Abstract
The ribosomal RNAs, along with their substrates the transfer RNAs, contain the most highly conserved nucleotides in all of biology. We have assembled a database containing structure-based alignments of sequences of the small-subunit rRNAs from organisms that span the entire phylogenetic spectrum, to identify the nucleotides that are universally conserved. In its simplest (bacterial and archaeal) forms, the small-subunit rRNA has ∼1500 nt, of which we identify 140 that are absolutely invariant among the 1961 species in our alignment. We examine the positions and detailed structural and functional interactions of these universal nucleotides in the context of a half century of biochemical and genetic studies and high-resolution structures of ribosome functional complexes. The vast majority of these nucleotides are exposed on the subunit interface surface of the small subunit, where the functional processes of the ribosome take place. However, only 40 of them have been directly implicated in specific ribosomal functions, such as contacting the tRNAs, mRNA, or translation factors. The roles of many other invariant nucleotides may serve to constrain the positions and orientations of those nucleotides that are directly involved in function. Yet others can be rationalized by participation in unusual noncanonical tertiary structures that may uniquely allow correct folding of the rRNA to form a functional ribosome. However, there remain at least 50 nt whose universal conservation is not obvious, serving as a metric for the incompleteness of our understanding of ribosome structure and function.
Keywords: 16S rRNA, RNA conservation, protein synthesis, ribosomal RNA, ribosome
INTRODUCTION
Following completion of the first sequences of the large 16S and 23S ribosomal RNAs (Brosius et al. 1978, 1980), it became clear that it might be possible to determine their secondary structures by using comparative sequence analysis, following the example of Fox and Woese (1975) on the small 5S rRNA. In this approach, compensating base changes between the rRNAs of different species provide evidence for their Watson–Crick complementarity. The resulting secondary structure models for 16S rRNA deduced in this way (Woese et al. 1980; Noller and Woese 1981; Stiegler et al. 1981; Zwieb et al. 1981; Gutell et al. 1985; Cannone et al. 2002) were supported by chemical and enzymatic probing experiments (Moazed et al. 1986) and ultimately confirmed by X-ray structures of ribosomal subunits and complete ribosomes (Ban et al. 2000; Schluenzen et al. 2000; Wimberly et al. 2000; Yusupov et al. 2001; Korostelev et al. 2006; Selmer et al. 2006).
A stumbling block to deducing secondary structure from comparative sequence analysis was the occurrence of highly conserved or even invariant nucleotides, which had first been detected in Woese et al.’s (1975) RNase T1 oligonucleotide catalogs from the 16S and 18S rRNAs (Woese et al. 1975). The discovery of universally conserved nucleotides drew attention to their possible functional importance, suggesting that the ribosomal RNAs were not simply structural scaffolds. Although it had been imagined that these bases might be conserved to create recognition sites for binding of ribosomal proteins, their susceptibility to attack in the intact ribosome by single-strand-specific chemical probes such as kethoxal (Noller 1974; Moazed et al. 1986) indicated just the opposite—that these nucleotides were exposed, and thus available for participation in ribosome function, a heterodox suggestion at the time (Noller and Chaires 1972; Noller 1991; Noller et al. 1992).
Together with the invariant bases of tRNA, those of the rRNAs must represent the most ancient nucleotides in all of biology. With the revolutionary advances in rapid DNA sequencing in recent years, coupled with the unmatched power of ribosomal RNA as a tool in phylogenetic research, several million complete sequences of ribosomal RNAs have been determined, stretching across the entire phylogenetic spectrum (Quast et al. 2013; Cole et al. 2014). Here, focusing on the rRNA of the small ribosomal subunit, the 16S-like rRNAs, we ask the following questions: Which bases are truly invariant, and why? The answer must be that mutation of any of these bases is ultimately a lethal event over the evolutionary time span of the organism. Can we rationalize their universal conservation in light of the wealth of information about ribosome structure and function that has emerged over the past half century? The extent to which this is successful can thus be taken as a metric for how well we currently understand the ribosome.
After identifying the universally conserved nucleotides, we examine their positions and detailed structural and functional interactions in high-resolution X-ray and cryo-EM structures of ribosome functional complexes. Except where noted, our reference structure throughout is the recent 2.0 Å cryo-EM structure of the E. coli 70S ribosome containing mRNA and tRNAs bound to the A, P, and E sites (Watson et al. 2020). Additional structures have provided the contact sites for elongation factors EF-G (Gao et al. 2009; Brilot et al. 2013; Zhou et al. 2014; Carbone et al. 2021; Petrychenko et al. 2021; Rundlet et al. 2021) and EF-Tu (Schmeing et al. 2009), initiation factors (Hussain et al. 2016) and release factors (Korostelev et al. 2008).
We limit our focus to the 16S rRNAs of bacteria, archaea, and the cytoplasmic 18S rRNAs of eukarya. The 1961 sequences in our alignment (the CRW alignment set) (https://crwsite.chemistry.gatech.edu/DAT/3A/Summary/index.php) cover essentially the entire phylogenetic spectrum, including 1345 bacteria, 519 eukarya, and 97 archaea (Fig. 1). It contains representatives from all of the main phylogenetic branches of each of these three domains. Much higher sequence variation is seen in the rRNAs of mitochondria, which is particularly extreme in the animal mitochondria, in which some small subunit rRNAs are missing more than half of the RNA of their bacterial counterparts (Gutell et al. 1985; Okimoto et al. 1992; Cannone et al. 2002). Chloroplast rRNAs, which are clearly related to those of cyanobacteria, also show greater variation, although much less so than mitochondria (Gutell et al. 1985; Cannone et al. 2002). We have confined our study exclusively to the rRNAs of free-living organisms, given our poor understanding of the reasons for the higher sequence variation observed for the rRNAs of the organelles and endosymbionts. Also, because of the lack of a comparably rich database for the post-transcriptionally modified nucleotides of the rRNAs, we are unable to address their conservation in similar depth. In any case, direct RNA sequence analysis has indicated that these modifications are generally less conserved (Woese et al. 1975). For sake of brevity, we shall often refer to both the 16S and 18S rRNAs from the small ribosomal subunit as “16S” rRNAs.
FIGURE 1.
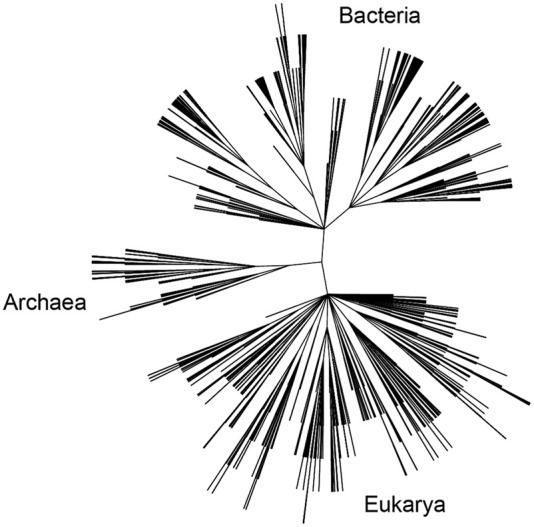
Phylogenetic distribution of small-subunit rRNAs used for analysis. This dendrogram was generated from the list of organisms in our sequence alignment using the NCBI Taxonomy Browser (https://www.ncbi.nlm.nih.gov/Taxonomy/CommonTree/wwwcmt.cgi). The three principal domains of life are indicated, representing sequences from the 1345 bacteria, 519 eukarya, and 97 archaea in the CRW alignment used in this work.
RESULTS AND DISCUSSION
Derivation of conservation values
Although there are now hundreds of thousands of rRNA sequences available in public databases such as the very extensive SILVA database (Quast et al. 2013), we have resisted the temptation to increase the number of sequences in our alignment set because of the danger of introducing errors. In identifying universally conserved nucleotides, even a single error can eliminate a position from classification as universal. Errors arise from four main sources: (1) sequencing errors in the databases (Hugenholtz and Huber 2003; Ashelford et al. 2005, 2006; Liu et al. 2012), (2) errors in sequence alignment, (3) the presence of nonannotated introns in rRNA entries in the databases (Jackson et al. 2002), and (4) the presence of rRNA pseudogenes in the databases—inactive (recessive) lethal mutant forms of rRNA that might be tolerated in a single copy of a 16S rRNA gene against a background of multiple wild-type copies. Unfortunately, pseudogenes are not readily identified in the course of high-throughput DNA sequencing, from which most rRNA sequences are derived.
We have attempted to exclude sequences that contain suspected errors or pseudogenes, such as those positions for which there is a single variant among the 1961 sequences in our alignment. Since this degree of variation is similar to the level of occurrence of sequencing errors, we searched for the presence of multiple independent sequences for the identical organism in the SILVA rRNA database (Quast et al. 2013) and aligned them with our CRW sequence set. If the replicate sequences agreed with the consensus base at that position, the outlier base was excluded from further analysis as a likely sequencing error or a silent mutation in an rRNA pseudogene. If there were one or more independent sequences containing the outlier base, we scored the position as near-universal. In spite of these efforts, the list of 140 invariant positions may well represent an underestimate of the true total.
Another crucial step is accurate alignment of the sequences. In our view, sequence alignment implies that the aligned nucleotides are three-dimensional structural counterparts of each other in ribosomes from different organisms. This was originally inferred, in an iterative process, from their positions in their respective secondary structures (Woese et al. 1980, 1983). Our alignment set is based on that developed by the Comparative RNA web (CRW) site (Cannone et al. 2002), which contains a smaller, carefully curated collection of representative rRNA sequences. Alignment errors are minimized in the CRW database by using a structure-based alignment that takes advantage of common 16S rRNA secondary structure features, as well as islands of highly conserved sequences (Gutell et al. 2002). Although the completeness of the secondary structures obtained by comparative sequence analysis evolved over many years and originally contained some errors in detail (Gutell et al. 2002), their accuracy can now be verified directly from the increasing number of high-resolution three-dimensional X-ray and cryo-EM structures of ribosomes
We find the conservation value (C) to be a useful parameter for quantification of the extent of conservation of individual nucleotide positions. It is obtained from the following expression, derived from information theory (Gutell et al. 1985; MS Waterman and HF Noller, unpubl.):
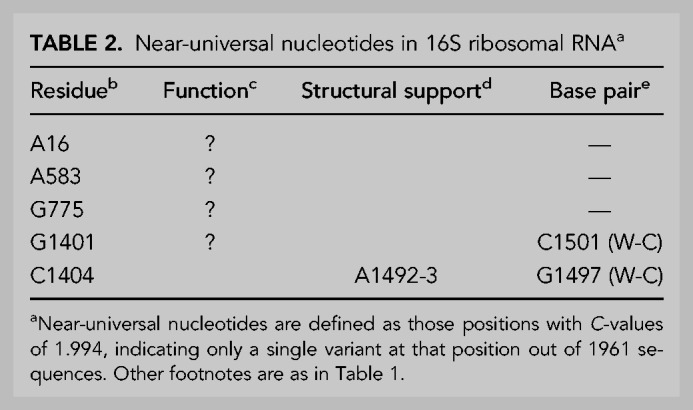
where Pi is the frequency of occurrence of the most common base i (A,C,G or U) at a given position in the RNA sequence and PΔ is the frequency of deletions at that position. For an invariant position, the conservation value C is then 2.000. Figure 2 shows the distribution of C-values for the 1542-nt positions. The prominent peak corresponding to positions with C-values at or near 2.000 gives a clear indication of the extraordinary conservation of bases at specific positions in the 16S rRNAs. Figure 3 shows an expanded view of the distribution of C-values between 1.900 and 2.000. It can be seen that it is dominated by the peak at C = 2.000, falling off abruptly at lower C-values. In our alignment, we identify 140 invariant (C = 2.000) positions out of the 1542-nt residues in our reference E. coli 16S rRNA sequence; these positions are listed in Table 1. We focus here on these universally conserved nucleotide positions. We also identified five true near-universal positions with C-values of 1.994, indicating a single confirmed variant sequence for each of these five positions (Table 2).
FIGURE 2.
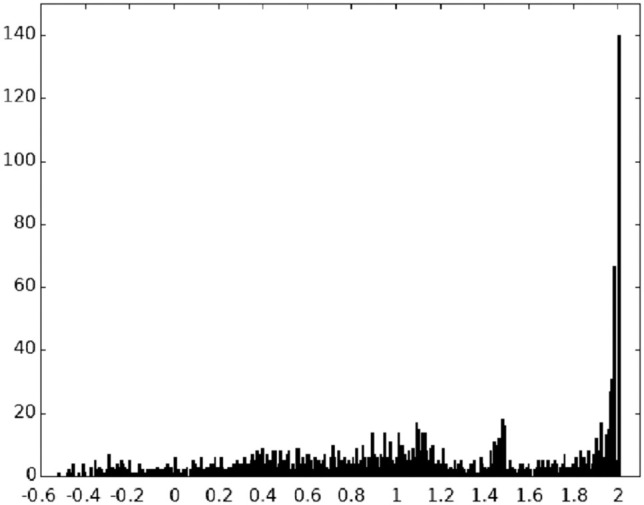
Distribution of C-values in 16S rRNAs. The number of sequences scoring each given C (conservation) value is shown for the 1961 sequences in our 16S rRNA database representing organisms across the entire phylogenetic spectrum. Note that the C-value 2.000 corresponds to bases that are universally conserved.
FIGURE 3.
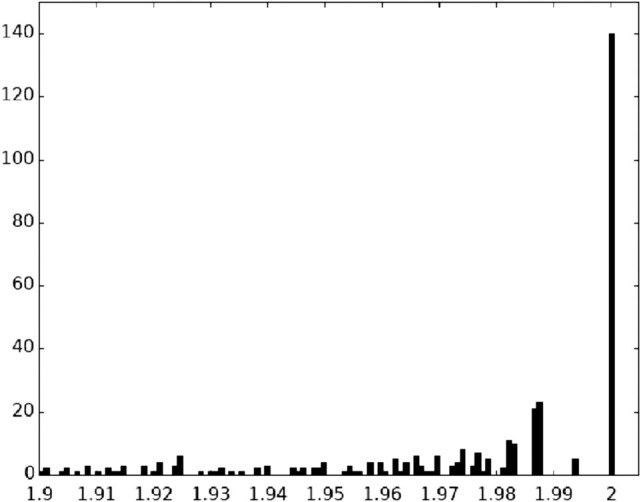
Distribution of C-values between 1.9 and 2.0. An expanded view of the distribution of the highest C-values shown in Figure 1. The most prominent peak, which has the maximum possible phylogenetic conservation value of C = 2.000, indicates that there are 140 positions that are invariant across the phylogenetic spectrum represented in the1961aligned sequences in our database.
Table 1.
Universal nucleotides in 16S ribosomal RNAa
TABLE 2.
Near-universal nucleotides in 16S ribosomal RNAa
Locations of the invariant nucleotides in the secondary structure of 16S rRNA
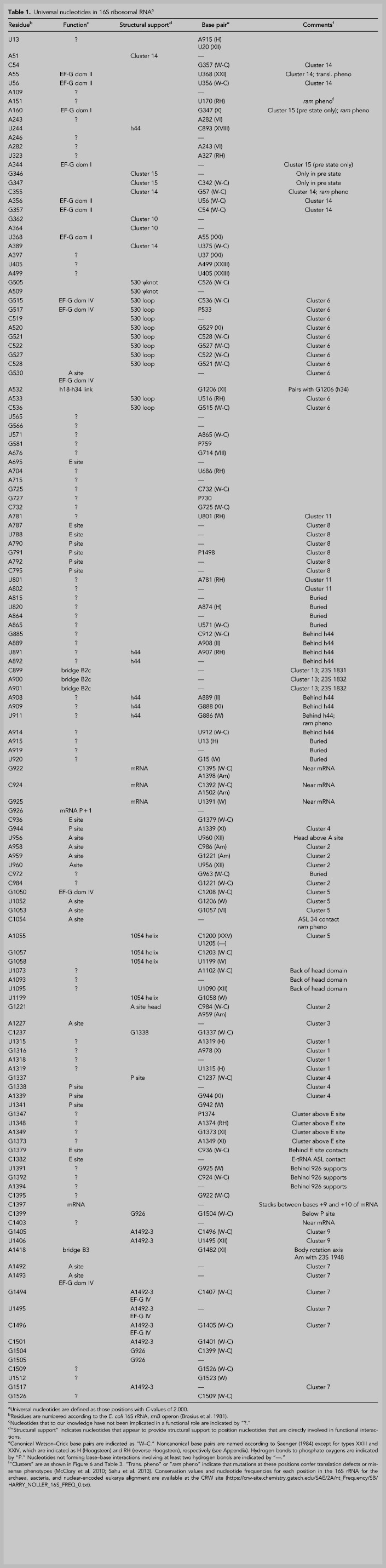
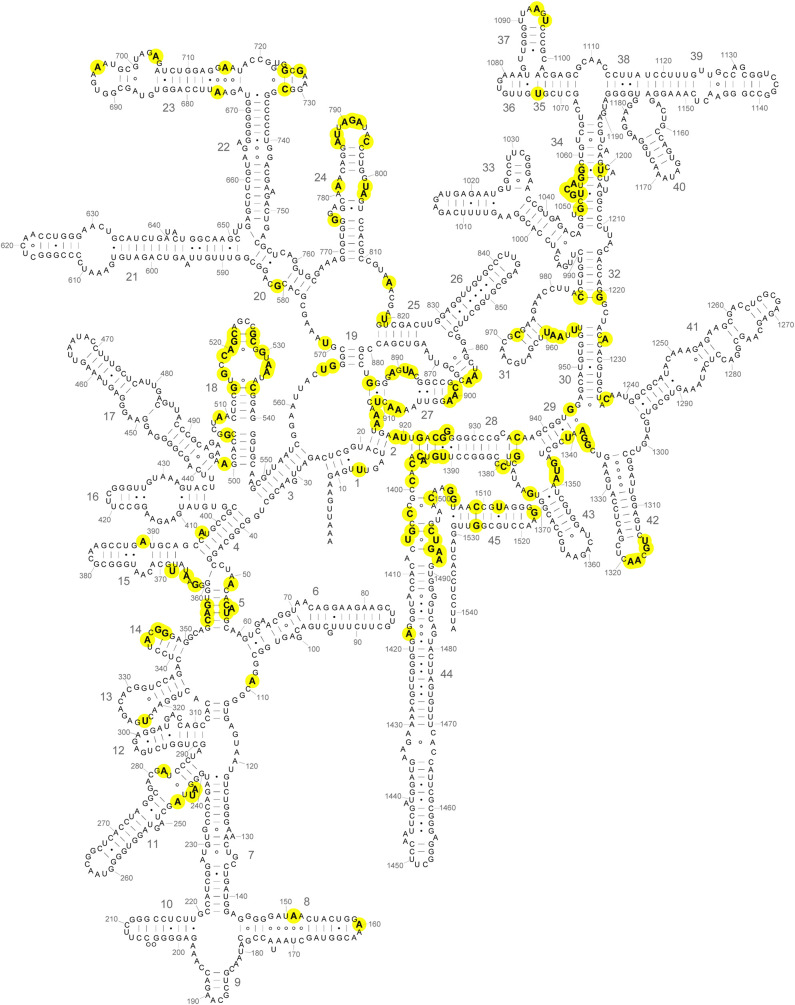
Figure 4 shows the positions of the invariant (C = 2.000) nucleotides highlighted on the secondary structure of the E. coli 16S rRNA (Woese et al. 1980; Gutell et al. 1985; Cannone et al. 2002). They are distributed widely in the secondary structure, with examples in all four secondary structural domains. Most notable are clusters in or around helices h5, h18, h24, h27, h28, h31, h34, h42, and h44 (Fig. 4). Many of these had been identified with ribosomal functions, such as the binding sites for tRNA in the A site (h18 and h44) and P site (h24, h28, h31, and h42) in early chemical probing studies (Moazed and Noller 1986, 1990), and subsequently shown in crystal structures of functional complexes to make direct contacts with the tRNAs (Yusupov et al. 2001; Korostelev et al. 2006; Selmer et al. 2006; Jenner et al. 2010). The universal clusters are discussed in detail below. Nearly all of the invariant nucleotides are found in nonhelical regions of the secondary structure. Notable exceptions can be seen in helices h5 and h28.
FIGURE 4.
Positions of universal bases in the secondary structure of 16S rRNA. Universally conserved (C = 2.000) bases are indicated in boldface on a diagram of the secondary structure of the E. coli 16S rRNA (Cannone et al. 2002; this work). Nucleotides are numbered at every ten positions; large numbers indicate the numbering for the 45 different helical elements. Several prominent universal clusters can be seen, many of which correspond to functional sites of the small ribosomal subunit (Fig. 6).
Positions of invariant nucleotides in the three-dimensional structure
Figure 5 shows the positions of the 140 universally conserved nucleotides in the three-dimensional structure of 16S rRNA in the 30S subunit from the recent 2.0 Å resolution structure of the E. coli ribosome (Watson et al. 2020). Most striking is the clustering of invariants on the subunit interface side of the RNA, particularly near the cleft between the head and body domains of the small ribosomal subunit. It is clear that invariants are nearly absent on the solvent face of the RNA (Fig. 5A,B). When they are viewed in the context of the complete 30S subunit, with the ribosomal proteins in place (Fig. 5C,D), two additional points become evident. First, almost no invariants on the interface side are covered by proteins, leaving the vast majority fully exposed; one of the few exceptions is a small cluster at the top of the head domain, which is covered by proteins S14 and S19 (cf. Fig. 5A,C). Second, the rare invariant nucleotides on the solvent face of the RNA are covered by protein (Fig. 5D). Thus, the vast majority of the universally conserved nucleotides are located on the interface surface of the 30S subunit, and exposed to the solvent, consistent with their likely roles in ribosome function.
FIGURE 5.
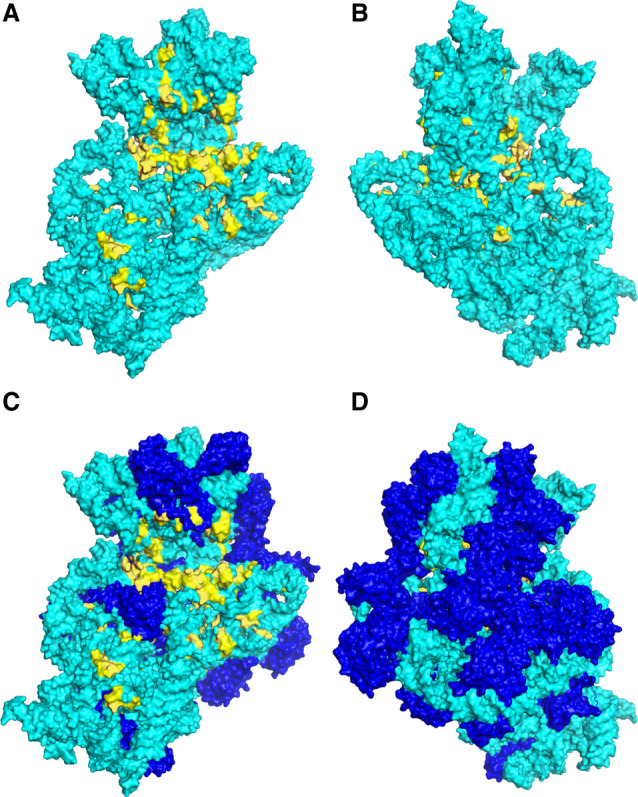
Three-dimensional positions of the universal nucleotides in 16S rRNA. The positions of the invariant nucleotides (C = 2.000) are shown in yellow in the context of the 2.0 Å resolution cryo-EM structure of the 16S rRNA (A,B) and the 30S ribosomal subunit (C,D) in the E. coli 70S ribosome (Watson et al. 2020). The 16S rRNA (cyan) and proteins (blue) are shown in surface rendering. (A,C) Subunit interface view. (B,D) Solvent view. Many of the invariants are clustered at sites known to participate in ribosome function, as indicated in Figure 6 and Table 3. Almost no invariant nucleotides are exposed on the solvent face of 16S rRNA.
The majority of the 15 most prominent clusters of invariant nucleotides shown in Figure 6 can be assigned to ribosomal functions (Table 3). These include the A and P sites for the tRNAs, as noted above, and the binding sites for the mRNA and the initiation, elongation and termination factors. However, it is unclear in many cases why only certain specific bases are compatible with these functions. Moreover, there are three main clusters (clusters 1, 10, and 11) (Table 3) whose functional importance remains unknown. There are many examples of participation of invariant nucleotides in noncanonical base pairing interactions, which may uniquely enable critical folding schemes (discussed below). We refer to these noncanonical pairs by the numbering system used by Saenger (1984), examples of which are provided in the Appendix for reference.
FIGURE 6.
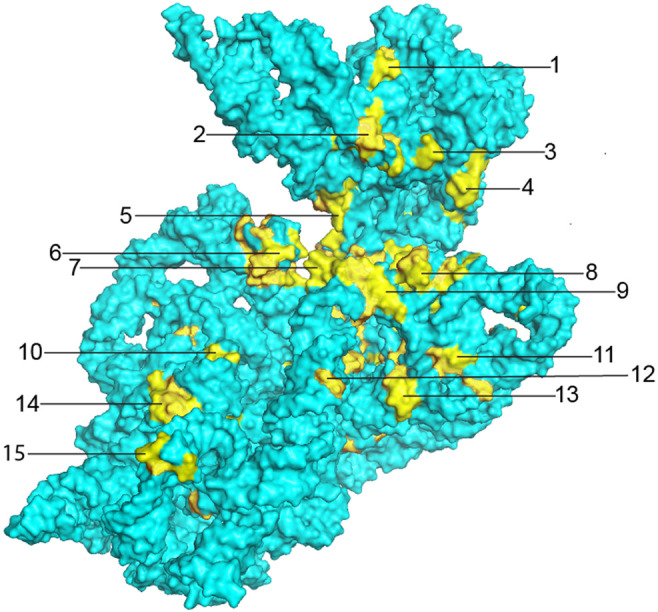
Universal clusters. Clusters of invariant nucleotides are indicated by their numbers. Many of the clusters can be assigned to ribosomal functions, as presented in Table 3. These include the binding sites for A-site (clusters 2, 3, 5, 6, and 7) and P-site (clusters 4, 8, and 9) tRNAs (Moazed and Noller 1990; Ogle et al. 2001; Yusupov et al. 2001; Selmer et al. 2006), sites of interaction with elongation factor EF-G (clusters 5, 6, 7, 14, and 15) (Gao et al. 2009; Brilot et al. 2013; Zhou et al. 2013) and sites of contact with the 50S subunit at intersubunit bridges B2c and B3 (clusters 12 and 13) (Yusupov et al. 2001; Selmer et al. 2006).
TABLE 3.
Universal clustersa
In cases where functional sites of 16S rRNA have been well characterized, the clusters of universal bases can be rationalized. The A- and P-site tRNA anticodon stem–loops bind in the cleft formed between the head and body domains of the 30S subunit; a prominent clustering of universal bases surrounds this cleft (clusters 4–9) (Fig. 6), which also forms the mRNA channel (clusters 6 and 7) (Fig. 7). The invariant G530, A1492, A1493, G926, and C1054 are among the few 16S rRNA bases that make direct contact with the mRNA (Fig. 7); their individual interactions and functional roles are described in further detail below. Positioning of the mRNA is mainly established by codon–anticodon interaction with the A- and P-site tRNAs, which are in turn held by their respective tRNA binding sites. Two additional clusters form the 16S rRNA components of intersubunit bridges B2c and B3, both of which contact elements of 23S rRNA in the 50S subunit (clusters 12 and 13) (Fig. 6; Yusupov et al. 2001; Selmer et al. 2006). Coupled translocation of mRNA and tRNA is catalyzed by elongation factor EF-G, which contacts 16S rRNA at five different clusters of universal nucleotides (clusters 5, 6, 7, 14, and 15) (Fig. 6; Gao et al. 2009; Brilot et al. 2013; Zhou et al. 2013), three of which (5, 6, and 7) also interact with the mRNA and A-site tRNA.
FIGURE 7.
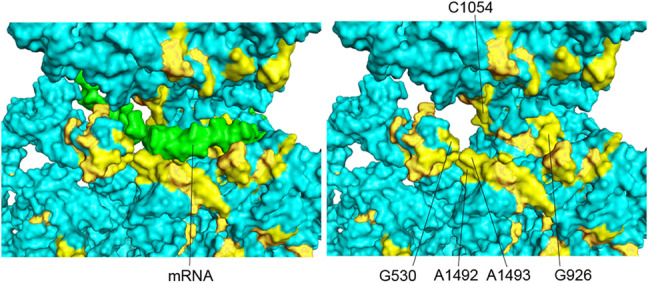
The mRNA channel. In this view, the 16SrRNA is tilted relative to the view in Figure 6, to show the position of the mRNA (green) and the invariant nucleotides flanking the mRNA. The mRNA is removed in the right-hand panel to reveal the universally conserved G530, A1492, A1493, G926, and C1054, which are among the few 16S rRNA bases that directly contact the mRNA.
Invariant nucleotides of the 30S A site
The tRNA binding sites of the 30S subunit have been studied most extensively. The 30S A site, also known as the decoding site, acts to enforce Watson–Crick pairing between the codon and anticodon of the incoming aminoacyl-tRNA (Ogle et al. 2001; Demeshkina et al. 2012). The four bases contacting the A-site tRNA are the aforementioned G530, C1054, A1492, and A1493 (Fig. 8), all of which are invariant. These nucleotides form a simple, compact structure that interacts with the minor groove of the codon–anticodon helix (Ogle et al. 2001; Demeshkina et al. 2012). A1493 and A1492 make type I and type II A-minor interactions with the base pairs formed between the first and second bases of the A-site codon and positions 36 and 35 of the tRNA anticodon, respectively, while G530 and C1054, which pack against the base pair formed between the third (wobble) position of the mRNA and position 34 of the anticodon (Fig. 8), make less extensive contact with the wobble base pair, explaining the variation in pairing that is allowed in the third codon position (Ogle et al. 2001). The substructure created by G530, C1054, A1492, and A1493 forms a sort of steric cage that restricts binding uniquely to codon–anticodon duplexes that have Watson–Crick geometry, particularly in the first and second positions, even when they are mispaired (Demeshkina et al. 2012).
FIGURE 8.
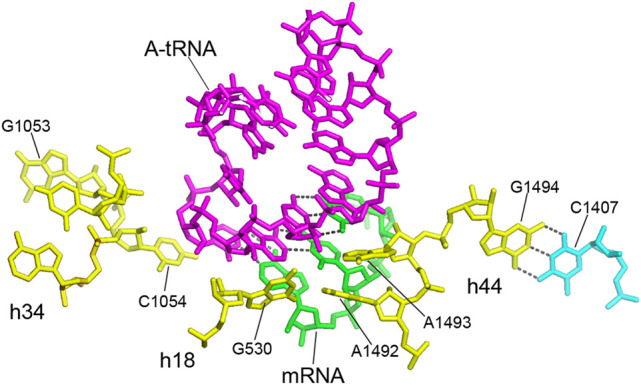
The 30S A site—the decoding site. The central participants are the universally conserved G530, C1054, A1492, and A1493 of 16S rRNA, which contact the codon–anticodon duplex. A1492 and A1493 in helix h44 form A-minor type interactions with the minor groove of the helix formed between the codon of the mRNA (green) and the anticodon of the incoming aminoacyl-tRNA (magenta); G530 in h18 and C1054 in h34 pack against the wobble nucleotide at the bottom of the anticodon (Ogle et al. 2001; Jenner et al. 2010; Watson et al. 2020). These 4 nt form a simple three-dimensional RNA cage that enforces Watson–Crick pairing of the anticodon with the A-site codon (Ogle et al. 2001; Demeshkina et al. 2012). The adjacent invariant nucleotide G1494 forms a tertiary Watson–Crick base pair with the conserved C1407 (C = 1.987). This interaction likely helps to fix the positions of the crucial A1492 and A1493. The noncanonical A532–G1206 base pair (see Fig. 10) forms a connection between h18 and h34, which positions C1054 and G530 in proximity to each other at the bottom of the anticodon as shown here, connecting the A-site elements of the head and body domains of the 30S subunit (see Figs. 10, 12). Universal 16S rRNA nucleotides are shown in yellow.
Most interestingly, an adjacent fourth nucleotide, G1494, is also invariant, but contacts neither the mRNA nor tRNA (Fig. 8). Instead, it forms a tertiary Watson–Crick base pair with C1407, which is not universal, but nevertheless highly conserved (C = 1.987). The likely role of G1494 is to help fix the positions of the crucial A1492 and A1493. This suggests that a second reason for universal conservation is not because of direct participation in ribosome function, but in supporting a precise stereochemical framework for the nucleotides that do make functional interactions. We shall see numerous further examples of invariant nucleotides of this type.
Helix h18: the 530 loop
Early indications of the importance of the 530 loop (the hairpin loop of helix h18) came from chemical probing studies, which showed that the invariant G530 was among the most chemically reactive, and therefore accessible, bases in the ribosome (Noller 1974) and that it was protected, along with A1492 and A1493, from chemical probes by A-site tRNA (Moazed and Noller 1986, 1990). High-resolution crystal structures of complexes of the 30S subunit and 70S ribosome bound with A-site tRNA or its anticodon stem–loop showed that G530 indeed contacts the minor groove of the A-site codon–anticodon duplex (Ogle et al. 2001; Demeshkina et al. 2012). The 530 loop has a complex fold containing a strong pseudoknot interaction formed between the 6-nt bulge loop around position 510 and the hairpin loop around position 525 (Woese and Gutell 1989). This creates an unusual backbone geometry, from which, somewhat surprisingly, solely the two universal bases G530 and A532 project from the structure (Fig. 9). Both bases are implicated in functional roles.
FIGURE 9.
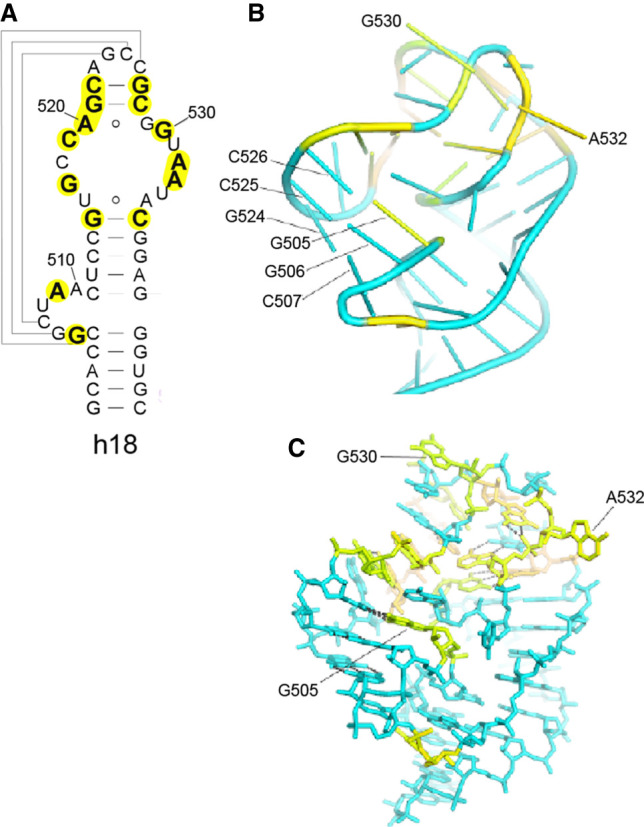
The 530 loop—a key component of the A site. (A,B) A pseudoknot structure formed by pairing of residues 507–509 and 524–526 in helix h18 (A) (Woese and Gutell 1989) creates a stacked coaxial 5-bp all-G–C helix (B) that stabilizes an unusual distortion of the RNA backbone of the hairpin loop of helix h18. (C) Note that the universal G530 and G532 are the sole bases that project from the pseudoknot, where their positions are fixed by numerous tertiary interactions involving conserved nucleotides, including the type XI A520–G529 pair and the reverse Hoogsteen U513–A535 pair. Pairing of A532 with G1206 connects the A-site elements h18 and h34 (Figs. 10, 12).
The universal A532 links the A-site elements of the head and body domains
The universal A532 is located in the hairpin loop of helix h18 close to G530. In the E. coli 70S ribosome complex (Watson et al. 2020), A532 makes a type XI A–G pair with G1206 in the minor groove of helix h34 in the 30S head domain (Fig. 10). Although G1206 is not universal, it is nevertheless highly conserved (C = 1.987) and forms a G–U wobble pair with the invariant U1052. This results in formation of an A–G–U base triple connecting helices h18 and h34, which creates the sole noncovalent connection between the head and body domains of 16S rRNA (Fig. 10). This interhelical base triple is stabilized by being sandwiched between two additional base triples in helix h34 (Fig. 11). One of these is an A–C–U triple formed from H-bonding of the bulged universal U1205 to the noncanonical type XXIV A1055–C1200 bp (Fig. 11B). The second is a G–G–C triple formed by binding of the bulged universal G1053 to the major groove edge of the G1057–C1203 Watson–Crick pair, creating a noncanonical type VI G1053–G1057 pair (Fig. 11A).
FIGURE 10.
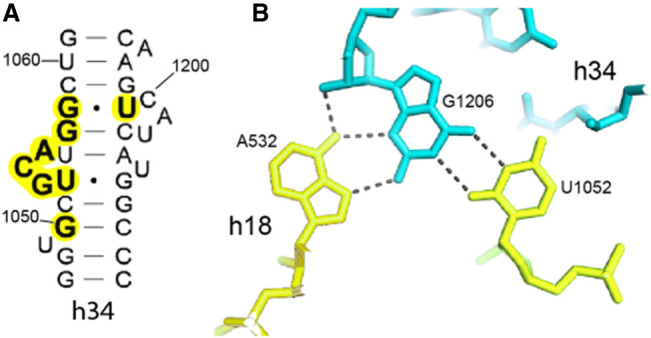
A base triple connecting helix h34 to helix h18. (A) Helix h34 contains three single-base and two 3-base bulge loops. (B) A532, which projects from the h18 pseudoknot (Fig. 9), makes a type XI A–G pair with G1206 in the minor groove of helix h34 (Fig. 12B). This interaction creates the sole noncovalent connection between h18 in the body domain and h34 of the head domain of the 30S subunit, two principal elements of the 30S A site.
FIGURE 11.
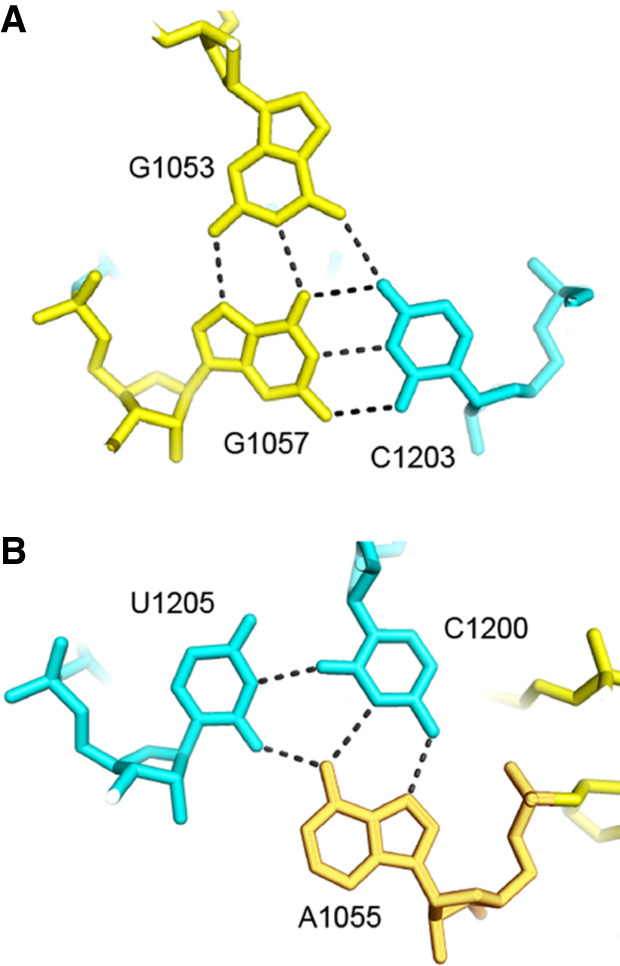
Base triples in helix h34 that sandwich the h18–h34 base triple. The base triple containing A532 (Fig. 13) is stabilized by stacking of 1053–1057–1203 (A) and 1055–1205–1200 (B) base triples in helix h34 on its two surfaces.
Surprisingly, the highly irregular and complex secondary structure of h34, which contains five bulges and three stacked base triples, creates a structure that at first glance resembles an A-form RNA helix (Fig. 12A). The interactions involving the universal bases converge on one face of the helical structure at the point of extrusion of the invariant C1054, the sole base that is not stacked inside the helix. The extruded C1054 stacks on the wobble nucleotide 34 in the anticodon loop of the aminoacyl-tRNA in the decoding site, where it forms the main interaction between the 30S head domain and the A-site tRNA (Jenner et al. 2010; Watson et al. 2020). In addition, we see again that the role of many of the peripheral universally conserved nucleotides is to create a structure that is responsible for accurate positioning of nucleotides that are directly involved in functional interactions.
FIGURE 12.
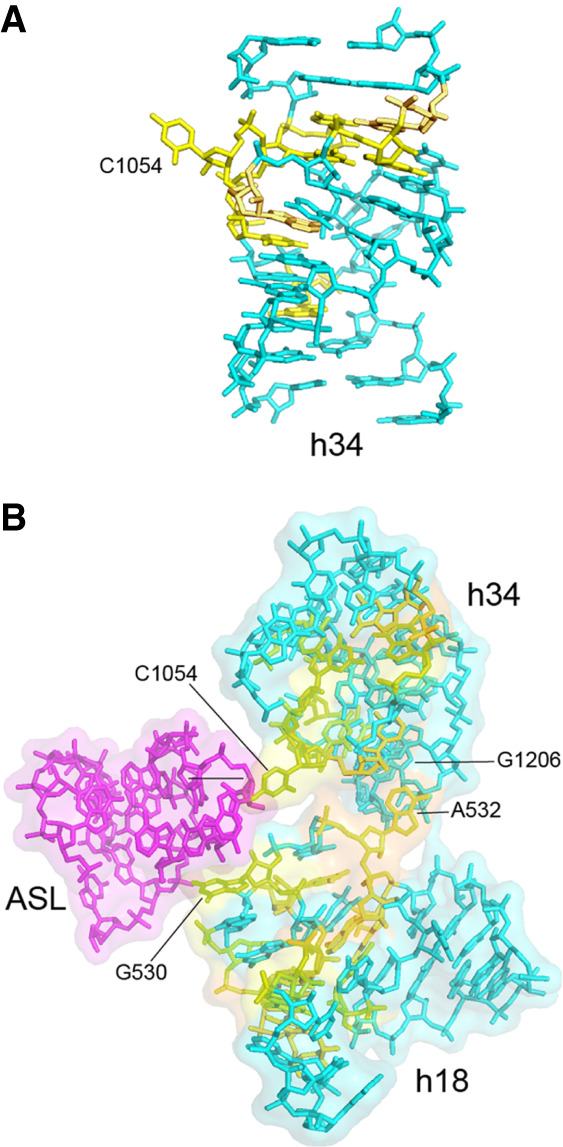
Helices h18 and h34 combine to position A-site tRNA contacts. (A) In spite of its three single-base bulges and two 3-base bulge loops (Fig. 10), C1054 is the sole base that projects from helix h34, stabilized by numerous flanking tertiary interactions involving universally conserved nucleotides (Fig. 11). (B) Formation of the noncanonical A532–G1206 tertiary base pair (Fig. 10) positions G530 and C1054 to contact the anticodon loop of the A-site tRNA (magenta).
The noncanonical A532–G1206 pair brings helices h18 and h34 together in a way that juxtaposes the two critical A-site bases G530 and C1054 into close proximity with one another (Fig. 12B). Mutations in either of these two bases confer strong A-site-related phenotypes (Powers and Noller 1990; Chernoff et al. 1996; Pagel et al. 1997; Abdi and Fredrick 2005; McClory et al. 2010). As expected, the A532–G1206 interaction is disrupted in ribosomes trapped in intermediate states of translocation in which large-scale rotation of the head domain is observed (Zhou et al. 2014). Thus, disruption of the tertiary A532–G1206 base pair must precede translocation of the A-site codon–anticodon duplex to the P site.
Invariant nucleotides of the 30S P site
Another critical function is positioning the first nucleotide of the P-site codon of the mRNA, notably during recognition of the start codon during initiation, which sets the translational reading frame. The invariant G926, which forms a single-base bulge in helix h18 (Fig. 14), makes a strong, polar bifurcated hydrogen bond to phosphate +1 of the mRNA, in which both the N1 and N2 positions of the guanine donate H-bonds to the OP2 phosphate oxygen (Fig. 13; Korostelev et al. 2006; Jenner et al. 2010). The invariant G1505 stacks on G926, restraining the orientation of the χ torsion angle of its guanine base for optimal interaction with the mRNA phosphate (Fig. 13).
FIGURE 14.
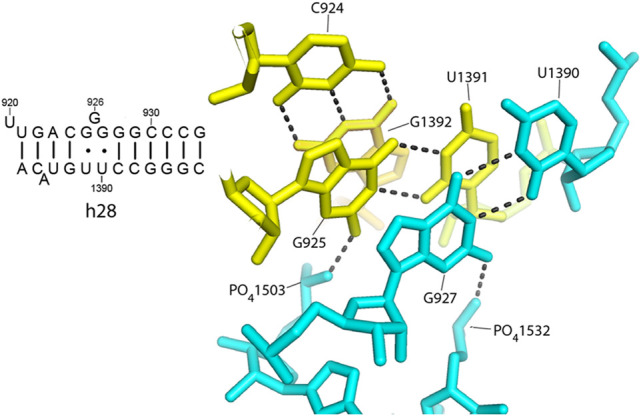
The role of the G–U pairs flanking G926. The G925–U1391 and G927–U1390 pairs project their two-amino group into the minor groove of helix 28, where they H-bond to phosphates 1503 and 1532, respectively, likely stabilizing the position of the bulged G926.
FIGURE 13.
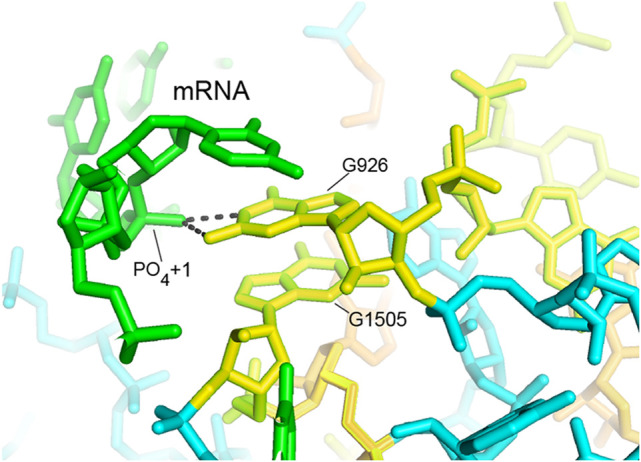
G926 H-bonds to phosphate +1 of the P-site codon of the mRNA. Both the N1 and N2 positions of the invariant bulged base G926 form hydrogen bonds with the OP2 oxygen of the phosphate group of nucleotide +1 of the mRNA, corresponding to the first position of the P-site codon (Korostelev et al. 2006; Jenner et al. 2010).
The bulged G926 is flanked by two conserved G–U wobble pairs (Fig. 14). The G925–U1391 pair is invariant, while the G927–U1390 pair is present in ∼75% of sequences. These flanking G–U pairs help to fix the position of G926 by creating tertiary interactions with surrounding elements of 16S rRNA (Fig. 14). Formation of these conserved G–U wobble pairs projects the N2 amino groups of their guanine bases into the minor groove of helix 28, where they both form hydrogen bonds with backbone phosphates of conserved nucleotides in the 3′-minor domain. Thus, the two-amino group of G925 H-bonds to the phosphate of the conserved A1503 (C = 1.926), a base that intercalates into the mRNA between positions −1 and −2 (Zhou et al. 2013); and G927 H-bonds to the phosphate of U1532 (C = 1.886) (Fig. 14). The G925–U1391 pair is further stabilized by stacking on the conserved G924–C1392 Watson–Crick pair. The strong conservation of all of these bases appears to be driven by the importance of constraining the position of the bulged G926 and thereby the position of phosphate +1 of the mRNA. H-bonding of the two-amino groups of guanines with phosphates of both mRNA and rRNA plays a prominent role here.
The most crucial element of the 30S P site is the tandem pair of the invariant nucleotides G1338 and A1339, which are directly involved in binding P-site tRNA. These bases were initially identified as elements of the P site in chemical probing experiments (Moazed and Noller 1986, 1990). The importance of G1338 for P-site tRNA binding was then demonstrated by modification-interference studies (von Ahsen and Noller 1995). Both G1338 and A1339 bind to the minor groove of the anticodon stem of the P-site tRNA (Fig. 15), forming type II and type I A-minor-like interactions, respectively. A-minor interactions are particularly strong when binding to the minor groove at G–C pairs, which are found at the 29–41 and 30–40 bp of the anticodon stem in all initiator tRNAs (Mangroo et al. 1995). Mutational studies have provided evidence that these two universal bases help to discriminate the initiator Met tRNA from elongator Met tRNA by virtue of the presence of these G–C pairs (Lancaster and Noller 2005). Thus, A-minor interactions play essential roles in the binding of tRNA to both the A and P sites of the small ribosomal subunit.
FIGURE 15.
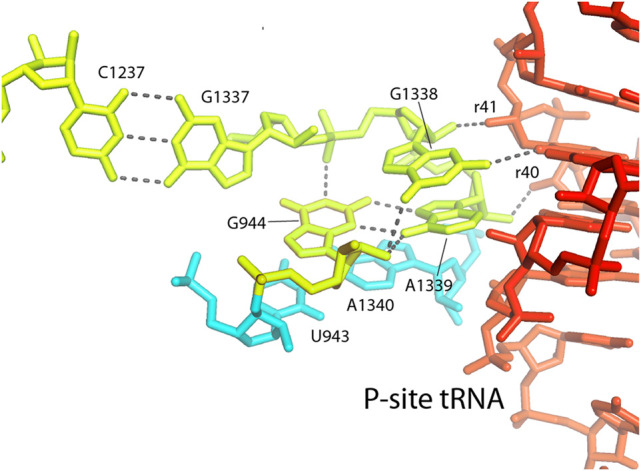
G1338 and A1339 contact the P-site tRNA. The universally conserved G1338 and A1339 contact the minor groove of the anticodon stem of the P-site tRNA at base pairs 29–41 and 30–40. The flanking universal nucleotide G1337 forms a tertiary Watson–Crick base pair with C1237, and the nearly universal G944 forms a compact hydrogen-bonded network (Fig. 11), both of which help to fix the three-dimensional positions and orientations of G1338 and A1339.
Another important role for G1338 and A1339 is in movement of tRNA from the 30S P site to the E site during EF-G-catalyzed translocation. The crystal structure of a chimeric hybrid-state translocation intermediate showed that in the rotated state of the 30S head domain, the P-site tRNA is bound to the head through its interactions with G1338 and A1339 (Zhou et al. 2014) exactly as in the nonrotated state (Carter et al. 2001; Korostelev et al. 2006; Selmer et al. 2006) . The interactions of the head domain with the P-tRNA anticodon stem shown in Figure 15 are thus preserved during its trajectory from the P site to the E site, following disruption of its contacts with P-site elements of the body domain.
Once again, invariant bases are also found adjacent to this important functional site (Fig. 15). Immediately upstream is the universal G1337, which forms a tertiary Watson–Crick pair with the universal C1237 (Fig. 15). In addition, the universal G944 forms an unusual single-nucleotide hydrogen-bonded clamp that fixes the relative positions and orientations of G1338 and A1339 (Fig. 16). The G944 network includes interactions with both ribose and phosphate backbone elements and a type XI noncanonical A–G pair between G944 and A1339 (Fig. 16). Thus, nearby invariant bases help to position G1338 and A1339 by both Watson–Crick and noncanonical base pairing as well as tertiary interactions with the RNA backbone
FIGURE 16.
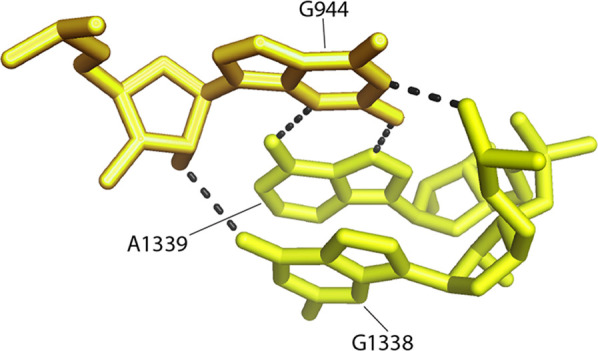
G944 forms a single-nucleotide clamp to fix the orientations of G1338 and A1339. The conserved nucleotide G944 (C = 1.994) forms a compact H-bonded network involving base, ribose, and phosphate moieties, fixing the relative positions of G1338 and A1339.
Contacts with elongation factor EF-G
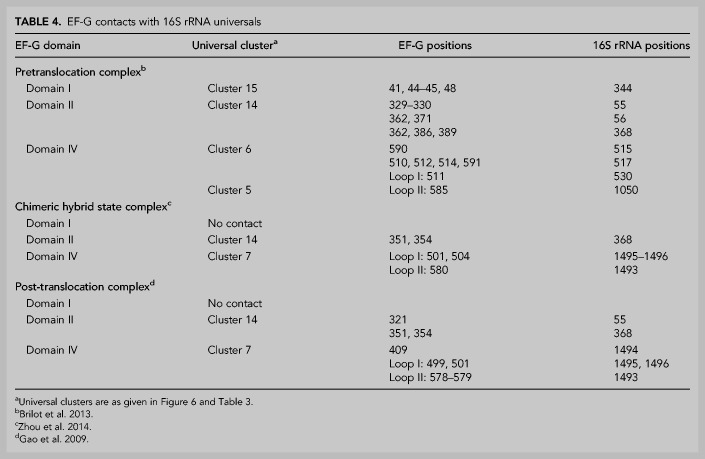
Following peptide bond formation, the GTPase elongation factor EF-G catalyzes the coupled translocation of mRNA and tRNAs from the A to P to E sites. This involves large-scale structural movements in the ribosome, including rotation of the body and head domains of the 30S subunit; elements of 16S rRNA are involved in both rotational movements (Mohan et al. 2014; Belardinelli et al. 2016; Noller et al. 2017). Five clusters (clusters 5, 6, 7, 14, and 15) of universally conserved nucleotides in 16S rRNA are contacted by domains I, II, and IV of elongation factor EF-G (Table 4; Fig. 6). In cryo-EM and crystal structures of complexes containing EF-G trapped in intermediate states of translocation (Gao et al. 2009; Brilot et al. 2013; Zhou et al. 2014; Carbone et al. 2021; Petrychenko et al. 2021; Rundlet et al. 2021), EF-G contacts different overlapping sets of these five clusters (Fig. 17). These interactions are summarized in Table 4.
TABLE 4.
EF-G contacts with 16S rRNA universals
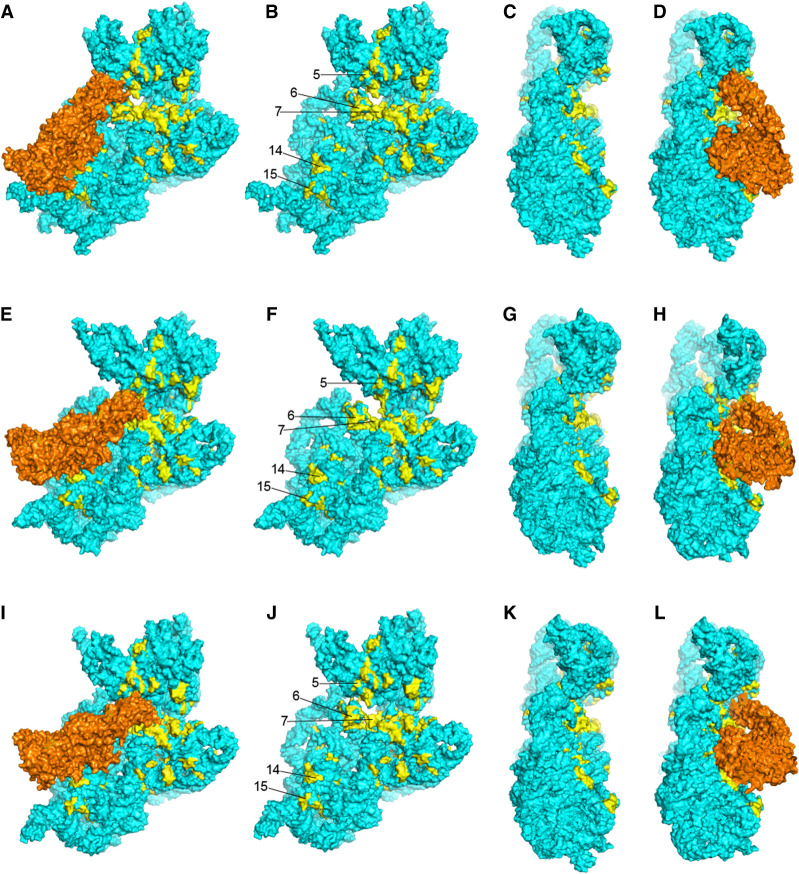
FIGURE 17.
Contacts between EF-G and universal clusters in 16S rRNA. Different binding orientations and contacts of EF-G in the cryo-EM structure of a pretranslocation complex (A–D) (Brilot et al. 2013), crystal structure of a chimeric hybrid-state complex (E–H) (Zhou et al. 2014), and crystal structure of a post-translocation complex (I–L) (Table 3; Gao et al. 2009). EF-G is shown in orange; coloring of 16S rRNA is as in Figures 5 and 6. Ribosomal proteins, mRNAs, and tRNAs are not shown.
In the cryo-EM structure of a pretranslocation complex (Brilot et al. 2013), domain I of EF-G contacts universal cluster 15 (Table 4; Fig. 17A,D), but not in the chimeric-hybrid state (Fig. 17E,H; Zhou et al. 2014) or classical-state (Fig. 17I,L; Gao et al. 2009) EF-G complexes, where domain I of EF-G is displaced by more than 20 Å from cluster 15. Domains II and IV of EF-G contact universal clusters in all EF-G complexes, but the interactions differ in detail for both EF-G domains. For domain II, there are distinct but overlapping sets of contacts with cluster 14 in 16S rRNA (Fig. 17; Table 4). EF-G appears to pivot around its domain II interactions between the three states. In the pretranslocation complex (Brilot et al. 2013) the very conserved Arg362 of EF-G contacts the universal U368, a bulged base in helix h15 that forms a tertiary Watson–Crick pair with the bulged universal A55 in helix h5 (Fig. 18). In the chimeric-hybrid (Zhou et al. 2014) and classical-state (Gao et al. 2009) EF-G complexes, contact shifts from Arg 362 to a similar packing arrangement with Arg 354 (Fig. 19). These interactions represent a rare instance of contact between a protein and the base moiety of a universally conserved nucleotide, although the contact does not involve any H-bonding. Disruption of the tertiary A55–U368 base pair by mutation of A55 to U affects the elongation phase of translation (Sahu et al. 2013). Whether this contact between EF-G and a conserved tertiary base pair that seems important for stabilizing a long-range interaction in the 5′ domain of 16S rRNA has implications for the structural dynamics of 16S rRNA is an open question. Moreover, it remains unclear why this base pair is universally conserved.
FIGURE 18.
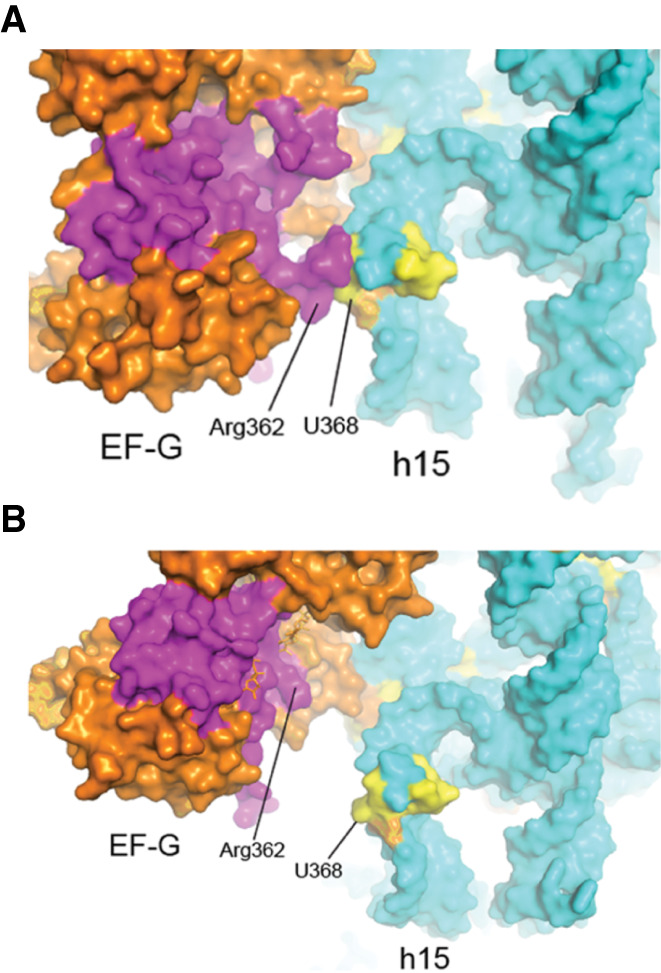
Interaction of domain I of EF-G with 16S rRNA. Domain I (magenta) of EF-G contacts universal cluster 14 (yellow) of 16S rRNA in the (A) pretranslocation complex (Brilot et al. 2013) but not in the (B) post-translocation (Gao et al. 2009) or chimeric hybrid-state (Zhou et al. 2014) complexes (Table 4).
FIGURE 19.
Domain II of EF-G contacts the conserved U368–A55 base pair. The main contact by domain II (magenta) with the universal U368 is from the conserved Arg 362.
Domain IV of EF-G, which forms a superdomain with domain V, flexes relative to the less dynamic cluster of domains I, II, and III during translocation (Gao et al. 2009; Brilot et al. 2013; Ramrath et al. 2013; Zhou et al. 2014; Carbone et al. 2021; Petrychenko et al. 2021; Rundlet et al. 2021). The tip of domain IV moves from cluster 6 in the pretranslocation state complex to cluster 7 in the chimeric-hybrid and post-translocation complexes (Figs. 6, 17; Table 3). In the pretranslocation state, the tip of domain IV is wedged between the loop of helix h18 at the decoding site and helix h34 in the 30S head domain (Fig. 17A–D). It contacts the RNA backbone at the universal positions 515 and 517 and the invariant G530, which, along with C1054, A1492, and A1493, contributes to the functional core of the decoding site, as described above. If these interactions influence the structure of the 530 loop, they would be prime candidates for triggering EF-G-induced conformational changes that lead to the release of the codon–anticodon duplex from the decoding site, a proposed rate-limiting step for translocation (Khade and Joseph 2011).
In the chimeric-hybrid state (Zhou et al. 2014) and classical-state post-translocation (Gao et al. 2009) complexes, the tip of domain IV moves into contact with all three 16S rRNA elements of the 30S A site: helices h18 and h44 in the body domain and h34 in the head domain (Figs. 17E–L, 20). Domain IV now occupies the position of the displaced anticodon stem–loop of the A-site tRNA (Ramrath et al. 2013; Zhou et al. 2014). As in the pretranslocation complex, all contacts by EF-G are with the 16S rRNA backbone, most notably at the universally conserved positions 1494, 1495, and 1496. In the transition from the chimeric-hybrid to post-translocation states, the tip of domain IV moves further toward the 30S P site (Fig. 17I–L). An ∼20° rotation of the head domain between the latter two states (Ramrath et al. 2013; Zhou et al. 2014) results in a shift of head domain interactions from A-site- to P-site-specific contacts. In contrast to the previous post-translocation complex, trapped in the presence of fusidic acid (Gao et al. 2009), recent time-resolved cryo-EM structures (Carbone et al. 2021) show no bound EF-G in the post-translocation state, suggesting that in the absence of antibiotic EF-G dissociates from the ribosome in the transition from the chimeric-hybrid to post-translocation states. Much is yet to be learned about the complex structural dynamics and sequence of events in EF-G-catalyzed translocation.
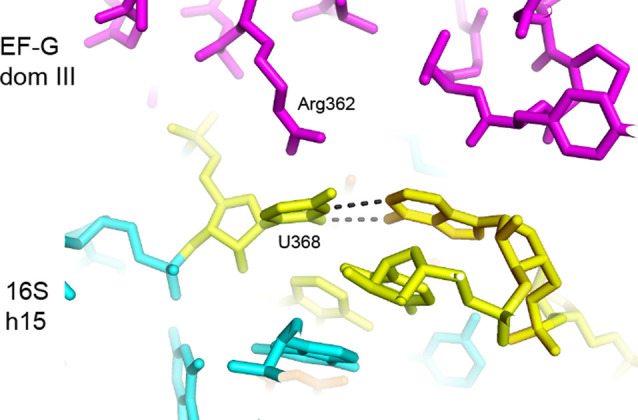
FIGURE 20.
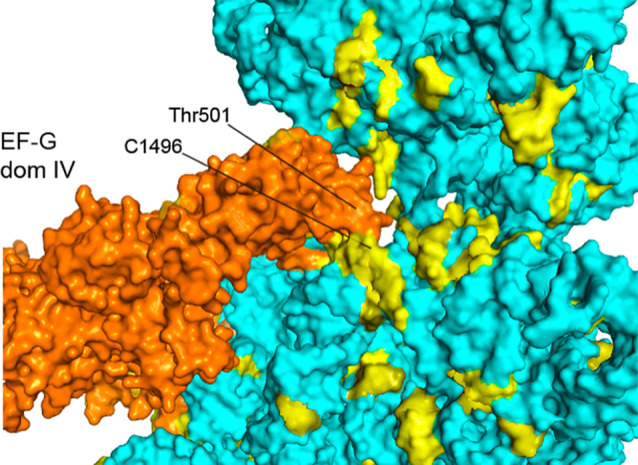
Interactions between domain IV of EF-G and universal A-site nucleotides. Loop I at the tip of domain IV of EF-G (Thr501) contacts universally conserved nucleotides adjacent to the A site (A1496) in the chimeric-hybrid state (Table 4; Zhou et al. 2014).
Some unexplained universal clusters
Cluster 13 is made up of universally conserved bases in helix h27 between positions 885 and 911 that are buried behind helix h44 on the interface side of the 30S subunit (Fig. 21). They form a complementary surface to the minor groove of helix h44 on the opposite face from A1418, which forms bridge B3 near the axis of intersubunit rotation (Fig. 21). The roles of these rare buried universals are unknown. Their location suggests that they may play some as-yet unknown role in influencing intersubunit rotation.
FIGURE 21.
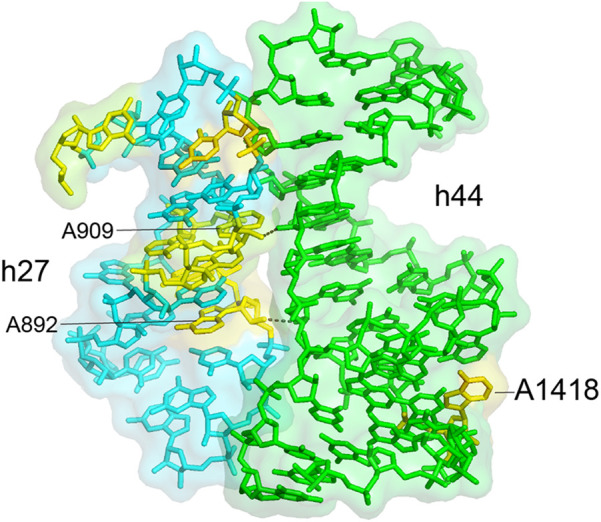
Universal nucleotides in helix h27 pack against the inside of helix h44. The cluster of conserved bases in helix h27 form a complementary surface to the minor groove of helix h44 on the opposite face from A1418, which forms intersubunit bridge B3 at the axis for intersubunit rotation.
Another of the few unexplained clusters of universally conserved nucleotides is seen at the top of the head domain (Cluster 1 in Figs. 6, 22). To our knowledge, no known function has been ascribed to this region of the 16S rRNA, which is remote from any known sites of interaction with functional ligands or with the 50S subunit. Moreover, when the structures of the ribosomal proteins are superimposed on the 16S rRNA, this universal cluster is completely masked (Figs. 5C, 24). The universal nucleotides in this cluster form an intricate three-dimensional network built around the conserved hairpin loop of helix h42 (Fig. 23). Interestingly, the bases are all oriented internally, rather than projecting outward as do almost all other universals. Two noncanonical base pairs, the U1315–A1319 reverse Hoogsteen pair and the A978–A1360 type V A–A pair form the core of the network, while G1316 makes a hydrogen-bonded contact with phosphate 1319.
FIGURE 22.
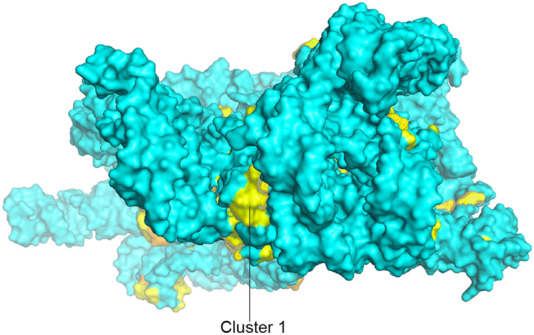
The head domain cluster (cluster 1). A view of 16S rRNA from the top of the head domain, rotated 90° around the X-axis from the view in Figure 5A, showing the cluster of invariant nucleotides on the top of the domain.
FIGURE 24.
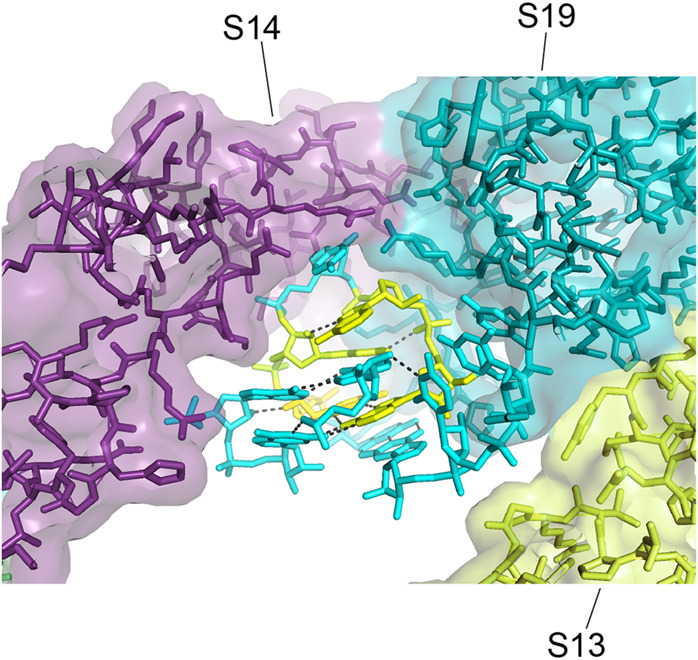
Interactions between proteins S14 and S19 and cluster 1. A cleft is formed between proteins s14 and S19 that wraps around one side of the head domain cluster. The proteins contact the RNA backbone of the network; no contact is made by either protein with any of the invariant bases.
FIGURE 23.
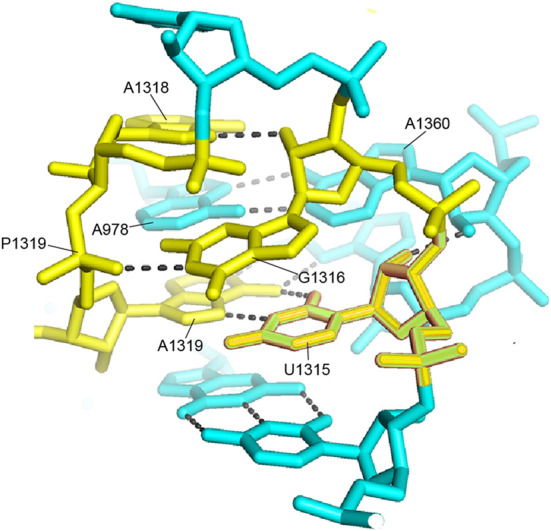
A network of universals in cluster 1. Four invariant nucleotides form a network around the noncanonical A1319–U1315 reverse Hoogsteen pair and the A978–A1360 type V A–A pair in the hairpin loop of helix h42. Note that the bases are virtually inaccessible inside the network, which is remote from any known functional site.
This network appears to stabilize a complex three-way tertiary interaction between the hairpin loops of h42 (positions 1315–1319) and h43 (positions 1360–1361) involving A978 from the connecting loop between helices h31 and h32. The overall structure is surrounded on one side by proteins S14 and S19, which contact each other precisely at their contact with the RNA network (Fig. 24). This is one of the few examples of a group of universally conserved nucleotides that is contacted by a ribosomal protein. In spite of the presence of four universal nucleotides, there is no contact of any of the respective bases with either of the proteins. The RNA is bound via van der Waals and ionic interactions to a complementary surface formed in a cleft between the two proteins. It is thus not obvious why these particular bases are universally conserved across the phylogenetic spectrum. In particular it is difficult to understand why A1318, which makes only a single H-bond with ribose 1316, is invariant.
Involvement of invariant nucleotides in unusual structural motifs
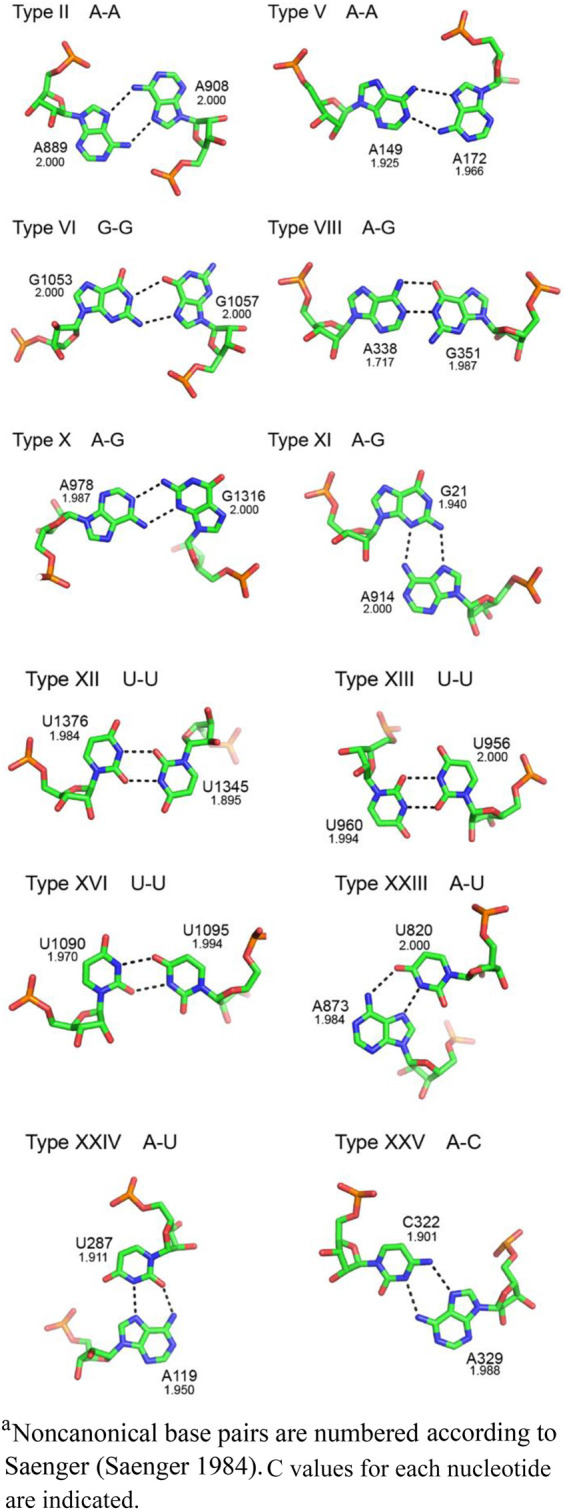
A further potential explanation for universal conservation of some bases in rRNA is that they are involved in unique structural motifs that can only be formed by particular RNA bases, and that these motifs are essential for the functional activity of the rRNA or for its folding and/or assembly. Inspection of the structural contexts of the universal bases indeed reveals that many of them participate in unusual RNA structural motifs, several of which are involved in RNA folds at the functional sites described above (Figs. 10–16). The universal conservation of other bases may be explained by their participation in noncanonical base–base interactions that are uniquely able to create a critical fold in the rRNA. Examples of the noncanonical base pairs found in 16S rRNA are provided in the Appendix, using the Saenger (1984) numbering system.
It was noted many years ago that adenine is strikingly overrepresented among non-base-paired bases (Gutell et al. 1985, 2000), and we can now see that this extends to the universals. Among the 140 universal bases, 60 (or 43%) are adenines. In a review of 16S rRNA structure (Noller 2005), it was noted that the so-called A-minor structural motifs (Cate et al. 1996; Nissen et al. 2001) are widespread in 16S rRNA, as they are in 23S rRNA (Nissen et al. 2001; Bokov and Steinberg 2009). A-minor interactions are formed by hydrogen bonding between the base and ribose moieties of adenosines with the base and ribose moieties in the minor groove of an RNA helix. Nissen et al. (2001) identified three classes of A-minor interactions, of which type I interactions involve the most extensive H-bonding. A prominent example is shown in Figure 25, where the invariant A959 forms a type I A-minor base triple with the invariant G1221–C984 base pair in helix h32, where the N1, N3, and 2′-OH positions of A959 form H-bonds with groups on the minor groove side of the G–C pair. We have seen that A-minor interactions play essential roles in binding tRNA to both the A and P sites of the small subunit (Figs. 8, 15).
FIGURE 25.
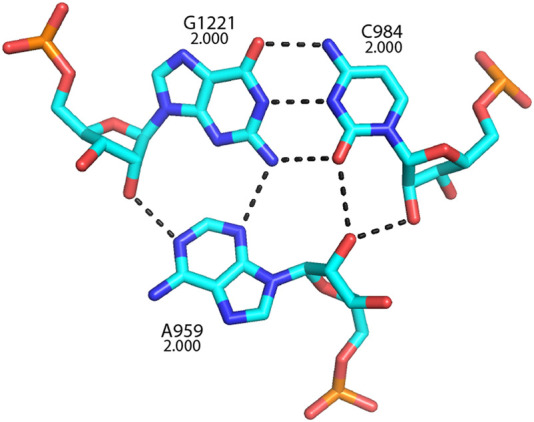
A universally conserved type I A-minor base triple in 16S rRNA. Conservation values are indicated for each of the three bases. This structure likely supports A-site elements in the head domain of the 30S subunit.
Yet another type of minor-groove interaction is shown in Figure 26, which shows a base quadruple formed between two Watson–Crick base pairs, in which the N6 and N7 positions of the adenine in an A–U pair form a Type XI noncanonical A–G pair by H-bonding to the N2 and N3 positions on the minor groove side of the guanine in a G–C pair. Note that two of the bases, C54 and A55 are consecutive in sequence, stabilizing a sharp turn in the RNA backbone by formation of the quadruple.
FIGURE 26.
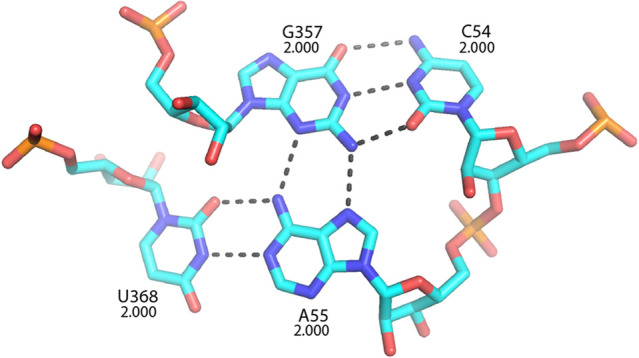
A conserved base quadruple. Two conserved Watson–Crick pairs form a quadruple via the noncanonical type XI A55–G357 pair. Since C54 and A55 are consecutive in sequence, formation of the quadruple stabilizes a sharp turn in the RNA backbone. This quadruple forms the core of cluster 14, which contacts elongation factors EF-G and EF-Tu.
Conclusions
Our analysis shows that 140 nt of the 16S-like rRNAs (including the eukaryotic 18S rRNAs) are universally conserved. This seems an enormous number of nucleotides whose mutation would have eliminated the existence of any free-living organism on our planet, regardless of the vast diversity of selective pressures over >3.5 billion years. Why is it impossible for any organism to live and reproduce with a substitution in any one of these 140 nt? Biochemical, genetic and structural studies over the past half century have led to the accumulation of strong evidence for the direct involvement of specific bases in ribosomal functional sites—a total of ∼40, by our count. These include 24 nt for the A, P and E tRNA binding sites (Table 1). Many, but by no means all of these interactions, depend on contacts that are base-specific, such as the A-minor interactions between A1492 and A1493 with the minor groove of the codon–anticodon duplex (Fig. 8), and the strong H-bonding interaction between G926 and the mRNA at phosphate +1 of the P-site codon (Fig. 13) and several other examples of G-phosphate H-bonding (Figs. 14–16, 23). In contrast, virtually none of the contacts between universal nucleotides and elongation factors EF-G and EF-Tu involve base-specific interactions. Instead, conservation of bases in these contact sites appears to be constrained by preservation of both noncanonical and Watson–Crick pairing that support the arrangement of unique RNA backbone features that are recognized by the elongation factors. The remaining functional bases are located at two of the 12 intersubunit bridges. Bridge B3 is located near the axis of intersubunit rotation—the pivot point for the ∼10° rotation of the 30S subunit relative to the 50S subunit. Universal conservation of A1418 of 16S rRNA preserves its ability to form a type I A-minor interaction with the minor groove of helix H71 of 23S rRNA in the 50S subunit. In contrast, the conserved nucleotides positioned at bridge B2c make only backbone contacts with 23S rRNA. This leaves 100 nt whose universal conservation does not appear to be due to direct participation in ribosome function.
Some 51 universal nucleotides are positioned near the above functional sites, contacting them directly or indirectly in ways that suggest that they might provide structural support to fix the positions and orientations of those nucleotides that are directly involved in function. This may amount essentially to formation of the RNA structures surrounding the functional sites. Alternatively, some of these nucleotides may be involved in fine tuning or stabilization of the local conformations of functional bases. Examples of bases involved in these kinds of structural support are shown for the A site in Figures 10–12 and for the P site in Figures 13–16.
With 91 universal nucleotides implicated directly or indirectly in ribosomal function, at least 49 remain unexplained (Table 1). One possible explanation for their universal conservation is that they might be involved in unusual noncanonical base–base interactions, such as those shown in Figures 25 and 26. Indeed, many of these unexplained universals make noncanonical interactions, including five guanines that are involved in H-bonding to phosphates (Table 1). This begs the further question as to why these noncanonical interactions must be invariant. We suggest that there are at least two possibilities. One possibility is that these interactions are essential for assembling an active ribosome, and that unusual base pairings are somehow uniquely able to achieve this, for example by avoiding the “alternative conformer hell” described by Uhlenbeck (1995). A second possibility is that noncanonical pairing may in some cases support the structural dynamics of the ribosome. The many conformational rearrangements underlying ribosome function, including, most notably, translocation, may depend on flexibility in specific parts of the ribosome structure that do not have obvious connections to particular functions.
One interesting finding is that almost no invariant nucleotides are found in the ribosomal protein binding sites (Fig. 5), with the notable exception of cluster 1, which forms a contact site for proteins S14 and S19 (Figs. 22–24). And although clusters of invariants are found in the contact sites for the elongation factors (Table 4; Fig. 17), virtually none of the actual bases directly contact the factors. These observations are in keeping with our view that the ribosome is fundamentally an RNA-based molecular machine; they also support the possibility that the basic ribosomal mechanisms were in place before the arrival of proteins as we know them in the evolution of life on our planet.
The questions raised here provide ample indications of the incompleteness of our present understanding of ribosome structure and function. One obvious approach to answering them would be to target these unexplained invariant positions for mutational analysis, testing their effects in vivo and in vitro. And with the newly achieved capabilities and speed of high-resolution cryo-EM structure determination, the detailed effects of these mutations on ribosome structure and assembly could also be examined. It is also important to remember that our analysis has focused exclusively on the 16S-like rRNAs, so a full assessment of our knowledge will require a similar study of the 23S-like rRNAs, which are nearly twice the size. In conclusion, although we have witnessed enormous progress in understanding this remarkable biological machine over the last half century, it is clear that much remains shrouded in mystery.
Appendix 1:
Examples of noncanonical base pair classesa
ACKNOWLEDGMENTS
This work was supported by grant number R35-GM118156 (to H.F.N.) and grant number R01-GM067317 (to R.R.G.) from the National Institutes of Health. We thank Laura Lancaster for a critical reading of the manuscript and Sharon Sussman and Laura Lancaster for expert help with rendering molecular graphics figures. This paper is dedicated to the memory of our long-time colleague and friend Carl Woese, who was the first to recognize the profound importance of the invariant nucleotides of ribosomal RNA to life on our planet.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079019.121.
REFERENCES
- Abdi NM, Fredrick K. 2005. Contribution of 16S rRNA nucleotides forming the 30S subunit A and P sites to translation in Escherichia coli. RNA 11: 1624–1632. 10.1261/rna.2118105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol 71: 7724–7736. 10.1128/AEM.71.12.7724-7736.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. 2006. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl Environ Microbiol 72: 5734–5741. 10.1128/AEM.00556-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920. 10.1126/science.289.5481.905 [DOI] [PubMed] [Google Scholar]
- Belardinelli R, Sharma H, Caliskan N, Cunha CE, Peske F, Wintermeyer W, Rodnina MV. 2016. Choreography of molecular movements during ribosome progression along mRNA. Nat Struct Mol Biol 23: 342–348. 10.1038/nsmb.3193 [DOI] [PubMed] [Google Scholar]
- Bokov K, Steinberg SV. 2009. A hierarchical model for evolution of 23S ribosomal RNA. Nature 457: 977–980. 10.1038/nature07749 [DOI] [PubMed] [Google Scholar]
- Brilot AF, Korostelev AA, Ermolenko DN, Grigorieff N. 2013. Structure of the ribosome with elongation factor G trapped in the pretranslocation state. Proc Natl Acad Sci 110: 20994–20999. 10.1073/pnas.1311423110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J, Palmer ML, Kennedy PJ, Noller HF. 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci 75: 4801–4805. 10.1073/pnas.75.10.4801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J, Dull TJ, Noller HF. 1980. Complete nucleotide sequence of a 23S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci 77: 201–204. 10.1073/pnas.77.1.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J, Dull TJ, Sleeter DD, Noller HF. 1981. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol 148: 107–127. 10.1016/0022-2836(81)90508-8 [DOI] [PubMed] [Google Scholar]
- Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, et al. 2002. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3: 2. 10.1186/1471-2105-3-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone C, Loveland A, Gamper H, Hou Y-M, Demo G, Korostelev A. 2021. Time-resolved cryo-EM visualizes ribosomal translocation with EF-G and GTP. Nat Commun 12: 7236. 10.1038/s41467-021-27415-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AP, Clemons WM Jr, Brodersen DE, Morgan-Warren RJ, Hartsch T, Wimberly BT, Ramakrishnan V. 2001. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science 291: 498–501. 10.1126/science.1057766 [DOI] [PubMed] [Google Scholar]
- Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. 1996. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science 273: 1678–1685. 10.1126/science.273.5282.1678 [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Newnam GP, Liebman SW. 1996. The translational function of nucleotide C1054 in the small subunit rRNA is conserved throughout evolution: genetic evidence in yeast. Proc Natl Acad Sci 93: 2517–2522. 10.1073/pnas.93.6.2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM. 2014. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42: D633–D642. 10.1093/nar/gkt1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G. 2012. A new understanding of the decoding principle on the ribosome. Nature 484: 256–259. 10.1038/nature10913 [DOI] [PubMed] [Google Scholar]
- Fox GE, Woese CR. 1975. 5S RNA secondary structure. Nature 256: 505–507. 10.1038/256505a0 [DOI] [PubMed] [Google Scholar]
- Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. 2009. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 326: 694–699. 10.1126/science.1179709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell RR, Weiser B, Woese CR, Noller HF. 1985. Comparative anatomy of 16-S-like ribosomal RNA. Prog Nucleic Acid Res Mol Biol 32: 155–216. 10.1016/S0079-6603(08)60348-7 [DOI] [PubMed] [Google Scholar]
- Gutell RR, Cannone JJ, Shang Z, Du Y, Serra MJ. 2000. A story: unpaired adenosine bases in ribosomal RNAs. J Mol Biol 304: 335–354. 10.1006/jmbi.2000.4172 [DOI] [PubMed] [Google Scholar]
- Gutell RR, Lee JC, Cannone JJ. 2002. The accuracy of ribosomal RNA comparative structure models. Curr Opin Struct Biol 12: 301–310. 10.1016/S0959-440X(02)00339-1 [DOI] [PubMed] [Google Scholar]
- Hugenholtz P, Huber T. 2003. Chimeric 16S rDNA sequences of diverse origin are accumulating in the public databases. Int J Syst Evol Microbiol 53: 289–293. 10.1099/ijs.0.02441-0 [DOI] [PubMed] [Google Scholar]
- Hussain T, Llacer JL, Wimberly BT, Kieft JS, Ramakrishnan V. 2016. Large-scale movements of IF3 and tRNA during bacterial translation initiation. Cell 167: 133–144.e113. 10.1016/j.cell.2016.08.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S, Cannone J, Lee J, Gutell R, Woodson S. 2002. Distribution of rRNA introns in the three-dimensional structure of the ribosome. J Mol Biol 323: 35–52. 10.1016/S0022-2836(02)00895-1 [DOI] [PubMed] [Google Scholar]
- Jenner LB, Demeshkina N, Yusupova G, Yusupov M. 2010. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat Struct Mol Biol 17: 555–560. 10.1038/nsmb.1790 [DOI] [PubMed] [Google Scholar]
- Khade PK, Joseph S. 2011. Messenger RNA interactions in the decoding center control the rate of translocation. Nat Struct Mol Biol 18: 1300–1302. 10.1038/nsmb.2140 [DOI] [PubMed] [Google Scholar]
- Korostelev A, Trakhanov S, Laurberg M, Noller HF. 2006. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell 126: 1065–1077. 10.1016/j.cell.2006.08.032 [DOI] [PubMed] [Google Scholar]
- Korostelev A, Asahara H, Lancaster L, Laurberg M, Hirschi A, Zhu J, Trakhanov S, Scott WG, Noller HF. 2008. Crystal structure of a translation termination complex formed with release factor RF2. Proc Natl Acad Sci 105: 19684–19689. 10.1073/pnas.0810953105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster L, Noller HF. 2005. Involvement of 16S rRNA nucleotides G1338 and A1339 in discrimination of initiator tRNA. Mol Cell 20: 623–632. 10.1016/j.molcel.2005.10.006 [DOI] [PubMed] [Google Scholar]
- Liu LY, Li S, Hu N, He Y, Pong R, Lin D, Lu L, Law M. 2012. Comparison of next-generation sequencing systems. J Biomed Biotechnol 2012: 251364. 10.1201/b16568-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangroo D, Wu XQ, RajBhandary UL. 1995. Escherichia coli initiator tRNA: structure-function relationships and interactions with the translational machinery. Biochem Cell Biol 73: 1023–1031. 10.1139/o95-109 [DOI] [PubMed] [Google Scholar]
- McClory SP, Leisring JM, Qin D, Fredrick K. 2010. Missense suppressor mutations in 16S rRNA reveal the importance of helices h8 and h14 in aminoacyl-tRNA selection. RNA 16: 1925–1934. 10.1261/rna.2228510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D, Noller HF. 1986. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell 47: 985–994. 10.1016/0092-8674(86)90813-5 [DOI] [PubMed] [Google Scholar]
- Moazed D, Noller HF. 1990. Binding of tRNA to the ribosomal A and P sites protects two distinct sets of nucleotides in 16S rRNA. J Mol Biol 211: 135–145. 10.1016/0022-2836(90)90016-F [DOI] [PubMed] [Google Scholar]
- Moazed D, Stern S, Noller HF. 1986. Rapid chemical probing of conformation in 16S ribosomal RNA and 30S ribosomal subunits using primer extension. J Mol Biol 187: 399–416. 10.1016/0022-2836(86)90441-9 [DOI] [PubMed] [Google Scholar]
- Mohan S, Donohue JP, Noller HF. 2014. Molecular mechanics of 30S subunit head rotation. Proc Natl Acad Sci 111: 13325–13330. 10.1073/pnas.1413731111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. 2001. RNA tertiary interaction in the large ribosomal subunit: the A-minor motif. Proc Natl Acad Sci 98: 4899–4903. 10.1073/pnas.081082398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller HF. 1974. Topography of 16S RNA in 30S ribosomal subunits. Nucleotide sequences and location of sites of reaction with kethoxal. Biochemistry 13: 4694–4703. 10.1021/bi00720a003 [DOI] [PubMed] [Google Scholar]
- Noller HF. 1991. Ribosomal RNA and translation. Annu Rev Biochem 60: 191–227. 10.1146/annurev.bi.60.070191.001203 [DOI] [PubMed] [Google Scholar]
- Noller HF. 2005. RNA structure: reading the ribosome. Science 309: 1508–1514. 10.1126/science.1111771 [DOI] [PubMed] [Google Scholar]
- Noller HF, Chaires JB. 1972. Functional modification of 16S ribosomal RNA by kethoxal. Proc Natl Acad Sci 69: 3113–3118. 10.1073/pnas.69.11.3115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller HF, Woese CR. 1981. Secondary structure of 16S ribosomal RNA. Science 212: 403–411. 10.1126/science.6163215 [DOI] [PubMed] [Google Scholar]
- Noller HF, Hoffarth V, Zimniak L. 1992. Unusual resistance of peptidyl transferase to protein extraction procedures. Science 256: 1416–1419. 10.1126/science.1604315 [DOI] [PubMed] [Google Scholar]
- Noller HF, Lancaster L, Mohan S, Zhou J. 2017. Ribosome structural dynamics in translocation: yet another functional role for ribosomal RNA. Q Rev Biophys 50: e12. 10.1017/S0033583517000117 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Brodersen DE, Clemons WM, Tarry MJ, Carter AP, Ramakrishnan V. 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292: 897–902. 10.1126/science.1060612 [DOI] [PubMed] [Google Scholar]
- Okimoto R, Macfarlane JL, Clary DO, Wolstenholme DR. 1992. The mitochondrial genomes of two nematodes, Caenorhabditis elegans and Ascaris suum. Genetics 130: 471–498. 10.1093/genetics/130.3.471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel FT, Zhao SQ, Hijazi KA, Murgola EJ. 1997. Phenotypic heterogeneity of mutational changes at a conserved nucleotide in 16 S ribosomal RNA. J Mol Biol 267: 1113–1123. 10.1006/jmbi.1997.0943 [DOI] [PubMed] [Google Scholar]
- Petrychenko V, Peng B-Z, Schwarzer A, Peske F, Rodnina M, Fischer N. 2021. Structural mechanism of GTPase-powered ribosome-tRNA movement. Nat Commun 12: 5933. 10.1038/s41467-021-26133-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T, Noller HF. 1990. Dominant lethal mutations in a conserved loop in 16S rRNA. Proc Natl Acad Sci 87: 1042–1046. 10.1073/pnas.87.3.1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramrath DJ, Lancaster L, Sprink T, Mielke T, Loerke J, Noller HF, Spahn CM. 2013. Visualization of two transfer RNAs trapped in transit during elongation factor G-mediated translocation. Proc Natl Acad Sci 110: 20964–20969. 10.1073/pnas.1320387110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundlet EJ, Holm M, Schacherl M, Natchiar SK, Altman RB, Spahn CMT, Myasnikov AG, Blanchard SC. 2021. Structural basis of early translocation events on the ribosome. Nature 595: 741–745. 10.1038/s41586-021-03713-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenger W. 1984. Principles of nucleic acid structure. Springer-Verlag, New York. [Google Scholar]
- Sahu B, Khade PK, Joseph S. 2013. Highly conserved base A55 of 16S ribosomal RNA is important for the elongation cycle of protein synthesis. Biochemistry 52: 6695–6701. 10.1021/bi4008879 [DOI] [PubMed] [Google Scholar]
- Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, et al. 2000. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell 102: 615–623. 10.1016/S0092-8674(00)00084-2 [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FV, Weir JR, Ramakrishnan V. 2009. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 326: 688–694. 10.1126/science.1179700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. 2006. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313: 1935–1942. 10.1126/science.1131127 [DOI] [PubMed] [Google Scholar]
- Stiegler P, Carbon P, Ebel JP, Ehresmann C. 1981. A general secondary-structure model for procaryotic and eucaryotic RNAs from the small ribosomal subunits. Eur J Biochem 120: 487–495. 10.1111/j.1432-1033.1981.tb05727.x [DOI] [PubMed] [Google Scholar]
- Uhlenbeck OC. 1995. Keeping RNA happy. RNA 1: 4–6. [PMC free article] [PubMed] [Google Scholar]
- von Ahsen U, Noller HF. 1995. Identification of bases in 16S rRNA essential for tRNA binding at the 30S ribosomal P site. Science 267: 234–237. 10.1126/science.7528943 [DOI] [PubMed] [Google Scholar]
- Watson ZL, Ward FR, Meheust R, Ad O, Schepartz A, Banfield JF, Cate JH. 2020. Structure of the bacterial ribosome at 2 Å resolution. Elife 9: e60482. 10.7554/eLife.60482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberly BT, Brodersen DE, Clemons WM Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. 2000. Structure of the 30S ribosomal subunit. Nature 407: 327–339. 10.1038/35030006 [DOI] [PubMed] [Google Scholar]
- Woese CR, Gutell RR. 1989. Evidence for several higher order structural elements in ribosomal RNA. Proc Natl Acad Sci 86: 3119–3122. 10.1073/pnas.86.9.3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Fox GE, Zablen L, Uchida T, Bonen L, Pechman K, Lewis BJ, Stahl D. 1975. Conservation of primary structure in 16S ribosomal RNA. Nature 254: 83–86. 10.1038/254083a0 [DOI] [PubMed] [Google Scholar]
- Woese CR, Magrum LJ, Gupta R, Siegel RB, Stahl DA, Kop J, Crawford N, Brosius J, Gutell R, Hogan JJ, et al. 1980. Secondary structure model for bacterial 16S ribosomal RNA: phylogenetic, enzymatic and chemical evidence. Nucleic Acids Res 8: 2275–2293. 10.1093/nar/8.10.2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Gutell RR, Gupta R, Noller HF. 1983. Detailed analysis of the higher-order structure of 16S-like ribosomal ribonucleic acids. Microbiol Rev 47: 621–669. 10.1128/mr.47.4.621-669.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov M, Yusupova G, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292: 883–896. 10.1126/science.1060089 [DOI] [PubMed] [Google Scholar]
- Zhou J, Lancaster L, Donohue JP, Noller HF. 2013. Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science 340: 1236086. 10.1126/science.1236086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lancaster L, Donohue JP, Noller HF. 2014. How the ribosome hands the A-site tRNA to the P site during EF-G-catalyzed translocation. Science 345: 1188–1191. 10.1126/science.1255030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb C, Glotz C, Brimacombe R. 1981. Secondary structure comparisons between small subunit ribosomal RNA molecules from six different species. Nucleic Acids Res 9: 3621–3640. 10.1093/nar/9.15.3621 [DOI] [PMC free article] [PubMed] [Google Scholar]