Abstract
It remains unclear whether immune responses following natural infection can be sustained or potentially prove critical for long-term immune protection against SARS-CoV-2 reinfection. Here, we systematically mapped the phenotypic landscape of SARS-CoV-2-specific immune responses in peripheral blood samples of convalescent patients with COVID-19 by single-cell RNA sequencing. The relative percentage of the CD8 + effector memory subset was increased in both convalescent moderate and severe cases, but NKT-CD160 and marginal zone B clusters were decreased. Innate immune responses were attenuated reflected by decreased expression of genes involved in interferon-gamma, leukocyte migration and neutrophil mediated immune response in convalescent COVID-19 patients. Functions of T cell were strengthened in convalescent COVID-19 patients by clear endorsement of increased expression of genes involved in biological processes of regulation of T cell activation, differentiation and cell–cell adhesion. In addition, T cell mediated immune responses were enhanced with remarkable clonal expansions of TCR and increased transition of CD4 + effector memory and CD8 + effector-GNLY in severe subjects. B cell immune responses displayed complicated and dual functions during convalescence of COVID-19, providing a novel mechanism that B cell activation was observed especially in moderate while humoral immune response was weakened. Interestingly, HLA class I genes displayed downregulation while HLA class II genes upregulation in both T and B cell subsets in convalescent individuals. Our results showed that innate immunity was declined but SARS-CoV-2-specific T cell responses were retained even strengthened whereas complicated and dual functions of B cells, including declined humoral immunity were presented at several months following infections.
Keywords: COVID-19, SARS-CoV-2, Single cell RNA sequencing, Immune response, T/B cell receptor repertoire, Bioinformatics analysis
1. Introduction
The coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, is an ongoing pandemic infectious disease, with more than 225 million confirmed infections and more than 4.6 million deaths according to the statistics of World Health Organization (WHO) until Sep 16th, 2021 (https://covid19.who.int/). At present, there is no effective drugs and vaccines against SARS-CoV-2, and the excessive inflammation associated with severe COVID-19 can lead to respiratory failure, septic shock, and ultimately, death [1], [2]. The total mortality rate of COVID-19 is 0.5%-3.5%, even as high as ∼ 10% in specific regions [1], [2]. The majority of COVID-19 patients exhibit mild to moderate symptoms, up to 15% progress to severe pneumonia and approximately 5% eventually develop acute respiratory distress syndrome (ARDS) and/or multiple organ failure [3]. Higher fatality rates have been observed in elderly individuals with comorbidities and those who are immunocompromised [4], [5], [6].
The capacity to mount a protective adaptive immune response plays a decisive role if viral propagation can be suppressed early or if the virus will massively destroy tissues with high angiotensin-converting enzyme 2 (ACE2) expression, ultimately leading to a severe inflammatory response with multiple organ damage, specifically in the lungs and the kidneys [7], [8], [9]. It will therefore be critical for public health reasons to determine whether individuals with moderate or severe COVID-19 develop robust immunity against SARS-CoV-2. Furthermore, there is considerable interest in understanding how immune responses act to control acute infection and provide protection from reinfection, which provide us a potential prognostic and diagnostic markers and to design effective therapeutic interventions for COVID-19 severe patients.
Global efforts are currently underway to map the determinants of immune protection against SARS-CoV-2. Up to now whether convalescent COVID-19 patients generate protection against SARS-CoV-2 reinfection is not solidly evidenced. Recent research has shown that SARS-CoV-2 infection generates near-complete protection against rechallenge in rhesus macaques [10]. However, there is very limited evidence of reinfection of recovered COVID-19 patients [11]. Further work is therefore urgently required to define the mechanisms that underlie these observations and evaluate the durability of protective immune responses elicited by primary infection with SARS-CoV-2. Most related studies of immune protection against COVID-19 have focused on induction of neutralizing antibodies against SARS-CoV-2 [12], [13], [14], [15]. Longitudinal studies on humoral immunity in COVID-19 as well as studies in convalescent subjects indicate that humoral immunity is often short lived and that most SARS-CoV-2 antibodies exhibit limited somatic hypermutation [13], [16], [17], [18]. Multiple studies have demonstrated that the level of neutralizing antibodies declines over time and may even lead to loss of detectable virus-specific antibodies in a substantial proportion of individuals [19], [20]. In line with antibody, memory B cell responses tend to be short lived after infection with SARS-CoV-1 [21], [22]. In contrast, memory T cell responses can persist for many years [22], [23], [24]. SARS-CoV-2-specific T cells have been identified in humans [25], [26]. SARS-CoV-2-specific T cells were even detectable in antibody-seronegative exposed family members and convalescent individuals with a history of asymptomatic and mild COVID-19, which revealed that SARS-CoV-2 elicits broadly directed and functionally replete memory T cell responses, suggesting that natural exposure or infection may prevent recurrent episodes of severe COVID-19 [27]. Recent studies identified that functional SARS-CoV-2-specific T-cell responses are retained at about six months following infection [27], [28], [29].
Single-cell RNA sequencing (scRNA-seq) has emerged as powerful tools to dissect the immune responses under various conditions at the finest resolution, and has been applied to COVID-19 studies [30], [31], [32], [33], [34]. Zhang et al. implemented scRNA-seq to obtain an unbiased and comprehensive transcriptome atlas of the immunological responses in PBMCs from COVID-19 patients, including moderate, severe and convalescent patients and found that most cell types in COVID-19 patiets showed a strong interferon-α response and an overall acute inflammatory response while highly cytotoxic effector T cell subsets including CD4 + effector-GNLY (granulysin), CD8 + effector-GNLY and NKT CD160 were intensive expanded in convalescent moderate patients [35]. Cao et al. reported that they rapid and defficient idenditification of SARS-CoV-2-neutralizing antibodies achieved by scRNA-seq and VDJ sequening of antigen-binding B cells from convalescent COVID-19 patients [36]. Our previous study applied scRNA-seq to analyze 284 samples from 196 COVID-19 patients and controls to create a comprehensive immune landscape. We found that lymphopenia and active T and B cell responses coexist and are shaped by age and sex. More importantly, SARS-CoV-2 infects diverse epithelial and immune cells, inducing distinct responses. Furthermore, cytokine storms with systemic calprotectin are associated with COVID-19 severity [37].
However, it remains unclear whether natural infections with SARS-CoV-2 generate long-lasting immune responses and how innate, T and B cell responses changes in a long-term post recovery and whether potentially prove critical for long-term immune protection against SARS-CoV-2 reinfection. Hubei Province of China was the very first field hit by SARS-CoV-2. Accordingly, all patients who experienced the longest phase of convalescence following COVID-19 reside here or neighborhood. To address this knowledge gap, we characterized SARS-CoV-2-specific innate, T and B cells in donors at four months following primary infection by scRNA-seq.
In this study, we systematically performed a scRNA-seq analysis to depict the immune landscape of convalescent COVID-19 patients. We found innate immune responses are attenuated in convalescent severe patients. Morerover, T cell mediated immune responses are retained even strengthened during COVID-19 convalescence while B cell immune responses display complicated and dual functions in convalescent severe patients. There were some unique IGV genes in severe cases which may assist the design of therapeutics and vaccines.
2. Results
2.1. Integrated analysis of scRNA-seq data of peripheral immune cells from convalescent patients with COVID-19.
We collected samples between 3.5 and 4.2 months following first symptom onset or between 0 and 3.3 months following closest discharge. The demographics and clinical features of the patients are listed in Table 1 . The laboratory findings of enrolled patients are shown in Supplementary Table 1. Thirteen convalescent patients with COVID-19 were classified into two clinical conditions: convalescent patients with moderate COVID-19 (n = 5) and convalescent patients with severe COVID-19 (n = 8) (Table 1; Supplementary Table 1). The five patients with moderate COVID-19 profiled were four males and one female, aged 28 to 39 years, at an average age of 35.4 years. The eight patients with severe COVID-19 profiled were six males and two females, aged 29 to 53 years, at an average age of 38.9 years. The healthy donors were four males and one female, aged 27 to 30 years, at an average age of 28.8 years. All healthy donors were also confirmed without SARS-CoV-2 infection by real-time RT-PCR. All the patients were confirmed between Jan 18th, 2020 and Feb 1st, 2020. Four of thirteen samples were collected from ventilated COVID-19 patients who were diagnosed with acute respiratory distress syndrome (ARDS; Supplementary Table 1). No patients received azithromycin, which has potential immunomodulatory effects, prior to sampling (Supplementary Table 1). No patients received Remdesivir during hospitalization prior to sampling.
Table 1.
Demographics, sample characteristics and disease course of convalescent patients with COVID-19.
|
ID of Samples |
Donors |
Age (Years) |
Sex |
Nucleic acid testing of SARS-CoV-2 |
Clinical classification |
Initial symptom onset |
Admission level |
Ventilated/ ARDS? |
Immunomodulators prior to blood draw |
PaO2/FiO2 on (ICU) admission |
SaO2 on (ICU) admission |
PaO2 on (ICU) admission |
Months from initial symptom onset to blood draw |
Months from closest discharge to blood draw |
Clinical outcome |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1 | Healthy donor-1 | 28 | M | Negative | Health | asymptomatic | |||||||||
| S2 | Healthy donor-2 | 30 | M | Negative | Health | asymptomatic | |||||||||
| S3 | Healthy donor-3 | 30 | M | Negative | Health | asymptomatic | |||||||||
| S4 | Healthy donor-4 | 27 | M | Negative | Health | asymptomatic | |||||||||
| S5 | Healthy donor-5 | 29 | M | Negative | Health | asymptomatic | |||||||||
| S6 | Convalescent patients with moderate COVID-19–1 |
28 | M | Positive | Mild | Fever | Floor | No | No | 4.2 | 2.5 | Discharged home |
|||
| S7 | Convalescent patients with moderate COVID-19–2 |
34 | F | Positive | Mild | Muscle soreness | Floor | No | No | 3.9 | 1.4 | Discharged home |
|||
| S8 | Convalescent patients with moderate COVID-19–3 |
38 | M | Positive | Mild | Fever | Floor | No | No | 3.9 | 2.3 | Discharged home |
|||
| S9 | Convalescent patients with moderate COVID-19–4 |
39 | M | Positive | Mild | Pulmonary shadow | Floor | No | No | 4 | 1.4 | Discharged home |
|||
| S10 | Convalescent patients with moderate COVID-19–5 |
38 | M | Positive | Mild | Dry cough | Floor | No | No | 3.8 | 3.3 | Discharged home |
|||
| S11 | Convalescent patients with severe COVID-19–1 |
29 | M | Positive | Severe | Fever | Floor | No | No | 106 | 90 | 74.00 | 4.1 | 3 | Discharged home |
| S12 | Convalescent patients with severe COVID-19–2 |
36 | M | Positive | Severe | Fever | ICU | Yes | No | 173 | 92 | 57 | 3.7 | 2.6 | Discharged home |
| S13 | Convalescent patients with severe COVID-19–3 |
28 | M | Positive | Severe | Fever | Floor | No | No | 203 | 90 | 74 | 4.1 | 3 | Discharged home |
| S14 | Convalescent patients with severe COVID-19–4 |
40 | M | Positive | Severe | Fever | Floor | No | No | 182 | 92 | 74.00 | 3.9 | 3.2 | Discharged home |
| S15 | Convalescent patients with severe COVID-19–5 |
36 | M | Positive | Severe | Fever | ICU | Yes | No | 107 | 46 | 28 | 3.5 | 0 | Discharged home |
| S16 | Convalescent patients with severe COVID-19–6 |
53 | M | Positive | Severe | Fever | ICU | Yes | No | 200 | 92 | 59 | 4.1 | 2.9 | Discharged home |
| S17 | Convalescent patients with severe COVID-19–7 |
36 | F | Positive | Severe | Fever | ICU | Yes | No | 130 | 78 | 43.00 | 4 | 2.2 | Discharged home |
| S18 | Convalescent patients with severe COVID-19–8 |
53 | F | Positive | Severe | Fever | Floor | No | No | 179 | 93 | 59.00 | 4 | 1.6 | Discharged home |
There were no significant differences in the absolute number of leukocytes, neutrophils, monocytes and platelets counts between moderate and severe group. However, the severe group exhibited a significantly lower absolute lymphocyte count and albumin than the moderate group (P = 0.003 and P = 0, respectively) (Supplementary Table 1). These findings were in accordance with the results of previous study (PMID: 32641700). All the patients were shown positive in SARS-CoV-2 RNA testing, but 60% (3 of 5) and 50% (4 of 8) of patients were seropositive in moderate and severe respectively. Interestingly, however, we found no patients were seropositive on the days performed SARS-CoV-2 RNA testing (Supplementary Table 2).
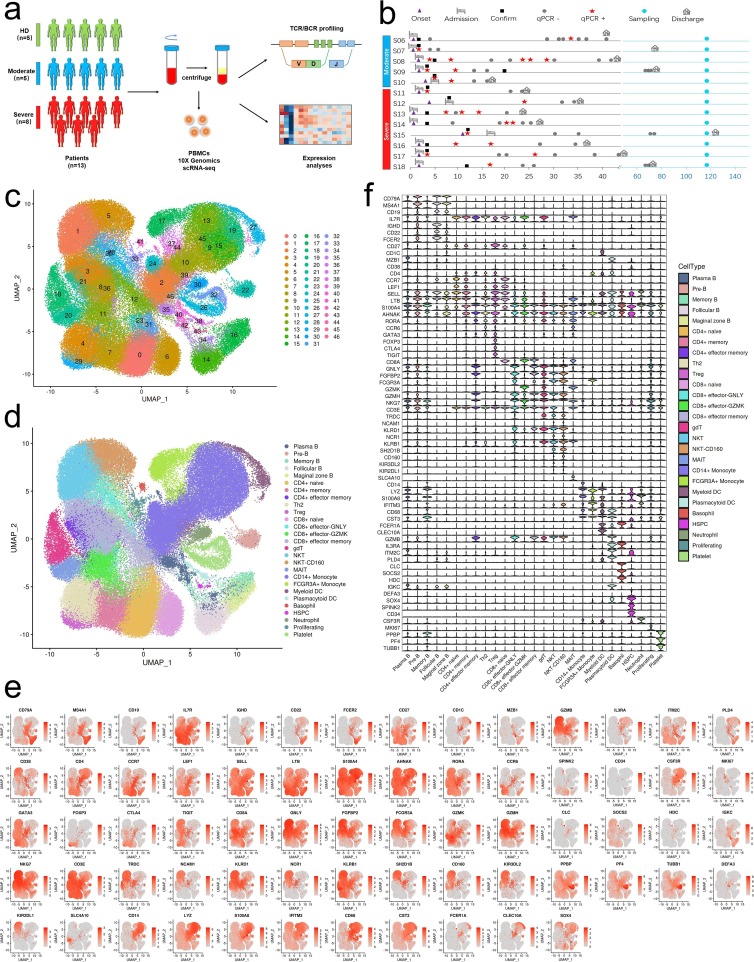
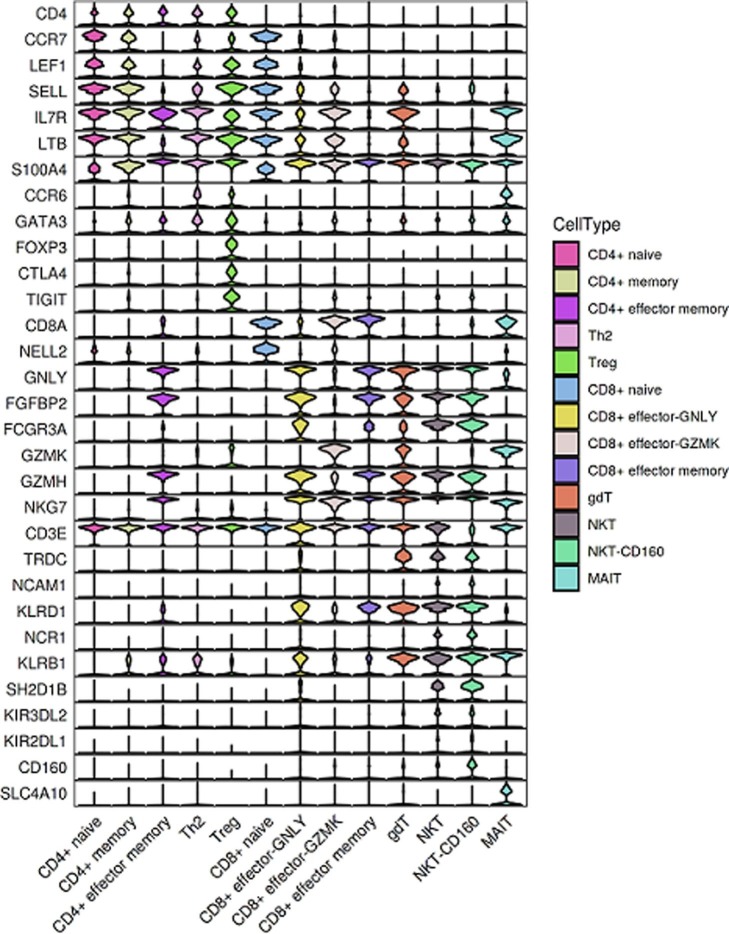
All samples were subjected to scRNA-seq based on the 10X Genomics 5′ sequencing platform to generate both the gene expression and T cell receptor (TCR) or B cell receptor (BCR) data of peripheral blood mononuclear cells (PBMCs) from the subjects e.g. convalescent patients with moderate COVID-19 (n = 5) and convalescent patients with severe COVID-19 (n = 8) and 5 healthy donors (HDs; n = 5) (Fig. 1 a and b). The scRNA-seq raw data were analyzed by a unified analysis pipeline, including the kallisto and bustools programs, to obtain the gene expression data of individual cells and by the CellRanger program to obtain TCR and BCR sequences. We applied a common set of stringent quality control criteria to ensure that the selected data were from single and live cells and that their transcriptomic phenotypes were comprehensively characterized. We abandoned one sample (S02 in HD group) due to the unsatisfactory health records. A total of 122,726 high-quality single cells were ultimately obtained from 17 samples, with an average of 7,219 cells per sample (Supplementary Table 3). After the unified single-cell analysis pipeline (see Methods), 11,7102 cells were used for identification of cell types (Supplementary Table 3). Among these (11,7102) cells, 27,781 cells (23.72%) were from the healthy donors, 30,447 cells (26%) were from the convalescent moderate condition and 58,874 cells (50.3%) were from the convalescent severe conditions. All high-quality cells were integrated into an unbatched and comparable dataset and subjected to principal component analysis after correction for read depth and mitochondrial read counts (Supplementary Table 4). Using graph-based clustering of uniform manifold approximation and projection (UMAP), we identified 47 major cell types (Supplementary Table 5) and captured 27 clusters according to the expression of canonical gene markers (Fig. 1c-f, Supplementary Table 5). These cell types included CD14 (+) monocyte (CD14 + Monocyte; CD14 + LYZ + S100A8 + ), FCGR3A (+) monocyte (FCGR3A + Monocyte; FCGR3A + IFITM3 + CD68 + ), myeloid dendritic cells (Myeloid DC; CD1C + CST3 + FCER1A + CLEC10A + ), plasmacytoid dendritic cells (Plasmacytoid DC; GZMB + IL3RA + ITM2C + PLD4 + ), basophiles (Basophile; CLC + SOCS2 + HDC + ), CD34(+) hematopoietic stem and progenitor cells (HSPC; DEFA3 + LYZ + SOX4 + SPINK2 + CD34 + ), neutrophils (Neutrophil; CSF3R + ), naïve CD4(+) T cells (CD4 + naïve; CD4 + CCR7 + LEF1 + SELL + ), CD4(+) T cell memory (CD4 + memory; CD4 + IL7R + LTB + S100A4 + ), CD4(+) effector memory T cells (CD4 + effector memory; CD4 + AHNAK + RORA + S100A4 + ), T helper 2 (Th2; CD4 + CCR6 + GATA3 + ), regulatory T (Treg) cells (Treg; CD4 + FOXP3 + CTLA4 + TIGIT + ), naïve CD8(+) T cells (CD8 + naïve; CD8A + CCR7 + LEF1 + NELL2 + ), CD8(+) effector-GNLY (CD8 + effector-GNLY; CD8A + GNLY + FGFBP2 + FCGR3A + ), CD8(+) effector- GZMK (CD8 + effector-GZMK; CD8A + GZMK + ), CD8(+) effector memory T cells (CD8 + effector memory; CD8A + FGFBP2 + GZMH + NKG7 + ), gammadeltaT cells (gdT; CD3E + TRDC + CD8A-CD4-), natural killer T (NKT) cells (NKT; NCAM1 + KLRD1 + NCR1 + KLRB1 + SH2D1B + KIR3DL2 + KIR2DL1 + CD8A + ), CD160 (+) natural killer T (NKT) cells (NKT-CD160; NCAM1 + KLRD1 + NCR1 + KLRB1 + CD160 + SH2D1B + KIR3DL2 + KIR2DL1 + CD8A + ), mucosal-associated invariant T (MAIT) cells (MAIT; CD3E + SLC4A10 + ), pre-B cells (Pre B; CD79A + MS4A1 + CD19 + IL7R + CD34-), marginal zone B cells (Marginal zone B; CD79A + MS4A1 + CD19 + CD1C + ), follicular B cells (Follicular B; CD79A + MS4A1 + CD19 + CD22 + FCER2 + CD27-), plasma B cells (Plasma B; CD79A + MZB1 + CD38 + IGKC + ), memory B cells (Memory B; CD79A + MS4A1 + CD27 + IGHD-), Proliferating (MKI67 + ), Platelet (PPBP + PF4 + TUBB1 + ).
Fig. 1.
Study design and single-cell transcriptional profiling of PBMCs from HDs and convalescent COVID-19 patients. a, Study design and single-cell transcriptional profiling of PBMCs from HDs and convalescent COVID-19 patients. A schematic showing the overall study design. The scRNA-seq was applied to PBMCs across three conditions and the output data were used for TCR and BCR profiling and expression analyses. b, Timeline of the course of disease for 13 patients infected with SARS-CoV-2 and enrolled in our study. qPCR indicates PCR testing for SARS-CoV-2 nucleic acids. qPCR positive indicates nasopharyngeal swab samples that were positive for SARS-CoV-2 nucleic acids. Healthy donors are colored in green, convalescent patients with moderate COVID-19 are colored in blue and convalescent severe patients are colored in reds. The color bars on the left represent conditions with the same color as in a. c, Quality control and cell populations identified. A total of 17 PBMC samples with HDs (n = 4), and convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8), were sequenced and obtained a total of 122,726 high-quality single cells. After quality control, 117,102 cells were obtained from 122,726 of PBMCs, 47 clusters of cells were identified by UMAP. Each dot corresponds to a single cell, colored according to cell type. d, 27 Cellular populations identified. The 47 clusters of cells in Fig. 1c were identified further by cell types. The UMAP projection of 117,102 single cells from HDs (n = 4) and convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8), showing 27 clusters with the respective labels. The 27 clusters were identified and showed with UMAP. Each dot corresponds to a single cell, colored according to cell type. e, Canonical cell markers were used to label clusters by cell identity as represented in the UMAP plot. Data are colored according to expression levels and the legend is labeled in log scale. f, Canonical cell markers were used to label clusters by cell identity in 27 clusters as represented in the violin plot. Y-axis represents log2 (normalized count + 1).
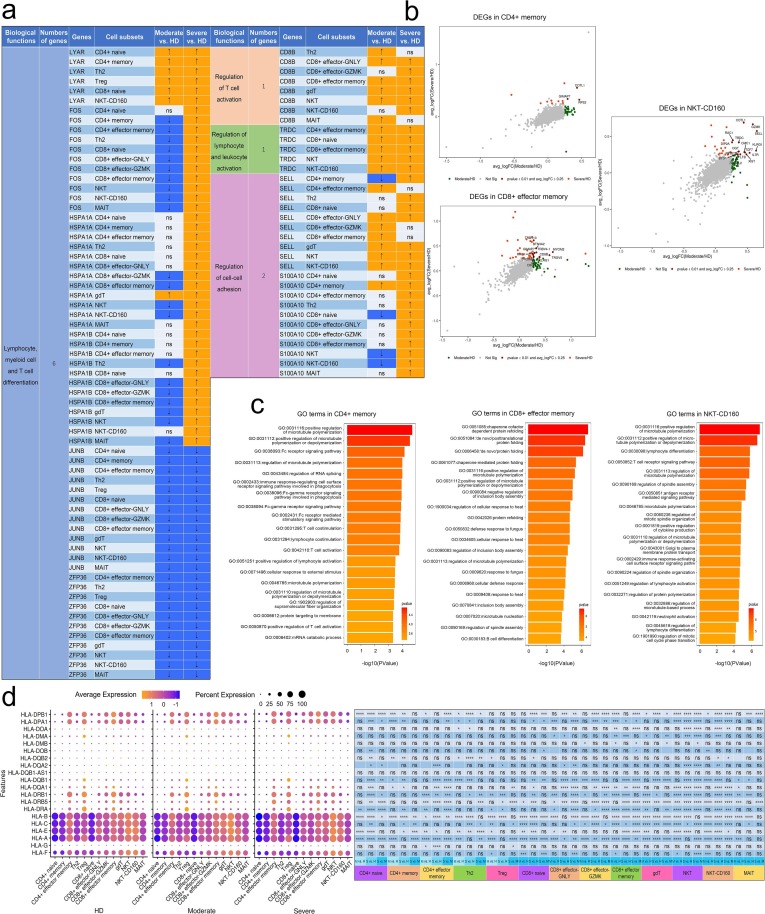
2.2. Phenotype characteristics of cell types in convalescent patients with moderate and severe COVID-19.
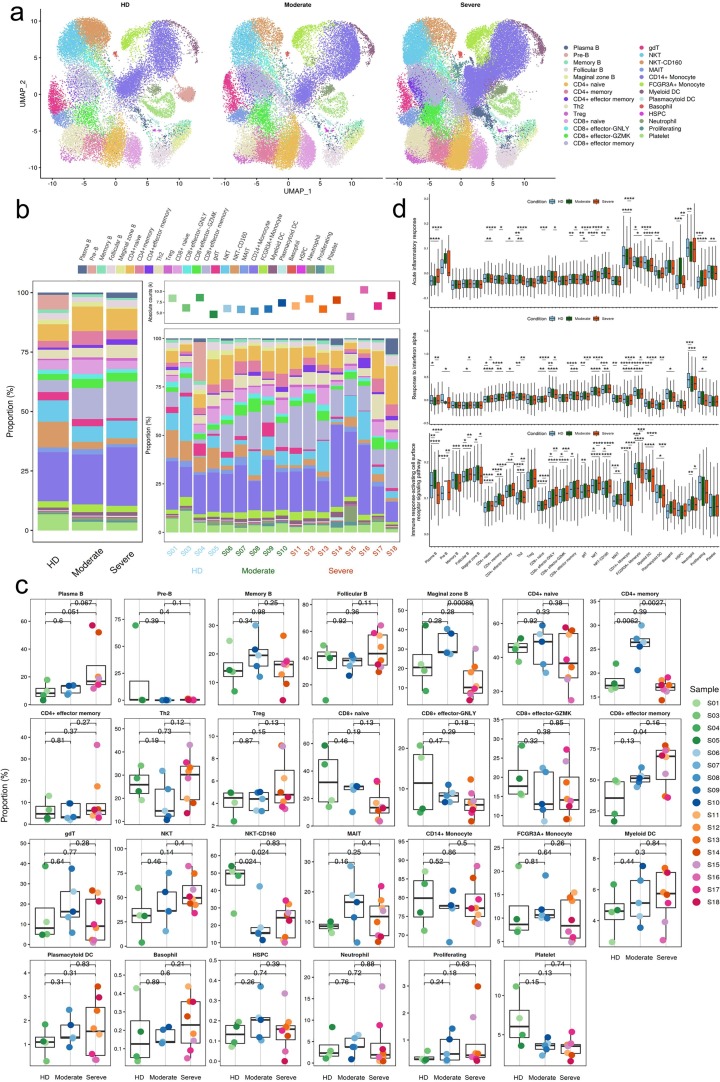
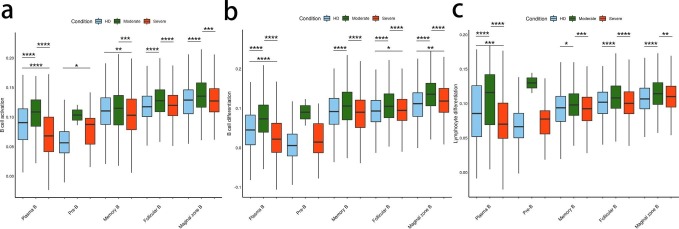
To show the differences in cell composition across two conditions, e.g. convalescent patients with moderate or severe COVID-19, and to compare with that in healthy donors, the relative percentage of the 27 major cell types in the PBMCs of each individual were calculated on the basis of scRNA-seq data. Notable differences could be observed in the immune compositions of healthy donors and the convalescent COVID-19 patients based on UMAP (Fig. 2 a–c). The relative percentage of of CD4 + memory cluster was increased significantly in convalescent moderate COVID-19 patients compared with HD (P = 0.0062) and decreased significantly in convalescent severe COVID-19 patients compared with moderate (P = 0.0027). The relative percentage of the CD8 + effector memory cell cluster increased significantly with disease severity (HD vs. convalescent severe patients, P = 0.04). In contrast, the relative percentage of the NKT-CD160 cell cluster was decreased significantly (HD vs. convalescent moderate patients, P = 0.024; HD vs. convalescent severe patients, P = 0.024). Compared with convalescent moderate patients, Marginal zone B cluster was decreased significantly in convalescent severe patients (P = 0.000089).
Fig. 2.
Differences in cell composition between HDs and convalescent COVID-19 patients. a, The UMAP projection of the HD, convalescent patients with moderate COVID-19 or with severe COVID-19. Each dot corresponds to a single cell, colored according to cell type. b, Average proportion of each cell type derived from HDs (n = 4) and convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8). The left picture depicts average proportion of each cell type derived from three groups, HDs, convalescent patients with moderate COVID-19 and convalescent patients with severe COVID-19. The calculation method is: (the number of specific cell cluster in one group) ⁄ (the number of total cells in one group). The dot plot in the upper panel of the right picture shows the sum of the absolute counts of cell subsets in the PBMCs of each sample, and the bottom bar plot shows cell compositions at a single sample level. The calculation method is: (the number of specific cell cluster in one sample) ⁄ (the number of total cells in one sample). c, Proportions of each cell type in each sample colored by donor of origin. Description of calculation method of cell ratio as follows: B cells included Pre-B, Marginal zone B, Follicular B, Plasma B and Memory B. The percentage of each cell cluster is calculated as: (the number of specific cell cluster in one sample) ⁄ (the number of B cells in one sample); CD4+ T cells included CD4+ naive, CD4+ memory, CD4+ effector memory, Th2, and Treg. The percentage of each cell cluster is calculated as: (the number of specific cell cluster in one sample) ⁄ (the number of CD4+ T cells in one sample); CD8+ T cells include CD8+ Naive, CD8+ effector-GNLY, CD8+ effector-GZMK, CD8+ effector memory, and the percentage of each cluster is calculated as: (the number of specific cell cluster in one sample) ⁄ (the number of CD8+ T cells in one sample); NKT/gdT cells include gdT, NKT, NKT-CD160, and MAIT, and the percentage of each cluster is calculated as: (the number of specific cell cluster in one sample) ⁄ (the number of (NKT/gdT) cells in one sample); Myeloid cells include CD14+ Monocyte, FCGR3A+ Monocyte, Myeloid DC, Plasmacytoid DC, and Neutrophile. The percentage of each cluster is calculated as: (the number of specific cell cluster in one sample) ⁄ (the number of Myeloid cells in one sample); Other cells include Basophil, HSPC, Proliferating, and Platelet, and the percentage of each cluster is calculated as: (the number of specific cell cluster in one sample) ⁄ (the number of total cells in one sample (117,102 cells)). The x axes correspond to the status of each patient e.g. HDs, convalescent patients with moderate COVID-19 and convalescent patients with severe COVID-19. Shown are adjusted P values by unpaired two-sided t-test. d, Box plots of the cell state score using genes from three GO biological process terms (Acute inflammatory responses, Responses to interferon alpha and Immune responses-activating cell surface receptor signaling pathway) across cell types derived from HDs (n = 4) and convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8). For each GO pathway, the AddModuleScore (Seurat v3.2.0, default parameter) function was used to calculate the cell stat scores of of genes from the GO pathway in each cell. Box plots of are drawn according to the three groups of HDs, convalescent patients with moderate COVID-19 and convalescent patients with severe COVID-19, and the cell stat scores of the cells from three groups are compared. Conditions are shown in different colors. The vertical axis represents median values, with whiskers extending to the farthest data point within a maximum of 1.5 × interquartile range. All differences with *P < 0.01 are indicated. **P < 0.001; ***P < 0.0001; using unpaired two-sided T-test. Shown are adjusted P values by unpaired two-sided t-test.
Next, to explore the antiviral and pathogenic immune responses during infection, we analysed expression levels of three important Gene Ontology (GO) biological process terms e.g. acute inflammatory response (GO0002526), response to interferon (IFN)-α (GO0035455) and immune response-activating cell surface receptor signaling pathway (GO0002429) in major cell types across three conditions. We found that the acute inflammatory response was significantly upregulated in CD8 + effector memory with disease severity. This response in NKT and NKT-CD160 significantly upregulated in moderate and significantly downregulated in severe. In contrast, it was significantly downregulated in Myeloid DC in moderate and significantly upregulated in severe. This response in CD14 Monocyte and FCGR3A + Monocyte significantly upregulated in severe (Fig. 2d; Supplementary Table 6). The response to IFN-α was significantly upregulated in CD4 + naïve, CD8 + naïve and NKT-CD160 with disease severity. In contrast, it was significantly downregulated in CD8 + effector-GNLY and Neutrophil. In gdT and NKT, it was significantly upregulated in moderate and significantly downregulated in severe. In contrast, in CD14 + Monocyte and FCGR3A + Monocyte it was significantly downregulated in moderate and significantly upregulated in severe (Fig. 2d; Supplementary Table 6). The immune response-activating cell surface receptor signaling pathway was found significantly upregulated in Follicular B, CD4 + naïve, CD4 + effector memory, CD8 + naïve, CD8 + effector-GNLY and CD8 + effector-GZMK with disease severity. But it was found significantly upregulated in CD4 + memory, Th2 and CD14 Monocyte in moderate and significantly downregulated in severe. In contrast, it was found significantly downregulated in NKT and NKT-CD160 in moderate and significantly upregulated in severe (Fig. 2d; Supplementary Table 6).
2.3. Attenuated innate immune responses were observed in convalescent COVID-19 patients.
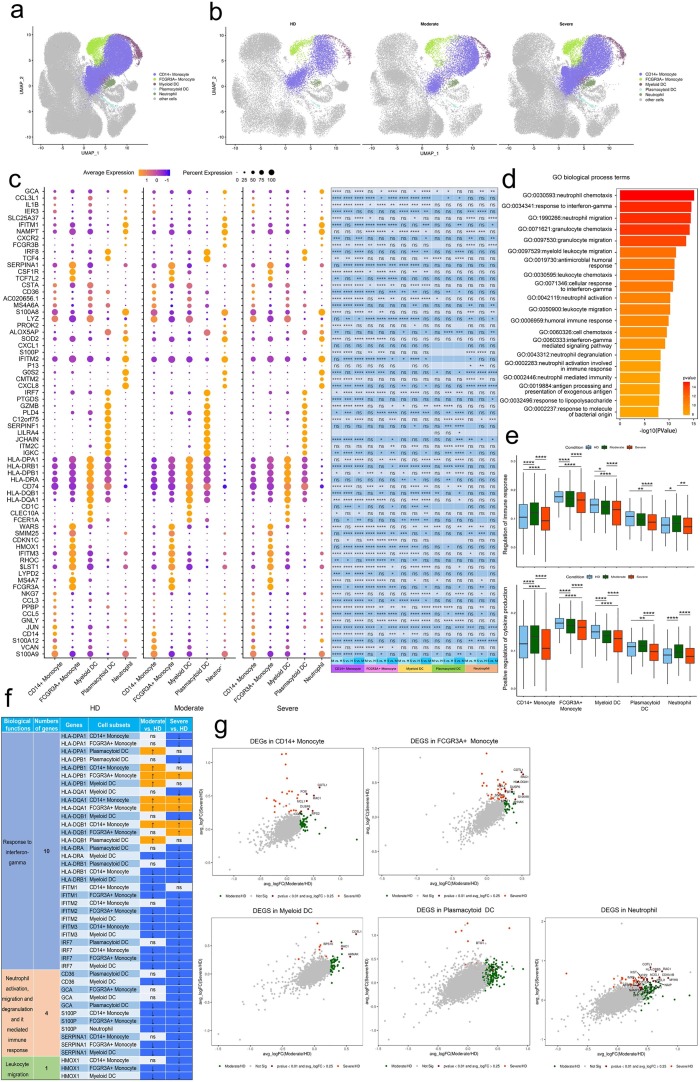
To further probe the transcriptomic changes of innate immune cells post infection with SARS-CoV-2, we compared the expression patterns of the convalescent moderate or convalescent severe condition with that of the HD in cell subtypes relevant to innate immunity. Using UMAP, we identified 5 major cell types according to the expression of canonical gene markers. These cell types included CD14 + Monocyte, FCGR3A + Monocyte, Myeloid DC, Plasmacytoid DC and Neutrophil (Fig. 3 a, b).
Fig. 3.
Characterization of innate immune cells in individuals across three conditions. a, The UMAP projection of innate immune cell subsets including Monocytes, DCs and neutrophils. Other cluster of cells are labeled as other cells. Each dot corresponds to a single cell, colored according to cell type. b, The UMAP projection of monocytes, DCs and neutrophils derived from HDs (n = 4) and convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8). Each dot corresponds to a single cell, colored according to cell type. c, Dot plot depicting gene expression and percent of expression of cells of top 70 differentially expressed genes in monocytes, DCs and neutrophils derived from HDs (n = 4) and convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8). The right panel shows differential expression significance between groups. The top 10 DEGs in each cluster are identified according to the average log foldchange and summarized the top 10 DEGs of all cell clusters from HDs, convalescent patients with moderate COVID-19 and convalescent patients with severe COVID-19 and a total of 70 DEGs are obtained. The size of the dot represents the percent of expression of cells, and color represents the expression levels after log scale. Statistical analysis was performed using Wilcoxon rank sum test. The right panel depicted the differential expression significance by across tree groups, HDs, convalescent patients with moderate COVID-19 and convalescent patients with severe COVID-19. Statistical analysis was performed using unpaired two-sided t-test. d, Gene enrichment analyses of the DEGs in Fig. 3c in the Biological Process (BP) database in Gene Ontology (GO). GO terms are labeled with name and ID and sorted by − log10 (P value). The top 20 enriched GO terms are shown. e, Box plots of the cell state score using genes from two GO biological process terms (Regulation of innate immune responses and Positive regulation of cytokine production) across clusters of monocytes, DCs and neutrophils derived from HDs (n = 4) and convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8). For each GO pathway, the AddModuleScore (Seurat v3.2.0, default parameter) function was used to calculate the cell stat scores of of genes from the GO pathway in each cell. Box plots of are drawn according to the two groups of HDs, convalescent patients with moderate COVID-19 and convalescent patients with severe COVID-19, and the cell stat scores of the cells from three groups are compared. Conditions are shown in different colors. The vertical axis represents median values, with whiskers extending to the farthest data point within a maximum of 1.5 × interquartile range. All differences with *P < 0.01 are indicated. **P < 0.001; ***P < 0.0001; using unpaired two-sided t-test. Shown are adjusted P values by unpaired two-sided t-test. f, Expression pattern of DEGs relevant to specific biological functions in regulating innate immunity during convalescence of COVID-19. DEGs relevant to specific biological functions in regulating innate immunity were identified from Fig. 3c and expression pattern of these DEGs in cell subsets were shown. Up arrow was correspondence to upregulation and down arrow was correspondence to downregulation. ns represented as no significance. g. Scatter-plot showing DEGs in monocytes, DCs and neutrophils derived from HDs (n = 4) and convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8) calculated by Markers (Seurat v3.2.0, default parameters) function. Each red dot denotes an individual gene with P < 0.01 and average log2 fold change greater than 0.25. The plots with the gene symbols are the genes which are significantly differentially expressed both when convalescent patients with moderate COVID-19 (n = 5) groups compared with HDs (n = 4) groups and convalescent patients with severe COVID-19 (n = 8) groups compared with HDs (n = 4) groups. Orange dots indicate the genes which are only significantly differentially expressed when convalescent patients with severe COVID-19 (n = 8) groups compared with HDs (n = 4) groups. Green dots indicate the genes which are only significantly differentially expressed when convalescent patients with moderate COVID-19 (n = 5) groups compared with HDs (n = 4) groups.
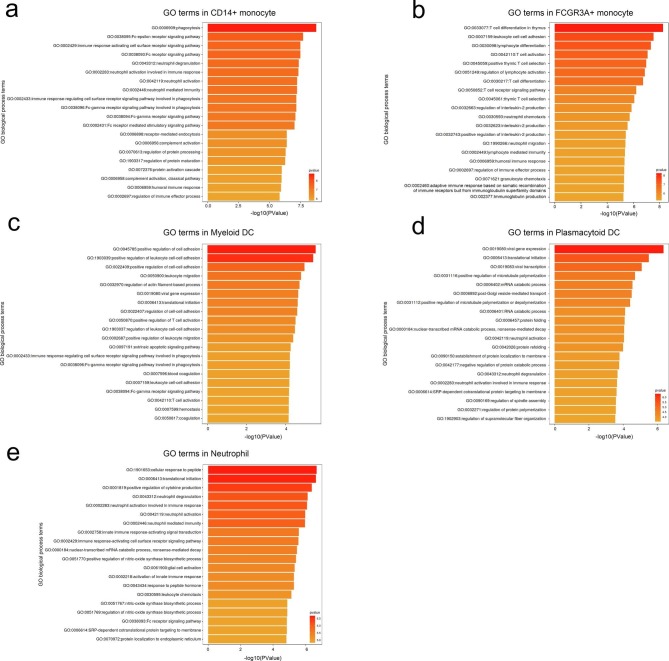
We analyzed differentially expressed genes (DEGs) from innate immune cell types like monocytes, dendritic cells and neutrophils from convalescent COVID-19 patients relative to healthy donors and used these genes to identify enriched Gene Ontology. We found that 70 genes related to innate immune responses were significantly differentially expressed across disease conditions (Fig. 3c; Supplementary Table 7 and 8). We found that significantly DEGs were involved in neutrophil response including neutrophil chemotaxis (GO: 0030593), neutrophil migration (GO: 1990266), neutrophil activation (GO: 0042119), neutrophil degranulation (GO:0043312), neutrophil activation involved in immune response (GO: 0002283) and neutrophil mediated immunity (GO: 0002446), such interferon-gamma response as response to interferon-gamma (GO:0034341), cellular response to interferon-gamma (GO:0071346) and interferon-gamma-mediated signaling pathway (GO:0060333) and such humoral response as antimicrobial humoral response (GO:0019730) and humoral immune response (GO:0006959) (Fig. 3d).
To unbias elucidate the status of innate cell subsets, which always reflected by specific mode of gene expression, from two kinds of convalescent COVID-19 patients, we deeply explored the expression pattern of DEGs involved in top 20 GO terms based on classification of their functions. We classified these genes into three categories including (1) response to interferon-gamma, (2) leukocyte migration and (3) neutrophil activation, migration and degranulation and immune response they mediated. We found substantial amounts of genes regarding response to interferon-gamma were downregulated in several innate cell subsets with the exception of some HLA class II molecules like HLA-DQA1, HLA-DQB1 and HLA-DPB1, which were upregulated in CD14 + Monocytes and FCGR3A + Monocytes from convalescent patients with COVID-19. Downregulation of these kinds of genes were interferon-stimulated genes (ISGs) e.g. IFITM1, IFITM2, IFITM3, IRF7 and HLA class II genes HLA-DRA and HLA-DRB1 in CD14 + Monocytes, FCGR3A + Monocytes, Myeloid DCs and Plasmacytoid DCs from convalescent patients with COVID-19. The gene, HMOX1, involved in leukocyte migration, was alsodecreased its’ expression in CD14 + Monocytes, FCGR3A + Monocytes and Myeloid DCs from convalescent patients with COVID-19. With the same trend, multiple genes, SERPINA1 and S100P, which regulate neutrophil activation, migration and degranulation and it mediated immune response, were downregulated in CD14 + Monocytes, FCGR3A + Monocytes, Neutrophils and Myeloid DCs and GCA and CD36 were only downregulated in Myeloid DCs and Plasmacytoid DCs from convalescent patients with COVID-19 (Fig. 3c, d, f; Supplementary Table 7 and 8).
We also found that CD14 + Monocytes, FCGR3A + Monocytes, Myeloid DCs and Plasmacytoid DCs significantly downregulated regulation of innate immune response and positive regulation of cytokine production in severe patients compared to HD group (Fig. 3e). These results suggest that innate immune responses including neutrophil response and interferon-gamma response were attenuated in convalescent COVID-19 patients. Taken together, innate cell subsets decreased expression of genes involved in interferon-gamma, leukocyte migration and neutrophil mediated immune response in convalescent COVID-19 patients.
To further characterize the functions of DEGs in innate immune cell subsets, we performed analyses CD14 + Monocyte, FCGR3A + Monocyte, Myeloid DC, Plasmacytoid DC and Neutrophil of convalescent moderate or severe patients in comparison with those of HDs. Total DEGs in 5 innate immune cell subsets from convalescent moderate or severe patients in comparison with those of HDs were shown in Supplementary Table 9 and the DEGs shared by both comparisons were presented in Fig. 3g. We found DEGs were involved in phagocytosis, neutrophil mediated immune response and immune response-activating cell surface receptor signaling pathway in CD14 + Monocyte, T cell activation, differentiation and T cell receptor signaling pathway and leukocyte cell–cell adhesion in FCGR3A + Monocyte, regulation of T and leukocyte cell–cell adhesion in Myeloid DC, viral transcription and gene expression in Plasmacytoid DC, and neutrophil mediated immune response and regulation of cytokine production in Neutrophil in convalescent COVID-19 patients (Supplementary Fig. 1).
2.4. Strengthened T cell and lymphocyte activation, differentiation and cell–cell adhesion in convalescent COVID-19 patients
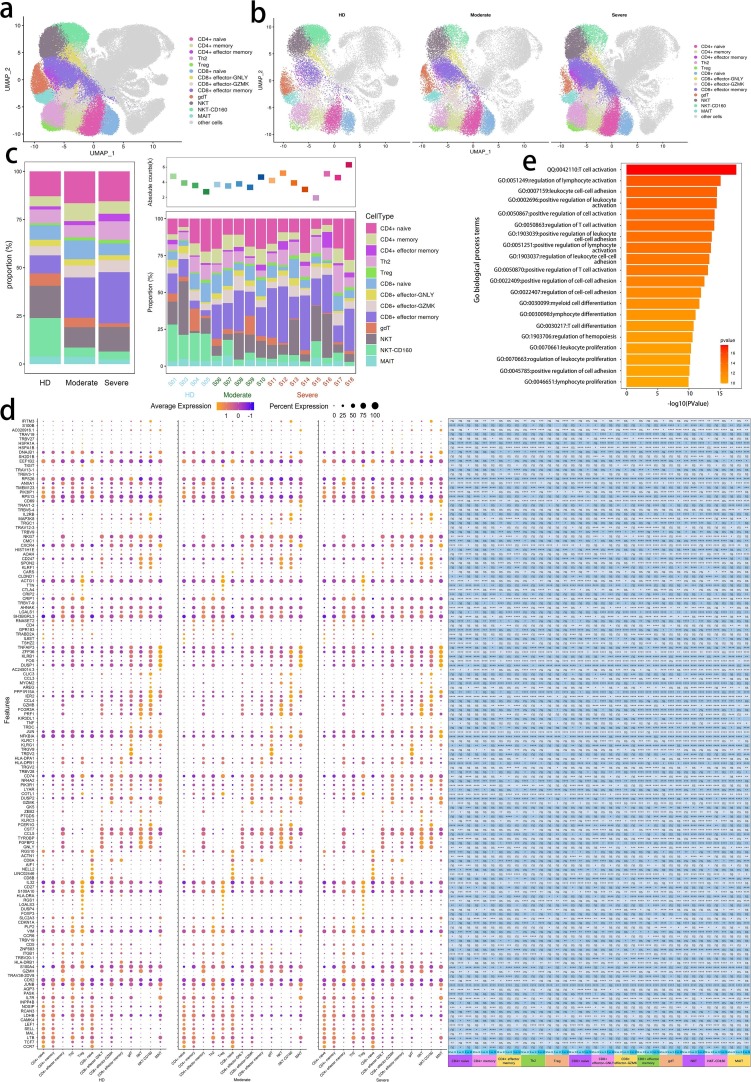
To characterize changes in individual T cell subsets after SARS-CoV-2 infection among the subjects across three conditions, we subclustered T cells from PBMCs and obtained 13 subsets according to the expression and distribution of canonical T cell markers using UMAP. These cell types included CD4 + naïve, CD4 + memory, CD4 + effector memory, Th2, Treg, CD8 + naïve, CD8 + effector-GNLY, CD8 + effector-GZMK, CD8 + effector memory, gdT, NKT, NKT-CD160 and MAIT (Fig. 4 a–c; Supplementary Fig. 2).
Fig. 4.
Characterization of T cells in individuals across three conditions a, The UMAP projection of T cells. Each dot corresponds to a single cell, colored according to cell type. The T cell subsets dimensionality reduction map includes CD4 + T, CD8 + T, gdT, NKT and MAIT, and other cluster of cells are labeled as other cells. b, The UMAP projection of T cells derived from HDs (n = 4) and convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8). Each dot corresponds to a single cell, colored according to cell type. c, Average proportion of each cell type derived from HDs (n = 4) and convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8). The left picture depicted average proportion of each T cell type derived from three groups, HDs, convalescent patients with moderate COVID-19 and convalescent patients with severe COVID-19. The calculation method is: (the number of specific cell cluster in one group) ⁄ (the number of total cells in one group). The dot plot in the upper panel of the right picture shows the sum of the absolute counts of T cell subsets in the PBMCs of each sample, and the bottom bar plot shows T cell compositions at a single sample level. The calculation method is: (the number of specific T cell cluster in one sample) ⁄ (the number of total cells in one sample). d, Dot plot depicting gene expression and percent of expression of cells of top 142 differentially expressed genes in T cells derived from HDs (n = 4) and convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8). The right panel shows differential expression significance between groups. The top 10 DEGs in each cluster are identified according to the average log foldchange and summarized the top 10 DEGs of all cell clusters from HDs, convalescent patients with moderate COVID-19 and convalescent patients with severe COVID-19 and a total of 142 DEGs are obtained. The size of the dot represents the percent of expression of cells, and color represents the expression levels after log scale. Statistical analysis was performed using Wilcoxon rank sum test. The right panel depicts the differential expression significance by across tree groups, HDs, convalescent patients with moderate COVID-19 and convalescent patients with severe COVID-19. Statistical analysis was performed using unpaired two-sided t-test. e, Gene enrichment analyses of 142 DEGs in Fig. 4d in the Biological Process (BP) database in Gene Ontology (GO). GO terms are labeled with name and ID and sorted by − log10 (P value). The top 20 enriched GO terms are shown.
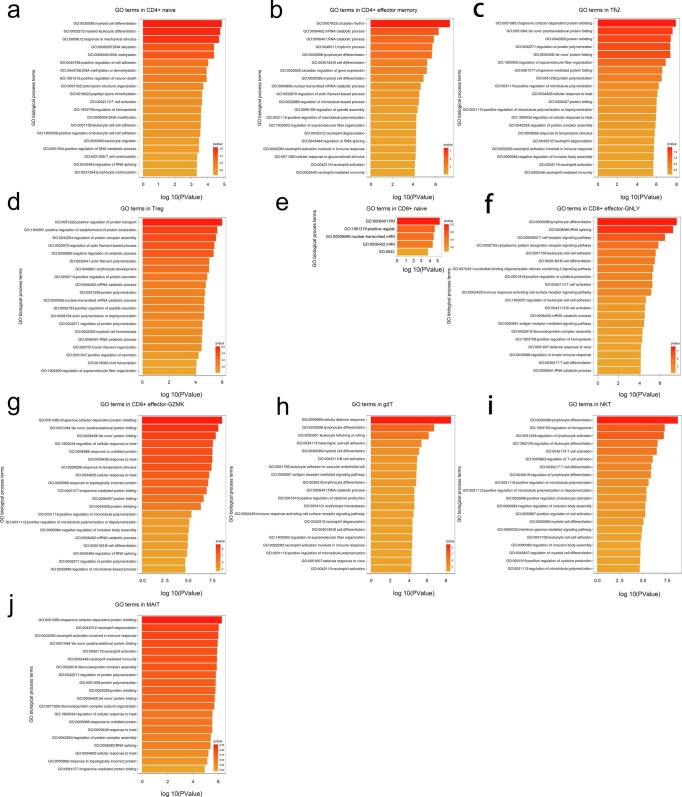
It demonstrated that 142 genes related to T cell responses were significantly differentially expressed across disease conditions (Fig. 4d; Supplementary Table 10 and 11). We found that significantly DEGs were involved in T cell and lymphocyte activation, differentiation and proliferation including T cell activation (GO:0042110), positive regulation of T cell activation (GO:0050870), regulation of T cell activation (GO:0050863), T cell differentiation (GO:0030217), regulation of lymphocyte activation (GO:0051249), positive regulation of lymphocyte activation (GO:0051251), lymphocyte differentiation (GO:0030098) and lymphocyte proliferation (GO:0046651), cell–cell adhesion such as leukocyte cell–cell adhesion (GO:0007159), positive regulation of leukocyte cell–cell adhesion (GO:1903039), regulation of leukocyte cell–cell adhesion (GO:1903037), positive regulation of cell–cell adhesion (GO:0022409), regulation of cell–cell adhesion (GO:0022407) and positive regulation of cell adhesion (GO:0045785) and myeloid cell responses including myeloid cell differentiation (GO:0030099),regulation of hemopoiesis (GO:1903706) and regulation of myeloid cell differentiation (GO:0045637) (Fig. 4e).
Same as analysis procedure of DEGs in innate cell subsets, we firstly classified DEGs in T cell subsets into four categories including (1) regulation of T cell activation, (2) regulation of lymphocyte and leukocyte activation (3) T cell, lymphocyte and myeloid cell differentiation and (4) regulation of cell–cell adhesion and then deeply explored the expression pattern of these genes involved in top 20 GO terms based on the classification. It depicted enhanced functions of activation, differentiation and adhesion of multiple T cell compartments from convalescent patients with COVID-19. Genes involved in biological processes of regulation of T cell activation and regulation of lymphocyte and leukocyte activation remarkably increased their expression from both convalescent patients with moderate and severe COVID-19. We found that CD8B were upregulated in CD8 + effector-GNLY, CD8 + effector memory, gdT and NKT and TRDC were upregulated in CD4 + effector memory, CD8 + naïve, CD8 + effector memory, NKT and NKT-CD160. SELL and S100A10 were upregulated in nearly more than 10 T cell subsets in convalescent patients with severe COVID-19. With the similar trend, genes, FOS, HSPA1A and HSPA1B, relating to biological process of T cell, lymphocyte and myeloid cell differentiation, were highly expressed in more than 10 T cell subsets in convalescent patients with severe COVID-19 while expression of Ly-1 antibody reactive clone (LYAR) was raised in CD4 + naïve, CD4 + memory, Th2, Treg, CD8 + naïve and NKT-CD160 from both kinds of convalescent patients. However, we observed a prominent feature that JUNB and ZFP36 declined in a wide range of T cell subsets from both kinds of convalescent subjects. The previous studies revealed that JUNB inhibits CD4 + T cell by promoting Th17 cell [38], [39] and ZFP36 RBPs in restraining T cell expansion and effector functions [40] (Fig. 5 a; Supplementary Table 10 and 11). Combined with the above results, we summarize that functions of T cell were retained even strengthened in convalescent COVID-19 patients by clear endorsement of increased expression of genes involved in biological processes of regulation of T cell activation, regulation of lymphocyte and leukocyte activation, T cell, lymphocyte and myeloid cell differentiation and regulation of cell–cell adhesion.
Fig. 5.
DEGs in T cells in individuals across three conditions. a, Expression pattern of DEGs relevant to specific biological functions in regulating T cell immunity during convalescence of COVID-19. DEGs relevant to specific biological functions in regulating innate immunity were identified from Fig. 4d and expression pattern of these DEGs in cell subsets were shown. Up arrow was correspondence to upregulation and down arrow was correspondence to downregulation. ns represented as no significance. b, Scatter-plot showing DEGs in T cell subsets (CD4 + Memory, CD8 + Effector memory and NKT-CD-160) derived from HDs (n = 4) and convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8) calculated by Markers (Seurat v3.2.0, default parameters) function. Each red dot denotes an individual gene with P < 0.01 and average log2 fold change greater than 0.25. The plots with the gene symbols are the genes which are significantly differentially expressed both when convalescent patients with moderate COVID-19 compared with HDs groups and convalescent patients with severe COVID-19 compared with HDs. Orange dots indicate the genes which are only significantly differentially expressed when convalescent patients with severe COVID-19 compared with HDs. Green dots indicate the genes which are only significantly differentially expressed when convalescent patients with moderate COVID-19 compared with HDs. c, Gene enrichment analyses of the DEGs in Fig. 4d in the Biological Process (BP) database in Gene Ontology (GO). GO terms are labeled with name and ID and sorted by − log10 (P value). The top 20 enriched GO terms are shown. d, Dot plot depicts percent expression and average expression of all detected HLA genes in T cells derived from HDs (n = 4) and convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8). The size of the dot represents the expression percentage of T cell clusters, and data color represents the expression levels after log scale. The right panel shows differential expression significance between the three groups with unpaired two-sided t-test.
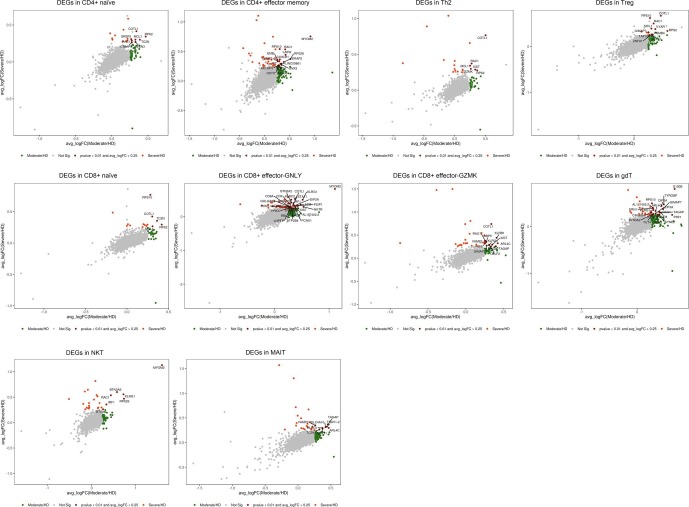
To further characterize the functions of DEGs in 13 T cell subsets, we performed analyses of T cells from moderate or severe convalescent patients in comparison with those of HDs. Total DEGs from moderate or severe convalescent patients in comparison with those of HDs were shown in Supplementary Table 12 and the DEGs shared by both comparisons were presented in Fig. 5b and Supplementary Fig. 3. We found DEGs were involved in myeloid cell and myeloid leukocyte differentiation in CD4 + naïve, regulation of microtubule polymerization and Fc receptor signaling pathway in CD4 + memory (Fig. 5c), mRNA and RNA catabolic process in CD4 + effector memory, regulation of protein transport, localization and complex assembly in Treg, RNA catabolic process in CD8 + naïve, T cell activation and T cell receptor signaling pathway in CD8 + effector-GNLY, posttranslational protein folding in Th2, CD8 + effector-GZMK and CD8 + effector memory (Fig. 5c), cellular defense response and lymphocyte differentiation in gdT, T cell activation and regulation of hemopoiesis in NKT, T cell receptor signaling pathway and antigen receptor-mediated signaling pathway in NKT-CD160 (Fig. 5c), and neutrophil activation involved in immune response in MAIT in convalescent COVID-19 patients (Fig. 5c; Supplementary Fig. 4).
Classical HLA class I genes e.g. HLA-A and HLA-B were downregulated in CD4 + naïve, CD4 + memory, CD4 + effector memory, Th2, Treg, CD8 + naïve, CD8 + effector-GNLY, CD8 + effector-GZMK, CD8 + effector memory, gdT, NKT and NKT-CD160 subsets and HLA-C was were downregulated in CD4 + memory, Th2, CD8 + naïve, CD8 + effector-GZMK, CD8 + effector memory, gdT, while Non-classical HLA class I genes e.g. HLA-E was also downregulated in CD4 + naïve, CD4 + memory, CD4 + effector memory, Th2, CD8 + naïve, CD8 + effector-GNLY, CD8 + effector memory, NKT and NKT-CD160 subsets of convalescent severe COVID-19 patients (Fig. 5d and Supplementary Table 13). However, three genes e.g. HLA-DPA1, HLA-DPB1 and HLA-DQB1 encoding HLA class II molecules were upregulated in CD4 + naïve, CD4 + memory, Th2, CD8 + naïve, CD8 + effector-GNLY, CD8 + effector-GZMK, CD8 + effector memory, NKT and NKT-CD160 subsets and other HLA class II molecules, e.g. HLA-DQA1 and HLA-DRB5 were upregulated in CD4 + effector memory, CD8 + effector-GNLY, CD8 + effector-GZMK, CD8 + effector memory, NKT and NKT-CD160 subset of convalescent severe COVID-19 patients (Fig. 5d and Supplementary Table 13).
2.5. Clonal expansion in T cells and preferred usage of V(D)J genes in convalescent COVID-19 patients.
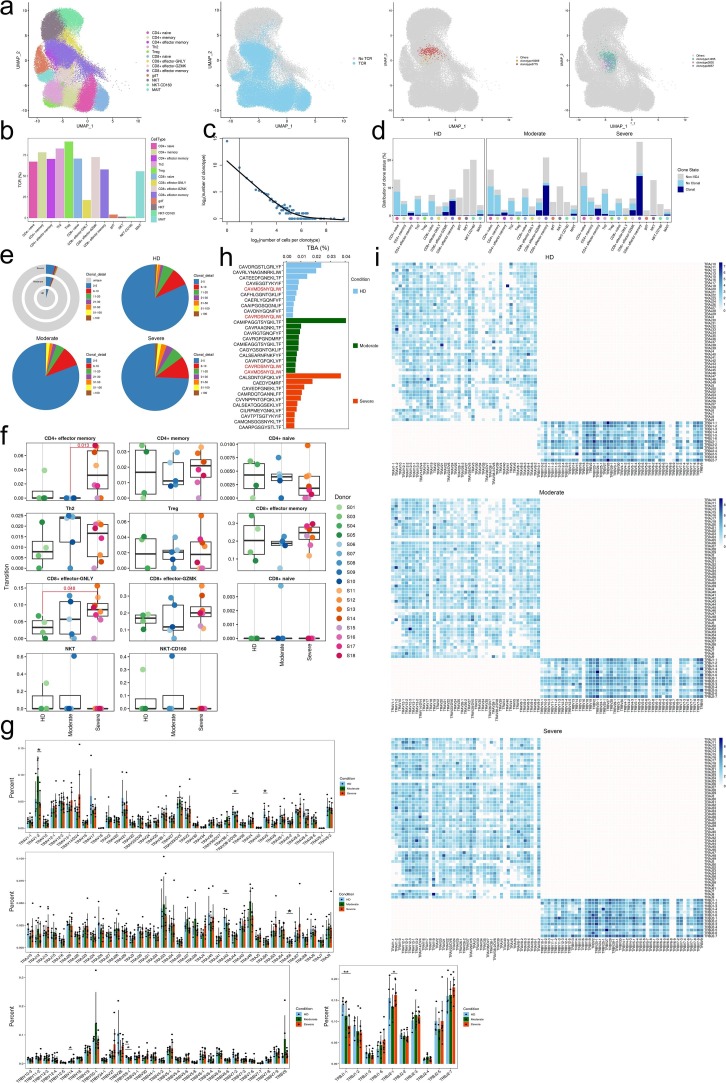
Next, to gain insight into the clonal relationship among individual T cells and usage of V(D)J genes across three conditions, we reconstructed TCR sequences from the TCR sequencing. Briefly, there were more than 75% of cells in CD4 + memory, Th2 and Treg subsets, more than 70% of cells in CD4 + naïve, CD4 + effector memory, CD8 + naïve and CD8 + effector-GNLY subsets, more than 50% of cells in CD8 + effector memory and MAIT subsets with matched TCR sequences, except for the three γδT, NKT and NKT-CD160 subsets (Fig. 6 a, b). Compared to the HDs, increased clonal expansion was found in patients with COVID-19 (Fig. 6c–e). The clonal expansion in the severe condition was higher than that of the HD and moderate conditions. Meanwhile, the clonal expansions (clonal size 50–100) were observed both in moderate and severe condition whereas clonal expansions (clonal size > 100) were only observed in severe condition (Fig. 6e), revealing that clonal expansion of effector T cells might be active in severe patients. We surveyed different degrees of clonal expansion among T cell subsets (Fig. 6c and d). CD4 + effector memory, CD8 + effector-GZMK and CD4 + effector memory showed high proportions of clonal cells both in patients increased with disease severity (Fig. 6d), suggesting that effector T cells underwent dynamic state transitions. We used STARTRAC to analyze the expansion and transition of T cell subsets. The results showed that CD4 + effector memory and CD8 + effector-GNLY increased transition in severe than those in moderate and HD respectively (Fig. 6f; Supplementary Table 14).
Fig. 6.
Expanded TCR clones and selective usage of V(D)J genes. a, The UMAP projection of T cells derived from PBMCs. Clusters are denoted by colors labeled with inferred cell types (first picture from left), TCR detection, T-cell expressing TCR with blue dots indicate cells with TCR and gray dots indicate cells without TCR (second picture from left), TCR clonotype number with copy number greater than 50 in one cell cluster (third picture from left) and TCR clonotype number with copy number greater than 50 in different cell clusters (fourth picture from left). b, Percentage of TCR cells in each T cell cluster. Bar plots showing the percentage of TCR detection in each T cell cluster. c, The association between the number of T cell clones and the number of cells per clonotype. The dashed line separates nonclonal and clonal cells. The x-axis is log2 (the number of cells per clonotype), and the y-axis is the number of cells clonotype. The dashed line at x = 1 represents the clonotype of TCR with nonclonal on the left and clonal cells on the right. The regression curve algorithm is LOESS fitting. d, The distribution of TCR clonal state of different T cell clusters of HDs (n = 4), convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8) groups. Clonotype copy number greater than 2 is clonal while clonotype copy number equal to 1 is nonclonal, and no clonotype represent non-TCR is marked as Non-VDJ. e, The clonal status percentage of T cells (top left) and percentage of different levels of clonal T cells (top right, bottom left and bottom right) across T cells derived from HDs (n = 4) and convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8). f, TCR clonal expansion and transition level was detected using Startrac.run (STARTRAC v0.1.0, default parameters) in samples among HDs (n = 4), convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8). Pairwise comparative analysis was performed. Adjusted P-values smaller than 0.05 are indicated (unpaired two-sided t-test). g, Usage of some TRAV (first), TRAJ (second), TRBV (bottom left) and TRBJ (bottom right) genes across T cells derived from HDs (n = 4) and convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8) by donor. The average percentages of the genes in each condition are showed with the height of the column. Higher column represents more gene appears in the group. Pairwise comparative analysis was performed with statistical analysis of Wilcoxon Rank Sum test. P < 0.05 was considered significant difference between two groups. Error bars represent ± s.e.m. for 4 HDs and 13 convalescent patients. h, The top ten CDR3 usages of protein sequences of TRA/TRB are shown across groups of HDs (n = 4), convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8). Each bar is colored by condition identity. Shared CDR3 sequences are in a red font. i, TRA/B rearrangement differences across three conditions of HDs (n = 4), convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8). The colors indicate the log2((usage count) + 1) of specific V-J gene pairs. Darker color represents more gene combination appear.
To study the dynamics and gene preference of TCRs in COVID-19 patients and HDs, we compared the usage of V(D)J genes across three conditions. The top 10 complementarity determining region 3 (CDR3) sequences were different across three conditions. The moderate and HD shared two CDR3 sequences. The usage percentage of the top 10 CDR3 sequences in the HD condition was lower and more balanced compared to moderate and severe conditions (Fig. 6h). Of note, we discovered a different usage of V(D)J genes with decreased diversity in patients with COVID-19, which was more pronounced in TRA genes (Fig. 6i).
We found that convalescent COVID-19 severe patients exhibit a preferred TCRVα-region bias toward TRAJ13, TRAJ23, TRAJ4, TRAJ45, TRAJ53 and TCRJ-region bias toward TRAJ57 and TRAV12-1, TRAV13-1, TRAV14-DV4, TRAV26-1, TRAV35, TRAV4 and TRAV83 gene segments, respectively (Fig. 6g and Supplementary Table 15). Meanwhile, convalescent COVID-19 severe patients shown a preferred TCRVβ-region bias toward TRBJ1-6 and TRBJ2-7 TRVB27, TRVB30, TRVB5-6 and TRVB6-1 (Fig. 6g and Supplementary Table 15).
To study biased V(D)J rearrangements of the TCR, we compared the usage of V(D)J genes across three conditions (Fig. 6g and Supplementary Table 15). We observed more specific V(D)J usage in the severe patients compared with the other two groups, indicating that T cells might have undergone unique and specific V(D)J rearrangements in severe COVID-19 patients (Fig. 6g, i). We also discovered paired genes of TRAV1-2/TRAJ33 in all HDs and COVID-19 patients (Fig. 6i). However, there were some unique V-J pairs in severe group. The shared V-J pairs e.g. TRAV27/TRAJ2-5, TRAV19/TRAJ8, TRAV19/TRAJ6, TRAV19/TRAJ57, TRAV21/TRAJ48, TRAV8-3/TRAJ43, TRAV19/TRAJ8, TRAV38-2/DV8/TRAJ40, TRAV14/DV4/TRAJ36 and TRAV12-1/TRAJ8 were only found in in severe group (Fig. 6i). In brief, increased T cell clonality and skewed usage of the TRAV and TRAJ genes in severe subjects indicate that infection with SARS-CoV-2 is closely associated with V(D)J rearrangements in T cells of the hosts. Notably, selective usage of dominant genes, especially TRAV27/TRAJ2-5, TRAV19/TRAJ8, TRAV19/TRAJ6, TRAV19/TRAJ57, TRAV21/TRAJ48, TRAV8-3/TRAJ43, TRAV19/TRAJ8, TRAV38-2/DV8/TRAJ40, TRAV14/DV4/TRAJ36 and TRAV12-1/TRAJ8 in severe patients, may facilitate the design of vaccines or therapeutics.
2.6. Dual functions of B cell subsets in convalescent COVID-19 patients
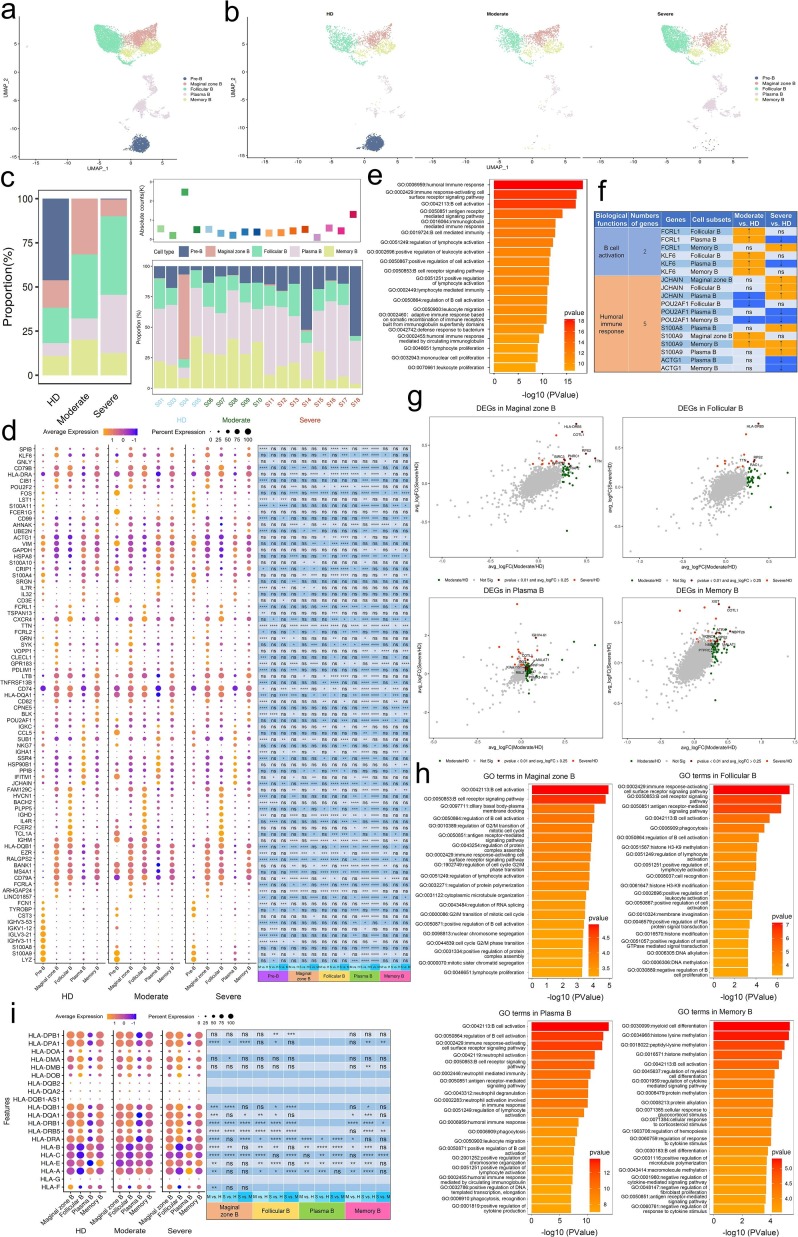
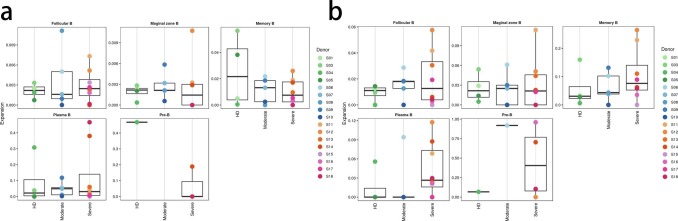
To trace the dynamic changes of different B subtypes, we subclustered B cells into five subsets according to the expression and distribution of canonical B cell markers. We identified Pre B, Marginal zone B, Follicular B, Plasma B and Memory B (Fig. 7 a and b and Supplementary Figs. 5 and 6). Notably, the proportions of plasma B and Follicular B subsets increased in convalescent severe COVID-19 patients in comparison with those of HDs. In contrast, the proportion of memory B cells decreased in convalescent severe COVID-19 patients (Fig. 7c).
Fig. 7.
Characterization of immunological features of B cell subsets in individuals across three conditions. a, The UMAP projection of B cell subsets. Each dot corresponds to a single cell, colored according to cell type. b, The UMAP projection of B cell subsets derived from HDs (n = 4) and convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8). The UMAP projection were used and each dot corresponds to a single cell. c, Bar plot showing B cell compositions at the single sample level. Average proportion of each cell type derived from HDs (n = 4) and convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8). The left picture depicted average proportion of each B cell type derived from three groups, HDs, convalescent patients with moderate COVID-19 and convalescent patients with severe COVID-19. The calculation method is: (the number of specific cell cluster in one group) ⁄ (the number of total cells in one group). The dot plot in the upper panel of the right picture shows the sum of the absolute counts of B cell subsets in the PBMCs of each sample, and the bottom bar plot shows T cell compositions at a single sample level. The calculation method is: (the number of specific B cell cluster in one sample) ⁄(the number of total cells in one sample). d, Dot plot depicted gene expression and percent of expression of cells of top 82 differentially expressed genes in B cells derived from HDs (n = 4) and convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8). The right panel shows differential expression significance between groups. The top 10 DEGs in each cluster are identified according to the average log foldchange and summarized the top 10 DEGs of all cell clusters from HDs, convalescent patients with moderate COVID-19 and convalescent patients with severe COVID-19 and a total of 82 DEGs are obtained. The size of the dot represents the percent of expression of cells, and color represents the expression levels after log scale. Statistical analysis was performed using Wilcoxon rank sum test. The right panel depicts the differential expression significance by across tree groups, HDs, convalescent patients with moderate COVID-19 and convalescent patients with severe COVID-19. Statistical analysis was performed using unpaired two-sided t-test. e, Gene enrichment analyses of 82 DEGs in Fig. 7d in the Biological Process (BP) database in Gene Ontology (GO). GO terms are labeled with name and ID and sorted by − log10 (P value). The top 20 enriched GO terms are shown. f, Expression pattern of DEGs was relevant to specific biological functions in regulating B cell immunity during convalescence of COVID-19. DEGs was relevant to specific biological functions in regulating innate immunity were identified from Fig. 7d and expression pattern of these DEGs in cell subsets were shown. Up arrow was correspondence to upregulation and down arrow was correspondence to downregulation. ns represented as no significance. g. Scatter-plot showing DEGs in B cell subsets derived from HDs (n = 4) and convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8) calculated by Markers (Seurat v3.2.0, default parameters) function. Each red dot denotes an individual gene with P < 0.01 and average log2 fold change greater than 0.25. The plots with the gene symbols are the genes which are significantly differentially expressed both when convalescent patients with moderate COVID-19 compared with HDs groups and convalescent patients with severe COVID-19 compared with HDs. Orange dots indicate the genes which are only significantly differentially expressed when convalescent patients with severe COVID-19 compared with HDs. Green dots indicate the genes which are only significantly differentially expressed when convalescent patients with moderate COVID-19 compared with HDs. h, Gene enrichment analyses of the DEGs in Fig. 7d in the Biological Process (BP) database in Gene Ontology (GO). GO terms are labeled with name and ID and sorted by − log10 (P value). The top 20 enriched GO terms are shown. i, Dot plot depicts percent expression and average expression of all detected HLA genes in B cells derived from HDs (n = 4) and convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8). The size of the dot represents the expression percentage of B cell clusters, and data color represents the expression levels after log scale. The right panel shows differential expression significance between the three groups with unpaired two-sided t-test.
To further investigate diverse transcriptomic changes in B cells post SARS-CoV-2 infection, we compared the expression profiles of B cells of the moderate or severe condition to those of the HD condition. It demonstrated that 82 genes related to B cell responses were significantly differentially expressed across disease conditions (Fig. 7d; Supplementary Table 16 and 17). We found that DEGs were involved in humoral immune response (GO:0006959), B cell activation (GO:0042113; GO:0050864) and cell surface signaling pathway including immune response-activating cell surface receptor signaling pathway (GO:0002429), antigen receptor-mediated signaling pathway (GO:0050851) and B cell receptor signaling pathway (GO:0050853) (Fig. 7e).
Humoral immunity plays an important role against viral infection. Then we further unbiasly mapped the status of B cell subtypes, using the same analysis procedures as innate and T cells, by evaluating the expression pattern of DEGs involved in top 20 GO terms based on classification of their functions. We classified these genes into two categories e.g. B cell activation and humoral immune response. Two genes, Kruppel-like factor 6 (KLF6) and Fc receptor-like 1 (FcRL1), relevant to B cell activation, were observed to be upregulated in B cells from moderate convalescent COVID-19 patients, with KLF6 increased in Follicular B and Plasma B whereas FcRL1 increased in Follicular B, Plasma B and Memory B. It is noteworthy that FcRL1 was upregulated in Memory B only from severe convalescent COVID-19 patients. A study showed that FcRL1 enhances B cell activation and function [41]. In another panel of genes relevant to humoral immune response, we found two molecules S100A8 and S100A9 upregulated in Plasma B only from severe subjects. However, two genes showed an opposite status, revealed S100A9 were upregulated and POU2AF1 were downregulated in Memory B both from moderate and severe convalescent COVID-19 patients. POU2AF1 and ACTG1 demonstrated a decreased expression in Memory B and Plasma B from moderate subjects. In contrary, JCHAIN was upregulated in Marginal zone B, Follicular B and Plasma B from severe convalescent subjects (Fig. 7d, e, f; Supplementary Table 16 and 17).
We also found that Plasma B, Follicular B, Memory B and Marginal zone B significantly downregulated B cell activation (GO: 0042113), B cell differentiation (GO: 0030183) and lymphocyte differentiation (GO: 0030098) in severe patients compared to HD group (Supplementary Fig. 6). These results suggest that B cell activation and differentiation responses including neutrophil response and interferon-gamma response were attenuated in convalescent COVID-19 patients. To further characterize the functions of DEGs in B cells, we performed analyses B cells of moderate or severe convalescent patients in comparison with those of HDs. Total 82 DEGs from moderate or severe convalescent patients in comparison with those of HDs were shown in Supplementary Table 18 and the DEGs shared by both comparisons were presented in Fig. 7g. We found DEGs were involved in neutrophil activation involved in immune response in Plasma B, regulation of mRNA processing in Pre B, B cell receptor signaling pathway in Follicular B in convalescent COVID-19 patients (Fig. 7h).
Classical HLA class I genes HLA-A and HLA-B were significantly downregulated in Marginal zone B, Follicular B, Plasma B and Memory B subsets from both moderate or severe convalescent patients and HLA-C was were significantly upregulated in Marginal zone B, Follicular B, Plasma B and Memory B subsets from moderate convalescent patients but downregulated in Marginal zone B, Plasma B and Memory B subsets from severe convalescent patients. Non-classical HLA class I genes HLA-DPA1, HLA-DPB1, HLA-DQB1, HLA-DRA and HLA-DRB1 were significantly upregulated in Marginal zone B and Memory B subsets from severe convalescent patients while HLA-DPA1, HLA-DQB1 and HLA-DRB1 were significantly downregulated in Follicular B subsets from severe convalescent patients. Furthermore, we found that HLA-DRB5 was significantly upregulated in Marginal zone B, Follicular B and Memory B subsets from both moderate or severe convalescent patients (Fig. 7i; Supplementary Table 19).
2.7. Expanded B cells and specific rearrangements of V(D)J genes in severe patients.
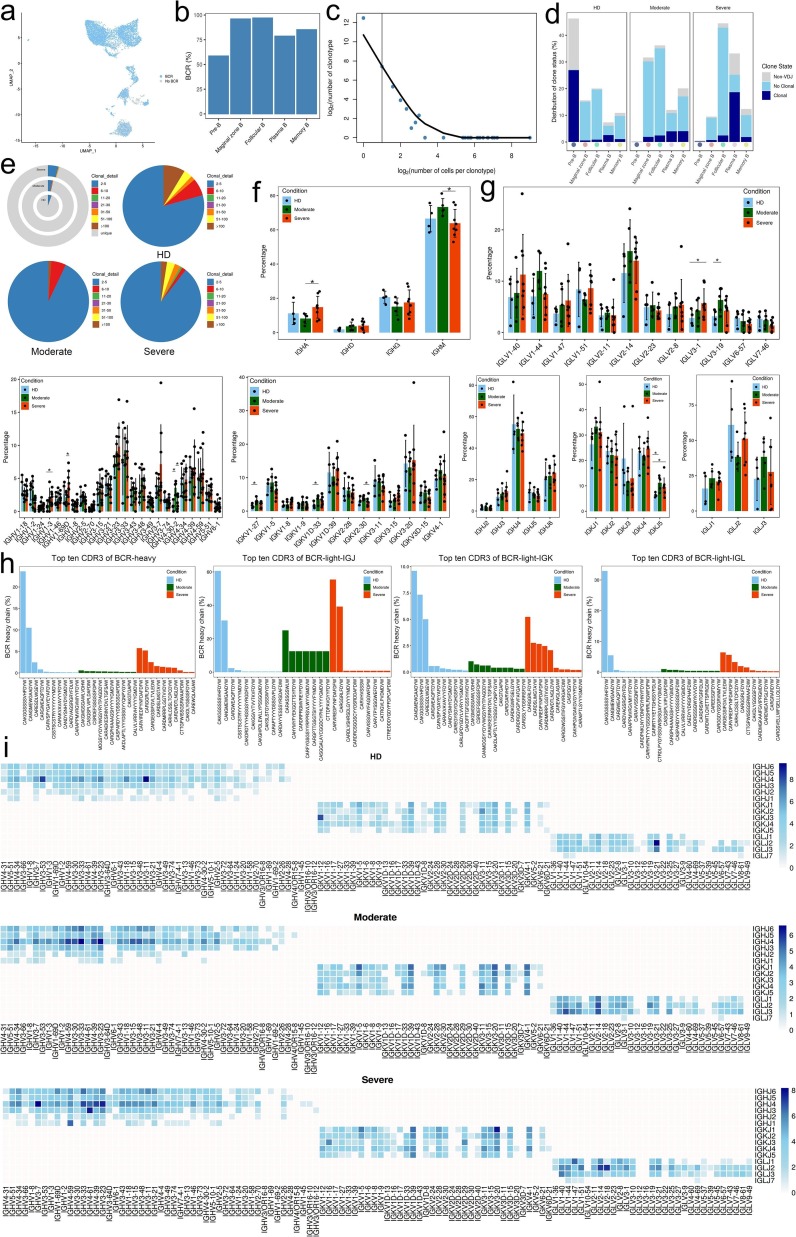
Next, to gain insight into the clonal relationship among individual B cells and usage of V(D)J genes across three conditions, we reconstructed BCR sequences from the BCR sequencing. Briefly, more than 75% of BCRs were detected in Marginal zone B, Follicular B, Plasma B and Memory B subsets (Fig. 8 a, b). Compared to the HDs, it shown an inconsistently tendency in clonal expansion between T and B cell subsets. Although the clonal expansion was obviously detected in Plasma B subsets from severe convalescent patients, in other B cell subsets e.g. Marginal zone B, Follicular B and Memory B, the clonal expansion level was not significantly increased from both moderate or severe convalescent patients, even weakened in later patients (Fig. 8c–d).
Fig. 8.
Expanded BCR clones and selective usage of V(D)J genes. a, The UMAP of B cells derived from PBMCs. The UMAP of B cells is colored based on BCR detection. B cells expressing BCR with blue dots indicate cells with BCR and gray dots indicate cells without BCR. b, Bar plot showing percentage of B cells expressing BCR detection in each B cell cluster. c, The association between the number of B cell clones and the number of cells per clonotype. The dashed line separates nonclonal and clonal cells. The x-axis is log2 (the number of cells per clonotype), and the y-axis is the number of cells clonotype. The dashed line at x = 1 represents the clonotype of BCR with nonclonal on the left and clonal cells on the right. The regression curve algorithm is LOESS fitting. d, Bar plot showing the distribution of the clone state of B cells in each cluster of HDs (n = 4), convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8) groups. Clonotype copy number greater than 2 is clonal while clonotype copy number equal to 1 is nonclonal, and no clonotype represent non-BCR is marked as Non-VDJ. e, The clonal status percentage of B cells (top left) and percentage of different levels of clonal B cells (top right, bottom left and bottom right) across B cells derived from HDs (n = 4) and convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8). f, Bar plot showing the percentages of IGHA, IGHD, IGHG and IGHM in each condition, with error bars representing ± s.e.m. for HDs (n = 4) and 13 patients, convalescent patients with moderate COVID-19 (n = 5) and convalescent patients with severe COVID-19 (n = 8). Higher column represents more gene appears in the group. All differences with P < 0.05 are indicated and were analyzed using unpaired two-sided t-test. g, Usage of some IGHV (top left), IGHJ (top right), IGKV (middle left), IGKJ (middle right), IGLV (bottom left) and IGLJ (bottom right) genes across conditions of HDs (n = 4) and convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8). The average percentages of the genes in each condition are showed with the height of the column. Conditions are shown in different colors. Error bars represent ± s.e.m. for 4 HDs and 13 patients. Higher column represents more gene appears in the group. All differences with P < 0.05 are indicated and were analyzed using unpaired two-sided t-test. Error bars represent ± s.e.m. for 4 HDs and 13 convalescent patients. h, The top ten CDR3 usages of protein sequences of heavy chain and light chain are shown across groups of HDs (n = 4), convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8). Each bar is colored by condition identity. Shared CDR3 sequences are in a red front. i, Heat maps showing IGH/K/L rearrangement differences across conditions of HDs (n = 4) and convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8). Colors indicate the log2((usage count) + 1) of specific V-J gene pairs. Darker color represents more gene combination appear.
Meanwhile, clonal expansions (clonal size greater than 100) were decreased both in moderate and severe condition, especially in moderate group (Fig. 8e), indicating that convalescent COVID-19 patients might have a faded clonal expansion of effector B cells. We also used STARTRAC to analyze the expansion and transition of B cell subsets. The results showed that there was no significant difference in expansion and transition among subjects (Supplementary Fig. 7a and b; Supplementary Table 20).
Next, we explored the distribution of IgA, IgD, IgG and IgM (IgE not detected) in each patient at moderate or severe convalescent conditions, respectively. In most patients, IgM was the predominant immunoglobulin. Compared to moderate convalescent patients, the abundance of IgA was significantly decreased, whereas IgM increased in severe ones (Fig. 8f). The top 10 CDR3 sequences were differently distributed across three conditions (Fig. 8h). To study biased V(D)J rearrangements of the BCR, we compared the usage of V(D)J genes across three conditions. We observed more specific V(D)J usage in the severe condition compared with the other two groups, indicating that B cells might have undergone unique and specific V(D)J rearrangements in severe COVID-19 patients (Fig. 8g and i). We also found comprehensive usage of IGHJ4, IGKJ1 and IGLJ2 in all HDs and patients, but the paired genes among IGHV/IGHJ4, IGKV/IGKJ1 and IGLV/IGLJ2 were different in patients compared with HDs. The shared V-J pairs IGHV3-33/IGHJ4, IGHV4-39/IGHJ4, IGHV3-23/IGHJ4, IGKV1-5/IGKJ1, IGKV1D-39/IGKJ1, IGKV3-20/IGKJ1 and IGLV2-14/IGLJ2 were detected in both moderate and severe patients whereas IGHV3-11/IGHJ4 and IGLV3-21/IGLJ2 were only observed in HDs, respectively. However, it attracted our attention that there were unique V-J pairs in severe group, such as IGHV4-59/IGHJ1, IGHV4-61/IGHJ3 and IGLV2-18/IGLJ2. similarly with T cells, increased B cell clonality and skewed usage of the IGHV and IGKJ genes in severe COVID-19 patients suggest that SARS-CoV-2 infection is closely associated with V(D)J rearrangements in B cells of the host. Notably, selective usage of dominant IGV genes, especially IGHV3-7/IGHJ4, IGHV3-33/IGHJ4, IGHV4-61/IGHJ4, IGHV4-39/IGHJ4, IGHV3-23/IGHJ4, IGLV2-14/IGLJ2, IGLV2-18/IGLJ2, IGKV1-5/IGKJ1, IGKV1D-39/IGKJ1, IGKV3-20/IGKJ1, IGHV4-59/IGHJ1 and IGHV4-61/IGHJ3 in severe patients, may facilitate the design of vaccines.
3. Discussion
Infection with SARS-CoV-2 causes a range of clinical manifestations, from asymptomatic or mild infection to severe COVID-19 that requires hospitalization [5], [42], [43]. Host immune response against acute infection with SARS-CoV-2 not only plays an antiviral role, but also simultaneous cause pathogenic injury of multiple organs and tissues, especially in the lungs of patients, which determines the disease severity, progression and outcome [35]. Currently, there are no protective therapeutics and vaccines available against SARS-CoV-2. It will be critical to determine whether individuals after several months following natural infection develop robust innate or adaptive immunity or potentially prove critical for long-term immune protection against SARS-CoV-2 reinfection [20]. There is not enough evidence show that a person is protected against SARS-CoV-2 after primary infection. It remains unclear that whether individual’s immunity to SARS-CoV-2 is long-term or temporary.
Recently, Nevada State Public Health Laboratory in the United States reported the first individual in global to have sever symptomatic reinfection with SARS-CoV-2. This patient aged 25 years old, with severe COVID-19, had the shortest interval between two diagnoses with only 48 days. During the second infection, the patient needed continuous oxygen inhalation [44]. Why are these patients infected again followed with more sever symptom, and are these cases rare or common still need to be answered. At present, the pathogenic mechanism of the secondary SARS-CoV-2 infection is still unclear.
Studies have reported the characteristics of signatures of neutralizing antibodies or T cell immune responses in COVID-19 patients or convalescent individuals, which have provide us understanding of the potential pathogenesis of SARS-CoV-2 infection and immune functions post infection [9], [23], [25], [27], [34], [35], [45], [46]. To date, however, there have been few studies to provide integrated scenario to depict SARS-CoV-2-specific innate, T and B cell immune responses and their role in convalescent stage by scRNA-seq, although cellular and molecular immune responses of virus-specific T cells have been analyzed in COVID-19 progression [34], [47], [48], [49]. To fill this knowledge gap, here we used scRNA-seq to obtain an unbiased and comprehensive visualization of the SARS-CoV-2-specific innate, T and B cell immunological responses in peripheral blood samples of healthy donors and convalescent patients with COVID-19, including moderate and severe individuals who donated blood 4 months after primary infection. We found innate immune responses are attenuated in COVID-19 convalescent severe patients based on the results that numerous genes regarding response to interferon-gamma were remarkable downregulated in several innate cellular subtypes. ISGs, the key effectors of innate immunity, such as IFITM1, IFITM2, IFITM3 and IRF7 displayed downregulation in monocytes and dendritic cells from the patients. Innate immunity is the primary line of defense against the invasion of pathogenic microorganisms. The main cells involved in innate immunity are monocytes-macrophages, dendritic cells, granulocytes, NK cells and NKT cells [50]. Some studies have shown that 12 h and 24 h after SARS-COV-2 infection, the levels of monocytes and NK cells increased significantly, while granulocytes decreased significantly, which further confirmed that coronavirus infection caused a strong immune response [51]. Type I interferon system is an important part of innate immunity. One previous study demonstrated that SARS-CoV-2 robustly triggered expression of numerous IFN-stimulated genes (ISGs) [52]. Zhang et al. found that IFN and acute inflammatory responses were significantly upregulated in both monocytes and NK cells after SARS-CoV-2 infection, especially in severe patients [35]. Researchers also found that SARS-CoV-2 induces overt but delayed IFN responses and this virus perturbs host innate immune response via both its structural and nonstructural proteins [53]. Interferon-induced transmembrane proteins (IFITM1,2,3) are a family of intracellular antiviral proteins that can inhibit the replication of many viruses [54]. Recent studies have shown that IFITM1, IFITM2 and IFITM3 can inhibit SARS-COV-2 virus infection, but it is interesting that IFITM3 can not only inhibit virus endocytosis, but also enhance virus fusion on the cell membrane and enhance virus infection [55]. We found that the expression of IFITM family was downregulated during the recovery period in patients with moderate and severe recovery compared with the healthy group. Interferon regulatory factor 7 (IRF7) is a member of the IRF family, which can mediate the production of type I interferon. Transcriptional analysis of human alveolar adenocarcinoma cells infected with SARS-COV-2 showed that IRF7 was highly expressed and positively correlated with the body's response to the virus [56]. Our study found that the expression of IRF7 was down-regulated in moderate and severe convalescent patients, and the above results suggested that the interferon response was down-regulated in convalescent patients. Severe patients with SARS-COV-2 infection are often accompanied by neutropenia. On the one hand, neutrophils clear part of SARS-COV-2 virus by phagocytosis and degranulation, on the other hand, persistent infiltration of neutrophils can enhance immune response and further release more chemokines and cytokines [1]. A large number of neutrophil infiltrating was also found in the lungs of dead patients with SARS-COV-2 infection [57]. Our study found that neutrophil-related genes such as CD36, GCA, S100P, SERPINA1 were significantly down-regulated in patients with moderate to severe recovery. The expression of HOXM1, a gene related to leukocyte migration, is also significantly downregulated. Our results show that the innate immunity of patients with moderate and severe recovery from SARS-C0V-2 infection is suppressed.
T cells and B cells play critical roles in clearing viral infections, and immunological T and B cell memory generated after infection is acting fundamental role in protecting the host from severe disease upon re-exposure [45]. Cellular immunity mainly refers to T cell-mediated immunity, which is the most effective defense method for intracellular clearance of pathogenic microorganisms [58]. Our results showed that T cell mediated immune responses are retained even strengthened during COVID-19 convalescence. The substantive evidence were genes involved in regulation of T cell activation and regulation of lymphocyte and leukocyte activation remarkably increased their expression from both convalescent patients with moderate and severe COVID-19. We found that CD8B, encoding surface glycoprotein on T cells, were upregulated in CD8 + effector-GNLY, CD8 + effector memory, gdT and NKT. Previous studies revealed that CD8β depletion can induce virus reactivation in simian immunodeficiency virus-infected rhesus macaques which provided compelling evidence for direct antiviral role of CD8β + T cells [59]. In our study, we found that CD8B genes regulating T cell activation were significantly upregulated in moderate and severe convalescent patients. It has aroused our great concern that both key regulators JUNB, an activator protein-1 (AP-1) factor, and ZFP36, an RNA-binding protein, declined in a wide range of T cell subsets from both kinds of convalescent subjects. JUNB can regulate the homeostasis of Treg cells. Tregs lacking JUNB from mice can produce abnormal activation of T helper cells, which eventually leads to multiple organ autoimmunity [60]. It is necessary for JUNB to exert its effect on T helper 17 (Th17) cells in chronically infected mice [38]. The previous study revealed that JUNB inhibits CD4 + T cell by promoting Th17 cell, which contributes to both immunity and autoimmunity [38]. The authors shown that JunB acted as an obligatory regulator of Th17 cell stability that promotes the development of inflammatory Th17 cells and restricts flexibility towards alternative effector and regulatory programs. ZFP36 plays an important role in regulating immune response. One study revealed that depletion of ZFP36 in vivo accelerated T cell responses to acute viral infection and enhanced anti-viral immunity [40]. Our study found that ZFP36 was significantly downregulated in moderate and severe recovery patients, further revealing that T cell function was activated in convalescent patients. In patients with chronic hepatitis C and non-alcoholic steatohepatitis, the expression of HSAP1A is upregulated and positively correlated with the severity of hepatitis. Transgenic mice overexpressing HSPA1A showed a strong ability to protect against liver injury, indicating that HSPA1A can reduce hepatocyte death [61]. The expression of HSPA1B is downregulated in stroke patients, and mice that knock out HSPA1B can develop thrombus earlier, indicating that HSPA1B can be used as a target for stroke prevention [62]. Our study shows that in most patients with moderate and severe recovery, the expression of HSPA1A is up-regulated, suggesting that the body shows an antiviral response to SARS-COV-2 infection. SELL and S100A10 are cell adhesion-related molecules. SELL is significantly upregulated in patients with type I diabetes [63]. Mice that knock out S100A10 are more sensitive to E. coli-induced sepsis and endotoxin-induced lethal shock [64]. Our study revealed that SELL and S100A10 were significantly overexpressed in moderately and severely recovered patients. In this study, we also took advantage of an analytical framework named as STARTRAC [65] to quantitatively characterize the dynamic properties of various T cell subsets and found CD4 + effector memory and CD8 + effector-GNLY increased transition in severe than those in moderate and HD respectively. We summarize that functions of T cell were retained even strengthened in convalescent COVID-19 patients by clear endorsement of increased expression of genes involved in regulation of T cell activation, regulation of lymphocyte and leukocyte activation, T cell, lymphocyte and myeloid cell differentiation and regulation of cell–cell adhesion or of decreased of genes in restricting T cell responses. Studies also found that SARS-CoV-2 specific T cells displayed a highly activated cytotoxic phenotype during the acute phase of COVID-19 and displayed polyfunctional T cells responses during the convalescent-phase. One study demonstrated the cytotoxicity and exhaustion scores of different effector state T cell subsets across four conditions, including moderate, severe and convalescence and heatthy donors. The authors found that highly cytotoxic effector T cell subsets, such as CD4 + effector-GNLY (granulysin), CD8 + effector-GNLY and NKT CD160, was associated with convalescence in moderate patients [35]. Meanwhile, the CD4 + effector-GZMK, CD8 + effector-GZMK and NKT CD160 clusters showed higher exhaustion scores than those of the other subsets. Within these highly exhausted subsets, heatthy donors all had the lowest exhaustion scores, whereas severe patients showed the most exhausted state [35]. Besides, SARS-CoV-2-specific T cells were detectable in antibody-seronegative individuals [27]. In addition, other researchers also reported a decrease levels of lymphocytes and peripheral T cells in COVID-19 patients [1], [66], [67]. Relevant research results show that T cells will be attracted from the blood and enter infected area to control the infection. And the increased exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severity of the COVID-19 patients [68]. Although the patient’s response will be impaired, COVID-19 recovered patients developed coronavirus-specific memory T cells.
Humoral immunity mainly refers to the mechanism by which effector B cells produce antibodies to protect the body [69]. Patients recovering from SARS-COV-2 infection still show high titers of IgG antibodies2 weeks after discharge [26]. Ourprevious study reported that the median duration of IgM and IgA antibody detection was 5 (IQR, 3–6) days, while IgG was detected 14 (IQR, 10–18) days after symptom onset [70]. However, another study showed that within 6 to 10 weeks of SARS-COV-2 infection, the level of (RBD)-specific IgG specific to the receptor domain decreased [71], indicating that B lymphocytes have dual functions during the recovery phase. A recent study reported key roles of Fc receptor-like 1 (FCRL1), a newly identified BCR co-receptor specifically expressed on the surface of B cells, in antibody production unveiled by experiment showing that FcRL1-deficient mice exhibited markedly impaired formation of extrafollicular plasmablasts and germinal centers, along with decreased antibody production upon antigen stimulation [41]. In our study, we found that the expression of FCRL1, a gene related to B cell activation, was significantly increased in patients with moderate recovery, suggesting that B cell function was activated. S100A8 and S100A9 are a large number of calcium-binding proteins in the cytoplasm of neutrophils, often forming S100A8/A9 heterodimers, called calcitonin [72]. The level of S100A8/A9 in severe patients with SARS-COV-2 infection is significantly higher than that in mild or healthy controls [73]. Our study found that S100A8 and S100A9 was highly expressed in patients during the recovery period. Researchers reported that severe SARS-CoV-2 infections blunt the germinal center response, which is likely to dampen the generation of long-lived antibody responses [74]. The defective Bcl-6 + T follicular helper cells (TFH) differentiation is the reason for the failure establishment of germinal center reaction [74]. This is the common characteristic of related coronaviruses causing severe infection in humans such as SARS and MERS. The above results suggest that B cell immune responses display complicated and dual functions in convalescent severe patients. As we all know, SARS-COV-2 virus shows more antigenic epitope variability [75], vaccine doesn’t solve all the problems, but T cells are activated in both moderate and severe patients and convalescent patients, which is a new direction of treatment. T cells and B cells are significantly reduced in the lung tissues of patients infected with COVID-19, and TFH and germinal centers almost cannot be found in the lymph nodes of these patients. This is believed to be related to the low levels of IgM and IgG in the dead [76]. In addition, 10% of COVID-19 infected patients have not detected antibodies, and medical staff generally lack antibodies, suggesting that after SARS-CoV-2 infection, it is unlikely to produce durable protective antibodies against the virus [77]. We actually found no patients were seropositive on the days performed SARS-CoV-2 RNA testing (Supplementary Table 2).
We have not deciphered BCR/TCR responses in COVID-19 thus far. Therefore, a more profound insight into the nature of beneficial versus detrimental BCR/TCR responses is critically needed. On the one hand, this will facilitate better prognostication in patients with risk factors and improve the monitoring of immunity to SARS-CoV-2 in recovered individuals. On the other hand, this knowledge may be used to enhance treatment (e.g., by cloning therapeutic monoclonal antibodies derived from immune repertoires of recovered patients) and, most important, to determine the optimal T cell engagement strategies for vaccines [9]. A previous study showed that individuals with COVID-19 or a history of COVID-19 developed antibodies to SARS-CoV-2, including patients with major disturbances in B cell/Ig levels at active disease, including high IgA and normal or low IgG and IgM amounts as well as B lymphocytosis [9]. Two VH genes VH3-38 and VH3-53 were concentrated and enriched in severe patients compared with mild patients [78]. But in our results, we found that there were unique V-J pairs in severe group, such as IGHV4-59/IGHJ1, IGHV4-61/IGHJ3 and IGLV2-18/IGLJ2. For recovery patients, the ratio of (IgA + IgG + IgE) to (IgD + IgM) increased in early recovery patients, but decreased with the extension of recovery time, indicating that B cell activity decreased in convalescent patients. This is consistent with our research. Our study shows that IgA decreased significantly and IgM was significantly higher in severe recovery patients than in moderate recovery patients, and B cells may undergo a unique and specific V (D) J rearrangement in severe patients. Our research provides a direction for the development of vaccines and the preparation of antibodies. In early recovery patients, the expansion of T cells was significantly higher than that in healthy controls, and the most significant clone was TRAV8-6-TRAJ45:TRAV7-8-TRBJ2-1. In addition, the cloned CD8 + CTL also had extensive antiviral effects. These results showed that the clonal expansion of T cells in convalescent patients was helpful to eliminate viral infection [79]. Our study found that T cell clones increased significantly both in moderately and severely recovered patients, and CD4 + effector memory, CD8 + effector GZMK and CD8 + effector memory all increased with the aggravation of the disease. We also found that V (D) J rearrangement was more specific in severe cases. In short, T cell clones increased in convalescent patients, which is very important to resist virus re-invasion.
One of the major limitations of this dataset is the paucity of available blood samples from a small cohort of convalescent patients with COVID-19. This material is difficult to obtain since patients with early disease. Another limitation of this study is lacking study on time course sampling.
In conclusion, we systematically and unbiasly mapped the phenotypic landscape of SARS-CoV-2-specific immune responses in fresh peripheral blood samples of 4 healthy donors and 13 convalescent patients with COVID-19 from Hubei province of China, including moderate and severe cases, by single-cell RNA sequencing. Our results highlighted that innate immunity was attenuated but T cell responses were retained even strengthened while B cell with a dual-function, together with the findings of TCR/BCR dataset, which provided insights into evaluation of possibility of reinfection of exposed individuals with COVID-19 and facilitation to design of effective therapeutics and vaccines.
4. Materials and methods
4.1. Patient cohort and clinical characteristics
Thirteen patients with COVID-19 were admitted at the Huanggang Hospital of Traditional Chinese Medicine, Hubei Province, P. R. China and enrolled in the study from 10 March to 15 May 2020. The categorization of three clinical groups was based on Guidelines for Diagnosis and Treatment of Corona Virus Disease 2019 issued by the National Health Commission of China (7th edition). The moderate group included non-pneumonia and mild pneumonia cases. The severe group included severe patients who met one of the following criteria: (1) Respiratory distress, respiratory rate ≥ 30 breathsmin − 1; (2) Pulse oxygen saturation (SpO2) ≤ 93% without inhalation of oxygen support at quiet resting state; (3) Arterial partial pressure of oxygen (PaO2)/oxygen concentration (FiO2) ≤ 300 mmHg; (4) Computed tomography (CT) image shows there is more than 50% increase of lung infltrating change within 24 to 48 h. The convalescent patients met the discharge criteria as follows: afebrile for more than 3d, resolution of respiratory symptoms, substantial improvement of chest CT images and two consecutive negative RT-qPCR tests for viral RNA in respiratory tract swab samples obtained at least 24 h apart. Four critical-illed patients who were grouped in the severe condition in this study required mechanical ventilation and exhibited respiratory failure, septic shock, and/or multiple organ dysfunction/ failure that required monitoring and treatment in the intensive care unit (ICU). This study strictly follows the principles according to the Declaration of Helsinki, with written informed consents obtained from all participants before sample collection according to regular principles. Ethical approvals were gained from the Ethics Committees of Huanggang Hospital of Traditional Chinese Medicine, Province, China and written informed consents or telephone call permissions were obtained from each patient or their guardians. The clinical data and disease course of the 13 patients are shown in Table 1 and Supplementary Table 1, respectively.
4.2. Sample collection
The peripheral blood mononuclear cells (PBMCs) were isolated using standard density gradient centrifugation and then used for 10x single-cell RNA-seq. Peripheral venous blood sample were obtained on 15th May 2020 of 13 patients with COVID-19 and 5 HDs. The samples were placed into vacutainer tubes and centrifuged at 400g for 5 min at 4 °C. Plasma samples were collected and stored at − 80 °C until use. For each sample, cell viability exceeded 90%.
4.3. Single cell RNA library preparation and sequencing
Using a Single Cell 5′ Library and Gel Bead kit (10X Genomics, 1000006) and Chromium Single Cell A Chip kit (10X Genomics, 120236), the cell suspension (300–600 living cells per ml as determined by Count Star) was loaded onto a Chromium single cell controller (10X Genomics) to generate single-cell gel beads in the emulsion (GEMs) according to the manufacturer’s protocol. Briefly, single cells were suspended in PBS containing 0.04% BSA. Approximately 10,000 cells were added to each channel and approximately 5,000 target cells were recovered. Captured cells were lysed and the released RNA was barcoded through reverse transcription in individual GEMs. Reverse transcription was performed on a S1000TM Touch Thermal Cycler (Bio Rad) at 53 °C for 45 min, followed by 85 °C for 5 min and a hold at 4 °C. Complementary DNA was generated and amplified, after which, quality was assessed using an Agilent 4200 (performed by CapitalBio Technology). According to the manufacturer’s introduction, scRNA-seq libraries were constructed using a Single Cell 5′ Library and Gel Bead kit, Single Cell V(D)J Enrichment kit, Human T Cell (1000005) and a Single Cell V(D)J Enrichment kit, Human B Cell (1000016). The libraries were sequenced using an Illumina Novaseq6000 sequencer with a paired-end 150-bp (PE150) reading strategy (performed by CapitalBio Technology, China).
4.4. Single-cell RNA-seq data processing
Single-cell sequencing data were aligned and quantified using CellRanger (Version 3.1.0) against the GRCh38 human reference genome downloaded from 10x Genomics official website. Preliminary counts were then used for downstream analysis. Quality control was applied to cells based on three metrics step by step: the total UMI counts, number of detected genes and proportion of mitochondrial gene counts per cell. Specifically, cells with less 200 detected genes and more than 10% mitochondrail gene counts were filtered. After quality control, a total of 117,102 cells were remained. We normalized the UMI counts with the NormalizeData() function implemented in the R package Seurat Version3.2.0. Then the logarithmic normalized counts were used for the downstream analysis.
4.5. Cell type annotation and cluster markers identification
To detect the most variable genes used for harmony algorithm, we performed variable gene selection separately for each sample. A consensus list of 2,000 variable genes was then formed by selecting the genes with the greatest recovery rates across samples, with ties broken by random sampling. Next, we calculate a PCA matrix with 20 components using such informative genes and then feed this PCA matrix into RunPCA() function implemented in R package Seurat. The resulting matrix was used to build nearest neighbor graph using FindNeighbors() function implemented in R package Seurat. Such nearest neighbor graph was then used to find clusters by Louvain algorithm [80] using FindClusters() function implemented in R package Seurat. The cluster-specific marker genes were identified using the FindAllMarkers() function implemented in R package Seurat.
The first round of clustering (resolution = 2) identified 23 major cell types including CD14 + Monocyte cells, FCGR3A + Monocyte cells, Myeloid DC cells, Plasmacytoid DC cells, Basophil cells, HSPC, Neutrophil cells, Proliferating cells, Platelet, CD4 + naive cells, CD4 + memory cells, CD4 + effector memory cells, Th2 cells, Treg cells, CD8 + naive cells, CD8 + effector-GNLY cells, CD8 + effector-GZMK cells, CD8 + effector memory cells, gdT cells, NKT cells, NKT-CD160 cells, MAIT cells and B cells. To identify clusters within B major cell type, we performed a second round of clustering on B cells. The procedure of the second round of clustering is the same as first round and we identified 5B sub cell types including Plasma B cells, Pre-B cells, Memory B cells, Follicular B cells and Marginal zone B cells. Thus, we identified 27 cell typs in total.
4.6. TCR and BCR analysis
TCR/BCR sequences were assembled and quantified following Cell Ranger (Version 3.1.0) vdj protocol against GRCh38 reference genome. Assembled contigs labeled as low-confidence, non-productive were discarded. To identify TCR clonotype for each T cell, only cells with at least one TCR α-chain (TRA) and one TCR β-chain (TRB) were remained. For a given T cell, if there are two or more α or β chains assembled, the highest expression level (UMI or reads) α or β chains was regarded as the dominated α or β chain in the cell. Each unique dominated α-β pair (CDR3 nucleotide sequences and rearranged VDJ genes included) was defined as a clonotype. T cells with exactly the same clonotype constituted a T cell clone. BCR clonotypes were identified similar to TCR. Only cells with at least one heavy chain (IGH) and one light chain (IGL or IGK) were kept. For a given B cell, if there are two or more IGH or IGL/IGK assembled, the highest expression level (UMI or reads) IGH or IGL/IGK was defined as the dominated IGH or IGL/IGK in the cell. Each unique dominated IGH-IGL/IGK pair (CDR3 nucleotide sequences and rearranged VDJ genes) was defined as a clonotype. B cells with exactly the same clonotype constituted a B cell clone. 34,242 T cells with TCR information and 8,087B cells with BCR information were used to perform the STARTRAC analysis as previously described [65]. STARTRAC-expa was used to quantified the potential clonal expansion level. TCR/BCR diversity was calculated as Shannon’s entropy shown below:
The pi represents the frequency of a given TCR/BCR clone among all T/B cells with TCR/BCR identified.
4.7. T-test Analysis
To further assess how different patients’ phenotypes and their potential interactions influence cell type proportions, we performed unpaired two-sided T-test on cell type proportions and on diversity of BCR/TCR based on different patient phenotypes. Interactions between variables were regarded as significantly associated with cell type proportions when FDR < 0.05.
4.8. Differential expression and GO term enrichment analysis
To investigate the impact of virus infection on cells, we identify differential expressed genes by performing two-sided unpaired Wilcoxon tests on all the expressed genes (expressed in at least 10% cells in either group of cells). Differential highly expressed genes of each group were shown in the scatter plots. Based on these genes, enriched GO terms were then acquired for each group of cells using R package clusterProfiler [81].
4.9. Functional module analysis
We used cell scores to evaluate the degree to which individual cells expressed a certain predefined expression functional gene set. The cell scores were initially based on the average expression of the genes from the predefined gene set in the respective cell. For a given cell i and a gene set j [28], the cell score SCj(i) quantifying the relative expression of Gj in cell i as the average relative expression (Er) of the genes in Gj compared to the average relative expression of a control gene set (Gjcont): SCj(i) = average(Er(Gj,i)) - average (Er(Gjcount,i)). The control gene set was randomly selected based on aggregate expression levels bins, which yield a comparable distribution of expression levels and over size to that of the considered gene set. The AddModuleScore() function in Seurat was used to implement the method with default settings.
4.10. HLA family gene expression analysis
The mean normalized expression levels of genes of each sample were used to represent their relative expression levels among the different cell clusters.
4.11. Statistics
The statistical tools, methods and threshold for each analysis are explicitly described with the results or detailed in the Figure Legends or Methods sections.
Ethics approval and consent to participate
Ethical approvals were gained from the Ethics Committees of Huanggang Hospital of Traditional Chinese Medicine, Province, China and written informed consents or telephone call permissions were obtained from each patient or their guardians.
Consent for publication
The manuscript was approved by all authors for publication. I would like to declare on behalf of my co-authors that the work described was original research that has not been published previously, and not under consideration for publication elsewhere, in whole or in part. All the authors listed have approved the manuscript that is enclosed.
Availability of data and materials
The datasets used or analysed during the current study are available from the corresponding author on reasonable request.
Funding
This work was supported by Natural Science Foundation of Guangdong Province, China (Grant No.: 2021A1515012429 to Dr. Pingsen Zhao) and Research Project for Outstanding Scholar of Yuebei People's Hospital (RS202001 to Dr. Pingsen Zhao)
CRediT authorship contribution statement
Pingsen Zhao: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing-original draft, Writing-review & editing. Jiahua Zou: Investigation, Resources, Methodology, Software. Fan Zhou: Data curation, Investigation, Methodology, Software. Yanyan Zhu: Data curation, Investigation, Methodology, Software. Qibin Song: Investigation, Resources, Methodology, Software. Dongdong Yu: Investigation, Resources, Methodology, Software. Xiangpan Li: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Writing-original draft, Writing-review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Analytical BioSciences for their help in analyzing this dataset. We thank CapitalBio Technology Inc. for their help in performing 10X Genomics scRNA-seq.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.intimp.2022.108767.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
Supplementary figure 1.
GO terms of DEGs in monocytes, DCs and neutrophils in individuals across three conditions. a-e, DEGs with Benjamini–Hochberg adjusted P value (two-sided unpaired Mann–Whitney U -test) ≤0.01 and average log2 fold change ≥0.25 in the moderate/HD and severe/HD comparisons in CD14+ Monocytes, FCGR3A+ Monocytes, Myeloid DCs, Plasmacytoid DCs and Neutrophils in individuals across three conditions.
Supplementary figure 2.
Violin plots showing the expression distribution of selected canonical T cell markers in the 13 clusters. The rows represented clusters and the columns represent selected marker genes.
Supplementary figure 3.
GO terms of DEGs in T cell subsets in individuals across three conditions. a-j, DEGs with Benjamini–Hochberg adjusted P value (two-sided unpaired Mann–Whitney U -test) ≤0.01 and average log2 fold change ≥0.25 in the moderate/HD and severe/HD comparisons in CD4+ naive, CD4+ memory, CD4+ effector memory, Th2, Treg, CD8+ naive, CD8+ effector-GNLY, CD8+ effector-GZMK, CD8+ effector memory, gdT, NKT, NKT-CD160 and MAIT in individuals across three conditions.
Supplementary figure 4.
GO terms of DEGs in T cells in individuals across three conditions. GO terms are labeled with name and ID and sorted by −log10 (P value). The top 20 enriched GO terms are shown.
Supplementary figure 5.
Violin plots showing the expression distribution of selected canonical B cell markers in the 5 clusters. The rows represented clusters and the columns represent selected marker genes.
Supplementary figure 6.
BCR clonal expansion and transition level was detected using Startrac.run (STARTRAC v0.1.0, default parameters) in samples among HDs (n = 4), convalescent patients with moderate COVID-19 (n = 5), convalescent patients with severe COVID-19 (n = 8). Pairwise comparative analysis was performed. Adjusted P-values smaller than 0.05 are indicated (unpaired two-sided t-test).
Supplementary figure 7.
The laboratory findings of enrolled convalescent patients with moderate COVID-19.
Detailed timeline of nucleic acid testing results for throat samples along with the IgM and IgG responses.
Counts of cells were used for identification of cell types.
Summary of Cell subtyping characterization after quality control of 117,102 cells.
Datasets of 47 major cell types identified.
Immune response-activating cell surface receptor signaling pathway (GO:0002429).
Statistical analysis of 70 DEGs in 5 innate immune cell subsets.
Average expression of 70 DEGs in 5 innate immune cell subsets.
Datasets of total DEGs in 5 innate immune cell subsets from convalescent moderate or severe patients in comparison with those of HDs.
Statistical analysis of 142 DEGs in 13 T cell subsets.
Average expression of 142 DEGs in 13 T cell subsets.
Datasets of total DEGs in 13 T cell subsets from convalescent moderate or severe patients in comparison with those of HDs.
Datasets of average expression of HLA genes in 13 T cell subsets from convalescent moderate or severe patients in comparison with those of HDs.
Datasets of expansion and transition of T cell subsets analyzed by STARTRAC.
Statistical analysis of usage of TRAV, TRAJ, TRBV and TRBJ genes across tree conditions.
Statistical analysis of 82 DEGs in 5 B cell subsets.
Average expression of 82 DEGs in 5 B cell subsets.
Datasets of total DEGs in 5 B cell subsets from convalescent moderate or severe patients in comparison with those of HDs.
Datasets of average expression of HLA genes in 4 B cell subsets from convalescent moderate or severe patients in comparison with those of HDs.
Datasets of expansion and transition of B cell subsets analyzed by STARTRAC.
References
- 1.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., Du B., Li L.J., Zeng G., Yuen K.Y., Chen R.C., Tang C.l., Wang T., Chen P.Y., Xiang J., Li S.Y., Wang J.L., Liang Z.J., Peng Y.X., Wei L., Liu Y., Hu Y.-H., Peng P., Wang J.M., Liu J.Y., Chen Z., Li G., Zheng Z.J., Qiu S.Q., Luo J., Ye C.J., Zhu S.Y., Zhong N.S. Clinical characteristics of Coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 3.Cao X. COVID-19: immunopathology and its implications for therapy. Nat. Rev. Immunol. 2020;20(5):269–270. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang D., Hu B.o., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in wuhan, china. JAMA. 2020;323(11):1061. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y.i., Zhang L.i., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L.i., Xie J., Wang G., Jiang R., Gao Z., Jin Q.i., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet (London, England) 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in china: summary of a report of 72 314 cases from the chinese center for disease control and prevention. JAMA. 2020;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 7.McKechnie J.L., Blish C.A. The innate immune system: fighting on the front lines or fanning the flames of COVID-19? Cell Host Microbe. 2020;27(6):863–869. doi: 10.1016/j.chom.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puelles V.G., Lütgehetmann M., Lindenmeyer M.T., Sperhake J.P., Wong M.N., Allweiss L., Chilla S., Heinemann A., Wanner N., Liu S., Braun F., Lu S., Pfefferle S., Schröder A.S., Edler C., Gross O., Glatzel M., Wichmann D., Wiech T., Kluge S., Pueschel K., Aepfelbacher M., Huber T.B. Multiorgan and renal tropism of SARS-CoV-2. N. Engl. J. Med. 2020;383(6):590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schultheiß C., Paschold L., Simnica D., Mohme M., Willscher E., von Wenserski L., Scholz R., Wieters I., Dahlke C., Tolosa E., Sedding D.G., Ciesek S., Addo M., Binder M. Next-generation sequencing of T and B cell receptor repertoires from COVID-19 patients showed signatures associated with severity of disease. Immunity. 2020;53(2):442–455.e4. doi: 10.1016/j.immuni.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandrashekar A., Liu J., Martinot A.J., McMahan K., Mercado N.B., Peter L., Tostanoski L.H., Yu J., Maliga Z., Nekorchuk M., Busman-Sahay K., Terry M., Wrijil L.M., Ducat S., Martinez D.R., Atyeo C., Fischinger S., Burke J.S., Slein M.D., Pessaint L., Van Ry A., Greenhouse J., Taylor T., Blade K., Cook A., Finneyfrock B., Brown R., Teow E., Velasco J., Zahn R., Wegmann F., Abbink P., Bondzie E.A., Dagotto G., Gebre M.S., He X., Jacob-Dolan C., Kordana N., Li Z., Lifton M.A., Mahrokhian S.H., Maxfield L.F., Nityanandam R., Nkolola J.P., Schmidt A.G., Miller A.D., Baric R.S., Alter G., Sorger P.K., Estes J.D., Andersen H., Lewis M.G., Barouch D.H. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science (New York, NY) 2020;369(6505):812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirkcaldy R.D., King B.A., Brooks J.T. COVID-19 and postinfection immunity: limited evidence, many remaining questions. JAMA. 2020;323(22):2245–2246. doi: 10.1001/jama.2020.7869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hotez P.J., Corry D.B., Strych U., Bottazzi M.E. COVID-19 vaccines: neutralizing antibodies and the alum advantage. Nat. Rev. Immunol. 2020;20(7):399–400. doi: 10.1038/s41577-020-0358-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., Hägglöf T., Oliveira T.Y., Viant C., Hurley A., Hoffmann H.-H., Millard K.G., Kost R.G., Cipolla M., Gordon K., Bianchini F., Chen S.T., Ramos V., Patel R., Dizon J., Shimeliovich I., Mendoza P., Hartweger H., Nogueira L., Pack M., Horowitz J., Schmidt F., Weisblum Y., Michailidis E., Ashbrook A.W., Waltari E., Pak J.E., Huey-Tubman K.E., Koranda N., Hoffman P.R., West A.P., Rice C.M., Hatziioannou T., Bjorkman P.J., Bieniasz P.D., Caskey M., Nussenzweig M.C. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584(7821):437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.E. Seydoux, L.J. Homad, A.J. MacCamy, K.R. Parks, N.K. Hurlburt, M.F. Jennewein, N.R. Akins, A.B. Stuart, Y.H. Wan, J. Feng et al., Characterization of neutralizing antibodies from a SARS-CoV-2 infected individual. bioRxiv : the preprint server for biology 2020. [DOI] [PMC free article] [PubMed]
- 15.Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A., van Kuppeveld F.J.M., Haagmans B.L., Grosveld F., Bosch B.J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11(1):2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., Bentlage A.E.H., van Haaren M.M., Guerra D., Burger J.A., Schermer E.E., Verheul K.D., van der Velde N., van der Kooi A., van Schooten J., van Breemen M.J., Bijl T.P.L., Sliepen K., Aartse A., Derking R., Bontjer I., Kootstra N.A., Wiersinga W.J., Vidarsson G., Haagmans B.L., Ward A.B., de Bree G.J., Sanders R.W., van Gils M.J. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science (New York, NY) 2020;369(6504):643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolf K.W., Baumgart K., Traut W. Cytology of Lepidoptera. III. Giant cysts: a morphological trait of apyrene spermatogenesis in an Ephestia kuehniella strain. Gamete Res. 1988;20(3):353–364. doi: 10.1002/mrd.1120200310. [DOI] [PubMed] [Google Scholar]
- 18.Woloshin S., Patel N., Kesselheim A.S. False negative tests for SARS-CoV-2 infection - challenges and implications. N. Engl. J. Med. 2020;383(6):e38. doi: 10.1056/NEJMp2015897. [DOI] [PubMed] [Google Scholar]
- 19.Ibarrondo F.J., Fulcher J.A., Goodman-Meza D., Elliott J., Hofmann C., Hausner M.A., Ferbas K.G., Tobin N.H., Aldrovandi G.M., Yang O.O. Rapid decay of anti-SARS-CoV-2 antibodies in persons with mild Covid-19. N. Engl. J. Med. 2020;383(11):1085–1087. doi: 10.1056/NEJMc2025179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Long Q.-X., Tang X.-J., Shi Q.-L., Li Q., Deng H.-J., Yuan J., Hu J.-L., Xu W., Zhang Y., Lv F.-J., Su K., Zhang F., Gong J., Wu B.o., Liu X.-M., Li J.-J., Qiu J.-F., Chen J., Huang A.-L. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26(8):1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 21.Channappanavar R., Fett C., Zhao J., Meyerholz D.K., Perlman S., Sandri-Goldin R.M. Virus-specific memory CD8 T cells provide substantial protection from lethal severe acute respiratory syndrome coronavirus infection. J. Virol. 2014;88(19):11034–11044. doi: 10.1128/JVI.01505-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang F., Quan Y., Xin Z.-T., Wrammert J., Ma M.-J., Lv H., Wang T.-B., Yang H., Richardus J.H., Liu W., Cao W.-C. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J. Immunol. 2011;186(12):7264–7268. doi: 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- 23.Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., Chia W.N., Chen M.-C., Wang L.-F., Ooi E.E., Kalimuddin S., Tambyah P.A., Low J.-H., Tan Y.-J., Bertoletti A. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584(7821):457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 24.Yang L.-T., Peng H., Zhu Z.-L., Li G., Huang Z.-T., Zhao Z.-X., Koup R.A., Bailer R.T., Wu C.-Y. Long-lived effector/central memory T-cell responses to severe acute respiratory syndrome coronavirus (SARS-CoV) S antigen in recovered SARS patients. Clin. Immunol. (Orlando, Fla) 2006;120(2):171–178. doi: 10.1016/j.clim.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., Marrama D., de Silva A.M., Frazier A., Carlin A.F., Greenbaum J.A., Peters B., Krammer F., Smith D.M., Crotty S., Sette A. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ni L., Ye F., Cheng M.-L., Feng Y.u., Deng Y.-Q., Zhao H., Wei P., Ge J., Gou M., Li X., Sun L., Cao T., Wang P., Zhou C., Zhang R., Liang P., Guo H., Wang X., Qin C.-F., Chen F., Dong C. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020;52(6):971–977.e3. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sekine T., Perez-Potti A., Rivera-Ballesteros O., Strålin K., Gorin J.-B., Olsson A., Llewellyn-Lacey S., Kamal H., Bogdanovic G., Muschiol S., Wullimann D.J., Kammann T., Emgård J., Parrot T., Folkesson E., Rooyackers O., Eriksson L.I., Henter J.-I., Sönnerborg A., Allander T., Albert J., Nielsen M., Klingström J., Gredmark-Russ S., Björkström N.K., Sandberg J.K., Price D.A., Ljunggren H.-G., Aleman S., Buggert M., Akber M., Berglin L., Bergsten H., Brighenti S., Brownlie D., Butrym M., Chambers B., Chen P., Jeannin M.C., Grip J., Gomez A.C., Dillner L., Lozano I.D., Dzidic M., Tullberg M.F., Färnert A., Glans H., Haroun-Izquierdo A., Henriksson E., Hertwig L., Kalsum S., Kokkinou E., Kvedaraite E., Loreti M., Lourda M., Maleki K., Malmberg K.-J., Marquardt N., Maucourant C., Michaelsson J., Mjösberg J., Moll K., Muva J., Mårtensson J., Nauclér P., Norrby-Teglund A., Medina L.P., Persson B., Radler L., Ringqvist E., Sandberg J.T., Sohlberg E., Soini T., Svensson M., Tynell J., Varnaite R., Kries A.V., Unge C. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183(1):158–168.e14. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.J.L. Ziwei Li, Hui Deng, Xuecheng Yang, Hua Wang, Xuemei Feng, Gennadiy Zelinskyy, Mirko Trilling, Kathrin Sutter, Lu Mengji, Ulf Dittmer, Baoju Wang, Dongliang Yang, Xin Zheng, Jia Liu, SARS-CoV-2-specific T cell memory is long-lasting in the majority of convalsecent COVID-19 individuals. bioRxiv : the preprint server for biology 2020.
- 29.J. Zuo, H. Pearce, K. Verma, H.M. Long, J. Begum, F. Aiano, Z. Amin-Chowdhury, B. Hallis, L. Stapley, R. Borrow, E. Linley, S. Ahmad, B. Parker, A. Horsley, G. Amirthalingam, K. Brown, M.E. Ramsay, S. Ladhani, P. Moss, Robust SARS-CoV-2-specific T-cell immunity is maintained at 6 months following primary infection. bioRxiv : the preprint server for biology 2020.
- 30.Chua R.L., Lukassen S., Trump S., Hennig B.P., Wendisch D., Pott F., Debnath O., Thürmann L., Kurth F., Völker M.T., Kazmierski J., Timmermann B., Twardziok S., Schneider S., Machleidt F., Müller-Redetzky H., Maier M., Krannich A., Schmidt S., Balzer F., Liebig J., Loske J., Suttorp N., Eils J., Ishaque N., Liebert U.G., von Kalle C., Hocke A., Witzenrath M., Goffinet C., Drosten C., Laudi S., Lehmann I., Conrad C., Sander L.-E., Eils R. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020;38(8):970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- 31.Guo C., Li B., Ma H., Wang X., Cai P., Yu Q., Zhu L., Jin L., Jiang C., Fang J., Liu Q., Zong D., Zhang W., Lu Y., Li K., Gao X., Fu B., Liu L., Ma X., Weng J., Wei H., Jin T., Lin J., Qu K. Single-cell analysis of two severe COVID-19 patients reveals a monocyte-associated and tocilizumab-responding cytokine storm. Nat. Commun. 2020;11(1) doi: 10.1038/s41467-020-17834-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He J., Cai S., Feng H., Cai B., Lin L., Mai Y., Fan Y., Zhu A., Huang H., Shi J., Li D., Wei Y., Li Y., Zhao Y., Pan Y., Liu H.e., Mo X., He X.i., Cao S., Hu F., Zhao J., Wang J., Zhong N., Chen X., Deng X., Chen J. Single-cell analysis reveals bronchoalveolar epithelial dysfunction in COVID-19 patients. Protein Cell. 2020;11(9):680–687. doi: 10.1007/s13238-020-00752-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., Liu L., Amit I., Zhang S., Zhang Z. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26(6):842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 34.Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martínez-Colón G.J., McKechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T., Simpson L.J., Grant P., Subramanian A., Rogers A.J., Blish C.A. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26(7):1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J.-Y., Wang X.-M., Xing X., Xu Z., Zhang C., Song J.-W., Fan X., Xia P., Fu J.-L., Wang S.-Y., Xu R.-N., Dai X.-P., Shi L., Huang L., Jiang T.-J., Shi M., Zhang Y., Zumla A., Maeurer M., Bai F., Wang F.-S. Single-cell landscape of immunological responses in patients with COVID-19. Nat. Immunol. 2020;21(9):1107–1118. doi: 10.1038/s41590-020-0762-x. [DOI] [PubMed] [Google Scholar]
- 36.Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X.u., Zheng Y., Geng C., Chai X., He R., Li X., Lv Q.i., Zhu H., Deng W., Xu Y., Wang Y., Qiao L., Tan Y., Song L., Wang G., Du X., Gao N., Liu J., Xiao J., Su X.-D., Du Z., Feng Y., Qin C., Qin C., Jin R., Xie X.S. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients' B Cells. Cell. 2020;182(1):73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xianwen Ren W.W., Fan X., Hou W., Bin S., Cai P., Li J., Liu Y., Tang F., Fan Zhang Y., Yang J.H., Ma W., He J., Wang P., Cao Q., Chen F., Chen Y., Cheng X., Deng G., Deng X., Ding W., Feng Y., Gan R., Guo C., Guo W., He S., Jiang C., Liang J., Li Y.-min., Lin J., Ling Y., Liu H., Liu J., Liu N., Liu S.-Q., Luo M., Ma Q., Song Q., Sun W., Wang G., Wang F., Wang Y., Wen X., Qian W., Gang X., Xie X., Xiong X., Xing X., Hao X., Yin C., Dongdong Y., Kezhuo Y., Yuan J., Zhang B., Zhang P., Zhang T., Zhao J., Zhao P., Zhou J., Zhou W., Zhong S., Zhong X., Zhang S., Zhu L., Zhu P., Zou B., Zou J., Zuo Z., Bai F., Huang X., Zhou P., Jiang Q., Huang Z., Bei J.-X., Wei L., Bian X.-W., Liu X., Cheng T., Li X., Zhao P., Wang F.-S., Wang H., Bing S., Zhang Z., Kun Q., Wang X., Chen J., Jin R., Zhang Z. COVID-19 immune features revealed by a large-scale single cell transcriptome atlas. Cell. 2021 doi: 10.1016/j.cell.2021.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carr T.M., Wheaton J.D., Houtz G.M., Ciofani M. JunB promotes Th17 cell identity and restrains alternative CD4(+) T-cell programs during inflammation. Nat. Commun. 2017;8(1):301. doi: 10.1038/s41467-017-00380-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hasan Z., Koizumi S.I., Sasaki D., Yamada H., Arakaki N., Fujihara Y., Okitsu S., Shirahata H., Ishikawa H. JunB is essential for IL-23-dependent pathogenicity of Th17 cells. Nat. Commun. 2017;8:15628. doi: 10.1038/ncomms15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore M.J., Blachere N.E., Fak J.J., Park C.Y., Sawicka K., Parveen S., Zucker-Scharff I., Moltedo B., Rudensky A.Y., Darnell R.B. ZFP36 RNA-binding proteins restrain T cell activation and anti-viral immunity. eLife. 2018;7 doi: 10.7554/eLife.33057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao X., Xie H., Zhao M., Ahsan A., Li X., Wang F., Yi J., Yang Z., Wu C., Raman I., Li Q.-Z., Kim T.J., Liu W., Raman I., et al. Fc receptor-like 1 intrinsically recruits c-Abl to enhance B cell activation and function. Sci. Adv. 2019;5(7) doi: 10.1126/sciadv.aaw0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L.i., Tai Y., Bai C., Gao T., Song J., Xia P., Dong J., Zhao J., Wang F.-S. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8(4):420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matthay M.A., Aldrich J.M., Gotts J.E. Treatment for severe acute respiratory distress syndrome from COVID-19. Lancet Respir. Med. 2020;8(5):433–434. doi: 10.1016/S2213-2600(20)30127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kam Y.-W., Ahmed M.Y., Amrun S.N., Lee B., Refaie T., Elgizouli K., Fong S.-W., Renia L., Ng L.FP. Systematic analysis of disease-specific immunological signatures in patients with febrile illness from Saudi Arabia. Clin. Transl. Immunol. 2020;9(8) doi: 10.1002/cti2.v9.810.1002/cti2.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cañete P.F., Vinuesa C.G. COVID-19 makes B cells forget, but T cells remember. Cell. 2020;183(1):13–15. doi: 10.1016/j.cell.2020.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D., Dejnirattisai W., Rostron T., Supasa P., Liu C., et al. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21(11):1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schulte-Schrepping J., Reusch N., Paclik D., Baßler K., Schlickeiser S., Zhang B., Krämer B., Krammer T., Brumhard S., Bonaguro L., et al. Severe COVID-19 is marked by a dysregulated Myeloid cell compartment. Cell. 2020;182(6):1419–1440.e1423. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu L., Yang P., Zhao Y., Zhuang Z., Wang Z., Song R., Zhang J., Liu C., Gao Q., Xu Q., Wei X., Sun H.-X., Ye B., Wu Y., Zhang N., Lei G., Yu L., Yan J., Diao G., Meng F., Bai C., Mao P., Yu Y., Wang M., Yuan Y., Deng Q., Li Z., Huang Y., Hu G., Liu Y., Wang X., Xu Z., Liu P., Bi Y., Shi Y.i., Zhang S., Chen Z., Wang J., Xu X., Wu G., Wang F.-S., Gao G.F., Liu L., Liu W.J. Single-cell sequencing of peripheral mononuclear cells reveals distinct immune response landscapes of COVID-19 and influenza patients. Immunity. 2020;53(3):685–696.e3. doi: 10.1016/j.immuni.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meckiff B.J., Ramírez-Suástegui C., Fajardo V., Chee S.J., Kusnadi A., Simon H., Eschweiler S., Grifoni A., Pelosi E., Weiskopf D., Sette A., Ay F., Seumois G., Ottensmeier C.H., Vijayanand P. Imbalance of regulatory and cytotoxic SARS-CoV-2-reactive CD4(+) T cells in COVID-19. Cell. 2020;183(5):1340–1353.e16. doi: 10.1016/j.cell.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee S., Channappanavar R., Kanneganti T.-D. Coronaviruses: Innate immunity, inflammasome activation, inflammatory cell death, and cytokines. Trends Immunol. 2020;41(12):1083–1099. doi: 10.1016/j.it.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yao Z., Zheng Z., Wu K.e., Junhua Z. Immune environment modulation in pneumonia patients caused by coronavirus: SARS-CoV, MERS-CoV and SARS-CoV-2. Aging. 2020;12(9):7639–7651. doi: 10.18632/aging.103101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou Z., Ren L., Zhang L.i., Zhong J., Xiao Y., Jia Z., Guo L.i., Yang J., Wang C., Jiang S., Yang D., Zhang G., Li H., Chen F., Xu Y.u., Chen M., Gao Z., Yang J., Dong J., Liu B.o., Zhang X., Wang W., He K., Jin Q.i., Li M., Wang J. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27(6):883–890.e2. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z., Wang C., Wang Y., Li L.i., Ren L., Guo F., Zhao Z., Zhou Z., Xiang Z., Wang J. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11(1) doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yánez D.C., Ross S., Crompton T. The IFITM protein family in adaptive immunity. Immunology. 2020;159(4):365–372. doi: 10.1111/imm.13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi G., Kenney A.D., Kudryashova E., Zani A., Zhang L., Lai K.K., Hall-Stoodley L., Robinson R.T., Kudryashov D.S., Compton A.A., et al. Opposing activities of IFITM proteins in SARS-CoV-2 infection. Embo J. 2020:e106501. doi: 10.15252/embj.2020106501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prasad K., Khatoon F., Rashid S., Ali N., AlAsmari A.F., Ahmed M.Z., Alqahtani A.S., Alqahtani M.S., Kumar V. Targeting hub genes and pathways of innate immune response in COVID-19: A network biology perspective. Int. J. Biol. Macromol. 2020;163:1–8. doi: 10.1016/j.ijbiomac.2020.06.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barnes B.J., Adrover J.M., Baxter-Stoltzfus A., Borczuk A., Cools-Lartigue J., Crawford J.M., Daßler-Plenker J., Guerci P., Huynh C., Knight J.S., Loda M., Looney M.R., McAllister F., Rayes R., Renaud S., Rousseau S., Salvatore S., Schwartz R.E., Spicer J.D., Yost C.C., Weber A., Zuo Y.u., Egeblad M. Targeting potential drivers of COVID-19: Neutrophil extracellular traps. J. Exp. Med. 2020;217(6) doi: 10.1084/jem.20200652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saravia J., Raynor J.L., Chapman N.M., Lim S.A., Chi H. Signaling networks in immunometabolism. Cell Res. 2020;30(4):328–342. doi: 10.1038/s41422-020-0301-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McBrien J.B., Wong A.K.H., White E., Carnathan D.G., Lee J.H., Safrit J.T., Vanderford T.H., Paiardini M., Chahroudi A., Silvestri G., Simon V. Combination of CD8β depletion and Interleukin-15 superagonist N-803 induces virus reactivation in simian-human immunodeficiency virus-infected, long-term ART-treated rhesus macaques. J. Virol. 2020;94(19) doi: 10.1128/JVI.00755-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koizumi S.-I., Sasaki D., Hsieh T.-H., Taira N., Arakaki N., Yamasaki S., Wang K.e., Sarkar S., Shirahata H., Miyagi M., Ishikawa H. JunB regulates homeostasis and suppressive functions of effector regulatory T cells. Nat. Commun. 2018;9(1) doi: 10.1038/s41467-018-07735-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Levada K., Guldiken N., Zhang X., Vella G., Mo F.-R., James L.P., Haybaeck J., Kessler S.M., Kiemer A.K., Ott T., Hartmann D., Hüser N., Ziol M., Trautwein C., Strnad P. Hsp72 protects against liver injury via attenuation of hepatocellular death, oxidative stress, and JNK signaling. J. Hepatol. 2018;68(5):996–1005. doi: 10.1016/j.jhep.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Allende M., Molina E., Guruceaga E., Tamayo I., González-Porras J.R., Gonzalez-López T.J., Toledo E., Rabal O., Ugarte A., Roldán V., Rivera J., Oyarzabal J., Montes R., Hermida J. Hsp70 protects from stroke in atrial fibrillation patients by preventing thrombosis without increased bleeding risk. Cardiovasc. Res. 2016;110(3):309–318. doi: 10.1093/cvr/cvw049. [DOI] [PubMed] [Google Scholar]
- 63.Jin Y., Sharma A., Carey C., Hopkins D., Wang X., Robertson D.G., Bode B., Anderson S.W., Reed J.C., Steed R.D., Steed L., She J.-X. The expression of inflammatory genes is upregulated in peripheral blood of patients with type 1 diabetes. Diabetes Care. 2013;36(9):2794–2802. doi: 10.2337/dc12-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lou Y., Han M., Liu H., Niu Y., Liang Y., Guo J., Zhang W., Wang H. Essential roles of S100A10 in Toll-like receptor signaling and immunity to infection. Cell. Mol. Immunol. 2020;17(10):1053–1062. doi: 10.1038/s41423-019-0278-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang L., Yu X., Zheng L., Zhang Y., Li Y., Fang Q., Gao R., Kang B., Zhang Q., Huang J.Y., Konno H., Guo X., Ye Y., Gao S., Wang S., Hu X., Ren X., Shen Z., Ouyang W., Zhang Z. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature. 2018;564(7735):268–272. doi: 10.1038/s41586-018-0694-x. [DOI] [PubMed] [Google Scholar]
- 66.Wong R.S., Wu A., To K.F., Lee N., Lam C.W., Wong C.K., Chan P.K., Ng M.H., Yu L.M., Hui D.S., et al. Haematological manifestations in patients with severe acute respiratory syndrome: retrospective analysis. BMJ (Clin. Res. Ed.) 2003;326(7403):1358–1362. doi: 10.1136/bmj.326.7403.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cui W., Fan Y., Wu W., Zhang F., Wang J.-Y., Ni A.-P. Expression of lymphocytes and lymphocyte subsets in patients with severe acute respiratory syndrome. Clin. Infect. Dis.: An Official Publication Infect. Dis. Soc. Am. 2003;37(6):857–859. doi: 10.1086/378587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zheng H.-Y., Zhang M.i., Yang C.-X., Zhang N., Wang X.-C., Yang X.-P., Dong X.-Q., Zheng Y.-T. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell. Mol. Immunol. 2020;17(5):541–543. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nutt S.L., Hodgkin P.D., Tarlinton D.M., Corcoran L.M. The generation of antibody-secreting plasma cells. Nat. Rev. Immunol. 2015;15(3):160–171. doi: 10.1038/nri3795. [DOI] [PubMed] [Google Scholar]
- 70.Guo L.i., Ren L., Yang S., Xiao M., Chang D.e., Yang F., Dela Cruz C.S., Wang Y., Wu C., Xiao Y., Zhang L., Han L., Dang S., Xu Y., Yang Q.-W., Xu S.-Y., Zhu H.-D., Xu Y.-C., Jin Q.i., Sharma L., Wang L., Wang J. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin. Infect. Dis.: An Official Publication Infect. Dis. Soc. Am. 2020;71(15):778–785. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beaudoin-Bussières G., Laumaea A., Anand S.P., Prévost J., Gasser R., Goyette G., Medjahed H., Perreault J., Tremblay T., Lewin A., Gokool L., Morrisseau C., Bégin P., Tremblay C., Martel-Laferrière V., Kaufmann D.E., Richard J., Bazin R., Finzi A., Ho D.D., Goff S.P. Decline of humoral responses against SARS-CoV-2 spike in convalescent individuals. mBio. 2020;11(5) doi: 10.1128/mBio.02590-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang S., Song R., Wang Z., Jing Z., Wang S., Ma J. S100A8/A9 in Inflammation. Front. Immunol. 2018;9:1298. doi: 10.3389/fimmu.2018.01298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Silvin A., Chapuis N., Dunsmore G., Goubet A.-G., Dubuisson A., Derosa L., Almire C., Hénon C., Kosmider O., Droin N., Rameau P., Catelain C., Alfaro A., Dussiau C., Friedrich C., Sourdeau E., Marin N., Szwebel T.-A., Cantin D., Mouthon L., Borderie D., Deloger M., Bredel D., Mouraud S., Drubay D., Andrieu M., Lhonneur A.-S., Saada V., Stoclin A., Willekens C., Pommeret F., Griscelli F., Ng L.G., Zhang Z., Bost P., Amit I., Barlesi F., Marabelle A., Pène F., Gachot B., André F., Zitvogel L., Ginhoux F., Fontenay M., Solary E. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell. 2020;182(6):1401–1418.e18. doi: 10.1016/j.cell.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kaneko N., Kuo H.-H., Boucau J., Farmer J.R., Allard-Chamard H., Mahajan V.S., Piechocka-Trocha A., Lefteri K., Osborn M., Bals J., Bartsch Y.C., Bonheur N., Caradonna T.M., Chevalier J., Chowdhury F., Diefenbach T.J., Einkauf K., Fallon J., Feldman J., Finn K.K., Garcia-Broncano P., Hartana C.A., Hauser B.M., Jiang C., Kaplonek P., Karpell M., Koscher E.C., Lian X., Liu H., Liu J., Ly N.L., Michell A.R., Rassadkina Y., Seiger K., Sessa L., Shin S., Singh N., Sun W., Sun X., Ticheli H.J., Waring M.T., Zhu A.L., Alter G., Li J.Z., Lingwood D., Schmidt A.G., Lichterfeld M., Walker B.D., Yu X.G., Padera R.F., Pillai S. Loss of bcl-6-expressing T Follicular Helper cells and germinal centers in COVID-19. Cell. 2020;183(1):143–157.e13. doi: 10.1016/j.cell.2020.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Forni D., Cagliani R., Pontremoli C., Mozzi A., Pozzoli U., Clerici M., Sironi M. Antigenic variation of SARS-CoV-2 in response to immune pressure. Mol. Ecol. 2020 doi: 10.1111/mec.15730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Duan Y.-q., Xia M.-H., Ren L., Zhang Y.-F., Ao Q.-L., Xu S.-P., Kuang D., Liu Q., Yan B., Zhou Y.-w., Chu Q., Liu L., Yang X.-P., Wang G.-P. Deficiency of Tfh Cells and Germinal Center in Deceased COVID-19 Patients. Curr Med Sci. 2020;40(4):618–624. doi: 10.1007/s11596-020-2225-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.D. Matthew, S.A. Beasley, Fiona M. Gracey, Ruban Kannan, S.R. Avisa Masarati, Madhara Udawela Premaratne, Rebecca E. Wood, Shereen Jabar, Nicole L. Church, Thien-Kim Le, Dahna Makris, Bradley K. McColl, Ben R. Kiefel, Antibodies that potently inhibit or enhance SARS-CoV-2 spike protein-ACE2 interaction isolated from synthetic single-chain antibody libraries. bioRxiv : the preprint server for biology 2020.
- 78.M.W. Michael Mor, Joel Alter, Modi Safra, Elad Chomsky, Smadar Hada-Neeman, Ksenia Polonsky, Cameron J. Nowell, Alex E. Clark, Anna Roitburd-Berman, Noam Ben Shalom, Michal Navon, Dor Rafael, Hila Sharim, Evgeny Kiner, Eric Griffis, Jonathan M. Gershoni, Oren Kobiler, Sandra Lawrynowicz Leibel, Oren Zimhony, Aaron F. Carlin, Gur Yaari, Moshe Dassau, Meital Gal-Tanamy, David Hagin, Ben A. Croker, Natalia T. Freund, Multi-Clonal Live SARS-CoV-2 In Vitro Neutralization by Antibodies Isolated from Severe COVID-19 Convalescent Donors. bioRxiv : the preprint server for biology 2020.
- 79.Wen W., Su W., Tang H., Le W., Zhang X., Zheng Y., Liu X., Xie L., Li J., Ye J., et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020;6:31. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Traag V.A., Waltman L., van Eck N.J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 2019;9(1):5233. doi: 10.1038/s41598-019-41695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The laboratory findings of enrolled convalescent patients with moderate COVID-19.
Detailed timeline of nucleic acid testing results for throat samples along with the IgM and IgG responses.
Counts of cells were used for identification of cell types.
Summary of Cell subtyping characterization after quality control of 117,102 cells.
Datasets of 47 major cell types identified.
Immune response-activating cell surface receptor signaling pathway (GO:0002429).
Statistical analysis of 70 DEGs in 5 innate immune cell subsets.
Average expression of 70 DEGs in 5 innate immune cell subsets.
Datasets of total DEGs in 5 innate immune cell subsets from convalescent moderate or severe patients in comparison with those of HDs.
Statistical analysis of 142 DEGs in 13 T cell subsets.
Average expression of 142 DEGs in 13 T cell subsets.
Datasets of total DEGs in 13 T cell subsets from convalescent moderate or severe patients in comparison with those of HDs.
Datasets of average expression of HLA genes in 13 T cell subsets from convalescent moderate or severe patients in comparison with those of HDs.
Datasets of expansion and transition of T cell subsets analyzed by STARTRAC.
Statistical analysis of usage of TRAV, TRAJ, TRBV and TRBJ genes across tree conditions.
Statistical analysis of 82 DEGs in 5 B cell subsets.
Average expression of 82 DEGs in 5 B cell subsets.
Datasets of total DEGs in 5 B cell subsets from convalescent moderate or severe patients in comparison with those of HDs.
Datasets of average expression of HLA genes in 4 B cell subsets from convalescent moderate or severe patients in comparison with those of HDs.
Datasets of expansion and transition of B cell subsets analyzed by STARTRAC.
Data Availability Statement
The datasets used or analysed during the current study are available from the corresponding author on reasonable request.