Summary
High atmospheric humidity levels profoundly impact host-pathogen interactions in plants by enabling the establishment of an aqueous living space that benefits pathogens. The effectors HopM1 and AvrE1 of the bacterial pathogen Pseudomonas syringae have been shown to induce an aqueous apoplast under such conditions. However, the mechanisms by which this happens remain unknown. Here, we show that HopM1 and AvrE1 work redundantly to establish an aqueous living space by inducing a major reprogramming of the Arabidopsis thaliana transcriptome landscape. These effectors induce a strong abscisic acid (ABA) signature, which promotes stomatal closure, resulting in reduced leaf transpiration, and water-soaking lesions. Furthermore, these effectors preferentially increase ABA accumulation in guard cells, which control stomatal aperture. Notably, a guard-cell specific ABA transporter, ABCG40, is necessary for HopM1 induction of water-soaking lesions. This study provides molecular insights into a chain of events of stomatal manipulation that create an ideal microenvironment to facilitate infection.
Graphical Abstract
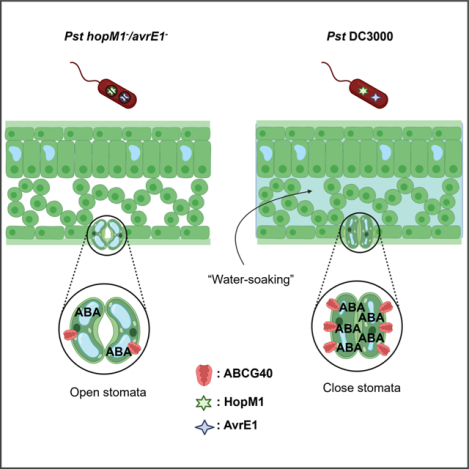
eTOC Blurb
Roussin-Léveillée et al. demonstrate that two Pseudomonas syringae effector proteins induce abscisic acid (ABA) biosynthesis and signaling in Arabidopsis. Although stomatal opening early in infection allows access to the leaf apoplast, they show that ABA induces stomatal closure in later stages, leading to water soaking lesions that benefit the bacteria.
Introduction
Many biotrophic pathogens secrete proteins, known as effectors, that manipulate their host to their benefit. These effectors are delivered, in many cases, to the cytoplasm of their host’s cells via diverse mechanisms. Bacterial pathogens deliver such effectors via a needle-like structure, known as the type-3 secretion system (T3SS). In a compatible host-pathogen interaction, effector activity results in effector-triggered susceptibility (ETS) to infection. A conserved and essential strategy used by microbial invaders is to employ effectors to subvert the host immune system (Macho, 2016; Toruño et al., 2016). However, this aspect of ETS is not sufficient for optimal pathogen growth as full pathogenicity is not restored to effector-less pathogens in mutant plants that are near completely compromised in their immune system (Xin et al., 2016). This indicates that effectors are required to induce physiological microenvironments favorable to pathogen growth in addition to inhibiting plant defenses.
Recent reports indicate that an important aspect of ETS involves the establishment of an ideal microenvironment in the apoplast (Bezrutczyk et al., 2018; Cohn et al., 2014; Cox et al., 2017; Xin et al., 2016). Elevated humidity levels in the environment are widely acknowledged as a key contributing factor to disease development in the field. During infection, such conditions have been shown to allow microbial pathogens to induce an aqueous living space in the apoplast, referred as “water-soaking lesions”. In addition to suppressing host defenses (Hauck et al., 2003; Lozano-Durán et al., 2014; Xin et al., 2015), the widely conserved effectors HopM1 and AvrE1 were previously identified as the main inducers of water-soaking lesions in an Arabidopsis-Pseudomonas pathosystem (Xin et al., 2016). HopM1 has been shown to interact with the regulator of endocytic trafficking MIN7 (Nomura, 2006). It has also been reported that HopM1 triggers proteaphagy, whereas the selective autophagy receptor NBR1 antagonizes HopM1-induced water-soaking (Üstün et al., 2018). A functional homolog of AvrE1 from Pantoea stewartii, WstE, interacts with the protein phosphatase 2A B’ (PP2A) subunit in maize (Jin et al., 2016). However, the molecular and physiological basis for the ability of HopM1 and AvrE1 to induce water-soaking lesions remain elusive.
Stomata are formed by specialized cells that mediate gas exchange and water transpiration in plants. Many plant pathogens gain access to the leaf interior through these same structures. Upon sensing of microbe-associated molecular patterns (MAMPs), stomata close, thereby preventing pathogen entry (Melotto et al., 2006). This phenomenon, known as stomatal immunity, is the first line of defense in MAMP-triggered immunity (MTI) and is often targeted by pathogens. Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) produces a phytotoxin, known as coronatine (COR), that induces a re-opening of closed stomata, allowing the pathogen access to the host apoplastic environment (Melotto et al., 2006). Although stomatal behavior during the early stages of infection has been the subject of numerous studies, their role in late stages is still unclear.
In this study, we have investigated the molecular and physiological basis for HopM1/AvrE1 induction of an aqueous microenvironment in the apoplast. Transcriptional profiling of Arabidopsis thaliana during an infection revealed that HopM1 and AvrE1 are functionally redundant in triggering abscisic acid (ABA) biosynthesis and signaling pathways. We show that ABA accumulation induced by HopM1 and AvrE1 triggers stomatal closure after an initial opening by coronatine and that manipulation of stomata has a dramatic effect on pathogenesis. Our study identifies a molecular and physiological explanation behind humidity-driven pathogenesis through the manipulation of ABA signaling.
Results
Water-soaking effectors induce a major reprogramming of the Arabidopsis transcriptome and induce ABA pathways
To characterize the role of water-soaking effectors, we profiled the transcriptome of A. thaliana inoculated with Pst using RNA-sequencing (RNA-Seq). We included hopM1− and avrE1− single and double mutants of Pst, as these two effectors have been reported to be required for the induction of water-soaked lesions (Xin et al., 2016). Leaves were syringe-infiltrated with a low dose of inoculum (1 × 106 CFU/ml) and tissues harvested during the Pst DC3000 exponential growth phase at 36 hpi, as determined by bacterial growth curve dynamics (Figure S1A and S1B). This time point was selected to provide insights into the transcriptional profile of Arabidopsis during the active growth phase of the pathogen, when we expect effector proteins to be highly active. Transcriptional responses induced by Pst were highly influenced by the presence of HopM1 and AvrE1 (Figure 1A, 1B and 1C). A principal component analysis of the RNA-Seq data revealed a close transcriptional relationship between Pst DC3000, hopM1− and avrE1− infected plants, but a more distant relationship to the hopM1−/avrE1− (h−/a−) infected plants (Figure 1B). The number of differentially expressed genes (DEGs) in Arabidopsis Col-0 plants infected by Pst DC3000 was strikingly higher than in their h−/a− infected counterparts (9623 vs 4906, respectively) (Figure 1C and S1C–G). We also compared the transcriptome of plants infected with Pst h−/a− to a previously described dataset from plants treated with the MTI-inducing peptide flg22 (Winkelmüller et al., 2021) to compare HopM1/AvrE1 transcriptional reprogramming in contrast to the response of an Arabidopsis immune response. Although these datasets were generated from different experimental conditions, we nonetheless find that they result in distinct transcriptional signatures. That is, less than half of the genes upregulated by the flg22 response were also induced by Pst h−/a− and there was only a very minor overlap in downregulated genes (Figure S1G). This suggests that Pst effectors other than HopM1 and AvrE1 contribute to countering the transcriptional reprogramming associated with an MTI response.
Figure 1. Water-soaking effectors induce a major transcriptional reprogramming and increase ABA biosynthesis.
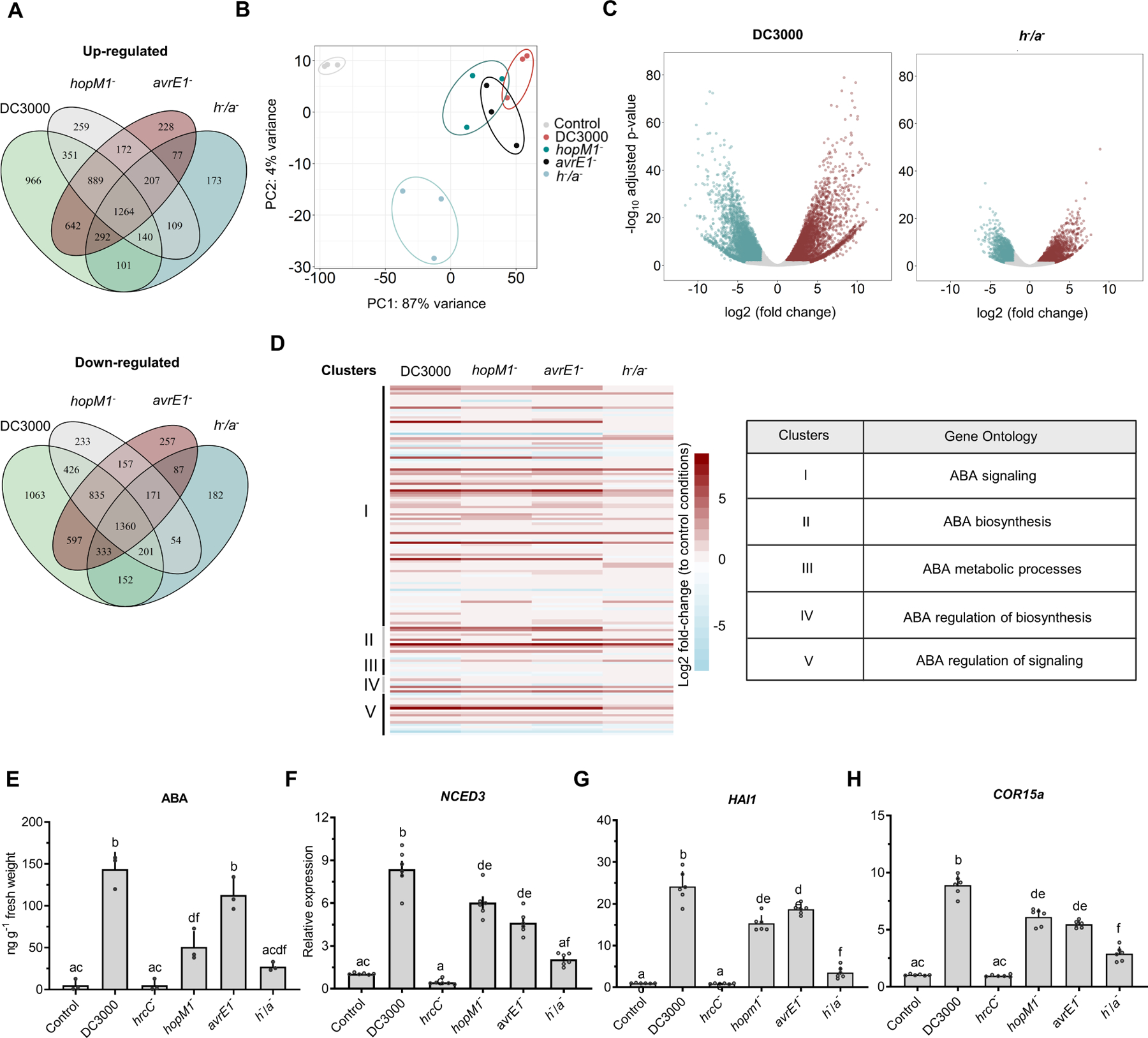
(A) Venn diagrams displaying specific and shared differentially expressed genes (DEGs) in A. thaliana Col-0 challenged with Pst DC3000, hopM1−, avrE1− and hopM1−/avrE1− (h−/a−) (1 × 106 CFU/ml) at 36 hours post inoculation (hpi). (B) Principal component (PC) analysis showing expression changes of transcriptomic data from the conditions described in a, as indicated. (C) Volcano plot representing DEGs in Arabidopsis plants infected with Pst DC3000 or h−/a− (1 × 106 CFU/ml at 36 hpi). Blue, red and gray dots represent significantly down-regulated, upregulated and not significantly altered DEGs, respectively. (D) Heat map representing expression differences of genes with gene ontologies involved in ABA-related processes, as indicated, upon infection of Arabidopsis plants with the indicated Pst strains. (E) ABA quantification in Arabidopsis Col-0 leaves infected with 0.5 × 108 CFU/ml of the indicated Pst strains by UPLC-MS at 24 hpi. (F) Relative expression of the ABA biosynthesis marker gene NCED3 in Arabidopsis Col-0 plants infiltrated with the indicated Pst strains (1 × 108 CFU/ml) at 24 hpi measured by real-time quantitative PCR (RT-qPCR). (G-H) Relative expression of the ABA signaling pathway marker genes HAI1 (G) and COR15a (H) in Arabidopsis Col-0 plants inoculated as in (E). Data are represented as mean of total experimental replicates ± SEM. Different letters indicate statistically significant differences (adjusted P < 0.01, one-way ANOVA).
Gene ontology (GO) analysis associated with the transcriptional response of Arabidopsis to infection by Pst DC3000, hopM1−, avrE1− and h−/a− indicated that HopM1 and AvrE1 induce a gene expression pattern related to an ABA response (Figure 1D, Table S1). ABA signaling is a well-known response to water stress and can impact multiple regulatory pathways. Thus, we investigated changes in ABA responses mediated by HopM1 and AvrE1. Using ultra performance liquid chromatography-mass spectrometry (UPLC-MS), we observed an increase in ABA levels in plants infected with Pst DC3000, compared to uninfected plants, but not with h−/a− (Figure 1E). In concordance with this, A. thaliana plants infected with Pst DC3000, hopM1− and avrE1− single mutants, but not with h−/a− double mutant, showed significantly higher expression levels of the ABA biosynthesis marker gene NCED3 than mock infected plants (Figure 1F). Furthermore, inoculation with Pst DC3000 resulted in an increase in expression of known components of the ABA signaling pathway, HAI1 (negative regulator of ABA signaling; ~24-fold increase) and COR15a (cold- and ABA-regulated gene; ~9-fold increase) (Baker et al., 1994; Yoshida et al., 2006). In contrast, the h−/a− double mutant showed only a moderate increase (~2.5-fold for both HAI1 and COR15a) compared to control plants, while the hopM1− and avrE1− single mutants showed an intermediate level of expression (at least half the level of induction induced by DC3000) (Figure 1G and 1H). Importantly, this ABA signature was observed at 24 hpi, at a timepoint when bacterial densities do not significantly differ between WT and mutant strains lacking HopM1 and/or AvrE1 (Figure 2F). Together, these results indicate a role for water-soaking effectors in increasing ABA accumulation and subsequent responses.
Figure 2. Pseudomonas syringae water-soaking effectors induce stomatal closure in the late stages of infection.
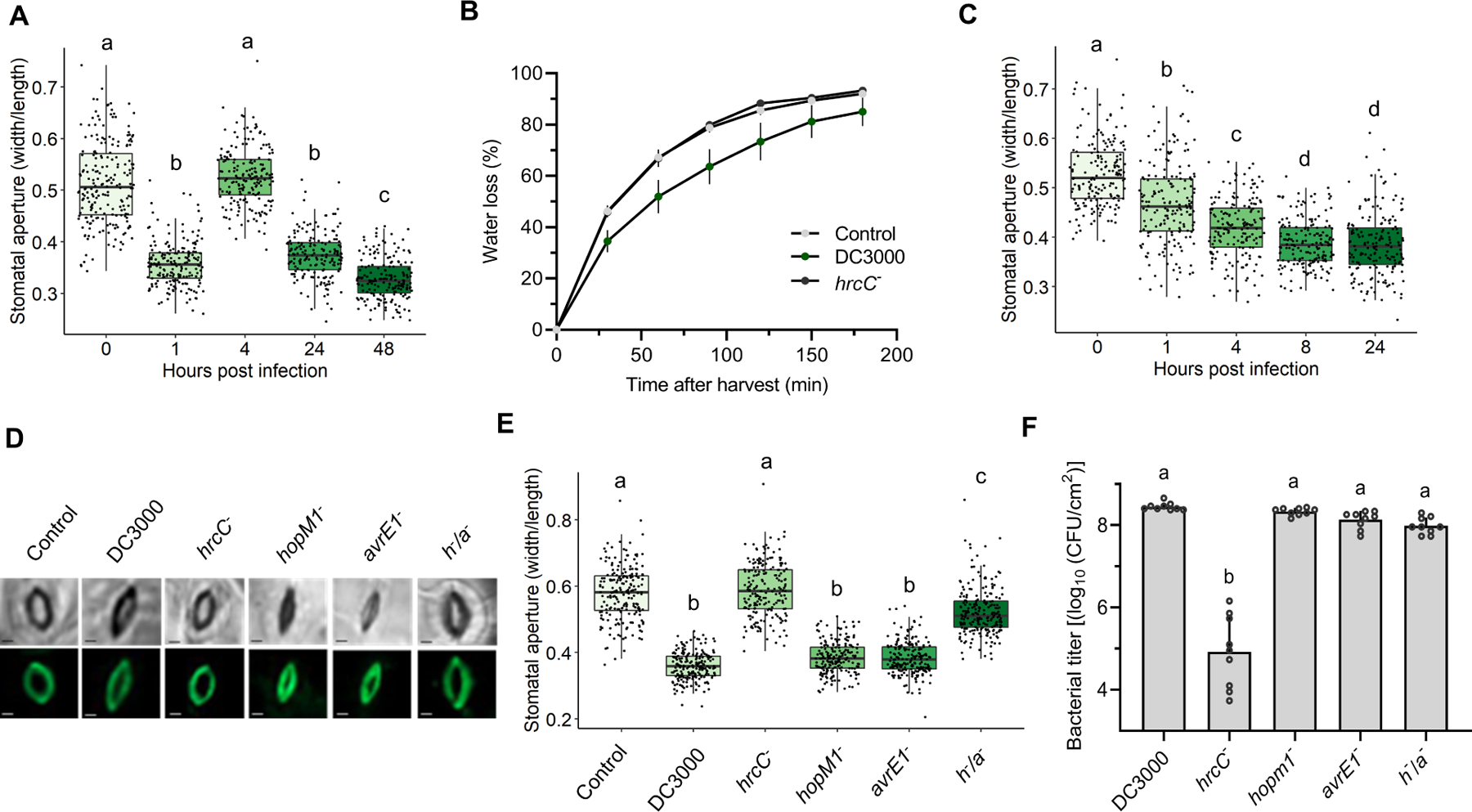
(A) Arabidopsis Col-0 plants were dip-inoculated in a Pst DC3000 bacterial suspension (2 × 108 CFU/ml) containing 0.025% Silwet L-77. Epifluorescence microscopy images were used to measure stomatal apertures (n > 150) at the indicated hours post infection (hpi). (B) Water loss in Arabidopsis Col-0 plants inoculated as in a with mock (control), WT (DC300), or T3SS defective (hrcC−) strains at 24 hpi. Leaves were detached from plants and stomata were fixed in stomatal fixation solution at the beginning of water loss measurement. (C) Stomatal aperture measurements of Arabidopsis Col-0 plants syringe-infiltrated with Pst DC3000 (1 × 108 CFU/ml) measured over time (n > 150 stomata). (D) Microscopy images of abaxial sides of four-week-old A. thaliana Col-0 leaves infiltrated with MgCl2 10 mM (control), Pst DC3000, hrcC−, hopM1−, avrE1− and h/a− (1 × 108 CFU/ml), as indicated, at 24 hpi. Upper panel: bright field, lower panel: GFP field to visualize Rhodamine 6G fluorescence. Bar scales represent 2 μm. (E) Stomatal aperture measurements of Arabidopsis Col-0 plants inoculated as in D at 24 hpi. (F) Bacterial counts found in plants analyzed in (D) and (E). Data are represented as median values, with minimum and maximum values indicated (A, C, E) or as means of total experimental replicates, ± SEM (B, F). Different letters indicate statistically significant differences (adjusted P < 0.01, one-way ANOVA).
HopM1 and AvrE1 induce stomatal closure following leaf entry
Under high levels of humidity, Pst DC3000 induces clear water-soaking lesions, which are crucial for its virulence (Xin et al., 2016). Since stomata mediate gas exchange, including loss of water vapour, between the leaf apoplast and the extracellular environment, we hypothesized that Pst DC3000 might induce stomatal closure following leaf entry to maintain high internal levels of humidity. As stomata respond rapidly to changes in relative humidity settings, we optimized a stomatal fixation method to prevent stomatal movement during analyses (Eisele et al., 2016). Briefly, we found that treatment of leaves with a solution containing 4% formaldehyde allowed for the processing of large numbers of leaves in a context where stomatal aperture does not change upon transfer of leaves from experimental to analytical conditions (Figure S2A–E).
We investigated A. thaliana stomatal responses to Pst DC3000 under elevated relative humidity over the three-day course of an infection. Plants dip inoculated with Pst DC3000 displayed the well-documented phenomenon of stomatal closure within one hour of inoculation, followed by re-opening of stomata after four hours. Interestingly, stomata were closed at the 24 hours post inoculation (hpi) timepoint in Pst DC3000 inoculated plants, suggesting a dynamic manipulation of stomatal movement (Figure 2A). In agreement with this, leaves infected with Pst DC3000 underwent slower water loss than control plants (Figure 2B). These phenomena required the bacterial type-three secretion system (T3SS) as a Pst mutant lacking this system (Pst hrcC−) did not affect stomatal aperture or water loss (Figure 2B, 2D and 2E). Stomatal closure was also observed in Arabidopsis plants that were syringe-infiltrated with Pst DC3000, although no reopening was observed at 4 hpi (Figure 2C).
Given the importance of HopM1 and AvrE1 in inducing ABA responses, we tested the importance of these effectors in inducing stomatal closure. Interestingly, Arabidopsis plants infected with Pst hopM1− and avrE1− single mutants displayed closed stomata at 24 hpi, whereas those infected with the h−/a− double mutant show stomatal aperture values similar to uninfected or hrcC- infected plants (Figure 2D and 2B). The differences in aperture opening cannot be due to differences in bacterial density as bacterial levels in the apoplast are not significantly different at this timepoint (Figure 2F). To further validate the role of these two effectors in inducing stomatal closure, we used transgenic Arabidopsis lines expressing HopM1 and AvrE1 under the control of a dexamethasone (DEX)-inducible promoter. Upon DEX treatment, HopM1 and AvrE1 expressing lines displayed significant stomatal closure compared to wild-type and non-DEX-treated plants, as well as induction of NCED3 expression (Figure S3A–B). These results suggest that Pst has evolved a strategy to initially open stomata to facilitate entry into the apoplast, after which it induces stomatal closure to maintain high levels of humidity in its environment.
ABA-mediated stomatal closure as the primary mechanism behind water-soaking lesions
Control of gas exchange by stomata is primarily orchestrated by the phytohormone ABA (Kollist et al., 2019). Interestingly, ABA pathway manipulation by microbial invaders is also known to contribute to susceptibility in many plant pathosystems (Fan et al., 2009). Since ABA is known to be exploited by Pst DC3000 to increase its virulence (de Torres-Zabala et al., 2007), we hypothesized that this enhanced susceptibility to infection might be caused by the pathogen’s ability to induce water-soaking lesions in an ABA-dependant manner. To test this, we inoculated WT Arabidopsis and an aba2–1 mutant, which is known to produce considerably less ABA (Rook et al., 2001). The induction of water-soaked lesions by Pst was completely abolished in the aba2–1 mutant at 24 hpi (Figure 3A). Likewise, Pst inoculation did not result in stomatal closure or in the production of ABA in the aba2–1 mutant (Figure 3B and 3G) and leaves from aba2–1 plants showed the same degree of water loss in the presence or absence of bacteria (Figure 3C). Importantly, the levels of bacteria were the same at the timepoints tested, ruling out bacterial density as a factor in these observations (Figure 3D). Furthermore, it is reported that bacterial levels of Pst DC3000 in ABA biosynthesis or signaling mutants are similar to the levels of Pst h−/a− in WT Arabidopsis (Hu et al., 2022). To strengthen the link between ABA and water-soaking lesions, we co-infiltrated Pst with fluridon, which has been shown to interfere with carotenoid and ABA biosynthesis (Gamble & Mullet, 1986). We found that co-infiltrating the pathogen with fluridon prevented Pst from causing water-soaking lesions through stomatal closure, supporting a role for ABA in inducing stomatal closure during an infection (Figure 3H–I).
Figure 3. Abscisic acid biosynthesis is required for humidity-driven pathogenesis in Arabidopsis.
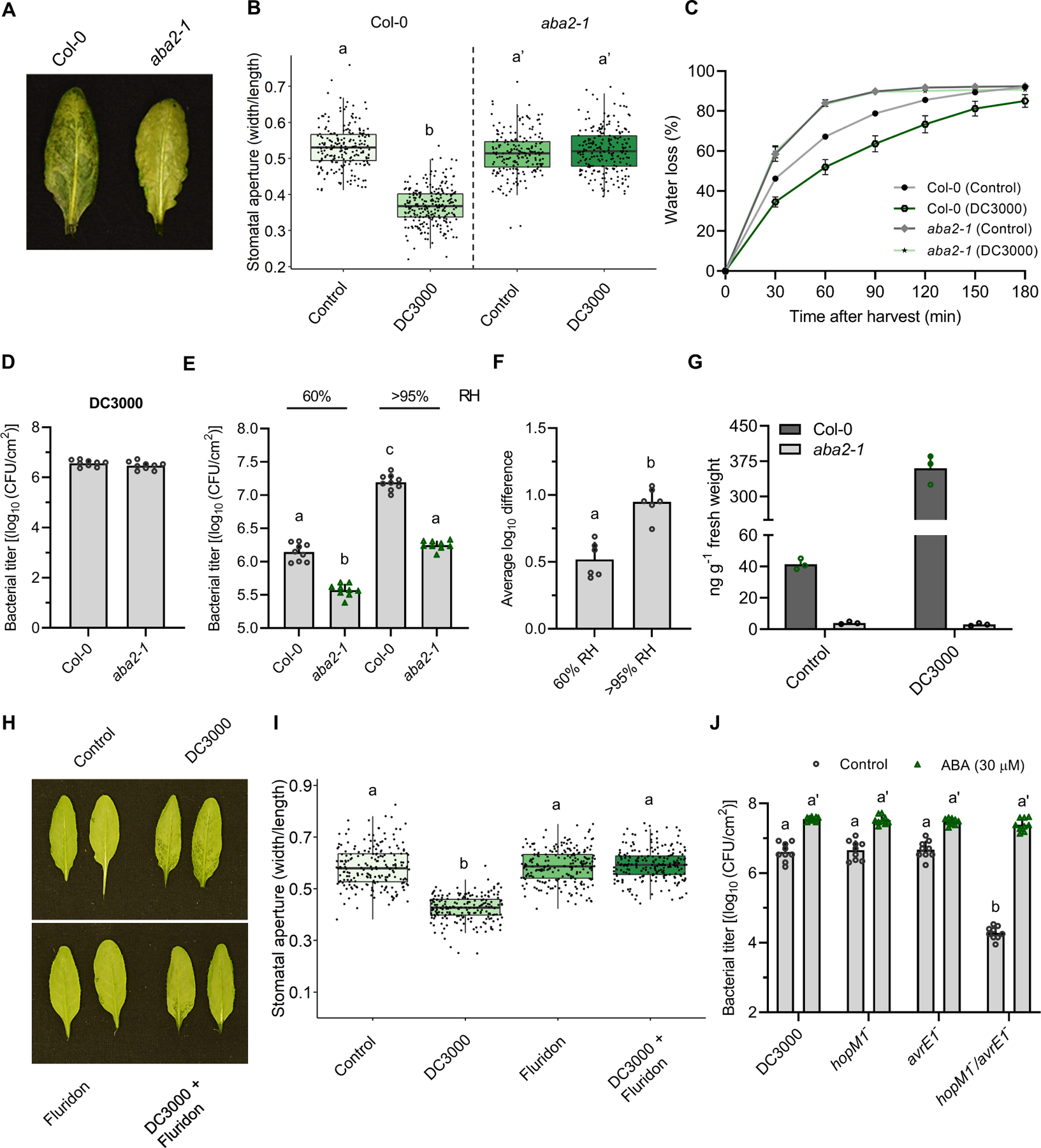
(A) Leaves of Col-0 Arabidopsis and the aba2–1 mutant were infiltrated with Pst DC3000 (1 × 108 CFU/ml) and plants grown at 95% humidity. Photos taken at 24 hpi. (B) Stomatal aperture measurements of Col-0 and the aba2–1 mutant inoculated with Pst DC3000 under the same conditions as in (A) (n > 150 stomata). (C) Water loss was measured in leaves of Arabidopsis Col-0 and aba2–1 mutant plants inoculated with 10 mM MgCl2 (control) or Pst DC3000 (1 × 108 CFU/ml). Leaves were detached at 24 hpi and stomata fixed in stomatal fixation solution at the beginning of water loss measurements. (D) Bacterial titres measured in plants analyzed in (A) at 24hpi. (E) Col-0 and aba2–1 mutant plants were infected with Pst DC3000 (1 × 105 CFU/ml) and kept under either 60% or >95% RH levels. Bacterial titers were quantified 3 days post infection. (F) Average log difference in bacterial titers between WT and aba2–1 plants assayed under the conditions as in (E). (G) Quantification of ABA in Arabidopsis Col-0 and aba2–1 mutant plants mock inoculated (control) or inoculated with Pst DC3000 (1 × 108 CFU/ml), as measured by UPLC-MS at 24 hpi. (H) Water-soaking lesion phenotypes in Arabidopsis leaves syringe-inoculated with MgCl2 10 mM (control), Pst DC3000 (1 × 108 CFU/ml), fluridon (100 μM), or both Pst DC3000 and fluridon. (I) Stomatal aperture measurements from leaves described in (H). (J) ABA-mediated rescue of the hopM1−/avrE1− double mutant. Arabidopsis Col-0 plants were infected with the indicated Pst mutants (1 × 105 CFU/ml) followed by treatment, at 2, 24, and 48 hpi, with either ABA (30 μM) or control solution (0.5% ethanol). Plants were treated at 2, 24 and 48 hpi. Treatments were carried out by spraying each solution on the abaxial and adaxial part of each plant. Bacterial titers were evaluated at 3 dpi. Data are represented as median values, with minimum and maximum values indicated (B, I) or as mean of total experimental replicates, ± SEM (C-–G, and J). Different letters indicate statistically significant differences, P < 0.0005, two-tailed Student’s t-test (B, F) or ANOVA (E, I, J).
To better understand the role of ABA in humidity-driven pathogenesis, we infected WT and aba2–1 mutant plants with Pst and maintained two different humidity levels over the course of the infection. Differences in bacterial levels between Col-0 and aba2–1 infected plants were greater under elevated humidity conditions as compared to ambient humidity levels, suggesting that ABA contributes to humidity-driven pathogenesis (Figure 3E and 3F). Next, we questioned whether treating plants with ABA would affect the growth rate of the h−/a− mutant. A significant difference in bacterial growth at three dpi was observed between the h−/a− double mutant in Arabidopsis as compared to DC3000, as well as hopM1− and avrE1− single mutants, as expected (Figure 3J). However, treatment of plants with ABA resulted in a complete rescue of the double mutant growth rate to the same levels as Pst DC3000 (Figure 3J). We suggest that ABA’s ability to rescue the h−/a− mutant growth deficiency is due to its ability to induce stomatal closure, which in turn results in water-soaking lesions (Figure S4A–D) that are beneficial to the bacteria. As such, ABA appears to be both necessary and sufficient for the induction of stomatal closure.
Coronatine does not prevent ABA-mediated stomatal closure
Pst produces coronatine in the early stages of an infection to re-open closed stomata to gain access to the apoplastic environment (Melotto et al., 2006). We questioned whether the amount of COR produced by Pst in planta was sufficient to antagonize ABA-induced stomatal closure once the pathogen has entered the apoplast. We measured in planta levels of COR and ABA, as well as several other phytohormones, in Arabidopsis plants infected by Pst DC3000 over a time course of 24 hours under two different humidity settings (Figure S5A–F). Approximate concentrations of these molecules in planta were calculated based on the amount of water found in Arabidopsis leaves, as determined by measuring fresh and dry weight in leaves of plants grown at 95% humidity (Figure 4A and 4B). ABA and COR were then infiltrated into Arabidopsis Col-0 leaves at concentrations approximating those found in vivo during a Pst-Arabidopsis interaction. As expected, we found that at concentrations found in Pst-infected leaves, COR induced stomatal opening and ABA was sufficient to induce stomatal closure and water soaking in the absence of bacteria. However, at physiologically relevant concentrations, COR did not prevent ABA from inducing stomatal closure or from inducing water soaking lesions (Figure 4C and 4D). Furthermore, a Pst mutant deficient in the biosynthesis of COR, DB29 (Brooks et al., 2004), displayed no difference in terms of water-soaking symptoms or in its ability to induce stomatal closure, compared to wild-type (Figure S5G–I). Thus, although COR and ABA have been shown to have opposite effects on stomatal behavior in early stages of infection, the concentration levels found in the later stage of an infection result in stomatal closure.
Figure 4. COR levels found in vivo do not prevent ABA from inducing stomatal closure and water-soaking lesions.
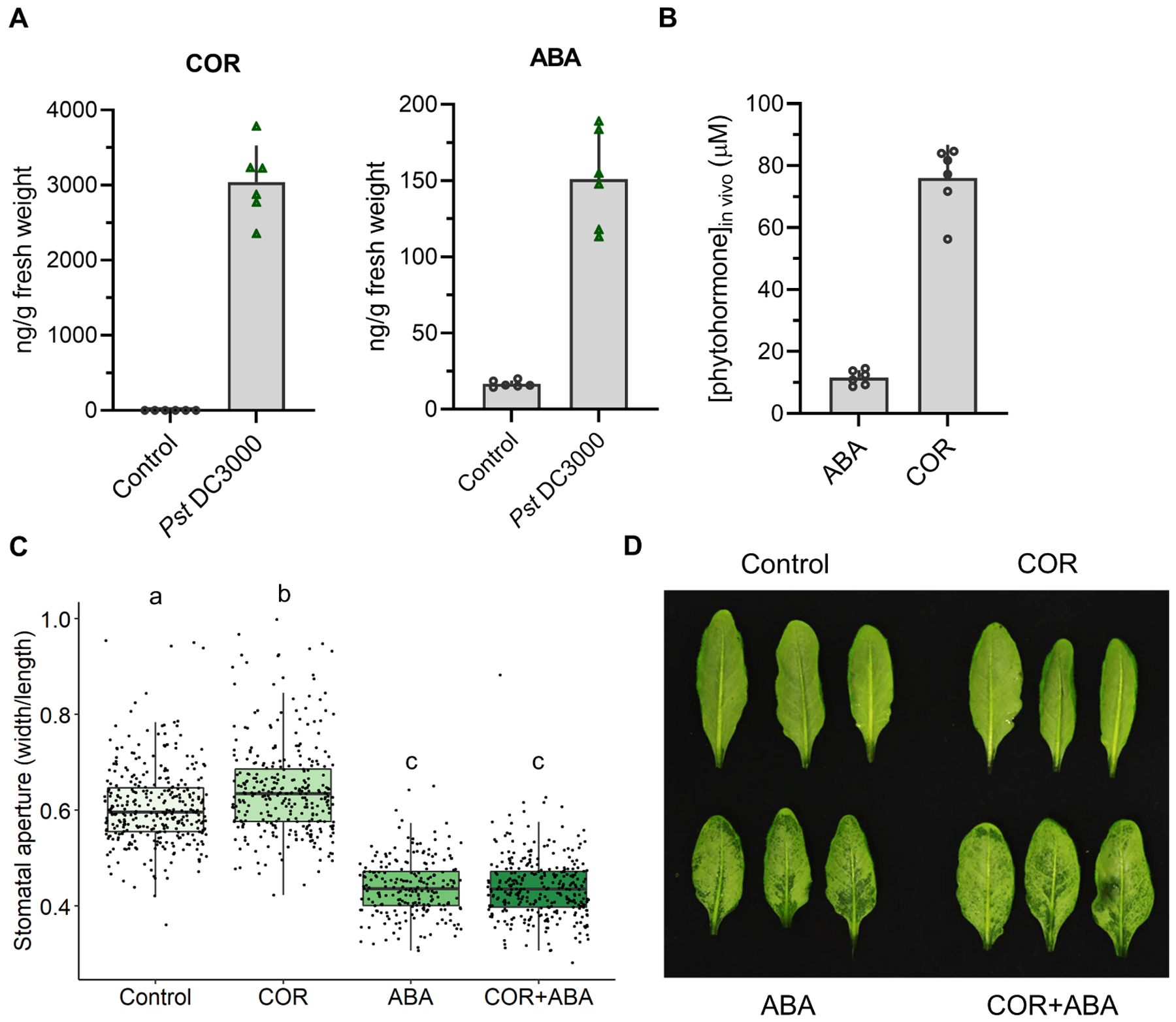
(A) COR and ABA levels found in planta during a Pst DC3000 infection. COR and ABA levels were quantified by UPLC-MS in Arabidopsis Col-0 leaves mock inoculated (control) or infiltrated with Pst DC3000 (5 × 107 CFU/ml) at 24 hpi. (B) Average calculated concentration of ABA and COR found in Arabidopsis leaves infiltrated with Pst DC3000 (5 × 107 CFU/ml) at 24 hpi (n = 6 plants from three independent experimental replicates). (C) Stomatal apertures of Arabidopsis Col0 leaves infiltrated with ABA (10 μM) and/or COR (80 μM), as measured from epifluorescence images from stomata-fixed leaves at 8 hours post treatment. (D) Appearance of water-soaking lesions in leaves described in (C). Data are represented as mean of total experimental replicates ± SEM (A, B), or as median values, with minimum and maximum values indicated (C). Different letters indicate statistically significant differences, P < 0.05, one-way ANOVA.
Importance of late stomatal closure in bacterial pathogenesis
The non-virulent Pst strain hrcC− is unable to grow well in the apoplastic environment of Arabidopsis. We therefore questioned whether forcing stomatal closure could re-establish virulence to Pst hrcC−. To test this, we established a protocol whereby Arabidopsis plants were subjected to a series of treatments to affect stomatal movement either before or after inoculation with Pst DC3000 or Pst hrcC− (Figure 5A). Treatments included H2O alone, ABA or Tert-butylhydroquinone (TBHQ). TBHQ is an aromatic compound derived from hydroquinone, and has been shown to induce stomatal closure in an ABA-independent manner (Toh et al., 2018). We find that, like ABA, TBHQ treatment induces water soaking lesions at high humidity, as well as stomatal closure, suggesting that the latter is sufficient for the former (Figure S4E and S4G).
Figure 5. A role for late stomatal closure in bacterial pathogenesis.
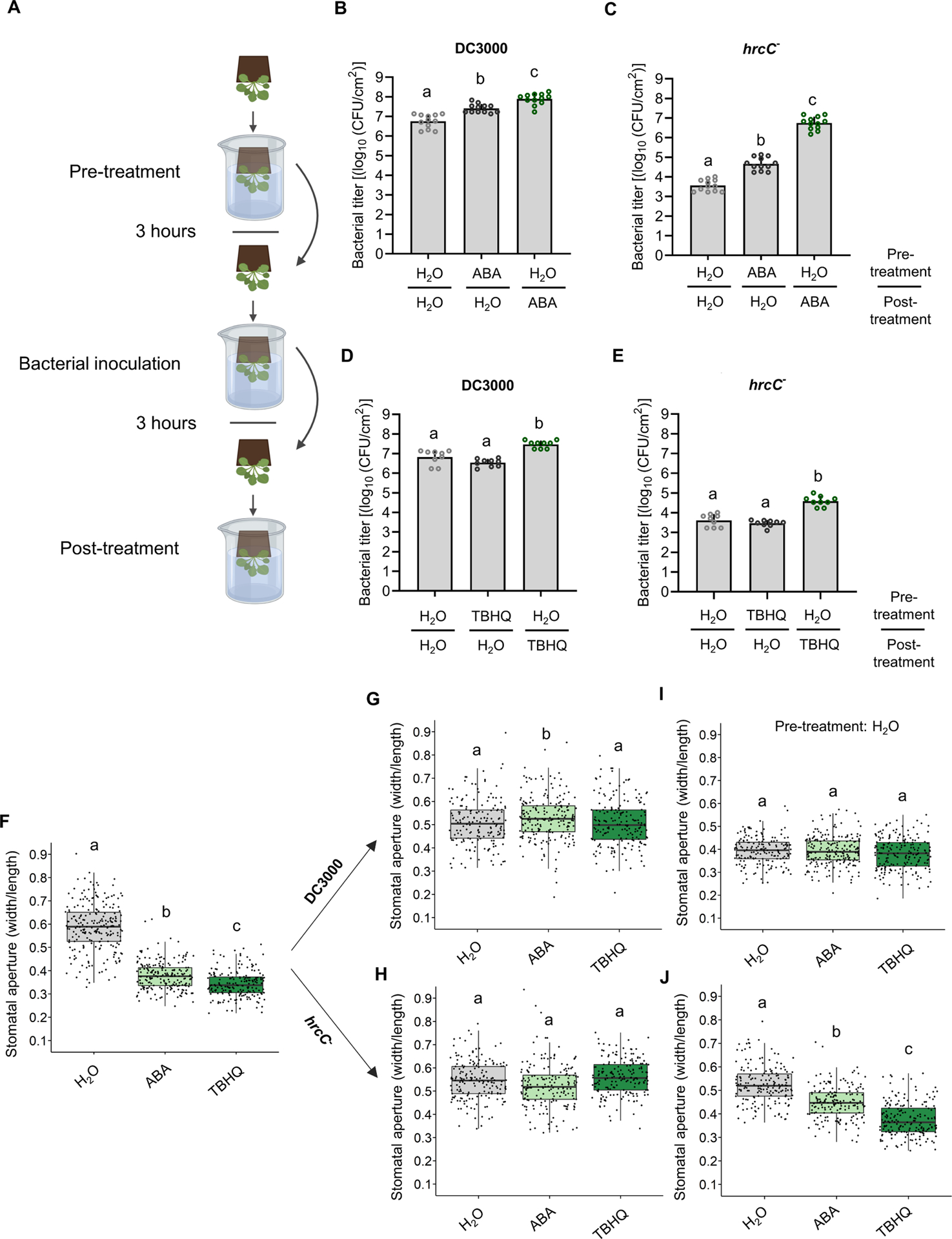
(A) Schematic diagram depicting the experimental design setup to evaluate the importance of pre- vs post-infection stomatal closure. Created with BioRender. (BE) All plants were subjected to three dip treatments. In the Pre-treatment, plants were first dipped in a solution containing either 0.05% ethanol plus 0.025% Silwet L-77 in water (H2O) or the same solution also containing either ABA (30 μM) or TBHQ (100 μM). Three hours after pre-treatment, plants were dip-inoculated with a solution of 2 × 108 CFU/ml of Pst DC3000 or hrcC−, as indicated. All plants were subsequently dip-inoculated in H2O, ABA or TBHQ solutions, as in the pretreatment (Post-treatment). (B-E) Bacterial titers were evaluated 3 days post infection (dpi). Bacterial titers are shown from plants subjected to different combinations of pre- and post-treatments, as indicated, for DC3000 (B and D) or hrcC− (C and E). Data are represented as mean of total experimental replicates, ± SEM. (F-J) Stomatal apertures measured in WT Arabidopsis plants treated as in (B-E). Apertures were measured three hours after pre-treatment with H2O, ABA or TBHQ (F). (G-H) Plants were pre-treated with H2O, ABA or TBHQ, as indicated, followed by bacterial inoculation. Stomatal apertures were measured three hours after inoculation with Pst DC3000 (G) or with hrcC− (H). (I-J) Plants were pretreated with H2O only, followed by inoculation with Pst DC3000 (I) or with hrcC− (J) and Post-treatment with H2O, ABA or TBHQ, as indicated. Stomatal apertures were measured three hours after post-treatment. Data are represented as median values, with minimum and maximum values indicated (F-J). Different letters indicate statistically significant differences, P < 0.05, one-way ANOVA.
We found that pre-treatment of Arabidopsis plants with ABA three hours prior to bacterial dip-inoculation resulted in enhanced growth of both Pst DC3000 and Pst hrcC−, compared to plants pre-treated with H2O (Figure 5B and 5C). However, when Arabidopsis plants were instead treated with ABA three hours after bacterial inoculation, this enhanced growth was more dramatic, with Pst hrcC− growing at levels nearing those of Pst DC3000 in untreated plants (Figure 5B and 5C).
Since ABA has been reported to dampen immune signaling (de Torres Zabala et al., 2009; Mine et al., 2017), we sought to test the effect of inducing stomatal closure without affecting immunity. Arabidopsis plants treated with TBHQ prior to pathogen challenge did not show any difference in proliferation of Pst DC3000 or Pst hrcC− three days post inoculation (Figure 5D and 5E). However, plants that were treated with TBHQ three hours post pathogen inoculation displayed enhanced growth of both Pst DC3000 and Pst hrcC− (Figure 5D and 5E). As TBHQ did not alter the protection provided by a flg22 pre-treatment (Figure S4F), we suggest that TBHQ does not affect immune signaling, but rather, augments bacterial growth by inducing stomatal closure and causing an aqueous apoplastic environment.
The interpretation of the above results depends on the actual behavior of stomates in this set of experiments. As such, we measured the stomatal apertures of plants treated as above. As expected, pretreatment with ABA or TBHQ resulted in stomatal closure (Figure 5F). Interestingly, both Pst DC3000 and Pst hrcC− induced a reopening of stomata three hours after dip-inoculation, regardless of ABA or TBHQ pre-treatment (Figure 5G and 5H). This suggests that the stomatal reopening capacity of the pathogen overcomes the action of ABA and TBHQ in this situation. In plants pretreated with H2O alone, post treatments with H2O, ABA or TBHQ three hours after DC3000 inoculation all resulted in similar stomatal closure activity (Figure 5I). This is presumably because the action of DC3000 effectors is sufficient to induce stomatal closure. In contrast, post-treatment of plants inoculated with Pst hrcC− with H2O alone did not result in stomatal closure, whereas ABA and TBHQ treatments did (Figure 5J). Thus, stomatal closure following leaf entry appears to be an important element in bacterial pathogenesis in addition to immune suppression.
HopM1 induction of water-soaking lesions requires the guard cell-specific ABA transporter ABCG40
As guard cells respond rapidly to environmental and hormonal stimuli, we assessed whether HopM1 and AvrE1 increase ABA accumulation ubiquitously or in a more localized manner. To assess this question, we used Arabidopsis transgenic lines expressing ABAleon2.1, a recombinant protein reporter that undergoes FRET between mTurquoise and cpV173 fluorophores in the absence of ABA (Waadt et al., 2014). We took advantage of the molecular quenching that occurs between the mTurquoise and cpV173 fluorophores in the presence of ABA to directly visualize ABA concentration changes in planta. We found that mTurquoise emission rates were significantly higher in plants infiltrated with Pst DC3000, hopM1− and avrE1− as compared to those infiltrated with hrcC−, h−/a− and mock-inoculated controls (Figure 6A). These observations suggest that cpV173 is less able to quench mTurquoise emission signal due to a higher quantity of ABA present in these cells. Interestingly, the mTurquoise emission appeared to be much greater in guard cells compared to pavement cells (Figure 6B and 6C).
Figure 6. Pseudomonas syringae enhances ABA accumulation in guard cells and requires the ABA transporter ABCG40 to induce water-soaking lesions.
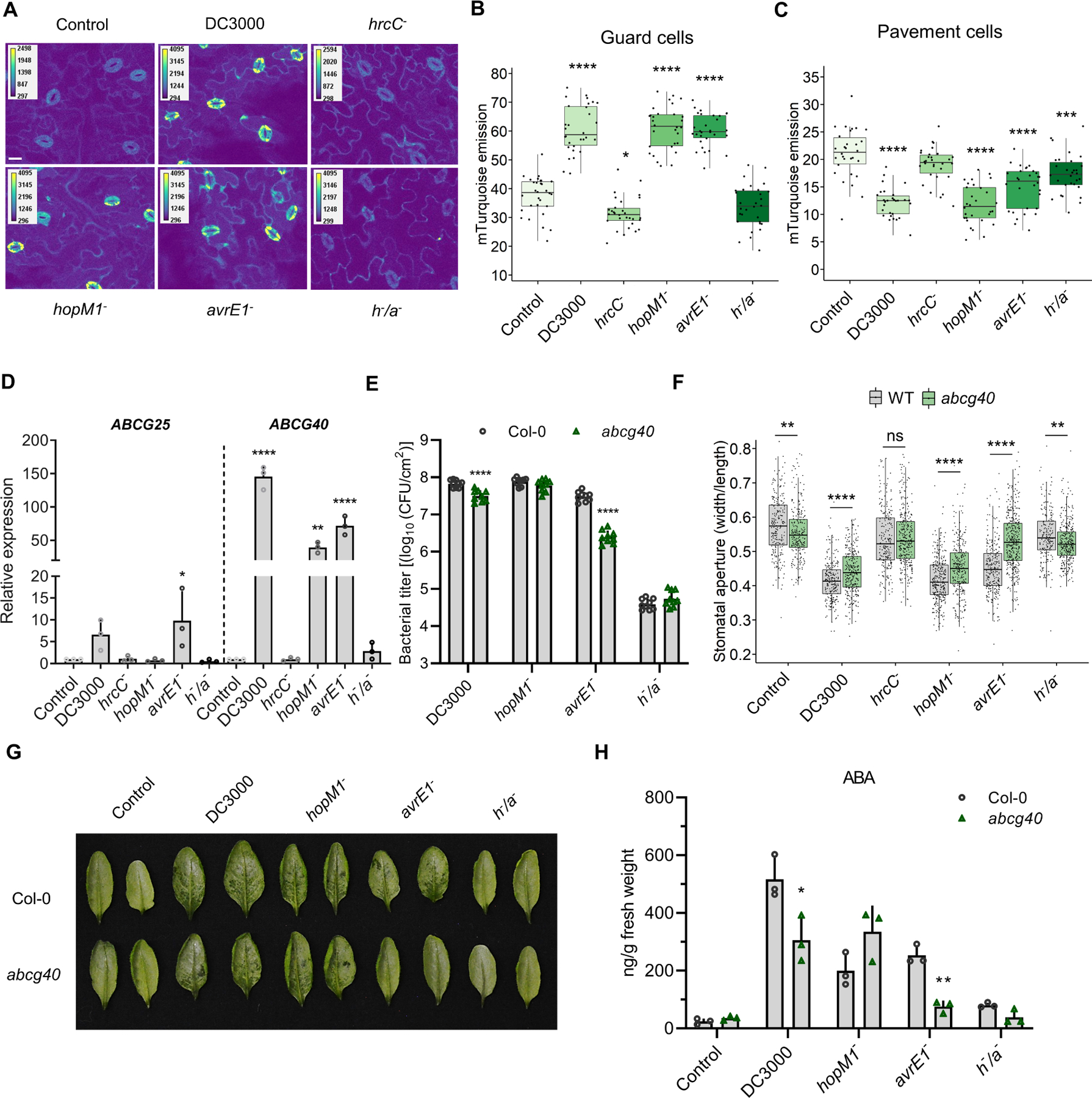
(A) Confocal microscopy images of Arabidopsis Col-0 leaves expressing the ABAleon2.1 reporter, mock inoculated (control) or inoculated by syringe with different Pst strains, as indicated. Images represent the emission of mTurquoise following excitation to evaluate the quenching of cpV173 fluorescence in the presence of ABA. Leaves were infected with 1 × 108 CFU/ml of bacteria and harvested at 24 hpi before being mounted on microscopy slides. Bar scale represents 20 μm. (B-C) mTurquoise emission in guard cells (B) and pavement cells (C) of plants infected and harvested as in (A). Each data point represents mean of mTurquoise emission in specific cells of one image (n = 30 images/condition). (D) Relative expression at 24 hpi of the guard cell ABA transporters involved in export (ABCG25) or import (ABCG40) in leaves inoculated with the indicated Pst strains (1 × 108 CFU/ml). (E) Bacterial titers in Col-0 and abcg40 mutant plants inoculated with 1 × 105 CFU/ml of the indicated Pst strains at 3 dpi. (F) Stomatal aperture measurements in wild-type and abcg40 mutant plants infected with the indicated Pst strains at 24 hpi (1 × 108 CFU/ml). (G) Water-soaking phenotypes in Col-0 and abcg40 mutants evaluated in (F). (H) ABA quantification in Col-0 and abcg40 mutant plants at 24 hpi with the indicated Pst strains (1 × 108 CFU/ml). Data are represented as median values, with minimum and maximum values indicated (B, C) or as mean ± SEM (D, E, H). *P < 0.05, **P < 0.05, ***P < 0.005, ****P < 0.0005, one-way ANOVA.
Plant cells are equipped with a variety of ABA transporters involved in either ABA export or import, often expressed in a cell-type and/or tissue-specific manner (Kuromori et al., 2018). We explored the possibility that Pst DC3000 might increase ABA accumulation in guard cells by increasing the expression of a guard-cell specific ABA transporter. We found that inoculation of plants with Pst DC3000, Pst hopM1−, Pst avrE1−, induced a dramatic increase in the expression levels of ABCG40, which is involved in stomatal ABA import (Kang et al., 2010) (Figure 6D). In contrast, transcripts encoding the non-tissue-specific ABA exporter ABCG25 were not upregulated by Pst infection to the same extent as ABCG40 (Figure 6D). Bacterial growth assays showed that WT Pst DC3000 grew somewhat less in the abcg40 mutant compared to WT, whereas Pst hopM1− growth was unaffected (Figure 6E). In contrast, Pst avrE1− displayed a more dramatic growth deficiency in abcg40 plants, compared to Col-0 (Figure 6E). Consistent with this, Pst avrE1− can no longer induce stomatal closure or water-soaking lesions in the abcg40 mutant, whereas Pst DC3000 and Pst hopM1− show only minor difference between WT and abcg40 plants in these assays (Figure 6F and 6G). In combination with the fact that only the h−/a− double mutant is affected in guard cell ABA accumulation (Figure 6A and 6B), this suggests a certain redundancy between HopM1 and AvrE1 in ABA induction and its concentration in guard cells. Interestingly, in the absence of ABCG40, Pst avrE1− induces almost no accumulation of ABA in contrast to Pst hopM1− (Figure 6H). Thus, since HopM1 cannot compensate for a lack of AvrE1 in the abcg40 mutant, this suggests that the effects of HopM1 on ABA-associated virulence are mediated at least in part through ABCG40.
Discussion
High humidity following consecutive rains is a factor well known to contribute to disease development in the field (Aung et al., 2018). Such conditions are often associated with the induction of water-soaking lesions, which are thought to be crucial for virulence (Xin et al., 2016). Homologues of HopM1 and AvrE1 are present in the genomes of a variety of bacterial pathogens, and functional homologues of AvrE1 have been identified in oomycete pathogens (Deb et al., 2018; Degrave et al., 2015). Here, we provide evidence that HopM1 and AvrE1 induce stomatal closure upon pathogen entry, which prevents transpiration, resulting in an accumulation of water in the apoplast. Through transcriptome analysis, we show that HopM1 and AvrE1 act redundantly in affecting the Arabidopsis transcriptome (Figure 1A). Among the most prominent differences in the transcriptional signatures induced by WT Pst versus Pst lacking HopM1 and AvrE1, we found an important induction of genes associated with ABA-related pathways (Figure 1D). Some transcriptional changes between WT and mutant strains could be due to the differences in levels of bacteria present in the leaf at the time of sampling (36h). However, ABA-related gene expression, as well as ABA levels, are clearly increased at earlier time points when bacterial levels are similar (Figure 1E–H, 2F). It has been previously reported that transgenic expression of the Pst effector AvrPtoB leads to increased ABA biosynthesis in Arabidopsis (de Torres-Zabala et al., 2007). However, we found that expression levels of the ABA biosynthesis marker gene NCED3 was unaltered in Arabidopsis plants infected with an avrPtoB mutant or with an effector-less strain in which AvrPto and AvrPtoB have been reintegrated (Figure S3C–D). Interestingly, ABA has also been implicated in the induction of water soaking lesions in rice, and rice mutants with reduced ABA synthesis show decreased growth of Xanthomonas bacteria, likely due to increased transpiration rates (Peng et al., 2019; D. Zhang et al., 2019). Thus, manipulating the ABA pathway to mediate stomatal closure may be a widely used strategy employed by pathogens to induce water-soaking lesions in plants. At the same time, although stomatal closure induced by ABA or TBHQ is sufficient to induce water soaking and increase pathogenicity (Figure 5, S4), additional mechanisms undoubtedly contribute to creating aqueous apoplastic environments, such as promoting water intake in the apoplast (Schwartz et al., 2017). Whether such mechanisms are dependant on ABA or not remains to be elucidated.
Considering that many bacterial pathogens have evolved to overcome stomatal immunity by producing diverse toxins, such as COR and SyrA, manipulating ABA to close stomata while producing COR might seem counterintuitive (Melotto et al., 2006; Schellenberg et al., 2010). The actions of such toxins are presumably required to allow pathogens access to the leaf interior only in the early stages of infection and would be detrimental to water-soaking induction at later stages. Indeed, Pst DC3000 can reopen stomata previously closed by ABA or TBHQ (Figure 5G–H). Thus, at three hours post treatment, stomatal opening activity appears to dominate. The interplay between COR and ABA at this early phase remains to be elucidated however, as COR is barely detectable in planta at this timepoint (de Torres Zabala et al., 2009) (Figure S5E). Nonetheless, by measuring the levels of COR and ABA present in late-stage infected leaves, we find that physiologically relevant levels of ABA could override COR-induced stomatal closure. This would be consistent with a chain of events wherein COR allows the pathogen to gain access to the apoplast during the early stage of an infection, but whose stomatal opening activity is subsequently countered by ABA after an eventual accumulation of T3SS effectors in infected tissues. At the same time, COR appears to have stomate-independent roles in virulence by interfering with immune signaling in the post-invasive stages (Brooks et al., 2005; Mine et al., 2017; Zheng et al., 2012). However, with respect to stomata, in later stages ABA signaling would appear to predominate over COR signaling, effectively closing the doors previously opened by COR.
Growth levels of Pst hrcC− in Arabidopsis plants treated post-infection with ABA reached those of Pst DC3000 in untreated plants (Figure 5B and 5C). When stomatal closure and water-soaking was induced with TBHQ, a similar effect was seen on Pst hrcC− growth, but to a lesser extent (Figure 5D and 5E). We interpret the greater effect of ABA on bacterial growth as being consistent with the ability of ABA to interfere with immune signaling in addition to inducing water-soaking lesions (de Torres Zabala et al., 2009).
We found that ABA appears to accumulate to a greater degree in guard cells than pavement cells in infected Arabidopsis leaves (Figure 6A–E). One possible explanation for this would be that HopM1 and AvrE1 might be delivered in a cell-type specific manner by the pathogen to induce ABA specifically in stomata. Alternatively, this could be due to a concentration of ABA in guard cells through the activity of ABA transporters. ABCG40, also known as PDR12, has previously been identified as an ABA transporter specifically expressed in guard cells (Kang et al., 2010). We found that Pst DC3000 induces an increase of ABCG40 expression levels in a HopM1 and AvrE1-dependent manner. Moreover, Pst avrE1, but not Pst hopM1−, was unable to cause stomatal closure or water-soaking lesions in an abcg40 mutant plant. This suggests that HopM1 requires the presence of ABCG40 to induce stomatal closure. Interestingly, ABCG40 has been identified as an important contributor to plant immunity alongside PEN3 against Botrytis cinerea via camalexin secretion (He et al., 2019). Thus, ABCG40 might have different functionalities in host-pathogen interactions depending on the type of interaction. Another explanation for the observation of specific ABA accumulation in guard cells may be related to global control of ABA movement and homeostasis. It has been proposed that during a drought response, less ABA is imported into mesophyll cells, whereas its accumulation reaches high levels in guard cells, possibly due to the activity of ABCG40 (Kang et al., 2010; Y. Zhang et al., 2021). Therefore, HopM1 and AvrE1 modulation of ABA accumulation in guard cells could be attributable to the fact that they may prompt a drought-like transcriptional response during an infection. Indeed, it has been reported that the Pst transcriptional signature resembles what is observed during a drought stress (Gupta & Senthil-Kumar, 2017). Nonetheless, regardless of the redundancy of both effectors in inducing ABCG40 expression, the requirement for ABCG40 for HopM1-induced disease development appears to represent a divergence in the functions of these two effectors.
Previous studies have identified MIN7 and PP2A subunits as HopM1 and AvrE1 interactors, respectively (Jin et al., 2016; Nomura, 2006). Arabidopsis min7 mutants have previously been reported to display partial water-soaking lesions following infiltration with Pst h−/a− (Xin et al., 2016). The Arabidopsis min7 mutant is highly sensitive to changes in growth conditions (Nomura et al., 2011), and under the growth conditions used in this study for stomatal analyses, infection with Pst h/a− did not lead to water-soaking lesions in the min7 mutant. In these conditions, min7 mutants showed no significant differences in either stomatal closure or in ABA accumulation following infection with Pst DC3000, h−/a− or hrcC− when compared to wild-type plants (Figure S6A and S6B). Interestingly, however, min7 mutant plants display an ABA hypersensitive phenotype (Figure S6C). Furthermore, in growth conditions where min7 mutants display partial water-soaking lesions upon infection with Pst h−/a−, although there was no significant difference in the accumulation of NCED3 transcripts between plants infected with Pst DC3000 or h−/a− at 24 h post-infection, intermediate NCED3 transcript levels were observed at 7 h post-infection in min7 plants (Figure S6D and S6E). Because HopM1 targets multiple host proteins (Nomura 2006), the partial water-soaking phenotype of the min7 mutant indicates that MIN7 is involved in a water-soaking step that does not lead to full-scale ABA induction. Water soaking induced by Pst h−/a− may be due in part to the fact that min7 mutants are hypersensitive to ABA (Figure S6C) and future research is needed to determine which other HopM1 host targets contribute, additively or synergistically, to full-scale water-soaking.
The interactions between several PP2A subunits and AvrE1 could also be linked to increased ABA signaling. Indeed, PP2A catalytic subunits have been associated with negative regulation of ABA signaling (Hu et al., 2014; Luo et al., 2006; Pernas et al., 2007). At the same time, it has been reported that AvrE1 targets Type One Protein Phosphatases (TOPPs), which are also negative regulators of ABA signaling (Hu et al., 2022). Ablation of TOPPs leads to enhanced water-soaking and stomatal closure, even in the absence of the pathogen, and recues the virulence phenotype of Pst h−/a−. These results further highlight the important role for ABA production and signaling in the establishment of an aqueous living space for the pathogen.
In conclusion, our data provide a comprehensive portrait of stomatal behavior during an infection and highlight the role of stomatal movement manipulation in bacterial pathogenesis. How HopM1 and AvrE1 induce the expression of ABA biosynthesis and transporter genes remains to be investigated. By understanding how pathogens induce water-soaking lesions, we anticipate that broad disease resistance in plants may be achieved by interfering with these mechanisms.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Peter Moffett (peter.moffett@usherbrooke.ca).
Materials availability
This study did not generate new unique reagents.
Data code availability
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
This paper analyzes existing, publicly available data (GSE115991) from (Winkelmüller et al., 2021).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Bacterial and virus strains | ||
| Pst DC13000 wild-type | (Cuppels, 1986) | N/A |
| Pst hopM1 − | (Badel et al. 2003) | CUCPB5368 |
| Pst avrE1 − | (Badel et al. 2006) | CUCPB5374 |
| Pst hopM1 − /avrE1 − | (Badel et al. 2006) | CUCPB5377 |
| Pst hrcC − | (Wei et al. 2000) | N/A |
| Pst DB29 | (Brooks et al. 2004) | N/A |
| Pst avrPtoB | (Lin and Martin, 2005) | N/A |
| Pst D28E + AvrPtoB | (Cunnac et al. 2011) | CUCPB6012 |
| Biological samples | ||
| Promix Premium Potting Mix | PremierTech Inc. | N/A |
| SUREMIX | Michigan Grower Products Inc. | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Abscisic acid | Millipore-Sigma | CAT#C8115 |
| Coronatine | Millipore-Sigma | CAT#45511 |
| Jasmonic acid | Millipore-Sigma | CAT#14631 |
| Fluridon | Millipore-Sigma | CAT#45511 |
| Water suitable for HPLC | Millipore-Sigma | CAT#270753 |
| Methanol | Millipore-Sigma | CAT#34860 |
| Rhodamine 6G | Millipore-Sigma | CAT#R4127 |
| Formaldehyde | Millipore-Sigma | CAT#252549 |
| Dexamethasone | Millipore-Sigma | CAT#D4902 |
| tert-Butylhydroquinone | Millipore-Sigma | CAT#112941 |
| QIAzol Reagent | QIAGEN | CAT#79306 |
| Advanced Q-PCR Mastermix With Supergreen LO-ROX | Wisent Bioproducts | CAT#800–431-UL |
| SuperScript™ II Reverse Transcriptase | ThermoFisher Sci. | CAT#18064022 |
| Ambion™DNase I (RNase-free) | ThermoFisher Sci. | CAT#AM2222 |
| Jasmonic acid - isoleucine | Cayman Chemical | CAT#10740 |
| Abscisic acid-d6 | Cayman Chemical | CAT#29093 |
| Critical commercial assays | ||
| NEBNext® Multiplex Oligos for Illumina | New England BioLabs | E7335L |
| Deposited data | ||
| GEO Submission (RNA-Seq) | GSE186836 | |
| Experimental models: Cell lines | ||
| Experimental models: Organisms/strains | ||
| A. thaliana: Col-0 wild-type | TAIR | CS70000 |
| A. thaliana: aba2–1 ethylmethane sulfonate mutant | TAIR | CS156 |
| A. thaliana: abcg40 TDNA insertion mutant | TAIR | SALK_148565C |
| A. thaliana: min7 TDNA insertion mutant | (Nomura et al. 2006) | N/A |
| Oligonucleotides | ||
| All primers are listed in Table S3 | N/A | |
| Recombinant DNA | ||
| Software and algorithms | ||
| R statistical environment | https://www.r-project.org/ | N/A |
| CutAdapt | (Martin, 2005) | N/A |
| featureCounts | (Liao et al. 2014) | N/A |
| DESEQ2 | (Love et al. 2014) | N/A |
| Galaxy | (Blankenberg et al. 2010) | N/A |
| PANTHER | (Mi et al. 2013) | N/A |
| REVIGO | (Supek et al. 2011) | N/A |
| Other | ||
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Plant material and growth conditions
The Arabidopsis thaliana (L.) Heynh. ecotype Columbia (Col-0) was used in this study. The T-DNA insertional mutants (Alonso et al., 2003) CS156 (AtABA2) and SALK_148565C (AtABCG40) were obtained from the Arabidopsis Biological Resource Center (ABRC, https://abrc.osu.edu/ /).
Arabidopsis thaliana plants were grown in Promix™ Premium potting mix (PremierTech, Rivière-du-Loup, QC) in Sherbrooke or in “Arabidopsis Mix” soil (equal parts of SUREMIX [Michigan Grower Products Inc., Galesburg, MI], medium vermiculate and perlite) at Michigan State University and Duke University in growth chambers with 12 hours light/dark photoperiod, with relative humidity of approximately 60% at 21°C. Nicotiana benthamiana and Solanum lycopersicum plants were grown in BM6 soil (Berger) in growth chambers under with 16 hours light/8 hours dark photoperiod, with relative humidity of approximately 60% at 23°C.
Four to five-week-old Arabidopsis plants were used for bacterial disease assay and stomatal aperture measurement assay. Six to seven-week-old Nicotiana and Solanum plants were used for stomatal aperture measurement assay.
Bacterial strains
Pseudomonas syringae pv. tomato DC3000 strains were from laboratory stocks and originated from studies described in the key resources table.
METHOD DETAILS
Stomatal aperture assay
Leaves were cut at the base of the petiole and immediately immersed in stomatal fixation solution (formaldehyde 4%, rhodamine 6G 0.5 μM) for 1 min to stop stomatal movement and excess of solution was removed by blotting. A quarter of each leaf was cut with a razor blade and stomata observed by epifluorescence microscopy (see Figure S2A). Stomatal aperture was measured by using the software OMERO. Between 150 to 400 stomata were measured for each data point. Data collection and analysis was performed by using double-blinded standards to avoid bias.
For analysis of stomata in infected leaves, plants were kept at room humidity levels for two hours to allow infiltrated apoplastic liquid to dissipate following bacterial infiltration. Once apoplastic water evaporated, plants were domed and kept at 95% relative humidity (RH) levels to allow development of disease symptoms, unless stated otherwise.
Bacterial disease assay
Pst DC3000 and mutant strains were cultured overnight at 28°C in Luria-Bertani (LB) media containing 50 mg/L of rifampicin (and/or other appropriate antibiotics if necessary). On the day of the infection, fresh LB media was inoculated with 0.5 mL of the overnight culture and bacteria collected when OD600 reaches between 0.8–1. Bacteria were centrifuged at 4000 g for 10 minutes and the pellet resuspended in MgCl2 10 mM. Bacterial density was adjusted to 0.2 (~ 1 × 108 CFU/ml) prior to further dilutions.
For syringe infiltration, bacterial suspensions were infiltrated directly into the apoplast. Infiltrated plants were kept under ambient humidity levels for 1–2 h to allow water to evaporate and let plant leaves return to a pre-infiltration appearance, unless stated otherwise. Thereafter, plants were domed with a plastic unit to maintain high humidity (>95% RH), unless stated otherwise. For dip-inoculation method, plants were dipped in the bacterial suspension of OD600 = 0.2, with 0.025% Silwet L-77 added. Plants were domed as mentioned previously once surface liquid had evaporated to allow disease to develop.
Bacterial populations were monitored by harvesting infected Arabidopsis leaves, surface sterilizing in ethanol 80% for one minute and rinsing in sterile water twice. Leaf disks were taken from three leaves from the same plant (one per leaf; total of three leaf disks) using a cork borer (0.58 mm in diameter) and ground in sterile MgCl2 10 mM. This served as biological replicates. Three biological replicates were performed for each biological experiment. Colony-forming units were determined by making serial dilutions (100–10−6) and plating on LB plates containing 50 mg/L of rifampicin. Each dilution was plated in three technical replicates. Experiments were at least three times for biological repeatability.
Water loss assay
Arabidopsis leaves were detached at the base of the petiole and weighed over time to quantify water loss as described previously (Takahashi et al., 2018).
Chemical treatments
To assess the potential of ABA and TBHQ in inducing water-soaking lesions, plants were either infiltrated, sprayed or dipped with/in an ABA (30 μM) or TBHQ (100 μM) solution. For spraying and dipping experiments, Silwet L-77 (0.025%) was added to each solution.
Inhibition of the ABA biosynthesis pathway was made possible by syringe infiltrating fluridon (10 μM) in Arabidopsis leaves.
RNA extraction and qPCR
RNA was extracted from flash-frozen, ground leaf tissue followed with QIAZOL (QIAGEN) reagents followed by on-column DNase treatment (QIAGEN), according to the manufacturer’s protocol. RNA purity was assessed with a spectrophotometer and quality by gel electrophoresis. cDNA was generated by using 5X All-In-One RT Master Mix (ABM).
Quantitative real-time PCR was performed with a Bio-Rad CFX96 machine. Each reaction was composed to 1X Wisent Advanced SYBR Master Mix, specific primers and a 1:20 dilution of 500 ng of cDNA stock. Amplification cycle setups will be as follow: 2 min at 95°C; 40 cycles of 6 seconds at 95°C and 30 seconds at 60°C. Melting curves were verified at the end of the 40 cycles for the confirmation of primer specificities. All reactions were performed in three technical and biological replicates. Three biological replicates were performed to confirm significance and repeatability. Average Cq values were normalized by ΔΔCT formula against the indicated reference gene ACT2. Primers used in this study are listed in Table S3.
RNA-sequencing and data analysis
RNA integrity was evaluated by an Agilent Bioanalyzer 2100 with the Eukaryote Total RNA Nano Series II. cDNA libraries were generated with NEBNext® Multiplex Oligos for Illumina®, according to the manufacturer’s protocol. cDNA libraries were sequenced by RNA-seq at the Université de Sherbrooke RNomics Platform using an Illumina NextSeq 500 system with 43-bp strand-specific paired-end read. Approximately 20 million reads were generated per sample.
Reads quality was assessed using FastQC and low-quality sequences removed by using cutadapt with a quality cutoff of 30. The resulting reads were mapped onto the Arabidopsis thaliana genome (TAIR10) using RNA STAR. Mapped reads were counted using featureCounts. Differential gene expression analysis was performed by using the DESeq2 package. A cutoff of q-value <0.01 and absolute log2 fold change > 1 was applied to identity DEGs. The R function plotPCA was used for principal component analysis. GO analysis was performed with the R package for PANTHER/REVIGO. Differentially expressed genes (DEG) are listed in Tables S2.
Phytohormone extraction and quantification
Fully expanded four weeks-old Arabidopsis leaves were harvested and weighed for fresh weight calculation and immediately flash-freeze in liquid nitrogen. Tissues were ground with a plastic pestle and phytohormones extracted overnight using 0.5–1 ml of ice-cold extraction buffer (methanol: water (80:20 v/v), 0.1% formic acid, 0.1 g/L butylated hydroxytoluene and 100 nM ABA-d6 as an internal standard). Extracted phytohormones were filtered using centrifugal filter units.
Filtered extracts were quantified using an Acquity Ultra Performance Liquid Chromatography system (Waters Corporation, Milford, MA) as described previously (Huot et al., 2017) with some modifications. Briefly, capillary, cone and extractor voltage were set at 3.5 kV, 25 V and 5 V, respectively. Desolvation and cone gas will be set to a flow rate of 600 L/hrs and 50 L/hrs, respectively. Selected ion monitoring was conducted in the negative ES mode for all processed analytes (SA, SAG, ABA, JA, JA-Ile, COR). Analyte responses based on peak area integrations relative to internal standard was determined by using QuanLynx 4.1 software (Waters, Milford, MA). All analytes were quantified based on their standard curve to calculate sample concentration (nM), which was converted to ng using the molecular weight of each specific compound and the extraction volume used. All data was normalized to initial fresh weight in grams.
ABA visualization by molecular quenching
Four-week-old Arabidopsis leaves expressing the ABAleon2.1 construct were harvested with forceps and a quarter of the leaf cut with a razor blade before being mounted on a microscopy slide to avoid any major veins. mTurquoise was excited at a wavelength of 440 nm and emission was monitored at 476 nm. Images were acquired on a FV3000 Olympus confocal microscope. Images were analyzed and quantified using ImageJ software [National Institutes of Health (NIH)].
QUANTIFICATION AND STATISTICAL ANALYSIS
All experiments were carried out at least three times with at least three biological replicates for each experiment. Statistical significances correspond to Tukey’s HSD test, Student’s T-test, One-way or Two-way ANOVA as indicated in the respective figure legends. Statistical analyses were performed using the GraphPad Prism 8.4.3 software.
For bacterial titer quantification, dots in figures represent nine plants from three independent experimental replicates. For stomatal aperture assays, each dot in figures represent one stomate. At least 50 stomates were quantified per biological replicate and carried out three times (> 150 stomates/experiment). For phytohormone and COR quantification, data points represent three biological replicates from a representative experimental replicate.
Supplementary Material
Table S1: Differentially expressed genes between Arabidopsis plants infected with WT and water-soaking mutant strains of Pst DC3000 compared to control. Related to Figure 1A.
Table S2: Differentially expressed genes associated with ABA pathways in Arabidopsis plants infected with water-soaking mutant strains of Pst DC3000 compared to control. Related to Figure 1D.
Table S3: Oligonucleotides used in this study. Related to Key Resources Table.
Highlights.
Pseudomonas syringae HopM1 and AvrE1 markedly affect the Arabidopsis transcriptome
HopM1 and AvrE1 induce stomatal closure by inducing ABA biosynthesis and signaling
Bacterial manipulation of ABA pathways is required to cause water-soaking lesions
Induction of water-soaking by HopM1 involves the Arabidopsis ABA transporter ABCG40
Acknowledgements
We would like to thank all members of the Moffett and He labs for their critical inputs into this study. We particularly thank Adam Seroka for his help setting the UPLC-MS experiments during C.R.-L. research visit to the He lab and to all He lab members for insightful discussions. We are grateful to Xiu-Fang Xin, Yezhou Hu and Yanxia Ding (Chinese Academy of Sciences, Shanghai) for valuable discussions and sharing of unpublished data. We would also like to thank Dr. Alan Collmer (Cornell University) for providing the hopM1−, avrE1− and hopM1−/avrE1− mutants used in this study. This study was supported by a National Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant and a Fonds de Recherche du Québec – Nature et Technologies (FRQNT) Team Grant to P.M., as well as by a grant from the United States National Institute of Health (NIH) 1R01AI155441to S.Y.H. C.R.-L. was supported by a VoiceAge excellence fellowship and by an international internship program from the FRQNT.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest
The authors declare no competing interests.
Inclusion and diversity statement
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, … Ecker JR (2003). Genome-Wide Insertional Mutagenesis of Arabidopsis thaliana. Science, 301(5633), 653–657. 10.1126/science.1086391 [DOI] [PubMed] [Google Scholar]
- Aung K, Jiang Y, & He SY (2018). The role of water in plant-microbe interactions. The Plant Journal, 93(4), 771–780. 10.1111/tpj.13795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badel JL, Nomura K, Bandyopadhyay S, Shimizu R, Collmer A, He SY, 2003. Pseudomonas syringae pv. tomato DC3000 HopPtoM (CEL ORF3) is important for lesion formation but not growth in tomato and is secreted and translocated by the Hrp type III secretion system in a chaperone-dependent manner. Mol Microbiol 49, 1239–51. doi: 10.1046/j.1365-2958.2003.03647.x. [DOI] [PubMed] [Google Scholar]
- Badel JL, Shimizu R, Oh HS, Collmer A, 2006. A Pseudomonas syringae pv. tomato avrE1/hopM1 mutant is severely reduced in growth and lesion formation in tomato. Mol Plant Microbe Interact 19, 99–111. doi: 10.1094/MPMI-19-0099. [DOI] [PubMed] [Google Scholar]
- Baker SS, Wilhelm KS, & Thomashow MF (1994). The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Molecular Biology, 24(5), 701–713. 10.1007/BF00029852 [DOI] [PubMed] [Google Scholar]
- Bezrutczyk M, Yang J, Eom J-S, Prior M, Sosso D, Hartwig T, Szurek B, Oliva R, Vera-Cruz C, White FF, Yang B, & Frommer WB (2018). Sugar flux and signaling in plant-microbe interactions. The Plant Journal, 93(4), 675–685. 10.1111/tpj.13775 [DOI] [PubMed] [Google Scholar]
- Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J, 2010. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol 19, 1–21. doi: 10.1002/0471142727.mb1910s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DM, Bender CL, & Kunkel BN (2005). The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic aciddependent defences in Arabidopsis thaliana. Molecular Plant Pathology, 6(6), 629–639. 10.1111/j.1364-3703.2005.00311.x [DOI] [PubMed] [Google Scholar]
- Brooks DM, Hernández-Guzmán G, Kloek AP, Alarcón-Chaidez F, Sreedharan A, Rangaswamy V, Peñaloza-Vázquez A, Bender CL, & Kunkel BN (2004). Identification and Characterization of a Well-Defined Series of Coronatine Biosynthetic Mutants of Pseudomonas syringae pv. Tomato DC3000. Molecular Plant-Microbe Interactions®, 17(2), 162–174. 10.1094/MPMI.2004.17.2.162 [DOI] [PubMed] [Google Scholar]
- Cohn M, Bart RS, Shybut M, Dahlbeck D, Gomez M, Morbitzer R, Hou B-H, Frommer WB, Lahaye T, & Staskawicz BJ (2014). Xanthomonas axonopodis Virulence Is Promoted by a Transcription Activator-Like Effector–Mediated Induction of a SWEET Sugar Transporter in Cassava. Molecular Plant-Microbe Interactions®, 27(11), 1186–1198. 10.1094/MPMI-06-140161-R [DOI] [PubMed] [Google Scholar]
- Cox KL, Meng F, Wilkins KE, Li F, Wang P, Booher NJ, Carpenter SCD, Chen L-Q, Zheng H, Gao X, Zheng Y, Fei Z, Yu JZ, Isakeit T, Wheeler T, Frommer WB, He P, Bogdanove AJ, & Shan L (2017). TAL effector driven induction of a SWEET gene confers susceptibility to bacterial blight of cotton. Nature Communications, 8(1), 15588. 10.1038/ncomms15588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunnac S, Chakravarthy S, Kvitko BH, Russell AB, Martin GB, Collmer A, 2011. Genetic disassembly and combinatorial reassembly identify a minimal functional repertoire of type III effectors in Pseudomonas syringae. Proc Natl Acad Sci U S A 15, 2975–80. doi: 10.1073/pnas.1013031108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuppels DA, 1986. Generation and Characterization of Tn5 Insertion Mutations in Pseudomonas syringae pv. tomato. Appl Environ Microbiol 51, 323–327. doi: 10.1128/aem.51.2.323-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torres Zabala M, Bennett MH, Truman WH, & Grant MR (2009). Antagonism between salicylic and abscisic acid reflects early host-pathogen conflict and moulds plant defence responses. The Plant Journal, 59(3), 375–386. 10.1111/j.1365-313X.2009.03875.x [DOI] [PubMed] [Google Scholar]
- de Torres-Zabala M, Truman W, Bennett MH, Lafforgue G, Mansfield JW, Rodriguez Egea P, Bögre L, & Grant M (2007). Pseudomonas syringae pv. Tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. The EMBO Journal, 26(5), 1434–1443. 10.1038/sj.emboj.7601575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb D, Mackey D, Opiyo SO, & McDowell JM (2018). Application of alignment-free bioinformatics methods to identify an oomycete protein with structural and functional similarity to the bacterial AvrE effector protein. PLOS ONE, 13(4), e0195559. 10.1371/journal.pone.0195559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degrave A, Siamer S, Boureau T, & Barny M-A (2015). The AvrE superfamily: Ancestral type III effectors involved in suppression of pathogen associated molecular pattern-triggered immunity. Molecular Plant Pathology, 16(8), 899–905. 10.1111/mpp.12237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisele JF, Fäßler F, Bürgel PF, & Chaban C (2016). A Rapid and Simple Method for Microscopy-Based Stomata Analyses. PLOS ONE, 11(10), e0164576. 10.1371/journal.pone.0164576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Hill L, Crooks C, Doerner P, & Lamb C (2009). Abscisic Acid Has a Key Role in Modulating Diverse Plant-Pathogen Interactions. Plant Physiology, 150(4), 1750–1761. 10.1104/pp.109.137943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble PE, & Mullet JE (1986). Inhibition of carotenoid accumulation and abscisic acid biosynthesis in fluridone-treated dark-grown barley. European Journal of Biochemistry, 160(1), 117–121. 10.1111/j.14321033.1986.tb09947.x [DOI] [PubMed] [Google Scholar]
- Gupta A, & Senthil-Kumar M (2017). Transcriptome changes in Arabidopsis thaliana infected with Pseudomonas syringae during drought recovery. Scientific Reports, 7(1), 9124. 10.1038/s41598-017-09135-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck P, Thilmony R, & He SY (2003). A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proceedings of the National Academy of Sciences, 100(14), 8577–8582. 10.1073/pnas.1431173100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Xu J, Wang X, He X, Wang Y, Zhou J, Zhang S, & Meng X (2019). The Arabidopsis Pleiotropic Drug Resistance Transporters PEN3 and PDR12 Mediate Camalexin Secretion for Resistance to Botrytis cinerea. The Plant Cell, 31(9), 2206–2222. 10.1105/tpc.19.00239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R, Zhu Y, Shen G, & Zhang H (2014). TAP46 Plays a Positive Role in the ABSCISIC ACID INSENSITIVE5-Regulated Gene Expression in Arabidopsis. Plant Physiology, 164(2), 721–734. 10.1104/pp.113.233684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Ding Y, Cai B, Qin X, Wu J, Yuan M, Wan S, Zhao Y, Xin X-F. (2022). Bacterial effectors manipulate plant ABA signaling and stomatal movement for creation of an aqueous apoplast. Cell Host & Microbe. [DOI] [PubMed] [Google Scholar]
- Huot B, Castroverde CDM, Velásquez AC, Hubbard E, Pulman JA, Yao J, Childs KL, Tsuda K, Montgomery BL, & He SY (2017). Dual impact of elevated temperature on plant defence and bacterial virulence in Arabidopsis. Nature Communications, 8(1), 1–12. 10.1038/s41467-017-01674-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Ham JH, Hage R, Zhao W, Soto-Hernández J, Lee SY, Paek SM, Kim MG, Boone C, Coplin DL, & Mackey D (2016). Direct and Indirect Targeting of PP2A by Conserved Bacterial Type-III Effector Proteins. PLOS Pathogens, 12(5), e1005609. 10.1371/journal.ppat.1005609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Hwang J-U, Lee M, Kim Y-Y, Assmann SM, Martinoia E, & Lee Y (2010). PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proceedings of the National Academy of Sciences, 107(5), 2355–2360. 10.1073/pnas.0909222107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollist H, Zandalinas SI, Sengupta S, Nuhkat M, Kangasjärvi J, & Mittler R (2019). Rapid Responses to Abiotic Stress: Priming the Landscape for the Signal Transduction Network. Trends in Plant Science, 24(1), 25–37. 10.1016/j.tplants.2018.10.003 [DOI] [PubMed] [Google Scholar]
- Kuromori T, Seo M, & Shinozaki K (2018). ABA Transport and Plant Water Stress Responses. Trends in Plant Science, 23(6), 513–522. 10.1016/j.tplants.2018.04.001 [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W, 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–30. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Lin NC, Martin GB, 2005. An avrPto/avrPtoB mutant of Pseudomonas syringae pv. tomato DC3000 does not elicit Pto-mediated resistance and is less virulent on tomato. Mol Plant Microbe Interact 18, 43–51. doi: 10.1094/MPMI-18-0043. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S, 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Durán R, Bourdais G, He SY, & Robatzek S (2014). The bacterial effector HopM1 suppresses PAMP-triggered oxidative burst and stomatal immunity. New Phytologist, 202(1), 259–269. 10.1111/nph.12651 [DOI] [PubMed] [Google Scholar]
- Luo J, Shen G, Yan J, He C, & Zhang H (2006). AtCHIP functions as an E3 ubiquitin ligase of protein phosphatase 2A subunits and alters plant response to abscisic acid treatment. The Plant Journal, 46(4), 649–657. 10.1111/j.1365-313X.2006.02730.x [DOI] [PubMed] [Google Scholar]
- Macho AP (2016). Subversion of plant cellular functions by bacterial type-III effectors: Beyond suppression of immunity. New Phytologist, 210(1), 51–57. 10.1111/nph.13605 [DOI] [PubMed] [Google Scholar]
- Martin M, 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, & He SY (2006). Plant Stomata Function in Innate Immunity against Bacterial Invasion. Cell, 126(5), 969–980. 10.1016/j.cell.2006.06.054 [DOI] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Casagrande JT, Thomas PD, 2013. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 8, 1551–66. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mine A, Berens ML, Nobori T, Anver S, Fukumoto K, Winkelmüller TM, Takeda A, Becker D, & Tsuda K (2017). Pathogen exploitation of an abscisic acid- and jasmonate-inducible MAPK phosphatase and its interception by Arabidopsis immunity. Proceedings of the National Academy of Sciences, 114(28), 7456–7461. 10.1073/pnas.1702613114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura K (2006). A Bacterial Virulence Protein Suppresses Host Innate Immunity to Cause Plant Disease. Science, 313(5784), 220–223. 10.1126/science.1129523 [DOI] [PubMed] [Google Scholar]
- Nomura K, Mecey C, Lee Y-N, Imboden LA, Chang JH, & He SY (2011). Effector-triggered immunity blocks pathogen degradation of an immunityassociated vesicle traffic regulator in Arabidopsis. Proceedings of the National Academy of Sciences, 108(26), 10774–10779. 10.1073/pnas.1103338108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z, Hu Y, Zhang J, Huguet-Tapia JC, Block AK, Park S, Sapkota S, Liu Z, Liu S, & White FF (2019). Xanthomonas translucens commandeers the host rate-limiting step in ABA biosynthesis for disease susceptibility. Proceedings of the National Academy of Sciences, 116(42), 20938–20946. 10.1073/pnas.1911660116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernas M, García-Casado G, Rojo E, Solano R, & Sánchez-Serrano JJ (2007). A protein phosphatase 2A catalytic subunit is a negative regulator of abscisic acid signalling1: PP2Ac is involved in ABA signalling. The Plant Journal, 51(5), 763–778. 10.1111/j.1365-313X.2007.03179.x [DOI] [PubMed] [Google Scholar]
- Rook F, Corke F, Card R, Munz G, Smith C, & Bevan MW (2001). Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. The Plant Journal, 26(4), 421–433. 10.1046/j.1365-313X.2001.2641043.x [DOI] [PubMed] [Google Scholar]
- Schellenberg B, Ramel C, & Dudler R (2010). Pseudomonas syringae Virulence Factor Syringolin A Counteracts Stomatal Immunity by Proteasome Inhibition. Molecular Plant-Microbe Interactions®, 23(10), 1287–1293. 10.1094/MPMI-04-10-0094 [DOI] [PubMed] [Google Scholar]
- Schwartz AR, Morbitzer R, Lahaye T, & Staskawicz BJ (2017). TALEinduced bHLH transcription factors that activate a pectate lyase contribute to water soaking in bacterial spot of tomato. Proceedings of the National Academy of Sciences, 114(5), E897–E903. 10.1073/pnas.1620407114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T, 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6, E21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi F, Suzuki T, Osakabe Y, Betsuyaku S, Kondo Y, Dohmae N, Fukuda H, Yamaguchi-Shinozaki K, & Shinozaki K (2018). A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nature, 556(7700), 235–238. 10.1038/s41586-018-0009-2 [DOI] [PubMed] [Google Scholar]
- Toh S, Inoue S, Toda Y, Yuki T, Suzuki K, Hamamoto S, Fukatsu K, Aoki S, Uchida M, Asai E, Uozumi N, Sato A, & Kinoshita T (2018). Identification and Characterization of Compounds that Affect Stomatal Movements. Plant and Cell Physiology, 59(8), 1568–1580. 10.1093/pcp/pcy061 [DOI] [PubMed] [Google Scholar]
- Toruño TY, Stergiopoulos I, & Coaker G (2016). Plant-Pathogen Effectors: Cellular Probes Interfering with Plant Defenses in Spatial and Temporal Manners. Annual Review of Phytopathology, 54(1), 419–441. 10.1146/annurev-phyto-080615-100204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Üstün S, Hafrén A, Liu Q, Marshall RS, Minina EA, Bozhkov PV, Vierstra RD, & Hofius D (2018). Bacteria Exploit Autophagy for Proteasome Degradation and Enhanced Virulence in Plants. The Plant Cell, 30(3), 668–685. 10.1105/tpc.17.00815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Hitomi K, Nishimura N, Hitomi C, Adams SR, Getzoff ED, & Schroeder JI (2014). FRET-based reporters for the direct visualization of abscisic acid concentration changes and distribution in Arabidopsis. ELife, 3, e01739. 10.7554/eLife.01739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Plovanich-Jones A, Deng WL, Jin QL, Collmer A, Huang HC, He SY, 2000. The gene coding for the Hrp pilus structural protein is required for type III secretion of Hrp and Avr proteins in Pseudomonas syringae pv. tomato. Proc Natl Acad Sci U S A 29, 2247–52. doi: 10.1073/pnas.040570097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelmüller TM, Entila F, Anver S, Piasecka A, Song B, Dahms E, Sakakibara H, Gan X, Kułak K, Sawikowska A, Krajewski P, Tsiantis M, Garrido-Oter R, Fukushima K, Schulze-Lefert P, Laurent S, Bednarek P, & Tsuda K (2021). Gene expression evolution in pattern-triggered immunity within Arabidopsis thaliana and across Brassicaceae species. The Plant Cell, 33(6), 1863–1887. 10.1093/plcell/koab073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin X-F, Nomura K, Aung K, Velásquez AC, Yao J, Boutrot F, Chang JH, Zipfel C, & He SY (2016). Bacteria establish an aqueous living space in plants crucial for virulence. Nature, 539(7630), 524–529. 10.1038/nature20166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin X-F, Nomura K, Ding X, Chen X, Wang K, Aung K, Uribe F, Rosa B, Yao J, Chen J, & He SY (2015). Pseudomonas syringae Effector Avirulence Protein E Localizes to the Host Plasma Membrane and Down-Regulates the Expression of the NONRACE-SPECIFIC DISEASE RESISTANCE1/HARPIN-INDUCED1-LIKE13 Gene Required for Antibacterial Immunity in Arabidopsis. Plant Physiology, 169(1), 793–802. 10.1104/pp.15.00547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Nishimura N, Kitahata N, Kuromori T, Ito T, Asami T, Shinozaki K, & Hirayama T (2006). ABA-Hypersensitive Germination3 Encodes a Protein Phosphatase 2C (AtPP2CA) That Strongly Regulates Abscisic Acid Signaling during Germination among Arabidopsis Protein Phosphatase 2Cs. Plant Physiology, 140(1), 115–126. 10.1104/pp.105.070128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Tian C, Yin K, Wang W, & Qiu J-L (2019). Postinvasive Bacterial Resistance Conferred by Open Stomata in Rice. Molecular Plant-Microbe Interactions®, 32(2), 255–266. 10.1094/MPMI-06-18-0162-R [DOI] [PubMed] [Google Scholar]
- Zhang Y, Kilambi HV, Liu J, Bar H, Lazary S, Egbaria A, Ripper D, Charrier L, Belew ZM, Wulff N, Damodaran S, Nour-Eldin HH, Aharoni A, Ragni L, Strader L, Sade N, Weinstain R, Geisler M, & Shani E (2021). ABA homeostasis and long-distance translocation are redundantly regulated by ABCG ABA importers. Science Advances, 7(43), eabf6069. 10.1126/sciadv.abf6069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Spivey NW, Zeng W, Liu P-P, Fu ZQ, Klessig DF, He SY, & Dong X (2012). Coronatine Promotes Pseudomonas syringae Virulence in Plants by Activating a Signaling Cascade that Inhibits Salicylic Acid Accumulation. Cell Host & Microbe, 11(6), 587–596. 10.1016/j.chom.2012.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Differentially expressed genes between Arabidopsis plants infected with WT and water-soaking mutant strains of Pst DC3000 compared to control. Related to Figure 1A.
Table S2: Differentially expressed genes associated with ABA pathways in Arabidopsis plants infected with water-soaking mutant strains of Pst DC3000 compared to control. Related to Figure 1D.
Table S3: Oligonucleotides used in this study. Related to Key Resources Table.


