Abstract
Our understanding of the spatiotemporal regulation of eukaryotic gene expression has recently been greatly stimulated by the findings that many of the regulators of chromatin, transcription, and RNA processing form biomolecular condensates often assembled through liquid-liquid phase separation. Increasing number of reports suggest that these condensates functionally regulate gene expression, largely by concentrating the relevant biomolecules in the liquid-like micro-compartments. However, it remains poorly understood how the physicochemical properties, especially the material properties, of the condensates regulate gene expression activity. In this review, we discuss current data on various nuclear condensates and their biophysical properties with the underlying molecular interactions, and how they may functionally impact gene expression at the level of chromatin organization and activities, transcription, and RNA processing.
Introduction
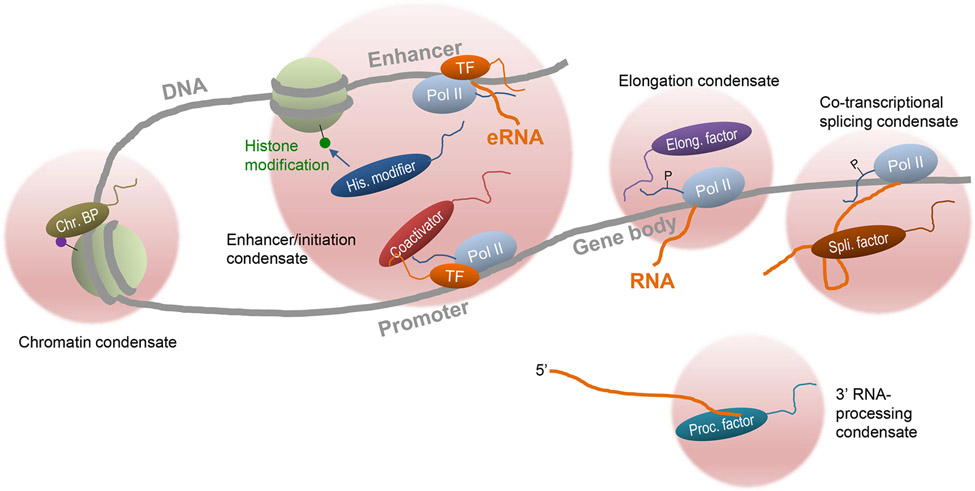
Regulation of the genetic information flow is at the very heart of cell’s activities. Dysregulation of gene expression underlies numerous human diseases including cancer and developmental disorders. While gene expression includes the entire process from DNA sequences to the final functional proteins, we in this review choose to focus on the early steps in gene expression that occur in the nucleus of eukaryotic cells, namely, chromatin regulation, transcription, and RNA processing. In nucleus, the long chromatin polymer undergoes high-order organization that can impact gene expression. For a typical protein-encoding gene, upon cell signaling, activated transcription factors (TFs) bind to specific DNA sequences at gene promoter and enhancer that are induced to interact with each other, and recruit chromatin modification and remodeling proteins to make the local genomic sites more accessible. The recruited general transcription machinery including RNA polymerase initiates transcription at the core promoter, but often pauses until further signal to enter productive elongation state. Nascent RNA transcripts, while still attached to the local chromatin, are almost immediately bound by many RNA-binding proteins which carry out extensive RNA processing including splicing (Figure 1). The mature transcripts are then exported to cytoplasm for translation to make proteins.
Figure 1. Protein condensates that regulate different steps of gene expression in the nucleus.
A highly simplified diagram for protein condensates at various locations in the nucleus that regulate different aspects of gene expression. Chromatin is the central organizer of these protein condensates and itself is in part organized by condensation. The number of proteins in the condensates are minimized for simplicity. Not shown here for simplicity, multiple genomic sites are often co-located in the condensates for co-regulation.
Much like all other cellular processes, the highly ordered and coordinated steps of gene expression process require the involved molecules to be at the right location in the three-dimensional nuclear space with the right concentration at the right time. A fundamental strategy cell uses to achieve spatiotemporal control of biochemistry, including gene expression, is forming membraneless micro-compartments, also termed biomolecular condensates [1], that enrich specific molecules. These condensates are often assembled by weak and multivalent intermolecular interactions between the same (homotypic) or different (heterotypic) molecules, thereby effectively separating these molecules from the solvent phase, a process called liquid-liquid phase separation (LLPS) [2]. It is being increasingly appreciated that the liquid-like condensed state is likely a fundamental state of cellular proteins along with the native state and the amyloid state (a solid-like condensed state), with distinct biological functions associated with each state [3]. Biomolecular condensation has multiple functions across different size scales. The most prominent function at the molecular level is the regulation of biochemical process rates through a number of mechanisms, including concentration, sequestration, and exclusion of specific components for a biochemical process [4].
In an integrated view, different types of high-order assemblies including biomolecular condensates have a structural and dynamic continuum, with very different physicochemical properties that profoundly impact the biological outcomes [5]. The material states of biomolecular condensates can range from highly dynamic liquid to less dynamic and highly viscous liquid or gels, and to non-dynamic and solid-like aggregates or fibers, with no clear-cut boundaries between these states (Figure 2). Compared to solid, the defining characteristic of liquid is its rapid molecular rearrangement. This allows molecules to rapidly find and collide with each other, which facilitates their interactions and subsequently the possible chemical reactions [2]. Proteins have evolved to have proper sequence elements that allow them to adopt optimal material properties to fulfill their specific biological roles. For example, a number of endogenous condensates adopt non-dynamic and solid-like states that are compatible with their specific biological functions [6], sometimes in response to certain environmental conditions [7]. However, aberrant material properties of biomolecular condensates are linked to human disease, most notably the neurodegenerative disease [8, 9]. Molecular composition is another important property of the condensates [10, 11]. Indeed, the endogenous nuclear condensates are always composed of many macromolecules and largely driven by the thermodynamics of the heterotypic interactions of these different molecules [12]. As discussed in [11], we have only rudimentary understanding of the mechanisms that control what molecules are incorporated into or excluded from the condensates, and at what concentration and stoichiometry if incorporated.
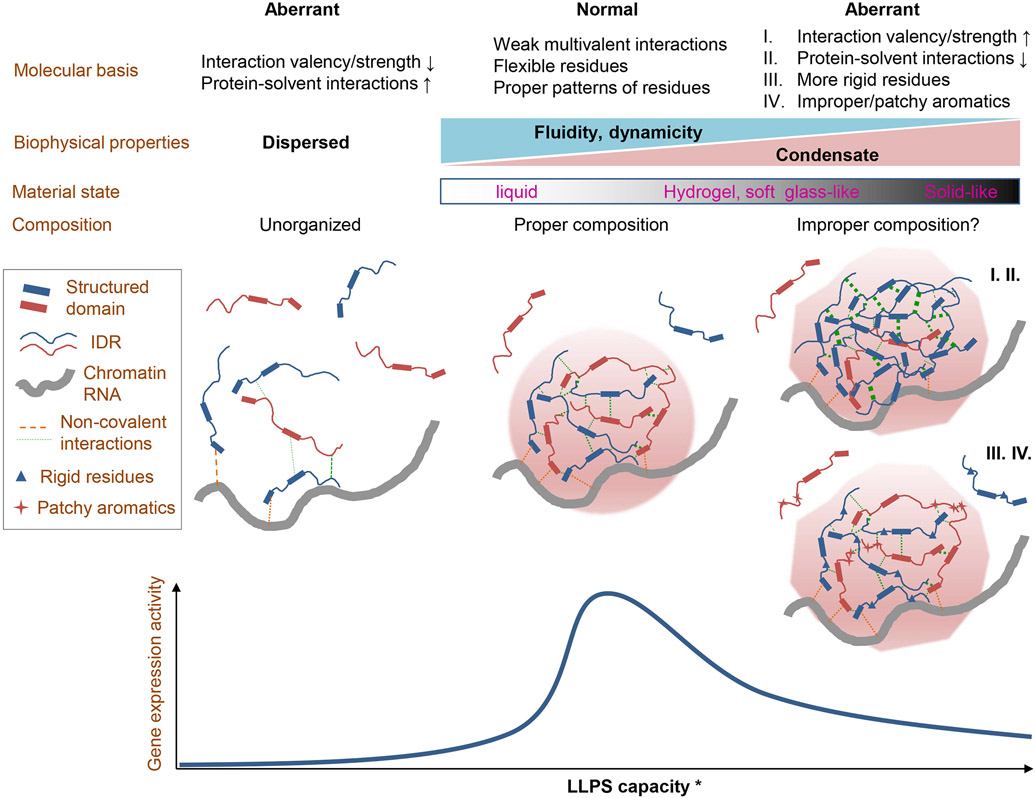
Figure 2. Molecular basis of protein condensate properties in gene regulation.
This figure shows the molecular basis for the material and composition properties of nuclear protein condensates in regulation of gene expression, with a few worthy notes:
a. Proteins in red and blue are drawn here to show that cellular condensates are mostly heterotypic, and the grey curve line shows that chromatin and RNAs are major nuclear polymer structures that organize and profoundly affect these condensates in gene regulation.
b. Each protein is interacting with multiple other proteins to form system-spanning network in the condensates.
c. Both structured domains and IDRs can be engaged in interactions that contribute to condensation.
d. In the diagram for scenarios I and II, the thickened dashed lines show enhanced intermolecular interactions. The increase in the homotypic interactions among the blue proteins may result in reduced co-partition of the red proteins (improper composition) in the co-condensates.
e. * While the x axis shows that increase in LLPS capacity often (I, II) correlates with the transition to a more solid-like state, other molecular changes, such as more rigid residues and patchy aromatics (III, IV), do not necessarily change LLPS capacity and can still harden the condensates.
A large number of excellent reviews have covered the fundamental concept of LLPS [2, 13, 14], biomolecular condensates [1, 15], and their biological regulation and function [4, 8, 16-20]. Biomolecular condensates exist in a very diverse range of locations in cells, including the nucleus [21]. Many nuclear condensate play critical roles in regulation of gene expression, from chromatin organization and activity, transcription, to various steps in RNA processing [22, 23]. Here we will only focus on those nuclear condensates involved in the RNA polymerase II (Pol II)-associated gene expression events and regulation (Figure 1), and also discuss the biophysical properties of these condensates in relation to their function in gene regulation. Though not discussed in this review, nucleolus is a paradigm of phase separated nuclear condensates with heterotypic protein-RNA composition and different phases of physicochemical properties to function in the expression and biogenesis of ribosomal RNAs [24, 25].
Determinants of the biophysical properties of biomolecular condensates
The material properties of condensates, including liquidity, viscosity, and elasticity, are determined by multiple factors including the primary amino acid sequences of the scaffold and client proteins [26]. The sequence determinants of protein condensation are under active studies and still largely unclear [27, 28]. One of the most recognizable features of many phase separating proteins in literature, especially for those involved in nuclear gene regulation events, is the long intrinsically disordered regions (IDRs), parts on proteins that lack well-folded structures [21]. IDRs are very abundant in human proteome, and especially abundant in nuclear proteins, with >70% to be highly disordered, many of which are involved in gene expression processes [29, 30].
Is having IDR equivalent to phase separation of the protein? This is a very practical question for many biologists who may be considering their favorite proteins in phase separation. There are many examples of phase separation driven by multivalent interactions of proteins with structured modular domains [1, 31, 32]. IDRs are not always associated with phase separation, but rather have abundant other roles in biology, mainly through their flexible interaction capacity [33]. Sequence that bestows a disordered state does not automatically mediates intermolecular interactions and high-order assembly, but the lack of strong engagement of the amino acid side chains in binding to other residues on the same molecule does make many of them more exposed and readily available for interactions with other molecules. This often provides the key element for phase separation, multivalency, which allows each molecule to engage in interactions with multiple different molecules to form system-spanning network [14], as illustrated in Figure 2. The major driving forces of IDR-mediated LLPS are usually relatively weak and short-lived, and include electrostatic interactions, π–π interactions, cation–π interactions, hydrogen bonding, and hydrophobic interactions (Figure 3) [1, 21, 34]. The weak interactions of IDRs promote break-reform cycles with either the same and different molecules. Furthermore, the “fuzzy” state, i.e. the large amplitude of conformational fluctuations, of IDRs allow them to rapidly sample interactions with other molecules. These features of interactions form the basis of molecular dynamics and liquidity of the condensates [35]. Indeed, perturbations or mutations that increase molecular interactions are often associated with enhanced propensity in phase separation and also reduced dynamicity, shifting the material state to a more solid-like state (Figure 2 and Table 1).
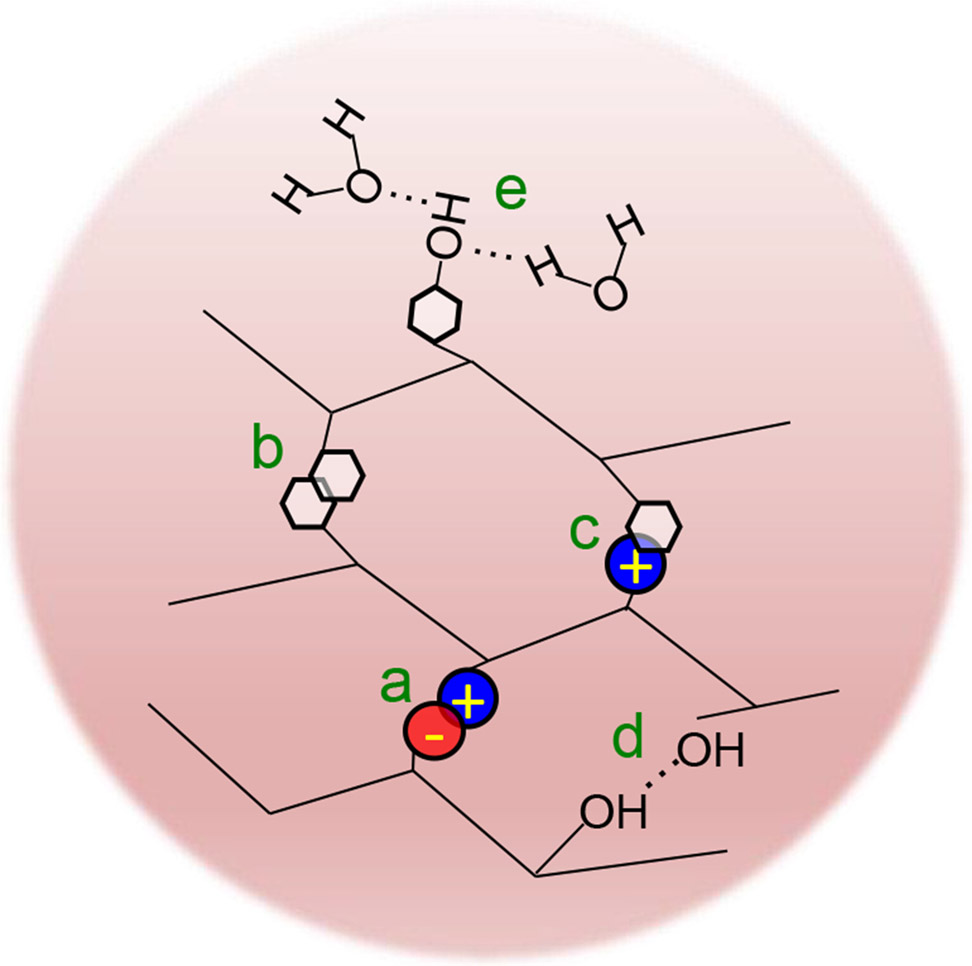
Figure 3. Major molecular interactions mediated by IDRs that control the assembly and biophysical properties of protein condensates.
These interactions include (a) electrostatic interactions between oppositely charged amino acids, (b) π–π interactions mainly between (but not limited to) aromatic rings, (c) cation–π interactions between positively charged and aromatic amino acids, (d) hydrogen bonding between hydroxyl groups of different protein molecules, and (not shown) hydrophobic interactions. (e) shows hydrogen bonding between the hydroxyl group of the protein Tyr and water molecules, and this interaction may play a role in regulating the liquidity and dynamics of the condensates.
Table 1. Properties of protein condensates regulate gene expression.
This table shows four prominent reports in which altered condensate properties are linked to activities in regulating gene expression.
| Protein | ER | HOXD13 | AKAP95 | UTX |
|---|---|---|---|---|
| Function | Transcription factor | Transcription factor | Splicing regulator | Chromatin modulator |
| Disease connection | Synpolydactyly | Tumorigenesis | Tumor suppression | |
| Mut/var/perturb | Ligand stimulation | Alanine expansion | Tyr to Phe | His to Tyr Y chromosome homolog |
| Methods to determine material properties | FRAP 1,6-Hexanediol sensitivity |
Morphology FRAP |
Morphology FRAP, RICS |
Morphology FRAP, RICS |
| Liquidity-function |
|
|
|
|
| Composition | ? | Reduced co-activator amount | ? | ? |
| Reference | [84] | [101] | [108] | [78] |
Under a given condition, the protein sequence elements, including composition and pattern of the amino acids, determine the biophysical characteristics of intermolecular interactions (including those with solvent) in the condensates and the dynamics of the connecting regions on the molecules [5]. These molecular characteristics profoundly impact the material properties of the condensates (Figure 2) [14, 27, 36]. The affinity and valency of the interactions among the proteins, which may be viewed effectively as associative polymers, are a major factor for the condensate dynamics, with increased interaction strength generally resulting in reduced dynamics. The pattern of the interacting residues is also important. For example, for an IDR with a fixed number of adhesive aromatic residues (thus fixed valency), IDR with evenly distributed aromatics gives rise to liquid droplets but IDR with highly patchy or clustered aromatics gives rise to solid-like aggregates [36]. While the linkers between the interacting elements do not greatly contribute to the intermolecular interaction strength, their ability to undergo rapid conformational exchange is an important factor for the condensate dynamics [5]. Moreover, the linkers can adopt either expanded or compact conformation due to their different effective solvation volume (or excluded volume), and can thus affect the condensate properties [14, 37]. In line with the conformational flexibility of the glycine-associated peptide bond, glycine maintains, while serine and glutamine oppose, the liquidity of condensates formed by FUS, all without much impact on condensation propensity [27, 38]. It is unclear if these amino acids control material properties of other protein condensates in the same way, and what other amino acids may play a role in regulating condensate liquidity without impacting condensation propensity.
Other factors, including post-translational modifications and other cellular factors, also regulate the material states of condensates, through ultimately tuning the molecular interaction valency and/or strength. They are beyond the protein primary sequences and thus provide rich means of biological regulation of condensate properties. Post-translational modifications elicit diverse effects on protein phase separation by altering their steric, hydrophobic, or electrostatic properties [16, 39]. Methylation of arginine in the FUS structured C-terminal region weakens the cation-phi interaction with the tyrosine in the IDR, and reduces FUS LLPS propensity and gelation of FUS condensates [40]. RNA-binding deficient TDP-43 forms a unique condensate structure composed of liquid spherical shells with liquid cores, and HSP70 family chaperons help maintain the liquidity of shells and cores [41]. Molecular chaperones including the co-chaperones for HSP70 and small heat shock proteins have also been recently shown to modulate the phase separation propensity of FUS and inhibit the liquid-to-solid/fiber transition of the FUS condensates [42-44]. Nuclear import receptors suppress LLPS and fibrillization of a number of RNA-binding proteins including FUS [38, 45-50]. The material properties of heterotypic condensates are also affected by protein network structure dictated by the relative abundance of each factors and their different interaction strengths, as increase in DAXX concentration in the SPOP-DAXX co-condensates results in a dominance of the weaker DAXX-DAXX interactions over the stronger SPOP-DAXX interactions, and the consequent transition from a gel-like state to a liquid-like state [51].
Chromatin-related condensates
As the starting point of the whole gene expression process, chromatin is a physical platform that is organized at least in part by the phase separation principle and meanwhile a central organizer of numerous nuclear condensates involved in gene expression (Figure 1) [52-54]. Our understanding of chromatin structural organization has been constantly evolving. Current data suggest that regional chromatin may be organized and regulated by LLPS, as short-ranged nucleosomal arrays can form histone tail-driven phase separated liquid-like droplets that recapitulate certain important chromatin regulations including de-condensation by histone acetylation [55]. Chromatin as a long nucleosomal polymer in the nucleus, however, behaves more like a solid or hydro-gel than liquid, with limited DNA molecular mobility in both heterochromatin and euchromatin regions, possibly constrained by the summed strong DNA-histone tail interactions [56]. The solid-like state of chromatin thus provides a scaffold, which not only provides mechanical stability to the genome and nucleus, but also may promote the formation of dynamic condensates of numerous chromatin-associated proteins. On the other hand, LLPS of these proteins also plays an active role in promoting the formation of the different chromatin regions. In particular, a number of studies have shown that constitutive heterochromatin formation is promoted by LLPS of several key heterochromatin-associated proteins, including HP1 alone or together with the H3K9 methylation writer protein, and nuclear matrix protein SAFB in cooperation with major satellite RNAs [57-61]. Another key component of constitutive heterochromatin, MeCP2, also forms phase separated liquid-like condensates. MeCP2 LLPS selectively concentrates heterochromatin cofactors, is promoted by methylated DNA, and is impaired by Rett syndrome-causing mutations in MeCP2 [62, 63]. Facultative heterochromatin compaction, linked to gene silencing, may also be promoted by condensation of the associated proteins, including the polycomb group proteins. CBX2, a component of PRC1, forms liquid condensates that can concentrate DNA and nucleosomes. Residues that are important for nucleosome compaction are also important for CBX2 condensation [64, 65]. Engineered PRC1 condensation induced by light-activated nucleation can read and write histone modification marks, which subsequently induce and maintain chromatin compaction [66]. Sustained X-chromosome heterochromatinization and inactivation was recently shown to require the formation of a heteromeric condensate containing the long non-coding RNA Xist and a number of Xist RNA-binding proteins through protein-RNA and protein-protein interactions [67]. In addition, LLPS of histones themselves, especially the linker histone H1, may also contribute to organization of repressed chromatin [68, 69].
Chromatin is also known to form higher-order structures including topologically associating domains (TADs) and other more dynamic domains, such as promoter-enhancer, promoter-promoter, or enhancer-enhancer loops that are intimately associated with transcriptional regulation [70]. Some of these higher-order structures are maintained by key proteins including cohesin and CTCF. Purified cohesin and DNA complexes form liquid-like condensates in a DNA length-dependent manner, suggesting a phase separation mechanism based on DNA-cohesin-DNA bridge, which may contribute to cohesin-mediated chromatin organization [71]. CTCF forms dynamic nuclear clusters to enhance its target search efficiency by locally concentrating CTCF and to regulate chromatin looping, though the nature of these clusters seems less clear [72-74]. Engineered and genomically targeted liquid nuclear condensates upon light activation-mediated oligomerization in a model system can mechanically sense and restructure the local genomic regions [75].
Active enhancers are associated with H3K4me1 and H3K27ac mainly catalyzed by the KMT2C/D (MLL3/4) branch of the SET1/MLL complexes and p300 acetyltransferase, respectively. MLL4 forms phase separated nuclear condensates that are dependent on a polyQ track in its prion-like domain, and participates and promotes the enhancer condensate formation that will be discussed below. Loss-of-function mutation of MLL4 found in MLL4-associated Kabuki syndrome increases the clustering of PRC1 and chromatin compaction, and causes nuclear mechanical stress by affecting nuclear architecture [76]. P300 and TFs form dynamic co-condensates through interactions of their IDRs. Functionally, the co-condensation promotes the acetylation activity of p300 and subsequent recruitment of coactivators, leading to enhanced transcriptional activation. Moreover, the p300-TF co-condensation can modulate transcriptional bursting kinetics [77]. KDM6A/UTX, a histone H3K27 demethylase often associated with enhancers, also forms phase separated liquid-like condensates mediated by its central long IDR. This property underlies its activities in suppressing tumor and regulating embryonic stem cell differentiation. The co-condensates formed by MLL4 and UTX enhance the H3K4 mono-methylation activity of MLL4, and also regulate the high-order chromatin interaction. These results indicate a crucial role of UTX condensation in cancer and developmental biology through regulation of chromatin organization, histone modifications, and transcription [78]. KDM7A, a demethylase for histone H3K9 and H3K27 demethylation, forms unique ring-shaped condensates with chromatin wrapping around them [79].
Transcriptional condensates
Transcriptional activation involves condensation of basically all categories of proteins that are central to transcription, including Pol II [80, 81], TFs [82-84], coactivators [76, 85-89], and elongation factors [90], highlighting a fundamental role of biomolecular condensation in transcriptional regulation as proposed (Figure 1) [85].
Gene-specific transcription is directed by TFs that have structured DNA binding domains that bind to cognate DNA sequences and also unstructured activation domains that recruit co-activators. TFs often undergo LLPS mediated by the disordered activation domains, either by itself thus acting as a driver in the formation of transcriptional condensates, or by acting as a client (e.g. OCT4) that is incorporated into the co-activator condensates through the interaction between the TF activation domain and the co-activator. Transcriptional co-activators, including BRD4, MED1, YAP/TAZ, and BRD3, form phase-separated condensates in vitro and in living cells [86-89]. These co-activators all possess IDRs that mediated LLPS, except TAZ, which relies on its coiled-coil domain for condensation [88]. These co-activator condensates are often localized at enhancers, especially super-enhancers, compartmentalize TFs and Pol II, and function to regulate specific target gene expression [86-89]. The Pol II C-terminal domain (CTD), consisting of many heptapeptide repeats and disordered, directly binds to the hydrogel formed by IDRs of the FET protein family (FUS, EWS, and TAF15) in a phosphorylation-sensitive manner [80]. Hypophosphorylated Pol II CTD undergoes LLPS, which mediates Pol II clustering at active genes for transcription initiation [91]. Engineered light-driven TAF15 condensates enhance Pol II recruitment and clustering, which in turn enhances the condensation of TAF15 as a positive feedback mechanism and activates local transcription [92]. Combined with other advanced imaging studies in living cells [82, 93], these studies together show that TFs, co-activators, and Pol II form dynamic hubs through the extensive and very often IDR-mediated homotypic and heterotypic protein interactions to drive transcription initiation.
In accord with the notion above that chromatin is not a passive platform but a central and active organizer of nuclear condensates, DNA sequences with high TF binding valency and affinity provide a nucleation mechanism that cooperatively promotes the transcriptional condensate formation at enhancers and the following transcription activation. The strong interactions between DNA and the structured DNA-binding domains of TFs actively contribute to the transcriptional condensate formation driven by weak-multivalent interactions among the protein IDRs (Figure 1) [94]. As a transcription-related example here that IDRs play roles beyond phase separation, extended IDRs of certain TFs guide promoter selection by weak and multiple interactions of the long IDRs with the genomic sites [95]. RNAs can also play a role in the transcriptional condensate formation. BRD3 condensate formation is promoted by a long non-coding RNA DIGIT [89]. Upon estrogen signaling, multiple activated TFs and co-activators assemble together into phase-separated dynamic condensates at induced enhancers in a manner dependent on the enhancer RNA (eRNA), and induces spatial proximity of the activated enhancers to turn on transcription [84].
LLPS also plays an important role in transcription elongation. A key step in elongation is the phosphorylation of the Pol II CTD by kinases including p-TEFb, which consists of CDK9 and cyclin T1. This phosphorylation event is promoted by LLPS of cyclin T1 through a histidine-rich domain on cyclin T1 and Pol II recruitment into the cyclin T1 condensates [81]. Promoter-proximal pause and release of Pol II is another important regulatory mechanism of transcription. Release is mediated by transition of p-TEFb sequestered in an inactive complex to that in the active super-elongation complex (SEC). Components in the SEC complex, ENL and AFF4, can dynamically extract and concentrate cyclin T1 from the inactive complex and form the SEC condensates through heterotypic interactions, and function to release the paused Pol II to a productive elongation stage for rapid gene expression [90]. Pol II CTD phosphorylation also directs the partition of Pol II from the transcriptional initiation condensates filled with Mediator to the splicing condensates filled with splicing factors for transcription elongation and co-transcriptional RNA processing [96]. Moreover, both in vitro and in vivo data suggest that RNA produced from transcription acts as a negative feedback mechanism for transcription through regulating the transcriptional condensates. The low amount of RNA products of transcription initiation promotes the formation of the co-activator condensates by electrostatic interaction-mediated complex coacervation, but the high level of RNA produced in transcription elongation can dissolve the co-activator condensates and thus dampen the transcription process [97].
These transcriptional condensates have important implications in disease. Many forms of malignancies are caused by protein fusion of portions, often including IDRs, of these transcriptional regulators. The EWS-FLI1 fusion protein, through the LLPS property of the EWSR1 IDR, recruits and retargets the BAF chromatin remodeling complex to tumor-specific enhancers in the genome, thereby driving oncogenic gene expression programs in Ewing sarcoma [98]. It is likely that fusions involving the other FET family proteins, FUS and TAF15, also promote cancer in a similar way. The LLPS property of the IDR in the NUP98-HOXA9 fusion enhances genomic occupancy of the fusion protein, induces aberrant chromatin looping for proto-oncogenes, and promotes oncogenic transcriptional activation, thereby driving leukemogenesis [99]. The fusion of ENL with MLL in leukemia can enhance the SEC condensation, and this may play a role in promoting the oncogenic gene expression program orchestrated by the MLL-ENL fusion [90]. Recurrent small insertion and deletion mutations in the ENL chromatin reader domain found in Wilms tumor enhance ENL condensation in an IDR-dependent manner, thereby enriching transcription regulatory factors and activating oncogenic target gene expression [100]. As will be further discussed below, repeat expansion in the IDRs of a number of TFs promote LLPS but also alter the physicochemical properties of transcription condensates, leading to dysregulated transcriptional program and inherited human disorders [101].
RNA processing condensates
Pre-mRNA splicing is initiated and regulated by a large number of RNA-binding proteins binding to the nascent RNA transcripts often still attached to chromatin. Current data suggest that co-transcriptional splicing might occur in nuclear speckles, a dynamic nuclear membraneless organelle highly enriched in splicing factors and actively transcribing Pol II [102, 103]. Consistent with RNA-binding proteins being the most enriched proteins that can adopt a hydrogel-like state identified in b-isox-mediated aggregation assay [104], RNA-binding proteins have been extensively shown to undergo LLPS mediated by their IDRs and often further regulated by RNA binding [105]. LLPS is thus expected to play a very important role in RNA processing (Figure 1).
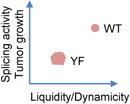
A number of splicing-regulatory proteins have been shown to form biomolecular condensates in order to be functionally active. The Rbfox family of splicing regulators form higher-order assembly mediated by a tyrosine-rich low-complexity sequence, and such assembly is essential for the normal function of Rbfox in alternative splicing [106]. Multiple hnRNP family proteins, known as important splicing regulators, also form higher-order assemblies that require Gly-Tyr repeat-rich IDRs. Interestingly, the mammalian-specific alternative exons are particularly enriched with these IDR-encoding regions, suggesting that the IDR-mediated condensation of these splicing regulators may expand their gene regulatory capacity in mammals [107]. AKAP95 is a zinc-finger RNA-binding protein that can play a positive role in tumorigenesis in part through regulating cancer-related pre-mRNA splicing. AKAP95 undergoes LLPS, which is abolished by mutations of IDR tyrosine to serine or alanine (YS or YA) and restored by replacing its IDR with condensation-capable IDR from irrelevant proteins. AKAP95 LLPS is important for its activity in regulating splicing and tumorigenesis, as its YS and YA mutants abolish these biological functions [108].
RNA 3′-end processing is also regulated by LLPS (Figure 1). Arabidopsis RNA-binding protein FCA forms phase-separated dynamic nuclear condensates, a property that is promoted by a coiled-coil protein FLL2. Their co-condensates compartmentalize many necessary proteins involved in RNA 3’-end processing and thus enhance RNA polyadenylation at specific sites [109].
Material properties of nuclear condensates affecting gene regulation
As mentioned above, reaction kinetics is affected by the interaction rate of the reactants, which is determined by the local concentrations and diffusion rates of the reactants. The most obvious, and possibly most important function, of forming condensate-like micro-compartments is to greatly increase the local molecular concentrations [4, 110, 111]. Diffusion rates of the reactants are linked to the material properties of the condensates, but only a few studies have touched on the question whether material properties of these protein condensates play a role in gene regulation (Table 1).
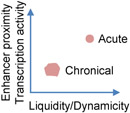
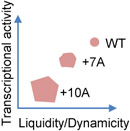
Phase-separated protein condensates in vitro commonly undergo transition from a liquid-like to a solid-like state and lose dynamics, a process known as aging or maturation [1]. Aging of liquid condensates can happen through either gelation that involves extensive physical crosslinks or turning into a glassy state with unchanged elasticity but increased viscosity [17, 112]. Condensate aging can be induced by strong light stimulation of engineered proteins in cells [113], but is otherwise less commonly observed in homeostatic cells, likely due to many cellular factors actively maintaining the condensate properties. In response to estrogen signaling, the physical properties of the eRNA-promoted enhancer condensates can alter depending on the stimulation duration. Acute stimulation results in dynamic, liquid-like, and 1,6-Hexanediol-sensitive enhancer condensates, close spatial proximity of enhancers, and higher transcriptional activity. However, prolonged stimulation leads to less dynamic, solid-like, and 1,6-Hexanediol-resistant condensates, lack of enhancer proximity, and reduced transcriptional activity (Table 1), consistent with the time-dependent in vitro aging of the purified enhancer condensates together with eRNA. eRNA appears to have differential effect on dynamics of different components of the condensates, and it is unclear how eRNA affects the aging process [84]. These data suggest that the dynamicity and liquidity of the enhancer condensates may be important for transcriptional activity, and, more provocatively, may help chromatin interaction through the liquid droplet surface tension as proposed in a model system [75]. It will also be interesting to study how the material properties transition in this system, by gelation or turning into a glassy state [17].
SEC components not only promote the formation of heterotypic condensates with p-TEFb, but also profoundly regulate the material properties of the condensates [90]. Strikingly, the intracellular condensates of ENL and CDK9 appear to be in non-dynamic and solid-like condensates until liquefied by the addition of AFF4. The dynamics of AFF4 is also promoted by ENL, suggesting that these SEC components together maintain the dynamic liquid-like SEC condensates [90]. Though functional evidence is lacking, it is reasonable to speculate that these dynamic condensate properties support the SEC-dependent rapid transcription activation.
In the aforementioned study of AKAP95 condensation [108], the authors show that Tyr to Phe mutation (YF) in the IDR, also impairs the activity in regulating splicing and tumorigenesis. However, opposite to the YS or YA mutations, YF enhances AKAP95 condensation ability, and shifts the material state of the condensates toward a less dynamic and more solid-like state (Table 1). The substantially reduced diffusion rate in the YF condensates may reduce the splicing reaction kinetics. It should be pointed out that it is unclear if active splicing reactions occur in the AKAP95 condensates, though these condensates often co-localize with elongating Pol II and thus like sites of active co-transcriptional splicing. A requirement of dynamic condensates is also consistent with them being the storage place for splicing-related factors, in that dynamic release of these factors can be also important for rapidly providing the factors to splicing reactions occurring elsewhere. A balance of Tyr and Phe residue number in a protein IDR may be important in achieving the proper material states and optimal biological activity, as reflected by the conservation and mutual swap of these two residues in human and mouse AKAP95 IDR [108].
Tyr provides a stronger driver force of phase separation than Phe for certain RNA binding proteins including FUS, suggesting that the hydroxyl group on the Tyr aromatic ring enhances the molecular interactions for phase separation of those proteins [27]. It remains unclear why YF mutation enhances AKAP95 LLPS and reduces the condensate dynamicity, but we may compare it to a similar example, in which the two (S/G)Y(S/G) motifs in FUS low complexity domain form reversible fibril cores [114]. Structural studies show that the first core is stabilized by intra- or inter-molecular hydrogen bonding of the tyrosine hydroxyl group with another Tyr or Ser, while the second core contains water molecules that form hydrogen bond with FUS Tyr58. Disruption of this hydrogen bond by mutation to Phe causes the second core to form irreversible fibrils [114]. This study suggests that, depending on the specific positional context in the protein, the hydroxyl group of Tyr can engage in hydrogen bonding with either protein or solvent molecules in the condensates, resulting in opposite effects on LLPS ability and the material properties of the condensates (Figure 3). We thus speculate that the tyrosine residues in AKAP95 IDR may form hydrogen bond with water to keep the condensates in a more liquid-like state, while the YF mutant loses the hydrogen-bonding with water and the condensates tend to form irregular aggregates.
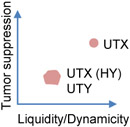
Another example of gene expression activity affected by biophysical properties of condensates comes from the aforementioned study of UTX condensation in cancer regulation [78]. UTX IDR is, surprisingly, severely depleted of aromatic residues known to promote LLPS. Mutation of the top enriched amino acid in that region (His) to Tyr (HY) greatly enhances UTX condensation, reduces the molecular dynamics and diffusion rate in the condensates, and significantly impairs the tumor suppressive activity of UTX. In addition, UTY, the Y-chromosome homolog of UTX, is a weaker tumor suppressor than UTX, at least in part due to the differential properties of its IDR condensates. UTY IDR has substantially more Tyr and Phe than UTX IDR, and also abundant clusters of oppositely charged residues not found in UTX IDR, both features known to enhance phase separation. Indeed, UTY IDR has enhanced LLPS propensity and also markedly reduced condensate dynamics (Table 1). Both the artificial mutation and the natural variation suggest that the UTX IDR may have evolved a proper sequence composition and pattern, such as avoiding aromatics and clusters of oppositely charged residues, for a balanced condensation propensity and material states to ensure its optimal physiological activities [78]. This may also partially explain why Tyr and Arg, both known for their strong capacity of driving phase separation [27], are not significantly enriched in human proteins that are predicted to drive droplet formation [115].
Mutations/variations that affect the condensate material state usually have less impact on the biological activities than those that disrupt condensate formation [78, 108], suggesting that getting the relevant molecules in locally high concentration is probably the primary function of the condensates, and maintaining the proper material state (and likely other properties including correct composition) is an additional but also very important determinant of the activities. We do note that the condensates of these mutations/variations still maintain a low level of fluidity and dynamicity, and thus it remains possible that a completely non-dynamic solid state of the condensates will abolish all activities.
Complications of condensate properties in gene regulation
It is possible that specific biological activities in gene expression in cells may be differentially impacted by the altered condensate material states. A phosphomimetic substitution, S48E, in the globular domain of the RNA-binding protein TDP-43 was shown to disrupt the protein polymerization. S48E also reduces the TDP-43 phase separation propensity and markedly fluidizes the formed condensates. This mutation reduces the activity of TDP-43 in regulating alternative splicing of a minigene construct [116]. Substitution mutations of a single glycine residue to different amino acids in TDP-43 was found to have different impacts on phase separation including the propensity and condensate material properties, and also have different effects on its activity in excluding alternative exon of a minigene construct [117]. G335A increases phase separation ability and also greatly reduces the condensate fluidity, yet slightly enhanced the minigene exon exclusion [117]. Another study shows that a TDP-43 mutant deficient in phase separation can still mediate endogenous splicing [118]. These studies suggest that the physiological dependence of gene expression on either the phase separation propensity or the condensate material states are likely influenced by multiple biological contexts.
These complications reflect how little we understand the relationship between the condensates and biological processes in the cell, which is discussed as follows. (1) As LLPS propensity increase is often correlated with reduced liquidity, the effect of the mutation or perturbation on the biological activity may be dependent on whether it pushes the properties toward or away from the balanced “sweet spot”. (2) While reduced molecular dynamics and diffusion rate in the hardened condensates can restrict the interactions of reactants inside the condensates and thus slow down reaction kinetics, they may have differential impacts on other factors of the reactions [26]. Material properties of condensates may affect the interaction time and stability of certain molecules, and also the network mesh size that may selectively impact certain molecules, thus making it difficult to predict the outcomes. (3) We often do not have a clear idea where exactly the reaction occurs with regard to the condensates. The notion of condensates acting as reaction crucible may leave an impression that the reaction occurs inside the condensates. Advanced microscopy shows that actively transcribing Pol II is located at the surface, not inside, of both the light-activated TAF15 condensates [92] and the condensates formed by the activated endogenous YAP co-activator [87]. Histone H2B ubiquitylation on gene-body nucleosomes probably occurs at the catalytic shell wrapping around the core condensate structure [119]. (4) We are ignorant on the detailed properties of the substructures in most small condensates in live cell nucleus, and thus far from being able to consistently explain the impact of the condensate properties on biological activities. (5) The intrinsic material properties of the protein condensates seen in vitro often cannot fully reflect the mobility measured in live cells, considering the extensive interactions of the nuclear proteins with chromatin and/or RNA, the resident long polymers in the nucleus. Pol II CTD length negatively impacts Pol II dynamics in nucleus likely by enhancing its association with the chromatin scaffold [91], which is rather immobile [56]. RNA-binding mutation of AKAP95 also seems to increase the liquidity and dynamics of AKAP95 nuclear condensates ([108] and unpublished observations), suggesting that RNA-tethering restrains AKAP95 mobility.
Other properties of condensates, such as molecular composition, can also affect the condensate-mediated gene expression activity. Repeat expansions in the IDRs of many transcription factors, such as HOXD13, can enhance their LLPS capacity and also alter the condensate properties, including decreasing dynamics and perturbing the composition of the condensates (termed “unblending”) (Figure 2 and Table 1) [101]. Transcription factors need to work together with co-activators to induce proper transcriptional program, therefore, the reduced amount of the Mediator and other co-activators incorporated into the TF condensates impairs transcription and ultimately lead to certain human disorders [101]. These data also suggest the importance of balanced homotypic and heterotypic interactions for the appropriate properties and thus function of the condensates, as the alanine repeat expansion in HOXD13 IDR enhances the homotypic interactions at the expense of the HOXD13-Mediator heterotypic interactions, and leads to less active transcriptional condensates with hardened material properties and improper composition [101]. We also note the mutual impact and interplay between the material and composition properties of the condensates. On one hand, the hardened condensates of the Ala-expanded HOXD13 disfavor the co-partition of co-activators [101]. On the other hand, changing the composition of the heterotypic condensates can affect the network structure and the material states of the condensates [51].
It is worth noting that it is challenging to draw a solid conclusion on a causal role of a specific material property or any other properties of the condensates in the associated biological outcomes. Just like the general difficulty for establishing causality in most biological research, multiple aspects of the condensate properties are intrinsically linked and it may not be possible to individually perturb one property without affecting some of the others, including some unappreciated properties of biomolecular condensates. For example, mutations that enhance the inter-molecular interactions can increase the phase separation propensity (reduction in saturation concentration), but also tends to reduce the molecular dynamics and harden the condensates (Figures 2 and Table 1) [40, 78, 101, 108, 116], and meanwhile may alter the compositional properties of the condensates [101]. While certain mutations may preferentially impact condensate material state with little effect on saturation concentration [27, 38], it remains unknown if other properties, such as condensate composition, are affected in cells. It is also hard to clearly interpret how condensation-enhancing mutations may differentially affect gene expression programs through differential impacts on different physicochemical properties. Do the condensation-enhancing mutations in ENL chromatin-reader domain [100] also affect the biophysical properties and composition of the condensates and how might those changes contribute to the transcriptional outcomes?
Conclusions and Perspectives
While a clustered distribution of numerous gene-regulatory proteins in the nucleus have been observed and studied for decades [120-123], the recent boom in phase separation research has injected new enthusiasm into the studies of spatiotemporal regulation of genome organization and gene expression. In our opinion, this does not merely provide a different perspective in understanding these structures, but also propels major advancement and revelations through some important approaches. One such approach is the in vitro demonstration that certain purified proteins have the intrinsic ability to form condensates, thus likely to be the “driver” (and probably biologically meaningful) of the nuclear foci formation rather than being passively dragged into the foci in cells. More importantly, dissections and manipulations of the protein sequence elements and other cellular factors greatly help reveal the functionality of many of these nuclear foci, as well as the activity and mechanism of the protein itself. The key molecular activities of certain nuclear proteins are quite enigmatic until LLPS sheds new light on them. There are a number of epigenetic enzymes whose catalytic activities appear to be far from sufficient to account for their biological roles [124]. UTX, for example, is a histone H3K27 demethylase whose demethylase activity is often dispensable for UTX to regulate many biological processes. UTX IDR-mediated LLPS is now found to be an underlying property for its activities in regulating tumor and stem cell differentiation [78]. The findings that the repeat expansions in TF IDRs alter the transcriptional condensate properties have brought critical and mechanistic understanding of otherwise perplexing pathologies [101].
There is no doubt that many more nuclear proteins will be shown in the coming years to form condensates that play a role in gene regulation. It will be of great interest to understand (1) how the physicochemical properties of these nuclear condensates are determined by protein-intrinsic and extrinsic factors, (2) how different cellular conditions (such as cell differentiation stages) and disease mutations may affect the physicochemical properties of the condensates [125], and (3) how these condensate properties regulate the associated biochemical activities and the biological functions.
What molecular “grammars”, if any, on the primary sequence level do proteins (including the nuclear proteins involved in gene expression) adopt, in order to maintain a balance of propensity in forming condensates and keeping the condensates in proper material states with proper molecular composition? We have too few examples at this stage to draw any generalizable rules. As we discussed above, such balance appears to be important for the optimal biochemical activities and biological functions of the proteins.
A great deal of research has focused on the role of IDRs on individual proteins in regulating the nuclear condensates. Future research will need to spend much more effort on (a) the interplay of IDRs and structured domains of these proteins, and also (b) the interplay of different molecules in modulating the condensate assembly, disassembly, and their physicochemical properties, in relationship with the function. These two aspects may also be interconnected, as the structured domains may regulate the condensation of full-length protein either through direct effect (without another molecule) or through interacting with another molecule in the condensates. Dimerization or oligomerization of a structured domain, such as shown by TDP-43, enhances LLPS driven by IDR-mediated interaction network and reduces the condensate fluidity [116]. Cation-π interaction between arginine on the structured domain and tyrosine of the IDR on FUS is a major driving force for FUS LLPS and profoundly shapes the material properties of the condensates [40]. An indirect effect of structured domain on LLPS is commonly seen in the enhanced TF condensation with DNA by multivalent interactions of structured DNA-binding domain of TFs with DNA sequences [94].
Multiple approaches will be necessary to answer the fundamental question of how condensates and their properties functionally regulate gene expression. Loss-of-function approach using physiologically relevant in vivo systems is an indispensable and major approach. This approach typically involves mutagenesis of endogenous gene sequences that encode proteins regions or residues critical for condensate properties, followed by functional analysis in gene expression. However, mechanistic dissection is not easy with this approach and indirect effects can be difficult to exclude. One important approach in mechanistic studies of gene expression has been in vitro biochemical dissection of chromatin modifications and remodeling, transcription, and RNA processing, using “homogenized” nuclear extracts and reconstituted systems with purified factors. Homogenization here refers to the loss of cellular structure at a rather large size scale, but it is unclear how much of the mesoscale condensates are still preserved or lost, and thus unclear whether the biochemical activities seen in these systems are aided by any condensates. Biochemical reconstitution with fluorescently trackable factors will allow manipulation of condensates formation and properties to be directly linked to the functional readouts in the system. Certain new approaches, especially the light-activated condensation in live cells, allow direct effects to be revealed by acute and site-specific manipulation of condensates followed by functional readouts in situ [66, 77, 92]. These can be useful complementary methods to understand the direct effects of condensation and condensate properties on gene expression.
Looking forward, we are just starting to appreciate the functional relevance of the numerous nuclear condensates and their properties in gene regulation, and will surely encounter new surprises along our journeys toward a better understanding of how genetic information flow is controlled by these small droplets. These studies may also suggest intervention approaches to modulate, including selectively soften and harden, the material and other properties of these nuclear condensates in disease treatment.
Acknowledgements
Work in authors’ laboratory is supported by the NIH (1 R21 CA257936-01), Department of Defense (BC190343), American Cancer Society Research Scholar Award (128609-RSG-15-166-01-DMC), and the Leukemia and Lymphoma Society Scholar Award. The authors are grateful to Rohit Pappu for stimulating discussions.
References
- [1].Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nature reviews Molecular cell biology. 2017;18:285–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hyman AA, Weber CA, Julicher F. Liquid-liquid phase separation in biology. Annual review of cell and developmental biology. 2014;30:39–58. [DOI] [PubMed] [Google Scholar]
- [3].Fuxreiter M, Vendruscolo M. Generic nature of the condensed states of proteins. Nature cell biology. 2021;23:587–94. [DOI] [PubMed] [Google Scholar]
- [4].Lyon AS, Peeples WB, Rosen MK. A framework for understanding the functions of biomolecular condensates across scales. Nature reviews Molecular cell biology. 2021;22:215–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wu H, Fuxreiter M. The Structure and Dynamics of Higher-Order Assemblies: Amyloids, Signalosomes, and Granules. Cell. 2016;165:1055–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Woodruff JB, Hyman AA, Boke E. Organization and Function of Non-dynamic Biomolecular Condensates. Trends in biochemical sciences. 2018;43:81–94. [DOI] [PubMed] [Google Scholar]
- [7].Franzmann TM, Jahnel M, Pozniakovsky A, Mahamid J, Holehouse AS, Nüske E, et al. Phase separation of a yeast prion protein promotes cellular fitness. Science (New York, NY). 2018;359. [DOI] [PubMed] [Google Scholar]
- [8].Alberti S, Dormann D. Liquid-Liquid Phase Separation in Disease. Annual review of genetics. 2019;53:171–94. [DOI] [PubMed] [Google Scholar]
- [9].Mathieu C, Pappu RV, Taylor JP. Beyond aggregation: Pathological phase transitions in neurodegenerative disease. Science (New York, NY). 2020;370:56–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, et al. Compositional Control of Phase-Separated Cellular Bodies. Cell. 2016;166:651–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ditlev JA, Case LB, Rosen MK. Who's In and Who's Out-Compositional Control of Biomolecular Condensates. Journal of molecular biology. 2018;430:4666–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Riback JA, Zhu L, Ferrolino MC, Tolbert M, Mitrea DM, Sanders DW, et al. Composition-dependent thermodynamics of intracellular phase separation. Nature. 2020;581:209–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bergeron-Sandoval LP, Safaee N, Michnick SW. Mechanisms and Consequences of Macromolecular Phase Separation. Cell. 2016;165:1067–79. [DOI] [PubMed] [Google Scholar]
- [14].Choi JM, Holehouse AS, Pappu RV. Physical Principles Underlying the Complex Biology of Intracellular Phase Transitions. Annual review of biophysics. 2020;49:107–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Science (New York, NY). 2017;357:eaaf4382. [DOI] [PubMed] [Google Scholar]
- [16].Snead WT, Gladfelter AS. The Control Centers of Biomolecular Phase Separation: How Membrane Surfaces, PTMs, and Active Processes Regulate Condensation. Molecular cell. 2019;76:295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Alberti S, Hyman AA. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nature reviews Molecular cell biology. 2021;22:196–213. [DOI] [PubMed] [Google Scholar]
- [18].Boeynaems S, Alberti S, Fawzi NL, Mittag T, Polymenidou M, Rousseau F, et al. Protein Phase Separation: A New Phase in Cell Biology. Trends in cell biology. 2018;28:420–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Uversky VN. Protein intrinsic disorder-based liquid-liquid phase transitions in biological systems: Complex coacervates and membrane-less organelles. Advances in colloid and interface science. 2017;239:97–114. [DOI] [PubMed] [Google Scholar]
- [20].Mitrea DM, Kriwacki RW. Phase separation in biology; functional organization of a higher order. Cell communication and signaling : CCS. 2016;14:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sabari BR, Dall'Agnese A, Young RA. Biomolecular Condensates in the Nucleus. Trends in biochemical sciences. 2020;45:961–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sabari BR. Biomolecular Condensates and Gene Activation in Development and Disease. Developmental cell. 2020;55:84–96. [DOI] [PubMed] [Google Scholar]
- [23].Peng L, Li EM, Xu LY. From start to end: Phase separation and transcriptional regulation. Biochimica et biophysica acta Gene regulatory mechanisms. 2020;1863:194641. [DOI] [PubMed] [Google Scholar]
- [24].Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, et al. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell. 2016;165:1686–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lafontaine DLJ, Riback JA, Bascetin R, Brangwynne CP. The nucleolus as a multiphase liquid condensate. Nature reviews Molecular cell biology. 2021;22:165–82. [DOI] [PubMed] [Google Scholar]
- [26].Schuster BS, Regy RM, Dolan EM, Kanchi Ranganath A, Jovic N, Khare SD, et al. Biomolecular Condensates: Sequence Determinants of Phase Separation, Microstructural Organization, Enzymatic Activity, and Material Properties. The journal of physical chemistry B. 2021;125:3441–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wang J, Choi JM, Holehouse AS, Lee HO, Zhang X, Jahnel M, et al. A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell. 2018;174:688–99.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chong S, Mir M. Towards Decoding the Sequence-Based Grammar Governing the Functions of Intrinsically Disordered Protein Regions. Journal of molecular biology. 2020:166724. [DOI] [PubMed] [Google Scholar]
- [29].Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. Journal of molecular biology. 2004;337:635–45. [DOI] [PubMed] [Google Scholar]
- [30].Frege T, Uversky VN. Intrinsically disordered proteins in the nucleus of human cells. Biochemistry and biophysics reports. 2015;1:33–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhu G, Xie J, Kong W, Xie J, Li Y, Du L, et al. Phase Separation of Disease-Associated SHP2 Mutants Underlies MAPK Hyperactivation. Cell. 2020;183:490–502.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Martin EW, Holehouse AS. Intrinsically disordered protein regions and phase separation: sequence determinants of assembly or lack thereof. Emerging topics in life sciences. 2020;4:307–29. [DOI] [PubMed] [Google Scholar]
- [34].Vernon RM, Chong PA, Tsang B, Kim TH, Bah A, Farber P, et al. Pi-Pi contacts are an overlooked protein feature relevant to phase separation. eLife. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Fuxreiter M Fuzzy protein theory for disordered proteins. Biochemical Society transactions. 2020;48:2557–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Martin EW, Holehouse AS, Peran I, Farag M, Incicco JJ, Bremer A, et al. Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science (New York, NY). 2020;367:694–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Harmon TS, Holehouse AS, Rosen MK, Pappu RV. Intrinsically disordered linkers determine the interplay between phase separation and gelation in multivalent proteins. eLife. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Niaki AG, Sarkar J, Cai X, Rhine K, Vidaurre V, Guy B, et al. Loss of Dynamic RNA Interaction and Aberrant Phase Separation Induced by Two Distinct Types of ALS/FTD-Linked FUS Mutations. Molecular cell. 2020;77:82–94.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bah A, Forman-Kay JD. Modulation of Intrinsically Disordered Protein Function by Post-translational Modifications. The Journal of biological chemistry. 2016;291:6696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Qamar S, Wang G, Randle SJ, Ruggeri FS, Varela JA, Lin JQ, et al. FUS Phase Separation Is Modulated by a Molecular Chaperone and Methylation of Arginine Cation-π Interactions. Cell. 2018;173:720–34.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Yu H, Lu S, Gasior K, Singh D, Vazquez-Sanchez S, Tapia O, et al. HSP70 chaperones RNA-free TDP-43 into anisotropic intranuclear liquid spherical shells. Science (New York, NY). 2021;371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Gu J, Liu Z, Zhang S, Li Y, Xia W, Wang C, et al. Hsp40 proteins phase separate to chaperone the assembly and maintenance of membraneless organelles. Proceedings of the National Academy of Sciences of the United States of America. 2020;117:31123–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Liu Z, Zhang S, Gu J, Tong Y, Li Y, Gui X, et al. Hsp27 chaperones FUS phase separation under the modulation of stress-induced phosphorylation. Nature structural & molecular biology. 2020;27:363–72. [DOI] [PubMed] [Google Scholar]
- [44].Boczek EE, Fürsch J, Jawerth L, Jahnel M, Niedermeier ML, Ruer-Gruß M, et al. HspB8 prevents aberrant phase transitions of FUS by chaperoning its folded RNA binding domain. bioRxiv. 2021:2021.04.13.439588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Baade I, Hutten S, Sternburg EL, Pörschke M, Hofweber M, Dormann D, et al. The RNA-binding protein FUS is chaperoned and imported into the nucleus by a network of import receptors. The Journal of biological chemistry. 2021:100659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Guo L, Fare CM, Shorter J. Therapeutic Dissolution of Aberrant Phases by Nuclear-Import Receptors. Trends in cell biology. 2019;29:308–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Guo L, Kim HJ, Wang H, Monaghan J, Freyermuth F, Sung JC, et al. Nuclear-Import Receptors Reverse Aberrant Phase Transitions of RNA-Binding Proteins with Prion-like Domains. Cell. 2018;173:677–92.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hofweber M, Hutten S, Bourgeois B, Spreitzer E, Niedner-Boblenz A, Schifferer M, et al. Phase Separation of FUS Is Suppressed by Its Nuclear Import Receptor and Arginine Methylation. Cell. 2018;173:706–19.e13. [DOI] [PubMed] [Google Scholar]
- [49].Springhower CE, Rosen MK, Chook YM. Karyopherins and condensates. Current opinion in cell biology. 2020;64:112–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Yoshizawa T, Ali R, Jiou J, Fung HYJ, Burke KA, Kim SJ, et al. Nuclear Import Receptor Inhibits Phase Separation of FUS through Binding to Multiple Sites. Cell. 2018;173:693–705.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Schmit JD, Bouchard JJ, Martin EW, Mittag T. Protein Network Structure Enables Switching between Liquid and Gel States. Journal of the American Chemical Society. 2020;142:874–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Feric M, Misteli T. Phase Separation in Genome Organization across Evolution. Trends in cell biology. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Palikyras S, Papantonis A. Modes of phase separation affecting chromatin regulation. Open biology. 2019;9:190167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Erdel F, Rippe K. Formation of Chromatin Subcompartments by Phase Separation. Biophysical journal. 2018;114:2262–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Gibson BA, Doolittle LK, Schneider MWG, Jensen LE, Gamarra N, Henry L, et al. Organization of Chromatin by Intrinsic and Regulated Phase Separation. Cell. 2019;179:470–84.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Strickfaden H, Tolsma TO, Sharma A, Underhill DA, Hansen JC, Hendzel MJ. Condensed Chromatin Behaves like a Solid on the Mesoscale In Vitro and in Living Cells. Cell. 2020;183:1772–84.e13. [DOI] [PubMed] [Google Scholar]
- [57].Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, et al. Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature. 2017;547:236–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Sanulli S, Trnka MJ, Dharmarajan V, Tibble RW, Pascal BD, Burlingame AL, et al. HP1 reshapes nucleosome core to promote heterochromatin phase separation. Nature. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH. Phase separation drives heterochromatin domain formation. Nature. 2017;547:241–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Huo X, Ji L, Zhang Y, Lv P, Cao X, Wang Q, et al. The Nuclear Matrix Protein SAFB Cooperates with Major Satellite RNAs to Stabilize Heterochromatin Architecture Partially through Phase Separation. Molecular cell. 2019. [DOI] [PubMed] [Google Scholar]
- [61].Wang L, Gao Y, Zheng X, Liu C, Dong S, Li R, et al. Histone Modifications Regulate Chromatin Compartmentalization by Contributing to a Phase Separation Mechanism. Molecular cell. 2019. [DOI] [PubMed] [Google Scholar]
- [62].Li CH, Coffey EL, Dall'Agnese A, Hannett NM, Tang X, Henninger JE, et al. MeCP2 links heterochromatin condensates and neurodevelopmental disease. Nature. 2020;586:440–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Wang L, Hu M, Zuo MQ, Zhao J, Wu D, Huang L, et al. Rett syndrome-causing mutations compromise MeCP2-mediated liquid-liquid phase separation of chromatin. Cell research. 2020;30:393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Tatavosian R, Kent S, Brown K, Yao T, Duc HN, Huynh TN, et al. Nuclear condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase separation. The Journal of biological chemistry. 2019;294:1451–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Plys AJ, Davis CP, Kim J, Rizki G, Keenen MM, Marr SK, et al. Phase separation of Polycomb-repressive complex 1 is governed by a charged disordered region of CBX2. Genes & development. 2019;33:799–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Eeftens JM, Kapoor M, Brangwynne CP. Epigenetic memory as a time integral over prior history of Polycomb phase separation. bioRxiv. 2020:2020.08.19.254706. [Google Scholar]
- [67].Pandya-Jones A, Markaki Y, Serizay J, Chitiashvili T, Mancia Leon WR, Damianov A, et al. A protein assembly mediates Xist localization and gene silencing. Nature. 2020;587:145–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Shakya A, Park S, Rana N, King JT. Liquid-Liquid Phase Separation of Histone Proteins in Cells: Role in Chromatin Organization. Biophysical journal. 2020;118:753–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Turner AL, Watson M, Wilkins OG, Cato L, Travers A, Thomas JO, et al. Highly disordered histone H1–DNA model complexes and their condensates. Proceedings of the National Academy of Sciences. 2018;115:11964–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Misteli T The Self-Organizing Genome: Principles of Genome Architecture and Function. Cell. 2020;183:28–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Ryu JK, Bouchoux C, Liu HW, Kim E, Minamino M, de Groot R, et al. Bridging-induced phase separation induced by cohesin SMC protein complexes. Science advances. 2021;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Hansen AS, Amitai A, Cattoglio C, Tjian R, Darzacq X. Guided nuclear exploration increases CTCF target search efficiency. Nature chemical biology. 2020;16:257–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Zirkel A, Nikolic M, Sofiadis K, Mallm JP, Brackley CA, Gothe H, et al. HMGB2 Loss upon Senescence Entry Disrupts Genomic Organization and Induces CTCF Clustering across Cell Types. Molecular cell. 2018;70:730–44.e6. [DOI] [PubMed] [Google Scholar]
- [74].Hansen AS, Hsieh TS, Cattoglio C, Pustova I, Saldaña-Meyer R, Reinberg D, et al. Distinct Classes of Chromatin Loops Revealed by Deletion of an RNA-Binding Region in CTCF. Molecular cell. 2019;76:395–411.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Shin Y, Chang YC, Lee DSW, Berry J, Sanders DW, Ronceray P, et al. Liquid Nuclear Condensates Mechanically Sense and Restructure the Genome. Cell. 2018;175:1481–91.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Fasciani A, D'Annunzio S, Poli V, Fagnocchi L, Beyes S, Michelatti D, et al. MLL4-associated condensates counterbalance Polycomb-mediated nuclear mechanical stress in Kabuki syndrome. Nature genetics. 2020;52:1397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Ma L, Gao Z, Wu J, Zhong B, Xie Y, Huang W, et al. Co-condensation between transcription factor and coactivator p300 modulates transcriptional bursting kinetics. Molecular cell. 2021;81:1682–97 e7. [DOI] [PubMed] [Google Scholar]
- [78].Shi B, Li W, Song Y, Wang Z, Ju R, Ulman A, et al. UTX condensation underlies its tumour-suppressive activity. Nature. 2021;597:726–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Ming H, Wang Q, Zhang Y, Ji L, Cheng L, Huo X, et al. The nuclear bodies formed by histone demethylase KDM7A. Protein & cell. 2021;12:297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, Mirzaei H, et al. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell. 2013;155:1049–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Lu H, Yu D, Hansen AS, Ganguly S, Liu R, Heckert A, et al. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature. 2018;558:318–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Chong S, Dugast-Darzacq C, Liu Z, Dong P, Dailey GM, Cattoglio C, et al. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science (New York, NY). 2018;361:aar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Boija A, Klein IA, Sabari BR, Dall'Agnese A, Coffey EL, Zamudio AV, et al. Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell. 2018;175:1842–55.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Nair SJ, Yang L, Meluzzi D, Oh S, Yang F, Friedman MJ, et al. Phase separation of ligand-activated enhancers licenses cooperative chromosomal enhancer assembly. Nature structural & molecular biology. 2019;26:193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A Phase Separation Model for Transcriptional Control. Cell. 2017;169:13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Sabari BR, Dall'Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science (New York, NY). 2018;361:aar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Cai D, Feliciano D, Dong P, Flores E, Gruebele M, Porat-Shliom N, et al. Phase separation of YAP reorganizes genome topology for long-term YAP target gene expression. Nature cell biology. 2019;21:1578–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Lu Y, Wu T, Gutman O, Lu H, Zhou Q, Henis YI, et al. Phase separation of TAZ compartmentalizes the transcription machinery to promote gene expression. Nature cell biology. 2020;22:453–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Daneshvar K, Ardehali MB, Klein IA, Hsieh FK, Kratkiewicz AJ, Mahpour A, et al. lncRNA DIGIT and BRD3 protein form phase-separated condensates to regulate endoderm differentiation. Nature cell biology. 2020;22:1211–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Guo C, Che Z, Yue J, Xie P, Hao S, Xie W, et al. ENL initiates multivalent phase separation of the super elongation complex (SEC) in controlling rapid transcriptional activation. Science advances. 2020;6:eaay4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Boehning M, Dugast-Darzacq C, Rankovic M, Hansen AS, Yu T, Marie-Nelly H, et al. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nature structural & molecular biology. 2018. [DOI] [PubMed] [Google Scholar]
- [92].Wei MT, Chang YC, Shimobayashi SF, Shin Y, Strom AR, Brangwynne CP. Nucleated transcriptional condensates amplify gene expression. Nature cell biology. 2020;22:1187–96. [DOI] [PubMed] [Google Scholar]
- [93].Cho WK, Spille JH, Hecht M, Lee C, Li C, Grube V, et al. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science (New York, NY). 2018;361:412–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Shrinivas K, Sabari BR, Coffey EL, Klein IA, Boija A, Zamudio AV, et al. Enhancer Features that Drive Formation of Transcriptional Condensates. Molecular cell. 2019;75:549–61.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Brodsky S, Jana T, Mittelman K, Chapal M, Kumar DK, Carmi M, et al. Intrinsically Disordered Regions Direct Transcription Factor In Vivo Binding Specificity. Molecular cell. 2020;79:459–71.e4. [DOI] [PubMed] [Google Scholar]
- [96].Guo YE, Manteiga JC, Henninger JE, Sabari BR, Dall'Agnese A, Hannett NM, et al. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature. 2019;572:543–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Henninger JE, Oksuz O, Shrinivas K, Sagi I, LeRoy G, Zheng MM, et al. RNA-Mediated Feedback Control of Transcriptional Condensates. Cell. 2021;184:207–25.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Boulay G, Sandoval GJ, Riggi N, Iyer S, Buisson R, Naigles B, et al. Cancer-Specific Retargeting of BAF Complexes by a Prion-like Domain. Cell. 2017;171:163–78.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Ahn JH, Davis ES, Daugird TA, Zhao S, Quiroga IY, Uryu H, et al. Phase separation drives aberrant chromatin looping and cancer development. Nature. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Wan L, Chong S, Xuan F, Liang A, Cui X, Gates L, et al. Impaired cell fate through gain-of-function mutations in a chromatin reader. Nature. 2020;577:121–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Basu S, Mackowiak SD, Niskanen H, Knezevic D, Asimi V, Grosswendt S, et al. Unblending of Transcriptional Condensates in Human Repeat Expansion Disease. Cell. 2020;181:1062–79.e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Spector DL, Lamond AI. Nuclear speckles. Cold Spring Harbor perspectives in biology. 2011;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Quinodoz SA, Ollikainen N, Tabak B, Palla A, Schmidt JM, Detmar E, et al. Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. Cell. 2018;174:744–57.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Mittag T, Parker R. Multiple Modes of Protein-Protein Interactions Promote RNP Granule Assembly. Journal of molecular biology. 2018;430:4636–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Ying Y, Wang XJ, Vuong CK, Lin CH, Damianov A, Black DL. Splicing Activation by Rbfox Requires Self-Aggregation through Its Tyrosine-Rich Domain. Cell. 2017;170:312–23.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Gueroussov S, Weatheritt RJ, O'Hanlon D, Lin ZY, Narula A, Gingras AC, et al. Regulatory Expansion in Mammals of Multivalent hnRNP Assemblies that Globally Control Alternative Splicing. Cell. 2017;170:324–39.e23. [DOI] [PubMed] [Google Scholar]
- [108].Li W, Hu J, Shi B, Palomba F, Digman MA, Gratton E, et al. Biophysical properties of AKAP95 protein condensates regulate splicing and tumorigenesis. Nature cell biology. 2020;22:960–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Fang X, Wang L, Ishikawa R, Li Y, Fiedler M, Liu F, et al. Arabidopsis FLL2 promotes liquid-liquid phase separation of polyadenylation complexes. Nature. 2019;569:265–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Zhang Y, Narlikar GJ, Kutateladze TG. Enzymatic Reactions inside Biological Condensates. Journal of molecular biology. 2020:166624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Peeples W, Rosen MK. Mechanistic dissection of increased enzymatic rate in a phase-separated compartment. Nature chemical biology. 2021;17:693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Jawerth L, Fischer-Friedrich E, Saha S, Wang J, Franzmann T, Zhang X, et al. Protein condensates as aging Maxwell fluids. Science (New York, NY). 2020;370:1317–23. [DOI] [PubMed] [Google Scholar]
- [113].Shin Y, Berry J, Pannucci N, Haataja MP, Toettcher JE, Brangwynne CP. Spatiotemporal Control of Intracellular Phase Transitions Using Light-Activated optoDroplets. Cell. 2017;168:159–71.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Luo F, Gui X, Zhou H, Gu J, Li Y, Liu X, et al. Atomic structures of FUS LC domain segments reveal bases for reversible amyloid fibril formation. Nature structural & molecular biology. 2018;25:341–6. [DOI] [PubMed] [Google Scholar]
- [115].Hardenberg M, Horvath A, Ambrus V, Fuxreiter M, Vendruscolo M. Widespread occurrence of the droplet state of proteins in the human proteome. Proceedings of the National Academy of Sciences of the United States of America. 2020;117:33254–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Wang A, Conicella AE, Schmidt HB, Martin EW, Rhoads SN, Reeb AN, et al. A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing. The EMBO journal. 2018;37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Conicella AE, Dignon GL, Zerze GH, Schmidt HB, D'Ordine AM, Kim YC, et al. TDP-43 α-helical structure tunes liquid-liquid phase separation and function. Proceedings of the National Academy of Sciences of the United States of America. 2020;117:5883–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Schmidt HB, Barreau A, Rohatgi R. Phase separation-deficient TDP43 remains functional in splicing. Nature communications. 2019;10:4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Gallego LD, Schneider M, Mittal C, Romanauska A, Gudino Carrillo RM, Schubert T, et al. Phase separation directs ubiquitination of gene-body nucleosomes. Nature. 2020;579:592–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].van Steensel B, Brink M, van der Meulen K, van Binnendijk EP, Wansink DG, de Jong L, et al. Localization of the glucocorticoid receptor in discrete clusters in the cell nucleus. Journal of cell science. 1995;108 ( Pt 9):3003–11. [DOI] [PubMed] [Google Scholar]
- [121].Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends in genetics : TIG. 2011;27:295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Spector DL. Macromolecular domains within the cell nucleus. Annual review of cell biology. 1993;9:265–315. [DOI] [PubMed] [Google Scholar]
- [123].Papantonis A, Cook PR. Transcription factories: genome organization and gene regulation. Chemical reviews. 2013;113:8683–705. [DOI] [PubMed] [Google Scholar]
- [124].Morgan MAJ, Shilatifard A. Reevaluating the roles of histone-modifying enzymes and their associated chromatin modifications in transcriptional regulation. Nature genetics. 2020;52:1271–81. [DOI] [PubMed] [Google Scholar]
- [125].Tsang B, Pritišanac I, Scherer SW, Moses AM, Forman-Kay JD. Phase Separation as a Missing Mechanism for Interpretation of Disease Mutations. Cell. 2020;183:1742–56. [DOI] [PubMed] [Google Scholar]