (Southern Biotech Cat. #6061-05) diluted 1:500 in blocking solution, was added to all wells and incubated for 1 h at room temperature. Plates were washed 3 times with T-PBS and 3 times with 1X PBS, and 100 μL of 1-step Ultra TMB-ELISA substrate solution (Thermo Fisher Scientific) was added to all wells. The reaction was stopped after 5 min using 100 μL of 1M HCl, and the plates were analyzed at a wavelength of 490 nm using a microtiter plate reader (BioTek).
Quantification and statistical analysis
Statistical significance was assigned when P values were < 0.05 using GraphPad Prism version 9.3. Tests, number of animals, median and geometric mean values, and statistical comparison groups are indicated in the Figure legends. Analysis of weight change was determined by two-way ANOVA. Changes in infectious virus titer, viral RNA levels, or serum antibody responses were compared to unvaccinated or mRNA-control immunized animals and were analyzed by one-way ANOVA with a multiple comparisons correction, unpaired t-test, or Mann-Whitney test, dependent on the number of comparison and the distribution of the data.
Acknowledgments
We thank Florian Krammer and Ali Ellebedy for the plasmid and recombinant S protein, respectively. This study was supported by the NIH (R01 AI157155 and U01 AI151810), NIAID Centers of Excellence for Influenza Research and Response [CEIRR] contracts 75N93021C00014 and 75N93021C00016, and the Collaborative Influenza Vaccine Innovation Centers [CIVIC] contract 75N93019C00051. It was also supported, in part, by the National Institutes of Allergy and Infectious Diseases Center for Research on Influenza Pathogenesis (HHSN272201400008C) and the Japan Program for Infectious Diseases Research and Infrastructure (JP21wm0125002) from the Japan Agency for Medical Research and Development (AMED).
Author contributions
T.L.B., B.Y., B.W., C.-Y.L., L.A.V., G.B., and T.L.D. performed mouse and hamster experiments. T.L.B., B.Y., and T.L.D. quantified virus titers in collected tissues. K.S., A.J., and B.W. determined viral load by real-time quantitative RT-PCR. Y.K. and P.J.H. isolated, expanded, and sequenced the B.1.621 virus. T.L.D. and L.A.V. performed the virus neutralization assays. M.S.D., A.C.M.B., T.L.D., L.B.T., and B.Y. analyzed the data. A.C.M.B., B.Y., and M.S.D. performed the statistical analysis. A.C.M.B. and M.S.D. had unrestricted access to all data. A.C.M.B. and M.S.D. wrote the first draft of the manuscript, and all authors reviewed and edited the final version. All authors agreed to submit the manuscript, read and approved the final draft, and take full responsibility of its content.
Declaration of interests
The A.C.M.B. laboratory has received unrelated funding support in sponsored research agreements from AI Therapeutics, GreenLight Biosciences, Inc., and Nano Targeting & Therapy Biopharma, Inc. The A.C.M.B. laboratory has received funding support from AbbVie, Inc., for the commercial development of SARS-CoV-2 mAbs. M.S.D. is a consultant for Inbios, Vir Biotechnology, Senda Biosciences, and Carnival Corporation and is on the Scientific Advisory Boards of Moderna and Immunome. The M.S.D. laboratory has received unrelated funding support in sponsored research agreements from Vir Biotechnology, Kaleido, and Emergent BioSolutions and past support from Moderna not related to these studies. S.M.E. and D.K.E. are employees of and shareholders in Moderna, Inc.
Published: April 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.medj.2022.03.009.
Supplemental information
References
- 1.Bajema K.L., Dahl R.M., Prill M.M., Meites E., Rodriguez-Barradas M.C., Marconi V.C., Beenhouwer D.O., Brown S.T., Holodniy M., Lucero-Obusan C., et al. Effectiveness of COVID-19 mRNA vaccines against COVID-19-associated hospitalization - five veterans affairs medical Centers, United States, February 1-August 6, 2021. MMWR Morb Mortal Wkly Rep. 2021;70:1294–1299. doi: 10.15585/mmwr.mm7037e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grannis S.J., Rowley E.A., Ong T.C., Stenehjem E., Klein N.P., DeSilva M.B., Naleway A.L., Natarajan K., Thompson M.G., VISION Network Interim estimates of COVID-19 vaccine effectiveness against COVID-19-associated emergency department or urgent Care clinic encounters and hospitalizations among adults during SARS-CoV-2 B.1.617.2 (Delta) variant predominance - Nine States, June-August 2021. MMWR Morb Mortal Wkly Rep. 2021;70:1291–1293. doi: 10.15585/mmwr.mm7037e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tenforde M.W., Self W.H., Naioti E.A., Ginde A.A., Douin D.J., Olson S.M., Talbot H.K., Casey J.D., Mohr N.M., Zepeski A., et al. Sustained effectiveness of Pfizer-BioNTech and Moderna vaccines against COVID-19 associated hospitalizations among adults - United States, March-July 2021. MMWR Morb Mortal Wkly Rep. 2021;70:1156–1162. doi: 10.15585/mmwr.mm7034e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fowlkes A., Gaglani M., Groover K., Thiese M.S., Tyner H., Ellingson K., HEROES-RECOVER Cohorts Effectiveness of COVID-19 vaccines in preventing SARS-CoV-2 infection among frontline workers before and during B.1.617.2 (Delta) variant Predominance - eight U.S. Locations, December 2020-August 2021. MMWR Morb Mortal Wkly Rep. 2021;70:1167–1169. doi: 10.15585/mmwr.mm7034e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nanduri S., Pilishvili T., Derado G., Soe M.M., Dollard P., Wu H., Li Q., Bagchi S., Dubendris H., Link-Gelles R., et al. Effectiveness of Pfizer-BioNTech and Moderna vaccines in preventing SARS-CoV-2 infection among nursing home residents before and during widespread circulation of the SARS-CoV-2 B.1.617.2 (Delta) variant - national healthcare safety network, March 1-August 1, 2021. MMWR Morb Mortal Wkly Rep. 2021;70:1163–1166. doi: 10.15585/mmwr.mm7034e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., Gilchuk P. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y., Liu J., Plante K.S., Plante J.A., Xie X., Zhang X., et al. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature. 2022;602:294–299. doi: 10.1038/s41586-021-04245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laiton-Donato K., Franco-Muñoz C., Álvarez-Díaz D.A., Ruiz-Moreno H.A., Usme-Ciro J.A., Prada D.A., Reales-González J., Corchuelo S., Herrera-Sepúlveda M.T., Naizaque J., et al. Characterization of the emerging B.1.621 variant of interest of SARS-CoV-2. Infect. Genet. Evol. 2021;95:105038. doi: 10.1016/j.meegid.2021.105038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt F., Weisblum Y., Rutkowska M., Poston D., DaSilva J., Zhang F., Bednarski E., Cho A., Schaefer-Babajew D.J., Gaebler C., Caskey M. High genetic barrier to SARS-CoV-2 polyclonal neutralizing antibody escape. Nature. 2021;600:512–516. doi: 10.1038/s41586-021-04005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fratev F. The R346K mutation in the Mu variant of SARS-CoV-2 alters the interactions with monoclonal antibodies from class 2: a free energy of perturbation study. J. Chem. Inf. Model. 2022;62:627–631. doi: 10.1101/2021.10.12.463781. [DOI] [PubMed] [Google Scholar]
- 12.Suthar M.S., Arunachalam P.S., Hu M., Trisal N.R.M., Raeber O., Chinthrajah S., et al. Durability of immune responses to the BNT162b2 mRNA vaccine. Med. 2022;3:25–27. doi: 10.1101/2021.09.30.462488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tada T., Zhou H., Dcosta B.M., Samanovic M.I., Cornelius A., Herati R.S., Mulligan M.J., Landau N.R. Neutralization of Mu and C.1.2 SARS-CoV-2 variants by vaccine-elicited antibodies in individuals with and without previous history of infection. Preprint at bioRxiv. 2021 doi: 10.1101/2021.10.19.463727. [DOI] [Google Scholar]
- 14.Uriu K., Kimura I., Shirakawa K., Takaori-Kondo A., Nakada T.A., Kaneda A., Nakagawa S., Sato K. Ineffective neutralization of the SARS-CoV-2 Mu variant by convalescent and vaccine sera. Preprint at bioRxiv. 2021 doi: 10.1101/2021.09.06.459005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L., Wang L., Kainulainen M.H., Jiang N., Di H., Bonenfant G., Mills L., Currier M., Shrivastava-Ranjan P., Calderon B.M., et al. Differential neutralization and inhibition of SARS-CoV-2 variants by antibodies elicited by COVID-19 mRNA vaccines. Preprint at bioRxiv. 2021 doi: 10.1101/2021.11.24.469906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Messali S., Bertelli A., Campisi G., Zani A., Ciccozzi M., Caruso A., Caccuri F. A cluster of the new SARS-CoV-2 B.1.621 lineage in Italy and sensitivity of the viral isolate to the BNT162b2 vaccine. J. Med. Virol. 2021;93:6468–6470. doi: 10.1002/jmv.27247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rathnasinghe R., Jangra S., Cupic A., Martínez-Romero C., Mulder L.C., Kehrer T., Yildiz S., Choi A., Mena I., De Vrieze J., Aslam S. The N501Y mutation in SARS-CoV-2 spike leads to morbidity in obese and aged mice and is neutralized by convalescent and post-vaccination human sera. Preprint at medRxiv. 2021 doi: 10.1101/2021.01.19.21249592. [DOI] [Google Scholar]
- 18.Gu H., Chen Q., Yang G., He L., Fan H., Deng Y.Q., Wang Y., Teng Y., Zhao Z., Cui Y., et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen R.E., Winkler E.S., Case J.B., Aziati I.D., Bricker T.L., Joshi A., Darling T.L., Ying B., Errico J.M., Shrihari S., et al. In vivo monoclonal antibody efficacy against SARS-CoV-2 variant strains. Nature. 2021;596:103–108. doi: 10.1038/s41586-021-03720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y., Hu G., Wang Y., Ren W., Zhao X., Ji F., Zhu Y., Feng F., Gong M., Ju X., Zhu Y. Functional and genetic analysis of viral receptor ACE2 orthologs reveals a broad potential host range of SARS-CoV-2. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2025373118. e2025373118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bricker T.L., Darling T.L., Hassan A.O., Harastani H.H., Soung A., Jiang X., Dai Y.N., Zhao H., Adams L.J., Holtzman M.J., et al. A single intranasal or intramuscular immunization with chimpanzee adenovirus-vectored SARS-CoV-2 vaccine protects against pneumonia in hamsters. Cell Rep. 2021;36:109400. doi: 10.1016/j.celrep.2021.109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmitz A.J., Turner J.S., Liu Z., Zhou J.Q., Aziati I.D., Chen R.E., Joshi A., Bricker T.L., Darling T.L., Adelsberg D.C., et al. A vaccine-induced public antibody protects against SARS-CoV-2 and emerging variants. Immunity. 2021;54:2159–2166.e2156. doi: 10.1016/j.immuni.2021.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sia S.F., Yan L.M., Chin A.W., Fung K., Choy K.T., Wong A.Y., Kaewpreedee P., Perera R.A., Poon L.L., Nicholls J.M., Peiris M. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su W., Sia S.F., Schmitz A.J., Bricker T.L., Starr T.N., Greaney A.J., Turner J.S., Mohammed B.M., Liu Z., Choy K.T., Darling T.L. Neutralizing monoclonal antibodies that target the spike receptor binding domain confer Fc receptor-independent protection against SARS-CoV-2 infection in Syrian hamsters. mBio. 2021;12 doi: 10.1128/mBio.02395-21. e02395–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorman M.J., Patel N., Guebre-Xabier M., Zhu A.L., Atyeo C., Pullen K.M., Loos C., Goez-Gazi Y., Carrion R., Tian J.H., et al. Fab and Fc contribute to maximal protection against SARS-CoV-2 following NVX-CoV2373 subunit vaccine with Matrix-M vaccination. Cell Rep. Med. 2021;2:100405. doi: 10.1016/j.xcrm.2021.100405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zohar T., Loos C., Fischinger S., Atyeo C., Wang C., Slein M.D., Burke J., Yu J., Feldman J., Hauser B.M., et al. Compromised humoral functional evolution tracks with SARS-CoV-2 mortality. Cell. 2020;183:1508–1519.e1512. doi: 10.1016/j.cell.2020.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bos R., Rutten L., van der Lubbe J.E.M., Bakkers M.J.G., Hardenberg G., Wegmann F., Zuijdgeest D., de Wilde A.H., Koornneef A., Verwilligen A., et al. Ad26 vector-based COVID-19 vaccine encoding a prefusion-stabilized SARS-CoV-2 Spike immunogen induces potent humoral and cellular immune responses. NPJ Vaccin. 2020;5:91. doi: 10.1038/s41541-020-00243-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alter G., Yu J., Liu J., Chandrashekar A., Borducchi E.N., Tostanoski L.H., McMahan K., Jacob-Dolan C., Martinez D.R., Chang A., et al. Immunogenicity of Ad26.COV2.S vaccine against SARS-CoV-2 variants in humans. Nature. 2021;596:268–272. doi: 10.1038/s41586-021-03681-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sadoff J., Le Gars M., Shukarev G., Heerwegh D., Truyers C., de Groot A.M., Stoop J., Tete S., Van Damme W., Leroux-Roels I., et al. Interim results of a phase 1-2a trial of Ad26.COV2.S covid-19 vaccine. N. Engl. J. Med. 2021;384:1824–1835. doi: 10.1056/NEJMoa2034201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stephenson K.E., Le Gars M., Sadoff J., de Groot A.M., Heerwegh D., Truyers C., Atyeo C., Loos C., Chandrashekar A., McMahan K., et al. Immunogenicity of the Ad26.COV2.S vaccine for COVID-19. Jama. 2021;325:1535–1544. doi: 10.1001/jama.2021.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fiege J.K., Block K.E., Pierson M.J., Nanda H., Shepherd F.K., Mickelson C.K., Stolley J.M., Matchett W.E., Wijeyesinghe S., Meyerholz D.K., Vezys V. Mice with diverse microbial exposure histories as a model for preclinical vaccine testing. Cell Host Microbe. 2021;29:1815–1827. doi: 10.1016/j.chom.2021.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyakawa K., Jeremiah S.S., Kato H., Ryo A. Neutralizing efficacy of vaccines against the SARS-CoV-2 Mu variant. Preprint at medRxiv. 2021 doi: 10.1101/2021.09.23.21264014. [DOI] [Google Scholar]
- 33.Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenke K., Meade-White K., Letko M., Clancy C., Hansen F., Liu Y., Okumura A., Tang-Huau T.L., Li R., Saturday G., et al. Defining the Syrian hamster as a highly susceptible preclinical model for SARS-CoV-2 infection. Emerg Microb. Infect. 2020;9:2673–2684. doi: 10.1080/22221751.2020.1858177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hassan A.O., Case J.B., Winkler E.S., Thackray L.B., Kafai N.M., Bailey A.L., McCune B.T., Fox J.M., Chen R.E., Alsoussi W.B., et al. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell. 2020;182:744–753.e4. doi: 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zang R., Gomez Castro M.F., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., Liu Z., Brulois K.F., Wang X., Greenberg H.B., et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5:eabc3582. doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nelson J., Sorensen E.W., Mintri S., Rabideau A.E., Zheng W., Besin G., Khatwani N., Su S.V., Miracco E.J., Issa W.J., et al. Impact of mRNA chemistry and manufacturing process on innate immune activation. Sci. Adv. 2020;6:eaaz6893. doi: 10.1126/sciadv.aaz6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hassett K.J., Benenato K.E., Jacquinet E., Lee A., Woods A., Yuzhakov O., Himansu S., Deterling J., Geilich B.M., Ketova T., et al. Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol. Ther. Nucleic Acids. 2019;15:1–11. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stadlbauer D., Amanat F., Chromikova V., Jiang K., Strohmeier S., Arunkumar G.A., Tan J., Bhavsar D., Capuano C., Kirkpatrick E., et al. SARS-CoV-2 seroconversion in humans: a detailed protocol for a serological assay, antigen production, and test setup. Curr. Protoc. Microbiol. 2020;57:e100. doi: 10.1002/cpmc.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Asthagiri Arunkumar G., Jurczyszak D., Polanco J., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winkler E.S., Bailey A.L., Kafai N.M., Nair S., McCune B.T., Yu J., Fox J.M., Chen R.E., Earnest J.T., Keeler S.P., et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020;21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ying B., Whitener B., VanBlargan L.A., Hassan A.O., Shrihari S., Liang C.Y., et al. Protective activity of mRNA vaccines against ancestral and variant SARS-CoV-2 strains. Science Translational Medicine. 2022;14(630) doi: 10.1101/2021.08.25.457693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.VanBlargan L.A., Adams L.J., Liu Z., Chen R.E., Gilchuk P., Raju S., Smith B.K., Zhao H., Case J.B., Winkler E.S., et al. A potently neutralizing SARS-CoV-2 antibody inhibits variants of concern by utilizing unique binding residues in a highly conserved epitope. Immunity. 2021;54:2399–2416.e6. doi: 10.1016/j.immuni.2021.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chu D.K.W., Pan Y., Cheng S.M., Hui K.P., Krishnan P., Liu Y., Ng D.Y., Wan C.K., Yang P., Wang Q., Peiris M. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper and are available from the corresponding author upon request. This paper does not include original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.