Abstract
BACKGROUND.
Current clinical tools have limited accuracy in differentiating patients with localized prostate cancer who are at risk of recurrence from patients with indolent disease. We aimed to identify a gene expression signature that jointly with clinical variables could improve upon the prediction of clinical recurrence after RP for patients with stage T2 PCa.
METHODS.
The study population includes consented patients who underwent a radical retropubic prostatectomy (RP) and bilateral pelvic lymph node dissection at the University of Southern California in the PSA-era (1988–2008). We used a nested case-control study of 187 organ-confined patients (pT2N0M0): 154 with no recurrence (“controls”) and 33 with clinical recurrence (“cases”). RNA was obtained from laser capture microdissected malignant glands representative of the overall Gleason score of each patient. Whole genome gene expression profiles (29,000 transcripts) were obtained using the Whole Genome DASL HT platform (Illumina, Inc). A gene expression signature of PCa clinical recurrence was identified using stability selection with elastic net regularized logistic regression. Three existing datasets generated with the Affymetrix Human Exon 1.0ST array were used for validation: Mayo Clinic (MC, n = 545), Memorial Sloan Kettering Cancer Center (SKCC, n = 150), and Erasmus Medical Center (EMC, n = 48). The areas under the ROC curve (AUCs) were obtained using repeated fivefold cross-validation.
RESULTS.
A 28-gene expression signature was identified that jointly with key clinical variables (age, Gleason score, pre-operative PSA level, and operation year) was predictive of clinical recurrence (AUC of clinical variables only was 0.67, AUC of clinical variables, and 28-gene signature was 0.99). The AUC of this gene signature fitted in each of the external datasets jointly with clinical variables was 0.75 (0.72–0.77) (MC), 0.90 (0.86–0.94) (MSKCC), and 0.82 (0.74–0.91) (EMC), whereas the AUC for clinical variables only in each dataset was 0.72 (0.70–0.74), 0.86 (0.82–0.91), and 0.76 (0.67–0.85), respectively.
CONCLUSIONS.
We report a novel gene-expression based classifier identified using agnostic approaches from whole genome expression profiles that can improve upon the accuracy of clinical indicators to stratify early stage localized patients at risk of clinical recurrence after RP.
Keywords: gene expression, stage II, clinical recurrence, stability selection
INTRODUCTION
In the United States, it is estimated that one in six men will be diagnosed with prostate cancer (PCa) in their lifetime and approximately 80% are diagnosed with tumors confined to the prostate. Most localized PCa tumors are indolent and will never become aggressive during a patient’s lifetime. However, to date, over half of localized PCa patients have undergone radical prostatectomy (RP) as their primary treatment choice [1]. Despite treatment, approximately 20% of these patients may continue to experience a rising PSA level after surgery (biochemical recurrence) of which 20–30% will develop metastasis and PCa-related death [2,3]. Therefore, distinguishing men diagnosed with localized disease but still at risk of progression from men with localized PCa who will not progress is a pressing priority in the clinical treatment of PCa. This would allow for early identification of men with truly indolent tumors who can avoid RP and enroll in active surveillance and those with aggressive tumors who will benefit more from definitive treatment and/or more aggressive and earlier interventions.
Current prognostic tools to determine risk of progression use clinical variables, such as Gleason score, stage, pre-operative PSA level, but these variables have limited predictive accuracy [4–7]. Tumor biomarkers used jointly with existing clinical variables have been shown to provide additional information to accurately differentiate aggressive and indolent primary PCa tumors [8–11]. Previous studies indicate that within tumors histologically classified as non-aggressive, subsets of cells present at the time of diagnosis may harbor gene expression profiles characteristic of cells with metastatic potential that can be predictive of clinical recurrence [12–20]; however, few of these profiles have been adopted in the clinic for further validation [13,14,17]. Moreover, few of these studies utilized whole genome gene expression data [14,20] and/or utilized tissue microdissection to account for tumor heterogeneity in RNA sampling [12,18,20]. Therefore, there is still a need for novel tumor biomarkers that can help improve prediction of prostate cancer recurrence upon clinical variables.
We report a novel gene expression-based profile that improves the prediction of PCa clinical recurrence over clinical variables alone. This predictive signature was identified using whole genome expression data (over 27,000 coding transcripts and over 1,500 non-coding transcripts) obtained from microdissected malignant prostate glands from 187 prostate cancer patients diagnosed with organ-confined disease (stage T2a-T2c), treated with radical prostatectomy at the USC Norris Comprehensive Cancer Center, Department of Urology, who either developed clinical recurrence, or remained disease free after a comparable follow-up time.
MATERIALS AND METHODS
Patient Population
This study included patients diagnosed with organ-confined disease (pT2) who underwent a radical retropubic prostatectomy and bilateral pelvic lymph node dissection (RRP/PLND) at the University of Southern California from 1988 to 2008 (within the PSA-era at the institution) (n = 2,646). After surgery, patients who consented to enroll in this patient database were followed every 4–6 months in year 1, every 6 months in years 2 and 3, and once annually afterward. During the visits, patients received a physical examination, had a serum PSA measurement, and chest x-ray. Bone scans were also completed if there were signs of progression, such as an increase in PSA levels. Biochemical recurrence (BCR) was defined as a detectable PSA level based on the era-specific assay’s detectability limit, verified by two consecutive increased PSA tests, with 3–4 months in-between blood draws [21]. Patients were defined as having clinical recurrence (CR) after the detection of recurrent local or distant disease by imaging. All specimens from radical prostatectomies were assessed using consistent pathological reporting.
Patients from this cohort were selected for this nested case control study. Eligible participants did not have lymph node involvement (N0), but had formalin-fixed paraffin-embedded (FFPE) prostatectomy tissue available for processing, and also had available clinical and follow-up data. Among the 2,552 eligible patients, there were 2,359 patients who had no evidence of disease after surgery (NED), 147 with biochemical/PSA recurrence only (BCR), and 46 with clinical recurrence (CR). Since some BCR patients do not ever experience metastatic disease, in order to enrich for genes predictive of truly aggressive PCa in this study, we compared NED and CR patients to determine a gene expression signature predictive of aggressive disease. Our final sample set included 154 NED patients and 33 with CR (those with tissue blocks available for microdissection).
Laser Capture Microdissection and RNA Extraction
All FFPE prostatectomy tissue sections were reviewed by an expert pathologist (Dr. Andy Sherrod) with the primary goal of determining the densest region of tumor to capture as much tumor RNA as possible, which usually translates into the region of higher Gleason score/grade of the tumor. In order to enrich for malignant glands and avoid contamination with stromal tissue or non-malignant glands, a laser capture microdissection (LCM) microscope (Arcturus® Laser Capture Microdissection, Model Veritas; Applied Biosystems by Life Technologies, Foster City, CA) was used to microdissect malignant prostate glands. Tumor sections cut at 5 microns were lightly stained with hematoxylin and eosin prior to microdissection. RNA extraction was performed using the Qiagen AllPrep DNA/RNA FFPE kit (Qiagen, Valencia, CA).
Gene Expression Microarray
Genome-wide gene expression profiles were generated for all samples, 50–200 ng RNA each, using the Whole-Genome DASL-HT Assay (Illumina, Inc.) [22]. The HumanHT-12 v4 BeadChip was used to detect the following transcripts using the RNA from the tumor samples: 27,253 coding transcripts (well-established annotations), 426 coding transcripts (provisional annotations), 1,580 non-coding transcripts (well-established annotations), and 26 non-coding transcripts (provisional annotations) (Illumina; Whole-Genome DASL ® HT Assay for Expression Profiling in FFPE Samples; Data Sheet: RNA Analysis, 2010). For quality control purposes, 20% of samples were included as duplicates and an even number of cases and controls were run on the same array chip. Technical replicates that were used to measure the variation induced by the processing of samples showed very good reproducibility (r2 = 0.97). Three of the samples processed had low sensitivities (~2,000–4,000 genes detected) based on the P-value thresholds and therefore were not included in further analyses.
Pre-Processing of Gene Expression Data
All pre-processing of data and subsequent analyses were performed using R and Bioconductor [23]. Control probes and sample probes were used to preprocess (normalization and background correction) and to assess quality control using Bioconductor’s lumi and limma packages. A specific pre-processing package (neqc) allowed for non-parametric background correction followed by quantile normalization using both control and sample probes [24]. This method provides the optimal compromise between precision and bias that occurs when using algorithms in preprocessing. We further considered and adjusted for possible batch effects by chip array during the microarray processing using ComBat [25]. The adjustment also took into account any batch effects by shipment since each shipment of RNA samples sent to Illumina involved the use of several BeadChips.
Differential Gene Expression
We identified differentially expressed genes (DEGs) between tumors of NED patients (n = 154) and CR patients (n = 33) using the empirical Bayes moderated t-test [26], which was applied on the entire set of ~29,000 features, adjusting for age (coded as a continuous variable), pre-operative PSA level (continuous), pathologic Gleason score (≤6, 7, 8–10), neoadjuvant hormone therapy (no, yes), operation year (continuous), and surgical margin status (positive, negative). Multiple testing correction was done by calculating the False Discovery Rate (FDR) using the Benjamini–Hochberg method [27].
Pathway Analyses
Using the resulting DEGs obtained from the analysis, GeneOntology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed using WebGestalt software (WEB-based GEne SeT AnaLysis Toolkit) (Nashville, TN). [28–30] Pathway analyses were completed for all differentially expressed genes (DEGs) and separately for genes that had higher expression in tumors of CR patients compared to NED patients (hereafter, referred to as up-regulated genes) and for genes that had lower expression in tumors of CR patients compared to NED patients (hereafter, referred to as down-regulated genes). All WebGestalt gene enrichment analyses were done using the following parameters: Benjamini–Hochberg correction for multiple testing, minimum of four genes within each category, and a significance level of multiple tested corrected P < 0.05. Ingenuity Pathway Analysis (IPA) was also used to gain further insight into the gene networks, canonical pathways, diseases, and functions associated with the genes, and upstream regulators using the DEGs.
Identification of Gene Signatures Predictive of Aggressive PCa
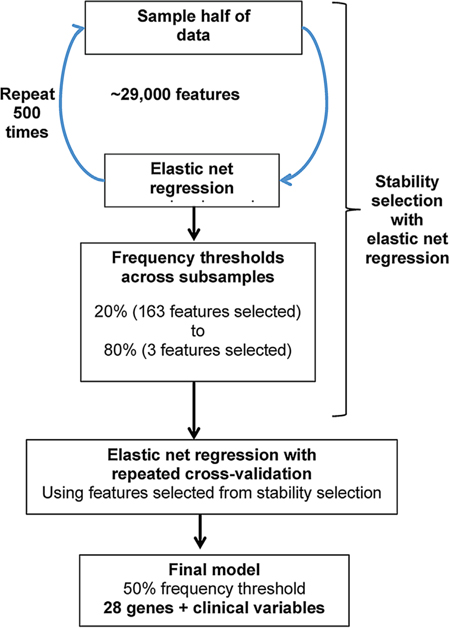
Using the pre-processed list of 29,000 targets, we identified a multivariate risk prediction model for PCa clinical recurrence using stability selection with elastic net-regularized logistic regression [31]. We used the R Bioconductor package caret to calibrate the optimal tuning parameter, using elastic net with repeated 10-fold cross-validation, and settled on an α = 0.2 as this would maximize the AUC (area under the curve) estimate while allowing for inclusion of as many possible features while maintaining good prediction. Using this tuning parameter, we implemented stability selection using 500 subsamples using the package glmnet in R, with each subsample having half of the sample size (original sample size n = 187). We used different frequency thresholds from 20 to 80% to determine the most predictive features. The different models identified were evaluated by estimating the average AUC across repeated (10 times) fivefold cross-validation. The following clinical variables were force-included in all models: Gleason, PSA, year of operation, and age at surgery. In addition, we also included use of neo-adjuvant androgen deprivation therapy (NADT), as a possible variable for selection, although not forced-included. A flowchart of the statistical analyses undertaken is shown in Figure 1.
Fig. 1.
Analytical plan for predictive model development.
In Silico Validation of Gene Signature
For validation of the identified models, we used three external datasets from three different studies that used whole-genome gene expression of PCa tumors. These datasets were: (i) from the Mayo Clinic (MC) [14]; (ii) Memorial Sloan-Kettering Cancer Center (MSKCC) [32]; and (iii) Erasmus Medical Center (EMC) [33]. Genomic and clinical data for these studies were obtained through the National Center of Biotechnology Information (NCBI) database repository for genomic data, Gene Expression Omnibus (GEO) (series accession numbers: GSE46691, GSE21032, and GSE41410). All three studies used the Affymetrix Human Exon 1.0 ST array to obtain gene expression data. This array consists of ~1.4 million probe sets, with approximately four probes per exon and about 40 probes per gene. In order to perform validation using these datasets, all probes and expression profiles corresponding to the genes in our predictive gene signature were extracted. Partek® Genomics Suite (Partek Inc., St. Louis, MO), was used to extract the raw data (Affymetrix CEL files) from GEO and was normalized through standard robust multi-array average (RMA) method and background correction for Affymetrix arrays. In order to ensure that all possible probes with good reliability were included in the validation, extended and full annotations were obtained for all probes pertaining to genes in the model. Probes from the full probeset annotation were used for final validation, since probe intensity distributions among extended and full probes are almost indistinguishable [34]. Using corresponding expression data from the patient population from each of the studies, repeated fivefold cross-validation using elastic net (α = 0.2 and no standardization of the probe variables) was performed for validation. To determine the best prediction of a parsimonious model, the average AUC across all cross-validation runs was obtained using the LASSO penalty parameter set as one standard error above the detected minimum penalty, which indicates the lowest cross-validation error. Genes for all the possible predictive models generated from the stability selection (frequency threshold 20–80%) were assessed using the probe and clinical data available for each dataset.
RESULTS
Whole-genome gene expression profiles were generated for 154 patients who had no evidence of disease (NED) following surgery after at least 2 years of follow-up, and 33 patients who experienced clinical recurrence of disease, with local or distal metastasis detected (CR). Compared to NED patients, CR patients had higher Gleason score (Gleason 8–10, 36% CR vs. 16% NEDs, P = 0.01), and more had neoadjuvant hormonal therapy prior to surgery (24% CR vs. 4% NEDs, P = 0.001) (Table I). CR patients were also more likely to be classified as high-risk according to the D’Amico risk classification using available diagnostic data prior to surgery. The median follow-up time was 9.55 years for NED patients and 5.83 years until clinical recurrence for CR patients. There were no differences in racial/ethnic distribution between the NED and CR patients, with the majority of patients being non-Hispanic White (89% and 88%, respectively).
TABLE I.
Clinical Characteristics of Localized Prostate Cancer Patients
| Controls (NED) | Cases (CR) | P-value | |
|---|---|---|---|
|
| |||
| n = 154 | n = 33 | ||
| Age | |||
| Median | 64 | 66 | 0.056 |
| IQR | 56–69 | 62–71 | |
| Range | 44–82 | 45–76 | |
| PSA before surgery (ng/ml) | |||
| Median | 7.16 | 7.4 | 0.17 |
| IQR | 4.81–10.40 | 5.60–13.0 | |
| Range | 1.1–120 | 2.2–65.80 | |
| Pathologic Gleason score | |||
| ≤6 | 56 (36) | 5 (15) | 0.01 |
| (3 + 4) or (2 + 5) | 60 (39) | 11 (33) | |
| (4 + 3) or (5 + 2) | 14 (9) | 5 (15) | |
| 8–10 | 24 (16) | 12 (36) | |
| Surgical margin status | |||
| Negative | 119 (77) | 24 (73) | 0.651 |
| Positive | 35 (23) | 9 (27) | |
| Race/ethnicity | |||
| Non-Hispanic White | 137 (89) | 29 (88) | 0.255 |
| Hispanic | 12 (8) | 1 (3) | |
| African-American | 4 (3) | 2 (6) | |
| Asian/PI | 1 (1) | 1 (3) | |
| Clinical stage | |||
| cT1 | 105 (68) | 17 (52) | 0.089 |
| cT2 | 48 (31) | 15 (45) | |
| cT3 | 1 (1) | 1 (3) | |
| Pathologic stage | |||
| T2a | 10 (6) | 3 (9) | 0.819 |
| T2b | 9 (6) | 1 (3) | |
| T2c | 134 (87) | 29 (88) | |
| T2 with unknown laterality | 1 | 0 | |
| Prostatectomy year | |||
| 07/1988–07/1994 | 56 (37) | 18 (55) | 0.093 |
| 07/1994–03/2005 | 90 (59) | 13 (39) | |
| 03/2005–06/2008 | 6 (4) | 2 (6) | |
| D’Amico risk groups (Those with available clinical data: Gleason, stage, PSA) | |||
| Low | 50 (40) | 2 (8) | <0.001 |
| Intermediate | 60 (48) | 11 (42) | |
| High | 15 (12) | 13 (50) | |
| Neoadjuvant hormonal therapy | |||
| No | 148 (96) | 25 (76) | 0.001 |
| Yes | 6 (4) | 8 (24) | |
| Radiation therapy | |||
| No | 135 (88) | 26 (79) | 0.177 |
| Yes | 19 (12) | 7 (21) | |
| Adjuvant hormone therapy | |||
| No | 151 (98) | 33 (100) | |
| Yes | 3 (2) | 0 (0) | |
| Median follow-up time (IQR) | 9.55 (6.61–15.25) | 5.83 (4.18–8.69) | |
NED, patients with “no evidence of disease” after radical prostatectomy; CR, patients with clinical recurrence after radical prostatectomy.
Identification of Differentially Expressed Genes (DEGs)
There were 184 differentially expressed features/probes, which represent a total of 172 differentially expressed genes (DEGs), in comparisons of tumors from CR and NED patients (Supplementary Table SI). All but 10 genes had a fold change of >1.2 with FDR-corrected P-values < 0.05.
Gene Set Enrichment Analyses
We did enrichment analyses for GeneOntology (GO) categories for all DEGs between tumors of CR patients and NED patients and identified eight functional categories enriched for DEGs that related to bioenergetics, with the three terminal categories being: NADP binding (four genes), oxidoreductase activity acting on the CH-OH group of donors with NAD or NADP as acceptor (six genes), and growth factor binding (five genes) (Supplementary Fig. S1a, Table SII). When considering DEGs with lower expression in tumors of CR patients compared to NED patients, we identified a similar pattern with seven enriched GO categories, with the three terminal categories being: coenzyme binding (six genes), oxidoreductase activity acting on CH-OH group of donors, NAD or NADP as acceptor (five genes), peptide binding (five genes) (Supplementary Fig. S1b, Table SII). There were two enriched GO categories for DEGs with higher expression in tumors of CR patients compared to tumors of NED patients, and the one terminal category included five genes in growth factor binding (Supplementary Fig. S1c). Further analyses using KEGG annotation identified three pathways enriched among all DEGs: metabolic pathways (12 DEGs), regulation of actin cytoskeleton (four DEGs), and pathways in cancer (four DEGs) (Supplementary Table SIII).
Disease-enrichment analyses showed enrichment of DEGs annotated to 27 disease categories, with the highest enrichment, based on the ratio of observed number of genes over the expected (R), found in the “fractures, bones” category (four genes). Among these categories, nine were directly related to cancer, including “prostatic neoplasms” (four DEGs: REPS2, CTBP2, LDAH, ANO7) (Supplementary Table SIV). We summarize in Table II all 33 DEGs identified as part of the terminal enriched pathways through GO and KEGG annotations, and genes found to be enriched as part of “prostatic neoplasms.” Among these 33 DEGs, eight were previously reported to be involved in PCa tumorigenesis (ADI1, ANO7, BMPR2, CTBP2, DCXR, LDAH, NRP1, REPS2) and 17 were reported to be involved in tumorigenesis of other cancers.
TABLE II.
DEGs Identified as Part of Enriched Pathways Identified With Gene Ontology and KEGG
| Gene | Main functions | Reported role in PCa | Reported in other cancers | Enrichment category | Expression in CR compared to NED | References |
|---|---|---|---|---|---|---|
| ACADSB | Fatty acid metabolism, mitochondrial localization, determines vitamin D levels | Metabolic (KEGG) | Lower | [82] | ||
| ADI1 | Methionine salvage | Progression | Metabolic (KEGG) | Higher | [35–37] | |
| AGPAT2 | Phospholipid synthesis | Osteosarcoma | Metabolic (KEGG) | Higher | [64] | |
| ALDH1A2 | Retinoic acid synthesis | Metabolic (KEGG) | Lower | [83] | ||
| ANO7 | Ion channel transporter, cell–cell interaction, glucose transport | Grade | Prostatic neoplasia (DE) | Lower | [39,40] | |
| ARHGEF6 | G protein | Actin/cytoskeleton reg. (KEGG) | Higher | |||
| BAIAP2 | Angiogenesis inhibitor, insulin receptor TK, filopodia formation, cell mobility, cancer cell growth | Actin/cytoskeleton regulation (KEGG) | Lower | [84] | ||
| BMPR2 | BMP receptor, cell invasion | Cell invasion | Growth factor binding (GO) | Higher | [85] | |
| CDKN2B | Cell proliferation | Several cancers | Pathways in cancer (KEGG) | Higher | [86,87] | |
| CHPT1 | Phosphatidylcholine metabolism | Breast | Metabolic (KEGG) | Higher | [65] | |
| COLIA2 | Collagen chain, cell proliferation and cell migration | Bladder | Growth factor binding (GO) | Higher | [60] | |
| CTBP2 | Transcriptional repressor, cell proliferation | Progression, cell proliferatio | Breast, esophagus, ovarian | Oxidoreductase activity (GO); pathways in cancer (KEGG); | Lower | [45–47] |
| Prostatic neoplasm (DE) | ||||||
| CYB5A | Metabolism ferric Hb metabolism, autophagy | Pancreatic | Oxidoreductase activity (GO) | Lower | [55] | |
| DCXR | Glucose metabolism | Biomarker | Liver, melanoma | Oxidoreductase activity (GO); metabolic (KEGG) | Lower | [48–50] |
| DECR1 | Fatty acid metabolism, mitochondrial localization | Binding (GO) | Lower | |||
| DUOX1 | Hydrogen peroxide producer, cell migration, antimicrobial defense | Binding (GO) | Higher | [59] | ||
| ESM1 | Endothelial cells factor | Breast and gastric | Growth factor binding (GO) | Higher | [61,88] | |
| FMO5 | Trimethylamine metabolism, determines mtDNA levels | Binding (GO) | Lower | [44] | ||
| GLDC | Glycine metabolism, mitochondrial localization | Lung | Metabolic (KEGG) | Higher | [66] | |
| GRHPR | Pyruvate metabolism | Liver | Oxidoreductase activity (GO); binding (GO); metabolic (KEGG) | Lower | [56] | |
| LAMA3 | Base membrane formation, cell migration | Gastric | Pathways in cancer (KEGG) | Higher | [89] | |
| LDAH | Cholesterol homeostasis | Risk | Prostatic neoplasia (DE) | Lower | [53,54] | |
| MAT2B | SAM biosynthesis, growth, tumorigenesis | Oxidoreductase activity (GO); metabolic (KEGG) | Higher | [58] | ||
| ME2 | Metabolism, mitochondrial localization | Melanoma | Oxidoreductase activity (GO) | Lower | [57] | |
| MYH9 | Cytokinesis, cell motility, cell shape, cell migration | Head and neck | Actin/cytoskeleton reg. (KEGG) | Higher | [69] | |
| NADSYN1 | Metabolism (NAD synthesis, calcium), determines VitD levels | Metabolic (KEGG) | Lower | [90,91] | ||
| NRP1 | Cell migration, cell survival, angiogenesis | Relapse | Bladder | Growth factor binding (GO) | Higher | [51,52] |
| PIAS4 | Autophagy | Pancreas | Pathways in cancer(KEGG) | Higher | [92,93] | |
| REPS2 | Cell proliferation, vesicular trafficking, endocytosis | Progression | Prostatic neoplasia (DE) | Lower | [41–43] | |
| RHBDF2 | Protease | Ovarian | Growth factor binding (GO) | Higher | [63] | |
| SRM | Spermidine synthesis, cell growth | Metabolic (KEGG) | Lower | |||
| SYNJ1 | Synaptic transmissions, membrane trafficking | Metabolic (KEGG) | Higher | [94] | ||
| TMSB4X | Cell proliferation, cell migration, differentiation, anti-apoptotic, cancer stem cells | Glioblastoma, CRC, breast, ovarian, lung, kidney, uterine | Actin/cytoskeleton reg. (KEGG) | Lower | [70–72] |
DEG, differentially expressed gene; CR, clinical recurrence patients; NED, no evidence of disease patients; CRC, colorectal cancer.
In order to identify candidate chromosomal deletions or amplifications directly responsible for differential gene expression, we analyzed whether there were specific genomic locations enriched for DEGs, as this may identify chromosomal deletions or amplifications. When all DEGs were considered, four cytogenetic bands were identified: (i) Chr3q: 10 DEGs; (ii) Chr12q: 11 DEGs; (iii) Chr18q: 6 DEGs, and (iv) Chr7q22: 4 DEGs. No chromosomal regions were found to be enriched when considering only upregulated or down-regulated DEGs (Supplementary Table SV).
We next used enrichment analysis to identify candidate miRNAs that may regulate the expression of DEGs. For all DEGs, there were 27 significantly enriched miRNA. Among them, we identified MIR-506 as a putative regulator of 13 DEGs and MIR-181, as a putative regulator of 11 DEGs. All other identified DEGs were identified as putative regulators of 4–7 DEGs. For DEGs with higher expression in CR patients compared to NED patients, there were 12 enriched miRNAs, of which nine overlapped with the miRNA found for all DEGs. For DEGs with lower expression in CR patients compared to NED patients, there were four enriched miRNAs that overlapped with the miRNA found for all DEGs (Supplementary Table VI).
Additional analysis included exploring transcription factor (TF) binding sites associated with the regulation of the genes in the lists of DEGs. For the list of all DEGs, there were 117 significantly enriched TF binding sites, 32 for DEGs with higher regulation among tumors of CR patients compared to tumors of NED patients, and 36 for DEGs with lower expression in the same group comparison (Supplementary Table SVII)
Finally, we investigated the associated small molecule targets given the set of DEGs. For all DEGs, seven categories were found, which adenine being the one with most targets [7], followed by NADH [6], and adenosine [5]. For DEGs with higher expression in tumors of CR patients compared to tumors of NED patients, we identified only one small molecule (glycine), and four for DEGs with lower expression in the same group comparison (Supplementary Table SVIII).
Gene Network Analyses
We used IPA software (IngenuitySystems) to analyze functional relationships among DEGs. Each generated network includes a score based on the negative log of the P-value calculated from the Fisher’s Exact t-test used to indicate the likelihood of the network generated by random chance alone. The three IPA networks identified for all DEGs with a score >40 include: (i) embryonic development, organismal survival, cell death, and survival; (ii) organismal development, cancer, organismal injury, and abnormalities; (iii) cell morphology, cellular development, cellular growth, and proliferation. For each of these networks, we noted the following main “regulatory hubs” (genes predicted to regulate multiple DEGs): NF-kb (network 1), ERK and ERK1 (network 2), Akt (network 3) (Supplemental Fig. S2). The one network identified for DEGs with lower expression in tumors among CR patients versus tumors of NED patients with a score >40 was cellular assembly and organization, cellular compromise, cellular movement. The three networks identified for DEGs with higher expression in the same comparison include: (i) cancer, organismal injury, and abnormalities, reproductive system disease; (ii) molecular transport, nucleic acid metabolism, small molecular biochemistry; (iii) digestive system development and function, organ morphology, organismal development (Supplemental Fig. S2).
Gene Expression Signature Predictive of Aggressive PCa
Using stability selection with elastic net regression at different thresholds (20–80%), we identified eight different models. The model with 28 genes identified at 50% frequency threshold showed the highest AUC after repeated cross-validation and therefore was selected as the most predictive model (Table III). AUCs for the genes obtained at thresholds of 60–80% while including all clinical variables ranged from 0.80 to 0.92. After repeated cross-validation, a model with only clinical variables (Gleason score, PSA, operation year, and age at diagnosis) predicted CR with an AUC of 0.60 whereas the same model with the addition of the 28 genes increased the AUC to 0.97. A heat map of the 28 gene signature contrasting cases and control is shown in Supplemental Figure S3.
TABLE III.
Genes Included in the Prostate Cancer Recurrence Predictive Model
| Rank | Gene symbol | Gene name | Gene function | Cytogenetic band | Fold change (CR:NED) | FDR adjusted P-value | Relevance to PCa and other cancers | References |
|---|---|---|---|---|---|---|---|---|
| 4 | ABCC11 | ATP-binding cassette transporter, sub-family C, member 11 | Transporter, drug resistance | 16q12.1 | 2.387 | 0.0311 | Breast and liver | [95,96] |
| 13 | ABLIM1 | Actin binding LIM protein 1 | Cytoskeleton, cell adhesion | 10q25 | −2.576 | 0.0248 | [97] | |
| 3 | ADRA2C | Alpha-2-adrenergic receptor | Adrenergic receptor, neurotransmitter release | 4p16.3 | −2.503 | 0.0928 | Breast and colorectal | [98,99] |
| 8 | CLEC4F | C-type lectin domain family 4, member F | Glycolipids presentation (Kupffer cells) | 2p13.3 | 2.962 | 0.0441 | ||
| 6 | CPVL | Carboxypeptidase, vitellogenic-like | Post-translational protein modification | 7p15.1 | 2.213 | 0.2197 | [100] | |
| 26 | DUOX1 | Dual oxidase 1 | Hydrogen peroxide producer, cell migration, antimicrobial defense | 15q15.3 | 3.143 | 0.0086 | [59] | |
| 24 | DUOXA1 | Dual oxidase maturation factor 1 | Hydrogen peroxide producer, DU0X1 activator, cell adhesion | 15q21.1 | 2.511 | 0.0423 | [101] | |
| 14 | EDARADD | EDAR-associated death domain | Death domain containing protein | 1q24.3 | 3.013 | 0.0136 | ||
| 17 | F10 | Coagulation factor X | Coagulation factor | 13q34 | −2.099 | 0.2283 | ||
| 19 | GLDC | Glycine dehydrogenase | Metabolism (glycine), mitochondrial localization | 9p22 | 3.147 | 0.0430 | Lung | [66] |
| 15 | GPR81 | G protein-coupled receptor-81 | Lactate receptor, g-protein | 12q24.31 | −2.851 | 0.0193 | Multiple | [102] |
| 18 | KCNA3 | Potassium voltage-gated channel, shaker-related subfamily, member 3 | Potassium channel | 1p13.3 | 2.375 | 0.0774 | Prostate | [103] |
| 20 | KCNQ2 | Potassium voltage-gated channel, KQT-like subfamily, member 2 | Potassium channel | 20q13.3 | −2.481 | 0.1202 | ||
| 25 | LDAH | Chromosome 2 open reading frame 43 | Metabolism (cholesterol) | 2p24.1 | −2.623 | 0.0338 | Prostate | [53,54] |
| 23 | MB | Myoglobin | Oxygen storage and diffusion | 22q13.1 | −2.15 | 0.1821 | Breast | [104] |
| 5 | MMP11 | Matrix metalloproteinase-11 | Extracellular matrix breakdown | 22q11.23 | 3.422 | 0.0006 | Prostate and breast | [105–107] |
| 16 | MYBPC1 | Myosin binding protein C | Myosin binding, muscle | 12q23.2 | −2.754 | 0.0020 | ||
| 1 | NKX2–1 | NK2 homeobox 1 | Thyroid function, morphogenesis | 14q13 | 4.279 | 0.0248 | Endometrial | [108] |
| 28 | NPR3 | Natriuretic peptide receptor C/guanylate cyclase C | Natriuretic receptor | 5p14-p13 | −2.533 | 0.1410 | Medulloblastoma | [109] |
| 9 | OAS2 | 2–5-oligoadenylate synthetase 2 | viral infection response | 12q24.2 | −2.369 | 0.0971 | Prostate | [110] |
| 27 | PCA3 | Prostate cancer antigen 3 | LncRNA | 9q21.2 | −2.019 | 0.2026 | Prostate | [82] |
| 12 | PCBP3 | Poly(rc) binding protein 3 | Post-transcriptional activator | 21q22.3 | 2.093 | 0.1432 | ||
| 10 | PGC | Progastricsin | Digestive enzyme | 6p21.1 | −3.937 | 0.0181 | Prostate, gastric | [111,112] |
| 21 | RAPGEF1 | Rap guanine nucleotide exchange factor (GEF) 1 | Apoptosis, cell transformation | 9q34.3 | 2.249 | 0.1536 | GI and ovarian | [113,114] |
| 22 | TUBB2B | Tubulin, beta 2B class IIb | Microtubules | 6p25 | 2.181 | 0.1354 | ||
| 2 | UPK1A | Uroplakin 1A | Growth, motility | 19q13.3 | 2.58 | 0.0928 | Gastric, CRC, esophageal, bladder | [115–117] |
| 11 | UPK3B | Uroplakin 3B | Apical plaques in urothelium | 7q11.2 | 2.245 | 0.0811 | ||
| 7 | ZYG11A | Zyg-11 family member A, cell cycle regulator | Cell cycle | 1p32.3 | 4.314 | 0.0002 |
DEG, differentially expressed gene; CR, clinical recurrence patients; NED, no evidence of disease patients; FDR, false discovery rate; GI, gastrointestinal; CRC, colorectal cancer.
In Silico Validation of the 28-Gene Signature
To further assess the predictive ability of the 28-gene signature, we identified three external datasets with whole genome data that included appropriate outcomes (MC, MSKCC, and EMC), which we used for in silico validation. Across all datasets, the 28-gene signature improved upon the predictive ability (AUC) of the available clinical variables alone. Specifically, using the MC dataset, the 28-gene model with Gleason score yielded an AUC = 0.75, a 3% increase above AUC = 0.72 in the model with only Gleason score (no other clinical variables were available). Using the MSKCC expression data, the 28-gene model with clinical variables without any missing data (age at diagnosis, race/ethnicity, neo-adjuvant, and adjuvant treatment status) obtained an AUC = 0.90, a 4% improvement over clinical variables alone with AUC = 0.86. With the EMC dataset, the 28-gene model with the available clinical variables (Gleason score and pathologic T stage) yielded an AUC = 0.82, a 6% improvement over clinical variables only with an AUC = 0.76 (Table IV).
TABLE IV.
In Silico Validation of 28-Gene Signature Using Three External Datasets
| MC | MSKCC | EMC | |
|---|---|---|---|
| Tissue used for gene expression and clinical outcomes | 333 PM (NED + BCR) versus 212 PM (CR) | 131 PM versus 19 tissues from MET lesions | 39 PM (non-CR) versus 9 PM (CR) |
| AUC (95%CI) | AUC (95%CI) | AUC (95%CI) | |
| USC 28 gene model + clinical variables | 0.75 (0.72–0.77) | 0.90 (0.86–0.94) | 0.82 (0.74–0.91) |
| Clinical variables only* | 0.72 (0.70–0.74) | 0.86 (0.82–0.91) | 0.76 (0.67–0.85) |
PM, primary tumors; NED, no evidence of disease (no recurrence patients); CR, clinical recurrence; MET, metastasis tissue; MC, Mayo Clinic; MSKCC, Memorial Sloan-Kettering Cancer Center; EMC, Erasmus Medical Center; AUC, area under the curve; CI, confidence interval.
Clinical variables in model: MC—Gleason score only; MSKCC—age at diagnosis, race/ethnicity, neo-adjuvant treatment, and adjuvant treatment for all patients (no missing data); EMC—pathologic stage and Gleason score (no missing data).
DISCUSSION
In this study, we report the identification of a 28-gene expression-based signature that improves the predictive ability of clinical recurrence upon a model with clinical variables alone, in both the original dataset and in three independent datasets. This signature was identified using whole genome gene expression of 187 resected tumors from radical prostatectomy patients diagnosed with organ-confined prostate cancer (stage pT2N0M0) with extensive follow-up. Our dataset included 154 prostate tumors from patients with no evidence of disease (NED) after surgery and 33 tumors from patients who experienced clinical recurrence (CR) after surgery. The results from this study provide novel data that with further validation may contribute to more accurate assessment of prognosis and thus aid in appropriate treatment decisions among patients diagnosed with early stage organ-confined localized disease.
When comparing whole genome expression profiles of tumors of CR patients to NED patients, we identified 172 statistically significant differentially expressed genes (DEGs). Among these DEGs, we observed enrichment in several key pathways defined by molecular function, biological function, and/or disease association. These enriched pathways include various metabolic pathways, including methionine and glucose metabolism, several localizing to the mitochondria, cell proliferation, cell motility and migration, and membrane trafficking. Altogether, these enriched pathways included 33 of the 172 DEGs. Among these 33 DEGs, there were eight genes previously reported to be relevant for prostate carcinogenesis: ANO7, ADI1, BMPR2, CTBP2, DCXR, LDAH, NPR1, and REPS2. ADI1 and ANO7 are two androgen-responsive genes reported to play a suppressor role in PCA progression and tumor invasion [35–37], and to inversely correlate with Gleason grade inversely correlate with Gleason grade [38–40], respectively. REPS2 has also been reported to be involved in PCa cell proliferation and to be down-regulated during PCa progression [41–43]. Consistent with these functions, we found reduced expression of ADI1, ANO7, and REPS2 in CR tumors compared to NED tumors. BMPR2, which we found to be up-regulated in CR patients compared to NED, has been reported to play a role in PCa cell invasion [44]. CTBP2 is reported to have two isoforms with different functions, was found to promote PCa cell proliferation and progression, and to also act as a transcriptional repressor [45–47]. Herein, we found this gene to have reduced expression in CR compared to NED tumors, which is in contrast to what has been reported previously for PCa [45]. DCXR has been reported to be a biomarker of PCa [48] in addition to playing a role in hepatocellular carcinoma [49] and melanocyte lesions [50]. We observed that CR tumors had lower levels of DCXR expression compared to NED. NRP1, which we also found to have higher expression in CR tumors, has been reported to participate in cell migration and survival and in predicting bladder cancer progression and prostate cancer relapse [51,52]. Finally, we observed lower expression of LDAH, a gene reported to associate with PCa risk [53,54].
Based on our GO analysis of the DEGs, there was enrichment for three main molecular function categories: oxidoreductase activity, NADP binding, and growth factor binding with the first two categories being mostly driven by down-regulated genes, and the latter being mostly driven by up-regulated genes in CR compared to NED patients. Among DEGs identified as part of the oxidoreductase activity, two were previously discussed genes, CTBP2 and DCXR, as well as four additional genes involved in several metabolic reactions, CYB5A (ferrous hemoglobin metabolism), GRHPR (pyruvate metabolism), MAT2B (S-adenosyl methionine biosynthesis), and ME2 (malic acid metabolism). CYP5A, GRHPR, and ME2 expression have been reported to be de-regulated in pancreatic cancer, liver cancer, and melanoma, respectively [55–57]. MAT2B participates in the recruitment of MEK and ERK during tumorigenesis in several cancers [58]; consistent with this function we observed overexpression of this gene in CR tumors compared to NED tumors.
Among genes enriched in the binding molecular function category were the previously discussed GRHPR as well as DUOX1, DECR1, and FMO5. DUOX1 is a hydrogen peroxide producer and participates in cell migration and antimicrobial defense [59]. DECR1 and FMO5 are related to mitochondrial function, the former participates in fatty acid metabolism in the mitochondria and the second one was reported to associate with mitochondrial DNA (mtDNA) levels [44]. There were five genes enriched in the growth factor binding GO category: COL1A2, ESM1, RHBDF2 and the two previously discussed BMPR2 and NRP1 genes. COL1A2 encodes for a type I collagen chain reported to associate with proliferation and migration of breast cancer cells [60]; consistent with this role, we observed this gene had higher expression in CR compared to NED tumors. ESM1 is an endothelial-specific factor reported to be a possible biomarker for gastric and breast cancer and associated with breast cancer invasiveness [61,62]. RHBDF2 is a protease reported to associate with ovarian cancer progression [63].
KEGG pathway enrichment analyses identified metabolism, pathways in cancer, and regulation of actin and cytoskeleton as being enriched for DEGS. Among DEGs in the metabolic pathways, there were four genes previously discussed (ADI1, CCXR, GRHPR, MAT2B) and eight additional genes. Among them, two participate in metabolic reactions that take place in the mitochondria: ACAFSB (fatty acid metabolism), GLDC (glycine metabolism); and five others participate in various metabolic pathways: AGPAT2 (phospholipid synthesis), ALDH1A2 (retinoic acid synthesis), CHPT1 (phosphatidylcholine metabolism), NADSYN1 (NAD synthesis), and SRM (spermidine synthesis). Finally, we identified SYNJ1, which is involved in membrane trafficking. Three of these 12 genes have been previously reported to be associated with tumorigenesis of osteosarcoma (AGPAT2) [64], breast cancer (CHPT1) [65], and lung cancer (GLDC) [66]. Within this enriched KEGG category, we highlight the cysteine and methionine metabolism pathway, involving MAT2B, SRM, and ADI1, which have been shown to associate with cancer development [67,68].
Among genes identified as part of the KEGG Pathways in Cancer was CTBP2, previously discussed, CDKN2B, involved in cell proliferation and reported to associate with several cancers [66], LAMA3, which participates in base membrane formation and cell migration and was reported to associate with gastric cancer [66], and PIAS4, which participates in autophagy and was reported to associate with pancreas cancer [66]. Among genes enriched in the KEGG actin/cytoskeleton regulation pathway were two genes that participate in cell migration: MYH9 and TMSBX4 both previously reported to associate with head and neck cancer [69] and several cancer types [70–72], respectively. In addition, this pathway was enriched by one G-protein gene (ARHGEF6) and a gene that participates in cell mobility and angiogenesis (BAIAP2).
The networks generated from the pathway analysis of the DEGs identified several central genes that act as “hubs” in these pathways and have been reported to be involved in cancer development and progression. Among them were NF-κβ, VEGF, ERK1 (MAPK3), and Tgfβ, Akt (protein kinase B), PI3 K, Ras, and EGFR. NF-κβ, EGFR has been reported to be involved in several cancers, including prostate cancer where its increased signaling has been reported to be involved in stem-like human prostate tumor-initiating cells and progression of disease [73–75]. De-regulation of VEGF, TGFβ, EGFR, and PI3 K/Akt have been reported as important steps in cancer invasion and metastasis, including prostate cancer [76–78]. Alterations of the Ras oncogene and MAPK3 have been reported to play an important role in the progression of prostate cancer cells to androgen resistance [79,80]. Similarly, our miRNA enrichment analyses identified an enrichment of DEGs that are regulated by many miRNAs known to be associated with prostate cancer progression [81].
We validated the predictive performance of our 28-gene model in three separate datasets that included whole-genome expression profiles that were obtained using a different platform than the one used in this study. Using data from all three datasets, the 28-gene model improved upon the prediction of clinical recurrence over models that included clinical variables only, with improvements ranging from 3 to 6%. Of the three datasets, the most comparable to our study design was the one from the Mayo Clinic. The AUC obtained for the 28-gene signature including Gleason score, which was the only clinical variable publicly available in the Mayo Clinic dataset, (AUC = 0.75; 0.72–0.77), was identical to the AUC reported by investigators from the Mayo Clinic and GenomeDx, for their 22-gene signature (AUC = 0.75; 0.68–0.83). We observed a comparable improvement in AUC when comparing the model with only clinical variables to the model that included both clinical and genetic components [14]. A key difference between our model and the GenomeDx one is that ours was identified among purely organ-confined prostate cancer patients with stage T2, although we note that the majority were stage T2C. In contrast, this Mayo clinic dataset included patients with stages higher than stage II, had a dissimilar distribution of pathological stage between cases and controls, and included lymph node positive patients.
Our study has several strengths. One, the use of microdissected tissue to enrich for malignant glands representative of the tumor’s Gleason grade, which ensured the obtainment of gene expression profiles not contaminated with non-malignant tissue. Second, we used tumors from a well-annotated cohort with extensive and active follow-up. Third, we focused our analyses to organ-confined cancer patients of stage pT2, which allowed for comparisons between cases and controls without biases introduced by differences in stage distribution, and the identification of gene profiles representative of very early PCa. Finally, we used rigorous statistical methods to minimize variability in order to capture the most predictive genes of metastatic disease. Among the limitations of this study is the possibility that tumor heterogeneity within the prostate may not have allowed for proper sampling of the foci most representative of the tumor’s potential to progress. This fact would introduce misclassification in our sample, and given that this misclassification would be non-differential with respect to case or control status (as both are equally likely to show heterogeneity), it could bias our findings toward the null. Therefore, we are more likely to have missed important associations between expression profiles and clinical recurrence rather that reporting inflated associations. We also acknowledge the relatively modest number of metastatic cases (n = 33). In spite of the large size of our cohort, few stage II patients experience a clinical recurrence. We sought to address this by validating our model in a dataset with higher numbers of clinical metastatic patients and obtained promising results that show an improvement of prediction of our model compared to clinical variables only. Lastly, for the original discovery of the 28-gene signature, given the modest number of patients with recurrence, we used cross-validation to estimate the AUC. Given that we used the same dataset used for discovery, our estimated AUC is likely an overestimate due to overfitting. However, the external validation datasets confirm that a model with clinical variables and the 28-gene signature is more predictive than clinical variables alone. Therefore, additional validation studies will be needed to confirm our findings and to obtain more accurate estimates of predictive accuracy. Moreover, future validation studies are needed to determine the utility of this signature to predict adverse pathology and risk of cancer progression at the time of initial biopsy.
CONCLUSIONS
In summary, we report a 28-gene model that used in conjunction with clinical variables improves the prediction of clinical recurrence among early stage localized PCa. This model, once validated in additional external databases, may aid clinicians in identifying patients with early localized disease at high risk of recurrence who may benefit from more aggressive treatments at the time of radical prostatectomy.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Derek Gruter for his assistance with laser capture microdissection of the prostatectomy specimens, Mo-li Chen and Alex Trena for their help with providing the prostatectomy sections for microdissection, and Tracy Campanelli and Jie Cai for assistance with clinical variables.
Grant sponsor: National Institute of Health (NIH); Grant number: TL1RR031992; Grant sponsor: Mirage Foundation; Grant sponsor: National Cancer Institute; Grant number: P30CA014089.
Jian-Bing Fan and Mariana C. Stern co-directed this work.
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of this article at the publisher’sweb-site.
REFERENCES
- 1.Cooperberg MR, Broering JM, Kantoff PW, Carroll PR. Contemporary trends in low risk prostate cancer: Risk assessment and treatment. J Urol 2007;178(3 Pt 2):S14–S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loeb S, Feng Z, Ross A, Trock BJ, Humphreys EB, Walsh PC. Can we stop prostate specific antigen testing 10 years after radical prostatectomy? J Urol 2011;186(2):500–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pound CR, Partin AW, Eisenberger MA, Chan DW, Pearson JD, Walsh PC. Natural history of progression after PSA elevation following radical prostatectomy. JAMA 1999;281(17):1591–1597. [DOI] [PubMed] [Google Scholar]
- 4.D’Amico AV, Whittington R, Malkowicz SB, Schultz D, Blank K, Broderick GA, Tomaszewski JE, Renshaw AA, Kaplan I, Beard CJ, Wein A. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. JAMA 1998;280(11):969–974. [DOI] [PubMed] [Google Scholar]
- 5.Cooperberg MR, Pasta DJ, Elkin EP, Litwin MS, Latini DM, Du Chane J, Carroll PR. The University of California, San Francisco Cancer of the Prostate Risk Assessment score: A straightforward and reliable preoperative predictor of disease recurrence after radical prostatectomy. J Urol 2005;173(6): 1938–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kattan MW, Eastham JA, Stapleton AM, Wheeler TM, Scardino PT. A preoperative nomogram for disease recurrence following radical prostatectomy for prostate cancer. J Natl Cancer Inst 1998;90(10):766–771. [DOI] [PubMed] [Google Scholar]
- 7.Partin AW, Mangold LA, Lamm DM, Walsh PC, Epstein JI, Pearson JD. Contemporary update of prostate cancer staging nomograms (Partin Tables) for the new millennium. Urology 2001;58(6):843–848. [DOI] [PubMed] [Google Scholar]
- 8.Donovan MJ, Hamann S, Clayton M, Khan FM, Sapir M, Bayer-Zubek V, Fernandez G, Mesa-Tejada R, Teverovskiy M, Reuter VE, Scardino PT, Cordon-Cardo C. Systems pathology approach for the prediction of prostate cancer progression after radical prostatectomy. J Clin Oncol 2008;26(24):3923–3929. [DOI] [PubMed] [Google Scholar]
- 9.Glinsky GV, Glinskii AB, Stephenson AJ, Hoffman RM, Gerald WL. Gene expression profiling predicts clinical outcome of prostate cancer. J Clin Invest 2004;113(6):913–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Talantov D, Jatkoe TA, Bohm M, Zhang Y, Ferguson AM, Stricker PD, Kattan MW, Sutherland RL, Kench JG, Wang Y, Henshall SM. Gene based prediction of clinically localized prostate cancer progression after radical prostatectomy. J Urol 2010;184(4):1521–1528. [DOI] [PubMed] [Google Scholar]
- 11.Stephenson AJ, Smith A, Kattan MW, Satagopan J, Reuter VE, Scardino PT, Gerald WL. Integration of gene expression profiling and clinical variables to predict prostate carcinoma recurrence after radical prostatectomy. Cancer 2005;104(2):290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheville JC, Karnes RJ, Therneau TM, Kosari F, Munz JM, Tillmans L, Basal E, Rangel LJ, Bergstralh E, Kovtun IV, Savci-Heijink CD, Klee EW, Vasmatzis G. Gene panel model predictive of outcome in men at high-risk of systemic progression and death from prostate cancer after radical retropubic prostatectomy. J Clin Oncol 2008;26(24):3930–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuzick J, Swanson GP, Fisher G, Brothman AR, Berney DM, Reid JE, Mesher D, Speights VO, Stankiewicz E, Foster CS, Møller H, Scardino P, Warren JD, Park J, Younus A, Flake DD, Wagner S, Gutin A, Lanchbury JS, Stone S. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: A retrospective study. Lancet Oncol 2011;12(3):245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erho N, Crisan A, Vergara IA, Mitra AP, Ghadessi M, Buerki C, Bergstralh EJ, Kollmeyer T, Fink S, Haddad Z, Zimmermann B, Sierocinski T, Ballman KV, Triche TJ, Black PC, Karnes RJ, Klee G, Davicioni E, Jenkins RB. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS ONE 2015;8(6):e66855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakagawa T, Kollmeyer TM, Morlan BW, Anderson SK, Bergstralh EJ, Davis BJ, Asmann YW, Klee GG, Ballman KV, Jenkins RB. A tissue biomarker panel predicting systemic progression after PSA recurrence post-definitive prostate cancer therapy. PLoS ONE 2008;3(5):e2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Penney KL, Sinnott JA, Fall K, Pawitan Y, Hoshida Y, Kraft P, Stark JR, Fiorentino M, Perner S, Finn S, Calza S, Flavin R, Freedman ML, Setlur S, Sesso HD, Andersson SO, Martin N, Kantoff PW, Johansson JE, Adami HO, Rubin MA, Loda M, Golub TR, Andren O, Stampfer MJ, Mucci LA. MRNA expression signature of Gleason grade predicts lethal prostate cancer. J Clin Oncol 2011;29(17):2391–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cullen J, Rosner IL, Brand TC, Zhang N, Tsiatis AC, Moncur J, Ali A, Chen Y, Knezevic D, Maddala T, Lawrence HJ, Febbo PG, Srivastava S, Sesterhenn IA, McLeod DG. A biopsy-based 17-gene genomic prostate score predicts recurrence after radical prostatectomy and adverse surgical pathology in a racially diverse population of men with clinically low- and intermediate-risk prostate cancer. Eur Urol 2015;68(1):123–131. [DOI] [PubMed] [Google Scholar]
- 18.Kosari F, Munz JM, Savci-Heijink CD, Spiro C, Klee EW, Kube DM, Tillmans L, Slezak J, Karnes RJ, Cheville JC, Vasmatzis G. Identification of prognostic biomarkers for prostate cancer. Clin Cancer Res 2008;14(6):1734–1743. [DOI] [PubMed] [Google Scholar]
- 19.Sboner A, Demichelis F, Calza S, Pawitan Y, Setlur SR, Hoshida Y, Perner S, Adami HO, Fall K, Mucci LA, Kantoff PW, Stampfer M, Andersson SO, Varenhorst E, Johansson JE, Gerstein MB, Golub TR, Rubin MA, Andren O. Molecular sampling of prostate cancer: A dilemma for predicting disease progression. BMC Med Genomics 2010;3:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, McDonald C, Thomas R, Dhir R, Finkelstein S, Michalopoulos G, Becich M, Luo JH. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol 2004;22(14):2790–2799. [DOI] [PubMed] [Google Scholar]
- 21.Dorin RP, Lieskovsky G, Fairey AS, Cai J, Daneshmand S. Outcomes after radical prostatectomy for patients with clinical stages T1-T2 prostate cancer with pathologically positive lymph nodes in the prostate-specific antigen era. Urologic Oncol 2013;31(8):1441–1447. [DOI] [PubMed] [Google Scholar]
- 22.April C, Klotzle B, Royce T, Wickham-Garcia E, Boyaniwsky T, Izzo J, Cox D, Jones W, Rubio R, Holton K, Matulonis U, Quackenbush J, Fan JB. Whole-genome gene expression profiling of formalin-fixed, paraffin-embedded tissue samples. PLoS ONE 2009;4(12):e8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol 2004;5(10):R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi W, Oshlack A, Smyth GK. Optimizing the noise versus bias trade-off for Illumina whole genome expression Bead-Chips. Nucleic Acids Res 2015;38(22):e204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen C, Grennan K, Badner J, Zhang D, Gershon E, Jin L, Liu C. Removing batch effects in analysis of expression microarray data: An evaluation of six batch adjustment methods. PLoS ONE 2015;6(2):e17238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004;3(Article3) Epub 2004 Feb 12. [DOI] [PubMed] [Google Scholar]
- 27.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodol). 1995;57(1):289–300. [Google Scholar]
- 28.Kirov S, Ji R, Wang J, Zhang B. Functional annotation of differentially regulated gene set using WebGestalt: A gene set predictive of response to ipilimumab in tumor biopsies. Methods Mol Biol 2014;1101:31–42. [DOI] [PubMed] [Google Scholar]
- 29.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis toolkit (WebGestalt): Update. Nucleic Acids Res 2013;41(Web Server issue):W77–W83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang B, Kirov S, Snoddy J. WebGestalt: An integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res 2005;33(Web Server issue):W741–W748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meinshausen N, Buehlmann P. Stability selection. J R Stat Soc Ser B-Stat Method 2010;72:417–473. [Google Scholar]
- 32.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, Antipin Y, Mitsiades N, Landers T, Dolgalev I, Major JE, Wilson M, Socci ND, Lash AE, Heguy A, Eastham JA, Scher HI, Reuter VE, Scardino PT, Sander C, Sawyers CL, Gerald WL. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010;18(1):11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boormans JL, Korsten H, Ziel-van der Made AJ, van Leenders GJ, de Vos CV, Jenster G, Trapman J. Identification of TDRD1 as a direct target gene of ERG in primary prostate cancer. Int J Cancer 2013;133(2):335–345. [DOI] [PubMed] [Google Scholar]
- 34.Robinson MD, Speed TP. A comparison of Affymetrix gene expression arrays. BMC Bioinf 2007;8:449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oram SW, Ai J, Pagani GM, Hitchens MR, Stern JA, Eggener S, Pins M, Xiao W, Cai X, Haleem R, Jiang F, Pochapsky TC, Hedstrom L, Wang Z. Expression and function of the human androgen-responsive gene ADI1 in prostate cancer. Neoplasia 2007;9(8):643–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oram S, Jiang F, Cai X, Haleem R, Dincer Z, Wang Z. Identification and characterization of an androgen-responsive gene encoding an aci-reductone dioxygenase-like protein in the rat prostate. Endocrinology 2004;145(4):1933–1942. [DOI] [PubMed] [Google Scholar]
- 37.Uekita T, Gotoh I, Kinoshita T, Itoh Y, Sato H, Shiomi T, Okada Y, Seiki M. Membrane-type 1 matrix metalloproteinase cytoplasmic tail-binding protein-1 is a new member of the Cupin superfamily. A possible multifunctional protein acting as an invasion suppressor down-regulated in tumors. J Biol Chem 2004;279(13):12734–12743. [DOI] [PubMed] [Google Scholar]
- 38.Das S, Hahn Y, Walker DA, Nagata S, Willingham MC, Peehl DM, Bera TK, Lee B, Pastan I. Topology of NGEP, a prostate-specific cell:cell junction protein widely expressed in many cancers of different grade level. Cancer Res 2015;68(15):6306–6312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohsenzadegan M, Madjd Z, Asgari M, Abolhasani M, Shekarabi M, Taeb J, Shariftabrizi A. Reduced expression of NGEP is associated with high-grade prostate cancers: A tissue microarray analysis. Cancer Immunol Immunother 2013;62(10):1609–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bera TK, Das S, Maeda H, Beers R, Wolfgang CD, Kumar V, Hahn Y, Lee B, Pastan I. NGEP a gene encoding a membrane protein detected only in prostate cancer and normal prostate. Proc Natl Acad Sci USA 2004;101(9): 3059–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oosterhoff JK, Penninkhof F, Brinkmann AO, Anton Grootegoed J, Blok LJ. REPS2/POB1 is downregulated during human prostate cancer progression and inhibits growth factor signalling in prostate cancer cells. Oncogene 2003;22(19):2920–2925. [DOI] [PubMed] [Google Scholar]
- 42.Oosterhoff JK, Kuhne LC, Grootegoed JA, Blok LJ. EGF signalling in prostate cancer cell lines is inhibited by a high expression level of the endocytosis protein REPS2. Int J Cancer J Int du Cancer 2015;113(4):561–567. [DOI] [PubMed] [Google Scholar]
- 43.Penninkhof F, Grootegoed JA, Blok LJ. Identification of REPS2 as a putative modulator of NF-kappaB activity in prostate cancer cells. Oncogene 2004;23(33):5607–5615. [DOI] [PubMed] [Google Scholar]
- 44.Lopez S, Buil A, Souto JC, Casademont J, Martinez-Perez A, Almasy L, Soria JM. A genome-wide association study in the genetic analysis of idiopathic thrombophilia project suggests sex-specific regulation of mitochondrial DNA levels. Mitochondrion 2014;18:34–40. [DOI] [PubMed] [Google Scholar]
- 45.Debiais-Delpech C, Godet J, Pedretti N, Bernard FX, Irani J, Cathelineau X, Cussenot O, Fromont G. Expression patterns of candidate susceptibility genes HNF1beta and CtBP2 in prostate cancer: Association with tumor progression. Urologic Oncol 2015;32(4):426–432. [DOI] [PubMed] [Google Scholar]
- 46.Takayama K, Suzuki T, Fujimura T, Urano T, Takahashi S, Homma Y, Inoue S. CtBP2 modulates the androgen receptor to promote prostate cancer progression. Cancer Res 2014;74(22):6542–6553. [DOI] [PubMed] [Google Scholar]
- 47.Zhang C, Gao C, Xu Y, Zhang Z. CtBP2 could promote prostate cancer cell proliferation through c-Myc signaling. Gene 2014;546(1):73–79. [DOI] [PubMed] [Google Scholar]
- 48.Cho-Vega JH, Tsavachidis S, Do KA, Nakagawa J, Medeiros LJ, McDonnell TJ. Dicarbonyl/L-xylulose reductase: A potential biomarker identified by laser-capture microdissection-micro serial analysis of gene expression of human prostate adenocarcinoma. Cancer Epidemiol Biomarkers Prev 2007;16(12):2615–2622. [DOI] [PubMed] [Google Scholar]
- 49.Liu S, Ma L, Huang W, Shai Y, Ji X, Ding L, Liu Y, Yu L, Zhao S. Decreased expression of the human carbonyl reductase 2 gene HCR2 in hepatocellular carcinoma. Cell Mol Biol Lett 2006;11(2):230–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cho-Vega JH, Vega F, Schwartz MR, Prieto VG. Expression of dicarbonyl/L-xylulose reductase (DCXR) in human skin and melanocytic lesions: Morphological studies supporting cell adhesion function of DCXR. J Cutan Pathol 2007;34(7):535–542. [DOI] [PubMed] [Google Scholar]
- 51.Cheng W, Fu D, Wei ZF, Xu F, Xu XF, Liu YH, Ge JP, Tian F, Han CH, Zhang ZY, Zhou LM. NRP-1 expression in bladder cancer and its implications for tumor progression. Tumour Biol 2014;35(6):6089–6094. [DOI] [PubMed] [Google Scholar]
- 52.Talagas M, Uguen A, Garlantezec R, Fournier G, Doucet L, Gobin E, Marcorelles P, Volant A, DE Braekeleer M. VEGFR1 and NRP1 endothelial expressions predict distant relapse after radical prostatectomy in clinically localized prostate cancer. Anticancer Res 2013;33(5):2065–2075. [PubMed] [Google Scholar]
- 53.Takata R, Akamatsu S, Kubo M, Takahashi A, Hosono N, Kawaguchi T, Tsunoda T, Inazawa J, Kamatani N, Ogawa O, Fujioka T, Nakamura Y, Nakagawa H. Genome-wide association study identifies five new susceptibility loci for prostate cancer in the Japanese population. Nature Genet 2010;42(9):751–754. [DOI] [PubMed] [Google Scholar]
- 54.Long QZ, Du YF, Ding XY, Li X, Song WB, Yang Y, Zhang P, Zhou JP, Liu XG. Replication and fine mapping for association of the C2orf43, FOXP4, GPRC6A and RFX6 genes with prostate cancer in the Chinese population. PLoS ONE 2015;7(5):e37866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giovannetti E, Wang Q, Avan A, Funel N, Lagerweij T, Lee JH, Caretti V, van der Velde A, Boggi U, Wang Y, Vasile E, Peters GJ, Wurdinger T, Giaccone G. Role of CYB5A in pancreatic cancer prognosis and autophagy modulation. J Natl Cancer Inst 2014;106(1):djt346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pan Y, Ni R, Deng Q, Huang X, Zhang Y, Lu C, Li F, Huang D, He S, Chen B. Glyoxylate reductase/hydroxypyruvate reductase: A novel prognostic marker for hepatocellular carcinoma patients after curative resection. Pathobiology 2013;80(3):155–162. [DOI] [PubMed] [Google Scholar]
- 57.Chang YL, Gao HW, Chiang CP, Wang WM, Huang SM, Ku CF, Liu GY, Hung HC. Human mitochondrial NAD(P) (+)-dependent malic enzyme participates in cutaneous melanoma progression and invasion. J Invest Dermatol 2015;135(3):807–815. [DOI] [PubMed] [Google Scholar]
- 58.Peng H, Dara L, Li TW, Zheng Y, Yang H, Tomasi ML, Tomasi I, Giordano P, Mato JM, Lu SC. MAT2B-GIT1 interplay activates MEK1/ERK 1 and 2 to induce growth in human liver and colon cancer. Hepatology 2013;57(6):2299–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hristova M, Veith C, Habibovic A, Lam YW, Deng B, Geiszt M, Janssen-Heininger YM, van der Vliet A. Identification of DUOX1-dependent redox signaling through protein S-glutathionylation in airway epithelial cells. Redox Biol 2014;2:436–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mori K, Enokida H, Kagara I, Kawakami K, Chiyomaru T, Tatarano S, Kawahara K, Nishiyama K, Seki N, Nakagawa M. CpG hypermethylation of collagen type I alpha 2 contributes to proliferation and migration activity of human bladder cancer. Int J Oncol 2009;34(6):1593–1602. [DOI] [PubMed] [Google Scholar]
- 61.Lv Z, Fan Y, Chen H, Zhao D. Endothelial cell-specific molecule-1: a potential serum marker for gastric cancer. Tumour Biol 2014;35(10):10497–10502. [DOI] [PubMed] [Google Scholar]
- 62.Rhee JJ, Sampson L, Cho E, Hughes MD, Hu FB, Willett WC. Comparison of methods to account for implausible reporting of energy intake in epidemiologic studies. Am J Epidemiol 2015;181(4):225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wojnarowicz PM, Provencher DM, Mes-Masson AM, Tonin PN. Chromosome 17q25 genes, RHBDF2 and CYGB, in ovarian cancer. Int J Oncol 2012;40(6):1865–1880. [DOI] [PubMed] [Google Scholar]
- 64.Rastegar F, Gao JL, Shenaq D, Luo Q, Shi Q, Kim SH, Jiang W, Wagner ER, Huang E, Gao Y, Shen J, Yang K, He BC, Chen L, Zuo GW, Luo J, Luo X, Bi Y, Liu X, Li M, Hu N, Wang L, Luther G, Luu HH, Haydon RC, He TC. Lysophosphatidic acid acyltransferase beta (LPAATbeta) promotes the tumor growth of human osteosarcoma. PLoS ONE 2015;5(12):e14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ghosh A, Akech J, Mukherjee S, Das SK. Differential expression of cholinephosphotransferase in normal and cancerous human mammary epithelial cells. Biochem Biophys Res Commun 2002;297(4):1043–1048. [DOI] [PubMed] [Google Scholar]
- 66.Zhang WC, Shyh-Chang N, Yang H, Rai A, Umashankar S, Ma S, Soh BS, Sun LL, Tai BC, Nga ME, Bhakoo KK, Jayapal SR, Nichane M, Yu Q, Ahmed DA, Tan C, Sing WP, Tam J, Thirugananam A, Noghabi MS, Pang YH, Ang HS, Mitchell W, Robson P, Kaldis P, Soo RA, Swarup S, Lim EH, Lim B. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell 2012;148(1–2):259–272. [DOI] [PubMed] [Google Scholar]
- 67.Stern PH, Wallace CD, Hoffman RM. Altered methionine metabolism occurs in all members of a set of diverse human tumor cell lines. J Cell Physiol 1984;119(1):29–34. [DOI] [PubMed] [Google Scholar]
- 68.Townsend DM, Tew KD, Tapiero H. The importance of glutathione in human disease. Biomed Pharmacother 2003;57(3–4):145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.MYH9 regulates p53 stability and is a tumor suppressor in SCC. Cancer Discovery 2014;4(3):OF22. [DOI] [PubMed] [Google Scholar]
- 70.Wirsching HG, Krishnan S, Florea AM, Frei K, Krayenbuhl N, Hasenbach K, Reifenberger G, Weller M, Tabatabai G. Thymosin beta 4 gene silencing decreases stemness and invasiveness in glioblastoma. Brain 2014;137(Pt 2):433–448. [DOI] [PubMed] [Google Scholar]
- 71.Kang YJ, Jo JO, Ock MS, Chang HK, Lee SH, Ahn BK, Baek KW, Choi YH, Kim WJ, Leem SH, Cha HJ. Thymosin beta4 was upregulated in recurred colorectal cancers. J Clin Pathol 2014;67(2):188–190. [DOI] [PubMed] [Google Scholar]
- 72.Ji YI, Lee BY, Kang YJ, Jo JO, Lee SH, Kim HY, Kim YO, Lee C, Koh SB, Kim A, Lee JY, Jung MH, Ock MS, Cha HJ. Expression patterns of thymosin beta4 and cancer stem cell marker CD133 in ovarian cancers. Pathol Oncol Res: POR 2013;19(2):237–245. [DOI] [PubMed] [Google Scholar]
- 73.Jin R, Yi Y, Yull FE, Blackwell TS, Clark PE, Koyama T, Smith JA Jr., Matusik RJ. NF-kappaB gene signature predicts prostate cancer progression. Cancer Res 2014;74(10):2763–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rajasekhar VK, Studer L, Gerald W, Socci ND, Scher HI. Tumour-initiating stem-like cells in human prostate cancer exhibit increased NF-kappaB signalling. Nat Commun 2011;2:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen CD, Sawyers CL. NF-kappa B activates prostate-specific antigen expression and is upregulated in androgen-independent prostate cancer. Mol Cell Biol 2002;22(8):2862–2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.de Brot S, Ntekim A, Cardenas R, James V, Allegrucci C, Heery DM, Bates DO, Odum N, Persson JL, Mongan NP. Regulation of vascular endothelial growth factor in prostate cancer. Endocr Relat Cancer 2015;22(3):R107–R123. [DOI] [PubMed] [Google Scholar]
- 77.Jiang WG, Sanders AJ, Katoh M, Ungefroren H, Gieseler F, Prince M, Thompson SK, Zollo M, Spano D, Dhawan P, Sliva D, Subbarayan PR, Sarkar M, Honoki K, Fujii H, Georgakilas AG, Amedei A, Niccolai E, Amin A, Ashraf SS, Ye L, Helferich WG, Yang X, Boosani CS, Guha G, Ciriolo MR, Aquilano K, Chen S, Azmi AS, Keith WN, Bilsland A, Bhakta D, Halicka D, Nowsheen S, Pantano F, Santini D. Tissue invasion and metastasis: Molecular, biological and clinical perspectives. Semin Cancer Biol 2015. [DOI] [PubMed] [Google Scholar]
- 78.Mimeault M, Batra SK. Frequent gene products and molecular pathways altered in prostate cancer- and metastasis-initiating cells and their progenies and novel promising multitargeted therapies. Mol Med 2011;17(9–10):949–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Whitaker HC, Neal DE. RAS pathways in prostate cancer— mediators of hormone resistance?. Curr Cancer Drug Targets 2010;10(8):834–839. [DOI] [PubMed] [Google Scholar]
- 80.Edwards J, Bartlett JM. The androgen receptor and signal-transduction pathways in hormone-refractory prostate cancer. Part 2: Androgen-receptor cofactors and bypass pathways. BJU Int 2005;95(9):1327–1335. [DOI] [PubMed] [Google Scholar]
- 81.Jackson BL, Grabowska A, Ratan HL. MicroRNA in prostate cancer: Functional importance and potential as circulating biomarkers. BMC Cancer 2014;14:930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Luo Y, Gou X, Huang P, Mou C. The PCA3 test for guiding repeat biopsy of prostate cancer and its cut-off score: A systematic review and meta-analysis. Asian J Androl 2014;16(3):487–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Amory JK, Arnold S, Lardone MC, Piottante A, Ebensperger M, Isoherranen N, Muller CH, Walsh T, Castro A. Levels of the retinoic acid synthesizing enzyme aldehyde dehydrogenase-1A2 are lower in testicular tissue from men with infertility. Fertil Steril 2014;101(4):960–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu PS, Jong TH, Maa MC, Leu TH. The interplay between Eps8 and IRSp53 contributes to Src-mediated transformation. Oncogene 2010;29(27):3977–3989. [DOI] [PubMed] [Google Scholar]
- 85.Breen MJ, Moran DM, Liu W, Huang X, Vary CP, Bergan RC. Endoglin-mediated suppression of prostate cancer invasion is regulated by activin and bone morphogenetic protein type II receptors. PLoS ONE 2015;8(8):e72407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lara-Riegos JC, Ortiz-Lopez MG, Pena-Espinoza BI, Montufar-Robles I, Pena-Rico MA, Sanchez-Pozos K, Granados-Silvestre MA, Menjivar M. Diabetes susceptibility in mayas: Evidence for the involvement of polymorphisms in HHEX, HNF4alpha, KCNJ11, PPARgamma, CDKN2A/2B, SLC30A8, CDC123/CAMK1D, TCF7L2, ABCA1 and SLC16A11 genes. Gene 2015;565(1):68–75. [DOI] [PubMed] [Google Scholar]
- 87.Li WQ, Pfeiffer RM, Hyland PL, Shi J, Gu F, Wang Z, Bhattacharjee S, Luo J, Xiong X, Yeager M, Deng X, Hu N, Taylor PR, Albanes D, Caporaso NE, Gapstur SM, Amundadottir L, Chanock SJ, Chatterjee N, Landi MT, Tucker MA, Goldstein AM, Yang XR. Genetic polymorphisms in the 9p21 region associated with risk of multiple cancers. Carcinogenesis 2014;35(12):2698–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Roudnicky F, Poyet C, Wild P, Krampitz S, Negrini F, Huggenberger R, Rogler A, Stohr R, Hartmann A, Provenzano M, Otto VI, Detmar M. Endocan is upregulated on tumor vessels in invasive bladder cancer where it mediates VEGF-A-induced angiogenesis. Cancer Res 2013;73(3):1097–1106. [DOI] [PubMed] [Google Scholar]
- 89.Bizama C, Benavente F, Salvatierra E, Gutierrez-Moraga A, Espinoza JA, Fernandez EA, Roa I, Mazzolini G, Sagredo EA, Gidekel M, Podhajcer OL. The low-abundance transcriptome reveals novel biomarkers, specific intracellular pathways and targetable genes associated with advanced gastric cancer. Int J Cancer 2015;134(4):755–764. [DOI] [PubMed] [Google Scholar]
- 90.Jorde R, Svartberg J, Joakimsen RM, Grimnes G. Associations between polymorphisms related to calcium metabolism and human height: The Tromso Study. Ann Hum Genet 2012;76(3):200–210. [DOI] [PubMed] [Google Scholar]
- 91.Ahn J, Yu K, Stolzenberg-Solomon R, Simon KC, McCullough ML, Gallicchio L, Jacobs EJ, Ascherio A, Helzlsouer K, Jacobs KB, Li Q, Weinstein SJ, Purdue M, Virtamo J, Horst R, Wheeler W, Chanock S, Hunter DJ, Hayes RB, Kraft P, Albanes D. Genome-wide association study of circulating vitamin D levels. Hum Mol Genet 2010;19(13):2739–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chien W, Lee KL, Ding LW, Wuensche P, Kato H, Doan NB, Poellinger L, Said JW, Koeffler HP. PIAS4 is an activator of hypoxia signalling via VHL suppression during growth of pancreatic cancer cells. Br. J Cancer 2013;109(7):1795–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Naidu SR, Lakhter AJ, Androphy EJ. PIASy-mediated Tip60 sumoylation regulates p53-induced autophagy. Cell Cycle 2012;11(14):2717–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Drouet V, Lesage S. Synaptojanin 1 mutation in Parkinson’s disease brings further insight into the neuropathological mechanisms. Biomed Res Int 2014;2014:289728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yamada A, Ishikawa T, Ota I, Kimura M, Shimizu D, Tanabe M, Chishima T, Sasaki T, Ichikawa Y, Morita S, Yoshiura K, Takabe K, Endo I. High expression of ATP-binding cassette transporter ABCC11 in breast tumors is associated with aggressive subtypes and low disease-free survival. Breast Cancer Res Treat 2015;137(3):773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Borel F, Han R, Visser A, Petry H, van Deventer SJ, Jansen PL, Konstantinova P. Adenosine triphosphate-binding cassette transporter genes up-regulation in untreated hepatocellular carcinoma is mediated by cellular microRNAs. Hepatology 2012;55(3):821–832. [DOI] [PubMed] [Google Scholar]
- 97.Chambers KF, Pearson JF, Pellacani D, Aziz N, Guzvic M, Klein CA, Lang SH. Stromal upregulation of lateral epithelial adhesions: Gene expression analysis of signalling pathways in prostate epithelium. J Biomed Sci 2011;18:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Powe DG, Voss MJ, Habashy HO, Zanker KS, Green AR, Ellis IO, Entschladen F. Alpha- and beta-adrenergic receptor (AR) protein expression is associated with poor clinical outcome in breast cancer: An immunohistochemical study. Breast Cancer Res Treat 2011;130(2):457–463. [DOI] [PubMed] [Google Scholar]
- 99.Lee H, Flaherty P, Ji HP. Systematic genomic identification of colorectal cancer genes delineating advanced from early clinical stage and metastasis. BMC Med Genomics 2013;6:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hu C, Zhang R, Yu W Wang J, Wang C, Pang C, Ma X, Bao Y, Xiang K, Jia W. CPVL/CHN2 genetic variant is associated with diabetic retinopathy in Chinese type 2 diabetic patients. Diabetes 2011;60(11):3085–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ostrakhovitch EA, Li SS. NIP1/DUOXA1 expression in epithelial breast cancer cells: Regulation of cell adhesion and actin dynamics. Breast Cancer Res Treat 2010;119(3):773–786. [DOI] [PubMed] [Google Scholar]
- 102.Roland CL, Arumugam T, Deng D, Liu SH, Philip B, Gomez S, Burns WR, Ramachandran V, Wang H, Cruz-Monserrate Z, Logsdon CD. Cell surface lactate receptor GPR81 is crucial for cancer cell survival. Cancer Res 2014;74(18):5301–5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fraser SP, Grimes JA, Diss JK, Stewart D, Dolly JO, Djamgoz MB. Predominant expression of Kv1.3 voltage-gated K+ channel subunit in rat prostate cancer cell lines: Electrophysiological, pharmacological and molecular characterisation. Pflugers Arch 2003;446(5):559–571. [DOI] [PubMed] [Google Scholar]
- 104.Meller S, Bicker A, Montani M, Ikenberg K, Rostamzadeh B, Sailer V, Wild P, Dietrich D, Uhl B, Sulser T, Moch H, Gorr TA, Stephan C, Jung K, Hankeln T, Kristiansen G. Myoglobin expression in prostate cancer is correlated to androgen receptor expression and markers of tumor hypoxia. Virchows Arch 2014;465(4):419–427. [DOI] [PubMed] [Google Scholar]
- 105.Min KW, Kim DH, Do SI, Pyo JS, Kim K, Chae SW, Sohn JH, Oh YH, Kim HJ, Choi SH, Choi YJ, Park CH. Diagnostic and prognostic relevance of MMP-11 expression in the stromal fibroblast-like cells adjacent to invasive ductal carcinoma of the breast. Ann Surg Oncol 2013;20(Suppl 3):S433–S442. [DOI] [PubMed] [Google Scholar]
- 106.Roscilli G, Cappelletti M, De Vitis C, Ciliberto G, Di Napoli A, Ruco L, Mancini R, Aurisicchio L. Circulating MMP11 and specific antibody immune response in breast and prostate cancer patients. J Transl Med 2014;12:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nonsrijun N, Mitchai J, Brown K, Leksomboon R, Tuamsuk P. Overexpression of matrix metalloproteinase 11 in Thai prostatic adenocarcinoma is associated with poor survival. Asian Pac J Cancer Prev: APJCP 2013;14(5):3331–3335. [DOI] [PubMed] [Google Scholar]
- 108.Ervine A, Leung S, Gilks CB, McCluggage WG. Thyroid transcription factor-1 (TTF-1) immunoreactivity is an adverse prognostic factor in endometrioid adenocarcinoma of the uterine corpus. Histopathology 2014;64(6):840–846. [DOI] [PubMed] [Google Scholar]
- 109.Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, Bouffet E, Clifford SC, Hawkins CE, French P, Rutka JT, Pfister S, Taylor MD. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol: Off J Am Soc Clin Oncol 2011;29(11):1408–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kazma R, Mefford JA, Cheng I, Plummer SJ, Levin AM, Rybicki BA, Casey G, Witte JS. Association of the innate immunity and inflammation pathway with advanced prostate cancer risk. PLoS ONE 2015;7(12):e51680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Antunes AA, Reis ST, Leite KR, Real DM, Sousa-Canavez JM, Camara-Lopes LH, Dall’Oglio MF, Srougi M. PGC and PSMA in prostate cancer diagnosis: Tissue analysis from biopsy samples. Int Braz J Urol 2013;39(5):649–656. [DOI] [PubMed] [Google Scholar]
- 112.Su B, Gao L, Baranowski C, Gillard B, Wang J, Ransom R, Ko HK, Gelman IH. A genome-wide RNAi screen identifies FOXO4 as a metastasis-suppressor through counteracting PI3 K/AKT signal pathway in prostate cancer. PLoS ONE 2015;9(7):e101411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Che YL, Luo SJ, Li G, Cheng M, Gao YM, Li XM, Dai JM, He H, Wang J, Peng HJ, Zhang Y, Li WY, Wang H, Liu B, Linghu H. The C3G/Rap1 pathway promotes secretion of MMP-2 and MMP-9 and is involved in serous ovarian cancer metastasis. Cancer Lett 2015;359(2):241–249. [DOI] [PubMed] [Google Scholar]
- 114.Samuelsson J, Alonso S, Ruiz-Larroya T, Cheung TH, Wong YF, Perucho M. Frequent somatic demethylation of RAPGEF1/C3G intronic sequences in gastrointestinal and gynecological cancer. Int J Oncol 2011;38(6):1575–1577. [DOI] [PubMed] [Google Scholar]
- 115.Lund-Johansen P The hemodynamics of the aging cardiovascular system. J Cardiovasc Pharmacol 1988;12(Suppl 8):S20–S30. [PubMed] [Google Scholar]
- 116.Kong KL, Kwong DL, Fu L, Chan TH, Chen L, Liu H, Li Y, Zhu YH, Bi J, Qin YR, Law SY, Guan XY. Characterization of a candidate tumor suppressor gene uroplakin 1A in esophageal squamous cell carcinoma. Cancer Res 2010;70(21):8832–8841. [DOI] [PubMed] [Google Scholar]
- 117.Olsburgh J, Harnden P, Weeks R, Smith B, Joyce A, Hall G, Poulsom R, Selby P, Southgate J. Uroplakin gene expression in normal human tissues and locally advanced bladder cancer. J Pathol 2003;199(1):41–49. [DOI] [PubMed] [Google Scholar]
- 118.Boormans JL, Korsten H. Ziel-van der made AJ, van leenders GJ, de vos CV, Jenster G, Trapman J. Identification of TDRD1 as a direct target gene of ERG in primary prostate cancer. Int J Cancer 2015;133(2):335–345. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.