Abstract
The impact of genetics on physiology and sports performance is one of the most debated research aspects in sports sciences. Nearly 200 genetic polymorphisms have been found to influence sports performance traits, and over 20 polymorphisms may condition the status of the elite athlete. However, with the current evidence, it is certainly too early a stage to determine how to use genotyping as a tool for predicting exercise/sports performance or improving current methods of training. Research on this topic presents methodological limitations such as the lack of measurement of valid exercise performance phenotypes that make the study results difficult to interpret. Additionally, many studies present an insufficient cohort of athletes, or their classification as elite is dubious, which may introduce expectancy effects. Finally, the assessment of a progressively higher number of polymorphisms in the studies and the introduction of new analysis tools, such as the total genotype score (TGS) and genome-wide association studies (GWAS), have produced a considerable advance in the power of the analyses and a change from the study of single variants to determine pathways and systems associated with performance. The purpose of the present study was to comprehensively review evidence on the impact of genetics on endurance- and power-based exercise performance to clearly determine the potential utility of genotyping for detecting sports talent, enhancing training, or preventing exercise-related injuries, and to present an overview of recent research that has attempted to correct the methodological issues found in previous investigations.
Keywords: Sports performance, Professional athlete, Genes, Innate factors, Heritability
Introduction
Research focused on ascertaining the effect of genetics on exercise traits has been progressively growing in the last few years, providing information about associations between some genetic polymorphisms and endurance- and power-based exercise performance, among other sports disciplines (Ahmetov and Fedotovskaya 2015; Ahmetov et al. 2021). Although the research in this field is extensive, there is still controversy and debate about the magnitude of the potential effect of genetics on sports performance and about how the influence of positive/negative heritable traits can be overcome by training and diet (Pickering et al. 2019a; Sarzynski et al. 2017; Stephan 2012). Some of this debate is based on the limitations and methodological issues of the research that presents the evidence on this topic (Breitbach et al. 2014). In this regard, studies investigating the effect of genetics on endurance and power sports performance may be confounded by the low sample sizes, incorrect categorization of elite athletes, the lack of measurement of valid exercise performance traits, and the obtaining of outcomes based only upon cross-sectional observations of genotype frequencies between athlete and non-athlete populations (Varillas Delgado et al. 2019, 2020; Varillas-Delgado et al. 2021). Additionally, elite athletes have undergone intensive training and field testing/selection prior to being studied and investigations rarely contain samples of athletes in which prior training has been standardized. In fact, in most of the studies that controlled the training stimuli, the duration of the training was short, and it was oriented to the improvement of fitness rather than to the achievement of high-performance aims (Issurin 2008, 2019). Although it is acknowledged that the standardization of training is unfeasible when studying elite athletes, particularly in studies with large sample sizes and in individual sports, the current evidence may limit the generalizations that can be generated from results of studies in the field of genetics and sports performance (Guth and Roth 2013; Bouchard 2012; Montgomery et al. 1998). As a result, the conclusions provided in the current studies remain approximate, without providing conclusive data, despite the high amount of human and economic resources invested, to clarify how genetics impact sports performance.
Interested readers are directed to review articles on the different links that some genetic polymorphisms and profiles have with sports performance (Joyner and Coyle 2008; Pickering et al. 2019a; Ostrander et al. 2009; Lippi et al. 2010; Storz et al. 2015; Tucker et al. 2013), according to phenotypes in endurance (Ahmetov et al. 2016; Ghosh and Mahajan 2016; Zhou 2014; Vellers et al. 2018; Collins et al. 2016) and power sports (Ghosh and Mahajan 2016; Eynon et al. 2013). There are also interesting and recent review articles documenting the advances made in exercise performance, fitness, and genomics (Wang et al. 2013; Pitsiladis et al. 2013; Sarzynski et al. 2016; Loos et al. 2015a).
The growing knowledge about the role of these genes in the improvement of sports performance and the advances in gene therapy make them likely targets for gene doping approaches (López et al. 2020; Baoutina et al. 2007). Several strategies are currently used in gene therapy. Adeno-associated, virus-derived vectors, lentiviral vectors and CRISPR/Cas9 gene editing are used as gene therapy approaches in several models, such as hematopoietic stem cell gene therapy (Lamsfus-Calle et al. 2020), cochlear gene therapy (György and Maguire 2018), and gene therapy targeting Schwann cells for demyelinating neuropathies (Sargiannidou et al. 2020). These rapid advances in molecular biology techniques open up many possibilities for the development of methods for gene doping detection (López et al. 2020).
However, the purpose of this review is to comprehensively appraise evidence on the impact of genetics on exercise performance to clearly determine if it supports the utility of genotyping to detect sports talent, enhance training or prevent exercise-related injuries, with a specific focus on endurance and power exercise.
Sports performance
Genetic approximations in sports performance
It is well accepted that some innate factors play a key role in sports performance and related phenotypes, such as power, strength, aerobic capacity, flexibility, coordination, and temperament (Naureen et al. 2020). Despite a relatively strong influence of heritability on the probability of becoming an elite athlete (> 70%, depending on the sports discipline) (De Moor et al. 2007) or on the values of phenotypes typically associated with performance (Simoneau and Bouchard 1995; Alonso et al. 2014), the search for genetic variants contributing to a greater predisposition to success in certain sports disciplines has been a challenging task (Holdys et al. 2013; Tucker et al. 2013; Maciejewska-Skrendo et al. 2019b). Sports genomics is a new scientific discipline focusing on the organization and functioning of genetics in elite athletes (Ahmetov and Fedotovskaya 2015). The era of sports genomics began in the 2000s with the discovery of the first genetic markers associated with sports performance; angiotensin-converting enzyme (ACE) (Jones et al. 2002), α-actinin 3 (ACTN3) (Yang et al. 2003), adenosine monophosphate deaminase 1 (AMPD1) (Rubio et al. 2005) and peroxisome proliferator-activated receptor gamma co-activator 1-alpha (PGC1α) (Lucia et al. 2005). With genotyping, sequencing and the use of the deoxyribonucleic acid (DNA) microarray analysis availability, a large number of genetic studies have been published with associations in candidate genes variants with elite athlete status (Ahmetov and Fedotovskaya 2015; Varillas Delgado et al. 2019; Pickering and Kiely 2020). To date, the affirmation that genetic variability contributes to interindividual responses to or during exercise, or to the likelihood of becoming an excellent athlete, is well supported, but when this affirmation is translated to determining which polymorphisms contribute to the phenotype of a champion, the evidence is weaker due to the low number of replication studies. Additionally, genetic predisposition is not the only factor for athletes to become champions and it is necessary to consider multiple environmental and epigenetic variables that confer the final phenotype of elite athlete (Tanisawa et al. 2020).
Several single-nucleotide polymorphisms (SNPs) have been associated with elite endurance and power performance in the last few years, and the polygenic trait is the current paradigm in elite performance, with minor contributions of each variant to the athletic phenotypes, like ACE insertion/deletion (I/D) (rs4646994), ACTN3 c.1729C > T (rs1815739), angiotensinogen (AGT) c.4072 T > C (rs699), AMPD1 c.34C > T (rs17602729), homeostatic iron regulator (HFE) c.187C > G (rs1799945) and c.845G > A (rs1800562), interleukin-6 (IL6) c.-174C > G (rs1800795), endothelial nitric oxide synthase 3 (NOS3) c.-786 T > C (rs1799983) and c.894G > T (rs1799983) have been linked to traits of elite endurance athletes. The polymorphisms in the peroxisome proliferator-activated receptor alpha (PPARα) intron 7 G > C (rs4253778), peroxisome proliferator-activated receptor gamma (PPARδ) c.294 T > C (rs2016520), polypeptide N-acetylgalactosaminyltransferase-like 6 (GALNTL6) last intron C > T (rs558129) and mitochondrial uncoupling protein 2 (UCP2) exon 4 C > T (p.Ala55Val; rs660339) have also been associated with elite performance, but the findings are less consistent (Eynon et al. 2013; Ruiz et al. 2010; Maciejewska-Skrendo et al. 2019b; Ben-Zaken et al. 2019; Díaz Ramírez et al. 2020; Moir et al. 2019). A recent review revealed that, to date, more than 69 genetic markers have been associated with power athlete status (Ahmetov et al. 2021).
A fundamental aspect that may influence the link between sports performance and genetics is the field of nutrigenomics and nutrigenetics, which are experimental approaches that use genetic testing technologies to examine the role of individual genetic differences in adapting an athlete's response to nutritional interventions (Mathers 2017; Reddy et al. 2018). This is important when assessing the influence of genetics on sports performance because an individual's dietary and supplement strategies can influence his/her sports performance (Guest et al. 2019; Thomas et al. 2016). Personalized nutrition in elite athletes based on genetic profile aims to optimize health, body composition, and sports performance (Saunders et al. 2019). Sport dietitians have been adept at implementing additional scrutiny of the dietary recommendations in all general population dietary guidelines to accommodate various sporting populations (Guest et al. 2019; Pickering and Kiely 2018). Genetic differences are known to impact absorption, metabolism, uptake, utilization and excretion of nutrients, and food bioactives may affect several metabolic pathways (Görman et al. 2013). However, to date, the investigations that consider the influence of nutritional factors when analyzing the link between genetics and exercise performance are scarce (Guest et al. 2019). In future, it will be necessary to discover the genetic modifiers of various dietary factors that impact an athlete's nutritional status, body composition and sports performance to optimize these nutritional factors.
Genetics and endurance performance
The ability to perform endurance exercise is associated with the aerobic metabolism or the capacity of using oxygen to produce energy, strongly supported by the enhanced mitochondrial function, gene expression and enzyme activity of elite athletes (Lippi et al. 2010). In a wider perspective, the capacity to perform endurance exercise is influenced by several central factors, relating to muscle and cardiovascular function (Al-Khelaifi et al. 2020; Ahmetov et al. 2015). These include the proportion of slow-twitch fibers in skeletal muscle and factors such as maximal cardiac output which underlies the maximal rate of oxygen consumption (VO2max). Regular endurance exercise training induces major adaptations in skeletal muscle, favoring metabolic consequences of the adaptations of skeletal muscle to endurance exercise. Examples are the slower utilization of muscle glycogen and blood glucose, a greater reliance on fat oxidation as a fuel during exercise, and less lactate production at a given exercise intensity, an enlargement in cardiac dimension, and an increase in blood volume. All of which favors a greater filling of the ventricles and a consequent larger stroke volume and cardiac output (Hellsten and Nyberg 2015), playing an important role in the large increase in the ability to perform prolonged strenuous exercise in response to endurance exercise training (Holloszy and Coyle 1984; Davies and Thompson 1979); but not all of them may be mainly determined by genetics (Varillas-Delgado et al. 2021). Still, it is well accepted that most endurance-related phenotypes are under strong genetic influence, with the length, duration, type, intensity and age of initiation of the training stimulus as additional contributors (Levine 2008). Some of the 40–50% variance in the proportion of slow-twitch fibers in human muscles seems genetically determined (Simoneau and Bouchard 1995), while VO2max and aerobic power also have high heritability (Moir et al. 2019; Miyamoto-Mikami et al. 2018; Zadro et al. 2017). Interestingly, gains in VO2max present a large interindividual variation even in response to standardized exercise training, and it is estimated that – 50% of the change in VO2max with aerobic training is associated with innate factors (Sarzynski et al. 2017). However, to date, the influence of genetics on other phenotypes highly associated with endurance performance such as exercise intensity or VO2 at blood lactate threshold are non-existent, despite the utility of these variables to predict endurance performance has been recognized for several decades (Brooks 1985).
Endurance athlete status remains the most-studied trait because up to 100 genetic variants have been associated with a genetic predisposition (Table 1) (Miyamoto-Mikami et al. 2018; Ahmetov et al. 2021), but still much research is needed to clearly pinpoint the genetic variants that enable better endurance performance or better adaptation to endurance training. This will be a difficult task for future years because of the complexity of all systemic and molecular phenomena associated with endurance performance (Lundby et al. 2017).
Table 1.
Genetic markers for endurance athlete status (Ahmetov et al. 2021)
| Gene | Full name | Locus | Polymorphism | Endurance-related allele, %a |
|---|---|---|---|---|
| ACE | Angiotensin I converting enzyme | 17q23.3 | Alu I/D | I, 53.0 |
| ACTN3 | Actinin alpha 3 | 11q13.1 | rs1815739 C > T | T, 40.1 |
| ADRB2 | Adrenoceptor beta 2 | 5q31-q32 | rs1042713 G > A | A, 47.6 |
| AGTR2 | Angiotensin II receptor type 2 | Xq22-q23 | rs11091046 A > C | C, 52.9 |
| AQP1 | Aquaporin 1 | 7p14 | rs1049305 C > G | C, 54.2 |
| AMPD1 | Adenosine monophosphate deaminase 1 | 1p13 | rs17602729 C > T | C, 96.2 |
| BDKRB2 | Bradykinin receptor B2 | 14q32.1-q32.2 | + 9/-9 (exon 1) | − 9, NA |
| CK-MM | Creatine kinase M-type | 19q13.32 | rs8111989 A > G | A, 66.0 |
| COL5A1 | Collagen type V alpha 1 chain | 9q34.2-q34.3 | rs12722 C > T | T, 34.9 |
| CYP2D6 | Cytochrome P450 family 2 subfamily D member 6 | 22q13.1 | rs3892097 G > A | G, 85.3 |
| FTO | FTO alpha-ketoglutarate dependent dioxygenase | 16q12.2 | rs9939609 T > A | T, 66.0 |
| GABPB1 | GA binding protein transcription factor subunit beta 1 | 15q21.2 | rs12594956 A > C | A, 58.7 |
| rs7181866 A > G | G, 11.1 | |||
| GALNTL6 | Polypeptide N-acetylgalactosaminyltransferase 6 | 4q34.1 | rs558129 T > C | C, 76.8 |
| GSTP1 | Glutathione S-transferase Pi 1 | 11q13.2 | rs1695 A > G | G, 35.3 |
| HFE | Homeostatic iron regulator | 6p21.3 | rs1799945 C > G | G, 7.3 |
| HIF1α | Hypoxia inducible factor 1 subunit alpha | 14q23.2 | rs11549465 C > T | C, 92.7 |
| MCT1 | Monocarboxylate transporter 1 | 1p12 | rs1049434 A > T | T, 67.7 |
| MtDNA loci | Mitochondrial DNA | MtDNA | MtDNA haplogroups | H, NA |
| Unfavourable: K, NA | ||||
| MYBPC3 | Myosin binding protein C3 | 11p11.2 | rs1052373 A > G | G, 52.4 |
| NFATC4 | Nuclear factor of activated T cells 4 | 14q11.2 | rs2229309 G > C | G, 72.0 |
| NFIA-AS2 | NFIA antisense RNA 2 | 1p31.3 | rs1572312 C > A | C, 91.7 |
| NOS3 | Nitric oxide synthase 3 | 7q36 | rs2070744 T > C | T, 76.6 |
| PPARα | Peroxisome proliferator-activated receptor alpha | 22q13.31 | rs4253778 G > C | G, 72.6 |
| PPARGC1α | Peroxisome proliferator-activated receptor, gamma, coactivator 1 alpha | 4p15.1 | rs8192678 G > A | G, 73.4 |
| PARGC1β | Peroxisome proliferator-activated receptor, gamma, coactivator 1 beta | 5q32 | rs7732671 G > C | C, 11.0 |
| PPP3R1 | Protein phosphatase 3 regulatory subunit B, alpha | 2p15 | Promoter 5I/5D | 5I, NA |
| RBFOX1 | RNA binding fox-1 homolog 1 | 16p13.3 | rs7191721 G > A | G, 61.4 |
| SPEG | Striated muscle enriched protein kinase | 2q35 | rs7564856 G > A | G, 46.4 |
| TFAM | Transcription factor A, mitochondrial | 10q21 | rs1937 G > C | C, 8.8 |
| TSHR | Thyroid stimulating hormone receptor | 14q31 | rs7144481 T > C | C, 15.7 |
| UCP2 | Uncoupling protein 2 | 11q13 | rs660339 C > T | T, 41.6 |
| UCP3 | Uncoupling protein 3 | 11q13 | rs1800849 C > T | T, 18.6 |
| VEGFA | Vascular endothelial growth factor A | 6p12 | rs2010963 G > C | C, 32.6 |
| VEGFR2 | Vascular endothelial growth factor receptor 2 | 4q11-q12 | rs1870377 T > A | A, 21.2 |
aThe frequency of endurance-related allele in the general population is shown (1000 Genomes project, n = 2504) (Ahmetov et al. 2021)
Genetics, endurance performance and cardiovascular disease
Several genes have been associated with endurance sports and phenotypes in terms of muscle functioning (ACTN3 and the c.1729C > T polymorphism) or blood pressure regulation (ACE and I/D polymorphism) with substantially high replication in multiple cohorts. Specifically, several studies have found that elite endurance athletes with the TT genotype in the ACTN3 or with the DD genotype in the ACE are more frequent than in control populations of untrained individuals or have higher values of variables associated to endurance exercise performance than their counterparts with TT/CT or II/ID genotypes (Ma et al. 2013; McAuley et al. 2021; Vaughan et al. 2016; Miyamoto-Mikami et al. 2018); however, other studies have not found any effect of the ACTN3 and ACE genotypes on endurance exercise performance (Papadimitriou et al. 2018; Mägi et al. 2016). Additionally, the genetic component for the observed variability of muscle fiber types in humans is ~ 40–50%, indicating that muscle fiber-type composition is determined equally by genotype and environment (Ahmetov et al. 2012). Several polymorphisms of genes involved in the calcineurin-NFAT pathway, mitochondrial biogenesis, glucose and lipid metabolism, cytoskeletal function, hypoxia and angiogenesis, and circulatory homeostasis have been associated with fiber-type composition (Ahmetov et al. 2012; Eynon et al. 2011a; Maciejewska-Skrendo et al. 2019a; Mustafina et al. 2014). Many of these gene variants in muscle have been associated with sports performance and the elite athlete status, as well as with metabolic and cardiovascular diseases (Houweling et al. 2018). Genetic variants associated with fiber-type proportions have relevant implications for the understanding of muscle performance (Mustafina et al. 2014; Hughes et al. 2011; Ahmetov et al. 2012).
VO2max is a strong prognostic factor of morbidity and mortality from all causes and, particularly, from cardiovascular disease (CVD) (Lavie et al. 2019). Numerous genetic association studies have been performed for other CVD risk factors, such as poor diet, high blood pressure and cholesterol, stress, smoking and obesity, revealing new therapeutic targets (Montefiori et al. 2018). VO2max has a large genetic component but association studies are still lacking (Bye et al. 2020). Identification of genes associated with VO2max would lead to a better understanding of this complex trait between VO2max and CVD associated with endurance performance (Bouchard et al. 2000; Rico-Sanz et al. 2004; Ahmetov et al. 2015; Al-Khelaifi et al. 2020). However, to date, studies are limited in size and employ prespecified genomic associations, limiting the discovery of new genetic markers (Bye et al. 2020). A large-scale systematic screening of genetic variants associated with directly measured VO2max in the athlete population is still needed (Bray et al. 2009). So far, the lack of studies directly measuring VO2max in high-performance athletes has limited the possibilities to associate with this phenotype.
Genetics and interindividual difference in response to aerobic training
The response to an exercise intervention is often described, with the assumption that average is a typical response for most groups of athlete populations. However, the phenomenon of “high responders” and “low responders” following a standardized training intervention may provide helpful insights into mechanisms of training adaptation and methods of individualized training prescription (Fyfe et al. 2014). Genetics makes an important contribution to individual variation in training responses, like change in body mass index (Bae et al. 2007), body composition (Duoqi et al. 2015), improvement in insulin activity for glucose utilization during exercise (Weiss et al. 2005), a favorable response to endurance training for diastolic blood pressure (DBP) and mean arterial pressure (MAP) (Jayewardene et al. 2016; Rankinen et al. 2000), and improvement of VO2max following endurance exercise training (Timmons et al. 2010). The association between genotype and training response has been supported using heritability estimates; however, recent studies have shown variation in some training responses to specific SNPs (Mann et al. 2014; He et al. 2007, 2010; Rice et al. 2012; Thomaes et al. 2011). Heritability is often expressed through the influence of innate factors on the pre-training phenotype, with some parameters showing a hereditary effect on the pre-training phenotype but not on the subsequent training response (Mann et al. 2014). Individual variation in response to standardized training that cannot be explained by genetic influences may be related to the characteristics of the training program, compliance with the program or differences in lifestyle factors among individuals (Mann et al. 2014; Ahtiainen et al. 2020; Zubair et al. 2019). Interestingly, the level of recovery between exercise sessions in endurance athletes has recently been shown to be related to liver-metabolizing genes, such as Cytochrome P450 2D6 (CYP2D6) and other genes encoding the family of G-glutathione transferases (Varillas Delgado et al. 2019), suggesting that future studies should associate genetic variants with the capacity of recovery by scavenging free radicals, or the neutralization of pH (Lewis et al. 2015). In this context, a recent study of interest by Varillas et al. (Varillas Delgado et al. 2019) presented the influence of polymorphisms in P450 cytochrome gene (i.e., c.506-1G > A polymorphism in the CYP2D6; formerly 1846G > A: rs3892097) and the glutathione S-transferase family (“null polymorphism” in GSTM, GSTP p.Ile105Val polymorphism (rs1695) and “null polymorphism” GSTP), and they found that the frequency of “favorable” variants in these genes was different among endurance athletes compared to a control population, which was confirmed by different scores in a total genotype score (TGS) model. Such information on these hepatic variables would be of interest to promote genetic scoring through various genes involved in recovery metabolism.
Some individuals may develop chronic fatigue and even maladaptation, due to imbalance between overall stress and recovery, contributing to variation in pre–post training responses (Doma et al. 2019; Mann et al. 2014). Training response can be modulated by the timing and composition of dietary intake, and hence nutritional factors could also potentially contribute to this individual variation (Close et al. 2016; van Loon and Tipton 2013). For example, acute ingestion of the phytochemical caffeine is a dietary factor that increases endurance performance with considerable interindividual differences (Guest et al. 2021). Future genetic studies that associate caffeine ergogenicity, genetics and interindividual responses may help to enhance the use of caffeine supplementation to improve sports performance in a more individualized manner (Gutiérrez-Hellín and Varillas-Delgado 2021). Finally, a certain amount of individual variation in responses to training may also be attributed to measurement error, a factor that should be accounted for wherever possible in future studies, to clarify and quantify the role of these factors with the ultimate goal of improving the prediction of training responses to endurance exercise and to provide further insight into the mechanisms of aerobic-based training adaptations (Fig. 1).
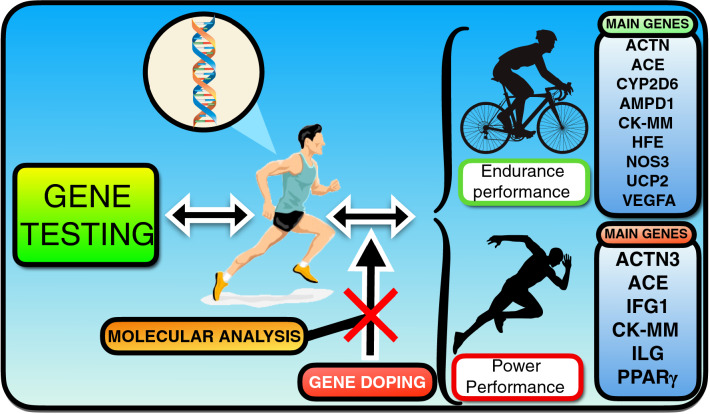
Fig. 1.
Schematic of genetic differences in power and endurance performance to identify talent for sports based on DNA testing. Knowledge of genetics facilitates gene doping, using the rapid advances in molecular biology techniques for the future development of methods to detect this doping modality. ACE angiotensin-converting enzyme, ACTN3 α-actinin 3, AMPD1 adenosine monophosphate deaminase 1, CK-MM creatine kinase isoenzyme MM, CYP2D6 cytochrome P450 family 2 subfamily D member 6, HFE homeostatic iron regulator 6, IGF1 insulin-like growth factor 1, IL6 interleukin 6, NOS3 endothelial nitric oxide synthase 3, PPARγ Peroxisome proliferator-activated receptor gamma, UCP2 uncoupling protein 2, VEGFA vascular endothelial growth factor A
Genetics and power performance
Genetics strongly influences the ability of skeletal muscle to produce force at a high velocity, crucial for success in power and sprint-based sports (Eynon et al. 2013; Ruiz et al. 2010).
To date, the genetic profile of elite athletes still remains poorly characterized, and it is likely that each SNP makes a limited contribution to an elite athletic phenotype (Ahmetov et al. 2016). The genetic profile in elite-level power/sprint performance is different from that of elite endurance performance (Eynon et al. 2011b; Maciejewska-Skrendo et al. 2019b; Buxens et al. 2011). The metabolic and biochemical phenotypes are polar opposites to perform in endurance or power/sprints endurance events (Eynon et al. 2013). Elite endurance performance requires sustained muscular contraction over a long period of time, using a high VO2max and other traditional endurance capabilities involved in the mitochondrial respiratory chain (Miyamoto-Mikami et al. 2018; Hagberg et al. 2001; Greggio et al. 2017). However, short-distance sprint and power performance require high speed and forceful muscle contraction, depending on the anaerobic pathways, using intramuscular stores of creatine phosphate (CP) and adenosine triphosphate (ATP) as the main substrate for energy production (Spencer and Gastin 2001; Praagh 2007; Sousa et al. 2017).
Recent studies have investigated the link of the myostatin gene (MSTN) with exercise performance. Within the sequence of this gen, the p.K153R polymorphism has been deemed as a genetic variant to influence skeletal muscle phenotypes as the MSTN RR variant is more frequent top level endurance runners than in national-level counterparts (Ben-Zaken et al. 2015c). Additionally, the combination of the R allele in the MSTN gene and T allele in the interferon gamma-1 (IGF-1) c.-1245C > T (rs35767) polymorphism The IGF c.1245C > T (rs35767) has been associated with higher circulating IGF-I levels, greater muscle mass and improved endurance performance (Ben-Zaken et al. 2017). Also, the MSTN c.163G > A (p (rs180565) polymorphism plays an important role for the stability of MSTN propeptide inhibitory activity (Ferrell et al. 1999), showing significantly different allele frequencies in Caucasians and African Americans, making it possible to explain the difference in muscle mass and strength among power athletes in these ethnicities. These allelic variants in MSTN provide markers for examining the association between the myostatin gene and interindividual variation in muscle mass and differences in loss of muscle mass with aging (Ferrell et al. 1999). The MSTN polymorphism could be a potential candidate that can positively affect the skeletal muscle phenotype after power exercise. However, previous myostatin SNPs studies mainly observed chronic effects such as muscle hypertrophy after exercise related to ethnicity (Li et al. 2014; Kostek et al. 2009), and there was no study about acute response such as muscle damage.
The genetic influence on elite power/sprint performance has received less scientific attention unlike the genetics of endurance performance. Only a few studies have characterized the associations between genetic variants and elite power/sprint performance (Table 2) (Ruiz et al. 2010; Eynon et al. 2013; Ben-Zaken et al. 2015b, 2019; Ahmetov et al. 2016, 2021; Pickering et al. 2019b). Several studies have reported that muscle power and muscle strength are important for power/sprint performance, involving a strong genetic influence (Calvo et al. 2002; Thomis et al. 1998a, b; Suchomel et al. 2018).
Table 2.
Genetic markers for power athlete status (Ahmetov et al. 2021)
| Gene | Full name | Locus | Polymorphism | Power-related allele, %a |
|---|---|---|---|---|
| ABHD17C | Abhydrolase domain containing 17C | 15q25.1 | rs7165759 G > A | A, 27.2 |
| ACTG1 | Actin gamma 1 | 17q25.3 | rs6565586 T > A | A, 45.3 |
| ACTN3 | Actinin alpha 3 | 11q13.1 | rs1815739 C > T | C, 59.9 |
| ADCY3 | Adenylate cyclase 3 | 2p23.3 | rs10203386 T > A | T, 52.0 |
| ADPGK | ADP dependent glucokinase | 15q24.1 | rs4776614 C > G | C, 53.8 |
| AGT | Angiotensinogen | 1q42.2 | rs699 T > C | C, 70.5 |
| ANGPT2 | Angiopoietin 2 | 8p23.1 | rs890022 G > A | A, 10.0 |
| ARPP21 | CAMP regulated phosphoprotein | 21 3p22.3 | rs1513475 T > C | C, 37.1 |
| BCDIN3D | Bicoid interacting 3 domain containing RNA methyltransferase | 12q13.12 | rs12367809 C > T | C, 75.7 |
| CK-MM | Creatine kinase, M-type | 19q13.32 | rs8111989 A/G | G, 34.0 |
| CNTFR | Ciliary neurotrophic factor receptor | 9p13.3 | rs41274853 C > T | T, 17.9 |
| CRTAC1 | Cartilage acidic protein 1 | 10q24.2 | rs563296 G > A | G, 55.2 |
| DHODH | Dihydroorotate dehydrogenase (quinone) | 16q22.2 | rs12599952 G > A | A, 43.1 |
| GALNTL6 | Polypeptide N-acetylgalactosaminyltransferase like 6 | 4q34.1 | rs558129 C > T | T, 23.2 |
| GBE1 | 1,4-alpha-glucan branching enzyme 1 | 3p12.2 | rs9877408 A > G | A, 42.7 |
| GBF1 | Golgi brefeldin a resistant guanine nucleotide exchange factor 1 | 10q24.32 | rs2273555 G > A | A, 59.6 |
| HIF1α | Hypoxia inducible factor 1 alpha | 14q21-q24 | rs11549465 C > T | T, 7.3 |
| IGF1 | Insulin like growth factor 1 | 12q23.2 | rs35767 C > T | T, 30.4 |
| IL6 | Interleukin 6 | 7p21 | rs1800795 C > G | G, 85.9 |
| ITPR1 | Inositol 1, 4, 5-triphosphate receptor type 1 | 3p26.1 | rs901850 G > T | T, 11.2 |
| KIF1B | Kinesin family member 1B | 1p36.22 | rs11121542 G > A | G, 92.1 |
| LRPPRC | Leucine-rich pentatricopeptide repeat cassette | 2p21 | rs10186876 A > G | A, 67.9 |
| MLN | Motilin | 6p21.31 | rs12055409 A/G | G, 51.7 |
| MMS22L | Methyl methanesulfonate-sensitivity protein 22-like | 6q16.1 | rs9320823 T > C | T, 31.1 |
| MTHFR | Methylenetetrahydrofolate reductase | 1p36.3 | rs1801131 A > C | C, 24.9 |
| NPIPB6 | Nuclear pore complex interacting protein family member B6 | 16p12.1 | rs2726036 A > C | A, 70.4 |
| PHACTR1 | Phosphate and actin regulator 1 | 6p24.1 | rs6905419 C > T | C, 70.9 |
| PLEKHB1 | Pleckstrin homology domain containing B1 | 11q13.4 | rs7128512 A > G | G, 74.9 |
| PPARAα | Peroxisome proliferator-activated receptor alpha | 22q13.31 | rs4253778 G > C | C, 27.4 |
| PPARγ | Peroxisome proliferator-activated receptor gamma | 3p25.2 | rs1801282 C > G | G, 7.0 |
| PPARGC1α | Peroxisome proliferator-activated receptor, gamma, coactivator 1 alpha | 4p15.2 | rs8192678 G > A | A, 26.6 |
| R3HDM1 | R3H domain containing 1 | 2q21.3 | rs6759321 G > T | T, 80.7 |
| RASGRF1 | Ras protein specific guanine nucleotide releasing factor 1 | 15q25.1 | rs1521624 C > A | A, 56.9 |
| RMC1 | Regulator of MON1-CCZ1 | 18q11.2 | rs303760 C > T | C, 82.9 |
| SLC39A8 | Solute carrier family 39-member 8 | 4q24 | rs13135092 A > G | A, 97.5 |
| TFAP2D | Transcriptional factor AP-2 delta | 6p12.3 | rs56068671 G > T | T, 6.1 |
| ZKSCAN5 | Zinc finger with KRAB and SCAN domains 5 | 7q22.1 | rs3843540 T > C | C, 40.2 |
| ZNF608 | Zinc finger protein 608 | 5q23.2 | rs4626333 G > A | G, 76.6 |
aThe frequency of power-related allele in the general population is shown (1000 Genomes project, n = 2504). (Ahmetov et al. 2021)
A previous study by Ruiz et al. (Ruiz et al. 2010) using six polymorphisms in candidate genes (ACE, ACTN3, AGT, GDF-8, IL6, and NOS3) and adding the influence of all variants in a TGS model, identified a polygenic profile to distinguish elite power athletes from both endurance athletes and nonathletic individuals, reducing the possibility of finding individuals with a “perfect” TGS, while increasing the predictive accuracy of the model. Moreover, it must be kept in mind that, beyond genetic endowment or complex gene–gene and gene–environment interactions, there are other numerous contributors to the “complex trait” of being an athletic champion, such as motivation, socioeconomic factors, or simply opportunity (Ruiz et al. 2010). Hence, the possession of a high TGS for power performance does not guarantee success in power-based sports.
The inter-individual difference is the ergogenic response to acute caffeine intake has been linked to variants in the cytochrome P450 1A2 (CYP1A2) gene, specifically to the c.-163C > A (rs762551) polymorphism. Although there is evidence to support that athletes with the AA genotype in this gene obtain greater benefits from acute caffeine intake (Guest et al. 2018), most evidence suggests that AA and C-allele carriers equally benefit from caffeine supplementation in several exercise and sports scenarios (Del Coso et al. 2012; Muñoz et al. 2020; Puente et al. 2018). However, to date, most evidence suggests that the ergogenic effect of caffeine on neuromuscular performance may be independent of the CYP1A2 genotype (Barreto et al. 2021).
Genetics and muscle performance
Mammalian skeletal muscle is an extremely heterogeneous tissue which is essential for its function. Differences in the mechanical and energetic properties of isolated mammalian slow-twitch and fast-twitch muscle fibers have been well documented (Lippi et al. 2010; Schiaffino and Reggiani 2011). Slow-twitch fibers are slower, less powerful and more economical in terms of energy at force generation than fast-twitch muscle fibers. Furthermore, the peak efficiency of slow-twitch muscle fibers occurs at slower shortening speeds than that of fast-twitch muscle fibers (Bottinelli and Reggiani 2000). This heterogeneity covers all possible aspects of muscle contractile function and is directed at optimizing the contractile responses and performing different motor tasks, minimizing fatigue. Endurance capacity has been related to a predominance of slow-twitch fibers (> 50%) (Wilson et al. 2012), whereas fast-twitch fibers are related to power and speed capacity (Fitts et al. 1991). Power and sprint athletes have a high proportion of fast-twitch muscle fibers with low oxidative capacity compared to endurance athletes (Bergh et al. 1978).
Several polymorphisms in target genes codifying muscle proteins have been linked to muscle performance: The creatine kinase isoenzyme MM (CK-MM) gene encodes the cytosolic muscle isoform of creatine kinase responsible for the rapid regeneration of ATP during intensive muscle contraction and plays a vital role in the energy homeostasis of muscle cells (Echegaray and Rivera 2001). Within the numerous polymorphisms located in the CK-MM gene (Chen et al. 2017), the c.*800A > G polymorphism (rs8111989), located in the 3 'untranslated region of the gene has been previously associated with the status of elite athlete (Eider et al. 2015; Lucía et al. 2005; Rivera et al. 1997b; Chen et al. 2017). Overall, the distribution of the AA genotype and the A allele is more frequent in elite athletes (Grealy et al. 2015; Zhou et al. 2006), while the A allele can even affect the increase in aerobic power induced by endurance training (Rivera et al. 1997a). Further investigation is warranted, in this is as a creatine kinase and is a central controller of cellular energy homeostasis. In this case, the study of the influence of variants in the CK-MM gene on exercise performance should be amplified by investigating other forms of creatine kinase as the mitochondrial isoenzyme of creatine kinase (MtCK) (Schlattner et al. 2006). Specifically, because MtCK interferes with the regulation of oxidative phosphorylation, as well as mitochondrial permeability transition (Calbet et al. 2020) and polymorphisms in the gene codifying this protein may produce different response to situations of energy stress (e.g., endurance exercise).
The ACTN3 gene codifies α-actinin-3, a component of the contractile machinery in fast skeletal muscle fibers in mammals with a highly conserved sequence (Eynon et al. 2013; Lippi et al. 2010). The ACTN3 RR variant is nearly always present in elite power athletes, whereas homozygosity in the 577X allele, associated with a premature stop codon that produces complete α-actinin-3 deficiency, is prevalent in elite endurance athletes, such as marathon runners and rowers (Yang et al. 2003; Shang et al. 2010; Gutiérrez-Hellín et al. 2021; Ben-Zaken et al. 2015a). The ACTN3 XX variant shows reduced fast fiber diameter, increased activity of multiple enzymes in the aerobic metabolic pathway, altered contractile properties and enhanced recovery from fatigue (Broos et al. 2012). The presence of α-actinin-3 has a globally beneficial effect on the function of skeletal muscle in generating forceful contractions at high velocity, related to increased sprint performance. Independent studies, however, failed to demonstrate a significant association between this ACTN3 polymorphism and ultra-endurance performance (Lucia et al. 2006; Houweling et al. 2018; Baltazar-Martins et al. 2020).
Myosin light chain kinase, a calcium-calmodulin-dependent multi-functional enzyme, plays a critical role in the regulation of smooth muscle contraction (Del Coso et al. 2016). Polymorphisms in the gene that codifies the myosin light chain kinase (MYLK) (C49T and C37885A) have been associated to the level of exercise induced muscle damage. CA heterozygotes for the C37885A polymorphism presented lower reductions in leg muscle power and lower increases in serum creatine kinase after a competitive marathon than CA runners (Del Coso et al. 2016). However, just the contrary has been also suggested with CA individuals presenting higher markers of muscle damage than CC after elbow flexion eccentric exercise (Clarkson et al. 2005). Also, individuals who were TT homozygotes for C49T had a significantly greater increase in creatine kinase and myoglobin after exercise when compared with CT and CC counterparts (Huang et al. 2018; Shen et al. 2015). Collectively, it seems that the variations in the sequence of the MYLK gene may contribute to skeletal muscle’s properties to endure muscle contraction. Still, more research is needed to warrant the identification of MYLK variants that produce beneficial muscle phenotypes and to determine if exercise training may overcome the potential influence of the variants in the MYLK gene on muscle performance.
The ACE gene has two alleles: the deletion (D) allele of the human ACE is habitually associated with higher activity of the enzyme angiotensin-converting enzyme than the insertion (I) allele in both tissue (Yan et al. 2018) and serum (Zhang et al. 2017). This genetic variation might be associated with many heritable traits, including skill parameters, and physiological and sports performance (Moran et al. 2006; Scott et al. 2005), showing an increased frequency of the ACE I allele in elite endurance athletes (Myerson et al. 1999; Nazarov et al. 2001; Amir et al. 2007) and increased frequency of the ACE D allele in elite sprint and power performance (Myerson et al. 1999; Costa et al. 2009). The mechanism underlying the association of the D allele with power-oriented, anaerobic sports is probably mediated through differences in skeletal muscle strength gain, since a greater training-related increase in quadriceps muscle strength has been associated with the D allele, while the I allele may influence endurance performance through improvements in substrate delivery and the efficiency of skeletal muscle, with subsequent conservation of energy stores (Moran et al. 2006; Nazarov et al. 2001; Lippi et al. 2010).
The AMP deaminase 1 gene is an important regulator of energy metabolism in the muscle fiber that catalyzes the irreversible deamination of adenosine monophosphate (AMP) to inosine monophosphate (IMP) and is one component of the purine nucleotide cycle (Van den Berghe et al. 1992). Subjects with the TT genotype in c.34C > T in the gene that codifies this protein (AMPD1) have diminished exercise capacity and cardiorespiratory responses to exercise when compared to C-allele carriers (Varillas Delgado et al. 2020; Rico-Sanz et al. 2003). Moreover, carriers of the T allele have a limited training response of ventilatory phenotypes during maximal exercise (Rico-Sanz et al. 2003; Rubio et al. 2005; Gineviciene et al. 2014) and a reduced submaximal aerobic capacity (Gineviciene et al. 2014; Lucia et al. 2009).
The combination of various polymorphisms has recently been studied to help in the understanding of muscle performance. In this case, several studies show the association of ACE and ACTN3 is a better explanation of muscle performance. A recent study by Wagle et al. (Wagle et al. 2021) suggested the potential combined influence of ACTN3 RR and ACE DD polymorphisms on isometric and dynamic strength testing. This study may serve as a framework to generate hypotheses regarding the effect of genetics on performance, suggesting that specific genetic profiles in these polymorphisms might influence human physical performance (Ma et al. 2013; Papadimitriou et al. 2016, 2018; Eynon et al. 2009). This current knowledge of ACE and ACTN3 should be expanded in future studies of cohorts of athletes with the other genes involved in muscle performance; CK-MM, MYLK and AMPD1 using TGS methodology to show the additive or synergistic influence of these polymorphisms in sports performance in both endurance and power modalities.
Gene doping
Athletes have long been using substances to improve their sports performance (De Rose 2008; Cantelmo et al. 2020). The use of doping substances had the purpose of boosting performance in various parts of the ancient world, and some strategies persist nowadays (Verroken 2000; Palmi et al. 2019; La Gerche and Brosnan 2018). With the evolution of knowledge about the organism’s physiological dynamics, new ways have emerged of achieving enhanced performance through gene manipulation. There is the possibility of manipulating human genetic material and regulating gene expression to increase or decrease the production of certain enzymes and other proteins associated with processes that are key for human performance (Cantelmo et al. 2020; Unal and Ozer Unal 2004). The genetic material could be manipulated using normal or genetically modified cells including transferring nucleic acid polymers or their analogues (Brzeziańska et al. 2014; Heuberger and Cohen 2019). This gene manipulation arose with the advent of gene therapy, which has been practiced and improved for the treatment of severe human diseases over the last 30 years (Baoutina et al. 2013).
Increased athletic performance through genetic manipulation could be achieved with the development of the clustered regularly interspaced short palindromic repeats with DNA endonuclease Cas9 (CRISPR/Cas9) methodology (Pokrywka et al. 2013). To restrain this practice, the World Anti-Doping Agency (WADA) added “gene doping” to the “Prohibited List” in 2004. However, the 2018 version expanded the gene doping ban by adding to the list of banned substances and methods any gene editing agents designed to alter genome sequences and/or the transcriptional or epigenetic regulation of gene expression. Nevertheless, to date, it is unclear how WADA will enforce such rules (Thevis et al. 2019) or if it has the capacity to detect any type of gene modification with the current methodology for obtaining samples.
Gene therapy was defined by Haisma et al 2006 as the transfer of genetic material to human cells for treatment or prevention of a disease. In this regard, the genetic materials that can be transferred are DNA, ribonucleic acid (RNA) or genetically altered cells (Haisma and de Hon 2006). This therapy still poses numerous limitations and risks, such as the patient’s immune response and consequent treatment rejection (Haisma and de Hon 2006; Gaffney and Parisotto 2007; Cantelmo et al. 2020). To date, no athlete has yet been formally accused of using gene therapy as a form of doping, maybe because there is no official confirmatory test for this therapy, which is still in the early stages of development (Paßreiter et al. 2020). Indeed, until recently, there has been no efficient gene-editing system until the discovery of the CRISPR technique (Jinek et al. 2012), which shows severe side effects related to gene therapy (van der Gronde et al. 2013). To date, we can only speculate which genes would be the best candidates for gene doping according to their potential to boost sports performance, as in the case of those that encode erythropoietin (EPO), myostatin blockers, the insulin-like growth factor (IGF-1), growth hormone (GH), vascular endothelial growth factor (VEGF), fibroblast growth factor (FGF), ACTN3, cytosolic phosphoenolpyruvate carboxykinase (PEPKC), and peroxisome proliferator-activated receptor-δ (PPARδ) (Cantelmo et al. 2020; Azzazy et al. 2005; Haisma and de Hon 2006; Hakimi et al. 2007; van der Gronde et al. 2013).
Of special interest is the success of the BNT162b2 mRNA-based vaccine against the SARS-CoV-2 virus, a promising recent development in the regulation of transgene expression (Wang et al. 2020) with ramifications to other research fields. In fact, there is evidence of the success of viral EPO gene transfer using adenovirus, adeno-associated virus, and retrovirus in rodents and non-human primates (Neuberger et al. 2012; Segura et al. 2007) with the purpose of increasing red blood cell production and hemoglobin concentration to improve endurance performance, providing experimental data to support the potential use of gene therapy employing adenovirus or retrovirus vectors as an effective and difficult to control form of gene doping.
Both the scientific and the sports communities have yet to reach a consensus regarding banning or regulating gene therapy in sports and, consequently, gene doping. Much still needs to be studied, developed, and discussed, but what seems to be decisive to resolve this question is to enforce some limits. Defining which conduct should be discouraged or regulated, elaborating effective and applicable rules in the scientific environment, and being able to detect undesirable behaviors with the purpose of exposing and punishing those responsible for these doping methods, are policies that still need to be developed in terms of gene doping (Solomon et al. 2009; Cantelmo et al. 2020).
The future in genetics and sports performance
Most current knowledge in sports genetics has been generated from candidate gene analysis using small sample sizes (typically, a few hundred) (Wang et al. 2013), presenting new candidate genes every year in various physiological pathways that increase knowledge of sports performance (Varillas Delgado et al. 2019). Candidate gene association studies have often produced inconclusive results. Therefore, the vast majority of the candidate genes for sporting performance discovered to date are not strongly associated with phenotypes of interest (Wang et al. 2013), except for a few exceptions, such as ACTN3 and ACE. Genetics and other innate factors represent just a portion of the likelihood of becoming an excellent athlete, but at the same time, genetics provides useful insights, as sports performance can be ultimately defined as polygenic traits intimately associated with the characteristics of the sport (Lippi et al. 2010). It is commonly accepted that there will be many genes involved in sports performance (Ahmetov and Fedotovskaya 2015; Ahmetov et al. 2016).
Exercise training regulates the expression of genes encoding various enzymes in muscle and other tissues, and genetic research in sports will help to clarify several aspects of human biology and physiology, such as RNA- and protein-level regulation under specific circumstances (Lippi et al. 2010). Although an individual’s potential for excelling in endurance or power performance can be partly predicted on specific genetic variants, the contribution of complex gene–gene interactions, environmental factors and epigenetic mechanisms are also important contributors to the “complex trait” of being an athletic champion (Buxens et al. 2011). A combination of the fields of genomics, epigenomics and transcriptomics along with improved bioinformatic tools, in addition to precise phenotyping, is required for future research to understand the inter-relations of exercise physiology, sports performance and susceptibility to diseases (Ehlert et al. 2013; Eynon et al. 2011b).
DNA polymorphisms (1% frequency or greater) (Takahashi and Tajima 2017) and rare DNA mutations (< 1% frequency) (Sloan et al. 2018) can generally be classified as genetic markers associated with endurance, power and strength (or combined power/strength). It should be noted that other possible athlete statuses involving coordination and flexibility have still not been studied. Although most of the genetic variants associated with sports performance seem to be implicated in musculoskeletal and cardiopulmonary functions, genes associated with enhanced functions of the central nervous system (CNS) may also contribute to elite athletic traits (Kitazawa et al. 2021). The significance of a particular sport-related genetic marker is based on several criteria, such as the type of the polymorphism (stop loss/gain, frameshift, missense, synonymous, 3′/5′-UTR, intronic, non-coding RNA, 5′/3′-near gene, intergenic, etc.) (Ergun and Oztuzcu 2016; Calabrese et al. 2020; Hon et al. 2017; Ahmetov et al. 2016).
Despite the obvious role of genetics in athletic performance, there is little unequivocal evidence in support of a specific genetic variant with a major effect on a relevant sports performance phenotype. This is certain at least across the normal range of human trait distributions because sports performance phenotypes are complex traits and fundamentally polygenic (numerous genes with small effects) (Loos et al. 2015b; Sarzynski et al. 2016; Muniesa et al. 2010; Lippi et al. 2010), or because researchers failed to take into consideration the full range of environmental effects, or both (Rice et al. 1997; Aaltonen et al. 2020; Ghosh and Mahajan 2016). It is very important to note that each DNA locus can probably explain a very small proportion of the phenotypic variance (0.1–1%) (Timmons et al. 2010; Lippi et al. 2010). Therefore, very large sample sizes are needed to detect associations, and various combinatorial approaches should be used.
Considering the low predictive capacity of findings with SNPs, researchers have turned to TGS, whereby several elite athlete-associated SNPs are combined into a single polygenic score. This method was developed by (Williams and Folland 2008). (Ruiz et al. 2010; Ruiz et al. 2009) and recently Varillas et al. (Varillas Delgado et al. 2019, 2020) utilized this approach with elite endurance and power athletes. Furthermore, whilst a TGS may help in discriminating between athlete and non-athlete populations, Santiago et al. (Santiago et al. 2010) demonstrated that, in a group of rowers, it did not distinguish between different levels of performance (i.e., World vs. National medalists). An earlier study by (Hughes et al. 2011) demonstrates that there is considerable similarity in polygenic scores in humans (athletes and non-athletes alike) when a low number of SNPs are used, such that again, this approach would likely have limited real world specificity and sensitivity. Lastly, the configuration of TGS to differentiate between populations of athletes usually confers the same score on each SNP while the individual influence of each gene on the status of elite athlete is likely uneven, with some genetic variants exerting a greater influence than others. For this reason, the weighted-total genotype score (w-TGS) model presented by (Varillas Delgado et al. 2020) is a score of genes involved in energy and iron metabolism, suggesting that the relative weighting of the influence of each SNP may add further value in the development of increasingly accurate TGS. As such, further work should seek to identify additional SNPs associated with elite athlete status across a variety of sporting phenotypes and replicate existing SNPs (Pickering et al. 2019a). New genotyping/sequencing technologies are developing rapidly, allowing a more thorough search across the whole human genome (Ng and Kirkness 2010; Boulygina et al. 2020). The genome-wide association studies (GWAS) represent a promising and productive way to study sports-related genotypes in the future (Boulygina et al. 2020; Al-Khelaifi et al. 2020; Ahmetov et al. 2015). This new approach involves rapidly scanning > 5 million markers in complete sets of DNA, using microchips from many individuals to find DNA polymorphisms associated with a particular trait (Ahmetov et al. 2016; Dehghan 2018; Flanagan 2015). One of the advantages of GWAS is that it is unbiased concerning genomic structure and previous knowledge of the trait (hypothesis-free), in contrast to candidate gene studies, where knowledge of the trait is used to identify candidate loci contributing to the trait of interest (Ahmetov et al. 2016; Wang et al. 2013; Al-Khelaifi et al. 2020; Flanagan 2015). However, the use of hypothesis-free models in GWAS contributes to the identification of genetic variants linked to exercise and endurance performance with no mechanistic bases to support how the variant may modify a specific trait. Despite this limitation, GWAS facilitated by high-throughput genotyping technologies have been enormously successful in identifying single-nucleotide polymorphisms (SNPs) that are associated with complex traits (Gallagher and Chen-Plotkin 2018; Visscher et al. 2017). For this reason, GWAS studies should be able to aid in resolving the low predictive capacity of current genetic research (Al-Khelaifi et al. 2019; Eynon et al. 2011b). The systematic review present by Ahmetov et al. shows 41 markers identified in the last 2 years by performing GWAS on African American, Jamaican, Japanese, and Russian athletes, indicating that GWAS represent a promising and productive way to study sports-related genotypes (Ahmetov et al. 2016). Of note, 31 genetic markers have shown positive associations with athlete status in at least 2 studies and 12 of them in 3 or more studies. Conversely, the significance of 29 markers was not replicated in at least 1 study, raising the possibility that several findings might be false positives. These results open up a path for future research including multicenter GWAS and whole-genome sequencing in large cohorts of athletes with further validation and replication, contributing to the discovery of large numbers of the causal genetic variants (mutations and DNA polymorphisms) that would partly explain the heritability of athlete status and related phenotypes (Al-Khelaifi et al. 2020; Ahmetov et al. 2015).
To date, there are serious and well-placed concerns about the use of genetic information, either alone or in combination with existing measures, for talent identification in sport (Webborn et al. 2015; Guth and Roth 2013). It is generally considered that, in sporting contexts, genetic testing should not be carried out on athletes younger than 18 years of age (Pickering et al. 2019a; Webborn et al. 2015). According to the current state of knowledge, no child or young athlete should be exposed to direct-to-consumer (DTC) genetic testing to define or alter training or for talent identification aimed at selecting gifted children or adolescents. Large-scale collaborative projects may help to develop a stronger scientific foundation on these issues in the future. There remains a lack of universally accepted guidelines and legislation for DTC testing regarding all forms of genetic testing and not just for talent identification (Webborn et al. 2015). It is advocated that talent identification and development programs should be dynamic and interconnected taking into consideration maturity status and the potential to develop rather than excluding children at an early age, and more representative real-world tasks should be developed and employed in a multidimensional design to increase the efficacy of talent identification and development programs (Vaeyens et al. 2008).
According to a recent review by Joyner (Joyner 2019), we are still a long way from developing a complete understanding of the genetic influence on sports performance, and in the future, we might be able to model it using field tests or even with data from real competitions, which could be more predictive of an athlete’s performance that current approaches by SNP and TGS studies. Limitations in the methodologies used in studies of genetic association and sports performance could be overcome using GWAS with larger sample sizes and by increasing the internal and external validity of the associations between genetics and sport-specific phenotypes. In addition, the use of algorithms through machine learning-based model for prediction, as in the detection of diseases (Heo et al. 2019; Kalafi et al. 2019; Uddin et al. 2019), would give us answers to the doubts that still exists in the field of genetics and sports performance for the detection of talents based on DNA testing. Still, it is important to mention that the use of DNA testing will never outperform the prediction capacity of a field test, but it can be used together with field and laboratory testing to enhance the likelihood of detecting sports talents or athletes more prone to injury.
Conclusion
This narrative review discusses the current evidence on the impact of genetics on endurance- and power-based exercise performance to clearly determine the utility of genotyping to detect sports talent, enhance training or prevent exercise-related injuries. The comprehensiveness of this review is aided by the explanation of the most common methods used to assess the influence of genetics on sports performance. From an applied perspective, the ideas discussed in this review suggest that talent/sports performance phenotype identification based on DNA testing is likely to be of limited value at present, and that field testing, which is essentially a higher-order ‘bioassay’, is likely to remain a key element of talent identification in both the near and foreseeable future (Webborn et al. 2015). Nevertheless, the rhythm of inclusion of new technologies to assess DNA variants, the reduction of costs for genetic testing and the cooperation of research groups to produce large-scale sample sizes, will help to enhance the predictive capacity of genetic testing in the future, at least in sports where a clear endurance vs. power phenotype is strongly associated with overall performance.
Abbreviations
- ACE
Angiotensin-converting enzyme
- ACTN3
α-actinin 3
- AGT
Angiotensinogen
- AMPD1
Adenosine monophosphate deaminase 1
- CK-MM
Creatine kinase isoenzyme MM
- CYP1A2
Cytochrome P450 1A2
- DNA
Deoxyribonucleic acid
- GWAS
Genome-wide association studies
- HFE
Homeostatic iron regulator
- IL6
Interleukin-6
- MYLK
Myosin light chain kinase
- NOS3
Endothelial nitric oxide synthase 3
- PGC1α
Peroxisome proliferator-activated receptor gamma coactivator 1-alpha
- RNA
Ribonucleic acid
- TGS
Total genotype score
- VO2max
Maximal rate of oxygen consumption
Author contributions
DV-D and JdC had the idea for the article, DV-D, EM, JG-H, MA and AM performed the literature search, and all authors contributed to write and/or critically revised the work. All authors read and approved the final manuscript.
Funding
This review has not been funded.
Declarations
Conflict of interest
On behalf of all the authors, the corresponding author states that there is no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Aaltonen S, Latvala A, Jelenkovic A, Rose RJ, Kujala UM, Kaprio J, Silventoinen K. Physical activity and academic performance: genetic and environmental associations. Med Sci Sports Exerc. 2020;52(2):381–390. doi: 10.1249/mss.0000000000002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmetov II, Fedotovskaya ON. Current progress in sports genomics. Adv Clin Chem. 2015;70:247–314. doi: 10.1016/bs.acc.2015.03.003. [DOI] [PubMed] [Google Scholar]
- Ahmetov II, Vinogradova OL, Williams AG. Gene polymorphisms and fiber-type composition of human skeletal muscle. Int J Sport Nutr Exerc Metab. 2012;22(4):292–303. doi: 10.1123/ijsnem.22.4.292. [DOI] [PubMed] [Google Scholar]
- Ahmetov I, Kulemin N, Popov D, Naumov V, Akimov E, Bravy Y, Egorova E, Galeeva A, Generozov E, Kostryukova E, Larin A, Mustafina L, Ospanova E, Pavlenko A, Starnes L, Żmijewski P, Alexeev D, Vinogradova O, Govorun V. Genome-wide association study identifies three novel genetic markers associated with elite endurance performance. Biol Sport. 2015;32(1):3–9. doi: 10.5604/20831862.1124568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmetov II, Egorova ES, Gabdrakhmanova LJ, Fedotovskaya ON. genes and athletic performance: an update. Med Sport Sci. 2016;61:41–54. doi: 10.1159/000445240. [DOI] [PubMed] [Google Scholar]
- Ahmetov II, Hall ECR, Semenova EA, Pranckevičienė E, Ginevičienė V. (2021) Advances in sports genomics. In: Advances in Clinical Chemistry. Elsevier. 10.1016/bs.acc.2021.07.004 [DOI] [PubMed]
- Ahtiainen JP, Sallinen J, Häkkinen K, Sillanpää E. Inter-individual variation in response to resistance training in cardiometabolic health indicators. Scand J Med Sci Sports. 2020;30(6):1040–1053. doi: 10.1111/sms.13650. [DOI] [PubMed] [Google Scholar]
- Al-Khelaifi F, Diboun I, Donati F, Botrè F, Abraham D, Hingorani A, Albagha O, Georgakopoulos C, Suhre K, Yousri NA, Elrayess MA. Metabolic GWAS of elite athletes reveals novel genetically-influenced metabolites associated with athletic performance. Sci Rep. 2019;9(1):19889. doi: 10.1038/s41598-019-56496-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khelaifi F, Yousri NA, Diboun I, Semenova EA, Kostryukova ES, Kulemin NA, Borisov OV, Andryushchenko LB, Larin AK, Generozov EV, Miyamoto-Mikami E, Murakami H, Zempo H, Miyachi M, Takaragawa M, Kumagai H, Naito H, Fuku N, Abraham D, Hingorani A, Donati F, Botrè F, Georgakopoulos C, Suhre K, Ahmetov II, Albagha O, Elrayess MA. Genome-wide association study reveals a novel association between MYBPC3 gene polymorphism, endurance athlete status, aerobic capacity and steroid metabolism. Front Genet. 2020;11:595. doi: 10.3389/fgene.2020.00595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso L, Souza E, Oliveira M, do Nascimento L, Dantas P. Heritability of aerobic power of individuals in northeast Brazil. Biol Sport. 2014;31(4):267–270. doi: 10.5604/20831862.1120933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir O, Amir R, Yamin C, Attias E, Eynon N, Sagiv M, Meckel Y. The ACE deletion allele is associated with israeli elite endurance athletes. Exp Physiol. 2007;92(5):881–886. doi: 10.1113/expphysiol.2007.038711. [DOI] [PubMed] [Google Scholar]
- Azzazy HM, Mansour MM, Christenson RH. Doping in the recombinant era: strategies and counterstrategies. Clin Biochem. 2005;38(11):959–965. doi: 10.1016/j.clinbiochem.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Bae JS, Kang BY, Lee KO, Lee ST. Genetic variation in the renin-angiotensin system and response to endurance training. Med Princ Pract. 2007;16(2):142–146. doi: 10.1159/000098368. [DOI] [PubMed] [Google Scholar]
- Baltazar-Martins G, Gutiérrez-Hellín J, Aguilar-Navarro M, Ruiz-Moreno C, Moreno-Pérez V, López-Samanes Á, Domínguez R, Del Coso J. Effect of ACTN3 genotype on sports performance, exercise-induced muscle damage, and injury epidemiology. Sports (Basel) 2020 doi: 10.3390/sports8070099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baoutina A, Alexander IE, Rasko JE, Emslie KR. Potential use of gene transfer in athletic performance enhancement. Mol Ther. 2007;15(10):1751–1766. doi: 10.1038/sj.mt.6300278. [DOI] [PubMed] [Google Scholar]
- Baoutina A, Coldham T, Fuller B, Emslie KR. Improved detection of transgene and nonviral vectors in blood. Hum Gene Ther Methods. 2013;24(6):345–354. doi: 10.1089/hgtb.2013.128. [DOI] [PubMed] [Google Scholar]
- Barreto G, Grecco B, Merola P, Reis CEG, Gualano B, Saunders B. Novel insights on caffeine supplementation, CYP1A2 genotype, physiological responses and exercise performance. Eur J Appl Physiol. 2021;121(3):749–769. doi: 10.1007/s00421-020-04571-7. [DOI] [PubMed] [Google Scholar]
- Ben-Zaken S, Eliakim A, Nemet D, Rabinovich M, Kassem E, Meckel Y. ACTN3 polymorphism: comparison between elite swimmers and runners. Sports Med Open. 2015;1(1):13. doi: 10.1186/s40798-015-0023-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Zaken S, Meckel Y, Nemet D, Eliakim A. Genetic score of power-speed and endurance track and field athletes. Scand J Med Sci Sports. 2015;25(2):166–174. doi: 10.1111/sms.12141. [DOI] [PubMed] [Google Scholar]
- Ben-Zaken S, Meckel Y, Nemet D, Rabinovich M, Kassem E, Eliakim A. Frequency of the MSTN Lys(K)-153Arg(R) polymorphism among track and field athletes and swimmers. Growth Horm IGF Res. 2015;25(4):196–200. doi: 10.1016/j.ghir.2015.04.001. [DOI] [PubMed] [Google Scholar]
- Ben-Zaken S, Meckel Y, Nemet D, Eliakim A. The combined frequency of IGF and myostatin polymorphism among track & field athletes and swimmers. Growth Horm IGF Res. 2017;32:29–32. doi: 10.1016/j.ghir.2016.12.002. [DOI] [PubMed] [Google Scholar]
- Ben-Zaken S, Eliakim A, Nemet D, Meckel Y. Genetic variability among power athletes: the stronger vs. the faster. J Strength Cond Res. 2019;33(6):1505–1511. doi: 10.1519/jsc.0000000000001356. [DOI] [PubMed] [Google Scholar]
- Bergh U, Thorstensson A, Sjödin B, Hulten B, Piehl K, Karlsson J. Maximal oxygen uptake and muscle fiber types in trained and untrained humans. Med Sci Sports. 1978;10(3):151–154. [PubMed] [Google Scholar]
- Bottinelli R, Reggiani C. Human skeletal muscle fibres: molecular and functional diversity. Prog Biophys Mol Biol. 2000;73(2–4):195–262. doi: 10.1016/s0079-6107(00)00006-7. [DOI] [PubMed] [Google Scholar]
- Bouchard C, Rankinen T, Chagnon YC, Rice T, Pérusse L, Gagnon J, Borecki I, An P, Leon AS, Skinner JS, Wilmore JH, Province M, Rao DC (2000) Genomic scan for maximal oxygen uptake and its response to training in the HERITAGE Family Study. J Appl Physiol (1985) 88 (2):551–559. 10.1152/jappl.2000.88.2.551 [DOI] [PubMed]
- Bouchard C. Genomic predictors of trainability. Exp Physiol. 2012;97(3):347–352. doi: 10.1113/expphysiol.2011.058735. [DOI] [PubMed] [Google Scholar]
- Boulygina EA, Borisov OV, Valeeva EV, Semenova EA, Kostryukova ES, Kulemin NA, Larin AK, Nabiullina RM, Mavliev FA, Akhatov AM, Andryushchenko ON, Andryushchenko LB, Zmijewski P, Generozov EV, Ahmetov II. Whole genome sequencing of elite athletes. Biol Sport. 2020;37(3):295–304. doi: 10.5114/biolsport.2020.96272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray MS, Hagberg JM, Pérusse L, Rankinen T, Roth SM, Wolfarth B, Bouchard C. The human gene map for performance and health-related fitness phenotypes: the 2006–2007 update. Med Sci Sports Exerc. 2009;41(1):35–73. doi: 10.1249/mss.0b013e3181844179. [DOI] [PubMed] [Google Scholar]
- Breitbach S, Tug S, Simon P. Conventional and genetic talent identification in sports: will recent developments trace talent? Sports Med. 2014;44(11):1489–1503. doi: 10.1007/s40279-014-0221-7. [DOI] [PubMed] [Google Scholar]
- Brooks GA. Anaerobic threshold: review of the concept and directions for future research. Med Sci Sports Exerc. 1985;17(1):22–34. [PubMed] [Google Scholar]
- Broos S, Malisoux L, Theisen D, Francaux M, Deldicque L, Thomis MA. Role of alpha-actinin-3 in contractile properties of human single muscle fibers: a case series study in paraplegics. PLoS One. 2012;7(11):e49281. doi: 10.1371/journal.pone.0049281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzeziańska E, Domańska D, Jegier A. Gene doping in sport - perspectives and risks. Biol Sport. 2014;31(4):251–259. doi: 10.5604/20831862.1120931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxens A, Ruiz JR, Arteta D, Artieda M, Santiago C, Gonzalez-Freire M, Martinez A, Tejedor D, Lao JI, Gomez-Gallego F, Lucia A. Can we predict top-level sports performance in power vs endurance events? a genetic approach. Scand J Med Sci Sports. 2011;21(4):570–579. doi: 10.1111/j.1600-0838.2009.01079.x. [DOI] [PubMed] [Google Scholar]
- Bye A, Klevjer M, Ryeng E, Silva G, Moreira JBN, Stensvold D, Wisløff U. Identification of novel genetic variants associated with cardiorespiratory fitness. Prog Cardiovasc Dis. 2020;63(3):341–349. doi: 10.1016/j.pcad.2020.02.001. [DOI] [PubMed] [Google Scholar]
- Calabrese C, Davidson NR, Demircioğlu D, Fonseca NA, He Y, Kahles A, Lehmann KV, Liu F, Shiraishi Y, Soulette CM, Urban L, Greger L, Li S, Liu D, Perry MD, Xiang Q, Zhang F, Zhang J, Bailey P, Erkek S, Hoadley KA, Hou Y, Huska MR, Kilpinen H, Korbel JO, Marin MG, Markowski J, Nandi T, Pan-Hammarström Q, Pedamallu CS, Siebert R, Stark SG, Su H, Tan P, Waszak SM, Yung C, Zhu S, Awadalla P, Creighton CJ, Meyerson M, Ouellette BFF, Wu K, Yang H, Brazma A, Brooks AN, Göke J, Rätsch G, Schwarz RF, Stegle O, Zhang Z. Genomic basis for RNA alterations in cancer. Nature. 2020;578(7793):129–136. doi: 10.1038/s41586-020-1970-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calbet JAL, Martín-Rodríguez S, Martin-Rincon M, Morales-Alamo D. An integrative approach to the regulation of mitochondrial respiration during exercise: focus on high-intensity exercise. Redox Biol. 2020;35:101478. doi: 10.1016/j.redox.2020.101478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo M, Rodas G, Vallejo M, Estruch A, Arcas A, Javierre C, Viscor G, Ventura JL. Heritability of explosive power and anaerobic capacity in humans. Eur J Appl Physiol. 2002;86(3):218–225. doi: 10.1007/s004210100522. [DOI] [PubMed] [Google Scholar]
- Cantelmo RA, da Silva AP, Mendes-Junior CT, Dorta DJ. Gene doping: present and future. Eur J Sport Sci. 2020;20(8):1093–1101. doi: 10.1080/17461391.2019.1695952. [DOI] [PubMed] [Google Scholar]
- Chen C, Sun Y, Liang H, Yu D, Hu S. A meta-analysis of the association of CKM gene rs8111989 polymorphism with sport performance. Biol Sport. 2017;34(4):323–330. doi: 10.5114/biolsport.2017.69819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson PM, Hoffman EP, Zambraski E, Gordish-Dressman H, Kearns A, Hubal M, Harmon B, Devaney JM ( 2005) ACTN3 and MLCK genotype associations with exertional muscle damage. J Appl Physiol 99(2):564–569. 10.1152/japplphysiol.00130.2005 [DOI] [PubMed]
- Close GL, Hamilton DL, Philp A, Burke LM, Morton JP. New strategies in sport nutrition to increase exercise performance. Free Radic Biol Med. 2016;98:144–158. doi: 10.1016/j.freeradbiomed.2016.01.016. [DOI] [PubMed] [Google Scholar]
- Collins M, O’Connell K, Posthumus M. Genetics of Musculoskeletal Exercise-Related Phenotypes. Med Sport Sci. 2016;61:92–104. doi: 10.1159/000445242. [DOI] [PubMed] [Google Scholar]
- Costa AM, Silva AJ, Garrido ND, Louro H, de Oliveira RJ, Breitenfeld L. Association between ACE D allele and elite short distance swimming. Eur J Appl Physiol. 2009;106(6):785–790. doi: 10.1007/s00421-009-1080-z. [DOI] [PubMed] [Google Scholar]
- Davies CT, Thompson MW. Aerobic performance of female marathon and male ultramarathon athletes. Eur J Appl Physiol Occup Physiol. 1979;41(4):233–245. doi: 10.1007/bf00429740. [DOI] [PubMed] [Google Scholar]
- De Rose EH. Doping in athletes–an update. Clin Sports Med. 2008;27(1):107–130. doi: 10.1016/j.csm.2007.10.001. [DOI] [PubMed] [Google Scholar]
- De Moor MH, Spector TD, Cherkas LF, Falchi M, Hottenga JJ, Boomsma DI, De Geus EJ. Genome-wide linkage scan for athlete status in 700 British female DZ twin pairs. Twin Res Hum Genet. 2007;10(6):812–820. doi: 10.1375/twin.10.6.812. [DOI] [PubMed] [Google Scholar]
- Dehghan A. Genome-wide association studies. Methods Mol Biol. 2018;1793:37–49. doi: 10.1007/978-1-4939-7868-7_4. [DOI] [PubMed] [Google Scholar]
- Del Coso J, Muñoz-Fernández VE, Muñoz G, Fernández-Elías VE, Ortega JF, Hamouti N, Barbero JC, Muñoz-Guerra J. Effects of a caffeine-containing energy drink on simulated soccer performance. PLoS One. 2012;7(2):e31380. doi: 10.1371/journal.pone.0031380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Coso J, Valero M, Lara B, Salinero JJ, Gallo-Salazar C, Areces F. Myosin light chain kinase (MLCK) gene influences exercise induced muscle damage during a competitive marathon. PLoS One. 2016;11(8):e0160053. doi: 10.1371/journal.pone.0160053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz Ramírez J, Álvarez-Herms J, Castañeda-Babarro A, Larruskain J, Ramírez de la Piscina X, Borisov OV, Semenova EA, Kostryukova ES, Kulemin NA, Andryushchenko ON, Larin AK, Andryushchenko LB, Generozov EV, Ahmetov II, Odriozola A. The GALNTL6 gene rs558129 polymorphism is associated with power performance. J Strength Cond Res. 2020;34(11):3031–3036. doi: 10.1519/jsc.0000000000003814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma K, Deakin GB, Schumann M, Bentley DJ. Training considerations for optimising endurance development: an alternate concurrent training perspective. Sports Med. 2019;49(5):669–682. doi: 10.1007/s40279-019-01072-2. [DOI] [PubMed] [Google Scholar]
- Duoqi Z, Qing H, Yang H, Yanchun L, Yi X, Li W. Association between KCNJ11 gene E23K polymorphism and body composition together with its response to endurance training. Open Biomed Eng J. 2015;9:121–125. doi: 10.2174/1874120701509010121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echegaray M, Rivera MA. Role of creatine kinase isoenzymes on muscular and cardiorespiratory endurance: genetic and molecular evidence. Sports Med. 2001;31(13):919–934. doi: 10.2165/00007256-200131130-00003. [DOI] [PubMed] [Google Scholar]
- Ehlert T, Simon P, Moser DA. Epigenetics in sports. Sports Med. 2013;43(2):93–110. doi: 10.1007/s40279-012-0012-y. [DOI] [PubMed] [Google Scholar]
- Eider J, Ahmetov II, Fedotovskaya ON, Moska W, Cieszczyk P, Zarebska A, Czubek Z, Klocek T, Stepien-Slodkowska M, Maciejewska-Karlowska A, Sawczuk M. CKM gene polymorphism in Russian and Polish rowers. Genetika. 2015;51(3):389–392. doi: 10.7868/s0016675815030029. [DOI] [PubMed] [Google Scholar]
- Ergun S, Oztuzcu S. Sequence-based analysis of 5'UTR and coding regions of CASP3 in terms of miRSNPs and SNPs in targetting miRNAs. Comput Biol Chem. 2016;62:70–74. doi: 10.1016/j.compbiolchem.2016.04.003. [DOI] [PubMed] [Google Scholar]
- Eynon N, Alves AJ, Yamin C, Sagiv M, Duarte JA, Oliveira J, Ayalon M, Goldhammer E, Meckel Y. Is there an ACE ID—ACTN3 R577X polymorphisms interaction that influences sprint performance? Int J Sports Med. 2009;30(12):888–891. doi: 10.1055/s-0029-1238291. [DOI] [PubMed] [Google Scholar]
- Eynon N, Ruiz JR, Meckel Y, Morán M, Lucia A. Mitochondrial biogenesis related endurance genotype score and sports performance in athletes. Mitochondrion. 2011;11(1):64–69. doi: 10.1016/j.mito.2010.07.004. [DOI] [PubMed] [Google Scholar]
- Eynon N, Ruiz JR, Oliveira J, Duarte JA, Birk R, Lucia A. Genes and elite athletes: a roadmap for future research. J Physiol. 2011;589(Pt 13):3063–3070. doi: 10.1113/jphysiol.2011.207035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eynon N, Hanson ED, Lucia A, Houweling PJ, Garton F, North KN, Bishop DJ. Genes for elite power and sprint performance: ACTN3 leads the way. Sports Med. 2013;43(9):803–817. doi: 10.1007/s40279-013-0059-4. [DOI] [PubMed] [Google Scholar]
- Ferrell RE, Conte V, Lawrence EC, Roth SM, Hagberg JM, Hurley BF. Frequent sequence variation in the human myostatin (GDF8) gene as a marker for analysis of muscle-related phenotypes. Genomics. 1999;62(2):203–207. doi: 10.1006/geno.1999.5984. [DOI] [PubMed] [Google Scholar]
- Fitts RH, McDonald KS, Schluter JM. The determinants of skeletal muscle force and power: their adaptability with changes in activity pattern. J Biomech. 1991;24(Suppl 1):111–122. doi: 10.1016/0021-9290(91)90382-w. [DOI] [PubMed] [Google Scholar]
- Flanagan JM. Epigenome-wide association studies (EWAS): past, present, and future. Methods Mol Biol. 2015;1238:51–63. doi: 10.1007/978-1-4939-1804-1_3. [DOI] [PubMed] [Google Scholar]
- Fyfe JJ, Bishop DJ, Stepto NK. Interference between concurrent resistance and endurance exercise: molecular bases and the role of individual training variables. Sports Med. 2014;44(6):743–762. doi: 10.1007/s40279-014-0162-1. [DOI] [PubMed] [Google Scholar]
- Gaffney GR, Parisotto R. Gene doping: a review of performance-enhancing genetics. Pediatr Clin North Am. 2007;54(4):807–822. doi: 10.1016/j.pcl.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Gallagher MD, Chen-Plotkin AS. The post-GWAS era: from association to function. Am J Hum Genet. 2018;102(5):717–730. doi: 10.1016/j.ajhg.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Mahajan PB. Can genotype determine the sports phenotype? a paradigm shift in sports medicine. J Basic Clin Physiol Pharmacol. 2016;27(4):333–339. doi: 10.1515/jbcpp-2015-0090. [DOI] [PubMed] [Google Scholar]
- Gineviciene V, Jakaitiene A, Pranculis A, Milasius K, Tubelis L, Utkus A. AMPD1 rs17602729 is associated with physical performance of sprint and power in elite Lithuanian athletes. BMC Genet. 2014;15:58. doi: 10.1186/1471-2156-15-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görman U, Mathers JC, Grimaldi KA, Ahlgren J, Nordström K. Do we know enough? A scientific and ethical analysis of the basis for genetic-based personalized nutrition. Genes Nutr. 2013;8(4):373–381. doi: 10.1007/s12263-013-0338-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grealy R, Herruer J, Smith CL, Hiller D, Haseler LJ, Griffiths LR. Evaluation of a 7-gene genetic profile for athletic endurance phenotype in ironman championship triathletes. PLoS One. 2015;10(12):e0145171. doi: 10.1371/journal.pone.0145171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greggio C, Jha P, Kulkarni SS, Lagarrigue S, Broskey NT, Boutant M, Wang X, Conde Alonso S, Ofori E, Auwerx J, Cantó C, Amati F. Enhanced respiratory chain supercomplex formation in response to exercise in human skeletal muscle. Cell Metab. 2017;25(2):301–311. doi: 10.1016/j.cmet.2016.11.004. [DOI] [PubMed] [Google Scholar]
- Guest N, Corey P, Vescovi J, El-Sohemy A. Caffeine, CYP1A2 genotype, and endurance performance in athletes. Med Sci Sports Exerc. 2018;50(8):1570–1578. doi: 10.1249/mss.0000000000001596. [DOI] [PubMed] [Google Scholar]
- Guest NS, Horne J, Vanderhout SM, El-Sohemy A. Sport nutrigenomics: personalized nutrition for athletic performance. Front Nutr. 2019;6:8. doi: 10.3389/fnut.2019.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guest NS, VanDusseldorp TA, Nelson MT, Grgic J, Schoenfeld BJ, Jenkins NDM, Arent SM, Antonio J, Stout JR, Trexler ET, Smith-Ryan AE, Goldstein ER, Kalman DS, Campbell BI. International society of sports nutrition position stand: caffeine and exercise performance. J Int Soc Sports Nutr. 2021;18(1):1. doi: 10.1186/s12970-020-00383-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guth LM, Roth SM. Genetic influence on athletic performance. Curr Opin Pediatr. 2013;25(6):653–658. doi: 10.1097/MOP.0b013e3283659087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Hellín J, Varillas-Delgado D. Energy drinks and sports performance, cardiovascular risk, and genetic associations; future prospects. Nutrients. 2021 doi: 10.3390/nu13030715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Hellín J, Baltazar-Martins G, Aguilar-Navarro M, Ruiz-Moreno C, Oliván J, Del Coso J. Effect of ACTN3 R577X genotype on injury epidemiology in elite endurance runners. Genes (Basel) 2021 doi: 10.3390/genes12010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- György B, Maguire CA. Extracellular vesicles: nature’s nanoparticles for improving gene transfer with adeno-associated virus vectors. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2018;10(3):e1488. doi: 10.1002/wnan.1488. [DOI] [PubMed] [Google Scholar]
- Hagberg JM, Moore GE, Ferrell RE. Specific genetic markers of endurance performance and VO2max. Exerc Sport Sci Rev. 2001;29(1):15–19. doi: 10.1097/00003677-200101000-00004. [DOI] [PubMed] [Google Scholar]
- Haisma HJ, de Hon O. Gene doping. Int J Sports Med. 2006;27(4):257–266. doi: 10.1055/s-2006-923986. [DOI] [PubMed] [Google Scholar]
- Hakimi P, Yang J, Casadesus G, Massillon D, Tolentino-Silva F, Nye CK, Cabrera ME, Hagen DR, Utter CB, Baghdy Y, Johnson DH, Wilson DL, Kirwan JP, Kalhan SC, Hanson RW. Overexpression of the cytosolic form of phosphoenolpyruvate carboxykinase (GTP) in skeletal muscle repatterns energy metabolism in the mouse. J Biol Chem. 2007;282(45):32844–32855. doi: 10.1074/jbc.M706127200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Hu Y, Feng L, Bao D, Xi Y, Wen L, Lucia A. Relationship between TFAM gene polymorphisms and endurance capacity in response to training. Int J Sports Med. 2007;28(12):1059–1064. doi: 10.1055/s-2007-965064. [DOI] [PubMed] [Google Scholar]
- He ZH, Hu Y, Li YC, Bao DP, Ruiz JR, Lucia A. Polymorphisms in the calcineurin genes are associated with the training responsiveness of cardiac phenotypes in Chinese young adults. Eur J Appl Physiol. 2010;110(4):761–767. doi: 10.1007/s00421-010-1558-8. [DOI] [PubMed] [Google Scholar]
- Hellsten Y, Nyberg M. Cardiovascular adaptations to exercise training. Compr Physiol. 2015;6(1):1–32. doi: 10.1002/cphy.c140080. [DOI] [PubMed] [Google Scholar]
- Heo J, Yoon JG, Park H, Kim YD, Nam HS, Heo JH. Machine learning-based model for prediction of outcomes in acute stroke. Stroke. 2019;50(5):1263–1265. doi: 10.1161/strokeaha.118.024293. [DOI] [PubMed] [Google Scholar]
- Heuberger J, Cohen AF. Review of WADA prohibited substances: limited evidence for performance-enhancing effects. Sports Med. 2019;49(4):525–539. doi: 10.1007/s40279-018-1014-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdys J, Gronek P, Kryściak J, Stanisławski D. Genetic variants of uncoupling proteins-2 and -3 in relation to maximal oxygen uptake in different sports. Acta Biochim Pol. 2013;60(1):71–75. doi: 10.18388/abp.2013_1953. [DOI] [PubMed] [Google Scholar]
- Holloszy JO, Coyle EF. Adaptations of skeletal muscle to endurance exercise and their metabolic consequences. J Appl Physiol Respir Env Exerc Physiol. 1984;56(4):831–838. doi: 10.1152/jappl.1984.56.4.831. [DOI] [PubMed] [Google Scholar]
- Hon CC, Ramilowski JA, Harshbarger J, Bertin N, Rackham OJ, Gough J, Denisenko E, Schmeier S, Poulsen TM, Severin J, Lizio M, Kawaji H, Kasukawa T, Itoh M, Burroughs AM, Noma S, Djebali S, Alam T, Medvedeva YA, Testa AC, Lipovich L, Yip CW, Abugessaisa I, Mendez M, Hasegawa A, Tang D, Lassmann T, Heutink P, Babina M, Wells CA, Kojima S, Nakamura Y, Suzuki H, Daub CO, de Hoon MJ, Arner E, Hayashizaki Y, Carninci P, Forrest AR. An atlas of human long non-coding RNAs with accurate 5’ ends. Nature. 2017;543(7644):199–204. doi: 10.1038/nature21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houweling PJ, Papadimitriou ID, Seto JT, Pérez LM, Coso JD, North KN, Lucia A, Eynon N. Is evolutionary loss our gain? the role of ACTN3 p.Arg577Ter (R577X) genotype in athletic performance, ageing, and disease. Hum Mutat. 2018;39(12):1774–1787. doi: 10.1002/humu.23663. [DOI] [PubMed] [Google Scholar]
- Huang J, Gao N, Wang S, Milewicz DM, Kamm KE, Stull JT. Genetic approaches to identify pathological limitations in aortic smooth muscle contraction. PLoS One. 2018;13(3):e0193769. doi: 10.1371/journal.pone.0193769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes DC, Day SH, Ahmetov II, Williams AG. Genetics of muscle strength and power: polygenic profile similarity limits skeletal muscle performance. J Sports Sci. 2011;29(13):1425–1434. doi: 10.1080/02640414.2011.597773. [DOI] [PubMed] [Google Scholar]
- Issurin V. Block periodization versus traditional training theory: a review. J Sports Med Phys Fit. 2008;48(1):65–75. [PubMed] [Google Scholar]
- Issurin VB. Biological background of block periodized endurance training: a review. Sports Med. 2019;49(1):31–39. doi: 10.1007/s40279-018-1019-9. [DOI] [PubMed] [Google Scholar]
- Jayewardene AF, Mavros Y, Gwinn T, Hancock DP, Rooney KB. Associations between CD36 gene polymorphisms and metabolic response to a short-term endurance-training program in a young-adult population. Appl Physiol Nutr Metab. 2016;41(2):157–167. doi: 10.1139/apnm-2015-0430. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A, Montgomery HE, Woods DR. Human performance: a role for the ACE genotype? Exerc Sport Sci Rev. 2002;30(4):184–190. doi: 10.1097/00003677-200210000-00008. [DOI] [PubMed] [Google Scholar]
- Joyner MJ. Genetic approaches for sports performance: how far away are we? Sports Med. 2019 doi: 10.1007/s40279-019-01164-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyner MJ, Coyle EF. Endurance exercise performance: the physiology of champions. J Physiol. 2008;586(1):35–44. doi: 10.1113/jphysiol.2007.143834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalafi EY, Nor NAM, Taib NA, Ganggayah MD, Town C, Dhillon SK. Machine learning and deep learning approaches in breast cancer survival prediction using clinical data. Folia Biol (Praha) 2019;65(5–6):212–220. [PubMed] [Google Scholar]
- Kitazawa H, Hasegawa K, Aruga D, Tanaka M. Potential genetic contributions of the central nervous system to a predisposition to elite athletic traits: state-of-the-art and future perspectives. Genes (Basel) 2021 doi: 10.3390/genes12030371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostek MA, Angelopoulos TJ, Clarkson PM, Gordon PM, Moyna NM, Visich PS, Zoeller RF, Price TB, Seip RL, Thompson PD, Devaney JM, Gordish-Dressman H, Hoffman EP, Pescatello LS. Myostatin and follistatin polymorphisms interact with muscle phenotypes and ethnicity. Med Sci Sports Exerc. 2009;41(5):1063–1071. doi: 10.1249/MSS.0b013e3181930337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Gerche A, Brosnan MJ. Drugs in sport—a change is needed, but what? Heart Lung Circ. 2018;27(9):1099–1104. doi: 10.1016/j.hlc.2018.04.302. [DOI] [PubMed] [Google Scholar]
- Lamsfus-Calle A, Daniel-Moreno A, Ureña-Bailén G, Raju J, Antony JS, Handgretinger R, Mezger M. Hematopoietic stem cell gene therapy: the optimal use of lentivirus and gene editing approaches. Blood Rev. 2020;40:100641. doi: 10.1016/j.blre.2019.100641. [DOI] [PubMed] [Google Scholar]
- Lavie CJ, Ozemek C, Carbone S, Katzmarzyk PT, Blair SN. Sedentary behavior, exercise, and cardiovascular health. Circ Res. 2019;124(5):799–815. doi: 10.1161/circresaha.118.312669. [DOI] [PubMed] [Google Scholar]
- Levine BD. VO2max: what do we know, and what do we still need to know? J Physiol. 2008;586(1):25–34. doi: 10.1113/jphysiol.2007.147629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis NA, Howatson G, Morton K, Hill J, Pedlar CR. Alterations in redox homeostasis in the elite endurance athlete. Sports Med. 2015;45(3):379–409. doi: 10.1007/s40279-014-0276-5. [DOI] [PubMed] [Google Scholar]
- Li X, Wang SJ, Tan SC, Chew PL, Liu L, Wang L, Wen L, Ma L. The A55T and K153R polymorphisms of MSTN gene are associated with the strength training-induced muscle hypertrophy among Han Chinese men. J Sports Sci. 2014;32(9):883–891. doi: 10.1080/02640414.2013.865252. [DOI] [PubMed] [Google Scholar]
- Lippi G, Longo UG, Maffulli N. Genetics and sports. Br Med Bull. 2010;93:27–47. doi: 10.1093/bmb/ldp007. [DOI] [PubMed] [Google Scholar]
- Loos RJ, Hagberg JM, Perusse L, Roth SM, Sarzynski MA, Wolfarth B, Rankinen T, Bouchard C. Advances in exercise, fitness, and performance genomics in 2014. Med Sci Sports Exerc. 2015;47(6):1105–1112. doi: 10.1249/mss.0000000000000645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos RJ, Hagberg JM, Pérusse L, Roth SM, Sarzynski MA, Wolfarth B, Rankinen T, Bouchard C. Advances in exercise, fitness, and performance genomics in 2014. Med Sci Sports Exerc. 2015;47(6):1105–1112. doi: 10.1249/mss.0000000000000645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López S, Meirelles J, Rayol V, Poralla G, Woldmar N, Fadel B, Figueiredo M, Costa Padilha MD, de Aquino Neto FR, Gualberto Pereira HM, Pizzatti L. Gene doping and genomic science in sports: where are we? Bioanalysis. 2020;12(11):801–811. doi: 10.4155/bio-2020-0093. [DOI] [PubMed] [Google Scholar]
- Lucia A, Gómez-Gallego F, Santiago C, Bandrés F, Earnest C, Rabadán M, Alonso JM, Hoyos J, Córdova A, Villa G, Foster C. ACTN3 genotype in professional endurance cyclists. Int J Sports Med. 2006;27(11):880–884. doi: 10.1055/s-2006-923862. [DOI] [PubMed] [Google Scholar]
- Lucia A, Martin MA, Esteve-Lanao J, San Juan AF, Rubio JC, Olivan J, Arenas J. C34T mutation of the AMPD1 gene in an elite white runner. BMJ Case Rep. 2009 doi: 10.1136/bcr.07.2008.0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucía A, Gómez-Gallego F, Chicharro JL, Hoyos J, Celaya K, Córdova A, Villa G, Alonso JM, Barriopedro M, Pérez M, Earnest CP. Is there an association between ACE and CKMM polymorphisms and cycling performance status during 3-week races? Int J Sports Med. 2005;26(6):442–447. doi: 10.1055/s-2004-821108. [DOI] [PubMed] [Google Scholar]
- Lundby C, Montero D, Joyner M. Biology of VO(2) max: looking under the physiology lamp. Acta Physiol (oxf) 2017;220(2):218–228. doi: 10.1111/apha.12827. [DOI] [PubMed] [Google Scholar]
- Ma F, Yang Y, Li X, Zhou F, Gao C, Li M, Gao L. The association of sport performance with ACE and ACTN3 genetic polymorphisms: a systematic review and meta-analysis. PLoS One. 2013;8(1):e54685. doi: 10.1371/journal.pone.0054685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejewska-Skrendo A, Buryta M, Czarny W, Król P, Stastny P, Petr M, Safranow K, Sawczuk M. The polymorphisms of the peroxisome-proliferator activated receptors’ alfa gene modify the aerobic training induced changes of cholesterol and glucose. J Clin Med. 2019 doi: 10.3390/jcm8071043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejewska-Skrendo A, Cieszczyk P, Chycki J, Sawczuk M, Smolka W. Genetic markers associated with power athlete status. J Hum Kinet. 2019;68:17–36. doi: 10.2478/hukin-2019-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mägi A, Unt E, Prans E, Raus L, Eha J, Veraksitš A, Kingo K, Kõks S. The association analysis between ACE and ACTN3 genes polymorphisms and endurance capacity in young cross-country skiers: longitudinal study. J Sports Sci Med. 2016;15(2):287–294. [PMC free article] [PubMed] [Google Scholar]
- Mann TN, Lamberts RP, Lambert MI. High responders and low responders: factors associated with individual variation in response to standardized training. Sports Med. 2014;44(8):1113–1124. doi: 10.1007/s40279-014-0197-3. [DOI] [PubMed] [Google Scholar]
- Mathers JC. Nutrigenomics in the modern era. Proc Nutr Soc. 2017;76(3):265–275. doi: 10.1017/s002966511600080x. [DOI] [PubMed] [Google Scholar]
- McAuley ABT, Hughes DC, Tsaprouni LG, Varley I, Suraci B, Roos TR, Herbert AJ, Kelly AL. The association of the ACTN3 R577X and ACE I/D polymorphisms with athlete status in football: a systematic review and meta-analysis. J Sports Sci. 2021;39(2):200–211. doi: 10.1080/02640414.2020.1812195. [DOI] [PubMed] [Google Scholar]
- Miyamoto-Mikami E, Zempo H, Fuku N, Kikuchi N, Miyachi M, Murakami H. Heritability estimates of endurance-related phenotypes: a systematic review and meta-analysis. Scand J Med Sci Sports. 2018;28(3):834–845. doi: 10.1111/sms.12958. [DOI] [PubMed] [Google Scholar]
- Moir HJ, Kemp R, Folkerts D, Spendiff O, Pavlidis C, Opara E. Genes and elite marathon running performance: a systematic review. J Sports Sci Med. 2019;18(3):559–568. [PMC free article] [PubMed] [Google Scholar]
- Montefiori LE, Sobreira DR, Sakabe NJ, Aneas I, Joslin AC, Hansen GT, Bozek G, Moskowitz IP, McNally EM, Nóbrega MA. A promoter interaction map for cardiovascular disease genetics. Elife. 2018 doi: 10.7554/eLife.35788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery HE, Marshall R, Hemingway H, Myerson S, Clarkson P, Dollery C, Hayward M, Holliman DE, Jubb M, World M, Thomas EL, Brynes AE, Saeed N, Barnard M, Bell JD, Prasad K, Rayson M, Talmud PJ, Humphries SE Human gene for physical performance. Nature. 1998;393(6682):221–222. doi: 10.1038/30374. [DOI] [PubMed] [Google Scholar]
- Moran CN, Vassilopoulos C, Tsiokanos A, Jamurtas AZ, Bailey ME, Montgomery HE, Wilson RH, Pitsiladis YP. The associations of ACE polymorphisms with physical, physiological and skill parameters in adolescents. Eur J Hum Genet. 2006;14(3):332–339. doi: 10.1038/sj.ejhg.5201550. [DOI] [PubMed] [Google Scholar]
- Muniesa CA, Gonzalez-Freire M, Santiago C, Lao JI, Buxens A, Rubio JC, Martin MA, Arenas J, Gomez-Gallego F, Lucia A. World-class performance in lightweight rowing: is it genetically influenced? a comparison with cyclists, runners and non-athletes. Br J Sports Med. 2010;44(12):898–901. doi: 10.1136/bjsm.2008.051680. [DOI] [PubMed] [Google Scholar]
- Muñoz A, López-Samanes Á, Aguilar-Navarro M, Varillas-Delgado D, Rivilla-García J, Moreno-Pérez V, Del Coso J. Effects of CYP1A2 and ADORA2A genotypes on the ergogenic response to caffeine in professional handball players. Genes (Basel) 2020 doi: 10.3390/genes11080933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafina LJ, Naumov VA, Cieszczyk P, Popov DV, Lyubaeva EV, Kostryukova ES, Fedotovskaya ON, Druzhevskaya AM, Astratenkova IV, Glotov AS, Alexeev DG, Mustafina MM, Egorova ES, Maciejewska-Karłowska A, Larin AK, Generozov EV, Nurullin RE, Jastrzębski Z, Kulemin NA, Ospanova EA, Pavlenko AV, Sawczuk M, Akimov EB, Danilushkina AA, Zmijewski P, Vinogradova OL, Govorun VM, Ahmetov II. AGTR2 gene polymorphism is associated with muscle fibre composition, athletic status and aerobic performance. Exp Physiol. 2014;99(8):1042–1052. doi: 10.1113/expphysiol.2014.079335. [DOI] [PubMed] [Google Scholar]
- Myerson S, Hemingway H, Budget R, Martin J, Humphries S, Montgomery H (1999) Human angiotensin I-converting enzyme gene and endurance performance. J Appl Physiol (1985) 87 (4):1313–1316. 10.1152/jappl.1999.87.4.1313 [DOI] [PubMed]
- Naureen Z, Perrone M, Paolacci S, Maltese PE, Dhuli K, Kurti D, Dautaj A, Miotto R, Casadei A, Fioretti B, Beccari T, Romeo F, Bertelli M. Genetic test for the personalization of sport training. Acta Biomed. 2020;91(13-s):e2020012. doi: 10.23750/abm.v91i13-S.10593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarov IB, Woods DR, Montgomery HE, Shneider OV, Kazakov VI, Tomilin NV, Rogozkin VA. The angiotensin converting enzyme I/D polymorphism in Russian athletes. Eur J Hum Genet. 2001;9(10):797–801. doi: 10.1038/sj.ejhg.5200711. [DOI] [PubMed] [Google Scholar]
- Neuberger EW, Jurkiewicz M, Moser DA, Simon P. Detection of EPO gene doping in blood. Drug Test Anal. 2012;4(11):859–869. doi: 10.1002/dta.1347. [DOI] [PubMed] [Google Scholar]
- Ng PC, Kirkness EF. Whole genome sequencing. Methods Mol Biol. 2010;628:215–226. doi: 10.1007/978-1-60327-367-1_12. [DOI] [PubMed] [Google Scholar]
- Ostrander EA, Huson HJ, Ostrander GK. Genetics of athletic performance. Annu Rev Genom Hum Genet. 2009;10:407–429. doi: 10.1146/annurev-genom-082908-150058. [DOI] [PubMed] [Google Scholar]
- Palmi I, Berretta P, Tini A, Ricci G, Marinelli S. The unethicality of doping in sports. Clin Ter. 2019;170(2):e100–e101. doi: 10.7417/ct.2019.2117. [DOI] [PubMed] [Google Scholar]
- Papadimitriou ID, Lucia A, Pitsiladis YP, Pushkarev VP, Dyatlov DA, Orekhov EF, Artioli GG, Guilherme JP, Lancha AH, Jr, Ginevičienė V, Cieszczyk P, Maciejewska-Karlowska A, Sawczuk M, Muniesa CA, Kouvatsi A, Massidda M, Calò CM, Garton F, Houweling PJ, Wang G, Austin K, Druzhevskaya AM, Astratenkova IV, Ahmetov II, Bishop DJ, North KN, Eynon N. ACTN3 R577X and ACE I/D gene variants influence performance in elite sprinters: a multi-cohort study. BMC Genom. 2016;17:285. doi: 10.1186/s12864-016-2462-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadimitriou ID, Lockey SJ, Voisin S, Herbert AJ, Garton F, Houweling PJ, Cieszczyk P, Maciejewska-Skrendo A, Sawczuk M, Massidda M, Calo CM, Astratenkova IV, Kouvatsi A, Druzhevskaya AM, Jacques M, Ahmetov II, Stebbings GK, Heffernan S, Day SH, Erskine R, Pedlar C, Kipps C, North KN, Williams AG, Eynon N. No association between ACTN3 R577X and ACE I/D polymorphisms and endurance running times in 698 Caucasian athletes. BMC Genom. 2018;19(1):13. doi: 10.1186/s12864-017-4412-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paßreiter A, Thomas A, Grogna N, Delahaut P, Thevis M. First steps toward uncovering gene doping with CRISPR/Cas by identifying SpCas9 in plasma via HPLC-HRMS/MS. Anal Chem. 2020;92(24):16322–16328. doi: 10.1021/acs.analchem.0c04445. [DOI] [PubMed] [Google Scholar]
- Pickering C, Kiely J. Are the current guidelines on caffeine use in sport optimal for everyone? inter-individual variation in caffeine ergogenicity, and a move towards personalised sports nutrition. Sports Med. 2018;48(1):7–16. doi: 10.1007/s40279-017-0776-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering C, Kiely J. Can genetic testing predict talent? a case study of 5 elite athletes. Int J Sports Physiol Perform. 2020;16(3):429–434. doi: 10.1123/ijspp.2019-0543. [DOI] [PubMed] [Google Scholar]
- Pickering C, Kiely J, Grgic J, Lucia A, Del Coso J. Can genetic testing identify talent for sport? Genes (Basel) 2019 doi: 10.3390/genes10120972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering C, Suraci B, Semenova EA, Boulygina EA, Kostryukova ES, Kulemin NA, Borisov OV, Khabibova SA, Larin AK, Pavlenko AV, Lyubaeva EV, Popov DV, Lysenko EA, Vepkhvadze TF, Lednev EM, Leońska-Duniec A, Pająk B, Chycki J, Moska W, Lulińska-Kuklik E, Dornowski M, Maszczyk A, Bradley B, Kana-Ah A, Cięszczyk P, Generozov EV, Ahmetov II. A genome-wide association study of sprint performance in elite youth football players. J Strength Cond Res. 2019;33(9):2344–2351. doi: 10.1519/jsc.0000000000003259. [DOI] [PubMed] [Google Scholar]
- Pitsiladis Y, Wang G, Wolfarth B, Scott R, Fuku N, Mikami E, He Z, Fiuza-Luces C, Eynon N, Lucia A. Genomics of elite sporting performance: what little we know and necessary advances. Br J Sports Med. 2013;47(9):550–555. doi: 10.1136/bjsports-2013-092400. [DOI] [PubMed] [Google Scholar]
- Pokrywka A, Kaliszewski P, Majorczyk E, Zembroń-Łacny A. Genes in sport and doping. Biol Sport. 2013;30(3):155–161. doi: 10.5604/20831862.1059606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praagh EV. Anaerobic fitness tests: what are we measuring? Med Sport Sci. 2007;50:26–45. doi: 10.1159/000101074. [DOI] [PubMed] [Google Scholar]
- Puente C, Abián-Vicén J, Del Coso J, Lara B, Salinero JJ. The CYP1A2 -163C>A polymorphism does not alter the effects of caffeine on basketball performance. PLoS One. 2018;13(4):e0195943. doi: 10.1371/journal.pone.0195943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankinen T, Rice T, Pérusse L, Chagnon YC, Gagnon J, Leon AS, Skinner JS, Wilmore JH, Rao DC, Bouchard C. NOS3 Glu298Asp genotype and blood pressure response to endurance training: the HERITAGE family study. Hypertension. 2000;36(5):885–889. doi: 10.1161/01.hyp.36.5.885. [DOI] [PubMed] [Google Scholar]
- Reddy VS, Palika R, Ismail A, Pullakhandam R, Reddy GB. Nutrigenomics: opportunities and challenges for public health nutrition. Indian J Med Res. 2018;148(5):632–641. doi: 10.4103/ijmr.IJMR_1738_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice T, Després JP, Pérusse L, Gagnon J, Leon AS, Skinner JS, Wilmore JH, Rao DC, Bouchard C. Segregation analysis of abdominal visceral fat: the HERITAGE Family Study. Obes Res. 1997;5(5):417–424. doi: 10.1002/j.1550-8528.1997.tb00664.x. [DOI] [PubMed] [Google Scholar]
- Rice TK, Sarzynski MA, Sung YJ, Argyropoulos G, Stütz AM, Teran-Garcia M, Rao DC, Bouchard C, Rankinen T. Fine mapping of a QTL on chromosome 13 for submaximal exercise capacity training response: the HERITAGE family study. Eur J Appl Physiol. 2012;112(8):2969–2978. doi: 10.1007/s00421-011-2274-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico-Sanz J, Rankinen T, Joanisse DR, Leon AS, Skinner JS, Wilmore JH, Rao DC, Bouchard C. Associations between cardiorespiratory responses to exercise and the C34T AMPD1 gene polymorphism in the HERITAGE family study. Physiol Genom. 2003;14(2):161–166. doi: 10.1152/physiolgenomics.00165.2002. [DOI] [PubMed] [Google Scholar]
- Rico-Sanz J, Rankinen T, Rice T, Leon AS, Skinner JS, Wilmore JH, Rao DC, Bouchard C. Quantitative trait loci for maximal exercise capacity phenotypes and their responses to training in the HERITAGE family study. Physiol Genom. 2004;16(2):256–260. doi: 10.1152/physiolgenomics.00035.2003. [DOI] [PubMed] [Google Scholar]
- Rivera MA, Dionne FT, Simoneau JA, Pérusse L, Chagnon M, Chagnon Y, Gagnon J, Leon AS, Rao DC, Skinner JS, Wilmore JH, Bouchard C. Muscle-specific creatine kinase gene polymorphism and VO2max in the HERITAGE family study. Med Sci Sports Exerc. 1997;29(10):1311–1317. doi: 10.1097/00005768-199710000-00006. [DOI] [PubMed] [Google Scholar]
- Rivera MA, Dionne FT, Wolfarth B, Chagnon M, Simoneau JA, Pérusse L, Boulay MR, Gagnon J, Song TM, Keul J, Bouchard C. Muscle-specific creatine kinase gene polymorphisms in elite endurance athletes and sedentary controls. Med Sci Sports Exerc. 1997;29(11):1444–1447. doi: 10.1097/00005768-199711000-00009. [DOI] [PubMed] [Google Scholar]
- Rubio JC, Martin MA, Rabadan M, Gomez-Gallego F, San Juan AF, Alonso JM, Chicharro JL, Perez M, Arenas J, Lucia A (2005) Frequency of the C34T mutation of the AMPD1 gene in world-class endurance athletes: does this mutation impair performance? J Appl Physiol (1985) 98 (6):2108– 2112. 10.1152/japplphysiol.01371.2004 [DOI] [PubMed]
- Ruiz JR, Gomez-Gallego F, Santiago C, Gonzalez-Freire M, Verde Z, Foster C, Lucia A. Is there an optimum endurance polygenic profile? J Physiol. 2009;587(Pt 7):1527–1534. doi: 10.1113/jphysiol.2008.166645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz JR, Arteta D, Buxens A, Artieda M, Gomez-Gallego F, Santiago C, Yvert T, Moran M, Lucia A (2010) Can we identify a power-oriented polygenic profile? J Appl Physiol (1985) 108 (3):561–566. 10.1152/japplphysiol.01242.2009 [DOI] [PubMed]
- Santiago C, Ruiz JR, Muniesa CA, Gonzalez-Freire M, Gomez-Gallego F, Lucia A. Does the polygenic profile determine the potential for becoming a world-class athlete? Insights from the sport of rowing. Scand J Med Sci Sports. 2010;20(1):e188–194. doi: 10.1111/j.1600-0838.2009.00943.x. [DOI] [PubMed] [Google Scholar]
- Sargiannidou I, Kagiava A, Kleopa KA. Gene therapy approaches targeting schwann cells for demyelinating neuropathies. Brain Res. 2020;1728:146572. doi: 10.1016/j.brainres.2019.146572. [DOI] [PubMed] [Google Scholar]
- Sarzynski MA, Loos RJ, Lucia A, Pérusse L, Roth SM, Wolfarth B, Rankinen T, Bouchard C. Advances in exercise, fitness, and performance genomics in 2015. Med Sci Sports Exerc. 2016;48(10):1906–1916. doi: 10.1249/mss.0000000000000982. [DOI] [PubMed] [Google Scholar]
- Sarzynski MA, Ghosh S, Bouchard C. Genomic and transcriptomic predictors of response levels to endurance exercise training. J Physiol. 2017;595(9):2931–2939. doi: 10.1113/jp272559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders B, El-Sohemy A, Derave W. Editorial: personalized sport and exercise nutrition. Front Nutr. 2019;6:139. doi: 10.3389/fnut.2019.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol Rev. 2011;91(4):1447–1531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- Schlattner U, Tokarska-Schlattner M, Wallimann T. Mitochondrial creatine kinase in human health and disease. Biochim Biophys Acta. 2006;1762(2):164–180. doi: 10.1016/j.bbadis.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Scott RA, Moran C, Wilson RH, Onywera V, Boit MK, Goodwin WH, Gohlke P, Payne J, Montgomery H, Pitsiladis YP. No association between angiotensin converting enzyme (ACE) gene variation and endurance athlete status in Kenyans. Comp Biochem Physiol A Mol Integr Physiol. 2005;141(2):169–175. doi: 10.1016/j.cbpb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Segura J, Fillat C, Andreu D, Llop J, Millan O, de la Torre BG, Nikolovski Z, Gomez V, Andreu N, Pinyot A, Castelo R, Gispert JD, Pascual JA. Monitoring gene therapy by external imaging of mRNA: pilot study on murine erythropoietin. Ther Drug Monit. 2007;29(5):612–618. doi: 10.1097/FTD.0b013e31811f3af6. [DOI] [PubMed] [Google Scholar]
- Shang X, Huang C, Chang Q, Zhang L, Huang T. Association between the ACTN3 R577X polymorphism and female endurance athletes in China. Int J Sports Med. 2010;31(12):913–916. doi: 10.1055/s-0030-1265176. [DOI] [PubMed] [Google Scholar]
- Shen K, Ramirez B, Mapes B, Shen GR, Gokhale V, Brown ME, Santarsiero B, Ishii Y, Dudek SM, Wang T, Garcia JG. Structure-function analysis of the non-muscle myosin light chain kinase (nmMLCK) Isoform by NMR spectroscopy and molecular modeling: influence of MYLK variants. PLoS One. 2015;10(6):e0130515. doi: 10.1371/journal.pone.0130515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoneau JA, Bouchard C. Genetic determinism of fiber type proportion in human skeletal muscle. Faseb j. 1995;9(11):1091–1095. doi: 10.1096/fasebj.9.11.7649409. [DOI] [PubMed] [Google Scholar]
- Sloan DB, Broz AK, Sharbrough J, Wu Z. Detecting rare mutations and dna damage with sequencing-based methods. Trends Biotechnol. 2018;36(7):729–740. doi: 10.1016/j.tibtech.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon LM, Mordkoff DS, Noll RC. Physical enhancement of human performance: is law keeping pace with science? Gend Med. 2009;6(1):249–258. doi: 10.1016/j.genm.2009.04.008. [DOI] [PubMed] [Google Scholar]
- Sousa FAB, Vasque RE, Gobatto CA. Anaerobic metabolism during short all-out efforts in tethered running: Comparison of energy expenditure and mechanical parameters between different sprint durations for testing. PLoS One. 2017;12(6):e0179378. doi: 10.1371/journal.pone.0179378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer MR, Gastin PB. Energy system contribution during 200- to 1500-m running in highly trained athletes. Med Sci Sports Exerc. 2001;33(1):157–162. doi: 10.1097/00005768-200101000-00024. [DOI] [PubMed] [Google Scholar]
- Stephan DA. Genetic determinants of athletic performance. Recent Pat DNA Gene Seq. 2012;6(3):175–179. doi: 10.2174/187221512802717376. [DOI] [PubMed] [Google Scholar]
- Storz JF, Bridgham JT, Kelly SA, Garland T., Jr Genetic approaches in comparative and evolutionary physiology. Am J Physiol Regul Integr Comp Physiol. 2015;309(3):R197–214. doi: 10.1152/ajpregu.00100.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchomel TJ, Nimphius S, Bellon CR, Stone MH. The importance of muscular strength: training considerations. Sports Med. 2018;48(4):765–785. doi: 10.1007/s40279-018-0862-z. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Tajima F. The amount of DNA polymorphism when population size changes linearly. Genes Genet Syst. 2017;92(1):55–57. doi: 10.1266/ggs.16-00055. [DOI] [PubMed] [Google Scholar]
- Tanisawa K, Wang G, Seto J, Verdouka I, Twycross-Lewis R, Karanikolou A, Tanaka M, Borjesson M, Di Luigi L, Dohi M, Wolfarth B, Swart J, Bilzon JLJ, Badtieva V, Papadopoulou T, Casasco M, Geistlinger M, Bachl N, Pigozzi F, Pitsiladis Y. Sport and exercise genomics: the FIMS 2019 consensus statement update. Br J Sports Med. 2020;54(16):969–975. doi: 10.1136/bjsports-2019-101532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevis M, Kuuranne T, Geyer H. Annual banned-substance review: Analytical approaches in human sports drug testing. Drug Test Anal. 2019;11(1):8–26. doi: 10.1002/dta.2549. [DOI] [PubMed] [Google Scholar]
- Thomaes T, Thomis M, Onkelinx S, Fagard R, Matthijs G, Buys R, Schepers D, Cornelissen V, Vanhees L. A genetic predisposition score for muscular endophenotypes predicts the increase in aerobic power after training: the CAREGENE study. BMC Genet. 2011;12:84. doi: 10.1186/1471-2156-12-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DT, Erdman KA, Burke LM. American college of sports medicine joint position statement nutrition and athletic performance. Med Sci Sports Exerc. 2016;48(3):543–568. doi: 10.1249/mss.0000000000000852. [DOI] [PubMed] [Google Scholar]
- Thomis MA, Beunen GP, Maes HH, Blimkie CJ, Van Leemputte M, Claessens AL, Marchal G, Willems E, Vlietinck RF. Strength training: importance of genetic factors. Med Sci Sports Exerc. 1998;30(5):724–731. doi: 10.1097/00005768-199805000-00013. [DOI] [PubMed] [Google Scholar]
- Thomis MA, Beunen GP, Van Leemputte M, Maes HH, Blimkie CJ, Claessens AL, Marchal G, Willems E, Vlietinck RF. Inheritance of static and dynamic arm strength and some of its determinants. Acta Physiol Scand. 1998;163(1):59–71. doi: 10.1046/j.1365-201x.1998.00344.x. [DOI] [PubMed] [Google Scholar]
- Timmons JA, Knudsen S, Rankinen T, Koch LG, Sarzynski M, Jensen T, Keller P, Scheele C, Vollaard NB, Nielsen S, Akerström T, MacDougald OA, Jansson E, Greenhaff PL, Tarnopolsky MA, van Loon LJ, Pedersen BK, Sundberg CJ, Wahlestedt C, Britton SL, Bouchard C (2010) Using molecular classification to predict gains in maximal aerobic capacity following endurance exercise training in humans. J Appl Physiol (1985) 108 (6):1487–1496. 10.1152/japplphysiol.01295.2009 [DOI] [PMC free article] [PubMed]
- Tucker R, Santos-Concejero J, Collins M. The genetic basis for elite running performance. Br J Sports Med. 2013;47(9):545–549. doi: 10.1136/bjsports-2013-092408. [DOI] [PubMed] [Google Scholar]
- Uddin S, Khan A, Hossain ME, Moni MA. Comparing different supervised machine learning algorithms for disease prediction. BMC Med Inform Decis Mak. 2019;19(1):281. doi: 10.1186/s12911-019-1004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unal M, Ozer Unal D. Gene doping in sports. Sports Med. 2004;34(6):357–362. doi: 10.2165/00007256-200434060-00002. [DOI] [PubMed] [Google Scholar]
- Vaeyens R, Lenoir M, Williams AM, Philippaerts RM. Talent identification and development programmes in sport : current models and future directions. Sports Med. 2008;38(9):703–714. doi: 10.2165/00007256-200838090-00001. [DOI] [PubMed] [Google Scholar]
- Van den Berghe G, Bontemps F, Vincent MF, Van den Bergh F. The purine nucleotide cycle and its molecular defects. Prog Neurobiol. 1992;39(5):547–561. doi: 10.1016/0301-0082(92)90006-z. [DOI] [PubMed] [Google Scholar]
- van der Gronde T, de Hon O, Haisma HJ, Pieters T. Gene doping: an overview and current implications for athletes. Br J Sports Med. 2013;47(11):670–678. doi: 10.1136/bjsports-2012-091288. [DOI] [PubMed] [Google Scholar]
- van Loon LJ, Tipton KD. Concluding remarks: nutritional strategies to support the adaptive response to prolonged exercise training. Nestle Nutr Inst Workshop Ser. 2013;75:135–141. doi: 10.1159/000345862. [DOI] [PubMed] [Google Scholar]
- Varillas Delgado D, Telleria Orriols JJ, Martin Saborido C. Liver-metabolizing genes and their relationship to the performance of elite Spanish male endurance athletes; a prospective transversal study. Sports Med Open. 2019;5(1):50. doi: 10.1186/s40798-019-0227-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varillas Delgado D, Tellería Orriols JJ, Monge Martín D, Del Coso J. Genotype scores in energy and iron-metabolising genes are higher in elite endurance athletes than in nonathlete controls. Appl Physiol Nutr Metab. 2020;45(11):1225–1231. doi: 10.1139/apnm-2020-0174. [DOI] [PubMed] [Google Scholar]
- Varillas-Delgado D, Tellería Orriols JJ, Del Coso J. Genetic profile in genes associated with cardiorespiratory fitness in elite spanish male endurance athletes. Genes. 2021 doi: 10.3390/genes12081230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan D, Brogioli M, Maier T, White A, Waldron S, Rittweger J, Toigo M, Wettstein J, Laczko E, Flück M. The Angiotensin converting enzyme insertion/deletion polymorphism modifies exercise-induced muscle metabolism. PLoS One. 2016;11(3):e0149046. doi: 10.1371/journal.pone.0149046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellers HL, Kleeberger SR, Lightfoot JT. Inter-individual variation in adaptations to endurance and resistance exercise training: genetic approaches towards understanding a complex phenotype. Mamm Genome. 2018;29(1–2):48–62. doi: 10.1007/s00335-017-9732-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verroken M. Drug use and abuse in sport. Baillieres Best Pract Res Clin Endocrinol Metab. 2000;14(1):1–23. doi: 10.1053/beem.2000.0050. [DOI] [PubMed] [Google Scholar]
- Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, Yang J. 10 Years of GWAS discovery: biology, function, and translation. Am J Hum Genet. 2017;101(1):5–22. doi: 10.1016/j.ajhg.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagle JP, Carroll KM, Cunanan AJ, Wetmore A, Taber CB, DeWeese BH, Sato K, Stuart CA, Stone MH. Preliminary investigation into the effect of ACTN3 and ACE polymorphisms on muscle and performance characteristics. J Strength Cond Res. 2021;35(3):688–694. doi: 10.1519/jsc.0000000000002809. [DOI] [PubMed] [Google Scholar]
- Wang G, Padmanabhan S, Wolfarth B, Fuku N, Lucia A, Ahmetov II, Cieszczyk P, Collins M, Eynon N, Klissouras V, Williams A, Pitsiladis Y. Genomics of elite sporting performance: what little we know and necessary advances. Adv Genet. 2013;84:123–149. doi: 10.1016/b978-0-12-407703-4.00004-9. [DOI] [PubMed] [Google Scholar]
- Wang F, Kream RM, Stefano GB. An evidence based perspective on mRNA-SARS-CoV-2 vaccine development. Med Sci Monit. 2020;26:e924700. doi: 10.12659/msm.924700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webborn N, Williams A, McNamee M, Bouchard C, Pitsiladis Y, Ahmetov I, Ashley E, Byrne N, Camporesi S, Collins M, Dijkstra P, Eynon N, Fuku N, Garton FC, Hoppe N, Holm S, Kaye J, Klissouras V, Lucia A, Maase K, Moran C, North KN, Pigozzi F, Wang G. Direct-to-consumer genetic testing for predicting sports performance and talent identification: consensus statement. Br J Sports Med. 2015;49(23):1486–1491. doi: 10.1136/bjsports-2015-095343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss EP, Kulaputana O, Ghiu IA, Brandauer J, Wohn CR, Phares DA, Shuldiner AR, Hagberg JM. Endurance training-induced changes in the insulin response to oral glucose are associated with the peroxisome proliferator-activated receptor-gamma2 Pro12Ala genotype in men but not in women. Metabolism. 2005;54(1):97–102. doi: 10.1016/j.metabol.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Williams AG, Folland JP. Similarity of polygenic profiles limits the potential for elite human physical performance. J Physiol. 2008;586(1):113–121. doi: 10.1113/jphysiol.2007.141887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JM, Loenneke JP, Jo E, Wilson GJ, Zourdos MC, Kim JS. The effects of endurance, strength, and power training on muscle fiber type shifting. J Strength Cond Res. 2012;26(6):1724–1729. doi: 10.1519/JSC.0b013e318234eb6f. [DOI] [PubMed] [Google Scholar]
- Yan X, Dvir N, Jacques M, Cavalcante L, Papadimitriou ID, Munson F, Kuang J, Garnham A, Landen S, Li J, O'Keefe L, Tirosh O, Bishop DJ, Voisin S, Eynon N (2018) ACE I/D gene variant predicts ACE enzyme content in blood but not the ACE, UCP2, and UCP3 protein content in human skeletal muscle in the Gene SMART study. J Appl Physiol (1985) 125 (3):923–930. 10.1152/japplphysiol.00344.2018 [DOI] [PubMed]
- Yan X, Dvir N, Jacques M, Cavalcante L, Papadimitriou ID, Munson F, Kuang J, Garnham A, Landen S, Li J, O'Keefe L, Tirosh O, Bishop DJ, Voisin S. Eynon N (2018) ACE I/D gene variant predicts ACE enzyme content in blood but not the ACE, UCP2, and UCP3 protein content in human skeletal muscle in the Gene SMART study. J Appl Physiol. 1985;125(3):923–930. doi: 10.1152/japplphysiol.00344.2018. [DOI] [PubMed] [Google Scholar]
- Yang N, MacArthur DG, Gulbin JP, Hahn AG, Beggs AH, Easteal S, North K. ACTN3 genotype is associated with human elite athletic performance. Am J Hum Genet. 2003;73(3):627–631. doi: 10.1086/377590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadro JR, Shirley D, Andrade TB, Scurrah KJ, Bauman A, Ferreira PH. The beneficial effects of physical activity: Is it down to your genes? a systematic review and meta-analysis of twin and family studies. Sports Med Open. 2017;3(1):4. doi: 10.1186/s40798-016-0073-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YF, Wang H, Cheng Q, Qin L, Tang NL, Leung PC, Kwok TC. The relationship between angiotensin-converting enzyme (ACE) insertion (I)/deletion (D) polymorphism, serum ACE activity and bone mineral density (BMD) in older Chinese. J Renin Angiotensin Aldosterone Syst. 2017;18(1):1470320316688345. doi: 10.1177/1470320316688345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W. Advances in the genetics of exercise performance. Yi Chuan. 2014;36(4):301–308. [PubMed] [Google Scholar]
- Zhou DQ, Hu Y, Liu G, Gong L, Xi Y, Wen L. Muscle-specific creatine kinase gene polymorphism and running economy responses to an 18-week 5000-m training programme. Br J Sports Med. 2006;40(12):988–991. doi: 10.1136/bjsm.2006.029744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubair N, Conomos MP, Hood L, Omenn GS, Price ND, Spring BJ, Magis AT, Lovejoy JC. Genetic Predisposition impacts clinical changes in a lifestyle coaching program. Sci Rep. 2019;9(1):6805. doi: 10.1038/s41598-019-43058-0. [DOI] [PMC free article] [PubMed] [Google Scholar]