Abstract
Repeat expansions in the C9orf72 gene are a common cause of amyotrophic lateral sclerosis and frontotemporal lobar degeneration, two devastating neurodegenerative disorders. One of the proposed mechanisms of GGGGCC repeat expansion is their translation into non-canonical dipeptide repeats, which can then accumulate as aggregates and contribute to these pathologies. There are five different dipeptide repeat proteins (polyGA, polyGR, polyPR, polyPA and polyGP), some of which are known to be neurotoxic.
In the present study, we used BioID2 proximity labelling to identify the interactomes of all five dipeptide repeat proteins consisting of 125 repeats each. We identified 113 interacting partners for polyGR, 90 for polyGA, 106 for polyPR, 25 for polyPA and 27 for polyGP. Gene Ontology enrichment analysis of the proteomic data revealed that these target interaction partners are involved in a variety of functions, including protein translation, signal transduction pathways, protein catabolic processes, amide metabolic processes and RNA-binding. Using autopsy brain tissue from patients with C9orf72 expansion complemented with cell culture analysis, we evaluated the interactions between polyGA and valosin containing protein (VCP). Functional analysis of this interaction revealed sequestration of VCP with polyGA aggregates, altering levels of soluble valosin-containing protein. VCP also functions in autophagy processes, and consistent with this, we observed altered autophagy in cells expressing polyGA. We also observed altered co-localization of polyGA aggregates and p62 in cells depleted of VCP.
All together, these data suggest that sequestration of VCP with polyGA aggregates contributes to the loss of VCP function, and consequently to alterations in autophagy processes in C9orf72 expansion disorders.
Keywords: ALS, C9orf72, VCP, autophagy
Using BioID2 proximity labelling, Božič et al. identify the interactomes of all five dipeptide repeat proteins associated with C9orf72 repeat expansion. Functional analysis of the interaction between polyGA and VCP reveals sequestration of VCP with polyGA aggregates, resulting in partial loss of VCP function.
Introduction
Amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD) are related neurodegenerative diseases. Mutations in >30 different genes have been linked to ALS and FTLD, including a hexanucleotide GGGGCC repeat expansion (G4C2 repeat) in the C9orf72 gene, which is the most common genetic cause of both ALS and FTLD. Patients with a G4C2 expansion have hundreds, or even thousands, of these repeats, while healthy individuals typically have between two and 23 repeats.1
There are three proposed mechanisms of how the G4C2 repeats cause these diseases. The first is the loss of function of the C9orf72 gene due to haploinsufficiency.2,3 The second is the production of sense and antisense repeat RNA, which can form RNA foci known to bind and sequester RNA-binding proteins.2,4,5 The third proposed mechanism is the translation of repeat RNA by repeat-associated non-ATG translation, resulting in the production of dipeptide repeat proteins (DPRs) that can accumulate in aggregates and contribute to these pathologies. There are five different DPRs, as polyGA, polyGR and polyGP from the sense strand, and polyPA, polyPR and again polyGP from the antisense strand. These are associated with variably toxicities.6-9
Studies have shown that expression of arginine-containing DPRs (i.e. polyGR, polyPR) is toxic in human cell line models and in Drosophila and mouse models.10–12 Although the molecular pathomechanisms of polyGR- and polyPR-induced neuronal toxicity are not fully understood, they are believed to involve cytotoxicity induced by inhibition of translation,13 by nuclear dysfunction, and disruption of nucleocytoplasmic transport.10,14 In addition, many independent studies have demonstrated the cellular toxicity of polyGA in various models,9,15,16 but again with only partially described mechanisms of this pathogenesis. PolyGA is highly prone to aggregation, and can therefore form large cytoplasmic inclusions, which then induce endoplasmic reticulum stress,16 affect nucleocytoplasmic transport proteins and proteasomal degradation.15,17 In contrast, polyGP and polyPA have been reported to be non-toxic.9,14
To gain deeper insight into the molecular mechanisms of DPR-induced cytotoxicity, we performed proximity labelling proteomics for all five of these DPRs and validated the most significant ones. Furthermore, we functionally evaluated the interactions between VCP and polyGA.
Materials and methods
Antibodies
The following antibodies were sourced and used in this study: Myc-tagged mouse monoclonal antibody (9B11; Cell Signalling Technology; #2276), Myc-tagged rabbit polyclonal antibody (Proteintech; #16286–1-AP), histone H3 (trimethyl Lys9) monoclonal antibody (6F12-H4; Novus Biologicals; #NBP1-30141), GAPDH rabbit polyclonal antibody (Proteintech; #10494–1-AP), VCP mouse monoclonal antibody (Proteintech; #60316–1-Ig), B23/NPM1 mouse monoclonal antibody (Proteintech; #60096–1-Ig), NMT1 rabbit polyclonal antibody (Proteintech; #11546–1-AP), RPL23A rabbit polyclonal antibody (Proteintech; #16386–1-AP), MAP4 rabbit polyclonal antibody (Proteintech; #11229–1-AP), ribosomal protein L17 mouse monoclonal antibody (C-8; Santa Cruz Biotechnology; #sc-515904), hnRNPc mouse monoclonal antibody (Santa Cruz Biotechnology; #sc-32308), NOP2 mouse monoclonal antibody (Santa Cruz Biotechnology; #sc-398884), SRP54 mouse monoclonal antibody (Santa Cruz Biotechnology; #sc393855), RRS1 mouse monoclonal antibody (Santa Cruz Biotechnology; #sc-515462), p62 mouse monoclonal antibody (Abcam; #ab 56416), LC3B rabbit monoclonal antibody (Abcam; #ab192890), ubiquitin rabbit monoclonal antibody (Abcam; #ab 134953), VCP rabbit polyclonal antibody (Genetex; # 101089), GA mouse monoclonal antibody (Millipore, # MABN 889), VCP rabbit recombinant antibody (Abcam; #ab 109240), MAP2 chicken polyclonal antibody (Abcam; #ab 5392), SYP mouse monoclonal antibody (Santa Cruz Biotechnology; #sc-17750), HRP goat anti-mouse antibody (Jackson ImmunoResearch; #115–035-068), HRP goat anti-rabbit antibody (Jackson ImmunoResearch; #111–035-045), Alexa Fluor 488 goat anti-rat antibody (Thermo Fisher Scientific; #A11006), Alexa Fluor 546 goat anti-rabbit antibody (Thermo Fisher Scientific; #A11010), Alexa Fluor 555 goat anti-mouse antibody (Thermo Fisher Scientific; #A21424), Alexa Fluor 488 goat anti-rabbit antibody (Thermo Fisher Scientific; #A11008), Alexa Fluor 647 goat anti-rabbit antibody (Thermo Fisher Scientific; #A21071) and rabbit Anti-TAR DNA-Binding Protein 43 (TDP-43) (CosmoBio; #CAC-TIP-PTD-MO1).
Generation of plasmids
The PolyGA, polyGP, polyGR, polyPA and polyPR sequences (as 125 repeats) were a kind gift from Youn-Bok Lee (King's College London, UK.) and were synthesized by GeneArt (Thermo Fisher Scientific), as described previously.17 Each protein sequence of each dipeptide was converted to a DNA sequence using a codon-optimization procedure. In this way, possible effects due to hexanucleotide repeats at the RNA level could be excluded.9 Detailed information about generation of BioID and GFP plasmids are described in the Supplementary material.
Cell culture and transfection
Human embryonic kidney 293T (HEK293T) cells were cultured in Dulbecco’s modified Eagle medium with high glucose, as GlutaMAX supplement (Thermo Fisher Scientific) with the addition of 10% foetal bovine serum (Thermo Fisher Scientific) and 1% penicillin–streptomycin (Thermo Fisher Scientific). For the large-scale pull-down experiments, the cells were plated the day before transfection at 3 × 106 cells per 100-mm plate. The cells were transfected using PolyJetTM (SignaGen Laboratories), according to the manufacturer’s instructions. Neuroblastoma cells SH-SY5Y FlpIn were cultured in DMEM/F12 medium 1 : 1 (Sigma) supplemented with 10% foetal bovein serum (FBS; tetracycline-free) and penicillin–streptomycin solution in a CO2 incubator (5%) at 37°C and 95% air humidity. The primary rat neurons [in vitro Day 7 (DIV7)] were cultured in Neurobasal media with 2% B-27, 1% GlutaMAX and 1% penicillin–streptomycin (all from Invitrogen) with 2% horse serum (Sigma-Aldrich). Cultures were incubated at 37°C with 5% CO2.
For cell fractionation and whole-cell lysate preparation, the cells were plated the day before transfection at 5 × 105 cells/well in six-well plates. The cells were transfected using Lipofectamine 3000 (Thermo Scientific), according to the manufacturer’s instructions. For the autophagic flux experiment, cells were treated with 100 µM bafilomycin A1 (Merck) for 4 h. For VCP silencing, the VCP siRNA (Santa Cruz Biotechnology; # sc-37187) and control siRNA (Santa Cruz Biotechnology; # sc-37007) were transfected using PepMute siRNA Transfection Reagent (SignaGene Laboratories; #SL100566), according to the manufacturer’s instructions. Primary rat cortical neurons (DIV7) were transfected with GA and EGFP plasmids with Lipofectamine 2000 (Thermo Scientific) using 250 ng DNA. Two days later, cells were fixed with 4% PFA for 15 min and ICC was performed.
FlpIn SH-SY5Y DPR cells generation
SH-SY5Y-TR-FRT cell line was generated as described before.18 FlpIn SH-SY5Y-TR-FRT cells containing a single FRT insertion site were used for the generation of an inducible SH-SY5Y-TR-FRT-mScarletI-DPR cell line. Gene sequences of PolyGA, polyGP, polyPA and polyPR were cut out of the pEGFP constructs using KpnI and HindIII restriction enzymes (both Thermo Scientific) and polyGR construct were cut out of a pcDNA5-myc plasmid by KpnI and EcoRV (Thermo Scientific) enzymes. All inserts were ligated into a pcDNA5-FRT-TO-mScarlet vector. The newly constructed plasmids were sequenced to ensure the correct sequence. SH-SY5Y-TR-FRT cells were cotransfected with pcDNA5-FRT-TO-mScarletI-GA, pcDNA5-FRT-TO-mScarletI-GR, pcDNA5-FRT-TO-mScarletI-PR, pcDNA5-FRT-TO-mScarletI-GP or pcDNA5-FRT-TO-mScarletI-PA and pOG44 vectors at 1:9 ratio using Lipofectamine 3000 and a stable SH-SY5Y-TR-FRT-mScarletI-GA, SH-SY5Y-TR-FRT-mScarletI-GR, SH-SY5Y-TR-FRT-mScarletI-PR, SH-SY5Y-TR-FRT-mScarletI-GP and SH-SY5Y-TR-FRT-mScarletI-PA cell lines were established by hygromycin (110 μg/μl) selection and single cell cloning. For production of DPRs, the FlpIn SH-SY5Y DPR cell lines were induced by 1 μg/ml of doxycycline (Sigma-Aldrich).
Diferentiation of FlpIn SH-SY5Y DPR cell lines
All five FlpIn SH-SY5Y DPR cell lines were differentiated into neuron cells. Cells were plated onto 24-well plates with poly-L-lysine-coated coverslips to achieve 50–70% confluence. The next day cells were washed with phosphate-buffered saline (PBS) and the differentiation media containing DMEM/F12 medium supplemented with 0.5% FBS (tetracycline-free), penicillin–streptomycin and 10 µM retinoic acid was added. On Day 4 the differentiation media was changed and new differentiation media containing DMEM/F12 medium supplemented with 0.5% FBS (tetracycline-free), penicillin–streptomycin, 10 µM retinoic acid and 50 ng/ml of BDNF was added. On Day 6, cells were fixed and ICC was performed.
Subcellular fractionation of cells and tissue
Cell fractionation was performed as described previously by Prpar Mihevc etal.19 Briefly, the cells were grown in six-well plates and transfected as described. After 24 h they were lysed in cold cell lysis buffer [50 mM Tris, pH 7.4, 10 mM NaCl, 0.5% Igepal Ca-630 (Sigma-Aldrich), 0.25% Triton X-100, protease inhibitor cocktail (Roche)] and centrifuged at 3000g for 5 min at 4°C. The soluble supernatants were transferred to fresh tubes and centrifuged at 16 100g for 10 min at 4°C. The first pellets were washed three times in cell lysis buffer, resuspended in 1× Laemmli buffer, sonicated, boiled for 5 min and centrifuged again. The supernatants obtained represented the insoluble fractions. Tissue fractionation was performed using Subcellular Protein Fractionation kits for tissues (Thermo Scientific; # 87790), according to the manufacturer’s instructions, frontal cortex, frozen tissue, n = 3 for normal controls and n = 3 from C9orf72 familial ALS. All the tissues were processed at the same time.
Immunoblotting
Further details of immunoblotting and autophagy analyses protocols can be found in the Supplementary material.
BioID2 pull-down assay
The BioID2 pull-down assays were performed as described previously.20 Briefly, for the large-scale pull-down assays, 24 h after transfection, 50 µM biotin was added to the cells in 100-mm plates. After a 16-h incubation with biotin, the cells were washed three times in Dulbecco's PBS, and lysed in 600 µl of lysis buffer [50 mM Tris, pH 7.4, 500 mM NaCl, 0.2% sodium dodecyl sulphate, 1 mM dithiothreitol, protease inhibitor cocktail (Roche)]. When the cells were collected, Triton X-100 was added to 2% final concentration. After sonication (UP200S; Hielscher Ultrasound Technology) an equal volume of cold Tris pH 7.4 was added to the lysates. The lysates were centrifuged at 16 500g for 10 min at 4°C, and the supernatant was collected and incubated with 100 µl Dynabeads MyOne Streptavidin T1 (Thermo Fisher Scientific) overnight on a rotating mixer at 4°C. After incubation, the beads were collected using a magnetic stand and washed twice with 1.5 ml 2% (w/v) sodium dodecyl sulphate. During each washing step, the samples were incubated on a rotating mixer for 8 min, followed by collection of the beads on a magnetic stand. Then the beads were washed three times with wash buffer 1 [0.1% (w/v) deoxycholic acid, 1% (w/v) Triton X-100, 1 mM EDTA, 500 mM NaCl, 50 mM HEPES, pH 7.5], then three times with wash buffer 2 [0.5% (w/v) deoxycholic acid, 0.5% (w/v) NP-40, 1 mM EDTA, 250 mM LiCl, 10 mM Tris, pH 7.4] and once with 50 mM Tris, pH 7.4. Finally, 20% of the samples was saved for future analysis, and the remaining 80% was resuspended in 50 µl 50 mM NH4HCO3 for mass spectrometry.
Immunofluorescence—co-localization analysis
The cells were grown on poly-L-lysine-coated coverslips in 24-well plates at a density of 1 × 105 cells/well. Twenty-four hours after transient transfection using PolyJet (SignaGen Laboratories), according to the manufacturer’s instructions, the cells were fixed using 4% paraformaldehyde in PBS or absolute methanol (chilled at −20°C) for 15 min, and permeabilized with 0.1% Triton X-100 in PBS for 5 min. After three washing steps with PBS, the coverslips were blocked in blocking buffer [10% goat serum (Euroclone) in PBS, containing 0.1% Tween] at room temperature for 30 min. Then the coverslips were incubated with the primary antibodies in 1% BSA in PBS, containing 0.1% Tween overnight at 4°C. After three washes in PBS, the coverslips were incubated with Alexa Fluor 488- or 647-conjugated secondary anti-rabbit or anti-mouse antibodies (Cell Signaling Technology) in 1% BSA in PBS, containing 0.1% Tween, for 1 h at room temperature. Alternatively, checking for biotinylated proteins in cells, on fixation and washing, coverslips were incubated with ATTO-488 streptavidin (ATTO-Tech; #AD488-61). After additional washing steps, the nuclei were counterstained with DAPI and the coverslips were mounted on glass slides using ProLong Gold (Thermo Fisher Scientific). Images were acquired using an inverted laser scanning microscope (LSM 710; Zeiss) with an oil immersion objective (Plan-Apochromat 63×/1.4 NA M27) with immersion oil (Carl Zeiss), using the ZEN 2011 image software (Carl Zeiss).
In situ proximity ligation assay
The insitu proximity ligation assays were performed according to the manufacturer’s instructions (Duolink PLA, Sigma-Aldrich). After fixation and blocking steps, the coverslips with cells were incubated with a Myc-tagged rabbit polyclonal antibody (Proteintech; #16286–1-AP) and the VCP mouse monoclonal antibody (Proteintech; #60316–1-Ig), overnight at 4°C. For the Myc-tagged-VCP interaction proximity ligation assay probe, anti-mouse minus was used for VCP and anti-rabbit plus was used for the Myc tag. The probes were diluted (Duolink insitu antibody diluent) and incubated for 1 h at 37°C in a humidified chamber. Ligation and amplification were performed using detection reagent orange (Duolink). Controls were prepared in the absence of one or both of the antibodies. After additional washing steps, the nuclei were counterstained with DAPI and the coverslips were mounted on glass slides (ProLong Gold; Thermo Fisher Scientific) followed by image acquisition.
Mass spectrometry
All of the samples were subjected to on-bead digestion. Beads in 50 µl NH4HCO3 buffer were diluted with 100 µl Tris-HCL, pH 7.8 and digested overnight at 37°C with 1 µg trypsin. After acidifying to a final 1% formic acid, the samples were desalted on C-18 cartridges (SOLA HRP) and dried under vacuum centrifugation. The samples were resuspended in 50 µl 100 mM triethylammonium bicarbonate (TEAB). Peptide quantification was carried out using colorimetric peptide quantification kit (Pierce) with TEAB diluted to 5 mM. According to these, 15.6 µg peptide was labelled with the tandem mass tags (TMT) as follows, as triplicates fo all samples: BioID + TMT126; GP + TMT127N; GA + TMT127C; PA + TMT128N; GR + TMT128C; PR + TMT129N; and two pools as 1/18Pool TMT 129C and 1% Pool (1% of each sample) TMT 130N. The samples were separated on a 50-cm spray column (ES803; EASY, Thermo Fisher Scientific) and analysed on a Lumos platform (Dionex Ultimate 3000/Orbitrap Fusion; Thermo Fisher Scientific), as described previously.21 The data were acquired using the MultiNotch MS3 method22 with the parameters as deposited.23
The liquid chromatography–tandem mass spectrometry data were analysed using Proteome Discoverer 2.2. The proteins were identified using Sequest HT, against a reviewed Uniprot Homo sapiens database (retrieved June 2018). The search parameters included: mass tolerances, 10 ppm for precursor and 0.5 Da fragment mass tolerance; TMT10plex (K), acetylation (protein N-term) and oxidation (M) were set as dynamic modifications, and TMT10plex (N-term) and alkylation (C) as static modifications; up to two missed cleavages with trypsin; and the results were filtered to 1% [e discovery rate stringency based on a target-decoy database strategy (percolator)].24 The triplicate sample sets were quantified and the fold-change enrichment of proteins were determined relative to the negative control (BioID), which represented any changed association of the identified proteins with the DPR substrates.
Bioinformatics
The significantly enriched Gene Ontology (GO) terms for the DPR interactors were searched for using the DAVID bioinformatics tool (DAVID Bioinformatics Resources 6.8; available at https://david.ncifcrf.gov/ accessed 20 September 2020) and GOrilla (gene ontology and enrichment analysis and visualization tool) available at http://cbl-gorilla.cs.technion.ac.il/, accessed 18 September 2020. For analysis of enriched GO ontologies (e.g. cellular compartmentation, molecular functions, biological processes) of the various DPR interaction partners (specific DPR versus pooled control), the interacting partners used were only those where more than two unique peptides were obtained using mass spectrometry, and where the fold-change compared to the control had at least doubled (Fc > 2), and P < 0.05 determined. In addition, a search for functional association of VCP within each DPR’s candidate set, has been performed by BioMine tool (https://biomine.ijs.si/, accessed 18 July 2021).
Human post-mortem tissue
The human post-mortem brain (i.e. cortex, hippocampus) samples fixed in buffered formalin were obtained from the archives of the Department of (Neuro)Pathology (Amsterdam UMC, University of Amsterdam, The Netherlands), n = 6 C9orf72 ALS-FTD patients, and n = 5 age-matched controls. For western blot analysis frozen tissue samples from cortex [n = 3 C9orf72 ALS-FTD patients, n = 3 controls were used from the archives of the Department of (Neuro) Pathology (Amsterdam UMC, University of Amsterdam, The Netherlands) and the Institute of Neuropathology (Aachen, Germany)]. Both the control and the ALS cases were selected from a retrospective searchable neuropathological database that was reviewed independently by two neuropathologists (E.A. and Dirk Troost), which included cases with consent for post-mortem brain/spinal cord autopsy and use of the autopsy tissue and their medical records for research purposes. All of the C9orf72 ALS-FTD patients suffered from clinical signs and symptoms of lower and upper motor neuron disease, with the eventual involvement of the cortex and brainstem motor nuclei. All of the patients had fulfilled the diagnostic criteria for ALS.25 The control participants included in the present study were adult individuals without any history of neurological diseases, based on their last clinical evaluation. The clinical details of the cases used in this study are provided in Supplementary Table 1.
Diaminobenzidine staining
Further details on diaminobenzidine staining can be found in the Supplementary material.
Immunofluorescence of cortex and hippocampal tissue sections
Double immunofluorescence staining was performed as described.27–29 In brief, deparaffinized tissue sections were heated in citrate buffer, pH 6 (Dako), for 20 min in a pressure cooker. Sections were blocked with 10% normal goat serum (Life Technologies), for 1 h at room temperature, and incubated with the required primary antibody (dilutions: VCP, 1:50; GA, 1:100; GR, 1:100), at 4°C overnight. After washing in TBST for 10 min, the sections were incubated with Alexa conjugated secondary antibody (dilution, 1: 500 in PBS) at room temperature for 2 h. Sections were washed in TBST (2 × 10 min) and stained for 10 min with 0.1% Sudan Black in 80% ethanol, to suppress endogenous lipofuscin auto-fluorescence. Finally, the sections were washed for 5 min in TBST and mounted with Vectashield mounting medium (Vector Laboratories) containing DAPI. Images were obtained with a laser scanning confocal microscope (LSM 700; Zeiss) using the 40× and 63× objectives (Zeiss). Images were acquired by averaging four scans per area of interest resulting in an image size of 1024 × 1024 pixels. The laser intensity was kept constant for all of the sections examined and images were analysed using Adobe Photoshop CS5 and ZEN (Blue edition) 2009 software.
Ethics statement
The human post-mortem tissue samples were used in compliance with the Declaration of Helsinki. The studies were approved by the Ethical Committees of the Medical Faculty, RWTH Aachen (EK 127/18) and of the Academic Medical Center, Amsterdam (W11 073).
Statistical analysis
Data are presented as means ± standard deviation. Prism software (GraphPad Software, USA) was used for the statistical analysis, using either unpaired or paired Student’s t-tests or one-way ANOVA with Dunnett’s or ANOVA with Tukey’s multiple comparisons test. The use of each specific test is depicted in the figure legends. P-values <0.05 were considered statistically significant.
Data availability
All of the data are available on reasonable request. The liquid chromatography–tandem mass spectrometry data are available via ProteomeXchange with identifier PXD024120.
Results
Characterization of the DPR interactome in HEK293 cells using BioID2 proximity-dependent labelling
To determine which proteins interact with the DPRs in living cells, the five different DPRs were conjugated to biotin ligase, which can biotinylate proteins, based on proximity.30 DPRs with 125 repeats of GA, GP, GR, PA and PR were conjugated at the C -terminus with the Myc-tagged birA enzyme and an intermediate 13-amino-acid-long linker was included for GA, GP and GR, and a 20-amino-acid-long linker for PA and PR (Fig. 1A). The DNA sequence encoding the codon optimized DPRs with an ATG start that allowed protein-induced effects to be tested, thus ruling out the negative effects of RNA mediated by G4C2 repeats.
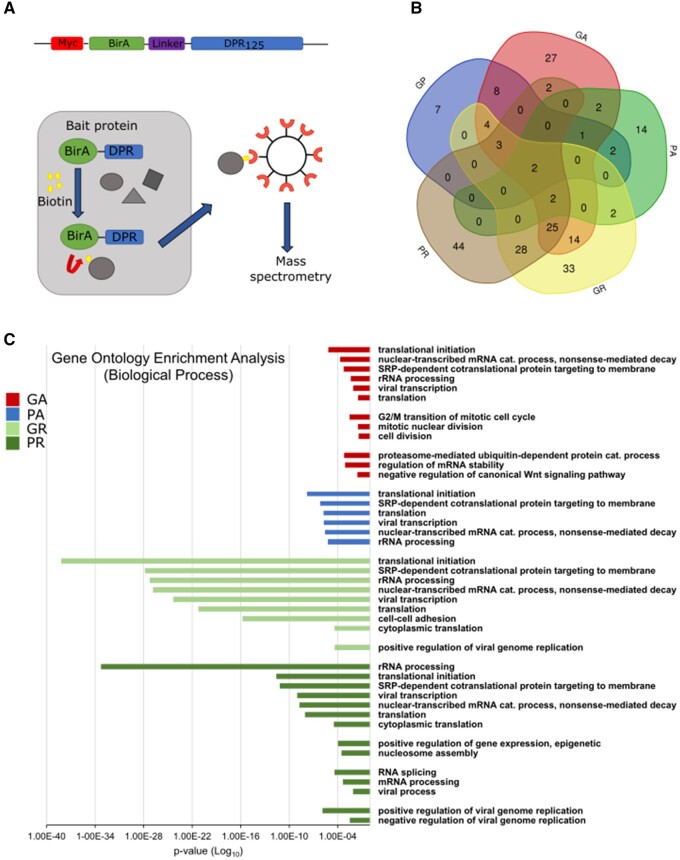
Figure 1.
Interactome of all five DPRs using BioID2 pull-down. (A) Schematic representation of constructs and pull-down experiments. The five DPR constructs consisted of 125 DPRs. (B) Venn diagram illustrating the overlap between the total interactors of the five DPRs after proteomic analysis, for Quant ratio versus control >2.0. The Venn diagram was prepared using the online tool provided by VIB and Ghent University (http://bioinformatics.psb.ugent.be/webtools/Venn/). (C) GO enrichment analysis for the interactors with Quant ratio versus BioID >2.0, showing the involvement of DPR interactors in different biological processes. There was no enrichment in the biological processes for polyGP interactors. P-values were derived via the DAVID algorithm.
The constructs were transiently transfected into HEK293T cells. The localization and biotinylation efficiency of the overexpressed DPRs were determined for each experiment separately, by immunoblotting and immunocytochemistry (Supplementary Fig. 1). Supplementary Fig. 1A shows polyGA mainly localized as distinct perinuclear aggregates, as reported previously.31,32 PolyGP and polyPA were soluble and evenly distributed in the cell cytoplasm, similar to the control BioID2. PolyGR also distributed in the cell cytoplasm and showed accumulation in the nucleoli of certain cells. PolyPR was predominantly in the form of nucleolar aggregates, as reported previously.31,33 Staining of biotinylated proteins (with streptavidin-Atto 488) showed the level of co-localization of the overexpressed DPRs with the biotinylated protein interactors (Supplementary Fig. 1B).
After pull-down purification of the biotinylated protein candidates, liquid chromatography–mass spectrometry proteomic analysis was performed to identify the interactors, which included three biological replicates. Samples of cells transfected with the Myc-BioID vector served as controls. In total, 220 proteins bound to different DPRs with two or more unique peptides and a fold-change >2.0 (Quant ratio versus control; Supplementary Table 2A) were revealed. We identified 27 interactors for polyGP and 25 for polyPA. The number of interactors was higher in the case of the hydrophobic polyGA, where 90 interacting proteins were identified, similar to polyPR and polyGR, with 106 and 113, respectively (Fig. 1B). This is consistent with the notion that polyGP, polyPA and polyGA are all uncharged; however, unlike polyGA, the first two DPRs have a flexible coil structure and are unable to aggregate by themselves.33–36 Consistent with their structures, polyGP and polyPA possibly interact with fewer intracellular proteins when compared to other DPR species and are thus less toxic.33 Only two of the proteins identified were interactors of all of these five different DPRs (Fig. 1B), namely nucleophosmin (NPM1) and nascent polypeptide-associated complex subunit α and muscle-specific form (NACA). Previous studies have shown that polyPR co-localizes with NPM1.33,37,38 For polyGA, 27 of the 90 interactors were unique to polyGA, while 25 of the interactors were also interactors of polyGR and polyPR (Fig. 1B).
DPR protein interactors are involved in various biological processes
To identify overrepresented biological processes and molecular functions for our hit list, we performed GO enrichment analysis using the DAVID Functional Annotation Tool.39,40 For the GO analyses, the increased number of interacting partners, including proteins with one or more unique peptides and a fold-change >2.0 was used (Quant ratio versus control, Supplementary Table 2B). The GO analysis for each set of interactors showed enrichment in proteins involved in the biological processes of translational initiation, translation, rRNA processing, SRP-dependent cotranslational protein targeting of membranes and nuclear-transcribed mRNA catabolic processes (Fig. 1C). For polyGA, the interactors were also seen to be involved in mitotic nuclear division, and proteasome-mediated ubiquitin-dependent catabolic processes. For polyPR, there was enrichment in proteins involved in nucleosome assembly and RNA splicing, in agreement with the nucleolar localization of overexpressed polyPR (Fig. 1C). GO analysis by cellular compartments confirmed the identified location of interactors in the nucleus, nucleolus and ribosomes, with the levels of significance higher for polyGR/PR interactors (Supplementary Fig. 2A). Of note, the last is consistent with previous co-immunoprecipitation studies.13,33,41,42
When the interactors of all five of DPRs were compared according to their molecular functions, they shared RNA-binding function. In addition, for polyGA and polyPR, there were interactors involved in ATP binding (Supplementary Fig. 2B).
Validation of protein interactors by immunoblotting and immunocytochemistry
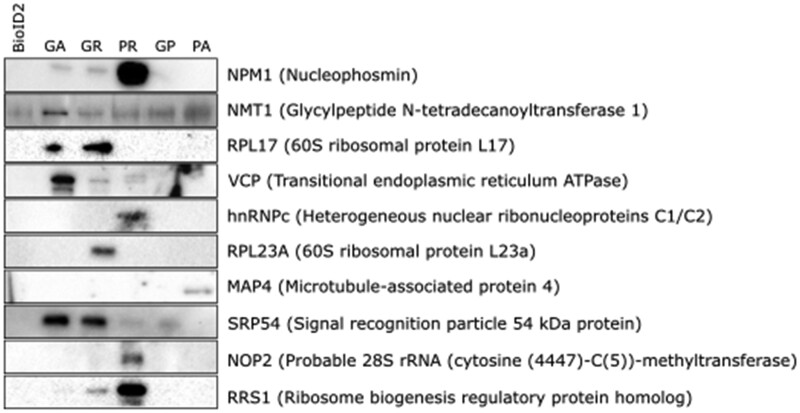
To validate the proteomic data on DPR interactors obtained, pull-down samples 27 specific interactors (characterized by the most significant fold-change and/or link to ALS) were tested and 12 that worked for western blot were quantified for enrichment. In all, 10 of protein interaction candidates were confirmed to interact with: polyGA: NPM1, NMT1, RPL17, VCP and SRP54; polyGR: NPM1, NMT1, RPL17, VCP, RPL23A, SRP54 and RRS1; polyPR: NPM1, HNRNPC, SRP54, NOP2 and RRS1; polyGP: NMT1 and SRP54; polyPA: MAP4, whereas two of them USP5 and ANXA5 did not show enrichment in polyGA samples (Fig. 2 and Supplementary Fig. 3). Immunocytochemistry analysis of co-localization and/or redistribution of interactors showed significant co-localization only for polyGA and the VCP (Fig. 3A). Co-localization or redistribution was not observed for NPM1, NMT1, RPL17, HNRNPC, RPL23A, MAP4, SRP54, NOP2 and RRS1, due to the diffuse distribution of the soluble proteins all over the cell.
Figure 2.
DPR interactors validated by immunoblotting. Representative immunoblotting showing validation of the 10 indicated DPR interactors (out of 27) in pull-down assays after BioID.
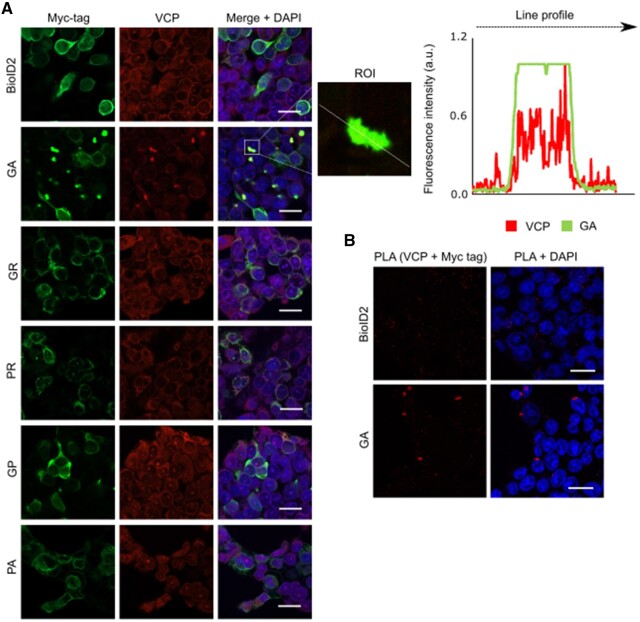
Figure 3.
Cellular localization of DPRs and VCP in HEK293 cells. (A) HEK293 cells transfected with the DPR constructs, revealed using an anti-Myc antibody (green) and endogenous VCP (red). VCP co-localized with polyGA, but not with the other DPRs. The line profile plot indicates the intensity distribution of the green and red channels through the white line in the magnified view of the region of interest (ROI) in the merged panel (middle). Scale bars = 20 µm. (B) Representative proximity ligation assay (PLA) using anti-Myc and anti-VCP antibodies revealed interactions between polyGA and VCP (red dots).
Moreover, the analysis of the interacting protein candidates with the VCP within each DPR’s candidate set, was performed in BioMine (https://biomine.ijs.si/) (Supplementary Fig. 4). There it is shown that among the set of polyGA identified protein interactors are indeed a lot of them that directly functionally associate with the VCP pathway.
VCP co-localizes with polyGA aggregates
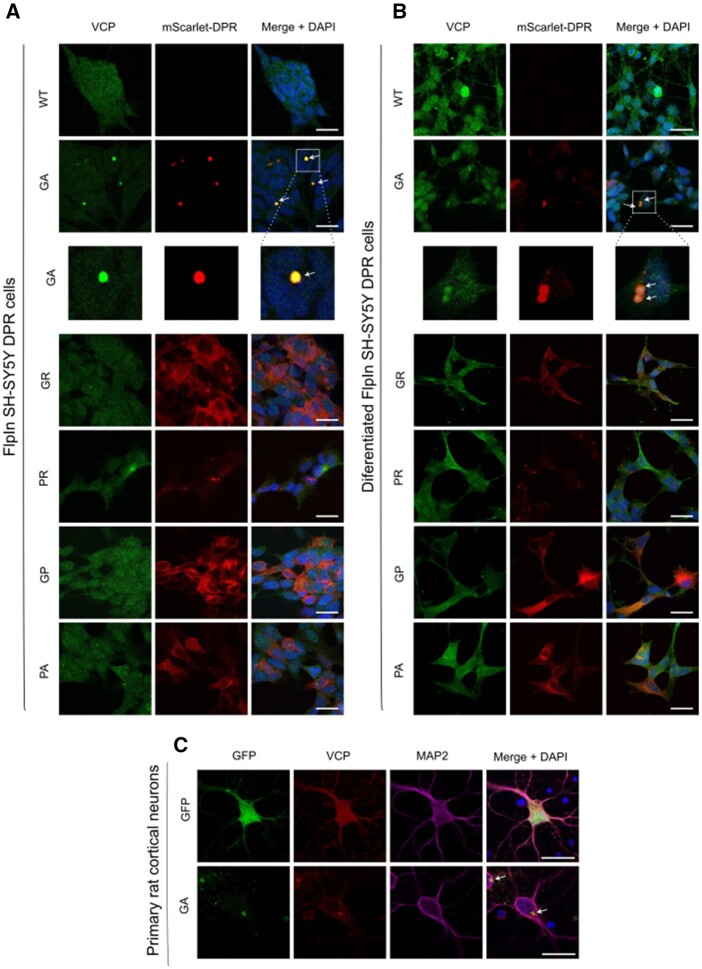
To validate the interactions between the DPRs and VCP in cells, Myc-tagged DPR constructs were expressed in HEK293 cells and stained for Myc and endogenous VCP (Fig. 3A). VCP showed co-localization with polyGA aggregates, but not with the other DPRs. The co-localization of VCP and polyGA was confirmed using line profile plots (Fig. 3A). This interaction between VCP and polyGA was additionally confirmed in HEK293 cells using in situ proximity ligation assays (Fig. 3B). Furthermore, we confirmed co-localization of polyGA and VCP in FlpIn SH-SY5Y GA cells (Fig. 4A) and their differentiated neuronal derivative stably expressing polyGA (Fig. 4B and Supplementary Fig. 5), and in primary rat cortical neurons transfected with GFP-GA expression plasmids (Fig. 4C).
Figure 4.
Cellular localization of DPRs and VCP in FlpIn SH-SY5Y DPR cell lines and their differentiated neuronal derivatives. (A) FlpIn SH-SY5Y DPR cell lines activated with doxycycline for 24 h (red) and endogenous VCP (green). VCP co-localized with polyGA, but not with the other DPRs. The enlarged images represent co-localized VCP in GA aggregates. Scale bars = 20 µm. (B) Differentiated FlpIn SH-SY5Y DPR activated with doxycycline for 24 h (red) and endogenous VCP (green). VCP co-localized with polyGA, but not with the other DPRs. The enlarged images represent co-localized VCP in GA aggregates. Scale bars = 20 µm. (C) Primary rat cortical neurons (DIV7) transfected with EGFP-GA and EGFP plasmids (green). The cells were stained with VCP (red) and MAP2 (purple) antibody and images were taken by Nikon A1R confocal. Scale bar = 20 µm.
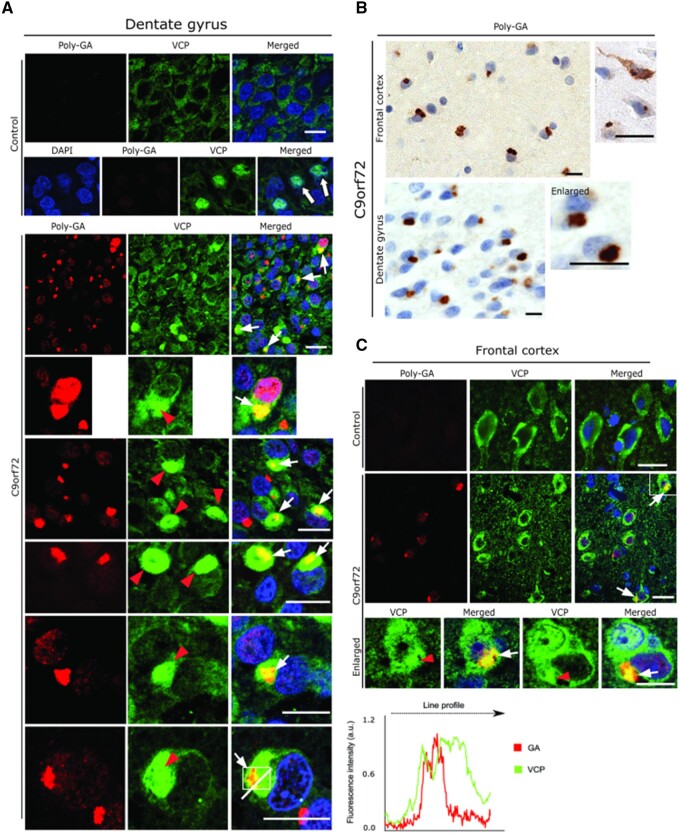
We next sought to validate the cell culture results in autopsy brain tissue obtained from C9orf72 ALS-FTD patients, in comparison to non-neuropathological (‘normal’) controls. Serial sections from the frontal cortex and hippocampus (Fig. 5A and B) were analysed here, where abundant polyGA aggregates were detected using a polyGA antibody (Fig. 5B).
Figure 5.
Neuropathological analysis of VCP in cortex and hippocampus of C9orf72 ALS/FTLD patients. (A) Representative double immunofluorescence labelling using antibodies against VCP and polyGA in the dentate gyrus from normal controls (top) and from C9orf72 ALS and FTLD patients (bottom). Note the large globular aggregates of VCP (red arrowheads) that are co-localized with polyGA aggregates (white arrows). The line profile shows co-localization of GA and VCP in one of the aggregates. (B) Representative DAB immunohistochemistry of frontal cortex (top) and hippocampus (bottom) from C9orf72 ALS and FTLD patients using an antibody against polyGA. Note the abundant polyGA aggregates in the cortical and dentate gyrus neurons. (C) Representative double immunofluorescence labelling using antibodies against VCP and polyGA in the frontal cortex. Note the large globular aggregates of VCP that are co-localized with polyGA aggregates in the cortical neurons (white arrows). Paraffin sections; scale bars = 15 µm.
Co-immunolabelling using anti-VCP and anti-GA antibodies revealed distinct patterns of immunoreactivity, where >70% of the cortical and hippocampal dentate gyrus neurons showed presence of large globular aggregates of VCP (Fig. 5A and C, red arrowheads; bottom enlarged panels). Such large globular aggregates of VCP appeared to actually decorate the polyGA aggregates and were found to be often co-localized (Fig. 5A and C, white arrows), which were later confirmed by line profile obtained from one of the representative dentate gyrus neuron harbouring both VCP and GA aggregate (Fig. 5A, right). Further examination of these C9orf72 patients hippocampal dentate gyrus neurons revealed that such VCP inclusions, which decorates GA aggregates were also co-aggregating with pTDP-43 aggregates (Supplementary Fig. 6A). Interestingly, in many instance GA aggregates appeared in the core of pTDP-43 inclusions (Supplementary Fig. 6B). In contrast, co-immunolabelling using the VCP antibody together with GR antibody did not yield such significant co-localization in the cortical and hippocampal dentate gyrus neurons (data not shown). The cortical and the dentate gyrus neurons from the normal control brain tissue showed a basal level of cytoplasmic (top panel) as well as nuclear immunoreactivity of VCP in many neurons (arrows in control panel; Fig. 5A), and they were devoid of polyGA aggregates (Fig. 5A and C).
PolyGA sequesters VCP
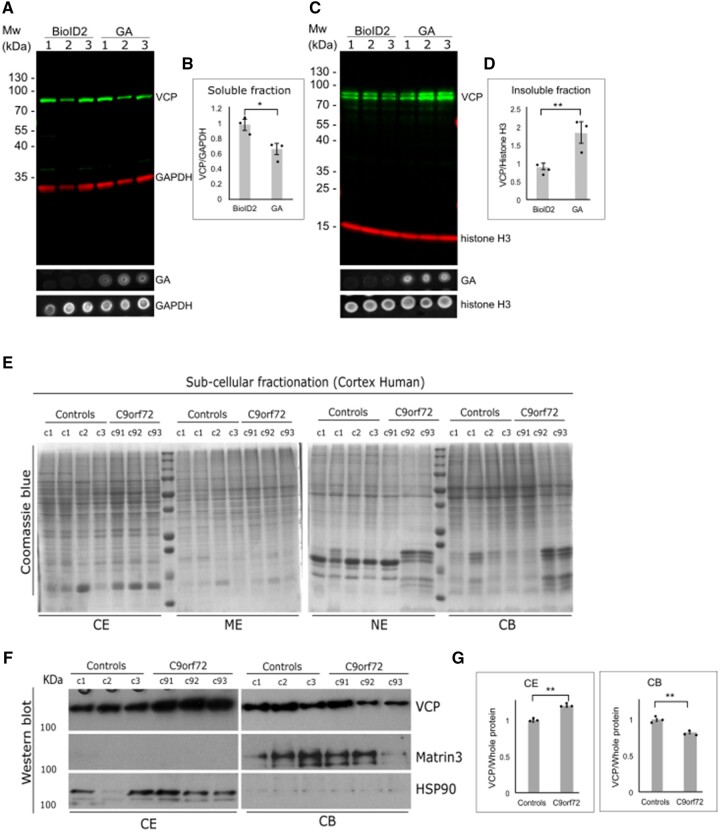
To understand the pathophysiology of this VCP–polyGA interaction in both cell culture and in C9orf72 cases, immunoblotting for VCP was performed with soluble/cytosolic fractions and insoluble/nuclear fractions. Here, we analysed VCP in three independent lysates of HEK293 cells transfected with the BioID2 construct, and three lysates of HEK293 cells transfected with the BioID- polyGA construct. Figure 6A and B both show lower VCP levels in the soluble fraction. In contrast, the VCP content was higher in the insoluble fraction of polyGA-expressing cells (Fig. 6C and D). The level of GA expression was tested by dot blots (Fig. 6A and C). There was also sequestration of VCP within the polyGA aggregates (Figs 3A, 4 and 5), with these large insoluble aggregates formed by polyGA explaining the higher VCP levels in the insoluble fraction.
Figure 6.
PolyGA sequesters VCP into GA aggregates. (A) HEK293 cells were transfected with BioID or polyGA, incubated for 24 h, and then the soluble fraction was extracted. Representative immunoblotting with anti-VCP antibodies (green) and anti-GAPDH antibodies (loading control; red). The dot blot shows expression level of polyGA in soluble fraction. (B) Quantification of immunoblotting as in A. Data are means ± SEM (n = 3 biological repeats). *P ˂ 0.05 (unpaired t-test). (C) HEK293 cells were transfected with BioID or polyGA, incubated for 24 h, and then the insoluble fraction was extracted. Immunoblotting with anti-VCP antibodies (green) and anti-histone H3 antibodies (loading control; red). The dot blot shows expression level of polyGA in insoluble fraction. (D) Quantification of immunoblotting as in C. Data are means ± SEM (n = 3 biological repeats). **P ˂ 0.01 (unpaired t-test). (E) Fractionation of brain tissue of three control samples (control) and three samples from C9orf72 patients (C9). The Coomassie blue gels show loading of total proteins. (F) Immunoblotting shows the amounts of VCP in the cytoplasmic extract (CE) and chromatin-bound extract (CB) fractions. MATR3 and HSP90 represent fractionation control for chromatin-bound and cytoplasmic extracts, respectively. (G) Quantification of immunoblotting as in F, normalized for total protein. Data are means ± SEM (n = 3 normal control and n = 3 C9orf72 ALS-FTD). **P ˂ 0.001 (unpaired t-test). ME = membrane extract; NE = nuclear extract.
To support these findings in C9orf72 ALS-FTD, subcellular fractionation was performed using frozen human brain tissue (frontal cortex) from three normal controls as well as from three C9orf72 ALS-FTD patients. To avoid ambiguity and to maintain the consistency, we used the frozen tissue from the same brain samples that were used for the immunohistochemical analysis (Fig. 5) and also used the same VCP antibody for the western blot analysis. These subcellular fractions obtained from the brain lysates were defined as the cytoplasmic extract, membrane extract, nuclear extract, chromatin-bound and extract.
We used cytoplasmic and nuclear chromatin-bound extracts for further analysis. The immunoblotting performed on these fractions showed an elevated level of VCP only in the cytoplasmic extract fractions from C9orf72 ALS-FTD patients; however, in contrast the nuclear chromatin-bound fractions showed a significantly reduced levels of VCP (Fig. 6F and G), but no change of VCP was observed in other nuclear extract fraction (not shown) in C9orf72 ALS-FTD patients’ brain compared to the normal controls (Fig. 6E–G).
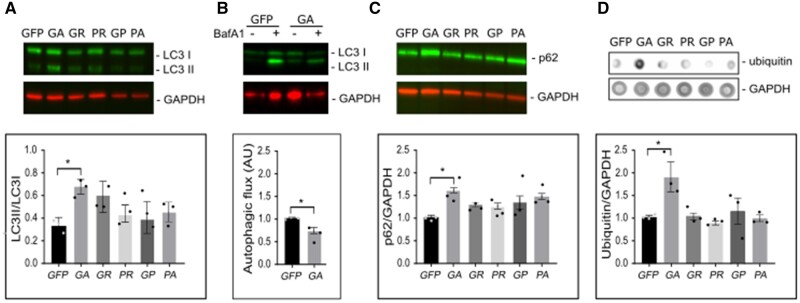
Overexpression of polyGA, but not other DPRs, leads to accumulation of autophagic markers LC3, p62 and ubiquitin
Several recent reports have suggested VCP as a key mediator of autophagy processes, in addition to its major role in mediating UPS mediated degradation44–47 and displaying disaggregase activity on Tau neurofibrillary tangles formation.48 Since we observed that VCP is sequestered specifically with the polyGA aggregates rather than other DPRs, the next logical step was to determine the autophagy status in these cells overexpressing the DPRs, including polyGA. Autophagy can be monitored at different stages by several well-established methods, along with assays based on lipidation of LC3I to LC3II, and the ratio of which is indicative of altered autophagy (by increased autophagy flux due increased autophagosome formation or decreased autophagy flux due decreased autophagosome turnover).43,49 Western blot analysis performed on the lysates of HEK293 cells transfected with the DPRs showed higher levels of LC3II (reflected by increased LC3II/LC3I ratio) specifically in the cells overexpressing polyGA, and not in cells expressing the other DPRs (Fig. 7A). Furthermore, the autophagic flux was measured for control cells and cells that expressed GA in the presence of bafilomycin A (Fig. 7B). Autophagic flux is measured as the difference between LC3 II intensity in bafilomycin treated cells and LC3 II intensity of bafilomycin untreated cells. Here, a significantly lower autophagic flux was observed in cells expressing GA. Consistent with this, increased levels of the autophagy substrate p62 were also observed, indicating inhibited autophagy in the cells overexpressing polyGA.50,51 In agreement with the findings that expression of polyGA can lead to accumulation of ubiquitylated proteins, significantly higher levels of ubiquitin conjugates were observed in dot blot analysis, again, specific to the polyGA-overexpressing cells (Fig. 7D). The decreased autophagic flux and the increase in LC3 II and p62 determine inhibition of autophagy52 in the late stage. Overall, thus our data suggest that polyGA aggregates impair late-stage autophagy.
Figure 7.
Expression of polyGA results in accumulation of LC3, p62 and ubiquitin in HEK293 cells. HEK293 cells were transfected with the GFP-DPR constructs, and analysed 24 h later. (A) Representative immunoblotting showing higher ratio between LC3 II and LC3 I in samples expressing polyGA. Bottom, quantification of immunoblotting. (B) Representative immunoblotting showing lower autophagic flux in cells expressing polyGA. Bottom: Quantification of immunoblotting. (C) Representative immunoblotting showing levels of p62. Bottom: Quantification of immunoblotting. (D) Representative dot blot for levels of ubiquitin. Bottom: Quantification of dot blots. Quantification data are means ± SEM (n = 3 biological repeats). *P ˂ 0.05 (B: unpaired t-tests, A, C and D: one-way ANOVA with Dunnett’s multiple comparisons test). Baf A1 = bafilomycin A1.
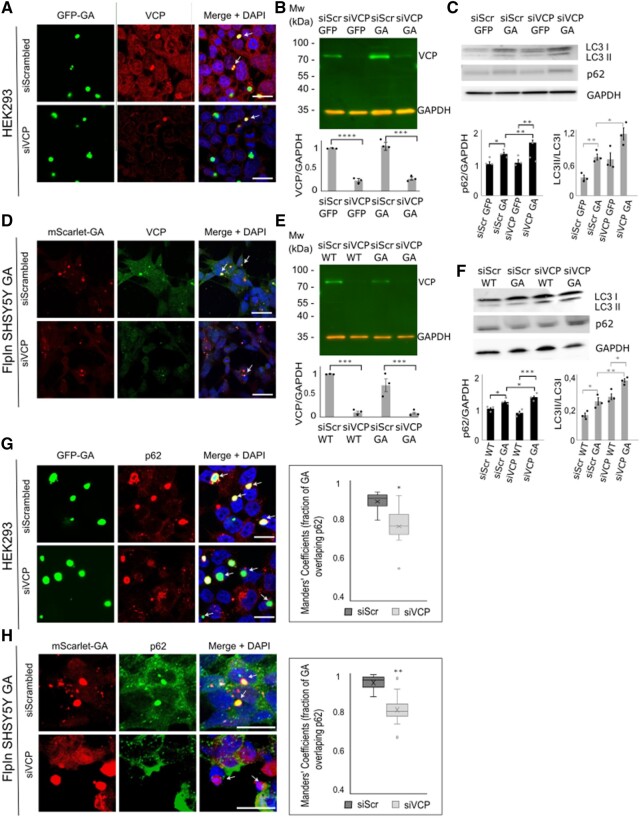
Silencing of VCP decreases the level of co-localization of polyGA and p62
Prompted by the observed accumulation of p62 in polyGA-overexpressing cells, and also with co-localization of VCP and polyGA aggregates, we next performed silencing of VCP to further investigate the functional significance of this VCP–polyGA interaction on p62 localization and autophagy status. HEK293 cells were cotransfected with the siRNA control (siScrambled) and the EGFP-polyGA construct, or with the siRNA sequence against VCP (siVCP) and the EGFP-polyGA construct. Almost 70% silencing of VCP was achieved in cells cotransfected with siVCP and EGFP-polyGA construct compared with control cells transfected with siScrambled RNA and EGFP-polyGA (Fig. 8A and B), whereas nearly 80% silencing of VCP was achieved in FlpIn SH-SY5Y-GA cells (Fig. 8D and E). Immunocytochemistry of these cells confirmed the lower signals for VCP immunoreactivity (Fig. 8A and D). Interestingly, although the levels of VCP were rather low (Fig. 8B and E), VCP was still found to be sequestered within GA aggregates (Fig. 8A and D, bottom panels, arrows). Loss of VCP function and/or defects in VCP can lead to impaired autophagy and cause accumulation of LC3 and p62.53 Consistent with this, we observed increased levels of LC3 by western blot analysis in VCP-depleted cells (Fig. 8C and F), indicating abnormal or inhibited autophagy/autophagosome accumulation. We observed increased accumulation of LC3 and p62 in cells that were VCP-deficient and expressed EGFP-polyGA, versus control cells that were VCP-deficient but expressed only EGFP (Fig. 8C). The same accumulation of LC3 and p62 was observed in FlpIn SH-SY5Y GA cells on VCP silencing (Fig. 8F). These data suggest that polyGA aggregates can sequester the remaining VCP and enhance the accumulation of LC3 and p62. Furthermore, we analysed co-localization of polyGA aggregates with the autophagosome marker p62 (Fig. 8G) in HEK293 cells cotransfected with siScrambled and EGPF-polyGA or siVCP and EGFP-polyGA, and in siScrambled FlpIn SH-SY5Y GA or siVCP FlpIn SH-SY5Y GA cells (Fig. 8H). Here, in the cells with endogenous VCP expression, most polyGA aggregates co-localized with p62 (Fig. 8G and H, top panel, arrows). In contrast, co-localization of polyGA aggregates and p62 appeared decreased in the cells deficient in VCP (Fig. 8G and H, bottom panel). PolyGA/p62 co-localization was quantified by the overlap between polyGA and p62 according to Manders’ overlap coefficients. Analysis of 15 frames from three independent experiments revealed significantly reduced overlap of polyGA with p62 in the VCP-silenced cells (Fig. 8G and H). Given the function of VCP as a mediator of selective autophagy,54 we would suggest here that VCP recognizes polyGA as its substrate and guides it to autophagy.
Figure 8.
VCP deficiency prevents co-localization of polyGA and p62. (A) Representative immunofluorescence of HEK293 cells cotransfected with GFP-GA (green) with siScrambled or siVCP constructs (as indicated) and stained with anti-VCP antibodies (red) and for nuclear DAPI (blue). Arrows show co-localization of polyGA and VCP. Scale bars = 20 µm. (B) Representative immunoblotting showing efficient silencing of VCP (green) in HEK293 cells cotransfected with siScrambled and GFP/GFP-GA or siVCP and GFP/GFP-GAconstructs. Red, GAPDH loading control. Quantification of immunoblotting is also shown. Data are means ± SEM (n = 3 biological repeats). ***P ˂ 0.001 (ANOVA with Tukey’s multiple comparisons test). (C) Representative immunoblotting showing higher LC3II/LC3I ratio and accumulation of p62 protein in HEK293 cells cotransfected with siVCP and GFP-GA. Quantification of immunoblotting is also shown. Data are means ± SEM (n = 3 biological repeats). *P ˂ 0.05 (ANOVA with Tukey’s multiple comparisons test). (D) Representative immunofluorescence of FlpIn SH-SY5Y GA cells (red) transfected with with siScrambled or siVCP constructs (as indicated) and stained with anti-VCP antibodies (red) and for nuclear DAPI (blue). Arrows show co-localization of polyGA and VCP. Scale bars = 20 µm. (E) Representative immunoblotting showing efficient silencing of VCP (green) in FlpIn SH-SY5Y GA and FlpIn SH-SY5Y cells transfected with siScrambled or siVCP. Red, GAPDH loading control. Quantification of immunoblotting is also shown. Data are means ± SEM (n = 3 biological repeats). ***P ˂ 0.001 (ANOVA with Tukey’s multiple comparisons test). (F) Representative immunoblotting showing a higher LC3II/LC3I ratio and accumulation of p62 protein in FlpIn SH-SY5Y GA cells transfected with siVCP. Quantification of immunoblotting is also shown. Data are means ± SEM (n = 3 biological repeats). *P ˂ 0.05 (ANOVA with Tukey’s multiple comparisons test). (G) Representative immunofluorescence of HEK293 cells cotransfected with GFP-GA (green) and siScrambled or siVCP constructs, stained with anti-p62 antibodies (red) and for nuclear DAPI (blue). Arrows: co-localization of polyGA and p62 (yellow) in the top panels; no co-localization in the bottom panels. Scale bars = 20 µm. Right: Quantification of Manders' coefficients for GA overlapping with p62, as shown on the left. Data are means ± SEM from three independent experiments n = 3 (five frames measured for every experiment). *P ˂ 0.05 (two-tailed paired t-tests). (H) Representative immunofluorescence of FlpIn SH-SY5Y GA cells transfected siScrambled or siVCP constructs, stained with anti-p62 antibodies (green) and for nuclear DAPI (blue). Arrows: co-localization of polyGA and p62 (yellow) in the top panels; no co-localization in the bottom panels. Scale bars = 20 µm. Right: Quantification of Manders' coefficients for GA overlapping with p62, as shown on the left. Data are means ± SEM from three independent experiments n = 3 (five frames measured for every experiment). **P ˂ 0.01 (two-tailed paired t-tests).
Discussion
Here, we identified the interactome of all five of these DPRs harbouring 125 pathogenic repeats using proximity-dependent labelling with BioID2. BioID is a unique method to screen for physiologically relevant protein interactions that occur in living cells. The method is unique and has several advantages over other methods for screening protein interactions, especially when searching for interactions with insoluble or membrane-associated proteins. Other advantages include the identifying weak and/or transient interactions and screening for interactions in a relatively natural cellular environment.30
A previous study on the interactome of DPRs (of only 50 repeats) using HEK293 cells overexpressing GFP-DPR constructs and immunoprecipitation with an anti-GFP nanobody, identified 130 interacting partners for polyGR, 147 for polyPR, 14 for polyGA and none for polyPA and polyGP.33 In contrast, in the present BioID2 study, we identified interacting partners for all five of these DPRs, more for polyGA than seen by Lee et al.33 and Moens et al.13
Previous proteomic studies of the DPR interactome have focused primarily on the polyPR and polyGR interactomes, as these have been shown to be the most cytotoxic.10–12,37 Consistent with previous studies, we have confirmed here that polyPR- and polyGR-interacting partners are involved in translation, RNA processing and regulation of gene expression.13,33,41,42 Since polyGA has shown toxicity in some models,15,17,55 and GA aggregates are the most abundant of these DPRs in patient tissue56,57 (see also Fig. 4A), it is important to identify and understand the polyGA-interacting partners. We identified 90 different interacting proteins for polyGA, including proteins that participate in translation, nuclear-transcribed mRNA catabolic processes, the mitotic cell cycle and proteasome-mediated ubiquitin-dependent protein catabolic processes. Five interactors are seen in common with the polyGA-interactors identified by Moens et al.,13 namely ACACA, HSPA8, MCCC1, PC and RPS27A, although there are no matches with the top 20 polyGA-interactors reported by May et al.58 However, in agreement with May et al.,58 we both detected components of the ubiquitin-proteasome system as major polyGA-interactors. Among the other interacting proteins, we identified VCP as a strong polyGA interactor. C9orf72 ALS/FTD patients probably express all DPRs rather than a single specific DPR.9,59 Moreover it was demonstrated that DPRs may not be translated as single dipeptide units but may be translated in combinations due to ribosomal frameshifting and the term ‘chimeric DPRs’ was ascribed to these putative DPRs.60 So to actually understand how DPRs contribute to disease, future research should necessarily focus on disease models that express multiple DPRs, ideally at physiologically relevant levels. To further address the issue that some of our identified interacting proteins appear to be associated with a particular subcellular localization and this way influence the interpretation of our in vitro results, it is important to recognize that the greatest challenge facing C9orf72 ALS/FTD research today is to reconcile the apparent discrepancy between the pattern of DPR pathology in human post-mortem tissues and in different animal and in vitro models. Although polyGR and polyPR are significantly more hydrophilic and less prone to aggregation than polyGA, they are nevertheless readily transported into the nucleus in vitro and in vivo in patients,6,10,31,56,61 despite the general belief that DPRs are mainly localized in the cytoplasm. The demonstrated localization of polyGR and polyPR DPRs in the nucleolus, cytoplasm and even mitochondria12,33,35,38,62 is consistent with the results of our pathway analyses of the identified interacting proteins. Post-mortem studies also indicated the localization polyPR in intranuclear aggregates,12 supporting the notion that DPR-positive inclusions in C9orf72-diseases may be localized in the nucleolus in addition to the cytoplasm.63,64 In this regard, we believe that our newly generated FlpIn SH-SY5Y DPR cell lines, which stably express DPRs localized in distinct subcellular localization, may prove to be a valuable model.
The insitu structure of polyGA aggregates did not reveal an abundance of VCP.65 Interestingly, however, here we showed significant co-localization of VCP with polyGA aggregates in HEK293 cells, and in cortical and hippocampal dentate gyrus neurons from C9orf72 ALS and FTLD patients. Of note, consistent with the previous reports, we also confirmed that VCP positive TDP-43 inclusions were also abundantly present in our samples together with a central GA aggregate core. These results are intriguing; however, they warrant a more detailed understanding of the role of VCP specifically in the degradation of aggregate-prone proteins for the maintenance of the neuronal proteostasis in ALS-FTLD.66–68 Moreover, we would suggest that polyGA sequesters VCP and probably modulates proteins involved in nucleocytoplasmic transport,15 as well as the transport factor UNC119.57 Expression of polyGA leads to accumulation of ubiquitinated conjugates and the p62 and LC3 proteins, which suggests proteotoxic stress resulting in impairment of protein degradation pathways. The lower autophagic flux further explains that in cells expressing polyGA, autophagy is impaired in its late stage. The present data are in line with previous reports showing accumulation of p62 and ubiquitin after polyGA expression,15,57 and impaired autophagy on loss of the C9ORF72 protein, as well as following DPR expresssion.69 In addition, the two known VCP cofactors, namely NPLOC4/NPL4 (NPL4 homolog, ubiquitin recognition factor) and UFD1 (Ubiquitin recognition factor in ER associated degradation 1)70 were found together with the VCP adaptor protein UBXN1 (UBX Domain protein 1)71 as shown in our Supplementary Table 2A and Supplementary Fig. 4, suggesting a possible interaction with GA (although not significant). VCP, in combination with these adaptors, has the role of identifying ubiquitylated proteins and targeting them for degradation by the proteasome or by autophagy. The ATP-dependent protein unfolding activity of VCP is dependent on NPLOC4-UFD1, ATP hydrolysis and substrate ubiquitination.72 Since this is shown to be impaired in cells with mutant or silenced VCP,70,73 it is likely that GA induced sequestration of VCP may similarly affect aggresome formation and cell clearance mechanism.
Impairment of VCP function or VCP deficiency might lead to impaired autophagy by inhibiting the fusion of autophagosomes with lysosomes.52 Consistent with this, we observed significantly lower VCP levels in the soluble fraction after polyGA expression, which suggests that sequestration of VCP within polyGA aggregates leads to a partial loss of VCP function, and contributes to the polyGA pathology. These data are supported by the impairment of autophagy in its late stage similar to impairment of autophagy when VCP is deficient. In line with this, we consistently observed lower levels of VCP in subcellular fractions obtained from cortical lysates of C9orf72 patients. Moreover, consistent with the report that PolyGA aggregates efficiently co-localized with p62,42,57,65 we observed that the overlap of polyGA with p62 was significantly lower in VCP-depleted cells. Of note, VCP recognizes poly-ubiquitinated proteins via adaptor subunits to direct them to the proteasome74,75; however, emerging evidence suggests that VCP may also act as a crucial mediator of selective autophagy.53,75,76 Based on the evidence presented here, it is therefore reasonable to propose that VCP is involved in the pathophysiology in C9orf72 ALS and FTLD by mediating selective autophagy of polyGA aggregates.
Supplementary Material
Acknowledgements
The mass spectrometry data were generated in the Discovery Proteomics Facility headed by Roman Fischer, as part of the TDI biological mass spectrometry laboratory under Benedikt M. Kessler. We acknowledge the team that contributed to the establishment of the Dutch ALS Tissue Bank, as well as the team that contributed to the collection of ALS tissue samples (Prof. Dr D. Troost, Prof. Dr M. de Visser, Dr A. J. van der Kooi, Dr J. Raaphorst) and J. Anink (AMC, Amsterdam) for providing technical support. We also thank Dr Istvan Katona (Institute of Neuropathology, Aachen) for providing one frozen control cortical tissue.
Funding
This study was supported by the Slovenian Research Agency (ARRS, grants J3-8201, P4-0127, N3-0141, J7-9399, J3-9263 and Z3-7307) and ICGEB (CRP/SVN19-03). E.A. is supported by the ALS Stichting (grant ‘The Dutch ALS Tissue Bank’). The neuropathological work-up at the Institute of Neuropathology is supported by the German Research Foundation (DFG; WE 1406/16–1).
Competing interests
The authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
Glossary
- ALS
amyotrophic lateral sclerosis
- DPR
dipeptide repeat
- FTLD
frontotemporal lobar degeneration
References
- 1. Renton AE, Majounie E, Waite A, et al. ; ITALSGEN Consortium . A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. DeJesus-Hernandez M, Mackenzie IR, Boeve BF, et al. . Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72(2):245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kovanda A, Zalar M, Šket P, Plavec J, Rogelj B.. Anti-sense DNA d(GGCCCC)n expansions in C9ORF72 form i-motifs and protonated hairpins. Sci Rep. 2015;5:17944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gendron TF, Belzil VV, Zhang YJ, Petrucelli L.. Mechanisms of toxicity in C9FTLD/ALS. Acta Neuropathol. 2014;127(3):359–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Česnik AB, Darovic S, Mihevc SP, et al. . Nuclear RNA foci from C9ORF72 expansion mutation form paraspeckle-like bodies. J Cell Sci. 2019;132(5):jcs224303. [DOI] [PubMed] [Google Scholar]
- 6. Zu T, Liu Y, Bañez-Coronel M, et al. . RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci USA. 2013;110(51):E4968–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gendron TF, Bieniek KF, Zhang YJ, et al. . Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 2013;126(6):829–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mori K, Arzberger T, Grässer FA, et al. . Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol. 2013;126(6):881–893. [DOI] [PubMed] [Google Scholar]
- 9. Lee YB, Baskaran P, Gomez-Deza J, et al. . C9orf72 poly GA RAN-translated protein plays a key role in amyotrophic lateral sclerosis via aggregation and toxicity. Hum Mol Genet. 2017;26(24):4765–4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kwon I, Xiang S, Kato M, et al. . Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science. 2014;345(6201):1139–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mizielinska S, Grönke S, Niccoli T, et al. . C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science. 2014;345(6201):1192–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wen X, Tan W, Westergard T, et al. . Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron. 2014;84(6):1213–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moens TG, Niccoli T, Wilson KM, et al. . C9orf72 arginine-rich dipeptide proteins interact with ribosomal proteins in vivo to induce a toxic translational arrest that is rescued by eIF1A. Acta Neuropathol. 2019;137(3):487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Freibaum BD, Lu Y, Lopez-Gonzalez R, et al. . GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015;525(7567):129–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang YJ, Gendron TF, Grima JC, et al. . C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nat Neurosci. 2016;19(5):668–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang YJ, Jansen-West K, Xu YF, et al. . Aggregation-prone c9FTD/ALS poly(GA) RAN-translated proteins cause neurotoxicity by inducing ER stress. Acta Neuropathol. 2014;128(4):505–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schludi MH, May S, Grässer FA, et al. . Distribution of dipeptide repeat proteins in cellular models and C9orf72 mutation cases suggests link to transcriptional silencing [published correction appears in Acta Neuropathol. 2015 Oct;130(4):557-8]. Acta Neuropathol. 2015;130(4):537–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Motaln H, Čerček U, Recek N, Česnik Mozetič BA, Rogelj M.. B. Cold atmospheric plasma induces stress granule formation via an eIF2α-dependent pathway. Biomater Sci. 2020;7(19):5293–5305. [DOI] [PubMed] [Google Scholar]
- 19. Prpar Mihevc S, Baralle M, Buratti E, Rogelj B.. TDP-43 aggregation mirrors TDP-43 knockdown, affecting the expression levels of a common set of proteins. Sci Rep. 2016;6:33996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim DI, Jensen SC, Noble KA, et al. . An improved smaller biotin ligase for BioID proximity labeling. Mol Biol Cell. 2016;27(8):1188–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Davis S, Charles PD, He L, Mowlds P, Kessler BM, Fischer R.. Expanding proteome coverage with charge ordered parallel ion analysis (CHOPIN) combined with broad specificity proteolysis. J Proteome Res. 2017;16(3):1288–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McAlister GC, Nusinow DP, Jedrychowski MP, et al. . MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal Chem. 2014;86(14):7150–7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vizcaíno JA, Csordas A, Del-Toro N, et al. . 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44(22):11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Käll L, Canterbury JD, Weston J, Noble WS, MacCoss MJ.. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4(11):923–925. [DOI] [PubMed] [Google Scholar]
- 25. Ludolph A, Drory V, Hardiman O, et al. ; WFN Research Group on ALS/MND . A revision of the El Escorial criteria - 2015. Amyotroph Lateral Scler Front Degener. 2015;16(5-6):291–292. [DOI] [PubMed] [Google Scholar]
- 26. Goswami A, Jesse CM, Chandrasekar A, et al. . Accumulation of STIM1 is associated with the degenerative muscle fibre phenotype in ALS and other neurogenic atrophies. Neuropathol Appl Neurobiol. 2015;41(3):304–318. [DOI] [PubMed] [Google Scholar]
- 27. Dreser A, Vollrath JT, Sechi A, et al. . The ALS-linked E102Q mutation in Sigma receptor-1 leads to ER stress-mediated defects in protein homeostasis and dysregulation of RNA-binding proteins. Cell Death Differ. 2017;24(10):1655–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Marrone L, Drexler HCA, Wang J, et al. . FUS pathology in ALS is linked to alterations in multiple ALS-associated proteins and rescued by drugs stimulating autophagy. Acta Neuropathol. 2019;138 (1):67–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yamoah A, Tripathi P, Sechi A, et al. . Aggregates of RNA binding proteins and ER chaperones linked to exosomes in granulovacuolar degeneration of the Alzheimer’s disease brain. J Alzheimer’s Dis. 2020;75(1):139–156. [DOI] [PubMed] [Google Scholar]
- 30. Roux KJ, Kim DI, Burke B, May DG.. BioID: A screen for protein-protein interactions. Curr Protoc Protein Sci. 2018;91(1):19.23.1–19.23.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Callister JB, Ryan S, Sim J, Rollinson S, Pickering-Brown SM.. Modelling C9orf72 dipeptide repeat proteins of a physiologically relevant size. Hum Mol Genet. 2016;25(23):5069–5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cristofani R, Crippa V, Vezzoli G, et al. . The small heat shock protein B8 (HSPB8) efficiently removes aggregating species of dipeptides produced in C9ORF72-related neurodegenerative diseases. Cell Stress Chaperones. 2018;23(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee KH, Zhang P, Kim HJ, et al. . C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell. 2016;167(3):774–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schmitz A, Marques JP, Oertig I, Maharjan N, Saxena S.. Emerging perspectives on dipeptide repeat proteins in C9ORF72 ALS/FTD. Front Cell Neurosci. 2021;18:637548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lopez-Gonzalez R, Lu Y, Gendron TF, et al. . Poly(GR) in C9ORF72-related ALS/FTD compromises mitochondrial function and increases oxidative stress and DNA damage in iPSC-derived motor neurons. Neuron. 2016;92(2):383–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Freibaum BD, Taylor JP.. The role of dipeptide repeats in C9ORF72-related ALS-FTD. Front Mol Neurosci. 2017;13:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang YJ, Guo L, Gonzales PK, et al. . Heterochromatin anomalies and double-stranded RNA accumulation underlie C9orf72 poly(PR) toxicity. Science. 2019;363(6428):eaav2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. White MR, Mitrea DM, Zhang P, et al. . C9orf72 Poly(PR) dipeptide repeats disturb biomolecular phase separation and disrupt nucleolar function. Mol Cell. 2019;74(4):713–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang DW, Sherman BT, Lempicki RA.. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huang DW, Sherman BT, Tan Q, et al. . DAVID bioinformatics resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35(Web Server issue):W169–W175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hartmann H, Hornburg D, Czuppa M, et al. . Proteomics and C9orf72 neuropathology identify ribosomes as poly-GR/PR interactors driving toxicity. Life Sci Alliance. 2018;1(2):e201800070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lin Y, Mori E, Kato M, et al. . Toxic PR poly-dipeptides encoded by the C9orf72 repeat expansion target LC domain polymers. Cell. 2016;167(3):789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mizushima N. Autophagy: Process and function. Genes Dev. 2007;21(22):2861–2873. [DOI] [PubMed] [Google Scholar]
- 44. Lei Y, Klionsky DJ.. New functions of a known autophagy regulator: VCP and autophagy initiation. Autophagy. 2021;17(5):1063–1064. May [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sun H, Wang R, Liu Y, Mei H, Liu X, Peng Z.. USP11 induce resistance to 5-fluorouracil in colorectal cancer through activating autophagy by stabilizing VCP. J Cancer. 2021;12(8):2317–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wrobel L, Hill SM, Ashkenazi A, Rubinsztein DC.. VCP/p97 modulates PtdIns3P production and autophagy initiation. Autophagy. 2021;17(4):1052–1053. Apr [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mengus C, Neutzner M, Bento ACPF, et al. . VCP/p97 cofactor UBXN1/SAKS1 regulates mitophagy by modulating MFN2 removal from mitochondria. Autophagy. Published online 9 May 2021. 10.1080/15548627.2021.1922982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Darwich NF, Phan JM, Kim B, et al. . Autosomal dominant VCP hypomorph mutation impairs disaggregation of PHF-tau. Science. 2020;370(6519):eaay8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rubinsztein DC, Cuervo AM, Ravikumar B, et al. . In search of an “autophagomometer”. Autophagy. 2009;5(5):585–589. [DOI] [PubMed] [Google Scholar]
- 50. Komatsu M, Waguri S, Koike M, et al. . Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131(6):1149–1163. [DOI] [PubMed] [Google Scholar]
- 51. Waguri S, Komatsu M.. Chapter 9 biochemical and morphological detection of inclusion bodies in autophagy-deficient mice. In: Klionsky DJ, ed. Autophagy in disease and clinical applications. Methods Enzymol. 2009;453:181–196. [DOI] [PubMed] [Google Scholar]
- 52. Klionsky DJ, Abdalla FC, Abeliovich H, et al. . Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8(4):445–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ju JS, Fuentealba RA, Miller SE, et al. . Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J Cell Biol. 2009;187(6):875–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bento AC, Bippes CC, Kohler C, Hemion C, Frank S, Neutzner A.. UBXD1 is a mitochondrial recruitment factor for p97/VCP and promotes mitophagy. Sci Rep. 2018;8(1):12415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schludi MH, Becker L, Garrett L, et al. . Spinal poly-GA inclusions in a C9orf72 mouse model trigger motor deficits and inflammation without neuron loss. Acta Neuropathol. 2017;134(2):241–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mori K, Weng SM, Arzberger T, et al. . The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339(6125):1335–1338. [DOI] [PubMed] [Google Scholar]
- 57. Saberi S, Stauffer JE, Jiang J, et al. . Sense-encoded poly-GR dipeptide repeat proteins correlate to neurodegeneration and uniquely co-localize with TDP-43 in dendrites of repeat-expanded C9orf72 amyotrophic lateral sclerosis. Acta Neuropathol. 2018;135(3):459–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. May S, Hornburg D, Schludi MH, et al. . C9orf72 FTLD/ALS-associated Gly-Ala dipeptide repeat proteins cause neuronal toxicity and Unc119 sequestration. Acta Neuropathol. 2014;128(4):485–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. He F, Flores BN, Krans A, et al. . The carboxyl termini of RAN translated GGGGCC nucleotide repeat expansions modulate toxicity in models of ALS/FTD. Acta Neuropathol Commun. 2020;4:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. McEachin ZT, Gendron TF, Raj N, et al. . Chimeric peptide species contribute to divergent dipeptide repeat pathology in c9ALS/FTD and SCA36. Neuron. 2020;107(2):292–305.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ash PE, Bieniek KF, Gendron TF, et al. . Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77(4):639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Frottin F, Schueder F, Tiwary S, et al. . The nucleolus functions as a phase-separated protein quality control compartment. Science. 2019;365(6451):342–347. [DOI] [PubMed] [Google Scholar]
- 63. Baborie A, Griffiths TD, Jaros E, et al. . Accumulation of dipeptide repeat proteins predates that of TDP-43 in frontotemporal lobar degeneration associated with hexanucleotide repeat expansions in C9ORF72 gene. Neuropathol Appl Neurobiol. 2015;41(5):601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vatsavayai SC, Yoon SJ, Gardner RC, et al. . Timing and significance of pathological features in C9orf72 expansion-associated frontotemporal dementia. Brain. 2016;139(Pt 12):3202–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Guo Q, Lehmer C, Martínez-Sánchez A, et al. . In situ structure of neuronal C9orf72 poly-GA aggregates reveals proteasome recruitment. Cell. 2018;172(4):696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mackenzie IR, Frick P, Grässer FA, et al. . Quantitative analysis and clinico-pathological correlations of different dipeptide repeat protein pathologies in C9ORF72 mutation carriers. Acta Neuropathol. 2015;130(6):845–861. [DOI] [PubMed] [Google Scholar]
- 67. Mackenzie IR, Arzberger T, Kremmer E, et al. . Dipeptide repeat protein pathology in C9ORF72 mutation cases: Clinico-pathological correlations. Acta Neuropathol. 2013;126(6):859–879. [DOI] [PubMed] [Google Scholar]
- 68. Nonaka T, Masuda-Suzukake M, Hosokawa M, et al. . C9ORF72 dipeptide repeat poly-GA inclusions promote intracellular aggregation of phosphorylated TDP-43. Hum Mol Genet. 2018;27(15):2658–2670. [DOI] [PubMed] [Google Scholar]
- 69. Boivin M, Pfister V, Gaucherot A, et al. . Reduced autophagy upon C9ORF72 loss synergizes with dipeptide repeat protein toxicity in G4C2 repeat expansion disorders. EMBO J. 2020;39(4):e100574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Blythe EE, Gates SN, Deshaies RJ, Martin A.. Multisystem proteinopathy mutations in VCP/p97 increase NPLOC4·UFD1L binding and substrate processing. Structure. 2019;27(12):1820–1829.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Pan M, Zheng Q, Yu Y, et al. . Seesaw conformations of Npl4 in the human p97 complex and the inhibitory mechanism of a disulfiram derivative. Nat Commun. 2021;12(1):121–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mukkavalli S, Klicksten JA, Oritz B, Juo P, Raman M.. The p97–UBXN1 complex regulates aggresome formation. J Cell Sci. 2021;134 (7):jcs254201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ganji R, Mukkavalli S, Somanji F, Raman M.. The VCP-UBXN1 complex mediates triage of ubiquitylated cytosolic proteins bound to the BAG6 complex. Mol Cell Biol. 2018;38(13):e00154–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Meyer H, Weihl CC.. The VCP/p97 system at a glance: Connecting cellular function to disease pathogenesis. J Cell Sci. 2014;127(18):3877–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kobayashi H, Shoji K, Kiyokawa K, Negishi L, Tomari Y.. VCP machinery mediates autophagic degradation of empty argonaute. Cell Rep. 2019;28(5):1144–1153. [DOI] [PubMed] [Google Scholar]
- 76. Papadopoulos C, Kirchner P, Bug M, et al. . VCP/p97 cooperates with YOD 1, UBXD 1 and PLAA to drive clearance of ruptured lysosomes by autophagy. EMBO J. 2017;36(2):135–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data are available on reasonable request. The liquid chromatography–tandem mass spectrometry data are available via ProteomeXchange with identifier PXD024120.