Abstract
Amyotrophic lateral sclerosis is a devastating disease characterized primarily by motor system degeneration, with clinical evidence of cognitive and behavioural change in up to 50% of cases. Amyotrophic lateral sclerosis is both clinically and biologically heterogeneous. Subgrouping is currently undertaken using clinical parameters, such as site of symptom onset (bulbar or spinal), burden of disease (based on the modified El Escorial Research Criteria) and genomics in those with familial disease. However, with the exception of genomics, these subcategories do not take into account underlying disease pathobiology, and are not fully predictive of disease course or prognosis.
Recently, we have shown that resting-state EEG can reliably and quantitatively capture abnormal patterns of motor and cognitive network disruption in amyotrophic lateral sclerosis. These network disruptions have been identified across multiple frequency bands, and using measures of neural activity (spectral power) and connectivity (comodulation of activity by amplitude envelope correlation and synchrony by imaginary coherence) on source-localized brain oscillations from high-density EEG. Using data-driven methods (similarity network fusion and spectral clustering), we have now undertaken a clustering analysis to identify disease subphenotypes and to determine whether different patterns of disruption are predictive of disease outcome.
We show that amyotrophic lateral sclerosis patients (n = 95) can be subgrouped into four phenotypes with distinct neurophysiological profiles. These clusters are characterized by varying degrees of disruption in the somatomotor (α-band synchrony), frontotemporal (β-band neural activity and γl-band synchrony) and frontoparietal (γl-band comodulation) networks, which reliably correlate with distinct clinical profiles and different disease trajectories. Using an in-depth stability analysis, we show that these clusters are statistically reproducible and robust, remain stable after reassessment using a follow-up EEG session, and continue to predict the clinical trajectory and disease outcome.
Our data demonstrate that novel phenotyping using neuroelectric signal analysis can distinguish disease subtypes based exclusively on different patterns of network disturbances. These patterns may reflect underlying disease neurobiology. The identification of amyotrophic lateral sclerosis subtypes based on profiles of differential impairment in neuronal networks has clear potential in future stratification for clinical trials. Advanced network profiling in amyotrophic lateral sclerosis can also underpin new therapeutic strategies that are based on principles of neurobiology and designed to modulate network disruption.
Keywords: amyotrophic lateral sclerosis, EEG, resting-state, clustering, subphenotyping
Dukic et al. report that ALS patients can be subgrouped into four phenotypes with distinct neurophysiological profiles. Using in-depth stability analyses, they show that the clusters are reproducible, remain stable after reassessment, and continue to predict the clinical trajectory and disease outcome.
Introduction
Amyotrophic lateral sclerosis (ALS) is a heterogeneous neurodegenerative disorder that primarily affects the motor system, causing varying degrees of upper and lower motor neuron dysfunction,1 with additional involvement of extra-motor regions2 presenting as cognitive and/or behavioural impairment that overlaps with frontotemporal dementia (FTD).3,4 The ALS population is clinically heterogeneous both in presentation and prognosis, and with variability in underlying disease pathobiology.5 Current clinical phenotypes are based on the predominant site of symptom onset (spinal, bulbar and respiratory), family history (sporadic and familial) and relative degree of upper and lower motor neurone involvement (lower and upper motor predominant). In addition, patients with ALS are often categorized on the basis of their survival period (short, average and long).5 Quantitative measurements that correlate with the clinical subgroups have been sought using structural and functional MRI,6 PET7,8 and neurophysiological (EEG and EMG) data.9-12
Additional refinements in clinical phenotyping in ALS include the interrogation of behavioural subphenotypes,13 data from early clinical consultation to determine ranges of survival probability14 and genomic characterization. At least 30 identified genes and three main pathophysiological processes (i.e. RNA biology, protein turnover and axonal transport) have been associated with ALS.15 Taken together, these observations, along with the absence of a clear correlation between ALS-associated genes, and highly distinctive molecular neuropathological and clinical subtypes,16 provide evidence that ALS can no longer be considered as a single disease with a singular pathophysiology and clinical course.
Current imaging and neurophysiology evidence suggests that differential disruption of neural networks, underpinned by biological pathology and genetic factors,17,18 is likely to reflect heterogeneous clinical presentations. This heterogeneity cannot be fully discerned using existing clinical tools, such as the revised ALS Functional Rating Scale (ALSFRS-R),19 which measures motor decline, and the Edinburgh Cognitive and Behavioural ALS screen (ECAS),20 which screens for cognitive and behavioural change.21 Though validated as a primary outcome measure in clinical trials, the ALSFRS-R is ordinal, semiquantitative and the subscales within the instrument are subject to floor and ceiling effects.22
Technological improvements in neuro-electro-physiological measures, and more specifically EEG, can provide additional insights into functional changes associated with different neurodegenerative diseases at a network level.21 Using this approach with task-based paradigms, we have shown changes implicating dysfunction of the frontoparietal network.9,23,24 Furthermore, we have shown that resting-state EEG, which can provide distinct measures that reflect different processes in the brain,25 can quantitatively capture both motor and cognitive networks affected in ALS. More specifically, using sensor-space analysis, we have found resting-state EEG changes that are correlated with structural changes in MRI10 and in line with other EEG studies.26–28 In a follow-up study using advanced source-space analysis, we further delineated dysfunctional networks and corroborated the findings with both structural MRI and clinical data.29
Here, we hypothesize that patient subgroups can be identified based on patterns of network disruption that could be used to reveal potentially different responses to therapy and thus, should be monitored and studied as complementary profiling measures. We show how the EEG measures of activity and connectivity in the brain networks provide the information for forming stable clusters of ALS patients and the distinct neurophysiological profiles associated with these patient clusters.
Materials and methods
Ethical approval
Approval was obtained from the ethics committee of Beaumont Hospital, Dublin, Ireland (reference: 13/102) and the Tallaght Hospital/St. James's Hospital Joint Research Ethics Committee (reference: 2014 Chairman’s Action 7) for St James’s Hospital, Dublin, Ireland. The experimental procedure conformed to the Declaration of Helsinki. All participants provided written informed consent before taking part in the experiments.
Participants
Patient recruitment
Patients with ALS were recruited from the National ALS clinic in Beaumont Hospital Dublin. Healthy controls included neurologically normal, age-matched individuals recruited from an existing population-based control bank.
Inclusion criteria
All ALS patients were within the first 18 months from diagnosis and fulfilled the revised El Escorial diagnostic criteria for Possible, Probable or Definite ALS.30 All patients underwent cognitive screening and were classified according to the revised Strong Criteria.31
Exclusion criteria
Patients diagnosed with primary lateral sclerosis, progressive muscular atrophy, flail arm/leg syndromes, previous transient ischaemic attacks, multiple sclerosis, stroke, epilepsy, seizure disorder, brain tumours, structural brain abnormalities, other neurodegenerative conditions and other medical morbidities, such as human immunodeficiency virus, were excluded.
The demographic profiles
A total of 95 ALS patients: 70 with spinal onset [male/female: 52/18; mean ± standard deviation (SD) age: 59 ± 12 years], 21 patients with bulbar onset (male/female: 14/7; age: 60 ± 11) and four patients with respiratory onset (male/female: 3/1; age: 62 ± 4) were included, along with 77 healthy controls (male/female: 29/48; age: 60 ± 11). Five patients (male/female: 2/3; age: 70 ± 9) were diagnosed as ALS-FTD (based on the Strong criteria)31 and 11 patients (male/female: 6/5; age: 61 ± 6) had the hexanucleotide repeat expansion in the C9orf72 gene. Patients and controls were matched for age (Mann–Whitney U-test, P = 0.73), but not for gender (Fisher’s exact test, P < 0.001).
Experiment
Experimental paradigm
The experiment was resting-state with eyes open, divided into three 2-min recording blocks, allowing for rest between blocks and to ensure patients remained awake. Subjects were seated in a comfortable chair, asked to relax, while they fixated their gaze at the letter X (6 × 8 cm) printed on an A4 sheet of paper placed ∼1 m in front of them.
EEG data
EEG data with 128 channels were collected using the BioSemi Active Two system (BioSemi B.V.) and sampled at 512 Hz, after a lowpass anti-aliasing filter (0–104 Hz) was applied by the acquisition hardware. Additional filtering was applied during the analysis. Recordings were conducted in a dedicated laboratory with a Faraday cage isolation at St. James’s Hospital, Dublin. Besides the initial recording session for 95 ALS and 77 healthy controls, 36 ALS patients had one follow-up EEG session after 4–6 months.
Disease severity and neuropsychology data
The scores from ALSFRS-R (n = 88),19 ECAS (n = 72)20 and Beaumont behavioural inventory (BBI, n = 87)32 were used to provide clinical profiles of clusters based on neurophysiological patterns. All clinical subscores were either normalized or standardized: ALSFRS-R subscores were normalized by dividing by the maximum possible value in each subscore and subtracting it from one; ECAS subscores were z-score standardized using age and education matched normative data from an Irish population33,34 and multiplied by minus one; and BBI score was normalized by dividing by the maximum possible value. This ensured that all subscores had the same direction of change, wherein an increased subscore means an increased impairment in the corresponding function.
In addition to this, King’s staging (n = 84),35 which assesses the disease burden in patients in stages from one (single region involved) to four (ventilatory support and/or gastrostomy), was used.
Data analysis
EEG data
The preprocessing and processing procedures were identical to those described in our cross-sectional study.29 Briefly, we have applied the linearly constrained minimum variance beamformer36 on the bandpassed (1–97 Hz) EEG data to obtain time-varying signals originating from 90 brain regions (excluding the cerebellum) based on the automated anatomical labelling atlas (Supplementary material).37 Using the 90 source-reconstructed signals, we estimated normalized spectral power (with respect to total spectral power), comodulation (amplitude envelope correlation) and synchrony (imaginary coherence). Spectral power was estimated for each region, while comodulation and synchrony were estimated between all pairs of brain regions resulting in a 90 × 90 symmetrical connectivity matrix, wherein 4005 (90 × 89/2) connectivity features were unique. All three measures were estimated in six frequency bands: δ (2–4 Hz), θ (5–7 Hz), α (8–13 Hz), β (14–30 Hz) and γ (γl: 31–47 Hz, γh: 53–97 Hz). The analysis resulted in three groups of EEG features: spectral power (90 × 6 = 540 features), comodulation (4005 × 6 = 24 030) and synchrony (24 030). This analysis was applied on both healthy control and ALS data (Supplementary material).
Clustering
Without previous knowledge of the EEG features that distinguish one ALS patient from the other, an unsupervised clustering approach was chosen and applied on all available EEG features. First, the similarity network fusion (SNF) method38 was used for combining and preparing the high-dimensional dataset and subsequently the spectral clustering39 was used for inference of the clusters.
For preparation of the data before the clustering, each EEG feature was z-scored. Three patient similarity matrices (one for each group of EEG features) based on the Euclidean distance, were constructed with multiple Gaussian kernels40 and fused into one similarity matrix using the SNF method. The SNF method iteratively updates each matrix with the information from the other matrices, thus fusing in the complementary information. To ensure that the irrelevant associations between patients emerging from the accumulated noise over many features are removed, the fused similarity matrix was denoised using the network enhancement method.41 Finally, subgrouping of patients was undertaken using spectral clustering.39 For additional information, see the Supplementary material. This clustering pipeline was selected based on the non-parametric and robust nature of these methods pertinent for clustering, especially in combining the EEG measures, which served to avoid finding clusters that heavily depend on specific mathematical assumptions or individual data values.
Statistical significance of clusters
The optimal number of patient subgroups (k = 2, …, 7) was chosen using a statistical approach applied on the eigengap and rotation cost indices,42 which are based on the eigenvalues and eigenvectors in the spectral clustering method, respectively. The biggest eigengap and the smallest rotation cost indicate the optimal number of clusters in the dataset (Supplementary material).
Taking a conservative approach, a statistical procedure that tests whether the two indices are likely to give such high (eigengap) and low (rotation cost) values under a null hypothesis of no actual clusters in data (i.e. homogenous data) was applied. In our Monte Carlo procedure, the null hypothesis was that the data come from the same dataset but with randomly permuted values within each EEG feature. The clustering was performed on the permuted dataset and the two indices were calculated. This procedure was repeated 5000 times to obtain the empirical null distributions and the P-values.43 In addition to this, bootstrapping was used to estimate the non-null distribution of the indices, statistical power at α = 0.05 (1 − β0.05) and Cliff’s delta (a non-parametric measure of effect size).44 All the statistical measures were calculated for each clustering solution, k = 2, …, 7.
Neurophysiological profiles
For each participant, 90 ‘EEG networks’ were defined based on the networks that are known to be activated at rest45 and affected in ALS.9,29,46,47 Namely, the 90 networks were constructed by separately averaging spectral power, comodulation and synchrony in five anatomical networks (somatomotor, frontotemporal, frontoparietal, default mode and salience) and in six frequency bands (3 × 5 × 6 = 90 Supplementary material). For each combination (i.e. ‘EEG network)’ an AUC (area under the curve of the receiver operating characteristics curve) was used to make the comparison between each ALS cluster and the control data. To further infer statistical significance in the multidimensional space and to control for multiple comparisons (q = 0.1, false discovery rate, FDR),29 empirical Bayesian inference (EBI)43 was applied on the AUC test statistics. This statistical tool exploits both the original (non‐null) observations and null‐permuted data to estimate the probability density function of the data and null, respectively.
The obtained AUC values were then used to determine brain networks that are strongly and exclusively associated with each of the identified clusters. An EEG network was considered as a potential and exclusive characteristic of a cluster if it was statistically significant compared to controls, and unique or directionally opposite in its change compared to other clusters (Supplementary material). Here, we reported the most characteristic EEG network that is affected for each cluster. Additionally, for each EEG network, the statistical difference between clusters (χ2-statistic, Kruskal–Wallis one-way ANOVA) was tested, while accounting for multiple comparisons (q = 0.05, FDR),48,49 and a Monte Carlo permutation procedure was applied to estimate the associated statistical power (1 − β0.05).
Additionally, complete brain maps for each EEG measure and frequency band were obtained in a similar manner using AUC and EBI between each cluster and the control data. These maps were then masked using P-values from Kruskal–Wallis one-way ANOVA to distinguish the EEG abnormalities that are shared by all identified clusters and those specific to each cluster.
Clinical profiles
Clinical profiles of subgroups were compared using the subscores of motor (ALSFRS-R), cognitive (ECAS) and behavioural (BBI) dysfunction. Significant difference of scores across the clusters was tested using Kruskal–Wallis one-way ANOVA. In addition to this, associations between the identified EEG clusters and known clinical factors (type of initial diagnosis, site of disease onset and C9orf72 gene status) that could influence our findings were tested using the Fisher’s exact test. Survival probability was analysed using the Kaplan–Meier method, wherein patients that were alive at the time of analysis were right-censured and survival was measured from the time of the reported symptom onset. The log rank test was used for testing of the difference between the survival curves.
Cluster validation
For a reliable interpretation of the derived clusters, we have assessed the accuracy, robustness and stability of clusters mathematically as well as experimentally. Specifically, four validation approaches were implemented using: a different clustering method (the Louvain method for community detection),50 a classification approach and by inspecting the reassignment of patients under small perturbations of data and when using a single follow-up EEG assessment (after 4–6 months following the initial session, n = 36 ALS patients). These validation methods are detailed in the Supplementary material.
Clustering using clinical data
To assess whether the derived EEG clusters simply recapitulate the subtypes that can be derived directly from the clinical data, the clustering procedure was applied on n = 60 patients with the complete clinical dataset. A fused similarity matrix was constructed from three similarity matrices based on 12 ALSFRS-R, five ECAS and one BBI subscores (n = 18 subscores in total). The optimal number of clusters was determined using the statistical approach as in the main analysis. Furthermore, the accuracy and the robustness of the clustering solution was evaluated using the same procedures that were described previously.
Additionally, for comparison with our identified EEG clusters, we inspected the clinical profiles of ALS subgroups that are based on four stages of King’s staging system.
Data availability
The data that support the findings of this study are available from the corresponding author on reasonable request from qualified investigators, and are subject to the approvals by Data Protection Officer and The Office of Corporate Partnership and Knowledge Exchange in Trinity College Dublin.
Results
EEG measures identify four clusters of amyotrophic lateral sclerosis patients
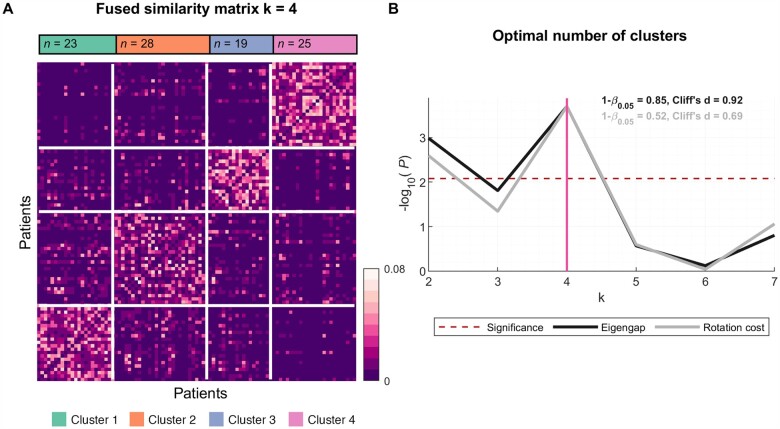
Four distinct clusters were identified based on analysis of spectral EEG patterns of neural activity and connectivity. As assessed by eigengap and rotation cost indices (Fig. 1), the solution of four clusters had high statistical power (0.85 and 0.52, respectively) and a large to medium (0.92 and 0.69, Cliff’s d, respectively) effect size, suggesting reproducible findings. The demographics of the identified clusters are shown in Table 1.
Figure 1.
EEG measures identify four ALS clusters: Fused similarity matrix and the optimal number of ALS clusters. (A) Fused similarity matrix of ALS patients is sorted based on the clusters, which were identified using spectral clustering. (B) At k = 4, both measures reflecting the optimal number of clusters (eigengap, black; rotation cost, grey) reach the highest significance (P < 0.008, Bonferroni corrected; red dashed line) with statistical power (1 − β0.05) 0.85 and 0.52, and effect size (Cliff’s d) 0.92 and 0.69, respectively. The number of patients in clusters 1–4 are n = 23, 28, 19 and 25, respectively.
Table 1.
Breakdown of cluster characteristics
| Group | n | Gender Male/female |
Age (years) | Disease duration (months) | Site of onset (S/B/T) |
Diagnosis (ALS/ALS-FTD) |
C9orf72 (+/−) |
|---|---|---|---|---|---|---|---|
| All | 95 | 69/26 | 59.2 ± 11.6 | 21.9 ± 17.5 | 70/21/4 | 90/5 | 11/84 |
| Cluster 1 | 23 | 14/9 | 61.0 ± 12.7 | 21.3 ± 16.8 | 17/5/1 | 23/0 | 0/23 |
| Cluster 2 | 28 | 22/6 | 56.6 ± 13.0 | 25.7 ± 24.3 | 23/2/3 | 28/0 | 3/25 |
| Cluster 3 | 19 | 14/5 | 58.5 ± 11.5 | 17.8 ± 8.9 | 14/5/0 | 16/3 | 2/17 |
| Cluster 4 | 25 | 19/6 | 60.7 ± 9.0 | 22.8 ± 20.2 | 16/9/0 | 23/2 | 6/19 |
Disease duration: time interval between the estimated symptom onset and the EEG recording; site of onset: spinal/bulbar/thoracic (S/B/T); C9orf72: presence (+) or absence (−) of the repeat expansion in C9orf72; age and disease duration: mean ± SD.
EEG clusters show distinct neurophysiological profiles
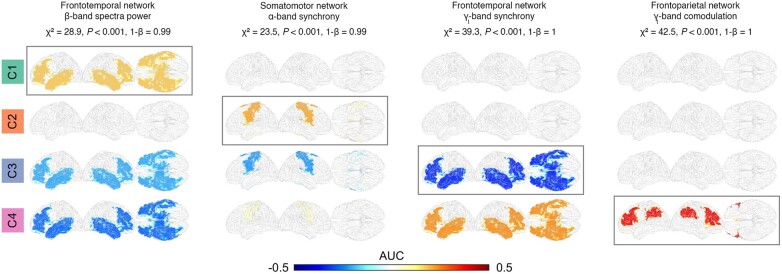
Analysis of neurophysiological profiles of the four clusters based on EEG measures revealed evidence of distinctly impaired neural networks for each cluster (Fig. 2). For example, cluster 1 shows a characteristic increase in β-band spectral power in the frontotemporal network, whereas clusters 3 and 4 show decreased power in the same network. Similarly, cluster 2 shows a characteristic increase in α-band synchrony in the somatomotor network, cluster 3 decrease in γl-band synchrony in the frontotemporal network and cluster 4 increase in γl-band comodulation in the frontoparietal network. The Kruskal–Wallis one-way ANOVA showed that the four networks vary significantly across clusters (P < 0.001, FDR).
Figure 2.
Distinct neurophysiological profiles of ALS clusters. For each cluster, a unique neurophysiological change (brain network, frequency band and EEG measure) was identified using AUC statistics estimated between the ALS clusters and control data (Supplementary material). The networks vary significantly across clusters in all four cases (Kruskal–Wallis one-way ANOVA, P < 0.001, FDR). The potential effects of age and gender on the identified changes were rejected based on the linear model analysis (Supplementary material). AUC = area under the receiver operating characteristic curve centred around zero; positive values indicate an increase, whereas negative values indicate a decrease compared to healthy controls.
The EEG abnormalities associated with all four clusters were identified as increased comodulation (δ- to α-band oscillations) and decreased synchrony (δ- to β-band) in the somatomotor and frontotemporal brain regions (Supplementary Fig. 1).
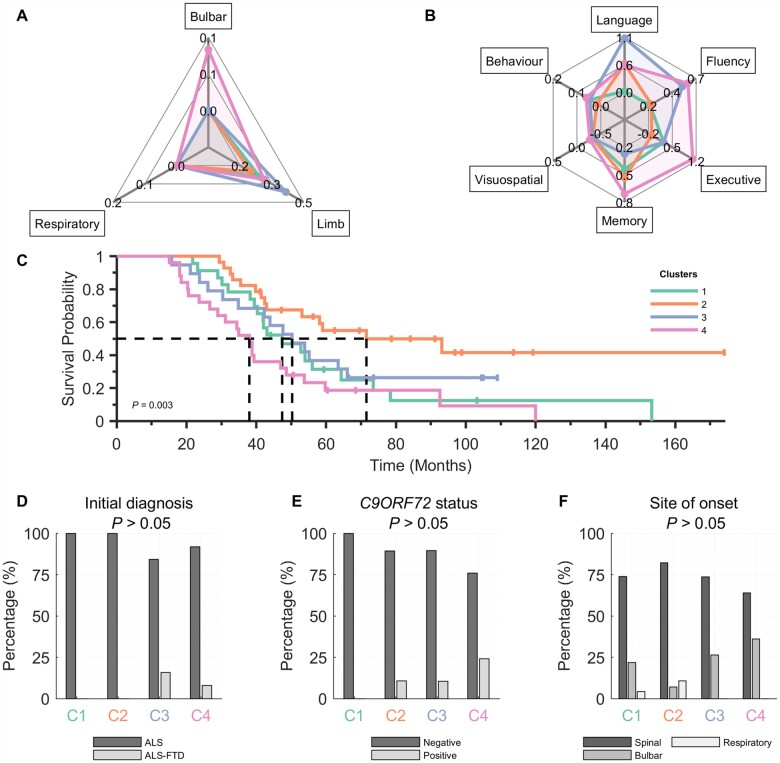
EEG clusters have concordant clinical and neurophysiological profiles
The analysis of clinical profiles using the functional scores shows clinical characteristics of each cluster (Fig. 3A and B, see also Supplementary Fig. 2). Although none of the clinical scores vary significantly across the clusters (P > 0.05, FDR), the changes are concordant with altered neurophysiological profiles. More specifically, cluster 1 (which has a uniquely increased β-band spectral power in the frontotemporal network) shows moderate limb and mild verbal fluency, executive and memory dysfunction, but no apparent change in the language domain; cluster 2 (which has a uniquely increased α-band spectral power in the somatomotor network) is characterized by mild impairment of limb and verbal fluency, and moderate language and memory impairment, with preservation of executive domain; cluster 3 (which has a uniquely decreased γl-band synchrony in the frontotemporal network) was characterized by marked impairment of limb, language and verbal fluency; cluster 4 (which has a uniquely increased γl-band comodulation frontoparietal network) was primarily characterized by impairments in bulbar function, verbal fluency, executive and memory. None of the clusters has notable impairment in the visuospatial domain, whereas all but cluster 2 exhibited mild aspects of behavioural impairment.
Figure 3.
Clinical profiles of ALS clusters derived from EEG measures are concordant with the neurophysiological profiles. The four EEG clusters (colour-coded) suggest different trends in functional/clinical scores in different domains: (A) normalized ALSFRS-R (bulbar, limb and respiratory) and (B) z-scored ECAS (language, fluency, executive, memory and visuospatial) and normalized BBI (behaviour) score are all non-significant (P > 0.05, FDR). (C) Kaplan–Meier survival curves corresponding to the ALS clusters. (D–F) Clinical characteristics. Clinical subscores (A and B) are all normalized or standardized, see the ‘Materials and methods’ section. Note that there are in total: five ALS-FTD, 11 C9orf72-positive and four respiratory-onset patients. Statistical tests: Kruskal–Wallis one-way ANOVA (A and B), logrank test (C) and Fisher’s exact test (D–F); all FDR corrected.
In addition to clinical subphenotypes, the clusters were associated with significant differences in overall survival (log rank χ2 = 13.84; P = 0.003). The survival probability curves (Fig. 3C) show that cluster 4 has the shortest survival (median: ∼3 years), whereas cluster 2 has the longest survival (∼6 years).
Although the associations between the clusters and commonly used clinical stratification parameters (type of initial diagnosis, site of disease onset and C9orf72 gene status; Fig. 3D–F) are not significant (P > 0.05, FDR), the results are consistent with clinical profiles of clusters. Specifically, clusters 3 and 4 (which have the greatest degree of impairment across most cognitive subscores; Fig. 3B) included all patients with the initial diagnosis of ALS-FTD (3/19 and 2/25; ALS-FTD/total). Furthermore, cluster 4 has the highest proportion of C9orf72-positive patients (6/25), compared to clusters 2 and 3 (3/28 and 2/19).
There were no significant between-group differences in disease duration, King’s staging, age, gender or riluzole usage, which could have affected EEG measures and the reported results (Supplementary Fig. 2). Additionally, we demonstrated that King’s staging cannot explain the clusters identified by EEG networks nor how the progression patterns differ in the EEG clusters (Supplementary Fig. 3). The potential effects of age and gender on the identified changes in neurophysiological profiles were tested and rejected based on the linear model analysis (Supplementary material).
Patient clusters show stability across multiple tests
Further analysis revealed that each cluster has high accuracy, robustness and remained stable at reassessment. Specifically, the clustering solution based on the Louvain community detection method converged to the same number of clusters (k = 4) and had a very high overlap with the spectral clustering solution from the main analysis, wherein only seven patients were assigned differently. Furthermore, the estimated clustering accuracy reached 89%, and the analysis of robustness showed that in the presence of data perturbation 82% of the cluster labels remain stable (both tests are conservatively quantified by the average adjusted Rand index, which controls for chance level). Last, using the longitudinal dataset (n = 36, with one follow-up EEG measurement 4–6 months after the initial recording session), the overall cluster (re)assignment is 72% (P < 0.001, Fisher’s exact test; Fig. 4), showing an experimental stability of the discovered clusters.
Figure 4.
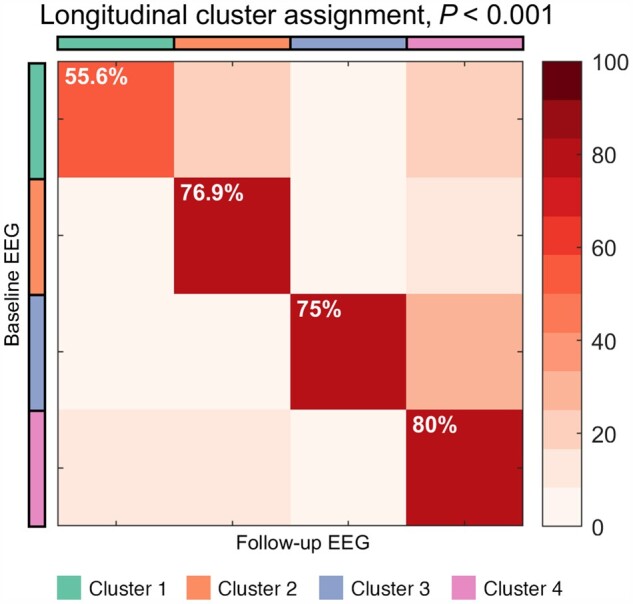
Clusters show high stability at reassessment. The overall stability is 72% and statistically significant (P < 0.001, Fisher’s exact test). Total number of patients with a follow-up (mean ± standard deviation: 5.1 ± 1.8 months after the initial recording session) is n = 36, wherein 9, 13, 4 and 10 patients belong to clusters 1–4, respectively.
Clustering based solely on clinical data does not identify stable subgroups
Using the same methodology, all the clinical measures were combined and underwent statistical analysis of the indices that estimate the optimal number of clusters. No significant clusters were identified, demonstrating that commonly applied clinical determinants were not driving the neurophysiological clustering data (Supplementary material and Supplementary Fig. 4).
Discussion
We have shown that analysis of network disturbance using multidimensional quantitative EEG can identify subgroups within ALS that are not discoverable using standard clinical assessment tools. Each of the subgroups, identified by data-driven clustering, demonstrates a distinct neurophysiological profile that in turn recapitulates a different combination of clinical attributes. These neurophysiological profiles are stable at reassessment and are associated with different prognostic outcomes.
Identified EEG clusters characterize distinct brain network impairments
Clinical heterogeneity has emerged as a major obstacle in understanding the pathophysiology of neurodegenerative diseases. This has implications for drug development as clinical stratification parameters remain relatively insensitive as predictors of disease progression and survival. While it is not surprising that the network disruptions that characterize our identified clusters do not strongly correlate or overlap with the commonly defined clinical phenotypic subtypes of disease, our results are in alignment with the observations from previous studies. For instance, cluster 4 in this study has the highest proportion of patients with C9orf72 expansion, which is known to implicate frontotemporal, temporoparietal and subcortical MRI6,51 and EEG9 changes, and is frequently associated with cognitive and behavioural impairment.52 Accordingly, in our study the neurophysiological profile of this cluster is characterized by the distinctive abnormal changes in γl-band comodulation within the frontoparietal network (also commonly known as central executive network), while the clinical profile of this cluster shows marked dysfunction in the verbal fluency, executive and memory domain. Similarly, this cluster has the highest proportion of bulbar patients, in which MRI studies have shown extensive thinning in frontotemporal, temporoparietal and subcortical brain regions.6 Furthermore, while cluster 4 has the highest proportion of patients with C9orf72 expansion, which is associated with both ALS and FTD, clusters 3 and 4 include all the ALS-FTD patients. Consistent with other studies,53 these two EEG phenotypes show the lowest survival probability in our analysis. Considering the presence of notably increased dysfunction in cognitive and behavioural profile of these two clusters, these ALS patients are likely to have clinical features that align with the FTD-side of the ALS-FTD spectrum.31 Interestingly, C9orf72 patients did not form one separate cluster, suggesting diverging network impairments caused by the same genetic mutation. These findings confirm a complex and heterogeneous nature of the variables (e.g. gene mutation status and presence/absence of FTD) currently used in ALS classification systems. By contrast, subphenotypes derived from EEG measures transcend traditional classification systems of ALS patients and characterize distinct brain networks affected in each subgroup.
Our findings are consistent with previous neuro-electro-magnetic studies in ALS. For example, a recent resting-state magnetoencephalography (MEG) connectivity study reports increased broadband comodulation in the posterior parts of the brain.54 Additionally, studies investigating brain network topology using graph theory, showed diverging MEG γ-synchrony (as assessed by phase lag index)55 affecting global brain patterns56 and increased EEG γ-synchrony (as assessed by partial directed coherence)57 patterns in the frontal networks.46 These resting-state findings are in line with the identified connectivity patterns in clusters 3 and 4.
The neurophysiological profiles of clusters 1 and 2 point to the characteristic changes in the β-band frontotemporal and α-band motor network, respectively, while the corresponding clinical subscores in the language, verbal fluency and motor domains indicate relative preservation of these functions. These abnormal network activations could be attributed either to the topological resilience or active compensation mechanisms that are unique to each cluster,58,59 or likely, to subtle impairments to which current clinical tools are not sufficiently sensitive.60–62
Our work emphasizes that not all cluster-specific patterns may be identifiable when ALS patients are compared to controls as a single group. This is due to due to the difference in the patterns of impairment between different clusters. The identified β-band power changes suggest two diverging patterns, which could explain the contradictory findings between an MEG study63 that reported an increased cortical β-desynchronization in ALS patients and EEG studies that reported decreased27–29 or no difference.64 Additionally, the findings in resting-state studies investigating brain network topology using graph theory, show globally increased EEG α-synchrony (as assessed by partial directed coherence)46 and increased α-band comodulation mostly in the central brain regions.65 Furthermore, our findings support the relevance of γ-oscillations in ALS (Supplementary material).
Clinical relevance
We have shown that clusters based on patterns of disruption in brain networks are associated with reproducible aggregates of clinical attributes and rate of disease progression, confirming the clinical relevance of our findings. EEG-based subphenotypes with superior statistical power do not recapitulate phenotypes that can be found using clinical data or burden of disease (e.g. King’s staging). This indicates that these neurophysiologic patterns provide additional information to that which is discerned by clinical evaluation alone. The EEG-based clusters are statistically robust with distinct patterns, whereas the clinical scores alone could not form meaningful significant clusters. A more in-depth analysis that further explores associations between EEG and clinical observations, would require larger and detailed clinical and genomic datasets.
The identification of such stable subtypes with high statistical power has significant biological and clinical implications. Our findings could contribute to modification of the existing stratification system, which is purely based on the clinical observations. In fact, simulated analysis resulting in high classification accuracy (89%) of new patients—where individual patients are classified to clusters—suggests the potential of our clustering approach to render clinically meaningful findings on an individual patient level. While the underlying neurobiological processes that determine these patterns or network disruption cannot be discerned at this point, the stability of the clusters could reflect premorbid patterns of network function and integrity.
Analysis of cluster stability using follow-up data shows that for many patients in our dataset the cluster assignment does not change. This stability further supports that our findings are based on characteristic pathological changes that are reasonably stable over a period spanning several months. Notwithstanding, future studies with more systematic inclusion of the disease stages and the analysis of longitudinal evolution of clusters (over multiple follow-ups) are warranted.
Limitations
This study is limited by its single-site nature. Alternative solutions with more than four clusters are likely to exist, especially if additional more sophisticated neurophysiological measures are included in the clustering analysis. Notwithstanding these limitations, our conservative validation analyses of clustering solutions show that the findings are both robust and reproducible.
Translation of our findings into a clinical setting will require medical-grade equipment with equal or lower number of EEG electrodes (e.g. 32 or 19 from the 10–20 system), which warrants an additional validation study. While this could reduce the preparation time, it should be approached with caution.66 Studies showed that electrode arrays with <32 sensors lead to severe mislocalizations.67 Moreover, our neurophysiological profiles include γ-band findings, and in this context, decreasing the number of electrodes might have a negative effect on our ability to capture these oscillations.68,69 Nevertheless, since localization accuracy starts to plateau from 64 channels,67,70 a medical-grade 64-channel system could be considered as a candidate for future translational steps.
Conclusion
Our findings have shown for the first time that EEG measures of neural activity and connectivity can be used to reproducibly group ALS patients into subphenotypes with distinct clinical patterns and neurophysiological signatures. Replication of our findings in an independent population with additional clinical and genomic data will be required to further understand the neurobiological factors that underpin these different patterns of network disruption. The demonstration that each cluster is associated with a different disease trajectory and outcome opens a new path towards the discovery of quantitative biomarkers of disease heterogeneity.
Taken together, our results highlight the strengths of using EEG data in identifying ALS subtypes that have distinct clinical and neurophysiological profiles. The identification of data-driven ALS subtypes based on patterned changes in neuronal networks can facilitate the identification of targeted therapies that are effective across the subtype. The development of reliable biomarkers to identify subtypes will also provide much needed prognostic information for patient stratification.
Supplementary Material
Acknowledgements
We would like to thank the students and staff at Trinity Biomedical Sciences Institute and Beaumont Hospital who facilitated this study. We would like to acknowledge the assistance and support of the Wellcome—HRB Clinical Research Facility at St. James’s Hospital in providing a dedicated environment for the conduct of high-quality clinical research activities. We would like to thank all the patients and their families who contributed their time for this research.
Funding
Funding for this study was provided by the Health Research Board of Ireland (HRA-POR-2013–246; MRCG-2018–02), the Irish Motor Neurone Disease Research Foundation (IceBucket Award; MRCG-2018–02), Irish Research Council (Government of Ireland Postdoctoral Research Fellowship GOIPD/2015/213 to B.N. and Government of Ireland Postdoctoral Postgraduate Scholarship GOIPG/2017/1014 to R.M.) and Science Foundation Ireland (16/ERCD/3854). The additional data analytic aspects of the study were supported by the ALS Association (multi-year grant 20-IIA-546). Marjorie Metzger is supported by the Thierry Latran Foundation (Project award to O.H.). Peter Bede and the Computational Neuroimaging Group are supported by the Health Research Board of Ireland (Emerging Investigator Award HRB-EIA-2017–019), the Irish Institute of Clinical Neuroscience (IICN) - Novartis Ireland research grant, The Iris O’Brien Foundation and The Perrigo clinician-scientist research fellowship. Muthuraman Muthuraman is supported by the German Collaborative Research (DFG-CRC-1193 and CRC-TR-128).
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
Glossary
- ALS
amyotrophic lateral sclerosis
- ALSFRS-R
ALS Functional Rating Scale-Revised
- BBI
Beaumont Behavioural Inventory
- ECAS
Edinburgh cognitive and behavioural ALS screen
- FTD
frontotemporal dementia
References
- 1. Hardiman O, Van Den Berg LH, Kiernan MC.. Clinical diagnosis and management of amyotrophic lateral sclerosis. Nat Rev Neurol. 2011;7(11):639–649. [DOI] [PubMed] [Google Scholar]
- 2. van Es MA, Hardiman O, Chio A, et al. . Amyotrophic lateral sclerosis. Lancet. 2017;390(10107):2084–2098. [DOI] [PubMed] [Google Scholar]
- 3. Pender N, Pinto-Grau M, Hardiman O.. Cognitive and behavioural impairment in amyotrophic lateral sclerosis. Curr Opin Neurol. 2020;33(5):649–654. [DOI] [PubMed] [Google Scholar]
- 4. Lomen-Hoerth C, Anderson T, Miller B.. The overlap of amyotrophic lateral sclerosis and frontotemporal dementia. Neurology. 2002;59(7):1077–1079. [DOI] [PubMed] [Google Scholar]
- 5. Al-Chalabi A, Hardiman O, Kiernan MC, Chiò A, Rix-Brooks B, van den Berg LH.. Amyotrophic lateral sclerosis: Moving towards a new classification system. Lancet Neurol. 2016;15(11):1182–1194. [DOI] [PubMed] [Google Scholar]
- 6. van der Burgh HK, Westeneng H-J, Walhout R, et al. . Multimodal longitudinal study of structural brain involvement in amyotrophic lateral sclerosis. Neurology. 2020;94(24):e2592–e2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cistaro A, Pagani M, Montuschi A, et al. . The metabolic signature of C9ORF72-related ALS: FDG PET comparison with nonmutated patients. Eur J Nucl Med Mol Imaging. 2014;41(5):844–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pagani M, Chiò A, Valentini MC, et al. . Functional pattern of brain FDG-PET in amyotrophic lateral sclerosis. Neurology. 2014;83(12):1067–1074. [DOI] [PubMed] [Google Scholar]
- 9. McMackin R, Dukic S, Broderick M, et al. . Dysfunction of attention switching networks in amyotrophic lateral sclerosis. NeuroImage Clin. 2019;22:101707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nasseroleslami B, Dukic S, Broderick M, et al. . Characteristic increases in EEG connectivity correlate with changes of structural MRI in amyotrophic lateral sclerosis. Cereb Cortex. 2019;29(1):27–41. [DOI] [PubMed] [Google Scholar]
- 11. Baumann F, Henderson RD, Ridall PG, Pettitt AN, McCombe PA.. Use of Bayesian MUNE to show differing rate of loss of motor units in subgroups of ALS. Clin Neurophysiol. 2012;123(12):2446–2453. [DOI] [PubMed] [Google Scholar]
- 12. Geevasinga N, Menon P, Howells J, Nicholson GA, Kiernan MC, Vucic S.. Axonal ion channel dysfunction in C9orf72 familial amyotrophic lateral sclerosis. JAMA Neurol. 2015;72(1):49–57. [DOI] [PubMed] [Google Scholar]
- 13. Burke T, Pinto-Grau M, Lonergan K, et al. . A Cross-sectional population-based investigation into behavioral change in amyotrophic lateral sclerosis: Subphenotypes, staging, cognitive predictors, and survival. Ann Clin Transl Neurol. 2017;4(5):305–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ganesalingam J, Stahl D, Wijesekera L, et al. . Latent cluster analysis of ALS phenotypes identifies prognostically differing groups. PLoS ONE. 2009;4(9):e7107. Rubinsztein DC, ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mathis S, Goizet C, Soulages A, Vallat JM, Masson GL.. Genetics of amyotrophic lateral sclerosis: A review. J Neurol Sci. 2019;399:217–226. [DOI] [PubMed] [Google Scholar]
- 16. Ravits J, Appel S, Baloh RH, et al. . Deciphering amyotrophic lateral sclerosis: What phenotype, neuropathology and genetics are telling us about pathogenesis. Amyotroph Lateral Scler Front Degener. 2013;14 (Suppl 1):5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mendelsohn AI, Dasen JS, Jessell TM.. Divergent hox coding and evasion of retinoid signaling specifies motor neurons innervating digit muscles. Neuron. 2017;93(4):792–805.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bikoff JB, Gabitto MI, Rivard AF, et al. . Spinal inhibitory interneuron diversity delineates variant motor microcircuits. Cell. 2016;165(1):207–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cedarbaum JM, Stambler N, Malta E, et al. . The ALSFRS-R: A revised ALS functional rating scale that incorporates assessments of respiratory function. J Neurol Sci. 1999;169(1-2):13–21. [DOI] [PubMed] [Google Scholar]
- 20. Abrahams S, Newton J, Niven E, Foley J, Bak TH.. Screening for cognition and behaviour changes in ALS. Amyotroph Lateral Scler Frontotemporal Degener. 2014;15(1-2):9–14. [DOI] [PubMed] [Google Scholar]
- 21. McMackin R, Muthuraman M, Groppa S, et al. . Measuring network disruption in neurodegenerative diseases: New approaches using signal analysis. J Neurol Neurosurg Psychiatry. 2019;90(9):1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Franchignoni F, Mora G, Giordano A, Volanti P, Chiò A.. Evidence of multidimensionality in the ALSFRS-R Scale: A critical appraisal on its measurement properties using Rasch analysis. J Neurol Neurosurg Psychiatry. 2013;84(12):1340–1345. [DOI] [PubMed] [Google Scholar]
- 23. Iyer PM, Mohr K, Broderick M, et al. . Mismatch negativity as an indicator of cognitive sub-domain dysfunction in amyotrophic lateral sclerosis. Front Neurol. 2017;8:395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McMackin R, Dukic S, Costello E, et al. . Localization of brain networks engaged by the sustained attention to response task provides quantitative markers of executive impairment in amyotrophic lateral sclerosis. Cereb Cortex. 2020;30(9):4834–4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Engel AK, Gerloff C, Hilgetag CC, Nolte G.. Intrinsic coupling modes: Multiscale interactions in ongoing brain activity. Neuron. 2013;80(4):867–886. [DOI] [PubMed] [Google Scholar]
- 26. Deligani RJ, Hosni SI, Borgheai SB, et al. . Electrical and hemodynamic neural functions in people with ALS: An EEG-fNIRS resting-state study. IEEE Trans Neural Syst Rehabil Eng. 2020;28(12):3129–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bizovičar N, Dreo J, Koritnik B, Zidar J.. Decreased movement-related beta desynchronization and impaired post-movement beta rebound in amyotrophic lateral sclerosis. Clin Neurophysiol. 2014;125(8):1689–1699. [DOI] [PubMed] [Google Scholar]
- 28. Riva N, Falini A, Inuggi A, et al. . Cortical activation to voluntary movement in amyotrophic lateral sclerosis is related to corticospinal damage: Electrophysiological evidence. Clin Neurophysiol. 2012;123(8):1586–1592. [DOI] [PubMed] [Google Scholar]
- 29. Dukic S, McMackin R, Buxo T, et al. . Patterned functional network disruption in amyotrophic lateral sclerosis. Hum Brain Mapp. 2019;40(16):4827–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ludolph A, Drory V, Hardiman O, et al. ; WFN Research Group on ALS/MND . A revision of the El Escorial criteria - 2015. Amyotroph Lateral Scler Front Degener. 2015;16(5-6):291–292. [DOI] [PubMed] [Google Scholar]
- 31. Strong MJ, Abrahams S, Goldstein LH, et al. . Amyotrophic lateral sclerosis - frontotemporal spectrum disorder (ALS-FTSD): Revised diagnostic criteria. Amyotroph Lateral Scler Front Degener. 2017;18(3-4):153–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Elamin M, Pinto-Grau M, Burke T, et al. . Identifying behavioural changes in ALS: Validation of the Beaumont Behavioural Inventory (BBI). Amyotroph Lateral Scler Front Degener. 2017;18(1-2):68–73. doi: 10.1080/21678421.2016.1248976 [DOI] [PubMed] [Google Scholar]
- 33. Costello E, Lonergan K, Madden C, et al. . Equivalency and practice effects of alternative versions of the Edinburgh Cognitive and Behavioral ALS Screen (ECAS). Amyotroph Lateral Scler Front Degener. 2020;21(1-2):86–91. [DOI] [PubMed] [Google Scholar]
- 34. Pinto-Grau M, Burke T, Lonergan K, et al. . Screening for cognitive dysfunction in ALS: Validation of the Edinburgh Cognitive and Behavioural ALS Screen (ECAS) using age and education adjusted normative data. Amyotroph Lateral Scler Front Degener. 2017;18(1-2):99–106. [DOI] [PubMed] [Google Scholar]
- 35. Balendra R, Jones A, Jivraj N, et al. . Estimating clinical stage of amyotrophic lateral sclerosis from the ALS Functional Rating Scale. Amyotroph Lateral Scler Front Degener. 2014;15(3-4):279–284. [DOI] [PubMed] [Google Scholar]
- 36. Van Veen BD, Van Drongelen W, Yuchtman M, Suzuki A.. Localization of brain electrical activity via linearly constrained minimum variance spatial filtering. IEEE Trans Biomed Eng. 1997;44(9):867–880. [DOI] [PubMed] [Google Scholar]
- 37. Tzourio-Mazoyer N, Landeau B, Papathanassiou D, et al. . Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15(1):273–289. [DOI] [PubMed] [Google Scholar]
- 38. Wang B, Mezlini AM, Demir F, et al. . Similarity network fusion for aggregating data types on a genomic scale. Nat Methods. 2014;11(3):333–337. [DOI] [PubMed] [Google Scholar]
- 39. Ng A, Jordan M, Weiss Y.. On spectral clustering: Analysis and an algorithm. Adv Neural Inf Process Syst. 2002;14. [Google Scholar]
- 40. Wang B, Zhu J, Pierson E, Ramazzotti D, Batzoglou S.. Visualization and analysis of single-cell RNA-seq data by kernel-based similarity learning. Nat Methods. 2017;14(4):414–416. [DOI] [PubMed] [Google Scholar]
- 41. Wang B, Pourshafeie A, Zitnik M, et al. . Network enhancement as a general method to denoise weighted biological networks. Nat Commun. 2018;9(1):3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zelnik-Manor L, Perona P.. Self-tuning spectral clustering. In: Proceedings of the 17th International Conference on Neural Information Processing Systems. NIPS’04. MIT Press; 2004:1601–1608. [Google Scholar]
- 43. Nasseroleslami B. An implementation of empirical Bayesian inference and non-null bootstrapping for threshold selection and power estimation in multiple and single statistical testing. bioRxiv. [Preprint] doi: 10.1101/342964 [DOI] [Google Scholar]
- 44. Cliff N. Dominance statistics: Ordinal analyses to answer ordinal questions. Psychol Bull. 1993;114(3):494–509. [Google Scholar]
- 45. Thomas Yeo BT, Krienen FM, Sepulcre J, et al. . The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106(3):1125–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Iyer PM, Egan C, Pinto-Grau M, et al. . Functional connectivity changes in resting-state EEG as potential biomarker for amyotrophic lateral sclerosis. PLoS ONE. 2015;10(6):e0128682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Blain-Moraes S, Mashour GA, Lee H, Huggins JE, Lee U.. Altered cortical communication in amyotrophic lateral sclerosis. Neurosci Lett. 2013;543:172–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Benjamini Y, Hochberg Y.. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57(1):289–300. [Google Scholar]
- 49. Benjamini Y, Krieger AM, Yekutieli D.. Adaptive linear step-up procedures that control the false discovery rate. Biometrika. 2006;93(3):491–507. [Google Scholar]
- 50. Blondel VD, Guillaume JL, Lambiotte R, Lefebvre E.. Fast unfolding of communities in large networks. J Stat Mech Theory Exp. 2008;2008(10):P10008. [Google Scholar]
- 51. Bede P, Bokde ALW, Byrne S, et al. . Multiparametric MRI study of ALS stratified for the C9orf72 genotype. Neurology. 2013;81(4):361–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Byrne S, Elamin M, Bede P, et al. . Cognitive and clinical characteristics of patients with amyotrophic lateral sclerosis carrying a C9orf72 repeat expansion: A population-based cohort study. Lancet Neurol. 2012;11(3):232–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Beeldman E, Raaphorst J, Twennaar MK, De Visser M, Schmand BA, De Haan RJ.. The cognitive profile of ALS: A systematic review and meta-analysis update. J Neurol Neurosurg Psychiatry. 2016;87(6):611–619. [DOI] [PubMed] [Google Scholar]
- 54. Proudfoot M, Colclough GL, Quinn A, et al. . Increased cerebral functional connectivity in ALS: A resting-state magnetoencephalography study. Neurology. 2018;90(16):e1418–e1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stam CJ, Nolte G, Daffertshofer A.. Phase lag index: Assessment of functional connectivity from multi channel EEG and MEG with diminished bias from common sources. Hum Brain Mapp. 2007;28(11):1178–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sorrentino P, Rucco R, Jacini F, et al. . Brain functional networks become more connected as amyotrophic lateral sclerosis progresses: A source level magnetoencephalographic study. NeuroImage Clin. 2018;20:564–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Baccalá LA, Sameshima K.. Partial directed coherence: A new concept in neural structure determination. Biol Cybern. 2001;84(6):463–474. [DOI] [PubMed] [Google Scholar]
- 58. Anthony M, Lin F.. A systematic review for functional neuroimaging studies of cognitive reserve across the cognitive aging spectrum. Arch Clin Neuropsychol. 2018;33(8):937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rittman T, Borchert R, Jones S, et al. ; Genetic Frontotemporal Dementia Initiative (GENFI) . Functional network resilience to pathology in presymptomatic genetic frontotemporal dementia. Neurobiol Aging. 2019;77:169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vucic S, Nicholson GA, Kiernan MC.. Cortical hyperexcitability may precede the onset of familial amyotrophic lateral sclerosis. Brain. 2008;131(6):1540–1550. [DOI] [PubMed] [Google Scholar]
- 61. Gregory JM, McDade K, Bak TH, et al. . Executive, language and fluency dysfunction are markers of localised TDP-43 cerebral pathology in non-demented ALS. J Neurol Neurosurg Psychiatry. 2020;91(2):149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. McMackin R, Dukic S, Costello E, et al. . Cognitive network hyperactivation and motor cortex decline correlate with ALS prognosis. Neurobiol Aging. 2021;104:57–70. [DOI] [PubMed] [Google Scholar]
- 63. Proudfoot M, Rohenkohl G, Quinn A, et al. . Altered cortical beta-band oscillations reflect motor system degeneration in amyotrophic lateral sclerosis. Hum Brain Mapp. 2017;38(1):237–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fraschini M, Demuru M, Hillebrand A, et al. . EEG functional network topology is associated with disability in patients with amyotrophic lateral sclerosis. Sci Rep. 2016;6(1):38653–38657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fraschini M, Lai M, Demuru M, et al. . Functional brain connectivity analysis in amyotrophic lateral sclerosis: An EEG source-space study. Biomed Phys Eng Express. 2018;4(3):037004. [Google Scholar]
- 66. Michel CM, Brunet D.. EEG source imaging: A practical review of the analysis steps. Front Neurol. 2019;10:325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Michel CM, Murray MM, Lantz G, Gonzalez S, Spinelli L, Grave De Peralta R.. EEG source imaging. Clin Neurophysiol. 2004;115(10):2195–2222. [DOI] [PubMed] [Google Scholar]
- 68. Zelmann R, Lina JM, Schulze-Bonhage A, Gotman J, Jacobs J.. Scalp EEG is not a blur: It can see high frequency oscillations although their generators are small. Brain Topogr. 2014;27(5):683–704. [DOI] [PubMed] [Google Scholar]
- 69. Kuhnke N, Schwind J, Dümpelmann M, Mader M, Schulze-Bonhage A, Jacobs J.. High frequency oscillations in the ripple band (80–250 Hz) in scalp EEG: Higher density of electrodes allows for better localization of the seizure onset zone. Brain Topogr. 2018;31(6):1059–1072. [DOI] [PubMed] [Google Scholar]
- 70. Schomer DL, Lopes da Silva FH, Michel CM, He B.. EEG mapping and source imaging. In: Schomer DL, Lopes da Silva FH.. Niedermeyer's Electroencephalography: Basic Principles, Clinical Applications, and Related Fields. 7 ed. Oxford University Press; 2017. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author on reasonable request from qualified investigators, and are subject to the approvals by Data Protection Officer and The Office of Corporate Partnership and Knowledge Exchange in Trinity College Dublin.