Abstract
Amyotrophic lateral sclerosis is a late-onset adult neurodegenerative disease, although there is mounting electrophysiological and pathological evidence from patients and animal models for a protracted preclinical period of motor neuron susceptibility and dysfunction, long before clinical diagnosis. The key molecular mechanisms linked to motor neuron vulnerability in amyotrophic lateral sclerosis have been extensively studied using transcriptional profiling in motor neurons isolated from adult mutant superoxide dismutase 1 mice. However, neonatal and embryonic motor neurons from mutant superoxide dismutase 1 mice show abnormal morphology and hyperexcitability, suggesting preceding transcriptional dysregulation. Here, we used RNA sequencing on motor neurons isolated from embryonic superoxide dismutase 1G93A mice to determine the earliest molecular mechanisms conferring neuronal susceptibility and dysfunction known in a mouse model of amyotrophic lateral sclerosis. Transgenic superoxide dismutase 1G93A mice expressing the spinal motor neuron homeobox HB9:green fluorescent protein reporter allowed unambiguous identification and isolation of motor neurons using fluorescence-activated cell sorting. Gene expression profiling of isolated motor neurons revealed transcriptional dysregulation in superoxide dismutase 1G93A mice as early as embryonic Day 12.5 with the majority of differentially expressed genes involved in RNA processing and α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate-mediated glutamate receptor signalling. We confirmed dysregulation of the α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate receptor Subunit 2, at transcript and protein levels, in embryonic superoxide dismutase 1G93A mouse motor neurons and human motor neurons derived from amyotrophic lateral sclerosis patient induced pluripotent stem cells harbouring pathogenic superoxide dismutase 1 mutations. Mutant superoxide dismutase 1 induced pluripotent stem cell-derived motor neurons showed greater vulnerability to α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate-mediated excitotoxicity, consistent with α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate receptor Subunit 2 downregulation. Thus, α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate receptor Subunit 2 deficiency leading to enhanced α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate receptor signalling, calcium influx, hyperexcitability, and chronic excitotoxicity is a very early and intrinsic property of spinal motor neurons that may trigger amyotrophic lateral sclerosis pathogenesis later in life. This study reinforces the concept of therapeutic targeting of hyperexcitability and excitotoxicity as potential disease-modifying approaches for amyotrophic lateral sclerosis.
Keywords: AMPA receptor, amyotrophic lateral sclerosis, motor neurons, transcriptomics, RNA processing
Zanganeh et al. report the earliest known transcriptional dysregulation in motor neurons from embryonic mutant superoxide dismutase 1 mice and amyotrophic lateral sclerosis patients. They conclude that glutamate receptor excitotoxicity and RNA processing defects are early determinants of cell vulnerability in amyotrophic lateral sclerosis.
Graphical Abstract
Graphical Abstract.
Introduction
Amyotrophic lateral sclerosis (ALS) is an adult-onset neurodegenerative disease that is caused by the loss of both upper and lower motor neurons. ALS results in rapidly progressive physical disability, leading to fatal paralysis for which there is no effective treatment or cure.1,2 The majority of ALS cases are sporadic (SALS), whereas the remaining 10% are inherited (familial ALS or FALS). Dominant mutations in the superoxide dismutase 1 (SOD1) gene account for 20% of FALS and transgenic mice expressing mutant SOD1 remain the most widely used, best understood and accepted mouse model of ALS.3
Although ALS typically presents in mid-to-late life, there are multiple lines of evidence suggesting the existence of a protracted preclinical period before symptom onset in ALS patients and mouse models. First in patients, asymptomatic SOD1 mutation carriers exhibited both a reduction in motor unit number estimation using electromyography, indicative of preclinical motor neuron loss.4 Increased cortical hyperexcitability detected by threshold tracking transcranial magnetic stimulation which reflects upper motor neuron dysfunction also occurs in presymptomatic SOD1 mutation carriers which progress to ALS.5 In SOD1G93A mice that develop symptom onset at postnatal Day 90 (P90), it is well established that peripheral synapses of spinal motor neurons degenerate as early as P456 which was subsequently predated to P25,7 preceding distal axon loss at P80 and cell body loss at P120.6 At a subcellular level, Golgi apparatus fragmentation at P308 and mitochondrial swelling at P149 are early features of spinal motor neuron pathology in SOD1G93A mice. At a synaptic level, dendritic pathology and increased synaptic neurotransmission in spinal motor neurons occur at ∼P8 in SOD1G93A mice;10 however, the molecular basis of this hyperexcitability remains unclear. Finally, at a molecular level, mTOR activation at P15, mutant SOD1 aggregation at P711 and endoplasmic reticulum (ER) stress at P512 are the earliest reported dysfunctions in spinal motor neurons in SOD1G93A mice.
Collectively, these findings in ALS patients and mouse models provide evidence of cell vulnerability and dysfunction before symptom onset in motor neurons and suggest the existence of a long-lasting disease process that may commence years, or potentially even decades, before clinical deficits. In support of this, Eisen et al.13 recently proposed that ALS onset may even date back to the perinatal period of life and conception, based on the embryonic expression of ubiquitous ALS-linked genes. These authors suggested that a combination of genetic, epigenetic and exogenous factors, coupled with metabolic and oxidative stress associated with early motor neuron development, may render motor neurons inherently vulnerable, thereby increasing the risk of ALS later in life. Indeed, there is evidence for abnormalities in spinal motor neurons cultured from embryonic SOD1G93A mice shown by enhanced hyperexcitability14 and axonal transport deficits,15 although these are cell culture experiments and evidence is lacking in vivo.
We, therefore, sought to test this provocative hypothesis using an unbiased transcriptomic approach in spinal motor neurons of embryonic SOD1G93A mice. While gene expression profiling of isolated motor neurons has been extensively performed in mutant SOD1 mice uncovering potential candidate disease pathways,12,16–18 these studies have been restricted to adult (P30–P120) or early postnatal (P5) mice using laser-capture microdissection and microarray analysis. Here, we devised a novel methodology to isolate enriched populations of spinal motor neurons from embryonic SOD1G93A mice. We coupled fluorescence-activated cell sorting (FACS)-based isolation and purification of motor neurons from SOD1G93A mice co-expressing a well-validated fluorescent motor neuron reporter, homeobox HB9 (HB9):green fluorescent protein (GFP),19 with highly sensitive transcriptional profiling to identify early changes in gene expression in embryonic wild-type (WT) and SOD1G93A mice. A distinct gene expression profile was identified as early as embryonic Day (E) 12.5 in SOD1G93A mice. We identified significant changes in genes associated with excitotoxicity and RNA processing and validated the involvement of these molecular mechanisms at transcriptional and protein levels. Importantly, these findings were confirmed in induced pluripotent stem cell (iPSC)-derived spinal motor neurons from SOD1-linked ALS patients carrying three distinct mutations which accordingly showed greater vulnerability to excitotoxicity. Our study identifies dysregulation of α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate (AMPA) receptor signalling and RNA processing genes as very early, intrinsic and key contributors linked to motor neuron susceptibility in ALS in utero.
Materials and methods
Animals
All animal procedures were performed in accordance with the guidelines of the Australian National Health and Medical Research Council’s published Code of Practice for the use of animals in research and were approved by the Florey Institute of Neuroscience and Mental Health Animal Ethics Committee (permit number: 16-026). Transgenic HB9:GFP mice [B6.Cg-Tg(Hlxb9-GFP)1Tmj/J, stock no. 005029]19 and SOD1G93A mice [B6.Cg-Tg(SOD1*G93A)1Gur/J line; stock no. 004435; RRID:IMSR_JAX:004435] were obtained from the Jackson Laboratory (Bar Harbor, ME, USA) and maintained on C57BL/6J backgrounds. SOD1G93A male and HB9:GFP female mice were crossed to generate SOD1G93A; HB9:GFP+/− and HB9:GFP+/− mice, which were subsequently crossed to generate experimental animals: SOD1G93A; HB9:GFP+/+ and HB9:GFP+/+ mice. HB9:GFP+/+ mice were henceforth called WT for this study. Mice were maintained under standard conditions of 12 h light/dark cycles with access to food and water ad libitum. Mice were time-mated overnight, and embryonic (E) Day 0.5 was designated following visualization of a vaginal plug the next morning.
DNA extraction and genotyping
Tissue for DNA extraction was obtained from embryo tail. DNA was extracted using a rapid genotyping method using red extract-N-AmpTM Tissue PCR Kit (Sigma–Aldrich; 254-457-8). Two μl of diluted DNA was added to the master mix containing 100 nM forward and reverse primers, nuclease-free water and 5 μl of SsoAdvanced universal SYBR Green Supermix (BioRad; 1725274). Samples were then genotyped on a real-time PCR machine (BioRad; CFX96), targeting human SOD1 using forward (5′-CATCAGCCCTAATCCATCTGA-3′) and reverse primers (5′-CGCGACTAACAATCAAAGTGA-3′). The thermal cycling condition was set as the following: 2 min denaturation at 95°C, annealing at 95°C for 5 s and extension at 60°C for 5 s for 40 cycles.
Isolation of spinal motor neurons
Pregnant mice were deeply anaesthetized with 3% isoflurane inhalation (Delvet, Seven Hills, NSW, Australia) and administered sodium pentobarbital (11 mg/kg 0.1 ml, i.p.). Lumbar spinal cords were microdissected from HB9:GFP and SOD1G93A HB9:GFP embryos at E12.5 in chilled Leibovitz’s L-15 medium (Gibco™; 11415064). Littermate lumbar spinal cords of the same genotype were pooled (2–3 per genotype) and dissociated into a single-cell suspension using a neural tissue dissociation kit (Miltenyi Biotec; 130-094-802) according to the manufacturer’s instructions. Cell viability and density were determined by trypan blue exclusion counting. Cells were resuspended in 500 μl of HBSSCa2+ Mg2+ containing 1% (v/v) bovine serum albumin (Sigma–Aldrich; 9048-46-8) and the cell suspension filtered using a 40 μm cell strainer. Sorting was conducted on a FACS AriaTM Fusion Flow Cytometer (Becton Dickinson) using 85 μm nozzle size, 45 psi sheath pressure, 1.0 flow rate and 488 nm optical path/laser. Dead cells and doublets were excluded using side scatter and forward scatter parameters based on DAPI staining and gating for the collection of GFP+ cells was set using GFP− tissue. Sorted GFP+ cells were directly collected in RLT lysis buffer (Qiagen; 74004) and the lysates were stored at −80°C.
RNA extraction and qRT-PCR
Total RNA was isolated from cell lysates using a RNeasy Micro kit (Qiagen; 74004), including an on-column DNase I (Qiagen) digestion step according to the manufacturer’s instructions. RNA quantity and integrity were assessed by a 5200 Fragment Analyzer system (Agilent Technologies Ltd). First-strand cDNA was synthesized from 50 ng of total RNA using a reverse transcription reagent kit according to the manufacturer (PrimeScriptTM RT; Takara; RR037A). Primers for each candidate gene were designed using Primer320 and are shown in Table 1. Real-time qPCR was carried out using SYBR® Premix Ex TaqTM II (Takara; RR820L) and 10 μM primer with 4–5 biological replicates for each group. Relative gene expression normalized to mouse Gapdh was determined using the ΔΔCt method.21 Droplet digital PCR and restriction fragment length polymorphism (RFLP) analyses are described in Supplementary Tables 1 and 2.
Table 1.
List of primers used in this study for quantitative RT-PCR
| Gene | Forward primer (5′–3′) | Reverse primer (5′–3′) |
|---|---|---|
| Adarb1 | AGTGAGGGTCTTCAGTTGCA | TGCATGAGGGGAGGAAAAGT |
| Flrt2 | AAATGGGCCACAGTCTCGTA | CGGTAGTTGAACGCATCCAG |
| Gfap | TAAGCTAGCCCTGGACATCG | GCCTTCTGACACGGATTTGG |
| Gria1 | TGCTTTGTCACAACTCACGG | ATGGCGTACACTCCTTTGGA |
| Gria2 | AATACCCTGGAGCACACACA | CCTCTGCTTCCGAAGATTGC |
| Gria3 | ACAAAGCCCTCCTGATCCTC | GTGAAGAACCACCAAACCCC |
| Gria4 | TGTTGGGAAGCACGTCAAAG | TCGTCACCATGGGCGTATTA |
| Mbp | TACCCTGGCTAAAGCAGAGC | CAGGGAGCCATAATGGGTAG |
| Mnx1 | GTTGGAGCTGGAACACCAGT | CTTTTTGCTGCGTTTCCATT |
| Pemt | TGCTTTTGAACATCCTCCGC | CTGGACAGCACAAACACGAA |
| Slc17a7 (Vglut1) | CTGGGGTCCTTGTGCAGTAT | GTATTTGCGCTCCTCCTCAG |
| Gas5 | GGTTTAAGTGGACGCAGCAA | CCATCACAGAGGTCCACACT |
| Isac2 | CTTTCCCAGGCTTTTGACCC | TGCAGCCTGAGGAATTCTGA |
| Malat1 | CCAGTTTCCCCAGCTTTTCC | TACTGGCTGCATCAAGGTGA |
| Meg3 | CGGAGGACACTTGGACTCTT | TCGATGGAGAAGAGCGAGTC |
| Pnisr | CATCTCCAGCTCCCCAAGAA | GCGCATCTTTAGCCACGTAA |
| Ankr16 | GATGCAATTCAGTGTGGCCA | CTTCATCCTGCCCAGTGACT |
| Mir124-1 | TGGGAGGGGTGCATTGTTAT | TGGGAAGGGGATCTGGTAGA |
| ND4L | GCTCCATACCAATCCCCATCA | TCTGTTCCGTACGTGTTTGA |
| Cpne6 | GACTTCATCGGCGAGTTCAC | CTTGTCTCGGTACTTGGGGT |
| Ankrd23 | CTGGTGAACAAGCTGCTGG | TTGTCTTGTGCGTTGACCTG |
| Adamts10 | AGTGCCTGTGAGACCATTGA | GTGACTCAGGGACAGGTTCA |
| Appl2 | CACGACCTGTCAATGGCAAA | CGTTGAGTGCGCAGTAGTAC |
| Rmst | ACTGAAGAGTACAACCGCCA | GCCTTATTGCTGCTCCTGTC |
| Gapdh | GGTGCTGAGTATGTCGTGGA | GTGGTTCACACCCATCACAA |
| Human-GRIA2 | CAAACAGCTTCGCAGTCACT | TGAAGGAGACGTGGAGTGTT |
| Human-ADAR2 | TGTACATCAGCACCTCTCCC | TTTTGGTCCGTAGCTGTCCT |
| Human 18S | GTAACCCGTTGAACCCCATT | CCATCCAATCGGTAGTAGCG |
| Human GAS5 | GAGCAAGCCTAACTCAAGCC | CCCAAGCAAGTCATCCATGG |
| Human ISCA2 | TCAGAATTCCTCAGGCTGCA | GCCCCACCCTGTTCAAATAC |
| Human MALAT1 | GGCTCAGTTGCGTAATGGAA | GAATTCGATCACCTTCCGCC |
| Human MEG3 | ATCATCCGTCCACCTCCTTG | CAGTGAGTGGCTGCTTTGTA |
| Human PNSIR | AGATGCTGAAGGAGGGGATG | AACTGGGGATGGACTTCTGG |
| Human ANKR16 | CACATTGCCAGTCGAGAAGG | CCTTGACTGCCTCCAAATGG |
RNA-seq library preparation and transcriptome analysis
RNA-seq libraries were prepared using the NeuGen Ovation® SoLo kit (0407-32) and subjected to 150 bp, paired-end sequencing on an Illumina HiSeq 2000 sequencer with four independent biological replicates analysed per group (four litters/collections per genotype). Reads were aligned to a reference mouse genome (mm10) and human genome (hg38) for identification of the human SOD1 transgene, both obtained from the University of California Santa Cruz using HISAT2 (2.1.0), under default parameters, with the exception of reporting only unique reads mapping to a unique location on the reference genome.22 The relative abundance of transcripts was measured using the FeatureCounts function in the Subread package.23 Differential expression analysis and principal component analysis (PCA) were conducted with DeSeq2 in RStudio (3.6.0).24 Gene ontology and pathway analysis were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) functional tool25 and Ingenuity Pathway Analysis (IPA; QIAGEN), respectively.
Immunohistochemistry
Embryonic lumbar spinal cords were fixed in 4% (w/v) paraformaldehyde (PFA) in 0.1 M phosphate buffer (PB) and picric acid at pH 7.2 for 2 h. Spinal cords were transferred to a 20% (w/v) sucrose in 0.1 M PB overnight at 4°C. Samples were embedded in OCT compound (Pro-SciTech, Queensland, Australia) and snap-frozen in isopentane on dry ice. Spinal cords were cross-sectioned at a thickness of 20 μm using a cryostat and placed on glass slides in series of 1:6 for immunostaining. For immunohistochemistry, the sections were rinsed with phosphate buffer saline (PBS) and blocked with 10% (v/v) normal donkey serum diluted in PBS with 0.3% (v/v) Triton X-100 for 1 h. The sections were incubated overnight at room temperature with primary antibodies. Primary antibodies were as follows: chicken anti-GFP (1:1000; Abcam; AB13970), rabbit anti-NeuN (1:1000; Abcam; AB104225), goat anti-ChAT (1:500; Abcam; AB34419) and guinea pig anti-AMPA receptor 2 subunit (GluA2) (1:500; Alomone Labs; AGP-073). On the following day, the sections were incubated in a secondary antibody (1:250) for 2 h at room temperature. Secondary antibodies were as follows: donkey, anti-chicken Alexa Fluor® 488 (Jackson ImmunoResearch Labs; 703-545-155), donkey, anti-guinea pig Alexa Fluor® 594 F(ab’)2 (Jackson ImmunoResearch Labs; 706-586-148) and anti-rabbit Alexa Fluor® 647 (Jackson ImmunoResearch Labs; 711-605-152). The sections were cover slipped using fluorescence mounting medium (DAKO; S3023). The slides were left to dry overnight in the dark at room temperature before imaging.
Microscopy
Images were captured using an LSM 780 Zeiss confocal laser scanning microscope under 40× magnification. Quantification of GluA2 signal intensity was determined in Zen Blue software (ZEN lite, Zeiss) by measuring signal intensity of HB9:GFP-positive motor neurons. Five sections of L4 lumbar spinal cord (equivalent to 10 ventral horns) were selected at equally spaced intervals per animal with four mice per genotype. To compare signal intensity between different samples, the images were taken at the same excitation light intensity, exposure time and gain settings.
Generation of iPSC-derived motor neurons
Patient iPSC lines containing SOD1G85S, SOD1L144F or SOD1I114T mutations and control line 11b26 were either generated in-house or kindly donated by Dr Kevin Eggan (Harvard Stem Cell Institute, MA, USA) and differentiated into spinal motor neurons as previously described.27 At Day 28, motor neurons were defined as mature by the expression of ChAT and Tuj1. RNA was harvested from mature motor neurons using RNeasy Micro kit (Qiagen; 74004) or the cells were fixed for immunocytochemistry.
Immunocytochemistry
Cells were fixed for 10 min in 4% (w/v) PFA and washed twice in DPBS (ThermoFisher Scientific). Cells were blocked in 10% (v/v) normal donkey serum with 0.1% (v/v) Triton-X 100 in 0.1 M PBS and incubated overnight at 4°C in the following primary antibodies: goat anti-ChAT (1:500, Millipore; AB144P), chicken anti-β III tubulin (1:1000, Abcam; AB41489) and rabbit anti-GluA2 (1:500; Alomone Labs; AGC-005). Cells were incubated in the following secondary antibodies in the same diluent (1:400), for 2 h at room temperature; anti-goat Alexa Fluor®-488 (Jackson ImmunoResearch Labs; 705-545-147), anti-rabbit Cy™-550 (Jackson ImmunoResearch Labs; 711-165-152), anti-chicken Alexa Fluor®-647 (Jackson ImmunoResearch Labs; 703-605-155); followed by 15 min incubation in 1 μg/ml Hoechst 33342 (LifeTech; H1399). Cells were imaged at 20× magnification on Zeiss fluorescence microscope (Carl Zeiss AG, Germany). GluA2 expression was quantified using Zen Blue software (ZEN lite, Zeiss) by manual tracing of the cell body of motor neurons, using a minimum of 50 cells per biological replicate (n = 3 biological replicates per line). The intensity values were then subtracted from the background signal. The background signal was calculated as the mean of the pixels outside of a cell. To compare signal intensity between different lines, the images were taken at the same excitation light intensity, exposure time and gain settings.
Excitotoxicity assay
Cryopreserved Day 19 motor neurons were seeded on laminin-521-coated 96-well plates at 1 × 105 cells/well and cultured for 24 h in BrainPhys™ medium supplemented with 0.5 mM retinoic acid, 0.1 mM purmorphamine, 0.1 mM compound E, 20 ng/ml IGF1, 20 ng/ml CNTF1, 20 ng/ml BDNF and 10 mM Y-27632. An assay was performed on Day 28 motor neurons. Live cells were labelled with 2 μM Calcein Red™ (AAT Bioquest; 21900) and 0.02% (w/v) Pluronic® F-127 according to the manufacturer’s instructions at 37°C for 30 min and imaged using a Zeiss Axio fluorescence microscope (Zeiss AG, Germany) under 10× magnification. Cells were then treated with 100 μM of AMPA (Tocris Bioscience; 0254) for 24 h. Following treatment, cells were labelled again with Calcein Red™ and imaged. Cells were counted after treatment using Zen Blue image analysis software (ZEN lite, Zeiss) by counting two fields of view per well.
Statistical analyses
All quantitative data are represented as means ± standard error of the means (SEMs). Statistical significance testing was performed using an unpaired Student’s t-test for comparison of two groups or one-way ANOVA with Dunnett’s multiple comparisons test for three or greater groups using GraphPad Prism 7.0 (GraphPad, USA).
Data availability
The RNA-seq data generated from this study have been deposited in GEO accession number GSE142654.
Results
Transcriptional profiling of spinal motor neurons of embryonic Day 12.5 SOD1G93A mice
To date, all gene expression profiling studies of motor neurons in ALS have been conducted in post-mortem clinical endstage patient samples or postnatal often adult mouse models. To identify the earliest molecular pathways in SOD1G93A mice that may underlie cell vulnerability to ALS, the transcriptional profile of lumbar spinal cord motor neurons was evaluated at E12.5. E12.5 represents motor neuron birth in the spinal cord, when motor neuron progenitors have exited the cell cycle and moved to their final position along motor pools.28,29 To specifically profile motor neurons in the spinal cord, HB9:GFP reporter mice expressing GFP under control of the motor neuron-specific homeodomain transcription factor HB9 promoter19 were crossed with SOD1G93A mice to generate HB9:GFP controls (henceforth called WT) and SOD1G93A; HB9:GFP mice (henceforth called SOD1G93A). Lumbar spinal cords were dissected, pooled within genotypes, dissociated into a single-cell suspension and GFP+ motor neurons isolated using FACS (Fig. 1A). The percentage of GFP+ cells isolated from WT and SOD1G93A embryos was 15 ± 2.6 and 15 ± 2.2% (mean ± SEM), respectively, when gated against GFP− cells from spinal cord (Fig. 1B).
Figure 1.
Gene expression profiling of spinal motor neurons of embryonic SOD1G93A mice. (A) Representative images of HB9:GFP-SOD1G93A transgenic mice at E12.5 and immunolabelling of HB9:GFP in a transverse section of spinal cord. Scale bar, 50 μm. Lumbar spinal cords were dissociated into single-cell suspension and motor neurons were isolated using FACS. (B) Bar graph representing the percentage of GFP+ cells in SOD1G93A and WT samples. n = 4 biological replicates. (C) RNA was extracted from the sorted motor neurons, and purity of samples was examined using qRT-PCR by testing the expression for motor neuron, interneuron, oligodendrocyte and astrocyte gene markers. n = 4–11 biological replicates. (D) RNA sequencing reads were aligned to the human genome to evaluate the expression level of human SOD1 in samples. (E) Quantification of number of reads mapped to human SOD1. n = 6 biological replicates. (F) Assessment of sample purity using RNA sequencing normalized read counts for the expression of neuronal and glial marker genes. n = 12 biological replicates. (G) Clustering of gene expression profiles of spinal motor neurons from WT and SOD1G93A mice were compared by PCA. Data represent mean ± SEM, unpaired student t-test, n = 4 biological replicates. *P < 0.01, **P < 0.001, ***P < 0.0001, ****P < 0.00001; GFPNeg, GFP-negative; GFPPos, GFP-positive.
To confirm the enrichment of motor neurons in the GFP+ cell fraction, high integrity RNA (RNA integrity number >9) was extracted from GFP+, GFP− and unsorted cell populations from WT embryos (Supplementary Fig. 1). Real-time qPCR was conducted to test for selected neuronal and non-neuronal populations. Mnx1 (HB9), the key marker for motor neurons, was significantly enriched (16 ± 3.3-fold) in the GFP+ fraction, relative to the unsorted cell fraction (P < 0.0001). The interneuron marker Ptf1a was significantly depleted (0.8 ± 0.05-fold) in the positive fraction (P < 0.05) and enriched in the negative fraction (1.3 ± 0.4-fold), relative to unsorted cells (P < 0.01). No significant change was observed in the expression of Vglut1 expressed by interneurons. The expression of the oligodendrocyte marker Mbp was significantly depleted (1.6 ± 0.1-fold) in both the positive and negative fractions, relative to the unsorted sample (P < 0.00001), whereas the astrocytic gene Gfap was significantly depleted in the positive fraction (1.0 ± 0.06, P < 0.0001) and negative fraction (0.06 ± 0.08, P < 0.0001) (Fig. 1C).
To enable unbiased gene expression profiling of motor neurons, RNA sequencing was conducted on GFP+ cells sorted from WT controls and SOD1G93A embryos. Whole transcriptome cDNA libraries were subject to 150 bp paired-end sequencing at an average read depth of 60 million reads per sample. Motor neurons isolated from SOD1G93A embryos showed a 30,000 ± 3,000-fold increase in human SOD1 expression, compared with WT (P < 0.0001) as expected (Fig. 1D–E). Consistent with the qPCR findings, normalized RNA-seq reads showed an enrichment of motor neuron-specific transcripts Snp25 (3237 ± 119-fold), Islet1 (1651 ± 136-fold) and Mnx1 (3713 ± 279-fold). A corresponding depletion of transcripts specific for microglia, oligodendrocytes, Schwann cells, astrocytes and vascular endothelial cells was also detected (Fig. 1F). Following validation of sample purity, examination of the global gene expression profile by PCA showed the samples segregated into two distinct clusters corresponding to genotypes (Fig. 1G), indicating a subtle difference in the motor neuron transcriptome between WT and SOD1G93A mice at E12.5.
Identification of potential mechanisms involved in susceptibility to ALS
Differential expression analysis was conducted to identify significantly dysregulated genes in motor neurons at E12.5. Significant changes (corrected for multiple testing) in 21 genes were observed at E12.5 in SOD1G93A motor neurons compared with WT (q value <0.05) (Table 2). Hierarchical clustering of differentially expressed genes indicated the highest similarity between the replicates of each genotype (Fig. 2A). Classification of the differentially expressed genes based on their known molecular function revealed that the majority of genes were involved in RNA processing (43%). The remaining genes were classified as developmental-related (19%), mitochondrial (19%), AMPA receptor signalling (10%) and unknown genes (9%) (Fig. 2B).
Table 2.
Significantly differentially expressed genes in SOD1G93A motor neurons at E12.5
| Gene symbol | Fold changea | Adj P-value | Entrez ID |
|---|---|---|---|
| Rmst | −0.80 | 4.76 × 10−02 | 110333 |
| Gria2 | −0.78 | 1.78 × 10−04 | 14800 |
| mt-Rnr2 | −0.76 | 3.88 × 10−02 | 17725 |
| Appl2 | −0.75 | 4.76 × 10−02 | 216190 |
| Pnisr | −0.73 | 3.88 × 10−02 | 66625 |
| ND6 | −0.72 | 2.88 × 10−02 | 17722 |
| Isca2 | −0.72 | 4.76 × 10−02 | 74316 |
| mt-Rnr1 | −0.70 | 2.44 × 10−04 | 17724 |
| Adamts10 | −0.70 | 3.44 × 10−02 | 224697 |
| Mir124-1 | −0.66 | 3.88 × 10−02 | 268755 |
| Gas5 | −0.65 | 1.17 × 10−02 | 14455 |
| Meg3 | −0.63 | 2.78 × 10−05 | 17263 |
| Ankrd16 | −0.62 | 2.62 × 10−04 | 320816 |
| Malat1 | −0.59 | 2.07 × 10−05 | 72289 |
| Mir10b | −0.55 | 3.42 × 10−03 | 387144 |
| ND4L | −0.54 | 2.40 × 10−09 | 17720 |
| Ankrd23 | −0.48 | 4.76 × 10−02 | 78321 |
| Cpne6 | −0.40 | 3.64 × 10−02 | 12891 |
| Scarna9 | −0.38 | 5.85 × 10−03 | 100216535 |
| Rnu1b2 | −0.34 | 4.48 × 10−02 | 19845 |
| Flrt2 | 1.54 | 2.44 × 10−04 | 399558 |
Negative value represents downregulation.
Figure 2.
Deconstructing dysregulated pathways of early pathogenesis in ALS. (A) Heat map shows the relative expression levels of differentially expressed genes, along with the hierarchical clustering of genes and samples. Expression levels used in the heatmap were calculated by mean of the log2 normalized values of genes. Visual representation of downregulation and upregulation is shown. (B) Classification of the differentially expressed genes based on their known molecular function. (C) Enriched biological function and (D) molecular function identified by gene ontology analysis (DAVID) using significantly differentially expressed genes (P < 0.01). (E and F) Network analysis using significantly differentially expressed genes (P < 0.01) determined by IPA.
Gene ontology analysis for biological and molecular processes using a less stringent significance threshold (not corrected for multiple testing) was conducted to capture a broader set of dysregulated genes and pathways (P < 0.01, Supplementary Table 3) and showed highly significant enrichment of RNA processing pathways (Fig. 2C–D). Consistent with gene ontology analysis, network analysis using the enlarged list of differentially expressed genes identified significant pathways centred around processing of RNA (Fig. 2E) and mRNA transcription (expression of RNA; P < 0.01) (Fig. 2F).
AMPA receptor expression is dysregulated in SOD1G93A mouse motor neurons at E12.5
Validation of significantly changed genes at E12.5 was conducted on a separate batch of GFP+ isolated motor neurons from WT and SOD1G93A mice using real-time qPCR where primers could be designed (16 of 21 genes). A significant decrease in expression was identified in the RNA processing gene Pnisr (−0.2 ± 0.06, P = 0.04) (Fig. 3A); no significant changes in the mitochondrial gene was validated (Fig. 3B); nervous system development-related genes Flrt2 (−0.5 ± 0.1, P = 0.003), Pemt (−0.1 ± 0.04, P = 0.01) and Adamts10 (−0.2 ± 0.06, P = 0.02) (Fig. 3C) and the AMPA receptor gene Gria2 (GluA2; −0.2 ± 0.09, P = 0.01); related Mir124-1 (−0.2 ± 0.07, P = 0.03) (Fig. 3D), and no significant changes in unknown gene category were identified (Fig. 3E).
Figure 3.
Validation of differentially expressed genes using qRT-PCR. (A–E) qPCR verification of the expression of genes involved in biological process identified as enriched by GO analysis compared with WT control. (F) Fold change expression of Ca2+-permeable AMPAR subunit Gria1, Gria3 and Gria4 mRNAs, relative to WT motor neurons at E12.5. (G) Relative expression of Adarb1 mRNA in SOD1G93A motor neurons at E12.5. (H) Schema showing the position of the fully complementary miR-124 target site at the 5′-end of the mouse Gria2, 3′-UTR. The seed region of miR-124 is shown. Data represent mean ± SEM, unpaired student t-test, n = 5–7 biological replicates, *P < 0.05.
Gria2 encodes GluA2 which reduces Ca2+ permeability of AMPA receptors when incorporated within the heteromeric receptor. Deficiency of the GluA2 Subunit of AMPA receptor in motor neurons is associated with Ca2+-mediated excitotoxity in ALS.30 As excitotoxicity is one of the most important processes implicated in ALS pathogenesis, Gria2 was selected as an interesting target for further validation. Evidence of mRNA dysregulation in other components of AMPA receptors was then examined in SOD1G93A mouse motor neurons. No significant changes in expression of other AMPA receptor subunits Gria1, Gria3 or Gria4 were detected at E12.5 (Fig. 3F). However, a significant decrease in the expression of the Gria2 editing enzyme Adar2 (Adarb1) gene (0.13 ± 0.06, P = 0.01) was identified using qRT-PCR (Fig. 3G).
Interestingly, Gria2 was shown as a potential target of transcriptional regulation by the significantly changed RNA processing gene miR124-1 in our study (by Targetscan and miRanda). The 3′-UTR of Gria2 contains a highly conserved miR-12-1 target site (Fig. 3H), and it has been previously shown that miR-124-mediated regulation of Gria2 produces rapid changes in Gria2 expression.31,32
To further confirm the involvement of a potential Gria2 and Adar2 network, droplet digital PCR was used. A significant downregulation of Gria2 (19 ± 6 copies per μl, P < 0.01) and Adar2 (3 ± 1.3 copies per μl, P < 0.01) transcript numbers were identified in SOD1G93A motor neurons compared with WT (Supplementary Fig. 2A–C). Enzymatic editing of Gria2 transcripts by Adar2 regulates its permeability to Ca2+ influx. Gria2 has been reported to be less efficiently edited in motor neurons due to downregulation of Adar2 in symptomatic SALS patients and mouse models.33,34 Hence, RFLP PCR was used to detect Gria2 mRNA post-transcriptional editing using a Bbv1 endonuclease in SOD1G93A motor neurons at E12.5.35 Despite downregulation of Adar2, SOD1G93A motor neurons exhibited normal Gria2 editing at E12.5 (Supplementary Fig. 2D).
GluA2 is downregulated in motor neurons of embryonic SOD1G93A mice
To determine if GluA2 protein was reduced in motor neurons of SOD1G93A mice, embryonic spinal cords were subjected to immunohistochemistry at E12.5 (Fig. 4A–J) and E17.5 (Fig. 4K–T). GluA2 levels were significantly reduced in GFP + lumbar motor neurons in SOD1G93A mice, compared with WT motor neurons at E12.5 (P < 0.05) (Fig. 4U). Examination of E17.5 spinal cord showed that this significant reduction in GluA2 protein levels persisted throughout embryonic development (P < 0.05) (Fig. 4V). Furthermore, GFP intensity was not reduced in spinal cords at E12.5 or E17.5 and there was no loss of motor neurons at E17.5 (Supplementary Fig. 3), confirming GluA2 downmodulation in motor neurons. In addition, GluA2 signal was not downregulated in GFP-NeuN+ cells, suggesting a motor neuron-specific property (Supplementary Fig. 3). Thus, GluA2 downregulation is a very early and sustained event in vulnerable motor neurons from SOD1G93A mice.
Figure 4.
Expression of GluA2 in spinal cords of embryonic SOD1G93A mice. Cross-sections of lumbar spinal cord from WT (HB9:GFP; WT) and SOD1G93A (SOD1G93A; HB9:GFP) mice at (A–J) E12.5 and (K–T) E17.5. Double-immunolabelling for GFP, GluA2 and NeuN (Neuronal nuclei). Plots represent quantification analysis of GluA2 signal intensity in HB9:GFP motor neurons at (U) E12.5 and (V) E17.5. Data represent mean ± SEM, unpaired student t-test performed on n = 4 biological replicates, ∼50 neurons analysed per biological replicate, *P < 0.05. Scale bars 50 μm.
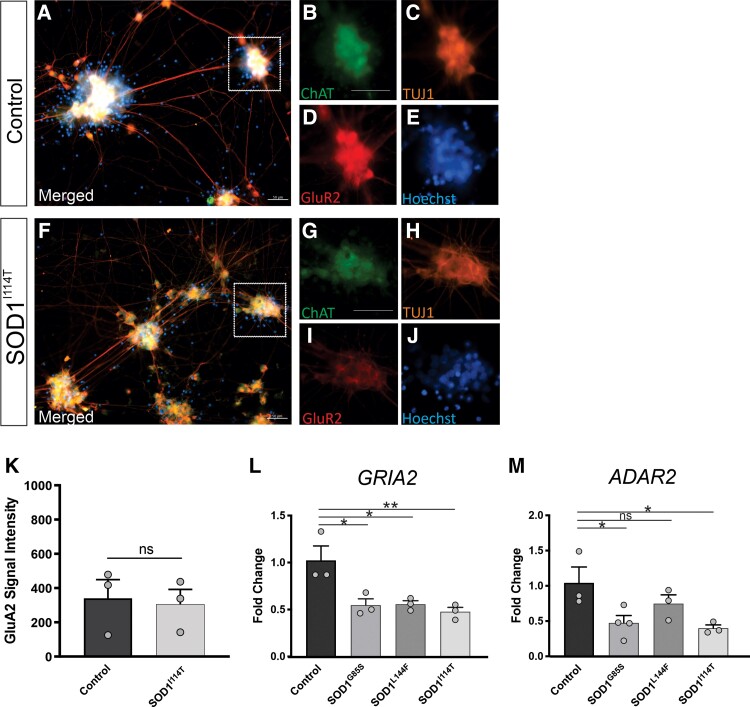
AMPA receptor subunit dysregulation occurs in ALS-linked SOD1 patient iPSC-derived motor neurons
To confirm our findings from the SOD1G93A mouse model, we cultured motor neurons from SOD1-linked ALS patient iPSCs. We used lines carrying different SOD1 point mutations generated in-house or previously established.26 In 28-day-old motor neurons, there was a trend for reduced GluA2 levels in mutant SOD1 motor neurons compared with control cells by immunocytochemistry, although not statistically significant (Fig. 5A–K), most likely due to the variation in differentiated human motor neurons, in contrast to motor neurons isolated from transgenic SOD1G93A mice which are more homogeneous. However, GRIA2 expression assessed by qPCR was significantly reduced in SOD1G85S (0.5 ± 0.1-fold, P < 0.01), SOD1L144F (0.5 ± 0.1-fold, P < 0.01) and SOD1I114T (0.6 ± 0.1-fold, P < 0.001), compared with control motor neurons (Fig. 5L). A significant reduction in ADAR2 expression was observed in SOD1G85S (0.6 ± 0.2-fold, P < 0.05) and SOD1I114T (0.6 ± 0.2-fold, P < 0.05), but not in SOD1L144F motor neurons, compared with control (Fig. 5M). Again, despite downregulation of ADAR2, mutant SOD1 motor neurons revealed normal GRIA2 editing (Supplementary Fig. 4). Hence, dysregulation of AMPA receptor signalling genes occurs in motor neurons of both SOD1-linked ALS patients and mice.
Figure 5.
Expression of GRIA2 and ADAR2 in iPSC motor neurons derived from ALS patients with SOD1 mutations and healthy control lines. Representative images of iPSC mature motor neurons derived from (A–E) healthy control line and (F–J) SOD1I114T line, immunolabelled with ChAT, GluA2 and TUJ1, counterstained with Hoechst. (K) Plot represents quantification analysis of GluA2 signal intensity in iPSC motor neurons. Data represent mean ± SEM, unpaired student t-test performed on n = 3 biological replicates, 50 neurons analysed per biological replicate. (L) Fold change expression of GRIA2 in SOD1 lines, compared with healthy control line determined by qRT-PCR. (M) Fold change expression of ADAR2 in SOD1 lines, compared with healthy control line determined by qRT-PCR. Data represent mean ± SEM, n = 3 biological replicates, one-way ANOVA with Dunnett's multiple comparison test, *P < 0.01, **P < 0.005. Scale bars 50 μm.
To functionally validate the impact of GRIA2 downregulation on survival of SOD1 patient-derived motor neurons, we next performed an excitotoxicity assay. Excitotoxicity was induced using the AMPA receptor-selective agonist AMPA. Exposure to AMPA resulted in higher levels of cell death in mutant SOD1 motor neurons (SOD1G85S, 46%; SOD1L144F, 48% and SOD1I114T, 44.5%), compared with control motor neurons (21.6%) (Fig. 6A–I). These findings suggest that SOD1 patient-derived motor neurons show increased vulnerability to AMPA-induced excitotoxicity through reduced GRIA2 expression which increases Ca2+ permeability.
Figure 6.
AMPA-induced excitotoxicity in SOD1 patient-derived motor neurons. Excitotoxity was induced in Day 28 motor neurons by 100 μM AMPA treatment for 24 h and cell viability was assessed by calcein red staining. Representative images of Calcein Red stained motor neurons derived from (A–B) healthy control or (C–H) mutant SOD1 lines before and after 24 h treatment with AMPA. (I) The bar graph represents percentage of cell death of each line following AMPA treatment. Data represent mean ± SEM, n = 3 biological replicates. One-way ANOVA with Dunnett’s multiple comparison test, *P < 0.01. Scale bars 50 μm.
RNA processing gene dysregulation in ALS-linked SOD1 patient iPSC-derived motor neurons
Dysregulated RNA processing genes were validated using qPCR in motor neurons from SOD1-linked ALS patient iPSCs. The expression of ANKRD16 and GAS5 genes was significantly reduced in SOD1G85S (0.5 ± 0.08-fold, P < 0.0005, 0.7 ± 0.1-fold, P < 0.0001, respectively), SOD1L144F (0.3 ± 0.08-fold, P < 0.009, 0.4 ± 0.06-fold, P < 0.0006, respectively) and SOD1I114T (0.6 ± 0.08-fold, P < 0.0002, 0.6 ± 0.06-fold, P < 0.0001, respectively), compared with control motor neurons (Fig. 7A–B). PNISR was downregulated in SOD1I114T (0.7 ± 0.2-fold, P < 0.04) but not in SOD1G85S and SOD1L144F motor neurons (Fig. 7C). A significant reduction in ISCA2 expression was also observed in SOD1G85S (0.6 ± 0.1-fold, P < 0.01) and SOD1I114T (0.7 ± 0.1-fold, P < 0.01) but not in SOD1L144F motor neurons (Fig. 7D). The expression of MEG3 was highly elevated in SOD1I114T (2036 ± 271-fold, P < 0.0002) but was not changed in SOD1G85S and SOD1L144F motor neurons (Fig. 7E). Finally, MALAT1 expression was not affected in mutant SOD1 lines, compared with control (Fig. 7F). Thus, dysregulation of RNA processing genes in motor neurons of SOD1G93A mice also occurs in SOD1-linked ALS patients.
Figure 7.
Expression of RNA processing genes in iPSC motor neurons derived from ALS patients with SOD1 mutations and healthy control lines. Fold change expression of (A) ANKRD16, (B) GAS5, (C) PNISR, (D) ISCA2, (E) MEG3 and (F) MALAT1 in SOD1 mutant lines, compared with healthy control line determined by qRT-PCR. Data represent mean ± SEM, n = 3 biological replicates. One-way ANOVA with Dunnett’s multiple comparison test, *P < 0.01, **P < 0.005, ***P < 0.0005, ****P < 0.0001. ns, not significant.
Discussion
Gene expression profiling studies of mouse models of ALS have focussed largely on adult symptomatic and presymptomatic16–18 or early postnatal ages.12 Although these studies provide molecular insights into potential mechanisms of motor neuron dysfunction and degeneration, they lack resolution of early molecular mechanisms that confer cell vulnerability and are likely to precede cell dysfunction. In this study, using a novel approach with a HB9:GFP reporter mouse, FACS and RNA sequencing, we have characterized the molecular signature of motor neurons from embryonic SOD1G93A mice. The approach we have used has several key advantages over previous gene expression profiling studies in ALS: (i) we have analysed gene expression changes specifically in motor neurons without glial contamination; (ii) we have assessed fetal gene expression changes and (iii) RNA sequencing allows unbiased screening of gene expression in SOD1G93A motor neurons.
To our knowledge, we present the first evidence for motor neuron gene dysregulation at E12.5 in a mouse model of ALS. This is a critical test of the hypothesis proposed by Eisen et al.13 and our findings provide support for motor neuron transcriptional dysregulation very early in the ALS pathogenic cascade. In a longitudinal microarray study by Saxena et al.12, ER stress markers and the unfolded protein response (UPR) in SOD1G93A mice were upregulated from P5. Our transcriptomic data reveal no dysregulation of genes involved in ER stress or protein misfolding management pathways, supporting the hypothesis that these pathways are in fact secondary events in ALS pathogenesis. This is corroborated by recent findings that genetic ablation of the key UPR sensor, PERK and downstream mediators GADD34 or CHOP, does not ameliorate the phenotype of SOD1G93A mice.36 Results from our study showed that most of the significantly changed genes (q < 0.05) were involved in RNA processing, followed by development-related genes, mitochondrial genes and AMPA receptor signalling.
The key finding of the present study was the downregulation of GluA2 subunit of AMPA receptors, at both transcript and protein levels, in embryonic motor neurons of SOD1G93A mice. Decreased expression of GluA2 may potentially confer selective cell vulnerability by allowing Ca2+ influx into motor neurons, leading to glutamate-mediated excitotoxicity,37 a proposed central pathogenic mechanism and treatment target in ALS. This was confirmed by the increased susceptibility of mutant SOD1 ALS patient iPSC-derived motor neurons to AMPA-induced excitotoxicity, which also demonstrated GluA2 downregulation. Our finding of dysregulated AMPA receptor subunit expression in SOD1-linked ALS patient-derived motor neurons is also consistent with motor neurons from C9orf72-linked ALS patients,38 suggesting a common early disease pathway in ALS. Indeed, genetic GluA2 deficiency or overexpression strongly modifies disease progression in SOD1G93A mice.37,39 Further, GluA2 deficiency promotes mutant SOD1 misfolding and aggregation in spinal cords of SOD1G93A mice39 and our findings place GluA2 dysregulation upstream of mutant SOD1 aggregation and cellular homeostatic responses, such as ER stress. Finally, riluzole which attenuates excitotoxicity remains the forefront approved drug for ALS, arguing for the primacy of excitotoxicity in ALS pathophysiology.
The reduced Ca2+ permeability of GluA2-containing AMPA receptors is due to the presence of arginine at position 586 (Q/R site) of the GluA2 protein, instead of the genetically encoded glutamine.40 This arginine residue at the Q/R site is introduced by the editing of Gria2 pre-mRNA, which is mediated by adenosine deaminase acting on RNA 2 (Adar2) and is critical to the survival of motor neurons.41,42 Here, we identified downregulation of Adar2, an enzyme that edits Gria2 transcripts, in motor neurons of SOD1G93A mice, although we did not detect inefficient editing of Gria2 in spinal motor neurons of embryonic SOD1G93A mice. Downregulation of ADAR2 and inefficient Gria2 RNA editing at Q/R site have been shown to occur in motor neurons of almost all ALS patients.43–45 However, RNA editing deficiencies at the Q/R site were not observed in SOD1G93A mice,46 concordant with our findings. Interestingly, selective Adar2 knockout in motor neurons leads to ALS-like symptoms and pathology in mice,47 supporting both Gria2 and Adar2 as susceptibility factors for motor neurons. Our study significantly advances these findings by placing Gria2 and Adar2 perturbations in motor neurons very early in the pathogenic cascade of ALS. We also demonstrated Gria2 and Adar2 transcript downregulation in iPSC-derived motor neurons from ALS patient carrying SOD1 point mutations which further validated our findings since these cells have a fetal transcriptome similar to mouse motor neurons at E12.5. Thus, transcriptional dysregulation in SOD1G93A mice shows high concordance with ALS patient-derived motor neurons, supporting the preclinical validity and usefulness of mutant SOD1 mice.
In this study, we also found Mir-124-1 downregulation in embryonic SOD1G93A motor neurons and computational identification of a conserved miR-124-1 site in the 3′-UTR of Gria2 gave rise to the hypothesis that Mir-124-mediated regulation of Gria2 could produce rapid changes in GluA2 expression. However, our results conflict with previous published reports of an inverse correlation of Mir124-1 and Gria2 levels.48 How Mir-124 regulates the expression of Gria2 in motor neurons and the exact molecular link to mutant SOD1 remains to be elucidated.
A novel finding of our study was dysregulation of RNA processing genes, particularly Pnisr, in mutant SOD1 motor neurons at E12.5 as confirmed by qPCR. Further studies are required to uncover the exact functional role and the downstream effect of Pnisr dysregulation in SOD1G93A mice. Dysregulation of RNA processing genes was further validated using qPCR in ALS patient iPSC-derived motor neurons. A significant downregulation of ANKRD16, GAS5, PNISR and ISCA2 genes were identified in SOD1 lines relative to controls, consistent with RNA-seq findings. RNA dysregulation is a key contributor to ALS pathogenesis. The major ALS-causative gene mutations in TDP-43, FUS and C9orf72 are involved in aspects of RNA metabolism processes, such as mRNA transcription, alternative splicing, RNA transport, mRNA stabilization and miRNA biogenesis.49 Evidence of RNA processing dysregulation in presymptomatic SOD1G93A mice has not been shown previously; however, using a combination of laser-capture microdissection and microarray analysis, downregulation of mRNA processing genes in SOD1G93A motor neurons at P90 was identified.18 Our findings, therefore, support an early and key role of RNA processing gene dysregulation in SOD1-linked ALS.
In summary, this study provides the earliest evidence of transcriptional dysregulation occurring in affected motor neurons of SOD1G93A mice in utero, supporting a potential perinatal origin of ALS as proposed by others.13 Our data highlight dysregulation of AMPA receptor Ca2+ permeability and RNA processing as very early and intrinsic events in the pathogenic cascade of mutant SOD1 (Fig. 8). Based on our evidence for AMPA receptor dysregulation in embryonic motor neurons of SOD1G93A mice and SOD1-linked ALS patient-derived motor neurons, we propose that aberrant Ca2+-mediated influx and signalling are proximal and key events, leading to downstream consequences such as hyperexcitability, excitotoxicity, ER stress and protein aggregation which may account for selective motor neuron death in ALS.
Figure 8.
Temporal sequence of major dysregulated cellular mechanisms in spinal motor neurons of SOD1G93A mice. In SOD1G93A mice, the onset of clinical symptoms occurs at postnatal (P) Day 90. The key pathological, subcellular and molecular events previously reported are summarized in the above timeline. In this study, we identified the earliest known dysregulated mechanisms as shown at embryonic (E) Day 12.5 that may confer motor neuron vulnerability, involving dysfunction of RNA processing, mitochondria and AMPA receptor-mediated excitotoxicity. Mito, mitochondria; NMJ, neuromuscular junction.
Supplementary Material
Acknowledgements
The authors thank Dr Kevin Eggan for the ALS patient iPSC lines, Dr Vickie Perreau, Brittany Cuic and Nayomi Wanniarachchillage for their expert technical assistance and acknowledge the support of the Flow Cytometry Facility at the Murdoch Children’s Research Institute and Micromon Genomics Facility.
Abbreviations
- ADAR2 =
adenosine deaminase acting on RNA 2
- ALS =
amyotrophic lateral sclerosis
- AMPA =
α-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate
- E =
embryonic day
- ER =
endoplasmic reticulum
- FACS =
fluorescence-activated cell sorting
- GluA2 =
AMPA receptor 2 subunit
- GFP =
green fluorescent protein
- Gria2 =
glutamate ionotropic receptor AMPA type Subunit 2
- HB9 =
homeobox HB9
- iPSC =
induced pluripotent stem cell
- mito =
mitochondrial
- P =
postnatal day
- PCA =
principal component analysis
- RFLP =
restriction fragment length polymorphism
- SEM =
standard error of the mean
- SOD1 =
superoxide dismutase 1
- WT =
wild-type
Funding
P.F.Z. was supported by a University of Melbourne International Research Scholarship. C.R.B. was supported by an Australian National Health and Medical Research Council Early Career Fellowship. B.J.T. was supported by an Australian National Health and Medical Research Council—Australian Research Council (NHMRC-ARC) Dementia Research Leadership Fellowship (1137024). S.K.B. was supported by an Australian NHMRC-ARC Dementia Research Development Fellowship (1110040). This research was funded by Motor Neurone Disease Research Australia (MND Benalla Act to D’feet MND Research Grant), FightMND Drug Screening Programme and the Stafford Fox Medical Research Foundation (B.J.T.). The Florey Institute of Neuroscience and Mental Health acknowledges the strong support from the Victorian Government and in particular the funding from the Operational Infrastructure Support Grant.
Competing interests
The authors declare no competing interests associated with this work.
Supplementary material
Supplementary material is available at Brain Communications online.
References
- 1. Taylor JP, Brown RH Jr, Cleveland DW. Decoding ALS: From genes to mechanism. Nature. 2016;539(7628):197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cook C, Petrucelli L. Genetic convergence brings clarity to the enigmatic red line in ALS. Neuron. 2019;101(6):1057–1069. [DOI] [PubMed] [Google Scholar]
- 3. Turner BJ, Talbot K. Transgenics, toxicity and therapeutics in rodent models of mutant SOD1-mediated familial ALS. Prog Neurobiol. 2008;85(1):94–134. [DOI] [PubMed] [Google Scholar]
- 4. Aggarwal A, Nicholson G. Detection of preclinical motor neurone loss in SOD1 mutation carriers using motor unit number estimation. J Neurol Neurosurg Psychiatry. 2002;73(2):199–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vucic S, Nicholson GA, Kiernan MC. Cortical hyperexcitability may precede the onset of familial amyotrophic lateral sclerosis. Brain J Neurol. 2008;131(Pt 6):1540–1550. [DOI] [PubMed] [Google Scholar]
- 6. Fischer LR, Culver DG, Tennant P, et al. Amyotrophic lateral sclerosis is a distal axonopathy: Evidence in mice and man. Exp Neurol. 2004;185(2):232–240. [DOI] [PubMed] [Google Scholar]
- 7. Gould TW, Buss RR, Vinsant S, et al. Complete dissociation of motor neuron death from motor dysfunction by Bax deletion in a mouse model of ALS. J Neurosci. 2006;26(34):8774–8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mourelatos Z, Gonatas NK, Stieber A, Gurney ME, Dal Canto MC. The Golgi apparatus of spinal cord motor neurons in transgenic mice expressing mutant Cu, Zn superoxide dismutase becomes fragmented in early, preclinical stages of the disease. Proc Natl Acad Sci U S A. 1996;93(11):5472–5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bendotti C, Calvaresi N, Chiveri L, et al. Early vacuolization and mitochondrial damage in motor neurons of FALS mice are not associated with apoptosis or with changes in cytochrome oxidase histochemical reactivity. J Neurol Sci. 2001;191(1–2):25–33. [DOI] [PubMed] [Google Scholar]
- 10. Fogarty MJ, Mu EWH, Lavidis NA, Noakes PG, Bellingham MC. Motor areas show altered dendritic structure in an amyotrophic lateral sclerosis mouse model. Front Neurosci. 2017;11:609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Saxena S, Roselli F, Singh K, et al. Neuroprotection through excitability and mTOR required in ALS motoneurons to delay disease and extend survival. Neuron. 2013;80(1):80–96. [DOI] [PubMed] [Google Scholar]
- 12. Saxena S, Cabuy E, Caroni P. A role for motoneuron subtype-selective ER stress in disease manifestations of FALS mice. Nat Neurosci. 2009;12(5):627–636. [DOI] [PubMed] [Google Scholar]
- 13. Eisen A, Kiernan M, Mitsumoto H, Swash M. Amyotrophic lateral sclerosis: A long preclinical period? J Neurol Neurosurg Psychiatry. 2014;85(11):1232–1238. [DOI] [PubMed] [Google Scholar]
- 14. Kuo JJ, Schonewille M, Siddique T, et al. Hyperexcitability of cultured spinal motoneurons from presymptomatic ALS mice. J Neurophysiol. 2004;91(1):571–575. [DOI] [PubMed] [Google Scholar]
- 15. Kieran D, Hafezparast M, Bohnert S, et al. A mutation in dynein rescues axonal transport defects and extends the life span of ALS mice. J Cell Biol. 2005;169(4):561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lobsiger CS, Boillee S, Cleveland DW. Toxicity from different SOD1 mutants dysregulates the complement system and the neuronal regenerative response in ALS motor neurons. Proc Natl Acad Sci U S A. 2007;104(18):7319–7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Perrin FE, Boisset G, Docquier M, Schaad O, Descombes P, Kato AC. No widespread induction of cell death genes occurs in pure motoneurons in an amyotrophic lateral sclerosis mouse model. Hum Mol Genet. 2005;14(21):3309–3320. [DOI] [PubMed] [Google Scholar]
- 18. Ferraiuolo L, Heath PR, Holden H, Kasher P, Kirby J, Shaw PJ. Microarray analysis of the cellular pathways involved in the adaptation to and progression of motor neuron injury in the SOD1 G93A mouse model of familial ALS. J Neurosci. 2007;27(34):9201–9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wichterle H, Lieberam I, Porter JA, Jessell TM. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110(3):385–397. [DOI] [PubMed] [Google Scholar]
- 20. Untergasser A, Cutcutache I, Koressaar T, et al. Primer3–new capabilities and interfaces. Nucleic Acids Res. 2012;40(15):e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37(8):907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liao Y, Smyth GK, Shi W. The subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41(10):e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. [DOI] [PubMed] [Google Scholar]
- 26. Boulting GL, Kiskinis E, Croft GF, et al. A functionally characterized test set of human induced pluripotent stem cells. Nat Biotechnol. 2011;29(3):279–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Du Z-W, Chen H, Liu H, et al. Generation and expansion of highly pure motor neuron progenitors from human pluripotent stem cells. Nat Commun. 2015;6(1):6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lu DC, Niu T, Alaynick WA. Molecular and cellular development of spinal cord locomotor circuitry. Front Mol Neurosci. 2015;8:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nornes HO, Carry M. Neurogenesis in spinal cord of mouse: An autoradiographic analysis. Brain Res. 1978;159(1):1–16. [DOI] [PubMed] [Google Scholar]
- 30. Wright A, Vissel B. The essential role of AMPA receptor GluR2 subunit RNA editing in the normal and diseased brain. Front Mol Neurosci. 2012;5:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gascon E, Lynch K, Ruan H, et al. Alterations in microRNA-124 and AMPA receptors contribute to social behavioral deficits in frontotemporal dementia. Nat Med. 2014;20(12):1444–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smith BN, Ticozzi N, Fallini C, et al. Exome-wide rare variant analysis identifies TUBA4A mutations associated with familial ALS. Neuron. 2014;84(2):324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kwak S, Kawahara Y. Deficient RNA editing of GluR2 and neuronal death in amyotropic lateral sclerosis. J Mol Med (Berl). 2005;83(2):110–120. [DOI] [PubMed] [Google Scholar]
- 34. Yamashita T, Kwak S. The molecular link between inefficient GluA2 Q/R site-RNA editing and TDP-43 pathology in motor neurons of sporadic amyotrophic lateral sclerosis patients. Brain Res. 2014;1584:28–38. [DOI] [PubMed] [Google Scholar]
- 35. Paschen W, Schmitt J, Uto A. RNA editing of glutamate receptor subunits GluR2, GluR5 and GluR6 in transient cerebral ischemia in the rat. J Cereb Blood Flow Metab. 1996;16(4):548–556. [DOI] [PubMed] [Google Scholar]
- 36. Dzhashiashvili Y, Monckton CP, Shah HS, Kunjamma RB, Popko B. The UPR-PERK pathway is not a promising therapeutic target for mutant SOD1-induced ALS. Neurobiol Dis. 2019;127:527–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Van Damme P, Braeken D, Callewaert G, Robberecht W, Van Den Bosch L. GluR2 deficiency accelerates motor neuron degeneration in a mouse model of amyotrophic lateral sclerosis. J Neuropathol Exp Neurol. 2005;64(7):605–612. [DOI] [PubMed] [Google Scholar]
- 38. Selvaraj BT, Livesey MR, Zhao C, et al. C9ORF72 repeat expansion causes vulnerability of motor neurons to Ca2+-permeable AMPA receptor-mediated excitotoxicity. Nat Commun. 2018;9(1):347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tateno M, Sadakata H, Tanaka M, et al. Calcium-permeable AMPA receptors promote misfolding of mutant SOD1 protein and development of amyotrophic lateral sclerosis in a transgenic mouse model. Hum Mol Genet. 2004;13(19):2183–2196. [DOI] [PubMed] [Google Scholar]
- 40. Verdoorn TA, Burnashev N, Monyer H, Seeburg PH, Sakmann B. Structural determinants of ion flow through recombinant glutamate receptor channels. Science. 1991;252(5013):1715. [DOI] [PubMed] [Google Scholar]
- 41. Burnashev N, Monyer H, Seeburg PH, Sakmann B. Divalent ion permeability of AMPA receptor channels is dominated by the edited form of a single subunit. Neuron. 1992;8(1):189–198. [DOI] [PubMed] [Google Scholar]
- 42. Sommer B, Köhler M, Sprengel R, Seeburg PH. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67(1):11–19. [DOI] [PubMed] [Google Scholar]
- 43. Hideyama T, Yamashita T, Aizawa H, et al. Profound downregulation of the RNA editing enzyme ADAR2 in ALS spinal motor neurons. Neurobiol Dis. 2012;45(3):1121–1128. [DOI] [PubMed] [Google Scholar]
- 44. Kawahara Y, Ito K, Sun H, Aizawa H, Kanazawa I, Kwak S. Glutamate receptors: RNA editing and death of motor neurons. Nature. 2004;427(6977):801. [DOI] [PubMed] [Google Scholar]
- 45. Aizawa H, Hideyama T, Yamashita T, et al. Deficient RNA-editing enzyme ADAR2 in an amyotrophic lateral sclerosis patient with a FUS(P525L) mutation. J Clin Neurosci. 2016;32:128–129. [DOI] [PubMed] [Google Scholar]
- 46. Kawahara Y, Sun H, Ito K, et al. Underediting of GluR2 mRNA, a neuronal death inducing molecular change in sporadic ALS, does not occur in motor neurons in ALS1 or SBMA. Neurosci Res. 2006;54(1):11–14. [DOI] [PubMed] [Google Scholar]
- 47. Hideyama T, Yamashita T, Suzuki T, et al. Induced loss of ADAR2 engenders slow death of motor neurons from Q/R site-unedited GluR2. J Neurosci. 2010;30(36):11917–11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ho VM, Dallalzadeh LO, Karathanasis N, et al. GluA2 mRNA distribution and regulation by miR-124 in hippocampal neurons. Mol Cell Neurosci. 2014;61:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Butti Z, Patten SA. RNA dysregulation in amyotrophic lateral sclerosis. Front Genet. 2019;9:712. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data generated from this study have been deposited in GEO accession number GSE142654.