Summary
Mucormycosis is a lethal and emerging disease that has lacked a genetic model fulfilling both high virulence and the possibility of performing stable and reproducible gene manipulation by homologous recombination (HR). Here, we developed a new methodology to successfully perform HR in Rhizopus microsporus. We isolated an uracil auxotrophic recipient strain and optimized the critical steps in the genetic transformation of this fungus. This was followed by an adaptation of a plasmid-free CRISPR-Cas9 system coupled with microhomology repair templates. We reproducibly generated stable mutants in the genes leuA and crgA, encoding a 3-isopropylmalate dehydratase and an ubiquitin ligase, respectively. Our new genetic model showed that mutations in the gene pyrF, a key virulence gene in several bacterial and fungal pathogens, correlated with an avirulent phenotype in an immunocompetent murine host. This was reverted by gene complementation, showing the broad possibilities of our methodology.
Keywords: mucormycosis, Mucorales, homologous recombination, genetic model, Rhizopus microsporus, CRISPR, virulence, pyrF, leuA, crgA
Graphical abstract
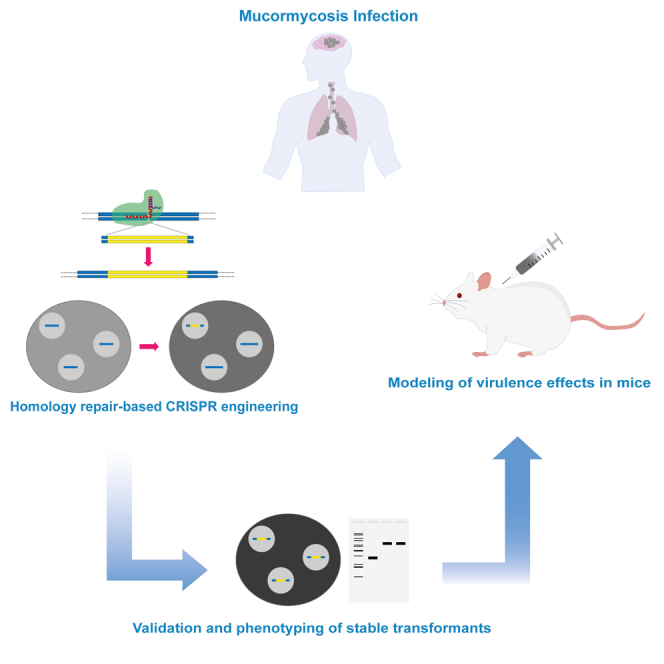
Highlights
-
•
Isolates and characterizes a mutagen-free recipient R. microsporus strain
-
•
Establishes and optimizes the conditions for protoplast transformation
-
•
Establishes a CRISPR-based procedure to generate stable homokaryotic mutant strains
-
•
Tests the virulence of the generated mutant strains in immunocompetent mice
Motivation
Due to increasing cases of the fatal fungal disease mucormycosis, including cases associated with COVID-19 patients, there is an urgent need to improve genetically tractable models of the causative fungal species. Genetic manipulation tools are available in Mucor lusitanicus, but this species shows low virulence and is therefore not the ideal model. Our goal was to develop tools for genetic manipulation of Rhizopus microsporus, because it is a virulent and frequently causative agent of mucormycosis and has also been used to model interactions between Mucorales and endobacteria. With our methodology, which combines the use of CRISPR-Cas9 and microhomology DNA templates, we have achieved stable targeted integrations by homologus recombination. This has allowed us to analyze the first visual phenotypes in R. microsporus caused by gene disruption and to discover a possible role for the pyrF gene in virulence. Considering these newly developed genetic tools, we propose R. microsporus as a model to study virulence and molecular genetics in the mucormycosis field.
Lax et al. establish a procedure for genetic manipulation of the fungus Rhizopus microsporus, one of the main causal agents of mucormycosis. Stable homologous recombination enables editing through CRISPR-Cas9 and microhomology repair templates. Stable, homokaryotic mutants are generated and tested with virulence assays in immunocompromised mice.
Introduction
The Mucorales is a group of early-diverging fungi with many distinct and unique features that has been scarcely studied due to the reluctance to be genetically transformed (Obraztsova et al., 2004). The interest in the study of Mucorales has increased because of the renewed emergence of the fungal infectious disease known as mucormycosis. Mucormycosis is a lethal disease caused by several mucoralean species, being the most frequent among the genus Rhizopus, followed by Mucor and Lichtheimia (Alvarez et al., 2009; Cornely et al., 2019; Roden et al., 2005). In the past, mucormycosis was considered a rare infection affecting immunosuppressed and otherwise compromised patients. However, new clinical reports and improvements in the correct diagnosis of mucormycosis have shown an emerging increase in the number of cases (Chayakulkeeree et al., 2006; Kontoyiannis, 2017). Indeed, the increased incidence of mucormycosis in COVID-19 patients associated with steroid treatment has arisen the concerns of the scientific and clinical community to treat infections caused by the named black fungus (John et al., 2021; Veisi et al., 2021). More importantly, an escalating number of mucormycosis cases has been reported in healthy patients without known predisposing diseases (Prakash and Chakrabarti, 2019; Sridhara et al., 2005). Mucormycosis has mortality rates that can reach 90% in the cases of bloodstream-disseminated infection (Hassan and Voigt, 2019; Jeong et al., 2019). These high mortality rates are mainly due to the innate antifungal drug resistance of Mucorales, which leaves clinicians with a few poorly effective treatments against mucormycosis (Caetano et al., 2019; Caramalho et al., 2017; Cornely et al., 2019; Dannaoui, 2017; Luo et al., 2013; Maurer et al., 2015). In addition, Mucorales can rapidly acquire new antifungal drug resistance through an exclusive RNAi-based mechanism to quickly and temporally generate resistant epimutants (Calo et al., 2014). In this sense, most of the current studies in Mucorales are focused on investigating new genes, pathways, methodologies, and virulence factors that might be the targets for future antifungal developments against mucormycosis (Binder et al., 2018; Gebremariam et al., 2019; Lax et al., 2020; López-Fernández et al., 2018; López-Muñoz et al., 2018; Navarro-Mendoza et al., 2018; Pérez-Arques et al., 2019, 2021a, 2021b; Trieu et al., 2017).
In the study of mucormycosis, the difficulty of the genetic transformation and manipulation of the Mucorales species has a remarkable exception: the fungus Mucor lusitanicus, previously known as Mucor circinelloides f. lusitanicus (Wagner et al., 2020). Hundreds of genes have been successfully interrupted or replaced by homologous recombination (HR) in this fungus. In addition, other genetic tools such as plasmid transformation, RNAi induction, genetic complementation, directed mutagenesis, and gene tagging are also available in M. lusitanicus (Navarro-Mendoza et al., 2019; Nicolas et al., 2003; Trieu et al., 2015, 2017). These tools have allowed the genetic study of many cellular processes in Mucorales, including the light responses, the RNAi mechanism, and more recently, the Mucorales pathogenesis. Unfortunately, this fungal model shows very low virulence, and infection assays only work in few specific murine strains after strong immunosuppression (Li et al., 2011) or drug-induced diabetes (Valle-Maldonado et al., 2020). In this sense, the genetic manipulation reluctance of Mucorales is limiting the research community to study mucormycosis.
Our purpose in this work is to overcome this limitation by creating a new methodology that allows stable and reproducible genetic manipulation in a Mucoral different from the Mucor species. We selected Rhizopus microsporus as a candidate model because it is one of the most frequent causal agents of mucormycosis and a well-known model to study the interaction of Mucorales with endobacteria (Mondo et al., 2017; Prakash and Chakrabarti, 2019; Skiada et al., 2011). We have optimized all the critical steps for transforming and achieving stable and reproducible genetic disruption of R. microsporus genes. Thus, we interrupted the gene leuA, creating a leucine auxotrophic strain, and crgA, which resulted in a pleiotropic phenotype that affected development of aerial hyphae and melanin overaccumulation, resembling the pleiotropic phenotype observed in the M. lusitanicus crgA mutant, suggesting conservation of the crgA function in Mucorales. Finally, we established a virulence assay that uses immunocompetent mice that revealed that uracil biosynthesis is essential for full virulence in R. microsporus. This work may be a milestone in the study of mucormycosis, as it provides a new study model that combines complete virulence robustness and capacity for proper genetic manipulation.
Results
Spontaneous uracil auxotrophic strain isolation
The first step for developing a transformation and gene disruption procedure for R. microsporus was to isolate a recipient strain with a reduced mutational burden by selecting spontaneous uracil auxotrophs. The complementation of uracil auxotrophy has been extensively used as a selective marker in the transformation of other fungi, including Mucorales (Gutiérrez et al., 2011; Ibragimova et al., 2020; Ibrahim et al., 2007; Nosheen et al., 2021). To select spontaneous mutants in uracil biosynthesis (Figure S1A), spores of the wild-type (WT) strain ATCC 11559 were inoculated in liquid-rich media (YPG) supplemented with 5-Fluoroorotic acid (5-FOA) and with uridine. The uracil biosynthesis pathway converts 5-FOA into 5-fluorouracil, a toxic compound that inhibits growth. After 48 h of growth, the cells were plated in solid YPG media, also supplemented with 5-FOA and uridine. Twenty 5-FOA-resistant colonies were isolated (R1–R20) and inoculated in both minimal medium (YNB) with and without uridine to check uracil auxotrophy. Isolates that showed strong growth in 5-FOA and no-growth in YNB without uridine were selected as uracil auxotrophs (Figures S1A and S1B). Mutations in either the pyrG gene encoding the orotidine 5′-phosphate decarboxylase (JGI: 279857) or the pyrF gene encoding the orotate phosphoribosyl transferase (JGI: 231940) block the uracil biosynthesis (Boeke et al., 1987; Bruni et al., 2019; Ibragimova et al., 2020). Sequencing of both genes from four different auxotrophs (R1, R3, R5, and R8) revealed that two of them (R3 and R5) had a mutation in the pyrF gene, but not in the pyrG gene. The other two, R1 and R8, have no mutations in the pyrG or the pyrF genes, suggesting the presence of mutations in other genes involved in the uracil metabolism. The R3 and R5 strains carry a nucleotide substitution that results in a nonsynonymous change in position 73 (K73E) (Figure S1C), suggesting that they both derive from the same initial auxotroph. Position 73 is part of the enzyme active site (AYKG) that interacts with the substrate α-D-5-phosphoribosyl-1-pyrophosphate (PRPP) (Figures S1D and S1E) (Scapin et al., 1995), and its mutation in the Salmonella typhimurium enzyme produced a 50–100-fold decrease in Kcat and an 8–12-fold increase in the PRPP KM (Ozturk et al., 1995). Based on these results, we selected the R3, thereafter named UM1, as the recipient strain for further experiments.
Optimized conditions for protoplast generation
A transformation protocol for Mucorales requires to determine the optimal germination stage to enhance the action of the lytic enzymes and the survival of the resulting protoplasts. Fresh spores of R. microsporus were grown in YPG medium supplemented with uridine at different temperatures in a rotary shaker (Figure S2). Spores swelled to approximately double their size, from 4.90 ± 0.62 to 9.39 ± 0.98 μm, and generally sprouted 1–3 germ tubes per spore in the early stages. The germination stage is optimal when spores are completely swollen, and they start emitting the germ tubes (Figure S2A, framed in blue) (Nicolás et al., 2018). The time required for germination decreased as the temperature increased (Figure S2). Although germination was faster at 37°C (between 3 and 3.5 h), the viability of the protoplasts was more affected at this temperature than 26°C and 33°C, as determined by direct protoplast observation and preliminary viability tests (Figure S2B). Therefore, 33°C and 4.5 h were selected as the germination conditions for further experiments. Following the procedures established in other fungi, we tested increasing concentrations of a combination of lysing enzymes from Trichoderma harzianum and chitosanase from Streptomyces griseus until convenient lysis was observed, when cells were subjected to a hypotonic shock (de Bekker et al., 2009; Gruber et al., 1990; Gutiérrez et al., 2011). We concluded that with a combination of 3 mg of lysing enzymes and 0.0008 units of chitosanase per 1 × 107 spores, the cell wall degradation was optimal, and the protoplast viability was not exceedingly affected, as we ascertained in the next steps.
Electroporation and efficient transformation of R. microsporus protoplasts
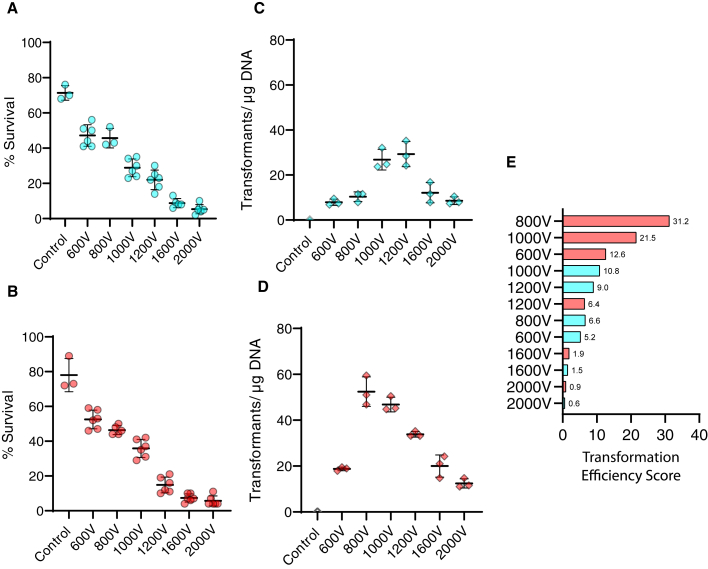
To establish the electroporation parameters, protoplasts of the uracil auxotroph UM1 were transformed with a self-replicative plasmid containing R. microsporus pyrF gene (pMAT1819). We tested two different electroporation methods: exponential decay pulse (ED) and time constant pulse (TC). ED starts with the set voltage and rapidly decays (exponentially) to zero. This method was successfully applied to M. lusitanicus and several other fungal species (Escoffre et al., 2009; Gutiérrez et al., 2011). On the other hand, the TC method supposes a continuous pulse of a set voltage during a set time (Escoffre et al., 2009). The survival rate and the transformation efficiency were tested at 600, 800, 1000, 1200, 1600, and 2000 V for both ED and TC pulses. For TC pulses, time was set to 5 milliseconds (ms), because preliminary tests with different time pulses showed that the highest transformation efficiency was in the range of 5–7 ms (Figure S3). For ED pulses, capacitance and resistance were set to 25 μF and 400 Ω, respectively, following the well-established protocol for M. lusitanicus (Gutiérrez et al., 2011). The results showed decreased survival rates for both ED and TC pulses as the voltage increased (Figures 1A and 1B). Survival rates ranged from 40% to 60% (compared with the negative control) at lower voltages (600–800 V) to ∼10% for higher voltages (1600–2000 V). In ED pulses, the highest transformation rates were obtained at 1000–1200 V with approximately 30 transformants/μg of DNA, whereas higher and lower voltages produced less than 20 transformants/μg of DNA (Figure 1C).
Figure 1.
Transformation efficiency and survival rate of electroporated protoplast of the UM1 strain
(A and B) The survival rate of the UM1 protoplast after electroporation after different ED (A) and TC pulses (B). Data is represented as mean ± SD. Each point corresponds to a different plate with 200 protoplasts (2 plates/cuvette).
(C and D) Transformation rate obtained after ED (C) and TC pulses (D). Data is represented as mean ± SD. Each point corresponds to a different electroporation cuvette.
(E) Transformation efficiency score was obtained as the product of the average transformants/μg of DNA and the average survival rate (normalized considering 1 as the value of the survival rate of the negative control).
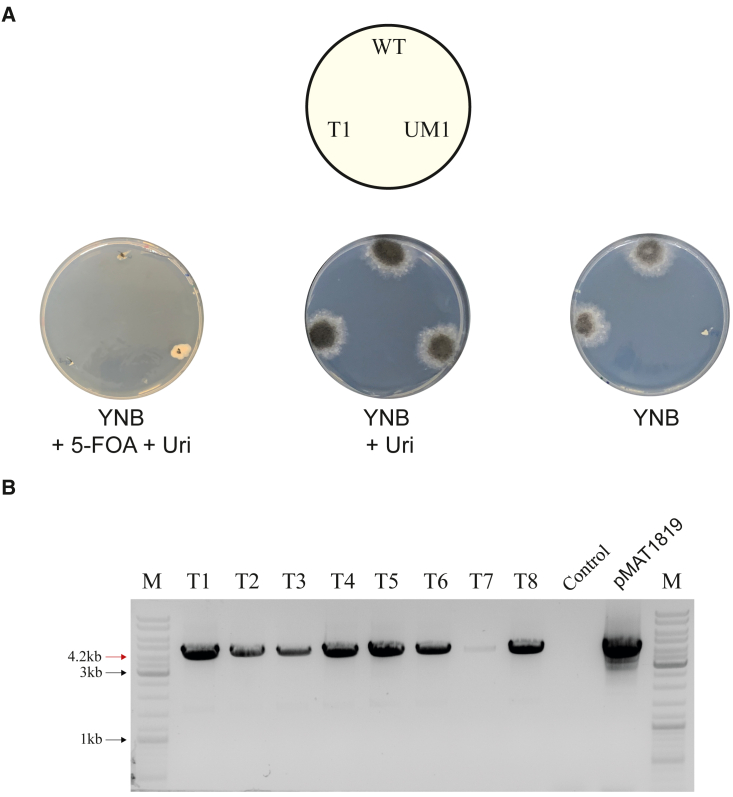
Interestingly, for TC pulses, a higher transformation rate was obtained, especially at 800 V and 1000 V, which nearly doubled the best of the ED pulses with approximately 50 transformants/μg of DNA (Figure 1D). Analysis of the transformation efficiency scores revealed that the 800 V TC pulse was the best condition. The transformants obtained were able to grow without uridine supplementation in minimal media but unable to grow in the presence of 5-FOA, like the WT strain and unlike the recipient strain UM1 (Figure 2A). The presence of the plasmid in the transformants, verified by PCR analysis in eight randomly selected transformants (Figure 2B), confirmed the success of this transformation method.
Figure 2.
Characterization of the transformants generated with the pMAT1819 plasmid
(A) Growth of the R. microsporus wild type strain ATCC 11559 (WT), UM1 strain, and a UM1 transformant-carrying plasmid pMAT1819 that complemented uracil auxotrophy (T1) on YNB medium supplemented with 5-FOA (3 g/L) and uridine (200 mg/L) (left), YNB medium supplemented with uridine (200 mg/L) (middle), and YNB medium (right).
(B) DNA was extracted from eight randomly selected transformants (T1–T8) and amplified by PCR using the M13_F and M13_R primers that bind to the vector sequence flanking the pyrF gene in plasmid pMAT1819. PCR amplification with the same primers of DNA from the untransformed UM1 strain and the purified plasmid served as negative and positive controls, respectively. Red arrow points to the expected amplification fragment. Black arrows point to the indicated bands of the marker.
Stable disruption of crgA and leuA genes by CRISPR-Cas9 and HR
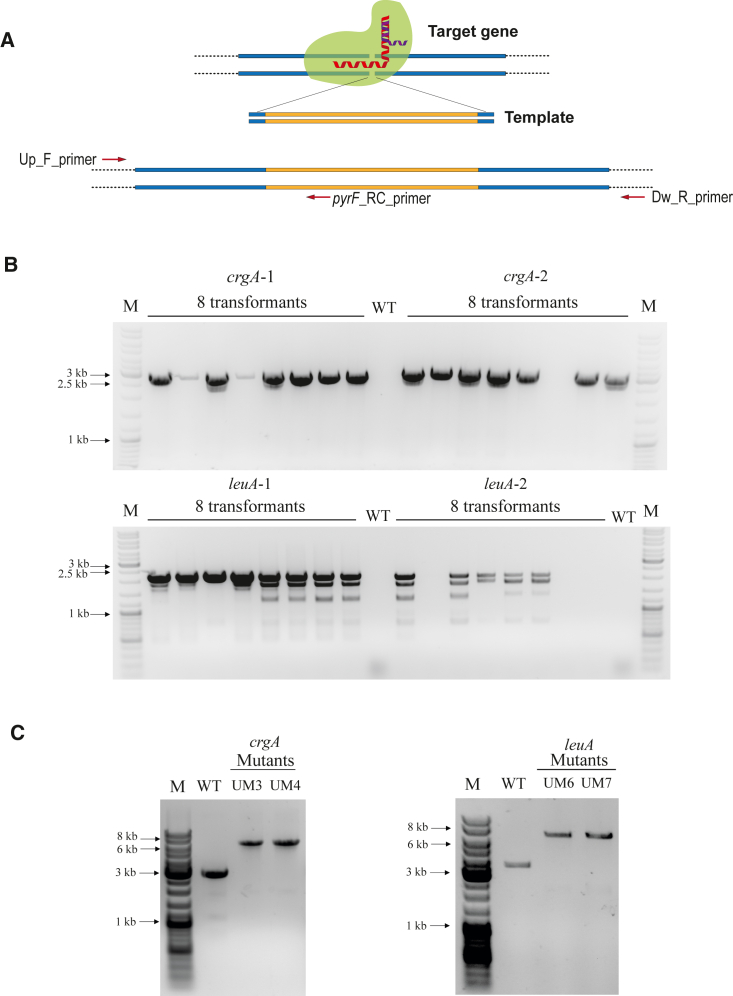
Once R. microsporus transformation using a plasmid was achieved, we set up a procedure to disrupt target genes using a plasmid-free CRISPR-Cas9 system and HR-mediated DNA repair (Figure 3A) with microhomology templates (from 30 to 50 base pairs [bp]). These templates were generated by PCR amplification of the pyrF locus with primers that included short tails corresponding to sequences flanking the cleavage site of a guided Cas9. For this approach, we reduced the pyrF gene length from 4.2 to 3.4 kb because the transformation rate was unaffected in transformations with self-replicative plasmids.
Figure 3.
Disruption of crgA and leuA genes
(A) Strategy followed to disrupt target genes with the combination of gRNA-Cas9 complex and a designed DNA template. Positions of the primers used to check gene disruption and homokaryosis are also shown.
(B) Gene disruption validation by PCR using the Up_F_primer and the pyrF_RC_primer for eight randomly selected transformants obtained with crgA_gRNA1 + crgATemplate1 (expected fragment of 2.8 kb), crgA_gRNA2 + crgATemplate2 (expected fragment of 2.9 kb), leuA_gRNA1 + leuATemplate1 (expected fragment of 2.1 kb), and leuA_gRNA2 + leuATemplate2 (expected fragment of 2.2 kb). DNA from the R. microsporus wild-type strain ATCC 11559 was used as a negative control (WT).
(C) PCR fragments smaller than the ones expected in some lanes of the leuA transformants corresponded to nonspecific PCR products. (C) PCR amplification of the crgA and leuA locus of the indicated transformants using the Up_F_primer and the Dw_R_primer. Expected PCR DNA fragments from WT and disrupted crgA locus were 3.1 kb and 6.6 kb in length, respectively. For leuA, expected PCR products from WT and disrupted locus were 3.6 kb and 7.1 kb in length, respectively. Black arrows point to the indicated bands of the marker.
We tested this strategy by disrupting two different genes: crgA (JGI: 226533) and leuA (JGI: 228594). crgA is the predicted homolog to the M. lusitanicus crgA gene, which encodes a ubiquitin ligase that represses carotenogenesis, sporangiophore length, and sporulation by inactivating an activator of the light response of this fungus (Navarro et al., 2013; Nicolas et al., 2008; Silva et al., 2008). For its part, leuA is the predicted homolog to M. lusitanicus leuA gene that codes for a 3-isopropylmalate dehydratase, involved in leucine biosynthesis (Roncero et al., 1989). We designed two different guide RNAs (gRNAs) (gRNA1 and gRNA2) for each gene, targeting different sequences, and the corresponding HR templates (Table S1). These templates consisted of the 3.4-kb pyrF gene flanked by 38-bp sequences complementary to the 5′ and 3′ regions adjacent to the predicted Cas9 cleavage site. In vitro assembled ribonucleoprotein (RNP) complexes formed by the Cas9 and a gRNA, together with the corresponding templates, were used to transform the UM1 strain. The transformation efficiency was reduced compared with plasmid transformation and varied among the different combinations of gRNA and templates (Table 1). In vitro cleavage activity assays of each gRNA-Cas9 and template combination revealed that transformation efficiency correlates with in vitro cleavage efficiencies (Figure S4). Thus, when the Cas9 loaded with leuA_gRNA1 was incubated with a leuA fragment, the amount of uncut DNA was higher than when leuA_gRNA2 was used. Correspondingly, this second gRNA yielded approximately double the number of transformants/μg of DNA (Table 1). To confirm a proper pyrF integration at the target site, we analyzed the targeted loci of 16 randomly selected transformants obtained from each transformation (32 for each gene) by PCR using the gene-specific Up_F_primer and pyrF_R_primer. Thus, a total of 64 transformants were checked for integration in the targeted regions (Figures 3A and 3B show representative gels with eight transformants for each gRNA). The correct integration ranges from 62.5% to 96.9% depending on the gRNA (Table 1), proving the high reliability of this method to perform targeted integration and gene disruption. As R. microsporus has multinucleated spores, initial transformants were expected to be heterokaryons, and therefore, two initial transformants for the disruption of crgA and leuA genes (UM3 and UM4 for crgA and UM6 and UM7 for leuA) were grown for 4–5 vegetative cycles in minimal medium, and the targeted loci were analyzed by PCR using the corresponding Up_F_primers and the Dw_R_primers. The absence of the PCR products corresponding to the WT crgA and leuA loci and the only presence of PCR products 3.4 kb larger evidenced that all transformants were homokaryons for the disruption (Figure 3C). Furthermore, this result also confirmed that the integration was stable and maintained through the successive cycles of vegetative growth.
Table 1.
Results of transformations for crgA and leuA genes using CRISPR-Cas9
| crgA_g1 crgA_Template1 | crgA_g2 crgA_Template2 | leuA_g1 leuA_Template1 | leuA_g2 leuA_Template2 | |
|---|---|---|---|---|
| Transformants/μg DNA | 23.32 ± 5.17 | 32.89 ± 2.66 | 13.82 ± 1.15 | 27.76 ± 0.19 |
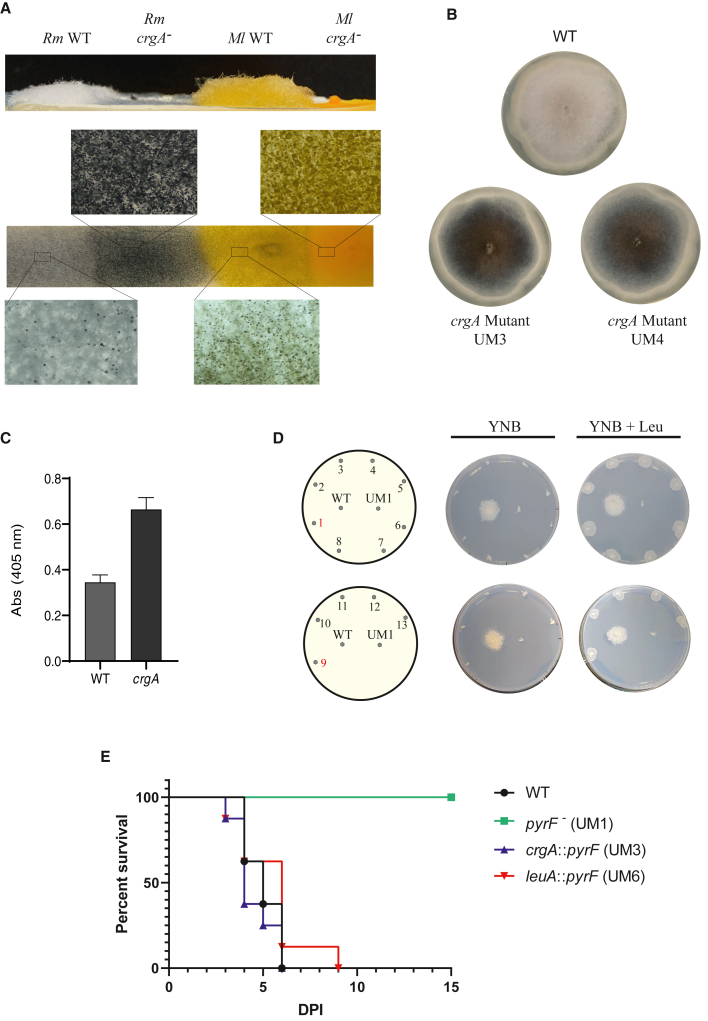
crgA mutants were affected in aerial mycelia development and melanin synthesis, while leuA disruption caused leucine auxotrophy
The phenotype of the mutants was analyzed to elucidate the function of crgA and leuA in R. microsporus. The radial growth and spore production of two different crgA mutants (UM3 and UM4), one from each gRNA + template combination, was similar to the WT strain (Figure S5). However, the mutants showed a clear defective development of aerial mycelium similar to the crgA null mutant of M. lusitanicus (Figure 4A). The mutant M. lusitanicus also shows a dark-yellow color caused by the overproduction of beta carotene (Navarro et al., 2001; Nicolas et al., 2008), which cannot be observed in R. microsporus because it lacks colored carotenoids. However, R. microsporus crgA mutants showed a dark brown color that could be associated with melanin overaccumulation (Figures 4A and 4B). Thus, we analyzed the melanin levels in extracts of the WT and mutants by measuring absorbance (Gupta and Chattoo, 2007; Saha et al., 2020). The absorbance at 405 nm was 1.92-fold higher in the crgA mutant than in the WT strain (Figure 4C), suggesting crgA mutant overaccumulated melanin. Regarding the phenotype of leuA mutants, we plated all the mutants confirmed by PCR in minimal YNB medium with and without leucine supplementation. Unlike the WT strain, leuA mutants were unable to grow without the presence of leucine (Figure 4D), evidencing the loss of gene function.
Figure 4.
Phenotypic effects of crgA- and leuA-targeted disruption
(A) Aerial mycelium and mycelial color of R. microsporus WT strain, an R. microsporus crgA mutant, M. lusitanicus WT strain (CBS 277.49), and an M. lusitanicus crgA mutant.
(B) Growth of R. microsporus wild-type strain (WT) and two crgA mutants (72 h, YPG media).
(C) Analysis of melanin production measured by its absorbance at 405 nm (p value of 0.0008).
(D) leuA mutants (1–8 from gRNA1+Template1 and 9–13 from gRNA2+Template2) after 48 h of growth on YNB media supplemented with leucine (right) and without leucine (left). Mutant growth is compared with the R. microsporus wild-type strain (WT) and the uridine auxotrophic strain UM1. UM6 (1) and UM7 (9) mutants are highlighted in red.
(E) Survival assays of mutant strains generated from R. microsporus. The survival assays were made in groups of eight immunocompetent Swiss mice, which were injected with 1 × 106 spores of the mutants pyrF− (UM1, green), crgA:pyrF (UM3, blue), and leuA::pyrF (UM6, red). In addition, the control WT strain was also injected (black). The survival rate of the mutants crgA:pyrF and leuA::pyrF and the WT strain were compared with the avirulent mutant strain pyrF− by a Mantel-Cox test (p value < 0.0001).
Testing the pathogenic potential of genetically modified strains from R. microsporus. Endogenous uracil production is essential for virulence
The capacity to genetically modify R. microsporus opened a whole range of genetic engineering possibilities in this fungus. However, our main goal was to generate a new genetic model to study mucormycosis. Thus, we tested the mutants crgA:pyrF (UM3) and leuA::pyrF (UM6) in a survival assay using a murine model in which the fungal spores were intravenously injected (Figure 4E). Instead of using immunosuppressed mice as with M. lusitanicus, we employed the common murine strain Swiss without immunosuppression treatment. Spores of the mutants in crgA and leuA genes caused rapid death of all mice between 6 and 7 days post-injection, similarly to the WT strain. However, spores of the pyrF mutant did not kill any mice, showing a complete avirulent phenotype. The virulent phenotype of the crgA and leuA mutants demonstrated that the lack of function of pyrF was utterly rescued in these mutants by the ectopic expression of the pyrF integrated in those loci, indicating that gene complementation was also successful in the genetic model of R. microsporus. It is worthy of noting that our results showed that genetically modified strains of R. microsporus could be easily tested for their pathogenic potential. Other mucormycosis models require immunosuppression before and during the infection assays or the use of neutropenic strains; thus, our strategy proposes a cost-effective and efficient virulence assay method.
Discussion
Genetic transformation of Mucorales has always been an obstacle hampering the study of these early-diverging fungi. Several attempts of genetic transformation of different species did not thrive in the long term. One example is Rhizopus delemar, another highly frequent isolated causal agent from mucormycosis patients (Prakash and Chakrabarti, 2019). An attempt to disrupt the gene ftr1, coding for a ferroxidase involved in the high-affinity iron uptake by double crossover HR generates only one heterokaryotic mutant strain that failed to segregate homokaryons (Ibrahim et al., 2010). Other studies used the CRISPR-Cas9 system to induce directed mutagenesis in a selective marker by the nonhomologous end-joining repair mechanism after a double-strand break caused by the guided Cas9 enzyme. This CRISPR-Cas9 system was used in R. delemar and Lichtheimia corymbifera, another frequent causal agent of mucormycosis, to mutate genes pyrF and pyrG, respectively, selecting for 5-FOA resistance (Bruni et al., 2019; Ibragimova et al., 2020). The main drawback of this strategy is that the mutagenesis can be exclusively directed to genes with a selective method such as the 5-FOA resistance, which makes it impossible to widen it to the whole genome. Another strategy to genetically manipulate Mucorales is the transformation with self-replicative plasmids, first achieved in M. lusitanicus and, later, in R. delemar and L. corymbifera (Benito et al., 1995; Bruni et al., 2019; Ibragimova et al., 2020). These plasmids have been used to manipulate gene expression through the RNAi mechanism (Bruni et al., 2019; Ibragimova et al., 2020; Ibrahim et al., 2010; Soliman et al., 2021). The main problem of the self-replicative plasmids is that they do not replicate in all the nuclei of multinucleated and coenocytic hyphae. During nuclei segregation and sporulation, only a variable proportion of the descendant spores contains some nucleus-carrying plasmids (Nicolas et al., 2003). Moreover, among the subpopulation that receives the plasmid, only those with a high number of plasmid copies can trigger the RNAi mechanism (Nicolas et al., 2003).
Stable and reproducible gene disruption by HR in Mucorales has been only achieved in Mucor, particularly M. lusitanicus (Navarro et al., 2001). Consequently, other derived genetic tools were also developed in this fungus, such as gene overexpression (Pérez-Arques et al., 2021a), complementation (Trieu et al., 2015), and protein tagging (Navarro-Mendoza et al., 2019). During the last decade, this fungus has served as the unique model with a wide repertoire of genetic engineering tools in the urgent search for new target genes that are involved in Mucorales pathogenesis (Lax et al., 2020; Li et al., 2011; Trieu et al., 2017; Navarro-Mendoza et al., 2018; Pérez-Arques et al., 2019, 2021b; López-Fernández et al., 2018; Patiño-Medina et al., 2018, 2019). However, M. lusitanicus shows reduced virulence, and survival assays can only be done in the specific mouse strains following rigorous immunosuppression (Li et al., 2011).
In this scenario, understanding of Mucorales pathogenesis urgently requires a genetic model that unifies the genetic amenability of M. lusitanicus and high virulence. Here, we developed a new genetic model of mucormycosis based on R. microsporus and the methodology to transform it and generate stable mutants in target genes. First, we developed a new procedure to obtain spontaneous uracil auxotrophic strains that resulted in the isolation of a strain mutated in the pyrF gene that can be used as a recipient in transformation experiments. As this strain is a spontaneous mutant, it is expected that it carries a low mutational burden, reducing the likelihood that additional mutations in the genome negatively affect fitness and virulence, which could interfere with genetic studies of new potential virulence factors.
Since the implementation of M. lusitanicus as a model for HR-mediated genetic manipulations in Mucorales, the methodology to transform this species has been optimized several times (Gutiérrez et al., 2011; van Heeswijck and Roncero, 1984; Nicolás et al., 2018; Trieu et al., 2017; Vellanki et al., 2018), providing knowledge about the critical steps that must be carefully taken to ensure a successful transformation. In this work, those essential steps were optimized for R. microsporus, finding that this fungus requires specific conditions in all the critical steps. Thus, R. microsporus germinates faster than M. lusitanicus at higher temperatures, which might be related to its higher virulence (Bhabhra and Askew, 2005). After germination, the digestion of R. microsporus needs higher concentrations of lysing enzymes and chitosanase than M. lusitanicus, indicating a more robust cell wall. The cell wall differences might also be behind the improvement in the electroporation efficiency after changing the pulse method. Finally, the use of a plasmid-free CRISPR-Cas9 system definitively ensured HR, which is a highly specific gene targeting system in fungi (Abdallah et al., 2018). We showed that a template DNA containing upstream and downstream homologous tails of only 38 bp was enough to disrupt the target gene (Yuzbashev et al., 2015). This represents an advantageous manner of generating the recombinant constructs for HR, because they can be produced by PCR using affordable long primers with the corresponding saving of time and effort, compared with the construction of DNA fragments with longer arms (Abdallah et al., 2017; Yuzbashev et al., 2015). By using this methodology, we generated homokaryotic and stable mutants in two genes of R. microsporus. The gene leuA, ortholog of the M. lusitanicus leuA gene, was selected to generate a second auxotrophic strain that will be helpful in future studies requiring the functional analysis of two genes. The second gene, crgA, was selected because of its pleiotropic and striking visual phenotype in mutants of its putative ortholog in M. lusitanicus. The lack of crgA in M. lusitanicus induces the overaccumulation of carotenoids, resulting in dark-yellow mycelium (Navarro et al., 2001; Nicolás-Molina et al., 2008). This visual phenotype in M. lusitanicus is pronounced by the lack of aerial sporangiophores, a phenotype also found in the crgA mutants of R. microsporus. R. microsporus does not accumulate colored carotenoids, but disruption of crgA provokes black-colored mycelia after sporulation as a result of an overaccumulation of melanin. Carotenoids and melanin are secondary metabolites produced after the initial growth phase. The fact that melanin is overaccumulated in crgA mutants suggests that crgA has evolved in R. microsporus to regulate melanin biosynthesis in a similar manner as carotenoids biosynthesis in M. lusitanicus (Silva et al., 2008).
The lack of aerial sporangiophores and the overaccumulation of melanin are the first visual phenotypes obtained by gene disruption in a Mucoral other than M. lusitanicus. However, the results of the survival assays made in this work are even more relevant for the future impact of R. microsporus as a new genetic model for the study of mucormycosis. The WT strain, the spontaneous pyrF auxotrophic strain, and the two generated mutants showed expected results for typical performance in a murine survival assay. The WT strain killed the mice rapidly and without the necessity of a previous and continued immunosuppression treatment, an important advantage over M. lusitanicus. Despite the pleiotropic effect provoked by disruption of crgA in R. microsporus, the virulence of the crgA mutant suggests that none of the regulatory pathways controlled by this gene are involved in the pathogenic potential of Mucorales. The lack of melanin correlates with a decreased virulence in other Mucorales (Soliman et al., 2020), but its overaccumulation has not been analyzed. Full virulence of the crgA mutant suggests that melanin overaccumulation does not increase virulence, although we cannot discard a compensatory effect on virulence by the diverse pathways altered in the crgA mutant. Regarding the mutants in the gene leuA, the virulent phenotype was equal to the WT strain, results that are similar to those previously observed for leucine auxotroph in M. lusitanicus (Li et al., 2011), confirming that this auxotrophy does not affect virulence in Mucorales. However, auxotrophy for uracil provoked a complete avirulent phenotype in R. microsporus. These findings are consistent with the result in other fungi, such as Aspergillus fumigatus and Candida albicans, whose uracil auxotrophs also present reduced virulence (Bain et al., 2006; Brand et al., 2004; D'Enfert et al., 1996). Likely, the free uridine/uracil in the tissues of the host is not enough to keep a regular fungal growth (D'Enfert et al., 1996). The tandem of controls, positive and negative, corresponding to the WT and uracil auxotrophic strains, will represent an invaluable tool for future virulence analyses in R. microsporus.
In summary, we have achieved stable and efficient HR-mediated genetic modification in R. microsporus using a plasmid-free CRISPR-Cas9 method. It is first achieved in a highly prevalent causal agent of mucormycosis and represents the second mucoralean genus that can be genetically modified stably and efficiently. Mucormycosis is currently a lethal, emerging, virulent, and drug-resistant infectious disease, and this new genetic model will represent a landmark in the study of the molecular aspect of this disease. Moreover, we propose to export the methodology developed in this work to other mucoralean species that are models in research fields different from mucormycosis, extending the genetic and molecular studies to the whole order of Mucorales.
Limitations of the study
The methodology presented in this work allows for genetic transformation and targeted integration by HR combined with the use of the CRISPR-Cas9 technology. This has required a deep optimization of every step of the process. Some of those steps are highly specific for R. microsporus. For that reason, in order to apply this procedure to any other related fungal species, these specific steps should be adapted in consequence. The possibility of targeted integration of DNA templates opens broad possibilities for the development of multiple genetic tools. Some factors such as the maximum size of DNA fragments that can be used as templates remain to be further characterized.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Escherichia coli Dh5α | Thermo Fisher Scientific | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Uridine | Merck KGaA | Cat#U3003 |
| L-Leucine | Merck KGaA | Cat#L8000 |
| 5 Fluoroorotic acid monohydrate (5-FOA) | Apollo Scientific | Cat#PC4054 |
| EDTA | Merck KGaA | Cat#EDS |
| Tris | Merck KGaA | Cat#T1503 |
| Niacine (nicotinc acid) | Merck KGaA | Cat#N0761 |
| Thiamine | Merck KGaA | Cat#T1270 |
| D-Sorbitol | Merck KGaA | Cat#S1876 |
| Lysing Enzymes | Merck KGaA | Cat#L412 |
| Chitosanase | Merck KGaA | Cat#C9830 |
| DMSO | Merck KGaA | Cat#D8418 |
| SDS | Merck KGaA | Cat#L3771 |
| RNase A | Merck KGaA | Cat#R6513 |
| Phenol | Merck KGaA | Cat#P1037 |
| Chloroform | Merck KGaA | Cat#C2432 |
| Isoamyl alcohol | Merck KGaA | Cat#309435 |
| Ammonium acetate | Merck KGaA | Cat#A1542 |
| Ethanol | Merck KGaA | Cat#51976 |
| Hydrochloric acid | Merck KGaA | Cat#H1758 |
| Critical commercial assays | ||
| Alt-R™ CRISPR-Cas9 crRNA | IDT | See Table S1 |
| Alt-R™ CRISPR-Cas9 tracrRNA | IDT | Cat#1072533 |
| Alt-R™ S.p. Cas9 Nuclease V3 | IDT | Cat#1081058 |
| Deposited data | ||
| Mucoromycotina genomes | Joint Genome Institute Mycocosm | https://mycocosm.jgi.doe.gov/mucoromycotina/mucoromycotina. info.html |
| R. microsporus pyrG | Joint Genome Institute Mycocosm | R. microsporus Protein ID 279857 |
| R. microsporus pyrF | Joint Genome Institute Mycocosm | R. microsporus Protein ID 231940 |
| R. microsporus leuA | Joint Genome Institute Mycocosm | R. microsporus Protein ID 228594 |
| R. microsporus crgA | Joint Genome Institute Mycocosm | R. microsporus Protein ID 226533 |
| Experimental models: Organisms/strains | ||
| Rhizopus microsporus ATCC 11559 | Kindly provided by Dr. Teresa E. Pawlowska (Cornell University) | N/A |
| Rhizopus microsporus UM1: pyrF- | This Study | N/A |
| Rhizopus microsporus UM3: crgA::pyrF | This Study | N/A |
| Rhizopus microsporus UM4: crgA::pyrF | This Study | N/A |
| Rhizopus microsporus UM6: leuA::pyrF | This Study | N/A |
| Rhizopus microsporus UM7: leuA::pyrF | This Study | N/A |
| Mucor lusitanicus CBS 277.49 | von Arx and Schipper, 1978 | N/A |
| Mucor lusitanicus MU223: crgAΔ::pyrG | Silva et al., 2008 | N/A |
| Oligonucleotides | ||
| Oligonucleotides | This Study | See Table S1 |
| Recombinant DNA | ||
| pBluescript II SK+ | Agilent | Cat#212205 |
| pMAT1819 | This Study | N/A |
| Software and algorithms | ||
| Snapgene v5.1.0 | Snapgene Software | https://www.snapgene.com/ |
| EuPaGDT | Peng and Tarleton, 2015 | http://grna.ctegd.uga.edu/ |
| NIS Elements Software | Nikon | https://www.microscope.healthcare.nikon.com/products/software/nis-elements |
| ImageJ v1.51m9 | Schneider et al., 2012 | |
| Zeiss Zen v2.5 | Zeiss | https://www.zeiss.com/microscopy/us/products/microscope-software/zen.html#introduction |
| Jmol | http://jmol.sourceforge.net/ | |
| Mol∗ | http://molstar.org | |
| GraphPad Prism v8.0.2 | Graphpad Software | https://www.graphpad.com/ |
| IBM SPSS Statistic | IBM Software | https://www.ibm.com/analytics/spss-statistics-software |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and fulfilled by the lead contact, Victoriano Garre (vgarre@umes.es).
Materials availability
All plasmids and fungal strains generated in this study are available from the lead contact without restriction.
Experimental model and subject details
All the fungal strains used and generated in this work derive from Rhizopus microsporus ATCC 11559 and M. circinelloides CBS277.49. Uridine and leucine auxotrophies were checked in the minimal media yeast nitrogen base, YNB (Lasker and Borgia, 1980). When specified, media was supplemented with uridine (200 mg/L) or leucine (20 mg/L). Transformants of the auxotrophic strain UM1 strain with the pyrF complementation plasmid or the pyrF template used in CRISPR/Cas9 disruption experiments were grown in Minimal Media with Casamino acids (MMC) (Nicolas et al., 2007). Electroporated protoplasts were resuspended in ice-cold YPG media + 0.5 M Sorbitol for 90 min and then centrifugated at 800 rpm and resuspended in YNB media +0.5 M Sorbitol. The resuspended protoplasts were plated in MMC + 0.5 M Sorbitol media to select transformants. All strains were grown at 30°C. Escherichia coli strain DH5α (Thermo Fisher Scientific) was used for all cloning experiments.
Methods details
Protoplast generation and electroporation
A total of 1x107 spores/mL were inoculated in 25 mL liquid YPG media and preincubated overnight at 4°C in YPG media supplemented with 200 mg/L of uridine. The culture was incubated at 33°C for approximately 4 hours and 30 min at 250 rpm in a rotary shaker. When spores reached the optimal germination stage, the culture was centrifuged, and the spores were washed twice with PS buffer (1X PBS and 0.5M Sorbitol) and resuspended in 5 mL of PS buffer. To digest the cell wall, 3 mg of lysing enzymes from Trichoderma harzianum (Merck KGaA, Darmstadt, Germany) and 0.0008 units of Chitosanase from Streptomyces griseus (Merck KGaA, Darmstadt, Germany) were added to the resuspended germinules. The digestion was incubated at 30°C for 90 min at 60 rpm in a rotary shaker. Protoplasts were washed twice with ice-cold 0.5 M sorbitol and transferred to 0.2 cm electroporation cuvettes (Fisher Scientific, FB102). Up to 20 μl of pMAT1819 (1- 4 μg of DNA) or gRNA-Cas9 complex (see gRNA-Cas9 assembly and in vitro activity tests) and up to 20 μl of purified template DNA (0,5 - 4 μg of DNA) were added and mixed with 200 μl of protoplasts. After the electroporation pulses, applied with Gene Pulser XCell (Biorad, Hercules, California, USA), protoplasts were rapidly resuspended in YPG + 0.5 M sorbitol. When survival rates of the protoplasts were analyzed, 10μl of pulsed protoplasts were diluted in 90 μl of 0.5 M sorbitol, and an aliquot was counted in a Neubauer chamber to determine concentration. Then, two hundred protoplasts were plated on minimal media (MMC) supplemented with uridine (200 mg/L) (two plates/cuvette). As a control, two hundred protoplasts that did not undergo electroporation were also plated in the same medium. The same procedure was followed to determine the effect of germination temperature on protoplasts viability. pMAT1819 plasmid was used in CRISPR/Cas9 disruption experiments as a positive control of the transformation/electroporation efficiency, while no DNA was added in negative controls.
Transformation efficiency score was obtained as the product of the average transformants/μg of DNA and the average survival rate (normalized considering 1 as the value of the survival rate of the negative control).
Nucleic acids manipulation, amplification, and plasmid construction
DNA from R. microsporus was extracted following a chloroform/phenol protocol. Briefly, mycelia were quickly frozen in liquid nitrogen and then ground using mortar and pestle. 200 to 500 mg of the resulting powder was treated with Extraction Buffer (Tris-HCl 200 mM, EDTA 100 mM and 1% SDS), RNAse A (10 mg/mL), and 1 mL of phenol pH 8.0. After mixing and centrifuge, the aqueous phase was washed three times using 1/2 volume of phenol pH 8 and 1/2 volume of isoamyl alcohol:chloroform (24:1). The aqueous phase was treated again with 1 volume of isoamyl alcohol:chloroform and then transferred to a different tube and mixed with 1/10 volume of 5 M ammonium acetate and 1 volume of ice-cold ethanol. The DNA pellet was washed with 1 mL of 80% ethanol. The supernatant was discarded, and the DNA was dried at room temperature for 10 min. Finally, DNA was resuspended with ultrapure water. All PCRs amplifications were performed with the Herculase II Fusion DNA polymerase (Agilent, Santa Clara, California, USA), adapting annealing temperature to each primer pair following the manufacturer's recommendations. DNA templates and fragments for crRNAs in in vitro test were purified using GeneJet PCR Purification Kit (Thermo Scientific).
To construct the plasmid pMAT1819 to complement pyrF mutation, we first PCR amplified the 4.2-kb pyrF locus from the wild-type DNA using the primers pyrF_F_EcoRI and pyrF_R_XhoI, which have the indicated restriction sites at their 5′ end (Table S1). The pyrF fragment in this plasmid included a 2357-bp sequence upstream the translation start codon and 1010-bp sequence downstream the stop codon of the gene to ensure that the construction contained the promoter and terminator of the pyrF gene. The pBluescript SK(+) vector was double digested using the EcoRI and XhoI restriction enzymes, and the 4.2-kb pyrF fragment was ligated at room temperature using T4 ligase (Thermo Scientific, Waltham, Massachusetts, USA). Electrocompetent cells of Escherichia coli (DH5α) were transformed and plated in LB media supplemented with ampicillin (100 mg/L). The plasmid was extracted from colonies using the GeneJet Plasmid Miniprep Kit (Thermo Scientific, Waltham, Massachusetts, USA) and checked by digestion followed to electrophoresis and sequenced with pyrF_F_SEQ and pyrF_R_SEQ (Table S1). Sequencing was performed with a 3500 Genetic Analyzer (Thermo Scientific, Waltham, Massachusetts, USA).
Selection of uracil auxotrophic mutants
After 4-5 days of growth in solid YPG media, 1x107 spores were collected, washed, and inoculated in 200 mL of liquid YPG media supplemented with 3 g/L of 5-FOA and 200 mg/L uridine. After 48 hours of growth at 30°C in a rotary shaker (250 rpm), the complete culture was centrifugated, and the pellet was plated on solid YPG media plates, also supplemented with 3 g/L of 5-FOA and 200 of mg/L uridine. After 24 hours, resistant colonies were isolated and plated again on solid YPG media supplemented with 3 g/L of 5-FOA and 200 mg/L of uridine. The colonies showing a robust growth were plates on solid YNB media with and without uridine (200 mg/L). DNA from the colonies resistant to 5-FOA and unable to grow without uridine was extracted. The pyrG gene and the pyrF gene were PCR amplified using the primers pyrF_F_SEQ and pyrF_R_SEQ (pyrF) and pyrG_F_SEQ and pyrG_R_SEQ (pyrG) (Table S1) and sequenced. The sequences of the pyrG and pyrF genes were aligned to the wild-type sequences available at JGI MycoCosm portal (https://mycocosm.jgi.doe.gov) (Lastovetsky et al., 2016) using SnapGene® software (from GSL Biotech; available at snapgene.com).
gRNA – Cas9 assembly and in vitro activity tests
Targeted gene disruption and in vitro activity tests required the ribonucleoprotein (gRNA – Cas9) complex assembly. Alt-R™ CRISPR-Cas9 crRNA, Alt-R™ CRISPR-Cas9 tracrRNA, and Alt-R™ S.p. Cas9 Nuclease were purchased from IDT (https://www.idtdna.com/site/order/oligoentry/index/crispr). The tracrRNA and crRNA were assembled to generate de gRNA following the manufacturer's recommendations and the protocol developed for Aspergillus fumigatus (Abdallah et al., 2017). Briefly, we used a 33 μM equimolecular mix of crRNA, tracRNA (common to every crRNA), and Nuclease-Free Duplex Buffer (IDT). The mix was incubated at 95°C for 5 min and cooled down at room temperature. 1.5 μl per electroporation cuvette of gRNA were mixed with 0.075 μg of Cas9 and 11 μl of PBS 1X. The mix was left for 5-10 min at room temperature before the electroporation to allow ribonucleoprotein complex assembly. 20-nt protospacer sequences were designed using the genome of R. microsporus (https://mycocosm.jgi.doe.gov) (Lastovetsky et al., 2016) and the tool EuPaGDT (Eukaryotic Pathogen CRISPR guide RNA/DNA Design Tool) with recommended default settings (Peng and Tarleton, 2015). The crRNAs were purchased from IDT (https://eu.idtdna.com/site/order/oligoentry/index/crispr, IDT, Coralville, IA, USA) (Table S1). For in vitro activity assays, gRNA (crRNA and tracRNA) and the ribonucleoprotein complex were assembled following the same procedure mentioned above. Subsequently, the gRNA, Cas9, and 100-400 ng of DNA were incubated at 30°C for 2 hours. The DNA corresponding to the wild-type loci were PCR amplified, using the Up_F_primer and the Dw_R_primer (Table S1), and purified using the Gene Jet PCR Purification Kit (Thermo Scientific, Waltham, Massachusetts, USA). Negative controls contained the same mix except for the crRNA. The in vitro reaction was inactivated (65°C for 20 min), and the cleavage products were separated by agarose gel electrophoresis (0.7%).
Phenotypic characterization of crgA and leuA mutants
For radial growth and spore production analysis, a drop with 1x103 spores was inoculated in the center of a plate. For radial growth, diameter measures were taken after 24, 48, and 72 hours. For spore production, a chunk of agar of 1cm2 (one per plate, three for each strain and time) was transferred to a 50 ml tube containing 10mL of PBS 1X. The tube was vigorously vortexed to release spores. Spores were counted in a Neubauer chamber to determine the concentration, and total spore production was calculated. For total melanin quantification, 20 mg of lyophilized biomass was crushed and resuspended in 0.5 mL of a 1 N NaOH solution + 10 % DMSO. Samples were incubated at 80°C for 1 hour and vortex every 5-10 min. After centrifugation, the absorbance of the supernatants was measured at 405 nm (Gupta and Chattoo, 2007; Saha et al., 2020). For leucine auxotrophy analysis, small pieces of mycelium were plated in minimal YNB and YNB media supplemented with leucine (20 mg/l), and the growth (or its absence) observed after 48 hours.
Microscopy imaging, data representation, and statistical analysis
For the germination experiments, 15 μl of spores or germinated spores were placed on a slide and covered with a coverslip. Images of germinated spores were acquired with a Nikon Eclipse 80i microscopy equipped with a Nikon DS-Ri2 camera and processed with NIS Elements Software and ImageJ (Schneider et al., 2012). Microscopy images showing the mycelia and growth differences (Figure 4) were obtained using a Stemi 305 Zeiss microscope (integrated camera). Schematic illustrations of the orotate phosphoribosyl transferase structure were obtained using the available structure of the S. typhimurium enzyme (PDB: 1OPR, (Ozturk et al., 1995; Scapin et al., 1995), using Jmol (http://jmol.sourceforge.net/) and Mol∗ (http://molstar.org).
Survival assays
Male Swiss mice weighing 30 g and one-month-old (Animal Facilities Services, University of Murcia, Spain) were used as an animal model for virulence assays. The mice were kept in groups of 8 individuals per cage. The infection was initiated one week later by forming the groups, letting a stable hierarchy and no conflict in the group. Mice were injected intravenously by retroorbital injection of 1·106 spores (Chang and Heitman, 2019). Isoflurane anesthetic was used during the procedure to ensure animal welfare, and the mice were monitored after the procedure until full recovery from the anesthesia. Mice were maintained in established conditions with free food, autoclaved water, and continuous ventilation. The animal welfare was checked twice a day for 15 days, and mice following the discomfort criteria were euthanized in a CO2 chamber. Survival rates were plotted in a Kaplan–Meier curve (GraphPad Prism), and differences were considered statistically significant with a p-value ≤ 0.05 in a Mantel–Cox test. To guarantee the welfare of the animals and the ethics of any procedure related to animal experimentation, all the experiments performed in this work comply with the Guidelines of the European Union Council (Directive 2010/63/EU) and the Spanish RD 53/2013. Experiments and procedures were supervised and approved by the University of Murcia Animal Welfare and Ethics Committee and the Council of Water, Agriculture, Farming, Fishing and Environment of Murcia (Consejería de Agua, Agricultura, Ganadería, Pesca y Medio Ambiente de la CARM), Spain (authorization number REGA ES300305440012).
Quantification and statistical analysis
For the graphical representation of data, GraphPad Prism version 8.0.2 for Windows was used (GraphPad Software, San Diego, California USA, www.graphpad.com). Radial growth and spore production results were expressed as mean ± S.E. The data were analyzed with the software IBM SPSS Statistic for Mac [IBM Corp (2014) Version 23.0.; https://www.ibm.com/SPSS-Statistics/]. An ANOVA of a single factor was used to determine statistically significant differences between the wild type strain and the mutants, assuming a significance level of 95% (P-value < 0.05), followed by the Tuckey or Games Howell tests, according to the homogeneity of the variables.
Acknowledgments
This work was supported by the Fundación Séneca-Agencia de Ciencia y Tecnología de la Región de Murcia, Spain (20897/PI/18). The student C.L. was funded by Spanish MICIU (FPU17/05814).
Author contributions
Methodology, C.L., M.I.N.-M., and C.P.-A.; investigation, formal analysis, visualization, writing – original draft, and writing – review & editing, C.L.; funding acquisition and resources, E.N.; conceptualization, supervision, project administration, funding acquisition, and writing – review & editing, F.E.N. and V.G.
Declaration of interests
The authors declare no competing interests.
Published: December 6, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2021.100124.
Contributor Information
Francisco Esteban Nicolás, Email: fnicolas@um.es.
Victoriano Garre, Email: vgarre@um.es.
Supplemental information
Data and code availability
•All data reported in this paper will be shared by the lead contact upon request.
•This paper does not report original code.
•Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abdallah Q.A., Ge W., Fortwendel J.R. A simple and universal system for gene manipulation in Aspergillus fumigatus: In vitro-assembled cas9-guide RNA ribonucleoproteins coupled with microhomology repair templates. mSphere. 2017;2:e00446. doi: 10.1128/mSphere.00446-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdallah Q.A., Souza A.C.O., Martin-vicente A., Ge W., Fortwendel J.R. Whole-genome sequencing reveals highly specific gene targeting by in vitro assembled cas9-ribonucleoprotein complexes in Aspergillus fumigatus. Fungal Biol. Biotechnol. 2018;5:1–8. doi: 10.1186/s40694-018-0057-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez E., Sutton D.A., Cano J., Fothergill A.W., Stchigel A., Rinaldi M.G., Guarro J. Spectrum of zygomycete species identified in clinically significant specimens in the United States. J. Clin. Microbiol. 2009;47:1650–1656. doi: 10.1128/JCM.00036-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain J.M., Stubberfield C., Gow N.A.R. Ura-status-dependent adhesion of Candida albicans mutants. FEMS Microbiol. Lett. 2006;204:323–328. doi: 10.1111/j.1574-6968.2001.tb10905.x. [DOI] [PubMed] [Google Scholar]
- Benito E.P., Campuzano V., Lôpez-Matas M.A., De Vicente J.I., Eslava A.P. Isolation, characterization and transformation, by autonomous replication, of Mucor circinelloides OMPdecase-deficient mutants. Mol. Gen. Genet. 1995;248:126–135. doi: 10.1007/BF02190793. [DOI] [PubMed] [Google Scholar]
- Bhabhra R., Askew D.S. Thermotolerance and virulence of Aspergillus fumigatus: Role of the fungal nucleolus. Med. Mycol. 2005;43 doi: 10.1080/13693780400029486. S87–93. [DOI] [PubMed] [Google Scholar]
- Binder U., Navarro-Mendoza M.I., Naschberger V., Bauer I., Nicolas F.E., Pallua J.D., Lass-Flörl C., Garre V. Generation of a Mucor circinelloides reporter strain—A promising new tool to study antifungal drug efficacy and mucormycosis. Genes (Basel) 2018;9:613. doi: 10.3390/genes9120613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J.D., Trueheart J., Natsoulis G., Fink G.R. [10] 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Brand A., MacCallum D.M., Brown A.J.P., Gow N.A.R., Odds F.C. Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaryot. Cell. 2004;3:900–909. doi: 10.1128/EC.3.4.900-909.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruni G.O., Zhong K., Lee S.C., Wang P. CRISPR-Cas9 induces point mutation in the mucormycosis fungus Rhizopus delemar. Fungal Genet. Biol. 2019;124:1–7. doi: 10.1016/j.fgb.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caetano L.A., Faria T., Springer J., Loeffler J., Viegas C. Antifungal-resistant Mucorales in different indoor environments. Mycology. 2019;10:75–83. doi: 10.1080/21501203.2018.1551251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo S., Shertz-Wall C., Lee S.C., Bastidas R.J., Nicolas F.E., Granek J.A., Mieczkowski P., Torres-Martinez S., Ruiz-Vazquez R.M., Cardenas M.E., et al. Antifungal drug resistance evoked via RNAi-dependent epimutations. Nature. 2014;513:555–558. doi: 10.1038/nature13575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caramalho R., Tyndall J.D.A., Monk B.C., Larentis T., Lass-Flörl C., Lackner M. Intrinsic short-tailed azole resistance in mucormycetes is due to an evolutionary conserved aminoacid substitution of the lanosterol 14α-demethylase. Sci. Rep. 2017;7:15898. doi: 10.1038/s41598-017-16123-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Z., Heitman J. Drug-resistant epimutants exhibit organ-specific stability and induction during murine infections caused by the human fungal pathogen Mucor circinelloides. mBio. 2019;10 doi: 10.1128/mBio.02579-19. e02579–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chayakulkeeree M., Ghannoum M.A., Perfect J.R. Zygomycosis: The re-emerging fungal infection. Eur. J. Clin. Microbiol. Infect. Dis. 2006;25:215–229. doi: 10.1007/s10096-006-0107-1. [DOI] [PubMed] [Google Scholar]
- Cornely O.A., Alastruey-Izquierdo A., Arenz D., Chen S.C.A., Dannaoui E., Hochhegger B., Hoenigl M., Jensen H.E., Lagrou K., Lewis R.E., et al. Global guideline for the diagnosis and management of mucormycosis: An initiative of the European confederation of medical mycology in cooperation with the mycoses study group education and research consortium. Lancet Infect. Dis. 2019;19:e405–e421. doi: 10.1016/S1473-3099(19)30312-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bekker C., Wiebenga A., Aguilar G., Wösten H.A.B. An enzyme cocktail for efficient protoplast formation in Aspergillus niger. J. Microbiol. Methods. 2009;76:305–306. doi: 10.1016/j.mimet.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Dannaoui E. Antifungal resistance in mucorales. Int. J. Antimicrob. Agents. 2017;50:617–621. doi: 10.1016/j.ijantimicag.2017.08.010. [DOI] [PubMed] [Google Scholar]
- D’Enfert C., Diaquin M., Delit A., Wuscher N., Debeaupuis J.P., Huerre M., Latge J.P. Attenuated virulence of uridine-uracil auxotrophs of Aspergillus fumigatus. Infect. Immun. 1996;64:4401–4405. doi: 10.1128/iai.64.10.4401-4405.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escoffre J.M., Portet T., Wasungu L., Teissié J., Dean D., Rols M.P. What is (still not) known of the mechanism by which electroporation mediates gene transfer and expression in cells and tissues. Mol. Biotechnol. 2009;41:286–295. doi: 10.1007/s12033-008-9121-0. [DOI] [PubMed] [Google Scholar]
- Gebremariam T., Alkhazraji S., Soliman S.S.M., Gu Y., Jeon H.H., Zhang L., French S.W., Stevens D.A., Edwards J.E., Filler S.G., et al. Anti-CotH3 antibodies protect mice from mucormycosis by prevention of invasion and augmenting opsonophagocytosis. Sci. Adv. 2019;5:eaaw1327. doi: 10.1126/sciadv.aaw1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber F., Visser J., Kubicek C.P., de Graaff L.H. The development of a heterologous transformation system for the cellulolytic fungus Trichoderma reesei based on a pyrG-negative mutant strain. Curr. Genet. 1990;18:71–76. doi: 10.1007/BF00321118. [DOI] [PubMed] [Google Scholar]
- Gupta A., Chattoo B.B. A novel gene MGA1 is required for appressorium formation in Magnaporthe grisea. Fungal Genet. Biol. 2007;44:1157–1169. doi: 10.1016/j.fgb.2007.02.014. [DOI] [PubMed] [Google Scholar]
- Gutiérrez A., López-García S., Garre V. High reliability transformation of the basal fungus Mucor circinelloides by electroporation. J. Microbiol. Methods. 2011;84:442–446. doi: 10.1016/j.mimet.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Hassan M.I.A., Voigt K. Pathogenicity patterns of mucormycosis: Epidemiology, interaction with immune cells and virulence factors. Med. Mycol. 2019;57:S245–S256. doi: 10.1093/mmy/myz011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibragimova S., Szebenyi C., Sinka R., Alzyoud E.I., Homa M., Vágvölgyi C., Nagy G., Papp T. CRISPR-Cas9-based mutagenesis of the mucormycosis-causing fungus Lichtheimia corymbifera. Int. J. Mol. Sci. 2020;21:1–11. doi: 10.3390/ijms21103727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim A.S., Gebermariam T., Fu Y., Lin L., Husseiny M.I., French S.W., Schwartz J., Skory C.D., Edwards J.E., Spellberg B.J. The iron chelator deferasirox protects mice from mucormycosis through iron starvation. J. Clin. Invest. 2007;117:2649–2657. doi: 10.1172/JCI32338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim A.S., Gebremariam T., Lin L., Luo G., Husseiny M.I., Skory C.D., Fu Y., French S.W., Edwards J.E., Jr., Spellberg B. The high affinity iron permease is a key virulence factor required for Rhizopus oryzae pathogenesis. Mol. Microbiol. 2010;77:587–604. doi: 10.1111/j.1365-2958.2010.07234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong W., Keighley C., Wolfe R., Lee W.L., Slavin M.A., Kong D.C.M., Chen S.C.-A. The epidemiology and clinical manifestations of mucormycosis: A systematic review and meta-analysis of case reports. Clin. Microbiol. Infect. 2019;25:26–34. doi: 10.1016/j.cmi.2018.07.011. [DOI] [PubMed] [Google Scholar]
- John T.M., Jacob C.N., Kontoyiannis D.P. When uncontrolled diabetes mellitus and severe COVID-19 converge: The perfect storm for mucormycosis. J. Fungi. 2021;7:298. doi: 10.3390/jof7040298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontoyiannis D.P. Antifungal resistance: An emerging reality and a global challenge. J. Infect. Dis. 2017;216:S431–S435. doi: 10.1093/infdis/jix179. [DOI] [PubMed] [Google Scholar]
- Lasker B.A., Borgia P.T. High-frequency heterokaryon formation by Mucor racemosus. J. Bacteriol. 1980;141:565–569. doi: 10.1128/jb.141.2.565-569.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lastovetsky O.A., Gaspar M.L., Mondo S.J., LaButti K.M., Sandor L., Grigoriev I.V., Henry S.A., Pawlowska T.E. Lipid metabolic changes in an early divergent fungus govern the establishment of a mutualistic symbiosis with endobacteria. Proc. Natl. Acad. Sci. USA. 2016;113:15102–15107. doi: 10.1073/pnas.1615148113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lax C., Pérez-arques C., Navarro-mendoza M.I., Cánovas-márquez J.T., Tahiri G., Pérez-ruiz J.A., Osorio-concepción M., Murcia-flores L., Navarro E., Garre V., et al. Genes, pathways, and mechanisms involved in the virulence of mucorales. Genes (Basel) 2020;11:317. doi: 10.3390/genes11030317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.H., Cervantes M., Springer D.J., Boekhout T., Ruiz-Vazquez R.M., Torres-Martinez S.R., Heitman J., Lee S.C. Sporangiospore size dimorphism is linked to virulence of Mucor circinelloides. PLoS Pathog. 2011;7:e1002086. doi: 10.1371/journal.ppat.1002086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Fernández L., Sanchis M., Navarro-Rodríguez P., Nicolás F.E., Silva-Franco F., Guarro J., Garre V., Navarro-Mendoza M.I., Pérez-Arques C., Capilla J. Understanding Mucor circinelloides pathogenesis by comparative genomics and phenotypical studies. Virulence. 2018;9:707–720. doi: 10.1080/21505594.2018.1435249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Muñoz A., Nicolás F.E., García-Moreno D., Pérez-Oliva A.B., Navarro-Mendoza M.I., Hernández-Oñate M.A., Herrera-Estrella A., Torres-Martínez S., Ruiz-Vázquez R.M., Garre V., et al. An adult zebrafish model reveals that mucormycosis induces apoptosis of infected macrophages. Sci. Rep. 2018;8:12802. doi: 10.1038/s41598-018-30754-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G., Gebremariam T., Lee H., French S.W., Wiederhold N.P., Patterson T.F., Filler S.G., Ibrahim A.S. Efficacy of liposomal amphotericin B and posaconazole in intratracheal models of murine mucormycosis. Antimicrob. Agents Chemother. 2013;57:3340–3347. doi: 10.1128/AAC.00313-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer E., Binder U., Sparber M., Lackner M., Caramalho R., Lass-Flörl C. Susceptibility profiles of amphotericin B and posaconazole against clinically relevant Mucorales species under hypoxic conditions. Antimicrob. Agents Chemother. 2015;59:1344–1346. doi: 10.1128/AAC.04424-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondo S.J., Lastovetsky O.A., Gaspar M.L., Schwardt N.H., Barber C.C., Riley R., Sun H., Grigoriev I.V., Pawlowska T.E. Bacterial endosymbionts influence host sexuality and reveal reproductive genes of early divergent fungi. Nat. Commun. 2017;8:1843. doi: 10.1038/s41467-017-02052-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro E., Lorca-Pascual J., Quiles-Rosillo M., Nicolás F., Garre V., Torres-Martínez S., Ruiz-Vázquez R. A negative regulator of light-inducible carotenogenesis in Mucor circinelloides. Mol. Genet. Genomics. 2001;266:463–470. doi: 10.1007/s004380100558. [DOI] [PubMed] [Google Scholar]
- Navarro E., Peñaranda A., Hansberg W., Torres-Martínez S., Garre V. A White Collar 1-like protein mediates opposite regulatory functions in Mucor circinelloides. Fungal Genet. Biol. 2013;52:42–52. doi: 10.1016/j.fgb.2012.12.003. [DOI] [PubMed] [Google Scholar]
- Navarro-Mendoza M.I., Pérez-Arques C., Murcia L., Martínez-García P., Lax C., Sanchis M., Capilla J., Nicolás F.E., Garre V. Components of a new gene family of ferroxidases involved in virulence are functionally specialized in fungal dimorphism. Sci. Rep. 2018;8:7660. doi: 10.1038/s41598-018-26051-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Mendoza M.I., Pérez-Arques C., Panchal S., Nicolás F.E., Mondo S.J., Ganguly P., Pangilinan J., Grigoriev I.V., Heitman J., Sanyal K., et al. Early diverging fungus Mucor circinelloides lacks centromeric histone CENP-A and displays a mosaic of point and regional centromeres. Curr. Biol. 2019;29:3791–3802.e6. doi: 10.1016/j.cub.2019.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas F.E., Torres-Martínez S., Ruiz-Vázquez R.M. Two classes of small antisense RNAs in fungal RNA silencing triggered by non-integrative transgenes. EMBO J. 2003;22:3983–3991. doi: 10.1093/emboj/cdg384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas F.E., de Haro J.P., Torres-Martinez S., Ruiz-Vazquez R.M. Mutants defective in a Mucor circinelloides dicer-like gene are not compromised in siRNA silencing but display developmental defects. Fungal Genet. Biol. 2007;44:504–516. doi: 10.1016/j.fgb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Nicolas F.E., Calo S., Murcia-Flores L., Garre V., Ruiz-Vazquez R.M., Torres-Martinez S. A RING-finger photocarotenogenic repressor involved in asexual sporulation in Mucor circinelloides. FEMS Microbiol. Lett. 2008;280:81–88. doi: 10.1111/j.1574-6968.2007.01044.x. [DOI] [PubMed] [Google Scholar]
- Nicolás F.E., Navarro-Mendoza M.I., Pérez-Arques C., López-García S., Navarro E., Torres-Martínez S., Garre V. Methods in Molecular Biology. Humana Press Inc.; 2018. Molecular tools for carotenogenesis analysis in the mucoral Mucor circinelloides; pp. 221–237. [DOI] [PubMed] [Google Scholar]
- Nicolás-Molina F.E., Navarro E., Ruiz-Vázquez R.M. Lycopene over-accumulation by disruption of the negative regulator gene crgA in Mucor circinelloides. Appl. Microbiol. Biotechnol. 2008;78:131–137. doi: 10.1007/s00253-007-1281-5. [DOI] [PubMed] [Google Scholar]
- Nosheen S., Yang J., Naz T., Nazir Y., Ahmad M.I., Fazili A.B.A., Li S., Mustafa K., Song Y. Annotation of AMP-activated protein kinase genes and its comparative transcriptional analysis between high and low lipid producing strains of Mucor circinelloides. Biotechnol. Lett. 2021;43:193–202. doi: 10.1007/s10529-020-02990-2. [DOI] [PubMed] [Google Scholar]
- Obraztsova I.N., Prados N., Holzmann K., Avalos J., Cerdá-Olmedo E. Genetic damage following introduction of DNA in Phycomyces. Fungal Genet. Biol. 2004;41:168–180. doi: 10.1016/j.fgb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Ozturk D.H., Dorfman R.H., Scapin G., Sacchettini J.C., Grubmeyer C. Locations and functional roles of conserved lysine residues in Salmonella typhimurium orotate phosphoribosyltransferase. Biochemistry. 1995;34:10755–10763. doi: 10.1021/bi00034a007. [DOI] [PubMed] [Google Scholar]
- Patiño-Medina J.A., Maldonado-Herrera G., Pérez-Arques C., Alejandre-Castañeda V., Reyes-Mares N.Y., Valle-Maldonado M.I., Campos-García J., Ortiz-Alvarado R., Jácome-Galarza I.E., Ramírez-Díaz M.I., et al. Control of morphology and virulence by ADP-ribosylation factors (Arf) in Mucor circinelloides. Curr. Genet. 2018;64:853–869. doi: 10.1007/s00294-017-0798-0. [DOI] [PubMed] [Google Scholar]
- Patiño-Medina J.A., Reyes-Mares N.Y., Valle-Maldonado M.I., Jácome-Galarza I.E., Pérez-Arques C., Nuñez-Anita R.E., Campos-García J., Anaya-Martínez V., Ortiz-Alvarado R., Ramírez-Díaz M.I., et al. Heterotrimeric G-alpha subunits Gpa11 and Gpa12 define a transduction pathway that control spore size and virulence in Mucor circinelloides. PLoS One. 2019;14:e0226682. doi: 10.1371/journal.pone.0226682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng D., Tarleton R. (2015). EuPaGDT: a web tool tailored to design CRISPR guide RNAs for eukaryotic pathogens. Microb Genomics. 2015;1 doi: 10.1099/mgen.0.000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Arques C., Navarro-Mendoza M.I., Murcia L., Lax C., Martínez-García P., Heitman J., Nicolás F.E., Garre V. Mucor circinelloides thrives inside the phagosome through an Atf-mediated germination pathway. mBio. 2019;10:1–15. doi: 10.1128/mBio.02765-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Arques C., Navarro-Mendoza M.I., Murcia L., Navarro E., Garre V., Nicolás F.E. The RNAi mechanism regulates a new Exonuclease gene involved in the virulence of mucorales. Int. J. Mol. Sci. 2021;22:2282. doi: 10.3390/ijms22052282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Arques C., Navarro-Mendoza M.I., Murcia L., Lax C., Sanchis M., Capilla J., Navarro E., Garre V., Nicolás F.E. A mucoralean white collar-1 photoreceptor controls virulence by regulating an intricate gene network during host interactions. Microorganisms. 2021;9:459. doi: 10.3390/microorganisms9020459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash H., Chakrabarti A. Global epidemiology of mucormycosis. J. Fungi. 2019;5:26. doi: 10.3390/jof5010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden M.M., Zaoutis T.E., Buchanan W.L., Knudsen T.A., Sarkisova T.A., Schaufele R.L., Sein M., Sein T., Chiou C.C., Chu J.H., et al. Epidemiology and outcome of zygomycosis: A review of 929 reported cases. Clin. Infect. Dis. 2005;41:634–653. doi: 10.1086/432579. [DOI] [PubMed] [Google Scholar]
- Roncero M.I.G., Jepsen L.P., Strøman P., van Heeswijck R. Characterization of a leuA gene and an ARS element from Mucor circinelloides. Gene. 1989;84:335–343. doi: 10.1016/0378-1119(89)90508-8. [DOI] [PubMed] [Google Scholar]
- Saha P., Ghosh S., Roy-Barman S. MoLAEA regulates secondary metabolism in Magnaporthe oryzae. mSphere. 2020;5:e00936. doi: 10.1128/mSphere.00936-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scapin G., Ozturk D.H., Grubmeyer C., Sacchettini J.C. The crystal structure of the orotate phosphoribosyltransferase complexed with orotate and α-D-5-Phosphoribosyl-1-pyrophosphate. Biochemistry. 1995;34:10744–10754. doi: 10.1021/bi00034a006. [DOI] [PubMed] [Google Scholar]
- Schneider Caroline A., Rasband Wayne S., Eliceiri Kevin W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva F., Navarro E., Peñaranda A., Murcia-Flores L., Torres-Martínez S., Garre V. A RING-finger protein regulates carotenogenesis via proteolysis-independent ubiquitylation of a White Collar-1-like activator. Mol. Microbiol. 2008;70:1026–1036. doi: 10.1111/j.1365-2958.2008.06470.x. [DOI] [PubMed] [Google Scholar]
- Skiada A., Pagano L., Groll A., Zimmerli S., Dupont B., Lagrou K., Lass-Florl C., Bouza E., Klimko N., Gaustad P., et al. Zygomycosis in Europe: Analysis of 230 cases accrued by the registry of the European confederation of medical mycology (ECMM) working group on zygomycosis between 2005 and 2007. Clin. Microbiol. Infect. 2011;17:1859–1867. doi: 10.1111/j.1469-0691.2010.03456.x. [DOI] [PubMed] [Google Scholar]
- Soliman S.S.M., Hamdy R., Elseginy S.A., Gebremariam T., Hamoda A.M., Madkour M., Venkatachalam T., Ershaid M.N., Mohammad M.G., Chamilos G., et al. Selective inhibition of Rhizopus eumelanin biosynthesis by novel natural product scaffold-based designs caused significant inhibition of fungal pathogenesis. Biochem. J. 2020;477:2489–2507. doi: 10.1042/BCJ20200310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliman S.S.M., Baldin C., Gu Y., Singh S., Gebremariam T., Swidergall M., Alqarihi A., Youssef E.G., Alkhazraji S., Pikoulas A., et al. Mucoricin is a ricin-like toxin that is critical for the pathogenesis of mucormycosis. Nat. Microbiol. 2021;6:313–326. doi: 10.1038/s41564-020-00837-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhara S.R., Paragache G., Panda N.K., Chakrabarti A. Mucormycosis in immunocompetent individuals: An increasing trend. J. Otolaryngol. 2005;34:402–406. doi: 10.2310/7070.2005.34607. [DOI] [PubMed] [Google Scholar]
- Trieu T.A., Calo S., Nicolás F.E., Vila A., Moxon S., Dalmay T., Torres-Martínez S., Garre V., Ruiz-Vázquez R.M. A non-canonical RNA silencing pathway promotes mRNA degradation in basal fungi. PLoS Genet. 2015;11:e1005168. doi: 10.1371/journal.pgen.1005168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu T.A., Navarro-Mendoza M.I., Perez-Arques C., Sanchis M., Capilla J., Navarro-Rodriguez P., Lopez-Fernandez L., Torres-Martinez S., Garre V., Ruiz-Vazquez R.M., et al. RNAi-based functional genomics identifies new virulence determinants in mucormycosis. PLoS Pathog. 2017;13:e1006150. doi: 10.1371/journal.ppat.1006150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heeswijck R., Roncero M.I.G. High frequency transformation of Mucor with recombinant plasmid DNA. Carlsberg Res. Commun. 1984;49:691. [Google Scholar]
- Valle-Maldonado M.I., Patiño-Medina J.A., Pérez-Arques C., Reyes-Mares N.Y., Jácome-Galarza I.E., Ortíz-Alvarado R., Vellanki S., Ramírez-Díaz M.I., Lee S.C., Garre V., et al. The heterotrimeric G-protein beta subunit Gpb1 controls hyphal growth under low oxygen conditions through the protein kinase A pathway and is essential for virulence in the fungus Mucor circinelloides. Cell. Microbiol. 2020;22:e13236. doi: 10.1111/cmi.13236. [DOI] [PubMed] [Google Scholar]
- Veisi A., Bagheri A., Eshaghi M., Rikhtehgar M.H., Rezaei Kanavi M., Farjad R. Rhino-orbital mucormycosis during steroid therapy in COVID-19 patients: A case report. Eur. J. Ophthalmol. 2021 doi: 10.1177/11206721211009450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellanki S., Navarro-Mendoza M.I., Garcia A., Murcia L., Perez-Arques C., Garre V., Nicolas F.E., Lee S.C. Mucor circinelloides: Growth, maintenance, and genetic manipulation. Curr. Protoc. Microbiol. 2018;49:e53. doi: 10.1002/cpmc.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arx J.A., Schipper M.A.A. The CBS fungus collection. Adv. Appl. Microbiol. 1978;24:215–236. doi: 10.1016/s0065-2164(08)70641-5. [DOI] [PubMed] [Google Scholar]
- Wagner L., Stielow J.B., de Hoog G.S., Bensch K., Schwartze V.U., Voigt K., Alastruey-Izquierdo A., Kurzai O., Walther G. A new species concept for the clinically relevant Mucor circinelloides complex. Persoonia. 2020;44:67–97. doi: 10.3767/persoonia.2020.44.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzbashev T.V., Larina A.S., Vybornaya T.V., Yuzbasheva E.Y., Gvilava I.T., Sineoky S.P. Repetitive genomic sequences as a substrate for homologous integration in the Rhizopus oryzae genome. Fungal Biol. 2015;119:494–502. doi: 10.1016/j.funbio.2015.02.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
•All data reported in this paper will be shared by the lead contact upon request.
•This paper does not report original code.
•Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.