Abstract
The adsorption process of SARS-CoV-2 Omicron spike protein to the nano-gold colloid surfaces was examined by monitoring the surface plasmon resonance (SPR) band shift of gold-nano particles ranging between diameters of d = 10–100 nm. The externally changed pH between 3 and 11 at 24.5 ± 0.4 °C initiated a reversible formation of the gold colloid aggregates, where formation/deformation of the aggregates were monitored by red/blue shift of the peak of the SPR band. There was no sign of reversible aggregation for d = 10, 15, and 20 nm gold colloids. A clear undulation of the peak shift corresponding to pH hopping between pH ~3 and ~11 was confirmed for colloidal d > 30 nm. This degree of the reversibility was compared to previously reported SARS-CoV-2 Alpha spike protein coated gold colloids. It was concluded that Omicron spike protein possesses a similar low affinity for gold nano particle d < 20 nm and possesses the higher affinity to the gold nanoparticles of d > 30 nm. However, the Omicron spike protein conformation was presumed to be more denatured compared to the SARS-CoV-2 Alpha spike protein. Our finding suggested Omicron spike protein was more acid labile/flexible.
Keywords: Omicron, SARS-CoV-2, Spike protein, Gold nano-particles, Protein folding, Protein corona, Reversible aggregation, SPR (Surface plasmon resonance) band
1. Introduction
The SARS-CoV-2, also known as the coronavirus, has undergone natural mutation and antigenic variation over time [1], [2], and multiple highly concerning variants have been reported, such as, the Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), and Delta (B.1.617.2) coronavirus [3]. The Omicron variant (B.1.1.529) had 32 mutations of the spike protein, presenting a serious increase of pathogenicity compared to the previous variants. Relative to Alpha, Beta, and Delta SARS-CoV-2 variants, Omicron has a 5.5–11 times higher mutation rate in the receptor-binding motif (RBM) [4], [5]. The S1 subunit spike protein contains a receptor binding domain (RBD) composed of 319–541 residues. The reported mutations for Omicron in RBD region are: G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, and Y505H [6].
Among all the mutations, the crucial mutations in the RBM of the Omicron variant are considered to be T478K, E484A, Q493R and N501Y [7], since these mutations are considered to increase the affinity to the hACE2 receptor binding [5]. Because a transformation to a hydrophilic amino acid (E: Glutamic acid) to a hydrophobic amino acid (A: Alanine) in 484 position, N: Asparagine to Y: Tyrosine in 501 position [4], Q: Glutamine to R: Arginine in 493 position, and T: Threonine to K: Lysine in 478 position, and the T478K mutation also occurred in the delta variant. Our group investigated the behavior of the cell fusion protein with respect to the molecular machinery function and the affinity of SARS-CoV-2 Alpha spike protein over gold-nano colloids. As proven in more serious infection rates, the several critical mutations in S1 unit suggesting significantly different mechanical functions can be found in the spike protein of the Omicron variant.
The adsorption of proteins over the nano-particles has been known as protein corona, and there have been many important applications in biomaterial sciences, the biomedical field, and immunology. The protein corona plays a major role in forming nano-aggregates. In order to characterize the aggregation of the bio-nano particles, understanding the conformation of a forming protein corona is invaluable to design new bio-nano-materials [1], [6], [8], [9]. A layer of protein corona was divided into two regimes: hard and soft. The designation of hard and soft corona is dependent on the relative strength of the binding energy. The hard designation is given to the regime of the protein corona with stronger binding energy [2]. With a significant amount of interest paid towards proteins associated with antibodies, our research group has been investigating the protein corona, of amyloidogenic peptides, to the gold nano-particles in order to identify the intermediates of fibrillogenesis (amyloids). We prepared the amyloidogenic peptide coated gold nano-particles and controlled the conformational change by externally varying the pH, making peptides unfold in acidic conditions and refold in basic conditions. Since the segments used for the adsorption over gold nano-particles won’t be critical for protein networking, the investigation of the aggregation process would focus on the segments responsible for protein-protein interaction.
We conducted the characterization of nano-size dependent proteins of corona by examining the SARS-CoV-2 Alpha spike protein through the formation of pH dependent gold colloid aggregates [3], [10]. We found that an affinity was significantly more noticeable for gold colloids with a diameter (d) of 30 nm and larger. There was indication of a reversible transition between folded and unfolded conformations of RBD, possibly dimerizing the S protein through the RBD. The highly acidic condition (e.g., pH ~3) is speculated to prepare the denatured conformation of an S-protein before initiating the binding to the hACE2 and also is considered to provide a hint of the conformation after endocytosis of Alpha SARS-CoV-2 to achieve a cell fusion process [11], [12], [13], [14], [15].
2. Experimental
Since the preparation of gold colloid coated with spike protein was described in previous reports [3], [10], [16], the sample preparations will be briefly described here. A total of nine different sizes of gold colloidal particles with conventional diameters of (d) = 10 (9.8 ± 1), 15 (15.2 ± 1.5), 20 (19.7 ± 1.1), 30 (30.7 ± 1.3), 40 (40.6 ± 1.1), 50 (51.5 ± 4), 60 (60 ± 1.0), 80 (80 ± 1.0), and 100 (99.5 ± 1.3) nm were used (Ted Pella Inc., Redding, California, USA), and their shapes were confirmed to be spherical [17]. The SARS-CoV-2 Spike trimer, His Tag (B.1.1.529/Omicron) was purchased from Acro Biosystems ACRO Biosystems (Newark, Delaware, USA; Catalog # SPN-C52Hz), and the molecular weight was 138.2 kDa. Since Omicron has almost an identical molecular weight as that of Alpha, the prepared stock solution and concentration was equivalent to that for Alpha spike protein [3], [10]. The fixed concentration of Omicron S-protein (~100 picomole) was mixed with each gold nano-particles with the ratio of [S-protein] / [Gold Colloid] ranging from ~5 to ~3400.
We examined the shift of the Surface Plasmon Resonance (SPR) band of S protein coated nano-gold colloids as a function of the change of pH at 24.5 ± 0.4 °C by utilizing Cary 5000 Model UV–VIS–NIR Spectrophotometer (Agilent Technologies Inc., Santa Clara, CA, USA). Two different schemes of pH changes were used: (1) a gradual change to the pH ~3 with the addition of hydrochloric acid (HCl), and (2) pH hop between acidic at pH 3.5 ± 0.6 (pH ~3) and basic condition around pH 10.7 ± 0.5 (pH ~11) by inserting HCl and sodium hydroxide (NaOH) with pre-tested amount to maintain pH ~3 and pH~11, respectively. In both schemes, each spectrum was processed with the component of the band expressed by a Gaussian profile by Peak Fit function of OriginPro 2018b (OriginLab Corporation, Northampton, MA, USA), and the spectrum area weight average peak position of the spectrum in the region between 480 nm and 850 nm was extracted [16].
3. Results
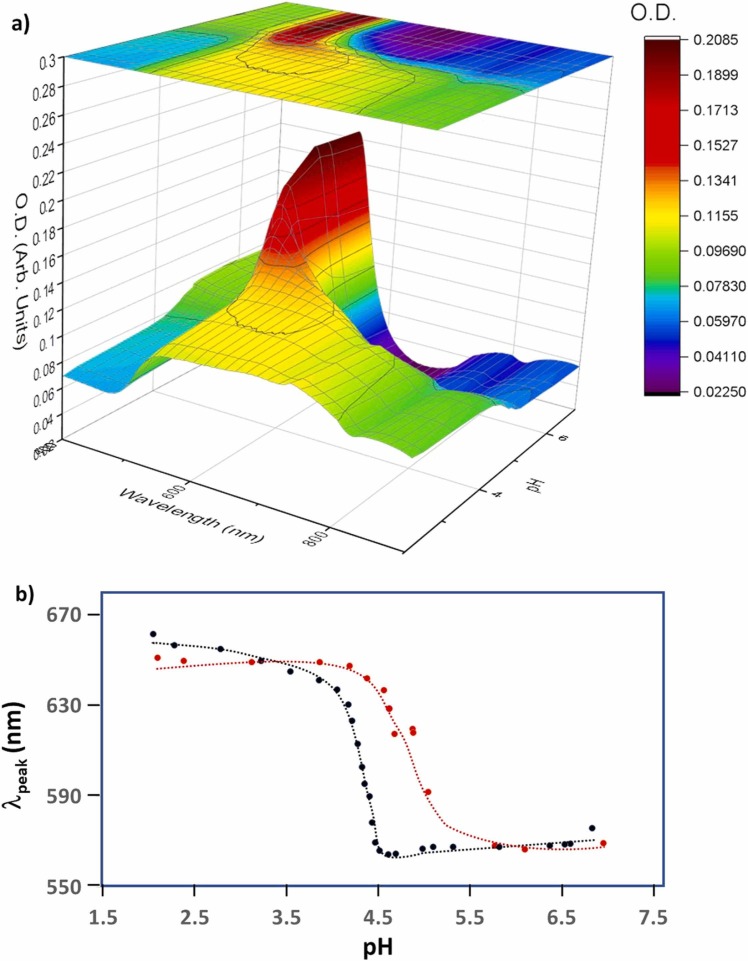
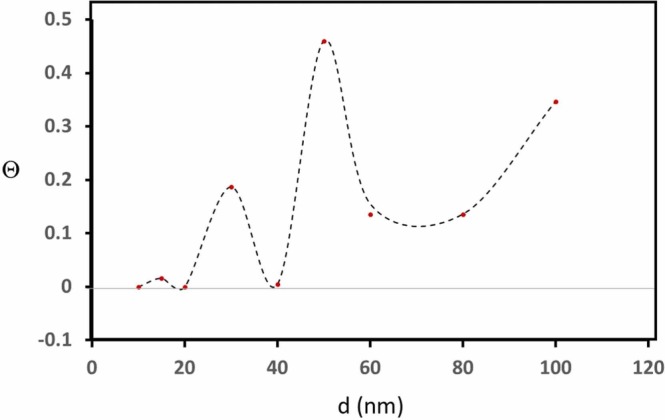
The gradual pH change down to ~pH 3 reflected on the gradual red shift of SPR band as shown in Fig. 1a, represented for the case of Omicron coated gold 50 nm particles. As the trace of the average peak band position, in the white dotted line exhibited a sigmoidal curve as shown in Fig. 1b. This sigmoidal feature was analyzed by the Boltzmann model and the inflection point (pHo) and the tangential slope at pHo, pHo (1), where dpH = (λmax – λmin)/4λpeak (1) [18], as shown in Table 1a, Table 1b. The difference of pHo between Omicron protein coated gold colloids and bare gold colloids, ΔpHo, were postulated to reflect the change of the surface property due to the adsorption of Omicron spike protein over the gold surface. The best plausible correlation, <R2 > = 0.46, was observed between 1/dpH (∝ λ(1)) and ΔpHo (> 0) and excluding ΔpHo for d = 60 nm. This relationship was used to extract the protein coverage fraction, Θ as shown in Table 2. The relatively low correlation shows that an assumption of a linear relationship between dpH and ΔpHo did not stand for these systems. The extracted Θ values were reproduced by conducting the geometric simulation reported previously. Because of the physical similarities to SARS-CoV-2 Alpha spike protein, the initial geometry of a prolate was expressed with that optimized to SARS-CoV-2 spike protein; a = 5.3 nm and b = 8.5 nm with a “spiking-out” orientation of the protein from the particle surface [16]. The axial lengths were optimized to match with extracted Θ for each gold colloid of diameter d (conventional size) and reported diameter size d was used for the fit as shown in Fig. 2. A unique point in Omicron protein was that much less contribution of the secondary layer was observed for an entire range of gold colloid diameters and here the analysis for d = 10, 15, and 20 nm were excluded due to a sign of no adsorption, and d = 60 nm due to a sign of involvement of a different factor which did not follow a linear relationship between dpH and ΔpH. While the average contribution of the second layer in amyloidogenic peptides coated gold colloids was ~40%, and that for Alpha was ~ 30%, for Omicron s-protein only d = 50 nm (Θ-2nd ~40%) exhibited a significant contribution from the 2nd layer. Quite interestingly, the sigmoidal fit was not successful for d = 60 nm for both Alpha and Omicron indicating the Z-shape sigmoidal curve. In Table 1a, the values for Alpha are those after the adjustment for d = 60 nm and showed around <R2 > ~ 0.7 before the adjustment, which was like the current work for Omicron showing <R2 > ~0.89 in contrast to the average value of 0.97. The general trend in pHo was an increase as the d increases and the trend was like what we observed for Alpha. In Fig. 3, the summary and comparison of ΔpHo was shown to justify the effect of the mutation to the effect to pH where protein conformation change took place. While a general trend was roughly the same, a striking feature was seen in the d = ~20 nm where Omicron shows strongly negative value implying that the negative value was a sign of the desorption of protein from the gold surface. Fig. 4.
Fig. 1.
a) The SPR band for visible range (400–900 nm) as a function of pH. The contour implying the is shown by a white dotted line on the surface contour map for Omicron S-protein mixed with 50 nm gold nano particles. b) The plot of vs. pH for both Omicron (red) and Alpha (black) S-protein mixed with 50 nm gold particles and the optimized fit by Boltzmann model is shown by the dotted line as a guide. Each data point exclude the deviations of measurements. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 1a.
The summary of Boltzmann plots for sigmoidal features for SARS-CoV-2 Omicron spike protein and Alpha spike proteins for each gold colloidal size from this study and compared to our previous report [10].
| Omicron | d (nm) | A1 | A2 | pHo | dpH | <R2 > |
|---|---|---|---|---|---|---|
| 10 | 666(4) | 581(1) | 1.92(2) | 0.13(2) | 0.98498 | |
| 15 | 610(3) | 575(1) | 3.10(8) | 0.34(5) | 0.99081 | |
| 20 | 689(9) | 575.7(3) | 1.83(9) | 0.55(3) | 0.99924 | |
| 30 | 652(2) | 576(2) | 4.40(1) | 0.07(1) | 0.98441 | |
| 40 | 666(6) | 572(6) | 4.33(8) | 0.31(9) | 0.94079 | |
| 50 | 650(2) | 567(3) | 4.92(4) | 0.25(4) | 0.98252 | |
| 60 | 733(30) | 544(33) | 4.5(4) | 0.8(4) | 0.896 | |
| 80 | 679(1) | 580(2) | 4.50(2) | 0.15(1) | 0.99552 | |
| 100 | 697(4) | 617(4) | 4.60(5) | 0.20(4) | 0.95608 | |
| Avg. | 671(34) | 576(19) | 3.8(12) | 0.3(2) | 0.97(3) | |
| Alpha | d (nm) | A1 | A2 | pHo | dpH | <R2 > |
| 10 | 630(2) | 585.4(5) | 3.27(2) | 0.21(2) | 0.9799 | |
| 15 | 650(2) | 582.4(7) | 3.54(3) | 0.31(2) | 0.9902 | |
| 20 | 643(2) | 585.2(5) | 3.33(4) | 0.37(3) | 0.99 | |
| 30 | 644(2) | 569.8(9) | 4.34(2) | 0.20(2) | 0.9888 | |
| 40 | 661(1) | 574.4(9) | 4.33(2) | 0.22(2) | 0.9947 | |
| 50 | 651(2) | 567(2) | 4.28(1) | 0.092(1) | 0.9803 | |
| 60 | 668(3) | 562(2) | 4.180(7) | 0.051(8) | 0.9815 | |
| 80 | 684(1) | 571.5(7) | 4.47(1) | 0.26(1) | 0.9978 | |
| 100 | 698.5(9) | 617.2(9) | 4.85(2) | 0.26(2) | 0.995 | |
| Avg. | 659(22) | 579(16) | 4.1(6) | 0.22(10) | 0.989(7) |
Table 1b.
The summary of pHo and ΔpHo for SARS-CoV-2 Omicron spike protein and Alpha spike proteins for each gold colloidal size from this study and compared to our previous report [10].
| Omicron |
Alpha |
|||
|---|---|---|---|---|
| d (nm) | pHo | ΔpHo | pHo | ΔpHo |
| 10 | 1.92(2) | –1.48 | 3.27(2) | –0.13 |
| 15 | 3.10(8) | 0.03 | 3.54(3) | 0.47 |
| 20 | 1.83(9) | –1.87 | 3.33(4) | –0.37 |
| 30 | 4.40(1) | 0.35 | 4.34(2) | 0.29 |
| 40 | 4.33(8) | 0.01 | 4.33(2) | 0.01 |
| 50 | 4.92(4) | 0.86 | 4.28(1) | 0.22 |
| 60 | 4.5(4) | 0.29 | 4.180(7) | –0.03 |
| 80 | 4.50(2) | 0.254 | 4.47(1) | 0.224 |
| 100 | 4.60(5) | 0.650 | 4.85(2) | 0.90 |
| Average | 3.8(12) | –0.10 (93) | 4.1(6) | 0.18(37) |
Table 2.
The predicted Θ based on the linear correlation between ΔpHo and 1/dpH.
| d (nm) | Θ |
|---|---|
| 15 | 0.016 |
| 30 | 0.187 |
| 40 | 0.005 |
| 50 | 0.459 |
| 60 | 0.136 |
| 80 | 0.136 |
| 100 | 0.347 |
| Avg. | 0.18(17) |
Fig. 2.
The observed surface coverage fraction Θ as a function of d (nm) shown in red dots. The simulated values were shown by the dotted curve. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
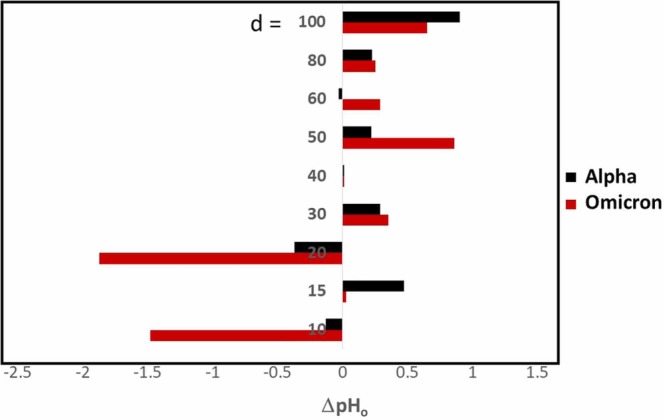
Fig. 3.
The comparison of ΔpHo as a function of gold colloid size (d) nm) for Omicron (red) and Alpha (black). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 4.
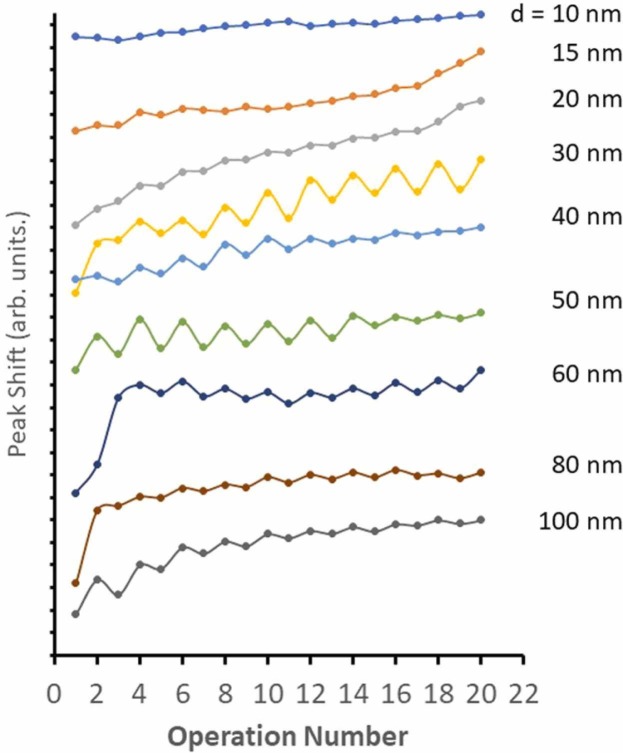
The trend of shift in SPR band, , as a function of operation number, n, where pH ~7 at n = 1, pH ~10 for odd n (red tick marks), and pH ~3 for even n (blue tick marks) for SARS-CoV-2 Omicron spike protein coated gold colloid for d = 10, 15, 20, 30, 40, 50, 60, 80, and 100 nm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
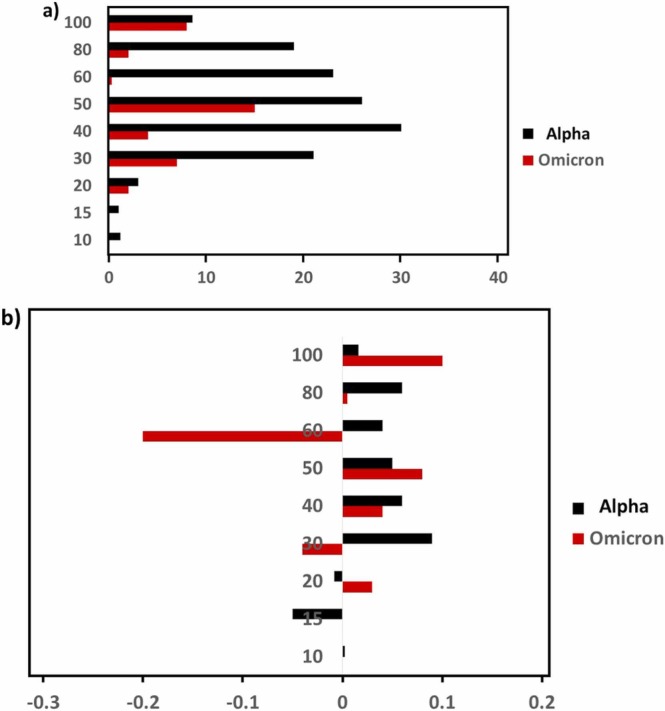
Except for d = 30, 40, 80, and 100 nm, ΔpHo was significantly different between two spike proteins. Especially, d = 10 and 20 showed a drastic decrease for Omicron and confirming the no adsorption of Omicron s-protein at those diameters. A huge increase for 50 nm and 60 nm shows a drastic surface character change and likely higher coverage. In summary, as seen in Fig. 2, the Θ was not a monotonic function of d as seen in the Alpha or amyloidogenic peptides.
Under the pH hopping scheme, the undulation of was obviously observed for the Omicron protein gold colloid of d > 30 nm. A remarkable feature indicating the reversible aggregation/de-aggregation was observed in the system for d > 30 nm, and all parameters were listed in Table 3. There was no sign of reversible process for d = 10, 15, and 20 nm and the parameters D and E in Eq. (1) were not obtained. Here, the pH value at the n (operational value) = 1 has an initial pH at around 7. For even operational numbers, it indicates a pH at around 3 and for odd operational numbers > 3 the pH’s were set at around 11. The analytical formula was applied to explain this behavior and was concluded that there was a reversible aggregation/disaggregation of gold nano-colloid mediated by Omicron protein at the interface. The undulation feature was explained by Eq. (1) [19].
| (1) |
Table 3.
The summary of the parameters utilized to describe the reversible aggregations formations for SARS-CoV-2 Omicron spike protein for the various gold colloidal sizes. In below, those parameters reported for SARS-CoV-2 Alpha spike proteins9 are reproduced as a comparison. A comparison of the parameters D and E for Omicron and Alpha are shown in Fig. 6a and b.
| Omicron | ||||||
|---|---|---|---|---|---|---|
| d (nm) | A (nm) | B (nm) | C (nm) | D (nm) | E | <R2 > |
| 10 | 582(2) | 2(1) | 0.8(2) | – | – | 0.91415 |
| 15 | 587(2) | 0.03(3) | 2.5(4) | – | – | 0.988 |
| 20 | 575(4) | 13(3) | 0.68(6) | 2(2) | 0.03(11) | 0.93464 |
| 30 | 581(5) | 27(5) | 0.04(4) | 7(2) | -0.04(2) | 0.97604 |
| 40 | 622(4) | 5(3) | 0.8(1) | 4(2) | 0.04(6) | 0.94479 |
| 50 | 629(4) | 5(3) | 0.6(2) | 15(2) | 0.08(2) | 0.94302 |
| 60 | 560(14) | 70(16) | 0.14(6) | 0.3(16) | -0.2(3) | 0.77487 |
| 80 | 581(2) | 62(3) | 0.153(9) | 2(1) | 0.005(41) | 0.99323 |
| 100 | 621(5) | 22(5) | 0.45(6) | 8(3) | 0.10(6) | 0.97158 |
| Alpha | ||||||
| d (nm) | A (nm) | B (nm) | C (nm) | D (nm) | E | <R2 > |
| 10 | 580(1) | 0.7(5) | 1.1(2) | 1.2(9) | 0.002(70) | 0.929 |
| 15 | 581(1) | 0.16(6) | 1.8(1) | 1.0(5) | -0.05(4) | 0.988 |
| 20 | 583(3) | 11(2) | 0.72(5) | 3(1) | 0.008(33) | 0.992 |
| 30 | 593(4) | 15(3) | 0.48(6) | 21(2) | 0.09(2) | 0.982 |
| 40 | 596(10) | 6(6) | 0.9(3) | 30(7) | 0.06(3) | 0.898 |
| 50 | 590(12) | 8(7) | 0.8(3) | 26(7) | 0.05(3) | 0.889 |
| 60 | 574(8) | 11(6) | 0.7(1) | 23(4) | 0.04(2) | 0.948 |
| 80 | 592(5) | 28(4) | 0.45(4) | 19(3) | 0.06(2) | 0.985 |
| 100 | 635(1) | 5.5(8) | 0.70(4) | 8.6(6) | 0.016(7) | 0.995 |
An initial peak position at neutral pH(i.e., (n = 1)) is given by A – D, and the parameters B and C show the average wave peak position shift as pH varies between pH ~3 and pH ~10. The parameters D represents a degree of reversibility amplitude, where (n = even) – (n = odd), and E is a damping (E < 0) or amplifying (E > 0) factor for the repetitive undulation. A reversible process was interpreted to be the stage of gold nano particle dispersed and aggregated. The aggregation of the gold colloid was presumed to be due to the mediation of the Omicron protein over the gold colloid surface. The obvious reversible process was observed for the gold colloid over diameter of 30 nm. Opposed to that, Alpha possessed a bi-structural undulation (i.e., two different amplitude sections), all pH hop features were single component undulation. In summary, the sign of adsorption was not observed for d = 10, 15, and 20 nm based on the reversible aggregation/de-aggregation process observed in the pH hop experiment. The case for d = 50 and d = 100 nm in Fig. 5 demonstrates that the amplitude of reversible process (parameter D) was reduced in the case of Omicron and the damping factor (parameter E) was significantly larger for Omicron.
Fig. 6.
The comparison of D and E parameters in Eq. (1) between Omicron (red) and Alpha (black) spike protein. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 5.
The trend of shift in SPR band, , as a function of operation number, n, for SARS-CoV-2 Omicron (red) and Alpha (black) spike protein coated gold colloid for a) d = 10, b) d = 50, and c) d = 100 nm. In order to make a fair comparison, the scale for this plot for was set the same.
4. Discussions
Omicron showed more correlation in nano-size dependence in D (or E) parameter(s) and ΔpHo. It indicates that Omicron spike protein may possess more significant nano-size dependent affinity and the adsorbed conformation was more sensitive to the acidic condition. In Θ simulation, a protein was approximated to have a prolate shape. Based on the studies with Alpha s-protein and near negligible mutations on the C-terminal side, the adsorption on the nano-gold surface took place at the C-terminal side. We explained the relatively lower Θ value by the gyration of the prolates with relatively larger Sd (distance between two adjacent prolates) [20]. The relatively larger Sd value was considered to be an outcome of repulsive interaction between adsorbed proteins. The repulsive forces must originate from an electrostatic repulsion between δ + (δ–) and δ + (δ–) in dipole-dipole interaction [20]. Among notable mutations in RBD of Omicron, T478K may support the above assumption, since polar uncharged Threonine (T) was mutated into positively charged Lysine (K), which exhibit more electro-positive condition.
There was no evidence of colloidal surface change due to the adsorption of the proteins [16], [17]. A recent unpublished report utilizing TEM of Alpha spike protein revealed that the adsorption orientation was confirmed to be “quasi” spiking-out orientation at pH 7–11 with a layer thickness of 6 nm [21], [22]. While there is not confirmation of a clear spiking-out orientation, it would support that the peptide forms a corona on the gold colloidal surface. If the spiking-out orientation was squeezed due to the pull of δ– surface of the gold colloid, it would make sense that the layer thickness is smaller than 10 nm, which was a rough ball park figure of the length of the spike protein [23]. Also, there was a possibility that the full length of a spike-protein could not be resolved by TEM, especially if the edge of the spike protein was less dense. Remarkably, there was a clear conformational change at pH 3 exhibiting almost sheet – like layers with thickness of ~2 nm. Thus, we have a confirmation of conformation change of a spike protein as a function of pH change. This also strongly suggests a conformation of β-pleated sheet forming from the unfolded peptide conformations.
Since the major mutations in the Omicron variant occur in the RBD, the significant differences observed between Alpha and Omicron coated gold colloids further supported that the adsorption site was on the C-terminal side and the RBD site was facing toward the outside. Otherwise, the major RBD mutations would not be expected to result in the significant changes observed in nano-size dependent aggregation processes. However, a limitation of the current study is the inferred direction of the RBD of the S-protein, and we are in the process of studying the effect of an insertion of hACE2 to the spike protein coated gold colloid to better elucidate S-protein orientation [24]. The binding of the hACE2 to RBD is expected to cause a cleavage of the RBD followed by the formation of pre-hairpin by an extension of protomers of fusion proteins [25], [26], [27], [28], [29], [30], [31], [32], while the hACE2 has no effect if the RBD side was used for the adsorption contact site. As we continue our investigation, a sub-microscale local conformational change of the form of adsorbed or networking proteins or segments of it will be probed by SERS Raman imaging technique under various pH and with/without hACE2 as a function of the nano-size to investigate the general role of fusion process involving s-protein adsorbed over the gold-nanoparticles.
The clear evidence of nano-size dependence affinity was observed for d = 10, 15, and 20 nm in both pH gradience and pH hop scheme, showing that the Omicron S-protein had no binding affinity for those sizes. This was also implied in Alpha S-protein and confirming that the size of S-protein is comparable to those sizes, and they may require more than the areas provided by spherical area of those gold colloids due to either protein-protein repulsive interaction or requirement of degree of freedom in mobility. With a rough estimation, the active range for S-protein to adsorb on the nano surface needs a ± 15 nm range. General consensus of the length of a spike protein is roughly 10 nm long [23]. The contacting point of the protein to the cell membrane was secured with envelope and membrane protein, and it may significantly reduce the mobility of S-protein over the cell surface. On the other hand, the gold colloidal surface possesses more drastic electrostatic conditions from surface plasmons, which provides a δ– surface charge field. If the field created by surface plasmon is considered to be more instantaneous or non-stationary, it would induce more degree of mobility to the dipole over the surface (i.e., gyration motion of a prolate) resulting in more mobility in the gold colloid surface compared to the case of S-protein placed over the membrane.
The current work demonstrated a good example of a formation of the protein corona for a different nano-size and the corresponding surface property in response to an external pH change. The hard protein corona is characterized as those proteins possessing the higher affinity. The indication of the reduced involvement of the 2nd layer, which has the lower affinity, the present work shows that S-protein corona was mainly formed with the hard protein corona. The property of S-protein hard core under the lower pH was, therefore, revealed to be less resilient compared to that of Alpha. If the RBD side is assumed to be responsible for an aggregation process, then one, or more, of the major mutations K, A, R, or Y from T478K, E484A, Q493R, N501Y is inferred to be causing the peptide to show less reversibility in aggregation (i.e., completely denatured or unfolded by acidic solution environment). Our studies on Aβ1–40 coated gold colloid showed that Lysine was easily impacted by the acid insertion to act as a pivotal point for unfolding (or folding) conformational change.
Popularly accepted Verma effect [33], [34], [35] developed for characterizing protein corona describes the first initiation of soft corona formation with proteins of the weaker affinity being replaced by strong corona of the higher affinity under the diffusion process. The current work revealed the aggregation process was interpreted to monitor the interaction between the hard corona (corresponding to the 1st layer in this work) and the soft corona (corresponding to the 2nd layer in this work) as initiated by the pH change. The observed reversible aggregation process proves there was a control and pH dependency in the conformation of the hard protein corona. Because reversible amplitude, D parameter, was relatively low, it could conclude soft-hard corona interaction was stronger for the Omicron variant than the earlier Alpha variant.
5. Conclusions
The spectroscopic features of pH dependent SPR band shift was utilized to evaluate the affinity of SARS-CoV-2 Omicron spike protein to the nano-gold colloids. There were consistent results that were observed in both Omicron and Alpha spike proteins. For example, relatively high affinity was confirmed for the gold colloid of diameter, d > 30 nm. No monotonic relationship between surface coverage ratio Θ and gold nano-colloid diameter, d, was observed. While no conclusive evidence of S-protein orientation was available, it is reasonable to assume Omicron spike protein took “spiking-out” adsorption orientation, which was the same conclusion drawn for Alpha spike protein. Compared to Alpha spike protein, Omicron spike protein was found to be more easily denatured by acidic condition resulting in a larger damping factor and smaller reversibility amplitude (i.e., difference of the average peak position between basic ad acidic condition). This increased acid-induced denaturation suggests the Omicron S-protein might be more responsive to the intracellular acidification. The investigation of pH induced reversible aggregation process provided the insights of the formation of soft and hard protein corona, and the Omicron variant was concluded to possess stronger of and hard protein corona interaction prohibiting the spike protein to conduct reversible folding-unfolding conformation.
CRediT authorship contribution statement
K.Y., T.L., J.S., and A.I. developed, performed, and analyzed the simulations. K. Y. designed the research, analyzed data, and wrote the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Wolfram J., Yang Y., Shen J., Moten A., Chen C., Shen H., Ferrari M., Zhao Y. The nano-plasma interface: Implications of the protein corona. Colloids Surf. B. 2014;124:17–24. doi: 10.1016/j.colsurfb.2014.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhdanov V.P., Kasemo B. Monte Carlo simulation of the kinetics of protein adsorption. Protein.: Struct., Funct. Genet. 1998;30:177–182. [PubMed] [Google Scholar]
- 3.Yokoyama K., Ichiki A. Nano-size dependence in the adsorption by the SARS-CoV-2 spike protein over gold colloid. Colloids Surf. A: Physicochem. Eng. Asp. 2021;615 doi: 10.1016/j.colsurfa.2021.126275. 126275-112681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weissman D., Alameh M.G., de Silva T., Collini P., Hornsby H., Brown R., LaBranche C.C., Edwards R.J., Sutherland L., Santra S., Mansouri K., Gobeil S., McDanal C., Pardi N., Hengartner N., Lin P.J.C., Tam Y., Shaw P.A., Lewis M.G., Boesler C., Şahin U., Acharya P., Haynes B.F., Korber B., Montefiori D.C. D614G spike mutation increases SARS CoV-2 susceptibility to neutralization. Cell Host Microbe. 2021;29:23–31. doi: 10.1016/j.chom.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wrapp D., De Vlieger D., Corbett K.S., Torres G.M., Wang N., Van Breedam W., Roose K., van Schie L., Hoffmann M., Pohlmann S., Graham B.S., Callewaert N., Schepens B., Saelens X., McLellan J.S. Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell. 2020;181:1004–1015. doi: 10.1016/j.cell.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rahman M., Laurent S., Tawil N., Yahia L.H., Mahmoudi M. Springer; 2013. Protein-Nanoparticle Interactions, The Bio-nano Interface; pp. 21–44. [Google Scholar]
- 7.Yi C., Sun X., Ye J., Ding L., Liu M., Yang Z., Lu X., Zhang Y., Ma L., Gu W., Qu A., Xu J., Shi Z., Ling Z., Sun B. Key residues of the receptor binding motif in the spike protein of SARS-CoV-2 that interact with ACE2 and neutralizing antibodies. Cell Mol. Immunol. 2020;17:621–630. doi: 10.1038/s41423-020-0458-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hooshmand N., Thoutam A., Anikovskiy M., Labouta H.I., El-Sayed M. Localized surface plasmon resonance as a tool to study protein corona formation on nanoparticles. J. Phys. Chem. C. 2021;125:24765–24776. [Google Scholar]
- 9.Lundqvist M., Sethson I., Jonsson B.-H. Protein adsorption onto silica nanoparticles: Conformational changes depend on the particles’ curvature and the protein stability. Langmuir. 2004;20:10639–10647. doi: 10.1021/la0484725. [DOI] [PubMed] [Google Scholar]
- 10.Yokoyama K., Ichiki A. Spectroscopic investigation on the affinity of SARS-CoV-2 spike protein to gold nano-particles. Colloid Interface Sci. Commun. 2021;40:100356–100362. doi: 10.1016/j.colcom.2020.100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Francés-Monerris A., Hognon C.C., Miclot T., García-Iriepa C., Iriepa I., Terenzi A., Grandemange S.P., Barone G., Marazzi M., Monari A. Molecular basis of SARS-CoV-2 infection and rational design of potential antiviral agents: Modeling and simulation approaches. J. Proteome Res. 2020;19:4291–4315. doi: 10.1021/acs.jproteome.0c00779. [DOI] [PubMed] [Google Scholar]
- 12.Yuan M., Wu N.C., Zhu X., Lee C.-C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirchdoerfer R.N., Wang N., Pallesen J., Wrapp D., Turner H.L., Cottrell C.A., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci. Rep. 2018;8:15701–15711. doi: 10.1038/s41598-018-34171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malik Y.A. Properties of coronavirus and SARS-CoV-2. Malays. J. Pathol. 2020;42:3–11. [PubMed] [Google Scholar]
- 16.Yokoyama K., Brown K., Shevlin P., Jenkins J., D’Ambrosio E., Ralbovsky N., Battaglia J., Deshmukh I., Ichiki A. Examination of adsorption orientation of amyloidogenic peptides over nano-gold colloidal particles’ surfaces. Surf. Int. J. Mol. Sci. 2019;20:5354–5380. doi: 10.3390/ijms20215354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yokoyama K., Cho H., Cullen S.P., Kowalik M., Briglio N.M., Hoops H.J., Zhao Z., Carpenter M.A. Microscopic investigation of reversible nanoscale surface size dependent protein conjugation. Int. J. Mol. Sci. 2009;10:2348–2366. doi: 10.3390/ijms10052348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yokoyama K., Ichiki A. In: Advances in Chemical Research. Taylor J.C., editor. NOVA Science Publisher; Hauppauge, NY, USA: 2020. Oligomerization and Adsorption Orientation of Amyloidogenic Peptides over Nano-gold Colloidal Particle Surfaces; pp. 139–194. [Google Scholar]
- 19.Yokoyama K. In: Nanoscale Spectroscopy with Applications. Musa S.M., editor. CRC Press-Taylor and Francis Group LLC; 2013. Controlling Reversible Self-assembly Path of Amyloid Beta Peptide over Gold Colloidal Nanoparticle’s Surface; pp. 279–304. [Google Scholar]
- 20.Yokoyama K., Ichiki A. A model for nano-scale spherical surface coverage and protein corona formation by amyloidogenic peptides. Int. J. Biocomput. Nano Technol. 2022;1:21–28. [Google Scholar]
- 21.Yokoyama K. American Chemical Society National MeetingSan Diego Convention Center; San Diego, CA, USA: 2022. Adsorption orientation of amyloidogenic peptides over nano-gold colloid surface. [Google Scholar]
- 22.Yokoyama K. American Chemical Society National MeetingSan Diego Convention Center; San Diego, CA, USA: 2022. Characterization of SARS-CoV-2 Spike Protein at Nano-scale Interface. [Google Scholar]
- 23.Bar-On Y.M., Flamholz A., Phillips R., Milo R. SARS-CoV-2 (COVID-19) by the numbers. eLife x. 2020;9:1–15. doi: 10.7554/eLife.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.K. Yokoyama, T. Lam, J. Santariello, Z.C. Lin, Reversible aggregation process of SARS-CoV-2 Alpha spike protein over gold colloid nano particles and its effect by ACE2, (to be submitted).
- 25.Verdecchia P., Reboldi G., Cavallini C., Mazzotta G., Angeli F. ACE-inhibitors, angiotensin receptor blockers and severe acute respiratory syndrome caused by coronavirus. G Ital. Cardiol. 2020;21:321–327. doi: 10.1714/3343.33127. [DOI] [PubMed] [Google Scholar]
- 26.Turner A.J., Tipnis S.R., Guy J.L., Rice G.I., Hooper N.M. ACEH/ACE2 is a novel mammalian metallocarboxypeptidase and a homologue of angiotensin-converting enzyme insensitive to ACE inhibitors. Can. J. Physiol. Pharmacol. 2002;80:346–353. doi: 10.1139/y02-021. [DOI] [PubMed] [Google Scholar]
- 27.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vickers C., Hales P., Kaushik V., Dick L., Gavin J., Tang J., Godbout K., Parsons T., Baronas E., Hsieh F., Acton S., Patane M., Nichols A., Tummino P. Hydrolysis of biological peptides by human angiotensin-converting enzyme-related carboxypeptidase. J. Biol. Chem. 2002;277:14838–14843. doi: 10.1074/jbc.M200581200. [DOI] [PubMed] [Google Scholar]
- 29.Katovich M.J., Grobe J.L., Huentelman M., Raizada M.K. Angiotensin-converting enzyme 2 as a novel target for gene therapy for hypertension. Exp. Physiol. 2005;90:299–305. doi: 10.1113/expphysiol.2004.028522. [DOI] [PubMed] [Google Scholar]
- 30.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tipnis S.R., Hooper N.M., Hyde R., Karran E., Christie G., Turner A.J. A Human homolog of angiotensin-converting enzyme, cloning and functional expression as a captoril-insensitive carboxypeptidase. J. Biol. Chem. 2000;275:33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- 32.Choudhury A., Mukherjee S. In silico studies on the comparative characterization of the interactions of SARS‐CoV‐2 spike glycoprotein with ACE‐2 receptor homologs and human TLRs. J. Med. Virol. 2020:1–9. doi: 10.1002/jmv.25987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verma A., Simard J.M., Rotello V.M. Effect of ionic strength on the binding of a-chymotrypsin to nanoparticle receptors. Langmuir. 2004;20:4178–4181. doi: 10.1021/la036183v. [DOI] [PubMed] [Google Scholar]
- 34.Verma A., Uzun O., Hu Y., Hu Y., Han H.-S., Watson N., Chen S., Irvine D.J., Stellacci F. Surface-structure-regulated cell-membrane penetration by monolayer-protected nanoparticles. Nat. Mater. 2008;7:588–595. doi: 10.1038/nmat2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verma A., Stellacci F. Effect of surface properties on nanoparticle-cell interaction. Small. 2010;6:12–21. doi: 10.1002/smll.200901158. [DOI] [PubMed] [Google Scholar]