Abstract
Differentiation of multi-potent mesenchymal stromal cells (MSCs) is directed by the activities of lineage-specific transcription factors and co-factors. A subset of these proteins controls the accessibility of chromatin by recruiting histone acetyl transferases or deacetylases that regulate acetylation of the N-termini of H3 and H4 histone proteins. Bromodomain (BRD) proteins recognize these acetylation marks and recruit the RNA pol II containing transcriptional machinery. Our previous studies have shown that Brd4 is required for osteoblast differentiation in vitro. Here, we investigated the role of Brd4 on endochondral ossification in C57BL/6 mice and chondrogenic differentiation in cell culture models. Conditional loss of Brd4 in the mesenchyme (Brd4 cKO, Brd4fl/fl: Prrx1-Cre) yields smaller mice that exhibit alteration in endochondral ossification. Importantly, abnormal growth plate morphology and delayed long bone formation is observed in juvenile Brd4 cKO mice. One week old Brd4 cKO mice have reduced proliferative and hypertrophic zones within the physis and exhibit a delay in the formation of the secondary ossification center. At the cellular level, Brd4 function is required for chondrogenic differentiation and maturation of both ATDC5 cells and immature mouse articular chondrocytes. Mechanistically, Brd4 loss suppresses Sox9 levels and reduces expression of Sox9 and Runx2 responsive endochondral genes (e.g., Col2a1, Acan, Mmp13 and Sp7/Osx). Collectively, our results indicate that Brd4 is a key epigenetic regulator required for normal chondrogenesis and endochondral ossification.
Keywords: Brd4, Epigenetics, Genetic Animal Model, Growth Plate, Histone, Limb Patterning
1. INTRODUCTION
Skeletal development in vertebrates occurs with precise timing and regulation [1] and begins in humans during the second month of development in utero and continues until age twenty five [2]. Skeletogenesis is initiated by the migration of mesenchymal stromal/stem cells (MSCs) that condense into primordial fibrous scaffolds [3] to support bone formation via intramembranous or endochondral ossification [4]. Intramembranous ossification generates bones of the skull and clavicles by direct conversion of condensed MSCs into osteoblasts which begin to secrete an osseous matrix and calcify the fibrous template [5]. The majority of the bones in the skeleton are formed via endochondral ossification where MSCs differentiate into chondrocytes within the physis [6], which serves as a transition zone between cartilage and newly calcified bone matrix [7].
Separate layers within the growth plate are characterized by distinct cell types and matrix proteins [8], including a reserve or germinal zone that harbors immature resting chondrocyte progenitors, a proliferative zone where chondrocytes align and divide into a columnar arrangement, and a hypertrophic zone where chondrocytes increase in size and remodel the surrounding matrix [9]. Chondrocyte differentiation is driven by several transcription factors, including the Sox trio (i.e., Sox9, Sox6 and Sox5) [10], that support expression of collagen type II (Col2a1) and aggrecan (Acan) to form the cartilaginous condensations that serve as a template for new bone formation. Mature hypertrophic chondrocytes remodel the collagen type II matrix by upregulating transcription factors Runx2 and Osx/Sp7 that mediate production of matrix metalloproteinase 13 (Mmp13) and type X collagen (Col10a1). Subsequent apoptosis of hypertrophic chondrocytes and migration of osteoblasts into the resulting voids initiates formation of calcifying osteoid tissue to form the primary ossification center [11]. This cycle of chondrocyte proliferation, hypertrophy, and osteoblast invasion occurs concomitantly with the advancing chondrocyte pool at the end of the bone, which leaves a mineralized matrix behind and supports the formation of a secondary ossification center at the end of long bones [12].
Different stages of growth plate development and chondrocyte maturation are controlled by epigenetic mechanisms that support unique gene expression profiles and overall cell function [13–20]. Skeletal genes are suppressed during very early stages of embryogenesis because genes are rendered inaccessible by formation of dense heterochromatin. Remodeling of heterochromatin is mediated by dynamic post-translational modifications of histone tails that support transcriptional activation of skeletal genes. Our group and others have focused on the dynamic equilibrium between histone lysine methylation and acetylation in the context of skeletal development [16, 17, 21–29].
Studies on the biological relevance of histone tail acetylation have been of particular interest as this mark induces euchromatin formation and is indicative of active transcription [30, 31]. Acetylation of histone proteins is recognized by bromodomain (BRD) containing proteins that mediate a direct link with RNA polymerase II activity to stimulate skeletal gene expression. Previous studies have shown that osteoblast differentiation is controlled by Brd2 [32] and Brd4 [33–37]. These two proteins are closely related and contain both a bromodomain and extra terminal domain (i.e., BET proteins). Therefore, these proteins bind acetylated lysine residues via the bromodomain and also interact with additional transcription factors and epigenetic regulators via the extra terminal domain [38]. The dual function of these proteins provides an ideal mechanism to serve as a molecular bridge between histone tail acetylation and downstream effectors such as transcription factors and RNA polymerase II in skeletal cells.
The clinical relevance of BRD4/Brd4 is evident from missense mutations in human BRD4 that have been linked to skeletal abnormalities including flat feet, short stature, brachydactyly, and clinodactyly [39]. In addition, copy number variations in BRD4 have been associated with craniofacial abnormalities such as oral clefts [40]. At the molecular level, BRD4 is of particular interest, because it is a relatively stable protein (half-life > 24h) [41] that is linked to transcriptional initiation and super-enhancer formation [42, 43]. The present study investigates the role of Brd4 in the developing skeleton, as well as during chondrocyte differentiation and maturation. The main findings are that conditional ablation of Brd4 in the mesenchyme (Prrx1-Cre: Brd4f l/f l mice) causes a significant skeletal phenotype characterized by shortened long bones and abnormal growth plate morphology. Furthermore, our results indicate that Brd4 is required for proper differentiation and function of primary and immortalized chondrocytes.
2. METHODS
2.1. Animal Welfare
All animal studies were conducted according to guidelines provided by the National Institutes of Health and the Institute of Laboratory Animal Resources, National Research Council. The Mayo Clinic Institutional Animal Care and Use Committee approved all animal studies. Animals were housed in an accredited facility under a 12-h light/dark cycle and provided water and food (PicoLab Rodent Diet 20, LabDiet) ad libitum.
2.2. Conditional Deletion of Brd4 in the Mesenchyme
Mice containing the Brd4f l/f l transgene harboring two loxP sites flanking the third exon of Brd4 genomic DNA were kindly provided by Dr. Anup Dey and Dr. Keiko Ozato (National Institutes of Health, Bethesda, MD) [44]. Brd4 was conditionally deleted in the mesenchyme by crossing the Brd4f l/f l line with transgenic mice expressing Cre recombinase under control of the Prrx1 enhancer [45]. All mice are on the C57BL/6 genetic background. Genotyping primer pairs utilized are shown (Suppl. Table 1). Skeletal tissues of postnatal mice (one, three, eight, and sixteen weeks of age) were harvested and fixed in 10% neutral buffered formalin for 48 hours. Tissues were transferred to 70% ethanol for storage and downstream analysis, including micro-computed tomography (µCT), radiography (X-ray), histology, and immunohistochemistry (IHC). Limb buds (DNA and protein analysis) and tails (genotyping) from 14.5 days post coitum (dpc) embryos were harvested on ice and stored in liquid nitrogen and processed as described.
2.3. Genomic DNA analysis
Genomic DNA was extracted using aDNA extraction kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. For qPCR analysis, 50ng of genomic DNA was amplified with the QuantiTect SYBR-Green PCR Kit (Qiagen) and a CFX384 real time system machine (Bio-Rad, Hercules, CA) using the following conditions: 10 min 95°C followed by 45 cycles of 30 sec at 95°C, 30 sec at 60°C and 30 sec at 72°C. Primer pairs targeting the analyzed genes are listed (Suppl. Table 1). PCR assays were performed in triplicate and expression of Brd4 exon three primer sets (excised by Cre recombinase activity) were normalized to the levels of Brd4 exon six primer sets (not excised by Cre recombinase activity) by the 2ΔΔCt method.
2.4. Whole Mount Staining
Whole mount staining of neonatal mice (day 1) was performed as previously described [25]. Briefly, skeletons were dissected and fixed in ethanol overnight. Cartilage was stained with 0.2% Alcian blue (Sigma, St. Louis, MO) bones were stained with 75 µg/ml alizarin red (Sigma). Images of long bones were obtained using a Wild M420 macroscope (Wild Heerbrugg, Heerbrugg, Switzerland) and ProGres C3 camera (Jenoptik, Jena, Germany).
2.5. µCT Analysis
Bone architecture of femora and vertebrae from 3-week-old mice was evaluated using microcomputed Tomography (µCT) analysis. Femora were scanned in 70% ethanol on a μCT35 scanner (Scanco Medical AG, Basserdorf, Switzerland) at 10-μm voxel size using an energy setting of 70 kVp and an integration time of 300 ms. Bone parameters were computed using the manufacturer’s software [46]. Measurements of femoral trabecular bone quality were taken within the region spanning the equivalent of 20% to 30% of total bone length measured from the distal end of the femur. This window was chosen because 20% of total bone length was sufficient to set a starting point proximal to the growth plate and proceeding to 30% of bone length reached the diaphysis in all bones. Measurements of femoral cortical bone quality were taken at the level equivalent to 50% of total length in all bones. Measurements of the growth plate were obtained using Bruker CTAn micro-CT software (Bruker, Kontich, Belgium). Vertebrae were scanned on a SkyScan model 1276 scanner Bruker) with a pixel size of 8 µm with an energy source voltage of 55 kV, a rotation step of 0.2 degrees and exposure of 473 ms. 3D images were reconstructed used NRecon software (Bruker) and analysis and measurements were computed using DataViewer and CTan Software (Bruker).
2.6. Radiography
Radiographic images were obtained using a cabinet X-ray (Faxitron, Tucson, AZ) at 25kV for 8 seconds of exposure. Measurements of radiographs were made using ImageJ software [47].
2.7. Assessment of femoral growth rates
X-ray images were taken of femora collected from one, three, and eight week old male mice. Measurements of each bone were taken by drawing a straight line from the greater trochanter to the patellar groove using ImageJ software analysis. After plotting the data, linear regression analysis was performed for each individual line segment (1 to 3 weeks and 3 to 8 weeks). The slope of the line was interpreted as the growth rate for each time period. A two-way ANOVA test (alpha = 0.05) was utilized to compare the three groups. The time variable accounted for 73.36% of the variation observed (p < 0.001). The genotype variable (reported on the graph) accounted for 18.08% of the variation observed (p < 0.001).
2.8. Histological analysis
Femora from one and three week old mice were decalcified in 15% EDTA for seven days. Complete decalcification was ensured by radiography. Decalcified bones were then paraffin-embedded, sectioned, and stained with Weigert’s Hematoxylin, 0.4% Aqueous Fast Green, and 0.125% Safranin-O as described [23, 25].
2.9. Immature mouse articular chondrocytes (iMAC) isolation
iMACs were collected from five day-old wild-type C57BL/6J mice under IACUC approved protocol. Briefly, hind limbs were disarticulated, and soft tissues removed. Knee and ankle joints were dissected and preserved in cold DMEM media. Cartilage pieces were digested in 3ml of digestion media (3mg/ml collagenase II in DMEM) for one hour at 37°C (8 cartilage pieces per 3ml digestion media). After one hour, tissues and digestion medium were mixed by pipetting ~40 times before removing digestion media. 3ml of 0.5mg/ml digestion media was then added to the remaining cartilage pieces and incubated overnight at 37°C. The next day, tissues and digestion medium were mixed by pipetting ~40 times before filtering through a cell strainer. Cells were counted and micromasses formed as described below.
2.10. ATDC5 and iMAC cell culture
ATDC5 and iMAC cells were maintained and expanded in DMEM media (Gibco, 4.5g/L D-Glucose, 4.5g/L L-Glutamine, no Sodium Pyruvate) supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals, Flowery Branch, GA), 100U/ml Penicillin (Gibco), and 100µg/ml Streptomycin (Gibco). For differentiation assays, base DMEM media was supplemented with 2% FBS, 100U/ml Penicillin (Gibco, Grand Island, NY), and 100µg/ml Streptomycin (Gibco) plus the chondrogenic differentiation cocktail [final concentration 50µg/ml ascorbic acid (Sigma), 10mM β-glycerol phosphate (Sigma), and 1X Insulin-Transferrin-Selenium (Gibco)]. Differentiation assays were performed in 3D micromass cultures. To form micromasses, 10µl of media containing 200,000 cells were pipetted onto the tissue culture plastic. Micromasses were then incubated without media at 37°C for one hour. The edge of the well was lined with media to prevent desiccation of the micromasses. After one hour, wells were flooded with 3ml of culture media without differentiation cocktail. Two days later, media was changed, and differentiation cocktail was introduced. Media was changed every three days for the remainder of the experiment.
2.11. mRNA isolation
At indicated time points, cells were lysed using TRI-Reagent (Zymo Research, Irvine, CA) and RNA was isolated using the Direct-zol RNA isolation kit (Zymo Research). Purified RNA was quantified, and quality tested using a NanoDrop 2000 spectrophotometer (Thermo Fischer Scientific, Waltham, MA).
2.12. RT-qPCR
RNA was isolated as described above and reverse transcribed into cDNA using the Promega Reverse Transcription kit and protocol (Promega, Madison, WI). Gene expression was quantified using real-time quantitative PCR with QuantiTect SYBR Green PCR Kit (Qiagen) and the CFX384 Real-Time System (Bio-Rad). Transcript levels were quantified using the 2ΔΔCt method and normalized to the housekeeping gene Gapdh (set at 100). Primer pairs utilized in our studies are shown (Suppl. Table 1).
2.13. RNA-sequencing
We have previously characterized the mRNA transcriptome of differentiating iMAC cultures by RNA-sequencing (RNA-seq) [23]. Normalized gene counts for paired-end reads were obtained using a standard RNA-seq pipeline [33] in which MAPRSeq was used to obtain expression values for each gene that normalized to 1 million reads and corrected for gene length (Fragments of reads Per Kilobase pair per Million mapped reads, FPKM). This data set has been deposited in the Gene Expression Omnibus (GEO) database (Accession # GSE97118). We accessed control samples from days three, seven, and fourteen to characterize expression of key chondrogenic genes.
2.14. +JQ1 treatment
The compound +JQ1 (Cayman Chemicals, Ann Arbor, MI) was dissolved in DMSO and added to culture media at indicated time points. Vehicle control groups were supplemented with equivalent concentration of DMSO when appropriate.
2.15. siRNA-mediated knockdown of Brd4
ATDC5s were plated at 10,000 cells/cm2 in DMEM media (Gibco, 4.5g/L D-Glucose, 4.5g/L L-Glutamine, no Sodium Pyruvate) supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals), 100U/ml Penicillin (Gibco), and 100µg/ml Streptomycin (Gibco). The next day, cells were transfected with siRNAs using Lipofectamine RNAiMAX (Thermo Fisher) transfection reagent in standard MEM alpha media. Dharmacon smart-pool siRNAs (GE Healthcare, Chicago, IL) targeting Brd4 (Cat#: L-041493–00) as well as non-targeting control (Cat#: D-001810–10-20) were used at a final concentration of 20 nM. Cells were cultured in the presence of siRNAs overnight. The next day, ATDC5s were trypsinized and formed into micromasses as described above. Two days after formation of micromasses, chondrogenic differentiation cocktail [final concentration 50µg/ml ascorbic acid (Sigma), 10mM β-glycerol phosphate (Sigma), and 1X Insulin-Transferrin-Selenium (Gibco)] was added to the cells. Media was changed every three days for the remainder of the experiment.
2.16. Western blotting
Western blotting was performed as previously described [25]. Cells were lysed in RIPA buffer at indicated time points while E14.5 limb buds were lysed in 3% SDS and 125 mM Tris-HCl (pH = 6.8) using plastic pestles (Grainger Cat# 6HAY3). Proteins were visualized using SuperSignal West Femto Reagent (Thermo Fischer Scientific) and imaged using the ChemiDoc Imaging System (Bio-Rad). Antibodies and concentrations used for western blotting were as follows: Brd4 (Bethyl Labs: A301–985A, 1:2,000) and Gapdh (Cell Signaling: 51745, 1:5,000).
2.17. MTS Assay
MTS assay (Promega) was assessed according to the manufacturer’s protocol at indicated time points. Enzymatic activity was calculated from absorbance at 490nm determined using a SpectraMAX Plus spectrophotometer (Molecular Devices, San Jose, CA).
2.18. Alcian Blue Staining
At indicated time points, media was removed, and cells were washed with PBS. Cells were then fixed in 10% NBF for ten minutes. NBF was removed, cells were washed with H2O, and stained with 0.5% Alcian Blue Stain (Alcian Blue 8GX dye, Sigma) for two hours. Stain was removed and wells were washed two times with 100% ethanol. Lastly, wells were washed twice with H2O. Plates were scanned and staining was quantified by ImageJ software [47].
2.19. Alizarin Red Staining
At indicated time points, media was removed, and cells were washed with PBS. Cells were then fixed in 10% NBF. After one hour, NBF was removed, cells were washed with PBS and stained with 2% Alizarin Red (Thermo Fisher) for ten minutes. Stain was removed and wells were washed five times with H2O. Plates were scanned and staining was quantified by ImageJ software [47].
2.20. Statistics
Where appropriate, data are plotted as boxplots indicating the median, interquartile range, and minimum and maximum values in each dataset. Individual measurements are represented by a single data point within the figures. When comparing two groups, statistical analysis was performed using an unpaired, two-tailed Student’s t-test. In the case of multiple groups, an analysis of variance (ANOVA) test was performed. If the ANOVA p-value was less than 0.05, subsequent pairwise comparisons were performed using a Tukey multiple comparisons test. The in vivo study outcomes were compared between the two or three experimental groups (CON, HET, and cKO) using unpaired Student’s t-test (two groups) or one-factor ANOVA (three groups). Separate analyses were performed for each outcome. When the overall ANOVA p-value was less than 0.05, pairwise comparisons were performed between the experimental groups using a Tukey multiple comparisons test. The overall type-I error rate was set at α = 0.05. Significance is noted in the figures.
3. RESULTS
3.1. Mesenchymal loss of Brd4 causes a reduction in overall body size
To understand the role of acetylation and acetyl recognition in skeletal development, we investigated the contribution of Brd4 during endochondral ossification in vivo. Whole-body knockout of Brd4 has been shown to be embryonic lethal [48]. Therefore, we utilized the Prrx1-Cre driver system to eliminate Brd4 expression in the limb bud [45]. Crossing of Prrx1-Cre mice with Brd4 transgene mice (Brd4f l/f l) [44] generated control (CON, Brd4wt/wt: Prrx1-Cre), heterozygous (HET, Brd4wt/f l: Prrx1-Cre) and conditional knockout (cKO, Brd4f l/f l: Prrx1-Cre) animals. Brd4 cKO mice are viable at birth but are substantially smaller in size. The small overall stature is evident at three (Fig. 1A&B) and sixteen (Fig. 1C&D) weeks of age. Specifically, photographic imaging of gross anatomy and X-ray assessment reveals shortened long bones of front and hind limbs. In support, whole mount staining of front legs further reveals the shortened long bones in one day old Brd4 cKO pups (Fig. 1E). The growth impairment is also quantified by a significant reduction in overall body weight at several time points (Fig. 1F). It is important to note that the HET mice showed no significant phenotypic changes when compared to control mice. Together, these initial observations demonstrate that Brd4 is required for proper skeletal formation.
Figure 1. Mesenchymal loss of Brd4 results in smaller mice.
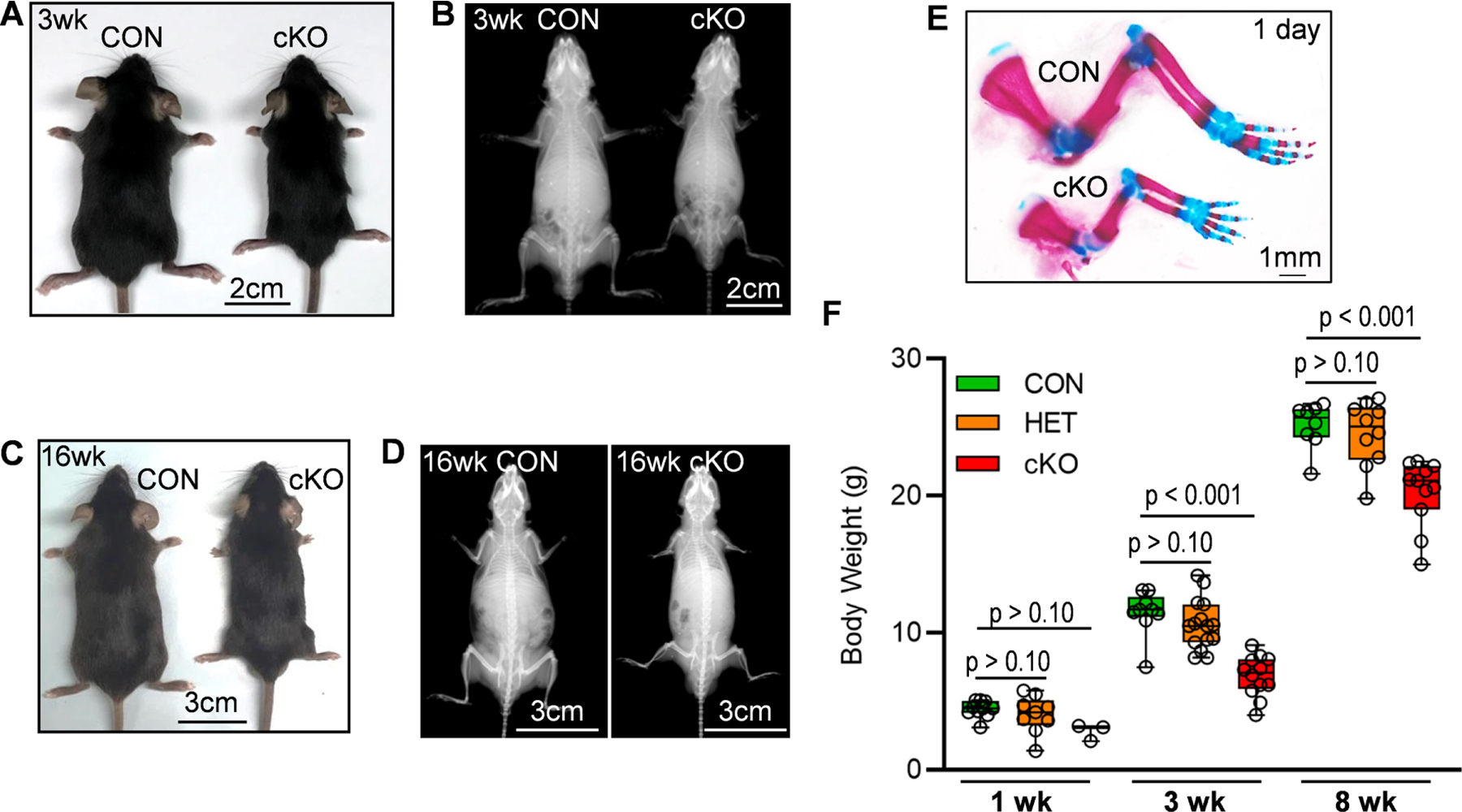
Brd4 CON, (Brd4wt/wt: Prrx1-Cre), HET (Brd4wt/f l: Prrx1-Cre), and cKO (Brd4f l/f l: Prrx1-Cre) mice were assessed at several postnatal time points. Photograph (A) and X-ray radiograph (B) of three week old male mice. Photograph (C) and X-ray (D) of sixteen week old male mice. Whole mount staining (front legs) of one day old male pups (E). Total body weight of male mice at one, three, and eight weeks (F). Boxplots indicate the median, interquartile range, and the minimum and maximum value in each dataset. Individual mice are represented by a single point on the graph (1 Week: CON (n = 10), HET (9), cKO (n = 3); 3 Weeks: CON (n = 9), HET (15), cKO (n = 12); 8 Weeks: CON (n = 8), HET (n = 10), cKO (n = 11). A one-way ANOVA was performed across all groups in panel F and demonstrated significant variation within the dataset (p < 0.001). A follow-up multiple comparisons test was performed using the Tukey method (correct for multiple comparisons) to compare the means of each group one-to-one. P-values shown in panel E represent the results from the comparison between the indicated group and the CON group at the same age.
To assess the recombination efficiency of the Cre-LoxP system in this mouse model, conditional ablation of Brd4 was assessed on limb buds of 14.5 dpc embryos. Quantitative PCR reveal robust and significant recombination of Brd4 alleles in the limb buds of cKO mice as quantified by primer pairs targeting the floxed site within exon 3 (Primer Set #1, 86% reduction; Primer Set #2, 82% reduction) (Fig. 2A). Primer pairs targeting exon 6 of Brd4 were used for normalization. As a result of recombination and removal of the ATG translational start site of Brd4 [49], we also note a significant reduction (82%) of Brd4 protein in the limb buds of 14.5 dpc embryos (Fig. 2B&C). These data indicate that genetic recombination occurs in this model and results in a reduction in Brd4 protein levels within the limb bud.
Figure 2. Efficient deletion of Brd4 by Prrx1-Cre recombinase.
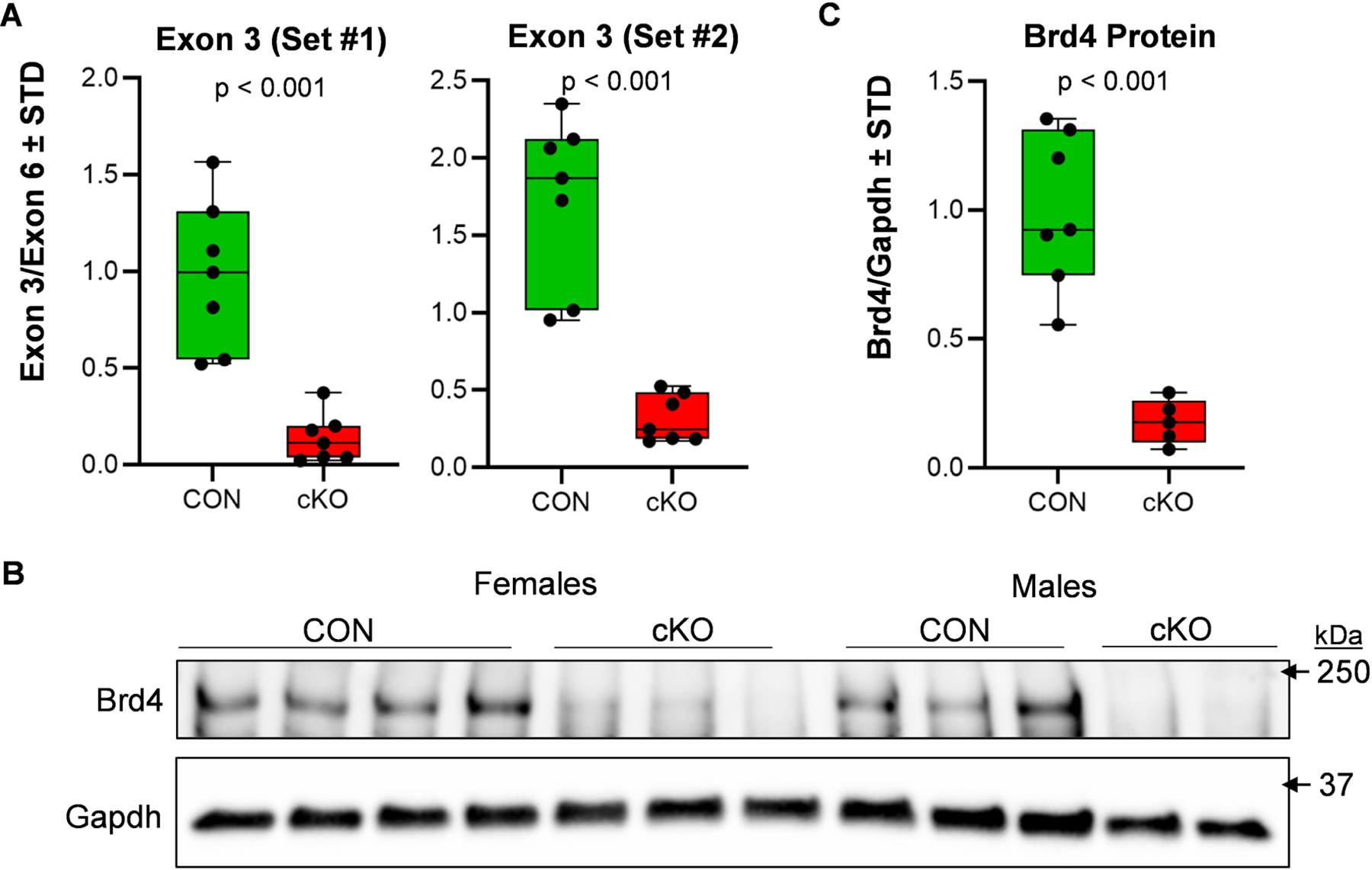
Limb buds were collected from CON (Brd4f l/wt and Brd4f l/f l: Prrx1-Cre negative) and cKO (Brd4f l/f l: Prrx1-Cre positive) 14.5 dpc embryos. Quantitative PCR analysis was conducted on DNA isolated from the limb buds. Both male and female embryos were included in the analysis. Deletion of exon three was assessed using two distinct primer pairs targeting the floxed region within exon 3 and evaluated in relation to non-targeted exon six (n = 7) (A). Western blotting for Brd4 in limb bud tissues of 14.5 dpc embryos (B) and quantification when normalized to Gapdh (n = 7 and 5) (C). Boxplots indicate the median, interquartile range, and the minimum and maximum value in each dataset. Individual mice are represented by a single point on the graph. P-values shown on the graphs represent the results of an unpaired, two-tailed t-test between CON and cKO.
3.2. Mesenchymal ablation of Brd4 impairs long bone development
To further assess long bone phenotypes, femora from three week old female mice were assessed by X-ray analysis (Fig. 3A&B) and microcomputed tomography (Fig. 3C&D). Strikingly, the femora of cKO animals are ~40% shorter than those of the CON mice. Furthermore, µCT analysis reveals an increase in bone volume fraction (BV/TV) and total mineral density (TMD) of trabecular bone in cKO mice (Suppl. Fig. 1A). We also examined trabecular bone parameters of L5 vertebrae from three week old mice. While trabecular bone parameters are retained, Brd4 depletion results in shorted vertebrae (Suppl. Fig. 1B). To gain a better understanding of how Brd4 loss affects the development of long bones over time, we collected femora from male CON, HET, and cKO mice at one, three, and eight weeks of age and assessed bone length by X-ray analysis and linear regression (Fig. 3E). The femora of cKO mice exhibit a significantly reduced growth rate between one and three weeks of age. Interestingly, the growth rate in cKO femora is similar to that of the CON group between three and eight weeks of age. Collectively, these results indicate that the detrimental effects of Brd4 loss are most prominent during rapid long bone growth and skeletal alterations persist at skeletal maturity.
Figure 3. Long bone alterations in mice lacking mesenchymal Brd4 expression.
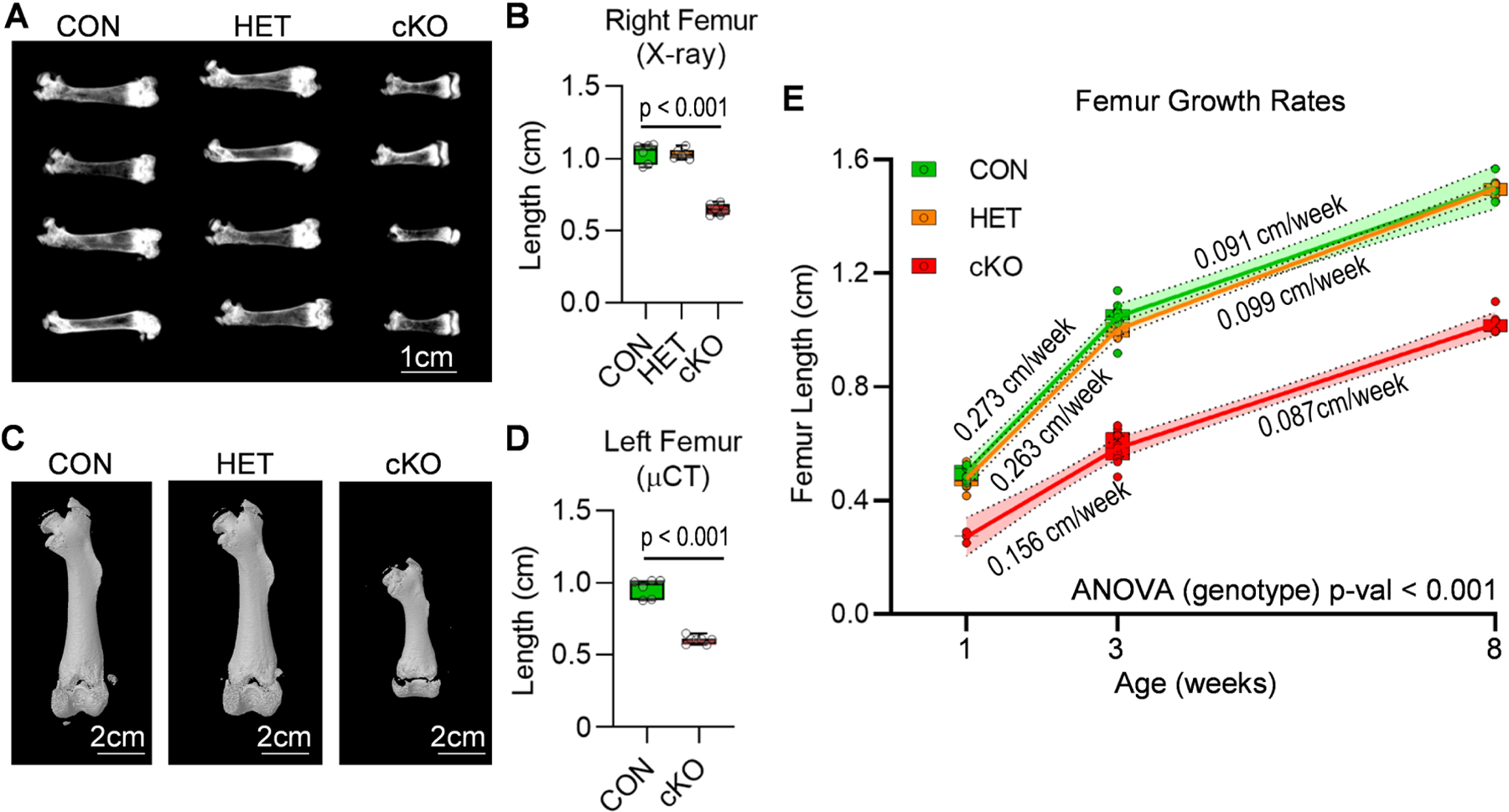
CON (Brd4wt/wt: Prrx1-Cre), HET (Brd4wt/f l: Prrx1-Cre), and cKO (Brd4f l/f l: Prrx1-Cre) female mice were assessed by X-ray and µCT analysis. X-ray radiographs (A) and length quantification (n = 6) (B) of femora collected from three week old female mice. Micro-computed tomography (µCT) reconstructions (C) and length quantification (n = 6 and 7) (D) of femora collected from three week old female mice. Measurements of femora lengths obtained from X-ray images from one, three, and eight week old male mice (1 Week: CON (n = 7), HET (n = 7), cKO (n = 3); 3 Weeks: CON (n = 9), HET (n = 7), cKO (n = 9); 8 Weeks: CON (n = 3), HET (7), cKO (n = 7). (E). Linear regression analysis was performed for each individual line segment (1 to 3 weeks and 3 to 8 weeks). The slope of the line (growth rate) is indicated on the graph, and the 95% confidence interval is indicated by the shaded region. Boxplots indicate the median, interquartile range, and the minimum and maximum value in each dataset. Individual mice are represented by a single point on the graph. P-value shown in panel B represents the results of a one-way ANOVA followed by a multiple comparisons test that was performed using the Tukey method (correct for multiple comparisons) to compare the means of the cKO and CON groups. P-value shown in panel D represent the results from an unpaired, two-tailed t-test between CON and cKO. For panel E, a two-way ANOVA was performed across the three groups. The time variable accounted for 73.36% of the variation observed (p < 0.001). The genotype variable (reported on the graph) accounted for 18.08% of the variation observed (p < 0.001).
3.3. Mice lacking Brd4 in the mesenchyme exhibit alterations in endochondral ossification
The non-mineralized surfaces within the growth plate region of femora appear disproportionally larger in Brd4 cKO mice when compared to CON mice (Fig. 4A). To further investigate this observation, the depth of non-mineralized area within growth plate was measured at fifteen cross sections throughout coronal and sagittal planes within the growth plate area (Fig. 4E). An increase in the depth of non-mineralized surface within the growth plate in cKO mice is noted (Fig. 4B), which is further exacerbated when accounting for difference in femur lengths (Fig. 4C). Brd4 cKO mice also exhibit a significant reduction in secondary ossification center (SOC) area (Fig. 4D). To investigate individual growth plate zones, trichrome staining was conducted on right femora collected from one and three week old male mice. Abnormal growth plate morphology is observed at one (Fig. 4F and Suppl. Fig. 2A) and three (Suppl. Fig. 2B) weeks of age in the cKO mice. Similar to the µCT-analysis, we observed that SOC formation is severely delayed in the cKO mice when compared to CON and HET mice. At one week of age, the SOC is completely absent in the cKO mice. A significant reduction is observed in proliferative and hypertrophic zones of cKO mice (Fig. 4G&H). Together, these data demonstrate that alterations in the physis in cKO mice may result from impaired progression of growth plate chondrocytes from the resting phase into proliferative and hypertrophic states.
Figure 4. Growth plate alterations in mice lacking mesenchymal Brd4 expression.
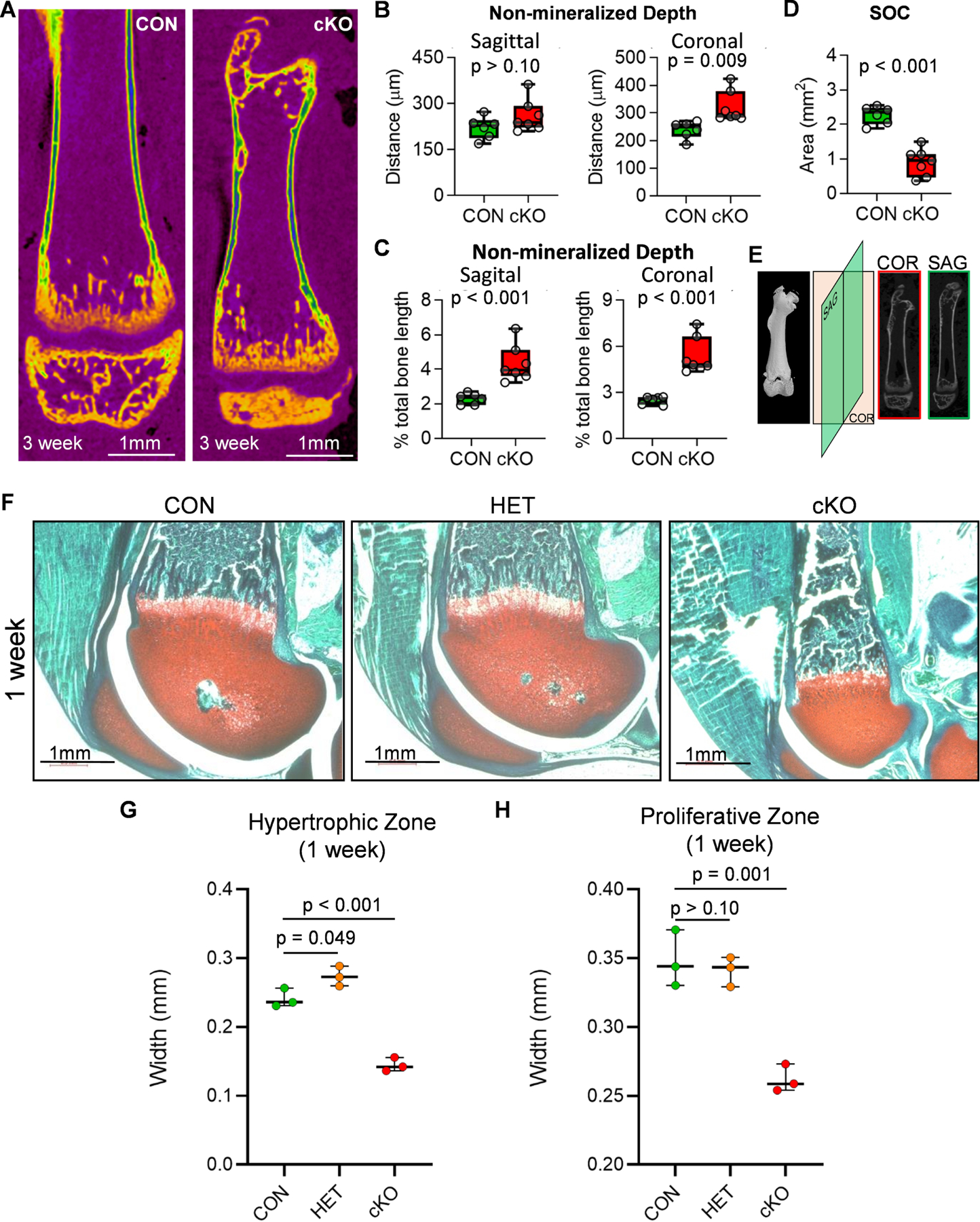
Distal femora of three week old CON (Brd4wt/wt: Prrx1-Cre) and cKO (Brd4f l/f l: Prrx1-Cre) female mice were assessed by µCT analysis (n = 6 and 7) (A-E). µCT reconstructions of femora collected from three week old female mice (A). Non-mineralized surface within the growth plate area measured throughout the sagittal (left) and coronal (right) planes (B). Each point represents an average of fifteen measurements taken for each mouse femur. Quantification of non-mineralized surface within growth plate area as a percentage of total femur length obtained from µCT images (C). Area of secondary ossification center (SOC) determined from medial slice of µCT images (D). Schematic depicting orientation of cross sections used to measure non-mineralized surface within the growth plate area of distal femora (E). Trichrome staining of distal femora derived from one week old CON (Brd4wt/wt: Prrx1-Cre), HET (Brd4wt/f l: Prrx1-Cre), and cKO (Brd4f l/f l: Prrx1-Cre) male mice (F). Red = cartilage, blue/green = bone, brown = nuclei. Measurements of hypertrophic (G) and proliferative (H) growth plate zones from trichrome stained sections of one week old male femora (n = 3). Individual measurements are represented by a single point on the graph for each mouse. Boxplots indicate the median, interquartile range, and the minimum and maximum value in each dataset. P-values shown in panels B-D represent the results from unpaired, two-tailed Student’s t-test between CON and cKO groups on each graph. P-values shown in panels G and H represent the results of a one-way ANOVA followed by a multiple comparisons test performed using the Tukey method (correct for multiple comparisons) to compare the means of the indicated groups.
3.4. Transcriptional profiling of immature mouse articular chondrocytes (iMACs) during chondrogenic differentiation
To further investigate the impact of Brd4 loss on chondrocyte differentiation, we utilized immature mouse articular chondrocytes (iMACs) as an ex vivo model system for chondrocyte maturation and hypertrophy. We first assessed the gene expression profile of iMACs using both real-time quantitative PCR (RT-qPCR) and RNA-sequencing (RNA-seq) to ensure these cells recapitulate chondrocyte maturation. iMACs were collected from five day-old wild-type C57BL/6J mice and cultured in three-dimensional micromass (3D-µmass) conditions. During a 14-day chondrogenic differentiation time course, a reduction in early chondrogenic transcription factors (Sox5, Sox6, and Sox9) and an increase in hypertrophic transcription factors (Runx2 and Osx/Sp7) is observed by both RNA-seq (Suppl. Fig. 3A) and RT-qPCR (Suppl. Fig. 3B). In addition, a significant reduction in the expression of early chondrogenic markers (Col2a1, Acan, and Comp) is observed over time by RNA-seq (Suppl. Fig. 3C) and RT-qPCR (Suppl. Fig. 3D). Expression patterns of genes associated with late-stage/hypertrophic chondrocytes (Col10a1, Mmp13, and Ihh) were also evaluated. An increase in these late-stage chondrocyte markers is observed by both RNA-seq (Suppl. Fig. 3E) and RT-qPCR (Suppl. Fig. 3F) over time. Lastly, we analyzed expression levels of genes associated with osteogenic differentiation (Bglap, Ibsp, and Col1a1), because these markers are elevated in the end-stages of hypertrophic chondrogenesis. Similar to hypertrophic chondrocyte markers, increased expression of these markers is observed over the differentiation time course by RNA-seq (Suppl. Fig. 3G) and RT-qPCR (Suppl. Fig. 3H). In sum, transcriptional profiling indicates that iMACs are transitioning from an early-stage chondrogenic phenotype to a more mature/hypertrophic state over the differentiation time course.
3.5. Inhibition of Brd4 with the small molecule +JQ1 prevents maturation of iMACs
To understand the contribution of Brd4 on chondrocyte hypertrophy, we exposed cells to +JQ1, a small molecule inhibitor that binds competitively to bromo-domains and prevents BET proteins from binding acetylated lysine residues [50], at different time points during chondrogenic differentiation of iMACs (Fig. 5A&B). As in the iMAC validation experiments (Suppl. Fig. 3), a decrease in expression over time of early chondrogenic transcription factors (Fig. 5C) and matrix proteins (Fig. 5D) is observed. Short-term treatment with +JQ1 (100 nM) for the first three days of differentiation had little effect on the expression of these early chondrogenic markers (Fig 5C&D, blue squares). Continuous (Fig. 5C&D, red triangle) and late administration (Fig. 5C&D, green diamond) had a more pronounced effect but may be under-appreciated as expression of these genes is suppressed by the differentiation process itself. Interestingly, the effects of Brd4 inhibition are more pronounced on the expression levels of late stage chondrogenic transcription factors (Fig. 5E) and genes that are typically up-regulated during the differentiation process of iMACs (Fig. 5F&G). Continuous treatment with +JQ1 severely impaired activation of the late stage chondrocyte markers (Fig. 5E–G, red triangle), and late-stage treatment has an acutely significant effect on gene expression (Fig. 5E–G, green diamond). For statistical evaluation of the impact of +JQ1 treatment throughout the chondrogenic differentiation time course, a mixed-effects model (REML) analysis was performed for each graph (alpha = 0.05). The p-value reported for the fixed effects was then assessed. For all datasets, the time variable had a statistically significant impact with a p-value < 0.001. The p-value calculated for the “treatment group” variable is reported on each graph. Interestingly, the “treatment” variable was found to have a significant impact in most genes that demonstrated increased expression throughout the time course. A follow-up multiple comparisons test was performed using the Tukey method (correct for multiple comparisons) to provide a comparison of each treatment group (Suppl. Table 2). These in vitro results support the in vivo observation that Brd4 is important for chondrocyte maturation and hypertrophy. Interestingly, we also note that Brd4 inhibition in later stages of maturation can drastically alter gene expression of maturing chondrocytes.
Figure 5. +JQ1 treatment prevents maturation of iMACs.
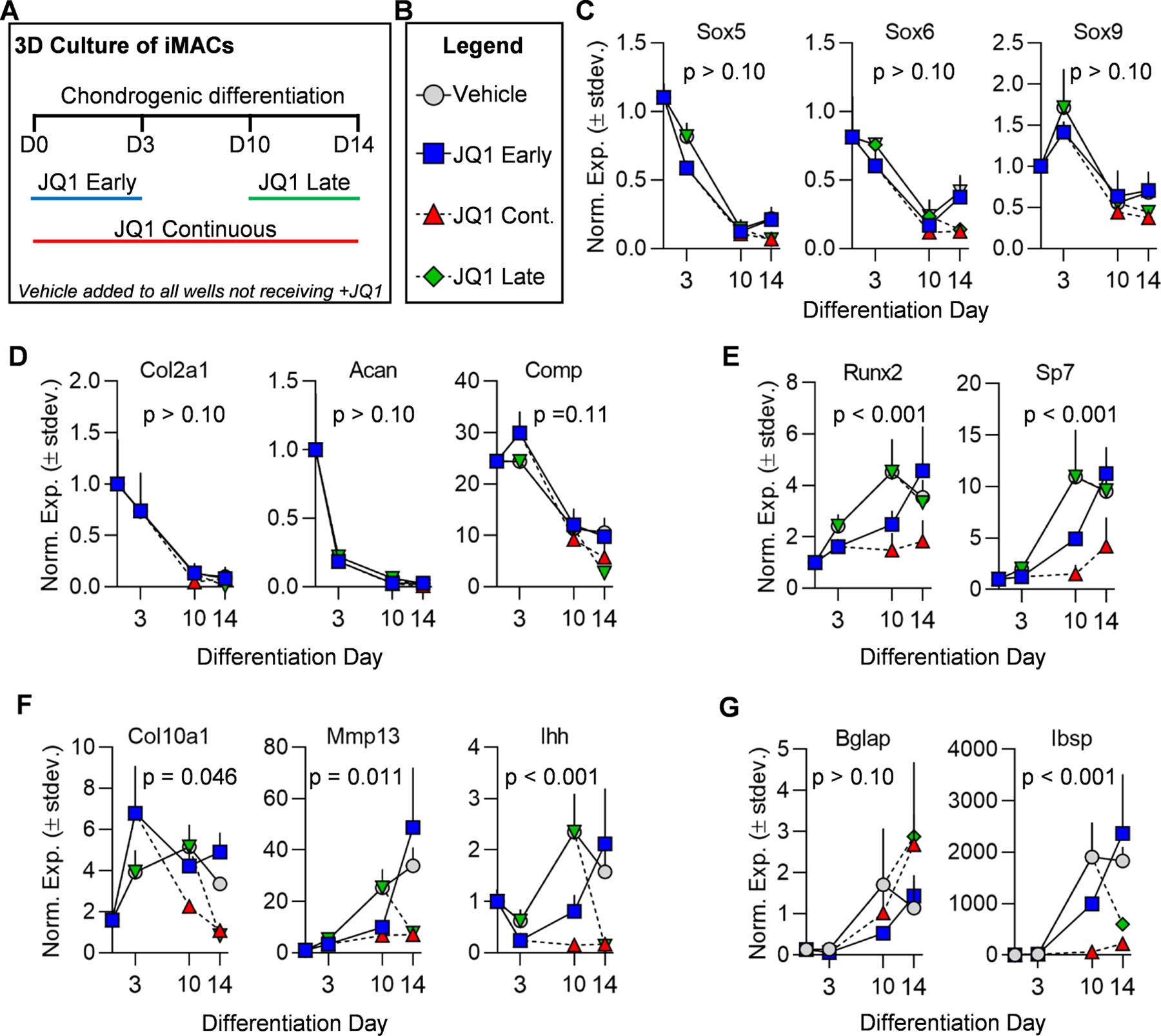
iMACs were collected from five day-old wild-type C57BL/6J mice and cultured in three-dimensional micromass (3D-µmass) conditions in chondrogenic differentiation media. Schematic of treatment regimen with vehicle (DMSO) control or 100nm +JQ1 (A). Legend indicating treatment groups in subsequent graphs (B). RT-qPCR analysis of chondrogenic transcription factors (C), early chondrogenic markers (D), hypertrophic/osteogenic transcription factors (E), hypertrophic markers (F), and hypertrophic/osteogenic markers (G). Experiments were performed in n = 3 biological replicates and the mean is indicated by each point on the graph. Error bars represent standard deviation and statistical significance is indicated in each panel. A mixed-effects model (REML) analysis was performed for each graph (alpha = 0.05). The p-value reported for the fixed effects was then assessed. For all datasets, the time variable had a statistically significant impact with a p-value < 0.001. The p-value calculated for the “treatment group” variable is reported on each graph.
3.6. Knockdown of Brd4 suppresses expression of key transcription factors in ATDC5 cells
Because +JQ1 inhibits other BET family members, we employed genetic based approaches to assess Brd4 function in chondrocytes. We initially performed 3D studies in which Brd4f l/f l iMACs were transduced with adenoviral vectors containing GFP (Ad-GFP) or Cre recombinase (Ad-Cre). These studies were limited in interpretation because even high Ad-Cre titers (multiplicity of infection = 1,000) only modestly reduce Brd4 protein levels (data not shown). We thus employed siRNA-mediated knockdown of Brd4 in ATDC5 cells, a well-established in vitro model of chondrogenic differentiation. Similar to iMACs, ATDC5 cells were assessed in the presence of chondrogenic medium in a 3D-µmass system (Fig. 6A). At day two post-transfection, robust knockdown of Brd4 at both the protein (Fig. 6B) and mRNA (Fig. 6C) levels is noted while cell viability as measured by MTS activity assay (Fig. 6D) and expression of cell proliferation markers (Fig. 6E) is not affected by Brd4 loss. Of interest, mRNA expression levels of key chondrogenic transcription factors, including Sox5, Sox9, and Sp7, are significantly reduced two days after the initial transfection of Brd4 siRNA (Fig. 6F). Taken together, these findings suggest that loss of Brd4 may disrupt early activation of the chondrogenic transcriptional program.
Figure 6. Depletion of Brd4 prevents early transcription factor expression in ATDC5 cells.
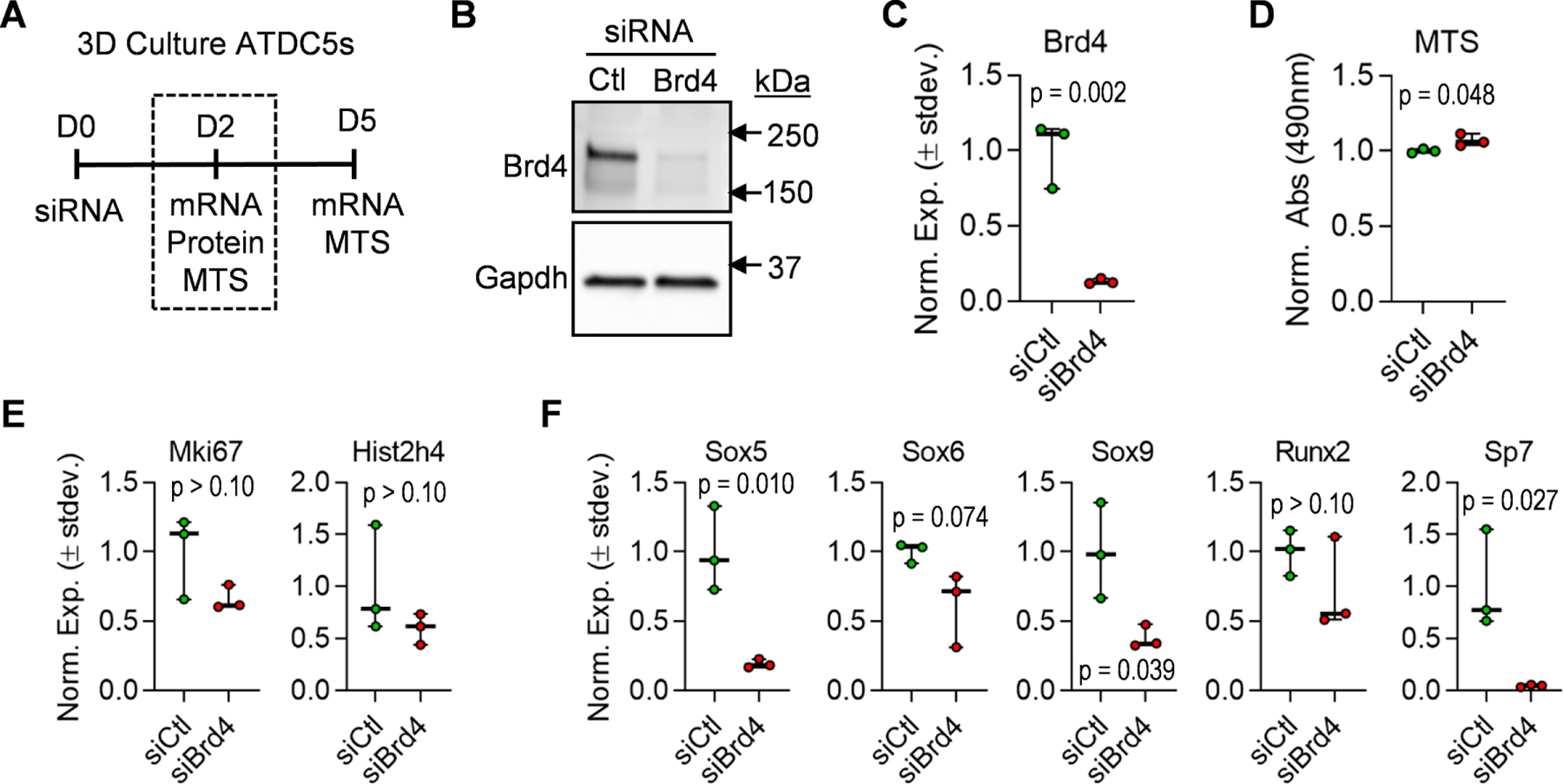
ATDC5 cells were transfected with non-targeting siRNA (siCtl) or Brd4-targeting siRNA (siBrd4) and cultured in three-dimensional micromass cultures in chondrogenic differentiation media. Schematic of siRNA transfection experiment and data collection points (A). Western blot analysis performed on protein lysates collected two days after transfection (B). RT-qPCR analysis on mRNA isolated two days post-transfection (C). MTS assay conducted two days after transfection (D). RT-qPCR analysis of cell cycle genes (E) and transcription factors performed on mRNA isolated two days post-transfection. Boxplots indicate the median, interquartile range, and the minimum and maximum value in each dataset (n = 3). Individual biological replicates are represented by a single point on the graph. P-values indicate results from unpaired, two-tailed Student’s t-test between siCtl and siBrd4 groups.
3.7. Knockdown of Brd4 prevents maturation of ATDC5 cells
To understand the downstream impact of Brd4 knockdown, we assessed the differentiated state of ATDC5 cells five days after transfection (Fig. 7A). Except for Sp7/Osx and in contrast to cells transfected for just two days (Fig. 6F), expression levels of transcription factors involved in chondrogenesis are similar between control and Brd4-targeting siRNAs (Fig. 7B). These data suggest that the early effects of Brd4 loss on transcription factor gene expression levels are not permanent, likely due to the diminishing effects of Brd4 targeting siRNA five days post-transfection (compare Fig. 6C to Fig. 7C). However, a significant impact on early chondrocyte markers (Col2a1, Acan, Comp) is observed in differentiating ATDC5 cells (Fig. 7D). We also demonstrate a reduction in the expression of Mmp13, a late-stage hypertrophic marker (Fig. 7E). Importantly, knockdown of Brd4 and the subsequent changes in gene expression resulted in a significant reduction in glycosaminoglycan content indicated by Alcian Blue staining (Fig. 7F) as well as a decrease in matrix mineralization as evidenced by a reduction in Alizarin Red staining (Fig. 7G). Together, these results indicate that Brd4 is required for activation of the early chondrogenic transcriptional network necessary for driving chondrocyte maturation and extracellular matrix production.
Figure 7. Brd4 knock-down prevents maturation of ATDC5 cells.
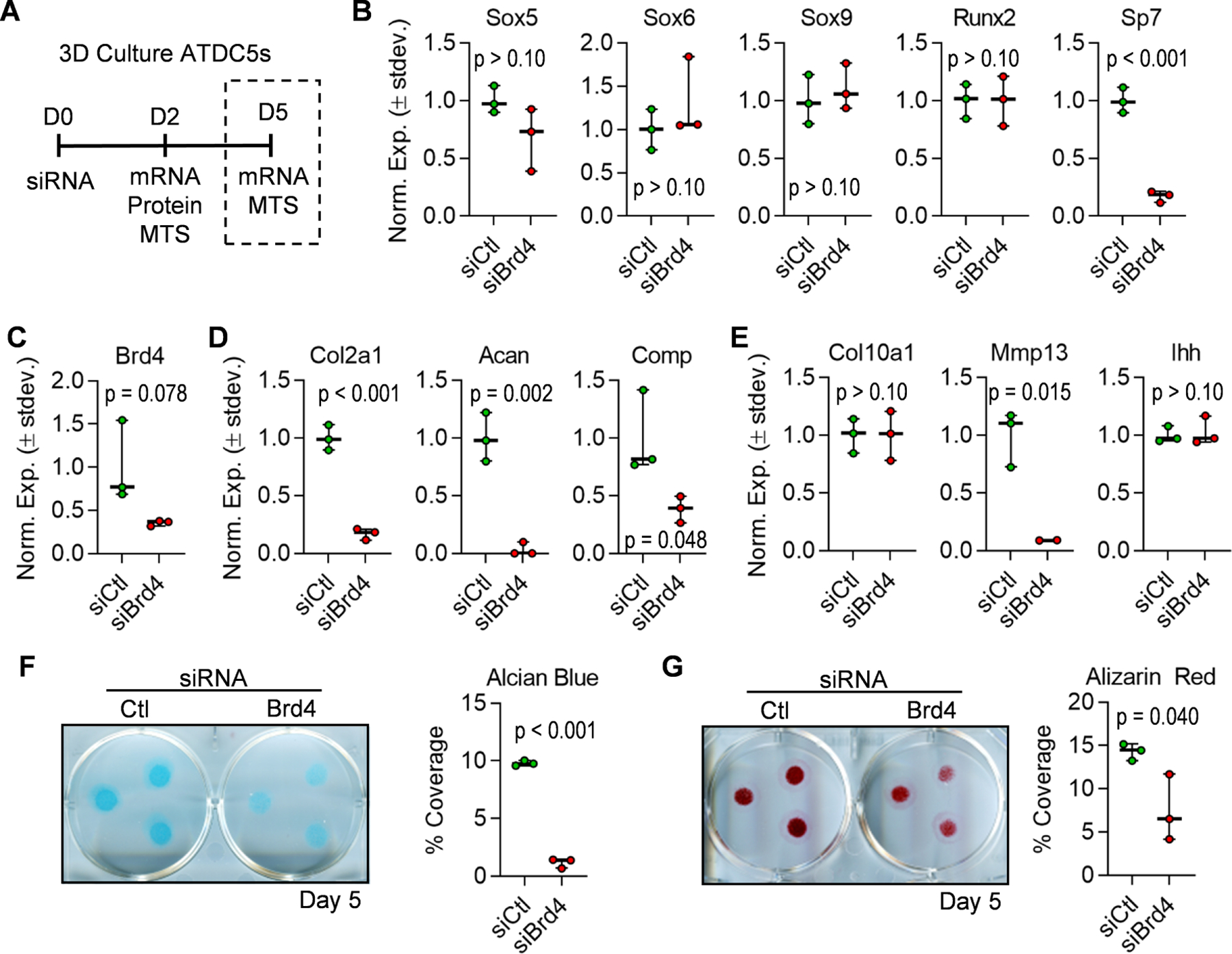
ATDC5 cells were transfected with non-targeting siRNA (siCtl) or Brd4-targeting siRNA (siBrd4) and cultured in three-dimensional micromass cultures in chondrogenic differentiation media. Schematic of siRNA transfection experiment and data collection points (A). RT-qPCR analysis of transcription factors (B), Brd4 (C), early chondrogenic genes (D), and hypertrophic genes (E) performed on mRNA isolated five days post-transfection. Alcian Blue staining and quantification performed five days after induction of chondrogenic differentiation (F). Alizarin red staining and quantification performed five days after induction of chondrogenic differentiation (G). For quantification of staining, percent coverage of a 1.13cm2 circle was measured for each micromass (n = 3 micromasses per well). Boxplots indicate the median, interquartile range, and the minimum and maximum value in each dataset (n = 3). Individual biological replicates are represented by a single point on the graph. P-values indicate results from unpaired, two-tailed Student’s t-test between siCtl and siBrd4 groups.
4. DISCUSSION
In this study, we establish that Brd4 is required for proper skeletal development in vivo. This conclusion is based on a transgenic mouse model in which Brd4 was conditionally ablated in the mesenchyme (Brd4f l/f l: Prrx1-Cre) [44, 45]. DNA assessment by PCR analysis reveals a significant reduction (82 to 85%) in the floxed region of the Brd4 transgene in limb buds. These findings are supported by the observation that the Prrx1-Cre reporter cassette is active within the limb bud at 9.5 dpc [45]. In this transgenic model, the floxed region lies within the region of exon 3 containing the ATG start codon. As such, we observe a significant reduction in Brd4 protein levels in limb buds of the Brd4f l/f l: Prrx1-Cre mice.
It is immediately apparent that the Brd4 knockout mice are smaller and have strikingly shorter limbs when compared to control mice. To better understand the cause of these skeletal abnormalities, we conducted X-ray and µCT imaging, whole mount staining, in vivo histological analyses, and ex vivo cell culture experiments. Assessment of long bone development using femora of three week old mice revealed a significant reduction in femora length upon Brd4 loss in the mesenchyme. It has been established that Prxx1-Cre is mainly restricted to the mesenchyme and a subset of the craniofacial mesenchyme [45]. While our assessment revealed a shortening of vertebrae by ~11% at three weeks of age upon Brd4 depletion, the long bones are reduced by ~44% in the same cohorts. Corroborating this finding, a drastic difference in long bones are observed at skeletal maturity (16 weeks) while the body length of the mice is very similar between controls and Brd4 depleted mice. Interestingly, long bone cortical thickness remains unchanged, but an increase in trabecular parameters including BV/TV and trabecular TMD is observed in cKO mice, which may be an area of research worth exploring in the future. Similar skeletal patterning is observed clinically in patients diagnosed with Schmid Metaphyseal Chondrodysplasia [51, 52]. In these patients, craniofacial patterning and overall bone quality remains normal, but the long bones fail to develop to full length. Interestingly, a subtype of Metaphyseal Chondrodysplasia, Metaphyseal Dysplasia with Maxillary Hypoplasia with or without Brachydactyly (MDMHB) [53], has been linked to a gain of function mutation in Runx2 [54–56].
We also assessed the growth rate of femora in cKO mice at one, three, and eight weeks of age. A significant reduction in the rate of growth is observed between one and three weeks of age, a time in development when elongation of long bones occurs at a rapid rate. However, there is no appreciable difference in growth rates in the cKO mice between weeks three and eight, a time of slower bone elongation. This phenotypic finding also recapitulates the Schmid Metaphyseal Chondrodysplasia phenotype as patients often appear asymptomatic until the age of two when long bones begin to grow rapidly [52]. These findings suggest that Brd4 knockout is most detrimental to the process of endochondral ossification during times of rapid growth when proper growth plate function is critical.
These observations of reduced overall length and growth rate reduction in femora indicated growth plate dysfunction in the Brd4 knockout animals. To further investigate, we performed histological and µCT analysis on distal femora with a specific focus on the growth plate region. In doing so, we observe a significant difference in the gross anatomy of the growth plate region in the cKO animals characterized by an enlarged resting chondrocyte region, narrowed proliferative and hypertrophic zones, and delayed formation of the secondary ossification center. This abnormal growth plate phenotype indicates that ablation of Brd4 in the mesenchyme alters the ability of chondrocytes to progress through the maturation process required for proper endochondral ossification. In support, whole mount staining of one day old pups reveals a significant long bone phenotype in Brd4 cKO mice. Thus, the skeletal changes are induced before birth and the observed phenotype in growing mice is an accumulation of deficiencies that are started in utero.
To examine the role of Brd4 during chondrogenic differentiation, we used both ex vivo (iMACs) and in vitro (ATDC5) 3D chondrogenic cell culture models in which Brd4 function was reduced using BET inhibitor, transient RNA silencing and/or Cre recombination of the Brd4 cKO alleles. Each of these approaches has specific technical limitations unique to each culture model and interpretations are dependent on the biological context of each model (e.g., primary chondrocytes versus teratoma derived cells). Yet, within these constraints, our experiments provide interpretable data that increases our understanding of Brd4 function. In both the iMAC and ATDC5 systems, we observed that addition of the BET inhibitor +JQ1 or the addition of Brd4-targeting siRNAs significantly impaired chondrogenic gene expression and differentiation. Specifically, in the iMAC cell culture model, we note a significant reduction in Sox9 target genes after BET protein inhibition. Of note, differentiation of chondrocytes from mesenchymal progenitors is driven by upregulated expression of the transcription factor Sox9 [57]. To further validate drug-based studies, we utilized the ATDC5 cell line, an established model of chondrogenic differentiation that is derived from mouse teratomas [58]. siRNA-mediated knockdown of Brd4 results in a significant reduction in Sox9 expression early in the differentiation time course that is accompanied by a reduction in Sox9 target genes Col2a1 and Acan [59]. The reduction in expression of Sox9 and its downstream targets suggests an inability to activate the chondrogenic-specific transcriptional network in the absence of Brd4 activity. These observations support previous findings which have shown that Brd4 inhibition alters Sox9 expression levels and protein stability [60]. Additionally, previous Sox9 transgenic mouse models have demonstrated severe effects on skeletal development [61]. Although the phenotype in the Brd4 knockout model is not as severe, these studies collectively suggest a regulatory relationship between Brd4 and Sox9 that remains to be further explored. In addition, cooperation of Runx2 and Brd4 to activate target genes has been previously demonstrated by our group and others in the context of osteogenic differentiation [33, 35]. The present study supports these previous findings and suggest a Brd4-Runx2 interaction. Of note, we observe additional evidence for the interaction of Brd4 and Runx2 in the +JQ1 iMAC ex vivo experiments as the expression of Runx2 target genes Mmp13, Ihh, and Ibsp [62] are markedly reduced with +JQ1 treatment.
Several groups have demonstrated that Brd4 interacts with numerous other lineage-specific transcription factors to activate the transcriptional networks required to drive adipogenesis and myogenesis [44, 63, 64]. Together, our findings in chondrocytes support these previous observations that Brd4 interacts with lineage-specific transcription factors. Specific to chondrocyte differentiation and hypertrophy, our current and recently published studies [33] suggest a model in which Brd4 supports Sox9 expression to facilitate the expression of early chondrogenic genes and this epigenetic regulator may also interacts with Runx2 during later stages of maturation to activate hypertrophic genes [33, 35].
The phenotype observed in the Brd4 cKO mice is consistent with our in vitro observation that loss of Brd4 delays maturation of chondrocytes. In the developing long bones, rapid chondrocyte proliferation, maturation, and hypertrophy are crucial to facilitate proper endochondral ossification. When Brd4 is ablated in the mesenchyme, chondrocytes in cKO mice can no longer effectively progress through the developmental stages of hypertrophy resulting in impaired endochondral ossification and ultimately shorter long bones. Our studies show that Brd4 is a key regulator of chondrogenic differentiation and hypertrophy and that loss of Brd4 in the mesenchymal lineage results in abnormal long bone development. This phenotype recapitulates some of the skeletal abnormalities observed in human clinical cases of Metaphyseal Chondrodysplasias.
BET protein inhibitors are gaining popularity as therapeutic agents in the field of cancer and orthopedic regenerative medicine [36, 65]. The present study illustrates the importance of the bromodomain-contain protein Brd4 during bone development. Before these inhibitors are widely adopted, it is important to consider the potential impact of systemic and local administration of these pharmacological agents on endochondral ossification in the developing skeleton.
Supplementary Material
Highlights.
Depletion of Brd4 in the mesenchyme causes changes in skeletal formation.
Mesenchymal Brd4 loss induces abnormal growth plate morphology.
Loss of Brd4 in the mesenchyme impairs long bone formation.
Brd4 function is required for chondrogenic differentiation and maturation.
5. ACKNOWLEDGMENTS
We thank our Mayo Clinic colleagues and members of the Orthopedic Research Laboratories, including Drs. Jennifer Westendorf, Wenchun Qu, and David Deyle for stimulating discussions and sharing reagents. We also thank Dr. Anup Dey and Dr. Keiko Ozato at the National Institutes of Health (Bethesda, MD) for providing the conditional knockout mouse model central to these studies. We acknowledge the assistance from the Biomaterials Characterization and Quantitative Histomorphometry Core Facility as well as the X-ray Imaging (XRI) Core Facility at Mayo Clinic.
6. FUNDING
All studies were done at Mayo Clinic. This publication was made possible through an intramural award from Mayo Clinic (Career Development Award in Orthopedics Research to AD) and the National Institute of Arthritis and Musculoskeletal and Skin Diseases (R01 AR049069 to AJvW). We also thank William and Karen Eby for generous philanthropic support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURES
ANL performed consulting with funds directed to research for Globus, Medtronic, Orthopediatrics, and Depuy. ANL holds a patent on anterior vertebral body tethering cord. Other authors have no disclosures to report.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available in Gene Expression Omnibus (GEO) database at https://www.ncbi.nlm.nih.gov/geo/, Accession # GSE97118.
8 REFERENCES
- [1].Kobayashi T, Kronenberg HM, Overview of skeletal development, Methods Mol Biol 1130 (2014) 3–12. [DOI] [PubMed] [Google Scholar]
- [2].Breeland G, Menezes RG, Embryology, Bone Ossification, StatPearls Publishing, Available from: https://www.ncbi.nlm.nih.gov/books/NBK539718/, 2019. [PubMed] [Google Scholar]
- [3].Hall BK, Miyake T, The membranous skeleton: the role of cell condensations in vertebrate skeletogenesis, Anat Embryol (Berl) 186(2) (1992) 107–24. [DOI] [PubMed] [Google Scholar]
- [4].Berendsen AD, Olsen BR, Bone development, Bone 80 (2015) 14–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Long F, Building strong bones: molecular regulation of the osteoblast lineage, Nat Rev Mol Cell Biol 13(1) (2011) 27–38. [DOI] [PubMed] [Google Scholar]
- [6].Aghajanian P, Mohan S, The art of building bone: emerging role of chondrocyte-to-osteoblast transdifferentiation in endochondral ossification, Bone Res 6 (2018) 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kronenberg HM, Developmental regulation of the growth plate, Nature 423(6937) (2003) 332–6. [DOI] [PubMed] [Google Scholar]
- [8].Marino R, Growth plate biology: new insights, Current opinion in endocrinology, diabetes, and obesity 18(1) (2011) 9–13. [DOI] [PubMed] [Google Scholar]
- [9].Cooper KL, Oh S, Sung Y, Dasari RR, Kirschner MW, Tabin CJ, Multiple phases of chondrocyte enlargement underlie differences in skeletal proportions, Nature 495(7441) (2013) 375–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lefebvre V, Dvir-Ginzberg M, SOX9 and the many facets of its regulation in the chondrocyte lineage, Connect Tissue Res 58(1) (2017) 2–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mackie EJ, Ahmed YA, Tatarczuch L, Chen KS, Mirams M, Endochondral ossification: how cartilage is converted into bone in the developing skeleton, Int J Biochem Cell Biol 40(1) (2008) 46–62. [DOI] [PubMed] [Google Scholar]
- [12].Kozhemyakina E, Lassar AB, Zelzer E, A pathway to bone: signaling molecules and transcription factors involved in chondrocyte development and maturation, Development 142(5) (2015) 817–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Allis CD, Jenuwein T, The molecular hallmarks of epigenetic control, Nat Rev Genet 17(8) (2016) 487–500. [DOI] [PubMed] [Google Scholar]
- [14].Gibney ER, Nolan CM, Epigenetics and gene expression, Heredity (Edinb) 105(1) (2010) 4–13. [DOI] [PubMed] [Google Scholar]
- [15].Hata K, Epigenetic regulation of chondrocyte differentiation, Japanese Dental Science Review 51(4) (2015) 105–113. [Google Scholar]
- [16].van Wijnen AJ, Westendorf JJ, Epigenetics as a New Frontier in Orthopedic Regenerative Medicine and Oncology, J Orthop Res 37(7) (2019) 1465–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Dudakovic AJ van Wijnen, Epigenetic Control of Osteoblast Differentiation by Enhancer of Zeste Homolog 2 (EZH2), Current Molecular Biology Reports 3(2) (2017) 94–106. [Google Scholar]
- [18].Ferguson GB, Van Handel B, Bay M, Fiziev P, Org T, Lee S, Shkhyan R, Banks NW, Scheinberg M, Wu L, Saitta B, Elphingstone J, Larson AN, Riester SM, Pyle AD, Bernthal NM, Mikkola HK, Ernst J, van Wijnen AJ, Bonaguidi M, Evseenko D, Mapping molecular landmarks of human skeletal ontogeny and pluripotent stem cell-derived articular chondrocytes, Nat Commun 9(1) (2018) 3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Paradise CR, Galeano-Garces C, Galeano-Garces D, Dudakovic A, Milbrandt TA, Saris DBF, Krych AJ, Karperien M, Ferguson GB, Evseenko D, Riester SM, van Wijnen AJ, Larson AN, Molecular characterization of physis tissue by RNA sequencing, Gene 668 (2018) 87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Emons J, Dutilh BE, Decker E, Pirzer H, Sticht C, Gretz N, Rappold G, Cameron ER, Neil JC, Stein GS, van Wijnen AJ, Wit JM, Post JN, Karperien M, Genome-wide screening in human growth plates during puberty in one patient suggests a role for RUNX2 in epiphyseal maturation, The Journal of endocrinology 209(2) (2011) 245–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Khani F, Thaler R, Paradise CR, Deyle DR, Kruijthof-de Julio M, Galindo M, Gordon JA, Stein GS, Dudakovic A, van Wijnen AJ, Histone H4 Methyltransferase Suv420h2 Maintains Fidelity of Osteoblast Differentiation, J Cell Biochem 118(5) (2017) 1262–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Dudakovic A, Evans JM, Li Y, Middha S, McGee-Lawrence ME, van Wijnen AJ, Westendorf JJ, Histone deacetylase inhibition promotes osteoblast maturation by altering the histone H4 epigenome and reduces Akt phosphorylation, J Biol Chem 288(40) (2013) 28783–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Camilleri ET, Dudakovic A, Riester SM, Galeano-Garces C, Paradise CR, Bradley EW, McGee-Lawrence ME, Im HJ, Karperien M, Krych AJ, Westendorf JJ, Larson AN, van Wijnen AJ, Loss of histone methyltransferase Ezh2 stimulates an osteogenic transcriptional program in chondrocytes but does not affect cartilage development, J Biol Chem 293(49) (2018) 19001–19011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Feigenson M, Shull LC, Taylor EL, Camilleri ET, Riester SM, van Wijnen AJ, Bradley EW, Westendorf JJ, Histone Deacetylase 3 Deletion in Mesenchymal Progenitor Cells Hinders Long Bone Development, J Bone Miner Res 32(12) (2017) 2453–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Dudakovic A, Camilleri ET, Xu F, Riester SM, McGee-Lawrence ME, Bradley EW, Paradise CR, Lewallen EA, Thaler R, Deyle DR, Larson AN, Lewallen DG, Dietz AB, Stein GS, Montecino MA, Westendorf JJ, van Wijnen AJ, Epigenetic Control of Skeletal Development by the Histone Methyltransferase Ezh2, J Biol Chem 290(46) (2015) 27604–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Dudakovic A, Camilleri ET, Paradise CR, Samsonraj RM, Gluscevic M, Paggi CA, Begun DL, Khani F, Pichurin O, Ahmed FS, Elsayed R, Elsalanty M, McGee-Lawrence ME, Karperien M, Riester SM, Thaler R, Westendorf JJ, van Wijnen AJ, Enhancer of zeste homolog 2 (Ezh2) controls bone formation and cell cycle progression during osteogenesis in mice, J Biol Chem 293(33) (2018) 12894–12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hemming S, Cakouros D, Codrington J, Vandyke K, Arthur A, Zannettino A, Gronthos S, EZH2 deletion in early mesenchyme compromises postnatal bone microarchitecture and structural integrity and accelerates remodeling, FASEB journal : official publication of the Federation of American Societies for Experimental Biology 31(3) (2017) 1011–1027. [DOI] [PubMed] [Google Scholar]
- [28].Hemming S, Cakouros D, Vandyke K, Davis MJ, Zannettino AC, Gronthos S, Identification of Novel EZH2 Targets Regulating Osteogenic Differentiation in Mesenchymal Stem Cells, Stem Cells Dev 25(12) (2016) 909–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Carpio LR, Bradley EW, McGee-Lawrence ME, Weivoda MM, Poston DD, Dudakovic A, Xu M, Tchkonia T, Kirkland JL, van Wijnen AJ, Oursler MJ, Westendorf JJ, Histone deacetylase 3 supports endochondral bone formation by controlling cytokine signaling and matrix remodeling, Science signaling 9(440) (2016) ra79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Grunstein M, Histone acetylation in chromatin structure and transcription, Nature 389(6649) (1997) 349–52. [DOI] [PubMed] [Google Scholar]
- [31].Sabari BR, Zhang D, Allis CD, Zhao Y, Metabolic regulation of gene expression through histone acylations, Nat Rev Mol Cell Biol 18(2) (2017) 90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Galea GL, Paradise CR, Meakin LB, Camilleri ET, Taipaleenmaki H, Stein GS, Lanyon LE, Price JS, van Wijnen AJ, Dudakovic A, Mechanical strain-mediated reduction in RANKL expression is associated with RUNX2 and BRD2, Gene: X 5 (2020) 100027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Paradise CR, Galvan ML, Kubrova E, Bowden S, Liu E, Carstens MF, Thaler R, Stein GS, van Wijnen AJ, Dudakovic A, The epigenetic reader Brd4 is required for osteoblast differentiation, J Cell Physiol 235(6) (2020) 5293–5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lamoureux F, Baud’huin M, Rodriguez Calleja L, Jacques C, Berreur M, Rédini F, Lecanda F, Bradner JE, Heymann D, Ory B, Selective inhibition of BET bromodomain epigenetic signalling interferes with the bone-associated tumour vicious cycle, Nat Commun 5 (2014) 3511. [DOI] [PubMed] [Google Scholar]
- [35].Najafova Z, Tirado-Magallanes R, Subramaniam M, Hossan T, Schmidt G, Nagarajan S, Baumgart SJ, Mishra VK, Bedi U, Hesse E, Knapp S, Hawse JR, Johnsen SA, BRD4 localization to lineage-specific enhancers is associated with a distinct transcription factor repertoire, Nucleic Acids Res 45(1) (2017) 127–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Baud’huin M, Lamoureux F, Jacques C, Rodriguez Calleja L, Quillard T, Charrier C, Amiaud J, Berreur M, Brounais-LeRoyer B, Owen R, Reilly GC, Bradner JE, Heymann D, Ory B, Inhibition of BET proteins and epigenetic signaling as a potential treatment for osteoporosis, Bone 94 (2017) 10–21. [DOI] [PubMed] [Google Scholar]
- [37].Jacques C, Lavaud M, Georges S, Tesfaye R, Baud’huin M, Lamoureux F, Ory B, BET bromodomains’ functions in bone-related pathologies, Epigenomics 12(2) (2020) 127–144. [DOI] [PubMed] [Google Scholar]
- [38].Taniguchi Y, The Bromodomain and Extra-Terminal Domain (BET) Family: Functional Anatomy of BET Paralogous Proteins, Int J Mol Sci 17(11) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Jin HS, Kim J, Kwak W, Jeong H, Lim GB, Lee CG, Identification of a Novel Mutation in BRD4 that Causes Autosomal Dominant Syndromic Congenital Cataracts Associated with Other Neuro-Skeletal Anomalies, PLoS One 12(1) (2017) e0169226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Cao Y, Li Z, Rosenfeld JA, Pursley AN, Patel A, Huang J, Wang H, Chen M, Sun X, Leung TY, Cheung SW, Choy KW, Contribution of genomic copy-number variations in prenatal oral clefts: a multicenter cohort study, Genet Med 18(10) (2016) 1052–5. [DOI] [PubMed] [Google Scholar]
- [41].Badreldin AA, Bagheri L, Zhang B, Larson AN, van Wijnen AJ, Relative mRNA and protein stability of epigenetic regulators in musculoskeletal cell culture models, Gene 766 (2021) 145032. [DOI] [PubMed] [Google Scholar]
- [42].Devaiah BN, Gegonne A, Singer DS, Bromodomain 4: a cellular Swiss army knife, J Leukoc Biol 100(4) (2016) 679–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA, Selective inhibition of tumor oncogenes by disruption of super-enhancers, Cell 153(2) (2013) 320–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lee JE, Park YK, Park S, Jang Y, Waring N, Dey A, Ozato K, Lai B, Peng W, Ge K, Brd4 binds to active enhancers to control cell identity gene induction in adipogenesis and myogenesis, Nat Commun 8(1) (2017) 2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Logan M, Martin JF, Nagy A, Lobe C, Olson EN, Tabin CJ, Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer, Genesis 33(2) (2002) 77–80. [DOI] [PubMed] [Google Scholar]
- [46].Bouxsein ML, Boyd SK, Christiansen BA, Guldberg RE, Jepsen KJ, Muller R, Guidelines for assessment of bone microstructure in rodents using micro-computed tomography, J Bone Miner Res 25(7) (2010) 1468–86. [DOI] [PubMed] [Google Scholar]
- [47].Schneider CA, Rasband WS, Eliceiri KW, NIH Image to ImageJ: 25 years of image analysis, Nat Methods 9(7) (2012) 671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Houzelstein D, Bullock SL, Lynch DE, Grigorieva EF, Wilson VA, Beddington RS, Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4, Mol Cell Biol 22(11) (2002) 3794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Devaiah BN, Case-Borden C, Gegonne A, Hsu CH, Chen Q, Meerzaman D, Dey A, Ozato K, Singer DS, BRD4 is a histone acetyltransferase that evicts nucleosomes from chromatin, Nature structural & molecular biology 23(6) (2016) 540–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE, Selective inhibition of BET bromodomains, Nature 468(7327) (2010) 1067–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Richmond CM, Savarirayan R, Schmid Metaphyseal Chondrodysplasia, in: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A (Eds.), GeneReviews((R)), University of Washington, Seattle, Seattle (WA), 1993. [PubMed] [Google Scholar]
- [52].Al Kaissi A, Ghachem MB, Nabil NM, Kenis V, Melchenko E, Morenko E, Grill F, Ganger R, Kircher SG, Schmid’s Type of Metaphyseal Chondrodysplasia: Diagnosis and Management, Orthop Surg 10(3) (2018) 241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Halal F, Picard JL, Raymond-Tremblay D, de Bosset P, Metaphyseal dysplasia with maxillary hypoplasia and brachydactyly, Am J Med Genet 13(1) (1982) 71–9. [DOI] [PubMed] [Google Scholar]
- [54].Moffatt P, Ben Amor M, Glorieux FH, Roschger P, Klaushofer K, Schwartzentruber JA, Paterson AD, Hu P, Marshall C, Fahiminiya S, Majewski J, Beaulieu CL, Boycott KM, Rauch F, Metaphyseal dysplasia with maxillary hypoplasia and brachydactyly is caused by a duplication in RUNX2, Am J Hum Genet 92(2) (2013) 252–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Avela K, Hirvinen H, Ben Amor M, Rauch F, Metaphyseal dysplasia with maxillary hypoplasia and brachydactyly in a Finnish woman: first confirmation of a duplication in RUNX2 as pathogenic variant, Eur J Med Genet 57(11–12) (2014) 617–20. [DOI] [PubMed] [Google Scholar]
- [56].Al-Yassin A, Calder AD, Harrison M, Lester T, Lord H, Oldridge M, Watkins S, Keen R, Wakeling EL, A three-generation family with metaphyseal dysplasia, maxillary hypoplasia and brachydactyly (MDMHB) due to intragenic RUNX2 duplication, Eur J Hum Genet 26(9) (2018) 1288–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Ohba S, He X, Hojo H, McMahon AP, Distinct Transcriptional Programs Underlie Sox9 Regulation of the Mammalian Chondrocyte, Cell Rep 12(2) (2015) 229–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Yao Y, Wang Y, ATDC5: an excellent in vitro model cell line for skeletal development, J Cell Biochem 114(6) (2013) 1223–9. [DOI] [PubMed] [Google Scholar]
- [59].Han Y, Lefebvre V, L-Sox5 and Sox6 drive expression of the aggrecan gene in cartilage by securing binding of Sox9 to a far-upstream enhancer, Mol Cell Biol 28(16) (2008) 4999–5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Hong SH, You JS, SOX9 is controlled by the BRD4 inhibitor JQ1 via multiple regulation mechanisms, Biochem Biophys Res Commun 511(4) (2019) 746–752. [DOI] [PubMed] [Google Scholar]
- [61].Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B, The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6, Genes Dev 16(21) (2002) 2813–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Komori T, Runx2, an inducer of osteoblast and chondrocyte differentiation, Histochem Cell Biol 149(4) (2018) 313–323. [DOI] [PubMed] [Google Scholar]
- [63].Sakurai N, Inamochi Y, Inoue T, Hariya N, Kawamura M, Yamada M, Dey A, Nishiyama A, Kubota T, Ozato K, Goda T, Mochizuki K, BRD4 regulates adiponectin gene induction by recruiting the P - TEFb complex to the transcribed region of the gene, Sci Rep 7(1) (2017) 11962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Gilmour J, Assi SA, Noailles L, Lichtinger M, Obier N, Bonifer C, The Co-operation of RUNX1 with LDB1, CDK9 and BRD4 Drives Transcription Factor Complex Relocation During Haematopoietic Specification, Sci Rep 8(1) (2018) 10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Doroshow DB, Eder JP, LoRusso PM, BET inhibitors: a novel epigenetic approach, Ann Oncol 28(8) (2017) 1776–1787. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available in Gene Expression Omnibus (GEO) database at https://www.ncbi.nlm.nih.gov/geo/, Accession # GSE97118.


