Abstract
Translation termination, which liberates a nascent polypeptide from the ribosome specifically at stop codons, must occur accurately and rapidly. We established single-molecule fluorescence assays to track the dynamics of ribosomes and two requisite release factors (eRF1 and eRF3) throughout termination using an in vitro-reconstituted yeast translation system. We found that the two eukaryotic release factors bind together to recognize stop codons rapidly and elicit termination via a tightly regulated, multi-step process that resembles tRNA selection during translation elongation. Because the release factors are conserved from yeast to humans, the molecular events that underlie yeast translation termination are likely broadly fundamental to eukaryotic protein synthesis.
One Sentence Summary:
Direct visualization of eukaryotic translation termination reveals the dynamics of termination at stop codons.
Protein synthesis concludes when a translating ribosome encounters a stop codon at the end of an open reading frame, triggering recruitment of two factors to liberate the nascent polypeptide: eukaryotic release factor 1 (eRF1), a tRNA-shaped protein that decodes the stop codon in the ribosomal aminoacyl-tRNA site (A site) and cleaves the peptidyl-tRNA bond (1, 2, 3), and eukaryotic release factor 3 (eRF3), a GTPase that promotes eRF1 action (4, 5, 6). After translation termination, the ribosome, peptidyl-tRNA site (P-site) tRNA, and mRNA are released via recycling (4, 7, 8). Despite decades of study, the order and timing of molecular events that drive translation termination remain unclear, as multistep processes are difficult to assess using traditional approaches. A cohesive understanding of translation termination and its underlying steps, central to normal translation, would also support the treatment of diseases caused by premature stop codons, which include cystic fibrosis, muscular dystrophy, and hereditary cancers (9). As premature stop codons cause 11% of all heritable human diseases (10), stop-codon readthrough therapeutics have immense clinical potential (9, 11).
Direct tracking of release factor dynamics
Here we used an in vitro-reconstituted yeast translation system (12) and single-molecule fluorescence spectroscopy to track eukaryotic release factor dynamics and termination directly. We reasoned that ribosomes translating mRNAs with very short open reading frames would provide the simplest system for detailed analysis of the discrete sub-steps of termination. Ribosome complexes were programmed with Met (M-Stop) or Met-Phe (M-F-Stop) mRNAs, achieved via incubation with purified Met-tRNAMeti / initiation factors / elongation factors and tRNAs (as appropriate), and then reacted with saturating amounts of eRF1 and eRF3 (13). Peptide release from both M-Stop and M-F-Stop ribosome complexes occurred at similar rates as a longer tetrapeptide (M-F-K-K-Stop) programmed ribosome complex (Fig. 1A and S1A–B), and also matched the rate previously characterized for tripeptide programmed ribosome complexes (4, 5). To monitor eRF1 and eRF3 binding to ribosomes in real time, we labeled both proteins specifically with fluorescent dyes (Fig. S1C–D) and established that both labeled proteins exhibited wild-type peptide release activity (Fig. 1B and S1E–F). Association of eRF1 with the ribosome was monitored by Förster resonance energy transfer (FRET) between 60S subunits labeled with Cy3 (FRET donor) on the C-terminus of uL18 (14) and eRF1 labeled with Cy5 (FRET acceptor) on the N-terminus. Structural models place these termini ~50 Å apart when eRF1 is bound in the A site (Fig. 1C) (3, 14, 15). Next, Cy3-labeled ribosomal complexes programmed with Met in the P site and either UAA or UUC in the A site were combined with Cy5-eRF1 and unlabeled eRF3, and FRET was monitored at equilibrium using total internal reflection fluorescence (TIRF) Microscopy. eRF1-60S FRET was only observed when a stop codon was in the A site (Fig. 1D and S1G–H), demonstrating the specificity of the FRET signal for proper eRF1 association mediated by a stop codon.
Fig. 1:
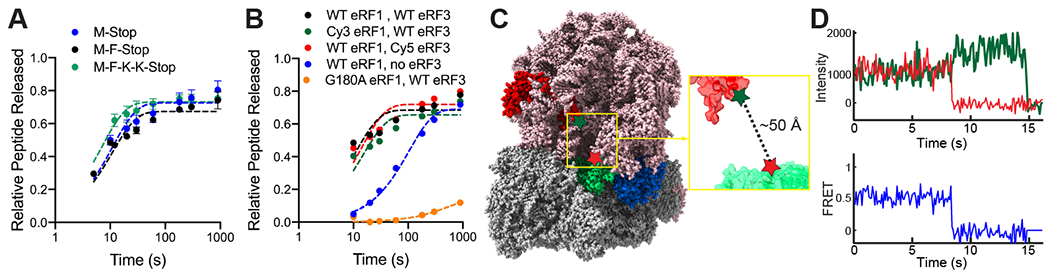
Bulk biochemical and single-molecule studies of termination. (A) Peptides are liberated at similar rates from ribosomes translating a variety of model mRNAs. (B) Wild-type and labeled release factors liberate peptides from ribosomes; catalytically-dead eRF1 (orange) is inactive. (C) Structural modeling (PDB ID: 5LZT (15)) suggests labeled eRF1 (green, Cy5-labeled at red star) would FRET with ribosomes (red, Cy3-labeled at green star) upon binding to A site. (D) FRET observed with Cy3-eRF1 and Cy5-60S by TIRF microscopy.
We leveraged this FRET-based binding signal to probe the roles of eRF1 and eRF3 in translation termination. We first prepared 80S ribosomal complexes programmed on 5’-biotinylated M-Stop mRNAs with Cy5-labeled 60S subunits; these complexes were tethered to neutravidin-coated zero-mode waveguide (ZMW) surfaces (16, 17). Upon start of real-time data acquisition, Cy3-eRF1, excess GTP, and unlabeled eRF3 were added to ZMWs, and Cy3-eRF1 and Cy5-60S fluorescence within individual ZMWs were monitored by excitation with a 532-nm laser (Fig. 2A).
Fig. 2:
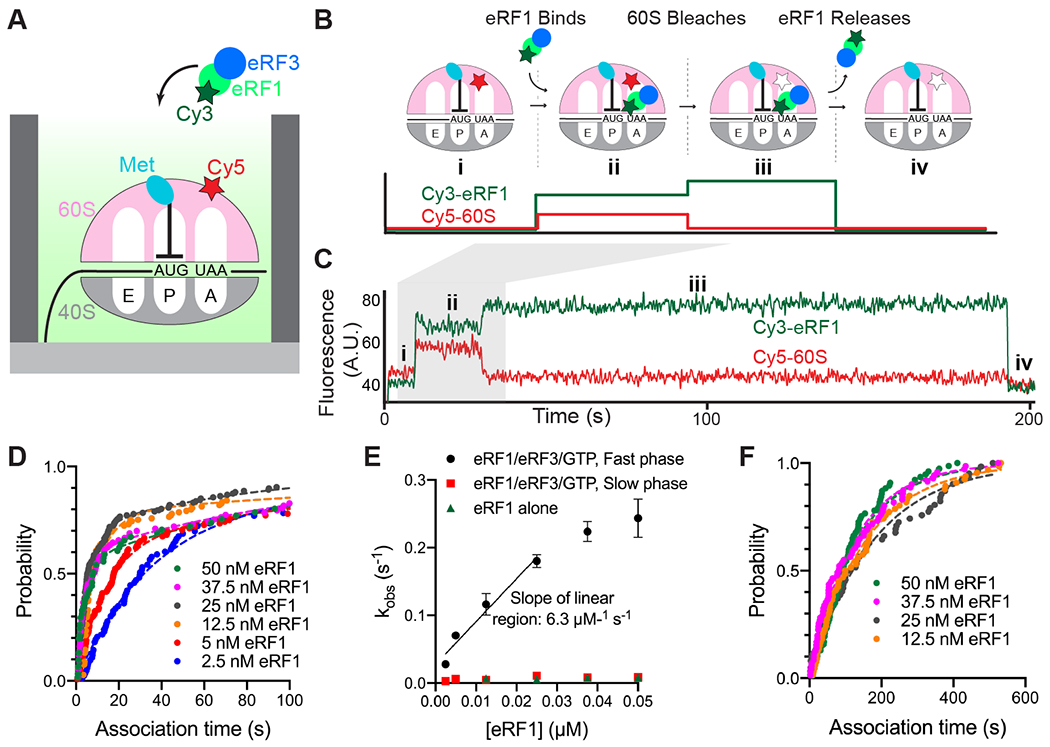
eRF3 promotes fast binding of eRF1 to ribosomes halted at stop codons. (A) Legend. (B) Assay schematic. (C) Example of fast binding of eRF1 (Cy3, green) to M-Stop ribosomes (Cy5, red) observed in the presence of eRF3. (D) Binding of eRF1 to M-Stop ribosomes is fast and concentration-dependent in the presence of eRF3; arrival time distributions were fit to a double-exponential model. (E) Observed rates of eRF1 binding to M-Stop ribosomes (kobs) with and without eRF3; the plateau observed with eRF1/eRF3/GTP fast phase coincides with the rate of sample mixing in ZMWs (17). (F) Binding of eRF1 to M-Stop ribosomes is slow and eRF1 concentration-independent without eRF3; arrival time distributions were fit to an exponential model.
Rapid, concentration-dependent eRF1 binding to the ribosomal A site was detected upon delivery of the release factors (Fig. 2B–C and S2A–C). Association kinetics were fit to a double-exponential function with a dominant (56 – 83%) eRF1 concentration-dependent fast phase with a pseudo-second order rate constant of 6.3 ± 3.9 μM−1 s−1 (95% CI; Fig. 2D–E and S2B); a minor (17 – 44%) slow phase, which did not vary with eRF1 concentration, was also observed (e.g. kobs = ~0.009 s−1; Fig. 2E and S2B). Conversely, eRF1 bound very slowly to these same complexes in the absence of eRF3 (e.g. kobs = ~0.008 s−1; Fig. 2E–F and S2D–E); the rate constant for this eRF3-independent binding was similar to the slow phase observed with eRF3, and also was unaffected by eRF1 concentration. In all cases, eRF1 binding events were long-lived (e.g. τ = 227 ± 13 s, Fig. S2B and S2E), and prolonged detection is likely limited by dye photobleaching lifetime (Fig. S2F). Notably, in the presence of eRF3, the rapid eRF1 binding observed here is similar to the rate of Phe-tRNAPhe ternary complex binding to its cognate A-site codon under similar conditions (9.0 ± 0.4 μM−1 s−1; Fig. S2G). These results indicate that eRF1 binding, which would otherwise be limited by a slow event, is rapid enough to compete with tRNAs for A-site occupancy when assisted by eRF3.
We next tracked eRF3 dynamics directly, independent of eRF1, to establish a baseline understanding of its interaction with the ribosome. We used a previously established inter-ribosomal subunit FRET signal to confirm 80S complex formation (14), and monitored dye-labeled eRF3 dynamics via fluorescent bursts that occur upon factor binding to immobilized ribosomes. Ribosomes, Cy3-labeled on uL18 and Cy5-labeled on uS19 (yielding FRET upon 80S formation), were programmed with 5’-biotinylated M-Stop mRNAs and tethered to ZMWs. Next, Cy5-eRF3 and GTP were added to ZMWs and were illuminated with 532- and 642-nm lasers. After an initial phase of FRET, typified by rapid 40S-Cy5 photobleaching, brief bursts of additional Cy5 signal were observed that marked binding and dissociation of eRF3 (Fig. 3A–B). eRF3 binding was concentration-dependent (Fig. 3C), and association kinetics were fit to an exponential function with a pseudo-second order rate constant of 0.4 ± 0.2 μM−1 s−1 (95% CI; Fig. S3A–B). eRF3 resided briefly on the ribosome (τ = 0.15 ± 0.01 s; Fig. 3D), and the dwell times between eRF3 binding events varied with its concentration (Fig. S3B), consistent with a bimolecular association reaction. Inclusion of GTP analogs, GDP, or a GTPase-deficient eRF3 mutant (H348E (4)) did not markedly impact its association or dissociation rates (~2-fold or less; Fig. 3D and S3C) suggesting that this binding cycle occurs independent of the eRF3 nucleotide-bound state or GTP hydrolysis.
Fig. 3:
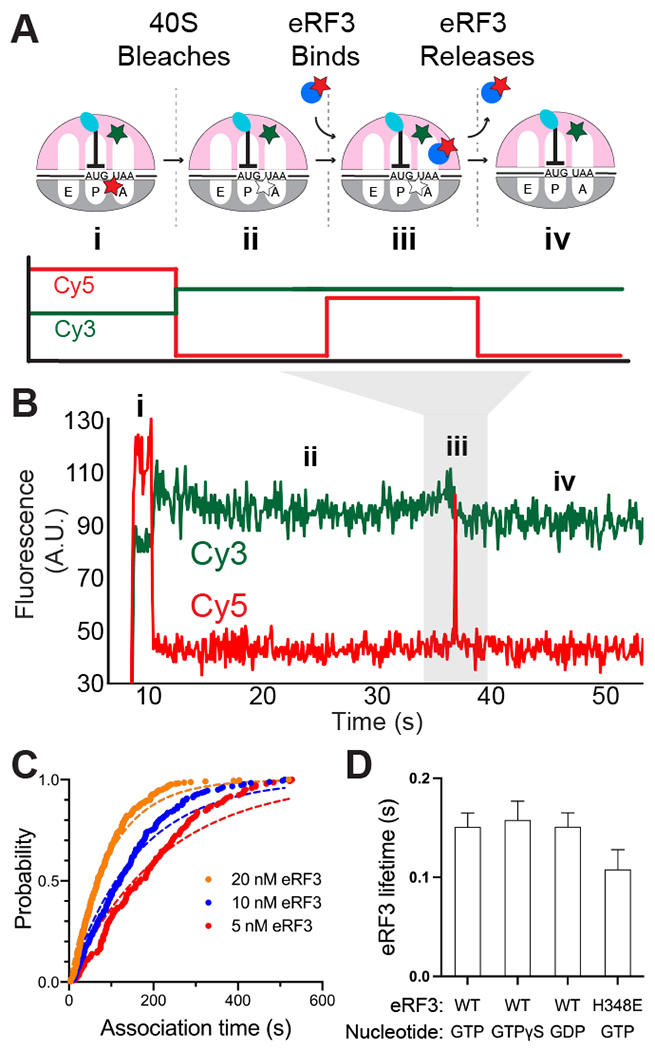
Observing eRF3 dynamics in ZMWs. (A) Assay schematic. (B) Example of eRF3 binding to M-Stop ribosomes. (C) Binding of eRF3 to M-Stop ribosomes is concentration-dependent; arrival time distributions were fit to an exponential model. (D) GTP hydrolysis by eRF3 is not required for its release from the ribosome in the absence of eRF1.
Two distinct models could explain how eRF3 promotes fast association of eRF1 with ribosomes halted at stop codons. eRF3 may first bind to ribosomes, triggering rearrangements that favor subsequent association of eRF1 with ribosomes. Alternatively, eRF3 may act as a chaperone, directly delivering eRF1 to ribosomes (5). To distinguish between these models, we performed single-molecule experiments similar to those described above but now simultaneously tracking fluorescent eRF1 and eRF3. We observed concurrent binding of the two factors to M-Stop ribosomes (Fig. 4A–B). While we also observed eRF1 and/or eRF3 binding individually to ribosomes in these experiments–unsurprising since the release factors can each bind alone to ribosomes and are at sub-saturating concentrations–the likelihood of such independent binding events occurring simultaneously was very low (<0.1%), allowing us to rule out that co-arrivals happen primarily by random chance (see Methods). Analysis of Cy5 (eRF1) and Cy3.5 (eRF3) fluorescence intensities, aligned to the beginning of apparent co-association events (“post-synchronization”), further demonstrates that eRF3 binding to a stop codon-halted ribosome is transient, whereas eRF1 resides longer on the ribosome (Fig. 4C). Omission of GTP decreased the number of observed co-binding events by 17-fold, confirming that eRF1, eRF3, and GTP bind the ribosome together as a pre-formed ternary complex (Fig. S4A). Ternary complex association kinetics were fit to a double-exponential function, yielding a dominant fast phase rate that was dependent upon eRF1 concentration and a pseudo-second order rate constant of 2.6 ± 5.1 μM−1 s−1 (95% CI; Fig. 4D and S4B–D). In contrast to the dynamics of eRF3 in absence of eRF1 (where eRF3 lifetime had little dependence on GTP hydrolysis, ranging from 0.11 – 0.16 s; Fig 3C), here GTP accelerated eRF3 release from ribosomes by 8-fold compared to experiments performed with the more slowly hydrolyzed analog GTPγS (0.3 ± 0.1 s with GTP vs. 2.5 ± 0.1 s with GTPγS; Fig. 4E and S4E–F). Substitution of wild-type eRF3 with a GTPase-deficient mutant similarly slowed its release from the ribosome by 5-fold (1.6 ± 0.1 s; Fig. 4E and S4E–F). Thus, eRF3 is a chaperone that delivers eRF1 to ribosomes halted at stop codons, and eRF3 departure from the ribosome is partly governed by its GTPase activity.
Fig. 4:
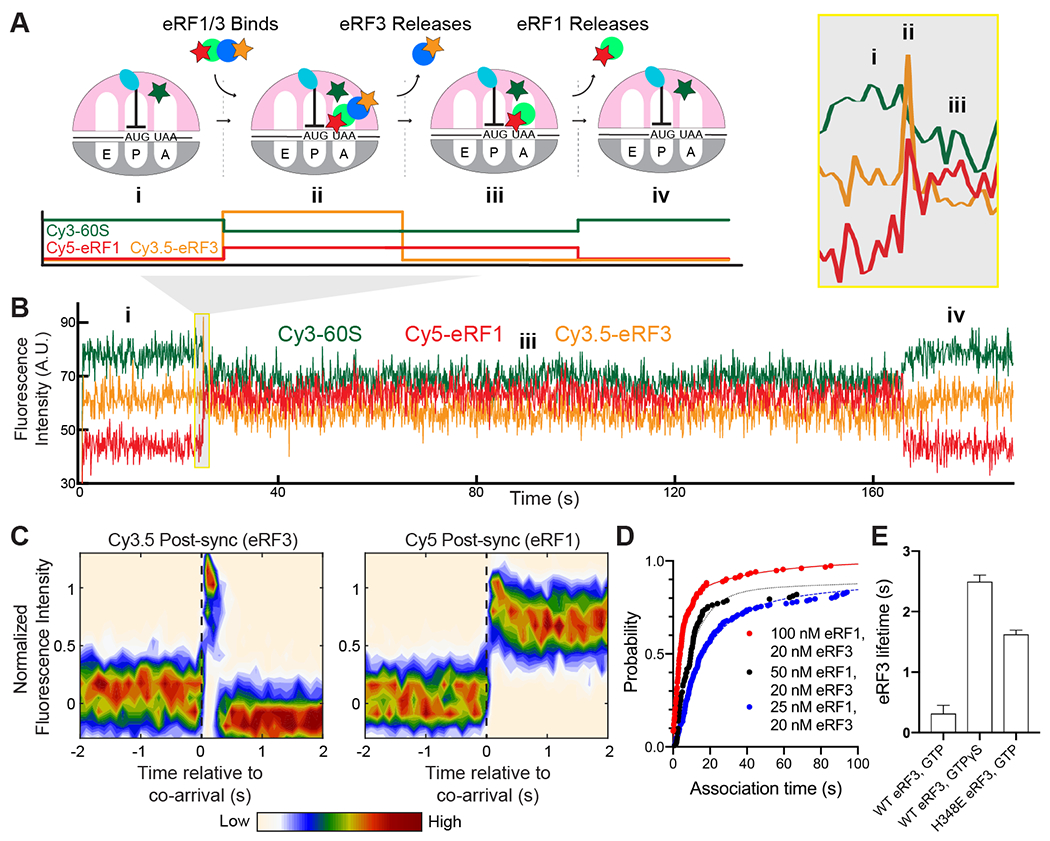
eRF3 delivers eRF1 quickly to ribosomes halted at stop codons. (A) Assay schematic. (B) Example of simultaneous binding of eRF1 (Cy5, red) and eRF3 (Cy3.5, yellow) to M-Stop ribosomes (Cy3, green). (C) Post-synchronization plot of fluorescence changes observed upon simultaneous binding of eRF1 and eRF3 (denoted as a dashed, black vertical line). (D) Simultaneous binding of eRF1 and eRF3 to M-Stop ribosomes is fast and concentration-dependent; arrival time distributions were fit to a double-exponential model. (E) GTP hydrolysis by eRF3 accelerates its release from the ribosome in the presence of eRF1.
Real-time monitoring of translation termination
We next sought to understand the timing and regulation of peptidyl-tRNA ester bond hydrolysis catalyzed by eRF1. Peptide hydrolysis triggers rapid rearrangement of P-site tRNA from a classical (P/P) to a hybrid (P/E) state (18), and we hypothesized this rearrangement could be tracked via FRET between labeled P-site tRNA and A-site-bound eRF1 (~34 Å separation before vs. ~50 Å after rearrangement; Fig. 5A). To test this, we tethered to ZMWs ribosomes programmed on an M-Stop mRNA with Cy3-labeled Met-tRNAMeti (FRET donor) in the P site, added catalytically-inactive Cy5-eRF1 (G180A, FRET acceptor), unlabeled eRF3, and GTP, and illuminated with a 532-nm laser. As expected, high FRET was observed between the classical-state tRNA and eRF1 (μ = 0.63, σ = 0.10; Fig. 5B). Next, we repeated the assay but added puromycin, a drug that cleaves the peptidyl-tRNA bond, and indeed observed lower efficiency FRET between the now-hybrid-state tRNA and catalytically-inactive eRF1 (μ = 0.53, σ = 0.10; Fig. 5B). Therefore, peptidyl-tRNA bond status can be deduced by monitoring P-site tRNA conformation with respect to eRF1 through this FRET signal.
Fig. 5:
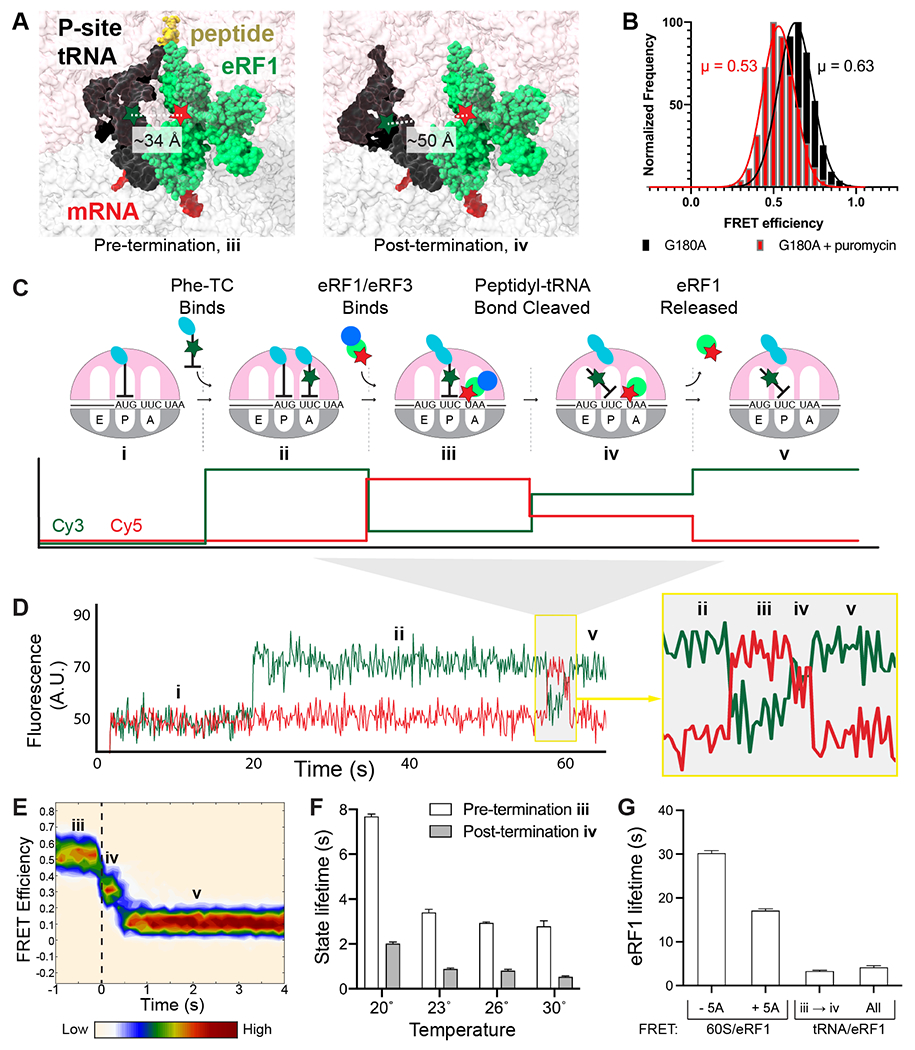
Tracking peptidyl-tRNA bond hydrolysis. (A) Model of FRET between eRF1 (Cy5, red star) and P-site tRNA (Cy3, green star) before and after termination. Left, pre-termination (modeled by 40S alignment, PDB IDs: 5LZU and 5LZT (15)). Right, post-termination (modeled by 40S alignment, PDB IDs: 5LZU and 3J77 (15, 20)). (B) FRET observed between P-site Cy3-Met-tRNAi and G180A Cy5-eRF1 (black); addition of puromycin (red) yields lower FRET as expected. (C) Assay schematic. (D) Example of FRET observed with Cy3-Phe (green) and Cy5-eRF1 (red). (E) Post-synchronization plot of FRET efficiency observed before and after peptidyl-tRNA bond hydrolysis (marked by a dashed, black vertical line). (F) Pre- and post-termination state lifetimes observed at different temperatures. (G) Dependence of eRF1 lifetime on eIF5A and detection methods; eRF1 lifetimes observed via tRNA/eRF1 FRET either represent all binding events (all) or only those resulting in termination (iii → iv).
We used this FRET signal to correlate eRF1 dynamics with peptidyl-tRNA bond hydrolysis in real time. Ribosomes programmed on 5’-biotinylated M-F-Stop mRNAs were tethered to ZMWs and illuminated with a 532-nm laser. We then added a mixture of Cy3-labeled Phe-tRNAPhe (FRET donor), Cy5-eRF1 (FRET acceptor), excess eRF3, elongation factors, and eIF5A (an accessory factor that accelerates elongation and termination (19)). Fast tRNA binding, denoted by high Cy3 signal, was observed soon after factor addition and persisted until translocation and subsequent eRF1 binding occurred (Fig. 5C–D and S5A). Association of eRF1 initially resulted in a high-FRET signal (μ = 0.67, σ = 0.07; herein referenced as “pre-termination state”; Fig. S5B–C) and was followed by a lower-FRET signal (μ = 0.46, σ = 0.11; “post-termination state”; Fig. 5C–E and S5B). Finally, eRF1 was released from the ribosome, resulting in restoration of high Cy3 signal (Fig. 5C–E). Critically, substitution of eRF1 with an inactive mutant reduced the number of observed high- to lower-FRET transitions by ~7-fold (Fig. S5D–E), which closely matched the relative extent of peptide release observed in bulk with either wild-type or G180A eRF1 (Fig. 1B and S1E–F). P-site tRNA/eRF1 FRET is also specific for stop codon recognition, as replacement of the UAA stop codon with near-cognate UAU completely eliminated these FRET transitions (Fig. S5D).
We used this tRNA/eRF1 FRET assay to characterize the kinetics of peptidyl-tRNA bond hydrolysis, focusing on ribosomal events before and after termination. Pre-termination state lifetimes of eRF1 on the 80S ribosome fit well to a two-step, irreversible kinetic model with termination occurring in 2.8 s at 30 °C (95% CI: 2.6 – 3.0 s; Fig. 5F and S5F–G). eRF1 dissociation kinetics, fit to a single step model by a single exponential, revealed that eRF1 is released quickly after termination in 0.5 s (95% CI: 0.5 – 0.6 s, Fig. 5F and S5G–H); eRF1 lifetime was unaffected by laser power variation, showing that its lifetime is not limited by dye photobleaching, Fig. S5I). Further support that peptidyl-tRNA bond hydrolysis favors eRF1 release was observed with catalytically-inactive eRF1, which resided 6-fold longer on ribosomes (Fig. S5J). Peptidyl-tRNA bond cleavage also hindered rebinding of eRF1 (Fig. S5K), demonstrating that termination decreases the affinity of eRF1 for ribosomes. The long eRF1 lifetime observed in our prior measurements (Fig. 2 and 4) is attributable to differences in detection methods and the omission of eIF5A from those assays, as eIF5A does not affect eRF1 association rates but does enhance termination and eRF1 release rates (Fig. 5G and S6) (19). Peptidyl-tRNA bond hydrolysis rates increased with temperature from 20 – 30 °C (Fig. 5F, S5G, and S7A–C), and subsequent Eyring and Gibbs analyses revealed that termination is regulated by a step with a significant energetic barrier (Fig. S7D); consistent with this notion, prior structural work demonstrated that eRF1 undergoes a large-scale conformational change (termed “accommodation”) to render it catalytically active (3, 15), and the tRNA rearrangement tracked by this assay would also depend upon transition of the ribosome from a classical to a hybrid state (20). Release of eRF1 from the ribosomal A site is also energetically costly (Fig. 5F and S7E), likely due to extensive interactions that anchor eRF1 to the stop codon (3). Termination proceeded at a similar rate when the UAA stop codon was changed to either UAG or UGA (Fig. S8), suggesting that a common mechanism is employed at all three stop codons. We therefore observed an ordered series of events at stop codons, with eRF1 eliciting peptidyl-tRNA bond cleavage within ~3 s of ribosomal association, followed by rapid eRF1 release.
Stop-codon readthrough effectors slow termination dynamics
We next explored the impact of cis-acting mRNA elements on termination. Prior work uncovered a class of 3’ untranslated region (“UTR”) mRNA sequences that promote stop-codon readthrough (21, 22), yet it was unclear if these elements function in part by inhibiting termination. Indeed, insertion of a 6-nucleotide sequence that promotes stop-codon readthrough with 30% efficiency (CAAUUA) into the 3’ UTR of M-F-Stop mRNAs lengthened pre-termination state duration by 2-fold (5.9 s, 95% CI: 5.2 – 7.0 s with CAAUUA vs. 2.9 s, 95% CI: 2.7 – 3.0 s without CAAUUA; Fig. 6A and S9A–B). Insertion of other sequences that promote readthrough at lower efficiencies also lengthened the pre-termination state, albeit to lesser degrees (Fig. 6A and S9A–B). Thus, sequences that enhance stop-codon readthrough hinder peptidyl-tRNA bond cleavage by eRF1.
Fig. 6:
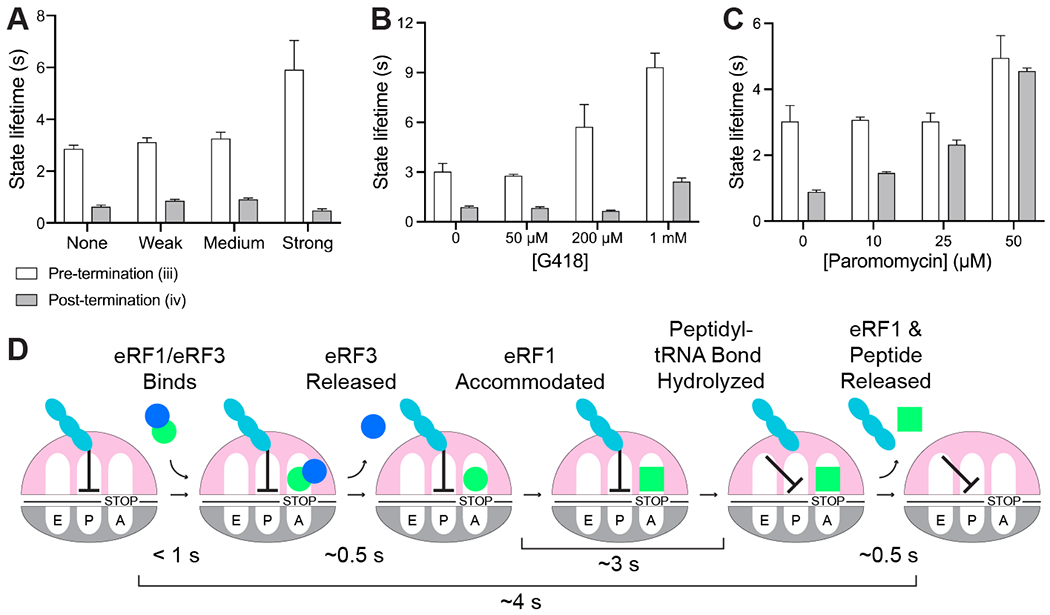
Termination is regulated by release factors, 3’ UTR mRNA sequence, and small molecules. (A) Insertion of 3’ UTR mRNA sequences known to promote stop-codon readthrough (“Weak” = CAAAGA, 10% efficiency; “Medium” = CAAUCA, 20% efficiency; “Strong” = CAAUUA, 30% efficiency) hinder termination; eRF1 release is unaffected. (B-C) The aminoglycosides G418 (B) and paromomycin (C) slow termination and eRF1 release. (D) Order and timing of events in eukaryotic termination.
The aminoglycosides paromomycin and G418 also promote stop-codon readthrough (23), primarily by stabilizing near-cognate tRNA in the A site. Paromomycin also inhibits bacterial termination (24), yet its effects on eukaryotic termination were not deeply characterized (25). G418 was recently identified as a eukaryotic termination inhibitor via bulk biochemical studies with a mixed mammalian system, however its mode of action remains unclear (26). We therefore applied our suite of assays to examine the effects of these drugs on eukaryotic termination. Addition of 1 mM G418 (EC50 of ~2 mM (27)) lengthened the pre-and post-termination state duration (by 2- and 3-fold, respectively; Fig. 6B and S9C–E). Similarly, 50 μM paromomycin (EC50 of ~35 μM (27)) increased the pre-termination state duration (by nearly 2-fold; Fig. 6C and S9C–E), and lengthened post-termination state duration (by 5-fold; Fig. 6C and S9C–E). A titration of paromomycin revealed concentration-dependent effects of the drug on post-termination state duration (Fig. 6C and S9C–E). Simultaneous tracking of eRF1/3 dynamics (as described in Fig. 4) revealed that 50 μM paromomycin slowed eRF1/eRF3 co-binding to the ribosome (by more than 2-fold; Fig. S9F). Together, these studies demonstrate that stop-codon readthrough effectors hinder numerous facets of termination, thus uncovering additional nodes to target with potential therapeutics.
Discussion
Whereas prior work broadly described the roles of eRF1 and eRF3 in regulating eukaryotic termination (2, 4, 5, 19, 28), here we directly monitored the kinetics of individual sub-steps to obtain a higher-resolution view of this essential process (Fig. 6D). First, a pre-bound ternary complex of eRF1, eRF3, and GTP rapidly binds to a ribosome halted at a stop codon (Fig. 2 and 4). eRF3 appears to unlock eRF1 conformation to facilitate fast ribosomal binding, as the association of eRF1 alone is slow and governed by an eRF1 concentration-independent event (Fig. 2D–E); consistent with this notion, prior structural studies demonstrated that the predominant conformation of eRF1 free in solution is incompatible with ribosomal binding (Fig. S10) (3, 15, 29, 30). Next, eRF3 hydrolyzes GTP to promote its own release (Fig. 4D), which permits the rearrangement of eRF1 to an active conformation (15). Accommodated eRF1 then rapidly cleaves the peptidyl-tRNA bond, triggering ribosomal intersubunit rotation, movement of the deacylated P-site tRNA to a P/E hybrid state and ejection of both eRF1 and the liberated peptide (Fig. 5). Indeed, eRF1 release was slowed by small molecules that hinder ribosomal rotation (G418 and paromomycin, Fig. 6B–C) (27), which further shows the interdependence of these events. Direct tracking of peptidyl-tRNA bond hydrolysis further uncovered how small molecules and mRNA sequences inhibit discrete steps in termination to promote stop-codon readthrough (Fig. 6A–C). With the critical caveats that termination may also be influenced by unidentified nascent chain dynamics and other trans-acting factors, we propose that the termination mechanisms described here are fundamental to eukaryotic translation, as the release factors are widely conserved from yeast to humans (Fig. S11) (1, 6).
To assess the physiological relevance of our in vitro results, we compared them to ribosome profiling measurements that report A-site occupancy via the ratio of short (21 nt: empty A site) versus long (28 nt: occupied A site) ribosome-protected footprints (RPFs). Prior studies suggested that binding of eRF1 to ribosomes is not rate-limiting for termination, as long RFPs significantly outnumber short RFPs at stop codons (31). We confirmed this hypothesis through direct observation of termination sub-steps (Fig. 6D), which demonstrated that release factor binding is indeed faster than subsequent ribosomal events. The finding that termination (~4 s) is fast relative to initiation ~20 – 60 s (32, 33), albeit somewhat slower than elongation (0.05 – 1.4 s per codon (33)), suggests the existence of an intricate choreography that prevents the accumulation of ribosomes at stop codons. Consistent with this, ribosomal profiling in eRF1-depleted cells revealed a dramatic increase in queueing of ribosomes at stop codons (31).
Eukaryotic termination differs strikingly from the mechanisms described previously for bacterial termination, in which the bacterial namesake of eRF3, RF3, drives departure of eRF1-like factors (RF1/2) from ribosomes (34). Instead, eukaryotic termination more closely resembles translation elongation (5), in which bacterial EF-Tu and eukaryotic eEF1A assist in the selection of proper tRNAs via a tightly-regulated, multi-step process. In eukaryotic termination, eRF3 (itself an EF-Tu/eEF1A homolog) in complex with GTP quickly delivers eRF1 (a tRNA-shaped protein) to a stop codon in the A site (Fig. 4A), similar to the rapid association of eEF1A/tRNA/GTP ternary complex with a sense codon in the A site. eRF3 hydrolyzes GTP to promote its release from the ribosome and facilitate eRF1 accommodation (Fig. 4D) (15), just as EF-Tu and eEF1A hydrolyze GTP to accelerate their ejection and favor tRNA accommodation (35, 36). Thus, the similarities between elongation and eukaryotic termination are not only limited to factor architecture, but also include the molecular choreography of these processes.
The fidelity of translation elongation is driven in part by kinetic proofreading, in which EF-Tu/eEF1A preferentially rejects non-cognate tRNAs in two sequential steps to boost overall accuracy (35, 36, 37). While the basis of termination fidelity is unknown, we consider kinetic proofreading an intriguing model. eRF3 is essential for termination fidelity, as its inclusion boosts specificity by 2,600-fold (28). Here we showed eRF3 conformationally unlocks and delivers eRF1 to ribosomes (Fig. 2 and 4) and facilitates eRF1 accommodation in an eRF3 GTPase-dependent manner (Fig. 4), thus providing eRF3 with multiple opportunities to favor genuine stop codons. Further study of termination sub-step kinetics at cognate and near-cognate stop codons will reveal whether proofreading governs eukaryotic termination fidelity.
Mutations that introduce a premature stop codon pose a unique challenge for therapeutic intervention. These mutations trigger premature termination, liberating an incomplete polypeptide, and the defective mRNAs are further degraded via nonsense-mediated decay (NMD) (38). To achieve effective therapeutic readthrough of premature stop codons, elongation, termination, and NMD must all be carefully tuned to avoid widespread misregulation of gene expression while still eliciting enough readthrough to alleviate disease. Thus, termination and NMD inhibitors may prove most useful as adjuvants, lengthening the kinetic window for drug-mediated readthrough of premature stop codons. Extension of these single-molecule assays to monitor stop-codon readthrough and NMD will provide the quantitative tools necessary to evaluate combination therapies, paving the way to effective treatments.
Supplementary Material
Acknowledgements:
We thank Christopher Lapointe, Christine Preston, and Amanda Gilliam Valentic for editing assistance, and also Puglisi and Green lab members for discussion and input.
Funding:
This work was supported by the A.P. Giannini Foundation (Postdoctoral Fellowship, MRL), Cystic Fibrosis Foundation (PUGLISI20GO, JDP), and NIGMS (R37GM059425, RG and R01GM113078 and R01GM51266, JDP).
Footnotes
Competing interests: None declared.
Data and materials availability:
All data are available in the main text or the supplementary materials. Relevant expression constructs are available upon request.
References and Notes
- 1.Frolova L et al. , A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature 372, 701–703 (1994). [DOI] [PubMed] [Google Scholar]
- 2.Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV, In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell 125, 1125–1136 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Brown A, Shao S, Murray J, Hegde RS, Ramakrishnan V, Structural basis for stop codon recognition in eukaryotes. Nature 524, 493–496 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shoemaker CJ, Green R, Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc. Natl. Acad. Sci. USA 108, E1392–1398 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eyler DE, Wehner KA, Green R, Eukaryotic release factor 3 is required for multiple turnovers of peptide release catalysis by eukaryotic release factor 1. J. Biol. Chem 288, 29530–29538 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhouravleva G et al. , Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 14, 4065–4072 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heuer A et al. , Structure of the 40S-ABCE1 post-splitting complex in ribosome recycling and translation initiation. Nat. Struct. Mol. Biol 24, 453–460 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Becker T et al. , Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature 482, 501–506 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keeling KM, Xue X, Gunn G, Bedwell DM, Therapeutics based on stop codon readthrough. Annu. Rev. Genom. Hum. Genet 15, 371–394 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mort M, Ivanov D, Cooper DN, Chuzhanova NA, A meta-analysis of nonsense mutations causing human genetic disease. Hum. Mutat 29, 1037–1047 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Bordeira-Carrico R, Pego AP, Santos M, Oliveira C, Cancer syndromes and therapy by stop-codon readthrough. Trends Mol. Med 18, 667–678 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Acker MG, Kolitz SE, Mitchell SF, Nanda JS, Lorsch JR, Reconstitution of yeast translation initiation. Methods Enzymol. 430, 111–145 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Methods and materials are available as supplementary online materials.
- 14.Wang J et al. , eIF5B gates the transition from translation initiation to elongation. Nature 573, 605–608 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shao S et al. , Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes. Cell 167, 1229–1240.e1215 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uemura S et al. , Real-time tRNA transit on single translating ribosomes at codon resolution. Nature 464, 1012–1017 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J et al. , High-throughput platform for real-time monitoring of biological processes by multicolor single-molecule fluorescence. Proc. Natl. Acad. Sci. USA 111, 664–669 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flis J et al. , tRNA Translocation by the Eukaryotic 80S Ribosome and the Impact of GTP Hydrolysis. Cell Rep 25, 2676–2688.e2677 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schuller AP, Wu CC, Dever TE, Buskirk AR, Green R, eIF5A Functions Globally in Translation Elongation and Termination. Mol. Cell 66, 194–205.e195 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Svidritskiy E, Brilot AF, Koh CS, Grigorieff N, Korostelev AA, Structures of yeast 80S ribosome-tRNA complexes in the rotated and nonrotated conformations. Structure 22, 1210–1218 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Namy O, Hatin I, Rousset JP, Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep 2, 787–793 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Namy O et al. , Identification of stop codon readthrough genes in Saccharomyces cerevisiae. Nucleic Acids Res. 31, 2289–2296 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wangen JR, Green R, Stop codon context influences genome-wide stimulation of termination codon readthrough by aminoglycosides. eLife 9, e52611 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Youngman EM, Cochella L, Brunelle JL, He S, Green R, Two distinct conformations of the conserved RNA-rich decoding center of the small ribosomal subunit are recognized by tRNAs and release factors. Cold Spring Harb Symp Quant Biol 71, 545–549 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Eyler DE, Green R, Distinct response of yeast ribosomes to a miscoding event during translation. RNA 17, 925–932 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Susorov D, Egri S, Korostelev AA, Termi-Luc: a versatile assay to monitor full-protein release from ribosomes. RNA 26, 2044–2050 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prokhorova I et al. , Aminoglycoside interactions and impacts on the eukaryotic ribosome. Proc Natl Acad Sci U S A 114, E10899–e10908 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.G. Indrisiunaite, Uppsala University, Digital Comprehensive Summaries of Uppsala Dissertations from the Faculty of Science and Technology (2019). [Google Scholar]
- 29.Song H et al. , The crystal structure of human eukaryotic release factor eRF1--mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 100, 311–321 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Cheng Z et al. , Structural insights into eRF3 and stop codon recognition by eRF1. Genes Dev. 23, 1106–1118 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu CC, Zinshteyn B, Wehner KA, Green R, High-Resolution Ribosome Profiling Defines Discrete Ribosome Elongation States and Translational Regulation during Cellular Stress. Mol. Cell 73, 959–970.e955 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shah P, Ding Y, Niemczyk M, Kudla G, Plotkin JB, Rate-limiting steps in yeast protein translation. Cell 153, 1589–1601 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chu D et al. , Translation elongation can control translation initiation on eukaryotic mRNAs. EMBO J. 33, 21–34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Freistroffer DV, Pavlov MY, MacDougall J, Buckingham RH, Ehrenberg M, Release factor RF3 in E.coli accelerates the dissociation of release factors RF1 and RF2 from the ribosome in a GTP-dependent manner. EMBO J. 16, 4126–4133 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pape T, Wintermeyer W, Rodnina MV, Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J. 17, 7490–7497 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pape T, Wintermeyer W, Rodnina M, Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J. 18, 3800–3807 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loveland AB, Demo G, Korostelev AA, Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading. Nature 584, 640–645 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He F, Jacobson A, Upf1p, Nmd2p, and Upf3p regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs. Mol. Cell. Biol 21, 1515–1530 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yin J et al. , Genetically encoded short peptide tag for versatile protein labeling by Sfp phosphopantetheinyl transferase. Proc. Natl. Acad. Sci. USA 102, 15815–15820 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gutierrez E et al. , eIF5A promotes translation of polyproline motifs. Mol. Cell 51, 35–45 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shoemaker CJ, Eyler DE, Green R, Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science 330, 369–372 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tesina P et al. , Molecular mechanism of translational stalling by inhibitory codon combinations and poly(A) tracts. The EMBO journal 39, e103365–e103365 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J et al. , Structural basis for the transition from translation initiation to elongation by an 80S-eIF5B complex. Nature Communications 11, 5003 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mitkevich VA et al. , Termination of translation in eukaryotes is mediated by the quaternary eRF1*eRF3*GTP*Mg2+ complex. The biological roles of eRF3 and prokaryotic RF3 are profoundly distinct. Nucleic Acids Res. 34, 3947–3954 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kononenko AV et al. , GTP-dependent structural rearrangement of the eRF1:eRF3 complex and eRF3 sequence motifs essential for PABP binding. Nucleic Acids Res. 38, 548–558 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ho B, Baryshnikova A, Brown GW, Unification of Protein Abundance Datasets Yields a Quantitative Saccharomyces cerevisiae Proteome. Cell Syst 6, 192–205.e193 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M, Systematic identification of pathways that couple cell growth and division in yeast. Science 297, 395–400 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Johnson AG et al. , RACK1 on and off the ribosome. RNA (New York, N.Y.) 25, 881–895 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Korobov V. OV, Chemical Kinetics with Mathcad and Maple. (Springer-Verlag/Wien, 2011). [Google Scholar]
- 50.Petrov A, Grosely R, Chen J, O’Leary SE, Puglisi JD, Multiple Parallel Pathways of Translation Initiation on the CrPV IRES. Mol. Cell 62, 92–103 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marshall RA, Dorywalska M, Puglisi JD, Irreversible chemical steps control intersubunit dynamics during translation. Proc Natl Acad Sci U S A 105, 15364–15369 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aitken CE, Puglisi JD, Following the intersubunit conformation of the ribosome during translation in real time. Nat. Struct. Mol. Biol 17, 793–800 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Juette MF et al. , Single-molecule imaging of non-equilibrium molecular ensembles on the millisecond timescale. Nat. Methods 13, 341–344 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bronson JE, Fei J, Hofman JM, Gonzalez RL Jr., Wiggins CH, Learning rates and states from biophysical time series: a Bayesian approach to model selection and single-molecule FRET data. Biophys. J 97, 3196–3205 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choi J, Puglisi JD, Three tRNAs on the ribosome slow translation elongation. Proc. Natl. Acad. Sci. USA 114, 13691 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steinert HS, Rinnenthal J, Schwalbe H, Individual basepair stability of DNA and RNA studied by NMR-detected solvent exchange. Biophys. J 102, 2564–2574 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rinnenthal J, Klinkert B, Narberhaus F, Schwalbe H, Direct observation of the temperature-induced melting process of the Salmonella fourU RNA thermometer at base-pair resolution. Nucleic Acids Res. 38, 3834–3847 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Magis C et al. , T-Coffee: Tree-based consistency objective function for alignment evaluation. Methods Mol Biol 1079, 117–129 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Pettersen EF et al. , UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saini P, Eyler DE, Green R, Dever TE, Hypusine-containing protein eIF5A promotes translation elongation. Nature 459, 118–121 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park MH, Cooper HL, Folk JE, Identification of hypusine, an unusual amino acid, in a protein from human lymphocytes and of spermidine as its biosynthetic precursor. Proc. Natl. Acad. Sci. USA 78, 2869–2873 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the supplementary materials. Relevant expression constructs are available upon request.


